- 1Graduate School, Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 2Tianjin Rehabilitation Center, Tianjin, China
- 3Department of Pathology, Tianjin Union Medical Center, Tianjin, China
- 4Nankai University School of Medicine, Nankai University, Tianjin, China
- 5Department of Thyroid Surgery, Tianjin Union Medical Center, Tianjin, China
Wnt/β-catenin signaling is a delicate and complex signal transduction pathway mediated by multiple signaling molecules, which plays a significant role in regulating human physiology and pathology. Abnormally activated Wnt/β-catenin signaling pathway plays a crucial role in promoting malignant tumor occurrence, development, recurrence, and metastasis, particularly in cancer stem cells. Studies have shown that the Wnt/β-catenin signaling pathway controls cell fate and function through the transcriptional and post-translational regulation of omics networks. Therefore, precise regulation of Wnt/β-catenin signaling as a cancer-targeting strategy may contribute to the treatment of some malignancies. SUMOylation is a post-translational modification of proteins that has been found to play a major role in the Wnt/β-catenin signaling pathway. Here, we review the complex regulation of Wnt/β-catenin signaling by SUMOylation and discuss the potential targets of SUMOylation therapy.
Introduction
The wingless and Int-1 (Wnt)/β-catenin signaling pathway is a key cascade in embryonic development, self-renewal of organisms, cell proliferation, differentiation (1–3), and usually involves in various cancers, such as colorectal cancer (4), breast cancer (5), gastric cancer (6), ovarian cancer (7), pancreatic cancer (8), prostate cancer (9), leukemia (10), and melanoma (11). Abnormal activation of this pathway plays an essential role in chemoradiotherapy resistance (12). In addition, Wnt/β-catenin has emerged as a critical regulator of cancer stem cells (CSCs), which is considered one of the root causes of cancer recurrence and metastasis because of their heterogeneity and plasticity (13). Furthermore, the Wnt/β-catenin can crosstalk with other signaling pathways including Notch, FGF, Hedgehog, and TGF-β/BMP signaling cascades to form a signaling network to regulate the survival and progression of cancer cells (14–16). Post-translational modifications (PTMs) of proteins, including phosphorylation, acetylation, ubiquitination, and SUMOylation, can regulate the function of proteins, determine the active state and subcellular location of proteins, and dynamically interact with other proteins related to carcinogenesis and progression (17–20). Conceptually, it is an efficient way to treat cancer involved the hijacking of PTMs of the key molecules in Wnt/β-catenin. SUMOylation of proteins is an important mechanism in cellular responses to environmental stress (21, 22). Recent reports based on proteomic studies have identified many SUMOylated substrates that play important roles in the development and progression of cancer. Proteins associated with the Wnt/β-catenin pathway have been identified as SUMOylated substrates, and evidences suggested that the initiation and progression of cancers depended on the function of the SUMOylation (23). This suggests the possibility that strictly regulated self-renewal mediated by Wnt signaling in cancer cells may be disturbed by the SUMOylation pathway to allow more malignant proliferation. Hence, some studies have indicated that targeting the SUMOylation modification in Wnt/β-catenin might be a strategy for the treatment cancer (24).
Following the first member of the Wnt family identified over the last four decades (25), several recent studies have focused on the Wnt/β-catenin signaling pathway (26, 27). This has led to an improved understanding of the multilayered mechanisms of this pathway transduction proceeds as well as the molecular mechanism regulating this pathway (28). Signaling is initiated when the Wnt ligand binds to the Frizzled receptor on the cell membrane and the LDL receptor-associated protein 5/6 (LRP5/6) co-receptor. This receptor can induce phosphorylation of dishevelment (DVL) and recruitment of Axam to destroy the destructive complex (29). This destructive complex include adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK-3β), Axin, and casein kinase 1 (CK1), which promote ubiquitination-dependent degradation of phosphorylated β-catenin (30). Dephosphorylated β-catenin is then translocated to the nucleus where it initiates intracellular signal transduction. In the nucleus, β-catenin can bind to members of the transcription factor (T-cell factor/lymphoid enhancer factor TCF/LEF) family and recruit transcription coactivators and CREB binding protein (CBP) to the transcription of the Wnt target genes (31–33). β-catenin in the nucleus plays key regulatory roles in cancer cell target genes, including c-Myc (34), CD44 (35), Lgr5 (25, 36), and axis inhibition protein 2 (Axin2) (37). The C-terminal binding protein (CtBP) can bind to various transcription factors and play a bidirectional regulatory role for the transcription of Wnt target gene (38, 39).
SUMOylation of proteins is a process in which small ubiquitin-like modifier (SUMO) proteins bind to substrates covalently or non-covalently under the catalysis of SUMO proteases (40). SUMO proteins in mammalian cells present four SUMO isoforms: SUMO1, SUMO2/3, and SUMO4 (41). Although SUMO2 and SUMO3 cannot be distinguished by antibodies as they are very similar, SUMO1 is quite divergent. Furthermore, the expression levels of SUMO2/3 exceed SUMO1 in human cells (42). The SUMO4 precursor possess proline at amino acid 90, which seems to limit its processing to yield a mature form (43). Despite this difference, all SUMO proteins are activated and conjugated by the same SUMO proteases in an orderly manner. SUMO proteases are composed of three types of enzymes: SUMO-activating enzymes (E1), SUMO-conjugating enzymes (E2), and SUMO-specific ligases (E3) (44). E1s are composed of two subunits: SAE1/Aos1 and SAE2/Uba2 (45). There is only one E2, UBC9, among mammals (46). With the advancement of SUMOylation research, an increasing number of E3s have been identified, including the PIAS family (PIAS1, PIASx, PIAS3, and PIASy), RanBP2, and the human polycomb 2 (Pc2) (47, 48). The SUMO enzymes and proteins are predominantly enriched in the nucleus. However, they have also been detected in other subcellular locations (cytoplasm, Golgi apparatus, and plasma membrane) (49). Three distinct but not mutually exclusive mechanisms determine whether proteins can be SUMOylated (40). Firstly, substrates can directly interact with Ubc9 (SUMO consensus site-directed SUMOylation). Secondly, the substrate possesses the SUMO interaction motif (SIM) (SIM-dependent SUMOylation). Thirdly, proteins are recognized by a SUMO E3 ligase, which is accessible to Ubc9 (E3 ligase-dependent SUMOylation). Although all SUMO isoforms use the same enzymatic mechanism, the underlying mechanism determining their specificity remains unclear. SUMOylation of proteins is a multi-step enzymatic cascade reaction in which the first step of the SUMOylation modification is the maturation of SUMO proteins by SUMO-specific proteases (50). Mature SUMO proteins are then directly activated by SUMO E1 via ATP hydrolysis. Consequently, upon interaction of the charged E1 enzyme with E2, SUMO is transferred from E1 to E2, forming an E2-SUMO thioester. Finally, SUMO is linked to the lysine residues of target proteins via an isopeptide bond, with or without stimulation by the ligase enzyme (E3). SUMOylation of proteins provide a platform for enhancing protein-protein interactions (51, 52).
DeSUMOylation is the process of removing the SUMO from SUMOylated substrates (53, 54). The proteins that perform the function of removing SUMO are mainly two different enzymes: SUMO-specific proteases and SUMO-targeted ubiquitin ligases (STUbLs) (44). The main function of SUMO-specific proteases is to promote maturation of and activate the SUMO protein. DeSUMOylation can also remove SUMO protein from the substrate protein and inhibit SUMOylation of the substrates. The bidirectional action of the enzyme may depend on the output of the integrated signal in the environment in which the enzyme is located. With the discovery of sentrin-specific protease 1 (SENP1) (55), more SUMO-specific proteases have been discovered, including SENP1-SENP3 and SENP5-SENP7 (56). Subsequently, three novel SUMO-specific proteases have been identified in humans: desulphurized isopeptidase 1 (DeSI1), desulphurized isopeptidase 2 (DeSI2), and ubiquitin-specific protease-like 1(USPL1) (57, 58). Compared to the SENP protease, the three novel SUMO-specific proteases have a unique binding sequence pattern. In addition, STUbls, a family of functionally conserved enzymes, can negatively regulate SUMOylation levels. They catalyze the addition of ubiquitin to proteins previously modified by SUMOylation and promote protein degradation, illustrating that SUMO can indirectly promote protein degradation, such as RNF4 and Arkadia (RNF111) (59). The sub-cellular localization and activity of these enzymes in response to different stimuli are effective mechanisms to adjust cellular SUMOylation levels (60, 61).
SUMOylation and deSUMOylation remain in a dynamic equilibrium, allowing cells to respond rapidly to changing external and internal pressures and stimuli (44, 62). SUMOylation and deSUMOylation are involved in cellular processes such as metabolism, DNA repair, and signal transduction (40, 63). Dysregulation of SUMOylation and deSUMOylation can lead to severe defects in cell proliferation and genomic stability. In this review, we systematically describe the crosstalk between Wnt/β-catenin signaling and the SUMOylation modification and briefly emphasize its therapeutic potential for treating cancer. The detail regulatory mechanisms between SUMOylation and deSUMOylation are shown in Figure 1.
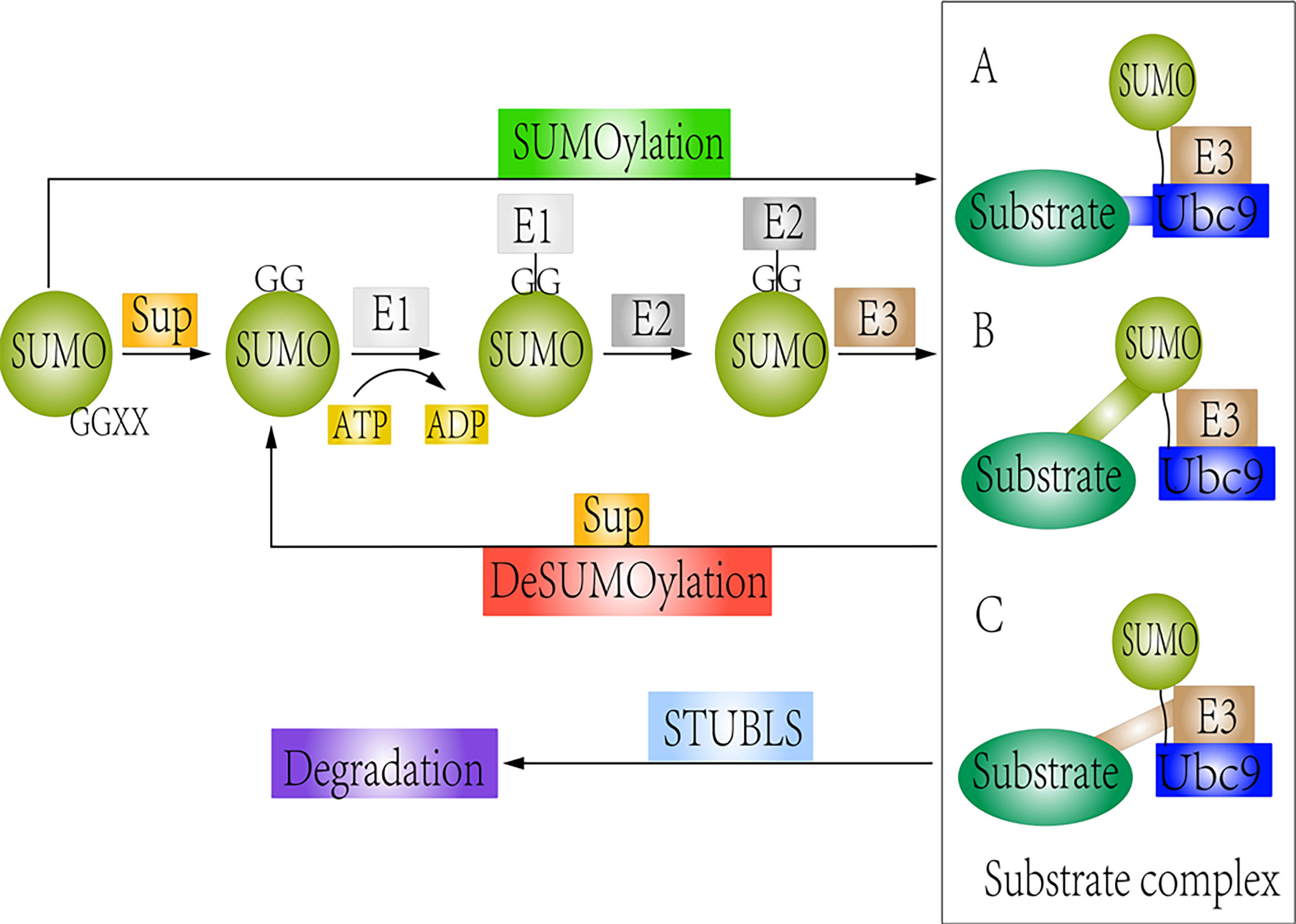
Figure 1 Schematic representation of SUMOylation and deSUMOylation. The first step is maturation of the SUMO protein. Then, mature SUMO is directly activated by SUMO E1 via ATP hydrolysis. Upon interaction of the charged E1 enzyme with E2, SUMO is transferred from E1 to E2. Finally, SUMO links to substrates by different forms: (A) Substrates directly interact with the Ubc9 (SUMO consensus site–directed SUMOylation); (B) Substrates possess the SUMO interaction motif (SIM) (SIM-dependent SUMOylation). (C) Proteins are recognized by a SUMO E3 ligase, which can be accessible to the charged Ubc9 (E3 ligase-dependent SUMOylation). However, SUMO-specific proteases could remove the SUMO protein from the substrate. STUbls promote degradation of SUMOylated substrate.
SUMOylation is Essential for Nuclear Import of β-Catenin
β-catenin is an evolutionarily conserved molecule that plays crucial roles in a variety of developmental and homeostatic processes (64). Aberrant activation of β-catenin is associated with cancer. More specifically, β-catenin is not only a structural component of the adhesion junctions of cadherins, but also a key nuclear effector of Wnt/β-catenin signaling (64). The Wnt/β-catenin pathway is ultimately centered on the PTM of β-catenin to maintain its abundance and regulate its subcellular localization (28). Accumulated β-catenin can translocate from the cytoplasm to the nucleus and participate in the transcription of multiple target genes. Nuclear translocation of β-catenin is regulated by multiple PTMs, including phosphorylation, acetylation, ubiquitination, and SUMOylation (65–68). However, several studies demonstrated that β-catenin had no recognizable nuclear localization signal (NLS) that could be transported to the nucleus by SUMOylation (67). To date, the mechanism of the interaction between β-catenin and the SUMOylation modification is not completely clear. All the studies mentioned above are based on interactions with other proteins, and there are no reports on whether β-catenin itself has SUMOylated modification sites to bind SUMO.
RanBP2 is part of the nuclear pore complex (NPC) and acts as a SUMO E3 enzyme, which helps macromolecular proteins that lack nuclear localization signals to enter the nucleus (69). It promotes the formation of the TCF-4-SUMO1-RanGAP1-Ubc9-RanBP2 complex by enhancing the SUMOylation of TCF-4 and the hydrolysis of RanGTP by RanGAP1, which binds to β-catenin and promotes its nuclear translocation (70). Another study demonstrated that total β-catenin and nuclear β-catenin expression were significantly decreased and phosphorylated β-catenin expression was increased after Uba2, an important component of the E1 activating enzyme, was knocked out (71). Briefly, β-catenin is able to enter the nucleus through SUMOylation of interacting proteins. However, the phosphorylation level of β-catenin in the cytoplasm increases when SUMOylation is inhibited, which promotes ubiquitination-dependent degradation of β-catenin. The SUMOylated modification appears to be the “ON/OFF” switch that regulates the nuclear import of β-catenin.
Jiang et al. demonstrated that the deSUMOylation activity of SENP2 was important for β-catenin stability. SENP2, a deSUMOylation enzyme, was downregulated in hepatocellular carcinoma (HCC) tissues (72). The study found that the stability of β-catenin was significantly reduced, whereas overexpression of SENP2 inhibited the growth and colony formation of HCC cells (72). Choi et al. found that SUMOylation excluded TBL1-TBLR1 from the nuclear hormone receptor inhibitor (NCoR) complex and increased recruitment of the TBL1-TBLR1/β-catenin complex to the Wnt target gene promoter, resulting in the expression of the Wnt signal target gene. In contrast, SENP1 reduces the formation of the TBL1-TBLR1/β-catenin complex, resulting in the inhibition of β-catenin-mediated transcription (73).
These studies suggest that the dynamic balance between SUMOylation and deSUMOylation can maintain the stability and transcriptional activity of β-catenin. The TCF-4-SUMO1-RanGAP1-Ubc9-RanBP2 complex can be used as a target to prevent the SUMO modification of β-catenin. β-catenin without SUMO modification cannot enter into the nuclear and play the role in the initiation and development of cancer.
SUMOylation is a Vital Regulator of Kinase Activity, Protein Stability, and Nuclear Localization of GSK-3β
GSK-3β is a ubiquitously expressed, broadly specific, serine/threonine kinase. Therefore, GSK-3β regulates a number of intracellular signaling pathways and is involved in a variety of biological events, including autophagy, cell apoptosis, and cancer progression (74–77). This is the axis of the classical Wnt signaling pathway (78). GSK-3β promotes β-catenin phosphorylation, leading to ubiquitin-mediated proteolysis. Active GSK-3β is considered a cancer suppressor as it promotes the destruction of several oncogenic proteins [e.g., β-catenin, c-Myc, and myeloid cell leukemia sequence 1 (MCL-1)] (79). Moreover, GSK-3β has also been found to exhibit carcinogenic properties, as it up-regulates pathways critical for cancer cell survival and drug resistance (80).
The SUMOylation of GSK-3β can occur in vitro or in vivo and is critical for GSK-3β function (81). GSK-3β has been identified as a potential SUMO substrate in proteomic studies, and the lysine residue K292 within the activation ring of GSK-3β has been identified as a SUMO receptor site. SUMOylation is a positive regulator of kinase activity, protein stability, and nuclear localization of GSK-3β. In addition, mutation of the SUMOylation sites in GSK-3β reduces its kinase activity and stability, significantly increasing cell survival (81). L. Eun Jeoung, H, et al. evaluated that the effects of GSK-3β SUMOylation on β-catenin stability and Wnt signaling and confirmed that GSK-3β SUMO mutant (K292R) increased the cell survival rate compared to the wild type GSK-3β (81). However, direct evidence is not sufficient to prove this. Wetzel et al. identified the lysine residue K730 in human ZO-2 as a potential SUMO-modification site directly bound to SUMO1 (82). SUMOylated ZO-2 binds directly to GSK-3β and promotes the kinase activity of GSK-3β. It is inhibited in the presence of SENP1, but not by inactivated SENP1 protein (82). SUMOylation of GSK-3β and upregulation of specific SUMO E3 enzyme of GSK-3β in the tumor microenvironment can inhibit the growth of malignant tumor, which may be used as a potential target for cancer treatment.
Activation of TCF/LEF Could be Regulated by SUMOylation
Constitutive activation of TCF/LEF is a key downstream effector of the Wnt/β-catenin pathway and is often observed in lung, breast, and colorectal cancers (83). The human TCF/LEF family consists of four homologous members: TCF-1, LEF-1, TCF-3, and TCF-4 (84). TCF-4 is found to be conjugated to SUMO at an endogenous level (70). The SUMOylation site of TCF-4 is Lys297. When Lys297 is mutated to arginine, activation of TCF-4 is inhibited. SUMOylation of TCF-4 can be amplified by PIASy, a SUMO E3 enzyme, and suppressed by Axam, a deSUMOylation enzyme (70). PIASy enhances TCF-4/β-catenin-dependent transcriptional activity, whereas Axam inhibits the SUMOylation of TCF-4. Conversely, lowering Axam protein levels by siRNA resulted in increased SUMOylation of TCF-4 and activation of TCF-4 (70). These results suggest that SUMOylation of TCF-4 is involved in β-catenin-dependent and TCF-4-mediated gene expression in the Wnt signaling. LEF1 is a transcription factor and can activate β-catenin-dependent transcription (85). S. Sachdev, L. et al. identified that LEF1 could be covalent modified by SUMO at Lys25 and Lys 267 (86). PIASy, as a novel interaction partner of LEF1, could significantly increase SUMOylation of LEF1. However, LEF1 activity is effectively inhibited after co-expression of PIASy with LEF1, suggesting that SUMOylated LEF1 inhibits the activation of Wnt/β-catenin pathway (86).
SUMOylation of Axin has No Effect on β-catenin Rather Than JNK
Axin is a multi-domain protein that interacts with several other proteins and acts as a negative regulator of Wnt signaling by down-regulating β-catenin levels (87, 88). Axin is found to interact with SUMO1 and three SUMO E3 ligase enzymes, PIAS1, PIASx-β, and PIASy (89). Six amino acids in the C-terminus of Axin interact with SUMO. The mutation of the six amino acid residues significantly reduces the expression of SUMOylated Axin (89). Both the mutation-type and wild-type Axin can decrease the transcriptional activity of LEF1 and destabilize β-catenin. SUMOylation of Axin has no effect on Wnt/β-catenin signaling. In contrast, removing any SUMO sites on Axin could reduce JNK activation (89). This indicates that SUMOylation of the same protein will have different effects on different interacting proteins, which depends on the subcellular location of the protein and the crosstalk between other signaling pathways. SUMOylation modification of Axin can effectively cause apoptosis via the JNK pathway rather than the Wnt/β-catenin pathway.
Axam and DeSUMOylation
Axam (also called SENP2) belongs to the ubiquitin-like protease 1 (Ulp1) cysteine protease family, which possesses hydrolase and isopeptidase activities of the SUMO-specific proteases (90). Axam usually localizes to the nucleoplasmic side of NPC by non-covalent binding with SUMO (91). A recent study showed that Axam was down-regulated in bladder cancer cells and HCC tissues (92). Additionally, Axam is identified as a novel Axin-binding protein that inhibits Wnt signaling by inhibiting the binding of DVL to Axin (93). Axam loses its deSUMOylation activity when its catalytic domain is mutated, which restores β-catenin activity. These results indicate that Axam acts as a deSUMOylation enzyme and is involved in negative regulation of the Wnt signaling pathway (94). Another study identified a new SUMO-specific protease called XSENP1 in Xenopus laevis that inhibited the expression of Wnt signal transduction target genes in a manner similar to that of Axam (95). Usually, Axam cannot be used as an independent therapeutic target for cancer treatment because of its non-specific deSUMOylation activity.
The Self-Acetylation and SUMOylation of CBP Compete With Each Other
CBP is a histone acetyl transferase and transcription cofactor that regulates the expression of genes involved in a wide range of cellular processes (96). CBP has attracted great interest as a promising new epigenetic target for a variety of diseases, including malignancies, since its discovery in 2006 (97). Studies showed that CBP could be covalently modified by SUMO1 in vitro and in vivo (98, 99). This covalent modification occurs at lysine residues 999, 1034, and 1057 of the CBP protein. CBP SUMOylation can be reversed by the overexpression of SENP2, a SUMO-specific protease (98). SUMO modification negatively regulates CBP transcriptional activity by recruiting the death domain-associated protein (Daxx) (98). Because CBP does not bind to DNA on its own, SUMO modification inhibits the intrinsic transactivation of CBP, and decreases the activity of Wnt signaling pathway. SUMO1 binding the ZZ domain in CBP is identified using nuclear magnetic resonance (NMR) spectroscopy (100). This SUMO site represents a unique SUMO interaction epitope that is spatially opposed to the epitope observed in a typical SIM. The presence of the ZZ domain in CBP enhances SUMOylation (100). Another study showed that the bromodomain (BRD) and plant homeodomain (PHD) domains of CBP, in addition to the ZZ domain, were the sites of interaction between SUMO1 and Ubc9, further suggesting that CBP might act as a SUMO E3 ligase enzyme to promote intramolecular SUMOylation of adjacent cycle regulatory domain 1 (CRD1) (99). The competition between self-acetylation and SUMOylation of lysine residues in CRD1 determines that CBP responds to various signals as a transcriptional activator or as a suppressor (99). The roles of the different modifications (self-acetylation and SUMOylation) of CBP in Wnt/β-catenin pathway need to clarify in the future.
SUMOylation of CtBP1 Profoundly Affects its Subcellular Localization and Transcriptional Activity
Mammalian CtBPs (CtBP1 and CtBP2) bind to various transcription factors and play a bidirectional regulatory role as a gene-specific activator and repressor of Wnt target gene transcription (39, 101). In earlier investigation, CtBPs played its role in transcriptional repression (102). However, CtBP can directly co-activate TCF4/LEF at key sites of the target genes to promote CSC self-renewal in CRCs (38). Lin et al. reported that SUMOylation of CtBP1 occurred at a single lysine residue (Lys428) of CtBP1, which was promoted by PIAS1 and PIASx-β. SUMOylation of CtBP1 substantially affected its subcellular localization and transcriptional activity (103). CtBP1 is transferred from the nucleus to the cytoplasm and loses its transcriptional activity after mutation at the SUMOylated Lys428 site (103, 104). Pc2 is a SUMO E3 ligase enzyme and significantly amplifies CtBP SUMOylation (105–107). Riefler et al. demonstrated an increase in nuclear accumulation of CtBP1 after binding to SUMO (108). The decreased SUMOylation of CtBP1 can result in the cytoplasmic retention of CtBP1 (108). These studies suggest that SUMOylation plays a critical role in nuclear retention of CtBP and positively regulating the activity of Wnt signaling pathway. Notably, CtBP2, a close homolog of CtBP1, lacks SUMOylation sites and is not modified by SUMO1. In addition, SUMOylation of friend of GATA1 could promote the interaction with members of the CtBP family, particularly CtBP1 (109).
SUMOylation of Smad3/4
Wnt/β-catenin and Smad3/4 can crosstalk with each other (110). In the cytoplasm, Smad3 directly promotes the nuclear translocation of β-catenin (111), which regulates the expression of different target genes. Several studies have shown that inactivation of Smad4 can promote self-renewal of CSCs (112). As a cancer suppressor, Smad4 is involved in Wnt-Kras-mediated inhibition of colorectal cancer. The loss of Smad4 and activation of Wnt signaling can create conditions favorable to the fate of stem cells and enable them to re-enter the cell cycle (113). Freeman et al. demonstrated that reduced levels of Smad4 correlated with increased levels of β-catenin mRNA (114). Smad3/4 is a target of PTMs, such as phosphorylation, ubiquitination, SUMOylation, and ADP ribosylation. However, the effect of PTM of Smad3/4 on Wnt signaling, particularly SUMOylation, has been poorly reported.
Recent data indicated that SUMO1 could bind to the C-terminal domain of Smad3, which was increased in the presence of PIASy, resulting in its translocation to the cytoplasm in vivo and in vitro (115). The cytoplasmic Smad3 can promote the nuclear translocation of β-catenin and activate Wnt signaling pathway. The co-expression of PIASy and SUMO1 not only affects the subcellular localization of Smad3 but also affects the binding activity of Smad3 to DNA. PIASy can regulate TGF-β/Smad3-mediated signaling by stimulating SUMOylation and nuclear output of Smad3 (116). SUMO1 and SUMO2/3 could bind to Smad4 in vitro and in vivo (117, 118). SUMOylation affects the stability, transcriptional activity, and subcellular localization of Smad4 (119). The major SUMOylation site in Smad4 is located in its linker fragment at Lys159, with an additional site at Lys113 in the MH-1 domain. Double mutations at these two sites enhance Smad4 stability (120). SUMOylation of Smad4 is amplified by PIAS1, PIASx-α, PIASx-β, and PIASy (118). Overexpression of SUMO1 and Ubc9 inhibits TGF-β reactivity reporter genes, whereas co-transfection with SUMO1 protease-1 (SuPr-1) increases TGF-β recovery (117). In addition, direct fusion of Smad4 with mutant sites (K113R and K159R) and SUMO1 effectively inhibits its transcriptional activity (117). Lee et al. reported that Smad4 could be modified by SUMO2/3 (but not SUMO1) to further activate TGF-β/Smad signaling in mesangial cells under high glucose conditions (121). Liu et al. demonstrated that ginkgolic acid (GA), a SUMO1 inhibitor, could inhibit carcinogenesis and cancer progression in oral squamous cell carcinoma by inhibiting Smad4 SUMOylation in a time- and concentration-dependent manner (122). The transcriptional activity of SUMOylationed Smad4 can be stimulated or inhibited in different cells at different niches, which may lead to different transcriptional effects.
CD44 and SUMOylation
CD44 is a member of the transmembrane glycoprotein family (123), and involves in several physiological and pathological processes, including hematopoiesis, inflammation, and cancer (124, 125). CD44, as a CSC marker, is a key target gene of the Wnt signaling pathway (35, 126), and can positively regulate Wnt signaling by regulating the localization and activation of LRP6 (127). Although few studies have focused on the PTM of CD44, it has been confirmed that CD44 was regulated by SUMOylation (128). This is supported by the fact that the expression of MMP14 and CD44 is reduced and the migration of cells in basal breast cancer is reduced by knockdown of Ubc9 and PIAS1, which suggests that SUMOylation has a significant effect on the stability of CD44 (128, 129). At the same time, small molecule inhibitors of SUMO in primary colorectal cancers could inhibit the mRNA levels and protein expression of CD44, and the former decreased more significantly in vitro (130). However, Huang, X et al. proved that deficiency of Ubc9 could lead to an increase in CD44+ stem cells, which promoted the proliferation and migration of bladder cancer cells (131). These seemingly contradictory outcomes indicate that different mechanisms of SUMOylation regulating CD44 expression exist in different cell tumor microenvironments. A clearer insight of the molecular environment and the different cellular components that elicit differential results of CD44 will be essential to understand the effects of the SUMOylation modification on renewal in CSCs. A schematic representation of the crosstalk between SUMOylation and Wnt/β-catenin pathway is shown in Figure 2.
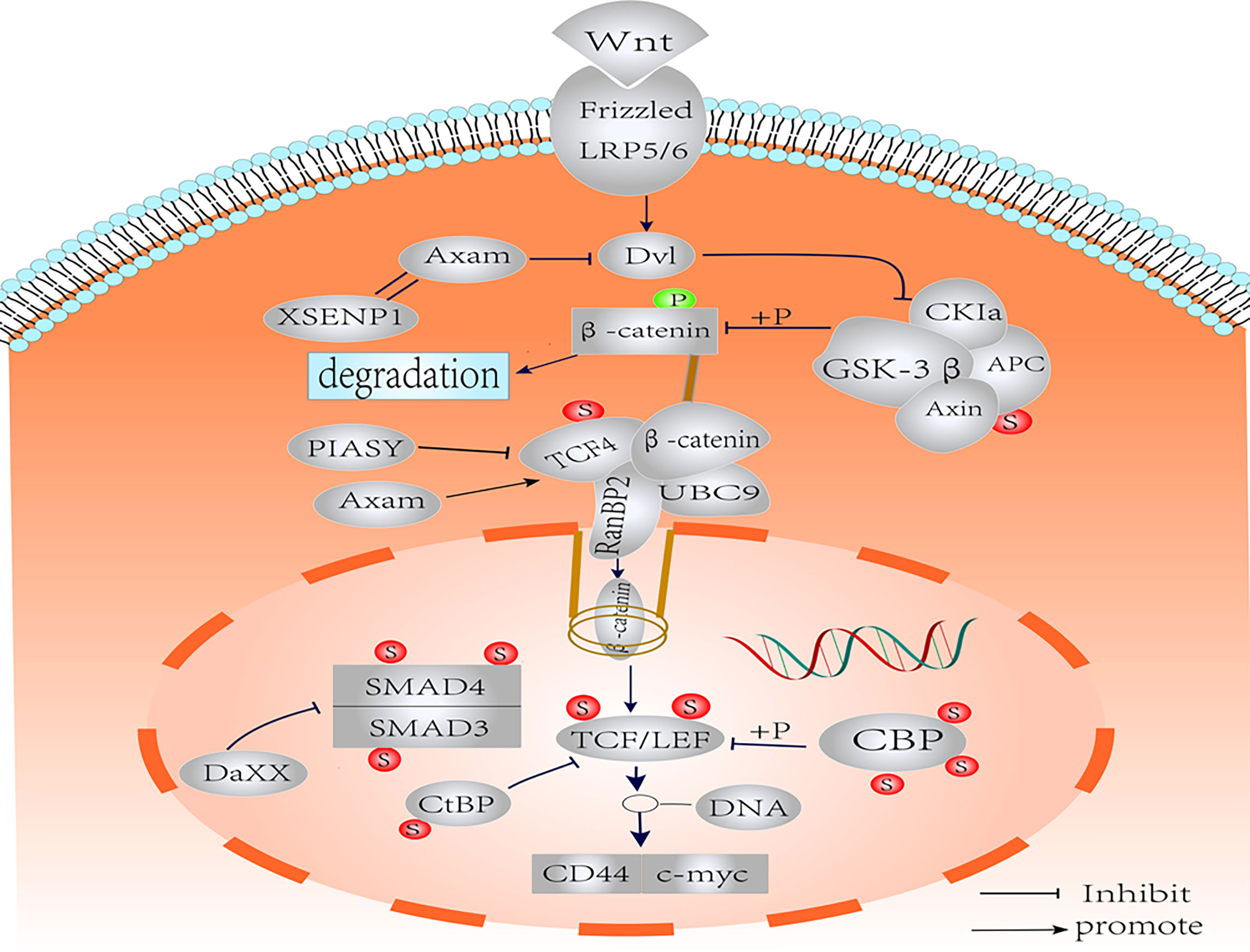
Figure 2 Schematic representation of the SUMOylation on the components of Wnt/β-catenin pathways. Nuclear translocation of β-catenin is regulated by SUMO. The SUMOylation of TCF-4 increases the activity of TCF-4. SUMOylated TCF promotes the binding of β-catenin to RanBP2 of the nuclear pore complex into the nucleus. SUMOylation of GSK-3β improves kinase activity and protein stability. The SUMOylation of LEF1 and its co-expression with PIASy resulted in effective inhibition of LEF1 activity. The SUMOylation of Axin had no effect on Wnt signaling. Axam acts as deSUMOylated enzyme to downregulate β-catenin and implicates the negative regulation of Wnt signaling pathway. SUMO modification negatively regulates CBP transcriptional activity by recruiting Daxx. The CBP may act as the SUMO E3 ligase enzyme, which promotes intramolecular SUMOylation of adjacent CRD1 domains. Binding of CtBP1 to SUMO increases nuclear accumulation. Smad3 SUMOylation is enhanced in the presence of PIASy, resulting in translocation to the cytoplasm. Under the influence of PIASy, PIAS1, PIASxα, and PIASxβ, SUMOylation reduces the stability and transcriptional activity of Smad4, but the effect on Wnt/β-catenin is not clear.
Wnt/β-Catenin Signaling Regulates SUMOylation
Several studies have reported that Wnt/β-catenin pathway could regulate the SUMOylation of other proteins (132, 133). More recently, Satow et al. showed that Wnt proteins regulated SUMOylation of ZIC5 (132). Wnt activity could promote the binding of SUMO to lysine residues in the highly conserved (ZF-NC) domain of ZIC5, which was critical for neural ridge development in mice. Interestingly, after treating the HEK293T cells with the GSK-3β inhibitor LiCl, the proportion of SUMOylated ZIC5 increased, whereas the ability of ZIC5 to inhibit TCF-dependent transcription decreased in a time-dependent manner (132). This study reveals the importance of SUMOylation of the ZF-NC domain of ZIC5 and the activity of Wnt in neural crest development in mice. It has also observed that targeting global SUMOylation or Wnt may have potential side effects, and a more precise location and time of intervention of SUMOylation need to studied in detail. Furthermore, another study showed that β-catenin interacted with promyelocytic leukemia protein (PML) transcript variant IV and disrupted PML nuclear body (NB) formation by inhibiting SUMOylation of PML transcript variant IV mediated by RanBP2 (133). Furthermore, Picard et al. reported that a striking decrease of SUMOylated estrogen receptor (ER) β appeared after the inhibition of GSK-3β expression. The SUMOylation modification of ERβ regulated by GSK-3β was associated with the stabilization and transcriptional activity of ERβ (134). ERβ could interfere with ERα-mediated oncogenic proliferation of breast cancer cell (135).
Conclusion
Genome-wide mapping techniques and proteomics have further promoted the study of dynamic changes in Wnt signaling networks and transcription complexes. These SUMO families have been found to effectively integrate into the Wnt signaling pathway, playing a key role in cancer self-renewal. Just as normal crosstalk between Wnt signaling and the SUMOylation modification may promote self-renewal of stem cells and organization, aberrant crosstalk may be tumorigenic in different systems. Finally, the influence of the SUMOylation modification on Wnt signaling appears to extend to CSCs, the most primitive cancer cells. As Wnt/β-catenin signaling seems to be required for stem cells and cancer cells, the ability of the SUMOylation modification to modulate Wnt signaling either positively or negatively may be of therapeutic relevance. Thus, understanding the interference of the SUMOylation modification on Wnt signaling may allow us to better control cancer cells when the treatment effect is unsatisfactory. SUMOylation on the key proteins involved in Wnt/β-catenin signaling pathway and their corresponding effects on the activity of Wnt/β-catenin signaling pathway have been showed in Table 1. Targeting SUMOylation modification of proteins has been proposed for the treatment of cancer. Several researchers have proposed improving the efficiency of cancer therapy by modifying targeted cancer pathways using SUMO, including small-molecule inhibitors. GA, a small-molecule SUMO inhibitor, could inhibit the growth of basal breast cancer xenografts in mice (128). Liu et al. demonstrated that GA could inhibit carcinogenesis and cancer progression in oral squamous cell carcinoma by inhibiting Smad4 SUMOylation in a time- and concentration-dependent manner (122). However, SUMOylation modification is also subject to a series of precise regulations, and has different binding forms with target proteins. In addition, tumor microenvironment can also regulate the SUMOylation modification to exert different functions. At present, most of the studies about SUMOylation modification were finished in vitro and in vivo, cell and animal experiments were difficult to simulate the microenvironment of human malignant tumors. Furthermore, while we focused on the interaction between SUMO and different Wnt members, the effects of SUMOylation modification on proteins other than Wnt signaling pathway cannot be ignored.
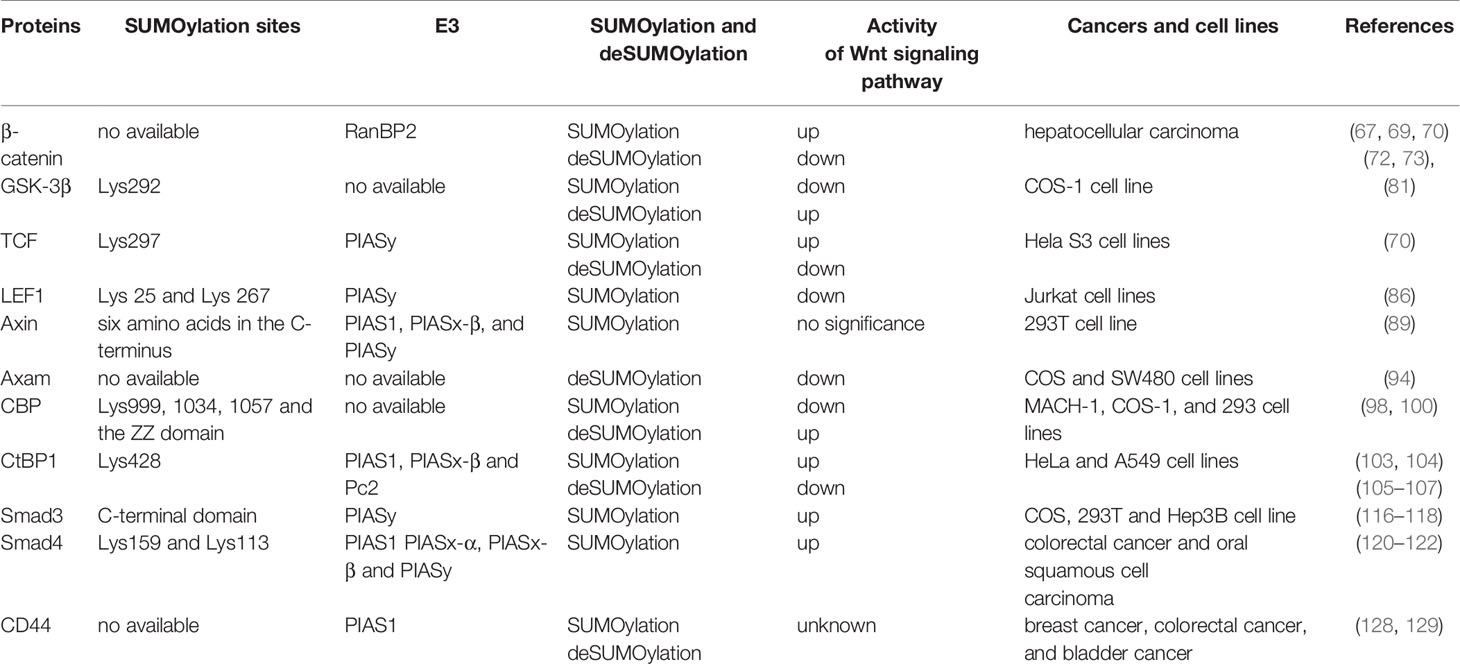
Table 1 SUMOylation on the key proteins involved in Wnt/β-catenin signaling pathway and their corresponding effects on the activity of Wnt/β-catenin signaling pathway.
Wnt/β-catenin pathway components undergo SUMOylation at different reaction levels. Owing to constantly evolving technology, the stage in which the SUMOylation modification may influence Wnt signaling in cancer cells may extend beyond the examples illustrated above. This pathway regulation is further complicated by the fact that the SUMO family is involved in many component modifications and is subject to crosstalk with other PTMs. SUMOylation of the same protein may result in opposite regulation of different downstream effector proteins. The same protein in cancer cells signals multiple dynamic changes in response to internal and external environmental changes in different cancer niches. The challenge for the future is that small-molecule inhibitors of SUMO in different stages of Wnt/β-catenin should be more targeted and spatio-temporally specific to achieve a precise impact on cancer stem cells under macro conditions.
Author Contributions
SZ and MG designed the paper; contributed to manuscript writing; and approved the manuscript before submission. LF, XuY and MZ contributed to manuscript writing; and approved the manuscript before submission. XiY and YN and LF collected literatures and approved the manuscript before submission. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants from the National Science Foundation of China (#82173283 and #82103088), and Foundation of committee on science and technology of Tianjin (#21JCZDJC00230, #2020036, #20JCYBJC01230). The funders had no roles in the design of the study, data collection, analysis and interpretation, or decision to write and publish the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge Editage service for the manuscript language edit.
Abbreviations
PTM, post-translational modification; CSCs, cancer stem cells; LRP5/6,LDL receptor-associated protein 5/6; DVL, dishevelment; APC, adenomatous polyposis coli; GSK-3β, glycogen synthase kinase 3β; TCF,T-cell factor; LEF, lymphoid enhancer factor; SUMO, Small Ubiquitin-like Modifier; CBP, CREB binding protein; CtBP, C-terminal binding protein; SIM, SUMO interaction motif; Ubc9,ubiquitin conjugating enzyme; StUbls, SUMO-targeted ubiquitin ligase; SENP1,sentrin-specific protease 1; DeSI1, desulphurized isopeptidase 1; USPL1, ubiquitin-specific protease-like 1; NPC, the nuclear pore complex; GA, ginkgolic acid.
References
1. Soleas JP, D'Arcangelo E, Huang L, Karoubi G, Nostro MC, McGuigan AP, et al. Assembly of Lung Progenitors Into Developmentally-Inspired Geometry Drives Differentiation via Cellular Tension. Biomaterials (2020) 254:120128. doi: 10.1016/j.biomaterials.2020.120128
2. Salik B, Yi H, Hassan N, Santiappillai N, Vick B, Connerty P, et al. Targeting RSPO3-LGR4 Signaling for Leukemia Stem Cell Eradication in Acute Myeloid Leukemia. Cancer Cell (2020) 38:263–78.e6. doi: 10.1016/j.ccell.2020.05.014
3. Choi BR, Cave C, Na CH, Sockanathan S. GDE2-Dependent Activation of Canonical Wnt Signaling in Neurons Regulates Oligodendrocyte Maturation. Cell Rep (2020) 31:107540. doi: 10.1016/j.celrep.2020.107540
4. Tang Q, Chen J, Di Z, Yuan W, Zhou Z, Liu Z, et al. TM4SF1 Promotes EMT and Cancer Stemness via the Wnt/β-Catenin/SOX2 Pathway in Colorectal Cancer. J Exp Clin Cancer Res (2020) 39:232. doi: 10.1186/s13046-020-01690-z
5. Xu X, Zhang M, Xu F, Jiang S. Wnt Signaling in Breast Cancer: Biological Mechanisms, Challenges and Opportunities. Mol Cancer (2020) 19:165. doi: 10.1186/s12943-020-01276-5
6. Tian S, Peng P, Li J, Deng H, Zhan N, Zeng Z, et al. SERPINH1 Regulates EMT and Gastric Cancer Metastasis via the Wnt/β-Catenin Signaling Pathway. Aging (Albany NY) (2020) 12:3574–93. doi: 10.18632/aging.102831
7. Wang Y, Zhao G, Condello S, Huang H, Cardenas H, Tanner EJ, et al. Frizzled-7 Identifies Platinum-Tolerant Ovarian Cancer Cells Susceptible to Ferroptosis. Cancer Res (2021) 81:384–99. doi: 10.1158/0008-5472.CAN-20-1488
8. Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X, et al. LncRNA PVT1 Promotes Gemcitabine Resistance of Pancreatic Cancer via Activating Wnt/β-Catenin and Autophagy Pathway Through Modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 Axes. Mol Cancer (2020) 19:118. doi: 10.1186/s12943-020-01237-y
9. Murillo-Garzón V, Kypta R. WNT Signalling in Prostate Cancer. Nat Rev Urol (2017) 14:683–96. doi: 10.1038/nrurol.2017.144
10. Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-Catenin Pathway is Required for the Development of Leukemia Stem Cells in AML. Science (2010) 327:1650–3. doi: 10.1126/science.1186624
11. Spranger S, Bao R, Gajewski TF. Melanoma-Intrinsic β-Catenin Signalling Prevents Anti-Tumour Immunity. Nature (2015) 523:231–5. doi: 10.1038/nature14404
12. Bugter JM, Fenderico N, Maurice MM. Mutations and Mechanisms of WNT Pathway Tumour Suppressors in Cancer. Nat Rev Cancer (2021) 21:5–21. doi: 10.1038/s41568-020-00307-z
13. Tayama S, Motohara T, Narantuya D, Li C, Fujimoto K, Sakaguchi I, et al. The Impact of EpCAM Expression on Response to Chemotherapy and Clinical Outcomes in Patients With Epithelial Ovarian Cancer. Oncotarget (2017) 8:44312–25. doi: 10.18632/oncotarget.17871
14. Katoh M, Katoh M. WNT Signaling Pathway and Stem Cell Signaling Network. Clin Cancer Res (2007) 13:4042–5. doi: 10.1158/1078-0432.CCR-06-2316
15. Rheinbay E, Suvà ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, et al. An Aberrant Transcription Factor Network Essential for Wnt Signaling and Stem Cell Maintenance in Glioblastoma. Cell Rep (2013) 3:1567–79. doi: 10.1016/j.celrep.2013.04.021
16. Kandyba E, Leung Y, Chen YB, Widelitz R, Chuong CM, Kobielak K. Competitive Balance of Intrabulge BMP/Wnt Signaling Reveals a Robust Gene Network Ruling Stem Cell Homeostasis and Cyclic Activation. Proc Natl Acad Sci U S A (2013) 110:1351–6. doi: 10.1073/pnas.1121312110
17. Cui Y, Ma W, Lei F, Li Q, Su Y, Lin X, et al. Prostate Tumour Overexpressed-1 Promotes Tumourigenicity in Human Breast Cancer via Activation of Wnt/β-Catenin Signalling. J Pathol (2016) 239:297–308. doi: 10.1002/path.4725
18. Lettini G, Sisinni L, Condelli V, Matassa DS, Simeon V, Maddalena F, et al. TRAP1 Regulates Stemness Through Wnt/β-Catenin Pathway in Human Colorectal Carcinoma. Cell Death Differ (2016) 23:1792–803. doi: 10.1038/cdd.2016.67
19. Li H, Zhang W, Yan M, Qiu J, Chen J, Sun X, et al. Nucleolar and Spindle Associated Protein 1 Promotes Metastasis of Cervical Carcinoma Cells by Activating Wnt/β-Catenin Signaling. J Exp Clin Cancer Res (2019) 38:33. doi: 10.1186/s13046-019-1037-y
20. Liang CJ, Wang ZW, Chang YW, Lee KC, Lin WH, Lee JL. SFRPs Are Biphasic Modulators of Wnt-Signaling-Elicited Cancer Stem Cell Properties Beyond Extracellular Control. Cell Rep (2019) 28:1511–25.e5. doi: 10.1016/j.celrep.2019.07.023
21. Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 Modified by SUMO-2/3 Promotes Gastric Cancer Progression and Regulates mRNA M5c Methylation. Cell Death Dis (2021) 12:842. doi: 10.1038/s41419-021-04127-3
22. Seeler JS, Dejean A. SUMO and the Robustness of Cancer. Nat Rev Cancer (2017) 17:184–97. doi: 10.1038/nrc.2016.143
23. Du L, Li YJ, Fakih M, Wiatrek RL, Duldulao M, Chen Z, et al. Role of SUMO Activating Enzyme in Cancer Stem Cell Maintenance and Self-Renewal. Nat Commun (2016) 7:12326. doi: 10.1038/ncomms12326
24. Mojsa B, Tatham MH, Davidson L, Liczmanska M, Branigan E, Hay RT. Identification of SUMO Targets Associated With the Pluripotent State in Human Stem Cells. Mol Cell Proteomics (2021) 20:100164. doi: 10.1016/j.mcpro.2021.100164
25. Nusse R, Varmus HE. Many Tumors Induced by the Mouse Mammary Tumor Virus Contain a Provirus Integrated in the Same Region of the Host Genome. Cell (1982) 31:99–109. doi: 10.1016/0092-8674(82)90409-3
26. Duchartre Y, Kim YM, Kahn M. The Wnt Signaling Pathway in Cancer. Crit Rev Oncol Hematol (2016) 99:141–9. doi: 10.1016/j.critrevonc.2015.12.005
27. Polakis P. Wnt Signaling in Cancer. Cold Spring Harb Perspect Biol (2012) 4: a008052. doi: 10.1101/cshperspect.a008052
28. Steinhart Z, Angers S. Wnt Signaling in Development and Tissue Homeostasis. Development (2018) 145:146589. doi: 10.1242/dev.146589
29. Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, Toyoshima K, et al. Axin, an Inhibitor of the Wnt Signalling Pathway, Interacts With Beta-Catenin, GSK-3beta and APC and Reduces the Beta-Catenin Level. Genes Cells (1998) 3:395–403. doi: 10.1046/j.1365-2443.1998.00198.x
30. Kimelman D, Xu W. Beta-Catenin Destruction Complex: Insights and Questions From a Structural Perspective. Oncogene (2006) 25:7482–91. doi: 10.1038/sj.onc.1210055
31. Mosimann C, Hausmann G, Basler K. Beta-Catenin Hits Chromatin: Regulation of Wnt Target Gene Activation. Nat Rev Mol Cell Biol (2009) 10:276–86. doi: 10.1038/nrm2654
32. Teo JL, Kahn M. The Wnt Signaling Pathway in Cellular Proliferation and Differentiation: A Tale of Two Coactivators. Adv Drug Delivery Rev (2010) 62:1149–55. doi: 10.1016/j.addr.2010.09.012
33. Veeman MT, Axelrod JD, Moon RT. A Second Canon. Functions and Mechanisms of Beta-Catenin-Independent Wnt Signaling. Dev Cell (2003) 5:367–77. doi: 10.1016/S1534-5807(03)00266-1
34. Madan B, Harmston N, Nallan G, Montoya A, Faull P, Petretto E, et al. Temporal Dynamics of Wnt-Dependent Transcriptome Reveal an Oncogenic Wnt/MYC/ribosome Axis. J Clin Invest (2018) 128:5620–33. doi: 10.1172/JCI122383
35. Wei CY, Zhu MX, Yang YW, Zhang PF, Yang X, Peng R, et al. Downregulation of RNF128 Activates Wnt/β-Catenin Signaling to Induce Cellular EMT and Stemness via CD44 and CTTN Ubiquitination in Melanoma. J Hematol Oncol (2019) 12:21. doi: 10.1186/s13045-019-0711-z
36. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of Stem Cells in Small Intestine and Colon by Marker Gene Lgr5. Nature (2007) 449:1003–7. doi: 10.1038/nature06196
37. Valenta T, Lukas J, Korinek V. HMG Box Transcription Factor TCF-4's Interaction With CtBP1 Controls the Expression of the Wnt Target Axin2/Conductin in Human Embryonic Kidney Cells. Nucleic Acids Res (2003) 31:2369–80. doi: 10.1093/nar/gkg346
38. Patel J, Baranwal S, Love IM, Patel NJ, Grossman SR, Patel BB. Inhibition of C-Terminal Binding Protein Attenuates Transcription Factor 4 Signaling to Selectively Target Colon Cancer Stem Cells. Cell Cycle (2014) 13:3506–18. doi: 10.4161/15384101.2014.958407
39. Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-Terminal-Binding Protein Directly Activates and Represses Wnt Transcriptional Targets in Drosophila. EMBO J (2006) 25:2735–45. doi: 10.1038/sj.emboj.7601153
40. Flotho A, Melchior F. Sumoylation: A Regulatory Protein Modification in Health and Disease. Annu Rev Biochem (2013) 82:357–85. doi: 10.1146/annurev-biochem-061909-093311
41. Geiss-Friedlander R, Melchior F. Concepts in Sumoylation: A Decade on. Nat Rev Mol Cell Biol (2007) 8:947–56. doi: 10.1038/nrm2293
42. Saitoh H, Hinchey J. Functional Heterogeneity of Small Ubiquitin-Related Protein Modifiers SUMO-1 Versus SUMO-2/3. J Biol Chem (2000) 275:6252–8. doi: 10.1074/jbc.275.9.6252
43. Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic Interplay of the SUMO and ERK Pathways in Regulating Elk-1 Transcriptional Activity. Mol Cell (2003) 12:63–74. doi: 10.1016/S1097-2765(03)00265-X
44. Chang HM, Yeh ETH. SUMO: From Bench to Bedside. Physiol Rev (2020) 100:1599–619. doi: 10.1152/physrev.00025.2019
45. Kunadis E, Lakiotaki E, Korkolopoulou P, Piperi C. Targeting Post-Translational Histone Modifying Enzymes in Glioblastoma. Pharmacol Ther (2021) 220:107721. doi: 10.1016/j.pharmthera.2020.107721
46. Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, et al. Ubc9 Sumoylation Regulates SUMO Target Discrimination. Mol Cell (2008) 31:371–82. doi: 10.1016/j.molcel.2008.05.022
47. Lao M, Shi M, Zou Y, Huang M, Ye Y, Qiu Q, et al. Protein Inhibitor of Activated STAT3 Regulates Migration, Invasion, and Activation of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis. J Immunol (2016) 196:596–606. doi: 10.4049/jimmunol.1403254
48. Wotton D, Merrill JC. Pc2 and SUMOylation. Biochem Soc Trans (2007) 35:1401–4. doi: 10.1042/BST0351401
49. Zhao X. SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes. Mol Cell (2018) 71:409–18. doi: 10.1016/j.molcel.2018.07.027
50. Johnson ES. Protein Modification by SUMO. Annu Rev Biochem (2004) 73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118
51. Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-Binding Motif That Recognizes SUMO-Modified Proteins. Proc Natl Acad Sci U S A (2004) 101:14373–8. doi: 10.1073/pnas.0403498101
52. Song J, Zhang Z, Hu W, Chen Y. Small Ubiquitin-Like Modifier (SUMO) Recognition of a SUMO Binding Motif: A Reversal of the Bound Orientation. J Biol Chem (2005) 280:40122–9. doi: 10.1074/jbc.M507059200
53. Barysch SV, Stankovic-Valentin N, Miedema T, Karaca S, Doppel J, Nait Achour T, et al. Transient Desumoylation of IRF2BP Proteins Controls Early Transcription in EGFR Signaling. EMBO Rep (2021) 22:e49651. doi: 10.15252/embr.201949651
54. Cui CP, Wong CC, Kai AK, Ho DW, Lau EY, Tsui YM, et al. SENP1 Promotes Hypoxia-Induced Cancer Stemness by HIF-1α Desumoylation and SENP1/HIF-1α Positive Feedback Loop. Gut (2017) 66:2149–59. doi: 10.1136/gutjnl-2016-313264
55. Gong L, Millas S, Maul GG, Yeh ET. Differential Regulation of Sentrinized Proteins by a Novel Sentrin-Specific Protease. J Biol Chem (2000) 275:3355–9. doi: 10.1074/jbc.275.5.3355
56. Yeh ET, Gong L, Kamitani T. Ubiquitin-Like Proteins: New Wines in New Bottles. Gene (2000) 248:1–14. doi: 10.1016/S0378-1119(00)00139-6
57. Schulz S, Chachami G, Kozaczkiewicz L, Winter U, Stankovic-Valentin N, Haas P, et al. Ubiquitin-Specific Protease-Like 1 (USPL1) is a SUMO Isopeptidase With Essential, non-Catalytic Functions. EMBO Rep (2012) 13:930–8. doi: 10.1038/embor.2012.125
58. Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, et al. DeSUMOylating Isopeptidase: A Second Class of SUMO Protease. EMBO Rep (2012) 13:339–46. doi: 10.1038/embor.2012.3
59. Sriramachandran AM, Meyer-Teschendorf K, Pabst S, Ulrich HD, Gehring NH, Hofmann K, et al. Arkadia/RNF111 is a SUMO-Targeted Ubiquitin Ligase With Preference for Substrates Marked With SUMO1-Capped SUMO2/3 Chain. Nat Commun (2019) 10:3678. doi: 10.1038/s41467-019-11549-3
60. Su YF, Yang T, Huang H, Liu LF, Hwang J. Phosphorylation of Ubc9 by Cdk1 Enhances SUMOylation Activity. PLoS One (2012) 7:e34250. doi: 10.1371/journal.pone.0034250
61. Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, et al. The Nuclear Pore Regulates GAL1 Gene Transcription by Controlling the Localization of the SUMO Protease Ulp1. Mol Cell (2013) 51:807–18. doi: 10.1016/j.molcel.2013.08.047
62. Wang Y, Dasso M. SUMOylation and Desumoylation at a Glance. J Cell Sci (2009) 122:4249–52. doi: 10.1242/jcs.050542
63. Garvin AJ, Morris JR. SUMO, a Small, But Powerful, Regulator of Double-Strand Break Repair. Philos Trans R Soc Lond B Biol Sci (2017) 372: 20160281. doi: 10.1098/rstb.2016.0281
64. Valenta T, Hausmann G, Basler K. The Many Faces and Functions of β-Catenin. EMBO J (2012) 31:2714–36. doi: 10.1038/emboj.2012.150
65. Ge X, Jin Q, Zhang F, Yan T, Zhai Q. PCAF Acetylates {Beta}-Catenin and Improves its Stability. Mol Biol Cell (2009) 20:419–27. doi: 10.1091/mbc.e08-08-0792
66. Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. The EDD E3 Ubiquitin Ligase Ubiquitinates and Up-Regulates Beta-Catenin. Mol Biol Cell (2011) 22:399–411. doi: 10.1091/mbc.e10-05-0440
67. Shitashige M, Satow R, Honda K, Ono M, Hirohashi S, Yamada T. Regulation of Wnt Signaling by the Nuclear Pore Complex. Gastroenterology (2008) 134:1961–71. doi: 10.1053/j.gastro.2008.03.010
68. Stamos JL, Weis WI. The β-Catenin Destruction Complex. Cold Spring Harb Perspect Biol (2013) 5:a007898. doi: 10.1101/cshperspect.a007898
69. He Y, Yang Z, Zhao CS, Xiao Z, Gong Y, Li YY, et al. T-Cell Receptor (TCR) Signaling Promotes the Assembly of RanBP2/RanGAP1-SUMO1/Ubc9 Nuclear Pore Subcomplex via PKC-θ-Mediated Phosphorylation of Rangap1. eLife (2021) 10:e67123. doi: 10.7554/eLife.67123.sa2
70. Yamamoto H, Ihara M, Matsuura Y, Kikuchi A. Sumoylation is Involved in Beta-Catenin-Dependent Activation of Tcf-4. EMBO J (2003) 22:2047–59. doi: 10.1093/emboj/cdg204
71. Li J, Sun X, He P, Liu WQ, Zou YB, Wang Q, et al. Ubiquitin-Like Modifier Activating Enzyme 2 Promotes Cell Migration and Invasion Through Wnt/β-Catenin Signaling in Gastric Cancer. World J Gastroenterol (2018) 24:4773–86. doi: 10.3748/wjg.v24.i42.4773
72. Jiang QF, Tian YW, Shen Q, Xue HZ, Li K. SENP2 Regulated the Stability of β-Catenin Through WWOX in Hepatocellular Carcinoma Cell. Tumour Biol (2014) 35:9677–82. doi: 10.1007/s13277-014-2239-8
73. Choi HK, Choi KC, Yoo JY, Song M, Ko SJ, Kim CH, et al. Reversible SUMOylation of TBL1-TBLR1 Regulates β-Catenin-Mediated Wnt Signaling. Mol Cell (2011) 43:203–16. doi: 10.1016/j.molcel.2011.05.027
74. Cadigan KM, Liu YI. Wnt Signaling: Complexity at the Surface. J Cell Sci (2006) 119:395–402. doi: 10.1242/jcs.02826
75. Doble BW, Woodgett JR. GSK-3: Tricks of the Trade for a Multi-Tasking Kinase. J Cell Sci (2003) 116:1175–86. doi: 10.1242/jcs.00384
76. Kikuchi A, Yamamoto H, Kishida S. Multiplicity of the Interactions of Wnt Proteins and Their Receptors. Cell Signal (2007) 19:659–71. doi: 10.1016/j.cellsig.2006.11.001
77. Mancinelli R, Carpino G, Petrungaro S, Mammola CL, Tomaipitinca L, Filippini A, et al. Multifaceted Roles of GSK-3 in Cancer and Autophagy-Related Diseases. Oxid Med Cell Longev (2017) 2017:4629495. doi: 10.1155/2017/4629495
78. Gandillet A, Park S, Lassailly F, Griessinger E, Vargaftig J, Filby A, et al. Heterogeneous Sensitivity of Human Acute Myeloid Leukemia to β-Catenin Down-Modulation. Leukemia (2011) 25:770–80. doi: 10.1038/leu.2011.17
79. Ding Q, He X, Xia W, Hsu JM, Chen CT, Li LY, et al. Myeloid Cell Leukemia-1 Inversely Correlates With Glycogen Synthase Kinase-3beta Activity and Associates With Poor Prognosis in Human Breast Cancer. Cancer Res (2007) 67:4564–71. doi: 10.1158/0008-5472.CAN-06-1788
80. Nagini S, Sophia J, Mishra R. Glycogen Synthase Kinases: Moonlighting Proteins With Theranostic Potential in Cancer. Semin Cancer Biol (2019) 56:25–36. doi: 10.1016/j.semcancer.2017.12.010
81. Eun Jeoung L, Sung Hee H, Jaesun C, Sung Hwa S, Kwang Hum Y, Min Kyoung K, et al. Regulation of Glycogen Synthase Kinase 3beta Functions by Modification of the Small Ubiquitin-Like Modifier. Open Biochem J (2008) 2:67–76. doi: 10.2174/1874091X00802010067
82. Wetzel F, Mittag S, Cano-Cortina M, Wagner T, Krämer OH, Niedenthal R, et al. SUMOylation Regulates the Intracellular Fate of ZO-2. Cell Mol Life Sci (2017) 74:373–92. doi: 10.1007/s00018-016-2352-5
83. Cadigan KM, Waterman ML. TCF/LEFs and Wnt Signaling in the Nucleus. Cold Spring Harb Perspect Biol (2012) 4:a007906. doi: 10.1101/cshperspect.a007906
84. Hoppler S, Kavanagh CL. Wnt Signalling: Variety at the Core. J Cell Sci (2007) 120:385–93. doi: 10.1242/jcs.03363
85. Song J, Xie C, Jiang L, Wu G, Zhu J, Zhang S, et al. Transcription Factor AP-4 Promotes Tumorigenic Capability and Activates the Wnt/β-Catenin Pathway in Hepatocellular Carcinoma. Theranostics (2018) 8:3571–83. doi: 10.7150/thno.25194
86. Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a Nuclear Matrix-Associated SUMO E3 Ligase, Represses LEF1 Activity by Sequestration Into Nuclear Bodies. Genes Dev (2001) 15:3088–103. doi: 10.1101/gad.944801
87. Mao J, Wang J, Liu B, Pan W, Farr GH 3rd, Flynn C, et al. Low-Density Lipoprotein Receptor-Related Protein-5 Binds to Axin and Regulates the Canonical Wnt Signaling Pathway. Mol Cell (2001) 7:801–9. doi: 10.1016/S1097-2765(01)00224-6
88. Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, et al. Axin, a Negative Regulator of the Wnt Signaling Pathway, Directly Interacts With Adenomatous Polyposis Coli and Regulates the Stabilization of Beta-Catenin. J Biol Chem (1998) 273:10823–6. doi: 10.1074/jbc.273.18.10823
89. Rui HL, Fan E, Zhou HM, Xu Z, Zhang Y, Lin SC. SUMO-1 Modification of the C-Terminal KVEKVD of Axin is Required for JNK Activation But has No Effect on Wnt Signaling. J Biol Chem (2002) 277:42981–6. doi: 10.1074/jbc.M208099200
90. Ihara M, Koyama H, Uchimura Y, Saitoh H, Kikuchi A. Noncovalent Binding of Small Ubiquitin-Related Modifier (SUMO) Protease to SUMO is Necessary for Enzymatic Activities and Cell Growth. J Biol Chem (2007) 282:16465–75. doi: 10.1074/jbc.M610723200
91. Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO Modification Pathway Localize to Filaments of the Nuclear Pore Complex. Mol Cell Biol (2002) 22:6498–508. doi: 10.1128/MCB.22.18.6498-6508.2002
92. Tan MY, Mu XY, Liu B, Wang Y, Bao ED, Qiu JX, et al. SUMO-Specific Protease 2 Suppresses Cell Migration and Invasion Through Inhibiting the Expression of MMP13 in Bladder Cancer Cells. Cell Physiol Biochem (2013) 32:542–8. doi: 10.1159/000354458
93. Kadoya T, Kishida S, Fukui A, Hinoi T, Michiue T, Asashima M, et al. Inhibition of Wnt Signaling Pathway by a Novel Axin-Binding Protein. J Biol Chem (2000) 275:37030–7. doi: 10.1074/jbc.M005984200
94. Kadoya T, Yamamoto H, Suzuki T, Yukita A, Fukui A, Michiue T, et al. Desumoylation Activity of Axam, a Novel Axin-Binding Protein, is Involved in Downregulation of Beta-Catenin. Mol Cell Biol (2002) 22:3803–19. doi: 10.1128/MCB.22.11.3803-3819.2002
95. Yukita A, Michiue T, Fukui A, Sakurai K, Yamamoto H, Ihara M, et al. XSENP1, a Novel Sumo-Specific Protease in Xenopus, Inhibits Normal Head Formation by Down-Regulation of Wnt/beta-Catenin Signalling. Genes Cells (2004) 9:723–36. doi: 10.1111/j.1356-9597.2004.00757.x
96. He ZX, Wei BF, Zhang X, Gong YP, Ma LY, Zhao W. Current Development of CBP/p300 Inhibitors in the Last Decade. Eur J Med Chem (2021) 209:112861. doi: 10.1016/j.ejmech.2020.112861
97. Cui Y, Wu X, Lin C, Zhang X, Ye L, Ren L, et al. AKIP1 Promotes Early Recurrence of Hepatocellular Carcinoma Through Activating the Wnt/β-Catenin/CBP Signaling Pathway. Oncogene (2019) 38:5516–29. doi: 10.1038/s41388-019-0807-5
98. Kuo HY, Chang CC, Jeng JC, Hu HM, Lin DY, Maul GG, et al. SUMO Modification Negatively Modulates the Transcriptional Activity of CREB-Binding Protein via the Recruitment of Daxx. Proc Natl Acad Sci U S A (2005) 102:16973–8. doi: 10.1073/pnas.0504460102
99. Park S, Stanfield RL, Martinez-Yamout MA, Dyson HJ, Wilson IA, Wright PE. Role of the CBP Catalytic Core in Intramolecular SUMOylation and Control of Histone H3 Acetylation. Proc Natl Acad Sci U S A (2017) 114:E5335–e5342. doi: 10.1073/pnas.1703105114
100. Diehl C, Akke M, Bekker-Jensen S, Mailand N, Streicher W, Wikström M. Structural Analysis of a Complex Between Small Ubiquitin-Like Modifier 1 (SUMO1) and the ZZ Domain of CREB-Binding Protein (CBP/p300) Reveals a New Interaction Surface on SUMO. J Biol Chem (2016) 291:12658–72. doi: 10.1074/jbc.M115.711325
101. Chinnadurai G. CtBP Family Proteins: More Than Transcriptional Corepressors. Bioessays (2003) 25:9–12. doi: 10.1002/bies.10212
102. Sewalt RG, Gunster MJ, van der Vlag J, Satijn DP, Otte AP. C-Terminal Binding Protein is a Transcriptional Repressor That Interacts With a Specific Class of Vertebrate Polycomb Proteins. Mol Cell Biol (1999) 19:777–87. doi: 10.1128/MCB.19.1.777
103. Lin X, Sun B, Liang M, Liang YY, Gast A, Hildebrand J, et al. Opposed Regulation of Corepressor CtBP by SUMOylation and PDZ Binding. Mol Cell (2003) 11:1389–96. doi: 10.1016/S1097-2765(03)00175-8
104. Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. Acetylation by P300 Regulates Nuclear Localization and Function of the Transcriptional Corepressor Ctbp2. J Biol Chem (2006) 281:4183–9. doi: 10.1074/jbc.M509051200
105. Kagey MH, Melhuish TA, Powers SE, Wotton D. Multiple Activities Contribute to Pc2 E3 Function. EMBO J (2005) 24:108–19. doi: 10.1038/sj.emboj.7600506
106. Kagey MH, Melhuish TA, Wotton D. The Polycomb Protein Pc2 is a SUMO E3. Cell (2003) 113:127–37. doi: 10.1016/S0092-8674(03)00159-4
107. Chinnadurai G. Transcriptional Regulation by C-Terminal Binding Proteins. Int J Biochem Cell Biol (2007) 39:1593–607. doi: 10.1016/j.biocel.2007.01.025
108. Riefler GM, Firestein BL. Binding of Neuronal Nitric-Oxide Synthase (nNOS) to Carboxyl-Terminal-Binding Protein (CtBP) Changes the Localization of CtBP From the Nucleus to the Cytosol: A Novel Function for Targeting by the PDZ Domain of nNOS. J Biol Chem (2001) 276:48262–8. doi: 10.1074/jbc.M106503200
109. Snow JW, Kim J, Currie CR, Xu J, Orkin SH. Sumoylation Regulates Interaction of FOG1 With C-Terminal-Binding Protein (CTBP). J Biol Chem (2010) 285:28064–75. doi: 10.1074/jbc.M109.096909
110. Labbé E, Letamendia A, Attisano L. Association of Smads With Lymphoid Enhancer Binding Factor 1/T Cell-Specific Factor Mediates Cooperative Signaling by the Transforming Growth Factor-Beta and Wnt Pathways. Proc Natl Acad Sci U S A (2000) 97:8358–63. doi: 10.1073/pnas.150152697
111. Zhang M, Wang M, Tan X, Li TF, Zhang YE, Chen D. Smad3 Prevents Beta-Catenin Degradation and Facilitates Beta-Catenin Nuclear Translocation in Chondrocytes. J Biol Chem (2010) 285:8703–10. doi: 10.1074/jbc.M109.093526
112. Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the Common Smad Mediator of Transforming Growth Factor-Beta Family Signaling. J Biol Chem (2003) 278:27853–63. doi: 10.1074/jbc.M301755200
113. Perekatt AO, Shah PP, Cheung S, Jariwala N, Wu A, Gandhi V, et al. SMAD4 Suppresses WNT-Driven Dedifferentiation and Oncogenesis in the Differentiated Gut Epithelium. Cancer Res (2018) 78:4878–90. doi: 10.1158/0008-5472.CAN-18-0043
114. Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL, et al. Smad4-Mediated Signaling Inhibits Intestinal Neoplasia by Inhibiting Expression of β-Catenin. Gastroenterology (2012) 142:562–71.e2. doi: 10.1053/j.gastro.2011.11.026
115. Imoto S, Sugiyama K, Muromoto R, Sato N, Yamamoto T, Matsuda T. Regulation of Transforming Growth Factor-Beta Signaling by Protein Inhibitor of Activated STAT, PIASy Through Smad3. J Biol Chem (2003) 278:34253–8. doi: 10.1074/jbc.M304961200
116. Imoto S, Ohbayashi N, Ikeda O, Kamitani S, Muromoto R, Sekine Y, et al. Sumoylation of Smad3 Stimulates its Nuclear Export During PIASy-Mediated Suppression of TGF-Beta Signaling. Biochem Biophys Res Commun (2008) 370:359–65. doi: 10.1016/j.bbrc.2008.03.116
117. Long J, Wang G, He D, Liu F. Repression of Smad4 Transcriptional Activity by SUMO Modification. Biochem J (2004) 379:23–9. doi: 10.1042/bj20031867
118. Ohshima T, Shimotohno K. Transforming Growth Factor-Beta-Mediated Signaling via the P38 MAP Kinase Pathway Activates Smad-Dependent Transcription Through SUMO-1 Modification of Smad4. J Biol Chem (2003) 278:50833–42. doi: 10.1074/jbc.M307533200
119. Shimada K, Suzuki N, Ono Y, Tanaka K, Maeno M, Ito K. Ubc9 Promotes the Stability of Smad4 and the Nuclear Accumulation of Smad1 in Osteoblast-Like Saos-2 Cells. Bone (2008) 42:886–93. doi: 10.1016/j.bone.2008.01.009
120. Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. Activation of Transforming Growth Factor-Beta Signaling by SUMO-1 Modification of Tumor Suppressor Smad4/DPC4. J Biol Chem (2003) 278:18714–9. doi: 10.1074/jbc.M302243200
121. Zhou X, Gao C, Huang W, Yang M, Chen G, Jiang L, et al. High Glucose Induces Sumoylation of Smad4 via SUMO2/3 in Mesangial Cells. BioMed Res Int (2014) 2014:782625. doi: 10.1155/2014/782625
122. Liu K, Wang X, Li D, Xu D, Li D, Lv Z, et al. Ginkgolic Acid, a SUMO-1 Inhibitor, Inhibits the Progression of Oral Squamous Cell Carcinoma by Alleviating SUMOylation of SMAD4. Mol Ther Oncolytics (2020) 16:86–99. doi: 10.1016/j.omto.2019.12.005
123. Orian-Rousseau V, Ponta H. Perspectives of CD44 Targeting Therapies. Arch Toxicol (2015) 89:3–14. doi: 10.1007/s00204-014-1424-2
124. Erfani E, Roudi R, Rakhshan A, Sabet MN, Shariftabrizi A, Madjd Z. Comparative Expression Analysis of Putative Cancer Stem Cell Markers CD44 and ALDH1A1 in Various Skin Cancer Subtypes. Int J Biol Markers (2016) 31:e53–61. doi: 10.5301/jbm.5000165
125. Ponta H, Sherman L, Herrlich PA. CD44: From Adhesion Molecules to Signalling Regulators. Nat Rev Mol Cell Biol (2003) 4:33–45. doi: 10.1038/nrm1004
126. Walter RJ, Sonnentag SJ, Munoz-Sagredo L, Merkel M, Richert L, Bunert F, et al. Wnt Signaling is Boosted During Intestinal Regeneration by a CD44-Positive Feedback Loop. Cell Death Dis (2022) 13:168. doi: 10.1038/s41419-022-04607-0
127. Schmitt M, Metzger M, Gradl D, Davidson G, Orian-Rousseau V. CD44 Functions in Wnt Signaling by Regulating LRP6 Localization and Activation. Cell Death Differ (2015) 22:677–89. doi: 10.1038/cdd.2014.156
128. Bogachek MV, Chen Y, Kulak MV, Woodfield GW, Cyr AR, Park JM, et al. Sumoylation Pathway is Required to Maintain the Basal Breast Cancer Subtype. Cancer Cell (2014) 25:748–61. doi: 10.1016/j.ccr.2014.04.008
129. De Andrade JP, Lorenzen AW, Wu VT, Bogachek MV, Park JM, Gu VW, et al. Targeting the SUMO Pathway as a Novel Treatment for Anaplastic Thyroid Cancer. Oncotarget (2017) 8:114801–15. doi: 10.18632/oncotarget.21954
130. Bogachek MV, Park JM, De Andrade JP, Lorenzen AW, Kulak MV, White JR, et al. Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem Cell Rep (2016) 7:1140–51. doi: 10.1016/j.stemcr.2016.11.001
131. Huang X, Tao Y, Gao J, Zhou X, Tang S, Deng C, et al. UBC9 Coordinates Inflammation Affecting Development of Bladder Cancer. Sci Rep (2020) 10:20670. doi: 10.1038/s41598-020-77623-9
132. Ali RG, Bellchambers HM, Warr N, Ahmed JN, Barratt KS, Neill K, et al. WNT-Responsive SUMOylation of ZIC5 Promotes Murine Neural Crest Cell Development, Having Multiple Effects on Transcription. J Cell Sci (2021) 134:256792. doi: 10.1242/jcs.256792
133. Satow R, Shitashige M, Jigami T, Fukami K, Honda K, Kitabayashi I, et al. β-Catenin Inhibits Promyelocytic Leukemia Protein Tumor Suppressor Function in Colorectal Cancer Cells. Gastroenterology (2012) 142:572–81. doi: 10.1053/j.gastro.2011.11.041
134. Picard N, Caron V, Bilodeau S, Sanchez M, Mascle X, Aubry M, et al. Identification of Estrogen Receptor β as a SUMO-1 Target Reveals a Novel Phosphorylated Sumoylation Motif and Regulation by Glycogen Synthase Kinase 3β. Mol Cell Biol (2012) 32:2709–21. doi: 10.1128/MCB.06624-11
Keywords: sumoylation, Wnt, β-catenin, t-cell factor, lymphoid enhancer factor
Citation: Fan L, Yang X, Zheng M, Yang X, Ning Y, Gao M and Zhang S (2022) Regulation of SUMOylation Targets Associated With Wnt/β-Catenin Pathway. Front. Oncol. 12:943683. doi: 10.3389/fonc.2022.943683
Received: 14 May 2022; Accepted: 07 June 2022;
Published: 30 June 2022.
Edited by:
Shoumin Zhu, University of Miami Health System, United StatesReviewed by:
Hailong Wu, Shanghai University of Medicine and Health Sciences, ChinaLei Chen, Capital Medical Univerisity, China
Zhangyuan Yin, Broad Institute, United States
Copyright © 2022 Fan, Yang, Zheng, Yang, Ning, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiwu Zhang, emhhbmdzaGl3dTY2NkBhbGl5dW4uY29t
†These authors have contributed equally to this work
 Linlin Fan
Linlin Fan Xudong Yang
Xudong Yang Minying Zheng
Minying Zheng Xiaohui Yang
Xiaohui Yang Yidi Ning
Yidi Ning Ming Gao
Ming Gao Shiwu Zhang
Shiwu Zhang