
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 25 July 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.911316
This article is part of the Research Topic Targeting the Wnt/β-catenin Signaling Pathway in Cancer View all 10 articles
Early diagnosis and prognosis prediction of non-small cell lung cancer (NSCLC) have been challenging. Signaling cascades involving the Wingless-type (WNT) gene family play important biological roles and show prognostic value in various cancers, including NSCLC. On this basis, this study aimed to investigate the significance of WNTs in the prognosis and tumor immunity in NSCLC by comprehensive analysis. Expression and methylation levels of WNTs were obtained from the ONCOMINE, TIMER, and UALCAN. The dataset obtained from The Cancer Genome Atlas (TCGA) was utilized for prognostic analysis. cBioPortal was used to perform genetic alterations and correlation analysis of WNTs. R software was employed for functional enrichment and pathway analysis, partial statistics, and graph drawing. TRRUST was used to find key transcription factors. GEPIA was utilized for the analysis of expression, pathological staging, etc. Correlative analysis of immune infiltrates from TIMER. TISIDB was used for further immune infiltration validation analysis. Compared with that of normal tissues, WNT2/2B/3A/4/7A/9A/9B/11 expressions decreased, while WNT3/5B/6/7B/8B/10A/10B/16 expressions increased in lung adenocarcinoma (LUAD); WNT2/3A/7A/11 expressions were lessened, while WNT2B/3/5A/5B/6/7B/10A/10B/16 expressions were enhanced in squamous cell lung cancer (LUSC). Survival analysis revealed that highly expressed WNT2B and lowly expressed WNT7A predicted better prognostic outcomes in LUAD and LUSC. In the study of immune infiltration levels, WNT2, WNT9B, and WNT10A were positively correlated with six immune cells in LUAD; WNT1, WNT2, and WNT9B were positively correlated with six immune cells in LUSC, while WNT7B was negatively correlated. Our study indicated that WNT2B and WNT7A might have prognostic value in LUAD, and both of them might be important prognostic factors in LUSC and correlated to immune cell infiltration in LUAD and LUSC to a certain extent. Considering the prognostic value of WNT2B and WNT7A in NSCLC, we validated their mRNA and protein expression levels in NSCLC by performing qRT-PCR, western blot, and immunohistochemical staining on NSCLC pathological tissues and cell lines. This study may provide some direction for the subsequent exploration of the prognostic value of the WNTs and their role as biomarkers in NSCLC.
Lung cancer has a high incidence rate, accounting for 11.4% of cases and 18% of cancer mortality worldwide. By 2020, it has been the second most common cancer and the leading cause of death worldwide. There was little progress in its early screening and survival (1, 2). Non-small cell lung cancer (NSCLC) is the most common histological subtype in lung cancer patients, about 85% of these patients. It can be mainly divided into lung adenocarcinoma (LUAD), squamous cell lung cancer (LUSC), and large cell lung carcinoma, of which LUAD and LUSC are the most common (3, 4). For NSCLC therapeutic targets, molecular therapy, immune checkpoint inhibition, and lung cancer vaccination have made many advances (5–7). Nonetheless, lung cancer still has a 5-year survival rate lower than all other cancer types (8).
The Wingless-type (WNT) protein family consists of many cysteine-rich secreted glycoproteins, with a molecular weight of about 40 kDa, and 19 WNT genes have been identified in the human genome (9). WNT binds to members of the Frizzled (Fzd) receptor family on the cell surface to activate downstream effectors inside the cell, activating signal transduction cascades through three different pathways in the organism, i.e., the canonical WNT pathway, the non-canonical WNT/planar cell polarity (PCP) pathway, and the non-canonical WNT/calcium pathway (10). At the cellular level, WNT proteins participate in the regulation of cell proliferation, cell morphology, cell motility, and cell fate, playing an extensive role in organism development; moreover, dysregulation of WNT signaling also works during tumor formation (11, 12). WNT signaling serves as a molecular rheostat in organisms and participates in multiple physiological processes, including embryonic development, lineage commitment, adult stem cell homeostasis, and tissue regeneration (12). The transmission of WNT signaling and the generation and secretion of WNT ligands involve specialized and complex mechanisms (11–13). Conduction of WNT signaling has been demonstrated in many types of cancer (12), including but not limited to colorectal cancer (14), liver cancer (15), prostate cancer (16), skin cancer (17), melanoma (18) and lung cancer (12, 13). The WNT pathway plays an important biological role and represents prognostic value in NSCLC, and β-catenin produced by downstream activation of WNT ligands is indispensable in NSCLC development (12, 13, 19). Activation by WNT ligands in the canonical WNT pathway increases the concentrations of β-catenin in the cytoplasm, which binds to transcription factors of the T-cell factor (TCF)/Lymphoid enhancer-binding factor (LEF) family in the nucleus so as to drive transcription of WNT/β-catenin target genes. Some genomic data also suggest that an important component in lung cancer development is intrinsic WNT signaling within cancer cells (12). However, comprehensive studies on the expression and prognosis of the WNT family in NSCLC or its subtypes are lacking.
Our study used a publicly available tumor database to perform bioinformatics analysis of WNT gene family expression, prognosis, and immune infiltration in LUAD and LUSC so as to find reliable biomarkers and detailed prognosis of the WNT family in NSCLC.
ONCOMINE (https://www.ONCOMINE.org) is an online microarray database that allows differential expression analysis in many types of cancer (20). In this study, we compared the mRNA transcript expression differences of WNT genes by using Student’s t-test in this database, and the cut-off values of p value and fold-change were as follows: p value of 1E-4, fold-change of 2, gene rank of 10%.
TIMER (https://cistrome.shinyapps.io/timer/) is an online tool that can systematically assess the infiltration level of immune cells in different cancer types and their clinical impact (21). In the “DiffExp” module, the expression level distribution of WNTs was displayed using boxplots, and the statistical significance of differential expression was assessed using the Wilcoxon test. Scatter plots representing the correlation of infiltration levels between WNT genes and the six immune cells were obtained by the “Gene” module. Then, by using the “Survival” module, the clinical correlation between WNTs and tumor immunity was explored by Cox regression analysis, including risk ratio and statistical significance. The level of tumor invasion between WNTs and tumors with different somatic copy number alterations was compared using the Wilcoxon rank-sum test in the “SCNA” module defined by GISTIC2.0. The heatmap of the correlation between WNTs and immune scores was created using the R software v4.0.3 package pheatmap (v1.0.12). The lollipop chart using the R software v4.0.3 package GSVA (v1.34.0) built-in algorithm ssGSEA to verify the relationship between WNTs expression and immune cells. The dataset used contained mRNA sequence data from 516 lung adenocarcinomas and 501 squamous cell lung cancers from TCGA tumors.
UALCAN (http://ualcan.path.uab.edu) is a comprehensive network resource that is taken from publicly available cancer genomics data (TCGA, MET500, and CPTAC) (22). In this study, the mRNA transcript expression and methylation data of WNT gene family were obtained by using the expression analysis module and methylation level module of UALCAN.
GEPIA (http://gepia.cancer-pku.cn) is a web-based data mining platform with large RNA sequencing data from TCGA and GTEx (23). “Pathological Stage Plot” in the “ExpressionDIY” function is used for the correlation study of the pathological stage of WNTs in NSCLC patients. “Multiple Gene Comparison” module was used to obtain WNTs relative expression data, and matrix plots were created by the R v4.0.3 software package ggplot2 (v.3.3.5). Gene expression correlation analysis was performed in the “Correlation” module.
From the TCGA dataset (https://portal.gdc.com), to obtain raw counts and corresponding clinical information of RNA sequencing data of NSCLC tumors. For Kaplan-Meier (KM) curves, p values and hazard ratios (HR) with 95% confidence intervals (CI) were derived by log-rank test and univariate Cox proportional hazards regression, p < 0.05 was considered statistically significant. The above analysis method was created by the R software v4.0.3 package ggplot2 (v.3.3.5). The survival analysis results were verified by the Kaplan-Meier plotter (https://kmplot.com/) (24), the microarray database included caBIG, GEO, and TCGA. We obtained KM survival plots with hazard ratios and log-rank p values by univariate Cox regression analysis. For prognostic models of prognosis-related genes and clinical factors, univariate and multivariate Cox proportional hazards analyses were first performed, with risk scores including age, gender, pTNM stage of the tumor, and expression levels of prognosis-related genes. Based on the univariate and multivariate Cox regression analysis results, variables with significant differences in prognosis were extracted to construct a nomogram and predict the 1-year, 3-year, and 5-year survival rates of NSCLC patients. The above analysis was created using the R software v4.0.3 packages forestplot (v2.0.0) and rms (v6.2-0).
The portal (https://www.cbioportal.org/) simplifies molecular profiling data from cancer tissues and cell lines into easily understood genetic, epigenetic, gene expression and proteomics events. It contains large cancer genomics datasets and has functions such as visualization, download and analysis (25). Based on the TCGA database, we analyzed 586 lung adenocarcinoma samples (TCGA, Firehose Legacy) and 511 squamous cell lung cancer samples (TCGA, Firehose Legacy) obtained from cBioPortal. mRNA expression z-scores (RNA Seq V2 RSEM) enter a z-score threshold ± 2.0, and protein expression z-scores (RPPA) enter a z-score threshold ± 2.0. Pearson’s correlation coefficient and p value were obtained by the “Co-expression” module, and the correlation heatmap between genes was constructed using the R software v4.0.3 package pheatmap (v1.0.12).
GO enrichment analysis examined the function of the WNT gene family from three levels: Biology Process, Molecular Function, and Cellular Component. KEGG is then used for pathway analysis. Histogram is implemented by R software v4.0.3 package clusterprofiler (v3.18.1) (26), the threshold is set as follows: p value cut-off of 0.05, q value cut-off of 0.05, minGSSize = 5, maxGSSize = 5000, The p value correction method uses Benjamini-Hochberg (BH). Protein-protein interaction (PPI) networks were constructed by STRING and Cytoscape. STRING database (https://string-db.org/) is designed to collect, score, and integrate all publicly available sources of protein-protein interaction information, supplemented by computational predictions (27). We constructed a protein interaction network of WNTs by the “Multipleproteins” function in STRING and performed GO and KEGG analyses.
TRRUST (https://www.grnpedia.org/trrust/) is a TF-target interaction database based on text mining, including 8444 regulatory interactions of 800 transcription factors (TFs) in humans (28). A list of genes is entered on this website to query key TFs.
TISIDB (http://cis.hku.hk/TISIDB/index.php) is a web portal containing multiple heterogeneous data on tumor and immune system interactions (29). We used TISIDB to reveal the relations between the abundance of tumor-infiltrating lymphocytes (TILs) and the expression of WNTs.
During January and March 2022, we collected 20 pairs of NSCLC tissues and adjacent normal tissues from the Third Xiangya Hospital of Central South University. These tissues were used to detect the expression levels of WNTs mRNA by quantitative real-time PCR (qRT-PCR). The Ethics Committee of the Third Xiangya Hospital of Central South University has approved the study. The approval number is I-22054.
Human normal lung epithelial cell line (BEAS-2B), LUAD cell line (PC9) and LUSC cell line (NCI-H520) were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA). All cell lines were cultured in DMEM (Gibco, Carlsbad, CA) supplemented with 10% FBS (Gibco) and 1% pen/strep (Gibco) at 5% CO2 and 37°C.
According to the manufacturer’s instructions, total RNA was isolated from tissues or cells using Trizol reagent (TaKaRa, Japan). One microgram of RNA was reverse transcribed into cDNA using the Revert Aid First Strand cDNA Synthesis Kit (Thermo, USA). Quantitative RT-PCR was then performed with Pro Taq HS Premix Probe qPCR Kit (Accurate, Hunan, China). The amplification program consisted of one cycle of predenaturation at 95°C for 5 min, 37 cycles of denaturation at 95°C for 30 s, annealing at 61°C for 30 s, and extension at 72°C for 10 min. The GAPDH gene was used as an endogenous control gene for normalizing the expression of target genes. Each sample was analyzed in triplicate. Primer sequences are shown in Supplementary Table S1.
RIPA lysis buffer (Thermo Fisher Scientific, USA) containing protease and phosphatase inhibitors was used to lyse tissues. Protein lysates were separated by SDS-PAGE gels (Thermo Fisher Scientific, USA), blotted onto PVDF membrane (Roche, Switzerland) for analysis and incubated at 4°C overnight with the following primary antibodies: anti-WNT2B antibody (1:1000 dilution; Bioss, China), anti-WNT7A antibody (1:1000 dilution; Bioss, China), and anti-GAPDH antibody (1:2000 dilution; Cell Signaling Technology, USA). The results of the western blot analyses were performed with Image J software.
Immunohistochemistry was performed in 20 NSCLC tissues and adjacent normal tissues. Paraffin-embedded sections were stained to determine the expression level of proteins. Sections were incubated overnight at 4°C with anti-WNT2B antibody (1:100 dilution; Bioss, China), or anti-WNT7A antibody (1:100 dilution; Bioss, China). After washing with phosphate-buffered saline (PBS), the slides were incubated with a goat anti-rabbit IgG secondary antibody conjugated to fluorescein isothiocyanate (ZSDB-BIO, China) for 30 mins. They were washed with PBS and then incubated with an antifade reagent (Invitrogen, USA). Finally, staining was observed using an Olympus CX41 fluorescence microscope (Olympus, Japan). The results of the analyses were performed with Image J software.
Members of the WNT gene family and their respective specific chromosomal regions are presented in Table 1. We first examined the mRNA transcription of 19 WNT genes in lung cancer by the ONCOMINE database, as can be seen from Figure 1A. The detailed expression results in lung cancer are shown in Table 2, in which the expression levels of WNT2/2B/3A/7A/11 in lung cancer were significantly lower than those in normal tissues, while those of WNT3/5A/7B expressions were significantly higher, with altered expression levels of WNT5A and WNT7A in LUSC and the others are in LUAD.
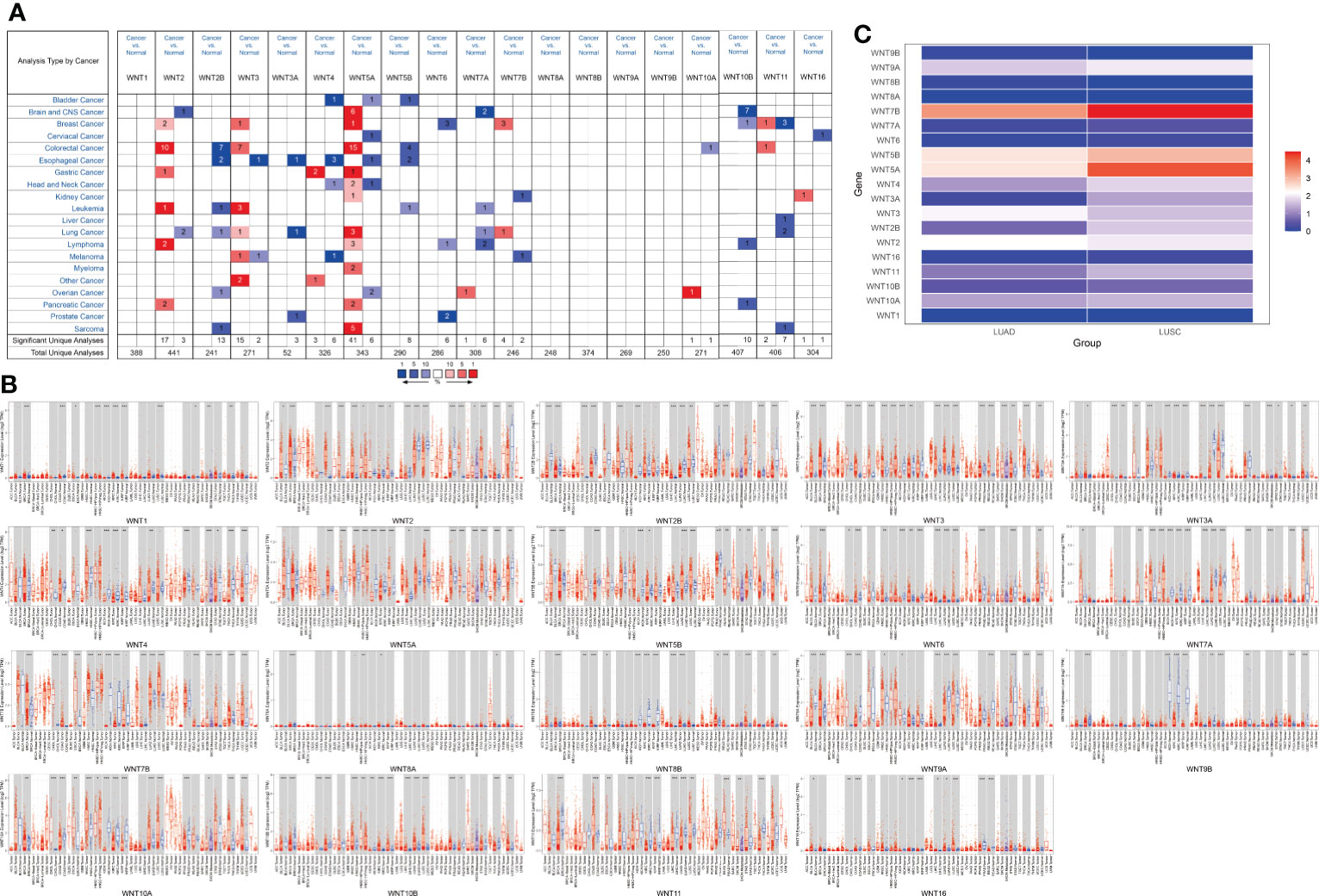
Figure 1 The mRNA transcription levels of the WNT gene family in LUAD and LUSC. (A) The mRNA transcription levels of WNT gene family in lung cancer (ONCOMINE). Red for overexpression, blue for downregulated expression. (B) Differential expression of WNT gene family in different tumor tissues and normal tissues (TIMER). Statistical significance of differential expression was assessed using the Wilcoxon test (p value significant codes: 0 ≤ *** < 0.001 ≤ ** < 0.01 ≤ * < 0.05 ≤. < 0.1). (C) The expression matrix plots for WNT gene family in LUAD and LUSC. Indicates the relative expression level among WNT genes.
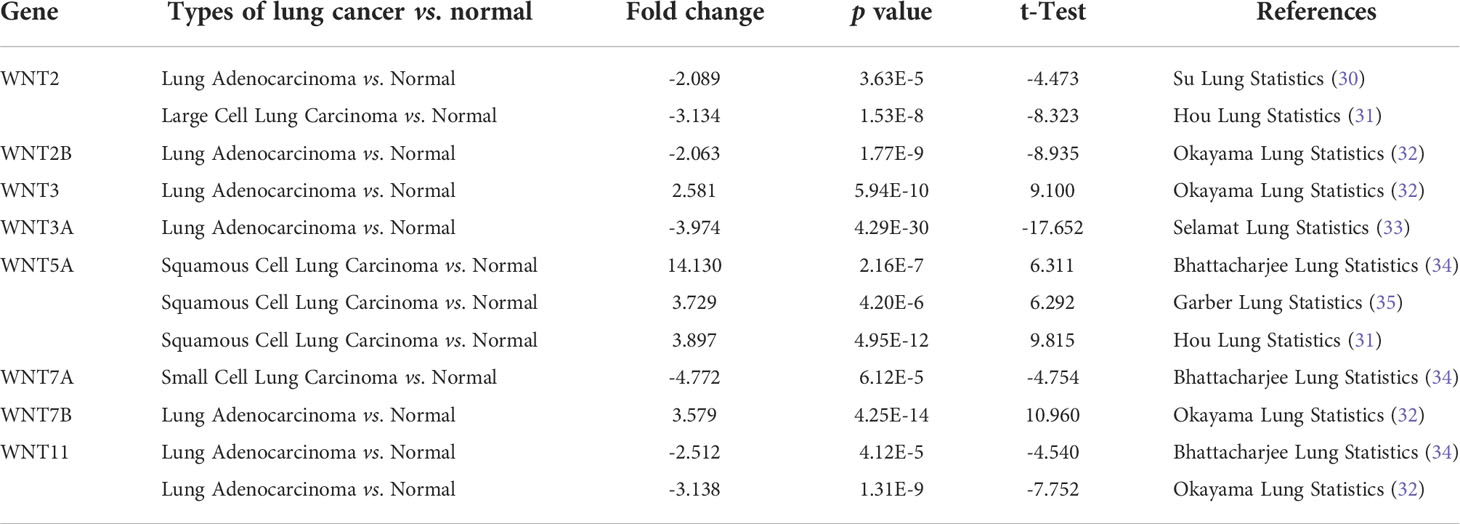
Table 2 The significant changes of WNTs mRNA expression in transcription level between different types of Lung cancer.
Additionally, we used TIMER to assess differences in expression levels of the WNT gene family in various cancers and normal tissues. As shown in Figure 1B, in LUAD, WNT3/5B/6/7B/8B/10A/10B/16 expressions significantly increased, WNT2/2B/3A/4/7A/9A/9B/11 expressions significantly decreased, and the results of WNT1 and WNT5A presented no statistical significance. In LUSC, the expressions of WNT2B/3/5A/5B/6/7B/10A/10B/16 were significantly promoted, while the expressions of WNT1/2/3A/7A/9A/9B/11 were significantly inhibited; the results of WNT4 and WNT8B were not statistically significant in LUSC, and WNT8A expression showed no statistical significance in LUAD and LUSC. Figure 1C showed the relative expression levels of WNT genes in LUAD and LUSC. Among the 19 WNT genes, WNT7B presented the highest relative expression level in LUAD, while the relative expression level of WNT5A/5B/7B was higher in LUSC than in LUAD, consistent with the above expression results.
Moreover, UALCAN was used to explore the expression differences of the WNT gene family in normal tissues and in LUAD and LUSC, with the results shown in Figure 2A. In LUAD, the expression of WNT2/2B/3A/4/7A/9A/9B/11 was lower than that in normal tissues, while the expressions of WNT1/3/5B/6/7B/8B/10A/10B/16 were higher than that in normal tissues, presenting statistical significance; WNT5A and WNT8A results were not statistically significant; WNT2/3A/7A/11 were less expressed in LUSC than in normal tissues, while WNT2B/3/4/5A/5B/6/7B/8B/10A/10B/16 showed the opposite circumstance with statistical significance; WNT1/8A/9A/9B expressions were not statistically significant in LUSC. In summary, according to the comparison between databases, LUAD represented lessened WNT2/2B/3A/4/7A/9A/9B/11 expressions and enhanced WNT3/5B/6/7B/8B/10A/10B/16 expressions compared with normal tissues; in contrast, LUSC presented prohibited WNT2/3A/7A/11 expressions and promoted WNT2B/3/5A/5B/6/7B/10A/10B/16 expressions.

Figure 2 Compared with normal tissues, the WNT gene family’s mRNA expression and methylation levels in LUAD and LUSC (UALCAN). (A) The mRNA transcription levels of the WNT gene family in LUAD and LUSC. (B) The methylation level of the WNT gene family in LUAD and LUSC. p < 0.05 was considered statistically significant.
In addition, UALCAN was employed to explore the methylation levels of WNTs in LUAD and LUSC, with the results depicted in Figure 2B. In both LUAD and LUSC, the methylation levels of WNT1/2/2B/3/3A/6/7A/9B/10A/10B/16 were significantly improved, while those of WNT8B and WNT11 were significantly diminished. For WNT8A and WNT9A, their methylation levels were significantly boosted in LUAD but declined in LUSC. The methylation level of WNT5B significantly dropped in LUSC but not in LUAD, and that of WNT7B significantly went up in LUSC but not in LUAD.
This section first analyzed the correlation between the WNT gene and different pathological stages of LUAD and LUSC. As shown in Figure 3A, WNT2, WNT2B, and WNT11 were significantly correlated with pathological stages of LUAD patients, while WNTs presented no statistical significance in the correlation with LUSC (Figure 3B), indicating that the mRNA expression of WNT2, WNT2B, and WNT11 was significantly correlated with individual cancer stages of LUAD patients.
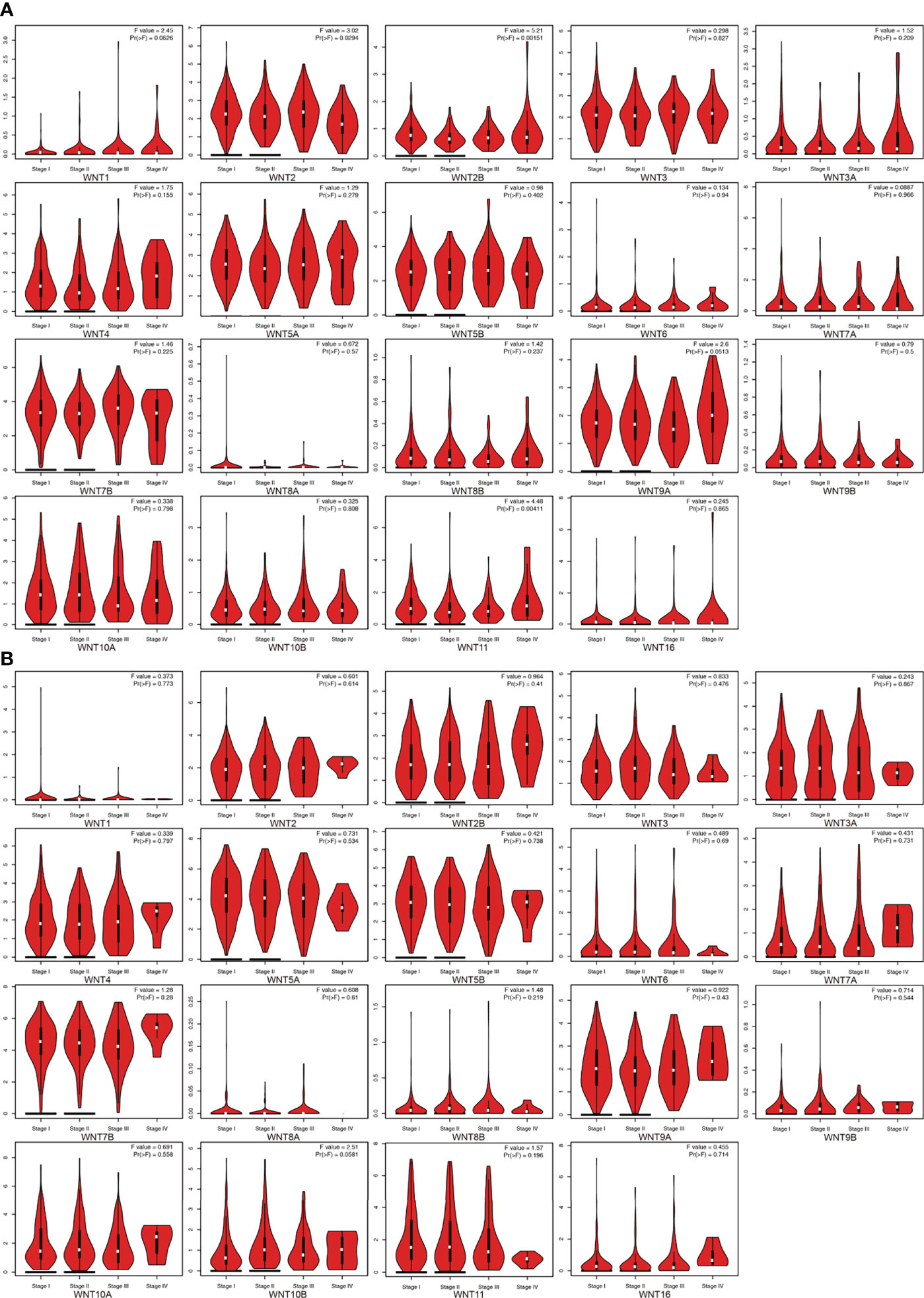
Figure 3 The mRNA expression of WNT gene family in different pathological stages of LUAD and LUSC (GEPIA). (A) Correlation between WNT gene family and pathological stage in patients with LUAD. (B) Correlation between WNT gene family and pathological stage in patients with LUSC.
Next, data were extracted from The Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.com). Raw counts and corresponding clinical information of RNA sequencing data were obtained from 516 LUAD patients and 501 LUSC patients for survival analysis. The clinical information of corresponding patients is shown in Supplementary Table S2. Figures 4A, B showed the overall survival (OS) curves in LUAD and LUSC. In LUAD patients, higher WNT2B expression and lower WNT7A expression were associated with a better prognosis. Similarly, higher expression of WNT2B and WNT8B and lower expression of WNT7A predicted better prognostic outcomes in LUSC patients. In addition, WNT8A expression was zero in more than half of the samples of LUAD and LUSC, which was thus not presented in this section. Then, the survival outcomes of WNTs in NSCLC were validated by the Kaplan-Meier plotter (Supplementary Figure 1), and higher WNT2B expression and lower WNT7A expression were associated with better prognostic outcomes in LUAD and LUSC, compatible with our results in TCGA. However, the results of WNT8B in LUSC were not significant, and thus WNT2B and WNT7A in LUAD and LUSC were selected for further analysis.
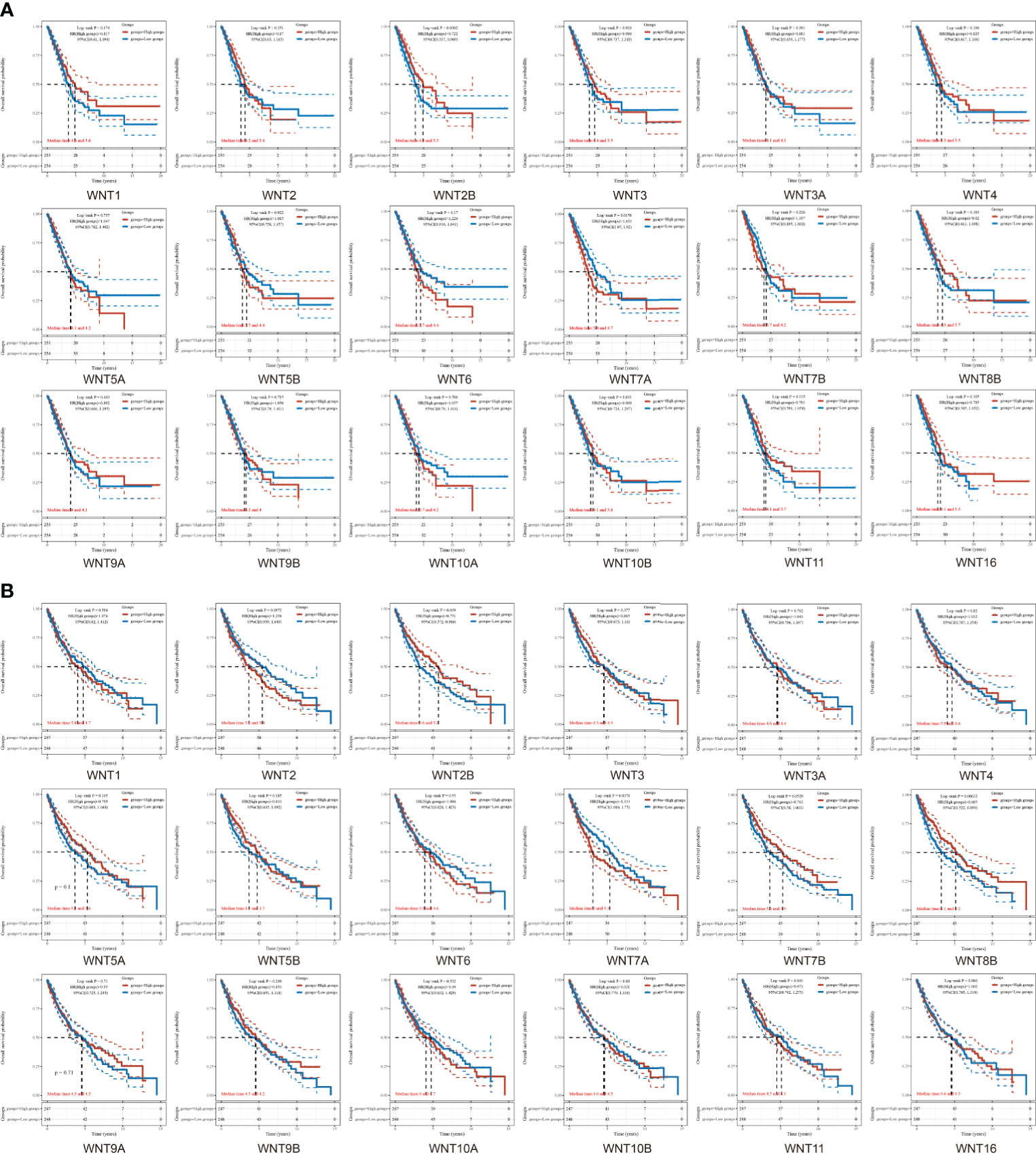
Figure 4 The overall survival (OS) curves of WNTs in LUAD and LUSC. (A) Kaplan-Meier survival analysis of WNTs in LUAD. (B) Kaplan-Meier survival analysis of WNTs in LUSC.
For the prognosis-related genes obtained from the survival analysis, a prognostic model was constructed (Figures 5A-H) to explore their clinical impact on the disease process of patients. For the univariate (Figure 5A) and multivariate (Figure 5B) analysis of LUAD, WNT7A and pTNM stage were significantly different variables; in contrast, WNT7A was not statistically significant for the prediction of the LUAD process (Figure 5C). The univariate (Figure 5E) and multivariate (Figure 5F) analyses of LUSC indicated that WNT7A, WNT2B, and pTNM stage were significantly different variables. Figure 5G illustrated that WNT7A and WNT2B were statistically significant in predicting the LUSC process. Taken together, our results showed that WNT2B and WNT7A might have prognostic value in LUAD, and both of them might be independent prognostic factors for LUSC patients.
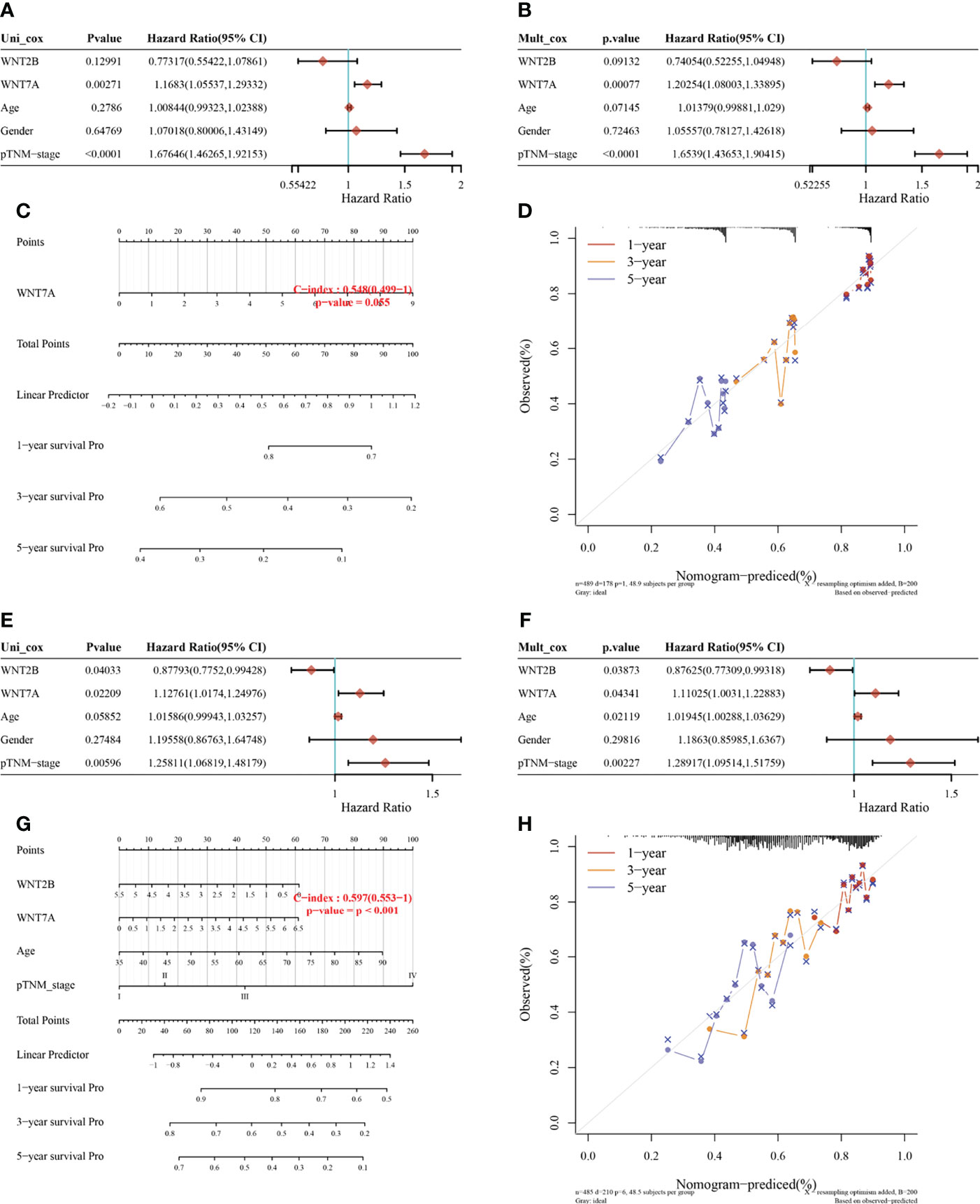
Figure 5 Univariate and multivariate Cox regression analysis of WNT gene expression and clinical characteristics and the nomogram predicting 1-y, 3-y, and 5-y survival in patients with LUAD and LUSC. (A, B, E, F) Hazard ratio and p value of constituents involved in univariate and multivariate Cox regression in LUAD (A, B) and LUSC (E, F). (C, G) Nomogram to predict the 1-y, 3-y and 5-y overall survival of LUAD (C) and LUSC (G) patients. (D, H) Calibration curve for the overall survival nomogram model in the discovery group. The dashed diagonal line represents the ideal nomogram, and the red, orange and blue represent the 1-y, 3-y and 5-y observed nomograms. (D) for LUAD. (H) for LUSC.
Through the cBioPortal online tool, we obtained the LUAD (TCGA, Firehose Legacy, http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/LUAD/20160128/) and the LUSC (TCGA, Firehose Legacy, http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/LUSC/20160128/) datasets from TCGA to analyze the genetic alterations and associations of WNT genes in NSCLC patients. As shown in Figures 6A, B, the WNT gene family represented different degrees of mutations in LUAD and LUSC. The mutation rate of WNT9A was the highest (11%) in LUAD (Figure 6A), while that of WNT5B was the highest (10%) in LUSC (Figure 6B). mRNA High and mRNA Low were the most common genetic changes in LUAD and LUSC.
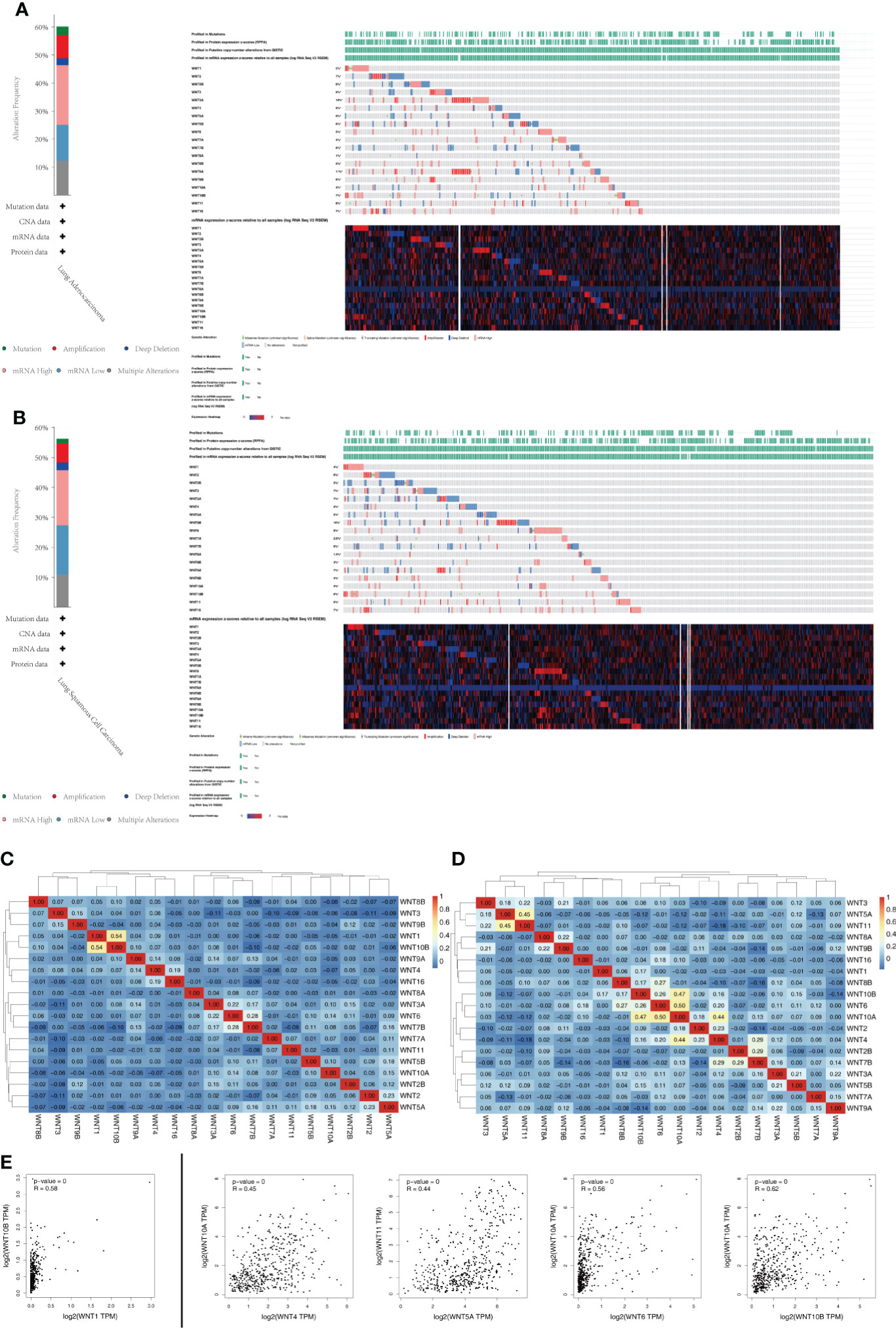
Figure 6 Genetic alterations and association analysis of the WNT gene family in NSCLC patients. (A) Genetic alterations of the WNT Gene Family in LUAD (cBioPortal). (B) Genetic alterations of the WNT Gene Family in LUSC (cBioPortal). (C, D) Correlation analysis heatmap between different WNT genes in LUAD (C) and LUSC (D). (E) Correlation analysis between different WNT genes (GEPIA).
In addition, Pearson’s correlation coefficients of relevant genes were obtained by the analysis of mRNA expression (RNA Seq V2 RSEM) samples (Figures 6C, D) and visualized by R software. A moderate correlation was observed between WNT1 and WNT10B (p value = 2.07e-40) in LUAD (Figure 6C), as well as between WNT4 and WNT10A (p value = 8.53e-25), WNT5A and WNT11 (p value = 5.96e-26), WNT6 and WNT10A (p value = 1.99e-32), and WNT10A and WNT10B (p value = 3.81e-29) in LUSC (Figure 6D). A total of 19 members of the WNT gene family were analyzed by the Correlation Analysis module of GEPIA, and similar correlation expression results were obtained, as shown in Figure 6E. In LUAD, WNT1 was moderately correlated with WNT10B (R = 0.58, p value = 0). However, In LUSC, moderate correlation was observed between WNT4 and WNT10A (R = 0.45, p value = 0), WNT5A and WNT11 (R = 0.44, p value = 0), and WNT6 and WNT10A (R = 0.56, p value = 0); moreover, a strong correlation was observed between WNT10A and WNT10B (R = 0.62, p value = 0).
To further predict the functional role of the WNT gene family, we performed GO enrichment and KEGG pathway analysis of WNT gene family members with differential expression. Figure 7A showed that, in Biological Process, WNT genes were mainly enriched in canonical WNT signaling pathway, cellular response to retinoic acid, pattern specification process, regulation of animal organ morphogenesis, regionalization, immune system development, developmental induction, cell fate commitment, positive regulation of myeloid leukocyte cytokine production involved in immune response, etc. In the part of Molecular Function (Figure 7B), WNTs were mainly enriched in signaling receptor binding, molecular function regulator, receptor regulator activity, receptor ligand activity, G protein-coupled receptor binding, Fzd binding, protein domain specific binding, cytokine activity, regulatory region nucleic acid binding, co-receptor binding, etc. Additionally, WNT genes in Cellular Component (Figure 7C) were mainly enriched in extracellular region, Golgi apparatus part, vesicle membrane, cytoplasmic vesicle membrane, endoplasmic reticulum lumen, endocytic vesicle, endocytic vesicle membrane, Golgi lumen, collagen-containing extracellular matrix, WNT signalosome, etc. Subsequently, KEGG pathway enrichment analysis (Figure 7D) identified that WNT genes were mainly enriched in Signaling pathways regulating pluripotency of stem cells, mTOR signaling pathway, Hippo signaling pathway, WNT signaling pathway, Basal cell carcinoma, Breast cancer, Gastric cancer, Cushing syndrome, Hepatocellular carcinoma, melanogenesis, etc.
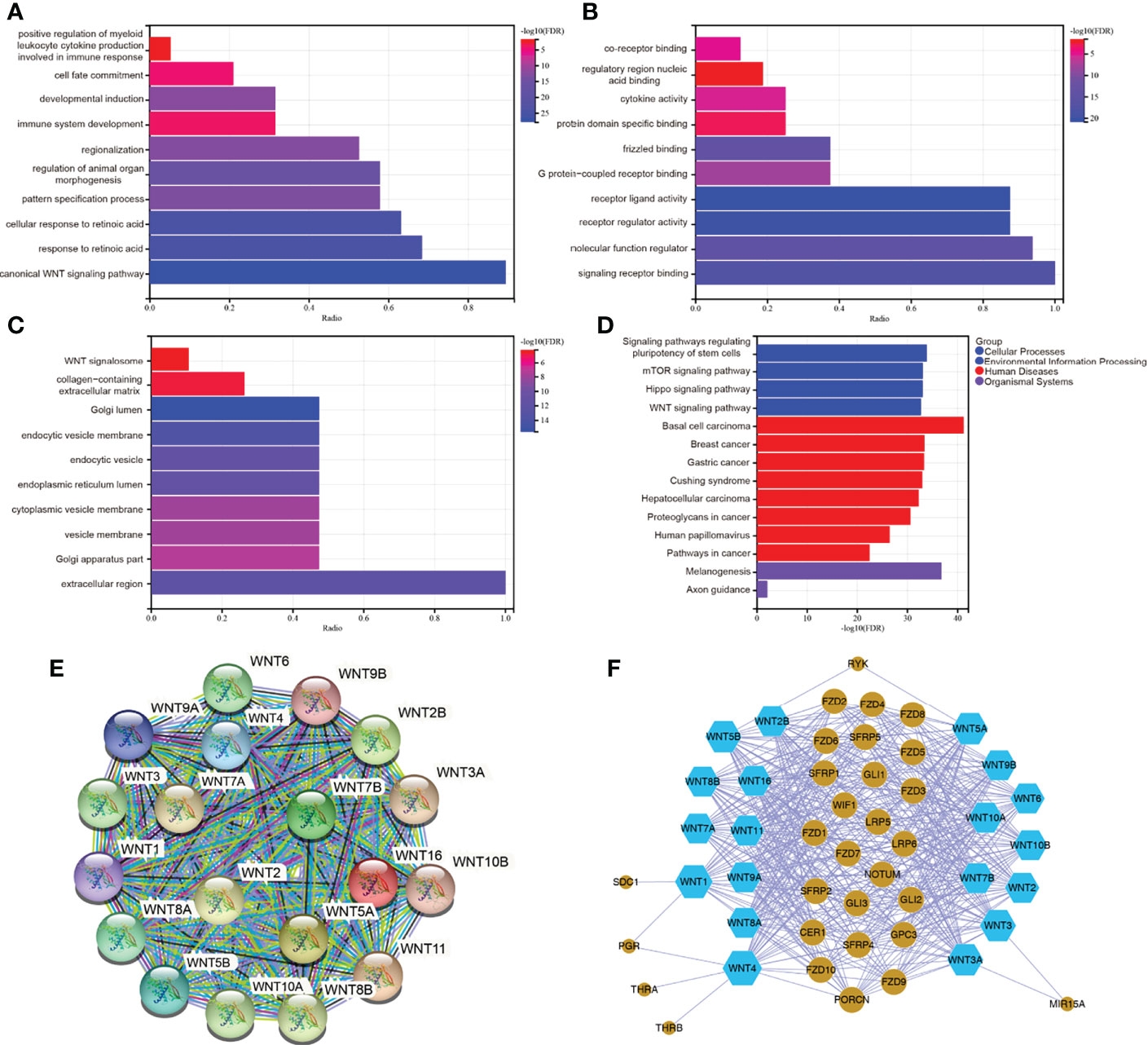
Figure 7 (A–D) GO and KEGG enrichment analysis of WNT gene family. (E, F) Protein-protein interaction network of WNT gene family. (E) for STRING. (F) for Cytoscape.
The PPI network analysis graph was constructed by STRING (Figure 7E) and Cytoscape (Figure 7F). STRING contained 19 nodes and 171 edges, PPI enrichment p value < 1.0e-16. As can be seen in Figure 7F, members of the WNT gene family had a close relationship with the Fzd receptor family, lipoprotein receptor-related protein (LRP) family, secreted Fzd-related proteins (SFRPs), and WNT inhibitory protein (WIF) family. Taken together, the functional enrichment and pathway analysis results for WNTs were as we know. The biological functions of the WNT gene family were mainly related to the canonical and non-canonical WNT signaling pathways, G-protein-coupled receptor binding, immune system development, and partial regulation of immune response.
Compared with that of normal tissues, the transcriptional expression of WNTs was significantly altered in both LUAD and LUSC patients. Since transcription factors were involved in the human transcription initiation, we used the TRRUST database to explore the key transcription factors of WNT genes in humans. Among 19 WNT genes, WNT2, WNT2B, WNT3, WNT3A, WNT4, WNT5A, WNT7B, and WNT11 were included in the TRRUST database. We found that PITX2 was a key transcription factor associated with WNT gene regulation (Table 3).
Through the TIMER database, we explored the correlation between the WNT gene and immune cell infiltration (Figure 8). In LUAD, WNT2, WNT9B, and WNT10A represented a significantly positive correlation with six immune cells, including B cell, CD4+ T cell, CD8+ T cell, dendritic cell, macrophage, and neutrophil; WNT3 and WNT8B were positively correlated with tumor purity, while WNT1/2/2B/6/7A/7B/9A/9B/10A/10B/16 were negatively correlated with tumor purity. In LUSC, WNT1, WNT2, WNT7B, and WNT9B were significantly correlated with six immune cells, among which WNT1, WNT2, and WNT9B were positively correlated, and WNT7B was negatively correlated. However, the effect of WNTs on tumor purity in LUSC was slightly poorer than that in LUAD, and WNT3/5A/8B/11 were positively correlated with tumor purity, while WNT1/2/4/6/7A/98/10A were negatively correlated with tumor purity. For WNT2B and WNT7A, which might have prognostic value in LUAD and LUSC, as can be seen, in LUAD, WNT2B was positively correlated with all immune infiltration cells except CD8+ T cell; WNT7A was positively correlated with CD4+ T cell, macrophage, neutrophil and dendritic cell. In LUSC, WNT2B was negatively correlated with CD8+ T cell, neutrophil and dendritic cell; WNT7A was significantly correlated with B cell, CD4+ T cell and macrophage, among which CD4+ T cell and macrophage were positively correlated, and B cell was negatively correlated. Subsequently, we validated the association of WNT2B and WNT7A with immune cell infiltration by the TISIDB database. Figures 9A, C represented the relationship between WNT2B and WNT7A and tumor-infiltrating lymphocytes (TILs) in different cancer types. Figure 9B showed that, in LUAD and LUSC, except for CD4+ T cell and neutrophil, the correlation between WNT2B and other TILs infiltration levels was the same as the results obtained in TIMER. Similarly, Figure 9D represented that except for CD4+ T cell and B cell, the correlation between WNT7A and other TILs infiltration levels was the same as the results obtained in TIMER. Additionally, based on the dataset containing mRNA sequence data from TCGA, we constructed a heatmap of the correlation between WNTs and immune scores (Supplementary Figure 2), as well as a lollipop chart (Supplementary Figure 3) implemented by the ssGSEA algorithm to show the correlation between WNT2B and WNT7A and the level of immune cell infiltration, and these results are generally compatible with ours. Next, using a Cox proportional hazards model in TIMER, we explored the clinical relevance of WNTs with LUAD and LUSC tumor immunity, with the results shown in Tables 4 and 5. As can be seen, B cell (p < 0.001), WNT2B (p < 0.05), WNT3A (p < 0.05), WNT6 (p < 0.05) and WNT7A (p < 0.05) were significantly associated with clinical outcomes in LUAD (Table 4) but not in LUSC (Table 5).
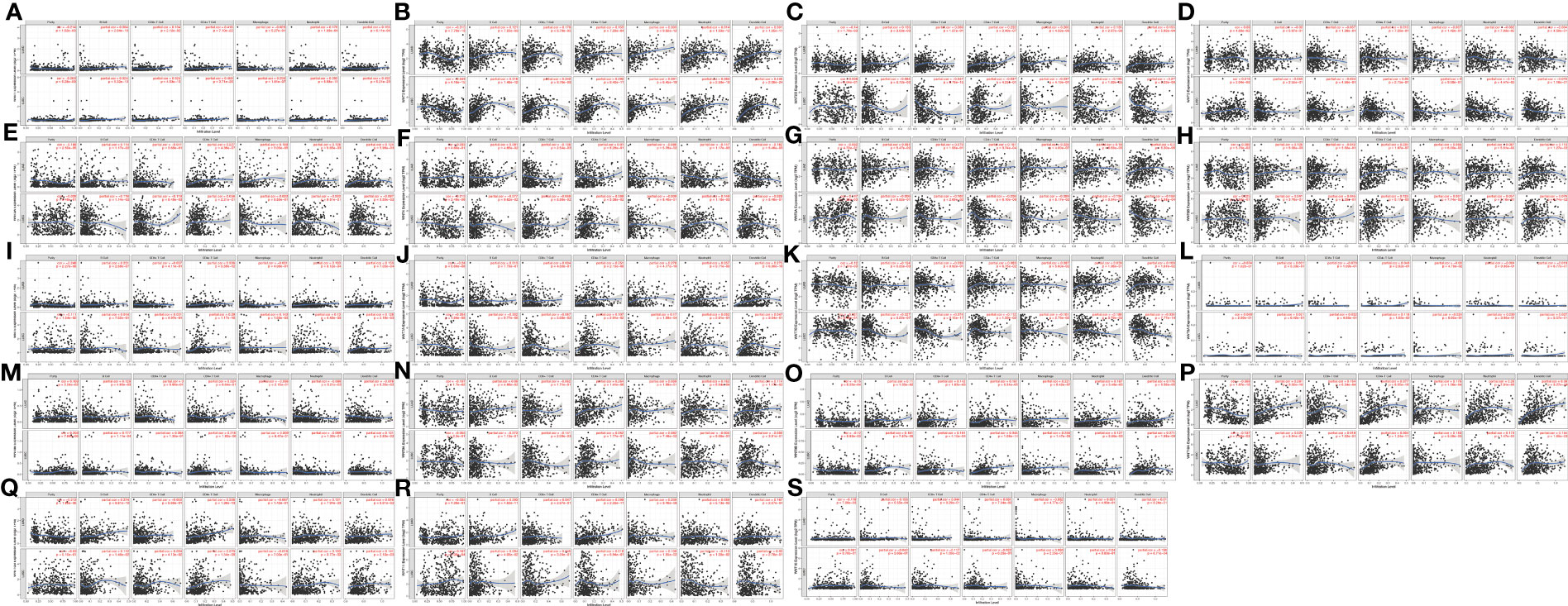
Figure 8 Correlation of WNT genes with immune cell infiltration in LUAD and LUSC (TIMER). (A) WNT1. (B) WNT2. (C) WNT2B. (D) WNT3. (E) WNT3A. (F) WNT4. (G) WNT5A. (H) WNT5B. (I) WNT6. (J) WNT7A. (K) WNT7B. (L) WNT8A. (M) WNT8B. (N) WNT9A. (O) WNT9B. (P) WNT10A. (Q) WNT10B. (R) WNT11. (S) WNT16.

Figure 9 The association of WNT2B and WNT7A with immune cell infiltration (TISIDB). (A, C) The landscape of relationship between WNT2B (A) and WNT7A (C) expression and TILs in different types of cancer (red is positive correlated and blue is negative correlated). (B) Relationship between WNT2B and immune infiltration levels of B cell, CD4+ T cell, macrophage, neutrophil, dendritic cell in LUAD, and CD8+ T cell, neutrophil, dendritic cell in LUSC. (D) Relationship between WNT7A and immune infiltration levels of CD4+ T cells, macrophage, neutrophil, dendritic cell in LUAD, and B cell, CD4+ T cell, macrophage in LUSC.
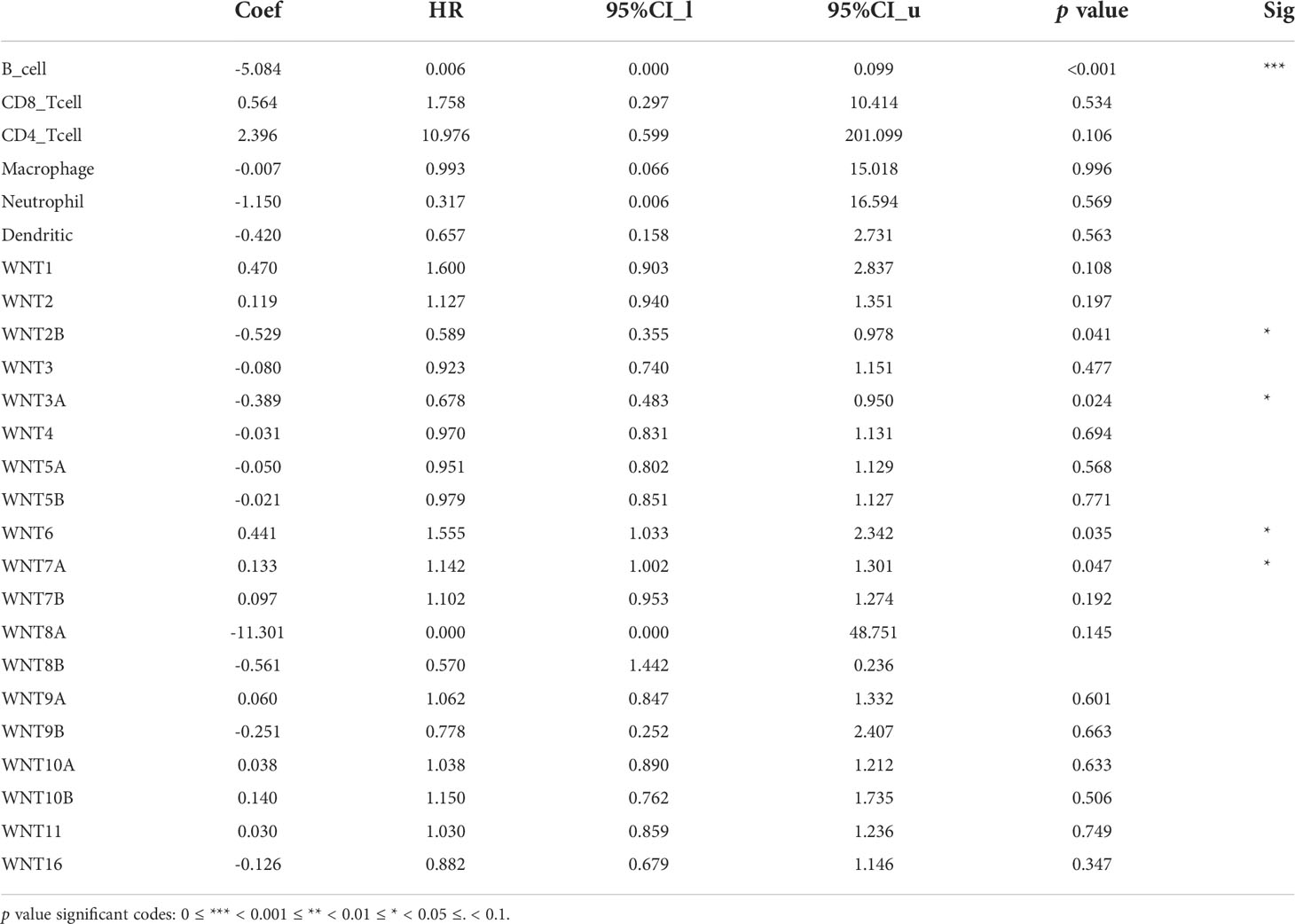
Table 4 The Cox proportional hazard model of WNT genes with tumor-infiltrating immune cells in LUAD patients.

Table 5 The Cox proportional hazard model of WNT genes with tumor-infiltrating immune cells in LUSC patients.
Moreover, for the 19 genes of WNTs, we compared the tumor infiltration level with different degrees of somatic copy number alterations (SCNA) in LUAD and LUSC patients, with the results illustrated in Figure 10. We found that all significant copy number change types appeared in arm-level deletion, arm-level gain, deep deletion, and high amplification. In LUAD, WNT7B and WNT8A were significantly correlated with deep deletion and arm-level deletion of six immune cells, respectively, and WNT3A and WNT9A were significantly correlated with high amplification of six immune cells. In LUSC, WNT4, WNT5A, WNT7A, and WNT8B were significantly correlated with arm-level deletion of six immune cells; WNT3 and WNT9B were significantly correlated with the arm-level gain of six immune cells; WNT7B was significantly correlated with high amplification of six immune cells. For WNT2B and WNT7A, WNT2B is significantly correlated with arm-level deletion of CD8+ T Cell, CD4+ T cell, macrophage, neutrophil and dendritic cell in LUAD; in LUSC, WNT2B showed a significant correlation with CNA levels in all immune cells except for CD8+ T cells. Similarly, WNT7A showed a significant correlation with CNA levels of all immune cells in LUAD except for CD8+ T Cell; and WNT7A showed a significant correlation with CNA levels of all immune cells in LUSC.
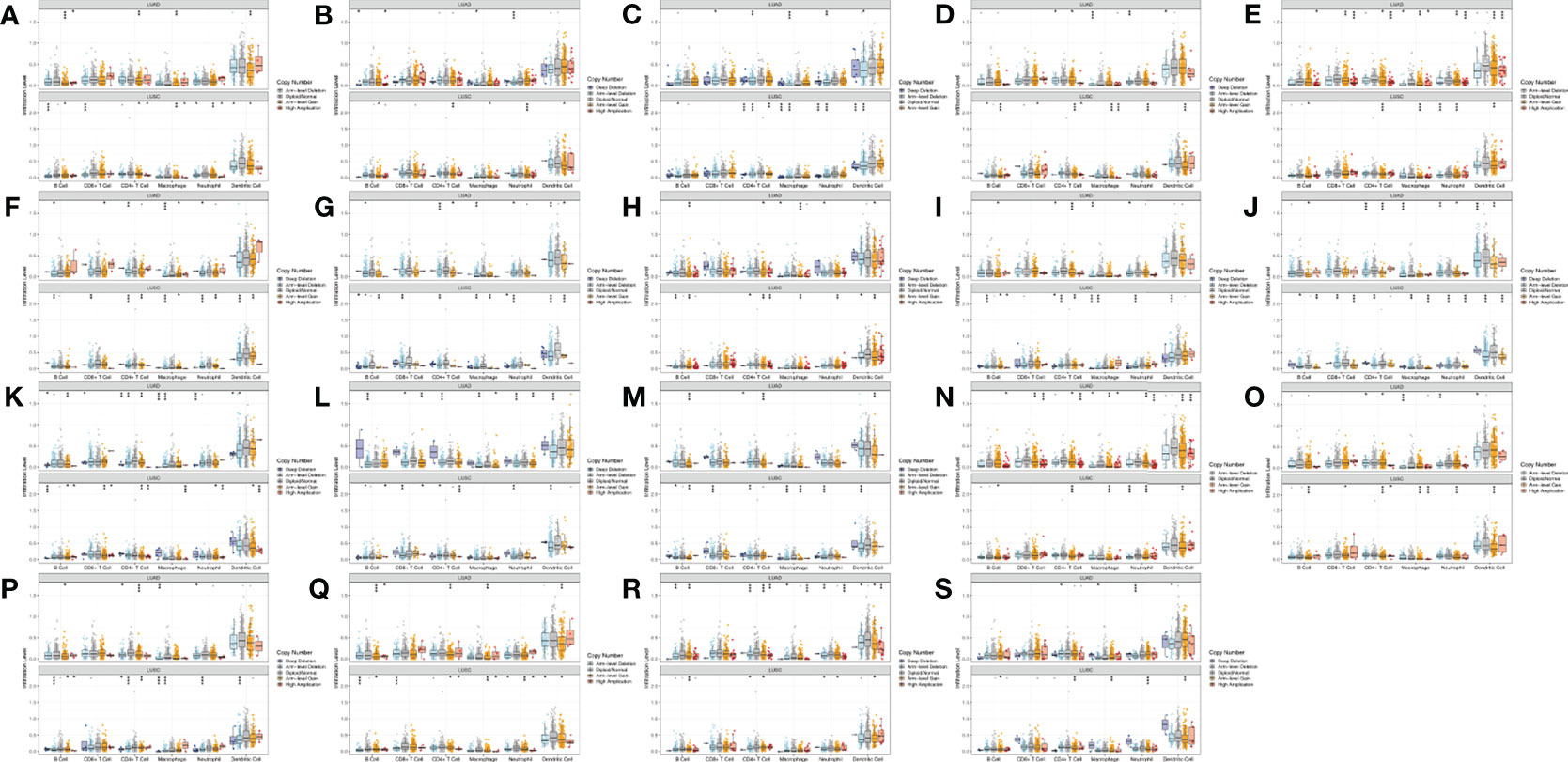
Figure 10 Comparison of tumor infiltration levels among tumors with different SCNA for WNTs in LUAD and LUSC samples (p value significant codes: 0 ≤ *** < 0.001 ≤ ** < 0.01 ≤ * < 0.05 ≤. < 0.1). (A) WNT1. (B) WNT2. (C) WNT2B. (D) WNT3. (E) WNT3A. (F) WNT4. (G) WNT5A. (H) WNT5B. (I) WNT6. (J) WNT7A. (K) WNT7B. (L) WNT8A. (M) WNT8B. (N) WNT9A. (O) WNT9B. (P) WNT10A. (Q) WNT10B. (R) WNT11. (S) WNT16.
To validate the mRNA expression levels of WNT2B and WNT7A in adjacent normal tissues, LUAD tissues, and LUSC tissues, qRT-PCR was performed (Figure 11). The results indicated that the expressions of WNT2B and WNT7A were downregulated in tissue samples from 10 LUAD cases. Moreover, WNT2B expression was upregulated, while WNT7A expression was downregulated in tissue samples from 10 LUSC cases (Figure 11A). In addition, compared with normal lung epithelial cell line (BEAS-2B), WNT2B and WNT7A were significantly underexpressed in the LUAD cell line (PC9), WNT2B was significantly highly expressed in the LUSC cell line (NCI-H520), and WNT7A was significantly underexpressed in LUSC cell line (NCI-H520) (Figure 11B). These results were consistent with the results of bioinformatics analysis. Next, the protein expression levels of WNT2B and WNT7A were validated by western blot (Figures 11C–F) and immunohistochemical staining (Figures 11G–J), which were consistent with the mRNA expression levels.
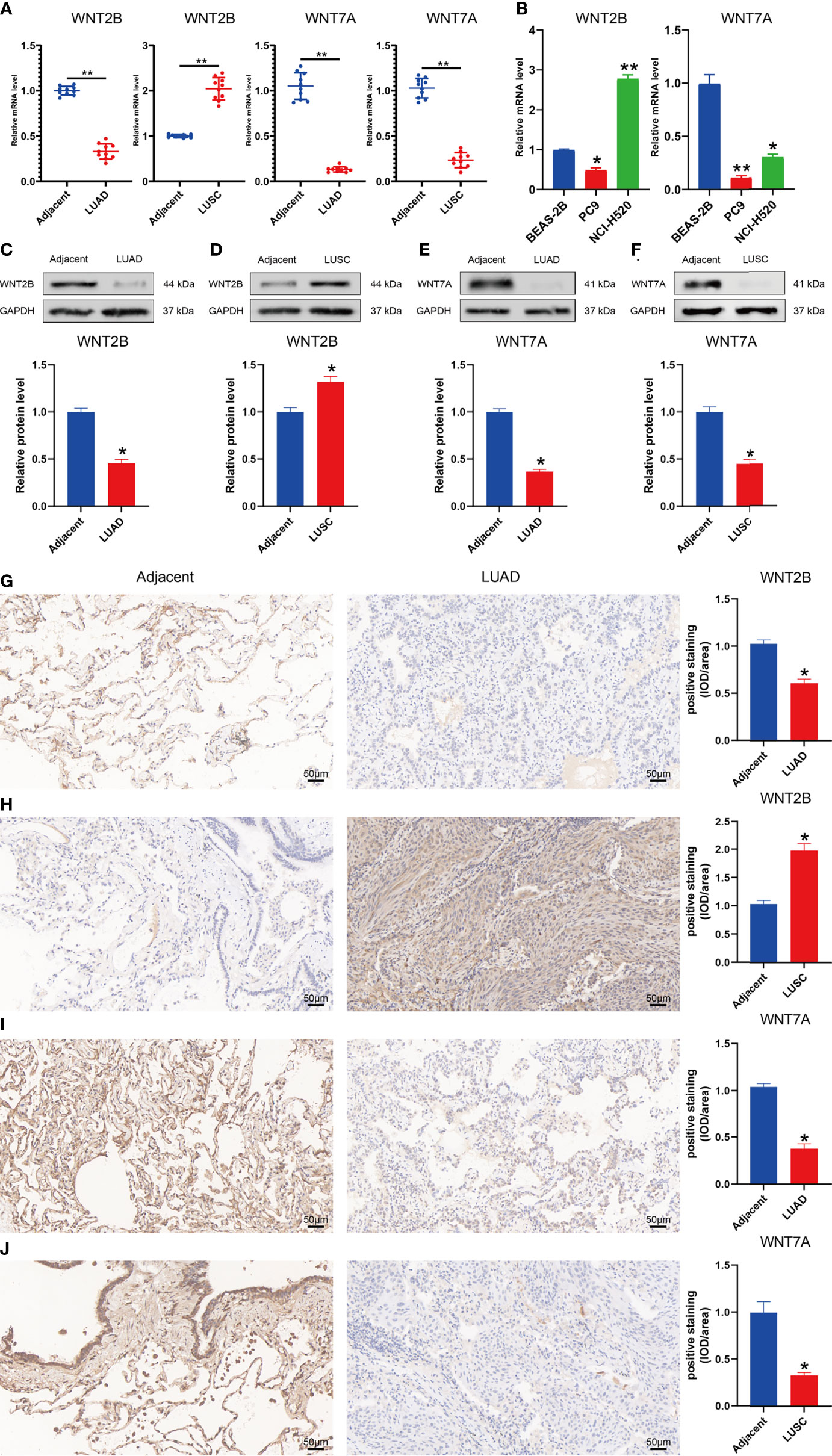
Figure 11 Validation of mRNA and protein expression levels of WNT2B and WNT7A in NSCLC. (A) Comparison of mRNA expression levels of WNTs in 20 paired NSCLC tissues and adjacent normal tissue samples by qRT-PCR. (B) Comparison of mRNA expression levels of WNTs in LUAD cell line (PC9) and LUSC cell line (NCI-H520) with human normal lung epithelial cell line (BEAS-2B). (C-F) The western blot analyses. (G–J) The immunohistochemical staining in 20 NSCLC tissues and adjacent normal tissues.
At the beginning of this study, we obtain the complete mRNA expression of WNT in LUAD and LUSC through three databases. In LUAD patients, WNT2/2B/3A/4/7A/9A/9B/11 expressions decrease, and WNT3/5B/6/7B/8B/10A/10B/16 expressions increase. In contrast, in LUSC patients, WNT2/3A/7A/11 expressions are decreased, and WNT2B/3/5A/5B/6/7B/10A/10B/16 expressions are increased. As in our findings, WNT2 is overexpressed in NSCLC (37). A higher expression of WNT2 was associated with a worse survival prognosis in some studies (38). In addition, anti-WNT2 antibodies could induce specific apoptosis in NSCLC by inhibiting WNT signaling (37), making anti-WNT2 monoclonal antibodies one of the strategies for treating lung cancer. Robert A Winn et al. carried out cell experiments and found that the mRNA expression level of WNT7A was significantly reduced in NSCLC cells and primary tumors (39), consistent with our down-regulation found.
In addition, according to the methylation analysis of the UALCAN database, in LUAD, the methylation levels of WNT1/2/2B/3/3A/6/7A/8A/9A/9B/10A/10B/16 are significantly enhanced, while those of WNT8B and WNT11 are significantly reduced; in LUSC, the methylation levels of WNT1/2/2B/3/3A/4/6/7A/7B/9B/10A/10B/16 are significantly promoted, while those of WNT5B/8A/8B/9A/11 are significantly inhibited. Methylation of WNTs has been demonstrated to show a partial correlation with tumor development (40). Tae-Hyung Kim et al. investigated a correlation between WNT7A promoter methylation and loss of E-cadherin expression by detecting WNT7A promoter methylation in NSCLC patients (40). E-cadherin loss was closely related to the poor prognosis and survival of many cancers (41), suggesting that promoter methylation-induced WNT7A loss might be an important prognostic factor in NSCLC. The 3p25 is the chromosome location of the WNT7A (Table 1). Cytogenetic analysis reveals that 3p25 is a common genetic deletion site in lung cancer, and similar regions also include 3p21.3 and 3p14-cen (42). The elevated methylation levels of WNT7A may be associated with its loss of expression (43).
In the next prognostic value and survival analysis, we first explore the WNT expression at different pathological stages of LUAD and LUSC patients. With the LUAD development, the WNT2 expression decreases as a whole, reaching the lowest level when the tumor progresses to stage IV; the expression of WNT2B presents no obvious change but tends to decrease with the disease progression, of which a few patients show a sudden rise in the expression in stage IV; WNT11 showed an increase in the expression with the tumor development. The results indicate that WNT2/2B/11 may be able to predict the tumor progression of LUAD patients. Next, we obtain the prognostic outcomes shown by the WNT gene with different degrees of expression in NSCLC patients by KM survival curves, and the survival results are also validated by another open database. In our study, highly expressed WNT2B and lowly expressed WNT7A predict better prognosis in LUAD and LUSC patients. After regression analysis and screening, the prediction results of WNT7A during the LUAD are not significant enough, according to the significant differential results obtained by univariate and multivariate Cox analyses of RNA-seq data from LUAD samples in TCGA. Despite this, we believe that WNT2B and WNT7A may act the important prognostic values in LUAD and LUSC, based on the significant differences shown in KM curves. The nomogram provides some guidance for WNT7A and WNT2B in predicting disease progression and survival in LUSC patients clinically.
WNT7A bound to Fzd9 and promoted epithelial differentiation of NSCLC cells by activating the cJun N-terminal kinases (JNKs) pathway (44). Moreover, the low expression of WNT7A in NSCLC might play a role in lung cancer progression through its effect on E-cadherin transcription (45). WNT7A could also induce hypertrophy of muscle fibers by driving activation of the AKT/mTOR signaling pathway (46). In addition, WNT7A acted as an upstream signaling molecule of the WNT canonical pathway (47). Its overexpression could transcriptionally activate WNT target genes through the WNT/β-catenin signaling pathway, thus leading to cancer development, or it could bind to some WNT receptors to promote the malignant phenotype of cancer (48), partially corroborating our findings. Some studies have suggested that the up-regulated expression of WNT2B promoted the malignant behavior of NSCLC (49, 50). After methodological comparison, we believe that the reason for the difference in the above results with our study may be relevant to the different cell types of NSCLC. That is, the previous studies focus on all NSCLC patients; in contrast, in this study, the prognostic analysis for WNT2B is based on the clinical samples of LUAD and LUSC patients. In addition, in our results, the transcriptional expression of WNT2B is divergent in LUAD and LUSC patients, implying the different roles of WNT2B in tumor progression in LUAD and LUSC. More research is required to explore the lung cancer field where WNT2B is located, which may be a breakthrough point in subsequent studies. WNT8B is one of the typical WNT ligands, transcriptionally regulated by zinc finger transcription factor 191 (ZNF 191) and playing an important role in hepatocellular carcinoma (HCC) proliferation through the canonical WNT pathway (serving as a new prognostic marker for HCC) (51). Few related studies were found on WNT8B in lung cancer, which may provide some directions for future research. It is worth mentioning that in the existing studies, WNT7A and WNT2B played different roles in the prognosis of different cancer types, which may be attributed to their various mechanisms of action in diverse cancer cell lines and the activated signaling pathways. More critically, our study is the first to use clinical samples to link the WNT expression with different cell types in LUAD and LUSC, providing more insights and evidence for the subsequent research of the WNT gene family in the prognosis of LUAD and LUSC patients. We look forward to more subsequent extensive studies of WNTs in different cell types of NSCLC.
Immediately afterward, we explore the WNT gene mutation in LUAD and LUSC by cBioPortal. All WNT genes are detected to show different degrees of mutation, of which mRNA High and mRNA Low are the most common genetic changes in LUAD and LUSC. The mutations presented by WNTs showed significant phenotypes in mice, Caenorhabditis elegans, and Drosophila (10). Heparin-sulfated forms of proteoglycans (HSPG) were involved in the WNT transport and played a role in stabilizing WNT proteins or assisting their transmission between cells. In Drosophila, loss of HSPG or mutations in genes encoding enzymes that modify them could lead to the generation of phenotypes similar to wingless mutants (9). Since it is associated with the biological role of stem cells (9), WNT pathway mutations are very common in cancer, especially in tissues relying on WNT for self-repair (11). However, mutations in WNT pathway components frequently appeared in colorectal cancer (19, 52). Although some studies have shown that mutations in β-catenin and its pathway components caused downstream activation of WNT signaling in LUAD (53), this mutation was still rare in lung cancer (13, 54). Analysis of RNA-seq and miRNA-seq data from several NSCLC patients suggested that alterations in WNT9A might impact cancer progression by affecting the interference between lncRNAs, miRNAs, and mRNAs (55). WNT5B signaled through the non-canonical WNT signaling pathway, which was usually used as an antagonist of the canonical WNT signaling pathway and played a role in the proliferation, differentiation, and migration of cells (56). Pairwise WNT genes presenting tight links in correlation analysis might play a synergistic or similar role in cancer progression. And both WNT5A and WNT11 act through the non-canonical WNT/calcium signaling pathway. It was found that when the mRNA of WNT5A or WNT11 was injected into one-cell zebrafish embryos, both of the mRNA of WNT5A or WNT11 could double the frequency of calcium transients in the enveloping layer of the blastodisc (57). Similar to WNT11 (58, 59), WNT5A has also been shown to help to inhibit cell migration and invasiveness in cancer cell lines (60).
According to the functional enrichment and pathway analysis results, we can see that WNT genes are highly enriched in WNT signaling pathways, immune system development and Fzd receptor/G protein-coupled receptor binding, etc. These functions are highly correlated with the action mode of the WNT gene family in the body. For example, overexpression of WNT4, WNT5A, and WNT11 has been found to play a role in early embryo development through the non-canonical WNT signaling pathway (57, 61). As an important regulator of cell growth and differentiation, WNT5A has also been demonstrated to participate in the tumor growth inhibition through the non-canonical WNT/calcium signaling pathway (62). WNT11 acted as a non-canonical WNT pathway ligand and similarly activated the WNT/calcium signaling pathway (63). WNT signaling might also serve as an essential regulator of stem cell proliferation or self-renewal selection in the body. For example, WNT3A promoted the self-renewal of hematopoietic stem cells in vitro (9). It turned out that there was overexpression of some WNT pathway components and active WNT signaling in most NSCLC (13). Mark Shapiro et al. performed immunoblot analysis on tumor and normal lung tissues from several patients with stage I NSCLC after resection, confirming that activation of the WNT pathway was significantly associated with high tumor recurrence rates (64).
It is well-known that transcription factors play an important role in the transcription process in humans. Therefore, we query key WNT-associated transcription factors in humans. Paired-like homeodomain transcription factor 2 (PITX2) is one of the bicoid/paired-like homeobox gene family, which is located at position 4q25 of the chromosome and is a transcription factor involved in the anterior structure development (65). PITX2 could promote cancer progression by activating the WNT gene and regulating the WNT/β-catenin pathway (36). In addition, PITX2 was also found to be a target gene of LEF1 (66). During WNT/β-catenin pathway regulation, PITX2 could activate specific growth regulatory genes, such as cyclin D1, cyclin D2 and c-Myc Kioussi (67). Jing Luo et al. investigated the involvement of PITX2 in the WNT/β-catenin pathway in LUAD, showing that PITX2 was a potential up-regulated oncogene in LUAD, and PITX2 was overexpressed in LUAD and relevant to poor prognosis. They also found that PITX2 could activate the WNT/β-catenin pathway by enhancing WNT3A transcription, thereby exerting an oncogenic effect (68).
After this, we explore the association between the WNT gene family and six immune cells, including B cell, CD4+ T Cell, CD8+ T Cell, dendritic cell, macrophage, and neutrophil. The prognosis-related genes WNT2B and WNT7A are significantly correlated with immune cell infiltration and the infiltration level of different SCNAs in both LUAD and LUSC. WNT signaling has been demonstrated to regulate immune cell function, significantly inhibit the maturation and differentiation of T cells and dendritic cells (69), and alter the differentiation of CD4+ cells and CD8+ effector T cells (70, 71). WNT signaling plays an important role in the tumor immune microenvironment, and WNT signaling can promote naive T cell maturation and control the proliferation and survival of progenitor B-cells and B-1 cells (72, 73). Moreover, WNT signaling can also increase the level of β-catenin in malignant cells, which leads to the subsistence of Regulatory T cells (Tregs), differentiation of CD4 + T cells into Th17 subtypes and secretion of IL10 and IL12 in dendritic cells (74). In addition, the canonical WNT signaling pathway was identified as one of the important immune evasion-related oncogenic pathway signals (75). Therefore, relevant WNT signaling pathways have been increasingly recognized as a potential target for cancer therapy. NFAT is a transcription factor regulated by the calcium/calmodulin-dependent protein phosphatase calcineurin and a downstream factor of WNT5A, localizing in the cytoplasm of resting T cells (76, 77). NFAT could enhance the expression of IL-2, an important mediator of graft-versus-host disease, under the stimulation of T cells, while cyclosporine A (CsA) acted as an immunosuppressive agent by reducing the nuclear accumulation of NFAT and then IL-2 expression (78). However, in the presence of CsA, T cell treatment with WNT5A resulted in nuclear accumulation of NFAT, enabling WNT5A to lead to CsA resistance through the WNT/heterotrimeric GTP-binding protein pathway (57). Through the B cell proliferation in WNT5A-deficient mice, WNT5A was found to inhibit B cell proliferation through WNT/calcium to act as a tumor suppressor (62). Immune escape has been a key in cancer research in recent decades, and the advent of treatments such as molecular therapy and immune checkpoint inhibition for cancer has also shed light on the conquest of lung cancer (5–7). Overall, based on the results in TIMER and TISIDB, our results suggest that there may be a degree of correlation between WNT2B and WNT7A and the level of immune cell infiltration. Based on limitations such as study objectives and experimental samples, etc, we did not conduct further experiments to verify the relationship between WNTs and immune cells, therefore, this field still needs to be explored through a large number of experiments. Furthermore, because of new drug resistance mechanisms, gene polymorphisms, and other reasons, many studies are still expected to support the research and treatment methods in this field.
Finally, we validate the mRNA and protein expression levels of WNT2B and WNT7A in NSCLC by qRT-PCR, western blot, and immunohistochemical staining, obtaining consistent experimental results with our analysis, further increasing the credibility and accuracy of our conclusions.
In summary, our study provides some insights into the WNT gene family in NSCLC patients about differential expression profile, potential prognostic value, immune infiltration level, etc. Nevertheless, this study still has some limitations. First, we did not perform the gene knockdown or overexpression of WNT2B and WNT7A to investigate the effects of WNT2B and WNT7A on prognostically relevant functions such as proliferation and migration of NSCLC, which will be the research focus of our team in the next phase. Second, based on existing studies, the role of some WNT genes in cancer conduction is not exhaustive, and the detailed mechanism of these genes still needs numerous subsequent studies. Our research is expected to provide further exploration direction and reference significance for subsequent research in this field.
This paper, for the first time, explored the comprehensive situation of the WNT gene family in LUAD and LUSC by bioinformatics methods and the differences of the WNT gene family in LUAD and LUSC fields. According to the survival analysis results, WNT2B and WNT7A might have prognostic value in LUAD, and both of them might be important prognostic factors for LUSC patients. In addition, WNT2B and WNT7A might show some correlation with immune cell infiltration of LUAD and LUSC, but their specific prognosis and immune infiltration-related mechanisms need further experimental validation. This study may provide some direction for the subsequent exploration of the biomarker or prognostic value of the WNT gene family in NSCLC.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the ethics committee of the Third Xiangya Hospital of Central South University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
WL and MT designed the study. JW and QY prepared the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Hunan Province Natural Science Foundation (No. 2021JJ40939, No. 2022JJ40253), the Scientific research project of Hunan Health Commission (No. 202203014949, No. 202102041763, No. 20200985), the Changsha Municipal Natural Science Foundation (No. kq2014267), and the Hunan Cancer Hospital Climb Plan (No. 2020QH001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.911316/full#supplementary-material
1. Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available from: https://gco.iarc.fr/today.
2. Schwartz AG, Cote ML. Epidemiology of lung cancer. Adv Exp Med Biol (2016) 893:21–41. doi: 10.1007/978-3-319-24223-1_2
3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc (2008) 83(5):584–94. doi: 10.4065/83.5.584
4. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, et al. Diagnosis of lung cancer in small biopsies and cytology: Implications of the 2011 international association for the study of lung Cancer/American thoracic Society/European respiratory society classification. Arch Pathol Lab Med (2013) 137(5):668–84. doi: 10.5858/arpa.2012-0263-RA
5. Sankar K, Gadgeel SM, Qin A. Molecular therapeutic targets in non-small cell lung cancer. Expert Rev Anticancer Ther (2020) 20(8):647–61. doi: 10.1080/14737140.2020.1787156
6. Rafei H, El-Bahesh E, Finianos A, Nassereddine S, Tabbara I. Immune-based therapies for non-small cell lung cancer. Anticancer Res (2017) 37(2):377–87. doi: 10.21873/anticanres.11330
7. Steendam CM, Dammeijer F, Aerts J, Cornelissen R. Immunotherapeutic strategies in non-small-cell lung cancer: The present and the future. Immunotherapy (2017) 9(6):507–20. doi: 10.2217/imt-2016-0151
8. Bade BC, Dela Cruz CS. Lung cancer 2020: Epidemiology, etiology, and prevention. Clinics Chest Med (2020) 41(1):1–24. doi: 10.1016/j.ccm.2019.10.001
9. Logan CY, Nusse R. The wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol (2004) 20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126
10. Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes Dev (1997) 11(24):3286–305. doi: 10.1101/gad.11.24.3286
11. Clevers H, Nusse R. Wnt/B-catenin signaling and disease. Cell (2012) 149(6):1192–205. doi: 10.1016/j.cell.2012.05.012
12. Parsons MJ, Tammela T, Dow LE. Wnt as a Driver and Dependency in Cancer. Cancer discovery (2021) 11(10):2413–29. doi 10.1158/2159-8290.Cd-21-0190
13. Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst (2014) 106(1):djt356. doi: 10.1093/jnci/djt356
14. Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/B-catenin signaling pathway in colorectal cancer. Biomed Pharmacother (2019) 110:473–81. doi: 10.1016/j.biopha.2018.11.082
15. He S, Tang S. Wnt/B-catenin signaling in the development of liver cancers. Biomed Pharmacother (2020) 132:110851. doi: 10.1016/j.biopha.2020.110851
16. Gounari F, Signoretti S, Bronson R, Klein L, Sellers WR, Kum J, et al. Stabilization of beta-catenin induces lesions reminiscent of prostatic intraepithelial neoplasia, but terminal squamous transdifferentiation of other secretory epithelia. Oncogene (2002) 21(26):4099–107. doi: 10.1038/sj.onc.1205562
17. Tan SH, Barker N. Wnt signaling in adult epithelial stem cells and cancer. Prog Mol Biol Trans Sci (2018) 153:21–79. doi: 10.1016/bs.pmbts.2017.11.017
18. Gajos-Michniewicz A, Czyz M. Wnt Signaling in Melanoma. Int J Mol Scie (2020) 21(14):4852. doi 10.3390/ijms21144852
19. Mazieres J, He B, You L, Xu Z, Jablons DM. Wnt signaling in lung cancer. Cancer Lett (2005) 222(1):1–10. doi: 10.1016/j.canlet.2004.08.040
20. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Oncomine: A cancer microarray database and integrated data-mining platform. Neoplasia (2004) 6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2
21. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. Timer: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77(21):e108–e10. doi: 10.1158/0008-5472.Can-17-0307
22. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
24. Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS One (2013) 8(12):e82241. doi: 10.1371/journal.pone.0082241
25. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal (2013) 6(269):pl1–pl. doi: 10.1126/scisignal.2004088
26. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An r package for comparing biological themes among gene clusters. Omics (2012) 16(5):284–7. doi: 10.1089/omi.2011.0118
27. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. String V11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res (2019) 47(D1):D607–d13. doi: 10.1093/nar/gky1131
28. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. Trrust V2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res (2018) 46(D1):D380–d6. doi: 10.1093/nar/gkx1013
29. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. Tisidb: An integrated repository portal for tumor-immune system interactions. Bioinformatics (Oxford England) (2019) 35(20):4200–2. doi: 10.1093/bioinformatics/btz210
30. Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, et al. Selection of Ddx5 as a novel internal control for q-Rt-Pcr from microarray data using a block bootstrap re-sampling scheme. BMC Genomics (2007) 8:140. doi: 10.1186/1471-2164-8-140
31. Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One (2010) 5(4):e10312. doi: 10.1371/journal.pone.0010312
32. Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in alk-positive and Egfr/Kras/Alk-negative lung adenocarcinomas. Cancer Res (2012) 72(1):100–11. doi: 10.1158/0008-5472.Can-11-1403
33. Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mrna expression. Genome Res (2012) 22(7):1197–211. doi: 10.1101/gr.132662.111
34. Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mrna expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A (2001) 98(24):13790–5. doi: 10.1073/pnas.191502998
35. Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A (2001) 98(24):13784–9. doi: 10.1073/pnas.241500798
36. Basu M, Roy SS. Wnt/B-catenin pathway is regulated by Pitx2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, skov-3. J Biol Chem (2013) 288(6):4355–67. doi: 10.1074/jbc.M112.409102
37. You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, et al. Inhibition of wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene (2004) 23(36):6170–4. doi: 10.1038/sj.onc.1207844
38. Huang C, Ma R, Xu Y, Li N, Li Z, Yue J, et al. Wnt2 promotes non-small cell lung cancer progression by activating Wnt/B-catenin pathway. Am J Cancer Res (2015) 5(3):1032–46.
39. Winn RA, Marek L, Han SY, Rodriguez K, Rodriguez N, Hammond M, et al. Restoration of wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-Mediated growth inhibition and promotion of cell differentiation. J Biol Chem (2005) 280(20):19625–34. doi: 10.1074/jbc.M409392200
40. Yang YL, Chen MW, Xian L. Prognostic and clinicopathological significance of downregulated e-cadherin expression in patients with non-small cell lung cancer (Nsclc): A meta-analysis. PLoS One (2014) 9(6):e99763. doi: 10.1371/journal.pone.0099763
41. Wong SHM, Fang CM, Chuah LH, Leong CO, Ngai SC. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit Rev Oncol Hematol (2018) 121:11–22. doi: 10.1016/j.critrevonc.2017.11.010
42. Hibi K, Takahashi T, Yamakawa K, Ueda R, Sekido Y, Ariyoshi Y, et al. Three distinct regions involved in 3p deletion in human lung cancer. Oncogene (1992) 7(3):445–9.
43. Tennis MA, Vanscoyk MM, Wilson LA, Kelley N, Winn RA. Methylation of Wnt7a is modulated by Dnmt1 and cigarette smoke condensate in non-small cell lung cancer. PLoS One (2012) 7(3):e32921. doi: 10.1371/journal.pone.0032921
44. Heasley LE, Winn RA. Analysis of Wnt7a-stimulated jnk activity and cjun phosphorylation in non-small cell lung cancer cells. Methods Mol Biol (Clifton NJ) (2008) 468:187–96. doi: 10.1007/978-1-59745-249-6_14
45. Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, et al. Wnt7a induces e-cadherin in lung cancer cells. Proc Natl Acad Sci U S A (2003) 100(18):10429–34. doi: 10.1073/pnas.1734137100
46. Schmidt M, Poser C, von Maltzahn J. Wnt7a counteracts cancer cachexia. Mol Ther Oncolytics (2020) 16:134–46. doi: 10.1016/j.omto.2019.12.011
47. Liu Y, Meng F, Xu Y, Yang S, Xiao M, Chen X, et al. Overexpression of Wnt7a is associated with tumor progression and unfavorable prognosis in endometrial cancer. Int J Gynecol Cancer (2013) 23(2):304–11. doi: 10.1097/IGC.0b013e31827c7708
48. Kirikoshi H, Katoh M. Expression of Wnt7a in Human Normal Tissues and Cancer, and Regulation of Wnt7a and Wnt7b in Human Cancer. Int J Oncol (2002) 21(4):895–900. doi 10.3892/ijo.21.4.895
49. Wang B, Sun L, Li J, Jiang R. Mir-577 suppresses cell proliferation and epithelial-mesenchymal transition by regulating the Wnt2b mediated Wnt/B-catenin pathway in non-small cell lung cancer. Mol Med Rep (2018) 18(3):2753–61. doi: 10.3892/mmr.2018.9279
50. Wu Y, Cheng K, Liang W, Wang X. Lncrna Rpph1 promotes non-small cell lung cancer progression through the mir-326/Wnt2b axis. Oncol Lett (2020) 20(4):105. doi: 10.3892/ol.2020.11966
51. Liu Y, Wu D, Cheng H, Chen L, Zhang W, Zou L, et al. Wnt8b, transcriptionally regulated by Znf191, promotes cell proliferation of hepatocellular carcinoma Via wnt signaling. Cancer Sci (2021) 112(2):629–40. doi: 10.1111/cas.14738
52. Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of sfrp genes allows constitutive wnt signaling in colorectal cancer. Nat Genet (2004) 36(4):417–22. doi: 10.1038/ng1330
53. Juan J, Muraguchi T, Iezza G, Sears RC, McMahon M. Diminished wnt -> B-catenin -> c-myc signaling is a barrier for malignant progression of Brafv600e-induced lung tumors. Genes Dev (2014) 28(6):561–75. doi: 10.1101/gad.233627.113
54. Ueda M, Gemmill RM, West J, Winn R, Sugita M, Tanaka N, et al. Mutations of the beta- and gamma-catenin genes are uncommon in human lung, breast, kidney, cervical and ovarian carcinomas. Br J Cancer (2001) 85(1):64–8. doi: 10.1054/bjoc.2001.1863
55. Zheng C, Li X, Qian B, Feng N, Gao S, Zhao Y, et al. The lncrna myocardial infarction associated transcript-centric competing endogenous rna network in non-small-cell lung cancer. Cancer Manag Res (2018) 10:1155–62. doi: 10.2147/cmar.S163395
56. Suthon S, Perkins RS, Bryja V, Miranda-Carboni GA, Krum SA. Wnt5b in physiology and disease. Front Cell Dev Biol (2021) 9:667581. doi: 10.3389/fcell.2021.667581
57. Kohn AD, Moon RT. Wnt and calcium signaling: Beta-Catenin-Independent pathways. Cell Calcium (2005) 38(3-4):439–46. doi: 10.1016/j.ceca.2005.06.022
58. Ouko L, Ziegler TR, Gu LH, Eisenberg LM, Yang VW. Wnt11 signaling promotes proliferation, transformation, and migration of Iec6 intestinal epithelial cells. J Biol Chem (2004) 279(25):26707–15. doi: 10.1074/jbc.M402877200
59. Toyama T, Lee HC, Koga H, Wands JR, Kim M. Noncanonical Wnt11 inhibits hepatocellular carcinoma cell proliferation and migration. Mol Cancer Res (2010) 8(2):254–65. doi: 10.1158/1541-7786.Mcr-09-0238
60. Syed Khaja AS, Helczynski L, Edsjö A, Ehrnström R, Lindgren A, Ulmert D, et al. Elevated level of Wnt5a protein in localized prostate cancer tissue is associated with better outcome. PloS One (2011) 6(10):e26539. doi: 10.1371/journal.pone.0026539
61. Du SJ, Purcell SM, Christian JL, McGrew LL, Moon RT. Identification of distinct classes and functional domains of wnts through expression of wild-type and chimeric proteins in xenopus embryos. Mol Cell Biol (1995) 15(5):2625–34. doi: 10.1128/mcb.15.5.2625
62. Liang H, Chen Q, Coles AH, Anderson SJ, Pihan G, Bradley A, et al. Wnt5a inhibits b cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell (2003) 4(5):349–60. doi: 10.1016/s1535-6108(03)00268-x
63. Veeman MT, Axelrod JD, Moon RT. A second canon. functions and mechanisms of beta-Catenin-Independent wnt signaling. Dev Cell (2003) 5(3):367–77. doi: 10.1016/s1534-5807(03)00266-1
64. Shapiro M, Akiri G, Chin C, Wisnivesky JP, Beasley MB, Weiser TS, et al. Wnt pathway activation predicts increased risk of tumor recurrence in patients with stage I nonsmall cell lung cancer. Ann Surg (2013) 257(3):548–54. doi: 10.1097/SLA.0b013e31826d81fd
65. Logan M, Pagán-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell (1998) 94(3):307–17. doi: 10.1016/s0092-8674(00)81474-9
66. Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, et al. Identification of a Wnt/Dvl/Beta-catenin –> Pitx2 pathway mediating cell-Type-Specific proliferation during development. Cell (2002) 111(5):673–85. doi: 10.1016/s0092-8674(02)01084-x
67. Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, et al. The Wnt/Beta-Catenin–>Pitx2 pathway controls the turnover of Pitx2 and other unstable mrnas. Mol Cell (2003) 12(5):1201–11. doi: 10.1016/s1097-2765(03)00407-6
68. Luo J, Yao Y, Ji S, Sun Q, Xu Y, Liu K, et al. Pitx2 enhances progression of lung adenocarcinoma by transcriptionally regulating Wnt3a and activating Wnt/B-catenin signaling pathway. Cancer Cell Int (2019) 19:96. doi: 10.1186/s12935-019-0800-7
69. Staal FJ, Luis TC, Tiemessen MM. Wnt signalling in the immune system: Wnt is spreading its wings. Nat Rev Immunol (2008) 8(8):581–93. doi: 10.1038/nri2360
70. Hong Y, Manoharan I, Suryawanshi A, Majumdar T, Angus-Hill ML, Koni PA, et al. B-catenin promotes regulatory T-cell responses in tumors by inducing vitamin a metabolism in dendritic cells. Cancer Res (2015) 75(4):656–65. doi: 10.1158/0008-5472.Can-14-2377
71. Fu C, Liang X, Cui W, Ober-Blöbaum JL, Vazzana J, Shrikant PA, et al. B-catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of Cd8+ T cells through regulation of il-10. Proc Natl Acad Sci U S A (2015) 112(9):2823–8. doi: 10.1073/pnas.1414167112
72. Rothenberg EV, Moore JE, Yui MA. Launching the T-Cell-Lineage developmental programme. Nat Rev Immunol (2008) 8(1):9–21. doi: 10.1038/nri2232
73. Osugui L, de Roo JJ, de Oliveira VC, Sodré ACP, Staal FJT, Popi AF. B-1 cells and b-1 cell precursors prompt different responses to wnt signaling. PLoS One (2018) 13(6):e0199332. doi: 10.1371/journal.pone.0199332
74. Patel S, Alam A, Pant R, Chattopadhyay S. Wnt signaling and its significance within the tumor microenvironment: Novel therapeutic insights. Front Immunol (2019) 10:2872. doi: 10.3389/fimmu.2019.02872
75. Pai SG, Carneiro BA, Mota JM, Costa R, Leite CA, Barroso-Sousa R, et al. Wnt/Beta-catenin pathway: Modulating anticancer immune response. J Hematol Oncol (2017) 10(1):101. doi: 10.1186/s13045-017-0471-6
76. Rao A, Luo C, Hogan PG. Transcription factors of the nfat family: Regulation and function. Annu Rev Immunol (1997) 15:707–47. doi: 10.1146/annurev.immunol.15.1.707
77. Saneyoshi T, Kume S, Amasaki Y, Mikoshiba K. The Wnt/Calcium pathway activates nf-at and promotes ventral cell fate in xenopus embryos. Nature (2002) 417(6886):295–9. doi: 10.1038/417295a
Keywords: NSCLC, LUAD, LUSC, WNT gene family, prognosis, immune infiltration
Citation: Wang J, Yang Q, Tang M and Liu W (2022) Validation and analysis of expression, prognosis and immune infiltration of WNT gene family in non-small cell lung cancer. Front. Oncol. 12:911316. doi: 10.3389/fonc.2022.911316
Received: 02 April 2022; Accepted: 28 June 2022;
Published: 25 July 2022.
Edited by:
Simone Patergnani, University of Ferrara, ItalyCopyright © 2022 Wang, Yang, Tang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Liu, bGl1d2VpeHkzQGNzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.