
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 17 March 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.855996
This article is part of the Research TopicCancer Stem Cell Differentiation: A Realistic Potential Therapeutic Option?View all 10 articles
 Zeyu Wang1†
Zeyu Wang1† Shasha Zhao2†
Shasha Zhao2† Xiaolin Lin3†
Xiaolin Lin3† Guanglong Chen4†
Guanglong Chen4† Jiawei Kang5
Jiawei Kang5 Zhongping Ma6
Zhongping Ma6 Yiming Wang6
Yiming Wang6 Zhi Li4*
Zhi Li4* Xiuying Xiao3*
Xiuying Xiao3* Aina He7*
Aina He7* Dongxi Xiang8*
Dongxi Xiang8*Organoids well recapitulate organ-specific functions from their tissue of origin and remain fundamental aspects of organogenesis. Organoids are widely applied in biomedical research, drug discovery, and regenerative medicine. There are various cultivated organoid systems induced by adult stem cells and pluripotent stem cells, or directly derived from primary tissues. Researchers have drawn inspiration by combination of organoid technology and tissue engineering to produce organoids with more physiological relevance and suitable for translational medicine. This review describes the value of applying organoids for tumorigenesis modeling and tumor vaccination. We summarize the application of organoids in tumor precision medicine. Extant challenges that need to be conquered to make this technology be more feasible and precise are discussed.
Organoids are three-dimensional cell complexes with a particular spatial structure cultured in vitro (1, 2). It is amplified and maintains certain structural and functional features of their source tissue (3). Organoids develop from stem-like cells or initiating cells including embryonic stem cells (ESCs) (4, 5), adult stem cells (ASCs) (6), induced pluripotent stem cells (iPSCs), and progenitor cells (7–9). ESCs are cells selected from the intraembryonic cell mass or obtained by inhibiting primordial germ cells in vitro, which has the ability of multidirectional differentiation (10, 11). ASCs are undifferentiated cells existing in various differentiated tissues that are responsible for repair and regeneration after tissue injury (5). Progenitor cells can repair and regenerate following tissue damage (12). In this review, we demonstrate organoid platforms derived from primary tissues, ASCs or iPSCs, summarize the organoid bioengineering advancement, and describe the possibility of applying organoids for carcinogenesis modeling (Figure 1). Current challenges about the broad application of organoids are also discussed.
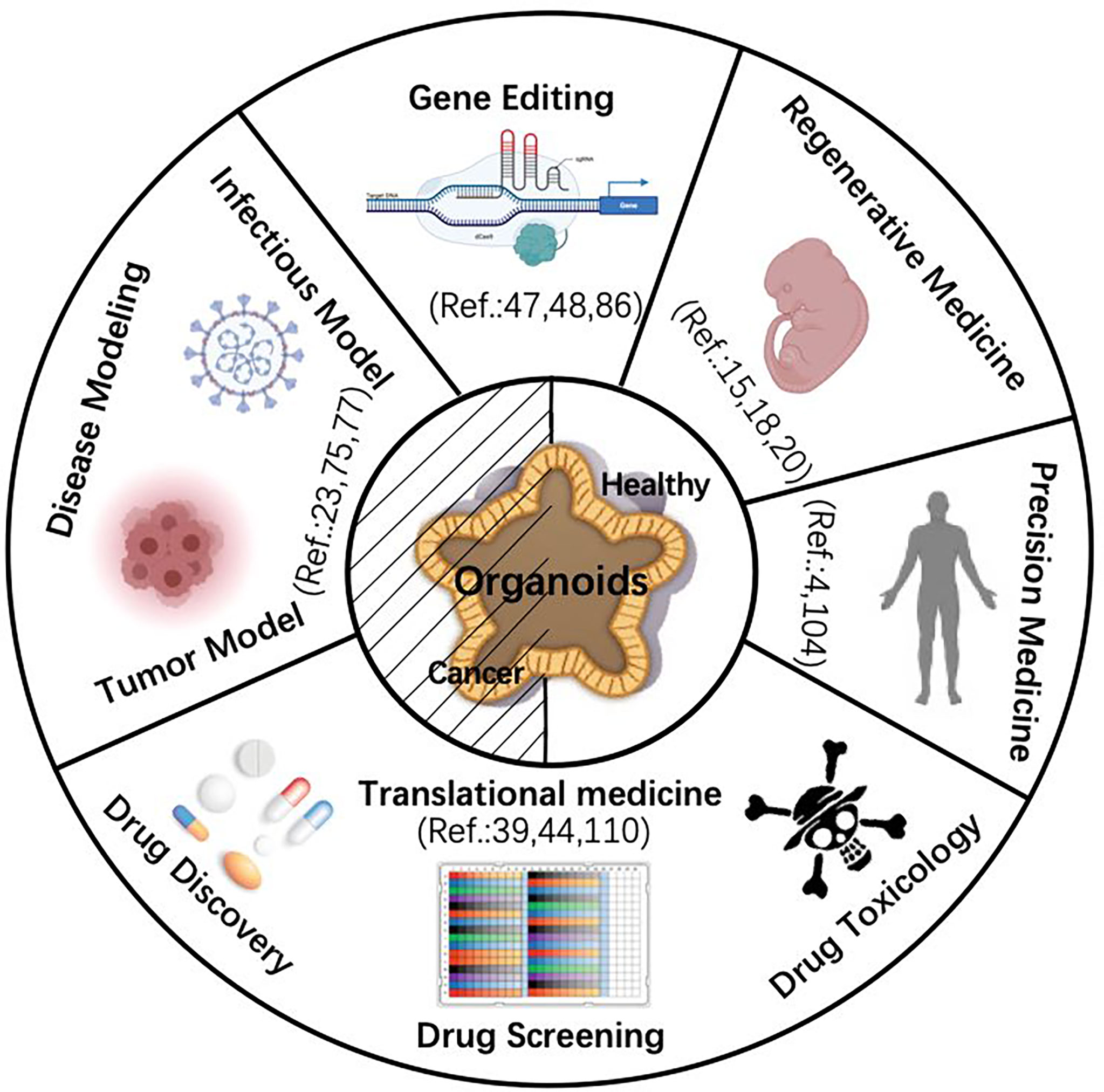
Figure 1 Schematic diagram of organoid application. The center of the diagram shows that both normal and cancer organoids share the five application fields indicated by the peripheral parts. [Note: e.g., Ref. 4 refers to reference (4)]. Images are adapted from Servier Medical Art (https://smart.servier.com).
Organoid culture became a major technological advance in 2009. Hans Clevers reconstructed a suitable niche better for maintaining intestinal stem cells in vitro (13). This system contains a key conditioned medium supplemented with multiple growth factors including Wnt agonists R-spondin-1 or -3, Wnt3a, epidermal growth factor (EGF), and noggin. Mouse primary intestine cells were embedded into hydrogel [i.e., extracellular matrix (ECM)] followed by gut organoid formation with villus structure owing to the self-renewal capacity of intestinal stem cells, which has pioneered the field of organoid cancer biology (13). The system guaranteed continuous proliferation of organoids, not only ensured stability and purification of mouse genome but also had an advantage of amplification (14). Research fellows have cultivated diverse organoid systems derived from tissues with epithelial origination, such as bladder, colon, rectum, endometrium, fallopian tube, kidney, liver, lung, esophagus, oral mucosa, pancreas, prostate, salivary gland, skin epidermis, stomach, and taste buds (15–25). Researchers have also generated organoids from normal cells in the urinary tract and bronchial lavage, rather than parenchymatous organs (26, 27). Organoids derived from ASCs maintain phenotypic and genetic stability, better reflecting their primary tissue genome (28, 29). Genomic mutation in organoids can be investigated by immunohistochemistry staining, whole-exome sequencing (WES), and RNA sequencing (30). Organoid is driven by the inherent ability of stem cells themselves (13). Their self-assembly ability allows organoids to produce functional mature “organs” by precise spatial and temporal order (31, 32). Similarly, organoids run self-assembly processes in vitro by changing cytokine constituents of culture media, simulating organoid differentiation and maturation (14, 33, 34). Gabriel et al. (35) observed that brain organoids assembled optic vesicles, including primitive corneal epithelial and lens-like cells. This study confirmed that the self-assembly process was carried out via a multistep process in the early stage of organoid genesis.
Organoid culture varies from different tissues of origin; several organs require more efforts to establish a stable condition (i.e., heart and immune organs) (36–38). It has been studied to generate complex and highly structured cardiac organs by embedding human iPSCs into matrix glue, regulating small molecule directional cardiac differentiation through the biphasic Wnt pathway (39). The lacrimal secretion of neurotransmitters by lacrimal gland organoids was verified by orthotopic transplantation in mice (40).
The inherent self-organizing of stem cells does not signify that organoid might form fine tissue under any condition (41). This process emphasizes that fate guides organoid to develop into mimic-tissues in a highly environment-dependent manner (42). Those established organoid-forming approaches have considerable defects: when cultured for too long, stem cells would uncontrollably develop into a circular cystic closed structure, with a short life span and non-physiological shape, resulting in inconsistency between organoid and organs in anatomy and physiology (43, 44). To solve this issue, bioengineering cultivated organoids into a variety of biomaterials that can promote their better proliferation, precise differentiation, and exact function (43, 45, 46). Tissue bioengineering uses bioactive substances to regenerate or repair tissues through in vitro construction (Figure 2) (47). It is accomplished by controlling the process of organoids and establishing the next generation with high physiological correlation (48). Researchers at the EPFL Institute have constructed an intestinal geometric scaffold with hydrogel, providing an appropriate place to guide organoid to form a true intestinal organ (49). In this method, stem cells were cultured in scaffolds simulating the surface of natural tissue and then combined into microfluidic chips. Due to their inherent self-organization, organoids grew on tubular scaffolds and self-organized to form intestines (47). Organoids would gradually form continuous cell layers with recess structure and villous-like domain and form “mini-intestines” in vitro, which maintain the same functional features as primary organs in vivo (48, 50, 51). Improved methods have been used for organoid generation (i.e., stomach, liver, kidney, etc.) (52–55).
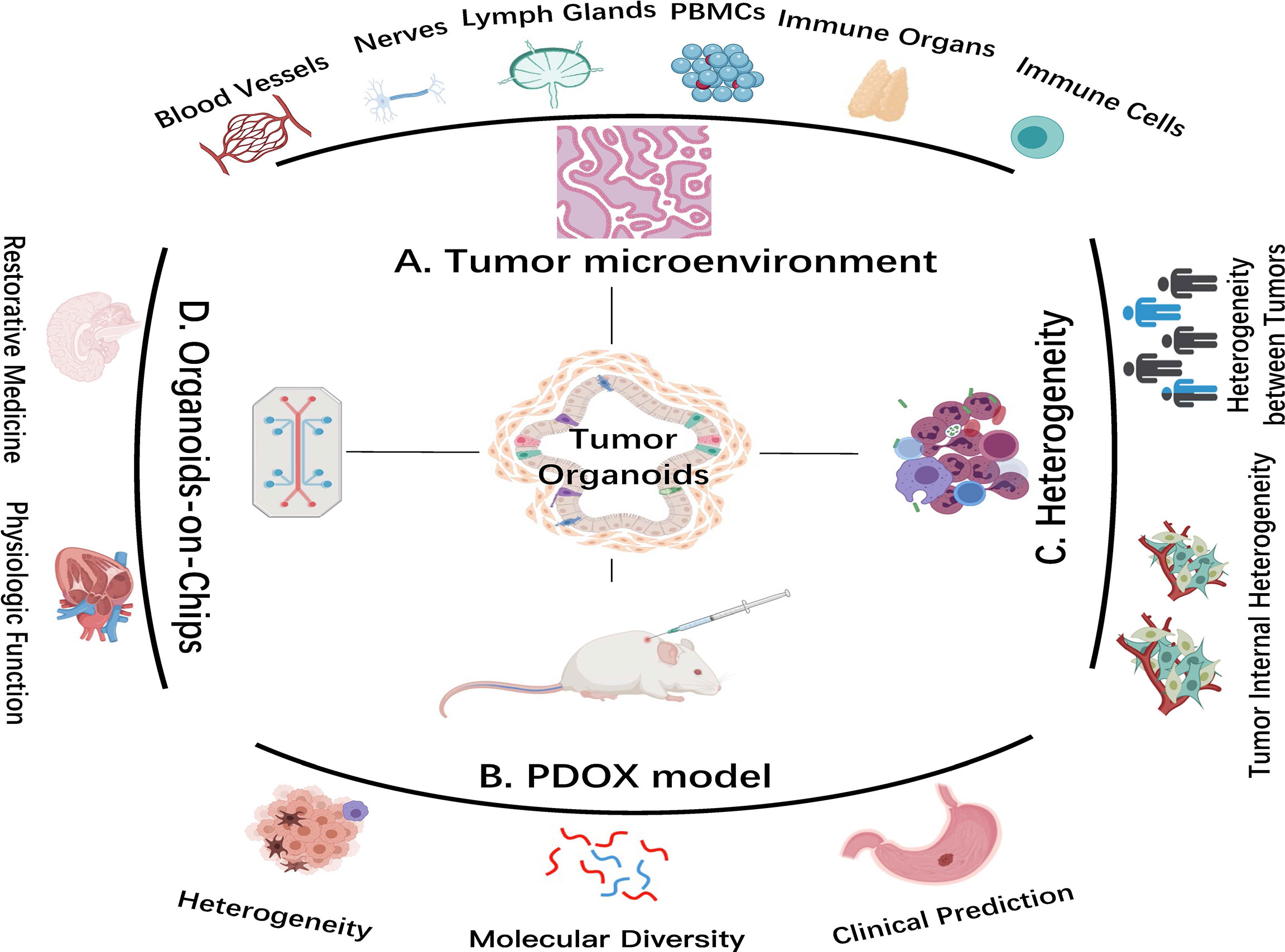
Figure 2 Advanced organoid technology in cancer research. (A) Coculture system of tumor organoids with components of the tumor microenvironment (TME). (B) Patient-derived organoid xenograft (PDOX) model. The PDOX model indicates tumor heterogeneity and molecular diversity and predicts clinical treatment in vivo. (C) Tumor organoids better reflect tumor heterogeneity. (D) Organoids-on-Chips system. This system combines organoid and tissue engineering to highly replicate physiological function in vitro and to facilitate regenerative medical development. Images are adapted from Servier Medical Art (https://smart.servier.com).
Microfluidic approach and organoid chip integrate mechanical and physiological parameters, expanding the usage of organoids for recapitulating the physiological function of their source organs (Figure 2) (37). Using organ setting on a chip, a single gastric organoid has been successfully established (56). Not singly but in pairs, the use of lumen flow and pressure circulation to induce peristaltic movement showed the feasibility of integrating engineering in organoid culture (57). Coincidentally, kidney organoids on chips were exposed to shear stress by applying fluid flow, thereby mimicking the kidney environment in vivo (58). The existence of fluid flow not only promoted maturation of renal organoids but also was conducive to formation of vascular network with perfusion lumen. Creative combination of organoids and tissue engineering allows organoid growth, which may overcome space constraints and promote shape-guided morphogenesis and physiology. Furthermore, researchers used micro fabricated cell rejection microporous (an apparatus for culturing organoids using tissue engineering) to culture and monitor homogeneous liver organoids from dissociated human iPSCs (54). Organoid bioengineering improves experimental replication, optimizes model quality, and realizes clinical transformation. Decembrini et al. (59) have proven that production of retinal organoids was accelerated and standardized in the best physicochemical microenvironment. A biomimetic hydrogel composed of circular bottom microwell arrays and optimized media formulation not only facilitated mouse ESC formation but also aggregated retinal organoids in a stereotypical manner, leading to an unlimited source of retinal neurons (59). Combining organoid culture with tissue engineering, organ devices accurately control organoid growth, while organoids simulate real physiology more in line with the situation in vivo (60–62).
Compared with traditional two-dimensional culture of tumor cells, tumor organoids maintain better cell heterogeneity, retain tumor characteristics, show less loss of tumor niche components, and provide a more authentic environment for clinical treatment (63, 64). In contrast to patient-derived tumor xenografts (PDXs), the success rate of tumor organoid construction is way higher (50%–90% vs. 10%–30%), organoids are maintained for a longer time with lower cost, so it is easy to gene editing and large-scale drug screening (65–68). There are two kinds of organoid construction techniques, one is derived from differentiation of iPSCs (16) and the other is directly derived from tumor tissues (69). Constructing tumor organoids from iPSCs largely depends on tumor types, and the culture operation is more complex (70). Tumor organoids obtained by iPSC differentiation lose the complexity of the tumor microenvironment (TME) (71). The more common organoid culture method is to directly use primary tissues, supplemented by cytokines, tumor matrix, and other components (70, 72). Tumor organoids such as colorectal, breast, pancreatic, prostate, liver, and gastric cancers have been successfully constructed (73–77). Researchers have obtained a diverse collection of tumor organoids with different characteristics through in vitro culture to constitute a living organoid biobank (78). Through histochemical observation of the morphology of tumor organoids, their internal structure was like that of primary tissue (1, 4, 6). Simultaneously, genome and single-cell sequencing was carried out to explore the variation between organoids and primary tumors in the gene mutation spectrum (79).
Organoids maintain tumor heterogeneity within and among tumors (80). Patient-derived organoids (PDOs) maintains the diversity and complexity of tumor origin in terms of cell hetereogeniety, histology, gene mutation, transcription spectrum, and even metabolism (39, 81, 82). Organoids not only reflect characteristics of primary tumors, but also exert advantages for explorations of tumorigenesis, cell communication, epigenetics, and invasion (34, 77).
Due to various sources of tumor samples and slightly discrepant culture system among laboratories, it is not easy to duplicate the experimental findings, and this hinders further verification and reference of obtained data. A study conducted by Dr. Manel Esteller, director of the Josep Carreras leukemia Institute (IJC), used epigenetic Infinium MethylationEPIC BeadChip (EPIC), a microarray chip from Illumina that interrogates more than 850,000 CpG sites to analyze DNA methylation status of 25 human cancer organoids (83). This data indicated that tumor organoids highly sustain biological properties and heterogeneity of tumor tissue in situ.
Tumor originates from accumulation of gene mutation in normal somatic cells; while not all mutations have access to induce tumorigenesis, tolerance of different tissues to the same mutation is widely divergent. Numerous cell and animal experiments have clarified key factors and decisive mechanisms initiated from gene mutation to tumorigenesis, fully understanding that such process is artificial due to the failure of monitoring and intervening the earliest process of tumor development. Innovative organoid system makes it possible to understand the transformation process from normal tissue to tumor (84). The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 system has revolutionized genetic engineering, allowing gene editing of normal organoids by inducing oncogenes to obtain tumor organoids and track the carcinogenic process from initial to advanced stage (85–87). Mutated tumor-related genes KRAS, CDKN2A, TP53, and SMAD4 in normal pancreatic organoids eventually developed to a state recapitulating pancreatic ductal adenocarcinoma (80, 88, 89). Lannagan et al. (90) found that mutated intestinal organoids could grow and reproduce without relying on any exogenous cytokines. When transplanting organoids with four mutations into mice, these organoids induced by colorectal cancer were invasive (90). This approach can amplify the effects of driving mutations in the same genetic background. Organoids from normal tissues mimic tumor pathogenesis by continuously introducing cancer-driven mutations (91, 92). Combination of organoid system and CRISPR/Cas9 strategy is powerful to study the origin of tumor (93, 94).
It is generally believed that cancer is a progressive disease caused by accumulation of abnormal gene mutation. These genetic anomalies include tumor suppressor/oncogene mutations and chromosome abnormalities (95). It is increasingly clear that tumors can be induced by epigenetic changes. Epigenetics refers to heritable change of gene function without a change of genetic material, eventually leading to different phenotypes (96). It mainly includes DNA methylation, histone modification, non-coding RNA regulation, and chromatin structure reconstruction. Potential tumor suppressor genes are inhibited or silenced at the transcriptional level by DNA methylation, promoting malignant transformation from normal cells (97, 98). It has begun to dig influence and underlying mechanisms that modulate methylation and chromatin states of tumor suppressor gene or tumor oncogene during tumor formation (96, 99). Epigenetic changes bring abundant precancerous cell expansion. They first occur in precancerous cells, determining subsequent genetic changes that promote malignant transformation and tumor cell clonal expansion (98, 99). Aloia et al. (100) have demonstrated that bile duct cells underwent epigenetic modification of genome-wide DNA methylation during tissue damage and subsequent organoid construction. This work that was inspired by those epigenetic changes of organoids from normal to malignant cell transition was able to determine subsequent uncontrollable modification of genetic machinery, eventually leading to malignant occurrence and development (95, 98, 100).
TME is an internal condition for the production and survival of tumor cells, including immune and inflammatory cells, fibroblasts, vascular endothelial cells (101). Bidirectional communication between tumor cells and TME possesses an indispensable position in tumor promotion (36). Tumor-infiltrating lymphocytes (TILs) are heterogeneously composed of different lymphocytes; their phenotypic and functional characteristics largely correlate with interaction with tumor cells (102). Researchers cultured tumor organoids through air–liquid interface to reproduce TME in vitro. Air-PDOs successfully retained inherent fibrous matrix and a variety of immune cell components of primary tumor tissue (103). Through integrated culture, the in situ tumor essence and matrix were reserved, including functional TILs. By continuously coculturing cancer organoids and peripheral blood monocytes (PBMCs) in the presence of T cell-stimulating growth factors, antigen-specific cytotoxic T cells were selected and amplified in about half of total samples (Figure 2). T cells expanded from adjacent healthy epithelial tissues resulted in undisturbed organoid expansion without a significant level of organoid cytotoxicity (104).
Cancer-associated fibroblasts (CAFs) secrete miscellaneous cytokines, chemokines, and growth factors to create a conducive microenvironment for tumor progression (105). The prominent role of CAFs is considered to shape stem cell niche to cultivate cancer stem cells (CSCs), while two-dimensional culture of cell lines is far from satisfactory in summarizing general characteristics of CSCs (106). The first coculture model of CAFs and organoids was established in pancreatic cancer, followed by in liver, colorectal, prostate, esophageal, and breast cancers (107–110). Recently, it is reported that organoids and CAFs from diethylnitrosamine (DEN)-induced mouse liver tumors can be cultured together; CAFs promoted organoid growth through paracrine signals. Cotransplantation of CAFs with liver tumor organoids promoted tumor growth in xenotransplantation models; CAFs may not regulate the efficiency of organoid initiation but accelerated its growth (111).
The deficiency of blood vessels in the organoid system brings challenges (112). One study emphasized that endothelial cells can be replaced by adaptable angiogenic cells to form a perfusion plastic vascular plexus for restrictive synthetic semipermeable membrane by organ chip system, and this provided a physiological platform for tumor organoid vascularization (113). This minimized hypoxia in tumor organoids provided a ponder over influence and mechanism of vascular endothelial cells on tumor occurrence and development (Figure 2) (112, 114).
Tumors are generally heterogeneous, and there is no fixed treatment for all types of tumors, which makes precision medicine a new direction for tumor therapy, that is, patients with different stages of tumor are entitled to discrepant treatment schemes (115, 116). Tumor organoids replicate complex signaling pathways and cell-to-cell relationship and more accurately reflect tumor genetic features (79). Organoids become a personalized treatment channel with its unique experimental advantages—high level of physiological relevance and convenience of in vitro operation (4, 65, 117).
Organoid technology for precision medicine refers to drug screening in vitro in PDOs and formulation of individual medication (32, 85). Tumor organoids largely maintain heterogeneity between source tumors and different patients, individual morphology and scale of organoids remain such uniform (3, 38, 118). These organoids not only remove confusing variables that may be introduced from animal models but also provide greater complexity than homogenized cell cultures (1, 44, 66). The valuable tumor model established by patients’ iPSCs is able to understand tumor pathogenesis and disease progression (3, 9). The established living biobank of tumor organoids has infinite predictive value for determining the distinct drug response of patients. In turn, it gives access to large-scale drug screening (63, 119). Tumor organoids swiftly select the optimal therapeutic schedule for patients, accompanied by reducing side effects and tumor recurrence (24, 70, 120). They significantly shorten the preclinical test period, working as a key slot in drug discovery, providing a large amount of biological data and a high-quality platform (6, 33). With the progress and standardization of organoid culture, the accessibility of tumor organs will be more widely realized (25, 30, 121).
Tumor organoid-based drug screening integrated with next-generation sequencing (NGS) is conducive to oncotherapy, which is combined with clinical treatment to form complementarity (122–124). NGS detects genetic mutations of patients at source and provides drug treatment options, but it alone does not guarantee clinical efficacy (125–128). As a distinct supplement, organoid is advantageous to well investigate this uncertainty (6, 65, 117). Patients with epidermal growth factor receptor amplification were usually treated with cetuximab guided by NGS, yet this consequence was overturned by the organoid system, which was consistent with actual clinical situation (129). Organoid drug screening further picks more effective approaches on the basis of sequencing to give conclusive recommendations and practical biology evidence for patients (20). This technology clarifies particular therapeutics for precision medicine and determines whether a particular group of patients is not suitable for a therapeutic (3, 70).
Organoid culture along with NGS and single-cell sequencing (scRNA-seq) is good to hunt for therapeutic targets and discover mutation-associated neoantigens (MANAs) for fresh targeted remedy or tumor vaccination (Figure 3) (130–134). With continuous innovation and improvement of benchwork for appraising tumor MANAs, such as the recently emerging NeoScreen technology, more previously hidden tumor antigen epitopes have been identified (135–138). Based on bioinformatics data analysis, MANAs reflecting individual disease are unearthed by a machine learning algorithm (138, 139). Using an organoid system, Demmers et al. (140) recently considered that human leukocyte antigen (HLA) class I peptide expression among different clonal cells from the same colorectal cancer patient was variability and its widespread difference in cloning specificity was generally common. By linking organoid proteomics and HLA peptide ligandomics, they discovered that tumor-specific ligands derived from DNA damage and tumor suppressor proteins were remarkably presented in tumor cells, which might be consistent with defunction of their cytoprotective effect. In general, their data demonstrated heterogeneous HLA peptide expression in an individual patient and presumed that a promising multipeptide tumor vaccine may be a feasible option to minimize immune escape risk.
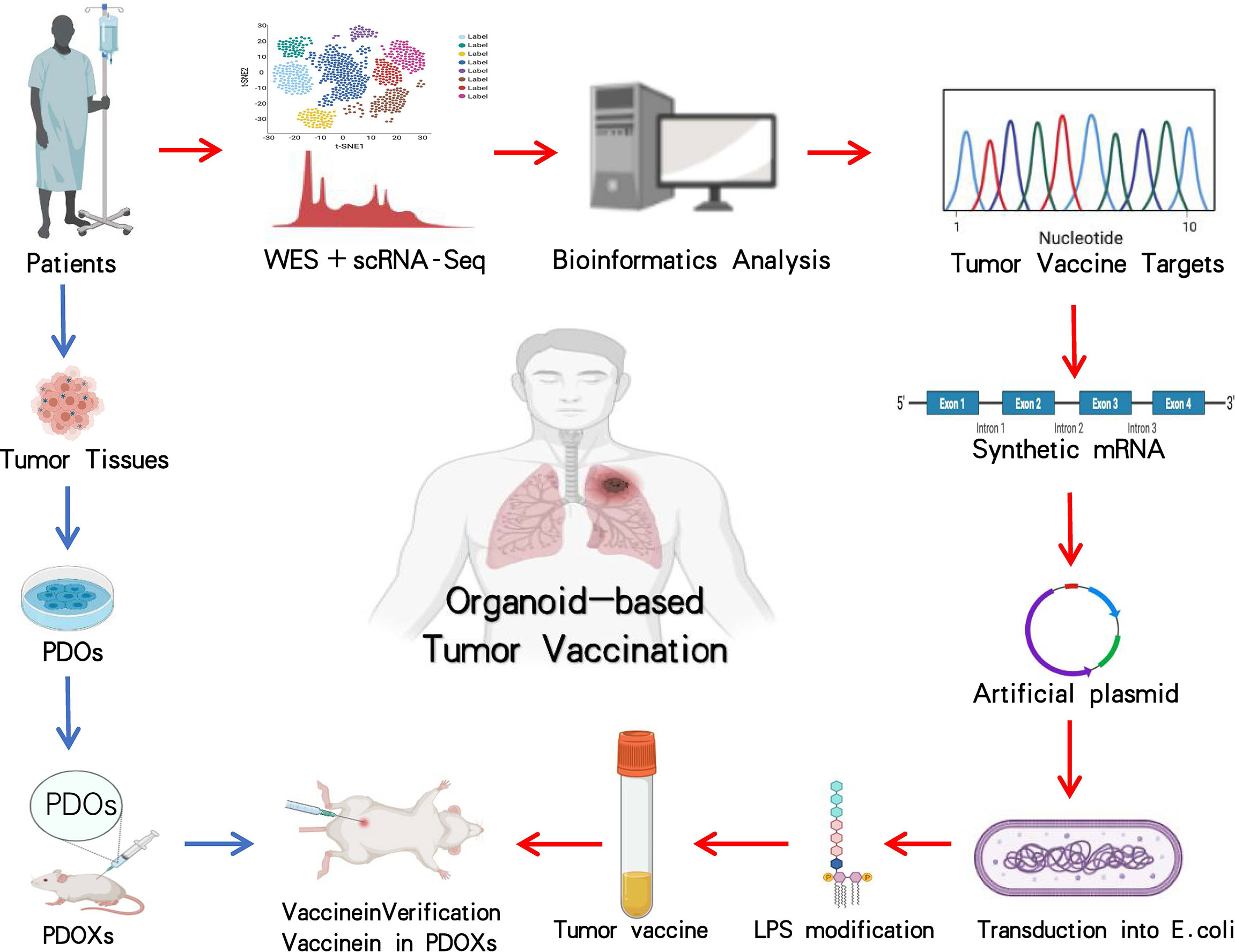
Figure 3 Application and investigation of organoid-based tumor vaccination. The workflow with blue arrows represents the patient-derived organoid xenograft (PDOX) model, and processes with red arrows indicate how tumor vaccine is generated. The entire steps are to verify the efficacy of tumor vaccination by using the PDOX model. Images are adapted from Servier Medical Art (https://smart.servier.com).
Taking certain mRNAs encoding MANAs as available templates, individualized tumor vaccines against MANAs are artificially synthesized, stimulating tumor-specific T-cell production for the sake of diminishing tumor cells (141–147). These tumor vaccines are theoretically proven to be potential therapeutics for accurate oncotherapy (146, 148, 149). Organoids have been implanted during the development and testing process of vaccines against pathogenic microorganisms including bacteria and viruses (141, 150–152). Researchers developed mini tonsils in vitro from surgery tissues and added coronavirus disease 2019 (COVID-19) candidate vaccines into the culture system, a predominant tool to verify vaccine efficacy, to observe whether tonsil organoids incite immune responses, specific immunocytes, and antibodies against viral surface spike proteins (153). It is believed that in the near future, organoids might be exploited in research and development of tumor vaccines (140, 142, 147, 149). PDOs with target gene mutations and autologous or non-autologous immune cells are cocultured. The constructed tumor vaccine candidates are subjected to the coculture system to detect tumor-specific T-cell production and tumor vaccine (145, 148, 154, 155). This emerging platform would be applied to the development of tumor vaccines on a large scale (Figure 3) (3, 36, 37, 65). Tumor organoids provide attractive options for tumor vaccine and are expected to be a unique utensil for tumor precision medicine (146, 153, 156).
Tumor chemoprevention generally refers to inhibition of tumor and vascular cell proliferation, which is expected to maintain its efficacy through the whole stage of chemotherapy for patients (157). It is required to develop advanced approaches for better tumor chemoprevention. The organoid model, a three-dimensional culture platform for primary cells, paves the way of evolution for tumor chemoprevention and provides a practical tool for tumor treatment. A study demonstrated that an A Disintegrin and Metalloprotease 10 inhibitor prevented glioma stem cells from integrating into brain organoids, similar to the mouse xenotransplantation outcome. Temozolomide and Adriamycin treatment reduced the size of tumor organoids by about 30% and 80%, respectively, but had no effect on normal nerve cell organoids. It is worth evaluating the reactivity of glioblastoma multiforme tumor and healthy brain cells when exposed to therapeutics by using the organoid system. They confirmed the biological correlation between organoids and clinical data, providing a basis for high-throughput drug screening and determining the most effective drug timely (158). One additional study indicated that the combination of extracellular signal-regulated kinase 1/2 (ERK1/2) inhibition and autophagy process reduced liver metastases in mice with pancreatic ductal adenocarcinoma PDO transplantation. Over the past decade, vitamin D3 has aroused great interest as a chemoprophylactic agent, especially for treating neoplasms from the digestive system (159). A recent comparative study reported that PDOs revealed the homeostatic effect of vitamin D3 on human intestinal mucosa (160). The organoid technique offers a promising preclinical model for evaluating chemotherapeutic efficacy, a more in-depth mechanistic insight into tumor biology and an organoid biobank system for exploring optimal tumor chemoprevention (161).
Current challenges of organoid application in the clinics include low reproducibility and presence of mixed cell populations, limiting strict experimental examination (38, 42, 77). We have described that the combination of biomaterials and organoids may address the abovementioned deficiencies. Astrocytes and neurons were generated in depart, and they were combined with microplates in the desired proportion and size. This approach accelerated maturation of astrocytes and allowed chemical or genetic manipulation of any cell type before coculture (162, 163). However, how to select certain types of cells as targets for transgenic or epigenetic manipulation, drug delivery, or local extracellular modification is still a bench-side issue (158, 159). Nanoparticles generated from polymer or liposomes are utilized as encapsulation carriers for astrocytes and other cells (164). In another innovative application of nanomedicine, iPSC-derived neural cells and the astrocyte neuron coculture system have been applied as a screening platform to evaluate neurotoxicity. Toxicity is a major risk for nanomedicine; the safety of any nanoparticles needs to be tested prior to application in vivo (165). An alternative way to reduce toxicity is to use exosomes as a source of nanocarriers, which can increase cell type-specific targeting and enhance their functionality by engineering surface antibodies. Verification of the toxicity of nanomaterials in organoids requires much further attention (158, 159, 164).
Organoid technology has made breakthroughs in regenerative medicine and tumor biology. Organoids have a wide range of potential applications, while they still face technical drawbacks (117, 118). Most of the existing organoids are derived from epithelial cells; non-epithelial-originated organoids are hard to be established and maintained for a long purpose (i.e., primary glioblastoma) (121). Coculture of tumor organoids with immune cells is a predictable model for cell therapy, but the culture condition requires further optimization. Different tumor types and individual patients maintain heterogeneity; it is difficult to accurately simulate about its dynamic adaptation (81). The classical organoid culture system contains non-human animal products, such as ECM and hydrogel (Matrigel or BME); these may bring undiscovered effects to the organoid biology. Further organoid-bioengineering strategy and conditioned medium developed with artificial substrates may solve the abovementioned limitations (166, 167). The organoid technology potentiates more accurate tumor recapitulation than cell lines and the PDX model (20, 168). The organoid culture system is good to predict drug sensitivity, tumor promotion, and tumor vaccination (119, 169). Having the in vivo modeling accessibility and developing application, organoid technology is expected to have a profound impact on both bench and bedside for precision medicine (84, 120).
ZW, SZ, and XL wrote the draft. ZL XX, and AH reviewed the article. GC and JK collected data. ZM and YW provided study materials. DX was responsible for review conception, design, and revision. All authors have read and agreed to the published version of the article.
This research was funded by the Natural Science Foundation of China (82173358 to AH and 32170924 to DX) and the Shanghai Municipal Committee of Science and Technology (21140901600 to DX).
ZM and YW served as lab technicians for Shanghai OneTar Biomedicine Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Artegiani B, Clevers H. Use and Application of 3D-Organoid Technology. Hum Mol Genet (2018) 27:R99–R107. doi: 10.1093/hmg/ddy187
2. McCusker D. Cellular Self-Organization: Generating Order From the Abyss. Mol Biol Cell (2020) 31:143–8. doi: 10.1091/mbc.E19-04-0207
3. Pasch CA, Favreau PF, Yueh AE, Babiarz CP, Gillette AA, Sharick JT, et al. Patient-Derived Cancer Organoid Cultures to Predict Sensitivity to Chemotherapy and Radiation. Clin Cancer Res (2019) 25:5376–87. doi: 10.1158/1078-0432.CCR-18-3590
4. Clevers HC. Organoids: Avatars for Personalized Medicine. Keio J Med (2019) 68(4):95. doi: 10.2302/kjm.68-006-ABST
5. Liu H, Zhang Y, Zhang YY, Li YP, Hua ZQ, Zhang CJ, et al. Human Embryonic Stem Cell-Derived Organoid Retinoblastoma Reveals a Cancerous Origin. Proc Natl Acad Sci USA (2020) 117:33628–38. doi: 10.1073/pnas.2011780117
6. Clevers H. Modeling Development and Disease With Organoids. Cell (2016) 165:1586–97. doi: 10.1016/j.cell.2016.05.082
7. Bershteyn M, Kriegstein AR. Cerebral Organoids in a Dish: Progress and Prospects. Cell (2013) 155:19–20. doi: 10.1016/j.cell.2013.09.010
8. Karagiannis P, Takahashi K, Saito M, Yoshida Y, Okita K, Watanabe A, et al. Induced Pluripotent Stem Cells and Their Use in Human Models of Disease and Development. Physiol Rev (2019) 99:79–114. doi: 10.1152/physrev.00039.2017
9. Ohnuki M, Takahashi K. Present and Future Challenges of Induced Pluripotent Stem Cells. Philos Trans R Soc Lond B Biol Sci (2015) 370:20140367. doi: 10.1098/rstb.2014.0367
10. Lim WF, Inoue-Yokoo T, Tan KS, Lai MI, Sugiyama D. Hematopoietic Cell Differentiation From Embryonic and Induced Pluripotent Stem Cells. Stem Cell Res Ther (2013) 4:71. doi: 10.1186/scrt222
11. Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, et al. Tissue-Specific Mutation Accumulation in Human Adult Stem Cells During Life. Nature (2016) 538:260–4. doi: 10.1038/nature19768
12. Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, et al. Unlimited In Vitro Expansion of Adult Bi-Potent Pancreas Progenitors Through the Lgr5/R-Spondin Axis. EMBO J (2013) 32:2708–21. doi: 10.1038/emboj.2013.204
13. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro Without a Mesenchymal Niche. Nature (2009) 459:262–5. doi: 10.1038/nature07935
14. Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, et al. Sustained In Vitro Intestinal Epithelial Culture Within a Wnt-Dependent Stem Cell Niche. Nat Med (2009) 15:701–6. doi: 10.1038/nm.1951
15. Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, et al. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell (2018) 23:787–93.e786. doi: 10.1016/j.stem.2018.11.016
16. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed Differentiation of Human Pluripotent Stem Cells Into Intestinal Tissue In Vitro. Nature (2011) 470:105–9. doi: 10.1038/nature09691
17. Fan YY, Davidson LA, Chapkin RS. Murine Colonic Organoid Culture System and Downstream Assay Applications. Methods Mol Biol (2019) 1576:171–81. doi: 10.1007/7651_2016_8
18. Muzio L, Consalez GG. Modeling Human Brain Development With Cerebral Organoids. Stem Cell Res Ther (2013) 4(6):154. doi: 10.1186/scrt384
19. Elbadawy M, Usui T, Mori T, Tsunedomi R, Hazama S, Nabeta R, et al. Establishment of a Novel Experimental Model for Muscle-Invasive Bladder Cancer Using a Dog Bladder Cancer Organoid Culture. Cancer Sci (2019) 110:2806–21. doi: 10.1111/cas.14118
20. Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology (2016) 150:1098–112. doi: 10.1053/j.gastro.2015.12.042
21. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-Term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett's Epithelium. Gastroenterology (2011) 141:1762–72. doi: 10.1053/j.gastro.2011.07.050
22. Fujii M, Sato T. Somatic Cell-Derived Organoids as Prototypes of Human Epithelial Tissues and Diseases. Nat Mater (2021) 20:156–69. doi: 10.1038/s41563-020-0754-0
23. Date S, Sato T. Mini-Gut Organoids: Reconstitution of the Stem Cell Niche. Annu Rev Cell Dev Biol (2015) 31:269–89. doi: 10.1146/annurev-cellbio-100814-125218
24. Wang J, Li X, Chen H. Organoid Models in Lung Regeneration and Cancer. Cancer Lett (2020) 475:129–35. doi: 10.1016/j.canlet.2020.01.030
25. Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid Culture Systems for Prostate Epithelial and Cancer Tissue. Nat Protoc (2016) 11:347–58. doi: 10.1038/nprot.2016.006
26. Yousef Yengej FA, Jansen J, Rookmaaker MB, Verhaar MC, Clevers H. Kidney Organoids and Tubuloids. Cells (2020) 9(6):1326. doi: 10.3390/cells9061326
27. Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Bottinger L, Klay D, et al. Long-Term Expanding Human Airway Organoids for Disease Modeling. EMBO J (2019) 38(4):e100300. doi: 10.15252/embj.2018100300
28. Lancaster MA, Knoblich JA. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science (2014) 345:1247125. doi: 10.1126/science.1247125
29. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature (2013) 501:373–9. doi: 10.1038/nature12517
30. Dutta D, Heo I, Clevers H. Disease Modeling in Stem Cell-Derived 3d Organoid Systems. Trends Mol Med (2017) 23:393–410. doi: 10.1016/j.molmed.2017.02.007
31. Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, et al. Divergent Routes Toward Wnt and R-Spondin Niche Independency During Human Gastric Carcinogenesis. Cell (2018) 174:856–69.e817. doi: 10.1016/j.cell.2018.07.027
32. Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. The Hope and the Hype of Organoid Research. Development (2017) 144:938–41. doi: 10.1242/dev.150201
33. Turco MY, Gardner L, Hughes J, Cindrova-Davies T, Gomez MJ, Farrell L, et al. Long-Term, Hormone-Responsive Organoid Cultures of Human Endometrium in a Chemically Defined Medium. Nat Cell Biol (2017) 19:568–77. doi: 10.1038/ncb3516
34. Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence During Disease Progression. Cell Stem Cell (2018) 22:454–67.e456. doi: 10.1016/j.stem.2017.12.009
35. Gabriel E, Albanna W, Pasquini G, Ramani A, Josipovic N, Mariappan A, et al. Human Brain Organoids Assemble Functionally Integrated Bilateral Optic Vesicles. Cell Stem Cell (2021) 28:1740–57.e1748. doi: 10.1016/j.stem.2021.07.010
36. Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in Immunological Research. Nat Rev Immunol (2020) 20:279–93. doi: 10.1038/s41577-019-0248-y
37. Sun W, Luo Z, Lee J, Kim HJ, Lee K, Tebon P, et al. Organ-On-a-Chip for Cancer and Immune Organs Modeling. Adv Healthc Mater (2019) 8:e1801363. doi: 10.1002/adhm.201801363
38. Rossi G, Manfrin A, Lutolf MP. Progress and Potential in Organoid Research. Nat Rev Genet (2018) 19:671–87. doi: 10.1038/s41576-018-0051-9
39. Xu H, Lyu X, Yi M, Zhao W, Song Y, Wu K. Organoid Technology and Applications in Cancer Research. J Hematol Oncol (2018) 11:116. doi: 10.1186/s13045-018-0662-9
40. Bannier-Helaouet M, Post Y, Korving J, Trani Bustos M, Gehart H, Begthel H, et al. Exploring the Human Lacrimal Gland Using Organoids and Single-Cell Sequencing. Cell Stem Cell (2021) 28:1221–32.e1227. doi: 10.1016/j.stem.2021.02.024
41. Florian S, Iwamoto Y, Coughlin M, Weissleder R, Mitchison TJ. A Human Organoid System That Self-Organizes to Recapitulate Growth and Differentiation of a Benign Mammary Tumor. Proc Natl Acad Sci USA (2019) 116:11444–53. doi: 10.1073/pnas.1702372116
42. Semertzidou A, Brosens JJ, McNeish I, Kyrgiou M. Organoid Models in Gynaecological Oncology Research. Cancer Treat Rev (2020) 90:102103. doi: 10.1016/j.ctrv.2020.102103
43. Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering Stem Cell Organoids. Cell Stem Cell (2016) 18:25–38. doi: 10.1016/j.stem.2015.12.005
44. Saengwimol D, Rojanaporn D, Chaitankar V, Chittavanich P, Aroonroch R, Boontawon T, et al. Three-Dimensional Organoid Model Recapitulates Tumorigenic Aspects and Drug Responses of Advanced Human Retinoblastoma. Sci Rep (2018) 8:15664. doi: 10.1038/s41598-018-34037-y
45. Takebe T, Wells JM. Organoids by Design. Science (2019) 364(6444):956–9. doi: 10.1126/science.aaw7567
46. Cruz-Acuna R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, et al. Synthetic Hydrogels for Human Intestinal Organoid Generation and Colonic Wound Repair. Nat Cell Biol (2017) 19:1326–35. doi: 10.1038/ncb3632
47. Grimm D, Egli M, Kruger M, Riwaldt S, Corydon TJ, Kopp S, et al. Tissue Engineering Under Microgravity Conditions-Use of Stem Cells and Specialized Cells. Stem Cells Dev (2018) 27:787–804. doi: 10.1089/scd.2017.0242
48. Rahmani S, Breyner NM, Su HM, Verdu EF, Didar TF. Intestinal Organoids: A New Paradigm for Engineering Intestinal Epithelium In Vitro. Biomaterials (2019) 194:195–214. doi: 10.1016/j.biomaterials.2018.12.006
49. Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, et al. Homeostatic Mini-Intestines Through Scaffold-Guided Organoid Morphogenesis. Nature (2020) 585:574–8. doi: 10.1038/s41586-020-2724-8
50. Sato T, Clevers H. Growing Self-Organizing Mini-Guts From a Single Intestinal Stem Cell: Mechanism and Applications. Science (2013) 340(6137):1190–4. doi: 10.1126/science.1234852
51. Li VSW. Modelling Intestinal Inflammation and Infection Using 'Mini-Gut' Organoids. Nat Rev Gastroenterol Hepatol (2021) 18:89–90. doi: 10.1038/s41575-020-00391-4
52. Mahe MM, Aihara E, Schumacher MA, Zavros Y, Montrose MH, Helmrath MA, et al. Establishment of Gastrointestinal Epithelial Organoids. Curr Protoc Mouse Biol (2013) 3:217–40. doi: 10.1002/9780470942390.mo130179
53. Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, et al. Oncogenic Transformation of Diverse Gastrointestinal Tissues in Primary Organoid Culture. Nat Med (2014) 20:769–77. doi: 10.1038/nm.3585
54. Liu H, Wang Y, Cui K, Guo Y, Zhang X, Qin J. Advances in Hydrogels in Organoids and Organs-On-a-Chip. Adv Mater (2019) 31:e1902042. doi: 10.1002/adma.201902042
55. Mitaka T, Ooe H. Characterization of Hepatic-Organoid Cultures. Drug Metab Rev (2010) 42:472–81. doi: 10.3109/03602530903492020
56. Park SE, Georgescu A, Huh D. Organoids-On-a-Chip. Science (2019) 364(6444):960–5. doi: 10.1126/science.aaw7894
57. Sugihara K, Yamaguchi Y, Usui S, Nashimoto Y, Hanada S, Kiyokawa E, et al. A New Perfusion Culture Method With a Self-Organized Capillary Network. PLoS One (2020) 15:e0240552. doi: 10.1371/journal.pone.0240552
58. Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, et al. Flow-Enhanced Vascularization and Maturation of Kidney Organoids In Vitro. Nat Methods (2019) 16:255–62. doi: 10.1038/s41592-019-0325-y
59. Decembrini S, Hoehnel S, Brandenberg N, Arsenijevic Y, Lutolf MP. Hydrogel-Based Milliwell Arrays for Standardized and Scalable Retinal Organoid Cultures. Sci Rep (2020) 10:10275. doi: 10.1038/s41598-020-67012-7
60. He J, Zhang X, Xia X, Han M, Li F, Li C, et al. Organoid Technology for Tissue Engineering. J Mol Cell Biol (2020) 12:569–79. doi: 10.1093/jmcb/mjaa012
61. Tan Q, Choi KM, Sicard D, Tschumperlin DJ. Human Airway Organoid Engineering as a Step Toward Lung Regeneration and Disease Modeling. Biomaterials (2017) 113:118–32. doi: 10.1016/j.biomaterials.2016.10.046
62. Garreta E, Kamm RD, Chuva de Sousa Lopes SM, Lancaster MA, Weiss R, Trepat X, et al. Rethinking Organoid Technology Through Bioengineering. Nat Mater (2021) 20:145–55. doi: 10.1038/s41563-020-00804-4
63. Tuveson D, Clevers H. Cancer Modeling Meets Human Organoid Technology. Science (2019) 364(6444):952–5. doi: 10.1126/science.aaw6985
64. Ji DB, Wu AW. Organoid in Colorectal Cancer: Progress and Challenges. Chin Med J (Engl) (2020) 133:1971–7. doi: 10.1097/CM9.0000000000000882
65. Aboulkheyr Es H, Montazeri L, Aref AR, Vosough M, Baharvand H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol (2018) 36:358–71. doi: 10.1016/j.tibtech.2017.12.005
66. Bleijs M, van de Wetering M, Clevers H, Drost J. Xenograft and Organoid Model Systems in Cancer Research. EMBO J (2019) 38:e101654. doi: 10.15252/embj.2019101654
67. Rowe RG, Daley GQ. Induced Pluripotent Stem Cells in Disease Modelling and Drug Discovery. Nat Rev Genet (2019) 20:377–88. doi: 10.1038/s41576-019-0100-z
68. Shinozawa T, Kimura M, Cai Y, Saiki N, Yoneyama Y, Ouchi R, et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology (2021) 160:831–46.e810. doi: 10.1053/j.gastro.2020.10.002
69. Maru Y, Hippo Y. Current Status of Patient-Derived Ovarian Cancer Models. Cells (2019) 8(5):505. doi: 10.3390/cells8050505
70. Wang Z, Wang SN, Xu TY, Miao ZW, Su DF, Miao CY. Organoid Technology for Brain and Therapeutics Research. CNS Neurosci Ther (2017) 23:771–8. doi: 10.1111/cns.12754
71. Smith RC, Tabar V. Constructing and Deconstructing Cancers Using Human Pluripotent Stem Cells and Organoids. Cell Stem Cell (2019) 24:12–24. doi: 10.1016/j.stem.2018.11.012
72. Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements During Tumorigenesis. Cell Stem Cell (2016) 18:827–38. doi: 10.1016/j.stem.2016.04.003
73. Xie BY, Wu AW. Organoid Culture of Isolated Cells From Patient-Derived Tissues With Colorectal Cancer. Chin Med J (Engl) (2016) 129:2469–75. doi: 10.4103/0366-6999.191782
74. Baker LA, Tiriac H, Clevers H, Tuveson DA. Modeling Pancreatic Cancer With Organoids. Trends Cancer (2016) 2:176–90. doi: 10.1016/j.trecan.2016.03.004
75. Jain T, Dudeja V. The War Against Pancreatic Cancer in 2020 - Advances on All Fronts. Nat Rev Gastroenterol Hepatol (2021) 18:99–100. doi: 10.1038/s41575-020-00410-4
76. Xu AT, Tong JL, Ran ZH. Organoids Derived From Digestive Tract, Liver, and Pancreas. J Dig Dis (2016) 17:3–10. doi: 10.1111/1751-2980.12305
77. Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grutzmann K, et al. Human Gastric Cancer Modelling Using Organoids. Gut (2019) 68:207–17. doi: 10.1136/gutjnl-2017-314549
78. Kawasaki K, Toshimitsu K, Matano M, Fujita M, Fujii M, Togasaki K, et al. An Organoid Biobank of Neuroendocrine Neoplasms Enables Genotype-Phenotype Mapping. Cell (2020) 183:1420–35.e1421. doi: 10.1016/j.cell.2020.10.023
79. Driehuis E, van Hoeck A, Moore K, Kolders S, Francies HE, Gulersonmez MC, et al. Pancreatic Cancer Organoids Recapitulate Disease and Allow Personalized Drug Screening. Proc Natl Acad Sci USA (2019) 116(52):26580–90. doi: 10.1073/pnas.1911273116
80. Hu X, Zhang L, Li Y, Ma X, Dai W, Gao X, et al. Organoid Modelling Identifies That DACH1 Functions as a Tumour Promoter in Colorectal Cancer by Modulating BMP Signalling. EBioMedicine (2020) 56:102800. doi: 10.1016/j.ebiom.2020.102800
81. Huang B, Trujillo MA, Fujikura K, Qiu M, Chen F, Felsenstein M, et al. Molecular Characterization of Organoids Derived From Pancreatic Intraductal Papillary Mucinous Neoplasms. J Pathol (2020) 252:252–62. doi: 10.1002/path.5515
82. Jin RU, Mills JC. Tumor Organoids to Study Gastroesophageal Cancer: A Primer. J Mol Cell Biol (2020) 12:593–606. doi: 10.1093/jmcb/mjaa035
83. Joshi R, Castro De Moura M, Pineyro D, Alvarez-Errico D, Arribas C, Esteller M. The DNA Methylation Landscape of Human Cancer Organoids Available at the American Type Culture Collection. Epigenetics (2020) 15:1167–77. doi: 10.1080/15592294.2020.1762398
84. Kretzschmar K. Cancer Research Using Organoid Technology. J Mol Med (Berl) (2021) 99:501–15. doi: 10.1007/s00109-020-01990-z
85. Fan H, Demirci U, Chen P. Emerging Organoid Models: Leaping Forward in Cancer Research. J Hematol Oncol (2019) 12:142. doi: 10.1186/s13045-019-0832-4
86. Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, et al. Fast and Efficient Generation of Knock-in Human Organoids Using Homology-Independent CRISPR-Cas9 Precision Genome Editing. Nat Cell Biol (2020) 22:321–31. doi: 10.1038/s41556-020-0472-5
87. Hendriks D, Artegiani B, Hu H, Chuva de Sousa Lopes S, Clevers H. Establishment of Human Fetal Hepatocyte Organoids and CRISPR-Cas9-Based Gene Knockin and Knockout in Organoid Cultures From Human Liver. Nat Protoc (2021) 16:182–217. doi: 10.1038/s41596-020-00411-2
88. Paes Dias M, Tripathi V, van der Heijden I, Cong K, Manolika EM, Bhin J, et al. Loss of Nuclear DNA Ligase III Reverts PARP Inhibitor Resistance in BRCA1/53BP1 Double-Deficient Cells by Exposing ssDNA Gaps. Mol Cell (2021) 81(22):4692–708.e9. doi: 10.1016/j.molcel.2021.09.005
89. Nguyen AT, Lee SY, Chin HJ, Le QV, Lee D. Kinase Activity of ERBB3 Contributes to Intestinal Organoids Growth and Intestinal Tumorigenesis. Cancer Sci (2020) 111:137–47. doi: 10.1111/cas.14235
90. Lannagan TRM, Lee YK, Wang T, Roper J, Bettington ML, Fennell L, et al. Genetic Editing of Colonic Organoids Provides a Molecularly Distinct and Orthotopic Preclinical Model of Serrated Carcinogenesis. Gut (2019) 68:684–92. doi: 10.1136/gutjnl-2017-315920
91. Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, Tapia P, et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell (2019) 24:927–43.e926. doi: 10.1016/j.stem.2019.04.017
92. Ballabio C, Anderle M, Gianesello M, Lago C, Miele E, Cardano M, et al. Modeling Medulloblastoma In Vivo and With Human Cerebellar Organoids. Nat Commun (2020) 11:583. doi: 10.1038/s41467-019-13989-3
93. Li H, Dai W, Xia X, Wang R, Zhao J, Han L, et al. Modeling Tumor Development and Metastasis Using Paired Organoids Derived From Patients With Colorectal Cancer Liver Metastases. J Hematol Oncol (2020) 13:119. doi: 10.1186/s13045-020-00957-4
94. Takeda H, Kataoka S, Nakayama M, Ali MAE, Oshima H, Yamamoto D, et al. CRISPR-Cas9-Mediated Gene Knockout in Intestinal Tumor Organoids Provides Functional Validation for Colorectal Cancer Driver Genes. Proc Natl Acad Sci USA (2019) 116:15635–44. doi: 10.1073/pnas.1904714116
95. Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov (2022) 12:31–46. doi: 10.1158/2159-8290.CD-21-1059
96. Stener-Victorin E, Deng Q. Epigenetic Inheritance of Polycystic Ovary Syndrome - Challenges and Opportunities for Treatment. Nat Rev Endocrinol (2021) 17:521–33. doi: 10.1038/s41574-021-00517-x
97. Jeffries MA. The Development of Epigenetics in the Study of Disease Pathogenesis. Adv Exp Med Biol (2020) 1253:57–94. doi: 10.1007/978-981-15-3449-2_2
98. Ushijima T, Clark SJ, Tan P. Mapping Genomic and Epigenomic Evolution in Cancer Ecosystems. Science (2021) 373(6562):1474–9. doi: 10.1126/science.abh1645
99. Carter B, Zhao K. The Epigenetic Basis of Cellular Heterogeneity. Nat Rev Genet (2021) 22:235–50. doi: 10.1038/s41576-020-00300-0
100. Aloia L, McKie MA, Vernaz G, Cordero-Espinoza L, Aleksieva N, van den Ameele J, et al. Epigenetic Remodelling Licences Adult Cholangiocytes for Organoid Formation and Liver Regeneration. Nat Cell Biol (2019) 21:1321–33. doi: 10.1038/s41556-019-0402-6
101. Koliaraki V, Prados A, Armaka M, Kollias G. The Mesenchymal Context in Inflammation, Immunity and Cancer. Nat Immunol (2020) 21:974–82. doi: 10.1038/s41590-020-0741-2
102. Lin B, Du L, Li H, Zhu X, Cui L, Li X. Tumor-Infiltrating Lymphocytes: Warriors Fight Against Tumors Powerfully. BioMed Pharmacother (2020) 132:110873. doi: 10.1016/j.biopha.2020.110873
103. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell (2018) 175:1972–88.e1916. doi: 10.1016/j.cell.2018.11.021
104. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of Tumor-Reactive T Cells by Co-Culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell (2018) 174:1586–98.e1512. doi: 10.1016/j.cell.2018.07.009
105. Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In Search of Definitions: Cancer-Associated Fibroblasts and Their Markers. Int J Cancer (2020) 146:895–905. doi: 10.1002/ijc.32193
106. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat Rev Cancer (2020) 20:174–86. doi: 10.1038/s41568-019-0238-1
107. Tsai S, McOlash L, Palen K, Johnson B, Duris C, Yang Q, et al. Development of Primary Human Pancreatic Cancer Organoids, Matched Stromal and Immune Cells and 3D Tumor Microenvironment Models. BMC Cancer (2018) 18:335. doi: 10.1186/s12885-018-4238-4
108. Chakrabarti J, Koh V, So JBY, Yong WP, Zavros YA. Preclinical Human-Derived Autologous Gastric Cancer Organoid/Immune Cell Co-Culture Model to Predict the Efficacy of Targeted Therapies. J Vis Exp (2021) 173:e61443. doi: 10.3791/61443
109. Naruse M, Ochiai M, Sekine S, Taniguchi H, Yoshida T, Ichikawa H, et al. Re-Expression of REG Family and DUOXs Genes in CRC Organoids by Co-Culturing With CAFs. Sci Rep (2021) 11:2077. doi: 10.1038/s41598-021-81475-2
110. Xu R, Zhou X, Wang S, Trinkle C. Tumor Organoid Models in Precision Medicine and Investigating Cancer-Stromal Interactions. Pharmacol Ther (2021) 218:107668. doi: 10.1016/j.pharmthera.2020.107668
111. Liu J, Li P, Wang L, Li M, Ge Z, Noordam L, et al. Cancer-Associated Fibroblasts Provide a Stromal Niche for Liver Cancer Organoids That Confers Trophic Effects and Therapy Resistance. Cell Mol Gastroenterol Hepatol (2021) 11:407–31. doi: 10.1016/j.jcmgh.2020.09.003
112. Palikuqi B, Nguyen DT, Li G, Schreiner R, Pellegata AF, Liu Y, et al. Adaptable Haemodynamic Endothelial Cells for Organogenesis and Tumorigenesis. Nature (2020) 585:426–32. doi: 10.1038/s41586-020-2712-z
113. Skardal A, Murphy SV, Devarasetty M, Mead I, Kang HW, Seol YJ, et al. Multi-Tissue Interactions in an Integrated Three-Tissue Organ-on-a-Chip Platform. Sci Rep (2017) 7:8837. doi: 10.1038/s41598-017-08879-x
114. Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, et al. Engineering of Human Brain Organoids With a Functional Vascular-Like System. Nat Methods (2019) 16:1169–75. doi: 10.1038/s41592-019-0586-5
115. Weil AR. Precision Medicine. Health Aff (Millwood) (2018) 37:687. doi: 10.1377/hlthaff.2018.0520
116. Dagogo-Jack I, Shaw AT. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat Rev Clin Oncol (2018) 15:81–94. doi: 10.1038/nrclinonc.2017.166
117. Li Y, Tang P, Cai S, Peng J, Hua G. Organoid Based Personalized Medicine: From Bench to Bedside. Cell Regener (2020) 9:21. doi: 10.1186/s13619-020-00059-z
118. The Promise of Organoids and Embryoids. Nat Mater (2021) 20:121. doi: 10.1038/s41563-021-00926-3
119. Abugomaa A, Elbadawy M, Yamawaki H, Usui T, Sasaki K. Emerging Roles of Cancer Stem Cells in Bladder Cancer Progression, Tumorigenesis, and Resistance to Chemotherapy: A Potential Therapeutic Target for Bladder Cancer. Cells (2020) 9(1):235. doi: 10.3390/cells9010235
120. Fatehullah A, Tan SH, Barker N. Organoids as an In Vitro Model of Human Development and Disease. Nat Cell Biol (2016) 18:246–54. doi: 10.1038/ncb3312
121. Andreatta F, Beccaceci G, Fortuna N, Celotti M, De Felice D, Lorenzoni M, et al. The Organoid Era Permits the Development of New Applications to Study Glioblastoma. Cancers (Basel) (2020) 12(11):3303. doi: 10.3390/cancers12113303
122. Narasimhan V, Wright JA, Churchill M, Wang T, Rosati R, Lannagan TRM, et al. Medium-Throughput Drug Screening of Patient-Derived Organoids From Colorectal Peritoneal Metastases to Direct Personalized Therapy. Clin Cancer Res (2020) 26:3662–70. doi: 10.1158/1078-0432.CCR-20-0073
123. Krall N, Superti-Furga G, Vladimer GI. Patient-Derived Model Systems and the Development of Next-Generation Anticancer Therapeutics. Curr Opin Chem Biol (2020) 56:72–8. doi: 10.1016/j.cbpa.2020.01.002
124. Aberle MR, Burkhart RA, Tiriac H, Olde Damink SWM, Dejong CHC, Tuveson DA, et al. Patient-Derived Organoid Models Help Define Personalized Management of Gastrointestinal Cancer. Br J Surg (2018) 105:e48–60. doi: 10.1002/bjs.10726
125. Morganti S, Tarantino P, Ferraro E, D'Amico P, Viale G, Trapani D, et al. Complexity of Genome Sequencing and Reporting: Next Generation Sequencing (NGS) Technologies and Implementation of Precision Medicine in Real Life. Crit Rev Oncol Hematol (2019) 133:171–82. doi: 10.1016/j.critrevonc.2018.11.008
126. Yang F, Anekpuritanang T, Press RD. Clinical Utility of Next-Generation Sequencing in Acute Myeloid Leukemia. Mol Diagn Ther (2020) 24:1–13. doi: 10.1007/s40291-019-00443-9
127. Doostparast Torshizi A, Wang K. Next-Generation Sequencing in Drug Development: Target Identification and Genetically Stratified Clinical Trials. Drug Discovery Today (2018) 23:1776–83. doi: 10.1016/j.drudis.2018.05.015
128. Tripathi P, Singh J, Lal JA, Tripathi V. Next-Generation Sequencing: An Emerging Tool for Drug Designing. Curr Pharm Des (2019) 25:3350–7. doi: 10.2174/1381612825666190911155508
129. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science (2018) 359:920–6. doi: 10.1126/science.aao2774
130. Skora AD, Douglass J, Hwang MS, Tam AJ, Blosser RL, Gabelli SB, et al. Generation of MANAbodies Specific to HLA-Restricted Epitopes Encoded by Somatically Mutated Genes. Proc Natl Acad Sci USA (2015) 112:9967–72. doi: 10.1073/pnas.1511996112
131. Danilova L, Anagnostou V, Caushi JX, Sidhom JW, Guo H, Chan HY, et al. The Mutation-Associated Neoantigen Functional Expansion of Specific T Cells (MANAFEST) Assay: A Sensitive Platform for Monitoring Antitumor Immunity. Cancer Immunol Res (2018) 6:888–99. doi: 10.1158/2326-6066.CIR-18-0129
132. Alcazer V, Bonaventura P, Tonon L, Wittmann S, Caux C, Depil S. Neoepitopes-Based Vaccines: Challenges and Perspectives. Eur J Cancer (2019) 108:55–60. doi: 10.1016/j.ejca.2018.12.011
133. Zhang W, Yin Q, Huang H, Lu J, Qin H, Chen S, et al. Personal Neoantigens From Patients With NSCLC Induce Efficient Antitumor Responses. Front Oncol (2021) 11:628456:628456. doi: 10.3389/fonc.2021.628456
134. Caushi JX, Zhang J, Ji Z, Vaghasia A, Zhang B, Hsiue EH, et al. Transcriptional Programs of Neoantigen-Specific TIL in Anti-PD-1-Treated Lung Cancers. Nature (2021) 596:126–32. doi: 10.1038/s41586-021-03752-4
135. Arnaud M, Chiffelle J, Genolet R, Navarro Rodrigo B, Perez MAS, Huber F, et al. Sensitive Identification of Neoantigens and Cognate TCRs in Human Solid Tumors. Nat Biotechnol (2021). doi: 10.1038/s41587-021-01072-6
136. Peng M, Mo Y, Wang Y, Wu P, Zhang Y, Xiong F, et al. Neoantigen Vaccine: An Emerging Tumor Immunotherapy. Mol Cancer (2019) 18(1):128. doi: 10.1186/s12943-019-1055-6
137. Gopanenko AV, Kosobokova EN, Kosorukov VS. Main Strategies for the Identification of Neoantigens. Cancers (Basel) (2020) 12(10):2879. doi: 10.3390/cancers12102879
138. Roudko V, Greenbaum B, Bhardwaj N. Computational Prediction and Validation of Tumor-Associated Neoantigens. Front Immunol (2020) 11:27:27. doi: 10.3389/fimmu.2020.00027
139. Richters MM, Xia H, Campbell KM, Gillanders WE, Griffith OL, Griffith M. Best Practices for Bioinformatic Characterization of Neoantigens for Clinical Utility. Genome Med (2019) 11:56. doi: 10.1186/s13073-019-0666-2
140. Demmers LC, Kretzschmar K, Van Hoeck A, Bar-Epraïm YE, van den Toorn HWP, Koomen M, et al. Single-Cell Derived Tumor Organoids Display Diversity in HLA Class I Peptide Presentation. Nat Commun (2020) 11(1):5338. doi: 10.1038/s41467-020-19142-9
141. Roth C, Cantaert T, Colas C, Prot M, Casademont I, Levillayer L, et al. A Modified mRNA Vaccine Targeting Immunodominant NS Epitopes Protects Against Dengue Virus Infection in HLA Class I Transgenic Mice. Front Immunol (2019) 10:1424:1424. doi: 10.3389/fimmu.2019.01424
142. Wang Y, Zhang L, Xu Z, Miao L, Huang L. mRNA Vaccine With Antigen-Specific Checkpoint Blockade Induces an Enhanced Immune Response Against Established Melanoma. Mol Ther (2018) 26(2):420–34. doi: 10.1016/j.ymthe.2017.11.009
143. Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, et al. Neoantigen Vaccine Generates Intratumoral T Cell Responses in Phase Ib Glioblastoma Trial. Nature (2019) 565:234–9. doi: 10.1038/s41586-018-0792-9
144. Hu Z, Leet DE, Allesøe RL, Oliveira G, Li S, Luoma AM, et al. Personal Neoantigen Vaccines Induce Persistent Memory T Cell Responses and Epitope Spreading in Patients With Melanoma. Nat Med (2021) 27(3):515–25. doi: 10.1038/s41591-020-01206-4
145. Li L, Goedegebuure SP, Gillanders WE. Preclinical and Clinical Development of Neoantigen Vaccines. Ann Oncol (2017) 28(suppl_12):xii11–7. doi: 10.1093/annonc/mdx681
146. Blass E, Ott PA. Advances in the Development of Personalized Neoantigen-Based Therapeutic Cancer Vaccines. Nat Rev Clin Oncol (2021) 18(4):215–29. doi: 10.1038/s41571-020-00460-2
147. Stanton SE, Gad E, Corulli LR, Lu H, Disis ML. Tumor-Associated Antigens Identified Early in Mouse Mammary Tumor Development Can Be Effective Vaccine Targets. Vaccine (2019) 37:3552–61. doi: 10.1016/j.vaccine.2019.05.024
148. Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, et al. Exploiting the Mutanome for Tumor Vaccination. Cancer Res (2012) 72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722
149. Song Q, Zhang CD, Wu XH. Therapeutic Cancer Vaccines: From Initial Findings to Prospects. Immunol Lett (2018) 196:11–21. doi: 10.1016/j.imlet.2018.01.011
150. Marrazzo P, Cricca M, Nastasi C. Are the Organoid Models an Invaluable Contribution to ZIKA Virus Research? Pathogens (2021) 10(10):1233. doi: 10.3390/pathogens10101233
151. Liu X, Mondal AM. Conditional Cell Reprogramming for Modeling Host-Virus Interactions and Human Viral Diseases. J Med Virol (2020) 92:2440–52. doi: 10.1002/jmv.26093
152. Ahammad I, Lira SS. Designing a Novel mRNA Vaccine Against SARS-CoV-2: An Immunoinformatics Approach. Int J Biol Macromol (2020) 162:820–37. doi: 10.1016/j.ijbiomac.2020.06.213
153. Wagar LE, Salahudeen A, Constantz CM, Wendel BS, Lyons MM, Mallajosyula V, et al. Modeling Human Adaptive Immune Responses With Tonsil Organoids. Nat Med (2021) 27:125–35. doi: 10.1038/s41591-020-01145-0
154. Pulendran B, Davis MM. The Science and Medicine of Human Immunology. Science (2020) 369(6511):eaay4014. doi: 10.1126/science.aay4014
155. Wisnewski AV, Redlich CA, Liu J, Kamath K, Abad QA, Smith RF, et al. Immunogenic Amino Acid Motifs and Linear Epitopes of COVID-19 mRNA Vaccines. PLoS One (2021) 16:e0252849. doi: 10.1371/journal.pone.0252849
156. Lu L, Jiang J, Zhan M, Zhang H, Wang QT, Sun SN, et al. Targeting Tumor-Associated Antigens in Hepatocellular Carcinoma for Immunotherapy: Past Pitfalls and Future Strategies. Hepatology (2021) 73:821–32. doi: 10.1002/hep.31502
157. Walczak K, Marciniak S, Rajtar G. Cancer Chemoprevention - Selected Molecular Mechanisms. Postepy Hig Med Dosw (Online) (2017) 71(0):149–61. doi: 10.5604/01.3001.0010.3799
158. Scuto M, Ontario ML, Salinaro AT, Caligiuri I, Rampulla F, Zimbone V, et al. Redox Modulation by Plant Polyphenols Targeting Vitagenes for Chemoprevention and Therapy: Relevance to Novel Anti-Cancer Interventions and Mini-Brain Organoid Technology. Free Radic Biol Med (2022) 179:59–75. doi: 10.1016/j.freeradbiomed.2021.12.267
159. Scuto M, Trovato Salinaro A, Caligiuri I, Ontario ML, Greco V, Sciuto N, et al. Redox Modulation of Vitagenes via Plant Polyphenols and Vitamin D: Novel Insights for Chemoprevention and Therapeutic Interventions Based on Organoid Technology. Mech Ageing Dev (2021) 199:111551. doi: 10.1016/j.mad.2021.111551
160. Umezawa S, Higurashi T, Komiya Y, Arimoto J, Horita N, Kaneko T, et al. Chemoprevention of Colorectal Cancer: Past, Present, and Future. Cancer Sci (2019) 110:3018–26. doi: 10.1111/cas.14149
161. Gairola K, Gururani S, Bahuguna A, Garia V, Pujari R, Dubey SK. Natural Products Targeting Cancer Stem Cells: Implications for Cancer Chemoprevention and Therapeutics. J Food Biochem (2021) 45:e13772. doi: 10.1111/jfbc.13772
162. Rickner H, Jiang L, Hong R, Wolozin B, Cheng C. Single Cell Transcriptomic Profiling of Neurodegeneration Mediated by Tau in a Novel 3D Neuron-Astrocyte Coculture Model. Alzheimers Dement (2021) 17 Suppl 2:e058551. doi: 10.1002/alz.058551
163. Flannagan K, Stopperan JA, Troutwine BM, Lysaker CR, Strope T, Draper J, et al. Mitochondrial Phenotypes in iPSC AD Models. Alzheimers Dement (2021) 17(Suppl 2):e058489. doi: 10.1002/alz.058489
164. Wang S, Wang D, Duan Y, Zhou Z, Gao W, Zhang L. Cellular Nanosponges for Biological Neutralization. Adv Mater (2021) e2107719. doi: 10.1002/adma.202107719
165. Yang K, Yang Z, Yu G, Nie Z, Wang R, Chen X. Polyprodrug Nanomedicines: An Emerging Paradigm for Cancer Therapy. Adv Mater (2021) 34(6):e2107434. doi: 10.1002/adma.202107434
166. Giobbe GG, Crowley C, Luni C, Campinoti S, Khedr M, Kretzschmar K, et al. Extracellular Matrix Hydrogel Derived From Decellularized Tissues Enables Endodermal Organoid Culture. Nat Commun (2019) 10:5658. doi: 10.1038/s41467-019-13605-4
167. Gjorevski N, Lutolf MP. Synthesis and Characterization of Well-Defined Hydrogel Matrices and Their Application to Intestinal Stem Cell and Organoid Culture. Nat Protoc (2017) 12:2263–74. doi: 10.1038/nprot.2017.095
168. O'Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, et al. Transplantation of Engineered Organoids Enables Rapid Generation of Metastatic Mouse Models of Colorectal Cancer. Nat Biotechnol (2017) 35:577–82. doi: 10.1038/nbt.3837
Keywords: organoids, tumor-initiating cell, tumorigenesis, precision medicine, tumor vaccine
Citation: Wang Z, Zhao S, Lin X, Chen G, Kang J, Ma Z, Wang Y, Li Z, Xiao X, He A and Xiang D (2022) Application of Organoids in Carcinogenesis Modeling and Tumor Vaccination. Front. Oncol. 12:855996. doi: 10.3389/fonc.2022.855996
Received: 16 January 2022; Accepted: 17 February 2022;
Published: 17 March 2022.
Edited by:
Claudio Tabolacci, National Institute of Health (ISS), ItalyReviewed by:
Barbara Breznik, National Institute of Biology (NIB), SloveniaCopyright © 2022 Wang, Zhao, Lin, Chen, Kang, Ma, Wang, Li, Xiao, He and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Li, bGl6aGlwdzMxNkAxNjMuY29t; Xiuying Xiao, eGlhb3hpdXlpbmcyMDAyQDE2My5jb20=; Aina He, YW5uYV8xMTg4QDEyNi5jb20=; Dongxi Xiang, ZHhpYW5nQHNoc211LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.