- 1Laboratory of Cell Signal Transduction, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Henan University, Kaifeng, China
- 2School of Life Sciences, Henan University, Kaifeng, China
- 3Department of Basic Sciences Research, Shaukat Khanum Memorial Cancer Hospital and Research Centre (SKMCH&RC), Lahore, Pakistan
- 4Department of Medical Laboratory Technology, The University of Haripur, Haripur, Pakistan
Pseudogenes have been considered as non-functional genes. However, peptides and long non-coding RNAs produced by pseudogenes are expressed in different tumors. Moreover, the dysregulation of pseudogenes is associated with cancer, and their expressions are higher in tumors compared to normal tissues. Recent studies show that pseudogenes can influence the liquid phase condensates formation. Liquid phase separation involves regulating different epigenetic stages, including transcription, chromatin organization, 3D DNA structure, splicing, and post-transcription modifications like m6A. Several membrane-less organelles, formed through the liquid phase separate, are also involved in the epigenetic regulation, and their defects are associated with cancer development. However, the association between pseudogenes and liquid phase separation remains unrevealed. The current study sought to investigate the relationship between pseudogenes and liquid phase separation in cancer development, as well as their therapeutic implications.
Introduction
Cancer remains a global health threat, and its impact on human being has been intense. Excluding melanoma skin cancer, there will be an estimated 19.3 million cancer diagnoses and around 10 million deaths worldwide in 2020 (1). The intricacy of cancer growth pathways, such as changes in the cellular microenvironment, environmental variables, and aberrant gene/epigenetic expressions, influences cancer management challenges. The epigenetic expression has been linked to the development of liquid-liquid phase separation (LLPS). LLPS mediates the epigenetic expression by providing the adaptability of the cellular microenvironment according to the cellular stress (2). LLPS is formed by the interaction between RNAs-RNAs, RNAs-protein, ribonucleoprotein, proteins-proteins, and non-coding RNAs interaction, which subsequently results in the formation of dynamic condensates (3–5).
Pseudogenes have been regarded as defective duplicates of coding genes that lack a functioning gene product for decades (junk DNA). However, using advanced technologies such as RNA sequencing and proteomics analysis, the findings indicate that pseudogenes can influence gene expression by competing with miRNA for the parental gene targets, being translated into peptides, and being transcribed into long non-coding RNAs that participate in other cellular functions (6, 7). The recent tremendous breakthrough in research on cancer has explored the link between pseudogenes and cancer progression. For instance, using cancer proteomics datasets, 1970 novel peptides of pseudogenes were found in tumor tissues, where some pseudogenes-encoded peptides are tumor-specific as pseudogene RHOXF1P3 is upregulated up to 16 folds in breast cancer (8). Moreover, peptide-encoded non-coding RNAs are linked to cancer formation, with 2044 unique peptides detected in tumor samples and 426 novel peptides found in healthy tissues (8). Along the same lines, Cx43 pseudogene (ΨCx43), a pseudogene of connexin43 (Cx43), which is a gap junction protein, is highly expressed in several cancer cell lines but not in normal cell lines (9). Table 1 summarises the correlation between dysregulated pseudogenes and cancer cell proliferation, migration, and poor prognosis.
Apart from their roles in cancer development, pseudogenes affect the formation of liquid phase separation, and various findings show that LLPS affects different epigenetic expression levels. As a result, the abnormal expression of pseudogenes and the formation of LLPS plays a significant role in cancer development. The current study sought to sift through the existing literature to assess the relationship between pseudogenes and LLPS and cancer formation, which may provide a new perspective on approaching cancer development in the future.
LLPS and Epigenetic Expression Stages
Current research focuses on epigenetic expression as a result of the dysregulation of several variables that contribute to cancer formation. Among these factors, LLPS influences numerous cellular events and subsequently affects different epigenetic expression stages, including chromatin organization, histone modification, transcription factors activation, RNA splicing, non-coding RNAs metabolism, and m6A modification.
LLPS and Chromatin Organization
Genomic DNA is wrapped around histone proteins, forming the more compact and dense complex known as chromatin. This organization regulates various nuclear processes and controls histone modifications or other chromatin-binding proteins (24). The existing literature shows that the nucleosomal array mediates the LLPS, where histone1 enhances the phase separation and leads to the aggregates’ formation, while histone acetylation suppresses the condensate formation (25).
Mechanically, MeCP2 competes with linker histone H1 and compacts the nucleosomal array to maintain the chromatin structure. A recent study shows that MeCP2 promotes chromatin condensates by inducing the DNA methylation and LLPS on nucleosomal arrays (26). In the same vein, heterochromatin-binding protein HP1 serves as a transcriptional repressor by binding to methylated lysine 9 residue of histone H3 and assists the chromatin cohesion, promoting aggregation formation in the nuclear state (27). Additionally, numerous membrane-free organelles generated by LLPS (such as paraspeckles and splicing speckles) interact with chromatin via their long non-coding RNAs (28).
To efficiently package the genome, the chromatin is settled in three dimensional (3D) structure in the nuclear loci. The study shows that the variation in 3D chromatin structure promotes tumorigenesis (29). Furthermore, the evidence points out the impact of LLPS on the 3D structure, where the suppression of liquid phase separate by 1,6-hexanediol compromises the 3D structure organization in living cells (30–33). Given that histone modification and three-dimensional structure are connected with liquid phase separation, and non-coding RNAs in membraneless organelles interact with chromatin structure, LLPS plays a critical role in chromatin organization.
LLPS and Transcription Factors
Transcription factors are proteins that can bind to DNA sequences to regulate the rate of mRNA transcription process (34). To achieve transcription precision, the transcriptional factors activate LLPS to concentrate the super-enhancers, enrich the transcriptional factors, and bind the RNA polymerase II. For instance, a transcriptional co-activator, Yes-associated protein (YAP) mediates phase separate condensates formation in the nucleus. Yu et al., 2021 reported that the interferon-gamma induces cancer drug resistance by promoting the nuclear translocation and phase different condensate of YAP, and the disruption of YAP condensates suppresses the tumor growth and promote immune responses (35). Along the same lines, the RNA polymerase II is recruited in phase separation condensates during the initiation of the transcription process; then, the formed aggregate assists the CycT1 to phosphorylate the carboxy-terminal domain of RNA polymerase II, which subsequently promotes the transcription elongation of polymerase II. Co-activator MED1 and bromodomain-containing proteins like BRD4 promotes the phase separation at the super-enhancers site (which is dominated by Nanog, Sox, and Oct4) to augment the transcriptional efficacy (36).
The study supports the function of transcription factors in LLPS, demonstrating that synthetic transcription factor aggregates upregulate gene expression up to five fold in various mammalian cell lines and an in vivo model (37). TAZ, a transcriptional co-activator with PDZ binding motif, is increased in more than 20% of breast cancers and promotes the growth and spread of cancer cells. TAZ coordinates transcriptional responses by condensing its DNA-binding cofactors and co-activators via the LLPS. The deletion of the TAZ coiled-coil domain hinders the formation of LLPS and the ability of the LLPS to begin the expression of its specific genes of interest (38). Based on these findings, LLPS plays a significant role in assuring transcription accuracy by aggregating the various transcription factors.
Splicing and LLPS
Alternative RNA splicing is the main step in gene expression regulation that allows the production of different messenger RNAs of varied functions from the same gene. Aberrant alternative RNA splicing involves in pathophysiology leading to various diseases, including cardiovascular diseases, immunopathological diseases, neurological diseases, and cancer (39).
The mRNA splicing mediates cellular developmental processes by regulating the liquid phase separation formation. For instance, Embryo defective 1579, which regulates the gene transcription and mRNA splicing, induces the formation of the condensate in vitro and in vivo, and its suppression affects the global gene expression and mRNA splicing as well (40). Similarly, Ubiquitin Specific Peptidase (USP42), which involves the deubiquitination process, uses its C-terminal disordered domain to drive the phase separation of spliceosome components in regulating the various mRNA splicing events. USP42 integrates the spliceosome component PLRG1 into nuclear speckles, and its inhibition affects the splicing process, which results in cancer development (41). To the same extent, alternative splicing drives the phase separation of heterogeneous ribonucleoprotein D-like (hnRNPDL), which is known to act as a transcriptional regulator. The study reveals that a mutation of the C-terminal disordered domain of hnRNPDL promotes the formation of the aggregates and affects the splicing products (42). Kawachi et al. show that splicing of the large exons is associated with phase separation of transcription factors, where the depletion of splicing factors, such as hnRNP K and SRSF3 disrupt the condensate assemblies (43). Le et al. demonstrate that a nuclear protein known as A Kina Anchoring Protein 95 (AKAP95), mediates different cellular events, including histone modification, cell-signaling pathways, and RNA splicing can induce phase liquid separate-like aggregates. The AKAP95 requires the LLPS to control the transcription and RNA splicing effectively, and its defect in the biophysical property and aggregates formation is associated with cancer development (44). Thus, LLPS is an essential regulator of the splicing process, and its aberrant production causes malignancy.
Role of m6A in LLPS Process
m6A is the most prevalent mRNA modification, accounting for 25% of all mRNAs. m6A affects the mRNA’s placement, translation, and degradation, depending on the included transcript. To achieve these mRNA metabolism processes accurately, m6A is assisted by its readers. The m6A readers such as YTHDF1, YTHDF2, and YTHDF3 can undergo LLPS, and this phase separation process depends on the abundance of m6A (45). For instance, YTHDF2 can recruit the m6A -containing transcript into P-bodies for being degraded (46). Moreover, Translocated in LipoSarcoma/Fused, a nuclear RNA-binding that forms the membrane-less aggregates, is affected by various m6A modifications in mediating the liquid phase separate condensates (47). Lee et al. show that the m6A modification regulates gene expression through liquid phase separate intervention. Furthermore, they reveal that m6A on enhancer RNAs display the highly active enhancers and recruit m6A reader (YTHDC1) to liquid phase separate into aggregates formation, which co-mixes with BRD4. This phase-separated condensate of m6A-enhancer RNA and YTHDC1 reveals the importance of enhancer RNA modification in the LLPS and gene expression control processes (48).
Apart from its role in mediating LLPS, the dysregulation of LLPS due to the defect in m6A expression is associated with cancer. The m6A is required for YTHDC1 to undergo LLPS and form nuclear aggregates, where the number of nuclear YTHDC1-m6A aggregates is higher in acute myeloid leukemia than in normal hematopoietic stem cells (49). Based on the aforementioned evidence, m6A modification and other epigenetic mediators like chromatin organization and splicing may collaborate to influence the RNA transcript regulation/destiny and mediate the liquid-liquid phase separation process. Their dysregulation contributes to cancer development (Figure 1). As the current study fouses on m6A, future studies could explore the impact of other RNA modifications to LLPS formation.
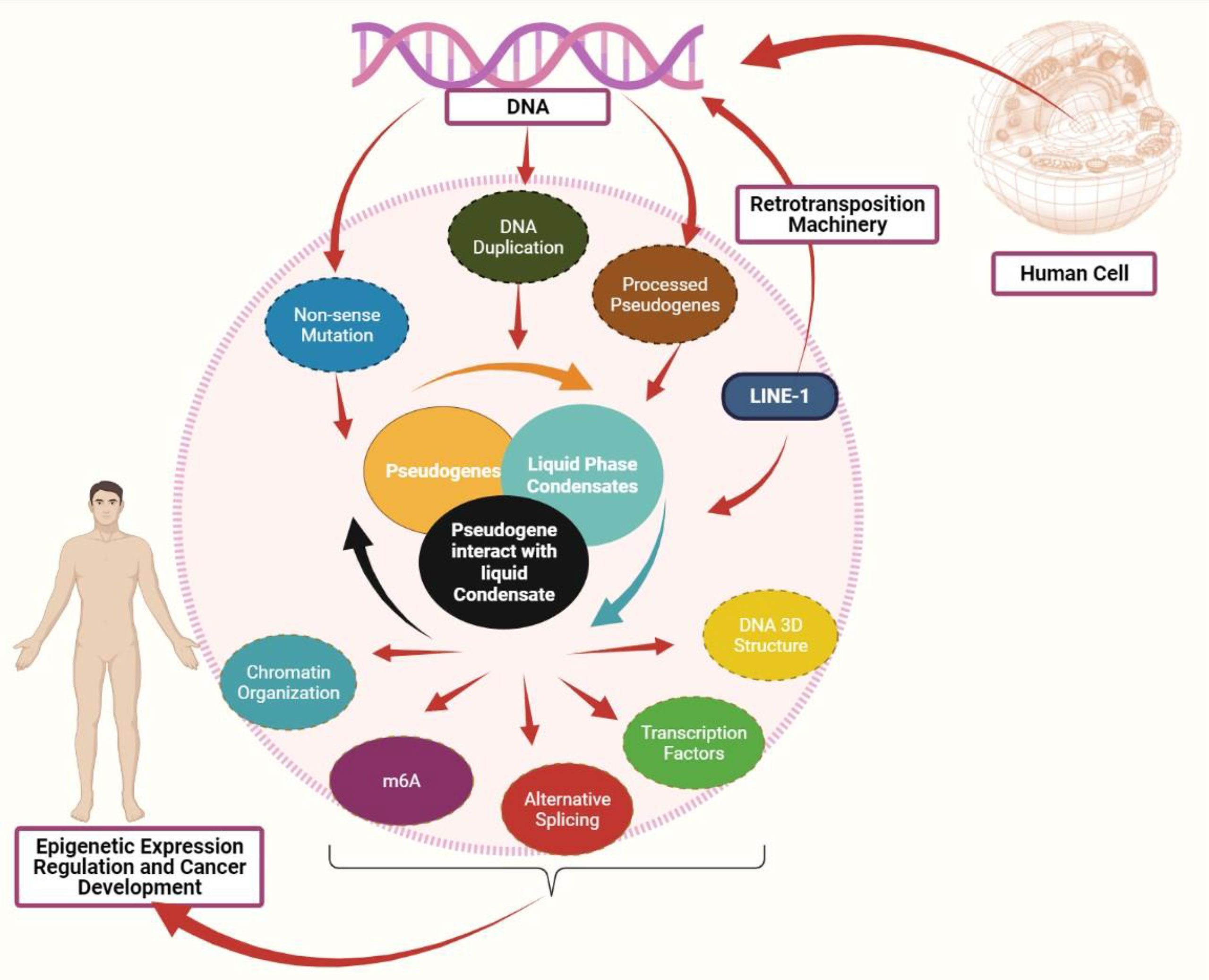
Figure 1 Interaction between pseudogenes and LLPS to mediate cancer: Pseudogenes can be produced in three processes: pseudogenes due to nonsense mutation, pseudogene due to DNA duplication, and processed pseudogenes as the result of retrotransposition machinery. Both LINE-1 and pseudogenes can mediate the liquid phase condensates formation. A defect in pseudogenes expression may affect the liquid phase separate process, eventually affecting the epigenetic mediators, including chromatin organization, DNA 3D structure, transcription factors, alternative splicing, and m6A modification. Abnormal expression in these epigenetic mediators promotes cancer development.
Pseudogenes Formation
Pseudogenes have been considered as non-functional relatives of genes that have lost their protein-coding capability as they lack the necessary sequence required for transcription or translation. More than 10% of the human genome is characterized as pseudogenes and more than 2,075 human genes are denoted by at least one pseudogene. Pseudogenes are frequently generated by DNA duplication, nonsense mutation, or mRNA transcripts that undergo reverse transcription which leads to processed pseudogenes. More than 8,000 processed pseudogenes are associated with Long Interspersed Element-1 (LINE-1 or L1) retrotransposition machinery (50). LINE-1 mRNA achieves its task by using its peptides, namely ORF1p (a nucleic acid chaperone) and ORF2p (an endonuclease and reverse transcriptase). L1 retro-transposition uses its enzymes to reverse the target mRNA transcript and integrate it into the host genome, resulting in a processed pseudogene as a final product (50).
Role of Pseudogenes in LLPS
LLPS is referred as physical changes of a substance from one state into another, where the homogeneous substance de-mixes into two liquid phase states depending on the threshold of concentration. The factors which influence the threshold concentration of substance include pH, chaperons, ATP, temperature, and posttranscriptional/posttranslational modification (51, 52). Membrane-less organelles are formed by LLPS, once diverse macromolecules such as peptides, coding RNAs, and non-coding RNA connect due to the presence of various interactions such as ionic bonds, hydrogen bonds, and van der Waals forces (53).
The main question is whether non-coding RNAs like pseudogenes can mediate the liquid phase condensates formation. To obtain the answer based on the published literature, the authors first analyzed L1 retrotransposons’ impact on liquid phase separation. As L1 retrotransposons play a critical role in processed pseudogene formation, the crosstalk of L1 retrotransposons to LLPS can indirectly affect the processed pseudogenes formation. Several findings unveil the role of L1 retrotransposons in liquid phase separate formation. The recent study discloses that LINE-1 ORF1 proteins are among other RNA-binding proteins co-localized in stress granules, and LINE-1 ORF1 proteins collaborate with stress granules and processing bodies (P-bodies) to mediate the processed pseudogenes formation (54). L1 is also involved in developing P-bodies since ORF1p co-localizes with non-L1 mRNA that is elevated in P-body granules (55).
Recent study has revealed that LLPS is regulated by intrinsically disordered proteins (IDPs). These misfolded proteins lack a proper 3D structure, which help them to form membrane-less organelles (56, 57). The alpha-synuclein and beta-amyloid are common misfolded proteins in liquid phase aggregate formation (57). As science evolves, a recent study shows that pseudogenes influence the LLPS by mediating the aggregation of these misfolded proteins. For instance, pseudogene T04B2 regulates the accumulation of beta-amyloid and alpha-synuclein proteins leading to the formation of the aggregates, where the downregulation of pseudogene TO4B2 intensifies the beta-amyloid and alpha-synuclein protein condensates formation (58). This suggests that the interplay of pseudogenes and misfolded proteins controls the liquid phase separation process.
Besides, pseudogene ACTBP2 is associated with β-Amyloid (Aβ) depositionin mediating blood-brain barrier permeability, which reveals the interaction between pseudogene and β-Amyloid in regulating barrier permeability (59). Moreover, mRNA purification in P-bodies reveals that 89% is enriched with protein-coding RNAs compared to 67% of non-coding RNAs, with the percentage of pseudogene RNAs among non-coding RNAs co-localized in the P-bodies (60). Similarly, the transcriptomes of lysate granules and stress granules show the presence of various classes of non-coding RNAs, such as long non-coding RNA and pseudogene RNAs as well (61). Another recent study shows a high 7SK small nuclear pseudogene (RN7SKP9) enrichment in stress granules (62). In addition, the nucleolusformed by LLPS involves ribosome biogenesis, cell cycle, DNA damage, and sensing the stress response (63). By using fluorescence-activated cell sorting to isolate the nucleolus and deep sequencing to characterize the nucleolus-associated domains (NADs), pseudogenes are the most enriched in NADs among non-ribosomal RNAs gene, which reveals the impact of pseudogenes in influencing the 3D chromatin structure (64).
Moreover, the dysregulation of the liquid phase separately affects nucleolus formation and leads to different diseases like ribosomopathies, neurodegenerative disease, aging, and cancer (65). The aberrant expression of pseudogenes in NADs can lead to various illnesses, based on evidence that NADs include pseudogenes and the fact that liquid phase condensate impacts the nucleolus to mediate physiology and pathology.
Another membrane-less organelle that formed via liquid phase separation is the Cajal body with coilin protein as scaffold protein. Cajal bodies are involved in the cell cycle, cell proliferation, ribonucleoprotein, and telomerase production (66). A recent study shows that the pseudogenes of coilin, coilp1 is accumulated in the nucleus, with strong accumulation in the nucleolus. The same research shows that the protein produced by pseudogene coilp1 has the ability to bind to small Cajal body-specific RNA 2 (scaRNA2) and small Cajal body-specific RNA 9 (scaRNA9) (67). The scaRNA2 is highly overexpressed in colorectal cancer than normal tissues. The findings reveal that the overexpression of scaRNA2 competes with miR-342-3p by ending up with the high expression of epidermal growth factor receptor, leading to colorectal cancer chemo-resistance (68). Increased expression of scaRNA2 also promotes cell proliferation, migration, and invasion in cutaneous squamous cell carcinoma (69).
To summarise, the data indicates that pseudogenes may play a significant role in LLPS by engaging in various misfolded proteins and co-localizing with various membrane organelles such as the nucleolus, Cajal bodies, stress granules, and P-bodies.
Pseudogenes and LLPS in Cancer
According to the aforementioned findings, LLPS significantly impacts several epigenetic stages, which therefore may mediates cancer development. The dysregulation of different membrane-less organelles formed via LLPS, including P-body, Cajal body, and stress granules, promotes cancer progression and metastasis. For instance, the mammary epithelial cells treated with transforming growth factor-beta promote the P-body formation and EMT, while the inhibition of P-body formation suppresses the EMT (70). Bearss et al. show that the enhancer of mRNA decapping 3 (EDC3) regulates the cancer cells proliferation and invasion by upregulating the P-body maturation. The inhibition of Pim1 and 3 protein kinases (which phosphorylate the EDC3) obstructs the localization of EDC3 in the P-body (71). Besides, Beneventi et al. show the role of Cajal bodies in cancer progression, where small Cajal body-specific RNAs 15 (SCARNA15) regulate the alternative splicing by modulating the pseudouridylation of U2 spliceosomal RNA, which influences the suppressor tumor genes like p53 and ATRX. Suppression of SCARNA15 downregulates the p53 expression, followed by cancer cells proliferation (72). Moreover, Adjibade et al. show how the stress granules formation affects cancer drug resistance, where Lapatinib, a tyrosine kinase inhibitor in breast cancer treatment, induces stress granules formation and suppresses the translation initiation by targeting the translation initiation factor elF2α (73). These results reveals therefore the importance of liquid phase separation and related membraneless organelles in driving cancer growth.
Apart from the involvement of LLPS in the epigenetic stages, the recent studies also show the impact of pseudogenes in these cellular processes, including splicing, transcription factors regulation, and chromatin organization. For instance, the splicing of pseudogene CYP3AP1 to CYP3A7 induces the formation of CYP3A7.1L, which has different functional properties and distribute in tissue specifically than the parental CYP3A7 enzyme (74). In the line of transcriptional activities, a transcriptional factor, Foxo3, is regulated by the expression of Foxo3 pseudogene (75). Another interesting finding is that the effects of pseudogene on the structure of chromatin are implicated by computational and experimental evidence showing a connection between the modifications of DNA and histones, as well as chromatin remodeling (76). Similarly, mOct4P4 lncRNA interacts SUV39H1 (histone methyltransferase) and RNA binding protein FUS to target its parental Oct4 promoter heterochromatin formation (77). Moreover, the recent study shows that the methylation of parental genes and pseudogenes are different in a tissue-specific manner (78). These findings reveal the role of pseudogenes in different cellular processes (Figure 2).
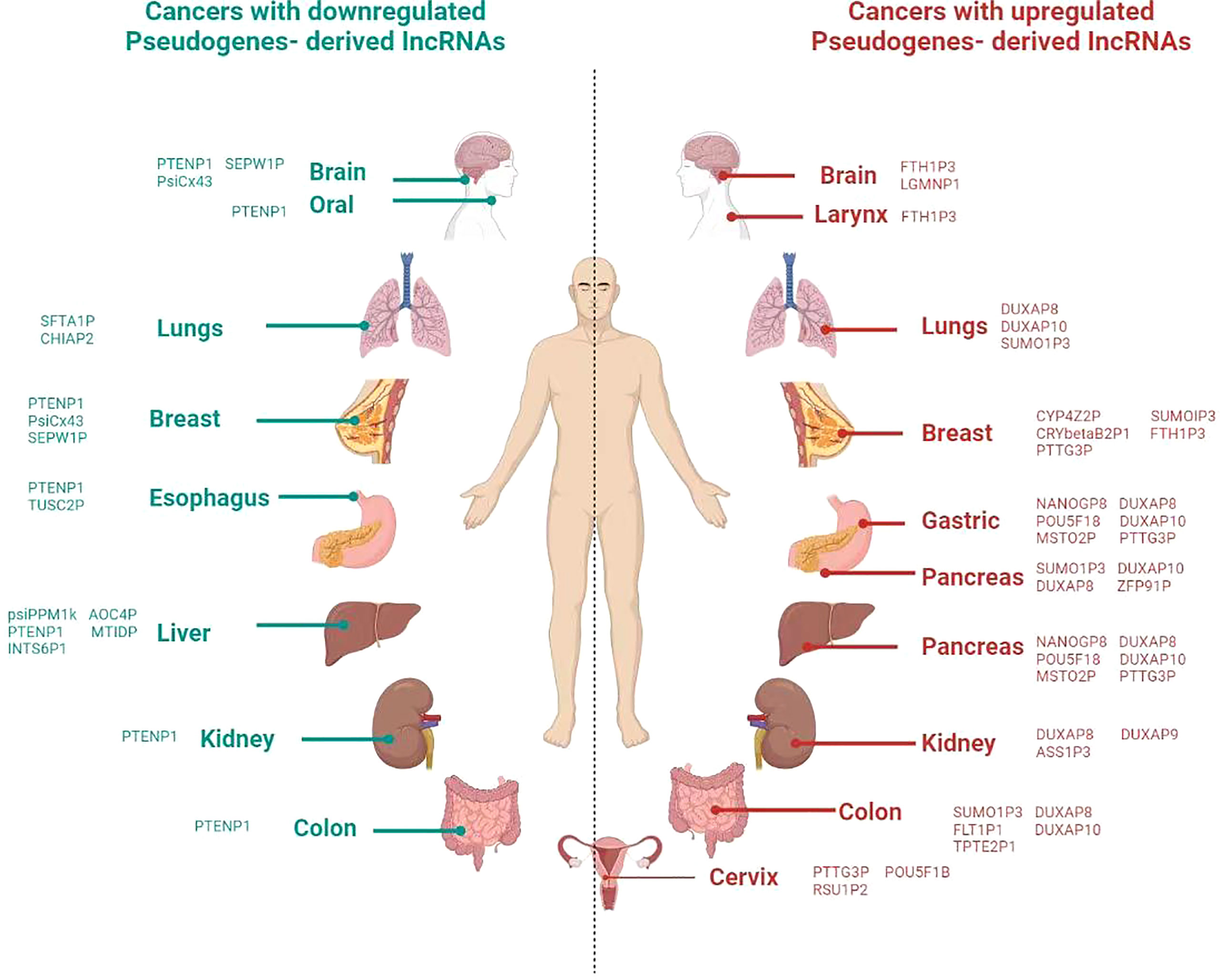
Figure 2 Overview of involvement of pseudogene in different cancer in the human body. Icons of body parts for the refrence are taken from different websources.
Based on the formation of pseudogenes, LINE-1 retrotransposons contribute a lot to produce the processed pseudogenes. Therefore, it is paramount to glance at the impact of these retrotransposons in cancer development before reaching the pseudogenes themselves. The literature shows that about half of all somatic cancers are associated with the integration of retrotransposons, and also LINE-1 retrotransposons compose about 17% of the entire DNA content (78). These evidence brought light to the hypothesis that overexpression LINE-1 retrotransposon might be considered a hallmark of many cancers. Mechanically, abnormal L1 integrations induces the loss of tumor suppressor genes or the amplification of oncogenes via breakage–fusion–bridge cycles. Pan-Cancer Analysis of Whole Genomes (PCAWG) project identified 19,166 somatically acquired retrotransposition events, which affected 35% of samples and LINE-1 variation is the most frequent in esophageal adenocarcinoma and the second in colorectal cancers (79). LINE-1 RNA is composed of two non-overlapping open reading frames, encoding two proteins, ORF1p and ORF2p. The expression level of ORF1p is 1000-10,000 times higher than ORF2p, which makes ORF2p less detectable in many cancers. The quantitative analysis shows that the LINE-1 ORF1p encoded peptides are highly overexpressed in uterine corpus endometrial carcinoma, ovarian cancer, and colon cancer compared to their respective normal samples (80).
Moreover, Cancer Genome Atlas (TCGA) analysis shows that expression levels of ORF1p bound mRNAs correlate with the expression of LINE-1 RNA in prostate cancer. In the same study, the findings show that ORF1p interacts with various non-LINE-1 mRNA targets, and these interactions were especially enriched to P-bodies in prostate cancer (55). According to the investigation of the cancer genome atlas and bioinformatics tools, the expression of ORF1p identified in the P-body is connected with the production of pseudogenes and the development of tumors (55). As the LINE-1 retrotransposition involves processed pseudogene formation, the recent study validates that retrotransposition promotes cancer growth. Retrotransposition is related to processed pseudogene insertion in small lung and colorectal cancers, where the pseudogenes are integrated into the promotor or first exon of the suppressor gene (81). Similarly, the retrocopy process is linked to structural variation in human genomes and that several prominent retrocopy insertions are present in malignancies, whereas it is absent in healthy persons (82).
Beta-amyloid fibrils and alpha-synuclein misfolded proteins interact with the pseudogene TO4B2. LLPS is known to be exacerbated by these misfolded proteins. Recent studies reveal the role of these misfolded proteins in cancer development. For instance, the accumulation of amyloid is considered as a part of the tumor microenvironment in glioma, and authors suggest that these accumulations might serve as diagnostic and therapeutic markers (83). The beta-amyloid also promotes the growth and migration of cancer cells by upregulating glial-specific fibrillary acidic protein (GFAP) expression and enhancing angiogenesis. The study also found that amyloid-beta is collected in breast cancer and that its high accumulation is related to high-grade breast cancer (84). The accumulation of amyloid-beta isassociated with the downregulation of tumor suppressor p53 in cancer (85).
To sum up, retrotransposition elements have a potential role in regulating LLPS and processed pseudogenes. Besides, the dysregulation of pseudogenes expression may affect the misfolded protein regulation. The abnormal retrotransposition elements may affect the pseudogene production, which affects the misfolded protein regulation, resulting in the disorganized liquid phase separate condensate, which can ultimately enhance tumorigenesis.
The findings from bioinformatics databases reveal that modifications and mutations in OCT4 pseudogenes are connected with low survival rate in cancer patient, indicating its potential for cancer prognosis (86). Besides, eight processed OCT4 pseudogenes, produced by the POU5F1 gene, exist in different cancer cell lines (87). Mutations in tumour suppressor genes, notably in the PTEN tumour suppressor gene, are thought to be a hallmark of cancer. Methylation of the PTENP1 pseudogene is significantly more prevalent in endometrial cancer than in normal tissues. Due to the possibility of competing for endogenous mRNAs, pseudogene methylation reduces transcription, resulting in the downregulation of the PTEN gene (88). Besides, some pseudogenes are cancer-specific, like CXADR-Ψ expression is upregulated in more than 25% of prostate cancer tissues and has no expression in normal tissues (89). For instance, the embryonic NANOG (NANOG1) gene, which is known as an essential regulator of pluripotency, its deregulation promotes cancer development. The recent study shows that NANOGP8 (NANOG-pseudogene) involves tumorigenesis, and single NANOG1-CRCs form spherical aggregates, indicating its potential in LLPS (90).
Apart from the role of pseudogenes at the transcription level, m6A on pseudogenes induce the gene expression accordingly. For instance, some RNAs of processed pseudogenes have more m6A levels than their equivalent protein-coding genes, and this modification promotes the degradation of RNA pseudogenes depending on the microRNAs’ involvement (91). Moreover, this modification assists the pseudogenes to mediate tumor growth, where m6A modified pseudogene HSPA7 regulates the immune responses in glioblastoma. The HSPA7 pseudogene triggers the development of YAP1 and LOX, which is followed by macrophage infiltration and the expression of SSP1 (92).
Taken together, the pseudogenes can influence gene expression and cancer development due to their presence in different epigenetic factors like LLPS, transcription process, m6A modification, regulating the misfolded proteins, and some of them are cancer-specific (Figure 2).
Clinical Utility of Pseudogenes and LLPS in Cancer Management
Several challenges have been identified in cancer management, including cancer relapse and chemo-resistance. It is known that the best treatment relies on accurate diagnosis. To overcome cancer therapy failure, new indicators could be investigated. Nevertheless, the new findings demonstrate that pseudogenes are elevated in cancer tissues than their normal counterparts, and their overexpressions are related to poor prognosis. For instance, pseudogene DUXAP10 is upregulated in different types of cancer (93). Besides, high expression of ANXA2 pseudogene induces a shorter overall survival in hepatocellular carcinoma patients (94). Similarly, increased expression of HSPB1P1 pseudogene is associated with poor prognosis in renal cell carcinoma (95). Thus, it reveals that pseudogenes can serve as novel markers in cancer diagnosis.
Additionally, a recent study discovered that LLPS-related genes are overexpressed in various malignancies, including ovarian epithelial carcinoma (96). Moreover, aberrant of LLPS is associated with cancer drug resistance in multiple myeloma. The mechanism behind this chemo-resistance is that the overexpression of histone methyltransferase NSD2 promotes the elevated steroid receptor coactivator-3 (SRC-3) by stimulating its aggregates formation via LLPS. Targeting this interaction using an inhibitor, SI-2, enhances the BTZ treatment functionality and overcomes this chemo-resistance (97). Along the same lines, the inhibition of core regulatory circuity, which interacts with super-enhancer to mediate LLPS at the transcriptional level, uses H3K27 demethylase inhibitor, GSK-J4, re-sensitizes the chemotherapy in osteosarcoma (98).
Following that, pseudogene can affect the liquid phase separation process. Abnormal pseudogene expression and LLPS formation can promote cancer growth, resulting in a low survival rate, a poor prognosis, and chemo-resistance (99). Therefore, impact of pseudogenes and LLPS may be used as biomarkers in cancer diagnosis, and targeting LLPS may give a novel therapeutic approach in near future.
Conclusions and Future Perspectives
Since the last few decades, much effort and research have been made to unravel the interplay of the small biomolecules involved in carcinogenesis. However, the connection between pseudogenes and the liquid phase remains fragmented. The purpose of this review is to examine the relationship between LLPS and pseudogenes and their potential impacts on cancer development. Based on the findings, pseudogenes considered junk DNA, are involved in different gene expression stages, including transcription process, post-transcription modifications, and regulating the liquid phase condensates formation.
The dynamicity of liquid phase separation (its property to mix and de-mix depending on cellular stress) greatly impacts gene epigenetic expression regulation. Its defect may result in different diseases, including infectious diseases, neurodegenerative disorders, and cancer. Since tumors may change morphology and respond in unpredictable ways, it’s critical to think about how pseudogenes (some of which are only found in tumor cells) can affect future cancer detection and treatment. Moreover, it will be interesting to explore the potential biomarkers based on LLPS in different diseases diagnosis and treatment. For instance, studies could investigate whether scaffold protein expression of the membraneless organelles can be used to differentiate cancer cells from normal cells. Furthermore, based on the fact that pseudogenes are considered as long non-coding RNAs, and their impact to LLPS is tremendous, it would be for paramount to assess the impact of microRNAs to the LLPS in the future.
Author Contributions
BN and FAK write the first draft, refined, edited, and revised the manuscript, designed the tables and figures. UAA, DW, and NF formatted and contributed to the materials organization and final editing of the manuscript. WW, WZ, and SJ conceptualized, designed and supervised the study. All authors listed have made a substantial, direct and intellectual contribution to the work and approved for publication.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31371386 SP.J).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
3D, Three dimensional; AKAP95, A Kina Anchoring Protein 95; Cx43, Connexin43; EDC3, Enhancer of mRNA decapping 3; hnRNPDL, Heterogeneous ribonucleoprotein D-like; IDPs, Intrinsically disordered proteins; LINE-1 or L1, Long Interspersed Element-1; LLPS, Liquid-liquid phase separation; NADs, Nucleolus associated domains; P-bodies, Processing bodies; PCAWG, Pan-Cancer Analysis of Whole Genomes; RN7SKP9, 7SK small nuclear pseudogene; scaRNA2, small Cajal body-specific RNA 2; TAZ, Transcriptional co-activator with PDZ binding motif; USP42, Ubiquitin Specific Peptidase; YAP, Yes-associated protein.
References
1. International Agency for Research on Cancer . Available at: https://gco.iarc.fr/today/data/factsheets/cancers/39-All-cancers-fact-sheet.pdf (Accessed November 19, 2021).
2. Harrison RES, Weng K, Wang Y, Peng Q. Phase Separation and Histone Epigenetics in Genome Regulation. Curr Opin Solid State Mater Sci (2021) 25(1):100892. doi: 10.1016/j.cossms.2020.100892
3. Liu M, Li H, Luo X, Cai J, Chen T, Xie Y, et al. A Comprehensive Database of RNAs Involved in Liquid-Liquid Phase Separation. 50(D1): D347–D355. doi: 10.1093/nar/gkab986
4. Guo Q, Shi X, Wang X. RNA and Liquid-Liquid Phase Separation. Noncoding RNA Res (2021) 6(2):92–9. doi: 10.1016/j.ncrna.2021.04.003
5. Nsengimana B, Khan FA, Ngowi EE, Zhou X, Jin Y, Jia Y, et al. Processing Body (P-Body) and its Mediators in Cancer. Mol Cell Biochem (2022) 477(4):1217–38. doi: 10.1007/s11010-022-04359-7
6. Brosch M, Saunders GI, Frankish A, Collins MO, Yu L, Wright J, et al. Shotgun Proteomics Aids Discovery of Novel Protein-Coding Genes, Alternative Splicing, and "Resurrected" Pseudogenes in the Mouse Genome. Genome Res (2011) 21(5):756–67. doi: 10.1101/gr.114272.110
7. Khan FA, Nsengimana B, Khan NH, Song Z, Ngowi EE, Wang Y, et al. Chimeric Peptides/Proteins Encoded by circRNA: An Update on Mechanisms and Functions in Human Cancers. Front Oncol (2022) 12:781270. doi: 10.3389/fonc.2022.781270
8. Xiang R, Ma L, Yang M, Zheng Z, Chen X, Jia F, et al. Increased Expression of Peptides From non-Coding Genes in Cancer Proteomics Datasets Suggests Potential Tumor Neoantigens. Commun Biol (2021) 4(1):496. doi: 10.1038/s42003-021-02007-2
9. Kandouz M, Bier A, Carystinos GD, Alaoui-Jamali MA, Batist G. Connexin43 Pseudogene is Expressed in Tumor Cells and Inhibits Growth. Oncogene (2004) 23(27):4763–70. doi: 10.1038/sj.onc.1207506
10. Lou W, Ding B, Fan W. High Expression of Pseudogene PTTG3P Indicates a Poor Prognosis in Human Breast Cancer. Mol Ther Oncolytics (2019) 14:15–26. doi: 10.1016/j.omto.2019.03.006
11. De Martino M, De Biase D, Forzati F, Credendino SC, Palma G, Barbieri A, et al. HMGA1-Pseudogene7 Transgenic Mice Develop B Cell Lymphomas. Sci Rep (2020) 10(1):7057. doi: 10.1038/s41598-020-62974-0
12. Chen X, Zhu H, Wu X, Xie X, Huang G, Xu X, et al. Downregulated Pseudogene CTNNAP1 Promote Tumor Growth in Human Cancer by Downregulating its Cognate Gene CTNNA1 Expression. Oncotarget (2016) 7(34):55518–28. doi: 10.18632/oncotarget.10833
13. Lian Y, Xiao C, Yan C, Chen D, Huang Q, Fan Y, et al. Knockdown of Pseudogene Derived From lncRNA DUXAP10 Inhibits Cell Proliferation, Migration, Invasion, and Promotes Apoptosis in Pancreatic Cancer. J Cell Biochem (2018) 119(4):3671–82. doi: 10.1002/jcb.26578
14. Esposito F, De Martino M, D'Angelo D, Mussnich P, Raverot G, Jaffrain-Rea ML, et al. HMGA1-Pseudogene Expression is Induced in Human Pituitary Tumors. Cell Cycle (2015) 14(9):1471–5. doi: 10.1080/15384101.2015.1021520
15. Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei C, et al. The Pseudogene DUXAP8 Promotes Non-Small-Cell Lung Cancer Cell Proliferation and Invasion by Epigenetically Silencing EGR1 and RHOB. Mol Ther (2017) 25(3):739–51. doi: 10.1016/j.ymthe.2016.12.018
16. Yue C, Liang C, Ge H, Yan L, Xu Y, Li G, et al. Pseudogene DUXAP10 Acts as a Diagnostic and Prognostic Marker and Promotes Cell Proliferation by Activating PI3K/AKT Pathway in Hepatocellular Carcinoma. Onco Targets Ther (2019) 12:4555–66. doi: 10.2147/OTT.S210623
17. Wei CC, Nie FQ, Jiang LL, Chen QN, Chen ZY, Chen X, et al. The Pseudogene DUXAP10 Promotes an Aggressive Phenotype Through Binding With LSD1 and Repressing LATS2 and RRAD in non Small Cell Lung Cancer. Oncotarget (2017) 8(3):5233–46. doi: 10.18632/oncotarget.14125
18. Ma H, Ma T, Chen M, Zou Z, Zhang Z. The Pseudogene-Derived Long non-Coding RNA SFTA1P Suppresses Cell Proliferation, Migration, and Invasion in Gastric Cancer. Biosci Rep (2018) 38(2):BSR20171193. doi: 10.1042/BSR20171193
19. He W, Yu Y, Huang W, Feng G, Li J. The Pseudogene DUXAP8 Promotes Colorectal Cancer Cell Proliferation, Invasion, and Migration by Inducing Epithelial-Mesenchymal Transition Through Interacting With EZH2 and H3k27me3. Onco Targets Ther (2020) 13:11059–70. doi: 10.2147/OTT.S235643
20. Ma HW, Xie M, Sun M, Chen TY, Jin RR, Ma TS, et al. The Pseudogene Derived Long non-Coding RNA DUXAP8 Promotes Gastric Cancer Cell Proliferation and Migration via Epigenetically Silencing PLEKHO1 Expression. Oncotarget (2016) 8(32):52211–24. doi: 10.18632/oncotarget.11075
21. Tian X, Song J, Zhang X, Yan M, Wang S, Wang Y, et al. MYC-Regulated Pseudogene HMGA1P6 Promotes Ovarian Cancer Malignancy via Augmenting the Oncogenic HMGA1/2. Cell Death Dis (2020) 11(3):167. doi: 10.1038/s41419-020-2356-9
22. Lian Y, Yang J, Lian Y, Xiao C, Hu X, Xu H. DUXAP8, a Pseudogene Derived lncRNA, Promotes Growth of Pancreatic Carcinoma Cells by Epigenetically Silencing CDKN1A and KLF2. Cancer Commun (Lond) (2018) 38(1):64. doi: 10.1186/s40880-018-0333-9
23. Zhang H, Chu K, Zheng C, Ren L, Tian R. Pseudogene DUXAP8 Promotes Cell Proliferation and Migration of Hepatocellular Carcinoma by Sponging MiR-490-5p to Induce BUB1 Expression. Front Genet (2020) 11:666. doi: 10.3389/fgene.2020.00666
24. Papin C, Le Gras S, Ibrahim A, Salem H, Karimi MM, Stoll I, et al. CpG Islands Shape the Epigenome Landscape. J Mol Biol (2021) 433(6):166659. doi: 10.1016/j.jmb.2020.09.018
25. Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell (2019) 179(2):470–484.e21. doi: 10.1016/j.cell.2019.08.037
26. Wang L, Hu M, Zuo MQ, Zhao J, Wu D, Huang L, et al. Rett Syndrome-Causing Mutations Compromise MeCP2-Mediated Liquid-Liquid Phase Separation of Chromatin. Cell Res (2020) 30(5):393–407. doi: 10.1038/s41422-020-0288-7
27. Qin W, Stengl A, Ugur E, Leidescher S, Ryan J, Cardoso MC, et al. Hp1β Carries an Acidic Linker Domain and Requires H3K9me3 for Phase Separation. Nucleus (2021) 12(1):44–57. doi: 10.1080/19491034.2021.1889858
28. Grosch M, Ittermann S, Shaposhnikov D, Drukker M. Chromatin-Associated Membraneless Organelles in Regulation of Cellular Differentiation. Stem Cell Rep (2020) 15(6):1220–32. doi: 10.1016/j.stemcr.2020.10.011
29. Xue Y, Yang Y, Tian H, Quan H, Liu S, Zhang L, et al. Computational Characterization of Domain-Segregated 3D Chromatin Structure and Segmented DNA Methylation Status in Carcinogenesis. 16(3): 699–716. doi: 10.1002/1878-0261.13127
30. Liu X, Jiang S, Ma L, Qu J, Zhao L, Zhu X, et al. Time-Dependent Effect of 1,6-Hexanediol on Biomolecular Condensates and 3D Chromatin Organization. Genome Biol (2021) 22(1):230. doi: 10.1186/s13059-021-02455-3
31. Ulianov SV, Velichko AK, Magnitov MD, Luzhin AV, Golov AK, Ovsyannikova N, et al. Suppression of Liquid-Liquid Phase Separation by 1,6-Hexanediol Partially Compromises the 3D Genome Organization in Living Cells. Nucleic Acids Res (2021) 49(18):10524–41. doi: 10.1093/nar/gkab249
32. Shi M, You K, Chen T, Hou C, Liang Z, Liu M, et al. Quantifying the Phase Separation Property of Chromatin-Associated Proteins Under Physiological Conditions Using an Anti-1,6-Hexanediol Index. Genome Biol (2021) 22(1):229. doi: 10.1186/s13059-021-02456-2
33. Itoh Y, Iida S, Tamura S, Nagashima R, Shiraki K, Goto T, et al. 1,6-Hexanediol Rapidly Immobilizes and Condenses Chromatin in Living Human Cells. Life Sci Alliance (2021) 4(4):e202001005. doi: 10.26508/lsa.202001005
34. D'OliveiraAlbanus R, Kyono Y, Hensley J, Varshney A, Orchard P, Kitzman JO, et al. Chromatin Information Content Landscapes Inform Transcription Factor and DNA Interactions. Nat Commun (2021) 12(1):1307. doi: 10.1038/s41467-021-21534-4
35. Yu M, Peng Z, Qin M, Liu Y, Wang J, Zhang C, et al. Interferon-γ Induces Tumor Resistance to Anti-PD-1 Immunotherapy by Promoting YAP Phase Separation. Mol Cell (2021) 81(6):1216–1230.e9. doi: 10.1016/j.molcel.2021.01.010
36. Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Co-Activator Condensation at Super-Enhancers Links Phase Separation and Gene Control. Science (2018) 361(6400):eaar3958. doi: 10.1126/science.aar3958
37. Schneider N, Wieland FG, Kong D, Fischer AAM, Hörner M, Timmer J, et al. Liquid-Liquid Phase Separation of Light-Inducible Transcription Factors Increases Transcription Activation in Mammalian Cells and Mice. Sci Adv (2021) 7(1):eabd3568. doi: 10.1126/sciadv.abd3568
38. Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, et al. Phase Separation of TAZ Compartmentalizes the Transcription Machinery to Promote Gene Expression. Nat Cell Biol (2020) 22(4):453–64. doi: 10.1038/s41556-020-0485-0
39. Zhang Y, Qian J, Gu C, Yang Y. Alternative Splicing and Cancer: A Systematic Review. Signal Transduct Target Ther (2021) 6(1):78. doi: 10.1038/s41392-021-00486-7
40. Zhang Y, Li Z, Chen N, Huang Y, Huang S. Phase Separation of Arabidopsis EMB1579 Controls Transcription, mRNA Splicing, and Development. PloS Biol (2020) 18(7):e3000782. doi: 10.1371/journal.pbio.3000782
41. Liu S, Wang T, Shi Y, Bai L, Wang S, Guo D, et al. USP42 Drives Nuclear Speckle mRNA Splicing via Directing Dynamic Phase Separation to Promote Tumorigenesis. Cell Death Differ (2021) 28(8):2482–98. doi: 10.1038/s41418-021-00763-6
42. Batlle C, Yang P, Coughlin M, Messing J, Pesarrodona M, Szulc E, et al. hnRNPDL Phase Separation Is Regulated by Alternative Splicing and Disease-Causing Mutations Accelerate Its Aggregation. Cell Rep (2020) 30(4):1117–1128.e5. doi: 10.1016/j.celrep.2019.12.080
43. Kawachi T, Masuda A, Yamashita Y, Takeda JI, Ohkawara B, Ito M, et al. Regulated Splicing of Large Exons is Linked to Phase-Separation of Vertebrate Transcription Factors. EMBO J (2021) 40(22):e107485. doi: 10.15252/embj.2020107485
44. Li W, Hu J, Shi B, Palomba F, Digman MA, Gratton E, et al. Biophysical Properties of AKAP95 Protein Condensates Regulate Splicing and Tumorigenesis. Nat Cell Biol (2020) 22(8):960–72. doi: 10.1038/s41556-020-0550-8
45. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. M6a Enhances the Phase Separation Potential of mRNA. Nature (2019) 571(7765):424–8. doi: 10.1038/s41586-019-1374-1
46. Wang J, Wang L, Diao J, Shi YG, Shi Y, Ma H, et al. Binding to M6a RNA Promotes YTHDF2-Mediated Phase Separation. Protein Cell (2020) 11(4):304–7. doi: 10.1007/s13238-019-00660-2
47. Yoneda R, Ueda N, Kurokawa R. M6a Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress. Int J Mol Sci (2021) 22(20):11014. doi: 10.3390/ijms222011014
48. Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, et al. Enhancer RNA M6a Methylation Facilitates Transcriptional Condensate Formation and Gene Activation. Mol Cell (2021) 81(16):3368–3385.e9. doi: 10.1016/j.molcel.2021.07.024
49. Zhang Z, Gerstein M. Large-Scale Analysis of Pseudogenes in the Human Genome. Curr Opin Genet Dev (2004) 14(4):328–35. doi: 10.1016/j.gde.2004.06.003
50. Feng X, Li H. Higher Rates of Processed Pseudogene Acquisition in Humans and Three Great Apes Revealed by Long-Read Assemblies. Mol Biol Evol (2021) 38(7):2958–66. doi: 10.1093/molbev/msab062
51. Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, et al. HSP70 Chaperones RNA-Free TDP-43 Into Anisotropic Intranuclear Liquid Spherical Shells. Science (2021) 371(6529):eabb4309. doi: 10.1126/science.abb4309
52. Lenard AJ, Hutten S, Zhou Q, Usluer S, Zhang F, Bourgeois BMR, et al. Phosphorylation Regulates CIRBP Arginine Methylation, Transportin-1 Binding and Liquid-Liquid Phase Separation. Front Mol Biosci (2021) 8:689687. doi: 10.3389/fmolb.2021.689687
53. Biswas A, Mallik BS. Dynamics of Ionic Liquid Through Intrinsic Vibrational Probes Using the Dispersion-Corrected DFT Functionals. J Phys Chem B (2021) 125(25):6994–7008. doi: 10.1021/acs.jpcb.1c04960
54. Goodier JL, Zhang L, Vetter MR, Kazazian HH Jr. LINE-1 ORF1 Protein Localizes in Stress Granules With Other RNA-Binding Proteins, Including Components of RNA Interference RNA-Induced Silencing Complex. Mol Cell Biol (2007) 27(18):6469–83. doi: 10.1128/MCB.00332-07
55. Briggs EM, McKerrow W, Mita P, Boeke JD, Logan SK, Fenyö D. RIP-Seq Reveals LINE-1 ORF1p Association With P-Body Enriched mRNAs. Mob DNA (2021) 12(1):5. doi: 10.1186/s13100-021-00233-3
56. Laaß K, Quiroz FG, Hunold J, Roberts S, Chilkoti A, Hinderberger D. Nanoscopic Dynamics Dictate the Phase Separation Behavior of Intrinsically Disordered Proteins. Biomacromolecules (2021) 22(2):1015–25. doi: 10.1021/acs.biomac.0c01768
57. Candelise N, Scaricamazza S, Salvatori I, Ferri A, Valle C, Manganelli V, et al. Protein Aggregation Landscape in Neurodegenerative Diseases: Clinical Relevance and Future Applications. Int J Mol Sci (2021) 22(11):6016. doi: 10.3390/ijms22116016
58. Shukla S, Srividya K, Nazir A. Not a Piece of Junk Anymore: Pseudogene T04B2.1 Performs non-Conventional Regulatory Role and Modulates Aggregation of α- Synuclein and β-Amyloid Proteins in C. Elegans. Biochem Biophys Res Commun (2021) 539:8–14. doi: 10.1016/j.bbrc.2020.12.029
59. Liu Q, Liu X, Zhao D, Ruan X, Su R, Shang X, et al. Pseudogene ACTBP2 Increases Blood-Brain Barrier Permeability by Promoting KHDRBS2 Transcription Through Recruitment of KMT2D/WDR5 in Aβ1-42 Microenvironment. Cell Death Discov (2021) 7(1):142. doi: 10.1038/s41420-021-00531-y
60. Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell (2017) 68(1):144–157.e5. doi: 10.1016/j.molcel.2017.09.003
61. Freibaum BD, Messing J, Yang P, Kim HJ, Taylor JP. High-Fidelity Reconstitution of Stress Granules and Nucleoli in Mammalian Cellular Lysate. J Cell Biol (2021) 220(3):e202009079. doi: 10.1083/jcb.202009079
62. Padrón A, Iwasaki S, Ingolia NT. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol Cell (2019) 75(4):875–887.e5. doi: 10.1016/j.molcel.2019.07.030
63. Engbrecht M, Mangerich A. The Nucleolus and PARP1 in Cancer Biology. Cancers (Basel) (2020) 12(7):1813. doi: 10.3390/cancers12071813
64. Pontvianne F, Carpentier MC, Durut N, Pavlištová V, Jaške K, Schořová Š, et al. Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. Thaliana Genome. Cell Rep (2016) 16(6):1574–87. doi: 10.1016/j.celrep.2016.07.016
65. Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The Nucleolus as a Multiphase Liquid Condensate. Nat Rev Mol Cell Biol (2021) 22(3):165–82. doi: 10.1038/s41580-020-0272-6
66. Yao Y, Tan HW, Liang ZL, Wu GQ, Xu YM, Lau ATY. The Impact of Coilin Nonsynonymous SNP Variants E121K and V145I on Cell Growth and Cajal Body Formation: The First Characterization. Genes (Basel) (2020) 11(8):895. doi: 10.3390/genes11080895
67. Poole AR, Enwerem II, Vicino IA, Coole JB, Smith SV, Hebert MD. Identification of Processing Elements and Interactors Implicate SMN, Coilin and the Pseudogene-Encoded Coilp1 in Telomerase and Box C/D scaRNP Biogenesis. RNA Biol (2016) 13(10):955–72. doi: 10.1080/15476286.2016.1211224
68. Zhang PF, Wu J, Wu Y, Huang W, Liu M, Dong ZR, et al. The lncRNA SCARNA2 Mediates Colorectal Cancer Chemo-Resistance Through a Conserved microRNA-342-3p Target Sequence. J Cell Physiol (2019) 234(7):10157–65. doi: 10.1002/jcp.27684
69. Zhang Z, Jia M, Wen C, He A, Ma Z. Long non-Coding RNA SCARNA2 Induces Cutaneous Squamous Cell Carcinoma Progression via Modulating miR-342-3p Expression. J Gene Med (2020) 22(12):e3242. doi: 10.1002/jgm.3242
70. Hardy SD, Shinde A, Wang WH, Wendt MK, Geahlen RL. Regulation of Epithelial-Mesenchymal Transition and Metastasis by TGF-β, P-Bodies, and Autophagy. Oncotarget (2017) 8(61):103302–14. doi: 10.18632/oncotarget.21871
71. Bearss JJ, Padi SK, Singh N, Cardo-Vila M, Song JH, Mouneimne G, et al. EDC3 Phosphorylation Regulates Growth and Invasion Through Controlling P-Body Formation and Dynamics. EMBO Rep (2021) 22(4):e50835. doi: 10.15252/embr.202050835
72. Beneventi G, Munita R, Cao Thi Ngoc P, Madej M, Cieśla M, Muthukumar S, et al. The Small Cajal Body-Specific RNA 15 (SCARNA15) Directs P53 and Redox Homeostasis via Selective Splicing in Cancer Cells. NAR Cancer (2021) 3(3):zcab026. doi: 10.1093/narcan/zcab026
73. Adjibade P, Simoneau B, Ledoux N, Gauthier WN, Nkurunziza M, Khandjian EW, et al. Treatment of Cancer Cells With Lapatinib Negatively Regulates General Translation and Induces Stress Granules Formation. PloS One (2020) 15(5):e0231894. doi: 10.1371/journal.pone.0231894
74. Rodriguez-Antona C, Axelson M, Otter C, Rane A, Ingelman-Sundberg M. A Novel Polymorphic Cytochrome P450 Formed by Splicing of CYP3A7 and the Pseudogene CYP3AP1. J Biol Chem (2005) 280(31):28324–31. doi: 10.1074/jbc.M502309200
75. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 Activity Promoted by non-Coding Effects of Circular RNA and Foxo3 Pseudogene in the Inhibition of Tumor Growth and Angiogenesis. Oncogene (2016) 35(30):3919–31. doi: 10.1038/onc.2015.460
76. Liu G, Feng F, Zhao X, Cai L. Nucleosome Organization Around Pseudogenes in the Human Genome. BioMed Res Int (2015) 2015:821596. doi: 10.1155/2015/821596
77. Schoeftner S, Scarola M, Benetti R. Chromatin Regulation at Parental Gene Promoters by Pseudogene Sense lncRNAs. Methods Mol Biol (2021) 2324:203–17. doi: 10.1007/978-1-0716-1503-4_13
78. Grunau C, Hindermann W, Rosenthal A. Large-Scale Methylation Analysis of Human Genomic DNA Reveals Tissue-Specific Differences Between the Methylation Profiles of Genes and Pseudogenes. Hum Mol Genet (2000) 9(18):2651–63. doi: 10.1093/hmg/9.18.2651
79. Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, et al. Mobile DNA in Cancer. Extensive Transduction of Nonrepetitive DNA Mediated by L1 Retrotransposition in Cancer Genomes. Science (2014) 345(6196):1251343. doi: 10.1126/science.1251343
80. Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, et al. Pan-Cancer Analysis of Whole Genomes Identifies Driver Rearrangements Promoted by LINE-1 Retrotransposition. Nat Genet (2020) 52(3):306–19. doi: 10.1038/s41588-019-0562-0
81. Cooke SL, Shlien A, Marshall J, Pipinikas CP, Martincorena I, Tubio JM, et al. Processed Pseudogenes Acquired Somatically During Cancer Development. Nat Commun (2014) 5:3644. doi: 10.1038/ncomms4644
82. Ewing AD, Ballinger TJ, Earl D, Broad Institute Genome Sequencing, Analysis Program and Platform, Harris CC, et al. Retrotransposition of Gene Transcripts Leads to Structural Variation in Mammalian Genomes. Genome Biol (2013) 14(3):R22. doi: 10.1186/gb-2013-14-3-r22
83. Zayas-Santiago A, Díaz-García A, Nuñez-Rodríguez R, Inyushin M. Accumulation of Amyloid Beta in Human Glioblastomas. Clin Exp Immunol (2020) 202(3):325–34. doi: 10.1111/cei.13493
84. Zayas-Santiago A, Martínez-Montemayor MM, Colón-Vázquez J, Ortiz-Soto G, Cirino-Simonet JG, Inyushin M. Accumulation of Amyloid Beta (Aβ) and Amyloid Precursor Protein (APP) in Tumors Formed by a Mouse Xenograft Model of Inflammatory Breast Cancer. 12(1):95-105. doi: 10.1002/2211-5463.13308
85. de Oliveira GAP, Cordeiro Y, Silva JL, Vieira TCRG. Liquid-Liquid Phase Transitions and Amyloid Aggregation in Proteins Related to Cancer and Neurodegenerative Diseases. Adv Protein Chem Struct Biol (2019) 118:289–331. doi: 10.1016/bs.apcsb.2019.08.002
86. Saha SK, Jeong Y, Cho S, Cho SG. Systematic Expression Alteration Analysis of Master Reprogramming Factor OCT4 and its Three Pseudogenes in Human Cancer and Their Prognostic Outcomes. Sci Rep (2018) 8(1):14806. doi: 10.1038/s41598-018-33094-7
87. Poursani EM, Mohammad Soltani B, Mowla SJ. Differential Expression of OCT4 Pseudogenes in Pluripotent and Tumor Cell Lines. Cell J (2016) 18(1):28–36. doi: 10.22074/cellj.2016.3984
88. Kovalenko TF, Morozova KV, Ozolinya LA, Lapina IA, Patrushev LI. The PTENP1 Pseudogene, Unlike the PTEN Gene, Is Methylated in Normal Endometrium, As Well As in Endometrial Hyperplasias and Carcinomas in Middle-Aged and Elderly Females. Acta Naturae (2018) 10(1):43–50. doi: 10.32607/20758251-2018-10-1-43-50
89. Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, et al. Expressed Pseudogenes in the Transcriptional Landscape of Human Cancers. Cell (2012) 149(7):1622–34. doi: 10.1016/j.cell.2012.04.041
90. Ibrahim EE, Babaei-Jadidi R, Saadeddin A, Spencer-Dene B, Hossaini S, Abuzinadah M, et al. Embryonic NANOG Activity Defines Colorectal Cancer Stem Cells and Modulates Through AP1- and TCF-Dependent Mechanisms. Stem Cells (2012) 30(10):2076–87. doi: 10.1002/stem.1182
91. Tan L, Cheng W, Liu F, Wang DO, Wu L, Cao N, et al. Positive Natural Selection of N6-Methyladenosine on the RNAs of Processed Pseudogenes. Genome Biol (2021) 22(1):180. doi: 10.1186/s13059-021-02402-2
92. Zhao R, Li B, Zhang S, He Z, Pan Z, Guo Q, et al. The N6-Methyladenosine-Modified Pseudogene HSPA7 Correlates With the Tumor Microenvironment and Predicts the Response to Immune Checkpoint Therapy in Glioblastoma. Front Immunol (2021) 12:653711. doi: 10.3389/fimmu.2021.653711
93. Yue C, Ren Y, Ge H, Yan L, Xu Y, Wang G, et al. Pseudogene DUXAP10 can be Used as a Diagnostic and Prognostic Biomarker in Human Cancers. J Cell Physiol (2019) 234(12):23685–94. doi: 10.1002/jcp.28937
94. Wang QS, Shi LL, Sun F, Zhang YF, Chen RW, Yang SL, et al. High Expression of ANXA2 Pseudogene ANXA2P2 Promotes an Aggressive Phenotype in Hepatocellular Carcinoma. Dis Markers (2019) 2019:9267046. doi: 10.1155/2019/9267046
95. Chen Z, Wang Z, Chen Z, Fu F, Huang X, Huang Z. Pseudogene HSPB1P1 Contributes to Renal Cell Carcinoma Proliferation and Metastasis by Targeting miR-296-5p to Regulate HMGA1 Expression. Cell Biol Int (2021) 45(12):2479–89. doi: 10.1002/cbin.11694
96. Qiu Y, Pan M, Chen X. A Liquid-Liquid Phase Separation-Related Gene Signature as Prognostic Biomarker for Epithelial Ovarian Cancer. Front Oncol (2021) 11:671892. doi: 10.3389/fonc.2021.671892
97. Liu J, Xie Y, Guo J, Li X, Wang J, Jiang H, et al. Targeting NSD2-Mediated SRC-3 Liquid-Liquid Phase Separation Sensitizes Bortezomib Treatment in Multiple Myeloma. Nat Commun (2021) 12(1):1022. doi: 10.1038/s41467-021-21386-y
98. Lu B, Zou C, Yang M, He Y, He J, Zhang C, et al. Pharmacological Inhibition of Core Regulatory Circuitry Liquid-Liquid Phase Separation Suppresses Metastasis and Chemoresistance in Osteosarcoma. Adv Sci (Weinh) (2021) 8(20):e2101895. doi: 10.1002/advs.202101895
Keywords: liquid phase separation, pseudogenes, RNA modification, epigenetic, cancer
Citation: Nsengimana B, Khan FA, Awan UA, Wang D, Fang N, Wei W, Zhang W and Ji S (2022) Pseudogenes and Liquid Phase Separation in Epigenetic Expression. Front. Oncol. 12:912282. doi: 10.3389/fonc.2022.912282
Received: 04 April 2022; Accepted: 13 June 2022;
Published: 08 July 2022.
Edited by:
Kevin J Ni, St George Hospital Cancer Care Centre, AustraliaReviewed by:
Pradeep Reddy Cingaram, University of Miami, United StatesCopyright © 2022 Nsengimana, Khan, Awan, Wang, Fang, Wei, Zhang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenqiang Wei, weiwq@henu.edu.cn; Weijuan Zhang, zwjuan1965@henu.edu.cn; Shaoping Ji, shaopingji@henu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Bernard Nsengimana
Bernard Nsengimana Faiz Ali Khan
Faiz Ali Khan Usman Ayub Awan
Usman Ayub Awan Dandan Wang1
Dandan Wang1 Shaoping Ji
Shaoping Ji