- 1Center of Stomatology, The Second Affiliated Hospital of Soochow University, Suzhou, China
- 2Department of Radiation Oncology, Suzhou Science & Technology Town Hospital, Suzhou, China
- 3Department of Oncology, The Second Affiliated Hospital of Soochow University, Suzhou, China
- 4Department of Radiotherapy & Oncology, The Second Affiliated Hospital of Soochow University, Suzhou, China
- 5Institute of Radiotherapy & Oncology, Soochow University, Suzhou, China
Background: Recent studies have highlighted the biomarker role of circulating miRNAs in oral squamous cell carcinoma (OSCC), indicating their potential application as early diagnostic markers for OSCC. However, the diagnostic results have proven inconclusive. This study was conducted to evaluate the diagnostic value of circulating miRNAs for OSCC diagnosis.
Methods: Eligible published studies were identified by a literature search carried out in several databases by using combinations of keywords associated with OSCC, circulating miRNAs, and diagnosis. The bivariate meta-analysis model was adopted to summarize the pooled parameters. Afterwards, we thoroughly explored the sources of heterogeneity after evaluating the risk of bias.
Results: A total of 60 studies focusing on 41 circulating miRNAs were included. The pooled sensitivity, specificity, and AUC were 0.75 (95%CI: 0.69-0.80), 0.76 (0.70-0.81), 0.82 (0.79-0.85), respectively. Subgroup analyses showed that miRNA combinations were more accurate than single miRNAs. Additionally, plasma may be a better matrix for miRNAs assays in OSCC diagnosis as the plasma-based miRNA assay had a higher level of diagnostic accuracy than serum-based miRNA assay. Subgroup analyses also suggested that using circulating miRNAs for OSCC diagnosis is more effective in Caucasians than in Asian ethnic groups. Finally, circulating miRNA assays based on large sample sizes have superior diagnostic accuracy than small sample sizes.
Conclusion: Circulating miRNAs might be applied as effective surrogate biomarkers for early diagnosis of OSCC. Nevertheless, future larger-scale prospective studies should be performed to enhance the diagnostic efficiency and investigate the miRNA combinations with more pronounced accuracy.
Introduction
Oral squamous cell carcinoma (OSCC), accounting for more than 90% of total oral cancer, is often diagnosed at advanced stages and is characterized by a low survival rate (1). Approximately, 50-70% of OSCC patients died within 5 years after they were diagnosed with OSCC due to frequent metastases to regional lymph nodes (2). Although surgical and medical treatments for OSCC have been rapidly developed, the therapeutic efficacy and the five-year survival rate remain unsatisfactory. Early detection of OSCC is vitally significant to enhance the survival outcome, as rates as high as 80-90% can be achieved in the early stages of OSCC patients (3). Currently, diagnosis of OSCC still remains a challenge as the procedures mainly depend on imaging and histological biopsy, which are invasive and uncomfortable (4). Although noninvasive screening tools have been employed, no alternative predictors have been proved effective (5). To this regard, there is a need to explore novel and effective biomarkers for more advancement to promote early detection of OSCC and improve the treatment options.
Over the last decades, microRNAs (miRNAs) have emerged as a new set of biomarkers which play a vital part in oral carcinogenesis (6). MiRNAs are a class of endogenous, non-coding, 18-25 nucleotide length single-strand RNAs that regulate the gene expression at the post-transcriptional level and are highly vital for cell growth and proliferation, mediating critical pathways involved in cancer initiation and progression (7). A plethora of reports have shown that abnormal expressions of specific miRNAs are associated with numerus human diseases, and miRNAs may function as oncogenes or tumor suppressors in various cancer types (8). So far, a significant number of studies have been performed to explore the biomarker role of miRNAs in different cancer types and evidence gathered has revealed the potential use of miRNAs as biomarkers in cancer detection and prognosis (9). Moreover, miRNAs are highly stable in rough conditions and their isolation and quantification have been found to be easy, convenient, and reproducible (10). Therefore, these unique properties make miRNAs as promising diagnostic and prognostic biomarkers in malignant tumors including OSCC which can start from non-invasive specimen collection avoiding postoperative pain.
As we all know, the initiation of OSCC may originate from a multistep accumulation of heterogeneous genetic alterations (11, 12). In the past decades, the expression patterns of miRNAs in OSCC have been recognized as new directions in the exploration of oral carcinogenesis (13). Recent reports have also shown a good association between specific miRNA expression and clinical parameters including tumor metastasis, relapse, and survival in OSCC patients (14, 15). Moreover, a number of studies have demonstrated that miRNAs play an important part during the initiation and progression of OSCC (16, 17). Prominently, miR-21 stands out to be frequently associated with OSCC and may serve as a potential biomarker for diagnosis, prognosis or therapeutic target. Therefore, circulating miRNAs may serve as useful biomarkers for early detection of OSCC (18). However, the diagnostic accuracy of miRNA profiles for OSCC was still inconsistent or even contradictory across different studies, which may be caused by different study designs, different sample sources, different sample sizes and different races.
In this study, we performed a comprehensive meta-analysis of previously published miRNA expression profiling studies to evaluate and summarize the clinical results in the literatures, regarding the potential application of circulating miRNAs in plasma or serum as biomarkers for OSCC diagnosis.
Materials and method
Guidelines and searches
This study was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guidelines. We have developed a comprehensive search strategy to retrieve all available studies from PubMed, Embase, and Web of Science by using different combinations of keywords including oral, tumor, carcinoma, cancer, squamous cell carcinoma, OSCC, microRNAs, miRNAs, miR-, diagnosis, diagnostic, detect, detection, screen, screening, sensitivity, specificity, and ROC curve. The database was searched from its inception up to April 25, 2022. In addition, the reference lists of relevant reviews were independently scanned to avoid missing any potential studies.
Eligibility criteria
To be eligible for enrollment in the analysis, the studies had to comply with the following inclusion criteria: (1) original studies focused on the diagnostic value of circulating miRNAs in OSCC; (2) the OSCC diagnosis was achieved based on histopathology as the reference test; (3) studies detected the expression of miRNAs in whole blood, plasma, or serum; (4) the studies provided sufficient data for further calculation, including parameters including specificity, sensitivity, and area under the receiver operating characteristic (ROC) curve. Studies were excluded according to these exclusion criteria as follows: (1) studies were published as review articles, meta-analysis, letters, commentaries, or abstracts; (2) studies failed to provide sufficient information to allow construction of two-by-two table; (3) studies were performed on species other than humans; (4) the language was non-English.
Data extraction and quality assessment
Two investigators (H.G. and Y.S.) independently reviewed the full texts of included studies and gathered baseline characteristic and clinical diagnostic data including (1) study characteristics: first author, year of study, and country; (2) patients’ demographic characteristics: ethnicity, number, mean age, and TNM stage; (3) miRNA features: miRNA signatures, detection methods, and sample types; (4) data used for further analyses: sensitivity and specificity or true positives (TP), true negatives (TN), false positives (FP), false negatives (FN), and area under the curve (AUC) values. In the present study, the qualities of studies were assessed independently by two reviewers using the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) criteria (19). If disagreements between the two reviewers occur, they will discuss together to achieve a consensus or consult with the third reviewer (Q.P.).
Statistical analysis
The bivariate meta-analysis model was adopted to summarize the sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR), respectively, together with their 95% confidence intervals (95% CIs) (20). Forest plots of summary statistics were constructed using the data from the enrolled studies. Afterwards, the Summary ROC (SROC) curves, which summarize the sensitivity and specificity of each enrolled study for the assessment of the overall diagnostic performance, were also established, while the AUC values were achieved from the constructed SROC curves (21). The Cochrane’s Q test and I2 statistic were applied to judge the presence of statistical heterogeneity across studies (22). P-value less than 0.05 and I2 value more than 50% indicated an obvious heterogeneity existing in the current study. When statistical heterogeneity existed between studies, subgroup analysis, meta-regression analysis and sensitivity analysis were adopted to seek the possible sources of heterogeneity (23). Moreover, the risk of publication bias of all the included diagnostic accuracy studies was measured by the Deeks’ funnel-plot asymmetry test (24). In our study, statistical analysis was undertaken using Stata 14.0 software. A p-value of <0.05 was considered statistically significant.
Results
Search results and characteristics of studies
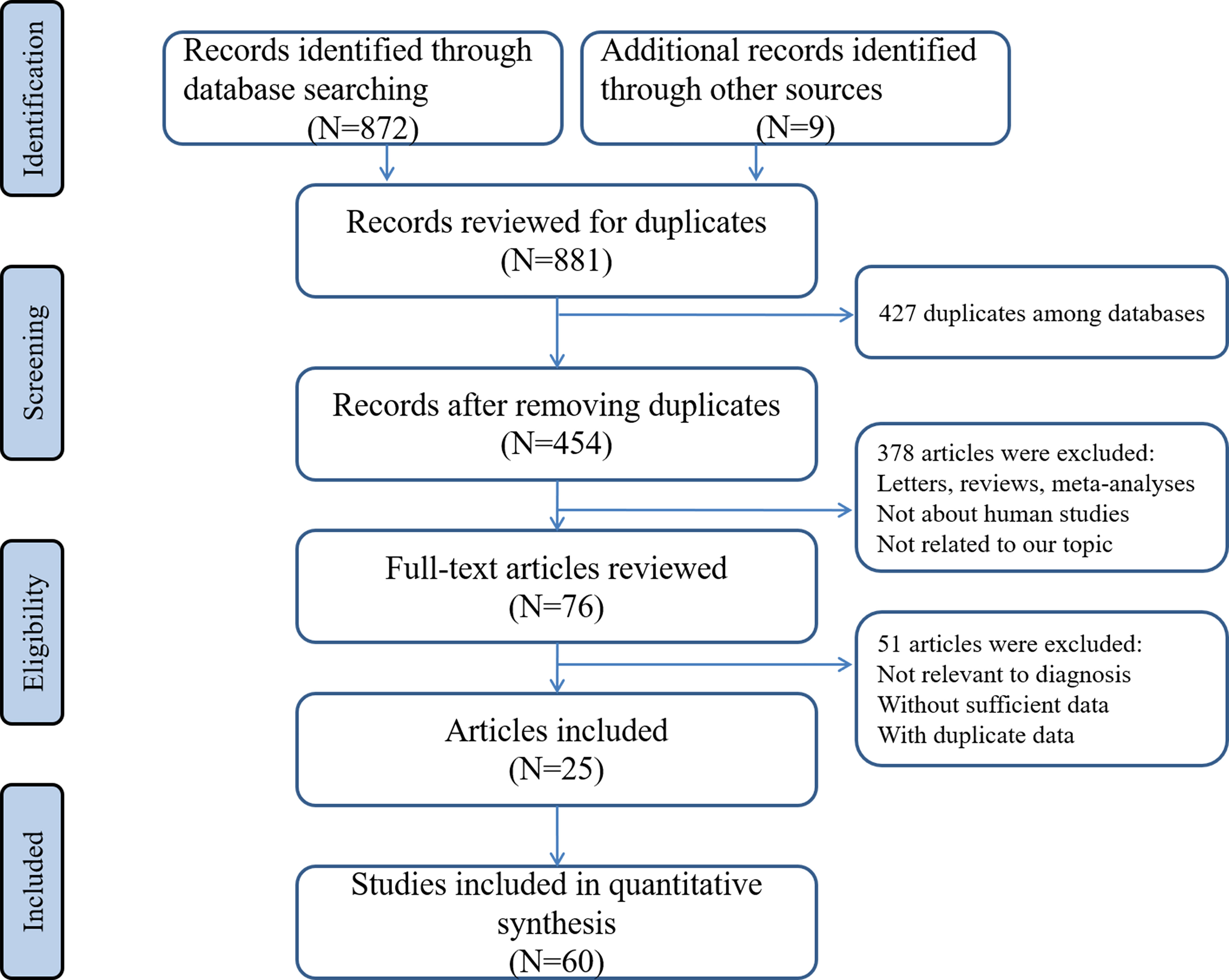
Through searching the electronic databases with different combinations of the above used keywords, avoiding obviously unrelated titles and abstracts, and after careful consideration of all collected studies, 25 articles involving 60 different miRNAs tests met all inclusion criteria and were included in the final analysis (25–49). A flowchart detailing the selection process was illustrated in Figure 1.
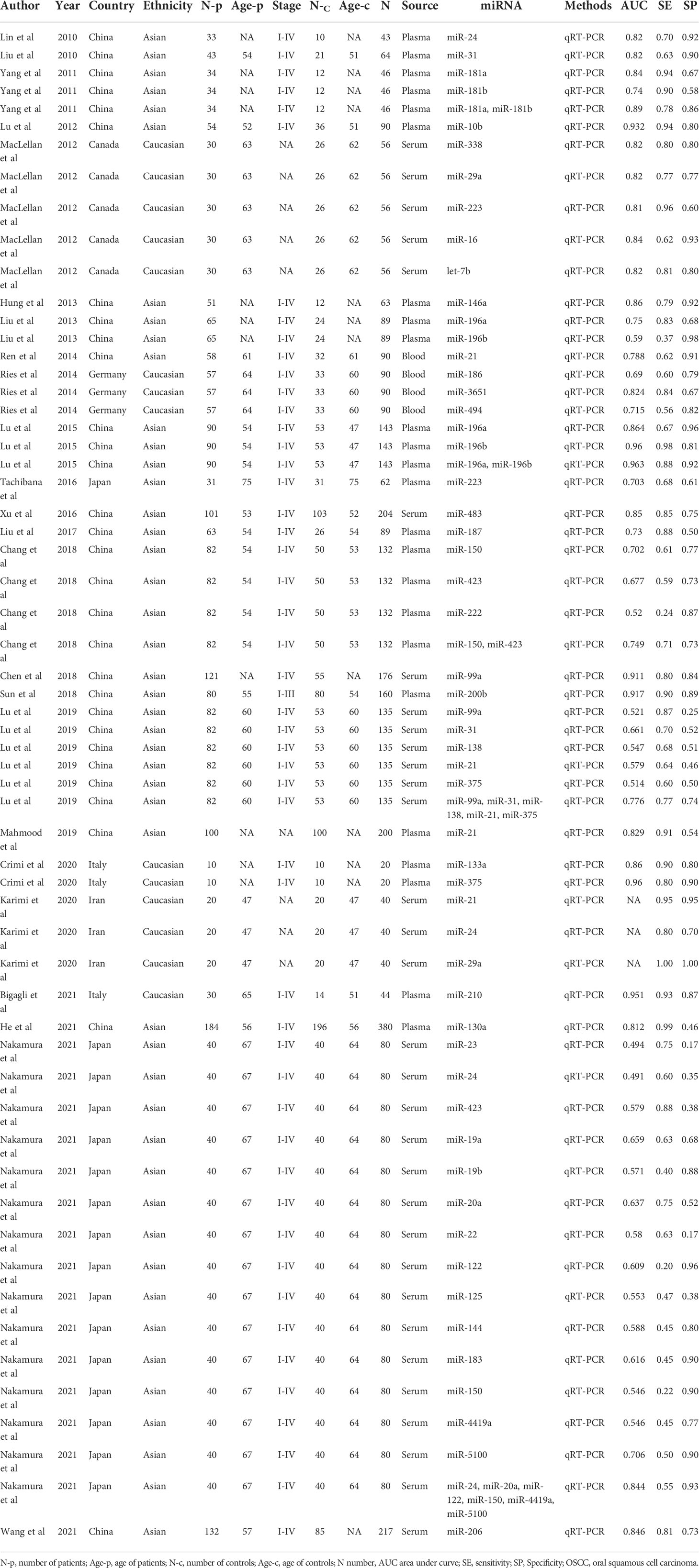
All the included studies were performed between 2010 and 2021. Thirty of the selected studies were from China, sixteen from Japan, five from Canada, three from Germany, three from Italy and three from Iran. The number of samples evaluated by each study ranged from 20 to 380. Concerning the type of biological specimen, 32 studies collected from serum, 24 studies from plasma, while 4 studies were collected from whole blood. A total of 55 studies focused on single miRNAs while 5 studies focused on miRNA combinations. All included studies used quantitative real-time reverse transcription-PCR (qRT-PCR) technique to detect the expression of miRNAs. According to the QUADAS-2 assessment tool, the overall analytical quality was acceptable. Details of the enrolled studies in the current study were presented at Table 1.
Diagnostic accuracy of circulating miRNAs for OSCC
The forest plots describing the pooled sensitivity and specificity for circulating miRNA profiling in distinguishing OSCC patients from normal participants were illustrated at Figure 2. Overall, the circulating miRNAs indicated good performances for the detection of OSCC with the pooled sensitivity of 0.75 (95% CI: 0.69-0.80) and the pooled specificity of 0.76 (0.70-0.81). Moreover, the combined PLR, NLR, and DOR were estimated to be 3.2 (2.5-4.0), 0.33 (0.27-0.41), and 9 (6-14), respectively. In addition, the SROC curve of circulating miRNAs in detecting OSCC was presented at Figure 3A, and the AUC was calculated to be 0.82 (0.79-0.85).
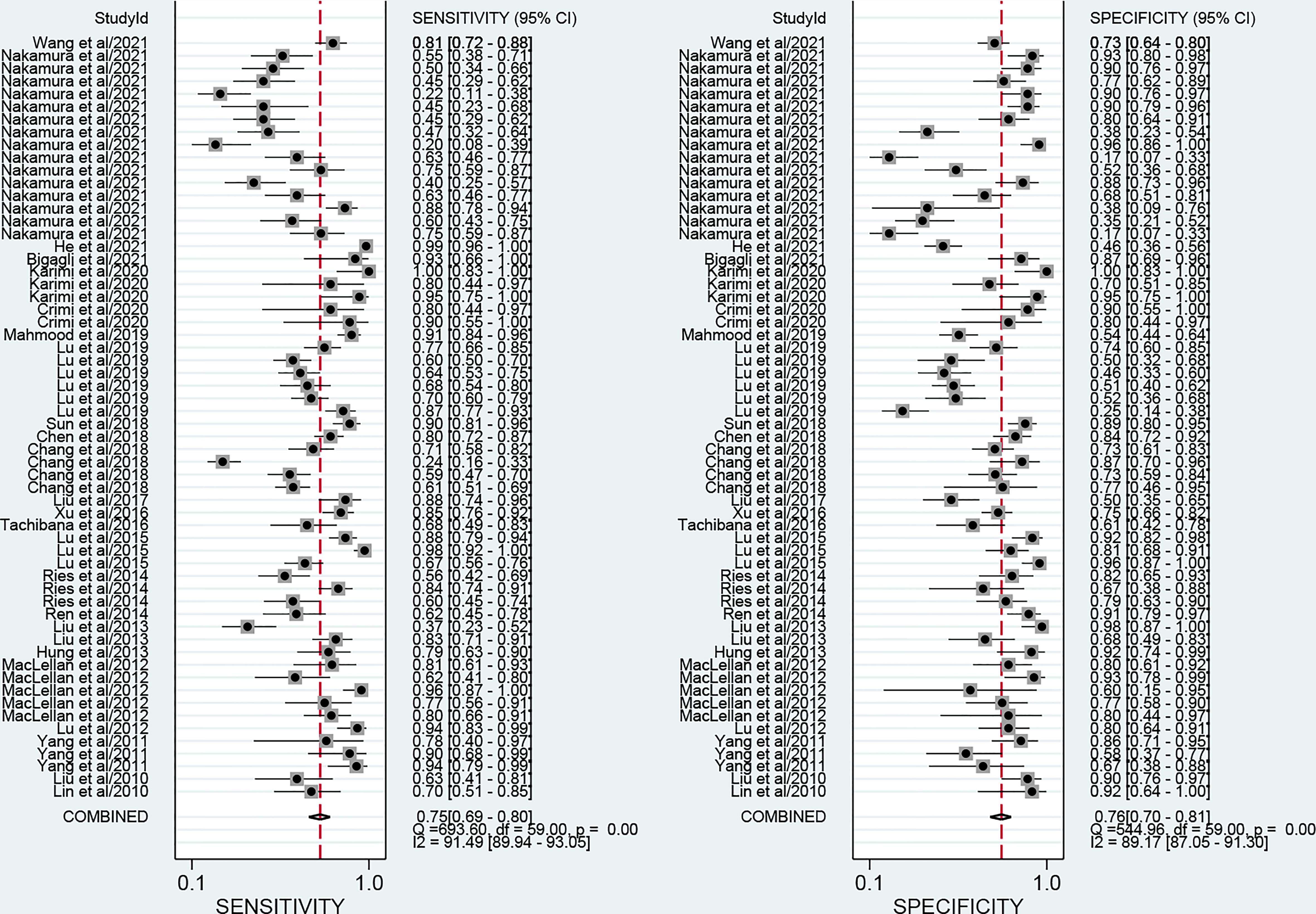
Figure 2 Forest plots of the pooled sensitivity and specificity for circulating miRNAs in detecting OSCC.
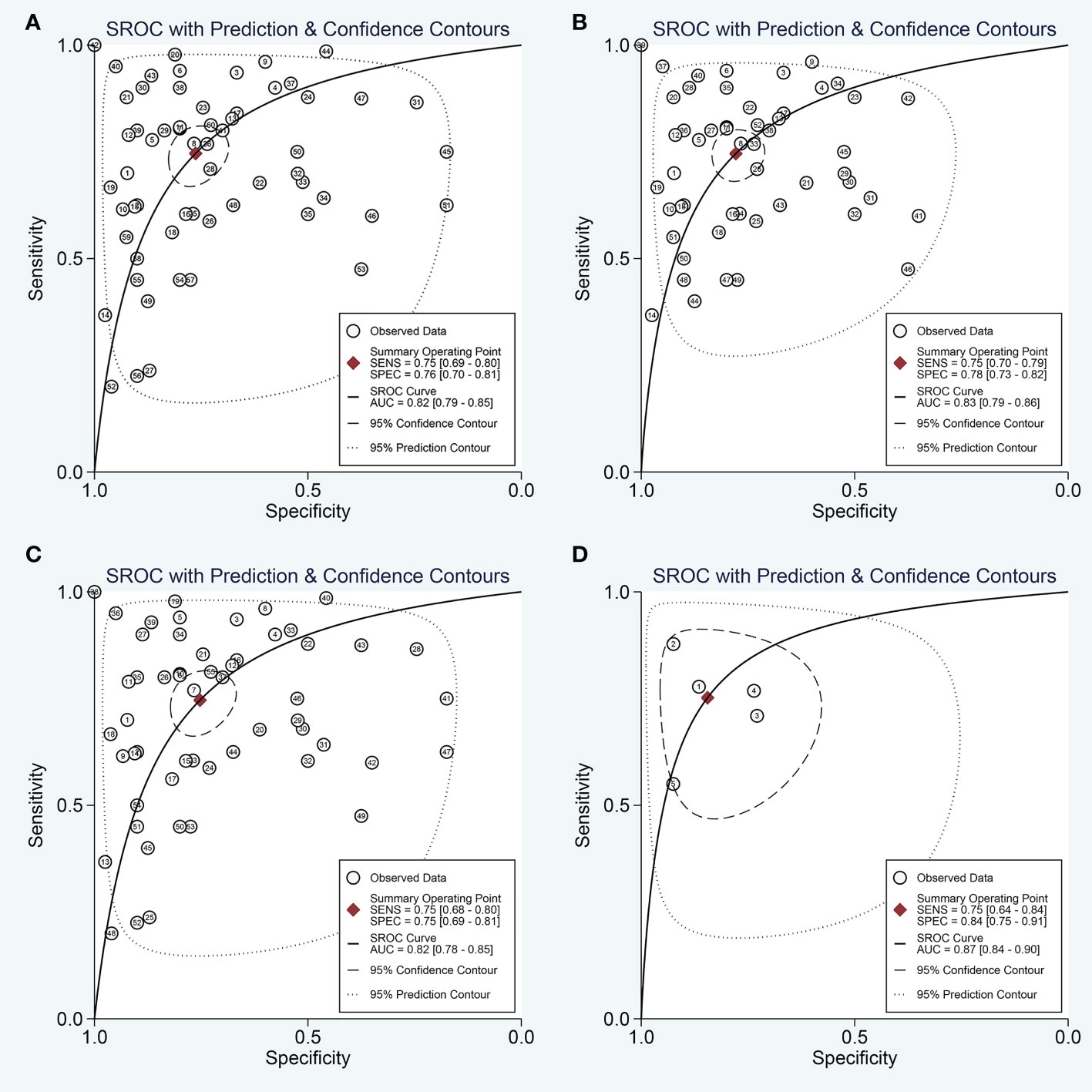
Figure 3 The SROC curve of the pooled analysis. (A) SROC curve of overall including the outliers for circulating miRNAs; (B) SROC curve of outliers excluded for circulating miRNAs; (C) SROC curve for circulating single miRNAs; (D) SROC curve for circulating combination miRNAs.
Then, the Fagan’s nomogram was utilized to confirm the clinical utility of the circulating miRNAs in OSCC. As illustrated in Figure 4, when the pre-test probability of OSCC was 20%, a positive measurement could improve the post-test probability of suffering cancer to 44%, while the post-test probability will reduce to 8% if a negative measurement happened.
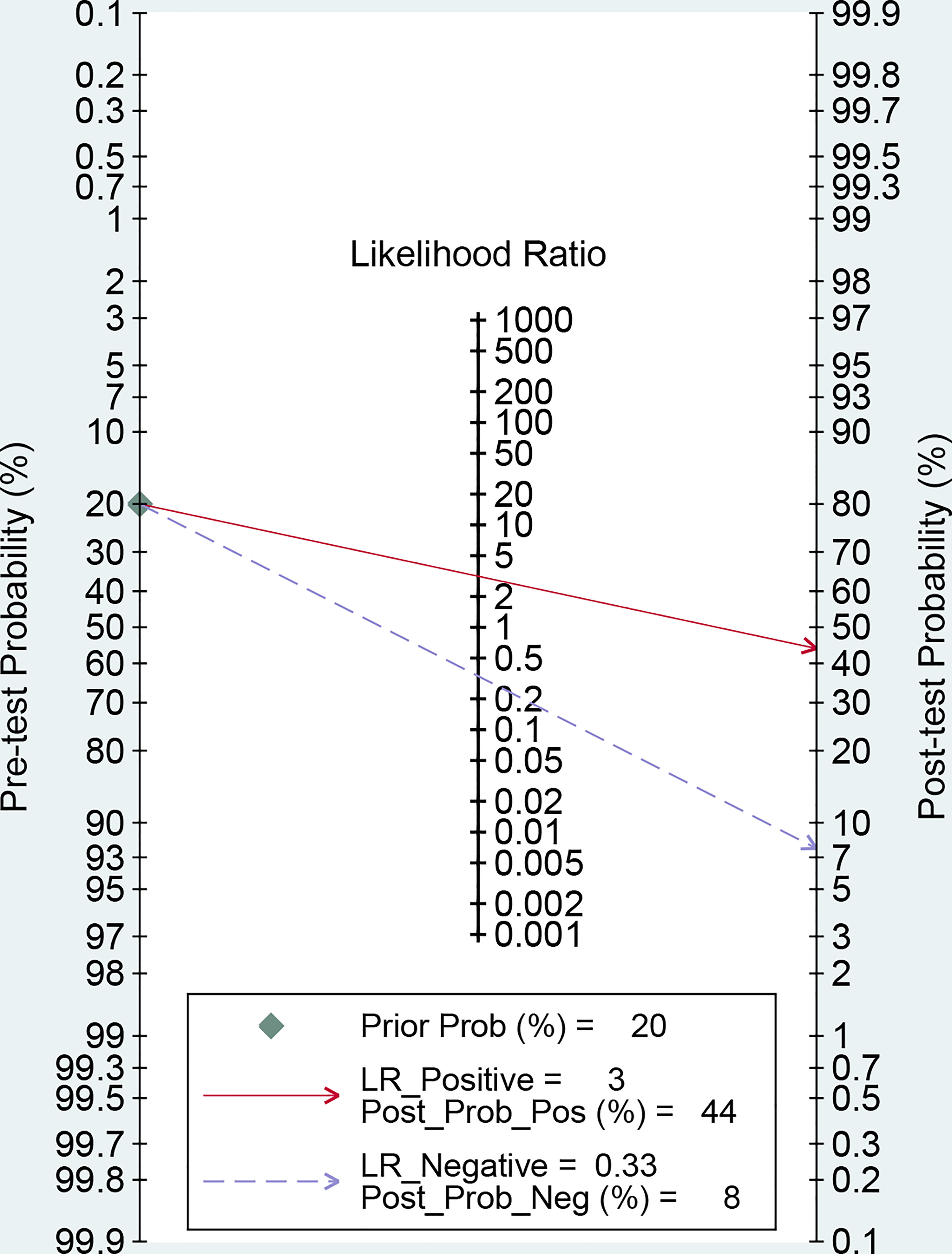
Figure 4 Fagan’s monogram evaluating the clinical utility of circulating miRNAs for differentiating OSCC patients.
The above parameters revealed moderate discriminative ability of the application of circulating miRNAs as biomarkers for the early diagnosis of OSCC.
Heterogeneity and subgroup analyses
In this study, the heterogeneity analysis using I2 suggested significantly higher heterogeneity in both pooled sensitivity (I2 = 91.49%; P<0.001) and pooled specificity (I2 = 89.17%; P<0.001). We first explored whether the heterogeneity may come from the threshold effect. The Spearman correlation coefficient between the sensitivity logarithm and (1-specificity) logarithm is calculated to be -0.24, and the P-value is 0.06, suggesting no obvious threshold effect.
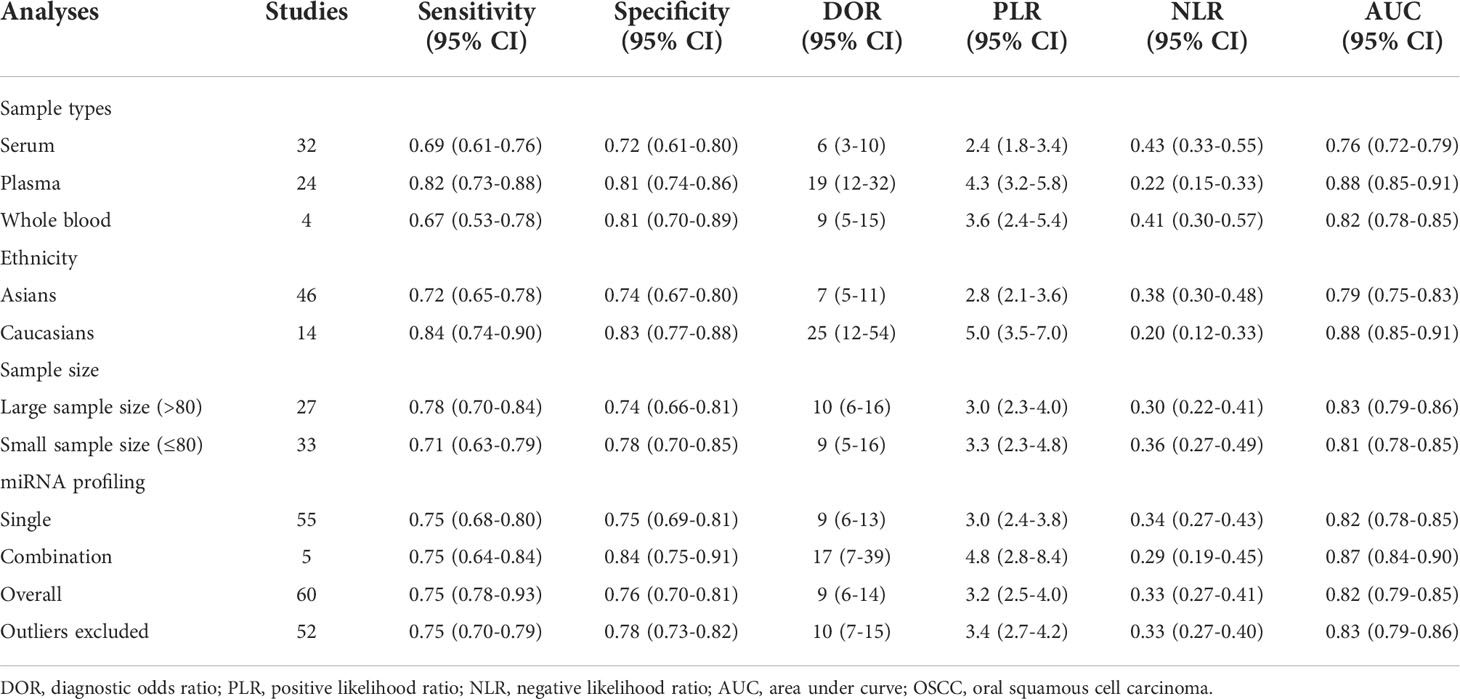
Next, the subgroup analyses were carried out. As exemplified by Table 2, miRNA combination testing achieved a more overall promising accuracy than single miRNA assay, with a similar sensitivity of 0.75 (0.64-0.84) versus 0.75 (0.68-0.80), a higher specificity of 0.84 (0.75-0.91) versus 0.75 (0.69-0.81), a higher AUC of 0.87 (0.84-0.90) versus 0.82 (0.78-0.85), respectively. The SROC curves of single miRNAs and combination miRNAs were plotted at Figures 3C, D, respectively. Following subgroup analysis depending on sample sources, the plasma-based miRNA assay exhibited a higher level of diagnostic accuracy than serum-based miRNA assay accompanied by a pooled sensitivity of 0.82 (0.73-0.88) versus 0.69 (0.61-0.76), specificity of 0.81 (0.74-0.86) versus 0.72 (0.61-0.80) and AUC of 0.88 (0.85-0.91) versus 0.76 (0.72-0.79). Moreover, the diagnostic accuracy based on ethnicity was also assessed through subgroup analysis. The results indicated that the sensitivity, specificity, and AUC of circulating miRNAs for OSCC detection in the Caucasian group were higher than those in the Asian group due to higher sensitivity: 0.84 (0.74-0.90) versus 0.72 (0.65-0.78), higher specificity: 0.83 (0.77-0.88) versus 0.74 (0.67-0.80), and higher AUC: 0.88 (0.85-0.91) versus 0.79 (0.75-0.83). In addition, we also conducted a subgroup analysis to identify whether sample size had a potential influence on the whole diagnostic value of circulating miRNAs. The stratified analysis by sample size suggested that the diagnostic value of miRNAs based on large sample sizes (N>80) might be more significant than it is for small sample sizes (N ≤ 80), with a pooled sensitivity of 0.78 (0.70-0.84) versus 0.71 (0.63-0.79), specificity of 0.74 (0.66-0.81) versus 0.78 (0.70-0.85) and AUC of 0.83 (0.79-0.86) versus 0.81 (0.78-0.85). The SROC curves of these subgroups were also plotted at Figure 5.
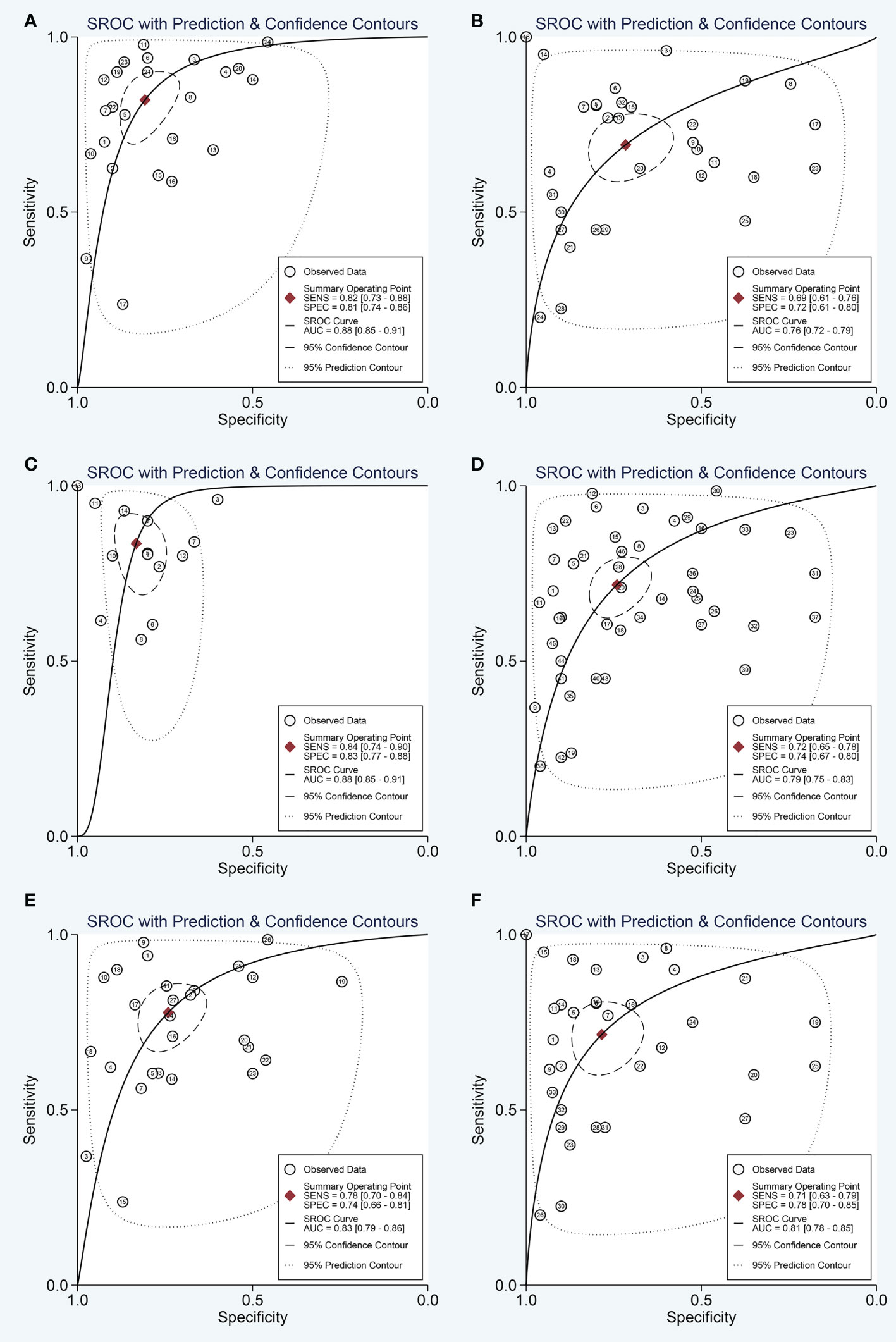
Figure 5 The SROC curve of the pooled analysis by some subgroups. (A) SROC curve for plasma miRNAs; (B) SROC curve for serum miRNAs; (C) SROC curve for circulating miRNAs based on Caucasian population; (D) SROC curve for circulating miRNAs based on Asian population; (E) SROC curve for circulating miRNAs based on large sample sizes; (F) SROC curve for circulating miRNAs based on small sample sizes.
Influence analysis and meta-regression
As heterogeneity observed in the present study, sensitivity analysis was performed (Figure 6). The influence analysis and outlier detection identified several outlier individual studies. After excluding seven outliers (34, 38, 41, 47, 48), the I2 for heterogeneity decreased both in sensitivity (from 91.49% to 84.30%) and specificity (from 89.17% to 84.31%). However, the overall results showed only minimal changes and were similar to the original ones as the pooled specificity of the overall study increased from 0.76 to 0.78, PLR increased from 3.2 to 3.4, DOR decreased from 9 to 10, and AUC increased from 0.82 to 0.83, while sensitivity and NLR had no changes. The SROC curve of outliers excluded for circulating miRNAs was plotted at Figure 3B. The above results suggested that our study was robust.
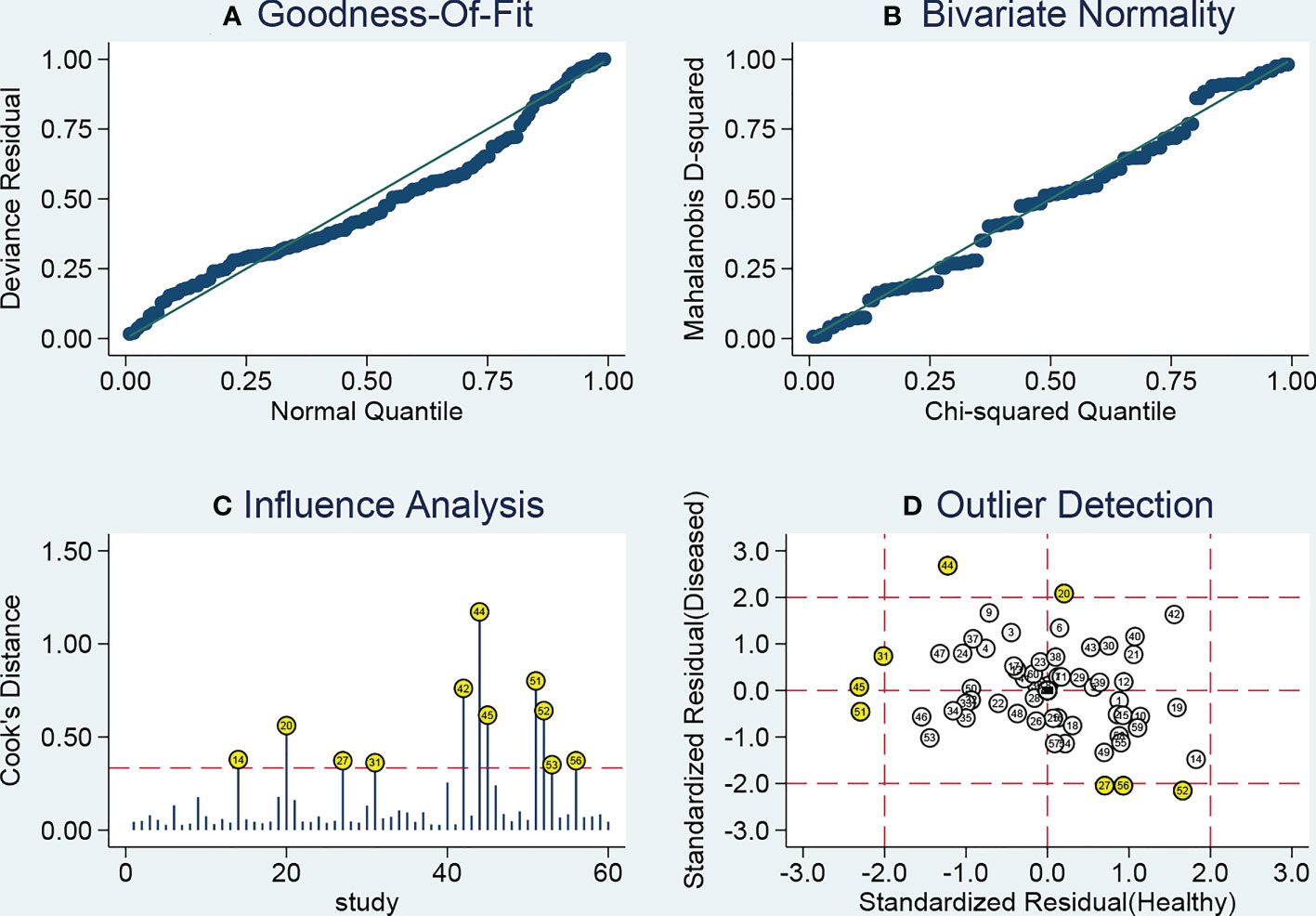
Figure 6 Sensitivity analysis results. (A) Goodness of fit; (B) bivariate normality; (C) influence analysis; (D) outlier detection.
Furthermore, a meta-regression analysis was applied to distinguish the potential sources of heterogeneity across studies by exploring research characteristics including specimen type, sample size, and ethnicity. The results suggested that ethnicity may be responsible for the heterogeneity while other factors reveled low likelihood of sources of inter-study heterogeneity.
Publication bias
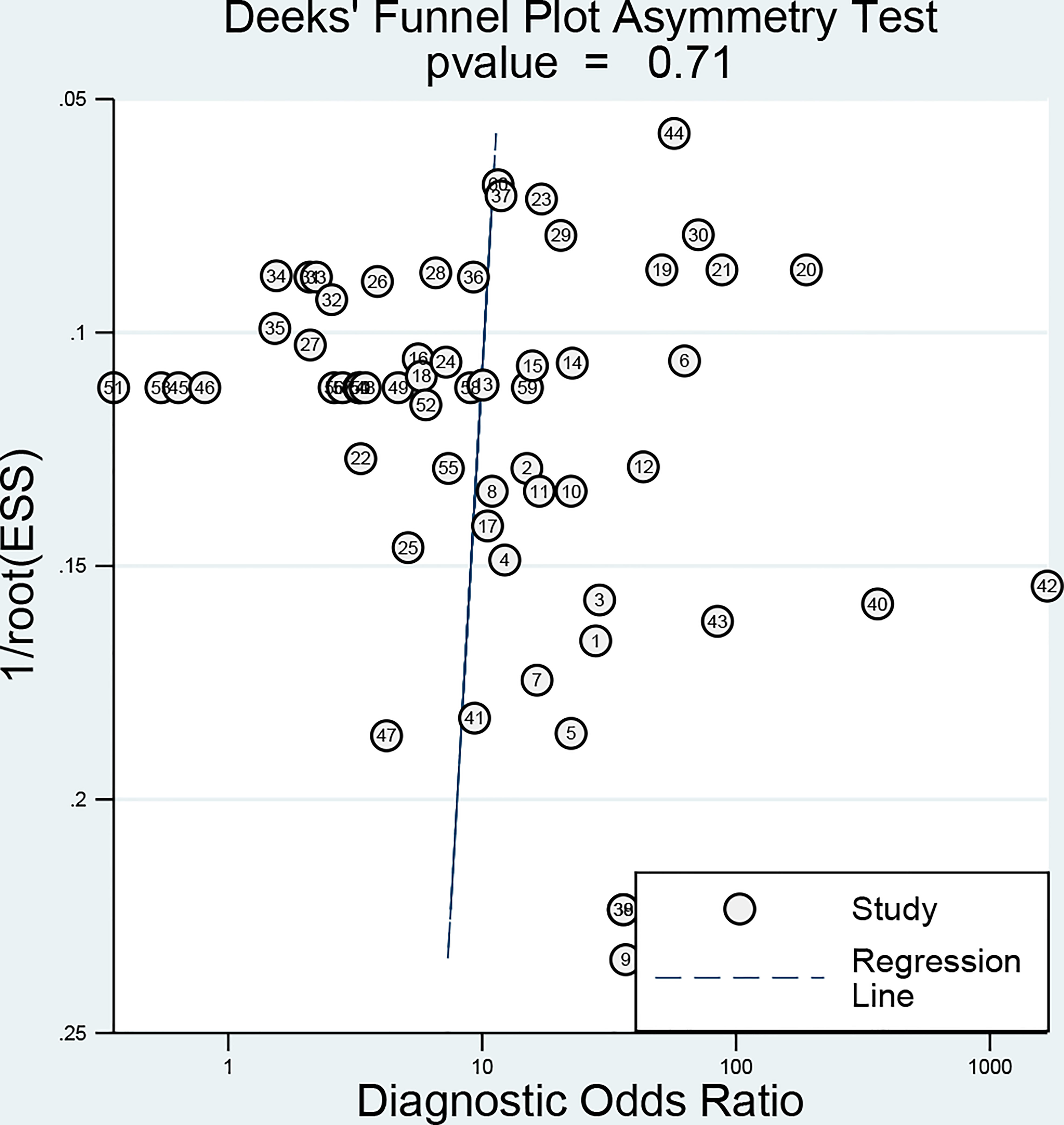
Deeks’ funnel-plot asymmetry test (Figure 7) was performed to investigate the potential publication bias. The assessment of publication bias indicated that the funnel plots were asymmetric with a slope coefficient P-value calculated to be 0.71 in the overall studies, indicating there was no obvious publication bias in the current study.
Discussion
Despite great advancement in the treatment modalities, the survival rate of patients with OSCC is still low. Early detection is of great clinical importance for the optimization of treatment strategies and improvement of long-term survival for OSCC patients. Although histopathological investigation is still recognized as the gold standard for clinicians, it is an invasive examination procedure that is temporally and spatially restricted and unable to indicate the whole molecular landscape of OSCC. Thus, it is indispensable to explore less invasive, reliable, and effective detection methods to enhance early OSCC detection. Currently, the miRNA detection in different biofluids has been explored as novel diagnostic biomarkers for OSCC. However, the clinical value of circulating miRNAs for OSCC diagnosis remains unclear. Due to the inconsistencies of previously published studies, we performed this comprehensive study to determine whether circulating miRNAs could be applied as a non-invasive and effective method in the early screening of OSCC.
To our knowledge, this is the most comprehensive study ever conducted on the role of circulating miRNAs in OSCC. A total of sixty studies were identified eligible and enrolled for the assessment of the diagnostic value of circulating miRNAs in OSCC. The overall analysis demonstrates moderate diagnostic accuracy as circulating miRNA profiling harbored a relatively high pooled diagnostic value in detecting OSCC, where the combined sensitivity and specificity were 0.75 and 0.76, respectively, corresponding with an AUC of 0.82. The stratified analysis by sample sources indicated that the plasma-based assay seemed to undergo a higher pooled sensitivity, specificity, and AUC than serum-based assay. Our results provided evidence that plasma may be a better matrix for diagnostic profiling of miRNAs in OSCC, which was also consistent with previous studies (50). Moreover, previous studies have suggested that genetic background may have some effect on the miRNA concentrations in body fluids or tumor lesions and the biomarker value of miRNAs varies among different ethnicities (51). In our pooled results by subgroup analysis, we found that the Caucasian population-based miRNA test harbored an overall higher accuracy compared than that of the Asians, suggesting that biomarker performance for circulating miRNAs in OSCC diagnosis may also be determined by different genetic background. Remarkably, the impact of genetic background on the diagnostic value of circulating miRNAs was also confirmed by a meta-regression analysis. Additionally, our previous study also reported that the diagnostic accuracies of miRNAs in different cancers were affected by sample sizes (52). More importantly, we observed a higher diagnostic accuracy in miRNA detection tests based on large sample size (n>80) than investigations with small sample sizes (n ≤ 80), which may suggest that future translational and clinical investigations concentrating on large-scale prospective studies are worth performing to validate the diagnostic efficacy of circulating miRNAs in OSCC.
Accumulating evidence described that tumorigenesis is a complex cell processes involving multiple miRNAs (53). For OSCC, it is a highly heterogeneous cancer with a complicated etiology and single miRNA is hard to diagnose OSCC with satisfactory performance (11). Combination biomarkers may be more comprehensive than single maker in dictating the complicated cancer evolutionary process and may be more powerful in cancer detection (54–56). There have been promising results yielded from studies that combination miRNAs had a higher level of diagnostic power than single miRNAs. However, since there were only five tests focusing on miRNA combination biomarkers in detecting OSCC, we could not answer the question that which and how many miRNAs should be combined together to enhance the diagnostic accuracy. This indicates that further research should be concentrated on the combined use of miRNA panels to investigate their effect on diagnostic accuracy.
There were 41 miRNAs utilized as diagnostic biomarkers involved in the present study, which may provide new insight into the early detection of OSCC. However, the exact role of miRNAs in the carcinogenesis of OSCC remains unclear. Recently, several researchers have demonstrated a functional role for miRNAs in the initiation and progression of OSCC, which may help us understand the potential biomarker role of miRNAs in OSCC. For example, Lin et al. reported that miR-31 played a pivotal part during the progression of OSCC through establishing a complicated network with its regulated genes and the signaling cascades such as EGF-AKT signaling axis, Hippo pathway, and Wnt signaling (57). Data from the study by Peng et al. revealed that downregulation of miR-130a could inhibit OSCC proliferation and metastasis by the Hippo-YAP pathway, which may provide potential target for OSCC therapy (58). Previous studies have also demonstrated that miR-133a plays tumor suppressive role in OSCC by inhibiting the Notch signaling pathway via binding to CTBP2 (59). Moreover, mechanism research has suggested that miR-222 affects OSCC cell proliferation, migration, invasion, and apoptosis by targeting CDKN1B (60). In addition, previous evidence has demonstrated that miR-144 inhibits tumorigenesis of OSCC by targeting ERO1L/STAT3 signaling pathway (61). Besides, available data suggest that the expression of miR-196a increased in OSCC cells against normal oral squamous cells and downregulation of miR-196a could inhibit the malignant biological processes of OSCC cells through targeting FOXO1 (62). In a word, the altered expressions of miRNAs and the involved pathways may contribute to the biological behaviors in OSCC, ultimately leading to tumorigenesis. However, as for more detailed mechanisms of miRNAs in OSCC, further studies are needed to make further investigations.
Since circulating miRNAs possess the distinctive advantages of tumor specificity, stable, extracted easily and non-invasive, they may be applied as perfect non-invasive biomarkers for cancer detection. Our study also demonstrated that circulating miRNAs may represent potentially promising biomarkers for the detection of OSCC. However, there is still a long way to go before they can be used in clinical practice in OSCC diagnosis. For example, different studies from different laboratories or testing platforms used diverse cut-off values and different normalization in the detection of miRNAs, which may potentially affect the diagnostic efficacy and generate heterogeneity and uncertainty. Therefore, a consensus in the scientific community should be indispensable to establish the optimum cut-off values and detection methods. Moreover, current studies on miRNAs detection in OSCC were isolated and the future exploration of miRNA biomarker for OSCC should be rooted in systematical and dynamical manner to develop integrative diagnostic models based on specific miRNAs and combinations with more appropriate and better prediction capacity. In addition, studies included in our studies are still far from enough and the size of available samples in the individual studies are still relatively small. Thus, large-scale and well-designed prospective randomized controlled studies are required to confirm the actual diagnostic value of circulating miRNA assays for OSCC and promote the clinical application. Moreover, we believe that large adequately designed prospective studies may help determine a specific miRNA type or specific miRNA combinations that may be more appropriate and better used in clinical decision-making for OSCC patients.
The present study has some limitations that must be acknowledged. First, we did observe significant heterogeneity because of discrepancies among the different studies. Although several common check methods of evaluating the risk of bias assessment including subgroup, meta-regression, and sensitivity analysis were performed, we were still unable to clarify the precise factors contributing to the significant heterogeneity. Second, since some clinical factors such as gender difference, age distribution, and TNM stage were not detailed in some studies, we could not carry out any other subgroup analysis based on them. Third, though we had planned to investigate the impact of cut-off value on heterogeneity by using the meta-regression analyses, this could not be accomplished due to insufficient data and the different standards in different enrolled diagnostic tests. Moreover, as discussed in the subgroup analyses and meta-regression analysis, ethnicity might be considered as a source of heterogeneity among individual studies. However, all the diagnostic miRNA tests were conducted on the basis of Asian and Caucasian population, whereas African population should have been enrolled. Finally, the history of tobacco and alcohol consumption may influence the study results. However, these data have not been provided by some of the included studies and we cannot perform such analysis. Regardless of these limitations, ours is the most comprehensive study that aggregated available data on circulating miRNAs and analyzed their application in the field of OSCC detection.
Conclusions
In conclusion, our results indicated that circulating miRNAs has a relatively high diagnostic accuracy in the detection of OSCC and might be applied in noninvasive screening tests for OSCC. In addition, combination miRNA biomarkers have exhibited a prominent advantage over single miRNAs in improving diagnostic accuracy of OSCC. However, further translational and clinical investigations based on large-scale prospective studies are necessary to validate the diagnostic efficacy of circulating miRNAs and promote their clinical application in OSCC.
Data availability statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Author contributions
HG, YS and ZF performed the computational analysis, and wrote the manuscript. YC, JY and YZ were responsible for the statistical analysis in meta-analysis part. QP conceived of the study, and took part in its design and coordination. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Suzhou Radiotherapy Clinical Medical Center (Szlcyxzx202103), Scientific Research Program for Young Talents of China National Nuclear Corporation (QP) and Leader Project of Clinical Technology Application Research for Jiangsu Gerontology (LR2021023).
Acknowledgments
We would like to thank the reviewers and editors for their constructive comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zini A, Czerninski R, Sgan-Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med (2010) 39(4):299–305. doi: 10.1111/j.1600-0714.2009.00845.x
2. Braakhuis BJ, Leemans CR, Visser O. Incidence and survival trends of head and neck squamous cell carcinoma in the Netherlands between 1989 and 2011. Oral Oncol (2014) 50(7):670–5. doi: 10.1016/j.oraloncology.2014.03.008
3. Chamoli A, Gosavi AS, Shirwadkar UP, Wangdale KV, Behera SK, Kurrey NK, et al. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol (2021) 121:105451. doi: 10.1016/j.oraloncology.2021.105451
4. Su YF, Chen YJ, Tsai FT, Li WC, Hsu ML, Wang DH, et al. Current insights into oral cancer diagnostics. Diagnostics (2021) 11(7):1287. doi: 10.3390/diagnostics11071287
5. Warnakulasuriya S, Kerr AR. Oral cancer screening: Past, present, and future. J Dent Res (2021) 100(12):1313–20. doi: 10.1177/00220345211014795
6. Dragomir MP, Knutsen E, Calin GA. Classical and noncanonical functions of miRNAs in cancers. Trends Genet (2022) 38(4):379–94. doi: 10.1016/j.tig.2021.10.002
7. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol (2019) 20(1):5–20. doi: 10.1038/s41580-018-0059-1
8. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery (2017) 16(3):203–22. doi: 10.1038/nrd.2016.246
9. Preethi KA, Selvakumar SC, Ross K, Jayaraman S, Tusubira D, Sekar D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol Cancer (2022) 21(1):54. doi: 10.1186/s12943-022-01525-9
10. Jet T, Gines G, Rondelez Y, Taly V. Advances in multiplexed techniques for the detection and quantification of microRNAs. Chem Soc Rev (2021) 50(6):4141–61. doi: 10.1039/D0CS00609B
11. Chai AWY, Lim KP, Cheong SC. Translational genomics and recent advances in oral squamous cell carcinoma. Semin Cancer Biol (2020) 61:71–83. doi: 10.1016/j.semcancer.2019.09.011
12. Prime SS, Cirillo N, Cheong SC, Prime MS, Parkinson EK. Targeting the genetic landscape of oral potentially malignant disorders has the potential as a preventative strategy in oral cancer. Cancer Lett (2021) 518:102–14. doi: 10.1016/j.canlet.2021.05.025
13. Osan C, Chira S, Nutu AM, Braicu C, Baciut M, Korban SS, et al. The connection between MicroRNAs and oral cancer pathogenesis: Emerging biomarkers in oral cancer management. Genes (2021) 12(12):1989. doi: 10.3390/genes12121989
14. Ghosh RD, Pattatheyil A, Roychoudhury S. Functional landscape of dysregulated MicroRNAs in oral squamous cell carcinoma: Clinical implications. Front Oncol (2020) 10:619. doi: 10.3389/fonc.2020.00619
15. Manzano-Moreno FJ, Costela-Ruiz VJ, Garcia-Recio E, Olmedo-Gaya MV, Ruiz C, Reyes-Botella C. Role of salivary MicroRNA and cytokines in the diagnosis and prognosis of oral squamous cell carcinoma. Int J Mol Sci (2021) 22(22):12215. doi: 10.3390/ijms222212215
16. Hsieh PL, Liao YW, Pichler M, Yu CC. MicroRNAs as theranostics targets in oral carcinoma stem cells. Cancers (2020) 12(2):340. doi: 10.3390/cancers12020340
17. Yete S, Saranath D. MicroRNAs in oral cancer: Biomarkers with clinical potential. Oral Oncol (2020) 110:105002. doi: 10.1016/j.oraloncology.2020.105002
18. Wang J, Lv N, Lu X, Yuan R, Chen Z, Yu J. Diagnostic and therapeutic role of microRNAs in oral cancer (Review). Oncol Rep (2021) 45(1):58–64. doi: 10.3892/or.2020.7854
19. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med (2011) 155(8):529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
20. Baragilly M, Willis BH. On estimating a constrained bivariate random effects model for meta-analysis of test accuracy studies. Stat Methods Med Res (2022) 31(2):287–99. doi: 10.1177/09622802211065157
21. Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surg (2005) 79(1):16–20. doi: 10.1016/j.athoracsur.2004.09.040
22. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327(7414):557–60. doi: 10.1136/bmj.327.7414.557
23. Ruppar T. Meta-analysis: How to quantify and explain heterogeneity? Eur J Cardiovasc Nurs (2020) 19(7):646–52. doi: 10.1177/1474515120944014
24. Lin L, Chu H. Quantifying publication bias in meta-analysis. Biometrics. (2018) 74(3):785–94. doi: 10.1111/biom.12817
25. Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS, Chang KW. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol (2010) 46(3):204–8. doi: 10.1016/j.oraloncology.2009.12.005
26. Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis (2010) 16(4):360–4. doi: 10.1111/j.1601-0825.2009.01646.x
27. Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH, Cheng HW, et al. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med (2011) 40(5):397–404. doi: 10.1111/j.1600-0714.2010.01003.x
28. Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH, Huang YC, et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res (Phila) (2012) 5(4):665–74. doi: 10.1158/1940-6207.CAPR-11-0358
29. Maclellan SA, Lawson J, Baik J, Guillaud M, Poh CF, Garnis C. Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med (2012) 1(2):268–74. doi: 10.1002/cam4.17
30. Hung PS, Liu CJ, Chou CS, Kao SY, Yang CC, Chang KW, et al. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PloS One (2013) 8(11):e79926. doi: 10.1371/journal.pone.0079926
31. Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW, Lin SC. miR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann Surg Oncol (2013) 20 Suppl 3:S406–414. doi: 10.1245/s10434-012-2618-6
32. Ren W, Qiang C, Gao L, Li SM, Zhang LM, Wang XL, et al. Circulating microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are promising novel biomarkers for detection of oral squamous cell carcinoma. Biomarkers. (2014) 19(7):590–6. doi: 10.3109/1354750X.2014.955059
33. Ries J, Vairaktaris E, Agaimy A, Kintopp R, Baran C, Neukam FW, et al. miR-186, miR-3651 and miR-494: potential biomarkers for oral squamous cell carcinoma extracted from whole blood. Oncol Rep (2014) 31(3):1429–36. doi: 10.3892/or.2014.2983
34. Lu YC, Chang JT, Huang YC, Huang CC, Chen WH, Lee LY, et al. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem (2015) 48(3):115–21. doi: 10.1016/j.clinbiochem.2014.11.020
35. Tachibana H, Sho R, Takeda Y, Zhang X, Yoshida Y, Narimatsu H, et al. Circulating miR-223 in oral cancer: Its potential as a novel diagnostic biomarker and therapeutic target. PloS One (2016) 11(7):e0159693. doi: 10.1371/journal.pone.0159693
36. Xu H, Yang Y, Zhao H, Yang X, Luo Y, Ren Y, et al. Serum miR-483-5p: A novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma. Tumour Biol (2016) 37(1):447–53. doi: 10.1007/s13277-015-3514-z
37. Liu CJ, Lin JS, Cheng HW, Hsu YH, Cheng CY, Lin SC. Plasma miR-187* is a potential biomarker for oral carcinoma. Clin Oral Investig (2017) 21(4):1131–8. doi: 10.1007/s00784-016-1887-z
38. Chang YA, Weng SL, Yang SF, Chou CH, Huang WC, Tu SJ, et al. A three-MicroRNA signature as a potential biomarker for the early detection of oral cancer. Int J Mol Sci (2018) 19(3):758. doi: 10.3390/ijms19030758
39. Chen L, Hu J, Pan L, Yin X, Wang Q, Chen H. Diagnostic and prognostic value of serum miR-99a expression in oral squamous cell carcinoma. Cancer biomark (2018) 23(3):333–9. doi: 10.3233/CBM-181265
40. Sun G, Cao Y, Wang P, Song H, Bie T, Li M, et al. miR-200b-3p in plasma is a potential diagnostic biomarker in oral squamous cell carcinoma. Biomarkers. (2018) 23(2):137–41. doi: 10.1080/1354750X.2017.1289241
41. Lu Z, He Q, Liang J, et al. miR-31-5p is a potential circulating biomarker and therapeutic target for oral cancer. Mol Ther Nucleic Acids (2019) 16:471–80. doi: 10.1016/j.omtn.2019.03.012
42. Mahmood N, Hanif M, Ahmed A, et al. Circulating miR-21 as a prognostic and predictive biomarker in oral squamous cell carcinoma. Pak J Med Sci (2019) 35(5):1408–12. doi: 10.12669/pjms.35.5.331
43. Crimi S, Falzone L, Gattuso G, Grillo CM, Candido S, Bianchi A, et al. Droplet digital PCR analysis of liquid biopsy samples unveils the diagnostic role of hsa-miR-133a-3p and hsa-miR-375-3p in oral cancer. Biology (2020) 9(11):379. doi: 10.3390/biology9110379
44. Emami N, Mohamadnia A, Mirzaei M, Bayat M, Mohammadi F, Bahrami N. miR-155, miR-191, and miR-494 as diagnostic biomarkers for oral squamous cell carcinoma and the effects of avastin on these biomarkers. J Korean Assoc Oral Maxillofac Surg (2020) 46(5):341–7. doi: 10.5125/jkaoms.2020.46.5.341
45. Karimi A, Bahrami N, Sayedyahossein A, Derakhshan S. Evaluation of circulating serum 3 types of microRNA as biomarkers of oral squamous cell carcinoma; a pilot study. J Oral Pathol Med (2020) 49(1):43–8. doi: 10.1111/jop.12959
46. Bigagli E, Locatello LG, Di Stadio A, Maggiore G, Valdarnini F, Bambi F, et al. Extracellular vesicles miR-210 as a potential biomarker for diagnosis and survival prediction of oral squamous cell carcinoma patients. J Oral Pathol Med (2022) 51(4):350–7. doi: 10.1111/jop.13263
47. He T, Guo X, Li X, Liao C, Wang X, He K. Plasma-derived exosomal microRNA-130a serves as a noninvasive biomarker for diagnosis and prognosis of oral squamous cell carcinoma. J Oncol (2021) 2021:5547911. doi: 10.1155/2021/5547911
48. Nakamura K, Hiyake N, Hamada T, Yokoyama S, Mori K, Yamashiro K, et al. Circulating microRNA panel as a potential novel biomarker for oral squamous cell carcinoma diagnosis. Cancers (Basel) (2021) 13(3):449. doi: 10.3390/cancers13030449
49. Wang L, Song H, Yang S. MicroRNA-206 has a bright application prospect in the diagnosis of cases with oral cancer. J Cell Mol Med (2021) 25(17):8169–73. doi: 10.1111/jcmm.16598
50. Peng Q, Shen Y, Lin K, Zou L, Shen Y, Zhu Y. Comprehensive and integrative analysis identifies microRNA-106 as a novel non-invasive biomarker for detection of gastric cancer. J Transl Med (2018) 16(1):127. doi: 10.1186/s12967-018-1510-y
51. Hu X, Peng Q, Zhu J, Shen Y, Lin K, Shen Y, et al. Identification of miR-210 and combination biomarkers as useful agents in early screening non-small cell lung cancer. Gene. (2020) 729:144225. doi: 10.1016/j.gene.2019.144225
52. Cai S, Ma J, Wang Y, Cai Y, Xie L, Chen X, et al. Biomarker value of miR-221 and miR-222 as potential substrates in the differential diagnosis of papillary thyroid cancer based on data synthesis and bioinformatics approach. Front Endocrinol (2021) 12:794490. doi: 10.3389/fendo.2021.794490
53. Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer (2018) 17(1):147. doi: 10.1186/s12943-018-0897-7
54. Peng Q, Zhang X, Min M, Zou L, Shen P, Zhu Y. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: a systematic review and meta-analysis. Oncotarget. (2017) 8(27):44893–909. doi: 10.18632/oncotarget.16488
55. Peng Q, Feng Z, Shen Y, Zhu J, Zou L, Shen Y, et al. Integrated analyses of microRNA-29 family and the related combination biomarkers demonstrate their widespread influence on risk, recurrence, metastasis and survival outcome in colorectal cancer. Cancer Cell Int (2019) 19:181. doi: 10.1186/s12935-019-0907-x
56. Peng Q, Shen Y, Lin K, Zou L, Shen Y, Zhu Y. Identification of microRNA-92a and the related combination biomarkers as promising substrates in predicting risk, recurrence and poor survival of colorectal cancer. J Cancer (2019) 10(14):3154–71. doi: 10.7150/jca.30306
57. Lin X, Wu W, Ying Y, Luo J, Xu X, Zheng L, et al. MicroRNA-31: a pivotal oncogenic factor in oral squamous cell carcinoma. Cell Death Discovery (2022) 8(1):140. doi: 10.1038/s41420-022-00948-z
58. Peng Y, Hu S, Zhang K, Wang Y, Rouzi M, Zhou D, et al. Downregulation of MicroRNA-130a inhibits oral squamous cell carcinoma proliferation and metastasis via the hippo-YAP pathway. Cancer Manag Res (2021) 13:4829–40. doi: 10.2147/CMAR.S287575
59. Liu W, Shi X, Wang B. microRNA-133a exerts tumor suppressive role in oral squamous cell carcinoma through the notch signaling pathway via downregulation of CTBP2. Cancer Gene Ther (2022) 29(1):62–72. doi: 10.1038/s41417-020-00289-y
60. Yang K, Chen Y, Cui Z, Chen H, Yang L, Zhao J, et al. MicroRNA-222-3p participates in the development of oral squamous cell carcinoma by targeting CDKN1B. J Oral Pathol Med (2020) 49(7):621–9. doi: 10.1111/jop.12986
61. Li X, Li Y, Jiang C, Chen L, Gan N. MicroRNA-144-3p inhibits tumorigenesis of oral squamous cell carcinoma by downregulating ERO1L. J Cancer (2020) 11(3):759–68. doi: 10.7150/jca.33267
Keywords: oral squamous cell carcinoma, microRNAs, biomarkers, diagnosis, circulating
Citation: Gao H, Shen Y, Feng Z, Cai Y, Yang J, Zhu Y and Peng Q (2022) The clinical implications of circulating microRNAs as potential biomarkers in screening oral squamous cell carcinoma. Front. Oncol. 12:965357. doi: 10.3389/fonc.2022.965357
Received: 09 June 2022; Accepted: 26 October 2022;
Published: 17 November 2022.
Edited by:
Wojciech Golusiński, Poznan University of Medical Sciences, PolandReviewed by:
Angela Pia Cazzolla, University of Foggia, ItalyAndrea Santarelli, Marche Polytechnic University, Italy
Copyright © 2022 Gao, Shen, Feng, Cai, Yang, Zhu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiliang Peng, cGVuZ3FpbGlhbmcxOTEyQDE2My5jb20=
†These authors have contributed equally to this work
 Huan Gao1†
Huan Gao1† Qiliang Peng
Qiliang Peng