- 1Jiangsu Cancer Hospital, Jiangsu Institute of Cancer Research, The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, China
- 2Research Division of Clinical Pharmacology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 3Department of Urology, Children’s Hospital of Nanjing Medical University, Nanjing, China
The roles of the microbiome in human beings have become clearer with the development of next-generation sequencing techniques. Several pieces of evidence showed strong correlations between the microbiome and human health and disease, such as metabolic disorders, infectious diseases, digestive system diseases, and cancers. Among these diverse microbiomes, the role of bacteria in human cancers, especially in cancer cells, has received extensive attention. Latest studies found that bacteria widely existed in cancers, mainly in cancer cells and immune cells. In this review, we summarize the latest advances in understanding the role of bacteria in human cancer cells. We also discuss how bacteria are transported into cancer cells and their physiological significance in cancer progression. Finally, we present the prospect of bacterial therapy in cancer treatment.
Introduction
Human microbiome is composed of bacteria, viruses, eukaryotic fungi, and protozoa (1). The roles of the microbiome in humans have become clearer with the development of next-generation sequencing techniques (2, 3). In healthy conditions, the human host and microbiome are symbiotic. The human host can provide a nutritious microenvironment for microbiome to help the human host with the metabolism and digestion process (4, 5). Several pieces of evidence showed strong correlations between the microbiome and human disease, such as metabolic disorders (6), infectious diseases (7), digestive system diseases (8), and cancers (9). Many microorganisms were related to the development of human cancers, such as bacteria and viruses, including human papillomavirus (HPV), hepatitis B virus (HBV), hepatitis C virus (HCV), Epstein–Barr virus (EBV) (9), and human endogenous retroviruses K (HERV-K) (10). There are three trillion bacterial members in human bodies, which can regulate the comprehensive interplay of physiological processes and disease susceptibilities (11). The high genetic diversity of bacteria encodes excellent mechanistic and metabolic competences that not only influence their own microbial niche but also regulate host tissue-specific and immune cell functions (12). Bacteria may also have health benefits, such as producing metabolites to fight cancers (13, 14). Butyrate, which is metabolized by intestinal bacteria, provides energy sources for colonocytes and suppresses inflammation and carcinogenesis by affecting immunity, gene expression, and epigenetic modification (14). Some bacteria, such as Lactococcus, Clostridia, Shigella, Bifidobacteria, Listeria, Vibrio, Salmonella, and Escherichia, have demonstrated significant potential for invasion and colonization in tumors sites, thereby leading to tumor clearance and retardation of neoplasm growth. Moreover, some bacteria such as Clostridia strains and Bifidobacterium longum are able to survive and colonize in the hypoxic condition of the tumor to destroy it (15, 16).
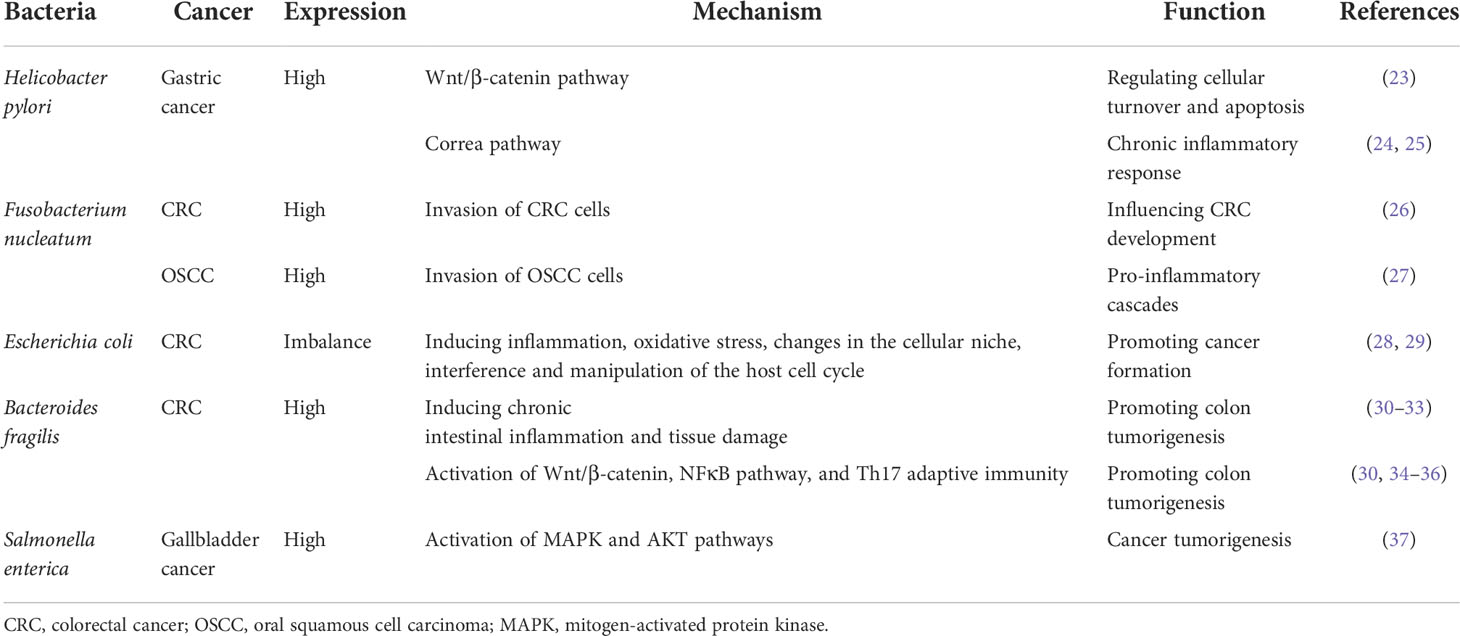
Recently, the role of bacteria in cancer development has drawn more attention (17). A well-established link between bacteria and human cancer is Helicobacter pylori and gastric cancer (18). The cytotoxin-associated gene A (CagA) produced by H. pylori is one of the first bacterial proteins identified to promote human cancer. CagA could activate the oncogenic signal transduction pathways, interfere with cell cycle and cell death, and increase the risk of gastric cancer (19, 20). Currently, only 11 organisms have been formally linked as potential causes of human cancers, including H. pylori (21), the only bacterium recognized by the International Agency for Research on Cancer (IARC). Despite the recent advances in microbiological and microbiome research, the protumorigenic microbe list by IARC has not been updated for more than one decade. Recent studies suggested that dozens of bacteria could modulate or contribute to cancers other than H. pylori (22), as shown in Table 1. Mechanically, bacteria can cause malignant tumors through the deleterious alterations in the physiological processes of the host, such as 1) chronic inflammation (38, 39), 2) antigen-driven lymph proliferation (40), 3) induction of hormones that increase the proliferation of epithelial cells (19, 20), 4) directly affecting oncogenesis through changing the cell transformation (41), or 5) interrupts the cellular signal by the production of carcinogenic metabolites or toxic substances, therefore, interfering with the regulation of cell growth (14, 42).
Bacteria were first discovered in human cancers about 100 years ago (43). All of the above studies described the role of extracellular bacteria in human cancers. The characterization of bacteria in human cancers has not been well studied due to their low biological expression. A recent report comprehensively analyzed the microorganisms in seven human cancers and demonstrated that the bacteria in cancers are primarily located in cells, including cancer cells and immune cells (44). Moreover, Livyatan et al. (45) also verified that bacteria were alive in cancer cells, but not bacterial components.
In this review, we summarize the latest progress in understanding the role of bacteria in human cancer cells and discuss how bacteria are transported into cancer cells and their physiological significance in cancer progression. Finally, we present the prospect of bacterial therapy in tumor treatment.
Bacteria inside human cancer cells
The microbiome of human cancer and its adjacent normal tissues was analyzed in more than 1,500 samples, and a bacterial catalog of seven different cancer types was generated (44). Cancer microbiome showed distinct microbial characteristics across different cancer types and even different subtypes (44). Visualization method showed that bacteria in cancers were mainly located in cancer and immune cells, not in the extracellular compartment (44, 45). Bacterial lipopolysaccharide (LPS) and lipoteichoic acid (LTA) were detected by immunohistochemistry (IHC) to label Gram-negative and Gram-positive bacteria, respectively (46). RNA fluorescence in situ hybridization (FISH), with a universal probe against bacterial 16S ribosomal RNA (rRNA), was used to detect bacterial RNA in cancer tissues (47). The examination of LPS, LTA, and bacterial 16S rRNA was performed in different cell types across 459, 427, and 354 cancer cores, respectively (44). It was found that LPS and 16S rRNA were mainly located in cancer cells and immune cells. In cancer cells, bacterial 16S rRNA was detected mostly in the cytoplasm, whereas LPS was detected in both the cytoplasm and the nucleus. LTA was only detected in macrophage cells but rarely in cancer cells and immune cells (44). To further verify the presence of bacteria inside cancer cells, the bacteria were found in close proximity to the nuclear membrane in four human breast cancer tissues by correlative light and electron microscopy (CLEM) (44). The bacteria were not detected in the nucleus, indicating that the appearance of LPS nuclear localization in some cancer cells was probably the staining of cytoplasmic perinuclear bacteria (44).
While positive FISH staining of bacterial 16S rRNA confirmed the diffused signal inside cancer cells, typical bacterial rods or cocci were rarely detected. Notably, no cell wall polymer LTA was detected in cancer cells, but many Gram-positive bacteria in human cancers were detected by 16S rDNA sequencing, suggesting that bacteria in human cancer cells may alter their envelope, perhaps result in a cell wall-deficient state, such as L-forms (48). Cell wall-deficient bacteria were only found inside cells (49, 50). In breast cancer, many intracellular cell wall-deficient bacteria were indeed found (44). Bacteria in the cytoplasm of breast cancer cells were also confirmed in the study by Fu et al. (51).
How are bacteria transported into human cancer cells?
There are more than 1,000 different species totaling 1,014 microorganisms of the human intestinal microbiome. The microbiome is involved in the important normal physiological activities of the intestine, such as energetic metabolism, proliferation of epithelial cells, and resistance of pathogens (52). Fusobacterium nucleatum is an opportunistic commensal anaerobe in the oral cavity, implicated in various forms of periodontal diseases. Outside the oral cavity, it is one of the most prevalent species in extraoral infections. F. nucleatum was highly expressed in colorectal cancer (CRC) and was related to CRC development (26), indicating that an oral pathogen associated with oral squamous cell carcinoma (OSCC) might also be related to distant cancers (26, 53). Additionally, F. nucleatum was also associated with liver metastasis, further suggesting its role in cancer development (54). F. nucleatum adheres to and invades endothelial and epithelial cells via FadA to get systemic transmission (27, 55). This leads to the internalization of pathogens and mediated the pro-inflammatory cascade by the induction of nuclear factor (NF)-κB (NFκB) and interleukin-6, constituting a possible way by which F. nucleatum invades OSCC cells (55, 56). FadA is highly conserved among F. nucleatum and was expressed on the bacterial surface (57). It can specifically bind to the EC5 region of E-cadherin on CRC cells to attach to and invade CRC cells, which was also demonstrated in HEK293 cells (58). F. nucleatum is also highly expressed in esophageal squamous cell carcinoma (ESCC) tissues (59) compared with their adjacent non-tumor tissues. Recently, it was also observed by transmission electron microscopy that it has the ability to invade ESCC cells (59). But how are bacteria transported into cancer cells was not reported in detail. Combined with previous reports, we speculated that bacteria might transfer to distant cancer cells by the digestive tract (direct diffusion), lymph, or blood pathway (60). The proposed mechanisms of how bacteria invade cancer cells are shown in Figure 1.
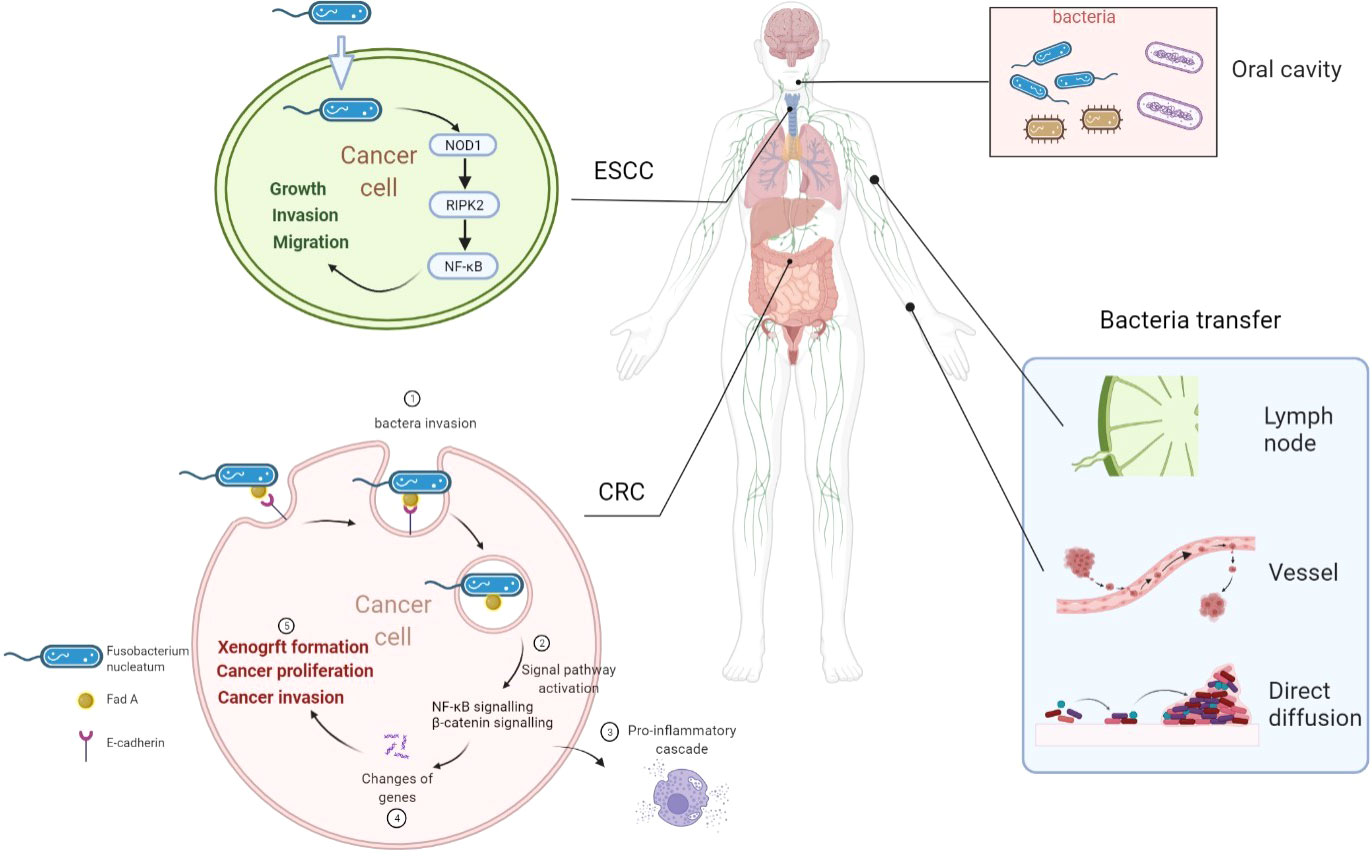
Figure 1 Overview of proposed mechanisms that Fusobacterium nucleatum invading cancer cells. F. nucleatum is a common bacterium in the oral cavity. F. nucleatum invades ESCC cells and CRC cells. F. nucleatum may transfer through lymph, vessel, or direct diffusion. F. nucleatum promotes ESCC progression by the NOD1/RIPK2/NFκB pathway. FadA is highly conserved among F. nucleatum and expressed on the bacterial surface. FadA binds E-cadherin on CRC cells to attach and invade CRC cells. Then, NFκB signaling and β-catenin signaling are activated to induce a pro-inflammatory cascade to promote CRC progression. ESCC, esophageal squamous cell carcinoma; CRC, colorectal cancer; Fn, Fusobacterium nucleatum; NOD1, nucleotide-binding oligomerization domain-containing protein 1; RIPK2, interacting serine threonine kinase 2; NFκB, nuclear factor-κB.
What roles do bacteria play in human cancer cells?
The study by Rubinstein et al. (58) demonstrated that FadA can bind to E-cadherin expressed on CRC cells and mediate F. nucleatum attachment and cell invasion. Then, FadA activates β-catenin signaling, leading to increased expression of transcription factors, oncogenes, Wnt genes, and inflammatory genes to promote CRC cell proliferation (58). Furthermore, FadA directly regulates Wnt and oncogene activation upon binding to CRC cells, but clathrin-mediated FadA internalization is also required for the inflammatory gene activation (58). In mice, CRC cells with F. nucleatum infection can increase their proliferation, invasive activity, and the ability to form xenograft cancers (58, 61). Another study demonstrated that F. nucleatum could activate toll-like receptor 4 signaling to MYD88, resulting in activation of the nuclear factor NFκB and elevated expression of miR21, which reduced the expression of the RAS GTPase RASA1 (61). In ESCC, F. nucleatum was associated with poor survival and promoted cancer cell growth, migration, and invasion. It can invade ESCC cells and induce the NFκB pathway through the nucleotide-binding oligomerization domain-containing protein 1 (NOD1)/receptor interacting serine threonine kinase 2 (RIPK2) pathway, leading to tumor progression. With the help of a murine spontaneous breast tumor model, it can be found that after clearing the bacteria in cancer cells, tumor weight was not affected, but lung metastasis decreased significantly. This shows that the intracellular bacteria are likely to affect the metastasis process rather than the growth of breast cancer (51). After bacteria invaded breast cancer cells, RhoA-ROCK signaling pathway can be activated to reorganize the cytoskeleton and help breast cancer cells resist the pressure from blood vessels and avoid damage during metastasis. Therefore, breast cancer cells carrying bacteria have a stronger ability to reach distant tissues for metastasis (51).
Bacteria in cancer treatment
The usefulness of eukaryotic and prokaryotic expression systems in delivering therapeutic payloads has been explored, including cytotoxic agents (62, 63), prodrug invertase (64, 65), immunomodulators (66), tumor matrix-targeting molecules (67, 68), and siRNA (68). Prokaryotic expression systems discussed above are the most commonly used methods. They encode targeted genes depending on bacterial prokaryotic plasmids (67, 69). Eukaryotic expression systems involve the transduction of host cells, such as immune cells or tumor cells, with the eukaryotic plasmids encoding the cDNA of target genes (70).
F. nucleatum is highly expressed in CRC tissues. Treatment with the antibiotic metronidazole can reduce F. nucleatum amount, cancer cell proliferation, and cancer growth in F. nucleatum-positive CRC (54). Therefore, bacteria may become therapeutic targets in cancer treatment. Mycobacterium bovis strain bacillus Calmette–Guérin (BCG) was first used in bladder perfusion therapy in the 1970s for the prevention and treatment of non-muscle-invasive bladder cancer (NMIBC). Intravesical instillation of BCG can significantly reduce cancer recurrence in NMIBC patients (71).
Despite being therapeutic targets in cancer treatment, bacteria can also regulate the effect of chemotherapy. Escherichia coli could influence the chemotherapy effect in gemcitabine and CB1954 by inducing drug resistance and activating cytotoxicity, separately (72, 73). As a cell taxon in pancreatic tumors, gammaproteobacteria can express an isoform of cytidine deaminase that inactivates gemcitabine, thereby reducing the concentration of gemcitabine in tumors and developing chemotherapy resistance (73). The number of F. nucleatum increased in CRC patients who relapsed after chemotherapy compared with those who did not relapse after chemotherapy (74). In CRC, F. nucleatum can biologically control chemotherapy resistance by coordinating TLR4-MyD88, miR18a and miR4802 and ULK1/ATG7 autophagy networks (74). This helps us to propose an important clinical question: are the conventional chemotherapy regimens including capecitabine plus oxaliplatin suitable for CRC patients with high expression of F. nucleatum? We suggest that chemotherapy regimens of patients with a high expression of F. nucleatum can be combined with anti-F. nucleatum treatment or autophagy inhibitors. A serious adverse reaction of irinotecan in cancer treatment is high intestinal toxicity. SN38, the active metabolite of irinotecan, is subsequently conjugated to SN38-G by hepatic UDP-glucuronyltransferase. SN38-G, an inactive metabolite of irinotecan, is excreted into the small intestine and then discharged from the body. The β-glucoronidases generated from intestinal bacteria can transform SN38-G into SN38, causing direct damage to the intestinal mucosa (75). Chemotherapy and associated mucosal damage may also affect the composition of the intestinal microbiome. Moreover, 5-fluorouracil (5-FU) can regulate the oral and fecal microbiome of laboratory animals by increasing Gram-negative anaerobic bacteria (76).
There was also an interaction between radiotherapy and ecological imbalance of intestinal flora. After radiotherapy, the imbalance of intestinal microbiota is characterized by the reduction of the abundance of symbiotic bifidobacteria, fecal bacilli, and Clostridium and the increase of Bacteroides and Enterococcus (77, 78). In vitro studies have shown that oral vancomycin induces the reduction of Gram-positive bacteria in the intestine and is associated with the enhanced efficacy of radiation therapy in melanoma, lung cancer, and cervical cancer models, which may possibly be through interferon-γ and CD8 T cell-dependent mechanisms (79). On the contrary, in the mouse model of breast cancer radiotherapy, antibiotics induced the decrease of Clostridium and the increase of intestinal yeast, promoting a macrophage-mediated tumor response (80). Intestinal bacteria may also be translocated through the damaged intestinal barrier, thereby regulating radiation toxicity, further contributing to uncontrolled intestinal immune response and tissue damage (81).
Compared with most other traditional drug delivery systems, bacteria have unique capabilities as drug carriers for cancer treatment. They can overcome physical barriers to target and accumulate in cancer tissues and initiate an anticancer immune response (82). The new discovery that bacteria exist in cancer cells (44) can promote the hypothesis that bacterial carriers can accurately deliver drugs to cancer cells.
In addition, bacteria can be genetically and chemically modified to produce and deliver anticancer agents to cancer tissues, thus improving the safety and effectiveness of cancer treatment while reducing the cytotoxicity to normal cells.
Conclusions and prospects
In this review, we briefly summarize the recent progress in understanding the development or correlation between bacteria and cancers and the future research approaches beneficial to this field, including carrying drugs to help kill cancer cells. Visualization method showed that bacteria in cancers were primarily located in cancer and immune cells, rather than in the extracellular compartment (44, 45). The bacteria were found in close proximity to the nuclear membrane in human breast cancer tissues (44). The correlation between F. nucleatum and CRC has been extensively demonstrated. Targeting this bacterium in the gastrointestinal tract and the potential development of bacteria-targeted therapy against F. nucleatum are promising research approaches. FadA could specifically bind to the EC5 region of E-cadherin on CRC cells to attach to and invade CRC cells (58). Then, FadA can activate the β-catenin signaling to promote CRC cell proliferation (58). F. nucleatum can also invade ESCC cancer cells and promote cancer progression through the NOD1/RIPK2/NFκB pathway. This is a specific study of how bacteria entered tumor cells and played their roles (Figure 1).
Studying the causal relationship and molecular interaction between bacteria and cancers promises to provide new clues for the development, progression, and treatment response of human cancers. In addition to trying to understand bacterial associations and causality in cancer, such research also faces arduous challenges related to sample allocation, processing, sequencing, and data analysis. Despite these challenges, the contribution of bacteria to cancer biology may occupy a central position in cancer research over the next decade, making more contributions to cancer diagnosis, patient stratification, and treatment.
Limitation
However, the way other bacteria enter cancer cells and how diseases affect tumor progression in which way still require further study. There are still many questions that need to be explored. What is the origin of bacteria in cancer? Does the composition of the bacteria change with cancer progression? Do bacteria “travel” with cancer cells to metastatic sites? Are there any bacteria at metastasis sites more relevant to the new location? We should focus on exploring and solving the above problems to better contribute to the treatment of cancers.
Author contributions
WZ and JW investigated and wrote the original draft. ZL and JW proposed the ideas and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Natural Science Foundation of Jiangsu Province (BK20201090), China Postdoctoral Science Foundation (2021M691338), and Young Scholars Fostering Fund of the First Affiliated Hospital of Nanjing Medical University (PY2021052).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Sci (New York NY) (2008) 320(5883):1647–51. doi: 10.1126/science.1155725
2. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature (2011) 473(7346):174–80. doi: 10.1038/nature09944
3. Human Microbiome Project Consortium. A framework for human microbiome research. Nature (2012) 486(7402):215–21. doi: 10.1038/nature11209
4. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
5. Xavier JB, Young VB, Skufca J, Ginty F, Testerman T, Pearson AT, et al. The cancer microbiome: Distinguishing direct and indirect effects requires a systemic view. Trends Cancer (2020) 6(3):192–204. doi: 10.1016/j.trecan.2020.01.004
6. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature (2013) 500(7464):541–6. doi: 10.1038/nature12506
7. Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ (Clinical Res ed) (2018) 360:j5145. doi: 10.1136/bmj.j5145
8. Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol (2018) 15(7):397–411. doi: 10.1038/s41575-018-0011-z
9. Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, et al. The human microbiome and cancer. Cancer Prev Res (Philadelphia Pa) (2017) 10(4):226–34. doi: 10.1158/1940-6207.Capr-16-0249
10. Salavatiha Z, Soleimani-Jelodar R, Jalilvand S. The role of endogenous retroviruses-K in human cancer. Rev Med Virol (2020) 30(6):1–13. doi: 10.1002/rmv.2142
11. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol (2016) 14(8):e1002533. doi: 10.1371/journal.pbio.1002533
12. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med (2018) 24(4):392–400. doi: 10.1038/nm.4517
13. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: The role of butyrate on colonic function. Alimentary Pharmacol Ther (2008) 27(2):104–19. doi: 10.1111/j.1365-2036.2007.03562.x
14. O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol (2016) 13(12):691–706. doi: 10.1038/nrgastro.2016.165
15. Fujimori M. Genetically engineeredbifidobacterium as a drug delivery system for systemic therapy of metastatic breast cancer patients. Breast Cancer (2006) 13(1):27–31. doi: 10.2325/jbcs.13.27
16. Roberts NJ, Zhang L, Janku F, Collins A, Bai RY, Staedtke V, et al. Intratumoral injection of clostridium novyi-nt spores induces antitumor responses. Sci Trans Med (2014) 6(249):249ra111. doi: 10.1126/scitranslmed.3008982
17. Lax AJ. Bacterial toxins and cancer--a case to answer? Nat Rev Microbiol (2005) 3(4):343–9. doi: 10.1038/nrmicro1130
18. Xue FB, Xu YY, Wan Y, Pan BR, Ren J, Fan DM, et al. Pylori infection with gastric carcinoma: A meta analysis. World J Gastroenterol (2001) 7(6):801–4. doi: 10.3748/wjg.v7.i6.801
19. Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-hydroxyguanine, an abundant form of oxidative DNA damage, causes G–-T and a–-C substitutions. J Biol Chem (1992) 267(1):166.
20. Smoot DT. How does helicobacter pylori cause mucosal damage? direct mechanisms. Gastroenterology (1997) 113(6-supp-S):S31. doi: 10.1016/s0016-5085(97)80008-x
21. IARC. Iarc monographs on the evaluation of carcinogenic risks to humans (2009). Available at: https://publications.iarc.fr/Book-And-Report-Series/Iarc-Monographs-On-The-Identification-Of-Carcinogenic-Hazards-To-Humans/.
22. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Sci (New York NY) (2021) 371(6536):eabc4552. doi: 10.1126/science.abc4552
23. Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology (2009) 136(6):1863–73. doi: 10.1053/j.gastro.2009.01.073
24. Correa P, Piazuelo MB. The gastric precancerous cascade. J Digestive Dis (2015) 13:2–9. doi: 10.1111/j.1751-2980.2011.00550.x
25. Correa P. A model for gastric cancer epidemiology. Lancet (London England) (1975) 306(7924):58–60. doi: 10.1016/s0140-6736(75)90498-5
26. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
27. Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. Fada from fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem (2007) 282(34):25000–9. doi: 10.1074/jbc.M611567200
28. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the human gut by e. coli and colorectal cancer risk. Clin Cancer Res (2014) 20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343
29. Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci (2017) 18(1):197. doi: 10.3390/ijms18010197
30. Cheng WT, Kantilal HK, Davamani F. The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malaysian J Med Sci (2020) 27(4):9–21.
31. Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/JCI72334
32. Chung L, Orberg ET, Geis AL, Chan JL, Fu K, Destefano Shields CE, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade Via targeting of colonic epithelial cells. Cell Host Microbe (2018) 23(3):421. doi: 10.1016/j.chom.2018.02.004
33. Sears CL. Enterotoxigenic bacteroides fragilis: A rogue among symbiotes. Clin Microbiol Rev (2009) 22(2):349. doi: 10.1128/CMR.00053-08
34. Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-myc expression and cellular proliferation. Gastroenterology (2003) 124(2):392–400. doi: 10.1053/gast.2003.50047
35. Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappab pathway. Infect Immun (2004) 72(10):5832–9. doi: 10.1128/IAI.72.10.5832-5839.2004
36. Kim JM, Lee JY, Kim YJ. Inhibition of apoptosis in bacteroides fragilis enterotoxin-stimulated intestinal epithelial cells through the induction of c-Iap-2. Eur J Immunol (2010) 38(8):2190–9. doi: 10.1002/eji.200838191
37. Scanu T, Spaapen R, Bakker J, Pratap C, Wu LE, Hofland I, et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe (2015) 17(6):763–74. doi: 10.1016/j.chom.2015.05.002
38. Buti L, Spooner E, Veen A, Rappuoli R, Covacci A, Ploegh HL. Helicobacter pylori cytotoxin-associated gene a (Caga) subverts the apoptosis-stimulating protein of P53 (Aspp2) tumor suppressor pathway of the host. Proc Natl Acad Sci (2011) 108(22):9238–43. doi: 10.1073/pnas.1106200108
39. Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R, Sasakawa C. Grb2 is a key mediator of helicobacter pylori caga protein activities. Mol Cell (2002) 10(4):745–55. doi: 10.1016/s1097-2765(02)00681-0
40. Suarez F, Lortholary O, Hermine O, Lecuit M. Infection-associated lymphomas derived from marginal zone b cells: A model of antigen-driven lymphoproliferation. Blood (2006) 107(8):3034–44. doi: 10.1182/blood-2005-09-3679
41. Ferrand J, Lehours P, Schmid-Alliana A, Mégraud F, Varon C. Helicobacter pylori infection of gastrointestinal epithelial cells in vitro induces mesenchymal stem cell migration through an nf-Kb-Dependent pathway. PLoS One (2011) 6(12):e29007. doi: 10.1371/journal.pone.0029007
43. Goodman B, Gardner H. The microbiome and cancer. J Pathol (2018) 244(5):667–76. doi: 10.1002/path.5047
44. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
45. Livyatan I, Nejman D, Shental N, Straussman R. Characterization of the human tumor microbiome reveals tumor-type specific intra-cellular bacteria. Oncoimmunology (2020) 9(1):1800957. doi: 10.1080/2162402x.2020.1800957
46. Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem (2002) 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414
47. Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16s rrna-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol (1990) 56(6):1919–25. doi: 10.1128/aem.56.6.1919-1925.1990
48. Klieneberger-Nobel E. Origin, development and significance of l-forms in bacterial cultures. J Gen Microbiol (1949) 3(3):434–43. doi: 10.1099/00221287-3-3-434
49. Errington J. L-form bacteria, cell walls and the origins of life. Open Biol (2013) 3(1):120143. doi: 10.1098/rsob.120143
50. Errington J. Cell wall-deficient, l-form bacteria in the 21st century: A personal perspective. Biochem Soc Trans (2017) 45(2):287–95. doi: 10.1042/bst20160435
51. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell (2022) 185(8):1356–72.e26. doi: 10.1016/j.cell.2022.02.027
52. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature (2012) 489(7415):242–9. doi: 10.1038/nature11552
53. Kim GW, Kim YS. Periodontitis is associated with an increased risk for proximal colorectal neoplasms. J Sci Rep (2019) 9(1):7528. doi: 10.1038/s41598-019-44014-8
54. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. J Sci (2017) 358(6369):1443–8. doi: 10.1126/science.aal5240
55. Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infection Immun (2000) 68(6):3140–6. doi: 10.1128/iai.68.6.3140-3146.2000
56. Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: Implication of oral bacteria in preterm birth. Infection Immun (2004) 72(4):2272–9. doi: 10.1128/iai.72.4.2272-2279.2004
57. Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriology (2005) 187(15):5330–40. doi: 10.1128/jb.187.15.5330-5340.2005
58. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-Cadherin/B-Catenin signaling Via its fada adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
59. Nomoto D, Baba Y, Liu Y, Tsutsuki H, Okadome K, Harada K, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression Via the Nod1/Ripk2/Nf-Kb pathway. Cancer Lett (2022) 530:59–67. doi: 10.1016/j.canlet.2022.01.014
60. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun (2020) 11(1):3259. doi: 10.1038/s41467-020-16967-2
61. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-Kb, and up-regulating expression of microrna-21. Gastroenterology (2017) 152(4):851–66.e24. doi: 10.1053/j.gastro.2016.11.018
62. Guan GF, Zhao M, Liu LM, Jin CS, Sun K, Zhang DJ, et al. Salmonella typhimurium mediated delivery of apoptin in human laryngeal cancer. Int J Med Sci (2013) 10(12):1639–48. doi: 10.7150/ijms.6960
63. Kimura H, Zhang L, Zhao M, Hayashi K, Tsuchiya H, Tomita K, et al. Targeted therapy of spinal cord glioma with a genetically modified salmonella typhimurium. Cell Proliferation (2010) 43(1):41–8. doi: 10.1111/j.1365-2184.2009.00652.x
64. Cheng CM, Lu YL, Chuang KH, Hung WC, Shiea J, Su YC, et al. Tumor-targeting prodrug-activating bacteria for cancer therapy. Cancer Gene Ther (2008) 15(6):393–401. doi: 10.1038/cgt.2008.10
65. Friedlos F, Lehouritis P, Ogilvie L, Hedley D, Davies L, Bermudes D, et al. Attenuated salmonella targets prodrug activating enzyme carboxypeptidase G2 to mouse melanoma and human breast and colon carcinomas for effective suicide gene therapy. Clin Cancer Res (2008) 14(13):4259–66. doi: 10.1158/1078-0432.ccr-07-4800
66. Loeffler M, Le'Negrate G, Krajewska M, Reed JC. Salmonella typhimurium engineered to produce Ccl21 inhibit tumor growth. Cancer Immunology Immunotherapy CII (2009) 58(5):769–75. doi: 10.1007/s00262-008-0555-9
67. Zuo SG, Chen Y, Wu ZP, Liu X, Liu C, Zhou YC, et al. Orally administered DNA vaccine delivery by attenuated salmonella typhimurium targeting fetal liver kinase 1 inhibits murine Lewis lung carcinoma growth and metastasis. Biol Pharm Bull (2010) 33(2):174–82. doi: 10.1248/bpb.33.174
68. Jia H, Li Y, Zhao T, Li X, Hu J, Yin D, et al. Antitumor effects of Stat3-sirna and endostatin combined therapies, delivered by attenuated salmonella, on orthotopically implanted hepatocarcinoma. Cancer Immunology Immunotherapy CII (2012) 61(11):1977–87. doi: 10.1007/s00262-012-1256-y
69. Berger E, Soldati R, Huebener N, Hohn O, Stermann A, Durmus T, et al. Salmonella Sl7207 application is the most effective DNA vaccine delivery method for successful tumor eradication in a murine model for neuroblastoma. Cancer Lett (2013) 331(2):167–73. doi: 10.1016/j.canlet.2012.12.026
70. Yoshimura K, Jain A, Allen HE, Laird LS, Chia CY, Ravi S, et al. Selective targeting of antitumor immune responses with engineered live-attenuated listeria monocytogenes. Cancer Res (2006) 66(2):1096–104. doi: 10.1158/0008-5472.can-05-2307
71. Gontero P, Bohle A, Malmstrom PU, O'Donnell MA, Oderda M, Sylvester R, et al. The role of bacillus calmette-guérin in the treatment of non-Muscle-Invasive bladder cancer. Eur Urol (2010) 57(3):410–29. doi: 10.1016/j.eururo.2009.11.023
72. Lehouritis P, Cummins J, Stanton M, Murphy CT, McCarthy FO, Reid G, et al. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci Rep (2015) 5:14554. doi: 10.1038/srep14554
73. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
74. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008
75. Alimonti A, Gelibter A, Pavese I, Satta F, Cognetti F, Ferretti G, et al. New approaches to prevent intestinal toxicity of irinotecan-based regimens. Cancer Treat Rev (2004) 30(6):555–62. doi: 10.1016/j.ctrv.2004.05.002
76. Von Bültzingslöwen I, Adlerberth I, Wold AE, Dahlén G, Jontell M. Oral and intestinal microflora in 5-fluorouracil treated rats, translocation to cervical and mesenteric lymph nodes and effects of probiotic bacteria. Oral Microbiol Immunol (2003) 18(5):278–84. doi: 10.1034/j.1399-302x.2003.00075.x.
77. Liu J, Liu C, Yue J. Radiotherapy and the gut microbiome: Facts and fiction. Radiat Oncol (2021) 16(1):9. doi: 10.1186/s13014-020-01735-9
78. Touchefeu Y, Montassier E, Nieman K, Gastinne T, Potel G, Bruley D, et al. Systematic review: The role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis – current evidence and potential clinical applications. Alimentary Pharmacol Ther (2014) 40(5):409–21. doi: 10.1111/apt.12878
79. Uribe-Herranz M, Rafail S, Beghi S, Gil-De-Gómez L, Facciabene A. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest (2019) 130(4):466–479. doi: 10.1172/JCI124332
80. Shiao SL, Kershaw KM, Limon JJ, You S. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell (2021) 39(1-12):1202–1213.e6. doi: 10.1016/j.ccell.2021.07.002
81. Al-Qadami G, Sebille YV, Le H, Bowen J. Gut microbiota: Implications for radiotherapy response and radiotherapy-induced mucositis. Expert Rev Gastroenterol Hepatol (2019) 13(5):485–496. doi: 10.1080/17474124.2019.1595586
Keywords: bacteria, cancer cells, Fusobacterium nucleatum (F.n), cancer treatment, colorectal cancer
Citation: Zhu W, Wang J-Z, Liu Z and Wei J-F (2022) The bacteria inside human cancer cells: Mainly as cancer promoters. Front. Oncol. 12:897330. doi: 10.3389/fonc.2022.897330
Received: 16 March 2022; Accepted: 14 July 2022;
Published: 12 August 2022.
Edited by:
Massimo Fantini, Precision Biologics, Inc., United StatesReviewed by:
Shahid Umar, University of Kansas Medical Center, United StatesJimmy Thomas Efird, The University of Newcastle, Australia
Copyright © 2022 Zhu, Wang, Liu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhixian Liu, bGl1emhpeGlhbkBuam11LmVkdS5jbg==; Ji-Fu Wei, d2VpamlmdUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Wei Zhu
Wei Zhu Jing-Zi Wang
Jing-Zi Wang Zhixian Liu
Zhixian Liu Ji-Fu Wei
Ji-Fu Wei