
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 19 July 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.896633
Despite the advancement in research methodologies and technologies for cancer research, there is a high rate of anti-cancer drug attrition. In this review, we discuss different conventional and modern approaches in cancer research and how human-centric models can improve on the voids conferred by more traditional animal-centric models, thereby offering a more reliable platform for drug discovery. Advanced three-dimensional cell culture methodologies, along with in silico computational analysis form the core of human-centric cancer research. This can provide a holistic understanding of the research problems and help design specific and accurate experiments that could lead to the development of better cancer therapeutics. Here, we propose a new human-centric research roadmap that promises to provide a better platform for cancer research and drug discovery.
Cancer arises when tumor cells start invading nearby tissues, leading to the disruption of tissue homeostasis. Common treatment strategies against cancer include surgery, chemotherapy, radiotherapy, and immunotherapy, depending upon the stage and the nature of cancer. Currently, every sixth person in the world dies of cancer, which is second only to the cardiovascular diseases (1). Despite advancements in early detection and novel therapeutics, cancer is still the second largest cause of human death (1). Globally, around 19.3 million new cancer cases were estimated, and 10 million deaths for the year 2020 (2). This is primarily attributed to our lack of understanding of the complexity of the disease. Some of the common complexities arise from heterogeneity within the cancer tissues and the development of therapeutic resistance during and after the treatment (3–6). Cancer is a genetically heterogeneous disease; i.e. cellular heterogeneity exists within a tumor population. Tumor heterogeneity suggests that it contains more than one cell types, which exhibit differential ability to proliferate, migrate, maintain stemness, and response to therapy (4). The stem cell-like population within the tumor are inherently more resistant to therapy [due to over-expression of specific drug neutralizing/exporting proteins, and anti-death proteins (7, 8)] and less accessible to drug molecules, whereas simultaneously also contributing to tumor heterogeneity. Therapy resistance is the result of either over-proliferation of cell types that are inherently resistant or may be acquired progressively during treatment. The sub-population of cancer cells generally coexist with immune and other non-cancerous cells in a complex ecosystem called tumor microenvironment. In addition to the intra-tumor cellular heterogeneity, the cancer cells co-exist with different non-cancerous cells like the Cancer associated fibroblasts (CAFs), endothelial cells, and immune cells (like macrophages, microglia, and lymphocytes); and the non-cellular components like extracellular matrix components (ECM) (fibronectin, laminin, collagen, hyaluronan, integrin etc.) to form a complex three dimensional (9) ecosystem called tumor microenvironment(TME) (5, 10). Different cells in the TME can cross-talk and release factors which can modify ECM. The chemical and physical signals triggered by ECM have been shown to regulate the tumor progression and therapeutic resistance by modulating cancer heterogeneity, clonal evolution, epithelial-mesenchymal transition, invasion, migration, neovascularization, and metastasis (9, 11, 12). Remodelling of endothelial cells, cross-talk between CAFs and tumor cells, and paracrine signalling by Tumor associated macrophages, all as a part of TME have been shown to promote cancer progression (13). Furthermore, intra-tumoral mechanisms of metabolite communications between different cell types act in a symbiotic way to promote tumor metabolism, maintenance and growth (14). In the last few years, researchers have discovered that TME mediate therapeutic resistance by regulating drug availability (13) and interstitial fluid pressure (15). Because of the complexities, even combinational therapy is unable to kill all the cancer cells, leading to disease relapse. Together, these factors play a pivotal role in tumor progression and therapy resistance (4–6, 16, 17), thus making it challenging to design targeted therapy. However, recent developments of the three dimensional (3D) in vitro cell culture methodologies and platforms provide immense scope to exploit the biology of tumor through better mimic of the TME (18, 19).
Despite substantial funding in research and development globally, less than 85% of the drugs showing encouraging signs in the pre-clinical cancer models succeed in the early clinical trials. and less than half of those passing through Phase 3 clinical trials make it for licencing (20). In fact, roughly 1 in 10,000 pre-clinical compounds reaches the market (21). Because of such a low success rate, prices of anti-cancer drugs touch an unaffordable range. There are many anticancer drugs at present which cost more than $10,000 (per year per patient) and the supporting claim pharma companies make to justify these costs is the high drug attrition rates at phase 2 and phase 3 of clinical trials (21–23). One of the main reasons for high cancer drug attrition is their reduced efficacy in humans compared to animal models (21, 23). This points out our lack of understanding of the complexity of the disease, which may be due to the way chemotherapeutic drugs are tested preclinically before being brought in the purview of the clinical trial. Heavy dependence on animal models for pre-clinical studies is perhaps one of the important reasons for the current setback. A better understanding of drug dynamics inside the human body and their interaction with different cell types will help decrease the intensity of the problem in hand. Thus, it becomes essential not to consider cancer from only tumor cell-centric perspective but rather as an ecosystem containing a plethora of cells, acting as a protective shelter for tumor cells. The use of human-centric models may help in mitigating the limitations posed by animal-centric research models. Its cumulative understanding will be useful to go beyond our current limitations and prepare new generation drugs with better efficacy. Traditional ways of performing preclinical research primarily depend on human cell lines (2D in vitro), and non-human animals, such as rodents and primates (in vivo). Historically, animal-centric research has been heavily relied on for understanding cancer and testing new anti-cancer drugs because of the conservation of basic biological principles and the evolution of modern genetic tools. Unfortunately, the traditional animal-based strategies for cancer research have not been able to deliver as per the expectations. Animal research has its own limitations and has not been the best of the approaches to depend upon as it fails to reliably mimic human cancer models, leading to high drug attrition rates (21, 24). In recent years, there has been an increased focus on developing more human-centric methods for enhancing drug predictivity and ensuring human-predictive drug efficacy. Due to the known limitations of the existing systems of drug-testing, it becomes essential to explore varieties of model systems relevant to humans, including human-specific models for improving drug efficacy, reducing drug costs, and enhancing quicker market reach.
In this review, we summarize the current and traditional ways of cancer research, including 2D cell culture and the use of various animal models. Thereafter, we look upon the avenues and opportunities which are presented by the advanced human-based cancer models such as modern 3D (three-dimensional) culture, organ-on-a-chip, systems biology approaches, and in silico models, which promise to go past the limitations posed by the animal-centric research models. We further discuss a future roadmap of cancer research, which focuses on the application of different advanced human-centric models and explores their scope to replace animal models.
The traditional in vitro cell culture models use two dimensional (2D) techniques, employing isolated immortal cell lines originally obtained from people with cancer (25) and now banked and commercially available. The first cell line to be established was the HeLa cell line, which was obtained in 1951 from Henrietta Lacks, a 31yr old cervical cancer patient (26). 2D models are still largely used for many experiments that involve deciphering molecular mechanisms and drug discovery. However, there are limitations with these models, leading to partly accurate results and predictions. Although the 2D cell culture model does not resemble the actual tumor and overlooks the complex cellular architecture of cancer cells and tumor microenvironment, it is still the easiest and most economical way to perform large scale screening and pilot studies. In real world. cell division, maintenance of cellular homeostasis, and cell death all occur in a three-dimensional environment within the complex tissue architecture in higher organisms. Experiments with cells growing on a two-dimensional plastic surface cannot mimic the inherent intricacies of cell-cell interactions, cell-matrix interactions, and biomolecule availability and drug dose response (hormesis) (27). This led to the development of different techniques, trying to mimic the 3D tissue microenvironment, and these 3D approaches are being actively used to study diseases like diabetes (28, 29), cardiomyopathy (30), and cancer (31). Culture of cells in 3D, both with and without scaffolds, is revolutionizing biological research (32). The first research with cells in 3D dates to more than a century back when bacteria were clamped in a hanging drop (33). Later, this technique was employed to grow the first tissue in a scaffold-free manner (34). 3D cell culture models relevant to human disease started coming to the limelight since the 1980s and have been advancing since then. 3D cancer models resemble the tumor architecture more closely (35) and can mimic the cellular crosstalk in the different co-culture systems (36). Different cancer and non-cancer cell lines are grown in 3D scaffolds to resemble tumor microenvironment. Cytotoxicity testing, modelling of cancer stem cells, and epithelial to mesenchymal transition is better represented in the 3D culture system when compared to conventional 2D culture (31, 37). However, the major limiting factors of scaffold-dependent 3D cell culture models are its inability to mimic the biomolecular circulation and the controlled availability (which is physiologically aided by vasculature). Further, these scaffold-dependent models fail to mimic the appropriate biophysical extracellular structures/cues as necessary to tissues. To date, conventional 2D and 3D in vitro models fail to fully replicate the complex dynamic system operating within a tumor (38).
Other notable advancements in cancer research are the organoid and spheroid 3D cell culture systems. Both Spheroid and organoid models are 3D cell-culture models but with marked differences. A spheroid model is the simpler of the two, formed by of simple 3D aggregation of cells and does not contain any extracellular matrix or hydrogel scaffold and thus cultured as free floating aggregates. It can be developed from any cancerous or non-cancerous cell line or primary cells (by using a non-adherent plate). On the other hand, an organoid is a complex 3D structure of cells which is often supported by extracellular matrix or hydrogel scaffold and specific growth factors (32, 39) and is developed using stem cells primarily derived from the human healthy to tumor tissues. The cells in the spheroid predominantly has stem cell like property but, an organoid contains more differentiated cells (aided by scaffold and growth factors) and closely resembles the tissue architecture and function (40), which can be cryopreserved (41). Organoids grown with this method acquire a micro-anatomy, which is similar to the native tissue by self-organization and spatial orientation (42) and helps retain heterogeneity of the original tumor (19). Patient-derived organoid models have been generated with liver, colorectal, pancreatic, and prostate cancer (19). Gene-editing to introduce mutation and co-culture techniques to mimic different cellular interactions have also been recently possible using organoid models in cancer (43). The organoid models have advantages over the regular 3D model in simulating tumor-specific differential growth patterns like quiescence. On the other hand, the spheroid culture presents another 3D model, which resembles the 3D structure of a tumor and is established with low technical difficulty and cost. These spheroids loosely resemble the architecture of the native tumor and can also be co-cultured with other cells to mimic the exact tumor microenvironment (44). They have been shown to be more reliable for in vitro drug screening (45) and for modelling drug-resistance (46) than conventional 2D culture.
Features like hypoxic gradient and nutrition gradient can be mimicked more realistically in these advanced 3D models in comparison to the native tumor (47). Over the years, with the advancement in the knowledge of stem cells and the usage of patient-derived tumor organoid models, these advanced 3D models hold key promise in the field of personalized medicine (48–50). These characteristics make them more relevant compared to in vivo animal models, being not only cheaper but also lacking the ethical conflicts of the latter (51). Recently, there has been significant progress in the application of microfluidic-based devices to create models of human organs, specifically for drug development and bio-modelling. This is called the organ-on-a-chip/organ chip model, which accounts for the constant circulation of media, mimicking the human vascular system. Scientists have been successful in developing organ-chip models of intestines, blood-brain barrier, bone marrow, lungs, liver, kidney, and skin and using them as effective preclinical models in cancer, and other diseases (52). The organ chip model recapitulates tissue-tissue interface, and multicellular architecture very closely and helps modulate local molecular, chemical, and biophysical parameters in a precisely controlled manner (53). Tumor-chip models have been shown to closely resemble tumor stroma and are beneficial for modelling multi-organ metastasis (19). The organ-on-a-chip model has been used to study important cancer hallmarks like angiogenesis, migration, and invasion in a precise manner (53). Further, 3D based microfluidic research shows applications in the study involving drug testing, metastasis, and drug resistance (54). Although organ-chips are by far the most advanced of the in vitro technology systems, and successfully mimic organ-level physiology, this technology is still in its infancy and is likely to require more validation in different contexts with in vivo human system before we move to a paradigm shift in our methodologies for basic and translational cancer research.
Rodents are the main animal used as in vivo models to test drug efficacy. The xenograft mouse model is one of the most prominently used mouse models to model cancer and test drugs in which in most cases commercial human cancer cell lines are inserted within the immunocompromised mice (55). This strategy has been perceived as the gold standard for the preclinical drug testing and elucidating molecular mechanisms in cancer. However, these animal models have significant limitations. Since the model organisms are not genetically identical to humans, the tumors do not reflect the exact human condition and thus makes it difficult to predict drug efficacy and behavior. Furthermore, because the immune system is compromised, complex cellular interactions between the immune cells and the cancerous cells cannot be studied. Considering that we now know the role of immune cells in cancer cell survival and progression (56), these might not be the appropriate way to model human cancers (55). Furthermore, these cells which have been grown for many generations in plastic, may not be very ideal to recapitulate tumor close to being original. More recently, animal models have been used for patient-derived xenograft models (PDX) and genetically engineered models (GEM). In the PDX models, the primary tumor cells are taken directly from patients to develop a tumor in immunocompromised mice. It has added advantages of mimicking the heterogeneity of parental cancer. However, it also has the disadvantage of using immunocompromised animals (57) and that patient-derived cells have been shown to undergo animal-specific modifications (58), besides being expensive and time consuming. There have been several attempts to develop GEM to mimic the original human tumors (59). The mutations are either made in somatic or germline cells to model the tumor. Recent progress in gene editing tools has advanced this field considerably. These models are supposed to be superior to conventional models (59). However, the major problem of these models is that they do not incorporate human cells, and they are not cost-effective. An alternative to rodents, invertebrate (Drosophila) and fishes (zebrafish) have been used less prominently as a model for therapeutic research because of their lesser resemblance to the human genome. Naturally occurring tumors in canines and felines, are also used in cancer research. Although they have the advantage of naturally occurring tumors (60) and longer life span when compared to rodents, they have their own limitations. They do not exactly mimic human physiology. Additionally, because of the lack of scope of genetic modifications and the extra demand for time and efforts, lack of sufficient numbers, these models do not promise to be the most ideal for cancer research (61).
Besides the widely used traditional cell culture and animal models, human-specific models for cancer research offer exciting avenues for preclinical testing. Human tissues have been used in oncology research for a long time (62). The ongoing advancement in tissue and cell culture reveal their promise for overcoming the common problems of animal-based research. One promising aspect is the primary cell culture from patient samples, where tumors from the patients are directly used and cultured on a dish, maintaining the inherent cellular heterogeneity. This technique is more suitable for drug testing and modelling chemoresistance (63). In comparison to conventional scaffold-based 3D cell culture models, the spheroid and organoid models of the primary tumor are far better representative of the original human tumor and can be used for better cancer modelling. A more recent trend is to utilize the tumor cells in a co-culture system with other cells of the tumor stroma, such as fibroblast and immune cells. These models can be used for high throughput drug-screening assays and have been successfully tried in a 96-well culture system (45, 64, 65). Alongside, organ chip model has been successful in taking care of fluid circulation and biomechanics of the tissues very closely. These microfluidic devices perhaps provide the best platforms along with the advanced 3D models to build a better and more complete cancer model which can closely resemble the parental tumors (53, 66). The most advanced form of human tissue reconstruction is 3D bioprinting which is now applied to model different types of cancer. This field combines the principles of biomaterials and tissue engineering with the delicacy of cell biology to provide a printable tissue that can be used for organ transplantation, drug discovery, and personalized medicine (67). These models have shown promising results in mimicking native tissue microenvironment and complexities (68–70). 3D bio-printed cancer models showed greater cell survival, protein expression patterns that resemble host cancer, and higher chemoresistance to anticancer drugs similar to host. They can also retain features that closely resembled host tumor characteristics like tumor heterogeneity, necrotic cores, and microenvironment. These methods have helped to better understand cancer formation, progression, and response to anticancer therapies. Since animal models differ physiologically from human cancer, a bio-printed cancer model with human cells can be a better representative. These models are also able to mimic the effects of metastasis which is a significant advancement in the fields of in vitro cancer modelling. Bioprinted cancer models have been shown to provide effective reconstructions of host cell and cell-matrix interaction patterns. These models were not only able to simulate metastasis, but the local chemical signalling profile was also found to be similar to the host. The current limitation of this technique is that the cells can only be cultured for a limited period. The expansion of human cell biobanks will help in the supply of a variety of patient’s cancer cells (of different grades and types) to research institutes, which currently have limited accessibility. We discussed this further in the last section (Future roadmap and perspective).
Data generated from OMICs-based research approaches (such as genomics, transcriptomics, proteomics, and metabolomics) have added greater value to the field of cancer research as it has in any other biomedical field. The ‘OMICs’ generally refers to the analysis of structure, function, and origin of biomolecules derived from high throughput studies, along with their implication in the applied fields of biology (71). OMICs based studies tend to generate vast amounts of data, and a systematic approach is needed to store, access, and analyse them using various tools. Advances in the field of molecular and computational biology have enabled us to perform genomic, proteomic, and transcriptomic analyses from the patient samples or primary cell culture (72, 73). Databases generated from “Omics” studies certainly provide a platform for biologists to access them from different sources and integrate it in their research (74, 75). Although some of these databases are behind a paywall or require specific permission, most of these are in the public domain and are made freely available for cancer researchers. Some of the publicly available database document information regarding the human disease, mutations, expression of proteins, and response to drugs (e.g., TCGA Database) while other databases enlist similar information regarding cell lines (e.g., CCLE). Databases are also available to explore other potential areas like epigenetic modification, miRNA regulation, and detailed mutation analysis (76). Some of the relevant databases are listed in Table 1, nevertheless describing them in detail is beyond the scope of this article. As we have discussed earlier regarding the therapeutic challenges posed by tumor heterogeneity, drug resistance, and drug mistargeting, it becomes vital to analyse OMICs data to understand disease progression, identify molecular markers, and response to therapy (98). The main use of cancer databases is to enable the generation of scientific hypotheses from the available datasets. These hypotheses can vary from the expression of a certain gene to the efficiency of certain drugs. In certain cases, these hypotheses can be tested using scientific tools, like cBioportal, which gives information about associated mutations, responsible protein function, and their association with survival. Alongside, proteomic databases are being used for developing diagnostics, identifying mechanisms, and designing treatment strategies. The main purposes of these databases are to save time by providing information that might be acquired from traditional wet lab and animal experiments, whereby avoiding repetition and duplication of experiments Moreover, many of these databases are based on integrated human and cell line experimental data and are carefully curated. Therefore, these databases can provide a starting platform where we can get information similar to that obtained with animal models and help to avoid further or duplicate use of animals. With data being generated at a high pace and their huge scope of application, one must admit that above-mentioned databases have been underutilized and can be used more broadly and efficiently.
The major advantage of many of the above mentioned repositories is that many of them contain patients’ data and thus are useful for accurate, human-relevant predictions. Increasingly, more data sets are being validated using other supporting experiments and have slowly started getting global acceptance. These approaches have the potential not only to complement but also to reduce many of the currently used wet-lab experimental models in an inexpensive manner. They open several new possibilities in the field of personalized medicine. However, a major limitation of these databases is their sheer volume and difficulty in handling them. The in silico models, which apply sophisticated algorithms, help in handling voluminous data of different cancer databases, to advance scientific understanding. Experience in computational analysis and specialized computational labs capable of handling big data are necessary to utilize the full potential of these databases. Therefore, a systematic approach is taken to analyse these data and model different outcomes. These can be built based on several approaches such as statistical models, network-based models, and tissue-based models (99). Examples of these models are provided in Table 2.
A new in silico approach to drug development is based on AI (artificial intelligence). Machine learning and deep learning methods are employed to improve the above models (109). These models can automatically access most of the available data and draw a conclusion using the AI approach to design lead compounds, refine existing drugs, validate them on artificial disease models and predict the emergence of drug resistance (110). Some software include modelling options. For e.g., Cytoscape is a free software package that provides user-friendly visualization of these open-source data (111). It may be noted that several of the primary databases also integrate these modelling features or allow the users to model the available data according to their needs. Though we have described these platforms primarily as cancer databases, it must be noted that several of these databases serve a dual role of data repository as well as a platform to analyse these data and model cancer. The field is evolving rapidly, and very soon AI-based platforms will be available as a complete package. These packages may be able to offer a more user friendly approach to analysing the databases and thus, supplementing cancer research. Advanced AI platforms, in combination with superior 3D cancer models, have the potential to replace unnecessary animal experiments as they more human relevant besides being faster, resource saving, effective and reproducible.
Presently, there is a need to integrate different human-based interdisciplinary research methodologies from the perspective of a holistic understanding of disease mechanisms, prediction, and treatment. 2D cell culture can still serve as a model for large scale pilot drug screening studies or mechanistic studies until 3D cell culture is made more economical and workable. However, after initial screening, different 3D cell culture models can play a big role in narrowing down potential drugs or to reveal specific molecular insights into biological mechanisms. These techniques promise better and more accurate results than conventional 2D cell culture models. Preliminary analysis using widely available human OMICs data alongside or in prior can help in better predictions and minimise the need for additional laboratory experiments. Simple co-culture experiments using 3D setup can aid groups studying the tumor microenvironment to understand complex interactions of cancer and non-cancer cells, which can be integrated with organ chip or the printed organ model to mimic actual organ with high precision. Publicly available datasets can be used for basic to extensive in silico analysis to understand the etiology, design drugs, and predict disease outcomes. For non-computational or wet-lab researchers, several software tools are publicly available and being developed in a way that can easily be handled by them for basic yet important analysis. While few research groups can independently perform and integrate both wet-lab and computational analysis, there is a need for scientists to collaborate and form large interdisciplinary groups, get people from diverse backgrounds under one umbrella to solve difficult and important problems. The use of systems biology approaches by the amalgamation of different human-specific research approaches (by integrating diverse research groups) promises to save time, resources, and provide specificity by reducing errors. Scaling of human biobanks would be vital for aiding human-centric cancer research. Its objective is not only to aid in personalized therapy but also provide human tissues for basic and translational research. Few existing cancer biobanks are listed here (112). With evolving modern in vitro culture methodologies, the cancer biobank can provide human cells/tissues to research labs to understand molecular mechanisms, intracellular communications, regulation of microenvironment, besides being used for high-throughput screening and personalized therapeutics. These can potentially more precisely tap into areas and scope of research which was not possible with animal models.
Biobanks represent the core for the development of future human-centric research. Cancer tissue biobanks need both vertical and horizontal expansion. For the latest in vitro technologies to be of the highest value, a framework has to be in place which ensures rapid, affordable tissue distribution from the biobanks to places (labs and institutes) that do not have easy access. An increase in collaboration both at the level of institute and nation will aid faster development. Policy changes aiming at expanding the cancer biobanks and making them accessible to wider research institutions, whereby promoting projects involving collaborations with hospitals will enhance and widen the scope of human-centric research. Minimizing extensive paperwork for setting up hospital collaborations without compromising on good research practice would be a welcoming step in this regard. Alongside, development of accessible, more economical and advanced cell culture techniques and availability of user-friendly, free computational tools with increased availability of human tissue/cell repositories, surely has the potential to replace animal-centric models that have not been successful enough for long. The proposed roadmap of the human-centric cancer research models as an alternative to the animal-centric model is demonstrated in Figure 1.
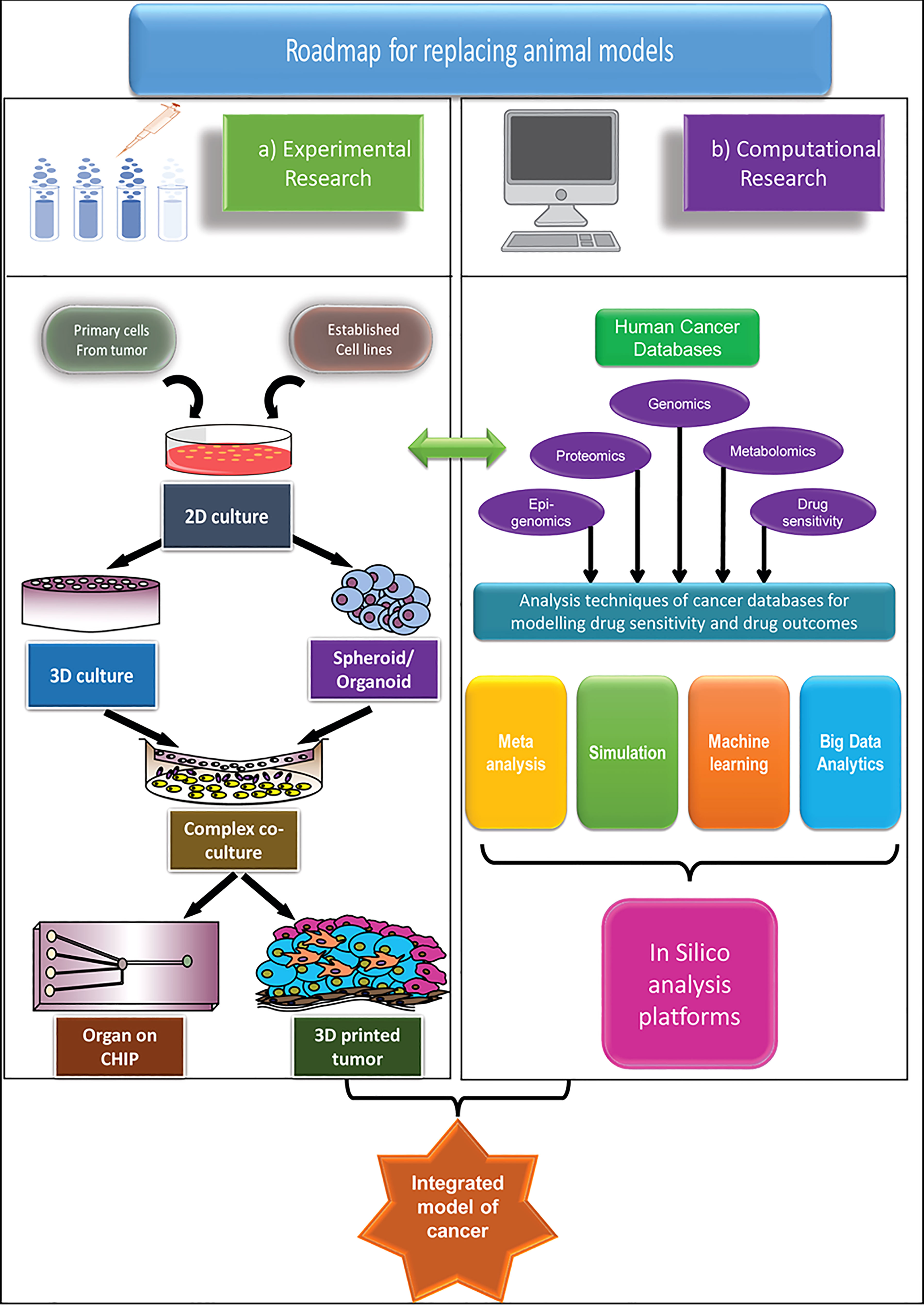
Figure 1 An integrated roadmap to apply advanced human-centric research approaches and potentially replace animal models in cancer research, both for basic and applied research. Two main approaches are considered for replacement of animal models (A) Experimental model and (B) Computational model. The experimental models are mainly based on the in vitro cell culture platforms, which grows in complexity from simple 2D models to the extremely sophisticated organ-on-chip, or 3D printed tumour models to match the specific microenvironment of primary cancer. The computational models take advantage of the different human-based databases and apply different computational methods to model cancer and its outcome. While both methods are advancing significantly, only by combining the two models, we can hope to truly predict the outcome of the cancer therapeutics.
We must realize that developing human-centric methods are essential for studying heterogeneous human diseases like cancer, where disease progression depends upon multiple components. With significant advancements in both wet-lab and dry lab technologies, it gives us the opportunity to work with human cells, thereby moving close to understanding the disease. These advanced set-ups will provide greater leads in understanding the disease both from the mechanistic and therapeutic perspective. We envisage that the human-centric advancements will add great value in reducing cancer-related death by adding specificity to treatment against cancer and specifically drug resistance. We need to shift from a cultural reliance on animal models to more human-centric models which certainly seems to be a relevant and an economical substitute. The integration of computational biology with the modern in vitro human-based cell culture techniques has the potential to replace the necessity for animal experiments. The merits and demerits of all the different models in cancer research are described in Table 3.

Table 3 Potential and limitations of different cancer research models (including in vitro, in vivo and in silico models).
Overall, the whole community including academic and industrial partners as well as their regulators need to show more confidence in using human-based advanced models and explore avenues to better exploit these models for improved drug-predictability, leading to lesser drug attrition, faster and efficient drug development, leading to higher clinical success rates. Collaborative efforts of cancer biologists, computational biologists, and data mining specialists can lead to a better interpretation of these data. Government policy makers and universities should promote such collaborative endeavours by opening new interdisciplinary centers. Funding must be raised from numerous sources to support and increase the reach of such platforms. Although several agencies and charitable institutes are supporting and encouraging funding for non-animal projects, a funding thrust from government and more private agencies can significantly promote the usage and validations using advanced in vitro models.
AP: Conceptualization, literature search, writing, proof reading, framework design. SD: Literature search, writing, conceptualization, figure drawing, proof reading; CD: Literature search, writing, conceptualization, proof reading; MM: Conceptualization, Proof Reading. All authors contributed to the article and approved the submitted version.
The review article receives funding from the “Human Society International”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Humane Society International/India (for their financial support for this project, and Biomed 21 collaboration for discussions and suggestions around the manuscript and editorial suggestions) Ashray Hastha Trust, and CSIR-Centre for Cellular and Molecular Biology. We also extend thanks to the host institutes, National Center for Biological Sciences (TIFR), Bengaluru and Indian Institute of Technology Kharagpur, Kharagpur and Anant National University.
1. Naghavi M, Abajobir AA, Abbafati C, Abbas KM, Abd-Allah F, Abera SF, et al. Global, Regional, and National Age-Sex Specifc Mortality for 264 Causes of Death, 1980-2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet (2017) 390(10100):1151–210. doi: 10.1016/S0140-6736(17)32152-9
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv Pharm Bull (2017) 7:339–48. doi: 10.15171/apb.2017.041
4. Dagogo-Jack I, Shaw AT. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat Rev Clin Oncol (2018) 15:81–94. doi: 10.1038/nrclinonc.2017.166
5. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
6. Burrell RA, Swanton C. Tumour Heterogeneity and the Evolution of Polyclonal Drug Resistance. Mol Oncol (2014) 8:1095–111. doi: 10.1016/j.molonc.2014.06.005
7. Dean M, Fojo T, Bates S. Tumor Stem Cells and Drug Resistance. Nat. Rev. Cancer (2005) 5 (4):275–84. doi: 10.1038/nrc1590
8. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, et al. Cancer Stem Cells (CSCs) in Drug Resistance and Their Therapeutic Implications in Cancer Treatment. Stem Cells Int (2018) 2018. doi: 10.1155/2018/5416923
9. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor Microenvironment Complexity and Therapeutic Implications at a Glance. Cell Communication Signaling (2020) 18(59):1–19. doi: 10.1186/s12964-020-0530-4
10. Balkwill FR, Capasso M, Hagemann T. The Tumor Microenvironment at a Glance. J Cell Sci (2012) 125(23):5591–6. doi: 10.1242/jcs.116392
11. Walker C, Mojares E, Del Río Hernández A. Role of Extracellular Matrix in Development and Cancer Progression. Int J Mol Sci (2018) 19(10):3028. doi: 10.3390/ijms19103028
12. Ungefroren H, Sebens S, Seidl D, Lehnert H, Hass R. Interaction of Tumor Cells With the Microenvironment. Cell Communication Signaling (2011) 9(1):18. doi: 10.1186/1478-811X-9-18
13. Trédan O, Galmarini CM, Patel K, Tannock IF. Drug Resistance and the Solid Tumor Microenvironment. J Natl Cancer Inst (2007) 99(19):1441–54. doi: 10.1093/jnci/djm135
14. Lyssiotis CA, Kimmelman AC. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol (2017) 27:863–75. doi: 10.1016/j.tcb.2017.06.003
15. Klemm F, Joyce JA. Microenvironmental Regulation of Therapeutic Response in Cancer. Trends Cell Biol (2015) 25(4):198–213. doi: 10.1016/j.tcb.2014.11.006
16. Roy S, Trinchieri G. Microbiota: A Key Orchestrator of Cancer Therapy. Nat Rev Cancer (2017) 17(5):271–85. doi: 10.1038/nrc.2017.13
17. Sounni NE, Noel A. Targeting the Tumor Microenvironment for Cancer Therapy. Clin Chem (2013) 59:85–93. doi: 10.1373/clinchem.2012.185363
18. Sleeboom JJF, Amirabadi HE, Nair P, Sahlgren CM, Den Toonder JMJ. Metastasis in Context: Modeling the Tumor Microenvironment With Cancer-on-a-Chip Approaches. Dis Model Mech (2018) 11(3). doi: 10.1242/dmm.033100
19. Fan H, Demirci U, Chen P. Emerging Organoid Models: Leaping Forward in Cancer Research. J Hematol Oncol (2019) 12:142. doi: 10.1186/s13045-019-0832-4
20. Mak IWY, Evaniew N, Ghert M. Lost in Translation: Animal Models and Clinical Trials in Cancer Treatment. Am J Trans Res (2014) 6(2):114–8. doi: 1943-8141/AJTR1312010
21. Moreno L, Pearson AD. How can Attrition Rates be Reduced in Cancer Drug Discovery? Expert Opin Drug Discovery (2013) 8(4):363–8. doi: 10.1517/17460441.2013.768984
22. Workman P, Draetta GF, Schellens JHM, Bernards R. How Much Longer Will We Put Up With 100,000 Cancer Drugs? Cell (2017) 168(4):579–83. doi: 10.1016/j.cell.2017.01.034
23. Hutchinson L, Kirk R. High Drug Attrition Rates - Where are We Going Wrong? Nat Rev Clin Oncol (2011) 8:189–90. doi: 10.1038/nrclinonc.2011.34
24. Adams DJ. The Valley of Death in Anticancer Drug Development: A Reassessment. Trends Pharmacol Sci (2012) 33:173–80. doi: 10.1016/j.tips.2012.02.001
25. Gillet J-P, Varma S, Gottesman MM. The Clinical Relevance of Cancer Cell Lines. J Natl Cancer Inst (2013) 105(7):452–8. doi: 10.1093/jnci/djt007
26. Jordan BThe Legacy of Henrietta Lacks. Medecine/Sciences (2021) 37(12):1189–93. doi: 10.1051/medsci/2021181
27. Bhakta-Guha D, Efferth T. Hormesis: Decoding Two Sides of the Same Coin. Pharmaceuticals (2015) 8(4):865–83. doi: 10.3390/ph8040865
28. Dayem AA, Lee SB, Kim K, Lim KM, Jeon T, Cho SG. Recent Advances in Organoid Culture for Insulin Production and Diabetes Therapy: Methods and Challenges. BMB Rep (2019) 52:295–303. doi: 10.5483/BMBRep.2019.52.5.089
29. Rogal J, Zbinden A, Schenke-Layland K, Loskill P. Stem-Cell Based Organ-on-a-Chip Models for Diabetes Research. Adv Drug Delivery Rev (2018) 140:101–28. doi: 10.1016/j.addr.2018.10.010
30. Zuppinger C. 3d Cardiac Cell Culture: A Critical Review of Current Technologies and Applications. Front Cardiovasc Med (2019) 6. doi: 10.3389/fcvm.2019.00087
31. Lv D, Hu Z, Lu L, Lu H, Xu X. Three-Dimensional Cell Culture: A Powerful Tool in Tumor Research and Drug Discovery. Oncol Lett (2017) 14:6999–7010. doi: 10.3892/ol.2017.7134
32. Alhaque S, Themis M, Rashidi H. Three-Dimensional Cell Culture: From Evolution to Revolution. Philos Trans R Soc B: Biol Sci (2018) 373. doi: 10.1098/rstb.2017.0216
33. Culturing Life - Hannah Landecker - Google Books. Available at: https://books.google.co.in/books?hl=en&lr=&id=CCvjXK91T9QC&oi=fnd&pg=PP15&ots=sGBu_gE7f2&sig=AkBxJAcH0oTZmPqGuKNaIyWSlH0&redir_esc=y#v=onepage&q=hanging&f=false.
34. Harrison RG, Greenman MJ, Mall FP, Jackson CM. Observations of the Living Developing Nerve Fiber. Anat Rec (1907) 1(5):116–28. doi: 10.1002/ar.1090010503
35. Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell (2007) 130:601–10. doi: 10.1016/j.cell.2007.08.006
36. Xin X, Yang H, Zhang F, Yang ST. 3D Cell Coculture Tumor Model: A Promising Approach for Future Cancer Drug Discovery. Process Biochem (2019) 78:148–60. doi: 10.1016/j.procbio.2018.12.028
37. Melissaridou S, Wiechec E, Magan M, Jain MV, Chung MK, Farnebo L, et al. The Effect of 2D and 3D Cell Cultures on Treatment Response, EMT Profile and Stem Cell Features in Head and Neck Cancer 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. Cancer Cell Int (2019) 19(1):16. doi: 10.1186/s12935-019-0733-1
38. Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol (2016) 4:12. doi: 10.3389/fbioe.2016.00012
39. Lancaster MA, Knoblich JA. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science (2014) 345(6194). doi: 10.1126/science.1247125
40. Derby B. Printing and Prototyping of Tissues and Scaffolds. Science (2012) 338:921–6. doi: 10.1126/science.1226340
41. Gunti S, Hoke ATK, Vu KP, London NR. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers (2021) 13(4):874. doi: 10.3390/cancers13040874
42. Fang Y, Eglen RM. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov (2017) 22:456–72. doi: 10.1177/1087057117696795
43. Tuveson D, Clevers H. Cancer Modeling Meets Human Organoid Technology. Science (2019) 364:952–5. doi: 10.1126/science.aaw6985
44. Ishiguro T, Ohata H, Sato A, Yamawaki K, Enomoto T, Okamoto K. Tumor-Derived Spheroids: Relevance to Cancer Stem Cells and Clinical Applications. Cancer Sci (2017) 108(3):283–9. doi: 10.1111/cas.13155
45. Mittler F, Obeïd P, Rulina AV, Haguet V, Gidrol X, Balakirev MY. High-Content Monitoring of Drug Effects in a 3D Spheroid Model. Front Oncol (2017) 7(DEC):293. doi: 10.3389/fonc.2017.00293
46. Nunes AS, Barros AS, Costa EC, Moreira AF, Correia IJ. 3D Tumor Spheroids as In Vitro Models to Mimic In Vivo Human Solid Tumors Resistance to Therapeutic Drugs. Biotechnol Bioeng (2019) 116(1):206–26. doi: 10.1002/bit.26845
47. Langhans SA. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front Pharmacol (2018) 9:6. doi: 10.3389/fphar.2018.00006
48. Granat LM, Kambhampati O, Klosek S, Niedzwecki B, Parsa K, Zhang D. The Promises and Challenges of Patient-Derived Tumor Organoids in Drug Development and Precision Oncology. Anim Model Exp Med (2019) 2(3):150–61. doi: 10.1002/ame2.12077
49. Scuto M, Trovato Salinaro A, Caligiuri I, Ontario ML, Greco V, Sciuto N, et al. Redox Modulation of Vitagenes via Plant Polyphenols and Vitamin D: Novel Insights for Chemoprevention and Therapeutic Interventions Based on Organoid Technology. Mech Ageing Dev (2021) 199(2):111551. doi: 10.1016/j.mad.2021.111551
50. Sun CP, Lan HR, Fang XL, Yang XY, Jin KT. Organoid Models for Precision Cancer Immunotherapy. Front Immunol (2022) 13. doi: 10.3389/fimmu.2022.770465
51. Aparicio S, Hidalgo M, Kung AL. Examining the Utility of Patient-Derived Xenograft Mouse Models. Nat Rev Cancer (2015) 15(5):311–6. doi: 10.1038/nrc3944
52. Ronaldson-Bouchard K, Vunjak-Novakovic G. Organs-On-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell (2018) 22(3):310–24. doi: 10.1016/j.stem.2018.02.011
53. Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling Cancer in Microfluidic Human Organs-on-Chips. Nat Rev Cancer (2019). doi: 10.1038/s41568-018-0104-6
54. Andrei L, Kasas S, Ochoa Garrido I, Stanković T, Suárez Korsnes M, Vaclavikova R, et al. Advanced Technological Tools to Study Multidrug Resistance in Cancer. Drug Resist Updat (2020) 48:100658. doi: 10.1016/j.drup.2019.100658
55. Holen I, Speirs V, Morrissey B, Blyth K. In Vivo Models in Breast Cancer Research: Progress, Challenges and Future Directions. Dis Model Mech (2017) 10(4):359–71. doi: 10.1242/dmm.028274
56. Gonzalez H, Hagerling C, Werb Z. Roles of the Immune System in Cancer: From Tumor Initiation to Metastatic Progression. Genes Dev (2018) 32:1267–84. doi: 10.1101/gad.314617.118
57. Pompili L, Porru M, Caruso C, Biroccio A, Leonetti C. Patient-Derived Xenografts: A Relevant Preclinical Model for Drug Development. J Exp Clin Cancer Res (2016) 35(1):1–8. doi: 10.1186/s13046-016-0462-4
58. Ben-David U, Ha G, Tseng Y-Y, Greenwald NF, Oh C, Shih J, et al. Patient-Derived Xenografts Undergo Mouse-Specific Tumor Evolution. Nat Genet (2017) 49(11):1567–75. doi: 10.1038/ng.3967
59. Lampreht Tratar U, Horvat S, Cemazar M. Transgenic Mouse Models in Cancer Research. Front Oncol (2018) 8:268. doi: 10.3389/fonc.2018.00268
60. Cekanova M, Rathore K. Animal Models and Therapeutic Molecular Targets of Cancer: Utility and Limitations. Drug Design Dev Ther (2014) 8:1911–22. doi: 10.2147/DDDT.S49584
61. Grimm D. Can Clinical Trials on Dogs and Cats Help People? Science (2016). doi: 10.1126/science.aag0742
62. Reuben A, Gopalakrishnan V, Wagner HE, Spencer CN, Austin-Breneman J, Jiang H, et al. Working With Human Tissues for Translational Cancer Research. J Vis Exp (2015) 105. doi: 10.3791/53189
63. Kodack DP, Farago AF, Dastur A, Held MA, Dardaei L, Friboulet L, et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep (2017) 21(11):3298–309. doi: 10.1016/j.celrep.2017.11.051
64. Phan N, Hong JJ, Tofig B, Mapua M, Elashoff D, Moatamed NA, et al. A Simple High-Throughput Approach Identifies Actionable Drug Sensitivities in Patient-Derived Tumor Organoids. Commun Biol (2019) 2(1):1–11. doi: 10.1038/s42003-019-0305-x
65. Kondo J, Inoue M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells (2019) 8(5):470. doi: 10.3390/cells8050470
66. Shang M, Soon RH, Lim CT, Khoo BL, Han J. Microfluidic Modelling of the Tumor Microenvironment for Anti-Cancer Drug Development. Lab Chip (2019) 19(3):369–86. doi: 10.1039/C8LC00970H
67. Murphy SV, Atala A. 3D Bioprinting of Tissues and Organs. Nat Biotechnol (2014) 32:773–85. doi: 10.1038/nbt.2958
68. Langer EM, Allen-Petersen BL, King SM, Kendsersky ND, Turnidge MA, Kuziel GM, et al. Modeling Tumor Phenotypes In Vitro With Three-Dimensional Bioprinting. Cell Rep (2019) 26(3):608–23. doi: 10.1016/j.celrep.2018.12.090
69. Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S. Bioprinting for Cancer Research. Trends Biotechnol (2015) 33:504–13. doi: 10.1016/j.tibtech.2015.06.007
70. Belgodere JA, King CT, Bursavich JB, Burow ME, Martin EC, Jung JP. Engineering Breast Cancer Microenvironments and 3D Bioprinting. Front Bioeng Biotechnol (2018) 6. doi: 10.3389/fbioe.2018.00066
71. Hasin Y, Seldin M, Lusis A. Multi-Omics Approaches to Disease. Genome Biol (2017) 18(1):83. doi: 10.1186/s13059-017-1215-1
72. Sheynkman GM, Shortreed MR, Cesnik AJ, Smith LM. Proteogenomics: Integrating Next-Generation Sequencing and Mass Spectrometry to Characterize Human Proteomic Variation. Annu Rev Anal Chem (Palo Alto Calif) (2016) 9(1):521–45. doi: 10.1146/annurev-anchem-071015-041722
73. Wuest DM, Harcum SW, Lee KH. Genomics in Mammalian Cell Culture Bioprocessing. Biotechnol Adv (2012) 30(3):629–38. doi: 10.1016/j.biotechadv.2011.10.010
74. Pinu FR, Beale DJ, Paten AM, Kouremenos K, Swarup S, Schirra HJ, et al. Systems Biology and Multi-Omics Integration: Viewpoints From the Metabolomics Research Community. Metabolites (2019) 9(4). doi: 10.3390/metabo9040076
75. Gomez-Cabrero D, Abugessaisa I, Maier D, Teschendorff A, Merkenschlager M, Gisel A, et al. Data Integration in the Era of Omics: Current and Future Challenges. BMC Syst Biol (2014) 8 Suppl 2:I1. doi: 10.1186/1752-0509-8-S2-I1
76. Kagohara LT, Stein-O’Brien GL, Kelley D, Flam E, Wick HC, Danilova LV, et al. Epigenetic Regulation of Gene Expression in Cancer: Techniques, Resources and Analysis. Brief Funct Genomics (2018) 17(1):49–63. doi: 10.1093/bfgp/elx018
77. Jimenez CR, Zhang H, Kinsinger CR, Nice EC. The Cancer Proteomic Landscape and the HUPO Cancer Proteome Project. Clin Proteomics (2018) 15(1):4. doi: 10.1186/s12014-018-9180-6
78. Aebersold R, Bader GD, Edwards AM, Van Eyk JE, Kussmann M, Qin J, et al. The Biology/Disease-Driven Human Proteome Project (B/D-HPP): Enabling Protein Research for the Life Sciences Community. J Proteome Res (2013) 12:23–7. doi: 10.1021/pr301151m
79. Ellis MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK, et al. Connecting Genomic Alterations to Cancer Biology With Proteomics: The NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discovery (2013) 3(10):1108–12. doi: 10.1158/2159-8290.CD-13-0219
80. Edwards NJ, Oberti M, Thangudu RR, Cai S, McGarvey PB, Jacob S, et al. The CPTAC Data Portal: A Resource for Cancer Proteomics Research. J Proteome Res (2015) 14(6):2707–13. doi: 10.1021/pr501254j
81. Rivers RC, Kinsinger C, Boja ES, Hiltke T, Mesri M, Rodriguez H. Linking Cancer Genome to Proteome: NCI’s Investment Into Proteogenomics. Proteomics (2014) 14(23–24):2633–6. doi: 10.1002/pmic.201400193
82. Li J, Akbani R, Zhao W, Lu Y, Weinstein JN, Mills GB, et al. Explore, Visualize, and Analyze Functional Cancer Proteomic Data Using the Cancer Proteome Atlas. Cancer Res (2017) 77(21):e51–e54. doi: 10.1158/0008-5472.CAN-17-0369
83. The Cancer Genome Atlas Program - National Cancer Institute. Available at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
84. Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: Exploring the World’s Knowledge of Somatic Mutations in Human Cancer. Nucleic Acids Res (2015) 43(D1):D805–11. doi: 10.1093/nar/gku1075
85. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signal (2013) 6(269). doi: 10.1126/scisignal.2004088
86. Marx V. Drilling Into Big Cancer-Genome Data. Nat Methods (2013) 10(4):293–7. doi: 10.1038/nmeth.2410
87. Packer BR. SNP500Cancer: A Public Resource for Sequence Validation, Assay Development, and Frequency Analysis for Genetic Variation in Candidate Genes. Nucleic Acids Res (2006) 34(90001):D617–21. doi: 10.1093/nar/gkj151
88. Samur MK, Yan Z, Wang X, Cao Q, Munshi NC, Li C, et al. Canevolve: A Web Portal for Integrative Oncogenomics. PloS One (2013) 8(2). doi: 10.1371/journal.pone.0056228
89. He X, Chang S, Zhang J, Zhao Q, Xiang H, Kusonmano K, et al. MethyCancer: The Database of Human DNA Methylation and Cancer. Nucleic Acids Res (2008) 36(SUPPL. 1). doi: 10.1093/nar/gkm730
90. Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: A Database for Somatic Mutations Impacting microRNA Function in Cancer. Nucleic Acids Res (2013) 41(D1). doi: 10.1093/nar/gks1138
91. Liu C, Bai B, Skogerbø G, Cai L, Deng W, Zhang Y, et al. NONCODE: An Integrated Knowledge Database of Non-Coding RNAs. Nucleic Acids Res (2005) 33(DATABASE ISS.). doi: 10.1093/nar/gki041
92. DATABASES: Decoding the Noncode. Science (2005) 307(5708):329b–b. doi: 10.1126/science.307.5708.329b
93. Bulusu KC, Tym JE, Coker EA, Schierz AC, Al-Lazikani B. CanSAR: Updated Cancer Research and Drug Discovery Knowledgebase. Nucleic Acids Res (2014) 42(Database issue):D1040–7. doi: 10.1093/nar/gkt1182
94. Zhang J, Finney RP, Rowe W, Edmonson M, Sei HY, Dracheva T, et al. Systematic Analysis of Genetic Alterations in Tumors Using Cancer Genome WorkBench (CGWB). Genome Res (2007) 17(7):1111–7. doi: 10.1101/gr.5963407
95. Goldman M, Craft B, Swatloski T, Ellrott K, Cline M, Diekhans M, et al. The UCSC Cancer Genomics Browser: Update 2013. Nucleic Acids Res (2013) 41(D1). doi: 10.1093/nar/gks1008
96. Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): A Resource for Therapeutic Biomarker Discovery in Cancer Cells. Nucleic Acids Res (2013) 41(D1). doi: 10.1093/nar/gks1111
97. Wishart DS, Mandal R, Stanislaus A, Ramirez-Gaona M. Cancer Metabolomics and the Human Metabolome Database. Metabolites (2016) 6. doi: 10.3390/metabo6010010
98. Pavlopoulou A, Spandidos DA, Michalopoulos I. Human Cancer Databases (Review). Oncol Rep (2015) 33(1):3–18. doi: 10.3892/or.2014.3579
99. Jean-Quartier C, Jeanquartier F, Jurisica I, Holzinger A. In Silico Cancer Research Towards 3R. BMC Cancer (2018) 18(1). doi: 10.1186/s12885-018-4302-0
100. Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, et al. Ensembl 2019. Nucleic Acids Res (2019) 47(D1):D745–51. doi: 10.1093/nar/gky1113
101. Lee CM, Barber GP, Casper J, Clawson H, Diekhans M, Gonzalez JN, et al. UCSC Genome Browser Enters 20th Year. (2019). pp. 1–6.
102. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res (2017) 45(D1):D353–61. doi: 10.1093/nar/gkw1092
103. Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER Version 11: Expanded Annotation Data From Gene Ontology and Reactome Pathways, and Data Analysis Tool Enhancements. Nucleic Acids Res (2017) 45(D1):D183–9. doi: 10.1093/nar/gkw1138
104. Carbon S, Dietze H, Lewis SE, Mungall CJ, Munoz-Torres MC, Basu S, et al. Expansion of the Gene Ontology Knowledgebase and Resources: The Gene Ontology Consortium. Nucleic Acids Res (2017) 45(D1):D331–8. doi: 10.1093/nar/gkw1108
105. Xenarios I. DIP, the Database of Interacting Proteins: A Research Tool for Studying Cellular Networks of Protein Interactions. Nucleic Acids Res (2002) 30(1):303–5. doi: 10.1093/nar/30.1.303
106. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–13. doi: 10.1093/nar/gky1131
107. Sivakumaran S, Hariharaputran S, Mishra J, Bhalla US. The Database of Quantitative Cellular Signaling: Management and Analysis of Chemical Kinetic Models of Signaling Networks. Bioinformatics (2003) 19(3):408–15. doi: 10.1093/bioinformatics/btf860
108. Kumar P, Han BC, Shi Z, Jia J, Wang YP, Zhang YT, et al. Update of KDBI: Kinetic Data of Bio-Molecular Interaction Database. Nucleic Acids Res (2009) 37(SUPPL. 1):636–41. doi: 10.1093/nar/gkn839
109. LeBeau AP. Considerations for Implementing an Informatics System to Support Biologics Drug Discovery. Drug Discovery Today (2019) 24(1):42–5. doi: 10.1016/j.drudis.2018.09.016
110. Mak KK, Pichika MR. Artificial Intelligence in Drug Development: Present Status and Future Prospects. Drug Discovery Today (2019). doi: 10.1016/j.drudis.2018.11.014
111. Su G, Morris JH, Demchak B, Bader GD. Biological Network Exploration With Cytoscape 3. Curr Protoc Bioinforma (2014) 24(3):773–80. doi: 10.1002/0471250953.bi0813s47
Keywords: drug attrition, human-centric models, cancer research, drug discovery, animal-centric models
Citation: Parekh A, Das S, Das CK and Mandal M (2022) Progressing Towards a Human-Centric Approach in Cancer Research. Front. Oncol. 12:896633. doi: 10.3389/fonc.2022.896633
Received: 15 March 2022; Accepted: 21 June 2022;
Published: 19 July 2022.
Edited by:
Baljinder Singh, Post Graduate Institute of Medical Education and Research (PGIMER), IndiaReviewed by:
Aranzazu Villasante, Institute for Bioengineering of Catalonia (IBEC), SpainCopyright © 2022 Parekh, Das, Das and Mandal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aditya Parekh, YWRpdHlhcGFyZWtoMTAxQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.