- 1Department of Pharmacology, School of Pharmacy, Weifang Medical University, Weifang, China
- 2Department of Medicinal Chemistry, School of Pharmacy, Weifang Medical University, Weifang, China
- 3Department of Toxicology, School of Public Health, Qingdao University, Qingdao, China
Aminopeptidase N (APN, CD13) is closely associated with the development and progression of cancer. Previous studies suggested APN as a biomarker for cancer stem cells. APN inhibitors have been intensively evaluated as chemosensitizers for cancer treatments. In the present study, tetrahydro-β-carboline scaffold was introduced to the structure of APN inhibitors. The synthesized compounds showed potent enzyme inhibitory activities compared with Bestatin, an approved APN inhibitor, in cell-based enzymatic assay. In combination with chemotherapeutic drugs, representative APN inhibitor molecules D12, D14 and D16 significantly improved the antiproliferative potency of anticancer drugs in the in vitro tests. Further mechanistic studies revealed that the anticancer effects of these drug combinations are correlated with decreased APN expression, increased ROS level, and induction of cell apoptosis. The spheroid-formation assay and colony-formation assay results showed effectiveness of Paclitaxel-APN inhibitor combination against breast cancer stem cell growth. The combined drug treatment led to reduced mRNA expression of OCT-4, SOX-2 and Nanog in the cancer stem cells tested, suggesting the reduced stemness of the cells. In the in vivo study, the selected APN inhibitors, especially D12, exhibited improved anticancer activity in combination with Paclitaxel compared with Bestatin. Collectively, potent APN inhibitors were discovered, which could be used as lead compounds for tumor chemo-sensitization and cancer stem cell-based therapies.
Introduction
Aminopeptidase N (APN, also known as CD13) is a widely expressed type II membrane bound metalloprotease (1). It plays important roles in various cellular processes including cell migration, survival (2), viral uptake (3), angiogenesis (4), and autophagy (5). Overexpression of APN have been demonstrated in various cancers such as breast (6), ovarian (7), colorectal (8) and hepatocellular carcinoma (9). It is reported that the serum level of APN is correlated with tumor size, lymph node metastasis, and tumor metastasis (10). Therefore, serum APN expression and activity could be utilized as diagnostic and prognostic biomarkers for different types of cancers.
Inhibition of APN has been intensively evaluated for the treatment of cancers (11). A number of APN inhibitors have been designed and synthesized for targeting various cancer cellular events, including cell migration, cell growth and tumor angiogenesis (12). According to the chemical structures, different kinds of APN inhibitors have been developed, such as antibodies, peptides, and nonpeptide small molecules (13). Bestatin, a natural product extracted from Streptomyces olivoreticuli, is an approved APN inhibitor (14).
Overexpression of APN played important roles in the resistance to anticancer agents, anti-apoptosis of cancer cells, as well as relapse of cancers (5, 15). APN inhibitors, such as Bestatin, have been extensively studied for their abilities in enhancing radiation sensitivity and chemo-sensitivity in different types of cancers (16, 17). Moreover, APN has been identified as a functional marker for cancer stem cells (CSCs) in human cancers (18). Inhibition of APN suppressed the self-renewal and tumor-initiative abilities of CSCs (19). It is indicated that APN is a novel therapeutic target for the treatment of cancer in combination with traditional chemotherapy. Development of APN inhibitors for the enhancement of chemotherapy sensitivity could be a new strategy in cancer treatment.
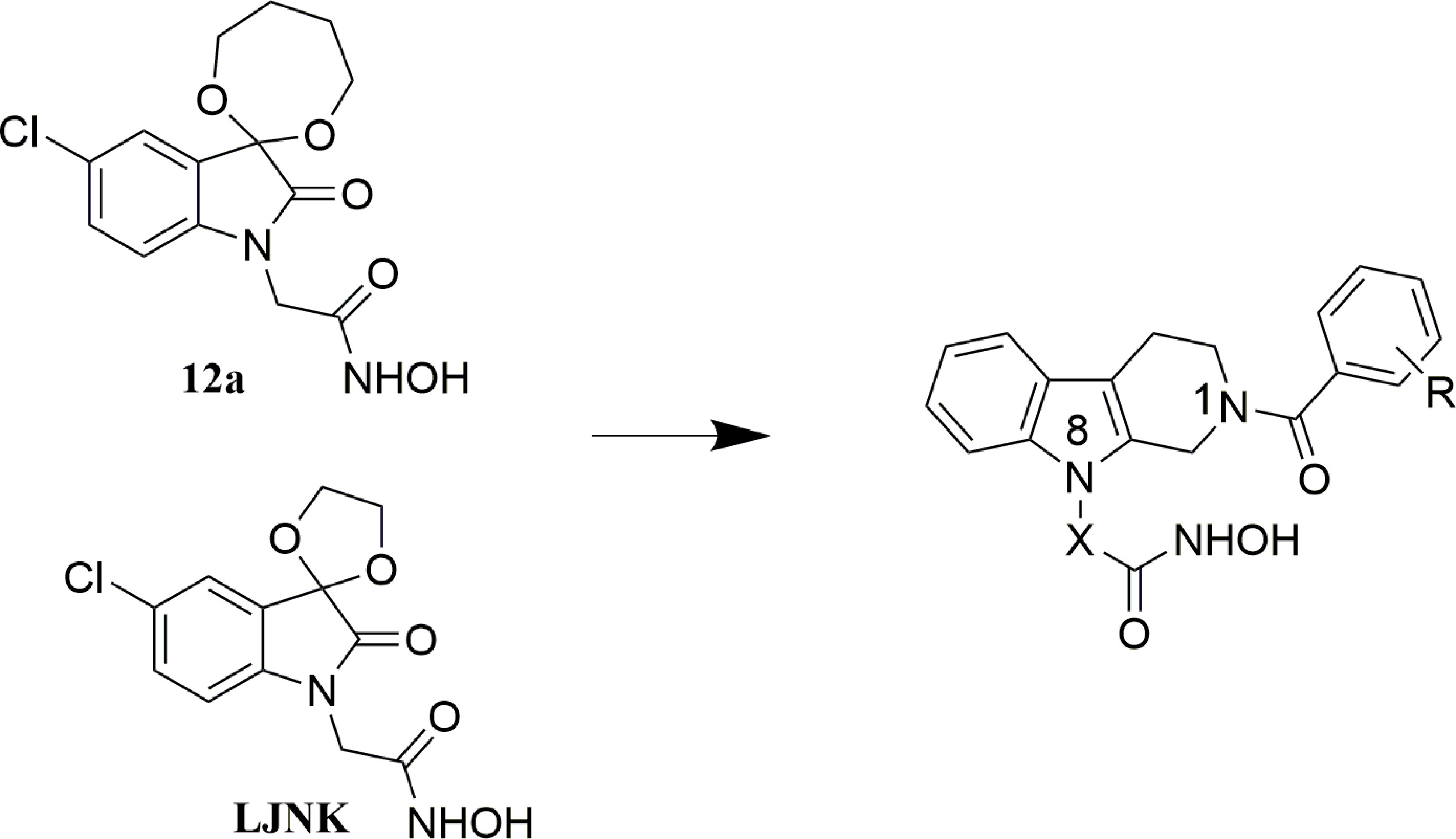
In our previous work, indoline-2,3-dione containing APN inhibitors have been developed with anticancer activities (20, 21). However, further development of these compounds was terminated due to lack of in vivo potency. To develop novel APN inhibitors as chemosensitizers, structural modification was performed on the existing compounds. Generally, a zinc binding group is needed for the chelation of zinc ion, which leads to the binding of inhibitors to the active site of APN. Fragments with various sizes and physicochemical properties were used to occupy different binding pockets in the active site. A linker is used to connect the former pharmacophores. To improve the binding affinity of target molecules in the active site, tetrahydro-β-carboline was utilized as a core fragment in the design of novel APN inhibitors (Figure 1). Substitutions were introduced by condensing different benzoyl chlorides to the N1-position in the carboline ring, and hydroxamic acid group with different linkers was introduced to N8-position as zinc binding group. Short linkers are usually selected in the design of APN inhibitors. In the present study, the contribution of both fatty acid and aromatic linkers was evaluated in the designed APN inhibitors. The synthesized molecules were evaluated using the enzymatic inhibition screening, in vitro antiproliferative test, breast CSC- based study and in vivo anticancer test.
Chemistry
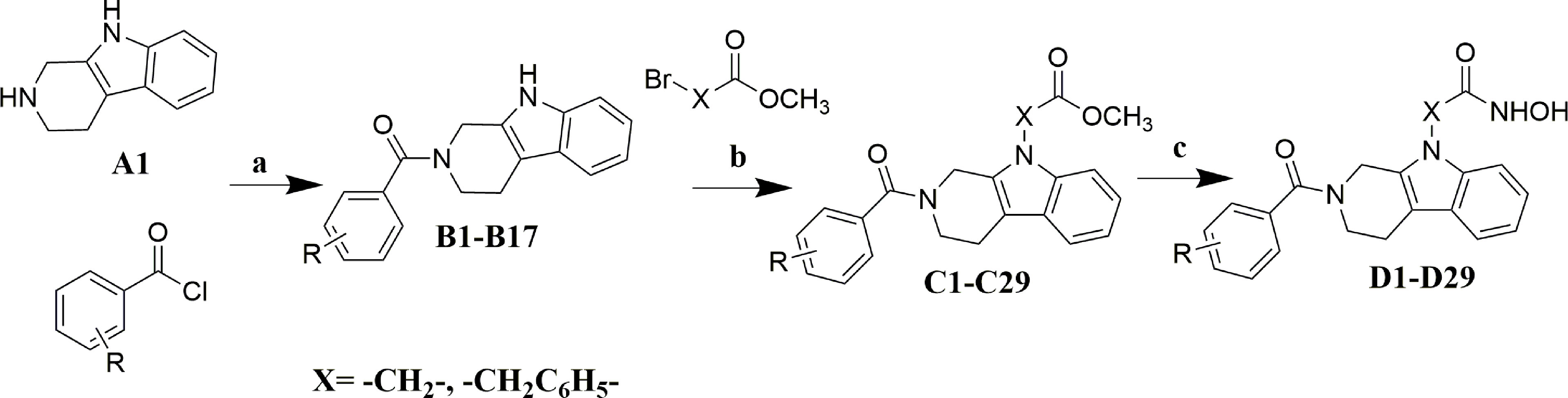
The synthesis of target compounds was described in Scheme 1. The target molecules were prepared by utilizing commercially available 2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole (A1) as the starting material. Firstly, compound A1 was condensed with substituted benzoyl chlorides to obtain intermediate B1-B17. Secondly, key intermediate C1-C29 were obtained by coupling of intermediate B1-B17 with methyl bromoacetate or methyl 4-(bromomethyl)benzoate. At last, target molecules D1-D29 were synthesized by treatment of intermediate C1-C29 with NH2OK in methanol.
Results and Discussions
Enzyme Inhibitory Activity of Synthesized Molecules
The substituted carboline structures are widely utilized in the design of anticancer molecules, such as histone deacetylase (HDAC6) inhibitors (22), kinesin spindle protein inhibitors, breast cancer resistance protein (ABCG2) inhibitors (23), phosphodiesterase 5 inhibitors (24), transforming growth factor beta (TGFβ) signaling pathway inhibitors (25), and bromodomain and extra terminal proteins (BET) inhibitors (26). The carboline moiety played important roles in the site occupation and hydrophobic interaction of the ligand-receptor complexes. On the other hand, the hydrophobic pocket in the active site of APN requires hydrophobic and bulky fragments for efficient binding. Therefore, in the current study, the tetrahydro-β-carboline scaffold was utilized for the occupation of target compounds to the hydrophobic pocket of APN.
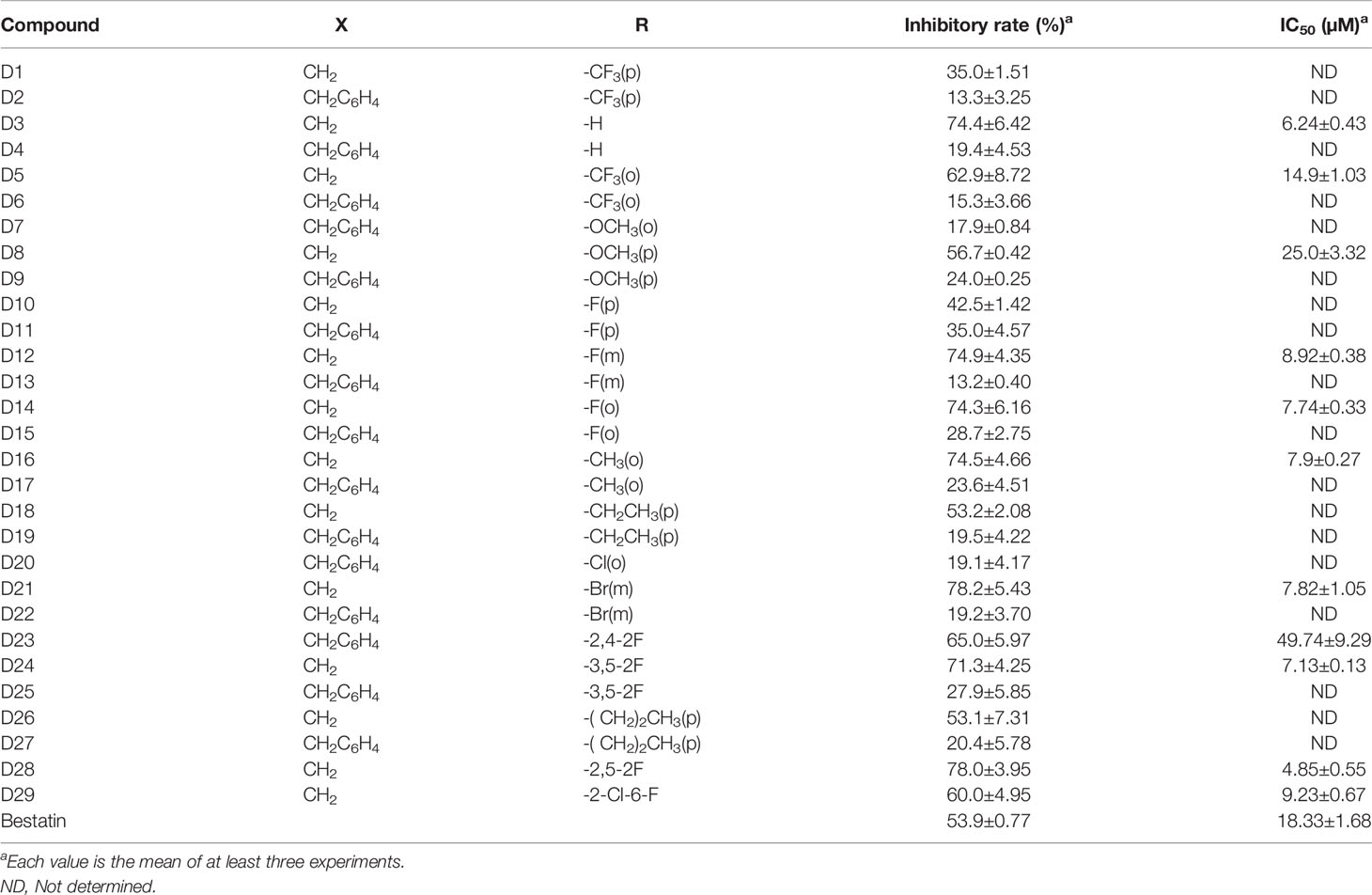
In the enzyme inhibitory assay, K562-CD13 monoclonal cells were used as APN enzyme source as described in our previous work (27). The derived target molecules were firstly screened at the concentration of 30 µM; then, compounds with inhibitory rate higher than 55% were further investigated for the IC50 value (Table 1 and Figures 2A, B). Notably, molecules with a short linker (CH2) between hydroxamic acid group and tetrahydro-β-carboline group exhibited better inhibitory activities than compounds with an aromatic linker (CH2C6H4). Several compounds exhibited increased activity compared with Bestatin (IC50 value 18.33 µM), such as D3 (IC50 value 6.24 µM), D14 (IC50 value 7.74 µM), D16 (IC50 value 7.90 µM), D21 (IC50 value 7.82 µM), D24 (IC50 value 7.13 µM), and D28 (IC50 value 4.85 µM). Comparing with D3, introduction of R groups by para substitution on the phenyl ring led to decreased activity as revealed in Table 1. Nevertheless, the fluorine substitution on the ortho and meta positions can maintain the inhibitory potency, as seen in D12, D14, D24 and D28. From the present structure activity relationship analysis, substitution on the ortho position of the phenyl ring could be beneficial to the inhibitory activity, and further structural modification could be performed by targeting this site.
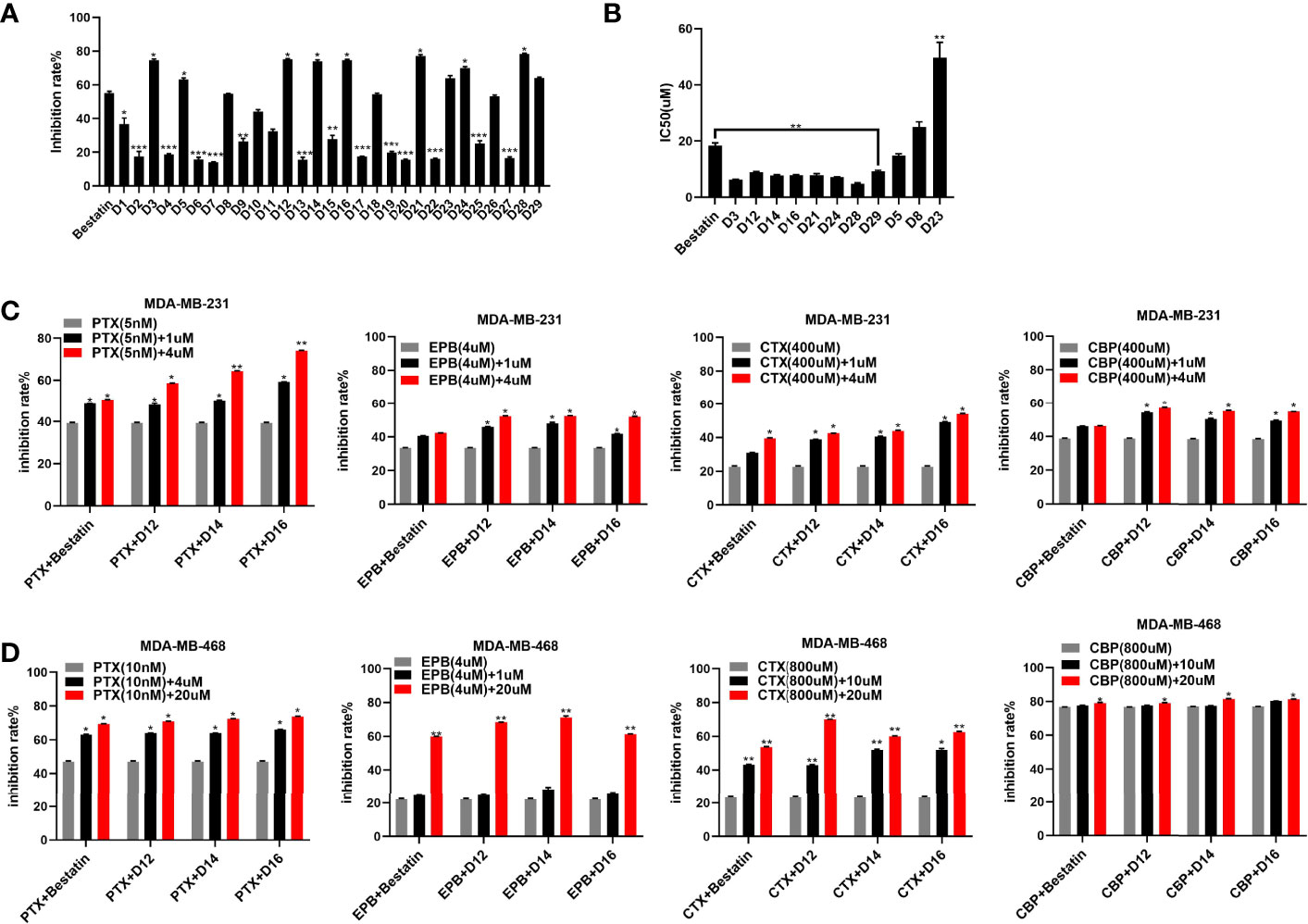
Figure 2 Enzyme inhibitory and antiproliferative activities of the derived APN inhibitors. a: APN enzyme inhibitory rates (A) of the synthesized compounds; (B) IC50 values of selected compounds; (C) antiproliferative activities of representative compounds in combination with chemotherapeutic drugs against MDA-MB-231 cells; (D) antiproliferative activities of representative compounds in combination with chemotherapeutic drugs against MDA-MB-468 cells; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Tumor Chemo-Sensitization Ability of Selected APN Inhibitors
APN has been revealed to be overexpressed in breast cancer and the APN expression is correlated with resistance to anticancer drugs, such as doxorubicin (EPB), in breast cancer cells (6, 28). The combination of APN inhibitor and paclitaxel (PTX), has been employed for the treatment of human breast cancer. In the current study, molecules with good APN inhibitory potency (D12, D14 and D16) were selected and used in combination with several chemotherapeutic drugs, such as PTX, EPB, cyclophosphamide (CTX) and carboplatin (CBP) to investigate its inhibitory effect on breast cancer. Human triple negative breast cancer cell (TNBC) lines MDA-MB-231 and MDA-MB-468 were utilized in the in vitro studies. The results revealed that the tested compounds improve the antiproliferative activities of chemotherapeutic drugs against both cell lines with superior potency than the approved APN inhibitor Bestatin (Figures 2C, D). It was also observed that the synthesized molecules enhanced chemotherapeutic drug sensitivity in a dose dependent manner. Therefore, further analysis was performed to examine the synergistic effects of the derived APN inhibitors and chemotherapeutic drugs.
APN Expression, ROS Level, and Apoptosis Analysis
PTX exhibited the highest inhibitory potency against both cell lines among the tested chemotherapeutic drugs. Moreover, APN inhibitor-induced chemo-sensitization had been frequently reported for PTX (17, 29). Therefore, the combination of PTX-APN inhibitor was selected in the subsequent studies. Enhanced APN expression was detected after treatment with the PTX relative to the untreated control, suggesting that PTX treatment up-regulated APN expression in both MDA-MB-231 and MDA-MB-468 cells (Figure 3A). Treatment with D12, D14, D16 or Bestatin in combination with PTX decreased APN expression in both cell lines compared with paclitaxel treatment alone. Compared to the control group, increased ROS levels were observed in both breast cancer cell lines following PTX treatment (Figure 3B). Co-treatment with selected APN inhibitors resulted in further increase of intracellular ROS levels than PTX treatment alone. The apoptotic results revealed that treatment of the tested breast cancer cell lines with APN inhibitors promoted the apoptosis-inducing effect of PTX (Figures 3C, D). Compared with Bestatin, the synthesized compounds D12, D14 and D16 in combination with PTX treatment induced higher apoptotic rate. These results suggested that the selected APN inhibitors can sensitize TNBC cells to PTX by decreasing APN expression, increasing ROS level and induction of apoptosis.
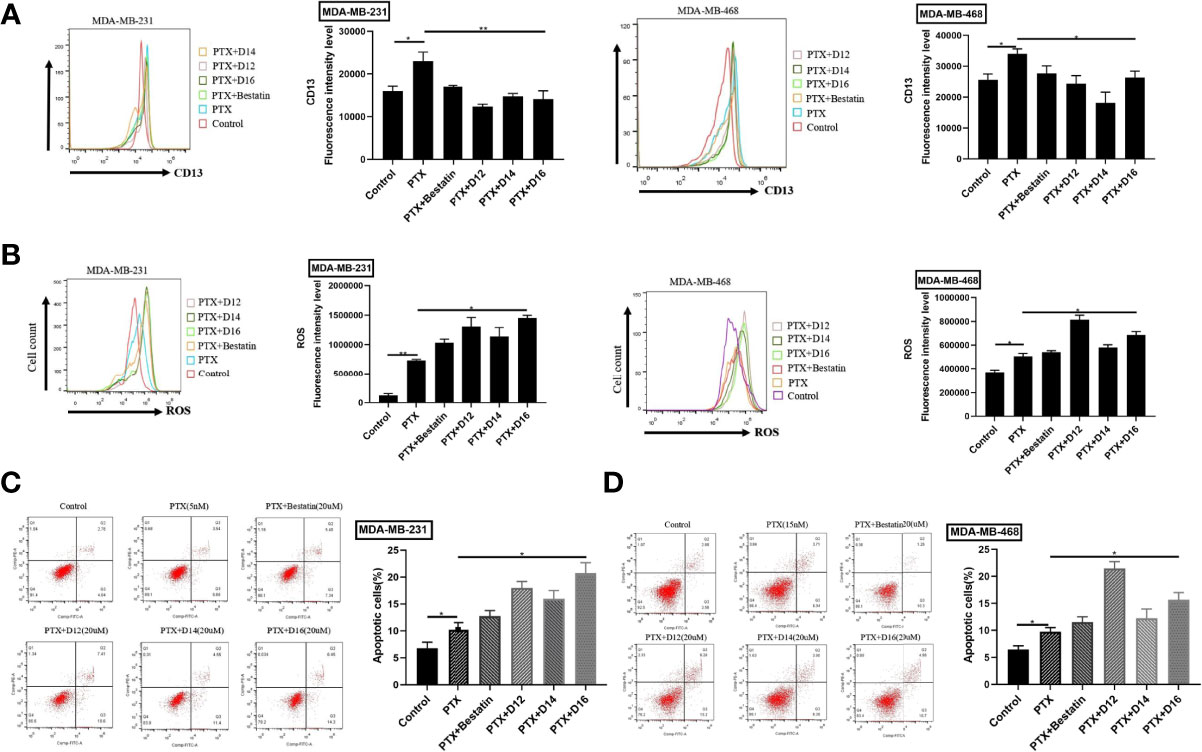
Figure 3 Combination of APN inhibitors with PTX decreased the expression CD13 (APN), increased ROS level, and promoted cell apoptosis. (A) CD13 (APN) expression regulated by various PTX-APN inhibitor combinations; (B) ROS level monitored by various PTX-APN inhibitor combinations; (C) MDA-MB-231 cell apoptosis induced by different PTX-APN inhibitor combinations; (D) MDA-MB-468 cell apoptosis induced by different PTX-APN inhibitor combinations; *P ≤ 0.05, **P ≤ 0.01.
Cancer Stemness Inhibitory Test
Cancer stem cells (CSCs) play an important role in the resistance of cancer cells to chemotherapeutic drugs. APN has been reported to be a therapeutic target in human cancer stem cells (18). Therefore, the CSC based test was performed in the present study. Colony-formation assay and spheroid-formation assay are two commonly used methods to assess the capacity of CSCs in vitro. In the spheroid-formation assay, MDA-MB-231 and MDA-MB-468 cells were dispersed into single cells, inoculated into low-adherence 6-well plates, and added with stem cell sphere medium. The stem cell spheres became visible after 5-6 days incubation. Stem cell sphere medium was added with 1nM of PTX in combination with 1 µM of D12, D14, D16 or Bestatin. After 7 days of incubation, significant decrease in the number of tumor spheres formed in both MDA-MB-231 and MDA-MB-468 cells was observed in the drug combination groups comparing with the control and PTX alone groups (Figures 4A, B).To evaluate the effect of PTX in combination with APN inhibitors on the colony formation ability of TNBC cells, MDA-MB-231 and MDA-MB-468 cells were inoculated with 3000 cells per well in a 6-well plate. After treating with 0.2 nM of PTX combined with 1µM of various APN inhibitors for 14 days, the drug combination groups exhibited reduced number of colonies compared with the control and single PTX treatment groups (Figures 4C, D). It is known that mRNA levels of the stem cell biomarkers OCT-4, SOX-2 and Nanog are associated with the characteristics of TNBC stem cells (30). Therefore, RT-PCR was performed on the TNBC cells to detect the mRNA levels of these markers. The result showed that the mRNA expression levels of OCT-4, SOX-2 and Nanog in the TNBC stem cell spheres significantly decreased in the combined treatment groups compared with the control and single PTX groups (Figures 4E, F). These results suggested that the synthesized APN inhibitors (D12, D14, D16) could inhibit the stemness of TNBC cells when combined with PTX, with a higher potency than Bestatin.
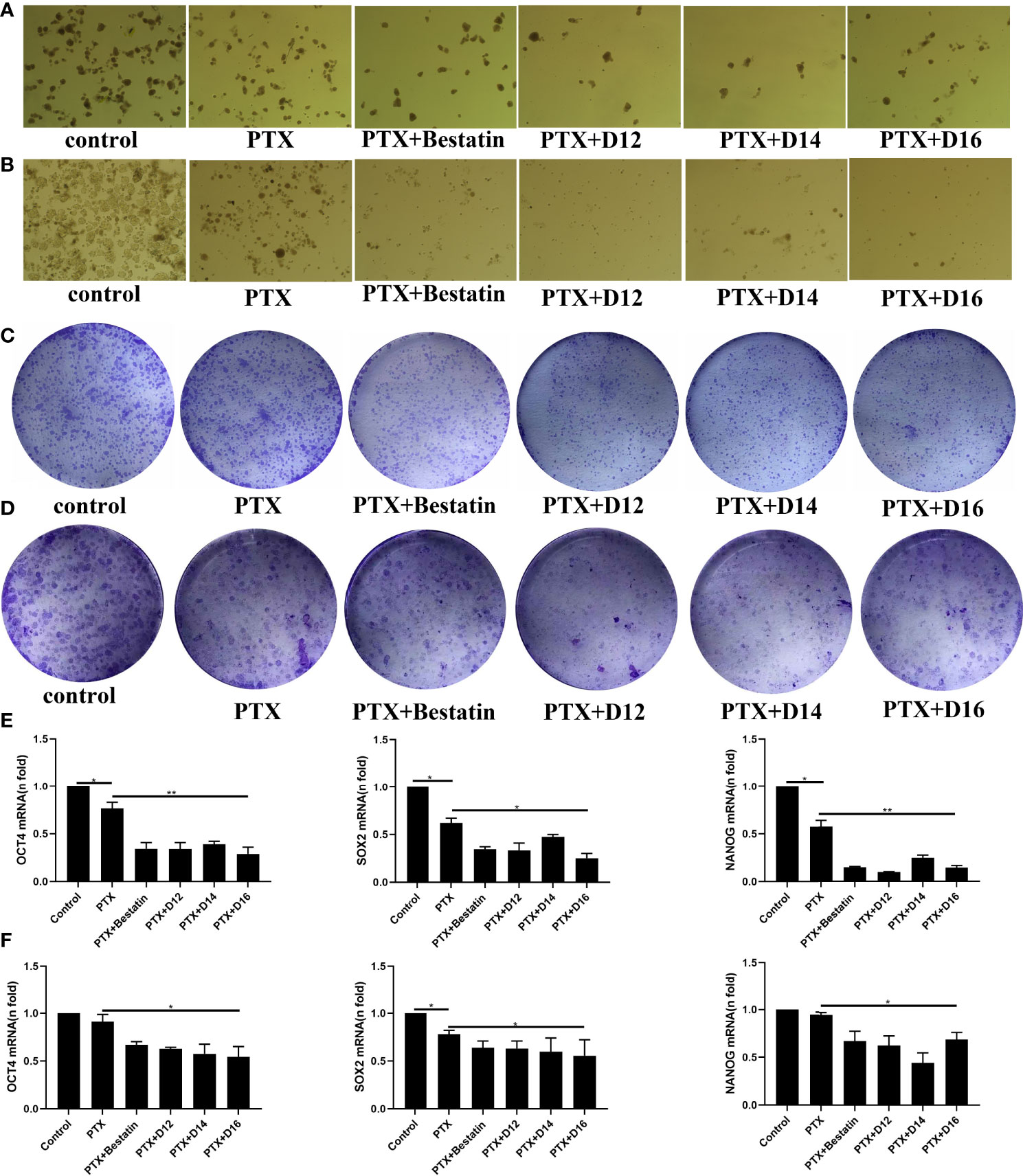
Figure 4 Effects of PTX-APN inhibitor combinations on spheroid and colony formation of tested TNBC cells, and on mRNA expression levels of OCT-4, SOX-2 and Nanog in the tested TNBC stem cells. (A) MDA-MB-231 cell spheroid formation affected by different PTX-APN inhibitor combinations; (B) MDA-MB-468 cell spheroid formation affected by different PTX-APN inhibitor combinations; (C) MDA-MB-231 cell colony formation affected by different PTX-APN inhibitor combinations; (D) MDA-MB-468 cell colony formation affected by different PTX-APN inhibitor combinations; (E) regulation of mRNA expression levels of OCT-4, SOX-2 and Nanog by different PTX-APN inhibitor combinations in MDA-MB-231 cells; (F) regulation of mRNA expression levels of OCT-4, SOX-2 and Nanog by different PTX-APN inhibitor combinations in MDA-MB-468 cells; *P ≤ 0.05, **P ≤ 0.01.
In vivo Anticancer Study
To evaluate the in vivo anticancer potency of the combination of PTX-APN inhibitor, cancer xenograft model was established by inoculation of luciferase- expressing MDA-MB-231 cells to female nude mice. About 1×107 MDA-MB-231 cells were injected subcutaneously into the right fat pad of the fourth mammary gland of the mice. The tumor growth status of mice in each group was observed with a small animal imaging device. Significant tumor size variations were observed among different groups (Figures 5A, B). Bestatin and the APN inhibitors (D12, D14, D16) improved the anticancer effect of PTX compared with the single PTX administration group (Figure 5C). Among the tested molecules, compound D12 in combination with PTX showed the best performance with the smallest tumor size, with an inhibitory rate of 54.02% compared with D14 (inhibitory rate of 31.32%), D16 (inhibitory rate of 14.15%) and Bestatin (inhibitory rate of 10.63%). It was also observed that the tested compounds inhibit the tumor growth without causing the loss in body weight (Figure 5D). Moreover, no clear signs of toxicity in liver and spleen were detected in the sacrificed mice. These results demonstrated that the derived APN inhibitors (especially D12) enhanced the anticancer effect of chemotherapeutic drug (PTX) compared with Bestatin in the in vivo animal model.
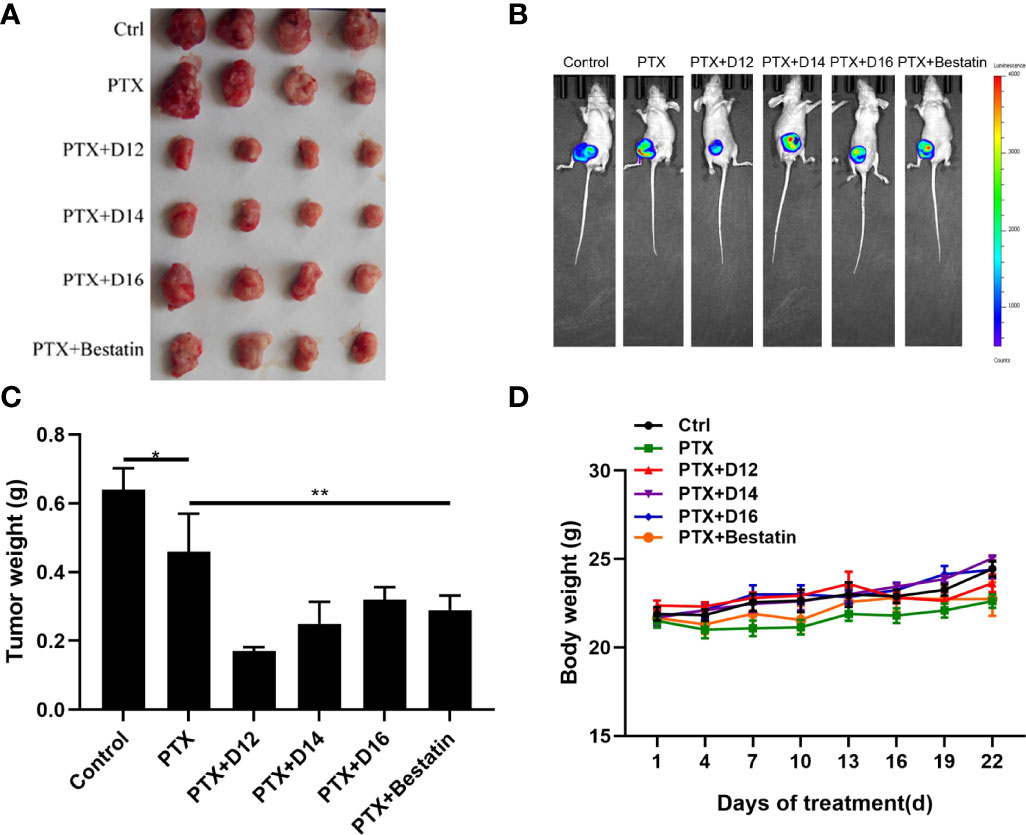
Figure 5 In vivo anticancer effects of the PTX-APN inhibitor combinations. (A) Dissected tumor tissues taken from mice after administration of various drug combinations; (B) representative in vivo imaging of tumors; (C) Tumor weight plot of different administration groups; (D) mice body weight plot of different administration groups; Data were taken as mean ± SEM (n = 4), *P ≤ 0.05, **P ≤ 0.01.
Conclusion
Chemotherapy is the standard treatment for various types of cancers, such as TNBC. However, adverse effects and development of multidrug resistance has led to the search for new drugs or combinatory drug regimens for cancer therapy. APN inhibitors have been investigated as chemosensitizers and used to enhance the efficacy of standard chemotherapy. In order to develop potent chemosensitizers for the cancer treatment, novel APN inhibitors with tetrahydro-β-carboline structure were designed and synthesized in the present study. The chemo-sensitization potential of the derived molecules was evaluated.
Based on the enzymatic assay, the synthesized compounds inhibit the bioactivity of APN with various potency. Several molecules with high enzyme inhibitory activities were selected for further anticancer analysis. In the in vitro study, compound D12, D14 and D16 synergistically improved the antiproliferative activity of chemotherapeutic drugs, such as PTX, EPB, CTX and CBP. PTX, with the highest inhibitory potency against the tested MDA-MB-231 and MDA-MB-468 cells, were evaluated in combination with representative APN inhibitors. It is revealed that the addition of APN inhibitors decreases APN expression level, enhances intracellular ROS production, and improves apoptotic rate in the TNBC cells. The subsequent TNBC stem cell-based studies showed that introduction of APN inhibitors (D12, D14 and D16) decreased the tumor sphere and colony formation abilities in the stem cells. The mRNA expression of stem cell markers OCT-4, SOX-2 and Nanog were decreased by combination of derived APN inhibitors to PTX. In the in vivo nude mouse model, the selected APN inhibitors, especially D12, improved the anticancer activity of PTX, as suggested by the observed smaller tumor size. Collectively, potent APN inhibitors were discovered, which can be used as lead compounds for the development of chemo-sensitizers in cancer treatment.
Materials and Methods
All commercially available starting materials, reagents and solvents were used without further purification. All reactions were monitored by TLC with 0.25 mm silica gel plates (60GF-254). UV light and ferric chloride were used to visualize the spots. 1H NMR and 13C NMR spectra were recorded on a Bruker DRX spectrometer at 500 MHz, using TMS as an internal standard. High-resolution mass spectra was performed in Weifang Medical University. A Thermo UltiMate 3000 High performance liquid chromatograph was used to measure the purity of the derived molecules, and all the target compounds achieved > 95% purity.
Preparation of B1 and its analogues: Derivatives B2-B17 were prepared as described for B1 (see below).
(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)(4-(trifluoromethyl)phenyl)methanone(B1).
To a solution of A1 (0.50 g, 2.9 mmol) in DCM 4-(trifluoromethyl)-benzoylchlorid (0.73 g, 3.5 mmol) and the followed Et3N (0.88 g, 8.7 mmol) were added dropwise at 0°C, and the mixture was stirred for 4 h. Then, the solvent was evaporated with the residue being taken up in EtOAc. The EtOAc solution was washed with saturated citric acid, NaHCO3 and saturated brine solution, dried over MgSO4, and evaporated under vacuum. The desired compound B1 (0.87 g, 87%) was derived by crystallization in EtOAc as white powder. HRMS m/z: 345.12012[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.93 (s, 1H), 7.86 (d, J = 8.0 Hz, 2H), 7.70 (d, J = 8.0 Hz, 2H), 7.40 (d, J = 8.0 Hz, 1H), 7.35-7.25 (m, 1H), 7.08-7.04 (m, 1H), 7.00-6.96 (m, 1H), 4.86-4.55 (m, 2H), 4.02-3,60 (m, 2H), 2.82-2.75 (m, 2H).
Phenyl(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B2).
HRMS m/z: 277.13254[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.94 (s, 1H), 7.48 (d, J = 4.0 Hz, 4H), 7.40 (d, J = 8.0 Hz, 1H), 7.34-7.26 (m, 1H), 7.05-7.04 (m, 1H), 6.99-6.96 (m, 1H), 4.83-4.60 (m, 2H), 3.99-3,63 (m, 2H), 2.75 (s, 2H).
(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)(2-(trifluoromethyl)phenyl)methanone (B3).
HRMS m/z: 345.11850[M+H]+. 1H-NMR (400 MHz, DMSO).δ 10.54 (s, 1H), 7.90-7.86 (m, 1H), 7.83-7.77 (m, 1H), 7.72-7.68 (m, 1H), 7.55-7.49 (m, 1H), 7.41-7.23 (m, 2H), 7.08-7.01 (m, 1H), 6.99-6.95 (m, 1H), 4.87 (dd, J = 94.0, 16.8 Hz, 1H), 4.41-4.22 (m, 1H), 3.47-3.42 (m, 2H), 2.81-2.62 (m, 2H).
(2-methoxyphenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B4).
HRMS m/z: 307.14319[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.61 (s, 1H), 7.46-7.32 (m, 3H), 7.25-7.20 (m, 1H), 7.18-6.95 (m, 3H), 4.84-4.37 (m, 2H), 3.80 (s, 2H), 3.72 (s, 1H), 3.50-3.45 (m, 2H), 2.79-2.62 (m, 2H).
(4-methoxyphenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B5).
HRMS m/z: 307.14322[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.84 (s, 1H), 7.46-7.45 (m, 2H), 7.44-7.39 (m, 1H), 7.30 (d, J = 8.0 Hz, 1H), 4.74 (s, 2H), 3.89-3.72 (m, 5H), 2.79-2.76 (m, 2H).
(4-fluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B6).
HRMS m/z: 295.12253[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.94 (s, 1H), 7.57-7.53 (m, 2H), 7.40 (d, J = 8.0 Hz, 1H), 7.34-7.30 (m, 3H), 7.07-7.03 (m, 1H), 6.99-6.96 (m, 1H), 4.82-4.61 (m, 2H), 3.98-3.63 (m, 2H), 2.77 (s, 2H).
(3-fluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B7).
HRMS m/z: 295.12314[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.92 (s, 1H), 7.57-7.51 (m, 1H), 7.41-7.39 (m, 1H), 7.36-7.30 (m, 4H), 7.07-7.04 (m, 1H), 6.99-6.96 (m, 1H), 4.83-4.59 (m, 2H), 3.99-3.62 (m, 2H), 2.75 (s, 2H).
(2-fluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B8).
HRMS m/z: 295.12253[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.96 (s, 1H), 7.58-7.53 (m, 1H), 7.49-7.25 (m, 5H), 7.07-7.04 (m, 1H), 6.99-6.96 (m, 1H), 4.89-4.50 (m, 2H), 4.03-3.56 (m, 2H), 2.82-2.69 (m, 2H).
(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)(o-tolyl)methanone (B9).
HRMS m/z: 291.14819[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.94 (s, 1H), 7.43-7.17 (m, 6H), 7.07-7.01 (m, 1H), 6.99-6.95 (m, 1H), 4.90 (d, J = 72.0 Hz, 2H), 4.40-4.24 (m, 2H), 3.82-3.47 (m, 2H), 2.82-2.63 (m, 2H), 2.27 (s, 2H), 2.15 (s, 1H).
(4-ethylphenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B10).
HRMS m/z: 305.16403[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.90 (s, 1H), 7.41-7.38 (m, 3H), 7.33-7.31 (m, 3H), 7.06-7.03 (m, 1H), 6.99-6.95 (m, 1H), 4.80-4.64 (m, 2H), 3.97-3.65 (m, 2H), 2.77 (s, 2H), 2.67 (q, J = 7.6 Hz, 2H),1.22 (t, J = 8.0 Hz, 3H).
(2-chlorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B11).
HRMS m/z: 311.09207[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.94 (s, 1H), 7.59-7.57 (m, 1H), 7.55-7.24 (m, 5H), 7.07-7.04 (m, 1H), 6.99-6.95 (m, 1H), 4.94-4.39 (m, 2H), 4.13-3.49 (m, 2H), 2.82-2.68 (m, 2H).
(3-bromophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B12).
HRMS m/z: 353.02856[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.92 (s, 1H), 7.71-7.70 (m, 1H), 7.69 (s, 1H), 7.66-7.41 (m, 3H), 7.39-7.26 (m, 1H), 7.07-7.03 (m, 1H), 6.99-6.95 (m, 1H), 4.83-4.59 (m, 2H), 3.98-3.62 (m, 2H), 2.75 (s, 2H).
(2,4-difluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B13).
HRMS m/z: 313.11172[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.93 (s, 1H), 7.58-7.50 (m, 1H), 7.44-7.19 (m, 4H), 7.08-7.02 (m, 1H), 6.99-6.95 (m, 1H), 4.87-4.50 (m, 2H), 4.02-3.56 (m, 2H), 2.81-2.69 (m, 2H).
(3,5-difluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B14).
HRMS m/z: 313.11337[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.93 (s, 1H), 7.44-7.26 (m, 5H), 7.08-7.02 (m, 1H), 6.99-6.96 (m, 1H), 4.83-4.59 (m, 2H), 3.98-3.59 (m, 2H), 2.80-2.75 (m, 2H).
(4-propylphenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B15).
HRMS m/z: 319.17862[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.94 (s, 1H), 7.41-7.37 (m, 3H), 7.31-7.29 (m, 3H), 7.06-7.03 (m, 1H), 6.99-6.95 (m, 1H), 4.81-4.62 (m, 2H), 3.97-3.33 (m, 2H), 2.76 (s, 2H), 2.61 (t, J = 8.0 Hz, 2H), 1.67-1.85 (m, 2H), 0.93 (t, J = 4.0 Hz, 3H).
(2,5-difluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B16).
HRMS m/z: 313.11273[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.97 (s, 1H), 7.43-7.38 (m, 4H), 7.35-7.27 (m, 1H), 7.08-7.03 (m, 1H), 7.00-6.96 (m, 1H), 4.88-4.52 (m, 2H), 4.04-3.57 (m, 2H), 2.76 (s, 2H), 2.82-2.71 (m, 2H).
(2-chloro-6-fluorophenyl)(1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl)methanone (B17).
HRMS m/z: 329.08441[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.61 (s, 1H), 7.59-7.52 (m, 1H), 7.49-7.27 (m, 4H), 7.09-7.03 (m, 1H), 7.00-6.96 (m, 1H), 4.94-4.45 (m, 2H), 4.11-3.55 (m, 2H), 2.83-2.69 (m, 2H).
Preparation of C1 and its analogues: Derivatives C2-C29 were prepared as described for C1 (see below).
Methyl 2-(2-(4-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9- yl)acetate (C1).
The solution of Cs2CO3 (1.17 g, 3.6 mmol) and B1 (0.40 g, 1.2 mmol) in DMF was firstly stirred under Ar2 and 0°C. After 30 min, methyl bromoacetate was added. The reaction solution was stirred at room temperature for 4 h. Then, the solvent was evaporated under vacuum. The residue was extracted with EtOAc (30 mL), washed with saturated NaHCO3 (3×30 mL), saturated brine solution (3×30 mL), dried over MgSO4, and evaporated under vacuum. The desired compound C1 (0.33 g, 66%) was derived by crystallization in EtOAc as powder. HRMS m/z: 417.14145[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.87 (d, J = 8.0 Hz, 2H), 7.73-7.65 (m, 2H), 7.46-7.37 (m, 2H), 7.14-7.11 (m, 1H), 7.07-7.03 (m, 1H), 5.15-4.93 (m, 2H), 4.83-4.54 (m, 2H), 4.00-3.50 (m, 2H), 3.71 (s, 3H), 2.78 (s, 2H).
Methyl 4-((2-(4-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol- 9-yl)methyl)benzoate (C2).
HRMS m/z: 493.17252[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.92 (d, J = 8.0 Hz, 1H), 7.86 (d, J = 8.0 Hz, 1H), 7.74-7.57 (m, 3H), 7.50-7.45 (m, 3H), 7.22-6.84 (m, 4H), 5.56-5.32 (m, 2H), 4.80-4.44 (m, 2H), 4.00 (s, 1H), 3.83 (s, 3H), 3.59 (s, 1H), 2.87-2.81 (m, 2H).
Methyl 2-(2-benzoyl-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C3).
HRMS m/z: 349.15399[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.50-7.40 (m, 7H), 7.14-7.11 (m, 1H), 7.07-7.04 (m, 1H), 5.15-4.92 (m, 2H), 4.80-4.60 (m, 2H), 3.98-3.51 (m, 5H), 2.80 (s, 2H).
Methyl 2-(2-benzoyl-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C4).
HRMS m/z: 425.18454[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.91-7.49 (m, 2H), 7.47-7.44 (m, 5H), 7.22 (s, 3H), 7.11 (t, J = 8.0 Hz, 1H), 7.05 (t, J = 8.0 Hz, 1H), 6.92 (s, 1H), 5.55-5.29 (m, 2H), 4.77-4.49 (m, 2H), 4.97-4.62 (m, 2H), 3.83 (s, 3H), 2.81 (s, 2H).
Methyl 2-(2-(2-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9- yl)acetate (C5).
HRMS m/z: 417.13935[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.86-7.67 (m, 3H), 7.58-7.56 (m, 1H), 7.48-7.33 (m, 2H), 7.14-7.10 (m, 1H), 7.09-7.03 (m, 1H), 5.15-4.67 (m, 4H), 4.38-3.86 (m, 1H), 3.72 (s, 2H), 3.49-3.36 (m, 2H), 2.83-2.61 (m, 2H).
Methyl 4-((2-(2-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol- 9-yl)methyl)benzoate (C6).
HRMS m/z: 493.17227[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.91-7.85 (m, 2H), 7.79-7.67 (m, 2H), 7.57-7.32 (m, 4H), 7.21 (d, J = 8.0 Hz, 1H), 7.12 (d, J = 8.0 Hz, 1H), 7.07-7.03 (m, 1H), 6.85 (d, J = 8.0 Hz, 1H), 5.57-5.35 (m, 2H), 5.07 (dd, J = 64.0, 16.0 Hz, 1H), 4.69-4.35 (m, 2H), 4.07-3.88 (m, 2H), 3.83 (s, 2H), 3.47-3.40 (m, 2H), 2.73-2.67 (m, 2H).
Methyl 4-((2-(2-methoxybenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzoate (C7).
HRMS m/z: 455.19534[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.94-7.73 (m, 2H), 7.50-7.21 (m, 5H), 7.14-7.00 (m, 4H), 6.93-6.81 (m, 1H), 5.55-5.21 (m, 2H), 4.83-4.32 (m, 2H), 4.23-4.16 (m, 1H), 3.86-3.62 (m, 6H), 3.46 (d, J = 8.0 Hz, 1H), 2.80-2.69 (m, 2H).
methyl 2-(2-(4-methoxybenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) acetate (C8).
HRMS m/z: 379.16446[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.45 (d, J = 8.0 Hz, 2H), 7.39 (d, J = 8.0 Hz, 1H), 7.13-7.12 (m, 1H), 7.06-7.01 (m, 2H), 5.08 (s, 2H), 4.72 (s, 2H), 3.82 (s, 3H), 3.66 (s, 2H), 2.81 (s, 2H).
Methyl 4-((2-(4-methoxybenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzoate (C9).
HRMS m/z: 455.19592[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.87 (s, 2H), 7.50-7.40 (m, 4H), 7.13-6.98 (m, 6H), 5.51 (s, 2H), 4.68 (s, 2H), 3.83 (s, 3H), 3.80-3.79 (m, 5H), 2.84-2.83 (m, 2H).
Methyl 2-(2-(4-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C10).
HRMS m/z: 367.14304[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.56 (s, 2H), 7.46-7.40 (m, 2H), 7.32 (t, J = 8.0 Hz, 2H), 7.14-7.10 (m, 1H), 7.07-7.03 (m, 1H), 5.15-4.94 (m, 2H), 4.78-4.60 (m, 2H), 3.95-3.62 (m, 5H), 2.80 (s, 2H).
Methyl 4-((2-(4-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl) benzoate (C11).
HRMS m/z: 465.15723[M+Na]+. 1H-NMR (400 MHz, DMSO). δ 7.91-7.75 (m, 2H), 7.55 (s, 1H), 7.50-7.45 (m, 2H),7.31-7.20 (m, 2H), 7.13-7.04 (m, 4H), 6.92 (s, 1H), 5.55-5.32 (m, 2H), 4.76-4.48 (m, 2H), 3.95 (s, 1H), 3.83 (s, 3H), 3.62 (s, 1H), 2.82 (s, 2H).
Methyl 2-(2-(3-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C12).
HRMS m/z: 367.14236[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.54 (d, J = 8.0 Hz, 1H), 7.45 (d, J = 8.0 Hz, 1H), 7.41-7.32 (m, 4H), 7.14-7.10 (m, 1H), 7.07-7.03 (m, 1H), 5.13-4.92 (m, 2H), 4.80-4.56 (m, 2H), 3.97-3.52 (m, 5H), 2.79 (s, 2H).
Methyl 4-((2-(3-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl) benzoate (C13).
HRMS m/z: 443.17584[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.93-7.73 (m, 2H), 7.55-7.32 (m, 5H), 7.21-7.04 (m, 4H), 6.92 (s, 1H), 5.56-5.31 (m, 2H), 4.77-4.47 (m, 2H), 3.96 (s, 1H), 3.83 (s, 3H), 3.60 (s, 1H), 2.81 (s, 2H).
Methyl 2-(2-(2-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C14).
HRMS m/z: 367.14447[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.55 (d, J = 8.0 Hz, 1H), 7.46 (d, J = 8.0 Hz, 1H), 7.41-7.34 (m, 4H), 7.14-7.11 (m, 1H), 7.07-7.04 (m, 1H), 5.16-4.94 (m, 2H), 4.81-4.57 (m, 2H), 3.97-3.53 (m, 5H), 2.79 (s, 2H).
Methyl 4-((2-(2-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl) benzoate (C15).
HRMS m/z: 443.17517[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.93-7.75 (m, 2H), 7.57-7.45 (m, 3H), 7.34-7.32 (m, 2H), 7.22-7.04 (m, 4H), 6.93 (s, 1H), 5.56-5.32 (m, 2H), 4.77-4.47 (m, 2H), 3.97 (s, 1H), 3.83 (s, 3H), 3.61 (s, 1H), 2.81 (s, 2H).
Methyl 2-(2-(2-methylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C16).
HRMS m/z: 363.16928[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.48-7.23 (m, 6H), 7.15-7.10 (m, 1H), 7.09-7.02 (m, 1H), 5.15-4.28 (m, 2H), 5.01-4.72 (m, 2H), 3.72-3.47 (m, 5H), 2.82-2.66 (m, 2H), 2.09 (s, 3H).
Methyl 4-((2-(2-methylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzoate (C17).
HRMS m/z: 439.20081[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.91-7.73 (m, 2H), 7.51-7.45 (m, 2H), 7.37-7.21 (m, 4H), 7.15-6.85 (m, 4H), 5.56-5.17 (m, 2H), 5.01-4.13 (m, 2H), 3.88 (s, 1H), 3.84 (t, J = 4.0 Hz, 2H), 3.75-3.47 (m, 2H), 2.84-2.69 (m, 2H), 2.22 (s, 2H), 1.19-1.16 (m, 1H).
Methyl 2-(2-(4-ethylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C18).
HRMS m/z: 377.18558[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.45 (d, J = 8.0 Hz, 1H), 7.40 (d, J = 8.0 Hz, 2H), 7.32 (d, J = 8.0 Hz, 2H), 7.13-7.10 (m, 1H), 7.07-7.03 (m, 1H), 5.13-4.96 (m, 2H), 4.95-4.76 (m, 2H), 3.70-3.64 (m, 5H), 2.79 (s, 2H), 2.69 (q, J = 7.6 Hz, 2H), 1.22 (t, J = 8.0 Hz, 3H).
Methyl 4-((2-(4-ethylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl) benzoate (C19).
HRMS m/z: 453.21667[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.91-7.76 (m, 2H), 7.50-7.45 (m, 2H), 7.38-7.20 (m, 4H), 7.14-6.90 (m, 4H), 5.55-5.32 (m, 2H), 4.75-4.55 (m, 2H), 3.95-3.65 (m, 5H), 2.82 (s, 2H), 2.65 (s, 2H), 1.20 (s, 3H).
Methyl 4-((2-(2-chlorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl) benzoate (C20).
HRMS m/z: 481.12839[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.94-7.73 (m, 2H), 7.58 (d, J = 8.0 Hz, 1H), 7.49-7.43 (m, 4H), 7.28-6.90 (m, 5H), 5.57 (s, 2H), 5.37-4.18 (m, 3H), 3.88 (s, 1H), 3.83 (s, 2H), 3.49-3.46 (m, 2H), 2.85-2.73 (m, 2H).
Methyl 2-(2-(2-bromobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C21).
HRMS m/z: 449.04633[M+Na]+. 1H-NMR (400 MHz, DMSO). δ 7.72-7.61 (m, 2H), 7.49-7.40 (m, 4H), 7.14-7.10 (m, 1H), 7.07-7.03 (m, 1H), 5.15-4.94 (m, 2H), 4.80-4.56 (m, 2H), 3.96-3.54 (m, 5H), 2.78 (s, 2H).
methyl 4-((2-(3-bromobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl) benzoate (C22).
HRMS m/z: 503.09595[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.93-7.65 (m, 3H), 7.53-7.45 (m, 4H), 7.25-7.19 (m, 2H), 7.13-7.10 (m, 1H), 7.05 (d, J = 8.0 Hz, 1H), 6.95 (s, 1H), 5.56-5.31 (m, 2H), 4.76-4.47 (m, 2H), 3.95-3.60 (m, 5H), 2.81 (s, 2H).
Methyl 4-((2-(2,4-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzoate (C23).
HRMS m/z: 461.16650[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.93-7.75 (m, 2H), 7.58-7.40 (m, 3H), 7.31 (q, J = 8.0 Hz, 1H), 7.24-7.20 (m, 2H), 7.14-7.03 (m, 2H), 6.98-6.92 (m, 1H), 5.55-5.35 (m, 2H), 4.81-4.36 (m, 2H), 3.99 (s, 1H), 3.85 (s, 1H), 3.82 (s, 2H), 3.58-3.55 (m, 1H), 2.84-2.76 (m, 2H).
Methyl 2-(2-(3,5-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) acetate (C24).
HRMS m/z: 385.13513[M+H5]+. 1H-NMR (400 MHz, DMSO). δ 7.46-7.40 (m, 3H), 7.35-7.26 (m, 2H), 7.18-7.10 (m, 1H), 7.07-7.03 (m, 1H), 5.15-4.96 (m, 2H), 4.80-4.55 (m, 2H), 3.96-3.56 (m, 5H), 2.78 (s, 2H).
Methyl 4-((2-(3,5-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzoate (C25).
HRMS m/z: 461.16635[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.93-7.74 (m, 2H), 7.50-7.38 (m, 2H), 7.25-6.94 (m, 7H), 5.56-5.34 (m, 2H), 4.76-4.43 (m, 2H), 3.95 (s, 1H), 3.83 (s, 3H), 3.59 (s, 1H), 2.80 (s, 2H).
Methyl 2-(2-(4-propylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetate (C26).
HRMS m/z: 391.20087[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.46-7.39 (m, 4H), 7.31-7.29 (m, 2H), 7.13-7.10 (m, 1H), 7.06-7.03 (m, 1H), 5.13-4.95 (m, 2H), 4.87-4.77 (m, 2H), 3.91-3.52 (m, 5H), 2.79 (s, 2H), 2.63-2.60 (m, 2H), 1.67-1.58 (m, 2H), 0.92 (t, J = 4.0 Hz, 3H).
Methyl 4-((2-(4-propylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzoate (C27).
HRMS m/z: 467.23260[M+H]+. 1H-NMR (400 MHz, DMSO). δ 7.91-7.76 (m, 2H), 7.47 (dd, J = 8.0, 7.6 Hz, 2H), 7.37-7.20 (m, 4H), 7.13-7.03 (m, 3H), 6.88 (s, 1H), 5.55-5.31 (m, 2H), 4.74-4.54 (m, 2H), 3.95 (s, 1H), 3.83 (s, 3H), 3.64 (s, 1H), 2.82 (s, 2H), 2.60 (s, 2H), 1.60 (s, 2H), 0.90 (s, 3H).
Methyl 2-(2-(2,5-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) acetate (C28).
HRMS m/z: 385.13501[M+H5]+. 1H-NMR (400 MHz, DMSO). δ 7.46-7.29 (m, 5H), 7.14-7.09 (m, 1H), 7.07-7.03 (m, 1H), 5.15-4.96 (m, 2H), 4.85-4.48 (m, 2H), 4.01-3.71 (m, 3H), 3.59-3.53 (m, 2H), 2.82-2.74 (m, 2H).
Methyl 2-(2-(2-chloro-6-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9- yl)acetate (C29).
HRMS m/z: 401.10547[M+H5]+. 1H-NMR (400 MHz, DMSO). δ 7.58-7.35 (m, 5H), 7.15-7.09 (m, 1H), 7.07-7.03 (m, 1H), 5.16-4.43 (m, 2H), 4.97-4.84 (m, 2H), 4.07-3.72 (m, 3H), 3.57-3.52 (m, 2H), 2.84-2.70 (m, 2H).
Preparation of D1and its’ analogues: D2-D29
N-hydroxy-2-(2-(4-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetamide (D1).
Compound C1 (0.14 g, 0.6 mmol) was dissolved in 14 mL of NH2OK methanol solution. After 2 h of stiring, the solvent was evaporated under vacuum. The residue was acidified with saturated citric acid, and then extracted with EtOAc for 3 times. The organic layers were combined, washed with brine and dried over MgSO4. The desired compound D1 (0.33 g, 54%) was derived by crystallization in EtOAc as white powder. Purity of 98.51%. HRMS (AP-ESI) m/z calcd for C21H18F3N3O3 [M+H]+ 417.13003, found: 418.13672[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.91 (d, J = 60.28 Hz, 1H), 9.02 (d, J = 46.6 Hz, 1H), 7.87 (d, J = 7.6 Hz, 2H), 7.72 (d, J = 7.6 Hz, 2H), 7.45-7.34 (m, 2H), 7.13 (t, J = 8.0 Hz, 1H), 7.03 (t, J = 8.0 Hz, 1H), 5.06-4.81 (m, 2H), 4.71-4.47 (m, 2H), 4.00-3.58 (m, 2H), 2.78 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 169.26, 164.80, 137.52, 132.47, 128.12, 126.73, 126.14, 123.16, 121.54, 119.60, 118.22, 115.33, 109.91, 107.50, 45.78, 44.13, 21.92 ppm. HPLC (λ = 210 nm): tR = 15.343 min (CH3CN/H2O, 45:55).
N-hydroxy-4-((2-(4-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)benzamide (D2).
Purity of 96.32%. HRMS (AP-ESI) m/z calcd for C27H22F3N3O3 [M+H]+ 493.16133, found: 494.16742[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.28 (s, 1H), 9.07 (s, 1H), 7.87-7.60 (m, 5H), 7.58-7.39 (m, 3H), 7.15-6.82 (m, 4H), 5.51-5.25 (m, 2H), 4.83-4.54 (m, 2H), 4.02-3.59 (m, 2H), 2.87-2.81 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 169.29, 164.33, 141.75, 140.72, 137.06, 132.29, 131.78, 130.09, 128.13, 127.85, 126.75 (d, J = 12.6 Hz), 126.43, 126.08, 121.83, 119.66, 118.43, 110.25, 107.83, 60.23, 46.17, 45.73, 21.95 ppm HPLC (λ = 210 nm): tR = 15.497 min (CH3CN/H2O, 45:55).
2-(2-benzoyl-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D3).
Purity of 95.50%. HRMS (AP-ESI) m/z calcd for C20H19N3O3 [M+H]+ 349.14264, found: 350.14944[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.96 (s, 1H), 9.03 (s, 1H), 7.49-7.40 (m, 7H), 7.12 (t, J = 7.6 Hz, 1H), 7.03 (t, J = 7.2 Hz, 1H), 5.04-4.70 (m, 4H), 3.97-3.62 (m, 2H), 2.78 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 168.76, 164.80, 137.28, 132.74, 13018, 129.03, 127.22, 126.76, 121.49, 119.57, 118.21, 109.88, 107.42, 45.92, 44.10, 22.03 ppm. HPLC (λ = 210 nm): tR = 14.070 min (CH3CN/H2O, 45:55).
4-((2-benzoyl-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D4).
Purity of 97.87%. HRMS (AP-ESI) m/z calcd for C26H23N3O3 [M+H]+ 425.17394, found: 426.1805[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.15 (s, 1H), 9.02 (s, 1H), 7.68-7.29 (m, 9H), 7.14-6.85 (m, 4H), 5.50-5.22 (m, 2H), 4.79-4.56 (m, 2H), 3.98-3.63 (m, 2H), 2.81 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 170.69, 164.34, 141.78, 137.04, 136.62, 132.20 (d, J = 28.1 Hz), 130.15, 129.00, 127.76, 127.20, 126.83, 126.72, 121.79, 119.63, 118.43, 110.21, 46.14, 45.79, 22.04 ppm. HPLC (λ = 210 nm): tR = 15.087 min (CH3CN/H2O, 45:55).
N-hydroxy-2-(2-(2-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetamide (D5).
Purity of 99.27%. HRMS (AP-ESI) m/z calcd for C21H18F3N3O3 [M+H]+ 417.13003, found: 418.13388[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.99 (s, 1H), 9.09 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.80 (t, J = 8.0 Hz, 1H), 7.74-7.67 (m, 1H), 7.58-7.57 (m, 1H), 7.47-7.33 (m, 2H), 7.12(q, J = 8.0 Hz, 1H), 7.03 (t, J = 8.0 Hz, 1H), 5.15-5.06 (m, 1H), 4.78-4.72 (m, 2H), 4.60-4.14 (m, 1H), 3.86-3.38 (m, 2H), 2.82-2.60 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 167.62, 164.75, 137.29, 135.43, 133.47 (d, J = 14.9 Hz), 132.19, 130.08 (d, J = 13.2 Hz), 127.76 (d, J = 12.1 Hz), 127.12 (d, J = 3.4 Hz), 126.65, 125.85-125.55 (m), 122.93, 121.54, 119.58, 118.27 (d, J = 11.6 Hz), 109.93, 107.20, 60.24, 45.31, 44.13, 21.46 ppm. HPLC (λ = 210 nm): tR = 14.690 min (CH3CN/H2O, 45:55).
N-hydroxy-4-((2-(2-(trifluoromethyl)benzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)benzamide (D6).
Purity of 97.92%. HRMS (AP-ESI) m/z calcd for C27H22F3N3O3 [M+H]+ 493.16133, found: 494.16602[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.18 (s, 1H), 9.05 (d, J = 9.6 Hz, 1H), 7.88-7.71 (m, 4H), 7.69-7.52 (m, 2H), 7.48-7.32 (m, 2H), 7.16-6.78 (m, 4H), 5.52-4.36 (m, 4H), 4.21-3.41 (m, 2H), 2.85-2.63 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 167.59, 164.40, 141.54 (d, J = 24.0 Hz), 137.06 (d, J = 5.0 Hz), 135.01 (d, J = 35.6 Hz), 133.38 (d, J = 24.5 Hz), 132.17 (d, J = 35.0 Hz), 131.40 (d, J = 20.0 Hz), 129.99(d, J = 32.3 Hz), 127.82 (d, J = 5.4 Hz), 127.46 (d, J = 25.4 Hz), 127.11 (d, J = 4.3 Hz), 126.88, 126.63(d, J = 5.2 Hz), 126.40, 125.87-125.55 (m), 121.84, 119.63 (d, J = 5.8 Hz), 118.46 (d, J = 7.5 Hz), 110.18 (d, J = 15.1 Hz), 108.1, 107.66, 60.23, 46.08 (d, J = 17.8 Hz), 45.06 (d, J = 34.0 Hz), 21.19 (d, J = 59.0 Hz) ppm. HPLC (λ = 210 nm): tR = 16.023 min (CH3CN/H2O, 45:55).
N-hydroxy-4-((2-(2-methoxybenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)benzamide (D7).
Purity of 98.27%. HRMS (AP-ESI) m/z calcd for C27H25N3O4 [M+H]+ 455.18451, found: 456.19095[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.15 (s, 1H), 9.01 (s, 1H), 7.70-7.54 (m, 2H), 7.50-7.30 (m, 3H), 7.22 (d, J = 8.0 Hz, 1H), 7.16-7.01 (m, 5H), 6.97-6.85 (m, 1H), 5.49-5.19 (m, 1H), 4.85-4.28 (m, 2H), 4.15-3.62 (m, 2H), 3.80-3.62 (m, 3H), 3.47 (d, J = 8.0 Hz, 2H), 2.80-2.69 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 167.87 (d, J = 37.5 Hz), 164.28 (d, J = 26.1 Hz), 155.29 (d, J = 34.8 Hz), 141.54 (d, J = 47.9 Hz), 137.02 (d, J = 7.6 Hz), 132.26 (d, J = 22.1 Hz), 132.03, 130.91, 128.05, 127.80, 126.92, 126.68 (d, J = 6.0 Hz), 126.26, 121.74, 121.16, 119.58, 118.41, 111.89, 110.15 (d, J = 5.9 Hz), 108.46, 107.93, 60.24, 55.92, 46.17, 44.95, 21.58 ppm. HPLC (λ = 210 nm): tR = 14.790 min (CH3CN/H2O, 45:55).
N-hydroxy-2-(2-(2-methoxybenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) acetamide (D8).
Purity of 98.39%. HRMS (AP-ESI) m/z calcd for C21H21N3O4 [M+H]+ 379.15321, found: 380.15985[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.92 (s, 1H), 9.02 (s, 1H), 7.47-7.43 (m, 3H), 7.38-7.31 (m, 1H), 7.12(t, J = 7.6 Hz, 1H), 7.05-7.01 (m, 3H), 4.83-4.65 (m, 4H), 3.82 (s, 3H), 3.73 (s, 2H), 2.80 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 170.53, 164.69, 160.83, 140.43, 137.22, 134.46, 132.92, 129.43, 128.58, 126.78, 123.40, 121.46, 119.55, 118.21, 114.23, 109.85, 107.74, 60.23, 55.74, 44.09, 21.87, 14.53 ppm. HPLC (λ = 210 nm): tR = 13.877 min (CH3CN/H2O, 45:55).
N-hydroxy-4-((2-(4-methoxybenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)benzamide (D9).
Purity of 95.09%. HRMS (AP-ESI) m/z calcd for C27H25N3O4 [M+H]+ 455.18451, 456.19104[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.15 (s, 1H), 9.01 (s, 1H), 7.65 (s, 2H), 7.50-7.43 (m, 4H), 7.15-6.99 (m, 6H), 5.45 (s, 2H), 4.72 (s, 2H), 3.81-3.73 (m, 5H), 2.83 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 171.74, 164.36, 160.79, 141.68, 137.03, 132.25, 129.35, 128.40, 127.74, 126.75, 121.77, 119.62, 118.43, 114.17, 110.19, 108.11, 55.71, 46.11, 44.52, 21.95 ppm. HPLC (λ = 210 nm): tR = 14.917 min (CH3CN/H2O, 45:55).
2-(2-(4-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D10).
Purity of 95.65%. HRMS (AP-ESI) m/z calcd for C20H18FN3O3 [M+H]+ 367.13322, found: 368.13782[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.98 (s, 1H), 9.08 (s, 1H), 7.58-7.56 (m, 2H), 7.45-7.40 (m, 2H), 7.34-7.32 (m, 2H), 7.12 (t, J = 7.6 Hz, 1H), 7.05-7.02 (m, 1H), 5.05-4.87 (m, 2H), 4.76-4.48 (m, 2H), 3.96-3.62 (m, 2H), 2.79 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 169.76, 164.56 (d, J = 36.6 Hz), 161.92, 137.22, 133.07, 129.93, 126.75, 121.51, 119.58, 118.22, 115.99 (d, J = 21.7 Hz), 109.87, 107.45.96, 44.10, 21.98 ppm. HPLC (λ = 210 nm): tR = 14.390 min (CH3CN/H2O, 45:55).
4-((2-(4-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide(D11).
Purity of 98.60%. HRMS (AP-ESI) m/z calcd for C26H22FN3O3 [M+H]+ 443.16452, found: 444.16885[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.18 (s, 1H), 9.04 (s, 1H), 7.69-7.55 (m, 3H), 7.49-7.31 (m, 4H), 7.13-7.03 (m, 3H), 9.04 (s, 2H), 5.50-5.25 (m, 2H), 4.78-4.57 (m, 2H), 3.97-3.63 (m, 2H), 2.82 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 169.79, 164.33, 161.87, 141.76, 137.05, 132.63 (d, J = 67.1 Hz), 132.00, 129.88 (d, J = 8.5 Hz), 127.77, 126.79, 126.71, 121.81, 119.64, 118.43, 115.96 (d, J = 21.4 Hz), 110.21, 107.92, 60.24, 46.12, 21.62 (d, J = 76.9 Hz), 14.56 ppm. HPLC (λ = 210 nm): tR = 15.263 min (CH3CN/H2O, 45:55).
2-(2-(3-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D12).
Purity of 97.51%. HRMS (AP-ESI) m/z calcd for C20H18FN3O3 [M+H]+ 367.13322, found: 368.13693[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.95 (s, 1H), 9.04 (s, 1H), 7.54 (s, 1H), 7.45-7.32 (m, 5H), 7.12 (t, J = 7.2, 1H), 7.05-7.02 (m, 1H), 5.05-4.46 (m, 4H), 3.97-3.60(m, 2H), 2.78 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 169.12, 164.73, 162.39 (d, J = 245.2 Hz), 139.06, 137.23, 132.52, 131.31, 126.73, 123.25, 121.51, 119.58, 118.23, 117.04 (d, J = 19.46 Hz), 114.30 (d, J = 22.17 Hz), 109.86, 107.44, 45.78, 44.10, 21.92 ppm. HPLC (λ = 210 nm): tR = 14.407 min (CH3CN/H2O, 45:55).
4-((2-(3-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide(D13).
Purity of 96.92%. HRMS (AP-ESI) m/z calcd for C26H22FN3O3 [M+H]+ 443.16452, found: 444.17139[M+H]+. 1H-NMR (400 MHz, DMSO).δ 11.15 (s, 1H), 9.01 (s, 1H), 7.69-7.55 (m, 3H), 7.53-7.45 (m, 2H), 7.34-7.20 (m, 3H), 7.19-7.03 (m, 2H), 6.88 (s, 2H), 5.50-5.24 (m, 2H), 4.79-4.54 (m, 2H), 3.98-3.61 (m, 2H), 2.81 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 169.14, 164.38, 162.35 (d, J = 246.2 Hz), 141.78, 138.90, 137.04, 132.31, 131.85, 131.29, 127.76, 126.84, 126.69, 123.26, 121.81, 119.63, 118.45, 117.03 (d, J = 21.0 Hz), 114.28 (d, J = 23.2 Hz), 110.21, 107.92, 60.24, 45.93 (d, J = 41.3 Hz), 21.60 (d, J = 71.3 Hz), 14.56 ppm. HPLC (λ = 210 nm): tR = 15.183 min (CH3CN/H2O, 45:55).
2-(2-(2-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D14).
Purity of 98.35%. HRMS (AP-ESI) m/z calcd for C20H18FN3O3 [M+H]+ 367.13322, found: HRMS m/z: 368.13776[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.87 (d, J =63.3 Hz, 1H), 8.99 (d, J = 48.9 Hz, 1H), 7.58-7.28 (m, 6H), 7.14-7.09 (m, 1H), 7.05-7.01 (m, 1H), 5.04-4.44 (m, 4H), 4.00-3.54 (m, 2H), 2.81 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 165.55, 164.74, 158.19 (d, J = 245.3 Hz), 137.25, 132.40, 132.01 (d, J = 8.2 Hz), 129.17, 126.64, 125.53 (d, J = 3.0 Hz), 124.86 (d, J = 18.1 Hz), 121.52, 119.58, 118.20, 116.38 (d, J = 21.0 Hz), 109.93, 107.28, 45.32, 44.13, 21.93 ppm. HPLC (λ = 210 nm): tR = 14.300 min (CH3CN/H2O, 45:55).
4-((2-(2-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D15).
Purity of 98.13%. HRMS (AP-ESI) m/z calcd for C26H22FN3O3 [M+H]+ 443.16452, found: 444.16870[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.17 (s, 1H), 9.04 (d, J = 13.6 Hz, 1H), 7.70-7.52 (m, 3H), 7.51-7.31 (m, 4H), 7.26-6.83 (m, 5H), 5.50-5.24 (m, 2H), 4.86-4.43 (m, 2H), 4.04-3.55 (m, 2H), 2.84-2.75 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 165.58, 164.40, 158.18 (d, J = 244.2 Hz), 141.54 (d, J =41.4 Hz), 137.06, 132.36, 132.03 (d, J = 8.0 Hz), 131.74, 128.98 (d, J = 30.4 Hz), 127.73 (d, J = 15.9 Hz), 126.90, 126.56 (d, J = 15.6 Hz), 125.52-124.65 (m), 121.84, 119.65, 118.47 (d, J = 8.1 Hz), 116.38 (d, J = 21.0 Hz), 110.20 (d, J = 8.7 Hz), 108.50, 107.76, 60.24, 46.12 (d, J = 14.4 Hz), 44.92 (d, J = 70.6 Hz), 21.52(d, J = 89.7 Hz) ppm. HPLC (λ = 210 nm): tR = 15.110 min (CH3CN/H2O, 45:55).
N-hydroxy-2-(2-(2-methylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetamide (D16).
Purity of 95.42%. HRMS (AP-ESI) m/z calcd for C21H21N3O3 [M+H]+ 363.15829, found: 364.16382[M+H]+. 1H NMR (400 MHz, DMSO). δ 10.90 (d, J = 71.4 Hz, 1H), 9.02 (d, J = 54.2 Hz, 1H), 7.47-7.16 (m, 6H), 7.14-7.11 (m, 1H), 7.05-7.01 (m, 1H), 5.11-4.71 (m, 3H), 4.51-4.39 (m, 1H), 3.73-3.47 (m, 2H), 2.82-2.66 (m, 2H), 2.25-2.13 (m, 3H). 13C-NMR (400 MHz, DMSO). δ 170.08, 164.75, 137.20 (d, J = 11.8 Hz), 134.22, 132.70, 130.73, 129.23, 126.70, 126.41, 125.95, 121.48, 119.55, 118.19, 109.91, 107.26, 44.85, 44.13, 21.92, 21.20, 19.06 ppm. HPLC (λ = 210 nm): tR = 14. 923 min (CH3CN/H2O, 45:55).
N-hydroxy-4-((2-(2-methylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl) methyl)benzamide (D17).
Purity of 95.30%. HRMS (AP-ESI) m/z calcd for C27H25N3O3 [M+H]+ 439.18959, found: 440.19635[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.20 (s, 1H), 9.03 (d, J = 9.2 Hz, 1H), 7.71-7.56 (m, 2H), 7.51-7.44 (m, 2H), 7.37-7.22 (m, 3H), 7.20-7.00 (m, 3H), 6.80-6.78 (d, J = 8.0 HZ, 2H), 5.51-5.02 (m, 3H), 4.98 -4.20 (m, 2H), 3.74-3.48 (m, 2H), 2.84-2.68 (m, 2H), 2.23 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 170.10, 164.38, 141.52 (d, J = 53.3 H), 137.03 (d, J = 4.4 Hz), 136.68, 133.92 (d, J = 62.5 Hz), 132.33, 132.04 (d, J = 5.3 HZ), 130.63 (d, J = 16.1 HZ), 129.11 (d, J = 26.9 HZ), 127.73 (d, J = 14.4 HZ), 126.78 (d, J = 18.4 Hz), 126.37 (d, J = 7.1 Hz), 125.75 (d, J = 46.3 Hz), 121.82 (d, J = 7.1 Hz), 119.61, 118.46 (d, J = 11.4 Hz), 110.18 (d, J = 7. Hz), 108.48, 107.71, 46.09 (d, J = 17.7 Hz), 44.81, 44.42, 21.53 (d, J = 83.4 Hz), 18.84 (d, J = 41.1 Hz) ppm. HPLC (λ = 210 nm): tR = 15.820 min (CH3CN/H2O, 45:55).
2-(2-(4-ethylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D18).
Purity of 97.92%. HRMS (AP-ESI) m/z calcd for C22H23N3O3 [M+H]+ 377.17394, found: 378.18045[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.96 (s, 1H), 9.06 (s, 1H), 7.45-7.41 (m, 4H), 7.34-7.32 (d, J = 7.6 Hz, 2H), 7.14-7.11 (m, 1H), 7.06-7.02 (m, 1H), 5.04-4.70 (m, 4H), 3.95-3.64 (m, 2H), 2.79 (s, 2H), 2.68 (dd, J = 15.2, 7.6 Hz, 2H), 1.23 (t, J = 7.6 Hz, 3H). 13C-NMR (400 MHz, DMSO). δ 170.74, 164.72, 146.06, 137.23, 134.03, 132.81, 128.32, 127.47, 126.77, 121.47, 119.56, 118.21, 109.87, 107.45.90, 44.10, 28.49, 22.04, 15.87 ppm. HPLC (λ = 210 nm): tR = 15.897 min (CH3CN/H2O, 45:55).
4-((2-(4-ethylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D19).
Purity of 98.88%. HRMS (AP-ESI) m/z calcd for C28H27N3O3 [M+H]+ 453.20524, found: 454.21204[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.15 (s, 1H), 9.01 (s, 1H), 7.67 (s, 2H), 7.50-7.46 (m, 2H), 7.44-7.39 (m, 3H), 7.13-6.85 (m, 5H), 5.50-5.26 (m, 2H), 4.77-4.61 (m, 2H), 3.97-3.65 (m, 2H), 2.82 (s, 2H), 2.68-2.65 (m, 2H), 1.21-1.16 (m, 3H). 13C-NMR (400 MHz, DMSO). δ 170.82, 164.38, 146.03, 141.74, 137.04, 133.91, 132.14, 128.29, 127.75, 127.41, 126.73, 121.78, 119.62, 118.42, 110.20, 108.02, 60.24, 46.12, 28.46, 22.08, 21.24, 15.82 ppm. HPLC (λ = 210 nm): tR = 17.653 min (CH3CN/H2O, 45:55).
4-((2-(2-chlorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D20).
Purity of 99.27%. HRMS (AP-ESI) m/z calcd for C26H22ClN3O3 [M+H]+ 459.13497, found: 460.13953[M+H]+. 1H-NMR (400 MHz, DMSO) δ 11.16 (s, 1H), 9.03 (d, J = 10.3 Hz, 1H), 7.69 (d, J = 8.0 HZ, 1H), 7.59-7.55 (m, 1H), 7.52-7.42 (m, 4H), 7.34-7.21 (m, 3H), 7.16-7.02 (m, 2H), 6.83 (d, J = 8.0 HZ, 1H), 5.51-5.15 (m, 2H), 4.96-4.25 (m, 2H), 3.87-3.48 (m, 2H), 2.85-2.73 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 167.07, 166.66, 137.06, 136.19 (d, J = 37.9 Hz), 131.71, 130.96 (d, J = 31.6 Hz), 129.67 (d, J = 21.1 Hz), 128.27 (d, J = 15.8 Hz), 127.71 (d, J = 16.3 Hz), 126.90, 126.54 (d, J = 15.9 Hz), 121.83, 119.64, 118.46 (d, J = 8.5 Hz), 110.17 (d, J = 12.5 Hz), 108.43, 107.74, 46.18, 44.60 (d, J = 65.3 Hz), 21.90, 21.10 ppm. HPLC (λ = 210 nm): tR = 14.690 min (CH3CN/H2O, 45:55).
2-(2-(3-bromobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D21).
Purity of 97.53%. HRMS (AP-ESI) m/z calcd for C20H18BrN3O3 [M+H]+ 427.05315, found: 428.05792[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.92 (d, J = 47.3 Hz, 1H), 9.03 (d, J = 38.0 Hz, 1H), 7.76-7.67 (m, 2H), 7.49-7.34 (m, 4H), 7.14-7.11 (m, 1H), 7.05-7.02 (m, 1H), 5.05-4.77 (m, 2H), 4.70-4.47 (m, 2H), 3.96-3.60 (m, 2H), 2.77 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 139.22, 138.25, 133.01, 131.39, 130.10, 126.72, 122.43, 121.52, 119.59, 118.25, 109.92, 107.34, 55.77, 44.11, 21.95 ppm. HPLC (λ = 210 nm): tR = 15.120 min (CH3CN/H2O, 45:55).
4-((2-(3-bromobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D22).
Purity of 97.45%. HRMS (AP-ESI) m/z calcd for C26H22BrN3O3 [M+H]+ 503.08445, found: 504.08893[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.15 (s, 1H), 9.01 (s, 1H), 7.68 (d, J = 8.4 HZ, 3H), 7.59 (s, 1H), 7.49-7.26 (m, 4H), 7.14-6.91 (m, 4H), 5.50-5.25 (m, 2H), 4.79-4.55 (m, 2H), 3.97-3.61 (m, 2H), 2.81 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 168.91, 164.39, 141.83, 138.94, 137.04, 132.97, 132.31, 131.83, 129.89, 127.77, 126.82, 126.68, 126.12, 122.26, 121.82, 119.63, 118.45, 110.21, 107.87, 108, 46.14, 45.77, 21.95 ppm. HPLC (λ = 210 nm): tR = 16.097 min (CH3CN/H2O, 45:55).
4-((2-(2,4-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D23).
Purity of 95.67%. HRMS (AP-ESI) m/z calcd for C26H21F2N3O3 [M+H]+ 461.15510, found: HRMS m/z: 462.16037[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.16 (s, 1H), 9.02 (s, 1H), 7.70-7.53 (m, 2H), 7.50-7.33 (m, 3H), 7.24-7.03 (m, 4H), 6.98-6.65 (m, 1H), 5.54-5.28 (m, 2H), 4.84-4.34 (m, 2H), 3.39-3.56 (m, 2H), 2.84 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 164.66 (d, J = 42.5 Hz), 161.99 (d, J = 12.3 Hz), 159.90 (d, J = 12.3 Hz), 157.44 (d, J = 12.7 Hz), 141.60 (d, J = 26.1 Hz), 137.06, 132.25 (d, J = 18.6 Hz), 131.68, 127.71(d, J = 18.6 Hz), 126.89, 126.55 (d, J = 15.0 Hz), 121.85, 121.32 (d, J = 18.5 Hz), 119.66, 118.48 (d, J = 7.8 Hz), 112.74 (d, J = 17.5 Hz), 110.23, 108.46, 107.76, 105.05 (d, J = 17.4 Hz), 46.09 (d, J = 18.2 Hz), 45.34, 44.61, 21.51 (d, J = 94.3 Hz) ppm. HPLC (λ = 210 nm): tR = 16.067 min (CH3CN/H2O, 45:55).
2-(2-(3,5-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D24).
Purity of 95.49%. HRMS (AP-ESI) m/z calcd for C20H17F2N3O3 [M+H]+ 385.12380, found: 386.12924[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.92 (d, J = 50.0 Hz, 1H), 9.03 (d, J = 39.1 Hz, 1H), 7.45-7.33 (m, 3H), 7.27-7.26 (m, 2H), 7.12 (t, J = 7.2 Hz, 1H), 7.05-7.02 (m, 1H), 5.05-4.78 (m, 2H), 4.70-4.49 (m, 2H), 3.96-3.59 (m, 2H), 2.77 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 167.91, 164.75, 161.51, 137.23, 137.50 (d, J = 40.0 Hz), 130.75, 126.70, 121.53, 119.59, 118.27, 110.71 (d, J = 26.5 Hz), 109.87, 107.48, 45.70, 44.11, 21.83 ppm. HPLC (λ = 210 nm): tR = 14.477 min (CH3CN/H2O, 45:55).
4-((2-(3,5-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)-N-hydroxybenzamide (D25).
Purity of 95.87%. HRMS (AP-ESI) m/z calcd for C26H21F2N3O3 [M+H]+ 461.15510, found: 462.15964[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.16 (s, 1H), 9.01 (s, 1H), 7.70-7.57 (m, 2H), 7.49-7.40 (m, 3H), 7.26-6.93 (s, 2H), 7.24-7.03 (m, 4H), 5.50-5.27 (m, 2H), 4.79-4.54 (m, 2H), 3.97-3.59 (m, 2H), 2.80 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 167.94, 164.37, 164.01 (d, J = 11.5 Hz), 161.54 (d, J = 12.0 Hz),141.77, 140.15, 137.06, 132.33, 131.62, 127.78, 126.85, 126.67, 121.83, 119.64, 118.46, 110.71 (d, J = 26.6 Hz), 110.21, 107.97, 105.60, 105.50 (d, J = 51.5 Hz), 60.24, 45.91 (d, J = 47.8 Hz), 21.55 (d, J = 64.1 Hz), 14.55 ppm. HPLC (λ = 210 nm): tR = 15. 933 min (CH3CN/H2O, 45:55).
N-hydroxy-2-(2-(4-propylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)acetamide (D26).
Purity of 98.35%. HRMS (AP-ESI) m/z calcd for C23H25N3O3 [M+H]+ 391.1859, found: 392.19434[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.97 (s, 1H), 9.07 (s, 1H), 7.45-7.40 (m, 4H), 7.31-7.30 (m, 2H), 7.12 (t, J = 8.0 Hz, 1H), 7.05-7.02 (m, 1H), 5.04-4.85 (m, 2H), 4.69-4.47 (m, 2H), 3.94-3.63 (m, 2H), 2.78 (s, 2H), 2.62 (t, J = 8.0 Hz, 2H), 1.68-1.58 (m, 2H), 0.92 (t, J = 8.0 Hz, 3H). 13C-NMR (400 MHz, DMSO). δ 170.81, 164.72, 144.47, 137.23, 134.05, 132.81, 128.88, 127.38, 126.77, 121.46, 119.55, 118.21, 109.87, 107.46, 60.23, 45.89, 44.10, 37.52, 24.37, 21.62 (d, J = 76.9 Hz), 14.14 ppm. HPLC (λ = 210 nm): tR = 16.550 min (CH3CN/H2O, 45:55).
N-hydroxy-4-((2-(4-propylbenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)methyl)benzamide (D27).
Purity of 99.31%. HRMS (AP-ESI) m/z calcd for C29H29N3O3 [M+H]+ 467.22089, found: 468.22525[M+H]+. 1H-NMR (400 MHz, DMSO). δ 11.15 (s, 1H), 9.01 (s, 1H), 7.67 (s, 2H), 7.49-7.43 (s, 2H), 7.38-7.29 (m, 3H), 7.12-6.85 (m, 5H), 5.49-5.24 (m, 2H), 4.77-4.59 (m, 2H), 3.96-3.64 (m, 2H), 2.82 (s, 2H), 2.60 (s, 2H), 1.60 (s, 2H), 0.90 (s, 3H). 13C-NMR (400 MHz, DMSO). δ 170.82, 164.34, 144.44, 141.75, 137.03, 133.93, 132.27, 132.13, 128.85, 127.75, 127.31, 126.78, 121.78, 119.62, 118.43, 110.19, 107.74, 60.24, 45.99 (d, J = 24.6 Hz),37.48, 24.34, 21.65 (d, J = 83.3 Hz), 14.56, 14.11 ppm. HPLC (λ = 210 nm): tR = 17.960 min (CH3CN/H2O, 45:55).
2-(2-(2,5-difluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D28).
Purity of 95.86%. HRMS (AP-ESI) m/z calcd for C20H17F2N3O3 [M+H]+ 385.12380, found: 386.13004[M+H]+. 1H-NMR (400 MHz, DMSO). δ 10.91 (d, J = 56.9 Hz, 1H), 9.03 (d, J = 43.9 Hz, 1H), 7.47-7.35 (m, 5H), 7.15-7.10 (m, 1H), 7.04 (t, J = 8.0 Hz, 1H), 5.05-4.83 (m, 2H), 4.71-4.48 (m, 2H), 4.00-3.57 (m, 2H), 2.74 (s, 2H). 13C-NMR (400 MHz, DMSO). δ 164.73, 164.31 (d, J = 29.9 Hz), 158.63 (d, J = 242.3 Hz), 154.36 (d, J = 240.8 Hz), 137.25, 132.19, 126.63, 126.20 (d, J = 10.8 Hz), 121.56, 118.28 (d, J = 8.8 Hz), 115.73 (d, J = 25.8 Hz), 109.89, 107.97, 107.33, 45.34, 44.13, 21.89, 21.06 ppm. HPLC (λ = 210 nm): tR = 14.470 min (CH3CN/H2O, 45:55).
2-(2-(2-chloro-6-fluorobenzoyl)-1,2,3,4-tetrahydro-9H-pyrido[3,4-b]indol-9-yl)-N-hydroxyacetamide (D29).
Purity of 99.09%. HRMS (AP-ESI) m/z calcd for C20H17ClFN3O3 [M+H]+ 401.09425, found: 402.09875[M+H]+. 1H NMR (400 MHz, DMSO). δ 10.91 (d, J = 65.9, 1H), 9.03 (dd, J = 50.4, 1H), 7.60-7.56 (m, 1H), 7.54-7.32 (m, 4H), 7.15-7.10 (m, 1H), 7.03 (t, J = 7.6 Hz, 1H), 4.99 (dd, J = 42.0, 16.8 Hz, 2H), 4.71-4.47 (m, 2H), 4.04-3.54 (m, 2H), 2.82-2.67 (m, 2H). 13C-NMR (400 MHz, DMSO). δ 164.73, 162.29, 158.58 (d, J = 247.4 Hz), 137.29, 132.25 (d, J = 9.6 Hz), 132.08, 131.01 (d, J = 6.1 Hz), 126.45 (d, J = 21.6 Hz), 124.78 (d, J = 23.3 Hz), 121.61, 119.62, 118.25, 115.50 (d, J = 21.2 Hz), 109.86 (d, J = 14.6 Hz), 107.91, 44.92, 44.17, 22.07, 21.15 ppm. HPLC (λ = 210 nm): tR = 16.050 min (CH3CN/H2O, 45:55).
Cell Culture
Human K562 myeloid lymphoblastoma cell line, human triple-negative breast cancer cell lines MDA-MB-231 and MDA-MB-468 were received from the Cell Bank of Shanghai (Shanghai in China) and maintained in (RPMI)-1640 supplemented with 10% fetal calf serum (FCS). The cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Ubenimex (Cat. B8385) and 2′, 7′-Dichlorofluorescin diaceta (DCFH-DA) were purchased from Sigma. Paclitaxel (PTX, Cat. SP8020) and doxorubicin (Epirubicin Hydrochloride, Cat. IE 0640) were purchased from Solarbio. Carboplatin (CBP, Cat.C-0219027) and Cyclophosphamide (CTX, Cat.C-0232450) were purchased from HEOWNS. All the compounds were dissolved in sterile dimethyl sulfoxide (DMSO) to constitute 20 mM stock agents and the aliquots were stored at -20°C.
Enzyme Activity Screening Assay
K562 cells with overexpression of APN were added into PBS for ultrasonic crushing, centrifuged for 10 minutes, and supernatant was taken as enzyme source and added into 96-well plate. Different concentrations of compounds were added subsequently, and 1.6 mM L-leucine-p-nitro anilide was used as substrate. After 30 minutes of reaction, the APN enzyme activity was measured at 405 nm absorbance. The inhibition rate of APN activity was analyzed by the formula (Od control OD tested)/Od control 100%.
MTT Assay
MDA-MB-231 and MDA-MB-468 cells were added into 96-well plates (5X103 cells/well) and treated with PTX, PTX+Bestatin, or PTX+ various compounds at 37°C, 5% CO2 for 48h. Then, 10μL of MTT solution (5 mg/mL) were added to each well. After incubation for 4h, the medium was carefully removed, and 100 μL DMSO was added to each well and fully dissolved in vibration for 10min. The optical density (OD) at 490 nm and 630 nm were measured with a microplate Spectrophotometer (M5, MD). IC50 values were obtained from at least 3 independent experiments.
Flow Cytometry of APN
The cells were seeded in 6-well plates (4 × 105 cells/well) and treated with tested compounds for 48 h. Then, cells were collected into 1.5 mL EP tubes, washed twice with PBS, and centrifuged at 2500rpm for 5minutes. Cells were resuspended with 100μlof PBS, then 2μL/tube of APN antibody was added, followed by incubation in dark for 30 minutes at 4°C. Then the cells were subjected to flow cytometry (FACSAria II, Becton-Dickinson) for the expression of APN.
Cellular Reactive Oxygen Species Detection
The cells were seeded in 6-well plates (4 × 105 cells/well) and treated with tested compounds for 48 h. Then, cells were resuspended with 500 μL of serum-free culture medium; after addition of10μMof DCFH-DA, cells were returned to the incubator for 30min. Finally, the cells were resuspended with PBS and subjected to flow cytometry.
Apoptosis Analysis
The cells were seeded in 6-well plates (3 × 105 cells/well) and treated with various compounds for 48 h. Then, cells were collected and washed twice with cold PBS. Annexin V-FITC staining solution was then added to the cells, and cells were incubated at 2-8°C for 15 minutes under dark conditions. Then, PI staining solution was added by gentle mix at 2-8°C. After 5 minutes of incubation under dark conditions, cells were tested with flow cytometry immediately.
Colony-Formation Assay
3000 cells were suspended in 2 mL medium, and seeded into the wells of six-well plate. After 12 hours of incubation, cells were treated with various compounds. Until each colony had more than 50 cells, this experiment terminated. Cells were washed twice with PBS and fixed with methanol for 10minutes. Then cells were stained with 1% crystal violet for 5minutes. Finally, colonies were washed and pictures were taken under a microscope (IX81, Olympus).
Spheroid-Formation Assay
MDA-MB-231 cells and MDA-MB-468 cells in logarithmic growth phase were collected, stem cell sphere culture medium without serum (containing 50X B-27Supplement,100X N-2 Supplement,20 μg/L epidermal growth factor (EGF), 20µg/L insulin-like growth factor (IGF-1), 10 μg/L fibroblast growth factor (b-FGF), 20X Knock Out SR, 2mM L-Glutamine, 5mg/L Heparin sodium, 1X PENICILLIN-STREPTOMYCIN and DMEM: F12 (1:1) medium was added for the re-suspension and fully blown into single-cell suspension. The cells were diluted to 5,000/2 mL, with stem cell sphere culture medium, and cultured in a 37°C, 5% CO2 cell incubator. The number of stem cell spheres were observed every day. After about five days, stem cell microspheres could be visually confirmed, then various compounds were added every two days for ten days.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
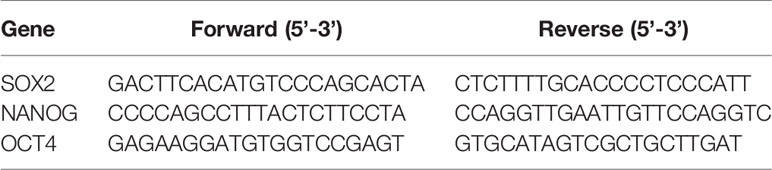
Total RNA was extracted with TRIzol reagent, and the RNA concentration was detected by a Nano Drop instrument (2000C, NanoDrop). Single-strand cDNA was reverse-transcribed with a Hi Fi Script cDNA Synthesis Kit (CWBIO, China) following the manufacturer-provided instruction. The cDNA was amplified with a SYBR Green mixture (CWBIO, China) and gene-specific primers (Table 2). The primers used for QRT-PCR were OCT-4, SOX-2 and Nanog. The relative gene expression was calculated with the 2-△△Ct method.
In vivo Imaging of Mice
Female Nude mice aged 3 weeks were purchased from Hunan SJA Experimental Animal Company and raised under SPF aseptic conditions. About 1×107 luciferase overexpressing MDA-MB-231 cells were injected subcutaneously into the right fat pad of the fourth mammary gland of each mouse. When the tumor volume reached100 mm3, the mice were randomly divided into six groups (n = 4): PBS group, PTX group, PTX+Bestatin group, PTX+D12 group, PTX+D14 group, PTX+D16 group. PBS group animals received PBS via intraperitoneal injection, while 10 mg/kg/3 days of PTX was administered to animals in the PTX group. 20 mg/kg/d of Bestatin, D12, D14, D16were administered in combination to10 mg/kg/3 days of PTX to the corresponding mice. After 3 weeks of administration, each mouse was intraperitoneally injected with 200uL of fluorescein substrate (d-fluorescein potassium salt, 150mg/kg), and then anesthetized with isoflurane. In vivo fluorescence imaging system (IVIS Spectrum, PerkinElmer) and SlideBook4.0 software were utilized for the result analysis. After the imaging analysis, mice were sacrificed and dissected for the detection of tumor size and calculation of liver/spleen indexes. All animal experiments were approved by the Animal Care and Use Committee of Weifang Medical University.
Statistical Analysis
All statistical analyses were performed with GraphPad Prism statistical software package. Quantitative data were presented as mean ± standard error of the mean (SEM). Statistical analysis was performed by one-way analysis of variance (ANOVA). When ANOVA returns significant results, post-hoc least significant different (LSD) tests were used be compare among the groups. P < 0.05 was considered statistically significant in all experiments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Committee of Weifang Medical University (ACUC-WFMU).
Author Contributions
LEZ designed the project. XX and YH performed the enzymatic screening. FL synthesized the molecules. LIZ, QH and HQ performed the in vitro and in vivo anticancer experiments. QJ, CF and WJ analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Science and technology support plan for youth innovation in universities of Shandong Province (Grant No. 2019KJM001), Natural Foundation of Shandong Province (Youth Found, Grant No. ZR2019QH005), National Natural Science Foundation of China (Youth Found, Grant No. 81803343).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.894842/full#supplementary-material
References
1. Taylor A. Aminopeptidases: Structure and Function. FASEB J (1993) 7(2):290–8. doi: 10.1096/fasebj.7.2.8440407
2. Wickstrom M, Larsson R, Nygren P, Gullbo J. Aminopeptidase N (CD13) as a Target for Cancer Chemotherapy. Cancer Sci (2011) 102(3):501–8. doi: 10.1111/j.1349-7006.2010.01826.x
3. Santiago C, Mudgal G, Reguera J, Recacha R, Albrecht S, Enjuanes L, et al. Allosteric Inhibition of Aminopeptidase N Functions Related to Tumor Growth and Virus Infection. Sci Rep-Uk (2017) 7:46045. doi: 10.1038/srep46045
4. Bhagwat SV, Lahdenranta J, Giordano R, Arap W, Pasqualini R, Shapiro LH. CD13/APN is Activated by Angiogenic Signals and is Essential for Capillary Tube Formation. Blood (2001) 97(3):652–9. doi: 10.1182/blood.V97.3.652
5. Zhao Y, Wu H, Xing X, Ma Y, Ji S, Xu X, et al. CD13 Induces Autophagy to Promote Hepatocellular Carcinoma Cell Chemoresistance Through the P38/Hsp27/CREB/ATG7 Pathway. J Pharmacol Exp Ther (2020) 374(3):512–20. doi: 10.1124/jpet.120.265637
6. Dixon J, Kaklamanis L, Turley H, Hickson ID, Leek RD, Harris AL, et al. Expression of Aminopeptidase-N (CD 13) in Normal Tissues and Malignant Neoplasms of Epithelial and Lymphoid Origin. J Clin Pathol (1994) 47(1):43–7. doi: 10.1136/jcp.47.1.43
7. van Hensbergen Y, Broxterman HJ, Hanemaaijer R, Jorna AS, van Lent NA, Verheul HM, et al. Soluble Aminopeptidase N/CD13 in Malignant and Nonmalignant Effusions and Intratumoral Fluid. Clin Cancer Res (2002) 8(12):3747–54.
8. Hashida H, Takabayashi A, Kanai M, Adachi M, Kondo K, Kohno N, et al. Aminopeptidase N is Involved in Cell Motility and Angiogenesis: Its Clinical Significance in Human Colon Cancer. Gastroenterology (2002) 122(2):376–86. doi: 10.1053/gast.2002.31095
9. Zhai MY, Yang ZX, Zhang CR, Li JP, Jia J, Zhou LY, et al. APN-Mediated Phosphorylation of BCKDK Promotes Hepatocellular Carcinoma Metastasis and Proliferation via the ERK Signaling Pathway. Cell Death Dis (2020) 11(5):396. doi: 10.1038/s41419-020-2610-1
10. Song YQ, Liu P, Wang CX, Zhang Q, Ge XC, Zhuang AS, et al. The Clinical Significance and Expression of TGF-Beta 1 and CD13 in Primary Lesion and Metastasis of Gastric Cancer. Int J Clin Exp Med (2017) 10(7):10643–51.
11. Amin SA, Adhikari N, Jha T. Design of Aminopeptidase N Inhibitors as Anti-Cancer Agents. J Med Chem (2018) 61(15):6468–90. doi: 10.1021/acs.jmedchem.7b00782
12. Zhang X, Fang H, Zhang J, Yuan Y, Xu W. Recent Advance in Aminopeptidase N (APN/CD13) Inhibitor Research. Curr Med Chem (2011) 18(32):5011–21. doi: 10.2174/092986711797535155
13. Schreiber CL, Smith BD. Molecular Imaging of Aminopeptidase N in Cancer and Angiogenesis. Contrast Media Mol Imaging (2018) 2018:5315172. doi: 10.1155/2018/5315172
14. Umezawa H, Aoyagi T, Suda H, Hamada M, Takeuchi T. Bestatin, an Inhibitor of Aminopeptidase B, Produced by Actinomycetes. J Antibiot (Tokyo) (1976) 29(1):97–9. doi: 10.7164/antibiotics.29.97
15. Zhang J, Fang C, Qu M, Wu H, Wang X, Zhang H, et al. CD13 Inhibition Enhances Cytotoxic Effect of Chemotherapy Agents. Front Pharmacol (2018) 9:1042. doi: 10.3389/fphar.2018.01042
16. Tsukamoto H, Shibata K, Kajiyama H, Terauchi M, Nawa A, Kikkawa F. Aminopeptidase N (APN)/CD13 Inhibitor, Ubenimex, Enhances Radiation Sensitivity in Human Cervical Cancer. BMC Cancer (2008) 8:74. doi: 10.1186/1471-2407-8-74
17. Yamashita M, Kajiyama H, Terauchi M, Shibata K, Ino K, Nawa A, et al. Involvement of Aminopeptidase N in Enhanced Chemosensitivity to Paclitaxel in Ovarian Carcinoma In Vitro and In Vivo. Int J Cancer (2007) 120(10):2243–50. doi: 10.1002/ijc.22528
18. Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, et al. CD13 Is a Therapeutic Target in Human Liver Cancer Stem Cells. J Clin Invest (2010) 120(9):3326–39. doi: 10.1172/JCI42550
19. Dou CH, Fang CY, Zhao Y, Fu XY, Zhang YF, Zhu DQ, et al. BC-02 Eradicates Liver Cancer Stem Cells by Upregulating the ROS-Dependent DNA Damage. Int J Oncol (2017) 51(6):1775–84. doi: 10.3892/ijo.2017.4159
20. Jin K, Zhang XP, Ma CH, Xu YY, Yuan YM, Xu WF. Novel Indoline-2,3-Dione Derivatives as Inhibitors of Aminopeptidase N (APN). Bioorgan Med Chem (2013) 21(9):2663–70. doi: 10.1016/j.bmc.2012.06.024
21. Hou JN, Jin K, Li J, Jiang YQ, Li XY, Wang XJ, et al. LJNK, an Indoline-2,3-Dione-Based Aminopeptidase N Inhibitor With Promising Antitumor Potency. Anti-Cancer Drug (2016) 27(6):496–507. doi: 10.1097/CAD.0000000000000351
22. Kalin JH, Butler KV, Akimova T, Hancock WW, Kozikowski AP. Second-Generation Histone Deacetylase 6 Inhibitors Enhance the Immunosuppressive Effects of Foxp3+ T-Regulatory Cells. J Med Chem (2012) 55(2):639–51. doi: 10.1021/jm200773h
23. Spindler A, Stefan K, Wiese M. Synthesis and Investigation of Tetrahydro-Beta-Carboline Derivatives as Inhibitors of the Breast Cancer Resistance Protein (Abcg2). J Med Chem (2016) 59(13):6121–35. doi: 10.1021/acs.jmedchem.6b00035
24. Mohamed HA, Girgis NM, Wilcken R, Bauer MR, Tinsley HN, Gary BD, et al. Synthesis and Molecular Modeling of Novel Tetrahydro-Beta-Carboline Derivatives With Phosphodiesterase 5 Inhibitory and Anticancer Properties. J Med Chem (2011) 54(2):495–509. doi: 10.1021/jm100842v
25. Zheng C, Fang YZ, Tong WG, Li GL, Wu HG, Zhou WB, et al. Synthesis and Biological Evaluation of Novel Tetrahydro-Beta-Carboline Derivatives as Antitumor Growth and Metastasis Agents Through Inhibiting the Transforming Growth Factor-Beta Signaling Pathway. J Med Chem (2014) 57(3):600–12. doi: 10.1021/jm401117t
26. Ran X, Zhao YJ, Liu L, Bai LC, Yang CY, Zhou B, et al. Structure-Based Design of Gamma-Carboline Analogues as Potent and Specific BET Bromodomain Inhibitors. J Med Chem (2015) 58(12):4927–39. doi: 10.1021/acs.jmedchem.5b00613
27. Liu H, Wang X, Yang H, Zhao Y, Ji S, Ma H, et al. New Method for Detecting the Suppressing Effect of Enzyme Activity by Aminopeptidase N Inhibitor. Chem Pharm Bull (Tokyo) (2019) 67(2):155–8. doi: 10.1248/cpb.c18-00667
28. Martinez JM, Prieto I, Ramirez MJ, Cueva C, Alba F, Ramirez M. Aminopeptidase Activities in Breast Cancer Tissue. Clin Chem (1999) 45(10):1797–802. doi: 10.1093/clinchem/45.10.1797
29. Ji S, Ma Y, Xing X, Ge B, Li Y, Xu X, et al. Suppression of CD13 Enhances the Cytotoxic Effect of Chemotherapeutic Drugs in Hepatocellular Carcinoma Cells. Front Pharmacol (2021) 12:660377. doi: 10.3389/fphar.2021.660377
Keywords: aminopeptidase N, inhibitor, cancer, chemosensitizer, tetrahydro-β-carboline
Citation: Xing X, Li F, Hu Y, Zhang L, Hui Q, Qin H, Jiang Q, Jiang W, Fang C and Zhang L (2022) Discovery of Novel Tetrahydro-β-carboline Containing Aminopeptidase N Inhibitors as Cancer Chemosensitizers. Front. Oncol. 12:894842. doi: 10.3389/fonc.2022.894842
Received: 12 March 2022; Accepted: 25 April 2022;
Published: 23 May 2022.
Edited by:
Jian-ye Zhang, Guangzhou Medical University, ChinaReviewed by:
Ashraf K. El-Damasy, Mansoura University, EgyptYingjie Zhang, Shandong University, China
Copyright © 2022 Xing, Li, Hu, Zhang, Hui, Qin, Jiang, Jiang, Fang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyan Fang, MTM4NTM2NjAzNDRAMTYzLmNvbQ==; Lei Zhang, bGVpemhhbmdjaGVtaWNhbEBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Xiaoyan Xing
Xiaoyan Xing Fahui Li2†
Fahui Li2† Lin Zhang
Lin Zhang Lei Zhang
Lei Zhang