
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 20 June 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.855807
The human programmed cell death 4 (PDCD4) gene was mapped at chromosome 10q24 and encodes the PDCD4 protein comprised of 469 amino acids. PDCD4 inhibits protein translation PDCD4 inhibits protein translation to suppress tumor progression, and its expression is frequently decreased in breast cancer. PDCD4 blocks translation initiation complex by binding eIF4A via MA-3 domains or by directly binding 5’ mRNA internal ribosome entry sites with an RNA binding domain to suppress breast cancer progression and proliferation. Numerous regulators and biological processes including non-coding RNAs, proteasomes, estrogen, natural compounds and inflammation control PDCD4 expression in breast cancer. Loss of PDCD4 expression is also responsible for drug resistance in breast cancer. HER2 activation downregulates PDCD4 expression by activating MAPK, AKT, and miR-21 in aromatase inhibitor-resistant breast cancer cells. Moreover, modulating the microRNA/PDCD4 axis maybe an effective strategy for overcoming chemoresistance in breast cancer. Down-regulation of PDCD4 is significantly associated with short overall survival of patients, which suggests that PDCD4 may be an independent prognostic marker for breast cancer.
Programmed cell death 4 (PDCD4) was first isolated as an antigen in proliferating cells that is bound by the monoclonal antibody Pr-28. Subsequently, the PDCD4 gene was identified in different species, including humans (1, 2), rats (3), and chicken (4). The human PDCD4 protein is comprised of 469 amino acids, and the protein sequence is a highly conserved from Drosophila to humans. The mouse and human PDCD4 proteins share approximately 92% identity in amino acid sequence. The human PDCD4 gene was mapped at chromosome 10q24 (5). PDCD4 has been demonstrated to be a protein translation inhibitor and a tumor suppressor, and consistently its expression frequently decreased in numerous types of malignancies including breast, colon, liver, lung, pancreatic, and skin cancers (6, 7). Loss of PDCD4 expression is associated with poor prognosis of patients with breast cancer (8, 9). In our review, we searched literatures in pubmed with the keywords PDCD4 and breast cancer in all fields. According to the contents of the target references, the article will focus on the function, regulation, and prognostic value of PDCD4 in breast cancer as well as its role in drug resistance.
Using yeast-two hybrid, PDCD4 was identified as a binding partner of translation initiation factor 4A (eIF4A) (10). The PDCD4-eIF4A interaction was further confirmed by mutation and crystallography (11–13). Crystallography of PDCD4-eIF4A showed that one molecule of PDCD4 interacts with two molecules of eIF4A through two MA-3 domains in PDCD4. The Glu210, Glu249, and Asp253 residues in the N-terminal segment of the MA-3 domain and the Glu373, Arg414, and Asp418 residues in the C-terminal segment form the ionic interaction with Arg110 and Arg161 of eIF4A (11, 12). Mutation of Glu249 to Lys, Asp253 to Ala, Asp414 to Lys and/or Asp418 to Ala in PDCD4 abolish its ability to bind eIF4A and result in loss of the ability to inhibit protein translation (13). This indicates that binding with eIF4A is required for the inhibition of translation by PDCD4. How does PDCD4-eIF4A binding inhibits protein translation? One possible mechanism is that PDCD4 inhibits protein translation through suppression of eIF4A’s helicase activity (10). eIF4A is an ATP-dependent RNA helicase. The function of eIF4A in protein translation initiation is to unwind the structured mRNA, allowing the translation initiation complex scan the mRNA from 5’ to 3’ end to locate the translation initiation codon (14). Therefore, inhibition of eIF4A activity by PDCD4 is expected to suppress protein translation, especially translation of mRNAs with complexly structured 5’untranslated regions (5’UTRs). PDCD4 was initially demonstrated to preferentially suppress translation of a luciferase mRNA with a stem loop-structured 5’UTR with free energy of -44.8 kcal/mol (13). This scenario was further confirmed by identification of the stress-activated-protein kinase interacting protein 1 (Sin1) and ribosomal S6 kinase 1 (S6K1) as PDCD4 targets, whose mRNAs have 5’UTRs that form secondary structures with free energies of -145 and -82.30 kcal/mol, respectively (15, 16). In agreement with inhibition of eIF4A by PDCD4, cells treated with an eIF4A inhibitor, silvestrol, had reduced the Sin1 protein abundance (16). It seems that the mRNAs that have 5’UTRs with a free energy greater than -44.8 kcal/mol are susceptible to translation inhibition by PDCD4. However, this notion requires identification and analysis of additional PDCD4 targets. In addition to inhibiting eIF4A activity, PDCD4 may inhibit translation of anti-apoptotic proteins such as Bcl-xl and XIAP by directly binding with their mRNAs at internal ribosome entry sites to block the translation initiation complex formation (17, 18). The mechanisms of translation inhibiton by PDCD4 are summarized in Figure 1.
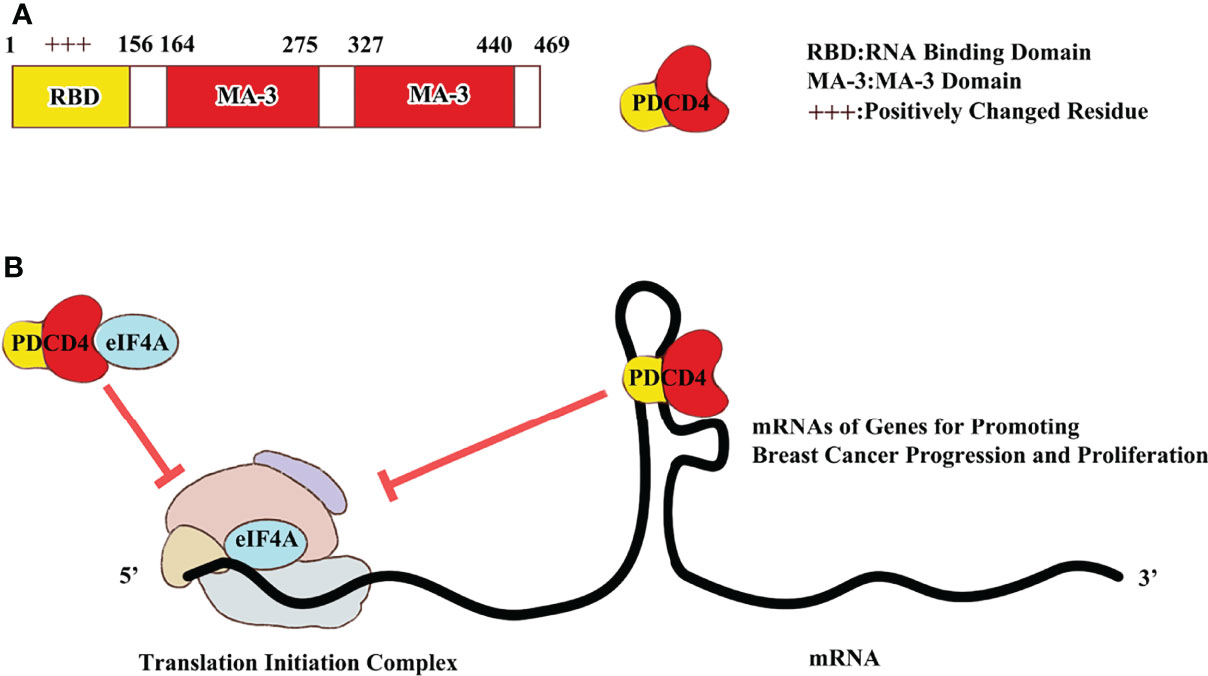
Figure 1 PDCD4 protein structure and mechanism to regulate translation initiation. (A) PDCD4 protein is 469 amino acid length with one RNA binding domain and two MA-3 domains. (B) PDCD4 protein could block translation initiation by either binding IRES in 5’ UTR or eIF4A in 48S translation initiation complex.
PDCD4 was initially suggested to be a tumor suppressor in transformation-resistant JB6 cells because the down-regulation of PDCD4 expression by antisense RNA resulted in a gain of transformation-susceptible phenotype (19). Conversely, overexpression of PDCD4 cDNA in the transformation-susceptible JB6 cells endowed resistance to tumor promoter-induced transformation (20). Mouse epidermal JB6 cells consist of three variants including transformation-susceptible, transformation-resistant, and transformed cells. Transformation-susceptible JB6 cells are sensitive to tumor promoter-induced transformation such as that induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment. By contrast, transformation-resistant JB6 cells are resistant to TPA-induced transformation. Later, several additional studies demonstrated that PDCD4 not only inhibits tumor promoter-induced transformation but also suppresses cancer cell proliferation, migration, invasion, and metastasis (for detail please see reviews (21, 22)). In breast cancer, PDCD4 has been shown to be a pro-apoptotic factor and inhibits cell growth (23, 24). Overexpression of PDCD4 reduces the rate of proliferation and promotes apoptosis in T-47D, MDA-MB-231, and MCF-7 breast cancer cells (25, 26). Conversely, down-regulation or repression of PDCD4 decreases apoptosis (27, 28). In addition to suppressing proliferation and cell growth, several studies also demonstrated that PDCD4 attenuates tumor invasion and metastasis. Immunohistochemical studies showed that PDCD4 expression was only slightly decreased in in situ ductal carcinoma samples but was markedly decreased in invasive ductal carcinoma samples (29). In addition, reverse phase protein arrays, a new technique to study the functional proteome in non-microdissected breast tumors (30), showed that loss of PDCD4 expression was significantly associated with lymph node metastasis in HER2- and ER-positive invasive breast carcinomas (31). These findings suggest that loss of PDCD4 expression promotes breast cancer invasion. This notion is further supported by in vitro studies showing that knockdown of PDCD4 boosts invasion of T47D cells (32). Recently, PDCD4 overexpression was demonstrated to suppress cell cycle progression from the G1 phase to the G2 phase in triple negative breast cancer (TNBC) cell line (33). Although it is clear that PDCD4 suppresses tumorigenesis in breast cancer, the underlying mechanism is still not fully understood. For example, PDCD4 inhibits invasion in breast cancer cells by reduction of inhibitors of metalloproteinases-2 (TIMP-2) expression (34, 35). TIMP-2 inhibits matrix metalloproteinase (MMP) activity to prevent degradation of the extracellular matrix and thereby impedes tumor cell migration and invasion (36). However, it is unknown of how PDCD4 inhibits TIMP-2 expression.
Currently, pieces of evidence suggest that PDCD4 may inhibit tumorigenesis through two possible mechanisms. As mentioned above, PDCD4 binds with eIF4A and inhibits its helicase activity. During translation initiation, eIF4A is required for the unwinding or mRNAs that have 5’UTRs with secondary structures. Usually, mRNAs with 5’UTRs that have secondary structures encode for growth factors, growth promotion genes, and proto-oncogenes (37). Thus, inhibition of eIF4A by PDCD4 is expected to cause suppression of cell proliferation, migration, and/or invasion. For example, PDCD4 inhibits Sin1 translation to attenuate cell invasion in colorectal carcinoma (16). Sin1 is a critical component of mammalian target of rapamycin complex 2 (mTORC2) that regulates Akt activation. Inhibition of Sin1 translation by PDCD4 results in reduction of Snail expression to suppress invasion via inactivation of Akt. In addition to the eIF4A-dependent mechanism, PDCD4 may repress tumorigenesis through an eIF4A-independent mechanism. Liwak et al. reported that PDCD4 directly binds with the mRNA of X-linked inhibitor of apoptosis (XIAP) at an internal ribosome binding site and inhibits XIAP translation by preventing the translation initiation complex formation (18). In addition, PDCD4 was reported to bind to rapamycin-insensitive companion of mTOR (Rictor), another key component of mTORC2, to repress mTORC2 activity (38), and this may regulate cell survival, migration, and invasion by inhibiting Akt phosphorylation. However, the role of PDCD4-Rictor binding in the suppression of tumorigenesis and the underlying mechanism needs further investigation.
Non-coding RNAs (NcRNAs) include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (CircRNAs). All of these have been reported to regulate the expression of PDCD4 and thereby control tumorigenesis in breast cancer.
miRNAs are small non-coding RNA molecules found in plants, animals and some viruses. They play critical roles in RNA silencing and post-transcriptional regulation of target genes. Among miRNAs, microRNA-21 (miR-21) was first identified to regulate PDCD4 expression (39). miR-21 is classified as an “oncomiR” and is commonly overexpressed in many types of solid cancers including breast cancer (40–42). Overexpression of miR-21 is implicated in various processes of tumorigenesis through the suppression of PDCD4 expression. These include the suppression of apoptosis (43, 44), promotion of cell proliferation and tumor growth (45, 46), and stimulation of invasion and metastasis (47, 48). Moreover, miR-21 significantly augmented PD-L1 expression and immune escape via PI3K/Akt pathway activation by targeting PDCD4 in breast cancer cells. According to this traits, Anti-PD-L1 antibody could enhance T cell immune responses and reduces resistance of breast cancer cells to radiotherapy in nude mice (49). Lu et al. found that the 3’-UTR of the PDCD4 mRNA contains a functional binding site for miR-21 (50). The PDCD4 mRNA level in MCF-7 cells did not change in the miR-21-transfected cells while the PDCD4 protein level decreased by approximately 50%, indicating that the reduction of PDCD4 protein is a result of post-transcriptional repression (50). Conversely. inhibition or down-regulation of miR-21 expression results in augmented PDCD4 expression (51). In addition to miR-21, miR-17-5p (52), miR-206 (53), miR-183-5p (54), miR-421 (28) and miR-424 (55) were reported to inhibit PDCD4 expression to influence the malignant behaviors of breast cancer cells (56).
lncRNAs are longer than 200 bp and comprise the most complex group of NcRNAs. lncRNAs contain miRNA binding sites and function as molecular sponges to effectively inhibit the expression of downstream genes targeted by miRNAs (57). A recent study showed that the expression of PDCD4-targeting lncRNA, PDCD4-AS1, is down-regulated during breast cancer progression, and its expression is positively correlated with PDCD4 level in breast cancer tissue (58, 59). Depletion of PDCD4-AS1 results in promotion of cell proliferation and migration in breast cancer cells. It is believed that PDCD4-AS1 stabilizes PDCD4 mRNA by forming an RNA duplex to attenuate the interaction between PDCD4 mRNA and RNA decay promoting factor HuR (60). Moreover, LncRNA GAS5 interacts with miR-21 via miRNA binding elements in breast cancer. Mediated by this sponge mechanism, GAS5 is involved in the upregulation of a number of mRNAs that encode tumor suppressor proteins such as PDCD4 (61). A similar mechanism that lncRNA SLC16A1-AS1 may serve as an internal sponge of miR-182 to augment PDCD4 has been reported recently. SLC16A1-AS1 is downregulated in TNBC and overexpression of SLC16A1-AS1 could suppress TNBC cell proliferation (33).
The circRNAs were initially identified from RNA viruses in 1976 and are connected at the 3’ and 5’ ends by exon or intron cyclization to form complete ring structures. CircRNAs have abundant binding sites for non-coding RNAs and thus act via absorption of non-coding RNAs like a sponge. CircRNAs affect the stability or the translation of target RNAs by competitively binding with miRNAs (62). For example, Hsa_circ_0053063 is a circRNA generated from several exons of hydroxyacyl CoA dehydrogenase trifunctional multienzyme complex subunit alpha (HADHA), and it inhibits cell viability, proliferation, and progression of breast cancer through stabilization of PDCD4 by targeting miR-330-3p (63). Similarly, Circ-NOL10 could suppress breast cancer carcinogenesis by upregulating PDCD4 via a circ-NOL10/miR-149-5p/miR-330-3p/miR-452-5p/PDCD4 pathway (64).
Exosomes are extracellular bilayer vesicles with diameters ranging from 30 to 100 nm that transfer a wealth of nucleic acids and proteins among cells under physiological and pathological conditions (65). Breast cancer cell-derived exosomes play an important role in promoting breast cancer bone metastasis, which is associated with the formation of a pre-metastatic niche via transferal of miR-21 to osteoclasts and consequent down-regulation of PDCD4 expression (66).
Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins as well as degrade misfolded and unneeded proteins. Proteins are tagged for degradation by a small protein called ubiquitin. Compromised proteasome complex assembly leads to reduced proteolytic activity and the accumulation of damaged and misfolded proteins, and this contributes to the pathogenesis in many diseases (67). Proteasomal degradation is another important mechanism that regulates PDCD4 expression. In response to mitogens such as TPA, PDCD4 is rapidly phosphorylated at Ser67 by the 70-kDa ribosomal protein 6 kinase (p70S6K). The Ser67 phosphorylated PDCD4 is recognized and bound by the ubiquitin E3 ligase β-transducin repeat-containing 1 (β-TRCP-1). The subsequent ubiquitination targets PDCD4 for proteasomal degradation and ultimately promotes tumor formation (68). Down-regulation of p70S6K by p70s6k shRNA increased PDCD4 expression in MDA-MB-231 cells (69). In addition to p70S6K, it was reported that the other p70S6K family member, p90 ribosomal S6 kinase (p90RSK), was able to phosphorylate PDCD4 at Ser76 and lead to PDCD4 degradation in several TNBC cell lines (70). Conversely, ubiquitin-specific protease 4 (USP4) can increase PDCD4 expression through deubiquitination and subsequent inhibition of its degradation in MCF-7 cells (71). A recent study showed that the S-phase kinase-associated protein 2 (SKP2) promotes PDCD4 phosphorylation at Ser67 and subsequently its proteasomal degradation, resulting in the promotion of breast cancer cell proliferation (72). Interestingly, PDCD4 was also reported to inhibit IGF1R/IR inhibitor-induced phosphorylation of p70S6K (15, 73), which suggests a feedback loop regulation between PDCD4 and p70S6K.
Accumulating evidence has shown that inflammation can promote all stages of tumorigenesis, including DNA damage, limitless replication, apoptosis evasion, sustained angiogenesis, self-sufficiency in growth signaling, insensitivity to anti-growth signaling, and tissue invasion/metastasis. An inflammatory tumor microenvironment may attenuate PDCD4 protein expression. For example, exposure of MCF-7 cells to conditioned medium from TPA-induced differentiated monocytes attenuate PDCD4 protein level. This conditioned medium contained proinflammatory cytokines such as tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), and IL-8, which stimulate PI3K–mTOR signaling pathway and thereby facilitate p70S6K-dependent proteasomal degradation of PDCD4 (74). In addition, activation of the Cyclooxygenase-2 (COX-2)/prostaglandin E2 (PGE2)/Interleukin-8 (IL-8) inflammatory pathway suppresses PDCD4 expression and promotes the invasion in MCF-7 cells (35). It also reported that proinflammatory cytokine transforming growth factor-β (TGF-β) stimulates miR-21 expression and thereby suppresses PDCD4 expression in MDA-MB-468 cells (75).
Lifetime estrogen exposure is widely accepted as a major risk factor for breast cancer development (76). Several epidemiological surveys indicate that supplementation with exogenous estrogen or a high level of endogenous estrogen significantly increase the incidence of breast cancer (77). In vitro studies also showed that dimerized estrogen binds with estrogen receptor (ER) and promotes cell proliferation and invasion of breast tumor cells (78). Natural estrogens include estradiol (E2), estrone (E) and estrotriol (E3), among which estradiol is the most bioactive form involved in breast tumorigenesis (79). Anti-estrogens are a class of drugs that prevent estrogens like E2 from mediating their biological effects in vivo. They act by blocking ER or by suppressing estrogen production. ICI-182780, an artificial anti-estrogen drug also named fulvestrant, can increase PDCD4 protein expression in ER-positive T-47D cells (25, 80). Interestingly, E2 treatment increases PDCD4 expression in MCF-7 cells by down-regulation of miR-21 in an ER-dependent manner (81). Further research proved that overexpression of ERα may up-regulate miR-21 to suppress PDCD4 expression in breast cancer cells (82). This interesting phenomenon can be interpreted as follows: if ER density is not increased, breast cancer cells may overexpress PDCD4 in the face of high estrogen levels, and if ER density is increased rising sensitivity to estrogen may decrease PDCD4 expression and thereby promote metastasis.
Sinomenine (Sino) is diffusely applied to heal rheumatoid arthritis and neuralgia. Gao et al. found that Sino exposure remarkably enhanced PDCD4 expression by inhibiting miR-29 in MDA-MB-231 cells (83). In addition, Gleditsia sinensis extract has been historically used in Chinese medicine and is considered one of the fundamental therapeutic herbs. The anti-breast cancer effect for Gleditsia sinensis probably results in upregulation of PDCD4 expression to promote cell apoptosis (84, 85). Curcumin is a constituent of the yellow powder extracted from the roots of Curcuma longa Linn. It is used to treat various diseases including hepatic disorders, anorexia, diabetic wounds, and rheumatism. Treatment with curcumin could down-regulates miR-21 expression and consistently up-regulates PDCD4 to promote apoptosis in breast cancer cell lines (86). Andrographolide is regarded as a “natural antibiotic” and is present in Andrographis paniculate (87), and it has shown to inhibit breast cancer growth and metastasis in vivo and to suppress cell proliferation, migration, and invasion in vitro. Mechanistically, andrographolide inhibited nuclear factor-kappa B (NF-κB) and elevates PDCD4 expression through suppression of miR-21-5p (88).
Retinoic acid receptors (RARs), which belong to steroid/thyroid hormone receptor gene family, are ligand-dependent transcription factors. They have been shown to inhibit the growth of breast cancer cells in vitro and in vivo. Afonja et al. treated T-47D and MCF-7 breast cancer cells with Am580 (a selective RARα agonist) and all-trans retinoic acid (ATRA, another RAR agonist derived from vitamin A retinoids) and found that PDCD4 expression was increased two- to five-fold. However, the detailed mechanism by which RAR agonists regulate PDCD4 expression is still unclear (25).
Human epidermal growth factor receptor-2 (HER2) is an oncogene, and its downstream signaling pathway plays important roles in the development and progression of certain aggressive types of breast cancer. HER2 antagonists (such as trastuzumab, pertuzumab and lapatinib) induce PDCD4 protein expression in T-47D cells, which express HER2 (25). Transfection of HER2 expression plasmid into MDA-MB-435 cells reduced PDCD4 expression by about 45% and concomitantly enhanced invasion and metastasis (89). Signal transducer and activator of transcription 3 (STAT3) was found to bind its response elements in the HER2 promoter to upregulate HER2 transcription in metastatic HER2-positive breast cancer. Moreover, STAT3 co-opts HER2 function by recruiting HER2 as its coactivator at the binding sites in the miR-21 promoter to enhance its expression, resulting in depression of PDCD4 (90).
The neuregulins (NRGs) comprise the largest subfamily of EGF-like polypeptide growth factors and act by binding to the ErbB/HER receptor tyrosine kinases. Breast cancer cells treated with NRGs have reduced phosphorylation of PDCD4 at Ser67, which results in increased PDCD4 stabilization due to avoidance of proteasomal degradation (91).
The chemical compound AC1MMYR2 (2,4-diamino-1, 3-diazinane-5-carbonitrile, also known as NSC211332) was identified as a specific small-molecule inhibitor of miR-21 that blocks the ability of Dicer to process pre-miR-21 to mature miR-21. AC1MMYR2 up-regulates the expression of PDCD4 to suppress proliferation and invasion in breast cancer (92).
In brief, various type of regulators control PDCD4 expression to influence hallmarks of breast cancer. Information on these regulatory factors is summarized in Table 1. Meanwhile, PDCD4 expression in different subtypes of breast cancer cell lines and tissues is summarized in Table 2.
Although various therapeutic agents such as tamoxifen, docetaxel, cisplatin, carboplatin, doxorubicin, gemcitabine, and mitoxantrone can improve overall survival and quality of life to some extent, patients eventually develop resistance to these treatments. The 5-year survival rate for patients with stage IV breast cancer is still as low as 20% (93). Thus, it is urgent to find new approaches that reduce drug resistance and improve the effectiveness of chemotherapy by understanding the relevant mechanisms of drug resistance in breast cancer. Increasing evidence suggests that up-regulation of PDCD4 in breast cancer may improve the sensitivity to systemic therapeutic drugs, especially for patients given neoadjuvant chemotherapy (preoperative chemotherapy where radical surgery is feasible), and thereby reduce tumor size, increase the chance of breast-conserving surgery and prolong lifespan.
Estrogens bind to ERs in the nucleus and promote their association with specific estrogen-response elements in the promoter region of target genes. This classic mechanism regulates the development of normal breast tissue and the progression of cancer cells (94). Based on the above mechanism, there are two main strategies to treat breast cancer with endocrine drugs (1): reduce the production of estrogen in the body; (2) competitively inhibit or destroy ERs. There are two main types of ERs in humans that have distinct functions: ERα and ERβ (95). ERα is expressed predominantly as a 66-kD transcript in breast cancer. ERβ has structural homology to ERα, particularly in their DNA-binding domains (95% amino acid identity) and in their ligand-binding domains (55% amino acid identity) (96). ERα is an important marker for breast cancer treatment as altered ERα signaling may cause resistance to endocrine therapy (97). By contrast, ERβ is proposed to inhibit ERα activity and may impair breast cancer cell proliferation by repressing the activation of the MAPK and PI3K signaling pathways (98, 99). ERβ agonists such as LY500307, ERB-041 and WAY200070 efficiently inhibit breast cancer cell growth and invasion. ERβ is now an interesting therapeutic target for patients with triple-negative breast cancer, who lack expression of ERα (100).
About 60%–70% of patients with breast cancer have ER-positive tumors (101). Therefore, inhibition of ER activity is the first therapeutic approach applied for patients with ER-positive breast cancer. Tamoxifen (TAM), a selective estrogen receptor modulator (SERM), is the most common drug for the treatment of ER-positive cancer (102). Nevertheless, approximately 40% of initially ERα-positive breast tumors become resistant to tamoxifen and other endocrine treatments (103). The resistance of ER antagonists may be due to: [1] overexpression of epidermal growth factor receptor (EGFR) and/or the oncogene HER2/neu/ErbB2 (104); [2] splice variants or point mutations in ER (105); [3] alterations in nuclear levels of ER coactivator or corepressor proteins, such as increased coactivator amplified in breast cancer 1 (AIB1) (106) or decreased corepressor nuclear receptor corepressor 1 (NCOR1) gene (107); [4] activation of the MAPK and PI3K/AKT signaling pathways (108, 109); and [5] increased expression of miR-221/222, which downregulate ER post-transcriptionally (110).
PDCD4 enhances sensitivity to several endocrine therapeutic drugs. Colburn and colleagues showed that down-regulation of PDCD4 by antisense RNA significantly reduced chemosensitivity to TAM in MCF-7 cells (111). In addition, PDCD4 is an independent predictive marker for resistance to TAM therapy in recurrent ER-positive breast cancer (112, 113). However, the mechanism through which PDCD4 enhances chemosensitivity to SERMs in breast cancer remains unclear.
Aromatase inhibitors (AIs) such as letrozole, anastrozole, and exemestane are used as first-line treatments for patients with post-menopausal breast cancer and block the peripheral conversion of androgens into estrogens and thereby reduce estrogen levels. Aromatase is the rate-limited enzyme for estrogen biosynthesis. As breast and ovarian cancer cells require estrogen to grow, AIs are administered to either block the production of estrogen or block the action of estrogen on receptors. Unfortunately, resistance to AI treatment is a significant clinical problem for a considerable number of patients with ER-positive breast cancer (114). Chen and his colleagues found that PDCD4 expression was down-regulated in AI-resistant breast cancer cells, and this down-regulation significantly correlated with activation of HER2 signaling in ER-positive breast tumors. HER2 downregulates PDCD4 expression by activating MAPK, AKT, and miR-21 in AI-resistant breast cancer cells. Overexpression of PDCD4 re-sensitizes AI-resistant breast cancer cells to AI, which suggests the potential role of PDCD4 in AI-resistant breast cancer (9).
PDCD4 is also associated with paclitaxel and doxorubicin resistance in breast cancer (115). Bourguignon et al. found that PDCD4 expression is down-regulated when MCF-7 breast cancer cells are treated with doxorubicin or paclitaxel (116). De Mattos et al. also reported that PDCD4 silencing by siRNAs enhances the resistance of SKBR3 breast cancer cells to paclitaxel and doxorubicin (117). In addition, it was suggested that the hyaluronan-CD44 interaction induced miR-21 production, and the consequent down-regulation of PDCD4 decreased apoptosis and increased the survival of MCF-7 cells and mediated resistance to both paclitaxel and doxorubicin treatment (116). Moreover, recent studies indicated that the miRNA/PDCD4 axis could modulate chemosensitivity in multiple resistant cancers (118–122). Taken together, this suggests that modulation of this axis may be an effective strategy for overcoming chemoresistance in breast cancer. Table 3 summarizes the relationship between PDCD4 and drug resistance by breast cancer as well as the possible related mechanisms.
A meta-analysis from 23 studies showed that down-regulation of PDCD4 was significantly associated with short overall survival of patients with breast cancer (123). Immunohistochemical staining showed that patients with nuclear PDCD4-positive (NPDCD4-positive) LumB-like tumors had better overall and disease-free survival rates compared to those with NPDCD4-positive LumA-like tumors. By contrast, loss of NPDCD4 expression increased the risk of disease recurrence and death in patients with LumB-like tumors (124). These findings suggest that PDCD4 may be an independent prognostic marker for breast cancer.
PDCD4 is an important suppressor of breast cancer tumorigenesis. Loss of PDCD4 promotes tumor cells proliferation, migration and invasion, and is associated with lymph node metastasis and worse disease-free survival in patients with ER- and HER2-positive breast cancer. This implicates PDCD4 as a possible prognostic marker. Moreover, PDCD4 has been shown to increase the sensitivity of breast cancer cells to endocrine therapy and chemotherapy, which render it as a relevant therapeutic target to overcome chemo-resistance. Thus, compounds that elevate PDCD4 expression are valuable agents for the combination of endocrine therapies or chemotherapies for breast cancer.
Taken together, the future research hotspot for the roles of PDCD4 in breast cancer will likely revolve around the following aspects: [1] the function PDCD4 as a biomarker for breast cancer diagnosis, prognosis, and drug-resistance; [2] the identification of novel upstream regulators and downstream target genes of PDCD4 and molecular biological mechanisms; [3] the development of small molecular and targeted drugs related to PDCD4.
JZ contributed to conception and design of the review. JZ and YCL organized the tables and the figure. QC wrote the first draft of the manuscript. HSY wrote one section (PDCD4 function) of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grant from the National Natural Science Foundation of China (No. 81402192).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.cn) for English language editing.
1. Matsuhashi HY S, Yatsuki H, Tsugita A, Hori K. Isolation of a Novel Gene Form a Human Cell Line With Pr-28 MAb Which Recognizes a Nuclear Antigen Involved in the Cell Cycle. Res Commun Biochem (1997) 1:109–20.
2. Azzoni L, Zatsepina O, Abebe B, Bennett IM, Kanakaraj P, Perussia B. Differential Transcriptional Regulation of CD161 and a Novel Gene, 197/15a, by IL-2, IL-15, and IL-12 in NK and T Cells. J Immunol (1998) 161(7):3493–500.
3. Goke A, Goke R, Knolle A, Trusheim H, Schmidt H, Wilmen A, et al. DUG is a Novel Homologue of Translation Initiation Factor 4G That Binds Eif4a. Biochem Biophys Res Commun (2002) 297(1):78–82. doi: j.pharmthera./s0006-291x(02)02129-0
4. Schlichter U, Kattmann D, Appl H, Miethe J, Brehmer-Fastnacht A, Klempnauer KH. Identification of the Myb-Inducible Promoter of the Chicken Pdcd4 Gene. Biochim Biophys Acta (2001) 1520(1):99–104. doi: j.pharmthera./s0167-4781(01)00252-4
5. Soejima H, Miyoshi O, Yoshinaga H, Masaki Z, Ozaki I, Kajiwara S, et al. Assignment of the Programmed Cell Death 4 Gene (PDCD4) to Human Chromosome Band 10q24 by in Situ Hybridization. Cytogenet Cell Genet (1999) 87(1-2):113–4. doi: 10.1159/000015408
6. Parsyan A. Translation and Its Regulation in Cancer Biology and Medicine. Netherlands: Springer (2014), ISBN: ISBN: 9789401790789. doi: 10.1007/978-94-017-9078-9
7. Korakiti AM, Moutafi M, Zografos E, Dimopoulos MA, Zagouri F. The Genomic Profile of Pregnancy-Associated Breast Cancer: A Systematic Review. Front Oncol (2020) 10:1773. doi: 10.3389/fonc.2020.01773
8. Qi L, Bart J, Tan LP, Platteel I, Sluis T, Huitema S, et al. Expression of miR-21 and its Targets (PTEN, PDCD4, TM1) in Flat Epithelial Atypia of the Breast in Relation to Ductal Carcinoma in Situ and Invasive Carcinoma [Research Support, Non-U.S. Gov't]. BMC Cancer (2009) 9:163. doi: 10.1186/1471-2407-9-163
9. Chen Z, Yuan YC, Wang Y, Liu Z, Chan HJ, Chen S. Down-Regulation of Programmed Cell Death 4 (PDCD4) is Associated With Aromatase Inhibitor Resistance and a Poor Prognosis in Estrogen Receptor-Positive Breast Cancer [Research Support, N.I.H., Extramural]. Breast Cancer Res Treat (2015) 152(1):29–39. doi: 10.1007/s10549-015-3446-8
10. Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The Transformation Suppressor Pdcd4 is a Novel Eukaryotic Translation Initiation Factor 4A Binding Protein That Inhibits Translation. Mol Cell Biol (2003) 23(1):26–37. doi: 10.1128/MCB.23.1.26-37.2003
11. Chang JH, Cho YH, Sohn SY, Choi JM, Kim A, Kim YC, et al. Crystal Structure of the Eif4a-PDCD4 Complex [Research Support, Non-U.S. Gov't]. Proc Natl Acad Sci U S A (2009) 106(9):3148–53. doi: 10.1073/pnas.0808275106
12. Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, et al. Structural Basis for Translational Inhibition by the Tumour Suppressor Pdcd4 [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't]. EMBO J (2009) 28(3):274–85. doi: 10.1038/emboj.2008.278
13. Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A Novel Function of the MA-3 Domains in Transformation and Translation Suppressor Pdcd4 is Essential for its Binding to Eukaryotic Translation Initiation Factor 4A. Mol Cell Biol (2004) 24(9):3894–906. doi: 10.1128/MCB.24.9.3894-3906.2004
14. Rogers GW Jr., Komar AA, Merrick WC. Eif4a: The Godfather of the DEAD Box Helicases [Research Support, U.S. Gov't, P.H.S. Review]. Prog Nucleic Acid Res Mol Biol (2002) 72:307–31. doi: j.pharmthera./S0079-6603(02)72073-4
15. Wang Q, Zhang Y, Zhu J, Zheng H, Chen S, Chen L, et al. IGF-1R Inhibition Induces MEK Phosphorylation to Promote Survival in Colon Carcinomas. Signal Transduct Target Ther (2020) 5(1):153. doi: 10.1038/s41392-020-0204-0
16. Wang Q, Zhu J, Wang YW, Dai Y, Wang YL, Wang C, et al. Tumor Suppressor Pdcd4 Attenuates Sin1 Translation to Inhibit Invasion in Colon Carcinoma. Oncogene (2017) 36(45):6225–34. doi: doi: 10.1038/onc.2017.228
17. Basu A, Sridharan S. Regulation of Anti-Apoptotic Bcl-2 Family Protein Mcl-1 by S6 Kinase 2. PLoS One (2017) 12(3):e0173854. doi: 10.1371/journal.pone.0173854
18. Liwak U, Thakor N, Jordan LE, Roy R, Lewis SM, Pardo OE, et al. Tumor Suppressor PDCD4 Represses Internal Ribosome Entry Site-Mediated Translation of Antiapoptotic Proteins and is Regulated by S6 Kinase 2. Mol Cell Biol (2012) 32(10):1818–29. doi: 10.1128/MCB.06317-11
19. Cmarik JL, Min H, Hegamyer G, Zhan S, Kulesz-Martin M, Yoshinaga H, et al. Differentially Expressed Protein Pdcd4 Inhibits Tumor Promoter-Induced Neoplastic Transformation [Research Support, U.S. Gov't, P.H.S.]. Proc Natl Acad Sci U S A (1999) 96(24):14037–42. doi: 10.1073/pnas.96.24.14037
20. Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 Suppresses Tumor Phenotype in JB6 Cells by Inhibiting AP-1 Transactivation. Oncogene (2003) 22(24):3712–20. doi: 10.1038/sj.onc.1206433
21. Matsuhashi S, Manirujjaman M, Hamajima H, Ozaki I. Control Mechanisms of the Tumor Suppressor PDCD4: Expression and Functions. Int J Mol Sci (2019) 20(9):2304. doi: 10.3390/ijms20092304
22. Wang Q, Yang HS. The Role of Pdcd4 in Tumour Suppression and Protein Translation. Biol Cell (2018). doi: 10.1111/boc.201800014
23. Lankat-Buttgereit B, Goke R. The Tumour Suppressor Pdcd4: Recent Advances in the Elucidation of Function and Regulation [Review]. Biol Cell (2009) 101(6):309–17. doi: 10.1042/BC20080191
24. Fan D, Zeng C, Wang S, Han J, Zhu L, Zhao H, et al. Forkhead Box P3 Promotes Breast Cancer Cell Apoptosis by Regulating Programmed Cell Death 4 Expression. Oncol letters (2020) 20(6):292. doi: 10.3892/ol.2020.12155
25. Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH. Induction of PDCD4 Tumor Suppressor Gene Expression by RAR Agonists, Antiestrogen and HER-2/Neu Antagonist in Breast Cancer Cells Evidence for a Role in Apoptosis. Oncogene (2004) 23(49):8135–45. doi: 10.1038/sj.onc.1207983
26. Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, et al. The Malignant Phenotype in Breast Cancer is Driven by Eif4a1-Mediated Changes in the Translational Landscape. Cell Death disease (2015) 6:e1603. doi: 10.1038/cddis.2014.542
27. Bitomsky N, Wethkamp N, Marikkannu R, Klempnauer KH. siRNA-Mediated Knockdown of Pdcd4 Expression Causes Upregulation of P21(Waf1/Cip1) Expression [Research Support, Non-U.S. Gov't]. Oncogene (2008) 27(35):4820–9. doi: 10.1038/onc.2008.115
28. Wang Y, Liu Z, Shen J. MicroRNA-421-Targeted PDCD4 Regulates Breast Cancer Cell Proliferation. Int J Mol Med (2019) 43(1):267–75. doi: 10.3892/ijmm.2018.3932
29. Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O. Alterations in the Expression of PDCD4 in Ductal Carcinoma of the Breast [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't]. Oncol Rep (2007) 18(6):1387–93.
30. Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-Microdissected Human Breast Cancers. Clin Proteomics (2010) 6(4):129–51. doi: 10.1007/s12014-010-9055-y
31. Meric-Bernstam F, Chen H, Akcakanat A, Do KA, Lluch A, Hennessy BT, et al. Aberrations in Translational Regulation are Associated With Poor Prognosis in Hormone Receptor-Positive Breast Cancer. Breast Cancer Res BCR. (2012) 14(5):R138. doi: 10.1186/bcr3343
32. Santhanam AN, Baker AR, Hegamyer G, Kirschmann DA, Colburn NH. Pdcd4 Repression of Lysyl Oxidase Inhibits Hypoxia-Induced Breast Cancer Cell Invasion [Research Support, N.I.H., Intramural]. Oncogene (2010) 29(27):3921–32. doi: 10.1038/onc.2010.158
33. Jiang B, Liu Q, Gai J, Guan J, Li Q. LncRNA SLC16A1-AS1 Regulates the miR-182/PDCD4 Axis and Inhibits the Triple-Negative Breast Cancer Cell Cycle. Immunopharmacol Immunotoxicol (2022) 22:1–18. doi: 10.1080/08923973.2022.2056482
34. Gonzalez-Villasana V, Nieves-Alicea R, McMurtry V, Gutierrez-Puente Y, Tari AM. Programmed Cell Death 4 Inhibits Leptin-Induced Breast Cancer Cell Invasion [Research Support, Non-U.S. Gov't]. Oncol Rep (2012) 27(3):861–6. doi: 10.3892/or.2011.1600
35. Nieves-Alicea R, Colburn NH, Simeone AM, Tari AM. Programmed Cell Death 4 Inhibits Breast Cancer Cell Invasion by Increasing Tissue Inhibitor of Metalloproteinases-2 Expression [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't]. Breast Cancer Res Treat (2009) 114(2):203–9. doi: 10.1007/s10549-008-9993-5
36. Strongin AY, Marmer BL, Grant GA, Goldberg GI. Plasma Membrane-Dependent Activation of the 72-kDa Type IV Collagenase is Prevented by Complex Formation With TIMP-2. J Biol Chem (1993) 268(19):14033–9. doi: j.pharmthera./S0021-9258(19)85205-5
37. De Benedetti A, Harris AL. Eif4e Expression in Tumors: Its Possible Role in Progression of Malignancies [Review]. Int J Biochem Cell Biol (1999) 31(1):59–72. doi: j.pharmthera./S1357-2725(98)00132-0
38. Bera A, Das F, Ghosh-Choudhury N, Kasinath BS, Abboud HE, Choudhury GG. microRNA-21-Induced Dissociation of PDCD4 From Rictor Contributes to Akt-IKKbeta-Mtorc1 Axis to Regulate Renal Cancer Cell Invasion. Exp Cell Res (2014) 328(1):99–117. doi: j.pharmthera./j.yexcr.2014.06.022
39. Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) Post-Transcriptionally Downregulates Tumor Suppressor Pdcd4 and Stimulates Invasion, Intravasation and Metastasis in Colorectal Cancer. Oncogene (2008) 27(15):2128–36. doi: 10.1038/sj.onc.1210856
40. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res (2005) 65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783
41. Diederichs S, Haber DA. Sequence Variations of microRNAs in Human Cancer: Alterations in Predicted Secondary Structure do Not Affect Processing. Cancer Res (2006) 66(12):6097–104. doi: 10.1158/0008-5472.CAN-06-0537
42. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc Natl Acad Sci U S A (2006) 103(7):2257–61. doi: 10.1073/pnas.0510565103
43. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res (2005) 65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137
44. Gao S, Tian H, Guo Y, Li Y, Guo Z, Zhu X, et al. miRNA Oligonucleotide and Sponge for miRNA-21 Inhibition Mediated by PEI-PLL in Breast Cancer Therapy [Research Support, Non-U.S. Gov't]. Acta biomaterialia (2015) 25:184–93. doi: j.pharmthera./j.actbio.2015.07.020
45. Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA Expression Abnormalities in Pancreatic Endocrine and Acinar Tumors are Associated With Distinctive Pathologic Features and Clinical Behavior. J Clin Oncol (2006) 24(29):4677–84. doi: 10.1200/JCO.2005.05.5194
46. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-Mediated Tumor Growth. Oncogene (2007) 26(19):2799–803. doi: 10.1038/sj.onc.1210083
47. Du J, Yang S, An D, Hu F, Yuan W, Zhai C, et al. BMP-6 Inhibits microRNA-21 Expression in Breast Cancer Through Repressing Deltaef1 and AP-1. Cell Res (2009) 19(4):487–96. doi: 10.1038/cr.2009.34
48. Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 Targets Tumor Suppressor Genes in Invasion and Metastasis. Cell Res (2008) 18(3):350–9. doi: 10.1038/cr.2008.24
49. Guo LM, Ding GF, Xu WC, Ge H, Jiang Y, Lu YF. Anti-PD-L1 Antibody Enhances T Cell Immune Responses and Reduces Resistance of Breast Cancer Cells to Radiotherapy. Oxid Med Cell Longev (2022) 2022:5938688. doi: 10.1155/2022/5938688
50. Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 Promotes Cell Transformation by Targeting the Programmed Cell Death 4 Gene. Oncogene (2008) 27(31):4373–9. doi: 10.1038/onc.2008.72
51. Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P, et al. Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker Cd133. Mol Ther (2019) 27(7):1252–61. doi: j.pharmthera./j.ymthe.2019.04.018
52. Jin YY, Andrade J, Wickstrom E. Non-Specific Blocking of miR-17-5p Guide Strand in Triple Negative Breast Cancer Cells by Amplifying Passenger Strand Activity. PLoS One (2015) 10(12):e0142574. doi: 10.1371/journal.pone.0142574
53. Lin CC, Sharma SB, Farrugia MK, McLaughlin SL, Ice RJ, Loskutov YV, et al. Kruppel-Like Factor 4 Signals Through microRNA-206 to Promote Tumor Initiation and Cell Survival. Oncogenesis (2015) 4:e155. doi: 10.1038/oncsis.2015.8
54. Cheng Y, Xiang G, Meng Y, Dong R. MiRNA-183-5p Promotes Cell Proliferation and Inhibits Apoptosis in Human Breast Cancer by Targeting the PDCD4. Reprod Biol (2016) 16(3):225–33. doi: j.pharmthera./j.repbio.2016.07.002
55. Zhang D, Shi Z, Li M, Mi J. Hypoxia-Induced miR-424 Decreases Tumor Sensitivity to Chemotherapy by Inhibiting Apoptosis [Research Support, Non-U.S. Gov't]. Cell Death Dis (2014) 5:e1301. doi: 10.1038/cddis.2014.240
56. Ghafouri-Fard S, Hussen BM, Mohaqiq M, Shoorei H, Baniahmad A, Taheri M, et al. Interplay Between Non-Coding RNAs and Programmed Cell Death Proteins. Front Oncol (2022) 12:808475. doi: 10.3389/fonc.2022.808475
57. Karreth FA, Pandolfi PP. ceRNA Cross-Talk in Cancer: When Ce-Bling Rivalries Go Awry. Cancer Discovery (2013) 3(10):1113–21. doi: 10.1158/2159-8290.CD-13-0202
58. Wang D, Wang Z, Zhang L, Sun S. LncRNA PDCD4-AS1 Alleviates Triple Negative Breast Cancer by Increasing Expression of IQGAP2 via miR-10b-5p. Transl Oncol (2021) 14(1):100958. doi: j.pharmthera./j.tranon.2020.100958
59. Eshghifar N, Rouhollah F, Barikrow N, Pouresmaeili F, Taheri M. The Role of Long Noncoding RNAs in Patients With Luminal A Invasive Breast Ductal Carcinoma. Pathol Res Pract (2021) 227:153645. doi: j.pharmthera./j.prp.2021.153645
60. Jadaliha M, Gholamalamdari O, Tang W, Zhang Y, Petracovici A, Hao Q, et al. A Natural Antisense lncRNA Controls Breast Cancer Progression by Promoting Tumor Suppressor Gene mRNA Stability. PLoS Genet (2018) 14(11):e1007802. doi: 10.1371/journal.pgen.1007802
61. Filippova EA, Fridman MV, Burdennyy AM, Loginov VI, Pronina IV, Lukina SS, et al. Long Noncoding RNA GAS5 in Breast Cancer: Epigenetic Mechanisms and Biological Functions. Int J Mol Sci (2021) 22(13):6810. doi: 10.3390/ijms22136810
62. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs Function as ceRNAs to Regulate and Control Human Cancer Progression. Mol Cancer. (2018) 17(1):79. doi: 10.1186/s12943-018-0827-8
63. Ji C, Hu J, Wang X, Zheng W, Deng X, Song H, et al. Hsa_circ_0053063 Inhibits Breast Cancer Cell Proliferation via Hsa_Circ_0053063/hsa-miR-330-3p/PDCD4 Axis. Aging (Albany NY). (2021) 13(7):9627–45. doi: 10.18632/aging.202707
64. Cai Y, Zhao X, Chen D, Zhang F, Chen Q, Shao CC, et al. Circ-NOL10 Regulated by MTDH/CASC3 Inhibits Breast Cancer Progression and Metastasis via Multiple miRNAs and PDCD4. Mol Ther Nucleic Acids (2021) 26:773–86. doi: j.pharmthera./j.omtn.2021.09.013
65. Barile L, Vassalli G. Exosomes: Therapy Delivery Tools and Biomarkers of Diseases. Pharmacol Ther (2017) 174:63–78. doi: j.pharmthera./j.pharmthera.02.020
66. Yuan X, Qian N, Ling S, Li Y, Sun W, Li J, et al. Breast Cancer Exosomes Contribute to Pre-Metastatic Niche Formation and Promote Bone Metastasis of Tumor Cells. Theranostics (2021) 11(3):1429–45. doi: 10.7150/thno.45351
67. Nassif ND, Cambray SE, Kraut DA. Slipping Up: Partial Substrate Degradation by ATP-Dependent Proteases. IUBMB Life (2014) 66(5):309–17. doi: 10.1002/iub.1271
68. Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-Mediated Degradation of PDCD4 Promotes Protein Translation and Cell Growth. Sciencer76 and lead to PDCD4 degradation in several TNBC cell lines(2006) 314(5798):467–71. doi: 10.1126/science.1130276
69. Akar U, Ozpolat B, Mehta K, Lopez-Berestein G, Zhang D, Ueno NT, et al. Targeting P70s6k Prevented Lung Metastasis in a Breast Cancer Xenograft Model. Mol Cancer Ther (2010) 9(5):1180–7. doi: 10.1158/1535-7163.MCT-09-1025
70. Cuesta R, Holz MK. RSK-Mediated Down-Regulation of PDCD4 is Required for Proliferation, Survival, and Migration in a Model of Triple-Negative Breast Cancer. Oncotarget (2016) 7(19):27567–83. doi: 10.18632/oncotarget.8375
71. Li Y, Jiang D, Zhang Q, Liu X, Cai Z. Ubiquitin-Specific Protease 4 Inhibits Breast Cancer Cell Growth Through the Upregulation of PDCD4. Int J Mol Med (2016) 38(3):803–11. doi: 10.3892/ijmm.2016.2685
72. Li C, Du L, Ren Y, Liu X, Jiao Q, Cui D, et al. SKP2 Promotes Breast Cancer Tumorigenesis and Radiation Tolerance Through PDCD4 Ubiquitination. J Exp Clin Cancer Res (2019) 38(1):76. doi: 10.1186/s13046-019-1069-3
73. Zhang Y, Wang Q, Chen L, Yang HS. Inhibition of P70s6k1 Activation by Pdcd4 Overcomes the Resistance to an IGF-1r/IR Inhibitor in Colon Carcinoma Cells. Mol Cancer Ther (2015) 14(3):799–809. doi: 10.1158/1535-7163.MCT-14-0648
74. Schmid T, Bajer MM, Blees JS, Eifler LK, Milke L, Rubsamen D, et al. Inflammation-Induced Loss of Pdcd4 is Mediated by Phosphorylation-Dependent Degradation [Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov't]. Carcinogenesis (2011) 32(10):1427–33. doi: 10.1093/carcin/bgr131
75. Davis BN, Hilyard AC, Lagna G, Hata A. SMAD Proteins Control DROSHA-Mediated microRNA Maturation. Nature (2008) 454(7200):56–61. doi: 10.1038/nature07086
76. Santen RJ, Boyd NF, Chlebowski RT, Cummings S, Cuzick J, Dowsett M, et al. Breast Cancer Prevention Collaborative G. Critical Assessment of New Risk Factors for Breast Cancer: Considerations for Development of an Improved Risk Prediction Model. Endocrine-related cancer (2007) 14(2):169–87. doi: 10.1677/ERC-06-0045
77. Travis RC, Key TJ. Oestrogen Exposure and Breast Cancer Risk. Breast Cancer Res BCR. (2003) 5(5):239–47. doi: 10.1186/bcr628
78. Chakravarty D, Nair SS, Santhamma B, Nair BC, Wang L, Bandyopadhyay A, et al. Extranuclear Functions of ER Impact Invasive Migration and Metastasis by Breast Cancer Cells. Cancer Res (2010) 70(10):4092–101. doi: 10.1158/0008-5472.CAN-09-3834
79. Turan VK, Sanchez RI, Li JJ, Li SA, Reuhl KR, Thomas PE, et al. The Effects of Steroidal Estrogens in ACI Rat Mammary Carcinogenesis: 17beta-Estradiol, 2-Hydroxyestradiol, 4-Hydroxyestradiol, 16alpha-Hydroxyestradiol, and 4-Hydroxyestrone. J Endocrinol (2004) 183(1):91–9. doi: 10.1677/joe.1.05802
80. DeFriend DJ, Howell A, Nicholson RI, Anderson E, Dowsett M, Mansel RE, et al. Investigation of a New Pure Antiestrogen (ICI 182780) in Women With Primary Breast Cancer. Cancer Res (1994) 54(2):408–14.
81. Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol Downregulates miR-21 Expression and Increases miR-21 Target Gene Expression in MCF-7 Breast Cancer Cells. Nucleic Acids Res (2009) 37(8):2584–95. doi: 10.1093/nar/gkp117
82. Klinge CM, Riggs KA, Wickramasinghe NS, Emberts CG, McConda DB, Barry PN, et al. Estrogen Receptor Alpha 46 is Reduced in Tamoxifen Resistant Breast Cancer Cells and Re-Expression Inhibits Cell Proliferation and Estrogen Receptor Alpha 66-Regulated Target Gene Transcription. Mol Cell Endocrinol (2010) 323(2):268–76. doi: 10.1016/j.mce.2010.03.013
83. Gao G, Liang X, Ma W. Sinomenine Restrains Breast Cancer Cells Proliferation, Migration and Invasion via Modulation of miR-29/PDCD-4 Axis. Artif Cells Nanomed Biotechnol (2019) 47(1):3839–46. doi: 10.1080/21691401.2019.1666861
84. Cai Y, Zhang C, Zhan L, Cheng L, Lu D, Wang X, et al. Anticancer Effects of Gleditsia Sinensis Extract in Rats Transplanted With Hepatocellular Carcinoma Cells. Oncol Res (2019) 27(8):889–99. doi: 10.3727/096504018X15482423944678
85. Yu J, Li G, Mu Y, Zhou H, Wang X, Yang P. Anti-Breast Cancer Triterpenoid Saponins From the Thorns of Gleditsia Sinensis. Nat Prod Res (2019) 33(16):2308–13. doi: 10.1080/14786419.2018.1443092
86. Pan W, Liu B, Gao X, Yu Z, Liu X, Li N, et al. A Graphene-Based Fluorescent Nanoprobe for Simultaneous Monitoring of miRNA and mRNA in Living Cells. Nanoscale (2018) 10(29):14264–71. doi: 10.1039/c8nr04106g
87. Patil R, Jain V. Andrographolide: A Review of Analytical Methods. J Chromatogr Sci (2021) 59(2):191–203. doi: 10.1093/chromsci/bmaa091
88. Li J, Huang L, He Z, Chen M, Ding Y, Yao Y, et al. Andrographolide Suppresses the Growth and Metastasis of Luminal-Like Breast Cancer by Inhibiting the NF-Kappab/miR-21-5p/PDCD4 Signaling Pathway. Front Cell Dev Biol (2021) 9:643525. doi: 10.3389/fcell.2021.643525
89. Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, et al. Up-Regulation of miR-21 by HER2/neu Signaling Promotes Cell Invasion. J Biol Chem (2009) 284(27):18515–24. doi: 10.1074/jbc.M109.006676
90. Venturutti L, Romero LV, Urtreger AJ, Chervo MF, Cordo Russo RI, Mercogliano MF, et al. Stat3 Regulates ErbB-2 Expression and Co-Opts ErbB-2 Nuclear Function to Induce miR-21 Expression, PDCD4 Downregulation and Breast Cancer Metastasis. Oncogene (2016) 35(17):2208–22. doi: 10.1038/onc.2015.281
91. Montero JC, Pandiella A. PDCD4 Limits Prooncogenic Neuregulin-ErbB Signaling. Cell Mol Life Sci (2021) 78(4):1799–815. doi: 10.1007/s00018-020-03617-5
92. Shi Z, Zhang J, Qian X, Han L, Zhang K, Chen L, et al. AC1MMYR2, an Inhibitor of Dicer-Mediated Biogenesis of Oncomir miR-21, Reverses Epithelial-Mesenchymal Transition and Suppresses Tumor Growth and Progression. Cancer Res (2013) 73(17):5519–31. doi: 10.1158/0008-5472.CAN-13-0280
93. Malhotra A, Jain M, Prakash H, Vasquez KM, Jain A. The Regulatory Roles of Long non-Coding RNAs in the Development of Chemoresistance in Breast Cancer. Oncotarget (2017) 8(66):110671–84. doi: 10.18632/oncotarget.22577
94. Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and Actions of Estrogens. N Engl J Med (2002) 346(5):340–52. doi: 10.1056/NEJMra000471
95. Hawkins MB, Thornton JW, Crews D, Skipper JK, Dotte A, Thomas P. Identification of a Third Distinct Estrogen Receptor and Reclassification of Estrogen Receptors in Teleosts. Proc Natl Acad Sci U S A. (2000) 97(20):10751–6. doi: 10.1073/pnas.97.20.10751
96. Enmark E, Gustafsson JA. Oestrogen Receptors - an Overview. J Intern Med (1999) 246(2):133–8. doi: 10.1046/j.1365-2796.1999.00545.x
97. Zhuang T, Zhu J, Li Z, Lorent J, Zhao C, Dahlman-Wright K, et al. P21-Activated Kinase Group II Small Compound Inhibitor GNE-2861 Perturbs Estrogen Receptor Alpha Signaling and Restores Tamoxifen-Sensitivity in Breast Cancer Cells. Oncotarget (2015) 6(41):43853–68. doi: 10.18632/oncotarget.6081
98. Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of Estrogen Receptor Beta on Gene Networks Regulated by Estrogen Receptor Alpha in Breast Cancer Cells. Endocrinology (2006) 147(10):4831–42. doi: 10.1210/en.2006-0563
99. Cotrim CZ, Fabris V, Doria ML, Lindberg K, Gustafsson JA, Amado F, et al. Estrogen Receptor Beta Growth-Inhibitory Effects are Repressed Through Activation of MAPK and PI3K Signalling in Mammary Epithelial and Breast Cancer Cells. Oncogene (2013) 32(19):2390–402. doi: 10.1038/onc.2012.261
100. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-Negative Breast Cancer: Challenges and Opportunities of a Heterogeneous Disease. Nat Rev Clin Oncol (2016) 13(11):674–90. doi: 10.1038/nrclinonc.2016.66
101. Burstein HJ, Temin S, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, et al. Adjuvant Endocrine Therapy for Women With Hormone Receptor-Positive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. J Clin Oncol (2014) 32(21):2255–69. doi: 10.1200/JCO.2013.54.2258
102. Ring A, Dowsett M. Mechanisms of Tamoxifen Resistance. Endocrine-related cancer (2004) 11(4):643–58. doi: 10.1677/erc.1.00776
103. Weihua Z, Saji S, Makinen S, Cheng G, Jensen EV, Warner M, et al. Estrogen Receptor (ER) Beta, a Modulator of ERalpha in the Uterus. Proc Natl Acad Sci U S A (2000) 97(11):5936–41. doi: 10.1073/pnas.97.11.5936
104. Dowsett M. Overexpression of HER-2 as a Resistance Mechanism to Hormonal Therapy for Breast Cancer. Endocrine-related cancer (2001) 8(3):191–5. doi: 10.1677/erc.0.0080191
105. Herynk MH, Fuqua SA. Estrogen Receptor Mutations in Human Disease. Endocr Rev (2004) 25(6):869–98. doi: 10.1210/er.2003-0010
106. Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 Functions as an E2F1 Coactivator to Promote Breast Cancer Cell Proliferation and Antiestrogen Resistance. Mol Cell Biol (2004) 24(12):5157–71. doi: 10.1128/MCB.24.12.5157-5171.2004
107. Girault I, Lerebours F, Amarir S, Tozlu S, Tubiana-Hulin M, Lidereau R, et al. Expression Analysis of Estrogen Receptor Alpha Coregulators in Breast Carcinoma: Evidence That NCOR1 Expression is Predictive of the Response to Tamoxifen. Clin Cancer Res (2003) 9(4):1259–66.
108. Fan P, Yue W, Wang JP, Aiyar S, Li Y, Kim TH, et al. Mechanisms of Resistance to Structurally Diverse Antiestrogens Differ Under Premenopausal and Postmenopausal Conditions: Evidence From In Vitro Breast Cancer Cell Models. Endocrinology (2009) 150(5):2036–45. doi: 10.1210/en.2008-1195
109. Masri S, Phung S, Wang X, Wu X, Yuan YC, Wagman L, et al. Genome-Wide Analysis of Aromatase Inhibitor-Resistant, Tamoxifen-Resistant, and Long-Term Estrogen-Deprived Cells Reveals a Role for Estrogen Receptor. Cancer Res (2008) 68(12):4910–8. doi: 10.1158/0008-5472.CAN-08-0303
110. Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 Confers Tamoxifen Resistance in Breast Cancer by Targeting p27Kip1. J Biol Chem (2008) 283(44):29897–903. doi: 10.1074/jbc.M804612200
111. Jansen AP, Camalier CE, Stark C, Colburn NH. Characterization of Programmed Cell Death 4 in Multiple Human Cancers Reveals a Novel Enhancer of Drug Sensitivity. Mol Cancer Ther (2004) 3(2):103–10.
112. De Marchi T, Liu NQ, Stingl C, Timmermans MA, Smid M, Look MP, et al. 4-Protein Signature Predicting Tamoxifen Treatment Outcome in Recurrent Breast Cancer. Mol Oncol (2016) 10(1):24–39. doi: 10.1016/j.molonc.2015.07.004
113. De Marchi T, Kuhn E, Dekker LJ, Stingl C, Braakman RB, Opdam M, et al. Targeted MS Assay Predicting Tamoxifen Resistance in Estrogen-Receptor-Positive Breast Cancer Tissues and Sera. J Proteome Res (2016) 15(4):1230–42. doi: 10.1021/acs.jproteome.5b01119
114. Mauri D, Pavlidis N, Polyzos NP, Ioannidis JPA. Survival With Aromatase Inhibitors and Inactivators Versus Standard Hormonal Therapy in Advanced Breast Cancer: Meta-Analysis. JNCI: J Natl Cancer Institute (2006) 98(18):1285–91. doi: 10.1093/jnci/djj357
115. Tao L, Wu YQ, Zhang SP. MiR-21-5p Enhances the Progression and Paclitaxel Resistance in Drug-Resistant Breast Cancer Cell Lines by Targeting PDCD4. Neoplasma (2019) 66(5):746–55. doi: 10.4149/neo_2018_181207N930
116. Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 Interaction With Protein Kinase C(epsilon) Promotes Oncogenic Signaling by the Stem Cell Marker Nanog and the Production of microRNA-21, Leading to Down-Regulation of the Tumor Suppressor Protein PDCD4, Anti-Apoptosis, and Chemotherapy Resistance in Breast Tumor Cells. J Biol Chem (2009) 284(39):26533–46. doi: 10.1074/jbc.M109.027466
117. De Mattos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovannetti E, Peg V, et al. MicroRNA-21 Links Epithelial-to-Mesenchymal Transition and Inflammatory Signals to Confer Resistance to Neoadjuvant Trastuzumab and Chemotherapy in HER2-Positive Breast Cancer Patients. Oncotarget (2015) 6(35):37269–80. doi: 10.18632/oncotarget.5495
118. Fu WF, Chen WB, Dai L, Yang GP, Jiang ZY, Pan L, et al. Inhibition of miR-141 Reverses Cisplatin Resistance in non-Small Cell Lung Cancer Cells via Upregulation of Programmed Cell Death Protein 4. Eur Rev Med Pharmacol Sci (2016) 20(12):2565–72.
119. Lei Y, Hu X, Li B, Peng M, Tong S, Zu X, et al. miR-150 Modulates Cisplatin Chemosensitivity and Invasiveness of Muscle-Invasive Bladder Cancer Cells via Targeting PDCD4 In Vitro. Med Sci monitor Int Med J Exp Clin Res (2014) 20:1850–7. doi: 10.12659/MSM.891340
120. Ning FL, Wang F, Li ML, Yu ZS, Hao YZ, Chen SS. MicroRNA-182 Modulates Chemosensitivity of Human non-Small Cell Lung Cancer to Cisplatin by Targeting PDCD4. Diagn pathol (2014) 9:143. doi: 10.1186/1746-1596-9-143
121. Wang YQ, Guo RD, Guo RM, Sheng W, Yin LR. MicroRNA-182 Promotes Cell Growth, Invasion and Chemoresistance by Targeting Programmed Cell Death 4 (PDCD4) in Human Ovarian Carcinomas. J Cell Biochem (2013) 114(7):1464–73. doi: 10.1002/jcb.24488
122. Zhao J, Cao J, Zhou L, Du Y, Zhang X, Yang B, et al. MiR-1260b Inhibitor Enhances the Chemosensitivity of Colorectal Cancer Cells to Fluorouracil by Targeting PDCD4/IGF1. Oncol letters (2018) 16(4):5131–9. doi: 10.3892/ol.2018.9307
123. Li JZ, Gao W, Ho WK, Lei WB, Wei WI, Chan JY, et al. The Clinical Association of Programmed Cell Death Protein 4 (PDCD4) With Solid Tumors and its Prognostic Significance: A Meta-Analysis. Chin J Cancer. (2016) 35(1):95. doi: 10.1186/s40880-016-0158-3
Keywords: PDCD4, breast cancer, translational control, drug resistance, tumor suppressor
Citation: Cai Q, Yang HS, Li YC and Zhu J (2022) Dissecting the Roles of PDCD4 in Breast Cancer. Front. Oncol. 12:855807. doi: 10.3389/fonc.2022.855807
Received: 16 January 2022; Accepted: 12 May 2022;
Published: 20 June 2022.
Edited by:
Roxana Schillaci, CONICET Instituto de Biología y Medicina Experimental (IBYME), ArgentinaReviewed by:
Brian D. Adams, Brain Institute of America, United StatesCopyright © 2022 Cai, Yang, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Zhu, emh1amlhbmdAcWlsdWhvc3BpdGFsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.