
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 13 April 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.845036
This article is part of the Research Topic Protein Kinase and Pseudokinase in Cancer Biology View all 7 articles
 Qianxu Jin1†
Qianxu Jin1† Jiahui Zhao2†
Jiahui Zhao2† Zijun Zhao1
Zijun Zhao1 Shiyang Zhang1
Shiyang Zhang1 Zhimin Sun3
Zhimin Sun3 Yunpeng Shi1
Yunpeng Shi1 Hongshan Yan1
Hongshan Yan1 Yizheng Wang1
Yizheng Wang1 Liping Liu2*
Liping Liu2* Zongmao Zhao1*
Zongmao Zhao1*Calcium/calmodulin-dependent protein ID (CAMK1D) is widely expressed in many tissues and involved in tumor cell growth. However, its role in gliomas has not yet been elucidated. This study aimed to investigate the roles of CAMK1D in the proliferation, migration, and invasion of glioma. Through online datasets, Western blot, and immunohistochemical analysis, glioma tissue has significantly lower CAMK1D expression levels than normal brain (NB) tissues, and CAMK1D expression was positively correlated with the WHO classification. Kaplan–Meier survival analysis shows that CAMK1D can be used as a potential prognostic indicator to predict the overall survival of glioma patients. In addition, colony formation assay, cell counting Kit-8, and xenograft experiment identified that knockdown of CAMK1D promotes the proliferation of glioma cells. Transwell and wound healing assays identified that knockdown of CAMK1D promoted the invasion and migration of glioma cells. In the above experiments, the results of overexpression of CAMK1D were all contrary to those of knockdown. In terms of mechanism, this study found that CAMK1D regulates the function of glioma cells by the PI3K/AKT/mTOR pathway. In conclusion, these findings suggest that CAMK1D serves as a prognostic predictor and a new target for developing therapeutics to treat glioma.
As the most frequent intracranial malignancy, glioma is characterized with infiltrative growth, high mortality, and easy recurrence (1–3). A clinical trial found that the 5-year survival rate of glioblastoma patients was less than 13% even after receiving the correct treatment including surgery, radiotherapy, and chemotherapy (4, 5). The treatment of glioma patients is becoming more standardized, but the prognosis of highly graded patients is still poor (6, 7). Due to the heterogeneity of glioma gene expression, identifying certain glioma-associated genes as therapeutic targets will help to develop more effective strategies for treating glioma (8, 9).
The development and occurrence of glioma is related to imbalanced expression and mutations of pro-tumor and anti-tumor genes, and therapies targeting molecular markers are a key research direction for the future (10). Previous studies have indicated that CAMK1D plays important roles in the CaMKK-CaMK1 signaling cascade triggered by calcium (11), regulates calcium-mediated granulocyte function and respiratory burst (12), and promotes basal dendrite growth of hippocampal neurons (13). Calmodulin (CaM) regulates the activity of a variety of proteins through binding to calcium, thereby regulating the function of relevant signaling pathways that control a variety of cellular functions (14). In addition, tumor-related studies have found that CAMK1D expression is higher in invasive breast cancer than in situ breast cancer and that overexpression of CAMK1D in breast epithelial cells promotes molecular and phenotypic alterations in epithelial–mesenchymal transition (EMT) (15). Overexpression of CAMK1D in lung adenocarcinoma cells promotes cell proliferation but inhibits vascular endothelial cell formation (16, 17). These findings suggest that CAMK1D plays critical roles in the pathogenesis of different cancers. Nevertheless, the clinical significance and function of CAMK1D in glioma remain unknown.
Phosphatidylinositol-3-kinase (PI3K) family is one of the kinases that specifically catalyze the hydroxyl phosphorylation of phosphatidylinositol and produce substances with second messenger effect. The signal pathway composed of PI3K and its downstream molecular signal protein kinase B (AKT)/rapamycin target protein (mTOR) is one of the most important intracellular signal pathways in mammals, which regulates important physiological functions such as cell cycle, protein synthesis, growth, and metabolism. Ribosomal p70S6 kinase (P70S6K) is one of the most characteristic downstream effector molecules of mTOR. The activation of PI3K pathway also increases the phosphorylation of P70S6K (18). It has been demonstrated that calmodulin affects autophagy in prostate cancer cells via AKT/mTOR (19) and also participates in PLCγ1-dependent ECM synthesis via the mTOR/P70S6K pathway (20). Thus, in this study, we determined if the PI3K/AKT/mTOR pathway plays an important role in CAMK1D regulation of tumor function.
This study focused on the function of CAMK1D in glioma. First, the relative level of CAMK1D in glioma and its relation with overall survival of glioma were analyzed using bioinformatics. We found that downregulation of CAMK1D was associated with poor prognosis in glioma patients. The role of CAMK1D in glioma cell proliferation, invasion, and migration was also explored. Finally, we investigated the potential mechanisms involved in CAMK1D-induced regulation of glioma.
Bioinformatics analysis was performed based on 6 glioma cohorts (CGGA, TCGA, Rembrandt, Gravendeel, Phillips and Kamoun) containing clinicopathological and gene expression data obtained from Gliovis (http://gliovis.bioinfo.cnio.es/). We also analyzed the pan-cancer data of CAMK1D in the Cancer Genome Atlas (TCGA), including 33 cancer types using UCSCXenaShiny (https://hiplot.com.cn/advance/ucscxena-shiny). Then, the association between clinical characteristics and CAMK1D expression was explored and visualized through the R package ggplot2. The high- and low-expression groups were distinguished by the median expression of CAMK1D. We subsequently performed survival analysis to compare the overall survival between the low-CAMK1D and high-CAMK1D groups through Survminer R packages.
To determine the exact mechanisms of CAMK1D in glioma, “limma” package in R was applied to detect the differentially expressed genes (DEGs). Then, KEGG (Kyoto Encyclopedia of Genes and Genomes), GSEA (Gene set enrichment analysis), and GO (Gene Ontology) were conducted to investigate potential activating biological function processes or pathways in the low-CAMK1D population.
A total of 80 glioma and normal tissues were obtained from a total of 80 patients treated in The Second Hospital of Hebei Medical University. All samples were confirmed by pathology. Patients who received radiotherapy or chemotherapy before surgery were excluded from this study. The Ethics Committee of The Second Hospital of Hebei Medical University approved this research. The ethics committee waived the requirement of written informed consent for participation.
A total of four glioma cell lines (U251, A172, LN229, and U87) and normal human astrocytes (NHAs) were obtained from Procell Life Science. The U251 cells were cultured in RPMI−1640 medium (Gibco™,11875093). The A172 and LN229 cells were cultured in DMEM (Gibco™, 10564011). The U87 cell lines were cultured in MEM (Gibco™, 11090081), while the NHA cells were cultured in astrocyte medium (AM, Sciencell™, #1801). All media were supplemented with 10% FBS (Gibco™, 10099141), 100 U/ml penicillin, and 100 μg/ml streptomycin (Pen-Strep Solution, BI, 2114091). Cells were incubated with 5% CO2 at a temperature of 37°C in a standard humidified incubator.
The CAMK1D−coding sequence was synthesized and cloned into the pcDNA 3.1 vector by Hanbio Biotechnology Co., Ltd. to construct a CAMK1D overexpression plasmid (pcCAMK1D). The blank pcDNA3.1 plasmid was used as negative control (NC) plasmid. The siRNA for silencing CAMK1D was designed and synthesized from Thermo Fisher Scientific. The targeting sequence for siCAMK1D was AAGAUGUAGGCAAUCACUCCG. Plasmids (8 μg/106 cells) or small interfering RNA (siRNA) (60 pmol/106 cells) were transfected in glioma cells using Lipofectamine 3000 (L3000015, Invitrogen), according to the product instruction. After 48–72 h, all cells were collected and used in the follow-up experiments.
Total RNA was extracted using TRIzol (Invitrogen) and then we used PrimeScript RT reagent (Takara) to reverse transcribe RNA (1 μg) into cDNA according to the instructions. Regarding RT-PCR, the ABI Prism 7500 RT-PCR system (Applied Biosystems) and the SYBR Premix ExTaq (Takara) were used. RT-PCR cycles included pre-denaturation for 30 s at 95°C, denaturation for 5 s at 95°C, annealing for 30 s at 60°C, and extension for 30 s at 72°C, 40 cycles in the last three steps. The primer sequences were as follows (5’−3’): CAMK1D, forward AATGGAGGGCAAAGGAGATGTGATG, and reverse GTA AGGTTTCTGGGCGAGGACTTC; GAPDH, forward GGAGCGAGATCCCTCCAAAAT, and reverse GGCTGTTGTCATACTTCTCATGG. The relative CAMK1D expression was calculated using the 2−ΔΔCq method.
Paraffin-embedded tissue specimens (4 μm thick) were sectioned, dewaxed, and rehydrated in gradient ethanol. Then, we add 3% H2O2 and soak for 10 min to remove endogenous peroxidase. The antigen retrieval was accomplished in 10 mM citrate at 95°C for 20 min. Slides were blocked in goat serum for 60 min and incubated with rabbit anti-CAMK1D antibodies (1:100; ab172618; Abcam) overnight at 4°C. After washes in phosphate buffered saline (PBS), goat anti-rabbit secondary antibody (sp-9001, Zhongshan Golden Bridge) was added for 60 min at room temperature. Subsequently, slides were incubated with horseradish peroxidase (HRP) (sp-9001, Zhongshan Golden Bridge). The slides were lightly counterstained with hematoxylin and dehydrated. Finally, images were captured with Leica DM2000 microscope.
Tissues and cells of human were lysed in radioimmunoprecipitation assay buffer (Beyotime) containing a protease and phosphatase inhibitor cocktail (K1015, APExBIO). The protein concentration was assayed by the BCA method. The extracted or enriched protein samples were separated by denaturing 10% SDS-polyacrylamide gel electrophoresis and transferred into polyvinylidene fluoride membranes. After being blocked by 5% bovine serum albumin (BSA) for 120 min at room temperature, the membranes were incubated with primary antibodies from abcam and Cell Signaling Technology against GAPDH (ab8245, 1:8,000), mTOR (ab2732, 1:1,500), AKT (ab18785, 1:1,000), p-mTOR (ab109268, 1:1,500), p-AKT (ab38449, 1:1,000), P70S6K (#2708, 1:1,000), p-P70S6K (#9205, 1:1,000), and CAMK1D (ab172618, 1:1,500) overnight at 4°C. After washing three times, the blots were further incubated with goat anti-rabbit IRDye 800CW preadsorbed secondary antibody (1:10,000; Abcam; ab216773). The images were detected with an Odyssey infrared imaging scanner (LI-COR, USA).
Cell samples were initially seeded at 100 μl of medium containing 5×103 cells/well on 96−well plates. Then, we measured the cell proliferation rate at 0, 1, 2, 3, and 4 days after transfection. Each well in the 96−well plate was supplemented with 10 μl of CCK-8 (Report) and incubated at 37°C for an additional 2 h. Eventually, the absorbance was measured at 450 nm of 96−well plates and was determined by the use of a strip reader (SpectraMax Plus 384).
Seeding of transfected cancer cells in the serum-free medium was performed in the upper chamber of Transwell (8−μm; BD Biosciences) precoated with 40 μl of Matrigel (to assess invasion) or non-coated (to assess migration). Then, 10% FBS containing culture medium was plated into the lower chamber. After incubating for 1 day, cells were removed on the upper chamber with cotton swabs, fixed with 4% paraformaldehyde, and stained in 0.1% crystal violet.
Cell samples of glioma were planted at 800 cells/well to the 6-well plates. Following 2 weeks of cell culture, samples were treated with 4% paraformaldehyde and Giemsa stain in succession, and then the total number of colonies was counted with ImageJ (version1.52p).
Cell samples of glioma were plated into the six-well plate for treatment and grew up to 90% confluency. Then, we created the scratch with a pipette tip on these cell monolayers. The scratch was photographed with the microscope (Olympus, Japan) at 0 h and 24 h at the same position. Finally, the width of the scratch was analyzed with ImageJ.
Glioma cells (5×106) were injected into the subcutis of athymic BALB/c nude mice (4 weeks, male). The length (L) and the width (W) of xenograft were estimated using a vernier caliper at 7, 14, 21, and 28 days. The xenograft volume was calculated by the equation: V = (W2 × L)/2. After 28 days, the mice were sacrificed and subcutaneous xenografts were collected, photographed, and weighed. The regulations of the Ethics Committee of the Second Hospital of Hebei Medical University were followed in animal experiments.
Each experiment was repeated thrice and exhibited as the mean values ± standard deviation (SD). All data were analyzed statistically by Prism 8 (GraphPad Inc, USA) and R software (version 3.6.3). Student’s t-test and one-way ANOVA were used in statistical analysis. The prognostic value of patients was shown using the Kaplan–Meier curve. Factors for overall survival were performed using Log-rank test and p-values less than 0.05 were judged as statistically significant difference between groups.
Pan-cancer analysis showed that CAMK1D expression levels were significantly different between multiple tumor tissue and adjacent tissues (or GTEx) (Figure 1A). Expression of CAMK1D was lower in BLCA, DLBC, GBM, LGG, PAAD, PRAD, SKCM, TGCT, THCA, and UCEC than in adjacent tissues, whereas CAMK1D expression levels were significantly higher in tumor of ACC, BRCA, CHOL, ESCA, KICH, KIRC, KIPR, LAML, LIHC, LUSC, LUAD, OV, PRAD, SARC, PCPG, STAD, UCS, and THYM than in adjacent tissues.
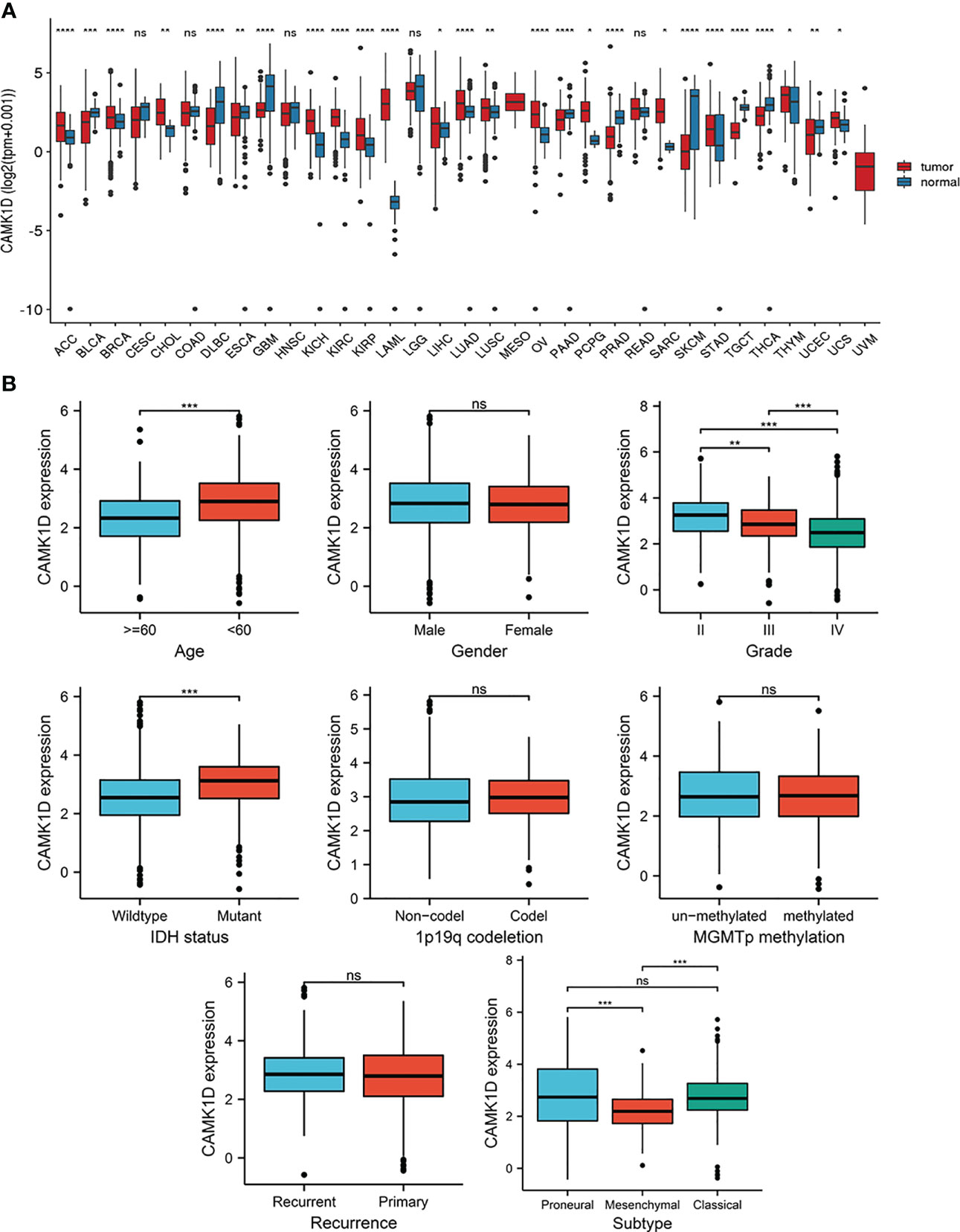
Figure 1 Pan-cancer analysis of CAMK1D expression and the relationship between the expression of CAMK1D and clinicopathological features. (A) CAMK1D mRNA expression in pan-cancer dataset of TCGA. (B) Expression of CAMK1D in different age, genders, WHO grades, IDH status, 1p19q codeletion, recurrence, MGMTp methylation, and subtypes of glioma. Please note the CAMK1D gene expression level changes in different tumor tissues. Furthermore, CAMK1D expression level was low in glioma tissue with higher WHO grade patients. ns, no significance; *p < 0.05 < **p < 0.01 < ***p < 0.001 < ****p < 0.0001.
To determine the role of CAMK1D in tumor pathogenesis and development, we studied the relationship between CAMK1D protein and clinicopathological features, including gender, age, recurrence, WHO grade, 1p19q codeletion, subtype, IDH status, and MGMTp methylation in the CGGA dataset. Our results show that the expression of CAMK1D in glioma tissues was decreased in aged and higher WHO grade patients. In addition, CAMK1D expression levels were different under different subtypes and different IDH mutations in different states (Figure 1B).
The patients were divided into low- and high-CAMK1D groups based on the median expression of CAMK1D. The survival analysis indicated that glioma patients with lower expression of CAMK1D in TCGA (HR: 0.33, 95% CI 0.25–0.44), CGGA (HR: 0.60, 95% CI 0.51–0.71), Rembrandt (HR: 0.54, 95% CI 0.43–0.68), Gravendeel (HR: 0.56, 95% CI 0.43–0.73), Kamoun (HR: 0.81, 95% CI 0.44–1.49), Phillips (HR: 0.43, 95% CI 0.26–0.73), Freije (HR: 1.04, 95% CI 0.62–1.73), and LeeY (HR: 0.85, 95% CI 0.63–1.14) exhibited significantly shorter survival time than patients with higher expression, while patients from the Kamoun cohort displayed the same trend with no statistical significance (Figure 2A). To show the reliability of our results, a meta-analysis was performed to gather the HR of the eight glioma datasets, and results also confirmed that low-CAMK1D patients displayed shorter OS time compared to high-CAMK1D patients (RR:0.5, 95% CI 0.52–0.63, Figure 2B).
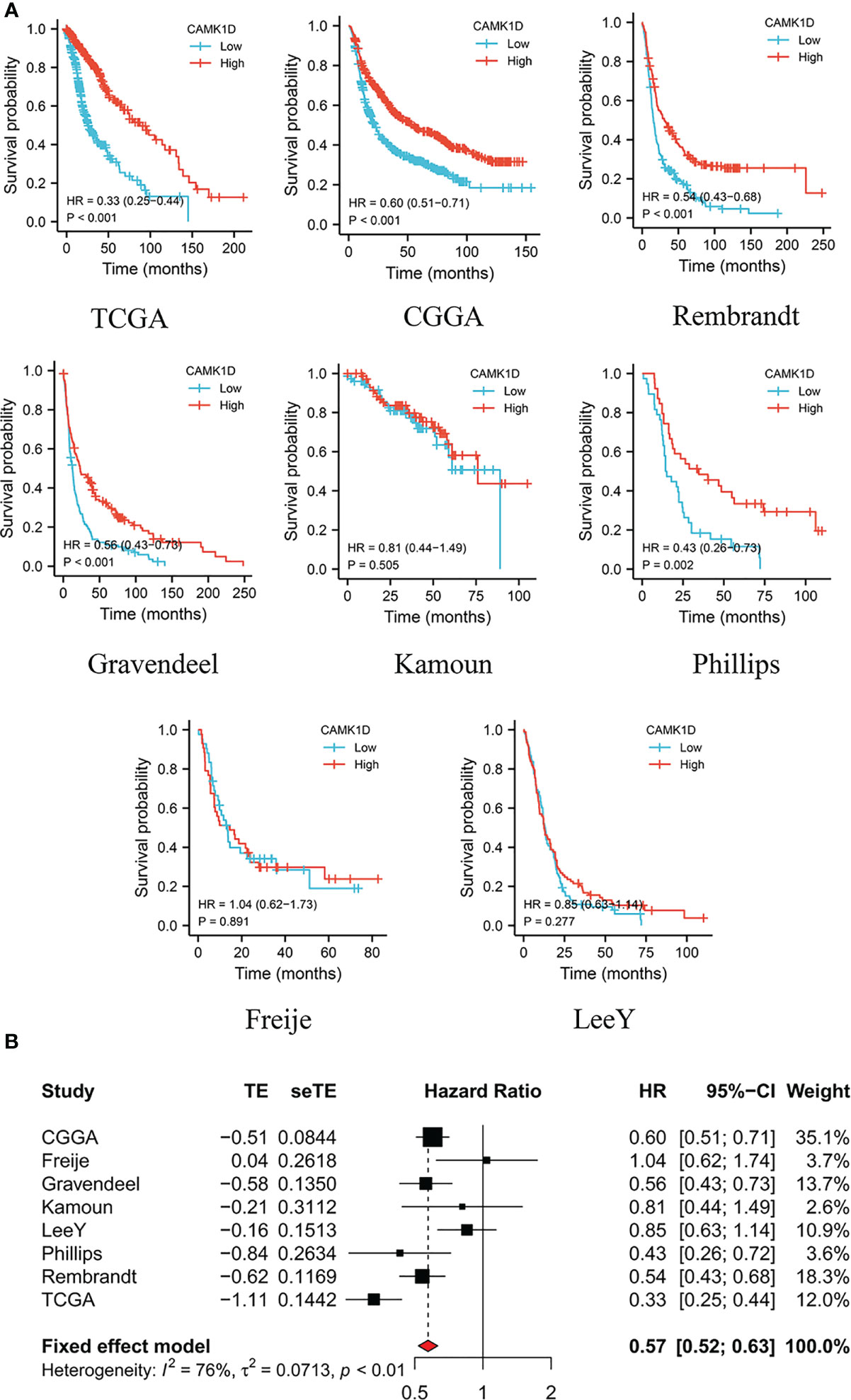
Figure 2 Decreased expression of CAMK1D predicts short survival time of glioma patients. (A) Survival curves of CAMK1D in various datasets of glioma showing that glioma patients with lower expression level of CAMK1D have shorter survival time than patients with higher expression levels. (B) The forest plot was presented in the RRs for glioma patients with high expression of CAMK1D compared to low expression.
To further explore the functions of CAMK1D in glioma, CAMK1D expression was estimated in glioma tissues compared with normal brain tissues using Western blot, IHC, and RT−qPCR. The results indicated that the expression of CAMK1D was strikingly downregulated in glioma tissues from patients at all 4 WHO grades compared with normal brain tissues (Figures 3A-C). The expression levels were decreased progressively as the grades increased. Similarly, the expressions of CAMK1D in 4 glioma cell lines were significantly lower than the NHA (Figures 3D, E). These results suggest that CAMK1D may be a cancer suppressor gene in glioma. The data of Kaplan–Meier survival were obtained from the medical records of glioma patients in The Second Hospital of Hebei Medical University hospital. According to the median cutoff of the expression of CAMK1D, the patients were separated into two groups. The result revealed that low CAMK1D expression was significantly related with the poor overall survival of glioma patients (Figure 3F).
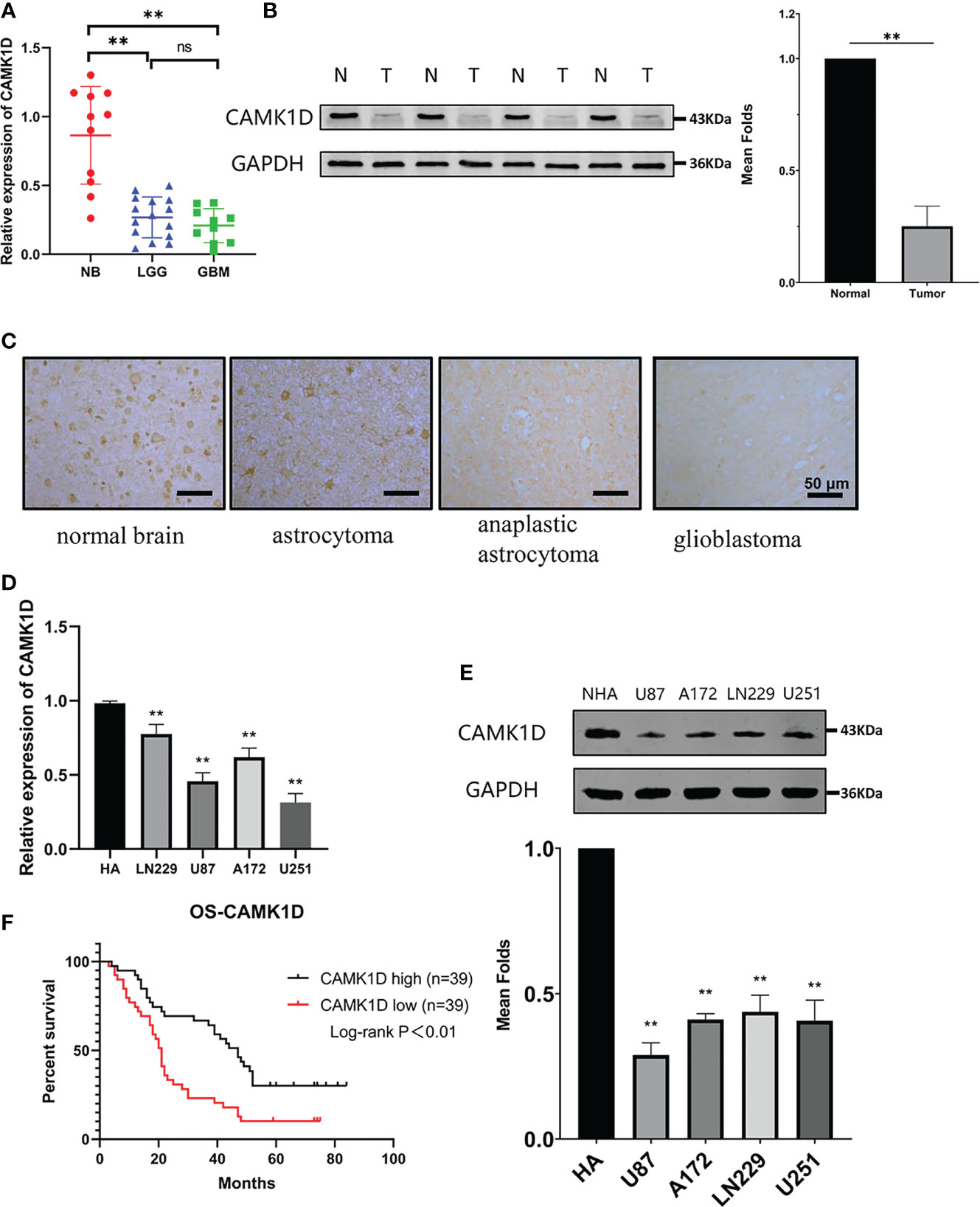
Figure 3 CAMK1D expression in tissues and cells. (A) CAMK1D mRNA expression was signficantly decreased in glioma samples compared with normal brain samples (NB = 11; LGG = 15; GBM = 10). (B) CAMK1D protein expression was significantly decreased in four glioma samples compared with four normal brain samples with Western blot analysis. (C) Immunohistochemistry showed that the expression of CAMK1D immunoreactivities were different in normal brain tissue, astrocytoma, anaplastic astrocytoma, and glioblastoma. (D, E) CAMK1D expression levels in HA cell lines and four glioma cell lines were determiend by RT-PCR and Western blot. Both mRNA and protein levels were signficantly decreased in the 4 glioma cell lines compared with HA cell lines. (F) Kaplan–Meier curves for overall survival of glioma patients based on expression of CAMK1D protein (78 glioma samples) showing that low-level CAMK1D expression was closely associated with the poor overall survival of glioma patients. ns, no significance; **p < 0.01.
We detected the expression levels of CAMK1D in U251 and U87 cells that were transfected with plasmid or siRNA for 2 days using RT−qPCR and Western blot. The mRNA and protein levels of CAMK1D were notably upregulated in glioma cells transfected with CAMK1D plasmids and were downregulated in glioma cells transfected with siCAMK1D, compared to the pcNC (transfected an empty vector) and siNC (transfected a scramble siRNA) (Figures 4A, B). As evidenced by CCK-8 assay and colony formation assay, CAMK1D overexpression significantly inhibited the proliferation of cells; on the contrary, CAMK1D knockdown resulted in the opposite (Figures 4C, D). Proliferation of cells were strengthened by CAMK1D knockdown compared with cells in the siNC group.
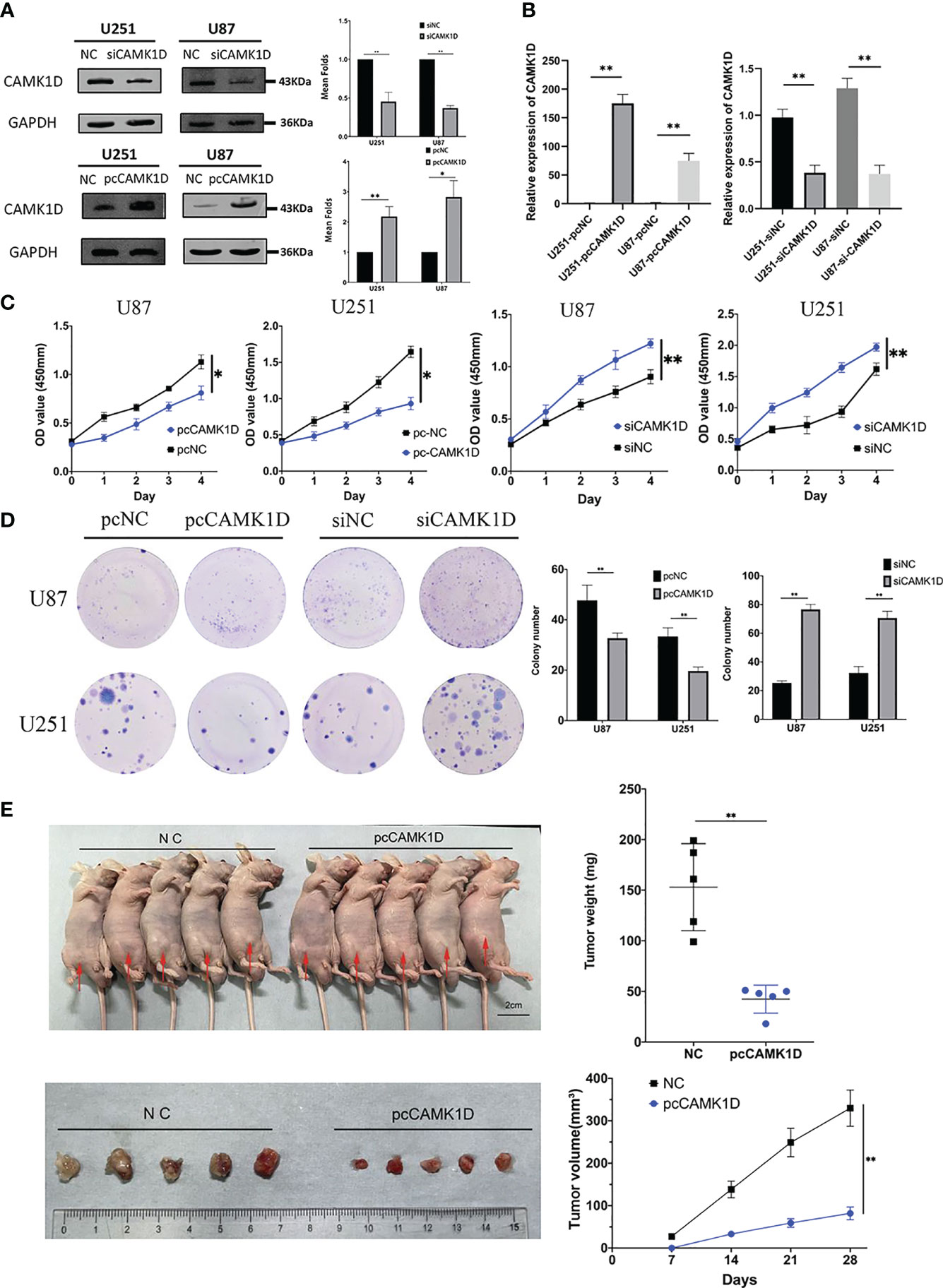
Figure 4 CAMK1D inhibits glioma cell proliferation. (A, B) The protein and mRNA levels of CAMK1D were able to be downregulated and overexpressed in glioma cells by siCAMK1D and paCAMK1D. (C) CCK8 assays measured the effect of CAMK1D on the growth of glioma cells. (D) Colony formation assay used to verify the influence of CAMK1D on glioma cell proliferation. (E) The tumor growth rate in vivo was inhibited after injection of U251 cells overexpressing CAMK1D. *p < 0.05; **p < 0.01.
Based on the experimental results in vitro, we inoculated pcCAMK1D and pcNC-treated U251 cells, subcutaneously into immunodeficient mice to establish a tumor transplantation model. The growth of tumors in two groups showed that tumors in the CAMK1D overexpression group grew significantly slower compared with the control group. Twenty-eight days after U251 cell injection, tumors were excised and weighed. The mean volume was 44.69 ± 18.64 mm3 in the CAMK1D overexpression group and 172.53 ± 73.32 mm3 in the control group. The mean mass weight was 42.40 ± 13.83 mg in the CAMK1D overexpression group and 153.00 ± 43.03 mg in the control group. There were significant differences in tumor weight and volume between the transfected and control groups (Figure 4E). These in vivo findings showed that CAMK1D inhibited the proliferative ability of U251 glioma cells, which was consistent with our findings in in vitro experiments.
To detect the role of CAMK1D in cell invasion and migration, we used scratch wound healing assays, transwell invasion, and migration assays. The results revealed that glioma cell invasion and migration were weakened by CAMK1D overexpression. Meanwhile, cell migration and invasion were strengthened by CAMK1D knockdown compared with cells in the siNC group (Figures 5A, B). The in vitro data suggest that CAMK1D represses glioma cell proliferation, invasion, and migration abilities.
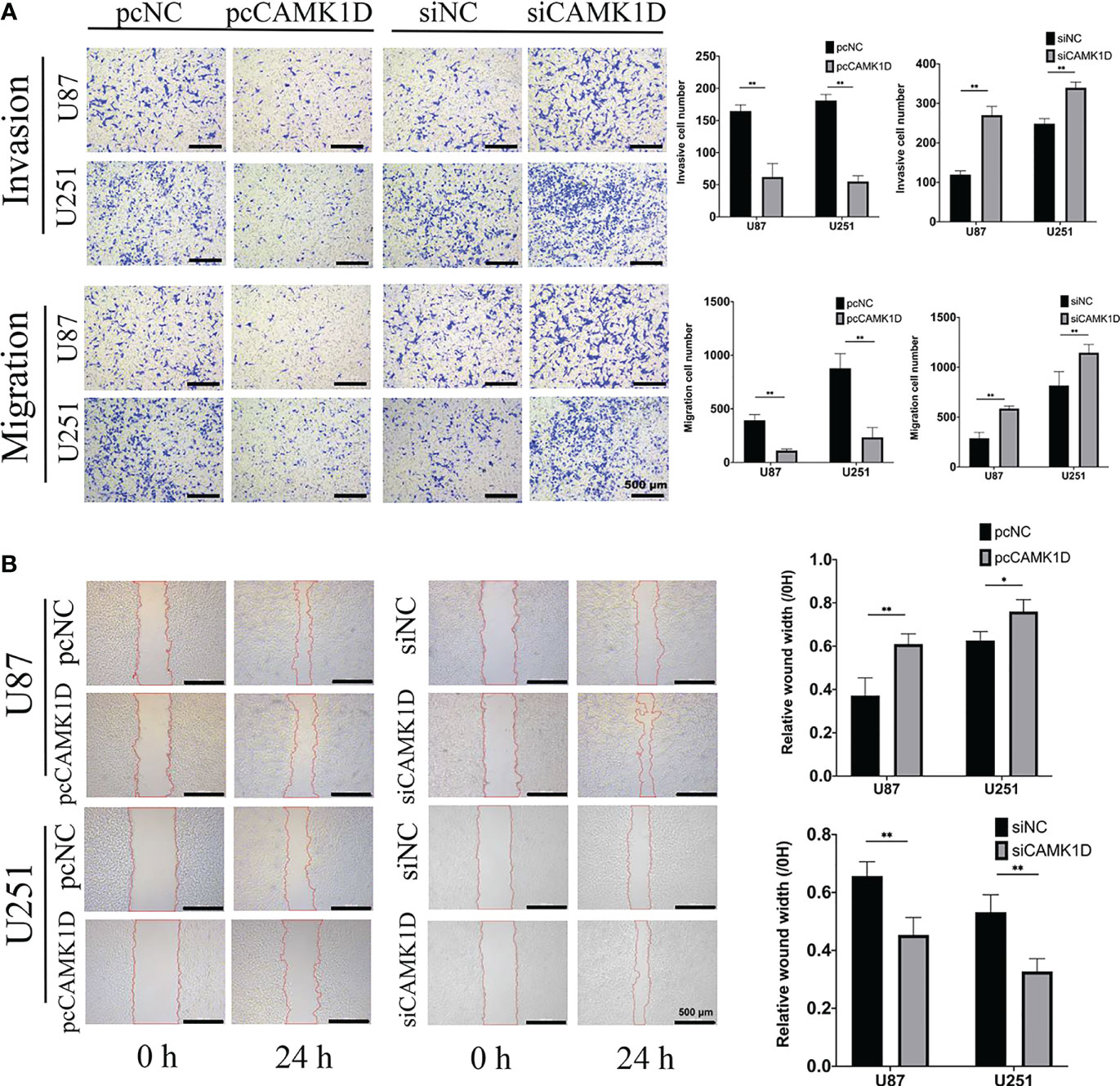
Figure 5 CAMK1D inhibits the invasion and migration of glioma cells. (A) Transwell assays revealed that CAMK1D overexpression reduced the invasion and migration of U87 and U251 glioma cells. The invasive cell numbers and migration cell numbers of U87 and U251 cell lines were significantly decreased by overexpression of CAMK1D with pcCAMK1D. (B) Wound healing assay showed that overexpression of CAMK1D reduced the relative wound width in U87 and U251 glioma cells. *p < 0.05; **p < 0.01.
To understand the potent mechanisms underlying the role of CAMK1D in regulating glioma proliferation, invasion, and migration, we analyzed DEGs between high and low CAMK1D groups in TCGA. Log2 fold-change (FC) > 2 and p-value less than 0.05 were recognized as screening crit eria, which contained 1,043 downregulated genes and 380 upregulated genes (Figure 6A). Enrichment analysis was performed, and the DEGs were mainly enriched in substrate-specific channel activity, dendrite membrane, regulation of immune effector process, neuroactive ligand–receptor interaction, and the PI3K/Akt/mTOR signal pathway (Figures 6B, C). Another 10 genes were identified by the STRING database as CAMK1D-related genes with significant interactions, including JAZF1, TSAN8, CDC123, THADA, CDKAL1, CALM3, CALM1, CREB1, CALM2, and NOS3. The PPI network of CAMK1D and CAMK1D-related genes was constructed and visualized by the STRING database (Figure 6D). Then, we used GSVA and GSEA methods to predict the potential carcinogenic pathway of CAMK1D. We found that many different signaling pathways were significantly related to CAMK1D expression (Figures 6E–G), such as EMT, glycolysis, p53 signaling pathways, and WNT signaling pathways. Notably, both analyses showed that the PI3K/AKT/mTOR signaling pathway were significantly enriched in tumor-related pathways, and the activation of this pathway was negatively correlated with CAMK1D expression (Figure 6H).
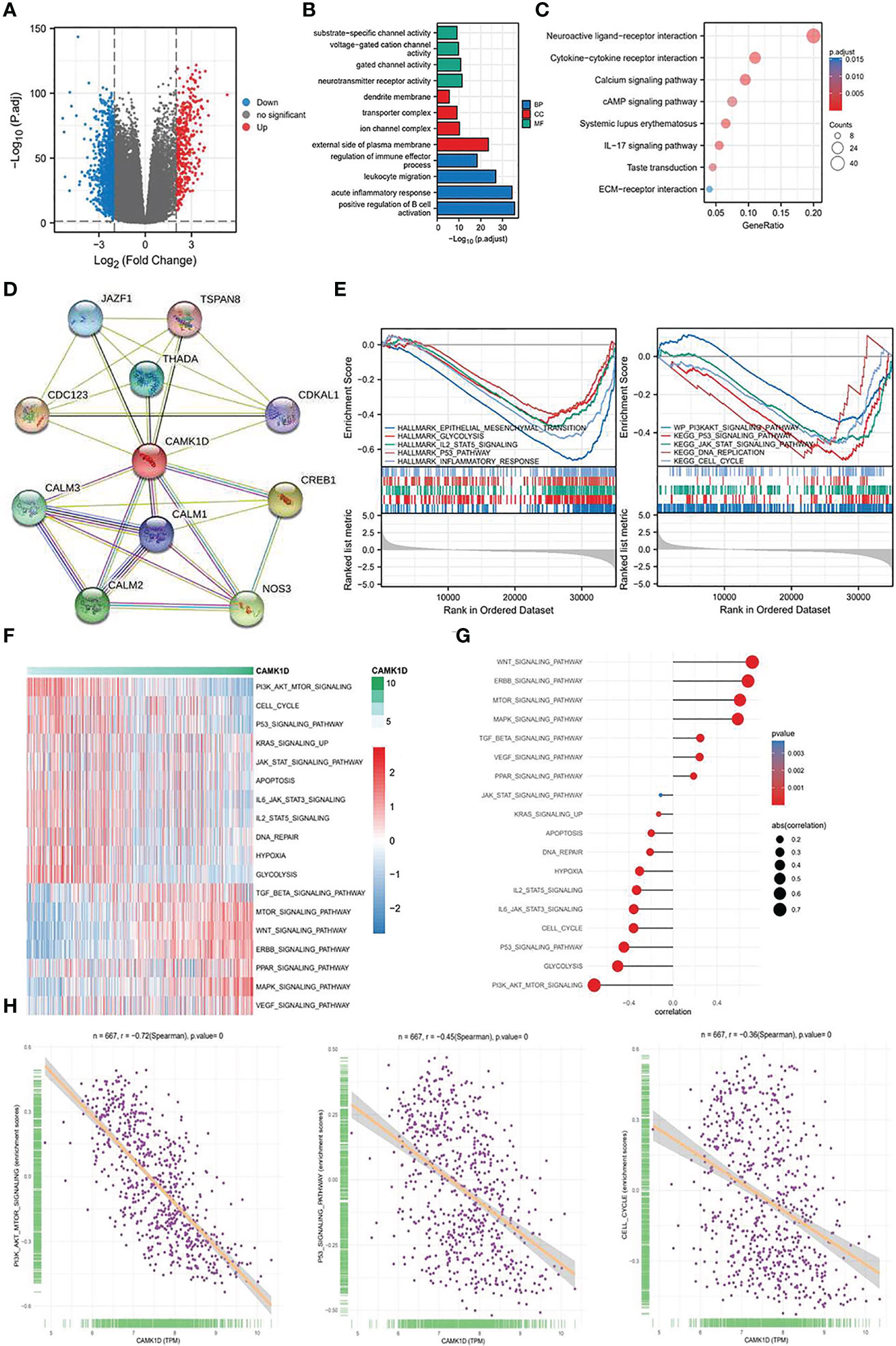
Figure 6 Determination of the biological mechanism and processes affected by CAMK1D. (A) The Volcano plot showed the DEGs in low- and high-CAMK1D groups. (B) Analysis of GO enrichment for the DEGs. (C) Analysis of KEGG pathway enrichment for the DEGs. (D) The PPI network of CAMK1D constructed by the STRING database. (E) GSEA between the low- and high-risk group (F) The heatmap showed the GSVA scores of 18 pathways in low- and high-risk populations. (G) The association graph between GSVA and risk scores of 18 pathways. (H) The correlation between the PI3K/AKT/mTOR signaling, P53 signaling, cell cycle, and the expression of CAMK1D.
Then, we determined the effect of CAMK1D overexpression on PI3K/AKT/mTOR pathway in glioma cells. Western blot analysis showed that overexpression of CAMK1D downregulated p-Akt, p-mTOR, and p-P70S6K, and decreased p-Akt/Akt, p-mTOR/mTOR, and p-P70S6K/P70S6K ratios in glioma cells (Figure 7A). Moreover, the involvement of this pathway in glioma cells was analyzed by applying PI3K/Akt/mTOR pathway inhibitor LY294002. By using CCK-8, transwell, and wound healing assay, we determined whether inhibition of the PI3K/AKT/mTOR signaling pathway by LY294002 reversed the inhibition of cell proliferation, invasion, and migration induced by CAMK1D overexpression. These results obtained from CCK-8, transwell, and wound healing assay all indicated that LY294002 reversed the promotion of cell proliferation, invasion, and migration by siCAMK1D (Figures 7B–E). These findings support the notion that CAMK1D regulates glioma cell proliferation, migration, and invasion through the PI3K/AKT/mTOR pathway.
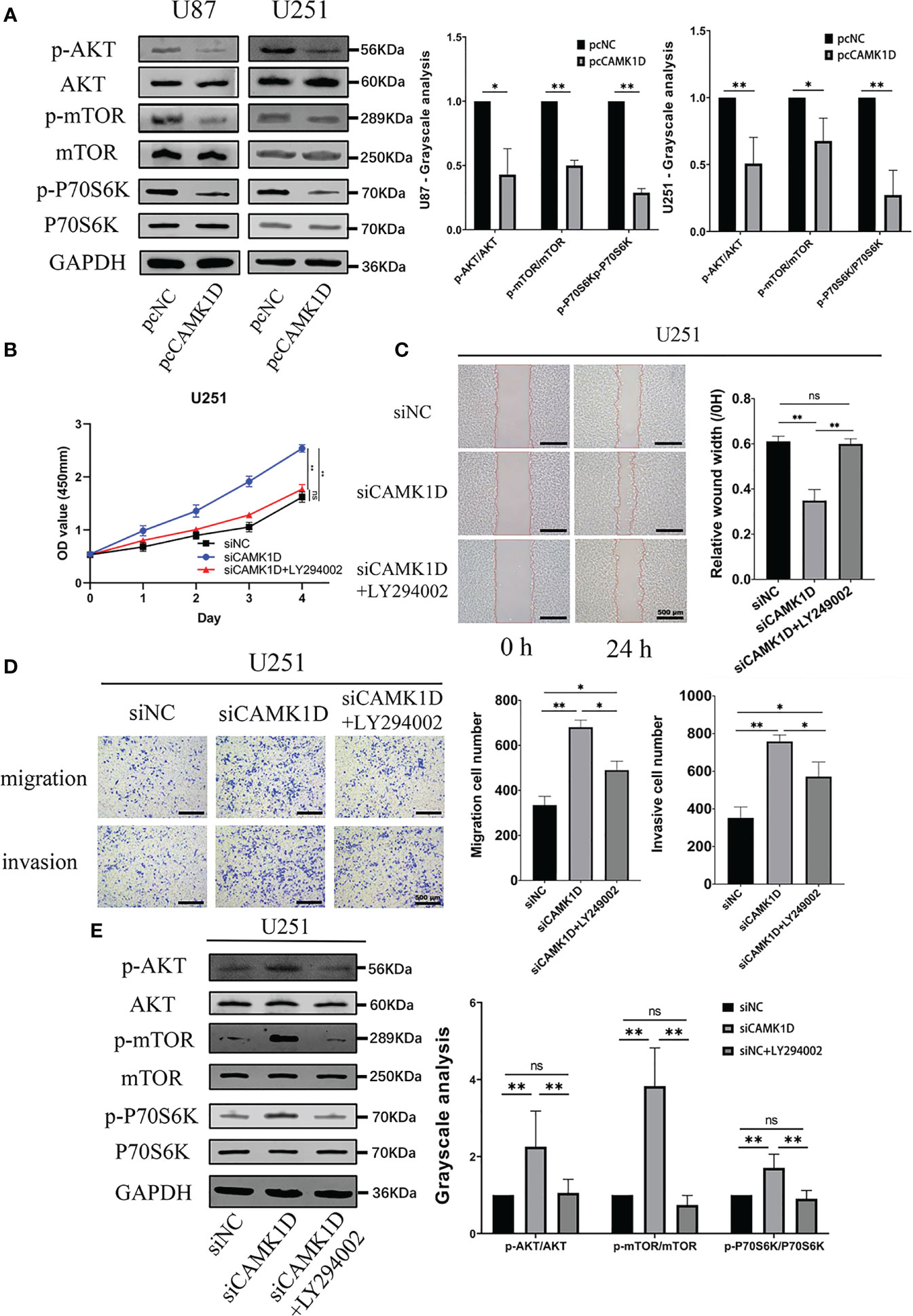
Figure 7 CAMK1D level changes on the PI3K/AKT/mTOR pathway. (A) Compared with the control, the expression of p-AKT and p-mTOR was significantly decreased after CAMK1D overexpression using pcCAMK1D plasmid construction in U87 and U251 cells. (B–D) LY294002 reversed the increases in cell proliferation, migration, and invasion induced by the decrease in CAMK1D using siCAMK1D in U87 and U251 cells. (E) LY294002 reversed the decrease of p-AKT and p-mTOR expression by siCAMK1D. ns, no significance; *p < 0.05; **p < 0.01.
CAMK1D is widely present in a variety of cell types and plays an important role in control of formation of synapses, growth cone movement and axon growth, aldosterone synthase expression, visual signaling processes, and the cell cycle (21–26). Based on bioinformatics analysis and screening, our study was the first to determine the role of CAMK1D in cell proliferation, migration, and invasion of glioma cells. We found that CAMK1D expression was significantly downregulated in glioma cell lines and glioma tissues from patients. However, based on the tumor expression data in the TCGA and GTEx databases, the expression level of CAMK1D in various tumors is not consistent with its corresponding normal tissues. The above findings suggest that CAMK1D probably plays different functions in different cancer types. In addition, CAMK1D expression in the database was associated with overall survival in patients with glioma. This implies that whether CAMK1D can be used as a prognostic marker for glioma patients needs to be studied in a larger collection of glioma specimens.
We found that overexpression of CAMK1D can inhibit the proliferation, migration, and invasion of glioma cells, whereas knockdown of camk1d using siRNA produced opposite effects. This is not identical to the previously reported function of CAMK1D in other cancers. For example, CAMK1D promotes the proliferation of breast cancer through CERB/CCND1 (15), inhibits the angiogenesis of lung adenocarcinoma by HH3 (17), enhances the resistance of multiple myeloma to T cells by phosphorylation of caspase-3 and caspase-6 (27), and inhibits the apoptosis of human choroidal trophoblastic cells (28). This reinforces the fact that camk1d plays different functions in different tumor types. The reason may be that the targeting mechanism of multifunctional kinases allows the same kinases in different cells to respond differently to stimulation and produce different functions (29–31). The function of CAMK1D-mediated depends on the different activation states and binding substrates in different cancers, thus affecting the function and signaling pathways involved in the substrates. Moreover, the different microenvironment of kinase will also affect its conformation and function. Thus, other functions of CAMK1D in glioma needs to be further studied.
To investigate the signaling pathway involved in the regulatory mechanism of CAMK1D in glioma, we predicted and determined that the PI3K/AKT/mTOR pathway was critically involved in the inhibitory effect of CAMK1D in glioma. It has been shown that abnormal activation of the PI3K/AKT/mTOR pathway is associated with many illnesses, including tumorigenesis (32, 33). In this study, we examined the protein levels of Akt/p-AKT, mTOR/p-mTOR, and P70S6K/p-P70S6k. We found that overexpression of camk1d significantly inhibited p−AKT, p-mTOR, and p-P70S6k. Block the PI3K/AKT/mTOR pathway in U251 cells using LY294002 reversed CAMK1D knockdown-mediated changes in cell function. These data suggest that CAMK1D inhibits cell proliferation, invasion, and migration by regulating the PI3K/AKT/mTOR signaling pathways.
Our findings were the first research report showing that CAMK1D overexpression inactivated the PI3K/AKT/mTOR signaling pathway. However, the limitation of this study is that we only studied the PI3K/Akt/mTOR signaling pathway. The regulation of CAMK1D in glioblastoma is a complex network involving multiple genes. In this study, we did not determine whether CERB or HH3 pathway was involved in CAMK1D overexpression-induced inhibition of glioma (15). In addition, we did not examine whether CAMK1D has other functions in glioma, such as reducing apoptosis, affecting EMT, and autophagy. Thus, we cannot rule out the possibility that other pathways are involved in the effect of CAMK1D in glioma, which deserves further investigation in our future studies. Since Ca2+ directly regulates CAMK1D activity (34, 35), Ca2+ dynamics, especially the frequency, amplitude, and duration of intracellular Ca2+ concentration changes, is a potential mechanism to regulate CAMK1D activity.
In summary, we found that CAMK1D promotes glioma cell proliferation, migration, and invasive processes through activation of the PI3K/AKT/mTOR signaling pathway. These findings suggest that CAMK1D expression is downregulated in glioma and can be used as a prognostic indicator for glioma patients.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Hospital of Hebei Medical University. The ethics committee waived the requirement of written informed consent for participation. The animal study was reviewed and approved by the Ethics Committee of the Second Hospital of Hebei Medical University.
QJ and JZ conducted experiments, analyzed and interpreted data, and wrote the original draft. ZJ Zhao performed the bioinformatics analysis. SZ, YS, and HY were involved in animal experiments. YW collected the samples and clinical data. ZS created tables, graphs, and figures. ZM Zhao and LL conceived the idea for the project, revised the paper, and received funding for the project. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (81870984); the National Key R & D Program Intergovernmental Cooperation on International Scientific and Technological Innovation of the Ministry of Science and Technology of China (2017YFE0110400); the Hebei Natural Science Foundation General Project—Beijing-Tianjin-Hebei Basic Research Cooperation Project (H2018206675); the Special Project for the Construction of Hebei Province International Science and Technology Cooperation Base (193977143D); the Government-funded Project on Training of outstanding Clinical Medical Personnel and Basic Research Projects of Hebei Province in the Year of 2017; and the Government-funded Project on Training of Outstanding Clinical Medical Personnel and Basic Research Projects of Hebei Province in the Year of 2019.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Molinaro AM, Taylor JW, Wiencke JK, Wrensch MR. Genetic and Molecular Epidemiology of Adult Diffuse Glioma. Nat Rev Neurol (2019) 15(7):405–17. doi: 10.1038/s41582-019-0220-2
2. Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro Oncol (2020) 22(12 Suppl 2):iv1–96. doi: 10.1093/neuonc/noaa200
3. Zhao Z, Wu Y, Wang Z, Xu J, Wang Y, Zhao Z. Establishment and Validation of Five Autophagy-Related Signatures for Predicting Survival and Immune Microenvironment in Glioma. Genes Genomics (2021) 44(1):79–95. doi: 10.1007/s13258-021-01172-2
4. Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. Jama (2017) 318(23):2306–16. doi: 10.1001/jama.2017.18718
5. Gu J, Wang J, You A, Li J, Zhang Y, Rao G, et al. MiR-137 Inhibits the Proliferation, Invasion and Migration of Glioma via Targeting to Regulate EZH2. Genes Genomics (2021) 43(10):1157–65. doi: 10.1007/s13258-021-01117-9
6. Gusyatiner O, Hegi ME. Glioma Epigenetics: From Subclassification to Novel Treatment Options. Semin Cancer Biol (2018) 51:50–8. doi: 10.1016/j.semcancer.2017.11.010
7. Wang H, Xu T, Huang Q, Jin W, Chen J, et al. Immunotherapy for Malignant Glioma: Current Status and Future Directions. Trends Pharmacol Sci (2020) 41(2):123–38. doi: 10.1016/j.tips.2019.12.003
8. Zachariah MA, Oliveira-Costa JP, Carter BS, Stott SL, Nahed BV. Blood-Based Biomarkers for the Diagnosis and Monitoring of Gliomas. Neuro Oncol (2018) 20(9):1155–61. doi: 10.1093/neuonc/noy074
9. Chen TC, da Fonseca CO, Schönthal AH. Intranasal Perillyl Alcohol for Glioma Therapy: Molecular Mechanisms and Clinical Development. Int J Mol Sci (2018) 19(12):3905. doi: 10.3390/ijms19123905
10. Lapointe S, Perry A, Butowski NA. Primary Brain Tumours in Adults. Lancet (2018) 392(10145):432–46. doi: 10.1016/S0140-6736(18)30990-5
11. Verploegen S, Lammers JW, Koenderman L, Coffer PJ. Identification and Characterization of CKLiK, a Novel Granulocyte Ca(++)/Calmodulin-Dependent Kinase. Blood (2000) 96(9):3215–23. doi: 10.1182/blood.V96.9.3215.h8003215_3215_3223
12. Verploegen S, Ulfman L, van Deutekom HW, van Aalst C, Honing H, Lammers JW, et al. Characterization of the Role of CaMKI-Like Kinase (CKLiK) in Human Granulocyte Function. Blood (2005) 106(3):1076–83. doi: 10.1182/blood-2004-09-3755
13. Kamata A, Sakagami H, Tokumitsu H, Owada Y, Fukunaga K, Kondo H, et al. Spatiotemporal Expression of Four Isoforms of Ca2+/calmodulin-Dependent Protein Kinase I in Brain and Its Possible Roles in Hippocampal Dendritic Growth. Neurosci Res (2007) 57(1):86–97. doi: 10.1016/j.neures.2006.09.013
14. Villalobo A, Berchtold MW. The Role of Calmodulin in Tumor Cell Migration, Invasiveness, and Metastasis. Int J Mol Sci (2020) 21(3):765. doi: 10.3390/ijms21030765
15. Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, Langerød A, et al. CAMK1D Amplification Implicated in Epithelial-Mesenchymal Transition in Basal-Like Breast Cancer. Mol Oncol (2008) 2(4):327–39. doi: 10.1016/j.molonc.2008.09.004
16. Lawson J, Dickman C, MacLellan S, Towle R, Jabalee J, Lam S, et al. Selective Secretion of microRNAs From Lung Cancer Cells via Extracellular Vesicles Promotes CAMK1D-Mediated Tube Formation in Endothelial Cells. Oncotarget (2017) 8(48):83913–24. doi: 10.18632/oncotarget.19996
17. Sui MH, Zhang WW, Geng DM, Sun DJ. CircPRKCI Regulates Proliferation, Migration and Cycle of Lung Adenocarcinoma Cells by Targeting miR-219a-5p-Regulated CAMK1D. Eur Rev Med Pharmacol Sci (2021) 25(4):1899–909. doi: 10.26355/eurrev_202102_25085
18. Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell (2012) 149(2):274–93. doi: 10.1016/j.cell.2012.03.017
19. Fan X, Zhou J, Yan X, Bi X, Liang J, Lu S, et al. Citrate Activates Autophagic Death of Prostate Cancer Cells via Downregulation CaMKII/AKT/mTOR Pathway. Life Sci (2021) 275:119355. doi: 10.1016/j.lfs.2021.119355
20. Zeng G, Cui X, Liu Z, Zhao H, Zheng X, Zhang B, et al. Disruption of Phosphoinositide-Specific Phospholipases Cγ1 Contributes to Extracellular Matrix Synthesis of Human Osteoarthritis Chondrocytes. Int J Mol Sci (2014) 15(8):13236–46. doi: 10.3390/ijms150813236
21. Rasmussen CD. Cloning of a Calmodulin Kinase I Homologue From Schizosaccharomyces Pombe. J Biol Chem (2000) 275(1):685–90. doi: 10.1074/jbc.275.1.685
22. Skelding KA, Rostas JA, Verrills NM. Controlling the Cell Cycle: The Role of Calcium/Calmodulin-Stimulated Protein Kinases I and II. Cell Cycle (2011) 10(4):631–9. doi: 10.4161/cc.10.4.14798
23. Nairn AC, Greengard P. Purification and Characterization of Ca2+/Calmodulin-Dependent Protein Kinase I From Bovine Brain. J Biol Chem (1987) 262(15):7273–81. doi: 10.1016/S0021-9258(18)48233-6
24. Wayman GA, Kaech S, Grant WF, Davare M, Impey S, Tokumitsu H, et al. Regulation of Axonal Extension and Growth Cone Motility by Calmodulin-Dependent Protein Kinase I. J Neurosci (2004) 24(15):3786–94. doi: 10.1523/JNEUROSCI.3294-03.2004
25. Condon JC, Pezzi V, Drummond BM, Yin S, Rainey WE. Calmodulin-Dependent Kinase I Regulates Adrenal Cell Expression of Aldosterone Synthase. Endocrinology (2002) 143(9):3651–7. doi: 10.1210/en.2001-211359
26. Jusuf AA, Sakagami H, Kikkawa S, Tereshima T. Expression of Beta Subunit 2 of Ca²+/Calmodulin-Dependent Protein Kinase I in the Developing Rat Retina. Kobe J Med Sci (2015) 61(4):E115–23.
27. Volpin V, Michels T, Sorrentino A, Menevse AN, Knoll G, Ditz M, et al. CAMK1D Triggers Immune Resistance of Human Tumor Cells Refractory to Anti-PD-L1 Treatment. Cancer Immunol Res (2020) 8(9):1163–79. doi: 10.1158/2326-6066.CIR-19-0608
28. Qin X, Liang Y, Guo Y, Liu X, Zeng W, Wu F, et al. Eukaryotic Initiation Factor 5A and Ca(2+)/Calmodulin-Dependent Protein Kinase 1D Modulate Trophoblast Cell Function. Am J Reprod Immunol (2018) 80(1):e12845. doi: 10.1111/aji.12845
29. Sang LJ, Ju HQ, Liu GP, Tian T, Ma GL, Lu YX, et al. LncRNA CamK-A Regulates Ca(2+)-Signaling-Mediated Tumor Microenvironment Remodeling. Mol Cell (2018) 72(1):71–83.e7. doi: 10.1016/j.molcel.2018.08.014
30. Racioppi L, Means AR. Calcium/calmodulin-Dependent Protein Kinase Kinase 2: Roles in Signaling and Pathophysiology. J Biol Chem (2012) 287(38):31658–65. doi: 10.1074/jbc.R112.356485
31. Selbert MA, Anderson KA, Huang QH, Goldstein EG, Means AR, Edelman AM, et al. Phosphorylation and Activation of Ca(2+)-Calmodulin-Dependent Protein Kinase IV by Ca(2+)-Calmodulin-Dependent Protein Kinase Ia Kinase. Phosphorylation of Threonine 196 Is Essential for Activation. J Biol Chem (1995) 270(29):17616–21. doi: 10.1074/jbc.270.29.17616
32. Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR Signaling Pathway in Ovarian Cancer: Biological and Therapeutic Significance. Semin Cancer Biol (2019) 59:147–60. doi: 10.1016/j.semcancer.2019.05.012
33. Zhang X, He X, Liu Y, Zhang H, Chen H, Guo S, et al. MiR-101-3p Inhibits the Growth and Metastasis of Non-Small Cell Lung Cancer Through Blocking PI3K/AKT Signal Pathway by Targeting MALAT-1. BioMed Pharmacother (2017) 93:1065–73. doi: 10.1016/j.biopha.2017.07.005
34. Rhoads AR, Friedberg F. Sequence Motifs for Calmodulin Recognition. FASEB J (1997) 11(5):331–40. doi: 10.1096/fasebj.11.5.9141499
Keywords: CAMK1D, glioma, cell proliferation, prognosis, PI3K/AKT/mTOR
Citation: Jin Q, Zhao J, Zhao Z, Zhang S, Sun Z, Shi Y, Yan H, Wang Y, Liu L and Zhao Z (2022) CAMK1D Inhibits Glioma Through the PI3K/AKT/mTOR Signaling Pathway. Front. Oncol. 12:845036. doi: 10.3389/fonc.2022.845036
Received: 29 December 2021; Accepted: 18 March 2022;
Published: 13 April 2022.
Edited by:
Jiancheng Hu, National Cancer Centre Singapore, SingaporeReviewed by:
David Akhavan, University of Kansas Medical Center, United StatesCopyright © 2022 Jin, Zhao, Zhao, Zhang, Sun, Shi, Yan, Wang, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongmao Zhao, enptNjkyMDE3QHNpbmEuY29t; Liping Liu, bGlwaW5nc2lzdGVyQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.