- Center for Uterine Cancer Diagnosis and Therapy Research of Zhejiang Province, Department of Obstetrics and Gynecology, The Second Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
Accumulated evidence has revealed that F-box protein, a subunit of SCF E3 ubiquitin ligase complexes, participates in carcinogenesis and tumor progression via targeting its substrates for ubiquitination and degradation. F-box proteins could be regulated by cellular signaling pathways and noncoding RNAs in tumorigenesis. Long noncoding RNA (lncRNA), one type of noncoding RNAs, has been identified to modulate the expression of F-box proteins and contribute to oncogenesis. In this review, we summarize the role and mechanisms of multiple lncRNAs in regulating F-box proteins in tumorigenesis, including lncRNAs SLC7A11-AS1, MT1JP, TUG1, FER1L4, TTN-AS1, CASC2, MALAT1, TINCR, PCGEM1, linc01436, linc00494, GATA6-AS1, and ODIR1. Moreover, we discuss that targeting these lncRNAs could be helpful for treating cancer via modulating F-box protein expression. We hope our review can stimulate the research on exploration of molecular insight into how F-box proteins are governed in carcinogenesis. Therefore, modulation of lncRNAs is a potential therapeutic strategy for cancer therapy via regulation of F-box proteins.
Introduction
F-box protein is a subunit in Skp1-Cullin1-F-box protein (SCF) E3 ubiquitin ligase complexes (1). It has been well documented that F-box proteins target their substrates via ubiquitination and proteasome degradation (2). It has been accepted that F-box proteins have 69 members in human genome (3). According to specific motifs in F-box proteins, these proteins are classified into three types: 10 FBXW proteins (WD40 repeat domains), and 22 FBXL proteins (leucine-rich repeat motifs), 37 FBXO proteins (other motifs) (4). Accumulated evidence demonstrated that F-box proteins participate in cancer initiation and progression via regulation of cell proliferation (5–7), apoptosis (8), motility and metastasis (9), cell cycle (10), EMT (11), cancer stem cells (12, 13), drug resistance (14, 15) and autophagy (16).
Noncoding RNAs have little or no protein coding capacity to encode proteins (17). Based on the lengths of nucleotides, noncoding RNAs are classified multiple types: long noncoding RNA (lncRNA, high than 200 bp), short noncoding RNA (17-30 bp) and mid-size noncoding RNA (31-200 bp) (18). It has been known that microRNA (miRNA) has approximately 22 nucleotides and targets gene expression via regulation of post-transcription (19, 20). Now, noncoding RNAs have been validated to critically participate in oncogenesis in various types of cancers (21–25). Not surprisingly, noncoding RNAs regulated numerous cellular biological processes and dysregulated noncoding RNAs lead to various diseases, including cancer (26–29). In recent years, accumulated evidence suggests that noncoding RNAs targets the expression of F-box proteins, leading to carcinogenesis and malignant progression. One review has well summarized the role of noncoding RNAs in regulation of F-box proteins in carcinogenesis (30). However, this review mainly described the role of microRNAs in governing the expression of F-box proteins. Here, we summarized the functions and mechanisms of lncRNAs in controlling F-box protein expression, leading to tumor development and progression.
LncRNAs regulate the expression of F-BOX proteins
Targeting FBXW family by lncRNAs
LncRNA SLC7A11-AS1 regulates β-TrCP1
SLC7A11-AS1 was downregulated in tumor tissues in patients with gastric cancer, and correlated with poor prognosis in these patients (31). Depletion of SLC7A11-AS1 contributed to tumor growth in cells and in mice via controlling the ASK1/p38/JNK signaling pathway in gastric cancer (31). In lung cancer cells, SLC7A11-AS1 facilitated tumor progression via suppressing miR-4775 and increasing the expression of TRAIP (32). SLC7A11-AS1 has been revealed to play a key role in chemoresistance in various types of cancers (33). Luo et al. found that SLC7A11-AS1 targeted miR-33a-5p and changed the expression of xCT, weakened cell growth, promoted ROS levels, and regulated cisplatin resistance in gastric cancer (34).
SLC7A11-AS1 was found to be significantly increased in PDAC tissues (35). PDAC cells with gemcitabine resistance have a high expression of SLC7A11-AS1, indicating that SLC7A11-AS1 could play an essential role in regulation of drug resistance. In fact, downregulation of SLC7A11-AS1 potentiated gemcitabine sensitivity in resistant PDAC cells and inhibited the PDAC stemness. In line with this, overexpression of SLC7A11-AS1 increased gemcitabine resistance via suppressing intracellular ROS levels by maintaining NRF2 stability (35). Mechanically, SLC7A11-AS1 can bind to the F-box motif of β-TrCP1 (also known as FBXW1), which blocks the ubiquitination and degradation of NRF2. Therefore, SLC7A11-AS1 attenuated β-TrCP-mediated degradation of NRF2, reduced ROS levels, and increased cancer stemness, which promoted gemcitabine resistance in PDAC (35). Hence, targeting SLC7A11-AS1 could overcome gemcitabine resistance to improve treatment benefits in PDAC patients.
LncRNA PCGEM1 regulates β-TrcP2
β-TrcP2, also known as FBXW11, has been characterized to take part in tumorigenesis (36). FBXW11 activated the β-catenin/TCF and NF-kappa B pathways and increased cell proliferation in lymphocytic leukemia (37). FBXW11 maintained stem-cell-like characters and enhanced liver metastasis via governing SIRT1 transcription in colorectal cancer (38). LncRNA PCGEM1 has been found to participate in the initiation and development of a variety of cancers via regulating several signaling pathways (39). LncRNA PCGEM1 expression was remarkably increased in cervical cancer specimens, which was associated with FIGO stage, lymph node metastasis, poor survival and distant metastasis in cervical cancer patients (40). PCGEM1 upregulation stimulated proliferation, invasion, migration, and cell cycle process and reduced apoptosis in cervical cancer cells (40). PCGEM1 can work as a ceRNA to sponge miR-182 and suppress its expression, leading to upregulation of FBXW11. Moreover, PCGEM1 can activate the NF-kappa B and β-catenin/TCF pathways, and this activation by PCGEM1 can be abrogated by knockdown of FBXW11 (40). Altogether, PCGEM1 exerted cervical cancer progression via modulation of miR-182 and FBXW11.
Several lncRNAs regulates FBXW7 expression
F-box and WD repeat domain containing 7 (FBXW7) is well studied and acts as one tumor suppressor gene in human carcinogenesis and tumor progression (41–43). One study identified that several lncRNAs are correlated with Fbxw7 deficiency in radiation-mediated thymic lymphoma (44). In mice with Fbxw7 deficiency, microarray dada from radiation-induced thymic lymphomas revealed that 372 lncRNAs are differentially expressed in tumor tissues. Among these lncRNAs, 170 lncRNAs were decreased, while 202 lncRNAs were increased in thymic lymphomas (44). Moreover, these FBXW7-associated lncRNAs were found to participate in DNA repair, cell cycle processes, lymphocyte activation and cell differentiation. Two lncRNAs (lncRNA position: 5119300, 5162836) were observed to be linked to Anxa2, Cecr2, Zeb1 and Zfp438 expressions, whereas one lncRNA (position: 182808654) was decreased and associated with Ampd1, Cd6, Clip1, Dap, Edaradd and Ptk2b (44). Furthermore, lncRNA A_30_P01032978 is correlated with poor disease free survival in patients with breast cancer (44). In this section, we will discuss how the several lncRNAs regulated the expression of FBXW7 in carcinogenesis.
LncRNA MT1JP regulates FBXW7
LncRNA MT1JP has been reported to be a tumor suppressor via promotion of the translation of p53 by interaction with TIAR (45). In retinoblastoma, MT1JP plays a tumor suppressive role via targeting Wnt/β-catenin signaling pathway (46). In breast cancer cells, MT1JP repressed oncogenesis and reversed cisplatin resistance through sponging miR-24-3p and inhibiting the Wnt/β-catenin (47). Consistently, MT1JP exhibited tumor suppressive functions via sponging miR-92-3p and targeting miR-214/RUNX3 axis in breast cancer cells (48, 49). In lung cancer cells, MT1JP blocked cell proliferation, migration and invasion through modulation of miR-423-3p/Bim axis (50). In glioma, MT1JP retarded tumor progression via competitively binding with miR-24 (51). In osteosarcoma cells, MT1JP was reported to increase the inhibitory function of miR-646 on FGF2 expression (52). In HCC cells, upregulation of MT1JP modulated cell apoptosis and migratory abilities via targeting miR-24-3p and regulating AKT, RUNX3 and p21 (53–55). Furthermore, MT1JP regulated miR-24-3p/Bcl2L2 signaling pathway and reduced lenvatinib sensitivity via suppression of apoptosis in HCC (56). Moreover, MT1JP upregulation abrogated the PTEN inactivation via miR-32 reduction in HCC cells (57).
In intrahepatic cholangiocarcinoma, MT1JP acted as a protective lncRNA via regulation of miR-18a-5p and FBP1 (58). MT1JP regulated miR-214-3p/RUNX3 signaling pathway and subsequently inhibited proliferation and migration of gastric cancer (59). Notably, low expression of MT1JP was related with poor prognosis in patients with gastric cancer (60). LncRNA MT1JP was downregulated in gastric cancer tissues compared with adjacent normal tissues (61). Gastric cancer patients had a better prognosis, who often have higher expression of MT1JP. In vitro experiment data showed that lncRNA MT1JP upregulation suppressed proliferation, invasion and migration and enhanced apoptosis of gastric cancer cells (61). In vivo data revealed that lncRNA MT1JP reduced tumor sizes and tumor metastasis. Mechanistical analysis demonstrated that lncRNA MT1JP sponged miR-92a-3p and upregulated FBXW7 in gastric cancer (61). Rescue experiments exhibited that downregulation of FBXW7 reversed MT1JP-induced inhibition of proliferation, invasion and migration in gastric cancer (61).
LncRNA TUG1 regulates FBXW7
Numerous studies have demonstrated the critical role of lncRNA Taurine upregulated gene 1 (TUG1) in cancer initiation and progression. LncRNA TUG1 underlined a tumor promotive property via impairing miR-421-mediated suppression of KDM2A and activating the ERK signaling in colorectal cancer cells (62). TUG1 suppressed cancer progression via targeting Siglec-15-mediated anti-immune activity in HCC (63). Moreover, TUG1 was reported to sponge miR-328-3p and increase the SRSF9 mRNA expression in HCC cells, leading to promotion of proliferation, invasion and migration (64). Xiu et al. found that TUG1 enhanced tumor malignant progression via binding with miR-516b-5p and increasing H6PD expression (65). Xia et al. reported that TUG1 stabilization by IGF2BP2 increased cisplatin resistance via targeting autophagy in colorectal cancer (66). One group identified that TUG1 governed the miR-320a/FOXQ1 axis and caused promotion of bladder tumor malignant phenotypes (67).
Sun et al. discovered that TUG1 increased chemoresistance and enhanced cancer stem cell behaviors via stabilizing GATA6 protein in colorectal cancer (68). TUG1 targeted AKT/mTOR signaling pathway via sponging miR-582-3p, which promoted ovarian cancer malignant behaviors (69). TUG1 sponged miR-29c-3p and upregulated the expression of VEGFA, which facilitated malignant phenotypes in stomach cancer (70). In addition, TUG1 competitively interacted with miR-29a and triggered the expression of IFITM3 in HCC cells (71). TUG1 promoted tumor progression and metastasis via modulating miR-140-3p and Annexin A8 axis in bladder cancer cells (72). Zhang et al. reported that TUG1 targeted miR-187-3p and TESC and modulated the NF-kappa B signaling pathway, which governed progression of pituitary adenoma (73). Li group reported that miR-199a-3p/MSI2 signaling pathway was involved in TUG1-mediated promotion of cell migration, invasion and proliferation in Ewing’s sarcoma (74). TUG1 upregulated the expression of XBP1 by sponging miR-498 in ESCC cells, which enhanced tumor metastasis and growth (75). One study revealed that TUG1 upregulated the expression of FBXW7 and induced FBXW7-triggered SIRT1 ubiquitination and degradation (76). Moreover, TUG1 compromised neuronal mitophagy via targeting TUG1/FBXW7 axis in cerebral ischemia and reperfusion injury (76). It is necessary to explore whether TUG1 regulates the expression of FBXW7 in carcinogenesis.
LncRNA FER1L4 regulates FBXW7
LncRNA Fer-1-like protein 4 (FER1L4) has been discovered to be involved in development of human cancer (77). Xia and colleagues found that FER1L4 knockdown suppressed cell growth and cell cycle progression via interacting with miR-372 and upregulating E2F1 expression in gliomas (78). In lung cancer cells, FER1L4 suppressed metastasis and growth and enhanced apoptosis via control of PI3K/AKT and p53 signaling pathways (79, 80). In osteosarcoma cells, FER1L4 regulated cell apoptosis and EMT via suppression of miR-18a-5p and promotion of SOCS5 and activation of PI3K/AKT pathway (81). In clear cell renal cell carcinoma (ccRCC) tissues, FER1L4 expression is higher than that in adjacent normal tissues (82). High expression of FER1L4 was linked to tumor grade, stage, metastasis and tumor aggressiveness and patient survival (82). In oral squamous cell carcinoma, FER1L4 facilitated tumor progression through regulation of miR-133a-5p/Prx1 axis (83). In colorectal cancer patients, FER1L4 expression levels were downregulated, while RB1 expression was upregulated. FER1L4 expression was associated with RB1 expression in colorectal cancer patients (84).
FER1L4 sponged miR-1273g-3p and increased the expression of PTEN and led to cell cycle arrest and metastasis suppression in colorectal cancer (85). One study showed that downregulation of FER1L4 inhibited the mRNA levels of RB1 in gastric cancer (86). Moreover, FER1L4 reduced cell growth via binding with miR-106a-5p and increased the expression of PTEN at both mRNA and protein levels in gastric cancer (87). Similarly, FER1L4 reduced growth, invasion, migration and metastasis by suppressing the Hippo-YAP signaling pathway in gastric cancer (88). Qiao et al. found that FER1L4 repressed cell proliferation and blocked cell cycle at G0/G1 phase as well as enhanced apoptosis via upregulation of PTEN in endometrial carcinoma (89). Furthermore, FER1L4 overexpression was correlated with favorable survival outcome in endometrial carcinoma patients (90). Ma et al. reported that FER1L4 decreased cell invasion and growth and promoted cell apoptosis and cell cycle arrest at G0/G1 phase in ESCC cells (91). In HCC cells, overexpression of FER1L4 attenuated cell migration and proliferation, increased apoptosis through targeting PI3K/AKT signaling pathway (92).
In ovarian cancer cells, FER1L4 upregulation reduced paclitaxel tolerance via modulation of the MAPK signaling pathway (93). The lower expression of lncRNA FER1L4 was observed in prostate cancer samples compared with normal prostate tissues (94). Early stage of prostate cancer patients had the higher expression of FER1L4 in prostate cancer specimens. Upregulation of FER1L4 decreased proliferation and increased apoptosis in prostate cancer cells via sponging miR-92a-3p and upregulating FBXW7 (94). Depletion of FBXW7 abrogated inhibition of cell proliferation caused by upregulation of FER1L4 in prostate cancer cells, indicating that FER1L4 exerted antitumor activities via miR-92a-3p/FBXW7 axis (94).
LncRNA TTN-AS1 targets FBXW7
LncRNA Titin-antisense RNA1 (TTN-AS1) has been reported to be involved in tumorigenesis in various type cancers, including esophageal squamous cell carcinoma (ESCC), cervical cancer, gastric cancer and lung cancer (95–98). Lin et al. reported that TTN-AS1 worked as an oncogene and was highly expressed in ESCC cells and tumor specimens, and overexpression of TTN-AS1 enhanced ESCC proliferation and metastasis (95). Mechanistically, TTN-AS1 competitively interacted with miR-133b and increased the expression of Snail1, leading to EMT cascade in ESCC cells (95). In addition, TTN-AS1 sponged miR-133b and increased the expression level of FSCN1 and resulted in invasion cascades in ESCC cells (95). Chen et al. reported that TTN-AS1 enhanced growth and metastasis of cervical cancer cells via regulation of miR-573/E2F3 axis (96). Dong et al. revealed that TTN-AS1 stimulated gastric cancer development via interacting with miR-376b-3p and KLF12 (97). Luo et al. observed that TTN-AS1 contributed to tumor progression via modulating PTEN/PI3K/AKT signaling pathway in lung adenocarcinoma (98). Similarly, TTN-AS1 activated cell invasion and migration via governing miR-4677-3p/ZEB1 axis in lung adenocarcinoma (99). In prostate cancer cells, TTN-AS1 reduced cell apoptosis and facilitated cell proliferation via binding with miR-193a-5p (100).
LncRNA TTN-AS1 sponged miR-134-5p and increased the expression of malignant brain tumor domain containing 1 (MBTD1), contributing to promotion of viability and drug resistance, inhibition of apoptosis in osteosarcoma cells (101). TTN-AS1 interacted with miR-376a-3p and subsequently upregulated KLF15, resulting in promotion of colorectal cancer progression (102). Cui et al. also reported that TTN-AS1 facilitated the cell invasion and growth through activation of miR-497-induced PI3K/AKT/mTOR pathway in colorectal cancer (103). Fang et al. found that TTN-AS1 enhanced invasion, EMT and cell growth via governing miR-139-5p/ZEB1 axis and miR-524-5p/RRM2 axis in breast cancer cells (104, 105). One group studied the role of TTN-AS1 in clear cell renal cell carcinoma and found that TTN-AS1 acted as a sponging RNA of miR-195 to increase the expression of cyclin D1 and promote tumor progression (106). It has been reported that lncRNA TTN-AS1 can sponge miR-15b-5p and regulate the expression of FBXW7 in ovarian cancer (107). The low expression of TTN-AS1 was found in ovarian cancer cells and tumor tissues. Upregulation of TTN-AS1 reduced proliferation and colony formation and stimulated apoptosis in ovarian cancer cells (107). Moreover, knockdown of FBXW7 attenuated the functions of TTN-AS1 upregulation on cell behaviors, suggesting that TTN-AS1 exerts its biological behaviors via upregulating FBXW7 in ovarian cancer cells (107).
LncRNA CASC2 targets FBXW7
LncRNA cancer susceptibility candidate 2 (CASC2) has been reported to serve as a tumor suppressor in carcinogenesis by sponging several miRNAs (108, 109). Upregulation of lncRNA CASC2 attenuated cell viability, induced apoptosis and affected autophagy via regulation of miR-19a and NF-kappa B signaling pathway in colon cancer (108). In line with this finding, lncRNA CASC2 enhance apoptosis and autophagy through targeting TRIM16 expression in colon cancer cells (110). CASC2 promoted berberine-mediated cytotoxicity via inhibition of Bcl2 in colorectal cancer (111). One group showed that CASC2 overexpression exhibited antitumor activities through sponging miR-24-3p in thyroid cancer (109). Another group reported that CASC2 increased radiotherapy sensitivity via sponging miR-155 in papillary thyroid cancer (112). Similarly, lncRNA CASC2 increased irradiation-triggered endoplasmic reticulum stress via regulation of PERK signaling pathway in NSCLC cells (113). In pancreatic cancer cells, lncRNA CASC2 increased the expression of PTEN and retarded cell metastasis via sponging miR-21 (114).
LncRNA CASC2 overexpression suppressed cell proliferation and tumor growth in mice in hepatocellular carcinoma (HCC) (115, 116). LncRNA CASC2 enhanced apoptosis and suppressed viability via targeting miR-24-3p in HCC cells (116). In TNF-related apoptosis-inducing ligand (TRAIL)-resistant HCC cells, CASC2 targeted miR-18a/receptor-interacting serine/threonine-protein kinase 1 (RIPK1) axis and the NF-kappa B pathway, whereas in TRAIL-sensitive cells, CASC2 affected miR-221/caspase-3 and miR-24/caspase-8 (115). In clinical tissues, HCC patients have lower expression of CASC2, which is associated with a poor overall survival rate (115, 117). Sun et al. observed that lncRNA CAS2 reduced cell viability, invasion and migratory activities via directly inhibiting miR-183 in HCC cells (118). Wang et al. reported that lncRNA CASC2 inhibited epithelial-mesenchymal transition (EMT) via targeting miR-367 and FBXW7 in HCC cells (117). Overexpression of lncRNA CASC2 repressed invasion and migration of HCC cells and suppressed EMT and blocked metastasis via sponging miR-367. In addition, FBXW7 was found to be a downstream target of miR-367 in HCC cells (117). Therefore, CASC2 regulates the expression of FBXW7 via regulation of miR-367 in HCC cells.
LncRNA MALAT1 targets FBXW7
LncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) has been known to be correlated with tumor metastasis in human cancer (119). MALAT1 expression was linked to the WHO grade, tumor size and poor survival in glioma patients (120). MALAT1 depletion increased proliferation of glioma stem cells and inhibited the expression of Nestin and Sox2, two stemness markers (121). MALAT1-mediated cell proliferation promotion was due to activation of ERK/MAPK signaling pathway in glioma cells (121). Han et al. found that MALAT1 downregulated MMP2 and blocked ERK/MAPK signaling pathway as well as exhibited tumor suppressive behaviors in glioma cells (122). Xiang et al. reported that knockdown of MALAT1 induced apoptosis via reduction of Cyclin D1 and Myc in U87 and U251 glioma cells (123). Studies showed that MALAT1 inhibited cell apoptosis and increased cell growth and activated autophagy via targeting miR-101 and derepressing Rap1B, RAB5A, ATG4D and STMN1 expression in glioma (124, 125).
MALAT1 was identified to recruit FBXW7 to stimulate the degradation of CRY2 and regulate trophoblast invasion and migration (126). MALAT1 was significantly downregulated in glioma samples and associated with tumor grade, tumor size and Karnofsky Performance status in glioma patients (127). MALAT1 repressed viability of glioma cells via suppressing miR-155 in vitro. Moreover, FBXW7 was identified as a key downstream molecule of miR-155 in glioma cells. Notably, FBXW7 mediated miR-155-triggered oncogenesis in U87 and SHG139 glioma cells. Strikingly, MALAT1 reduced cell viability by upregulation of FBXW7 expression due to downregulation of miR-155 (127). Hence, MALAT1 might be a potential therapeutic target for glioma.
LncRNA TINCR targets FBXW7
LncRNA terminal differentiation-induced lncRNA (TINCR) have been implicated in carcinogenesis and tumor progression (128). TINCR can reduce cell invasion and growth, and induce apoptosis through controlling the expression of miR-424-5p and LATS1 in cutaneous malignant melanoma (129). TINCR attenuated cell invasion and growth via targeting miR-210 and BTG in laryngeal squamous cell carcinoma (130). In HCC cells, TINCR enhanced cell invasion and growth via regulation of STAT3 pathway by binding to TCPTP (131). In breast cancer, TINCR governed cell metastatic ability and cell growth via regulating miR-761 and targeting OAS1 and EGFR (132–134).
In lung cancer tissues, TINCR expression levels were downregulated (135). In lung cancer cells, TINCR upregulation retarded cell invasion and proliferation via acting as a sponge of miR-544a. Moreover, FBXW7 was validated as a downstream target of miR-544a in lung cancer cells. In a rescue experiment, depletion of FBXW7 abrogated the suppression of TINCR on invasion and proliferation (135). Altogether, lncRNA TINCR performed anti-proliferative and invasive abilities in lung cancer cells through modulating miR-544a/FBXW7 axis. However, one study found that TINCR promoted tumor progression by BRAF-induced MAPK pathway in NSCLC (136). Therefore, further investigation is essential to determine the role of TINCR in lung cancer progression.
LncRNA MALAT1 targets FBXW8
MALAT1 has been validated to have a role in cancer diagnosis, prognosis and therapy (137). Emerging study has shown that MALAT1 can regulate the expression of FBXW8 in human cancer (138). FBXW8 has been reported to involve in cell growth and cell cycle progression in choriocarcinoma (139). Depletion of FBXW8 by siRNA transfection suppressed cell growth and induced cell cycle arrest at G2/M phase in choriocarcinoma JEG-3 cells (139). Overexpression of FBXW8 exhibited the opposite functions on cell growth and cell cycle. FBXW8 can regulate the expression of CDK1, CDK2, p27, Cyclin A and Cyclin B1 in choriocarcinoma cells (139). One study reported that miR-218 suppressed the cell proliferation via inhibition of FBXW8 in choriocarcinoma JEG-3 cells (140).
In choriocarcinoma cells, MALAT1 upregulation increased cell proliferation, while depletion of MALAT1 hindered cell growth (138). Moreover, MALAT1 exerted its biological behaviors via targeting miR-218 in choriocarcinoma cells. Depletion of MALAT1 reduced the tumor growth in vivo. What is more, FBXW8 was found to be a direct target of miR-218 and was involved in MALAT1-meidiated promotion of cell proliferation in choriocarcinoma (138). Hence, MALAT1 promoted cell proliferation via interaction with miR-218 and upregulation of FBXW8 in choriocarcinoma.
Targeting FBXO family by lncRNAs
Linc01436 regulates FBXO11
Linc01436 was reported to be controlled by E2F6 and served as a tumor promoter in NSCLC cells (141). Linc01436 worked as a miR-30a-3p sponge to increase the expression of EPAS1 in NSCLC, resulting in promotion of cell growth, invasion and migration in vitro and enhancement of tumor growth and tumor metastasis in mice (141).
Emerging evidence has revealed that linc01436 plays an oncogenic role in gastric cancer progression (142–144). Linc01436 repressed the expression of miR-585-3p and increased mitogen-activated protein kinase 1 (MAPK1) expression, which contributed to gastric cancer development (143). Similarly, linc01436 triggered gastric cancer progression through modulation of miR-513a-5p and apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) (144). The higher expression of linc01436 was observed in tumor tissues of gastric cancer patients and was associated with a poor survival in gastric cancer cases (142). Moreover, using in vitro experiments, knockdown of linc01436 retarded metastasis and blocked proliferation in BGC823 gastric cancer cells, while increased linc01436 promoted metastasis and proliferative activity in AGS gastric cancer cells (142). Mechanistically, miR-585 can bind to linc01463 and FBXO11, suggesting that linc01436 sponges miR-585 and inhibit it, leading to indirect promotion of FBXO11 expression in gastric cancer (142). Taken together, linc01463 targets miR-585/FBXO11 axis and subsequently promotes progression of gastric cancer.
LincRNA GATA6-AS1 regulates FBXO11
Xu et al. reported that lincRNA GATA6-AS1 regulated invasive and migratory capacities and viability via binding to miR-19a-5p and increasing TET2 in ovarian cancer cells (145). LincRNA GATA6-AS1 promoted GATA6 expression and controlled the behaviors of lung cancer cells (146). In lung cancer cells, lincRNA GATA6-AS1 suppressed cell proliferation and invasive ability (147). Using several approaches, including RNA sequencing dataset, RT-qPCR and TCGA data, one group found that GATA6-AS1 expression levels were downregulated in lung cancer tissues (147). Moreover, GATA6-AS1 overexpression increased the expression of FBXO11 and SP1 via sponging miR-324-5p, contributing to enhancement of invasion and proliferation in lung cancer cells. Furthermore, miR-324-5p overexpression abolished the effects of GATA6-AS1 upregulation in lung cancer (147). In a word, lincRNA GATA6-AS1 might regulate miR-324-5p/FBXO11 axis and facilitated lung cancer development.
LncRNA ODIR1 regulates FBXO25
FBXO25 has been reported to participate in cancer development and malignant behaviors (148, 149). Impairing PRKCD-FBXO25-HAX-1 signaling pathway led to lymphomagenesis and reduced the apoptotic reaction (150). FBXO25 facilitated cell invasion, migration and proliferation via regulation of YAP, cyclins, MMPs and β-catenin in NSCLC cells (148). Clinically, FBXO25 had the higher expression in the nucleus and cytoplasm of tumor tissues in lung cancer patients, and was associated with lymph node metastasis and TNM stage and overall survival (148). In cutaneous squamous cell carcinoma cells, FBXO25 increased cell growth and metastasis via binding with Oct-1, a Cyclin D1 repressor, and stabilization of Cyclin D1 (149). One study showed that lncRNA RP11-527N22.2, also known as osteogenic differentiation inhibitory lncRNA 1 (ODIR1), interacted with FBXO25 and promoted the destruction of FBXO25 protein by recruiting Cullin 3 (151). FBXO25 promoted H2BK120 ubiquitination and increased the trimethylation of H3K4 (H3K4me3), which increased osterix transcription and the expression of osteocalcin, osteopontin and ALP (151). In human umbilical cord-derived mesenchymal stem cells, downregulation of ODIR1 contributed to osteogenic differentiation, while upregulation of ODIR1 suppressed osteogenic differentiation (151). It is required to investigate the role of ODIR1-mediated FBXO25 disruption in oncogenesis and progression.
Linc00494 regulates FBXO32
FBXO32 promoter hypermethylation has been revealed to be linked to poor prognosis in patients with ovarian cancer (152). FBXO32 has been involved in carcinogenesis and tumor malignant behaviors. FBXO32 worked as an E3 ligase for PHPT1 ubiquitination, leading to reduction of PHPT1 accumulation, inactivation of the ERK/MAPK axis, which inhibited the proliferation of lung cancer cells (153). FBXO32 repressed tumorigenesis by targeting KLF4 for ubiquitination and proteasomal degradation in breast cancer (154). Linc00494 was predicted to bind with NF-kappa B1 by bioinformatics analysis in ovarian cancer cells (155). Dual-luciferase reporter assay, RIP and RNA pull-down confirmed the interaction between linc00494 and NF-kappa B1. Linc00494 increased the activity of NF-kappa B1 after their interaction. Moreover, NF-kappa B1 suppressed the transcription of FBXO32 via binding with the promoter region of FBXO32. Linc00494 upregulation accelerated the expression of NF-kappa B1 and caused invasion, migration and tumorigenesis in ovarian cancer cells. In consistent, upregulation of FBXO22 reversed the linc00494-mediated tumorgenicity in ovarian cancer (155). Strikingly, linc00494 expression levels were highly upregulated in ovarian cancer tissues, while FBXO32 has a lower expression in ovarian tumor specimens (155). In summary, linc00494 modulated NF-kappa B1 and FBXO32 and enhanced progression of ovarian cancer.
Conclusions and future perspectives
In conclusion, multiple lncRNAs have been reported to regulate the expression of several F-box proteins in tumorigenesis, including lncRNAs SLC7A11-AS1, MT1JP, TUG1, FER1L4, TTN-AS1, CASC2, MALAT1, TINCR, PCGEM1, linc01436, linc00494, GATA6-AS1, and ODIR1 (Figures 1 and 2). Modulation of these lncRNA expressions is a potential therapeutic strategy for cancer therapy via regulation of F-box proteins. Besides lncRNAs, miRNAs and circRNAs have also participated in modulation of F-box protein in carcinogenesis. It is necessary to note that several issues need to be addressed for clarifying the functions of lncRNAs in oncogenesis via targeting F-box proteins. For example, there are thousands of lncRNAs. However, only about a dozen lncRNAs were identified to regulate the expression of F-box proteins. More lncRNAs should be discovered, which modulate the F-box protein expression in cancer. Among the 69 F-box proteins, no lncRNA was discovered to target FBXL proteins in tumorigenesis. In addition, one lncRNA can target several F-box proteins. For example, MALAT1 targets both FBXW7 and FBXW8 in cancer cells. It is unclear whether MALAT1 targets two F-box proteins at the same time in carcinogenesis. Hence, further in-depth investigation is pivotal to determine whether regulation of F-box proteins by related lncRNAs is a therapeutic strategy for cancer treatment.
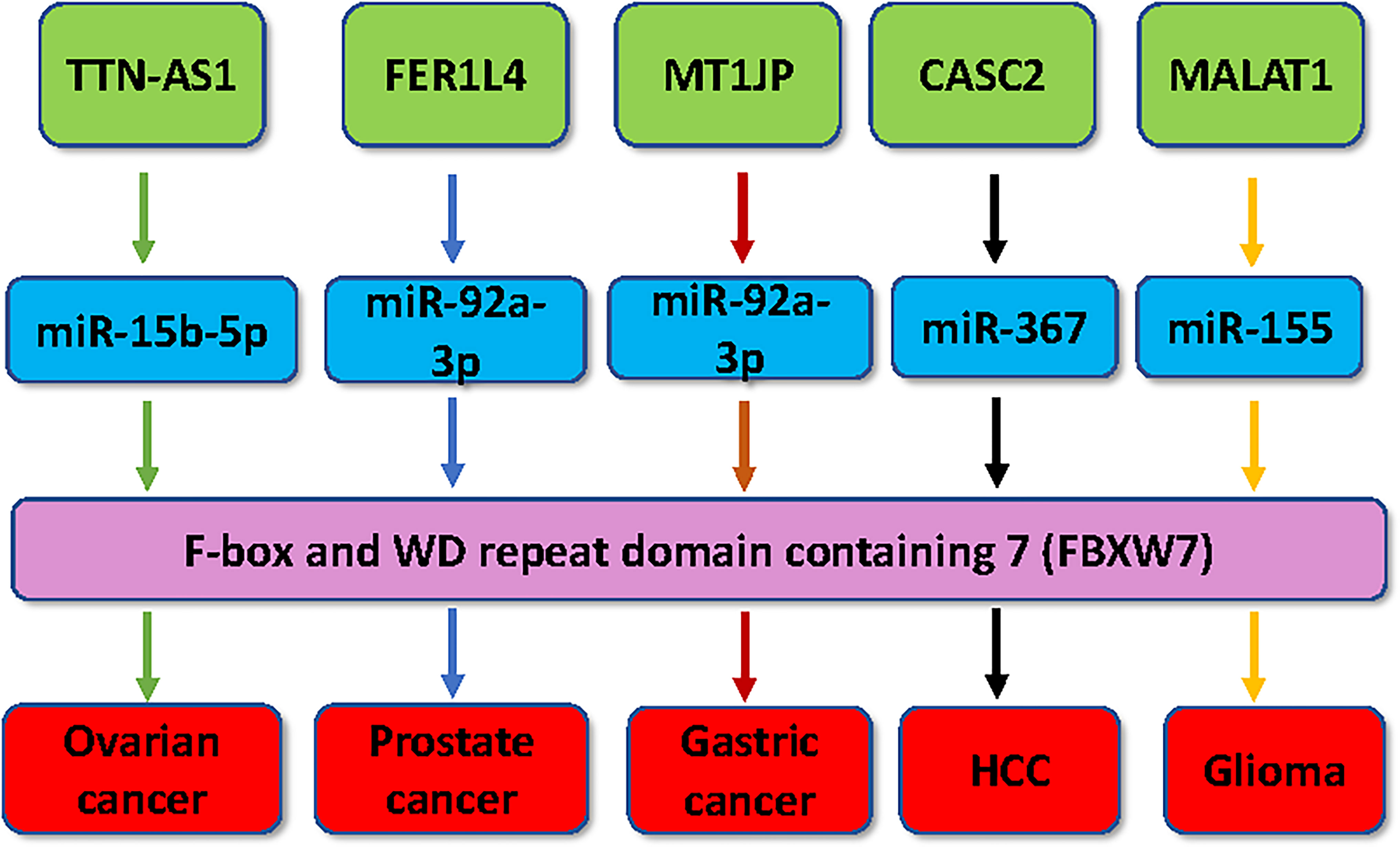
Figure 1 Multiple lncRNAs regulate the expression of FBXW7 in human cancer. Multiple lncRNAs, including MT1JP, FER1L4, TTN-AS1, CASC2 and MALAT1, have been demonstrated to regulate the expression of FBXW7 in tumorigenesis.
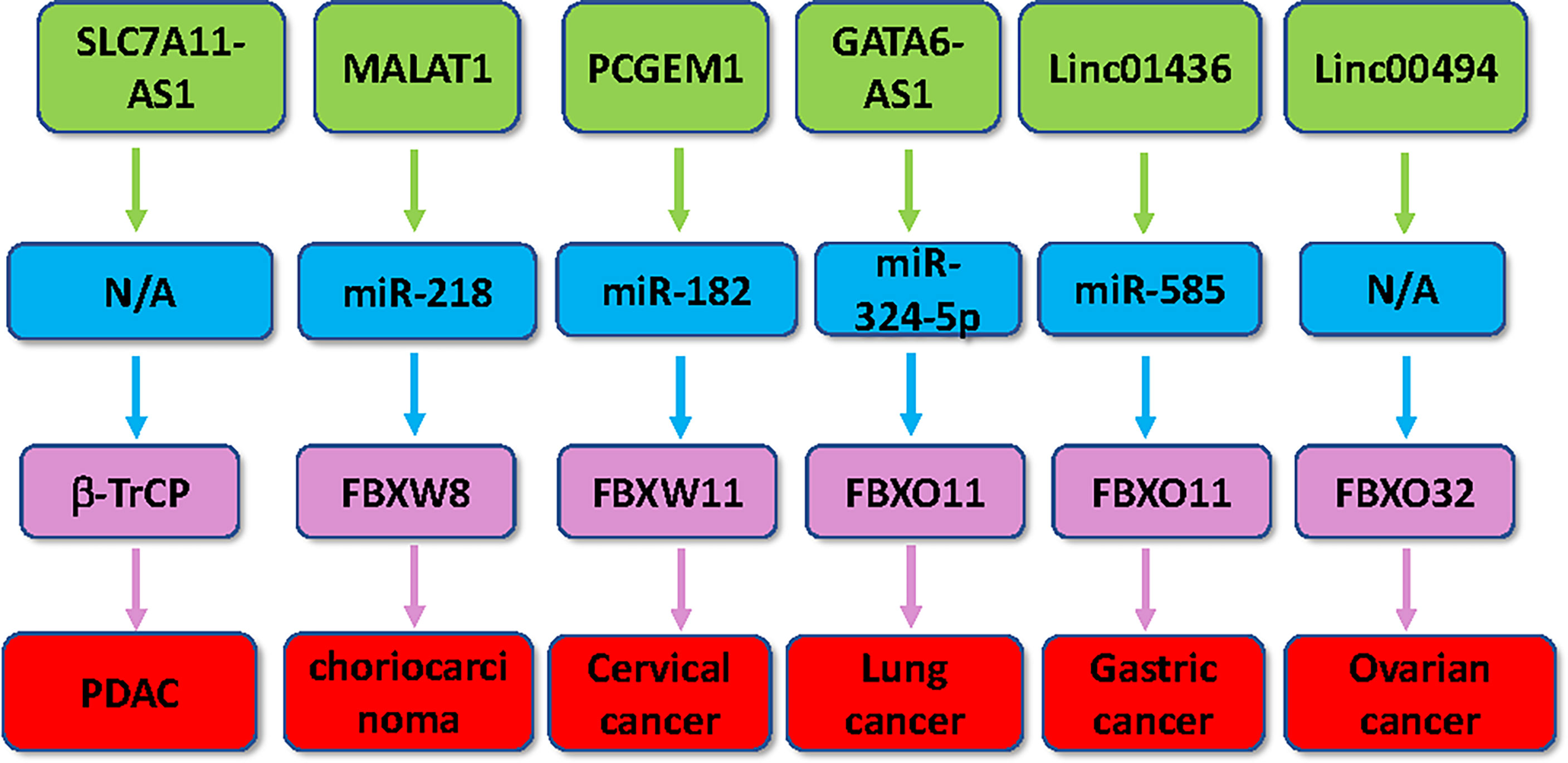
Figure 2 Multiple lncRNAs regulate the expression of F-box proteins in human cancer. Multiple lncRNAs, including SLC7A11-AS1, MALAT1, PCGEM1, linc01436, linc00494 and GATA6-AS1, regulate the expression of several F-box proteins in tumorigenesis.
Author contributions
LX drafted the original manuscript and made the figure. JC, MH, and JM edited the manuscript. ML edited this manuscript and supervised this study. All authors have read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by Natural Science Foundation of Zhejiang province (LY22H160033) and Wenzhou Medical University Basic Research Project KYYW202129.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CASC2, Cancer Susceptibility 2; FER1L4, Fer-1 Like Family Member 4; GATA6-AS1, GATA6 Antisense RNA 1; HCC, Hepatocellular carcinoma; LncRNA, Long noncoding RNA; MALAT1, Metastasis-associated lung adenocarcinoma transcript 1; MT1JP, Metallothionein 1J, pseudogene; NRF2, Nuclear factor erythroid-2-related factor 2; ODIR1, osteogenic differentiation inhibitory regulator 1; PCGEM1, Prostate cancer gene expression marker 1; PDAC, Pancreatic ductal adenocarcinoma; ROS, Reactive oxygen species; SLC7A11-AS1, Solute carrier family 7 member 11 antisense RNA 1; SCF, Skp1-Cullin1-F-box protein; STAT3, Signal transducer and activator of transcription 3; TCPTP, T-cell protein tyrosine phosphatase; TINCR, Terminal differentiation-induced non-coding RNA; TUG1, Taurine up-regulated 1; TTN-AS1, TTN Antisense RNA 1.
References
1. Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by f-box proteins. Nat Rev Mol Cell Biol (2013) 14:369–81. doi: 10.1038/nrm3582
2. Zheng N, Zhou Q, Wang Z, Wei W. Recent advances in SCF ubiquitin ligase complex: clinical implications. Biochim Biophys Acta (2016) 1866:12–22. doi: 10.1016/j.bbcan.2016.05.001
3. Wang Z, Liu P, Inuzuka H, Wei W. Roles of f-box proteins in cancer. Nat Rev Cancer (2014) 14:233–47. doi: 10.1038/nrc3700
4. Liu J, Chen T, Li S, Liu W, Wang P, Shang G. Targeting matrix metalloproteinases by E3 ubiquitin ligases as a way to regulate the tumor microenvironment for cancer therapy. Semin Cancer Biol Ahead Print (2022) S1044–579X(22):00138–9. doi: 10.1016/j.semcancer.2022.06.004
5. Gao J, Yang D, Cao R, Huang H, Ma J, Wang Z, et al. The role of Fbxo5 in the development of human malignant tumors. Am J Cancer Res (2022) 12:1456–64.
6. Wu L, Yu K, Chen K, Zhu X, Yang Z, Wang Q, et al. Fbxo45 facilitates pancreatic carcinoma progression by targeting USP49 for ubiquitination and degradation. Cell Death Dis (2022) 13(3):231. doi: 10.1038/s41419-022-04675-2
7. Wang Z, Dai X, Zhong J, Inuzuka H, Wan L, Li X, et al. SCF(beta-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun (2015) 6:10185. doi: 10.1038/ncomms10185
8. Randle SJ, Laman H. F-box protein interactions with the hallmark pathways in cancer. Semin Cancer Biol (2016) 36:3–17. doi: 10.1016/j.semcancer.2015.09.013
9. Yumimoto K, Akiyoshi S, Ueo H, Sagara Y, Onoyama I, Ueo H, et al. F-box protein FBXW7 inhibits cancer metastasis in a non-cell-autonomous manner. J Clin Invest (2015) 125:621–35. doi: 10.1172/JCI78782
10. Zheng N, Wang Z, Wei W. Ubiquitination-mediated degradation of cell cycle-related proteins by f-box proteins. Int J Biochem Cell Biol (2016) 73:99–110. doi: 10.1016/j.biocel.2016.02.005
11. Su J, Yin X, Zhou X, Wei W, Wang Z. The functions of f-box proteins in regulating the epithelial to mesenchymal transition. Curr Pharm Des (2015) 21:1311–7. doi: 10.2174/1381612821666141211144203
12. Song Y, Lin M, Liu Y, Wang ZW, Zhu X. Emerging role of f-box proteins in the regulation of epithelial-mesenchymal transition and stem cells in human cancers. Stem Cell Res Ther (2019) 10(1):124. doi: 10.1186/s13287-019-1222-0
13. Wang Z, Inuzuka H, Fukushima H, Wan L, Gao D, Shaik S, et al. Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep (2011) 13:36–43. doi: 10.1038/embor.2011.231
14. Yan L, Lin M, Pan S, Assaraf YG, Wang ZW, Zhu X. Emerging roles of f-box proteins in cancer drug resistance. Drug Resist Update (2020) 49:100673. doi: 10.1016/j.drup.2019.100673
15. Wang Z, Fukushima H, Gao D, Inuzuka H, Wan L, Lau AW, et al. The two faces of FBW7 in cancer drug resistance. Bioessays (2011) 33:851–9. doi: 10.1002/bies.201100101
16. Che X, Jian F, Wang Y, Zhang J, Shen J, Cheng Q, et al. FBXO2 promotes proliferation of endometrial cancer by ubiquitin-mediated degradation of FBN1 in the regulation of the cell cycle and the autophagy pathway. Front Cell Dev Biol (2020) 8:843. doi: 10.3389/fcell.2020.00843
17. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer (2018) 18:5–18. doi: 10.1038/nrc.2017.99
18. Goodall GJ, Wickramasinghe VO. RNA In cancer. Nat Rev Cancer (2021) 21:22–36. doi: 10.1038/s41568-020-00306-0
19. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discovery (2021) 20:629–51. doi: 10.1038/s41573-021-00219-z
20. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol (2019) 20:21–37. doi: 10.1038/s41580-018-0045-7
21. Liu J, Shang G. The roles of noncoding RNAs in the development of osteosarcoma stem cells and potential therapeutic targets. Front Cell Dev Biol (2022) 10:773038. doi: 10.3389/fcell.2022.773038
22. Liu F, Hu L, Pei Y, Zheng K, Wang W, Li S, et al. Long non-coding RNA AFAP1-AS1 accelerates the progression of melanoma by targeting miR-653-5p/RAI14 axis. BMC Cancer (2020) 20(1):258. doi: 10.1186/s12885-020-6665-2
23. Chen X, Liu Y, Liu H, Wang ZW, Zhu X. Unraveling diverse roles of noncoding RNAs in various human papillomavirus negative cancers. Pharmacol Ther (2022) 238:108188. doi: 10.1016/j.pharmthera.2022.108188
24. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer (2021) 21:446–60. doi: 10.1038/s41568-021-00353-1
25. Chen T, Liu J, Zhang H, Li J, Shang G. Long intergenic noncoding RNA00265 enhances cell viability and metastasis via targeting miR-485-5p/USP22 axis in osteosarcoma. Front Oncol (2022) 12:907472. doi: 10.3389/fonc.2022.907472
26. Zheng C, Chu M, Chen Q, Chen C, Wang ZW, Chen X. The role of lncRNA OIP5-AS1 in cancer development and progression. Apoptosis (2022) 27:311–21. doi: 10.1007/s10495-022-01722-3
27. Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer (2021) 20(1):116. doi: 10.1186/s12943-021-01406-7
28. Jiang W, Xia J, Xie S, Zou R, Pan S, Wang ZW, et al. Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Update (2020) 50:100683. doi: 10.1016/j.drup.2020.100683
29. Huarte M. The emerging role of lncRNAs in cancer. Nat Med (2015) 21:1253–61. doi: 10.1038/nm.3981
30. Lin M, Xu Y, Gao Y, Pan C, Zhu X, Wang ZW. Regulation of f-box proteins by noncoding RNAs in human cancers. Cancer Lett (2019) 466:61–70. doi: 10.1016/j.canlet.2019.09.008
31. Luo Y, Wang C, Yong P, Ye P, Liu Z, Fu Z, et al. Decreased expression of the long non-coding RNA SLC7A11-AS1 predicts poor prognosis and promotes tumor growth in gastric cancer. Oncotarget (2017) 8:112530–49. doi: 10.18632/oncotarget.22486
32. Liu Y, Fan X, Zhao Z, Shan X. LncRNA SLC7A11-AS1 contributes to lung cancer progression through facilitating TRAIP expression by inhibiting miR-4775. Onco Targets Ther (2020) 13:6295–302. doi: 10.2147/OTT.S253082
33. Xie W, Chu M, Song G, Zuo Z, Han Z, Chen C, et al. Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin Cancer Biol (2020) 83:303–18. doi: 10.1016/j.semcancer.2020.11.004
34. Luo Y, Xiang W, Liu Z, Yao L, Tang L, Tan W, et al. Functional role of the SLC7A11-AS1/xCT axis in the development of gastric cancer cisplatin-resistance by a GSH-dependent mechanism. Free Radic Biol Med (2022) 184:53–65. doi: 10.1016/j.freeradbiomed.2022.03.026
35. Yang Q, Li K, Huang X, Zhao C, Mei Y, Li X, et al. lncRNA SLC7A11-AS1 promotes chemoresistance by blocking SCF(beta-TRCP)-mediated degradation of NRF2 in pancreatic cancer. Mol Ther Nucleic Acids (2020) 19:974–85. doi: 10.1016/j.omtn.2019.11.035
36. Bi Y, Cui D, Xiong X, Zhao Y. The characteristics and roles of beta-TrCP1/2 in carcinogenesis. FEBS J (2021) 288:3351–74. doi: 10.1111/febs.15585
37. Wang L, Feng W, Yang X, Yang F, Wang R, Ren Q, et al. Fbxw11 promotes the proliferation of lymphocytic leukemia cells through the concomitant activation of NF-kappaB and beta-catenin/TCF signaling pathways. Cell Death Dis (2018) 9(4):427. doi: 10.1038/s41419-018-0440-1
38. Yao J, Yang J, Yang Z, Wang XP, Yang T, Ji B, et al. FBXW11 contributes to stem-cell-like features and liver metastasis through regulating HIC1-mediated SIRT1 transcription in colorectal cancer. Cell Death Dis (2021) 12(10):930. doi: 10.1038/s41419-021-04185-7
39. Su Y, Gu X, Zheng Q, Zhu L, Lu J, Li L. LncRNA PCGEM1 in human cancers: functions, mechanisms and promising clinical utility. Front Oncol (2022) 12:847745. doi: 10.3389/fonc.2022.847745
40. Zhang Q, Zheng J, Liu L. The long noncoding RNA PCGEM1 promotes cell proliferation, migration and invasion via targeting the miR-182/FBXW11 axis in cervical cancer. Cancer Cell Int (2019) 19:304. doi: 10.1186/s12935-019-1030-8
41. Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget (2014) 5:2000–15. doi: 10.18632/oncotarget.1859
42. Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH, et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett (2012) 586:1409–18. doi: 10.1016/j.febslet.2012.03.017
43. Fan J, Bellon M, Ju M, Zhao L, Wei M, Fu L, et al. Clinical significance of FBXW7 loss of function in human cancers. Mol Cancer (2022) 21(1):87. doi: 10.1186/s12943-022-01548-2
44. Snijders AM, Mao JH. Co-Expression network analysis of Fbxw7-associated lncrnas reveals their functions in radiation-induced thymic lymphoma. Insights Cancer Res (2016) 1:1–5.
45. Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G, et al. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget (2016) 7:15787–800. doi: 10.18632/oncotarget.7487
46. Bi LL, Han F, Zhang XM, Li YY. LncRNA MT1JP acts as a tumor inhibitor via reciprocally regulating wnt/beta-catenin pathway in retinoblastoma. Eur Rev Med Pharmacol Sci (2018) 22:4204–14. doi: 10.26355/eurrev_201807_15414
47. Zhu D, Zhang X, Lin Y, Liang S, Song Z, Dong C. MT1JP inhibits tumorigenesis and enhances cisplatin sensitivity of breast cancer cells through competitively binding to miR-24-3p. Am J Transl Res (2019) 11:245–56.
48. Wu H, Li S. Long non-coding RNA MT1JP exerts anti-cancer effects in breast cancer cells by regulating miR-92-3p. Gen Physiol Biophys (2020) 39:59–67. doi: 10.4149/gpb_2019039
49. Ouyang Q, Cui Y, Yang S, Wei W, Zhang M, Zeng J, et al. lncRNA MT1JP suppresses biological activities of breast cancer cells in vitro and in vivo by regulating the miRNA-214/RUNX3 axis. Onco Targets Ther (2020) 13:5033–46. doi: 10.2147/OTT.S241503
50. Ma J, Yan H, Zhang J, Tan Y, Gu W. Long-chain non-coding RNA (lncRNA) MT1JP suppresses biological activities of lung cancer by regulating miRNA-423-3p/Bim axis. Med Sci Monit (2019) 25:5114–26. doi: 10.12659/MSM.914387
51. Chen J, Lou J, Yang S, Lou J, Liao W, Zhou R, et al. MT1JP inhibits glioma progression via negative regulation of miR-24. Oncol Lett (2020) 19:334–42. doi: 10.3892/ol.2019.11085
52. Yang L, Liu G, Xiao S, Wang L, Liu X, Tan Q, et al. Long noncoding MT1JP enhanced the inhibitory effects of miR-646 on FGF2 in osteosarcoma. Cancer Biother Radiopharm (2020) 35:371–6. doi: 10.1089/cbr.2019.3328
53. Wu JH, Xu K, Liu JH, Du LL, Li XS, Su YM, et al. LncRNA MT1JP inhibits the malignant progression of hepatocellular carcinoma through regulating AKT. Eur Rev Med Pharmacol Sci (2020) 24:6647–56. doi: 10.26355/eurrev_202006_21651.
54. Shan QL, Chen NN, Meng GZ, Qu F. Overexpression of lncRNA MT1JP mediates apoptosis and migration of hepatocellular carcinoma cells by regulating miR-24-3p. Cancer Manag Res (2020) 12:4715–24. doi: 10.2147/CMAR.S249582
55. Mo W, Dai Y, Chen J, Liang L, Xu S, Xu X. Long noncoding RNA (lncRNA) MT1JP suppresses hepatocellular carcinoma (HCC) in vitro. Cancer Manag Res (2020) 12:7949–60. doi: 10.2147/CMAR.S253496
56. Yu T, Yu J, Lu L, Zhang Y, Zhou Y, Zhou Y, et al. MT1JP-mediated miR-24-3p/BCL2L2 axis promotes lenvatinib resistance in hepatocellular carcinoma cells by inhibiting apoptosis. Cell Oncol (Dordr) (2021) 44:821–34. doi: 10.1007/s13402-021-00605-0
57. Zhang S, Xu J, Chen Q, Zhang F, Wang H, Guo H. lncRNA MT1JP-overexpression abolishes the silencing of PTEN by miR-32 in hepatocellular carcinoma. Oncol Lett (2021) 22(2):604. doi: 10.3892/ol.2021.12865
58. Zhao W, Zhao J, Guo X, Feng Y, Zhang B, Tian L. LncRNA MT1JP plays a protective role in intrahepatic cholangiocarcinoma by regulating miR-18a-5p/FBP1 axis. BMC Cancer (2021) 21(1):142. doi: 10.1186/s12885-021-07838-0
59. Xu Y, Zhang G, Zou C, Zhang H, Gong Z, Wang W, et al. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem (2018) 46:2445–59. doi: 10.1159/000489651
60. Zhu C, Ma J, Li Y, Zhang Y, Da M. Low expression of long noncoding RNA MT1JP is associated with poor overall survival in gastric cancer patients: protocol for meta-analysis. Med (Baltimore) (2018) 97(21):e10394. doi: 10.1097/MD.0000000000010394
61. Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer (2018) 17(1):87. doi: 10.1186/s12943-018-0829-6
62. Liu W, Meng J, Su R, Shen C, Zhang S, Zhao Y, et al. SP1-mediated up-regulation of lncRNA TUG1 underlines an oncogenic property in colorectal cancer. Cell Death Dis (2022) 13(5):433. doi: 10.1038/s41419-022-04805-w
63. Ren Y, Lyu J, Guo Y, Yao Y, Hu L. Long noncoding RNA TUG1 inhibits tumor progression through regulating siglec-15-related anti-immune activity in hepatocellular carcinoma. J Immunol Res (2022) 2022:9557859. doi: 10.1155/2022/9557859
64. Liu Y, Mao X, Ma Z, Chen W, Guo X, Yu L, et al. Aberrant regulation of LncRNA TUG1-microRNA-328-3p-SRSF9 mRNA axis in hepatocellular carcinoma: a promising target for prognosis and therapy. Mol Cancer (2022) 21(1):36. doi: 10.1186/s12943-021-01493-6
65. Xiu C, Song R, Jiang J. TUG1 promotes retinoblastoma progression by sponging miR-516b-5p to upregulate H6PD expression. Transl Cancer Res (2021) 10:738–47. doi: 10.21037/tcr-19-1480
66. Xia C, Li Q, Cheng X, Wu T, Gao P, Gu Y. Insulin-like growth factor 2 mRNA-binding protein 2-stabilized long non-coding RNA taurine up-regulated gene 1 (TUG1) promotes cisplatin-resistance of colorectal cancer via modulating autophagy. Bioengineered (2022) 13:2450–69. doi: 10.1080/21655979.2021.2012918
67. Tan J, Liu B, Zhou L, Gao J, Wang XK, Liu Y, et al. LncRNA TUG1 promotes bladder cancer malignant behaviors by regulating the miR-320a/FOXQ1 axis. Cell Signal (2022) 91:110216. doi: 10.1016/j.cellsig.2021.110216
68. Sun J, Zhou H, Bao X, Wu Y, Jia H, Zhao H, et al. lncRNA TUG1 facilitates colorectal cancer stem cell characteristics and chemoresistance by enhancing GATA6 protein stability. Stem Cells Int (2021) 2021:1075481. doi: 10.1155/2021/1075481
69. Dai T, Liang J, Liu W, Zou Y, Niu F, Li M, et al. The miRNA mir-582-3p suppresses ovarian cancer progression by targeting AKT/MTOR signaling via lncRNA TUG1. Bioengineered (2021) 12:10771–81. doi: 10.1080/21655979.2021.2003662
70. Jin Y, Cao J, Hu X, Cheng H. Long noncoding RNA TUG1 upregulates VEGFA to enhance malignant behaviors in stomach adenocarcinoma by sponging miR-29c-3p. J Clin Lab Anal (2021) 35(12):e24106. doi: 10.1002/jcla.24106
71. Liu W, Feng Q, Liao W, Li E, Wu L. TUG1 promotes the expression of IFITM3 in hepatocellular carcinoma by competitively binding to miR-29a. J Cancer (2021) 12:6905–20. doi: 10.7150/jca.57477
72. Yuan JB, Gu L, Chen L, Yin Y, Fan BY. Annexin A8 regulated by lncRNA-TUG1/miR-140-3p axis promotes bladder cancer progression and metastasis. Mol Ther Oncolytics (2021) 22:36–51. doi: 10.1016/j.omto.2021.04.008
73. Zhang R, Yang F, Fan H, Wang H, Wang Q, Yang J, et al. Long non-coding RNA TUG1/microRNA-187-3p/TESC axis modulates progression of pituitary adenoma via regulating the NF-kappaB signaling pathway. Cell Death Dis (2021) 12(6):524. doi: 10.1038/s41419-021-03812-7
74. Li H, Huang F, Liu XQ, Liu HC, Dai M, Zeng J. LncRNA TUG1 promotes ewing’s sarcoma cell proliferation, migration, and invasion via the miR-199a-3p-MSI2 signaling pathway. Neoplasma (2021) 68:590–601. doi: 10.4149/neo_2021_201110N1198
75. Jin G, Yang Y, Tuo G, Wang W, Zhu Z. LncRNA TUG1 promotes tumor growth and metastasis of esophageal squamous cell carcinoma by regulating XBP1 via competitively binding to miR-498. Neoplasma (2020) 67:751–61. doi: 10.4149/neo_2020_190805N717
76. Xue LX, Chen SF, Xue SX, Liu PD, Liu HB. LncRNA TUG1 compromised neuronal mitophagy in cerebral ischemia/reperfusion injury by targeting sirtuin 1. Cell Biol Toxicol (2022). doi: 10.1007/s10565-022-09700-w
77. Mou J, Wang B, Liu Y, Zhao F, Wu Y, Xu W, et al. FER1L4A long non-coding RNA with multiple roles in the occurrence and development of tumors. Curr Pharm Des (2022). doi: 10.2174/1381612828666220324141016
78. Xia L, Nie D, Wang G, Sun C, Chen G. FER1L4/miR-372/E2F1 works as a ceRNA system to regulate the proliferation and cell cycle of glioma cells. J Cell Mol Med (2019) 23:3224–33. doi: 10.1111/jcmm.14198
79. Gao X, Wang N, Wu S, Cui H, An X, Yang Y. Long noncoding RNA FER1L4 inhibits cell proliferation and metastasis through regulation of the PI3K/AKT signaling pathway in lung cancer cells. Mol Med Rep (2019) 20:182–90. doi: 10.3892/mmr.2019.10219.
80. Ouyang L, Yang M, Wang X, Fan J, Liu X, Zhang Y, et al. Long noncoding RNA FER1L4 inhibits cell proliferation and promotes cell apoptosis via the PTEN/AKT/p53 signaling pathway in lung cancer. Oncol Rep (2021) 45:359–67. doi: 10.3892/or.2020.7861
81. Ye F, Tian L, Zhou Q, Feng D. LncRNA FER1L4 induces apoptosis and suppresses EMT and the activation of PI3K/AKT pathway in osteosarcoma cells via inhibiting miR-18a-5p to promote SOCS5. Gene (2019) 721:144093. doi: 10.1016/j.gene.2019.144093
82. Cox A, Tolkach Y, Kristiansen G, Ritter M, Ellinger J. The lncRNA Fer1L4 is an adverse prognostic parameter in clear-cell renal-cell carcinoma. Clin Transl Oncol (2020) 22:1524–31. doi: 10.1007/s12094-020-02291-0
83. Zhang N, Zeng L, Wang S, Wang R, Yang R, Jin Z, et al. LncRNA FER1L4 promotes oral squamous cell carcinoma progression via targeting miR-133a-5p/Prx1 axis. Onco Targets Ther (2021) 14:795–806. doi: 10.2147/OTT.S277351
84. Ostovarpour M, Khalaj-Kondori M, Ghasemi T. Correlation between expression levels of lncRNA FER1L4 and RB1 in patients with colorectal cancer. Mol Biol Rep (2021) 48:4581–9. doi: 10.1007/s11033-021-06488-6
85. Uboveja A, Satija YK, Siraj F, Saluja D. p73-regulated FER1L4 lncRNA sponges the oncogenic potential of miR-1273g-3p and aids in the suppression of colorectal cancer metastasis. iScience (2022) 25(2):103811. doi: 10.1016/j.isci.2022.103811
86. Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep (2014) 4:6088. doi: 10.1038/srep06088
87. Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep (2015) 5:13445. doi: 10.1038/srep13445
88. Xu J, Li N, Deng W, Luo S. Long noncoding RNA FER1L4 suppresses proliferation, invasion, migration and lymphatic metastasis of gastric cancer cells through inhibiting the hippo-YAP signaling pathway. Am J Transl Res (2020) 12:5481–95.
89. Qiao Q, Li H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem Biophys Res Commun (2016) 478:507–12. doi: 10.1016/j.bbrc.2016.06.160
90. Kong Y, Ren Z. Overexpression of LncRNA FER1L4 in endometrial carcinoma is associated with favorable survival outcome. Eur Rev Med Pharmacol Sci (2018) 22:8113–8. doi: 10.26355/eurrev_201812_16502
91. Ma W, Zhang CQ, Li HL, Gu J, Miao GY, Cai HY, et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci (2018) 22:2638–45. doi: 10.26355/eurrev_201805_14958
92. Wang X, Dong K, Jin Q, Ma Y, Yin S, Wang S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J Cell Biochem (2019) 120:6781–8. doi: 10.1002/jcb.27980
93. Liu S, Zou B, Tian T, Luo X, Mao B, Zhang X, et al. Overexpression of the lncRNA FER1L4 inhibits paclitaxel tolerance of ovarian cancer cells via the regulation of the MAPK signaling pathway. J Cell Biochem (2018) 120(5):7581–89. doi: 10.1002/jcb.28032
94. Huo W, Qi F, Wang K. Long non-coding RNA FER1L4 inhibits prostate cancer progression via sponging miR-92a-3p and upregulation of FBXW7. Cancer Cell Int (2020) 20:64. doi: 10.1186/s12935-020-1143-0
95. Lin C, Zhang S, Wang Y, Wang Y, Nice E, Guo C, et al. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin Cancer Res (2018) 24:486–98. doi: 10.1158/1078-0432.CCR-17-1851
96. Chen P, Wang R, Yue Q, Hao M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun (2018) 503:2956–62. doi: 10.1016/j.bbrc.2018.08.077
97. Dong MM, Peng SJ, Yuan YN, Luo HP. LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma (2019) 66:564–75. doi: 10.4149/neo_2018_180927N721
98. Luo J, Liu Z. Long non-coding RNA TTN-AS1 promotes the progression of lung adenocarcinoma by regulating PTEN/PI3K/AKT signaling pathway. Biochem Biophys Res Commun (2019) 514:140–7. doi: 10.1016/j.bbrc.2019.04.050
99. Zhong Y, Wang J, Lv W, Xu J, Mei S, Shan A. LncRNA TTN-AS1 drives invasion and migration of lung adenocarcinoma cells via modulation of miR-4677-3p/ZEB1 axis. J Cell Biochem (2019) 120:17131–41. doi: 10.1002/jcb.28973
100. Luo JF, Xu J, Zheng JZ. Long non-coding RNA TTN-AS1 promotes cell proliferation and inhibits cell apoptosis in prostatic cancer by sponging miR-193a-5p. Eur Rev Med Pharmacol Sci (2019) 23:7816–25. doi: 10.26355/eurrev_201909_18991
101. Fu D, Lu C, Qu X, Li P, Chen K, Shan L, et al. LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY) (2019) 11:8374–85. doi: 10.18632/aging.102325
102. Wang Y, Jiang F, Xiong Y, Cheng X, Qiu Z, Song R. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci (2020) 244:116936. doi: 10.1016/j.lfs.2019.116936
103. Cui Z, Han B, Wang X, Li Z, Wang J, Lv Y. Long non-coding RNA TTN-AS1 promotes the proliferation and invasion of colorectal cancer cells by activating miR-497-Mediated PI3K/Akt/mTOR signaling. Onco Targets Ther (2019) 12:11531–9. doi: 10.2147/OTT.S229104
104. Fang J, Huang C, Ke J, Li J, Zhang W, Xue H, et al. lncRNA TTN-AS1 facilitates proliferation, invasion, and epithelial-mesenchymal transition of breast cancer cells by regulating miR-139-5p/ZEB1 axis. J Cell Biochem (2020) 121(12):4772–84. doi: 10.1002/jcb.29700
105. Feng H, Wang Q, Xiao W, Zhang B, Jin Y, Lu H. LncRNA TTN-AS1 regulates miR-524-5p and RRM2 to promote breast cancer progression. Onco Targets Ther (2020) 13:4799–811. doi: 10.2147/OTT.S243482
106. Lin K, Chen H, Su C, Zhu H, Lai C, Shi Y. Long non-coding RNA TTN-AS1 serves as a competing endogenous RNA of miR-195 to facilitate clear cell renal cell carcinoma progression. Cancer Manag Res (2020) 12:3091–7. doi: 10.2147/CMAR.S249456
107. Miao S, Wang J, Xuan L, Liu X. LncRNA TTN-AS1 acts as sponge for miR-15b-5p to regulate FBXW7 expression in ovarian cancer. Biofactors (2020) 46:600–7. doi: 10.1002/biof.1622
108. Zhang P, Pan Y, Sun J, Pan G. Aberrant expression of LncRNA CASC2 mediated the cell viability, apoptosis and autophagy of colon cancer cells by sponging miR-19a via NF-kappaB signaling pathway. Int J Exp Pathol (2021) 102:163–71. doi: 10.1111/iep.12393
109. Li X, Sun J, Lou L, Fan X, Zhang W, Li Q. Overexpression of lncRNA CASC2 inhibits the tumorigenesis of thyroid cancer via sponging miR-24-3p. Am J Transl Res (2020) 12:6314–24.
110. Ju B, Liu Z, Nai C, Zhu X. Long non-coding RNA CASC2 induces apoptosis and autophagy in human colon cancer cells via modulation of TRIM16 expression. Am J Transl Res (2020) 12:2695–702.
111. Dai W, Mu L, Cui Y, Li Y, Chen P, Xie H, et al. Long noncoding RNA CASC2 enhances berberineinduced cytotoxicity in colorectal cancer cells by silencing BCL2. Mol Med Rep (2019) 20:995–1006. doi: 10.3892/mmr.2019.10326
112. Tao L, Tian P, Yang L, Guo X. lncRNA CASC2 enhances (131)I sensitivity in papillary thyroid cancer by sponging miR-155. BioMed Res Int (2020) 2020:7183629. doi: 10.1155/2020/7183629
113. Ding Z, Kang J, Yang Y. Long non-coding RNA CASC2 enhances irradiation-induced endoplasmic reticulum stress in NSCLC cells through PERK signaling. 3 Biotech (2020) 10(10):449. doi: 10.1007/s13205-020-02443-7
114. Zhang H, Feng X, Zhang M, Liu A, Tian L, Bo W, et al. Long non-coding RNA CASC2 upregulates PTEN to suppress pancreatic carcinoma cell metastasis by downregulating miR-21. Cancer Cell Int (2019) 19:18. doi: 10.1186/s12935-019-0728-y
115. Sun J, Xu H, Lei Z, Li Z, Zhu H, Deng Z, et al. The lncRNA CASC2 modulates hepatocellular carcinoma cell sensitivity and resistance to TRAIL through apoptotic and non-apoptotic signaling. Front Oncol (2021) 11:726622. doi: 10.3389/fonc.2021.726622
116. Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem (2018) 119:6391–7. doi: 10.1002/jcb.26479
117. Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer (2017) 16(1):123. doi: 10.1186/s12943-017-0702-z
118. Sun J, Liu L, Zou H, Yu W. The long non-coding RNA CASC2 suppresses cell viability, migration, and invasion in hepatocellular carcinoma cells by directly downregulating miR-183. Yonsei Med J (2019) 60(10):905–13. doi: 10.3349/ymj.2019.60.10.905
119. Malakoti F, Targhazeh N, Karimzadeh H, Mohammadi E, Asadi M, Asemi Z, et al. Multiple function of lncRNA MALAT1 in cancer occurrence and progression. Chem Biol Drug Des (2021). doi: 10.1111/cbdd.14006
120. Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol (2015) 36:3355–9. doi: 10.1007/s13277-014-2969-7
121. Han Y, Zhou L, Wu T, Huang Y, Cheng Z, Li X, et al. Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell Mol Neurobiol (2016) 36:1097–107. doi: 10.1007/s10571-015-0303-6
122. Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis (2016) 7:e2123. doi: 10.1038/cddis.2015.407
123. Xiang J, Guo S, Jiang S, Xu Y, Li J, Li L, et al. Silencing of long non-coding RNA MALAT1 promotes apoptosis of glioma cells. J Korean Med Sci (2016) 31:688–94. doi: 10.3346/jkms.2016.31.5.688
124. Li Z, Xu C, Ding B, Gao M, Wei X, Ji N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J Neurooncol (2017) 134:19–28. doi: 10.1007/s11060-017-2498-5
125. Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, et al. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun (2017) 492:480–6. doi: 10.1016/j.bbrc.2017.08.070
126. Wu L, Liu Q, Fan C, Yi X, Cheng B. MALAT1 recruited the E3 ubiquitin ligase FBXW7 to induce CRY2 ubiquitin-mediated degradation and participated in trophoblast migration and invasion. J Cell Physiol (2021) 236:2169–77. doi: 10.1002/jcp.30003
127. Cao S, Wang Y, Li J, Lv M, Niu H, Tian Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res (2016) 6:2561–74.
128. Sharma U, Barwal TS, Malhotra A, Pant N, Vivek Dey D, et al. Long non-coding RNA TINCR as potential biomarker and therapeutic target for cancer. Life Sci (2020) 257:118035. doi: 10.1016/j.lfs.2020.118035
129. Han X, Jia Y, Chen X, Sun C, Sun J. lncRNA TINCR attenuates the proliferation and invasion, and enhances the apoptosis of cutaneous malignant melanoma cells by regulating the miR4245p/LATS1 axis. Oncol Rep (2021) 46(5):238. doi: 10.3892/or.2021.8189
130. He G, Pang R, Han J, Jia J, Ding Z, Bi W, et al. TINCR inhibits the proliferation and invasion of laryngeal squamous cell carcinoma by regulating miR-210/BTG2. BMC Cancer (2021) 21(1):753. doi: 10.1186/s12885-021-08513-0
131. Tang C, Feng W, Bao Y, Du H. Long non-coding RNA TINCR promotes hepatocellular carcinoma proliferation and invasion via STAT3 signaling by direct interacting with T-cell protein tyrosine phosphatase (TCPTP). Bioengineered (2021) 12:2119–31. doi: 10.1080/21655979.2021.1930336
132. Zhang ML, Liu WW, Li WD. Imbalance of molecular module of TINCR-miR-761 promotes the metastatic potential of early triple negative breast cancer and partially offsets the anti-tumor activity of luteolin. Cancer Manag Res (2021) 13:1877–86. doi: 10.2147/CMAR.S288271
133. Lu D, Di S, Zhuo S, Zhou L, Bai R, Ma T, et al. The long noncoding RNA TINCR promotes breast cancer cell proliferation and migration by regulating OAS1. Cell Death Discovery (2021) 7(1):41. doi: 10.1038/s41420-021-00419-x
134. Wang Q, Liu J, You Z, Yin Y, Liu L, Kang Y, et al. LncRNA TINCR favors tumorigenesis via STAT3-TINCR-EGFR-feedback loop by recruiting DNMT1 and acting as a competing endogenous RNA in human breast cancer. Cell Death Dis (2021) 12(1):83. doi: 10.1038/s41419-020-03188-0
135. Liu X, Ma J, Xu F, Li L. TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. BioMed Pharmacother (2018) 99:9–17. doi: 10.1016/j.biopha.2018.01.049
136. Zhu ZJ, He JK. TINCR facilitates non-small cell lung cancer progression through BRAF-activated MAPK pathway. Biochem Biophys Res Commun (2018) 497:971–7. doi: 10.1016/j.bbrc.2018.02.059
137. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188502. doi: 10.1016/j.bbcan.2021.188502
138. Shi D, Zhang Y, Lu R, Zhang Y. The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. BioMed Pharmacother (2018) 97:543–50. doi: 10.1016/j.biopha.2017.10.083
139. Lin P, Fu J, Zhao B, Lin F, Zou H, Liu L, et al. Fbxw8 is involved in the proliferation of human choriocarcinoma JEG-3 cells. Mol Biol Rep (2011) 38:1741–7. doi: 10.1007/s11033-010-0288-7
140. Shi D, Tan Z, Lu R, Yang W, Zhang Y. MicroRNA-218 inhibits the proliferation of human choriocarcinoma JEG-3 cell line by targeting Fbxw8. Biochem Biophys Res Commun (2014) 450:1241–6. doi: 10.1016/j.bbrc.2014.06.094
141. Yuan S, Xiang Y, Wang G, Zhou M, Meng G, Liu Q, et al. Hypoxia-sensitive LINC01436 is regulated by E2F6 and acts as an oncogene by targeting miR-30a-3p in non-small cell lung cancer. Mol Oncol (2019) 13:840–56. doi: 10.1002/1878-0261.12437
142. Zhang Y, Yang G, He X, Chen S, Zhang F, Fang X. LINC01436, regulating miR-585 and FBXO11, is an oncogenic lncRNA in the progression of gastric cancer. Cell Biol Int (2020) 44:882–93. doi: 10.1002/cbin.11287
143. Xu Y, Dong M, Wang J, Zhao W, Jiao M. LINC01436 inhibited miR-585-3p expression and upregulated MAPK1 expression to promote gastric cancer progression. Dig Dis Sci (2021) 66:1885–94. doi: 10.1007/s10620-020-06487-w
144. Lu MD, Liu D, Li YX. LINC01436 promotes the progression of gastric cancer via regulating miR-513a-5p/APE1 axis. Onco Targets Ther (2020) 13:10607–19. doi: 10.2147/OTT.S257747
145. Xu H, Wang X, Zhang Y, Zheng W, Zhang H. GATA6-AS1 inhibits ovarian cancer cell proliferation and migratory and invasive abilities by sponging miR-19a-5p and upregulating TET2. Oncol Lett (2021) 22(4):718. doi: 10.3892/ol.2021.12979
146. Kang H, Ma D, Zhang J, Zhao J, Yang M. Long non-coding RNA GATA6-AS1 upregulates GATA6 to regulate the biological behaviors of lung adenocarcinoma cells. BMC Pulm Med (2021) 21(1):166. doi: 10.1186/s12890-021-01521-7
147. Wang Z, Pan L, Yang L, Lv P, Mai S, Wang Y. Long non-coding RNA GATA6-AS1 sponges miR-324-5p to inhibit lung cancer cell proliferation and invasion. Onco Targets Ther (2020) 13:9741–51. doi: 10.2147/OTT.S256336
148. Jiang GY, Zhang XP, Wang L, Lin XY, Yu JH, Wang EH, et al. FBXO25 promotes cell proliferation, invasion, and migration of NSCLC. Tumour Biol (2016) 37:14311–9. doi: 10.1007/s13277-016-5298-1
149. Kuzmanov A, Johansen P, Hofbauer G. FBXO25 promotes cutaneous squamous cell carcinoma growth and metastasis through cyclin D1. J Invest Dermatol (2020) 140:2496–504. doi: 10.1016/j.jid.2020.04.003
150. Baumann U, Fernandez-Saiz V, Rudelius M, Lemeer S, Rad R, Knorn AM, et al. Disruption of the PRKCD-FBXO25-HAX-1 axis attenuates the apoptotic response and drives lymphomagenesis. Nat Med (2014) 20:1401–9. doi: 10.1038/nm.3740
151. He S, Yang S, Zhang Y, Li X, Gao D, Zhong Y, et al. LncRNA ODIR1 inhibits osteogenic differentiation of hUC-MSCs through the FBXO25/H2BK120ub/H3K4me3/OSX axis. Cell Death Dis (2019) 10(12):947. doi: 10.1038/s41419-019-2148-2
152. Chou JL, Su HY, Chen LY, Liao YP, Hartman-Frey C, Lai YH, et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest (2010) 90:414–25. doi: 10.1038/labinvest.2009.138
153. Zhang N, Liao Y, Lv W, Zhu S, Qiu Y, Chen N, et al. FBXO32 targets PHPT1 for ubiquitination to regulate the growth of EGFR mutant lung cancer. Cell Oncol (Dordr) (2022) 45:293–307. doi: 10.1007/s13402-022-00669-6
154. Zhou H, Liu Y, Zhu R, Ding F, Wan Y, Li Y, et al. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene (2017) 36:3312–21. doi: 10.1038/onc.2016.479
Keywords: cancer, lncRNAs, F-box protein, treatment, oncogenesis, noncoding RNA
Citation: Xia L, Chen J, Huang M, Mei J and Lin M (2022) The functions of long noncoding RNAs on regulation of F-box proteins in tumorigenesis and progression. Front. Oncol. 12:963617. doi: 10.3389/fonc.2022.963617
Received: 07 June 2022; Accepted: 27 June 2022;
Published: 19 July 2022.
Edited by:
Aamir Ahmad, University of Alabama at Birmingham, United StatesReviewed by:
Mohammad Aatif, King Faisal University, Saudi ArabiaGuanning Shang, China Medical University, China
Copyright © 2022 Xia, Chen, Huang, Mei and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Lin, bGlubWlud211QDE2My5jb20=; d3psaW5taW5Ad211LmVkdS5jbg==
 Lu Xia
Lu Xia Jingyun Chen
Jingyun Chen Min Huang
Min Huang Min Lin
Min Lin