- 1S.H. Ho Urology Centre, Department of Surgery, The Chinese University of Hong Kong, Hong Kong SAR, China
- 2Department of Clinical Oncology, Tuen Mun Hospital, Hong Kong SAR, China
- 3Division of Urology, Department of Surgery, Tuen Mun Hospital, Hong Kong SAR, China
- 4Division of Urology, Department of Surgery, United Christian Hospital, Hong Kong SAR, China
- 5Division of Urology, Department of Surgery, Tseung Kwan O Hospital, Hong Kong SAR, China
- 6Department of Oncology, United Christian Hospital, Hong Kong SAR, China
- 7Department of Pathology, Hong Kong Sanatorium and Hospital, Hong Kong SAR, China
- 8Department of Clinical Oncology, State Key Laboratory of Translational Oncology, Sir YK Pao Centre for Cancer, Hong Kong Cancer Institute, The Chinese University of Hong Kong, Hong Kong SAR, China
- 9Comprehensive Oncology Centre, Hong Kong Sanatorium and Hospital, Hong Kong SAR, China
Background: In recent years, indications for genetic testing in prostate cancer (PC) have expanded from patients with a family history of prostate and/or related cancers to those with advanced castration-resistant disease, and even to early PC patients for determination of the appropriateness of active surveillance. The current consensus aims to provide guidance to urologists, oncologists and pathologists working with Asian PC patients on who and what to test for in selected populations.
Methods: A joint consensus panel from the Hong Kong Urological Association and Hong Kong Society of Uro-Oncology was convened over a series of 5 physical and virtual meetings. A background literature search on genetic testing in PC was performed in PubMed, ClinicalKey, EBSCOHost, Ovid and ProQuest, and three working subgroups were formed to review and present the relevant evidence. Meeting agendas adopted a modified Delphi approach to ensure that discussions proceed in a structured, iterative and balanced manner, which was followed by an anonymous voting on candidate statements. Of 5 available answer options, a consensus statement was accepted if ≥ 75% of the panelists chose “Accept Completely” (Option A) or “Accept with Some Reservation” (Option B).
Results: The consensus was structured into three parts: indications for testing, testing methods, and therapeutic implications. A list of 35 candidate statements were developed, of which 31 were accepted. The statements addressed questions on the application of PC genetic testing data and guidelines to Asian patients, including patient selection for germline testing, selection of gene panel and tissue sample, provision of genetic counseling, and use of novel systemic treatments in metastatic castration-resistant PC patients.
Conclusion: This consensus provides guidance to urologists, oncologists and pathologists working with Asian patients on indications for genetic testing, testing methods and technical considerations, and associated therapeutic implications.
1 Introduction
The incidence of prostate cancer (PC) has risen rapidly in Asia (1). In Hong Kong, PC was the third most commonly diagnosed male cancer in 2019, with the highest rate of increase in new cases among all cancers from the previous year (2).
Although genetic testing has traditionally not been performed in PC patients, there is growing evidence for its role in providing additional prognostic and therapeutic information for selected patient populations. Sequencing studies demonstrated that ~11.8% of metastatic PC (mPC) patients carry inherited DNA repair gene mutations based on germline testing (3), and ~23% of castration-resistant prostate cancer (CRPC) patients show DNA repair pathway aberrations on somatic testing (4). Currently, poly (adenosine diphosphate-ribose) polymerase inhibitor (PARPi) treatment is indicated for metastatic CRPC (mCRPC) patients who harbor homologous recombination repair (HRR) gene mutations, including BRCA1/2 or ATM, which showed an overall survival (OS) benefit in the PROfound trial (5, 6).
In recent years, indications for genetic testing in PC have expanded from patients with a family history (FH) of prostate and/or related cancers to those with advanced castration-resistant disease, and even to early PC patients for determination of the appropriateness of active surveillance (AS) (7). The latest international guidelines and consensuses provide guidance and perspectives on the indications and modes of genetic testing in PC, including relevant clinical scenarios where patient management could be affected by the identification of a pathogenic mutation. Depending on the indication for genetic testing, the selection of testing method and technique will likely vary, such as germline versus somatic testing, or tumor versus blood sampling. For somatic testing, when tumor tissue is not readily available, circulating tumor DNA (ctDNA) from blood plasma may provide a convenient alternative (8, 9).
Despite the availability of data and international guidelines in support of PC genetic testing, formidable barriers exist in applying the latest developments to an Asian setting. First, caution must be exercised in the direct application of Caucasian-based guidelines to Asian men with PC because Asian data for such testing are limited, and inter-ethnic differences in patterns of genetic mutations (10) and pharmacogenomics (11) are notable. Second, the lack of genetic counseling resources for hereditary conditions can deter physicians from suggesting germline testing to patients, even when a positive germline mutation (e.g. germline BRCA mutation) seems plausible. Third, an optimal testing panel will need to be defined, taking into consideration the given indication and setting.
To facilitate the development of strategies for genetic testing in PC in Hong Kong, an Asian setting, a local expert consensus was jointly organized by the Hong Kong Urological Association (HKUA) and Hong Kong Society of Uro-Oncology (HKSUO), which are the two most representative professional organizations for PC management in Hong Kong. The statements were derived from available literature and overseas guidelines, and supplemented with the expertise of panel members where evidence was limited.
2 Methods
2.1 Panel Formation and Meetings
The joint consensus panel was formed from 8 local experts: 4 urologists representing the HKUA, 3 oncologists representing the HKSUO, and 1 invited pathologist (Supplementary Table 1 and Appendix A). A series of 5 meetings were held for literature presentations, discussions and voting (Supplementary Table 2).
2.2 Topics and Literature Review
A literature review of 5 medical databases was conducted to retrieve major publications that were relevant to the present study: PubMed, ClinicalKey, EBSCOHost, Ovid and ProQuest. The basic keywords used were “genetic testing” and “prostate cancer”. Additional keywords were “BRCA”, “castration-resistant”, “chemotherapy”, “Delphi method”, “family history”, “germline”, “guideline”, “homologous recombination repair”, “liquid biopsy”, “metastatic”, “mismatch repair”, “PARP inhibitor” and “somatic.” A list of 47 major articles were selected for in-depth discussion, which comprised 30 large-scale studies, 2 meta-analyses, 6 major guidelines, 6 consensuses and 3 high-impact narrative reviews.
2.3 Modified Delphi Method
Where there is incomplete evidence, the Delphi method can be used to make estimations and predictions, determine collective values, and define foundational concepts (12). This method has been employed in recent genitourinary cancer consensuses (13–15). In this study, the following modified Delphi method was used to ensure that the panel discussions would proceed in a structured and iterative manner, contents would be well-balanced, and that participants could contribute fairly and equally: 1) for each of the 3 identified debate areas, a working subgroup was formed to review and present the latest literature evidence for deliberation; 2) each subgroup was also responsible for drafting a set of candidate consensus statements, which were further revised during the discussions, and compiled into the final voting statements; 3) each statement was discussed around the table twice, over two meetings; and 4) at the final meeting, panelists were instructed to vote anonymously on each statement’s “practicability of recommendation” in the locality, based on a set of predefined judgement criteria (Supplementary Table 3). A consensus statement was accepted if ≥ 75% of the panelists chose “Accept Completely” (Option A) or “Accept with Some Reservation” (Option B) (16). (Supplementary Table 4 contains the complete voting record.)
3 Results
Preliminary discussions converged on a “patient journey” approach to structuring the consensus (Figure 1). Moreover, 3 core debate areas were identified: 1) indications for genetic testing in PC; 2) testing methods and technical considerations; and 3) therapeutic implications. A total of 35 candidate statements were finalized for voting. The voting accepted 31 statements (Table 1); explanations for their acceptance are given below. Table 2 provides a brief summary of the consensus, organized by patient disease status. (Voting records, rejected statements, and the latter’s explanations can be found in Supplementary Tables 4, 5 and Appendix B, respectively.)
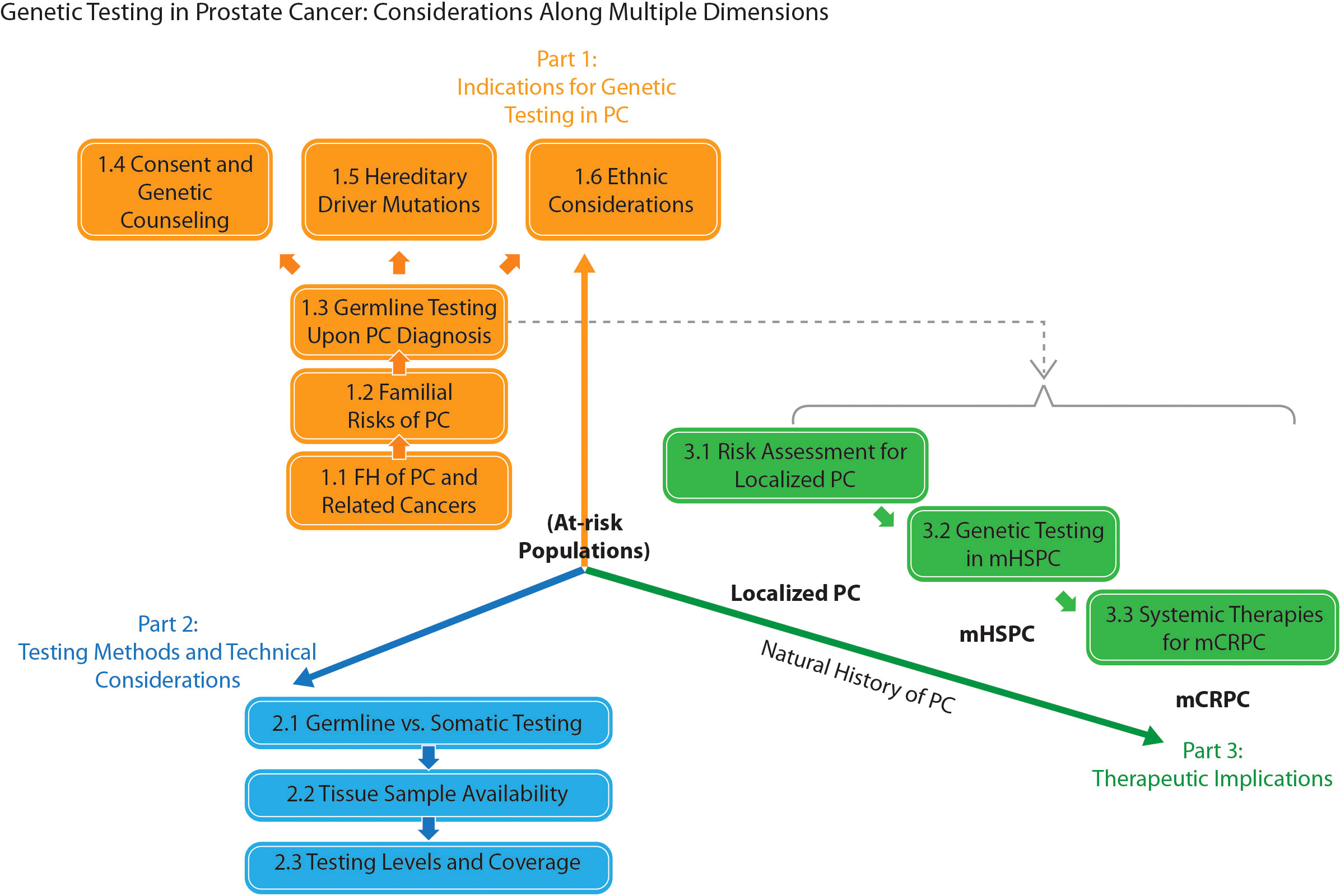
Figure 1 A “patient journey” schematic representation of the topic division for the consensus in prostate cancer (PC) genetic testing. FH, family history; mCRPC / mHSPC, metastatic castration-resistant / hormone-sensitive prostate cancer.
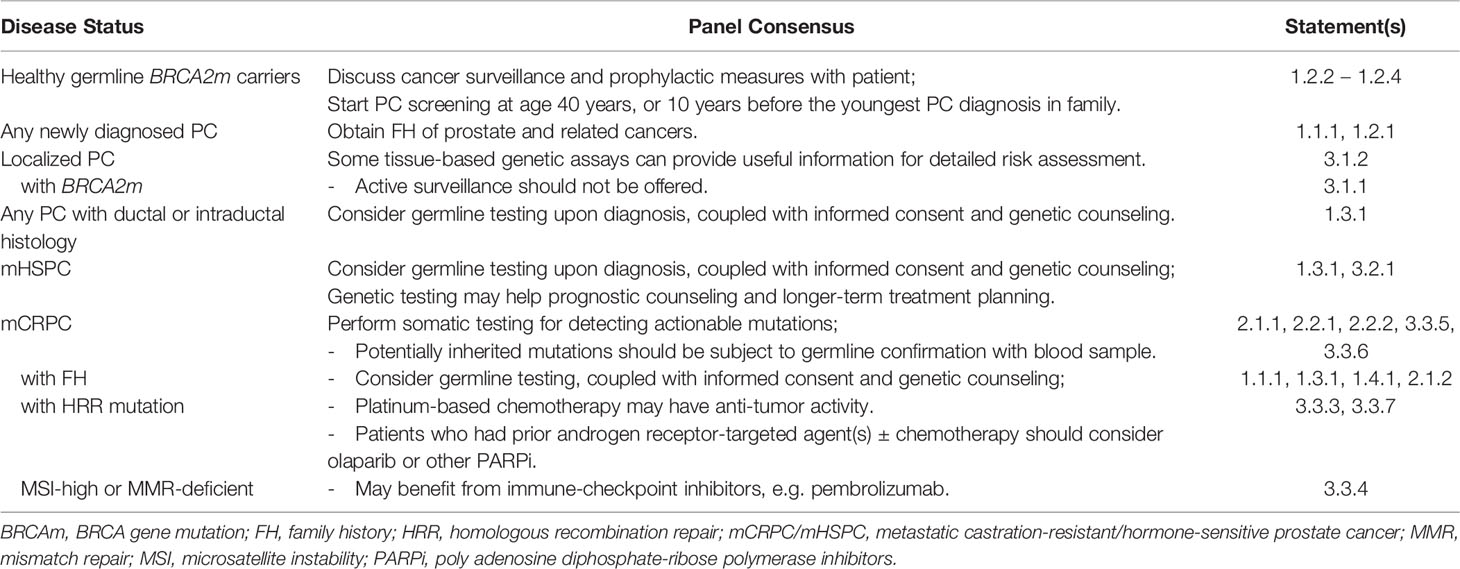
Table 2 Summary of panel consensus on genetic testing in prostate cancer (PC) according to disease status.
3.1 Part 1: Indications for Genetic Testing in PC
3.1.1 FH of PC and Related Cancers
The FH criteria (Statement 1.1.1 in Table 1) were modified from the National Comprehensive Cancer Network (NCCN)’s Prostate Cancer Early Detection guidelines (17) and Philadelphia Prostate Cancer Consensus 2019 (18), with considerations of the local age of PC onset (2). The exclusion of Grade Group 1 (i.e. low grade) PC followed that in the NCCN guidelines. Also, in a multicenter sequencing analysis of 82 mPC patients with pathogenic DNA repair gene mutations, only two had a Gleason score of ≤ 6 (3).
3.1.2 Familial Risks of PC
Taking FH is a definitive part of the clinical investigation of PC patients (1.2.1). In a U.S. survey of 132 urologists (21), 98% asked about FH of PC, but only some inquired about other malignancies associated with PC; e.g., 76%, 52% and 48% asked about breast, ovarian and colorectal cancers, respectively. Also, 78% and 62% of respondents were aware of the association between hereditary PC and BRCA2 mutations (BRCA2m) and BRCA1 mutations, respectively, but < 40% were aware of the association between hereditary PC and Lynch syndrome.
Through the increasing availability of germline testing, mutation carriers can be identified to facilitate early PC detection (1.2.2). Alternatively, patients with a personal history of related cancers may be referred for genetic testing and counselling through the relevant guideline recommendations, such as for male breast cancer (69). For germline mutation carriers, the associated personal and familial implications should be explained clearly, and should include associated cancer risks and ways to cope with potential stress and anxiety (see also Consent and genetic counseling).
For BRCA2m carriers (including those with PC), the NCCN Genetic/Familial High-Risk Assessment guidelines (70) recommend clinical breast examination starting at 35 years of age, pancreatic cancer screening (for those with FH of pancreatic cancer), and general melanoma risk management. For those with MLH1 mutations, the NCCN guidelines (71) recommend pancreatic cancer screening (for those with FH of pancreatic cancer), high-quality colonoscopy (every 1–2 years), and baseline esophagogastroduodenoscopy with random biopsy (every 3–5 years).
Although the usual age of PC onset may be later in Asian compared with Western populations (72), it is unclear whether this difference applies to mutation carriers. For example, the HOXB13 G84E mutation has been associated with early PC onset in Caucasian populations. Although the mutation is rarely detected in Asian men, alternative similar mutations have been reported, including G135E and G132E in Chinese and Japanese studies, respectively (73). Thus, further studies are needed to elucidate the prevalence and effects of these recurrent mutations on PC onset. In the 3-year interim report of IMPACT (22), a longitudinal study of 1,821 BRCA1/2 mutation carriers and 1,206 noncarriers, the median age of PC diagnosis was 61 years in the BRCA2m carrier arm and 64 years in the noncarrier arm (p = 0.04). In Hong Kong, most PC cases occur in patients over age 50 (2). Because of the increased aggressiveness of PC in BRCA2m carriers, early screening is recommended (1.2.3, 1.2.4).
3.1.3 Germline Testing Upon PC Diagnosis
The panel recommends germline testing in PC patients with metastatic disease (1.3.1), ductal or intraductal histology (1.3.2), or positive FH (1.3.3), because significantly increased rates of germline mutations have been reported in these populations. Studies suggest that germline mutation prevalence is much higher in men with mPC (11.8%) than those with localized PC (4.6%), or compared with healthy individuals (2–3%) (74). Also, PC patients with germline mutations were more likely to harbor intraductal/ductal histology than those without (48% vs. 12%, p < 0.01) (24), and ~50% of patients with prostatic ductal adenocarcinoma had DNA repair gene alterations (25).
3.1.4 Consent and Genetic Counseling
With regard to the manifold implications of germline testing, it is good clinical practice to require a detailed informed consent, in addition to the written consent required by the testing laboratory (1.4.1). Familial risk assessment should include a Bayesian analysis performed by a qualified genetic counselor. For germline testing indicated after somatic testing (19), prior consent may not be sufficient. The panel recommends discussing the following items during the consent process: testing purpose, panel choices, cost, potential implications on disease management, additional risks, health insurance, legal issues, and potential significance to family members (18).
Because of limitations in resources and training, urologists and oncologists may not be in the best position to provide specialized genetic counseling to patients. In Hong Kong, there exists a pressing need for professional genetic counselors in this area (1.4.2).
3.1.5 Hereditary Driver Mutations
In clinical practice, the number of genes selected for germline testing will depend on the choice of testing method and associated cost. For Asian patients, we suggest including the genes mentioned in Statement 1.5.1 in a multi-gene panel. Mutations in these genes possess potential prognostic and therapeutic significance (18, 19), and have been reported in studies involving Asian patients (33, 73). For example, mutations in the DNA mismatch repair (MMR) genes MLH1, MSH2, MSH6 and PMS2 have been reported in Chinese patients with Lynch syndrome, at 2–14% prevalence (75). We specifically did not include CHEK2 and HOXB13 because they appear to be rare in Chinese populations (76, 77) and, to our knowledge, no large study has reported on CDK12 germline mutations in Asian PC patients (see also Testing levels and coverage).
3.1.6 Ethnic Considerations
In Hong Kong, the Hong Kong Hereditary Breast Cancer Family Registry helps to archive the incidence of hereditary mutations for high-risk breast and ovarian cancers, including BRCA1/2 mutations (28). A similar registry is needed for hereditary PC (1.6.1). While variations exist between different regions of Asia (73), local studies in Hong Kong would be valuable, particularly the detection and classification of uncertain variants (34).
3.2 Part 2: Testing Methods and Technical Considerations
3.2.1 Germline Versus Somatic Testing
A landmark study on the genomics of advanced PC (4) showed that, among 150 mCRPC patients, 19/150 (12.7%) had BRCA2 loss, of which ~90% was biallelic. Eight (5.3%) affected individuals harbored pathogenic germline BRCA2m, with a later somatic event leading to biallelic loss. A subsequent study by the New York Memorial Sloan Kettering Cancer Center (78) demonstrated that, in 504 tumors from 451 PC patients, of whom 221 consented to germline testing, the incidence of any alteration in BRCA1/2, ATM and CHEK2 was 27%. These included germline mutations (BRCA2, 8.6%; BRCA1, 0.9%; ATM, 2.3%; CHEK2, 4.1%) and somatic mutations only (BRCA2, 7.7%; BRCA1, 0.9%; ATM, 4.5%; CHEK2, 0.9%). These studies suggest that germline mutations involving DNA repair genes account for almost half of all detected mutations through somatic testing of PC tissue.
Because both germline and somatic mutations are relevant to the molecular pathology of PC, genetic testing can proceed in two orders: 1) germline testing, after genetic counseling and informed consent (e.g. patients with FH); or 2) somatic testing of tumor tissue, after explaining to patients that any detected potential germline mutation may require further confirmatory testing (38). When an HRR or MMR mutation is detected, it will be important to undergo germline confirmation by testing a peripheral blood sample, so that proper genetic counseling and cascade testing can be offered to family members (2.1.1).
In a Hong Kong ovarian cancer consensus (28), opinion favored performing somatic testing before germline testing. In this order, the patient can decide whether to undergo testing solely for the purpose of informing treatment planning, without having to understand the potential hereditary implications in advance. Germline testing can be considered after treatment decision has been made. However, one disadvantage of this approach is that cascade testing of family members is more easily missed or forgotten (28).
A large-scale cross-sectional study (79) investigated the data of 3,607 men with a personal history of PC who underwent germline testing between 2013 and 2018. After examining the self-reported FH data, 37% of patients with positive detectable variants would not have been approved for germline testing using NCCN guidelines criteria. This suggests that DNA repair gene variants detected by somatic testing in PC should be considered for germline confirmation, irrespective of FH or Gleason score.
3.2.1.1 Large Genomic Rearrangements (LGRs)
LGRs are large, usually exon-level deletions, or other structural variations that span megabase segments of the human genome (39). A Hong Kong study (80) of 1,236 patients with high-risk hereditary breast and/or ovarian cancers showed that, among 120 deleterious BRCA mutations, 8 (6.7%) were LGRs involving BRCA1 (5/57, or 8.8%) and BRCA2 (3/63, or 4.8%). LGRs are not detectable by Sanger sequencing, and require multiple ligation-dependent probe amplification (MLPA) for detection (81). With the advancement of multi-gene next-generation sequencing (NGS) panels for germline testing, using molecular barcodes and specialized bioinformatics methods, large deletions have become detectable by coverage depth analysis, thus obviating routine MLPA.
Detection of LGRs in somatic testing using NGS on formalin-fixed paraffin-embedded (FFPE) tissue is challenging. Indeed, not all NGS methods are developed or optimized for the detection of copy number variants. For example, an inter-laboratory study (82) of FFPE tumor DNA samples showed that the large insertion BRCA1 exon13ins6kb, a known pathogenic variant, was not detected by any of the laboratories in the primary analysis. If a patient has FH, or is otherwise suspected of having inherited cancer, germline testing using a peripheral blood sample should be considered, even if NGS returned negative results (2.1.2). Various options of platforms and assays for PC genetic testing are available from different testing laboratories. Strengths and limitations of the available panels were discussed in Giri et al. (18).
3.2.2 Tissue Sample Availability
In the PROfound study, multi-gene NGS assays successfully sequenced and produced biomarker status outcomes in only 69% of tumor samples, and the success rate dropped to about 50% in tissue specimens that were stored for 5 or more years (41). This was comparable to those in recent trials involving somatic testing of tumor tissues. Major reasons for testing failure include a limited amount of tumor tissue collected during diagnostic biopsy, exhaustion of diagnostic material during histological examination, insufficient tumor content for genomic study, and suboptimal DNA quality and/or quantity due to DNA degradation during tissue fixation and storage (83).
The histopathologist is in a key position to optimize the success rate of multi-gene NGS assay in PC (2.2.1). FFPE samples for HRR gene mutation testing should contain sufficient cellularity (> 5,000 cells is equivalent to ~30 ng of DNA) to yield the required amount of DNA for testing (84). A minimum of neoplastic cell content is also required; i.e. tumor content should be at least 10–20% to enable reliable detection of somatic variants at > 5% allele frequency, and higher for detection of LGRs. Other recommendations on processing and storage of FFPE samples for DNA analysis in PC can be found in Gonzalez et al. (84).
3.2.3 Testing Levels and Coverage
Genomic profiling of PC is performed by multi-gene NGS panels that comprehensively cover all types of genetic variants; namely, single nucleotide variants, small insertion/deletions, copy number variants including large deletions, and gene fusions (2.3.1). To the clinician, the most important variants are actionable mutations that can guide treatment decisions. A real-world study (85) on clinical comprehensive genomic profiling in 3,476 clinically advanced PCs (1,660 primary tumors and 1,816 metastases) showed that potentially targetable genomic alterations were frequently identified in DNA damage repair, phosphatidylinositol-3-kinase (PI3K), and RAS/RAF/MEK pathways, among other genetic changes. Among the DNA damage repair pathways, homologous recombination defects (HRDs) were found in 23.4% of cases, Fanconi anemia gene defect in 4.8%, CDK12 abnormality in 5.6%, and MMR gene defects in 4.3%.
3.2.3.1 Ethnic Variations
Genetic mutations vary with ethnicity. The spectrum of mutations seen in Chinese PC patients differs from that observed in Caucasian populations. As the majority of participants in large-scale international studies were Western populations, some commonly reported mutations may, in fact, be rare in Chinese PC patients. For example, the most frequently reported mutation of the CHEK2 gene, c.1100delC (3), has not been found in Chinese studies. Likewise, while the HOXB13 gene is believed to be an uncommon cause of familial PC in European populations (86), the HOXB13 G84E hotspot missense mutation was not detected in a study of 1,123 patients from 18 centers across China (76). A study of breast and ovarian cancer patients from Southern China reported a unique set of recurrent or founder mutations of BRCA (87). Thus, annotation of genetic variants detected by multi-gene NGS panels should consider ethnic origins.
3.2.3.2 HRR Gene Mutations
The presence of HRR gene mutations in a mCRPC patient helps to predict response to PARPi treatment (6). Also, MMR gene mutations, which are associated with the microsatellite instability (MSI)-high (MSI-H) phenotype, increase susceptibility to immune checkpoint inhibitor (ICI) treatment, such as programmed death-1 (PD-1) blockade (88). For PC patients, DNA repair genes of interest include: HRR genes (most significant: BRAC2, BRCA1, ATM; others: ATR, BARD1, BRIP1, CHEK1, CHEK2, FAM175A, GEN1, MRE11A, PALB2, PPP2R2A, NBN, RAD31, RAD51B, RAD51C, RAD51D and RAD54L); MMR genes (MSH2, MSH6); Fanconi anemia genes (FANCA, FANCL); and CDK12 (3–5, 60). For mPC, the Philadelphia 2019 consensus (18) recommends large panels and somatic testing; the genes considered were BRCA1/2, HOXB13, CHEK2, ATM, NBN, MSH2, MSH6, MLH1, PMS2, BRIP1, TP53 and the Fanconi anemia genes.
For determining eligibility for PARPi therapy, it should be noted that gene-level testing alone, whether germline or somatic, cannot cover all HRDs (45) (2.3.2). Hence, in addition to HRR gene mutation testing, HRD testing by genomic signatures and scars, such as the genomic instability score or loss of heterozygosity (LOH) score from commercial laboratories may need to be considered, to better stratify patients and inform treatment selection (89). Efforts to standardize and harmonize these and other similar assays are needed. Novel techniques, including mutational signatures and functional assays for HRD testing, are currently under research (46).
3.2.3.3 MMR Gene Mutations
DNA MMR is a system to correct the erroneous insertion, deletion and mismatch of bases that can occur during DNA replication and recombination. Because the accumulation of mismatches leads to DNA disruption and cell death, MMR is essential for maintaining genomic stability. Major MMR genes in humans include MLH1, MSH2, MSH6 and PMS2.
A large amount of MMR defects can be characterized by a “MSI-H” phenotype. Conventionally, polymerase chain reaction (PCR) and capillary electrophoresis of two single-nucleotide repeat loci, and three multi-nucleotide repeat loci on paired normal tumor samples, are employed to determine MSI status (90) (2.3.3). Instability of one locus is termed “MSI-low (MSI-L),” instability of ≥ 2 loci is “MSI-H,” and “microsatellite-stable (MSS)” denotes that no loci have been affected.
Currently, MSI status is detected by NGS cancer genomic profiling, which is applicable to PC (47). Apart from MMR gene mutations, epigenetic changes such as MLH1 gene promoter hypermethylation can also cause sporadic or acquired MSI. Using a routine immunohistochemistry (IHC) panel of antibodies against MLH1, MSH2, MSH6 and PMS2, defective MMR status in PC may also be determined indirectly by the loss of MMR protein expression on tumor cells (91). The use of IHC is a reliable and cost-effective way of determining MMR status.
MMR defects are identified in ~5% of mPC, of which the most frequently mutated genes are MSH2 and MSH6; the majority (~75%) of these mutations occur at the somatic level and are not inherited (92). Nevertheless, as mentioned in Statement 2.1.1, MMR gene mutations detected by an NGS tumor panel should be subject to germline confirmation by testing a peripheral blood sample.
3.2.3.4 ICI Effect Biomarkers
There is growing evidence that tumor mutational burden (TMB), MSI status and programmed death ligand-1 (PD-L1) expression are biomarkers that can help to predict response to ICIs (2.3.4). In 2017, the US Food and Drug Administration (FDA) granted accelerated approval of the ICI pembrolizumab for treating patients with unresectable or metastatic MSI-H or MMR-deficient (dMMR) solid tumors who progressed on prior therapy with no satisfactory treatment alternative (88). This was also the FDA’s first tissue/site-agnostic approval for anti-cancer therapy. In 2020, a similar accelerated approval was granted for use of pembrolizumab in patients with advanced solid tumors that display high TMB, defined as ≥ 10 mutations/megabase in an FDA-approved test (93).
One caveat is that these biomarkers do not correlate well with one another. In a study of 11,348 cases across 26 types of cancer (49), 3.0%, 7.7% and 25.4% were MSI-H, TMB-high and PD-L1-positive, respectively. Of note, 30% of MSI-H cases were TMB-low, suggesting that the two biomarkers are not diagnostically interchangeable. This result might have been expected, because MSI focuses on specific genomic regions known to accumulate errors, whereas TMB covers the genome more broadly. Hence, MSI-H tumors are likely to be TMB-high, but not vice versa. In addition to genomic biomarkers (e.g. MSI and TMB), inflammatory biomarkers such as PD-L1 and T cell–inflamed gene expression profile (GEP) may also help to stratify patients by predicted clinical response (94). Because MSI prevalence in PC is low (~3%), and data for the reliability of IHC and PCR outside of Lynch syndrome are lacking, the European Society for Medical Oncology guideline recommends using NGS for MSI testing in PC patients (50).
3.2.3.5 Liquid Biopsy
In patients with advanced or mPC, especially in the castration-resistant state, it may be difficult to obtain tissue biopsy samples with a sufficient yield; for example, in those with bone-predominant metastases. Alternatively, using archival tissues from the primary prostatic biopsy presents significant limitations because such tissues may fail to reflect the latest tumor mutations, and the DNA yield and quality will be reduced, depending on storage duration and condition (see Tissue sample availability). Liquid biopsy using ctDNA is an emerging alternative method to capture tumor features (2.3.5). This DNA is shed into the bloodstream by degrading tumor cells, and its yield increases with disease burden and active progression (95). In a sequencing study of 72 genes from 45 ctDNA samples from mPC patients (9), prostate-specific antigen (PSA) concentrations did not differ in patients with or without detectable ctDNA, although patients with high PSA levels (> 370 ng/mL) all had detectable ctDNA. Consistent with previous studies, high ctDNA fractions were associated with poor prognosis and correlated with overall tumor burden (9).
In the PROfound study, BRCA and ATM mutations identified in ctDNA showed 81% positive percentage agreement and 92% negative percentage agreement with matched tumor tissues (8). Moreover, liquid biopsy may capture spatial tumor heterogeneity and temporal tumor evolution (96). Unlike a single tissue biopsy, liquid biopsy can integrate genomic alterations from more than one metastatic lesion and thereby survey any tumor heterogeneity in the patient, which may help to detect pathway alterations and resistance mechanisms (96).
3.3 Part 3: Indications for Genetic Testing in PC
3.3.1. Localized PC Patients
AS is a management option offered to patients with low-risk localized PC to defer curative-intent treatment in order to reduce over-treatment and treatment-related complications. This is usually applied to patients with International Society of Urological Pathology (ISUP) Grade Group 1, clinical stage T1c or T2a, PSA level < 10 ng/mL and PSA density < 0.15 ng/mL/cc (97). The presence of a germline BRCA2m is an independent, poor prognostic factor for localized PC, with increased risks of an aggressive phenotype and metastasis (98), shortened survival (99), and grade reclassification during AS (7). The Philadelphia consensus (18) encourages germline BRCA2 testing for AS discussions. In our opinion, current evidence does not warrant routine germline testing in all localized PC patients. However, we agree that patients with otherwise known germline mutations should not be offered AS, due to the higher risks of disease progression as mentioned above (3.1.1).
In localized PC patients, risk stratification usually includes consideration of PSA level, Gleason score, and clinical T-staging, each of which may have their own limitations. Genetic analysis can provide additional information on the molecular basis of the condition (3.1.2). Notable early events may include inactivation of tumor suppressors, cell cycle dysregulation, and rearrangement between androgen responsive genes (e.g. TMPRSS2) and the ETS transcription factor family of genes (most commonly ERG) (100). The Prolaris assay reports a cell cycle progression score that predicts the risk of cancer-specific mortality (52), biochemical recurrence after external beam radiotherapy (53), and outcomes after radical prostatectomy (54). The Decipher test predicts metastatic progression (55), including post-radical prostatectomy metastasis (56), cancer-specific mortality (101), and post-operative radiation sensitivity (102). Note that due to the low reported rates of genetic mutations in localized PC patients (see Germline testing upon PC diagnosis), our panel did not reach a consensus on genetic testing for driver mutations in this group (Supplementary Table 5 and Appendix B).
3.3.2 Genetic Testing in mHSPC
In addition to androgen deprivation therapy, there is growing evidence to support the use of chemotherapy (docetaxel) or an androgen receptor-signaling inhibitor to provide further survival benefit in mHSPC patients (103–105). In mHSPC patients receiving standard therapy, preliminary studies suggest that the presence of DNA repair gene mutations has not been associated with OS, progression-free survival (PFS) or response rate (58, 106), but was associated with early progression to CRPC (59). For patients with de novo mHSPC, genetic testing may provide additional information for prognostic counseling and longer-term treatment planning (3.2.1).
3.3.3 Systemic Therapies for mCRPC
We suggest that all mCRPC patients undergo genetic testing for actionable mutations (3.3.5), which may help to identify eligibility for systemic treatments, including PARPi (3.3.1–3.3.3), ICIs (3.3.4), and platinum-based chemotherapy (3.3.7). Due to potential resource limitations and psychological burden on the patient, somatic testing may initially be preferred (3.3.6; see also Germline versus somatic testing). It may be worth noting that eligibility to PARPi for PC patients, in terms of clinical trial inclusion or the approved indications (6, 107), has so far included both germline and somatic mutations.
3.3.3.1 PARPi
PARP is an enzyme that facilitates DNA repair by binding to the site of DNA damage and attracting DNA repair proteins (108). Inhibition of PARP leads to accumulation of DNA damage, genomic instability, and ultimately cell death – a mechanism that has been termed “synthetic lethality” (109). HRR mutations result in the utilization of more error-prone pathways, and thereby increase susceptibility to PARP inhibition.
PROfound (5) was a phase 3 open-label randomized trial comparing the PARPi olaparib with physician’s choice of a new hormonal agent (NHA; abiraterone or enzalutamide) in 387 mCRPC patients with ≥ 1 HRR mutation (Cohort A: BRCA1/2 or ATM mutations; Cohort B: 12 other prespecified genes), who progressed while receiving an NHA. In Cohort A, the primary endpoint of median radiographic PFS (rPFS) was significantly improved in the olaparib arm versus controls (7.4 vs. 3.6 months; hazard ratio [HR] = 0.34, p < 0.001); objective response rate (ORR) and time to pain progression were also significantly greater in the olaparib arm. In the overall population (Cohorts A and B), median rPFS was also significantly improved in the olaparib arm versus controls (5.8 vs. 3.5 months; HR = 0.49, p < 0.001). Encouragingly, the final median OS (61) also demonstrated a significant benefit in Cohort A versus the NHA arm (19.1 vs. 14.7 months; HR = 0.69, p < 0.02).
TRITON2 (60) was a phase 2 open-label trial of rucaparib in 115 mCRPC patients with BRCA mutation who progressed after one or two lines of androgen receptor-directed therapy and one taxane-based chemotherapy. The ORR (primary endpoint) was 43.5%, with median rPFS of ~9 months and confirmed PSA response rate of 54.8%.
3.3.3.2 ICIs
PC is recognized as poorly immunogenic, showing low levels of T-cell activation and high levels of immunosuppressive activities (110). The 2017 FDA approval of pembrolizumab for treating unresectable or metastatic MSI-H or dMMR solid tumors (88) was based on the KEYNOTE-158 results, which included 2 mCRPC patients, of whom one had a partial response and the other had stable disease for > 9 months (111). In the open-label phase 2 KEYNOTE-199 study of pembrolizumab in 258 patients with mCRPC regardless of MSI status (64), 9% achieved ≥ 50% PSA response, and 3–4% had an objective radiographic response. In a retrospective study of 1,033 mCRPC patients (65), of 11 MSI-H/dMMR patients who received ICI therapy, 6 (55%) had ≥ 50% PSA response and 4 showed radiographic response.
3.3.3.3 Platinum-Based Chemotherapy
Studies in breast and ovarian cancers have shown that patients with BRCA1/2 mutations are responsive to platinum-based chemotherapy (112). For PC patients, Cheng et al. (113) reported exceptional response to platinum-based chemotherapy in 3 patients with biallelic BRCA2 inactivation, despite disease progression on prior standard therapies. In a retrospective analysis of 141 mCRPC patients who received ≥ 2 doses of carboplatin and docetaxel (112), 6 of 8 (75%) of BRCA2m carriers experienced PSA declines > 50% within 12 weeks, compared with 17% of noncarriers (p < 0.001). In a multicenter retrospective analysis of 508 CRPC patients treated with platinum-based chemotherapy (67), those with DNA repair gene aberrations showed higher rates of PSA declines and soft tissue responses (vs. those without), although the differences were not statistically significance (p = 0.20 and p = 0.07, respectively). Among those with BRCA2m, large proportions demonstrated PSA declines of ≥ 50% or soft tissue responses (63.9% and 50% of 44 patients, respectively). Although the evidence is derived from retrospective studies, it has consistently suggested that platinum-based chemotherapy may have anti-tumor activity in mCRPC patients with HRR mutations (3.3.7).
4 Discussion
4.1 Further Practical Advice
Among the currently available testing methods, somatic testing using tumor tissue samples has the highest yield in detection of actionable genetic mutations, and may lead to new treatment options for the patient. However, it is important for clinicians to know the appropriate scope to test for, including HRR gene mutations (e.g. BRCA1/2, ATM, etc.), MMR gene mutations (e.g. MLH1, MSH2, MSH6, PMS2), presence of MSI, and TMB. Clinicians should note the following important technical aspects in genetic testing: 1) tumor testing success rates are reduced with longer storage times; 2) performing only somatic (tumor) testing may not necessarily reflect germline mutation status; 3) ctDNA is a convenient alternative for tumor testing; and 4) if a germline mutation was detected, proper genetic counseling provided by trained personnel is ideal, to inform the patient’s family members about hereditary risks. Cascade testing in PC can be challenging, as PC is not traditionally considered an inherited cancer, and is not usually an “index” cancer diagnosed in families with Lynch syndrome (114). Men may also be less likely than women to undergo testing (115).
4.2 Limitations
With emerging evidence and recommendations from international guidelines on the indications and scope of genetic testing for PC, this consensus aims to provide a local guide to urologists, oncologists and pathologists on who and what to test for in selected patients. In areas with limited data in Asian men, evidence from Caucasian studies and international guidelines were used for reference. Also, the term “Asian” is relatively broad, and the data mentioned here should not be taken as absolute values, but as reminders to clinicians of the ethnic and region-specific factors involved in treating Asian patients. More large-scale sequencing studies are warranted to identify ethnic-specific variants and better understand their pathophysiological effects.
5 Conclusion
This consensus provides guidance to urologists, oncologists and pathologists working with Asian patients on the indications for genetic testing, testing methods and technical considerations, and associated therapeutic implications.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
The panel meetings were co-chaired by DP and PC. All authors participated in the panel meetings, contributed to drafting the consensus statements and journal manuscript, and read and approved the final version.
Funding
This study did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors. Editorial assistance was provided by Best Solution, which was funded by the Hong Kong Society of Uro-Oncology.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.962958/full#supplementary-material
References
1. Kimura T, Egawa S. Epidemiology of Prostate Cancer in Asian Countries. Int J Urol (2018) 25(6):524–31. doi: 10.1111/iju.13593
2. Hong Kong Cancer Registry. Overview of Hong Kong Cancer Statistics of 2019. Hong Kong Hospital Authority (2021). Available at: https://www3.ha.org.hk/cancereg/pdf/overview/Overview%20of%20HK%20Cancer%20Stat%202019.pdf. Accessed January 25, 2022.
3. Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-Repair Gene Mutations in Men With Metastatic Prostate Cancer. N Engl J Med (2016) 375(5):443–53. doi: 10.1056/NEJMoa1603144
4. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell (2015) 161(5):1215–28. doi: 10.1016/j.cell.2015.05.001
5. de Bono JS, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med (2020) 382(22):2091–102. doi: 10.1056/NEJMoa1911440
6. U.S. Food & Drug Administration. FDA Approves Olaparib for HRR Gene-Mutated Metastatic Castration-Resistant Prostate Cancer (2020). Available at: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-olaparib-hrr-gene-mutated-metastatic-castration-resistant-prostate-cancer.
7. Carter HB, Helfand B, Mamawala M, Wu Y, Landis P, Yu H, et al. Germline Mutations in ATM and BRCA1/2 Are Associated With Grade Reclassification in Men on Active Surveillance for Prostate Cancer. Eur Urol (2019) 75(5):743–9. doi: 10.1016/j.eururo.2018.09.021
8. Chi KN, Barnicle A, Sibilla C, Lai Z, Corcoran C, Williams JA, et al. Concordance of BRCA1, BRCA2 (BRCA), and ATM Mutations Identified in Matched Tumor Tissue and Circulating Tumor DNA (ctDNA) in Men With Metastatic Castration-Resistant Prostate Cancer (mCRPC) Screened in the PROfound Study. J Clin Oncol (2021) 39(6_suppl):26–. doi: 10.1200/JCO.2021.39.6_suppl.26
9. Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst (2017) 109(12):djx118. doi: 10.1093/jnci/djx118
10. Mahal BA, Alshalalfa M, Kensler KH, Chowdhury-Paulino I, Kantoff P, Mucci LA, et al. Racial Differences in Genomic Profiling of Prostate Cancer. N Engl J Med (2020) 383(11):1083–5. doi: 10.1056/NEJMc2000069
11. Poon DMC, Chan K, Chan T, Cheung FY, Lam D, Lam M, et al. Ethnic Pharmacogenomic Differences in the Management of Asian Patients With Metastatic Prostate Cancer. Cancers (Basel) (2022) 14(2):407. doi: 10.3390/cancers14020407
12. Jorm AF. Using the Delphi Expert Consensus Method in Mental Health Research. Aust N Z J Psychiatry (2015) 49(10):887–97. doi: 10.1177/0004867415600891
13. Bessa A, Maclennan S, Enting D, Bryan R, Josephs D, Hughes S, et al. Consensus in Bladder Cancer Research Priorities Between Patients and Healthcare Professionals Using a Four-Stage Modified Delphi Method. Eur Urol (2019) 76(2):258–9. doi: 10.1016/j.eururo.2019.01.031
14. Saad F, Canil C, Finelli A, Hotte SJ, Malone S, Shayegan B, et al. Controversial Issues in the Management of Patients With Advanced Prostate Cancer: Results From a Canadian Consensus Forum. Can Urol Assoc J (2020) 14(4):E137–E49. doi: 10.5489/cuaj.6082
15. Tsiamis E, Millar J, Baxi S, Borg M, De Ieso P, Elsaleh H, et al. Development of Quality Indicators to Monitor Radiotherapy Care for Men With Prostate Cancer: A Modified Delphi Method. Radiother Oncol (2018) 128(2):308–14. doi: 10.1016/j.radonc.2018.04.017
16. Diamond IR, Grant RC, Feldman BM, Pencharz PB, Ling SC, Moore AM, et al. Defining Consensus: A Systematic Review Recommends Methodologic Criteria for Reporting of Delphi Studies. J Clin Epidemiol (2014) 67(4):401–9. doi: 10.1016/j.jclinepi.2013.12.002
17. National Comprehensive Cancer Network. Prostate Cancer Early Detection, Version 1.2021 — January 5, 2021. Available at: https://www.nccn.org/professionals/physician_gls/pdf/prostate_detection.pdf. Accessed June 3, 2021.
18. Giri VN, Knudsen KE, Kelly WK, Cheng HH, Cooney KA, Cookson MS, et al. Implementation of Germline Testing for Prostate Cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol (2020) 38(24):2798–811. doi: 10.1200/JCO.20.00046
19. National Comprehensive Cancer Network. Prostate Cancer, Version 2.2021 — February 17, 2021. Available at: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf. Accessed June 8, 2021.
20. Giri VN, Knudsen KE, Kelly WK, Abida W, Andriole GL, Bangma CH, et al. Role of Genetic Testing for Inherited Prostate Cancer Risk: Philadelphia Prostate Cancer Consensus Conference 2017. J Clin Oncol (2018) 36(4):414–24. doi: 10.1200/JCO.2017.74.1173
21. Loeb S, Byrne N, Walter D, Makarov DV, Wise DR, Becker D, et al. Knowledge and Practice Regarding Prostate Cancer Germline Testing Among Urologists: Gaps to Address for Optimal Implementation. Cancer Treat Res Commun (2020) 25:100212. doi: 10.1016/j.ctarc.2020.100212
22. Page EC, Bancroft EK, Brook MN, Assel M, Hassan Al Battat M, Thomas S, et al. Interim Results From the Impact Study: Evidence for Prostate-Specific Antigen Screening in BRCA2 Mutation Carriers. Eur Urol (2019) 76(6):831–42. doi: 10.1016/j.eururo.2019.08.019
23. So HC, Kwan JS, Cherny SS, Sham PC. Risk Prediction of Complex Diseases From Family History and Known Susceptibility Loci, With Applications for Cancer Screening. Am J Hum Genet (2011) 88(5):548–65. doi: 10.1016/j.ajhg.2011.04.001
24. Isaacsson Velho P, Silberstein JL, Markowski MC, Luo J, Lotan TL, Isaacs WB, et al. Intraductal/Ductal Histology and Lymphovascular Invasion Are Associated With Germline DNA-Repair Gene Mutations in Prostate Cancer. Prostate (2018) 78(5):401–7. doi: 10.1002/pros.23484
25. Schweizer MT, Antonarakis ES, Bismar TA, Guedes LB, Cheng HH, Tretiakova MS, et al. Genomic Characterization of Prostatic Ductal Adenocarcinoma Identifies a High Prevalence of DNA Repair Gene Mutations. JCO Precis Oncol (2019) 3):1–9. doi: 10.1200/po.18.00327
26. Pritzlaff M, Tian Y, Reineke P, Stuenkel AJ, Allen K, Gutierrez S, et al. Diagnosing Hereditary Cancer Predisposition in Men With Prostate Cancer. Genet Med (2020) 22(9):1517–23. doi: 10.1038/s41436-020-0830-5
27. Paller CJ, Antonarakis ES, Beer TM, Borno HT, Carlo MI, George DJ, et al. Germline Genetic Testing in Advanced Prostate Cancer; Practices and Barriers: Survey Results From the Germline Genetics Working Group of the Prostate Cancer Clinical Trials Consortium. Clin Genitourin Cancer (2019) 17(4):275–82 e1. doi: 10.1016/j.clgc.2019.04.013
28. Kwong A, Cheng KD, Hsue CV, Hui SK, Leung CR, Leung KA, et al. BRCA Mutation Testing for Ovarian Cancer in the Context of Available Targeted Therapy: Survey and Consensus of Hong Kong Specialists. Asia Pac J Clin Oncol (2019) 15 Suppl 2:20–31. doi: 10.1111/ajco.13116
29. Zayts O, Sarangi S, Thong MK, Chung BH, Lo IF, Kan AS, et al. Genetic Counseling/Consultation in South-East Asia: A Report From the Workshop at the 10th Asia Pacific Conference on Human Genetics. J Genet Couns (2013) 22(6):917–24. doi: 10.1007/s10897-013-9646-7
30. Doan DK, Schmidt KT, Chau CH, Figg WD. Germline Genetics of Prostate Cancer: Prevalence of Risk Variants and Clinical Implications for Disease Management. Cancers (Basel) (2021) 13(9):2154. doi: 10.3390/cancers13092154
31. Loeb S, Giri VN. Clinical Implications of Germline Testing in Newly Diagnosed Prostate Cancer. Eur Urol Oncol (2021) 4(1):1–9. doi: 10.1016/j.euo.2020.11.011
32. Sokolova AO, Cheng HH. Genetic Testing in Prostate Cancer. Curr Oncol Rep (2020) 22(1):5. doi: 10.1007/s11912-020-0863-6
33. Wei Y, Wu J, Gu W, Qin X, Dai B, Lin G, et al. Germline DNA Repair Gene Mutation Landscape in Chinese Prostate Cancer Patients. Eur Urol (2019) 76(3):280–3. doi: 10.1016/j.eururo.2019.06.004
34. Li D, Shi Y, Li A, Cao D, Su H, Yang H, et al. Retrospective Reinterpretation and Reclassification of BRCA1/2 Variants From Chinese Population. Breast Cancer (2020) 27(6):1158–67. doi: 10.1007/s12282-020-01119-7
35. Ren S, Wei GH, Liu D, Wang L, Hou Y, Zhu S, et al. Whole-Genome and Transcriptome Sequencing of Prostate Cancer Identify New Genetic Alterations Driving Disease Progression. Eur Urol (2018) 73(3):322–39. doi: 10.1016/j.eururo.2017.08.027
36. Tan DS, Mok TS, Rebbeck TR. Cancer Genomics: Diversity and Disparity Across Ethnicity and Geography. J Clin Oncol (2016) 34(1):91–101. doi: 10.1200/JCO.2015.62.0096
37. Wedge DC, Gundem G, Mitchell T, Woodcock DJ, Martincorena I, Ghori M, et al. Sequencing of Prostate Cancers Identifies New Cancer Genes, Routes of Progression and Drug Targets. Nat Genet (2018) 50(5):682–92. doi: 10.1038/s41588-018-0086-z
38. Cheng HH, Sokolova AO, Schaeffer EM, Small EJ, Higano CS. Germline and Somatic Mutations in Prostate Cancer for the Clinician. J Natl Compr Canc Netw (2019) 17(5):515–21. doi: 10.6004/jnccn.2019.7307
39. Jaratlerdsiri W, Chan EKF, Petersen DC, Yang C, Croucher PI, Bornman MSR, et al. Next Generation Mapping Reveals Novel Large Genomic Rearrangements in Prostate Cancer. Oncotarget (2017) 8(14):23588–602. doi: 10.18632/oncotarget.15802
40. Comanescu M, Annaratone L, D'Armento G, Cardos G, Sapino A, Bussolati G. Critical Steps in Tissue Processing in Histopathology. Recent Pat DNA Gene Seq (2012) 6(1):22–32. doi: 10.2174/187221512799303190
41. Hussain M, Corcoran C, Sibilla C, Fizazi K, Saad F, Shore N, et al. Tumor Genomic Testing for >4000 Men With Metastatic Castration-Resistant Prostate Cancer in the Phase III Trial PROfound (Olaparib). Clin Cancer Res (2022) 28(8):1518–1530. doi: 10.1158/1078-0432.Ccr-21-3940
42. Gilson C, Ingleby F, Gilbert DC, Parry MA, Atako NB, Ali A, et al. Genomic Profiles of De Novo High- and Low-Volume Metastatic Prostate Cancer: Results From a 2-Stage Feasibility and Prevalence Study in the Stampede Trial. JCO Precis Oncol (2020) 4):882–97. doi: 10.1200/po.19.00388
43. Paulo P, Maia S, Pinto C, Pinto P, Monteiro A, Peixoto A, et al. Targeted Next Generation Sequencing Identifies Functionally Deleterious Germline Mutations in Novel Genes in Early-Onset/Familial Prostate Cancer. PloS Genet (2018) 14(4):e1007355. doi: 10.1371/journal.pgen.1007355
44. Vlajnic T, Bubendorf L. Molecular Pathology of Prostate Cancer: A Practical Approach. Pathology (2021) 53(1):36–43. doi: 10.1016/j.pathol.2020.10.003
45. Sztupinszki Z, Diossy M, Krzystanek M, Borcsok J, Pomerantz M, Tisza V, et al. Prevalence of Homologous Recombination Deficiency (HRD)-Related Signatures Indicates That a Wider Range of Prostate Cancer Patients May Benefit From PARP-Inhibitor Therapy. Ann Oncol (2019) 30:vii14. doi: 10.1093/annonc/mdz413.051
46. Sztupinszki Z, Diossy M, Krzystanek M, Borcsok J, Pomerantz MM, Tisza V, et al. Detection of Molecular Signatures of Homologous Recombination Deficiency in Prostate Cancer With or Without BRCA1/2 Mutations. Clin Cancer Res (2020) 26(11):2673–80. doi: 10.1158/1078-0432.CCR-19-2135
47. Hempelmann JA, Lockwood CM, Konnick EQ, Schweizer MT, Antonarakis ES, Lotan TL, et al. Microsatellite Instability in Prostate Cancer by Pcr or Next-Generation Sequencing. J Immunother Cancer (2018) 6(1):29. doi: 10.1186/s40425-018-0341-y
48. Li K, Luo H, Huang L, Luo H, Zhu X. Microsatellite Instability: A Review of What the Oncologist Should Know. Cancer Cell Int (2020) 20(1):16. doi: 10.1186/s12935-019-1091-8
49. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite Instability Status Determined by Next-Generation Sequencing and Compared With Pd-L1 and Tumor Mutational Burden in 11,348 Patients. Cancer Med (2018) 7(3):746–56. doi: 10.1002/cam4.1372
50. Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, et al. Esmo Recommendations on Microsatellite Instability Testing for Immunotherapy in Cancer, and Its Relationship With Pd-1/Pd-L1 Expression and Tumour Mutational Burden: A Systematic Review-Based Approach. Ann Oncol (2019) 30(8):1232–43. doi: 10.1093/annonc/mdz116
51. Hartzfeld D, Berse B, Lowrance W, Dash A, Brawer M, Lawrence J, et al. Caring for Patients With Prostate Cancer Who Are BRCA Positive. Fed Pract (2016) 33(Suppl 1):46S–51S.
52. Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, Reid JE, et al. Prognostic Value of a Cell Cycle Progression Signature for Prostate Cancer Death in a Conservatively Managed Needle Biopsy Cohort. Br J Cancer (2012) 106(6):1095–9. doi: 10.1038/bjc.2012.39
53. Freedland SJ, Gerber L, Reid J, Welbourn W, Tikishvili E, Park J, et al. Prognostic Utility of Cell Cycle Progression Score in Men With Prostate Cancer After Primary External Beam Radiation Therapy. Int J Radiat Oncol Biol Phys (2013) 86(5):848–53. doi: 10.1016/j.ijrobp.2013.04.043
54. Bishoff JT, Freedland SJ, Gerber L, Tennstedt P, Reid J, Welbourn W, et al. Prognostic Utility of the Cell Cycle Progression Score Generated From Biopsy in Men Treated With Prostatectomy. J Urol (2014) 192(2):409–14. doi: 10.1016/j.juro.2014.02.003
55. Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and Validation of a Prostate Cancer Genomic Classifier That Predicts Early Metastasis Following Radical Prostatectomy. PloS One (2013) 8(6):e66855. doi: 10.1371/journal.pone.0066855
56. Klein EA, Yousefi K, Haddad Z, Choeurng V, Buerki C, Stephenson AJ, et al. A Genomic Classifier Improves Prediction of Metastatic Disease Within 5 Years After Surgery in Node-Negative High-Risk Prostate Cancer Patients Managed by Radical Prostatectomy Without Adjuvant Therapy. Eur Urol (2015) 67(4):778–86. doi: 10.1016/j.eururo.2014.10.036
57. Loeb S, Ross AE. Genomic Testing for Localized Prostate Cancer: Where Do We Go From Here? Curr Opin Urol (2017) 27(5):495–9. doi: 10.1097/MOU.0000000000000419
58. Chanza NM, Roodenbeke MTD, Desmyter L, Wilkin F, Badoer C, Vandernoot I, et al. Prevalence and Clinical Impact of BRCA1/2 Mutations in Patients With De Novo Metastatic Hormone-Sensitive Prostate Cancer (mHSPC). J Clin Oncol (2020) 38(6_suppl):44–. doi: 10.1200/JCO.2020.38.6_suppl.44
59. Vandekerkhove G, Struss WJ, Annala M, Kallio HML, Khalaf D, Warner EW, et al. Circulating Tumor DNA Abundance and Potential Utility in De Novo Metastatic Prostate Cancer. Eur Urol (2019) 75(4):667–75. doi: 10.1016/j.eururo.2018.12.042
60. Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in Men With Metastatic Castration-Resistant Prostate Cancer Harboring a BRCA1 or BRCA2 Gene Alteration. J Clin Oncol (2020) 38(32):3763–72. doi: 10.1200/JCO.20.01035
61. de Bono JS, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. 610O Final Overall Survival (OS) Analysis of PROfound: Olaparib Vs Physician's Choice of Enzalutamide or Abiraterone in Patients (Pts) With Metastatic Castration-Resistant Prostate Cancer (mCRPC) and Homologous Recombination Repair (HRR) Gene Alterations. Ann Oncol (2020) 31:S508. doi: 10.1016/j.annonc.2020.08.870
62. de Bono JS, Mehra N, Higano CS, Saad F, Buttigliero C, Mata M, et al. TALAPRO-1: A Phase II Study of Talazoparib (TALA) in Men With DNA Damage Repair Mutations (DDRmut) and Metastatic Castration-Resistant Prostate Cancer (mCRPC)—First Interim Analysis (IA). J Clin Oncol (2020) 38(6_suppl):119–. doi: 10.1200/JCO.2020.38.6_suppl.119
63. Smith MR, Sandhu SK, Kelly WK, Scher HI, Efstathiou E, Lara PN, et al. Pre-Specified Interim Analysis of Galahad: A Phase II Study of Niraparib in Patients (Pts) With Metastatic Castration-Resistant Prostate Cancer (mCRPC) and Biallelic DNA-Repair Gene Defects (DRD). Ann Oncol (2019) 30:v884–v5. doi: 10.1093/annonc/mdz394.043
64. Antonarakis ES, Piulats JM, Gross-Goupil M, Goh J, Ojamaa K, Hoimes CJ, et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II Keynote-199 Study. J Clin Oncol (2020) 38(5):395–405. doi: 10.1200/JCO.19.01638
65. Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol (2019) 5(4):471–8. doi: 10.1001/jamaoncol.2018.5801
66. Tucker MD, Zhu J, Berry WR, Ramalingam S, Zhang T, Harrison MR, et al. Pembrolizumab in Men With Heavily Treated Metastatic Castration-Resistant Prostate Cancer (mCRPC). J Clin Oncol (2019) 37(7_suppl):172. doi: 10.1200/JCO.2019.37.7_suppl.172
67. Schmid S, Omlin A, Higano C, Sweeney C, Martinez Chanza N, Mehra N, et al. Activity of Platinum-Based Chemotherapy in Patients With Advanced Prostate Cancer With and Without DNA Repair Gene Aberrations. JAMA Netw Open (2020) 3(10):e2021692. doi: 10.1001/jamanetworkopen.2020.21692
68. Mota JM, Barnett E, Nauseef JT, Nguyen B, Stopsack KH, Wibmer A, et al. Platinum-Based Chemotherapy in Metastatic Prostate Cancer With DNA Repair Gene Alterations. JCO Precis Oncol (2020) 4):355–66. doi: 10.1200/po.19.00346
69. Hassett MJ, Somerfield MR, Baker ER, Cardoso F, Kansal KJ, Kwait DC, et al. Management of Male Breast Cancer: Asco Guideline. J Clin Oncol (2020) 38(16):1849–63. doi: 10.1200/JCO.19.03120
70. National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2022 — August 11, 2021. Available at: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf. Accessed January 28, 2022.
71. National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Colorectal, Version 1.2021 — May 11, 2021. Available at: https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf. Accessed January 28, 2022.
72. Zhou CK, Check DP, Lortet-Tieulent J, Laversanne M, Jemal A, Ferlay J, et al. Prostate Cancer Incidence in 43 Populations Worldwide: An Analysis of Time Trends Overall and by Age Group. Int J Cancer (2016) 138(6):1388–400. doi: 10.1002/ijc.29894
73. Zhu Y, Wei Y, Zeng H, Li Y, Ng CF, Zhou F, et al. Inherited Mutations in Chinese Men With Prostate Cancer. J Natl Compr Canc Netw (2021) 20(1):54–62. doi: 10.6004/jnccn.2021.7010
74. Cheng H, Powers J, Schaffer K, Sartor O. Practical Methods for Integrating Genetic Testing Into Clinical Practice for Advanced Prostate Cancer. Am Soc Clin Oncol Educ Book (2018) 38:372–81. doi: 10.1200/EDBK_205441
75. Yuan Y, Zhu L, Xu D, Haixing J, Sun Y, Ding P-R, et al. The Prevalence of Germline Mutations in Chinese Colorectal Cancer Patients With Mismatch Repair Deficiency. J Clin Oncol (2018) 36(15_suppl):e13518–e. doi: 10.1200/JCO.2018.36.15_suppl.e13518
76. Dong B, Yang B, Li Y, Chen W, Li J, Xu Z, et al. Insights Into Chinese Prostate Cancer Germline Gene Mutation Profile: HOXB13 G84E Mutation Is Unsuitable for Genetic Testing. J Clin Oncol (2020) 38(15_suppl):e17515–e. doi: 10.1200/JCO.2020.38.15_suppl.e17515
77. Wu Y, Yu H, Zheng SL, Na R, Mamawala M, Landis T, et al. A Comprehensive Evaluation of CHEK2 Germline Mutations in Men With Prostate Cancer. Prostate (2018) 78(8):607–15. doi: 10.1002/pros.23505
78. Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol (2017) 1:PO.17.00029. doi: 10.1200/PO.17.00029
79. Nicolosi P, Ledet E, Yang S, Michalski S, Freschi B, O'Leary E, et al. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol (2019) 5(4):523–8. doi: 10.1001/jamaoncol.2018.6760
80. Kwong A, Chen J, Shin VY, Ho JC, Law FB, Au CH, et al. The Importance of Analysis of Long-Range Rearrangement of BRCA1 and BRCA2 in Genetic Diagnosis of Familial Breast Cancer. Cancer Genet (2015) 208(9):448–54. doi: 10.1016/j.cancergen.2015.05.031
81. Judkins T, Rosenthal E, Arnell C, Burbidge LA, Geary W, Barrus T, et al. Clinical Significance of Large Rearrangements in BRCA1 and BRCA2. Cancer (2012) 118(21):5210–6. doi: 10.1002/cncr.27556
82. Ellison G, Ahdesmaki M, Luke S, Waring PM, Wallace A, Wright R, et al. An Evaluation of the Challenges to Developing Tumor BRCA1 and BRCA2 Testing Methodologies for Clinical Practice. Hum Mutat (2018) 39(3):394–405. doi: 10.1002/humu.23375
83. Al-Kateb H, Nguyen TT, Steger-May K, Pfeifer JD. Identification of Major Factors Associated With Failed Clinical Molecular Oncology Testing Performed by Next Generation Sequencing (Ngs). Mol Oncol (2015) 9(9):1737–43. doi: 10.1016/j.molonc.2015.05.004
84. Gonzalez D, Mateo J, Stenzinger A, Rojo F, Shiller M, Wyatt AW, et al. Practical Considerations for Optimising Homologous Recombination Repair Mutation Testing in Patients With Metastatic Prostate Cancer. J Pathol Clin Res (2021) 7(4):311–25. doi: 10.1002/cjp2.203
85. Chung JH, Dewal N, Sokol E, Mathew P, Whitehead R, Millis SZ, et al. Prospective Comprehensive Genomic Profiling of Primary and Metastatic Prostate Tumors. JCO Precis Oncol (2019) 3:PO.18.00283. doi: 10.1200/PO.18.00283
86. Xu J, Lange EM, Lu L, Zheng SL, Wang Z, Thibodeau SN, et al. HOXB13 Is a Susceptibility Gene for Prostate Cancer: Results From the International Consortium for Prostate Cancer Genetics (ICPCG). Hum Genet (2013) 132(1):5–14. doi: 10.1007/s00439-012-1229-4
87. Kwong A, Ng EK, Wong CL, Law FB, Au T, Wong HN, et al. Identification of BRCA1/2 Founder Mutations in Southern Chinese Breast Cancer Patients Using Gene Sequencing and High Resolution DNA Melting Analysis. PloS One (2012) 7(9):e43994. doi: 10.1371/journal.pone.0043994
88. U.S. Food & Drug Administration. FDA Grants Accelerated Approval to Pembrolizumab for First Tissue/Site Agnostic Indication (2017). Available at: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnostic-indication.
89. Miller RE, Leary A, Scott CL, Serra V, Lord CJ, Bowtell D, et al. Esmo Recommendations on Predictive Biomarker Testing for Homologous Recombination Deficiency and PARP Inhibitor Benefit in Ovarian Cancer. Ann Oncol (2020) 31(12):1606–22. doi: 10.1016/j.annonc.2020.08.2102
90. Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: Development of International Criteria for the Determination of Microsatellite Instability in Colorectal Cancer. Cancer Res (1998) 58(22):5248–57.
91. Nava Rodrigues D, Rescigno P, Liu D, Yuan W, Carreira S, Lambros MB, et al. Immunogenomic Analyses Associate Immunological Alterations With Mismatch Repair Defects in Prostate Cancer. J Clin Invest (2018) 128(10):4441–53. doi: 10.1172/JCI121924
92. Sedhom R, Antonarakis ES. Clinical Implications of Mismatch Repair Deficiency in Prostate Cancer. Future Oncol (2019) 15(20):2395–411. doi: 10.2217/fon-2019-0068
93. U.S. Food & Drug Administration. FDA Approves Pembrolizumab for Adults and Children With Tmb-H Solid Tumors (2020). Available at: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors.
94. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-Tumor Genomic Biomarkers for Pd-1 Checkpoint Blockade-Based Immunotherapy. Science (2018) 362(6411):eaar3593. doi: 10.1126/science.aar3593
95. Gonzalez-Billalabeitia E, Conteduca V, Wetterskog D, Jayaram A, Attard G. Circulating Tumor DNA in Advanced Prostate Cancer: Transitioning From Discovery to a Clinically Implemented Test. Prostate Cancer Prostatic Dis (2019) 22(2):195–205. doi: 10.1038/s41391-018-0098-x
96. Russano M, Napolitano A, Ribelli G, Iuliani M, Simonetti S, Citarella F, et al. Liquid Biopsy and Tumor Heterogeneity in Metastatic Solid Tumors: The Potentiality of Blood Samples. J Exp Clin Cancer Res (2020) 39(1):95. doi: 10.1186/s13046-020-01601-2
97. Mottet NC P, van den Bergh RCN, Gillessen S, Grummet J, Henry AM, Lam TB, et al. Eau-Eanm-Estro-Esur-Siog Guidelines on Prostate Cancer. Arnhem, The Netherlands: EAU Guidelines Office (2020).
98. Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA Mutations Are Associated With Higher Risk of Nodal Involvement, Distant Metastasis, and Poor Survival Outcomes in Prostate Cancer. J Clin Oncol (2013) 31(14):1748–57. doi: 10.1200/JCO.2012.43.1882
99. Castro E, Goh C, Leongamornlert D, Saunders E, Tymrakiewicz M, Dadaev T, et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-Specific Survival After Radical Treatment for Localised Prostate Cancer. Eur Urol (2015) 68(2):186–93. doi: 10.1016/j.eururo.2014.10.022
100. Ross AE, D'Amico AV, Freedland SJ. Which, When and Why? Rational Use of Tissue-Based Molecular Testing in Localized Prostate Cancer. Prostate Cancer Prostatic Dis (2016) 19(1):1–6. doi: 10.1038/pcan.2015.31
101. Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, Marchionni L, et al. Tissue-Based Genomics Augments Post-Prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur Urol (2016) 69(1):157–65. doi: 10.1016/j.eururo.2015.05.042
102. Den RB, Yousefi K, Trabulsi EJ, Abdollah F, Choeurng V, Feng FY, et al. Genomic Classifier Identifies Men With Adverse Pathology After Radical Prostatectomy Who Benefit From Adjuvant Radiation Therapy. J Clin Oncol (2015) 33(8):944–51. doi: 10.1200/JCO.2014.59.0026
103. Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, et al. Abiraterone Plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med (2017) 377(4):352–60. doi: 10.1056/NEJMoa1704174
104. James ND, de Bono JS, Spears MR, Clarke NW, Mason MD, Dearnaley DP, et al. Abiraterone for Prostate Cancer Not Previously Treated With Hormone Therapy. N Engl J Med (2017) 377(4):338–51. doi: 10.1056/NEJMoa1702900
105. Sweeney CJ, Chen Y-H, Carducci M, Liu G, Jarrard DF, Eisenberger M, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. New Engl J Med (2015) 373(8):737–46. doi: 10.1056/NEJMoa1503747
106. Mateo J, Cheng HH, Beltran H, Dolling D, Xu W, Pritchard CC, et al. Clinical Outcome of Prostate Cancer Patients With Germline DNA Repair Mutations: Retrospective Analysis From an International Study. Eur Urol (2018) 73(5):687–93. doi: 10.1016/j.eururo.2018.01.010
107. U.S. Food & Drug Administration. FDA Grants Accelerated Approval to Rucaparib for BRCA-Mutated Metastatic Castration-Resistant Prostate Cancer (2020). Available at: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-rucaparib-BRCA-mutated-metastatic-castration-resistant-prostate.
108. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific Killing of BRCA2-Deficient Tumours With Inhibitors of Poly(ADP-Ribose) Polymerase. Nature (2005) 434(7035):913–7. doi: 10.1038/nature03443
109. Lord CJ, Ashworth A. PARP Inhibitors: Synthetic Lethality in the Clinic. Science (2017) 355(6330):1152–8. doi: 10.1126/science.aam7344
110. Sun BL. Immunotherapy in Treatment of Metastatic Prostate Cancer: An Approach to Circumvent Immunosuppressive Tumor Microenvironment. Prostate (2021) 81(15):1125–34. doi: 10.1002/pros.24213
111. Merck Sharp & Dohme Corp. Keytruda® (Pembrolizumab) [Prescribing Information]. U.S: Merck & Co., Inc (2020).
112. Pomerantz MM, Spisak S, Jia L, Cronin AM, Csabai I, Ledet E, et al. The Association Between Germline BRCA2 Variants and Sensitivity to Platinum-Based Chemotherapy Among Men With Metastatic Prostate Cancer. Cancer (2017) 123(18):3532–9. doi: 10.1002/cncr.30808
113. Cheng HH, Pritchard CC, Boyd T, Nelson PS, Montgomery B. Biallelic Inactivation of BRCA2 in Platinum-Sensitive Metastatic Castration-Resistant Prostate Cancer. Eur Urol (2016) 69(6):992–5. doi: 10.1016/j.eururo.2015.11.022
114. Haraldsdottir S, Hampel H, Wei L, Wu C, Frankel W, Bekaii-Saab T, et al. Prostate Cancer Incidence in Males With Lynch Syndrome. Genet Med (2014) 16(7):553–7. doi: 10.1038/gim.2013.193
Keywords: Asians, genetic counseling (MeSH), genetic testing, hereditary cancer syndromes, liquid biopsy, molecular targeted therapy, practice guideline (MeSH), prostate cancer
Citation: Chiu PKF, Lee EKC, Chan MTY, Chan WHC, Cheung MH, Lam MHC, Ma ESK and Poon DMC (2022) Genetic Testing and Its Clinical Application in Prostate Cancer Management: Consensus Statements from the Hong Kong Urological Association and Hong Kong Society of Uro-Oncology. Front. Oncol. 12:962958. doi: 10.3389/fonc.2022.962958
Received: 07 June 2022; Accepted: 24 June 2022;
Published: 18 July 2022.
Edited by:
Kouji Izumi, Kanazawa University, JapanReviewed by:
Lap Yin Ho, Independent Researcher, Hong Kong, SAR ChinaJeanny B. Aragon-Ching, Inova Schar Cancer Institute, United States
Copyright © 2022 Chiu, Lee, Chan, Chan, Cheung, Lam, Ma and Poon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darren M. C. Poon, bWNfcG9vbkBjbG8uY3Voay5lZHUuaGs=
 Peter K. F. Chiu
Peter K. F. Chiu Eric K. C. Lee2
Eric K. C. Lee2 Marco T. Y. Chan
Marco T. Y. Chan Darren M. C. Poon
Darren M. C. Poon