- Department of Oncology, The fifth Affiliated Hospital of Sun Yat-sen University, Zhuhai, China
Dysregulation of cysteine cathepsin protease activity is pivotal in tumorigenic transformation. However, the role of cathepsin protease in lung cancer remains unknown. Here, we analyzed GEO database and found that lung cancer presented high expression of cathepsin V (CTSV). We then performed immunohistochemistry assay in 73 paired lung cancer tissues and normal lung tissues and confirmed that CTSV is overexpressed in lung cancer and correlates with poor prognosis. The mass spectrometry experiment showed that the N-glycosylation locus of CTSV are N221 and N292, glycosylated CTSV (band 43 kDa) was particularly expressed in lung cancer samples and correlated with lymph node metastasis. Mechanistic studies showed that only glycosylated CTSV (43-kDa band) are secreted to extracellular matrix (ECM) and promoted the metastasis of lung cancer. Importantly, the Elisa detection in serum of 12 lung cancer patients and 12 healthy donors showed that the level of CTSV in serum distinguished lung cancer patients from healthy donors. Together, our findings reveal the clinical relevance of CTSV glycosylation and CTSV drives the metastasis of lung cancer, suggesting that the glycosylated CTSV in serum is a promising biomarker for lung cancer.
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide (1). The high lethality rate observed in lung cancer patients is related to the common diagnosis of advanced stage disease, which hinders curative treatment and indicates poor 5-year survival. Metastasis has been proven to be the primary cause of cancer-related death among lung cancer patients (2, 3). Known protooncogenes of lung cancer include EGFR, HER2, HER3, AKL/EML4, KRAS, BRAF, MET, NRAS, PIK3CA and ROS1, among others (4, 5). Lung cancer-related tumor suppressors include RBM5, FUS1, TP53, LKB1, SNPs, APC, etc. The upregulation of oncogenes and the mutation of tumor suppressor genes may both involved in the progression of lung cancer (6). In recent years, targeted therapeutics in the treatment of lung cancer would be a substantial advance, as such events represent new targets for promising therapeutic intervention (7–10). Consequently, understanding the molecular mechanisms of lung cancer pathogenesis and identifying potential therapeutic targets for the clinical management of lung cancer are imminent research objectives.
Recently study showed that released cathepsins increases tumor cell invasion and metastasis in lung cancer (11). CTSS degradation of nidogen-1 is strongly associated with NSCLC (12). CTSF was found to be downregulated in NSCLC samples and overexpression of CTSF was correlated with favorable prognosis of NSCLC (13). Also, CTSB was proved to be closely related to the pathogenesis of lung cancer in a mouse model (14). The regulation of cathepsin C and cathepsin H (CTSC and CTSH) can cause a split anergic state in NK cells (15). The cysteine proteinase cathepsin O (CTSO) decreases the protein levels of BRCA1 and ZNF423 by proteinase-mediated degradation to regulate the tamoxifen sensitivity of breast cancer (16). Mutation of cathepsin F (CTSF) is correlated with type B Kufs disease and Alzheimer’s disease (17, 18). Cathepsin W (CTSW) is an important host factor for the entry of influenza A virus into target cells from late endosomes (19). Cathepsin V (CTSV) has been shown to be a tumor metastasis-associated protease in colorectal cancer, breast carcinoma and thymic epithelial tumors and is correlated with poor outcomes (20–23). Yuki Niwa et al. demonstrated that the N-glycosylation of CTSV is crucial for its transportation to lysosomes and secretion (24). However, the detailed picture of the downstream molecular mechanism of CTSV is still unclear. It is of great importance to understand the crucial roles and individual functions of CTSV in disease from the perspectives of both basic science and pharmacological targeting.
In this study, we reported that the overexpression of CTSV in lung cancer patients are positively correlated with poor overall survival. CTSV was glycosylated at N221 and N292 sites and the glycosylation of CTSV is correlated with lymph node metastasis. Notably, serum (secreted) CTSV distinguished lung cancer patients from healthy donors, and glycosylated CTSV determined its secretion and pro-metastatic role in lung cancer. Our study reveals glycosylated CTSV holds promise for providing effective prognostic biomarker for lung cancer patients.
Methods and Materials
Human Tissue Samples
73 fresh-frozen paired samples of primary lung cancer tissues and normal lung tissues were obtained from the Oncology Department at The Fifth Affiliated Hospital of Sun Yat-sen University. All human sample studies were reviewed and approved by the Ethics Committee of Sun Yat-sen University Cancer Center (GZR2020-154), and informed written consent was obtained from all donors.
Cell Culture and Transfection
All cell lines A-549 (RRID : CVCL_0023), NCI-H1975 (RRID : CVCL_1511), NCI-H292 (RRID : CVCL_0455), HEK293T (RRID : CVCL_0063), NCI-H1993 (RRID : CVCL_1512) were purchase from ATCC and identified by short tandem repeat (STR) profiles from the China Center For Type Culture Collection (CCTCC, Wuhan University, China). All experiments were performed with mycoplasma-free cells. Cells were cultured with Dulbecco’s modified Eagle’s medium media (Thermo Scientific, USA) with 10% fetal bovine serum (FBS, Thermo Scientific, USA). HEK-293T cells were cultured in a six-well plate with complete culture medium the day before and transfected when the cell confluence was approximately 90%. We used a plasmid lentiviral packaging system, and the two packaging plasmids psPAX2 and PMD2.G (Thermo Scientific, USA) and the target gene were mixed at 3 μg:2 μg:1 μg. Then, polyethylenimine (PEI) was added to a quarter of the plasmid. Finally, 200 μL Opti-MEM medium was added to the mixture, and then the mixture was evenly added to the cells. After 6-8 hours, the solution was changed to complete medium. After 48 hours, the cell culture supernatant was collected and filtered with a 0.4-μm filter membrane to obtain virus solution.
Migration and Invasion Assays
The indicated lung cancer cell migration and invasion ability were detected by Transwell assay using 24-well Boyden chambers (BD Inc., USA) with 8-μm pores coated with (invasion) or without (migration) Matrigel. We seeded 5×104 A549 cells or 1×105 (NCI-H1975 and NCI-H292) cells per well on the Transwell inserts and incubated them in 300 μL of serum-free DMEM at 37°C in the top chambers for 12 hours and 24 hours, whereas DMEM containing 20% FBS was added to the lower chamber. Cells that traversed the inserts to the bottom chamber surface were fixed, stained and observed under phase-contrast microscopy.
Western Blotting
Briefly, cells were collected and analyzed in lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.1% SDS) supplemented with 1% protease inhibitor (Sigma-Aldrich, USA) after being gently washed 3 times with cold PBS, and then the lysate supernatant was obtained after centrifugation at 13,000 ×g and 4°C for 20 min. Protein (20 μg) was loaded and separated on 10% SDS-PAGE minigels, transferred onto PVDF membranes (Millipore, USA) and blocked with 5% blocking buffer for 1 hour at room temperature. The PVDF membranes were incubated with primary antibody overnight at 4°C with constant shaking, followed by incubation with horseradish peroxidase-conjugated secondary antibody (Promega, USA) for 1 hour at room temperature. The proteins were detected with an enhanced chemiluminescence (ECL) system (Bio-Rad, USA).
Protein Mass Spectrometry Analysis
Briefly, A549 and NCI-H1975 cells were transfected to express Flag-tagged CTSV. The cells were then lysed in NETN buffer containing 15 mmol/L NaF, 60 mmol/L β-glycerophosphate, and 1 mg/mL each of pepstatin A and aprotinin. The debris was removed and incubated with Flag-conjugated beads at 4°C for 4 hours. Then, the beads were washed with NETN buffer 5 times, the bound proteins were analyzed by SDS-PAGE, and protein mass spectrometry was performed by PTM BioLabs. The immunocomplexes were then washed 4 times with NETN buffer and determined by SDS-PAGE and Western blot.
Immunohistochemistry Staining
Lung cancer tissues were fixed in 4% paraformaldehyde for 16 hours at room temperature and then dehydrated in descending concentrations of ethanol. The tumor sections were incubated in 0.3% H2O2 solution at room temperature for 20 min and washed with PBS 3 times. Then, the cells were probed with monoclonal anti-CTSV (1:200) at 4°C overnight in a humidified container. After washing with PBS 3 times, the tissue sections were treated with goat anti-rabbit at room temperature for 2 hours. Streptavidin/peroxidase complex and diaminobenzidine were used for immunostaining, and hematoxylin was used for counterstaining.
Antibodies
Antibodies specific for CTSV (WB: Abcam Cat#ab166894, IHC: Theremofish Cat#PA5-47061, ELISA: Theremofish, Cat#PA5-112393, RRID : AB_2261304), Flag-Tag (Cell Signaling, Cat#14793S, RRID : AB_2797401), HA-Tag (Cell Signaling, Cat#3724S, RRID : AB_391833), Tubulin (Genetex, Cat#GTX112141, RRID : AB_1157911) HSP70 (Abcam, Cat#ab2787, RRID : AB_1874830) and GAPDH (Genetex, Cat#GTX100118, RRID : AB_2617427) were purchased from the indicated companies.
Concentration Medium Obtaining
The condition medium from indicated cultured cells were collected, and centrifuge at a centrifugal force of 2,000g per minute for 20 minutes, collected the supernatant for centrifuge at a centrifugal force of 10,000g per minute for 1 hour, collected the supernatant and repeated the centrifugation with 100,000g for 1 hour. Finally subjected the supernatant to concentration column and obtained that concentrated medium(CM).
Statistical Analysis
All data were obtained from two or three independent experiments and are presented as the mean ± SD. We conducted the analysis in version 7.00 of GraphPad Prism using the unpaired, two-tailed Student’s t-test module, where ns represents no statistical significance, * represents P value<0.05, ** represents P value<0.01, *** represents P value<0.001, and **** represents P value<0.0001. Graphs were drawn with GraphPad Prism software. Kaplan-Meier survival analyses were used to compare survival among lung cancer patients based on CTSV expression. Western blot bands were quantified with Image-Pro Plus, and the results were normalized to those for the control GAPDH. Statistically significance was defined as P<0.05.
Results
CTSV Is Upregulated in Lung Cancer and Correlated With Poor Survival
Cysteine cathepsins are pivotal in disease development and progression; they have been reported to be overexpressed in a number of cancers and to lead to increased cancer cell invasion and metastasis (25–27), however, many facets of CTSV activity in lung cancer remain uncharacterized. To explore the role that the cysteine cathepsin family plays in lung cancer pathogenesis, we investigated The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets and compared the gene expression of cathepsins family (CTSB, CTSC, CTSF, CTSH, CTSK, CTSL, CTSO, CTSS, CTSV, CTSW and CTSX) changes in lung cancer tissues and normal tissues. The RNA sequence data from the TCGA demonstrated that the expression of CTSW, CTSH, CTSO and CTSS was downregulated in lung cancer tissues; notably, only CTSV was found upregulated in lung cancer tissues (n=1037), approximately 2.6-fold higher than that in normal tissues (n=108) (Figures 1A, B), and no significant changes were observed in the remaining cathepsin family members. In addition, analysis of GEO datasets (GSE31210, GSE19188 and GSE3269) showed that lung cancer exhibits high CTSV levels (Supplementary Figures S1A–C). Next, we performed microarray gene expression profile analysis of lung cancer tissues from patients (n=73) with newly diagnosed lung cancer with paired adjacent normal tissues. Immunohistochemical staining of CTSV showed that CTSV expression in lung tumor tissues was higher than that in normal tissues (P<0.0001) (Figures 1C, D). We then analyzed the association of CTSV expression with the prognosis of 73 lung cancer patients. Our Kaplan-Meier analysis revealed that high expression of CTSV were correlated with poor overall survival (P=0.028) (Figure 1E). Further exploration of the Kaplan-Meier Plotter database (28) showed that elevated CTSV expression was correlated with poor survival in cancer patients (Figure 1F). Collectively, these results showed that CTSV is upregulated in lung cancer and is correlated with poor overall survival, implying the oncogenic role of CTSV.
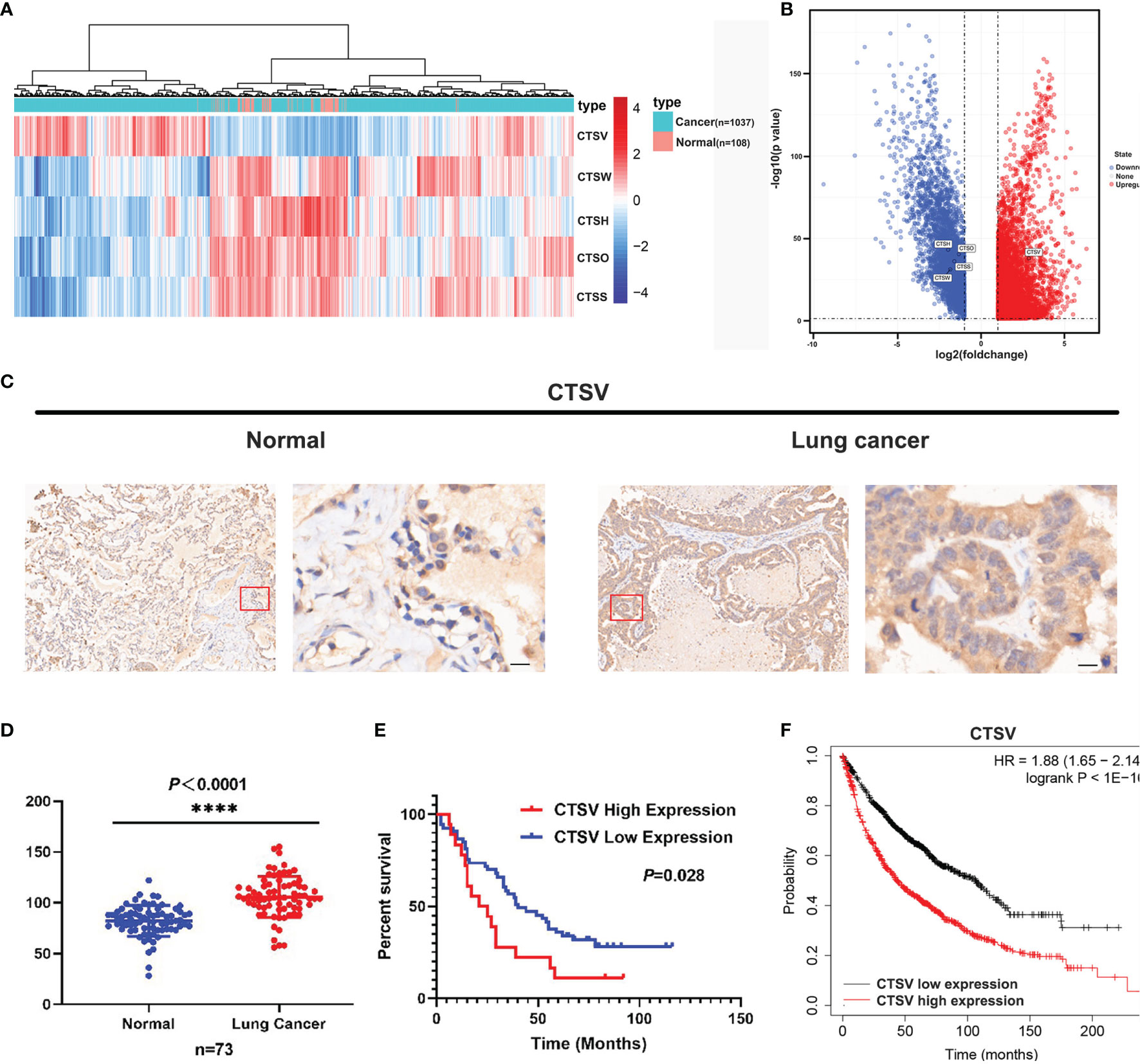
Figure 1 Lung cancer patients presented with high CTSV expression. (A), (B) Analysis of mRNA expression of cysteine cathepsin family members (11 members) in lung cancer tissue and normal tissue from the TCGA database. (C) Representative images of CTSV IHC staining of lung cancer tissue and normal tissue. Scale bars, 50 μm. (D) Expression of CTSV in 73 paired lung cancer normal tissue samples. (E) Kaplan-Meier survival curves based on CTSV expression in lung cancer tissues. (F) Analysis of lung cancer patients’ overall survival in the Kaplan-Meier Plotter database. ****P < 0.0001.
CTSV Is Glycosylated in Lung Cancer
While detecting the protein expression of CTSV in cancer cell lines, we noticed that the CTSV detection by Western blotting revealed more than one band, and the majority of the protein was found at 43 kDa. To determine whether the higher band is responsible for CTSV glycosylation, we treated NCI-H292 cells lysates with peptide-N-glycosidase F (PNGase F) and endoglycosidase (Endo H) to remove the structure of N-glycan and then analyzed the cell lysates by Western blotting. A significant reduction in 39- and 43-kDa CTSV and a remarkable increase in 37-kDa CTSV were observed (Figure 2A). The results generally revealed that glycosylated CTSV is present and that the higher band was indeed the glycosylated form of CTSV.
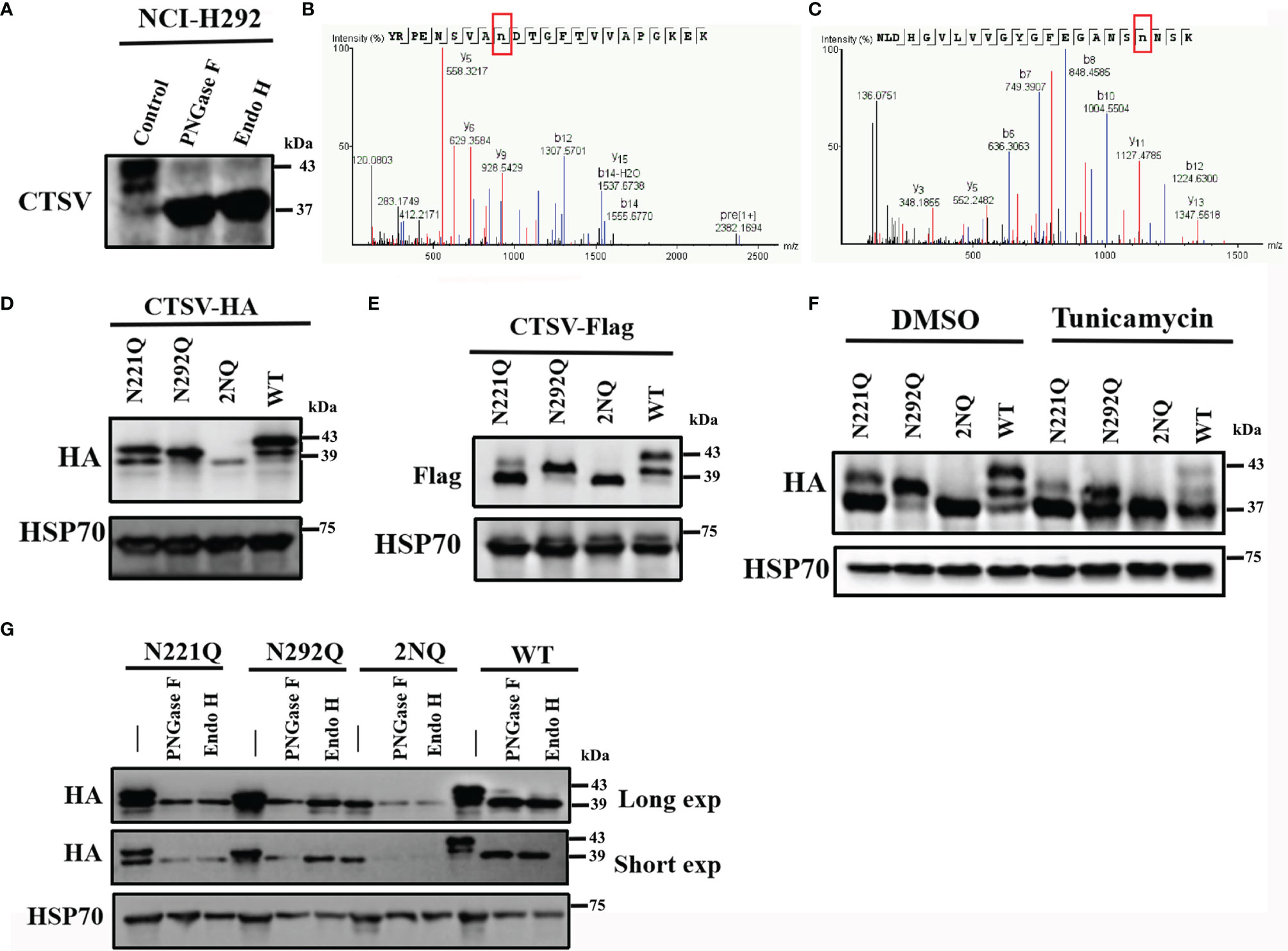
Figure 2 Identification of the glycosylation locus CTSV. (A) NCI-H292 cells lysates treated with PNGase F and Endo H were analyzed by Western blotting. (B, C) LC-MS/MS of the CTSV N221 and N292 glycosylation sites. (D, E) Cells transfected with the indicated plasmids with N221, N292 or both mutated sites were analyzed by Western blotting. (F) The indicated cells with N221Q, N292Q or both mutated sites were incubated with DMSO and tunicamycin (100ng/mL) and then analyzed by Western blotting. (G) Western blot analysis of the indicated stable cells lysates treated with PNGase F and Endo H with a long exposure time (60 sec) and short exposure time (20 sec).
Yuki Niwa et al. observed that cathepsin V is glycosylated at both Asn221 and Asn292 in human fibrosarcoma HT1080 cells (24). To further investigate the glycan locus in lung cancer cells, we used liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) to determine certain sites of glycosylation for CTSV. Glycopeptides carrying N-glycans were identified for 2 N-glycosylation sites, which is consistent with a previous study (Figures 2B, C). The substitution of asparagine (N) to glutamine (Q) (N221Q, N292Q or both [2NQ]), caused a certain level of decrease in glycosylation when compared with that in the wild-type (WT) cells (Figures 2D, E). When treated with the N-linked glycosylation inhibitor tunicamycin, the glycosylation of CTSV was completely suppressed (Figure 2F). Similarly, hemagglutinin (HA)-tagged CTSV had a molecular weight shift from its regular size when treated with Endo H and PNGase F (Figure 2G). Together, the results suggested that CTSV is exclusively N-glycosylated at N221 and N292 in lung cancer cells.
Glycosylation of CTSV Is Associated With Metastasis in Lung Cancer Patients
Next, we explored the clinical relevance of CTSV glycosylation in the lymph node metastasis of lung cancer. Western blot analysis of 24 paired lung samples showed that glycosylated CTSV was expressed at different levels in lung cancer tissues (Figures 3A–D). Importantly, three bands were observed in most lung cancer tissues but one or two bands were observed in normal tissues; therefore, we suspected that the glycosylation of CTSV might be associated with the progression of lung cancer. In this regard, we further assessed the relationship of glycosylation level with lymph node metastasis. Our data showed that the protein levels of the first and second bands of CTSV (the glycosylated forms of CTSV; bands at 43 and 39 kDa) were significantly higher in tumor tissues than in normal tissues, and no significant differences were observed in the third band (Figures 3E, G, I). In addition, we found that a higher protein level of the first glycosylation band (band 43 kDa) in lung cancer patients was correlated with lymph node metastasis (P=0.0173), while no significant changes were observed in the second and third bands (39 kDa and 37 kDa) of CTSV (Figures 3F, H, J and Supplementary Table 1). Furthermore, the data also showed that the protein level of total CTSV was significantly higher than that in normal tissues and was correlated with lymph node metastasis as well (Figures 3K, I). Together, the results suggested that the level of glycosylation CTSV (band 43 kDa) might be a more sensitive prognostic marker for lung cancer patients with metastasis.
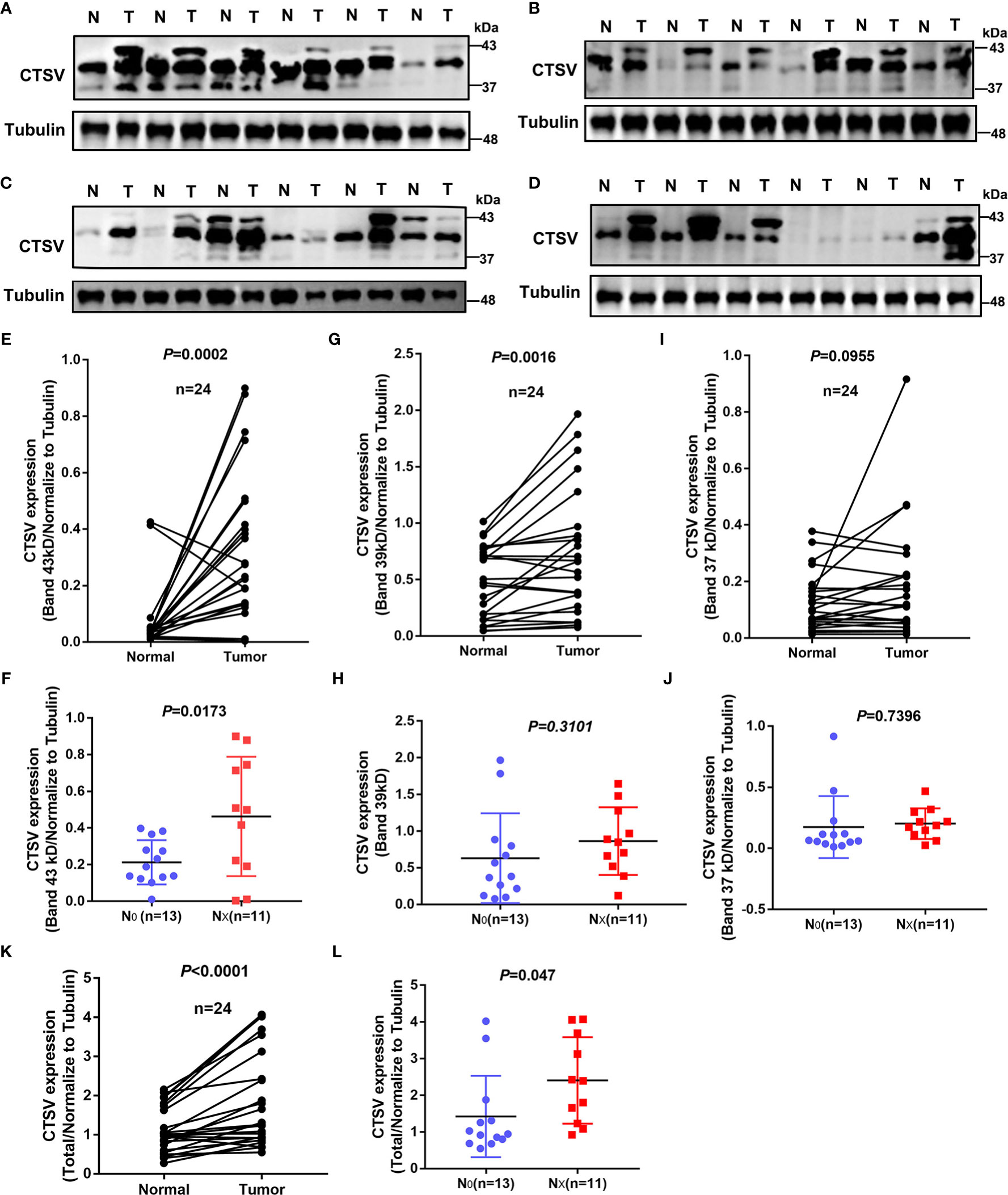
Figure 3 Glycosylation of CTSV is correlated with metastasis in lung cancer. (A–D) Western blot analysis of 24 pairs of lung specimens from lung cancer patients using anti-CTSV antibody. (E–I, K) Quantification of the Western blot bands by using ImageJ 6.0. (F–J, L) All patients were divided into the no lymph node metastasis group and the lymph node metastasis group, and CTSV expression was analyzed by Western blotting.
Glycosylation of CTSV Determine its Secretion and Metastasis of Lung Cancer Cells
CTSV acts as an intracellular proteolytic enzyme, we suspected that CTSV is secreted from the cells to the intercellular space. To this end, we collected concentrated medium (CM) with a concentration column (10kDa) from the condition medium of indicated cells, which removed microvesicles (MV) and exosome (Exo), and Western blotting was conducted to determine the protein level of CTSV and exosome markers. As a result, we have excluded MV and Exo from condition medium, and 43 kDa CTSV was detected in CM, but not in MV and Exo (Figures 4A, B). Additionally, CTSV overexpression led to higher protein levels of CTSV in CM (Figure 4C). We then performed migration and invasion assays in A549 cells pretreated with 5μg CM of CTSV-HA-medium or CTSV-HA-high cells(Overexpression effect). We found that the CM of the indicated cells promoted the migration and invasion of A549 cells (Figure 4D).
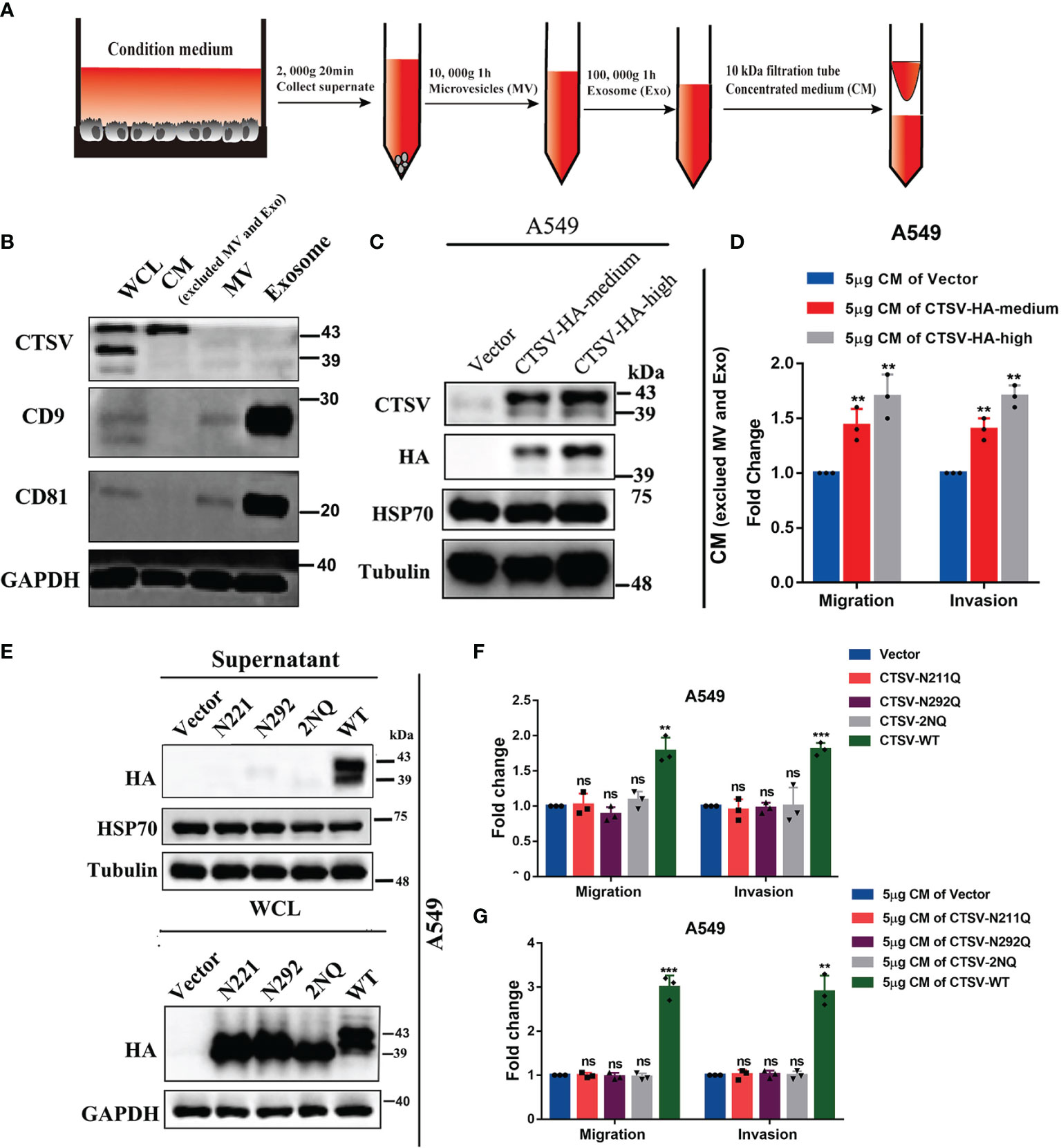
Figure 4 Mutation of glycosylation sites inhibits CTSV secretion and suppresses the metastasis of lung cancer cells. (A) Schematic diagrams of concentrated medium (CM) concentration. (B) Western blot analysis of CM using anti-CTSV, anti-CD9, anti-81 and anti-GAPDH antibodies. (C) Western blot analysis of the indicated stable A549 cells using anti-CTSV, anti-HA, anti-HSP70 and anti-Tubulin antibodies. (D) Migration and invasion assay of A549 cells pretreated with 5μg of CTSV-HA supernatant. (E) Western blot analysis of supernatant and whole cell lysate of A549 cells with N221Q, N292Q or both (2NQ) using anti-HA and anti-HSP70 antibodies. (F) Migration and invasion assay of A549 cells with N221Q, N292Q or both (2NQ). (G) Migration and invasion assay of A549 cells pretreated with 5μg of CTSV-N221Q, CTSV-N292Q or CTSV-2NQ supernatant. The results are shown as the mean ± SD of three biological independent experiments. **P < 0.01, ***P < 0.001 was analyzed by using the Student’s t-test. ns represent no significant change.
Glycosylation is vital for maintaining protein structure and function. Secreted cathepsins have emerged as potent effectors that modify the tumor microenvironment by degrading the ECM (29). However, the role of N-glycosylated CTSV in determining its pro-metastatic behavior remains unclear. To determine the contribution of glycosylation to CTSV distribution, we performed Western blotting assays in supernatant and whole cell lysate and found that N221Q, N292Q and 2NQ suppressed the secretion of CTSV from intracellular to extracellular matrix (Figure 4E). Next, overexpression of CTSV-N221Q, CTSV-N292Q, CTSV-2NQ or wild type CTSV in A549 cells, respectively, we found N221Q, N292Q, both [2NQ] CTSV did not change the migration and invasion ability, but wild-type CTSV significantly increased the migration and invasion of A549 cells (Figure 4F). Consistently, we performed migration and invasion assays in A549 cells pretreated with 5μg CM of CTSV-N221Q, CTSV-N292Q, CTSV-2NQ, or CTSV-WT cells, the data revealed that CM from the CTSV-N221, CTSV-N292 or CTSV-2NQ cells showed no significant change in migration and invasion, but greatly promoted cell migration and invasion in wild-type CM group (Figure 4G). Interestingly, different from a previous study (24), we found that both glycosylation sites N221 and N292 of CTSV, rather than N292 alone, are important for its secretion, this regulating its promoting effect on tumor metastasis in lung cancer. Collectively, these results supported that glycosylation of CTSV is required for secretion, thereafter determine the metastasis behavior of lung cancer cells.
Serum CTSV Distinguish Lung Cancer Patients From Healthy Donors
Our previous evidence support that glycosylated CTSV (band 43 kDa) is particularly expressed in lung cancer samples, and the first glycosylation band (band 43 kDa) of CTSV, which has been secreted outside of the cells, we reasoned that the glycosylated CTSV might serve as a more sensitive prognosis marker for lung cancer. To this end, we isolated serum from the plasma of lung cancer patients and healthy donors, the level of CTSV in circulating serum was significantly higher in patients with lung cancer than that in healthy donors (Figure 5). Together, the data suggested that the level of CTSV in serum distinguished lung cancer patients from healthy donors, revealing that the level of CTSV in serum (glycosylation CTSV) might be a biological marker for lung cancer.
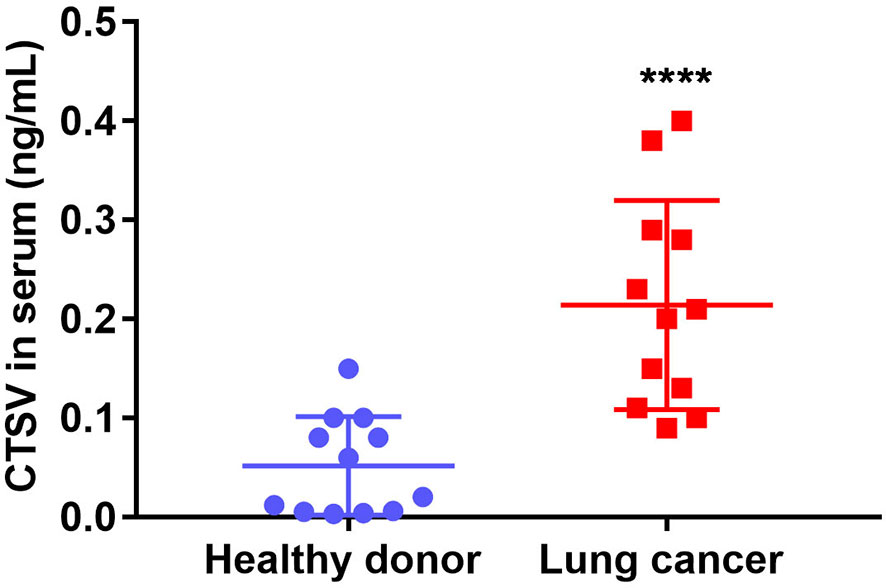
Figure 5 The level of CTSV on serum distinguishes patients with lung cancer from healthy donors. Elisa of human CTSV on serum in plasma samples from lung cancer patients and healthy donors. ****P < 0.0001 was analyzed by using the Student’s t-test.
Discussion
Cathepsins are vital acid proteolytic enzymes within lysosomes and therefore represent the crucial effector molecules of protein catabolism and autophagy (30, 31), which support the intense metabolic needs of proliferating cancer cells. In addition, cathepsins are integrated into almost all processes related to lysosomes, such as protein and lipid metabolism, protein degradation, antigen presentation, growth factor receptor cycling, stress signaling and lysosome-mediated cell death (29). They make critical contributions to cancer progression and dissemination in numerous cancer types.
Cysteine cathepsins are cysteine proteases that belong to the papain-like family; 11 cysteine cathepsins (B, C, F, H, K, L, O, S, V, W, and X) are encoded in the human genome (32). Cysteine cathepsins play crucial roles in regulating numerous physical processes, particularly proteolysis in the endolysosomal system and MHC II-mediated immune reactions (33, 34). Among others, these enzymes function differently in terms of localization and expression profile, tissue and cellular distribution, structural differences and modulation of enzyme activity. Although tightly controlled under physiological conditions, the dysregulation and overactivation of cathepsins in the extracellular milieu are marked hallmarks of numerous diseases, such as inflammation and inflammation-associated diseases, various types of cancer, bone disorders, rheumatoid arthritis and cardiovascular diseases (35–37). Therefore, cysteine cathepsins represent promising biological targets for therapeutic intervention.
Mutant HRAS upregulates both CTSB and CTSL and transforms mammary epithelial cells (38). CTSS expression in cancer cells is crucial for the promotion of colorectal cancer growth and progression (39). CTSZ promotes pancreatic neuroendocrine tumor growth and progression from both cancer cell-derived and tumor-associated macrophage-derived sources (40). However, the molecular mechanisms of cathepsins that lead to the malignancy of tumors remain unclear, which is amplified by the fact that members of the cathepsin superfamily have different roles in different types of cancer. Herein, our clinical data, based on RNA-seq, TCGA and GEO datasets, IHC and prognosis analysis, revealed that CTSV is a prognostic biomarker that is upregulated in lung cancer. The functional impact and mechanism of the dysregulation of CTSV in lung cancer remain to be determined.
N-glycosylation plays an important role in determining the structure and functions of proteins (41). The glycosylation of CTSV has been reported before in human fibrosarcoma HT1080 cells (24). Consistently, we showed that glycosylation of CTSV generally exists in lung cancer tissues and lung cancer cell lines, and protein mass spectrometry identified glycosylation at the N221 and N292 residues. Mutation of the N221 and N292 residues with Q residues, generating N221Q and N292Q, decreased the glycosylation of CTSV, thereby inhibited the secretion of CTSV, and remarkably suppressed the metastasis, invasion and migration of lung cancer cells. Importantly, we found that the glycosylation band (43 kDa) was positively correlated with lymph node metastasis, which suggests that the glycosylation of CTSV can serve as a risk factor for malignant progression of lung cancer. To further understand the role of CTSV in lung cancer, it is critical to dissect the complex processes by which upregulation of protease expression disrupts the homeostasis of normal tissue. The results that the CM of CTSV overexpression cells presented a stronger band confirmed our hypothesis. Our data provide mechanistic insight into this metastasis-driven issue.
In conclusion, our research showed that CTSV is upregulated in lung cancer and correlated with a poor prognosis, indicating that CTSV might be a prognostic biomarker for lung cancer. Herein, we confirmed that CTSV is N-glycosylated at N221 and N292, and its pro-metastatic behavior is glycosylation and extracellular secretion dependent. Notably, the level of CTSV in serum distinguished lung cancer patients from healthy donors and the glycosylated 43 kDa CTSV (secreted CTSV) is associated with lymph node metastasis. Collectively, our findings suggest that glycosylated CTSV could be a sensitive biomarker for lung cancer patients with metastasis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Sun Yat-sen University Cancer Center. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LY: Conceptualization, Methodology, Writing-Original draft preparation. QZ, YD: Methodology, Data curation, Software, Formal analysis. YQ: Visualization, Investigation. WY, YL: Resources, Validation, Supervision. LY, WY, YL: Writing,Reviewing and Editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.876245/full#supplementary-material
Abbreviations
CTSC, cathepsin C; CTSF, cathepsin F; CTSH, cathepsin H; CTSO, cathepsin O; CTSV, cathepsin V; CTSW, Cathepsin W; CM, Concentration medium; ECM, extracellular matrix; Endo H, endoglycosidase; Exo, exosome; HA, hemagglutinin; IHC, Immunohistochemistry staining; MS, Protein mass spectrometry; MV, microvesicles.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, et al. Cancer Screening in the United States, 2018: A Review of Current American Cancer Society Guidelines and Current Issues in Cancer Screening. CA Cancer J Clin (2018) 68(4):297–316. doi: 10.3322/caac.21446
3. T. National Lung Screening Trial Research. Lung Cancer Incidence and Mortality With Extended Follow-Up in the National Lung Screening Trial. J Thorac Oncol (2019) 14(10):1732–42. doi: 10.1016/j.jtho.2019.05.044
4. Oxnard GR, Binder A, Janne PA. New Targetable Oncogenes in Non-Small-Cell Lung Cancer. J Clin Oncol (2013) 31(8):1097–104. doi: 10.1200/JCO.2012.42.9829
5. Stella GM, Luisetti M, Pozzi E, Comoglio PM. Oncogenes in Non-Small-Cell Lung Cancer: Emerging Connections and Novel Therapeutic Dynamics. Lancet Respir Med (2013) 1(3):251–61. doi: 10.1016/S2213-2600(13)70009-2
6. Liu Y, Chen Q, Zhang JT. Tumor Suppressor Gene 14-3-3sigma is Down-Regulated Whereas the Proto-Oncogene Translation Elongation Factor 1delta Is Up-Regulated in Non-Small Cell Lung Cancers as Identified by Proteomic Profiling. J Proteome Res (2004) 3(4):728–35. doi: 10.1021/pr034127+
7. Hastings K, Yu HA, Wei W, Sanchez-Vega F, DeVeaux M, Choi J, et al. EGFR Mutation Subtypes and Response to Immune Checkpoint Blockade Treatment in Non-Small-Cell Lung Cancer. Ann Oncol (2019) 30(8):1311–20. doi: 10.1093/annonc/mdz141
8. Dai J, Lv J, Zhu M, Wang Y, Qin N, Ma H, et al. Identification of Risk Loci and a Polygenic Risk Score for Lung Cancer: A Large-Scale Prospective Cohort Study in Chinese Populations. Lancet Respir Med (2019) 7(10):881–91. doi: 10.1016/S2213-2600(19)30144-4
9. Li L, Ng SR, Colon CI, Drapkin BJ, Hsu PP, Li Z, et al. Identification of DHODH as a Therapeutic Target in Small Cell Lung Cancer. Sci Transl Med (2019) 11(517). doi: 10.1126/scitranslmed.aaw7852
10. Socinski MA, Obasaju C, Gandara D, Hirsch FR, Bonomi P, Bunn PA Jr., et al. Current and Emergent Therapy Options for Advanced Squamous Cell Lung Cancer. J Thorac Oncol (2018) 13(2):165–83. doi: 10.1016/j.jtho.2017.11.111
11. Kundu ST, Grzeskowiak CL, Fradette JJ, Gibson LA, Rodriguez LB, Creighton CJ, et al. TMEM106B Drives Lung Cancer Metastasis by Inducing TFEB-Dependent Lysosome Synthesis and Secretion of Cathepsins. Nat Commun (2018) 9(1):2731. doi: 10.1038/s41467-018-05013-x
12. Willumsen N, Bager CL, Leeming DJ, Bay-Jensen AC, Karsdal MA. Nidogen-1 Degraded by Cathepsin S Can Be Quantified in Serum and Is Associated With Non-Small Cell Lung Cancer. Neoplasia (2017) 19(4):271–8. doi: 10.1016/j.neo.2017.01.008
13. Song L, Wang X, Cheng W, Wu Y, Liu M, Liu R, et al. Expression Signature, Prognosis Value and Immune Characteristics of Cathepsin F in Non-Small Cell Lung Cancer Identified by Bioinformatics Assessment. BMC Pulm Med (2021) 21(1):420. doi: 10.1186/s12890-021-01796-w
14. Zhang J, Zhou B, Jin S, Huang Z, Ma B, Shao Q, et al. Mechanism of Action of Panax Notoginoside Against Lung Cancer in Mice Based on Response to CTSB Gene. BMC Complement Med Ther (2020) 20(1):367. doi: 10.1186/s12906-020-03159-0
15. Magister S, Tseng HC, Bui VT, Kos J, Jewett A. Regulation of Split Anergy in Natural Killer Cells by Inhibition of Cathepsins C and H and Cystatin F. Oncotarget (2015) 6(26):22310–27. doi: 10.18632/oncotarget.4208
16. Cairns J, Ingle JN, Wickerham LD, Weinshilboum R, Liu M, Wang L. SNPs Near the Cysteine Proteinase Cathepsin O Gene (CTSO) Determine Tamoxifen Sensitivity in ERalpha-Positive Breast Cancer Through Regulation of BRCA1. PloS Genet (2017) 13(10):e1007031. doi: 10.1371/journal.pgen.1007031
17. Di Fabio R, Moro F, Pestillo L, Meschini MC, Pezzini F, Doccini S, et al. Pseudo-Dominant Inheritance of a Novel CTSF Mutation Associated With Type B Kufs Disease. Neurology (2014) 83(19):1769–70. doi: 10.1212/WNL.0000000000000953
18. Bras J, Djaldetti R, Alves AM, Mead S, Darwent L, Lleo A, et al. Exome Sequencing in a Consanguineous Family Clinically Diagnosed With Early-Onset Alzheimer's Disease Identifies a Homozygous CTSF Mutation. Neurobiol Aging (2016) 46:236 e1–6. doi: 10.1016/j.neurobiolaging.2016.06.018
19. Edinger TO, Pohl MO, Yanguez E, Stertz S. Cathepsin W Is Required for Escape of Influenza A Virus From Late Endosomes. mBio (2015) 6(3):e00297. doi: 10.1128/mBio.00297-15
20. Wang CH, Wang LK, Wu CC, Chen ML, Kuo CY, Shyu RY, et al. Cathepsin V Mediates the Tazarotene-Induced Gene 1-Induced Reduction in Invasion in Colorectal Cancer Cells. Cell Biochem Biophys (2020) 78(4):483–94. doi: 10.1007/s12013-020-00940-3
21. Kiuchi S, Tomaru U, Ishizu A, Imagawa M, Kiuchi T, Iwasaki S, et al. Expression of Cathepsins V and S in Thymic Epithelial Tumors. Hum Pathol (2017) 60:66–74. doi: 10.1016/j.humpath.2016.09.027
22. Toss M, Miligy I, Gorringe K, Mittal K, Aneja R, Ellis I, et al. Prognostic Significance of Cathepsin V (CTSV/CTSL2) in Breast Ductal Carcinoma in Situ. J Clin Pathol (2020) 73(2):76–82. doi: 10.1136/jclinpath-2019-205939
23. Sereesongsaeng N, McDowell SH, Burrows JF, Scott CJ, Burden RE. Cathepsin V Suppresses GATA3 Protein Expression in Luminal A Breast Cancer. Breast Cancer Res (2020) 22(1):139. doi: 10.1186/s13058-020-01376-6
24. Niwa Y, Suzuki T, Dohmae N, Umezawa K, Simizu S. Determination of Cathepsin V Activity and Intracellular Trafficking by N-Glycosylation. FEBS Lett (2012) 586(20):3601–7. doi: 10.1016/j.febslet.2012.08.001
25. Sevenich L, Bowman RL, Mason SD, Quail DF, Rapaport F, Elie BT, et al. Analysis of Tumour- and Stroma-Supplied Proteolytic Networks Reveals a Brain-Metastasis-Promoting Role for Cathepsins. Nat Cell Biol (2014) 16(9):876–88. doi: 10.1038/ncb3011
26. Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, et al. Distinct Roles for Cysteine Cathepsin Genes in Multistage Tumorigenesis. Genes Dev (2006) 20(5):543–56. doi: 10.1101/gad.1407406
27. Akkari L, Gocheva V, Kester JC, Hunter KE, Quick ML, Sevenich L, et al. Distinct Functions of Macrophage-Derived and Cancer Cell-Derived Cathepsin Z Combine to Promote Tumor Malignancy via Interactions With the Extracellular Matrix. Genes Dev (2014) 28(19):2134–50. doi: 10.1101/gad.249599.114
28. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer. PloS One (2013) 8(12):e82241. doi: 10.1371/journal.pone.0082241
29. Olson OC, Joyce JA. Cysteine Cathepsin Proteases: Regulators of Cancer Progression and Therapeutic Response. Nat Rev Cancer (2015) 15(12):712–29. doi: 10.1038/nrc4027
31. Kondo Y, Kanzawa T, Sawaya R, Kondo S. The Role of Autophagy in Cancer Development and Response to Therapy. Nat Rev Cancer (2005) 5(9):726–34. doi: 10.1038/nrc1692
32. Vizovisek M, Fonovic M, Turk B. Cysteine Cathepsins in Extracellular Matrix Remodeling: Extracellular Matrix Degradation and Beyond. Matrix Biol (2019) 75-76:141–59. doi: 10.1016/j.matbio.2018.01.024
33. Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, et al. Cysteine Cathepsins: From Structure, Function and Regulation to New Frontiers. Biochim Biophys Acta (2012) 1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002
34. Turk V, Turk B, Turk D. Lysosomal Cysteine Proteases: Facts and Opportunities. EMBO J (2001) 20(17):4629–33. doi: 10.1093/emboj/20.17.4629
35. Turk B. Targeting Proteases: Successes, Failures and Future Prospects. Nat Rev Drug Discov (2006) 5(9):785–99. doi: 10.1038/nrd2092
36. Drag M, Salvesen GS. Emerging Principles in Protease-Based Drug Discovery. Nat Rev Drug Discov (2010) 9(9):690–701. doi: 10.1038/nrd3053
37. Turk B, Turk D, Turk V. Protease Signalling: The Cutting Edge. EMBO J (2012) 31(7):1630–43. doi: 10.1038/emboj.2012.42
38. Sloane BF, Moin K, Sameni M, Tait LR, Rozhin J, Ziegler G. Membrane Association of Cathepsin B Can Be Induced by Transfection of Human Breast Epithelial Cells With C-Ha-Ras Oncogene. J Cell Sci (1994) 107(Pt 2):373–84. doi: 10.1242/jcs.107.2.373
39. Small DM, Burden RE, Jaworski J, Hegarty SM, Spence S, Burrows JF, et al. Cathepsin S From Both Tumor and Tumor-Associated Cells Promote Cancer Growth and Neovascularization. Int J Cancer (2013) 133(9):2102–12. doi: 10.1002/ijc.28238
40. Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, et al. IL-4 Induces Cathepsin Protease Activity in Tumor-Associated Macrophages to Promote Cancer Growth and Invasion. Genes Dev (2010) 24(3):241–55. doi: 10.1101/gad.1874010
Keywords: CTSV, lung cancer, metastasis, glycosylation, prognosis
Citation: Yang L, Zeng Q, Deng Y, Qiu Y, Yao W and Liao Y (2022) Glycosylated Cathepsin V Serves as a Prognostic Marker in Lung Cancer. Front. Oncol. 12:876245. doi: 10.3389/fonc.2022.876245
Received: 15 February 2022; Accepted: 11 March 2022;
Published: 13 April 2022.
Edited by:
Aamir Ahmad, University of Alabama at Birmingham, United StatesReviewed by:
C Sykora, Australian National University, AustraliaGerald Schwan, University of Texas Southwestern Medical Center, United States
Copyright © 2022 Yang, Zeng, Deng, Qiu, Yao and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Yao, eWFvdml2aUAxNjMuY29t; Yifeng Liao, bGlhb3lmQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
 Lewei Yang
Lewei Yang Qi Zeng
Qi Zeng Yun Deng
Yun Deng Yeqing Qiu
Yeqing Qiu Wei Yao
Wei Yao Yifeng Liao
Yifeng Liao