- 1Department of Advanced Robotics, Istituto Italiano di Tecnologia, Genoa, Italy
- 2Unit of Otorhinolaryngology – Head and Neck Surgery, IRCCS Ospedale Policlinico San Martino, Genoa, Italy
- 3Department of Surgical Sciences and Integrated Diagnostics (DISC), University of Genoa, Genoa, Italy
- 4Unit of Otorhinolaryngology – Head and Neck Surgery, ASST Spedali Civili of Brescia, Brescia, Italy
- 5Department of Medical and Surgical Specialties, Radiological Sciences, and Public Health, University of Brescia, Brescia, Italy
- 6The BioRobotics Institute and Department of Excellence in Robotics and AI, Scuola Superiore Sant’Anna, Pisa, Italy
Introduction: Narrow Band Imaging (NBI) is an endoscopic visualization technique useful for upper aero-digestive tract (UADT) cancer detection and margins evaluation. However, NBI analysis is strongly operator-dependent and requires high expertise, thus limiting its wider implementation. Recently, artificial intelligence (AI) has demonstrated potential for applications in UADT videoendoscopy. Among AI methods, deep learning algorithms, and especially convolutional neural networks (CNNs), are particularly suitable for delineating cancers on videoendoscopy. This study is aimed to develop a CNN for automatic semantic segmentation of UADT cancer on endoscopic images.
Materials and Methods: A dataset of white light and NBI videoframes of laryngeal squamous cell carcinoma (LSCC) was collected and manually annotated. A novel DL segmentation model (SegMENT) was designed. SegMENT relies on DeepLabV3+ CNN architecture, modified using Xception as a backbone and incorporating ensemble features from other CNNs. The performance of SegMENT was compared to state-of-the-art CNNs (UNet, ResUNet, and DeepLabv3). SegMENT was then validated on two external datasets of NBI images of oropharyngeal (OPSCC) and oral cavity SCC (OSCC) obtained from a previously published study. The impact of in-domain transfer learning through an ensemble technique was evaluated on the external datasets.
Results: 219 LSCC patients were retrospectively included in the study. A total of 683 videoframes composed the LSCC dataset, while the external validation cohorts of OPSCC and OCSCC contained 116 and 102 images. On the LSCC dataset, SegMENT outperformed the other DL models, obtaining the following median values: 0.68 intersection over union (IoU), 0.81 dice similarity coefficient (DSC), 0.95 recall, 0.78 precision, 0.97 accuracy. For the OCSCC and OPSCC datasets, results were superior compared to previously published data: the median performance metrics were, respectively, improved as follows: DSC=10.3% and 11.9%, recall=15.0% and 5.1%, precision=17.0% and 14.7%, accuracy=4.1% and 10.3%.
Conclusion: SegMENT achieved promising performances, showing that automatic tumor segmentation in endoscopic images is feasible even within the highly heterogeneous and complex UADT environment. SegMENT outperformed the previously published results on the external validation cohorts. The model demonstrated potential for improved detection of early tumors, more precise biopsies, and better selection of resection margins.
Introduction
At present, the cornerstone of the otolaryngological clinical examination of the upper aero-digestive tract (UADT) is represented by endoscopy. Whether performed through the nose with flexible instrumentation or transorally by rigid telescopes, endoscopy, especially if coupled with high-definition (HD) technology, provides a detailed, magnified, and comprehensively enhanced vision of the UADT. Endoscopy enhancing filters (EEFs), such as Narrow Band Imaging (NBI) or the Storz Professional Image Enhancement System (SPIES), have been playing a fundamental role in the past decade, empowering conventional white light (WL) endoscopy by highlighting the submucosal and subepithelial neoangiogenic network associated with malignant transformation (1). By enhancing visualization of the cancer-related abnormal intrapapillary capillary loops, these “bioendoscopic” tools have been shown to provide better performance compared to standard WL endoscopy in the diagnosis of UADT carcinomas (2–5). Nowadays, EEFs like NBI are widely used in various head and neck subsites such as the larynx/hypopharynx (3, 6, 7), oropharynx (8, 9), nasopharynx (10–12), and oral cavity (13–15), where they play a fundamental role in detection, characterization, and delineation of superficial margins of malignant lesions. However, caution is needed in the analysis and interpretation of UADT videoendoscopies, especially in centers less experienced with these techniques. Even with EEFs, in fact, the detection and evaluation of vascular abnormalities is limited by the considerable heterogeneity in the appearance of squamous cell carcinomas (SCCs) of this area. Moreover, margins delineation can be challenging when mucosal vascularization is altered by other factors, such as inflammatory disease or previous (chemo)radiotherapy (16). Finally, several aspects hinder the large-scale implementation of EEFs during routine UADT endoscopic assessment, such as its intrinsic operator-dependent nature and the relatively steep learning curve needed to master this technique.
Artificial intelligence (AI) is a potentially powerful ally to support clinicians in this complex scenario, prompting our research group to envision the birth of “Videomics” as a new and promising field of application of such a tool in the diagnostic challenges of the UADT oncologic diseases (17). The term Videomics was coined to refer to computer vision and deep learning methods that are systematically used to process the unstructured video data obtained from diagnostic endoscopy to convert subjective assessment into objective findings. Parallelly, the use of AI in videoendoscopy, especially in the gastrointestinal field, has already become relevant in the literature and even on the market (18). When moving to the specific field of UADT, however, only a few studies have been published in the current literature, with most focusing on laryngeal endoscopy (19). Among all AI-powered methods, deep learning (DL) techniques based on convolutional neural networks (CNNs) are increasingly used in UADT videoendoscopy analysis for automatic disease detection (20–22), classification (23, 24), and segmentation (25). In fact, thanks to their unique architecture, CNNs provide improved performance compared to conventional computer vision and machine learning methods.
Image segmentation is typically used to locate objects in images by marking their specific contours and the area inside those. In computer vision, semantic segmentation is referred as the task of assigning each pixel in an image to a predefined set of classes. Within the different computer vision tasks, semantic segmentation is particularly interesting for UADT endoscopy. Indeed, the possibility offered by DL to automatically detect tumor boundaries, especially if coupled with EEFs imaging, would represent a valuable support in clinical practice. This could make the benefits of EEFs accessible to all physicians and contribute to improve their performance in tumor recognition and margins delineation. However, only a few studies have pursued automatic segmentation of UADT lesions so far (26–29) and, thus, further research is needed to progress this technology and advance it towards its clinical implementation.
In this paper, we describe a new CNN-based semantic segmentation model for videoendoscopy of the UADT, named SegMENT. This model was specifically developed for the identification and segmentation of UADT cancer in endoscopic video frames, with particular attention to laryngeal squamous cell carcinoma (LSCC), oral cavity squamous cell carcinoma (OCSCC) and oropharyngeal squamous cell carcinoma (OPSCC). The list of abbreviations used in the article is reported in Table 1.
Materials and Methods
Data Acquisition
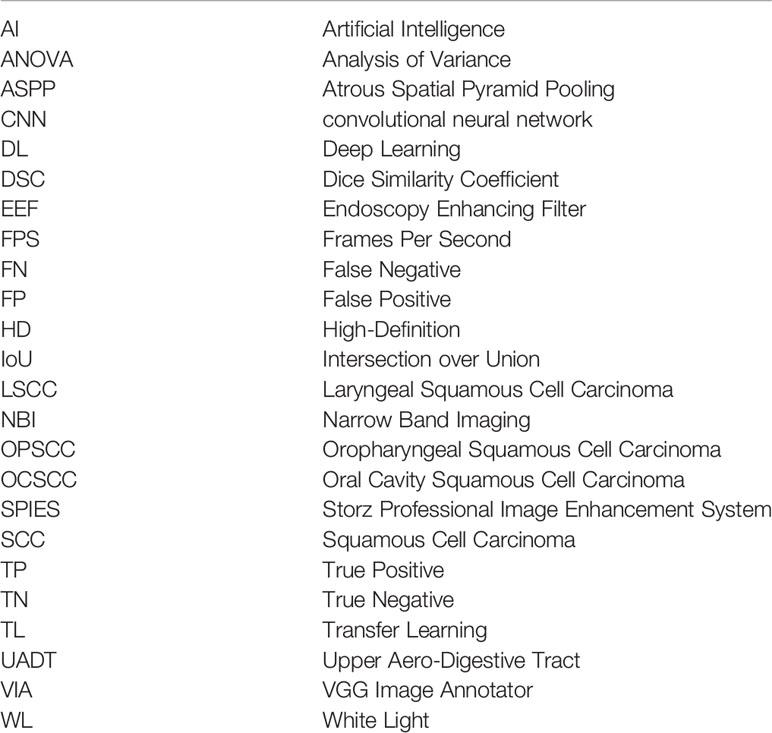
Recorded videoendoscopies of patients treated between 2014 and 2019 at the Unit of Otorhinolaryngology – Head and Neck Surgery of the IRCSS Ospedale Policlinico San Martino, Genoa, Italy, were retrospectively revised. Selection criteria comprised a pathology report positive for LSCC and the availability of at least one recorded videoendoscopy before treatment. Local Institutional Review Board approval was obtained (CER Liguria: 230/2019). All patients were first examined through transnasal videolaryngoscopy (HD Video Rhino-laryngoscope Olympus ENF-VH, Olympus Medical System Corporation, Tokyo, Japan) in the office before treatment. For those submitted to transoral laryngeal microsurgery, an additional intraoperative endoscopic evaluation was conducted using 0°, 30° or 70° telescopes coupled to a HD camera head connected to a Visera Elite CLV-S190 light source (Olympus Medical System Corporation). In both settings, a thorough examination was conducted under WL videoendoscopy, then switching to NBI.
From each of the collected videos, four expert physicians extracted, when available, one WL and one NBI frame. Frames were selected to be the most representative of the lesion and possibly offer a clear view of its boundaries. Priority in image selection was given to steady frames with few artifacts and blur. The extracted videoframes were then labeled by the same physicians using the VGG Image Annotator (VIA) 2.0 (https://www.robots.ox.ac.uk/~vgg/software/via/), an open-source web-based annotation software. The annotation process consisted in the manual segmentation of the neoplastic lesion borders: this was done by manually tracing its contour following the visible tumor margins identified both in WL and in NBI. The pixels comprised in the traced regions were classified as “LSCC” and no specific label was assigned for NBI or WL images. If multiple lesions were visible, multiple segmentations were carried out in order to select all the LSCC pixels in the image. If a physician was not completely sure about the correctness of the annotations, all four otolaryngologists revised them collectively. Finally, a senior surgeon (the author G.P.) checked all annotations and referred the inexact ones for collective revision. Figure 1 summarizes the data acquisition process.
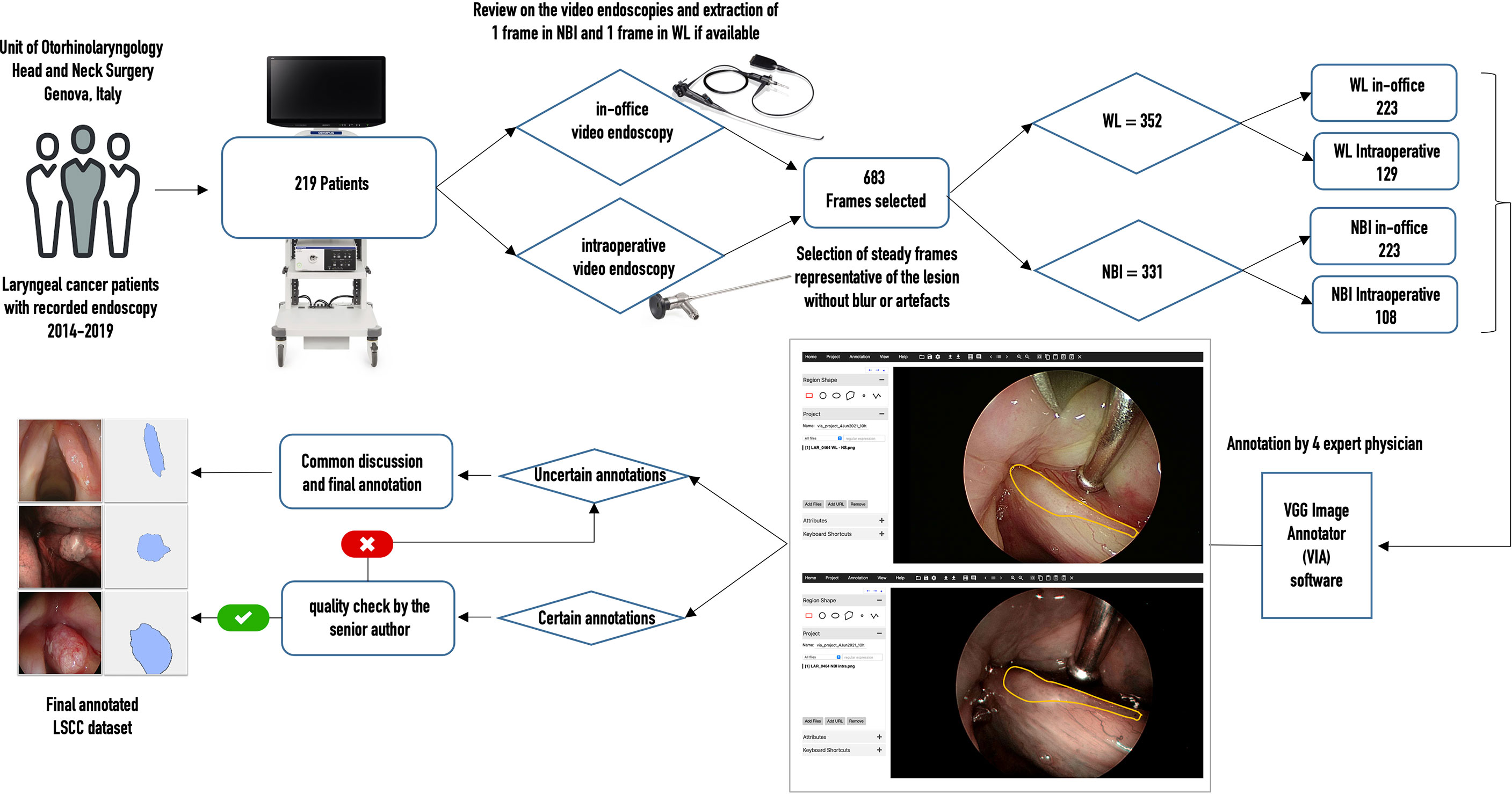
Figure 1 Flow chart of data acquisition for the laryngeal squamous cell carcinoma (LSCC) dataset. WL, white light; NBI, narrow band imaging.
Finally, two datasets of NBI endoscopic images were obtained from a previous study on automatic segmentation of OCSCC and OPSCC (26). The datasets included the corresponding ground-truth annotations, as previously described.
SegMENT Architecture
Figure 2 describes the architecture of SegMENT. This latter is based on concepts introduced by the DeepLabV3+ model (30), which were expanded and customized here for precise cancer segmentation in UADT videoendoscopies. The segMENT backbone was built on the Xception architecture (31), which was chosen for its high benchmark results on ImageNet (32), the largest dataset of natural images publicly available. Xception has a smaller number of parameters compared to the most popular CNN architectures like VGG16 and VGG19, but an almost equal number of parameters than Resnet50 and DensNet121 (33). The Xception backbone architecture is composed of two primary components: convolutional layers with pooling for feature extraction, and fully connected dense layers at the top of the network for classification. To customize this backbone network for segmentation tasks, we removed the network fully connected dense layers and maintained only the feature extraction layers. The functionalities to resize the input frames into 256x256 pixels were maintained. In addition, we used three-skip connections to get feature map outputs (of size 16×16, 32×32, and 64×64 pixels) from Xception backbone convolution layers. These were then merged using Atrous Spatial Pyramid Pooling (ASPP) blocks in the encoder part of our model.
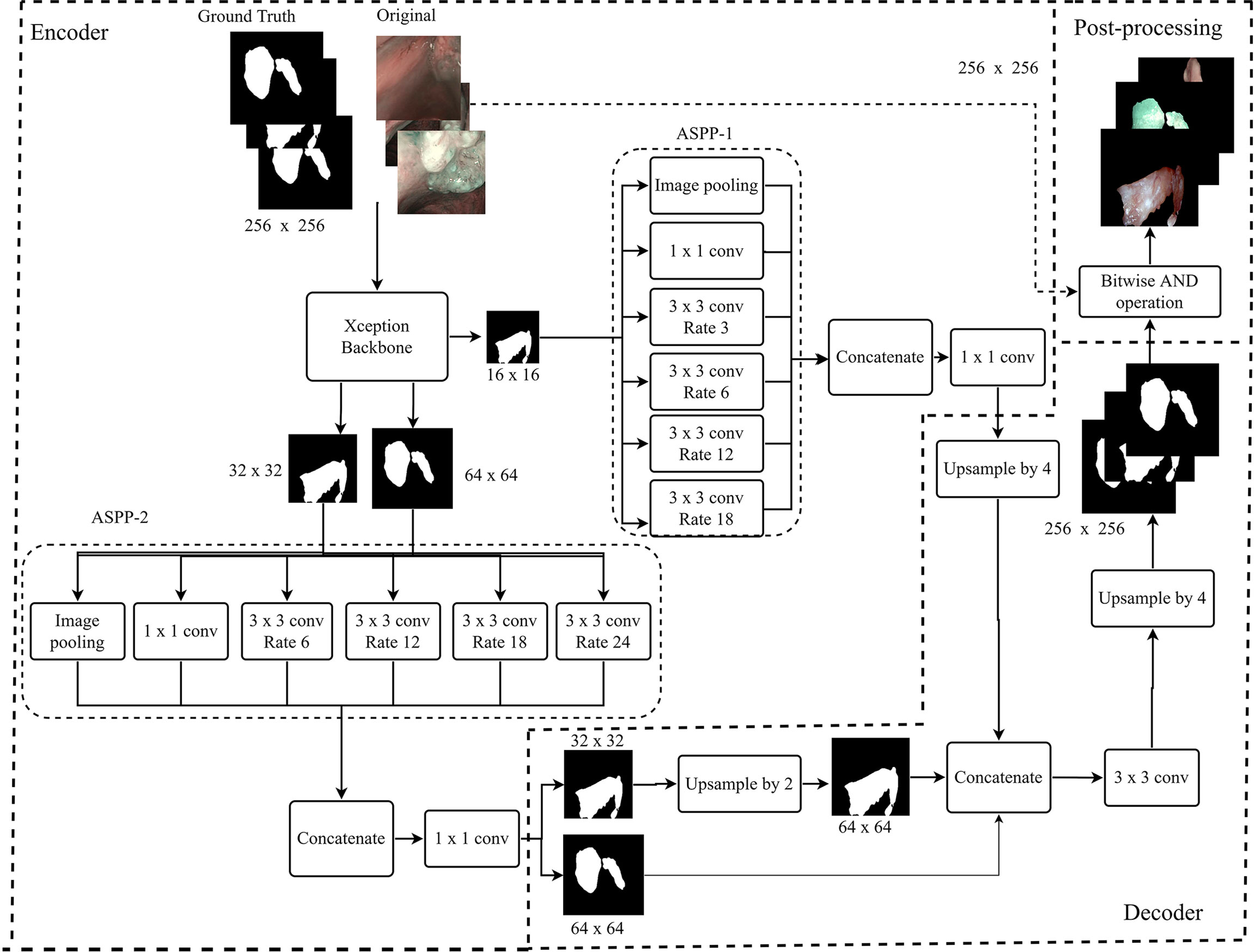
Figure 2 Workflow diagram of the proposed SegMENT semantic segmentation architecture. The Xception backbone is customized for segmentation tasks by eliminating the dense layers for classification while maintaining the feature extraction layers. The model encoder uses two ASPP blocks designed, respectively, for high- and low-level image features. The decoder section concatenates the information coming from the two ASPP blocks, producing segmentation masks.
Given the heterogeneous nature of UADT lesions in terms of dimension, form, and contour, we designed the encoder part of SegMENT to use two ASPP blocks. These can potentially contribute to increased segmentation accuracy. The ASPP block-1 is designed for high-level image features (shapes, tumor composition, etc.). It is fed with 16×16 pixels input images directly from the Xception backbone and contains 256 filters. ASPP block-1 comprises five rates of dilation convolution layers (1, 3, 6, 12, and 18), which were chosen given the small-scale input images. The ASPP block-2 is designed for low-level image features (edges, contours, texture) and accepts two scales of input images (32×32 and 64×64 pixels). It is also comprised of five rates of dilation convolution layers (1, 6, 12, 18, and 24), which are higher here because of the larger scale of the input images. The ASPP block-2 employs 48 filters for 64×64 pixels input images and 64 filters for 32×32 pixels input images. Finally, the convolutional layers of SegMENT use the Mish activation function (except for the last layers that use a sigmoid activation function). This activation function was selected to replace the traditional Relu activation function as it was shown to provide better performance (34). The decoder section accepts the encoder outputs with the three resized (16×16, 32×32, and 64×64 pixels) images. The output images of ASPP block-1 are four times up-sampled using the UpSampling2D layer and bilinear interpolation technique to yield 64×64 pixels images. The images from ASPP block-2 with 32×32 pixels are up-sampled two times. Next, 64×64 pixel images from ASPP block-2 are concatenated with the up-sampled 64×64 pixel images generated by ASPP block-1. This concatenation produces 64×64 pixels images. Afterward, a 2D convolutional layer with a kernel size of 3×3 and 256 filters is applied to these images. Again, images are further up-sampled four times to get 256×256 pixel images. Finally, the segmented tumor area is retrieved through a bitwise-AND operation, which generates an output image of 256x256 pixels.
Baseline Models
Three state-of-the-art baseline CNNs for semantic segmentation (i.e., UNet, ResUNet, and DeepLabv3+) were investigated and tested as part of a comparative study. Each segmentation model incorporates a backbone architecture for feature extraction. The backbones considered were VGG16, VGG19, MobileNetV2, ResNet50, ResNet101V2, DenseNet121, and Xception. Pre-trained backbone weights from the ImageNet dataset were used (32). The UNet network (35) is equipped with an encoding path that learns to encode texture descriptors and a decoding path that achieves the segmentation task. ResUNet (36) is a segmentation model based on the UNet architecture that implements residual units instead of plain neural units, in order to obtain good performance with fewer parameters. Finally, DeepLabV3+ (30) uses dilated separable convolutions and spatial pyramid pooling in a U-shaped architecture to produce accelerated inference times and reduced loss values. The training and testing of these baseline models were performed in the same environment and using the same data as SegMENT.
Ensemble Technique
Multiple ensemble techniques are described in the literature for decreasing segmentation errors and optimizing efficiency (37–39). Their utility becomes evident especially when the available training dataset for a new application area is small or highly heterogeneous, such as the case of the OCSCC and OPSCC datasets. In this work, we evaluated the value of using the weighted average ensemble approach (27) during the testing phase of the segmentation network. This technique implements a weighted ensemble of predictions from different models. Its integration into the proposed segmentation architecture is shown in Figure 3.
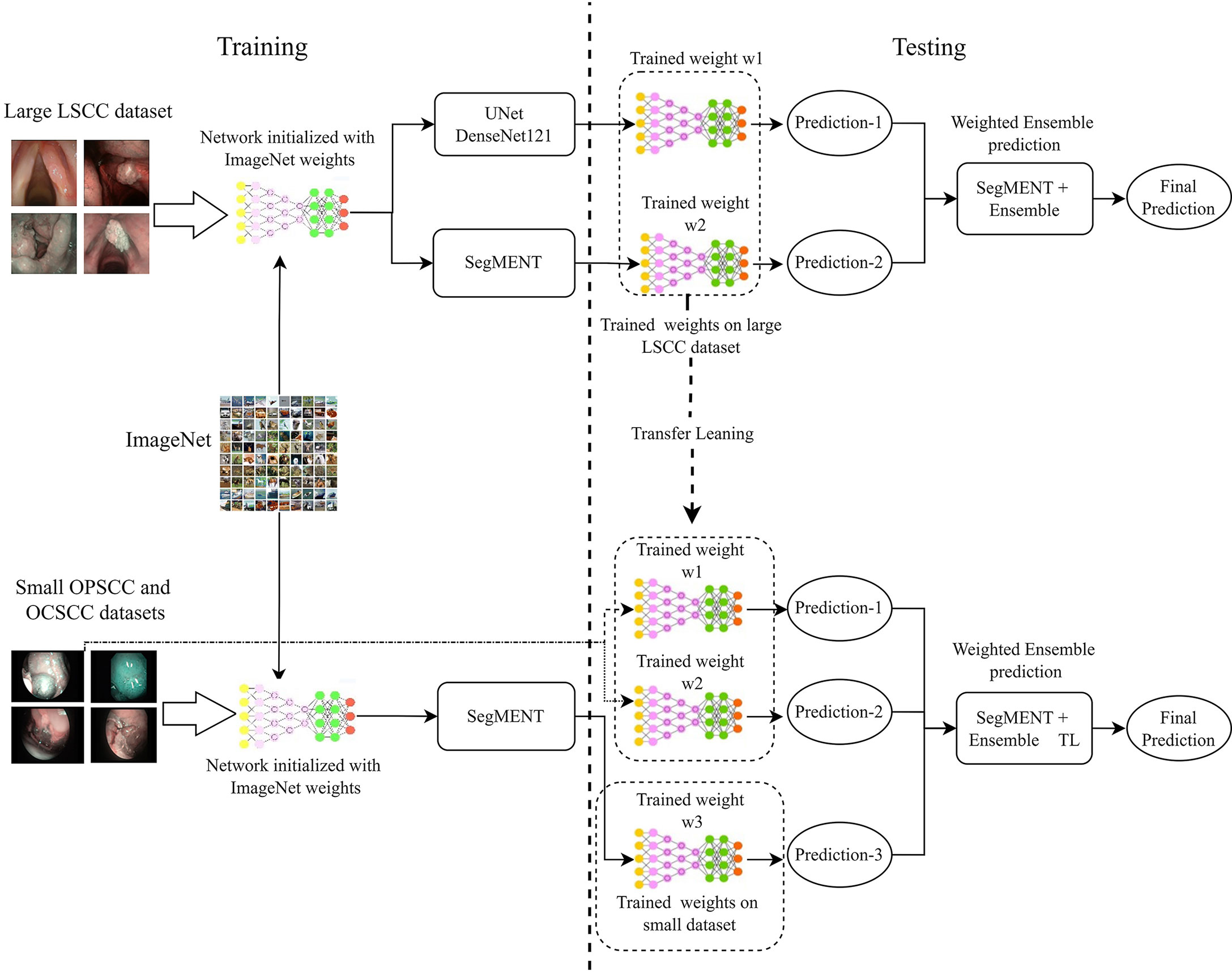
Figure 3 Diagram describing the training and testing of the models assessed in this work. Initially, the models are initialized with weights obtained from training on the ImageNet dataset. Next, the models are trained on the specific datasets of interest. During testing, the trained models provide segmentation predictions, and ensemble used to generate the final segmentations. In addition, in-domain transfer learning (TL) can be used to enhance the segmentation performance on small datasets using trained weights from other anatomical subsites. LSCC, laryngeal squamous cell carcinoma; OCSCC, oral cavity squamous cell carcinoma; OPSCC, oropharyngeal squamous cell carcinoma.
In LSCC segmentation, the predictions of the two best-performing models (SegMENT and UNet-DenseNet121) were combined to improve the accuracy of laryngeal cancer segmentation. Initially, the two models were independently trained on the LSCC dataset. Their predictions were then combined following the weighted average ensemble approach.
In OCSCC segmentation tasks, three different predictions were ensembled during testing. One was taken from a SegMENT model trained on the OCSCC dataset. The other two predictions were taken from SegMENT and UNet-DenseNet121 trained on the LSCC dataset. To produce the single final output prediction, the three output predictions were multiplied by the assigned weight values, which were obtained through a grid search technique.
The same strategy was used for OPSCC segmentation, with the difference that the first SegMENT model was trained on the OPSCC dataset. The other two predictions were taken from SegMENT and UNet-DenseNet121 models trained on the LSCC dataset, as before.
Training Parameters
For training of SegMENT, the Tverky loss function (40) was used. This loss function is used for highly unbalanced datasets. In our model, we used a learning rate of 0.001 with a batch size of 8 images per epoch during training. The learning rate decay was set to a factor of 0.1. If the training loss did not improve after four consecutive epochs of learning, the decay was slowed down. Data augmentation was used during training to increase the variability of the training dataset: flip, crop, translation, rotation, and scaling were applied, as well as hue, brightness, and contrast augmentation. A Tesla K80 GPU with 12 GB of memory and an Intel(R) Xeon(R) CPU running at 2.20 GHz with 13 GB of memory using Keras and a Tensorflow (41) back-end were used for all experiments.
Validation on LSCC Dataset
The first experiments were performed on the LSCC dataset, which was split into a training and a test sets using a respective split ratio of 80% and 20% with randomly selected images. Initially, the baseline models and SegMENT were trained and tested on the LSCC dataset starting from pre-trained weights obtained from ImageNet. Afterward, the weights of SegMENT and UNet-DenseNet121 were combined using the weighted ensemble method in the testing phase.
Validation on OCSCC and OPSCC Datasets
The OCSCC and OPSCC datasets were separately used to validate SegMENT on these different UADT sites. Each dataset was split into a training and a validation/test groups with a 70/30 percent split ratio based on a random image selection process.
SegMENT was first trained and tested separately on the OCSCC and OPSCC datasets, starting from ImageNet pre-trained weights without applying the ensemble technique. Following this, the described in-domain transfer learning (TL) method based on a weighted ensemble technique was used to assess potential performance improvements. We hypothesized that, compared to the standard TL provided by ImageNet, which is based on natural images (such as daily objects, and animals), a specific in-domain TL based on LSCC trained weights might enable better performance on images from other UADT regions. Therefore, the SegMENT ensemble model, incorporating features initially learned from the LSCC dataset, was tested on the OCSCC and OPSCC datasets during the testing phase of the segmentation framework.
Outcome Analysis
The outcomes of each DL model were evaluated by comparing the predicted segmentations with the manual annotations performed by expert physicians (i.e., the ground-truth segmentations). Standard evaluation metrics for semantic segmentation were used as previously reported (42). A classification of each pixel in the images as true positive (TP), true negative (TN), false positive (FP), or false negative (FN) was used to derive the evaluation metrics below.
● Accuracy: the percentage of pixels in the image that is correctly classified by the model.
● Precision (positive predictive value): the fraction of pixels that are true positives (correctly predicted pixels of the targeting class) among the total predicted pixels:
● Recall (sensitivity): the fraction of pixels that are true positives among the total ground truth segmented pixels:
● Dice similarity coefficient (DSC): represent the harmonic weight of Precision and Recall values (also called F1 score):
● Intersection over Union (IoU): the fraction of pixels that are true positives among the union of pixels that are positive predictions and belong to the target class (Figure 4).
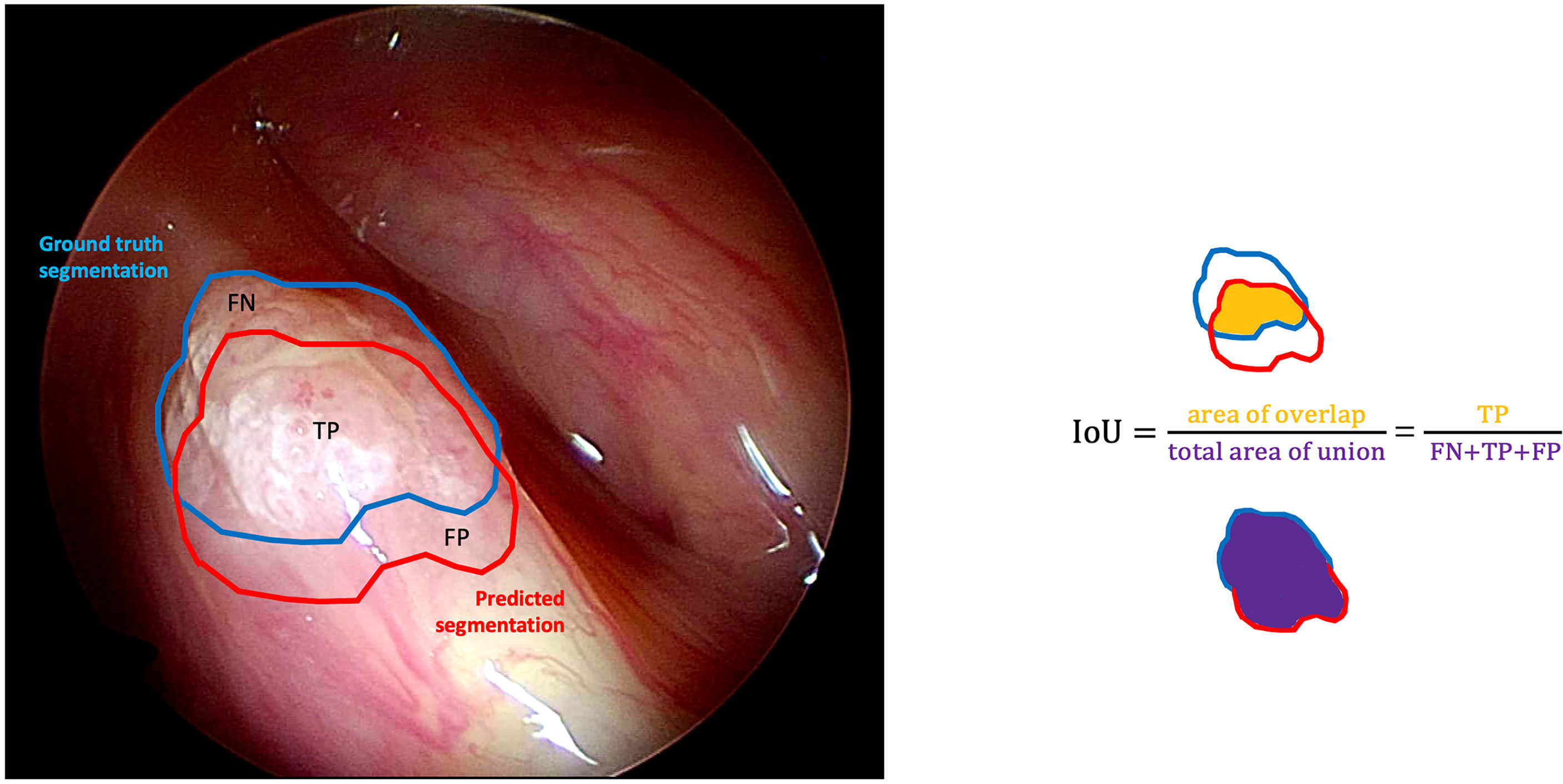
Figure 4 Graphical representation of the intersection over union (IoU) calculation on a white light right glottic cancer intraoperative videoframe. The boundary traced in light blue represents the ground truth segmentation provided by an expert clinician, while the red one represents the model prediction. The IoU is calculated by dividing the overlapping area (containing the true positive pixels) by the total area of union (encompassing the false negative, true positive, and false positive pixels).
Statistical Analysis
Differences in distributions of continuous variables among more than two independent groups were assessed with the analysis of variance (ANOVA) test. Post-hoc analysis was performed using Tukey’s multiple comparisons test to control for the inflated Type I error. A p<0.05 was considered significant. Data analysis was carried out using statistical functions (scipy.stat) and statistical models (statmodels v0.13.2) libraries in python (v3.9).
Results
Two hundred and nineteen patients with a mean age of 67.9 years (SD ± 11.8 years) were enrolled. Among these, 196 (89.4%) were males and 23 (10.6%) females. A total of 683 frames representing LSCC were extracted from videolaryngoscopies. Of those, 223 were in-office WL, 129 intraoperative WL, 223 in-office NBI, and 108 intraoperative NBI images. Figure 5 presents an overview of the final composition of the LSCC dataset.
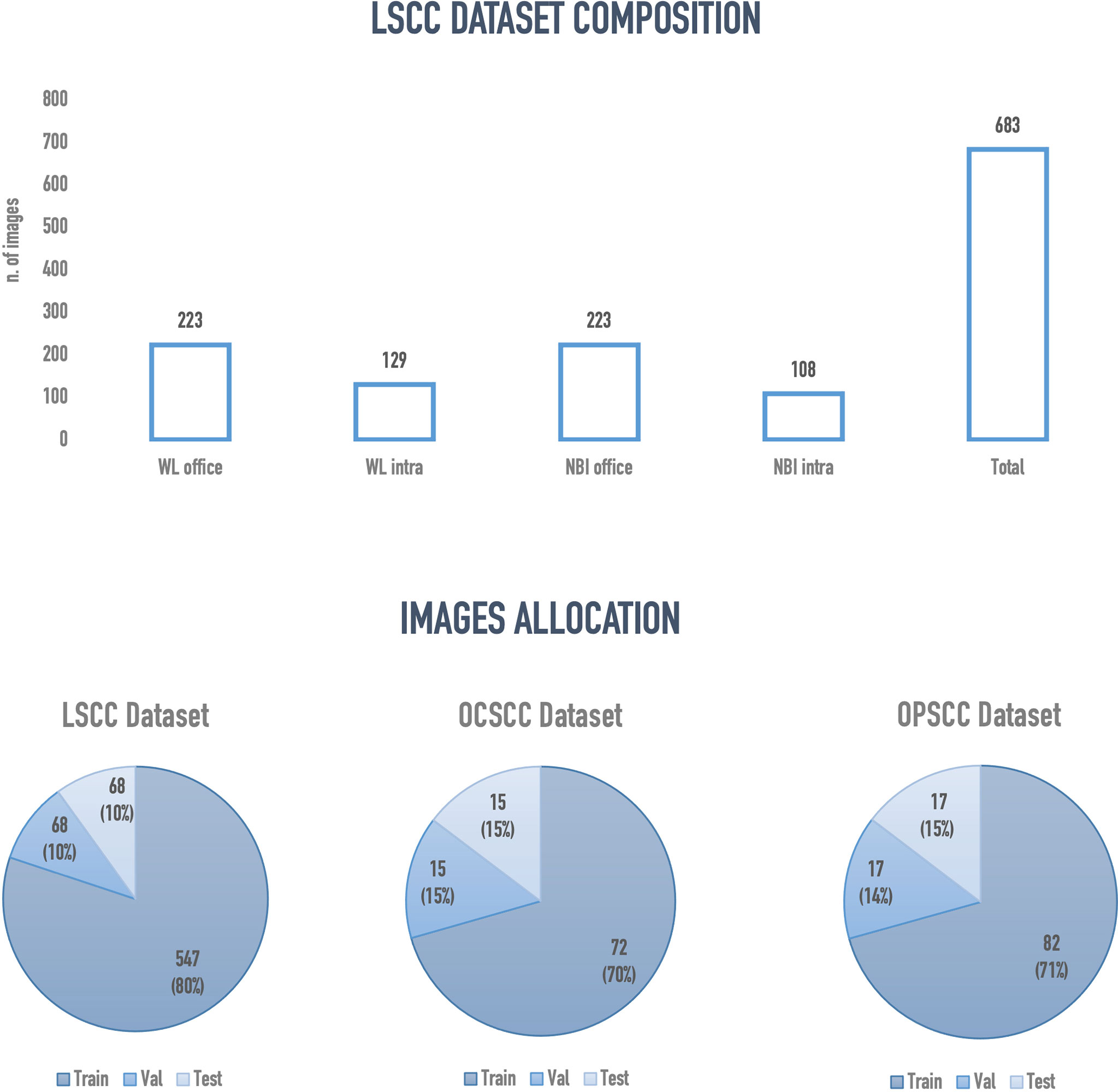
Figure 5 Overview of the final configuration of laryngeal squamous cell carcinoma (LSCC), oral cavity squamous cell carcinoma (OCSCC) and oropharyngeal squamous cell carcinoma (OPSCC) dataset. Val, validation; WL, white light; NBI, narrow band imaging; Intra, intraoperative endoscopy acquired image; Office, inoffice endoscopy acquired image.
The semantic segmentation models were trained on the LSCC dataset after random distribution of the images into a training set (547 images), a validation set (68 images), and a test set (68 images). During experiments, it was observed that the proposed SegMENT model outperformed the other state-of-the-art segmentation models. Among the baseline models, the UNet-DenseNet121 performed better than the other baseline models. Thus, the ensemble technique was used to integrate the training weights from this model with those from SegMENT, leading to better segmentation performance during testing. The performances of the models on the test set are shown in Table 2. The median values achieved by SegMENT with the ensemble technique on the LSCC dataset were: IoU=0.685, DSC=0.814, recall=0.951, precision=0.785, and accuracy=0.973. The boxplots showing the IoU and DSC score performances of SegMENT and the other state-of-the-art segmentation models during testing on the LSCC dataset are shown in Figures 6, 7. The processing rates of all base-line models ranged from 6.3 to 8.7 frames per second (fps), while the proposed ensemble model processed an average of 2.1 fps (taking 0.48 seconds to process a single frame). Examples of LSCC segmentation including ground-truth labels and the resulting automatic segmentations are shown in Figure 8.
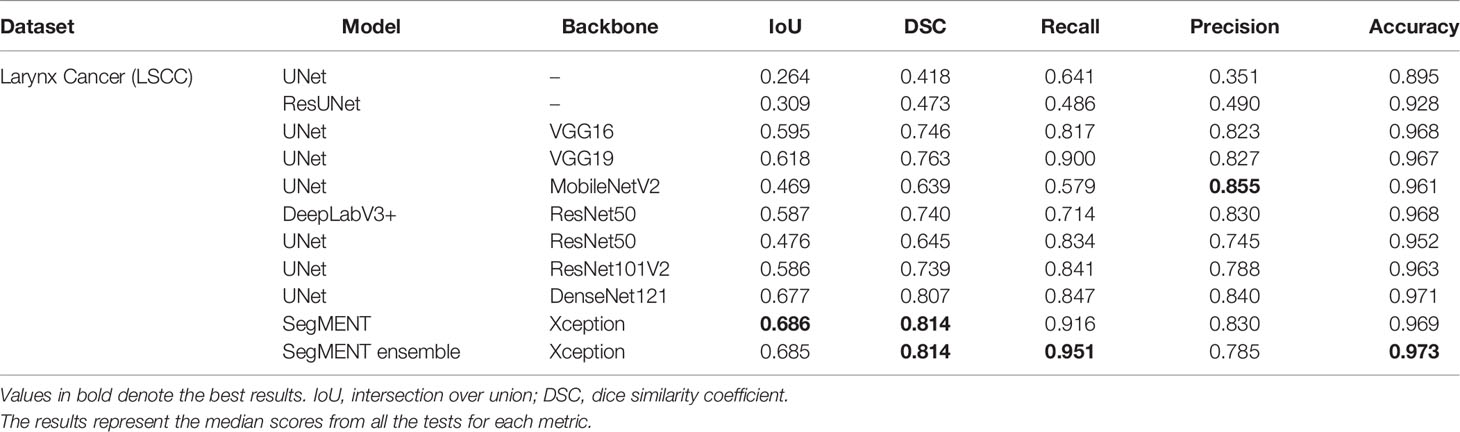
Table 2 Performance evaluation of different semantic segmentation models during testing on the laryngeal squamous cell carcinoma (LSCC) dataset.
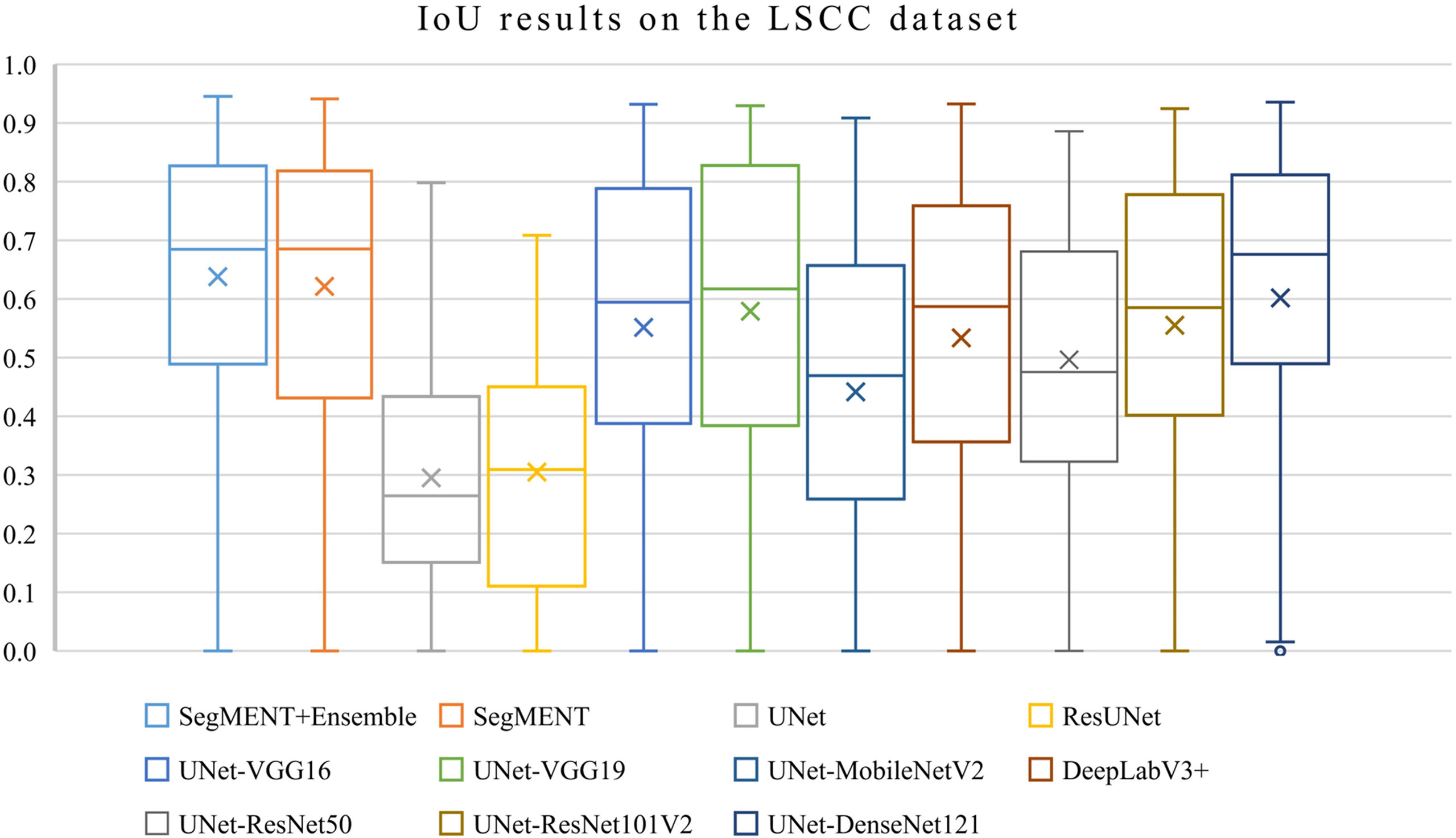
Figure 6 Boxplots of Intersection over Union (IoU) results from SegMENT ensemble and the other state-of-the-art segmentation models on the laryngeal squamous cell carcinoma (LSCC) dataset. The cross sign represents the mean value while the horizontal line inside the boxplot shows the median value.
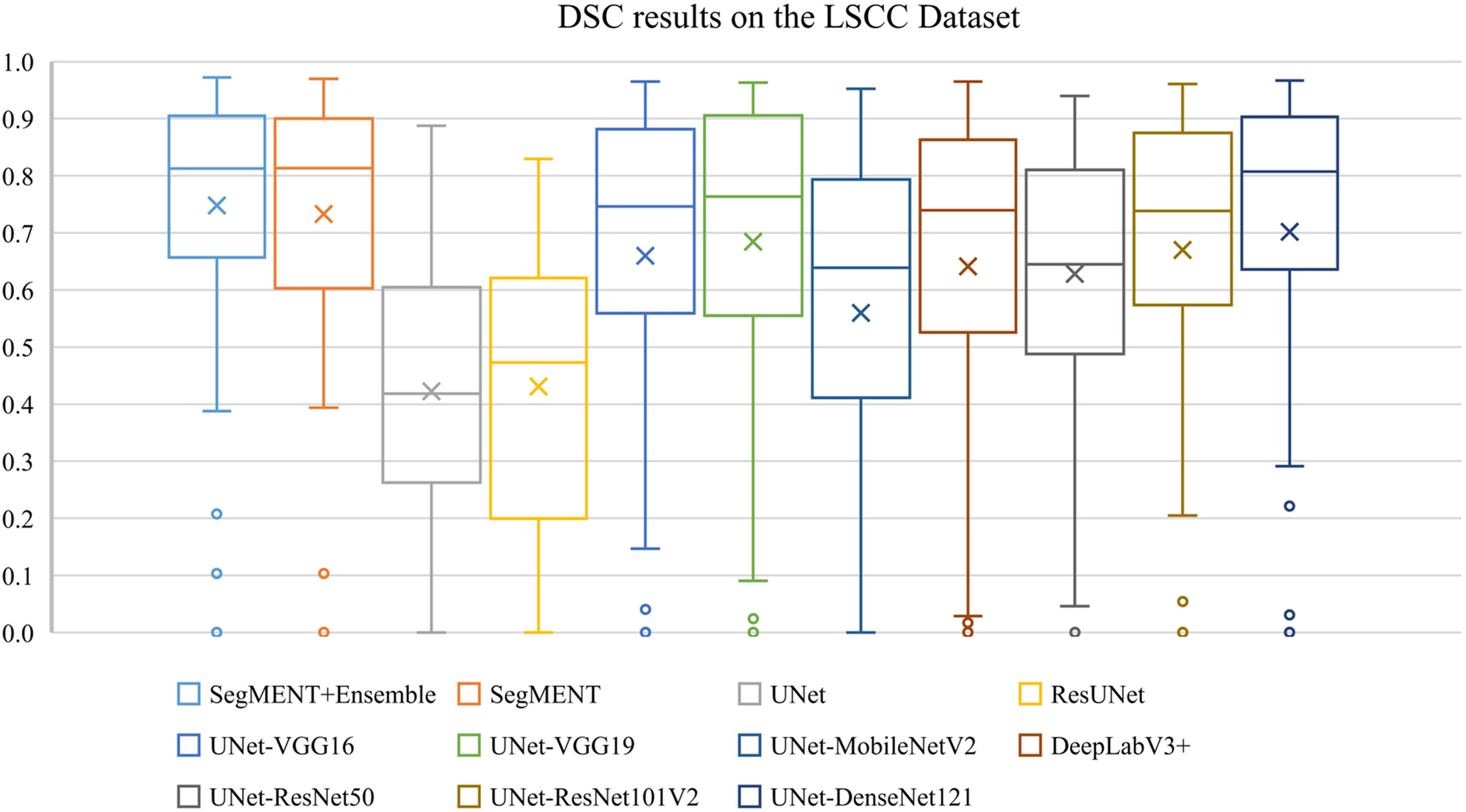
Figure 7 Boxplots of Dice similarity coefficient (DSC) results from SegMENT ensemble and the other state-of-the-art segmentation models on the laryngeal squamous cell carcinoma (LSCC) dataset. The cross sign represents the mean value, while the horizontal line inside the boxplot shows the median value.
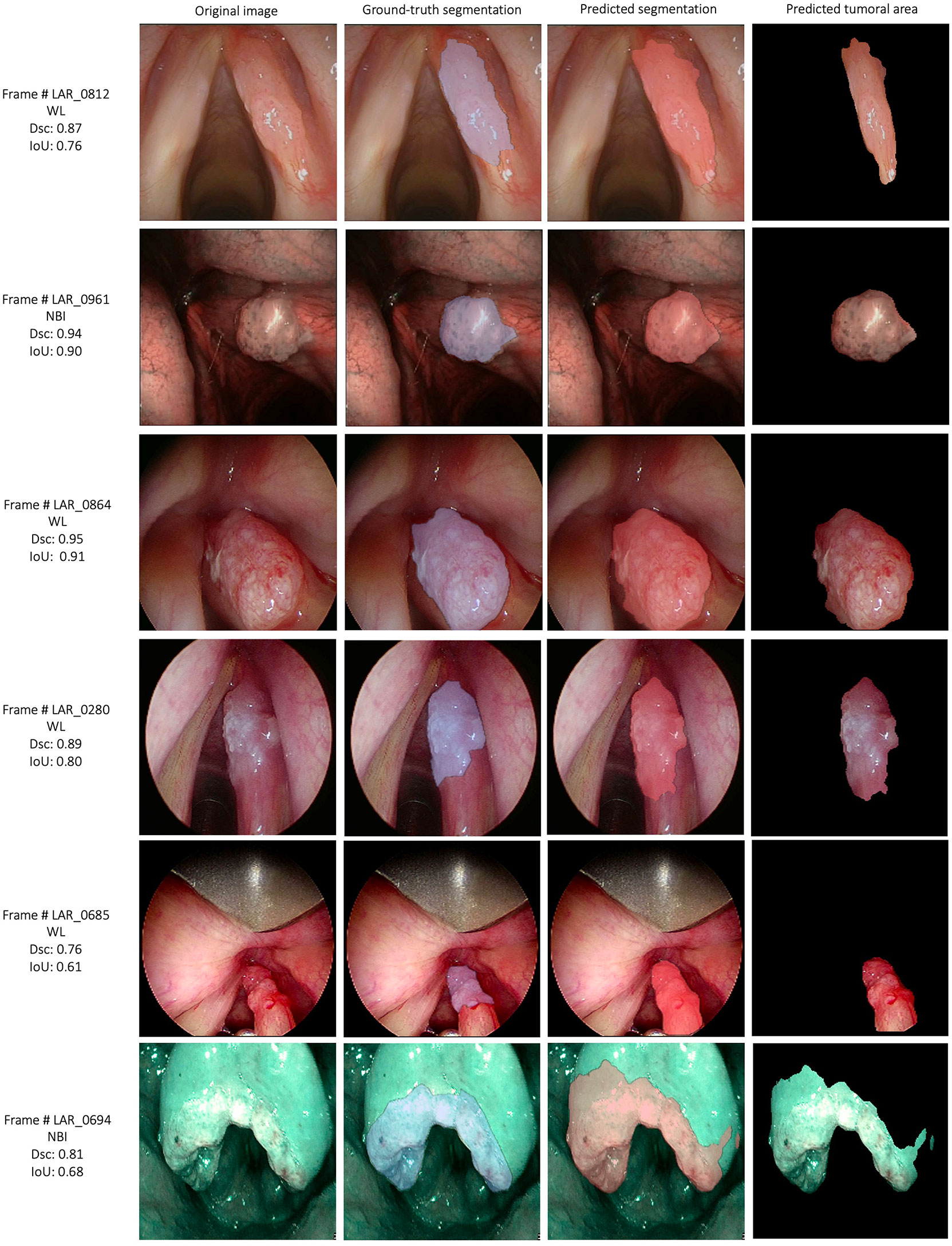
Figure 8 Examples of automatic segmentation results for the laryngeal squamous cell carcinoma dataset using SegMENT ensemble. DSC, dice similarity coefficient; IoU, intersection over union; WL, white light; NBI, Narrow Band Imaging.
When comparing results among all the models using ANOVA test, the differences were significant for each metric (p<0.001). When performing multiple comparisons on IoU, the SegMENT ensemble model achieved significantly better results than UNet, ResUNet, UNet-MobileNetV2, and UNet-ResNet50 CNNs (p=0.001, p=0.001, p=0.001, and p=0.03, respectively). Concerning DSC, the SegMENT ensemble model achieved a significantly better result compared to UNet, ResUNet, and UNet-MobileNetV2 CNNs (p=0.001, p=0.001, and p=0.001, respectively). Considering recall, the SegMENT ensemble model significantly outperformed UNet, ResUNet, UNet-VGG16, UNet-MobileNetV2, and DeepLabv3+CNNs (p=0.001, p=0.001, p=0.02, p=0.001, and p=0.001, respectively). For precision and accuracy values, the SegMENT ensemble model performed significantly better compared to UNet and ResUNet (p=0.001 and p=0.001, respectively).
The external validation cohorts comprised 102 images for OCSCC (72 for training, 15 for validation, and 15 for testing) and 116 images for OPSCC (82 for training, 17 for validation, and 17 for testing). Previously published outcomes (26) are compared to the proposed models performance in Table 3. However, while the images used were the same, it must be highlighted that it was not possible to perform the same exact image allocation in the training/testing cohorts as in the previous study. The SegMENT model pre-trained on ImageNet already performed better than the previous study CNNs on all metrics. The in-domain TL also helped to improve the results, especially for the OPSCC dataset. Indeed, the median metrics on the OCSCC and OPSCC datasets improved compared to the previously published by, respectively, 10.3% and 11.9% for DSC, 15.0% and 5.1% for recall, 17.0% and 14.7% for precision, and 4.1% and 10.3% for accuracy. The processing rate of our model was 3.9 fps on both the OCSCC and OPSCC datasets. Examples of segmentation of OCSCC and OPSCC frames displaying both the ground-truth labels and the resulting automatic segmentations are shown in Figure 9.
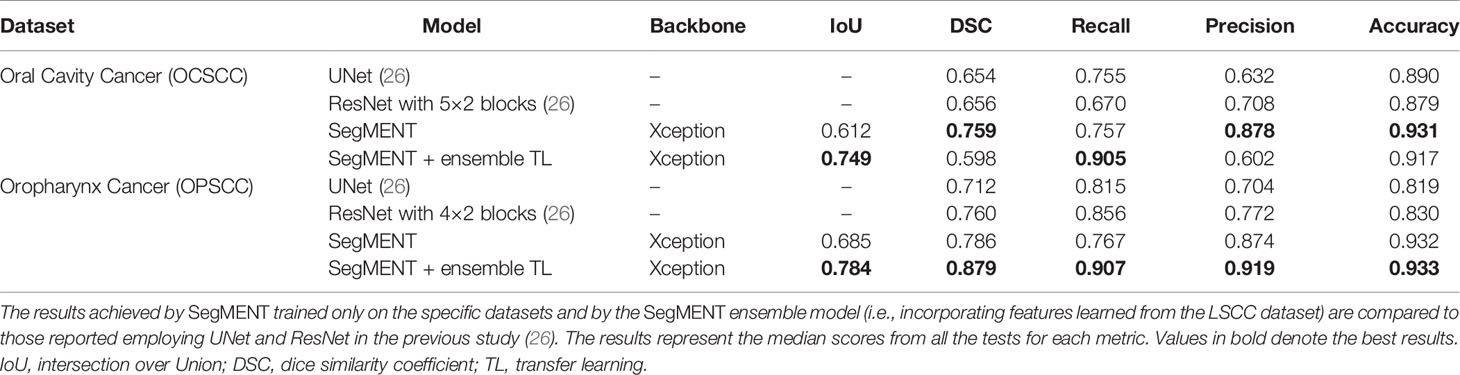
Table 3 Performance evaluation of models during testing on the oral cavity (OCSCC) and oropharynx squamous cell carcinoma (OPSCC) datasets.
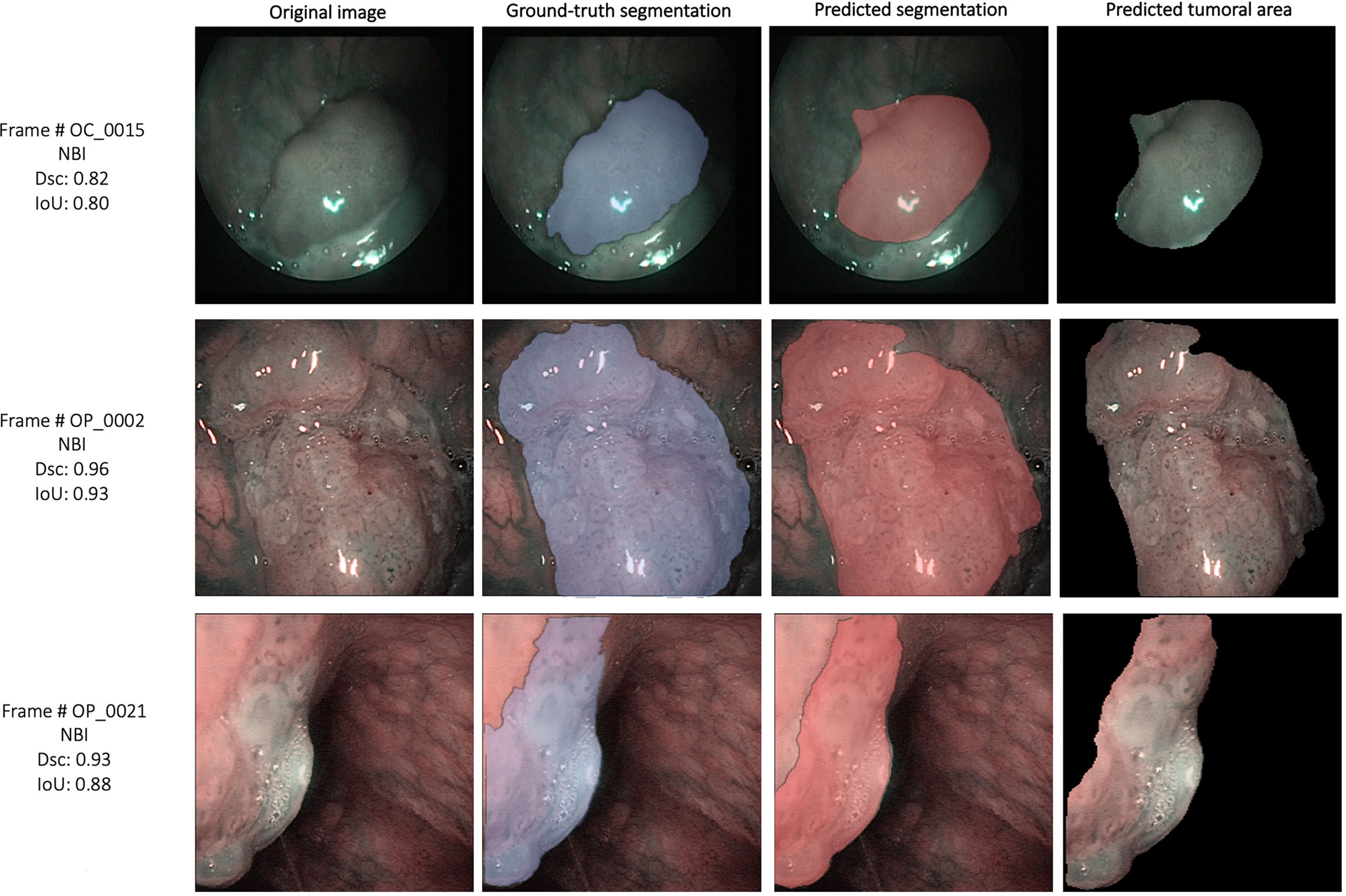
Figure 9 Examples of automatic segmentation results for the oral cavity and oropharyngeal squamous cell carcinoma datasets using SegMENT + ensemble in-domain transfer learning. DSC, Dice similarity coefficient; IoU, intersection over union; NBI, Narrow Band Imaging.
Discussion
The development of semantic segmentation AI models for medical image analysis is a field of study that is becoming increasingly widespread, especially for radiologic imaging (43–45). Conversely, the exploitation of these algorithms to investigate videoendoscopic images represents a sphere of research less explored in the literature, as demonstrated by the lack of proper terminology to indicate such a field of interest before our first proposal to identify it as “Videomics” (17). Notably, the vast majority of studies analyzing the applicability of DL-based semantic segmentation models in endoscopy come from the gastrointestinal field (46, 47). On the other hand, reports regarding the use of such algorithms in videoendoscopic examination of the UADT are scarce. This can be attributed to many factors: first, the insidious and diversified mucosal anatomy and a wide variety of lesions arising from this region are a challenge for AI-based image recognition models; second, while HD colonoscopy and esophago-gastroduodenoscopy have been routinely used in worldwide screening protocols for decades (48, 49), implementation of UADT endoscopic examination coupled with HD-videoendoscopy is more recent and less widespread, thus limiting the amount of data available for the application of AI technologies.
In this study, the authors present the promising results of a new CNN specifically designed for the automatic segmentation of UADT SCC. This new processing network was first tested on a dataset of LSCC images and subsequently validated on OCSCC and OPSCC images obtained from a previous study (26). The proposed model showed similar diagnostic outcomes for all three investigated sites, demonstrating good generalization capacity and, thus, the potential for using it in real-life clinical scenarios. Considering the tests performed on the LSCC dataset, the SegMENT ensemble model performed better than the state-of-the-art CNNs, especially considering the IoU and Dsc metrics, which are the most reliable and widely used for the evaluation of semantic segmentation models. These performances were maintained when the model was validated on the OCSCC and OPSCC cohorts, where SegMENT outperformed the results of the state-of-the-art models previously investigated on the same datasets (26). Notably, the adjunction of specific-TL features borrowed from the LSCC dataset allowed reaching even higher results, especially for the OPSCC dataset, compared to the basal SegMENT pre-trained on ImageNet. While most methods for medical image analysis employ TL from general natural images (e.g., ImageNet) (32) this strategy has been proven to be less effective compared to in-domain TL due to the mismatch in learned features between natural and medical images (50). Similarly, our results indicate that in-domain TL is a promising strategy for the processing of UADT videoendoscopies. Nonetheless, our findings should be further validated on larger datasets, as the results on the OCSCC dataset were improved less compared to OPSCC. These contradictory outcomes may be explained by the heterogeneous image composition of the OCSCC dataset. Indeed, endoscopic examination of the oral cavity often includes videoframes of the lip cutaneous surfaces, alveolar ridges, or teeth crowns. Therefore, these non-mucosal areas that differ markedly from the laryngeal and oropharyngeal endoscopic appearance may have contributed to confuse the model and decrease its performance. Moreover, the limited number of images included in the validation datasets might limit the effect of in-domain TL which we believe could lead to even better results if tested on more images.
The comparable results obtained by the proposed model for the LSCC (WL+NBI), the OCSCC (NBI only), and OPSCC (NBI only) datasets confirm the good generalization capacity of this model, which performs well regardless of the light source used to acquire images. Moreover, even without in-domain TL, the SegMENT ensemble model maintained its performance on the OCSCC and OPSCC datasets regardless of the small training datasets (13% and 15%, respectively, compared to the LSCC dataset). This finding possibly suggests that EEFs images, with their enhanced visual characteristics, may offer more information to the model during the training phase, thus helping to mitigate the shortage of images. Nevertheless, a prospective study comparing WL vs. EEFs cohorts is recommended to better investigate this finding.
To date, the present study represents the first attempt to validate a DL-based semantic segmentation model capable of achieving good results in the endoscopic assessment of SCC arising from the oral cavity, oropharynx, and larynx. The segmentation task has been seldomly applied to videoendoscopic images of the UADT, making this field of study innovative. Laves et al. previously tested different CNNs to automatically delineate different tissues and anatomical subsites on images obtained during intraoperative endoscopic evaluation of the glottis (27). The authors reported high mean values of IoU (84.7%) by segmenting every object in the image but did not focus on the annotation of cancer. Moreover, their dataset consisted of similar images obtained from only two patients, hence impairing a reasonable comparison with our results. A more specific paper on laryngeal lesion segmentation was published by Ji and colleagues (28). In this work, several CNN models were implemented to delineate glottic leucoplakias on a dataset of 649 images with segmentation metrics in line with our results (DSC=0.78 and IoU=0.66). Interestingly, the best processing performance achieved by their models was 5 fps which, together with the previous results of Paderno and colleagues (ranging within 8.7 and 16.9 fps) (26), were faster than the 3.9 fps processed by our proposed model. Notwithstanding, all these processing times are still far from real-time inferences (20-30 fps), meaning that different strategies and different CNNs must be explored in order to maintain high diagnostic performance while reaching real-time efficiency. Investigating a different UADT subsite, Li and colleagues employed a CNN to automatically segment endoscopic images of nasopharyngeal carcinoma (29). Their work included a large dataset of WL in-office endoscopic frames (30,396 images) obtained in a single tertiary-level institution. Of note, the only segmentation metric reported is mean DSC (0.75±0.26), which is comparable to the results of our model. Interestingly, the authors underlined the value of the proposed semantic segmentation algorithm as an instrument to perform a target biopsy in case of suspicious nasopharyngeal lesions. Indeed, the common presence of adenoid/lymphatic hyperplasia in this area burdened the performance of endoscopic biopsies taken in an office-based setting, thus resulting in considerable false negatives rates (29). The same issue is frequently encountered when performing endoscopy-driven biopsies of suspicious neoplasms of other UADT districts. Regarding the oropharynx, lesions arising from the base of tongue and amygdalo-glossal sulcus are frequently hidden by lymphatic tissue present in this area, and can sometimes be misinterpreted and confused with mucosal/lymphatic hyperplasia (26). Similarly, tumors in the oral cavity can nest in inflammatory lesions such as leucoplakias or lichen, which may also hinder the real target to biopsy. Regarding the larynx, the performance of incisional biopsies under WL has been largely questioned due to its low sensitivity (51). The introduction of EEFs, by allowing to better select the most suspicious area to target, represented an important step forward leading to a significant improvement in the performances of endoscopic biopsies (2, 52–54). Nevertheless, the difficulties encountered during human evaluation of such images burden the capillary application of EEFs, hence paving the way to the innovative application of AI for these tasks. Indeed, the use of trustworthy semantic segmentation DL models during endoscopy may automatically delineate the superficial area where to conduct the biopsy, even when facing heterogeneous and mystifying lesions such as SCCs of the UADT.
Additionally, automatic-segmentation models may find a field of application even intraoperatively for surgical guidance (55), or for driving tumor excision and provide an improved rate of negative surgical margins. Of note, the use of NBI during surgery for SCC of the oral cavity, oropharynx, and larynx has been already shown to be effective in these tasks (56–59), but we believe that AI-based tools, once rigorously validated, will represent a more precise and objective method for surgical margins sampling. Finally, pursuing the automatic segmentation in this field is expected to become increasingly relevant in the future not only for surgical practice but in other fields as well. In fact, semantic segmentation is paramount for establishing boundaries between objects in order to explain complex situations to computers. In the future, highly elaborated AI tools might be able to autonomously understand the relationships between different elements, even in a highly complex environment such as the UADT, and suggest meaningful clinical decisions to physicians.
The present work has limitations that must be acknowledged. First, the study conclusions are restricted by the small size of the datasets, especially the external validation cohorts, and by its retrospective design. To overcome such drawbacks, an enriched data collection will characterize our future projects in order to increase the dataset’s size. Additionally, data acquisition protocols will be implemented by gathering videos from different video sources, with the purpose of enhancing the generalization capability of the algorithm. Furthermore, the previously collected NBI images from the validation cohorts did not allow head-to-head comparison of WL vs. NBI. Moreover, as SegMENT was trained to recognize the appearance of lesions, the finding of diffuse and narrow intrapapillary capillary loops, typical of inflammatory diseases or radiotherapy, may lead to decreased lesion segmentation performance. To minimize this potential issue, future research should consider training CNN models using endoscopic images obtained from heterogeneous cohorts of patients, including those that were previously irradiated or are concomitantly affected by inflammatory diseases. Finally, the annotations performed by physicians were not cross-validated by other institutions, representing a bias that will be addressed in future studies.
Conclusions
This work represents the first multicentric validation of a DL-based semantic segmentation model applied on UADT videoendoscopic images of SCC. The model maintained reliable diagnostic performance analyzing both WL and NBI images from three distinct anatomical subsites. Ensemble strategies and in-domain transfer learning techniques demonstrated the potential to increase segmentation performance. Exploration of new CNNs should be carried out to pursue real-time clinical implementation, while further studies powered by a larger training dataset and larger external validation cohorts are needed before setting up clinical trials.
Data Availability Statement
The raw data analyzed during the current study are available from the corresponding author on reasonable request for research purposes only.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of IRCCS Ospedale Policlinico San Martino, Genova, Italy (CER Liguria). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
Conceptualization, MA, AI, CS, and LM; Data curation, MA, AI, CS, PB, GG, MV, VC, SL, LG, and AP; Formal analysis, MA, CS, and SL; Investigation, MA, AI, CS, PB, GG, MV, VC, and LM; Methodology, MA, AI, CS, LM, and SM; Supervision, SM, CP, GP, LG, and LM; Writing—original draft, CS, AI, MA, SM, and LM; Writing—review and editing, AI, CS, MA, SM, CP, AP, GP, and LM. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Piazza C, Del Bon F, Peretti G, Nicolai P. “Biologic Endoscopy”: Optimization of Upper Aerodigestive Tract Cancer Evaluation. Curr Opin Otolaryngol Head Neck Surg (2011) 19:67–76. doi: 10.1097/MOO.0B013E328344B3ED
2. Fiz I, Mazzola F, Fiz F, Marchi F, Filauro M, Paderno A, et al. Impact of Close and Positive Margins in Transoral Laser Microsurgery for Tis-T2 Glottic Cancer. Front Oncol (2017) 7:245. doi: 10.3389/fonc.2017.00245
3. Piazza C, Cocco D, De Benedetto L, Del Bon F, Nicolai P, Peretti G. Narrow Band Imaging and High Definition Television in the Assessment of Laryngeal Cancer: A Prospective Study on 279 Patients. Eur Arch Oto-Rhino-Laryngol 2009 2673 (2010) 267:409–14. doi: 10.1007/S00405-009-1121-6
4. Vilaseca I, Valls-Mateus M, Nogués A, Lehrer E, López-Chacón M, Avilés-Jurado FX, et al. Usefulness of Office Examination With Narrow Band Imaging for the Diagnosis of Head and Neck Squamous Cell Carcinoma and Follow-Up of Premalignant Lesions. Head Neck (2017) 39:1854–63. doi: 10.1002/hed.24849
5. Carta F, Sionis S, Cocco D, Gerosa C, Ferreli C, Puxeddu R. Enhanced Contact Endoscopy for the Assessment of the Neoangiogenetic Changes in Precancerous and Cancerous Lesions of the Oral Cavity and Oropharynx. Eur Arch oto-rhino-laryngol (2016) 273:1895–903. doi: 10.1007/s00405-015-3698-2
6. Ni X-G, He S, Xu Z-G, Gao L, Lu N, Yuan Z, et al. Endoscopic Diagnosis of Laryngeal Cancer and Precancerous Lesions by Narrow Band Imaging. J Laryngol Otol (2011) 125:288–96. doi: 10.1017/S0022215110002033
7. Arens C, Piazza C, Andrea M, Dikkers FG, Tjon Pian Gi REA, Voigt-Zimmermann S, et al. Proposal for a Descriptive Guideline of Vascular Changes in Lesions of the Vocal Folds by the Committee on Endoscopic Laryngeal Imaging of the European Laryngological Society. Eur Arch Oto-Rhino-Laryngol (2016) 273:1207–14. doi: 10.1007/s00405-015-3851-y
8. Piazza C, Cocco D, Del Bon F, Mangili S, Nicolai P, Majorana A, et al. Narrow Band Imaging and High Definition Television in Evaluation of Oral and Oropharyngeal Squamous Cell Cancer: A Prospective Study. Oral Oncol (2010) 46:307–10. doi: 10.1016/j.oraloncology.2010.01.020
9. Filauro M, Paderno A, Perotti P, Marchi F, Garofolo S, Peretti G, et al. Role of Narrow-Band Imaging in Detection of Head and Neck Unknown Primary Squamous Cell Carcinoma. Laryngoscope (2018) 128:2060–6. doi: 10.1002/LARY.27098
10. Madana J, Lim CM, Loh KS. Narrow Band Imaging of Nasopharynx to Identify Specific Features for Possible Detection of Early Nasopharyngeal Carcinoma. Head Neck (2015) 37:1096–101. doi: 10.1002/hed.23705
11. Vlantis AC, Woo JKS, Tong MCF, King AD, Goggins W, van Hasselt CA. Narrow Band Imaging Endoscopy of the Nasopharynx is Not More Useful Than White Light Endoscopy for Suspected Nasopharyngeal Carcinoma. Eur Arch oto-rhino-laryngol (2016) 273:3363–9. doi: 10.1007/s00405-016-3940-6
12. Vlantis AC, Wong EWY, Ng SK, Chan JYK, Tong MCF. Narrow Band Imaging Endoscopy of the Nasopharynx for Malignancy: An Inter- and Intraobserver Study. Laryngoscope (2019) 129:1374–9. doi: 10.1002/lary.27483
13. Vu A, Matias M, Farah C. Diagnostic Accuracy of Narrow Band Imaging for the Detection of Oral Potentially Malignant Disorders. Oral Dis (2015) 21:519–29. doi: 10.1111/ODI.12315
14. Deganello A, Paderno A, Morello R, Fior M, Berretti G, Del Bon F, et al. Diagnostic Accuracy of Narrow Band Imaging in Patients With Oral Lichen Planus: A Prospective Study. Laryngoscope (2021) 131:E1156–61. doi: 10.1002/LARY.29035
15. Kim DH, Kim SW, Lee J, Hwang SH. Narrow-Band Imaging for Screening of Oral Premalignant or Cancerous Lesions: A Systematic Review and Meta-Analysis. Clin Otolaryngol (2021) 46:501–7. doi: 10.1111/coa.13724
16. Wang WH, Lin YC, Chen WC, Chen MF, Chen CC, Lee KF. Detection of Mucosal Recurrent Nasopharyngeal Carcinomas After Radiotherapy With Narrow-Band Imaging Endoscopy. Int J Radiat Oncol Biol Phys (2012) 83:1213–9. doi: 10.1016/J.IJROBP.2011.09.034
17. Paderno A, Holsinger FC, Piazza C. Videomics: Bringing Deep Learning to Diagnostic Endoscopy. Curr Opin Otolaryngol Head Neck Surg (2021) 29:143–8. doi: 10.1097/MOO.0000000000000697
18. Kudo S-E, Mori Y, Abdel-Aal UM, Misawa M, Itoh H, Oda M, et al. Artificial Intelligence and Computer-Aided Diagnosis for Colonoscopy: Where do We Stand Now? Transl Gastroenterol Hepatol (2021) 6:64. doi: 10.21037/tgh.2019.12.14
19. Yao P, Usman M, Chen YH, German A, Andreadis K, Mages K, et al. Applications of Artificial Intelligence to Office Laryngoscopy: A Scoping Review. Laryngoscope (2021) 00:1–24. doi: 10.1002/lary.29886
20. Azam MA, Sampieri C, Ioppi A, Africano S, Vallin A, Mocellin D, et al. Deep Learning Applied to White Light and Narrow Band Imaging Videolaryngoscopy: Toward Real-Time Laryngeal Cancer Detection. Laryngoscope (2021) 00:1–9. doi: 10.1002/lary.29960
21. Tamashiro A, Yoshio T, Ishiyama A, Tsuchida T, Hijikata K, Yoshimizu S, et al. Artificial Intelligence-Based Detection of Pharyngeal Cancer Using Convolutional Neural Networks. Dig Endosc (2020) 32:1057–65. doi: 10.1111/den.13653
22. Inaba A, Hori K, Yoda Y, Ikematsu H, Takano H, Matsuzaki H, et al. Artificial Intelligence System for Detecting Superficial Laryngopharyngeal Cancer With High Efficiency of Deep Learning. Head Neck (2020) 42:2581–92. doi: 10.1002/hed.26313
23. Cho WK, Lee YJ, Joo HA, Jeong IS, Choi Y, Nam SY, et al. Diagnostic Accuracies of Laryngeal Diseases Using a Convolutional Neural Network-Based Image Classification System. Laryngoscope (2021) 131(11):2558–66. doi: 10.1002/lary.29595
24. Dunham ME, Kong KA, McWhorter AJ, Adkins LK. Optical Biopsy: Automated Classification of Airway Endoscopic Findings Using a Convolutional Neural Network. Laryngoscope (2022) 132 Suppl:S1–8. doi: 10.1002/lary.28708
25. Fehling MK, Grosch F, Schuster ME, Schick B, Lohscheller J. Fully Automatic Segmentation of Glottis and Vocal Folds in Endoscopic Laryngeal High-Speed Videos Using a Deep Convolutional LSTM Network. PloS One (2020) 15:1–29. doi: 10.1371/journal.pone.0227791
26. Paderno A, Piazza C, Del Bon F, Lancini D, Tanagli S, Deganello A, et al. Deep Learning for Automatic Segmentation of Oral and Oropharyngeal Cancer Using Narrow Band Imaging: Preliminary Experience in a Clinical Perspective. Front Oncol (2021) 11:626602. doi: 10.3389/fonc.2021.626602
27. Laves MH, Bicker J, Kahrs LA, Ortmaier T. A Dataset of Laryngeal Endoscopic Images With Comparative Study on Convolution Neural Network-Based Semantic Segmentation. Int J Comput Assist Radiol Surg (2019) 14:483–92. doi: 10.1007/s11548-018-01910-0
28. Ji B, Ren J, Zheng X, Tan C, Ji R, Zhao Y, et al. A Multi-Scale Recurrent Fully Convolution Neural Network for Laryngeal Leukoplakia Segmentation. BioMed Signal Process Control (2020) 59:1–9. doi: 10.1016/j.bspc.2020.101913
29. Li C, Jing B, Ke L, Li B, Xia W, He C, et al. Development and Validation of an Endoscopic Images-Based Deep Learning Model for Detection With Nasopharyngeal Malignancies 08 Information and Computing Sciences 0801 Artificial Intelligence and Image Processing. Cancer Commun (2018) 38:1–11. doi: 10.1186/s40880-018-0325-9
30. Firdaus-Nawi M, Noraini O, Sabri MY, Siti-Zahrah A, Zamri-Saad M, Latifah H. DeepLabv3+_Encoder-Decoder With Atrous Separable Convolution for Semantic Image Segmentation. Pertanika J Trop Agric Sci (2011) 34:137–43. doi: 10.48550/arXiv.1802.02611
31. Chollet F. Xception: Deep Learning With Depthwise Separable Convolutions, in: Proc - 30th IEEE Conf Comput Vis Pattern Recognition, CVPR 2017. IEEE: Institute of Electrical and Electonics Engineers (2017). pp. 1800–7. doi: 10.1109/CVPR.2017.195
32. Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. ImageNet: A Large-Scale Hierarchical Image Database, in: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE: Institute of Electrical and Electonics Engineers (2017). pp. 248–55. doi: 10.1109/CVPR.2009.5206848
33. Bianco S, Cadene R, Celona L, Napoletano P. Benchmark Analysis of Representative Deep Neural Network Architectures. IEEE Access (2018) 6:64270–7. doi: 10.1109/ACCESS.2018.2877890
34. Misra D. Mish: A Self Regularized Non-Monotonic Activation Function (2019). Available at: https://arxiv.org/abs/1908.08681v3 (Accessed February 2, 2022).
35. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect Notes Comput Sci (2015) 9351:234–41. doi: 10.1007/978-3-319-24574-4_28
36. Zhang Z, Liu Q, Wang Y. Road Extraction by Deep Residual U-Net. IEEE Geosci Remote Sens Lett (2018) 15:749–53. doi: 10.1109/LGRS.2018.2802944
37. Pham VT, Tran CM, Zheng S, Vu TM, Nath S. Chest X-Ray Abnormalities Localization via Ensemble of Deep Convolutional Neural Networks. Int Conf Adv Technol Commun (2021), 125–30. doi: 10.1109/ATC52653.2021.9598342
38. Mahendran N, Vincent DR, Srinivasan K, Chang CY, Garg A, Gao L, et al. Sensor-Assisted Weighted Average Ensemble Model for Detecting Major Depressive Disorder. Sensors (Switzerland) (2019) 19:1–16. doi: 10.3390/s19224822
39. Nourani V, Elkiran G, Abba SI. Wastewater Treatment Plant Performance Analysis Using Artificial Intelligence - An Ensemble Approach. Water Sci Technol (2018) 78:2064–76. doi: 10.2166/wst.2018.477
40. Abraham N, Khan NM. A Novel Focal Tversky Loss Function With Improved Attention U-Net for Lesion Segmentation. Proc - Int Symp BioMed Imaging (2019), 683–7. doi: 10.1109/ISBI.2019.8759329
41. Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. TensorFlow: A System for Large-Scale Machine Learning. 12th Symp Oper Syst Des Implement (2016), 265–83. doi: 10.1016/0076-6879(83)01039-3
42. Jha D, Ali S, Tomar NK, Johansen HD, Johansen D, Rittscher J, et al. Real-Time Polyp Detection, Localization and Segmentation in Colonoscopy Using Deep Learning. IEEE Access (2021) 9:40496–510. doi: 10.1109/ACCESS.2021.3063716
43. Qiu B, van der Wel H, Kraeima J, Glas HH, Guo J, Borra RJH, et al. Automatic Segmentation of Mandible From Conventional Methods to Deep Learning-A Review. J Pers Med (2021) 11:1–26. doi: 10.3390/jpm11070629
44. Wallner J, Schwaiger M, Hochegger K, Gsaxner C, Zemann W, Egger J. A Review on Multiplatform Evaluations of Semi-Automatic Open-Source Based Image Segmentation for Cranio-Maxillofacial Surgery. Comput Methods Prog BioMed (2019) 182:105102. doi: 10.1016/j.cmpb.2019.105102
45. Zhong Y, Yang Y, Fang Y, Wang J, Hu W. A Preliminary Experience of Implementing Deep-Learning Based Auto-Segmentation in Head and Neck Cancer: A Study on Real-World Clinical Cases. Front Oncol (2021) 11:638197. doi: 10.3389/fonc.2021.638197
46. Wu Z, Ge R, Wen M, Liu G, Chen Y, Zhang P, et al. ELNet:Automatic Classification and Segmentation for Esophageal Lesions Using Convolutional Neural Network. Med Image Anal (2021) 67:1–21. doi: 10.1016/j.media.2020.101838
47. Mendel R, Ebigbo A, Probst A, Messmann H, Palm C. Barrett’s Esophagus Analysis Using Convolutional Neural Networks. In: Maier-Hein geb. Fritzsche KH, Deserno TM, Lehmann Handels H, Tolxdorff T, editors. Bildverarbeitung Für Die Medizin 2017. Berlin, Heidelberg: Springer Berlin Heidelberg (2017). p. 80–5.
48. Winawer SJ, Zauber AG, Fletcher RH, Stillman JS, O’brien MJ, Levin B, et al. Guidelines for Colonoscopy Surveillance After Polypectomy: A Consensus Update by the US Multi-Society Task Force on Colorectal Cancer and the American Cancer Society. CA Cancer J Clin (2006) 56:143–5. doi: 10.3322/canjclin.56.3.143
49. Saito Y, Oka S, Kawamura T, Shimoda R, Sekiguchi M, Tamai N, et al. Colonoscopy Screening and Surveillance Guidelines. Dig Endosc (2021) 33:486–519. doi: 10.1111/den.13972
50. Alzubaidi L, Fadhel MA, Al-Shamma O, Zhang J, Santamaría J, Duan Y, et al. Towards a Better Understanding of Transfer Learning for Medical Imaging: A Case Study. Appl Sci (2020) 10:1–21. doi: 10.3390/app10134523
51. Cohen JT, Benyamini L. Transnasal Flexible Fiberoptic in-Office Laryngeal Biopsies-Our Experience With 117 Patients With Suspicious Lesions. Rambam Maimonides Med J (2014) 5:e0011. doi: 10.5041/RMMJ.10145
52. Galli J, Settimi S, Mele DA, Salvati A, Schiavi E, Parrilla C, et al. Role of Narrow Band Imaging Technology in the Diagnosis and Follow Up of Laryngeal Lesions: Assessment of Diagnostic Accuracy and Reliability in a Large Patient Cohort. J Clin Med (2021) 10:1–10. doi: 10.3390/jcm10061224
53. Nair D, Qayyumi B, Sharin F, Mair M, Bal M, Pimple S, et al. Narrow Band Imaging Observed Oral Mucosa Microvasculature as a Tool to Detect Early Oral Cancer: An Indian Experience. Eur Arch Oto-Rhino-Laryngol (2021) 278:3965–71. doi: 10.1007/s00405-020-06578-4
54. Carobbio ALC, Vallin A, Ioppi A, Missale F, Ascoli A, Mocellin D, et al. Application of Bioendoscopy Filters in Endoscopic Assessment of Sinonasal Schneiderian Papillomas. Int Forum Allergy Rhinol (2021) 11(6):1025–8. doi: 10.1002/alr.22760
55. Gong J, Holsinger FC, Noel JE, Mitani S, Jopling J, Bedi N, et al. Using Deep Learning to Identify the Recurrent Laryngeal Nerve During Thyroidectomy. Sci Rep (2021) 11:1–11. doi: 10.1038/s41598-021-93202-y
56. Garofolo S, Piazza C, Del Bon F, Mangili S, Guastini L, Mora F, et al. Intraoperative Narrow Band Imaging Better Delineates Superficial Resection Margins During Transoral Laser Microsurgery for Early Glottic Cancer. Ann Otol Rhinol Laryngol (2015) 124:294–8. doi: 10.1177/0003489414556082
57. Bertino G, Cacciola S, Fernandes WB, Fernandes CM, Occhini A, Tinelli C, et al. Effectiveness of Narrow Band Imaging in the Detection of Premalignant and Malignant Lesions of the Larynx: Validation of a New Endoscopic Clinical Classification. Head Neck (2015) 37:215–22. doi: 10.1002/HED.23582
58. Tirelli G, Piovesana M, Gatto A, Tofanelli M, Biasotto M, Boscolo Nata F. Narrow Band Imaging in the Intra-Operative Definition of Resection Margins in Oral Cavity and Oropharyngeal Cancer. Oral Oncol (2015) 51:908–13. doi: 10.1016/j.oraloncology.2015.07.005
Keywords: larynx cancer, oral cancer, oropharynx cancer, machine learning, endoscopy, laryngoscopy, computer vision, otorhinolaryngology
Citation: Azam MA, Sampieri C, Ioppi A, Benzi P, Giordano GG, De Vecchi M, Campagnari V, Li S, Guastini L, Paderno A, Moccia S, Piazza C, Mattos LS and Peretti G (2022) Videomics of the Upper Aero-Digestive Tract Cancer: Deep Learning Applied to White Light and Narrow Band Imaging for Automatic Segmentation of Endoscopic Images. Front. Oncol. 12:900451. doi: 10.3389/fonc.2022.900451
Received: 20 March 2022; Accepted: 26 April 2022;
Published: 01 June 2022.
Edited by:
Lorenzo Cobianchi, University of Pavia, ItalyReviewed by:
Jacopo Martellucci, Careggi University Hospital, ItalyJuan Verde, IHU Strasbourg, France
Copyright © 2022 Azam, Sampieri, Ioppi, Benzi, Giordano, De Vecchi, Campagnari, Li, Guastini, Paderno, Moccia, Piazza, Mattos and Peretti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudio Sampieri, Y2xhdWRpby5zYW1waWVyaUBvdXRsb29rLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Muhammad Adeel Azam1†
Muhammad Adeel Azam1† Claudio Sampieri
Claudio Sampieri Alessandro Ioppi
Alessandro Ioppi Pietro Benzi
Pietro Benzi Marta De Vecchi
Marta De Vecchi Valentina Campagnari
Valentina Campagnari Shunlei Li
Shunlei Li Alberto Paderno
Alberto Paderno Sara Moccia
Sara Moccia Cesare Piazza
Cesare Piazza Leonardo S. Mattos
Leonardo S. Mattos