- Department of Oncology, The First Affiliated Hospital of Nanchang University, Nanchang, China
Liver hepatocellular carcinoma (LIHC) seriously endangers the health and quality of life of individuals worldwide. Increasing evidence has underscored that the copper metabolism MURR1 domain (COMMD) family plays important roles in tumorigenesis. However, the specific role, biological function, mechanism and prognostic value of COMMD2 and its correlation with immune cell infiltration in LIHC remain unknown. In this study, we first determined the expression and prognostic potential of COMMD2 in human tumors using The Cancer Genome Atlas (TCGA) data and identified COMMD2 as a potential oncogene in LIHC. High COMMD2 expression was associated with pathological tumor stage and metastasis. Subsequently, noncoding RNAs (ncRNAs) upregulating COMMD2 expression were identified by performing expression, correlation, and survival analyses in combination. The CRNDE/LINC00511/SNHG17/HCG18-miR-29c-3p axis was identified as the most likely ncRNA-associated pathway upstream of COMMD2 in LIHC. Next, the expression profiles of COMMD2 and ncRNAs were validated in LIHC tissues and adjacent normal tissues. Furthermore, COMMD2 was significantly positively correlated with tumor immune cell infiltration, immune cell biomarkers, and immune checkpoint molecule expression. Importantly, COMMD2 potentially influenced prognosis by regulating immune cell infiltration in LIHC. Finally, COMMD2 was knocked down in LIHC cell lines using siRNAs for functional assays in vitro, resulting in suppressed cell proliferation and migration. In summary, our findings showed that the ncRNA-mediated upregulation of COMMD2 was associated with an unfavorable prognosis correlated with immune cell infiltration in LIHC.
Introduction
Liver hepatocellular carcinoma (LIHC) is the most common type of primary cancer in the liver and third leading cause of cancer-related mortality worldwide (1). Although substantial improvements have been made in LIHC therapy, particularly in molecular targeted therapy and immunotherapy (2, 3), the 5-year survival rate of LIHC is dismal because of its high heterogeneity, complex genetics and clinical features (4, 5). Thus, molecular biomarkers urgently need to be identified to improve the prognosis and develop novel therapeutic strategies for LIHC.
The copper metabolism MURR1 domain (COMMD) protein family comprises 10 members (COMMD1–COMMD10), all of which share a structurally conserved C-terminal motif and are implicated in regulating many biological processes through protein-protein interactions (6). COMMD proteins are frequently dysregulated in various cancers and are associated with cancer progression and metastasis (7–11). For example, decreased COMMD1 expression increased tumor invasion (12) and suppressed the sensitivity of ovarian cancer cells to cisplatin (13). COMMD7 promotes cell proliferation, migration, and invasion by upregulating NF-κB signaling (14) or CXCL10 (15). COMMD9 promotes the progression of non-small cell lung cancer by increasing TFDP1/E2F1 activation (8). However, the expression, biological function, possible mechanism and prognostic relevance of COMMD2 and its correlation with immune cell infiltration in human cancers, including LIHC, remain unknown.
In this study, we first performed expression profiling and survival analysis of COMMD2 in various human cancers. Additionally, COMMD2 expression was correlated with cancer stage, tumor grade, lymph node metastasis and the TP53 mutation status in LIHC. Next, the mechanism underlying the regulation of COMMD2 by noncoding RNAs (ncRNAs), including long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), through a competing endogenous RNA (ceRNA) network in LIHC was explored. Furthermore, we determined the correlations of COMMD2 with tumor-infiltrating immune cells, immune cell biomarkers, and immune checkpoint molecules in LIHC and analyzed the correlations between COMMD2 and infiltrating immune cells in the tumor microenvironment. Importantly, COMMD2 influenced the overall survival (OS) of LIHC patients through immune cell infiltration. Additionally, we performed a series of functional assays to further evaluate the effects of COMMD2 knockdown on LIHC cell proliferation and migration in vitro. Taken together, our findings suggest that the ncRNA-mediated upregulation of COMMD2 plays crucial roles in the development of LIHC by regulating immune cell infiltration.
Materials And Methods
Cell Culture and Transfection
The LIHC cell lines MHCC-97H and Huh-7 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). MHCC-97H and Huh-7 cells were cultured in DMEM (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2. COMMD2-targeting siRNA oligonucleotides were purchased from GenePharma (Shanghai, China). Cells were transfected with the indicated siRNAs using TurboFect transfection reagent (R0532; Thermo Scientific Scientific, Waltham, MA, United States). The siRNA sequences used in our study were as follows:
COMMD2-Homo-54: 5′-GGAAUUGUCCGAGGAGCAUTT-3′, COMMD2-Homo-222: 5′-GCAUGGUGUGGAAGGAUUATT-3′.
Real-Time Quantitative PCR
Total RNA was extracted from cells and clinical samples using the acid guanidine method with TRIzol reagent (Thermo Fisher Scientific), chloroform and isopropanol according to the manufacturer’s instructions. cDNA was obtained by reverse transcription using the PrimeScript™ RT reagent kit (Takara, Dalian, China). Real-time quantitative PCR (RT–qPCR) was performed using TB Green™ Premix Ex Taq II (Takara, Dalian, China) on an CFX96 Real-Time PCR Detection System (Bio-Rad, USA) and following minimum standard MIQE guidelines (16). The RT–qPCR cycling conditions were as follows: 95°C for 30 s, 42 cycles at 95°C for 5 s, and 60°C for 30 s. The melt curve stage was set as follows: 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. The primer sequences are shown in Table 1. The miRNA, mRNA and lncRNA levels were normalized to those of U6 or GAPDH. The relative expression level of mRNA from cells was calculated using the 2−ΔΔCT method and the miRNA, mRNA and lncRNA levels from clinical samples was assessed using the 2−ΔCt method.
Tissue Samples and Ethical Statement
Fresh LIHC specimens and adjacent normal tissue were obtained from 10 LIHC patients who were undergoing surgery at the First Affiliated Hospital of Nanchang University (Nanchang, China). The patients were treatment-naïve before surgery. This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University.
CCK8 Viability and Clone Formation Assays
Cells were seeded in 96-well plates at a density of 2 × 103 cells per well, after which 100 μl of FBS-free medium containing 10% CCK8 was added to each well and incubated for 2 h at 37°C. Next, the OD values at 450 nm were detected using a microplate reader (Thermo Scientific) for 5 days. For the clone formation assay, cells were seeded in 6-well plates at a density of 2 × 103 cells per well and cultured for 14 days. Next, the clones were fixed with methanol, stained with 1% crystal violet and counted.
Wound Healing Assay
Cells were seeded in 6-well plates and cultured in serum-free DMEM to 100% density, after which wounds were created by scratching with 10 μl pipette tips. Images were acquired at 0 and 24 h, and the wound areas were quantified using ImageJ software.
Cell Migration Assay
Cells were seeded on the upper transwell chamber at a density of 3 × 104 cells in 200 μl of serum-free culture medium, and 600 μl of medium containing 20% FBS was added to the lower chamber. After 48 h, the cells that migrated through the membranes were fixed with methanol, stained with 1% crystal violet and counted under a light microscope.
Data Acquisition and Processing
We downloaded LIHC-related mRNA-seq expression profiles from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.Cancer.gov/). Additionally, the expression profiling data of the arrays GSE55092 and GSE107170 (GPL570 sequencing platform) from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) were downloaded as validation datasets.
Immune Infiltration Analysis
TIMER (https://cistrome.shinyapps.io/timer/) is an online database used to comprehensively analyze tumor-infiltrating immune cells in various cancer types (17). The expression levels of COMMD2 in multiple cancers and their correlation with immune cell infiltration or immune checkpoint molecules in LIHC were analyzed using TIMER. CIBERSORT is a deconvolution algorithm based on gene expression that assesses the relative variations in immune cell infiltration (18). The immune infiltration levels of 22 immune cell types in patients in the TCGA-LIHC cohort, GSE55092 and GSE107170 were analyzed using the CIBERSORT algorithm. A p value <0.05 was considered to be statistically significant.
UALCAN Analysis
UALCAN (http://ualcan.path.uab.edu) is a comprehensive and interactive online tool that includes 31 cancer types from the TCGA database (19). In this study, COMMD2 mRNA expression in various cancer types and its association with survival prognosis were analyzed. Furthermore, UALCAN was used to analyze the associations of COMMD2 with clinicopathologic parameters, such as patient sex, patient age, cancer stage, tumor grade, nodal metastasis status and the TP53 mutation status in LIHC. A p value <0.05 was considered to be statistically significant.
GEPIA Database Analysis
GEPIA (http://gepia.cancer-pku.cn/) is an interactive analysis online tool for cancer and normal gene expression profiling using TCGA and Genotype-Tissue Expression (GTEx) data (20). GEPIA was used to determine and assess the expression and prognostic values of candidate lncRNAs in LIHC. Additionally, the correlations between COMMD2 and immune checkpoint molecules in LIHC were evaluated. A p value <0.05 was considered to be statistically significant.
Candidate miRNA Prediction
miRNAs binding upstream of COMMD2 were predicted using the TarBase (http://www.microrna.gr/tarbase), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw//), miRWalk (http://mirwalk.uni-hd.de/) and starBase (http://starbase.sysu.edu.cn/) databases. Only the predicted miRNAs that commonly appeared in more than two of the abovementioned databases were considered candidate miRNAs of COMMD2.
StarBase Database Analysis
starBase is an online database for exploring miRNA, lncRNA and RNA interaction networks (21). starBase was used to conduct the following correlation analyses in LIHC: miRNA-COMMD2, lncRNA-miR-29c-3p and lncRNA-COMMD2. The expression of candidate miRNAs in LIHC was also analyzed. Additionally, starBase was used to predict candidate lncRNAs that could potentially bind to miR-29c-3p. A p value <0.05 was considered statistically significant.
Kaplan-Meier Plotter Analysis
Kaplan-Meier plotter (http://kmplot.com/analysis/), an online database containing data on the relationships between gene or miRNA expression and clinical outcomes in more than 20 cancer types, was used to assess associations of miR-29c-3p with the survival of LIHC patients and the associations of COMMD2 with patient survival in various cancer types. OS and progression-free survival (PFS) with hazard ratios (HRs) with 95% confidence intervals (95% CIs) and log-rank p value were evaluated (22).
Statistical Analysis
The statistical analyses in this study were automatically performed using the above online databases. Paired Student’s t test was used to evaluate the differences in ncRNA and COMMD2 expression between the cancer and control groups. Student’s t test was used for comparisons. P<0.05 was considered statistically significant. * P<0.05 and ** P<0.01.
Results
Expression Levels of COMMD2 in Multiple Cancers
To investigate the possible roles of COMMD2 in tumorigenesis, its expression levels in tumor and normal tissue samples of multiple cancer types were analyzed using the TIMER database. COMMD2 was expressed at significantly higher levels in the tumor tissues of 10 various cancer types, including bladder cancer (BLCA), cervical squamous cell carcinoma (CESC), cholangiocarcinoma (CHOL), colorectal adenocarcinoma (COAD), esophageal cancer (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), LIHC, lung squamous cell carcinoma (LUSC) and stomach adenocarcinoma (STAD), than in the corresponding normal tissues. However, in breast cancer (BRCA), kidney chromophobe (KICH), lung adenocarcinoma (LUAD), prostate adenocarcinoma (PRAD) and thyroid cancer (THCA) tissues, COMMD2 expression was markedly lower than that in the corresponding normal samples (Figure 1A). To further evaluate COMMD2 expression in human cancers, the UALCAN database was evaluated, revealing that the COMMD2 expression levels in BLCA, CHOL, COAD, ESCA, GBM, HNSC, LIHC, LUSC and STAD tissues were significantly higher than those in the corresponding normal tissues (Figures 1B–J). However, compared with its levels in corresponding normal tissues, the COMMD2 expression levels in KICH, LUAD, PRAD and THCA tissues were obviously lower (Figures 1K–N). COMMD2 expression levels in BRCA and CESC tissues has no significantly difference than its levels in corresponding normal tissues (Figures 1O, P). Overall, COMMD2 expression was upregulated in BLCA, CHOL, COAD, ESCA, GBM, HNSC, LIHC, LUSC and STAD tissues and downregulated in KICH, LUAD, PRAD and THCA tissues, indicating that COMMD2 may crucially regulate carcinogenesis in these 13 types of cancer.
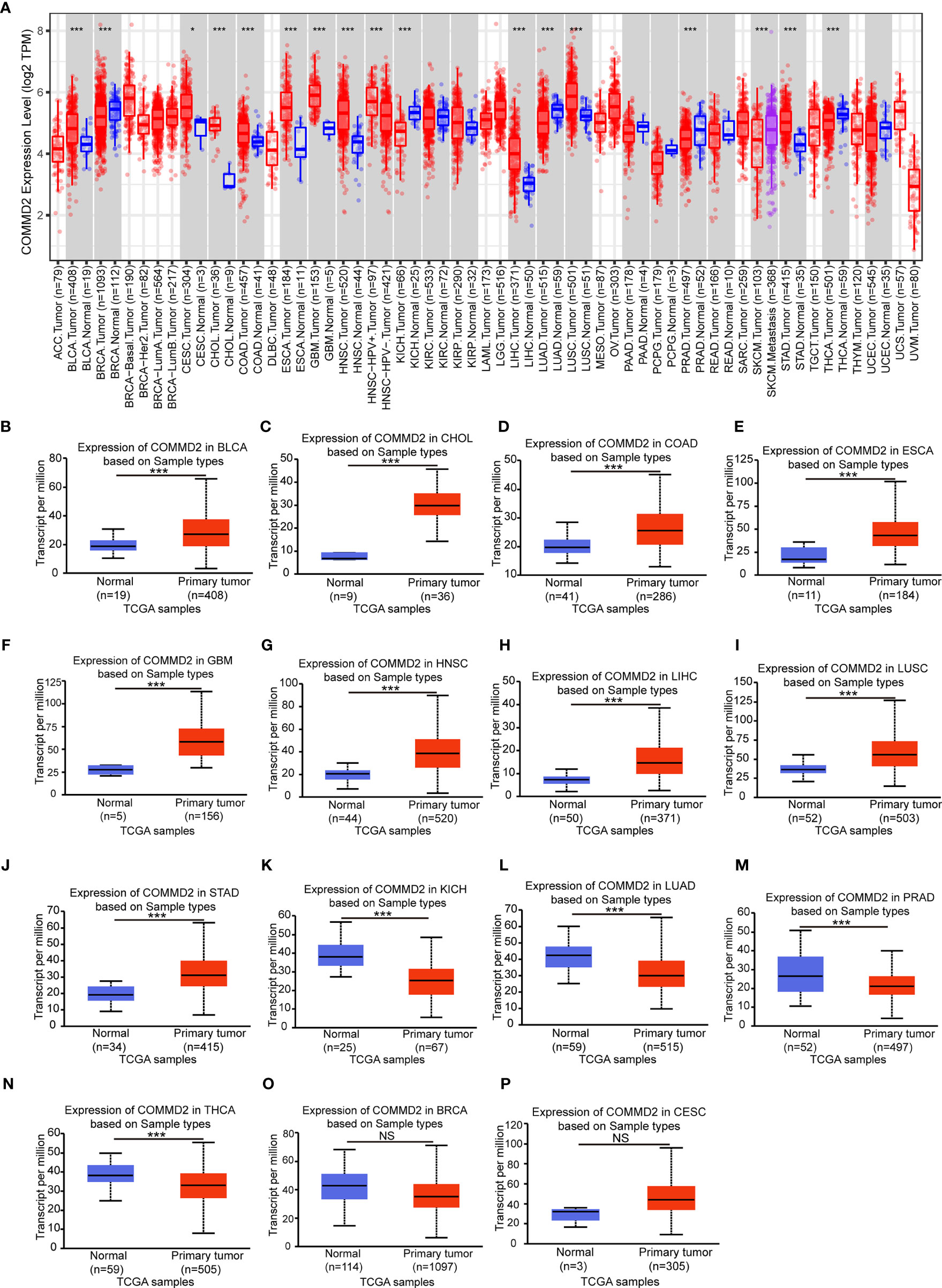
Figure 1 Expression levels of COMMD2 in different cancer types. (A) COMMD2 expression in different types of cancer was determined using the TIMER database. (B–M) COMMD2 expression in BLCA (B), CHOL (C), COAD (D), ESCA (E), GBM (F) HNSC (G), LIHC (H), LUSC (I), STAD (J), KICH (K), LUAD (L), PRAD (M), THCA (N), BRCA (O) and CESC (P) tissues compared with corresponding normal tissues as determined using the UALCAN database. *p value < 0.05, **p value < 0.01, ***p value < 0.001. NS, Not Significant.
Prognostic Value of COMMD2 in Various Human Cancers
The association between COMMD2 expression and prognosis was analyzed for various candidate types of cancer using the UALCAN database (Supplementary Figure 1). Notably, higher COMMD2 expression was significantly associated with a worse prognosis in LIHC (p= 0.0051, Supplementary Figure 1F), while higher expression of COMMD2 in CHOL indicated a better prognosis (p= 0.032, Supplementary Figure 1B). To further examine the prognostic potential of COMMD2 in different cancers, the Kaplan-Meier plotter database was used. Regarding OS, COMMD2 upregulation was associated with an unfavorable prognosis in LIHC (p= 0.00028; Figure 2D), but a higher level of COMMD2 was significantly associated with a positive prognosis in LUSC (p= 0.018; Figure 2F). Increased expression of COMMD2 was significantly correlated with short relapse-free survival (RFS) in BLCA, LIHC and STAD (Figures 2I, L, O). In other cancer types (Figures 2A–C, E, G–H, J–K, M–N, P), no significant correlations were found between COMMD2 and patient prognosis. In addition, the protein expression of COMMD2 was explored with the Human Protein Atlas (HPA) database. Similarly, COMMD2 was overexpressed in LIHC tissues compared with that in normal hepatic tissue. Higher COMMD2 expression was markedly correlated with a worse OS in LIHC patients (Supplementary Figures 2A, B). By combining the prognostic values from various databases, we concluded that COMMD2 might be an unfavorable prognostic biomarker for LIHC.
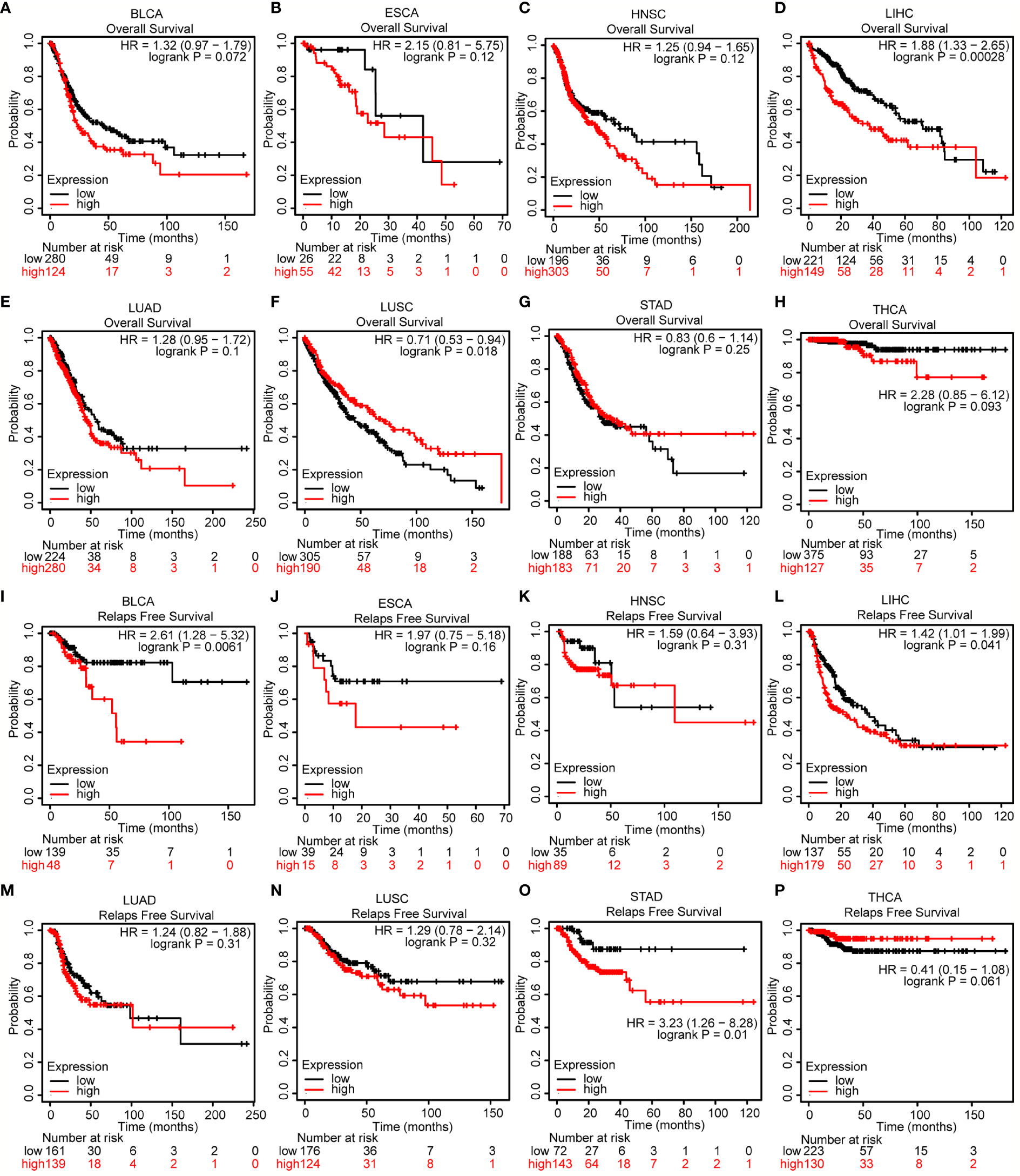
Figure 2 Prognostic value of COMMD2 in various human cancers. (A–H) Overall survival (OS) analysis of COMMD2 in BLCA (A), ESCA (B), HNSC (C), LIHC (D), LUAD (E), LUSC (F), STAD (G) and THCA (H). (I–P) Relapse-free survival (RFS) analysis of COMMD2 in BLCA (I), ESCA (J), HNSC (K), LIHC (L), LUAD (M), LUSC (N), STAD (O) and THCA (P) .
Association of COMMD2 Expression With the Clinicopathological Features of LIHC Patients
The relationships between COMMD2 expression and the clinicopathological parameters of LIHC patients, including patient sex, patient age, cancer stage, tumor grade, nodal metastasis status and the TP53 mutation status, were analyzed using UALCAN database. Regarding sex, COMMD2 expression was significantly upregulated in the LIHC tissues of both male and female patients compared with that in the normal tissues (Figure 3A). The COMMD2 level was significantly related to the age of the patient (Figure 3B) and remarkably correlated with cancer stage. Compared with that in normal tissues, COMMD2 expression was significantly higher in stage 1, stage 2, stage 3 and stage 4 cancers (Figure 3C). Concerning tumor grade, upregulation of COMMD2 expression was observed in grade 1, grade 2, grade 3, and grade 4 tumors, and COMMD2 expression increased as the pathological grade increased (Figure 3D). Moreover, COMMD2 expression was significantly related to the nodal metastasis status (Figure 3E). Furthermore, COMMD2 was expressed at a significantly higher level in the TP53 mutant than in the TP53 wild-type (Figure 3F). Additionally, to better understand the prognostic value of COMMD2 expression in LIHC, we explored the association between COMMD2 expression and clinical characteristics using the Kaplan-Meier database. High COMMD2 expression was significantly correlated with a poor OS in male and female patients with LIHC. Regarding the different tumor stages, upregulation of COMMD2 expression was associated with the a poor OS of patients with LIHC classified as stage 1 + 2, stage 2, and stage 2 + 3. A significant correlation between COMMD2 expression and a poor OS was observed in patients with American Joint Committee on Cancer (AJCC) stage T-2 and grade 2 LIHC. Additionally, high COMMD2 expression was significantly associated with an unfavorable OS for LIHC patients with microvascular invasion, patients who did not consume alcohol and patients without hepatitis. These results imply that COMMD2 expression possesses prognostic value in LIHC (Figure 3G).
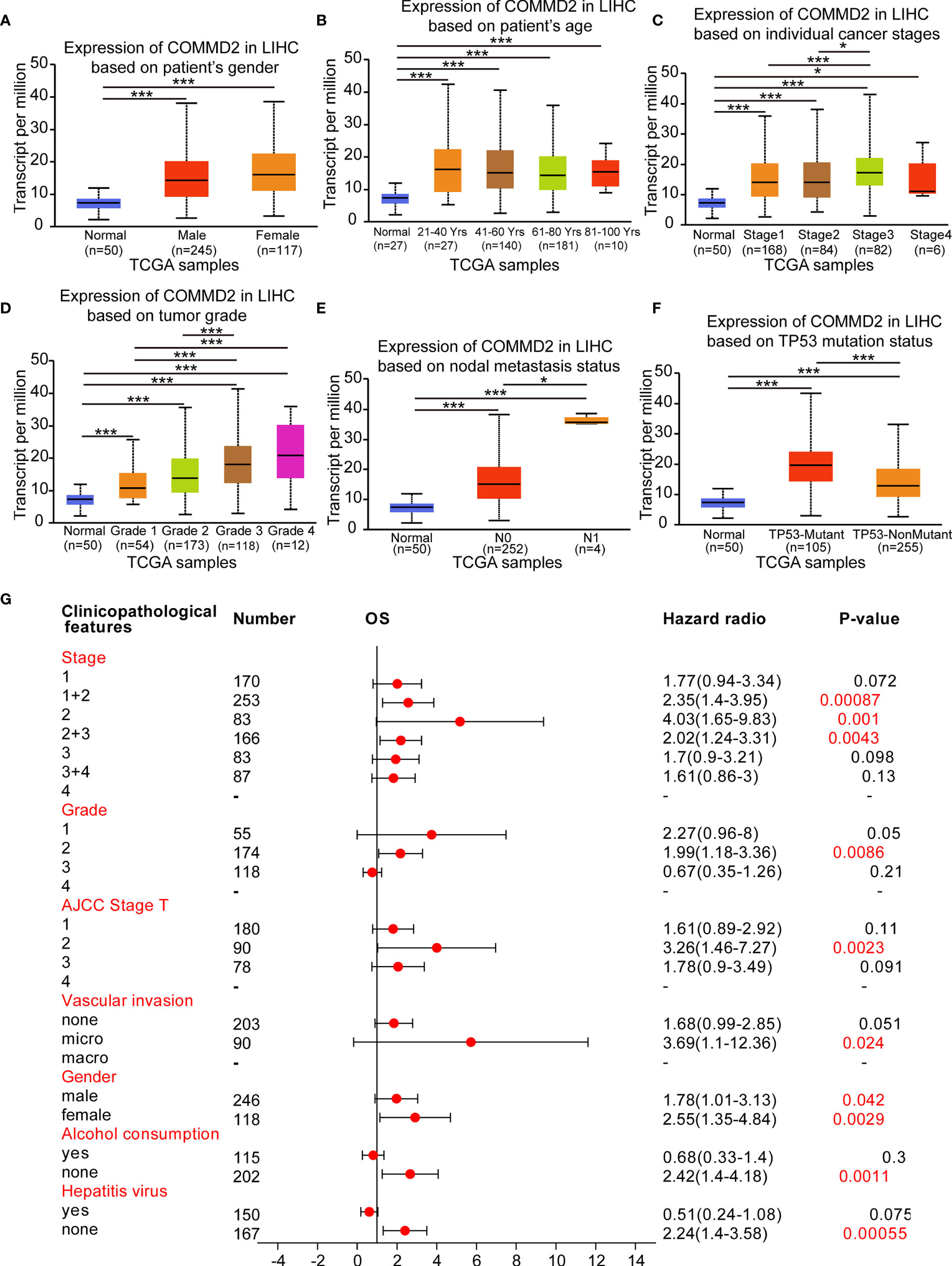
Figure 3 Association of COMMD2 expression with the clinicopathological features of LIHC patients. (A–F) Boxplot showing COMMD2 expression in normal individuals and LIHC patients based on clinicopathological features using the UALCAN database. Patient sex (A), patient age (B), cancer stage (C), tumor grade (D), nodal metastasis status (E) and TP53 mutation status parameters were analyzed (F). (G) Forest plot showing the association between COMMD2 expression and clinicopathological parameters of LIHC patients. *p < 0.05; ***p < 0.001.
Prediction and Analysis of miRNAs Upstream of COMMD2
Increasing evidence has shown that ncRNAs play key roles in the development of cancers by regulating gene expression. To determine whether COMMD2 is modulated by ncRNAs, the miRNAs binding upstream of COMMD2 were predicted by several target gene prediction programs, and 21 candidate miRNAs were ultimately identified (Figure 4A). The LIHC-associated miRNA-COMMD2 regulatory network was established using Cytoscape software (Figure 4B). According to the mechanism by which miRNAs generally negatively regulate downstream target genes, miRNAs and COMMD2 should be negatively correlated. Therefore, correlation analysis was conducted. COMMD2 was significantly negatively correlated with miR-29a-3p, miR-29b-3p and miR-29c-3p and positively correlated with miR-365b-5p in LIHC (Figure 4C). No significant relationships were observed between the expression of COMMD2 and other 17 predicted miRNAs. Finally, the expression and prognostic values of miR-29a-3p, miR-29b-3p, miR-29c-3p and miR-365b-5p in LIHC were determined. As presented in Figures 4D–G, miR-29a-3p, miR-29b-3p and miR-29c-3p were markedly downregulated in LIHC, but no significant upregulation of miR-365b-5p in LIHC was observed. Furthermore, only the high expression of miR-29c-3p was associated with a positive prognosis for LIHC patients (Figures 4H–K). Thus, miR-29c-3p might be the most promising miRNA that regulates COMMD2 in LIHC.
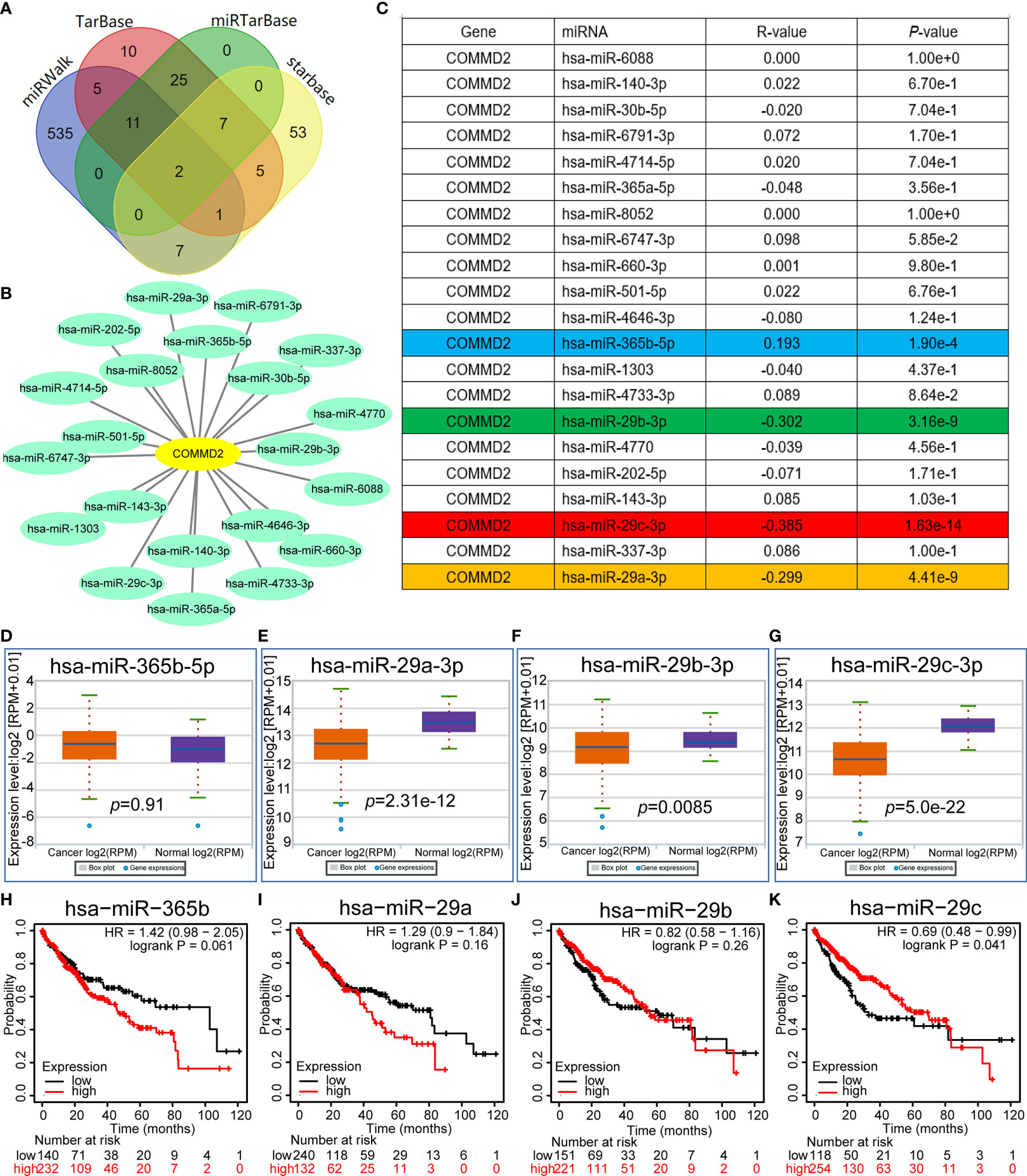
Figure 4 Prediction and analysis of miRNAs upstream of COMMD2. (A) miRNAs predicted to be upstream of COMMD2 using the miRWalk, TarBase, miRTarBase and starBase databases. (B) miRNA-COMMD2 regulatory network established using Cytoscape software. (C) Correlations between the predicted miRNAs and COMMD2 in LIHC as determined by the starBase database. (D–G) Expression of miR-365b-5p (D), miR-29a-3p (E), miR-29b-3p (F) and miR-29c-3p (G) in LIHC and normal tissue samples as determined by the starBase database. (H–K) Prognostic values of miR-365b-5p (H), miR-29a-3p (I), miR-29b-3p (J) and miR-29c-3p (K) in LIHC as assessed by Kaplan-Meier plotter.
Prediction and Analysis of LncRNAs Upstream of miR-29c-3p
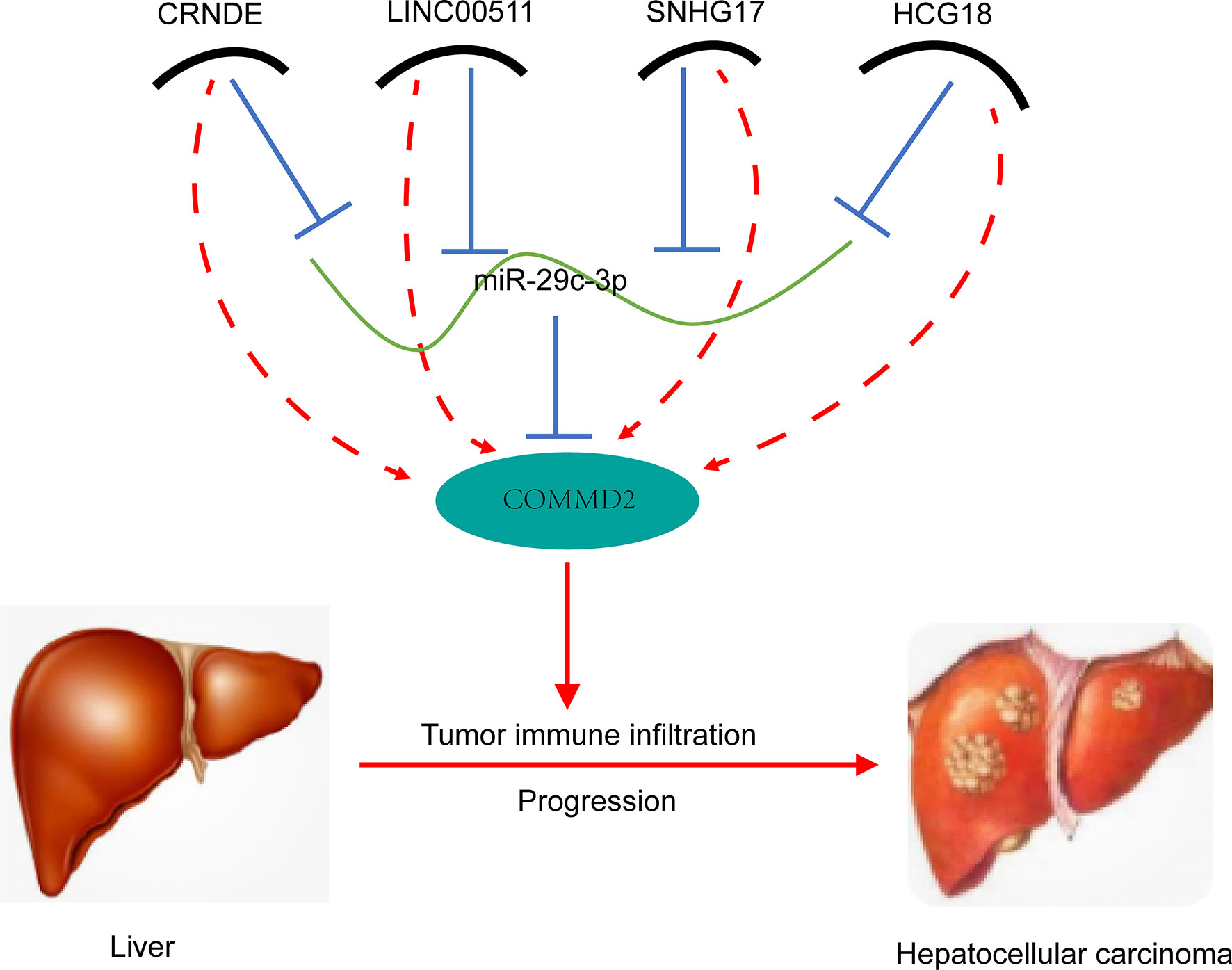
Next, the lncRNAs upstream of miR-29c-3p were predicted using the starBase database. Fifty-four possible lncRNAs were predicted, and an lncRNA-miR-29c-3p regulatory network was constructed using Cytoscape software (Supplementary Figure 3). Next, the expression of these lncRNAs in LIHC was determined using GEPIA. Among the 54 predicted lncRNAs, only CRNDE, LINC00511, SNHG17, and HCG18 were expressed at significantly higher levels in LIHC tissues than in normal tissues (Figures 5A–D). Subsequently, the associations between the four lncRNAs and prognosis of LIHC patients were evaluated. High expression of CRNDE, SNHG17, or HCG18 was significantly associated with both unfavorable OS and disease-free survival (DFS) in LIHC patients (Figures 5E–L). In addition, increased expression of LINC00511 indicated a poor OS. Additionally, to further confirm the expression of ncRNAs and COMMD2 in LIHC, RT-qPCR was performed in 10 pairs of fresh LIHC specimens and adjacent normal tissue. In accordance with our previous analytic data, miR-29c-3p was significantly decreased in LIHC tissue compared with those in adjacent normal tissue (Figure 6E). In contrast, the expression levels of COMMD2, CRNDE, LINC00511, SNHG17 and HCG18 were significantly increased in LIHC tissue compared with adjacent normal tissue (Figures 6A–D, F). Based on the known interactions of these ncRNAs in ceRNA networks, lncRNAs potentially promote mRNA expression by competitively binding to matched miRNAs. Thus, negative correlations between lncRNAs and miRNAs or positive correlations between lncRNAs and mRNAs should have been observed. Correlation analysis with the starBase database indicated a positive or negative relationship between each of the four lncRNAs, particularly SNHG17 and HCG18, and COMMD2 or miR-29c-3p (Figures 6H–O). By combining expression, survival, and correlation analysis, a CRNDE/LINC00511/SNHG17/HCG18-miR-29c-3p-COMMD2 ceRNA network was constructed (Figure 6G) and could potentially serve as a prognostic model in LIHC.
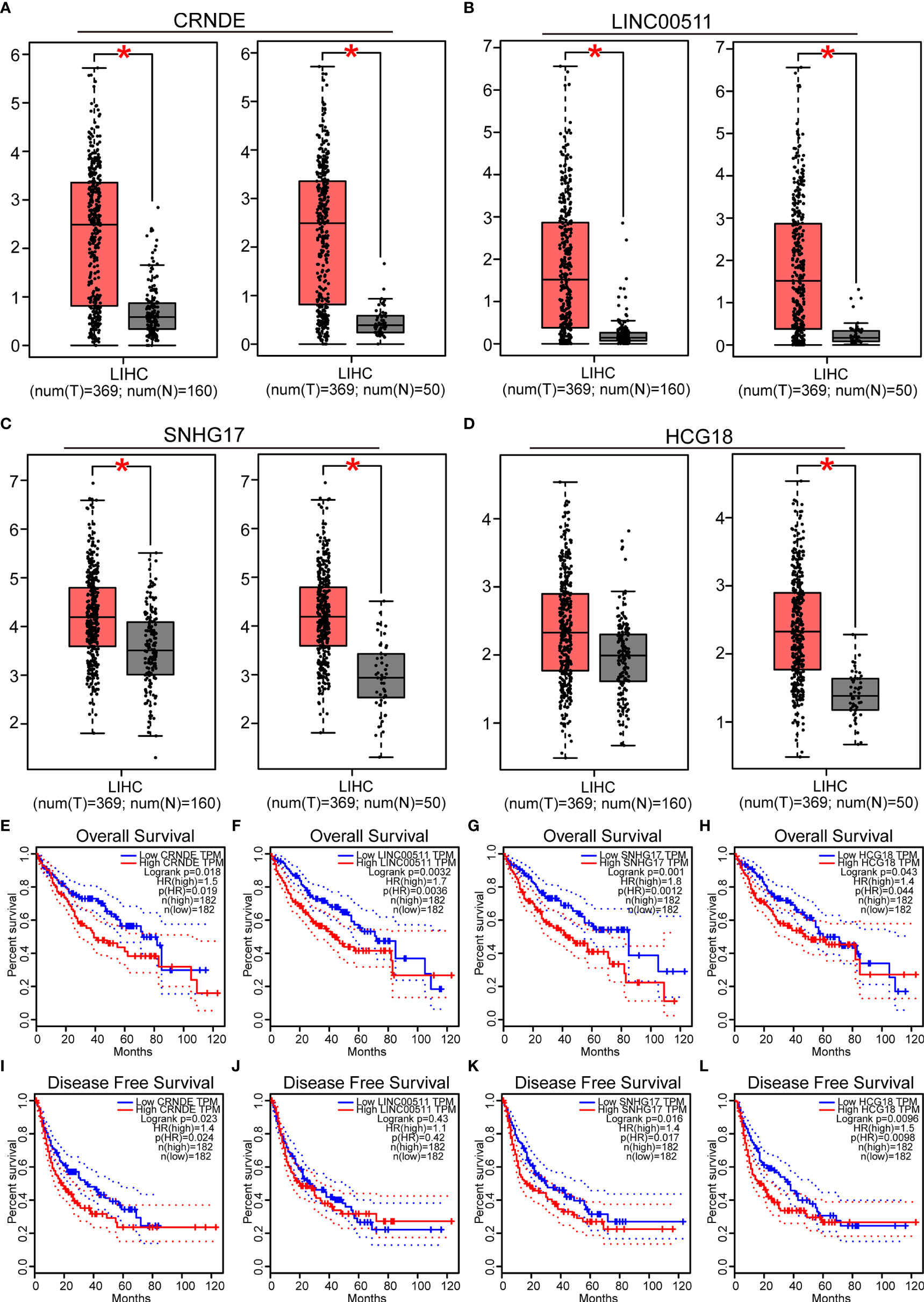
Figure 5 Prediction and analysis of lncRNAs upstream of miR-29c-3p. (A–D) Expression of CRNDE (A), LINC00511 (B), SNHG17 (C) and HCG18 (D) in LIHC data compared with that in “TCGA normal” or “TCGA and GTEx normal” data. (E–L) OS analysis of CRNDE (E), LINC00511 (F), SNHG17 (G) and HCG18 (H) in LIHC. RFS analysis of CRNDE (I), LINC00511 (J), SNHG17 (K) and HCG18 (L) in LIHC. ∗p value < 0.05.
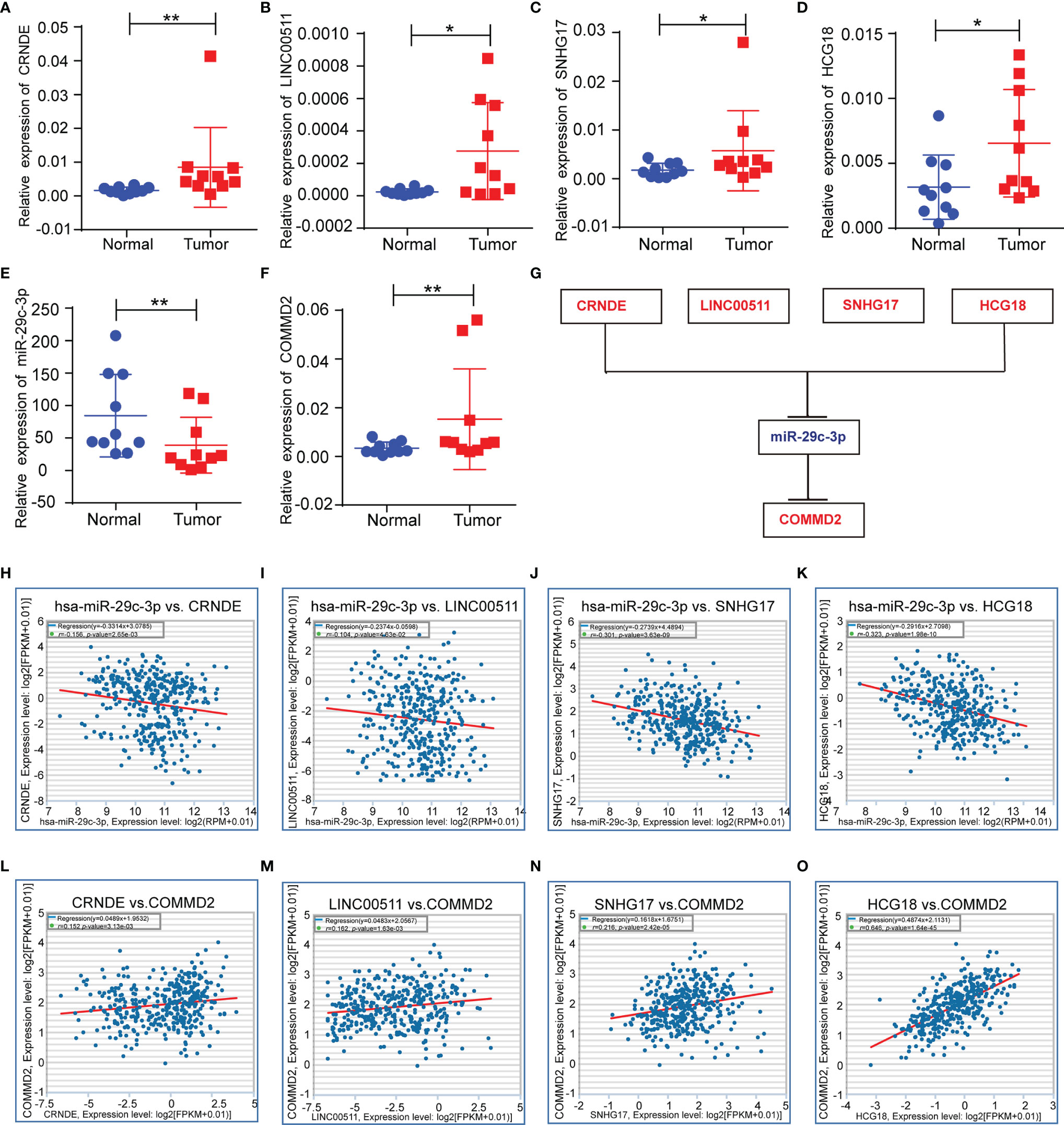
Figure 6 Correlations of lncRNAs with miR-29c-3p and COMMD2 in LIHC. (A–F) Expression of crucial ncRNAs and COMMD2 in fresh LIHC specimens and adjacent normal tissue as determined by RT-qPCR. (G) Schematic model of the ceRNA network. Red indicates upregulation and blue indicates downregulation. (H–K) Correlations of CRNDE (H), LINC00511 (I), SNHG17 (J) and HCG18 (K) with miR-29c-3p in LIHC. (L–O) Correlations of CRNDE (L), LINC00511 (M), SNHG17 (N) and HCG18 (O) with COMMD2 in LIHC. *p < 0.05; **p < 0.01.
Relationship Between COMMD2 and Immune Cell Infiltration in LIHC
COMMD2 is a member of the COMMD family, the members of which affect the prognosis of patients by participating in the inflammatory response and immune cell infiltration. Therefore, the relationships between COMMD2 and infiltrating immune cells were displayed using TIMER. Positive relationships were observed between the expression of COMMD2 and the infiltration of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Figures 7B–G). Moreover, COMMD2 copy number alterations could affect the infiltration level of six dominant immune cells, especially high amplification and arm-level deletion (Figure 7A). To further understand the association between COMMD2 expression and 22 immune cell types in the TCGA-LIHC cohort, we summarized the relative fractions of these immune cells in each LIHC patient using the CIBERSORT method (Figure 8A). Patients with high COMMD2 expression exhibited significantly higher proportions of M0 macrophages and neutrophils (P <0.05) and lower proportions of CD8 T cells (Figure 8B). Next, we used two gene expression omnibus (GEO) datasets to determine the above results and found that patients with high COMMD2 expression had a significant increase in the abundance of M0 macrophages, neutrophils, resting dendritic cells and resting mast cells and a significant decrease in the abundance of plasma cells and resting NK cells in GSE55092 (Figures 8C, D). In GSE107170, M0 macrophages, neutrophils, naïve B cells, activated memory CD4 T cells and resting dendritic cells increased and plasma cells decreased in patients with high COMMD2 expression (Figures 8E, F). Thus, high COMMD2 expression is significantly associated with higher proportions of M0 macrophages and neutrophils. All these findings suggested that COMMD2 is closely related to the level of immune infiltration, suggesting that COMMD2 may be involved in regulating LIHC tumor immunity.
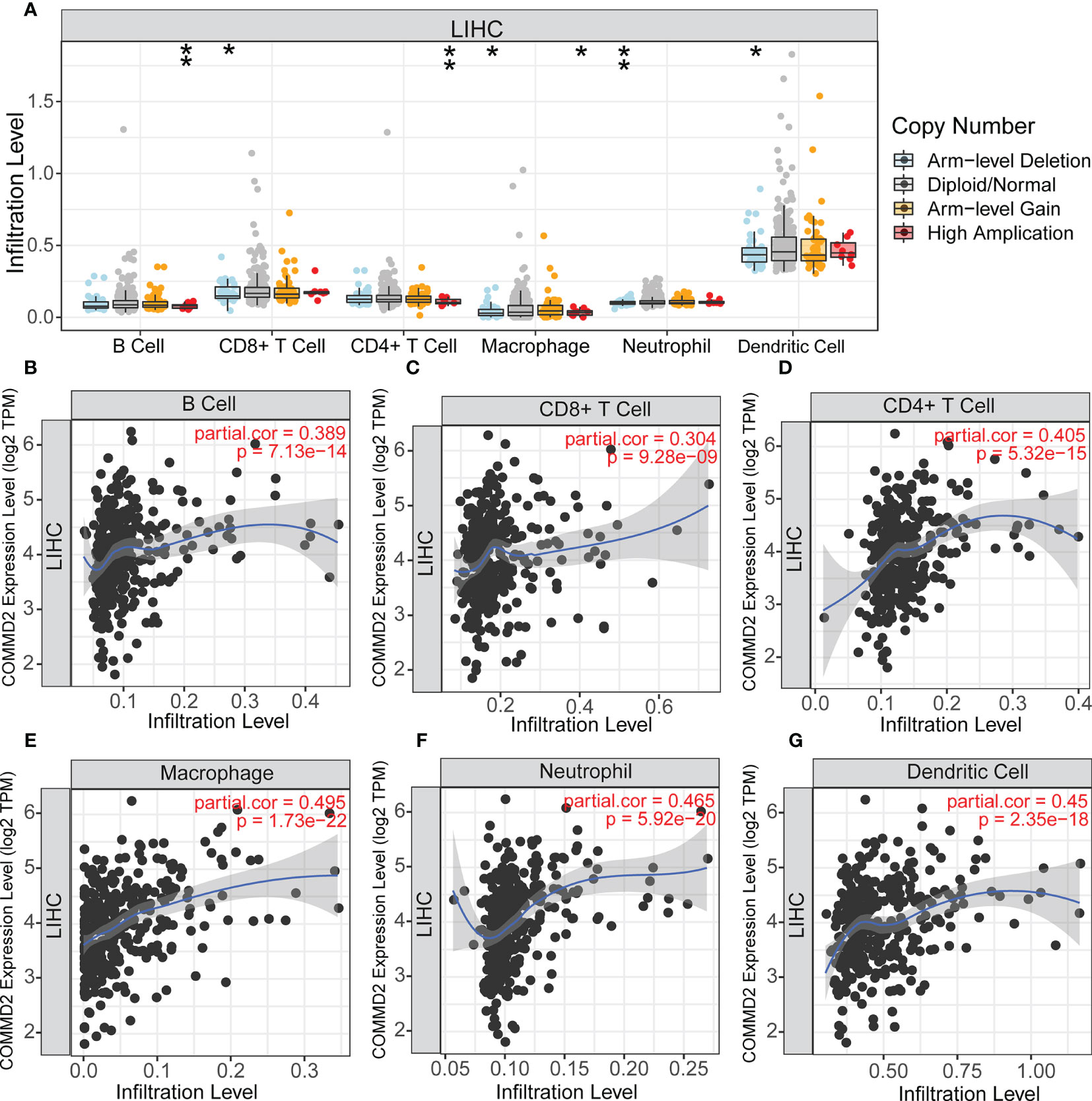
Figure 7 Relationship Between COMMD2 and immune cell infiltration in LIHC. (A) Relationship Between COMMD2 copy number alterations and the infiltration level of six dominant immune cells in LIHC. (B–G) Correlations of COMMD2 with infiltrating B cells (B), CD8+ T cells (C), CD4+ T cells (D), neutrophils (E), macrophages (F) and dendritic cells (G) in LIHC. *p < 0.05; **p < 0.01.
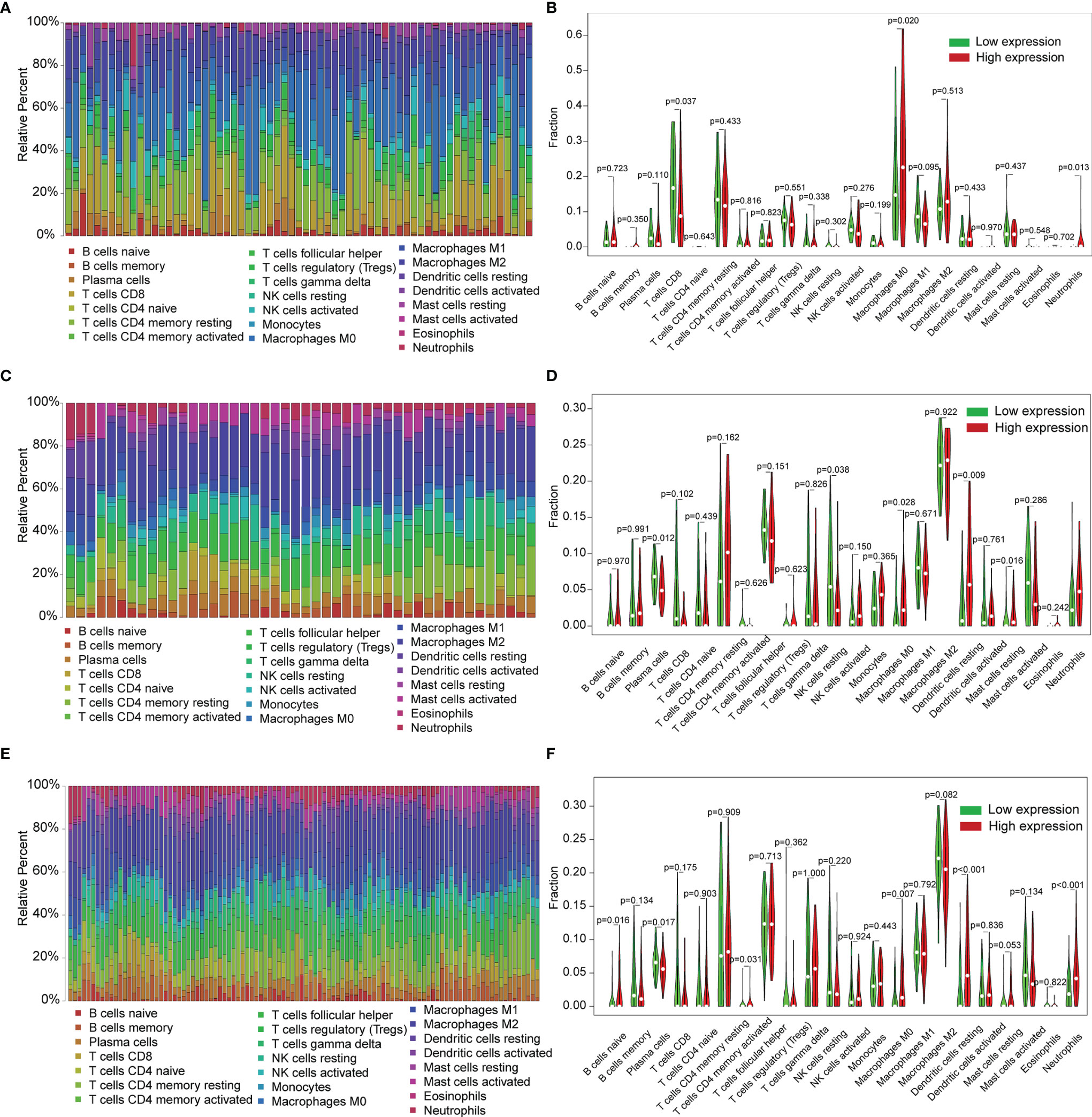
Figure 8 Relationship of COMMD2 expression with 22 immune cell types in LIHC based on CIBERSORT. (A, C, E) Relative fractions of 22 immune cell types in the TCGA-LIHC cohort (A), GSE55092 (C) and GSE107170 (E). (B, D, F) Violin plots of the difference in 22 immune cell types between patients with high and low COMMD2 expression from the TCGA-LIHC cohort (B), GSE55092 (D) and GSE107170 (F).
Prognostic Analysis of COMMD2 Expression Based on Immune Cells in LIHC
Because COMMD2 expression was significantly associated with immune cell infiltration and a poor prognosis in LIHC, we next investigated whether COMMD2 expression influences the OS of LIHC patients by regulating immune cell infiltration. We performed survival analyses of LIHC patients based on COMMD2 expression in related immune cell subgroups. As shown in Figure 9 and Supplementary Figure 4, different infiltration levels of B cells, CD4+ memory T cells, CD8+ T cells, macrophages, mesenchymal stem cells, regulatory T cells, type 1 T helper cells and type 2 T helper cells were found in LIHC patients, and those with high COMMD2 expression had a poor prognosis. These results indicate that COMMD2 may influence the OS of LIHC patients by regulating immune cell infiltration.
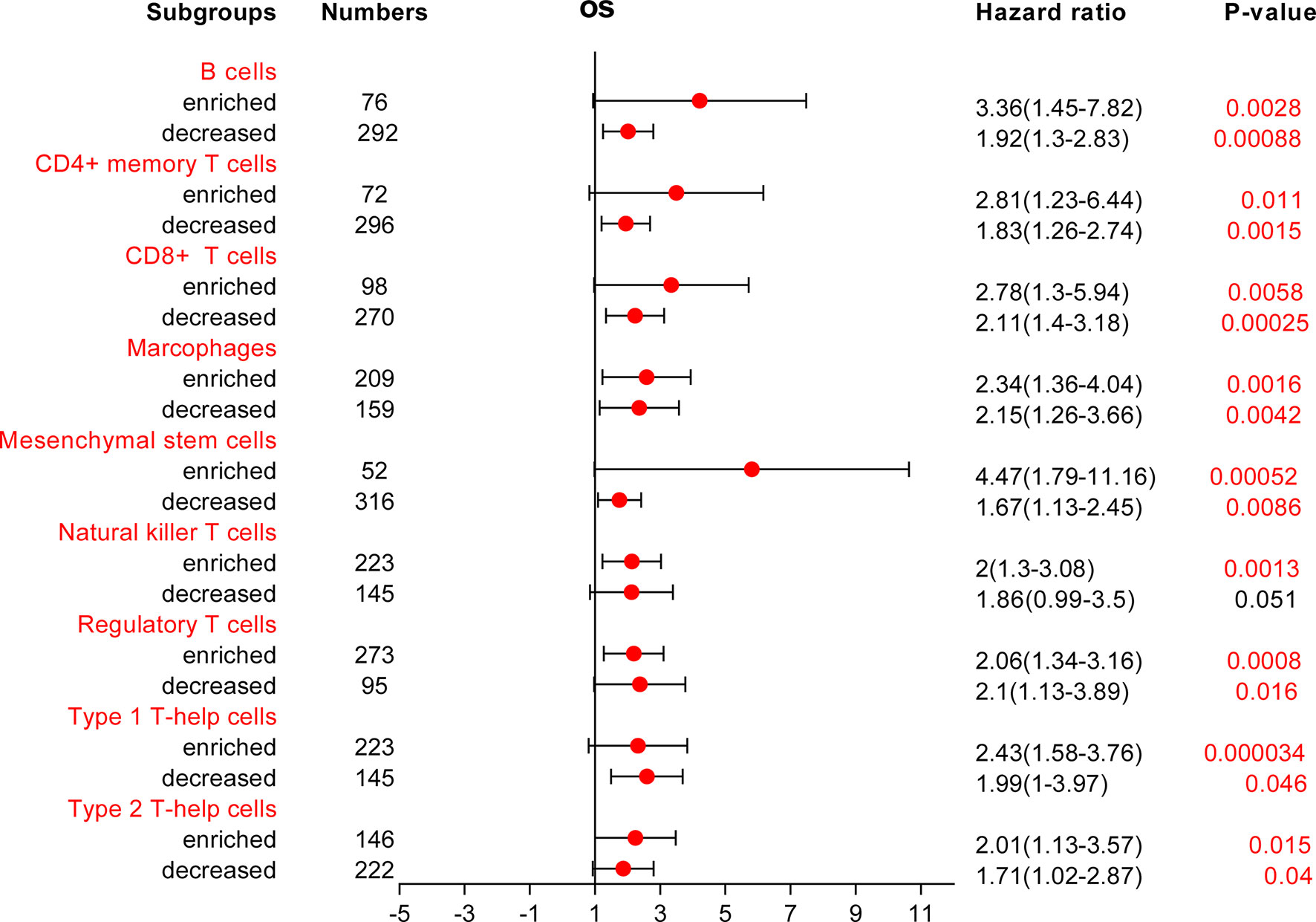
Figure 9 Forest plot of the prognostic value of COMMD2 based on different immune cell subgroups of LIHC patients.
Correlations of COMMD2 With Immune Cell Biomarkers in LIHC
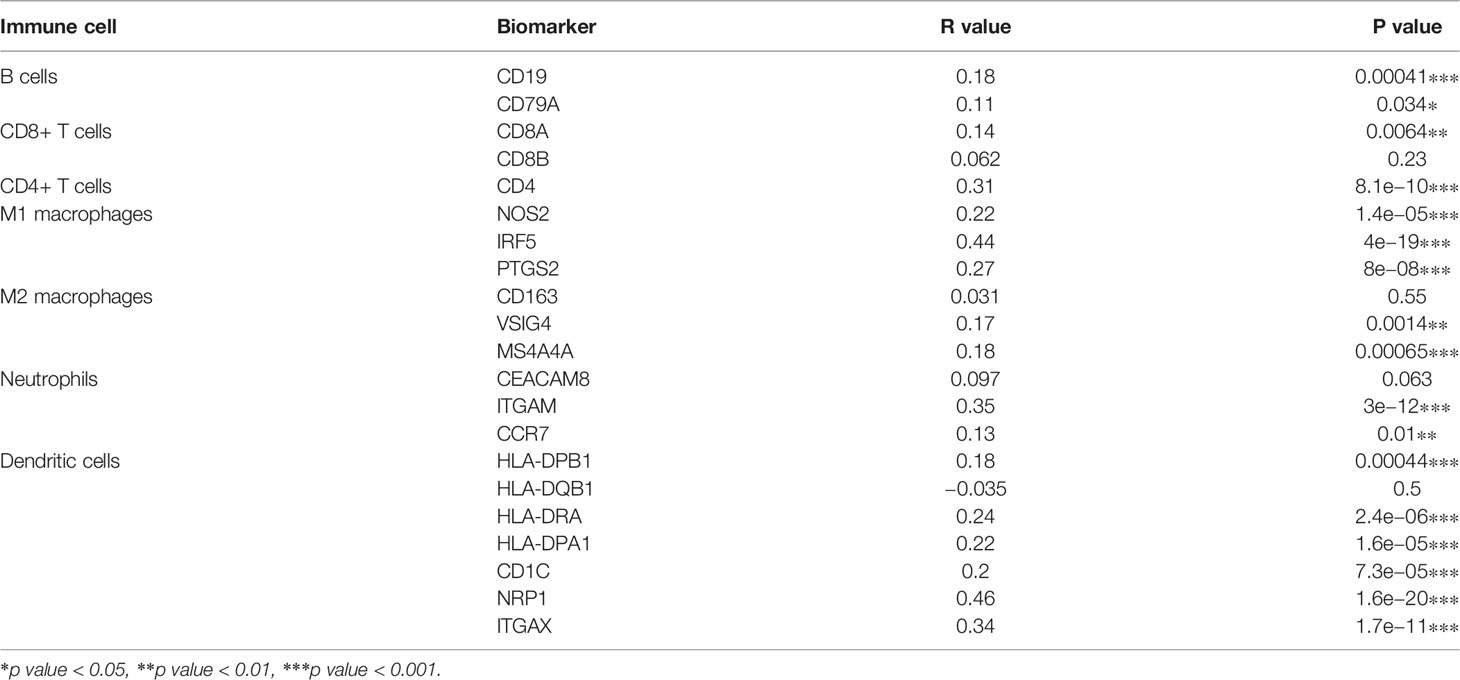
To further validate the role of COMMD2 in antitumor immunity, we explored the relationships between COMMD2 and immune cell biomarkers in LIHC using the GEPIA database. As listed in Table 2, COMMD2 was significantly positively correlated with most immune markers in various immune cell types, including B cells, CD8+ T cells, CD4+ T cells, M1 macrophages, M2 macrophages, neutrophils and dendritic cells. These results further support that COMMD2 is positively related to tumor immune cell infiltration in LIHC.
Correlations Between COMMD2 and Immune Checkpoint Molecules in LIHC
As crucial immune checkpoint molecules, programmed death-1 (PD1), programmed death-ligand 1 (PD-L1) and cytotoxic T ymphocyte antigen-4 (CTLA-4) play important roles in tumor immune escape. Based on the potential oncogenic role of COMMD2 in LIHC, the correlations of COMMD2 with PD1, PD-L1 and CTLA-4 were estimated. COMMD2 was significantly positively correlated with PD1, PD-L1 and CTLA-4 in LIHC (Figures 10A–C). Similar results were found using the GEPIA database, which revealed significant positive correlations of COMMD2 with PD1, PD-L1 or CTLA 4 in LIHC (Figures 10D–F). These results demonstrate that COMMD2 may be involved in tumor immune escape during LIHC tumorigenesis.
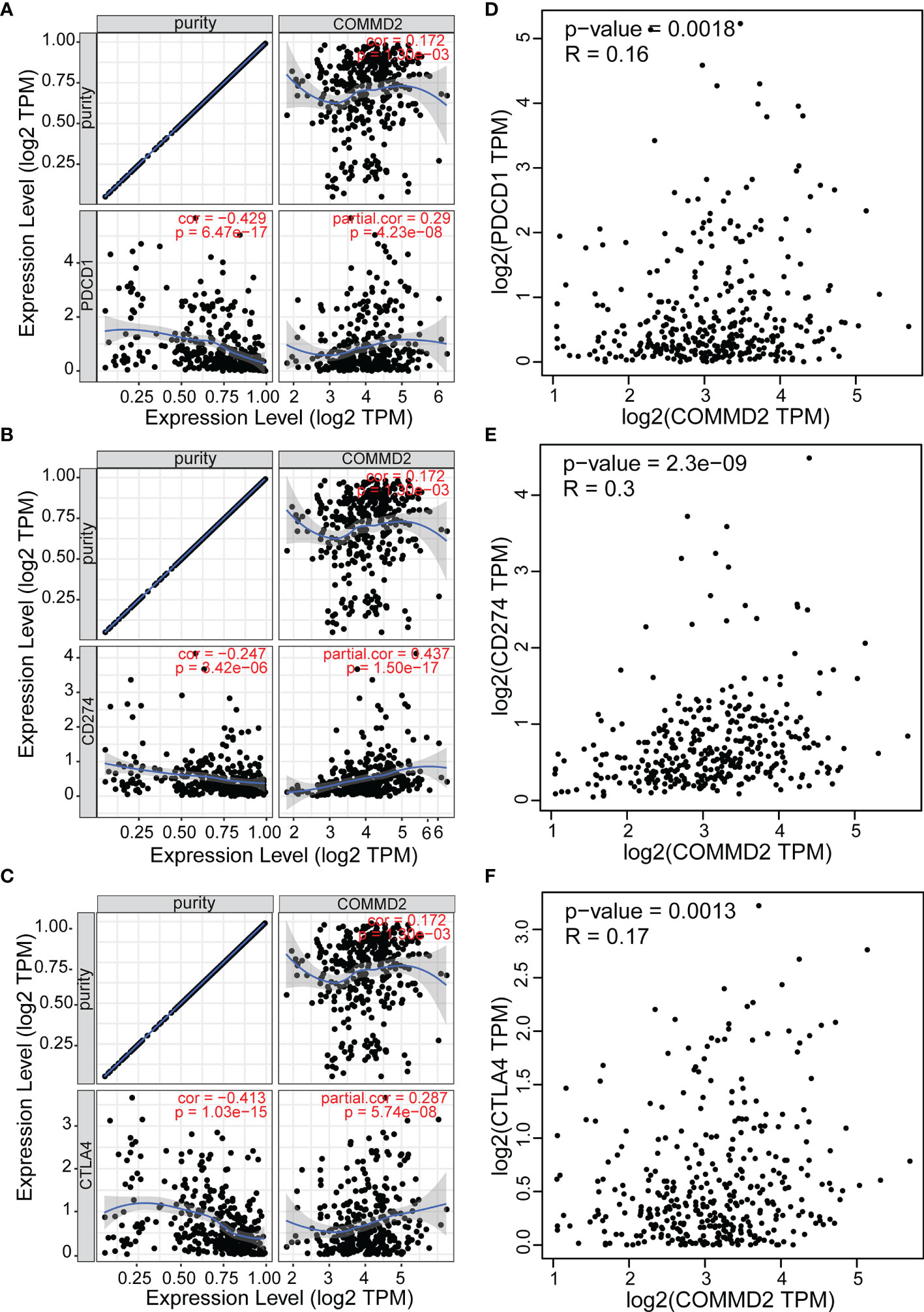
Figure 10 Correlations between COMMD2 and PD-1, PD-L1 and CTLA-4 in LIHC. (A-C) Correlations of COMMD2 with PD-1 (A), PD-L1 (B) and CTLA-4 (C) in LIHC adjusted for purity using TIMER. (D-F) Relationships of COMMD2 with PD-1 (D), PD-L1 (E) and CTLA-4 (F) in LIHC as determined using the GEPIA database.
Effects of COMMD2 Knockdown on the Proliferation and Migration of LIHC Cells In Vitro
To assess the function of COMMD2 in LIHC, we knocked down its expression in HuH-7 and MHCC97-H cells using siRNAs, and the silencing efficiency was determined by RT-qPCR (Figures 11A, B). CCK8 and colony formation assays were performed to explore the effect of COMMD2 knockdown on LIHC cell proliferation, revealing that the proliferation of HuH-7 and MHCC97-H cells was significantly decreased after COMMD2 downregulation (Figures 11C–F). Subsequently, to investigate the impacts of COMMD2 knockdown on LIHC cell migration ability, wound healing and transwell assays were performed, demonstrating that COMMD2 knockdown drastically decreased the migration ability of HuH-7 and MHCC97-H cells compared with that of control group cells (Figures 11G–K).
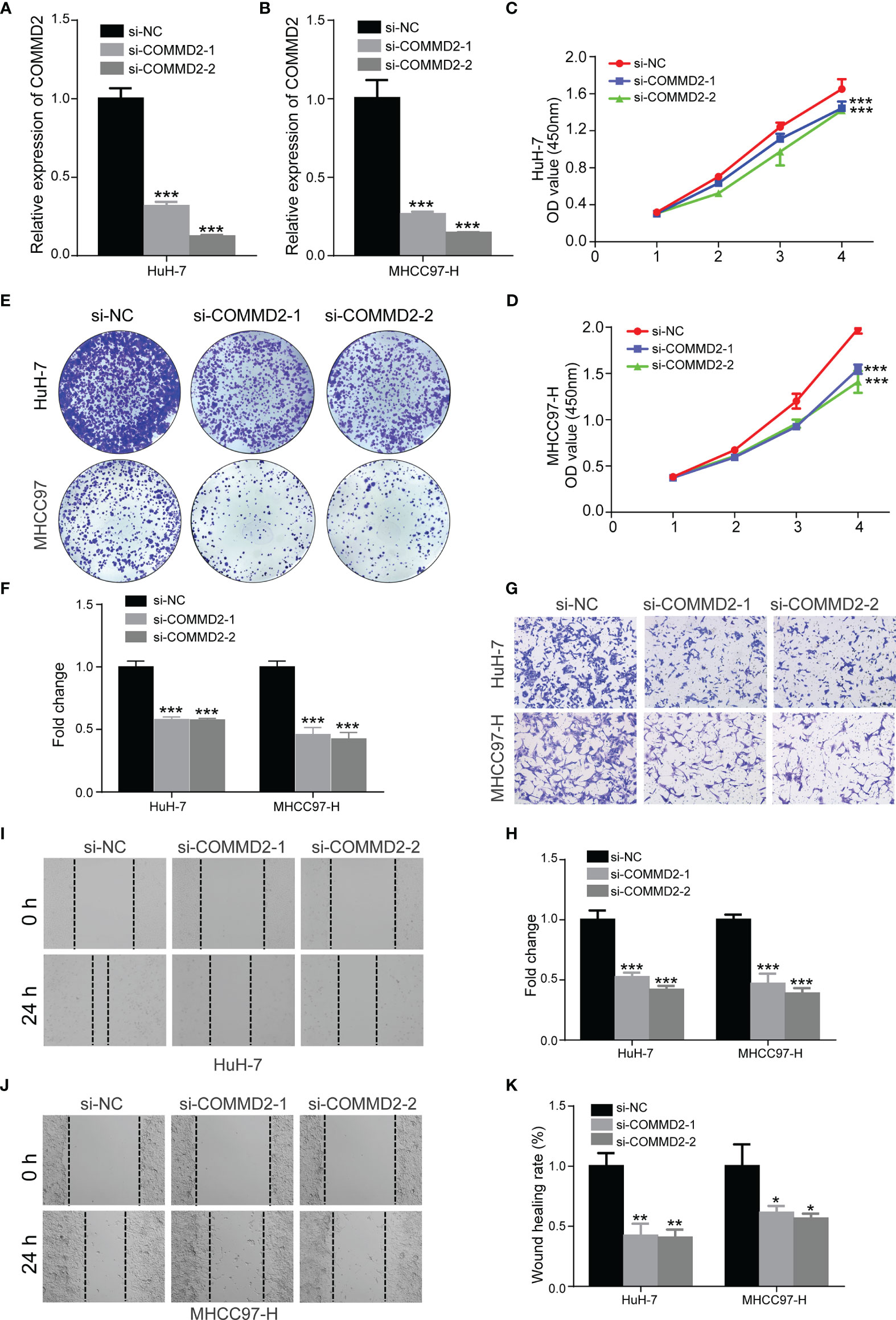
Figure 11 Effects of COMMD2 knockdown on the proliferation and migration of LIHC cells. (A, B) The efficiency of COMMD2 downregulation in HuH-7 (A) and MHCC97-H (B) cells after COMMD2 siRNA transfection was detected by RT-qRCR. (C–F) Effect of COMMD2 knockdown on the proliferation of HuH-7 and MHCC97-H cells as determined by CCK8 (C, D) and colony formation assays (E, F). (G–K) Effect of COMMD2 knockdown on the migration of HuH-7 and MHCC97-H cells as detected by transwell (G, H) and wound healing assays (I–K). Scale bar=50 μm. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
LIHC is the third leading cause of cancer-related mortality worldwide (1). Although various therapeutic strategies have been adopted for LIHC patients, their efficacies remain unsatisfactory (23). Identifying novel biomarkers of malignant LIHC is essential to identify new effective therapeutic targets and improve LIHC patient prognosis. Increasing evidence has demonstrated that COMMD proteins play key roles in the development and progression of multiple human cancers (12, 13), including LIHC. However, the underlying mechanisms and clinical value of COMMD2 and its correlation with immune cell infiltration in LIHC remain unknown.
In the present study, we first performed pancancer analysis of COMMD2 expression using the TIMER and UALCAN databases, and found that COMMD2 was abnormally expressed in the tumor tissues of 13 different cancer types compared with that in the corresponding normal tissues. Association analyses of COMMD2 with the survival of patients with candidate types of cancer indicated that high COMMD2 expression was associated with a poor prognosis in LIHC. Clinical association analyses demonstrated that increased COMMD2 expression was correlated with higher histological grade, more advanced clinical stage, lymph node metastasis and the TP53 mutation status in LIHC patients. Furthermore, COMMD2 knockdown suppresses LIHC cell proliferation and migration in vitro via a series of functional assays.
Previous studies have revealed that ncRNAs, particularly miRNAs, lncRNAs, and circular RNAs, are involved in the development and progression of tumors through gene regulation mechanisms involving ceRNA regulatory networks (24–27). To explore the upstream miRNAs that modulate COMMD2 expression, we used four prediction programs to predict miRNAs that potentially bind COMMD2 and ultimately identified 21 miRNAs. Most of these miRNAs play suppressor roles in LIHC. For example, miR−29b-3p regulates the TGF−β1 and p53 signaling pathways to inhibit the growth and induce the apoptosis of LIHC ascites H22 cells (28). miR-29a-3p inhibits cell proliferation and migration by targeting PTEN and thereby regulating the NF-kappaB pathway in LIHC (29). miR-29c-3p inhibits tumor progression by regulating the methylation of DNMT3B and LATS1 in LIHC (30). Among the 21 identified candidate miRNAs, only miR-29c-3p was expressed at a low level, which was negatively correlated with the high expression of COMMD2 and associated with a better prognosis for LIHC patients as determined by the combination of expression, correlation, and survival analyses. Thus, miR-29c-3p was selected as the most promising upstream miRNA of COMMD2. Previous studies also showed that miR-29c-3p inhibits LIHC proliferation (31).
According to the ceRNA hypothesis (32), the potential lncRNAs upstream of the miR-29c-3p/COMMD2 axis should be oncogenic lncRNAs in LIHC. Subsequently, lncRNAs upstream of the miR-29c-3p/COMMD2 axis were also predicted, ultimately identifying 54 possible lncRNAs. By performing expression, correlation, and survival analyses, CRNDE, LINC00511, SNHG17 and HCG18 were identified as the most promising upregulated lncRNAs. High expression of CRNDE, LINC00511, SNHG17 and HCG18, which have positive and negative relationships with COMMD2 and miR-29c-3p, respectively, was associated with a poor prognosis in LIHC. The four lncRNAs functioned as oncogenes in multiple tumors, including LIHC. For example, the lncRNA CRNDE facilitates the proliferation, invasion, migration and chemoresistance of LIHC (33–36). LINC00511 promotes malignant cell behaviors and correlates with prognosis in LIHC (37–39). The lncRNA SNHG17 promotes cell proliferation and migration and predicts a poor prognosis in LIHC (40, 41). The lncRNA HCG18 contributes to the progression of LIHC (42, 43). Thus, the CRNDE/LINC00511/SNHG17/HCG18-miR-29c-3p-COMMD2 axis regulates the development and progression of LIHC.
Numerous recent studies have confirmed that tumor immune cell infiltration influences tumor angiogenesis and the prognosis of patients with LIHC (44–48). Herein, we found that COMMD2 was significantly positively correlated with various infiltrating immune cells in LIHC, especially M0 macrophages and neutrophils by various database. Moreover, COMMD2 was closely related to immune markers of these tumor-infiltrating immune cells. Importantly, COMMD2 was shown to influence the OS of LIHC patients through immune cell infiltration. These findings indicate that tumor immune infiltration may partially explain the carcinogenic effect of COMMD2 in LIHC. In addition, immune checkpoint molecules, including PD-1, PD-L1, and CTLA-4, are associated with the prognosis of LIHC patients (49–51). Checkpoint inhibitors (CPIs) targeting PD-1, PD-L1, or CTLA-4 have led to clinical breakthroughs in oncological treatment (52–55). Thus, we also assessed the relationships between COMMD2 and immune checkpoint molecules. High COMMD2 expression was significantly linked to PD1, PD-L1 and CTLA-4 levels in LIHC, suggesting that targeting COMMD2 might enhance immunotherapeutic efficacy in LIHC.
However, some limitations in our study should be considered. First, our finding is mainly relies on public databases, more data and larger LIHC cohorts were required to validate its clinical suitability. Second, the role of COMMD2 in tumor immune infiltration needs to be further confirmed in vitro or in vivo. Finally, the carcinogenic mechanism of the CRNDE/LINC00511/SNHG17/HCG18-miR-29c-3p-COMMD2 axis in LIHC requires more functional studies to elucidate. Therefore, further investigations, including basic experiments and clinical trials, are needed to perform in the future.
In conclusion, our results indicate a carcinogenic effect of COMMD2 and its potential as a novel prognostic biomarker in LIHC. Furthermore, we further elucidated the underlying oncogenic mechanism of COMMD2 by constructing a CRNDE/LINC00511/SNHG17/HCG18-miR-29c-3p ceRNA network in LIHC (Figure 12). Additionally, our study showed that COMMD2 might play a cancer-promoting role by regulating tumor immune cell infiltration in patients with LIHC. Therefore, these findings provide a potentially valuable target for LIHC prognosis and immunotherapy.
Publisher’s Note
All the claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, editors and reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LZ and JX designed this work. WF conducted the experiments and collected the data. WF and YG analyzed the data. WF and LZ drafted the manuscript. JX revised the manuscript. All the authors approved the final version of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81760431 and 81860427) and the Natural Science Foundation of Jiangxi Province, China (grant number 20161BAB205243).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.853026/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib Versus Sorafenib in First-Line Treatment of Patients With Unresectable Hepatocellular Carcinoma: A Randomised Phase 3 non-Inferiority Trial. Lancet (2018) 391:1163–73. doi: 10.1016/S0140-6736(18)30207-1
3. Pinter M, Scheiner B, Peck-Radosavljevic M. Immunotherapy for Advanced Hepatocellular Carcinoma: A Focus on Special Subgroups. Gut (2021) 70:204–14. doi: 10.1136/gutjnl-2020-321702
4. Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, et al. Global Trends and Predictions in Hepatocellular Carcinoma Mortality. J Hepatol (2017) 67:302–9. doi: 10.1016/j.jhep.2017.03.011
5. Villanueva A. Hepatocellular Carcinoma. N Engl J Med (2019) 380:1450–62. doi: 10.1056/NEJMra1713263
6. Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, et al. COMMD Proteins, a Novel Family of Structural and Functional Homologs of MURR1. J Biol Chem (2005) 280:22222–32. doi: 10.1074/jbc.M501928200
7. Zoubeidi A, Ettinger S, Beraldi E, Hadaschik B, Zardan A, Klomp LW, et al. Clusterin Facilitates COMMD1 and I-kappaB Degradation to Enhance NF-kappaB Activity in Prostate Cancer Cells. Mol Cancer Res (2010) 8:119–30. doi: 10.1158/1541-7786.MCR-09-0277
8. Zhan W, Wang W, Han T, Xie C, Zhang T, Gan M, et al. COMMD9 Promotes TFDP1/E2F1 Transcriptional Activity via Interaction With TFDP1 in non-Small Cell Lung Cancer. Cell Signal (2017) 30:59–66. doi: 10.1016/j.cellsig.2016.11.016
9. Yang SS, Li XM, Yang M, Ren XL, Hu JL, Zhu XH, et al. FMNL2 Destabilises COMMD10 to Activate NF-kappaB Pathway in Invasion and Metastasis of Colorectal Cancer. Br J Cancer (2017) 117:1164–75. doi: 10.1038/bjc.2017.260
10. Zheng L, Liang P, Li J, Huang XB, Liu SC, Zhao HZ, et al. ShRNA-Targeted COMMD7 Suppresses Hepatocellular Carcinoma Growth. PloS One (2012) 7:e45412. doi: 10.1371/journal.pone.0045412
11. Yang M, Huang W, Sun Y, Liang H, Chen M, Wu X, et al. Prognosis and Modulation Mechanisms of COMMD6 in Human Tumours Based on Expression Profiling and Comprehensive Bioinformatics Analysis. Br J Cancer (2019) 121:699–709. doi: 10.1038/s41416-019-0571-x
12. van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E, et al. COMMD1 Disrupts HIF-1alpha/Beta Dimerization and Inhibits Human Tumor Cell Invasion. J Clin Invest (2010) 120(6):2119–30. doi: 10.1172/JCI40583
13. Fedoseienko A, Wieringa HW, Wisman GB, Duiker E, Reyners AK, Hofker MH, et al. Nuclear COMMD1 Is Associated With Cisplatin Sensitivity in Ovarian Cancer. PloS One (2016) 11(10):e0165385. doi: 10.1371/journal.pone.0165385
14. Zheng L, You N, Huang X, Gu H, Wu K, Mi N, et al. COMMD7 Regulates NF-kappaB Signaling Pathway in Hepatocellular Carcinoma Stem-Like Cells. Mol Ther Oncol (2019) 12:112–23. doi: 10.1016/j.omto.2018.12.006
15. You N, Li J, Huang X, Wu K, Tang Y, Wang L, et al. COMMD7 Promotes Hepatocellular Carcinoma Through Regulating CXCL10. BioMed Pharmacother (2017) 88:653–7. doi: 10.1016/j.biopha.2017.01.046
16. Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, et al. MIQE Précis: Practical Implementation of Minimum Standard Guidelines for Fluorescence-Based Quantitative Real-Time PCR Experiments. BMC Mol Biol (2010) 11:74. doi: 10.1186/1471-2199-11-74
17. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77:e108–10. doi: 10.1158/0008-5472.CAN-17-0307
18. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust Enumeration of Cell Subsets From Tissue Expression Profiles. Nat Methods (2015) 12:453–7. doi: 10.1038/nmeth.3337
19. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
20. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45:W98–W102. doi: 10.1093/nar/gkx247
21. Li JH, Liu S, Zhou H, Qu LH, Yang JH. Starbase V2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks From Large-Scale CLIP-Seq Data. Nucleic Acids Res (2014) 42:D92–97. doi: 10.1093/nar/gkt1248
22. Lanczky A, Gyorffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res (2021) 23(7):e27633. doi: 10.2196/27633
23. Deng Y, Zhu J, Liu Z, Huang M, Chang DW, Gu J. Elevated Systemic Inflammatory Responses, Factors Associated With Physical and Mental Quality of Life, and Prognosis of Hepatocellular Carcinoma. Aging (Albany NY) (2020) 12:4357–70. doi: 10.18632/aging.102889
24. Zhan M, He K, Xiao J, Liu F, Wang H, Xia Z, et al. LncRNA HOXA11-AS Promotes Hepatocellular Carcinoma Progression by Repressing miR-214-3p. J Cell Mol Med (2018) 22:3758–67. doi: 10.1111/jcmm.13633
25. Lou W, Ding B, Wang J, Xu Y. The Involvement of the Hsa_Circ_0088494-miR-876-3p-CTNNB1/CCND1 Axis in Carcinogenesis and Progression of Papillary Thyroid Carcinoma. Front Cell Dev Biol (2020) 8:605940. doi: 10.3389/fcell.2020.605940
26. Gao S, Ding B, Lou W. microRNA-Dependent Modulation of Genes Contributes to ESR1's Effect on ERalpha Positive Breast Cancer. Front Oncol (2020) 10:753. doi: 10.3389/fonc.2020.00753
27. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in Cancer: Possible Functions and Clinical Implications. J Med Genet (2015) 52:710–8. doi: 10.1136/jmedgenet-2015-103334
28. Liu YL, Yang WH, Chen BY, Nie J, Su ZR, Zheng JN, et al. Mir29b Suppresses Proliferation and Induces Apoptosis of Hepatocellular Carcinoma Ascites H22 Cells via Regulating TGFbeta1 and P53 Signaling Pathway. Int J Mol Med (2021) 48:157. doi: 10.3892/ijmm.2021.4990
29. Ma JH, Bu X, Wang JJ, Xie YX. MicroRNA-29-3p Regulates Hepatocellular Carcinoma Progression Through NF-kappaB Pathway. Clin Lab (2019) 65:801–6. doi: 10.7754/Clin.Lab.2018.181012
30. Wu H, Zhang W, Wu Z, Liu Y, Shi Y, Gong J, et al. miR-29c-3p Regulates DNMT3B and LATS1 Methylation to Inhibit Tumor Progression in Hepatocellular Carcinoma. Cell Death Dis (2019) 10:48. doi: 10.1038/s41419-018-1281-7
31. Lv T, Jiang L, Kong L, Yang J. MicroRNA29c3p Acts as a Tumor Suppressor Gene and Inhibits Tumor Progression in Hepatocellular Carcinoma by Targeting TRIM31. Oncol Rep (2020) 43:953–64. doi: 10.3892/or.2020.7469
32. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
33. Wang H, Ke J, Guo Q, Barnabo Nampoukime KP, Yang P, Ma K. Long non-Coding RNA CRNDE Promotes the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cells Through miR-217/MAPK1 Axis. J Cell Mol Med (2018) 22:5862–76. doi: 10.1111/jcmm.13856
34. Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, et al. LncRNA CRNDE Facilitates Epigenetic Suppression of CELF2 and LATS2 to Promote Proliferation, Migration and Chemoresistance in Hepatocellular Carcinoma. Cell Death Dis (2020) 11:676. doi: 10.1038/s41419-020-02853-8
35. Ji D, Jiang C, Zhang L, Liang N, Jiang T, Yang B, et al. LncRNA CRNDE Promotes Hepatocellular Carcinoma Cell Proliferation, Invasion, and Migration Through Regulating miR-203/ BCAT1 Axis. J Cell Physiol (2019) 234:6548–60. doi: 10.1002/jcp.27396
36. Chen L, Sun L, Dai X, Li T, Yan X, Zhang Y, et al. LncRNA CRNDE Promotes ATG4B-Mediated Autophagy and Alleviates the Sensitivity of Sorafenib in Hepatocellular Carcinoma Cells. Front Cell Dev Biol (2021) 9:687524. doi: 10.3389/fcell.2021.687524
37. Hu WY, Wei HY, Li KM, Wang RB, Xu XQ, Feng R. LINC00511 as a ceRNA Promotes Cell Malignant Behaviors and Correlates With Prognosis of Hepatocellular Carcinoma Patients by Modulating miR-195/EYA1 Axis. BioMed Pharmacother (2020) 121:109642. doi: 10.1016/j.biopha.2019.109642
38. Wang RP, Jiang J, Jiang T, Wang Y, Chen LX. Increased Long Noncoding RNA LINC00511 is Correlated With Poor Prognosis and Contributes to Cell Proliferation and Metastasis by Modulating miR-424 in Hepatocellular Carcinoma. Eur Rev Med Pharmacol Sci (2019) 23:3291–301. doi: 10.26355/eurrev_201904_17691
39. Peng X, Li X, Yang S, Huang M, Wei S, Ma Y, et al. LINC00511 Drives Invasive Behavior in Hepatocellular Carcinoma by Regulating Exosome Secretion and Invadopodia Formation. J Exp Clin Cancer Res (2021) 40:183. doi: 10.1186/s13046-021-01990-y
40. Ma T, Zhou X, Wei H, Yan S, Hui Y, Liu Y, et al. Long Non-Coding RNA SNHG17 Upregulates RFX1 by Sponging miR-3180-3p and Promotes Cellular Function in Hepatocellular Carcinoma. Front Genet (2020) 11:607636. doi: 10.3389/fgene.2020.607636
41. Zhu XM, Li L, Ren LL, Du L, Wang YM. LncRNA SNHG17 Predicts Poor Prognosis and Promotes Cell Proliferation and Migration in Hepatocellular Carcinoma. Eur Rev Med Pharmacol Sci (2021) 25:4219–27. doi: 10.26355/eurrev_202106_26127
42. Zou Y, Sun Z, Sun S. LncRNA HCG18 Contributes to the Progression of Hepatocellular Carcinoma via miR-214-3p/CENPM Axis. J Biochem (2020) 168:535–46. doi: 10.1093/jb/mvaa073
43. Zhang L, Wang Z, Li M, Sun P, Bai T, Wang W, et al. HCG18 Participates in Vascular Invasion of Hepatocellular Carcinoma by Regulating Macrophages and Tumor Stem Cells. Front Cell Dev Biol (2021) 9:707073. doi: 10.3389/fcell.2021.707073
44. Zhang J, Chang L, Zhang X, Zhou Z, Gao Y. Meta-Analysis of the Prognostic and Clinical Value of Tumor-Associated Macrophages in Hepatocellular Carcinoma. J Invest Surg (2021) 34:297–306. doi: 10.1080/08941939.2019.1631411
45. Fu J, Zhang Z, Zhou L, Qi Z, Xing S, Lv J, et al. Impairment of CD4+ Cytotoxic T Cells Predicts Poor Survival and High Recurrence Rates in Patients With Hepatocellular Carcinoma. Hepatology (2013) 58:139–49. doi: 10.1002/hep.26054
46. Yu S, Wang Y, Hou J, Li W, Wang X, Xiang L, et al. Tumor-Infiltrating Immune Cells in Hepatocellular Carcinoma: Tregs is Correlated With Poor Overall Survival. PloS One (2020) 15:e0231003. doi: 10.1371/journal.pone.0231003
47. Xu X, Tan Y, Qian Y, Xue W, Wang Y, Du J, et al. Clinicopathologic and Prognostic Significance of Tumor-Infiltrating CD8+ T Cells in Patients With Hepatocellular Carcinoma: A Meta-Analysis. Med (Baltimore) (2019) 98:e13923. doi: 10.1097/MD.0000000000013923
48. Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. Interaction Between Tumour-Infiltrating B Cells and T Cells Controls the Progression of Hepatocellular Carcinoma. Gut (2017) 66:342–51. doi: 10.1136/gutjnl-2015-310814
49. Gao Q, Wang XY, Qiu SJ, Yamato I, Sho M, Nakajima Y, et al. Overexpression of PD-L1 Significantly Associates With Tumor Aggressiveness and Postoperative Recurrence in Human Hepatocellular Carcinoma. Clin Cancer Res (2009) 15:971–9. doi: 10.1158/1078-0432.CCR-08-1608
50. Umemoto Y, Okano S, Matsumoto Y, Nakagawara H, Matono R, Yoshiya S, et al. Prognostic Impact of Programmed Cell Death 1 Ligand 1 Expression in Human Leukocyte Antigen Class I-Positive Hepatocellular Carcinoma After Curative Hepatectomy. J Gastroenterol (2015) 50:65–75. doi: 10.1007/s00535-014-0933-3
51. Chang H, Jung W, Kim A, Kim HK, Kim WB, Kim JH, et al. Expression and Prognostic Significance of Programmed Death Protein 1 and Programmed Death Ligand-1, and Cytotoxic T Lymphocyte-Associated Molecule-4 in Hepatocellular Carcinoma. APMIS (2017) 125:690–8. doi: 10.1111/apm.12703
52. Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab Versus Docetaxel for Previously Treated, PD-L1-Positive, Advanced non-Small-Cell Lung Cancer (KEYNOTE-010): A Randomised Controlled Trial. Lancet (2016) 387:1540–50. doi: 10.1016/S0140-6736(15)01281-7
53. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab Versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med (2015) 373:1627–39. doi: 10.1056/NEJMoa1507643
54. Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, et al. Atezolizumab Versus Docetaxel for Patients With Previously Treated non-Small-Cell Lung Cancer (POPLAR): A Multicentre, Open-Label, Phase 2 Randomised Controlled Trial. Lancet (2016) 387:1837–46. doi: 10.1016/S0140-6736(16)00587-0
55. Chae YK, Arya A, Iams W, Cruz MR, Chandra S, Choi J, et al. Current Landscape and Future of Dual Anti-CTLA4 and PD-1/PD-L1 Blockade Immunotherapy in Cancer; Lessons Learned From Clinical Trials With Melanoma and non-Small Cell Lung Cancer (NSCLC). J Immunother Cancer (2018) 6:39. doi: 10.1186/s40425-018-0349-3
Keywords: COMMD2, CRNDE, LINC00511, SNHG17, HCG18, miR-29c-3p, tumor immune infiltration, liver hepatocellular carcinoma
Citation: Fang W, Gan Y, Zhang L and Xiong J (2022) COMMD2 Upregulation Mediated by an ncRNA Axis Correlates With an Unfavorable Prognosis and Tumor Immune Infiltration in Liver Hepatocellular Carcinoma. Front. Oncol. 12:853026. doi: 10.3389/fonc.2022.853026
Received: 12 January 2022; Accepted: 28 March 2022;
Published: 29 April 2022.
Edited by:
Alessandro Passardi, Scientific Institute of Romagna for the Study and Treatment of Tumors (IRCCS), ItalyReviewed by:
Louis Charles Penning, Utrecht University, NetherlandsZiheng Wang, Affiliated Hospital of Nantong University, China
Copyright © 2022 Fang, Gan, Zhang and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Zhang, bmR5ZnkxMzA2QG5jdS5lZHUuY24=; Jianping Xiong, anB4aW9uZzA2MzBAb3V0bG9vay5jb20=
 Weidan Fang
Weidan Fang Yu Gan
Yu Gan Ling Zhang
Ling Zhang Jianping Xiong
Jianping Xiong