
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 03 March 2022
Sec. Thoracic Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.846536
This article is part of the Research Topic Prognostic Factors in Non-Small Cell Lung Cancer View all 37 articles
 Xiulin Jiang1†
Xiulin Jiang1† Lin Tang2†
Lin Tang2† Yixiao Yuan2†
Yixiao Yuan2† Juan Wang2†
Juan Wang2† Dahang Zhang2
Dahang Zhang2 Kebao Qian2
Kebao Qian2 William C. Cho3*
William C. Cho3* Lincan Duan2*
Lincan Duan2*Background: Hyaluronan-mediated motility receptor (HMMR) plays a pivotal role in cell proliferation in various cancers, including lung cancer. However, its function and biological mechanism in lung adenocarcinoma (LUAD) remain unclear.
Methods: Data on HMMR expression from several public databases were extensively analyzed, including the prognosis of HMMR in the Gene Expression Profiling Interactive Analysis (GEPIA) database. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed using DAVID and gene set enrichment analysis (GSEA) software. The correlation between HMMR expression and immune cell infiltration was analyzed in the Tumor Immune Estimation Resource (TIMER) database, and the gene and protein networks were examined using the GeneMANIA and STRING databases. Experimentally, the expression of HMMR in LUAD and lung cancer cell lines was determined using immunohistochemistry and quantitative RT-PCR assays. Besides, the function of HMMR on cancer cell proliferation and migration was examined using cell growth curve and colony formation, Transwell, and wound healing assays.
Results: In this study, we found that HMMR was elevated in LUAD and that its high expression was associated with poor clinicopathological features and adverse outcomes in LUAD patients. Furthermore, our results demonstrated that the expression of HMMR was positively correlated with immune cell infiltration and immune modulation. Interestingly, diverse immune cell infiltration affects the prognosis of LUAD. In the functional assay, depletion of HMMR significantly repressed the cancer cell growth and migration of LUAD. Mechanically, we found that that the DNA methylation/TMPO-AS1/let-7b-5p axis mediated the high expression of HMMR in LUAD. Depletion of TMPO-AS1 and overexpression of let-7b-5p could result in the decreased expression of HMMR in LUAD cells. Furthermore, we found that TMPO-AS1 was positively correlated with HMMR, yet negatively correlated with let-7b-5p expression in LUAD.
Conclusions: Our findings elucidated that the DNA methylation/TMPO-AS1/let-7b-5p axis mediated the high expression of HMMR, which may be considered as a biomarker to predict prognosis in LUAD.
Lung cancer is one of the main causes of cancer death and has brought huge public health burden worldwide (1). Lung cancer mainly consists of non-small cell lung cancer (NSCLC) and small cell lung cancer. NSCLCs include lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell carcinoma (1, 2). Owing to the lack of effective diagnostic markers, most patients with LUAD are diagnosed in the advanced stage and thus might miss the best treatment opportunities (2–4). Therefore, it is imperative to identify useful biomarkers for the treatment of lung cancer.
In a previous study, we developed a new method called cross-value association analysis (CVAA), which functions without a normalization and distribution assumption. We applied this method to large-scale pan-cancer transcriptome data generated by The Cancer Genome Atlas (TCGA) project and successfully discovered numerous new differentially expressed genes (DEGs). Hyaluronan-mediated motility receptor (HMMR) is one of these DEGs (5). HMMR, also named as RHAMM (receptor for hyaluronan-mediated motility), plays a pivotal role in cell proliferation (6). Studies have shown that HMMR was mainly expressed in the nervous system (7). Mutations in HMMR might cause neurodevelopmental defects (8). Studies also showed that a high HMMR expression has been associated with various cancers, such as breast cancer (9), colorectal cancer (10), stomach cancer (11), endometrial cancer (12), and prostate cancer (13). A previous study indicated that HMMR was essential to maintain the stemness of glioblastoma stem cells (14). He et al. found that HMMR was upregulated in bladder cancer and correlated with poor prognosis. Knockdown of HMMR significantly inhibited bladder cancer growth, invasion, epithelial-to-mesenchymal transition, and inactivation of the Wnt/β-catenin signaling pathway (15). In addition, Fan et al. found that the messenger RNA (mRNA) expression of HMMR was significantly increased in hepatocellular carcinoma (HCC) tissues and also correlated with the histologic grade, pathological stage, and survival status. Univariate and multivariate analyses indicated that HMMR is an independent predictive factor associated with overall survival (OS) in HCC. However, the biological function and potential mechanisms of HMMR in LUAD progression and immune response regulation remain to be elucidated.
In this study, we used Tumor Immune Estimation Resource (TIMER), Gene Expression Profiling Interactive Analysis (GEPIA), the Cancer Cell Line Encyclopedia (CCLE), and the Kaplan–Meier (KM) plotter database to explore the expression level, clinical significance, diagnosis, and prognostic value of HMMR in LUAD. Furthermore, we used the TIMER and GEPIA databases to examine the relationship between HMMR and tumor-infiltrating immune cells in the tumor microenvironment. Moreover, we further explored potential HMMR dysregulation via analysis of the upstream long non-coding RNAs (lncRNAs)/microRNAs (miRNAs). Finally, immunohistochemistry (IHC), Western blot, quantitative real-time PCR (qRT-PCR), growth curve, and colony formation, Transwell, and wound healing assays were used to study HMMR in LUAD progression. Our findings underline the vital role of HMMR in LUAD. Also, we provide an underlying mechanism of HMMR expression in potentially regulating the infiltration of immune cells, partly affecting the prognosis of LUAD.
TCGA-LUAD cohort data and the corresponding clinical information of 535 LUAD patients were downloaded from TCGA database (https://portal.gdc.cancer.gov/repository). LUAD patients were classified into low and high HMMR expression groups according to the median HMMR expression value. The gene expression profiles were normalized using the scale method provided in the “limma” R package. Data analysis was performed with R (version 3.6.3) and the ggplot2 (3.3.3) package. The expression data were normalized to transcripts per kilobase million (TPM) values before further analysis. In addition, a receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value of HMMR using the R packages pROC and ggplot2.
GEPIA (http://gepia.cancer-pku.cn/index.html) is a user-friendly web portal for gene expression analysis based on TCGA and GTEx data (16). In the current study, the CCLE (https://sites.broadinstitute.org/ccle) and GEPIA (http://gepia.cancer-pku.cn/) databases were used to analyze the expression and prognostic value of HMMR in pan-cancer. Furthermore, we used GEPIA to study the correlation between HMMR expression and pathological stage.
We used KM plotter (http://kmplot.com), an online database that contains gene expression data and the survival information of 3,452 clinical lung cancer patients, to analyze the prognostic value of HMMR in lung cancer (17). The patient samples were divided into two groups based on the median expression (high expression and low expression) to analyze the OS, with hazard ratios (HRs), 95% confidence intervals (95% CIs), and log-rank p-values.
TIMER (https://cistrome.shinyapps.io/timer/) is a comprehensive resource for systematic analysis of immune infiltrates across diverse cancer types. In this study, TIMER was used to examine the correlation between the somatic copy number alterations of HMMR and the immune cell infiltration levels of B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs). We also used the R package GSVA to quantify the LUAD immune infiltration of 24 tumor-infiltrating immune cells in tumor samples through single-sample gene set enrichment analysis (ssGSEA). According to the 509 gene signatures of 24 tumor-infiltrating lymphocytes (TILs) (18), comprising natural killer (NK) cells, T follicular helper (Tfh) cells, CD56bright NK cells, CD56dim NK cells, central memory CD4+ T cells, macrophages, cytotoxic cells, DCs, CD8+ B cells, effector memory T (Tem) cells, eosinophils, gamma delta T cells, activated DCs (aDCs), immature DCs (iDCs), mast cells, neutrophils, plasmacytoid DCs (pDCs), T helper cells, regulatory T cells (Tregs), type 1 T helper cells (Thp1), Th2, and Th17, the relative enrichment score of every immunocyte was quantified. The correlation between HMMR and the infiltration levels of immune cells was analyzed with Spearman’s correlation, and immune cells with different expression groups of HMMR were analyzed using the Wilcoxon rank-sum test.
Univariate and multivariate Cox regression analyses were performed to examine the prognostic value of HMMR in LUAD.
In the present study, we utilized the LinkedOmics database (http://www.linkedomics.org/login.php) to study the co-expression genes of HMMR in LUAD. The gene set “kegg.v6.2.symbols.gmt,” which served as a reference gene set, was downloaded from the Molecular Signatures Database (MSigDB) (http://software.broadinstitute.org/gsea/msigdb). We utilized the GSEA software and clusterProfiler package to perform the GO and KEGG enrichment analyses of the signaling pathways of HMMR in LUAD (19–21).
The GeneMANIA database (http://www.genemania.org) was utilized to construct the HMMR interaction network (22), and the STRING online database (https://string-db.org/) was employed to construct the protein–protein interaction (PPI) network of HMMR (23).
We used starBase (http://starbase.sysu.edu.cn/) to predict the potential upstream miRNAs of HMMR and to examine the expression, prognosis, and correlation between let-7b-5p and lncRNA. starBase was also used to predict the binding sites among the miRNAs, mRNAs, and lncRNAs (24). The lncLocator (www.csbio.sjtu.edu.cn/bioinf/lncLocator) is a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier, and CPC2 (http://cpc2.cbi.pku.edu.cn) is a fast and accurate coding potential calculator based on sequence intrinsic features (25, 26). In this study, lncLocator and CPC2 were used to explore the subcellular localization and the protein-coding ability of TMPO-AS1, respectively.
The human bronchial epithelial cell line (BEAS2B) and the LUAD cell lines were purchased from the Cell Bank of Kunming Institute of Zoology and cultured in bronchial epithelial cell growth medium (BEGM) (CC-3170; Lonza, Basel, Switzerland). The HEK-293T cell line was obtained from the American Type Culture Collection (ATCC). The lung cancer cell lines A549, H1299, and H1975 were purchased from Cobioer (Nanjing, China) with short tandem repeat (STR) document. A549, H1299, and H1975 cells were all cultured in RPMI 1640 medium (Corning, Corning, NY, USA) supplemented with 10% fetal bovine serum (cat. no. 10099141C; Gibco, Waltham, MA, USA) and 1% penicillin/streptomycin. HEK-293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning). The short hairpin RNA (shRNA) for HMMR was constructed using pLKO.1 vector. The shRNA for the HMMR primer sequences are as follows: HMMR shRNA#1: AAACAGCTGGAAGATGAAGAAGGAA; HMMR shRNA#2: CAGCTGGAAGATGAAGAAGGAAGAA.
The qRT-PCR assay was performed as described (27). For real-time reverse transcription PCR (RT-PCR) assay, the indicated cells were lysed with RNAiso Plus (cat. no. 108-95-2; Takara Bio, Beijing, China). Total RNA was extracted according to the manufacturer’s protocol and then reverse transcribed using the RT reagent kit. Real-time PCR was performed with the FastStart Universal SYBR Green Master Mix (cat. no. 04194194001; Roche, Basel, Switzerland; cat. no. FP411-02; TIANGEN Biotech, Beijing, China) using an Applied Biosystems 7500 machine. The primer sequences are as follows: HMMR-F: ATGATGGCTAAGCAAGAAGGC, HMMR-R: TTTTCCCTTGAGACTCTTCGAGA; β-actin-F: CTTCGCGGGCGACGAT, β-actin-R: CCATAGGAATCCTTCTGACC. Expression quantification was calculated with the 2−ΔΔCt method.
The cancer cell migration and invasion abilities were assessed with the Transwell assay (28). For the cell proliferation assay, the indicated cells were plated into 12-well plates at a density of 2 × 104. The cell numbers were subsequently counted each day using the automatic cell analyzer Countstar (IC1000; Shanghai RuiYu Biotech Co., Shanghai, China). For the colony formation assay, 500 cells were seeded in a six-well plate at 500 cells/well supplemented with 2 ml cell culture medium. The cell culture medium was changed every 3 days for 2–3 weeks. Indicated cells were fixed with 4% paraformaldehyde (PFA) and stained with 0.5% crystal violet.
The migration ability of the indicated cells was evaluated by the wound healing and Transwell assays. For the wound healing assay, cells were plated in six-well plates at 1 × 106 cells/well in 2 ml culture medium. After 24 h, a wound was scratched on the adherent cell monolayer with an Eppendorf tip. Wounds were imaged at 5–10 positions along each well. For the Transwell assay, 2 × 104 cells in serum-free medium were plated on uncoated insets and incubated using 24-well chemotaxis chambers (Corning cell culture inserts, 8-μm pore size). Fetal bovine serum (10%) was added into the bottom wells of the chambers as a chemoattractant. The cells were allowed to migrate through the membrane for an indicated time. Non-migrating cells were removed and cells migrating to the lower face were stained with cresyl violet (Sigma, St. Louis, MO, USA). Stained cells in the entire field were counted under an inverted microscope.
Western blotting and the immunohistochemistry staining assay were performed as described (29). Briefly, the cell lysates were collected, Western blot was performed, and the primary antibody (HMMR) was incubated overnight. The secondary antibody was also incubated. Finally, western blot develop using protein developing instrument conducted. The detail information of antibodies employ in our study are as follows: HMMR antibody (CST Group (HMMR, Rabbit mAb #87129, 1:1,000) and β-actin.
For the datasets from TCGA, statistical analyses were performed using R (v.3.6.3). The Wilcoxon rank-sum test and the chi-square test were used to estimate the association between HMMR and the clinicopathological characteristics. The Kaplan–Meier method was used to calculate the survival rates of LUAD patients. Cox univariate and multivariate analyses were performed to assess the correlation between clinical features and OS, disease-specific survival (DSS), and progression-free survival (PFS). GraphPad Prism 7.0 was used for statistical analysis of data regarding the function of HMMR. Student’s t-test evaluated the statistical significance between experimental groups, and multiple group comparisons were analyzed using one-way ANOVA. Values of *p < 0.05, **p < 0.01, and ***p < 0.001 were considered significant.
TIMER tools were utilized to examine the expression of HMMR in multifarious cancer. The results indicated that HMMR was elevated in pan-cancer, including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), LUAD, lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) (Figure 1A). To further verify the results, we used GEPIA database analysis and found that HMMR was significantly overexpressed in adrenocortical carcinoma (ACC), BLCA, BRCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), ESCA, glioblastoma multiforme (GBM), HNSC, LIHC, LUAD, LUSC, ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), READ, skin cutaneous melanoma (SKCM), STAD, thymoma (THYM), UCEC, and uveal melanoma (UVM) compared to matched normal tissues. HMMR showed low expression in acute myeloid leukemia (LAML) and TGCT (Figure 1B). Finally, we found that HMMR was highly expressed in diverse cancer cell lines, including LUAD cell lines (Figure 1C). Primarily, these results confirm that HMMR may play a crucial role in human cancer progression.
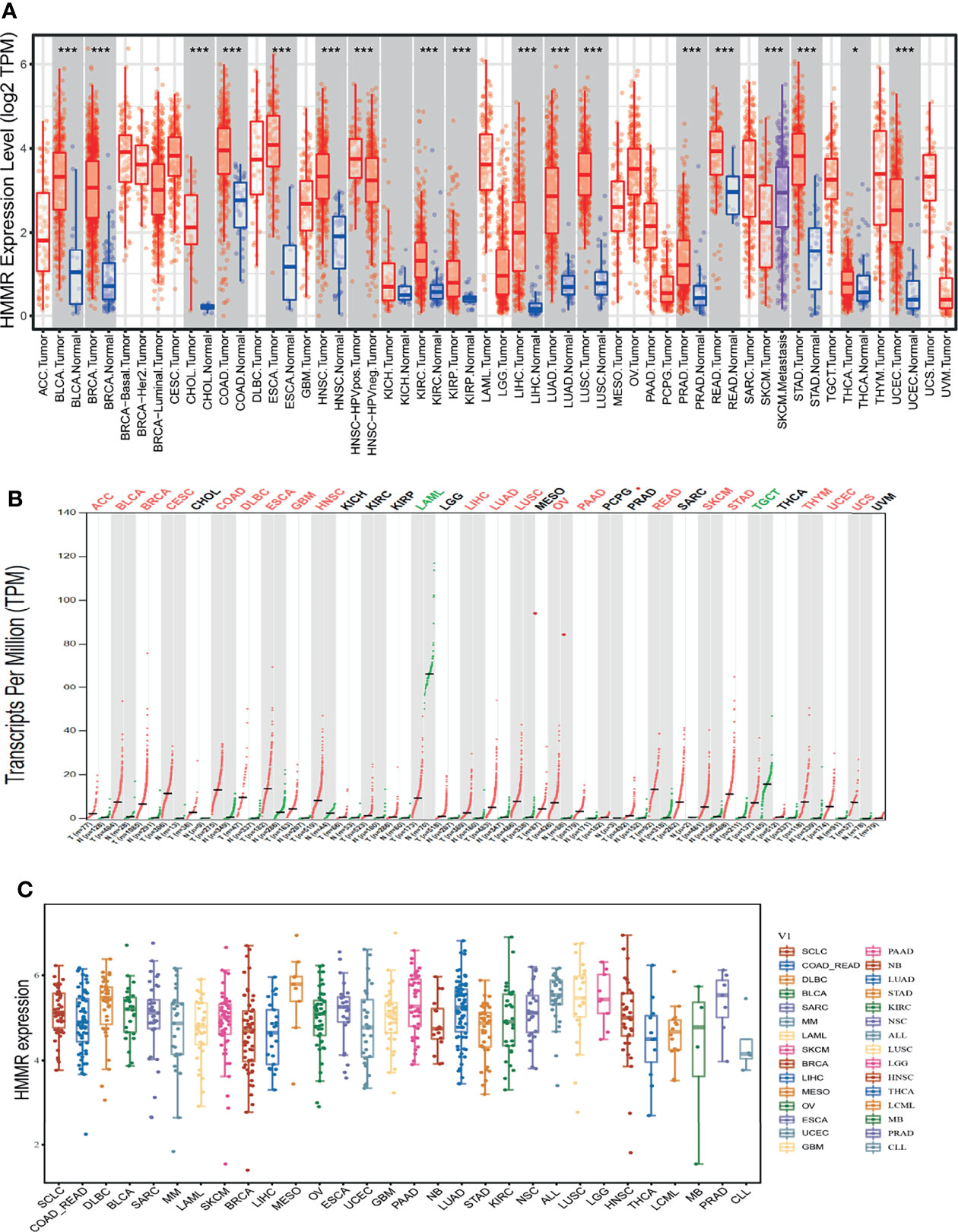
Figure 1 HMMR was highly expressed in pan-cancer. (A) The expression of HMMR in pan-cancers examined by TIMER tools. (B) The expression of HMMR in pan-cancers examined by GEPIA tools. (C) The expression of HMMR in pan-cancers cells lines examined by CCLE database. *P < 0.05; ***P < 0.001.
Given that HMMR was overexpressed in pan-cancer, we further analyzed its prognosis in pan-cancer. The results showed that a high HMMR expression was correlated with poor OS in ACC, COAD, chromophobe renal cell carcinoma (KICH), KIRC, KIRP, LGG, LIHC, LUAD, MESO, PAAD, PCPG, PRAD, THYM, and UVM (Supplementary Figure S1A), related to poor DSS in KIRP, LIHC, MESO, SARC, and THCA (Supplementary Figure S1B), and was associated with poor disease-free survival (DFS) in ACC, KICH, KIRC, KIRP, LGG, LIHC, LUAD, MESO, PRAD, THCA, and UVM (Supplementary Figure S1C). It was also linked to poor PFS in ACC, KICH, KIRC, KIRP, LGG, LIHC, LUAD, MESO, PCPG, and UVM (Supplementary Figure S1D). We also utilized GEPIA tools to examine the correlation between HMMR expression and the pathological stage of diverse cancer. Interestingly, we found that the expression of HMMR was markedly positive with the pathological stage of ACC, BRCA, ESCA, NHCS, KICH, KIRC, KIRP, LAML, LAML, LIHC, LUAD, and LUSC (Figures 2A–L). Taken together, these results suggest that HMMR may serve as an oncogene in human cancers.
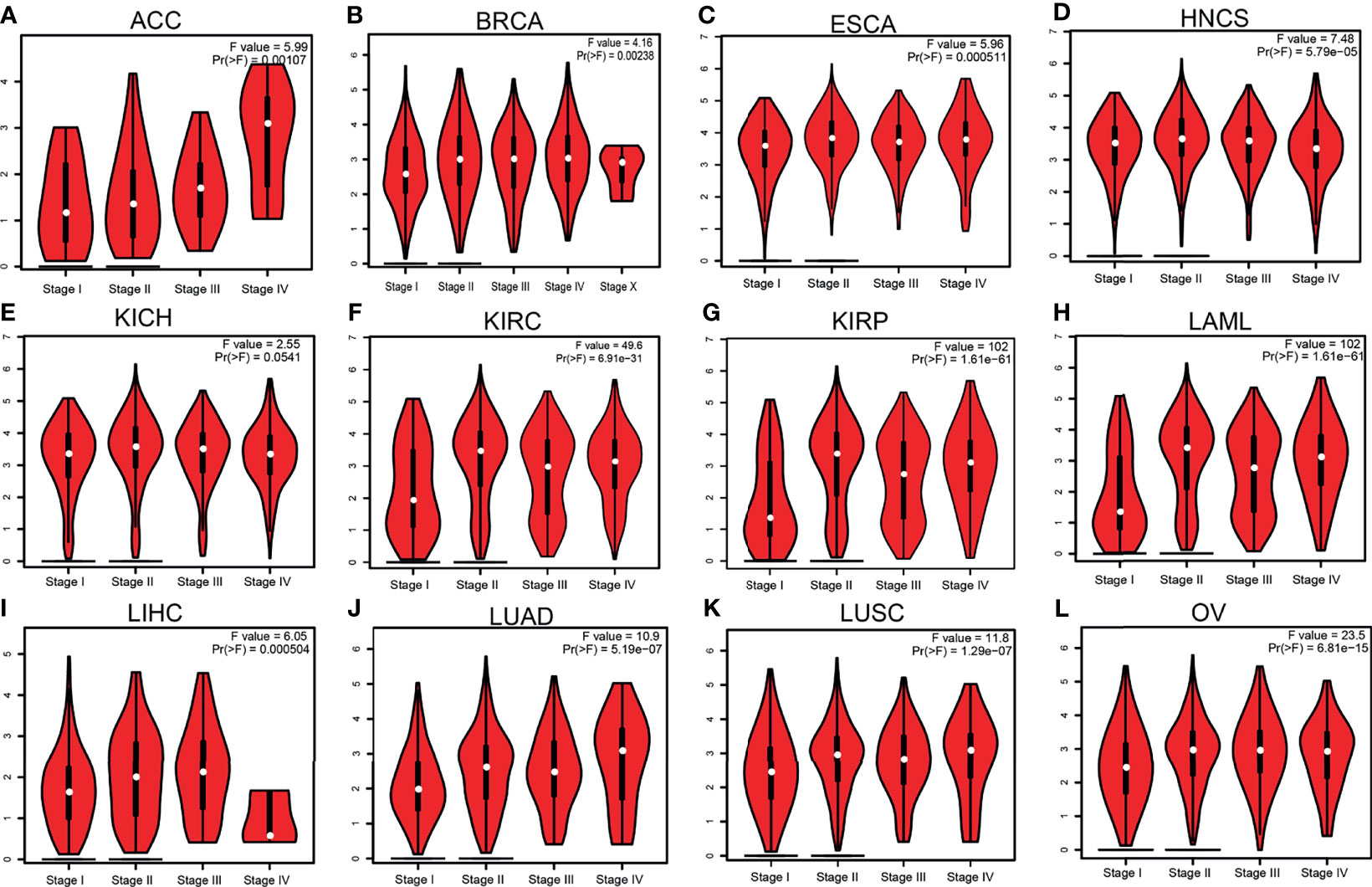
Figure 2 Pathological stage analysis of hyaluronan-mediated motility receptor (HMMR) in various human cancers. (A–K) Pathological stage of HMMR in adrenocortical carcinoma (ACC) (A), breast invasive carcinoma (BRCA) (B), esophageal carcinoma (ESCA) (C), head and neck squamous cell carcinoma (HNSC) (D), chromophobe renal cell carcinoma (KICH) (E), kidney renal clear cell carcinoma (KIRC) (F), kidney renal papillary cell carcinoma (KIRP) (G), acute myeloid leukemia (LAML) (H), liver hepatocellular carcinoma (LIHC) (I), lung adenocarcinoma (LUAD) (J), lung squamous cell carcinoma (LUSC) (K), and ovarian serous cystadenocarcinoma (OV) (L) examined using Gene Expression Profiling Interactive Analysis (GEPIA).
Previous reports have shown that cancer could accord to the molecular characteristics divided into different immune and molecular subtypes. Thus, we utilized the Tumor Immune System Interactions Database (TISIDB) to analyze the expression of HMMR in diverse immune and molecular subtypes of human cancer. Concerning the immune subtypes, the analysis results showed HMMR to have differential expression patterns in cancers (Supplementary Figure S2A). A quintessential example is LUAD, where HMMR was elevated in C1 and C2, while its expression was observed to be low in C3. With regard to the molecular subtypes, HMMR also displayed a distinctive expression pattern (Supplementary Figure S2B), in which it showed a high expression in C1 of LUAD, but decreased in C3. To summarize, our results indicate that the expression pattern of HMMR has tissue-dependent specificity.
We initially examined the expression of HMMR between tumor and normal tissues in LUAD using TCGA database. In paired samples, we found that HMMR expression was significantly higher in tissues than in adjacent normal groups (Figures 3A, B). This result was validated by the Gene Expression Omnibus (GEO) dataset (Supplementary Figure S3A). A further study found that a higher HMMR expression was correlated with adverse clinicopathological features in LUAD, including pathological stage (p < 0.001), TNM stage (p < 0.001), residual tumor (p < 0.001), primary therapy outcome (p < 0.001), and smoking status (p < 0.001). Additionally, the ROC analysis showed that the expression levels of HMMR in LUAD, LUSC, and LUAD were 0.970, 0.989, and 0.975, respectively (Figures 3C–J and Table 1). The ROC curve results were validated in the GEO dataset (Supplementary Figure S3B). Owing to HMMR overexpression being correlated with poor clinicopathological features, we then explored its prognostic value in LUAD. The results demonstrated that an elevated HMMR expression was associated with poor OS, DSS, and PFS (Figure 3K). This result was validated in the GEO dataset (Supplementary Figure S3C). The different subgroups, including TNM stage, age, pathological stage, primary therapy outcome, residual tumor, and smoking status, also correlated with poor prognosis (Figures 4A–H). Cox univariate and multivariate analyses indicated that TNM stage and HMMR expression were independent risk factors for LUAD patients resulting in adverse outcomes (Table 2).
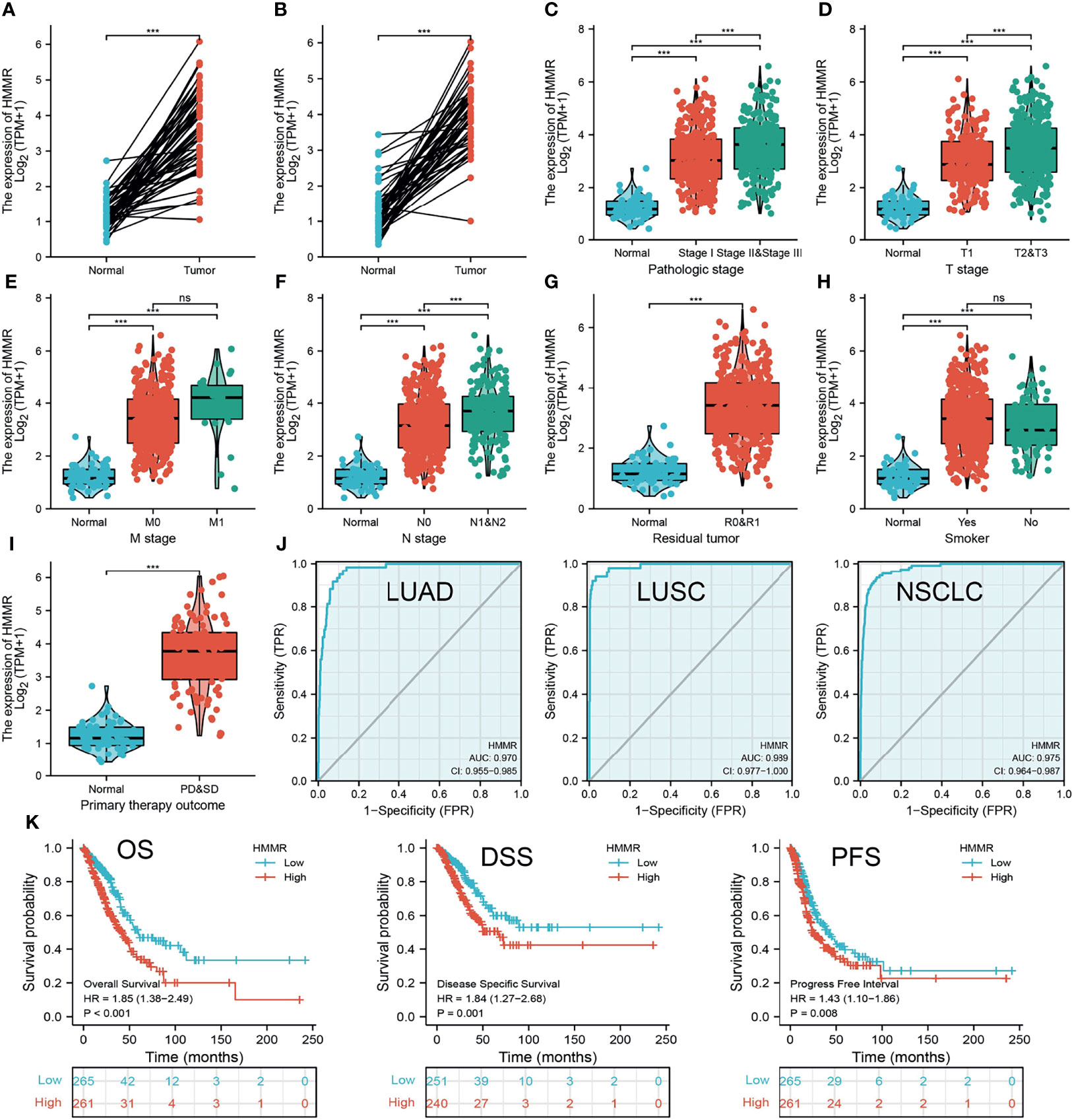
Figure 3 HMMR was up-regulated in LUAD. (A, B) The expression of HMMR in LUAD examine by TCGA databases. (C–I) The correlation between HMMR expression and clinical features in LUAD. (J) The ROC curve of HMMR in LUAD. (K) The prognosis of HMMR in LUAD examine by TCGA database. Overall survival (os), disease specific survival (DSS), progression-free survival (PFS), SD (stable disease), and PD (progressive disease). Ns, P > 0.05; ***P < 0.001.
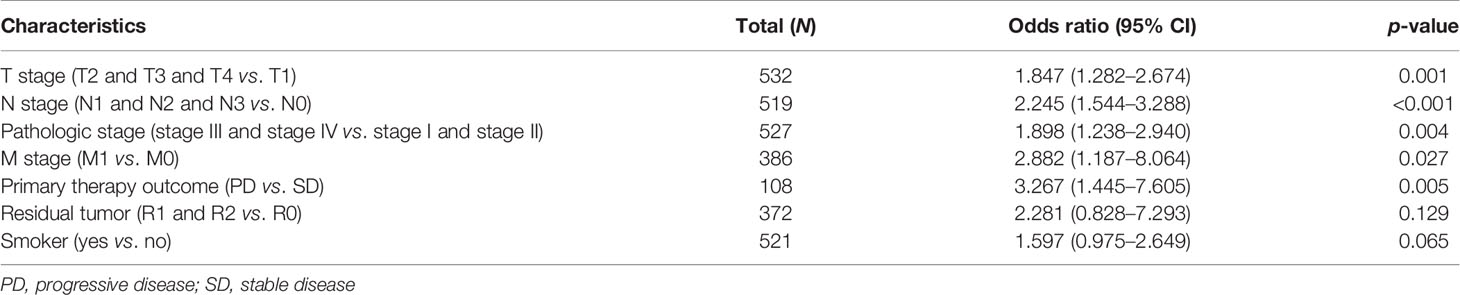
Table 1 Correlation between hyaluronan-mediated motility receptor (HMMR) expression and the clinicopathological characteristics in TCGA-LUAD dataset.
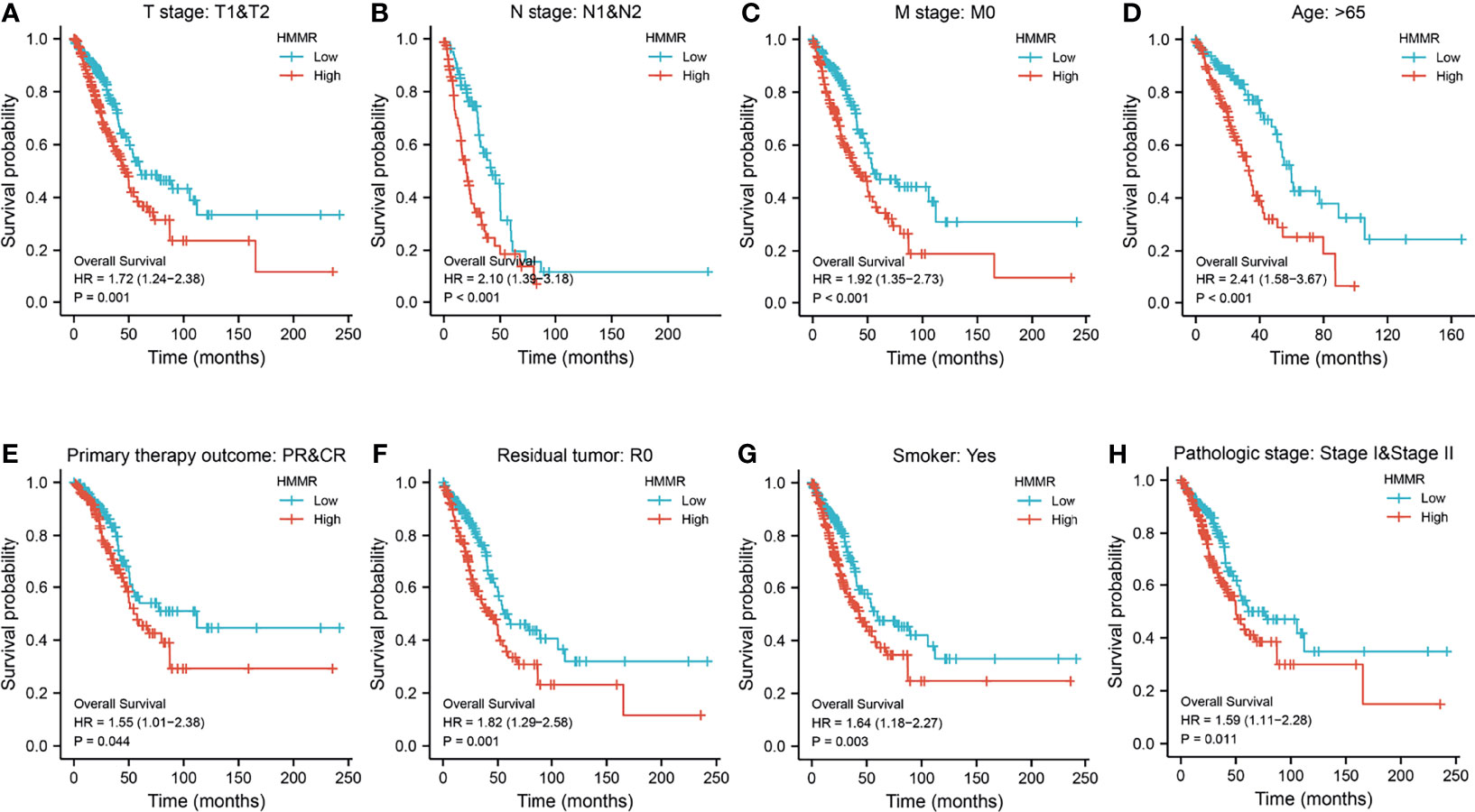
Figure 4 Prognosis of hyaluronan-mediated motility receptor (HMMR) based on different subgroups. (A–H) Prognosis of HMMR based on TNM stage, age, pathologic stage, and smoking status.
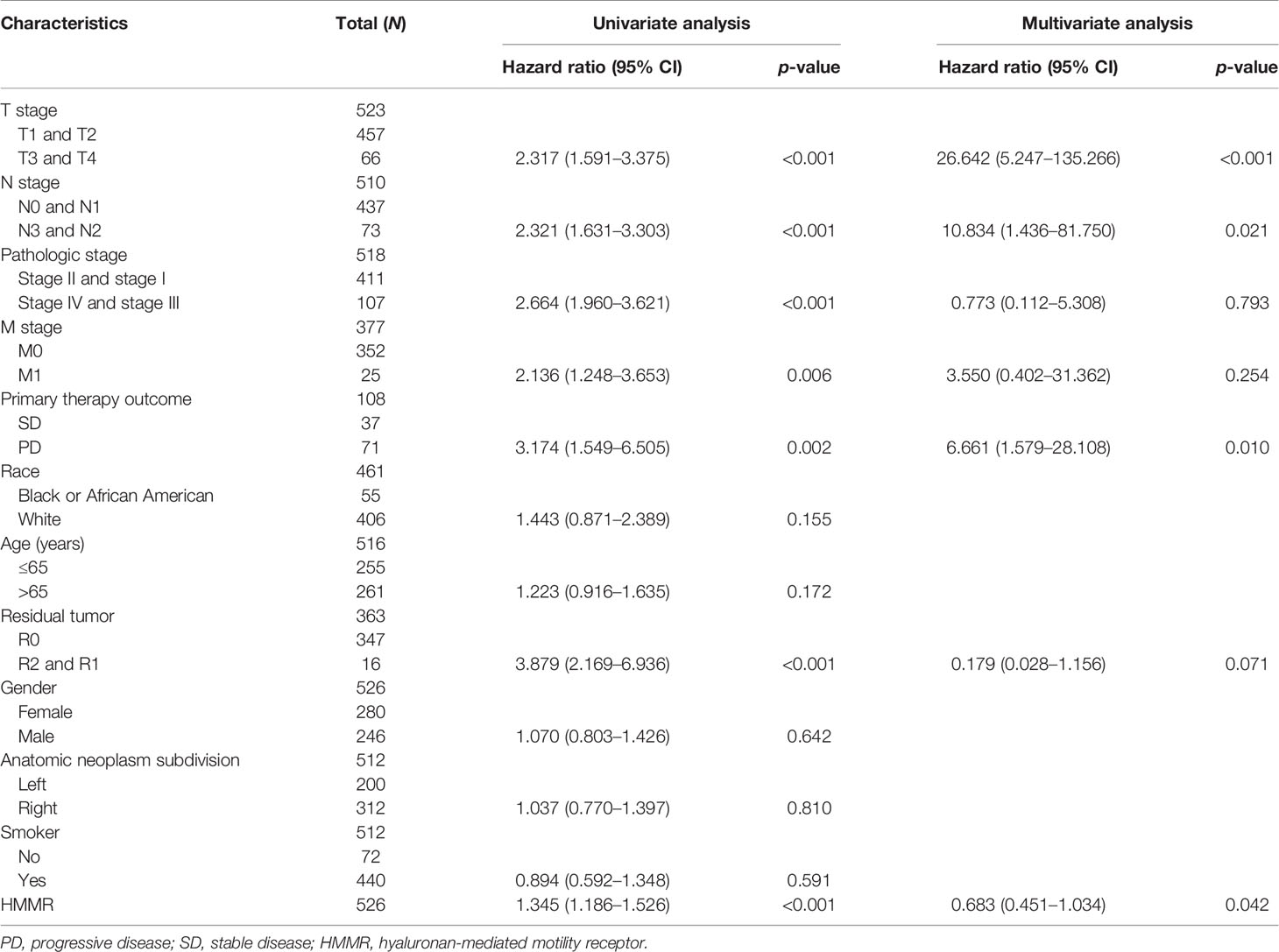
Table 2 Univariate regression and multivariate survival model of the prognostic covariates in patients with lung adenocarcinoma (LUAD).
To explore the gene mutation information of HMMR in LUAD, cBioportal was employed to perform comprehensive analysis of HMMR. The results showed that the mutation rate of HMMR reached 2.9% in LUAD (Supplementary Figure S4A). We also examined the mutation type and base mutation in LUAD and found that missense substitution and base G>A reached the highest mutation rate in LUAD (Supplementary Figures S4B, C). The results also displayed the mutation of HMMR in different LUAD molecular subtypes (Supplementary Figure S4D). At the DNA level, gain and diploid were the main drivers of the high expression of HMMR in LUAD (Supplementary Figure S4E). Overall, these results emphasize that the gene mutation of HMME may contribute to HMMR being elevated in LUAD.
To elucidate the biological functions of HMMR in LUAD, the LinkedOmics database was used to examine positive genes with HMMR. As shown in Figures 5A–C, the most positive gene (r > 0.7) is displayed in a heatmap. Subsequently, we performed GO and KEGG analyses using the top 100 co-expression genes. Biological processes mainly involved DNA replication, chromosome segregation, cell division, and protein localization (Figures 5D). For the KEGG enrichment results, the pathways mainly included cell cycle, P53 signaling pathway, non-small cell lung cancer, and FOXO signal pathways (Figures 5E). In addition, we analyzed the most relevant gene of HMMR using GeneMANIA. The results indicated that 20 genes were most relevant, with gene functions mainly involved in cell cycle (Figure 5F). We also used the STRING database to construct a PPI network. The PPI network of HMMR mainly included PLK4, CD44, AURKA, NEK2, CDK1, and FAM83D (Figure 5G).
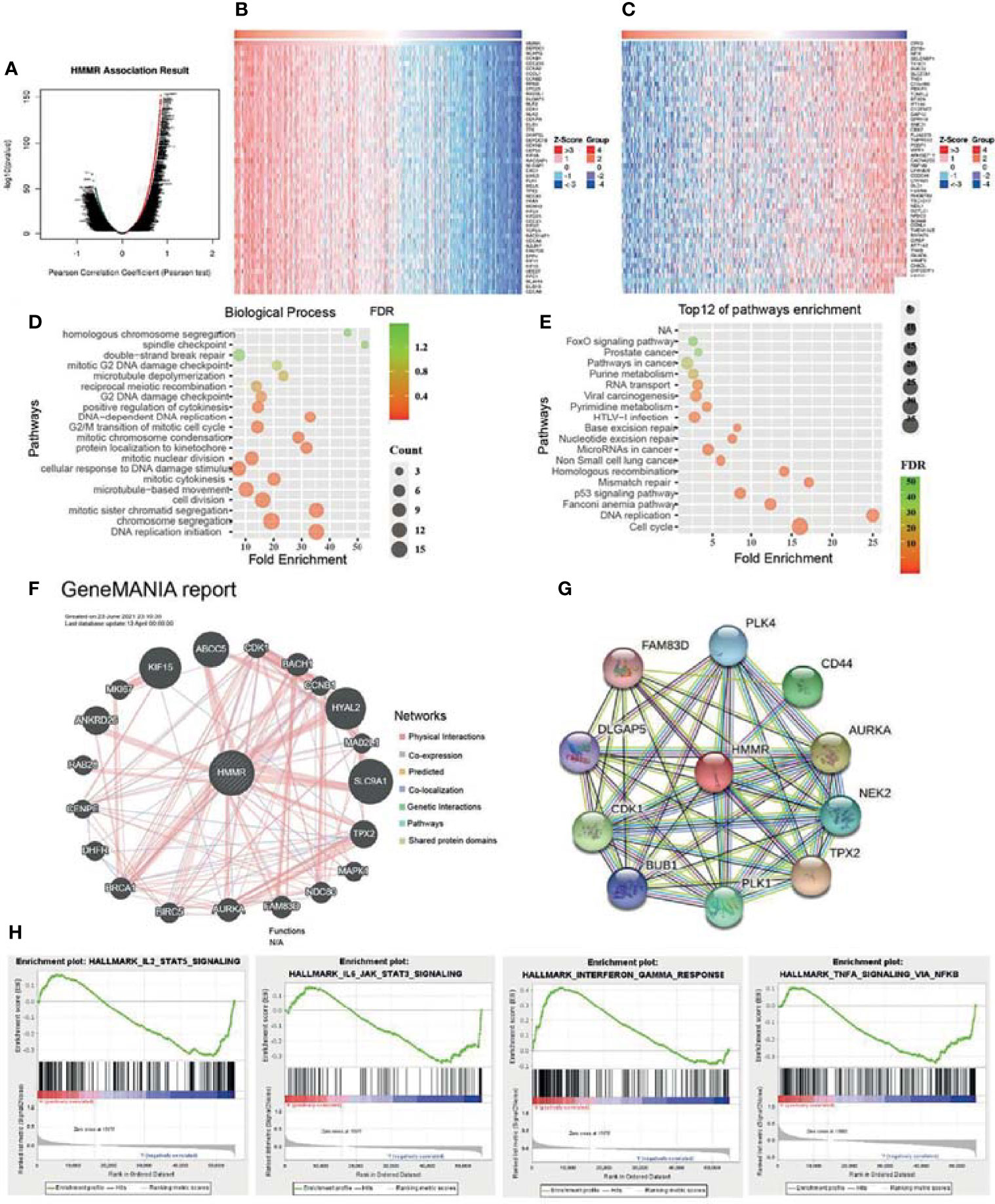
Figure 5 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of hyaluronan-mediated motility receptor (HMMR) in lung adenocarcinoma (LUAD). (A) Differentially expressed genes displayed in a volcano plot. (B, C) Positive and negative correlations with HMMR displayed in a heatmap. (D) Analysis of the biological process of HMMR. (E) Analysis of the KEGG pathways of HMMR. (F) Gene interaction network of HMMR constructed using GeneMANIA. (G) Construction of the protein–protein interaction network of HMMR using STRING. (H) Signaling pathways enriched using gene set enrichment analysis (GSEA) software.
To uncover the signaling pathways of HMMR in the progression of LUAD, we utilized the GSEA software to perform KEGG pathway enrichment. The analysis results showed that upregulation of HMMR expression was mainly involved in the IL2/STAT5 signaling pathway, IL6/JAK/STAT3 signaling pathway, interferon-γ response, and TNF-α signaling pathway (Figure 5H). These results suggest that HMMR plays a pivotal role in immune response regulation in LUAD.
The TIMER database was utilized to examine the relationship between HMMR expression and immune infiltration in LUAD. The results showed that the gene copy number change of HMMR significantly affects the immune infiltration levels of B cells, CD4+ T cells, CD8+ T cells, macrophage cells, neutrophils, and DCs in LUAD (Figure 6A). Furthermore, the TIMER database was used to examine the relationship between the HMMR level and immune infiltration levels in LUAD. The results showed that HMMR expression was markedly positively associated with B cells (r = 0.47, p = 7.69e−30), CD4+ T cells (r = 0.71, p = 1.71e−78), CD8+ T cells (r = 0.51, p = 1.22e−35), neutrophils (r = 0.77, p = 3.54e−103), macrophage cells (r = 0.44, p = 8.88e−26), and DCs (r = 0.84, p = 5.94e−139) in LUAD (Figure 6B). We also found that the expression of HMMR was markedly positively correlated with immune modulators, including CD274 (r = 0.34, p = 9.3e−15), CTLA4 (r = 0.556, p = 5), HAVCR2 (r = 0.71, p = 0), LAG3 (r = 0.39, p = 0), PDCD1 (r = 0.56, p = 0), TIGIT (r = 0.71, p = 0), and PDCD1LG2 (r = 0.67, p = 0) in LUAD (Figure 6C). These results demonstrate that HMMR plays crucial roles in the regulation of tumor immune infiltration in LUAD.
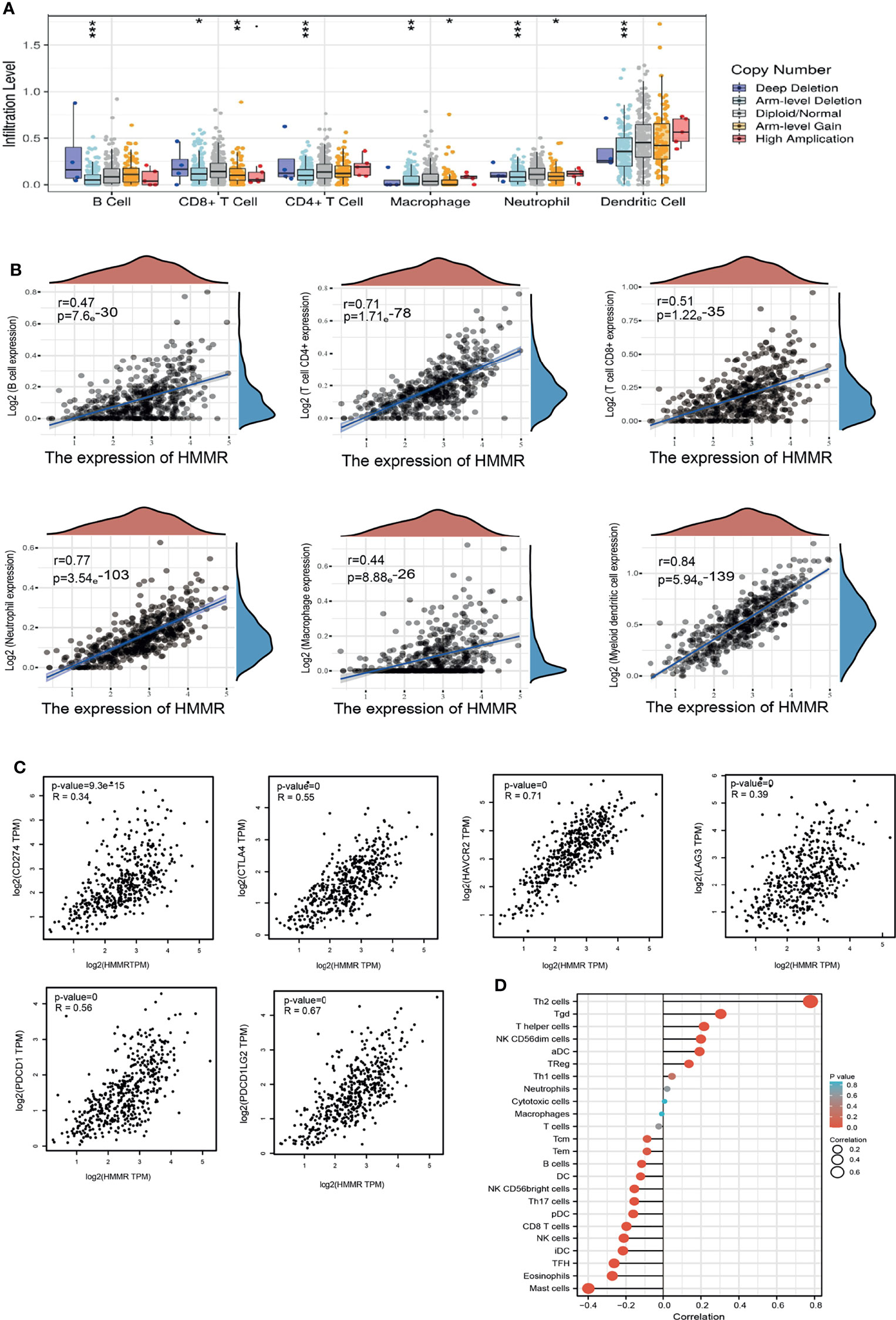
Figure 6 HMMR expression was associated with immune infiltration in LUAD. (A) The correlation between HMMR CNV and immune cells in LUAD examined by TIMER database. (B) The correlation between HMMR expression and immune infiltration in LUAD examined by TIMER database. (C) The correlation between HMMR expression and immune checkpoints related gene expression in LUAD. (D) The correlation between HMMR expression level and 24 immune cell types. *P < 0.05; **P < 0.01; *** < 0.001.
Moreover, ssGSEA with Spearman’s rank correlation was employed to measure the correlation between the expression of HMMR and the infiltration levels of 24 immune cell types. The results revealed that the expression of HMMR was markedly positively correlated with the infiltrating level of Th2 cells, gamma delta T cells, T helper cells, NK CD56dim cells, and aDCs and negatively correlated with the infiltrating levels of NK CD56bright cells, Th17 cells, pDCs, CD8 T cells, NK cells, iDCs, Tfh cells, eosinophils, and mast cells (Figure 6D).
Considering the significance of HMMR in immune regulation, we next explored the relationship between HMMR expression and diverse immune modulators, including tumor-infiltrating lymphocytes, immune stimulators, immune inhibitors, chemokines, receptors, and major histocompatibility complexes (MHCs) in LUAD. The analysis revealed that HMMR expression was positively correlated with the 28 tumor-infiltrating lymphocytes (Supplementary Figure S5A), 24 immune inhibitors (Supplementary Figure S5B), 45 immune stimulators (Supplementary Figure S5C), 21 MHCs (Supplementary Figure S5D), 41 chemokines (Supplementary Figure S5E), and 18 receptors (Supplementary Figure S5F) in LUAD. These findings indicate that HMMR plays an indispensable role in the regulation of immune response in LUAD.
The KM plotter database was utilized to explore the prognosis of HMMR based on the different immune cells in LUAD. It was found that the cohort with an elevated HMMR expression and decreased B cells, CD4+ T cells, CD8+ T cells, macrophages, NK T cells, and Tregs had poor prognosis (Figures 7A–L). These data indicate that diverse immune cell infiltration could significantly affect the prognosis of HMMR in LUAD.
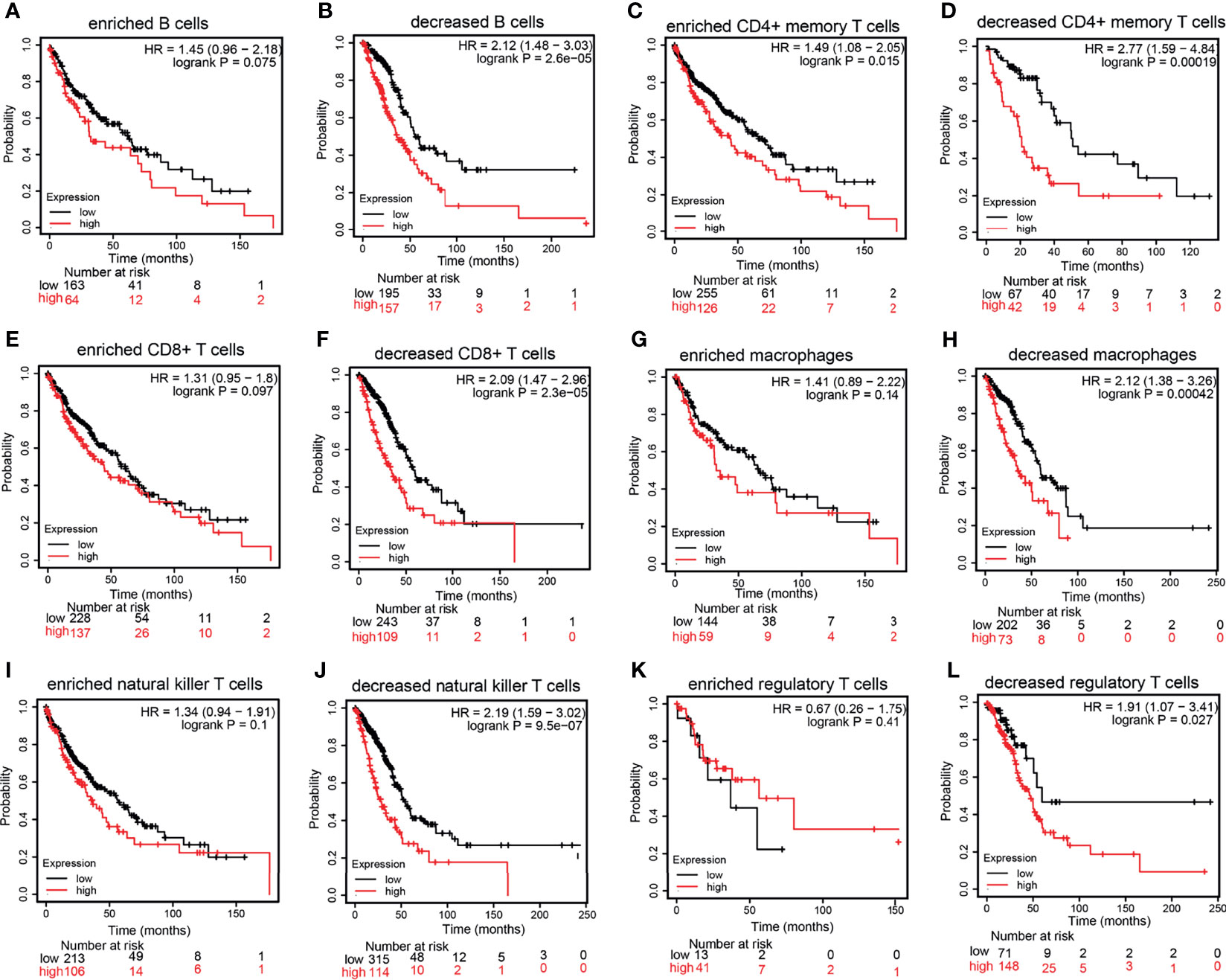
Figure 7 Prognostic potential of the expression of hyaluronan-mediated motility receptor (HMMR) in different tumors based on immune cells. (A–L) Relationship between HMMR expression and overall survival (OS) based on the immune cell subgroups examined using the Kaplan–Meier (KM) plotter.
MiRNAs play an important role in the modulation of gene expression. We further examined the upstream miRNAs that regulate HMMR expression in the progression of lung cancer. We utilized starBase and TargetScan to predict the potential miRNAs of HMMR. The results showed four miRNAs that bind with the 3′-UTR of HMMR, namely, let-7b-5p, hsa-miR-18a-5p, hsa-miR-33a-5p, and hsa-miR-369-3p. In the analysis of the correlation between these miRNAs and HMMR, only let-7b-5p was markedly negative with the HMMR expression in LUAD (Figures 8A, C). Subsequently, starBase was used to examine the target sites between HMMR and let-7b-5p (Figure 8B). A further study found that let-7b-5p was downregulated in LUAD (Figure 8D), and a low expression of let-7b-5p was related to poor prognosis and clinicopathological features. The ROC analysis showed that let-7b-5p expression in LUAD was 0.754 (Figures 8E–J). Additionally, the qRT-PCR assay examined the expression of let-7b-5p in LUAD cell lines, with the data indicating that it was decreased in LUAD cells compared to control cells (Figure 8K). To determine whether let-7b-5p affects the expression of HMMR, we overexpressed let-7b-5p in the A549 cell line and found that the mRNA and protein levels of HMMR were significantly reduced after the overexpression of let-7b-5p (Figures 8L, M). Collectively, these data imply that let-7b-5p may participate in the regulation of HMMR expression in LUAD.
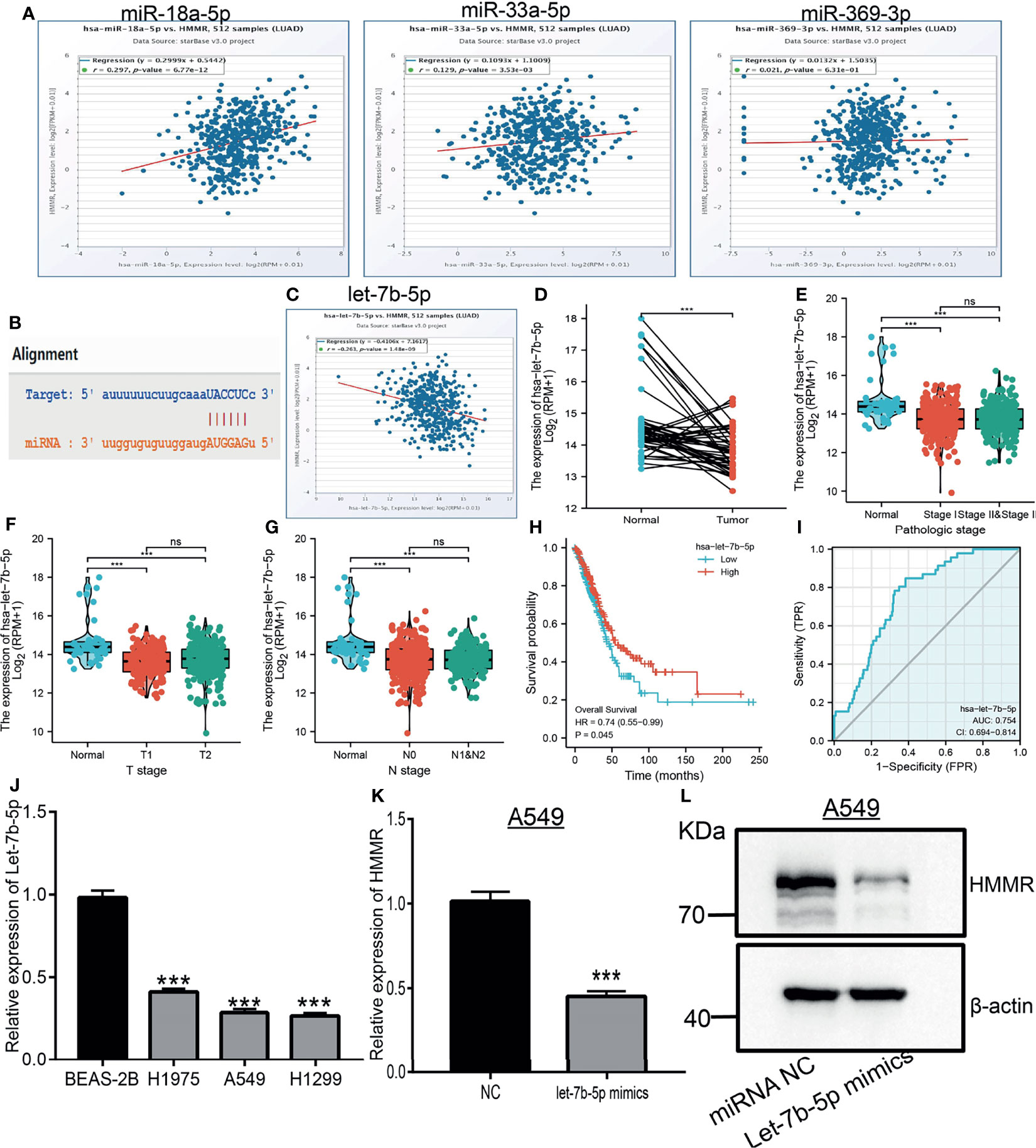
Figure 8 Predicted and analysis the upstream miRNAs of HMMR in LUAD. (A) The correlation between the HMMR expression and miRNA-18a-5p, miR-33a-5p and miR-369-3p analysis by starBase. (B) The target sites between HMMR and hsa-let-7b-5p were predicted by starBase. (C) The correlation between the HMMR expression and hsa-let-7b-5p analysis by starbase. (D) The expression of hsa-let-7b-5p in LUAD analysis by TCGA. (E–G) The correlation between hsa-let-7b-5p expression and clinical features in LUAD. (H) The prognosis of hsa-let-7b-5p in LUAD analysis by kmplot. (I) The ROC curve of hsa-let-7b-5p in LUAD. (J) The expression of hsa-let-7b-5p in LUAD cells analysis by employed qRT-PCR assay. (K) The expression of HMMR after overexpression of hsa-let-7b-5p in LUAD cells analysis by qRT-PCR assay. (L) The expression of HMMR after overexpression of hsa-let-7b-5p in LUAD Cells analysis by western blot assay. NC, Negative control. P > 0.05 (ns), ***P < 0.001, was considered significantly.
It has been shown that lncRNAs play crucial roles in the regulation of the expressions of miRNAs and mRNAs. The above findings showed that let-7b-5p may modulate the expression of HMMR via binding with its 3′-UTR. We next explored the upstream lncRNAs of let-7b-5p. starBase and lncBase were utilized to predict the potential lncRNAs that act as a miRNA sponge and control mRNA expression. By performing the related analysis, we obtained three lncRNAs—SNHG12, LINC02242, and TMPO-AS1—and further analyzed their correlation with hsa-let-7b-5p. The data indicated that SNHG12 and LINC02242 were positively correlated with let-7b-5p. The expression of TMPO-AS1 was not only negative with hsa-let-7b-5p but also positive with HMMR as a target gene of hsa-let-7b-5p (Figures 9A–C). In addition, we found that TMPO-AS1 was highly expressed in LUAD (Figures 9D–G). We performed localization and coding potential analysis using diverse public databases. The subcellular localization of TMPO-AS1 was determined employing lncLocator. The results indicated that TMPO-AS1 was mainly located in the cytoplasm (Figure 9H). We also examined the coding potential of TMPO-AS1 using the coding potential calculator and found that TMPO-AS1 does not possess a protein coding ability (Figure 9I). Finally, it was shown that depletion of TMPO-AS1 markedly reduced the expression of HMMR and upregulated the expression of hsa-let-7b-5p in A549 cells (Figure 9J).
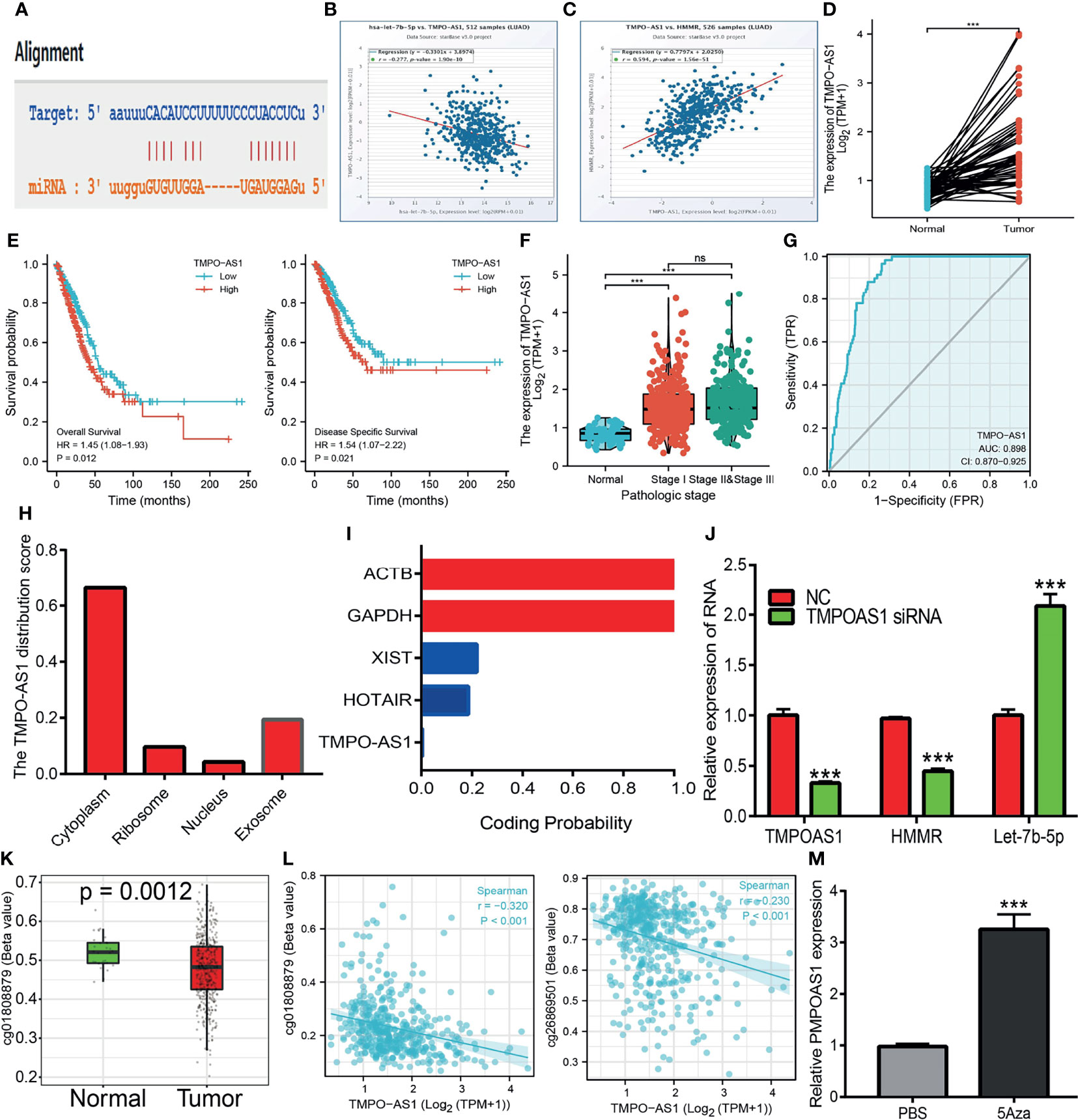
Figure 9 Predicted and analysis the upstream LncRNAs of let-7b-5p in LUAD. (A) The target sites between the TMPO-AS1 and hsa-let-7b-5p were predicted by starbase. (B) The correlation between hsa-let-7b-5p expression and TMPO-AS1 analysis by starbase. (C) The correlation between the HMMR expression and TMPO-AS1 analysis by starbase. (D) The expression of TMPO-AS1 in LUAD analysis by starbase. (E, F) The prognosis of TMPO-AS1 in LUAD analysis by kmplot. (G) The ROC curve of TMPO-AS1 in LUAD. (H) The subcellular localization of TMPO-AS1 analysis by the lncLocator tools. (I) The coding potential of TMPO-AS1 analysis by the coding potential calculator. (J) The HMMR and hsa-let-7b-5p expression after depletion of TMPO-AS1 in LUAD cells analysis by qRT-PCR assay. (K) The DNA methylation of TMPO-AS1 in LUAD. (L) The correlation between DNA methylation and expression of TMPO-AS1 in LUAD. (M) The expression of TMPO-AS1 in LUAD cells after treat with 5Aza examined by qRT-PCR assay. P >0.05 (ns), P < 0.001 (***), was considered significantly.
Upon exploration of the potential mechanism of TMPO-AS1 overexpression in LUAD, we further found that the DNA methylation of TMPO-AS1 was decreased and negatively correlated with its expression in LUAD. Treatment with 5-azacytidine, an inhibitor of DNA methyltransferases, resulted in the increased level of TMPO-AS1 in LUAD cells (Figures 9K–M). These results suggest that hypomethylation in the TMPO-AS1 promoter DNA results in the increased expression of this gene in LUAD.
To further determine the function of HMMR in LUAD progression, IHC and qRT-PCR assays were performed to examine the expression of HMMR in different LUAD tissues and cell lines. The results showed that HMMR was significantly elevated in lung cancer and in LUAD cells (Figures 10A, B), especially in A549 and H1299 cells. Subsequently, we knocked down HMMR in A549 and H1299 cells and used qRT-PCR and Western blot to examine the knockdown efficiency (Figures 10C, D). The growth curve and colony formation assay showed that depletion of HMMR inhibited the cell growth of LUAD cells (Figures 10E, F). The Transwell and wound healing assays also demonstrated that the knock down of HMMR inhibited the cell growth of LUAD cells (Figures 10G, H). These findings suggest that HMMR promotes the cell growth and migration of LUAD cells.
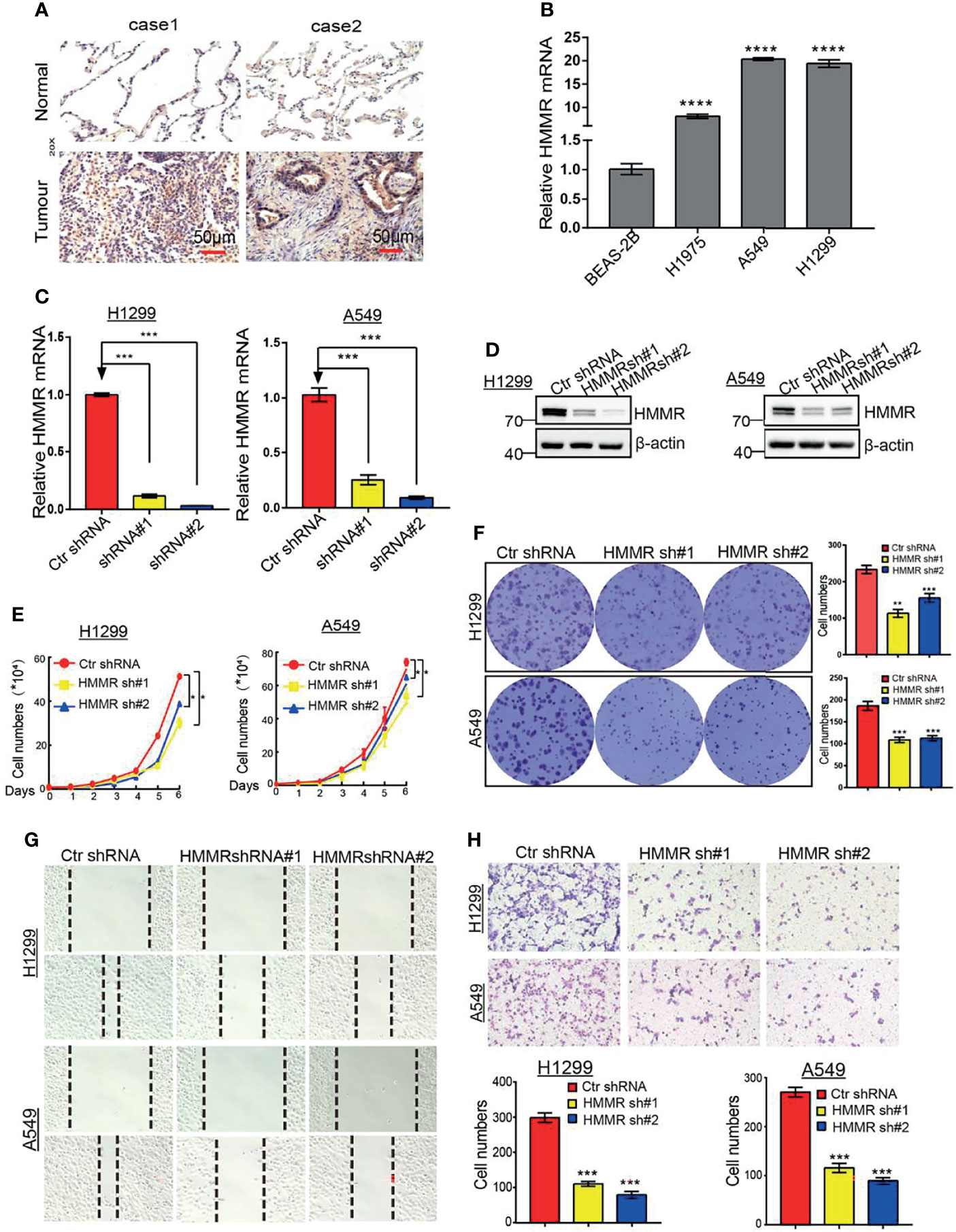
Figure 10 Depletion of HMMR inhibits growth and migration of LUAD cells in vivro. (A) IHC analysis of HMMR in LUAD. (B) The expression of HMMR in LUAD cell lines by qRT-PCR. (C, D) Establishment of HMMR knockdown in A549 and H1299 cell lines and verified by qRT-PCR and Western blot. (E) The growth curve assay was employed detect the proliferation of LUAD cells. (F) The colony formation assay was employed detect the proliferation of LUAD cells. (G) The transwell assay was employed detect the migration of LUAD cells, (H) The wound healing assay was employed detect the migration of LUAD cells. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***), P < 0.0001 (***) was considered significantly.
It has been reported that HMMR plays a crucial role in the progression of human cancers (14). For example, studies have shown that HMMR was elevated in glioblastoma and that a high expression of HMMR could boost the self-renewal of GBM stem cells (GSCs) (14). In breast cancer, elevated levels of HMMR were related to poor clinical outcomes (29). Presently, important evidence for the significance of HMMR in LUAD is still lacking.
In this study, we employed various databases to conduct a comprehensive analysis of the expression, prognosis, clinical significance, and biological function in LUAD. We found that HMMR was highly expressed in various cancer tissues, including LUAD. The high expression of HMMR was significantly correlated with poor clinicopathological features and adverse outcomes in LUAD. These results indicate that HMMR plays a central role in the progression and metastasis of LAUD. Our results are consistent with previous studies. HMMR was upregulated in some cancer tissues and was correlated with adverse clinicopathological features and poor prognosis (15, 30).
The results of the survival analysis confirmed that a higher HMMR expression was correlated with poor OS, DFS, and PFS in LUAD. Cox univariate and multivariate analyses indicated that the TNM stage and HMMR expression were independent risk factors for LUAD patients resulting in adverse outcomes. Consistent with previous studies, HMMR affects cancer cell proliferation and epithelial-to-mesenchymal transition and results in a poor prognosis (15, 31). Our findings strongly confirmed that HMMR can be used as a prognostic biomarker for LAUD.
Previous studies have shown that HMMR plays crucial roles in tissue homeostasis (7), neural development (7), and cancer progression (32). For example, it has been demonstrated that HMMR is highly expressed in GBM tumors and that its depletion impairs GSC self-renewal and inhibits the expressions of its markers and regulators. Furthermore, HMMR silencing suppresses GSC-derived tumor growth and extends the survival of mice bearing GSC xenografts. In this study, we found that HMMR mainly participated in cell cycle, p53 signaling pathway, non-small cell lung cancer, and FOXO signal pathway. We also utilized the STRING database to construct the PPI network, which mainly included PLK4, CD44, AURKA, NEK2, CDK1, and FAM83D. It has been shown that HMMR was elevated in LUAD and that its high expression was related to the tumor size and lymph node metastasis (33). A recent study has found that CD44 was able to elevate the expression of programmed death-ligand 1 (PD-L1) by regulating CD274 transcription, resulting in inhibition of the tumor-intrinsic function of PD-L1 (34). It has also been reported that alisertib, an inhibitor of AURKA, was able to treat mammary tumors when combined with PD-L1 blockade (35). NEK2 was reported to be elevated in lung cancer, regulated by EGFR mutation. The overexpression of NEK2 significantly promoted cell proliferation and induced cell cycle progression in LUAD cells (36). The above results indicate that HMMR may play a central regulatory role in cancer progression.
The GSEA pathway enrichment analysis found that HMMR may be associated with immune regulation and is involved in the IL2/STAT5 signaling pathway, IL6/JAK/STAT3 signaling pathway, interferon-γ response, and TNF-α signaling pathway. Therefore, we attempted to examine the correlation between the expression of HMMR and immune response. HMMR was previously found to modulate the tumor microenvironment (37, 38). By performing a correlation analysis, it was revealed that the expression of HMMR was associated with the immune infiltration of B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and DCs. Our results are consistent with previous studies (38). Chong et al. found that HMMR was overexpressed in renal cancer and affected the progression, prognosis, and immune microenvironment of renal cell carcinoma (39).
Kaplan Meier-Plotter analysis showed that up-regulation of HMMR and enriched in a variety of immune cells correlated with poor prognosis in LUAD. DCs can promote tumor metastasis by increasing Tregs and decreasing the cytotoxicity of CD8+ T cells (40). Previous studies have also confirmed that the proportions of CD8+ T cells and Tregs in LUAD patients were associated with adverse clinical outcomes (41). These results may explain the increased HMMR expression partly affecting the prognosis of LUAD patients partially via immune cell infiltration. These findings suggest that HMMR could be an immune-related biomarker in LUAD.
It has been well documented that lncRNAs and miRNAs play crucial roles in controlling gene expression. To elucidate the potential mechanism of HMMR overexpression in LUAD, we predicted and analyzed the upstream miRNAs of HMMR. We found that let-7b-5p, a known tumor suppressor gene, could modulate the expression of HMMR. Wang et al. found that let-7b-5p inhibited the cell proliferation of myeloma by regulating the expression of IGF1R (42). Subsequently, we found that the lncRNA TMPOAS1 acted as a miRNA sponge, inhibiting let-7b-5p and elevating HMMR expressions in LUAD. As a matter of fact, mounting evidence has demonstrated that TMPOAS1 plays an oncogenic role in the progression of cancer. For instance, Zhao et al. found that TMPO-AS1, via negative regulation of the expression of miR-383-5p, promoted lung adenocarcinoma progression (43). Similarly, Chen et al. reported that TMPO-AS1 promoted the proliferation and metastasis of LUAD cells by upregulating ERBB2 via sponging miR-204-3p (44). A previous study reported that the HCG18/miR-34a-5p/HMMR axis promoted the progression of lung adenocarcinoma (45). In this study, the TMPO-AS1/let-7b-5p/HMMR axis was identified as a potential regulatory pathway in LUAD. Finally, we found that the knockdown of HMMR significantly reduced the proliferation and migration ability of LUAD cells. Our results are consistent with previous studies.
This study improves our understanding of the correlation between HMMR and LUAD, but some limitations exist. Firstly, although we explored the correlation between HMMR and immune infiltration in LUAD patients, there is a lack of experiments validating the function of HMMR in the tumor microenvironment regulation of LUAD. Secondly, we uncovered that the depletion of HMMR inhibited the cell proliferation and cell migration of LUAD cells. However, the potential molecular mechanisms of HMMR in tumor growth and metastasis need to be explored in further studies. Thirdly, we did not conduct in vivo experiments to validate the function of HMMR in tumor metastasis and the tumor microenvironment regulation of LUAD. In the future, we will pay more attention to the function of HMMR in tumor metastasis and the tumor microenvironment regulation of LUAD.
In conclusion, our findings uncovered, for the first time, the biological function of HMMR in LUAD. The expression of HMMR in LUAD was correlated with poor prognosis was associated with patient pathological stage, TNM stage, residual tumor, primary therapy outcome, and smoking status. In addition, we also found that HMMR may play a vital role in regulating the immune microenvironment of LUAD and, thus, affect its progression. The upregulation of HMMR may be attributed to the TMPO-AS1/let-7b-5p axis. The knockdown of HMMR significantly reduced the proliferation and migration ability of LUAD cells. Exploring the role of HMMR in LUAD and its immune microenvironment will be helpful to better understand this cancer and could result in the identification of a new gene-targeted immunotherapy for LUAD. Therefore, HMMR can be used as a promising molecular predictor to evaluate the prognosis of LUAD patients and as a therapeutic target in the clinical detection of LUAD.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
XJ, LT, YY, and JW designed this work and performed related assays. DZ and KQ analyzed the data. WC and LD supervised and wrote the manuscript. All authors have read and approved the final version of the manuscript.
This work was supported by the National Nature Science Foundation of China (82160508), Yunnan Applied Basic Research Projects (YNWRMY-2019-067, 2019FE001), and Yunnan Province Specialized Training Grant for High-Level Healthcare Professionals (D-201614).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank support from the Department of Thoracic Surgery, The Third Affiliated Hospital of Kunming Medical University (Yunnan Tumor Hospital), Kunming, China.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.846536/full#supplementary-material
1. Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med (2020) 41(1):1–24. doi: 10.1016/j.ccm.2019.10.001
2. Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am (2016) 25(3):447–68. doi: 10.1016/j.soc.2016.02.003
3. Zeng Y, Shi Y, Xu L, Zeng Y, Cui X, Wang Y, et al. Prognostic Value and Related Regulatory Networks of MRPL15 in Non-Small-Cell Lung Cancer. Front Oncol (2021) 11:656172. doi: 10.3389/fonc.2021.656172
4. Liang R, Li X, Li W, Zhu X, Li C. DNA Methylation in Lung Cancer Patients: Opening a “Window of Life” Under Precision Medicine. BioMed Pharmacother (2021) 144:112202. doi: 10.1016/j.biopha.2021.112202
5. Li QG, He YH, Wu H, Yang CP, Pu SY, Fan SQ, et al. A Normalization-Free and Nonparametric Method Sharpens Large-Scale Transcriptome Analysis and Reveals Common Gene Alteration Patterns in Cancers. Theranostics (2017) 7(11):2888–99. doi: 10.7150/thno.19425
6. He Z, Mei L, Connell M, Maxwell CA. Hyaluronan Mediated Motility Receptor (HMMR) Encodes an Evolutionarily Conserved Homeostasis, Mitosis, and Meiosis Regulator Rather Than a Hyaluronan Receptor. Cells (2020) 9(4). doi: 10.3390/cells9040819
7. Casini P, Nardi I, Ori M. RHAMM mRNA Expression in Proliferating and Migrating Cells of the Developing Central Nervous System. Gene Expr Patterns (2010) 10(2-3):93–7. doi: 10.1016/j.gep.2009.12.003
8. Prager A, Hagenlocher C, Ott T, Schambony A, Feistel K. Hmmr Mediates Anterior Neural Tube Closure and Morphogenesis in the Frog Xenopus. Dev Biol (2017) 430(1):188–201. doi: 10.1016/j.ydbio.2017.07.020
9. Assmann V, Gillett CE, Poulsom R, Ryder K, Hart IR, Hanby AM. The Pattern of Expression of the Microtubule-Binding Protein RHAMM/IHABP in Mammary Carcinoma Suggests a Role in the Invasive Behaviour of Tumour Cells. J Pathol (2001) 195(2):191–6. doi: 10.1002/path.941
10. Zlobec I, Baker K, Terracciano LM, Lugli A. RHAMM, P21 Combined Phenotype Identifies Microsatellite Instability-High Colorectal Cancers With a Highly Adverse Prognosis. Clin Cancer Res (2008) 14(12):3798–806. doi: 10.1158/1078-0432.CCR-07-5103
11. Li H, Guo L, Li JW, Liu N, Qi R, Liu J. Expression of Hyaluronan Receptors CD44 and RHAMM in Stomach Cancers: Relevance With Tumor Progression. Int J Oncol (2000) 17(5):927–32. doi: 10.3892/ijo.17.5.927
12. Rein DT, Roehrig K, Schöndorf T, Lazar A, Fleisch M, Niederacher D, et al. Expression of the Hyaluronan Receptor RHAMM in Endometrial Carcinomas Suggests a Role in Tumour Progression and Metastasis. J Cancer Res Clin Oncol (2003) 129(3):161–4. doi: 10.1007/s00432-003-0415-0
13. Gust KM, Hofer MD, Perner SR, Kim R, Chinnaiyan AM, Varambally S, et al. RHAMM (CD168) is Overexpressed at the Protein Level and may Constitute an Immunogenic Antigen in Advanced Prostate Cancer Disease. Neoplasia (2009) 11(9):956–63. doi: 10.1593/neo.09694
14. Tilghman J, Wu H, Sang Y, Shi X, Guerrero-Cazares H, Quinones-Hinojosa A, et al. HMMR Maintains the Stemness and Tumorigenicity of Glioblastoma Stem-Like Cells. Cancer Res (2014) 74(11):3168–79. doi: 10.1158/0008-5472.CAN-13-2103
15. Yang D, Ma Y, Zhao P, Ma J, He C. HMMR is a Downstream Target of FOXM1 in Enhancing Proliferation and Partial Epithelial-to-Mesenchymal Transition of Bladder Cancer Cells. Exp Cell Res (2021) 408(2):112860. doi: 10.1016/j.yexcr.2021.112860
16. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–w102. doi: 10.1093/nar/gkx247
17. Győrffy B. Survival Analysis Across the Entire Transcriptome Identifies Biomarkers With the Highest Prognostic Power in Breast Cancer. Comput Struct Biotechnol J (2021) 19:4101–9. doi: 10.1016/j.csbj.2021.07.014
18. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity (2013) 39(4):782–95. doi: 10.1016/j.immuni.2013.10.003
19. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics (2012) 16(5):284–7. doi: 10.1089/omi.2011.0118
20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc Natl Acad Sci USA (2005) 102(43):15545–50. doi: 10.1073/pnas.0506580102
21. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46(D1):D956–d963. doi: 10.1093/nar/gkx1090
22. Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, et al. GeneMANIA Prediction Server 2013 Update. Nucleic Acids Res (2013) 41(Web Server issue):W115–22. doi: 10.1093/nar/gkt533
23. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–13. doi: 10.1093/nar/gky1131
24. Li JH, Liu S, Zhou H, Qu LH, Yang JH. Starbase V2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks From Large-Scale CLIP-Seq Data. Nucleic Acids Res (2014) 42(Database issue):D92–7. doi: 10.1093/nar/gkt1248
25. Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, et al. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res (2017) 45(W1):W12–6. doi: 10.1093/nar/gkx428
26. Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The Lnclocator: A Subcellular Localization Predictor for Long Non-Coding RNAs Based on a Stacked Ensemble Classifier. Bioinformatics (2018) 34(13):2185–94. doi: 10.1093/bioinformatics/bty085
27. Jiang LP, Fan SQ, Xiong QX, Zhou YC, Yang ZZ, Li GF, et al. GRK5 Functions as an Oncogenic Factor in Non-Small-Cell Lung Cancer. Cell Death Dis (2018) 9(3):295. doi: 10.1038/s41419-018-0299-1
28. Duan L, Shen H, Zhao G, Yang R, Cai X, Zhang L, et al. Inhibitory Effect of Disulfiram/copper Complex on Non-Small Cell Lung Cancer Cells. Biochem Biophys Res Commun (2014) 446(4):1010–6. doi: 10.1016/j.bbrc.2014.03.047
29. Wang C, Thor AD, Moore DH, Zhao Y, Kerschmann R, Stern R, et al. The Overexpression of RHAMM, a Hyaluronan-Binding Protein That Regulates Ras Signaling, Correlates With Overexpression of Mitogen-Activated Protein Kinase and is a Significant Parameter in Breast Cancer Progression. Clin Cancer Res (1998) 4(3):567–76.
30. Guo H, Fan Q. Identification of the HMMR Gene as a Diagnostic and Prognostic Biomarker in Hepatocellular Carcinoma Based on Integrated Bioinformatics Analysis. Evid Based Complement Alternat Med (2021) 2021:5970085. doi: 10.1155/2021/5970085
31. Heldin P, Basu K, Olofsson B, Porsch H, Kozlova I, Kahata K. Deregulation of Hyaluronan Synthesis, Degradation and Binding Promotes Breast Cancer. J Biochem (2013) 154(5):395–408. doi: 10.1093/jb/mvt085
32. Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM Functions in Ran-Dependent Microtubule Nucleation and Pole Formation During Anastral Spindle Assembly. Curr Biol (2004) 14(20):1801–11. doi: 10.1016/j.cub.2004.10.002
33. Zhou Q, Fan G, Dong Y. Polo-Like Kinase 4 Correlates With Greater Tumor Size, Lymph Node Metastasis and Confers Poor Survival in Non-Small Cell Lung Cancer. J Clin Lab Anal (2020) 34(4):e23152. doi: 10.1002/jcla.23152
34. Kong T, Ahn R, Yang K, Zhu X, Fu Z, Morin G, et al. CD44 Promotes PD-L1 Expression and Its Tumor-Intrinsic Function in Breast and Lung Cancers. Cancer Res (2020) 80(3):444–57. doi: 10.1158/0008-5472.CAN-19-1108
35. Yin T, Zhao ZB, Guo J, Wang T, Yang JB, Wang C, et al. Aurora A Inhibition Eliminates Myeloid Cell-Mediated Immunosuppression and Enhances the Efficacy of Anti-PD-L1 Therapy in Breast Cancer. Cancer Res (2019) 79(13):3431–44. doi: 10.1158/0008-5472.CAN-18-3397
36. Chen C, Peng S, Li P, Ma L, Gan X. High Expression of NEK2 Promotes Lung Cancer Progression and Drug Resistance and is Regulated by Mutant EGFR. Mol Cell Biochem (2020) 475(1-2):15–25. doi: 10.1007/s11010-020-03854-z
37. Willemen Y, Van den Bergh JM, Bonte SM, Anguille S, Heirman C, Stein BM, et al. The Tumor-Associated Antigen RHAMM (HMMR/CD168) is Expressed by Monocyte-Derived Dendritic Cells and Presented to T Cells. Oncotarget (2016) 7(45):73960–70. doi: 10.18632/oncotarget.12170
38. Spranger S, Jeremias I, Wilde S, Leisegang M, Stärck L, Mosetter B, et al. TCR-Transgenic Lymphocytes Specific for HMMR/Rhamm Limit Tumor Outgrowth In Vivo. Blood (2012) 119(15):3440–9. doi: 10.1182/blood-2011-06-357939
39. Xu F, Guan Y, Xue L, Huang S, Gao K, Yang Z, et al. The Effect of a Novel Glycolysis-Related Gene Signature on Progression, Prognosis and Immune Microenvironment of Renal Cell Carcinoma. BMC Cancer (2020) 20(1):1207. doi: 10.1186/s12885-020-07702-7
40. Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global Characterization of T Cells in Non-Small-Cell Lung Cancer by Single-Cell Sequencing. Nat Med (2018) 24(7):978–85. doi: 10.1038/s41591-018-0045-3
41. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA Circ-CPA4/let-7 miRNA/PD-L1 Axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in Non-Small Cell Lung Cancer (NSCLC). J Exp Clin Cancer Res (2020) 39(1):149. doi: 10.1186/s13046-020-01648-1
42. Xu H, Liu C, Zhang Y, Guo X, Liu Z, Luo Z, et al. Let-7b-5p Regulates Proliferation and Apoptosis in Multiple Myeloma by Targeting IGF1R. Acta Biochim Biophys Sin (Shanghai) (2014) 46(11):965–72. doi: 10.1093/abbs/gmu089
43. Mu X, Wu H, Liu J, Hu X, Wu H, Chen L, et al. Long Noncoding RNA TMPO-AS1 Promotes Lung Adenocarcinoma Progression and is Negatively Regulated by miR-383-5p. BioMed Pharmacother (2020) 125:109989. doi: 10.1016/j.biopha.2020.109989
44. Yu X, Lin Q, Liu F, Yang F, Mao J, Chen X. LncRNA TMPO-AS1 Facilitates the Proliferation and Metastasis of NSCLC Cells by Up-Regulating ERBB2 via Sponging miR-204-3p. Int J Immunopathol Pharmacol (2020) 34:2058738420958947. doi: 10.1177/2058738420958947
Keywords: hyaluronan-mediated motility receptor, non-small cell lung cancer, non-coding RNA, immune infiltration, cell proliferation
Citation: Jiang X, Tang L, Yuan Y, Wang J, Zhang D, Qian K, Cho WC and Duan L (2022) NcRNA-Mediated High Expression of HMMR as a Prognostic Biomarker Correlated With Cell Proliferation and Cell Migration in Lung Adenocarcinoma. Front. Oncol. 12:846536. doi: 10.3389/fonc.2022.846536
Received: 31 December 2021; Accepted: 24 January 2022;
Published: 03 March 2022.
Edited by:
Luciano Mutti, Temple University, United StatesReviewed by:
Chen Li, Free University of Berlin, GermanyCopyright © 2022 Jiang, Tang, Yuan, Wang, Zhang, Qian, Cho and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lincan Duan, ZHVhbm11bXVodW9zYW5AMTYzLmNvbQ==; William C. Cho, Y2hvY3NAaGEub3JnLmhr
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.