- 1Department of Oncology and Central Laboratory, The Affiliated Hospital of Chengde Medical University, Chengde, China
- 2Cancer Center, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, China
JC polyomavirus (JCPyV) belongs to the human polyomavirus family. Based on alternative splicing, the early region encodes the large and small T antigens, while the late region encodes the capsid structural proteins (VP1, VP2, and VP3) and the agnoprotein. The regulatory transcription factors for JCPyV include Sp1, TCF-4, DDX1, YB-1, LCP-1, Purα, GF-1, and NF-1. JCPyV enters tonsillar tissue through the intake of raw sewage, inhalation of air droplets, or parent-to-child transmission. It persists quiescently in lymphoid and renal tissues during latency. Both TGF-β1 and TNF-α stimulates JCPyV multiplication, while interferon-γ suppresses the process. The distinct distribution of caspid receptors (α-2, 6-linked sialic acid, non-sialylated glycosaminoglycans, and serotonin) determines the infection capabilities of JCPyV virions, and JCPyV entry is mediated by clathrin-mediated endocytosis. In permissive cells, JCPyV undergoes lytic proliferation and causes progressive multifocal leukoencephalopathy, while its DNA is inserted into genomic DNA and leads to carcinogenesis in non-permissive cells. T antigen targets p53, β-catenin, IRS, Rb, TGF-β1, PI3K/Akt and AMPK signal pathways in cancer cells. Intracranial injection of T antigen into animals results in neural tumors, and transgenic mice develop neural tumors, lens tumor, breast cancer, gastric, Vater’s, colorectal and pancreatic cancers, insulinoma, and hepatocellular carcinoma. Additionally, JCPyV DNA and its encoded products can be detected in the brain tissues of PML patients and brain, oral, esophageal, gastric, colorectal, breast, cervical, pancreatic, and hepatocellular cancer tissues. Therefore, JCPyV might represent an etiological risk factor for carcinogenesis and should be evaluated for early prevention, diagnosis, and treatment of cancers.
Introduction
JC polyomavirus (JCPyV) belongs to the human non-enveloped polyomavirus family in combination of SV40 and BK viruses. The genomic DNA homology between JCPyV and SV40 or BK viruses is 69% or 75%, respectively, showing their close evolutionary relationships (1). A serological study has indicated asymptomatic JCPyV infection in 46.1% of 1-month-old infants, 80.7% of 1- year-old infants, 85.9% of 2-year-old children, 85.5% of 3-year-old children, and about 90% of the adult population (2). As shown in Figure 1, JCPyV consists of a small, circular, double-stranded DNA genome of 5,130 base pairs and icosahedral capsids. The transcription of early and late coding regions occurs to produce small t and large T antigens by an interposed transcription control region. The late region encodes the capsid structural proteins (VP1, VP2, and VP3) by alternative splicing and a small regulatory protein (agnoprotein). T and t antigens are responsible for DNA replication, and the VP proteins for assemble with viral DNA to form virions (1). JCPyV may be activated for cell lysis under immunosuppressive conditions (e.g., HIV infection or the transplantation of bone marrow, liver, lung or kidney), and therefore is an established etiologic factor of demyelinating progressive multifocal leukoencephalopathy (PML) (3–7). Moreover, JCPyV could infect the enteric glia and cause chronic idiopathic intestinal pseudo-obstruction (8), or result in male lower urinary tract symptoms (9). The autoimmune diseases of JCPyV-associated brain syndromes include multiple sclerosis (MS), Crohn’s disease, and psoriasis, which were not previously considered as predisposing factors for PML (10). In non-permissive cells (i.e., cells that do not allow viral replication), JCPyV infection causes either abortive infection or malignant transformation (1) (Figure 2).
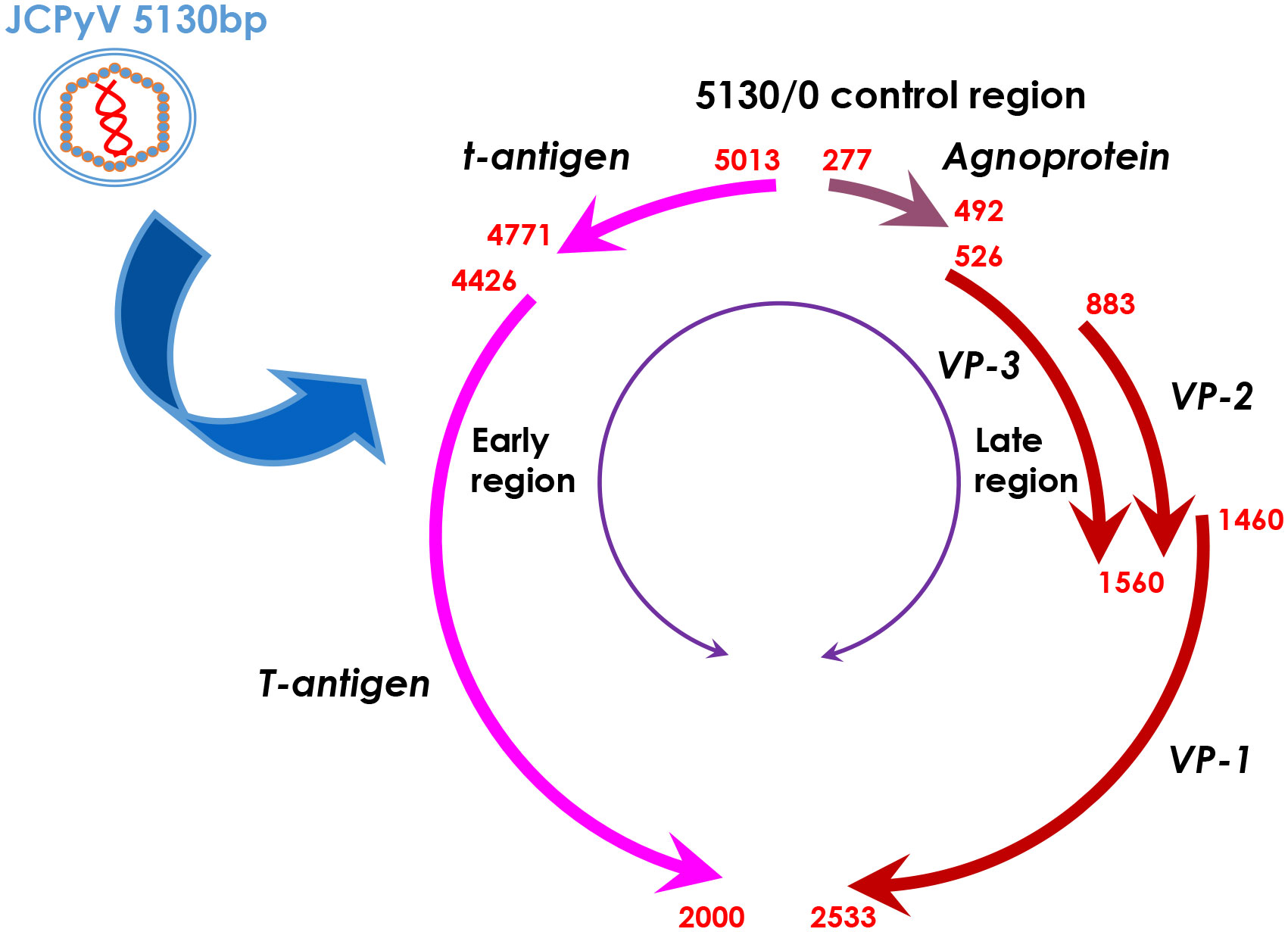
Figure 1 The genomic DNA structure of JCPyV. JCPyV has icosahedral capsids and small, circular, double-stranded genomic DNA of 5130 base pairs. It is composed of early and late coding regions, which are transcribed in opposite directions initiated by a transcriptional control region. The early region encodes both small and large T antigens by alternative splicing. The late region encodes the capsid structural proteins (VP1, VP2 and VP3) by alternative splicing and a small regulatory protein, agnoprotein.
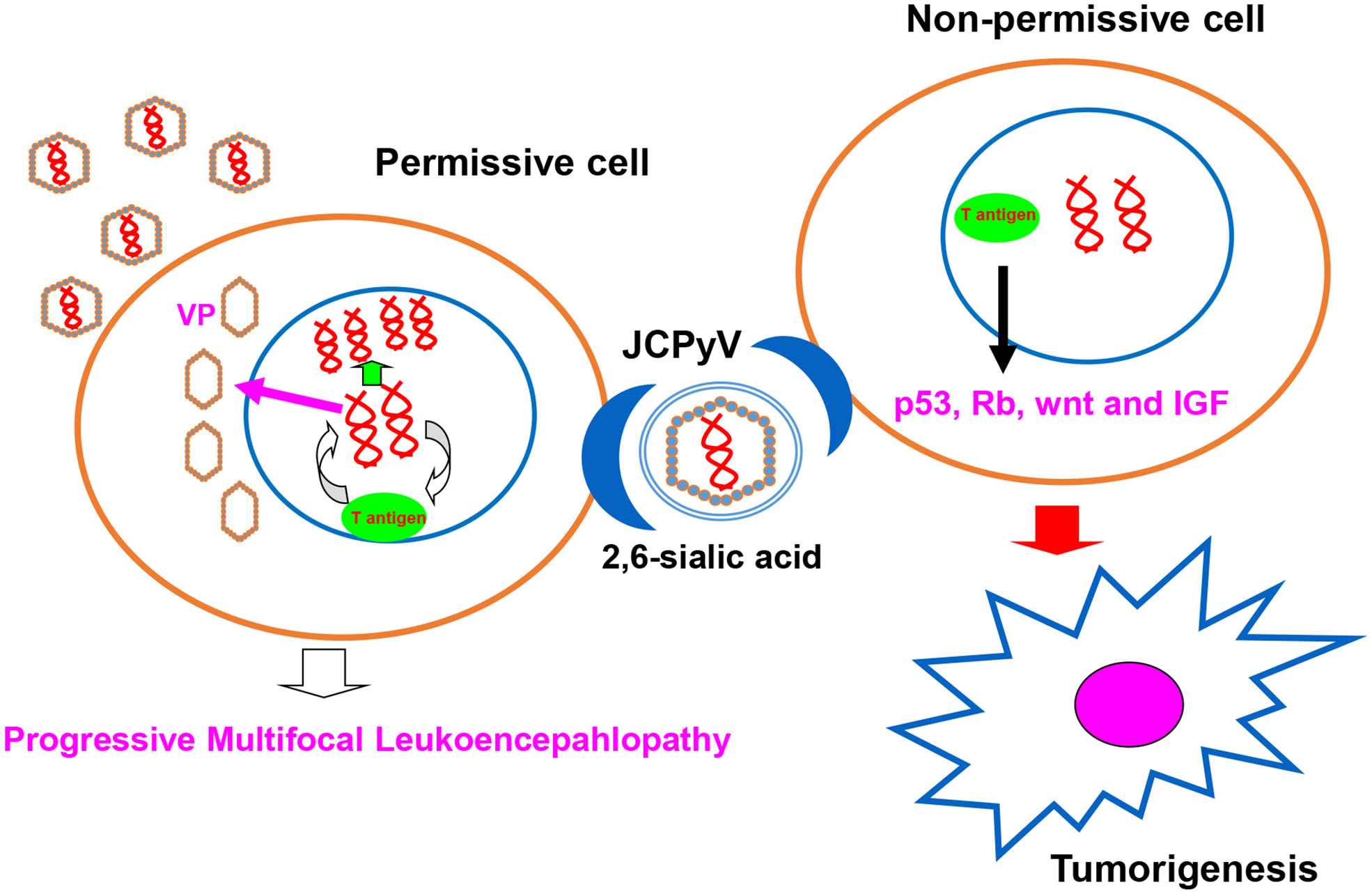
Figure 2 JCPyV infection outcome. JCPyV infection is initiated by its binding to JCPyV-sensitive cell surfaces. JCPyV capsids undergo endocytosis via capsid receptors (e.g., α 2, 6-linked sialic acid, non-sialylated glycosaminoglycans, and serotonin). In permissive cells, JCPyV may be activated for cell lysis and cause progressive multifocal leukoencephalopathy under immunosuppression (e.g., HIV infection, immunosuppressive drugs for organ transplantation, and cancer chemotherapy). In non-permissive cells, T antigen DNA from JCPyV can be inserted into genomic DNA, and T antigen can induce the malignant transformation of normal cells by targeting the p53, Rb, wnt, and IGF signal pathways.
The infection and replication of JCPyV
As shown in Figure 3, JCPyV enters the human body through the intake of raw sewage or the inhalation of air droplets, and persists quiescently in tonsillar lymphoid and renal tissues during latency (11). Parent-to-child transmission is also common for its propagation (12). After asymptomatic primary infection in childhood, the virus spreads through the bloodstream from the primary sites of infection to secondary sites (kidney and lymphoid tissues, peripheral blood leukocytes, and brain tissue) (13). JCPyV DNA replication occurs primarily in lymphoid and glial cells that contain transcription factors specific for JCPyV (14, 15).
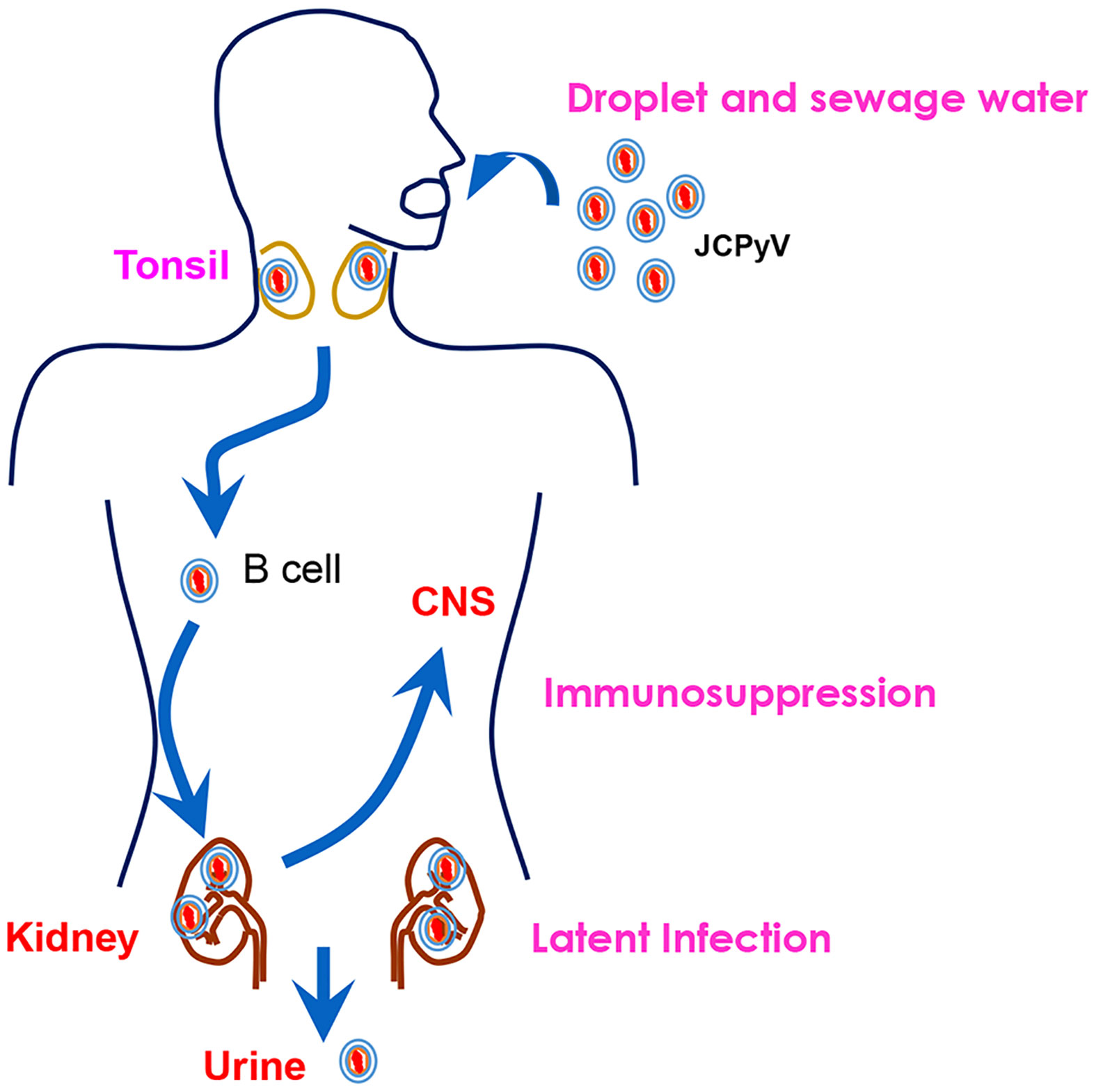
Figure 3 The natural history of JCPyV. JCPyV enters the human body through the intake of raw sewage or inhalation of air droplets. It is transported to the kidneys via B cells. It persists quiescently in the tonsil and renal tissues during latency. Upon immunosuppression, JCPyV enters the central nervous system (CNS) and undergoes lytic proliferation, resulting in the demyelinating brain disease, progressive multifocal leukoencephalopathy (PML). It can be excreted from the human body through the urine.
JCPyV infects human cells by the interaction of capsid VP proteins with receptors on JCPyV-sensitive cells, followed by endocytosis and nuclear transport of JCPyV virions. In the nucleus, the viral DNA is uncoated, initiating the transcription of the early region (16). The tissue-specific distribution of the VP receptors (α 2, 6-linked sialic acid, non-sialylated glycosaminoglycans, and serotonin) determines the different infection capabilities of JCPyV (17–21). JCPyV infection is dependent on the interactions between VP capsid proteins and asparagine N-linked sialic acids or the serotonin 5-hydroxytryptamine 2A receptor (5-HT2AR) on the cell surface. Treatment with an inhibitor of N-linked glycosylation (tunicamycin), 5HT2AR antagonists (ketanserin and ritanserin), or anti-5HT2aR antibody reduces JCPyV infection, while treatment with PNGase F to remove N-linked oligosaccharides does not influence JCPyV infection (18, 19, 22). VP1-composed virion-like particles (VLPs) can bind to sialoglycoproteins (α1 acid-glycoprotein, transferrin receptor, and fetuin) and glycolipids (gangliosides and lactosylceramide) (23). Exposure to either anti-VP1 antibody or sialidase to hydrolyze sialic acid residues can suppress viral entry into host cells. After interaction between capsid proteins and their receptors, JCPyV binds to caveolin-1 and undergoes eps15 and Rab5-GTPase-mediated internalization and clathrin-dependent endocytosis (24), which is facilitated by the interaction of β-arrestin with 5-hydroxytryptamine receptors (25).
After the entry of JCPyV into cells, TCF-4-T-antigen complex binds to the JCPyV promoter in U87-MG cells, increasing the ability of the T antigen to replicate viral DNA (26). LCP-1 also interacts with the lytic control element and differentially regulates T antigen expression (27). Glial factor 1 (GF-1) has homology with the central region of Sμbp-2 and can bind to the promoter B-regulatory domain of JCPyV (28). Purα interacts with T antigen to modulate T-antigen-mediated transcriptional activation, while the Purα-BAG-1 complex suppresses JCPyV DNA replication in glial cells (29, 30). The terminal core kinase of the MAPK cascade (MAPK-ERK) facilitates the transcription of the JCPyV by up-regulating the transcription factors downstream of the MAPK cascade (i.e., c-myc and SMAD4) and shuttling them to the nucleus (31), while SF2/ASF (splicing factor 2/alternative splicing factor) weakens the transcription and alternative splicing of JCPyV genes via direct interaction with the viral promoter (32), and retinoic acid-inducible gene I (RIG-I) and cGMP-AMP synthase negatively controls JCPyV replication in human astrocytes (33).
Moreover, HIV-1 induces cytokines that reactivate JCPyV to cause PML in the brain, suggesting a direct correlation between inflammatory cytokines and the susceptibility to JCPyV infection (34, 35). The treatment of glial cells with IFNα and IFNβ increases the endogenous levels of C/EBPβ-LIP, which inhibits basal and NF-κB-stimulated JCPyV transcription via the NF-κB-C/EBPβ-LIP -JCPyV DNA complex (36). Both TGF-β1 and TNF-α can stimulate JCPyV multiplication and increase the overall number of infected cells via the Smad and NF-κB pathways, respectively (37). Tat and Fast1 can cooperate with Smad2, 3, and 4 at the JCPyV DNA control region, stimulating its gene transcription in oligodendroglial cells (38). JCPyV infection significantly increases nuclear HIF-1α levels in glial cells, which binds to and activates the JCPyV early promoter via Smad3 and Smad4 (39). IL-1β dramatically increases JCPyV transcription in glial cells via NF-1 binding to the JCPyV enhancer region via the PKC pathways (40). However, interferon-γ inhibits JCPyV replication by down-regulating T antigen expression via Jak1 signaling (41).
According to recent literatures, topoisomerase I inhibitors (β-lapachone and topotecan) are found to inhibit JCPyV infection in neuroblastoma cells (42). Irisolidone, an isoflavone metabolite, negatively modulates JCPyV gene expression by suppressing Sp1 binding in glial cells (24). The Cdk inhibitor, R-roscovitine, suppresses the proliferation and production of JCPyV by inhibiting the phosphorylation of T antigen (43). Hexadecyloxypropyl- cidofovir suppresses JCPyV replication in fetal brain SVG cells (44). JCPyV infection can be suppressed by nocodazole, cytochalasin D, or acrylamide in glial cells (45). Moreover, O’Hara et al. (46) found that teriflunomide could inhibit JCPyV infection and propagation in choroid plexus epithelial cells and glial cells. PARP-1 inhibitor, 3-aminobenzamide, could significantly suppress JCPyV replication and spread (47). In contrast, both trichostatin A (TSA) and butyrate can activate the JCPyV promoter and hyperacetylation of this promoter in non-glial cells. The enhancer and Sp1 element upstream of the TATA box are necessary for TSA-mediated activation (48). Some reagents are expected to prevent the infection and replication of JCPyV in the future.
The functions of JCPyV-encoded proteins
T antigen
T antigen is a multifunctional and oncogenic phosphoprotein essential for viral DNA replication in G2-arrested cells via ATM- and ATR-mediated G2 checkpoint signaling (49). It binds to and breaks DNA to unwind the double helix and recruits helicase, ATPase, and polymerase (1, 50). T antigen primarily targets protein complexes that have PP4 and PP1 phosphatase, v-ATPase, and E3-ubiquitin ligase activities (51). Its N-terminal portion contains LXCXE and J domains, which are necessary for binding and inactivating the Rb family (52) and its N-terminal phosphorylation site at threonine 125 is critical to T-antigen-mediated replication via stabilizing T antigen, interaction with the Rb family members p107 and p130 and the release of E2F from RB-E2F complex (53). The origin-binding domain of T antigen contains a C-terminal pocket and interacts with the major groove of GAGGC sequences. The pocket residue increases T antigen expression, supporting JCPyV DNA replication (54).
Reportedly, AP-1 family (c-Fos and Jun) functionally interacts with T antigen, significantly diminishing T-antigen-mediated replication and transcription of JCPyV genes in glial cells. The c-Jun-binding domain for T antigen maps to the middle portion of the protein, while the T-antigen-binding domain for c-Jun is its basic-DNA binding region (55). In glial cells, T antigen interacts with Purα and serine/arginine-rich splicing factor 1 (SRSF1). T antigen promotes JCPyV gene expression by binding to the SRSF1 promoter and weakening SRSF1 transcription (56, 57). Purα and T antigen bind to the JCPyV early promoter via T-antigen, ameliorating SRSF1-mediated inhibition of JCPyV gene expression and replication (58). P53 can interact with T antigen, blocking viral DNA replication (59). However, neurofibromatosis type 2 could induce proteasomal degradation of the T-antigen and suppress T-antigen protein expression in glioblastoma cells, weakening T-antigen-mediated regulation of the JCPyV promoter (60), and LIP (liver inhibitory protein) expression also induced the degradation of JCPyV T antigen in transgenic mouse tumor cells (32, 61). The partner proteins modulate the biological functions and protein instability of T antigen, which is involved in carcinogenesis and subsequent progression.
Agnoprotein
The JCPyV agnoprotein shares 50–60% homology with those of BK and SV40 viruses; however, its carboxyl-terminal region is relatively unique. It is firstly detected on day 3 of JCPyV post-infection, and its levels increase until the late stage of infection, and responsible for virion release and viral propagation (62). Agnoprotein localizes to the endoplasmic reticulum (ER) early in infection and then the plasma membrane late in infection (63). Agnoprotein is 71 amino acids (8kDa) and stably forms dimers and oligomers through its hydrophobic Leu/Ile/Phe-rich (aa 28–39) domain (64). Residues Lys22 to Asp44 may be the transmembrane domain, and the disulfide bond at Cys40 may trigger oligomerization (65). Its basic amino acid residues at positions 8 and 9 determine its viroporin activity (63). In agnoprotein, the major amphipathic α-helix conformation spans amino acids 23–39 of the Leu/Ile/Phe-rich region, while the minor α-helix consists of Leu6 to Lys13 (66). Leu29 and Leu36 of the major amphipathic α-helix are at the dimer interface, keeping the spatial structure and protein stability (67). All three Phe residues are localized to this amphipathic α-helix and mediate protein folding and stability (68). Moreover, agnoprotein primarily targets 501 cellular proteins containing “coiled-coil” motifs. The agnoprotein- host interactions were involved in protein synthesis and degradation, cellular transport, and organelles, including mitochondria, ER-Golgi, and the nucleus. Among the agnoprotein partners, Rab11B, importin, and Crm-1 have been biochemically validated (68).
In nucleus, agnoprotein promotes T antigen binding to the viral origin with indirect interactions with DNA. It contains several potential phosphorylation sites (ser7, ser11, and thr21) that can be phosphorylated by PKC (69). Small t antigen (aa 82-124) also interacts with agnoprotein and PP2A, suppressing the PP2A-mediated dephosphorylation of agnoprotein and promoting JCPyV replication (70). The amino-terminal of agnoprotein can bind to YB-1 and reduce YB-1-mediated gene transcription (71). The interaction of p53 with agnoprotein can lead to p21 expression, causing G2/M arrest and sensitizing cells to cisplatin via chromosome fragmentation, micronuclei formation, and impaired double-strand DNA break repair activity by up-regulating the expression of the DNA repair proteins (e.g., Ku70 and Ku80) (72).
In cytosol, agnoprotein predominantly localizes to the perinuclear region of JCPyV-infected cells, and colocalizes with the cellular cytoskeletal protein tubulin (73), which is co-precipitated with phosphorylated agnoprotein (74). Suzuki et al. (75) also demonstrated that agnoprotein could directly interact with fasciculation and elongation protein zeta 1 (FEZ1) and microtubules. The interaction dissociated FEZ1 from the microtubules and inhibited FEZ1-facilitated neurite outgrowth. Saxena et al. (76) reported that the mitochondrial targeting sequence and dimerization domain of agnoprotein mediate mitochondrial localization, where agnoprotein decreased the respiration rate, mitochondrial membrane potential, and ATP production while increasing ROS production and Ca2+ uptake.
Caspids
Major coat protein VP1 couples with a minor coat protein (VP2 or VP3). VP2 and VP3 share DNA binding domain, VP1-binding domain, and nuclear localization signal (NLS). The 16 carboxy- terminal and 12 amino-terminal amino acids of VP1 are essential for the assembly of VLPs. Both minor coat proteins and the myristylation site on VP2 are important for properly packaging the genomic DNA of JCPyV (77). Furthermore, the cysteine residues of VP1 are dispensable for protein stability and oligomerization (78, 79). VP1 mediates VLP entry into the nucleus by importins α and β via its NLS (80). Point mutations in VP1 can influence virion binding to cellular glycan receptors and their recognition by polyomavirus-specific antibodies (81). Mutation 186G→C (Lys→Asp) in the VP1 gene could predispose MS patients undergoing treatment with natalizumab to PML (82). A deletion of the C-terminal 10 bp of VP1 is closely linked to lytic infection of granule cell neurons and atrophy in the cerebellum of an HIV/PML patient (83). VP1 mutations that are involved binding to sialic acid cell receptors favored PML onset (84). Hsp70 could interact with T antigen and VP2 or VP3, which accumulates in the nucleus of the infected cells and enhances viral DNA replication (85). VP2 binds to DNA through its DNA-binding domain between Lys332 and Lys336 (86). As for capsid expression, Ravichandran et al. (87) found that TGF-β1 activated MEK1/2 and subsequent phosphorylation of Smads, which bound to or increased binding to the JCPyV promoter for VP-1 synthesis.
The signal pathways of JCPyV
Multi-omics analysis has demonstrated that JCPyV-related carcinogenesis involves aberrant Forkhead box O, AMPK, p53, and PI3K/Akt signaling pathways. Moreover, T antigen can upregulate the expression of Akt, Rb, and survivin and downregulate p21 expression, indicating that it might activate the Akt/NF-κB/survivin pathway to block apoptosis and cause Rb hyper-expression and p21 hypo-expression for cell cycle progression (88). The upregulated proteins are involved in signaling through Cyclin-CDK, TGF-β receptor 1, fibroblast growth factor family receptor and platelet-derived growth factor receptor and the inflammatory responses mediated by Cox-2 (89). T antigen might interact with ribosomal proteins, various keratins, G proteins, apolipoproteins, ubiquitin-related proteins, CCAAT enhancer-binding proteins, β-catenin, RPL19, β-TRCP, and p53 in lens tumor cells (88). T antigen knockdown could suppress glycolysis, mitochondrial respiration, proliferation, migration, and invasion in lens tumor cells; however, it promoted apoptosis. T antigen can also activate the Akt/NF-κB/survivin pathway, producing an anti-apoptosis effect and causing Rb hyperexpression and p21 hypoexpression to mediate cell cycle progression (88). These findings suggest that the T antigen can aggravate the cellular phenotype, possibly by inactivating tumor suppressors, activating oncogenes, or disrupting metabolism and cell adhesion.
As shown in Figure 4, p53 interacts with T antigen to repress transcription from the JCPyV early promoter and JCPyV DNA replication in non-glial cells (90, 91). The interaction between p53 and T-antigen up-regulated the p53 downstream target protein, p21/WAF1 (92). Additionally, E2F-1 dissociated from the pRb-E2F-1 complex and stimulated S phase-specific genes following the formation of a pRb-T antigen complex or Rb phosphorylation (93). T antigen can bind to pRb2/p130, p107, and pRb/p105, activating the E2F transcription factor family and promoting entry into S phase (94). As IRS1 signal pathway, T antigen also induces the nuclear translocation of IRS-1, and IRS-1 interacts with T antigen, which is independent of IRS-1 tyrosine phosphorylation and blocked by IRS-1 serine phosphorylation (95). After T-antigen-mediated nuclear translocation, IRS-1 binds to Rad51 at the site of damaged DNA to direct DNA repair, causing accumulation of mutations in the affected cells (96). IRS-1-Rad51 nuclear interaction also sensitizes JCV T-antigen positive medulloblastoma cells to cisplatin and γ-irradiation (97). T-antigen requires the presence of a functional insulin-like growth factor I receptor (IGF-IR) for transformation of fibroblasts and for survival of medulloblastoma cell line. IGF-IR is phosphorylated in medulloblastoma biopsies and JCV T-antigen inhibits homologous recombination-directed DNA repair, causing accumulation of mutations. In Wnt- β-catenin pathway, the interaction between the central domain (residues 82–628) of T-antigen and the C-terminal residues of β-catenin (aa 695-781) increases β-catenin levels and its nuclear entry, resulting in the upregulation of its downstream genes (c-myc, VEGF, and Cyclin D1). T antigen binds to the F-box proteins β-transducin repeat-containing protein-1 and 2 (βTrCP1/2) and recruits Rac1 to form the T antigen-Rac1-β-catenin complex that suppresses the ubiquitin- dependent degradation of β-catenin by proteasomes (98–100). T antigen downregulates BAG-3 expression to inhibit apoptosis by blocking AP2 binding to the BAG3 promoter. Bag3 interacts with the T antigen, inducing its autophagic degradation (101). Additionally, T antigen binds to and activates the survivin promoter, upregulating its expression and mediating the nuclear translocation of suvivin via the T-antigen-survivin complex (102, 103). In contrast, T antigen can arrest G1, sustain G2, and block ROS induction and cytotoxicity during glucose deprivation. T antigen can also stimulate the expression of transaldolase-1 and hexokinase-2 (104).
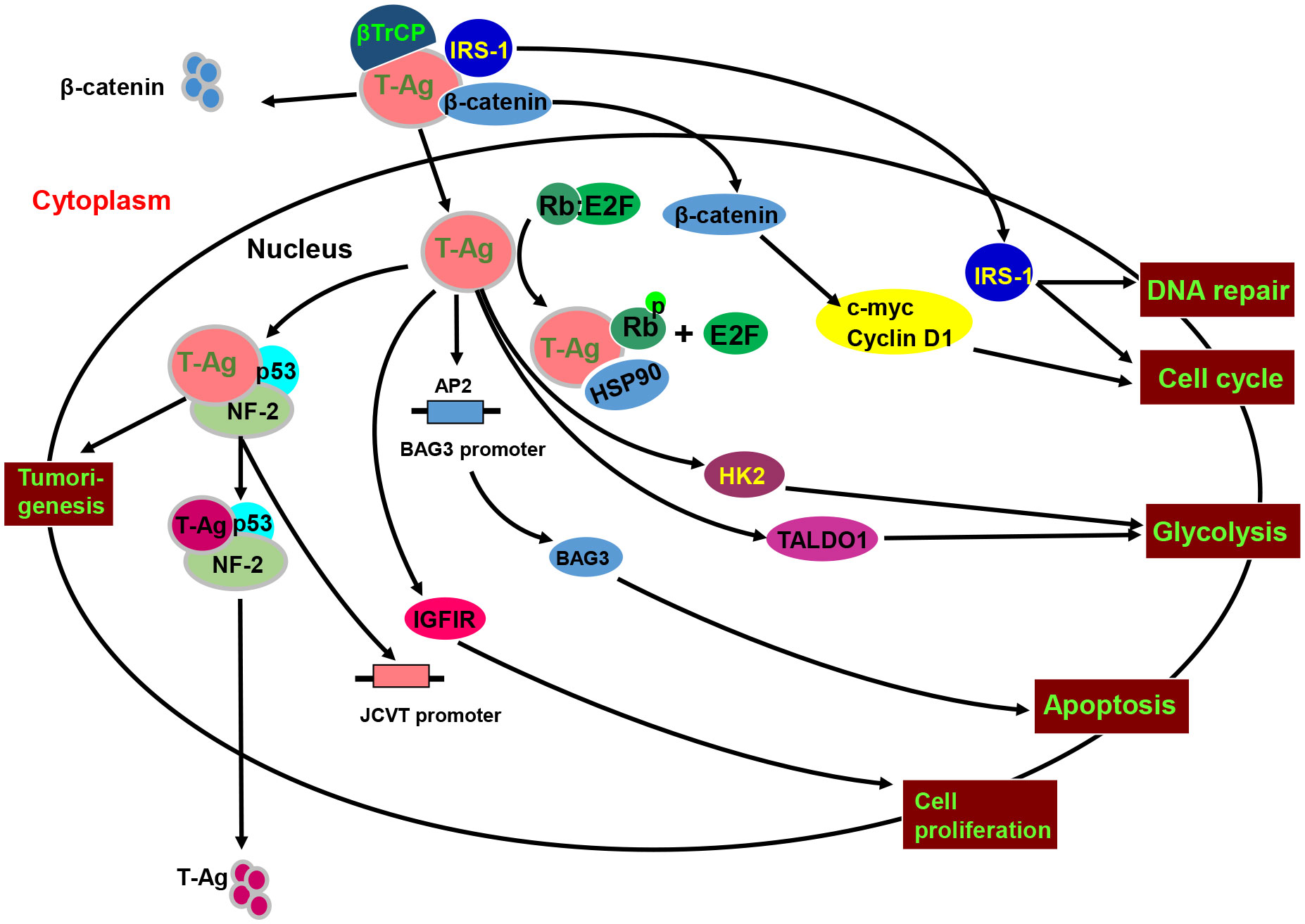
Figure 4 The biological function and signal pathways of the JCPyV T antigen. JCPyV T antigen binds to βTrCP1/2 protein to cause ubiquitin-mediated degradation of β-catenin, and binds to β-catenin to enhance its protein stability and facilitate its entry into the nucleus. Nuclear β-catenin promotes S-phase transformation by up-regulating c-myc and Cyclin D1 protein expression. T antigen interacts with p53 and neurofibromatosis-2 (NF-2) for the proteasome-mediated degradation of T antigen. The binding of T antigen to phosphorylated Rb protein results in the separation of Rb-E2F, resulting in an abnormal cell cycle. T antigen can promote the translocation of the insulin receptor substrate 1(IRS1) to the nucleus, induce cell cycle evolution, and participate in DNA repair. In addition, T antigen up-regulates the expression and phosphorylation of IRS1 and IGF1 receptor (IGF-1R), promoting cell proliferation and disrupting normal cell activity by binding to the transcription factor AP-1. The DNA binding domain of T antigen can bind to the AP2 sequence in the BAG3 promoter and CPG binding protein promoter of transcriptional regulatory methylation and regulate the expression of BAG3. T antigen can also stimulate the expression of transaldolase-1 (TALDO1) and hexokinase-2 (HK2) to induce glycolysis.
The association between JCPyV and carcinogenesis
In transformed cells, JCPyV can cause anchorage-dependent growth, rapid division, prolonged life span, increased ploidy, unstable multicentric chromosomes, centric and acentric rings, dysregulated genomic stability and DNA repair, and increased micronuclei formation (105–107). Intracranially inoculated JCPyV caused glioblastoma in juvenile owl monkeys (108), grade 3-4 astrocytoma in adult owl monkeys (109), undifferentiated neuroectodermal tumors in the cerebrums of newborn Sprague-Dawley rats (110), cerebellar medulloblastoma, plexus tumors, medulloblastoma, and thalamic gliomatosis in hamsters (111), and neuroblastoma in the abdominal cavity, pelvis, mediastinum, and neck region of Syrian hamsters (112). Padgett et al. (113) demonstrated that malignant brain tumors developed in Syrian golden hamsters during a 6.5-month observation period following intracerebral inoculation of different JCPyV strains into newborns. The Mad-2 strain caused cerebellar medulloblastomas, whereas the MAD-3 strain induced extracranial neuroblastomas. In contrast, the Mad-4 strain produced pineal gland and cerebellar tumors.
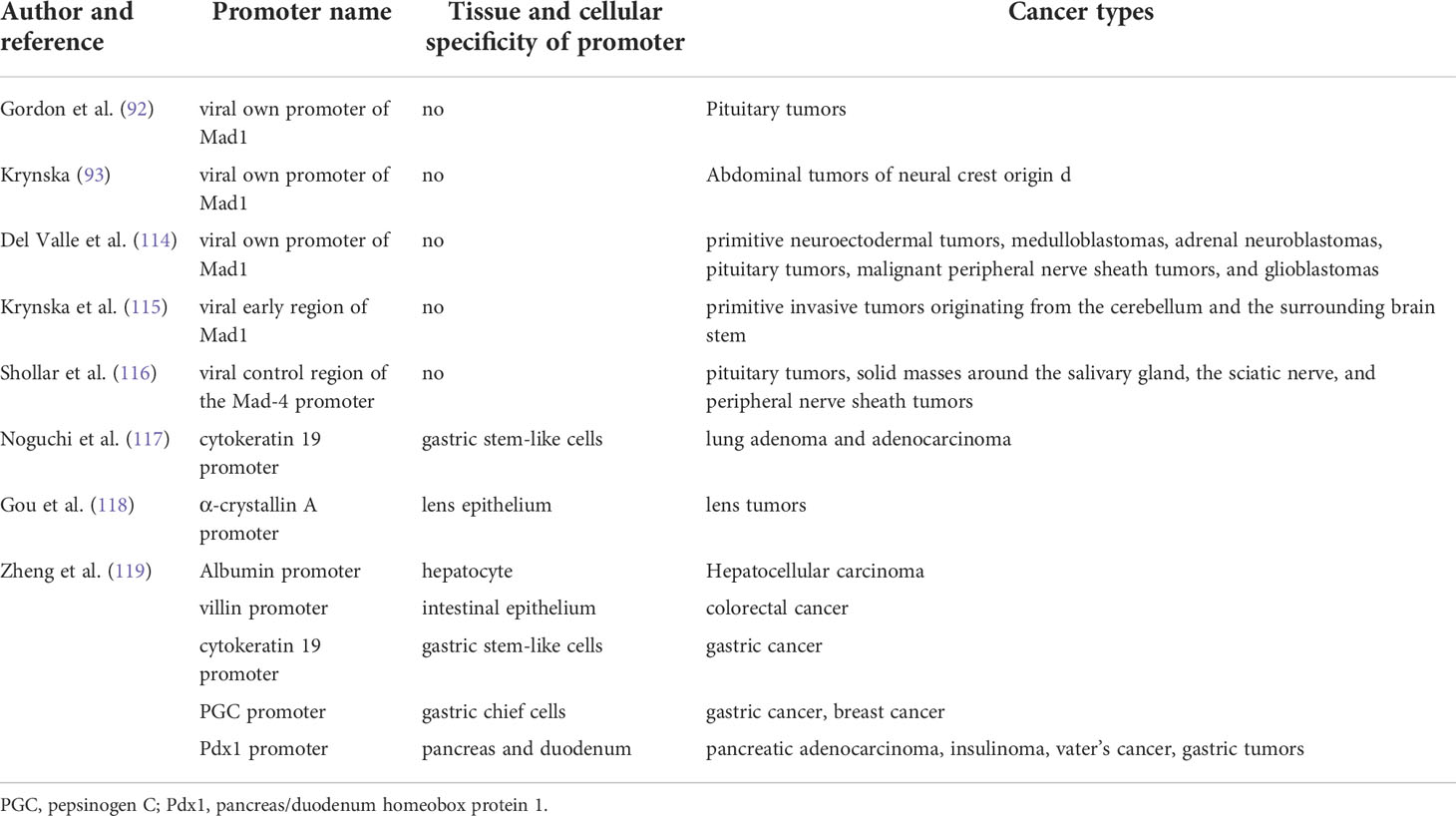
The spontaneous tumors in the transgenic mice of JCV T antigen can provide direct evidences for the oncogenic role of JCPyV as shown in Table 1. The transgenic mouse with the early encoding region of the archetype strain was generated using its own promoter and developed neural crest tumors, such as primitive neuroectodermal tumors, adrenal neuroblastomas, medulloblastomas, pituitary tumors, glioblastomas, and malignant peripheral nerve sheath tumors (114). Krynska et al. (115) established the same transgenic mice and observed primitive tumors originating from the cerebellum and adjacent brain stem that were grossly and histologically similar to human medulloblastoma and primitive neuroectodermal tumors. However, Gordon et al. (92) used the same promoter to generate transgenic mice overexpressing T antigen, which developed large, solid pituitary masses. Shollar et al. (116) established transgenic mice expressing T-antigen under the control of the Mad-4 promoter and observed pituitary tumors by one year of age. Krynska et al. (93) found that transgenic mice harboring T antigen could develop massive abdominal tumors of neural crest origin. In our group, a transgene with the K19 promoter was generated and pulmonary tumors with T-antigen, p53, and CK19 expression and EGFR mutation were observed (117). We also established T antigen-expressing transgenic mice using α-crystallin A and observed lens tumors that were positive for T antigen, N-cadherin, p53, and β-catenin. Enlarged eyeballs were observed, and the tumors invaded the brain (118). Additionally, we generated CAG-loxp-LaZ-loxp T antigen transgenic mice with T antigen activation induced using matching tissue-specific cre transgenic mice. Gastric poorly-differentiated carcinoma was observed in gastric stem-like and chief cells following T antigen overexpression. Moreover, spontaneous hepatocellular and colorectal cancers developed in Alb-cre (hepatocytes)/T antigen and villin-cre (intestinal epithelium)/T antigen transgenic mice. Gastric, colorectal, and breast cancer were observed in PGC (Pepsinogen C)-cre/T antigen mice. Pancreatic insulinoma and ductal adenocarcinoma, gastric adenoma, and duodenal cancer were detected in Pdx1-cre/T antigen mice. There was alternative splicing of T antigen mRNA in all target organs of these transgenic mice and various cells transfected with pEGFP-N1-T antigen. It has been suggested that the JCPyV T antigen might induce gastroenterological carcinogenesis in a cell-specific manner (119).
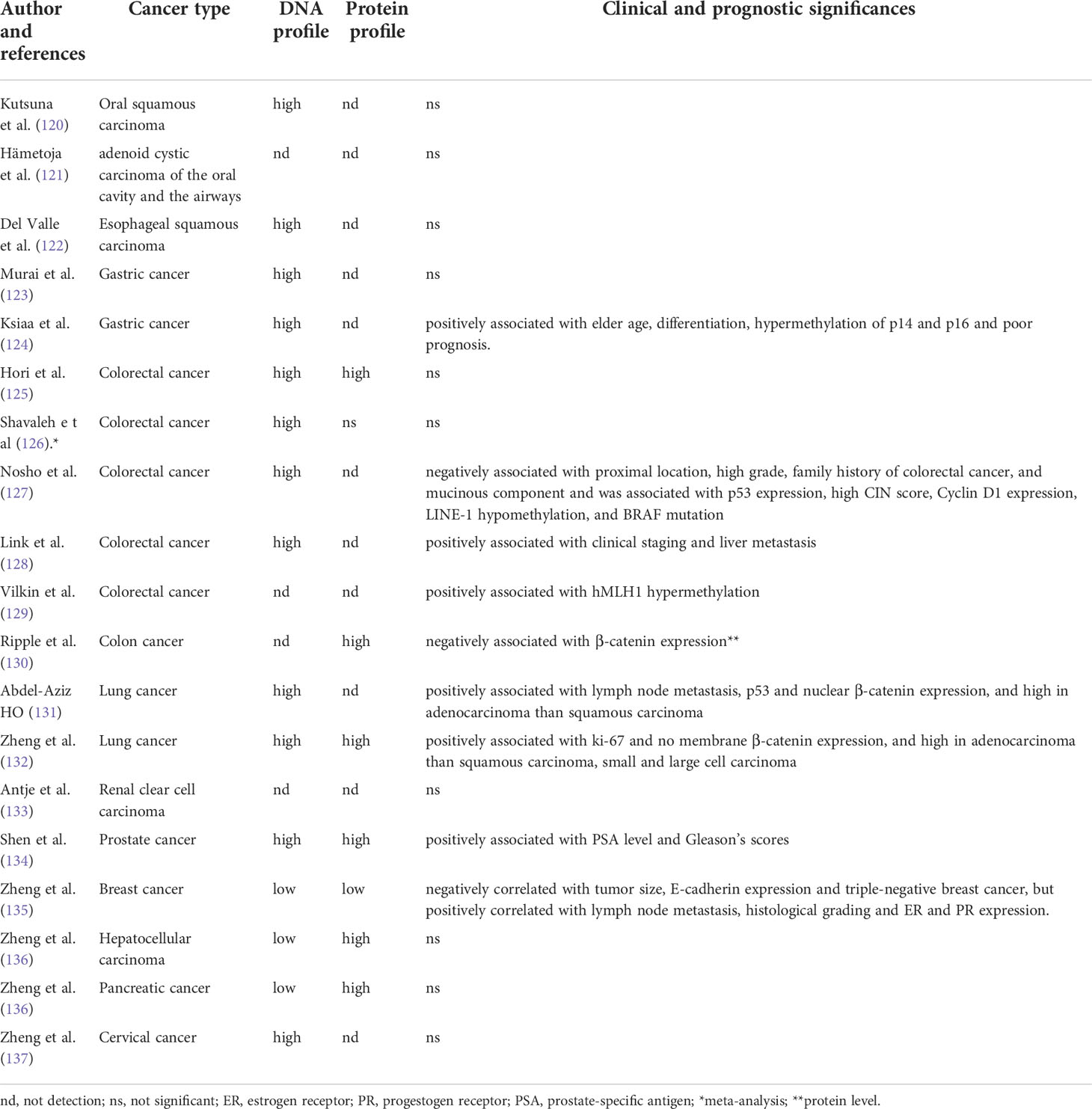
It is important to detect and compare JCPyV DNA in cancer and adjacent normal tissues using either molecular or morphological approach. JCPyV detection might determine the etiology for JCPyV-related cancer. The correlation of JCPyV T antigen with carcinogenesis and subsequent progression was summarized in Table 2. Although JCPyV DNA was found in ependymomas and choroid plexus papilloma (138), Kutsuna et al. (120) found that glossitis and tongue dysplasia had significantly lower copies of JCPyV than tongue cancer. They observed T antigen DNA and protein in the nuclei of tongue cancer cells but not in normal or dysplastic epithelia. JCPyV DNA and T antigen were found in adenoid cystic carcinomas samples of the trachea, paranasal sinuses, and oral cavity by PCR and immunohistochemistry respectively (121). JCPyV DNA was more frequently detected in esophageal carcinomas than in normal, benign, or premalignant esophageal samples (122). JCPyV T antigen load is also higher in gastric cancer than in normal mucosa (123). Indeed, its DNA and protein were detected in the nuclei of gastric cancer cells. Moreover, T-antigen DNA is correlated with differentiation and the methylation of p14 and p16 in this cancer (124).
In colorectal cancer, the positivity rate of T antigen is decreased from colorectal adenocarcinoma to adenoma to mucosa (125, 126). Nosho et al. (127) reported that T antigen could inactivate wild-type p53, resulting in chromosomal instability. It was positively correlated with p53 expression, p21 loss, nuclear β-catenin, LINE-1 hypo-methylation and hyper-expression, and low MSI (microsatellite instability) levels. Link et al. (128) found that T antigen enhanced the migration and invasion of colorectal cancer cells via Akt and MAPK signaling. Indeed, T antigen could be detected by IHC in primary colorectal cancers and their corresponding liver metastases. The interaction between T-antigen and β-catenin and the nuclear detection of β-catenin in T-antigen-positive colorectal cancer cells demonstrates dys-regulation of the Wnt pathway (15, 129). Ripple et al. (130) found that T antigen and β-catenin were co-localized in the nuclei of colorectal cancer cells, resulting in the activation of TCF4-dependent promoters and the transcription of TCF4 downstream targets (e.g. c-myc, VEGF and Cyclin D1).
In the respiratory cancer, the positivity rate for the JCPyV T antigen in the respiratory system is lower in normal lung tissue than in tumors; T antigen DNA is strongly observed in lung adenocarcinoma (131). One study found a lower JCPyV copy number in normal lung cancer than in lung tumors (132). Moreover, the copy number was lower in lung adenocarcinomas compared to squamous, small, or large cell carcinomas. Lung cancers with a high JCPyV copy number were characterized by high proliferation and low β-catenin-mediated cell adhesion (132).
In urinary tract neoplasms, JCPyV has also been detected in renal pelvic urothelial carcinoma and renal cell carcinoma (133). Shen et al. (134) found that 90.1% of the urothelial carcinomas and all the renal cell carcinomas that they evaluated were positive for JCPyV using nested PCR. Prostate cancer is more susceptible to JCPyV infection than benign prostate hyperplasia. Tumors with both high prostate-specific antigen levels and high Gleason scores were associated with a high risk of JCPyV infection.
In addition, we reported that the positivity rate and expression levels of T antigen were lower in breast cancer than in normal breast tissue (135), in line with hepatocellular and pancreatic cancer (136). T antigen DNA positivity was inversely associated with E-cadherin expression and triple-negative breast cancer but positively associated with lymph node involvement and ER and PR expression. JCPyV copies were negatively linked to tumor size and E-cadherin expression in breast cancer but positively associated with histological grading. Additionally, we for the first time found that JCPyV was less detectable in cervical epithelium than dysplasia and carcinoma (137). We also observed T antigen DNA and protein in hepatocellular, pancreatic, breast and cervical cancer cells using in situ PCR and immunohistochemistry (134–136).
Conclusions and perspective
JCPyV enters eukaryotic cells and is inserted into genomic DNA. It induces tumorigenesis with tissue specificity by targeting the p53, β-catenin, IRS, Rb, TGF-β1, PI3K/Akt, and AMPK signal pathways. Pathological examination and animal experiments have demonstrated that the JCPyV T antigen might induce tumorigenesis in neural and gastroenterological systems and breast. Thus, JCPyV might be an etiological risk factor for carcinogenesis and should be emphasized in tertiary prevention and treatment of cancer.
Because JCPyV infection rate reaches 80%, we should try our best to prevent the entry of JCPyV into the human body through the sewage and air droplet. In addition, it is better to block the endocytosis and nuclear transport of JCPyV virions by receptor antagonists. Finally, the agents to block the JCPyV infection or inhibit the JCPyV-related signal pathway should be developed to prevent and treat JCPyV-related cancers. In the future, we can realize the early diagnosis, finding and treatment of JCPyV-related cancers.
Author contributions
H-CZ conceived the review, and HX and CY-Z led its drafting and managed the editing of it. All authors contributed to the drafting of the review and approved the final manuscript.
Funding
This review was supported by Award for Liaoning Distinguished Professor, Natural Science Foundation of Hebei Province (21377772D) and National Natural Scientific Foundation of China (81672700).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zheng HC, Yan L, Cui L, Guan YF, Takano Y. Mapping the history and current situation of research on John Cunningham virus - a bibliometric analysis. BMC Infect Dis (2009) 9:28. doi: 10.1186/1471-2334-9-28
2. Elia F, Villani S, Ambrogi F, Signorini L, Dallari S, Binda S, et al. JC virus infection is acquired very early in life: Evidence from a longitudinal serological study. J Neurovirol (2017) 23:99–105. doi: 10.1007/s13365-016-0477-9
3. Rastogi A, Gulati N, Bihari C, Chaudhary A, Bansal K, Sasturkar S, et al. JC virus-related progressive multifocal leukoencephalopathy after living-donor liver transplant: A rare case. Exp Clin Transplant (2019) 17:414–7. doi: 10.6002/ect.2016.0242
4. Panchabhai TS, Choudhary C, Isada C, Folch E, Mehta AC. Progressive multifocal leukoencephalopathy in a lung transplant recipient: Isolation of John Cunningham (JC) virus from bronchoalveolar lavage. J Glob Infect Dis (2016) 8:51–4. doi: 10.4103/0974-777X.176150
5. Mischitelli M, Fioriti D, Anzivino E, Bellizzi A, Barucca V, Boldorini R, et al. Viral infection in bonerow transplants: is JC virus involved? J Med Virol (2010) 82:138–45. doi: 10.1002/jmv.21558
6. Querido S, Ormonde C, Adragão T, Costa I, Pessanha MA, Gomes P, et al. Outcomes of living kidney donor candidates and living kidney recipient dandidates with JC polyomavirus and BK polyomavirus viruria. Int J Nephrol (2021) 2021:8010144. doi: 10.1155/2021/8010144
7. Karalic D, Lazarevic I, Banko A, Cupic M, Jevtovic D, Jovanovic T. Analysis of variability of urinary excreted JC virus strains in patients infected with HIV and healthy donors. J Neurovirol (2018) 24:305–13. doi: 10.1007/s13365-017-0608-y
8. Selgrad M, De Giorgio R, Fini L, Cogliandro RF, Williams S, Stanghellini V, et al. JC virus infects the enteric glia of patients with chronic idiopathic intestinal pseudo-obstruction. Gut (2009) 58:25–32. doi: 10.1136/gut.2008.152512
9. Thomas S, Dunn CD, Campbell LJ, Strand DW, Vezina CM, Bjorling DE, et al. A multi-omic investigation of male lower urinary tract symptoms: Potential role for JC virus. PLoS One (2021) 16:e0246266. doi: 10.1371/journal.pone.0246266
10. Miskin DP, Koralnik IJ. Novel syndromes associated with JC virus infection of neurons and meningeal cells: no longer a gray area. Curr Opin Neurol (2015) 28:288–94. doi: 10.1097/WCO.0000000000000201
11. Bofill-Mas S, Girones R. Role of the envirionment in the transmission of JC virus. J Neurovirol (2003) 9 Suppl 1:54–8. doi: 10.1080/13550280390195306
12. Kunitake T, Kitamura T, Guo J, Taguchi F, Kawabe K, Yogo Y. Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. J Clin Microbiol (1995) 33:1448–51. doi: 10.1128/jcm.33.6.1448-1451.1995
13. Pietropaolo V, Prezioso C, Bagnato F, Antonelli G. John Cunningham Virus: an overview on biology and disease of the etiological agent of the progressive multifocal leukoencephalopathy. New Microbiol (2018) 41:179–86.
14. Major EO, Amemiya K, Tornatore CS, Houff SA, Berger JR. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev (1992) 5:49–73. doi: 10.1128/CMR.5.1.49
15. Atwood WJ, Amemiya K, Traub R, Harms J, Major EO. Interaction of the human polyomavirus, JCV, with human b-lymphocytes. Virology (1992) 190:716–23. doi: 10.1016/0042-6822(92)90909-9
16. Suzuki S, Sawa H, Komagome R, Orba Y, Yamada M, Okada Y, et al. Broad distribution of the JC virus receptor contrasts with marked cellular restriction of virus replication. Virology (2001) 286:100–12. doi: 10.1006/viro.2001.0972
17. Schaumburg C, O'Hara BA, Lane TE, Atwood WJ. Human embryonic stem cell-derived oligodendrocyte progenitor cells express the serotonin receptor and are susceptible to JC virus infection. J Virol (2008) 82:8896–9. doi: 10.1128/JVI.00406-08
18. Nukuzuma S, Nakamichi K, Nukuzuma C, Takegami T. Inhibitory effect of serotonin antagonists on JC virus propagation in a carrier culture of human neuroblastoma cells. Microbiol Immunol (2009) 53:496–501. doi: 10.1111/j.1348-0421.2009.00156.x
19. Maginnis MS, Haley SA, Gee GV, Atwood WJ. Role of n-linked glycosylation of the 5-HT2A receptor in JC virus infection. J Virol (2010) 84:9677–84. doi: 10.1128/JVI.00978-10
20. Dugan AS, Gasparovic ML, Atwood WJ. Direct correlation between sialic acidbinding and infection of cells by two human polyomaviruses (JC virus and BK virus). J Virol (2008) 82:2560–4. doi: 10.1128/JVI.02123-07
21. Eash S, Tavares R, Stopa EG, Robbins SH, Brossay L, Atwood WJ. Differential distribution of the JC virus receptor-type sialic acid in normal human tissues. Am J Pathol (2004) 164:419–28. doi: 10.1016/S0002-9440(10)63132-X
22. O'Hara BA, Atwood WJ. Interferon beta1-a and selective anti-5HT(2a) receptor antagonists inhibit infection of human glial cells by JC virus. Virus Res (2008) 132:97–103. doi: 10.1016/j.virusres.2007.11.002
23. Sawa H, Komagome R. The JC virus-like particle overlay assay. Methods Mol Biol (2005) 292:175–86.
24. Querbes W, Benmerah A, Tosoni D, Di Fiore PP, Atwood WJ. A JC virus-induced signal is required for infection of glial cells by a clathrin- and eps15-dependent pathway. J Virol (2004) 78:250–6. doi: 10.1128/JVI.78.1.250-256.2004
25. Mayberry CL, Soucy AN, Lajoie CR, DuShane JK, Maginnis MS. JC polyomavirus entry by clathrin-mediated endocytosis is driven by β-arrestin. J Virol (2019) 93:e01948–18. doi: 10.1128/JVI.01948-18
26. Enam S, Gan DD, White MK, Del Valle L, Khalili K. Regulation of humanneurotropic JCV in colon cancer cells. Anticancer Res (2006) 26:833–41.
27. Tada H, Khalili K. A novel sequence-specific DNA-binding protein, LCP-1, interacts with single-stranded DNA and differentially regulates early gene expression of the human neurotropic JC virus. J Virol (1992) 66:6885–92. doi: 10.1128/jvi.66.12.6885-6892.1992
28. Chen NN, Kerr D, Chang CF, Honjo T, Khalili K. Evidence for regulation of transcription and replication of the human neurotropic virus JCV genome by the human s(mu)bp-2 protein in glial cells. Gene (1997) 185:55–62. doi: 10.1016/S0378-1119(96)00630-0
29. Chang CF, Gallia GL, Muralidharan V, Chen NN, Zoltick P, Johnson E, et al. Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence- specific single-stranded DNA-binding protein puralpha. J Virol (1996) 70:4150 –6. doi: 10.1128/jvi.70.6.4150-4156.1996
30. Devireddy LR, Kumar KU, Pater MM, Pater A. BAG-1, a novel bcl-2-interactingprotein, activates expression of human JC virus. J Gen Virol (2000) 81:351–7.
31. DuShane JK, Berry CL, Wilczek MP, Nichols SL, Maginnis MS. JCPyV-induced MAPK aignaling activates transcription factors during infection. Int J Mol Sci (2019) 20:4779. doi: 10.3390/ijms20194779
32. Uleri E, Beltrami S, Gordon J, Dolei A, Sariyer IK. Extinction of tumor antigen expression by SF2/ASF in JCV-transformed cells. Genes Cancer (2011) 2:728–36. doi: 10.1177/1947601911424578
33. Chiang C, Dvorkin S, Chiang JJ, Potter RB, Gack MU. The small t antigen of JC virus antagonizes RIG-I-Mediated innate immunity by inhibiting TRIM25's RNA binding ability. mBio (2021) 12:e00620–21. doi: 10.1128/mBio.00620-21
34. Manley K, Gee GV, Simkevich CP, Sedivy JM, Atwood WJ. Microarray analysis of glial cells resistant to JCV infection suggests a correlation between viral infection and inflammatory cytokine gene expression. Virology (2007) 366:394–404. doi: 10.1016/j.virol.2007.05.016
35. Wollebo HS, Safak M, Del Valle L, Khalili K, White MK. Role for tumor necrosis factor-α in JC virus reactivation and progressive multifocal leukoencephalopathy. J Neuroimmunol (2011) 233:46–53. doi: 10.1016/j.jneuroim.2010.11.013
36. May D, Bellizzi A, Kassa W, Cipriaso JM, Caocci M, Wollebo HS. IFNα and β mediated JCPyV suppression through C/EBPβ-LIP isoform. Viruses (2021) 13:1937. doi: 10.3390/v13101937
37. Nukuzuma S, Nakamichi K, Kameoka M, Sugiura S, Nukuzuma C, Tasaki T, et al. TNF-α stimulates efficient JC virus replication in neuroblastoma cells. J Med Virol (2014) 86:2026–32. doi: 10.1002/jmv.23886
38. Stettner MR, Nance JA, Wright CA, Kinoshita Y, Kim WK, Morgello S, et al. SMAD proteins of oligodendroglial cells regulate transcription of JC virus early and late genes coordinately with the tat protein of human immunodeficiency virus type 1. J Gen Virol (2009) 90:2005–201. doi: 10.1099/vir.0.011072-0
39. Piña-Oviedo S, Khalili K, Del Valle L. Hypoxia inducible factor-1 alpha activation of the JCV promoter: role in the pathogenesis of progressive multifocal leukoencephalopathy. Acta Neuropathol (2009) 118:235–47. doi: 10.1007/s00401-009-0533-0
40. Kim SY, Choi EC, Woo Jo Y, Henson JW, Kim HS. Transcriptional activation of JC virus early promoter by phorbol ester and interleukin-1beta: Critical role of nuclear factor-1. Virology (2004) 327:60–9. doi: 10.1016/j.virol.2004.06.021
41. De-Simone FI, Sariyer R, Otalora YL, Yarandi S, Craigie M, Gordon J, et al. IFN-gamma inhibits JC virus replication in glial cells by suppressing T-antigen expression. PLoS One (2015) 10:e0129694. doi: 10.1371/journal.pone.0129694
42. Nukuzuma S, Nukuzuma C, Kameoka M, Sugiura S, Nakamichi K, Tasaki T, et al. CPT11 prevents virus replication in JCI cells persistently infected with JC polyomavirus. Microbiol Immunol (2017) 61:232–38. doi: 10.1111/1348-0421.12486
43. Orba Y, Sunden Y, Suzuki T, Nagashima K, Kimura T, Tanaka S, et al. Pharmacological cdk inhibitor r-roscovitine suppresses JC virus proliferation. Virology (2008) 370:173–83. doi: 10.1016/j.virol.2007.08.034
44. Jiang ZG, Cohen J, Shall LJ, Major EO. Hexadecyloxypropyl-cidofovir (CMX001) suppresses JC virus replication in human fetal brain SVG cell cultures. Antimicrob Agents Chemother (2010) 54:4723–32. doi: 10.1128/AAC.00837-10
45. Ashok A, Atwood WJ. Contrasting roles of endosomal pH and the cytoskeleton in infection of human glial cells by JC virus and simian virus 40. J Virol (2003) 77:1347–56. doi: 10.1128/JVI.77.2.1347-1356.2003
46. O'Hara BA, Gee GV, Haley SA, Morris-Love J, Nyblade C, Nieves C, et al. Teriflunomide inhibits JCPyV infection and spread in glial cells and choroid plexus epithelial cells. Int J Mol Sci (2021) 22:9809. doi: 10.3390/ijms22189809
47. Nukuzuma S, Kameoka M, Sugiura S, Nakamichi K, Nukuzuma C, Takegami T. Suppressive effect of PARP-1 inhibitor on JC virus replication in vitro. J Med Virol (2013) 85:132–7. doi: 10.1002/jmv.23443
48. Kim SY, Woo MS, Kim WK, Choi EC, Henson JW, Kim HS. Glial cell-specific regulation of the JC virus early promoter by histone deacetylase inhibitors. J Virol (2003) 77:3394–401. doi: 10.1128/JVI.77.6.3394-3401.2003
49. Orba Y, Suzuki T, Makino Y, Kubota K, Tanaka S, Kimura T, et al. Large T Antigen promotes JC virus replication in G2-arrested cells by inducing ATM- and ATR-mediated G2 checkpoint signaling. J Biol Chem (2010) 285:1544–54. doi: 10.1074/jbc.M109.064311
50. Ahye N, Bellizzi A, May D, Wollebo HS. The role of the JC virus in central nervous system tumorigenesis. Int J Mol Sci (2020) 21:6236. doi: 10.3390/ijms21176236
51. Saribas S, Safak M. A comprehensive proteomics analysis of the JC virus (JCV) large and small tumor antigen interacting proteins: Large T primarily targets the host protein complexes with v-ATPase and ubiquitin ligase activities while small t mostly associates with those having phosphatase and chromatin-remodeling functions. Viruses (2020) 12:1192. doi: 10.3390/v12101192
52. Bollag B, Kilpatrick LH, Tyagarajan SK, Tevethia MJ, Frisque RJ. JC virus T'135, T'136 and T'165 proteins interact with cellular p107 and p130 in vivo and influence viral transformation potential. J Neurovirol (2006) 12:428–42. doi: 10.1080/13550280601009553
53. Tyagarajan SK, Frisque RJ. Stability and function of JC virus large T antigen and t' proteins are altered by mutation of their phosphorylated threonine 125 residues. J Virol (2006) 80:2083–91. doi: 10.1128/JVI.80.5.2083-2091.2006
54. Meinke G, Phelan PJ, Kalekar R, Shin J, Archambault J, Bohm A, et al. Insights into the initiation of JC virus DNA replication derived from thecrystal structure of the T-antigen origin binding domain. PLoS Pathog (2014) 10:e1003966.
55. Kim J, Woolridge S, Biffi R, Borghi E, Lassak A, Ferrante P, et al. Members of the AP-1 family, c-jun and c-fos, functionally interact with JC virus early regulatory protein large T antigen. J Virol (2003) 77:5241–52. doi: 10.1128/JVI.77.9.5241-5252.2003
56. Craigie M, Regan P, Otalora YL, Sariyer IK. Molecular interplay between T-antigen and splicing factor, arginine/serine-rich 1 (SRSF1) controls JC virus gene expression in glial cells. Virol J (2015) 12:196. doi: 10.1186/s12985-015-0426-x
57. Sariyer IK, Khalili K. Regulation of human neurotropic JC virus replication by alternative splicing factor SF2/ASF in glial cells. PLoS One (2011) 6:e14630. doi: 10.1371/journal.pone.0014630
58. Sariyer IK, Sariyer R, Otte J, Gordon J. Pur-alpha induces JCV gene expression and viral replication by suppressing SRSF1 in glial cells. PLoS One (2016) 11:e0156819. doi: 10.1371/journal.pone.0156819
59. Staib C, Pesch J, Gerwig R, Gerber JK, Brehm U, Stangl A, et al. p53 inhibits JC virus DNA replication in vivo and interacts with JC virus large T-antigen. Virology (1996) 219:237–46. doi: 10.1006/viro.1996.0241
60. Beltrami S, Branchetti E, Sariyer IK, Otte J, Weaver M, Gordon J. Neurofibromatosis type 2 tumor suppressor protein, NF2, induces proteasome-mediated degradation of JC virus T-antigen in human glioblastoma. PLoS One (2013) 8:e53447.
61. Bellizzi A, White MK, Wollebo HS. Degradation of polyomavirus JC T-antigen by stress involves the LIP isoform of C/EBP. Cell Cycle (2015) 14:2075–9. doi: 10.1080/15384101.2015.1042631
62. Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J Virol (2001) 75:1476–86. doi: 10.1128/JVI.75.3.1476-1486.2001
63. Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, et al. The human polyoma JC virus agnoprotein acts as a viroporin. PloS Pathog (2010) 6:e1000801. doi: 10.1371/journal.ppat.1000801
64. Sami Saribas A, Abou-Gharbia M, Childers W, Sariyer IK, White MK, Safak M. Essential roles of Leu/Ile/Phe-rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology (2013) 443:161–76. doi: 10.1016/j.virol.2013.05.003
65. Hidaka K, Hojo K, Fujioka S, Nukuzuma S, Tsuda Y. Oligomerization of neutral peptides derived from the JC virus agnoprotein through a cysteine residue. Amino Acids (2015) 47:2205–13. doi: 10.1007/s00726-015-2004-3
66. Saribas AS, White MK, Safak M. Structure-based release analysis of the JC virus agnoprotein regions: a role for the hydrophilic surface of the major alpha helix domain in release. J Cell Physiol (2018) 233:2343–59. doi: 10.1002/jcp.26106
67. Coric P, Saribas AS, Abou-Gharbia M, Childers W, Condra JH, White MK, et al. Nuclear magnetic resonance structure of the human polyoma JC virus agnoprotein. J Cell Biochem (2017) 118:3268–80. doi: 10.1002/jcb.25977
68. Saribas AS, Datta PK, Safak M. A comprehensive proteomics analysis of JC virus agnoprotein-interacting proteins: Agnoprotein primarily targets the host proteins with coiled-coil motifs. Virology (2020) 540:104–18. doi: 10.1016/j.virol.2019.10.005
69. Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. J Virol (2006) 80:3893–903. doi: 10.1128/JVI.80.8.3893-3903.2006
70. Sariyer IK, Khalili K, Safak M. Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: Inhibition by small t antigen. Virology (2008) 375:464–79. doi: 10.1016/j.virol.2008.02.020
71. Safak M, Sadowska B, Barrucco R, Khalili K. Functional interaction between JC virus late regulatory agnoprotein and cellular y-box binding transcription factor, YB-1. J Virol (2002) 76:3828–38. doi: 10.1128/JVI.76.8.3828-3838.2002
72. Darbinyan A, Siddiqui KM, Slonina D, Darbinian N, Amini S, White MK, et al. Role of JC virus agnoprotein in DNA repair. J Virol (2004) 78:8593–600. doi: 10.1128/JVI.78.16.8593-8600.2004
73. Endo S, Okada Y, Orba Y, Nishihara H, Tanaka S, Nagashima K, et al. JC virus agnoprotein colocalizes with tubulin. J Neurovirol (2003) 9 Suppl 1:10–4. doi: 10.1080/13550280390195333
74. Okada Y, Endo S, Takahashi H, Sawa H, Umemura T, Nagashima K. Distribution and function of JCV agnoprotein. J Neurovirol (2001) 7:302–6. doi: 10.1080/13550280152537148
75. Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, Endo S, et al. Identification of FEZ1 as a protein that interacts with JC virus agnoprotein and microtubules: role of agnoprotein-induced dissociation of FEZ1 from microtubules in viral propagation. J Biol Chem (2005) 280:24948–56. doi: 10.1074/jbc.M411499200
76. Saxena R, Saribas S, Jadiya P, Tomar D, Kaminski R, Elrod JW, et al. Human neurotropic polyomavirus, JC virus, agnoprotein targets mitochondrion and modulates its functions. Virology (2021) 553:135–53. doi: 10.1016/j.virol.2020.11.004
77. Ou WC, Chen LH, Wang M, Hseu TH, Chang D. Analysis of minimal sequences on JC virus VP1 required for capsid assembly. J Neurovirol (2001) 7:298–301. doi: 10.1080/13550280152537139
78. Kobayashi S, Suzuki T, Igarashi M, Orba Y, Ohtake N, Nagakawa K, et al. Cysteine residues in the major capsid protein,Vp1, of the JC virus are important for protein stability and oligomer formation. PLoS One (2013) 8:e76668. doi: 10.1371/journal.pone.0076668
79. Gasparovic ML, Gee GV, Atwood WJ. JC virus minor capsid proteins Vp2 and Vp3 are essential for virus propagation. J Virol (2006) 80:10858–61. doi: 10.1128/JVI.01298-06
80. Qu Q, Sawa H, Suzuki T, Semba S, Henmi C, Okada Y, et al. Nuclear entry mechanism of the human polyomavirus JC virus-like particle: Role of importins and the nuclear pore complex. J Biol Chem (2004) 279:27735–42. doi: 10.1074/jbc.M310827200
81. Lauver MD, Lukacher AE. JCPyV VP1 mutations in progressive multifocal leukoencephalo- pathy: Altering tropismor mediating immune evasion? Viruses (2020) 12:1156. doi: 10.3390/v12101156
82. Flores J, Anguiano O, Rivas-Alonso V, González-Conchillos H, Pérez-Saldivar M, Sotelo J, et al. Mutations in the John Cunningham virus VP1 gene could predispose to the development of progressive multifocal leukoencephalopathy in multiple sclerosis patients undergoing treatment with natalizumab. Mult Scler Relat Disord (2021) 56:103266. doi: 10.1016/j.msard.2021.103266
83. Dang X, Vidal JE, Penalva de Oliveira AC, Simpson DM, Morgello S, Hecht JH, et al. JC virus granule cell neuronopathy is associated with VP1 c terminus mutants. J Gen Virol (2012) 93:175–83. doi: 10.1099/vir.0.037440-0
84. Gorelik L, Reid C, Testa M, Brickelmaier M, Bossolasco S, Pazzi A, et al. Progressive multifocal leukoencephalopathy (PML) development is associated with mutations in JC virus capsid protein VP1 that change its receptor specificity. J Infect Dis (2011) 204:103–14. doi: 10.1093/infdis/jir198
85. Saribas AS, Mun S, Johnson J, El-Hajmoussa M, White MK, Safak M. Human polyoma JC virus minor capsid proteins, VP2 and VP3, enhance large T antigen binding to the origin of viral DNA replication: Evidence for their involvement in regulation of the viral DNA replication. Virology (2014) 449:1–16. doi: 10.1016/j.virol.2013.10.031
86. Huang YL, Wang M, Ou WC, Fung CY, Chen LS, Chang D. Analysis of DNA-binding activity of the JC virus minor capsid protein VP2. J Neurovirol (2003) 9 Suppl1:21–4. doi: 10.1080/13550280390195289
87. Ravichandran V, Jensen PN, Major EO. MEK1/2 inhibitors block basal and transforming growth factor 1beta1-stimulated JC virus multiplication. J Virol (2007) 81:6412–8. doi: 10.1128/JVI.02658-06
88. Zheng HC, Xue H, Jin YZ, Jiang HM, Cui ZG. The oncogenic effects, pathways and target molecules of JC polyoma virus T antigen in cancer cells. Front Oncol (2022) 12:744886. doi: 10.3389/fonc.2022.744886
89. Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol (2003) 77:10638–44. doi: 10.1128/JVI.77.19.10638-10644.2003
90. Henson JW, Schnitker BL, Lee TS, McAllister J. Cell-specific activation of the glial-specific JC virus early promoter by large T antigen. J Biol Chem (1995) 270:13240–5. doi: 10.1074/jbc.270.22.13240
91. Kim HS, Woo MS. Transcriptional regulation of the glial cell-specific JC virus by p53. Arch Pharm Res (2002) 25:208–13. doi: 10.1007/BF02976565
92. Gordon J, Del Valle L, Otte J, Khalili K. Pituitary neoplasia induced by expression of human neurotropic polyomavirus, JCV, early genome in transgenic mice. Oncogene (2000) 19:4840–6. doi: 10.1038/sj.onc.1203849
93. Krynska B, Gordon J, Otte J, Franks R, Knobler R, DeLuca A, et al. Role of cell cycle regulators in tumor formation in transgenic mice expressing the human neurotropic virus, JCV, early protein. J Cell Biochem (1997) 67:223–30. doi: 10.1002/(SICI)1097-4644(19971101)67:2<223::AID-JCB7>3.0.CO;2-Z
94. Caracciolo V, Reiss K, Crozier-Fitzgerald C, De Pascali F, Macaluso M, Khalili K, et al. Interplay between the retinoblastoma related pRb2/p130 and E2F-4 and -5 in relation to JCV-TAg. J Cell Physiol (2007) 212:96–104. doi: 10.1002/jcp.21005
95. Lassak A, Del Valle L, Peruzzi F, Wang JY, Enam S, Croul S, et al. Insulin receptor substrate 1 translocation to the nucleus by the human JC virus T-antigen. J Biol Chem (2002) 277:17231–8. doi: 10.1074/jbc.M110885200
96. Reiss K, Khalili K, Giordano A, Trojanek J. JC virus large T-antigen and IGF-I signaling system merge to affect DNA repair and genomic integrity. J Cell Physiol (2006) 206:295–300. doi: 10.1002/jcp.20455
97. Trojanek J, Ho T, Croul S, Wang JY, Chintapalli J, Koptyra M, et al. IRS-1-Rad51 nuclear interaction sensitizes JCV T-antigen positive medulloblastoma cells to genotoxic treatment. Int J Cancer (2006) 119:539–48. doi: 10.1002/ijc.21828
98. Reviriego-Mendoza MM, Frisque RJ. Interaction and co-localization of JC virus large T antigen and the f-box protein β-transducin-repeat containing protein. Virology (2011) 410:119–28. doi: 10.1016/j.virol.2010.10.038
99. Gan DD, Khalili K. Interaction between JCV large T-antigen and beta-catenin. Oncogene (2004) 23:483–90. doi: 10.1038/sj.onc.1207018
100. Bhattacharyya R, Noch EK, Khalili K. A novel role of Rac1 GTPase in JCV T-antigen- mediated beta-catenin stabilization. Oncogene (2007) 26:7628–36. doi: 10.1038/sj.onc.1210576
101. Sariyer IK, Merabova N, Patel PK, Knezevic T, Rosati A, Turco MC, et al. Bag3-induced autophagy is associated with degradation of JCV oncoprotein, T-Ag. PloS One (2012) 7:e45000. doi: 10.1371/journal.pone.0045000
102. Piña-Oviedo S, Urbanska K, Radhakrishnan S, Sweet T, Reiss K, Khalili K, et al. Effects of JC virus infection on anti-apoptotic protein survivin in progressive multifocal leukoencephalopathy. Am J Pathol (2007) 170:1291–304. doi: 10.2353/ajpath.2007.060689
103. Del Valle L, Sweet T, Parker-Struckhoff A, Perez-Liz G, Piña-Oviedo S. JCPyV T-antigen activation of the anti-apoptotic survivin promoter-its role in the development of progressive multifocal leukoencephalopathy. Viruses (2020) 12:1253. doi: 10.3390/v12111253
104. Noch E, Sariyer IK, Gordon J, Khalili K. JC virus T-antigen regulates glucose metabolic pathways in brain tumor cells. PloS One (2012) 7:e35054. doi: 10.1371/journal.pone.0035054
105. Wright PJ, Bernhardt G, Major EO, di Mayorca G. Comparison of the serology, transforming ability, and polypeptide composition of human papovaviruses isolated from urine. J Virol (1976) 17:762–75. doi: 10.1128/jvi.17.3.762-775.1976
106. Takemoto KK, Kanda T. Lymphotropic papovavirus transformation of hamster embryo cells. J Virol (1984) 50:100–5. doi: 10.1128/jvi.50.1.100-105.1984
107. Darbinyan A, White MK, Akan S, Radhakrishnan S, Del Valle L, Amini S, et al. Alterations of DNA damage repair pathways resulting from JCV infection. Virology (2007) 364:73–86. doi: 10.1016/j.virol.2007.02.015
108. Major EO, Vacante DA, Traub RG, London WT, Sever JL. Owl monkey astrocytoma cells in culture spontaneously produce infectious JC virus which demonstrates altered biological properties. J Virol (1987) 61:1435–41. doi: 10.1128/jvi.61.5.1435-1441.1987
109. London WT, Houff SA, Madden DL, Fuccillo DA, Gravell M, Wallen WC, et al. Brain tumors in owl monkeys inoculated with a human polyomavirus (JC virus). Science (1978) 201:1246–9. doi: 10.1126/science.211583
110. Ohsumi S, Ikehara I, Motoi M, Ogawa K, Nagashima K, Yasui K. Induction of undifferentiated brain tumors in rats by a human polyomavirus (JC virus). Jpn J Cancer Res (1985) 76:429–31.
111. Nagashima K, Yasui K, Kimura J, Washizu M, Yamaguchi K, Mori W. Induction of brain tumors by a newly isolated JC virus (Tokyo-1 strain). Am J Pathol (1984) 116:455–63.
112. Varakis J, ZuRhein GM, Padgett BL, Walker DL. Induction of peripheral neuroblastomas in syrian hamsters after injection as neonates with JC virus, a human polyoma virus. Cancer Res (1978) 38:1718–22.
113. Padgett BL, Walker DL, ZuRhein GM, Varakis JN. Differential neurooncogenicity of strains of JC virus, a human polyoma virus, in newborn Syrian hamsters. Cancer Res (1977) 37:718–20.
114. Del Valle L, Khalili K. Induction of brain tumors by the archetype strain of human neurotropic JCPyV in a transgenic mouse model. Viruses (2021) 13:162. doi: 10.3390/v13020162
115. Krynska B, Otte J, Franks R, Khalili K, Croul S. Human ubiquitous JCV(CY)T-antigen gene induces brain tumors in experimental animals. Oncogene (1999) 18:39–46. doi: 10.1038/sj.onc.1202278
116. Shollar D, Del Valle L, Khalili K, Otte J, Gordon J. JCV T-antigen interacts with the neurofibromatosis type 2 gene product in a transgenic mouse model of malignant peripheral nerve sheath tumors. Oncogene (2004) 23:5459–67. doi: 10.1038/sj.onc.1207728
117. Noguchi A, Kikuchi K, Ohtsu T, Yoshiwara M, Nakamura Y, Miyagi Y, et al. Pulmonary tumors associated with the JC virus T-antigen in a transgenic mouse model. Oncol Rep (2013) 30:2603–8. doi: 10.3892/or.2013.2782
118. Gou WF, Zhao S, Shen DF, Yang XF, Liu YP, Sun HZ, et al. The oncogenic role of JC virus T antigen in lens tumors without cell specificity of alternative splicing of its intron. Oncotarget (2015) 6:8036–45. doi: 10.18632/oncotarget.3507
119. Zheng HC, Xue H EY, Sun HZ, Cui ZG. The oncogenic roles of John Cunningham virus T antigen in digestive epithelial cells with tissue specificity BMC Cancer in press.
120. Kutsuna T, Zheng H, Abdel-Aziz HO, Murai Y, Tsuneyama K, Furuta I, et al. High JC virus load in tongue carcinomas be a risk factor for tongue tumorigenesis. Virchows Arch (2008) 452:405–10. doi: 10.1007/s00428-007-0534-0
121. Hämetoja H, Hagström J, Haglund C, Bäck L, Mäkitie A, Syrjänen S. Polyomavirus JCPyV infrequently detectable in adenoid cystic carcinoma of the oral cavity and the airways. Virchows Arch (2019) 475:609–16. doi: 10.1007/s00428-019-02617-6
122. Del Valle L, White MK, Enam S, Piña Oviedo S, Bromer MQ, Thomas RM, et al. Detection of JC virus DNA sequences and expression of viral T antigen and agnoprotein in esophageal carcinoma. Cancer (2005) 103:516–27. doi: 10.1002/cncr.20806
123. Murai Y, Zheng HC, Abdel Aziz HO, Mei H, Kutsuna T, Nakanishi Y, et al. High JC virus load in gastric cancer and adjacent non-cancerous mucosa. Cancer Sci (2007) 98:25–31. doi: 10.1111/j.1349-7006.2006.00354.x
124. Ksiaa F, Ziadi S, Mokni M, Korbi S, Trimeche M. The presence of JC virus in gastric carcinomas correlates with patient's age, intestinal histological type and aberrant methylation of tumor suppressor genes. Mod Pathol (2010) 23:522–30. doi: 10.1038/modpathol.2009.184
125. Hori R, Murai Y, Tsuneyama K, Abdel-Aziz HO, Nomoto K, Takahashi H, et al. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Arch (2005) 447:723–30. doi: 10.1007/s00428-005-0014-3
126. Shavaleh R, Kamandi M, Feiz Disfani H, Mansori K, Naseri SN, Rahmani K, et al. Association between JC virus and colorectal cancer: Systematic review and meta-analysis. Infect Dis (Lond) (2020) 52:152–60. doi: 10.1080/23744235.2019.1692145
127. Nosho K, Shima K, Kure S, Irahara N, Baba Y, Chen L, et al. JC virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia (2009) 11:87–95. doi: 10.1593/neo.81188
128. Link A, Shin SK, Nagasaka T, Balaguer F, Koi M, Jung B, et al. JC virus mediates invasion and migration in colorectal metastasis. PLoS One (2009) 4:e8146. doi: 10.1371/journal.pone.0008146
129. Vilkin A, Niv Y. Association between hMLH1 hypermethylation and JC virus (JCV) infection in human colorectal cancer (CRC). Clin Epigenet (2011) 2:1–5. doi: 10.1007/s13148-010-0013-3
130. Ripple MJ, Parker Struckhoff A, Trillo-Tinoco J, Li L, Margolin DA, McGoey R, et al. Activation of c-myc and cyclin D1 by JCV T-antigen and β-catenin in colon cancer. PLoS One (2014) 9:e106257. doi: 10.1371/journal.pone.0106257
131. Abdel-Aziz HO, Murai Y, Hong M, Kutsuna T, Takahashi H, Nomoto K, et al. Detection of the JC virus genome in lung cancers: Possible role of the T-antigen in lung oncogenesis. Appl Immunohistochem Mol Morphol (2007) 15:394–400. doi: 10.1097/01.pai.0000213126.96590.64
132. Zheng H, Abdel Aziz HO, Nakanishi Y, Masuda S, Saito H, Tsuneyama K, et al. Oncogenic role of JC virus in lung cancer. J Pathol (2007) 212:306–15. doi: 10.1002/path.2188
133. Knöll A, Stoehr R, Jilg W, Hartmann A. Low frequency of human polyomavirus BKV and JCV DNA in urothelial carcinomas of the renal pelvis and renal cell carcinomas. Oncol Rep (2003) 10:487–91.
134. Shen C, Tung C, Chao C, Jou Y, Huang S, Meng M, et al. The differential presence of human polyomaviruses, JCPyV and BKPyV, in prostate cancer and benign prostate hypertrophy tissues. BMC Cancer (2021) 21:1141. doi: 10.1186/s12885-021-08862-w
135. Zheng HC EY, Cui ZG, Zhao S, Zhang Y. The oncogenic roles of JC virus T antigen in breast carcinogenesis. Front Mol Biosci (2021) 8:687444. doi: 10.3389/fmolb.2021.687444
136. Zheng HC, Yang N, Yun WJ, Xue H, Cui ZG. The oncogenic roles of JC polympavirus T antigen in hepatocellular and pancreatic cancers. Cancer J in press.
137. Zheng HC, Hu CY, Xue H, Yang N, Yun WJ, Cui ZG. The oncogenic roles of JC polyomavirus T antigen on cervical cancer. BMC Cancer in press.
Keywords: JC polyomavirus, cancer, oncogenesis, signal pathway, virus replication, virus infection
Citation: Zheng H-c, Xue H and Zhang C-y (2022) The oncogenic roles of JC polyomavirus in cancer. Front. Oncol. 12:976577. doi: 10.3389/fonc.2022.976577
Received: 23 June 2022; Accepted: 05 August 2022;
Published: 23 September 2022.
Edited by:
Bangshun He, Nanjing Medical University, ChinaCopyright © 2022 Zheng, Xue and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua-chuan Zheng, emhlbmdfaHVhY2h1YW5AaG90bWFpbC5jb20=
 Hua-chuan Zheng
Hua-chuan Zheng Hang Xue
Hang Xue Cong-yu Zhang2
Cong-yu Zhang2