- 1National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, The First Affiliated Hospital of Soochow University, Suzhou, China
- 2Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou, China
- 3Key Laboratory of Cell Differentiation and Apoptosis of Ministry of Education, Department of Pathophysiology, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai, China
Acute myeloid leukemia (AML) is an aggressive hematological malignancy that recurs in approximately 50% of cases. Elevated homing and uncontrolled expansion are characteristics of AML cells. Here, we identified that Fifth Ewing Variant (FEV) regulates the homing and expansion of AML cells. We found that FEV was re-expressed in 30% of primary AML samples and in almost all relapsed AML samples, and FEV expression levels were significantly higher in relapsed samples compared to primary samples. Interference of FEV expression in AML cell lines delayed leukemic progression and suppressed homing and proliferation. Moreover, FEV directly activated integrin subunit alpha 4 (ITGA4) transcription in a dose-dependent manner. Inhibition of integrin α4 activity with natalizumab (NZM) reduced the migration and colony-forming abilities of blasts and leukemic-initiating cells (LICs) in both primary and relapsed AML. Thus, our study suggested that FEV maintains the homing and expansion of AML cells by activating ITGA4 transcription and that targeting ITGA4 inhibits the colony-forming and migration capacities of blasts and LICs. Thus, these findings suggested that the FEV-ITGA4 axis may be a therapeutic target for both primary and relapsed AML.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematological malignancy in which immature cells accumulate and expand uncontrollably. Nearly 50% of patients relapse after induction chemotherapy (1, 2). Although new approaches have improved the prognosis of some patients, the majority of primary and relapsed AML patients still lack effective treatment (1–4). Therefore, there is an urgent need for novel targets or drugs to improve the outcomes of these patients.
Enhanced homing and migration abilities are features of AML cells, which rapidly home to bone marrow (BM) and hijack the normal hematopoietic niche to aid extensive expansion of leukemic initiating cells (LICs) (5–7). Cell-to-cell or cell-to-matrix interactions mediated by C-X-C Motif Chemokine Receptor 4 (CXCR4)- C-X-C Motif Chemokine Ligand 12 (CXCL12), integrins and CD44 signaling pathways have been reported to contribute to the homing of leukemic cells to the BM microenvironment (5–11). However, the upstream mechanism that activates the pathways of VLA-4 remains unknown. Here, we identified that fifth Ewing variant (FEV) regulates integrin signaling.
FEV (also known as PET1 in mammals) is an E26 transformation-specific transcription factor (12, 13). FEV was initially reported to regulate the synthesis of 5-hydroxytryptamine (5-HT), and FEV deficiency results in the differentiation arrest of the majority of 5-HT–producing neurons and a 70–80% decrease in 5-HT (14, 15). In recent years, FEV has been reported to be a functional regulator in the generation and self-renewal of embryo hematopoietic stem cells (HSCs) (13, 16). In our previous work, we reported that FEV is silenced in normal adult hematopoiesis and re-expressed in leukemias of prenatal origin, and FEV deficiency significantly impairs the leukemia-propagating capacity of LICs in AML patient-derived xenograft mice (17). However, the mechanism of how FEV modulates the reconstitution of LICs remains unknown.
In the present study, we demonstrated that FEV was re-expressed in 30% of primary and in almost all relapsed AML samples with high expression levels, and we found that FEV maintains the homing and expansion abilities of AML cells by directly activating integrin subunit alpha 4 (ITGA4) transcription. Integrin α4 blockade inhibited the migration and colony-forming abilities of blasts and LICs in both primary and relapsed AML. Our results demonstrated the role of the FEV-ITGA4 axis in homing and expansion maintenance, providing a potential therapeutic target for primary and relapsed AML.
Materials and Methods
Human Samples
Human bone marrow (BM) aspirates of patients with leukemia from 2018 to 2020 were obtained from the First Affiliated Hospital of Soochow University, and the studies were approved by the Medical Ethical Committees of the hospital in accordance with the Declaration of Helsinki protocol. All human participants signed a written informed consent form. In total, 69 AML samples containing 16 relapsed AML samples were included. The detailed information for the clinical samples is shown in Supplementary Tables 1–3. Mononuclear cells (MNCs) from the samples were obtained using Ficoll (Sigma–Aldrich, St. Louis, MO, USA) gradient centrifugation and frozen in liquid nitrogen.
Xenograft Model
NOD-SCID mice (aged 6–8 weeks) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd., Shanghai, China. The mice were bred under pathogen-free conditions at the Laboratory Animal Center of Soochow University. All animal experiments were performed in accordance with the protocols approved by the Experimental Animal Ethical Committee at Soochow University. The NOD-SCID mice were irradiated with 2.0 Gy of X-rays and subsequently treated with CD122 antibody at a dose of 200 μg per mouse via intraperitoneal injection. The mice were injected with genetically modified MV4-11 cells at a dose of 1 × 105 through the caudal vein.
Real-Time Quantitative Polymerase Chain Reaction
Total RNA was extracted using a TRIzol reagent in accordance with the manufacturer’s instructions and reversely transcribed. RT-qPCR was also performed using TB Green Premix Ex Taq II (Takara Bio, Otsu, Japan) in accordance with the manufacturer’s instructions. All experiments were performed in triplicate with ABI QuantStudio 3 Real-time PCR System (Applied Biosystems, MA, USA). The primer sequences are summarised in Supplementary Table 4. Differences in cDNA input were normalized to the ACTB expression levels. FEV positivity was defined in accordance with a previous report (17).
Chromatin Immunoprecipitation
ChIP assays were performed using the Magna ChIP A/G kit (Merck Millipore, Billerica, MA, USA) in accordance with the manufacturer’s instructions. MV4-11 cells with ectopically expressed flag-FEV were cross-linked in 1% formaldehyde and then sonicated to create soluble chromatin. Antibodies against flag (Sigma–Aldrich, St. Louis, MO, USA) were added to precipitate the DNA fragments. The recovered DNA was amplified using PCR or quantitative PCR.
Homing Assay
Homing assays were performed as previously reported (18). In this assay, 2 × 106 transduced MV4-11 cells were injected into sublethally irradiated NOD-SCID mice by the caudal vein. 16 hours later, bone marrow (BM) cells of the recipients were stained with human CD45-PC7 and analysed using a flow cytometer (ACEA Biosciences, California, USA). All antibodies were obtained from BD Bioscience.
RNA Sequencing
MV4-11 cells transduced with NC or shFEV were cultured for 3 days and sorted by BD FACS Melody (BD Biosciences, San Jose, CA, USA). The cells were collected and high-throughput RNA sequencing (RNA-seq) was performed by Illumina HiSeq 2500 (Illumina, San Diego, CA) at CapitalBio Corporation (Beijing, China). Genes with a P value < 0.05 and fold changes ≥ 1.5 were recognized as differentially expressed genes between the two samples (Dataset 1). Pathway analysis (q value ≤ 0.001) of these differentially expressed genes was conducted using KOBAS 2.0, and the analysed results are provided in Supplementary Table 5.
Statistical Analysis
The Mann–Whitney U test was used to estimate the differences in FEV expression. The definition of FEV positivity was based on a previous report (17). The FEV positivity was analyzed using the chi-squared test. The survival of xenograft mice was assessed using the Kaplan–Meier method and the log-rank test. Statistical analysis was conducted using SPSS 25.0 software (SPSS Inc., Chicago, USA) and GraphPad Prism software (version 8.4.1, GraphPad Software, San Diego, CA).
Results
FEV Is Re-Expressed in Primary and Relapsed AML With High Expression Levels
To validate FEV expression in AML, BM aspirates from 53 primary AML patients were examined using reverse transcription-polymerase chain reaction (RT–PCR), and BM mononuclear cells (MNCs) and CD34+ cells from healthy donors were also examined as controls. According to the definition of FEV positivity from a previous report (17), FEV was negative in all normal BM MNCs (14/14) and CD34+ cells (7/7), and 30.2% (16/53) of primary AML samples were FEV positive (Figure 1A, Supplementary Table 1). Next, FEV expression was validated using the Vizome database (19) (www.vizome.org), which indicated that 37.0% (67/181) of primary AML samples were FEV positive (Figure 1B).
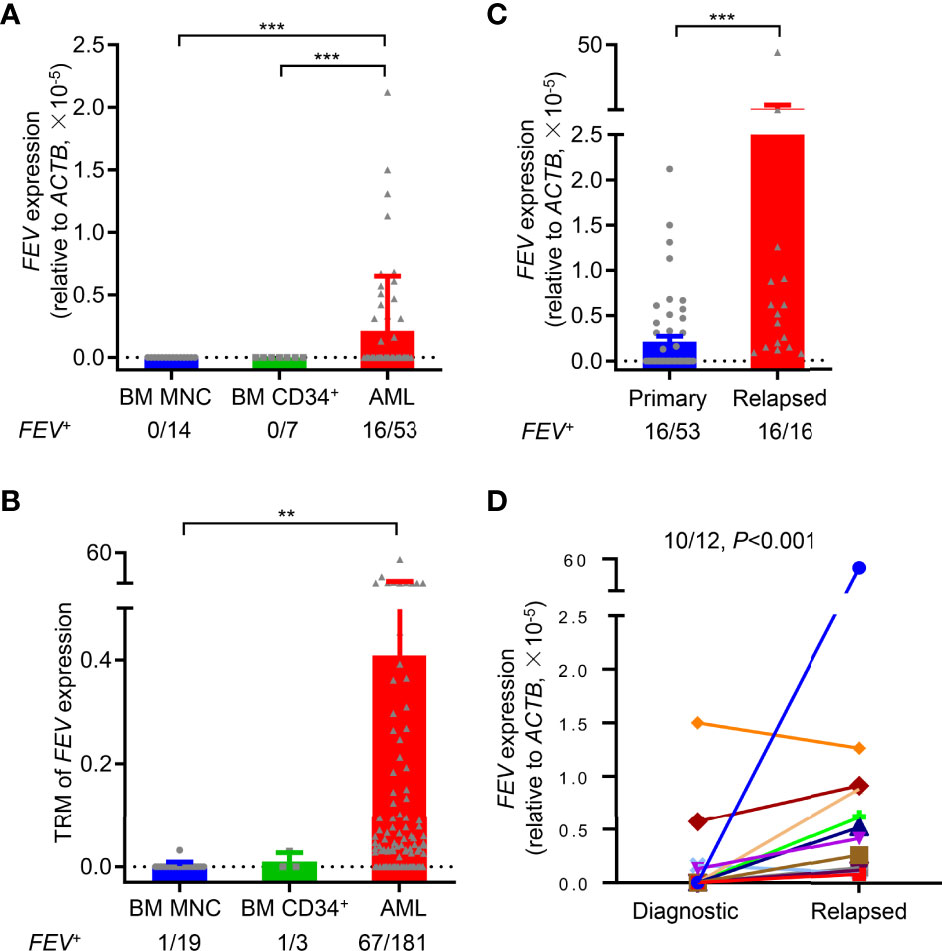
Figure 1 FEV is highly expressed in primary and relapsed AML. (A) FEV mRNA expression levels in primary AML samples, and BM MNCs and CD34+ cells from healthy donors were used as normal controls. (B) FEV mRNA expression levels were analyzed using the data from the Vizome database. (C) Comparison of FEV mRNA levels in samples from primary and relapsed AML. The proportion of FEV+ samples and relative mRNA level are indicated. Positivity for FEV expression was identified as previously described (17). (D) FEV mRNA expression levels in paired diagnostic and relapsed samples. FEV mRNA was significantly higher in 91.7% (11/12) of relapsed samples than in the diagnostic samples. **P < 0.01 and ***P < 0.001 (A–C), Mann–Whitney U test; (D), Wilcoxon test. All data are presented as the mean ± SD.
To explore whether FEV is preferentially expressed in AML patients, the clinical characteristics of the patients who were diagnosed and treated in our center were summarized. FEV was no preferentially expressed in age, sex, FAB subtypes, genetics (except for CEBPA) and cytogenetics. However, statistical analysis showed a worse risk category in FEV+ patients compared with the FEV- group (P=0.0017), according to the guideline of ELN risk stratification. Also, a significantly lower rate of complete remission was observed after two courses of induction chemotherapy in FEV+ group (12.5% vs. 94.6%, P<0.001, Supplementary Table 2). In addition, the FEV+ patients had unfavorable 2-year overall survival (OS) and relapse-free survival (RFS) than the FEV- patients (OS: 46.63% vs. 69.17%, P=0.020; RFS: 25.00% vs. 55.39%, P=0.030) (Supplementary Figures 1A, B). Therefore, we considered that FEV was preferentially expressed in aggressive and treatment-refractory subsets of AML.
BM samples from relapsed AML patients were also analyzed to further study FEV expression at different stages of AML progression, and their clinical characteristics were shown in Supplementary Table 3. All of the relapsed samples (16/16) were FEV positive, and the mRNA levels of FEV were significantly higher in relapsed samples than in primary samples (Figure 1C). However, 55.6% (5/9) of relapsed samples were FEV positive, and no significant difference in FEV expression was observed between primary and relapsed samples using data in the Vizome database, which may due to the small sample size (Supplementary Figure 1C). We next confirmed FEV expression in patients with paired diagnostic and relapsed samples. FEV expression was significantly higher in 83.3% (10/12) of relapsed samples than in the diagnostic samples (Figure 1D). These results indicated that FEV was re-expressed in 30% of primary AML samples and in almost all relapsed AML samples, and high FEV expression levels were determined in relapsed samples, suggesting that FEV may play a role in AML progression.
FEV Deficiency Inhibits the Colony-Forming Ability and Proliferation of AML Cells in Vitro
FEV expression in AML leukemic cell lines was examined by RT–PCR and immunoblotting (Supplementary Figures 2A, B), and FEV positively expression cell lines, MV4-11, THP-1 and KG-1 cells, were used to investigate the role of FEV in AML progression. FEV knockdown was achieved using lentiviral vector-driven interference shRNAs (shFEVs), and the most effective shRNAs (Supplementary Figure 2C) were selected as previously reported (17).
MV4-11 cells transduced with nonsilencing control (NC), FEV shRNA1 (sh1) or shRNA2 (sh2) were flow-sorted according to GFP positivity, and colony-forming cell (CFC) assay and cell proliferation assay were conducted. We found that FEV knockdown markedly reduced CFCs (Figure 2A). The cell count in the shFEV group was decreased after 2 days of culture (Figure 2B), which was confirmed by proliferation detection after 7 days of culture (Figure 2C). Reduced CFCs (Figures 2D, E) and inhibited proliferation (Figures 2F, G) were also observed in THP-1 or KG-1 cells transduced with shFEVs. The cell cycle status and apoptosis of AML cells were also evaluated. A slight increase in the frequency of G1 phase and a mild decrease in the frequency of S phase were observed in FEV-deficient cells, but no significant difference was found between the NC and shFEV (sh1) groups in terms of total Annexin V+ cells although a slight increase in the proportion of late apoptosis was observed. (Figures 2H–J), suggesting that FEV knockdown leads to proliferation inhibition with G1 arrest.
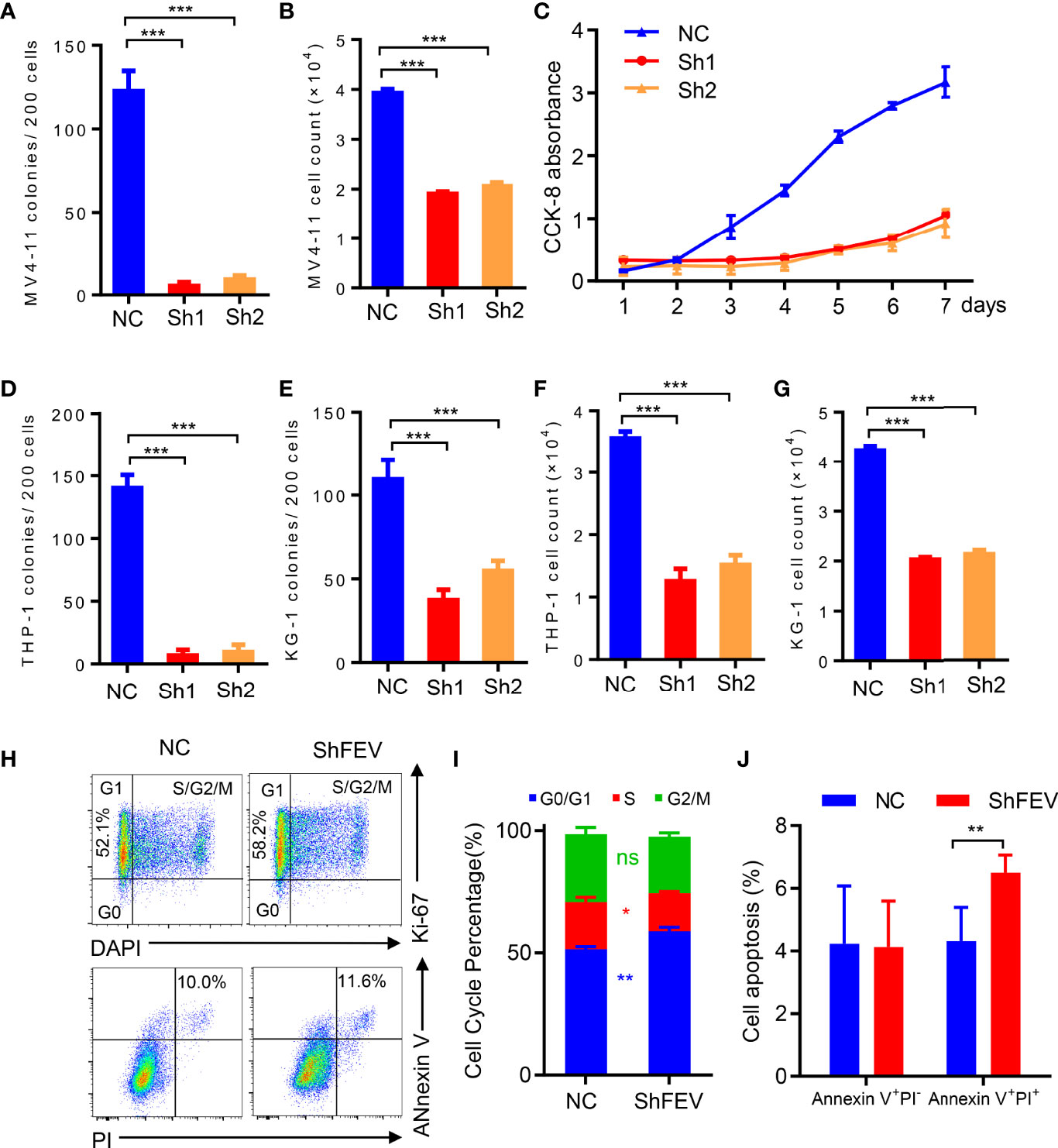
Figure 2 FEV deficiency inhibits the colony-forming ability and proliferation of AML cells in vitro. (A, D, E) Number of colonies formed by sorted MV4-11 (A), THP-1 (D) and KG-1 (E) cells transduced with nonsilencing control (NC), FEV shRNA1 (sh1) or shRNA2 (sh2) after 7 days. (B, F, G) Cell counts in sorted MV4-11 (B), THP-1 (F) and KG-1 (G) cells transduced with NC, sh1 or sh2 after 2 days of culture. (C) CCK-8 assay showing the effect of shFEV on proliferation over a period of 7 days for MV4-11 cells. (H) Representative flow cytometry images of the cell cycle and apoptosis of MV4-11 cells transduced with NC or shFEV at 2 days. (I, J) Statistical analysis of the percentage of cells in G1 phase (I) and Annexin V+ cells (J) in the NC and shFEV groups. The results are representative of at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 (Student’s t test). All data are presented as the mean ± SD.
FEV Deficiency Inhibits the Homing Ability of AML Cells in Vivo
MV4-11 cells transduced with NC or shFEV (sh1, GFP+) were transplanted into immunodeficient NOD-SCID mice treated with CD122 antibody (NS122 mice) (17, 20, 21). The mice that received FEV knockdown cells had extensively prolonged survival time compared to their NC counterparts (Figure 3A). FEV deficiency significantly reduced the engraftment of AML cells in the BM (Figures 3B–D), which was consistent with previous findings in leukemia with prenatal initiation (17). FEV knockdown also reduced the infiltration of leukemic cells in the spleen and liver (Figure 3D and Supplementary Figure 3A–C), indicating the impaired homing and migration abilities of the leukemic cells.
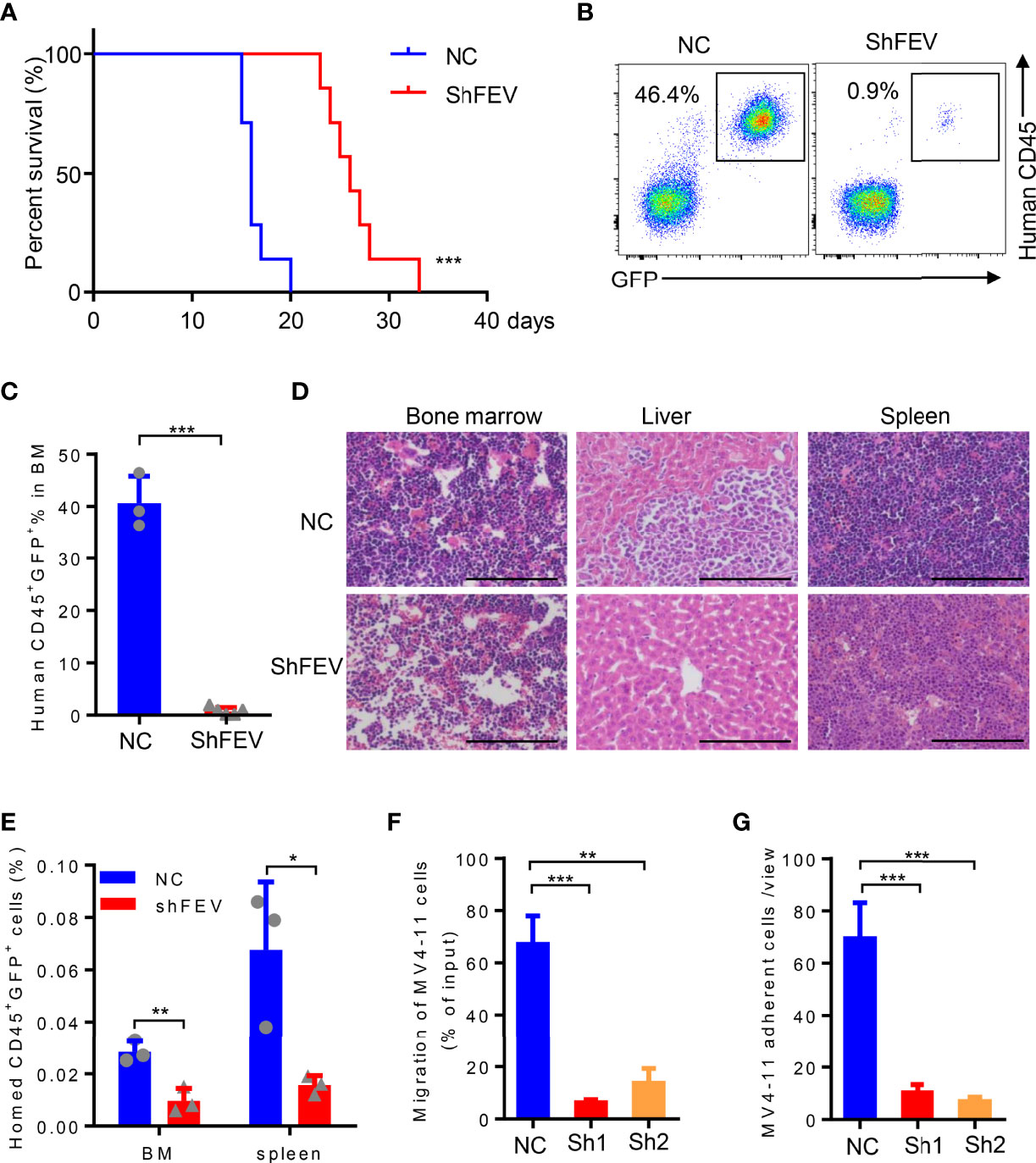
Figure 3 FEV deficiency inhibits the homing ability of AML cells in vivo. (A) Kaplan–Meier plot of disease-free survival of mice that received MV4-11 cells transduced with NC or shFEV (n=7). (B, C) Representative flow cytometry plot (B) and statistical analysis (C) of the percentage of engrafted cells in BM from mice that received NC (n=3) or shFEV MV4-11 cells after 15 days (n=5). (D) HE-stained sections (size bars = 50 μm) of BM, spleen and liver of mice that received NC or shFEV MV4-11 cells after 15 days. (E) Statistical analysis of the frequency of NC or shFEV cells homed to the BM and spleen at 16 h(n=3). (F) Frequency of migrating cells in the NC, sh1 and sh2 groups, which was normalized to the input control. (G) The average count of adherent MV4-11 cells in 10 random views. The results are representative of at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 (Student’s t test). All data are presented as the mean ± SD.
Given that enhanced homing and migration are the characteristics of leukemic cells, losing them may result in delayed leukemia progression (5–11). Therefore, the homing ability of leukemic cells after FEV knockdown was examined. Transduced MV4-11 cells were injected into sublethally irradiated NOD/SCID mice followed by analysis of human CD45+GFP+ cells in the spleen and BM 16 hours after the injection. The AML cells that homed to the BM or spleen were significantly decreased in the shFEV group (Figure 3E and Supplementary Figure 3D), indicating that FEV knockdown impairs the homing ability. In vitro transwell and adhesion assays supported that FEV knockdown decreased the frequency of migrated cells (Figure 3F and Supplementary Figure 3E) and those adherent to fibronectin (Figure 3G and Supplementary Figure 3F). Furthermore, the migration and adhesion abilities of FEV-deficient THP-1 or KG-1 cells were also inhibited (Supplementary Figures 3G–J).
Integrin Signaling Is Inhibited in FEV-Deficient AML Cells
MV4-11 cells transduced with NC or shFEV (sh1) were flow-sorted and subjected to gene transcriptome examination to investigate the underlying mechanisms. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis highlighted an intensive expression alteration of genes involved in cell adhesion molecules (CAMs), transcriptional misregulation in cancer and the PI3K-Akt signaling pathway (Figure 4A). Gene set enrichment analysis (GSEA) indicated that the FEV-deficient cells had a decreased enrichment of integrin cell surface interactions (Figure 4B) and cell adhesion molecules (Figure 4C). The mRNA and protein expression levels of genes related to the pathways were validated via RT–qPCR and western blot analyses. Key regulators of integrin signaling (integrin α4, phosphorylated RAC1, phosphorylated MAPK1, c-Jun and β-catenin) were downregulated (Figure 4D and Supplementary Figures 4A–D), indicating that the integrin signaling pathway may be downstream of FEV.
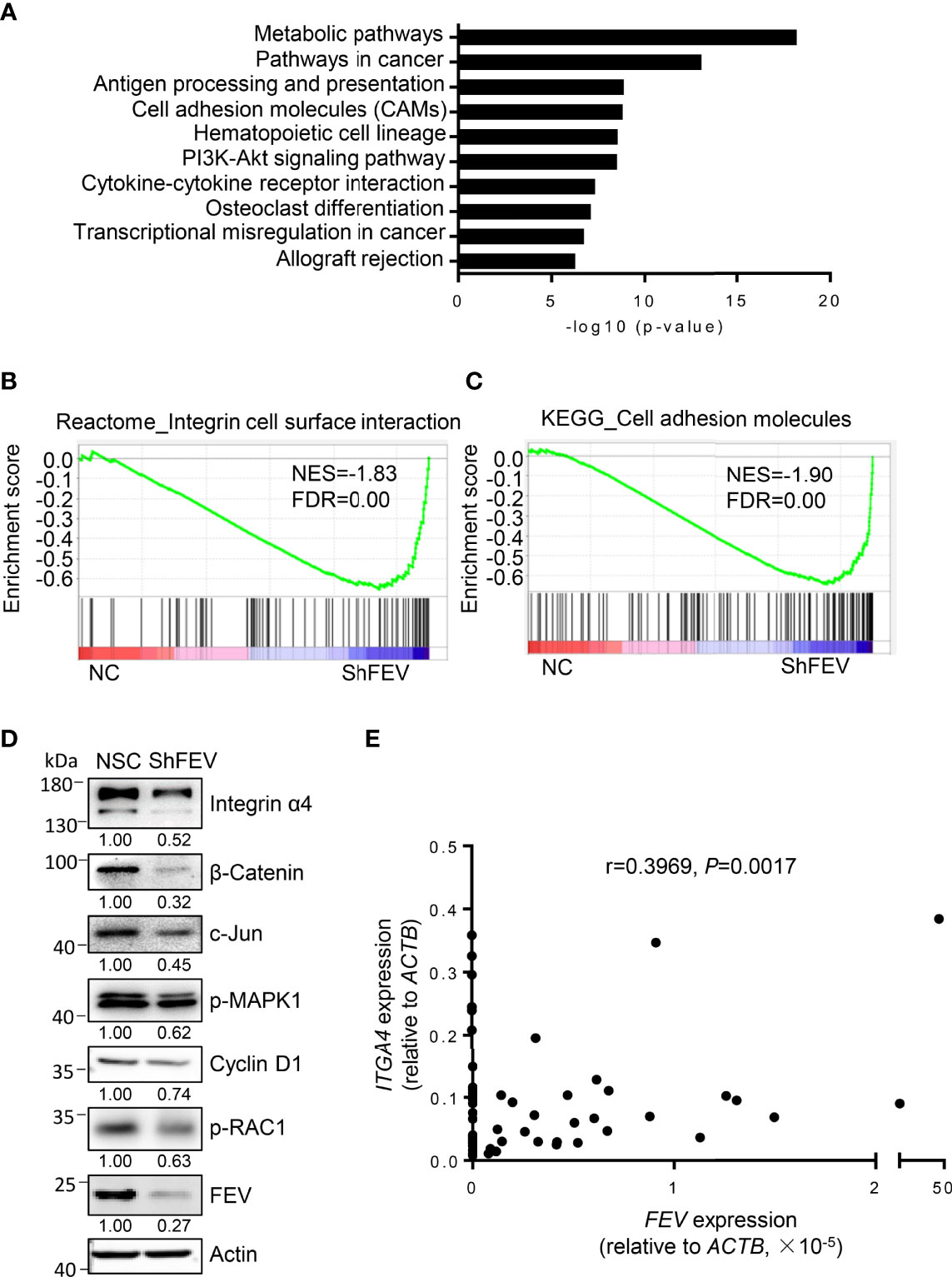
Figure 4 Integrin signaling is inhibited in FEV-deficient AML cells. (A) KEGG analysis highlighted the alteration of signaling pathways in MV4-11 cells transduced with NC or shFEV. (B, C) GSEA of selected signatures encoding the gene sets of integrin cell surface interaction (B) and cell adhesion molecules (C), which were differentially expressed between NC and shFEV cells. (D) Immunoblotting analysis of protein expression in the integrin signaling pathway. Relative protein levels were quantified using ImageJ software, and the numbers refer mean signals for three independent experiments. (E) Correlation analysis between FEV and ITGA4 expression in AML samples.
Given that integrins are key regulators of cell adhesion, migration and proliferation (22–25) and that ITGA4 is highly expressed in leukemic cells (26), ITGA4 may be a potential target of FEV. The correlation between FEV and ITGA4 expression in AML samples was analyzed, and a significant correlation between FEV and ITGA4 (Figure 4E) was observed. These results indicated that ITGA4 may be the target of FEV.
FEV Regulates AML Progression by Directly Activating ITGA4 Transcription
To study whether ITGA4 mediates FEV functions in AML progression, ITGA4 expression was interfered with by shRNAs (sh-ITGA4). Consistent with FEV knockdown, the number of CFCs was reduced (Supplementary Figure 5A), the proliferation of AML cells was inhibited (Supplementary Figure 5B) and the frequency of cells in G1 phase was increased (Supplementary Figure 5C) in the sh-ITGA4 group. Notably, cyclin D1 expression was decreased (Supplementary Figure 5D) in sh-ITGA4 cells, suggesting that ITGA4 may regulate the expansion of AML cells. In mice that received MV4-11 cells with ITGA4 interference, the survival time was extended (Supplementary Figure 5E), the reconstitution of leukemic cells was decreased (Supplementary Figure 5F), the infiltration of leukemic cells was decreased (Supplementary Figure 5G) and the frequency of cells homed to the BM or spleen was reduced (Supplementary Figure 5H). In vitro transwell and adhesion assays also exhibited a decrease in the proportion of migrated (Supplementary Figure 5I) and adhered (Supplementary Figure 5J) MV4-11 cells with ITGA4 interference.
ITGA4 was then ectopically expressed in FEV-knockdown MV4-11 cells, which were then subjected to in vitro and in vivo assays. Compared to the shFEV group, the number of colonies was significantly increased, the proliferation was significantly increased and G1 arrest was alleviated in the ITGA4-expressing shFEV group (Figure 5A and Supplementary Figures 6A, B). Migration and adhesion inhibition was also partly alleviated in ITGA4-expressing shFEV cells (Figures 5B, C). The mice that received ITGA4-expressing shFEV cells showed a significantly reduced survival time, which was comparable to that of their NC counterparts (Figure 5D). After ITGA4 expression, the frequency of infiltrated leukemic cells in organs was increased (Supplementary Figures 6C, D), and the homing arrest in shFEV cells to the BM and spleen was fully reversed (Figure 5E). Therefore, these findings indicated that ITGA4 is essential to FEV functions in expansion and homing of AML cells.
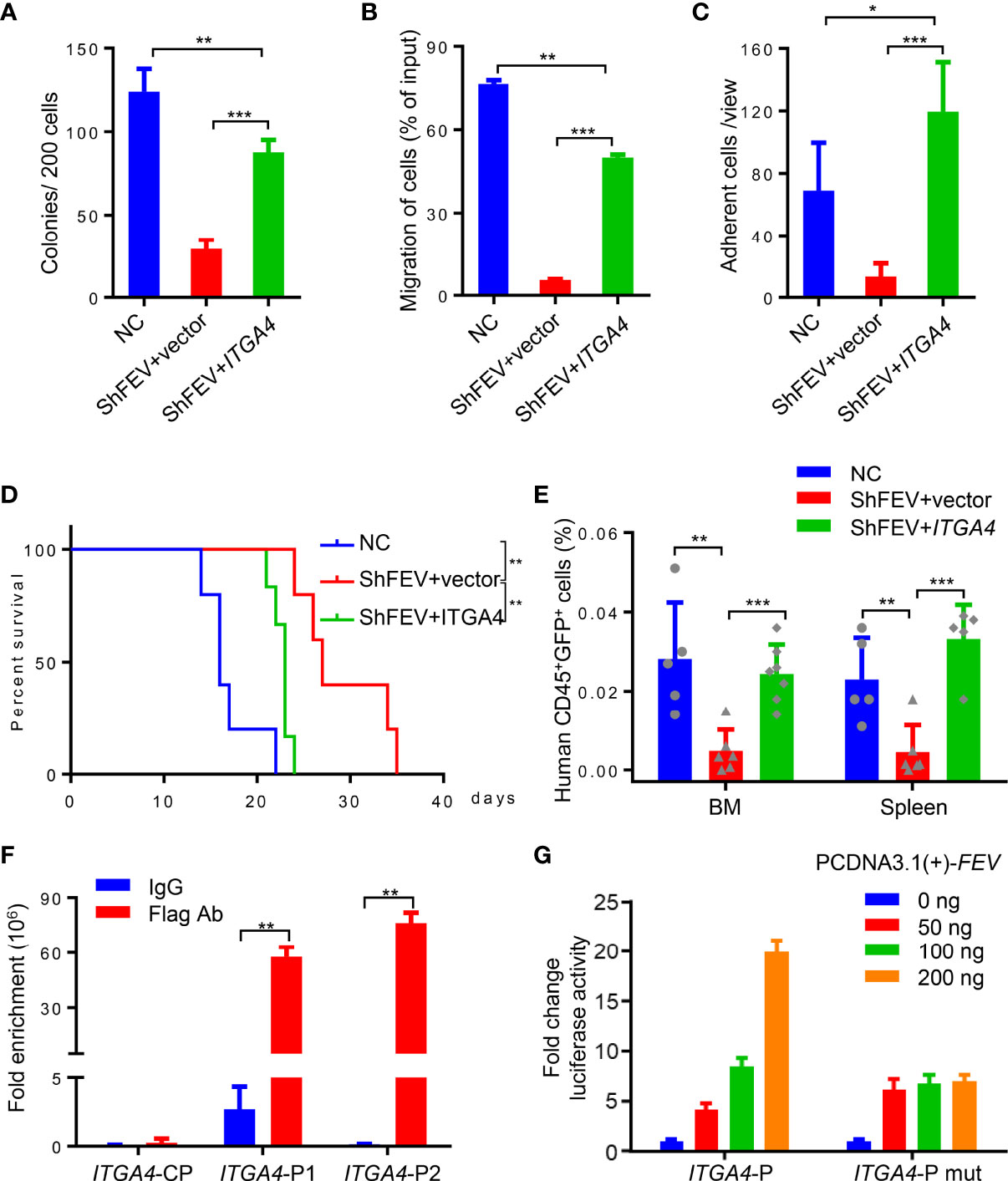
Figure 5 FEV regulates AML progression by directly activating ITGA4 transcription. (A) Number of colonies formed by NC and shFEV cells transduced with empty vector (shFEV+vector) or ectopically expressed ITGA4 (shFEV+ITGA4) after 7 days. (B) Frequency of migrated cells in the NC, shFEV+vector and shFEV+ITGA4 groups. (C) Average count of adherent cells in 10 random views. (D) Kaplan–Meier plot of disease-free survival of mice that received MV4-11 cells transduced with NC (n=5), shFEV+vector (n=5) or shFEV+ITGA4 (n=6). (E) Frequency of cells homed to the BM and spleen following intravenous injection in the NC (n=5), shFEV+vector (n=6) and shFEV+ITGA4 groups at 16 h (BM n=7; spleen n=5). (F) ChIP-qPCR analysis of FEV enrichment on the ITGA4 promoter. CP indicates the control sites without the FEV-binding site. P1 and P2 indicate the regions with conserved FEV-binding sites. (G) Dual luciferase reporter assays using ITGA4 promoter constructs that contain conserved (ITGA4-P) or mutated (ITGA4-P mut) FEV-binding sites cotransfected with the pCDNA3.1-FEV plasmid. The results are representative of at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 (Student’s t test). All data are presented as the mean ± SD.
Given that FEV is a transcription factor, the present study investigated whether FEV regulates the transcription of ITGA4. Two regions containing potential FEV-binding sites in the ITGA4 promotor were found by JASPAR (Supplementary Figures 6E, F). Chromatin immunoprecipitation (ChIP)-PCR assays with MV4-11 leukemic cells demonstrated that FEV binds to both regions (Supplementary Figure 6G). The amount of DNA fragments bound to the FEV protein was quantified by qPCR, and the results revealed that FEV preferred to bind to the P2 region (Figure 5F). The core binding sites in both regions were then mutated to TCTTCCCT to exclude nonspecific binding. A luciferase reporter assay in 293T cells was conducted using the promoter constructs of ITGA4 containing wild-type (ITGA4-P) or mutant FEV-binding sites (ITGA4-P mut). The results suggested that FEV positively regulated wild-type ITGA4 expression in a dose-dependent manner but not mutant ITGA4 (Figure 5G), indicating that ITGA4 is the direct target of FEV.
Blocking ITGA4 Activity Reduces the Colony-Forming, Migration and Adhesion Abilities of Blasts and LICs in Primary and Relapsed AML Patients
Because ITGA4 was identified as a downstream target of FEV in AML cells, we next investigated whether ITGA4 can be a target in the treatment of AML. Natalizumab (NZM), a therapeutic antibody approved by the U.S. Food and Drug Administration (FDA) for multiple sclerosis (27) and Crohn’s disease (28), was used to block the activity of integrin α4. Consistent with FEV knockdown, MV4-11 cells treated with NZM exhibited a decrease in the count of CFCs (Supplementary Figure 7A), migrated cells (Supplementary Figure 7B) and adhesion cells (Supplementary Figure 7C). After treatment with NZM, the proliferation of MV4-11 cells was inhibited, and an increased frequency of G1 phase cells was also found (Supplementary Figures 7D, E).
The samples of primary AML were thawed and treated with NZM and subjected to colony-forming, migration and adhesion assays. Thirteen primary AML samples were divided into FEV- and FEV+ groups according to FEV positivity. In FEV+ samples, NZM treatment reduced the number CFCs in the blasts (Supplementary Figure 8A) and inhibited the ability to migrate and adhere to fibronectin (Supplementary Figures 8B, C). However, no significant difference between IgG and NZM treatment was observed in the FEV- group, indicating that ITGA4 is a specific target of FEV.
Given that LICs are the main cause of relapse (7, 8, 29) and previous evidence has reported that the CD34+CD38−CD123+ putative LICs maintains high expression of integrin α4β1 (VLA-4) (30), we next determined whether LICs are eliminated by NZM treatment. CD34+CD38- LICs were sorted from 10 primary AML patients and treated with NZM. Consistent with blasts, LICs showed a reduced colony-forming capacity (Figure 6A), and the frequency of LICs that migrated or adhered to fibronectin was decreased after NZM treatment in the FEV+ group (Figures 6B, C). Moreover, no significant difference was observed in the FEV- group.
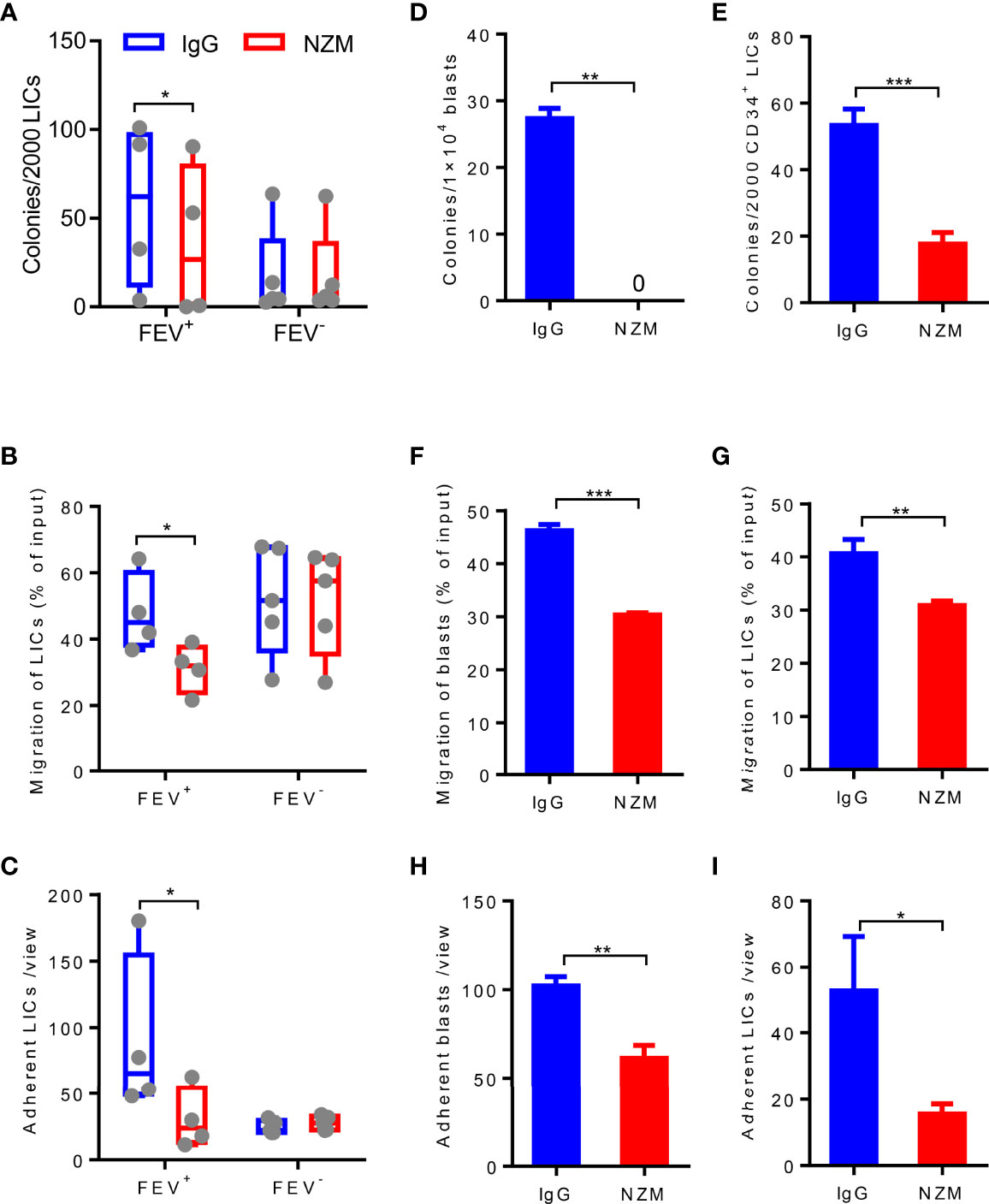
Figure 6 Blocking integrin α4 activity reduces the colony-forming, migration and adhesion abilities of blasts and LICs in primary and relapsed AML patients. (A) Number of colonies formed by LICs sorted from primary AML patients after IgG4 or NZM treatment after 14 days. (B) Frequency of migrated LICs from primary AML patients after 2 days of IgG4 or NZM treatment. (C) Average count of adherent LICs from primary AML patients per view. (D, E) Number of colonies formed by blasts (D) and LICs (E) from relapsed AML patients after IgG4 or NZM treatment. (F, G) Frequency of migrated blasts (F) and LICs (G) from relapsed AML patients after 2 days of IgG4 or NZM treatment. (H, I) Average count of adherent blasts (H) and LICs (I) per view from relapsed AML patients after IgG4 or NZM treatment. *P < 0.05, *P < 0.05, **P < 0.01 and ***P < 0.001 (A–C), paired t test; D-I, Student’s t test. All data are presented as the mean ± SD.
To determine whether NZM affects relapsed AML samples, blasts and LICs from relapsed AML patients were treated with NZM. The abilities of relapsed blasts or LICs to form colonies (Figures 6D, E), migrate (Figures 6F, G) and adhere (Figures 6H, I) were significantly inhibited after NZM treatment. These results suggested that ITGA4 may be a potential therapeutic target for relapsed AML.
In conclusion, FEV was re-expressed in 30% of primary and in almost all relapsed AML samples with high expression levels. FEV is functionally required for AML progression by regulating the homing and expansion of AML cells. ITGA4 is a direct target of FEV, and blockade of FEV-ITGA4-mediated homing and expansion may be a novel approach for primary and relapsed AML therapy.
Discussion
Approximately 40–45% of AML patients will achieve remission with current standard chemotherapy, but disease recurrence will appear in nearly 50% of these patients (1, 2). The prognosis of patients with relapsed AML is poor with no more than 10% overall survival in 3 years (1–4). Although new approaches, such as epigenetic drugs, leukemic antigen antibodies, inhibitors (for FLT3, IDH1, MDM2 and BCL2) and CAR-T therapy, have improved the outcome of some patients (2, 31, 32), a large number of primary or relapsed patients still lack effective treatment.
Here, we identified FEV as a regulator of AML progression and identified that the FEV-ITGA4 axis is involved in the homing and expansion of AML cells, which may be a potential therapeutic target for primary and relapsed AML.
In previous work, we reported that FEV is silenced in adult hematopoiesis and re-expressed in leukemias of prenatal origin, such as infants, children and young adults (<40 years); we also demonstrated that FEV is essential for leukemia propagation of LICs (17), but the mechanism is not clear. In the present study, we confirmed that FEV was re-expressed in approximately 30% of primary AML samples (our hospital, 30.2% FEV+; and Vizome database, 37.0% FEV+). The expression of FEV was associated with worse risk category, CR rate after 2 course of induction chemotherapy and prognosis. Most importantly, we observed that FEV was expressed in almost all relapsed AML samples, and the expression levels of FEV were higher in relapsed samples than in primary samples. We further demonstrated that FEV was required for AML progression. FEV deficiency markedly reduced the homing and expansion abilities of AML cells. These findings revealed the mechanism underlying the function of FEV in the propagation of LICs.
Leukemic cells are retained in the BM microenvironment with elevated homing and migration abilities, which are mediated by cell-to-cell or cell-to-matrix interactions. CXCR4-CXCL12 signaling, VLA4 signaling and CD44 signaling have been reported to contribute to the homing of leukemic cells to the BM microenvironment (5–11). However, the upstream signaling pathway remains unknown. In the present study, we found a significant correlation between FEV and ITGA4 (α4 subunit of VLA4) expression in AML samples, and ectopic ITGA4 expression elevated homing and expansion of shFEV MV4-11 cells and accelerated AML progression. ChIP and luciferase assays demonstrated that ITGA4 was a downstream target of FEV, which activated ITGA4 transcription by directly binding to the promoter in a dose-dependent manner. Thus, we identified a novel FEV-ITGA4 axis for homing and migration.
VLA4 is highly expressed in leukemic blasts and LICs (30), and it mediates the attachment of blasts and LICs to the ECM or stromal cells (5–11). High expression of VLA4 is associated with adverse outcomes in AML (26), chronic lymphocytic leukemia (33–35) and B-cell precursor acute lymphoblastic leukemia (36). Disruption of the adhesion of blasts within the microenvironment by a VLA-4 antibody or small molecular inhibitors sensitizes drug-resistant acute lymphoblastic leukemia (37–39) and AML (40, 41) to chemotherapy. In the present study, we showed that integrin α4 blockade with NZM resulted in reduced colony-forming, adhesion and migration abilities of blasts and LICs. NZM treatment also decreased the CFCs as well as the adhesion and migration of relapsed AML cells, although the account of colonies was less than that from fresh samples. Thus, these findings suggested that ITGA4 may be a potential target for both primary and relapsed AML. In agreement with Hsieh et al. (41, 42), blasts and LICs were not eradicated completely after NZM treatment; therefore, combination with cytotoxic chemotherapy agents or other targeted drugs may be a better approach.
In summary, our data provides mechanistic insight into the role of the FEV-ITGA4 axis in the progression of AML and the treatment application of NZM in primary and relapsed AML. Therefore, ITGA4 may be considered a therapeutic target for both primary and relapsed AML.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE166621.
Ethics Statement
The studies involving human participants were reviewed and approved by the Medical Ethical Committees of the First Affiliated Hospital of Soochow University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Experimental Animal Ethical Committee at Soochow University.
Author Contributions
JZ performed the experiments and wrote the manuscript. LQ and TW performed the experiments and analyzed the results. BZ and YF established the xenograft model. YL and MS collected the patient samples and clinical information. JA, DH, YX and DW discussed the data and contributed to the writing of the manuscript. TL supervised the project, designed the experiments and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Natural Science Foundation of China (81730003), the Excellent Youth Science Fund of Jiangsu Province (BK20211553), the Natural Science Foundation of China (81700139 and 81870120), the National Science and Technology Major Project (2017ZX09304021), the National Key R&D Program of China (2019YFC0840604 and 2017YFA0104502), the Key R&D Program of Jiangsu Province (BE2019798 and BE2019655), the Natural Science Fund of Jiangsu Province (BK20170360), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Jiangsu Medical Outstanding Talents Project (JCRCA2016002) and the Jiangsu Provincial Key Medical Center (YXZXA2016002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
All samples were from Jiangsu Biobank of Clinical Resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.890346/full#supplementary-material
References
1. Dohner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med (2015) 373(12):1136–52. doi: 10.1056/NEJMra1406184
2. Thol F, Ganser A. Treatment of Relapsed Acute Myeloid Leukemia. Curr Treat Options Oncol (2020) 21(8):66. doi: 10.1007/s11864-020-00765-5
3. Kantarjian H, Kadia T, DiNardo C, Daver N, Borthakur G, Jabbour E, et al. Acute Myeloid Leukemia: Current Progress and Future Directions. Blood Cancer J (2021) 11(2):41. doi: 10.1038/s41408-021-00425-3
4. Short NJ, Rytting ME, Cortes JE. Acute Myeloid Leukaemia. Lancet (2018) 392(10147):593–606. doi: 10.1016/s0140-6736(18)31041-9
5. Sipkins DA, Wei X, Wu JW, Runnels JM, Côté D, Means TK, et al. In Vivo Imaging of Specialized Bone Marrow Endothelial Microdomains for Tumour Engraftment. Nature (2005) 435(7044):969–73. doi: 10.1038/nature03703
6. Yamashita M, Dellorusso PV, Olson OC, Passegue E. Dysregulated Haematopoietic Stem Cell Behaviour in Myeloid Leukaemogenesis. Nat Rev Cancer (2020) 20(7):365–82. doi: 10.1038/s41568-020-0260-3
7. Konopleva MY, Jordan CT. Leukemia Stem Cells and Microenvironment: Biology and Therapeutic Targeting. J Clin Oncol (2011) 29(5):591–9. doi: 10.1200/JCO.2010.31.0904
8. Schepers K, Campbell TB, Passegué E. Normal and Leukemic Stem Cell Niches: Insights and Therapeutic Opportunities. Cell Stem Cell (2015) 16(3):254–67. doi: 10.1016/j.stem.2015.02.014
9. Witkowski MT, Kousteni S, Aifantis I. Mapping and Targeting of the Leukemic Microenvironment. J Exp Med (2020) 217(2):e20190589. doi: 10.1084/jem.20190589
10. Pollyea DA, Jordan CT. Therapeutic Targeting of Acute Myeloid Leukemia Stem Cells. Blood (2017) 129(12):1627–35. doi: 10.1182/blood-2016-10-696039
11. Medyouf H. The Microenvironment in Human Myeloid Malignancies: Emerging Concepts and Therapeutic Implications. Blood (2017) 129(12):1617–26. doi: 10.1182/blood-2016-11-696070
12. Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, et al. A New Member of the Ets Family Fused to Ews in Ewing Tumors. Oncogene (1997) 14(10):1159–64. doi: 10.1038/sj.onc.1200933
13. Liu F, Patient R. Genome-Wide Analysis of the Zebrafish Ets Family Identifies Three Genes Required for Hemangioblast Differentiation or Angiogenesis. Circ Res (2008) 103(10):1147–54. doi: 10.1161/CIRCRESAHA.108.179713
14. Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, et al. Pet-1 Ets Gene Plays a Critical Role in 5-Ht Neuron Development and Is Required for Normal Anxiety-Like and Aggressive Behavior. Neuron (2003) 37(2):233–47. doi: 10.1016/s0896-6273(02)01167-4
15. Lillesaar C, Tannhäuser B, Stigloher C, Kremmer E, Bally-Cuif L. The Serotonergic Phenotype Is Acquired by Converging Genetic Mechanisms Within the Zebrafish Central Nervous System. Dev Dyn (2007) 236(4):1072–84. doi: 10.1002/dvdy.21095
16. Wang L, Liu T, Xu L, Gao Y, Wei Y, Duan C, et al. Fev Regulates Hematopoietic Stem Cell Development Via Erk Signaling. Blood (2013) 122(3):367–75. doi: 10.1182/blood-2012-10-462655
17. Liu TH, Tang YJ, Huang Y, Wang L, Guo XL, Mi JQ, et al. Expression of the Fetal Hematopoiesis Regulator Fev Indicates Leukemias of Prenatal Origin. Leukemia (2017) 31(5):1079–86. doi: 10.1038/leu.2016.313
18. Zhang Y, Xia F, Liu X, Yu Z, Xie L, Liu L, et al. Jam3 Maintains Leukemia-Initiating Cell Self-Renewal Through Lrp5/Akt/B-Catenin/Ccnd1 Signaling. J Clin Invest (2018) 128(5):1737–51. doi: 10.1172/JCI93198
19. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional Genomic Landscape of Acute Myeloid Leukaemia. Nature (2018) 562(7728):526–31. doi: 10.1038/s41586-018-0623-z
20. Duan CW, Shi J, Chen J, Wang B, Yu YH, Qin X, et al. Leukemia Propagating Cells Rebuild an Evolving Niche in Response to Therapy. Cancer Cell (2014) 25(6):778–93. doi: 10.1016/j.ccr.2014.04.015
21. Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of Single Human Hematopoietic Stem Cells Capable of Long-Term Multilineage Engraftment. Science (2011) 333(6039):218–21. doi: 10.1126/science.1201219
22. Rettig MP, Ansstas G, DiPersio JF. Mobilization of Hematopoietic Stem and Progenitor Cells Using Inhibitors of Cxcr4 and Vla-4. Leukemia (2012) 26(1):34–53. doi: 10.1038/leu.2011.197
23. Williams DA, Rios M, Stephens C, Patel VP. Fibronectin and Vla-4 in Haematopoietic Stem Cell-Microenvironment Interactions. Nature (1991) 352(6334):438–41. doi: 10.1038/352438a0
24. Papayannopoulou T, Craddock C, Nakamoto B, Priestley GV, Wolf NS. The Vla4/Vcam-1 Adhesion Pathway Defines Contrasting Mechanisms of Lodgement of Transplanted Murine Hemopoietic Progenitors Between Bone Marrow and Spleen. Proc Natl Acad Sci USA (1995) 92(21):9647–51. doi: 10.1073/pnas.92.21.9647
25. Scott LM, Priestley GV, Papayannopoulou T. Deletion of Alpha4 Integrins From Adult Hematopoietic Cells Reveals Roles in Homeostasis, Regeneration, and Homing. Mol Cell Biol (2003) 23(24):9349–60. doi: 10.1128/mcb.23.24.9349-9360.2003
26. Matsunaga T, Takemoto N, Sato T, Takimoto R, Tanaka I, Fujimi A, et al. Interaction Between Leukemic-Cell Vla-4 and Stromal Fibronectin Is a Decisive Factor for Minimal Residual Disease of Acute Myelogenous Leukemia. Nat Med (2003) 9(9):1158–65. doi: 10.1038/nm909
27. Polman CH, O'Connor PW, Havrdova E, Hutchinson M, Kappos L, Miller DH, et al. A Randomized, Placebo-Controlled Trial of Natalizumab for Relapsing Multiple Sclerosis. N Engl J Med (2006) 354(9):899–910. doi: 10.1056/NEJMoa044397
28. Gordon FH, Lai CW, Hamilton MI, Allison MC, Srivastava ED, Fouweather MG, et al. A Randomized Placebo-Controlled Trial of a Humanized Monoclonal Antibody to Alpha4 Integrin in Active Crohn's Disease. Gastroenterology (2001) 121(2):268–74. doi: 10.1053/gast.2001.26260
29. Ho TC, LaMere M, Stevens BM, Ashton JM, Myers JR, O'Dwyer KM, et al. Evolution of Acute Myelogenous Leukemia Stem Cell Properties After Treatment and Progression. Blood (2016) 128(13):1671–8. doi: 10.1182/blood-2016-02-695312
30. Becker PS, Chien S, Roshal M, Wood B, Estey E, Papayannopoulou T, et al. Adhesion Receptor Expression by Acute Myeloid Leukemia (Aml) Bone Marrow Derived or Circulating Blasts and Aml Stem Cells: The Key to Overcoming Chemoresistance. Blood (2009) 114(22):2658. doi: 10.1182/blood.V114.22.2658.2658
31. Döhner H, Wei AH, Löwenberg B. Towards Precision Medicine for Aml. Nat Rev Clin Oncol (2021) 18(9):577–90. doi: 10.1038/s41571-021-00509-w
32. Rowe JM, Tallman MS. How I Treat Acute Myeloid Leukemia. Blood (2010) 116(17):3147–56. doi: 10.1182/blood-2010-05-260117
33. Gattei V, Bulian P, Del Principe MI, Zucchetto A, Maurillo L, Buccisano F, et al. Relevance of Cd49d Protein Expression as Overall Survival and Progressive Disease Prognosticator in Chronic Lymphocytic Leukemia. Blood (2008) 111(2):865–73. doi: 10.1182/blood-2007-05-092486
34. Majid A, Lin TT, Best G, Fishlock K, Hewamana S, Pratt G, et al. Cd49d Is an Independent Prognostic Marker That Is Associated With Cxcr4 Expression in Cll. Leuk Res (2011) 35(6):750–6. doi: 10.1016/j.leukres.2010.10.022
35. Strati P, Parikh SA, Chaffee KG, Achenbach SJ, Slager SL, Call TG, et al. Cd49d Associates With Nodal Presentation and Subsequent Development of Lymphadenopathy in Patients With Chronic Lymphocytic Leukaemia. Br J Haematol (2017) 178(1):99–105. doi: 10.1111/bjh.14647
36. Shalapour S, Hof J, Kirschner-Schwabe R, Bastian L, Eckert C, Prada J, et al. High Vla-4 Expression Is Associated With Adverse Outcome and Distinct Gene Expression Changes in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia at First Relapse. Haematologica (2011) 96(11):1627–35. doi: 10.3324/haematol.2011.047993
37. Hsieh YT, Gang EJ, Geng H, Park E, Huantes S, Chudziak D, et al. Integrin Alpha4 Blockade Sensitizes Drug Resistant Pre-B Acute Lymphoblastic Leukemia to Chemotherapy. Blood (2013) 121(10):1814–8. doi: 10.1182/blood-2012-01-406272
38. Ruan YS, Gang EJ, Kim H, Parekh C, Hisham A, Bhojwani D, et al. Ava4746, an Orally Available, Clinical Grade Antagonist of Integrin Alpha 4, Sensitizes Pre-B Cell Acute Lymphoblastic Leukemia to Chemotherapy. Blood (2016) 128(22):2765. doi: 10.1182/blood.V128.22.2765.2765
39. Härzschel A, Zucchetto A, Gattei V, Hartmann TN. Vla-4 Expression and Activation in B Cell Malignancies: Functional and Clinical Aspects. Int J Mol Sci (2020) 21(6):2206. doi: 10.3390/ijms21062206
40. Layani-Bazar A, Skornick I, Berrebi A, Pauker MH, Noy E, Silberman A, et al. Redox Modulation of Adjacent Thiols in Vla-4 by As101 Converts Myeloid Leukemia Cells From a Drug-Resistant to Drug-Sensitive State. Cancer Res (2014) 74(11):3092–103. doi: 10.1158/0008-5472.CAN-13-2159
41. Hsieh Y, Jiang E, Pham J, Kim H, Hisham A, Khazal S, et al. Vla4 Blockade in Acute Myeloid Leukemia. Blood (2013) 122(21):3944. doi: 10.1182/blood.V122.21.3944.3944
Keywords: FEV, AML (acute myeloid leukemia), ITGA4, homing, expansion
Citation: Zhang J, Qi L, Wang T, An J, Zhou B, Fang Y, Liu Y, Shan M, Hong D, Wu D, Xu Y and Liu T (2022) FEV Maintains Homing and Expansion by Activating ITGA4 Transcription in Primary and Relapsed AML. Front. Oncol. 12:890346. doi: 10.3389/fonc.2022.890346
Received: 05 March 2022; Accepted: 17 June 2022;
Published: 07 July 2022.
Edited by:
Tarik Moroy, Montreal Clinical Research Institute (IRCM), CanadaReviewed by:
Albrecht Reichle, University Medical Center Regensburg, GermanyChristian Kosan, Friedrich Schiller University Jena, Germany
Copyright © 2022 Zhang, Qi, Wang, An, Zhou, Fang, Liu, Shan, Hong, Wu, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianhui Liu, bGl1dGlhbmh1aUBzdWRhLmVkdS5jbg==; Yang Xu, eWFuZ3h1QHN1ZGEuZWR1LmNu; Depei Wu, d3VkZXBlaUBzdWRhLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Jubin Zhang
Jubin Zhang Lijuan Qi1,2†
Lijuan Qi1,2† Biqi Zhou
Biqi Zhou Yanglan Fang
Yanglan Fang Meng Shan
Meng Shan Dengli Hong
Dengli Hong Depei Wu
Depei Wu Yang Xu
Yang Xu Tianhui Liu
Tianhui Liu