- 1Department of Acupuncture and Moxibustion, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 2Department of Acupuncture and Moxibustion, Shanghai TCM-Integrated Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 3Department of Acupuncture and Moxibustion, Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 4Outpatient Department, Shanghai Research Institute of Acupuncture and Meridian, Shanghai, China
Sirtuin 3 (SIRT3), the nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase, acts as a metabolic modulator mainly located in mitochondria via regulating the process of the relevant biochemical processes by targeting crucial mediators. Recently, owing to its dual role in cancer, SIRT3 has attracted extensive attention. Cancer cells have different metabolic patterns from normal cells, and SIRT3-mediated metabolism reprogramming could be critical in the cancer context, which is closely related to the mechanism of metabolism reprogramming, metastasis, and chemoresistance in tumor cells. Therefore, it is crucial to elucidate the relevant pathological mechanisms and take appropriate countermeasures for the progression of clinical strategies to inhibit the development of cancer. In this review, existing available data on the regulation of cancer metabolism reprogramming, metastasis, and chemoresistance progression of SIRT3 are detailed, as well as the status quo of SIRT3 small molecule modulators is updated in the application of cancer therapy, aiming to highlight strategies directly targeting SIRT3-mediated tumor-suppressing and tumor-promoting, and provide new approaches for therapy application. Furthermore, we offer an effective evidence-based basis for the evolvement of potential personalized therapy management strategies for SIRT3 in cancer settings.
Introduction
Sirtuins are nicotinamide adenine dinucleotide (NAD+)-dependent class III histone deacetylases (HDACs) with homology to the saccharomyces cerevisiae silent information regulator 2 (Sir2) (1). Different from traditional HDACs, sirtuins deacetylate a series of substrates (2, 3). Moreover, as metabolic sensors, sirtuins connect their enzymatic activity with redox status and cellular energy through the co-factor NAD+ (1, 4). There are seven sirtuins, namely sirtuin 1-7 (SIRT1-7), that have been found in mammals. All sirtuins depend on NAD+, but they differ in location, types of post-translational modifications, and substrate affinity. Among these, SIRT3 has attracted much attention, as it exhibits the most powerful activity of mitochondrial deacetylase. SIRT3 could regulate a variety of mitochondrial proteins’ acetylation levels, such as metabolic enzymes and transcription factors, thereby playing a vital role in all kinds of cellular activities (5). Dysregulation of SIRT3 in cellular pathways may promote carcinogenesis and resistance to chemotherapy, which has been demonstrated in previous studies (6, 7). Thus, as a potential marker, SIRT3 expression could be identified in patients susceptible to metastasis or drug-resistant disease when cancer is initially diagnosed. Cancer remains a major health problem worldwide, and cancer metastasis is a principal consideration of cancer-related fatalities. In 2011, Hanahan and Weinberg summarized ten hallmarks of cancerous cells in Cell that include resisting cell death, enhancing invasion and metastasis, maintaining proliferative signaling, dysregulation of cellular energy, inducing angiogenesis, limitless replicative potential, evading immune destruction, gene instability and mutation, escaping growth inhibitors, and tumor-promoting inflammation (8). In fact, these ten cancer phenotypic features that differ from normal cells reflect altered metabolism or metabolism reprogramming of tumor cells. Despite recent advancements in cancer treatment, the chemotherapy resistance of neoplastic cells directly affects the prognosis of patients. Therefore, novel target-based avenues to enhance treatment effectiveness need to be explored.
Currently, owing to its dual role in cancer, SIRT3 has attracted lots of attention from researchers, particularly in the terms of regulating the metabolism reprogramming of tumor cells, involving cancer metastasis, drug resistance, and other aspects. Although reviews on the dual role of SIRT3 in cancer have been summarized and published, there is currently no systematic review on cancer metastasis and drug resistance about the role of SIRT3. In this review, we mainly pay close attention to how SIRT3 plays a dual role in regulating the cancer metabolism reprogramming, metastasis, and chemoresistance, as well as further summarize the SIRT3 small molecule modulators in the application of cancer therapy. Furthermore, we provide evidence to support the development of potential personalized therapy strategies for SIRT3 in cancer settings.
The Location and Structure of SIRT3
Seven mammalian sirtuins (SIRT1-7) have different flanking N-terminal and C-terminal extensions, which are in charge of the subcellular location of sirtuins. As a typical sirtuin, SIRT3 is a mitochondrial protein, although SIRT3 has also been confirmed to shift from the nucleus to the mitochondria under stress conditions (9, 10). Although all types of sirtuins are variable in length and sequence, they all have a catalytic core domain that is highly conserved with about 275 amino acids (11). The highly conserved enzymatic core contained in SIRT3 has two main domains, one is a large Rossmann fold domain and the other is a small zinc-binding domain. The large Rossmann fold domain shows the classic features of NAD+ binding sites and the specific features that recognize nicotinamide or ribose groups. The small zinc-binding domain can be served as an appealing and latent binding site for sirtuin inhibitors. The cleft between the two domains could be used as a docking site of acetylation substrates, where the NAD+ and peptide substrates containing acetyl-lysine enter and bind to the enzymes for deacetylation (12). In humans, SIRT3 is expressed as a full-length 44-kDa protein and is targeted to the cellular mitochondria proteins through its N-terminal location sequence. SIRT3 is expressed firstly in the cytoplasm as an inactive precursor of 399-amino acid (44-kDa). During the cellular stress, SIRT3 translocates to the mitochondrial matrix and is activated and stimulated by the mitochondrial matrix processing peptidase (MPP), which yields a comprising 257-amino acid residues (28-kDa) the short active form of SIRT3 (10, 13). Mitochondrial transport mainly relies on the N-terminal 100 amino acids, among which residues 1-25 play a vital role in mitochondrial targeting and proteolytic processing (13). Most studies support the localization of SIRT3 in mitochondria, but some studies have proved that SIRT3 may also exist in the nucleus, and it usually shows histone deacetylation activity and translocates to the mitochondria under stress conditions (10, 14). Hence, SIRT3 may autoregulate itself between the nucleus and the mitochondria. Despite the controversy, it can be concluded that SIRT3 mainly exerts a key role in the cellular mitochondria. The location and structural composition of SIRT3 is shown in Figure 1.
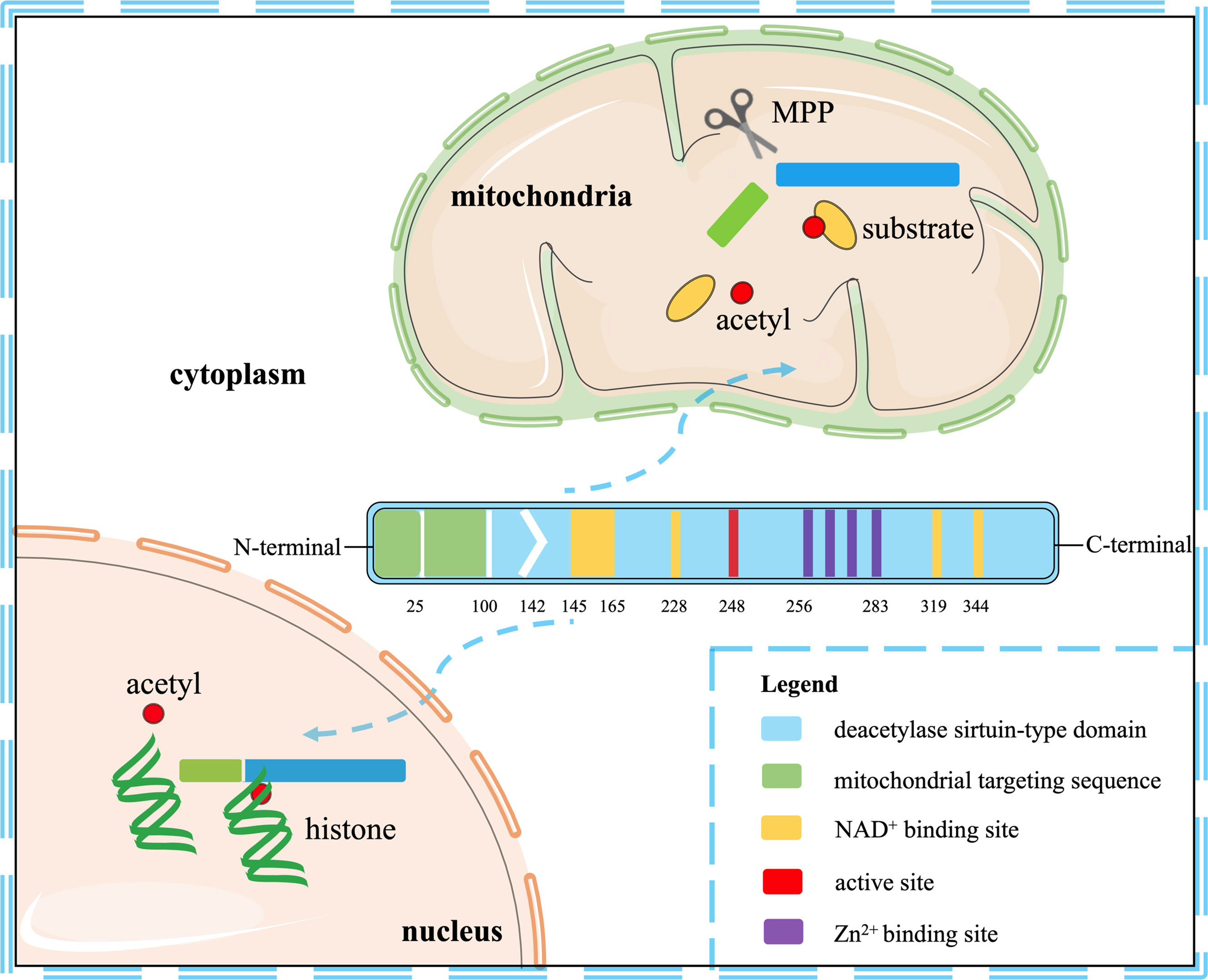
Figure 1 The location and structural composition of SIRT3. The conserved enzymatic core of SIRT3 contains an active site, four Zn2+ sites, four NAD+ sites, and a mitochondrial targeting sequence. SIRT3 is directed to the mitochondria through the targeting sequence, where it is cleaved by the mitochondrial MPP to produce mature proteins. As a deacetylase, it is primarily localized to mitochondria, but also shows histone deacetylation activity in the nucleus.
SIRT3 Deacetylation
The SIRT3 protein is mainly expressed in mitochondria-abundant tissues with metabolic activity, including the kidney, liver, brain, heart, and brown adipose tissue (15). As an important deacetylase in mitochondria, SIRT3 affects mitochondrial metabolism and function by modulating the acetylation level of the critical enzymes in mitochondrial metabolism to maintain the normal metabolic balance of cells (16). Mitochondria metabolism and function are highly regulated by protein acetylation. As a mitochondrial important and classic regulatory mechanism, acetylation could alter the protein-protein interactions network, affect complex stability, decide the location in mitochondria, and modulate enzyme activity. Acetylated proteins are richly expressed in the cells, such as the nucleus, mitochondria, and cytoplasm. Although mitochondrial proteins are highly acetylated leading to mitochondrial dysfunction, enzymes that regulate acetylation are rarely reported (17, 18). To be precise, the acetylation process is a biochemical reaction in which the acetyl group of acetyl-CoA is transferred to the amino acid residue of proteins with the help of acetyltransferase (19, 20). Deacetylation modification depends on the catalytic deacetylation function of deacetylase and serves as the reverse conversion of the acetylation reaction. The reaction process of NAD+-dependent SIRT3 deacetylation is rough as follows. The acetylated substrate and the NAD+ co-substrate combined with SIRT3 in the first place. Then, the acetyl group transfers from substrate to ADP-ribose moiety of NAD+, resulting in the production of bicyclic intermediates, which ultimately generate the deacetylated protein. Translocation in cells or conformational changes will occur after substrate deacetylation, which adjusts the activity of the substrate enzyme and regulates the reaction process involved by the enzyme (21, 22).
SIRT3 is mainly located in the mitochondria and functions as a vital role in regulating the glucose metabolism, tricarboxylic acid (TCA) cycle, oxidative phosphorylation (OXPHOS), reactive oxygen species (ROS) detoxification, electron transport chain (ETC), fatty acid (FA) metabolism, mitochondrial unfolded protein response (UPRmt), autophagy, etc. (23–25). SIRT3 could directly deacetylate pyruvate dehydrogenase (PDH) and glutamate oxaloacetate transaminase 2 (GOT2) to regulate glucose metabolism (26). As a multienzyme complex, PDH converts pyruvate to acetyl-Co, and the K336 site of its core subunit pyruvate dehydrogenase E1α (PDHA1) may be served as the potential deacetylation site of SIRT3 (27). The acetylation of GOT2 3K (K159, K185, K404) sites could improve the ability of the malate-aspartate shuttle, but GOT2 deacetylation through SIRT3 shows an inhibited function (28). In addition, SIRT3 could regulate the TCA cycle via deacetylating isocitrate dehydrogenase 2 (IDH2) and glutamate dehydrogenase (GDH). IDH2 is the rate-limiting enzyme of the TCA cycle to some certain extent in the case of SIRT3 low expression (29). The acetylation level of IDH2 at K413 and K360 sites is increased, and the decreased enzyme activity leads to the inability to participate in electronic transmission, thus obstructing the TCA cycle (30, 31). Similarly, SIRT3 deacetylates GDH and regulates the TCA cycle to promote amino acid oxidation, as well as one of the GDH acetylation sites, K527, has been reported (32, 33). Mitochondria produce ATP through OXPHOS, and SIRT3 deacetylates ETC subunits, such as complexes I–III and ATP synthase, to maintain cellular ATP levels (34, 35). Succinate dehydrogenase (SDH) is an enzyme that exists between the TCA cycle and the ETC (36). SIRT3 could directly deacetylate SDH A subunit, and the K179 site may be a potential deacetylation site (31). It is well known that the production of ROS is closely related to OXPHOS transferred electrons and mitochondrial energy metabolism, and SIRT3 can stabilize ROS levels by deacetylation of ETC proteins (37, 38). In addition, SIRT3 can directly deacetylate and activate antioxidant enzymes, such as IDH2 and superoxide dismutase 2 (SOD2), to protect cells from ROS (39, 40). SIRT3 can also deacetylate transcription factor forkhead box 3α (FOXO3α) to regulate the expression of antioxidant enzymes, and deacetylated sites of FOXO3α are mainly located at K271 and K290 (41, 42). Thus, activation of SIRT3 may contribute to ROS detoxification, thus avoiding oxidative stress damage. Moreover, SIRT3 plays an important role in regulating FA metabolism by deacetylation of long-chain-specific acyl-CoA dehydrogenase (LCAD) at K42, K318, and K322 sites (22, 43). In other aspects, as the primary coordinator of UPRmt, SIRT3 not only maintains protein homeostasis but also deacetylates the autophagy-related protein to repair defective autophagy, which is important for cell function and survival (44, 45). Consequently, as the key site of energy metabolism, mitochondria could regulate cellular metabolism and intracellular signal transduction to maintain homeostasis, and their imbalance indicates the arrival of diseases (46). Acting as a modulator of cell metabolism, SIRT3 displays tumor-suppressing and tumor-promoting features in cancer by regulating the acetylation level of multiple mitochondrial proteins (47). Although the most basic biological function of SIRT3 is the deacetylation of its substrate, cancer cell pathophysiology, metabolism reprogramming, metastasis, and chemoresistance are dependent on the regulation of SIRT3 deacetylation.
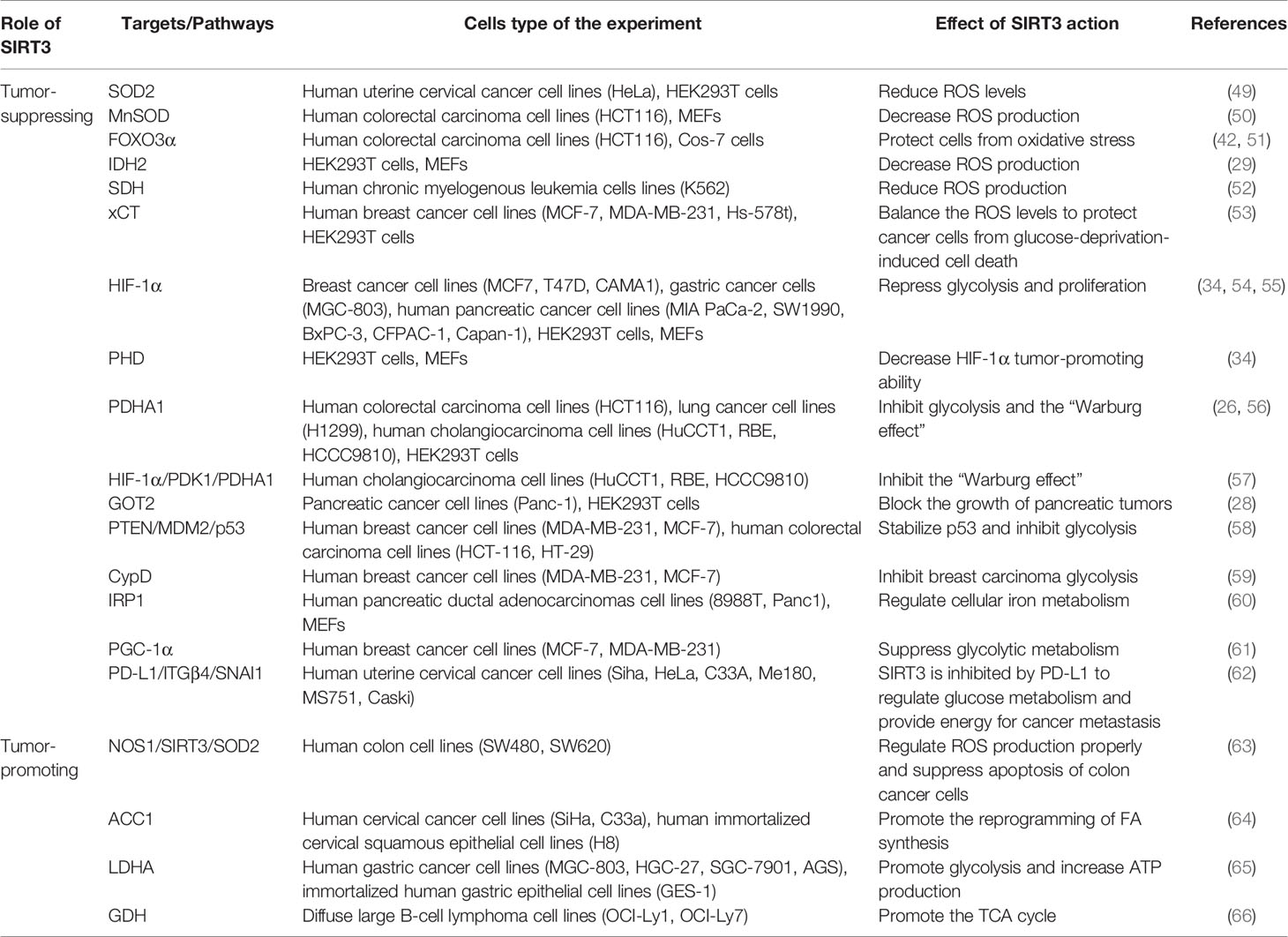
Regulation of Cancer Metabolism Reprogramming by SIRT3
The most striking feature of metabolism reprogramming of tumor cells is the “Warburg effect”, which is that most cancer cells are more dependent on aerobic glycolysis than OXPHOS. Glycolysis could provide a rapid energy supply and several beneficial conditions for the genesis and progress of the tumor microenvironment. To meet the needs of continued growth and proliferation, tumor cells provide themselves with more material and energy through metabolism reprogramming (48). The signaling functions of SIRT3 relevant to cancer metabolism reprogramming are shown in Figure 2 and Table 1.
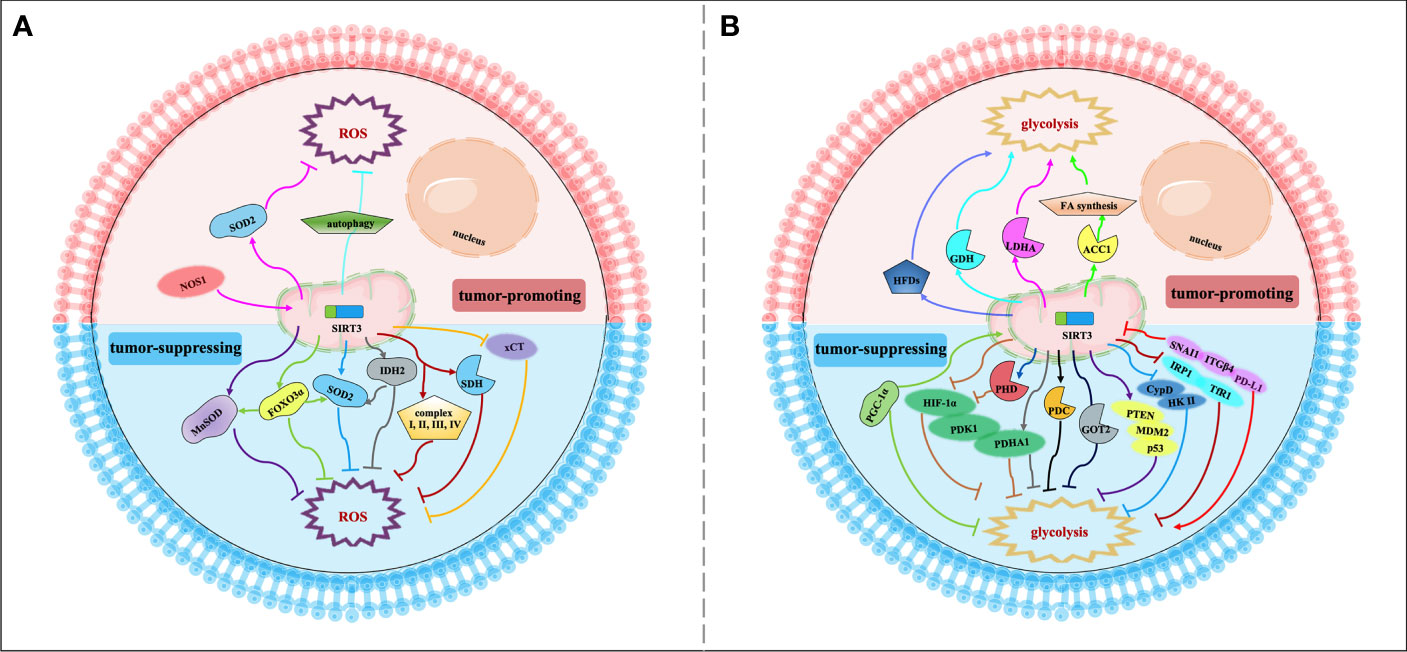
Figure 2 Signaling functions of SIRT3 relevant to cancer metabolism reprogramming. (A) SIRT3 regulates cancer metabolism reprogramming by modulating ROS levels. As a tumor-suppressing protein, SIRT3 could deacetylate and modulate the activity of a series of enzymes and transcription factors such as SOD2, MnSOD, FOXO3α, IDH2, SDH, and xCT to decrease the ROS levels and regulate tumor metabolism reprogramming. As a tumor-promoting protein, SIRT3 could regulate the NOS1/SIRT3/SOD2 pathway or mitochondrial autophagy to keep ROS at an appropriate level to promote tumorigenesis and prevent apoptosis. (B) SIRT3 regulates cancer metabolism reprogramming by modulating tumor cell glycolysis. As a tumor-suppressing protein, SIRT3 not only deacetylates and regulates the activity of various enzymes and proteins including HIF-1α, PHD, PDHA1, GOT2, CypD, IRP1, and PGC-1α, but also affects some signaling pathways such as HIF-1α/PDK1/PDHA, PTEN/MDM2/p53 pathway, CypD/HK II pathway, IRP1/TfR1 pathway, and PD-L1/ITGβ4/SNAI1 pathway, thereby inhibiting glycolysis and regulating tumor metabolism reprogramming. As a tumor-promoting protein, SIRT3 could deacetylate and activate ACC1, LDHA, GDH, and HFDs to promote glycolysis and increase the occurrence of cancer.
SIRT3 can modulate intracellular ROS levels to regulate tumor metabolism reprogramming (Figure 2A). Elevated oxidative stress caused by ROS is the most important feature of mitochondria reprogramming in cancer cells (67, 68). Cancer cells have higher levels of ROS compared with normal cells. ROS can oxidize specific intracellular chemical moieties to damage macromolecules, such as DNA, leading to genetic mutations and influencing the biochemical pathways of tumor cell proliferation and transformation (69). Although excessive accumulation of ROS levels may trigger cell death, a slight increase in ROS may benefit cell survival and growth. Thus, the role of ROS remains controversial (70, 71). SIRT3 could function as a tumor suppressor to limit ROS levels via the activation of antioxidant enzymes, such as superoxide dismutase (SODs). Dysregulation of the detoxification activity of antioxidant enzymes in tumors implies more oncogenic or therapeutic resistance phenotypes. As an antioxidant enzyme, SOD2 can be deacetylated by SIRT3 to remove ROS (49). SIRT3 can also target the mitochondrial enzyme manganese superoxide dismutase (MnSOD) to restore superoxide dismutase activity and decrease ROS production (50). SIRT3 could also deacetylate another important target, FOXO3α, to up-regulate antioxidant enzymes and protect cells from oxidative stress (42, 51). In addition, IDH2 can be directly deacetylated by SIRT3, thereby affecting SOD2-mediated mitochondrial ROS scavenging and cellular redox status (29). SIRT3 can also regulate the activity of complexes I-IV in the ETC to reduce ROS leakage from the complex, and improve the detoxification ability of oxidase, thus reducing ROS production (38, 50). For example, SIRT3 can catalyze the deacetylation of the SDH flavoprotein subunit, one of the Complex II subunits, and coordinate material oxidation and electron transport to reduce ROS production by modulating SDH activity (52). Recently, one study deserves attention that SIRT3 may play a protective role in the glucose-deprivation-induced cell death of breast cancer cells by regulating the xCT expression, also known as solute carrier family 7 member 11 (SLC7A11), to balance the ROS levels (53). In this regard, SIRT3 is considered a key scavenger of ROS in cells by playing a tumor-suppressing effect. On the contrary, SIRT3-mediated ROS removal may also contribute to the oncogenic effects. SIRT3 keeps ROS at an appropriate level to maintain proliferative and aggressive phenotypes that promote tumorigenesis and prevent apoptosis (72). For instance, mitochondrial neuronal nitric oxide synthase (nNOS, NOS1) could increase SIRT3 activity and thereby activating SOD2 to regulate ROS production properly, thus, suppressing apoptosis of colon cancer cells (63).
Similarly, SIRT3 may also regulate tumor metabolism reprogramming by modulating tumor cell glycolysis (Figure 2B). As a tumor suppressor, downregulation of SIRT3 may promote a Warburg phenotype (73). In breast cancer and gastric cancer, the loss of SIRT3 promotes ROS production, leading to hypoxia-inducible factor-1α (HIF-1α) stabilization. HIF-1α is a transcription factor that controls glycolytic gene expression, and SIRT3 mediates metabolism reprogramming by destabilizing HIF-1α to affect the progression of cancer (34, 54). In this regard, profilin1 (Pfn1) played an anti-cancer role in pancreatic cancer by improving the SIRT3 expression, resulting in the destabilization of HIF-1α to inhibit glycolytic (55). SIRT3 can also directly deacetylate prolyl hydroxylase (PHD) to regulate HIF-1α, thereby decreasing the stability of HIF-1α to inhibit glycolysis (34). In addition, SIRT3 could deacetylate and activate pyruvate dehydrogenase complex (PDC) and related component enzyme PDHA1 to inhibit glycolysis and decrease the “Warburg effect” (26, 56). Interestingly, SIRT3 inhibits the cholangiocarcinoma progression via inhibiting the “Warburg effect” through the HIF-1α/pyruvate dehydrogenase kinase 1 (PDK1)/PDHA1 downstream signaling pathway (57). In addition, SIRT3 plays a potential role in the deacetylation of GOT2. SIRT3 impairs the protein association between GOT2 and malate dehydrogenase 2 (MDH2), thereby inhibiting the activity of the malate–aspartate shuttle. As a restriction enzyme that regulates the glycolysis process, GOT2 is deacetylated by SIRT3, thus obstructing the transport of cytosolic nicotinamide adenine dinucleotide hydrogen (NADH) into the mitochondria to achieve the purpose of blocking the growth of pancreatic tumors (28). p53 is one of the tumor suppressors that slows down glycolysis and promotes OXPHOS (74). SIRT3 could inhibit glycolysis via stabilizing p53 in cancer cells, which was related to the phosphatase and tensin homolog (PTEN) and mouse double minute 2 (MDM2) (58). SIRT3 deacetylates and inactivates cyclophilin D (CypD) to impact the contact of the lactate metabolic enzyme hexokinase II (HK II) with mitochondria, further inhibiting breast carcinoma glycolysis (59). Besides, SIRT3 inhibits the aggregation of iron regulatory protein 1 (IRP1), and further reduces the transferrin receptor (TfR1) expression, thus, regulating cellular iron metabolism to affect the proliferation of pancreatic cancer cells (60). Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) regulates the cell survival and proliferation of cancer cells by controlling mitochondrial biogenesis and cellular metabolism. PGC-1α could suppress glycolytic metabolism by activating SIRT3 to moderate breast cancer cell proliferation (61). As a downstream effector factor of programmed death-ligand 1 (PD-L1), tumor suppressor SIRT3 is inhibited by PD-L1 and participates in integrin β4 (ITGB4)/snail family transcriptional repressor 1 (SNAI1) signaling pathway to regulate glucose metabolism, which provides energy to promote metastasis in cervical cancer (62). A recent study has shown that whey could target SIRT3 expression to induce metabolic dysfunctions and modulate the bioenergy characteristics of colon cancer cells (75). Of course, as a tumor promoter, SIRT3 also shows a dark side. In cervical cancer, SIRT3 significantly promotes the FA synthesis reprogramming by up-regulating acetyl-CoA carboxylase (ACC1) (64). In addition, as a key protein regulating anaerobic glycolysis, lactate dehydrogenase A (LDHA) could be deacetylated and activated by SIRT3 to promote glycolysis and increase ATP production in gastric cancer cells (65). Similarly, SIRT3 enhances the activity of GDH to promote the TCA cycle, thereby increasing the occurrence of lymphoma (66). Intriguingly, inhibition of SIRT3 expression may reduce the carcinogenic effect induced by high-fat diets (HFDs), suggesting that SIRT3 is served as an oncogene (76).
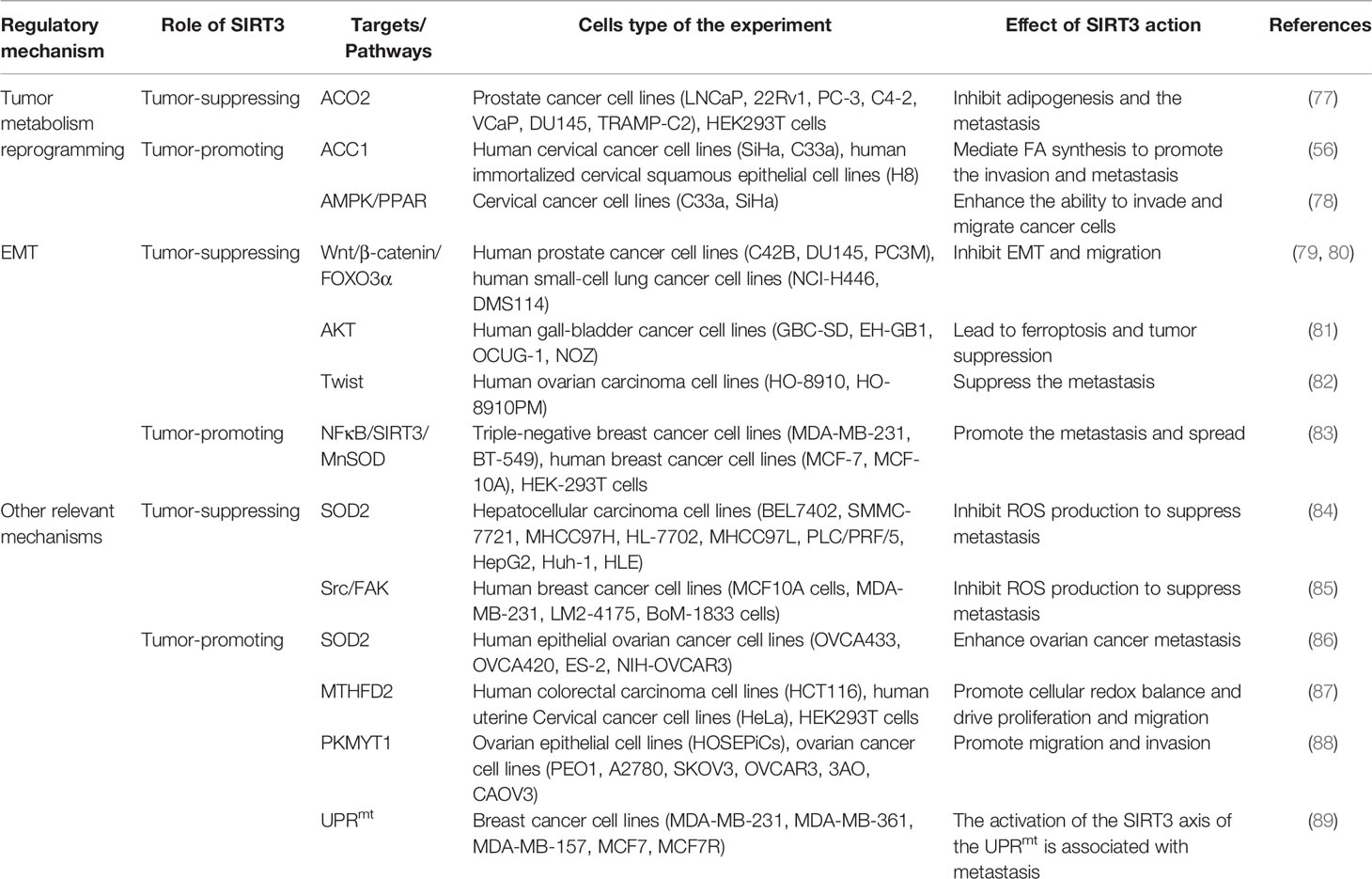
Regulation of Cancer Metastasis by SIRT3
The invasion and migration of tumor cells to surrounding tissues is the beginning and pivotal stage of cancer metastasis, which is a major principal consideration of cancer deaths. Regulation of metabolism reprogramming and EMT by SIRT3 can make a significant influence on cancer metastasis. Exploring the metabolic targets that drive invasive and metastatic cancers promises to be an effective therapeutic strategy. The signaling functions of SIRT3 relevant to cancer metastasis are shown in Figure 3 and Table 2.
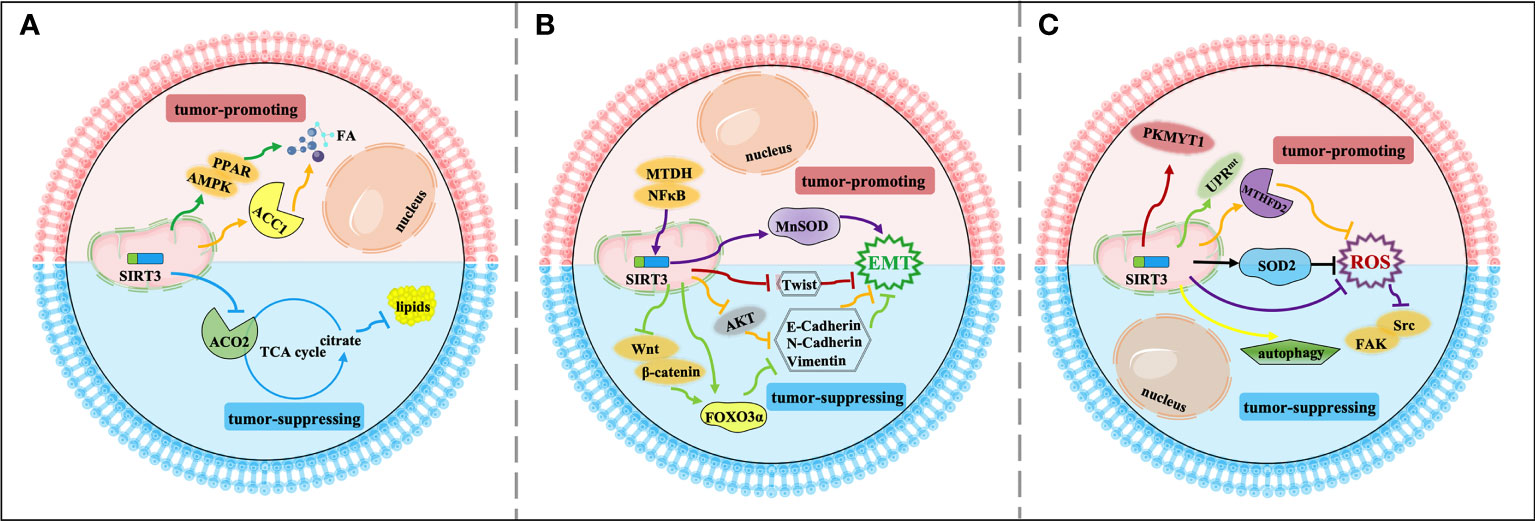
Figure 3 Signaling functions of SIRT3 relevant to cancer metastasis. (A) SIRT3 regulates cancer metastasis by modulating tumor metabolism reprogramming. As a tumor-suppressing protein, SIRT3 deacetylates and inactivates ACO2 to inhibit adipogenesis and cancer metastasis. As a tumor-promoting protein, SIRT3 deacetylates and activates ACC1 and the AMPK/PPAR pathway to mediate FA synthesis and promote cancer metastasis. (B) SIRT3 regulates cancer metastasis by modulating EMT. As a tumor-suppressing protein, SIRT3 up-regulates Twist and inhibits EMT-related markers such as E-cadherin, N-cadherin, and N-cadherin by affecting FOXO3α, AKT, and Wnt/β-catenin/FOXO3α pathway, thereby inhibiting EMT and cancer metastasis. As a tumor-promoting protein, SIRT3 regulates the NFκB/SIRT3/MnSOD pathway to promote EMT and cancer metastasis. (C) Other relevant mechanisms that SIRT3 participates in cancer metastasis. As a tumor-suppressing protein, SIRT3 regulates autophagy, and inhibits ROS production through SOD2 and the Src/FAK pathway, leading to suppression of cancer metastasis. As a tumor-promoting protein, SIRT3 deacetylates and regulates SOD2, MTHFD2, PKMYT1, and the SIRT3 axis of the UPRmt to promote cancer metastasis.
Firstly, metabolism reprogramming plays important role in tumor cell survival during the cancer metastasis process, which contributes to driving the successful formation of metastatic lesions (90). Studies have shown that changes in FA synthesis in cancer cells may offer selective advantages for the metastatic process (91, 92). The FA synthesis ability of tumor tissues with strong invasion ability is generally enhanced. Thus, for rapidly proliferating and strongly invasive tumor cells, there is high demand for FA (93). As a tumor suppressor, acetylation of aconitase (ACO2) could be reversibly regulated by SIRT3. Transcriptional repression of SIRT3 enhances the ACO2 activity to promote mitochondrial citrate synthesis and adipogenesis, further promoting the progression of aggressive prostate cancer to the bone (77). As a tumor promoter, SIRT3 promotes cervical cancer invasion and metastasis via up-regulating FA synthesis mediated through ACC1 (64). SIRT3 also activates the AMP-activated protein kinase (AMPK)/peroxisome proliferator-activated receptor (PPAR) pathway to effectively promote intracellular triglyceride synthesis. Increased FA synthesis of tumor cells enhances the invasion and infiltration in vitro and promotes the growth of primary tumors and distant metastases in vivo, which further enhances the ability of cervical cancer cells to invade and migrate (78, 94) (Figure 3A).
Moreover, SIRT3 may play a dual role in cancer metastasis by affecting epithelial-to-mesenchymal transition (EMT). EMT-mediated metastasis is when cancer cells spread from the primary tumor to adjacent tissues firstly, then spread and proliferate via the bloodstream and lymphatic system. From the perspective of mechanism, EMT regulates epithelial markers, mesenchymal markers, and transcriptional regulatory factors to play a dual role in cancer metastasis mainly manifested in down-regulation of E-cadherin and cytokeratin, up-regulation of N-cadherin, Vimentin, β-catenin, and Snail. SIRT3 plays a significant inhibitory role in the cancer metastasis process. SIRT3 regulates the EMT-related markers such as E-Cadherin, N-Cadherin, and Vimentin to suppress EMT, especially in prostate cancer and small‐cell lung cancer, which may be related to Wnt/β-catenin/FOXO3α pathway and protein kinase B (AKT, also known as PKB)-dependent ferroptosis (79–81). In ovarian cancer, as a key regulator of EMT, Twist is down-regulated by SIRT3 to suppress the metastasis of the tumor (82). Interestingly, SIRT3-mediated activation of MnSOD can promote EMT in cancer. SIRT3 could be mediated by metadherin (MTDH) to enhance its activity, and enhance EMT and invasion by participating in the NFκB/SIRT3/MnSOD pathway, ultimately promoting the metastasis and spread of triple-negative breast cancer (83) (Figure 3B).
Finally, in addition to regulating metabolism reprogramming and EMT, there are other potential mechanisms that SIRT3 participates in cancer metastasis. Down-regulation of SIRT3 promotes tumor metastasis ability, in which it plays a tumor suppressor role. ROS contributes to increased invasion and migration capacity, which is associated with the inhibition of SOD2 by mitochondrial Ca2+ uniporter (MCU)-dependent mitochondrial Ca2+ uptake (84). Moreover, down-regulation of SIRT3-mediated elevated ROS levels could promote the activation of the steroid receptor coactivator (Src)/focal adhesion kinase (FAK) signaling pathway to enhance cancer metastasis (85). SIRT3 also significantly promotes the expression of key proteins involved in autophagosome formation, thereby inducing proliferation and migration inhibition (95). However, as a tumor promoter, a recent study showed that SIRT3 could enhance ovarian cancer metastasis by rapidly up-regulating SOD2, which was a new piece of evidence of SIRT3’s role in promoting metastasis in cancer (86). SIRT3 can directly deacetylate the mitochondrial methylenetetrahydrofolate dehydrogenase/cyclohydrolase (MTHFD2) to promote cellular redox balance and drive colorectal cancer cell proliferation and diffusion (87). In ovarian cancer, SIRT3 enhances the ability of protein kinase membrane associated tyrosine/threonine 1 (PKMYT1) to promote cell migration and invasion, thus accelerating cancer metastasis (88). Furthermore, SIRT3 is a necessary component of the UPRmt required for breast cancer metastasis to secondary organs (89, 96), which further enriches our information on the dual role of SIRT3 in the regulation of cancer metastasis (Figure 3C).
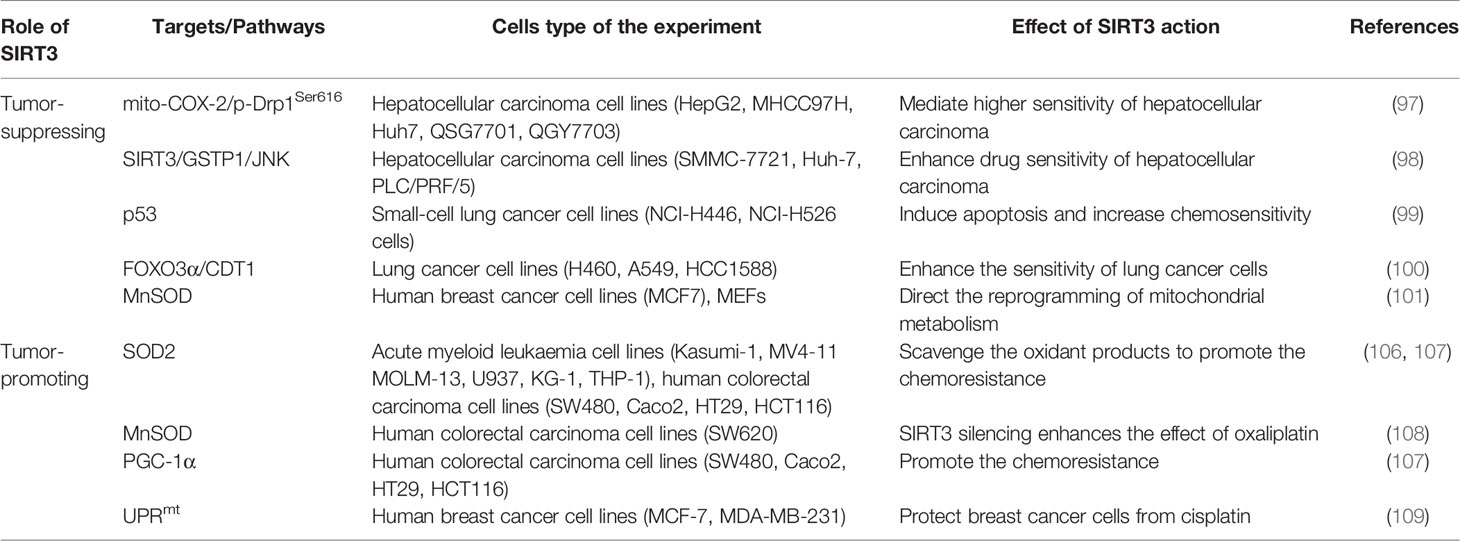
Regulation of Cancer Chemoresistance by SIRT3
Cancer drug resistance is the main factor leading to chemotherapy failure. Research on the mechanism of cancer chemoresistance is essential to establishing therapeutic strategies for drug-resistant tumors. Based on the dual role of SIRT3 in cancer, its involvement in cancer drug resistance is also bidirectional. The signaling functions of SIRT3 relevant to cancer chemoresistance are shown in Figure 4 and Table 3.
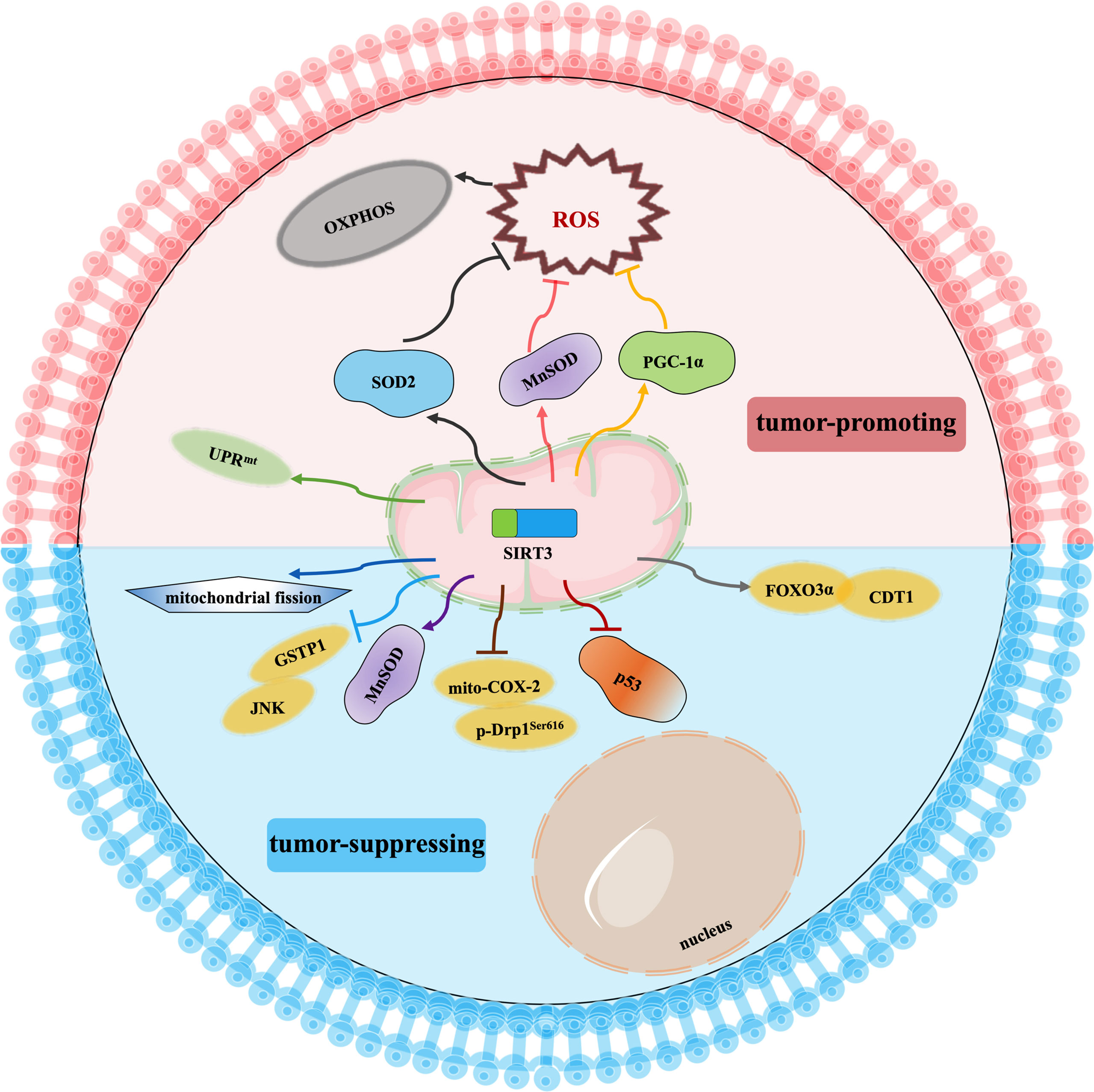
Figure 4 Signaling functions of SIRT3 relevant to cancer chemoresistance. As a tumor-suppressing protein, SIRT3 not only deacetylates MnSOD and p53 but also regulates the mito-COX-2/p-Drp1Ser616 pathway, SIRT3/GSTP1/JNK pathway, FOXO3α/CDT1 pathway, and mitochondrial fission to enhance chemotherapy drugs sensitivity of cancer. As a tumor-promoting protein, SIRT3 scavenges the oxidant products by deacetylating SOD2, MnSOD, and PGC-1α, and activates UPRmt to promote chemotherapy drugs resistance to cancer.
SIRT3 improves the sensitivity of cancer cells to chemotherapy to inhibit drug resistance. Chemotherapeutic agents, such as doxorubicin, cisplatin, oxaliplatin, and epirubicin, could inhibit the expression of SIRT3 in tumor cells. In hepatocellular carcinoma cells, SIRT3 sensitizes hepatocellular carcinoma cells to chemotherapy drugs via regulating mitochondrial cyclooxygenase-2 (mito-COX-2)/Drp1 with phosphorylation at serine 616 (p-Drp1Ser616) and glutathione S-transferase pi 1 (GSTP1)/c-Jun N-terminal kinase (JNK) signaling pathway (97, 98). In lung cancer, SIRT3 enhances the sensitivity of lung cancer cells to cisplatin by deacetylating mutant p53 in small-cell lung cancer and elevating the FOXO3α/chromatin licensing and DNA replication factor 1 (CDT1) axis in lung cancer (99, 100). In breast cancer, SIRT3 expression is reduced. Lowered expression of SIRT3 leads to an increase in MnSOD acetylation to direct the reprogramming of mitochondrial metabolism, thus, resulting in cisplatin and doxorubicin resistance (101). Moreover, SIRT3 enhances the sensitivity of ovarian cancer cells to cisplatin by promoting mitochondrial fission (106). Therefore, activation of SIRT3 protein may be a way to improve cancer treatment, especially to overcome the resistance of cancer cells to chemotherapy drugs.
Oppositely, SIRT3 also endows tumor cells with resistance to chemotherapy. Cancer cells exhibit enhanced ROS production and accumulation to give them an advantage in chemotherapy resistance, while ROS excess can lead to growth inhibition and apoptosis. Interestingly, SIRT3 could precisely reduce ROS production and result in cell survival (71, 107–109). When SIRT3 is highly expressed, tumor cells can balance ROS production, and become more resistant to chemotherapy drugs (71). Similarly, SIRT3 could deacetylate SOD2, and contribute to acute myeloid leukaemia chemoresistance via inhibiting the cytarabine-induced production of ROS to regulate mitochondrial OXPHOS (102, 103). In colorectal cancer, SIRT3 silencing not only reduces the antioxidant capacity of the cell by regulating MnSOD acetylation and increases cell death to enhance the effect of oxaliplatin but also regulates mitochondrial function under oxidative conditions through PGC-1α, thereby decreasing the chemoresistance to the colorectal cancer cells (103, 104). In this regard, SIRT3 inhibitors may be an attractive drug for the therapy of colorectal cancer in combination with cisplatin. In breast cancer, SIRT3 knockdown could enhance the effect of chemotherapy drugs. SIRT3 protects breast cancer cells from cisplatin by increasing UPRmt, suggesting that inhibition of SIRT3 is beneficial for enhancing chemotherapy sensitivity in breast cancer (105). SIRT3 silencing could produce an increment in ROS production and compromise the antioxidant response, thus, producing an enhancement of cisplatin-triggered apoptosis, which sensitizes breast cancer cells to cisplatin treatments (110).
In a word, the impact of SIRT3 on chemotherapy drug resistance varies with tumor type, which needs further study. It could be seen that SIRT3-mediated effects on mitochondrial metabolism reprogramming by regulating ROS levels are crucial for the chemoresistance of tumor cells. The selection of SIRT3 activators/inhibitors in combination with chemotherapeutic agents may offer a potential approach to cancer treatment to prevent the formation of a microenvironment conducive to tumor recurrence.
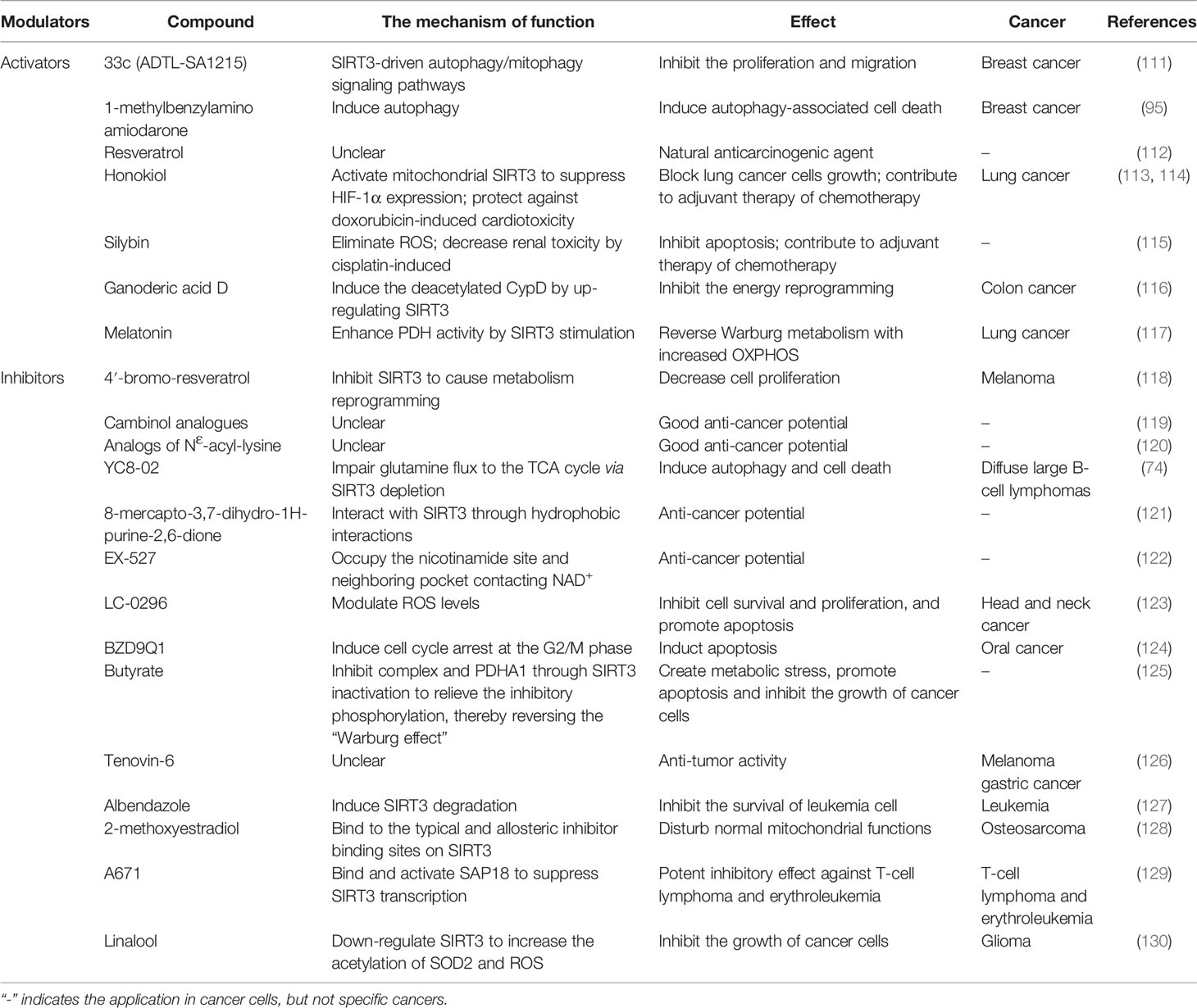
Perspectives for Therapy Targeting SIRT3 in Cancer
Existing studies support the dual role of SIRT3 in cancer development, suggesting that SIRT3 activators/inhibitors can be served as cancer treatments, which is a very attractive strategy for cancer therapy. Therefore, the development and application of targeted SIRT3 as a small molecule of anti-cancer drugs has been a hotspot for many researchers. In this section, we mainly discuss the therapeutic value of SIRT3 activators/inhibitors in cancer treatment (summarized in Table 4).
Application of SIRT3 Activators in Cancer
There are few reports about SIRT3 activators in cancer treatment. Compound 33c (ADTL-SA1215) may be the earliest SIRT3-specific small molecule activator in the current research field, which can regulate the SIRT3-driven autophagy/mitophagy signaling pathways to inhibit the proliferation and invasion of human breast cancer cells (111). 1-methylbenzylamino amiodarone (MA) is another SIRT3 novel small-molecule activator that induces autophagy-associated cell death in breast cancer (95). Although few SIRT3 activators have been reported, there are many active modulators of SIRT3 that can elevate SIRT3 expression. Most SIRT3 active modulators are derived from natural products, such as Resveratrol, Honokiol, Silybin, and Ganoderic acid D (GAD). As a natural anti-cancer agent, Resveratrol activates SIRT3 to regulate cancer at different stages, but the mechanism of action of Resveratrol is still unclear (112). As a natural phenolic compound, Honokiol has anti-cancer activity. Honokiol blocks the growth of lung cancer cells by activating SIRT3 to inhibit HIF-1α expression, and also be used as adjuvant chemotherapy to prevent doxorubicin-induced cardiotoxicity in tumors transplanted mice (113, 114). Silybin is another SIRT3 activator derived from natural plants. Activation of SIRT3 can reduce ROS production and inhibit apoptosis to protect kidney cells from death, and this protection can also be used in adjuvant chemotherapy by reducing cisplatin-induced nephrotoxicity (115). GAD is the main active component of Ganoderma lucidum, and GAD inhibits the energy metabolism reprogramming of colon cancer cells by activating SIRT3 to induce deacetylation of CypD (116). In addition, Melatonin can also activate SIRT3, then the SIRT3/PDH axis is stimulated to improve the ROS production to induce apoptosis of lung cancer cells (117).
Application of SIRT3 Inhibitors in Cancer
The development of SIRT3 inhibitors is much easier than the development of SIRT3 activators. Based on the structure and biochemical function of SIRT3, a variety of SIRT3 inhibitors have been developed and applied for cancer treatment, but it has been challenging to develop high-efficiency inhibitors with few side effects. On the one hand, the design of structural analogues based on acetylated substrates is the most common and effective design strategy. The small-molecule 4’‐bromo‐resveratrol (4’‐BR) has an inhibitory effect on SIRT3, and the reduced expression of SIRT3 could cause metabolism reprogramming, resulting in a significant anti-cancer effect in melanoma cells (118). Both Cambinol and NƐ-acyl-lysine analogs are all SIRT3 inhibitors with promising anti-cancer potential, but their anti-cancer mechanism remains unclear (119, 120). Another SIRT3-targeting inhibitor, YC8-02, induces autophagy and cell death by decreasing glutamine flux into the TCA cycle in diffuse large B-cell lymphomas cells (66). Lately, a novel SIRT3 inhibitor based on the 8-mercapto-3,7-dihydro-1H-purine-2,6-dione scaffold has been identified to inhibit SIRT3 through hydrophobic interactions, which provides new structural information for the development of effective SIRT3 inhibitors (121). On the other hand, the activity of SIRT3 could be decreased by NAD+ coenzyme inhibitors by inhibiting the progress of the deacetylate reaction. EX-527 has anti-cancer promise and inhibits the less active SIRT3 by holding the nicotinamide site and the adjacent pocket contacting NAD+ (122). LC-0296 is also considered a novel SIRT3 inhibitor because it is structurally an NAD+ inhibitor. LC-0296 inhibits cell survival and proliferation by regulating ROS and promoting head and neck cancer cell apoptosis (123).
Here, many other types of SIRT3 inhibitors have also been reported for cancer treatment. Compound BZD9Q1, as a SIRT3 inhibitor, elicits cytostatic effects by inducing cell cycle arrest in the G2/M phase to show promising anti-cancer properties against oral cancer (124). Butyrate acts as an inhibitor of SIRT3 to promote apoptosis of cancer cells via inducing high acetylation of PDHA1 to reverse the “Warburg effect” (125). Tenovin-6, a micromolar SIRT3 inhibitor, has anti-tumor ability in melanoma and gastric cancer cells, but its inhibitory effect remains unclear (126). Albendazole, an anthelmintic drug, is known as an inhibitor of leukemia cell survival by inducing SIRT3 degradation (127). 2-methoxyestradiol (2-ME) inhibits SIRT3 activity by combining with the binding site of canonical allosteric inhibitor, thereby killing osteosarcoma cells by disrupting normal mitochondrial function (128). Compound 3-O-chloroacetyl-gagamine (A671) binds and activates SIN3 Associated Protein 18 (SAP18) to inhibit the SIRT3 transcription, which has a good anti-cancer effect in T-cell lymphoma and erythroleukemia (129). Moreover, linalool inhibits the growth of glioma cells by stimulating SOD2-ROS signaling via SIRT3 inhibition (130).
Role of microRNA in SIRT3 Expression and Cancer Development
MicroRNA (miRNA) is a class of short non-coding RNA that performs its functions by regulating the expression of target genes at the stage of post-transcriptional. Dysregulation of miRNA has been supported to be closely related to carcinogenesis (131). Actually, miRNA is capable of controlling SIRT3 gene transcription in tumors to affect the progression of cancer (summarized in Table 5).
SIRT3 is a target of miR-1225-5p, miR-224, miR-494, miR-31, miR-708-5p, miR-421, miR-6858-5p, and miR-761. Notably, downregulation of miR-1225-5p and upregulation of miR-224 may promote tumor cell growth, proliferation, and metastasis via targeting SIRT3 directly in thyroid cancer and non-small cell lung cancer cells (132, 133). In hepatocellular carcinoma, the miR-494 induces endothelial to mesenchymal transition (EndMT) to promote cancer metastasis by targeting the SIRT3 signaling pathway (134). Furthermore, miR-31 directly targets SIRT3 to activate oxidative stress by disrupting mitochondrial activity in oral carcinoma (135). On the contrary, miR-708-5p and miR-421 inhibit the process of pancreatic ductal adenocarcinoma and gastric cancer by targeting SIRT3 to ablate the stimulatory activity on cell viability, invasion, and migration (136, 137). In the glioblastoma multiforme, the SIRT3 expression level could be decreased by miR-6858-5p mimic, which inhibits the growth-promoting AKT signaling pathway (138). In addition, miR-761 is closely associated with SIRT3-mediated chemical resistance, and changes in miR-761 abundance targeted SIRT3 to modulate the response of synovial sarcoma cells to pazopanib (139). Therefore, altered miRNA expression affects the signaling pathway of SIRT3 involved in cancer progression. The specific connection between miRNA and SIRT3 role, and whether miRNA can be used as an indicator of SIRT3-mediated related cancer development process should be further explored, which provides a potential basis for targeting SIRT3 to find new approaches in cancer therapy.
Discussion
Pathologic SIRT3 expression involves complex molecular signaling pathways and affects tumor metabolism, metastasis, and prognosis. As an important intracellular regulatory factor of energy metabolism, SIRT3 has the ability of metabolism reprogramming and contributes greatly to cancer fate. Available data have confirmed the dual role of SIRT3 in cancer, and the effects of SIRT3 are different at different stages of cancer development, although the study of such phenomena remained unsystematic.
Multiple studies have shown that the increase of mitochondrial ROS level is usually an important cause of the development of cancer. The role of SIRT3 in cancer is mainly to remove excess ROS through deacetylating and activating its substrates. SIRT3 regulates cell metabolism reprogramming, such as TCA, OXPHOS, and FA metabolism, to maintain mitochondria homeostasis, thereby affecting the fitness and survival of tumor cells. However, SIRT3 plays a carcinogenic role in some OXPHOS-addiction cancers, SIRT3 could promote glycolysis and tumorigenesis by maintaining ROS levels at an appropriate level, which depends on the cellular context. Interestingly, most studies have shown that SIRT3 could promote cancer metastasis, which may be closely related to the SIRT3-mediated tumor cell metabolism reprogramming and the EMT. Of course, SIRT3 can significantly inhibit cancer metastasis via inhibiting ROS levels, EMT, and the Src/FAK signaling pathway. Nowadays, the presence or absence of organ and tissue metastases is used as a predictor of patient survival, thus, using SIRT3 to predict cancer metastasis patient survival needs further study. Similarly, the impact of SIRT3 on cancer chemoresistance is also bidirectional. SIRT3-mediated effects on mitochondrial metabolism reprogramming by regulating ROS levels are crucial for the chemoresistance of tumor cells. Thus, the role of SIRT3 must not be generalized. The function of SIRT3 is different in different tumor tissues and normal tissues, which is related to the specificity of tumor cell type and the cellular context.
Importantly, SIRT3 is a potentially attractive molecular target based on its dual role in cancer. Several potential SIRT3 activators/inhibitors have been tested in different cell lines of cancer, but rarely in vivo. Further exploration of the biological function and structure of SIRT3 will contribute to the exploration of satisfactory with no side effects small-molecule modulators targeting SIRT3. However, the function of SIRT3 varies in different cancer types or at different stages of the same cancer type, which makes it relatively difficult for cancer therapy to target SIRT3. On the one hand, SIRT3 has a protective effect on most organs in the body, and inhibition of SIRT3 may affect the normal functions of other organs. On the other hand, activation of SIRT3 may also have unimaginable carcinogenic effects under certain contexts. Perhaps, combining SIRT3 activators/inhibitors with other drugs targeting a specific cancer type to design personalized therapy programs would be a potential treatment strategy. Thus, it will be a good choice to appropriately regulate SIRT3 in personalized therapy, so that it is in a dynamic balance, especially to overcome resistance to treatment.
In the future, further exploration of the dual role of SIRT3 in cancer progression should be done as follows. a) examining the role of SIRT3 in different cancer types or at different stages of the same cancer type and in both in vivo and in vitro settings; b) observing metabolism reprogramming changes that SIRT3-regulated is a driver or passenger of cancer progression; c) investigating the relationship between pathological expression of SIRT3 and specific predisposition to cancer metastasis; d) conducting of cohort studies using SIRT3 to predict cancer metastasis patient survival and establish prognosis evaluation systems; e) further developing of SIRT3 activators/inhibitors for therapeutic potential with no side effects to design personalized therapy programs. In conclusion, SIRT3 is a promising biomarker and plays an essential role in different cancer types. Exploring the function of SIRT3 in the field of cancer continues to enrich our knowledge bases. In addition, the personalized therapy combined application of SIRT3 small molecule modulators with the other drug targeting a specific cancer type has a promising future for the patients.
Author Contributions
QZ and JZ: wrote the original draft. FL, SG, LZ, and JL: reviewed and editing. The final submitted version was supervised and approved by QQ and YS. All authors contributed to the article and the final version was approved by all of them.
Funding
This work was supported by the Shanghai Sailing Program (No. 20YF1445800) and Shanghai Clinical Research Center for Acupuncture and Moxibustion (No. 20MC1920500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. North BJ, Verdin E. Sirtuins: Sir2-Related NAD-Dependent Protein Deacetylases. Genome Biol (2004) 5(5):224. doi: 10.1186/gb-2004-5-5-224
2. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional Silencing and Longevity Protein Sir2 Is an NAD-Dependent Histone Deacetylase. Nature (2000) 403(6771):795–800. doi: 10.1038/35001622
3. Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, et al. The Silencing Protein SIR2 and Its Homologs are NAD-Dependent Protein Deacetylases. Proc Natl Acad Sci USA (2000) 97(11):5807–11. doi: 10.1073/pnas.110148297
4. Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for Life-Span Extension by Calorie Restriction in Saccharomyces Cerevisiae. Science (2000) 289(5487):2126–8. doi: 10.1126/science.289.5487.2126
5. Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, et al. A Role for the Mitochondrial Deacetylase Sirt3 in Regulating Energy Homeostasis. Proc Natl Acad Sci USA (2008) 105(38):14447–52. doi: 10.1073/pnas.0803790105
6. Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and Cancer: Tumor Promoter or Suppressor? Biochim Biophys Acta (2011) 1816(1):80–8. doi: 10.1016/j.bbcan.2011.04.004
7. Xiong Y, Wang M, Zhao J, Han Y, Jia L. Sirtuin 3: A Janus Face in Cancer (Review). Int J Oncol (2016) 49(6):2227–35. doi: 10.3892/ijo.2016.3767
8. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
9. Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a Human SIR2 Homologue, is an NAD-Dependent Deacetylase Localized to Mitochondria. Proc Natl Acad Sci USA (2002) 99(21):13653–8. doi: 10.1073/pnas.222538099
10. Scher MB, Vaquero A, Reinberg D. SirT3 Is a Nuclear NAD+-Dependent Histone Deacetylase That Translocates to the Mitochondria Upon Cellular Stress. Genes Dev (2007) 21(8):920–8. doi: 10.1101/gad.1527307
11. Parihar P, Solanki I, Mansuri ML, Parihar MS. Mitochondrial Sirtuins: Emerging Roles in Metabolic Regulations, Energy Homeostasis and Diseases. Exp Gerontol (2015) 61:130–41. doi: 10.1016/j.exger.2014.12.004
12. Bruzzone S, Parenti MD, Grozio A, Ballestrero A, Bauer I, Del Rio A, et al. Rejuvenating Sirtuins: The Rise of a New Family of Cancer Drug Targets. Curr Pharm Des (2013) 19(4):614–23. doi: 10.2174/138161213804581954
13. Schwer B, North BJ, Frye RA, Ott M, Verdin E. The Human Silent Information Regulator (Sir)2 Homologue Hsirt3 Is a Mitochondrial Nicotinamide Adenine Dinucleotide-Dependent Deacetylase. J Cell Biol (2002) 158(4):647–57. doi: 10.1083/jcb.200205057
14. Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of Mouse Mitochondrial SIRT Proteins: Shift of SIRT3 to Nucleus by Co-Expression With SIRT5. Biochem Biophys Res Commun (2008) 366(1):174–9. doi: 10.1016/j.bbrc.2007.11.122
15. Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, et al. Biochemical Characterization, Localization, and Tissue Distribution of the Longer Form of Mouse SIRT3. Protein Sci (2009) 18(3):514–25. doi: 10.1002/pro.50
16. Liu L, Nam M, Fan W, Akie TE, Hoaglin DC, Gao G, et al. Nutrient Sensing by the Mitochondrial Transcription Machinery Dictates Oxidative Phosphorylation. J Clin Invest (2014) 124(2):768–84. doi: 10.1172/JCI69413
17. Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and Functional Diversity of Lysine Acetylation Revealed by a Proteomics Survey. Mol Cell (2006) 23(4):607–18. doi: 10.1016/j.molcel.2006.06.026
18. Finley LW, Haigis MC. Metabolic Regulation by SIRT3: Implications for Tumorigenesis. Trends Mol Med (2012) 18(9):516–23. doi: 10.1016/j.molmed.2012.05.004
19. Dancy BM, Cole PA. Protein Lysine Acetylation by P300/CBP. Chem Rev (2015) 115(6):2419–52. doi: 10.1021/cr500452k
20. Drazic A, Myklebust LM, Ree R, Arnesen T. The World of Protein Acetylation. Biochim Biophys Acta (2016) 1864(10):1372–401. doi: 10.1016/j.bbapap.2016.06.007
21. Shahbazian MD, Grunstein M. Functions of Site-Specific Histone Acetylation and Deacetylation. Annu Rev Biochem (2007) 76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114
22. Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, et al. Sirtuin 3 (SIRT3) Protein Regulates Long-Chain Acyl-CoA Dehydrogenase by Deacetylating Conserved Lysines Near the Active Site. J Biol Chem (2013) 288(47):33837–47. doi: 10.1074/jbc.M113.510354
23. Giralt A, Villarroya F. SIRT3, a Pivotal Actor in Mitochondrial Functions: Metabolism, Cell Death and Aging. Biochem J (2012) 444(1):1–10. doi: 10.1042/BJ20120030
24. Wang S, Zhang J, Deng X, Zhao Y, Xu K. Advances in Characterization of SIRT3 Deacetylation Targets in Mitochondrial Function. Biochimie (2020) 179:1–13. doi: 10.1016/j.biochi.2020.08.021
25. Zhang J, Xiang H, Liu J, Chen Y, He RR, Liu B. Mitochondrial Sirtuin 3: New Emerging Biological Function and Therapeutic Target. Theranostics (2020) 10(18):8315–42. doi: 10.7150/thno.45922
26. Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, et al. Tyr Phosphorylation of PDP1 Toggles Recruitment Between ACAT1 and SIRT3 to Regulate the Pyruvate Dehydrogenase Complex. Mol Cell (2014) 53(4):534–48. doi: 10.1016/j.molcel.2013.12.026
27. Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, et al. Sirt3 Regulates Metabolic Flexibility of Skeletal Muscle Through Reversible Enzymatic Deacetylation. Diabetes (2013) 62(10):3404–17. doi: 10.2337/db12-1650
28. Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, et al. SIRT3-Dependent GOT2 Acetylation Status Affects the Malate-Aspartate NADH Shuttle Activity and Pancreatic Tumor Growth. EMBO J (2015) 34(8):1110–25. doi: 10.15252/embj.201591041
29. Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 Protein Deacetylates Isocitrate Dehydrogenase 2 (IDH2) and Regulates Mitochondrial Redox Status. J Biol Chem (2012) 287(17):14078–86. doi: 10.1074/jbc.M112.355206
30. Zou X, Zhu Y, Park SH, Liu G, O’Brien J, Jiang H, et al. SIRT3-Mediated Dimerization of IDH2 Directs Cancer Cell Metabolism and Tumor Growth. Cancer Res (2017) 77(15):3990–9. doi: 10.1158/0008-5472.CAN-16-2393
31. Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, et al. Label-Free Quantitative Proteomics of the Lysine Acetylome in Mitochondria Identifies Substrates of SIRT3 in Metabolic Pathways. Proc Natl Acad Sci USA (2013) 110(16):6601–6. doi: 10.1073/pnas.1302961110
32. Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and Regulation Mechanisms for the Human Mitochondrial Sirtuins Sirt3 and Sirt5. J Mol Biol (2008) 382(3):790–801. doi: 10.1016/j.jmb.2008.07.048
33. Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 Homolog SIRT3 Regulates Global Mitochondrial Lysine Acetylation. Mol Cell Biol (2007) 27(24):8807–14. doi: 10.1128/MCB.01636-07
34. Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism Through HIF1α Destabilization. Cancer Cell (2011) 19(3):416–28. doi: 10.1016/j.ccr.2011.02.014
35. Vassilopoulos A, Pennington JD, Andresson T, Rees DM, Bosley AD, Fearnley IM, et al. SIRT3 Deacetylates ATP Synthase F1 Complex Proteins in Response to Nutrient- and Exercise-Induced Stress. Antioxid Redox Signal (2014) 21(4):551–64. doi: 10.1089/ars.2013.5420
36. Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, et al. Succinate Dehydrogenase Is a Direct Target of Sirtuin 3 Deacetylase Activity. PloS One (2011) 6(8):e23295. doi: 10.1371/journal.pone.0023295
37. Napolitano G, Fasciolo G, Venditti P. Mitochondrial Management of Reactive Oxygen Species. Antioxid (Basel) (2021) 10(11):1824. doi: 10.3390/antiox10111824
38. Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism During Stress. Cancer Cell (2010) 17(1):41–52. doi: 10.1016/j.ccr.2009.11.023
39. Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, et al. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss Under Caloric Restriction. Cell (2010) 143(5):802–12. doi: 10.1016/j.cell.2010.10.002
40. Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab (2010) 12(6):662–7. doi: 10.1016/j.cmet.2010.11.015
41. Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 Blocks the Cardiac Hypertrophic Response by Augmenting Foxo3α-Dependent Antioxidant Defense Mechanisms in Mice. J Clin Invest (2009) 119(9):2758–71. doi: 10.1172/JCI39162
42. Tseng AH, Shieh SS, Wang DL. SIRT3 Deacetylates FOXO3 to Protect Mitochondria Against Oxidative Damage. Free Radic Biol Med (2013) 63:222–34. doi: 10.1016/j.freeradbiomed.2013.05.002
43. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 Regulates Mitochondrial Fatty-Acid Oxidation by Reversible Enzyme Deacetylation. Nature (2010) 464(7285):121–5. doi: 10.1038/nature08778
44. Papa L, Germain D. SirT3 Regulates the Mitochondrial Unfolded Protein Response. Mol Cell Biol (2014) 34(4):699–710. doi: 10.1128/MCB.01337-13
45. Liu P, Huang G, Wei T, Gao J, Huang C, Sun M, et al. Sirtuin 3-Induced Macrophage Autophagy in Regulating NLRP3 Inflammasome Activation. Biochim Biophys Acta Mol Basis Dis (2018) 1864(3):764–77. doi: 10.1016/j.bbadis.2017.12.027
46. Annesley SJ, Fisher PR. Mitochondria in Health and Disease. Cells (2019) 8(7):680. doi: 10.3390/cells8070680
47. Chen Y, Fu LL, Wen X, Wang XY, Liu J, Cheng Y, et al. Sirtuin-3 (SIRT3), a Therapeutic Target With Oncogenic and Tumor-Suppressive Function in Cancer. Cell Death Dis (2014) 5(2):e1047. doi: 10.1038/cddis.2014.14
48. Martinez-Pastor B, Mostoslavsky R. Sirtuins, Metabolism, and Cancer. Front Pharmacol (2012) 3:22. doi: 10.3389/fphar.2012.00022
49. Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, et al. Tumour Suppressor SIRT3 Deacetylates and Activates Manganese Superoxide Dismutase to Scavenge ROS. EMBO Rep (2011) 12(6):534–41. doi: 10.1038/embor.2011.65
50. Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, et al. Sirt3-Mediated Deacetylation of Evolutionarily Conserved Lysine 122 Regulates MnSOD Activity in Response to Stress. Mol Cell (2010) 40(6):893–904. doi: 10.1016/j.molcel.2010.12.013
51. Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, et al. SIRT3 Interacts With the Daf-16 Homolog FOXO3a in the Mitochondria, as Well as Increases FOXO3a Dependent Gene Expression. Int J Biol Sci (2008) 4(5):291–9. doi: 10.7150/ijbs.4.291
52. Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of Succinate Dehydrogenase Activity by SIRT3 in Mammalian Mitochondria. Biochemistry (2010) 49(2):304–11. doi: 10.1021/bi901627u
53. Chen MC, Hsu LL, Wang SF, Hsu CY, Lee HC, Tseng LM. ROS Mediate xCT-Dependent Cell Death in Human Breast Cancer Cells Under Glucose Deprivation. Cells (2020) 9(7):1598. doi: 10.3390/cells9071598
54. Yang B, Fu X, Shao L, Ding Y, Zeng D. Aberrant Expression of SIRT3 Is Conversely Correlated With the Progression and Prognosis of Human Gastric Cancer. Biochem Biophys Res Commun (2014) 443(1):156–60. doi: 10.1016/j.bbrc.2013.11.068
55. Yao W, Ji S, Qin Y, Yang J, Xu J, Zhang B, et al. Profilin-1 Suppresses Tumorigenicity in Pancreatic Cancer Through Regulation of the SIRT3-Hif1α Axis. Mol Cancer (2014) 13:187. doi: 10.1186/1476-4598-13-187
56. Ozden O, Park SH, Wagner BA, Song HY, Zhu Y, Vassilopoulos A, et al. SIRT3 Deacetylates and Increases Pyruvate Dehydrogenase Activity in Cancer Cells. Free Radic Biol Med (2014) 76:163–72. doi: 10.1016/j.freeradbiomed.2014.08.001
57. Xu L, Li Y, Zhou L, Dorfman RG, Liu L, Cai R, et al. SIRT3 Elicited an Anti-Warburg Effect Through HIF1α/PDK1/PDHA1 to Inhibit Cholangiocarcinoma Tumorigenesis. Cancer Med (2019) 8(5):2380–91. doi: 10.1002/cam4.2089
58. Zhao K, Zhou Y, Qiao C, Ni T, Li Z, Wang X, et al. Oroxylin A Promotes PTEN-Mediated Negative Regulation of MDM2 Transcription via SIRT3-Mediated Deacetylation to Stabilize P53 and Inhibit Glycolysis in Wt-P53 Cancer Cells. J Hematol Oncol (2015) 8:41. doi: 10.1186/s13045-015-0137-1
59. Wei L, Zhou Y, Dai Q, Qiao C, Zhao L, Hui H, et al. Oroxylin A Induces Dissociation of Hexokinase II From the Mitochondria and Inhibits Glycolysis by SIRT3-Mediated Deacetylation of Cyclophilin D in Breast Carcinoma. Cell Death Dis (2013) 4(4):e601. doi: 10.1038/cddis.2013.131
60. Jeong SM, Lee J, Finley LW, Schmidt PJ, Fleming MD, Haigis MC. SIRT3 Regulates Cellular Iron Metabolism and Cancer Growth by Repressing Iron Regulatory Protein 1. Oncogene (2015) 34(16):2115–24. doi: 10.1038/onc.2014.124
61. Zu Y, Chen XF, Li Q, Zhang ST, Si LN. PGC-1α Activates SIRT3 to Modulate Cell Proliferation and Glycolytic Metabolism in Breast Cancer. Neoplasma (2021) 68(2):352–61. doi: 10.4149/neo_2020_200530N584
62. Wang S, Li J, Xie J, Liu F, Duan Y, Wu Y, et al. Programmed Death Ligand 1 Promotes Lymph Node Metastasis and Glucose Metabolism in Cervical Cancer by Activating Integrin β4/SNAI1/SIRT3 Signaling Pathway. Oncogene (2018) 37(30):4164–80. doi: 10.1038/s41388-018-0252-x
63. Wang Q, Ye S, Chen X, Xu P, Li K, Zeng S, et al. Mitochondrial NOS1 Suppresses Apoptosis in Colon Cancer Cells Through Increasing SIRT3 Activity. Biochem Biophys Res Commun (2019) 515(4):517–23. doi: 10.1016/j.bbrc.2019.05.114
64. Xu LX, Hao LJ, Ma JQ, Liu JK, Hasim A. SIRT3 Promotes the Invasion and Metastasis of Cervical Cancer Cells by Regulating Fatty Acid Synthase. Mol Cell Biochem (2020) 464(1-2):11–20. doi: 10.1007/s11010-019-03644-2
65. Cui Y, Qin L, Wu J, Qu X, Hou C, Sun W, et al. SIRT3 Enhances Glycolysis and Proliferation in SIRT3-Expressing Gastric Cancer Cells. PloS One (2015) 10(6):e0129834. doi: 10.1371/journal.pone.0129834
66. Li M, Chiang YL, Lyssiotis CA, Teater MR, Hong JY, Shen H, et al. Non-Oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell (2019) 35(6):916–931.e919. doi: 10.1016/j.ccell.2019.05.002
67. Yang Y, Karakhanova S, Hartwig W, D’Haese JG, Philippov PP, Werner J, et al. Mitochondria and Mitochondrial ROS in Cancer: Novel Targets for Anticancer Therapy. J Cell Physiol (2016) 231(12):2570–81. doi: 10.1002/jcp.25349
68. Breitenbach M, Rinnerthaler M, Hartl J, Stincone A, Vowinckel J, Breitenbach-Koller H, et al. Mitochondria in Ageing: There Is Metabolism Beyond the ROS. FEMS Yeast Res (2014) 14(1):198–212. doi: 10.1111/1567-1364.12134
69. Finkel T. Signal Transduction by Reactive Oxygen Species. J Cell Biol (2011) 194(1):7–15. doi: 10.1083/jcb.201102095
70. Panieri E, Santoro MM. ROS Homeostasis and Metabolism: A Dangerous Liason in Cancer Cells. Cell Death Dis (2016) 7(6):e2253. doi: 10.1038/cddis.2016.105
71. Galadari S, Rahman A, Pallichankandy S, Thayyullathil F. Reactive Oxygen Species and Cancer Paradox: To Promote or to Suppress? Free Radic Biol Med (2017) 104:144–64. doi: 10.1016/j.freeradbiomed.2017.01.004
72. Qiao A, Wang K, Yuan Y, Guan Y, Ren X, Li L, et al. Sirt3-Mediated Mitophagy Protects Tumor Cells Against Apoptosis Under Hypoxia. Oncotarget (2016) 7(28):43390–400. doi: 10.18632/oncotarget.9717
73. Liu H, Li S, Liu X, Chen Y, Deng H. SIRT3 Overexpression Inhibits Growth of Kidney Tumor Cells and Enhances Mitochondrial Biogenesis. J Proteome Res (2018) 17(9):3143–52. doi: 10.1021/acs.jproteome.8b00260
74. Puzio-Kuter AM. The Role of P53 in Metabolic Regulation. Genes Cancer (2011) 2(4):385–91. doi: 10.1177/1947601911409738
75. D’Onofrio N, Martino E, Balestrieri A, Mele L, Neglia G, Balestrieri ML, et al. SIRT3 and Metabolic Reprogramming Mediate the Antiproliferative Effects of Whey in Human Colon Cancer Cells. Cancers (Basel) (2021) 13(20):5196. doi: 10.3390/cancers13205196
76. Ahmed MA, O’Callaghan C, Chang ED, Jiang H, Vassilopoulos A. Context-Dependent Roles for SIRT2 and SIRT3 in Tumor Development Upon Calorie Restriction or High Fat Diet. Front Oncol (2019) 9:1462. doi: 10.3389/fonc.2019.01462
77. Sawant Dessai A, Dominguez MP, Chen UI, Hasper J, Prechtl C, Yu C, et al. Transcriptional Repression of SIRT3 Potentiates Mitochondrial Aconitase Activation to Drive Aggressive Prostate Cancer to the Bone. Cancer Res (2021) 81(1):50–63. doi: 10.1158/0008-5472.CAN-20-1708
78. Xiao JB, Ma JQ, Yakefu K, Tursun M. Effect of the SIRT3-AMPK/PPAR Pathway on Invasion and Migration of Cervical Cancer Cells. Int J Clin Exp Pathol (2020) 13(10):2495–501.
79. Li R, Quan Y, Xia W. SIRT3 Inhibits Prostate Cancer Metastasis Through Regulation of FOXO3A by Suppressing Wnt/β-Catenin Pathway. Exp Cell Res (2018) 364(2):143–51. doi: 10.1016/j.yexcr.2018.01.036
80. Wang X, Zeng Q, Li Z, Yang X, Xia W, Chen Z. Adjudin Synergizes With Paclitaxel and Inhibits Cell Growth and Metastasis by Regulating the Sirtuin 3-Forkhead Box O3a Axis in Human Small-Cell Lung Cancer. Thorac Cancer (2019) 10(4):642–58. doi: 10.1111/1759-7714.12976
81. Liu L, Li Y, Cao D, Qiu S, Li Y, Jiang C, et al. SIRT3 Inhibits Gallbladder Cancer by Induction of AKT-Dependent Ferroptosis and Blockade of Epithelial-Mesenchymal Transition. Cancer Lett (2021) 510:93–104. doi: 10.1016/j.canlet.2021.04.007
82. Dong XC, Jing LM, Wang WX, Gao YX. Down-Regulation of SIRT3 Promotes Ovarian Carcinoma Metastasis. Biochem Biophys Res Commun (2016) 475(3):245–50. doi: 10.1016/j.bbrc.2016.05.098
83. Neeli PK, Gollavilli PN, Mallappa S, Hari SG, Kotamraju S. A Novel MetadherinΔ7 Splice Variant Enhances Triple Negative Breast Cancer Aggressiveness by Modulating Mitochondrial Function via NFĸB-SIRT3 Axis. Oncogene (2020) 39(10):2088–102. doi: 10.1038/s41388-019-1126-6
84. Ren T, Zhang H, Wang J, Zhu J, Jin M, Wu Y, et al. MCU-Dependent Mitochondrial Ca2+ Inhibits NAD+/SIRT3/SOD2 Pathway to Promote ROS Production and Metastasis of HCC Cells. Oncogene (2017) 36(42):5897–909. doi: 10.1038/onc.2017.167
85. Lee JJ, van de Ven RAH, Zaganjor E, Ng MR, Barakat A, Demmers J, et al. Inhibition of Epithelial Cell Migration and Src/FAK Signaling by SIRT3. Proc Natl Acad Sci USA (2018) 115(27):7057–62. doi: 10.1073/pnas.1800440115
86. Kim YS, Gupta Vallur P, Jones VM, Worley BL, Shimko S, Shin DH, et al. Context-Dependent Activation of SIRT3 Is Necessary for Anchorage-Independent Survival and Metastasis of Ovarian Cancer Cells. Oncogene (2020) 39(8):1619–33. doi: 10.1038/s41388-019-1097-7
87. Wan X, Wang C, Huang Z, Zhou D, Xiang S, Qi Q, et al. Cisplatin Inhibits SIRT3-Deacetylation MTHFD2 to Disturb Cellular Redox Balance in Colorectal Cancer Cell. Cell Death Dis (2020) 11(8):649. doi: 10.1038/s41419-020-02825-y
88. Xuan ZH, Wang HP, Zhang XN, Chen ZX, Zhang HY, Gu MM. PKMYT1 Aggravates the Progression of Ovarian Cancer by Targeting SIRT3. Eur Rev Med Pharmacol Sci (2020) 24(10):5259–66. doi: 10.26355/eurrev_202005_21308
89. Kenny TC, Hart P, Ragazzi M, Sersinghe M, Chipuk J, Sagar MAK, et al. Selected Mitochondrial DNA Landscapes Activate the SIRT3 Axis of the UPRmt to Promote Metastasis. Oncogene (2017) 36(31):4393–404. doi: 10.1038/onc.2017.52
90. Pascual G, Domínguez D, Benitah SA. The Contributions of Cancer Cell Metabolism to Metastasis. Dis Model Mech (2018) 11(8):dmm032920. doi: 10.1242/dmm.032920
91. Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, et al. Emerging Roles of Lipid Metabolism in Cancer Metastasis. Mol Cancer (2017) 16(1):76. doi: 10.1186/s12943-017-0646-3
92. Nath A, Chan C. Genetic Alterations in Fatty Acid Transport and Metabolism Genes are Associated With Metastatic Progression and Poor Prognosis of Human Cancers. Sci Rep (2016) 6:18669. doi: 10.1038/srep18669
93. Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, et al. Targeting Metastasis-Initiating Cells Through the Fatty Acid Receptor CD36. Nature (2017) 541(7635):41–5. doi: 10.1038/nature20791
94. Okumura T, Ohuchida K, Sada M, Abe T, Endo S, Koikawa K, et al. Extra-Pancreatic Invasion Induces Lipolytic and Fibrotic Changes in the Adipose Microenvironment, With Released Fatty Acids Enhancing the Invasiveness of Pancreatic Cancer Cells. Oncotarget (2017) 8(11):18280–95. doi: 10.18632/oncotarget.15430
95. Zhang S, Zhang J, An Y, Zeng X, Qin Z, Zhao Y, et al. Multi-Omics Approaches Identify SF3B3 and SIRT3 as Candidate Autophagic Regulators and Druggable Targets in Invasive Breast Carcinoma. Acta Pharm Sin B (2021) 11(5):1227–45. doi: 10.1016/j.apsb.2020.12.013
96. Papa L, Germain D. Estrogen Receptor Mediates a Distinct Mitochondrial Unfolded Protein Response. J Cell Sci (2011) 124(Pt 9):1396–402. doi: 10.1242/jcs.078220
97. Che L, Wu JS, Du ZB, He YQ, Yang L, Lin JX, et al. Targeting Mitochondrial COX-2 Enhances Chemosensitivity via Drp1-Dependent Remodeling of Mitochondrial Dynamics in Hepatocellular Carcinoma. Cancers (Basel) (2022) 14(3):821. doi: 10.3390/cancers14030821
98. Tao NN, Zhou HZ, Tang H, Cai XF, Zhang WL, Ren JH, et al. Sirtuin 3 Enhanced Drug Sensitivity of Human Hepatoma Cells Through Glutathione S-Transferase Pi 1/JNK Signaling Pathway. Oncotarget (2016) 7(31):50117–30. doi: 10.18632/oncotarget.10319
99. Guo R, Li Y, Xue Y, Chen Y, Li J, Deng X, et al. SIRT3 Increases Cisplatin Sensitivity of Small-Cell Lung Cancer Through Apoptosis. Gene (2020) 745:144629. doi: 10.1016/j.gene.2020.144629
100. Cao Y, Li P, Wang H, Li L, Li Q. SIRT3 Promotion Reduces Resistance to Cisplatin in Lung Cancer by Modulating the FOXO3/CDT1 Axis. Cancer Med (2021) 10(4):1394–404. doi: 10.1002/cam4.3728
101. Gao Y, Zhu Y, Tran EL, Tokars V, Dean AE, Quan S, et al. MnSOD Lysine 68 Acetylation Leads to Cisplatin and Doxorubicin Resistance Due to Aberrant Mitochondrial Metabolism. Int J Biol Sci (2021) 17(5):1203–16. doi: 10.7150/ijbs.51184
102. Ma J, Liu B, Yu D, Zuo Y, Cai R, Yang J, et al. SIRT3 Deacetylase Activity Confers Chemoresistance in AML via Regulation of Mitochondrial Oxidative Phosphorylation. Br J Haematol (2019) 187(1):49–64. doi: 10.1111/bjh.16044
103. Paku M, Haraguchi N, Takeda M, Fujino S, Ogino T, Takahashi H, et al. SIRT3-Mediated SOD2 and PGC-1α Contribute to Chemoresistance in Colorectal Cancer Cells. Ann Surg Oncol (2021) 28(8):4720–32. doi: 10.1245/s10434-020-09373-x
104. Torrens-Mas M, Hernández-López R, Oliver J, Roca P, Sastre-Serra J. Sirtuin 3 Silencing Improves Oxaliplatin Efficacy Through Acetylation of MnSOD in Colon Cancer. J Cell Physiol (2018) 233(8):6067–76. doi: 10.1002/jcp.26443
105. Chen H, Zhang DM, Zhang ZP, Li MZ, Wu HF. SIRT3-Mediated Mitochondrial Unfolded Protein Response Weakens Breast Cancer Sensitivity to Cisplatin. Genes Genomics (2021) 43(12):1433–44. doi: 10.1007/s13258-021-01145-5
106. Hou L, Wang R, Wei H, Li S, Liu L, Lu X, et al. ABT737 Enhances Ovarian Cancer Cells Sensitivity to Cisplatin Through Regulation of Mitochondrial Fission via Sirt3 Activation. Life Sci (2019) 232:116561. doi: 10.1016/j.lfs.2019.116561
107. Sullivan LB, Chandel NS. Mitochondrial Reactive Oxygen Species and Cancer. Cancer Metab (2014) 2:17. doi: 10.1186/2049-3002-2-17
108. Hussain SP, Hofseth LJ, Harris CC. Radical Causes of Cancer. Nat Rev Cancer (2003) 3(4):276–85. doi: 10.1038/nrc1046
109. Sainz RM, Lombo F, Mayo JC. Radical Decisions in Cancer: Redox Control of Cell Growth and Death. Cancers (Basel) (2012) 4(2):442–74. doi: 10.3390/cancers4020442
110. Torrens-Mas M, Pons DG, Sastre-Serra J, Oliver J, Roca P. SIRT3 Silencing Sensitizes Breast Cancer Cells to Cytotoxic Treatments Through an Increment in ROS Production. J Cell Biochem (2017) 118(2):397–406. doi: 10.1002/jcb.25653
111. Zhang J, Zou L, Shi D, Liu J, Zhang J, Zhao R, et al. Structure-Guided Design of a Small-Molecule Activator of Sirtuin-3 That Modulates Autophagy in Triple Negative Breast Cancer. J Med Chem (2021) 64(19):14192–216. doi: 10.1021/acs.jmedchem.0c02268
112. Signorelli P, Ghidoni R. Resveratrol as an Anticancer Nutrient: Molecular Basis, Open Questions and Promises. J Nutr Biochem (2005) 16(8):449–66. doi: 10.1016/j.jnutbio.2005.01.017
113. Luo LX, Li Y, Liu ZQ, Fan XX, Duan FG, Li RZ, et al. Honokiol Induces Apoptosis, G1 Arrest, and Autophagy in KRAS Mutant Lung Cancer Cells. Front Pharmacol (2017) 8:199. doi: 10.3389/fphar.2017.00199
114. Pillai VB, Kanwal A, Fang YH, Sharp WW, Samant S, Arbiser J, et al. Honokiolan, an Activator of Sirtuin-3 (SIRT3) Preserves Mitochondria and Protects the Heart From Doxorubicin-Induced Cardiomyopathy in Mice. Oncotarget (2017) 8(21):34082–98. doi: 10.18632/oncotarget.16133
115. Li Y, Ye Z, Lai W, Rao J, Huang W, Zhang X, et al. Activation of Sirtuin 3 by Silybin Attenuates Mitochondrial Dysfunction in Cisplatin-Induced Acute Kidney Injury. Front Pharmacol (2017) 8:178. doi: 10.3389/fphar.2017.00178
116. Liu Z, Li L, Xue B. Effect of Ganoderic Acid D on Colon Cancer Warburg Effect: Role of SIRT3/cyclophilin D. Eur J Pharmacol (2018) 824:72–7. doi: 10.1016/j.ejphar.2018.01.026
117. Chen X, Hao B, Li D, Reiter RJ, Bai Y, Abay B, et al. Melatonin Inhibits Lung Cancer Development by Reversing the Warburg Effect via Stimulating the SIRT3/PDH Axis. J Pineal Res (2021) 71(2):e12755. doi: 10.1111/jpi.12755
118. George J, Nihal M, Singh CK, Ahmad N. 4’-Bromo-Resveratrol, a Dual Sirtuin-1 and Sirtuin-3 Inhibitor, Inhibits Melanoma Cell Growth Through Mitochondrial Metabolic Reprogramming. Mol Carcinog (2019) 58(10):1876–85. doi: 10.1002/mc.23080
119. Mahajan SS, Scian M, Sripathy S, Posakony J, Lao U, Loe TK, et al. Development of Pyrazolone and Isoxazol-5-One Cambinol Analogues as Sirtuin Inhibitors. J Med Chem (2014) 57(8):3283–94. doi: 10.1021/jm4018064
120. Chen D, Zheng W. Cyclic Peptide-Based Potent and Selective SIRT1/2 Dual Inhibitors Harboring Nϵ-Thioacetyl-Lysine. Bioorg Med Chem Lett (2016) 26(21):5234–9. doi: 10.1016/j.bmcl.2016.09.055
121. Han H, Li C, Li M, Yang L, Zhao S, Wang Z, et al. Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1h-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5. Molecules (2020) 25(12):2755. doi: 10.3390/molecules25122755
122. Gertz M, Fischer F, Nguyen GT, Lakshminarasimhan M, Schutkowski M, Weyand M, et al. Ex-527 Inhibits Sirtuins by Exploiting Their Unique NAD+-Dependent Deacetylation Mechanism. Proc Natl Acad Sci USA (2013) 110(30):E2772–81. doi: 10.1073/pnas.1303628110
123. Alhazzazi TY, Kamarajan P, Xu Y, Ai T, Chen L, Verdin E, et al. A Novel Sirtuin-3 Inhibitor, LC-0296, Inhibits Cell Survival and Proliferation, and Promotes Apoptosis of Head and Neck Cancer Cells. Anticancer Res (2016) 36(1):49–60.
124. Yeong KY, Nor Azizi MIH, Berdigaliyev N, Chen WN, Lee WL, Shirazi AN, et al. Sirtuin Inhibition and Anti-Cancer Activities of Ethyl 2-Benzimidazole-5-Carboxylate Derivatives. Medchemcomm (2019) 10(12):2140–5. doi: 10.1039/c9md00323a
125. Xu S, Liu CX, Xu W, Huang L, Zhao JY, Zhao SM. Butyrate Induces Apoptosis by Activating PDC and Inhibiting Complex I Through SIRT3 Inactivation. Signal Transduct Target Ther (2017) 2:16035. doi: 10.1038/sigtrans.2016.35
126. Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, et al. Discovery, In Vivo Activity, and Mechanism of Action of a Small-Molecule P53 Activator. Cancer Cell (2008) 13(5):454–63. doi: 10.1016/j.ccr.2008.03.004
127. Wang LJ, Lee YC, Huang CH, Shi YJ, Chen YJ, Pei SN, et al. Non-Mitotic Effect of Albendazole Triggers Apoptosis of Human Leukemia Cells via SIRT3/ROS/p38 MAPK/TTP Axis-Mediated TNF-α Upregulation. Biochem Pharmacol (2019) 162:154–68. doi: 10.1016/j.bcp.2018.11.003
128. Gorska-Ponikowska M, Kuban-Jankowska A, Eisler SA, Perricone U, Lo Bosco G, Barone G, et al. 2-Methoxyestradiol Affects Mitochondrial Biogenesis Pathway and Succinate Dehydrogenase Complex Flavoprotein Subunit A in Osteosarcoma Cancer Cells. Cancer Genomics Proteomics (2018) 15(1):73–89. doi: 10.21873/cgp.20067
129. Gajendran B, Varier KM, Liu W, Wang C, Sample KM, Zacksenhaus E, et al. A C21-Steroidal Derivative Suppresses T-Cell Lymphoma in Mice by Inhibiting SIRT3 via SAP18-Sin3. Commun Biol (2020) 3(1):732. doi: 10.1038/s42003-020-01458-3
130. Cheng Y, Dai C, Zhang J. SIRT3-SOD2-ROS Pathway is Involved in Linalool-Induced Glioma Cell Apoptotic Death. Acta Biochim Pol (2017) 64(2):343–50. doi: 10.18388/abp.2016_1438
131. Di Leva G, Garofalo M, Croce CM. MicroRNAs in Cancer. Annu Rev Pathol (2014) 9:287–314. doi: 10.1146/annurev-pathol-012513-104715
132. Wang S, Chen X, Zhang Z, Wu Z. MicroRNA-1225-5p Inhibits the Development and Progression of Thyroid Cancer via Targeting Sirtuin 3. Pharmazie (2019) 74(7):423–7. doi: 10.1691/ph.2019.9411
133. Zhang J, Han L, Yu J, Li H, Li Q. miR-224 Aggravates Cancer-Associated Fibroblast-Induced Progression of Non-Small Cell Lung Cancer by Modulating a Positive Loop of the SIRT3/AMPK/mTOR/HIF-1α Axis. Aging (Albany NY) (2021) 13(7):10431–49. doi: 10.18632/aging.202803
134. Zhang J, Zhu Y, Hu L, Yan F, Chen J. miR-494 Induces EndMT and Promotes the Development of HCC (Hepatocellular Carcinoma) by Targeting SIRT3/TGF-β/SMAD Signaling Pathway. Sci Rep (2019) 9(1):7213. doi: 10.1038/s41598-019-43731-4
135. Kao YY, Chou CH, Yeh LY, Chen YF, Chang KW, Liu CJ, et al. MicroRNA miR-31 Targets SIRT3 to Disrupt Mitochondrial Activity and Increase Oxidative Stress in Oral Carcinoma. Cancer Lett (2019) 456:40–8. doi: 10.1016/j.canlet.2019.04.028
136. Huang S, Guo H, Cao Y, Xiong J. MiR-708-5p Inhibits the Progression of Pancreatic Ductal Adenocarcinoma by Targeting Sirt3. Pathol Res Pract (2019) 215(4):794–800. doi: 10.1016/j.prp.2019.01.026
137. Ma J, Zhao G, Du J, Li J, Lin G, Zhang J. LncRNA FENDRR Inhibits Gastric Cancer Cell Proliferation and Invasion via the miR-421/SIRT3/Notch-1 Axis. Cancer Manag Res (2021) 13:9175–87. doi: 10.2147/CMAR.S329419
138. Wang C, Zhao Z, Qi Q, Wang J, Kong Y, Feng Z, et al. miR-6858 Plays a Key Role in the Process of Melatonin Inhibition of the Malignant Biological Behavior of Glioma. J Clin Neurosci (2021) 87:137–46. doi: 10.1016/j.jocn.2021.02.015
Keywords: sirtuin 3, cancer, metabolism reprogramming, metastasis, chemoresistance, activator, inhibitor, therapy
Citation: Zhao Q, Zhou J, Li F, Guo S, Zhang L, Li J, Qi Q and Shi Y (2022) The Role and Therapeutic Perspectives of Sirtuin 3 in Cancer Metabolism Reprogramming, Metastasis, and Chemoresistance. Front. Oncol. 12:910963. doi: 10.3389/fonc.2022.910963
Received: 09 April 2022; Accepted: 05 May 2022;
Published: 27 June 2022.
Edited by:
Jin-Ming Yang, University of Kentucky, United StatesReviewed by:
Dong Li, University of Kentucky, United StatesHamed Hemati, University of Kentucky, United States
Copyright © 2022 Zhao, Zhou, Li, Guo, Zhang, Li, Qi and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin Qi, cXFxaXFpbjI0NzhAMTI2LmNvbQ==; Yin Shi, Zmx5c3kwNjM2QDE2My5jb20=
†These authors have contributed equally to this work
 QingYi Zhao
QingYi Zhao Jing Zhou2†
Jing Zhou2† Sen Guo
Sen Guo Qin Qi
Qin Qi Yin Shi
Yin Shi