- 1Department of Radiation Oncology, The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou, China
- 2School of Computer Science and Artificial Intelligence, Changzhou University, Changzhou, China
- 3Key Laboratory of Computer Network and Information Integration, Southeast University, Nanjing, China
- 4Jiangsu Province Engineering Research Center of Medical Physics, Changzhou, China
- 5School of Electrical and Information Engineering, Changzhou Institute of Technology, Changzhou, China
- 6Laboratory of Image Science and Technology, Southeast University, Nanjing, China
- 7Centre de Recherche en Information Biomédicale Sino-francais, Rennes, France
- 8Jiangsu Provincial Joint International Research Laboratory of Medical Information Processing, Southeast University, Nanjing, China
Prostate cancer diagnosis is performed under ultrasound-guided puncture for pathological cell extraction. However, determining accurate prostate location remains a challenge from two aspects: (1) prostate boundary in ultrasound images is always ambiguous; (2) the delineation of radiologists always occupies multiple pixels, leading to many disturbing points around the actual contour. We proposed a boundary structure-preserving U-Net (BSP U-Net) in this paper to achieve precise prostate contour. BSP U-Net incorporates prostate shape prior to traditional U-Net. The prior shape is built by the key point selection module, which is an active shape model-based method. Then, the module plugs into the traditional U-Net structure network to achieve prostate segmentation. The experiments were conducted on two datasets: PH2 + ISBI 2016 challenge and our private prostate ultrasound dataset. The results on PH2 + ISBI 2016 challenge achieved a Dice similarity coefficient (DSC) of 95.94% and a Jaccard coefficient (JC) of 88.58%. The results of prostate contour based on our method achieved a higher pixel accuracy of 97.05%, a mean intersection over union of 93.65%, a DSC of 92.54%, and a JC of 93.16%. The experimental results show that the proposed BSP U-Net has good performance on PH2 + ISBI 2016 challenge and prostate ultrasound image segmentation and outperforms other state-of-the-art methods.
Introduction
Prostate cancer is the most common cancer among American men. The American Cancer Society estimated about 191,930 new cases of prostate cancer and about 33,330 deaths from prostate cancer in 2020 (1).
Ultrasound images can be applied for diagnosis and guide puncture and radiotherapy (2). The accurate delineation of the prostate boundary in ultrasound images is crucial for intraoperative navigation to help medical physicists operate, especially in ultrasound-guided puncture. Ultrasound-guided puncture as one of the monitoring means can reflect prostate deformation in real time.
In the era of deep learning, many convolutional neural network (CNN)-based segmentation approaches have been proposed for medical image segmentation. Fully convolutional network (FCN) is an innovative network for semantic segmentation (3). U-Net is the first CNN-based network with skip layers for biomedical image segmentation and is employed for biological microscopy images (4). 3D U-Net extends the typical U-Net architecture by replacing all 2D operations with their 3D counterparts and is applied in the volumetric segmentation of sparsely annotated volumetric images (5). U-Net++ is a new, more powerful architecture for medical image segmentation (6). The architecture of U-Net++ is a deeply supervised encoder–decoder network, where the encoder and decoder sub-networks are connected through a series of nested, dense skip pathways. The re-designed skip pathways aim at reducing the semantic gap between the feature maps of the encoder and decoder sub-networks. V-Net is employed for 3D image segmentation based on a volumetric fully convolutional neural network (FCNN) (7). The authors introduce a novel objective function based on Dice similarity coefficient (DSC). Progressive Dense V-net (PDV-Net) was employed as a 3D-CNN encoder for fast and automatic segmentation (8) to deal with situations where the number of foreground and background voxels has a strong imbalance. These U-Net architecture networks are useful for many modalities of medical image segmentation. However, they can hardly be used directly for ultrasound image segmentation because of the several defects of ultrasound imaging (e.g., attenuation, speckle, signal drop-out, low contrast, and signal shadowing).
Shape information and boundary information play a critical role in ultrasound image segmentation (9–14). Shape models bring essential information, particularly in prostate segmentation, because the anatomical structure of a healthy prostate is more likely an ellipse shape. Gong et al. (15) proposed a deformable super-ellipse model that drives shape evolution by an efficient and robust Bayesian segmentation algorithm. Badiei et al. (16) utilized image warping and ellipse fitting for prostate ultrasound segmentation. Shen et al. (17) used a statistical shape model that adopts normalized features to make prostate shape invariant to probe rotation. The experimental results of the aforementioned works showed that incorporating tissue shape information can improve segmentation accuracy. As a kind of shape model, the active shape model (ASM) can describe various shapes based on a mean position and variant modes (18). Hodge et al. extended ASM from 2D to 3D ultrasound prostate image segmentation (19). Some methods that combine ASM with other models have been proposed to improve the segmentation accuracy. Yan et al. proposed a discrete deformable model guided by partial ASM for transrectal ultrasound image segmentation (20). Bi et al. proposed a fast and accurate segmentation method using ASM with Rayleigh mixture model clustering for prostate ultrasound images (21).
Remarkably, authors started to combine shape information and deep learning. Mishra et al. proposed an FCNN with attention to boundaries conducted on the MICCAI 2011 IVUS challenge dataset resulting in a Dice index value of 0.91 (22). Chen et al. developed a new model based on deep learning, which takes into account boundary information (23). Murugesan et al. proposed a Psi-Net that joins shape and boundary into a multitask deep network that aids in ensuring the smoothness of segmentation prediction (24). Nguyen et al. proposed a consecutive deep encoder–decoder network combined with a boundary-emphasization data augmentation (25). Hou et al. proposed a robust 3D CNN with boundary correction (26). Soliman et al. proposed a novel CNN segmentation framework based on shape features described by the seventh-order Markov–Gibbs random field, which reached a high DSC of 98.37% ± 0.68% on 95 CT lung images (27). Hesse et al. improved U-Net segmentation by adding an active contour step to correct the imperfect ground-truth labels (28). Qin et al. combined superpixel and boundary information with CNNs for liver segmentation (29). Lee et al. proposed a novel image segmentation network for medical images with ambiguous boundaries (30). In summary, boundary information is helpful to improve segmentation accuracy by combining it with a neural network.
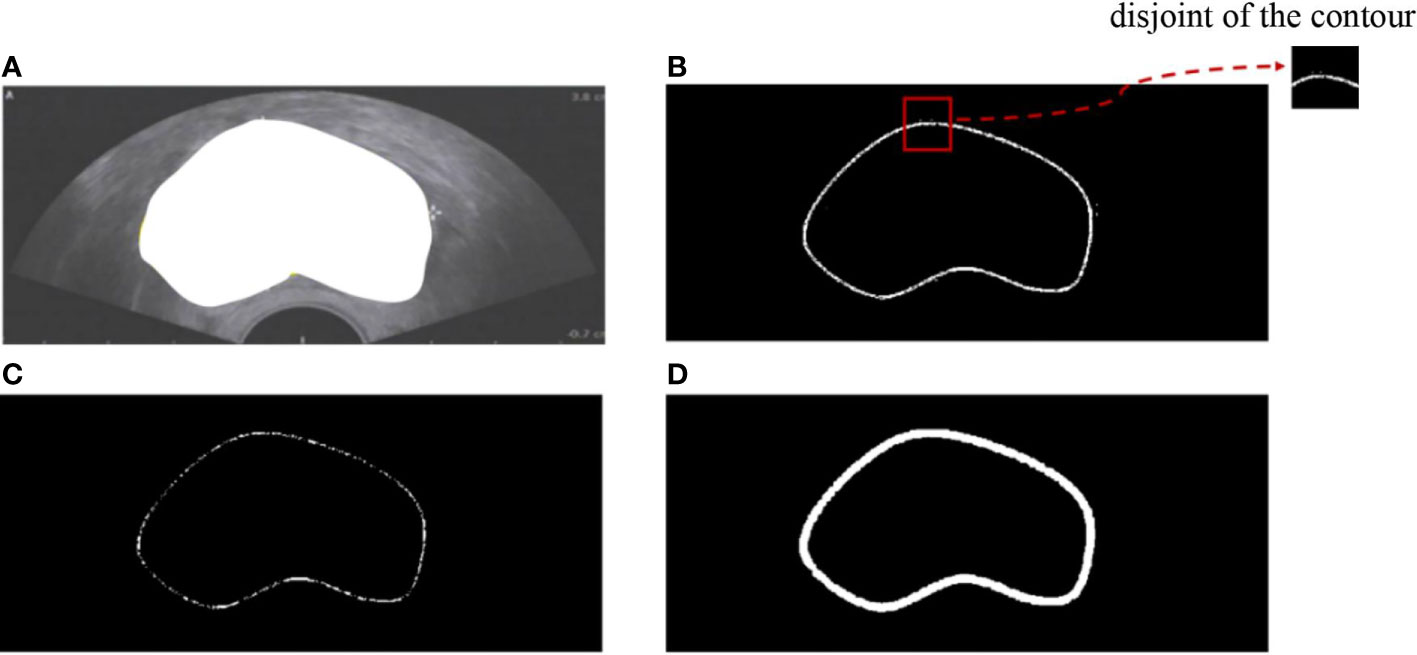
However, boundary representation remains a challenge in two aspects: (1) the contour of the existing dataset is implicit because the ground truth always provides semantic segmentation (Figure 1A), and (2) the actual contour is surrounded by many disturbing points (Figure 1E). In our work, we proposed a model that combines boundary information with deep learning for prostate ultrasound image segmentation. The contributions of our work are as follows: (1) we built an end-to-end neural network for prostate ultrasound segmentation; (2) an ASM-based method was utilized for boundary information extraction; (3) the key point selection models were easy to plug into any network. This paper is organized as follows. Section 2 presents the mathematical background of ASM. Section 3 describes the selection of the key points of initialization. Section 4 provides the experimental results, and Section 5 presents the concluding remarks.
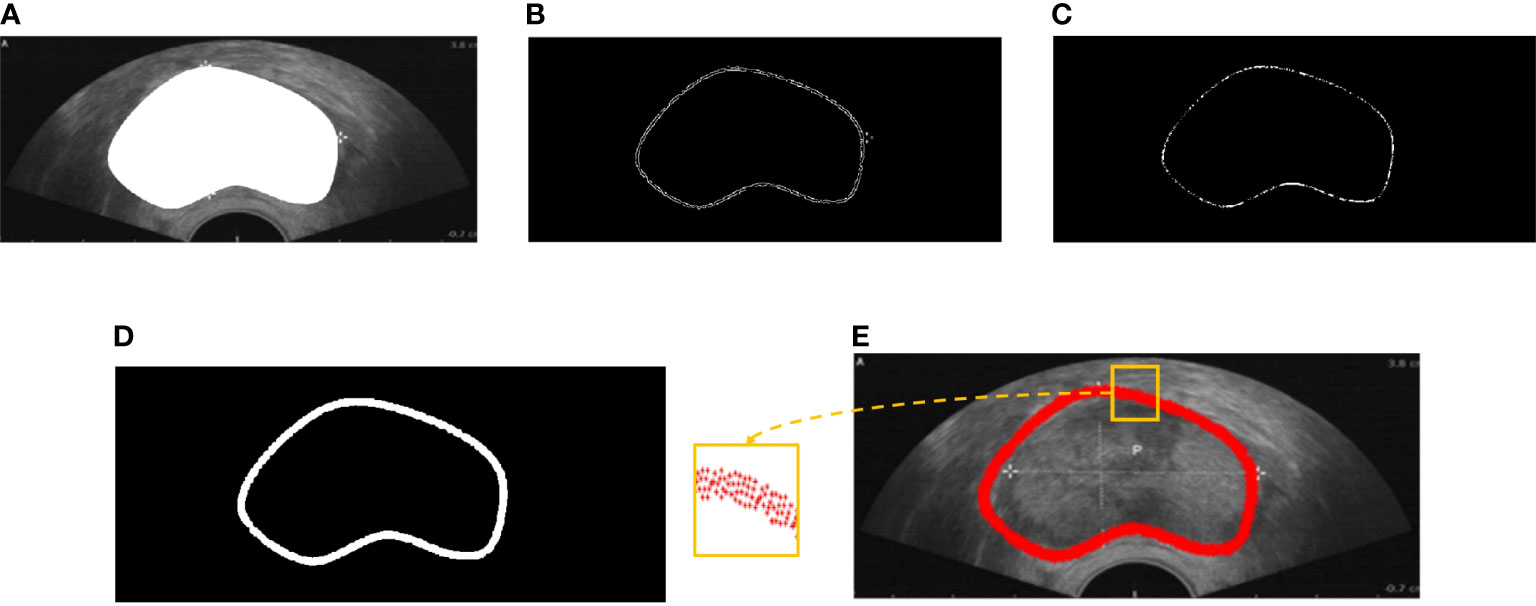
Figure 1 (A) Semantic segmentation of prostate ultrasound image. (B) Edge detection of the prostate. (C) Prostate boundary after erosion. (D) Erode prostate boundary after dilation. (E) Prostate contour.
Materials and methods
Materials
Two medical image segmentation datasets, namely, PH2 + ISBI 2016 Skin Lesion Challenge (31, 32) and our private dataset, were applied in the experiments. The PH2 + ISBI 2016 dataset, which is a publicly available dataset for evaluating skin lesion segmentation, includes 900 multi-size skin lesion images and 200 dermoscopic images. The private dataset is from the Second People’s Hospital of Changzhou Affiliated with Nanjing Medical University, which was approved by the Ethics Committee of the Second People’s Hospital of Changzhou. All subjects agreed to participate in the study and signed the informed consent. All data, including 100 prostate images, are desensitization data. The ground truth was based on the average delineation from three radiologists.
Methods
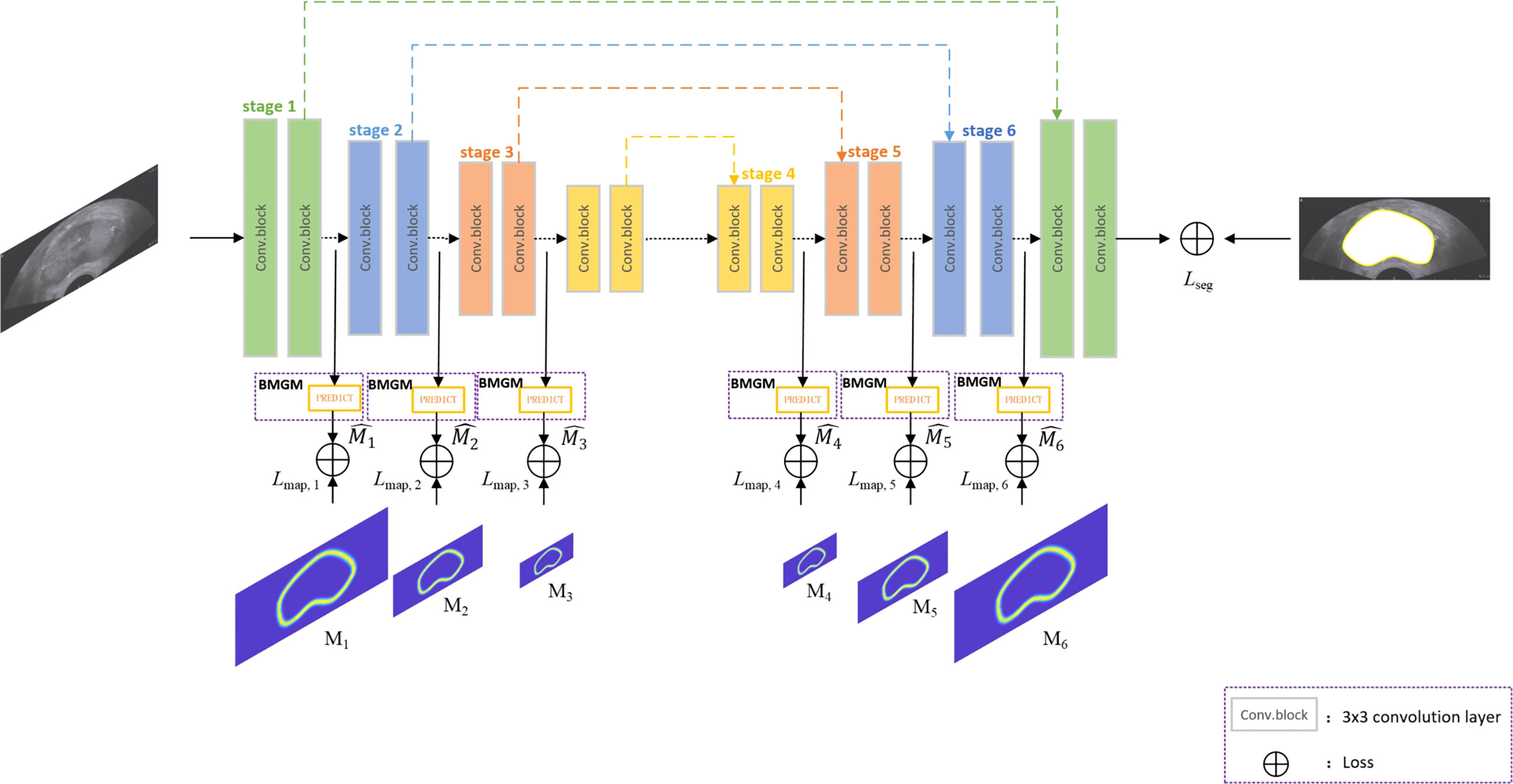
Figure 2 shows the overview of the proposed network architecture. It consists of one encoder–decoder network and a boundary map generation module (BMGM). The BMGM was utilized to incorporate boundary information in different scales of the image within the network.
Boundary map generation module
Figure 1E shows that the points extracted from semantic segmentation have two deficiencies: (1) in each dimension, several points make key points implicit; (2) the points are chaotic. Hence, we proposed a BMGM to alleviate this situation.
First, rough prostate boundary detection was processed, including binarization, edge extraction, and morphological operation. The adaptive threshold binarization of the Otsu algorithm and the edge extraction of the Canny algorithm were adopted for ultrasound images as shown in Figure 1B. A morphological operation consisting of erosion and dilation was conducted to remove the disturbing points and connect the isolated points (Figures 1C, D). Then, the rough boundary of the prostate was detected, and its pixel points formed the rough point set.
Second, prostate boundary finer was processed, including key point initialization and interpolation generation. Key point initialization is adopted to eliminate the disturbance of messy points. As shown in Figure 3A, the green circles denote only two points left on each vertical axis. Then, four salient points {kp11, kp12, kp13, kp14} were used as the initialization to demonstrate the prostate shape, where footnote 1 indicates the first iteration selection, 4 indicates the order of the points in the set (Figure 3B), and the red stars denote the key point set of the boundary. Then, interpolation generation was adopted to supplement the shape points to finish the finer contour detection. The interpolation needs to find the perpendicular direction of any two adjacent points. The normal direction of the perpendicular line connected to two points was achieved using Eq. (1):
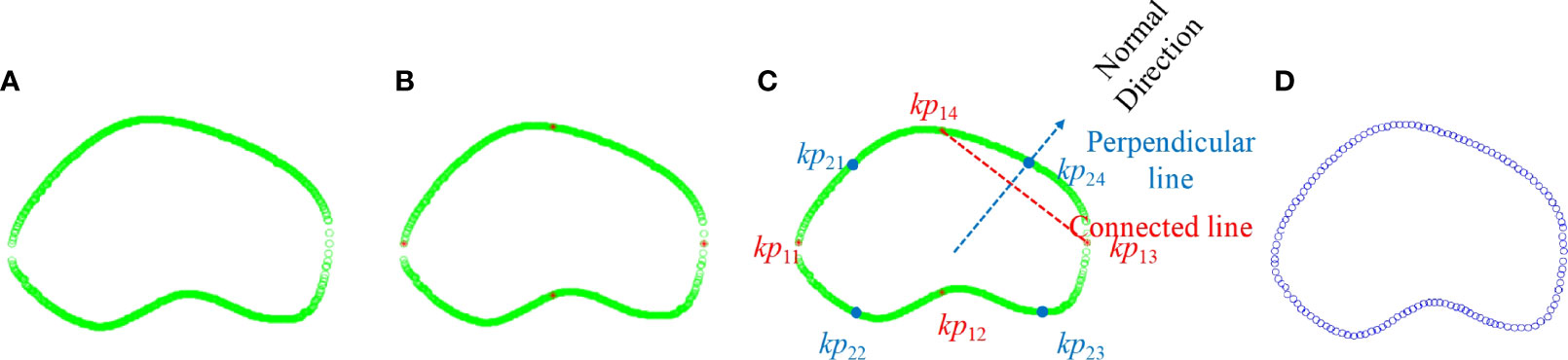
Figure 3 Illustration of candidate point selection and finer boundary detection. (A) Green—point selection after extra point removal; red—key points. (B) Detection of four salient points. (C) Interpolation of the first iteration. (D) Finer contour.
where Dxand Dy are the coordinates in the x-axis and y-axis, respectively; x1, y1 are the coordinates of point 1; and x2, y2 are the coordinates of point 2. Taking kp13 and kp14 as examples, the red line denotes the connected line, whereas the blue line denotes the perpendicular line. The point was interpolated through the candidate points. Euclidean distance was used to select the candidate points so that kp24 was added to the point set as shown in Figure 3C. Finally, we achieved the key points as Figure 3D shows.
Network Backbone
As an attractive structure-preserving method, Lee’s model (30) selects an optimal solution from a set of random sampling on the target contour, but the extracted points are not evenly distributed. Our proposed work generates the key points over the boundary map generation module, called the BMGM, to obtain a more uniform repartition of the key points along the contour. As the replacement of the random sampling module of Lee’s model, the BMGM is proposed to provide a more accurate information on the tissue boundary in our segmentation network. The BMGM module is utilized to evaluate the intermediate segmentation at the different stages of the network after the convolutional block. The prediction at different scales is used to restrain the segmentation stage. The prediction module was constructed directly by an upsampling block.
The network is a typical U-Net architecture that has encoder and decoder parts besides skip layers as shown in Figure 2. The encoder part is used for semantic feature extraction, and the decoder part helps with generator segmentation. The BMGM was placed after the convolution block. In the network, the BMGM was utilized to generate the predicted map to restrain the intermediate segmentation. The BMGM was used in the encoder and decoder parts. The BMGM at different scales was used to compare the stage with the ground-truth map.
We utilized cross-entropy to evaluate the stage with the ground truth and restrain the segmentation with the true boundary in different stages. The map loss Lmap is defined as:
where Mi is the boundary map generated by the ground truth, and is the map achieved by the network in the ith stage.
We also used cross-entropy to evaluate the segmentation result with the ground truth and restrain the segmentation with the true boundary. Segment loss Lseg is defined as:
where G is the ground truth, and S is the segmentation achieved by the network.
Total loss Ltotal is defined as follows:
Experimental implementation
BMGM implementation details
In the coarse boundary step, we detected the edge of the prostate based on Otsu’s algorithm. Then, all-one template size 2×2 for image erode and disk template size 5×5 for image dilate were implemented (Figure 4). Figure 4A depicts the semantic segmentation of the prostate. The edge of the prostate can be detected through image binarization as shown in Figure 4B. Figure 4B shows that more than two points along each dimension are disturbing points. Thus, image erosion was implemented to solve this issue. Figure 4 illustrates that the points were reduced along each dimension; however, the operation led to some disconnection along the contour. Image dilation was implemented to make contour connections as shown in Figure 4D. The closed contour is smooth and can be seen as the coarse contour of the prostate.
The proposed method was compared with the point selection method proposed in (30) to evaluate the proposed method qualitatively. We conducted experiments on 100 prostate ultrasound images. Four representative demos were used as Figure 5 shows. The first column represents the semantic segmentation responding to each ultrasound image. The second column is the point selection based on Lee’s method (30). As the figure shows, some overlap points are present inside the red box, and disconnection occurred along the boundary. The third column shows the points selected by the proposed method. The points have an equidistant distribution along the boundary and do not overlap.
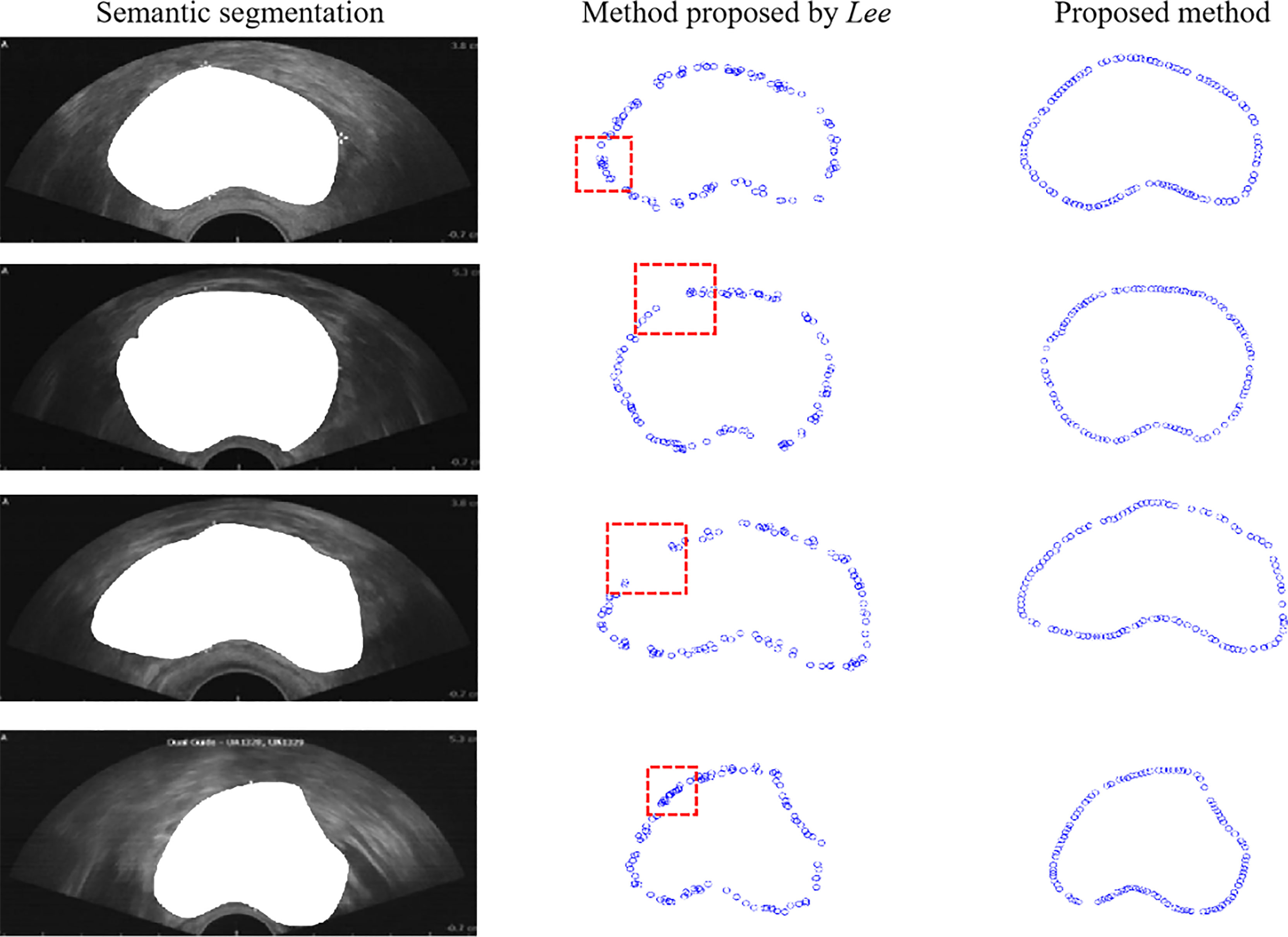
Figure 5 Comparison of boundary detection. The first column is the original image, the second column is the boundary detection based on Lee’s method, and the third column is the boundary detection based on the proposed method.
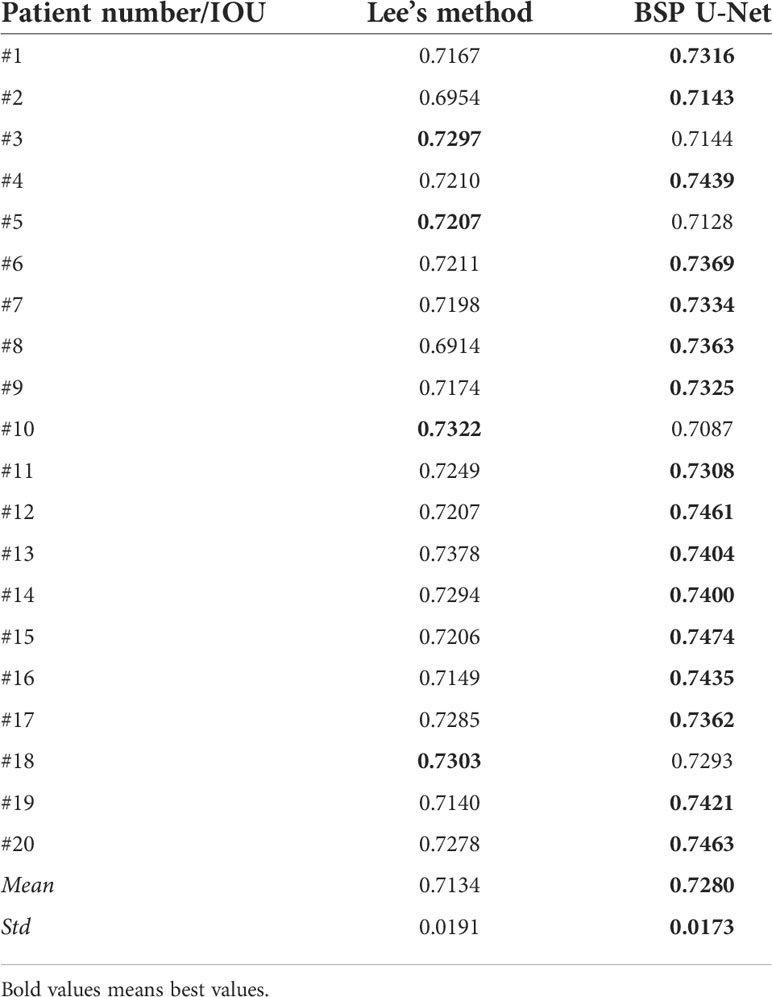
Furthermore, we used the intersection over union (IOU) to evaluate the proposed method quantitatively using Eq. (5), where SL denotes the region generated by the points based on Lee’s method and SP denotes the region generated by the points based on the proposed method. IOU equals the ratio of the overlap of SL and SP to the union of SL and SP. We calculated the mean and variance of 100 prostate ultrasound images, and representative images are shown in Table 1. Most key points generated based on the proposed method showed higher IOU than those generated by Lee’s method. Furthermore, the mean (0.7280) and variance (0.0173) of the proposed method were higher than those of Lee’s method (0.7134 and 0.0191, respectively).
BSP U-Net Implementation Details
All the experiments were implemented in TensorFlow 1.4.0 with an NVIDIA GeForce RTX 2080Ti GPU. We used a robust model training schema by 10-fold cross-validation and randomly shuffled all images. We trained the model by Adam optimization algorithm with an initial learning rate of 1e−4, a maximum epoch of 1,200, and a batch size of 8. The automatically saved optimal model was used to evaluate the testing set.
We evaluated the proposed method by the test set. We trained and tested other classic deep-learning based models, including U-Net (27), FCN (3), and Lee’s network (30), using the same dataset in the paper to evaluate the performance of our network objectively.
Discussion
Structure information is crucial for medical image segmentation within a deep learning network. Derived from the capability of ASM-based methods to capture the shape variability of the prostate within an ultrasound image, we employed ASM to generate a key point map from the delineated boundary based on physicians.
Though several methods are proposed to generate shape information, the demonstration of the prostate in ultrasound images is still a challenge because of the inhomogeneous images. Lee proposed a boundary key point selection algorithm for a selected point set to demonstrate the target object. In our work, we considered the prostate boundary as an ASM model to demonstrate an inhomogeneous prostate. ASM is applied to generate the key points of a prostate boundary to achieve more homogeneous tissue information, which is helpful for ultrasound image segmentation. This assumption was verified through experiments about plugging a key point selection module based on ASM in a CNN. The key point selection module based on ASM is simple to implement and conveniently attached to a network in a plug-and-play manner.
The proposed ASM-based method outperformed Lee’s method in terms of IOU. Compared with Lee’s method, the mean intersection over union (MIOU) of the proposed ASM-based method was improved by 1.3%, and the standard deviation declined by 9.4%. The larger mean IOU means higher accuracy, and the smaller standard deviation means more stability. The reason the proposed method showed better performance is that BMGM re-partitions the key points along the prostate contour. Compared with Lee’s method, BSP U-Net had better constraints on prostate shape.
In terms of segmentation results, the proposed ASM-based method’s key point map generation plugin (traditional U-net network) achieved considerable accuracy. We conducted experiments on two datasets. PH2 data: Compared with SCDRR, the segmentation accuracy of our method improved by 11.56% for DSC and 16.55% for Jaccard coefficient (JC) as Table 2 shown. Compared with JCLMM, the segmentation accuracy of our method improved by 15.80% for DSC. Compared with MSCA, the segmentation accuracy of our method improved by 17.62% for DSC and 22.47% for JC. Compared with SSLS, the segmentation accuracy of our method improved by 22.40% for DSC and 29.36% for JC. Compared with FCN, the segmentation accuracy of our method improved by 7.30% for DSC and 7.82% for JC. Compared with Bi, the segmentation accuracy of our method improved by 5.82% for DSC and 5.46% for JC. Compared with Lee, the segmentation accuracy of our method improved by 4.46% for DSC, and 5.08% for JC. Prostate ultrasound data: Compared with FCN, the segmentation accuracy of our method improved by 2.93% for pixel accuracy (PA), 4.84% for MIOU, 8.39% for DSC, and 5.16% for JC. Compared with U-Net, the segmentation accuracy of our method improved by 1.69% for PA, 2.95% for MIOU, 5.70% for DSC, and 3.27% for JC as Table 3 shown. Compared with Lee, the segmentation accuracy of our method improved by 0.84% for PA, 1.42% for MIOU, 2.05% for DSC, and 1.74% for JC. The reason is that the proposed method had a better performance in homogeneous tissue presentation to achieve more accurate segmentation results.
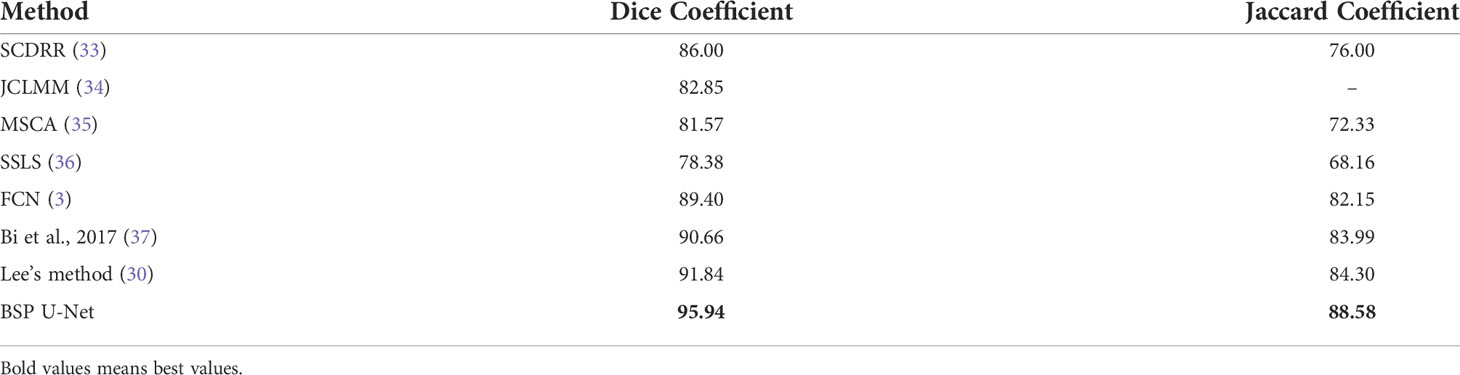
Table 2 Evaluation of U-Net, FCN, Lee’s network, and the proposed network on PH2 + ISBI 2016 challenge.

Table 3 Evaluation of U-Net, FCN, Lee’s network, and the proposed network on prostate ultrasound images.
Furthermore, we compared the performance of using a boundary mask (Figure 4A) and a boundary-preserving module (Figure 4D). The results are shown in Table 4. Compared with using a boundary mask, the segmentation accuracy of using boundary sample points improved by 10.56% for PA, 11.73% for MIOU, 5.39% for DSC, and 10.42% for JC. The reason is that boundary-preserving points are more adaptive to curve changes, which represent better constraints on prostate shape.

Table 4 Evaluation of using boundary mask and boundary-sampled points on prostate ultrasound images.
We also conducted experiments using boundary-preserving modules in the encoder layer, decoder layer, and both layers. The results are shown in Table 5. The performance of boundary preservation in both layers was better than that in the encoder layer; the segmentation accuracy improved by 2.14% for PA, 3.26% for MIOU, 3.28% for DSC, and 2.08% for JC. The boundary preservation performance in the encoder layer was better than that in the decoder layer; the segmentation accuracy improved by 2.57% for PA, 1.45% for MIOU, 1.44% for DSC, and 2.63% for JC. The reason is that the decoder layer also can obtain boundary information using boundary preservation in the encoder layer. The shape constraint is augmented into the information forward transmission.
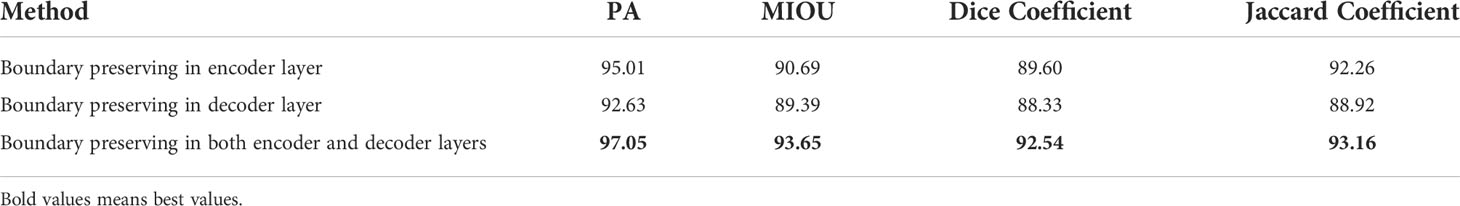
Table 5 Evaluation of using BSP U-Net in the encoder layer, decoder layer, and both layers on prostate ultrasound images.
This work focused on tumor and gland segmentation for ultrasound images with smooth and non-convex targets. The generation of key points would drop some points when encountering the bottleneck. In the future, we will conduct further research on tissues containing a bottleneck and to make our method effective on convex and non-smooth objects. In addition, considering the relatively small dataset of ultrasound images, the 10-fold cross-validation was adopted to evaluate the proposed method. In the future, for further evaluation of the proposed method, we will collect more kinds of ultrasound images and promote the proposed method for use in other organs, such as the thyroid, uterus, and liver.
Conclusion
BSP U-Net was proposed to obtain accurate prostate location during ultrasound-guided puncture. The ASM-based method was applied for key point selection in the segmentation of prostate ultrasound images from coarse to fine points. The BMGM is easy to plug into any network. The experimental results show that the proposed BSP U-Net has good performance on prostate ultrasound image segmentation in terms of several evaluation indexes.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://challenge.isic-archive.com/data/.
Author contributions
All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 62171125, No. 62141401), the 67th National Postdoctoral Program (Grant No. 2020M671277), the Natural Science Foundation of Jiangsu Province (Grant No. BK20190159), the Science and Technology Project of Changzhou City (Grant No. CE20215045), the Key Laboratory of Computer Network and Information Integration (Southeast University) of the Ministry of Education (Grant No. K93-9-2021-08), and the Key Medical Physics Laboratory of Changzhou (Grant No. CM20193005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. “American cancer society,” . Available at: http://www.cancer.org.
2. Sun H, Ni X, Yang J. Research status of ultrasound-guided radiotherapy for tumors. Chin J Radiat Oncol (2020) 29(04):317–20. doi: 10.3760/cma.j.cn113030-20181125-00014
3. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Conf Comput Vision Pattern Recognit (2015), 3431–40. doi: 10.1109/CVPR.2015.7298965
4. Ronneberger O, Fischer P, Brox T. (2015). U-Net: convolutional networks for biomedical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, . pp. 234–41. Springer
5. Çiçek Ö, Abdulkadir A, Lienkamp SS. (2016). 3d u-net: learning dense volumetric segmentation from sparse annotation, in: International Conference on Medical Image Computing and Computer Assisted Intervention, . pp. 424–32. Cham
6. Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. U-Net++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans Med Imag (2019) 39(6):1856–67. doi: 10.1109/TMI.2019.2959609
7. Milletari F, Navab N, Ahmadi SA. (2016). V-Net: Fully convolutional neural networks for volumetric medical image segmentation, in: 2016 Fourth International Conference on 3D Vision, . pp. 565–71. IEEE
8. Imran AAZ, Hatamizadeh A, Ananth SP, Ding X, Terzopoulos D, Tajbakhsh N. Automatic segmentation of pulmonary lobes using a progressive dense v-network. Deep Learn Med Imag Anal Multimo Learn Clin Dec Sup (2018), (10045):282–90. doi: 10.1007/978-3-030-00889-5_32
9. Nobel JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans Med Imag (2006) 25(8):987–1010. doi: 10.1109/TMI.2006.877092
10. Van Ginneken B, Frangi AF, Staal JJ, Ter Haar Romeny BM, Viergever MA. Active shape model segmentation with optimal features. IEEE Trans Med Imag (2002) 21(8):924–33. doi: 10.1109/TMI.2002.803121
11. Aarnink RG, Giesen RJB, Huynen AL, de la Rosette JJ, Debruyne FM, Wijkstra H. A practical clinical method for contour determination in ultrasonographic prostate images. Ultrasound Med Biol (1994) 20(8):705–17. doi: 10.1016/0301-5629(94)90028-0
12. Richard WD, Keen CG. Automated texture-based segmentation of ultrasound images of the prostate. Computer Med Imaging Graph (1996) 20(3):131–40. doi: 10.1016/0895-6111(96)00048-1
13. Choy MM, Jin JS. Morphological image analysis of left-ventricular endocardial borders in 2D echocardiograms. SPIE Proc Med Imag (1996) 2710:852–63. doi: 10.1117/12.237991
14. Lei T, Wang R, Wan Y, Zhang B, Meng H, Nandi AK. Medical image segmentation using deep learning: a survey. ArXiv Prep (2009), 13120. doi: 10.48550/arXiv.2009.13120
15. Gong L, Pathak SD, Haynor DR, Cho PS, Kim Y. Parametric shape modeling using deformable superellipses for prostate segmentation. IEEE Trans Med Imag (2004) 23(3):340–9. doi: 10.1109/TMI.2004.824237
16. Badiei S, Salcudean SE, Varah J, Morris WJ. Prostate segmentation in ultrasound images using image warping and ellipsoid fitting. Int Conf Med Imag Comp Comput Ass Interv (2006) 4191:17–24. doi: 10.1007/11866763_3
17. Shen D, Zhan Y, Davatzikos C. Segmentation of prostate boundaries from ultrasound images using statistical shape model. IEEE Trans Med Imag (2003) 22(4):539–51. doi: 10.1109/TMI.2003.809057
18. He P, Zheng J. Segmentation of tibia bone in ultrasound images using active shape models. Annu Int Conf IEEE Eng Med Biol Soc (2001) 3:2712–5. doi: 10.21236/ADA412425
19. Hodge AC, Fenster A, Downey DB, Ladak HM. Prostate boundary segmentation from ultrasound images using 2d active shape models: optimization and extension to 3d. Comput Methods Prog Biomed (2006) 84(2-3):99–113. doi: 10.1016/j.cmpb.2006.07.001
20. Yan P, Xu S, Turkbey B, Kruecker J. Discrete deformable model guided by partial active shape model for trus image segmentation. IEEE Trans Biomed Engineer (2010) 57(5):1158–66. doi: 10.1109/TBME.2009.2037491
21. Bi H, Jiang Y, Tang H, Yang G, Shu H, Dillenseger JL. Fast and accurate segmentation method of active shape model with rayleigh mixture model clustering for prostate ultrasound images. Comput Methods Prog Biomed (2020) 184:105097. doi: 10.1016/j.cmpb.2019.105097
22. Mishra D, Chaudhury S, Sarkar M, Soin AS. Ultrasound image segmentation: a deeply supervised network with attention to boundaries. IEEE Trans Biomed Engineer (2018) 66(6):1637–48. doi: 10.1109/TBME.2018.2877577
23. Chen X, Williams BM, Vallabhaneni SR, Czanner G, Williams R, Zheng Y. Learning active contour models for medical image segmentation. Proc IEEE Conf Comput Vision Pattern Recognit (2019), 11632–40. doi: 10.1109/CVPR.2019.01190
24. Murugesan B, Sarveswaran K, Shankaranarayana SM, Ram K, Joseph J, Sivaprakasam M. Psi-net: shape and boundary aware joint multi-task deep network for medical image segmentation. Annu Int Conf IEEE Eng Med Biol Soc (2019), 7223–6. doi: 10.1109/EMBC.2019.8857339
25. Nguyen NQ, Lee SW. Robust boundary segmentation in medical images using a consecutive deep encoder-decoder network. IEEE Acc (2019) 7:33795–808. doi: 10.1109/ACCESS.2019.2904094
26. Hou B, Kang G, Zhang N, Hu C. Robust 3D convolutional neural network with boundary correction for accurate brain tissue segmentation. IEEE Acc (2018) 6:75471–81. doi: 10.1109/ACCESS.2018.2882848
27. Soliman A, Shaffie A, Ghazal M, Gimel’farb G, Keynton R, EI-Baz A. A novel cnn segmentation framework based on using new shape and appearance features. IEEE Int Conf Imag Process (2018), 3488–92. doi: 10.1109/ICIP.2018.8451534
28. Hesse LS, Namburete AIL. Improving u-net segmentation with active contour based label correction. Springer Cham Annu Conf Med Imag Understanding Anal (2020), 69–81. doi: 10.1007/978-3-030-52791-4_6
29. Qin W, Wu J, Han F, Yuan Y, Zhao W, Ibragimov B. Superpixel-based and boundary-sensitive convolutional neural network for automated liver segmentation. Phys Med Biol (2018) 63(9):095017. doi: 10.1088/1361-6560/aabd19
30. Lee HJ, Kim JU, Lee S, Kim HG, Ro YM. Structure boundary preserving segmentation for medical image with ambiguous boundary. IEEE Conf Comput Vision Pattern Recognit (2020), 4817–26. doi: 10.1109/CVPR42600.2020.00487
31. Mendonca T, Ferreira PM, Marques JS, Marcal AR, Rozeira J. Ph2 - a dermoscopic image database for research and benchmarking. Annu Int Conf IEEE Eng Med Biol Soc (2012), 5437–40. doi: 10.1109/EMBC.2013.6610779
32. Gutman D, Codella NC, Celebi E, Helba B, Marchetti M, Mishra M. Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging, hosted by the international skin imaging collaboration. ArXiv Prep (2016), 1605.01397. doi: 10.1109/ISBI.2018.8363547
33. Bozorgtabar R, Abedini M, Garnavi R. Sparse coding based skin lesion segmentation using dynamic rule-based refinement. Springer Int Workshop Mach Learn Med Imag (2016), 254–61. doi: 10.1007/978-3-319-47157-0_31
34. Roy A, Pal A, Garain U. Jclmm: A finite mixture model for clustering of circular-linear data and its application to psoriatic plaque segmentation. Pattern Recognit (2017) 66:160–73. doi: 10.1016/j.patcog.2016.12.016
35. Bi L, Kim J, Ahn E, Feng D, Fulham M. (2016). Automated skin lesion segmentation via image-wise supervised learning and multi-scale superpixel based cellular automata, in: IEEE 13th International Symposium on Biomedical Imaging, . pp. 1059–62. IEEE
36. Ahn E, Bi L, Jung YH, Kim J, Li CY, Fulham M, et al. Automated saliency-based lesion segmentation in dermoscopic images. IEEE Eng Med Biol Soc (2015) 3009-3012. doi: 10.1109/EMBC.2015.7319025
Keywords: prostate ultrasound image segmentation, ASM-based key points selection, U-Net architecture, deep learning, shape prior
Citation: Bi H, Sun J, Jiang Y, Ni X and Shu H (2022) Structure boundary-preserving U-Net for prostate ultrasound image segmentation. Front. Oncol. 12:900340. doi: 10.3389/fonc.2022.900340
Received: 20 March 2022; Accepted: 30 June 2022;
Published: 28 July 2022.
Edited by:
Xiang Li, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Sekeun Kim, Yonsei University, South KoreaJerome Charton, Massachusetts General Hospital and Harvard Medical School, United States
Copyright © 2022 Bi, Sun, Jiang, Ni and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinye Ni, bnh5QG5qbXUuZWR1LmNu
 Hui Bi
Hui Bi Jiawei Sun4
Jiawei Sun4 Xinye Ni
Xinye Ni