- 1Graduate College, Heilongjiang University of Chinese Medicine, Harbin, China
- 2College of Life Science and Technology, Guangxi University, Nanning, China
- 3Department of Gastroenterology, The First Affiliated Hospital of Heilongjiang University of Chinese Medicine, Harbin, China
- 4Department of Gastroenterology, Heilongjiang Academy of Traditional Chinese Medicine, Harbin, China
- 5Department of Gynaecology, The First Affiliated Hospital of Heilongjiang University of Chinese Medicine, Harbin, China
Autophagy is a conserved cellular self-digesting process that degrades obsoleting proteins and cellular components and plays a crucial role in the tumorigenesis, metastasis, and drug resistance of various tumors such as gastric cancer (GC). As a hotspot in molecular biology, non-coding RNAs (ncRNAs) are involved in the regulation of multiple biological processes, such as autophagy. Increasing evidence indicate that various ncRNAs exert double roles in the initiation and progression of GC, either serve as oncogenes or tumor suppressors. Recent studies have shown that some ncRNAs could modulate autophagy activity in GC cells, which would affect the malignant transformation and drug resistance. Whether the function of ncRNAs in GC is dependent on autophagy is undefined. Therefore, identifying the underlying moleculr targets of ncRNAs in autophagy pathways and the role of ncRNA-regulated autophagy in GC could develop new treatment interventions for this disease. This review summarizes the autophagy process and its role in GC, and the regulatory mechanisms of ncRNAs, as well as focuses on the dual role of ncRNAs-mediated autophagy in GC, for the development of potential therapeutic strategies in GC patients.
Introduction
Gastric cancer (GC) is a gastrointestinal tumor accounting for nearly 15% of cancer-related deaths in the world, with high morbidity and high-grade malignancy (1). Surgical resection is a curative treatment for patients with early GC, whereas systemic chemotherapy provides an optimal therapeutic strategy for patients who have developed local or distant metastasis (2). Unfortunately, GC tumors increasingly become resistant to chemotherapy because of the intrinsic or acquired mechanisms of chemoresistance, which contributes to tumor metastasis and local recurrence (3). Autophagy is an evolutionarily conserved intracellular self-digesting process by which misfolded proteins and damaged organelles are degraded to regulate cell homeostasis (4). This process has become a hot topic in cancer research within recent years due to its involvement in tumorigenesis, metastasis, and chemoresistance (5). During tumor progression or chemotherapy-induced stress, obsolete organelles and useless proteins are recycled by autophagy to foster cancer cell growth and chemoresistance; conversely, removal of oncogenic substances via autophagy blocks tumorigenesis (6). Thus, the role of autophagy as a “friend” or “foe” in GC progression and chemoresistance is still controversial (7). It should be noted that autophagy comprises multiple autophagy-related genes (ATGs) and complexes that are indispensable to the formation of autophagosome, and that autophagy is intricately regulated by a variety of signaling pathways (8). Autophagic components and autophagy-related signaling molecules can be further modulated by non-coding RNAs (ncRNAs) at various levels, from transcriptional regulation to post-translational protein modification (9). Recent findings indicate that ncRNA-mediated autophagy is associated with malignant behaviors like proliferation, metastasis and chemoresistance that is extremely important in cancer development (10). Whether the dual role of autophagy in GC is determined by ncRNAs is still vague. Thus, it is crucial to clarify the regulative mechanisms of ncRNA on autophagy of GC. This review concisely summarizes the formation, signal regulation of autophagy and its paradoxical effects in GC, as well as the regulatory mechanisms of ncRNAs in the gene expression and protein modification. It emphasizes the latest research progression on the role of ncRNAs-regulated autophagy in GC, and discusses the effects of drug-mediated autophagy through regulating ncRNAs on GC treatment.
The process, pathways and roles of autophagy
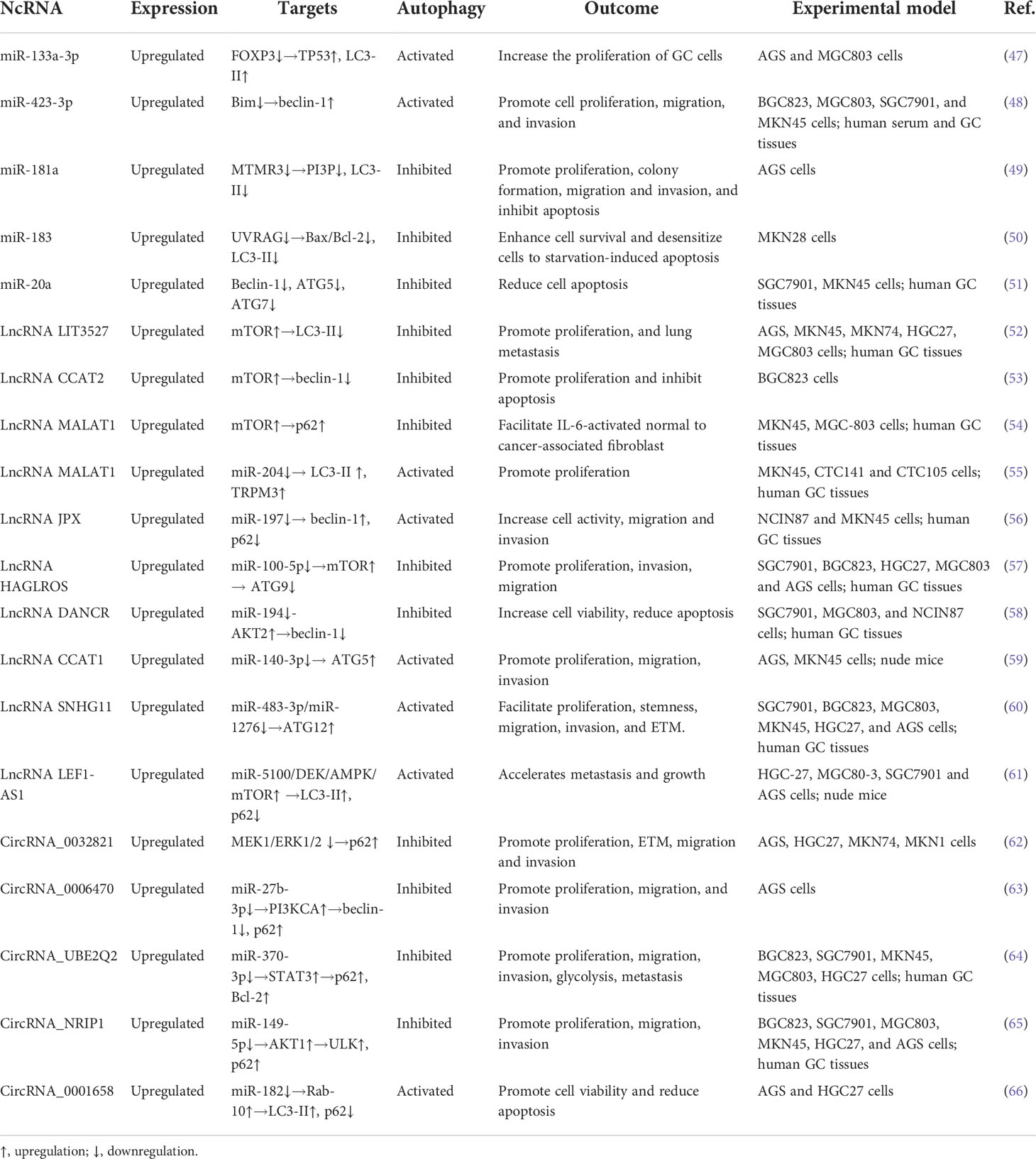
Autophagy can be generally classified into macroautophagy, microautophagy, and chaperone-mediated autophagy (11). Owing to its extensive and crucial roles in current research, macroautophagy is referred to as autophagy in this review. Cellular conditions like nutrient deficiency, hypoxia, and inflammation, can trigger autophagy signals in the affected cells (12). The process of autophagy consists of five steps: initiation and nucleation, elongation, maturation, fusion, and degradation. Each step of autophagy is regulated by ATGs and molecular complexes (Figure 1) (13). The inactivation of mammalian target of rapamycin (mTORC) complex induces the activation of a protein complex, composing of unc-51 like autophagy activating kinase (ULK1), ATG17, ATG13, and ATG101, which is recruited to form a pre-autophagosomal structure (PAS) that initiates the autophagy process. Inhibition of the mTOR/ULK1 signaling pathway activates autophagy signal in GC cells (14). This activated ULK1 complex phosphorylates the autophagy and beclin-1 regulator (AMBRA) protein that activates the class III phosphatidylinositol 3-kinase (PI3K) complex consisting of PI3KC3, beclin-1, ATG14 and vacuolar protein sorting 15 (VPS15), and produces phosphatidylinositol 3-phosphate (PI3P) at the endoplasmic reticulum (ER) (15, 16). PI3P subsequently attracts zinc-finger FYVE domain-containing protein 1 (ZFYVE1, also called DFCP1) and WD repeat domain phosphoinositide-interacting protein (WIPI2), which bind the ATG16L1 protein and make it accessible to recruit the ATG16L1/ATG5/ATG12 protein complex to the PAS (17). Reducing the expression of ATG16L1 in GC cells blocks the formation of ATG16L1/ATG5/ATG12 complex and thus suppress the occurrence of autophagy (18). This protein complex also combines with the ATG3/ATG7 complex to promote the binding of LC3-II, derived from the cleavage of microtubule associated protein light chain 3 (LC3) by cysteine protease ATG4, with phosphatidylethanolamine (PE) to form PE-conjugated LC3-II, which elicits the membrane sealing to form a mature autophagosome, and further to bind with cargo receptors such as p62 to select proteins or organelles (19). Then, the autophagosome fuses with the lysosome to form autolysosome. As the fusion completes, the autophagosome substances will be degraded and released from the autolysosome. These products of decomposition can be recycled for cellular anabolism and growth.
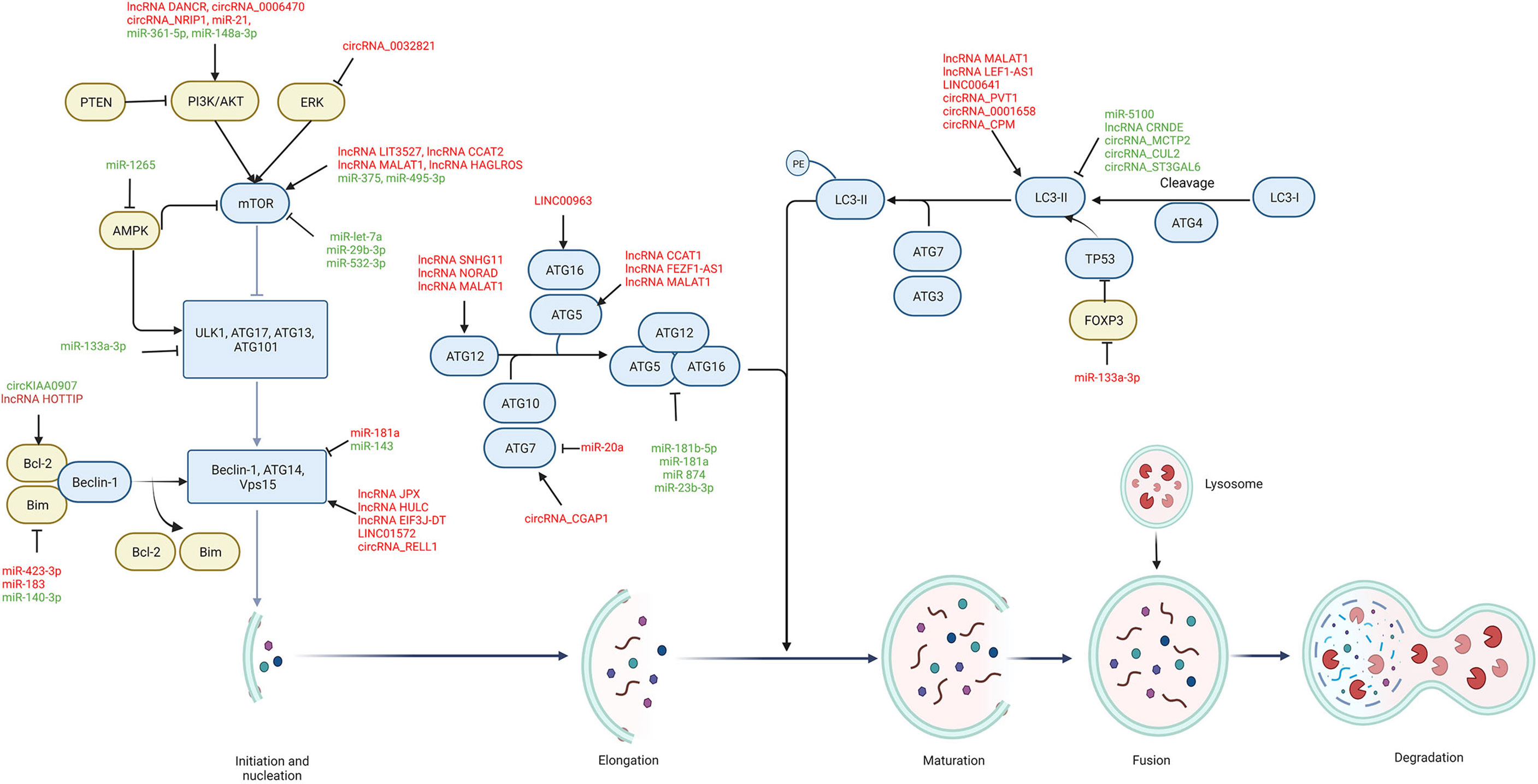
Figure 1 The regulatory role of ncRNAs on autophagy process in GC. Autophagic components including autophagy-related signaling pathways (yellow) and autophagic proteins (blue) are regulated by ncRNAs during each stage of autophagy, including initiation and nucleation, elongation, maturation, fusion, and degradation. Oncogenic ncRNAs (red) are highly expressed in GC and promote cancer progression and chemoresistance by modulating autophagic components, while tumor-suppressive ncRNAs (green) are downregulated in GC and their upregulation can suppress GC progression and chemoresistance through autophagy modulation. → indicates a promoting effect and ⊥ indicates an inhibitory effect.
Several signaling pathways are demonstrated to be involved in the regulation of autophagy. The inactivation of the mTOR signaling pathway is the central step to induce autophagy, which can be regulated by various signaling pathways, such as PI3K/AKT, adenosine monophosphate-activated protein kinase (AMPK), mitogen activated kinase-like protein (MAPK), and phosphatase and tensin homolog (PTEN) pathways (20). The PI3K phosphorylates phosphatidylinositol-4,5-bisphosphate to produce PI3P that induces Akt phosphorylation, which activates mTOR and results in the inhibition of autophagy in GC cells (21). Besides, cellular energy deficiency increases the level of AMPK that phosphorylates ULK1 to directly activate autophagy, as well as inactivates mTOR to indirectly induce autophagy (22). As an important regulating protein of the autophagic pathway, Bcl-2 binds to beclin-1 and thus blocks the formation of autophagosome. The c-Jun N-terminal kinase (JNK), a subtype of MAPK, phosphorylates Bcl-2, contributing to the dissociation of beclin-1 and the subsequent induction of autophagy (23). However, extracellular signal-regulated kinase (ERK), another subtype of MAPK, can be phosphorylated to block the Tuberous sclerosis complex 1 and 2 (TSC1/TSC2) complex and activate mTOR, thus leading to the suppression of autophagy. In addition, the presence of PTEN inhibits the PI3K/AKT signaling pathway (24). Consequently, inactivation of the AKT signaling pathway by PTEN facilitates autophagy activity.
Autophagy is believed to play dual roles in tumor progression that acts as a suppressor at early stages but as a promoter at the advanced stages of GC (6). For the tumor-inhibiting role, autophagy mitigates tumor initiation by removal of oncogenic substances and strengthens tumor immunosurveillance by interacting with factors involved in innate or adaptive immunity; moreover, excessive autophagy causes cellular energy depletion and ultimately leads to cell death, which is known as autophagic cell death (25). On the other hand, autophagy functions as a “recycling” process that digests useless and toxic components, and produces nutrients and essentials to maintain cell homeostasis, by which GC cells also survive under stress conditions such as hypoxia and chemotherapy (26).
Regulatory mechanisms of NcRNAs
NcRNAs mainly contain microRNA (miRNA), long non-coding RNA (lncRNA) and circular RNA (circRNA). MiRNAs are a kind of small endogenous, single-stranded ncRNAs with a length of 19–25 nucleotides. By pairing with the complementary sequences at the 3′-UTR of mRNAs of target genes, miRNAs suppress mRNAs translation or initiate their degradation (27). Depending on the degree of complementarity between miRNA and the target mRNA, different outputs can occur. The mRNA undergoes endonucleolytic cleavage when the complementarity is perfect, but, if the complementarity is not total, the target mRNA is translationally repressed, through interference with polyribosomes or by sequestering in cytoplasmic P-bodies (28). Thus, miRNAs function as master regulators that control the expression of genes, and are involved in the occurrence and progression of human diseases, such as tumors, neurodegenerative diseases, autoimmune diseases, and endocrine disorders (29). Of interest, the role of miRNAs in tumorigenesis is double-edged: acting as tumor suppressors by blocking the translation of mRNA of target genes that mediate differentiation, invasion, and malignance; conversely, playing oncogenic role via triggering the degradation of mRNA of tumor suppressor genes (30). Numerous studies have investigated the potential roles of miRNAs as a diagnostic or prognostic marker for cancer, and, as promising therapeutic agents to tackle cancer (31, 32).
LncRNAs are a class of transcripts of over 200 nucleotides, accounting for 80–90% of all ncRNAs, and no or limited protein-coding potential (33). The regulatory mechanisms of lncRNAs are determined by their structure. On the primary structure level, the functional activity of lncRNAs relies on Watson-Crick base pairing, promoting direct interactions with other RNA molecules. The secondary structures are shaped by base-pairing or ribose backbone interactions, that allow the formation of higher-order configurations with helices and hairpin loops. A third alternative mechanism of stabilization is circularization, leading to the formation of the circRNAs (34). There are three well-characterized action mechanisms of lncRNAs (1): interacting with DNA, chromatin-modifying enzymes, and transcription factors to regulate genetic transcription (2); sponging miRNAs from their target mRNAs or direct binding to mRNAs to influence translation, also known as a competing endogenous RNAs (ceRNAs) mechanism (3); serving as scaffolds for proteins to block their functions (35). LncRNAs have been demonstrated to widely participate in various physiological and pathological processes (36, 37). Some overexpressed lncRNAs in cancers are reported to promote tumor growth by regulating angiogenesis, migration and metastasis, while other downregulated lncRNAs are regarded as safeguards against cancer progression by inhibiting cell proliferation, inducing apoptosis, maintaining genomic stability, or promoting tumor suppressor expression (38).
CircRNAs are a kind of single-stranded covalently RNA molecules that form a closed loop through the link between 5’ and 3’ terminal nucleotide sequences, with the length of 100nt ~ over 4 kb (39). Thus, comparing with linear transcripts such as miRNAs and lncRNAs, circRNAs are more stable owing to lacking of 3ʹ and 5ʹ ends, suggesting a huge potential for diagnostic or prognostic markers of various diseases (40, 41). The exact action models of circRNAs are still unsolved, but they are believed to affect different biological processes in two manners: acting as sponges directly binding to miRNAs to prevent them from exerting biological functions; moreover, functioning as scaffolds for the protein complex assembly to suppress its functions (42). Moreover, circRNAs act as transcription modulators. It is assumed that circRNAs are nuclear limited, which is similar to the observation of nuclear limitation of linear RNAs containing conserved introns and forms a large number of posttranscriptional modulators. Thus, further suppression of circRNAs leads to a decrease in the expression of their parental genes (43). CircRNAs have become a research hotspot because of their association with disease progression, particularly in cancer (44, 45). As circRNAs regulate both the expression of oncogenes and tumor suppressors, they also play dual role in tumorigenesis (46).
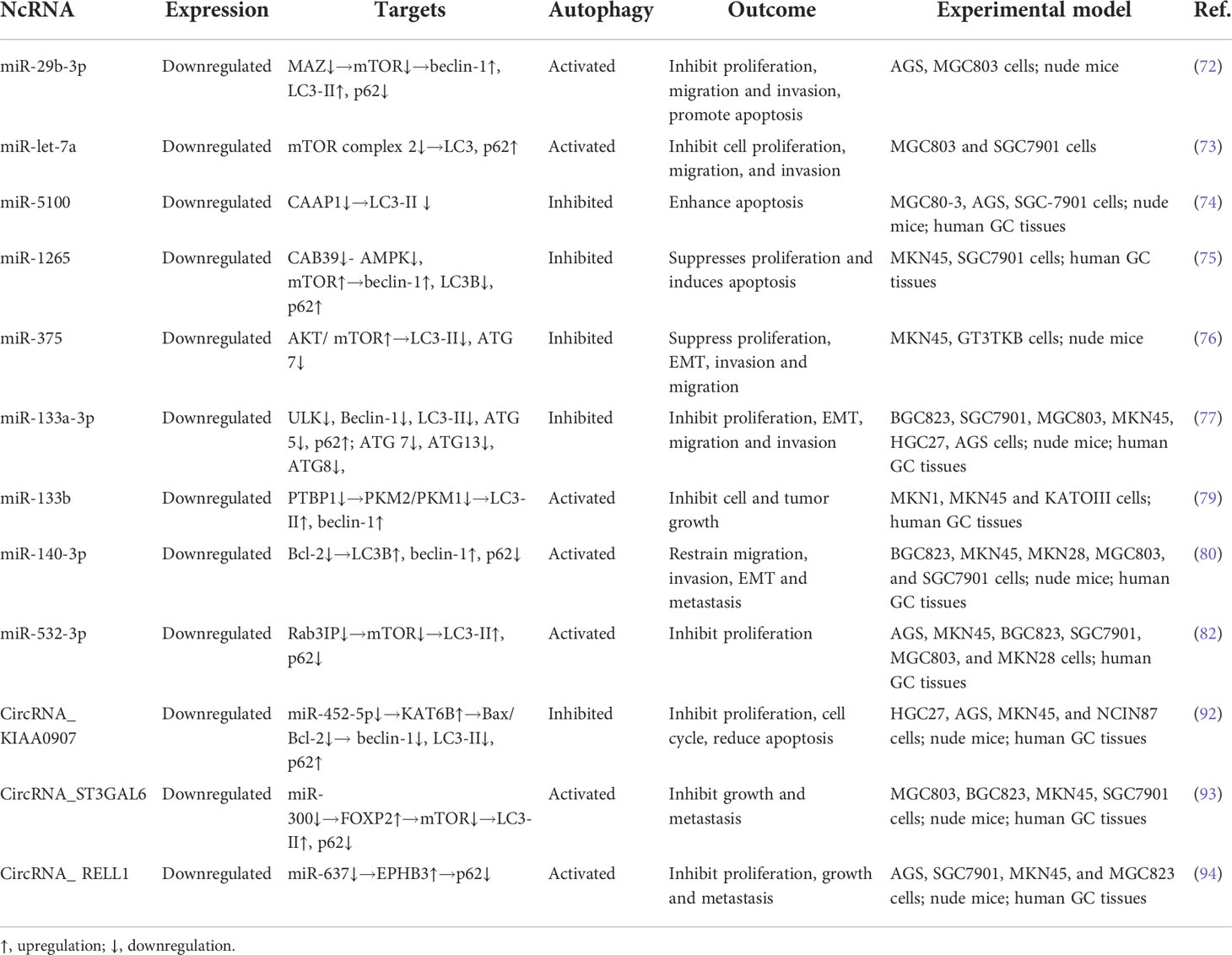
In conclusion, miRNAs directly target mRNAs to influence the gene translational process. Both lncRNAs and circRNAs can act as ceRNAs to sponge miRNAs, thus regulating the miRNAs-mediated gene epigenetic regulation. They also directly bind to proteins, suppressing proteins’ functions and blocking downstream signaling pathways (Figure 2). Dysfunction of ncRNAs is frequently emerged in cancer where ncRNAs function as both oncogenes and tumor suppressors. In this context, ncRNAs are capable of affecting various cellular processes, including autophagy, as they can regulate the expression of ATGs and autophagy-related signaling proteins. Thus, it can be speculated that the role of autophagy in tumors is can be further regulated by ncRNAs.
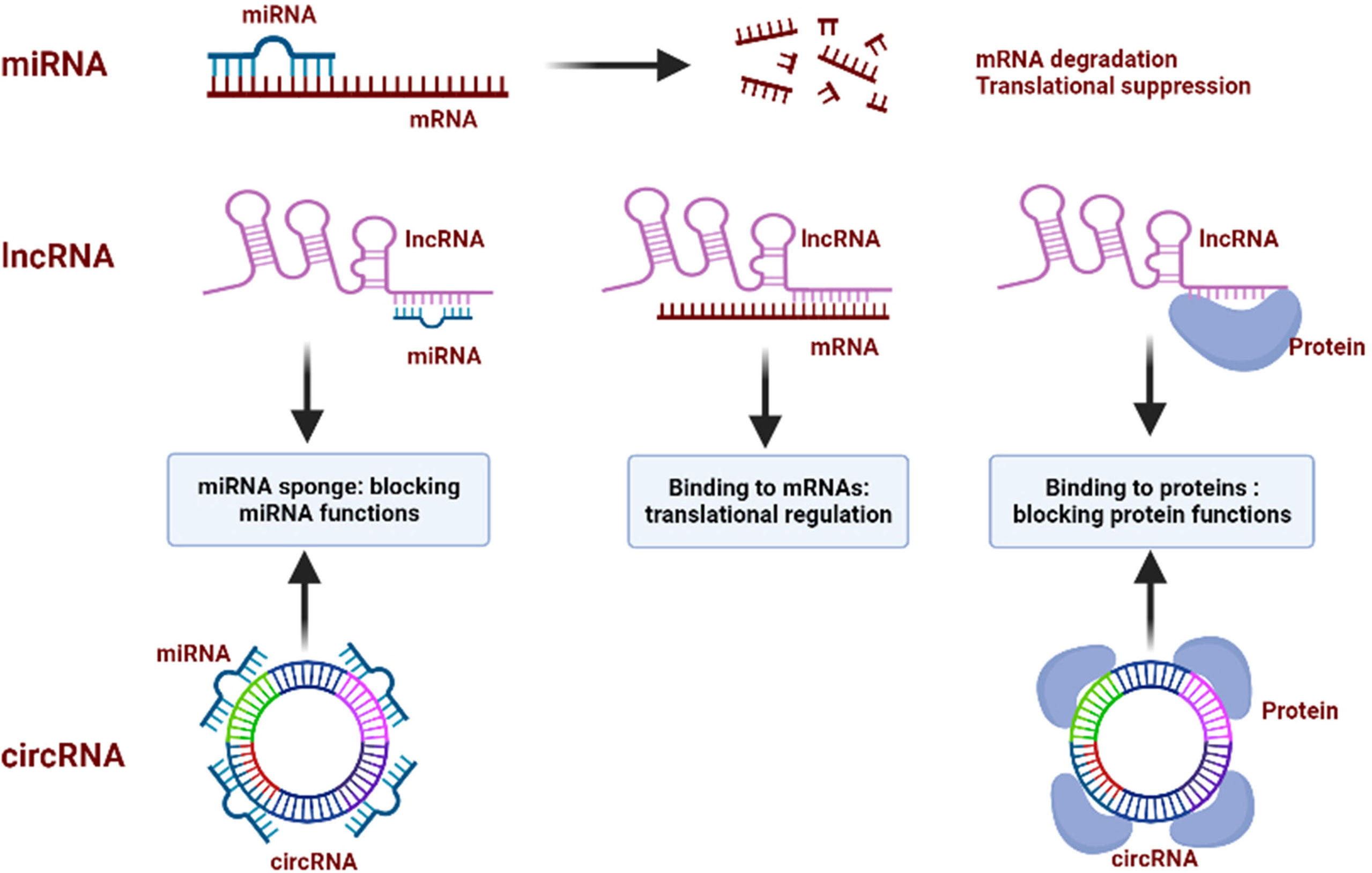
Figure 2 Regulatory mechanisms of ncRNAs. miRNAs bind to the target mRNAs and inhibit the function of mRNAs by inducing its degradation and blocking the translational process; lncRNAs sponge miRNAs to block their functions, bind to mRNAs to regulate their translation, and interact with proteins to repress their functions; circRNAs sponge miRNAs to block their functions, and bind to proteins to block their functions.
Roles of autophagy-regulating NcRNAs in GC
A total of 54 ncRNAs reported from experimental and clinical studies of GC are included to discuss their roles in regulating autophagy process in GC progression and chemoresistance (Tables 1–4). Mechanically, these autophagy-regulating ncRNAs modulate cellular autophagy by regulating ATGs and autophagy-related signaling pathways (Figure 1).
Tumor promoter
MiRNA molecules have been proved to directly regulate target genes to promote cancer cell proliferation in which autophagy is involved. For example, the miR-133a-3p/FOXP3 axis, playing an oncogenic role in GC cells, triggers the proliferation and autophagy via suppressing the expression of TP3, a tumor suppressor gene (47). Kong et al. illuminated that miR-423-3p, which indicated a poor prognosis in GC, was highly expressed in GC tissues and promoted cancer progression; however, this effect was blocked by knockdown of the ATG7, suggesting the oncogenic role of miR-423-3p via beclin-1-mediated autophagy (48). These findings indicate that oncogenic miRNAs trigger GC progression via inducing autophagy. However, miR-181a, highly expressed in both human GC tissues and cell lines, represses autophagy and thus leads to elevated migration and invasion via decreasing the expression of myotubularin related protein 3 (49), which contains PI3P phosphatases proteins that is required for the initiation of autophagy (67). Similarly, the ultraviolet radiation resistance-associated gene (UVRAG) is demonstrated to interact with Bcl-2 to further induce autophagy and GC cell death; moreover, miR-183 attenuates starvation-induced autophagy and apoptosis in GC cells by targeting the 3′-UTR region of UVRAG, suggesting an autophagy-related oncogenic role of miR-183 (50). As an oncogene, miR-20a is upregulated in GC and reduces both autophagy and apoptosis induced by the METase/SNHG5 axis (51). These results imply that oncogenic miRNAs also induce the malignant phenotype of GC via inhibiting both apoptosis and autophagy. H. pylori infection is believed to be the main risk factor for GC. Of interest, these bacteria induce the expression of miRNAs in host cells to facilitate GC progression via modulating autophagy. For instance, CagA released from H. pylori increases the expression of miR-543, which aggravates cell proliferation, migration, and invasion by impairing SIRT1-induced autophagy (68). Likewise, H. pylori-induced miR-30d overexpression in GC restrains autophagy to mediate H. pylori persistence and cancer progression through inhibiting the expression of ATG5, ATG12, and beclin-1 (69). Therefore, the infection-induced upregulation of miRNAs blocks autophagy and thus prevents H. pylori from autophagy-mediated clearance.
LncRNAs also promote GC progression by regulating autophagy. It was reported that lncRNA LIT3527 was highly expressed in GC, and its depletion drove cell autophagy and apoptosis via inhibiting AKT/ERK/mTOR signaling pathway, along with reduced proliferation, migration and lung metastasis of GC cells (52). Analogously, overexpressed lncRNA CCAT2 is observed in GC tissues; moreover, silencing of CCAT2 mediates apoptosis and autophagy, as well as the proliferation of BGC823 cells by suppressing the PI3K/mTOR activity (53). Intriguingly, upregulated lncRNA MALAT1 in GC cells protects IL-6 from autophagic degradation and thus aggravates IL-6-activated cancer-associated fibroblast conversion. Further mechanistic investigations explain that MALAT1 competitively interacts with ELAVL1 to destabilize PTEN mRNA because ELAVL1 bind a greater fraction of MALAT1 mRNA than PTEN mRNA, therefore blocking autophagy by activating the AKT/mTOR signaling pathway (54). Also, MALAT1 is demonstrated to function as a decoy of miR-204 to enhance GC cell proliferation but attenuate autophagy through downregulating the expression of LC3-II (55). Likewise, lncRNA JPX targets miR-197 to suppress beclin-1 expression while promoting p62 expression in GC cells, thus accelerating GC progression by regulating autophagy (56). New evidence shows that lncRNA HAGLROS, an indicator of a poor prognosis in GC patients, strengthens GC cell proliferation and maintains the malignant phenotype by inhibiting autophagy through competitively interacting with miR-100-5p to further activate mTORC1 and alleviate the expression of ATG9 (57). Besides, KLF5-induced lncRNA DANCR overexpression in GC elevates cancer cell viability while impeding autophagy through the miR-194/AKT2 axis (58). These results indicate that oncogenic lncRNAs mediate GC progression by suppressing autophagy through direct regulating autophagy-related pathways and acting as a sponge of miRNAs to indirectly inhibit the expression of ATGs. However, lncRNA CCAT1 promotes GC cell proliferation, migration and invasion, accompanied by increased autophagy activity by sponging miR-140-3p to restore the expression of ATG5 (59). High expression of lncRNA SNHG11 in GC also aggravates oncogenic autophagy by upregulating ATG12 expression through the miR-483-3p/miR-1276 axis (60). Similarly, overexpressed lncRNA LEF1-AS1 promotes the autophagy and malignant phenotype of GC by the miR-5100/DEK/AMPK/mTOR axis (61). Thus, lncRNAs also promote GC progression by inducing autophagy through sequestering miRNAs and enhancing ATGs expression.
Also, circRNAs play an oncogenic role in GC. It was reported that high circRNA_0032821 expression in patients with GC was associated with lymph node metastasis and poor prognosis. Further investigations unveiled that circRNA_0032821 facilitated GC cell proliferation, migration and invasion, but alleviated autophagy through activating the MEK1/ERK1/2 signaling pathway (62). Similarly, the elevated expression of circRNA_0006470 GC cells improved cell proliferation and migration through targeting miR-27b-3p to suppress autophagy. Mechanistic evaluation showed that circRNA_0006470 reduced the expression of LC3-II and beclin-1 but increased the expression of p62 by sponging miR-27b-3p to activate ROR1 and PI3KCA (63). Besides, the oncogenic circRNA_UBE2Q2 promoted glycolysis for GC cell invasion and metastasis through activating the miR-370-3p/STAT3 pathway (64). The AKT1/mTOR signaling pathway is also activated by circRNA_NRIP1, which inhibits autophagy but promotes GC progression by sponging miR-149-5p, indicating an underlying therapeutic target for GC (65). These studies suggest that oncogenic circRNAs in GC suppress autophagy by regulating autophagy-related pathways via targeting miRNAs. However, a recent study has reported that circRNA_0001658 promotes autophagy and GC cell viability through miR-182/Rab-10 axis (66).
It can be concluded that oncogenic ncRNAs are upregulated in GC tissues and promote malignant progression via activating or inhibiting autophagy. It is generally acknowledged that enhanced autophagy evokes GC progression in most cases owing to its cytoprotective feature (6). However, most oncogenic ncRNAs are related to low levels of autophagy, especially when ncRNAs function through regulating autophagy-related signaling pathways, including mTOR, PI3K, AKT, and ERK (Table 1). One explanation may be that oncogenic ncRNAs target these signaling pathways to block autophagic cell death. Hence, silencing these ncRNAs or targeting these downstream molecules may suppress tumorigenesis and progression by inducing autophagic cell death. Besides, it should be note that autophagy has been verified to interact with multiple biological processes associated with cancer progression, such as apoptosis, oxidative stress, DNA damage and repair (70, 71). Indeed, most ncRNAs exert their oncogenic role by inhibiting apoptosis and autophagy (Table 1). Thus, oncogenic ncRNAs in GC may mainly regulate malignant transformation processes with an additional autophagy suppression. It can be assumed that oncogenic ncRNAs-regulating autophagy in GC progression is complex due to its interaction with various cellular processes. However, the intrinsic interplay between autophagy and GC tumorigenesis has not been fully elaborated.
Tumor suppressor
Several miRNAs are downregulated in GC and act as tumor suppressor through regulating autophagy. Zhao et al. found that miR-29b-3p had a low expression in GC cells compared with normal human gastric mucosal epithelial cells, and that upregulation of miR-29b-3p induced apoptosis by inhibiting the autophagy-related protein MAZ, which can inhibit autophagy by activating the mTORC1 pathway (72). In addition, overexpression of miR-let-7a suppresses cell viability by enhancing autophagy in MGC803 and SGC7901 cells while downregulation of miR-let-7a reverses these effects. Further mechanistic investigations revealed that miR-let-7a regulated cellular autophagic levels by targeting Rictor and further affecting the AKT/mTOR activity (73). A similar study showed that miR-5100 promoted GC cell apoptosis while inhibiting autophagy by targeting caspase activity and apoptosis inhibitor 1, suggesting that downregulated miR-5100 was associated with the poor prognosis of GC patients (74). Coincidentally, high expression of miR-1265 is negatively related to tumor size in GC; moreover, it facilitates apoptosis but suppresses proliferation and autophagy by impairing the LKB1-STRAD-CAB39 complex and regulating the AMPK/mTOR signaling pathway (75). The AKT/mTOR pathway is also activated after upregulating miR-375 in GC cells, and autophagy is subsequently restrained, which inhibits GC cell migration and invasion both in vitro and in vivo (76). Furthermore, miR-133a-3p is found to target GABARAPL1 to block autophagy-mediated glutaminolysis, thereby repressing GC cell growth and metastasis (77). Similar effects are observed in Epstein-Barr virus-associated GC where miR-BART2-3p is elevated to attenuate autophagy and further reduce epithelial-mesenchymal transition (EMT) and cell migration by targeting ULK1 (78). These findings indicate that tumor-suppressive miRNAs inhibit GC progression by suppressing cytoprotective autophagy via targeting autophagy-related components. Additionally, overexpression of miR-133b in GC cells inhibits cell proliferation through eliciting autophagy by regulation of PKM isoforms; however, this inhibitory effect could be partially canceled when cells are administrated with 3-methyladenine, an autophagy inhibitor (79). As a tumor suppressor, miR-140-3p restrains EMT and metastasis in GC cells by suppressing Bcl-2 to further activate beclin-1-dependent autophagy (80). As a GTPase, RAB3IP is involved in the mTOR signaling pathway and regulates autophagy in cancer cells (81). A recent study has shown that miR-532-3p binds to the 3’UTR region of RAB3IP and inhibits its expression, which alleviates RAB3IP-mediated cell proliferation and autophagy inhibition (82). Of interest, H. pylori-induced miR-99b overexpression in GC tissues and BGC823 cells is shown to relieve bacterial loads and block cell proliferation but enhance autophagy by mTOR inhibition (83). Thus, miRNAs also function as tumor suppressor to inhibit GC malignant transformation by activating autophagy.
The downregulated lncRNAs in GC has been clarified to function as tumor suppressors. Mechanistically, these molecules are able to regulate autophagy and further affect GC progression. For example, overexpression of lncRNA FENDRR in GC represses tumor proliferation and invasion through the miR-421/SIRT3/Notch-1 axis (84). Further study revealed that FENDRR deteriorates cell apoptosis but inhibits autophagy via epigenetic suppression of ATG7 (85). These results suggest that lncRNAs promote GC cell apoptosis by targeting miRNAs, which may inhibit autophagy. The Notch-1 signaling pathway could be restrained by lncRNA SNHG1 in GC, and the cell proliferation is subsequently suppressed (86). However, depletion of SNHG1 alleviates apoptosis and autophagy by regulating miR-362-3p/JAK2/STAT3 pathway (87). Another deep impressed study demonstrated that lncRNA ADAMTS9-AS2 impeded GC progression and decreased the tumorigenicity of cancer stem cells through regulating SPOP, which may provide a novel target in the treatment of GC (88). Further mechanism analysis showed that ADAMTS9-AS2 downregulated the expression level of p62 and antiapoptotic protein Bcl-2 while upregulating the expression level of LC3-II, beclin-1, and the proapoptotic protein Bax by inactivating the PI3K/AKT/mTOR signaling pathway (89). In addition, upregulation of lncRNA DRAIC attenuates GC proliferation and metastasis via impairing the combination of UCHL5 and NFRKB and mediating the ubiquitination degradation of NFRKB (90). As a tumor suppressor, DRAIC can induce autophagy to maintain the cancer malignant phenotype via activating the AMPK-mTOR-S6K/ULK1 pathway (91). Therefore, lncRNAs seem to suppress GC progression by promoting autophagy, but there is lacking of direct evidence to support this point.
Also, low expressed circRNAs in GC have been implicated in tumor suppressor through regulating autophagy. Zhu et al. investigated the inhibitory effects of circRNA_KIAA0907 on migration and proliferation of GC cells. In their study, a remarkable KIAA0907 low expression was observed in GC compared with their adjacent normal tissue. KIAA0907 overexpression inhibited the cell cycle, proliferation, and autophagy, as well as promoted apoptosis. Molecular investigation demonstrated that KIAA0907 functioned as a specific sponge for miR-452-5p and further accelerated the expression of lysine acetyltransferase 6B. For in vivo efficacy assessment, a mouse GC model was used. KIAA0907 overexpression repressed proliferation of GC in mice. They also indicated that upregulation of KIAA0907 suppressed cell proliferation (92). Thus, this study demonstrates that circRNAs promote GC cell apoptosis but inhibit autophagy by targeting miRNAs. However, autophagy activation is also associated with the tumor-suppressive effect of circRNAs. A recent study has showed that circRNA-ST3GAL6 inhibits the malignant behaviors of GC by inducing apoptosis and autophagy through miR-300/FOXP2 axis (93). Likewise, circRNA-RELL1 blocks GC cell proliferation, invasion, migration via autophagy activation through the miR-637/EPHB3 axis (94).
In conclusion, various tumor suppressive ncRNAs are downregulated in GC cells compared with normal ones. Overexpression of these ncRNAs can inhibit cell proliferation and malignant transformation, accompanied with increased or reduced levels of autophagy. Modulation of autophagy by these ncRNAs is complicated due to they both target autophagy-related signaling pathways and ATGs (Table 2). On most occasions, tumor-suppressive ncRNAs inhibit cytoprotective autophagy. However, ncRNAs also induce pro-death autophagy to inhibit GC progression (80, 91). Overexpression of some tumor suppressor ncRNAs to block autophagy has become a potential therapeutic strategy in cancer via different molecular pathways (95). Thus, upregulation of tumor suppressive ncRNAs may be beneficial for GC treatment by inhibiting protective autophagy or facilitating autophagic cell death. Otherwise, autophagy induces cell death by crosstalk with non-apoptotic cell death pathways (96). It can be hypothesized that when tumor-suppressive ncRNAs activate nonapoptotic signals to provoke GC cell death, autophagy regulation may serve as an adaptive response to participate in ncRNAs-mediated tumor suppression. Thus, further clarifying the role of ncRNA-regulating autophagy is vital for GC treatment. In addition to ncRNAs dysfunction, the fluctuant autophagy level during GC progression may be associated with various stages of tumor development, histological and molecular subtypes of tumor, and other conditions.
Chemoresistance
Series of miRNAs are upregulated in GC and promote chemoresistance by regulating autophagy. A recent study has revealed that miR-3174 is highly expressed in GC cells and inhibits autophagic cell death by the ARHGAP10/mTORC1 axis, thus leading to cisplatin resistance (97). In another study performed by Gu et al., autophagy induction sensitizes GC cells to cisplatin, whereas its inhibition yields the opposite effects. Compared to parent cells, cisplatin-resistant GC cells express higher levels of miR-21. In GC cells transfected with miR-21 mimics, cisplatin resistance is restored via inhibiting autophagy by the PI3K/AKT/mTOR pathway; however, miR-21 suppressors confer GC cells to cisplatin sensitivity through activation of autophagy (98). These findings demonstrate that autophagy induction can be an effective therapeutic strategy in cisplatin-resistant GC. However, autophagy can counteract the chemotherapy-induced oxidative stress reaction. Oxidative stress-induced damage to cancer cells is a main model of action by chemotherapeutic drugs. Wang et al. showed that oxaliplatin-mediated oxidative stress could activate lncRNA NORAD, which further sponges miR-433-3p and stabilizes the ATG5-ATG12 complex, thereby enhancing the autophagy flux to alleviate oxidative stress, and ultimately leading to oxaliplatin resistance in GC (99). Thus, suppression of autophagy may be beneficial to inhibit oxaliplatin resistance in this context.
Some lncRNAs are overexpressed in GC tissues and induce cell resistance to the growth inhibition and apoptosis, which are mediated by chemotherapeutic agents through regulating autophagy. For instance, upregulated lncRNA FEZF1-AS1 in chemoresistant GC tissues drives chemoresistance of cancer cells to 5-FU through facilitating autophagy by directly targeting ATG5 (100). Similarly, lncRNA HULC, which indicates a poor prognosis in GC patients, induces autophagy and DDP resistance by inhibiting the ubiquitination of FoxM1 (101). Thus, silencing of lncRNA HULC elicits GC cell apoptosis induced by cisplatin (102). In addition, LINC00963, a new drug-resistant lncRNA, enhances proliferation and migration in oxaliplatin-resistant cells by negatively regulating miR-4458. Both downregulation of LINC00963 and upregulation of miR-4458 suppress autophagy by reducing the expression of ATG16, which indicates that targeting LINC00963 to block autophagy sensitizes GC cells to oxaliplatin (103). The miR188-3p/ATG14 axis is also targeted by lncRNA EIF3J-DT to activate autophagy and induce multiple chemotherapeutic drug resistance in GC cells (104). In cisplatin-resistant GC cells, LINC01572 is upregulated to induce chemoresistance, while depletion of LINC01572 promotes cisplatin sensitivity by inhibiting drug-induced autophagy through the miR-497-5p/ATG14 regulatory axis (105). A similar study unveiled that highly expressed LINC00641 in GC cells mediates oxaliplatin resistance via targeting miR-582-5p to regulate autophagy process (106). Besides, lncRNA MALAT1 functions as a ceRNA for miR-30b to sequester miR-30b from ATG5, thus inducing autophagy and cisplatin resistance (107). Of interest, MALAT1 causes poor disease-free survival and overall survival in patients who have received 5-FU-based adjuvant therapy; moreover, this lncRNA induces the resistance of GC cells to cisplatin, 5-FU and vincristine by triggering autophagy through sequestration of miR-23b-3p and then elevating ATG12 levels (108). Further mechanistic evaluation revealed that MALAT1 promotes GC cell proliferation and cisplatin resistance via the PI3K/AKT pathway (109). However, whether MALAT1-induced cisplatin resistance and autophagy is regulated by the PI3K/AKT pathway remains elusive. Additionally, overexpressed lncRNA HOTTIP in cisplatin-resistant GC cell line restrains autophagy and induces chemoresistance through sponging miR-216a-5p and further upregulating the Bcl-2 expression but decreasing the beclin-1 expression (110). These findings indicate that overexpressed lncRNAs in GC induce cell chemoresistance by inducing autophagy. However, when apoptosis is inhibited by lncRNAs to make GC cells become resistant to chemotherapeutic agents, autophagy might be suppressed as an additional effect.
CircRNAs are also upregulated in GC cells and contribute to chemoresistance by regulating autophagy. Yao and co-workers conducted a study on chemotherapeutic implication of circRNA_PVT1 in cisplatin-resistant GC cells. They showed that the knockdown of PVT1 expedited cisplatin sensitivity of GC via inhibiting autophagy and further promoting apoptosis and decreasing invasion. Molecular investigation demonstrated that PVT1 promoted cisplatin resistance and autophagy via the miR-30a-5p/YAP1 axis (111). Likewise, Ma et al. investigated the effects of miR-375 on circRNA_CGAP1-mediated apatinib chemoresistance of GC cells. In their study, a remarkable miR-3657 low expression was observed in GC tissues in comparison with their corresponding paracarcinoma samples. Silencing of CGAP1 inhibited apatinib-induced autophagy by stabilizing miR-3657 to augment its suppression on ATG7 expression; moreover, knockdown of CGAP1 sensitized GC cells to apatinib via autophagy inhibition in vitro and in vivo (112), suggesting that targeting CGAP1 may provide a novel target in the treatment of cisplatin-resistant GC. Also, overexpressed circRNA-CPM leads to the activation of autophagy and 5-FU chemoresistance in GC cells and tissues via the miR-21-3p/PRKAA2 axis (113). These results imply that overexpressed circRNAs in GC mediate chemoresistance by inducing autophagy, and that suppression of autophagy could improve chemotherapy efficacy in GC.
Collectively, chemoresistance-related ncRNAs are highly expressed in GC tissues and mediate the resistance to various chemotherapeutic drugs, along with various levels of autophagy. Of importance, most ncRNAs are associated with increased levels of autophagy, especially when ncRNAs directly upregulate ATGs (Table 3). Silencing these ncRNAs may alleviate the resistance of GC cells to chemotherapeutic agents by reducing protective autophagy. In fact, genetic interventions targeting ncRNAs such as miRNA mimics, miRNA sponges, anti-miRNA oligonucleotides are useful approaches to combat chemoresistance by regulating autophagy in cancer treatment (114). Thus, chemotherapies combined with suppression of autophagy via downregulating these ncRNAs or targeting ATGs may provide a promising therapeutic strategy for GC.
Chemosensitivity
Of importance, increasing researches have emphasized the role of miRNAs on enhancing the sensitivity of chemotherapeutic drugs in GC, in which the autophagic process is involved. MiR-181b-5p, regulated by the transcription factor CAGE, is downregulated in chemoresistant GC cells and reduces autophagic flux to promote the sensitivity to various anticancer drugs in these cells by targeting S1PR1 and further inhibiting the expression of beclin-1 (115). Similarly, overexpression of miR-495-3p is sufficient to reverse the multidrug resistant-cell to four chemotherapeutics and suppress the GC tumor growth, as miR-495-3p attenuates the process of autophagy via binding to GRP78 and thus activating mTOR (116). The PI3K/AKT/mTOR pathway is also regulated by miR-361-5p in GC to block autophagy and further to inhibit chemoresistance to docetaxel in vitro (117). In addition, upregulation of miR-181a in cisplatin-resistant SGC7901 cells expedites cisplatin sensitivity and reduces the growth of GC xenografts in vivo through negative regulating autophagy via targeting ATG5 (118). Similar to scenario observed in these cells, Du et al. have reported that miR-30a elevates the expression of P-gp and MDR1, reduces the expression of LC3-II, impedes autophagy, and reverses cisplatin resistance (119). Also, the overexpressed miR-874 in GC cells counteracts the multidrug resistance by directly downregulating the expression of ATG16L1 and thus blocking autophagy (18). Intriguingly, miR-148a-3p decreases the expression of A-kinase anchoring protein 1 and RAB12, a member of RAS oncogene family, and alleviates the inhibitory effects of RAB12 on mTORC1, thereby suppressing autophagy and reversing the resistance of GC cells to cisplatin (120). Of importance, miR-143 is regarded as an autophagy inhibitor to improve the efficacy of quercetin in GC cells via targeting GABARAPL1 (121). Moreover, miR-23b-3p is shown to inhibit autophagy, and reverses GC cell resistance to cisplatin, vincristine, and 5-FU by directly targeting ATG12 (122).These findings suggest that miRNAs induce chemosensitivity in GC though inhibiting autophagy. Thus, in GC with these downregulated miRNAs, chemotherapeutic drugs combined with autophagy inhibitors might provide a potential therapeutic strategy for this disease.
Additionally, lncRNAs have been demonstrated to be associated with chemosensitivity in GC by regulating autophagy. In a study performed by Zhang et al., decreased lncRNA CRNDE expression was related to the chemoresistance in GC cells, whereas its upregulation exhibited opposite effects. Further mechanistic investigations revealed that CRNDE increased proteasome ubiquitination-dependent degradation of SRSF6, a classical splicing factor. They unveiled that SRSF6 enhanced GC cellular autophagy by regulating alterative splicing of PICALM (S-to-L isoform switch), thereby mediating autophagy-induced resistance to oxaliplatin and 5-FU. Their results suggested that autophagy inhibited by the CRNDE/SRSF6 axis could be an effective therapeutic approach in chemoresistant GC (123). This finding implies that lncRNA CRNDE enhances chemosensitivity through suppressing autophagy. However, CRNDE derived from tumor-associated macrophages is demonstrated to promote GC cells proliferation and cisplatin resistance (124). The overexpressed CRNDE indicates a poor prognosis in GC patients; moreover, depression of CRNDE attenuates GC cell proliferation, migration and invasion by targeting miR-145 and affecting PI3K/AKT signaling pathways (125, 126). Therefore, the role of lncRNA CRNDE in GC is dependent on different cell types and conditions.
Furthermore, circRNAs can promote chemosensitivity in GC via autophagy regulation. Sun et al. investigated the effects of circRNA_MCTP2 on efficacy of chemotherapeutic agents in GC cells. In their study, a remarkable MCTP2 low expression was observed in cisplatin-resistant GC cells and tissues. MCTP2 overexpression inhibited proliferation and promoted apoptosis in GC cells. Most importantly, MCTP2 suppressed autophagy and sensitized GC cells to cisplatin by sponging miR-99a-5p and further upregulating the expression of MTMR3. For in vivo efficacy assessment, a GC mouse model was used. MCTP2 overexpression alleviated cisplatin resistance in vivo. They also proposed that upregulation of MCTP2 could be a promising therapeutic strategy for counteracting cisplatin resistance in GC (127). Similarly, overexpression of circRNA_CUL2 in cisplatin-resistant AGS and SGC7901 GC cells repressed malignant transformation in vitro and tumorigenicity in vivo, as CUL2 modulated tumor progression by sponging miR-142-3p to regulate ROCK2. Further investigation revealed that CUL2 enhanced the sensitivity of GC cells to cisplatin through miR-142-3p/ROCK2-mediated autophagy activation (128), since ROCK2 inhibition was associated with autophagy induction (129, 130). These results suggest that circRNAs can promote chemosensitivity in GC through modulating autophagy.
In sum, the expression level of chemosensitivity-related ncRNAs is depressed in GC tissues, and upregulation of these ncRNAs sensitizes GC cells to various chemotherapeutic drugs through inhibiting autophagy by both regulating autophagy-related signaling pathways and targeting ATGs (Table 4). The levels of some ncRNAs are negatively correlated with protective autophagy and drug resistance following chemotherapy, therefore rejuvenation of these downregulated ncRNAs in drug resistant GC cells may restore chemosensitivity by inhibiting autophagy. Moreover, ncRNA-based therapeutic approaches, including ncRNA-coated nanoparticles and ncRNA microinjection, are developed to enhance chemosensitivity (131). In this context, inhibition of cytoprotective autophagy by ncRNAs may pave the way to combat chemoresistance.
Conclusions
In this review, the role of ncRNAs-induced autophagy in the malignant transformation and drug resistance of GC are summarized in detail. NcRNAs are shown to serve as both oncogenes and tumor suppressors in GC. Of interest, NcRNA-regulated autophagy also perform a dual function in the progression and chemotherapy of GC. How the autophagy mechanism exerts a dual effect in the GC is unclear. It can be postulated that the role of autophagy is determined by its upstream modulators. Whether oncogenic ncRNAs or tumor-suppressive ncRNAs is dependent on various levels and functions of autophagy to affect GC progression and chemotherapeutic efficacy. Briefly, as tumor suppressors, ncRNAs inhibit cytoprotective autophagy during cainogenesis or induce autophagic cell death at the advanced stages of GC; whereas oncogenic ncRNAs trigger cytoprotective autophagy to facilitate tumorigenesis and progression of GC. The regulatory mechanisms of ncRNAs on autophagy is intricate. Noteworthy, oncogenic ncRNAs tends to regulate autophagy-related signaling pathways to inhibit autophagic cell death, and chemoresistance-related ncRNAs mainly increase the expression of ATGs to activate cytoprotective autophagy; moreover, upregulating the expression of ncRNAs that is originally repressed in tumor tissues promote tumor suppression and chemosensitivity through autophagy modulation via both regulating autophagy-related signaling pathways and ATGs. In this context, targeting oncogenic ncRNAs combined with autophagy inhibitors can be employed as a promising strategy to promote GC cells death and to sensitize them to chemotherapy. Thus, clarification of the regulatory role of ncRNAs on autophagy in GC development and chemoresistance is expected to strategize beneficial options in ncRNA-based therapies to eradicate this disease, as it will help contribute towards addressing key issues in GC drug resistance.
Author contributions
ZW wrote the manuscript. JL, JX, XY, and BW contributed to the writing and drafted the figures. WS and YZ critically reviewed the manuscript and contributed to the writing. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by the National Natural Science Foundation of China (81973601), TCM research projects of Heilongjiang Province (ZHY2020-041) and Postdoctoral funding of Heilongjiang Province (LBH-Z21218).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AMPK, adenosine monophosphate-activated protein kinase; miRNA, microRNA; AMBRA, autophagy and beclin-1 regulator; MAPK, mitogen activated kinase-like protein; ATGs, autophagy-related genes; ncRNA, non-coding RNA; CAAP1, caspase activity and apoptosis inhibitor 1; PI3K, phosphatidylinositol 3-kinase; ceRNA, competing endogenous RNA; PI3P. phosphatidylinositol 3-phosphate; circRNA, circular RNA; PAS, phosphatidylethanolamine; ERK, extracellular signal-regulated kinase; PE, pre-autophagosomal structure EMT; epithelial-mesenchymal transition PTEN phosphatase and tensin homolog ER endoplasmic reticulum TSC Tuberous sclerosis complex GC gastric cancer ULK1 unc-51 like autophagy activating kinase JNK c-Jun N-terminal kinase UVRAG, ultraviolet radiation resistance-associated gene; LC3, microtubule associated protein light chain 3; VPS15, vacuolar protein sorting 15; LncRNA, long non-coding RNA; WIPI2, WD repeat domain phosphoinositide-interacting protein; mTORC, mammalian target of rapamycin; ZFYVE1, zinc-finger FYVE domain-containing protein 1.
References
1. Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol (2020) 18(3):534–42. doi: 10.1016/j.cgh.2019.07.045
2. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/s0140-6736(20)31288-5
3. Shi WJ, Gao JB. Molecular mechanisms of chemoresistance in gastric cancer. World J Gastrointest Oncol (2016) 8(9):673–81. doi: 10.4251/wjgo.v8.i9.673
4. Morishita H, Mizushima N. Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol (2019) 35:453–75. doi: 10.1146/annurev-cellbio-100818-125300
5. Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: Recent advances and future directions. Cancer Discovery (2019) 9(9):1167–81. doi: 10.1158/2159-8290.CD-19-0292
6. Cao Y, Luo Y, Zou J, Ouyang J, Cai Z, Zeng X, et al. Autophagy and its role in gastric cancer. Clin Chim Acta (2019) 489:10–20. doi: 10.1016/j.cca.2018.11.028
7. Xu JL, Yuan L, Tang YC, Xu ZY, Xu HD, Cheng XD, et al. The role of autophagy in gastric cancer chemoresistance: Friend or foe? Front Cell Dev Biol (2020) 8:621428. doi: 10.3389/fcell.2020.621428
8. Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell (2019) 177(7):1682–99. doi: 10.1016/j.cell.2019.05.026
9. Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res (2018) 129:151–5. doi: 10.1016/j.phrs.2017.11.009
10. Yin Q, Feng W, Shen X, Ju S. Regulatory effects of lncRNAs and miRNAs on autophagy in malignant tumorigenesis. Biosci Rep (2018) 38(5):BSR20180516. doi: 10.1042/BSR20180516
11. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J (2017) 36(13):1811–36. doi: 10.15252/embj.201796697
12. Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev (2010) 90(4):1383–435. doi: 10.1152/physrev.00030.2009
13. Al-Bari MAA. A current view of molecular dissection in autophagy machinery. J Physiol Biochem (2020) 76(3):357–72. doi: 10.1007/s13105-020-00746-0
14. Chen Z, Zhang J, Dong C, Li D, Yin Y, Yu W, et al. TNFAIP8 regulates gastric cancer growth via mTOR-Akt-ULK1 pathway and autophagy signals. J Cell Mol Med (2021) 25(7):3361–70. doi: 10.1111/jcmm.16413
15. Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy (2013) 9(2):124–37. doi: 10.4161/auto.23323
16. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol (2013) 15(7):741–50. doi: 10.1038/ncb2757
17. Lystad AH, Carlsson SR, Simonsen A. Toward the function of mammalian ATG12-ATG5-ATG16L1 complex in autophagy and related processes. Autophagy (2019) 15(8):1485–6. doi: 10.1080/15548627.2019.1618100
18. Huang H, Tang J, Zhang L, Bu Y, Zhang X. miR-874 regulates multiple-drug resistance in gastric cancer by targeting ATG16L1. Int J Oncol (2018) 53(6):2769–79. doi: 10.3892/ijo.2018.4593
19. Zhang X, Wang L, Ireland SC, Ahat E, Li J, Bekier ME 2nd, et al. GORASP2/GRASP55 collaborates with the PtdIns3K UVRAG complex to facilitate autophagosome-lysosome fusion. Autophagy (2019) 15(10):1787–800. doi: 10.1080/15548627.2019.1596480
20. Paquette M, El-Houjeiri L, Pause A. mTOR pathways in cancer and autophagy. Cancers (2018) 10(1):18. doi: 10.3390/cancers10010018
21. Yu Y, Hou L, Song H, Xu P, Sun Y, Wu K. Akt/AMPK/mTOR pathway was involved in the autophagy induced by vitamin e succinate in human gastric cancer SGC-7901 cells. Mol Cell Biochem (2017) 424(1-2):173–83. doi: 10.1007/s11010-016-2853-4
22. Mao K, Klionsky DJ. AMPK activates autophagy by phosphorylating ULK1. Circ Res (2011) 108(7):787–8. doi: 10.1161/RES.0b013e3182194c29
23. Cheng CY, Tseng HH, Chiu HC, Chang CD, Nielsen BL, Liu HJ. Bovine ephemeral fever virus triggers autophagy enhancing virus replication via upregulation of the Src/JNK/AP1 and PI3K/Akt/NF-κB pathways and suppression of the PI3K/Akt/mTOR pathway. Vet Res (2019) 50(1):79. doi: 10.1186/s13567-019-0697-0
24. Cai J, Li R, Xu X, Zhang L, Lian R, Fang L, et al. CK1alpha suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat Cell Biol (2018) 20(4):465–78. doi: 10.1038/s41556-018-0065-8
25. Acevo-Rodriguez PS, Maldonado G, Castro-Obregon S, Hernandez G. Autophagy regulation by the translation machinery and its implications in cancer. Front Oncol (2020) 10:322. doi: 10.3389/fonc.2020.00322
26. Papaefthymiou A, Christodoulidis G, Koffas A, Doulberis M, Polyzos SA, Manolakis A, et al. Role of autophagy in gastric carcinogenesis. World J Gastrointest Oncol (2021) 13(10):1244–62. doi: 10.4251/wjgo.v13.i10.1244
27. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell (2009) 136(2):215–33. doi: 10.1016/j.cell.2009.01.002
28. Baptista B, Riscado M, Queiroz JA, Pichon C, Sousa F. Non-coding RNAs: Emerging from the discovery to therapeutic applications. Biochem Pharmacol (2021) 189:114469. doi: 10.1016/j.bcp.2021.114469
29. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery (2017) 16(3):203–22. doi: 10.1038/nrd.2016.246
30. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
31. Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology (2012) 143(1):35–47.e2. doi: 10.1053/j.gastro.2012.05.003
32. Daoud AZ, Mulholland EJ, Cole G, McCarthy HO. MicroRNAs in pancreatic cancer: biomarkers, prognostic, and therapeutic modulators. BMC Cancer (2019) 19(1):1130. doi: 10.1186/s12885-019-6284-y
33. Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol (2015) 22(1):5–7. doi: 10.1038/nsmb.2942
34. Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol (2019) 112:82–92. doi: 10.1016/j.molimm.2019.04.011
35. Liu QL, Zhang Z, Wei X, Zhou ZG. Noncoding RNAs in tumor metastasis: molecular and clinical perspectives. Cell Mol Life Sci (2021) 78(21-22):6823–50. doi: 10.1007/s00018-021-03929-0
36. Kong Y, Lu Z, Liu P, Liu Y, Wang F, Liang EY, et al. Long noncoding RNA: Genomics and relevance to physiology. Compr Physiol (2019) 9(3):933–46. doi: 10.1002/cphy.c180032
37. Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorg Chem (2019) 92:103214. doi: 10.1016/j.bioorg.2019.103214
38. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
39. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
40. Zhang Z, Yang T, Xiao J. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
41. Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer (2021) 20(1):13. doi: 10.1186/s12943-020-01298-z
42. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
43. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer (2020) 19(1):172. doi: 10.1186/s12943-020-01286-3
44. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther (2018) 187:31–44. doi: 10.1016/j.pharmthera.2018.01.010
45. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell (2019) 176(4):869–81.e13. doi: 10.1016/j.cell.2018.12.021
46. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer (2020) 6(4):319–36. doi: 10.1016/j.trecan.2020.01.012
47. Li JP, Zhang HM, Liu MJ, Xiang Y, Li H, Huang F, et al. miR-133a-3p/FOXP3 axis regulates cell proliferation and autophagy in gastric cancer. J Cell Biochem (2020) 121(5-6):3392–405. doi: 10.1002/jcb.29613
48. Kong P, Zhu X, Geng Q, Xia L, Sun X, Chen Y, et al. The microRNA-423-3p-Bim axis promotes cancer progression and activates oncogenic autophagy in gastric cancer. Mol Ther (2017) 25(4):1027–37. doi: 10.1016/j.ymthe.2017.01.013
49. Lin Y, Zhao J, Wang H, Cao J, Nie Y. miR-181a modulates proliferation, migration and autophagy in AGS gastric cancer cells and downregulates MTMR3. Mol Med Rep (2017) 15(5):2451–6. doi: 10.3892/mmr.2017.6289
50. Yuan Y, Zhang Y, Han L, Sun S, Shu Y. miR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int J Mol Med (2018) 42(6):3562–70. doi: 10.3892/ijmm.2018.3871
51. Xin L, Zhou LQ, Liu L, Yuan YW, Zhang HT, Zeng F. METase promotes cell autophagy via promoting SNHG5 and suppressing miR-20a in gastric cancer. Int J Biol Macromol (2019) 122:1046–52. doi: 10.1016/j.ijbiomac.2018.09.051
52. Wang B, Deng Y, Jin J, Wu Y, Shen L. Long noncoding RNA LIT3527 knockdown induces apoptosis and autophagy through inhibiting mTOR pathway in gastric cancer cells. J Cancer (2021) 12(16):4901–11. doi: 10.7150/jca.58185
53. Yu ZY, Wang Z, Lee KY, Yuan P, Ding J. Effect of silencing colon cancer-associated transcript 2 on the proliferation, apoptosis and autophagy of gastric cancer BGC-823 cells. Oncol Lett (2018) 15(3):3127–32. doi: 10.3892/ol.2017.7677
54. Wang Z, Wang X, Zhang T, Su L, Liu B, Zhu Z, et al. LncRNA MALAT1 promotes gastric cancer progression via inhibiting autophagic flux and inducing fibroblast activation. Cell Death Dis (2021) 12(4):368. doi: 10.1038/s41419-021-03645-4
55. Shao G, Zhao Z, Zhao W, Hu G, Zhang L, Li W, et al. Long non-coding RNA MALAT1 activates autophagy and promotes cell proliferation by downregulating microRNA-204 expression in gastric cancer. Oncol Lett (2020) 19(1):805–12. doi: 10.3892/ol.2019.11184
56. Han X, Liu Z. Long non−coding RNA JPX promotes gastric cancer progression by regulating CXCR6 and autophagy via inhibiting miR−197. Mol Med Rep (2021) 23(1):60. doi: 10.3892/mmr.2020.11698
57. Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer (2018) 17(1):6. doi: 10.1186/s12943-017-0756-y
58. Cheng Z, Liu G, Huang C, Zhao X. KLF5 activates lncRNA DANCR and inhibits cancer cell autophagy accelerating gastric cancer progression. NPJ Genom Med (2021) 6(1):75. doi: 10.1038/s41525-021-00207-7
59. Yang F, Peng ZX, Ji WD, Yu JD, Qian C, Liu JD, et al. LncRNA CCAT1 upregulates ATG5 to enhance autophagy and promote gastric cancer development by absorbing miR-140-3p. Dig Dis Sci (2021) 67(8):3725–41. doi: 10.1007/s10620-021-07187-9
60. Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. lncRNA SNHG11 promotes gastric cancer progression by activating the wnt/β-catenin pathway and oncogenic autophagy. Mol Ther (2021) 29(3):1258–78. doi: 10.1016/j.ymthe.2020.10.011
61. Zhang H, Wang J, Wang Y, Li J, Zhao L, Zhang T, et al. Long non-coding LEF1-AS1 sponge miR-5100 regulates apoptosis and autophagy in gastric cancer cells via the miR-5100/DEK/AMPK-mTOR axis. Int J Mol Sci (2022) 23(9):4787. doi: 10.3390/ijms23094787
62. Jiang Y, Zhang Y, Chu F, Xu L, Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int (2020) 20:74. doi: 10.1186/s12935-020-1151-0
63. Cui Y, Cao J, Huang S, Ye J, Huang H, Liao D, et al. circRNA_0006470 promotes the proliferation and migration of gastric cancer cells by functioning as a sponge of miR-27b-3p. Neoplasma (2021) 68(6):1245–56. doi: 10.4149/neo_2021_210222N235
64. Yang J, Zhang X, Cao J, Xu P, Chen Z, Wang S, et al. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell Death Dis (2021) 12(10):910. doi: 10.1038/s41419-021-04216-3
65. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer (2019) 18(1):20. doi: 10.1186/s12943-018-0935-5
66. Duan X, Yu X, Li Z. Circular RNA hsa_circ_0001658 regulates apoptosis and autophagy in gastric cancer through microRNA-182/Ras-related protein rab-10 signaling axis. Bioengineered (2022) 13(2):2387–97. doi: 10.1080/21655979.2021.2024637
67. Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett (2010) 584(7):1313–8. doi: 10.1016/j.febslet.2010.02.054
68. Shi Y, Yang Z, Zhang T, Shen L, Li Y, Ding S. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in helicobacter pylori CagA-associated gastric cancer. Cell Death Dis (2019) 10(9):625. doi: 10.1038/s41419-019-1859-8
69. Yang XJ, Si RH, Liang YH, Ma BQ, Jiang ZB, Wang B, et al. Mir-30d increases intracellular survival of helicobacter pylori through inhibition of autophagy pathway. World J Gastroenterol (2016) 22(15):3978–91. doi: 10.3748/wjg.v22.i15.3978
70. Gao L, Loveless J, Shay C, Teng Y. Targeting ROS-mediated crosstalk between autophagy and apoptosis in cancer. Adv Exp Med Biol (2020) 1260:1–12. doi: 10.1007/978-3-030-42667-5_1
71. Das S, Shukla N, Singh SS, Kushwaha S, Shrivastava R. Mechanism of interaction between autophagy and apoptosis in cancer. Apoptosis (2021) 26(9-10):512–33. doi: 10.1007/s10495-021-01687-9
72. Zhao X, Ye N, Feng X, Ju H, Liu R, Lu W. MicroRNA-29b-3p inhibits the migration and invasion of gastric cancer cells by regulating the autophagy-associated protein MAZ. Onco Targets Ther (2021) 14:3239–49. doi: 10.2147/ott.s274215
73. Fan H, Jiang M, Li B, He Y, Huang C, Luo D, et al. MicroRNA-let-7a regulates cell autophagy by targeting rictor in gastric cancer cell lines MGC-803 and SGC-7901. Oncol Rep (2018) 39(3):1207–14. doi: 10.3892/or.2018.6194
74. Zhang HM, Li H, Wang GX, Wang J, Xiang Y, Huang Y, et al. MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. Neoplasia (2020) 22(5):220–30. doi: 10.1016/j.neo.2020.03.001
75. Xu Z, Li Z, Wang W, Xia Y, He Z, Li B, et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett (2019) 449:226–36. doi: 10.1016/j.canlet.2019.02.026
76. Yuan KT, Li BX, Yuan YJ, Tan M, Tan JF, Dai WG, et al. Deregulation of MicroRNA-375 inhibits proliferation and migration in gastric cancer in association with autophagy-mediated AKT/mTOR signaling pathways. Technol Cancer Res Treat (2018) 17:1–11. doi: 10.1177/1533033818806499
77. Zhang X, Li Z, Xuan Z, Xu P, Wang W, Chen Z, et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res (2018) 37(1):320. doi: 10.1186/s13046-018-0993-y
78. Shi D, Zhang Y, Mao T, Liu D, Liu W, Luo B. MiR-BART2-3p targets unc-51-like kinase 1 and inhibits cell autophagy and migration in Epstein-Barr virus-associated gastric cancer. Virus Res (2021) 305:198567. doi: 10.1016/j.virusres.2021.198567
79. Sugiyama T, Taniguchi K, Matsuhashi N, Tajirika T, Futamura M, Takai T, et al. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci (2016) 107(12):1767–75. doi: 10.1111/cas.13091
80. Chen J, Cai S, Gu T, Song F, Xue Y, Sun D. MiR-140-3p impedes gastric cancer progression and metastasis by regulating BCL2/BECN1-mediated autophagy. Onco Targets Ther (2021) 14:2879–92. doi: 10.2147/ott.s299234
81. Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, et al. Ral and rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell (2014) 53(2):209–20. doi: 10.1016/j.molcel.2013.12.004
82. Guo W, Chen Z, Chen Z, Yu J, Liu H, Li T, et al. Promotion of cell proliferation through inhibition of cell autophagy signalling pathway by Rab3IP is restrained by MicroRNA-532-3p in gastric cancer. J Cancer (2018) 9(23):4363–73. doi: 10.7150/jca.27533
83. Yang L, Li C, Jia Y. MicroRNA-99b promotes helicobacter pylori-induced autophagyand suppresses carcinogenesis by targeting mTOR. Oncol Lett (2018) 16(4):5355–60. doi: 10.3892/ol.2018.9269
84. Ma J, Zhao G, Du J, Li J, Lin G, Zhang J. LncRNA FENDRR inhibits gastric cancer cell proliferation and invasion via the miR-421/SIRT3/Notch-1 axis. Cancer Manag Res (2021) 13:9175–87. doi: 10.2147/cmar.s329419
85. Zhao SP, Yu C, Yang MS, Liu ZL, Yang BC, Xiao XF. Long non-coding RNA FENDRR modulates autophagy through epigenetic suppression of ATG7 via binding PRC2 in acute pancreatitis. Inflammation (2021) 44(3):999–1013. doi: 10.1007/s10753-020-01395-7
86. Zhang Z, Wang H. lncRNA SNHG1 suppresses gastric cancer cell proliferation and promotes apoptosis via Notch1 pathway. J buon (2020) 25(1):302–7.
87. Zhou J, Li Z, Zhao Q, Wu T, Zhao Q, Cao Y. Knockdown of SNHG1 alleviates autophagy and apoptosis by regulating miR-362-3p/Jak2/stat3 pathway in LPS-injured PC12 cells. Neurochem Res (2021) 46(4):945–56. doi: 10.1007/s11064-020-03224-7
88. Wang F, Tang C, Xu D, Tang Y, Jiang Y, Gao X, et al. LncRNA ADAMTS9-AS2 suppresses the proliferation of gastric cancer cells and the tumorigenicity of cancer stem cells through regulating SPOP. J Cell Mol Med (2020) 24(8):4830–8. doi: 10.1111/jcmm.15161
89. Li H, Huang H, Li S, Mei H, Cao T, Lu Q. Long non-coding RNA ADAMTS9-AS2 inhibits liver cancer cell proliferation, migration and invasion. Exp Ther Med (2021) 21(6):559. doi: 10.3892/etm.2021.9991
90. Zhang Z, Hu X, Kuang J, Liao J, Yuan Q. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell Mol Biol Lett (2020) 25:29. doi: 10.1186/s11658-020-00221-0
91. Saha S, Zhang Y, Wilson B, Abounader R, Dutta A. The tumor-suppressive long noncoding RNA DRAIC inhibits protein translation and induces autophagy by activating AMPK. J Cell Sci (2021) 134(24):259306. doi: 10.1242/jcs.259306
92. Zhu L, Wang C, Lin S, Zong L. CircKIAA0907 retards cell growth, cell cycle, and autophagy of gastric cancer In vitro and inhibits tumorigenesis In vivo via the miR-452-5p/KAT6B axis. Med Sci Monit (2020) 26:e924160. doi: 10.12659/MSM.924160
93. Xu P, Zhang X, Cao J, Yang J, Chen Z, Wang W, et al. The novel role of circular RNA ST3GAL6 on blocking gastric cancer malignant behaviours through autophagy regulated by the FOXP2/MET/mTOR axis. Clin Transl Med (2022) 12(1):e707. doi: 10.1002/ctm2.707
94. Sang H, Zhang W, Peng L, Wei S, Zhu X, Huang K, et al. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation. Cell Death Dis (2022) 13(1):56. doi: 10.1038/s41419-021-04364-6
95. Liang S, Li X, Gao C, Zhang L. microRNA-based autophagy inhibition as targeted therapy in pancreatic cancer. BioMed Pharmacother (2020) 132:110799. doi: 10.1016/j.biopha.2020.110799
96. Miller DR, Cramer SD, Thorburn A. The interplay of autophagy and non-apoptotic cell death pathways. Int Rev Cell Mol Biol (2020) 352:159–87. doi: 10.1016/bs.ircmb.2019.12.004
97. Li B, Wang L, Li Z, Wang W, Zhi X, Huang X, et al. miR-3174 contributes to apoptosis and autophagic cell death defects in gastric cancer cells by targeting ARHGAP10. Mol Ther Nucleic Acids (2017) 9:294–311. doi: 10.1016/j.omtn.2017.10.008
98. Gu Y, Fei Z, Zhu R. miR-21 modulates cisplatin resistance of gastric cancer cells by inhibiting autophagy via the PI3K/Akt/mTOR pathway. Anticancer Drugs (2020) 31(4):385–93. doi: 10.1097/cad.0000000000000886
99. Wang J, Sun Y, Zhang X, Cai H, Zhang C, Qu H, et al. Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell Death Dis (2021) 12(1):90. doi: 10.1038/s41419-020-03368-y
100. Gui Z, Zhao Z, Sun Q, Shao G, Huang J, Zhao W, et al. LncRNA FEZF1-AS1 promotes multi-drug resistance of gastric cancer cells via upregulating ATG5. Front Cell Dev Biol (2021) 9:749129. doi: 10.3389/fcell.2021.749129
101. Xin L, Zhou Q, Yuan YW, Zhou LQ, Liu L, Li SH, et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol (2019) 145(10):2507–17. doi: 10.1007/s00432-019-03015-w
102. Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in human gastric cancer. J Med Biochem (2016) 35(2):137–43. doi: 10.1515/jomb-2015-0016
103. Hou M, Li C, Dong S. LINC00963/miR-4458 regulates the effect of oxaliplatin in gastric cancer by mediating autophagic flux through targeting of ATG16L1. Sci Rep (2021) 11(1):20951. doi: 10.1038/s41598-021-98728-9
104. Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang C, et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy (2021) 17(12):4083–101. doi: 10.1080/15548627.2021.1901204
105. Song Z, Jia N, Li W, Zhang XY. LINC01572 regulates cisplatin resistance in gastric cancer cells by mediating miR-497-5p. Onco Targets Ther (2020) 13:10877–87. doi: 10.2147/OTT.S267915
106. Hu Y, Su Y, Lei X, Zhao H, Wang L, Xu T, et al. LINC00641/miR-582-5p mediate oxaliplatin resistance by activating autophagy in gastric adenocarcinoma. Sci Rep (2020) 10(1):14981. doi: 10.1038/s41598-020-70913-2
107. Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagy−associated cisplatin resistance by regulating the microRNA−30b/autophagy−related gene 5 axis in gastric cancer. Int J Oncol (2019) 54(1):239–48. doi: 10.3892/ijo.2018.4609
108. YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer (2017) 16(1):174. doi: 10.1186/s12943-017-0743-3
109. Dai Q, Zhang T, Li C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag Res (2020) 12:1929–39. doi: 10.2147/CMAR.S243796
110. Zhao R, Zhang X, Zhang Y, Zhang Y, Yang Y, Sun Y, et al. HOTTIP predicts poor survival in gastric cancer patients and contributes to cisplatin resistance by sponging miR-216a-5p. Front Cell Dev Biol (2020) 8:348. doi: 10.3389/fcell.2020.00348
111. Yao W, Guo P, Mu Q, Wang Y. Exosome-derived circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis Via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm (2021) 36(4):347–59. doi: 10.1089/cbr.2020.3578
112. Ma L, Wang Z, Xie M, Quan Y, Zhu W, Yang F, et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis (2020) 11(3):169. doi: 10.1038/s41419-020-2352-0
113. Fang L, Lv J, Xuan Z, Li B, Li Z, He Z, et al. Circular CPM promotes chemoresistance of gastric cancer via activating PRKAA2-mediated autophagy. Clin Transl Med (2022) 12(1):e708. doi: 10.1002/ctm2.708
114. Lei Y, Chen L, Liu J, Zhong Y, Deng L. The MicroRNA-based strategies to combat cancer chemoresistance via regulating autophagy. Front Oncol (2022) 12:841625. doi: 10.3389/fonc.2022.841625
115. Yeon M, Kim Y, Pathak D, Kwon E, Kim DY, Jeong MS, et al. The CAGE-MiR-181b-5p-S1PR1 axis regulates anticancer drug resistance and autophagy in gastric cancer cells. Front Cell Dev Biol (2021) 9:666387. doi: 10.3389/fcell.2021.666387
116. Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D, et al. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis (2018) 9(11):1070. doi: 10.1038/s41419-018-0950-x
117. Tian L, Zhao Z, Xie L, Zhu J. MiR-361-5p suppresses chemoresistance of gastric cancer cells by targeting FOXM1 via the PI3K/Akt/mTOR pathway. Oncotarget (2018) 9(4):4886–96. doi: 10.18632/oncotarget.23513
118. Zhao J, Nie Y, Wang H, Lin Y. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene (2016) 576(2 Pt 2):828–33. doi: 10.1016/j.gene.2015.11.013
119. Du X, Liu B, Luan X, Cui Q, Li L. miR-30 decreases multidrug resistance in human gastric cancer cells by modulating cell autophagy. Exp Ther Med (2018) 15(1):599–605. doi: 10.3892/etm.2017.5354
120. Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J, et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett (2017) 410:212–27. doi: 10.1016/j.canlet.2017.09.035
121. Du F, Feng Y, Fang J, Yang M. MicroRNA-143 enhances chemosensitivity of quercetin through autophagy inhibition via target GABARAPL1 in gastric cancer cells. BioMed Pharmacother (2015) 74:169–77. doi: 10.1016/j.biopha.2015.08.005
122. An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, et al. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis (2015) 6(5):e1766. doi: 10.1038/cddis.2015.123
123. Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, et al. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer (2021) 20(1):6. doi: 10.1186/s12943-020-01299-y
124. Xin L, Zhou LQ, Liu C, Zeng F, Yuan YW, Zhou Q, et al. Transfer of LncRNA CRNDE in TAM-derived exosomes is linked with cisplatin resistance in gastric cancer. EMBO Rep (2021) 22(12):e52124. doi: 10.15252/embr.202052124
125. Hu CE, Du PZ, Zhang HD, Huang GJ. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145. Cell Physiol Biochem (2017) 42(1):13–21. doi: 10.1159/000477107
126. Du DX, Lian DB, Amin BH, Yan W. Long non-coding RNA CRNDE is a novel tumor promoter by modulating PI3K/AKT signal pathways in human gastric cancer. Eur Rev Med Pharmacol Sci (2017) 21(23):5392–8. doi: 10.26355/eurrev_201712_13925
127. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res (2020) 39(1):246. doi: 10.1186/s13046-020-01758-w
128. Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer (2020) 19(1):156. doi: 10.1186/s12943-020-01270-x
129. Shi J, Surma M, Yang Y, Wei L. Disruption of both ROCK1 and ROCK2 genes in cardiomyocytes promotes autophagy and reduces cardiac fibrosis during aging. FASEB J (2019) 33(6):7348–62. doi: 10.1096/fj.201802510R
130. Sun L, Ma Y, Zhang Z, Li X, Chen Y, Liu G, et al. ROCK2 regulates autophagy in the hippocampus of rats after subarachnoid hemorrhage. Neuroreport (2018) 29(18):1571–7. doi: 10.1097/wnr.0000000000001154
Keywords: autophagy, gastric cancer, non-coding RNA, tumorigenesis, chemoresistance
Citation: Wang Z, Liu J, Xie J, Yuan X, Wang B, Shen W and Zhang Y (2022) Regulation of autophagy by non-coding RNAs in gastric cancer. Front. Oncol. 12:947332. doi: 10.3389/fonc.2022.947332
Received: 18 May 2022; Accepted: 18 July 2022;
Published: 24 October 2022.
Edited by:
Dunfa Peng, University of Miami, United StatesReviewed by:
Kannappan Sriramajayam, University of Miami Health System, United StatesZhangjian Zhou, Second Affiliated Hospital of Xi’an Jiaotong University, China
Copyright © 2022 Wang, Liu, Xie, Yuan, Wang, Shen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Shen, aGxqd2VuanVhbnNoZW5AMTYzLmNvbQ==; Yang Zhang, eWFuZ3poYW5nODNAMTYzLmNvbQ==
 Zijian Wang1
Zijian Wang1 Xingxing Yuan
Xingxing Yuan Bingyu Wang
Bingyu Wang Wenjuan Shen
Wenjuan Shen Yang Zhang
Yang Zhang