
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 17 November 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.960072
This article is part of the Research TopicReviews in Molecular and Cellular OncologyView all 47 articles
Extracellular RNA (exRNA) is a special form of RNA in the body. RNA carries information about genes and metabolic regulation in the body, which can reflect the real-time status of cells. This characteristic renders it a biomarker for disease diagnosis, treatment, and prognosis. ExRNA is transported through extracellular vesicles as a signal medium to mediate communication between cells. Tumor cells can release more vesicles than normal cells, thereby promoting tumor development. Depending on its easy detection, the advantages of non-invasive molecular diagnostic technology can be realized. In this systematic review, we present the types, vectors, and biological value of exRNA. We briefly describe new methods of tumor diagnosis and treatment, as well as the difficulties faced in the progress of such research. This review highlights the groundbreaking potential of exRNA as a clinical biomarker.
Extracellular RNA (exRNA) is a class of nucleic acid molecules first discovered in serum and plasma (1, 2). Due to the extensive presence of RNA enzymes in the extracellular space and body fluids, experts previously deduced that exRNA could not be stable outside the cell. However, two studies (3, 4) demonstrated that RNA could be detected in microvesicles and exosomes; these structures are characterized by special membrane structures that permit the protection of the internal RNA from enzyme degradation. ExRNA can move along microvesicles and exosomes, acting as a signal molecule to affect neighboring cells or conduct long-distance regulation (5). Further, exRNA was found in almost all biological fluids, including blood, urine, and cerebrospinal fluid, and in the fluids collected in abdominal and pulmonary cavities (6–12). This is of great significance for the study of exRNA as a biomarker. A report showing a map of exRNA indicates the species carrier of exRNA in five human biological fluids: serum, plasma, cerebrospinal fluid, saliva, and urine (13). A model containing six exRNA types (CT1, CT2, CT3A, CT3B, CT3C, and CT4) indicates that each type of exRNA is associated with its carrier (vesicle and non-vesicle) (14). ExRNA transport carriers include extracellular vesicles (EVs), lipoprotein particles, and ribonucleoprotein. These exRNAs can be used as biomarkers to regulate important biological processes in the body and monitor the occurrence of various diseases (15). There is no doubt that exRNA has great potential in the field of biomarkers. If tumor biomarkers can be accurately located and detected on a non-invasive basis, exRNA may have groundbreaking impacts in the field of oncology.
EVs mainly have three forms: microvesicles, exosomes, and oncosomes. The difference between microvesicles and exosomes lies in the direction of membrane budding. Microvesicles are produced by the outward budding of the plasma membrane and released directly into the extracellular environment. In essence, exosomes are produced by the inward budding of the membrane. The first step is the inward budding of the membrane to form the multivesicular endosomes (MVEs). Then, intraluminal vesicles (ILVs) are formed in the lumen of MVEs. ESCRT components, ceramides, tetrasanins, and syntenins play a key role in this process. When the MVB membrane fuses with the plasma membrane, exosomes are released into the extracellular environment. There is also a class of specialized EVs released by tumor cells, which are called oncosomes. It is a large EV formed by cancer cells that carries several cancer-causing molecules. Studies have shown that the regulatory factors related to EV biogenesis are overexpressed in cancer, which may be related to the development, progression, and metastasis of cancer (16).
Other studies have found that the loading of extracellular RNA involves a complex sorting mechanism. RNA-binding protein (RBP) can not only bind RNA to prevent the degradation of exRNA but also play an essential role in the process of RNA targeting EVs. Some RBPs can also perform miRNA classification by recognizing specific RNA motifs. For example, hnRNPA2B1, SYNCRIP, and ANXA2 can mediate miRNA sorting and then induce the entry and localization of extracellular RNA to EVs. The regulation of miRNA sorting involves the expression level of miRNA, the sphingomyelin pathway, and the posttranscriptional modification of its 3’ terminus. The main components of miRISCs (mirNA-loaded RNA-induced silencing complex) such as Agos and GW182 can influence the sorting and loading process of miRNAs by associating with the endosomal pathway and then further mediate RNA silencing to affect the sorting and loading process of miRNA. The inhibition of neutral sphingomyelinase 2 and ceramide production can prevent multiple miRNAs from loading into EVs. In addition, uridylation and adenylation at the 3’ end of miRNAs are also associated with the release and retention of miRNAs in exosomes. When the EV-carrying information molecules meet the receptor cells, first of all, by directly triggering the membrane fusion mechanism, the contents of EVs can be directly released into the receptor cells, and finally, the transmission of information between cells can be completed. In addition, EVs can be ingested into recipient cells by corresponding endocytosis. EVs can also bind to receptors on the cell membrane, triggering intracellular signal transduction pathways, and then cascade information transmission process. When RNA is released from EVs, it can activate a series of biological functions, such as affecting protein translation (including promoting and inhibiting effects) and changing gene expression patterns (17, 18).
EVs can transport many types of RNA (19). These RNAs have specific biological functions and important clinical application values. In clinical disease surveillance, it is extremely important to read the RNA information in EVs. Giraldez et al. (20) developed a novel RNA sequencing method, Phospho-RNA-seq, which identifies thousands of mRNA/lncRNA fragments in plasma. This new RNA assay no longer misses critical transcriptome information and reveals the fact that the extracellular transcriptome in human biological fluids is more complex than previously known. Li et al. (21) conducted further studies on exRNA and updated the previously established exRNA database, Exorbase (www.exoRBase.org), to include more samples and data; this provides a more detailed detection and analysis for exLRs. Simultaneously, the RNA mapping program in human EVs/exosomes will be examined to 1) establish a standard protocol for the detection and analysis of exLRs and 2) construct the exLR mapping for common diseases. Uman et al. (22) proposed a natural mechanism for RNA exchange and cell-to-cell communication by using eukaryotes. EVs were used as RNA drug delivery vectors. However, the common vectors for RNA drug delivery are immunogenic or cytotoxic. The research team used human red blood cells to produce EVs for RNA therapy. Large numbers of RBCEVs deliver ASO to leukemia and breast cancer cells. The proliferation of tumor cells was reduced by the inhibition of miR-125b (22) (Figure 1). The further development of cancer-targeting peptides or antibody-coated RBCEVs could potentially target therapeutic RNA delivery to cancer cells and reduce side effects in normal tissues. As a potential biomarker for disease diagnosis, extracellular vesicle RNA is associated with many pathophysiological processes (23). The study of RNA in extracellular vesicles will advance new possibilities for disease surveillance and provide communities with more precise information about health and the disease status.
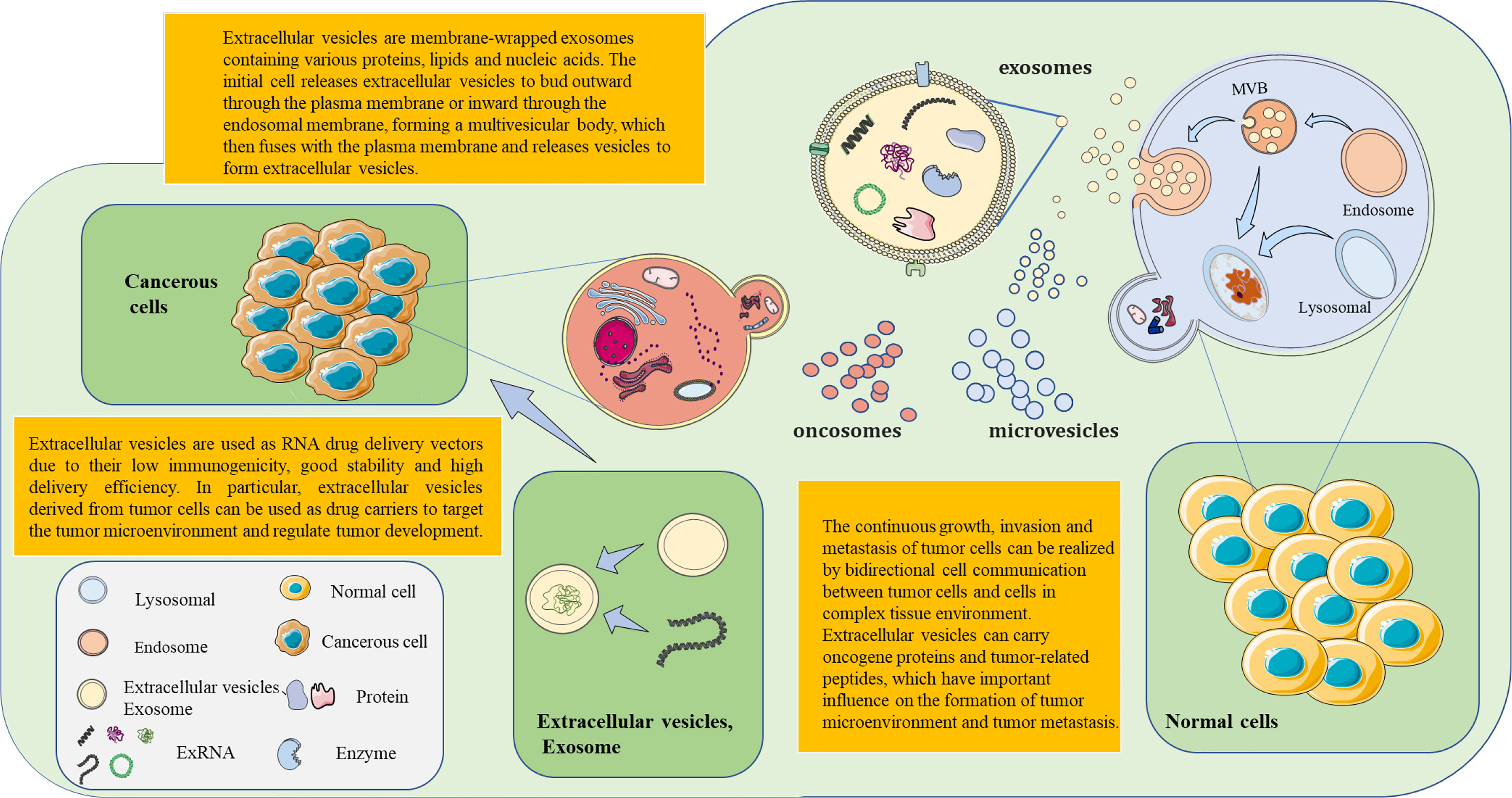
Figure 1 The formation of extracellular vesicles (EVs) and exosomes is based on continuous cell membrane renewal. EVs have three forms: microvesicles, exosomes, and tumor bodies. Microvesicles arise from the inward budding of the membrane. Exosomes are produced by the outward budding of the membrane. Tumor bodies are produced by tumor cells. The invagination of the cell membrane forms the endosome, which then assembles the components within the cell. This results in the formation of multivesicular bodies (MVBs). MVBs then fuse with the cell membrane. EVs within MVBs are released from the cell. EVs (exosomes) contain nucleic acids, lipids, and proteins from the source cell. These EVs, which carry information molecules, play an important role in the communication of information between cells. Based on the performance of exosomes as molecular carriers, therapeutic drugs (e.g., small interfering RNAs) are encapsulated in exosomes in vitro for targeted delivery to related cells. This opens a new horizon for novel precision therapy methods.
Exosomes are subtypes of extracellular vesicles secreted by various cell types such as stem cells, cancer cells, and immune cells. Faruqu et al. (24) successfully delivered siRNA and microRNA to target cells using exosomes. This study confirmed that exosomes can effectively deliver various biomolecules to recipient cells and play an important role in the delivery of anticancer and anti-inflammatory drugs. Due to the advantages of exosomes in drug therapy, the isolation of exosomes has become very important. The most popular method of exosome isolation is the precipitation of exosomes from the original material by hypervelocity centrifugation. With this method, protein coprecipitation usually occurs. If supercentrifugation is combined with density-based separation methods (such as the sucrose gradient), the content of impurities in isolated exosomes can be significantly reduced (25). Combining the three methods, hypercentrifugation, iodoxanol density-gradient centrifugation, and gel filtration can provide high quantities of exosomes for mass spectrometric analysis (26). However, it is important to note that the repeated centrifugation of samples may damage exosome vesicles, resulting in the poor quality of isolated exosomes. The ultrafiltration centrifugation method uses the interception of an ultrafiltration membrane with different relative molecular weights for selective separation. This method ensures the quality of exosomes, but the disadvantage is that exosome separation efficiency is low. Once exosomes block the filtration pores, the membrane life will be shortened.
In addition, adhesion between exosomes trapped on the membrane can also occur, reducing exosome yield (27). To utilize the specific markers on the surface of exosomes in vivo (28), researchers attempted to isolate exosomes using a magnetic bead-based immunoassay. In a clinical breast cancer biopsy study, a magnetic bead–based exosome immunoaffinity separation system was developed by using the high-affinity aptamer of CD63 protein. This system can effectively isolate exosomes from human breast cancer cell media (29). This assay fixed the captured receptor on the solid-phase carrier and targeted the exosome. Exosomes were further isolated and purified based on receptor–ligand affinity. However, this method poses challenges due to its low efficiency and potential errors considering the multistep purification processes. Based on this method, IBA Life Sciences developed an affinity traceless separation-based fragment antigen binding (FAB) method, which uses an affinity chromatography system for exosome separation without the use of magnetic beads. It works by utilizing the principle that double-stranded tags and FAB fragments (FABs) can reversibly bind and release exosomes. The application of this innovative method, with a simple and convenient purification process, has markedly improved exosome isolation as the separation reagents used in purification are reversible and have no impact on the biological activity and function of exosomes. Notably, exosomes can be isolated with high specificity and minimal contamination even in the setting of several interference factors in body fluid samples. In addition, functional materials and microfluidic chips have been used for exosome separation. This is a microfluidic exosome analysis platform based on a new GO/polydopamine (GO/PDA) nanointerface. Nanostructured GO/PDA interfaces greatly improve the efficiency of exosome immunocapture while effectively inhibiting non-specific exosome adsorption (30, 31). The innovation and development of exosome separation technology will greatly accelerate the application of exosome diagnosis and the treatment of disease (Table 1).
The exosome isolation strategies are different for different biological samples. The serum contains more exosomes than plasma because platelets release a certain amount of EVs during clotting. When studying the biology of platelets, serum samples are typically used. Plasma is recommended as a biological sample for exosome research. If exosomes in plasma are to be extracted, anticoagulants should be utilized to prevent clotting. The amount of exosomes in urine samples is lower than that in blood. Some studies have demonstrated that morning urine and midstream urine are more suitable biological samples for exosome extraction. The content of exosomes in the supernatant of the samples treated by eddy currents increased. Urine also contains specialized proteins, such as Tamm–Horsfall protein (THP). After inserting the reducing agent DTT, a large amount of EVs captured by THP is released, and the purity of exosomes can be effectively improved. In the case of breast-milk samples, the increase of dead cells after refrigeration interferes with exosome extraction. Therefore, exosomes will be extracted more efficiently by using fresher milk. Breast milk was centrifuged from high to low density, and then, the supernatant removed from fat and cells was taken to ensure further exosome extraction. If direct high-speed centrifugation is used, it is likely to cause the globulins in breast milk to agglomerate with extracellular vesicles, making the isolation of exosomes more difficult. Cerebrospinal fluid is the best biological fluid to judge the status of the central nervous system. For such small biological samples, filtration and molecular exclusion chromatography are usually more suitable.
Some studies have found EVs associated with tumor cells in the biological samples of cancer patients, carrying relevant carcinogenic factors and substances that can promote the occurrence and development of tumors. These factors have a profound impact on the invasion, immune escape, metastasis, and other aspects of cancer. For example, in the metastasis of breast cancer, macrophages can release EV-containing miR-223 to transmit the “invasion ability” of tumor cells from cell to cell. EV-RNA released by tumor cells can also damage the vascular endothelial barrier and cause secondary changes in endothelial permeability. Finally, it promotes the endothelial infiltration of cancer cells, leading to the enlargement of cancer cell spread, and even causes metastasis through the circulatory system. In addition, the role of EV-RNA from tumor cells in enhancing endothelial cell proliferation and angiogenesis should not be underestimated. The growth of tumor cells depends on the rich blood supply and nutrition. The pro-angiogenic effect of EV-RNA in tumor cells provides a good basis for tumor growth and metastasis to a certain extent. This essentially provides a driving force for tumor migration and invasion. In the process of cancer development, the communication of information is bidirectional, and there is mutual interference between tumor cells and normal cells. Normal cells can inhibit the proliferation of cancer cells and induce the apoptosis of cancer cells by releasing EVs rich in inhibitory miRNA, thus inhibiting the malignant phenotype of cancer cells. However, EV-RNA from certain normal cells may also help tumors grow and metastasize. For example, EV-miRNA from hypoxic mesenchymal stem cells (MSCs) can improve the survival rate of cancer cells and promote proliferation, migration, and invasion. When the gene expression of normal cells is regulated by EV-miRNA from tumor cells, a cancer-promoting effect appropriate to the biological function of cancer cells may occur, such as cancer-associated fibroblasts (CAFs) and mesenchymal stem cells in the tumor microenvironment. Once the balance between normal cells and cancer cells in the body is disrupted, cancer is endowed with the ability to metastasize and invade. The EV-RNA of cancer cells mediated the antitumor immune response by activating the function of immune cells, but it may also play an inhibitory effect to mediate the immune escape of the tumor. Under the interference of tumor EV-RNA, tumor-associated macrophages in the tumor microenvironment will change the cell phenotype and promote cancer progression. Studies have also shown that immune cells associated with hypoxia release EV-RNA, which can promote tumor growth and migration. In addition, the EV-RNA-mediated gene reprogramming of immune cells is one of the main reasons why the function of T cells is hindered, which promote the escape of cancer cells (16, 18).
Currently, the main clinical screening and diagnosis methods are endoscopies, imaging detection, and tumor marker detection (32). However, these methods are restricted to specific cases and are not sensitive enough. In the treatment of tumors, an accurate and early diagnosis often results in a more hopeful prognosis; this indicates the importance of primary diagnoses of tumors. Liquid biopsy is highly sensitive to trace tumor-derived nucleic acids and other markers, and miRNA, lncRNA, and mRNA can all be used as tumor-specific markers in the clinical application (33–35). This provides a new possibility for the study of the early or even ultra-early screening of tumors. In the identification of tumor-specific gene expression profiles, miRNA is often used as the preferred biomarker for detection. The miRNAs exist in blood or are encapsulated in exosomes and are not easily degraded. Specific serum miRNA expression levels can be used for the initial screening, diagnosis, and prognostic trackings of common tumors, such as lung cancer, breast cancer, and liver cancer. The scope of action of lncRNA includes almost all physiological and pathological processes, and it plays an important role in regulating gene silencing and gene expression. LncRNA has high stability in blood, and its high abundance compared with circulating tumor cells and cell-free DNA shows its potential as a more reliable tumor marker. In the current study, some lncRNAs that are closely related to the tumor diagnosis and prognosis have been detected in lung cancer, liver cancer, gastric cancer, and prostate cancer. Circulating lncRNA levels can reflect the existence of tumors in the body to a certain extent. This is of great significance for the early screening and diagnosis of tumors in the future.
Studies have shown that host cells or tumor cells are involved in tumor genesis, growth, invasion, and metastasis through special signal transduction modes (36, 37). During cell growth and development, the cell envelope invaginates to form multiple vesicles. After a series of physiological effects in the cell, it finally fuses with the cell membrane and then releases, forming exosomes containing a signal medium (38–42). EVs can mediate intercellular communication in different ways after secretion (Figure 1). Protein molecules or lipid ligands on the surface of exosomes directly activate receptors on the surface of target cells, generating signaling complexes and activating intracellular signaling pathways. The researchers engineered the surface of the cell membrane to drive the antiviral signal in the receptor breast cancer cell in a protein-free binding form, resulting in tumor growth (43, 44).
In addition, exosomes can fuse with target cells or enter the target cells through endocytosis, bringing their proteins, nucleic acids, lipids, and other active molecules into cells to regulate the function and biological behavior of cells. Lunavat et al. (45) determined the amount of EV RNAs released by cancer cells after treatment with verofenib. The increased expression of miR-211-5p was observed in EV secreted by verofenib-treated cells and tumor tissues from xenografted tumor patients. In conclusion, the treatment of verofenib can induce the upregulation of miR-211-5p in melanoma cells in vitro and in vivo. Tumor-derived exosomes contain components associated with tumor cell miRNAs and have demonstrated the ability to independently process precursor miRNAs into mature miRNAs (46). Roccaro et al. (47) collected many oncogenic proteins, cytokines, and kinases in exosomes isolated from bone marrow stromal cells in myeloma.
These tumor-associated miRNAs and proteins are transported by exosomes, creating an environment that favors tumor metastasis. Interestingly, certain properties of exosomes prompt novel possibilities in the treatment of cancer. Pi et al. (48) found that attaching antibody-like (i.e., Y-shaped) RNA nanoparticles to microvesicles can deliver effective RNA therapeutic agents such as small interfering RNA (siRNA) specifically to cancer cells. By using RNA nanotechnology, researchers have successfully generated extracellular vesicles capable of successfully targeting three types of cancer therapeutics in animal models. Further, its influence on the metabolism of tumor cells cannot be underestimated. Chen et al. (49) investigated the mechanism of the aerobic glycolysis of breast cancer tumor cells with a high quantity of macrophages. This study demonstrates that tumor-associated macrophages can transmit signals through EVs containing lncRNA. This signal enhances the aerobic glycolysis of breast cancer cells and improves the ability of antiapoptosis. The study also highlights the potential of lncRNAs as signal transducers, which propagate between immune cells and tumor cells by EVs and promote the aerobic glycolysis of tumor cells.
In summary, the growth, metastasis, and metabolism of tumors have a significant impact on the subsequent treatment of tumors. Fundamentally understanding the biological characteristics of tumor cells and further research on tumor biomarkers can significantly impact the treatment of tumors, bringing a renewed sense of hope to patients with tumors.
Cigarettes and environmental pollution are two major culprits of the high incidence of lung cancer. Lung cancer accounts for a high proportion of cancers, among which lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) are the main subtypes (50–52). Cheng et al. (53) performed a circRNA detection of LUSC and its adjacent normal tissues by using the ceRNA chip. Through coexpression analysis and qPCR verification, the target circRNA-circTp63 and the target gene FOXM1 of circTp63 were first identified. In further bioassay experiments, circTp63 was considered to promote the cell cycle progression by acting on FOXM1 and CENPA and CENPB downstream genes through miR-873-3p. It ultimately promotes the proliferation and tumor growth of LSC cells. Nigita et al. (54) combined miRNA sequencing data from 87 samples of non-small cell lung cancer (NSCLC) and 26 independent exosomes derived from plasma to investigate RNA editing. This experimental data suggest that there is an editing event in the dysregulated microRNA between the tumor and normal tissue in NSCLC. The fifth edit of miR-411-5p was significantly dysregulated in the tissues and plasma exosomes of patients with NSCLC. Hayashita et al. (55) found that miR17-92 was also associated with lung cancer, and this miRNA was overexpressed in most lung cancer cell lines. When exogenous MIR17-92 was introduced, the growth of lung cancer cells was significantly promoted. Liu et al. (56) performed differential lncRNA screening on spinal samples from patients with primary lung adenocarcinoma, patients with spinal metastases (SM) from lung adenocarcinoma, and normal controls. Linc00852 and MAPK pathways were associated with lung adenocarcinoma SM. In addition, in vivo, and in vitro experiments have shown that the target gene S100A9 of LINC00852 has a positive regulatory effect on the growth, migration, invasion, and metastasis of lung adenocarcinoma cells. S100A9 strongly activates P38 and Rek1/2 kinases and slightly activates JNK kinase phosphorylation in the MAPK pathway in A549 (human alveolar basal epithelial cells) and SPCA-1 cells, thereby promoting the progression and oncogenic ability of SM in lung adenocarcinoma. This suggests that SM intervention is a potential novel therapeutic target in the early stage of lung cancer. Further, evidence suggests that exRNA has a strong potential as a diagnostic marker for lung cancer. In future research on lung cancer, the detection of exRNA expression profiles in lung cancer tissue is expected to become the principal force of non-invasive detection methods (Table 2).
Esophageal squamous cell carcinoma (ESCC) is the sixth leading cause of cancer death worldwide (57, 58). To advance the study of non-invasive biomarker detection, Lin et al. (59) extracted and analyzed the G-Nchirna in exosomes from the tissues and saliva of mice and ESCC patients. The clinical potential of salivary exosomes G-Nchirna (SEG-Nchirna) as readily accessible biomarkers was assessed. The G-Nchirna of exosomes can be detected in ESCC cells and xenograft tumor models in nude mice. The tumor load was closely related to SEG-Nchirna levels. This study reveals that SEG-Nchirna can be used as a reliable biomarker to evaluate the initial detection, treatment response, and recurrence of ESGG (Table 2).
Gastric cancer (GC) is a malignant disease with high morbidity and fatality rates (60). Yuan et al. (61) measured the average expression of miR-551b-3p in patients with GC utilizing a quantitative reverse-transcription polymerase chain reaction experiment. In the study samples, the expression of miR-551b-3p was low in 64% of cases. Further analysis indicates that patients with low miR-551b-3p expression versus patients with high miR-551b-3p expression demonstrate varying prognoses. In addition, it was found that LncRNA SMARCC2 can act as an upstream regulator of Mir-551b-3p, thereby enhancing the proliferation and migration of GC cells by inhibiting the expression of MiR-551b-3p. Lu et al. (62) found that circ-Rangap1 was significantly upregulated in both the plasma exosomes and GC tissues of patients with GC. The high expression of circ-Rangap1 is closely related to TNM staging. The level of circ-Rangap1 in the plasma exosomes of patients with GC before surgery is upregulated, which enhances the migration and invasion ability of GC cells. Circ-Rangap1 may be a potential prognostic biomarker for GC and a therapeutic target for GC. Peritoneal dissemination was the most frequent metastatic method for GC. Hu et al. (63) found higher expression levels of miR-196, miR-92, and miR-1307 in the ascites-derived exosomes of GC patients. Moreover, the invasion of GC cells was closely related to the exosomes detected in the experiment. Taken together, these results demonstrate the significance of exRNA in the diagnosis, treatment, and prognosis of GC (Table 2).
The high morbidity and mortality of hepatocellular carcinoma (HCC) grants urgency to the development of more effective treatment programs for the disease (64–66). Zhang et al. (67) used ceRNA microchips to detect the expression of circRNA in the exosomes of HCC patients with high and low body fat percentages (BFRs). The results showed that the high expression of circ-BD in patients with high BFR played a certain role in promoting the tumor development of HCC. Wang et al. (68) studied the potential role of the circRNA of HCC exosomes in tumor cell migration and invasion, helping to clarify its potential mechanism. Signal transmission between cells with low or no metastatic potential and HCC cells with high metastatic potential can be achieved through exosomes. In another study, Cao et al. (69) selected HCC cells with non-metastatic (HepG2), low metastatic (97L), and highly metastatic (LM3) potential and sequenced the circRNAs in the exosomes. HCC cell–derived exosomes affected the expression of miR-21. By interfering with the expression process of downstream genes, it can regulate the physiological function of tumor cells (69). Liu et al. (70) found that endoplasmic reticulum–stressed HCC cells acted on macrophages by releasing exosomes containing specific miRNAs. It can also inhibit the function of T cells by affecting cell signal transduction pathways, leading to the escape of HCC cells from the immune monitoring system (Figure 2). From the direction of immunology, strengthening the immune monitoring function in the tumor microenvironment may establish new methods for the treatment of HCC (Table 2).
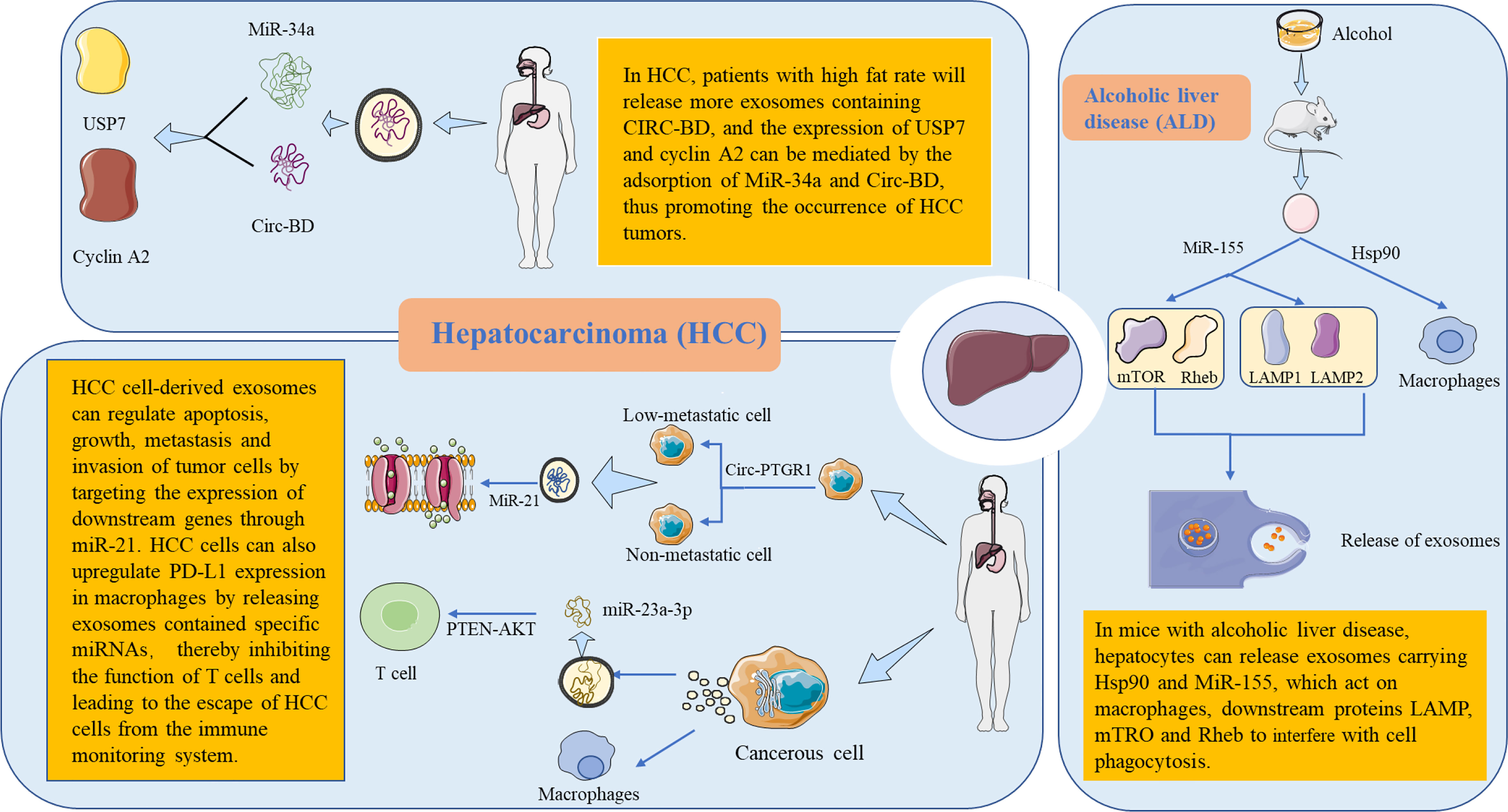
Figure 2 Metabolic functions and mechanisms of extracellular RNA (exRNA) in hepatocellular-derived exosomes. The liver is the body’s largest detoxification organ. In mice with alcoholic liver disease, liver cells interfere with the cellular function by releasing exRNA-carrying exosomes. HSP90 targets to macrophages and affects phagocytosis. MiR-155 acts on downstream proteins LAMP, MTRO, and RHEB, affecting autophagy through specific activation pathways and then interfering with the release of exosomes. In patients with hepatocellular carcinoma (HCC), it was found that patients with a high fat percentage released more exosomes containing circ-BD. Circ-BD was highly expressed in adipocyte exosomes. It promotes HCC tumorigenesis by absorbing miR-34a to mediate the expression of USP7 and cyclin A2. In the process of tumor cell invasion, non-metastatic and low-metastatic cells can obtain the ability of migration and invasion through exosomes containing circ-ptgr1. In addition, HCC cell–derived exosomes can increase the expression of miR-21, which regulates the apoptosis, growth, metastasis, and invasion of tumor cells by targeting the expression of downstream genes. Endoplasmic reticulum–stressed HCC cells raise the expression of PD-L1 in macrophages by releasing exosomes containing specific miRNAs. Subsequently, the miR-23a-PTEN-Akt pathway inhibits the function of T cells, leading to the escape of HCC cells from the immune monitoring system. PD-L1, programmed death-ligand 1.
In 2008, Kroh et al. first demonstrated the presence of miRNAs released from prostate cancer (PC) cells in the blood (71). The greatest clinical need in PC surveillance is to find reliable, non-invasive tools to distinguish between PC and benign prostate disease. Mitchell et al. (72) developed a “miRScore” based on serum levels of 14 miRNAs. Local and metastatic resistant prostate cancer (mCRPC), or low-grade and high-grade PCs, could be distinguished by analysis of free cell-free miRNAs. The results showed that some free acellular miRNAs were predictive in the differentiation between PC and benign prostatic hyperplasia (BPH). However, only a few miRNA biomarkers have been validated in multiple independent studies. Many other miRNAs have no definitive results and have shown conflicting results in some studies. The analysis of miRNA is thus regarded as a non-repeatable method. As research continues, cancer-derived EVs contain highly specific protocellular biomarkers and protect their RNA from RNA enzyme degradation in blood. Therefore, EV-based miRNA analysis may be superior to whole plasma/serum analysis. Endzelins et al. (73) determined the RNA isolated from plasma EV samples from 77 patients. The results showed that four miRNAs showed potential for the diagnosis of PC, and there were significant differences between the miRNAs encapsulated in EVs and the miRNA profiles in whole plasma. Only a small fraction of plasma acellular miRNAs was present in plasma EVs. MiR-375 was able to distinguish PC versus BPH in the whole plasma assay, while miR-200c-3p and miR21-5p were better in plasma EVs. The level of let-7a-5p in non-plasma EVs could distinguish PC patients with a Gleason score ≥8 vs. ≤6 (40), but plasma let-7a-5p had no diagnostic value. The prostate-specific antigen (PSA) test is currently recognized as the best method for early suspected prostate cancer. However, elevated PSA levels are not unique to prostate cancer and can also be caused by benign prostate disease. Therefore, in clinical practice, histopathology is generally obtained through the systemic needle biopsy of the prostate to make the definitive diagnosis (74–76). However, invasive procedures pose several challenges for patients. Non-invasive biomarker research is a pioneering field for the diagnosis and treatment of prostate cancer (Table 2).
Breast cancer in women surpassed lung cancer for the first time as the most common cancer globally in 2020, accounting for approximately 11.7% of incident cancer cases, according to the latest data on the global burden of cancer for 2020 released by the International Agency for Research on Cancer of the World Health Organization (77). Circulating miRNAs are formed by the nuclease degradation of RNA, which are then assembled by microvesicles and exosomes for transport. Zhao et al. (78) amplified miRNA by reverse-transcription PCR (RT-PCR) and analyzed miR-106a in blood by combining it with the miRNA microarray. It was found that the abnormal expression of miR-106a could be a potential biomarker for metastatic breast cancer. Liu et al. (79) designed a set of systematic screening methods for breast cancer–specific circRNAs and found that hsa_circ_001783 regulates tumor proliferation and metastasis through sponge miR-200C-3p. This study indicates that hsa_circ_001783 may be a novel prognostic marker and therapeutic target for breast cancer. The treatment of breast cancer has developed from simple surgical treatment to a combination of surgery, chemotherapy, radiotherapy, hormonal therapy, and/or targeted therapy. This makes breast cancer treatment more personalized and continuous. It is suspected that researching breast cancer at the molecular level will bring advantages for the screening, treatment, and prognosis of breast cancer, but this remains to be explored (Table 2).
Advanced ovarian cancer usually spreads to the visceral adipose tissue of the greater omentum (80, 81). Au et al. (82) identified and compared microRNA-21 from exosomes in ovarian cancer cells, cancer-associated adipocytes (CAAs), and cancer-associated fibroblasts (CAFs). Our results suggest that miR21 can metastasize from CAAs or CAFs to cancer cells and then inhibit ovarian cancer cell apoptosis and enable cancer cells to acquire drug resistance by binding to APAF1. In the retinal tumor microenvironment, exosomes secreted by adjacent stromal cells alter the malignant phenotype of metastatic ovarian cancer cells by the delivery of miR21. The inhibition of miR21 metastasis from a stroma has a promising application prospect in the treatment of metastatic and recurrent ovarian cancer. Endometrial cancer most often occurs in the endometrial epithelium of menopausal and postmenopausal women. Xu et al. (83) isolated circRNA in EVs from the serum samples of three patients with stage III endometrial cancer aged 50–60 years and three healthy subjects. A total of 209 circRNAs were upregulated, and 66 circRNAs were downregulated. The abnormal expression of two circRNAs, hsa_circ_0109046 and hsa_circ_0002577, was confirmed by RT-qPCR. These findings help predict the occurrence, metastasis, and prognosis of endometrial cancer (Table 2).
The intercellular communication between tumor and host microenvironment can be mediated by exosomes. Wei et al. (84) studied the cancer-derived exRNA by using glioblastoma (GBM) as a research model. The study provides a variety of categories of exRNA that can be used for biomarker discovery. At the same time, the miRNA with the greatest influence on GBM was also predicted. It provides additional possibilities for the study of biomarkers in cerebrospinal fluid. Wang et al. (85) screened circRNAs differentially expressed in GBM clinical samples by using the ceRNA microarray. The target circNT5E was identified by TargetScan prediction and an miRNA pulldown experiment. The phenotypic analysis in vivo and in vitro showed that AdARB2 can act on linear NT5E precursors and relieve ADAR-1’s inhibition of circnt5E transcription. The highly expressed circnt5E can be adsorbed to miRNA-422a to relieve its inhibition of downstream target genes. Then Akt, Smad2, and other signaling pathways are activated to promote the proliferation, migration, and invasion of GBM cells. In general, exRNA can regulate the occurrence and development of GBM through regulatory intervention on downstream target genes. This is a novel mechanism for the study of GBM (Table 2).
Multiple myeloma (MM) is a hematologic cancer caused by the abnormal expansion of plasma cells. MM is characterized by the malignant proliferation of monoclonal plasma cells and the secretion of large amounts of monoclonal immunoglobulins, which then inhibit the normal function of both polyclonal plasma cells and polyclonal immunoglobulins (86–88). Deng et al. (89) found that mesenchymal stem cell–derived exosomes promoted MM tumorigenesis by regulating the function of miR-15a/16 and Bcl-2 through lncRNA LINC00461. The expression profile analysis of lncRNA in bone marrow plasma cells showed that the dysregulation of lncRNA UCA1 was associated with the abnormal serum levels of albumin and M protein. In addition, lncRNA metastases–related MALAT1 overexpression was found in newly diagnosed MM patients compared with healthy individuals after treatment. This suggests that MALAT1 could be used as a marker to predict disease progression. The overexpression of specific lncRNA MSL1 was detected in 40% of MM samples. The knockdown of MSL1 by short hairpin RNAs significantly increased apoptosis in MM cell lines, suggesting that MSL1 may be a relevant therapeutic target. In conclusion, these data suggest that the changes in lncRNA expression are associated with the occurrence of MM (Table 2).
This review introduces the great potential of exRNA as a novel biomarker. Each study of the formation process of exRNA and the complex mechanism of its role in body diseases provides new possibilities for the detection and treatment of future diseases. At present, most clinical diagnoses of the disease still include invasive procedures, such as biopsies. Based on the special physiological characteristics of exRNA, researchers foresee much potential in utilizing exRNA for non-invasive diagnostic methods (5, 8). In addition, as studies continue to deepen, there is evidence that exRNA has a greater functional value. The use of extracellular vesicles to transport RNA drugs to treat diseases is increasingly widely studied (90). Whether in the detection, diagnosis, treatment, or prognosis of diseases, exRNA has highlighted a great advantage.
The study of exRNA could lead to the search for new diagnostic markers or new therapeutic targets. However, before translating its use into clinical practice, the mechanisms and pathways of exRNA treatment should be understood. It is still unknown how to introduce this biomarker into clinical disease surveillance (10, 91, 92). Further, interspecies exRNA transfer may occur through contact and feeding, opening a new entry point for the transboundary transfer of exRNA. Although information exchange and transmission serve as a bridge between species, it is not fully understood whether or how they play an important role in biological interactions (90, 93, 94).
At present, there has been the formation of an exRNA database (95–98). The existence of a wide spectrum of exRNA and its diversity suggests that it may be involved in important biological processes, such as the regulation of normal growth and development, as well as the occurrence of cancer and disease. The study of the mechanisms behind the regulatory functions of exRNA will provide new insights into the development of certain diseases (99, 100). However, the maturity of the new technology needs to go through a perfect development process, and the extensive scope of exRNA has brought some difficulties to its in-depth research. Exosome extraction and purification have a great impact on the study of exRNA to a large extent (101, 102). Therefore, from the study of exRNA to its practical application, it still needs to be verified in practice. It is believed that with the development of science and extensive research efforts, exRNA will bring widely impact science and medicine as a whole. The prospect of the clinical application of exRNA should be more anticipated.
XZ and ZL conceived the work. DW wrote and drafted the manuscript. TT, PL, ZL, and XZ discussed the manuscript. EE revised the manuscript. All authors read and approved the final version of the manuscript.
This work was supported partly by The National Natural Science Foundation of China (81972366); The Natural Science Foundation of Guangdong (2022A1515012606); The Shenzhen Science and Technology Innovation Commission projects (JCYJ20220818101808018, KCXFZ202110203002406).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kacian DL, Mills DR, Kramer FR, Spiegelman S. A replicating RNA molecule suitable for a detailed analysis of extracellular evolution and replication. Proc Natl Acad Sci U.S.A. (1972) 69:3038–42. doi: 10.1073/pnas.69.10.3038
2. Kamm RC, Smith AG. Nucleic acid concentrations in normal human plasma. Clin Chem (1972) 18:519–22. doi: 10.1093/clinchem/18.6.519
3. Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia (2006) 20:847–56. doi: 10.1038/sj.leu.2404132
4. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol (2007) 9:654–9. doi: 10.1038/ncb1596
5. Tosar JP, Witwer K, Cayota A. Revisiting extracellular RNA release, processing, and function. Trends Biochem Sci (2021) 46:438–45. doi: 10.1016/j.tibs.2020.12.008
6. Hildebrandt A, Kirchner B, Meidert AS, Brandes F, Lindemann A, Doose G, et al. Detection of atherosclerosis by small RNA-sequencing analysis of extracellular vesicle enriched serum samples. Front Cell Dev Biol (2021) 9:729061. doi: 10.3389/fcell.2021.729061
7. Hutchins E, Reiman R, Winarta J, Beecroft T, Richholt R, De Both M, et al. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci Data (2021) 8:276. doi: 10.1038/s41597-021-01056-w
8. Kim S, Jeon OH, Jeon YJ. Extracellular RNA: Emerging roles in cancer cell communication and biomarkers. Cancer Lett (2020) 495:33–40. doi: 10.1016/j.canlet.2020.09.002
9. Ning B, Huang Z, Youngquist BM, Scott JW, Niu A, Bojanowski CM, et al. Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma. Nat Nanotechnol (2021) 16:1039–44. doi: 10.1038/s41565-021-00939-8
10. O'Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA Delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol (2020) 21:585–606. doi: 10.1038/s41580-020-0251-y
11. Xue VW, Wong SCC, Song G, Cho WCS. Promising RNA-based cancer gene therapy using extracellular vesicles for drug delivery. Expert Opin Biol Ther (2020) 20:767–77. doi: 10.1080/14712598.2020.1738377
12. Zhou D, Gu J, Wang Y, Wu H, Cheng W, Wang Q, et al. Long non-coding RNA NEAT1 transported by extracellular vesicles contributes to breast cancer development by sponging microRNA-141-3p and regulating KLF12. Cell Biosci (2021) 11:68. doi: 10.1186/s13578-021-00556-x
13. Lasser C. Mapping extracellular RNA sheds lights on distinct carriers. Cell (2019) 177:228–30. doi: 10.1016/j.cell.2019.03.027
14. Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, et al. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell (2019) 177:463–477.e415. doi: 10.1016/j.cell.2019.02.018
15. Quinn JF, Patel T, Wong D, Das S, Freedman JE, Laurent LC, et al. Extracellular RNAs: development as biomarkers of human disease. J Extracell Vesicles (2015) 4:27495. doi: 10.3402/jev.v4.27495
16. Hu W, Liu C, Bi ZY, Zhou Q, Zhang H, Li LL, et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Mol Cancer (2020) 19:102. doi: 10.1186/s12943-020-01199-1
17. Corrado C, Barreca MM, Zichittella C, Alessandro R, Conigliaro A. Molecular mediators of RNA loading into extracellular vesicles. Cells (2021) 10:3355. doi: 10.3390/cells10123355
18. Redzic JS, Balaj L, van der Vos KE, Breakefield XO. Extracellular RNA mediates and marks cancer progression. Semin Cancer Biol (2014) 28:14–23. doi: 10.1016/j.semcancer.2014.04.010
19. Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D'Agostino VG. RNA Packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J Extracell Vesicles (2020) 10:e12043. doi: 10.1002/jev2.12043
20. Giraldez MD, Spengler RM, Etheridge A, Goicochea AJ, Tuck M, Choi SW, et al. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J (2019) 38:e101695. doi: 10.15252/embj.2019101695
21. Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem (2019) 65:798–808. doi: 10.1373/clinchem.2018.301291
22. Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun (2018) 9:2359. doi: 10.1038/s41467-018-04791-8
23. Chiabotto G, Gai C, Deregibus MC, Camussi G. Salivary extracellular vesicle-associated exRNA as cancer biomarker. Cancers (Basel) (2019) 11:891. doi: 10.3390/cancers11070891
24. Faruqu FN, Xu L, Al-Jamal KT. Preparation of exosomes for siRNA delivery to cancer cells. J Vis Exp (2018) 142:10.3791/58814. doi: 10.3791/58814
25. Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. Retrovirus-like gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell (2018) 172:262–274.e211. doi: 10.1016/j.cell.2017.12.022
26. Abramowicz A, Widlak P, Pietrowska M. Proteomic analysis of exosomal cargo: the challenge of high purity vesicle isolation. Mol Biosyst (2016) 12:1407–19. doi: 10.1039/C6MB00082G
27. Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, et al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell (2016) 165:1106–19. doi: 10.1016/j.cell.2016.04.015
28. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol (2015) 8:83. doi: 10.1186/s13045-015-0181-x
29. Song Z, Mao J, Barrero RA, Wang P, Zhang F, Wang T. Development of a CD63 aptamer for efficient cancer immunochemistry and immunoaffinity-based exosome isolation. Molecules (2020) 25:5585. doi: 10.3390/molecules25235585
30. He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip (2014) 14:3773–80. doi: 10.1039/C4LC00662C
31. Zhao Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip (2016) 16:489–96. doi: 10.1039/C5LC01117E
32. Tang Z, Li D, Hou S, Zhu X. The cancer exosomes: Clinical implications, applications and challenges. Int J Cancer (2020) 146:2946–59. doi: 10.1002/ijc.32762
33. Wallington-Beddoe CT, Mynott RL. Prognostic and predictive biomarker developments in multiple myeloma. J Hematol Oncol (2021) 14:151. doi: 10.1186/s13045-021-01162-7
34. Wang W, Han Y, Jo HA, Lee J, Song YS. Non-coding RNAs shuttled via exosomes reshape the hypoxic tumor microenvironment. J Hematol Oncol (2020) 13:67. doi: 10.1186/s13045-020-00893-3
35. Wu X, Kang M, Wang D, Zhu M, Hu Y, Zhang Y, et al. Heparan sulfate analogues regulate tumor-derived exosome formation that attenuates exosome functions in tumor processes. Int J Biol Macromol (2021) 187:481–91. doi: 10.1016/j.ijbiomac.2021.07.110
36. Tang Z, Xu Z, Zhu X, Zhang J. New insights into molecules and pathways of cancer metabolism and therapeutic implications. Cancer Commun (Lond) (2021) 41:16–36. doi: 10.1002/cac2.12112
37. Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci (2020) 10:31. doi: 10.1186/s13578-020-00396-1
38. Chen H, Wang L, Zeng X, Schwarz H, Nanda HS, Peng X, et al. Exosomes, a new star for targeted delivery. Front Cell Dev Biol (2021) 9:751079. doi: 10.3389/fcell.2021.751079
39. Gao Z, Pang B, Li J, Gao N, Fan T, Li Y. Emerging role of exosomes in liquid biopsy for monitoring prostate cancer invasion and metastasis. Front Cell Dev Biol (2021) 9:679527. doi: 10.3389/fcell.2021.679527
40. Li J, Gao N, Gao Z, Liu W, Pang B, Dong X, et al. The emerging role of exosomes in cancer chemoresistance. Front Cell Dev Biol (2021) 9:737962. doi: 10.3389/fcell.2021.737962
41. Prieto-Vila M, Yoshioka Y, Ochiya T. Biological functions driven by mRNAs carried by extracellular vesicles in cancer. Front Cell Dev Biol (2021) 9:620498. doi: 10.3389/fcell.2021.620498
42. Shi ZY, Yang XX, Malichewe C, Li YS, Guo XL. Exosomal microRNAs-mediated intercellular communication and exosome-based cancer treatment. Int J Biol Macromol (2020) 158:530–41. doi: 10.1016/j.ijbiomac.2020.04.228
43. Matei I, Kim HS, Lyden D. Unshielding exosomal RNA unleashes tumor growth and metastasis. Cell (2017) 170:223–5. doi: 10.1016/j.cell.2017.06.047
44. Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell (2017) 170:352–366. e313. doi: 10.1016/j.cell.2017.06.031
45. Lunavat TR, Cheng L, Einarsdottir BO, Olofsson Bagge R, Veppil Muralidharan S, Sharples RA, et al. BRAF(V600) inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc Natl Acad Sci U.S.A. (2017) 114:E5930–9. doi: 10.1073/pnas.1705206114
46. Batth IS, Mitra A, Manier S, Ghobrial IM, Menter D, Kopetz S, et al. Circulating tumor markers: harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann Oncol (2019) 30:1845. doi: 10.1093/annonc/mdz218
47. Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood (2009) 113:6669–80. doi: 10.1182/blood-2009-01-198408
48. Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol (2018) 13:82–9. doi: 10.1038/s41565-017-0012-z
49. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol (2019) 21:498–510. doi: 10.1038/s41556-019-0299-0
50. Xie S, Wu Z, Qi Y, Wu B, Zhu X. The metastasizing mechanisms of lung cancer: Recent advances and therapeutic challenges. BioMed Pharmacother (2021) 138:111450. doi: 10.1016/j.biopha.2021.111450
51. Ye Z, Huang Y, Ke J, Zhu X, Leng S, Luo H. Breakthrough in targeted therapy for non-small cell lung cancer. BioMed Pharmacother (2021) 133:111079. doi: 10.1016/j.biopha.2020.111079
52. Zhu X, Luo H, Xu Y. Transcriptome analysis reveals an important candidate gene involved in both nodal metastasis and prognosis in lung adenocarcinoma. Cell Biosci (2019) 9:92. doi: 10.1186/s13578-019-0356-1
53. Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun (2019) 10:3200. doi: 10.1038/s41467-019-11162-4
54. Nigita G, Distefano R, Veneziano D, Romano G, Rahman M, Wang K, et al. Tissue and exosomal miRNA editing in non-small cell lung cancer. Sci Rep (2018) 8:10222. doi: 10.1038/s41598-018-28528-1
55. Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res (2005) 65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352
56. Liu P, Wang H, Liang Y, Hu A, Xing R, Jiang L, et al. LINC00852 promotes lung adenocarcinoma spinal metastasis by targeting S100A9. J Cancer (2018) 9:4139–49. doi: 10.7150/jca.26897
57. Shiozaki A, Marunaka Y, Otsuji E. Roles of ion and water channels in the cell death and survival of upper gastrointestinal tract cancers. Front Cell Dev Biol (2021) 9:616933. doi: 10.3389/fcell.2021.616933
58. Xue W, Zheng Y, Shen Z, Li L, Fan Z, Wang W, et al. Involvement of long non-coding RNAs in the progression of esophageal cancer. Cancer Commun (Lond) (2021) 41:371–88. doi: 10.1002/cac2.12146
59. Lin Y, Dong H, Deng W, Lin W, Li K, Xiong X, et al. Evaluation of salivary exosomal chimeric GOLM1-NAA35 RNA as a potential biomarker in esophageal carcinoma. Clin Cancer Res (2019) 25:3035–45. doi: 10.1158/1078-0432.CCR-18-3169
60. Abadi AJ, Zarrabi A, Hashemi F, Zabolian A, Najafi M, Entezari M, et al. The role of SOX family transcription factors in gastric cancer. Int J Biol Macromol (2021) 180:608–24. doi: 10.1016/j.ijbiomac.2021.02.202
61. Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji R, et al. Molecular mechanisms of lncRNA SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett (2018) 418:84–96. doi: 10.1016/j.canlet.2018.01.032
62. Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett (2020) 471:38–48. doi: 10.1016/j.canlet.2019.11.038
63. Hu Y, Qi C, Liu X, Zhang C, Gao J, Wu Y, et al. Malignant ascites-derived exosomes promote peritoneal tumor cell dissemination and reveal a distinct miRNA signature in advanced gastric cancer. Cancer Lett (2019) 457:142–50. doi: 10.1016/j.canlet.2019.04.034
64. Lu G, Luo H, Zhu X. Targeting the GRP78 pathway for cancer therapy. Front Med (Lausanne) (2020) 7:351. doi: 10.3389/fmed.2020.00351
65. Turtoi M, Anghelache M, Bucatariu SM, Deleanu M, Voicu G, Safciuc F, et al. A novel platform for drug testing: Biomimetic three-dimensional hyaluronic acid-based scaffold seeded with human hepatocarcinoma cells. Int J Biol Macromol (2021) 185:604–19. doi: 10.1016/j.ijbiomac.2021.06.174
66. Zhu X, Zhang J, Fan W, Wang F, Yao H, Wang Z, et al. The rs391957 variant cis-regulating oncogene GRP78 expression contributes to the risk of hepatocellular carcinoma. Carcinogenesis (2013) 34:1273–80. doi: 10.1093/carcin/bgt061
67. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene (2019) 38:2844–59. doi: 10.1038/s41388-018-0619-z
68. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
69. Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, Xue P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer (2019) 18:148. doi: 10.1186/s12943-019-1075-2
70. Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, et al. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology (2019) 70:241–58. doi: 10.1002/hep.30607
71. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods (2010) 50:298–301. doi: 10.1016/j.ymeth.2010.01.032
72. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U.S.A. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
73. Endzelins E, Berger A, Melne V, Bajo-Santos C, Sobolevska K, Abols A, et al. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer (2017) 17:730. doi: 10.1186/s12885-017-3737-z
74. Gasi Tandefelt D, de Bono J. Circulating cell-free DNA: Translating prostate cancer genomics into clinical care. Mol Aspects Med (2020) 72:100837. doi: 10.1016/j.mam.2019.100837
75. Greene J, Baird AM, Lim M, Flynn J, McNevin C, Brady L, et al. Differential CircRNA expression signatures may serve as potential novel biomarkers in prostate cancer. Front Cell Dev Biol (2021) 9:605686. doi: 10.3389/fcell.2021.605686
76. Laraib U, Sargazi S, Rahdar A, Khatami M, Pandey S. Nanotechnology-based approaches for effective detection of tumor markers: A comprehensive state-of-the-art review. Int J Biol Macromol (2021) 195:356–83. doi: 10.1016/j.ijbiomac.2021.12.052
77. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Pineros M, Znaor A, et al. Cancer statistics for the year 2020: An overview. Int J Cancer (2021) 149:778–89. doi: 10.1002/ijc.33588
78. Zhao Q, Deng S, Wang G, Liu C, Meng L, Qiao S, et al. A direct quantification method for measuring plasma MicroRNAs identified potential biomarkers for detecting metastatic breast cancer. Oncotarget (2016) 7:21865–74. doi: 10.18632/oncotarget.7990
79. Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis (2019) 10:55. doi: 10.1038/s41419-018-1287-1
80. Fathi M, Barar J, Erfan-Niya H, Omidi Y. Methotrexate-conjugated chitosan-grafted pH- and thermo-responsive magnetic nanoparticles for targeted therapy of ovarian cancer. Int J Biol Macromol (2020) 154:1175–84. doi: 10.1016/j.ijbiomac.2019.10.272
81. Li K, Luo H, Luo H, Zhu X. Clinical and prognostic pan-cancer analysis of m6A RNA methylation regulators in four types of endocrine system tumors. Aging (Albany NY) (2020) 12:23931–44. doi: 10.18632/aging.104064
82. Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun (2016) 7:11150. doi: 10.1038/ncomms11150
83. Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics (2018) 10:187–97. doi: 10.2217/epi-2017-0109
84. Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun (2017) 8:1145. doi: 10.1038/s41467-017-01196-x
85. Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res (2018) 78:4812–25. doi: 10.1158/0008-5472.CAN-18-0532
86. Chen IC, Sethy B, Liou JP. Recent update of HDAC inhibitors in lymphoma. Front Cell Dev Biol (2020) 8:576391. doi: 10.3389/fcell.2020.576391
87. Tomaipitinca L, Russo E, Bernardini G. NK cell surveillance of hematological malignancies. therapeutic implications and regulation by chemokine receptors. Mol Aspects Med (2021) 80:100968. doi: 10.1016/j.mam.2021.100968
88. Yuan G, Huang Y, Yang ST, Ng A, Yang S. RGS12 inhibits the progression and metastasis of multiple myeloma by driving M1 macrophage polarization and activation in the bone marrow microenvironment. Cancer Commun (Lond) (2021) 42:60–4. doi: 10.1002/cac2.12228
89. Deng M, Yuan H, Liu S, Hu Z, Xiao H. Exosome-transmitted LINC00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microRNA/BCL-2 expression. Cytotherapy (2019) 21:96–106. doi: 10.1016/j.jcyt.2018.10.006
90. Born LJ, Harmon JW, Jay SM. Therapeutic potential of extracellular vesicle-associated long noncoding RNA. Bioeng Transl Med (2020) 5:e10172. doi: 10.1002/btm2.10172
91. Cabiati M, Salvadori C, Basta G, Del Turco S, Aretini P, Cecchettini A, et al. miRNA and long non-coding RNA transcriptional expression in hepatocellular carcinoma cell line-secreted extracellular vesicles. Clin Exp Med (2021) 22:245–55. doi: 10.1007/s10238-021-00744-6
92. Nikoobakht M, Shamshiripour P, Shahin M, Bouzari B, Razavi-Hashemi M, Ahmadvand D, et al. A systematic update to circulating extracellular vesicles proteome; transcriptome and small RNA-ome as glioma diagnostic, prognostic and treatment-response biomarkers. Cancer Treat Res Commun (2021) 30:100490. doi: 10.1016/j.ctarc.2021.100490
93. Gruner HN, McManus MT. Examining the evidence for extracellular RNA function in mammals. Nat Rev Genet (2021) 22:448–58. doi: 10.1038/s41576-021-00346-8
94. Wang BZ, Luo LJ, Vunjak-Novakovic G. RNA And protein delivery by cell-secreted and bioengineered extracellular vesicles. Adv Healthc Mater (2021) 11:e2101557. doi: 10.1002/adhm.202101557
95. Chiang SH, Thomas GA, Liao W, Grogan T, Buck RL, Fuentes L, et al. RNAPro*SAL: a device for rapid and standardized collection of saliva RNA and proteins. Biotechniques (2015) 58:69–76. doi: 10.2144/000114254
96. Hildebrandt A, Kirchner B, Nolte-'t Hoen ENM, Pfaffl MW. miREV: An online database and tool to uncover potential reference RNAs and biomarkers in small-RNA sequencing data sets from extracellular vesicles enriched samples. J Mol Biol (2021) 433:167070. doi: 10.1016/j.jmb.2021.167070
97. Li JR, Tong CY, Sung TJ, Kang TY, Zhou XJ, Liu CC. CMEP: a database for circulating microRNA expression profiling. Bioinformatics (2019) 35:3127–32. doi: 10.1093/bioinformatics/btz042
98. Rozowsky J, Kitchen RR, Park JJ, Galeev TR, Diao J, Warrell J, et al. exceRpt: A comprehensive analytic platform for extracellular RNA profiling. Cell Syst (2019) 8:352–357.e353. doi: 10.1016/j.cels.2019.03.004
99. Chen X, Rechavi O. Plant and animal small RNA communications between cells and organisms. Nat Rev Mol Cell Biol (2022) 23:185–203. doi: 10.1038/s41580-021-00425-y
100. Pin F, Beltra M, Garcia-Castillo L, Pardini B, Birolo G, Matullo G, et al. Extracellular vesicles derived from tumour cells as a trigger of energy crisis in the skeletal muscle. J Cachexia Sarcopenia Muscle (2022) 13:481–94. doi: 10.1002/jcsm.12844
101. Marchand V, Galvanin A, Motorin Y. Isolation, extraction and deep-sequencing analysis of extracellular RNAs (exRNAs) from human plasma. Methods Mol Biol (2021) 2300:165–82. doi: 10.1007/978-1-0716-1386-3_15
102. Poudyal RR, Meyer MO, Bevilacqua PC. Measuring the activity and structure of functional RNAs inside compartments formed by liquid-liquid phase separation. Methods Enzymol (2021) 646:307–27. doi: 10.1016/bs.mie.2020.06.010
Keywords: exRNA, exosome, circulating tumor cell, cell communication, biomarker
Citation: Wu D, Tao T, Eshraghian EA, Lin P, Li Z and Zhu X (2022) Extracellular RNA as a kind of communication molecule and emerging cancer biomarker. Front. Oncol. 12:960072. doi: 10.3389/fonc.2022.960072
Received: 02 June 2022; Accepted: 10 October 2022;
Published: 17 November 2022.
Edited by:
Young-Jun Jeon, Sungkyunkwan University, South KoreaReviewed by:
Kyungoh Jung, Chung-Ang University, South KoreaCopyright © 2022 Wu, Tao, Eshraghian, Lin, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zesong Li, lzssc@email.szu.edu.cn; Xiao Zhu, bioxzhu@yahoo.com
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.