
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 04 July 2022
Sec. Cancer Genetics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.943622
This article is part of the Research TopicHallmark of Cancer: Replicative ImmortalityView all 8 articles
 Peter Lansdorp1,2*
Peter Lansdorp1,2*The number of (TTAGGG)n repeats at the ends of chromosomes is highly variable between individual chromosomes, between different cells and between species. Progressive loss of telomere repeats limits the proliferation of pre-malignant human cells but also contributes to aging by inducing apoptosis and senescence in normal cells. Despite enormous progress in understanding distinct pathways that result in loss and gain of telomeric DNA in different cell types, many questions remain. Further studies are needed to delineate the role of damage to telomeric DNA, replication errors, chromatin structure, liquid-liquid phase transition, telomeric transcripts (TERRA) and secondary DNA structures such as guanine quadruplex structures, R-loops and T-loops in inducing gains and losses of telomere repeats in different cell types. Limitations of current telomere length measurements techniques and differences in telomere biology between species and different cell types complicate generalizations about the role of telomeres in aging and cancer. Here some of the factors regulating the telomere length in embryonic and adult cells in mammals are discussed from a mechanistic and evolutionary perspective.
Ever since it was observed that telomere repeats are lost with each DNA replication cycle in vitro (1) as well as with age in human tissues (2, 3) and that such losses are mechanistically linked to replicative senescence (4), the role of telomeres and telomerase in aging and cancer has been the subject of intensive research efforts (reviewed in (5–8)). As a result, the amount of relevant information can no longer be effectively communicated in a single review, posing a major challenge for researchers entering the field. This dilemma is by no means unique to the telomere field and this perspective does not pretend to solve it. Instead, it is hoped that a discussion of selected studies and observations will inspire discussion, insights and new experiments. For this purpose, a general discussion of the mechanisms involved in the elongation and the shortening of telomeres is followed by a discussion of factors and pathways that are implicated in regulating the number of telomere repeats at chromosome ends in embryonic and adult cells.
That complete replication of linear chromosomes could represent a problem was already proposed in the early 1970s (9, 10) and the “end-replication problem” is a now a well-recognized cause of telomere attrition (reviewed in (11). In most organisms with linear chromosomes the solution to the “end-replication problem” is extension of the 3’ end of DNA template strands by means of telomerase, a specialised reverse transcriptase (12, 13). However, telomerase is not the only solution to the “end replication problem”. For example, Drosophila melanogaster uses a transposition mechanism to replenish the DNA lost with each DNA replication cycle [reviewed in (14)].
Unfortunately, understanding the Yin and Yang at the telomere: the end replication problem and its primary solution, telomerase, only scratches the surface of why all chromosomes end up with a given number of telomeric repeats at any given point in time. While much progress has been made in the characterization of the molecules and processes involved in the loss and gain of telomere repeats at chromosome ends, many questions remain unsolved. Further work is needed to better understand the role of telomerase and its interaction with chromosome ends, the role of telomere chromatin (reviewed in (15) and the “shelterin” proteins that bind to telomeric DNA (reviewed in (16) in telomere function in different cell types during development and adult tissues. The “shelterin” complex is a set of related complexes that not only differ between cells but also function differently along the telomere at different stages of the cell cycle (reviewed in (7). Other areas that require further study are how cells respond to the DNA damage signals derived from critically short telomeres and how the composition of “shelterin” proteins is modulated by telomeric TERRA transcripts (reviewed in (17), R-loops and guanine quadruplex (G4) structures (reviewed in (18, 19).
Apart from differences between cell types, a complicating factor is that the regulation of telomerase and telomere length in various cell types is markedly different between short- and long-lived animals (reviewed in (5). Low levels of telomerase limit the replicative potential of somatic cells in long-lived animals including humans but not in relatively short-lived animals such as laboratory mice. Most likely, replication-dependent telomere erosion in somatic cells protects long-lived animals against tumors early in life be it at the expense of tissue regeneration and immune responses late in life. This idea, captured in the “telomere erosion in disposable soma (TEDS)” theory of human aging (5), provides the framework for the following discussion of telomere length regulation in human cells.
Next to the loss of telomeric DNA via the “end-replication problem”, telomeric DNA is also lost via the obligatory processing of chromosome ends following leading strand DNA replication (20). The resulting blunt ends must be processed in order to create a single strand 3’ overhang that is presumed to be present and required at every chromosome end. Apart from inevitable losses of telomeric DNA with each replication cycle, telomeric DNA can also be lost by other, less predictable mechanisms. Sporadic problems can arise following various types of damage to telomeric DNA but also when single stranded G-rich telomeric DNA is allowed to form secondary structures that interfere with replication or repair. In principle, all processes that disrupt duplex telomeric DNA can generate single stranded DNA which in turn can form secondary DNA structures known as guanine quadruples (G4) structures (21). Next to G4 structures, single stranded G-rich DNA, folded back into duplex telomeric DNA called T-loops, and RNA transcripts of telomeric DNA called TERRA, associated with telomeric DNA in structures called R-loops, can also cause stalling of the replication fork (22–24). In view of the variable length of TERRA and single stranded G-rich DNA telomere repeats, a variety of G4 structures could form at telomeres including stable hybrid G4 structures containing both RNA and DNA (25).
Unlike elsewhere in the genome, where a stalled replication fork can be rescued by a fork coming from the opposite direction, rescue by incoming forks is not expected at telomeres unless replication is initiated from origins within the telomere upon replication stress (26). Progression of the replication fork at telomeres can also be hampered by other types of DNA lesions including intra-strand crosslinks as well as tightly DNA-bound proteins in telomeric heterochromatin (11). Stalled replication forks can lead to fork collapse and breaks in telomeric DNA. When such breaks are not repaired by telomerase or recombination (27), large tracts of telomere repeats are lost.
Observations of telomeric DNA at individual chromosome ends in human and murine cells using quantitative fluorescence in situ hybridization (Q-FISH) documented heterogeneous telomere length distributions as well as sporadic loss of telomeric DNA (Figure 1) (5, 30–32). Importantly, in cells with low or undetectable telomerase levels such as human fibroblasts, the biological consequences of sporadic losses of telomeric DNA add to the inevitable loss of telomere repeats with each replication cycle to accelerate the replicative senescence or apoptosis of cells.
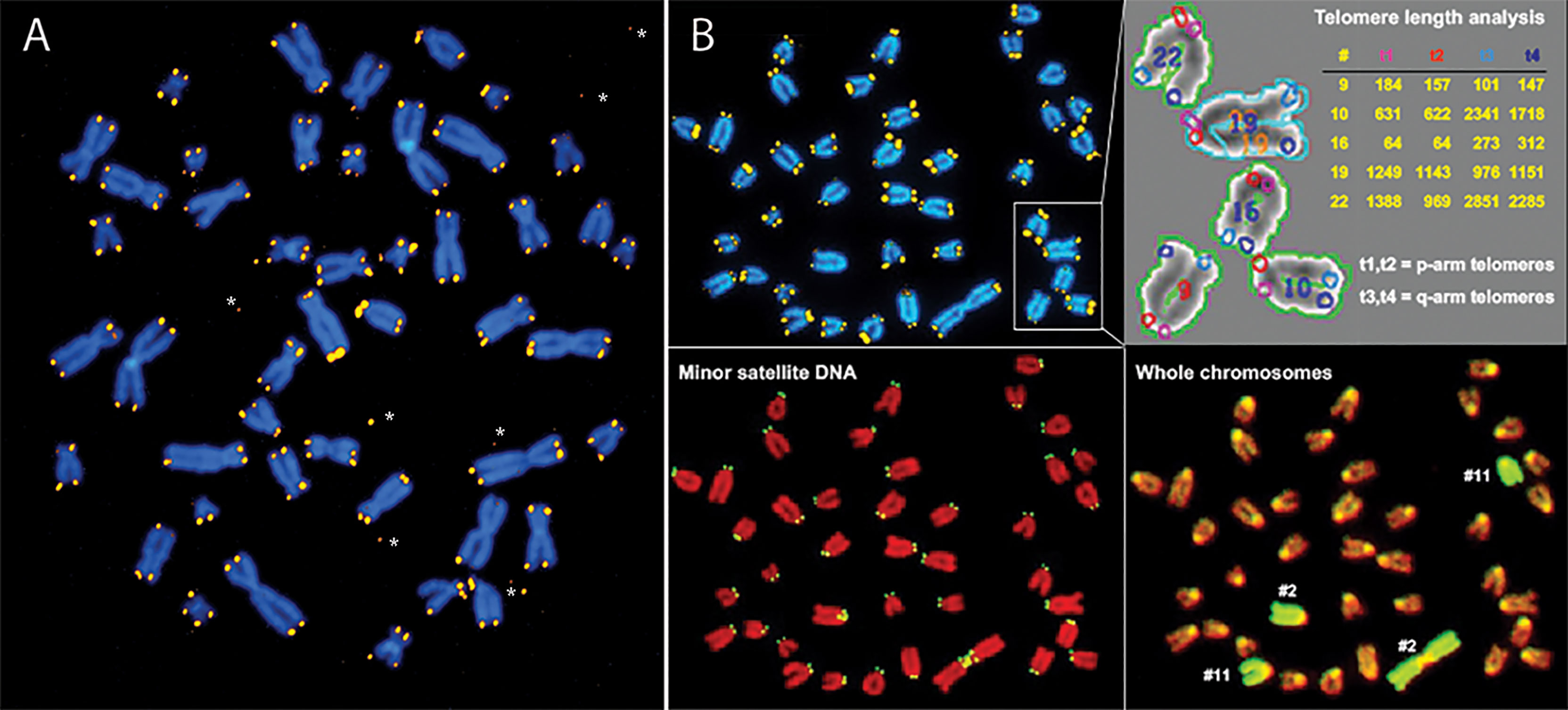
Figure 1 Q-FISH shows extreme variability in the length of telomere repeats in human (A) and mouse (B) chromosomes. (A) Human metaphase chromosomes stained with DAPI (blue) from a cultured fibroblast following hybridization with Cy3 labeled (CCCTAA)3 PNA (yellow). Note the large variation in telomere fluorescence intensity at individual chromosome ends, discrepancies in telomere fluorescence between sister chromatids and occasional telomere fluorescence spots outside chromosomes*. (B) Mouse metaphase chromosomes from a cultured skin fibroblast hybridized sequentially with a mixture of different fluorescent probes: first Cy3 labeled (CCCTAA)3 PNA (B, top left panel) and FITC labeled CGGCATTGTAGAACAGTG PNA specific for mouse minor satellite sequences (B, bottom left panel) followed by staining of DNA with DAPI, image acquisition and hybridization with FITC labeled chromosome paint probe specific for respectively chr2 and chr11 (B, bottom right panel). Telomere length was analyzed using the TFL-Telo software (B, top right panel) (28). Note the very short telomeres on the short arm of chr “16” and the very long telomeres on the long arm of chr “22” (arbitrary chromosome numbers and fluorescence intensity values). For details see (29, 30).
Most problems at telomeres probably arise or become apparent during DNA replication. Apart from the inevitable problems during replication discussed above several additional problems have been identified (reviewed in (11)). Lagging strand replication of telomeric DNA involves either the regular lagging strand DNA replication complex or a replication complex that is more specific to telomeres involving CTC1, STN1 and TEN1, a set of proteins known as the CST complex (7). The CST complex is similar but different from RPA, the protein complex that typically binds single strand DNA. The CST complex is required for “fill-in” synthesis of C-rich DNA from G-rich templates extended or produced by telomerase. Where or when lagging strand replication switches from RPA to CST during replication of telomeric DNA in the absence of telomerase is not known. CST protein levels could be important to suppress the formation of G4 structures from single stranded G-rich DNA at telomeres produced by telomerase or exposed during replication, transcription or recombination. The POT1 protein is another protein that binds to single stranded G-rich DNA to suppress the formation of G4 structures (33) and the full extent of its roles in telomere biology remains to be clarified (34, 35). In general, more studies are needed to better understand the role of DNA damage response pathways in telomere function. Such studies should elucidate the balance between proteins that suppress the formation of G4 DNA at telomeres and helicases such as RTEL1, BLM and FANCJ that are capable of unwinding G4 structures (36). Differences between cell types and the stochastic nature of G4 DNA formation provide significant challenges for such studies.
The switch between a protected telomere end and a chromosome end that signals DNA damage has many levels and components. Many of these components themselves have switch-like characteristics in that two alternate states can be present or selected. For example, it is possible that telomere damage signaling involves a liquid-liquid phase separation in the nucleus between heterochromatin and euchromatin (reviewed in (37–39). Both the position of telomeres in the nucleus and studies of telomere mobility provide some support for this possibility. Telomeres in human lymphocytes are not randomly distributed in the nucleus but appear positioned at the interface between euchromatin and heterochromatin (Figure 2). Interestingly, when the movement of telomeres in cultured mouse embryonic stem cells was studied using telomere binding protein tagged with green fluorescent protein (Venus-TRF1), it was observed that the faintest fluorescent spots showed the highest mobility. These findings are illustrated in Figure 3 (see also Supplementary Movies 1–3). More rapid movements of short telomeres were also observed with human cells (41). Together these observations suggest that a minimum number of telomeric repeats at chromosome ends could be required to anchor telomeres in heterochromatic areas of the nucleus. Such a location is expected to suppress transcription as well as activation of DNA damage signaling pathways.
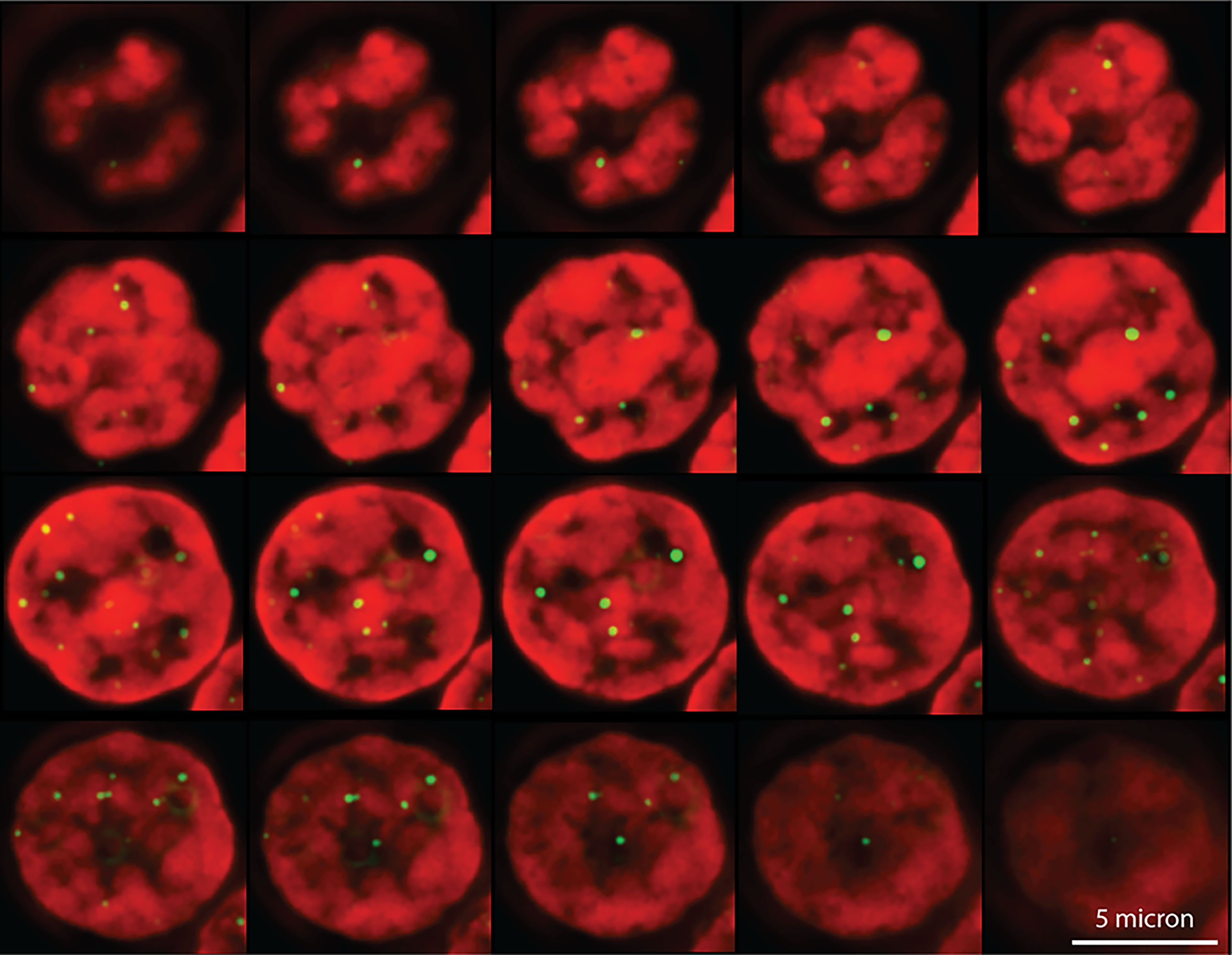
Figure 2 Telomeres in a human lymphocyte are not randomly distributed in the nucleus. Shown are optical sections through the interphase nucleus of a human T lymphocyte following formaldehyde fixation and fluorescence in situ hybridization with fluorescently labeled (CCCTAA)3 PNA (shown in yellow/green). DNA is counterstained with DAPI (shown in red). A stack of images, acquired at separate focal planes, was processed using deconvolution microscopy (40). Note that telomeres appear to cluster at the interface between DNA bright areas, presumably reflecting heterochromatin and DNA weakly stained areas, presumably representing euchromatin (Chavez and Lansdorp unpublished observations).
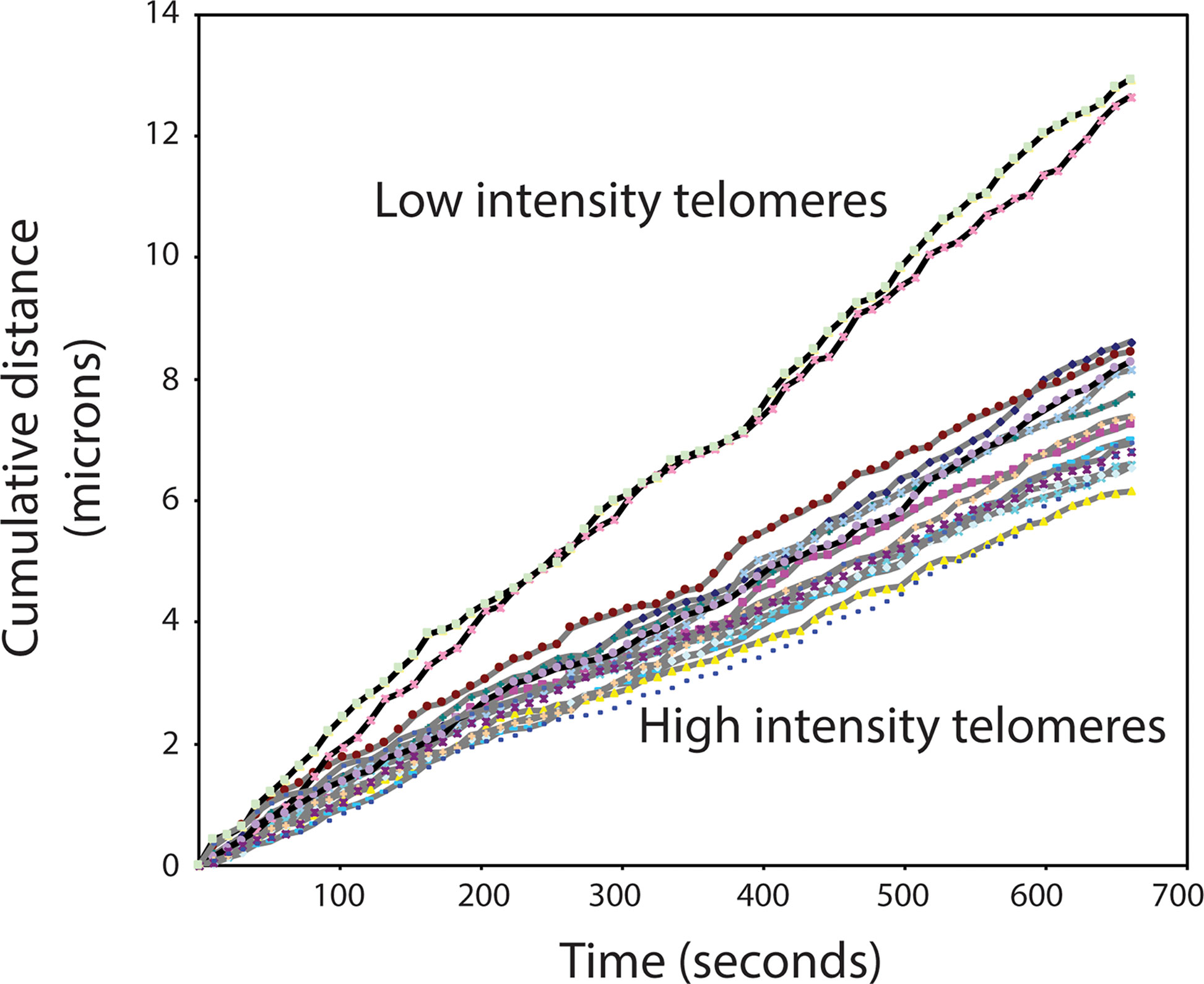
Figure 3 High mobility of very short telomeres in cultured mouse embryonic stem cells. Viable cells, tagged with Venus-TRF1, were imaged at a fixed position over 10 minutes. The position of individual fluorescent telomere spots was recorded every 10 seconds. Two categories of telomere spots were observed: low intensity spots and high intensity spots. The recorded position of each spot at each time interval was used to calculate the travel distance of individual telomeres. See Supplementary Information and Supplementary Movie 3 for details.
Liquid-liquid phase transitions are known to be important for the regulation of transcription and DNA repair (42, 43) and have also been implicated in the function of telomeres (44). Unfortunately, studies of the role of liquid-liquid phase separation in telomere function (and biology in general) are complicated by the difficulty to reconstitute relevant phase transitions in the laboratory. It is possible that the transition between duplex telomere repeats and G4 DNA also acts as a switch, perhaps facilitating liquid-liquid phase transitions and/or activating DNA damage signaling pathways (45, 46). Other switch events at telomeres that need further study include the increased mobility of histones in telomere chromatin (15), the switch from RPA to CST during telomere replication (reviewed in (7), the initiation and suppression of telomere transcription (reviewed in (47) and the role of subtelomeric DNA and associated proteins including boundary elements such as CTCF in subtelomeric chromatin and telomere function (reviewed in (48). Further studies of these factors and their interplay promise to yield interesting results.
The starting telomere length at the onset of life is presumably the telomere length present in the gametes from the parents. Little is known about the regulation of telomere length in oocytes of females throughout their reproductive lifespan. Perhaps all oocyte precursors express telomerase during embryonic development similar to spermatogonial stem cells in the mouse (49, 50). However, the average telomere length in human oocytes was reported to be around 1 kb shorter than in blastocysts, possibly reflecting loss of telomeric DNA during cell divisions of oocyte precursors in utero (51). Various factors implicated in regulating telomere length in embryos were recently reviewed (52). In males, the telomere length in sperm appears to slightly increase with age (53) and paternal age was found to correlate with telomere length in offspring (54). The most important events to shape the telomere length at birth and throughout life probably occur in the first few cell divisions of the fertilized oocyte when chromatin is completely reorganized (reviewed in (55)). First the dense chromatin in gametes is decompacted (56). The two-cell stage is characterized by demethylation and a unique gene expression profile that includes high expression of Zscan4 (57, 58). At this stage telomeres recombine, presumably to allow elongation and rescue of very short telomeres or damaged telomeres by break-induced replication (58, 59). Perhaps the telomere length in sperm chromosomes is the most important factor in adjusting the telomere length at the 2-cell stage. Suppression of G4 structures at telomeres could be particularly important when telomere chromatin is decompacted and telomerase is expressed at high levels. Undifferentiated human embryonic stem cells were reported to express high levels of G4 DNA (60). Telomere length adjustments at this stage could reflect a balance between single stranded G-rich DNA synthesized by telomerase, the CST proteins binding to such DNA, G4 DNA formation, G4 unwinding by helicase activity, C-strand “fill in” synthesis and various recombination processes. A major role for G4 DNA in the regulation of telomere length is supported by studies of mice. Laboratory mice, Mus musculus, with very long telomeres (>40 Kb) differ from Mus spretus mice with relatively short telomeres (~10 Kb) in RTEL1, the Regulator of Telomere Length helicase predicted to unwind guanine G4 structures at telomeres (29, 36, 61). More studies are needed to define the role of telomerase expression (62), primer sequestration, CST proteins and fill-in replication (63, 64) in the regulation of telomere length. Such studies should also clarify the role of factors suppressing the formation of G4 DNA at telomeres such as the CST complex (7), RAP1 (65) and POT1 (33) as well as helicases that can unwind G4 DNA structures that include, next to RTEL1, FANCJ (66) and BLM (67, 68). The protection of chromosome ends from DNA damage response pathways in early embryonic cells is very different from that in somatic cells (59, 69). How differences in end protection relate to telomere length adjustments will require further study. Such studies are complicated by the small numbers of relevant cells as well as the highly dynamic events within and between cell divisions during early embryogenic development.
Following telomere length adjustments at the 2-cell stage, telomerase expression in embryos may result in further elongation of telomeres. While the level of telomerase RNA appears to be the main limiting factor for telomerase activity in embryonic stem cells (70), it is possible that telomerase levels in blastocysts are further upregulated by low oxygen levels perhaps via induction of hTERT expression by the transcription factor hypoxia inducible factor 1 (HIF-1) alpha (71). The relatively high oxygen levels in typical tissue culture experiments (20%) do not match the much lower oxygen levels encountered by most cells in vivo and reducing oxygen levels during tissue culture increases the replicative potential of many cells in vitro (reviewed in (72). Culture of embryos at reduced oxygen levels could not only increase telomerase activity but also limit oxidative damage to telomeric DNA. Culture of embryos for assisted reproductive technologies should explore the effect of oxygen levels with some urgency given that reductions in telomere length prior to embryo implantation could have biological effects that only appear many years later. Future studies should also explore whether sex differences in telomere length and lifespan indeed reflect higher levels of dyskerin and telomerase in embryonic cells as was recently proposed (73). More generally, apart from overexpressing telomerase in cells, which seems neither feasible or advisable in vivo, measures that reduce exogenous damage to telomeric DNA represent perhaps the most effective ways to increase the replicative potential of cells in vitro and in vivo.
Telomerase levels are down-regulated in most human cells at some point during in utero growth and differentiation to limit the number of subsequent cell divisions. Both alternative splicing and transcriptional regulation of the telomerase transcriptase gene (hTERT) have been implicated in the silencing of telomerase activity (8, 74). However, details of the timing and degree of telomerase silencing during development and differences between specific cell types remain to be clarified. Downregulation of telomerase activity in somatic cells of long-lived animals was proposed to increase reproductive fitness by suppressing the growth of malignant tumors before reproduction (5). Several findings support this theory. Surprisingly, somatic mutations in dividing and non-dividing human tissues accumulate at a very similar rate of between 10-40 mutations per cell per year (75). In this study, cells of the germline were found to acquire only 1-2 mutations per year. Given that tumors arise by acquisition of mutations, one can wonder why DNA repair in somatic cells is not as effective as in cells of the germline. Apart from evolutionary arguments about the importance of germline versus somatic DNA in cells, the answer could involve a trade-off between the energy required to limit the acquisition of somatic mutations by improving DNA repair and other ways to suppress malignant growth prior to reproduction. Targeting telomerase expression levels in somatic cells to suppress tumors before reproduction could have provided a selective advantage that allowed lifespan to increase during evolution (5). Subsequent cell-type specific adjustments of telomerase levels could have allowed further increases in fitness. However, a requirement for tumor suppression by telomere shortening is that senescence or apoptosis is indeed enforced when telomeres become critically short. Given that most malignant tumors are deficient in p53, the Achilles heel of tumor suppression via telomere erosion in humans is perhaps loss of p53, resulting in failure to impose cell cycle arrest or apoptosis when telomeres are too short (6, 8, 76). Mechanistically, it seems possible that frequent loss of p53 on chr17p is related to the very short telomeres on that chromosome arm (77, 78). It is also tempting to speculate that multiple copies of p53 genes in long-lived animals such as elephants (79) reflect selection of more effective tumor suppression via DNA damage signals originating from short telomeres.
Next to the gradual loss of telomeric DNA with each cell division, sporadic loss of telomeric DNA also contributes to telomere shortening. Indeed, such losses are likely to underpin the strikingly heterogenous telomere length in human and murine cells. Sporadic truncation of telomeric DNA at specific chromosome ends was well-documented in cultured human fibroblasts (80). It is possible that the function of residual telomerase activity in most human somatic cells is not to prevent overall telomere shortening but to prevent premature senescence triggered by the sporadic loss of telomeric DNA (5). The need for such “telomere repair” by telomerase is expected to increase not only as a function of the overall telomere length but also as a function of the number of telomeres with damaged DNA. Differences between cells in the efficiency of telomere repair by telomerase argue against telomere shortening acting as a simple “mitotic clock” in all human somatic cells given that the number of cell divisions prior to replicative senescence could vary as a function of telomerase levels. Fibroblasts, with little if any telomerase activity, could encounter critical loss of telomeres and replicative senescence earlier than hematopoietic stem cells or T lymphocytes which have easily detectable telomerase activity (5). Telomerase activity could be regulated at many levels, ranging from regulation of gene expression and alternative splicing to variable processing of transcripts, proteins and assembly of the telomerase holoenzyme. Perhaps germinal center (GC) B cells represent the most extreme example of high telomerase activity in human somatic cells (81, 82). High levels of telomerase in GC B cells elongate telomeres relative to naive B cells, perhaps allowing some B cells to avoid the Hayflick limit altogether. Elongation of telomeres in GC B cells could reflect the need to enable more numerous replication cycles to support effective antibody production following clonal selection and affinity maturation. More effective antibody responses may have provided a selective advantage during evolution be it at the cost of increasing the risk of malignant transformation (5). This notion is compatible with observations indicating that B cell lymphomas in human adults are much more common than T cell lymphomas and that haploinsufficiency for telomerase causes pronounced defects in B cell responses (83). More generally, telomere-mediated limits in the proliferation of lymphocytes are likely to eventually compromise all different types of immune responses including those against other viruses and microorganisms (84) as well as tumor cells that can be recognized by the immune system (5). Of note, short telomere length in leukocytes was reported to correlate with poor outcome of COVID-19 infection irrespective of age (85).
The decline in telomere length over a human lifetime has been most clearly documented for leukocyte subsets using fluorescence in situ hybridization and flow cytometry or “flow FISH” (86). Rapid decline in telomere length is observed in the first few years of life in line with rapid cell divisions as well as a “mitotic clock” ticking in hematopoietic stem cells (87, 88). These findings were recently discussed in the context of the “Telomere Erosion in Disposable Soma” theory of human aging (5). The essence of this theory is that replicative senescence could be advantageous as a tumor suppressor mechanism early in life but disadvantageous late in life by limiting cell proliferation and tissue regeneration. Interestingly, differences in average telomere length and lifespan between males and females are correlated and perhaps reflect differences in embryonic telomerase levels (73). Given the role of oxidative damage to telomeric DNA in telomere shortening (89), further studies of the effect of oxidative stress on leukocyte telomere length dynamics are of interest. Paradoxically, chronic and acute oxidative stress appear to have opposite effects on telomere length in vivo. Exposure to high oxygen levels in deep sea divers was shown to result in transient elongation of telomeres in leukocytes (90). In this study the telomere length returned to below baseline levels after a year. Perhaps replacement of cells lost by damage from oxidative stress increased the turnover of primitive stem cells (with longer telomeres) which would otherwise have stayed dormant longer. It is possible that a similar “robbing Peter to pay Paul” principle applies to the reported elongation of leukocyte telomeres during space flight in pure oxygen (91). In general, further long-term studies of telomere length in relation to health outcomes are needed to develop a clearer picture of telomere-mediated senescence and regeneration defects in various diseases that are more prevalent in the elderly.
While much progress has been made in understanding various factors and pathways that play a role in regulating telomere length in different cell types many questions remain unsolved. Major variables are the length of telomeres in gametes and the processes at telomeres that occur between fertilization and embryo implantation. Variable suppression of telomerase levels in human somatic cells, ranging from little if any activity in fibroblasts to high levels in germinal center B cells, further complicate the picture. Whereas telomere shortening limits the proliferation of premalignant cells, it also limits immune responses. Variable suppression of telomerase in different cell types such as B and T lymphocytes could reflect ongoing selective forces. A major problem for the field is that all current telomere length measurements suffer from limitations (5, 92). Measurements of the average telomere length typically ignore variation in the length of telomere repeats at individual chromosomes which could be relevant for biological outcomes. Most techniques require thousands of cells and obscure differences between cells. Ideally, novel telomere length measurements should generate information about the average length as well as the distribution of telomere length in single cells. Single cell techniques are also needed to clarify the highly dynamic processes that result in adjustment of telomere length in the first few cell divisions after fertilization. With better telomere length measurements and more insight in telomere length regulation during development many of the current outstanding questions will be answered.
The author confirms being the sole contributor of this work and has approved it for publication.
Work in the Lansdorp laboratory is funded by a Program Project Grant (#1074) from the Terry Fox Research Institute, a Project Grant (#PJT-159787) from the Canadian Institutes of Health Research, and a grant (#40044) from the Canadian Foundation for Innovation and the Government of British Columbia.
The author is a founder and shareholder of Repeat Diagnostics Inc., a company specializing in clinical telomere length measurements.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This paper is dedicated to Mark Zijlmans and Elizabeth Chavez who are no longer with us. Both Mark and Liz made major contributions to some of the unpublished work described in this paper. I also thank Uwe Martens, Prakash Hande, Natalie Rufer, Tim Brümmendorf, Mike Schertzer, Gabriela Baerlocher, Irma Vulto, Geraldine Aubert and Katleen Lisaingo for unpublished work shown and discussed in this paper. I apologize to all my colleagues in and outside the telomere field whose relevant work was not mentioned. In most cases this is not by choice. Who can keep up with the ever-expanding universe of relevant data? Biology is more complex that most of us are willing to accept.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.943622/full#supplementary-material
1. Harley CB, Futcher AB, Greider CW. Telomeres Shorten During Ageing of Human Fibroblasts. Nature (1990) 345:458–60. doi: 10.1038/345458a0
2. Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere Reduction in Human Colorectal Carcinoma and With Ageing. Nature (1990) 346:866–8. doi: 10.1038/346866a0
3. de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, et al. Structure and Variability of Human Chromosome Ends. Mol Cell Biol (1990) 10:518–27. doi: 10.1128/mcb.10.2.518-527.1990
4. Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, et al. Extension of Life-Span by Introduction of Telomerase Into Normal Human Cells. Science (1998) 279:349–52. doi: 10.1126/science.279.5349.349
5. Lansdorp PM. Telomeres, Aging, and Cancer: The Big Picture. Blood (2022) 139:813–21. doi: 10.1182/blood.2021014299
6. Maciejowski J, De Lange T. Telomeres in Cancer: Tumour Suppression and Genome Instability. Nat Rev Mol Cell Biol (2017) 18:175–86. doi: 10.1038/nrm.2016.171
7. Lim CJ, Cech TR. Shaping Human Telomeres: From Shelterin and CST Complexes to Telomeric Chromatin Organization. Nat Rev Mol Cell Biol (2021) 22:283–98. doi: 10.1038/s41580-021-00328-y
8. Shay JW, Wright WE. Telomeres and Telomerase: Three Decades of Progress. Nat Rev Genet (2019) 20:299–309. doi: 10.1038/s41576-019-0099-1
9. Olovnikov AM. A Theory of Marginotomy. The Incomplete Copying of Template Margin in Enzymic Synthesis of Polynucleotides and Biological Significance of the Phenomenon. J Theor Biol (1973) 41:181–90. doi: 10.1016/0022-5193(73)90198-7
10. Watson JD. Origin of Concatemeric T7 DNA. Nat New Biol (1972) 239:197–201. doi: 10.1038/newbio239197a0
11. Bonnell E, Pasquier E, Wellinger RJ. Telomere Replication: Solving Multiple End Replication Problems. Front Cell Dev Biol (2021) 9:668171. doi: 10.3389/fcell.2021.668171
12. Greider CW, Blackburn EH. Identification of a Specific Telomere Terminal Transferase Activity in Tetrahymena Extracts. Cell (1985) 43:405–13. doi: 10.1016/0092-8674(85)90170-9
13. Ghanim GE, Fountain AJ, Van Roon AM, Rangan R, das R, Collins K, et al. Structure of Human Telomerase Holoenzyme With Bound Telomeric DNA. Nature (2021) 593:449–453. doi: 10.1038/s41586-021-03415-4
14. Servant G, Deininger PL. Insertion of Retrotransposons at Chromosome Ends: Adaptive Response to Chromosome Maintenance. Front Genet (2015) 6:358. doi: 10.3389/fgene.2015.00358
15. Soman A, Liew CW, Teo HL, Berezhnoy NV, Olieric V, Korolev N, et al. The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties. Nucleic Acids Res (2020) 48:5383–96. doi: 10.1093/nar/gkaa289
16. de Lange T. Shelterin-Mediated Telomere Protection. Annu Rev Genet (2018) 52:223–47. doi: 10.1146/annurev-genet-032918-021921
17. Fernandes RV, Feretzaki M, Lingner J. The Makings of TERRA R-Loops at Chromosome Ends. Cell Cycle (2021) 20:1745–59. doi: 10.1080/15384101.2021.1962638
18. Bryan TM. G-Quadruplexes at Telomeres: Friend or Foe? Molecules (2020) 25:3686. doi: 10.3390/molecules25163686
19. Rhodes D, Lipps HJ. G-Quadruplexes and Their Regulatory Roles in Biology. Nucleic Acids Res (2015) 43:8627–37. doi: 10.1093/nar/gkv862
20. Lingner J, Cooper JP, Cech TR. Telomerase and DNA End Replication: No Longer a Lagging Strand Problem? Science (1995) 269:1533–4. doi: 10.1126/science.7545310
21. Lipps HJ, Rhodes D. G-Quadruplex Structures: In Vivo Evidence and Function. Trends Cell Biol (2009) 19:414–22. doi: 10.1016/j.tcb.2009.05.002
22. Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian Telomeres End in a Large Duplex Loop. Cell (1999) 97:503–14. doi: 10.1016/S0092-8674(00)80760-6
23. Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 Dismantles T Loops and Counteracts Telomeric G4-DNA to Maintain Telomere Integrity. Cell (2012) 149:795–806. doi: 10.1016/j.cell.2012.03.030
24. Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, Lingner J. RAD51-Dependent Recruitment of TERRA lncRNA to Telomeres Through R-Loops. Nature (2020) 587:303–8. doi: 10.1038/s41586-020-2815-6
25. Xu Y, Kimura T, Komiyama M. Human Telomere RNA and DNA Form an Intermolecular G-Quadruplex. Nucleic Acids Symp Ser (Oxf) (2008), 52:169–70. doi: 10.1093/nass/nrn086
26. Drosopoulos WC, Deng Z, Twayana S, Kosiyatrakul ST, Vladimirova O, Lieberman PM, et al. TRF2 Mediates Replication Initiation Within Human Telomeres to Prevent Telomere Dysfunction. Cell Rep (2020) 33:108379. doi: 10.1016/j.celrep.2020.108379
27. Epum EA, Haber JE. DNA Replication: The Recombination Connection. Trends Cell Biol (2022) 32:45–57. doi: 10.1016/j.tcb.2021.07.005
28. Poon SS, Martens UM, Ward RK, Lansdorp PM. Telomere Length Measurements Using Digital Fluorescence Microscopy. Cytometry (1999) 36:267–78. doi: 10.1002/(SICI)1097-0320(19990801)36:4<267::AID-CYTO1>3.0.CO;2-O
29. Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, et al. Regulation of Murine Telomere Length by Rtel: An Essential Gene Encoding a Helicase-Like Protein. Cell (2004) 117:873–86. doi: 10.1016/j.cell.2004.05.026
30. Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, et al. Telomeres in the Mouse Have Large Inter-Chromosomal Variations in the Number of T2AG3 Repeats. Proc Natl Acad Sci U.S.A. (1997) 94:7423–8. doi: 10.1073/pnas.94.14.7423
31. Lansdorp PM, Verwoerd NP, Van De Rijke FM, Dragowska V, Little MT, Dirks RW, et al. Heterogeneity in Telomere Length of Human Chromosomes. Hum Mol Genet (1996) 5:685–91. doi: 10.1093/hmg/5.5.685
32. Mayer S, Bruderlein S, Perner S, Waibel I, Holdenried A, Ciloglu N, et al. Sex-Specific Telomere Length Profiles and Age-Dependent Erosion Dynamics of Individual Chromosome Arms in Humans. Cytogenet Genome Res (2006) 112:194–201. doi: 10.1159/000089870
33. Zaug AJ, Podell ER, Cech TR. Human POT1 Disrupts Telomeric G-Quadruplexes Allowing Telomerase Extension In Vitro. Proc Natl Acad Sci U.S.A. (2005) 102:10864–9. doi: 10.1073/pnas.0504744102
34. Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 Facilitates Telomere Elongation by Telomerase. Curr Biol (2003) 13:942–6. doi: 10.1016/S0960-9822(03)00339-7
35. Kelleher C, Kurth I, Lingner J. Human Protection of Telomeres 1 (POT1) is a Negative Regulator of Telomerase Activity In Vitro. Mol Cell Biol (2005) 25:808–18. doi: 10.1128/MCB.25.2.808-818.2005
36. Lansdorp P, Van Wietmarschen N. Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo. Genes (Basel) (2019) 10:870. doi: 10.3390/genes10110870
37. Asamitsu S, Shioda N. Potential Roles of G-Quadruplex Structures in RNA Granules for Physiological and Pathological Phase Separation. J Biochem (2021) 169:527–33. doi: 10.1093/jb/mvab018
38. Mehta S, Zhang J. Liquid-Liquid Phase Separation Drives Cellular Function and Dysfunction in Cancer. Nat Rev Cancer (2022) 22:239–52. doi: 10.1038/s41568-022-00444-7
39. Fuxreiter M, Vendruscolo M. Generic Nature of the Condensed States of Proteins. Nat Cell Biol (2021) 23:587–94. doi: 10.1038/s41556-021-00697-8
40. Mathog D, Hochstrasser M, Sedat JW. Light Microscope Based Analysis of Three-Dimensional Structure: Applications to the Study of Drosophila Salivary Gland Nuclei. I. Data Collection and Analysis. J Microsc (1985) 137:241–52. doi: 10.1111/j.1365-2818.1985.tb02582.x
41. Wang X, Kam Z, Carlton PM, Xu L, sedat JW, Blackburn EH. Rapid Telomere Motions in Live Human Cells Analyzed by Highly Time-Resolved Microscopy. Epigenet Chromatin (2008) 1:4. doi: 10.1186/1756-8935-1-4
42. Rippe K. Liquid-Liquid Phase Separation in Chromatin. Cold Spring Harb Perspect Biol (2022) 14:a040683. doi: 10.1101/cshperspect.a040683
43. Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, et al. Phase Separation of 53BP1 Determines Liquid-Like Behavior of DNA Repair Compartments. EMBO J (2019) 38:e101379. doi: 10.15252/embj.2018101379
44. King JT, Shakya A. Phase Separation of DNA: From Past to Present. Biophys J (2021) 120:1139–49. doi: 10.1016/j.bpj.2021.01.033
45. Zwang TJ, Tse ECM, Barton JK. Sensing DNA Through DNA Charge Transport. ACS Chem Biol (2018) 13:1799–809. doi: 10.1021/acschembio.8b00347
46. Lv L, Guo Z, Wang J, Wang E. G-Quadruplex as Signal Transducer for Biorecognition Events. Curr Pharm Des (2012) 18:2076–95. doi: 10.2174/138161212799958459
47. Azzalin CM, Lingner J. Telomere Functions Grounding on TERRA Firma. Trends Cell Biol (2015) 25:29–36. doi: 10.1016/j.tcb.2014.08.007
48. Beishline K, Vladimirova O, Tutton S, Wang Z, Deng Z, Lieberman PM. CTCF Driven TERRA Transcription Facilitates Completion of Telomere DNA Replication. Nat Commun (2017) 8:2114. doi: 10.1038/s41467-017-02212-w
49. Garbuzov A, Pech MF, Hasegawa K, Sukhwani M, Zhang RJ, Orwig KE, et al. Purification of GFRalpha1+ and GFRalpha1- Spermatogonial Stem Cells Reveals a Niche-Dependent Mechanism for Fate Determination. Stem Cell Rep (2018) 10:553–67. doi: 10.1016/j.stemcr.2017.12.009
50. Pech MF, Garbuzov A, Hasegawa K, Sukhwani M, Zhang RJ, Benayoun BA, et al. High Telomerase is a Hallmark of Undifferentiated Spermatogonia and is Required for Maintenance of Male Germline Stem Cells. Genes Dev (2015) 29:2420–34. doi: 10.1101/gad.271783.115
51. Turner S, Wong HP, Rai J, Hartshorne GM. Telomere Lengths in Human Oocytes, Cleavage Stage Embryos and Blastocysts. Mol Hum Reprod (2010) 16:685–94. doi: 10.1093/molehr/gaq048
52. Anifandis G, Samara M, Simopoulou M, Messini CI, Chatzimeletiou K, Thodou E, et al. Insights Into the Role of Telomeres in Human Embryological Parameters. Opinions Regarding IVF. J Dev Biol (2021) 9:49. doi: 10.3390/jdb9040049
53. Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, et al. Telomere Length Predicts Replicative Capacity of Human Fibroblasts. Proc Natl Acad Sci U.S.A. (1992) 89:10114–8. doi: 10.1073/pnas.89.21.10114
54. Unryn BM, Cook LS, Riabowol KT. Paternal Age is Positively Linked to Telomere Length of Children. Aging Cell (2005) 4:97–101. doi: 10.1111/j.1474-9728.2005.00144.x
55. Tardat M, Dejardin J. Telomere Chromatin Establishment and its Maintenance During Mammalian Development. Chromosoma (2018) 127:3–18. doi: 10.1007/s00412-017-0656-3
56. Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic Genome Architecture in the Nuclear Space: Regulation of Gene Expression in Three Dimensions. Nat Rev Genet (2007) 8:104–15. doi: 10.1038/nrg2041
57. Dan J, Rousseau P, Hardikar S, Veland N, Wong J, Autexier C, et al. Zscan4 Inhibits Maintenance DNA Methylation to Facilitate Telomere Elongation in Mouse Embryonic Stem Cells. Cell Rep (2017) 20:1936–49. doi: 10.1016/j.celrep.2017.07.070
58. Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, et al. Zscan4 Regulates Telomere Elongation and Genomic Stability in ES Cells. Nature (2010) 464:858–63. doi: 10.1038/nature08882
59. Markiewicz-Potoczny M, Lobanova A, Loeb AM, Kirak O, Olbrich T, Ruiz S, et al. TRF2-Mediated Telomere Protection is Dispensable in Pluripotent Stem Cells. Nature (2021) 589:110–5. doi: 10.1038/s41586-020-2959-4
60. Zyner KG, Simeone A, Flynn SM, Doyle C, Marsico G, Adhikari S, et al. G-Quadruplex DNA Structures in Human Stem Cells and Differentiation. Nat Commun (2022) 13:142. doi: 10.1038/s41467-021-27719-1
61. Awad A, Glousker G, Lamm N, Tawil S, Hourvitz N, Smoom R, et al. Full Length RTEL1 is Required for the Elongation of the Single-Stranded Telomeric Overhang by Telomerase. Nucleic Acids Res (2020) 48:7239–51. doi: 10.1093/nar/gkaa503
62. Rowland TJ, Dumbovic G, Hass EP, Rinn JL, Cech TR. Single-Cell Imaging Reveals Unexpected Heterogeneity of Telomerase Reverse Transcriptase Expression Across Human Cancer Cell Lines. Proc Natl Acad Sci U.S.A. (2019) 116:18488–97. doi: 10.1073/pnas.1908275116
63. Chen LY, Redon S, Lingner J. The Human CST Complex is a Terminator of Telomerase Activity. Nature (2012) 488:540–4. doi: 10.1038/nature11269
64. Zaug AJ, Lim CJ, Olson CL, Carilli MT, Goodrich KJ, Wuttke DS, et al. CST Does Not Evict Elongating Telomerase But Prevents Initiation by ssDNA Binding. Nucleic Acids Res (2021) 49:11653–65. doi: 10.1093/nar/gkab942
65. Traczyk A, Liew CW, Gill DJ, Rhodes D. Structural Basis of G-Quadruplex DNA Recognition by the Yeast Telomeric Protein Rap1. Nucleic Acids Res (2020) 48:4562–71. doi: 10.1093/nar/gkaa171
66. Sato K, Martin-Pintado N, Post H, Altelaar M, Knipscheer P. Multistep Mechanism of G-Quadruplex Resolution During DNA Replication. Sci Adv (2021) 7:eabf8653. doi: 10.1126/sciadv.abf8653
67. Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM Helicase Facilitates Telomere Replication During Leading Strand Synthesis of Telomeres. J Cell Biol (2015) 210:191–208. doi: 10.1083/jcb.201410061
68. Wu Y, Brosh RM Jr. G-Quadruplex Nucleic Acids and Human Disease. FEBS J (2010) 277:3470–88. doi: 10.1111/j.1742-4658.2010.07760.x
69. Ruis P, van Ly D, Borel V, Kafer GR, Mccarthy A, Howell S, et al. TRF2-Independent Chromosome End Protection During Pluripotency. Nature (2021) 589:103–9. doi: 10.1038/s41586-020-2960-y
70. Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Cancer-Associated TERT Promoter Mutations Abrogate Telomerase Silencing. Elife (2015) 4:e07918. doi: 10.7554/eLife.07918
71. Takahashi N, Davy PM, Gardner LH, Mathews J, Yamazaki Y, Allsopp RC. Hypoxia Inducible Factor 1 Alpha Is Expressed in Germ Cells Throughout the Murine Life Cycle. PLoS One (2016) 11:e0154309. doi: 10.1371/journal.pone.0154309
72. Otero-albiol D, Carnero A. Cellular Senescence or Stemness: Hypoxia Flips the Coin. J Exp Clin Cancer Res (2021) 40:243. doi: 10.1186/s13046-021-02035-0
73. Lansdorp PM. Sex Differences in Telomere Length, Lifespan, and Embryonic Dyskerin Levels. Aging Cell (2022) 21:e13614. doi: 10.1111/acel.13614
74. Penev A, Bazley A, Shen M, Boeke JD, Savage SA, Sfeir A. Alternative Splicing is a Developmental Switch for hTERT Expression. Mol Cell (2021) 81:2349–60.e6. doi: 10.1016/j.molcel.2021.03.033
75. Abascal F, Harvey LMR, Mitchell E, Lawson ARJ, Lensing SV, Ellis P, et al. Somatic Mutation Landscapes at Single-Molecule Resolution. Nature (2021) 593:405–10. doi: 10.1038/s41586-021-03477-4
76. Chin L, Artandi SE, Shen Q, Tam A, Lee SL, gottlieb GJ, et al. P53 Deficiency Rescues the Adverse Effects of Telomere Loss and Cooperates With Telomere Dysfunction to Accelerate Carcinogenesis. Cell (1999) 97:527–38. doi: 10.1016/S0092-8674(00)80762-X
77. Martens UM, Zijlmans JM, Poon SS, Dragowska W, Yui J, Chavez EA, et al. Short Telomeres on Human Chromosome 17p. Nat Genet (1998) 18:76–80. doi: 10.1038/ng0198-76
79. Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, et al. TP53 Copy Number Expansion is Associated With the Evolution of Increased Body Size and an Enhanced DNA Damage Response in Elephants. Elife (2016) 5:e11994. doi: 10.7554/eLife.11994
80. Martens UM, Chavez EA, Poon SS, Schmoor C, Lansdorp PM. Accumulation of Short Telomeres in Human Fibroblasts Prior to Replicative Senescence. Exp Cell Res (2000) 256:291–9. doi: 10.1006/excr.2000.4823
81. Weng NP, Granger L, Hodes RJ. Telomere Lengthening and Telomerase Activation During Human B Cell Differentiation. Proc Natl Acad Sci U.S.A. (1997) 94:10827–32. doi: 10.1073/pnas.94.20.10827
82. Martens UM, Brass V, Sedlacek L, Pantic M, Exner C, Guo Y, et al. Telomere Maintenance in Human B Lymphocytes. Br J Haematol (2002) 119:810–8. doi: 10.1046/j.1365-2141.2002.03910.x
83. Knudson M, Kulkarni S, Ballas ZK, Bessler M, Goldman F. Association of Immune Abnormalities With Telomere Shortening in Autosomal-Dominant Dyskeratosis Congenita. Blood (2005) 105:682–8. doi: 10.1182/blood-2004-04-1673
84. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-Reactive T Cells in Healthy Donors and Patients With COVID-19. Nature (2020) 587:270–4. doi: 10.1038/s41586-020-2598-9
85. Wang Q, Codd V, Raisi-Estabragh Z, Musicha C, Bountziouka V, Kaptoge S, et al. Shorter Leukocyte Telomere Length is Associated With Adverse COVID-19 Outcomes: A Cohort Study in UK Biobank. EBioMedicine (2021) 70:103485. doi: 10.1016/j.ebiom.2021.103485
86. Aubert G, Baerlocher GM, Vulto I, Poon SS, Lansdorp PM. Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes. PLoS Genet (2012) 8:e1002696. doi: 10.1371/journal.pgen.1002696
87. Baerlocher GM, Rice K, Vulto I, Lansdorp PM. Longitudinal Data on Telomere Length in Leukocytes From Newborn Baboons Support a Marked Drop in Stem Cell Turnover Around 1 Year of Age. Aging Cell (2007) 6:121–3. doi: 10.1111/j.1474-9726.2006.00254.x
88. Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a Mitotic Clock in Human Hematopoietic Stem Cells: Loss of Telomeric DNA With Age. Proc Natl Acad Sci U.S.A. (1994) 91:9857–60. doi: 10.1073/pnas.91.21.9857
89. Von Zglinicki T. Oxidative Stress Shortens Telomeres. Trends Biochem Sci (2002) 27:339–44. doi: 10.1016/S0968-0004(02)02110-2
90. Shlush LI, Skorecki KL, Itzkovitz S, Yehezkel S, Segev Y, Shachar H, et al. Telomere Elongation Followed by Telomere Length Reduction, in Leukocytes From Divers Exposed to Intense Oxidative Stress–Implications for Tissue and Organismal Aging. Mech Ageing Dev (2011) 132:123–30. doi: 10.1016/j.mad.2011.01.005
91. Garrett-Bakelman FE, Darshi M, Green SJ, Gur RC, Lin L, Macias BR, et al. The NASA Twins Study: A Multidimensional Analysis of a Year-Long Human Spaceflight. Science (2019) 364:eaau8650. doi: 10.1126/science.aau8650
Keywords: telomerase, telomere length measurements, telomere replication, telomere length regulation, development, quadruplex DNA, tumor suppression, lifespan
Citation: Lansdorp P (2022) Telomere Length Regulation. Front. Oncol. 12:943622. doi: 10.3389/fonc.2022.943622
Received: 13 May 2022; Accepted: 13 June 2022;
Published: 04 July 2022.
Edited by:
Susan M. Bailey, Colorado State University, United StatesReviewed by:
Sara Selig, Technion Israel Institute of Technology, IsraelCopyright © 2022 Lansdorp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Lansdorp, cGxhbnNkb3JAYmNjcmMuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.