
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 12 July 2022
Sec. Gynecological Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.941741
Background: Emerging evidence has demonstrated a close association between perturbations in vaginal microbiota composition in women and human papillomavirus (HPV) infection, cervical lesions, and cervical cancer (Ca); however, these findings are highly heterogeneous and inconclusive.
Aim: To perform a comprehensive systematic review of the global disturbance in the vaginal microbiota, specifically in women with HPV-associated cervical diseases, and to further conduct within- and across-disease comparisons.
Method: Twenty-two records were identified in a systematic literature search of PubMed, Web of Science, and Embase up to February 28, 2022. We extracted microbial changes at the community (alpha and beta diversity) and taxonomic (relative abundance) levels. Within- and across-disease findings on the relative abundance of taxonomic assignments were qualitatively synthesized.
Results: Generally, significantly higher alpha diversity was observed for HPV infection, cervical lesions, and/or cancer patients than in controls, and significant differences within beta diversity were observed for the overall microbial composition across samples. In within-disease comparisons, the genera Gardnerella, Megasphaera, Prevotella, Peptostreptococcus, and Streptococcus showed the greatest abundances with HPV infection; Sneathia and Atopobium showed inconsistent abundance with HPV infection, and Staphylococcus was observed in Ca. Across diseases, we find increased levels of Streptococcus and varying levels of Gardnerella were shared across HPV infections, high-grade squamous intraepithelial lesions, and Ca, whereas Lactobacillus iners varied depending on the HPV-related disease subtype.
Conclusions: This systematic review reports that vaginal microbiome disturbances are correlated to the depletion of Lactobacillus, enrichment of anaerobes, and increased abundance of aerobic bacteria in HPV infection and related cervical diseases. Moreover, L. iners may exert either protective or pathogenic effects on different HPV-related diseases.
Cervical cancer (Ca) remains the fourth most prevalent cancer in women worldwide, with 6,04,127 new cases just in 2020, and more than 3,41,831 deaths, accounting for nearly 8% of all female cancer-related deaths every year (1). This common infection-related neoplasm and its premalignant precursors are caused by high-risk human papillomavirus (HR-HPV) infections. HR-HPV persistence is a crucial contributor to the pathophysiology of cervical lesions and cancer, with preneoplastic lesions taking several years to develop (2). However, a major fraction of these patients undergo clinical HPV clearance within a few months, indicating that other potential cofactors may be involved in the progression of cervical carcinogeneses, such as vaginal microbiome disturbance.
Evidence has indicated a close relationship between the microbiome of the lower genital tract and gynecological diseases, such as polycystic ovary syndrome (PCOS), endometriosis, and HPV-related cervical diseases. Bacterial vaginosis-related (BV) microbes have been detected in PCOS and endometriosis in previous studies (3, 4). Epidemiological studies have identified relationships between vaginal dysbiosis and HPV-related diseases, particularly in BV, in relation to the risk factors involved in the initiation and progression of HPV infection (5–7). Advances in molecular microbiology have revealed perturbations in the vaginal microbiota composition in HPV-induced cervical diseases (8, 9), and cultivation-independent high-throughput sequencing has provided insights into the global patterns of vaginal microbiomes (10–12). Currently, a growing body of observational and interventional research has provided data for microbial characterization of the continuum of HPV-mediated cervical diseases. Three meta-analysis linked the epidemiological relationships among vaginal dysbiosis, HPV infection, and related cervical diseases, with case-control studies and observational investigations of vaginal dysbiosis-associated risk (6, 7). The remaining network meta-analysis of cross-sectional and longitudinal studies examined the risk of certain bacterial community types in relation to perturbations in the vaginal microbiota configuration (13). However, data have not been obtained to determine the amount and/or composition of vaginal microbiota that could potentially affect the progression of HPV infection to high-grade squamous intraepithelial lesions (HSIL) or carcinoma. In addition, the overlap of findings across the spectrum of HPV-related diseases has not yet been investigated.
Regarding specific taxa, the most common anaerobes were reported to be associated with HPV infection and cervical dysplasia (i.e., G. vaginals, Megasphaera, Prevotella, and Sneathia) (14), whereas Sneathia and Atopobium were found to be inconsistently abundant (15, 16). Other members (i.e., Streptococcus, Staphylococcus, Corynebacterium, Clostridium) have also been reported to be associated with cervical lesions and Ca (17–19). Additionally, the presence of cervical lesions and Ca among women has been correlated with an increase in Lactobacillus iners (13, 20) and a decrease in Lactobacillus jensenii and Lactobacillus crispatus (20). However, conflicting results have been obtained in terms of study- and method-related heterogeneity.
Therefore, we conducted an updated systematic review of vaginal microbiome studies to characterize the microbial disturbances in women with HPV infection, HPV persistence, cervical lesions, and cervical cancer. This review aims to synthesize the findings of previous studies and conduct comparisons between- and across-diseases.
The present review was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
A systematic literature search of PubMed, Web of Science, and Embase was carried out for articles published between January 01, 2000, and February 28, 2022. The keywords included HPV [all fields], HPV infection [all fields], human papillomavirus [all fields], cervical intraepithelial neoplasia (CIN) [all fields], cervical lesion [all fields], cervical dysplasia [all fields], cervical neoplasm [all fields], cervical cancer [all fields], cervical carcinoma [all fields], microbiome [all fields], microbiota [all fields], and flora [all fields].
The inclusion criteria for the study, were as follows (1): all observational studies (including case-control, cross-sectional, prospective, and retrospective cohort studies or interventional studies) (2); women with HPV infection (HPV+), HPV persistence, low-grade squamous intraepithelial lesions (LSIL), HSIL, and Ca – categorized according to the results of HPV testing and histology of cervical biopsy, compared to healthy controls (HPV-negative and cytology-negative women) or HPV-negative women (HPV-), cervical cytology was used for classification if histology was not reported (3); studies reporting microbiota analyses of vaginal samples based on high-throughput sequencing (4); studies with information on vaginal microbial alterations at the community level (alpha and beta diversity) and/or taxonomic levels (relative abundance of different microbial species).
Records were excluded if (1) they did not use high-throughput sequencing approaches (2), sequenced the microbiota from cervical or cervicovaginal swabs [because of anatomy-associated discrepancies in the microbiome (21)] (3), analyzed the microbiome in intestinal samples (4), were duplicates (5), were not published in English, or were review articles or conference abstracts.
Prospective or interventional studies reporting altered microbial composition without relevant baseline measurements were also excluded.
Study characteristics and microbiome quantification methods were extracted from the included references. Study characteristics include the following details: publication information, subject demographics, and clinical features, whereas the latter consists of a sequencing platform, sequencing target, DNA extraction, data analysis pipelines, and reference databases. As major outcomes of the analysis, we extracted microbial alterations at the community level (alpha and beta diversity) as well as taxonomic levels (relative abundance of different microbial species). Within- and across-disease findings for the relative abundance of taxonomic assignments were qualitatively synthesized and categorized as increased, decreased, or inconsistent abundancies. No findings were consistent, with less than 75% agreement between studies reporting this taxon (22).
We utilized the National Institutes of Health (NIH) National Health, Lung, and Blood Institute Study Quality Assessment Tool for Observational and Cross-sectional Studies to evaluate the internal validity and potential bias of the included studies. Study quality was rated as “good”, “fair, or poor” by two authors (MW and HL), with differences addressed through discussion.
A total of 3322 relevant records were obtained in the initial electronic search, including 1042 from PubMed, 912 from Web of Science, and 1368 from Embase. Six hundred ninety-nine studies were imported after removing duplicates and screening the titles and/or abstracts. Of those, studies were excluded due to non-vaginal samples (n=15), non-high-throughput sequencing approaches (n=2), no information on any change in taxa compared with HPV- (n=1), and longitudinal studies in the absence of controls (n=2) after assessing the full text (n=42). Ultimately, 22 studies were selected for the final analysis. A flowchart of this process is shown in Figure 1.
The 22 studies consisted of 17 case-control, three cross-sectional, and two longitudinal studies. Most studies (72.7%, 16/22) were conducted in Asia (China, Korea), five (22.7%) were performed in Western countries (USA, UK, Mexico, and Sweden), and one (4.5%) was performed in Africa (Nigeria). In the 17 case-control comparisons, six studies compared HPV+ or HPV persistence to the controls, ten studies compared women with cervical lesions and/or Ca to the controls, and 3 included women with HPV infection. Only one study compared ≥ HSIL with LSIL. Of these, one study grouped samples into LSIL, HSIL, and normal cytology groups based on observed cytology. In the two prospective studies, comparisons were performed between cases before treatment (local excisional treatment and neoadjuvant chemotherapy) and untreated controls. The detailed information is provided in Table 1.
Control samples were defined as healthy women in most studies, whereas in two studies the controls were defined as having normal cytology. The controls were HPV positive in one study.
The different microbiome analysis methods are shown in Table 2. Sequencing approaches included 16S rRNA (95.5%, 21/22) and metagenomics (4.5%, 1/22). Among the studies employing 16S sequencing, sequencing was predominantly performed using the Illumina MiSeq platform (52.4%, 11/21), followed by the HiSeq platform (33.3%, 7/21), the Ion Torrent PGM platform (9.5%, 2/21), and the Novaseq platform (4.8%, 1/21). Five different hypervariable regions were amplified, including V1-V2 (4.8%, 1/21), V1-V3 (4.8%, 1/21), V3 (9.5%, 2/21), V4 (42.9%, 9/21), and V3-V4 (33.3%, 7/21). However, one study did not provide a sequencing target. The bioinformatics pipelines used included QIIME (52.4%, 11/21), Mothur (19.0%, 4/21), QIIME and DATA2 (4.8%, 1/21), DATA2 (4.8%, 1/21), and USEARCH (4.8%, 1/21). The widely used reference databases for the taxonomic assignment were the SILVA (33.3%, 7/21), Greengenes (23.8%, 5/21), and Ribosomal Database Project (14.3%, 3/21) databases.
A total of 57 alpha-diversity assessments were performed across 21 studies (Figure 2) Reported metrics included measurements of richness (observed species/operational taxonomic units, Chao1, abundance-based coverage estimator; 43.9%, 25/57), richness and evenness (Shannon, Simpson, inverse Simpson; 51.7%, 30/58), biodiversity (Faith’s phylogenetic diversity, 1.8%, 1/57) and sequencing depth (Good’s coverage, 1.8%, 1/57). Most of the reported metrics were based on the Shannon index (36.8%, 21/57).
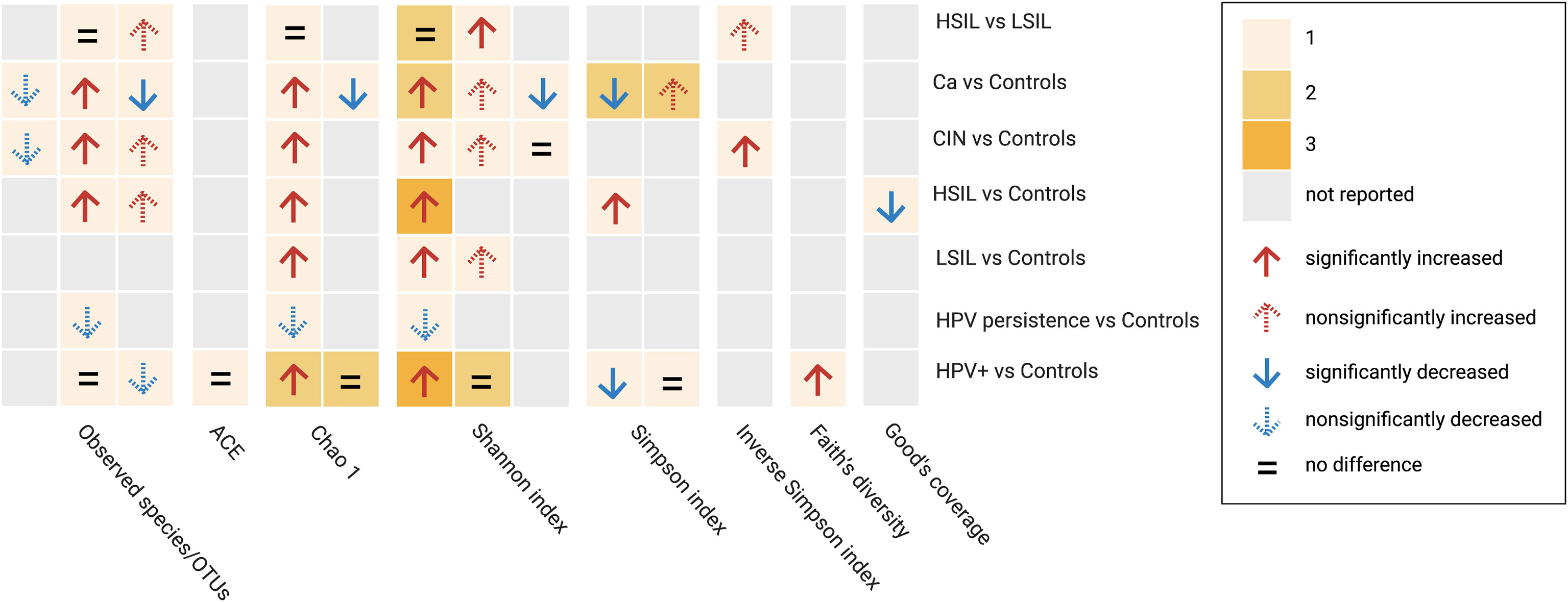
Figure 2 Alpha diversity assessments reported by studies between subgroup and controls based on different metrics. Cells are colored based on the number of assessments. HPV+, HPV-positive women; LSIL, low-grade squamous intraepithelial lesions; HSIL; high-grade squamous intraepithelial lesions; CIN, cervical intraepithelial neoplasia; Ca, cervical cancer; OTUs, operational taxonimic units.
When comparing HPV+ vs. controls or HPV−, five studies examined community richness using different metrics. Two studies indicated that there were no significant differences, two found significantly higher richness indices, whereas one found a non-significant decrease in richness indices. Regarding diversity, combining richness and evenness, significantly higher diversity was observed in three studies, significantly lower diversity was observed in one study, and no significant differences were observed in three studies. One study found a significant increase in Faith’s phylogenetic diversity.
When comparing HPV persistence vs. controls, lower richness indices were reported in two studies, and lower alpha diversity based on a combination of richness and evenness data was obtained in one study.
In comparisons between CIN and controls, two studies investigated higher richness and diversity by incorporating richness evenness indices in LSIL, four studies reported increases in both metrics in HSIL, and four studies reported a similar pattern in CIN. Among women with Ca, 4 studies reported differential richness data. Higher richness was reported in 2 studies, and lower richness was reported in the remaining studies. Differential diversity indices combining richness and evenness were reported in five studies, four of which reported higher diversity and two of which reported lower diversity. Four studies examined diversity based on a comparison between HSIL and LSIL, two of which reported no significant observations in terms of richness and/or evenness data, with the remaining studies providing higher indices. In addition, a significantly lower Good’s coverage was reported in one study comparing HSIL and controls.
Beta diversity comparisons were evaluated in 15 studies, with 13 studies comparing subgroup vs. controls, one study comparing combined subgroups vs. controls, and one study comparing within-subgroup results. Six of 13 studies identified significant differences between different samples, with 4 of 13 showing visually separated components with no significance, 2 finding no clear separation between groups, and 1 study without a report. In the across-group (controls excluded) comparisons, two studies reported no significant differences between subgroups (≥ HSIL vs. ≤ LSIL, HSIL vs. LSIL, and CIN vs. HPV+). One subgroup (HPV+ + LSIL + Ca) was significantly different from the controls. This study also showed a significant difference between the combined subgroup (LSIL + HSIL) and HPV infections. The beta diversity assessments are presented in Table 3.
Twenty-one studies have reported the relative abundance of vaginal microorganisms at the phylum, family, genus, and species levels. Between-subgroup comparisons of CIN were conducted relative to controls or in the order of abundance in subgroups reported by previous studies.
Figure 2 presents a summary of within- and across-disease comparisons for HPV-related diseases at the phylum, genus, and species levels. Taxa reported as increased or decreased in the subgroups of each study were combined. High inconsistencies were observed at different taxonomic levels in both comparisons. The following results were obtained for the taxa provided by at least two studies:
(1)In HPV+, the abundance of thirteen taxa increased or decreased: the nine with increased abundancies included the phyla Actinobacteria, Bacteroidetes, and Fusobacteria; the genera Gardnerella, Megasphaera, Peptostreptococcus, and Streptococcus; the species L. jensenii and Veillonella montpellierensis); the four taxa with a decrease in abundance included the phylum Firmicutes, the genus Lactobacillus, and the species L. crispatus and L. iners). Prevotella, Sneathia, Atopobium, Anaerococcus, and Gardnerella vaginalis were inconsistently identified (Figure 3).
(2)In HPV persistence, Lactobacillus numbers were decreased (Figure 3).
(3)In CIN, one increase in abundance in taxon (Sneathia) and two decrease in abundance in taxa (the phylum Firmicutes and the genus Lactobacillus) were observed in LSIL; five increased taxa (phylum Bacteroidetes, genera Prevotella, Streptococcus and Pseudomonas, and species L. iners), two decreased taxa (phylum Firmicutes and genus Lactobacillus), and three taxa with inconsistent numbers (phylum Actinobacteria, genera Gardnerella and Atopobium) were reported in HSIL (Figure 3).
(4)In Ca, the numbers of Streptococcus and Staphylococcus were increased, while Lactobacillus was decreased. Gardnerella and Megasphaera provided inconsistent data (Figure 3).
The differential numbers of some taxa were consistent between the subgroups. For example, higher Bacteroidetes was common to both HPV+ and HSIL; lower Firmicutes was common to HPV+, LSIL, and HSIL; higher Streptococcus was common to HPV+, HSIL, and Ca; lower Lactobacillus was common to all subgroups. The genera Gardnerella and Prevotella were inconsistently present in HPV+, HSIL, and Ca; Sneathia was inconsistent in HPV+ and LSIL; L. iners was inconsistent in HPV+ and HSIL (Figure 3).
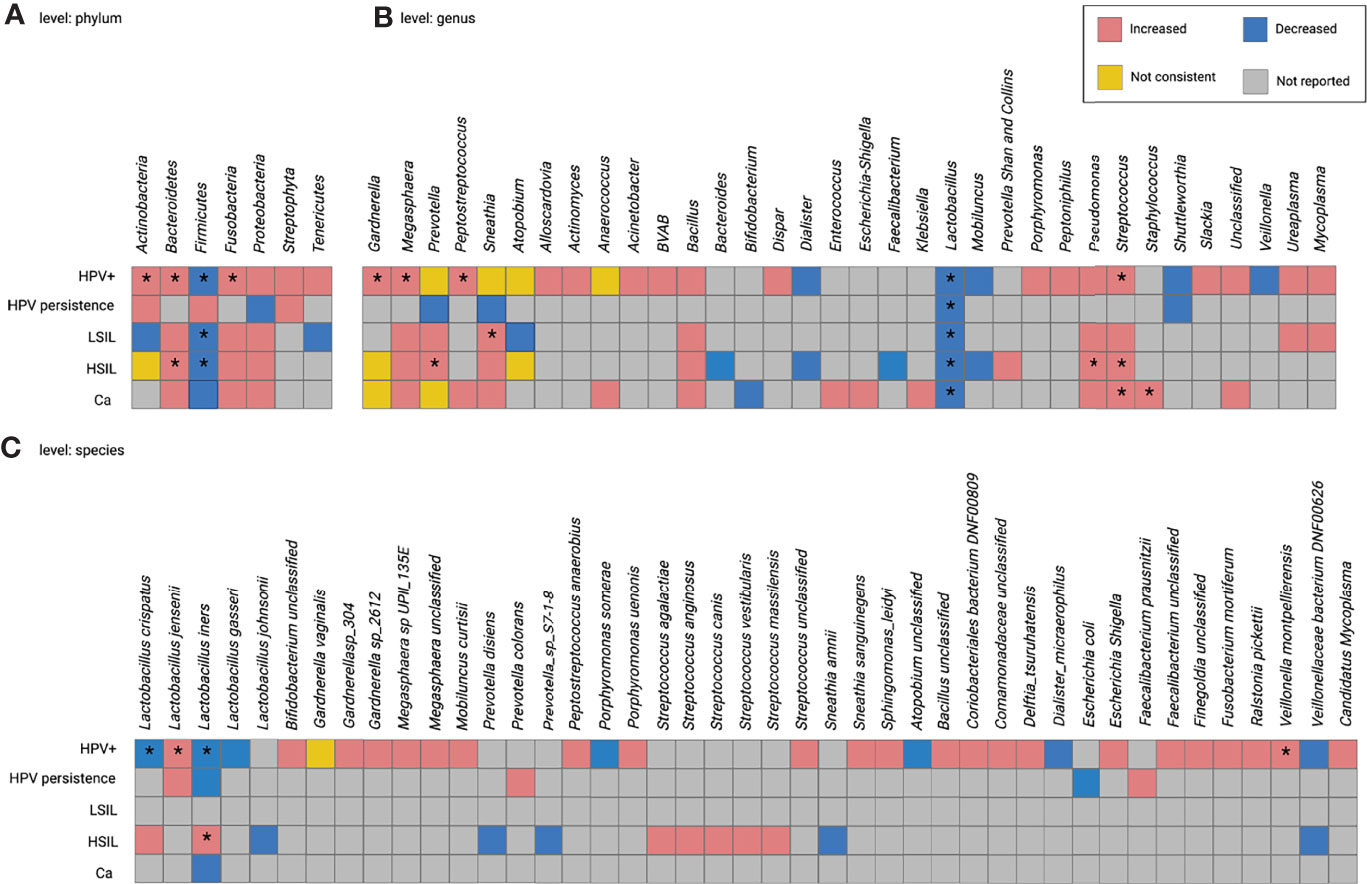
Figure 3 Within- and cross-disease changes of relative abundance of microbial taxa. *The relative abundance of taxa reported by more than 1 study. Not consistent, any finding with less than 75% agreement between studies reporting this taxon. HPV+, HPV-positive women; LSIL, low-grade squamous intraepithelial lesions; HSIL; high-grade squamous intraepithelial lesions; CIN, cervical intraepithelial neoplasia; Ca, cervical cancer.
Most studies were rated ‘fair’ (63.7%, 14/22). Only four studies were rated as ‘Good’ (18.2%, 4/22), which not only included study features, microbiome methods, and rich reporting of major outcomes but also considered several potential confounders in the reports. Four studies were rated as ‘poor’ quality (18.2%, 4/22). The major sources of potential bias were the lack of reporting of tools or methods for measuring outcomes, lack of detailed information on demographic or clinical characteristics, and lack of detailed definition of cases. The internal validity of the included studies is shown in Table 4.
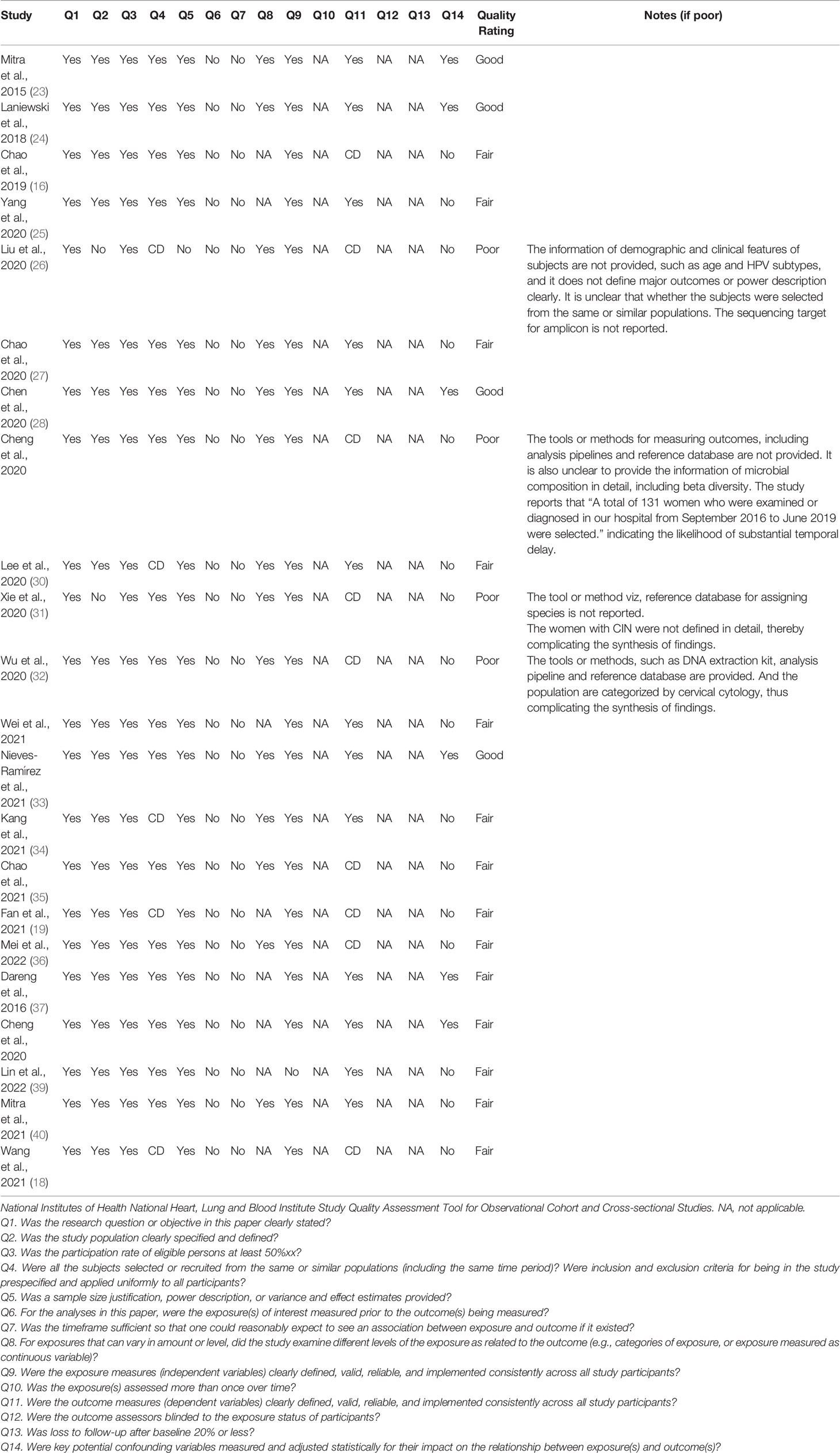
Table 4 Quality assessment of included studies using the Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies.
Our systematic review evaluated vaginal microbiome disturbances across a spectrum of HPV-associated diseases to analyze compositional alterations and perform within- and across-disease comparisons. A total of 22 studies were included in the meta-analysis. First, we outline the variances in both the study features and methodology. Next, a qualitative synthesis indicated that the alpha diversity was significantly higher than that of the controls, whereas the beta diversity showed a significant difference in the overall microbial composition across samples. The patterns of observed taxonomic changes revealed a large heterogeneity at the within- and across-disease levels. When considering alterations in abundance in at least two studies, we analyzed the alterations in consistent and inconsistent directions, within the disease. In addition, our results revealed an overlap across diseases in consistently and inconsistently altered taxa. The underlying mechanisms explaining the observed disturbances are discussed for both comparisons. The confounding effects of study characteristics and microbiome methodologies were also assessed.
Overall, our synthesis indicated a significant increase in richness and diversity in women with cervical lesions and cancer, but no difference was observed between HSIL and LSIL. Medium and large studies indicated a non-significant relationship between richness and/or evenness indices in women with HPV infection, suggesting that alpha diversity indices are of limited utility as a measurement of vaginal health, or for distinguishing between HPV infection and controls. A large number of intrinsic and extrinsic factors account for variations in diversity at the baseline, including race, age, menstrual cycles, sexual activities, contraception use, smoking, and diet, thereby complicating the measurement (33, 41–46).
However, reports with inconsistent data have been published. A few studies have reported a reduction in the richness and diversity of HPV infections. A similar pattern was observed in women with HPV persistence and Ca. These residual heterogeneities for certain diseases may be attributed to methodological effects and the study design. Alpha diversity is particularly affected by measurement errors in microbiome research, owing to the heavy bias of commonly used estimators (47). Although the number of sequences affects the alpha diversity estimates, several confounders in combination with data pre-processing may influence the data output (47, 48). Furthermore, richness metrics do not account for unobserved taxa or provide variance estimates (47). These multifactorial aspects may have contributed to the heterogeneous findings. It is worth noting that in one study investigating lower alpha diversity in Ca, a higher proportion of HPV+ women were a part of the control cohort, which may have resulted in less harmonization in the synthesis.
Regarding beta diversity, most studies have observed a significant cluster between the samples. In summary, two of the studies reported no significant differences between HSIL and LSIL, which is consistent with the pattern observed for alpha diversity.
Generally, when considering the differential taxa provided by more than two studies, we found that specific alterations occurred within the disease at the genus and species levels. The genus Staphylococcus was the only taxon enriched in Ca whereas, the genera Gardnerella, Megasphaera, Prevotella, Peptostreptococcus, and Streptococcus showed the most common increase in HPV infections, most of which were BV-related anaerobes, although they were also reported in HSIL and Ca. These associations are likely mediated by biological amine-induced oxidative stress since BV-related bacteria are linked to their production (14). Recent studies have highlighted the complex and bi-directional association between HPV and BV. Mechanistically, HPV inhibits basal and proinflammatory-induced host defense peptide expression by subverting the NF-κB and Wnt/β-catenin signaling cascades, thus leading to a significant reduction in the Lactobacillus species that rely on amino acid sources, which further promotes an imbalance in vaginal flora. In turn, oxidative stress resulting from BV infiltration promotes the progression of preneoplastic lesions (49).
Intriguingly, the genera Sneathia and Atopobium, two important, yet underappreciated pathogens, play controversial roles in HPV acquisition. Sneathia is the only microorganism enriched in the initiation and progression of cervical carcinogenesis (24) and arises as a consequence of the disease (14). Our analysis recapitulated that overrepresentation of Sneathia was reported in three studies comparing HPV infection and controls. Two studies reported depletion, one of which used only 16S rRNA amplicon pyrosequencing. Atopobium showed findings similar to those of our review. A plausible contribution to the pathomechanism may indicate the potential release of toxic products by adherent Sneathia to alter the characteristics of host tissue and directly mediate the effects on the cervical microenvironment (50). It is postulated that the enzyme, sialidase, facilitates the destruction of the mucus layer on the vaginal epithelium and entraps Atopobium (28). In addition, Prevotella bivia, as an early colonizer in BV, may pave the way for secondary colonizers like Atopobium and Sneathia (51). In contrast, a viral-driven over-reactive host immune response may lead to an observed decrease in the two pro-inflammatory genera (15). These findings represent a plausible mechanism to explain the contribution of Sneathia and Atopobium to the initiation of HPV infection. It is noteworthy that few studies have provided inconsistent abundance data on taxa, although such results may be more reflective of methodological aspects, rather than true intraspecific diversity.
Importantly, our observations implied that HPV-associated diseases share similar microbial alterations. Typically, we found an overlap between HPV infection, HSIL, and Ca in consistently and inconsistently altered taxa. Specifically, the anaerobe Gardnerella was enriched in HPV infection. An inconsistent change in HSIL was reported in two studies, which was also observed for Ca. These associations suggest that BV-related microbes may be considered indirect markers of sexual transmission of HPV, rather than promoters of progression to more severe lesions (52). Moreover, the genus Streptococcus was the only enriched taxon found in HPV infection, HSIL, and Ca, suggesting that enrichment of this genus may be characteristic of HPV-associated diseases, irrespective of the subtype. As described before, the genus Staphylococcus was the only taxon enriched in Ca. Consistently, 12 species were found in women with Ca, mainly Corynebacterium spp. and Staphylococcus spp., based on the identification of the cultivable aerobic bacterial microbiota (17). Notably, inflammation is crucial for the progression of cervical cancer to cancer (53, 54). A dysbiotic microbiome dominated by aerobic species can create an inflammatory environment that is favorable for tissue damage. It can also drive pathology by promoting immune evasion that favors tumor cell survival (17). Apart from BV-related anaerobes, the enrichment of other genera, such as Streptococcus and Staphylococcus, should be considered in the interpretation of vaginal microbiota during cervical carcinogenesis, which has been discussed in previous studies (52, 55). Our work strengthens this hypothesis by demonstrating that specific aerobic microorganisms may exert dominant functionality that is relevant to the progression of cervical lesions. Further studies are required to investigate the contribution of aerobic bacteria to cervical carcinogenesis.
Furthermore, L. iners, a strain that is the most common vaginal bacterium in healthy women (56), was found to be lower in HPV infection, HPV persistence, and Ca, but higher in HSIL. For example, decreased L. iners numbers have been linked to HPV infection, HPV persistence, and Ca (19, 27, 28). Alternatively, the presence of pre-cancer and cancer in women is associated with a high relative abundance of L. iners (20). This observation was partially confirmed here since two studies implicated increased numbers of L. iners in HSIL (30, 35). A plausible explanation is that species in the Lactobacillus genus play different roles in different contexts. The effect of L. iners on vaginal health may depend on specific community configuration. Genetically, this species can vary its gene expression when found within community state type (CST) IV-dominated communities (57). This intraspecific diversity may be a crucial determinant of structural stability by buffering the dominant Lactobacillus configuration against disturbances (58). A recent study indicated that the L. iners metabolite, lactate, could inhibit the proliferation and migration of cervical cancer cells (19). With respect to co-occurrence patterns, suppressed Lactobacillus species L. iners was positively associated with Gardnerella (59). BV-related microorganisms are more likely to be associated with HPV infection, thus, HPV infection may trigger BV establishment and promote the growth of L. iners, thereby promoting the progression of cervical preneoplastic lesions. However, there is little evidence regarding the potential contributors to HSIL. Similarly, one study detected a decrease in L. crispatus and L. gasseri in HPV infections, which are typically dominant in healthy vaginas (28). However, L. crispatus and L. jensenii were overrepresented in contrasting directions in HSIL and HPV infections, respectively (24, 28). Further analysis is required to determine the extent to which certain strains of Lactobacillus are protective or pathogenic in HPV-associated cervical diseases.
Among the multiple demographic and clinical features that may account for the extensive divergences across studies, existing evidence indicates that regional and control selection are dominant. The geographical region and the correlative effect of ethnicity can influence the configuration of the vaginal microbiome (60). Our findings indicate that some of the disturbances may be specific to Asian cohorts (i.e., a differential abundance of L. iners). At the same time, divergence was observed in the categorization of subgroups and controls, which is problematic in disentangling the panel of vaginal microbial disturbances observed in HPV infections and cervical neoplasms. For example, Laniewski et al. (24) reported a significant increase in L. crispatus in HSIL versus HPV infection. However, a significant increase in L. iners was observed in another group compared to that in the control group (34).
Despite advancements in bioinformatics techniques in this field, vaginal microbiota studies continue to face methodological challenges. Another source of bias is methodological variations across a range of laboratory processing methods (i.e., DNA extraction, sequencing target, and platform) and data preprocessing methods (i.e., reference database and quality filtration criteria for sequences). Our analyses may also suffer from the use of different sequencing platforms as well as differences in hypervariable regions since heterogeneity in evaluating microbial diversity has been reported (60). For example, five studies reported enriched Prevotella in women with HPV infection using 16S rRNA gene sequencing, although a decrease was observed in one study using 16S rRNA amplicon pyrosequencing. This observation was also similar to the inconsistent changes in the taxa Sneathia reported by 1 study. Technical and clinical factors across studies may increase the difficulty of comparing effect sizes, indicating the need for consistency among methodologies and encouraging data sharing with sufficient metadata. The VAginaL community state typE Nearest CentroId Assifier (VALENCIA) clustering tool (12) is a novel established resolution classification program for the assignment of vaginal microbiome patterns that may be a promising step toward methodologically reinforcing consistency and reproducibility.
Other covariates, including hormonal fluctuations, contraception use, temporal activity, and HPV subtypes, are also intimately correlated with perturbations in the vaginal microbiome (23, 33, 41, 61–63). There was evidence of an association between HPV 16/18 infection and specifically increased genera, viz., Gardnerella, Prevotella, and Atopobium (62); however, insufficient evidence was presented in our analysis. Thus, a rigorous collection of these factors and their careful consideration in assessments and interpretation should be employed by future research groups.
To our knowledge, this is the first systematic review analyzing vaginal microbiome disturbances across a spectrum of HPV-associated diseases globally, thus forming the basis for the identification of reproducible and generalizable potential biomarkers. This enabled us to further explore its clinical use in predicting the severity of HPV-related cervical diseases as a non-invasive diagnostic tool. Functional analyses have provided new insights into the roles of specific bacteria in cervical carcinogenesis. In particular, examining the role of Lactobacillus species may provide better rational targets for the advancement of novel probiotic-based prevention and treatment agents. It has been shown that maintenance of the vaginal microbiota and improvement of cervical epithelialization favors regression of cervical lesions through a prebiotic effect (64). However, the present study has some limitations. First, none of the selected studies provided microbial information for cervical or cervicovaginal samples, which prevented adequate comparison of the relationship between HPV-induced cervical disease and vaginal microbiota. The decision to remove records in these samples was dictated by knowledge of the anatomic potential and substantial site-associated discrepancies in the microbiome (21, 62). Second, the geographic distribution of the included studies indicates a high proportion of studies performed in China. This circles back to the link between regional variances and vaginal bacterial clusters, an imbalance that may have affected data synthesis in our analysis. Third, most studies had relatively small or moderate sample sizes, indicating that our results may still be underpowered. Fourth, given that the selected studies varied widely in terms of study design and bioinformatic analyses, meta-analysis of the selected studies seem complicated. Therefore, we performed qualitative systematic analyses rather than a meta-analysis. Further meta-analysis based on the harmonization of methodologies may provide robust and reproducible taxonomic changes. Finally, this systematic review aimed to integrate current analyses that commonly use 16S rRNA amplicon sequencing at the horizontal taxonomic level, rather than functional profiling. The generated evidence implies that local metabolic patterns correlated with BV include amino acid, dipeptide, polyamine, and ketone body pathways (55). Recent integrative work highlighted the close association of 3-hydroxybutyrate, macrophage migration inhibitory factors, pathobionts, and dysbiotic microbiota with cervical carcinogenesis (65). Relatively little is known about the sophisticated interactive mechanisms among vaginal microbiota, metabolites, and the host. Given the recognized functional redundancy (11), harnessing multi-omics techniques will provide a window on functional candidates or metabolites to clarify the contribution of host-microbe interplay in cervical tumorigenesis.
Going forward, definitive causality for the role of specific taxa in HPV clearance and cervical diseases will stem from a more molecular-related epidemiological study at a larger scale, as well as identification of the mechanisms involved in both clinical research and experimental animal research. It is important to consider individual- and disease-related covariates using multi-omics techniques. Moreover, elucidating the potential for therapeutic manipulations of the vaginal microbiota holds promise for improving outcomes in HPV clearance and cervical lesions.
This systematic review reports that, in HPV infection and related cervical diseases, in addition to an increase in anaerobes, an enrichment of aerobic bacteria may characterize vaginal microbiome disturbances. Lactobacillus iners may play either protective or pathogenic roles in HPV infections and cervical neoplasms. Hopefully, the altered microbial taxa established in this review can pave the way for further bacterial-driven causal studies on cervical carcinogenesis.
The original contributions presented in the study are included in the article material. Further inquiries can be directed to the corresponding author.
FX contributed to conception and design of the review. MW conducted literature search and drafted the first manuscript. HL and HY performed the data extraction. HL, HY, YY, CW, FT and AF made substantial contributions to writing the systematic review and revising it critically. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was funded by the Tianjin Municipal Science and Technology Commission Special Foundation for Science and Technology Major Projects in Control and Prevention of Major Diseases (Grant No. 18ZXDBSY00200), the General Project of the National Natural Science Foundation of China (Grant No. 82071674), the National Natural Science Foundation of China (Grant No. 82101705), the Natural Science Foundation of Tianjin Municipal Science and Technology (Grant No. 20JCYBJC00440), the Tianjin Health Science and Technology Project (Grant No. KJ20176; KJ20003), the Scientific Research Project of Tianjin Education Commission (Grant No. 2020KJ158), and Tianjin Key Medical Discipline (Specialty) Construction Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Mitra A, MacIntyre DA, Marchesi JR, Lee YS, Bennett PR, Kyrgiou M. The Vaginal Microbiota, Human Papillomavirus Infection and Cervical Intraepithelial Neoplasia: What do We Know and Where are We Going Next? Microbiome (2016) 4(1):58. doi: 10.1186/s40168-016-0203-0
3. Giampaolino P, Foreste V, Di Filippo C, Gallo A, Mercorio A, Serafino P, et al. Microbiome and PCOS: State-Of-Art and Future Aspects. Int J Mol Sci (2021) 22(4):2048. doi: 10.3390/ijms22042048
4. Oishi S, Mekaru K, Tanaka SE, Arai W, Ashikawa K, Sakuraba Y, et al. Microbiome Analysis in Women With Endometriosis: Does a Microbiome Exist in Peritoneal Fluid and Ovarian Cystic Fluid? Reprod Med Biol (2022) 21(1):e12441. doi: 10.1002/rmb2.12441
5. Gillet E, Meys JF, Verstraelen H, Bosire C, De Sutter P, Temmerman M, et al. Bacterial Vaginosis is Associated With Uterine Cervical Human Papillomavirus Infection: A Meta-Analysis. BMC Infect Dis (2011) 11:10. doi: 10.1186/1471-2334-11-10
6. Liang Y, Chen M, Qin L, Wan B, Wang H. A Meta-Analysis of the Relationship Between Vaginal Microecology, Human Papillomavirus Infection and Cervical Intraepithelial Neoplasia. Infect Agent Cancer (2019) 14:29. doi: 10.1186/s13027-019-0243-8
7. Brusselaers N, Shrestha S, van de Wijgert J, Verstraelen H. Vaginal Dysbiosis and the Risk of Human Papillomavirus and Cervical Cancer: Systematic Review and Meta-Analysis. Am J Obstet Gynecol (2019) 221(1):9–18 e8. doi: 10.1016/j.ajog.2018.12.011
8. Gao WJ, Weng JL, Gao YN, Chen XC. Comparison of the Vaginal Microbiota Diversity of Women With and Without Human Papillomavirus Infection: A Cross-Sectional Study. BMC Infect Dis (2013) 13. doi: 10.1186/1471-2334-13-271
9. Kovachev SM. Cervical Cancer and Vaginal Microbiota Changes. Arch Microbiol (2020) 202(2):323–7. doi: 10.1007/s00203-019-01747-4
10. Verstraelen H, Verhelst R, Claeys G, Temmerman M, Vaneechoutte M. Culture-Independent Analysis of Vaginal Microflora: The Unrecognized Association of Atopobium Vaginae With Bacterial Vaginosis. Am J Obstet Gynecol (2004) 191(4):1130–2. doi: 10.1016/j.ajog.2004.04.013
11. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal Microbiome of Reproductive-Age Women. Proc Natl Acad Sci U.S.A. (2011) 108 Suppl 1:4680–7. doi: 10.1073/pnas.1002611107
12. France MT, Ma B, Gajer P, Brown S, Humphrys MS, Holm JB, et al. VALENCIA: A Nearest Centroid Classification Method for Vaginal Microbial Communities Based on Composition. Microbiome (2020) 8(1):166. doi: 10.1186/s40168-020-00934-6
13. Norenhag J, Du J, Olovsson M, Verstraelen H, Engstrand L, Brusselaers N. The Vaginal Microbiota, Human Papillomavirus and Cervical Dysplasia: A Systematic Review and Network Meta-Analysis. BJOG (2020) 127(2):171–80. doi: 10.1111/1471-0528.15854
14. Kyrgiou M, Moscicki AB. Vaginal Microbiome and Cervical Cancer. Semin Cancer Biol (2022). doi: 10.1016/j.semcancer.2022.03.005
15. Wei ZT, Chen HL, Wang CF, Yang GL, Han SM, Zhang SL. Depiction of Vaginal Microbiota in Women With High-Risk Human Papillomavirus Infection. Front Public Health (2020) 8:587298. doi: 10.3389/fpubh.2020.587298
16. Chao XP, Sun TT, Wang S, Fan QB, Shi HH, Zhu L, et al. Correlation Between the Diversity of Vaginal Microbiota and the Risk of High-Risk Human Papillomavirus Infection. Int J Gynecol Cancer (2019) 29(1):28–34. doi: 10.1136/ijgc-2018-000032
17. Manzanares-Leal GL, Coronel-Martinez JA, Rodriguez-Morales M, Rangel-Cuevas I, Bustamante-Montes LP, Sandoval-Trujillo H, et al. Preliminary Identification of the Aerobic Cervicovaginal Microbiota in Mexican Women With Cervical Cancer as the First Step Towards Metagenomic Studies. Front Cell Infect Microbiol (2022) 12:838491. doi: 10.3389/fcimb.2022.838491
18. Wang Z, Xiao R, Huang J, Qin X, Hu D, Guo E, et al. The Diversity of Vaginal Microbiota Predicts Neoadjuvant Chemotherapy Responsiveness in Locally Advanced Cervical Cancer. Microb Ecol (2021). doi: 10.1007/s00248-021-01800-0
19. Fan Q, Wu Y, Li M, An F, Yao L, Wang M, et al. Lactobacillus Spp. Create a Protective Micro-Ecological Environment Through Regulating the Core Fucosylation of Vaginal Epithelial Cells Against Cervical Cancer. Cell Death Dis (2021) 12(12):1094. doi: 10.1038/s41419-021-04388-y
20. Yang X, Da M, Zhang W, Qi Q, Zhang C, Han S. Role of Lactobacillus in Cervical Cancer. Cancer Manage Res (2018) Volume 10:1219–29. doi: 10.2147/CMAR.S165228
21. Kim TK, Thomas SM, Ho MF, Sharma S, Reich CI, Frank JA, et al. Heterogeneity of Vaginal Microbial Communities Within Individuals. J Clin Microbiol (2009) 47(4):1181–9. doi: 10.1128/JCM.00854-08
22. Nikolova VL, Hall MRB, Hall LJ, Cleare AJ, Stone JM, Young AH. Perturbations in Gut Microbiota Composition in Psychiatric Disorders: A Review and Meta-Analysis. JAMA Psychiatry (2021) 78(12):1343–54. doi: 10.1001/jamapsychiatry.2021.2573
23. Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, et al. Cervical Intraepithelial Neoplasia Disease Progression is Associated With Increased Vaginal Microbiome Diversity. Sci Rep (2015) 5:16865. doi: 10.1038/srep16865
24. Łaniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, et al. Linking Cervicovaginal Immune Signatures, HPV and Microbiota Composition in Cervical Carcinogenesis in non-Hispanic and Hispanic Women. Sci Rep (2018) 8(1):7593. doi: 10.1038/s41598-018-25879-7
25. Yang Q, Wang Y, Wei X, Zhu J, Wang X, Xie X, et al. The Alterations of Vaginal Microbiome in HPV16 Infection as Identified by Shotgun Metagenomic Sequencing. Front Cell Infect Microbiol (2020) 10. doi: 10.3389/fcimb.2020.00286
26. Liu J, Luo M, Zhang Y, Cao G, Wang S. Association of High-Risk Human Papillomavirus Infection Duration and Cervical Lesions With Vaginal Microbiota Composition. Ann Transl Med (2020) 8(18):1161. doi: 10.21037/atm-20-5832
27. Chao X, Sun T, Wang S, Tan X, Fan Q, Shi H, et al. Research of the Potential Biomarkers in Vaginal Microbiome for Persistent High-Risk Human Papillomavirus Infection. Ann Transl Med (2020) 8(4):100. doi: 10.21037/atm.2019.12.115
28. Chen Y, Qiu X, Wang W, Li D, Wu A, Hong Z, et al. Human Papillomavirus Infection and Cervical Intraepithelial Neoplasia Progression are Associated With Increased Vaginal Microbiome Diversity in a Chinese Cohort. BMC Infect Dis (2020) 20(1):629. doi: 10.1186/s12879-020-05324-9
29. Cheng W, Xu F, Gao L, Liu J. The Correlation Between the Determination of Vaginal Micro-Ecological Composition and the Outcome of HPV Infection by High-Throughput Metagene Sequencing Information Technology on the Illumina Platform. J Infect Public Health (2020) 13(12):1961–6. doi: 10.1016/j.jiph.2020.05.024
30. Lee YH, Kang GU, Jeon SY, Tagele SB, Pham HQ, Kim MS, et al. Vaginal Microbiome-Based Bacterial Signatures for Predicting the Severity of Cervical Intraepithelial Neoplasia. Diagn (Basel) (2020) 10(12):1013. doi: 10.3390/diagnostics10121013
31. Xie Y, Feng Y, Li W, Zhan F, Huang G, Hu H, et al. Revealing the Disturbed Vaginal Micobiota Caused by Cervical Cancer Using High-Throughput Sequencing Technology. Front Cell Infect Microbiol (2020) 10:538336. doi: 10.3389/fcimb.2020.538336
32. Wu M, Gao J, Wu Y, Li Y, Chen Y, Zhao F, et al. Characterization of Vaginal Microbiota in Chinese Women With Cervical Squamous Intra-Epithelial Neoplasia. Int J Gynecol Cancer (2020) 30(10):1500–4. doi: 10.1136/ijgc-2020-001341
33. Nieves-Ramirez ME, Partida-Rodriguez O, Moran P, Serrano-Vazquez A, Perez-Juarez H, Perez-Rodriguez ME, et al. Cervical Squamous Intraepithelial Lesions Are Associated With Differences in the Vaginal Microbiota of Mexican Women. Microbiol Spectr (2021) 9(2):e0014321. doi: 10.1128/Spectrum.00143-21
34. Kang GU, Jung DR, Lee YH, Jeon SY, Han HS, Chong GO, et al. Potential Association Between Vaginal Microbiota and Cervical Carcinogenesis in Korean Women: A Cohort Study. Microorganisms (2021) 9(2):294. doi: 10.3390/microorganisms9020294
35. Chao X, Wang L, Wang S, Lang J, Tan X, Fan Q, et al. Research of the Potential Vaginal Microbiome Biomarkers for High-Grade Squamous Intraepithelial Lesion. Front Med (Lausanne) (2021) 8:565001. doi: 10.3389/fmed.2021.565001
36. Mei L, Wang T, Chen Y, Wei D, Zhang Y, Cui T, et al. Dysbiosis of Vaginal Microbiota Associated With Persistent High-Risk Human Papilloma Virus Infection. J Transl Med (2022) 20(1):12. doi: 10.1186/s12967-021-03201-w
37. Dareng EO, Ma B, Famooto AO, Adebamowo SN, Offiong RA, Olaniyan O, et al. Prevalent High-Risk HPV Infection and Vaginal Microbiota in Nigerian Women. Epidemiol Infect (2016) 144(1):123–37. doi: 10.1017/S0950268815000965
38. Cheng L, Norenhag J, Hu YOO, Brusselaers N, Fransson E, Ahrlund-Richter A, et al. Vaginal Microbiota and Human Papillomavirus Infection Among Young Swedish Women. NPJ Biofilms Microbiom (2020) 6(1):39. doi: 10.1038/s41522-020-00146-8
39. Lin W, Zhang Q, Chen Y, Dong B, Xue H, Lei H, et al. Changes of the Vaginal Microbiota in HPV Infection and Cervical Intraepithelial Neoplasia: A Cross-Sectional Analysis. Sci Rep (2022) 12(1):2812. doi: 10.1038/s41598-022-06731-5
40. Mitra A, MacIntyre DA, Paraskevaidi M, Moscicki AB, Mahajan V, Smith A, et al. The Vaginal Microbiota and Innate Immunity After Local Excisional Treatment for Cervical Intraepithelial Neoplasia. Genome Med (2021) 13(1):176. doi: 10.1186/s13073-021-00977-w
41. Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, et al. Temporal Dynamics of the Human Vaginal Microbiota. Sci Transl Med (2012) 4(132):132ra52. doi: 10.1126/scitranslmed.3003605
42. Lewis FMT, Bernstein KT, Aral SO. Vaginal Microbiome and Its Relationship to Behavior, Sexual Health, and Sexually Transmitted Diseases. Obstet Gynecol (2017) 129(4):643–54. doi: 10.1097/AOG.0000000000001932
43. Brotman RM, He X, Gajer P, Fadrosh D, Sharma E, Mongodin EF, et al. Association Between Cigarette Smoking and the Vaginal Microbiota: A Pilot Study. BMC Infect Dis (2014) 14:471. doi: 10.1186/1471-2334-14-471
44. Achilles SL, Austin MN, Meyn LA, Mhlanga F, Chirenje ZM, Hillier SL. Impact of Contraceptive Initiation on Vaginal Microbiota. Am J Obstet Gynecol (2018) 218(6):622.e1–.e10. doi: 10.1016/j.ajog.2018.02.017
45. Seo SS, Oh HY, Lee JK, Kong JS, Lee DO, Kim MK. Combined Effect of Diet and Cervical Microbiome on the Risk of Cervical Intraepithelial Neoplasia. Clin Nutr (2016) 35(6):1434–41. doi: 10.1016/j.clnu.2016.03.019
46. Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, et al. Differences in Vaginal Microbiome in African American Women Versus Women of European Ancestry. Microbiol (Reading) (2014) 160(Pt 10):2272–82. doi: 10.1099/mic.0.081034-0
47. Willis AD. Rarefaction, Alpha Diversity, and Statistics. Front Microbiol (2019) 10:2407. doi: 10.3389/fmicb.2019.02407
48. McGuinness AJ, Davis JA, Dawson SL, Loughman A, Collier F, O'Hely M, et al. A Systematic Review of Gut Microbiota Composition in Observational Studies of Major Depressive Disorder, Bipolar Disorder and Schizophrenia. Mol Psychiatry (2022) 24(4):1920–35. doi: 10.1038/s41380-022-01456-3
49. Lebeau A, Bruyere D, Roncarati P, Peixoto P, Hervouet E, Cobraiville G, et al. HPV Infection Alters Vaginal Microbiome Through Down-Regulating Host Mucosal Innate Peptides Used by Lactobacilli as Amino Acid Sources. Nat Commun (2022) 13(1):1076. doi: 10.1038/s41467-022-28724-8
50. Theis KR, Florova V, Romero R, Borisov AB, Winters AD, Galaz J, et al. Sneathia: An Emerging Pathogen in Female Reproductive Disease and Adverse Perinatal Outcomes. Crit Rev Microbiol (2021) 47(4):517–42. doi: 10.1080/1040841X.2021.1905606
51. Muzny CA, Laniewski P, Schwebke JR, Herbst-Kralovetz MM. Host-Vaginal Microbiota Interactions in the Pathogenesis of Bacterial Vaginosis. Curr Opin Infect Dis (2020) 33(1):59–65. doi: 10.1097/QCO.0000000000000620
52. Plisko O, Zodzika J, Jermakova I, Pcolkina K, Prusakevica A, Liepniece-Karele I, et al. Aerobic Vaginitis-Underestimated Risk Factor for Cervical Intraepithelial Neoplasia. Diagn (Basel) (2021) 11(1):97. doi: 10.3390/diagnostics11010097
53. Zhou ZW, Long HZ, Cheng Y, Luo HY, Wen DD, Gao LC. From Microbiome to Inflammation: The Key Drivers of Cervical Cancer. Front Microbiol (2021) 12:767931. doi: 10.3389/fmicb.2021.767931
54. Boccardo E, Lepique AP, Villa LL. The Role of Inflammation in HPV Carcinogenesis. Carcinogenesis (2010) 31(11):1905–12. doi: 10.1093/carcin/bgq176
55. Łaniewski P, Ilhan ZE, Herbst-Kralovetz MM. The Microbiome and Gynaecological Cancer Development, Prevention and Therapy. Nat Rev Urol (2020) 17(4):232–50. doi: 10.1038/s41585-020-0286-z
56. France M, Alizadeh M, Brown S, Ma B, Ravel J. Towards a Deeper Understanding of the Vaginal Microbiota. Nat Microbiol (2022) 7(3):367–78. doi: 10.1038/s41564-022-01083-2
57. France MT, Fu L, Rutt L, Yang H, Humphrys MS, Narina S, et al. Insight Into the Ecology of Vaginal Bacteria Through Integrative Analyses of Metagenomic and Metatranscriptomic Data. Genome Biol (2022) 23(1):66. doi: 10.1186/s13059-022-02635-9
58. Agashe D. The Stabilizing Effect of Intraspecific Genetic Variation on Population Dynamics in Novel and Ancestral Habitats. Am Nat (2009) 174(2):255–67. doi: 10.1086/600085
59. Li W, Ma ZS. Dominance Network Analysis of the Healthy Human Vaginal Microbiome Not Dominated by Lactobacillus Species. Comput Struct Biotechnol J (2020) 18:3447–56. doi: 10.1016/j.csbj.2020.10.033
60. Sirichoat A, Sankuntaw N, Engchanil C, Buppasiri P, Faksri K, Namwat W, et al. Comparison of Different Hypervariable Regions of 16S rRNA for Taxonomic Profiling of Vaginal Microbiota Using Next-Generation Sequencing. Arch Microbiol (2020) 203(3):1159–66. doi: 10.1007/s00203-020-02114-4
61. Carrillo-Ng H, Becerra-Goicochea L, Tarazona-Castro Y, Pinillos-Vilca L, Del Valle LJ, Aguilar-Luis MA, et al. Variations in Cervico-Vaginal Microbiota Among HPV-Positive and HPV-Negative Asymptomatic Women in Peru. BMC Res Notes (2021) 14(1):4. doi: 10.1186/s13104-020-05422-6
62. Zhang Z, Li T, Zhang D, Zong X, Bai H, Bi H, et al. Distinction Between Vaginal and Cervical Microbiota in High-Risk Human Papilloma Virus-Infected Women in China. BMC Microbiol (2021) 21(1):90. doi: 10.1186/s12866-021-02152-y
63. Huang X, Li C, Li F, Zhao J, Wan X, Wang K. Cervicovaginal Microbiota Composition Correlates With the Acquisition of High-Risk Human Papillomavirus Types. Int J Cancer (2018) 143(3):621–34. doi: 10.1002/ijc.31342
64. Lavitola G, Della Corte L, De Rosa N, Nappi C, Bifulco G. Effects on Vaginal Microbiota Restoration and Cervical Epithelialization in Positive HPV Patients Undergoing Vaginal Treatment With Carboxy-Methyl-Beta-Glucan. BioMed Res Int (2020) 2020:1–8. doi: 10.1155/2020/5476389
Keywords: HPV, human papillomavirus, cervical dysplasia, cervical cancer, microbiome, vaginal microbiota, systematic review
Citation: Wu M, Li H, Yu H, Yan Y, Wang C, Teng F, Fan A and Xue F (2022) Disturbances of Vaginal Microbiome Composition in Human Papillomavirus Infection and Cervical Carcinogenesis: A Qualitative Systematic Review. Front. Oncol. 12:941741. doi: 10.3389/fonc.2022.941741
Received: 11 May 2022; Accepted: 06 June 2022;
Published: 12 July 2022.
Edited by:
Stefano Restaino, Ospedale Santa Maria della Misericordia di Udine, ItalyReviewed by:
Carlo Ronsini, Università degli Studi della Campania “Luigi Vanvitelli”, ItalyCopyright © 2022 Wu, Li, Yu, Yan, Wang, Teng, Fan and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengxia Xue, eHVlZmVuZ3hpYUB0bXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.