- Department of Endoscopy, The First Affiliated Hospital of China Medical University, Shenyang, China
Worldwide, colorectal cancer (CRC) ranks as the third most common malignancy, and the second most deadly with nearly one million attributable deaths in 2020. Metastatic disease is present in nearly 25% of newly diagnosed CRC, and despite advances in chemotherapy, less than 20% will remain alive at 5 years. Epigenetic change plays a key role in the epithelial-to-mesenchymal transition (EMT), which is a crucial phenotype for metastasis and mainly includes DNA methylation, non-coding RNAs (ncRNAs), and N6-methyladenosine (m6A) RNA, seemingly valuable biomarkers in CRCs. For ncRNAs, there exists a “molecular sponge effect” between long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs). The detection of exosomes is a novel method in CRC monitoring, especially for predicting metastasis. There is a close relationship between exosomes and EMT in CRCs. This review summarizes the close relationship between epigenetic changes and EMT in CRCs and emphasizes the crucial function of exosomes in regulating the EMT process.
Introduction
In 2020, colorectal cancers (CRCs) have become significant public health problems worldwide, and their incidence has increased to twice in the top 10 cancers (1–3). Monitoring distant metastatic loci and recurrence in situ is the last and most crucial step for cancer treatment. Furthermore, it is the most common cause of concern and anxiety for patients with CRCs. Based on previous studies, the mortality of metastatic CRCs is much higher than that of primary CRCs, especially for liver metastasis. Metastatic cancers severely influence the 5-year survival rate and quality of life of patients (4). Therefore, it is pivotal to predict the metastasis and recurrence of CRCs in order to increase distal survival time and quality of life.
For CRC, epithelial–mesenchymal transition (EMT) is a vital phenotype in its metastasis. EMT is a reversible process that promotes tumor cells exfoliating into circulation and damaging the intercellular skeletal structure. These exfoliated tumor cells could reach distal cites and form metastatic loci. The transcription factors (TFs) SNAI1, SNAI2, Zeb1, Zeb2, and Twist, called EMT-TFs, and some non-coding RNAs (ncRNAs) can influence the EMT process to regulate the migration and infiltration of tumor cells (5, 6). EMT is a dynamic process that is affected by genetic, epigenetic, and immune environmental factors. N-cadherin, vimentin, matrix metalloproteinases (MMPs), E-cadherins, claudins, epithelial cell adhesion molecules (EpCAMs), and cytokeratins are common biomarkers for the detection of EMT or its reverse process, mesenchymal–epithelial transition (MET). EMT is an essential phenotype in the study of tumor metastasis.
The liquid biopsy (LB) technology is a novel method used to detect the genesis, progress of recurrence, and metastasis in CRC patients. Circulating tumor cells (CTCs), circulating tumor DNAs (ctDNAs), and cell-free RNAs are detectable biomarkers in serum (7). In addition, the detection of exosomes in the urine, cerebrospinal fluid, and saliva is applied widely and clinically. Detecting genetic materials has more perspective as nano-scale biological vesicles that are stable, suitable for storage, and valuable as both biomarkers and therapeutic nano-drugs, exosomes likely have a brighter future than cells (8).
Exosomes are tiny vesicles derived from tumor cells that contain DNA, ncRNAs, epigenetic modulation, and metabolites. Especially in comparison to obtaining tissue biopsies, liquid biopsies, including the relatively noninvasive venipuncture, or sampling other bodily fluids is often safer and more convenient. According to previous studies, exosomes play a pivotal role in predicting cancer metastasis. Comparably, exosome detection is safer and more convenient in malignant cancers, especially for CRCs. LB has shown considerable accuracy in CRC testing. Its sensitivity and specificity can reach up to 60% and 90%, respectively (9, 10). Therefore, this review summarized the relationship between epigenetic changes and EMT in CRCs and emphasized the association between exosomes and EMT.
Epigenetic Changes and the EMT Process in CRC
The reverse EMT process is characterized by the downregulation of E-cadherin, desmoplakin, claudins, and β-catenin and the upregulation of N-cadherin, vimentin, fibronectin, and Snail1/Snail2 mediate the migration and invasive metastasis of cancer cells (11–13). Whether to promote or inhibit is a question that can be answered using qualified biomarkers in EMT and reflects their influence on tumor migration, infiltration, or more aggressive properties. This process may not be complete; partial EMT is a novel phenomenon that has been recently put forward and proven to be more related to a malignant phenotype. It provides a warning that paying more attention to the regulators during the EMT process is more critical than just the results.
The chromosomal instability (CIN), microsatellite instability (MSI), and the CpG island (CGI) methylator phenotype (CIMP) pathways are three principle pathways involved in the genesis and development of CRCs. CIMP is related to the hypermethylation of promoters or the upstream regulator regions that usually contain CGIs, reported to be related to the detection of CRCs (14, 15). The definition of CIMP depends on the number of methylated loci on these CGIs; these also decide the survival outcomes and long-term prognosis of CRCs (16–18). The CIMP pathway could predict metastasis and recurrence via the detection of epigenetic biomarkers, such as DNA methylation, overexpression of ncRNAs, and N6-methyladenosine (m6A) modification. These epigenetic changes are regulated by genes and can be inherited by the next generation, but they cannot be transmitted into proteins to form phenotypes. Their influence is so strong that they have gained wide attention (19, 20). Several signaling pathways such as the Wnt signaling pathway, Raf/MEK/ERK pathway, and the NF-κB pathway function in CRCs. Especially for the Wnt signaling pathway, canonical Wnt signaling is frequently activated in the tumorigenesis of CRCs, with loss of the adenomatous polyposis coli (APC) genes and the translocation of β-catenin into the nucleus (21–23).
Regarding epigenetic changes, 5-methylcytosine (5mC) methylation for DNA and m6A modification for RNA are two epigenetic modifications affecting the transcription and expression of critical genes. In addition, ncRNA is another group that cannot be transcribed, but has high regulatory ability for coding genes. These two mechanisms can undoubtedly regulate the EMT process in CRC metastatic events.
DNA 5mC Methylation: A Robust Prognostic Biomarker in CRC
DNA methylation occurs when a methyl group is transferred to the carbon-5′ position of the cytosine base (5-methylcytosine) concentrated in CGIs under the catalytic action of DNA methyltransferase enzymes (24). The hypermethylation of 5-mC can regulate not only the transcription elements but also alternative splicing, which can modulate the production of ncRNAs (25–27). The methylation of promoters could weaken the transcription activity into messenger RNA (mRNA) and affect the function of the translated proteins. The intensity of their influence depends on the methylated sites on CGIs, which is differentially sensitive to the distance between the CpG sites among groups (28, 29).
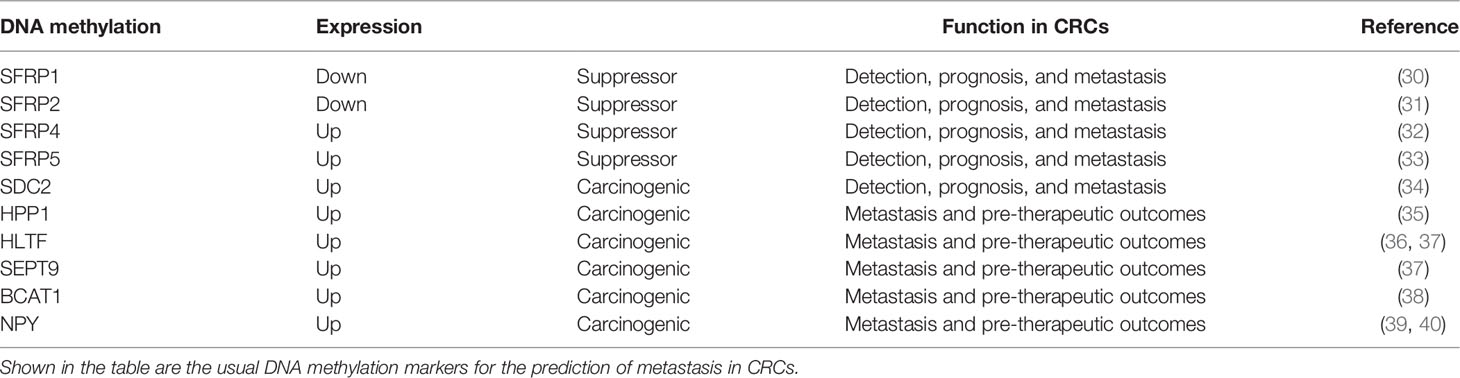
This review focuses on several common and proven effective DNA methylation indicators for CRCs, shown in Table 1. The secreted frizzled-related protein (SFRP) family is regarded as an antagonist of the Wnt signaling pathway for its similarity to frizzled (Fz) receptors with bidirectional function (41). SFRP1 and SFRP2 are expressed in stromal myofibroblasts and are downregulated from typical adjacent tumor (normal tissue adjacent to the tumor, NAT) tissues toward the tumor (30, 31, 42, 43). They are diagnostic and prognostic biomarkers and suppress the growth and metastasis of CRC tumor cells. SFRP4 and SFRP5 may also inhibit invasion in CRC, but the underlying mechanisms in regulating the signaling pathways are more complex by comparison (32, 33, 44). Additional studies focusing on the SFRPs family as it relates to CRC should be undertaken to clarify their overall role and potentially derive a novel therapeutic regimen. Different from that of SFRPs, SDC2 methylation is a carcinogenic biomarker in CTCs tested in patients with CRC (45). Hua et al. conducted a study to prove that the upregulation of SDC2 promotes cell proliferation, migration, and invasion; inhibits apoptosis; and activates and promotes EMT through mitogen-activated protein kinase (MAPK) signaling pathways (34). Apart from that of SFRPs and SDC2, the methylation of TAC1, SEPT9, HPP1, HLTF, and NPY has also been proven to be related to the recurrence and metastasis of CRCs (39). Moreover, the methylation of two factors, hyperplastic polyposis protein 1 (HPP1) and helicase-like transcription factor (HLTF), has prognostic value for therapy and metastatic location in CRC patients (35, 36, 46, 47). Several DNA methylation have been applied clinically with high sensitivity and specificity. Overall, the panels of methylated SEPT9 and SDC2 (48, 49), SEPT9 and OSMR (77.0%) (37), SEPT9 and OSMR (37), SEPT9 and TAC1 and HIC/CYCD2/VHL (50), APC/MGMT/RASSF2A/WIF1 (51), SYNE1 and FOXE1 (52), WIF1/NPY/PENK (53), THBD and C9orf50 (54), and the combinations of FAM123A/GLI3/PPP1R16B/SLIT3/TMEM90B (55) and MGMT/RASSF1A/SEPT9 (56) were reported to have diagnostic significance in early-stage CRCs and may be more promising than the use of single markers. The combinations of BCAT1/IKZF1 and BCAT1/IKZF1/IRF4 were exploited and were shown to perform better than individual markers (57, 58), with higher sensitivity (73.9%) and specificity (90.1%) and an area under the curve (AUC) of 0.82 (38, 59).
Methylated DNA analysis in exosomes has been reported for gastric cancer, prostate cancer, lymphoma, breast cancer, and gingivitis (60). Although an association between DNA methylation in vesicles and CRCs exists, no study on their relationship has been conducted. DNA methylation can regulate the expression of hypoxia-inducible factor (HIF), which can modulate the development and progression of hypoxic CRCs. It is a critical factor in the regulatory EMT process.
NcRNAs Regulate the EMT Process in CRC
NcRNAs are a class of small RNA fragments or circulars in the non-coding regions. Although an association between DNA methylation in vesicles and CRCs exists, no study on their relationship has been conducted. The ncRNAs in CTCs originate from three main pathways, namely, membrane-bound vesicles, apoptotic bodies, and RNA-binding proteins (RBPs), which have been elucidated in detail in a previous review (61). The ncRNAs can maintain highly stable in the bloodstream with the protection of exosomes or RBPs, such as argonaute-2 (AGO2) high-density lipoprotein (HDL), due to their small size and their ability to escape RNase-mediated degradation (62–66). NcRNAs include a number of RNA types, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs).
MiRNAs were the first discovered ncRNAs that can regulate the functions of genes and proteins. The sequence of miRNAs is short and can form complementary pairs with mRNAs. Most miRNAs are inhibitory and can lead to the degradation of mRNAs, which decreases the expressions of target proteins. Many cytoplasmic miRNAs have been discovered that can regulate expression of N-cadherin, Vimentin, MMPs, E-cadherin, Claudin, and EpCAM, thereby influencing the direction of EMT through molecular regulation in targeted pathways (67–69). Go et al. provided proof that the maternally expressed gene 3 differentially methylated region (MEG3-DMR) can control liver metastasis by regulating miRNAs, such as miR-655-3p, in the 14q32 cluster in CRCs (70, 71). This confirms that circulating miRNAs have an endothelial cell and blood component origin (19, 63, 72). A molecular classification based on epigenetically regulated gene expression profiles was established by Wang et al. to provide a better understanding of the epigenetic mechanisms underlying CRC heterogeneity using The Cancer Genome Atlas (TCGA) dataset and validated across Gene Expression Omnibus (GEO) datasets (73–78).
The upregulation or the downregulation of miRNAs hinges on regulating genes and epigenetic modifications. This variability, the targets and resultant outcomes are exemplified by the following: miR192-2 promoter hypermethylation was shown to modulate tumor metastasis in CRC by regulating the expression of SOX4 (80). MiR-31 has been reported to increase the sensitivity to 5-fluorouracil (5-FU) at an early stage (81). In the latest study, miR-31 has been proven to increase radiosensitivity in CRC cell lines through targeting STK40 (80, 82–85).
LncRNAs can promote or suppress the metastasis of tumor cells via upregulating or downregulating the process of EMT. LncRNAs have RNA sequences longer than 200 nucleotides that are not translated into proteins (86), so they are more stable than miRNAs (87). Similar to miRNAs, detection of expression or methylation of lncRNAs in CRC has been reported (88). Several lncRNAs, such as XIST, H19, and SPRY4-IT1, are related to EB1, 2/E-cadherin, and vimentin and can be detected from CTCs in plasma (89–91). The lncRNA H19 can promote CRC metastasis via inducing EMT (92). EMT promotes tumor cell growth, proliferation, and metastasis (93). The upregulation of the lncRNA TUG1 increases the risk of metastasis and further invasion in CRCs via activating the Wnt/β-catenin signaling pathway (94). The lncRNA MALAT1 is a predictive biomarker for metastasis by regulating the epigenetic model (95). Urothelial carcinoma-associated 1 (UCA1), which is produced by cancer-associated fibroblasts (CAFs) and released into serum, is associated with the metastasis of CRCs. The upregulation of UCA1 can change the expressions of the genes in EMT (96). The lncRNA BANCR is an oncogenic molecule that enhanced the aggressiveness of CRC metastasis (97). The TGF-β signaling pathway is a critical pathway in regulating cancer-associated expression. The lncRNA activated by TGF-β (LNCRNA-ATB) can suppress the expression of E-cadherin in EMT to promote CRC metastasis (98). The lncRNAs HOTAIR and HOXA-AS2 in plasma correlate with the invasion of CRCs via EMT (99, 100). The role of lncRNAs in serum is shown in Table 2.
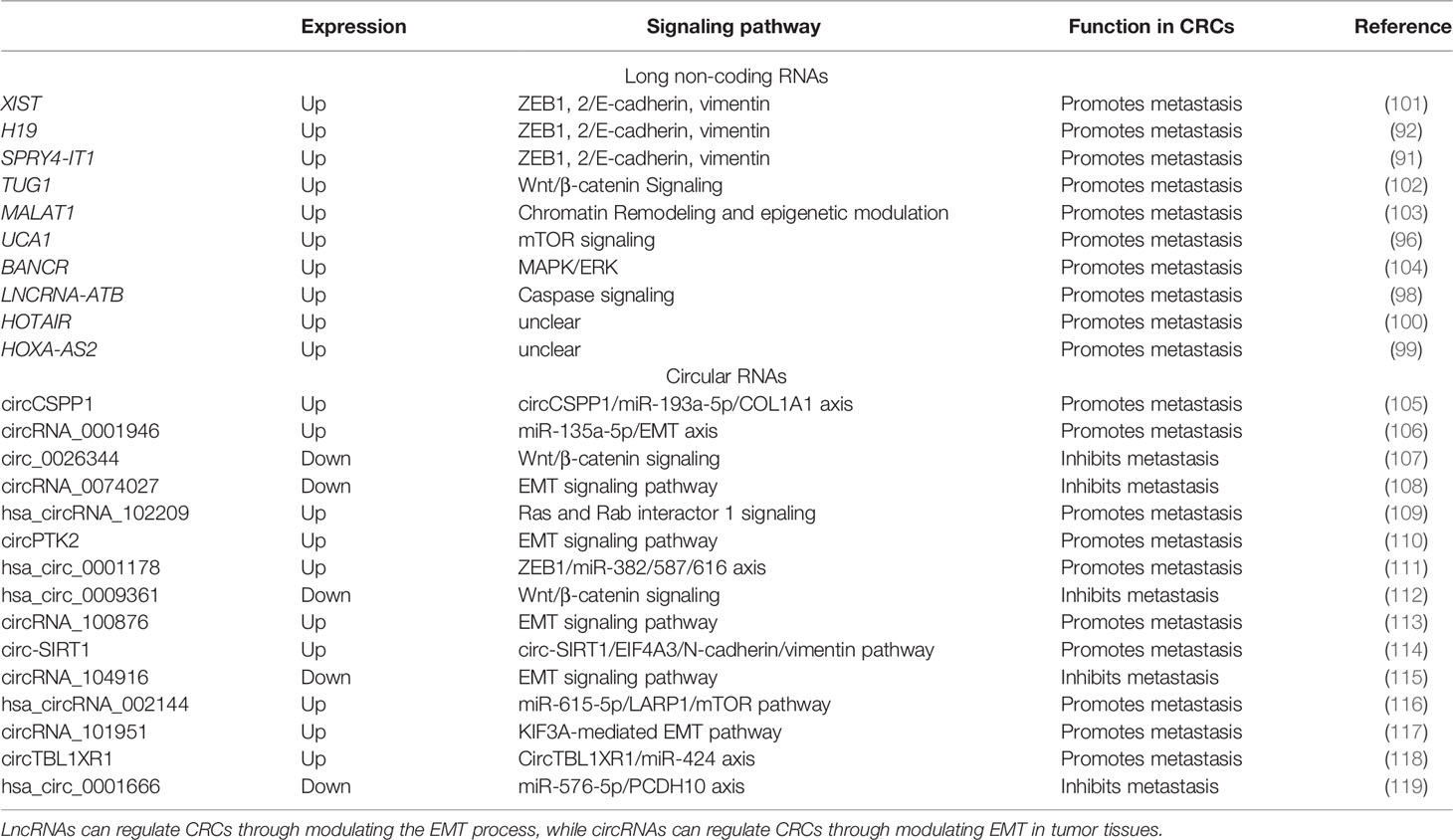
Table 2 Role of long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) associated with epithelial–mesenchymal transition (EMT) in colorectal cancers (CRCs).
CircRNAs are novel derivatives from the alternative splicing of lncRNAs that have small ring structures (120, 121). Compared to miRNAs and lncRNAs, circRNAs have shown their fantastic advantage in forecasting the metastasis of CRCs. By analyzing the expression profiles of circRNAs in the six pairs of CRC tissues and adjacent normal tissues, Chen et al. found that about two in three circRNAs were upregulated and one in three circRNAs was downregulated (122). In CRC tissues, the recovery of some circRNAs hints at the possibility of metastasis and recurrence of CRCs. CircRNA-1662 is an oncogenic circRNA that promoted CRC cell invasion and migration through control of EMT, and was reported to be a robust predictor for cancer metastasis (122). CircCSPP1 can promote CRC metastasis by regulating COL1A1, which is composed of type I collagen (105). CircRNA_0001946, hsa_circRNA_102209, circPTK2, hsa_circ_0001178, circRNA_100876, circ-SIRT1, hsa_circRNA_002144, circRNA_101951, and circTBL1XR can be overexpressed in CRC to promote metastasis and proliferation (106, 109–111, 113, 114, 116–118). On the other hand, circ_0026344, circRNA-0074027, hsa_circ_0009361, and hsa_circ_0001666 are downregulated in CRC tissues and suppress the metastasis of tumor cells (107, 108, 112, 115, 119). The elevation of circPTK2 in CRC CTCs increases the possibility of tumor metastasis (110). The hsa_circ_0001821 (circ3823) from both CRC tissues and sera may lead to tumor cell infiltration by binding with miR-30c-5p and its target to regulate the expression of TCF7 (123). The role of circRNAs in CRCs is shown in Table 2.
Interactions Between MiRNAs, LncRNAs, and CircRNAs—The “Sponge Effect”
There are reports that, at times, the outcomes of miRNA transcription and expression is less satisfactory than expected, which hints at the existence of miRNA inhibitors might be present in the tumor microenvironment of CRCs (124). Seitz et al. hypothesized this inhibitor as a “miRNA sponge” that competes with endogenous mRNAs for miRNAs (125). LncRNAs and circRNAs can regulate miRNAs from binding with the target genes by functioning as competing endogenous RNAs (ceRNAs), which can regulate the expressions of genes by competitively integrating with miRNAs (126). The “lncRNA/circRNAs/miRNA/mRNA/protein” pathway is also called the molecular sponge effect (126).
In contrast, miRNA modulation can regulate the expressions of lncRNAs as well. MiR-939-5p is a suppressive miRNA that can inhibit the invasion and metastasis of CRC cells by decreasing the level of LIMK2. LncRNA LINC00460 reduced the inhibition of miR-939-5p on LIMK2 and bound with miR-939-5p as a ceRNA (127). It is oncogenic through upregulation of annexin to promote the EMT process (128). The glycolysis-related lncRNA MIR17HG was found to competitively bind with miR-138-5p to increase glycolysis, which can promote CRC liver metastasis (129). The antisense lncRNA MCM3AP-AS1 sponged miR-193a-5p through upregulating SENP1 to promote the progression and migration of CRC (130). The lncRNA SNHG7 sponges miR-216b to promote the proliferation and liver metastasis of colorectal cancer through upregulating GALNT1 (131). In the cytoplasm, LINC00460 is a type of lncRNA that can regulate the expression of high mobility group A1 (HMGA1) of m6A modification under the catalytic function of methyltransferase 3 (METTL3) (132). The earliest found lncRNA, H19, functioned as a ceRNA that sequesters miR-138 and miR-200a and leads to the inhibition of vimentin and the protein expressions of ZEB1 and ZEB2 (90, 92). The regulatory pathway of lncRNAs on miRNAs is shown in Figure 1. Similarly, circRNAs can act as ceRNA sponges of miRNAs (133). Most circRNAs positively affect target genes and promote the metastasis of CRCs. A few circRNAs act as suppressors of tumor invasion and promotion. Circ_0026344, hsa_circ_0009361, and hsa_circ_0001666 are negative regulators of tumor proliferation and inhibit metastasis. The sponge mechanism of circRNAs on miRNAs is shown in Figure 2.
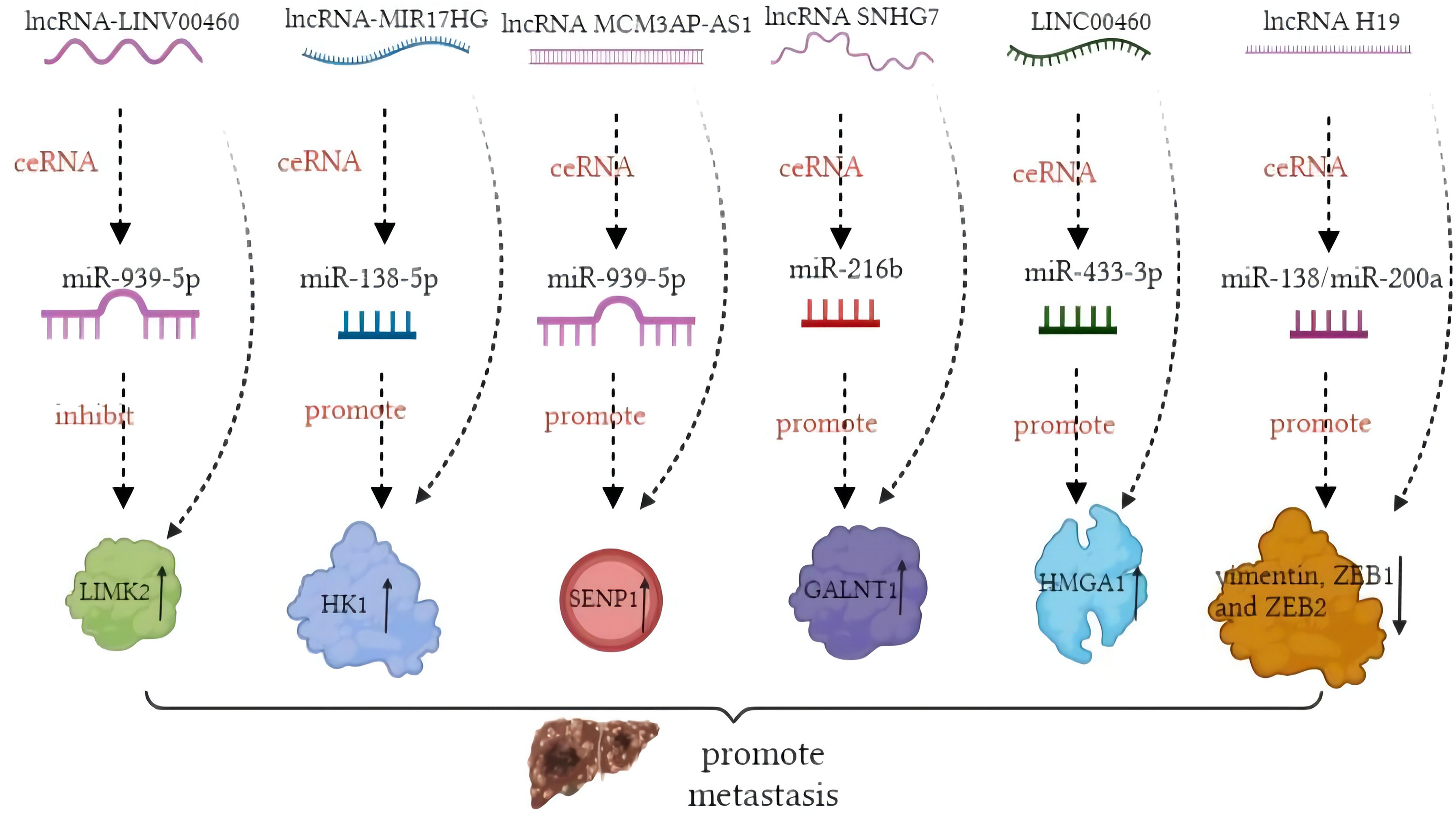
Figure 1 Mechanism of long non-coding RNAs (lncRNAs) acting as competing endogenous RNAs (ceRNAs) in colorectal cancer (CRC) metastasis.
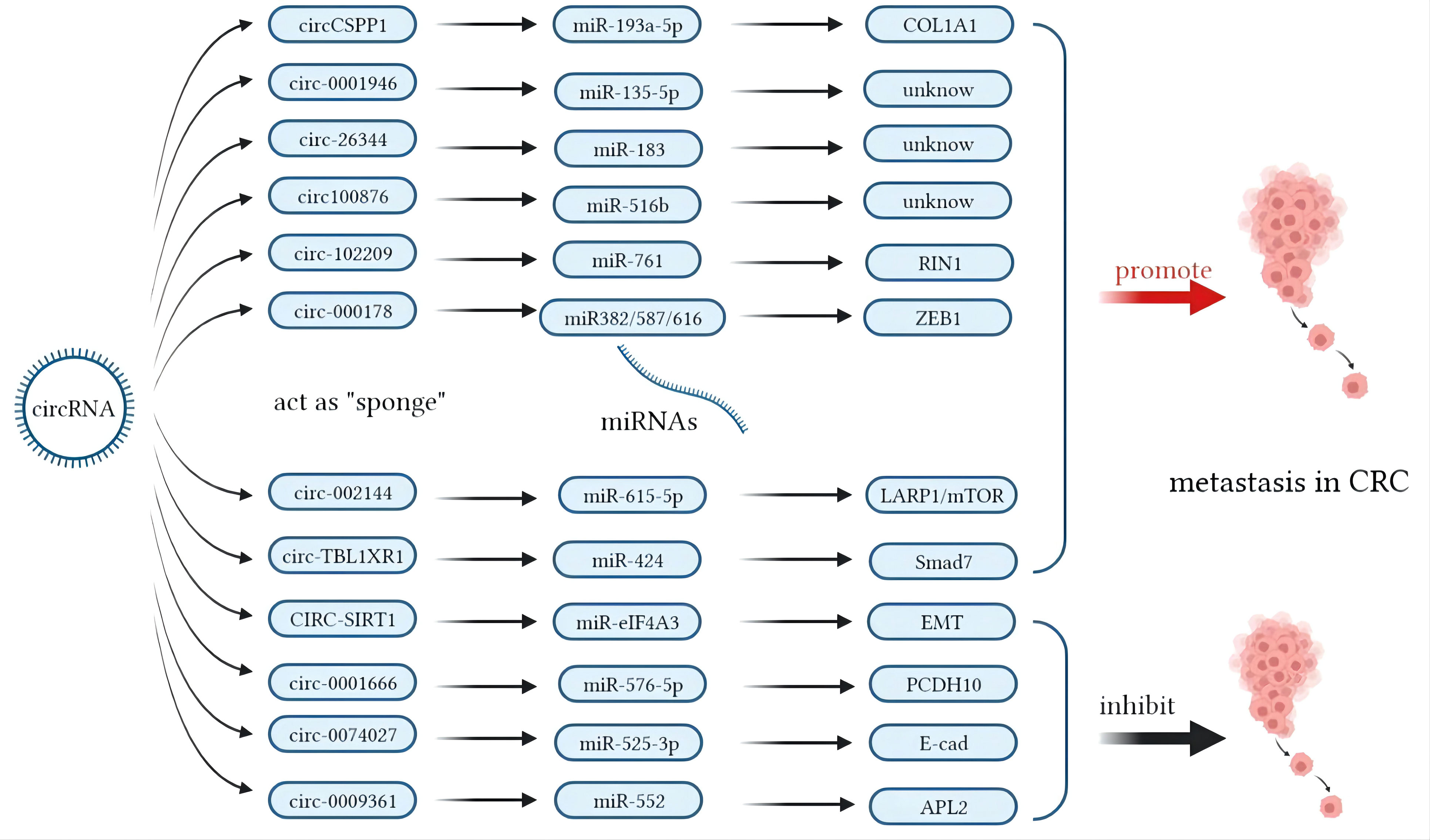
Figure 2 Mechanisms of circular RNAs (circRNAs) acting as competing endogenous RNAs (ceRNAs) in colorectal cancer (CRC) metastasis.
The Role of M6A Methylation of RNAs in EMT
M6A methylation is a novel and critical component of RNA modification related to the expressions of ncRNAs in epigenetics (134). M6A in RNA post-transcriptionally regulates mRNAs by affecting the splicing, export, stability, and translation of transcripts (135–138). Its methylation regulates miRNA synthesis, processing, and maturation, which are crucial in tumorigenesis and cancer progression (139). It has been reported that the m6A modification could alter its local RNA structure at the terminal loop region of primary miRNAs (pri-miRNAs) to promote their processing through nuclear transcripts and alternative splicing by modulating RALY binding (140–142). It is promising that m6A methylation can predict metastasis and the prognosis of therapy (143). The upregulation of HMGA1 of m6A modification in CRC cells enhances the probability of metastasis (132).
In total, 11 readers, 7 writers, and 2 erasers have already been found. The term “writers” often denotes methyltransferase as the beginning of m6A methylation, and different methyltransferases form a complex to gain more powerful catalytic ability. The (143)methyltransferase complex (MTC) comprises the m6A/METTL complex (MAC) and the m6A/METTL-associated complex (MACOM). MAC consists of METTL3 and METTL14, which can form stable heterodimers (144). Besides METTL3 and METTL14, WTAP is another writer that can participate in the formation of complex to regulate m6A. NcRNAs can affect the process of m6A by regulating the function of the “writer” complex. For example, METTL3 is the first discovered RNA methylation-related mRNA with a catalytic subunit and is upregulated in metastasis to promote cell migration and invasion in CRCs by regulating miR-1246 (145). Contrarily, METTL14 is regulated to suppress cell proliferation, invasion, and migration in CRCs via miR-375 (146–149). Furthermore, there exists mutual action between m6A modulation and ncRNAs. Prior studies demonstrated that knockdown of METTL3 downregulated the expression levels of miR-483, miR-676, miR-877 and circ1662-YAP1-SMAD3 axis in regulating CRC invasion (122, 142). The m6A modification of circRNA_103783 (also called circNSUN2) could increase export into the cytoplasm in the circNSUN2/IGF2BP2/HMGA2 ternary complex (150). In addition, the high expression of circNSUN2 predicts a more aggressive characteristic of CRC; thus, m6A modification enhances the metastasis risk of CRC cells (150).
Beyond the methyltransferases, the “readers” that can recognize m6A modulation sites, such as YTHDF1, IGF2BP1, IGF2BP3, and EIF3B (151), are also under the influence of ncRNAs. The tumor-suppressive miR-1266 promoted the occurrence and progression of CRCs by directly targeting FTO (152). Moreover, the expression of the lncRNA GAS5 can reversely regulate YAP1 via m6A (153). Surprisingly, the levels of m6A can positively increase with the increased concentrations of circRNAs (154). The upregulation of the lncRNA RP11 by m6A methylation can accelerate the spread of CRC cells through the upregulation of ZEB1 (155). Interestingly, it has been proven that m6A circRNAs are different from m6A mRNA sites. This indicates that disturbance in the m6A modification of mRNA is not equivalent to that of m6A circRNAs. Therefore, to abolish the effects of m6A modification, then the m6A of circRNAs must be considered as well (123, 154). Therefore, to abolish the effects of m6A modification, then the m6A of circRNAs must be considered as well. NcRNAs can regulate the critical enzymes as the downstream target molecules or combined partners. Feedback of the essential enzymes affects the m6A modulation of ncRNAs, and this process can change the cancer-related phenotype and lead to metastasis (155).
The Exosomes and EMT in CRC
What is an exosome? Exosomes are microvesicles with diameters from 30 to 150 nm. They are derived from normal intestinal cells, CRC cells, and other cancer-related stromal cells (156). Exosomes have various physiological functions and participate in multiple cancer-related signaling pathways. Exosomes derived from CRCs cells are associated with a metastatic phenotype in CRCs, such as migration, infiltration, and EMT. For exosomes, surface antigens act as biomarkers that contribute to recognition, and their components determine the tasks exosomes carry out. Exosomes are secreted not only from cancer cells but also from normal intestinal cells or other tumor-related cells. Hepatocytes and fibroblasts can also produce exosomes to enter into the blood, urine, or saliva (157, 158). NcRNAs excreted from exosomes play an important role in regulating the signaling pathways in CRC metastasis (159). Studies that excluded the interference of miRNAs and lncRNAs in cells via the addition of anti-miRNAs and exogenous exosomes containing miRNAs confirmed that ncRNAs negatively regulated the downstream target proteins or mRNA molecules to promote tumor metastasis via EMT (160–162). In contrast, EMT cancer cells can promote exosome secretion via increasing the permeability of vascular epithelial layers, and miR-27b-3p can regulate this process through the STAT3 pathway (163). There is a mutual regulation between exosomes and the EMT process, but most studies have focused on the influence of exosome transportation in EMT, as shown in Table 3. While the ncRNAs in exosomes can negatively regulate the target genes and promote tumor metastasis, this also hints that they may be brilliant therapeutic targets for the prevention of metastasis. Besides CRC cells, tumor-associated macrophages (TAMs) comprise another class involved in the regulation of exosome and regulate the EMT process in inflammatory or immune diseases (164). The effects being positive or negative depend on the inflammatory cells and cytokines that TAMs secrete. The miRNAs or lncRNAs wrapped in vesicles could affect the M1 or M2 polarization of TAMs (172).
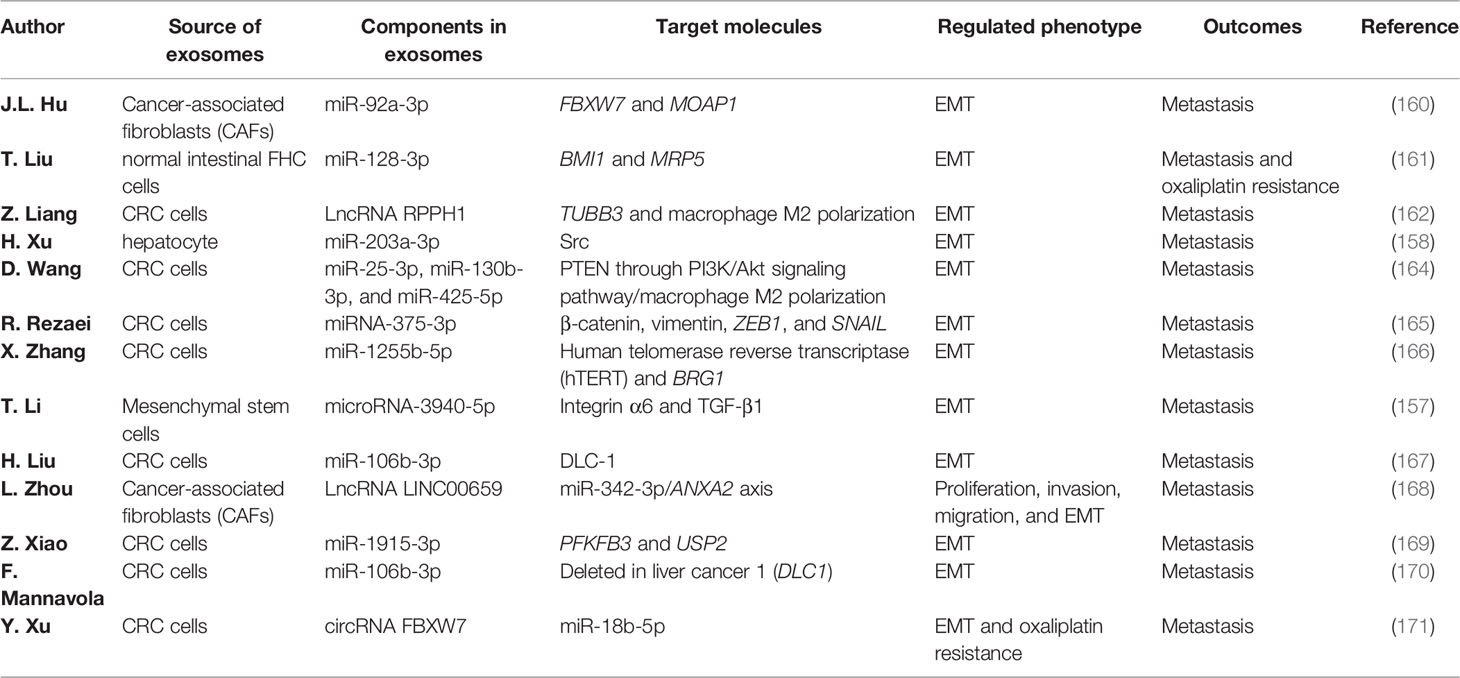
Table 3 Functions of exosomes in colorectal cancer (CRC) metastasis via regulating the epithelial–mesenchymal transition (EMT) process.
Conclusion
Cancer metastasis has always been a hot topic for scientists and clinicians, and discovering the risk factors and suitable therapeutic targets is a critical part of preventing metastasis. Epigenetic change is a popular context associated with the progression and metastasis of CRCs due to its variability and vulnerability. NcRNAs plays a crucial role in regulating epithelial tissues and the mesenchymal components. The dynamic process determines the tumor metastasis tendency. In this regulation, the sponge effect between lncRNAs/circRNAs/miRNAs/mRNA is critical. In addition, epigenetic modification is a crucial link. The 5mC methylation of DNA and the m6A methylation of RNA in CRCs could enhance the invasive and migration ability to promote metastasis. The mechanisms of the mutual effect between methylation and ncRNAs may provide a novel direction for future studies.
The EMT is a vital regulator in metastasis, while the mechanism and influence factor of EMT is still unclear. As mentioned before, these epigenetic changes are not independent but interacting. The methylation of promoters can regulate the process of alternative splicing and production of ncRNAs. The review interprets a crosstalk interaction in epigenetic changes from DNA methylation, ncRNAs, and m6A methylation. The function of epigenetic changes is powerful and complex in regulating mechanisms in CRC metastasis. At the same time, EMT is a cross point that the DNA methylation, m6A methylation, and ncRNAs can all affect CRC metastasis. They can upregulate or downregulate the biomarkers in EMT such as N-cadherin, Vimentin, MMPs, E-cadherin, Claudin, and EpCAM to control the progress of the epithelial transition.
Another novelty of the review is that the exosomes can regulate EMT as a carrier for transporting ncRNAs. The function of exosomes has gained widespread attention in cancers. The exosomes have been proved to be related to cancers detection. This review summarizes the effect of exosomes in CRC metastasis related to EMT. That indicates that exosomes are a vital factor in the tumor micro-environment. The tumor cells or other stromal cells can secret exosomes to regulate EMT. And this review indicates that DNA methylation, ncRNAs, m6A methylation, exosomes, and EMT can all act as potential therapeutic targets in CRC metastasis. The associated relationship and mutual interaction among them are elucidated thoroughly in this review. While prior studies have been published elucidating the mechanisms of exosomes and EMT, the field remains in its relative infancy, with the potential to discover important new therapeutic targets for CRC.
The detection of exosomes in LB is a novel method to predict metastasis in CRCs. The variety of stromal cells can produce exosomes to participate in the progression, proliferation, migration, and EMT in CRCs. Epigenetic changes, ncRNAs, and exosomes are not independent of each other; on the contrary, there exists an intrinsic interrelation between them. Different from previous studies, this review elaborated on the potential links between epigenetic changes, exosomes, and EMT in CRCs regarding metastasis. It provided novel sights for the study of probable mechanisms of metastasis in CRCs. There have already been several studies on the mechanisms of exosomes and EMT, but more studies are needed to discover new therapeutic targets for CRCs.
Author Contributions
MY and HZ designed the study design and wrote the main manuscript. MS contributed to the final proof. All authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Conteduca V, Sansonno D, Russi S, Dammacco F. Precancerous Colorectal Lesions (Review). Int J Oncol (2013) 43(4):973–84. doi: 10.3892/ijo.2013.2041
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Barnell EK, Kang Y, Barnell AR, Kruse KR, Fiske J, Pittz ZR, et al. Multitarget Stool RNA Test for Noninvasive Detection of Colorectal Neoplasias in a Multicenter, Prospective, and Retrospective Cohort. Clin Trans Gastroenterol (2021) 12(5):e00360. doi: 10.14309/ctg.0000000000000360
4. Govaert KM, van Kessel CS, Steller EJ, Emmink BL, Molenaar IQ, Kranenburg O, et al. Recurrence Location After Resection of Colorectal Liver Metastases Influences Prognosis. J Gastrointestinal Surg Off J Soc Surg Alimentary Tract (2014) 18(5):952–60. doi: 10.1007/s11605-014-2461-0
5. Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel Therapeutic Strategies: Targeting Epithelial–Mesenchymal Transition in Colorectal Cancer. Lancet Oncol (2021) 22(8):e358–68. doi: 10.1016/S1470-2045(21)00343-0
6. Rajamaki K, Taira A, Katainen R, Valimaki N, Kuosmanen A, Plaketti RM, et al. Genetic and Epigenetic Characteristics of Inflammatory Bowel Disease-Associated Colorectal Cancer. Gastroenterology (2021) 161(2):592–607. doi: 10.1053/j.gastro.2021.04.042
7. Palanca-Ballester C, Rodriguez-Casanova A, Torres S, Calabuig-Farinas S, Exposito F, Serrano D, et al. Cancer Epigenetic Biomarkers in Liquid Biopsy for High Incidence Malignancies. Cancers (2021) 13(12). doi: 10.3390/cancers13123016
8. Barault L, Amatu A, Siravegna G, Ponzetti A, Moran S, Cassingena A, et al. Discovery of Methylated Circulating DNA Biomarkers for Comprehensive Non-Invasive Monitoring of Treatment Response in Metastatic Colorectal Cancer. Gut (2018) 67(11):1995–2005. doi: 10.1136/gutjnl-2016-313372
9. Marcuello M, Duran-Sanchon S, Moreno L, Lozano JJ, Bujanda L, Castells A, et al. Analysis of A 6-Mirna Signature in Serum From Colorectal Cancer Screening Participants as Non-Invasive Biomarkers for Advanced Adenoma and Colorectal Cancer Detection. Cancers (2019) 11(10). doi: 10.3390/cancers11101542
10. Xie W, Xie L, Song X. The Diagnostic Accuracy of Circulating Free DNA for the Detection of KRAS Mutation Status in Colorectal Cancer: A Meta-Analysis. Cancer Med (2019) 8(3):1218–31. doi: 10.1002/cam4.1989
11. Yeung KT, Yang J. Epithelial-Mesenchymal Transition in Tumor Metastasis. Mol Oncol (2017) 11(1):28–39. doi: 10.1002/1878-0261.12017
12. Gurzu S, Turdean S, Kovecsi A, Contac AO, Jung I. Epithelial-Mesenchymal, Mesenchymal-Epithelial, and Endothelial-Mesenchymal Transitions in Malignant Tumors: An Update. World J Clin Cases (2015) 3(5):393–404. doi: 10.12998/wjcc.v3.i5.393
13. Ye Z, Zhou M, Tian B, Wu B, Li J. Expression of lncRNA-CCAT1, E-Cadherin and N-Cadherin in Colorectal Cancer and Its Clinical Significance. (2015).
14. Luo1 H, Zhao Q, Wei1 W, Zheng L. Circulating Tumor DNA Methylation Profiles Enable Early Diagnosis, Prognosis Prediction, and Screening for Colorectal Cancer. Sci Transl Med (2020) 12:eaax7533. doi: 10.1126/scitranslmed.aax7533
15. Øster B, Thorsen K, Lamy P, Wojdacz TK, Hansen LL, Birkenkamp-Demtro K, et al. Identification and Validation of Highly Frequent CpG Island Hypermethylation in Colorectal Adenomas and Carcinomas. Int J Cancer (2011) 129:2855–66. doi: 10.1002/ijc.25951
16. Singh MP, Rai S, Suyal S, Singh SK, Singh NK, Agarwal A, et al. Genetic and Epigenetic Markers in Colorectal Cancer Screening: Recent Advances. Expert Rev Mol Diagn (2017) 17(7):665–85. doi: 10.1080/14737159.2017.1337511
17. Kang KJ, Min BH, Ryu KJ, Kim KM, Chang DK, Kim JJ, et al. The Role of the CpG Island Methylator Phenotype on Survival Outcome in Colon Cancer. Gut Liver (2015) 9(2):202–7. doi: 10.5009/gnl13352
18. Shiovitz S, Bertagnolli MM, Renfro LA, Nam E, Foster NR, Dzieciatkowski S, et al. CpG Island Methylator Phenotype Is Associated With Response to Adjuvant Irinotecan-Based Therapy for Stage III Colon Cancer. Gastroenterology (2014) 147(3):637–45. doi: 10.1053/j.gastro.2014.05.009
19. Kaur S, Lotsari-Salomaa JE, Seppanen-Kaijansinkko R, Peltomaki P. MicroRNA Methylation in Colorectal Cancer. Adv Exp Med Biol (2016) 937:109–22. doi: 10.1007/978-3-319-42059-2_6
20. Katada S, Imhof A, Sassone-Corsi P. Connecting Threads: Epigenetics and Metabolism. Cell (2012) 148(1-2):24–8. doi: 10.1016/j.cell.2012.01.001
21. Najdi R, Holcombe RF, Waterman ML. Wnt Signaling and Colon Carcinogenesis: Beyond APC. J Carcinogenesis (2011) 10:5. doi: 10.4103/1477-3163.78111
22. Amelio I, Bertolo R, Bove P, Buonomo OC, Candi E, Chiocchi M, et al. Liquid Biopsies and Cancer Omics. Cell Death Discov (2020) 6(1):131. doi: 10.1038/s41420-020-00373-0
23. Butz H, Nofech-Mozes R, Ding Q, Khella HWZ, Szabo PM, Jewett M, et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell-Cell Communication in Renal Cell Carcinoma. Eur Urol Focus (2016) 2(2):210–8. doi: 10.1016/j.euf.2015.11.006
24. Holubekova V, Zubor P, Mendelova A. Epigenetic Regulation by DNA Methylation and miRNA Molecules in Cancer. (2017). doi: 10.2217/fon-2017-0363
25. Mazouji O, Ouhajjou A, Incitti R, Mansour H. Updates on Clinical Use of Liquid Biopsy in Colorectal Cancer Screening, Diagnosis, Follow-Up, and Treatment Guidance. Front Cell Dev Biol (2021) 9:660924. doi: 10.3389/fcell.2021.660924
26. Luo M, Yang X, Chen HN, Nice EC, Huang C. Drug Resistance in Colorectal Cancer: An Epigenetic Overview. Biochim Biophys Acta Rev Cancer (2021) 1876(2):188623. doi: 10.1016/j.bbcan.2021.188623
27. Ehrlich M. DNA Hypermethylation in Disease: Mechanisms and Clinical Relevance. Epigenetics (2019) 14(12):1141–63. doi: 10.1080/15592294.2019.1638701
29. Sancho E, Batlle E, Clevers H. Signaling Pathways in Intestinal Development and Cancer. Annu Rev Cell Dev Biol (2004) 20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805
30. Valcz G, Patai AV, Kalmar A, Peterfia B, Furi I, Wichmann B, et al. Myofibroblast-Derived SFRP1 as Potential Inhibitor of Colorectal Carcinoma Field Effect. PloS One (2014) 9(11):e106143. doi: 10.1371/journal.pone.0106143
31. Li H, Wang Z, Zhao G, Ma Y, Chen Y, Xue Q, et al. Performance of a MethyLight Assay for Methylated SFRP2 DNA Detection in Colorectal Cancer Tissue and Serum. Int J Biol Markers (2019) 34(1):54–9. doi: 10.1177/1724600818820536
32. Liu Y, Yu J, Xie Y, Li M, Wang F, Zhang J, et al. EZH2 Regulates Sfrp4 Expression Without Affecting the Methylation of Sfrp4 Promoter DNA in Colorectal Cancer Cell Lines. Exp Ther Med (2020) 20(5):33. doi: 10.3892/etm.2020.9160
33. Kirana C, Smith E. High Preoperative Levels of Circulating SFRP5 Predict Better Prognosis in Colorectal Cancer Patients. Future Oncol (2020) 16(31):2499–509. doi: 10.2217/fon-2020-0356
34. Hua R, Yu J, Yan X, Ni Q, Zhi X, Li X, et al. Syndecan-2 in Colorectal Cancer Plays Oncogenic Role via Epithelial-Mesenchymal Transition and MAPK Pathway. Biomed Pharmacother Biomed Pharmacother (2020) 121:109630. doi: 10.1016/j.biopha.2019.109630
35. Herbst A, Vdovin N, Gacesa S, Ofner A, Philipp A, Nagel D, et al. Methylated Free-Circulating HPP1 DNA Is an Early Response Marker in Patients With Metastatic Colorectal Cancer. Int J Cancer (2017) 140(9):2134–44. doi: 10.1002/ijc.30625
36. Helmer RA, Martinez-Zaguilan R, Kaur G, Smith LA, Dufour JM, Chilton BS. Helicase-Like Transcription Factor-Deletion From the Tumor Microenvironment in a Cell Line-Derived Xenograft Model of Colorectal Cancer Reprogrammed the Human Transcriptome-S-Nitroso-Proteome to Promote Inflammation and Redirect Metastasis. PLoS One (2021) 16:e0251132. doi: 10.17504/protocols.io.bs5xng7n
37. Yuan P, Cheng X, Wu X, Li L, Zhang L, Li Z, et al. OSMR and SEPT9: Promising Biomarkers for Detection of Colorectal Cancer Based on Blood-Based Tests. Trans Cancer Res (2016) 5(2):131–9. doi: 10.21037/tcr.2016.03.07
38. Young GP, Symonds EL, Nielsen HJ, Ferm L, Christensen IJ, Dekker E, et al. Evaluation of a Panel of Tumor-Specific Differentially-Methylated DNA Regions in IRF4, IKZF1 and BCAT1 for Blood-Based Detection of Colorectal Cancer. Clin Epigenet (2021) 13(1):14. doi: 10.1186/s13148-020-00999-y
39. Jensen LH, Olesen R, Petersen LN, Boysen AK, Andersen RF, Lindebjerg J, et al. NPY Gene Methylation as a Universal, Longitudinal Plasma Marker for Evaluating the Clinical Benefit From Last-Line Treatment With Regorafenib in Metastatic Colorectal Cancer. Cancers (2019) 11(11). doi: 10.3390/cancers11111649
40. Overs A, Flammang M, Hervouet E, Bermont L, Pretet JL, Christophe B, et al. The Detection of Specific Hypermethylated WIF1 and NPY Genes in Circulating DNA by Crystal Digital PCR Is a Powerful New Tool for Colorectal Cancer Diagnosis and Screening. BMC Cancer (2021) 21(1):1092. doi: 10.1186/s12885-021-08816-2
41. Cruciat CM, Niehrs C. Secreted and Transmembrane Wnt Inhibitors and Activators. Cold Spring Harbor Perspect Biol (2013) 5(3):a015081. doi: 10.1101/cshperspect.a015081
42. Silva AL, Dawson SN, Arends MJ, Guttula K, Hall N, Cameron EA, et al. Boosting Wnt Activity During Colorectal Cancer Progression Through Selective Hypermethylation of Wnt Signaling Antagonists. BMC Cancer (2014) 14:891. doi: 10.1186/1471-2407-14-891
43. Bartak BK, Kalmar A, Peterfia B, Patai AV, Galamb O, Valcz G, et al. Colorectal Adenoma and Cancer Detection Based on Altered Methylation Pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in Plasma Samples. Epigenetics (2017) 12(9):751–63. doi: 10.1080/15592294.2017.1356957
44. Huang D, Yu B, Deng Y, Sheng W, Peng Z, Qin W, et al. SFRP4 was Overexpressed in Colorectal Carcinoma. J Cancer Res Clin Oncol (2010) 136(3):395–401. doi: 10.1007/s00432-009-0669-2
45. Molnar B, Galamb O, Kalmar A, Bartak BK, Nagy ZB, Toth K, et al. Circulating Cell-Free Nucleic Acids as Biomarkers in Colorectal Cancer Screening and Diagnosis - An Update. Expert Rev Mol Diagn (2019) 19(6):477–98. doi: 10.1080/14737159.2019.1613891
46. Philipp AB, Nagel D, Stieber P, Lamerz R, Thalhammer I, Herbst A, et al. Circulating Cell-Free Methylated DNA and Lactate Dehydrogenase Release in Colorectal Cancer. (2014). doi: 10.1186/1471-2407-14-245
47. Ebert MP, Mooney SH, Tonnes-Priddy L, Lograsso J, Hoffmann J, Chen J, et al. Hypermethylation of the TPEF/HPP1 Gene in Primary and Metastatic Colorectal Cancers. Neoplasia (2005) 7(8):771–8. doi: 10.1593/neo.05235
48. Zhao G, Li H, Yang Z, Wang Z, Xu M, Xiong S, et al. Multiplex Methylated DNA Testing in Plasma With High Sensitivity and Specificity for Colorectal Cancer Screening. Cancer Med (2019) 8(12):5619–28. doi: 10.1002/cam4.2475
49. Chen Y, Wang Z, Zhao G, Sun C, Ma Y, Zhang L, et al. Performance of a Novel Blood-Based Early Colorectal Cancer Screening Assay in Remaining Serum After the Blood Biochemical Test. Dis Markers (2019) 2019:5232780. doi: 10.1155/2019/5232780
50. Melson J, Li Y, Cassinotti E, Melnikov A, Boni L, Ai J, et al. Commonality and Differences of Methylation Signatures in the Plasma of Patients With Pancreatic Cancer and Colorectal Cancer. Int J Cancer (2014) 134(11):2656–62. doi: 10.1002/ijc.28593
51. Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, et al. Aberrant Methylation of APC, MGMT, RASSF2A, and Wif-1 Genes in Plasma as a Biomarker for Early Detection of Colorectal Cancer. Clin Cancer Res Off J Am Assoc Cancer Res (2009) 15(19):6185–91. doi: 10.1158/1078-0432.CCR-09-0111
52. Melotte V, Yi JM, Lentjes MH, Smits KM, Van Neste L, Niessen HE, et al. Spectrin Repeat Containing Nuclear Envelope 1 and Forkhead Box Protein E1 Are Promising Markers for the Detection of Colorectal Cancer in Blood. Cancer Prev Res (2015) 8(2):157–64. doi: 10.1158/1940-6207.CAPR-14-0198
53. Roperch J-P, Incitti R, Forbin S, Bard F, Mansour H, Mesli F, et al. Aberrant Methylation of NPY, PENK, and WIF1 as a Promising Marker for Blood-Based Diagnosis of Colorectal Cancer. BMC Cancer (2013) 13(1). doi: 10.1186/1471-2407-13-566
54. Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H, et al. Genome-Scale Discovery of DNA-Methylation Biomarkers for Blood-Based Detection of Colorectal Cancer. PloS One (2012) 7(11):e50266. doi: 10.1371/journal.pone.0050266
55. Cho NY, Park JW, Wen X, Shin YJ, Kang JK, Song SH, et al. Blood-Based Detection of Colorectal Cancer Using Cancer-Specific DNA Methylation Markers. Diagnostics (2020) 11(1). doi: 10.3390/diagnostics11010051
56. Freitas M, Ferreira F, Carvalho S, Silva F, Lopes P, Antunes L, et al. A Novel DNA Methylation Panel Accurately Detects Colorectal Cancer Independently of Molecular Pathway. J Trans Med (2018) 16(1):45. doi: 10.1186/s12967-018-1415-9
57. Jedi M, Young GP, Pedersen SK, Symonds EL. Methylation and Gene Expression of BCAT1 and IKZF1 in Colorectal Cancer Tissues. Clin Med Insights Oncol (2018) 12:1179554918775064. doi: 10.1177/1179554918775064
58. Javierre BM, Rodriguez-Ubreva J, Al-Shahrour F, Corominas M, Graña O, Ciudad L, et al. Long-Range Epigenetic Silencing Associates With Deregulation of Ikaros Targets in Colorectal Cancer Cells. Mol Cancer Res (2011) 9(8):1139–51. doi: 10.1158/1541-7786.MCR-10-0515
59. Pedersen SK, Baker RT, McEvoy A, Murray DH, Thomas M, Molloy PL, et al. A Two-Gene Blood Test for Methylated DNA Sensitive for Colorectal Cancer. PloS One (2015) 10(4):e0125041. doi: 10.1371/journal.pone.0125041
60. Hur JY, Lee KY. Characteristics and Clinical Application of Extracellular Vesicle-Derived DNA. Cancers (2021) 13(15). doi: 10.3390/cancers13153827
61. Wang JJ, Wang X, Song YX, Zhao JH, Sun JX, Shi JX, et al. Circulating Noncoding RNAs Have a Promising Future Acting as Novel Biomarkers for Colorectal Cancer. Dis Markers (2019) 2019:2587109. doi: 10.1155/2019/2587109
62. Fernandez-Lazaro D, Garcia Hernandez JL, Garcia AC, Cordova Martinez A, Mielgo-Ayuso J, Cruz-Hernandez JJ. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer's Biomarkers. Diagnostics (2020) 10(4). doi: 10.3390/diagnostics10040215
63. Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res Off J Am Assoc Cancer Res (2017) 23(10):2391–9. doi: 10.1158/1078-0432.CCR-16-1676
64. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in Body Fluids–the Mix of Hormones and Biomarkers. Nat Rev Clin Oncol (2011) 8(8):467–77. doi: 10.1038/nrclinonc.2011.76
65. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 Complexes Carry a Population of Circulating microRNAs Independent of Vesicles in Human Plasma. Proc Natl Acad Sci USA (2011) 108(12):5003–8. doi: 10.1073/pnas.1019055108
66. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs Are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat Cell Biol (2011) 13(4):423–33. doi: 10.1038/ncb2210
67. Di Palo A, Siniscalchi C, Polito R, Nigro E, Russo A, Daniele A, et al. microRNA-377-3p Downregulates the Oncosuppressor T-Cadherin in Colorectal Adenocarcinoma Cells. Cell Biol Int (2021) 45(8):1797–803. doi: 10.1002/cbin.11605
68. Ye M, Huang T, Li J, Zhou C, Yang P, Ni C, et al. Role of CDH13 Promoter Methylation in the Carcinogenesis, Progression, and Prognosis of Colorectal Cancer: A Systematic Meta-Analysis Under PRISMA Guidelines. Medicine (2017) 96(4):e5956. doi: 10.1097/MD.0000000000005956
69. Duan BS, Xie LF, Wang Y. Aberrant Methylation of T-Cadherin Can Be a Diagnostic Biomarker for Colorectal Cancer. Cancer Genomics Proteomics (2017) 14(4):277–84. doi: 10.21873/cgp.20038
70. Oshima G, Poli EC, Bolt MJ, Chlenski A, Forde M, Jutzy JMS, et al. DNA Methylation Controls Metastasis-Suppressive 14q32-Encoded miRNAs. Cancer Res (2019) 79(3):650–62. doi: 10.1158/0008-5472.CAN-18-0692
71. Oshima G, Guo N, He C, Stack ME, Poon C, Uppal A, et al. In Vivo Delivery and Therapeutic Effects of a MicroRNA on Colorectal Liver Metastases. Mol Ther J Am Soc Gene Ther (2017) 25(7):1588–95. doi: 10.1016/j.ymthe.2017.04.005
72. He L, Hannon GJ. MicroRNAs: Small RNAs With a Big Role in Gene Regulation. Nat Rev Genet (2004) 5(7):522–31. doi: 10.1038/nrg1379
73. Wang X, Liu J, Wang D, Feng M, Wu X. Epigenetically Regulated Gene Expression Profiles Reveal Four Molecular Subtypes With Prognostic and Therapeutic Implications in Colorectal Cancer. Briefings Bioinf (2021) 22(4). doi: 10.1093/bib/bbaa309
74. Laiho P, Kokko A, Vanharanta S, Salovaara R, Sammalkorpi H, Jarvinen H, et al. Serrated Carcinomas Form a Subclass of Colorectal Cancer With Distinct Molecular Basis. Oncogene (2007) 26(2):312–20. doi: 10.1038/sj.onc.1209778
75. Kurozumi A, Goto Y, Okato A, Seki N. DNA Methylation and Dysregulation of miRNA in Cancer. Mol Oncol (2017) 6:281–96. doi: 10.1007/978-3-319-59786-7_10
76. Suzuki H, Takatsuka S, Akashi H, Yamamoto E, Nojima M, Maruyama R, et al. Genome-Wide Profiling of Chromatin Signatures Reveals Epigenetic Regulation of MicroRNA Genes in Colorectal Cancer. Cancer Res (2011) 71(17):5646–58. doi: 10.1158/0008-5472.CAN-11-1076
77. Croce CM. Causes and Consequences of microRNA Dysregulation in Cancer. Nat Rev Genet (2009) 10(10):704–14. doi: 10.1038/nrg2634
78. Lei L, An G, Zhu Z, Liu S, Fu Y, Zeng X, et al. C8orf48 Inhibits the Tumorigenesis of Colorectal Cancer by Regulating the MAPK Signaling Pathway. Life Sci (2021) 266:118872. doi: 10.1016/j.lfs.2020.118872
79. Ma Y, Zhang P, Yang J, Liu Z, Yang Z, Qin H. Candidate microRNA Biomarkers in Human Colorectal Cancer: Systematic Review Profiling Studies and Experimental Validation. Int J Cancer (2012) 130(9):2077–87. doi: 10.1002/ijc.26232
80. Zhang W, Zhu Y, Zhou Y, Wang J, Jiang P, Xue L. miRNA-31 Increases Radiosensitivity Through Targeting STK40 in Colorectal Cancer Cells. Asia-Pacific J Clin Oncol (2021). doi: 10.1111/ajco.13602
81. Wang CJ, Stratmann J, Zhou ZG, Sun XF. Suppression of microRNA-31 Increases Sensitivity to 5-FU at an Early Stage, and Affects Cell Migration and Invasion in HCT-116 Colon Cancer Cells. BMC Cancer (2010) 10:616. doi: 10.1186/1471-2407-10-616
82. Pinho SS, Reis CA. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat Rev Cancer (2015) 15(9):540–55. doi: 10.1038/nrc3982
83. Pucci M, Malagolini N, Dall'Olio F. Glycosyltransferase B4GALNT2 as a Predictor of Good Prognosis in Colon Cancer: Lessons From Databases. Int J Mol Sci (2021) 22(9). doi: 10.3390/ijms22094331
84. Kalluri R. The Biology and Function of Exosomes in Cancer. J Clin Invest (2016) 126(4):1208–15. doi: 10.1172/JCI81135
85. Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, et al. Circulating microRNA-203 Predicts Prognosis and Metastasis in Human Colorectal Cancer. Gut (2017) 66(4):654–65. doi: 10.1136/gutjnl-2014-308737
86. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long Noncoding RNAs With Enhancer-Like Function in Human Cells. Cell (2010) 143(1):46–58. doi: 10.1016/j.cell.2010.09.001
87. Han J, Rong LF, Shi CB, Dong XG, Wang J, Wang BL, et al. Screening of Lymph Nodes Metastasis Associated lncRNAs in Colorectal Cancer Patients. World J Gastroenterol (2014) 20(25):8139–50. doi: 10.3748/wjg.v20.i25.8139
88. Ye B, Li F, Chen M, Weng Y, Qi C, Xie Y, et al. A Panel of Platelet-Associated Circulating Long Non-Coding RNAs as Potential Biomarkers for Colorectal Cancer. Genomics (2022) 114(1):31–7. doi: 10.1016/j.ygeno.2021.11.026
89. Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, et al. Long Noncoding RNA XIST Expedites Metastasis and Modulates Epithelial-Mesenchymal Transition in Colorectal Cancer. Cell Death Dis (2017) 8(8):e3011. doi: 10.1038/cddis.2017.421
90. Chen SW, Zhu J, Ma J, Zhang JL, Zuo S, Chen GW, et al. Overexpression of Long Non-Coding RNA H19 is Associated With Unfavorable Prognosis in Patients With Colorectal Cancer and Increased Proliferation and Migration in Colon Cancer Cells. Oncol Lett (2017) 14(2):2446–52. doi: 10.3892/ol.2017.6390
91. Jin J, Chu Z, Ma P, Meng Y, Yang Y. Long non-Coding RNA SPRY4-IT1 Promotes Proliferation and Invasion by Acting as a ceRNA of miR-101-3p in Colorectal Cancer Cells. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2017) 39(7):1010428317716250. doi: 10.1177/1010428317716250
92. Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu J, et al. Long non-Coding RNA H19 Promotes Colorectal Cancer Metastasis via Binding to Hnrnpa2b1. J Exp Clin Cancer Res CR (2020) 39(1):141. doi: 10.1186/s13046-020-01619-6
93. Yagi T, Imaeda K, K E, Koyama H, Joh T. Glucagon Promotes Colon Cancer Cell Growth via Regulating AMPK and MAPK Pathways. Oncotarget (2018) 9:10650–64. doi: 10.18632/oncotarget.24367
94. Yue B, Liu C, Sun H, Liu M, Song C, Cui R, et al. A Positive Feed-Forward Loop Between LncRNA-CYTOR and Wnt/beta-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol Ther J Am Soc Gene Ther (2018) 26(5):1287–98. doi: 10.1016/j.ymthe.2018.02.024
95. Ji Q, Cai G, Liu X, Zhang Y, Wang Y, Zhou L, et al. MALAT1 Regulates the Transcriptional and Translational Levels of Proto-Oncogene RUNX2 in Colorectal Cancer Metastasis. Cell Death Dis (2019) 10(6):378. doi: 10.1038/s41419-019-1598-x
96. Jahangiri B, Khalaj-Kondori M, Asadollahi E, Sadeghizadeh M. Cancer-Associated Fibroblasts Enhance Cell Proliferation and Metastasis of Colorectal Cancer SW480 Cells by Provoking Long Noncoding RNA Uca1. J Cell Commun Signal (2019) 13(1):53–64. doi: 10.1007/s12079-018-0471-5
97. Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, et al. BRAF-Activated Long Non-Coding RNA Contributes to Colorectal Cancer Migration by Inducing Epithelial-Mesenchymal Transition. Oncol Lett (2014) 8(2):869–75. doi: 10.3892/ol.2014.2154
98. Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, et al. LncRNA-ATB Mediated E-Cadherin Repression Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Gastroenterol Hepatol (2016) 31(3):595–603. doi: 10.1111/jgh.13206
99. Tong G, Liu Y, Wu X, Wang S. Knockdown of HOXA-AS2 Suppresses Proliferation and Induces Apoptosis in Colorectal Cancer. (2017).
100. Wu XL, Lu RY, Wang LK, Wang YY, Dai YJ, Wang CY, et al. Long Noncoding RNA HOTAIR Silencing Inhibits Invasion and Proliferation of Human Colon Cancer LoVo Cells Via Regulating IGF2BP2. J Cell Biochem (2018).
101. Liu X, Cui L, Hua D. Long Noncoding RNA XIST Regulates miR-137-EZH2 Axis to Promote Tumor Metastasis in Colorectal Cancer. Oncol Res (2018) 27(1):99–106. doi: 10.3727/096504018X15195193936573
102. Shen X, Hu X, Mao J, Wu Y, Liu H, Shen J, et al. The Long Noncoding RNA TUG1 Is Required for TGF-Beta/TWIST1/EMT-Mediated Metastasis in Colorectal Cancer Cells. Cell Death Dis (2020) 11(1):65. doi: 10.1038/s41419-020-2254-1
103. Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-Coding RNA MALAT1 Promotes Tumour Growth and Metastasis in Colorectal Cancer Through Binding to SFPQ and Releasing Oncogene PTBP2 From SFPQ/PTBP2 Complex. Br J Cancer (2014) 111(4):736–48. doi: 10.1038/bjc.2014.383
104. Shen X, Bai Y, Luo B, Zhou X. Upregulation of lncRNA BANCR Associated With the Lymph Node Metastasis and Poor Prognosis in Colorectal Cancer. Biol Res (2017) 50(1):32. doi: 10.1186/s40659-017-0136-5
105. Wang Q, Shi L, Shi K, Yuan B, Cao G, Kong C, et al. CircCSPP1 Functions as a ceRNA to Promote Colorectal Carcinoma Cell EMT and Liver Metastasis by Upregulating Col1a1. Front Oncol (2020) 10:850. doi: 10.3389/fonc.2020.00850
106. Deng Z, Li X, Wang H, Geng Y, Cai Y, Tang Y, et al. Dysregulation of CircRNA_0001946 Contributes to the Proliferation and Metastasis of Colorectal Cancer Cells by Targeting MicroRNA-135a-5p. Front Genet (2020) 11:357. doi: 10.3389/fgene.2020.00357
107. Shen T, Cheng X, Liu X, Xia C, Zhang H, Pan D, et al. Circ_0026344 Restrains Metastasis of Human Colorectal Cancer Cells via miR-183. Artif Cells Nanomed Biotechnol (2019) 47(1):4038–45. doi: 10.1080/21691401.2019.1669620
108. Xiong G, Zhang J, Zhang Y, Pang X, Wang B, Zhang Y. Circular RNA_0074027 Participates in Cell Proliferation, Apoptosis and Metastasis of Colorectal Cancer Cells Through Regulation of Mir5253p. Mol Med Rep (2021) 23(5). doi: 10.3892/mmr.2021.11963
109. Li C, Zhou H. Circular RNA Hsa_circRNA_102209 Promotes the Growth and Metastasis of Colorectal Cancer Through miR-761-Mediated Ras and Rab Interactor 1 Signaling. Cancer Med (2020) 9(18):6710–25. doi: 10.1002/cam4.3332
110. Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (Hsa_Circ_0005273) as a Novel Therapeutic Target for Metastatic Colorectal Cancer. Mol Cancer (2020) 19(1):13. doi: 10.1186/s12943-020-1139-3
111. Rena C, Zhanga Z, Wang S, Zhu W, Zheng P, Wang W. Circular RNA Hsa_Circ_0001178 Facilitates the Invasion and Metastasis of Colorectal Cancer Through Upregulating ZEB1 via Sponging Multiple miRNAs. Biol Chem (2019) 401:487–96. doi: 10.1515/hsz-2019-0350
112. Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa_circ_0009361 Acts as the Sponge of miR-582 to Suppress Colorectal Cancer Progression by Regulating APC2 Expression. Clin Sci (2019) 133(10):1197–213. doi: 10.1042/CS20190286
113. Zhang J, Wang H, Wu K, Zhan F, Zeng H. Dysregulated circRNA_100876 Contributes to Proliferation and Metastasis of Colorectal Cancer by Targeting microRNA-516b (miR-516b). Cancer Biol Ther (2020) 21(8):733–40. doi: 10.1080/15384047.2020.1776075
114. Wang X, Liu S, Xu B, Liu Y, Kong P, Li C, et al. Circ-SIRT1 Promotes Colorectal Cancer Proliferation and EMT by Recruiting and Binding to Eif4a3. Analytical Cell Pathol (2021) 2021:5739769. doi: 10.1155/2021/5739769
115. Min L, Wang H, Zeng Y. CircRNA_104916 Regulates Migration, Apoptosis and Epithelial-Mesenchymal Transition in Colon Cancer Cells. Front Biosci (Landmark Ed) (2019) 24:819–32. doi: 10.2741/4753
116. Wu M, Kong C, Cai M, Huang W, Chen Y, Wang B, et al. Hsa_circRNA_002144 Promotes Growth and Metastasis of Colorectal Cancer Through Regulating miR-615-5p/LARP1/mTOR Pathway. Carcinogenesis (2021) 42(4):601–10. doi: 10.1093/carcin/bgaa140
117. Li Y, Pei F, Cao M. CircRNA_101951 Promotes Migration and Invasion of Colorectal Cancer Cells by Regulating the KIF3A-Mediated EMT Pathway. Exp Ther Med (2020) 19(5):3355–61.
118. Li N. CircTBL1XR1/miR-424 Axis Regulates Smad7 to Promote the Proliferation and Metastasis of Colorectal Cancer. J Gastrointestinal Oncol (2020) 11(5):918–31. doi: 10.21037/jgo-20-395
119. Zhou J, Wang L, Sun Q, Chen R, Zhang C, Yang P, et al. Et Al: Hsa_circ_0001666 Suppresses the Progression of Colorectal Cancer Through the miR-576-5p/PCDH10 Axis. Clin Trans Med (2021) 11(11):e565. doi: 10.1002/ctm2.565
120. Wang Y, Wang Z. Efficient Backsplicing Produces Translatable Circular mRNAs. Rna (2015) 21(2):172–9. doi: 10.1261/rna.048272.114
121. Li X, Yang L, Chen L-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell (2018) 71(3):428–42. doi: 10.1016/j.molcel.2018.06.034
122. Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W, et al. N6-Methyladenosine-Induced Circ1662 Promotes Metastasis of Colorectal Cancer by Accelerating YAP1 Nuclear Localization. Theranostics (2021) 11(9):4298–315. doi: 10.7150/thno.51342
123. Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 Contributes to Growth, Metastasis and Angiogenesis of Colorectal Cancer: Involvement of miR-30c-5p/TCF7 Axis. Mol Cancer (2021) 20(1):93. doi: 10.1186/s12943-021-01372-0
124. Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread Changes in Protein Synthesis Induced by microRNAs. Nature (2008) 455(7209):58–63. doi: 10.1038/nature07228
125. Seitz H. Redefining microRNA Targets. Curr Biol CB (2009) 19(10):870–3. doi: 10.1016/j.cub.2009.03.059
126. Thomson DW, Dinger ME. Endogenous microRNA Sponges: Evidence and Controversy. Nat Rev Genet (2016) 17(5):272–83. doi: 10.1038/nrg.2016.20
127. Zhang Y, Liu X, Li Q, Zhang Y. lncRNA LINC00460 Promoted Colorectal Cancer Cells Metastasis via miR-939-5p Sponging. Cancer Manage Res (2019) 11:1779–89. doi: 10.2147/CMAR.S192452
128. Hong W, Ying H, Lin F, Ding R, Wang W, Zhang M. lncRNA LINC00460 Silencing Represses EMT in Colon Cancer Through Downregulation of ANXA2 via Upregulating miR-433-3p. Mol Ther Nucleic Acids (2020) 19:1209–18. doi: 10.1016/j.omtn.2019.12.006
129. Zhao S, Guan B, Mi Y, Shi D, Wei P, Gu Y, et al. LncRNA MIR17HG Promotes Colorectal Cancer Liver Metastasis by Mediating a Glycolysis-Associated Positive Feedback Circuit. Oncogene (2021) 40(28):4709–24. doi: 10.1038/s41388-021-01859-6
130. Zhou M, Bian Z, Liu B, Zhang Y, Cao Y, Cui K, et al. Long Noncoding RNA MCM3AP-AS1 Enhances Cell Proliferation and Metastasis in Colorectal Cancer by Regulating miR-193a-5p/SENP1. Cancer Med (2021) 10(7):2470–81. doi: 10.1002/cam4.3830
131. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 Sponges miR-216b to Promote Proliferation and Liver Metastasis of Colorectal Cancer Through Upregulating GALNT1. Cell Death Dis (2018) 9(7):722. doi: 10.1038/s41419-018-0759-7
132. Hou P, Meng S, Li M, Lin T, Chu S, Li Z, et al. LINC00460/DHX9/IGF2BP2 Complex Promotes Colorectal Cancer Proliferation and Metastasis by Mediating HMGA1 mRNA Stability Depending on M6a Modification. J Exp Clin Cancer Res CR (2021) 40(1):52. doi: 10.1186/s13046-021-01857-2
133. Thomas LF, Saetrom P. Circular RNAs Are Depleted of Polymorphisms at microRNA Binding Sites. Bioinformatics (2014) 30(16):2243–6. doi: 10.1093/bioinformatics/btu257
134. Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic Insights Into M(6)A RNA Enzymes. Biochim Biophys Acta Gene Regul Mech (2019) 1862(3):222–9. doi: 10.1016/j.bbagrm.2018.10.014
135. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-Dependent M6a Demethylation Controls Splicing and Stability of Long 3'-UTR mRNAs in Male Germ Cells. Proc Natl Acad Sci USA (2018) 115(2):E325–33. doi: 10.1073/pnas.1717794115
136. Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN, et al. Regulation of Co-Transcriptional Pre-mRNA Splicing by M(6)A Through the Low-Complexity Protein hnRNPG. Mol Cell (2019) 76(1):70–81.e79. doi: 10.1016/j.molcel.2019.07.005
137. Zhang. YTHDC1 Mediates Nuclear Export of N6-Methyladenosine Methylated mRNAs. (2017). doi: 10.7554/eLife.31311
138. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
139. Yao L, Yin H, Hong M, Wang Y, Yu T, Teng Y, et al. RNA Methylation in Hematological Malignancies and Its Interactions With Other Epigenetic Modifications. Leukemia (2021) 35(5):1243–57. doi: 10.1038/s41375-021-01225-1
140. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
141. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine Marks Primary microRNAs for Processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
142. Sun L, Wan A, Zhou Z, Chen D, Liang H, Liu C, et al. RNA-Binding Protein RALY Reprogrammes Mitochondrial Metabolism via Mediating miRNA Processing in Colorectal Cancer. Gut (2021) 70(9):1698–712. doi: 10.1136/gutjnl-2020-320652
143. Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X, et al. Molecular Characterization and Clinical Relevance of M(6)A Regulators Across 33 Cancer Types. Mol Cancer (2019) 18(1):137. doi: 10.1186/s12943-019-1066-3
144. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
145. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 Promotes Metastasis of Colorectal Cancer via miR-1246/SPRED2/MAPK Signaling Pathway. J Exp Clin Cancer Res CR (2019) 38(1):393. doi: 10.1186/s13046-019-1408-4
146. Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, et al. METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther J Am Soc Gene Ther (2020) 28(2):599–612. doi: 10.1016/j.ymthe.2019.11.016
147. Xu L, L M, Wang M, Yan D, Feng G, An G. The Expression of microRNA-375 in Plasma and Tissue Is Matched in Human Colorectal Cancer. (2014).
148. Jepsen RK, N G, Klarskov LL, Christensen IJ, Riis LB, Høgdall E. Intra-Tumor Heterogeneity of microRNA-92a,microRNA-375 and microRNA-424 in Colorectal Cancer. Exp Mol Pathol (2016) 100:125–31. doi: 10.1016/j.yexmp.2015.12.004
149. Zaharie F, Muresan MS, Petrushev B, Berce C, Gafencu GA, Selicean S, et al. Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. J Gastrointestinal Liver Dis JGLD (2015) 24(4):435–43. doi: 10.15403/jgld.2014.1121.244.375
150. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-Methyladenosine Modification of Circnsun2 Facilitates Cytoplasmic Export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat Commun (2019) 10(1):4695. doi: 10.1038/s41467-019-12651-2
151. Kuai D, Zhu S, Shi H, Yang R, Liu T, Liu H, et al. Aberrant Expression of M(6)A mRNA Methylation Regulators in Colorectal Adenoma and Adenocarcinoma. Life Sci (2021) 273:119258. doi: 10.1016/j.lfs.2021.119258
152. SHEN X-P, L X, LU H, ZHOU C-X, ZHANG J-K, YU Q. Low Expression of microRNA-1266 Promotes Colorectal Cancer Progression via Targeting FTO. Eur Rev Med Pharmacol Sci (2018) 22:8220–6. doi: 10.26355/eurrev_201812_16516
153. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting With and Triggering YAP Phosphorylation and Degradation and Is Negatively Regulated by the M(6)A Reader YTHDF3. Mol Cancer (2019) 18(1):143. doi: 10.1186/s12943-019-1079-y
154. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-Wide Maps of M6a circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns That Are Distinct From mRNAs. Cell Rep (2017) 20(9):2262–76. doi: 10.1016/j.celrep.2017.08.027
155. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. M6a-Induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells Via Upregulation Zeb1. Mol Cancer (2019) 18(1). doi: 10.1186/s12943-019-1014-2
156. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The Biology, Function, and Applications of Exosomes in Cancer. Acta Pharm Sin B (2021) 11(9):2783–97. doi: 10.1016/j.apsb.2021.01.001
157. Li T, Wan Y, Su Z, Li J, Han M, Zhou C. Mesenchymal Stem Cell-Derived Exosomal microRNA-3940-5p Inhibits Colorectal Cancer Metastasis by Targeting Integrin Alpha6. Digestive Dis Sci (2021) 66(6):1916–27. doi: 10.1007/s10620-020-06458-1
158. Xu H, Lan Q, Huang Y, Zhang Y, Zeng Y, Su P, et al. The Mechanisms of Colorectal Cancer Cell Mesenchymal-Epithelial Transition Induced by Hepatocyte Exosome-Derived miR-203a-3p. BMC Cancer (2021) 21(1):718. doi: 10.1186/s12885-021-08419-x
159. Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA Biomarkers: Emerging Frontiers in Colorectal and Other Human Cancers. Expert Rev Mol Diagn (2016) 16(5):553–67. doi: 10.1586/14737159.2016.1156535
160. Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, et al. CAFs Secreted Exosomes Promote Metastasis and Chemotherapy Resistance by Enhancing Cell Stemness and Epithelial-Mesenchymal Transition in Colorectal Cancer. Mol Cancer (2019) 18(1):91. doi: 10.1186/s12943-019-1019-x
161. Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-Transmitted miR-128-3p Increase Chemosensitivity of Oxaliplatin-Resistant Colorectal Cancer. Mol Cancer (2019) 18(1):43. doi: 10.1186/s12943-019-0981-7
162. Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, et al. LncRNA RPPH1 Promotes Colorectal Cancer Metastasis by Interacting With TUBB3 and by Promoting Exosomes-Mediated Macrophage M2 Polarization. Cell Death Dis (2019) 10(11):829. doi: 10.1038/s41419-019-2077-0
163. Dou R, Liu K, Yang C, Zheng J, Shi D, Lin X, et al. EMT-Cancer Cells-Derived Exosomal miR-27b-3p Promotes Circulating Tumour Cells-Mediated Metastasis by Modulating Vascular Permeability in Colorectal Cancer. Clin Trans Med (2021) 11(12):e595. doi: 10.1002/ctm2.595
164. Wang D, Wang X, Si M, Yang J, Sun S, Wu H, et al. Exosome-Encapsulated miRNAs Contribute to CXCL12/CXCR4-Induced Liver Metastasis of Colorectal Cancer by Enhancing M2 Polarization of Macrophages. Cancer Lett (2020) 474:36–52. doi: 10.1016/j.canlet.2020.01.005
165. Rezaei R, Baghaei K, Amani D, Piccin A, Hashemi SM, Asadzadeh Aghdaei H, et al. Exosome-Mediated Delivery of Functionally Active miRNA-375-3p Mimic Regulate Epithelial Mesenchymal Transition (EMT) of Colon Cancer Cells. Life Sci (2021) 269:119035. doi: 10.1016/j.lfs.2021.119035
166. Zhang X, Bai J, Yin H, Long L, Zheng Z, Wang Q, et al. Exosomal miR-1255b-5p Targets Human Telomerase Reverse Transcriptase in Colorectal Cancer Cells to Suppress Epithelial-to-Mesenchymal Transition. Mol Oncol (2020) 14(10):2589–608. doi: 10.1002/1878-0261.12765
167. Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, et al. Colorectal Cancer-Derived Exosomal miR-106b-3p Promotes Metastasis by Down-Regulating DLC-1 Expression. Clin Sci (2020) 134(4):419–34. doi: 10.1042/CS20191087
168. Zhou L, Li J, Tang Y, Yang M. Exosomal LncRNA LINC00659 Transferred From Cancer-Associated Fibroblasts Promotes Colorectal Cancer Cell Progression via miR-342-3p/ANXA2 Axis. J Trans Med (2021) 19(1):8. doi: 10.1186/s12967-020-02648-7
169. Xiao Z, Liu Y, Li Q, Liu Q, Liu Y, Luo Y, et al. EVs Delivery of miR-1915-3p Improves the Chemotherapeutic Efficacy of Oxaliplatin in Colorectal Cancer. Cancer Chemother Pharmacol (2021) 88(6):1021–31. doi: 10.1007/s00280-021-04348-5
170. Mannavola F, Pezzicoli G, Tucci M. DLC-1 Down-Regulation via Exosomal miR-106b-3p Exchange Promotes CRC Metastasis by the Epithelial-to-Mesenchymal Transition. Clin Sci (2020) 134(8):955–9. doi: 10.1042/CS20200181
171. Xu Y, Qiu A, Peng F, Tan X, Wang J, Gong X. Exosomal Transfer of Circular RNA FBXW7 Ameliorates the Chemoresistance to Oxaliplatin in Colorectal Cancer by Sponging miR-18b-5p. Neoplasma (2021) 68(1):108–18. doi: 10.4149/neo_2020_200417N414
Keywords: colorectal cancer (CRC), epithelial to mesenchymal transition (EMT), exosomes, DNA methylation, non-coding RNAs (ncRNAs), N6-methyladenosine (m6A) RNA, metastasis
Citation: Yang M, Sun M and Zhang H (2022) The Interaction Between Epigenetic Changes, EMT, and Exosomes in Predicting Metastasis of Colorectal Cancers (CRC). Front. Oncol. 12:879848. doi: 10.3389/fonc.2022.879848
Received: 20 February 2022; Accepted: 01 April 2022;
Published: 30 May 2022.
Edited by:
Yongye Huang, Northeastern University, ChinaCopyright © 2022 Yang, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huijing Zhang, emhqMTIyM0Bob3RtYWlsLmNvbQ==
 Meiqi Yang
Meiqi Yang Mingjun Sun
Mingjun Sun Huijing Zhang
Huijing Zhang