
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 12 May 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.887532
This article is part of the Research Topic The Role of non-coding RNAs in Tumor Microenvironment and Metastasis of Gastrointestinal Cancers View all 9 articles
Colorectal cancer (CRC) is the second leading cause of cancer death and the third most prevalent malignancy. Colorectal tumors exchange information with the surrounding environment and influence each other, which collectively constitutes the tumor microenvironment (TME) of CRC. Many studies have shown that exosome-derived non-coding RNAs (ncRNAs) play important roles in various pathophysiological processes by regulating the TME of CRC. This review summarizes recent findings on the fundamental roles of exosomal ncRNAs in angiogenesis, vascular permeability, tumor immunity, tumor metabolism and drug resistance. Certainly, the in-depth understanding of exosomal ncRNAs will provide comprehensive insights into the clinical application of these molecules against CRC.
CRC is the third most common malignancy (1). Although data from the American Cancer Society showed a gradual decline in mortality rates for CRC, this trend hides a rise in mortality among young adults (2). With the death rate for young people with CRC rises, a better understanding of molecular pathogenesis and the exploration of sensitive surveillance tools for early diagnosis and therapy are critical.
CRC is widely regarded as a heterogeneous disease, and its pathogenesis involves multiple genetic changes and multiple pathways (3). Tumor heterogeneity causes differences between and within CRC, which also increases the difficulty in the treatment of CRC (4, 5). TME contributes significantly to this heterogeneity because it is the site of tumor cell formation and growth (6, 7). TME is an intricate system, composed of primary cancer cells, associated stromal and immune cells, which considerably affects the behavior of CRC cells at the primary tumor site as well as in metastatic lesions (8). More evidence indicates that exosomes may impact carcinogenesis and development as crucial players in the communication between tumor cells and surrounding components in TME (9, 10). According to a growing amount of scientific, lots of ncRNAs existing in exosomes play a regulatory role in various pathophysiological activities of CRC (11, 12). Furthermore, ncRNAs in exosomes can influence the malignant progression of tumorigenesis by a variety of ways, making them a research hotspot in recent years (10, 13).
In this review, we discussed the various functions and mechanisms of exosomal ncRNAs: microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA) in TME, thereby elucidating the possibility of exosomal ncRNAs being applied clinically to treat CRC.
Cancer progression is dependent on the capacity of tumor cells to establish a supportive TME (14). Studies have demonstrated that the microenvironment is involved in causing a condition of growth arrest in the tumor (15). At the same time, tumors fight against the normal microenvironment to overcome the anti-tumor pressure (15). While tumors communicate closely with the surrounding microenvironment, the biochemical signals in the microenvironment impact cell growth and tumorigenesis (16, 17). The normal microenvironment inhibits cancer progression in a steady state. On the contrary, when this steady-state structure is out of control, the microenvironment itself will send out tumor-promoting signals, promoting cell malignant transformation (18, 19).
TME is an important factor leading to the heterogeneity and targeted therapy of CRC (6, 20). The composition of TME is influenced by both tumor features and patient state, which impacts disease progression, responsiveness to cancer therapy, and survival prognosis (21). A lot of research shows that TME plays a crucial role in tumor growth, metastasis, and drug resistance (22). Components of the TME in CRC include colorectal tumor cells, blood vessels, fibroblasts, immune and inflammatory cells, the extracellular matrix, as well as many signaling molecules and pathways that impact the angiogenic response (18, 23, 24).
As the most abundant cell type in TME, CAFs regulate many aspects of tumorigenesis (25, 26). Studies have shown that cancer-associated fibroblasts (CAFs) contribute to CRC progression through immunosuppression, extracellular matrix (ECM) remodeling and promotion of epithelial mesenchymal transformation (EMT) (27). Colorectal tumor cells affect the recruitment of CAF precursors and induce the differentiation of normal fibroblasts into CAFs, promote tumor growth and maintain its malignant propensity (24). Suetsugu et al. found that colon metastatic tumor cells can recruit CAFs to metastatic sites and contribute to tumor progression (28). In addition, the TME shows great diversity in different types of cancer (29). In terms of immune cells in the TME of colon cancer, high tumor-associated macrophages (TAMs) are associated with fewer liver metastases (30). However, another study showed that high TAM is associated with a higher clinical stage in the TME of esophageal cancer (31). While high TAM is associated with poorly differentiated histology and lymph node metastasis in cholangiocarcinoma (32). Ugai et al. found the immunological microenvironment were different between moderate and advanced CRC patients. Moreover, lymphocytic response patterns, macrophages, and regulatory T cells in the TME were associated with patients’ age. Thus, immune cell profiles by age of diagnosis may help to explain the growth and progression of CRC in young people (33).
Tumor and stromal cells located in the TME can secrete both various soluble molecules and vesicles, including exosomes (24). Exosomes have been explored as key factors mediating cell-to-cell communication between tumor cells and the microenvironment, which are involved in various signaling pathways regulated in the TME. Hence, exosomes in the TME could be a promising therapeutic target for CRC therapy (34).
Exosomes were first described in the 1980s as membranous vesicles in reticulocytes (35). Initially, exosomes were considered cellular garbage, withal later studies have shown that exosomes can transfer genetic information to achieve cell-to-cell communication (36). The biogenesis of exosomes mainly involves three stages: first, the fusion of endocytic vesicles produces early endosomes (EEs), which encapsulate the cargo of endocytic cells that share certain biomolecules and membrane proteins; second, the late endosomes (LEs) are composed by the inward sprouting of the multivesicular body (MVB) membrane; finally, MVB can fuse with lysosomes or autophagosomes for degradation, which can also fuse with the plasma membrane to release the contained substances, namely exosomes (37, 38). They are encased in lipid bilayers and carry various biological molecules, such as RNA, DNA, proteins, glycans, and lipids (39). Many studies revealed that exosomes are rich in ncRNA (40). ncRNAs can bind to recipient target cells through exosome carriers to transmit information and change the gene expression and function of recipient cells, thus affecting cancer progression to a certain extent (34, 41).
Exosome-derived ncRNAs are involved in driver mutations and epigenetic modifications that drive various pathophysiological processes in CRC (42). A large number of studies on exosomes have shown that exosomal ncRNAs communicate throughout cancer and non-cancer cells and are closely involved in the occurrence and development of CRC (43, 44). Moreover, studies have shown that exosomes are much easier to take up by cancerous cells than other vesicles of an equal amount, indicating that exosomes have a higher selectivity for cancer targeting (45). We particularly focused on the roles of miRNAs, lncRNAs, and circRNAs in the TME of CRC (Table 1).
Since the discovery that exosomes carry genetic material inside, scientists have conducted extensive research on exosomes. Currently, the role of exosomal miRNAs in cancer is the most studied. miRNA is an endogenous short ncRNA sequence that binds to the 3′ untranslated region (UTR) of a target mRNA to suppress its production by degrading or repressing translation (60). As we know, miRNAs are capable of regulating different cellular processes (61). However, when some of these mechanisms are changed and disrupted miRNA expression, tumor growth deviates from its typical course of progression (61). Increasing evidence shows that miRNA-carrying exosomes released from immune cells, mesenchymal cells, and cancer cells in the TME can shuttle from donor cells to recipient cells, even being taken up by distant cells to alter gene expression (62, 63).
Exosomal miR-19a is enriched in the serum of CRC patients as well as associated with poor prognosis (64). Treating CRC-bearing mice with tumor-derived exosomal miR-34a significantly reduced tumor size and prolonged survival of CRC-bearing mice (65). Dai et al. found CRC cell-derived exosomal miR-10b transferred to fibroblast cells and directly inhibited PIK3CA expression, reduced PI3K/Akt/mTOR pathway activity, and boosted TGF-β and SM α-actin expression (46). Exosomal miR-16-5p from bone marrow-derived mesenchymal stem cells (BMSCs) acted on CRC cells, reduced CRC cell proliferation, migration and invasion by reducing ITGA2 (47). According to Tian et al., CRC-derived exosomal miR-221/222 transferred to hepatic stromal cells decreased serine protease inhibitor Kunitz type 1(SPINT1) expression to activate liver hepatocyte growth factor (HGF), which plays a vital role in the formation of pre-metastatic niche (PMN) effect, leading to CRC invasiveness (48). Moreover, CRC cell proliferation and invasion were inhibited by exosomal miR-22-3p from BMSCs, which inhibited the PI3K/AKT pathway by reducing RAP2B expression (49). CRC-derived exosomal miR-106b-3p promoted CRC cell invasiveness by targeting deleted in liver cancer-1(DLC-1) (50). CAFs-derived exosomal miR-17-5p targeted RUNX3 to increase TGF-β1 expression and activate the TGF-β signaling pathway. Furthermore, TGF-β1 in the TME activated CAFs, which in turn released additional exosomal miR-17-5p to CRC cells and resulted in a cancer-promoting feedback loop, finally promoting CRC aggressive phenotype (51). These findings suggested exosomal miRNAs expression is closely related to CRC progression.
lncRNAs have the characteristics of low expression, moderate sequence conservation, and high tissue-specific (66). What makes lncRNAs unique is that most of them are specifically expressed in certain conditions and tissues instead of having widespread roles (66, 67). Interestingly, lncRNAs can be preferentially sorted into exosomes, which are intercellular communication mediators and involved in CRC development (68).
According to great research findings, lncRNAs spread to cells via exosomes, shaping a favorable microenvironment for tumor cell growth (69). Compared with normal cell-derived exosomes, cancer cell-derived exosomes are enriched with specific lncRNAs, which further accelerate the malignant progression of cancer in recipient cells (61). Exosomal lncRNA MALAT1 secreted by metastatic CRC cells increased FUT4 expression and activated PI3K/Akt/mTOR to sponge miR-26a/26b in primary CRC cells, leading to CRC progression (52). CAFs-derived exosomes delivered LINC00659 to CRC cells and sponged miR-342-3p, which regulated ANXA2 for CRC cell proliferation, invasion and migration (53). Moreover, downregulation of lncRNA UCA1 in serum exosomes affected cell migration in CRC progression by controlling the ceRNA network (54). Through the miR-496/RAP2C axis, exosomal lncRNA NNT-AS1 promoted CRC cell proliferation, migration, and invasion (55). In addition, exosomal lnc CCAL and exosomal lnc CRNDE-h have been implicated in CRC progression (70, 71).
Circular RNA (circRNA) is an endogenous ncRNA created by exon back-splicing (72). One study calculated the ratio of back splicing to forward splicing product reads between cellular and cell-derived exosomes, the data obtained showed that the ratio of circRNA levels to linear RNA levels was approximately 6-fold higher in exosomes than in cells. This finding indicated that circRNAs are more present in exosomes than linear RNA (73). Duo et al. also came to the above conclusion by identifying KRAS mutant (DKO-1), mutant/wild-type (DLD-1) and wild-type (DKs-8) circRNA expression profiles in cells and exosomes (74). Moreover, numerous studies have shown that circRNAs are abundant and stable in exosomes and can translocate to nearby or distant cells and performed their functions (75, 76). Zhao et al. found that exosomal circ-ABCC1 from CRC cells promoted cell stemness, sphere formation, and metastasis by activating the Wnt/β-catenin pathway (56). Yang et al. found that exosomal circ_PTPRA inhibited CRC tumor growth by modulating the miR-671-5p/SMAD4 network (57). Through the miR-1305/TGF-β2/SMAD3 pathway, exosomal circCOG2 can transmit from cancer cells with high metastatic potential to cancer cells with low metastatic potential, promoting CRC proliferation, migration and invasion (58). Additionally, CRC cell-derived exosomal circEPB41L2 sponged miR-21-5p and miR-942-5p to inhibit proliferation and migration of CRC cells by regulating the PTEN/AKT signaling pathway (59).
The occurrence and progression of CRC require various complicated procedures (77). Metastasis is frequently the most dangerous and primary cause of CRC treatment failure which goes through a complex chain of events (78, 79). Abundant studies have shown that ncRNAs are significantly enriched in exosomes, which spread to the TME and remodel TME, ultimately leading to tumor metastasis, such as angiogenesis, vascular permeability, tumor immunity, tumor metabolism, drug resistance (Figure 1) (80–82). Additionally, drug resistance is closely linked to TME, exosomal ncRNAs play key roles in tumor cell adaptation to the TME and drug resistance (83). Based on the current studies, we discussed the role and mechanism of exosomal ncRNAs in TME of CRC in this section (Figure 2).
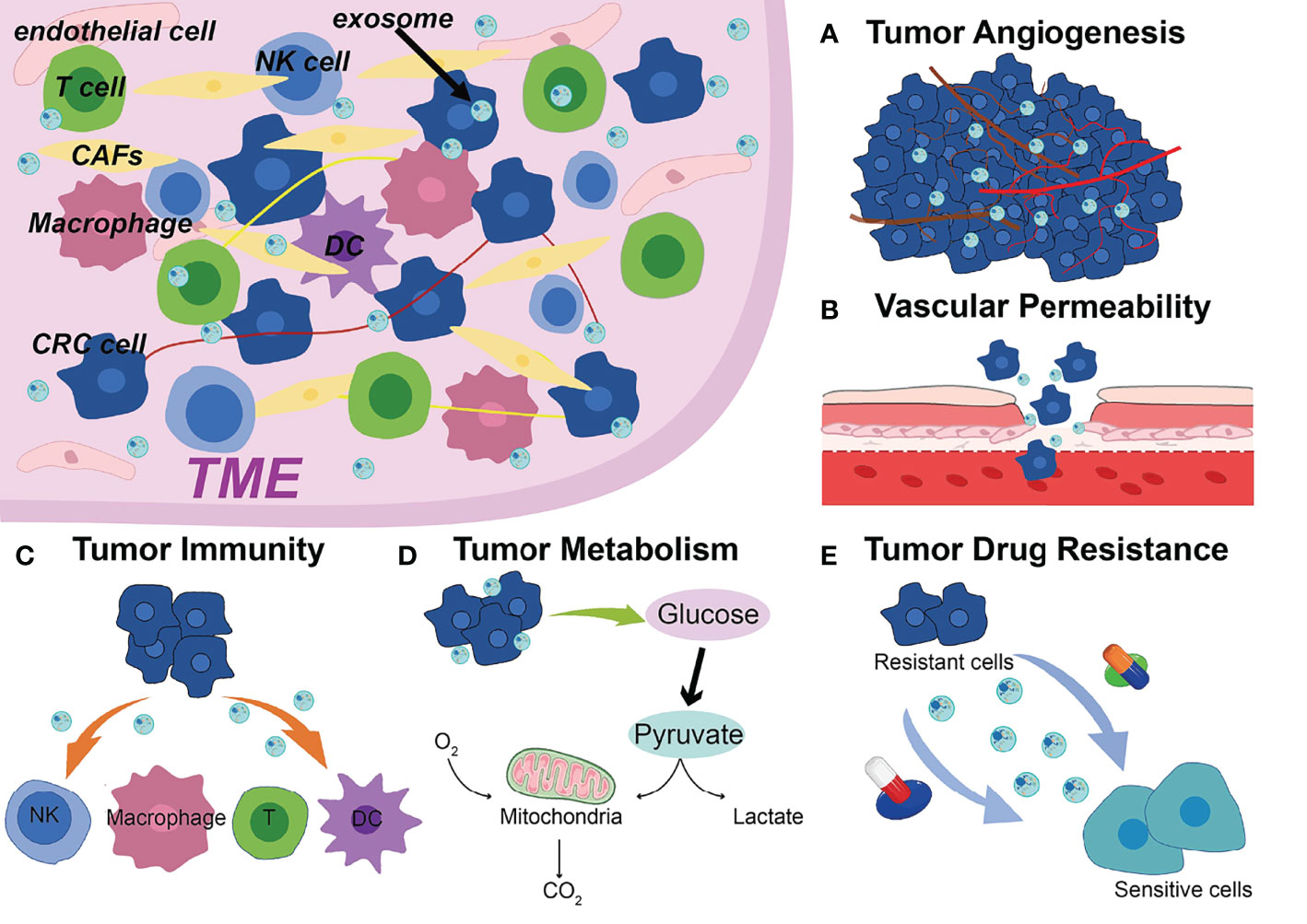
Figure 1 Exosomal ncRNAs contribute to the (A) tumor angiogenesis, (B) vascular permeability, (C) tumor immunity, (D) tumor metabolism, (E) drug resistance in the TME of CRC.
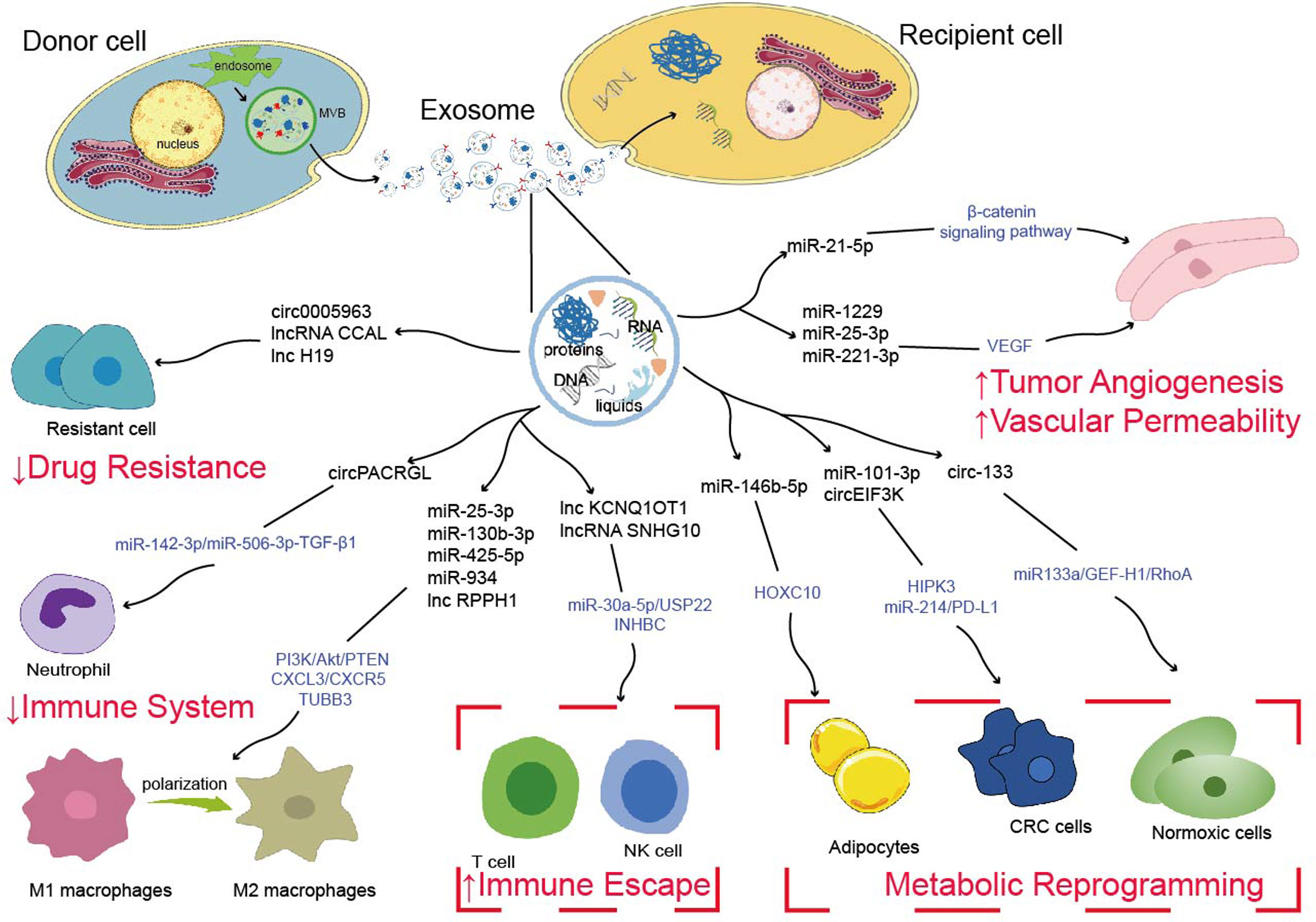
Figure 2 Possible mechanisms of exosome-derived ncRNAs to regulate tumor metastasis, tumor immunity, tumor metabolism and drug resistance.
One of the distinguishing characteristics of cancer is its ability to stimulate angiogenesis (84). In practice, TME needs to provide more oxygen and nutrients as the tumor continues to grow (23). During this process, TME sends signals to endothelial cells, causing an increase in the expression of mutiple angiogenic factors in order to form a new blood vessel (85). In particular, vascular endothelial growth factor (VEGF) is a powerful angiogenic factor that is widely recognized as a critical element in angiogenesis (86). VEGF regulates vessel formation by binding to VEGF receptors (VEGFRs)-1, -2, and -3, which are expressed on vascular endothelial cells (87). To create innovative anti-angiogenic medicines, it will be necessary to have a deeper knowledge of the cellular and molecular pathways that are involved in tumor angiogenesis.
Abnormal expression of exosomal ncRNAs impact cancer progression through regulating angiogenesis (88). He et al. found that miR-21-5p is propagated from CRC cells to recipient human umbilical vein endothelial cells (HUVECs) via exosomes, then activated β-catenin signaling pathway, increased downstream target expression, and regulated CRC angiogenesis and vascular permeability (89). In another study suggested significant upregulation of CRC cell-derived exosomal miR-1229 suppressed HIPK protein expression and activated the VEGF pathway of HUVECs. Moreover, exosomal miR-1229 inhibitor substantially inhibited tumor growth and angiogenesis as demonstrated by a nude mouse xenograft model (90). Zeng et al. showed that cancer-derived exosomal miR-25-3p drives CRC development by altering the expression of VEGFR2, ZO-1, occludin and Claudin5 in endothelial cells through targeting KLF2 and KLF4 (91). Likewise, the STAT3/VEGFR-2 signaling axis was activated by CRC cell-derived exosomal miR-221-3p, which promoted endothelial cells angiogenesis (92). Shang et al. discovered that miR-185-3p was significantly expressed in cancer cell-derived exosomes and overexpression of its target gene FOXO1 could reverse the increase in angiogenesis-related proteins caused by miR-183-5p in HMEC-1 cells (93). Chen et al. revealed that cancer cells derived exosomal miR-27b-3p trafficking into vascular endothelial cells reduced VE-Cad and p120 expression, increased vascular permeability in vivo, and eventually accelerated CRC metastasis (94). These studies demonstrate that exosomal ncRNAs affect angiogenesis and permeability to promote colorectal tumor metastasis.
Tumor-associated immune cells are a part of the TME, which play an important role in the TME and may have tumor-promoting or opposing effects (95, 96). Dunn et al. proposed the “cancer immunoediting” theory, which is based on the dual role of immunity in the complex interaction between tumor and host (97). In short, the immune system has the ability to prevent tumor development and progression while suppressing its occurrence and development (96, 97). However, some initial tumors can evade this attack and continue to develop in the host, increasing their chances of metastasis and recurrence (98). Interactions between immune cells and cancer cells, as well as exosome-related intercellular communication, are vital in tumor immune regulation, producing an immunosuppressive environment that promotes cancer development and progression (99).
Multiple studies demonstrate that exosomal ncRNAs mediate complex interactions between tumor and immune cells and induce changes in the expression of genes that regulate immunosuppression, confirming the functional significance of exosomal ncRNAs in immune modulation (100, 101). Another study showed that exosomal circPACRGL generated from CRC cells promoted neutrophil N1-N2 differentiation through the miR-142-3p/miR-506-3p-TGF-β1 axis (102). Moreover, the immune system normally responds to foreign antigens by promoting the proliferation and differentiation of cytotoxic T cells in TME (103). Xian et al. found that overexpression of exosomal lncRNA KCNQ1OT1 secreted by CRC cell affected cytotoxic T cells by regulating PD-L1 ubiquitination via miR-30a-5p/USP22, leading to immune escape (104). Similarly, exosomes ncRNAs can potentially serve as a bridge between tumor cells and NK cells, facilitating the exchange of information. For instance, exosomal lncRNA SNHG10 derived from CRC cells contributed to immune escape by suppressing NK cell function by upregulating INHBC expression as well (105).
Macrophages are innate immune cells that play a variety of roles in host defense and tissue homeostasis in TME (106). Exosomes have also been found to affect the pre-metastatic niche and promote cancer metastasis by altering the localization and function of tumor-associated macrophages (TAMs) through related mechanisms (11, 107, 108). Wang et al. found that when CXCL12/CXCR4 axis is activated, CRC cells produce exosomal miRNAs (miR-25-3p, miR-130b-3p, and miR-425-5p), which macrophages may take up and target PTEN through activation of PI3K/Akt signaling pathway, resulting in the M2 phenotypic transfer. Crucially, M2-polarized macrophages in turn secrete VEGF, which promotes CRC angiogenesis and liver metastases (109). Zhao et al. showed that tumor-derived exosomal miR-934-induced M2 macrophage polarization promotes CRC liver metastasis through activation of the CXCL13/CXCR5 axis (110). Meanwhile, another study found that CRC cell-derived exosomal lnc-RPPH1 promotes CRC cell metastasis and proliferation in vivo through mediating macrophage M2 polarization by binding to TUBB3 (111).
These studies confirmed that exosomal ncRNAs can alter the immunological microenvironment via influencing immune cell phenotypic.
In addition, reprogramming of energy metabolism is also seen as a hallmark of cancer (84). Glycolysis is the primary source of energy metabolism in tumors and obtains more sugar breakdown capacity, which can convert glucose into lactate to generate ATP, resulting in the formation of an acidic TME that is more conducive to cancer growth (112). Tao et al. found that exosomal miR-101-3p targeted and decreased HIPK3 expression in CRC cells, reduced mitochondrial membrane potential and produced reactive oxygen species (ROS), while increasing aerobic glycolysis and encouraging colorectal tumor development (113). Exosomal circ_0005963 from drug-resistant cells were delivered to drug-sensitive cells to inhibit glycolysis via the circ_0005963/miR-122/PKM2 pathway (114). Moreover, exosomes can mediate communication between cancer cells and adipocytes, which facilitate the transfer of nutrients such as lipids in the TME (115). Exosomal miR-146b-5p was released by cancer cells to promote browning of white adipose tissue (WATs) and increase lipolysis. Further study indicated that exosomal miR-146b-5p inhibited HOXC10 overexpression to increase WAT browning, reduce oxygen consumption and regulate lipolysis (116).
Furthermore, the nutritional status of oxygen has a substantial impact on cancer’s ability to use energy, and differences in energy storage between normoxic and hypoxic cells present various transfer potentials in the TME (117). Exosomal circ-133 generated from Hypoxic cells was transported into normoxic cells and promoted CRC metastasis by acting on the miR-133a/GEF-H1/RhoA axis (117). Yang et al. found that CAFs-derived exosomal circEIF3K was delivered to CRC cells and promoted CRC progression by regulating the miR-214/PD-L1 axis in the hypoxic microenvironment (118). According to these studies, targeting exosome cargoes that govern energy metabolism might provide a novel and successful method for cancer therapy.
Exosomal ncRNAs are involved not only in cancer progression and metastasis, but also in treatment resistance development (119). Obviously, one of the challenges in the treatment of tumor processes during chemoradiotherapy is the development of drug resistance. Exosomes present important mediators of intercellular communication, may contribute to the horizontal spread of drug resistance in heterogeneous cancer cell populations, which might make it impossible to treat many cancers effectively (120).
Methotrexate (MTX) is an antineoplastic drug that is widely used as standard chemotherapy in the treatment of various malignancies (121). Under MTX treatment, downregulation of the CDX2/HEPH axis by CAFs-derived exosomal miR-24-3p inhibitor accelerates the resistance of colon cancer cells to MTX (122). Recently, studies have demonstrated that exosomal miR-208b promotes regulatory T cells (Tregs) expansion by targeting PDCD4, and exosomal miR-208b upregulation was the most pronounced in oxaliplatin-resistant cells. That may be associated with reduced oxaliplatin-based chemosensitivity in CRC (123). Exosomal miR-208b can thus be employed as an oxaliplatin treatment response indicator. CAFs delivered exosomal lncRNA CCAL to cancer cells inhibited apoptosis in CRC cells, activated the β-catenin pathway, and promoted oxaliplatin resistance both in vitro and in vivo (70). Hon et al. found that exosomal circ-0000338 can transfer chemoresistance from FOLFOX-resistant cells to sensitive cells (124).
Furthermore, CRC patients who are undergoing chemotherapy commonly develop resistance to the drug 5-fluorouracil (5-FU) (125, 126). Similarly, overexpression of exosomal miR-181d-5p suppressed 5-FU sensitivity of CRC cells (127). According to Hu et al., exosomal miR-92a-3p was shown to be considerably higher in 5-FU/L-OHP resistant CRC patients than in 5-FU/L-OHP sensitive CRC patients. Further research found that CAFs-derived exosomal miR-92a-3p transfer to CRC cells then promoted the Wnt/β-catenin pathway, directly inhibited FBXW7 and MOAP1 to suppress mitochondrial apoptosis, which affects cancer growth and medication resistance (128). In addition, Ren et al. discovered that exosomal lncRNA H19 acted as a competitive endogenous RNA sponge for miR-141 to activate the Wnt/β-linked protein pathway, hence increasing resistance to oxaliplatin in CRC cells (129). These results implied that exosomal miRNAs might be as useful biomarkers for the development of CRC metastases and chemoresistance.
The above-mentioned evidence revealed that using exosomal ncRNAs to induce tumor cell resistance to anticancer drugs, cancer-associated cells were reprogrammed in the TME. We can determine that exosomal ncRNAs can also be potential therapeutic targets for overcoming drug resistance, thereby contributing to cancer patients.
CRC can be a preventable and treatable disease, and with current treatment, early diagnosis can greatly improve five-year survival rates (130). Biomarkers in bodily fluids can help identify the presence of cancer, metastasize and assess the response of treatment. Apparently, the methods of obtaining bodily fluids are significantly less invasive than traditional biopsies (131). Since exosomes can be detected in all bodily fluids and are produced by all cells, exosomes are great liquid biopsies that can be used to track the progression of sickness over time (37). Moreover, exosomal ncRNA is stable, abundant, reproducible and disease-specific, which lays the foundation for the early diagnosis of CRC (132, 133).
Min et al., found that miRNAs in exosomes, such as Let-7b-3p, miR-139-3p, miR-145-3p and miR150-3p, can be used as biomarkers for early diagnosis of CRC and have been validated in large cohorts (134). Besides, they had more discriminative power for multiple miRNA-binding diagnoses than for single miRNA-binding diagnoses (134). Moreover, high level of exosomal miR-19a expression in serum suggest the possibility of CRC recurrence (64). Pan et al. screened circ-0004771 as a potential diagnostic biomarker for CRC, and the ROC curve area of circ0004771 can distinguish benign intestinal diseases (BID), stage I/II CRC patients, and healthy controls (HCs) (135). Zhao et al. studied the exosomes taken from the plasma of CRC patients and discovered that the more severe the CRC patients, the lower the expression of miR-193a and the higher the expression of let-7g (136). Exosomal circR1 had specificity in CRC diagnosis and was associated with overall survival. In addition, the combination of exosomal circLPAR1, CEA, and CA19-9 enhanced the AUC value to 0.875 (137). As reported by Gao et al., CRC patients with high serum lncRNA 91H expression levels were generally at a higher risk of tumor recurrence or metastasis than other patients. Furthermore, CRC recurrence or metastasis can be predicted by detecting serum exosomal lncRNA 91H (138).
The above examples all demonstrate the potential and possibility of exosomal ncRNAs in various aspects, such as differential diagnosis, prognostic diagnosis, and staging characteristics.
Numerous studies have established that exosomes serve crucial biological functions in cancer growth. Therefore, specifically targeting exosomes may be a promising therapeutic approach. Interfering with CRC progression by modulating exosome expression or blocking its transport pathway may be a therapeutic strategy.
Exosomes, with their lipid bilayer structure, can delivery anti-tumor chemicals to target cells, so as to achieve the effect of treatment (139–141). Researchers have packaged interfering RNAs and chemical drugs into exosomes through various methods and targeted transport to specific cells (142, 143). This engineered exosome technology can achieve the purpose of treating disease areas or cells. Tian et al. targeted exosomes by intravenous injection to specifically deliver doxorubicin (DOX) to tumor tissue in nude mice to inhibit tumor growth (144). Bagheri et al. created a MUC1 aptamer-modified exosome for DOX administration and showed promising efficacy in reducing colorectal tumor development in vivo, laying the groundwork for the use of exosomes in preclinical cancer treatment (145). Asadirad et al. found that exosomes carrying miR-155 could stimulate bone marrow-derived DC cells to release inflammatory factors IL-12p70 and IFN-γto achieve anti-CRC effect (146). Furthermore, Zhan et al. designed exosomes as a nanoplatform capable of precisely delivering drugs and miR-21i to tumor cells, and animals treated with this technique showed increased tumor suppression without severe adverse effects (147). Liang et al. used exosome engineering to deliver the combination of miR-21 and the chemotherapeutic drug 5-FU to recipient cells, which effectively overcame drug resistance and increased the cytotoxicity of 5-FU-resistant colon cancer cells (148). With the continuous refinement and diversification of engineered exosome technology, the exosomal drug delivery system holds a lot of promise for use in the medical field as a natural nanoscale drug delivery platform.
Taken together, the potential applications of exosome-derived ncRNAs in cancer therapy are not limited to these aspects. Their roles in TME have great potential and application prospects in the future.
Exosome-derived ncRNAs play non-negligible roles in reshaping the CRC microenvironment. They serve as novel means of cell-to-cell crosstalk in TME, regulating signaling pathways that occupy their roles in cancer development and progression. Although research progress on functions and mechanisms of exosomal ncRNAs provide broad prospects for cancer diagnosis and therapeutic applications, the gap between their discovery and clinical practice cannot be ignored.
In the continuous research on the clinical application of exosomes, the technology of sensitive and accurate separation and detection of exosomes has not yet reached the expected level. Additionally, given the various advantages of exosomes, further research on how to maximize their loading efficiency is also required. The application of exosomes to cancer therapy will undoubtedly accelerate as more undiscovered areas are explored.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
XC and YZ were involved in the conception of the study. XC and MJ were involved in writing the article. JJ, ZZ and YZ critically revised the manuscript. All authors have read and approved the final manuscript.
This study was supported by The National Natural Science Foundation of China (grant no. 8217121619 to YZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors sincerely thank all participants involved in this study.
1. Keum N, Giovannucci E. Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat Rev Gastroenterol Hepatol (2019) 16(12):713–32. doi: 10.1038/s41575-019-0189-8
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
3. Fearon ER, Vogelstein B. A Genetic Model for Colorectal Tumorigenesis. Cell (1990) 61(5):759–67. doi: 10.1016/0092-8674(90)90186-i
4. Chen B, Scurrah CR, McKinley ET, Simmons AJ, Ramirez-Solano MA, Zhu X, et al. Differential Pre-Malignant Programs and Microenvironment Chart Distinct Paths to Malignancy in Human Colorectal Polyps. Cell (2021) 184(26):6262–80.e26. doi: 10.1016/j.cell.2021.11.031
5. Guo S, Ye Y, Liu X, Gong Y, Xu M, Song L, et al. Intra-Tumor Heterogeneity of Colorectal Cancer Necessitates the Multi-Regional Sequencing for Comprehensive Mutational Profiling. Cancer Manag Res (2021) 13:9209–23. doi: 10.2147/cmar.S327596
6. Li W, Li F, Zhang X, Lin H, Xu C. Insights Into the Post-Translational Modification and Its Emerging Role in Shaping the Tumor Microenvironment. Signal Transduct Target Ther (2021) 6(1):422. doi: 10.1038/s41392-021-00825-8
7. LeBleu VS. Imaging the Tumor Microenvironment. Cancer J (2015) 21(3):174–8. doi: 10.1097/ppo.0000000000000118
8. Wu Q, Zhang H, Sun S, Wang L, Sun S. Extracellular Vesicles and Immunogenic Stress in Cancer. Cell Death Dis (2021) 12(10):894. doi: 10.1038/s41419-021-04171-z
9. Leonard NA, Reidy E, Thompson K, McDermott E, Peerani E, Tomas Bort E, et al. Stromal Cells Promote Matrix Deposition, Remodelling and an Immunosuppressive Tumour Microenvironment in a 3d Model of Colon Cancer. Cancers (Basel) (2021) 13(23):5998. doi: 10.3390/cancers13235998
10. Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng J, et al. The Roles of Tumor-Derived Exosomes in Non-Small Cell Lung Cancer and Their Clinical Implications. J Exp Clin Cancer Res (2018) 37(1):226. doi: 10.1186/s13046-018-0901-5
11. Li W, Wang X, Li C, Chen T, Yang Q. Exosomal Non-Coding Rnas: Emerging Roles in Bilateral Communication Between Cancer Cells and Macrophages. Mol Ther (2021) 30(3):1036–53. doi: 10.1016/j.ymthe.2021.12.002
12. Rizk NI, Abulsoud AI, Kamal MM, Kassem DH, Hamdy NM. Exosomal-Long Non-Coding Rnas Journey in Colorectal Cancer: Evil and Goodness Faces of Key Players. Life Sci (2022) 292:120325. doi: 10.1016/j.lfs.2022.120325
13. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome Mediated Communication Within the Tumor Microenvironment. J Control Release (2015) 219:278–94. doi: 10.1016/j.jconrel.2015.06.029
14. Hui L, Chen Y. Tumor Microenvironment: Sanctuary of the Devil. Cancer Lett (2015) 368(1):7–13. doi: 10.1016/j.canlet.2015.07.039
15. Bakhshandeh S, Werner C, Fratzl P, Cipitria A. Microenvironment-Mediated Cancer Dormancy: Insights From Metastability Theory. Proc Natl Acad Sci USA (2022) 119(1):e2111046118. doi: 10.1073/pnas.2111046118
16. Del Prete A, Schioppa T, Tiberio L, Stabile H, Sozzani S. Leukocyte Trafficking in Tumor Microenvironment. Curr Opin Pharmacol (2017) 35:40–7. doi: 10.1016/j.coph.2017.05.004
17. Plaks V, Kong N, Werb Z. The Cancer Stem Cell Niche: How Essential Is the Niche in Regulating Stemness of Tumor Cells? Cell Stem Cell (2015) 16(3):225–38. doi: 10.1016/j.stem.2015.02.015
18. Han Z, Gong C, Li J, Guo H, Chen X, Jin Y, et al. Immunologically Modified Enzyme-Responsive Micelles Regulate the Tumor Microenvironment for Cancer Immunotherapy. Mater Today Bio (2022) 13:100170. doi: 10.1016/j.mtbio.2021.100170
19. Bissell MJ, Hines WC. Why Don't We Get More Cancer? A Proposed Role of the Microenvironment in Restraining Cancer Progression. Nat Med (2011) 17(3):320–9. doi: 10.1038/nm.2328
20. Kasprzak A. The Role of Tumor Microenvironment Cells in Colorectal Cancer (Crc) Cachexia. Int J Mol Sci (2021) 22(4):1565. doi: 10.3390/ijms22041565
21. Plundrich D, Chikhladze S, Fichtner-Feigl S, Feuerstein R, Briquez PS. Molecular Mechanisms of Tumor Immunomodulation in the Microenvironment of Colorectal Cancer. Int J Mol Sci (2022) 23(5):2782. doi: 10.3390/ijms23052782
22. Huang X, Pan J, Xu F, Shao B, Wang Y, Guo X, et al. Bacteria-Based Cancer Immunotherapy. Adv Sci (Weinh) (2021) 8(7):2003572. doi: 10.1002/advs.202003572
23. Weis SM, Cheresh DA. Tumor Angiogenesis: Molecular Pathways and Therapeutic Targets. Nat Med (2011) 17(11):1359–70. doi: 10.1038/nm.2537
24. Chen Y, Zheng X, Wu C. The Role of the Tumor Microenvironment and Treatment Strategies in Colorectal Cancer. Front Immunol (2021) 12:792691. doi: 10.3389/fimmu.2021.792691
25. Kochetkova M, Samuel MS. Differentiation of the Tumor Microenvironment: Are Cafs the Organizer? Trends Cell Biol (2021) 32(4):285–94. doi: 10.1016/j.tcb.2021.11.008
26. Linares J, Marín-Jiménez JA, Badia-Ramentol J, Calon A. Determinants and Functions of Cafs Secretome During Cancer Progression and Therapy. Front Cell Dev Biol (2020) 8:621070. doi: 10.3389/fcell.2020.621070
27. Chandra R, Karalis JD, Liu C, Murimwa GZ, Voth Park J, Heid CA, et al. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers (Basel) (2021) 13(24):6206. doi: 10.3390/cancers13246206
28. Suetsugu A, Osawa Y, Nagaki M, Saji S, Moriwaki H, Bouvet M, et al. Imaging the Recruitment of Cancer-Associated Fibroblasts by Liver-Metastatic Colon Cancer. J Cell Biochem (2011) 112(3):949–53. doi: 10.1002/jcb.23011
29. Yang M, McKay D, Pollard JW, Lewis CE. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res (2018) 78(19):5492–503. doi: 10.1158/0008-5472.Can-18-1367
30. Zhou Q, Peng RQ, Wu XJ, Xia Q, Hou JH, Ding Y, et al. The Density of Macrophages in the Invasive Front Is Inversely Correlated to Liver Metastasis in Colon Cancer. J Transl Med (2010) 8:13. doi: 10.1186/1479-5876-8-13
31. Hu JM, Liu K, Liu JH, Jiang XL, Wang XL, Yang L, et al. The Increased Number of Tumor-Associated Macrophage Is Associated With Overexpression of Vegf-C, Plays an Important Role in Kazakh Escc Invasion and Metastasis. Exp Mol Pathol (2017) 102(1):15–21. doi: 10.1016/j.yexmp.2016.12.001
32. Miura T, Yoshizawa T, Hirai H, Seino H, Morohashi S, Wu Y, et al. Prognostic Impact of Cd163+ Macrophages in Tumor Stroma and Cd8+ T-Cells in Cancer Cell Nests in Invasive Extrahepatic Bile Duct Cancer. Anticancer Res (2017) 37(1):183–90. doi: 10.21873/anticanres.11304
33. Ugai T, Väyrynen JP, Lau MC, Borowsky J, Akimoto N, Väyrynen SA, et al. Immune Cell Profiles in the Tumor Microenvironment of Early-Onset, Intermediate-Onset, and Later-Onset Colorectal Cancer. Cancer Immunol Immunother (2022) 71(4):933–42. doi: 10.1007/s00262-021-03056-6
34. Jan AT, Rahman S, Khan S, Tasduq SA, Choi I. Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal. Cells (2019) 8(2):99. doi: 10.3390/cells8020099
35. Harding C, Heuser J, Stahl P. Receptor-Mediated Endocytosis of Transferrin and Recycling of the Transferrin Receptor in Rat Reticulocytes. J Cell Biol (1983) 97(2):329–39. doi: 10.1083/jcb.97.2.329
36. Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, Biogenesis and Function. Nat Rev Immunol (2002) 2(8):569–79. doi: 10.1038/nri855
37. Kalluri R, LeBleu VS. The Biology, Function, and Biomedical Applications of Exosomes. Science (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
38. Chen Q, Li Y, Liu Y, Xu W, Zhu X. Exosomal Non-Coding Rnas-Mediated Crosstalk in the Tumor Microenvironment. Front Cell Dev Biol (2021) 9:646864. doi: 10.3389/fcell.2021.646864
39. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-To-Cell Communication. Nat Cell Biol (2019) 21(1):9–17. doi: 10.1038/s41556-018-0250-9
40. Corrado C, Barreca MM, Zichittella C, Alessandro R, Conigliaro A. Molecular Mediators of RNA Loading Into Extracellular Vesicles. Cells (2021) 10(12):3355. doi: 10.3390/cells10123355
41. Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell (2016) 164(6):1226–32. doi: 10.1016/j.cell.2016.01.043
42. Zhang F, Guo J, Zhang Z, Qian Y, Wang G, Duan M, et al. Mesenchymal Stem Cell-Derived Exosome: A Tumor Regulator and Carrier for Targeted Tumor Therapy. Cancer Lett (2022) 526:29–40. doi: 10.1016/j.canlet.2021.11.015
43. Bahrami A, Moradi Binabaj M, Gordon AF. Exosomes: Emerging Modulators of Signal Transduction in Colorectal Cancer From Molecular Understanding to Clinical Application. BioMed Pharmacother (2021) 141:111882. doi: 10.1016/j.biopha.2021.111882
44. Li Q, Wang D, Ding D, Feng Y, Hou R, Liu D, et al. The Role and Application of Exosomes in Gastric and Colorectal Cancer. Front Pharmacol (2021) 12:825475. doi: 10.3389/fphar.2021.825475
45. Yang J, Zhang L. The Roles and Therapeutic Approaches of Msc-Derived Exosomes in Colorectal Cancer. Clin Transl Oncol (2022). doi: 10.1007/s12094-021-02750-2
46. Dai G, Yao X, Zhang Y, Gu J, Geng Y, Xue F, et al. Colorectal Cancer Cell-Derived Exosomes Containing Mir-10b Regulate Fibroblast Cells Via the Pi3k/Akt Pathway. Bull Cancer (2018) 105(4):336–49. doi: 10.1016/j.bulcan.2017.12.009
47. Xu Y, Shen L, Li F, Yang J, Wan X, Ouyang M. Microrna-16-5p-Containing Exosomes Derived From Bone Marrow-Derived Mesenchymal Stem Cells Inhibit Proliferation, Migration, and Invasion, While Promoting Apoptosis of Colorectal Cancer Cells by Downregulating Itga2. J Cell Physiol (2019) 234(11):21380–94. doi: 10.1002/jcp.28747
48. Tian F, Wang P, Lin D, Dai J, Liu Q, Guan Y, et al. Exosome-Delivered Mir-221/222 Exacerbates Tumor Liver Metastasis by Targeting Spint1 in Colorectal Cancer. Cancer Sci (2021) 112(9):3744–55. doi: 10.1111/cas.15028
49. Wang Y, Lin C. Exosomes Mir-22-3p Derived From Mesenchymal Stem Cells Suppress Colorectal Cancer Cell Proliferation and Invasion by Regulating Rap2b and Pi3k/Akt Pathway. J Oncol (2021) 2021:3874478. doi: 10.1155/2021/3874478
50. Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, et al. Colorectal Cancer-Derived Exosomal Mir-106b-3p Promotes Metastasis by Down-Regulating Dlc-1 Expression. Clin Sci (Lond) (2020) 134(4):419–34. doi: 10.1042/cs20191087
51. Zhang Y, Wang S, Lai Q, Fang Y, Wu C, Liu Y, et al. Cancer-Associated Fibroblasts-Derived Exosomal Mir-17-5p Promotes Colorectal Cancer Aggressive Phenotype by Initiating a Runx3/Myc/Tgf-B1 Positive Feedback Loop. Cancer Lett (2020) 491:22–35. doi: 10.1016/j.canlet.2020.07.023
52. Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y, et al. Exosomal Malat1 Sponges Mir-26a/26b to Promote the Invasion and Metastasis of Colorectal Cancer Via Fut4 Enhanced Fucosylation and Pi3k/Akt Pathway. J Exp Clin Cancer Res (2020) 39(1):54. doi: 10.1186/s13046-020-01562-6
53. Zhou L, Li J, Tang Y, Yang M. Exosomal Lncrna Linc00659 Transferred From Cancer-Associated Fibroblasts Promotes Colorectal Cancer Cell Progression Via Mir-342-3p/Anxa2 Axis. J Transl Med (2021) 19(1):8. doi: 10.1186/s12967-020-02648-7
54. Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, et al. Lncrna Uca1, Upregulated in Crc Biopsies and Downregulated in Serum Exosomes, Controls Mrna Expression by Rna-Rna Interactions. Mol Ther Nucleic Acids (2018) 12:229–41. doi: 10.1016/j.omtn.2018.05.009
55. Yin H, Hu J, Ye Z, Chen S, Chen Y. Serum Long Non−Coding Rna Nnt−As1 Protected by Exosome Is a Potential Biomarker and Functions as an Oncogene Via the Mir−496/Rap2c Axis in Colorectal Cancer. Mol Med Rep (2021) 24(2):585. doi: 10.3892/mmr.2021.12224
56. Zhao H, Chen S, Fu Q. Exosomes From Cd133(+) Cells Carrying Circ-Abcc1 Mediate Cell Stemness and Metastasis in Colorectal Cancer. J Cell Biochem (2020) 121(5-6):3286–97. doi: 10.1002/jcb.29600
57. Yang Y, Yang N, Jiang J. Exosomal Circ_Ptpra Inhibits Tumorigenesis and Promotes Radiosensitivity in Colorectal Cancer by Enriching the Level of Smad4 Via Competitively Binding to Mir-671-5p. Cytotechnology (2022) 74(1):51–64. doi: 10.1007/s10616-021-00506-y
58. Gao L, Tang X, He Q, Sun G, Wang C, Qu H. Exosome-Transmitted Circcog2 Promotes Colorectal Cancer Progression Via Mir-1305/Tgf-B2/Smad3 Pathway. Cell Death Discovery (2021) 7(1):281. doi: 10.1038/s41420-021-00680-0
59. Jiang Z, Hou Z, Li L, Liu W, Yu Z, Chen S. Exosomal Circepb41l2 Serves as a Sponge for Mir-21-5p and Mir-942-5p to Suppress Colorectal Cancer Progression by Regulating the Pten/Akt Signalling Pathway. Eur J Clin Invest (2021) 51(9):e13581. doi: 10.1111/eci.13581
60. Hammond SM. An Overview of Micrornas. Adv Drug Delivery Rev (2015) 87:3–14. doi: 10.1016/j.addr.2015.05.001
61. Thakur A, Parra DC, Motallebnejad P, Brocchi M, Chen HJ. Exosomes: Small Vesicles With Big Roles in Cancer, Vaccine Development, and Therapeutics. Bioact Mater (2022) 10:281–94. doi: 10.1016/j.bioactmat.2021.08.029
62. Kahlert C, Kalluri R. Exosomes in Tumor Microenvironment Influence Cancer Progression and Metastasis. J Mol Med (Berl) (2013) 91(4):431–7. doi: 10.1007/s00109-013-1020-6
63. Garcia-Martin R, Wang G, Brandão BB, Zanotto TM, Shah S, Kumar Patel S, et al. Microrna Sequence Codes for Small Extracellular Vesicle Release and Cellular Retention. Nature (2022) 601(7893):446–51. doi: 10.1038/s41586-021-04234-3
64. Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal Microrna in Serum Is a Novel Biomarker of Recurrence in Human Colorectal Cancer. Br J Cancer (2015) 113(2):275–81. doi: 10.1038/bjc.2015.201
65. Hosseini M, Baghaei K, Hajivalili M, Zali MR, Ebtekar M, Amani D. The Anti-Tumor Effects of Ct-26 Derived Exosomes Enriched by Microrna-34a on Murine Model of Colorectal Cancer. Life Sci (2022) 290:120234. doi: 10.1016/j.lfs.2021.120234
66. Xu J, Bai J, Zhang X, Lv Y, Gong Y, Liu L, et al. A Comprehensive Overview of Lncrna Annotation Resources. Brief Bioinform (2017) 18(2):236–49. doi: 10.1093/bib/bbw015
67. Park EG, Pyo SJ, Cui Y, Yoon SH, Nam JW. Tumor Immune Microenvironment Lncrnas. Brief Bioinform (2021) 12(1):123. doi: 10.1093/bib/bbab504
68. Quinn JJ, Chang HY. Unique Features of Long Non-Coding Rna Biogenesis and Function. Nat Rev Genet (2016) 17(1):47–62. doi: 10.1038/nrg.2015.10
69. Nie H, Liao Z, Wang Y, Zhou J, He X, Ou C. Exosomal Long Non-Coding Rnas: Emerging Players in Cancer Metastasis and Potential Diagnostic Biomarkers for Personalized Oncology. Genes Dis (2021) 8(6):769–80. doi: 10.1016/j.gendis.2020.12.004
70. Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, et al. Long Noncoding Rna Ccal Transferred From Fibroblasts by Exosomes Promotes Chemoresistance of Colorectal Cancer Cells. Int J Cancer (2020) 146(6):1700–16. doi: 10.1002/ijc.32608
71. Sun J, Jia H, Bao X, Wu Y, Zhu T, Li R, et al. Tumor Exosome Promotes Th17 Cell Differentiation by Transmitting the Lncrna Crnde-H in Colorectal Cancer. Cell Death Dis (2021) 12(1):123. doi: 10.1038/s41419-020-03376-y
72. Chen LL. The Expanding Regulatory Mechanisms and Cellular Functions of Circular Rnas. Nat Rev Mol Cell Biol (2020) 21(8):475–90. doi: 10.1038/s41580-020-0243-y
73. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular Rna Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
74. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular Rnas Are Down-Regulated in Kras Mutant Colon Cancer Cells and Can Be Transferred to Exosomes. Sci Rep (2016) 6:37982. doi: 10.1038/srep37982
75. Li J, Zhang G, Liu CG, Xiang X, Le MTN, Sethi G, et al. The Potential Role of Exosomal Circrnas in the Tumor Microenvironment: Insights Into Cancer Diagnosis and Therapy. Theranostics (2022) 12(1):87–104. doi: 10.7150/thno.64096
76. Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. Circrnas and Exosomes: A Mysterious Frontier for Human Cancer. Mol Ther Nucleic Acids (2020) 19:384–92. doi: 10.1016/j.omtn.2019.11.023
77. Hamidi AA, Khalili-Tanha G, Nasrpour Navaei Z, Moghbeli M. Long Non-Coding Rnas as the Critical Regulators of Epithelial Mesenchymal Transition in Colorectal Tumor Cells: An Overview. Cancer Cell Int (2022) 22(1):71. doi: 10.1186/s12935-022-02501-5
78. Pietilä M, Ivaska J, Mani SA. Whom to Blame for Metastasis, the Epithelial-Mesenchymal Transition or the Tumor Microenvironment? Cancer Lett (2016) 380(1):359–68. doi: 10.1016/j.canlet.2015.12.033
79. Huang Z, Yang M, Li Y, Yang F, Feng Y. Exosomes Derived From Hypoxic Colorectal Cancer Cells Transfer Wnt4 to Normoxic Cells to Elicit a Prometastatic Phenotype. Int J Biol Sci (2018) 14(14):2094–102. doi: 10.7150/ijbs.28288
80. Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, et al. The Emerging Role of Exosome-Derived Non-Coding Rnas in Cancer Biology. Cancer Lett (2018) 414:107–15. doi: 10.1016/j.canlet.2017.10.040
81. Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging Role of Exosome-Derived Long Non-Coding Rnas in Tumor Microenvironment. Mol Cancer (2018) 17(1):82. doi: 10.1186/s12943-018-0831-z
82. Liu Y, Cao X. Characteristics and Significance of the Pre-Metastatic Niche. Cancer Cell (2016) 30(5):668–81. doi: 10.1016/j.ccell.2016.09.011
83. Taghvimi S, Vakili O, Soltani Fard E, Khatami SH, Karami N, Taheri-Anganeh M, et al. Exosomal Micrornas and Long Noncoding Rnas: Novel Mediators of Drug Resistance in Lung Cancer. J Cell Physiol (2022) 237(4):2095–106. doi: 10.1002/jcp.30697
84. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
85. Potente M, Gerhardt H, Carmeliet P. Basic and Therapeutic Aspects of Angiogenesis. Cell (2011) 146(6):873–87. doi: 10.1016/j.cell.2011.08.039
86. Tanaka S, Tatsuguchi A, Futagami S, Gudis K, Wada K, Seo T, et al. Monocyte Chemoattractant Protein 1 and Macrophage Cyclooxygenase 2 Expression in Colonic Adenoma. Gut (2006) 55(1):54–61. doi: 10.1136/gut.2004.059824
87. da Costa VR, Araldi RP, Vigerelli H, D'Ámelio F, Mendes TB, Gonzaga V, et al. Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications. Cells (2021) 10(10):2617. doi: 10.3390/cells10102617
88. Conti I, Varano G, Simioni C, Laface I, Milani D, Rimondi E, et al. Mirnas as Influencers of Cell-Cell Communication in Tumor Microenvironment. Cells (2020) 9(1):220. doi: 10.3390/cells9010220
89. He Q, Ye A, Ye W, Liao X, Qin G, Xu Y, et al. Cancer-Secreted Exosomal Mir-21-5p Induces Angiogenesis and Vascular Permeability by Targeting Krit1. Cell Death Dis (2021) 12(6):576. doi: 10.1038/s41419-021-03803-8
90. Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, et al. Exosomal Mir-1229 Derived From Colorectal Cancer Cells Promotes Angiogenesis by Targeting Hipk2. Int J Biol Macromol (2019) 132:470–7. doi: 10.1016/j.ijbiomac.2019.03.221
91. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer-Derived Exosomal Mir-25-3p Promotes Pre-Metastatic Niche Formation by Inducing Vascular Permeability and Angiogenesis. Nat Commun (2018) 9(1):5395. doi: 10.1038/s41467-018-07810-w
92. Dokhanchi M, Pakravan K, Zareian S, Hussen BM, Farid M, Razmara E, et al. Colorectal Cancer Cell-Derived Extracellular Vesicles Transfer Mir-221-3p to Promote Endothelial Cell Angiogenesis Via Targeting Suppressor of Cytokine Signaling 3. Life Sci (2021) 285:119937. doi: 10.1016/j.lfs.2021.119937
93. Shang A, Wang X, Gu C, Liu W, Sun J, Zeng B, et al. Exosomal Mir-183-5p Promotes Angiogenesis in Colorectal Cancer by Regulation of Foxo1. Aging (Albany NY) (2020) 12(9):8352–71. doi: 10.18632/aging.103145
94. Dou R, Liu K, Yang C, Zheng J, Shi D, Lin X, et al. Emt-Cancer Cells-Derived Exosomal Mir-27b-3p Promotes Circulating Tumour Cells-Mediated Metastasis by Modulating Vascular Permeability in Colorectal Cancer. Clin Transl Med (2021) 11(12):e595. doi: 10.1002/ctm2.595
95. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune Cells Within the Tumor Microenvironment: Biological Functions and Roles in Cancer Immunotherapy. Cancer Lett (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
96. Natua S, Dhamdhere SG, Mutnuru SA, Shukla S. Interplay Within Tumor Microenvironment Orchestrates Neoplastic RNA Metabolism and Transcriptome Diversity. Wiley Interdiscip Rev RNA (2021) 13(2):e1676. doi: 10.1002/wrna.1676
97. Dunn GP, Old LJ, Schreiber RD. The Three Es of Cancer Immunoediting. Annu Rev Immunol (2004) 22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803
98. Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New Insights Into Cancer Immunoediting and Its Three Component Phases–Elimination, Equilibrium and Escape. Curr Opin Immunol (2014) 27:16–25. doi: 10.1016/j.coi.2014.01.004
99. Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity (2018) 48(3):399–416. doi: 10.1016/j.immuni.2018.03.004
100. Deyell M, Garris CS, Laughney AM. Cancer Metastasis as a Non-Healing Wound. Br J Cancer (2021) 124(9):1491–502. doi: 10.1038/s41416-021-01309-w
101. Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-Derived Exosomes Regulate Expression of Immune Function-Related Genes in Human T Cell Subsets. Sci Rep (2016) 6:20254. doi: 10.1038/srep20254
102. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal Circpacrgl Promotes Progression of Colorectal Cancer Via the Mir-142-3p/Mir-506-3p- Tgf-B1 Axis. Mol Cancer (2020) 19(1):117. doi: 10.1186/s12943-020-01235-0
103. Liu J, Peng X, Yang S, Li X, Huang M, Wei S, et al. Extracellular Vesicle Pd-L1 in Reshaping Tumor Immune Microenvironment: Biological Function and Potential Therapy Strategies. Cell Commun Signal (2022) 20(1):14. doi: 10.1186/s12964-021-00816-w
104. Xian D, Niu L, Zeng J, Wang L. Lncrna Kcnq1ot1 Secreted by Tumor Cell-Derived Exosomes Mediates Immune Escape in Colorectal Cancer by Regulating Pd-L1 Ubiquitination Via Mir-30a-5p/Usp22. Front Cell Dev Biol (2021) 9:653808. doi: 10.3389/fcell.2021.653808
105. Huang Y, Luo Y, Ou W, Wang Y, Dong D, Peng X, et al. Exosomal Lncrna Snhg10 Derived From Colorectal Cancer Cells Suppresses Natural Killer Cell Cytotoxicity by Upregulating Inhbc. Cancer Cell Int (2021) 21(1):528. doi: 10.1186/s12935-021-02221-2
106. Chanmee T, Ontong P, Konno K, Itano N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers (Basel) (2014) 6(3):1670–90. doi: 10.3390/cancers6031670
107. Doak GR, Schwertfeger KL, Wood DK. Distant Relations: Macrophage Functions in the Metastatic Niche. Trends Cancer (2018) 4(6):445–59. doi: 10.1016/j.trecan.2018.03.011
108. Brown JM, Recht L, Strober S. The Promise of Targeting Macrophages in Cancer Therapy. Clin Cancer Res (2017) 23(13):3241–50. doi: 10.1158/1078-0432.Ccr-16-3122
109. Wang D, Wang X, Si M, Yang J, Sun S, Wu H, et al. Exosome-Encapsulated Mirnas Contribute to Cxcl12/Cxcr4-Induced Liver Metastasis of Colorectal Cancer by Enhancing M2 Polarization of Macrophages. Cancer Lett (2020) 474:36–52. doi: 10.1016/j.canlet.2020.01.005
110. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, et al. Tumor-Derived Exosomal Mir-934 Induces Macrophage M2 Polarization to Promote Liver Metastasis of Colorectal Cancer. J Hematol Oncol (2020) 13(1):156. doi: 10.1186/s13045-020-00991-2
111. Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, et al. Lncrna Rpph1 Promotes Colorectal Cancer Metastasis by Interacting With Tubb3 and by Promoting Exosomes-Mediated Macrophage M2 Polarization. Cell Death Dis (2019) 10(11):829. doi: 10.1038/s41419-019-2077-0
112. Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab (2016) 23(1):27–47. doi: 10.1016/j.cmet.2015.12.006
113. Tao L, Xu C, Shen W, Tan J, Li L, Fan M, et al. Hipk3 Inhibition by Exosomal Hsa-Mir-101-3p Is Related to Metabolic Reprogramming in Colorectal Cancer. Front Oncol (2021) 11:758336. doi: 10.3389/fonc.2021.758336
114. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-Delivered Circrna Promotes Glycolysis to Induce Chemoresistance Through the Mir-122-Pkm2 Axis in Colorectal Cancer. Mol Oncol (2020) 14(3):539–55. doi: 10.1002/1878-0261.12629
115. Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, et al. Exosomal Circrna Derived From Gastric Tumor Promotes White Adipose Browning by Targeting the Mir-133/Prdm16 Pathway. Int J Cancer (2019) 144(10):2501–15. doi: 10.1002/ijc.31977
116. Di W, Zhang W, Zhu B, Li X, Tang Q, Zhou Y. Colorectal Cancer Prompted Adipose Tissue Browning and Cancer Cachexia Through Transferring Exosomal Mir-146b-5p. J Cell Physiol (2021) 236(7):5399–410. doi: 10.1002/jcp.30245
117. Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, et al. Hypoxia Induced Exosomal Circrna Promotes Metastasis of Colorectal Cancer Via Targeting Gef-H1/Rhoa Axis. Theranostics (2020) 10(18):8211–26. doi: 10.7150/thno.44419
118. Yang K, Zhang J, Bao C. Exosomal Circeif3k From Cancer-Associated Fibroblast Promotes Colorectal Cancer (Crc) Progression Via Mir-214/Pd-L1 Axis. BMC Cancer (2021) 21(1):933. doi: 10.1186/s12885-021-08669-9
119. Seebacher NA, Krchniakova M, Stacy AE, Skoda J, Jansson PJ. Tumour Microenvironment Stress Promotes the Development of Drug Resistance. Antioxidants (Basel) (2021) 10(11):1801. doi: 10.3390/antiox10111801
120. Yun BD, Choi YJ, Son SW, Cipolla GA, Berti FCB, Malheiros D, et al. Oncogenic Role of Exosomal Circular and Long Noncoding Rnas in Gastrointestinal Cancers. Int J Mol Sci (2022) 23(2):930. doi: 10.3390/ijms23020930
121. Shakeran Z, Varshosaz J, Keyhanfar M, Mohammad-Beigi H, Rahimi K, Sutherland DS. Co-Delivery of Stat3 Sirna and Methotrexate in Breast Cancer Cells. Artif Cells Nanomed Biotechnol (2022) 50(1):29–39. doi: 10.1080/21691401.2022.2030746
122. Zhang HW, Shi Y, Liu JB, Wang HM, Wang PY, Wu ZJ, et al. Cancer-Associated Fibroblast-Derived Exosomal Microrna-24-3p Enhances Colon Cancer Cell Resistance to Mtx by Down-Regulating Cdx2/Heph Axis. J Cell Mol Med (2021) 25(8):3699–713. doi: 10.1111/jcmm.15765
123. Ning T, Li J, He Y, Zhang H, Wang X, Deng T, et al. Exosomal Mir-208b Related With Oxaliplatin Resistance Promotes Treg Expansion in Colorectal Cancer. Mol Ther (2021) 29(9):2723–36. doi: 10.1016/j.ymthe.2021.04.028
124. Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N. Extracellular Vesicle-Derived Circular Rnas Confers Chemoresistance in Colorectal Cancer. Sci Rep (2019) 9(1):16497. doi: 10.1038/s41598-019-53063-y
125. Xu F, Ye ML, Zhang YP, Li WJ, Li MT, Wang HZ, et al. Microrna-375-3p Enhances Chemosensitivity to 5-Fluorouracil by Targeting Thymidylate Synthase in Colorectal Cancer. Cancer Sci (2020) 111(5):1528–41. doi: 10.1111/cas.14356
126. Zhang Q, Liu RX, Chan KW, Hu J, Zhang J, Wei L, et al. Exosomal Transfer of P-Stat3 Promotes Acquired 5-Fu Resistance in Colorectal Cancer Cells. J Exp Clin Cancer Res (2019) 38(1):320. doi: 10.1186/s13046-019-1314-9
127. Pan S, Deng Y, Fu J, Zhang Y, Zhang Z, Qin X. N6−Methyladenosine Upregulates Mir−181d−5p in Exosomes Derived From Cancer−Associated Fibroblasts to Inhibit 5−Fu Sensitivity by Targeting Ncald in Colorectal Cancer. Int J Oncol (2022) 60(2):14. doi: 10.3892/ijo.2022.5304
128. Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, et al. Cafs Secreted Exosomes Promote Metastasis and Chemotherapy Resistance by Enhancing Cell Stemness and Epithelial-Mesenchymal Transition in Colorectal Cancer. Mol Cancer (2019) 18(1):91. doi: 10.1186/s12943-019-1019-x
129. Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, et al. Carcinoma-Associated Fibroblasts Promote the Stemness and Chemoresistance of Colorectal Cancer by Transferring Exosomal Lncrna H19. Theranostics (2018) 8(14):3932–48. doi: 10.7150/thno.25541
130. Durán-Vinet B, Araya-Castro K, Calderón J, Vergara L, Weber H, Retamales J, et al. Crispr/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer Through Exosomes Micro-Rna Detection: A Review. Cancers (Basel) (2021) 13(18):4640. doi: 10.3390/cancers13184640
131. Nabariya DK, Pallu R, Yenuganti VR. Exosomes: The Protagonists in the Tale of Colorectal Cancer? Biochim Biophys Acta Rev Cancer (2020) 1874(2):188426. doi: 10.1016/j.bbcan.2020.188426
132. He X, Qi Y, Zhang X, Liu X, Li X, Li S, et al. Current Landscape of Tumor-Derived Exosomal Ncrnas in Glioma Progression, Detection, and Drug Resistance. Cell Death Dis (2021) 12(12):1145. doi: 10.1038/s41419-021-04430-z
133. Umwali Y, Yue CB, Gabriel ANA, Zhang Y, Zhang X. Roles of Exosomes in Diagnosis and Treatment of Colorectal Cancer. World J Clin cases (2021) 9(18):4467–79. doi: 10.12998/wjcc.v9.i18.4467
134. Min L, Zhu S, Chen L, Liu X, Wei R, Zhao L, et al. Evaluation of Circulating Small Extracellular Vesicles Derived Mirnas as Biomarkers of Early Colon Cancer: A Comparison With Plasma Total Mirnas. J Extracell Vesicles (2019) 8(1):1643670. doi: 10.1080/20013078.2019.1643670
135. Pan B, Qin J, Liu X, He B, Wang X, Pan Y, et al. Identification of Serum Exosomal Hsa-Circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front Genet (2019) 10:1096. doi: 10.3389/fgene.2019.01096
136. Cho WC, Kim M, Park JW, Jeong SY, Ku JL. Exosomal Mir-193a and Let-7g Accelerate Cancer Progression on Primary Colorectal Cancer and Paired Peritoneal Metastatic Cancer. Transl Oncol (2021) 14(2):101000. doi: 10.1016/j.tranon.2020.101000
137. Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, et al. Exosomal Circlpar1 Functions in Colorectal Cancer Diagnosis and Tumorigenesis Through Suppressing Brd4 Via Mettl3-Eif3h Interaction. Mol Cancer (2022) 21(1):49. doi: 10.1186/s12943-021-01471-y
138. Gao T, Liu X, He B, Nie Z, Zhu C, Zhang P, et al. Exosomal Lncrna 91h Is Associated With Poor Development in Colorectal Cancer by Modifying Hnrnpk Expression. Cancer Cell Int (2018) 18:11. doi: 10.1186/s12935-018-0506-2
139. Zhao X, Wu D, Ma X, Wang J, Hou W, Zhang W. Exosomes as Drug Carriers for Cancer Therapy and Challenges Regarding Exosome Uptake. BioMed Pharmacother (2020) 128:110237. doi: 10.1016/j.biopha.2020.110237
140. Donoso-Quezada J, Ayala-Mar S, González-Valdez J. State-Of-the-Art Exosome Loading and Functionalization Techniques for Enhanced Therapeutics: A Review. Crit Rev Biotechnol (2020) 40(6):804–20. doi: 10.1080/07388551.2020.1785385
141. Shao J, Zaro J, Shen Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int J Nanomedicine (2020) 15:9355–71. doi: 10.2147/ijn.S281890
142. Huang X, Wu W, Jing D, Yang L, Guo H, Wang L, et al. Engineered Exosome as Targeted Lncrna Meg3 Delivery Vehicles for Osteosarcoma Therapy. J Control Release (2022) 343:107–17. doi: 10.1016/j.jconrel.2022.01.026
143. Zhang J, Ji C, Zhang H, Shi H, Mao F, Qian H, et al. Engineered Neutrophil-Derived Exosome-Like Vesicles for Targeted Cancer Therapy. Sci Adv (2022) 8(2):eabj8207. doi: 10.1126/sciadv.abj8207
144. Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, et al. A Doxorubicin Delivery Platform Using Engineered Natural Membrane Vesicle Exosomes for Targeted Tumor Therapy. Biomaterials (2014) 35(7):2383–90. doi: 10.1016/j.biomaterials.2013.11.083
145. Bagheri E, Abnous K, Farzad SA, Taghdisi SM, Ramezani M, Alibolandi M. Targeted Doxorubicin-Loaded Mesenchymal Stem Cells-Derived Exosomes as a Versatile Platform for Fighting Against Colorectal Cancer. Life Sci (2020) 261:118369. doi: 10.1016/j.lfs.2020.118369
146. Asadirad A, Baghaei K, Hashemi SM, Dehnavi S, Ghanbarian H, Mortaz E, et al. Dendritic Cell Immunotherapy With Mir-155 Enriched Tumor-Derived Exosome Suppressed Cancer Growth and Induced Antitumor Immune Responses in Murine Model of Colorectal Cancer Induced by Ct26 Cell Line. Int Immunopharmacol (2022) 104:108493. doi: 10.1016/j.intimp.2021.108493
147. Zhan Q, Yi K, Qi H, Li S, Li X, Wang Q, et al. Engineering Blood Exosomes for Tumor-Targeting Efficient Gene/Chemo Combination Therapy. Theranostics (2020) 10(17):7889–905. doi: 10.7150/thno.45028
Keywords: colorectal cancer, exosome, ncRNAs, tumor microenvironment, tumor biomarker
Citation: Chen X, Jia M, Ji J, Zhao Z and Zhao Y (2022) Exosome-Derived Non-Coding RNAs in the Tumor Microenvironment of Colorectal Cancer: Possible Functions, Mechanisms and Clinical Applications. Front. Oncol. 12:887532. doi: 10.3389/fonc.2022.887532
Received: 01 March 2022; Accepted: 19 April 2022;
Published: 12 May 2022.
Edited by:
Zhijie Xu, Central South University, ChinaReviewed by:
Chao Chen, Nanjing University of Chinese Medicine, ChinaCopyright © 2022 Chen, Jia, Ji, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjie Zhao, emhhb3lqQHFkdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.