- 1Department of Surgery, College of Medicine, University of Nigeria, Enugu, Nigeria
- 2Oncology Center, University of Nigeria Teaching Hospital, Enugu, Nigeria
- 3Department of Surgery, University of Abuja Teaching Hospital, Abuja, Nigeria
- 4Department of Surgery, University College Hospital, Ibadan, Nigeria
- 5Department of Pathology, National Hospital, Abuja, Nigeria
- 6Department of Epidemiology and Public Health, and Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore, MD, United States
- 7Department of Anatomic Pathology, Federal Medical Center, Abuja, Nigeria
- 8Department of Medical Microbiology, University College Hospital, Ibadan, Nigeria
- 9Department of Pathology, University College Hospital, Ibadan, Nigeria
- 10Institute of Human Virology Nigeria, Abuja, Nigeria
Breast cancer is now the commonest cancer in most sub-Saharan African countries. Few studies of the epidemiology and genomics of breast cancer and its molecular subtypes in these countries have been done. The African Female Breast Cancer Epidemiology (AFBRECANE) study, a part of the Human Heredity and Health in Africa (H3Africa) initiative, is designed to study the genomics and epidemiology of breast cancer and its molecular subtypes in Nigerian women. We link recruitment of breast cancer cases at study sites with population-based cancer registries activities to enable ascertainment of the incidence of breast cancer and its molecular subtypes. We use centralized laboratory processing to characterize the histopathological and molecular diagnosis of breast cancer and its subtypes using multiple technologies. By combining genome-wide association study (GWAS) data from this study with that generated from 12,000 women participating in our prospective cohort study of cervical cancer, we conduct GWAS of breast cancer in an entirely indigenous African population. We test associations between dietary intakes and breast cancer and focus on vitamin D which we measure using dietary intakes, serum vitamin D, and Mendelian randomization. This paper describes the AFBRECANE project, its design, objectives and anticipated contributions to knowledge and understanding of breast cancer.
Introduction
While the incidence of breast cancer has stabilized in high-income countries (HIC), it is rising in low income countries (LIC) that hitherto had low incidence (1). The WHO and the International agency for Research on Cancer (IARC) estimate that by the year 2040, there will be 3.06 million new cases of breast cancer every year and most of them will be in LIC (2). This is due to the epidemiological transition going on in these countries and the increase in the prevalence of known risk factors such as obesity, hormone replacement therapy (HRT), as well as increasing use of early detection and screening methods (3–7). Nevertheless, these factors do not account for all the increase in breast cancer incidence, while the specific contributions of some of them such as mammographic screening, hormone replacement treatments, and hormonal agents are hotly debated (5, 8–16).
In contrast to developed countries, there has been few systematic studies of breast cancer in sub-Saharan Africa. Studying the epidemiology and genomics of breast in developing countries like Nigeria will enable identification of critical environmental and genetic risk factors and improve our knowledge of the causes of its rising incidence. The AFBRECANE study is funded by the Office of the Director of the US National Institutes of Health (NIH) and the National Human Genome Research Institute (NHGRI) under the Human Heredity and Health in Africa (H3Africa) initiative for five years starting in November 2018. The project is designed to study the genetic and environmental risk factors of breast cancer and its subtypes in an indigenous African population.
The primary objectives of the AFBRECANE study are to determine the incidence and prevalence of total and molecular subtypes of breast cancer in Nigerian women and to characterize their secular trend in Nigeria. We achieve this through combination of case-control and cancer registration research methods through enrollment at our study sites and linkage with the Population Based Cancer Registries (PBCR) in the cities where participants are being enrolled in Nigeria. In this study, we link epidemiological risk factors’ data, cancer registry databases, clinical information and biorepository of tissues for breast cancer research in Nigeria. We engage multiple methods for determination of molecular subtypes of breast cancer in tissue biopsies and fine needle aspirates and evaluate determinants of discordance between the different methods.
Another objective of the AFBRECANE project is to conduct GWAS of breast cancer and its molecular subtypes to look for mutations associated with breast cancer and its subtypes in indigenous African populations. In order to efficiently achieve this objective, the project takes advantage of the GWAS data from H3Africa funded African Collaborative Center for Microbiome and Genomics Research (ACCME) project which enrolled women who volunteered for cervical cancer screening and were free of cancer at baseline. The use of an enriched microarray chip designed by the H3Africa Consortium for this GWAS study promises to increase the likelihood of detecting new variants that are significantly associated with breast cancer in this African population. We will use Mendelian Randomization to test associations between SNPs associated with serum vitamin D and breast cancer in Nigerian women.
The third objective of the AFBRECANE project is to evaluate associations between Nigerian diets and their food and nutrients composition, and breast cancer. Using our validated Food frequency questionnaire (FFQ), Food composition database (FCD), and Food picture book (FPB), we characterize the dietary intakes of our participants. Further, we specifically examine dietary and serum levels of vitamin of D, as well as use Mendelian Randomization (MR) of SNPs associated with serum levels of vitamin D, and risk of breast cancer in this population in Africa.
Methods And Analysis
Study Design and Rationale
In the AFBRECANE project, we establish clinical research sites in 3 cities in Nigeria and link these sites with the population-based cancer registries (PBCR) covering the population in these cities. This enables us to use cancer registration methods to ascertain the incidence of breast cancer and its molecular subtypes in this population. Further by enrolling population based controls, we evaluate the role of demographics, lifestyle, and diet in the epidemiology of incident breast cancer and its subtypes in Nigerian women (17–19).
Molecular Subtypes of Breast Cancer in Africa
Several studies have reported that the growth in incidence of breast cancer in developed countries was largely due to increases in incidence of Luminal types A and B (hormone receptors rich) breast cancers, while the incidence of other subtypes including triple negative breast cancer (TNBC) remained stable (8, 20). In a study using the SEER database, the proportion of Estrogen receptor (ER) positive and Progesterone receptor (PR) positive breast cancers among 40 to 69 years old women increased from 75.4% to 77.5% (p-value = 0.0002) and from 65.0% to 67.7% (p-value < 0.001) between 1992 and1998 (20). In contrast, several authors have suggested that the recent increase in breast cancer incidence in Africa is mostly due to rising prevalence of hormone-receptor poor tumors (21, 22). To date it has been difficult to test this hypothesis because of lack of population-based cancer registry (PBCR) and immunohistochemistry laboratories in most African countries (19). The AFBRECANE study resolves these challenges by working with the Nigerian National System of Cancer Registries and using a centralized laboratory for specimen analyses to evaluate secular differences in incidence of molecular subtypes of breast cancer (17–19).
Dietary Intakes and Breast Cancer Risk in Africa
The growing adoption of western lifestyles including dietary patterns has been touted as one of the risk factors for the rising incidence of breast cancer in African populations (16, 23–26). To date, there has been no robust study of the associations between dietary intakes and breast cancer in indigenous African populations (27). Very few of the studies done so far used validated Food Frequency Questionnaires (FFQ), the paradigmatic tool for nutrition epidemiology of complex diseases (28, 29). Nutrition epidemiology research also requires methods for obtaining information on portion sizes because of poor standardization of serving sizes in Africa, and food composition databases (FCD) of African foods to ensure correct computation of nutrient values. None of the studies of diets and breast cancer done in Africa till date has found strong associations between any African diet, their food and nutrient compositions, and the risk of breast cancer. In the AFBRECANE study, we use validated FFQ, FCD of Nigerian foods and a Food Picture Book (FPB) to obtain information on food intakes for studies of their association with breast cancer.
Vitamin D and Breast Cancer Risk
Studies from developed high-income countries have suggested that there are strong associations between dietary intake of vitamin D or serum levels of vitamin D and breast cancer risk and outcomes (30–34). This association may be stronger in premenopausal women, patients with advanced breast cancer and in triple negative breast cancer (35). Further, vitamin D status may explain some of the racial disparities in incidence and outcomes of breast cancer (36–40). African-Americans with low mean serum vitamin D levels had higher risk of breast cancer, compared to European Americans. Most Sub-Saharan Africans have low overall intake of vitamin D and recent studies have shown high prevalence of hypovitaminosis D in Africa populations, despite high levels of sun exposure (41). Skin tones, which may influence bioavailable vitamin D levels of Africans, also vary significantly and there has been no previous study of vitamin D and breast cancer that adjusted for skin tones. There has been no previous study of vitamin D and its association with breast cancer risk in indigenous Africans. The AFBRECANE study will fill this gap.
In addition to evaluating associations between vitamin D levels derived from nutrient levels in dietary intakes and serum vitamin D measurements, we also evaluate the association between vitamin D and breast cancer risk by Mendelian Randomization using a combination of SNPs as instrumental variables (IV). This overcomes the problems of reverse causation and confounders in assessing any causal relationships between vitamin D and breast cancer risk (42–51). 1,25-dihydroxyvitamin D (1,25(OH)2D), the natural ligand of the vitamin D receptor (VDR), is a transcription factor that is expressed in more than thirty-five tissues. More than 200 genes containing VDR-response elements and their expression is affected by 1,25(OH)2D. Mutations in one or more of these genes may affect the biological activity of vitamin D and its effect on disease occurrence. SNPs that are known to be associated with serum 25-hydroxyvitamin D levels would be used as instrumental variables to estimate the effect of vitamin D on breast cancer risk. Since SNP genotypes are determined at birth, they are not influenced by potentially confounding variables, and so the effect estimate from multiple regression analysis should be unaffected by reverse causation.
Genome-Wide Association Study (GWAS) of Breast Cancer in Indigenous Africans
Previous studies have described associations between some SNPs and breast cancer in African populations. Some of these SNPs have been reported in non-African ancestry populations while others appear to be specific to Africans (52). Although there have been several case-control GWAS of breast cancer in African ancestry populations, none has been solely of Africans living in Africa (53–80). In the previous studies, the genotyping array chips used were not designed to account for African populations. Given the genomic heterogeneity of Africans, it is probable that GWAS of breast cancer in indigenous African population using microarray chips enriched for African populations will identify more unique genomic loci associated with breast cancer and its molecular subtypes in African women. The AFBRECANE study provides an opportunity to conduct GWAS of breast cancer and its molecular subtypes in indigenous African population using chips enriched for genetic variants in African populations.
Participants
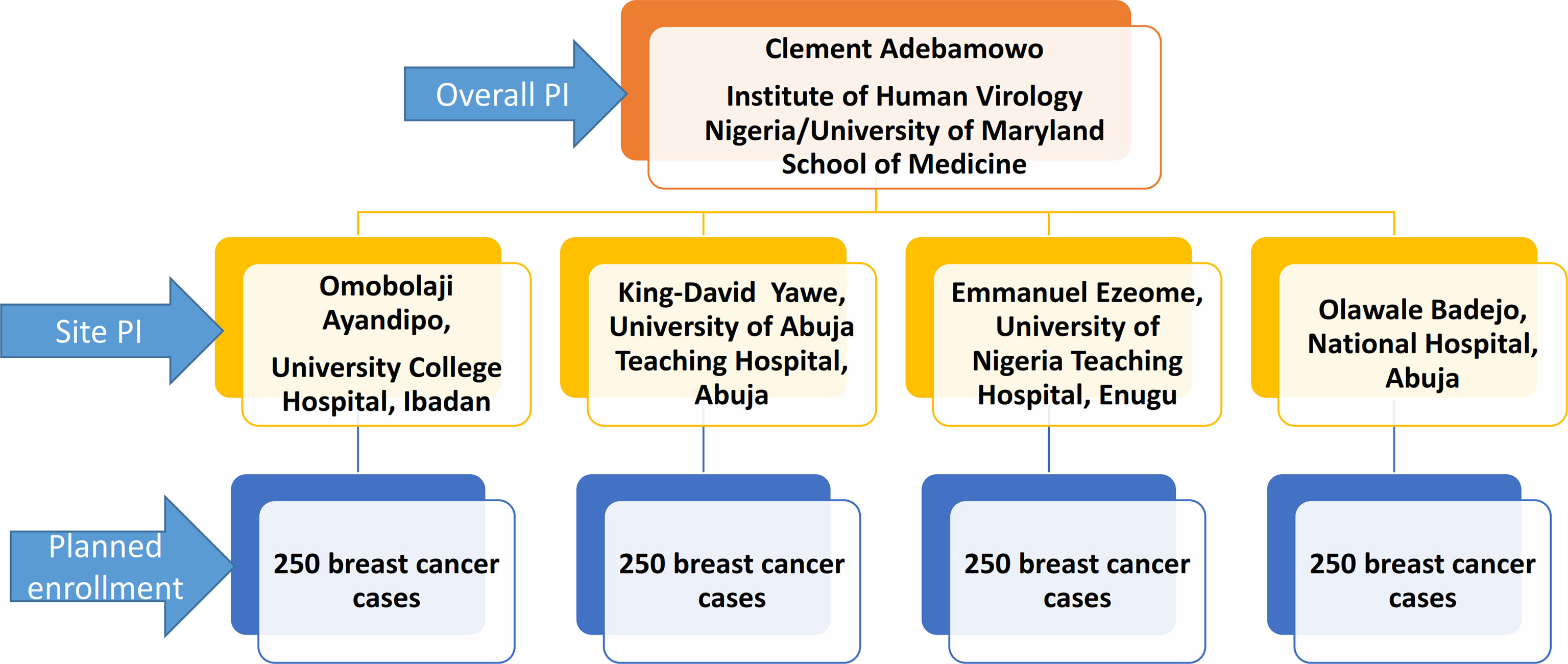
The cases in this study are women aged 18 years and older with clinical and histological diagnosis of breast cancer. A total of 1000 cases will be enrolled over five years from clinical sites across Nigeria. Enrollment commenced in November 2018 at National Hospital Abuja and University of Abuja Teaching Hospital Gwagwalada in central Nigeria, University College Hospital Ibadan in Western Nigeria and University of Nigeria Teaching Hospital, Enugu in Eastern Nigeria. The sites were selected based on their case volume, representation of different geographical zones of Nigeria as well as availability of population-based cancer registries in those areas. Figure 1 displays the organogram of the AFBRECANE study and how it is coordinated by the PI of the study. The research associates report cases enrolled to the cancer registrars in these cities to ensure more complete registration of breast cancer cases by the PBCR. Breast cancer patients identified by the cancer registrars are also invited to participate in the study. Breast cancer patients who opt not to participate in the study contribute to registry data and this enables us to compute the breast cancer incidence in the registries catchment areas. After obtaining informed consent from those willing to participate, research associates conduct confidential and structured face-to-face interviews in the language preferred by the participant. Patients who are too sick to participate or refused to give informed consent are excluded from the study.
For the GWAS component of this study, control participants are the ~12,000 women who are enrolled in the ACCME cohort study. These women had no history of any cancer at enrollment, were HIV negative by self-report and had given broad informed consent for future unspecified use genomic research. The profile of participants in the ACCME cohort has been previously described (81). To generate matching controls for epidemiological case-control analyses, we randomly select women in the ACCME cohort who are within +/- 2.5 years’ age of the breast cancer cases enrolled in the AFBRECANE study. Ethical clearance for this study was obtained from the National Health Research Ethics Committee of Nigeria and the Institutional Review Board (IRB) of the University of Maryland School of Medicine, Baltimore. Each patient gave broad informed consent for the research and the use of the data for future research.
Breast cancer cases are followed up at regular clinic appointments after enrollment until end of study or loss to follow-up. Participants data are stored in REDCap database with paper forms for back-up. The contact details of participants and their next of kin are collected and used for following up. Patients are required to inform the next of kin of their participation in this study and obtain consent of the next of kin so they can be contacted if we cannot reach the participant.
Research associates administer the AFBRECANE study questionnaire earlier developed for breast cancer studies in the Nigerian population. Data collected include socio-demographic data, lifestyle data, dietary intakes data, medical history, obstetrics, and gynecology history of participants (Table 1). The associates use our validated semi-quantitative FFQ assisted by the FPB to record participants dietary history (82, 83). They measure participants’ height, weight, waist and hip circumferences, and body shape information (somatotype) per standard protocol. All data are entered into REDCap databases (84, 85). Standard cancer registry methods were used to identify all population based diagnosis of incident breast cancer in the catchment areas of the two PBCR as previously described (18, 86, 87). The Nigerian National System of Cancer Registries Data Abstraction form are used to collect cancer registries data and the CanReg5® open source software is used to check the data and analyze the registries data.
The breast cancer cases are staged using the American Joint Committee of Cancer’s classification of Tumor, Node and Metastasis (TNM). All the patients receive clinical examination of the breasts, chest x-rays, abdominal ultrasound, and bone scans. Breast biopsies are done using core biopsy needles, excisional or incisional biopsies as indicated. One breast tumor sample is inserted into PAXgene® Tissue Fixative which ensures rapid penetration and fixation in 2 to 4 hours. The warm ischaemic time is recorded. The sample is inserted into PAXgene® Tissue Stabilizer to stabilize molecular contents and preserve morphology. A second sample are stored in -80°C. All samples were transported to the ACCME Laboratory at the Institute of Human Virology of Nigeria (IHVN), Abuja, Nigeria where two sets of slides are prepared.
Slides are prepared according to standard histology and immunohistochemistry procedures in the ACCME Laboratory, IHVN Abuja, Nigeria and the results are reported by the study pathologist. The estrogen, progesterone, and HER2 status of the tumors were graded using <1% positivity for cut-off as recommended by the American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP). Scores of 0, 1, 2, 3 are assigned to the tumors based on the intensity of staining. Additional molecular subtyping of the breast tumors is also done using RT-PCR methods to measure four breast somatic biomarkers, ESR1, PGR1, ERBB2, and MKi67 messenger RNAs (mRNAs), and one reference gene (CYFIP1) (88). The results of the molecular subtyping tests are compared and only concordant classifications are accepted. Slide images from discordant IHC tests are captured using Grundium Ocus®40 microscope scanners and evaluated at University of Maryland School of Medicine. Cases that are persistently discordant will be analyzed separately.
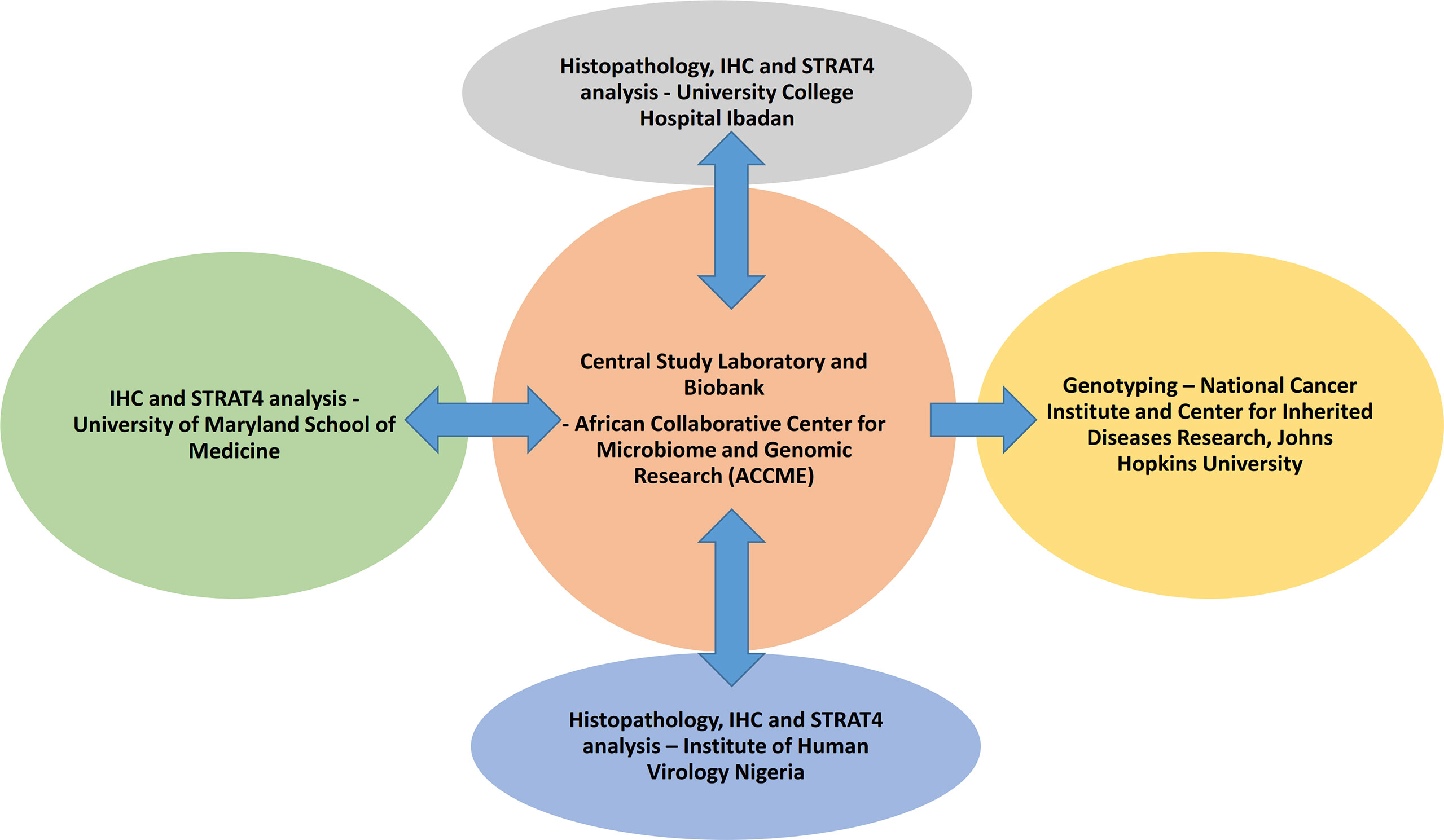
Blood samples are collected from the study participants and separated into plasma, buffy coat, and blot clots which are stored -80°C. Germline DNA were extracted from the buffy coat using Qiagen Qiacube HT® in the ACCME Laboratory in Nigeria. Plasma samples are used for standard chemistry, trace metals, lipids, and biomarkers, and blood clots are used for analysis of fatty acids. Figure 2 displays the components of the study conducted at different sites within and outside Nigeria.
Data Analysis And Quality Assurance
All data are entered into Redcap database hosted at the IHVN (84, 85). Research associates interview patients and enter epidemiological and clinical data into tablet computers. To improve quality of laboratory analyses and enhance reproducibility, we ensure that the study is done to a very high standard and implement several quality assurance activities (89). Use of Paxgene® tissue system for collection, handling, transport, and fixation of breast cancer samples reduces pre-analytic variation in tissue handling (90–92). All immunohistochemistry is done in a single central laboratory in Nigeria. We use the same hormone-receptor primary antibodies and clones [Estrogen Receptor Clone 6F11 Ready-To-Use Primary Antibody, Her2/Neu (SP3) Rabbit Monoclonal Antibody, and the Bond™ Ready-to-Use Primary Antibody for Progesterone Receptor (16)] as the Department of Pathology, University of Maryland School of Medicine (93). We keep detailed record of clones and lot numbers of the primary antibodies (94, 95).
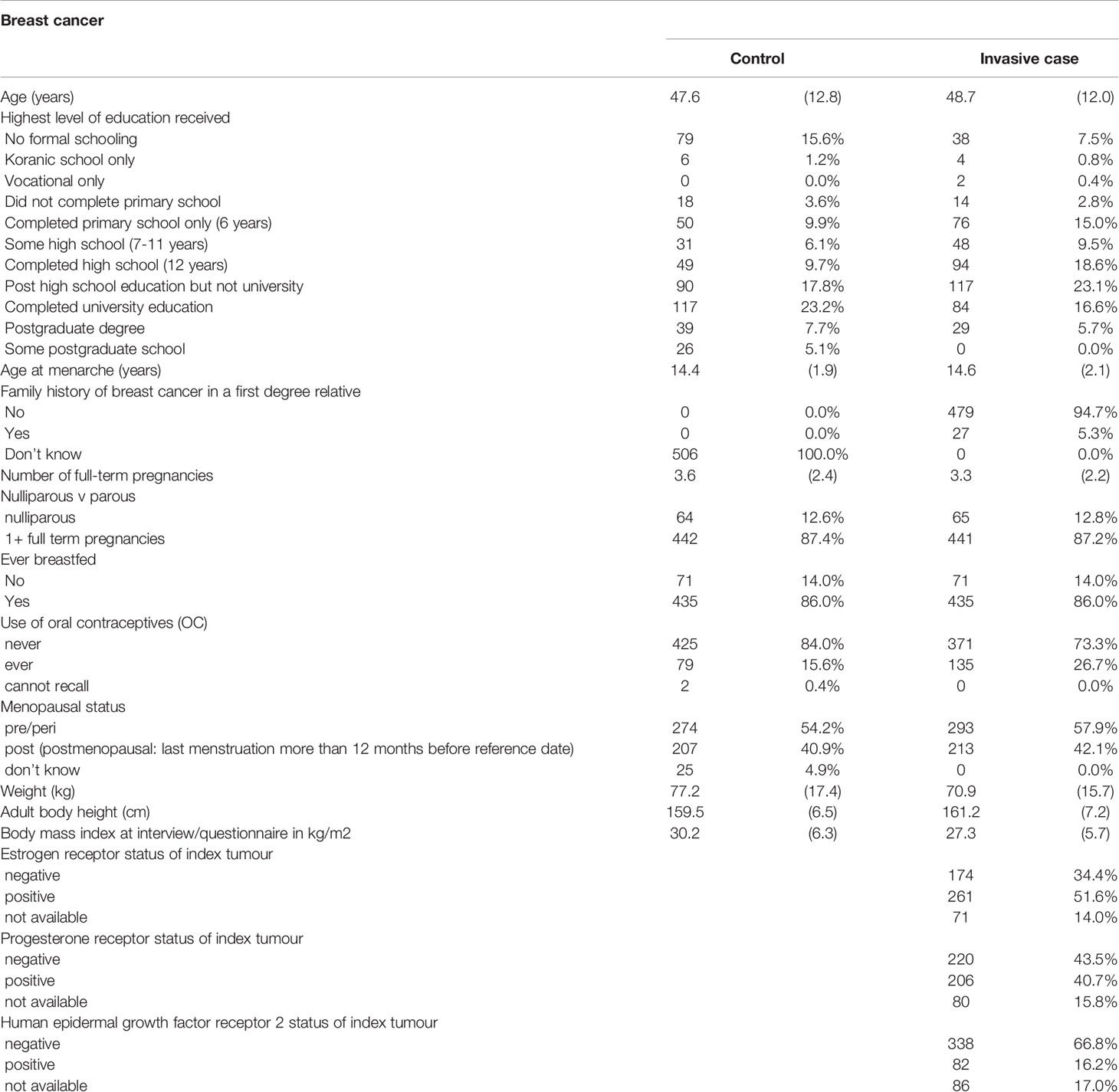
Recruitment of patients into the project started in November 2018. As of May 2021, we have enrolled 506 breast cancer cases and in Table 2, we present the analysis of their characteristics compared with matching controls from the ACCME cohort. The mean (SD) age of the controls was 47.6 (12.8) years while that of cases was 48.7 (12.0) years. Breast cancer cases had later age of onset of menarche, positive family history of breast cancer in a first degree relative, fewer full-term pregnancies, higher prevalence of nulliparity, history of use of oral contraceptives, were more likely to be pre-menopausal and had similar breast feeding history compared to controls. Breast cancer cases weighed less, were taller, and had lower BMI compared to controls. The molecular subtype patterns of the breast tumors are 51.6% ER positive, 34.4% ER negative, 40.7% PR positive, 43.5% PR negative, 66.8% HER2 positive and 16.2% HER2 negative.
Discussion
Scope of the Project and Future Directions
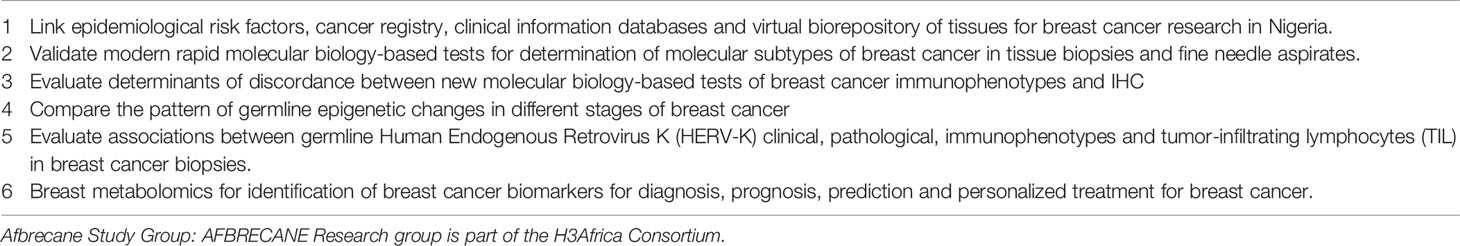
The AFBRECANE project will contribute to global knowledge about the epidemiological determinants of incident total and molecular breast cancer types, particularly of variants like triple negative breast cancer that have poor outcomes in indigenous African populations. The project will contribute to knowledge on the associations between indigenous African diets and breast cancer risk. The project focuses in a powerful way on the association between vitamin D and the risk of breast cancer by using different strategies to evaluate vitamin D intakes. Identification of genomic risks of breast cancer in African populations like Nigerians will clarify the roles of genomic variants that have been identified in non-African populations. Other research questions that the AFBRECANE study will address are listed on Table 3. The database of tissue samples, clinical details and genomic data collected in this cohort will help future genomic studies, data science studies and epidemiological studies of breast cancer in general and among Africans in particular. Future research will identify the functional relevance of the genes identified in this project and the results may lay the foundation for vitamin D prevention trials in breast cancer.
What Are the Main Strengths and Weaknesses?
AFBRECANE project is one of the few large case-controlled study of genomic data on breast cancer from solely indigenous Africa populations. The detailed collection of epidemiological, dietary and nutritional information in the database promises to give insight to the role of environmental and genomic variables in the development of breast cancer and how these have impacted the changing trend in breast cancer incidence both locally and globally. The detailed genomic characterization presents the potential for discovery of new genetic mutations and variants of known mutations that may be active within the African population given the genetic diversity of the African populations.
This is also the only breast cancer study in African populations that includes use of validated FFQ supported by FPB to capture portion size information and a native African FCD. The focus on the role of vitamin D and breast cancer risks includes three independent methods of characterizing vitamin D intakes and adjustment of skin tones which determines the impact of sun exposure on endogenous vitamin D levels. The project is therefore likely to contribute unique insights into understanding any associations between vitamin D and breast cancer in this population.
The main limitation of this cohort is that we plan to enroll 1,000 breast cancer cases, and this sample size may give adequate power for total breast cancer analysis but it may prove insufficient for exploration of some of our hypothesis including GWAS of molecular subtypes of breast cancer. We are enrolling participants from Nigeria only and this may impact the generalizability of our results to other parts of Africa.
Training and Capacity Development
Built into the AFBRECANE study are capacity development programs in Epidemiology, Data science, Genomics and Breast cancer research. Both pre-doctoral and post-doctoral students are currently working on different aspects of the project, including the data science, the sample analysis, and the epidemiological correlates.
Collaborations
A key component of the AFBRECANE project is the collaboration with other research projects in Africa and internationally. AFBRECANE project is part of the H3Africa consortium and is contributing data and biospecimen to the H3Africa repositories in accordance with the guidelines. We are also collaborating with the NCI on the Confluence project which aims to conduct GWAS of 300,000 breast cancer cases and 300,000 controls in order to uncover breast cancer genetic risk factors.
We currently collaborate with Cepheid Corporation to validate the STRAT4 GeneXpert test for molecular typing of breast cancer. STRAT4 uses GeneXpert technology to quantitatively classify the molecular subtypes of breast cancer as an alternative to traditional IHC testing. This will be responsive to the challenge of immunohistochemistry testing in Africa and improve access to these results for management and prognostication of breast cancer.
Can I Get Hold of the Data? Where Can I Find Out More?
The project data release plan will be in accordance with the H3Africa data storage and release policy (96). Access to the data is granted to researchers through the Database Access Committee (DBAC) of H3Afrca and investigators can access the database one year after the end of the project. Genotyping and phenotyping data will be deposited in dbGap (https://www.ncbi.nlm.nih.gov/gap/) More information on the data access protocol can be obtained at the H3Africa data archive @ https://www.h3abionet.org/resources/h3africa-archive. Specific inquiries and proposals for collaboration can be directed to the PI of the project. (cadebamowo@som.umaryland.edu).
Federal Capital Territory Abuja site group
1. Oge Ikwueme
2. Ayotunde Famooto
3. Tolu Gbolahan
4. Mary Ajayi
5. Mary-Favour Edet
6. Ayobami Ademola
7. Yinka Owoade
8. Faithful Nze
9. Temitayo Oladimeji
Enugu site Group
10. Robinson Ugwu
11. Mr. Agbalu Ifeanyi Sebastian
Ibadan site group
12. Temilola Yusuf
13. Tobi Oyediran
14. Adeola Akintola
15. Temitope Olayinka
16. Julius Adediji
17. Chibuzor Nkwodimmah
18. Bisola Famooto
19. Banke Ipadeola
Consultants
1. Prof. Emmanuel Ezeome
2. Prof. King-David Terna Yawe
3. Dr. Badejo Olawale Ayodele
4. Dr. Abike Fowotade
5. Dr. Omobolaji. O. Ayandipo
6. Dr. Gabriel Ogun
7. Dr. Izuu Achuzi
Cancer Registries Directors
1. Dr. Festus Igbinoba
2. Dr. Theresa Otu
3. Prof. Emmanuel Ezeome
Cancer Registrars
1. Chinyere Chukwubuike
2. Henry M. Kumai
3. Ann Okoroafor
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by: 1. Institutional Review Board, University of Maryland School of Medicine, Baltimore; 2. National Health Research Ethics Committee of Nigeria, Federal Ministry of Health, Nigeria. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: CA, AFBRECANE Research group. Data curation: EE, K-DY, OA, OB, BA, AF, GO, CA, AFBRECANE Research group. Formal analysis: EE, SA, and CA. Funding acquisition: EE, K-DY, MA, OB, SA, and CA. Methodology: EE, K-DY, OA, OB, SA, BA, AF, GO, CA, AFBRECANE Research group. Project administration: EE, K-DY, MA, AF, and CA. Visualization: EE, K-DY, OA, OB, SA, BA, AF, GO, and CA. Writing–original draft: EE, SA, and CA. All authors contributed to the article and approved the submitted version.
Funding
The project is supported by the African Female Breast Cancer Epidemiology (AFBRECANE, U01HG009784) and the African Collaborative Center for Microbiome and Genomics Research (ACCME U54HG006947) grants from the National Institute of Health Office of the Director/National Human Genome Research Institute. Additional support was received from the Maryland Department of Health’s Cigarette Restitution Fund, the University of Maryland School of Medicine Greenebaum Comprehensive Cancer Center Support Grant (National Cancer Institute Award Number: P30CA134274), and the American Cancer Society Institutional Research Grant (IRG-18-160-16).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The AFBRECANE team expresses their gratitude for the support and help from the IHVN African Collaborative Center for Microbiome and Genomics Research, IHVN Research Department and also Staff and collaborators in the project at the different recruitment sites and their Cancer Registries in Nigeria.
References
1. Xu S, Liu Y, Zhang T, Zheng J, Lin W, Cai J, et al. The Global, Regional, and National Burden and Trends of Breast Cancer From 1990 to 2019: Results From the Global Burden of Disease Study 2019. Front Oncol (2021) 11:689562. doi: 10.3389/fonc.2021.689562
2. IARC. Cancer Tomorrow 2021. Available at: https://gco.iarc.fr/tomorrow/graphic-isotype?type=0&type_sex=0&mode=population&sex=2&populations=900&cancers=20&age_group=value&apc_male=0&apc_female=0&single_unit=500000&print=0.
3. Parkin DM, Bray FI, Devesa SS. Cancer Burden in the Year 2000. Global picture Eur J Cancer (2001) 37 Suppl 8:S4–66. doi: 10.1016/S0959-8049(01)00267-2
4. Boyle P, Levin B. World Cancer Report 2008. Geneva: International Agency for Research on Cancer (2008). 2008.
5. Autier P, Boniol M, Gavin A, Vatten LJ. Breast Cancer Mortality in Neighbouring European Countries With Different Levels of Screening But Similar Access to Treatment: Trend Analysis of WHO Mortality Database. BMJ (2011) 343:d4411. doi: 10.1136/bmj.d4411
6. DeSantis CE, Bray F, Ferlay J, Lortet-Tieulent J, Anderson BO, Jemal A. International Variation in Female Breast Cancer Incidence and Mortality Rates. Cancer Epidemiol Biomarkers Prev (2015) 24(10):1495–506. doi: 10.1158/1055-9965.EPI-15-0535
7. Linos E, Spanos D, Rosner BA, Linos K, Hesketh T, Qu JD, et al. Effects of Reproductive and Demographic Changes on Breast Cancer Incidence in China: A Modeling Analysis. J Natl Cancer Inst (2008) 100(19):1352–60. doi: 10.1093/jnci/djn305
8. Valla M, Vatten LJ, Engstrom MJ, Haugen OA, Akslen LA, Bjorngaard JH, et al. Molecular Subtypes of Breast Cancer: Long-Term Incidence Trends and Prognostic Differences. Cancer Epidemiol Biomarkers Prev (2016) 25(12):1625–34. doi: 10.1158/1055-9965.EPI-16-0427
9. Russakoff DB, Hasegawa A. Generation and Application of a Probabilistic Breast Cancer Atlas. Med Image Comput Comput Assist Interv (2006) 9(Pt 2):454–61. doi: 10.1007/11866763_56
10. Glass AG, Lacey JV Jr., Carreon JD, Hoover RN. Breast Cancer Incidence, 1980-2006: Combined Roles of Menopausal Hormone Therapy, Screening Mammography, and Estrogen Receptor Status. J Natl Cancer Inst (2007) 99(15):1152–61. doi: 10.1093/jnci/djm059
11. Sant M, Francisci S, Capocaccia R, Verdecchia A, Allemani C, Berrino F. Time Trends of Breast Cancer Survival in Europe in Relation to Incidence and Mortality. Int J Cancer (2006) 119(10):2417–22. doi: 10.1002/ijc.22160
12. Sant M, Francisci S, Capocaccia R, Verdecchia A, Allemani C, Berrino F. Should We Use Incidence, Survival or Mortality to Assess Breast Cancer Trends in European Women? Nat Clin Pract Oncol (2006) 3(3):228–9. doi: 10.1038/ncponc0489
13. Boyle P, Ngoma T, Sullivan R, Brawley O. Cancer in Africa: The Way Forward. Ecancermedicalscience (2019) 13:953. doi: 10.3332/ecancer.2019.947
14. Boyle P, Autier P, Adebamowo C, Anderson B, Badwe RA, Ashton LP, et al. World Breast Cancer Report. Lyon: International Prevention Research Institute (2012).
15. Kantelhardt EJ, Muluken G, Sefonias G, Wondimu A, Gebert HC, Unverzagt S, et al. A Review on Breast Cancer Care in Africa. Breast Care (2015) 10(6):364–70. doi: 10.1159/000443156
16. Azubuike SO, Muirhead C, Hayes L, McNally R. Rising Global Burden of Breast Cancer: The Case of Sub-Saharan Africa (With Emphasis on Nigeria) and Implications for Regional Development: A Review. World J Surg Oncol (2018) 16(1):63. doi: 10.1186/s12957-018-1345-2
17. Jedy-Agba E, Curado MP, Ogunbiyi O, Oga E, Fabowale T, Igbinoba F, et al. Cancer Incidence in Nigeria: A Report From Population-Based Cancer Registries. Cancer Epidemiol (2012) 36(5):e271–8. doi: 10.1016/j.canep.2012.04.007
18. Jedy-Agba EE, Oga EA, Odutola M, Abdullahi YM, Popoola A, Achara P, et al. Developing National Cancer Registration in Developing Countries - Case Study of the Nigerian National System of Cancer Registries. Front Public Health (2015) 3:186. doi: 10.3389/fpubh.2015.00186
19. Adebamowo CA, Famooto A, Ogundiran TO, Aniagwu T, Nkwodimmah C, Akang EE. Immunohistochemical and Molecular Subtypes of Breast Cancer in Nigeria. Breast Cancer Res Treat (2008) 110(1):183–8. doi: 10.1007/s10549-007-9694-5
20. Li CI, Daling JR, Malone KE. Incidence of Invasive Breast Cancer by Hormone Receptor Status From 1992 to 1998. J Clin Oncol (2003) 21(1):28–34. doi: 10.1200/JCO.2003.03.088
21. Dickens C, Duarte R, Zietsman A, Cubasch H, Kellett P, Schuz J, et al. Racial Comparison of Receptor-Defined Breast Cancer in Southern African Women: Subtype Prevalence and Age-Incidence Analysis of Nationwide Cancer Registry Data. Cancer Epidemiol Biomarkers Prev (2014) 23(11):2311–21. doi: 10.1158/1055-9965.EPI-14-0603
22. Huo D, Ikpatt F, Khramtsov A, Dangou JM, Nanda R, Dignam J, et al. Population Differences in Breast Cancer: Survey in Indigenous African Women Reveals Over-Representation of Triple-Negative Breast Cancer. J Clin Oncol (2009) 27(27):4515–21. doi: 10.1200/JCO.2008.19.6873
23. Ferlay J SH, Bray F, Forman D, Mathers C, Parkin DM. Estimate of Worldwide Burden of Cancer in 2008. Globacom 2008. Int J Cancer (2008) 127:2893–917. doi: 10.1002/ijc.25516
24. Parkin DM, Nambooze S, Wabwire-Mangen F, Wabinga HR. Changing Cancer Incidence in Kampala, Uganda, 1991-2006. Int J Cancer (2010) 126(5):1187–95. doi: 10.1002/ijc.24838
25. Chokunonga EBM CZ, Nyakabau AM, Parkin DM. Trends in the Incidence of Cancer in the Black Population of Harare, Zimbabwe 1991–2010. Int J Cancer (2013) 133:721–9. doi: 10.1002/ijc.28063
26. Fregene A NL. Breast Cancer in Sub Saharan Africa:How Does it Relate to Breast Cancer in African-American Women. Cancer (2005) 103):1540–50. doi: 10.1002/cncr.20978
27. Bigman G, Otieno L, Adebamowo SN, Adebamowo C. Dietary Intake and Cancer in Sub-Saharan Africa: A Critical Review of Epidemiological Studies. Nutr Cancer (2022) 1–12. doi: 10.1080/01635581.2022.2032217
28. Jacobs I, Taljaard-Krugell C, Ricci C, Vorster H, Rinaldi S, Cubasch H, et al. Dietary Intake and Breast Cancer Risk in Black South African Women: The South African Breast Cancer Study. Br J Nutr (2019) 121(5):591–600. doi: 10.1017/S0007114518003744
29. Jordan I, Hebestreit A, Swai B, Krawinkel MB. Dietary Patterns and Breast Cancer Risk Among Women in Northern Tanzania: A Case-Control Study. Eur J Nutr (2013) 52(3):905–15. doi: 10.1007/s00394-012-0398-1
30. Garland CF, Gorham ED, Mohr SB, Garland FC. Vitamin D for Cancer Prevention: Global Perspective. Ann Epidemiol (2009) 19(7):468–83. doi: 10.1016/j.annepidem.2009.03.021
31. Bertone-Johnson ER. Vitamin D and Breast Cancer. Ann Epidemiol (2009) 19(7):462–7. doi: 10.1016/j.annepidem.2009.01.003
32. Shekarriz-Foumani R, Khodaie F. The Correlation of Plasma 25-Hydroxyvitamin D Deficiency With Risk of Breast Neoplasms: A Systematic Review. Iranian J Cancer Prev (2016) 9(3):e4469. doi: 10.17795/ijcp-4469
33. Yao S, Kwan ML, Ergas IJ, Roh JM, Cheng TM, Hong C, et al. Association of Serum Level of Vitamin D at Diagnosis With Breast Cancer Survival: A Case-Cohort Analysis in the Pathways Study. JAMA (2016) 3(3):351–7. doi: 10.1001/jamaoncol.2016.4188
34. Grant WB. Relation Between Prediagnostic Serum 25-Hydroxyvitamin D Level and Incidence of Breast, Colorectal and Other Cancers. J Photochem Photobiol B (2010) 101(2):130–6. doi: 10.1016/j.jphotobiol.2010.04.008
35. Yao S, Sucheston LE, Millen AE, Johnson CS, Trump DL, Nesline MK, et al. Pretreatment Serum Concentrations of 25-Hydroxyvitamin D and Breast Cancer Prognostic Characteristics: A Case-Control and a Case-Series Study. PloS One (2011) 6(2):e17251. doi: 10.1371/journal.pone.0017251
36. Grant WB. Vitamin D Status may Help Explain Racial Disparities in Breast Cancer Hospitalization Outcomes. Cancer Epidemiol (2016) 45:174. doi: 10.1016/j.canep.2016.09.008
37. Grant WB. Lower Vitamin D Status may Explain Racial Disparities in All-Cause Mortality Among Younger Commercially Insured Women With Incident Metastatic Breast Cancer. Breast Cancer Res Treat (2016) 159(1):173. doi: 10.1007/s10549-016-3916-7
38. Freedman DM, Looker AC, Chang SC, Graubard BI. Prospective Study of Serum Vitamin D and Cancer Mortality in the United States. J Natl Cancer Inst (2007) 99(21):1594–602. doi: 10.1093/jnci/djm204
39. Garland CF, Gorham ED, Mohr SB, Grant WB, Giovannucci EL, Lipkin M, et al. Vitamin D and Prevention of Breast Cancer: Pooled Analysis. J Steroid Biochem Mol Biol (2007) 103(3-5):708–11. doi: 10.1016/j.jsbmb.2006.12.007
40. Abbas S, Chang-Claude J, Linseisen J. Plasma 25-Hydroxyvitamin D and Premenopausal Breast Cancer Risk in a German Case-Control Study. Int J Cancer (2009) 124(1):250–5. doi: 10.1002/ijc.23904
41. Mogire RM, Mutua A, Kimita W, Kamau A, Bejon P, Pettifor JM, et al. Prevalence of Vitamin D Deficiency in Africa: A Systematic Review and Meta-Analysis. Lancet Global Health (2020) 8(1):e134–2. doi: 10.1016/S2214-109X(19)30457-7
42. Smith GD, Ebrahim S. 'Mendelian Randomization': Can Genetic Epidemiology Contribute to Understanding Environmental Determinants of Disease? Int J Epidemiol (2003) 32(1):1–22. doi: 10.1093/ije/dyg070
43. Burgess S, Dudbridge F, Thompson SG. Combining Information on Multiple Instrumental Variables in Mendelian Randomization: Comparison of Allele Score and Summarized Data Methods. Stat Med (2016) 35(11):1880–906. doi: 10.1002/sim.6835
44. Burgess S, Butterworth AS, Thompson JR. Beyond Mendelian Randomization: How to Interpret Evidence of Shared Genetic Predictors. J Clin Epidemiol (2016) 69:208–16. doi: 10.1016/j.jclinepi.2015.08.001
45. Burgess S, Small DS, Thompson SG. A Review of Instrumental Variable Estimators for Mendelian Randomization. Stat Methods Med Res (2015) 26(5):2333–55. doi: 10.1177/0962280215597579
46. Burgess S, Timpson NJ, Ebrahim S, Davey Smith G. Mendelian Randomization: Where Are We Now and Where are We Going? Int J Epidemiol (2015) 44(2):379–88. doi: 10.1093/ije/dyv108
47. Bowden J, Davey Smith G, Burgess S. Mendelian Randomization With Invalid Instruments: Effect Estimation and Bias Detection Through Egger Regression. Int J Epidemiol (2015) 44(2):512–25. doi: 10.1093/ije/dyv080
48. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, Consortium E-I. Using Published Data in Mendelian Randomization: A Blueprint for Efficient Identification of Causal Risk Factors. Eur J Epidemiol (2015) 30(7):543–52. doi: 10.1007/s10654-015-0011-z
49. Burgess S, Thompson SG. Multivariable Mendelian Randomization: The Use of Pleiotropic Genetic Variants to Estimate Causal Effects. Am J Epidemiol (2015) 181(4):251–60. doi: 10.1093/aje/kwu283
50. Davies NM, von Hinke Kessler Scholder S, Farbmacher H, Burgess S, Windmeijer F, Smith GD. The Many Weak Instruments Problem and Mendelian Randomization. Stat Med (2015) 34(3):454–68. doi: 10.1002/sim.6358
51. Burgess S, Daniel RM, Butterworth AS, Thompson SG, Consortium EP-I. Network Mendelian Randomization: Using Genetic Variants as Instrumental Variables to Investigate Mediation in Causal Pathways. Int J Epidemiol (2015) 44(2):484–95. doi: 10.1093/ije/dyu176
52. Chouchane KSSaL, Ibrahim M. Breast Cancer in African Populations. In: Rotimi C, editor. The Genetics of African Populations in Health and Diseases, 1st ed. United Kingdom: Cambridge University Press (2020). p. 199 – 216.
53. Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-Wide Association Studies Identify Four ER Negative-Specific Breast Cancer Risk Loci. Nat Genet (2013) 45(4):392–8e2. doi: 10.1038/ng.2561
54. Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A Common Variant at the TERT-CLPTM1L Locus is Associated With Estrogen Receptor-Negative Breast Cancer. Nat Genet (2011) 43(12):1210–4. doi: 10.1038/ng.985
55. Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, et al. Multiple Independent Variants at the TERT Locus are Associated With Telomere Length and Risks of Breast and Ovarian Cancer. Nat Genet (2013) 45(4):371–84e2. doi: 10.1038/ng.2566
56. Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-Scale Genotyping Identifies 41 New Loci Associated With Breast Cancer Risk. Nat Genet (2013) 45(4):353–61e2. doi: 10.1038/ng.2563
57. Siddiq A, Couch FJ, Chen GK, Lindström S, Eccles D, Millikan RC, et al. A Meta-Analysis of Genome-Wide Association Studies of Breast Cancer Identifies Two Novel Susceptibility Loci at 6q14 and 20q11. Hum Mol Genet (2012) 21(24):5373–84. doi: 10.1093/hmg/dds381
58. Couch FJ, Gaudet MM, Antoniou AC, Ramus SJ, Kuchenbaecker KB, Soucy P, et al. Common Variants at the 19p13.1 and ZNF365 Loci are Associated With ER Subtypes of Breast Cancer and Ovarian Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. Cancer Epidemiol Biomarkers Prev (2012) 21(4):645–57. doi: 10.1158/1055-9965.EPI-11-0888
59. Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, et al. Genome-Wide Association Analysis Identifies Three New Breast Cancer Susceptibility Loci. Nat Genet (2012) 44(3):312–8. doi: 10.1038/ng.1049
60. Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, et al. Novel Breast Cancer Susceptibility Locus at 9q31.2: Results of a Genome-Wide Association Study. J Natl Cancer Inst (2011) 103(5):425–35. doi: 10.1093/jnci/djq563
61. Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A Locus on 19p13 Modifies Risk of Breast Cancer in BRCA1 Mutation Carriers and is Associated With Hormone Receptor-Negative Breast Cancer in the General Population. Nat Genet (2010) 42(10):885–92. doi: 10.1038/ng.669
62. Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-Wide Association Study Identifies Five New Breast Cancer Susceptibility Loci. Nat Genet (2010) 42(6):504–7. doi: 10.1038/ng.586
63. Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A Multistage Genome-Wide Association Study in Breast Cancer Identifies Two New Risk Alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet (2009) 41(5):579–84. doi: 10.1038/ng.353
64. Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-Wide Association Study Identifies a New Breast Cancer Susceptibility Locus at 6q25.1. Nat Genet (2009) 41(3):324–8. doi: 10.1038/ng.318
65. Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly Discovered Breast Cancer Susceptibility Loci on 3p24 and 17q23.2. Nat Genet (2009) 41(5):585–90. doi: 10.1038/ng.354
66. Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common Variants on Chromosome 5p12 Confer Susceptibility to Estrogen Receptor-Positive Breast Cancer. Nat Genet (2008) 40(6):703–6. doi: 10.1038/ng.131
67. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A Genome-Wide Association Study Identifies Alleles in FGFR2 Associated With Risk of Sporadic Postmenopausal Breast Cancer. Nat Genet (2007) 39(7):870–4. doi: 10.1038/ng2075
68. Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-Wide Association Analysis of More Than 120,000 Individuals Identifies 15 New Susceptibility Loci for Breast Cancer. Nat Genet (2015) 47(4):373–80. doi: 10.1038/ng.3242
69. Huo D, Feng Y, Haddad S, Zheng Y, Yao S, Han YJ, et al. Genome-Wide Association Studies in Women of African Ancestry Identified 3q26.21 as a Novel Susceptibility Locus for Oestrogen Receptor Negative Breast Cancer. Hum Mol Genet (2016) 25(21):4835–46. doi: 10.1093/hmg/ddw305
70. Feng Y, Stram DO, Rhie SK, Millikan RC, Ambrosone CB, John EM, et al. A Comprehensive Examination of Breast Cancer Risk Loci in African American Women. Hum Mol Genet (2014) 23(20):5518–26. doi: 10.1093/hmg/ddu252
71. Chen F, Chen GK, Stram DO, Millikan RC, Ambrosone CB, John EM, et al. A Genome-Wide Association Study of Breast Cancer in Women of African Ancestry. Hum Genet (2013) 132(1):39–48. doi: 10.1007/s00439-012-1214-y
72. Zheng Y, Ogundiran TO, Adebamowo C, Nathanson KL, Domchek SM, Rebbeck TR, et al. Lack of Association Between Common Single Nucleotide Polymorphisms in the TERT-CLPTM1L Locus and Breast Cancer in Women of African Ancestry. Breast Cancer Res Treat (2012) 132(1):341–5. doi: 10.1007/s10549-011-1890-7
73. Huo D, Zheng Y, Ogundiran TO, Adebamowo C, Nathanson KL, Domchek SM, et al. Evaluation of 19 Susceptibility Loci of Breast Cancer in Women of African Ancestry. Carcinogenesis (2012) 33(4):835–40. doi: 10.1093/carcin/bgs093
74. Hou N, Zheng Y, Gamazon ER, Ogundiran TO, Adebamowo C, Nathanson KL, et al. Genetic Susceptibility to Type 2 Diabetes and Breast Cancer Risk in Women of European and African Ancestry. Cancer Epidemiol Biomarkers Prev (2012) 21(3):552–6. doi: 10.1158/1055-9965.EPI-11-0979
75. Fackenthal JD, Zhang J, Zhang B, Zheng Y, Hagos F, Burrill DR, et al. High Prevalence of BRCA1 and BRCA2 Mutations in Unselected Nigerian Breast Cancer Patients. Int J Cancer (2012) 131(5):1114–23. doi: 10.1002/ijc.27326
76. Zhang B, Fackenthal JD, Niu Q, Huo D, Sveen WE, DeMarco T, et al. Evidence for an Ancient BRCA1 Mutation in Breast Cancer Patients of Yoruban Ancestry. Familial Cancer (2009) 8(1):15–22. doi: 10.1007/s10689-008-9205-9
77. Huo D, Kim HJ, Adebamowo CA, Ogundiran TO, Akang EE, Campbell O, et al. Genetic Polymorphisms in Uridine Diphospho-Glucuronosyltransferase 1A1 and Breast Cancer Risk in Africans. Breast Cancer Res Treat (2008) 110(2):367–76. doi: 10.1007/s10549-007-9720-7
78. Garner CP, Ding YC, John EM, Ingles SA, Olopade OI, Huo D, et al. Genetic Variation in IGFBP2 and IGFBP5 is Associated With Breast Cancer in Populations of African Descent. Hum Genet (2008) 123(3):247–55. doi: 10.1007/s00439-008-0468-x
79. Fackenthal JD, Sveen L, Gao Q, Kohlmeir EK, Adebamowo C, Ogundiran TO, et al. Complete Allelic Analysis of BRCA1 and BRCA2 Variants in Young Nigerian Breast Cancer Patients. J Med Genet (2005) 42(3):276–81. doi: 10.1136/jmg.2004.020446
80. Gao Q, Tomlinson G, Das S, Cummings S, Sveen L, Fackenthal J, et al. Prevalence of BRCA1 and BRCA2 Mutations Among Clinic-Based African American Families With Breast Cancer. Hum Genet (2000) 107(2):186–91. doi: 10.1007/s004390000290
81. Adebamowo SN, Dareng EO, Famooto AO, Offiong R, Olaniyan O, Obende K, et al. Cohort Profile: African Collaborative Center for Microbiome and Genomics Research's (ACCME's) Human Papillomavirus (HPV) and Cervical Cancer Study. Int J Epidemiol (2017) 46(6):1745. doi: 10.1093/ije/dyx050
82. Akarolo-Anthony SN, Odubore FO, Yilme S, Aragbada O, Odonye G, Hu F, et al. Pattern of Dietary Carbohydrate Intake Among Urbanized Adult Nigerians. Int J Food Sci Nutr (2013) 64(3):292–9. doi: 10.3109/09637486.2012.746290
83. Akarolo-Anthony S, Odubore F, Yilme S, Aragbada O, Odonye G, Hu FB, et al. Pattern of Dietary Carbohydrate Intake Among Urbanized Adult Nigerians. Inter J Food Sci Nutr (2013) 64(3):292–3. doi: 10.3109/09637486.2012.746290
84. Franklin JD, Guidry A, Brinkley JF. A Partnership Approach for Electronic Data Capture in Small-Scale Clinical Trials. J Biomed Inf (2011) 44(Supple 1):S103–8. doi: 10.1016/j.jbi.2011.05.008
85. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap)—A Metadata-Driven Methodology and Workflow Process for Providing Translational Research Informatics Support. J Biomed Inf (2009) 42(2):377–81. doi: 10.1016/j.jbi.2008.08.010
86. Jensen OM, Parkin DM, MacLennan R, Muir CS, Skeet RG. Cancer Registration: Principles and Methods. Lyon, France: International Agency for Research on Cancer (1991).
87. Adebamowo C, Jedy-Agba E, Oga E, Osinubi P, Igbinoba F, Osubor G, et al. Creating a Nationwide Cancer Registration System to Support AIDS-Cancer Match Studies in Nigeria. Infect Agent Cancer (2012) 7(Suppl 1):P3. doi: 10.1186/1750-9378-7-S1-P3
88. Wu NC, Wong W, Ho KE, Chu VC, Rizo A, Davenport S, et al. Comparison of Central Laboratory Assessments of ER, PR, HER2, and Ki67 by IHC/FISH and the Corresponding mRNAs (ESR1, PGR, ERBB2, and MKi67) by RT-qPCR on an Automated, Broadly Deployed Diagnostic Platform. Breast Cancer Res Treat (2018) 172(2):327–38. doi: 10.1007/s10549-018-4889-5
89. Freedman LP, Inglese J. The Increasing Urgency for Standards in Basic Biologic Research. Cancer Res (2014) 74(15):4024–9. doi: 10.1158/0008-5472.CAN-14-0925
90. Groelz D, Sobin L, Branton P, Compton C, Wyrich R, Rainen L. Non-Formalin Fixative Versus Formalin-Fixed Tissue: A Comparison of Histology and RNA Quality. Exp Mol Pathol (2013) 94(1):188–94. doi: 10.1016/j.yexmp.2012.07.002
91. Bussolati G, Annaratone L, Maletta F. The Pre-Analytical Phase in Surgical Pathology. Recent results Cancer Res Fortschr der Krebsforschung Progres dans les recherches sur le Cancer (2015) 199:1–13. doi: 10.1007/978-3-319-13957-9_1
92. Bass BP, Engel KB, Greytak SR, Moore HM. A Review of Preanalytical Factors Affecting Molecular, Protein, and Morphological Analysis of Formalin-Fixed, Paraffin-Embedded (FFPE) Tissue: How Well do You Know Your FFPE Specimen? Arch Pathol Lab Med (2014) 138(11):1520–30. doi: 10.5858/arpa.2013-0691-RA
93. Baker M. Reproducibility Crisis: Blame it on the Antibodies. Nature (2015) 521(7552):274–6. doi: 10.1038/521274a
94. Hicks DG, Kushner L, McCarthy K. Breast Cancer Predictive Factor Testing: The Challenges and Importance of Standardizing Tissue Handling. J Natl Cancer Inst Monogr (2011) 2011(42):43–5. doi: 10.1093/jncimonographs/lgr003
95. Hicks DG, Boyce BF. The Challenge and Importance of Standardizing Pre-Analytical Variables in Surgical Pathology Specimens for Clinical Care and Translational Research. Biotechnic Histochem Off Publ Biol Stain Commission (2012) 87(1):14–7. doi: 10.3109/10520295.2011.591832
96. Consortium HA. H3Africa Consortium Data Access Release Policy 2020 [updated August 2020]. Available from: https://h3africa.org/wp-content/uploads/2020/06/H3Africa-Consortium-Data-Access-Release-Policy-April-2020.pdf.
Keywords: breast cancer, female, Africa, genomics, epidemiology
Citation: Ezeome ER, Yawe K-DT, Ayandipo O, Badejo O, Adebamowo SN, Achusi B, Fowotade A, Ogun G, AFBRECANE Research Group and Adebamowo CA (2022) The African Female Breast Cancer Epidemiology Study Protocol. Front. Oncol. 12:856182. doi: 10.3389/fonc.2022.856182
Received: 16 January 2022; Accepted: 08 March 2022;
Published: 13 April 2022.
Edited by:
Alireza Sadjadi, Tehran University of Medical Sciences, IranReviewed by:
Andreas Hadjisavvas, The Cyprus Institute of Neurology and Genetics, CyprusSophia S. Wang, City of Hope Beckman Research Institute/Comprehensive Cancer Center, United States
Copyright © 2022 Ezeome, Yawe, Ayandipo, Badejo, Adebamowo, Achusi, Fowotade, Ogun, AFBRECANE Research Group and Adebamowo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clement A. Adebamowo, Y2FkZWJhbW93b0Bzb20udW1hcnlsYW5kLmVkdQ==
†AFBRECANE Research group is part of the H3Africa Consortium
 Emmanuel R. Ezeome
Emmanuel R. Ezeome King-David T. Yawe3
King-David T. Yawe3 Sally N. Adebamowo
Sally N. Adebamowo Benerdin Achusi
Benerdin Achusi Clement A. Adebamowo
Clement A. Adebamowo