
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 08 February 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.841625
This article is part of the Research Topic RNA and RNA Modification in the Pathogenesis, Diagnosis and Treatment of Cancers View all 27 articles
Chemoresistance frequently occurs in cancer treatment, which results in chemotherapy failure and is one of the most leading causes of cancer-related death worldwide. Understanding the mechanism of chemoresistance and exploring strategies to overcome chemoresistance have become an urgent need. Autophagy is a highly conserved self-degraded process in cells. The dual roles of autophagy (pro-death or pro-survival) have been implicated in cancers and chemotherapy. MicroRNA (miRNA) is a class of small non-coding molecules that regulate autophagy at the post-transcriptional level in cancer cells. The association between miRNAs and autophagy in cancer chemoresistance has been emphasized. In this review, we focus on the dual roles of miRNA-mediated autophagy in facilitating or combating chemoresistance, aiming to shed lights on the potential role of miRNAs as targets to overcome chemoresistance.
Cancers with local organ invasion and distant metastasis often require systemic chemotherapy. Despite the newly developed therapeutic interventions such as immunotherapy, chemotherapy is still the most commonly applied treatment modality (1). In recent years, neoadjuvant chemotherapy has been included in the treatment guidelines of various solid tumors (2). However, after benefiting from the initial chemotherapeutic treatment, most patients will inevitably suffer from cancer relapse because of acquiring chemoresistance (3). Chemoresistance, a major cause of treatment failure and high mortality, remains a big challenge in clinics. Acquired drug resistance occurs after long-term chemotherapy, followed by devastating outcome (4), whereas intrinsic drug resistance exists without exposure to therapeutic drugs (5). It is reported that chemoresistance is responsible for more than ninety percent of cancer-related mortality (6). For instance, it has been documented that almost half of the patients diagnosed with metastatic colorectal cancer are resistant to 5-FU-based chemotherapy and their five-year survival rate is only slightly over 12% (7). Hence, there is an urgent need to elucidate the mechanism of chemoresistance and explore novel treatment strategies. After decades of works, several strategies to reverse chemoresistance have been proposed, including inhibition of P-glycoprotein (P-gp), combinational therapy, dosage enhancement, tumor microenvironment modulation and so on (8). Up to now, four generations of drug resistance reversal agents have been developed. The first generation of P-gp inhibitors such as verapamil and cyclosporin A can sensitize tumor to chemotherapeutic drugs only in vitro but not in vivo (9). The second generation of P-gp inhibitors such as S9788 and PSC833 also can’t be used clinically because it can inhibit cytochrome P4503A4 to bring about unpredictable toxicity and side effects (10). The third generation including tariquidar, laniquidar, zosuquidar and fourth generation including curcumin, andreia, tangeretin are under laboratory or clinical investigation and expected to be clinically used in the future (11). In addition, immunotherapy and targeted therapy are commonly used in clinic after chemoresistance occurs (12). Among these strategies, targeting autophagy to combat chemoresistance is gradually coming into sight.
Autophagy is an evolutionarily conserved process in which long-lived proteins, damaged organelles, or other cytoplasmic components are degraded and recycled to maintain energy homeostasis of cells (13). In 2016, Yoshinori Ohsumi was awarded the Nobel Prize for his contributions in elucidating the mechanism of autophagy, making autophagy a highlighted focus (14). Dysregulation of autophagy is involved in various pathological events such as cardiovascular disease (15), neurological disease (16), endocrine disorder (17), and especially cancers (18). Autophagy occurs frequently during chemotherapy, acting as either a pro-death or pro-survival process (19). The dual roles of autophagy in multi-drug resistance (MDR) have been described in our previously published review (13). On one hand, autophagy protects cancer cells from chemotherapeutic drugs to mediate drug resistance by eliminating damaged organelles and recycling degradation products. On the other hand, excessive autophagy can kill MDR cancer cells in which apoptosis pathways are inactive. Therefore, it is well recognized that autophagy is involved in chemoresistance in various types of cancers (20). The role of autophagy in chemoresistance is paradoxical and context-dependent, which needs comprehensive and systematic investigation.
MicroRNAs (miRNAs) are a class of small non-coding RNA with 19-25 nucleotides. They regulate gene expression by binding to the 3’-untranslated region (UTR) of target mRNAs to inhibit mRNA translation or facilitate mRNA degradation (21). Abnormal expression of miRNAs has been implicated in regulating cell proliferation, apoptosis, metastasis, migration, autophagy, and drug resistance in a large number of cancer types (22). Accumulating evidence indicated that miRNAs target some of the molecules in autophagic pathway thus resulting in chemoresistance or chemosensitivity during chemotherapy. Therefore, miRNAs could be promising targets for reversal of chemoresistance (23). Currently, miRNA-based therapies have been proposed. MiRNA mimics, miRNA sponges, anti-miRNA oligonucleotides, and small molecule inhibitors are promising strategies to modulate miRNAs (24). Miravirsen, the first miRNA-targeted drug, has been successfully tested in clinical Phase II trials for the treatment of hepatitis C (25). Miravirsen is a locked nucleic acid (LNA)-based antisense oligonucleotide targeting miR-122 (26). In the field of oncotherapy, MRX34, a liposomal miR-34a mimic, is the most advanced miRNA drug, which was designed to deliver miR-34a mimic to cancer cells for the treatment of several solid tumors (27). Additionally, novel miRNA-based drugs are being developed for the treatment of atherosclerosis (anti-miR-33a/b) (28), chronic heart failure (anti-miR-208, anti-miR-195) (29, 30), and other diseases.
In this review, we discussed the correlation between miRNAs and autophagy in chemoresistance/chemosensitivity, illustrated the current interventions targeting miRNA/autophagy axis to combat chemoresistance, aiming to provide novel insights from the perspective of miRNA-mediated autophagy for promoting chemotherapeutic efficacy.
Autophagy is initiated by the formation of double-membraned autophagic vesicles (AV) in response to a range of cellular stresses, including nutrient deprivation, hypoxia, organelle damage, and accumulation of reactive oxygen species (ROS) (31). The critical roles of autophagy in cell death, cell survival, metabolic adaptation, embryonic differentiation, immune surveillance and other biological processes have been verified (32). Therefore, dysregulation of autophagy has been implicated in various diseases such as Alzheimer’s disease, aging, microorganism infection, and multiple forms of cancers (33).
There are three types of autophagy, namely macroautophagy, microautophagy, and chaperone-mediated autophagy (34). Hereafter autophagy refers to macroautophagy, which is well understood and the mechanisms are established. In addition to general autophagy which functions in bulk degradation of cytoplasmic material, there exists selective autophagy targeting specific proteins or organelles such as mitochondria, endoplasmic reticulum (ER), bacteria, ribosomes, and ferritin (35). It is well accepted that autophagy is a multistep process involving approximately 30 autophagy-related genes (Atgs), that encode proteins executing the initiation of phagophore, AV maturation, and lysosomal fusion (36).
Mammalian target of rapamycin (mTOR) is at the upstream position of the autophagic process. mTOR consists of two distinct multiprotein complexes: mTORC1 and mTORC2 (37). As an environmental sensor, mTOR responds to intracellular and extracellular stressful conditions such as hypoxia, nutrient deprivation, or drug treatment (38). mTOR actively phosphorylates ATG, leading to the inhibition of autophagy under nutrient-rich conditions. In the case of nutrient deprivation, mTOR is inactivated and can no longer phosphorylate and inhibit the Unc-51-like kinase (ULK) complex, which consists of ULK family kinase, focal adhesion kinase interacting protein 200 kDa (FIP200), and ATG13. Dephosphorylated ULK1 dissociates from the mTOR complex and becomes active to trigger autophagosome membrane nucleation (39, 40). In addition to mTOR, 5’-AMP activated protein kinase (AMPK) also acts as a master regulator of energy stress to participate in the activation of ULK complex (41). Once activated, the ULK complex localizes to the phagophore and activates the Beclin1-Vacuolar protein sorting associated protein 34 (VPS34) complex, which contains VPS34, VPS15, Beclin1, and ATG14L (42). The VPS34 (a class III phosphatidylinositol 3-kinase, PI3K) complex generates phosphatidylinositol 3-phosphate (PI3P)-rich membranes most commonly derived from endoplasmic reticulum and Golgi complex (43). Elongation of phagophore membrane relies on two ubiquitin-like conjugation systems, the E1-like enzyme ATG7 and E2-like enzyme ATG10, which conjugate ATG5 to ATG12 (44). The E3 like enzyme ATG5-ATG12-ATG6L1 complex together with ATG7-ATG3 complex conjugate microtubule-associated protein 1 light chain 3 (LC3, ATG8) family members to phosphatidylethanolamine (PE) (45). The conversion of pro-LC3 to the active cytosolic isoform LC3-I requires the ATG4 family of cysteine proteases (46). Next, LC3-I is conjugated to PE to generate LC3-II, which is regarded as a key step of specific substrate recognition for selective degradation, therefore constructing cargo-loaded autophagosomes (47). In addition to serving as a marker for autophagosome, LC3 also acts as a docking site for cargo adaptors that bring autophagic cargo to the AVs. These adaptors such as SQSTM1 (p62) and neighbor of BRCA1 (NBR1) directly bind to proteins and organelles marked for autophagic degradation through NIX and FAM134B (48). Then, the double-membrane autophagosomes are degraded by fusing with lysosomes to form autolysosomes, that are regulated by Rab GTPases, SNARE, and HOPS complex (49, 50). During this process, the outer autophagosomal membrane is cleaved by ATG4, while the LC3-PE-conjugated inner membrane and the cytoplasmic contents were broken down by lysosomal proteases, thus recycling amino acids and other macromolecular building blocks (32).
The dual roles of autophagy in cancers have been largely demonstrated. Although autophagy may limit tumorigenesis in the earliest stage, accumulating evidence indicate that antophagy inhibition displays anti-proliferative effects in established cancers since antophagy can help cancer cells cope with hypoxia, nutrient deprivation or other cellular stresses (51). The basal level of autophagy plays a protective role against cancer through eliminating damaged organelles and proteins to maintain cellular homeostasis in normal cells (52). The abnormal autophagy contributes to the development of cancers. The first mouse model with deletion of autophagy gene was established to study the role of autophagy in tumorigenesis, and the results indicated that the deletion of BECN1 (gene symbol of Beclin1) increased the rate of spontaneous tumor formation compared with BECN1 wild-type (53). Depletion of the BECN1 is also observed in human breast, prostate, and ovarian cancers (54). Additionally, bax-interacting factor 1 (BIF-1) and UV radiation resistance-associated gene protein (UVRAG), which is related to Beclin1, was found to be absent or mutated in variety of cancer types (55, 56). However, a high basal-level of autophagy is observed in multiple established cancers, acting as a protective mechanism towards nutrient-stressed conditions (57). For example, in a Kras-driven lung cancer model, tumor cell growth and survival requires autophagy which plays a vital role in maintaining mitochondrial function (58). As a consequence, inhibition of cytoprotective autophagy in these cancers may result in tumor suppression (59). Additionally, a large body of literature has emerged and elucidated the role of autophagy induction to enable survival of cancer cells following chemotherapy or radiotherapy, indicating that autophagy is a key drug resistance mechanism in various cancer types (60).
Chemoresistance is a major cause of treatment failure, cancer relapse, and cancer metastasis (3). Drug resistance can be classified as resistance to either a single agent or multiple drugs with different structures and mechanisms of action (referring to MDR) (61). Mechanisms of cancer chemoresistance mainly include the following categories: (1) increased drug efflux by membrane transporters particularly ABC transporters (62), (2) reduced drug uptake by influx transporters such as solute carriers (63), (3) alterations in tumor microenvironment (TME), through secretion of multiple growth factors, chemokines, and cytokines by stromal and immune cells (64), (4) cancer stem cells, a class of tumor-triggering cells able to self-renew (65), (5) excessive DNA repair, which makes cancer cell survive and become tolerant to chemotherapeutic agents (66), (6) boosting drug metabolism mediated by glutathione S-transferase and cytochrome P450 enzymes (67, 68), (7) mutation in cancer-related genes including gain of function in oncogenes and loss of function in tumor suppressor genes (69), and (8) elevating adaptability by epigenetic and/or miRNA regulation (1).
The relationship between chemoresistance and autophagy has been studied for decades. It is known that resistance of cancer cells to chemotherapeutic agents is inevitable following prolonged exposure to drugs. This phenomenon may be partly mediated by induction of autophagy as a protective mechanism to cope with pressures during treatment (70). The autophagy triggered by chemotherapeutic drugs such as paclitaxel, epirubicin, or tamoxifen facilitates resistance of cancer cells to corresponding or multiple drugs (71–73). Various autophagy regulators and signaling pathways were confirmed to participate in this process. The mechanisms of metabolic-induced and therapeutic stress-induced autophagy might overlap in cancers. After chemotherapy is applied, nutrient and energy stress is amplified to increase autophagic flux. For example, after treatment with mTOR inhibitors, the transcription factor EB (TFEB)/transcription factor E3 (TFE3)/melanocyte inducing transcription factor (MITF) family can no longer be phosphorylated and translocate to nucleus, therefore activating transcription of the CLEAR network of genes to affect lysosome and autophagy (74). Another research demonstrated that the expression of S100A8 which is necessary for Beclin1-PI3KC3 complex formation is elevated to promote autophagy after adriamycin and vincristine treatment, contributing to drug resistance in leukemic cells (75). Moreover, ATG family members (76), bromodomain containing 4 (BRD4) (77), p53 (78), and ER stress-related genes (79) are also important factors involved in cytoprotective autophagy to mediate chemoresistance. Therefore, inhibition of such autophagy can re-sensitize resistant cancer cells to chemotherapeutic drugs. In recent years, the combination strategies of chemotherapeutic drugs and autophagy inhibitors have been proposed. Abundant basic and clinical research is ongoing. It is well established that genetic silencing of ATGs such as ATG5, ATG7, and Beclin1 blocks autophagy to sensitize drug resistant cells to therapeutic agents (80, 81). Chloroquine (CQ) and hydroxychloroquine (HCQ), which are clinically used for malaria treatment, are potent autophagy inhibitors through destroying lysosomes to prevent autophagosome degradation (82). Previous research has revealed that inhibition of autophagy by CQ sensitize vincristine-resistant gastric adenocarcinoma (83), epirubicin-resistant triple-negative breast cancer (84), sorafenib-resistant hepatocellular carcinoma (HCC) (85), cisplatin-resistant hypopharyngeal carcinoma (86), 5-fluorouracil-resistant gallbladder carcinoma (87) to chemotherapeutics. HCQ has been repurposed in numerous clinical trials either as a single agent or combined with therapeutic agents, some of which are in phase II studies (88). Lys05, a water-soluble analog of HCQ, displays stronger anticancer properties than HCQ both in vitro and in vivo (89). It can improve the efficiency of BRAF inhibitor against glioblastoma (90). Other autophagy-targeted compounds that are promising in combating chemoresistance include wogonin (91), SAR405 (92), tioconazole (93), 3-methyladenine (3-MA) (94) and others.
Paradoxically, while autophagy mainly acts as a pro-survival mechanism, excessive autophagy leads to a caspase-independent cell death called “type II programmed cell death” or “autophagic cell death”, which differs from apoptosis (95). In consequence, activation of such autophagy confers lethal effect on drug resistant cancer cells (96). Numerous studies have focused on identifying the novel agents that can effectively kill apoptosis-deficient cancer cells by inducing autophagic cell death. Since AKT/mTOR is the vital negative regulator of autophagy, the AKT/mTOR-associated autophagic cell death has gained a lot of attention. As a dual PI3K and mTOR inhibitor, NVP-BEZ235 was reported to sensitize osteosarcoma and urothelial cancer cells to cisplatin by activating autophagic flux independent of apoptosis (97, 98). Meanwhile, NVP-BEZ235 can also combat resistance to temozolomide and doxorubicin in glioma and neuroblastoma cells respectively (99, 100). Similarly, the Ganoderma microsporum immunomodulatory (GMI) protein targets AKT-mTOR-p70S6K pathway to reverse multidrug resistance by inducing pro-death autophagy in lung cancer (101). In addition to AKT/mTOR signaling pathway, the JNK activation and MCT1 inhibition also contributes to autophagic cell death, suggesting the possible autophagy-related targets to overcome chemoresistance (102, 103). Therefore it can be seen that autophagy demonstrates a role of pro-survival or pro-death to promote or suppress tumor growth, as well as mediate or combat chemoresistance. Inhibition of cytoprotective autophagy may enhance the sensitivity of cancer cells to chemotherapeutic agents, whereas induction of autophagic cell death can be used as an alternative cell death mechanism when the cells fail to undergo apoptosis. It is convinced that the role of autophagy is context- and tumor type-dependent, therefore clarifying the relationship between autophagy and chemoresistance is urgent and critical for improving the efficacy of chemotherapy.
MiRNAs are a class of small non-coding single-stranded RNA molecules with 19-25 nucleotides. They play fundamental roles in multiple biological processes through binding to the 3’-UTR of target mRNAs to accelerate mRNA degradation or terminate translation (104). MiRNAs are evolutionary conserved and found in a wide range of organisms (105). It is reported that more than 60% of human genes contain potential miRNA binding sites and approximately 10-40% of mRNAs are regulated by miRNAs (106). By post-transcriptional gene silencing, miRNAs regulate various cellular pathways including cell growth, differentiation, apoptosis, and homeostasis (107). MiRNA genes exist in both intergenic and intronic regions (108). They are transcribed into primary miRNA (pri-miRNA) with 5’ cap and a 3’ poly-A tail by RNA polymerase II (109). Meanwhile, RNA polymerase III is required for the transcription of some particular miRNAs (110). Following transcription, pri-miRNAs are processed in the nucleus by a core microprocessor complex including RNase III enzyme Drosha and its cofactor Pasha/DGCR8 to generate hairpin-structured premature-miRNAs (pre-miRNAs) with 60-70 nt (111). A single pri-miRNA transcript may generate more than one functional miRNA due to splicing (112). Then, Exportin-5 recognizes the 2-nucleotide overhang of the pre-miRNA and transports it from nucleus to the cytoplasm (113). In cytoplasm, the hairpin structure of pre-miRNAs is cleaved by DICER protein to form mature miRNAs, which are incorporated into an RNA-induced silencing complex (RISC) (114). Argonaute (AGO) proteins, the components of the RISC, guide mature miRNAs to specific target regions within mRNA transcripts, leading to mRNA degradation or translation blockage (106).
Dysregulation of miRNA often gives rise to multiple human diseases especially cancers. Abnormal expression of miRNAs is closely associated with cancer formation, progression, invasion, metastasis, and chemosensitivity (6). The complex roles of miRNAs as either tumor suppressors or oncogenes have been largely demonstrated. The correlation between miRNA-mediated autophagy and chemoresistance has been attracting a lot of interest. The fact that autophagy plays dual roles in chemoresistance provides a useful explanation on how miRNAs could reverse or facilitate chemoresistance through regulating autophagy. Since the protective mechanism of autophagy is the majority of cases to trigger tumor chemoresistance, inhibition of such autophagy by miRNAs may pave the way to combat chemoresistance. However, miRNA mediated-chemosensitivity by pro-death autophagy also possesses great value. Various tumor suppressive miRNAs are down-regulated in drug resistant cancer cells compared with sensitive ones, indicating that rejuvenation of these miRNAs may reverse chemoresistance by inhibiting protective autophagy or facilitating autophagic cell death. On the contrary, suppression of oncogenic miRNAs to modulate autophagy is another strategy for reversal of chemoresistance. In this section, we discuss the involvement of miRNAs in chemoresistance/chemosensitivity from different perspectives regulation of autophagy.
The process of autophagy is tightly regulated by ATGs, therefore targeting ATGs represent a promising strategy for reversal of drug resistance. Numerous ATGs are reported to be direct targets of miRNAs (Figure 1). For example, the 3’-UTR of ATG5 mRNA can be bound by miR-137 (115), miR-181a (116), miR-216b (117), miR-30a (118), and miR-153-3p (119) to facilitate ATG5 mRNA degradation, thus sensitizing various cancers, such as pancreatic cancer, gastric cancer, melanoma, chronic myelogenous leukemia, and non-small cell lung cancer (NSCLC), to chemotherapeutic agents by inhibiting protective autophagy. In addition to ATG5, Bcl-2 is a direct target of miR-153-3p in combating resistance to Imatinib in chronic myeloid leukemia (120). Similarly, miR-375 targets ATG7 (121) and ATG14 (122) to mediate chemosensitivity of HCC to sorafenib. The above studies indicated that a single miRNA can target different genes, likewise a specific gene is regulated by multiple miRNAs. Beclin-1, also known as ATG6, is a component of the PI3K complex which mediates vesicle-trafficking processes in autophagy (123). Targeting Beclin-1 by miR-409-3p (124), miR-17 (125), miR-216b (117), miR-17-5p (126), and miR-199a-5p (127) appears to reverse chemoresistance by inhibiting autophagy in different types of cancers. The interaction between miR-30 family and Beclin-1 has been revealed in recent years. It is reported that miR-30 or its homology miR-30a, miR-30a-5p binds to Beclin-1 mRNA to block autophagy-induced chemoresistance in chronic myeloid leukemia (118, 128), gastric cancer (129), osteosarcoma (130), small cell lung cancer (SCLC) (131) and other variety of cancers (132). A clinical study on Egyptian patients with chronic myeloid leukemia also confirmed this result (133). It was found that a single miRNA in different cancer types may share a common mechanism in mediating chemoresistance/chemosensitivity. Additionally, other ATGs are directly targeted by miRNAs, i.e. miR-541 targeting ATG 2A (134), miR-24-3p targeting ATG4A (135), miR-1 targeting ATG3 (136), miR-23b-3p and miR-200b targeting ATG12 (83, 137), miR-874 targeting ATG16L1 (138) and miR-34a targeting ATG4B (139). Therefore, overexpression of these miRNAs may result in sensitization of chemotherapeutic drugs in cancer treatment. Furthermore, miRNAs can also regulate ATGs indirectly. For instance, MiR-29c-3p targets FOXP1 to downregulate ATG14, leading to sensitization of ovarian cancer cells to cisplatin treatment by inhibiting autophagy (140).
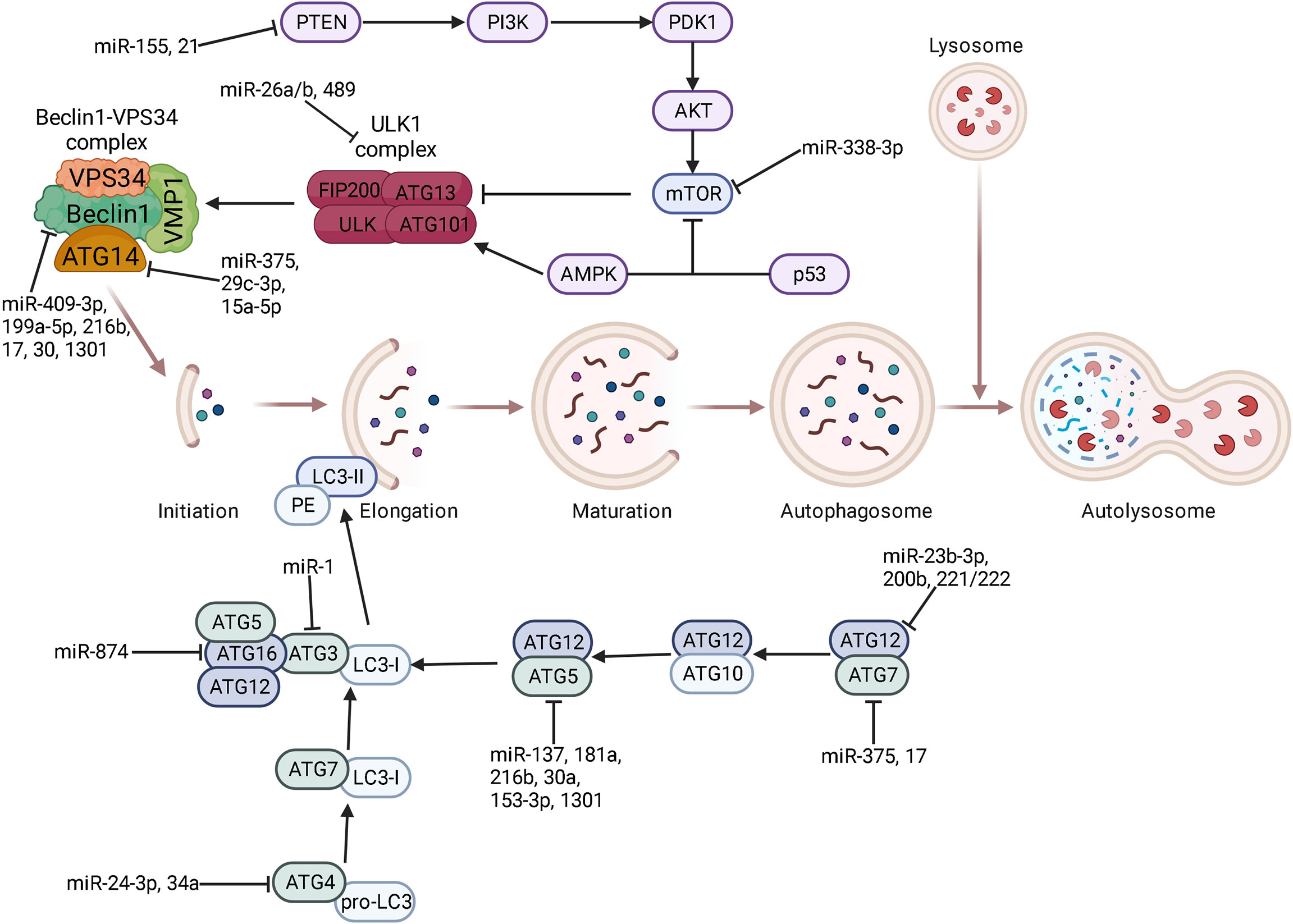
Figure 1 The regulatory role of miRNAs on each stage of autophagy. Core proteins and signaling pathways are related to each stage of autophagy including phagophore initiation and elongation, autophagosome maturation, and lysosomal fusion. Some key miRNAs target autophagy-related genes at the post-transcriptional level to participate in every stage of autophagy. ⊥ indicates an inhibitory effect and → indicates a promoting effect.
High mobility group box 1 (HMGB1) is a highly conserved DNA-binding nuclear protein which regulates various DNA-related activities such as replication, transcription, and repair (141). Abundant studies have confirmed the involvement of HMGB1 in multiple hallmarks of cancers, making HMGB1 a promising target to combat tumor progression, invasion, metastasis, and chemoresistance (142). As a key regulator of autophagy, HMGB1 promotes drug resistance of a number of cancers including osteosarcoma (143), lung adenocarcinoma (144), and leukemia (145) by activating protective autophagy following pharmacotherapy. Some investigators have attempted to reveal the association between miRNAs and HMGB1. They found that HMGB1 was targeted by miR-22, miR-218, miR-26a, miR-34a, miR-129-5p, and miR-142-3p to sensitize osteosarcoma, endometrial carcinoma, melanoma, retinoblastoma, breast cancer, NSCLC to chemotherapeutic agents through inhibiting autophagy (146–151). HMGN5 is another member of the HMG box family involved in oncogenesis and tumor progression. Meng and his colleagues conducted a series of experiments to elucidate the HMGN5-associated chemoresistance. Their work revealed that HMGN5-mediated autophagy contributes to chemoresistance in osteosarcoma. Targeting HMGN5 by miR-140-5p sensitizes osteosarcoma cells to chemotherapy, suggesting a potential application of miR-140-5p in the prognosis and treatment of chemoresistant cancers (152).
Other core autophagic components, regulators, or signaling pathways also associated with the mechanisms of miRNA-mediated chemosensitivity. For example, RAB family, the largest subfamily of Ras, consists of more than 60 small GTPases. RABs play essential roles in membrane traffic including autophagosome formation (153). Xu et al. found that high miR-541 expression potentiates the response of HCC to sorafenib treatment by targeting RAB1B (134). Additionally, miR-148a-3p inhibits the cytoprotective autophagy by suppressing RAB12 to enhance cisplatin cytotoxicity in gastric cancer (154). As a key initiator of autophagy, ULK1 is an attractive target for cancer treatment. The 3ʹ-UTR of ULK1 was reported to contain binding sites for miR-26a/b and miR-106a. Overexpression of miR-26a/b enhances the sensitivity of HCC to doxorubicin (Dox) and promotes apoptosis both in vitro and in vivo by inhibiting autophagy (155). Similarly, ectopic expression of miR-106a resulted in significant tyrosine kinase inhibitor (TKI)-induced cell death in lung adenocarcinoma compared to control transduced cells (156). Moreover, miR-489 overexpression inhibits ULK1 to suppress autophagy, thus sensitizing breast cancer cells to DOX (157). FOXO3a is a multifaceted transcription factor which guides autophagy directly or indirectly (158). A research by Zhou et al. revealed that FOXO3a is a direct downstream target of miR-223. Overexpression of miR-223 or agomiR-223 contributes to the enhancement of doxorubicin sensitivity in HCC (159). Furthermore, Wingless-type MMTV integration site family member 2 (WNT2) belongs to the WNT family which is evolutionarily conserved (160). The foremost roles of the Wnt/β-catenin signaling pathway in tumorigenesis and tumor progression have been well established especially in the aspects of cancer invasion and migration, whereas little is known about WNT and autophagy (161). Chen et al. found that overexpression of miR-199a/b-5p inhibits its direct target WNT2 and downstream signaling to influence autophagy formation, resulting in enhanced efficacy of Imatinib treatment in chronic myeloid leukemia (162). Overall, induction of autophagy following chemotherapy confers the survival mechanism of cancer cells. Overexpression of some tumor suppressor miRNAs to block autophagy has become a useful strategy to enhance chemosensitivity via different molecular pathways. See Table 1 for details.
The inactivation of apoptosis pathway following chemotherapy contributes to the development of drug resistance. Hence, alternative types of cell death to combat chemoresistance have attracted increasing attention. Opposite to cytoprotective autophagy, excessive autophagy promotes autophagic cell death during chemotherapy (183). It has been reported that some miRNAs trigger autophagic cell death in drug resistant cancers by repressing important upstream signals of autophagy pathway. These cases are few but of great significance. In an investigation into chemosensitivity of cervical carcinoma, Huang et al. found that miR-15a and miR-16 directly targets Rictor to attenuate the phosphorylation of mTORC1 and p70S6K. As a consequence, miR-15a/16 dramatically enhances chemotherapeutic efficacy of camptothecin towards cervical carcinoma partly due to autophagy-induced inhibition of cell proliferation (179). Similarly, another research revealed that in cisplatin-resistant NSCLC, downregulation of miR-181 correlates with reduced autophagy and apoptosis. MiR-181 overexpression restored LC3 and ATG5 protein by triggering PTEN/PI3K/AKT/mTOR signaling pathway, therefore promoting apoptosis in cisplatin-resistant NSCLC (180). These two studies emphasize mTOR as the key regulator in miRNA-induced autophagic cell death. In a study of miR-193b, it is reported that overexpression of miR-193b significantly enhances the cytotoxicity of 5-FU to oesophageal cancer cells, which is mediated by elevated autophagic flux rather than apoptosis. Although the exact targets of miR-193b are unknown, target prediction analysis suggests that stathmin 1 might be involved in this process (181). Additionally, another research demonstrated that miR-519a increased the sensitivity of glioblastoma to temozolomide through induction of autophagy by targeting STAT3/Bcl-2/Beclin-1 signaling pathway. These results provide an effective therapeutic strategy of drug combination for glioblastoma treatment (182). See Table 1 for details.
The expression of some autophagy inhibitory miRNAs was significantly increased in drug resistant cancer cells compared with their parental cells, indicating the possible mechanism of miRNAs-mediated chemoresistance by autophagy. Hence, silencing these oncogenic miRNAs may increase the sensitivity of drug resistant cancer cells to chemotherapeutic agents by inducing autophagic cell death. For example, miR-1301 promoted the proliferation of cisplatin-resistant ovarian cancer cells by inhibiting ATG5 and Beclin1, indicating that targeting miR-1301 is an effective approach to reverse cisplatin resistance by inducing autophagy (184). Another research demonstrated that miR-487b-5p at high level may be a potential biomarker of acquired Temozolomide resistance in lung cancer cells. MiR-487b-5p directly targets LAMP2 to block autophagy thus mediating Temozolomide resistance. In consequence, miR-487b-5p has been regarded as a chemotherapeutic target in the treatment of TMZ-resistant lung carcinoma by enhancing autophagy (185). Furthermore, inhibitions of miR-221/222 induced extended autophagy and cell death of multiple myeloma cells by enhanced autophagy via ATG12 and p27 upregulation (186). A recent study found that miR-15a-5p was overexpressed in chemoresistant acute myeloid leukemia (AML) patients compared with chemosensitive patients treated with daunorubicin and cytarabine. The upregulation of miR-15a-5p decreased daunorubicin-induced autophagy by targeting ATG9a, ATG14, GABARAPL1, and SMPD1, thus resulting in attenuating cell sensitivity to daunorubicin. This finding indicated that inhibition of miR-15a-5p may sensitize AML to daunorubicin by enhancing autophagy (187). MiR-21 contributes to the tamoxifen (TAM) and fulvestrant (FUL) resistance of breast cancer by inhibiting autophagy. Yu et al. found that silencing miR-21 increased the sensitivity of ER+ breast cancer cells to TAM or FUL by triggering autophagic cell death. Phosphatase and tensin homolog (PTEN) is the potential target of miR-21 to regulate autophagy by affecting downstream PI3K-AKT-mTOR signaling pathway (188). As mentioned above, PI3K-AKT-mTOR is the core negative regulator of autophagy (189). The activation of PI3K-AKT-mTOR by miR-21 reduces the efficacy of cisplatin on gastric cancer cells through autophagy inhibition (190). Similarly, a study by He et al. verified the role of miR-21 in mediating sorafenib resistance of HCC cells by inhibiting autophagy. Anti-miR-21 oligonucleotides re-sensitized sorafenib-resistant HCC cells by promoting autophagy via the PTEN/AKT signaling pathway (191). These studies show that the connections of miR-21 and PTEN-PI3K-AKT-mTOR have been well established in drug resistance of some cancer types. Meanwhile, the autophagy inhibition by miR-155 through PTEN-PI3K-AKT-mTOR signaling pathway to mediate adriamycin resistance of osteosarcoma has been proposed (192). To conclude, targeting miR-21 or miR-155 may restore PTEN to inhibit PI3K-AKT-mTOR signaling pathway, thus triggering autophagic cell death to overcome chemoresistance. Seca et al. found that autophagy enhancement by miR-21 inhibition decreases the expression of pro-survival genes such as Bcl-2, thus sensitizing leukemia cells to chemotherapeutic drugs (193). Furthermore, recent studies confirmed that the downstream biological effects following autophagy inhibition may benefit cancer survival. The suppression of autophagy by miR-3127-5p results in activation of STAT3 signaling pathway, which stimulates programmed death-ligand 1 (PD-L1) and subsequently mediates immune evasion of cancer cells (194). As a consequence, targeting miR-3127-5p to facilitate autophagy may aid in immune escape dismission and chemoresistance reversal. See Table 2 for details.
The levels of some miRNAs are positively correlated with cytoprotective autophagy and drug resistance following chemotherapy, therefore targeting these upregulated miRNAs in drug resistant cancer cells may restore chemosensitivity by inhibiting autophagy. MiR-138 was confirmed to be associated with glioblastoma cell survival and resistance to TMZ by inducing pro-survival autophagy which negatively correlates with BIM, the direct target of miR-138. Hence, targeting miR-138 may represent a novel strategy to overcome temozolomide resistance in glioblastoma by inhibiting autophagy (196). Interestingly, the role of miR-21 in autophagy regulation is controversial. Contrary to what is mentioned in previous section, miR-21-5p enhances pro-survival autophagic flux following inhibition of proteasome pathway to mediate drug resistance to topoisomerase inhibitors in colorectal cancer (CRC). Therefore, miR-21-5p could be a potential target for reversing drug resistance in CRC (195). Moreover, miR-338-3p confers resistance to 5-FU in p53 mutant colon cancer through mTOR downregulation-induced autophagy, indicating that targeting miR-338-3p is a promising strategy to overcome 5-FU resistance in p53 mutant colon cancer (199). Additionally, miR-140-5p and miR-155 promote the chemotherapy-induced autophagy to mediate drug resistance in osteosarcoma. Inositol 1,4,5-trisphosphate kinase 2 (IP3k2) was reported to be a direct target of miR-140-5p (197, 198). Similarly, miR-7-5p promotes autophagy via suppression of Bcl-2 to mediate cisplatin resistance in cervical cancer (200). MiR-223 directly targets FBXW7 thus promoting autophagy and rendering NSCLC cells resistant to cisplatin (201). Thus, targeting the above mentioned miRNAs could provide potential approach to combat chemoresistance. See Table 2 for details.
Since miRNAs play vital roles in chemoresistance and chemosensitivity via regulating autophagy, miRNA-based strategies including either miRNA inhibition or miRNA restoration have been proposed in cancer therapy. The rapid development of miRNA-based interventions, such as miRNA mimics, anti-miRNA oligonucleotides, miRNA sponges, and small molecule inhibitors has been witnessed in the last decade. Some of these agents are in different phases of clinical trials (202). The first miRNA-based therapy for cancer is MRX34, a miR-34 mimic designed to target Wnt signaling and tumor metastasis (203). In this section, we demonstrate some genetic or pharmacological interventions targeting miRNA to combat chemoresistance by modulating autophagy.
Isoliquiritigenin (ISL), a natural flavonoid isolated from the root of licorice, has been used for the treatment of inflammation, platelet aggregation, cancer, and cardiac injury for centuries (204). A research by Wang et al. revealed that ISL targets miR-25 to trigger autophagic cell death by increasing ULK1 expression in MCF-7/ADR cells, which provides evidence for ISL as a natural autophagy inducer to increase breast cancer chemosensitivity (205). Apigenin is a flavonoid with anti-proliferative properties against a broad spectrum of cancers (206). Apigenin can significantly upregulate miR-520b which targets ATG7 to block protective autophagy, thus sensitizing HCC cells to doxorubicin (207). Propofol is an intravenous sedative-hypnotic agent used in surgery. A growing number of studies have revealed the anti-tumor effect of propofol against different cancer types (208, 209). LncRNA MALAT1 targets miR-30e to facilitate autophagy via ATG5 upregulation. The downregulation of lncRNA MALAT1 by propofol results in inhibiting autophagy and promoting gastric cancer cells sensitive to cisplatin (210). A recent study demonstrated that rutin, the main component of Potentilla discolor Bunge, reverses sorafenib resistance by inhibiting autophagy through the BANCR/miRNA-590-5P/OLR1 axis in HCC (211). Furthermore, it is urgent to look for efficient miRNA delivery system for miRNA mimics that can’t enter cells efficiently on its own. Based on the novel drug delivery system, miR-375 and sorafenib were co-loaded into calcium carbonate nanoparticles with lipid coating (miR-375/Sf-LCC NPs). As an inhibitor of autophagy, miR-375 enhances cytotoxicity of sorafenib both in vitro and in vivo by targeting ATG7, thus producing potent anti-tumor effect to combat sorafenib resistance (121).
This review demonstrated that miRNAs, as epigenetic factors of autophagy, play a pivotal role in cancer chemoresistance. Various types of cancers develop resistance to chemotherapeutic drugs through complex regulatory mechanisms of miRNAs by targeting different genes at every stage of autophagy. Due to the paradoxical effects of autophagy in chemoresistance, there is an urgent need to understand the interactions between miRNA-mediated autophagy and chemoresistance, which may provide evidence for development of novel miRNA-based therapy. As mentioned above, altered expression of miRNAs can trigger chemoresistance or chemosensitivity through pro-death or pro-survival autophagy during chemotherapy. Hence, inhibiting miRNA function or restoring miRNA expression is a possible approach for combating chemoresistance (Figure 2). Genetic interventions targeting miRNAs such as miRNA mimics, miRNA sponges, anti-miRNA oligonucleotides are useful approaches (212). The pharmacological interventions such as small molecule compound or active ingredient can also be used to target miRNA to overcome chemoresistance. In addition, the upstream molecular pathway regulating miRNA/autophagy axis can also be the potential targets for chemoresistance reversal. LncRNAs, circRNAs, and proteins are the major upstream mediators of miRNA/autophagy axis (213, 214). The complex regulatory network of upstream factors on miRNA/autophagy axis necessitates further research.
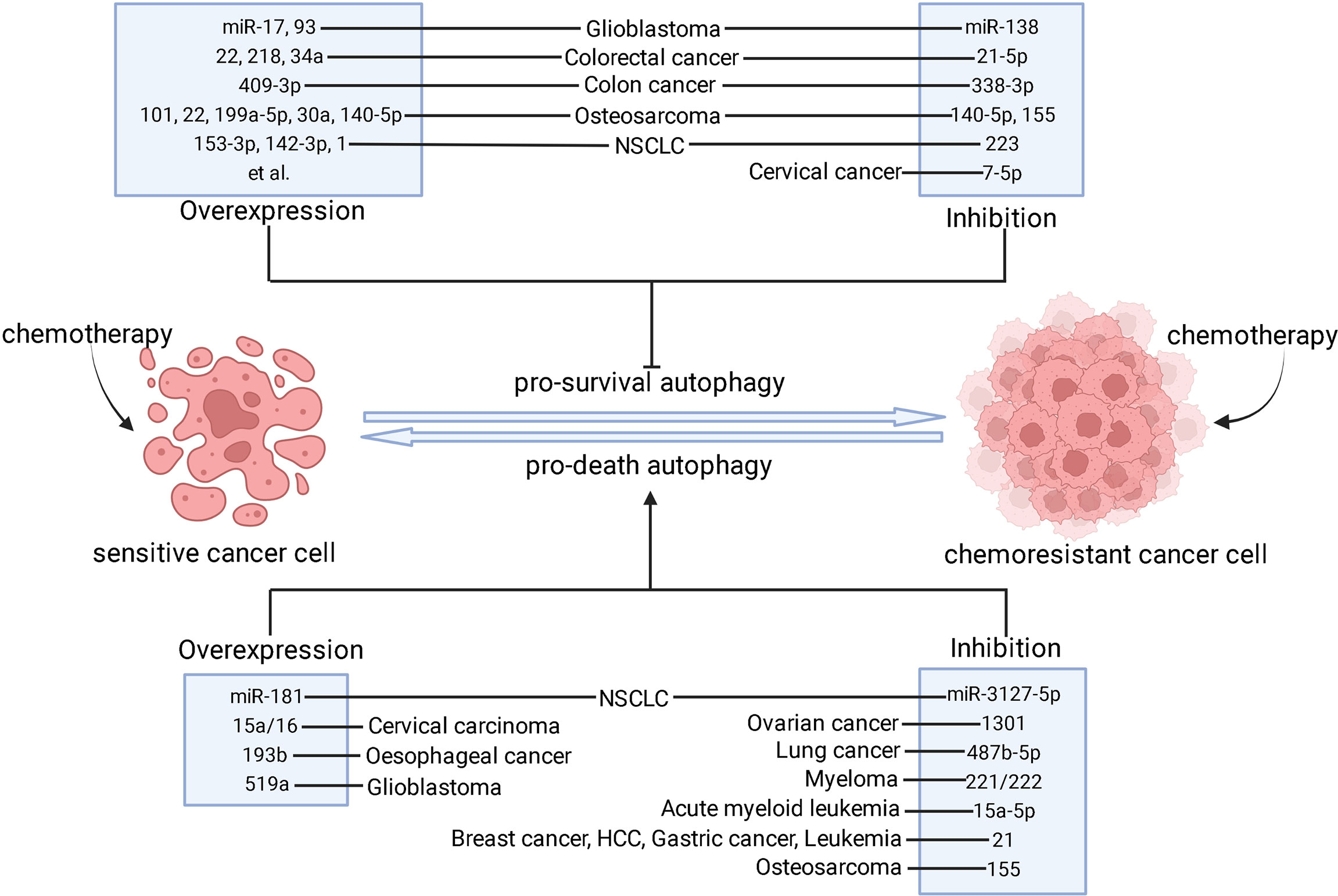
Figure 2 The strategies of modulating miRNAs to combat chemoresistance through autophagy. After chemotherapy is applied, sensitive cancer cells mainly undergo apoptotic cell death process whereas chemoresistant cancer cells fail to respond to chemotherapeutics. The pro-survival autophagy contributes to the development of chemoresistance. However, pro-death autophagy can be used as an alternative cell death mechanism in apoptosis-inactive cancer cells to re-sensitize them. Based on these facts, inhibition of pro-survival autophagy and induction of pro-death autophagy may result in chemoresistance reversal, which can be done by overexpression or inhibition of these miRNAs in different types of cancer. ⊥ indicates an inhibitory effect and → indicates a promoting effect.
MiRNA-based therapy as an adjuvant to immunotherapy and targeted therapy is highly feasible. MiRNA-based therapies may aid in the four principal cancer immunotherapy approaches including immune checkpoint blockade, cancer vaccines, cytokine therapy, and adoptive cell therapy (215). According to the study of Howell et al, the miR-31 inhibits CD8+ T cell function, leading to a substantial block to anti-tumor immunity. Hence, they proposed that miR-31 inhibitor combined with PD-1 inhibitor may prevent T cell from exhaustion and promote autoimmunity, thus displaying huge potential for cancer suppression (216). MiR-200 has also emerged as a potential therapeutic adjuvant for checkpoint inhibitors by acting on both immune and metastatic pathways via modulation of PD-L1 and EMT (217). Additionally, aberrant expression of miRNAs promotes resistance of different types of cancer to targeted therapy through multiple mechanisms. Therefore, the combination of miRNA-based therapy and targeted therapy may overcome the resistance of cancer cells to targeted drugs such as tyrosine kinase inhibitors and monoclonal antibody (218).
Recently, the delivery approaches for miRNAs including viral vector-, lipid-, inorganic material-, polymer-, cell-, and 3D scaffold-based approaches have emerged (219). The lipid-based delivery systems such as liposomes, lipid nanoparticles, and solid lipid nanoparticles (SLNs) have been widely used for introduction of miRNAs. With the development of nanoparticle delivery system, the introduction of miRNA turns out to be highly efficient in cancer therapeutics because these nano-miRNAs have a site specific action, which can deliver the miRNA or anti-miRNA directly to the transformed cells, thus reducing the unexpected toxicity in non-target cells (212). The viral vector delivery system also has high efficiency. However, the associated immunogenic responses and cytotoxicity limit the further application of these approaches respectively (220, 221). Currently, the safety concerns of miRNA therapy including off-target side-effects, toxicity, and carcinogenicity have become big challenges. Nowadays, less than 20 miR targeting molecules have entered clinical trials, and none progressed to phase III (219). Hence, further research is needed to promote the application value of miRNA therapy.
In conclusion, this review elucidated the microRNA-based strategies to combat cancer chemoresistance via regulating autophagy. We expect that patients will benefit from the improvement of chemotherapy efficacy through modulation of miR/autophagy axis in the future.
YZ and LD contributed to conception and design of the review. YL, LC, and JL drafted the manuscript. All authors contributed to the article and approved the submitted.
This work was supported by the National Natural Science Foundation of China (No. 81803790), National Natural Science Foundation of Guangdong (No. 2020A1515011090) and the Project of Administration of Traditional Chinese Medicine of Guangdong Province of China (Grant no. 20200511205949).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Jing Y, Liang W, Liu J, Zhang L, Wei J, Yang J, et al. Autophagy-Mediating microRNAs in Cancer Chemoresistance. Cell Biol Toxicol (2020) 36(6):517–36. doi: 10.1007/s10565-020-09553-1
2. Korde LA, Somerfield MR, Carey LA, Crews JR, Denduluri N, Hwang ES, et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J Clin Oncol (2021) 39(13):1485–505. doi: 10.1200/JCO.20.03399
3. Zheng HC. The Molecular Mechanisms of Chemoresistance in Cancers. Oncotarget (2017) 8(35):59950–64. doi: 10.18632/oncotarget.19048
4. Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E. The Challenge of Drug Resistance in Cancer Treatment: A Current Overview. Clin Exp Metastasis (2018) 35(4):309–18. doi: 10.1007/s10585-018-9903-0
5. Gottesman MM. Mechanisms of Cancer Drug Resistance. Annu Rev Med (2002) 53:615–27. doi: 10.1146/annurev.med.53.082901.103929
6. Si W, Shen J, Zheng H, Fan W. The Role and Mechanisms of Action of microRNAs in Cancer Drug Resistance. Clin Epigenet (2019) 11(1):25. doi: 10.1186/s13148-018-0587-8
7. Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, et al. Phase III Multicenter Randomized Trial of Oxaliplatin Added to Chronomodulated Fluorouracil-Leucovorin as First-Line Treatment of Metastatic Colorectal Cancer. J Clin Oncol (2000) 18(1):136–47. doi: 10.1200/JCO.2000.18.1.136
8. Panda M, Biswal BK. Cell Signaling and Cancer: A Mechanistic Insight Into Drug Resistance. Mol Biol Rep (2019) 46(5):5645–59. doi: 10.1007/s11033-019-04958-6
9. Palmeira A, Sousa E, Vasconcelos MH, Pinto MM. Three Decades of P-Gp Inhibitors: Skimming Through Several Generations and Scaffolds. Curr Med Chem (2012) 19(13):1946–2025. doi: 10.2174/092986712800167392
10. Nobili S, Landini I, Giglioni B, Mini E. Pharmacological Strategies for Overcoming Multidrug Resistance. Curr Drug Targets (2006) 7(7):861–79. doi: 10.2174/138945006777709593
11. Wu S, Fu L. Tyrosine Kinase Inhibitors Enhanced the Efficacy of Conventional Chemotherapeutic Agent in Multidrug Resistant Cancer Cells. Mol Cancer (2018) 17(1):25. doi: 10.1186/s12943-018-0775-3
12. Ji X, Lu Y, Tian H, Meng X, Wei M, Cho WC. Chemoresistance Mechanisms of Breast Cancer and Their Countermeasures. BioMed Pharmacother (2019) 114:108800. doi: 10.1016/j.biopha.2019.108800
13. Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, et al. Autophagy and Multidrug Resistance in Cancer. Chin J Cancer (2017) 36(1):52. doi: 10.1186/s40880-017-0219-2
14. Mizushima N. A Brief History of Autophagy From Cell Biology to Physiology and Disease. Nat Cell Biol (2018) 20(5):521–7. doi: 10.1038/s41556-018-0092-5
15. Saliba-Gustafsson P, Pedrelli M, Gertow K, Werngren O, Janas V, Pourteymour S, et al. Subclinical Atherosclerosis and Its Progression Are Modulated by PLIN2 Through a Feed-Forward Loop Between LXR and Autophagy. J Intern Med (2019) 286(6):660–75. doi: 10.1111/joim.12951
16. Lachance V, Wang Q, Sweet E, Choi I, Cai CZ, Zhuang XX, et al. Autophagy Protein NRBF2 has Reduced Expression in Alzheimer’s Brains and Modulates Memory and Amyloid-Beta Homeostasis in Mice. Mol Neurodegener (2019) 14(1):43. doi: 10.1186/s13024-019-0342-4
17. Guo X, Shi Y, Du P, Wang J, Han Y, Sun B, et al. HMGB1/TLR4 Promotes Apoptosis and Reduces Autophagy of Hippocampal Neurons in Diabetes Combined With OSA. Life Sci (2019) 239:117020. doi: 10.1016/j.lfs.2019.117020
18. Zhang Q, Wang X, Cao S, Sun Y, He X, Jiang B, et al. Berberine Represses Human Gastric Cancer Cell Growth In Vitro and In Vivo by Inducing Cytostatic Autophagy via Inhibition of MAPK/mTOR/p70S6K and Akt Signaling Pathways. BioMed Pharmacother (2020) 128:110245. doi: 10.1016/j.biopha.2020.110245
19. Kimmelman AC, White E. Autophagy and Tumor Metabolism. Cell Metab (2017) 25(5):1037–43. doi: 10.1016/j.cmet.2017.04.004
20. Ding R, Jin S, Pabon K, Scotto KW. A Role for ABCG2 Beyond Drug Transport: Regulation of Autophagy. Autophagy (2016) 12(5):737–51. doi: 10.1080/15548627.2016.1155009
21. Goradel NH, Mohammadi N, Haghi-Aminjan H, Farhood B, Negahdari B, Sahebkar A. Regulation of Tumor Angiogenesis by microRNAs: State of the Art. J Cell Physiol (2019) 234(2):1099–110. doi: 10.1002/jcp.27051
22. Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, et al. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer (2018) 9(19):3557–69. doi: 10.7150/jca.26350
23. Huang T, Xu Z, Cao L, Lian Y, Huang Y. Autophagy Regulation by microRNAs in Chemotherapy Resistance (Review). Oncol Rep (2020) 44(3):791–7. doi: 10.3892/or.2020.7655
24. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in Cancer: Rationale, Strategies and Challenges. Nat Rev Drug Discov (2010) 9(10):775–89. doi: 10.1038/nrd3179
25. Schmidt MF. miRNA Targeting Drugs: The Next Blockbusters? Methods Mol Biol (2017) 1517:3–22. doi: 10.1007/978-1-4939-6563-2_1
26. Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can Inhibit the Biogenesis of miR-122. Nucleic Acids Res (2014) 42(1):609–21. doi: 10.1093/nar/gkt852
27. Li WJ, Wang Y, Liu R, Kasinski AL, Shen H, Slack FJ, et al. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front Cell Dev Biol (2021) 9:640587. doi: 10.3389/fcell.2021.640587
28. Ono K. Functions of microRNA-33a/B and microRNA Therapeutics. J Cardiol (2016) 67(1):28–33. doi: 10.1016/j.jjcc.2015.10.017
29. Wang L, Qin D, Shi H, Zhang Y, Li H, Han Q. MiR-195-5p Promotes Cardiomyocyte Hypertrophy by Targeting MFN2 and FBXW7. BioMed Res Int (2019) 2019:1580982. doi: 10.1155/2019/1580982
30. Zhao X, Wang Y, Sun X. The Functions of microRNA-208 in the Heart. Diabetes Res Clin Pract (2020) 160:108004. doi: 10.1016/j.diabres.2020.108004
31. Russell RC, Yuan HX, Guan KL. Autophagy Regulation by Nutrient Signaling. Cell Res (2014) 24(1):42–57. doi: 10.1038/cr.2013.166
32. Towers CG, Wodetzki D, Thorburn A. Autophagy and Cancer: Modulation of Cell Death Pathways and Cancer Cell Adaptations. J Cell Biol (2020) 219(1):e201909033. doi: 10.1083/jcb.201909033
33. Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and Disease: Always Two Sides to a Problem. J Pathol (2012) 226(2):255–73. doi: 10.1002/path.3025
34. Chiang HL, Terlecky SR, Plant CP, Dice JF. A Role for a 70-Kilodalton Heat Shock Protein in Lysosomal Degradation of Intracellular Proteins. Science (1989) 246(4928):382–5. doi: 10.1126/science.2799391
35. Anding AL, Baehrecke EH. Cleaning House: Selective Autophagy of Organelles. Dev Cell (2017) 41(1):10–22. doi: 10.1016/j.devcel.2017.02.016
36. Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct Autophagosomal-Lysosomal Fusion Mechanism Revealed by Thapsigargin-Induced Autophagy Arrest. Mol Cell (2011) 42(6):731–43. doi: 10.1016/j.molcel.2011.04.024
37. Populo H, Lopes JM, Soares P. The mTOR Signalling Pathway in Human Cancer. Int J Mol Sci (2012) 13(2):1886–918. doi: 10.3390/ijms13021886
38. Tian T, Li X, Zhang J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int J Mol Sci (2019) 20(3):755. doi: 10.3390/ijms20030755
39. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-Dependent Mtorc1 Association With the ULK1-Atg13-FIP200 Complex Required for Autophagy. Mol Biol Cell (2009) 20(7):1981–91. doi: 10.1091/mbc.E08-12-1248
40. Torii S, Yoshida T, Arakawa S, Honda S, Nakanishi A, Shimizu S. Identification of PPM1D as an Essential Ulk1 Phosphatase for Genotoxic Stress-Induced Autophagy. EMBO Rep (2016) 17(11):1552–64. doi: 10.15252/embr.201642565
41. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR Regulate Autophagy Through Direct Phosphorylation of Ulk1. Nat Cell Biol (2011) 13(2):132–41. doi: 10.1038/ncb2152
42. Behrends C, Sowa ME, Gygi SP, Harper JW. Network Organization of the Human Autophagy System. Nature (2010) 466(7302):68–76. doi: 10.1038/nature09204
43. Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi Intermediate Compartment Is a Key Membrane Source for the LC3 Lipidation Step of Autophagosome Biogenesis. Elife (2013) 2:e00947. doi: 10.7554/eLife.00947
44. Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A Protein Conjugation System Essential for Autophagy. Nature (1998) 395(6700):395–8. doi: 10.1038/26506
45. Dikic I, Elazar Z. Mechanism and Medical Implications of Mammalian Autophagy. Nat Rev Mol Cell Biol (2018) 19(6):349–64. doi: 10.1038/s41580-018-0003-4
46. Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics Comparisons of Mammalian Atg4 Homologues Indicate Selective Preferences Toward Diverse Atg8 Substrates. J Biol Chem (2011) 286(9):7327–38. doi: 10.1074/jbc.M110.199059
47. Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, et al. Mechanism and Functions of Membrane Binding by the Atg5-Atg12/Atg16 Complex During Autophagosome Formation. EMBO J (2012) 31(22):4304–17. doi: 10.1038/emboj.2012.278
48. Moosavi MA, Djavaheri-Mergny M. Autophagy: New Insights Into Mechanisms of Action and Resistance of Treatment in Acute Promyelocytic Leukemia. Int J Mol Sci (2019) 20(14):3559. doi: 10.3390/ijms20143559
49. Nakamura S, Yoshimori T. New Insights Into Autophagosome-Lysosome Fusion. J Cell Sci (2017) 130(7):1209–16. doi: 10.1242/jcs.196352
50. Yu L, Chen Y, Tooze SA. Autophagy Pathway: Cellular and Molecular Mechanisms. Autophagy (2018) 14(2):207–15. doi: 10.1080/15548627.2017.1378838
51. Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov (2019) 9(9):1167–81. doi: 10.1158/2159-8290.CD-19-0292
52. Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 Regulates Autophagy. J Cell Biol (2010) 190(5):881–92. doi: 10.1083/jcb.200911078
53. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of Tumorigenesis by Heterozygous Disruption of the Beclin 1 Autophagy Gene. J Clin Invest (2003) 112(12):1809–20. doi: 10.1172/JCI20039
54. Shen Y, Li DD, Wang LL, Deng R, Zhu XF. Decreased Expression of Autophagy-Related Proteins in Malignant Epithelial Ovarian Cancer. Autophagy (2008) 4(8):1067–8. doi: 10.4161/auto.6827
55. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 Interacts With Beclin 1 Through UVRAG and Regulates Autophagy and Tumorigenesis. Nat Cell Biol (2007) 9(10):1142–51. doi: 10.1038/ncb1634
56. He S, Zhao Z, Yang Y, O’Connell D, Zhang X, Oh S, et al. Truncating Mutation in the Autophagy Gene UVRAG Confers Oncogenic Properties and Chemosensitivity in Colorectal Cancers. Nat Commun (2015) 6:7839. doi: 10.1038/ncomms8839
57. Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, et al. Transcriptional Control of Autophagy-Lysosome Function Drives Pancreatic Cancer Metabolism. Nature (2015) 524(7565):361–5. doi: 10.1038/nature14587
58. Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras Requires Autophagy to Maintain Oxidative Metabolism and Tumorigenesis. Genes Dev (2011) 25(5):460–70. doi: 10.1101/gad.2016311
59. Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy Is Critical for Pancreatic Tumor Growth and Progression in Tumors With P53 Alterations. Cancer Discov (2014) 4(8):905–13. doi: 10.1158/2159-8290.CD-14-0362
60. Rebecca VW, Amaravadi RK. Emerging Strategies to Effectively Target Autophagy in Cancer. Oncogene (2016) 35(1):1–11. doi: 10.1038/onc.2015.99
61. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer Drug Resistance: An Evolving Paradigm. Nat Rev Cancer (2013) 13(10):714–26. doi: 10.1038/nrc3599
62. Chun SY, Kwon YS, Nam KS, Kim S. Lapatinib Enhances the Cytotoxic Effects of Doxorubicin in MCF-7 Tumorspheres by Inhibiting the Drug Efflux Function of ABC Transporters. BioMed Pharmacother (2015) 72:37–43. doi: 10.1016/j.biopha.2015.03.009
63. International Transporter C, Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, et al. Membrane Transporters in Drug Development. Nat Rev Drug Discov (2010) 9(3):215–36. doi: 10.1038/nrd3028
64. Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi M, et al. The Role of Tumor Microenvironment in Chemoresistance: To Survive, Keep Your Enemies Closer. Int J Mol Sci (2017) 18(7):1586. doi: 10.3390/ijms18071586
65. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, et al. Cancer Stem Cells (CSCs) in Drug Resistance and Their Therapeutic Implications in Cancer Treatment. Stem Cells Int (2018) 2018:5416923. doi: 10.1155/2018/5416923
66. Chen QN, Wei CC, Wang ZX, Sun M. Long Non-Coding RNAs in Anti-Cancer Drug Resistance. Oncotarget (2017) 8(1):1925–36. doi: 10.18632/oncotarget.12461
67. Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 Pharmacogenetics and Cancer. Oncogene (2006) 25(11):1679–91. doi: 10.1038/sj.onc.1209377
68. Filomeni G, Turella P, Dupuis ML, Forini O, Ciriolo MR, Cianfriglia M, et al. 6-(7-Nitro-2,1,3-Benzoxadiazol-4-Ylthio)Hexanol, a Specific Glutathione S-Transferase Inhibitor, Overcomes the Multidrug Resistance (MDR)-Associated Protein 1-Mediated MDR in Small Cell Lung Cancer. Mol Cancer Ther (2008) 7(2):371–9. doi: 10.1158/1535-7163.MCT-07-0487
69. Zhu K, Liu Q, Zhou Y, Tao C, Zhao Z, Sun J, et al. Oncogenes and Tumor Suppressor Genes: Comparative Genomics and Network Perspectives. BMC Genomics (2015) 16:S8. doi: 10.1186/1471-2164-16-S7-S8
70. Mathew R, Karantza-Wadsworth V, White E. Role of Autophagy in Cancer. Nat Rev Cancer (2007) 7(12):961–7. doi: 10.1038/nrc2254
71. Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, et al. Macroautophagy Inhibition Sensitizes Tamoxifen-Resistant Breast Cancer Cells and Enhances Mitochondrial Depolarization. Breast Cancer Res Treat (2008) 112(3):389–403. doi: 10.1007/s10549-007-9873-4
72. Sun WL, Chen J, Wang YP, Zheng H. Autophagy Protects Breast Cancer Cells From Epirubicin-Induced Apoptosis and Facilitates Epirubicin-Resistance Development. Autophagy (2011) 7(9):1035–44. doi: 10.4161/auto.7.9.16521
73. Ajabnoor GM, Crook T, Coley HM. Paclitaxel Resistance Is Associated With Switch From Apoptotic to Autophagic Cell Death in MCF-7 Breast Cancer Cells. Cell Death Dis (2012) 3:e260. doi: 10.1038/cddis.2011.139
74. Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The Complex Relationship Between TFEB Transcription Factor Phosphorylation and Subcellular Localization. EMBO J (2018) 37(11):e98804. doi: 10.15252/embj.201798804
75. Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, et al. S100A8 Contributes to Drug Resistance by Promoting Autophagy in Leukemia Cells. PloS One (2014) 9(5):e97242. doi: 10.1371/journal.pone.0097242
76. Li X, Zhou Y, Li Y, Yang L, Ma Y, Peng X, et al. Autophagy: A Novel Mechanism of Chemoresistance in Cancers. BioMed Pharmacother (2019) 119:109415. doi: 10.1016/j.biopha.2019.109415
77. Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O’Prey J, Clark W, et al. Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol Cell (2017) 66(4):517–32.e519. doi: 10.1016/j.molcel.2017.04.027
78. Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, et al. Global Genomic Profiling Reveals an Extensive P53-Regulated Autophagy Program Contributing to Key P53 Responses. Genes Dev (2013) 27(9):1016–31. doi: 10.1101/gad.212282.112
79. Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic Reticulum Stress Triggers Autophagy. J Biol Chem (2006) 281(40):30299–304. doi: 10.1074/jbc.M607007200
80. Eum KH, Lee M. Targeting the Autophagy Pathway Using Ectopic Expression of Beclin 1 in Combination With Rapamycin in Drug-Resistant V-Ha-Ras-Transformed NIH 3T3 Cells. Mol Cells (2011) 31(3):231–8. doi: 10.1007/s10059-011-0034-6
81. Ge J, Chen Z, Huang J, Chen J, Yuan W, Deng Z, et al. Upregulation of Autophagy-Related Gene-5 (ATG-5) Is Associated With Chemoresistance in Human Gastric Cancer. PloS One (2014) 9(10):e110293. doi: 10.1371/journal.pone.0110293
82. Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L. Chloroquine and Hydroxychloroquine for Cancer Therapy. Mol Cell Oncol (2014) 1(1):e29911. doi: 10.4161/mco.29911
83. An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, et al. miR-23b-3p Regulates the Chemoresistance of Gastric Cancer Cells by Targeting ATG12 and HMGB2. Cell Death Dis (2015) 6:e1766. doi: 10.1038/cddis.2015.123
84. Chittaranjan S, Bortnik S, Dragowska WH, Xu J, Abeysundara N, Leung A, et al. Autophagy Inhibition Augments the Anticancer Effects of Epirubicin Treatment in Anthracycline-Sensitive and -Resistant Triple-Negative Breast Cancer. Clin Cancer Res (2014) 20(12):3159–73. doi: 10.1158/1078-0432.CCR-13-2060
85. Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW, et al. Targeting Autophagy Enhances Sorafenib Lethality for Hepatocellular Carcinoma via ER Stress-Related Apoptosis. Autophagy (2011) 7(10):1159–72. doi: 10.4161/auto.7.10.16818
86. Zhao XG, Sun RJ, Yang XY, Liu DY, Lei DP, Jin T, et al. Chloroquine-Enhanced Efficacy of Cisplatin in the Treatment of Hypopharyngeal Carcinoma in Xenograft Mice. PloS One (2015) 10(4):e0126147. doi: 10.1371/journal.pone.0126147
87. Liang X, Tang J, Liang Y, Jin R, Cai X. Suppression of Autophagy by Chloroquine Sensitizes 5-Fluorouracil-Mediated Cell Death in Gallbladder Carcinoma Cells. Cell Biosci (2014) 4(1):10. doi: 10.1186/2045-3701-4-10
88. Usman RM, Razzaq F, Akbar A, Farooqui AA, Iftikhar A, Latif A, et al. Role and Mechanism of Autophagy-Regulating Factors in Tumorigenesis and Drug Resistance. Asia Pac J Clin Oncol (2020) 17(3):193–208. doi: 10.1111/ajco.13449
89. Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. Phase I Clinical Trial and Pharmacodynamic Evaluation of Combination Hydroxychloroquine and Doxorubicin Treatment in Pet Dogs Treated for Spontaneously Occurring Lymphoma. Autophagy (2014) 10(8):1415–25. doi: 10.4161/auto.29165
90. Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, et al. A Phase I/II Trial of Hydroxychloroquine in Conjunction With Radiation Therapy and Concurrent and Adjuvant Temozolomide in Patients With Newly Diagnosed Glioblastoma Multiforme. Autophagy (2014) 10(8):1359–68. doi: 10.4161/auto.28984
91. Rong LW, Wang RX, Zheng XL, Feng XQ, Zhang L, Zhang L, et al. Combination of Wogonin and Sorafenib Effectively Kills Human Hepatocellular Carcinoma Cells Through Apoptosis Potentiation and Autophagy Inhibition. Oncol Lett (2017) 13(6):5028–34. doi: 10.3892/ol.2017.6059
92. Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, et al. A Highly Potent and Selective Vps34 Inhibitor Alters Vesicle Trafficking and Autophagy. Nat Chem Biol (2014) 10(12):1013–9. doi: 10.1038/nchembio.1681
93. Liu PF, Tsai KL, Hsu CJ, Tsai WL, Cheng JS, Chang HW, et al. Drug Repurposing Screening Identifies Tioconazole as an ATG4 Inhibitor That Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy. Theranostics (2018) 8(3):830–45. doi: 10.7150/thno.22012
94. Jiang K, Zhang C, Yu B, Chen B, Liu Z, Hou C, et al. Autophagic Degradation of FOXO3a Represses the Expression of PUMA to Block Cell Apoptosis in Cisplatin-Resistant Osteosarcoma Cells. Am J Cancer Res (2017) 7(7):1407–22.
95. Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy-Regulating microRNAs and Cancer. Front Oncol (2017) 7:65. doi: 10.3389/fonc.2017.00065
96. Anding AL, Baehrecke EH. Autophagy in Cell Life and Cell Death. Curr Top Dev Biol (2015) 114:67–91. doi: 10.1016/bs.ctdb.2015.07.012
97. Li JR, Cheng CL, Yang CR, Ou YC, Wu MJ, Ko JL. Dual Inhibitor of Phosphoinositide 3-Kinase/Mammalian Target of Rapamycin NVP-BEZ235 Effectively Inhibits Cisplatin-Resistant Urothelial Cancer Cell Growth Through Autophagic Flux. Toxicol Lett (2013) 220(3):267–76. doi: 10.1016/j.toxlet.2013.04.021
98. Huang JC, Cui ZF, Chen SM, Yang LJ, Lian HK, Liu B, et al. NVP-BEZ235 Synergizes Cisplatin Sensitivity in Osteosarcoma. Oncotarget (2018) 9(12):10483–96. doi: 10.18632/oncotarget.23711
99. Westhoff MA, Faham N, Marx D, Nonnenmacher L, Jennewein C, Enzenmuller S, et al. Sequential Dosing in Chemosensitization: Targeting the PI3K/Akt/mTOR Pathway in Neuroblastoma. PloS One (2013) 8(12):e83128. doi: 10.1371/journal.pone.0083128
100. Yu Z, Xie G, Zhou G, Cheng Y, Zhang G, Yao G, et al. NVP-BEZ235, a Novel Dual PI3K-mTOR Inhibitor Displays Anti-Glioma Activity and Reduces Chemoresistance to Temozolomide in Human Glioma Cells. Cancer Lett (2015) 367(1):58–68. doi: 10.1016/j.canlet.2015.07.007
101. Chiu LY, Hu ME, Yang TY, Hsin IL, Ko JL, Tsai KJ, et al. Immunomodulatory Protein From Ganoderma Microsporum Induces Pro-Death Autophagy Through Akt-mTOR-P70s6k Pathway Inhibition in Multidrug Resistant Lung Cancer Cells. PloS One (2015) 10(5):e0125774. doi: 10.1371/journal.pone.0125774
102. Shimizu S, Konishi A, Nishida Y, Mizuta T, Nishina H, Yamamoto A, et al. Involvement of JNK in the Regulation of Autophagic Cell Death. Oncogene (2010) 29(14):2070–82. doi: 10.1038/onc.2009.487
103. Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, et al. Mcl-1-Dependent Activation of Beclin 1 Mediates Autophagic Cell Death Induced by Sorafenib and SC-59 in Hepatocellular Carcinoma Cells. Cell Death Dis (2013) 4:e485. doi: 10.1038/cddis.2013.18
104. Pea A, Jamieson NB, Braconi C. Biology and Clinical Application of Regulatory RNAs in Hepatocellular Carcinoma. Hepatology (2021) 73:38–48. doi: 10.1002/hep.31225
105. Kim VN. MicroRNA Biogenesis: Coordinated Cropping and Dicing. Nat Rev Mol Cell Biol (2005) 6(5):376–85. doi: 10.1038/nrm1644
106. Dalmay T. Mechanism of miRNA-Mediated Repression of mRNA Translation. Essays Biochem (2013) 54:29–38. doi: 10.1042/bse0540029
107. Gebert LFR, MacRae IJ. Regulation of microRNA Function in Animals. Nat Rev Mol Cell Biol (2019) 20(1):21–37. doi: 10.1038/s41580-018-0045-7
108. Jansson MD, Lund AH. MicroRNA and Cancer. Mol Oncol (2012) 6(6):590–610. doi: 10.1016/j.molonc.2012.09.006
109. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA Genes Are Transcribed by RNA Polymerase II. EMBO J (2004) 23(20):4051–60. doi: 10.1038/sj.emboj.7600385
110. Borchert GM, Lanier W, Davidson BL. RNA Polymerase III Transcribes Human microRNAs. Nat Struct Mol Biol (2006) 13(12):1097–101. doi: 10.1038/nsmb1167
111. Luhur A, Chawla G, Wu YC, Li J, Sokol NS. Drosha-Independent DGCR8/Pasha Pathway Regulates Neuronal Morphogenesis. Proc Natl Acad Sci U.S.A. (2014) 111(4):1421–6. doi: 10.1073/pnas.1318445111
112. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The Nuclear RNase III Drosha Initiates microRNA Processing. Nature (2003) 425(6956):415–9. doi: 10.1038/nature01957
113. Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A High-Resolution Structure of the pre-microRNA Nuclear Export Machinery. Science (2009) 326(5957):1275–9. doi: 10.1126/science.1178705
114. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A Cellular Function for the RNA-Interference Enzyme Dicer in the Maturation of the Let-7 Small Temporal RNA. Science (2001) 293(5531):834–8. doi: 10.1126/science.1062961
115. Wang ZC, Huang FZ, Xu HB, Sun JC, Wang CF. MicroRNA-137 Inhibits Autophagy and Chemosensitizes Pancreatic Cancer Cells by Targeting ATG5. Int J Biochem Cell Biol (2019) 111:63–71. doi: 10.1016/j.biocel.2019.01.020
116. Zhao J, Nie Y, Wang H, Lin Y. MiR-181a Suppresses Autophagy and Sensitizes Gastric Cancer Cells to Cisplatin. Gene (2016) 576(2 Pt 2):828–33. doi: 10.1016/j.gene.2015.11.013
117. Luo M, Wu L, Zhang K, Wang H, Wu S, O’Connell D, et al. miR-216b Enhances the Efficacy of Vemurafenib by Targeting Beclin-1, UVRAG and ATG5 in Melanoma. Cell Signal (2018) 42:30–43. doi: 10.1016/j.cellsig.2017.09.024
118. Yu Y, Cao L, Yang L, Kang R, Lotze M, Tang D. microRNA 30A Promotes Autophagy in Response to Cancer Therapy. Autophagy (2012) 8(5):853–5. doi: 10.4161/auto.20053
119. Zhang W, Dong YZ, Du X, Peng XN, Shen QM. MiRNA-153-3p Promotes Gefitinib-Sensitivity in Non-Small Cell Lung Cancer by Inhibiting ATG5 Expression and Autophagy. Eur Rev Med Pharmacol Sci (2019) 23(6):2444–52. doi: 10.26355/eurrev_201903_17391
120. Li YL, Tang JM, Chen XY, Luo B, Liang GH, Qu Q, et al. MicroRNA-153-3p Enhances the Sensitivity of Chronic Myeloid Leukemia Cells to Imatinib by Inhibiting B-Cell Lymphoma-2-Mediated Autophagy. Hum Cell (2020) 33(3):610–8. doi: 10.1007/s13577-020-00367-1
121. Zhao P, Li M, Wang Y, Chen Y, He C, Zhang X, et al. Enhancing Anti-Tumor Efficiency in Hepatocellular Carcinoma Through the Autophagy Inhibition by miR-375/Sorafenib in Lipid-Coated Calcium Carbonate Nanoparticles. Acta Biomater (2018) 72:248–55. doi: 10.1016/j.actbio.2018.03.022
122. Yang S, Wang M, Yang L, Li Y, Ma Y, Peng X, et al. MicroRNA-375 Targets ATG14 to Inhibit Autophagy and Sensitize Hepatocellular Carcinoma Cells to Sorafenib. Onco Targets Ther (2020) 13:3557–70. doi: 10.2147/OTT.S247655
123. Zachari M, Ganley IG. The Mammalian ULK1 Complex and Autophagy Initiation. Essays Biochem (2017) 61(6):585–96. doi: 10.1042/EBC20170021
124. Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X, et al. miR-409-3p Sensitizes Colon Cancer Cells to Oxaliplatin by Inhibiting Beclin-1-Mediated Autophagy. Int J Mol Med (2016) 37(4):1030–8. doi: 10.3892/ijmm.2016.2492
125. Chatterjee A, Chattopadhyay D, Chakrabarti G. MiR-16 Targets Bcl-2 in Paclitaxel-Resistant Lung Cancer Cells and Overexpression of miR-16 Along With miR-17 Causes Unprecedented Sensitivity by Simultaneously Modulating Autophagy and Apoptosis. Cell Signal (2015) 27(2):189–203. doi: 10.1016/j.cellsig.2014.11.023
126. Chatterjee A, Chattopadhyay D, Chakrabarti G. miR-17-5p Downregulation Contributes to Paclitaxel Resistance of Lung Cancer Cells Through Altering Beclin1 Expression. PloS One (2014) 9(4):e95716. doi: 10.1371/journal.pone.0095716
127. Li Y, Jiang W, Hu Y, Da Z, Zeng C, Tu M, et al. MicroRNA-199a-5p Inhibits Cisplatin-Induced Drug Resistance via Inhibition of Autophagy in Osteosarcoma Cells. Oncol Lett (2016) 12(5):4203–8. doi: 10.3892/ol.2016.5172
128. Yu Y, Yang L, Zhao M, Zhu S, Kang R, Vernon P, et al. Targeting microRNA-30a-Mediated Autophagy Enhances Imatinib Activity Against Human Chronic Myeloid Leukemia Cells. Leukemia (2012) 26(8):1752–60. doi: 10.1038/leu.2012.65
129. Du X, Liu B, Luan X, Cui Q, Li L. miR-30 Decreases Multidrug Resistance in Human Gastric Cancer Cells by Modulating Cell Autophagy. Exp Ther Med (2018) 15(1):599–605. doi: 10.3892/etm.2017.5354
130. Xu R, Liu S, Chen H, Lao L. MicroRNA-30a Downregulation Contributes to Chemoresistance of Osteosarcoma Cells Through Activating Beclin-1-Mediated Autophagy. Oncol Rep (2016) 35(3):1757–63. doi: 10.3892/or.2015.4497
131. Yang X, Bai F, Xu Y, Chen Y, Chen L. Intensified Beclin-1 Mediated by Low Expression of Mir-30a-5p Promotes Chemoresistance in Human Small Cell Lung Cancer. Cell Physiol Biochem (2017) 43(3):1126–39. doi: 10.1159/000481754
132. Zou Z, Wu L, Ding H, Wang Y, Zhang Y, Chen X, et al. MicroRNA-30a Sensitizes Tumor Cells to Cis-Platinum via Suppressing Beclin 1-Mediated Autophagy. J Biol Chem (2012) 287(6):4148–56. doi: 10.1074/jbc.M111.307405
133. Khalil NA, Desouky MN, Diab IH, Hamed NAM, Mannaa HF. MicroRNA 30a Mediated Autophagy and Imatinib Response in Egyptian Chronic Myeloid Leukemia Patients. Indian J Hematol Blood Transfus (2020) 36(3):491–7. doi: 10.1007/s12288-019-01241-3
134. Xu WP, Liu JP, Feng JF, Zhu CP, Yang Y, Zhou WP, et al. miR-541 Potentiates the Response of Human Hepatocellular Carcinoma to Sorafenib Treatment by Inhibiting Autophagy. Gut (2020) 69(7):1309–21. doi: 10.1136/gutjnl-2019-318830
135. Pan B, Chen Y, Song H, Xu Y, Wang R, Chen L. Mir-24-3p Downregulation Contributes to VP16-DDP Resistance in Small-Cell Lung Cancer by Targeting ATG4A. Oncotarget (2015) 6(1):317–31. doi: 10.18632/oncotarget.2787
136. Hua L, Zhu G, Wei J. MicroRNA-1 Overexpression Increases Chemosensitivity of Non-Small Cell Lung Cancer Cells by Inhibiting Autophagy Related 3-Mediated Autophagy. Cell Biol Int (2018) 42(9):1240–9. doi: 10.1002/cbin.10995
137. Pan B, Feng B, Chen Y, Huang G, Wang R, Chen L, et al. MiR-200b Regulates Autophagy Associated With Chemoresistance in Human Lung Adenocarcinoma. Oncotarget (2015) 6(32):32805–20. doi: 10.18632/oncotarget.5352
138. Huang H, Tang J, Zhang L, Bu Y, Zhang X. miR-874 Regulates Multiple-Drug Resistance in Gastric Cancer by Targeting ATG16L1. Int J Oncol (2018) 53(6):2769–79. doi: 10.3892/ijo.2018.4593
139. Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z, Liu L, et al. Methylation-Induced Silencing of miR-34a Enhances Chemoresistance by Directly Upregulating ATG4B-Induced Autophagy Through AMPK/mTOR Pathway in Prostate Cancer. Oncol Rep (2016) 35(1):64–72. doi: 10.3892/or.2015.4331
140. Hu Z, Cai M, Zhang Y, Tao L, Guo R. miR-29c-3p Inhibits Autophagy and Cisplatin Resistance in Ovarian Cancer by Regulating FOXP1/ATG14 Pathway. Cell Cycle (2020) 19(2):193–206. doi: 10.1080/15384101.2019.1704537
141. Tang D, Kang R, Zeh HJ 3rd, Lotze MT. High-Mobility Group Box 1 and Cancer. Biochim Biophys Acta (2010) 1799(1-2):131–40. doi: 10.1016/j.bbagrm.2009.11.014
142. Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in Inflammation and Cancer. Annu Rev Immunol (2010) 28:367–88. doi: 10.1146/annurev.immunol.021908.132603
143. Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, et al. HMGB1 Promotes Drug Resistance in Osteosarcoma. Cancer Res (2012) 72(1):230–8. doi: 10.1158/0008-5472.CAN-11-2001
144. Pan B, Chen D, Huang J, Wang R, Feng B, Song H, et al. HMGB1-Mediated Autophagy Promotes Docetaxel Resistance in Human Lung Adenocarcinoma. Mol Cancer (2014) 13:165. doi: 10.1186/1476-4598-13-165
145. Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, et al. HMGB1-Induced Autophagy Promotes Chemotherapy Resistance in Leukemia Cells. Leukemia (2011) 25(1):23–31. doi: 10.1038/leu.2010.225
146. Li X, Wang S, Chen Y, Liu G, Yang X. miR-22 Targets the 3’ UTR of HMGB1 and Inhibits the HMGB1-Associated Autophagy in Osteosarcoma Cells During Chemotherapy. Tumour Biol (2014) 35(6):6021–8. doi: 10.1007/s13277-014-1797-0
147. Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang R, et al. MIR34A Regulates Autophagy and Apoptosis by Targeting HMGB1 in the Retinoblastoma Cell. Autophagy (2014) 10(3):442–52. doi: 10.4161/auto.27418
148. Ran X, Yang J, Liu C, Zhou P, Xiao L, Zhang K. MiR-218 Inhibits HMGB1-Mediated Autophagy in Endometrial Carcinoma Cells During Chemotherapy. Int J Clin Exp Pathol (2015) 8(6):6617–26.
149. Chen Y, Zhou X, Qiao J, Bao A. MiR-142-3p Overexpression Increases Chemo-Sensitivity of NSCLC by Inhibiting HMGB1-Mediated Autophagy. Cell Physiol Biochem (2017) 41(4):1370–82. doi: 10.1159/000467896
150. Shi Y, Gong W, Lu L, Wang Y, Ren J. Upregulation of miR-129-5p Increases the Sensitivity to Taxol Through Inhibiting HMGB1-Mediated Cell Autophagy in Breast Cancer MCF-7 Cells. Braz J Med Biol Res (2019) 52(11):e8657. doi: 10.1590/1414-431X20198657
151. Yu Y, Xiang N, Lin M, Huang JW, Zhang J, Cheng B, et al. miR- 26a Sensitizes Melanoma Cells To Dabrafenib Via Targeting HMGB1-Dependent Autophagy Pathways. Drug Des Devel Ther (2019) 13:3717–26. doi: 10.2147/DDDT.S225671
152. Meng Y, Gao R, Ma J, Zhao J, Xu E, Wang C, et al. MicroRNA-140-5p Regulates Osteosarcoma Chemoresistance by Targeting HMGN5 and Autophagy. Sci Rep (2017) 7(1):416. doi: 10.1038/s41598-017-00405-3
153. Chia WJ, Tang BL. Emerging Roles for Rab Family GTPases in Human Cancer. Biochim Biophys Acta (2009) 1795(2):110–6. doi: 10.1016/j.bbcan.2008.10.001
154. Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J, et al. MicroRNA-148a-3p Enhances Cisplatin Cytotoxicity in Gastric Cancer Through Mitochondrial Fission Induction and Cyto-Protective Autophagy Suppression. Cancer Lett (2017) 410:212–27. doi: 10.1016/j.canlet.2017.09.035
155. Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, et al. MiR-26 Enhances Chemosensitivity and Promotes Apoptosis of Hepatocellular Carcinoma Cells Through Inhibiting Autophagy. Cell Death Dis (2017) 8(1):e2540. doi: 10.1038/cddis.2016.461
156. Rothschild SI, Gautschi O, Batliner J, Gugger M, Fey MF, Tschan MP. MicroRNA-106a Targets Autophagy and Enhances Sensitivity of Lung Cancer Cells to Src Inhibitors. Lung Cancer (2017) 107:73–83. doi: 10.1016/j.lungcan.2016.06.004
157. Soni M, Patel Y, Markoutsa E, Jie C, Liu S, Xu P, et al. Autophagy, Cell Viability, and Chemoresistance Are Regulated By miR-489 in Breast Cancer. Mol Cancer Res (2018) 16(9):1348–60. doi: 10.1158/1541-7786.MCR-17-0634
158. Fitzwalter BE, Towers CG, Sullivan KD, Andrysik Z, Hoh M, Ludwig M, et al. Autophagy Inhibition Mediates Apoptosis Sensitization in Cancer Therapy by Relieving FOXO3a Turnover. Dev Cell (2018) 44(5):555–565 e553. doi: 10.1016/j.devcel.2018.02.014
159. Zhou Y, Chen E, Tang Y, Mao J, Shen J, Zheng X, et al. miR-223 Overexpression Inhibits Doxorubicin-Induced Autophagy by Targeting FOXO3a and Reverses Chemoresistance in Hepatocellular Carcinoma Cells. Cell Death Dis (2019) 10(11):843. doi: 10.1038/s41419-019-2053-8
160. Astudillo P. Wnt5a Signaling in Gastric Cancer. Front Cell Dev Biol (2020) 8:110. doi: 10.3389/fcell.2020.00110
161. Wu C, Zhuang Y, Jiang S, Liu S, Zhou J, Wu J, et al. Interaction Between Wnt/beta-Catenin Pathway and microRNAs Regulates Epithelial-Mesenchymal Transition in Gastric Cancer (Review). Int J Oncol (2016) 48(6):2236–46. doi: 10.3892/ijo.2016.3480
162. Chen PH, Liu AJ, Ho KH, Chiu YT, Anne Lin ZH, Lee YT, et al. microRNA-199a/B-5p Enhance Imatinib Efficacy via Repressing WNT2 Signaling-Mediated Protective Autophagy in Imatinib-Resistant Chronic Myeloid Leukemia Cells. Chem Biol Interact (2018) 291:144–51. doi: 10.1016/j.cbi.2018.06.006
163. Ren WW, Li DD, Chen X, Li XL, He YP, Guo LH, et al. MicroRNA-125b Reverses Oxaliplatin Resistance in Hepatocellular Carcinoma by Negatively Regulating EVA1A Mediated Autophagy. Cell Death Dis (2018) 9(5):547. doi: 10.1038/s41419-018-0592-z
164. Liu ZR, Song Y, Wan LH, Zhang YY, Zhou LM. Over-Expression of miR-451a can Enhance the Sensitivity of Breast Cancer Cells to Tamoxifen by Regulating 14-3-3zeta, Estrogen Receptor Alpha, and Autophagy. Life Sci (2016) 149:104–13. doi: 10.1016/j.lfs.2016.02.059
165. Yu X, Luo A, Liu Y, Wang S, Li Y, Shi W, et al. MiR-214 Increases the Sensitivity of Breast Cancer Cells to Tamoxifen and Fulvestrant Through Inhibition of Autophagy. Mol Cancer (2015) 14:208. doi: 10.1186/s12943-015-0480-4
166. Ueda S, Takanashi M, Sudo K, Kanekura K, Kuroda M. miR-27a Ameliorates Chemoresistance of Breast Cancer Cells by Disruption of Reactive Oxygen Species Homeostasis and Impairment of Autophagy. Lab Invest (2020) 100(6):863–73. doi: 10.1038/s41374-020-0409-4
167. Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D, et al. MicroRNA-495-3p Inhibits Multidrug Resistance by Modulating Autophagy Through GRP78/mTOR Axis in Gastric Cancer. Cell Death Dis (2018) 9(11):1070. doi: 10.1038/s41419-018-0950-x
168. Huang L, Hu C, Cao H, Wu X, Wang R, Lu H, et al. MicroRNA-29c Increases the Chemosensitivity of Pancreatic Cancer Cells by Inhibiting USP22 Mediated Autophagy. Cell Physiol Biochem (2018) 47(2):747–58. doi: 10.1159/000490027
169. Chang Z, Huo L, Li K, Wu Y, Hu Z. Blocked Autophagy by miR-101 Enhances Osteosarcoma Cell Chemosensitivity In Vitro. ScientificWorldJournal (2014) 2014:794756. doi: 10.1155/2014/794756
170. Meng CY, Zhao ZQ, Bai R, Zhao W, Wang YX, Sun L, et al. MicroRNA22 Regulates Autophagy and Apoptosis in Cisplatin Resistance of Osteosarcoma. Mol Med Rep (2020) 22(5):3911–21. doi: 10.3892/mmr.2020.11447
171. Wang P, Zhao ZQ, Guo SB, Yang TY, Chang ZQ, Li DH, et al. Roles of microRNA-22 in Suppressing Proliferation and Promoting Sensitivity of Osteosarcoma Cells via Metadherin-Mediated Autophagy. Orthop Surg (2019) 11(2):285–93. doi: 10.1111/os.12442
172. Zhang H, Tang J, Li C, Kong J, Wang J, Wu Y, et al. MiR-22 Regulates 5-FU Sensitivity by Inhibiting Autophagy and Promoting Apoptosis in Colorectal Cancer Cells. Cancer Lett (2015) 356(2 Pt B):781–90. doi: 10.1016/j.canlet.2014.10.029
173. Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Xie J, et al. Downregulation of YEATS4 by miR-218 Sensitizes Colorectal Cancer Cells to L-OHP-Induced Cell Apoptosis by Inhibiting Cytoprotective Autophagy. Oncol Rep (2016) 36(6):3682–90. doi: 10.3892/or.2016.5195
174. Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, et al. miR-34a Mediates Oxaliplatin Resistance of Colorectal Cancer Cells by Inhibiting Macroautophagy via Transforming Growth Factor-Beta/Smad4 Pathway. World J Gastroenterol (2017) 23(10):1816–27. doi: 10.3748/wjg.v23.i10.1816
175. Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, et al. microRNA-17 Regulates the Expression of ATG7 and Modulates the Autophagy Process, Improving the Sensitivity to Temozolomide and Low-Dose Ionizing Radiation Treatments in Human Glioblastoma Cells. Cancer Biol Ther (2013) 14(7):574–86. doi: 10.4161/cbt.24597
176. Huang T, Wan X, Alvarez AA, James CD, Song X, Yang Y, et al. MIR93 (microRNA -93) Regulates Tumorigenicity and Therapy Response of Glioblastoma by Targeting Autophagy. Autophagy (2019) 15(6):1100–11. doi: 10.1080/15548627.2019.1569947
177. Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, et al. MiRNA-30a-Mediated Autophagy Inhibition Sensitizes Renal Cell Carcinoma Cells to Sorafenib. Biochem Biophys Res Commun (2015) 459(2):234–9. doi: 10.1016/j.bbrc.2015.02.084
178. Li Y, Zhang G, Wu B, Yang W, Liu Z. miR-199a-5p Represses Protective Autophagy and Overcomes Chemoresistance by Directly Targeting DRAM1 in Acute Myeloid Leukemia. J Oncol (2019) 2019:5613417. doi: 10.1155/2019/5613417
179. Huang N, Wu J, Qiu W, Lyu Q, He J, Xie W, et al. MiR-15a and miR-16 Induce Autophagy and Enhance Chemosensitivity of Camptothecin. Cancer Biol Ther (2015) 16(6):941–8. doi: 10.1080/15384047.2015.1040963
180. Liu J, Xing Y, Rong L. miR-181 Regulates Cisplatin-Resistant Non-Small Cell Lung Cancer via Downregulation of Autophagy Through the PTEN/PI3K/AKT Pathway. Oncol Rep (2018) 39(4):1631–9. doi: 10.3892/or.2018.6268
181. Nyhan MJ, O’Donovan TR, Boersma AW, Wiemer EA, McKenna SL. MiR-193b Promotes Autophagy and Non-Apoptotic Cell Death in Oesophageal Cancer Cells. BMC Cancer (2016) 16:101. doi: 10.1186/s12885-016-2123-6
182. Li H, Chen L, Li JJ, Zhou Q, Huang A, Liu WW, et al. miR-519a Enhances Chemosensitivity and Promotes Autophagy in Glioblastoma by Targeting STAT3/Bcl2 Signaling Pathway. J Hematol Oncol (2018) 11(1):70. doi: 10.1186/s13045-018-0618-0
183. Jeong S, Kim BG, Kim DY, Kim BR, Kim JL, Park SH, et al. Cannabidiol Overcomes Oxaliplatin Resistance by Enhancing NOS3- and SOD2-Induced Autophagy in Human Colorectal Cancer Cells. Cancers (Basel) (2019) 11(6):781. doi: 10.3390/cancers11060781
184. Yu JL, Gao X. MicroRNA 1301 Inhibits Cisplatin Resistance in Human Ovarian Cancer Cells by Regulating EMT and Autophagy. Eur Rev Med Pharmacol Sci (2020) 24(4):1688–96. doi: 10.26355/eurrev_202002_20343
185. Bao L, Lv L, Feng J, Chen Y, Wang X, Han S, et al. miR-487b-5p Regulates Temozolomide Resistance of Lung Cancer Cells Through LAMP2-Medicated Autophagy. DNA Cell Biol (2016) 35(8):385–92. doi: 10.1089/dna.2016.3259
186. Xu J, Su Y, Xu A, Fan F, Mu S, Chen L, et al. miR-221/222-Mediated Inhibition of Autophagy Promotes Dexamethasone Resistance in Multiple Myeloma. Mol Ther (2019) 27(3):559–70. doi: 10.1016/j.ymthe.2019.01.012
187. Bollaert E, Claus M, Vandewalle V, Lenglez S, Essaghir A, Demoulin JB, et al. MiR-15a-5p Confers Chemoresistance in Acute Myeloid Leukemia by Inhibiting Autophagy Induced by Daunorubicin. Int J Mol Sci (2021) 22(10):5153. doi: 10.3390/ijms22105153
188. Yu X, Li R, Shi W, Jiang T, Wang Y, Li C, et al. Silencing of MicroRNA-21 Confers the Sensitivity to Tamoxifen and Fulvestrant by Enhancing Autophagic Cell Death Through Inhibition of the PI3K-AKT-mTOR Pathway in Breast Cancer Cells. BioMed Pharmacother (2016) 77:37–44. doi: 10.1016/j.biopha.2015.11.005
189. Choi AM, Ryter SW, Levine B. Autophagy in Human Health and Disease. N Engl J Med (2013) 368(7):651–62. doi: 10.1056/NEJMra1205406
190. Gu Y, Fei Z, Zhu R. miR-21 Modulates Cisplatin Resistance of Gastric Cancer Cells by Inhibiting Autophagy via the PI3K/Akt/mTOR Pathway. Anticancer Drugs (2020) 31(4):385–93. doi: 10.1097/CAD.0000000000000886
191. He C, Dong X, Zhai B, Jiang X, Dong D, Li B, et al. MiR-21 Mediates Sorafenib Resistance of Hepatocellular Carcinoma Cells by Inhibiting Autophagy via the PTEN/Akt Pathway. Oncotarget (2015) 6(30):28867–81. doi: 10.18632/oncotarget.4814
192. Wang L, Tang B, Han H, Mao D, Chen J, Zeng Y, et al. miR-155 Affects Osteosarcoma MG-63 Cell Autophagy Induced by Adriamycin Through Regulating PTEN-PI3K/AKT/mTOR Signaling Pathway. Cancer Biother Radiopharm (2018) 33(1):32–8. doi: 10.1089/cbr.2017.2306
193. Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, Vasconcelos MH. Targeting miR-21 Induces Autophagy and Chemosensitivity of Leukemia Cells. Curr Drug Targets (2013) 14(10):1135–43. doi: 10.2174/13894501113149990185
194. Tang D, Zhao D, Wu Y, Yao R, Zhou L, Lu L, et al. The miR-3127-5p/P-STAT3 Axis Up-Regulates PD-L1 Inducing Chemoresistance in Non-Small-Cell Lung Cancer. J Cell Mol Med (2018) 22(8):3847–56. doi: 10.1111/jcmm.13657
195. Chen JC, Hsieh YY, Lo HL, Li A, Chou CJ, Yang PM. In Vitro and In Silico Mechanistic Insights Into miR-21-5p-Mediated Topoisomerase Drug Resistance in Human Colorectal Cancer Cells. Biomolecules (2019) 9(9):467. doi: 10.3390/biom9090467
196. Stojcheva N, Schechtmann G, Sass S, Roth P, Florea AM, Stefanski A, et al. MicroRNA-138 Promotes Acquired Alkylator Resistance in Glioblastoma by Targeting the Bcl-2-Interacting Mediator BIM. Oncotarget (2016) 7(11):12937–50. doi: 10.18632/oncotarget.7346
197. Wei R, Cao G, Deng Z, Su J, Cai L. miR-140-5p Attenuates Chemotherapeutic Drug-Induced Cell Death by Regulating Autophagy Through Inositol 1,4,5-Trisphosphate Kinase 2 (IP3k2) in Human Osteosarcoma Cells. Biosci Rep (2016) 36(5):e00392. doi: 10.1042/BSR20160238
198. Chen L, Jiang K, Jiang H, Wei P. miR-155 Mediates Drug Resistance in Osteosarcoma Cells via Inducing Autophagy. Exp Ther Med (2014) 8(2):527–32. doi: 10.3892/etm.2014.1752
199. Han J, Li J, Tang K, Zhang H, Guo B, Hou N, et al. miR-338-3p Confers 5-Fluorouracil Resistance in P53 Mutant Colon Cancer Cells by Targeting the Mammalian Target of Rapamycin. Exp Cell Res (2017) 360(2):328–36. doi: 10.1016/j.yexcr.2017.09.023
200. Yang F, Guo L, Cao Y, Li S, Li J, Liu M. MicroRNA-7-5p Promotes Cisplatin Resistance of Cervical Cancer Cells and Modulation of Cellular Energy Homeostasis by Regulating the Expression of the PARP-1 and BCL2 Genes. Med Sci Monit (2018) 24:6506–16. doi: 10.12659/MSM.910969
201. Wang H, Chen J, Zhang S, Zheng X, Xie S, Mao J, et al. MiR-223 Regulates Autophagy Associated With Cisplatin Resistance by Targeting FBXW7 in Human Non-Small Cell Lung Cancer. Cancer Cell Int (2020) 20:258. doi: 10.1186/s12935-020-01284-x
202. Lei Y, Chen L, Zhang G, Shan A, Ye C, Liang B, et al. MicroRNAs Target the Wnt/betacatenin Signaling Pathway to Regulate Epithelialmesenchymal Transition in Cancer (Review). Oncol Rep (2020) 44(4):1299–313. doi: 10.3892/or.2020.7703
203. Zhang L, Liao Y, Tang L. MicroRNA-34 Family: A Potential Tumor Suppressor and Therapeutic Candidate in Cancer. J Exp Clin Cancer Res (2019) 38(1):53. doi: 10.1186/s13046-019-1059-5
204. Ye L, Gho WM, Chan FL, Chen S, Leung LK. Dietary Administration of the Licorice Flavonoid Isoliquiritigenin Deters the Growth of MCF-7 Cells Overexpressing Aromatase. Int J Cancer (2009) 124(5):1028–36. doi: 10.1002/ijc.24046
205. Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T, et al. MicroRNA-25 Regulates Chemoresistance-Associated Autophagy in Breast Cancer Cells, a Process Modulated by the Natural Autophagy Inducer Isoliquiritigenin. Oncotarget (2014) 5(16):7013–26. doi: 10.18632/oncotarget.2192
206. Sung B, Chung HY, Kim ND. Role of Apigenin in Cancer Prevention via the Induction of Apoptosis and Autophagy. J Cancer Prev (2016) 21(4):216–26. doi: 10.15430/JCP.2016.21.4.216
207. Gao AM, Zhang XY, Hu JN, Ke ZP. Apigenin Sensitizes Hepatocellular Carcinoma Cells to Doxorubic Through Regulating miR-520b/ATG7 Axis. Chem Biol Interact (2018) 280:45–50. doi: 10.1016/j.cbi.2017.11.020
208. Ou W, Lv J, Zou X, Yao Y, Wu J, Yang J, et al. Propofol Inhibits Hepatocellular Carcinoma Growth and Invasion Through the HMGA2-Mediated Wnt/beta-Catenin Pathway. Exp Ther Med (2017) 13(5):2501–6. doi: 10.3892/etm.2017.4253
209. Liu F, Qiu F, Fu M, Chen H, Wang H. Propofol Reduces Epithelial to Mesenchymal Transition, Invasion and Migration of Gastric Cancer Cells Through the MicroRNA-195-5p/Snail Axis. Med Sci Monit (2020) 26:e920981. doi: 10.12659/MSM.920981
210. Zhang YF, Li CS, Zhou Y, Lu XH. Propofol Facilitates Cisplatin Sensitivity via lncRNA MALAT1/miR-30e/ATG5 Axis Through Suppressing Autophagy in Gastric Cancer. Life Sci (2020) 244:117280. doi: 10.1016/j.lfs.2020.117280
211. Zhou M, Zhang G, Hu J, Zhu Y, Lan H, Shen X, et al. Rutin Attenuates Sorafenib-Induced Chemoresistance and Autophagy in Hepatocellular Carcinoma by Regulating BANCR/miRNA-590-5p/OLR1 Axis. Int J Biol Sci (2021) 17(13):3595–607. doi: 10.7150/ijbs.62471
212. Mishra S, Yadav T, Rani V. Exploring miRNA Based Approaches in Cancer Diagnostics and Therapeutics. Crit Rev Oncol Hematol (2016) 98:12–23. doi: 10.1016/j.critrevonc.2015.10.003
213. YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long Noncoding RNA MALAT1 Regulates Autophagy Associated Chemoresistance via miR-23b-3p Sequestration in Gastric Cancer. Mol Cancer (2017) 16(1):174. doi: 10.1186/s12943-017-0743-3
214. Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y, et al. Circhipk3 Promotes Oxaliplatin-Resistance in Colorectal Cancer Through Autophagy by Sponging miR-637. EBioMedicine (2019) 48:277–88. doi: 10.1016/j.ebiom.2019.09.051
215. Romano G, Kwong LN. Diagnostic and Therapeutic Applications of miRNA-Based Strategies to Cancer Immunotherapy. Cancer Metastasis Rev (2018) 37(1):45–53. doi: 10.1007/s10555-017-9716-7
216. Moffett HF, Cartwright ANR, Kim HJ, Godec J, Pyrdol J, Aijo T, et al. The microRNA miR-31 Inhibits CD8(+) T Cell Function in Chronic Viral Infection. Nat Immunol (2017) 18(7):791–9. doi: 10.1038/ni.3755
217. Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis Is Regulated via microRNA-200/ZEB1 Axis Control of Tumour Cell PD-L1 Expression and Intratumoral Immunosuppression. Nat Commun (2014) 5:5241. doi: 10.1038/ncomms6241
218. Ghasabi M, Mansoori B, Mohammadi A, Duijf PH, Shomali N, Shirafkan N, et al. MicroRNAs in Cancer Drug Resistance: Basic Evidence and Clinical Applications. J Cell Physiol (2019) 234(3):2152–68. doi: 10.1002/jcp.26810
219. Momin MY, Gaddam RR, Kravitz M, Gupta A, Vikram A. The Challenges and Opportunities in the Development of MicroRNA Therapeutics: A Multidisciplinary Viewpoint. Cells (2021) 10(11):3097. doi: 10.3390/cells10113097
220. Sadelain M, Papapetrou EP, Bushman FD. Safe Harbours for the Integration of New DNA in the Human Genome. Nat Rev Cancer (2011) 12(1):51–8. doi: 10.1038/nrc3179
Keywords: microRNA, chemoresistance, autophagy, cancer therapy, non-coding RNA
Citation: Lei Y, Chen L, Liu J, Zhong Y and Deng L (2022) The MicroRNA-Based Strategies to Combat Cancer Chemoresistance via Regulating Autophagy. Front. Oncol. 12:841625. doi: 10.3389/fonc.2022.841625
Received: 22 December 2021; Accepted: 13 January 2022;
Published: 08 February 2022.
Edited by:
Dong-Hua Yang, St. John’s University, United StatesReviewed by:
Jianyang Hu, City University of Hong Kong, Hong Kong SAR, ChinaCopyright © 2022 Lei, Chen, Liu, Zhong and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinqin Zhong, NjQxMjk2NDg0QHFxLmNvbQ==; Lijuan Deng, bGpkZW5nQGpudS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.