- 1Division of Radiotherapy and Imaging, The Institute of Cancer Research, Sutton, United Kingdom
- 2Sarcoma Unit, The Royal Marsden National Health Service (NHS) Foundation Trust, London, United Kingdom
- 3Division of Clinical Studies, The Institute of Cancer Research, London, United Kingdom
- 4Division of Molecular Pathology, The Institute of Cancer Research, Sutton, United Kingdom
A shift in radiology to a data-driven specialty has been unlocked by synergistic developments in imaging biomarkers (IB) and computational science. This is advancing the capability to deliver “virtual biopsies” within oncology. The ability to non-invasively probe tumour biology both spatially and temporally would fulfil the potential of imaging to inform management of complex tumours; improving diagnostic accuracy, providing new insights into inter- and intra-tumoral heterogeneity and individualised treatment planning and monitoring. Soft tissue sarcomas (STS) are rare tumours of mesenchymal origin with over 150 histological subtypes and notorious heterogeneity. The combination of inter- and intra-tumoural heterogeneity and the rarity of the disease remain major barriers to effective treatments. We provide an overview of the process of successful IB development, the key imaging and computational advancements in STS including quantitative magnetic resonance imaging, radiomics and artificial intelligence, and the studies to date that have explored the potential biological surrogates to imaging metrics. We discuss the promising future directions of IBs in STS and illustrate how the routine clinical implementation of a virtual biopsy has the potential to revolutionise the management of this group of complex cancers and improve clinical outcomes.
Introduction
The paradigm shift in radiology from an imaging-based to a data-driven specialty has been unlocked by synergistic developments in imaging biomarkers (IB) and computational science including artificial intelligence (AI) (1). This is propelling imaging to the forefront of oncology and advancing the capability to deliver “virtual biopsies” (2). Virtual biopsies are defined in this review as an imaging method, or collection of imaging methods, which provide similar information about the structure, function and pathology of a tissue to that obtained from histopathology. The ability to non-invasively probe tumour biology both spatially and temporally would fulfil the potential of imaging to inform management of complex tumours; improving diagnostic accuracy, providing new insights into inter- and intra-tumoral heterogeneity and individualised treatment planning and monitoring (3).
Neuroimaging has long served as the test bed for imaging innovation, afforded by the size and immobility of the brain, which allows for repeated acquisition of high quality images. These characteristics are shared with soft tissue sarcomas (STS). STS are often large masses located in the extremities and imaging is therefore not prone to motion and respiratory artefacts encountered in thoraco-abdominal imaging. Further, image-guided biopsy of STS of the peripheries is usually straightforward with low morbidity and can therefore serve as biological validation for IB studies. The primary treatment for local disease is surgery which also provides opportunities for radiology-pathology correlation, which is a crucial component of IB development as non-invasive surrogates for tumour biology (4, 5). Leveraging on these features, we should be recruiting the best innovators and technology to STS research. These tumours provide an ideal testing ground for translational research which can then be transferred to other cancer types but also present a significant clinical need for better diagnostics and treatments. STS are rare tumours of mesenchymal origin that account for 1% of all adult cancers with over 150 histological subtypes and notorious heterogeneity, both between patients and within a tumour itself (6). The combination of inter- and intra-tumoural heterogeneity and the rarity of the disease remain major barriers to effective treatments (7–10). Whilst initial local control is often achieved, distant recurrence is frequent and associated with a generally poor prognosis, with a median survival reported between 12 and 20 months (11–17). Without methods to tackle the vast heterogeneity, we fail to confidently stratify patient risk and identify patients most likely to benefit from treatment and in turn, continue to apply a “one size fits all” approach to therapy. As we advance our biological understanding of these sarcomas, combining this with innovative imaging tools and reliable IBs could dramatically alter the landscape of such a devastating disease.
Herein, we provide an overview of the long-standing challenges facing our ambition to advance treatment and improve clinical outcome for STS patients. We describe the advances in IBs and computational imaging in STS that can help overcome these challenges and the realising promise of “virtual biopsies”. We focus on the studies that have sought to correlate imaging with biology, the development of quantitative MRI parameters in STS and radiomics that has been applied to this group of cancers. We then discuss the challenges and future work that is required to discover, validate and translate IBs into the clinical setting for these patients.
Imaging Biomarkers in STS
With the rapid growth of IBs and computational image analysis capabilties in the past decade, radiological images are now increasingly recognised as valuable datasets that can provide complementary information to aid the diagnosis and management of patients with STS.
Improved imaging tools are required for STS tumours as is a detailed understanding of their relationship with histological changes which is vital for IB development. An IB is defined by the National Institutes of Health (NIH) as “characteristics that are objectively measured and evaluated as an indicator of normal biological processes, pathogenic process or biological responses to therapeutic intervention” (18, 19). IBs can be numerical (quantitative) or categorical (quantitative value or qualitative). An IB roadmap has been produced to accelerate the translation of IBs from conception into useful clinical tools (5). This includes two translational gaps that must be crossed. The first is to become medical research tools if the IB is confirmed to reliably test medical hypotheses, and the second as clinical decision-making tools if the IB is confirmed to be clinically useful and has been clinically validated (20). In order to bridge these translational gaps, IBs need to pass through a series of domains: discovery (Domain 1), validation (Domain 2) and qualification with ongoing technical validation (Domain 3), including technical validation (e.g. repeatability, reproducibility and availability) and biological and clinical validation (e.g. relationship with disease state, diagnostic, prognostic and predictive capabilities) (5).
In STS, IBs have the potential to provide non-invasive insights into tumour biology over time without sampling bias, including in relation to intra–tumoural heterogeneity (21, 22). Furthermore, they bypass any technical and clinical difficulties of biopsy for diagnosis and treatment monitoring, allowing improved patient risk stratification and treatment planning. The capabilities of multiparametric magnetic resonance imaging (MRI) present an attractive tool for quantitative IB development for clinical adoption (23). The ability to capture multiple parameters in a single scan, with the potential to explore multiple biological properties which contribute to intratumoural heterogeneity is highly appealing (3).
Quantitative MRI
Quantitative MRI is a multi-step process that requires image acquisition, reconstruction, segmentation and often mathematical modelling of parameters prior to feature extraction (Figure 1) (2). A semi-quantitative MRI parameter already in use in clinical practice is contrast-enhanced (CE-)MRI, where the difference in signal after intravenous injection of a gadolinium-based contrast agent is used as a surrogate for tumour vascular perfusion (22). Examples of fully quantitative parameters are Dixon MRI which enables fat content in tumours to be visualised and quantified and apparent diffusion coefficient (ADC) measurements from diffusion-weighted (DW)-MRI which maps tissue cellularity (22, 25–27). ADC is rapidly growing in use in oncology because of the widespread availability of DW-MRI on clinical scanners, ease of acquisition (not requiring additional equipment or injection of contrast agents), excellent repeatability and large number of biological and clinical validation studies (22, 27, 28). These factors make it an attractive IB, and as such, ADC has emerged as a useful quantitative IB in STS which has led to its incorporation in the European Organisation of Research and Treatment of Cancer (EORTC) guidance on standard of care MRI in STS (29–31).
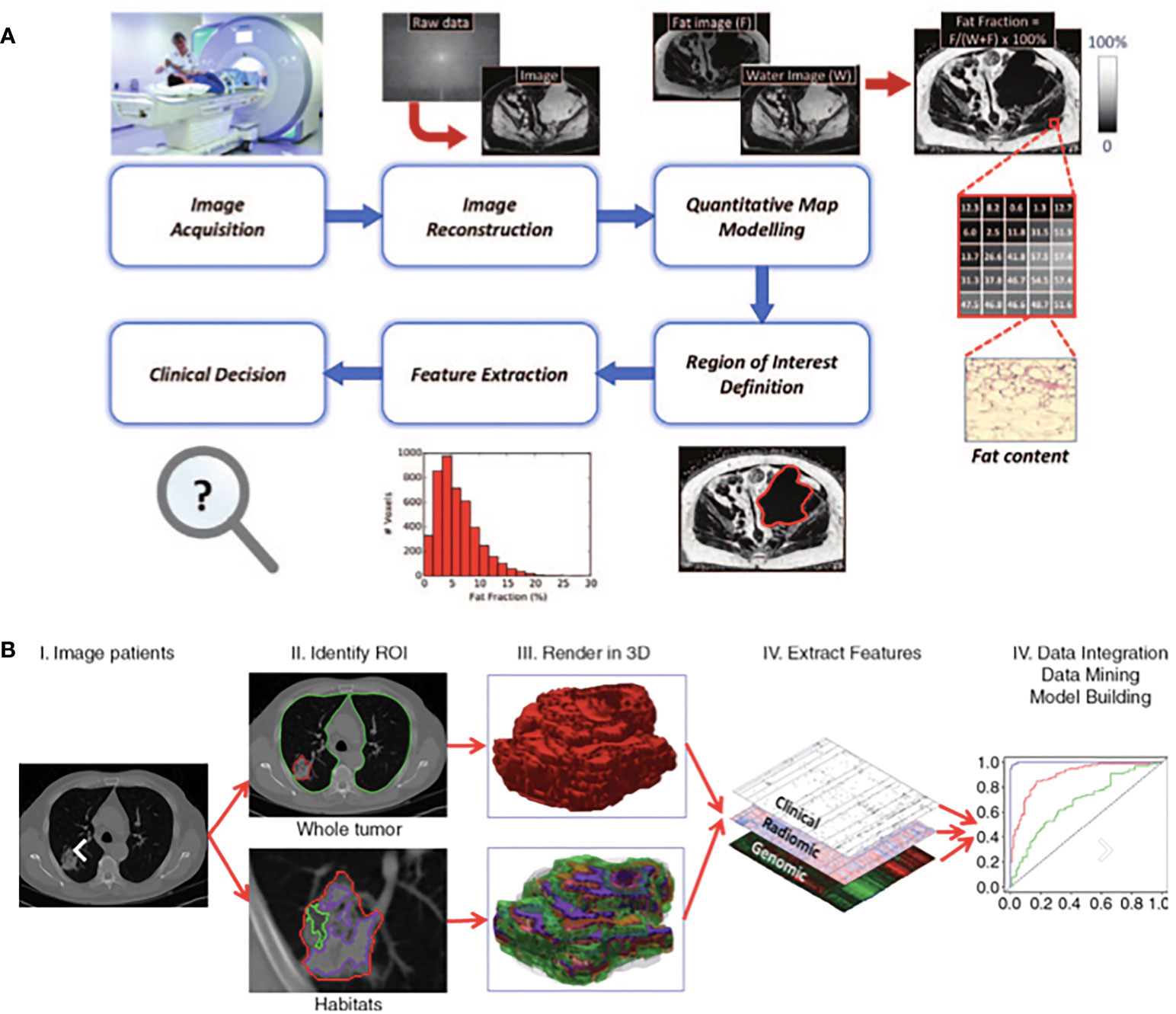
Figure 1 Exemplars of typical quantitative imaging in (A) and radiomic workflows in (B) taken from Blackledge et al. (22) and Gillies et al. (24) respectively. (A) Quantitative imaging Involves a detailed process of steps. Following image acquisition and image reconstruction, quantitative maps are developed either by the scanner or offline and these maps differ from the parent images as each voxel has an unit of measurement. Multiple regions of interest are often selected, features extracted accordingly to aid clinical decisions. (B) Radiomics involves a multi-step process. Following acquisition of high quality images, regions of interest and/or habitats are defined. These are reconstructed into 3D. Radiomic features are extracted and models are developed with correlation of these extracted features and pre-defined clinical outcomes of interest.
Radiomics
The emerging field of “radiomics” may also serve to identify IBs for use in STS. Radiomics describes the extraction of features from radiological images, converting them to mineable high-dimensional data and searching for correlations with defined clinical variables. It relies on the extraction of both semantic (radiology lexicon such as size and shape) and agnostic (quantitative descriptors such as texture and histogram) features, interrogating tumour morphology and behaviour on a deeper scale (32, 33). Correlative assessment of these features with clinical variables may drive generation of diagnostic, prognostic or predictive models for variables of interest, to act as complimentary non-invasive decision support tools (Figure 1) (24). Radiomics may identify an individual feature or set of features (a signature) that could be utilised as an IB. Radiomics is particularly attractive as it can explore and discover clinically relevant IBs without the need for new or complex imaging systems.
A typical radiomics workflow consists of multiple steps: high quality image acquisition, preprocessing of the images to ensure uniformity of scans, tumour segmentation and feature extraction. Extracted features are combined with other mineable data and correlations for clinical outcomes of interest are explored (24). Cancer types where radiomics has shown promise include lung cancer, glioblastoma and prostate cancer where radiomics has been used to distinguish benign from malignant tissue, measure the aggressiveness of tumours, aid diagnosis and selection of biopsy sites (24, 34–39). In addition, radiomics has identified imaging phenotypes suggestive of patient prognosis and predictions of treatment response (24, 34, 36–42). The use of radiomics in a highly heterogeneous malignancy like STS could deepen our understanding of the biology across the entire tumour volume, and its correlation with clinical outcomes. To date, preliminary radiomic studies in STS have focused on correlation of radiomic features with pathological grading and prognosis (43–47). Multiple studies have demonstrated an association of final histopathological grade in the surgical specimen on tissue examination with extractable radiomic features, including the ability to discriminate between tumours of low and high grade (43, 44, 47). In addition, radiomic features were predictive of distant metastases, early response to neoadjuvant chemotherapy and clinical outcome (46–50). The studies performed in STS remain limited to single centre cohorts with small patient numbers and imaging performed on single scanners. Radiomics requires large datasets to allow confident training of models and for robust conclusions to be drawn, and therefore, the rarity of STS and resultant small datasets has significantly hindered the progress of applying radiomics to “real life” clinical practice. Solutions for dealing with smaller datasets are emerging, such as using internal cross-validation to test radiomic models removing a dependence on large independent datasets (51). Overcoming this challenge would pave a bright future for radiomics in STS and its potential to supply unparalleled information about tumour heterogeneity and its impact on tumour behaviour and clinical outcome.
Unlocking the Potential of IBs With Computational Imaging
Quantitative MRI and radiomics require time-consuming processing and analysis and produce a vast amount of complex data which make interpretation challenging and prohibit translation as clinical decision making tools. Recently, AI is gaining traction within oncology as offering a complementary suite of technologies to drive uptake of IBs in clinical practice. Defined as the “development of computer systems able to perform tasks normally requiring human intelligence”, AI offers a solution to the time-consuming and complex image processing analyses required for some MRI parameters and for radiomics. It encompasses machine learning and its subset deep learning, each with multiple algorithms, and may prove an indispensable tool for complex data analysis (2, 52, 53). The integral radiomics workflow step of image delineation requires expert radiological input and is time consuming, particularly in the context of multiple tumour lesions or large, complex tumours. Deep learning can train and use algorithms for automated segmentation of tumours without the requirement for human intervention (54). In addition, it can analyse numeric data from predefined radiomic features as well as designing its own radiomics features from the direct analysis of images (55). This holds potential as a attractive tool, maximising the amount of “hidden data” that can be automatically extracted from radiological images, whilst addressing the real life constraints of time consuming segmentation and post processing.
Diagnosis
The current gold standard for diagnosis in STS remains histopathological examination of biopsy or surgical specimen tissue. Routinely, a pathologist will determine histological subtype, histological grading based on the three-tiered histopathological grading system of the French Federation of Cancer Centers Sarcoma Group (FNCLCC) and therapeutic response demonstated by percentage of remaining viable cells when required. If applicable to the suspected histological subtype, the pathologist will also routinely request ancillary tests to aid diagnosis. This includes immunohistochemistry (for example for proliferation index Ki67, specific proteins and immune cell staining), cytogenetics and molecular analysis for relevant alterations (56, 57). In addition to tumour size, grade by FNCLCC is the most important prognostic factor in STS (58–60). However, whilst core biopsy is the usual modality for obtaining tissue for diagnosis, it is unable to capture the full extent of heterogeneity due to sampling error, and therefore can have the potential to misrepresent histopathological grade (44, 61, 62). In particular, under-grading of STS on core biopsy is recognised (62). Furthermore, given the disease rarity and degree of heterogeneity, accurate diagnosis by non-specialist pathologists can be difficult. In fact, a previous study conducted across 3 European regions confirmed a concordance of only 56% in histopathological tissue examination by primary diagnosis and secondary opinion (63). The main discrepanices were namely histological grade (43%), histological type (24%), subtype (3%) and grade plus subtype or grade plus histological type (29%). A similar finding was reported in a further study which found a 71% concordance in diagnosis between primary and referral centres, with 16.4% major discrepancies and 11.8% minor discrepancies (64).This is complicated by the qualitative nature of histopathological measures used for diagnosis, where inter-observer discrepancies may lead to poor reproducibility and carries the potential to impact patient treatment planning. These findings support the drive to centralise sarcoma care into specialist centres.
Ultimately, we frequently fail to capture the heterogeneity within complex lesions like STS with tissue biopsy alone, and despite the substantial strides made in our biological understanding and management of these diseases, this remains incomplete for most subtypes. Imaging can overcome these challenges either by guiding precision biopsy of higher grade tissue, or by providing biological information or correcting underestimation of grade with direct IBs acting as a surrogate for underlying biological features. Furthermore, efforts to standardise the acquisition of scans will improve accuracy and consistency and could contribute to more robust clinical decision making (65). Below we discuss the evolution of IBs in STS over the past few years.
Conventional Imaging
The potential of biopsies to misrepresent grade in STS is well established (62). With regards to grading tumours, the primary goal of imaging has remained to achieve the equivalent of in vivo microscopy, or provide a complimentary diagnostic aide. In order to overcome the limitations of invasive core biopsies in STS, many MRI studies have sought to test the correlative power of imaging with tumour grade, comparing findings to histopathological examination of specimens based on FNCLCC. In 2008, Liu et al. correlated peripheral tumour growth pattern on MRI with histopathological grade in 59 STS patients, and found poorly defined margins in 60% of high grade tumours, versus well-defined margins in 60% of low grade tumours (66). This has been reinforced in a retrospective study by Zhao et al. where increased intensity of peritumoural contrast enhancement on imaging in higher grade tumours was the strongest independent indicator of histopathologically confirmed high grade STS (67). In addition, a study by Crombe et al. validated findings by Zhao et al. and demonstrated that the three following features of peritumoural enhancement, necrosis and heterogeneity in signal intensity on MRI could allow up-grading of underestimated tumour grade (61). This study also undertook survival analyses and determined that two out of three of these criteria were independent prognosticators of worse metastasis free survival and overall survival through uni- and multi-variable analysis (61).
Computed tomography (CT) sensitivity to detect tissue necrosis has also been utilised to demonstrate improved accuracy of grading in leiomyosarcoma (LMS) when combined with histopathological assessment of tissue, versus histopathology alone (68). Further to tumour grade, studies have explored the CT features of STS that could aid diagnosis, exploiting features such as fat content, margin status and presence of septations. An example of successful utilisation of these imaging parameters is in liposarcoma, a group of STS which can be frequently distinguished radiologically, without the requirement of a diagnostic biopsy (69). Contrast-enhanced CT, which is generally the standard investigation for retroperitoneal sarcomas, can be used for site confirmation and often tissue composition, and can confidently delineate well-differentiated liposarcoma (WDLPS) and angiomyolipoma (70, 71).
Positron Emission Tomography (PET)/CT
The value of PET/CT in assessing histological grade in STS has been explored. Using 18flurodeoxyglucose-positron emission tomography (18FDG-PET)/CT, the maximun standardised uptake value (SUVmax) of a lesion was demonstrated to correlate with the FNCLCC. In two meta-analyses, SUVmax was able to distinguish high-grade versus lower-grade STS such that this imaging technique could improve the diagnostic accuracy of core biopsies (72, 73). However, both meta-analyses reported poor methodological quality and lack of comparable parameters across studies, preventing confident conclusions to be drawn.
Quantitative MRI
Advancements in MRI capabilities has allowed the derivement of quantitative parameters that allow insight into biological features of tumours for example fat, vascularity and cellularity, as well as the spatial heterogeneity across an entire tumour. These parameters may represent translatable IBs that could improve accurate diagnosis. Although the multitude of potential parameters is expanding, some remain in early stages of exploration (Table 1). Exploiting MRI features of fat fraction and contrast sequences, liposarcomas can be distinguished from other STS, and provide information about the margin, shape, as well as internal architecture and properties (70, 75, 76). In a group of retroperitoneal sarcoma patients, MRI fat fraction strongly correlated with histopathological fat content on the surgical excision sample (22).
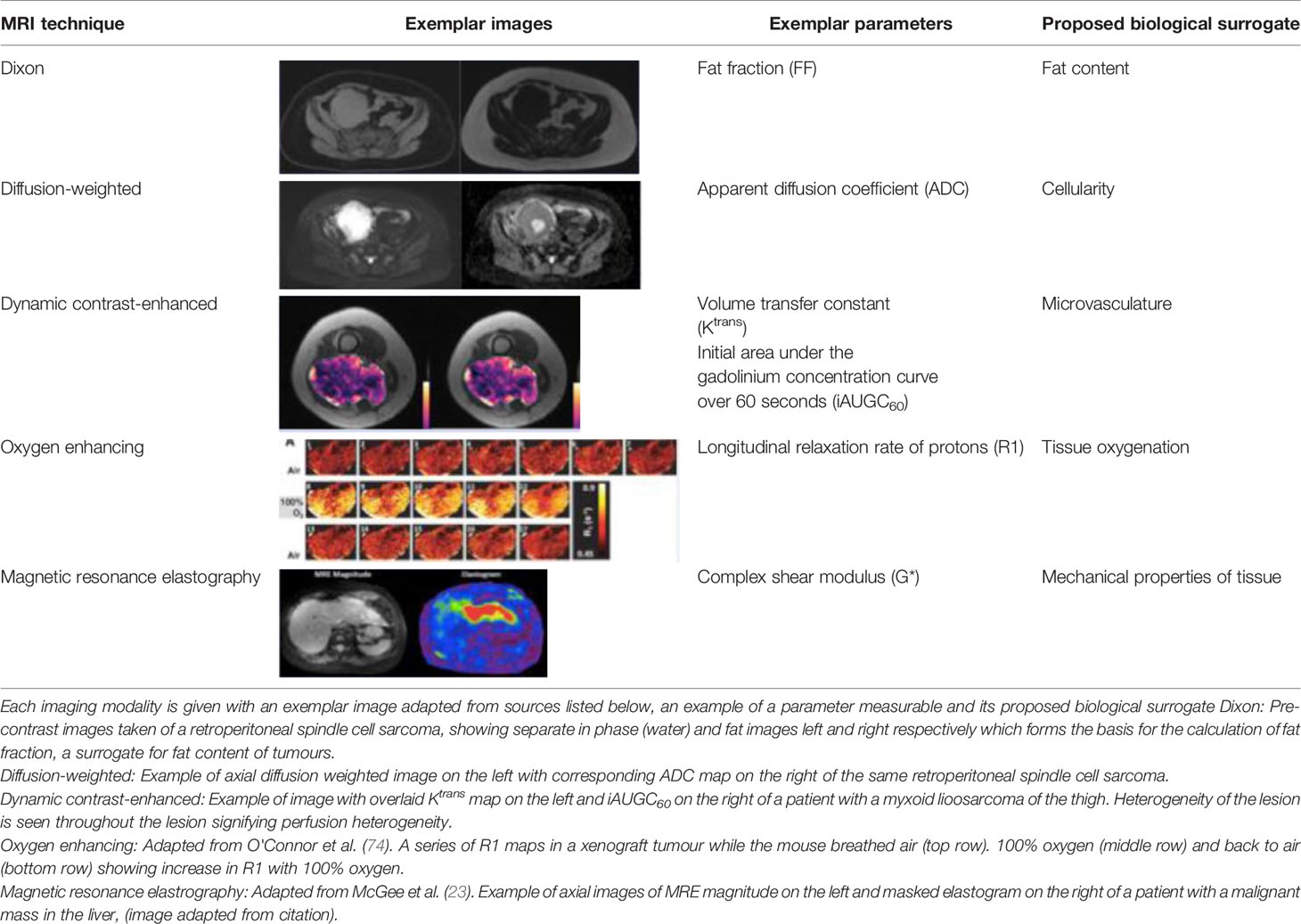
Table 1 Summary of the quantitative imaging techniques described in this paper and that represent potential IBs.
Schnapauff et al. in 2009 prospectively undertook DW-MRI in 30 patients prior to histological evaluation of regions of interest (ROIs), illustrating an inverse correlation of tumour cellularity and ADC, with low ADCs generated for areas of higher cellularity (77). This correlative relationship was not influenced by prior anti-cancer treatment. This finding was reproduced in two further retrospective studies, with lower mean ADC in higher grade sarcomas representative of the increased cellularity on histological analysis (78, 79). In a prospective study of 45 patients, the use of ADC to discriminate between malignant and benign soft tissue lesions was demonstrated. This study showed that malignant lesions had a significantly lower mean ADC value, further supporting the complimentary value of DW-MRI and ADC to soft tissue mass imaging (80). In addition, whilst most studies exploring ADC focus on the use of median or mean ADC values to objectively measure cellularity, this can fail to reflect baseline or post-treatment heterogeneity for any given tumour. Winfield et al. demonstrated ADC may be measured for various regions of interest on MRI, providing a deeper insight into ITH both within a region and across the entire tumour (22). An extension of DW-MRI is intravoxel incoherent motion (IVIM) which allows separation of perfusion from the true tissue diffusion. This allows calculation of quantitative parameters that reflect true tissue diffusivity (D), perfusion-related pseudodiffusion (D*) and perfusion fraction (f) which represents the contribution of water in capillaries (81). This could improve the accuracy of a quantitative parameter of diffusion by informing on the contribution of the microvascular circulation. Wu et al. demonstrated the combination of ADC and D were most useful in differentiating malignant and benign STS than ADC alone, and that the combination of ADC and f improved the differentiation between benign, intermediate and malignant STS (82). However, in Winfield et al’s study, the IVIM parameters exhibited poor repeatability when compared with ADC (22). A modification of DW-MRI is diffusion tensor imaging (DTI) and its advancement diffusion kurtosis imaging (DKI) (83, 84). These techniques add a quantitative assessment of the direction of diffusion of water molecules and may provide further diagnostic value by providing more structural information about lesions. DTI studies in muscoskeletal imaging are still in early stages but have been used to provide information about nerve fascicle visualisation for peripheral nerve sheath tumours and the integrity of axons (85–87). Similarly DKI studies remain evolutionary, however, have shown value in helping differentiate sarcoma from benign lesions and distinguishing recurrence from post surgical changes (88, 89).
Dynamic contrast-enhanced MRI (DCE-MRI) is being increasingly applied to obtain temporal information about tumour microvascularity and there have been a wealth of studies in the past few decades (90–94). It is well recognised that the tumour vascular system has an altered pathophysiology and role in tumourigenesis, supporting the diagnostic clinical application of DCE-MRI (95, 96). DCE-MRI utilises serial images capturing the arrival, duration and exit of a contrast agent in a tissue of interest, generating a signal intensity curve that may provide detailed information on both the physical and physiological properties of tissue in response to the agent (91, 97–99). The signal intensity curve and individual quantitative parameters such as the transport constant of the contrast agent from the blood plasma into the extracellular extravascular space (Ktrans), and the extracellular extravascular space volume (Ve) may inform on underlying tissue microvasculature (99). For example tumour areas of higher perfusion and permeability having a higher and quicker uptake of contrast compared with tumours areas of necrosis (100). An early study in STS has demonstrated that DCE-MRI can aid in discriminating between low and high grade tumours, with Ktrans differentiating grade I from grade III and grade II from grade III tumours (101). Combining Ktrans with two semi-quantitative parameters also derived from DCE-MRI, initial area under the gadolinium concentration-time curve (iAUC) and time to peak (TTP), had the highest diagnostic performance with an area under the curve (AUC) of 0.841.
Magnetic Resonance Spectroscopy (MRS) is a technique which can provide information about the biochemical processes without the requirement for invasive intravenous contrast agents. By detecting protons (1H-) containing metabolites in addition to water, this can provide information about the micro-metabolic environment within tissue. In STS, a prospective study used this technique to leverage on the measurement of choline which is an established marker for increased cell membrance turnover and can indicate malignancy. The authors showed that this can be useful in differentiating grade of muscoskeletal tumours (102). In addition, in a heterogeneous lesion like STS tumours, it might highlight more aggressive areas. However, this technique has poor sensitivity and specificity in STS and can be technically challenging is some anatomical sites such as extremities (103).
Radiomics
Recent efforts have centred on radiomic model development prediction of tumour grade given the crucial role of grade in preoperative diagnosis and treatment planning. Multiple studies have focused on identification of MRI-based radiomic features distinguishing low from high grade tumours (43, 45, 47, 78, 104). Using T2-weighted MRI images, Zhang et al. tested 5 radiomic features in a predictive model of STS grade in 35 patients with good accuracy (44). This was further explored by studies training machine learning algorithms based on radiomic features extracted from T2-weighted and/or T1-weighted MRI sequences to identify the optimal model of tumour grade prediction (43, 45). These studies used larger sample sizes (105 and 113 patients respectively) to form their complete cohort for training, testing and validation of their radiomic models, and achieved area under the curves (AUC) of over 0.9, demonstrating both feasibility and utility in their models for distinguishing grade (43, 45). In a study with the largest sample size to date, Navarro et al. retrospectively collected a training cohort of 148 patients to train a deep learning model comprising of features extracted from contrast-enhanced fat-saturated T1 and fat saturated T2 weighted images which was capable of distinguishing low from high grade tumours (105). They went on to externally validate this result in a cohort of 158 patients with AUCs of 0.75 and 0.76 for their T1 and T2 weighted radiomics models respectively. Corino et al. used DWI-MRI and it’s corresponding ADC maps in a group of 19 histologically confirmed STS patients for retrospective radiomic analysis to distinguish intermediate and high grade tumours. This study concluded that using a maximum of three features from first order statistics could achieve a model distinguishing grade with relatively good accuracy (78). Texture analysis tools can be applied to routine MR images to interrogate the inter-pixel relationships and grey level pattern of a ROI to provide a measure of heterogeneity (106, 107). This was utilised in a retrospective study of 29 patients by Meyer et al. to match Ki-67 index to discriminate between low and highly proliferating sarcomas (108). Ki-67 proliferation index is a widely used immunohistochemical test that reflects the activity of tumour cells, aiding the discrimination between low and high grade tumours, treatment response to neoadjuvant radiotherapy and can act as a predictor of clinical outcome in several cancers. Meyer et al. demonstrated several textural analyses correlated with Ki-67 index, namely run length and grey-level non uniformity (run length matrix features) had a positive correlation with Ki67 whilst sum average (grey-level histogram feature) and grey-level kurtosis (absolute gradient feature) had a negative correlation. Radiomics may be a valuable non-invasive tool for supplementing histological methods of distinguishing low and high grade STS, and may improve rates of undergrading tumours. However, MRI-based radiomic studies are usually retrospective, often limited to few imaging sequences and rarely externally validated, and therefore remain some way from constituting reliable and reproducible IBs. Despite its inferiority to MRI in soft tissue contrast, CT-based radiomics has also been explored in STS. Peeken et al. used three retrospective, independent cohorts for training, testing and validating (83, 87 and 51 patients respectively) a model to demonstrate feasibility of using CT-radiomics for grading STS, able to differentiate grade 3 from non-grade 3 tumours (47). There remains occasions, particularly for STS located within the retroperitoneum, when MRI is not conducted and therefore CT radiomics remains a useful avenue to explore. Whilst this study successfully validated its radiomics model, its AUC was 0.64, and therefore does not perform well enough to be a confident tool alongside the usual histopathological work-up for these patients.
Image Guided Biopsy Using Next Generation Imaging
Image guided biopsy could circumvent many issues related to tissue sampling, although this has undergone relatively little change for over 30 years (109). Biopsy usually takes place under CT or ultrasound (US) guidance alone, and whilst these modalities benefit from low cost, speed, and familiarity, they do not convey the same level of biological information as next generation imaging techniques (e.g. quantitative MRI).
In other tumour types including prostate cancer bone metastases, lymphoma and neuroendocrine tumours, functional imaging has been used to select the biopsy sites thought to be most deterministic in terms of biological behaviour, response to treatment and disease outcomes (110–112). Since tumour grade is known to be an important prognostic indicator of recurrence in sarcomas, this ‘precision biopsy’ approach could be adopted to provide more accurate grading and better inform treatment decisions e.g. alternative or adjuvant therapies (113). Practically speaking, a precision MRI biopsy requires either a direct ‘in-gantry’ approach, or some form of image fusion to leverage the benefits of the two imaging modalities - either ‘cognitive’ (visual estimation) or actual (software), the latter leveraging the benefits of two imaging modalities. Sampling tumours in multiple regions would afford the ability to capture intratumoural heterogeneity and can provide an accurate ground truth for imaging biomarker validation, with a view to “virtual biopsy” in the future. For example, biopsy in a single region may not capture the necrotic or myxoid elements, which are a key component of the FNCLCC grading scheme for adult sarcomas (114). A recent example of multiregional fusion biopsy has been reported by a group who sampled different regions of ovarian tumours using CT/US fusion, guided by radiomic habitats (115). Precision biopsy would also facilitate discovery of IBs with radiogenomic study of co-localised tissue samples, integrating data-rich radiological images with biological profiling inclusive of deeper histological and molecular metrics. There is research applying “-omics” approaches such as transcriptomics and proteomics to comprehensively profile STS and their functional status, particularly in the context of heterogeneity (116–118). Further, we are increasingly understanding the importance of the role the immune environment plays within these heterogeneous tumours, and its potential to inform on behaviour and clinical outcome of STS (119–122). Combining these powerful –omics platforms with next generation imaging and biopsy techniques could accelerate the discovery and validation of IBs in STS.
Treatment Planning and Therapeutic Monitoring
Aside from the search for IBs of histopathological phenomena, there is also a need for markers that surrogate biological features of lesions, particularly those of prognostic value that can allow better patient risk stratification. At present, the complexity of heterogeneity within STS lesions confounds our ability to individualise therapy for each patient. Upfront opportunities to identify patients at risk of relapse or poor outcome through IBs could lead to improved management strategies and clinical outcome. Furthermore, when positioned with other emerging tools such as dynamic prognostic nomograms, for example the Sarculator App (123) and PERsonalised SARComa care (PERSARC) (124), which model prognosis in STS patients using a number of clinicopathological factors, a clinical IB may improve the accuracy of these accessible and user-friendly applications for treatment planning.
Currently, radiotherapy in STS is primarily utilised in the neoadjuvant setting for tumours of the extremeties to reduce post-surgical local recurrence rates (125). This is generally preferred to radiation given in the adjuvant setting which can have long-term morbidity associated with the radiotherapy received (e.g. fractures, fibrosis and oedema) (126). A retrospective study also demonstrated that neoadjuvant radiotherapy achieved higher rates of negative surgical margins on resection of the tumour when compared with adjuvant radiotherapy and no radiotherapy (90% versus 75% and 80% respectively) (127). Unfortunately intratumoural heterogeneity causes variable treatment effect across a single tumour, wherein varying tissue composition will govern therapeutic, and up-front radiotherapy planning is difficult without improved biological information of the lesion. For example, it is known that hypoxic areas within tumours can result in chemo- and radioresistance (128, 129). Consequently a single STS tumour containing a number of hypoxic regions will fail to respond to radiotherapy in those hypoxic areas. Systemic agents may be used as neoadjuvant therapy to reduce the potential for metastasis, or in the advanced setting to reduce metastases and control symptoms (130). In high-risk STS, 50% of patients will develop advanced disease and chemotherapy forms the mainstay of management in the metastatic setting (131). The role of neoadjuvant chemotherapy is well established in certain subtypes such as rhabdomyosarcoma, however, its use in most sarcoma subtypes remains controversial (132). Available data from randomised controlled trials have significant limitations and the evidence base is insufficient (130, 133–138). In fact, the only randomised controlled trial investigating neoadjuvant chemotherapy and surgery with or without radiotherapy versus surgery with or without radiotherapy alone was not able to recruit target accrual and had a negative result (138). The addition of neoadjuvant chemotherapy was not proven to be more effective than surgery alone, with a 5-year disease free survival of 56% versus 52% respectively. Furthermore, when a histotype specific approach to neoadjuvant chemotherapy agent choice was investigated in a phase 3 multi-centre randomised controlled trial, there was no benefit observed over the standard chemotherapy regimen (133). In this study, local failure free survival at 46 months was 86% in the standard regimen group and 85% in the histotype-tailored chemotherapy group. Moreover, distant metastases free survival at 46 months was 74% for the standard chemotherapy group versus 45% for the histotype-tailored chemotherapy group. This suggests that therapeutic challenges in STS are in part related to disease rarity, however, also likely to be related to intratumoural heterogeneity. Without a greater understanding of the biology underlying this heterogeneity and the manner in which tumours may evolve in space and time, prediction of therapeutic response is difficult.
Furthermore, whilst serial biopsies theoretically serve to monitor therapy, this requires multiple invasive and sometimes technically challenging procedures, which remain prone to sampling error (125). In contrast, imaging affords non-invasive, global, and temporal assessment of tumours. However, radiological response to treatment is not reliably characterised with conventional size-based measurements on imaging in STS. At present, Response Evaluation Criteria for Solid Tumours (RECIST 1.1) is used to measure radiological response to therapy based on whether a tumour demonstrates shrinkage, an increase in size or no change in overall size on imaging scans (139, 140). In the neodjuvant setting there is a disparity between the therapy response measured radiologically and the histopathological response findings on tissue (141–143). Treatment induced necrosis on tissue examination is an independent marker of prognosis, and dimension-based imaging is a crude predictor of this (144, 145). Numerous radiological patterns of response can be seen and good responders following neoadjuvant therapy as defined by RECIST 1.1 may be rare (Figure 2) (141). A significant reduction in the volume of responding tumours following radiotherapy has been reported as low as 0%, and instead tumours may grow due to cystic transformation, haemorrhage and necrosis (pseudoprogression) (141, 142, 146, 147). In the phase 3 randomised controlled trial investigating the histotype approach to neoadjuvant chemotherapy, no patients achieved complete response and only 14% achieved a partial response with the rest being objectively measured as stable or progressive disease (133). A retrospective review conducted on patients with advanced sarcoma demonstrated that good responders (27% of the cohort defined as complete and partial responders) following neoadjuvant chemotherapy had comparable rates of relapse and overall survival to the non responders (148). A study by Delaney et al. demonstrated that in a prospectively recruited group of 48 STS patients treated with neoadjuvant chemotherapy and radiotherapy prior to surgery, 6 patients had progression as measured by RECIST 1.1 (149). However, on histological examination of the surgical specimen, 4 of the 6 patients had prominent necrosis (more than 90%) of the sample confirming good sensitivity to the neoadjuvant therapy received. Further, a previous study observed 31% of STS tumours increasing in size by more than 10%, with no association with a deterioration in clinical outcome (150). Efforts to establish alternative response criteria have been undertaken. The Choi criteria measures radiological response to therapy based on changes in tumour size and/or density (tumour attenuation measured in Hounsfield units (HU)) on computed tomography (CT) (151). Studies have demonstrated that density changes often preceed any dimensional response to the tyrosine kinase inhibitor imatinib in gastrointestinal stromal tumour, and therefore Choi criteria have a higher sensitivity to tyrosine kinase inhibitor benefit in these patients (151, 152). In other STS subtypes, Choi criteria have shown promise as superior predictors of histopathological response to cytotoxic chemotherapy, and a modified Choi criteria can allow assessment on MRI scans (153, 154). However, these criteria require appropriate software for postprocessing and the segmentations required impact on radiologist time. This has hindered validation, including reproducibility in larger prospective studies.
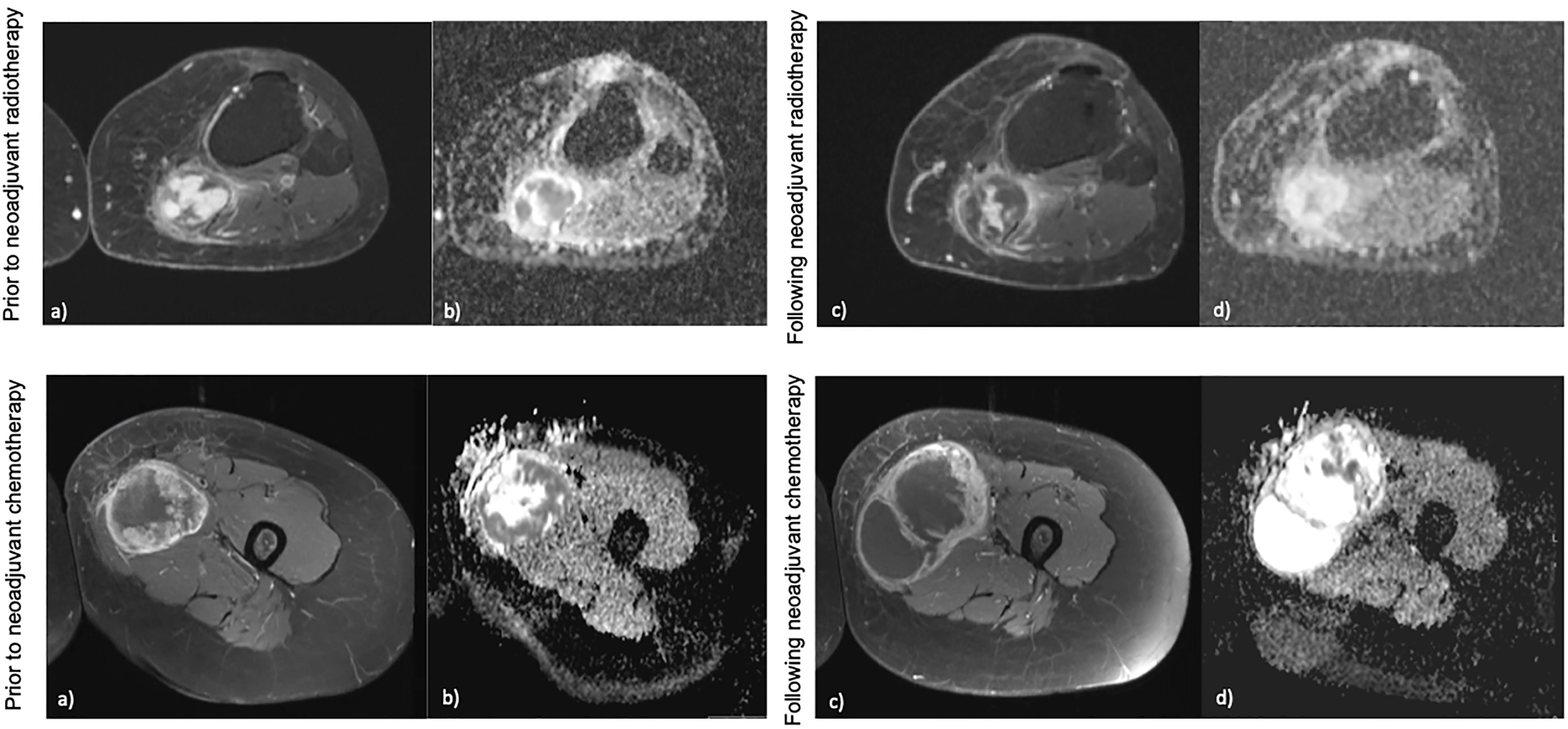
Figure 2 Examples of different radiological response following neoadjuvant therapy. Top: (A, B) demonstrate a hisloJogically confirmed myxofibrosarcoma in the calf prior to neoadjuvant radiotherapy on post-contrast imaging and ADC maps, (C, D) demonstrate the mass following neoadjuvant radiotherapy which shows no change in overall size, however a change in contrast signal and ADC can be seen. This would he stable by RECIST 1.1. Bottom: (A, B) demonstrate a biopsy confirmed ewing's sarcoma of the thigh prior to neoadjuvant chemotherapy on post-contrast imaging and ADC maps, (C, D) demonstrate the mass following neoadjuvant chemotherapy which has increased in size, however a change in ihe contrast signal and ADC within the mass can be seen. This mass would be measured as progression by RECIST 1.1 criteria.
To overcome the limitations of current therapy response criteria, the capacity to gain serial virtual biopsies by next generation imaging received as part of a patient’s pathway could improve treatment monitoring. If a robust IB can be developed, alternative and more precise monitoring of therapy is possible. Not only could this allow earlier alterations in therapeutic regimes for patients who are not responding, it would also improve dosing options and reduce the likelihood of severe adverse side effects. This IB may be used independently or again in concurrence with other therapy monitoring methods such as the use of tumour circulating-derived DNA(ctDNA) and tumour cell free DNA(ftDNA) (155) as markers of response as they translate into clinical practice. This in turn would bring us closer to the goal of personalised medicine and allow clinical decisions to be based of individualised tumour and patient characteristics that are achievable in a non-invasive and dynamic way.
PET/CT and PET/MRI
Studies involving 18F-FDG PET/CT have shown value in using the reduction in SUVmax following treatment to indicate possible therapeutic response even when overall tumour volume has not reduced (74, 156–158). Benz et al. demonstrated that a decrease in SUVmax of ≥ 35% from baseline to post-treatment scans was a sensitive predictor of histopathological response to neoadjuvant therapy in a group of 50 patients with resectable high grade STS (156). Schuetze et al. demonstrated a reduction in SUVmax of ≥ 40% was an independent predictor of improved prognosis (disease specific and overall survival) in 46 patients with high grade sarcoma receiving neoadjuvant chemotherapy (157). However, these PET/CT studies are limited to small patient numbers, single centres and are usually retrospective and therefore, confidence in SUVmax as a clinical IB will be dependent on ongoing prospective studies.
PET/MRI has also shown promise for monitoring of patients, adding the excellent soft tissue contrast possible with MRI to information obtained through PET. A study by Erfanian et al. reported improved accuracy in the detection of local recurrence of STS of 90% when compared to conventional MRI alone at 83% (159). Like PET/CT, progress for this imaging modality will rely on robust prospective studies. A further exciting avenue for PET research in STS is immuno-PET imaging as a potential non invasive and serial tool to monitor pre-selected immune markers or aid the selection of patients for immunotherapy. This remains in early stages, however, a study demonstrated feasibility of using a radiotracer and immuno-PET to detect and monitor programmed cell death protein 1 (PD1, an immune checkpoint protein) in humanised mouse models (160).
Quantitative MRI
Frequently ADC values change following radiotherapy signifying possible treatment response, even when overall tumour volume remains unchanged (22, 161–163). Winfield et al. also illustrated that in the same group of retroperitoneal STS, patient MRIs following radiotherapy demonstrated an increase in ADC, perhaps reflecting biological changes to a less cellular tumour suggestive of response. In this study, ADC was shown to have excellent repeatability, with a coefficient of variation (CoV) of 2.5% for median ADC (22). The repeatability of ADC adds to the confidence of its usage as a robust, standardised IB. However, it is important to note that at present, there remains no standardised method to measure ADC in STS, particularly with relation to therapeutic response (164). This can range from variations in the acquisition of diffusion-weighted images, the ROI selection process (for example 2D ROIs versus 3D volume ROIs) and the ADC parameter selected (for example minimum ADC versus mean ADC) (22, 77, 162). As such, this hinders our ability to standardise study findings and like other IBs, it also suffers from lack of reproducibility and validation efforts in prospectively designed studies.
DCE-MRI has also been used to demonstrate changes following therapy, suggestive of possible response (98, 165–168). Recently, in a subset of 10 patients with myxoid liposarcoma from the Dose Reduction in Preoperative Radiotherapy in Myxoid Liposarcomas (DOREMY) multicentre clinical trial, a higher baseline Ktrans was linked to response following neoadjuvant radiotherapy and may allow prediction of early response to radiotherapy (169). Although time consuming analysis and some limitations on anatomical coverage preclude routine clinical use, DCE-MRI represents an interesting avenue for further research into the STS vascular microenvironment and its role in therapeutic response.
Oxygen enhanced (OE-)MRI is an emerging technique that may offer insight into tumour hypoxia; a well-recognised negative prognostic factor in cancer (170–172). To date, there remains an unmet need to measure hypoxia through a biomarker, and MRI offers an attractive non-invasive and serial option (173). OE-MRI is capable of mapping and quantifying the spatial distribution of tumour oxygen delivery. By measuring the longitudinal relaxation rate of hydrogen nuclei (R1), which changes in relation to the dissolved oxygen concentration in tissue or plasma, OE-MRI can characterise well- and poorly-oxygenated tissue. Inhalation of hyperoxic gas results in delivery of excess oxygen to well-oxygenated tissue, however, given well- oxygenated tissue will have saturation of haemoglobin molecules with oxygen, the excess oxygen remains within the plasma and interstitial tissue fluid, increasing the tissue R1. Preclinical and clinical studies in renal cell carcinoma, glioma, prostate cancer and lung cancer have demonstrated the feasibility and accuracy of OE-MRI in mapping hypoxic subregions, including changes following radiotherapy (173–179). Whilst human studies in sarcoma are lacking, the early work by Cao-Pham et al. in rhabdomyosarcoma xenograft models showed that R1 as measured by OE-MRI is sensitive to changes in oxygenation (180). Further exploration in STS is required, however in a group of heterogeneous cancers, OE-MRI could optimise our treatment planning for these patients. Visualising areas of hypoxia across a tumour could aid radiotherapy planning, stratifying patients accordingly.
Similarly, magnetic resonance elastography (MRE) is gaining attention as a quantitative imaging method capable of assessing the mechanical properties of tissue. MRE informs on the viscoelastic properties of tissue by measuring the propagation of mechanical waves through tissue (181). Already a well-established technique in patients with chronic liver disease, there is a strong motivation to explore the applicability of MRE to other pathologies (182, 183). Preclinical studies have demonstrated that altered tissue “stiffness” (quantified using the complex shear modulus (G*) and related parameters) can be used to indicate malignant masses within brain parenchyma, and that changes in mechanical properties of a tumour may precede volume change following treatment indicative of response in colon cancer and non-Hodgkin’s lymphoma (181, 184–186). There are currently limited studies within STS, however, Pepin et al. demonstrated technical feasibility of MRE in a pilot study of 13 sarcoma patients, and possible utility in assessment of early response to radiation therapy (in 4 of these 13 patients) (187). Further studies are underway, and this may offer a further non-invasive method to interrogate deep and complex masses such as those within the retroperitoneum.
Radiomics
Much of the radiomic research in STS is focused on predicting therapeutic response and clinical outcome. In an externally validated study, Spraker et al. found that features extracted from T1-weighted MRI were independently associated with overall survival (OS), and a radiomics model combined with clinical features (age and grade) performed best (46). In a further study, the addition of T2-weighted MR radiomics to the clinical American Joint Committee on Cancer (AJCC) staging system into a nomogram improved the clinical net benefit for stratification of patients for OS, and was superior to using AJCC system alone (188). The same group went on to apply deep learning to the two cohorts (that had been slightly expanded), concluding that the addition of deep learning to their previous radiomic model yielded comparable prediction performance for OS. This finding was supported by a further nomogram constructed by Yan et al, demonstrating successful patient risk stratification based on progression free survival (PFS) when MR-based radiomics was used in addition to clinical factors, with an increased benefit to using AJCC staging system alone (189). In CT, the same study by Peeken et al. using radiomics to distinguish tumour grade in STS explored the predictive performance of radiomic features for OS, distant progression free survival (DPFS) and local progression free survival (LPFS). This study concluded that again, despite CT’s inferior soft tissue contrast, there was value in using a radiomics model to stratify patient risk based on their survival outcomes. This finding was successfully validated in an external cohort, and results were more consistent than using a clinical model (47).
In the context of response to therapy, delta-radiomics, the change in a given radiomics feature in a set of longitudinal images is of particular interest. Gao et al. used longitudinal DW-MRI images in 30 STS patients receiving neoadjuvant RT at three time points, to develop a predictive model of pathological therapeutic response (190). The model demonstrated that radiomic features alone were not sufficient to predict response, however, the inclusion of delta-radiomics features at mid- and post-therapy time points optimised prediction of response to RT. Delta-radiomics was also used for prediction of pathological response to neoadjuvant chemotherapy in a retrospective cohort of 65 STS patients. A radiomics model was developed based on longitudinal MRI scans taken at baseline and following two cycles of anthracycline-based therapy, prior to surgical excision from 50 of the 65 patients. The findings were then validated in the remaining 15 patients. Using 3 STS features of shape, heterogeneity and surrounding tissue, the study concluded an improved performance of the radiomics model when compared to RECIST 1.1, and a promising tool for the evaluation of early response following only two cycles of chemotherapy (50).
Multispectral Tissue Characterisation and Habitat Mapping
Previous studies have utilised quantitative parameters derived from diffusion weighted imaging, namely ADC, the transverse relaxation time (T2) and proton density (M0) to objectively segment tumours into distinct subregions and in relation to therapy (191–195). In a xenograft mouse model of radiation-induced fibrosarcoma (RIF-1), Henning et al. was able to segment tumours into regions based on two viable tumour groups and two non viable (necrotic) tissue groups which were confirmed on histopathological staining of a hypoxia marker (192). The group also analysed the viable tissue groups further and characterised them into well oxygenated radiosensitive and hypoxia and radioresistant groups (193). This is important ground work for methods of objectively characterising tissue based on quantitative markers for underlying biological properties. Building on this are habitat maps which incorporate multiple quantitative parameters obtained from numerous MRI sequences. These maps characterise tissue into distinct biological habitats based on their combination of multiparametric MRI parameters and provide insight into multiple aspects of biological heterogeneity and their interaction with one another (21). In a cohort of 30 retroperitoneal STS, automated segmentation of tumours into tissue subtypes based on fat fraction (FF), enhancement fraction (EF) and ADC, demonstrated distinct habitat maps and changes following radiotherapy suggestive of response, including when overall tumour volume remained the same or increased (Figure 3). These techniques may improve our understanding of the differing characteristics within this heterogeneous cancer and how distinct regions respond variably to the same treatment. In addition, it can illustrate results in a timely and clinician friendly manner that is not reliant on interpretation of numerical values, easing its transition into practice.
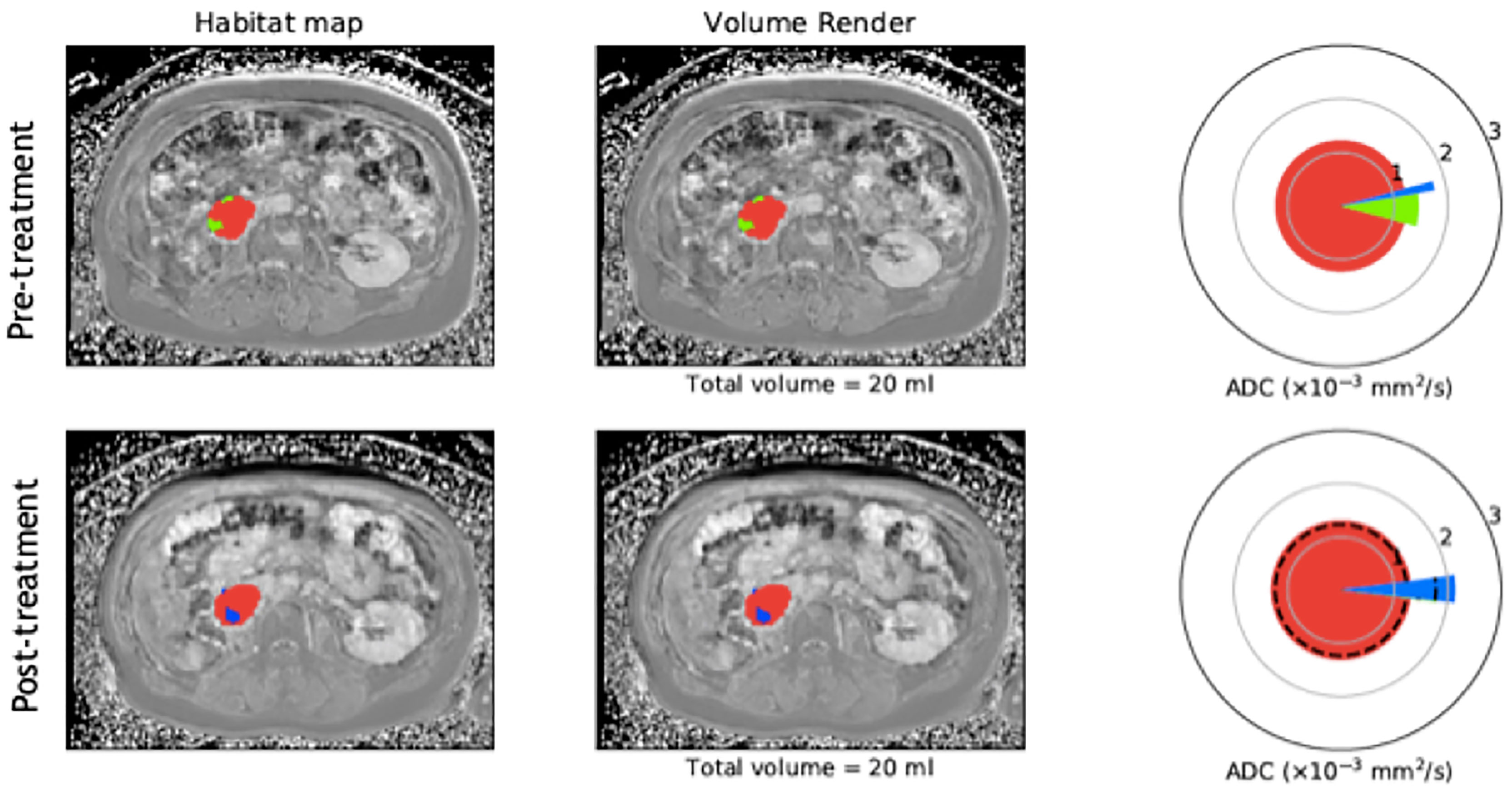
Figure 3 Example of habitat imaging as demonstrated by a supplementary figure provided by Blackedge et al. (21). Axial MRI scan taken pre and post-radiotherapy in a patient with a retropentoneal STS tumour. Habitat maps are overlaid on MRI scans acquired for this patient and volume renders shown. The different colours within the mass on MRI scans represent the different sub compartments assigned according to the qMRI parameters EF. FF and ADC. Tissue sub compartments with high ADC and low FF are represented in blue and may suggest necrotic or cystic tissue, low ADC and high EF in red and suggest cellular vascular tissue, and low ADC and low EF in green and suggest poorly vasculansed tissue. Spie charts show the average ADC for each tissue sub compartment as the illustrated radius of each segment, whilst the angular proportion represents the proportional volume of habitat class. Pre and post-treatment, the colours within the mass are shown to change, with loss of the green sub compartment (poorly vasculansed tissue) and increase in the blue sub compartment (necrotic/cystic tissue), signifying a change in tissue characteristics following therapy and possible response even when overall volume is unchanged. (Figure adapted from citation).
Ongoing Challenges and Future Directions
The integration of powerful imaging technology to inform on the biology of these heterogeneous tumours is needed in the clinical setting for this complex disease. Indeed, there is potential to enable a more personalised approach to patient management and uncover candidate therapeutic options. However, there are a number of challenges that need to be addressed prior to the successful implementation of “virtual biopsy” methods into practice. If the ultimate goal of quantitative and radiomic imaging techniques is the development of an IB, consideration of the steps of biomarker development is required (5). At present, despite a heavy investment into the search for IB discovery, most sarcoma work has faltered at the initial stage of technical (assay) validation (22).
Reproducibility and Complexiy of Data
Quantitative MRI sequences have a varying complexity of delivery, particularly those which require intricate post-processing of images, and therefore, make reliable and confident translation across radiology departments challenging. This is mirrored in the variation seen across different MRI scanners in image acquisition and measurement output, usually requiring adaptations at every centre and vendor. This weakens the reproducibility potential of quantitative MRI techniques and may oblige different centres to perform reproducibility/repeatability studies to conclude some clinical capability. As a result, most studies remain limited to a single centre using the same MRI machine. This hinders the progression of novel and promising quantitative MRI techniques through imaging biomarker validation. Similarly, a crucial step in the radiomics workflow is standardisation of acquired images to allow reliable image delineation and radiomic feature extraction, and this can pose significant challenges at multi-centre level. Each step in the radiomics workflow can in fact affect results, and similar to quantitative MRI, requires careful or expert processing of data. This makes reproducibility efforts challenging. To date, few studies have spanned multiple centres, and the majority of studies have not been tested in an independent external cohort. Attempts to provide standardised guidance for the translation of acquired imaging to high-throughput IBs are underway. The Image Biomarkers Standardization Initiative (IBSI), an independent international collaboration aims to provide detailed guidance on the mandatory steps for radiomic analyses, and Lambin et al. have defined the Radiomics Quality Score (RQS) to provide an objective estimate of the quality of the study (196, 197). Future studies may benefit from these to aid transition from proof of concept to real life clinically applicable tools.
Limited Cohort Size and External Validation
STS is a disease of rarity, and it’s diverse histological subtype profile confounds effort to accrue patient cohorts comparable to other cancer types. This can make assessment of the quality of findings within imaging studies challenging. In particular, radiomics is dependent on large datasets for reliable feature extraction and robust conclusions to be drawn from results. Furthermore, a crucial component of translating an IB is it’s independent validation in an external cohort, which is a recognised challenge in the sarcoma community. Whilst there are methods to deal with smaller datasets such as cross-validation, these still remain limited to the individuals studied and findings cannot be generalised to the rest of the population, not overcoming the issues pertaining to external validation (198).
A solution to this may be data-sharing. A powerful drive through the NIH’s Quantitative Imaging Network (QIN) and its leveraging on The Cancer Imaging Archive (TCIA) has demonstrated feasibility in data sharing of clinical images across various sites (199). This partnership holds promise to overcome challenges related to data-sharing, facilitating multi-site collaboration and support the development of IBs through validation (200–202). Builiding on this, there are efforts to harmonise quantitative MRI data across centres and develop a robust infrastructure to allow this, such as the National Cancer Imaging Translational Accelerator (NCITA) consortium which includes 9 UK centres aiming to develop a robust multicentre IB pipeline (65). This would provide opportunities for both multi-centre data to increase the size of initial datasets and external cohorts for the validation of discoverable IBs.
Retrospective Study Design
The challenges related to the low incidence and heterogeneity of the disease can in turn make prospective collection of data difficult. It can take multiple years to recruit a desired cohort size, and studies may be closed early upon failing to reach target accrual. As such, alongside the readily available retrospective data, many imaging studies within STS remain retrospective in nature. This carries risk of confounding and bias (such as selection or recall bias), and results in an inferior level of evidence. Furthermore, retrospective designs negate the ability to carry out repeatability tests for quantitative MRI and radiomic studies due to a lack of repeat scans performed around the time of the initial scan. Going forward, confident validation and translation of IBs is heavily dependent on prospective studies.
Within STS, a Cancer Research United Kingdom (CRUK) Sarcoma Accelerator consortium partnering centres across the UK, Spain and Italy aims to develop a multicentre, expansive data pipeline to develop clinical and biological models to further our understanding of sarcoma biology in patients with high risk STS undergoing perioperative management. This will involve prospective collection of imaging data across 5 years and spanning these multiple centres. This will greatly benefit IB discovery and validation in sarcoma, where limited patient numbers for studies will increasingly rely on close multi-centre collaborations and data-sharing to propel their progress to clinical translation.
Conclusion
The ambition to achieve non-invasive and global captures of tumour biology remains a priority in complex and heterogeneous malignancies such as STS. Whilst radiomics is promising, sub-optimal datasets and a lack of external validation mean this is some way from becoming a tool to direct patient care. However, we can now offer virtual biopsies in the research setting following the validation of a number of IBs in sarcomas, and which can be assimilated as habitat maps (Figure 4). The clinical impact of identifying these distinct imaging phenotypes is yet to be fully explored and as further IBs including radiomic signatures mature, they will also be incorporated into these models of heterogeneity. The next step is prospective clinical trials exploring habitat map-guided risk stratification, response assessment and adaptive therapies on patient outcomes. Translation to routine clinical care will also be ultimately reliant on improved proficiencies within radiology departments to seamlessly incorporate the necessary post-processing and data visualisation tools into routine workflow.
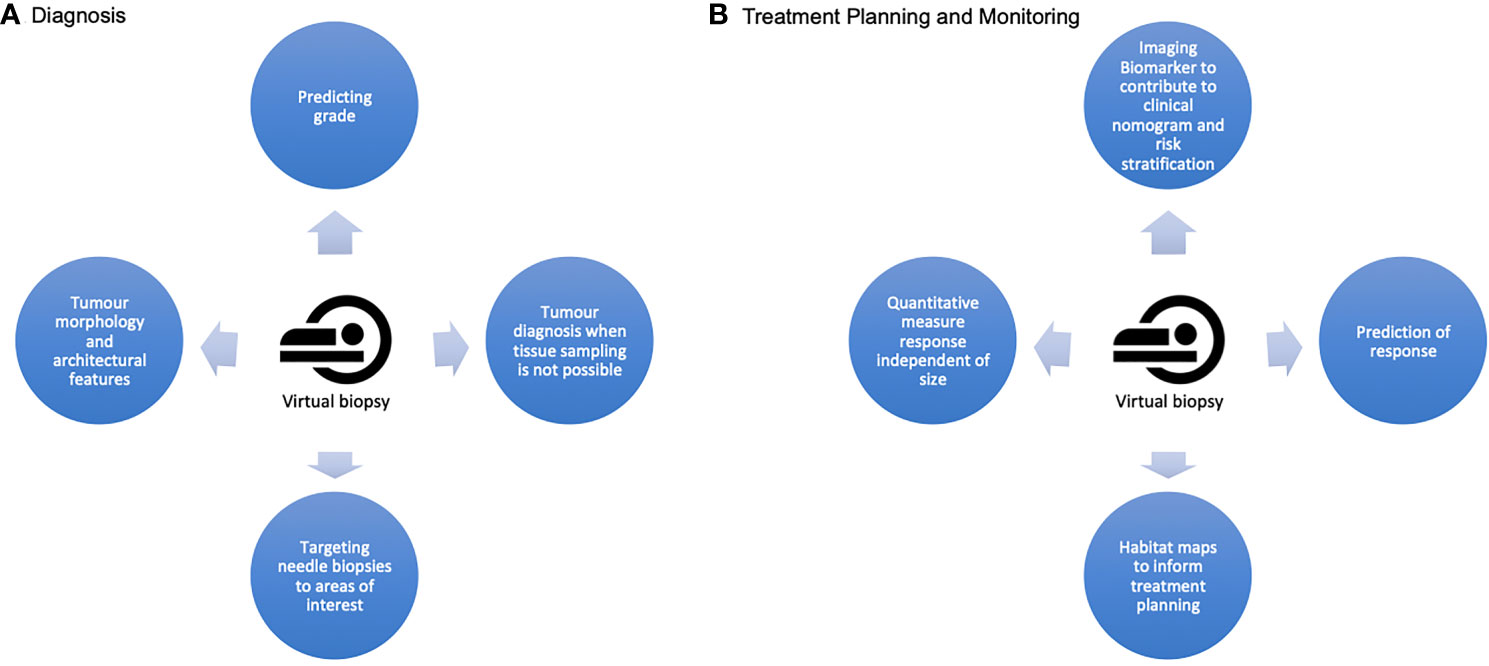
Figure 4 A summary of the potential applications of a virtual biopsy at the two crucial stages of a sarcoma patient's cane pathway (A) At diagnosis, a virtual biopsy can offer quantitative imaging biomarkers to improve accuracy of grading, complimentary architectural information about the lesion and information about diagnosis and grade when needle biopsy is not possible. It can also guide precision needle biopsy. (B) For treatment planning and monitoring, virtual biopsy offers the opportunities for improved patient risk stratification and prediction of therapeutic response. It can provide information assimilated in habitat maps to capture global and spatial heterogeneity for improved treatment planning, and provides the potential for serial quantitative, non invasive measures of response which are independent of tumour size.
Author Contributions
AA, EJ, JW, RJ, PH, and CM: literature search. AA, JW, MB, CM, and PH: figures. All authors contributed to manuscript revision and approved the final version.
Funding
This report is independent research funded by the National Institute for Health Research.
Conflict of Interest
RJ is the recipient of grants/research support from Merck Sharp & Dohme and GlaxoSmithKline. RJ is the recipient of consultation fees from Adaptimmune, Athenex, Blueprint Medicines, Clinigen, Eisai, Epizyme, Daichii, Deciphera, Immune Design, Lilly, Merck, PharmaMar and UpToDate.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge the support of the Wellcome Trust and the Royal Marsden/Institute of Cancer Research National Institute for Health Research Biomedical Research Centre.
References
1. Cullinane C, Solomon B, Hicks RJ. Imaging of Molecular Target Modulation in Oncology: Challenges of Early Clinical Trials. Clin Transl Imaging (2014) 2(1):5–12. doi: 10.1007/s40336-013-0047-6
2. Blackledge MD, Messiou C. Using Artificial Intelligence to Support the Adoption of Quantitative MRI Into Clinical Practice. RAD Mag (2018) 538(46):23–4.
3. Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL, Gillies RJ. Quantitative Imaging of Cancer in the Postgenomic Era: Radio(geno)mics, Deep Learning, and Habitats. Cancer (2018) 124:4633–49. John Wiley and Sons Inc. doi: 10.1002/cncr.31630
4. Kuo MD, Yamamoto S. Next Generation Radiologic-Pathologic Correlation in Oncology: Rad-Path 2.0. Am J Roentgenol (2011) 197(4):990–7. doi: 10.2214/AJR.11.7163
5. O’Connor JPB, Aboagye EO, Adams JE, Aerts HJWL, Barrington SF, Beer AJ, et al. Imaging Biomarker Roadmap for Cancer Studies. Nat Rev Clin Oncol (2017) 14(3):169–86. doi: 10.1038/nrclinonc.2016.162
6. Arifi S, Belbaraka R, Rahhali R, Ismaili N. Treatment of Adult Soft Tissue Sarcomas: An Overview. Rare Cancers Ther (2015) 3(1–2):69–87. doi: 10.1007/s40487-015-0011-x
7. Bovée JVMG, Hogendoorn PCW. Molecular Pathology of Sarcomas: Concepts and Clinical Implications. Virchows Arch (2010) 456:193–9. doi: 10.1007/s00428-009-0828-5
8. Dancsok AR, Asleh-Aburaya K, Nielsen TO. Oncotarget 7068 Www.Impactjournals.Com/Oncotarget Advances in Sarcoma Diagnostics and Treatment. Oncotarget (2017) 8(4):7068–93. doi: 10.18632/oncotarget.12548
9. Halcrow PW, Dancer M, Panteah M, Walden C, Ohm JE. Molecular Changes Associated With Tumor Initiation and Progression of Soft Tissue Sarcomas: Targeting the Genome and Epigenome. In: Progress in Molecular Biology and Translational Science. (Netherlands:Elsevier) B.V (2016). p. 323–80.
10. Jo VY, Fletcher CDM. WHO Classification of Soft Tissue Tumours: An Update Based on the 2013 (4th) Edition. Pathology (2014) 46(2):95–104. doi: 10.1097/PAT.0000000000000050
11. Ryan CW, Merimsky O, Agulnik M, Blay J-Y, Schuetze SM, Van Tine BA, et al. PICASSO III: A Phase III, Placebo-Controlled Study of Doxorubicin With or Without Palifosfamide in Patients With Metastatic Soft Tissue Sarcoma. J Clin Oncol (2016) 34(32):3898–905. doi: 10.1200/JCO.2016.67.6684
12. Tap WD, Wagner AJ, Schöffski P, Martin-Broto J, Krarup-Hansen A, Ganjoo KN, et al. Effect of Doxorubicin Plus Olaratumab vs Doxorubicin Plus Placebo on Survival in Patients With Advanced Soft Tissue Sarcomas: The ANNOUNCE Randomized Clinical Trial. JAMA (2020) 323(13):1266–76. doi: 10.1001/jama.2020.1707
13. Tap WD, Papai Z, Van Tine BA, Attia S, Ganjoo KN, Jones RL, et al. Doxorubicin Plus Evofosfamide Versus Doxorubicin Alone in Locally Advanced, Unresectable or Metastatic Soft-Tissue Sarcoma (TH CR-406/SARC021): An International, Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol (2017) 18(8):1089–103. doi: 10.1016/S1470-2045(17)30381-9
14. Seddon B, Strauss SJ, Whelan J, Leahy M, Woll PJ, Cowie F, et al. Gemcitabine and Docetaxel Versus Doxorubicin as First-Line Treatment in Previously Untreated Advanced Unresectable or Metastatic Soft-Tissue Sarcomas (GeDDiS): A Randomised Controlled Phase 3 Trial. Lancet Oncol (2017) 18(10):1397–410. doi: 10.1016/S1470-2045(17)30622-8
15. Judson I, Verweij J, Gelderblom H, Hartmann JT, Schöffski P, Blay J, et al. Results of a Randomised Phase III Trial (EORTC 62012) of Single Agent Doxorubicin Versus Doxorubicin Plus Ifosfamide as First Line Chemotherapy for Patients With Advanced or Metastatic Soft Tissue Sarcoma: A Survival Study by the Eortc Soft Tissue and Bone Sarcoma Group. Ann Oncol (2012) 23:ixe28. doi: 10.1016/S0923-7534(20)34351-9
16. Hendifar AE, Chawla SP, Leahy MG, Italiano A, Patel S, Santoro A, et al. Results of the Randomized Phase III Trial of Trabectedin (T) Versus Doxorubicin-Based Chemotherapy (DXCT) as First-Line Therapy in Patients (Pts) With Translocation-Related Sarcoma (TRS). J Clin Oncol (2013) 31(15_suppl):10517–7. doi: 10.1200/jco.2013.31.15_suppl.10517
17. Pautier P, Floquet A, Chevreau C, Penel N, Guillemet C, Delcambre C, et al. Trabectedin in Combination With Doxorubicin for First-Line Treatment of Advanced Uterine or Soft-Tissue Leiomyosarcoma (LMS-02): A non-Randomised, Multicentre, Phase 2 Trial. Lancet Oncol (2015) 16(4):457–64. doi: 10.1016/S1470-2045(15)70070-7
18. Adeniyi O, Aguel F, Agyeman A, Amur S, Bauri K, Canos D, et al. BEST (Biomarkers, EndpointS, and Other Tools) Resource. BEST ( Biomarkers, EndpointS, Other Tools ) Resour. Silver Spring (MD: Food and Drug Administration (US (2016). Bethesda (MD): National Institutes of Health (US).
19. Califf RM. Biomarker Definitions and Their Applications. Exp Biol Med (2018) 243(3):213. doi: 10.1177/1535370217750088
20. Sullivan DC, Obuchowski NA, Kessler LG, Raunig DL, Gatsonis C, Huang EP, et al. Metrology Standards for Quantitative Imaging Biomarkers1. Radiology (2015) 277(3):813–25. doi: 10.1148/radiol.2015142202
21. Blackledge MD, Winfield JM, Miah A, Strauss D, Thway K, Morgan VA, et al. Supervised Machine-Learning Enables Segmentation and Evaluation of Heterogeneous Post-Treatment Changes in Multi-Parametric MRI of Soft-Tissue Sarcoma. Front Oncol (2019) 9(SEP):941. doi: 10.3389/fonc.2019.00941
22. Winfield JM, Miah AB, Strauss D, Thway K, Collins DJ, DeSouza NM, et al. Utility of Multi-Parametric Quantitative Magnetic Resonance Imaging for Characterization and Radiotherapy Response Assessment in Soft-Tissue Sarcomas and Correlation With Histopathology. Front Oncol (2019) 9(APR). doi: 10.3389/fonc.2019.00280
23. McGee KP, Hwang KP, Sullivan DC, Kurhanewicz J, Hu Y, Wang J, et al. Magnetic Resonance Biomarkers in Radiation Oncology: The Report of AAPM Task Group 294. Med Phys (2021) 48(7):e697. doi: 10.1002/mp.14884
24. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More Than Pictures, They Are Data. Radiology (2016) 278(2):563–77. doi: 10.1148/radiol.2015151169
25. Ma J. Dixon Techniques for Water and Fat Imaging. J Magn Reson Imaging (2008) 28:543–58. doi: 10.1002/jmri.21492
26. Reeder S. Proton Density Fat-Fraction : A Standardized MR-Based Biomarker of Tissue Fat Concentration. J Magn Reson Imaging (2012) 1014:1011–4. doi: 10.1002/jmri.23741
27. Koh DM, Collins DJ. Diffusion-Weighted MRI in the Body: Applications and Challenges in Oncology. Am J Roentgenol (2007) 188(6):1622–35. doi: 10.2214/AJR.06.1403
28. Messina C, Bignone R, Bruno A, Bruno A, Bruno F, Calandri M, et al. Diffusion-Weighted Imaging in Oncology: An Update. Cancers (2020) 12:1–28. doi: 10.3390/cancers12061493
29. Malayeri AA, El Khouli RH, Zaheer A, Jacobs MA, Corona-Villalobos CP, Kamel IR, et al. Principles and Applications of Diffusion-Weighted Imaging in Cancer Detection, Staging, and Treatment Follow-Up. Radiographics (2011) 31(6):1773–91. doi: 10.1148/rg.316115515
30. Baliyan V, Das CJ, Sharma R, Gupta AK. Diffusion Weighted Imaging: Technique and Applications. World J Radiol (2016) 8(9):785. doi: 10.4329/wjr.v8.i9.785
31. Michoux NF, Ceranka JW, Vandemeulebroucke J, Peeters F, Lu P, Absil J, et al. Repeatability and Reproducibility of ADC Measurements: A Prospective Multicenter Whole-Body-MRI Study. Eur Radiol (2021) 31(7): 4514–27, doi: 10.1007/s00330-020-07522-0
32. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: The Process and the Challenges. Magn Reson Imaging (2012) 30(9):1234–48. doi: 10.1016/j.mri.2012.06.010
33. Wu J, Tha KK, Xing L, Li R. Radiomics and Radiogenomics for Precision Radiotherapy. J Radiat Res (2018) 59(January):i25–31. doi: 10.1093/jrr/rrx102
34. Nair VS, Gevaert O, Davidzon G, Napel S, Graves EE, Hoang CD, et al. Prognostic PET 18f-FDG Uptake Imaging Features are Associated With Major Oncogenomic Alterations in Patients With Resected non-Small Cell Lung Cancer. Cancer Res (2012) 72(15):3725–34. doi: 10.1158/0008-5472.CAN-11-3943
35. Zhou M, Hall L, Goldgof D, Russo R, Balagurunathan Y, Gillies R, et al. Radiologically Defined Ecological Dynamics and Clinical Outcomes in Glioblastoma Multiforme: Preliminary Results. Transl Oncol (2014) 7(1):5–13. doi: 10.1593/tlo.13730
36. Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, et al. Haralick Texture Analysis of Prostate MRI: Utility for Differentiating Non-Cancerous Prostate From Prostate Cancer and Differentiating Prostate Cancers With Different Gleason Scores. Eur Radiol (2015) 25(10):2840–50. doi: 10.1007/s00330-015-3701-8
37. Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas HA, et al. Automatic Classification of Prostate Cancer Gleason Scores From Multiparametric Magnetic Resonance Images. Proc Natl Acad Sci U S A (2015) 112(46):E6265–73. doi: 10.1073/pnas.1505935112
38. Chen CK, Wu HT, Chiou HJ, Wei CJ, Yen CH, Chang CY, et al. Differentiating Benign and Malignant Soft Tissue Masses by Magnetic Resonance Imaging: Role of Tissue Component Analysis. J Chin Med Assoc (2009) 72(4):194–201. doi: 10.1016/S1726-4901(09)70053-X
39. Sala E, Mema E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, et al. Unravelling Tumour Heterogeneity Using Next-Generation Imaging: Radiomics, Radiogenomics, and Habitat Imaging. Clin Radiol (2017) 72(1):3–10. doi: 10.1016/j.crad.2016.09.013
40. Tatli S, Gerbaudo VH, Mamede M, Tuncali K, Shyn PB, Silverman SG. Abdominal Masses Sampled at PET/CT-Guided Percutaneous Biopsy: Initial Experience With Registration of Prior PET/CT Images. Radiology (2010) 256(1):305–11. doi: 10.1148/radiol.10090931
41. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat Commun (2014) 5(1):1–9. doi: 10.1038/ncomms5644
42. Tomaszewski MR, Gillies RJ. The Biological Meaning of Radiomic Features. Radiology (2021) 298:505–16. doi: 10.1148/radiol.2021219005
43. Xu W, Hao D, Hou F, Zhang D, Wang H. Soft Tissue Sarcoma: Preoperative MRI-Based Radiomics and Machine Learning May Be Accurate Predictors of Histopathologic Grade. Am J Roentgenol (2020) 215(4):963–9. doi: 10.2214/AJR.19.22147
44. Zhang Y, Zhu Y, Shi X, Tao J, Cui J, Dai Y, et al. Soft Tissue Sarcomas: Preoperative Predictive Histopathological Grading Based on Radiomics of MRI. Acad Radiol (2018) 26(9):1262–8. doi: 10.1016/j.acra.2018.09.025
45. Wang H, Chen H, Duan S, Hao D, Liu J. Radiomics and Machine Learning With Multiparametric Preoperative MRI May Accurately Predict the Histopathological Grades of Soft Tissue Sarcomas. J Magn Reson Imaging (2020) 51(3):791–7. doi: 10.1002/jmri.26901
46. Spraker MB, Wootton LS, Hippe DS, Ball KC, Peeken JC, Macomber MW, et al. MRI Radiomic Features Are Independently Associated With Overall Survival in Soft Tissue Sarcoma. Adv Radiat Oncol (2019) 4(2):413–21. doi: 10.1016/j.adro.2019.02.003
47. Peeken JC, Bernhofer M, Spraker MB, Pfeiffer D, Devecka M, Thamer A, et al. CT-Based Radiomic Features Predict Tumor Grading and Have Prognostic Value in Patients With Soft Tissue Sarcomas Treated With Neoadjuvant Radiation Therapy. Radiother Oncol (2019) 135:187–96. doi: 10.1016/j.radonc.2019.01.004
48. Tian L, Zhang D, Bao S, Nie P, Hao D, Liu Y, et al. Radiomics-Based Machine-Learning Method for Prediction of Distant Metastasis From Soft-Tissue Sarcomas. Clin Radiol (2021) 76(2):158.e19–158.e25. doi: 10.1016/j.crad.2020.08.038
49. Vallières M, Freeman CR, Skamene SR, El Naqa I. A Radiomics Model From Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities. Phys Med Biol (2015) 60(14):5471. doi: 10.1088/0031-9155/60/14/5471
50. Crombé A, Périer C, Kind M, De Senneville BD, Le Loarer F, Italiano A, et al. T 2 -Based MRI Delta-Radiomics Improve Response Prediction in Soft-Tissue Sarcomas Treated by Neoadjuvant Chemotherapy. J Magn Reson Imaging (2019) 50(2):497–510. doi: 10.1002/jmri.26589
51. Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. Radiomics: The Facts and the Challenges of Image Analysis. Eur Radiol Exp (2018) 2(1):36. doi: 10.1186/s41747-018-0068-z
52. Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial Intelligence in Cancer Imaging: Clinical Challenges and Applications. CA Cancer J Clin (2019) 69(2):127–57. doi: 10.3322/caac.21552
53. Oren O, Gersh BJ, Bhatt DL. Artificial Intelligence in Medical Imaging: Switching From Radiographic Pathological Data to Clinically Meaningful Endpoints. Lancet Digit Health (2020) 2:e486–8. doi: 10.1016/S2589-7500(20)30160-6
54. Jiang H, Diao Z, Yao Y-D. Deep Learning Techniques for Tumor Segmentation: A Review. J Supercomput (2021) 78(2):1807–51. doi: 10.1007/s11227-021-03901-6
55. Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö. Radiomics With Artificial Intelligence: A Practical Guide for Beginners. Diagn Interv Radiol (2019) 25:485–95. doi: 10.5152/dir.2019.19321
56. Rubin BP, Cooper K, Fletcher CDM, Folpe AL, Gannon FH, Hunt JL, et al. Protocol for the Examination of Specimens From Patients With Tumors of Soft Tissue. Arch Pathol Lab Med (2010) 134(4):e31–9. doi: 10.5858/134.4.e31
57. Fisher C. Standards and Datasets for Reporting Cancers. (2017). London:The Royal College of Pathologists:
58. Coindre JM, Terrier P, Bui NB, Bonichon F, Collin F, Le Doussal V, et al. Prognostic Factors in Adult Patients With Locally Controlled Soft Tissue Sarcoma. A Study of 546 Patients From the French Federation of Cancer Centers Sarcoma Group. J Clin Oncol (1996) 14(3):869–77. doi: 10.1200/JCO.1996.14.3.869
59. Guillou L, Coindre JM, Bonichon F, Nguyen BB, Terrier P, Collin F, et al. Comparative Study of the National Cancer Institute and French Federation of Cancer Centers Sarcoma Group Grading Systems in a Population of 410 Adult Patients With Soft Tissue Sarcoma. J Clin Oncol (1997) 15(1):350–62. doi: 10.1200/JCO.1997.15.1.350
60. Coindre J-M, Terrier P, Guillou L, Le Doussal V, Oise Collin F, Ranchè D, et al. Predictive Value of Grade for Metastasis Development in the Main Histologic Types of Adult Soft Tissue Sarcomas A Study of 1240 Patients From the French Federation of Cancer Centers Sarcoma Group. Cancer (2001) 91(10):1914–26.
61. Crombé A, Marcellin PJ, Buy X, Stoeckle E, Brouste V, Italiano A, et al. Soft-Tissue Sarcomas: Assessment of MRI Features Correlating With Histologic Grade and Patient Outcome. Radiology (2019) 291(3):710–21. doi: 10.1148/radiol.2019181659
62. Schneider N, Strauss DC, Smith MJ, Miah AB, Zaidi S, Benson C, et al. The Adequacy of Core Biopsy in the Assessment of Smooth Muscle Neoplasms of Soft Tissues. Am J Surg Pathol (2017) 41(7):923–31. doi: 10.1097/PAS.0000000000000867
63. Ray-coquard I, Montesco MC, Coindre JM, Dei tos AP, Lurkin A, Ranchère-vince D, et al. Sarcoma: Concordance Between Initial Diagnosis and Centralized Expert Review in a Population-Based Study Within Three European Regions. Ann Oncol (2012) 23(9):2442. doi: 10.1093/annonc/mdr610
64. Thway K, Wang J, Mubako T, Fisher C. Histopathological Diagnostic Discrepancies in Soft Tissue Tumours Referred to a Specialist Centre: Reassessment in the Era of Ancillary Molecular Diagnosis. Sarcoma (2014) 2014:7. doi: 10.1155/2014/686902
65. McAteer MA, O’Connor JPB, Koh DM, Leung HY, Doran SJ, Jauregui-Osoro M, et al. Introduction to the National Cancer Imaging Translational Accelerator (NCITA): A UK-Wide Infrastructure for Multicentre Clinical Translation of Cancer Imaging Biomarkers. Br J Cancer (2021) 125(11):1462–5. doi: 10.1038/s41416-021-01497-5
66. Liu QY, Li HG, Chen JY, Liang BL. Correlation of MRI Features to Histopathologic Grade of Soft Tissue Sarcoma. Ai Zheng (2008) 27(8):856–60.
67. Zhao F, Ahlawat S, Farahani SJ, Weber KL, Montgomery EA, Carrino JA, et al. Can MR Imaging be Used to Predict Tumor Grade in Soft-Tissue Sarcoma? Radiology (2014) 272(1):192–201. doi: 10.1148/radiol.14131871
68. Mcaddy NC, Hallin M, Strauss D, Smith M, Hayes A, Yusuf S, et al. CT Imaging Improves Histopathological Grading of Retroperitoneal Leiomyosarcomas. Eur J Surg Oncol (2020) 46(2):288–92. doi: 10.1016/j.ejso.2019.10.007
69. Gamboa AC, Gronchi A, Cardona K. Soft-Tissue Sarcoma in Adults: An Update on the Current State of Histiotype-Specific Management in an Era of Personalized Medicine. CA Cancer J Clin (2020) 70(3):200–29. doi: 10.3322/caac.21605
70. Lahat G, Madewell JE, Anaya DA, Qiao W, Tuvin D, Benjamin RS, et al. Computed Tomography Scan-Driven Selection of Treatment for Retroperitoneal Liposarcoma Histologic Subtypes. Cancer (2009) 115(5):1081–90. doi: 10.1002/cncr.24045
71. Morosi C, Stacchiotti S, Marchianò A, Bianchi A, Radaelli S, Sanfilippo R, et al. Correlation Between Radiological Assessment and Histopathological Diagnosis in Retroperitoneal Tumors: Analysis of 291 Consecutive Patients at a Tertiary Reference Sarcoma Center. Eur J Surg Oncol (2014) 40(12):1662–70. doi: 10.1016/j.ejso.2014.10.005
72. Ioannidis JPA, Lau J. 8 F-FDG PET for the Diagnosis and Grading of Soft-Tissue Sarcoma: A Meta-Analysis.J Nucl Med (2003) 44(5):717–24
73. Bastiaannet E, Groen B, Jager PL, Cobben DCP, van der Graaf WTA, Vaalburg W, et al. The Value of FDG-PET in the Detection, Grading and Response to Therapy of Soft Tissue and Bone Sarcomas; a Systematic Review and Meta-Analysis. Cancer Treat Rev (2004) 30(1):83–101. doi: 10.1016/j.ctrv.2003.07.004
74. Ha SC, Oh JS, Roh JL, Moon H, Kim JS, Cho KJ, et al. Pretreatment Tumor SUVmax Predicts Disease-Specific and Overall Survival in Patients With Head and Neck Soft Tissue Sarcoma. Eur J Nucl Med Mol Imaging (2017) 44(1):33–40. doi: 10.1007/s00259-016-3456-8
75. Song T, Shen J, Liang BL, Mai WW, Li Y, Guo HC. Retroperitoneal Liposarcoma: MR Characteristics and Pathological Correlative Analysis. Abdom Imaging (2007) 32(5):668–74. doi: 10.1007/s00261-007-9220-6
76. El Ouni F, Jemni H, Trabelsi A, Ben Maitig M, Arifa N, Ben Rhouma K, et al. Liposarcoma of the Extremities: MR Imaging Features and Their Correlation With Pathologic Data. Orthop Traumatol Surg Res (2010) 96(8):876–83. doi: 10.1016/j.otsr.2010.05.010
77. Schnapauff D, Zeile M, Niederhagen MB, Fleige B, Tunn PU, Hamm B, et al. Diffusion-Weighted Echo-Planar Magnetic Resonance Imaging for the Assessment of Tumor Cellularity in Patients With Soft-Tissue Sarcomas. J Magn Reson Imaging (2009) 29(6):1355–9. doi: 10.1002/jmri.21755
78. Corino VDA, Montin E, Messina A, Casali PG, Gronchi A, Marchianò A, et al. Radiomic Analysis of Soft Tissues Sarcomas can Distinguish Intermediate From High-Grade Lesions. J Magn Reson Imaging (2018) 47(3):829–40. doi: 10.1002/jmri.25791
79. Chhabra A, Ashikyan O, Slepicka C, Dettori N, Hwang H, Callan A, et al. Conventional MR and Diffusion-Weighted Imaging of Musculoskeletal Soft Tissue Malignancy: Correlation With Histologic Grading. Eur Radiol (2019) 29(8):4485–94. doi: 10.1007/s00330-018-5845-9
80. Hassanien OA, Younes RL, Dawoud RM. Diffusion Weighted MRI of Soft Tissue Masses: Can Measurement of ADC Value Help in the Differentiation Between Benign and Malignant Lesions? Egypt J Radiol Nucl Med (2018) 49(3):681–8. doi: 10.1016/j.ejrnm.2018.04.008
81. Englund EK, Reiter DA, Shahidi B, Sigmund EE. Intravoxel Incoherent Motion Magnetic Resonance Imaging in Skeletal Muscle: Review and Future Directions. JMRI (2021) 55(4):988–1012. doi: 10.1002/jmri.27875
82. Wu H, Zhang S, Liang C, Liu H, Liu Y, Mei Y, et al. Intravoxel Incoherent Motion MRI for the Differentiation of Benign, Intermediate, and Malignant Solid Soft-Tissue Tumors. J Magn Reson Imaging (2017) 46(6):1611–8. doi: 10.1002/jmri.25733
83. Nucifora PGP, Verma R, Lee SK, Melhem ER. Diffusion-Tensor MR Imaging and Tractography: Exploring Brain Microstructure and Connectivity1. Radiology (2007) 245(2):367–84. doi: 10.1148/radiol.2452060445
84. Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional Kurtosis Imaging: The Quantification of Non-Gaussian Water Diffusion by Means of Magnetic Resonance Imaging. Mag Reson Med (2005) 53(6):1432–40. doi: 10.1002/mrm.20508
85. Schmidt M, Kasprian G, Amann G, Duscher D, Aszmann OC. Diffusion Tensor Tractography for the Surgical Management of Peripheral Nerve Sheath Tumors. Neurosurg Focus (2015) 39(3):E17. doi: 10.3171/2015.6.FOCUS15228
86. Mazal AT, Ashikyan O, Cheng J, Le LQ, Chhabra A. Diffusion-Weighted Imaging and Diffusion Tensor Imaging as Adjuncts to Conventional MRI for the Diagnosis and Management of Peripheral Nerve Sheath Tumors: Current Perspectives and Future Directions. Eur Radiol (2019) 29(8):4123–32. doi: 10.1007/s00330-018-5838-8
87. Chhabra A, Thakkar RS, Andreisek G, Chalian M, Belzberg AJ, Blakeley J, et al. Anatomic MR Imaging and Functional Diffusion Tensor Imaging of Peripheral Nerve Tumors and Tumorlike Conditions. Am J Neuroradiol (2013) 34(4):802–7. doi: 10.3174/ajnr.A3316
88. Steven AJ, Zhuo J, Melhem ER. Diffusion Kurtosis Imaging: An Emerging Technique for Evaluating the Microstructural Environment of the Brain. AJR Am J Roentgenol (2014) 202(1):W26–33. doi: 10.2214/AJR.13.11365
89. Hu P, Zhang S, Zhou Z. The Value of Bi-Exponential and non-Gaussian Distribution Diffusion-Weighted Imaging in the Differentiation of Recurrent Soft Tissue Neoplasms and Post-Surgical Changes. Ann Transl Med (2020) 8(21):1357–7. doi: 10.21037/atm-20-2025
90. Mayr NA, Yuh WTC, Magnotta VA, Ehrhardt JC, Wheeler JA, Sorosky JI, et al. Tumor Perfusion Studies Using Fast Magnetic Resonance Imaging Technique in Advanced Cervical Cancer: A New Noninvasive Predictive Assay. Int J Radiat Oncol Biol Phys (1996) 36(3):623–33. doi: 10.1016/S0360-3016(97)85090-0
91. Yankeelov TE, Gore JC. Dynamic Contrast Enhanced Magnetic Resonance Imaging in Oncology: Theory, Data Acquisition, Analysis, and Examples. Curr Med Imaging Rev (2009) 3(2):91. doi: 10.2174/157340507780619179
92. Preziosi P, Orlacchio A, Di Giambattista G, Di Renzi P, Bartolotti L, Fabiano A, et al. Enhancement Patterns of Prostate Cancer in Dynamic MRI. Eur Radiol (2003) 13(5):925–30. doi: 10.1007/s00330-002-1703-9
93. Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, et al. The Assessment of Antiangiogenic and Antivascular Therapies in Early-Stage Clinical Trials Using Magnetic Resonance Imaging: Issues and Recommendations. Br J Cancer (2005) 92(9):1599–610. doi: 10.1038/sj.bjc.6602550
94. Kuhl CK, Mielcareck P, Klaschik S, Leutner C, Wardelmann E, Gieseke J, et al. Dynamic Breast MR Imaging: Are Signal Intensity Time Course Data Useful for Differential Diagnosis of Enhancing Lesions?1. Radiology (1999) 211(1):101–10. doi: 10.1148/radiology.211.1.r99ap38101
95. Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell (2000) 100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9
96. Fang S, Yang Y, Xu N, Tu Y, Yin Z, Zhang Y, et al. An Update in Imaging Evaluation of Histopathological Grade of Soft Tissue Sarcomas Using Structural and Quantitative Imaging and Radiomics. J Magn Reson Imaging (2022) 55(5):1357–75. doi: 10.1002/jmri.27954
97. O’Connor JPB, Jackson A, Parker GJM, Roberts C, Jayson GC. Dynamic Contrast-Enhanced MRI in Clinical Trials of Antivascular Therapies. Nat Rev Clin Oncol (2012) 9(3):167–77. doi: 10.1038/nrclinonc.2012.2
98. Wang X, Jacobs M, Fayad L. Therapeutic Response in Musculoskeletal Soft Tissue Sarcomas: Evaluation by Magnetic Resonance Imaging. NMR Biomed (2011) 24(6):750. doi: 10.1002/nbm.1731
99. Gordon Y, Partovi S, Müller-Eschner M, Amarteifio E, Bäuerle T, Weber M-A, et al. Dynamic Contrast-Enhanced Magnetic Resonance Imaging: Fundamentals and Application to the Evaluation of the Peripheral Perfusion. Cardiovasc Diagn Ther (2014) 4(2):147. doi: 10.3978/j.issn2223-3652.2014.03.01
100. Lavini C, Buiter MS, Maas M. Reports in Medical Imaging Use of Dynamic Contrast Enhanced Time Intensity Curve Shape Analysis in MRI: Theory and Practice. Rep Med Imaging (2013), 6:71–82. doi: 10.2147/RMI.S35088
101. Li X, Wang Q, Dou Y, Zhang Y, Tao J, Yang L, et al. Soft Tissue Sarcoma: Can Dynamic Contrast-Enhanced (DCE) MRI be Used to Predict the Histological Grade? Skeletal Radiol (2020) 49(11):1829–38. doi: 10.1007/s00256-020-03491-z
102. Patni RS, Boruah DK, Sanyal S, Gogoi BB, Patni M, Khandelia R, et al. Characterisation of Musculoskeletal Tumours by Multivoxel Proton MR Spectroscopy. Skeletal Radiol (2017) 46(4):483–95. doi: 10.1007/s00256-017-2573-1
103. Wang CK, Li CW, Hsieh TJ, Chien SH, Liu GC, Tsai KB. Characterization of Bone and Soft-Tissue Tumors With in Vivo 1h MR Spectroscopy: Initial Results1. Radiology (2004) 232(2):599–605. doi: 10.1148/radiol.2322031441
104. Zhang Q-W, Gao Y-J, Zhang R-Y, Zhou X-X, Chen S-L, Zhang Y, et al. Personalized CT-Based Radiomics Nomogram Preoperative Predicting Ki-67 Expression in Gastrointestinal Stromal Tumors: A Multicenter Development and Validation Cohort. Clin Trans Med (2020) 9:12. doi: 10.1186/s40169-020-0263-4
105. Navarro F, Dapper H, Asadpour R, Knebel C, Spraker MB, Schwarze V, et al. Development and External Validation of Deep-Learning-Based Tumor Grading Models in Soft-Tissue Sarcoma Patients Using Mr Imaging. Cancers (Basel) (2021) 13(12):2866. doi: 10.3390/cancers13122866
106. Nalepa J, Szymanek J, Hayball MP, Brown SJ, Ganeshan B, Miles K. Texture Analysis for Identifying Heterogeneity in Medical Images. In: Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) Chem, Switzerland:Springer, vol. 8671. (2014). p. 446–53.
107. Larroza A, Bodí V, Moratal D. Texture Analysis in Magnetic Resonance Imaging: Review and Considerations for Future Applications. In: Assessment of Cellular and Organ Function and Dysfunction Using Direct and Derived MRI Methodologies. London:InTech (2016).
108. Meyer HJ, Renatus K, Höhn AK, Hamerla G, Schopow N, Fakler J, et al. Texture Analysis Parameters Derived From T1-And T2-Weighted Magnetic Resonance Images can Reflect Ki67 Index in Soft Tissue Sarcoma. Surg Oncol (2019) 30:92–7. doi: 10.1016/j.suronc.2019.06.006
109. Hopper KD. Percutaneous, Radiographically Guided Biopsy: A History. Radiology (1995) 196:329–33. doi: 10.1148/radiology.196.2.7617841
110. Padhani AR, Lecouvet FE, Tunariu N, Koh DM, De Keyzer F, Collins DJ, et al. Rationale for Modernising Imaging in Advanced Prostate Cancer. Eur Urol Focus (2017) 3:223–39. doi: 10.1016/j.euf.2016.06.018
111. Wang Z, Shi H, Zhang X, Pan J, Jin Z. Value of CT-Guided Percutaneous Needle Biopsy of Bone in the Diagnosis of Lymphomas Based on PET/CT Results. Cancer Imaging (2019) 19(1):42. doi: 10.1186/s40644-019-0230-8
112. Rozenblum L, Mokrane FZ, Yeh R, Sinigaglia M, Besson FL, Seban RD, et al. Imaging-Guided Precision Medicine in Non-Resectable Gastro-Entero-Pancreatic Neuroendocrine Tumors: A Step-by-Step Approach. Eur J Radiol (2020) 122:108743. doi: 10.1016/j.ejrad.2019.108743
113. Anaya DA, Lahat G, Wang X, Xiao L, Tuvin D, Pisters PW, et al. Establishing Prognosis in Retroperitoneal Sarcoma: A New Histology-Based Paradigm. Ann Surg Oncol (2009) 16(3):667–75. doi: 10.1245/s10434-008-0250-2
114. Neuville A, Chibon F, Coindre JM. Grading of Soft Tissue Sarcomas: From Histological to Molecular Assessment. Pathology (2014) 46(2):113–20. doi: 10.1097/PAT.0000000000000048
115. Beer L, Martin-Gonzalez P, Delgado-Ortet M, Reinius M, Rundo L, Woitek R, et al. Ultrasound-Guided Targeted Biopsies of CT-Based Radiomic Tumour Habitats: Technical Development and Initial Experience in Metastatic Ovarian Cancer. Eur Radiol (2021) 31(6):3765. doi: 10.1007/s00330-020-07560-8
116. Burns J, Wilding CP L, Jones R, H Huang P. Proteomic Research in Sarcomas – Current Status and Future Opportunities. Semin Cancer Biol (2020) 61:56–70. doi: 10.1016/j.semcancer.2019.11.003
117. Noujaim J, Payne LS, Judson I, Jones RL, Huang PH. Phosphoproteomics in Translational Research: A Sarcoma Perspective. Ann Oncol (2016) 27(5):787–94. doi: 10.1093/annonc/mdw030
118. Petitprez F, de Reyniès A, Chen TW-W, Sun C-M, Lacroix L, Adam J, et al. Immune Classification of Soft Tissue Sarcoma and Its Association With Molecular Characteristics, and Clinical Outcome. Ann Oncol (2018) 29:vi35. doi: 10.1093/annonc/mdy319.001
119. Sorbye SW, Kilvaer T, Valkov A, Donnem T, Smeland E, Al-Shibli K, et al. Prognostic Impact of Lymphocytes in Soft Tissue Sarcomas. PLos One (2011) 6(1):e14611. doi: 10.1371/journal.pone.0014611
120. Petitprez F, de Reyniès A, Keung EZ, Chen TWW, Sun CM, Calderaro J, et al. B Cells are Associated With Survival and Immunotherapy Response in Sarcoma. Nature (2020) 577(7791):556–60. doi: 10.1038/s41586-019-1906-8
121. Raj S, Miller LD, Triozzi PL. Addressing the Adult Soft Tissue Sarcoma Microenvironment With Intratumoral Immunotherapy. Sarcoma (2018) 2018:10. doi: 10.1155/2018/9305294
122. Dufresne A, Lesluyes T, Ménétrier-Caux C, Brahmi M, Darbo E, Toulmonde M, et al. Specific Immune Landscapes and Immune Checkpoint Expressions in Histotypes and Molecular Subtypes of Sarcoma. OncoImmunology (2020) 9(1):1792036. doi: 10.1080/2162402X.2020.1792036
123. Callegaro D, Miceli R, Bonvalot S, Ferguson PC, Strauss DC, van Praag VVM, et al. Development and External Validation of a Dynamic Prognostic Nomogram for Primary Extremity Soft Tissue Sarcoma Survivors. EClinicalMedicine (2019) 17:100215. doi: 10.1016/j.eclinm.2019.11.008
124. van Praag VM, Rueten-Budde AJ, Jeys LM, Laitinen M, Pollock R, Aston W, et al. A Prediction Model for Treatment Decisions in High-Grade Extremity Soft-Tissue Sarcomas: Personalised Sarcoma Care (PERSARC). Eur J Cancer (2017) :83:313–23. doi: 10.1016/j.ejca.2017.06.032
125. Gennaro N, Reijers S, Bruining A, Messiou C, Haas R, Colombo P, et al. Imaging Response Evaluation After Neoadjuvant Treatment in Soft Tissue Sarcomas: Where do We Stand? Crit Rev Oncol Hematol (2021) 160:103309. doi: 10.1016/j.critrevonc.2021.103309
126. Haas RL. Preoperative Radiotherapy in Soft Tissue Sarcoma: From General Guidelines to Personalized Medicine. Chin Clin Oncol (2018) 7(4):41. doi: 10.21037/cco.2018.05.02
127. Gingrich AA, Bateni SB, Monjazeb AM, Darrow MA, Thorpe SW, Kirane AR, et al. Neoadjuvant Radiotherapy Is Associated With R0 Resection and Improved Survival for Patients With Extremity Soft Tissue Sarcoma Undergoing Surgery: A National Cancer Database Analysis. Ann Surg Oncol (2017) 24(11):3252–63. doi: 10.1245/s10434-017-6019-8
128. Telarovic I, Wenger RH, Pruschy M. Interfering With Tumor Hypoxia for Radiotherapy Optimization. J Exp Clin Cancer Res (2021) 40(1):1–26. doi: 10.1186/s13046-021-02000-x
129. Gray LH, Conger AD, Ebert M, Hornsey S, Scott OCA. The Concentration of Oxygen Dissolved in Tissues at the Time of Irradiation as a Factor in Radiotherapy. Br J Radiol (2014) 26(312):638–48. doi: 10.1259/0007-1285-26-312-638
130. Pasquali S, Gronchi A. Neoadjuvant Chemotherapy in Soft Tissue Sarcomas: Latest Evidence and Clinical Implications. Ther Adv Med Oncol (2017) 9:415–29. doi: 10.1177/1758834017705588
131. Italiano A, Mathoulin-Pelissier S, Le Cesne A, Terrier P, Bonvalot S, Collin F, et al. Trends in Survival for Patients With Metastatic Soft-Tissue Sarcoma. Cancer (2011) 117(5):1049–54. doi: 10.1002/cncr.25538
132. Esnaola NF, Rubin BP, Baldini EH, Vasudevan N, Demetri GD, M Fletcher CD, et al. Response to Chemotherapy and Predictors of Survival in Adult Rhabdomyosarcoma. Annals of Surgery (2001) 234(2):215–23. doi: 10.1097/00000658-200108000-00012
133. Rizzoli O, Bologna I, Gronchi A, Ferrari S, Quagliuolo V, Broto JM, et al. Histotype-Tailored Neoadjuvant Chemotherapy Versus Standard Chemotherapy in Patients With High-Risk Soft-Tissue Sarcomas (ISG-STS 1001): An International, Open-Label, Randomised, Controlled, Phase 3, Multicentre Trial. Artic Lancet Oncol (2017) 18:812–34. doi: 10.1016/S1470-2045(17)30334-0
134. Haussmann J, Matuschek C, Bölke E, Tamaskovics B, Corradini S, Wessalowski R, et al. Comparison of Different Systemic Therapeutic Regimes in Resectable Soft-Tissue Sarcoma-Results of a Network Meta-Analysis. Cancers (Basel) (2021) 13(22):5631. doi: 10.3390/cancers13225631
135. Verma S, Younus J, Stys-Norman D, Haynes AE, Blackstein M. Meta-Analysis of Ifosfamide-Based Combination Chemotherapy in Advanced Soft Tissue Sarcoma. Cancer Treat Rev (2008) 34(4):339–47. doi: 10.1016/j.ctrv.2008.01.005
136. Grobmyer SR, Maki RG, Demetri GD, Mazumdar M, Riedel E, Brennan MF, et al. Neo-Adjuvant Chemotherapy for Primary High-Grade Extremity Soft Tissue Sarcoma. Ann Oncol (2004) 15(11):1667–72. doi: 10.1093/annonc/mdh431
137. Italiano A, Penel N, Robin YM, Bui B, Le Cesne A, Piperno-Neumann S, et al. Neo/adjuvant Chemotherapy Does Not Improve Outcome in Resected Primary Synovial Sarcoma: A Study of the French Sarcoma Group. Ann Oncol (2009) 20(3):425–30. doi: 10.1093/annonc/mdn678
138. Gortzak E, Azzarelli A, Buesa J, Bramwell VH, Coevorden Fv, Geel Av, et al. A Randomised Phase II Study on Neo-Adjuvant Chemotherapy for ‘High-Risk’ Adult Soft-Tissue Sarcoma. Eur J Cancer (2001) 37(9):1096–103. doi: 10.1016/S0959-8049(01)00083-1
139. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur J Cancer (2009) 45(2):228–47. doi: 10.1016/j.ejca.2008.10.026
140. Schwartz LH, Litière S, De Vries E, Ford R, Gwyther S, Mandrekar S, et al. RECIST 1.1—Update and Clarification: From the RECIST Committee. Eur J Cancer (2016) 62:132–7. doi: 10.1016/j.ejca.2016.03.081
141. Canter RJ, Martinez SR, Tamurian RM, Wilton M, Li CS, Ryu J, et al. Radiographic and Histologic Response to Neoadjuvant Radiotherapy in Patients With Soft Tissue Sarcoma. Ann Surg Oncol (2010) 17(10):2578–84. doi: 10.1245/s10434-010-1156-3
142. Roberge D, Skamene T, Nahal A, Turcotte RE, Powell T, Freeman C. Radiological and Pathological Response Following Pre-Operative Radiotherapy for Soft-Tissue Sarcoma. Radiother Oncol (2010) 97(3):404–7. doi: 10.1016/j.radonc.2010.10.007
143. Abu-Hijlih R. Correlation of Radiologic and Pathologic Response in Patients Receiving Neoadjuvant Radiotherapy for Soft Tissue Sarcoma. BioMed J Sci Tech Res (2018) 9(2):6973–7. doi: 10.26717/BJSTR.2018.09.001765
144. Bonvalot S, Wunder J, Gronchi A, Broto JM, Turcotte R, Rastrelli M, et al. Complete Pathological Response to Neoadjuvant Treatment Is Associated With Better Survival Outcomes in Patients With Soft Tissue Sarcoma: Results of a Retrospective Multicenter Study. Eur J Surg Oncol (2021) 47(8):2166–72. doi: 10.1016/j.ejso.2021.02.024
145. Eilber FC, Rosen G, Eckardt J, Forscher C, Nelson SD, Selch M, et al. Treatment-Induced Pathologic Necrosis: A Predictor of Local Recurrence and Survival in Patients Receiving Neoadjuvant Therapy for High-Grade Extremity Soft Tissue Sarcomas. J Clin Oncol (2001) 19(13):3203–9. doi: 10.1200/JCO.2001.19.13.3203
146. Wardelmann E, Haas RL, Bovée JVMG, Terrier P, Lazar A, Messiou C, et al. Evaluation of Response After Neoadjuvant Treatment in Soft Tissue Sarcomas; The European Organization for Research and Treatment of Cancer-Soft Tissue and Bone Sarcoma Group (EORTC-STBSG) Recommendations for Pathological Examination and Reporting. Eur J Cancer (2016) 53:84–95. doi: 10.1016/j.ejca.2015.09.021
147. Hong NJL, Hornicek FJ, Harmon DC, Choy E, Chen YL, Yoon SS, et al. Neoadjuvant Chemoradiotherapy for Patients With High-Risk Extremity and Truncal Sarcomas: A 10-Year Single Institution Retrospective Study. Eur J Cancer (2013) 49(4):875–83. doi: 10.1016/j.ejca.2012.10.002
148. Pisters PWT, Patel SR, Varma DGK, Cheng SC, Chen NP, Nguyen HTB, et al. Preoperative Chemotherapy for Stage IIIB Extremity Soft Tissue Sarcoma: Long-Term Results From a Single Institution. J Clin Oncol (2016) 15(12):3481–7. doi: 10.1200/JCO.1997.15.12.3481
149. DeLaney TF, Spiro IJ, Suit HD, Gebhardt MC, Hornicek FJ, Mankin HJ, et al. Neoadjuvant Chemotherapy and Radiotherapy for Large Extremity Soft-Tissue Sarcomas. Int J Radiat Oncol Biol Phys (2003) 56(4):1117–27. doi: 10.1016/S0360-3016(03)00186-X
150. Miki Y, Ngan S, Clark JCM, Akiyama T, Choong PFM. The Significance of Size Change of Soft Tissue Sarcoma During Preoperative Radiotherapy. Eur J Surg Oncol (2010) 36(7):678–83. doi: 10.1016/j.ejso.2010.05.021
151. Choi H, Charnsangavej C, Faria SC, Macapinlac HA, Burgess MA, Patel SR, et al. Correlation of Computed Tomography and Positron Emission Tomography in Patients With Metastatic Gastrointestinal Stromal Tumor Treated at a Single Institution With Imatinib Mesylate: Proposal of New Computed Tomography Response Criteria. J Clin Oncol (2007) 25:1753–9. doi: 10.1200/JCO.2006.07.3049
152. Benjamin RS, Choi H, Macapinlac HA, Burgess MA, Patel SR, Chen LL, et al. We Should Desist Using RECIST, at Least in GIST. J Clin Oncol (2007) 25(13):1760–4. doi: 10.1200/JCO.2006.07.3411
153. Stacchiotti S, Collini P, Messina A, Morosi C, Barisella M, Bertulli R, et al. High-Grade Soft-Tissue Sarcomas: Tumor Response Assessment–Pilot Study to Assess the Correlation Between Radiologic and Pathologic Response by Using RECIST and Choi Criteria. Radiology (2009) 251(2):447–56. doi: 10.1148/radiol.2512081403
154. Stacchiotti S, Verderio P, Messina A, Morosi C, Collini P, Llombart-Bosch A, et al. Tumor Response Assessment by Modified Choi Criteria in Localized High-Risk Soft Tissue Sarcoma Treated With Chemotherapy. Cancer (2012) 118(23):5857–66. doi: 10.1002/cncr.27624
155. Eastley NC, Ottolini B, Neumann R, Luo JL, Hastings RK, Khan I, et al. Circulating Tumour-Derived DNA in Metastatic Soft Tissue Sarcoma. Oncotarget (2018) 9(12):10549. doi: 10.18632/oncotarget.24278
156. Benz MR, Czernin J, Allen-Auerbach MS, Tap WD, Dry SM, Elashoff D, et al. FDG-PET/CT Imaging Predicts Histopathologic Treatment Responses After the Initial Cycle of Neoadjuvant Chemotherapy in High-Grade Soft-Tissue Sarcomas. Clin Cancer Res (2009) 15(8):2856–63. doi: 10.1158/1078-0432.CCR-08-2537
157. Schuetze SM, Rubin BP, Vernon C, Hawkins DS, Bruckner JD, Conrad EU, et al. Use of Positron Emission Tomography in Localized Extremity Soft Tissue Sarcoma Treated With Neoadjuvant Chemotherapy. Cancer (2005) 103(2):339–48. doi: 10.1002/cncr.20769
158. Eary JF, Conrad EU, O’Sullivan J, Hawkins DS, Schuetze SM, O’Sullivan F. Sarcoma Mid-Therapy [F-18]Fluorodeoxyglucose Positron Emission Tomography (FDG PET) and Patient Outcome. J Bone Joint Surg Am (2014) 96(2):152. doi: 10.2106/JBJS.M.00062
159. Erfanian Y, Grueneisen J, Kirchner J, Wetter A, Podleska LE, Bauer S, et al. Integrated 18f–FDG PET/MRI Compared to MRI Alone for Identification of Local Recurrences of Soft Tissue Sarcomas: A Comparison Trial. Eur J Nucl Med Mol Imaging (2017) 44(11):1823–31. doi: 10.1007/s00259-017-3736-y
160. Huang H, Zhu H, Xie Q, Tian X, Yang X, Feng F, et al. Evaluation of 124I-JS001 for Hpd1 Immuno-PET Imaging Using Sarcoma Cell Homografts in Humanized Mice. Acta Pharm Sin B (2020) 10(7):1321–30. doi: 10.1016/j.apsb.2020.02.004
161. Rutman AM, Kuo MD. Radiogenomics: Creating a Link Between Molecular Diagnostics and Diagnostic Imaging. Eur J Radiol (2009) 70(2):232–41. doi: 10.1016/j.ejrad.2009.01.050
162. Einarsdóttir H, Karlsson M, Wejde J, Bauer HCF. Diffusion-Weighted MRI of Soft Tissue Tumours. Eur Radiol (2004) 14(6):959–63. doi: 10.1007/s00330-004-2237-0
163. Dudeck O, Zeile M, Pink D, Pech M, Tunn PU, Reichardt P, et al. Diffusion-Weighted Magnetic Resonance Imaging Allows Monitoring of Anticancer Treatment Effects in Patients With Soft-Tissue Sarcomas. J Magn Reson Imaging (2008) 27(5):1109–13. doi: 10.1002/jmri.21358
164. Spinnato P, Kind M, Le Loarer F, Bianchi G, Colangeli M, Sambri A, et al. Soft Tissue Sarcomas: The Role of Quantitative MRI in Treatment Response Evaluation. Acad Radiol (2021) 29(7)1065–84. doi: 10.1016/j.acra.2021.08.007
165. Alic L, van Vliet M, Wielopolski PA, ten Hagen TLM, van Dijke CF, Niessen WJ, et al. Regional Heterogeneity Changes in DCE-MRI as Response to Isolated Limb Perfusion in Experimental Soft-Tissue Sarcomas. Contrast Media Mol Imaging (2013) 8(4):340–9. doi: 10.1002/cmmi.1528
166. Spratt DE, Arevalo-Perez J, Leeman JE, Gerber NK, Folkert M, Taunk NK, et al. Early Magnetic Resonance Imaging Biomarkers to Predict Local Control After High Dose Stereotactic Body Radiotherapy for Patients With Sarcoma Spine Metastases. Spine J (2016) 16(3):291–8. doi: 10.1016/j.spinee.2015.08.041
167. Huang W, Beckett BR, Tudorica A, Meyer JM, Afzal A, Chen Y, et al. Evaluation of Soft Tissue Sarcoma Response to Preoperative Chemoradiotherapy Using Dynamic Contrast-Enhanced Magnetic Resonance Imaging. Tomography (2016) 2(4):308–16. doi: 10.18383/j.tom.2016.00202
168. Xia W, Yan Z, Gao X. Volume Fractions of DCE-MRI Parameter as Early Predictor of Histologic Response in Soft Tissue Sarcoma: A Feasibility Study. Eur J Radiol (2017) 95:228–35. doi: 10.1016/j.ejrad.2017.08.021
169. Kousi E, Messiou C, Miah A, Orton M, Haas R, Thway K, et al. Descriptive Analysis of MRI Functional Changes Occurring During Reduced Dose Radiotherapy for Myxoid Liposarcomas. Br J Radiol (2021) 94(1126):20210310. doi: 10.1259/bjr.20210310
170. Nordsmark M, Alsner J, Keller J, Nielsen OS, Jensen OM, Horsman MR, et al. Hypoxia in Human Soft Tissue Sarcomas: Adverse Impact on Survival and No Association With P53 Mutations. Br J Cancer (2001) 84(8):1070–5. doi: 10.1054/bjoc.2001.1728
171. Nyström H, Jönsson M, Werner-Hartman L, Nilbert M, Carneiro A. Hypoxia-Inducible Factor 1α Predicts Recurrence in High-Grade Soft Tissue Sarcoma of Extremities and Trunk Wall. J Clin Pathol (2017) 70(10):879–85. doi: 10.1136/jclinpath-2016-204149
172. Smeland E, Kilvaer TK, Sorbye S, Valkov A, Andersen S, Bremnes RM, et al. Prognostic Impacts of Hypoxic Markers in Soft Tissue Sarcoma. Sarcoma (2012) 2012:10. doi: 10.1155/2012/541650
173. Connor O, Robinson JP, Waterton SP. Imaging Tumour Hypoxia With Oxygen-Enhanced MRI and BOLD MRI. Br J Radiol (2019) 92:20180642. doi: 10.1259/bjr.20180642
174. O’Connor JPB, Boult JKR, Jamin Y, Babur M, Finegan KG, Williams KJ, et al. Oxygen-Enhanced MRI Accurately Identifies, Quantifies, and Maps Tumor Hypoxia in Preclinical Cancer Models. Cancer Res (2016) 76(4):787–95. doi: 10.1158/0008-5472.CAN-15-2062
175. Salem A, Little RA, Latif A, Featherstone AK, Babur M, Peset I, et al. Oxygen-Enhanced MRI is Feasible, Repeatable, and Detects Radiotherapy-Induced Change in Hypoxia in Xenograft Models and in Patients With Non-Small Cell Lung Cancer. Clin Cancer Res (2019) 25(13):3818–29. doi: 10.1158/1078-0432.CCR-18-3932
176. Rodrigues LM, Howe FA, Griffiths JR, Robinson SP. Tumor R2* is a Prognostic Indicator of Acute Radiotherapeutic Response in Rodent Tumors. J Magn Reson Imaging (2004) 19(4):482–8. doi: 10.1002/jmri.20024
177. Zhou H, Hallac RR, Yuan Q, Ding Y, Zhang Z, Xie X-J, et al. Incorporating Oxygen-Enhanced MRI Into Multi-Parametric Assessment of Human Prostate Cancer. Diagnostics (2017) 7(3):48. doi: 10.3390/diagnostics7030048
178. Fan Q, Tang CY, Gu D, Zhu J, Li G, Wu Y, et al. Investigation of Hypoxia Conditions Using Oxygen-Enhanced Magnetic Resonance Imaging Measurements in Glioma Models. Oncotarget (2017) 8(19):31864. doi: 10.18632/oncotarget.16256
179. Ohno Y, Hatabu H, Takenaka D, Adachi S, Van Cauteren M, Sugimura K. Oxygen-Enhanced MR Ventilation Imaging of the Lung Prelimary Clinical Experience in 25 Subjects. Am J Roentgenol (2001) 177(1):185–94. doi: 10.2214/ajr.177.1.1770185
180. Cao-Pham TT, Tran LBA, Colliez F, Joudiou N, El Bachiri S, Grégoire V, et al. Monitoring Tumor Response to Carbogen Breathing by Oxygen-Sensitive Magnetic Resonance Parameters to Predict the Outcome of Radiation Therapy: A Preclinical Study. Int J Radiat Oncol (2016) 96(1):149–60. doi: 10.1016/j.ijrobp.2016.04.029
181. Manduca A, Bayly PJ, Ehman RL, Kolipaka A, Royston TJ, Sack I, et al. MR Elastography: Principles, Guidelines, and Terminology. Magn Reson Med (2021) 85:2377–90. doi: 10.1002/mrm.28627
182. Chen J, Yin M, Glaser KJ, Talwalkar JA, Ehman RL. MR Elastography of Liver Disease: State of the Art. Appl Radiol (2013) 42(4):5–12. doi: 10.37549/AR1982
183. Hoodeshenas S, Yin M, Venkatesh SK. Magnetic Resonance Elastography of Liver Current Update. Top Magn Reson Imaging (2018) 27:319–33. doi: 10.1097/RMR.0000000000000177
184. Pepin KM, McGee KP. Quantifying Tumor Stiffness With Magnetic Resonance Elastography the Role of Mechanical Properties for Detection, Characterization, and Treatment Stratification in Oncology. Top Magn Reson Imaging (2018) 27(5):353–62. doi: 10.1097/RMR.0000000000000181
185. Jugé L, Doan BT, Seguin J, Albuquerque M, Larrat B, Mignet N, et al. Colon Tumor Growth and Antivascular Treatment in Mice: Complementary Assessment With MR Elastography and Diffusion-Weighted MR Imaging. Radiology (2012) 264(2):436–44. doi: 10.1148/radiol.12111548
186. Jamin Y, Boult JKR, Li J, Popov S, Garteiser P, Ulloa JL, et al. Integrated Systems and Technologies Exploring the Biomechanical Properties of Brain Malignancies and Their Pathologic Determinants In Vivo With Magnetic Resonance Elastography. Cancer Res (2015) 75(7):1216–24. doi: 10.1158/0008-5472.CAN-14-1997
187. Pepin K, Grimm R, Kargar S, Howe BM, Fritchie K, Frick M, et al. Soft Tissue Sarcoma Stiffness and Perfusion Evaluation by MRE and DCE-MRI for Radiation Therapy Response Assessment: A Technical Feasibility Study. BioMed Phys Eng Express (2019) 5(4):1–18. doi: 10.1088/2057-1976/ab2175
188. Peeken JC, Spraker MB, Knebel C, Dapper H, Pfeiffer D, Devecka M, et al. Tumor Grading of Soft Tissue Sarcomas Using MRI-Based Radiomics. EBioMedicine (2019) 48:332–40. doi: 10.1016/j.ebiom.2019.08.059
189. Yan R, Hao D, Li J, Liu J, Hou F, Chen H, et al. Magnetic Resonance Imaging-Based Radiomics Nomogram for Prediction of the Histopathological Grade of Soft Tissue Sarcomas: A Two-Center Study. J Magn Reson Imaging (2021) 53(6):1683–96. doi: 10.1002/jmri.27532
190. Gao Y, Kalbasi A, Hsu W, Ruan D, Fu J, Shao J, et al. Treatment Effect Prediction for Sarcoma Patients Treated With Preoperative Radiotherapy Using Radiomics Features From Longitudinal Diffusion-Weighted MRIs. Phys Med Biol (2020) 65(17):175006. doi: 10.1088/1361-6560/ab9e58
191. Carano RAD, Ross AL, Ross J, Williams SP, Koeppen H, Schwall RH, et al. Quantification of Tumor Tissue Populations by Multispectral Analysis. Magn Reson Med (2004) 51(3):542–51. doi: 10.1002/mrm.10731
192. Henning EC, Azuma C, Sotak CH, Helmer KG. Multispectral Quantification of Tissue Types in a RIF-1 Tumor Model With Histological Validation. Part I. Magn Reson Med (2007) 57(3):501–12. doi: 10.1002/mrm.21161
193. Henning EC, Azuma C, Sotak CH, Helmer KG. Multispectral Tissue Characterization in a RIF-1 Tumor Model: Monitoring the ADC and T2 Responses to Single-Dose Radiotherapy. Part Ii. Magn Reson Med (2007) 57(3):513–9. doi: 10.1002/mrm.21178
194. Berry LR, Barck KH, Go MA, Ross J, Wu X, Williams SP, et al. Quantification of Viable Tumor Microvascular Characteristics by Multispectral Analysis. Magn Reson Med (2008) 60(1):64–72. doi: 10.1002/mrm.21470
195. Barck KH, Willis B, Ross J, French DM, Filvaroff EH, Carano RAD. Viable Tumor Tissue Detection in Murine Metastatic Breast Cancer by Whole-Body MRI and Multispectral Analysis. Magn Reson Med (2009) 62(6):1423–30. doi: 10.1002/mrm.22109
196. Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology (2020) 295(2):328–38. doi: 10.1148/radiol.2020191145
197. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, De Jong EEC, Van Timmeren J, et al. Radiomics: The Bridge Between Medical Imaging and Personalized Medicine. Nat Rev Clin Oncol (2017) 14(12):749–62. doi: 10.1038/nrclinonc.2017.141
198. Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J Radiol (2019) 20(7):1124. doi: 10.3348/kjr.2018.0070
199. Quantitative Imaging Network (QIN) | CIP Grant-supported Networks | Programs & Resources | Cancer Imaging Program (CIP). About the Quantitative Imaging Network (QIN) (2022). Available at: https://imaging.cancer.gov/programs_resources/specialized_initiatives/qin/about/default.htm#:~:text = The Quantitative Imaging Network %28QIN%29 promotes research%2C development,the overall goal of facilitating clinical decision making.
200. Kalpathy-Cramer J, Freymann JB, Kirby JS, Kinahan PE, Prior AFW. Quantitative Imaging Network: Data Sharing and Competitive Algorithm Validation Leveraging the Cancer Imaging Archive. Transl Oncol (2014) 7(1):147–52. doi: 10.1593/tlo.13862
201. Fedorov A, Beichel R, Kalpathy-Cramer J, Clunie D, Onken M, Riesmeier J, et al. Quantitative Imaging Informatics for Cancer Research. JCO Clin Cancer Inform (2020) 4(4):444–53. doi: 10.1200/CCI.19.00165
Keywords: sarcoma, MRI, radiomics, virtual biopsy, quantitative MRI (qMRI), radiology pathology correlation, soft tissue sarcoma (STS), imaging biomarker
Citation: Arthur A, Johnston EW, Winfield JM, Blackledge MD, Jones RL, Huang PH and Messiou C (2022) Virtual Biopsy in Soft Tissue Sarcoma. How Close Are We? Front. Oncol. 12:892620. doi: 10.3389/fonc.2022.892620
Received: 09 March 2022; Accepted: 31 May 2022;
Published: 01 July 2022.
Edited by:
Anastasia Constantinidou, University of Cyprus, CyprusReviewed by:
Chiara Giraudo, Università degli Studi di Padova, ItalyAmandine Crombe, Université de Bordeaux, France
Copyright © 2022 Arthur, Johnston, Winfield, Blackledge, Jones, Huang and Messiou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul H. Huang, UGF1bC5IdWFuZ0BpY3IuYWMudWs=; Christina Messiou, Q2hyaXN0aW5hLk1lc3Npb3VAcm1oLm5ocy51aw==
†These authors share senior authorship
 Amani Arthur
Amani Arthur Edward W. Johnston
Edward W. Johnston Jessica M. Winfield
Jessica M. Winfield Matthew D. Blackledge
Matthew D. Blackledge Robin L. Jones
Robin L. Jones Paul H. Huang
Paul H. Huang Christina Messiou1,2*†
Christina Messiou1,2*†