- 1School of Clinical Medicine, Shandong First Medical University & Shandong Academy of Medical Sciences, Jinan, China
- 2Central Laboratory, Linyi People’s Hospital, Linyi, China
- 3Department of Education and Teaching, Linyi People’s Hospital, Linyi, China
- 4Linyi Key Laboratory of Tumor Biology, Linyi, China
Circular RNAs (circRNAs) are non–protein-coding RNAs that have a circular structure and do not possess a 5` cap or 3` poly-A tail. Their structure is more stable than that of linear RNAs, and they are difficult to deform via hydrolysis. Advancements in measurement technology such as RNA sequencing have enabled the detection of circRNAs in various eukaryotes in both in vitro and in vivo studies. The main function of circRNAs involves sponging of microRNAs (MiRNAs) and interaction with proteins associated with physiological and pathological processes, while some circRNAs are involved in translation. circRNAs act as tumor suppressors or oncogenes during the development of many tumors and are emerging as new diagnostic and prognostic biomarkers. They also affect resistance to certain chemotherapy drugs such as imatinib. The objective of this review is to investigate the expression and clinical significance of circRNAs in hematological malignancies. We will also explore the effect of circRNAs on proliferation and apoptosis in hematological malignancy cells and their possible use as biomarkers or targets to determine prognoses. The current literature indicates that circRNAs may provide new therapeutic strategies for patients with hematologic malignancies.
Introduction
Circular RNAs (circRNAs) are noncoding RNAs formed by reverse splicing, with a downstream splice donor covalently linked to an upstream splice acceptor (1). circRNAs are classified into four types: exonic circRNAs (ecircRNAs), containing only postsplicing exons; circular intron RNAs (ciRNAs), formed from introns; exon-intron circRNAs (EIciRNAs), which are exon–intron circRNAs; and tRNA intronic circular RNAs (tricRNAs) (2, 3). ecircRNAs are abundant among mammalian transcripts, display strong conservation, and are highly expressive (4). ciRNAs are frequently detected in the nucleus and positively regulate parental gene expression (5). EIciRNAs are mainly localized in the nucleus and regulate the expression of parental genes in cis through positive feedback (6). The tricRNAs are stable and abundant, with typical characteristics of postnatal transcripts (7). Different isoforms of circRNAs can be produced from a single gene, such as CircSTAU2a and CircSTAU2b (8)
Currently, several mechanisms of circRNA biogenesis are known. Intron pairing-promoted cyclization (9), spliceosome-dependent lasso-driven cyclization (10), and facilitation of the 3’-end to 5’-end ligation of circRNAs by RNA binding proteins (RBPs) (11) are the most common biogenesis mechanisms of ecircRNAs and EIciRNAs. Furthermore, ciRNA formation is dependent on the intron, a GU-rich sequence in the lasso at the 5’ end, and an AC-rich sequence at the branch point (5). tricRNAs result from the intron ends generated by the pre-tRNA cleavage (12).
In 1976, circRNA was first recognized in plant viroids (13), and in 1979, circRNAs were first detected in human HeLa cells using electron microscopy (14). Subsequently, circRNAs were found in hepatitis D virus, which was the first instance that circRNAs were detected in an animal virus (15). Earlier, studies were not conducted on circRNAs because they were considered as minor products of RNA missplicing (16, 17). With the development of analytical techniques such as RNA sequencing and informatics tools, circRNAs have been detected in various eukaryotes (e.g., mice), as well as in human plasma and saliva (8, 10, 18–20).
Since the creation of the first genome-wide circRNA atlas in 2012 (21), various circRNA identification tools such as Find-CIRC, MapSplice, and CircRNAFisher have been constructed, and several circRNA databases including CircBase, Circpedia, and CircRNADb have been established (18). In addition, Dahl et al. obtained differential expression profiles that can distinguish between different B-cell malignancies using nano-string technology (22); and Issah et al. established the expression profile of N6-methyladenosine (m6A) circRNAs in AML patients by microarray analysis (23). Using these tools, the preliminary identification, characterization, and annotation of circRNAs can be achieved (18). These technologies have facilitated the advancement of circRNA research (18), and more than 10,000 human CircRNAs have been identified till date (8).
CircRNAs have a stable circular structure that lacks a 5` cap and 3` poly-A tail; therefore, it is difficult to deform circRNAs via hydrolysis (24). Meanwhile, the sequence of circRNAs is more strongly conserved and extensive than the corresponding linear RNAs. It has also been suggested in many studies that circRNAs display high specificity across various tissues (17). Furthermore, circRNAs can promote the transcription of parental genes by acting in cis- or trans- (25).
The main functions of circRNAs are as follows. Firstly, circRNAs are considered microRNA (miRNA) sponges (26). Mir-7 is one of the most widely known miRNAs as a target of circRNAs, both suppressing and inducing tumor progression (17). circRNA sponge for miR-7 (CIRS-7, also called CDR1as) has 74 conserved binding sites against mir-7 (4); the binding sites are predominantly located in the brain of placental mammals (27, 28), wherein they inhibit mir-7 and upregulate the expression of genes such as epidermal growth factor receptor (17). Secondly, circRNAs interact with proteins to influence protein expression, biogenesis, and pathophysiological processes (29). RBPs are a class of proteins that interact with RNA to regulate RNA maturation, translation, and other functions (29, 30). CircRNAs may bind to regulated RBPs and affect the expression of their target miRNAs (29). Next, even though the 5’ cap and 3’ poly-A tail required for mRNA translation are absent in circRNAs, they are translatable at sites such as the internal ribosome entry site (IRES) and m6A modification in their sequences, and many circRNAs have now been shown to produce proteins by translation (17, 31). The modification of circRNA by m6A contributes to the translation of circRNA (32). Recently, circRNAs have been found to play important roles in the proliferation and apoptosis of various tumors (33–36). The chromosome 9 and 22 translocations in chronic myelogenous leukemia (CML) produce the PML-RARα gene; similar chromosomal translocations in tumors are reported to produce fusion-circular RNAs (f-circRNAs) and facilitate the progression of diseases such as leukemia (37, 38). Moreover, circRNAs show potential as new therapeutic targets for and biomarkers of diseases (39). circPVT1 is considered to be a prognostic marker for gastric cancer (40). In recent years, increasing evidence has indicated that circRNAs play important roles in hematological tumor development and progression. These tumors have high circRNA content, are not easily degraded and are widely present in various body fluids; therefore, exploring circRNAs associated with hematologic malignancies as biomarkers or targets will create new possibilities for diagnosis, prognosis, and effective treatment. In this review, we summarize the expression of different types of circRNAs in hematological malignancies, describe their role in tumor cell growth and apoptosis. (Table 1).
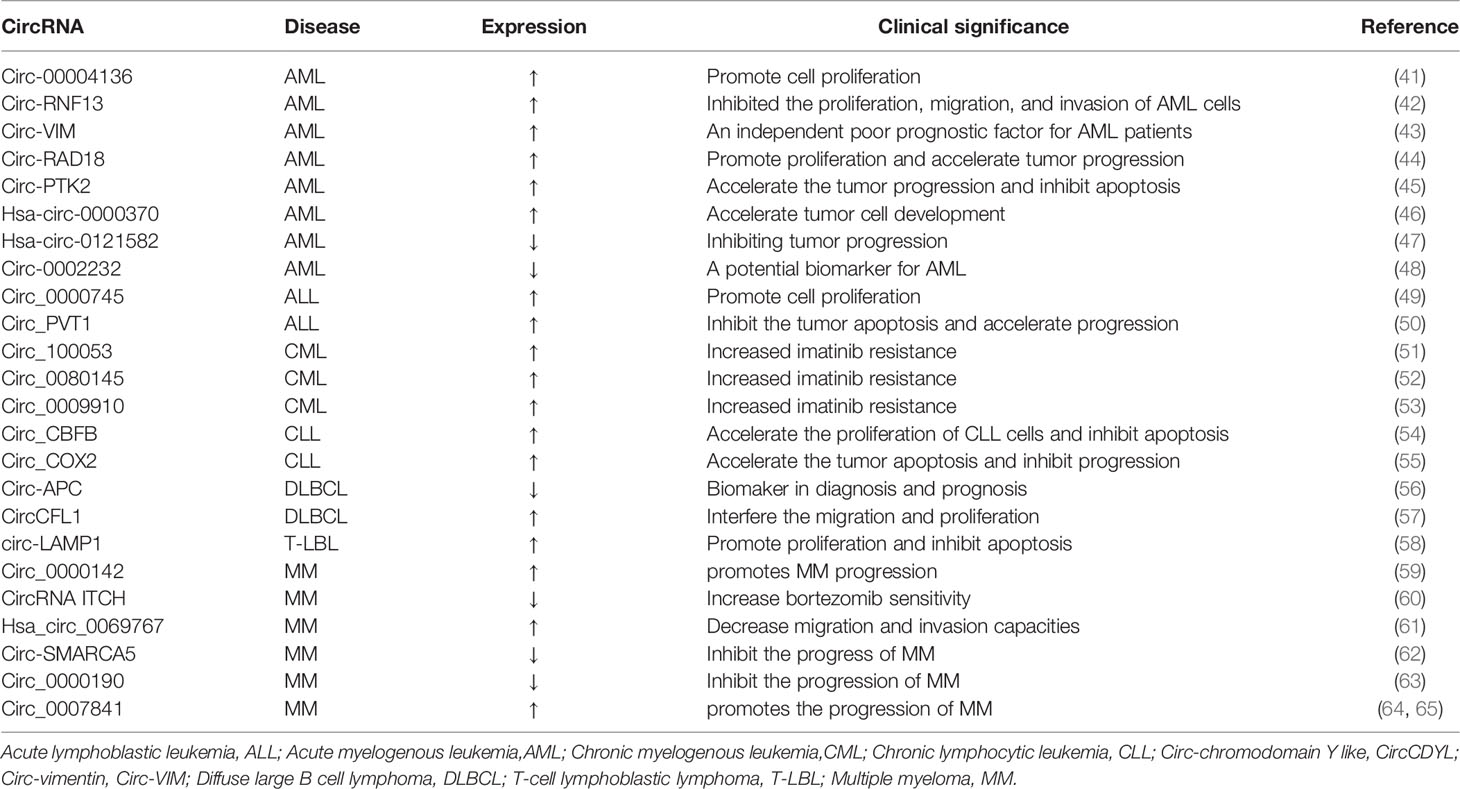
Table 1 Some abnormal circRNAs with confirmed functions in the universal category of hematological malignancies (↑: up-regulated;↓:down-regulated).
CircRNAs and Leukemia
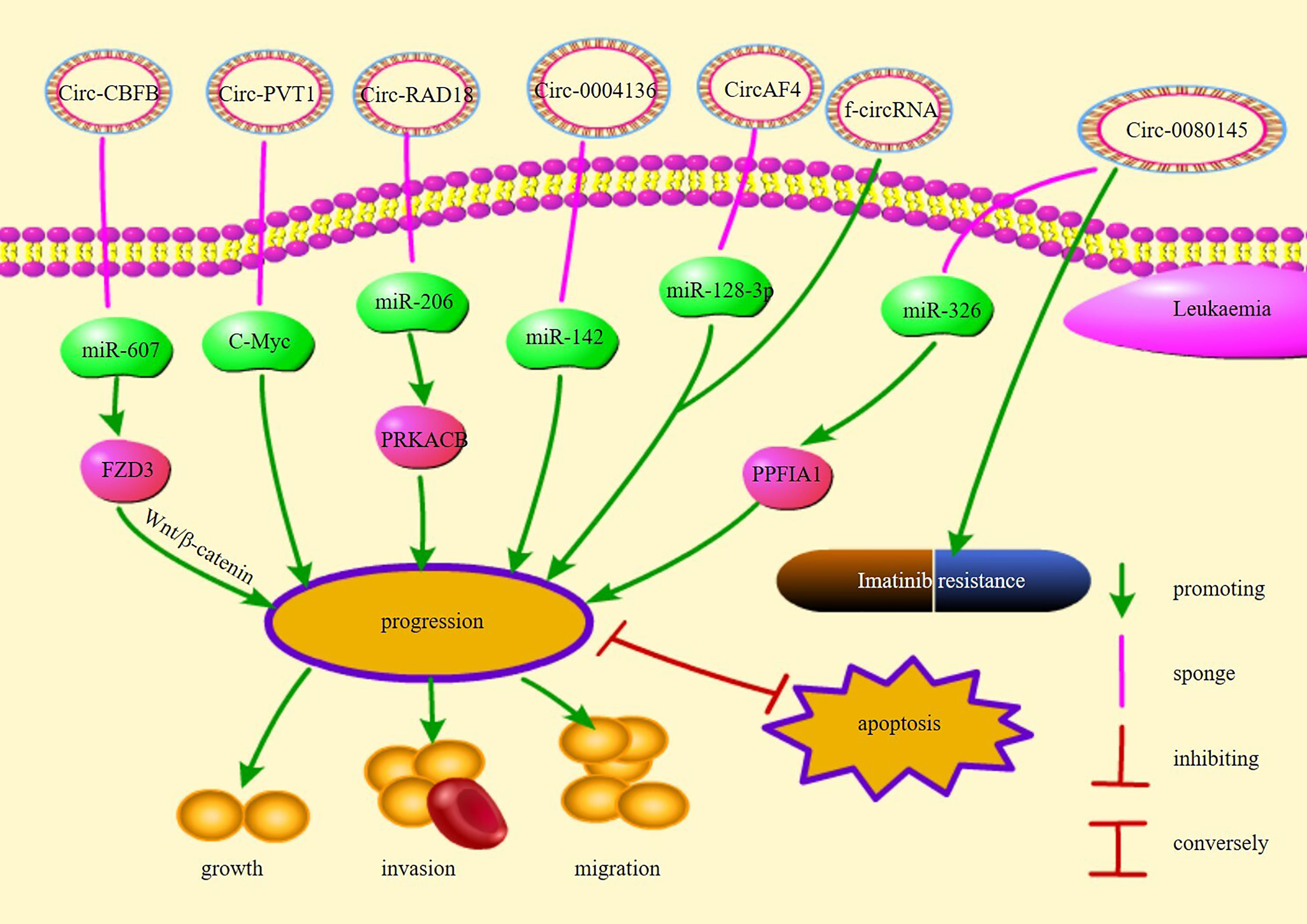
AML is a disease caused by malignant proliferation of myeloid cells in the bone marrow (45).Acute myelogenous leukemia (AML) is the most prevalent acute leukemia in adults (66). Although the diagnosis and treatment strategies of AML have been rapidly developed, the prognosis remains poor (44). With the advancement of analytical techniques, many circRNAs have been identified to be abnormally expressed in AML. In a study screening 273 circRNAs upregulated in AML, circ-00004136 was found to be significantly upregulated in pediatric AML, promoting leukemic cell proliferation via miR-142 sponging (41). circ-RNF13 was significantly upregulated in adult patients with AML and it inhibited AML cell proliferation, migration, and invasion by regulating miRNA-1224-5p expression (42). miRNA-1224-5p overexpression promotes early apoptosis of AML cells and arrests the cell cycle in the G1 phase (42). circ-vimentin expression has been found to be significantly upregulated and positively correlated with leukocyte counts and French–American–British (FAB) classifications. Furthermore, circ-vimentin overexpression is considered an independent and poor prognostic factor for significantly shorter overall survival (OS) and leukemia-free survival (LFS) in patients with AML (43). circ-RAD18 was found to be highly expressed in patients with AML, promotes AML cell proliferation, and accelerates tumor progression via miR-206/protein kinase CAMP-activated catalytic subunit beta (PRKACB) (44). PRKACB participates in various biological processes, including cell replication, gene transcription, and metabolism (44, 67). circRAD18 directly inhibits miR-206, whereas miR-206 inhibits PRKACB (44). circ-PTK2 affects the miR-330-5p/forkhead box M1(FOXM1) axis by binding to miR-330-5p, accelerating tumor progression, and inhibiting tumor apoptosis (45). circ-0009910, circ-0058058, and circ-DLEU2 are also overexpressed in AML, and their expression is negatively correlated with patient prognosis (67–69). AML patients with FMS-like tyrosine kinase 3 receptor (FLT3) mutations tend to have shorter survival rates and are more likely to experience relapse than patients without these mutations (70). hsa-circ-0000370 expression was found to significantly increase in patients who had AML with FLT3 internal tandem duplication and promoted tumor development by inhibiting miR-1299 expression (46). hsa-circ-100290 and hsa-circ-0079480 expression was also found to be elevated in AML, leading to the promotion of AML via the inhibition of miR-203 and miR-654-3p expression, respectively; hsa-circ-100290 and hsa-circ-0079480 knockdown induced apoptosis in AML cells (71, 72).
circRNA expression is downregulated in patients with AML. Lower circ-0002232 expression is correlated with increased patient survival and decreased phosphatase and tensin homolog deleted on chromosome 10(PTEN) expression (48). circ-0002232 might influence PTEN expression and AML progression via miR-92a-3p sponging (48). The expression of hsa-circ-0121582, a robust circRNA reverse-spliced from exon 1 to exon 7 in GSK3β, decreases in AML and inhibits tumor progression by binding to miR-224 and TET1 via the Wnt/β-catenin signaling pathway (47). Regardless of their expression in AML, circRNAs are expected to be developed as novel biomarkers or therapeutic targets for the diagnosis and treatment of AML, involved in assessing patient prognosis, early risk stratification, likelihood of recurrence, and treatment by targeting circRNA-miRNAs and related signaling pathway networks.
Acute lymphoblastic leukemia (ALL) is a malignant neoplastic disease in which lymphocytes proliferate abnormally in the bone marrow (73). ALL is more frequent in children, with adequate prognosis, while its prognosis in adults remains inadequate (49). circRNAs have the potential to serve as novel therapeutic targets to improve the prognosis of ALL patients. circ-0000745 and circ-PVT1 expression was found to be elevated in ALL (49, 50). circ-0000745 overexpression activates the ERK pathway, thereby promoting cell proliferation (49). In most cancers, MYC expression is elevated; high MYC expression indicates poor prognosis in patients with ALL (74). c-MYC is a member of the MYC family (75). The gene on chromosome 8q24.21 is named human plasmacytoma variant translocation 1 (PVT1), which is located 54 kb far downstream of MYC. circPVT1 is generated by cyclization of exon 2 of PVT1 (16, 76). High circPVT1 expression in ALL leads to the inhibition of c-MYC expression, in turn inhibited the apoptosis of ALL cells and promotion of cell proliferation (50). If the proliferation of ALL cells can be inhibited by targeting these circRNAs, then it is possible to improve the prognosis of ALL patients.
CML is a hematological malignancy caused by reciprocal translocation between the long arms of chromosomes 9 and 22 (77, 78). This translocation also creates the breakpoint cluster region (BCR) and Abelson (ABL) genes (77, 79). Imatinib is a first-line drug for CML (80). Drug resistance is a major obstacle to CML treatment (81). High circ-100053 and circ-0080145 levels are associated with imatinib resistance in CML (51, 52). Patients with high circ-100053 levels are more likely to develop resistance to imatinib than patients with low circ-100053 levels, thereby leading to worse prognosis (51). Moreover, circ-100053 is related to the BCR/ABL mutation status (51). circ-0080145 expression was found to be higher in patients who were resistant to imatinib than imatinib-sensitive patients. circ-0080145 knockdown increased imatinib-sensitivity of patients with CML, inhibited imatinib resistance and cell proliferation, and induced apoptosis of imatinib-resistant CML cells via miR-326 (circ_0080145 target) and protein alpha 1 (miR-326 target gene) (52). hsa-circ-0080145 was found to be highly expressed in CML cells with high proliferative capacity; furthermore, hsa-circ-0080145 directly inhibited miR296 (82). circBA9.3, a circRNA derived from BCR-ABL1, has been detected in some imatinib-resistant patients (83). circBA9.3 was found to be positively correlated with BCR-ABL1 expression, and the presence of BCR-ABL1 indicated poor prognosis. circBA9.3 has been reported to inhibit CML cell apoptosis and promote BCR-ABL1 protein expression (83). circ-0009910 accelerates CML resistance to imatinib via the miR-34a-5p/ULK1 axis (53). These findings suggest that circRNA has a significant impact on imatinib resistance, and may therefore serve as a therapeutic target for imatinib-resistant CML and potential biomarker for the diagnosis of CML.
Chronic lymphocytic leukemia (CLL) involves the malignant proliferation of CD5-positive monoclonal B lymphocytes (84, 85). circ-CBFB was found to be significantly overexpressed in CLL, which accelerated CLL cell proliferation and inhibited CLL cell apoptosis (54). circ-CBFB promotes FZD3 expression by inhibiting miR-607 via the activation of the Wnt/β-catenin pathway and acceleration of CLL progression (54). High mc-COX2 (mitochondrial genome–derived circRNA) expression was reported to be inversely correlated with prognosis while decreased mc-COX2 expression was found to affect mitochondrial function, inhibit cell proliferation, and induce apoptosis (55). Plasma circ-RPL15 was also found to promote CLL progression (86).
Chromosomal translocations may result in oncogenic fusion genes involved in the development of many tumors and are an important cause of leukemia (87). Guarnerio et al. demonstrated that f-circRNAs produced by chromosomal translocations associated with leukemia promote proliferation and disease progression and maintain the viability of leukemic cells, thereby increasing resistance to chemotherapy and reducing treatment efficacy (87). In addition, further insights into the origin of fusion gene chaperone circRNAs (FP-circRNAs) revealed that these FP-circRNAs have the potential to function as oncogenes in chromosomal translocation leukemias (88). For example, CircAF4 found in MLL leukemia originates from the partner of the MLL fusion gene AF4, promotes leukemogenesis, and represses MLL-AF4 gene expression through the sponge miR-128-3p (89). In summary, f-circRNAs and FP-CircRNAs may be considered a target for the diagnostic treatment of chromosomal translocation leukemia. Figure 1 illustrates the pathways of action and functions of circRNAs in leukemia.
CircRNAs and Lymphoma
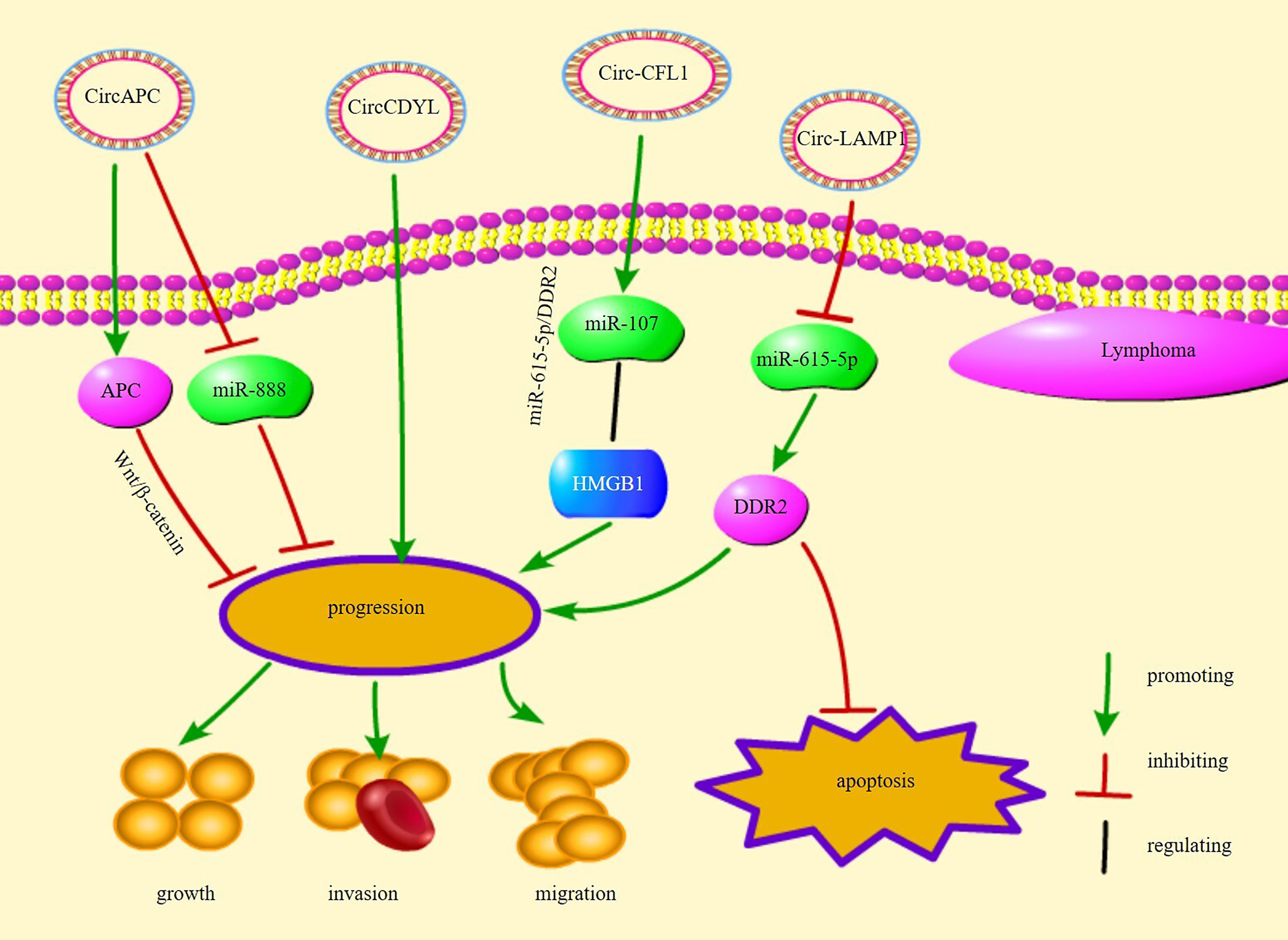
Lyphomas may be classified into various types, and the clinical presentation, treatment options, and prognosis vary based on the classification (90). T-cell lymphoblastic lymphoma (T-LBL) is a malignant tumor that arises from malignant thymocyte proliferation, leading to massive infiltration of immature T cells in the mediastinum and in lymphoid organs (90). Recently, circ-LAMP1 was found to be overexpressed in T-LBL tissues. circ-LAMP1 was reported to indirectly regulate discoidin domain receptor tyrosine kinase 2 (DDR2) expression by repressing miR-615-5p and to directly promote DDR2 expression and inhibit T-LBL cell apoptosis via the miR-615-5p/DDR2 pathway (58).
Diffuse large B-cell lymphoma (DLBCL) is the most common form of lymphoma in adults; approximately 30% of patients with DLBCL cannot be cured (91–93). It is a highly aggressive disease that originates in or outside the lymph nodes and is also caused by the transformation of other malignancies, such as CLL (92, 94, 95). circ-APC, a circRNA originating from the back-splicing of APC exon 7 to exon 14, sponges and decreases miR-888 expression or physically combines with the APC promoter to increase APC expression (56). circ-APC is a possible biomarker for DLBCL diagnosis and prognosis (56). APC inhibition decreases β-catenin aggregation in the nucleus, thereby suppressing the Wnt/β-catenin signaling pathway and inhibiting DLBCL growth (96). circCFL1 promotes the migration and proliferation of DLBCL cells (57). Using the dual-luciferase reporter gene system, it was confirmed that circCFL1 sponges its target gene miR-107 to reduce miR-107 expression, which binds to its target gene high mobility group box (HMGB)1 and reduces HMGB1 expression (57). HMGB1 enhances tumor invasion and metastasis and accelerates tumor growth (97). circCFL1 regulates the HMGB1 level via mir-107 and consequently promotes DLBCL cell migration and proliferation (57).
circ-chromodomain Y-like (circCDYL) is overexpressed in mantle cell lymphoma (MCL) and promotes MCL cell proliferation (98). Receiver operating characteristic curve analysis showed that circCDYL is a potential diagnostic biomarker for MCL (98). Overall, these circRNAs are involved in the proliferation, growth, and apoptosis of different types of lymphoma cells, implying that circRNAs are closely related to the progression of lymphoma and are potential therapeutic targets for lymphoma. Figure 2 illustrates several circRNA pathways of action and their functions in lymphomas.
CircRNAs and Multiple Myeloma
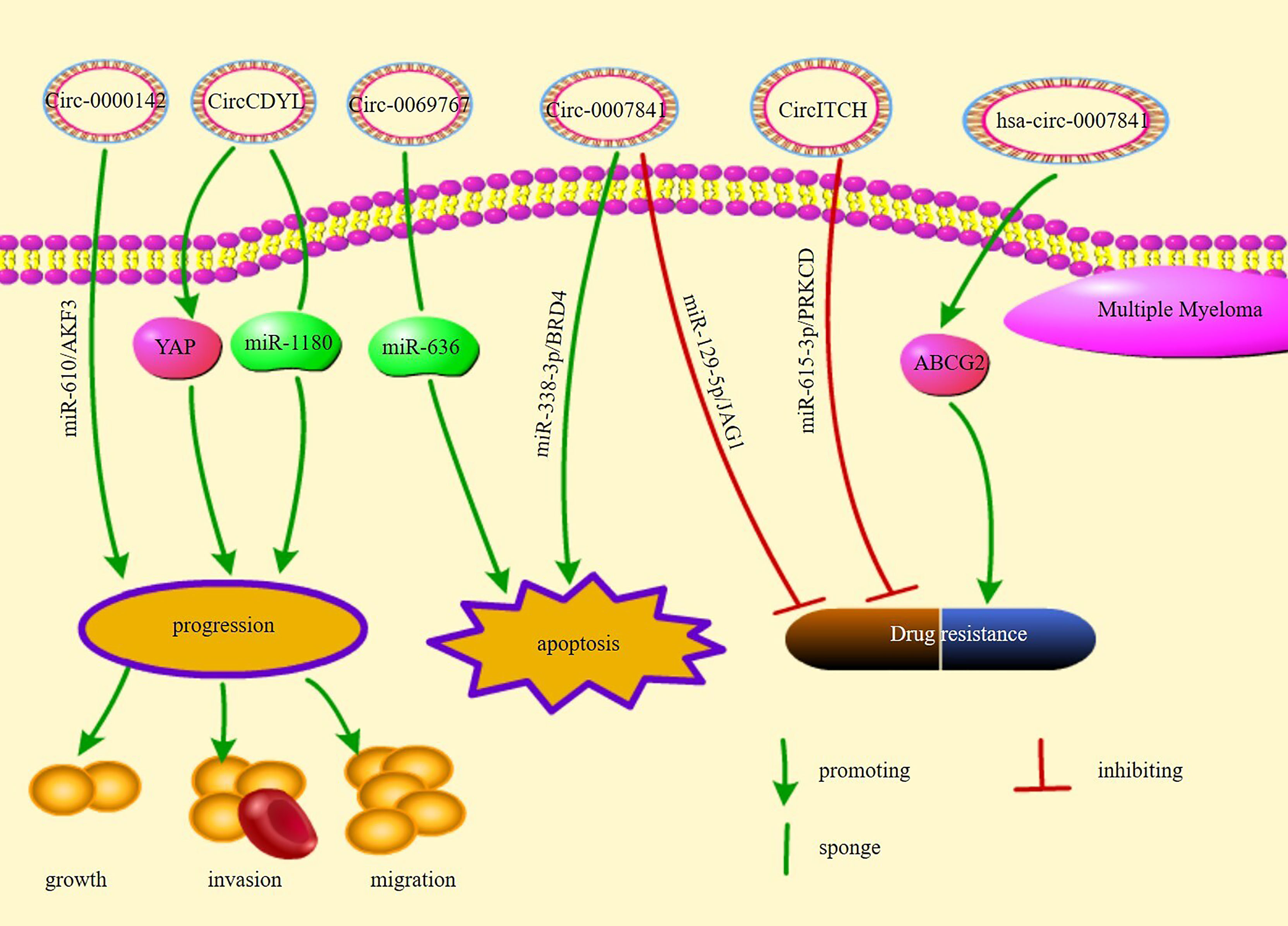
Multiple myeloma (MM) is the second most prevalent hematologic malignancy worldwide (99). It accounts for 1–2% of tumors worldwide and 2% cancer-related deaths (100). circRNAs play an important regulatory role in MM. Certain circRNAs are overexpressed in MM. Recently, it was shown that circ-CDYL is significantly overexpressed in both MM cells and plasma (101). circ-CDYL promotes the growth of MM cells by absorbing miR-1180 and increasing YAP expression (101). circ_0000142 was found to be overexpressed in patients with MM and was significantly correlated with higher International Staging System (ISS) and Durie-Salmon stages (59). circ_0000142 promoted MM cell proliferation, migration, and invasion and inhibited apoptosis by regulating miR-610/AKT3 expression (59). hsa-circRNA-101237 expression in the bone marrow is significantly upregulated in patients with MM, in MM cell lines, and in BTZ-resistant MM cells, especially those with recurrent/refractory disease (102). hsa-circRNA-101237 expression was found to significantly increase in patients who had MM with t (4, 14), t (14, 16), 13q14 deletion, P53 deletion, and 1q21 amplification; the hsa-circRNA-101237 expression was related to the prognosis, OS, and progression free survival (PFS) of patients with MM (102). circ_0007841 was overexpressed in MM cell lines and patients with MM and induced the activation of the PI3K/AKT signaling pathway via the miR-338-3p/bromodomain containing 4(BRD4) axis (64). circ_0007841 induced the cell cycle, cell growth, and metastasis and inhibited the apoptosis of MM cells via the miR-338-3p/BRD4 axis (64). The malignant behavior of MM cells is augmented by exosomes obtained from mesenchymal stromal cells (MSCs) via circ_0007841 (64). hsa-circ-0007841 was found to be overexpressed in MM drug-resistant cells and was associated with chromosomal aberrations, including 1q21, t (4:14), ATR, and IRF4 gene mutations (103, 104). hsa-circ-0007841 induces resistance in MM cells to doxorubicin by promoting ATP-binding cassette transporter G2 (ABCG2) (104). These studies suggest that overexpression of circRNAs promotes disease progression in MM, indicating their close association with its development, and nominate them as novel potential targets and biomarkers for the treatment of MM for diagnosis, and disease staging, among other uses.
Bortezomib (BTZ) is an effective drug for treating primary and relapsed MM (105). BTZ resistance remains a critical issue to overcome in the current MM treatment process (106). CircRNA itchy E3 ubiquitin protein ligase(circITCH) was found to decrease in the bone marrow of patients with MM, MM cell lines, and BTZ-resistant MM cells (60). Decreased circITCH expression is related to the poor prognosis of patients with MM (60). circITCH overexpression leads to the promotion of the sensitivity of BTZ-resistant MM cells to BTZ via the mir-615-3p/PRKCD pathway (60). circ_0007841 deletion reduces the resistance in MM cells to BTZ via the miR-129-5p/JAG1 axis (65). Therefore, targeting specific circRNAs may reduce the resistance of MM cells to BTZ and enhance the efficacy of the drug. In addition to BTZ, immunomodulatory drugs (IMids) such as lenalidomide and pomalidomide are crucial players in the treatment of MM (107). IMid-resistant cells affect the expression of circRNAs including CIRS-7, which is the most prominently affected circRNA. However, CIRS-7 knock-down does not reverse the drug sensitivity of MM cells. This suggests that CIRS-7 does not mediate the development of drug resistance in IMids (28).
Some circRNAs have been shown to have antitumor effects. circ-0069767 is overexpressed in MM, and its expression is correlated with longer PFS and OS (61). circ-0069767 overexpression inhibits the growth, migration, and invasion of MM cells and promotes apoptosis by sponging miR-636, which modulates KRAS expression (61). circ-MBYL2 was reported to decrease in the tissues and serum of patients with MM and was significantly related to advanced clinical stage and worse prognosis (108).Serum circ-MYBL2 analysis has been shown to be accurate for MM diagnosis (108). circ-MBYL2 overexpression inhibits the DNA synthesis in and cell cycle and growth of MM cells and decreases its viability by suppressing the binding of cyclin F to MYBL2, which inhibits the transcription of proliferation-related oncogenes (108). A study showed that circ_0000190 expression decreased but that of its target miR-767-5p increased in the bone marrow and peripheral blood of patients with MM (63). circ-0000190 suppressed cell cycle and growth and promoted the apoptosis of MM cells by regulating miR-767-5p/mitogen-activated protein kinase 4 (MAPK4) (63). Overexpression of Circ-0000190 arrests the cell cycle in G1 phase, and miR-767-5p promotes MM cell development by directly targeting MAPK4. Survival data analysis suggests that patients with high CIRC_0000190 expression have lower risk, and longer PFS and OS (63). circ-AMARCA5 expression has been found to decrease in MM and is negatively associated with beta-2-microglobulin (β2-MG) levels and ISS stage (62). circ-AMARCA5 overexpression was found to be related to improved complete response (CR), PFS, and OS and to suppress cell growth; meanwhile, it induced the apoptosis of MM cells by sponging miR-767-5p (62). In summary, patient prognosis may be improved by targeting circRNAs and their targets, and circRNAs display remarkable potential as biomarkers for MM.
Exosomes are secretory vesicles from the endosome (endosomal) with a diameter of about 40–160 nm (average diameter 100 nm) (109). They are widely distributed and abundant in body fluids (110) and can be secreted by most cells (111). Exosomes deliver nucleic acids including circRNAs, proteins, and other substances between cells, and regulate various pathophysiological activities (110, 112). CircDAP3, an exosomal CircRNA significantly decreased in essential thrombocythemia (ET), inhibited the conversion of K562 cells to megakaryocytes in in vitro studies, suggesting its possible involvement in the progression of ET (113). Peripheral neuropathy is the most prominent complication in MM (111), and is also one of the side effects of bortezomib (114). One of the circulating exosomal circRNAs exo-CircRNAs called ChR2:2744228-2744407+, which is upregulated in MM, has been found to potentially be involved in the induction of MM-induced peripheral neuropathy (111). Another serum exosomal CircRNA called CircMYC was shown to be significantly elevated in MM and suggested a poor prognosis. Expression was upregulated in relapsed refractory patients compared to primary patients, suggesting that it may be associated with high recurrence of MM. Additionally, expression was higher in BTZ-resistant patients compared to non-resistant patients, suggesting that CircMYC may be associated with BTZ resistance in patients (110). IgG MM interferes with a variety of cells in the tumor microenvironment by secreting CircHNRNPU via exosomes (115). Exosomal circ-ATP10A promotes angiogenesis in MM patients by targeting various miRNAs, such as hsa-miR-3620-3p, which is expected to improve the prognosis of patients (116). In summary, circRNAs secreted by exosomes can influence the course of MM, participate in the generation of MM complications, and interfere with the tumor microenvironment, providing a new entry point for the treatment of MM.
Figure 3 illustrates the pathways of action and functions of circRNAs in MM.
Discussion and Challenges
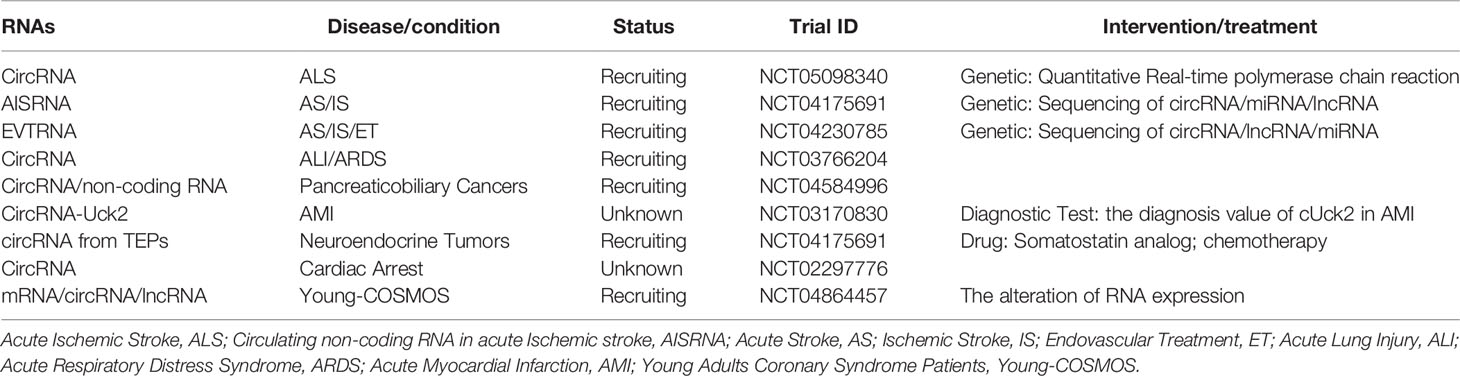
There is an increasing number of studies on circRNAs and hematologic malignancies. The expression of certain circRNAs, the interaction of many circRNAs with miRNAs, and the related signaling pathways are associated with growth, proliferation, invasion, and apoptosis of various hematological malignancies, and are also closely related to disease stage, OS, LFS, and PFS. In addition, the circRNAs participate in or manifest drug resistance, indicating their potential as therapeutic targets and biomarkers. Biomarkers are used for disease diagnosis, to determine disease staging, or to evaluate the safety and efficacy of new drugs or therapies in the target population (117). To become biomarkers or therapeutic targets, they must first have a high degree of specificity so that they can be targeted while reducing toxicity to normal cells; in addition, they must be sensitive and easy to detect and they must have a high degree of stability (118). Biomarkers and therapeutic targets need to undergo repeated screening, risk assessment, clinical trials, and prognostic analysis before they can be applied (117). CircRNAs in human blood (119) and other body fluids are abundant and easy to detect, indicating that CircRNAs are suitable as biomarkers or therapeutic targets. However, the specificity of circRNAs as diagnostic markers is still unclear, and their sensitivity as therapeutic targets is also unknown. Since a single circRNA may be aberrantly expressed in different diseases. For example, abnormal expression of circPVT1 can be detected in diseases such as gastric cancer and ALL (40, 50), so the diagnostic specificity of a single circRNA is insufficient. However, cumulative diagnosis may be achieved by combining multiple circRNAs or including corresponding target miRNAs, etc. Most of the current studies primarily focus on the interaction between circRNAs and miRNAs. Further mechanisms of action of circRNAs, as well as the case of multiple circRNAs that demonstrate abnormal expression in the same disease, require more investigation. Interactions between circRNAs are yet to be fully elucidated; a lack of sufficient clinical studies limit the applications these circRNAs. We have concluded the current clinical trials in Table 2.
Conclusion
In this review, we summarized the progress of circRNA research in hematologic malignancies. circRNAs have the potential to become new diagnostic markers and therapeutic targets for various diseases. Although further research on circRNAs is still required, circRNA-based therapeutic approaches may be a promising strategy for the treatment of hematologic malignancies in the future.
Author Contributions
JD designed the study and wrote the manuscript. LW edited and revised the manuscript. All authors have read and approved the submitted version.
Funding
This work was supported by grants from the key research project program of Shandong Province (2018GSF118035), the Medical Health Science and Technology Development Plan of Shandong Province (2017–462), the Affiliated Hospital Development Fund of Xuzhou Medical University (XYFM2020016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
circRNAs, circular RNAs; miRNA, microRNA; ecircRNAs, exonic circRNAs; ciRNAs, circular intron RNAs; EIciRNAs, exon-intron circRNAs; tricRNAs, tRNA intronic circular RNAs; RBPs, RNA binding proteins; m6A, N6-methyladenosine; CIRS-7,circRNA sponge for miR-7;IRES,internal ribosome entry site; f-circRNAs, fusion-circular RNAs; AML, Acute myelogenous leukemia; FAB, French–American–British; LFS, leukemia-free survival; PRKACB, protein kinase CAMP-activated catalytic subunit beta; FOXM1,forkhead box M1;FLT3,FMS-like tyrosine kinase 3 receptor;PTEN, phosphatase and tensin homolog deleted on chromosome 10; ALL, Acute lymphoblastic leukemia;PVT1, plasmacytoma variant translocation 1;CML, Chronic myelogenous leukemia; BCR, breakpoint cluster region; ABL, Abelson;CLL Chronic lymphocytic leukemia; FP-circRNAs,fusion gene chaperone circRNAs; T-LBL, T-cell lymphoblastic lymphoma; DLBCL, Diffuse large B-cell lymphoma; HMGB, high mobility group box; circCDYL, circ-chromodomain Y-like;MCL, mantle cell lymphoma; MM, Multiple myeloma; BTZ, Bortezomib; ISS, International Staging System; circITCH, circRNA itchy E3 ubiquitin protein ligase; IMids, immunomodulatory drugs; PFS, progression free survival; BRD4,bromodomain containing 4; MSCs, mesenchymal stromal cells; ABCG2,ATP-binding cassette transporter G2; MAPK4,mitogen-activated protein kinase 4; β2-MG,beta-2-microglobulin; CR, complete response; ET, essential thrombocythemia.
References
1. Qian L, Yu S, Chen Z, Meng Z, Huang S, Wang P. The Emerging Role of Circrnas and Their Clinical Significance in Human Cancers. Biochim Biophys Acta Rev Cancer (2018) 1870(2):247–60. doi: 10.1016/j.bbcan.2018.06.002
2. Zhou X, Zhan L, Huang K, Wang X. The Functions and Clinical Significance of Circrnas in Hematological Malignancies. J Hematol Oncol (2020) 13(1):138. doi: 10.1186/s13045-020-00976-1
3. Zhang Z, Yang T, Xiao J. Circular Rnas: Promising Biomarkers for Human Diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
4. Chen I, Chen CY, Chuang TJ. Biogenesis, Identification, and Function of Exonic Circular Rnas. Wiley Interdiscip Rev RNA (2015) 6(5):563–79. doi: 10.1002/wrna.1294
5. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding Rnas. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
6. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular Rnas Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
7. Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. Molecular Determinants of Metazoan Tricrna Biogenesis. Nucleic Acids Res (2019) 47(12):6452–65. doi: 10.1093/nar/gkz311
8. Bonizzato A, Gaffo E, Te Kronnie G, Bortoluzzi S. Circrnas in Hematopoiesis and Hematological Malignancies. Blood Cancer J (2016) 6(10):e483. doi: 10.1038/bcj.2016.81
9. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of Intron Sequences Reveals Hallmarks of Circular Rna Biogenesis in Animals. Cell Rep (2015) 10(2):170–7. doi: 10.1016/j.celrep.2014.12.019
10. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular Rnas Are Abundant, Conserved, and Associated With Alu Repeats. RNA (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
11. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. Fus Affects Circular Rna Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat Commun (2017) 8:14741. doi: 10.1038/ncomms14741
12. Noto JJ, Schmidt CA, Matera AG. Engineering and Expressing Circular Rnas Via Trna Splicing. RNA Biol (2017) 14(8):978–84. doi: 10.1080/15476286.2017.1317911
13. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids Are Single-Stranded Covalently Closed Circular Rna Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc Natl Acad Sci U S A (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
14. Hsu MT, Coca-Prados M. Electron Microscopic Evidence for the Circular Form of Rna in the Cytoplasm of Eukaryotic Cells. Nature (1979) 280(5720):339–40. doi: 10.1038/280339a0
15. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The Hepatitis Delta (Delta) Virus Possesses a Circular Rna. Nature (1986) 323(6088):558–60. doi: 10.1038/323558a0
16. Palcau AC, Canu V, Donzelli S, Strano S, Pulito C, Blandino G. Circpvt1: A Pivotal Circular Node Intersecting Long Non-Coding-Pvt1 and C-Myc Oncogenic Signals. Mol Cancer (2022) 21(1):33. doi: 10.1186/s12943-022-01514-y
17. Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on Circular Rnas and New Insights Into Their Roles in Cancer. Comput Struct Biotechnol J (2021) 19:910–28. doi: 10.1016/j.csbj.2021.01.018
18. Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, et al. The Bioinformatics Toolbox for Circrna Discovery and Analysis. Brief Bioinform (2021) 22(2):1706–28. doi: 10.1093/bib/bbaa001
19. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded Identification and Characterization of Mammalian Circular Rnas. Genome Biol (2014) 15(7):409. doi: 10.1186/s13059-014-0409-z
20. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular Rnas. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
21. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular Rnas Are the Predominant Transcript Isoform From Hundreds of Human Genes in Diverse Cell Types. PLoS One (2012) 7(2):e30733. doi: 10.1371/journal.pone.0030733
22. Dahl M, Daugaard I, Andersen MS, Hansen TB, Gronbaek K, Kjems J, et al. Enzyme-Free Digital Counting of Endogenous Circular Rna Molecules in B-Cell Malignancies. Lab Invest (2018) 98(12):1657–69. doi: 10.1038/s41374-018-0108-6
23. Issah MA, Wu D, Zhang F, Zheng W, Liu Y, Chen R, et al. Expression Profiling of N(6)-Methyladenosine Modified Circrnas in Acute Myeloid Leukemia. Biochem Biophys Res Commun (2022) 601:137–45. doi: 10.1016/j.bbrc.2022.02.087
24. Perez de Acha O, Rossi M, Gorospe M. Circular Rnas in Blood Malignancies. Front Mol Biosci (2020) 7:109. doi: 10.3389/fmolb.2020.00109
25. Shen T, Han M, Wei G, Ni T. An Intriguing Rna Species–Perspectives of Circularized Rna. Protein Cell (2015) 6(12):871–80. doi: 10.1007/s13238-015-0202-0
26. Verduci L, Strano S, Yarden Y, Blandino G. The Circrna-Microrna Code: Emerging Implications for Cancer Diagnosis and Treatment. Mol Oncol (2019) 13(4):669–80. doi: 10.1002/1878-0261.12468
27. Hansen TB, Kjems J, Damgaard CK. Circular Rna and Mir-7 in Cancer. Cancer Res (2013) 73(18):5609–12. doi: 10.1158/0008-5472.CAN-13-1568
28. Jakobsen T, Dahl M, Dimopoulos K, Gronbaek K, Kjems J, Kristensen LS. Genome-Wide Circular Rna Expression Patterns Reflect Resistance to Immunomodulatory Drugs in Multiple Myeloma Cells. Cancers (Basel) (2021) 13(3):365. doi: 10.3390/cancers13030365
29. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular Rna-Protein Interactions: Functions, Mechanisms, and Identification. Theranostics (2020) 10(8):3503–17. doi: 10.7150/thno.42174
30. Hentze MW, Castello A, Schwarzl T, Preiss T. A Brave New World of Rna-Binding Proteins. Nat Rev Mol Cell Biol (2018) 19(5):327–41. doi: 10.1038/nrm.2017.130
31. Sinha T, Panigrahi C, Das D, Chandra Panda A. Circular Rna Translation, a Path to Hidden Proteome. Wiley Interdiscip Rev RNA (2022) 13(1):e1685. doi: 10.1002/wrna.1685
32. Jamal M, Song T, Chen B, Faisal M, Hong Z, Xie T, et al. Recent Progress on Circular Rna Research in Acute Myeloid Leukemia. Front Oncol (2019) 9:1108. doi: 10.3389/fonc.2019.01108
33. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. Circhipk3 Promotes Colorectal Cancer Growth and Metastasis by Sponging Mir-7. Cell Death Dis (2018) 9(4):417. doi: 10.1038/s41419-018-0454-8
34. Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li D, et al. Increased Expression of Circrna_102231 in Lung Cancer and Its Clinical Significance. Biomed Pharmacother (2018) 102:639–44. doi: 10.1016/j.biopha.2018.03.084
35. Liu Y, Yang Y, Wang Z, Fu X, Chu XM, Li Y, et al. Insights Into the Regulatory Role of Circrna in Angiogenesis and Clinical Implications. Atherosclerosis (2020) 298:14–26. doi: 10.1016/j.atherosclerosis.2020.02.017
36. Shi Y, Jia X, Xu J. The New Function of Circrna: Translation. Clin Transl Oncol (2020) 22(12):2162–9. doi: 10.1007/s12094-020-02371-1
37. Babin L, Piganeau M, Renouf B, Lamribet K, Thirant C, Deriano L, et al. Chromosomal Translocation Formation Is Sufficient to Produce Fusion Circular Rnas Specific to Patient Tumor Cells. iScience (2018) 5:19–29. doi: 10.1016/j.isci.2018.06.007
38. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic Circular Rnas Arise From Chromosomal Translocations. Cancer Discovery (2016) 6(6):OF20. doi: 10.1158/2159-8290.CD-RW2016-068
39. Chen B, Huang S. Circular Rna: An Emerging Non-Coding Rna as a Regulator and Biomarker in Cancer. Cancer Lett (2018) 2018):41–50. doi: 10.1016/j.canlet.2018.01.011
40. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular Rna Profile Identifies Circpvt1 as a Proliferative Factor and Prognostic Marker in Gastric Cancer. Cancer Lett (2017) 388:208–19. doi: 10.1016/j.canlet.2016.12.006
41. Yuan DM, Ma J, Fang WB. Identification of Non-Coding Rna Regulatory Networks in Pediatric Acute Myeloid Leukemia Reveals Circ-0004136 Could Promote Cell Proliferation by Sponging Mir-142. Eur Rev Med Pharmacol Sci (2019) 23(21):9251–8. doi: 10.26355/eurrev_201911_19417
42. Zhang R, Li Y, Wang H, Zhu K, Zhang G. The Regulation of Circrna Rnf13/Mirna-1224-5p Axis Promotes the Malignant Evolution in Acute Myeloid Leukemia. BioMed Res Int (2020) 2020:5654380. doi: 10.1155/2020/5654380
43. Yi YY, Yi J, Zhu X, Zhang J, Zhou J, Tang X, et al. Circular Rna of Vimentin Expression as a Valuable Predictor for Acute Myeloid Leukemia Development and Prognosis. J Cell Physiol (2019) 234(4):3711–9. doi: 10.1002/jcp.27145
44. Wang Y, Guo T, Liu Q, Xie X. Circrad18 Accelerates the Progression of Acute Myeloid Leukemia by Modulation of Mir-206/Prkacb Axis. Cancer Manag Res (2020) 12:10887–96. doi: 10.2147/CMAR.S277432
45. Yi L, Zhou L, Luo J, Yang Q. Circ-Ptk2 Promotes the Proliferation and Suppressed the Apoptosis of Acute Myeloid Leukemia Cells Through Targeting Mir-330-5p/Foxm1 Axis. Blood Cells Mol Dis (2021) 86:102506. doi: 10.1016/j.bcmd.2020.102506
46. Zhang L, Bu Z, Shen J, Shang L, Chen Y, Wang Y. A Novel Circular Rna (Hsa_Circ_0000370) Increases Cell Viability and Inhibits Apoptosis of Flt3-Itd-Positive Acute Myeloid Leukemia Cells by Regulating Mir-1299 and S100a7a. Biomed Pharmacother (2020) 122:109619. doi: 10.1016/j.biopha.2019.109619
47. Chen JJ, Lei P, Zhou M. Hsa_Circ_0121582 Inhibits Leukemia Growth by Dampening Wnt/B-Catenin Signaling. Clin Trans Oncol (2020) 22(12):2293–302. doi: 10.1007/s12094-020-02377-9
48. Su X-Y, Zhao Q, Ke J-M, Wu D-H, Zhu X, Lin J, et al. Circ_0002232 Acts as a Potential Biomarker for Aml and Reveals a Potential Cerna Network of Circ_0002232/Mir-92a-3p/Pten. Cancer Manage Res (2020) 12:11871–81. doi: 10.2147/cmar.S278499
49. Liu X, Zhou C, Li Y, Deng Y, Lu W, Li J. Upregulation of Circ-0000745 in Acute Lymphoblastic Leukemia Enhanced Cell Proliferation by Activating Erk Pathway. Gene (2020) 751:144726. doi: 10.1016/j.gene.2020.144726
50. Hu J, Han Q, Gu Y, Ma J, McGrath M, Qiao F, et al. Circular Rna Pvt1 Expression and Its Roles in Acute Lymphoblastic Leukemia. Epigenomics (2018) 10(6):723–32. doi: 10.2217/epi-2017-0142
51. Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z. High Circ_100053 Predicts a Poor Outcome for Chronic Myeloid Leukemia and Is Involved in Imatinib Resistance. Oncol Res (2019). doi: 10.3727/096504018X15412701483326
52. Che H, Ding H, Jia X. Circ_0080145 Enhances Imatinib Resistance of Chronic Myeloid Leukemia by Regulating Mir-326/Ppfia1 Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3600
53. Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L, et al. Circ_0009910 Promotes Imatinib Resistance Through Ulk1-Induced Autophagy by Sponging Mir-34a-5p in Chronic Myeloid Leukemia. Life Sci (2020) 243:117255. doi: 10.1016/j.lfs.2020.117255
54. Xia L, Wu L, Bao J, Li Q, Chen X, Xia H, et al. Circular Rna Circ-Cbfb Promotes Proliferation and Inhibits Apoptosis in Chronic Lymphocytic Leukemia Through Regulating Mir-607/Fzd3/Wnt/Beta-Catenin Pathway. Biochem Biophys Res Commun (2018) 503(1):385–90. doi: 10.1016/j.bbrc.2018.06.045
55. Wu Z, Sun H, Wang C, Liu W, Liu M, Zhu Y, et al. Mitochondrial Genome-Derived Circrna Mc-Cox2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol Ther Nucleic Acids (2020) 20:801–11. doi: 10.1016/j.omtn.2020.04.017
56. Hu Y, Zhao Y, Shi C, Ren P, Wei B, Guo Y, et al. A Circular Rna From Apc Inhibits the Proliferation of Diffuse Large B-Cell Lymphoma by Inactivating Wnt/Beta-Catenin Signaling Via Interacting With Tet1 and Mir-888. Aging (Albany NY) (2019) 11(19):8068–84. doi: 10.18632/aging.102122
57. Chen X, Xie X, Zhou W. Circcfl1/Mir-107 Axis Targeting Hmgb1 Promotes the Malignant Progression of Diffuse Large B-Cell Lymphoma Tumors. Cancer Manag Res (2020) 12:9351–62. doi: 10.2147/CMAR.S263222
58. Deng L, Liu G, Zheng C, Zhang L, Kang Y, Yang F. Circ-Lamp1 Promotes T-Cell Lymphoblastic Lymphoma Progression Via Acting as a Cerna for Mir-615-5p to Regulate Ddr2 Expression. Gene (2019) 701:146–51. doi: 10.1016/j.gene.2019.03.052
59. Liu F, Wang YL, Wei JM, Huang ZD. Upregulation of Circ_0000142 Promotes Multiple Myeloma Progression by Adsorbing Mir-610 and Upregulating Akt3 Expression. J Biochem (2020) 169(3):327–36. doi: 10.1093/jb/mvaa106
60. Liu J, Du F, Chen C, Li D, Chen Y, Xiao X, et al. Circrna Itch Increases Bortezomib Sensitivity Through Regulating the Mir-615-3p/Prkcd Axis in Multiple Myeloma. Life Sci (2020) 262:118506. doi: 10.1016/j.lfs.2020.118506
61. Chen F, Wang X, Fu S, Wang S, Fu Y, Liu Z, et al. Effect of the Up-Regulation of Circular Rna Hsa_Circ_0069767 Derived From C-Kit on the Biological Behavior of Multiple Myeloma Cells. Cancer Manag Res (2020) 12:11321–31. doi: 10.2147/CMAR.S259393
62. Liu H, Wu Y, Wang S, Jiang J, Zhang C, Jiang Y, et al. Circ-Smarca5 Suppresses Progression of Multiple Myeloma by Targeting Mir-767-5p. BMC Cancer (2019) 19(1):937. doi: 10.1186/s12885-019-6088-0
63. Feng Y, Zhang L, Wu J, Khadka B, Fang Z, Gu J, et al. Circrna Circ_0000190 Inhibits the Progression of Multiple Myeloma Through Modulating Mir-767-5p/Mapk4 Pathway. J Exp Clin Cancer Res (2019) 38(1):54. doi: 10.1186/s13046-019-1071-9
64. Wang Y, Lin Q, Song C, Ma R, Li X. Circ_0007841 Promotes the Progression of Multiple Myeloma Through Targeting Mir-338-3p/Brd4 Signaling Cascade. Cancer Cell Int (2020) 20:383. doi: 10.1186/s12935-020-01475-6
65. Wang Y, Lin Q, Song C, Ma R, Li X. Depletion of Circ_0007841 Inhibits Multiple Myeloma Development and Btz Resistance Via Mir-129-5p/Jag1 Axis. Cell Cycle (2020) 19(23):3289–302. doi: 10.1080/15384101.2020.1839701
66. Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular Biomarkers in Acute Myeloid Leukemia. Blood Rev (2017) 31(1):63–76. doi: 10.1016/j.blre.2016.08.005
67. Wu DM, Wen X, Han XR, Wang S, Wang YJ, Shen M, et al. Role of Circular Rna Dleu2 in Human Acute Myeloid Leukemia. Mol Cell Biol (2018) 38(20):e00259-18. doi: 10.1128/MCB.00259-18
68. Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z. Silencing of Circ_0009910 Inhibits Acute Myeloid Leukemia Cell Growth Through Increasing Mir-20a-5p. Blood Cells Mol Dis (2019) 75:41–7. doi: 10.1016/j.bcmd.2018.12.006
69. Zhang T, Zhou Y, Guan J, Cheng H. Circ_0058058 Knockdown Inhibits Acute Myeloid Leukemia Progression by Sponging Mir-4319 to Regulate Eif5a2 Expression. Cancer Biother Radiopharm (2021). doi: 10.1089/cbr.2020.4170
70. Levis M, Perl AE. Gilteritinib: Potent Targeting of Flt3 Mutations in Aml. Blood Adv (2020) 4(6):1178–91. doi: 10.1182/bloodadvances.2019000174
71. Fan H, Li Y, Liu C, Liu Y, Bai J, Li W. Circular Rna-100290 Promotes Cell Proliferation and Inhibits Apoptosis in Acute Myeloid Leukemia Cells Via Sponging Mir-203. Biochem Biophys Res Commun (2018) 507(1-4):178–84. doi: 10.1016/j.bbrc.2018.11.002
72. Hu Q, Gu Y, Chen S, Tian Y, Yang S. Hsa_Circ_0079480 Promotes Tumor Progression in Acute Myeloid Leukemia Via Mir-654-3p/Hdgf Axis. Aging (Albany NY) (2020) 13(1):1120–31. doi: 10.18632/aging.202240
73. Malard F, Mohty M. Acute Lymphoblastic Leukaemia. Lancet (2020) 395(10230):1146–62. doi: 10.1016/s0140-6736(19)33018-1
74. Baluapuri A, Wolf E, Eilers M. Target Gene-Independent Functions of Myc Oncoproteins. Nat Rev Mol Cell Biol (2020) 21(5):255–67. doi: 10.1038/s41580-020-0215-2
75. Butler DSC, Cafaro C, Putze J, Wan MLY, Tran TH, Ambite I, et al. A Bacterial Protease Depletes C-Myc and Increases Survival in Mouse Models of Bladder and Colon Cancer. Nat Biotechnol (2021) 39(6):754–64. doi: 10.1038/s41587-020-00805-3
76. Ghetti M, Vannini I, Storlazzi CT, Martinelli G, Simonetti G. Linear and Circular Pvt1 in Hematological Malignancies and Immune Response: Two Faces of the Same Coin. Mol Cancer (2020) 19(1):69. doi: 10.1186/s12943-020-01187-5
77. Baccarani M, Castagnetti F, Gugliotta G, Rosti G, Soverini S, Albeer A, et al. The Proportion of Different Bcr-Abl1 Transcript Types in Chronic Myeloid Leukemia. Int Overview Leukemia (2019) 33(5):1173–83. doi: 10.1038/s41375-018-0341-4
78. Agarwal P, Isringhausen S, Li H, Paterson AJ, He J, Gomariz A, et al. Mesenchymal Niche-Specific Expression of Cxcl12 Controls Quiescence of Treatment-Resistant Leukemia Stem Cells. Cell Stem Cell (2019) 24(5):769–84.e6. doi: 10.1016/j.stem.2019.02.018
79. Asnafi AA, Deris Zayeri Z, Shahrabi S, Zibara K, Vosughi T. Chronic Myeloid Leukemia With Complex Karyotypes: Prognosis and Therapeutic Approaches. J Cell Physiol (2019) 234(5):5798–806. doi: 10.1002/jcp.27505
80. Vener C, Banzi R, Ambrogi F, Ferrero A, Saglio G, Pravettoni G, et al. First-Line Imatinib Vs Second- and Third-Generation Tkis for Chronic-Phase Cml: A Systematic Review and Meta-Analysis. Blood Adv (2020) 4(12):2723–35. doi: 10.1182/bloodadvances.2019001329
81. Lu Z, Xu N, He B, Pan C, Lan Y, Zhou H, et al. Inhibition of Autophagy Enhances the Selective Anti-Cancer Activity of Tigecycline to Overcome Drug Resistance in the Treatment of Chronic Myeloid Leukemia. J Exp Clin Cancer Res (2017) 36(1):43. doi: 10.1186/s13046-017-0512-6
82. Liu J, Kong F, Lou S, Yang D, Gu L. Global Identification of Circular Rnas in Chronic Myeloid Leukemia Reveals Hsa_Circ_0080145 Regulates Cell Proliferation by Sponging Mir-29b. Biochem Biophys Res Commun (2018) 504(4):660–5. doi: 10.1016/j.bbrc.2018.08.154
83. Pan Y, Lou J, Wang H, An N, Chen H, Zhang Q, et al. Circba9.3 Supports the Survival of Leukaemic Cells by Up-Regulating C-Abl1 or Bcr-Abl1 Protein Levels. Blood Cells Mol Dis (2018) 73:38–44. doi: 10.1016/j.bcmd.2018.09.002
84. Hallek M. Chronic Lymphocytic Leukemia: 2017 Update on Diagnosis, Risk Stratification, and Treatment. Am J Hematol (2017) 92(9):946–65. doi: 10.1002/ajh.24826
85. Hallek M, Shanafelt TD, Eichhorst B. Chronic Lymphocytic Leukaemia. Lancet (2018) 391(10129):1524–37. doi: 10.1016/s0140-6736(18)30422-7
86. Wu Z, Sun H, Liu W, Zhu H, Fu J, Yang C, et al. Circ-Rpl15: A Plasma Circular Rna as Novel Oncogenic Driver to Promote Progression of Chronic Lymphocytic Leukemia. Leukemia (2020) 34(3):919–23. doi: 10.1038/s41375-019-0594-6
87. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic Role of Fusion-Circrnas Derived From Cancer-Associated Chromosomal Translocations. Cell (2016) 165(2):289–302. doi: 10.1016/j.cell.2016.03.020
88. Lin Z, Long F, Zhao M, Zhang X, Yang M. The Role of Circular Rnas in Hematological Malignancies. Genomics (2020) 112(6):4000–8. doi: 10.1016/j.ygeno.2020.06.051
89. Huang W, Fang K, Chen TQ, Zeng ZC, Sun YM, Han C, et al. Circrna Circaf4 Functions as an Oncogene to Regulate Mll-Af4 Fusion Protein Expression and Inhibit Mll Leukemia Progression. J Hematol Oncol (2019) 12(1):103. doi: 10.1186/s13045-019-0800-z
90. Kroeze E, Loeffen JLC, Poort VM, Meijerink JPP. T-Cell Lymphoblastic Lymphoma and Leukemia: Different Diseases From a Common Premalignant Progenitor? Blood Adv (2020) 4(14):3466–73. doi: 10.1182/bloodadvances.2020001822
91. Wang L, Li LR, Young KH. New Agents and Regimens for Diffuse Large B Cell Lymphoma. J Hematol Oncol (2020) 13(1):175. doi: 10.1186/s13045-020-01011-z
92. Pasqualucci L, Dalla-Favera R. Genetics of Diffuse Large B-Cell Lymphoma. Blood (2018) 131(21):2307–19. doi: 10.1182/blood-2017-11-764332
93. Li S, Young KH, Medeiros LJ. Diffuse Large B-Cell Lymphoma. Pathology (2018) 50(1):74–87. doi: 10.1016/j.pathol.2017.09.006
94. He MY, Kridel R. Treatment Resistance in Diffuse Large B-Cell Lymphoma. Leukemia (2021) 35(8):2151–65. doi: 10.1038/s41375-021-01285-3
95. Ennishi D, Hsi ED, Steidl C, Scott DW. Toward a New Molecular Taxonomy of Diffuse Large B-Cell Lymphoma. Cancer Discovery (2020) 10(9):1267–81. doi: 10.1158/2159-8290.CD-20-0174
96. Fodde R, Brabletz T. Wnt/Beta-Catenin Signaling in Cancer Stemness and Malignant Behavior. Curr Opin Cell Biol (2007) 19(2):150–8. doi: 10.1016/j.ceb.2007.02.007
97. Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. Hmgb1 and Rage in Inflammation and Cancer. Annu Rev Immunol (2010) 28(1):367–88. doi: 10.1146/annurev.immunol.021908.132603
98. Mei M, Wang Y, Wang Q, Liu Y, Song W, Zhang M. Circcdyl Serves as a New Biomarker in Mantle Cell Lymphoma and Promotes Cell Proliferation. Cancer Manage Res (2019) 11:10215–2. doi: 10.2147/cmar.S232075
99. Federico C, Alhallak K, Sun J, Duncan K, Azab F, Sudlow GP, et al. Tumor Microenvironment-Targeted Nanoparticles Loaded With Bortezomib and Rock Inhibitor Improve Efficacy in Multiple Myeloma. Nat Commun (2020) 11(1):6037. doi: 10.1038/s41467-020-19932-1
100. Poh C, Keegan T, Rosenberg AS. Second Primary Malignancies in Multiple Myeloma: A Review. Blood Rev (2021) 46:100757. doi: 10.1016/j.blre.2020.100757
101. Chen F, Wang X, Fu S, Wang S, Fu Y, Zhang J, et al. Circular Rna Circ-Cdyl Sponges Mir-1180 to Elevate Yes-Associated Protein in Multiple Myeloma. Exp Biol Med (Maywood) (2020) 245(11):925–32. doi: 10.1177/1535370220918191
102. Liu X, Tang H, Liu J, Wang X. Hsa_Circrna_101237: A Novel Diagnostic and Prognostic Biomarker and Potential Therapeutic Target for Multiple Myeloma. Cancer Manag Res (2020) 12:2109–18. doi: 10.2147/CMAR.S241089
103. Gao M, Li C, Xiao H, Dong H, Jiang S, Fu Y, et al. Hsa_Circ_0007841: A Novel Potential Biomarker and Drug Resistance for Multiple Myeloma. Front Oncol (2019) 9:1261. doi: 10.3389/fonc.2019.01261
104. Song Y, Hu N, Song X, Yang J. Hsa_Circ_0007841 Enhances Multiple Myeloma Chemotherapy Resistance Through Upregulating Abcg2. Technol Cancer Res Treat (2020) 19:1533033820928371. doi: 10.1177/1533033820928371
105. Tibullo D, Giallongo C, Romano A, Vicario N, Barbato A, Puglisi F, et al. Mitochondrial Functions, Energy Metabolism and Protein Glycosylation Are Interconnected Processes Mediating Resistance to Bortezomib in Multiple Myeloma Cells. Biomolecules (2020) 10(5):696. doi: 10.3390/biom10050696
106. Gonsalves WI, Milani P, Derudas D, Buadi FK. The Next Generation of Novel Therapies for the Management of Relapsed Multiple Myeloma. Future Oncol (2017) 13(1):63–75. doi: 10.2217/fon-2016-0200
107. Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, et al. Cereblon Is a Direct Protein Target for Immunomodulatory and Antiproliferative Activities of Lenalidomide and Pomalidomide. Leukemia (2012) 26(11):2326–35. doi: 10.1038/leu.2012.119
108. Yu S, Ai L, Wei W, Pan J. Circrna Circ-Mybl2 Is a Novel Tumor Suppressor and Potential Biomarker in Multiple Myeloma. Hum Cell (2021) 34(1):219–28. doi: 10.1007/s13577-020-00441-8
109. Kalluri R, LeBleu VS. The Biology, Function, and Biomedical Applications of Exosomes. Science (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
110. Luo Y, Gui R. Circulating Exosomal Circmyc Is Associated With Recurrence and Bortezomib Resistance in Patients With Multiple Myeloma. Turk J Haematol (2020) 37(4):248–62. doi: 10.4274/tjh.galenos.2020.2020.0243
111. Zhang Y, Pisano M, Li N, Tan G, Sun F, Cheng Y, et al. Exosomal Circrna as a Novel Potential Therapeutic Target for Multiple Myeloma-Related Peripheral Neuropathy. Cell Signal (2021) 78:109872. doi: 10.1016/j.cellsig.2020.109872
112. Babin L, Andraos E, Fuchs S, Pyronnet S, Brunet E, Meggetto F. From Circrnas to Fusion Circrnas in Hematological Malignancies. JCI Insight (2021) 6(21):e151513. doi: 10.1172/jci.insight.151513
113. Wang Q, Yu G, He H, Zheng Z, Li X, Lin R, et al. Differential Expression of Circular Rnas in Bone Marrow-Derived Exosomes From Essential Thrombocythemia Patients. Cell Biol Int (2021) 45(4):869–81. doi: 10.1002/cbin.11534
114. Richardson PG, Xie W, Mitsiades C, Chanan-Khan AA, Lonial S, Hassoun H, et al. Single-Agent Bortezomib in Previously Untreated Multiple Myeloma: Efficacy, Characterization of Peripheral Neuropathy, and Molecular Correlations With Response and Neuropathy. J Clin Oncol (2009) 27(21):3518–25. doi: 10.1200/JCO.2008.18.3087
115. Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao S, et al. A Novel Protein Encoded by Circhnrnpu Promotes Multiple Myeloma Progression by Regulating the Bone Marrow Microenvironment and Alternative Splicing. J Exp Clin Cancer Res (2022) 41(1):85. doi: 10.1186/s13046-022-02276-7
116. Yu M, Yu J, Zhang Y, Sun X, Sun R, Xia M, et al. A Novel Circrna-Mirna-Mrna Network Revealed Exosomal Circ-Atp10a as a Biomarker for Multiple Myeloma Angiogenesis. Bioengineered (2022) 13(1):667–83. doi: 10.1080/21655979.2021.2012553
117. Henry NL, Hayes DF. Cancer Biomarkers. Mol Oncol (2012) 6(2):140–6. doi: 10.1016/j.molonc.2012.01.010
118. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of Circrnas in the Tumour Microenvironment. Mol Cancer (2020) 19(1):14. doi: 10.1186/s12943-019-1125-9
Keywords: circRNAs, hematological malignancies, biomarkers, oncogenes, proliferation, apoptosis
Citation: Du J, Jia F and Wang L (2022) Advances in the Study of circRNAs in Hematological Malignancies. Front. Oncol. 12:900374. doi: 10.3389/fonc.2022.900374
Received: 20 March 2022; Accepted: 23 May 2022;
Published: 20 June 2022.
Edited by:
Simona Soverini, University of Bologna, ItalyReviewed by:
Sabrina Strano, Hospital Physiotherapy Institutes (IRCCS), ItalyGuillermo Aquino-Jarquin, Hospital Infantil de México Federico Gómez, Mexico
Copyright © 2022 Du, Jia and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Wang, d2FuZ2xqNzMwQDE2My5jb20=; Feiyu Jia, QzEzMjkwMjA5ODY2QDE2My5jb20=
 Jingyi Du
Jingyi Du Feiyu Jia3*
Feiyu Jia3* Lijuan Wang
Lijuan Wang