- 1Department of Biotechnology and Bioinformatics, Faculty of Life Sciences, Jagadguru Sri Shivarathreeshwara (JSS) Academy of Higher Education and Research (JSSAHER), Mysuru, India
- 2Department of General Surgery, Adichunchanagiri Institute of Medical Sciences, Mandya, India
- 3Department of Medical Oncology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India
- 4Department of Microbiology, Faculty of Life Sciences, Jagadguru Sri Shivarathreeshwara (JSS) Academy of Higher Education and Research (JSSAHER), Mysuru, India
- 5School of Agro-Industry, Faculty of Agro-Industry, Chiang Mai University, Chiang Mai, Thailand
- 6Cluster of Agro Bio-Circular-Green Industry (Agro BCG), Chiang Mai University, Chiang Mai, Thailand
- 7Centre of Excellence in Molecular Biology and Regenerative Medicine (CEMR), Department of Biochemistry, JSS Medical College, JSS Academy of Higher Education and Research (JSSAHER), Mysuru, India
- 8Department of Plant and Soil Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
Cancers are known to have multifactorial etiology. Certain bacteria and viruses are proven carcinogens. Lately, there has been in-depth research investigating carcinogenic capabilities of some bacteria. Reports indicate that chronic inflammation and harmful bacterial metabolites to be strong promoters of neoplasticity. Helicobacter pylori-induced gastric adenocarcinoma is the best illustration of the chronic inflammation paradigm of oncogenesis. Chronic inflammation, which produces excessive reactive oxygen species (ROS) is hypothesized to cause cancerous cell proliferation. Other possible bacteria-dependent mechanisms and virulence factors have also been suspected of playing a vital role in the bacteria-induced-cancer(s). Numerous attempts have been made to explore and establish the possible relationship between the two. With the growing concerns on anti-microbial resistance and over-dependence of mankind on antibiotics to treat bacterial infections, it must be deemed critical to understand and identify carcinogenic bacteria, to establish their role in causing cancer.
Introduction
Cancer, one of the leading causes of morbidity and mortality in the world, is characterized by the uncontrolled growth of cells with potential to metastasize. Problems arise when these cancerous cells, carrying mutagenic DNA, turn into tumors (1). The World Health Organization (WHO) estimates that ~10 million deaths occurred due to cancer in 2020 alone (2), twice the number of global COVID-19 related deaths in the same year. Numerous causes of cancer have been identified, with enormous interlink between environmental and genetic factors (3). The alterations occurring in the genetic makeup are known to be influenced by various external factors mostly related to lifestyle, such as alcohol, tobacco abuse, and exposure to sunlight (3). In 2018, roughly 19% and 2% of cancers worldwide had been attributed to tobacco and alcohol intake respectively (4). Interestingly, microbial infections have also been recognized to potentially cause cancer(s) (5–7). According to a 2021 report of the International Agency for Research on Cancer (IARC), there were 2.2 million cancer cases globally related to microbial infection(s), caused by Helicobacter pylori, Human papillomavirus (HPV), Hepatitis B virus (HBV), Hepatitis C virus (HCV), and Schistosoma haematobium (8).
Traditionally, bacteria have not been considered as a significant etiologic factor for cancer. Though an infectious cause was suspected in the 16th century, the relationship between bacteria and cancer was not very clear due to many reasons. One such example is the varied duration between the onset of infection and the diagnosis of cancer, making it difficult to single out (9). Cancer causing bacteria modulate a variety of immune responses which are believed to play a role in tumor progression. However, the very mechanism of carcinogenesis by bacteria is yet to be elucidated. Notwithstanding, the bacteria-associated factors that may influence neoplasm are not well understood. Carcinogenesis is also influenced by the duration of infection (acute or chronic infections). While epidemiological evidence suggests a reduced risk of cancer in case of acute infections, persistent infections may increase the risk (10). The neoplastic potential of bacterial infections is reported to be influenced by various factors, such as the host immune response, presence of the bacterial toxin, etc. (11). A few bacterial infections are known to promote inflammatory responses amounting to mutagenesis (12), whereas the others are observed to impede the host cell signaling pathways (13). In addition, bacteria interact with host cell(s) and modulate their cell adhesion and cytoskeletal functions (13). This complex network in which a bacteria can possibly promote oncogenesis includes modified cell proliferation and death, alteration of the immune response, and change in the host metabolic processes (14). Recent findings have confirmed the vitality of inflammation in tumor growth promotion, with a direct causal relationship between the two (15, 16). Infection, persistent irritation, and inflammation, in combination, contribute to the development of cancer. In 2011, amongst other cancer hallmarks, tumor-promoting inflammation was highlighted as an enabling trait (17). Furthermore, non-steroidal anti-inflammatory drug use was linked to a lower chance of acquiring various tumors and a lower mortality rate, emphasizing the importance of inflammation in neoplastic transformations (18). Carcinogenesis and inflammation are both highly complicated processes relying independently on multiple signaling mechanisms. Advances in inflammation research have revealed a link between the inflammatory processes and neoplastic transformations, tumor growth, as well as the development of metastases and recurrences (16). The tumor microenvironment, predominantly regulated by inflammatory cells, has now been recognized as an essential participant in the neoplastic process, supporting the proliferation, survival, and migration events. Additionally, innate immune signaling molecules, such as selectins, chemokines, and their receptors, have been co-opted by tumor cells for the purposes of invasion, migration, and metastasis (15).
Surprisingly, the principal global research focus has been limited to establishing the nonspecific mechanisms of carcinogenesis by different microorganisms, including inflammation and toxic bacterial metabolites, rather than understanding the cancer-causing potential of any specific microbe. Helicobacter pylori, alongside other bacteria such as Chlamydia trachomatis, Propionibacterium acnes, and Fusobacterium nucleatum have been studied for their associations with cancer (Table 1.1). Though many hypotheses have been proposed based on findings from in vivo research, the function of persistent inflammation in bacterial oncogenesis has been most widely researched. In addition, specific bacterial virulence factors aiding infection establishment have been examined for their role in oncogenesis. The current review focuses on the various pathways examined in bacterial oncogenesis, taking into account the most widely researched bacterial infection models.
Helicobacter pylori (previously Campylobacter pylori)
In Gastric Adenocarcinoma
Helicobacter pylori (H. pylori) is the first bacterium to be termed carcinogenic by the IARC in 1994 (19). Its infection and relevance with respect to gastric adenocarcinoma are the best studied amongst all cancer causing bacteria. It is an excellent example of cancer caused by bacteria via the inflammatory mechanism. It was estimated that nearly one-fifth of all cancers worldwide are due to infections, and H. pylori could be implicated in more than 50% of the gastric cancer cases reported (38). The gram-negative bacterium H. pylori colonize the stomach and, despite being present on normal stomach epithelial cells, can result in an infection accompanied with inflammation, which, once established, can last for decades (39, 40). This substantiates the role of H. pylori as a potent risk factor that can increase the probability of cancer incidence.
An extensive study conducted by the EUROGAST study group found a statistically significant relationship between the incidence of gastric cancer, death rate, and the presence of anti-H. pylori antibodies in the serum in 13 countries which is also supported by studies from other researchers (41, 42). In several supporting studies, elevated serum IgG levels were found against H. pylori suggesting an infection even without isolation of the causative organism (43, 44). In up to 5% of patients per year, persistent inflammation of the superficial portion of the gastric mucosa is documented to evolve into chronic atrophic gastritis characterized by an advanced cancerous lesion (45, 46). The cancer risk rises 9-fold with substantial atrophy (47). Despite the absence of H. pylori in areas with atrophic gastritis, it has often been identified in non-atrophic regions of the same stomach (48).
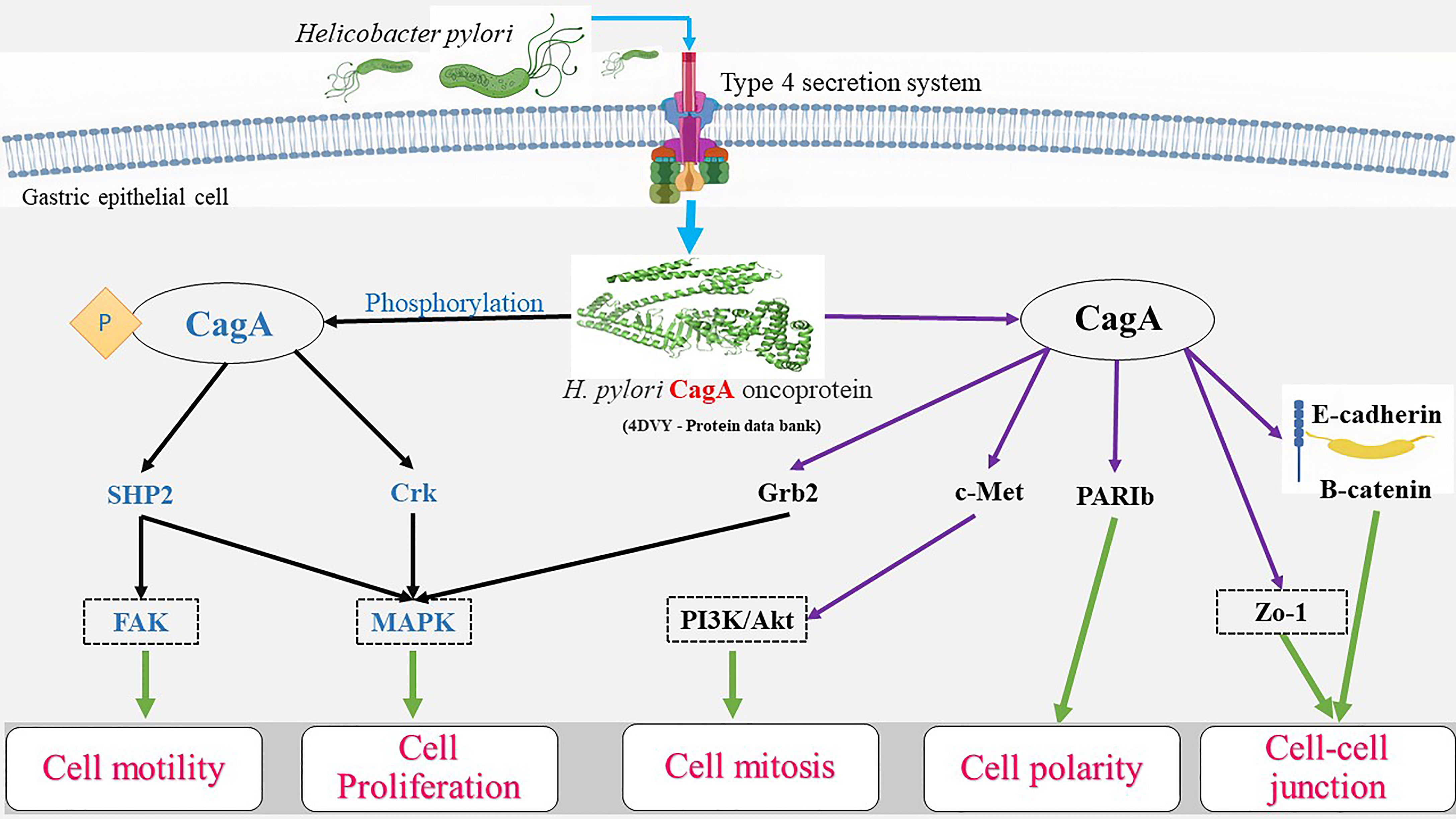
The probability of malignancy is relatively high with exposure to H. pylori infection, as persistent inflammation induces superficial gastritis (19).Oncogenesis promoted by the gram-negative, microaerophilic, spiral bacterium includes several factors comprising cytotoxin-associated gene A (CagA), vacuolating cytotoxin A (VacA), and ROS interactions. Promoters, such as the CagA, VacA, and CagY genes, lead to a higher proliferation of cells or affect gene expression and cell differentiation (49). H. pylori is believed to reside in the host for prolonged periods worsening inflammation translating into an increased chance of errors during DNA replication in proportion to cell proliferation, resulting in a cycle of damage, repair, proliferation, and eventually cancer.
Oxidative stress: Greater damage due to oxidative stress is linked to H. pylori infection in gastric cells (50). The consequences of oxidative stress upon gastric cells are documented via changes observed in the lipid and protein expressions and biomolecular damage (51, 52). Upon infection with H. pylori, the epithelial cells of the stomach release ROS, nitric oxide, and chemokines that triggered the production of proinflammatory cytokines, such as interleukin-8 (IL-8), which have been identified as effectors of the inflammatory role in the induction and promotion of the oncogenic process(es) (53, 54). There is also release of interleukin-6 (IL-6), an anti-apoptotic factor, which plays a crucial role in triggering critical signaling pathways, including the activation of JAK, STAT3, PI3K, MAPK, and AMPK ultimately leading to inflammation (55). H. pylori induce the Signal Transducer and Activator of Transcription 3 (STAT3) protein activation via ROS generation leading to increased expression of the interleukins -6 (IL-6) and -11 (IL-11) (56, 57). The induction of Type 1 T helper (Th-1) cellular response results in the activation of cytokines including gamma interferons (IFN-γ), and interleukin-1 (IL-1) among others, thus resulting in inflammation, loss of healthy host cells, and compensatory cell proliferation (58, 59). With a rising rate of proliferation, errors during replication and accumulation of mutations result from oxygen-free radical accumulation. 8-hydroxy-2’-deoxyguanine (8HdG), an end product of oxidative damage by ROS, leads to transversion of guanine to thiamine in the DNA (59–61). Some reports suggest that the mucosal surface of patients with infection had a higher percentage of 8HdG than those lacking the infection. The levels of this marker are found to be proportional to the infection, as the infection subsides, the 8HdG levels also return to nil, speculating the mutagenic nature of both the bacterium and its metabolite (62). Thence, the inflammation theory of H. pylori-induced-oncogenesis may be assumed true. Nonetheless, the other theories, including the activation of the mitogenic transduction pathway, have not been ruled out (63).
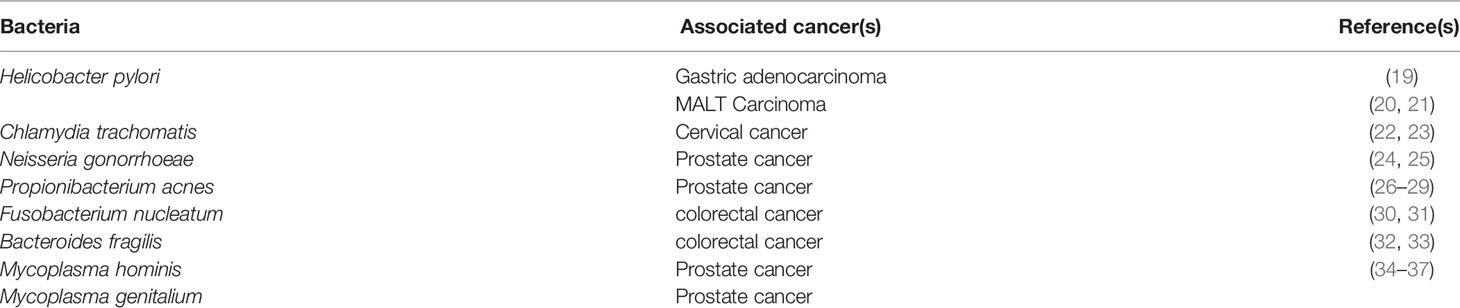
CAG PAI: Pathogenicity Island(s) (PAI), a part of the genome carrying virulence genes in pathogenic bacteria, are often absent in non-pathogenic isolates of the same bacteria. The PAI, first described in 1996, was reportedly obtained by the bacteria via horizontal transfer, and based on its presence, strains have been categorized into either the very virulent type 1 or the mildly virulent type 2 strains (64). The function of cytotoxin-associated gene pathogenicity island (CAG PAI) of H. pylori has been identified as one of the virulence factors in gastric cancer (65). The PAI is responsible for a type 4 secretion system that enables the insertion of CagA protein into the host cells (59, 66). CagA is made of 5 different amino acids, Glu-Pro-Ile-Tyr-Ala, together named EPIYA, occurring either as the EPIYA-D motifs or the multiple EPIYA-C phosphorylation sites, which are associated risk factors for gastric cancer or peptic ulcer disease (PUD) (67).
Once the CagA protein is transferred to the epithelial cells, interaction with host cell proteins, in both phosphorylation-dependent and independent manner, leads to the activation of various signaling pathways involved in cell elongation and scattering, eventually causing responses of the carcinogenic nature (68). Once internalized, CagA can also produce an inflammatory response leading to the release of cytokines such as the IL-8 and -6 via activation of the nuclear factor kappa B (NF-κB) (69, 70). Inside the cell, phosphorylation occurs by means of Src and Abl kinases (71), and the phosphorylated CagA activates Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2), further activating the extracellular signal-regulated kinase (ERK) pathway increasing its activation time with phosphatidylinositol 3-kinase (PI3K), leading to the reorganization of actin, and cellular elongation (72). The phosphorylated CagA interacts with the Src homology 2 (SH2) domains of SHP2, C-terminal Src kinase (CSK), growth factor receptor-bound protein 2 (Grb2), and CT10 regulator of kinase (CRK) proteins (73) containing protein tyrosine phosphatases (PTPs). Thereby causing activation of many tumorigenic signaling cascades by CagA, such as the Ras/Raf/Mitogen-activated protein kinase/ERK kinase (MEK)/extracellular-signal-regulated kinase (RAS/ERK), canonical Wnt pathway (WNT/β-catenin), Janus kinases/signal transducer, and activator of transcription (JAK/STAT), phosphatidylinositol 3-kinase/RAC-alpha serine/threonine-protein kinase (PI3K/AKT), and others along with the inhibition of tumor suppressors such as the tumor protein p53 and ultimately lead to a mitogenic response which is achieved by activation of the PI3K/AKT and ERK/mouse double minute 2 homolog (MDM2) pathways (74–76).
The CagA+ strain infection has been known to cause strong inflammation and damage to the gastric tissues (77, 78). It is noteworthy that these oncogenes were activated only by the positive strains of H. pylori (79). The proto-oncogene tyrosine-protein kinase activity is also inhibited by CagA, resulting in the dephosphorylation of tyrosine (80). Similar results were observed due to a defeat in the induction of cell retraction notwithstanding, the signaling molecules responsible have not been identified yet (81). The kinases are phosphorylated in the nucleus, thus triggering the transcription of E-26-like protein-1 (Elk-1) (82), which binds to the serum response factor and subsequently to the serum response elements and stimulates the oncogenic c-Fos and c-Jun upregulation (83, 84). Together, these genes express the activator protein-1 (AP-1) transcription factor, thereby promoting the expression of other late genes responsible for cell proliferation (85). The AP-1 transcription factor activates the transcription of cyclin D (86). In turn, augmented cyclin D activity results in the ultimate release of E2F transcription factors, which via cyclin E upregulation prompts the entry into the S-phase (87, 88).
CagA was found to trigger anti-apoptotic responses due to interaction with the p53 protein and thereby causing mutagenesis (89). A significant number of factors and pathways, including the kinases Akt and ERK, anti-apoptotic factors of the B-cell lymphoma family, including MCL-1, BCL-2, and BCL-Xl, were reportedly modulated by CagA (90–93). Furthermore, other proapoptotic factors are majorly involved in the downregulation of autophagy and increase of inflammation, such as Bcl-2-like protein 11 (BIM), BCL2 associated agonist of cell death (BAD), and the apoptosis regulatory SIVA1 are suppressed (91). Recently, the Siva1 protein was identified as a possible factor downregulated by CagA via the PI3K/Akt pathway to cause apoptosis inhibition alongside DNA damage (94). the apoptosis-stimulating protein of p53 2 (ASPP2), a critical CagA target and another tumor suppressor found in humans, aids in the survival of the CagA-positive H. pylori in the lumen. Notwithstanding, the molecular basis mediating disruption of gastric epithelial cell-polarity observed in the above event and subsequent oncogenesis is yet to be fully understood (95). Various mechanisms of CagA mediated gastric carcinogenesis have been summarized in Table 1.2.
In addition, a few other studies have observed the effects of non-phosphorylated CagA in host cells contributing to pathogenesis. Once inside the gastric cells, non-phosphorylated CagA interacts with E-cadherin leading to the disassociation of E-cadherin and β-catenin complex, amounting to the latter accumulation cytoplasm and nucleus (99). Zonula occludens-1 (ZO-1) and Junctional adhesion molecules (JAMs) interact with CagA and E-cadherin, resulting in junctional instability as well as β-catenin activation (100). Disruption of apical-junction complex (AJC) clubbed with a loss of cell polarity is achieved via translocation and activation of beta-catenin (100). It is known to target E-cadherin, tyrosine-protein kinase Met (c-Met), and kinase partitioning defective 1b (PAR1b) or microtubule affinity-regulating kinase 2 (MARK2), resulting in inflammation and mitogenesis (100, 101). The β-catenin and T-cell factor complexes formed trigger the expression of genes that encode cyclin D1 and cellular myelocytomatosis oncogene (c-Myc), leading to abnormal cell proliferation (102). Non-phosphorylated CagA also brings about alternations in cell motility and proliferation by binding to GRB/SOS/RAS and activation of Raf/MEK/Erk pathway, joining with ZO-1 and JAM-A tight junction proteins. The effects of phosphorylated as well as non-phosphorylated CagA in gastric neoplasm has been illustrated in Figure 1.
The number of CagA-positive H. pylori strains varies greatly among geographic regions. While almost all variants can be found in the East Asia, there are less than half prevalent in the west (103). The CagA-positive strains of H. pylori have been classified as the East Asian and the Western types based on the 3’ end region made of repeating sequences containing EPIYA phosphorylation site. Where, the former constituted EPIYA-A and EPIYA-B segments, and the latter contained EPIYA-C and EPIYA-D, respectively (104). EPIYA-D type segments were found to have more remarkable in vitro SHP-2 binding ability (104). In the transgenic mice model, the carcinogenic potential of CagA has been questioned concerning the positive and negative species of H. pylori, highlighting CagA as a potential oncoprotein (105). It is widely accepted that CagA-positive H. pylori are related to a greater risk of gastric cancer, however, the same outcome has not been seen in CagA-negative H. pylori (106). Notwithstanding, irrespective of the strain used, researchers failed to induce gastric cancer in the Mongolian gerbil model (107, 108).
VacA: Vacuolating cytotoxin (VacA) and those proteins linked with the outer membrane of H. pylori are involved in the process of vacuolation and ulcer formation (109). VacA, secreted by the type 5 secretion system in all isolates of H. pylori, is present in the mitochondria and affects its functions (110, 111). Initially formed as a 140kDa precursor, it matures to become an 88 kDa protein comprising p33 and p55 (112). The p55 domain is mainly responsible for the building of cell surface receptor proteins such as the tyrosine phosphatase (RPTP), epidermal growth factor (EGF), sphingomyelin, and fibronectin (113), while the p33 domain forms a channel of 6 subunits of VacA to facilitate chloride transport. This protein can separate the tight junction of gastric epithelial cells, thereby crossing the barrier (114). Once bound to the cell, VacA enters it by a mechanism independent of clathrin (115, 116). Many cell-surface components such as the RPTP-α (117), RPTP-β (118), various lipids (117), heparin sulphate (119), sphingomyelin (120), as well as Integrin beta chain-2 (integrin β2; CD18) on T cells (120) are targeted by VacA. Notwithstanding, the roles played by these factors in VacA uptake remain unidentified. Vacuolation of cells, disruption of apoptosis and lysosomal functions are some of the most important alterations caused by VacA cytotoxicity (121). Vacuole formation is achieved by means of in vitro endosomal compartment(s) disruption (122).
VacA activates akt via phosphatidylinositol 3-kinase dependent phosphorylation of glycogen synthase kinase – 3 beta (GSK3β) (123). Akt phosphorylation and activation are achieved via two protein kinases 3-Phosphoinositide-dependent kinase - 1 (PDK-1) and mammalian target of rapamycin complex 2 (mTORC2) (124). In VacA affected cells, inhibition of Rapamycin complex 1 (mTORC1) signaling positively regulates autophagy as well as affects the host cell metabolism and stress signaling (125). Cell death occurs via the Unc-51like autophagy activating kinase – 1 (ULK1) complex, using the low-density lipoprotein (LDL) receptors (125). Hence, Akt phosphorylation inhibits GSK3β and subsequent proliferation and survival (126, 127). GSK3β phosphorylates β-catenin in a cytoplasmic complex constituting auxin, adenomatous polyposis coli (APC) protein, and β-catenin in the absence of the ligand (128). The phosphorylated β-catenin is then ubiquitinated and destroyed by the proteasome (129). GSK3β remains inactivated in the presence of VacA, causing β-catenin accumulation in the cytoplasm (130). The β-catenin protein serves as a transcription factor coactivator, T cell factor, and lymphoid enhancer factor upon entering the nucleus to activate transcription of the β-catenin-dependent genes such as the cyclin D1 gene, CCND1, whose overexpression is linked to cancer (102). β-catenin signaling pathway is affected by VacA, presumably having an oncogenic role (131). The association of VacA and CagA in anti-apoptotic signaling may be one of the highly effective strategies of the bacterium to protect itself from the gastric niche and the human immune defense (112). In vivo studies involving Mongolian gerbil, models have observed apoptotic loss of pit cells by H. pylori and decreased apoptosis leading to hyperplasia and colonization mediated by CagA via MAP kinase protein (132). H. pylori can cause genomic instability in the gastric cells through epigenetic pathways (133). Previous in vitro studies have documented the induction of breakage in DNA strands by irrespective of CagA Presence in strains (134). Other studies have found that CAG PAI resultant products may have a crucial role in the accumulation of DNA strand breaks in the infected gastric cells (135). It was also hypothesized that host-bacterium interaction was responsible for DNA double-strand breaks, postulating that treatment and elimination of H. pylori may show reduced gastric cancer risk (136). The overall effect of virulence factors and inflammation on gastric epithelial cells is summarized in Figure 2.
In Gastric Mucosa-Associated Lymphoid Tissue (MALT) Carcinoma
The only human malignancies in which the etiological function of a specific bacterial infection has been broadly established are gastric adenocarcinoma and MALT lymphoma. As many as half of all MALT lymphoma cases are reportedly occurring in the stomach, and H. pylori were found to be prevalent in 90% of gastric MALT lymphoma tissues (20, 21). Given the morphological similarities between the follicles amongst gastric MALT lymphoma tissue(s) and those affected by H. pylori, a high incidence and direct relation were suspected between H. pylori and MALT lymphoma (21). A connection can thus be established between H. pylori and gastric MALT lymphoma (137).
As a result of repeated stimulation with H. pylori antigens, chronic infections lead to the formation of MALT in the stomach mucosa as they stimulate specific T-cells, marking the early stages of oncogenesis (138, 139). The assistance of tumor-infiltrating T-cells is essential for the development of MALT lymphoma in vitro (90). Tumor-infiltrating T-cells promote the proliferation of B cells when stimulated by H. pylori (90). Cytokine and CD-40 mediated cell signaling have been observed mandatory for lymphoma formation (90). In MALT lymphoma cells, the B-cell attracting chemokine 1 (BCA-1) and its receptor C-X-C motif chemokine receptor 5 (CXCR5) are augmented, which regulate B-cells and promote the production of the inflammation-causing interleukin-8 (IL-8) (140).
Translocated by the type 4 secretion system, CagA along with the SHP-2 stimulates B-cells via p38 kinase (105), bringing about B-cell proliferation via the control of endoplasmic reticulum kinases 1 and 2 (ERK 1 and 2) (105, 141). Due to phosphorylation and lowering of SHP-2, CagA promotes H. pylori-associated gastric neoplasm formation (142) in murine models. In addition, apoptosis of B-cells can be blocked due to the accumulation of p43 in the presence of CagA (141, 143). Alternations in the p53 suppressor gene influence the grade of lymphoma formed (144). Interference with antigen presentation of B-cells is brought about by VacA, affecting cell proliferation (145, 146). Molecular studies have shown changes in methylation of DNA at cysteine and guanine nucleotides which can subdue the tumor suppressor genes. Another contributing factor is the CpG island methylator phenotype found in 60% of MALT lymphomas due to H. pylori infection (147). Notwithstanding, growing chromosomal aberrations may enable MALT lymphomas to exist without an H. pylori infection (148).
Epidemiological studies reveal that the host factors such as the amount of salt intake also surged the degree of infection and frequency of cancer (149, 150). Experimental studies suggest synergistic effects of salt on lesions (151), wherein increased salt consumption leads to an augmented expression of CagA (152). These findings shed light on how H. pylori avoid inducing excessive cellular damage while maintaining long-term colonization. As observed, activation of cell proliferating signaling pathways was initiated by CagA and VacA. Nonetheless, further studies may be required to study and understand the effects of the inactivation of the above pathways in designing new therapeutic targets.
Treatment or Elimination of H. pylori
Antibiotics and proton pump inhibitors (PPIs) are commonly used in the event of H. pylori infection. Clarithromycin triple therapy with clarithromycin and amoxicillin, bismuth quadruple therapy with bismuth and tetracycline, and concomitant therapy with clarithromycin and amoxicillin in combination with a PPI and metronidazole constitute the recommended antibiotic regimens (153).
Meta-analyses of trials have resulted in reduced incidences of gastric cancer with the eradication of the bacteria (154, 155). In another similar trial, follow-up led to the reduced incidence of cancerous lesions after the eradication (156). Yet another trial comprising follow-up after eight years observed a 50% reversal of atrophic gastritis in the bacteria-eradicated patients (157). notwithstanding, the examination of available data indicates that no trials or studies have demonstrated a significant rise or decline in the incidence of cancer post-eradication after the infection is past the atrophic gastritis stage. However, eradication could undoubtedly prevent the development of precancerous lesions (158). This may be indicative of one clinical benefit that eradication at earlier stages of infection could be helpful. The prevailing notion is that eradicating infection before the dysplasia stage could be of benefit (159). Most meta-analyses have noted neither enough evidence nor data to claim any association between H. pylori and MALT carcinoma (155, 160). Similar observations have been made in individual studies, where MALT lymphomas were unresponsive to H. pylori eradication therapy (161). Once H. pylori infection was removed, 83% of the lymphomas were seen to be regressed (137). However, other studies have reported that the eradication therapy could be effective in long-term outcomes for H. pylori-induced-MALT lymphomas regardless of the infection stage (162, 163). Indicating the need for a more solid substantiation to link the bacteria’s eradication and cancer regression (164).
Chlamydia trachomatis
Chlamydia trachomatis, an intracellular, obligate, Gram-negative bacterium, is known to cause Chlamydia. While many discovered serovars of this species are known to infect different organ systems, the Serovars A-C instigate infection in the eyes, and serovars D through H colonize the genital tract. As of 2018, Cervical cancer is responsible for nearly 8% of cancer-related mortality, ranking 4th for both incidence and mortality (165). Many in vitro and in vivo studies reported an association between cervical neoplasm and chlamydial infection (22, 23). Implicated in a heightened risk of uncontrolled cervical cell growth, the presence of the chlamydial infection has also been associated with increased cancer incidence (166–169). Notwithstanding, the association remains controversial as various other reports indicated no alliance between the infection and the development of cancer (170, 171).
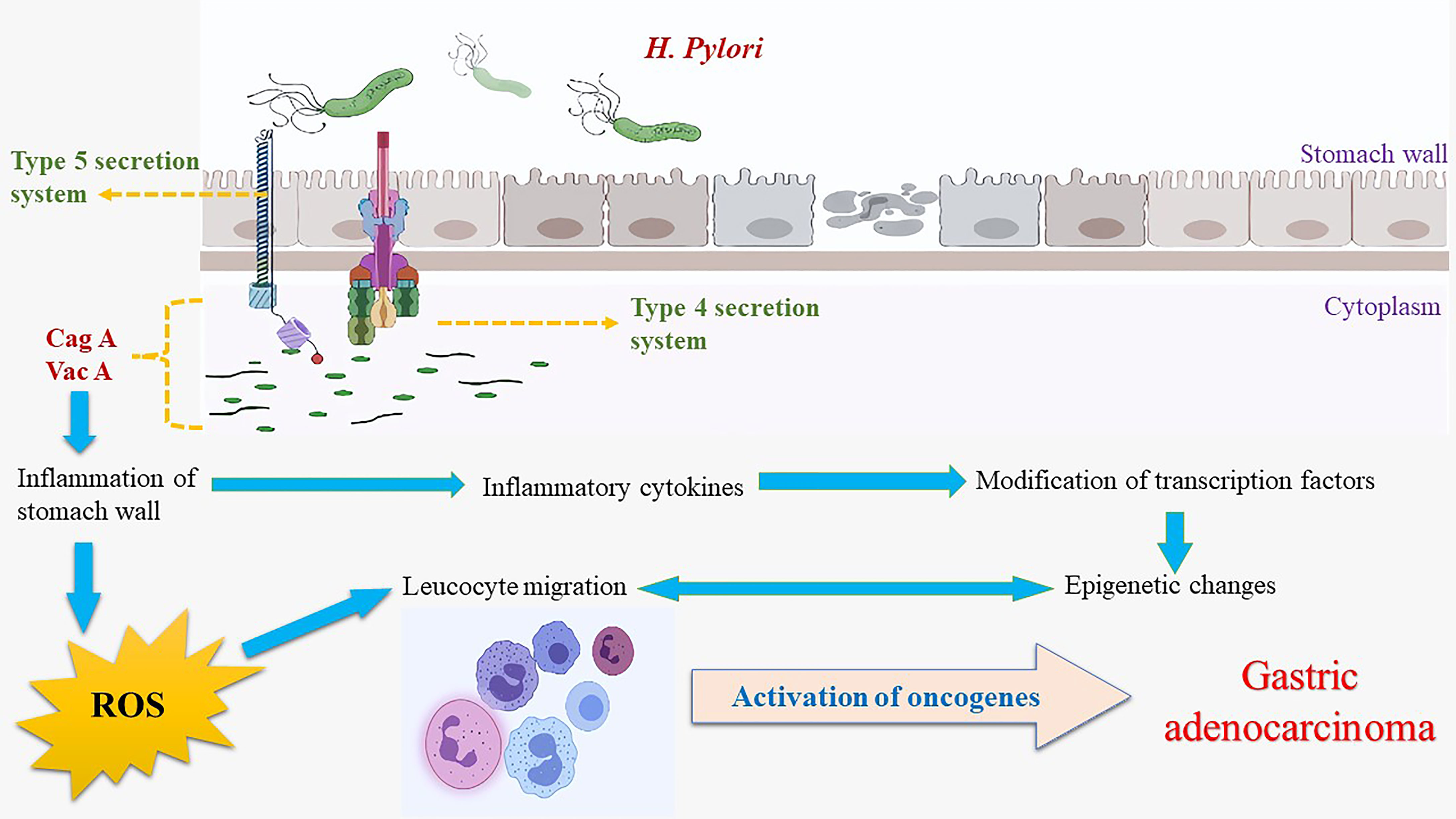
While the process of oncogenesis is yet unclear, it is hypothesized to arise from persistent inflammation and metaplasia (172), particularly through the squamous cell metaplasia. C. trachomatis has been known to cause cancer which may develop over years or decades (172, 173). As an intracellular pathogen, these bacteria can only multiply inside a host cell by dodging the immune system via prevention of phagolysosome formation (174), thereby affecting major histocompatibility complex (MHC) induced antigen expression (175) and its anti-apoptotic properties (176). C. trachomatis infection has been documented to modify the transcription of genes responsible for cell differentiation, cell death, and transcription factor(s) expression (177). Chronic inflammatory response(s), modified metabolite production, the amplified activity of cytokines, and decreased cell-mediated immunity contribute to mutagenesis by facilitating uncontrolled, multipolar mitosis and injury to DNA repair systems amounting to accumulation of aberrant DNA and thereby cancer (178, 179). In vitro studies evaluating the effects of C. trachomatis on apoptosis, inhibition observed unaffected DNA synthesis in the infected cells, which could undergo regular mitosis at any point of infection, linking it to a heightened risk of malignancy (180).
The apoptosis inhibition caused by C. trachomatis infection reasons the occurrence of neoplasm (181). Another mechanism through which apoptosis inhibition occurs is via mitochondrial cytochrome C inhibition (182). However, three different pathways of achieving this have been theorized. Firstly, by the inhibition of upstream activities controlling mitochondrial function via production of anti-apoptotic factors (183, 184). Secondly, Bcl-2 or Bcl-2-like molecule expression may prevent the activation of caspase and cytochrome c production (181) although, the expression of Bcl-2 does not guarantee blockage of apoptosis (185). Lastly, pertaining speculations indicate the involvement of other anti-apoptotic factors which are yet to be identified or understood (181).
Tyrosine phosphorylation of host cell proteins involved in signal transduction pathways is upregulated during C. trachomatis infection (177, 186–188). In addition, carcinogenic components of the Ras-Raf-MEK-ERK pathway are observed to be activated by the bacterium along with the ROS production for survival (189–191). The p62 knockdown was found not to affect host cells or autophagy during early infection, notwithstanding, in the later stages of infection, autophagy was affected by p61 silencing as seen in vitro (192). Thereby, it may be deduced that p62 has a significant role in bacterium-induced autophagy, providing the necessary supportive data and theoretical basis for further study into bacterial pathogenesis. The plasmid-encoded protein Pgp3 inhibits apoptosis with PI3K/AKT signaling pathway activation, MDM2 (murine double minute 2) phosphorylation, and nuclear entry, as well as p53 degradation (193). In HeLa cells, Pgp3-induced inhibition of apoptosis was hindered, suggesting that the PI3K/AKT pathway had a critical role MDM2-p53 axis in Pgp3 anti-apoptotic activity. Nonetheless, the precise molecular targets and pathways are due to be further identified (193).
The pORF5 plasmid protein plays a crucial role in mitochondrial autophagy and apoptosis by upregulation of knockdown high mobility group box 1 (HMGB1) which may be necessary to C. trachomatis in modulating mitophagy whose specific upstream and downstream signaling pathways remain unknown, hence establishing growth (194). Further, 3-phosphoinositide-dependent protein kinase one signaling is evoked by the infection leading to the stabilization and phosphorylation of MYC (195). MYC- PDPK1 signaling activates the hexokinase of host II (HKII), which is moved into the mitochondria. It was found that the prevention of HKII interaction with mitochondria with the use of exogenous peptides triggered the apoptosis of infected cells in a manner similar to inhibition of either PDPK1 or MYC, resulting in disruption of intracellular development of the bacteria (195). The target of the MYC-PDPK1-HKII-axis could be considered a novel scheme in overcoming therapeutic resistance to the infection (195).
Centrosomes and centrosome segregation defects were produced in excess during C. trachomatis infection as a result of multipolar cell division, promoting genetic instability (178). Various in vitro studies have documented that chlamydial infection led to incremented multinucleation of host cells, directly linked to neoplastic transformation (196). Defects in the mitotic spindle pole were due to heightened supernumerary centrosomes, amounting to apoptosis activation resistance in the cell division cycle and subsequently leading to oncogenesis (178, 180, 181, 197). Furthermore, centrosome amplification and segregation defects in the chromosomes were suggested to promote instability (178). Trigger of supernumerary centrosome production and chromosome segregation defects, multipolar mitosis, chromosome instability promotion, and multinucleation lead to the malignant transformation and subsequent tumor development (178, 180, 198).
Chlamydial Heat Shock Protein
Heat shock protein-60 (HSP60), a protein-folding protein, is found in the cytoplasm of cells (199). The HSP-60, similar to GroEL of Escherichia coli, can induce inflammation. It was proposed that the C. trachomatis HSP60 may serve as a risk factor for oncogenesis by the mediation of apoptosis (199). The host cells affected by the chlamydial HSP60 are highly susceptible to oncogene expression for survival, continued proliferation, and eventually malignancy (199). Contradicting this theory, Capello, 1990 (200) proposed the presence of anti-chlamydial HSP60 antibodies providing immunity against cancer. Reports indicate that copious amounts of HSP60 are produced by C. trachomatis during infectious stages (201). Some tumors were found to present HSP60 on their surface, bringing about antibodies towards their epitopes in an attempt to induce an anti-tumor response (202). This surplus of chlamydial HSP60 seen in the cytoplasm and the host cell membrane during a long-standing infection promotes activation of immune cells against the protein, followed by endocytosis (203). These endocytosed proteins bind to toll-like receptors (TLRs), resulting in the activation of signaling networks responsible for the proliferation of host cells (204, 205). Protein-mediated anti-apoptotic activity via the formation of a complex with Bax and Bak proteins to cut the outer membrane of mitochondria has been documented (206).
A higher incidence of cervical cancer was directly linked to the increased anti-chlamydial heat shock protein antibodies (202). Some studies concluded that the HSP, with anti-apoptotic properties, was blamed for chronic inflammation (207–209). Airenne 2002 and Carratelli 2000 (23, 210) observed that the heat-labile component C. pneumoniae is released during infection validates HSP60 as a risk for cancer. The risk of infection increases with time, right from the moment of serum sampling to cancer diagnosis, similar to the serological studies of H. pylori in gastric cancer (209). It has also been noted that the different serotypes of C. trachomatis show variable risks, and the serotypes B, D, E, G, I, and J have been linked to an increased risk of squamous cell cancer (211).
Emphasizing the need to consider and identify possible cofactors responsible for enhancing cervical carcinogenesis (212). While HPV infections are prominently linked to cervical cancer, reports suggest that only a fraction of these infections are responsible for oncogenesis (213). Much evidence also stipulates that the risk of HPV acquisition and persistence is raised with C. trachomatis infection (214). C. trachomatis infection history and HPV have been linked in two recent studies, thus confirming the hypothesis of C. trachomatis being a cofactor (214). Furthermore, C. trachomatis may have a suggested role in aiding HPV in the carcinogenesis via MMP-9/RECK imbalance during cervical inflammation as a part of the infection (215).
Recent meta-analyses have evaluated the use of azithromycin vs. doxycycline and found doxycycline to be more effective in treating C. trachomatis infection (216). Notwithstanding, the Centre for Disease Control (CDC) recommended treatment regimen for chlamydial infection includes doxycycline, azithromycin, or levofloxacin (217). Therefore, finding more screening techniques and treatment options is deemed necessary for those affected with C. trachomatis. The effects of C. trachomatis on cervical cells are summarized in Figure 3.
Fusobacterium nucleatum
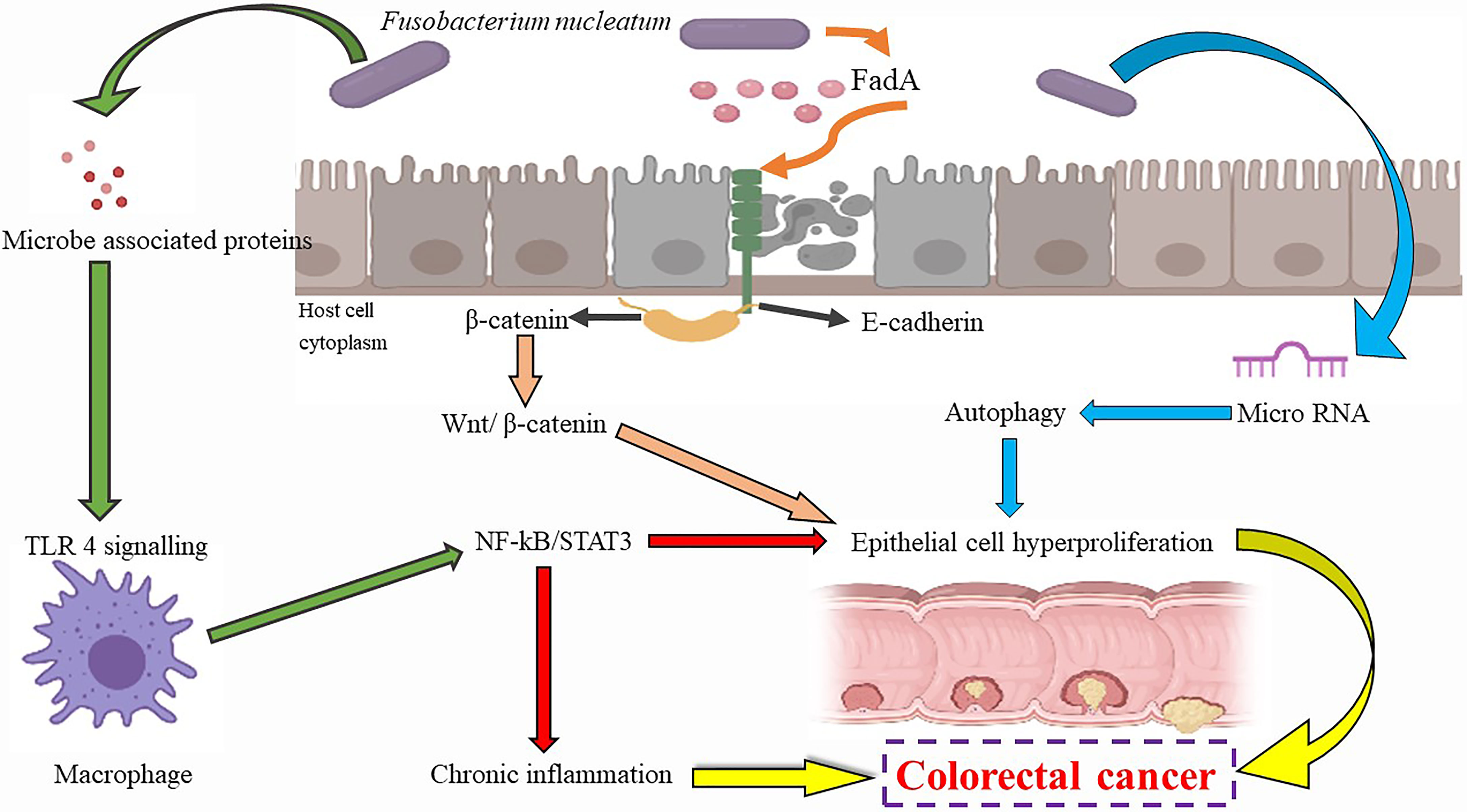
Fusobacterium nucleatum is a gram-negative, non-sporing bacterium capable of forming biofilms, commonly known for causing teeth infection (218). Speculations prevail that it brings about inflammation and invasive infections via hematogenous dissemination from the oral cavity to the colon (219–221). Many researchers have hypothesized an association between F. nucleatum and colorectal carcinogenesis (CRC) (30, 31) and considered the bacterium a risk factor for cancer progression. Recent meta-analyses and independent studies have found significantly raised levels of F. nucleatum during CRC incidents (222–231). Several other studies have also speculated that F. nucleatum may synergistically promote CRC with other bacteria such as the Streptococcus spp. and Campylobacter spp (232, 233). In an International ColoCare Study, it was observed in non-treated patients that varying levels of the bacterium were found at the tumor sites, indicating its use as a possible prognostic and diagnostic marker in the management of CRC (234).
The bacterium also aids neoplastic transformation via obstruction of anti-tumorigenic immunity by recruiting lymphocytes that infiltrate the tumor as well as activating immune checkpoints such as the T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) and Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), which aid in the inhibition of apoptosis (235–238). Generation of optimum microenvironment and activation of β-catenin signaling are some mechanisms by which F. nucleatum is involved in cancer progression (239–241). Recruitment of pro-inflammatory immune cells occurs due to the ROS-rich microenvironment (242, 243). Inflammation is worsened by the NKp46 receptor of natural killer cells in the presence of F. nucleatum, which prompts the release of TNF-α (244). Generation of the proinflammatory neoplastic microenvironment, higher rate of cell proliferation via Wnt/β-catenin signal activation, and signaling of NF-kB by TLR4 (31, 227, 240, 245) are some of the alternate mechanisms proposed for oncogenesis caused by F. nucleatum. Several in vitro studies have associated a higher prevalence of the bacteria with activation of oncogenic molecular cascades, including the instability of microsatellites, genetic mutations of BRAF, CHD7, CHD8, and TP53 CpG island methylator phenotype (246, 247).
The lipopolysaccharides, FadA, and Fap23 molecules present on the bacterium’s surface have been found to instigate oncogenesis (231, 248). Stimulation of malignant cell growth was found occurring due to β-catenin signal induction and tumorigenic gene expression via the virulence factor FadA (31, 239). The RadD adhesin, an arginine-instable adhesin, was reported to aid in the bacterial attachment and invasion into host cells, apart from aiding biofilm formation (31, 249). FadA, a virulence protein, monitors the bacterial entrance into host cells by activating the inflammatory and carcinogenic signals to induce growth in cells (250, 251). 2 forms of FadA, the pre-FadA and mFadA have been identified, which as the pre-FadA-mFadA complex are essential for the function as mentioned above (250, 252). FadA affects E-cadherin and β-cadherin, stimulating the T-cell factors and ultimately resulting in the expression of oncogenes, inflammation, and proliferation (31, 253).
Loss of E-cadherin alteration of the Wnt signaling pathway is an essential process in mesenchymal transition (254). In the Wnt pathway, β-catenin is responsible for the downregulation of E-cadherin, leading to the mesenchymal transition (255). Usually existing as a complex at the epithelial surface, β-catenin and E-cadherin are separated and migrated to the nucleus (227), which results in the alternation and deregulation of the Wnt signaling pathway, leading to tumor formation. Furthermore, the levels of the FadA gene in colorectal tissues of infected patients have been observed to be elevated and associated with inflammatory genes (256), hence substantiating the claims that virulence factors of F. nucleatum have a possible carcinogenic effect. In an in vivo study, the surface galactose-binding lectin, Fap2, was found to mediate F. nucleatum recruitment to the CRC cells (251). Furthermore, a polysaccharide D-galactose- β-N-acetyl-D-galactosamine (Gal-GalNAc) has been found in CRC tissues, which bind to FAP2, leading to the enrichment of the bacterium (249). A new miRNA-mediated pathway has been hypothesized by which F. nucleatum can affect the host cells and cancer (257). Researchers observed the enlarged tumor rate and decreased survival rates when APC/- mice were fed with F. nucleatum. In addition, the co-culture of F. nucleatum in the CRC cell lines led to increased cell proliferation in vitro and in vivo (257). With the theory of miRNA deregulation in colorectal cancer, many researchers studied miRNA expression in the exposed cell lines (245, 258). The engagement of fusobacterial lipopolysaccharide by toll-like receptors led to the induction of miR21, leading to the activation of RAS-MAPK signaling through the miR21 target RasGTPase enzyme (245, 258).
The putative mechanism of F. nucleatum and its effects are summarized in Figure 4. Markers of inflammation such as IL-8 and IL-6 TNF-α have been elevated in case of infection (259). The adhesion of cells, autophagic flux, and anti-tumor activities of immune cells are affected by F. nucleatum, apart from decreasing the activity of T cells in adaptive immunity by affecting the G1 phase (260). While F. nucleatum is noted to promote cell proliferation by modulating E-cadherin and β-catenin pathways, increasing miRNA-21 expression. On the contrary, in the human gingival fibroblasts, F. nucleatum prevents cell proliferation and induces cell death by activating the AKT and NF-kB signaling pathways (260). This bacterium has also been found to reduce chemotherapeutic effects in CRC due to activation of TLR4/NF-κB pathways (261, 262). All these findings together suggest an important role of F. nucleatum in cancer initiation. The presence of β-lactamase in a few strains may make these organisms resistant to penicillin, indicating that the anaerobic antibiotics such as metronidazole or clindamycin may be the drug of choice in the treatment of this infection (263, 264).
Bacteroides fragilis
Bacteroides fragilis are non-spore-forming, Gram-negative, anaerobic bacteria constituting two different classes, i.e., non-toxigenic B. fragilis (NTBF) and enterotoxigenic B. fragilis (ETBF), based on their ability to produce biofilm and the presence of the gene for zinc-dependent metalloprotease, B. fragilis toxin (BFT) (32, 265, 266). The infliction of tight junctions and increase in intestinal permeability caused by BFT may be necessary for inflammation of the intestine and, further, in neoplastic transformation (32, 33). In vitro tests on HT29/C1 cells with BFT treatment revealed a decline in membrane-associated E-cadherin initiated the nuclear localization of ß-catenin, which further induced translation of c-myc and continuous cell proliferation (267). This ability of BFT to affect the epithelial cells has led to many researchers concluding that the ETBF may contribute towards CRC (266–268). Long-term colonization by ETBF in the intestine results in chronic inflammation stimulation due to activation of STAT3, which leads to increased IL-17 production responsible for prolonged inflammation in the intestine (269). BFT modulates signaling pathways and is responsible for ROS production, leading to mutagenesis and cleavage of E-cadherin (266, 270). It can activate β-catenin signaling and induce IL-8 production in epithelial cells (268). Additionally, being biofilm producers, ETBF degrades E-cadherin in cells, causes the production of IL-6, and activates STAT3 pathways, enhancing cell proliferation. Indicating that biofilms are associated with neoplastic development in the colon (271). While the ETBF through biofilm can induce cancer, the NTBF cannot harm the intestinal tract (272). The ETBF promoting colorectal carcinogenesis, upregulation of JMJD2B, a histone demethylase, via TLR4-NFAT5-dependent pathway is caused by the ETBF promoting colorectal carcinogenesis (273).
Further, BFT has been documented to trigger the production of COX-2, which releases prostaglandin E2 (PGE2), which causes inflammation and controls cell proliferation via control of signaling pathways. Hence, COX-2 plays a vital role in colon carcinogenesis via angiogenesis promotion, stem cell formation, inhibition of apoptosis, increasing metastatic potential, and promotion of cell proliferation (266, 274–278). The serum COX-2 levels have also been used as a biomarker in CRC patients, indicating aggressive growth and higher mortality rates compared to normal individuals (279–281). Downregulation of miR-149-3p by ETBF was found to promote PHF5A-mediated RNA alternative splicing of KAT2A in CRC cells (282).
Via secretion of chemokine IL-17 along with other cell surface receptors, activated through induction of NF-kB pathways, BFT establishes a pro-carcinogenic signaling relay in ETBF-associated carcinogenesis (283). Chemokine motif ligand 3 is a macrophage inflammatory protein with CCR5 as the receptor. CCR5 plays a vital role in invasion and metastasis via inflammatory factors and tumor-associated genes to regulate NF-kB (284). Some studies have found that BFT promotes and may be necessary for the proliferation of colorectal cancer due to the acceleration of CCl-3 molecular pathways (285).
Significant associations have been established between the presence of ETBF and colorectal cancer, however, additional research is required to determine other factors affecting their relationship. Due to the presence of β-lactamase, the ETBF is resistant to penicillin. Antibiotics such as cefoxitin and clindamycin have little susceptibility towards the bacterium, while piperacillin/tazobactam, meropenem, and metronidazole are known to be more effective (286).
Neisseria gonorrhoeae
Neisseria gonorrhoeae (N. gonorrhoeae), the causative organism of gonorrhea, is a gram-negative, facultative intracellular pathogen. A history of infection with N. gonorrhoeae has been suggested to be associated with a higher incidence of prostate cancer risk, as reported by a few meta-analyses (24, 25). In 2018, prostate cancer was the second most frequent form of cancer in men worldwide (287, 288). Gonorrheal infection is one of the most common causes of prostate cancer (289).
Although the exact molecular mechanisms in oncogenesis are unclear, chronic and repeated infections of this bacterium have been associated with prostate cancer (290). The duration of infection has been observed to be directly proportionate to a higher risk of cancer (291). Following the infection, a persistent inflammatory phase is induced in the prostate. The bacteria attach to the epithelial cell surface made possible by the type IV pili, the unique appendages on the bacterial surface (292, 293). Once attached, the host cell signaling events occur, eliciting induction of the anti-apoptotic activities (294, 295). A large number of cytokines and chemokines (interleukins 6 and 8) are secreted following the damage due to inflammatory cells that promote oncogenesis (296). Pathological examinations have revealed proliferative atrophy with inflammation which may be a precursor lesion to cancer (296, 297).
N. gonorrhoeae can evade the autophagy pathways of host cells during later stages of invasion, which allows a small population of the bacteria to thrive for a prolonged duration and show exocytosis. This may be due to the modulation of autophagy pathway repressor mTORC1 and inhibition of autophagosome maturation and lysosomal fusion (298). Amphiregulin, a protein capable of inhibiting the growth of cancerous cells, is downregulated by N. gonorrhoeae during the G1 phase of the cell cycle alongside cyclin degradation (299). It may be noted here that the levels of cyclins were previously measured to identify mechanistic pathways (299). Several other factors such as the ribonuclease L, hereditary prostate cancer 1, and toll-like receptor have also been studied for their role in the development of cancer (300). The double-stranded DNA breaks have also been observed due to the N. gonorrhoeae infection, along with the downregulation of p53 (301). In addition, the bacteria produce increased levels of restriction endonucleases during an active infection, ultimately resulting in mutagenesis, which is evidently observed in the form of longer and impaired M-phase of spindle assembly, formation of micronuclei, and lagging of chromosomes (302).
On the contrary, the evidence as mentioned above has been disputed by several studies that could not find a correlation between infection and cancer (303, 304). One of the main challenges associated with the treatment of N. gonorrhoeae infection is the development of antimicrobial resistance to standard drugs, including cephalosporins, macrolides, and tetracyclines (305) suggesting the requirement of intensive screening, prevention, or cure for men with gonorrhoea (306).
Cutibacterium acnes (Formerly Propionibacterium acnes)
Cutibacterium acnes (C. acnes) is a gram-positive anaerobic bacillus commonly found in the follicles of the skin. In men, a higher prevalence of pro-inflammatory C. acnes has been associated with prostate cancer (26–29). In studies conducted using in situ hybridization, clusters of C. acnes in 50% of patients with prostate cancer were documented (307). Nonetheless, as a common skin commensal, the presence of C. acnes has been regarded as contamination (308). On the contrary, sequence typing of the bacteria indicated them to be urogenital pathogens and not skin commensals (309). Reports suggest that some species of the bacterium have cytotoxic and hemolytic properties (310) and are also noted for the extensive immunomodulatory character (311), revealing factors that interfere with virulence and host tissue (312). The bacterium showcased a wide range of virulence factors, including enzymes such as lipases, proteases, and chemotactic factors for immune cells (313). Reports also indicate the promotion of innate immune cells, including macrophages which release cytokines such as tumor necrosis factor, Il-1, 6, 8, and 12 (27, 314, 315). By upregulating the vascular endothelial growth factor (VEGF), IL-17 release contributes to the activation of malignant cell proliferation and the production of new blood cells (316). Further, the disbalance in IL-17 and regulatory T-cells (Treg) cells in tumors could aggravate oncogenesis by inducing immunosuppression (315, 317).
High antibody titers against C. acnes were observed in men with benign prostatic hyperplasia, indicating the existence of infection and inflammation (318). While the infection affects cell proliferation leading to transformation (319), inflammation elicits oncogenesis by enhanced mutagenesis, cell replication, and angiogenesis (320). An increased Th1-type immune response is observed in the site of infection as a result of inflammation (320, 321) which harms the neoplastic process. However, in prostate cancer, proliferation-promoting Th2-type of response was seen (322). Increased nuclear factor-kappa B (NF-κB) activity in tumors, due to increased IkappaB kinase (IB kinase) activity (323), lead to increased expression of several genes known to be crucial for cancer development and progression (324). The levels of serum inflammatory cytokines 1,6, and 8 were found to rise alongside intensified IL-6 secretion (314, 325). IL-6 triggered the JAK signaling pathway, which in turn activates STAT3, whose repeated stimulation was seen to enhance cell proliferation and eventually cancer (326). A prolonged C. acnes infection triggered the production of reactive oxygen species (ROS) in cells as well as the influx of immune cells such as macrophages to the infection site along with inhibition of apoptosis (327). Oncogenesis was suggested to be favored by this combination (319).
FOXM1 (Forkhead box M1), a transcription factor linked with cell proliferation and involved in tumorigenesis achieved via promoting cell progression into S and M phases, was downregulated during the C. acnes infection (328). FOXM1 also facilitated the recombination and repair of double-stranded DNA during breaks, maintenance of stability by control of Aurora B kinase, Cyclin B1, and Centromere protein F. It is speculated that FOXM1 downregulation could lead to mutagenesis, notwithstanding more information is yet to be obtained in this regard (328). Other researchers have proposed the role of androgen levels in cancer development (329), but the signaling pathways remain unclear as of now (330). In murine models, the in vivo inoculation of C. acnes showed an inflammatory response eliciting cell damage (331, 332). In vivo evidence in the mice model found the prostate cancer generation and following chronic infection of up to 2 months, accompanied by a rise in proliferation and decreased androgen receptor levels (309, 333) and validating the inflammatory theory (332). C acnes infection may be treated with antibiotics including (β-lactams, quinolones, clindamycin, or rifampicin, despite increasing evidence of resistance towards these classes. Further, treatment could require surgical intervention to completely eradicate the bacteria (334).
“Mycoplasma” Species
Since the initial hints of an association between Mycoplasmas and oncogenesis, many studies have attempted to understand its oncogenic properties and its direct or indirect role in the onset of cancer or its progression (335–337). There is not much evidence to support current proposed mechanisms for Mycoplasma-induced oncogenesis. Improving diagnosis and tracking of Mycoplasma infections in patients is necessary to improve available data linking infection with pathological and clinical outcomes. Mycoplasma infections can be identified in a timely manner in patients by identifying antibodies induced in the host following infection (338–340).
Yet to be proven, Mycoplasma pneumoniae has been suspected as a probable cause of leukaemia since the mid-20th century. The meta-analyses of various cancer studies revealed the possible involvement of Mycoplasma species in oncogenic processes (341). Gliomas, Hodgkin’s, along with non-Hodgkin’s lymphoma, head and neck cancer, as well as cervical cancer have all been linked to Mycoplasma spp (338, 342–344). During the first examination of the etiology and role of venereal diseases in prostate oncogenesis in the 1950s, persistent inflammation and atrophy were suggested as probable processes resulting in the development of prostate cancer (345–348). Mycoplasmas are commonly present in the male urogenital tract, with the most prevalent species being Mycoplasma hominis and Mycoplasma genitalium (349–351). Current research has looked at the function of mycoplasmas in prostate cancer development. Due to the chronic infections with mycoplasma species in oncogenic cases, their involvement in oncogenesis has been strongly suggested (34–37).
Although altered inflammatory pathways, along with disruption of cell division and DNA repair, have been viewed as possible causes for cancer initiation, the exact mechanisms for cancer formation by mycoplasmas remain unclear (352, 353). Mycoplasmas cause long-term infection and develop immune escape mechanisms by modifying the inflammatory response (353). Infections with mycoplasmas lead to chronicity by a range of strategies that undermine the immune response, including degradation of immune effector molecules, cell invasion, molecular mimicry, antigen variation, and biofilm development, besides inflammatory regulation (354). As a means of immune evasion, the invasion of the host cell may result in the production of proteins that modulate critical cellular processes such as apoptosis and DNA repair. sAs a result of these modifications, the likelihood of aberrant cell development and oncogenicity increases (355). Some Mycoplasmas (notably M. fermentans, M. penetrans, and M. hyorhinis) have been reported to have oncogenic potential due to their ability to cause phenotypic changes in the cells in addition to inducing DNA aberrations (356). Long-term infections with Mycoplasmas are also connected to the instability of chromosomes and neoplastic modifications such as decreased cell adherence, spindle shape, and multilayer growth in cell cultures (357). Although a direct link between Mycoplasma infection and cancer formation remains still explored, the epidemiologic and observational data strongly imply the greater risk of cancer development with Mycoplasma spp. infections (355).
The NLRP3 inflammasome, a protein complex that controls the production of pro-inflammatory cytokines, including IL-1 and IL-18, is also involved in cancer development and spread (358). Many in vitro carcinogenic models to demonstrate the cancer-causing abilities of mycoplasmas, have been developed. Mutagenesis, disruption of the cell cycle checkpoints, apoptosis, and altered cell growth signals have been observed to be caused by mycoplasmas (357, 359). As a result, researchers have hypothesized that chronic mycoplasma infections can cause genetic instability and DNA aberrations as a result of mitogenic and apoptotic effects, eventually leading to tumor formation (357, 359, 360). M. fermentans and M. penetrans demonstrated their capability of oncogenesis promotion in murine CH3 cells showing mutagenic properties via the c-Ras and c-myc genes (352, 361). Such cells have been observed to accumulate mutagens and eventually mutate their DNAs due to altered methylation of DNA (362). Mycoplasma DnaK, a chaperone protein from the HSP-70 family, binds to and inhibits the catalytic activity of poly adenosine diphosphate-ribose polymerase (PARP)-1, a protein involved in the detection and repair of DNA damage. It also binds to USP10, an important p53 regulator, compromising p53 stability and anti-cancer potential. Mycoplasma-associated carcinogenic activity, mediated via the suppression of DNA repair and p53, may initiate some cancers, albeit not always in later stages (363). The same has been demonstrated in vivo in mouse models via lymphomagenesis (364). NF-κB activation for the inhibition of p53 is a proposed mechanism of oncogenesis, as demonstrated in the murine model (357). In the tissues of mammals, a lipoprotein present on the surface membrane, P37, is known to play a role in adhesion (351) via association with epidermal growth factor receptor 2 (365, 366). M. hyorhinis produces the p37 protein, which can promote cancer cell invasion in a dose-dependent manner and blocked by monoclonal antibodies specific for p37. Because p37 makes prostate cancer more aggressive, the molecular events it causes could be a therapeutic target (365). Mycoplasma infection functions as a p53-suppressing oncogene that collaborates with Ras in cell transformation, implying that mycoplasma’s carcinogenic and mutagenic effects are due to its inhibition of p53 tumor suppressor activity (367), which has been demonstrated in several Mycoplasma strains, ultimately resulting in downregulation of apoptosis of the damaged cells. Similar in vitro studies performed on human cell lines showed malignancy in prostrate cells, cervical cells, and bronchial cells (365, 368, 369), while other in vivo studies have also concluded the species of Mycoplasma to promote oncogenesis (367).
Investigation of small cell lung cancer and mycoplasma association revealed a considerably high mycoplasma presence in the patients compared to healthy groups, which led the researchers to speculate a multistage oncogenesis pathway directed by the bacteria (370), however, more research in this line is required for elucidating its exact role. It was hypothesized that the association between mycoplasmas and renal cell carcinoma mechanism was a persistent infection in the kidneys which stimulated oxidative stress (371).
Although in vitro and in vivo studies have suggested the involvement of Mycoplasmas in oncogenesis, more studies are needed to decipher its specific role in oncogenesis, diving into cellular and molecular mechanisms involved in the neoplastic transformation. Any role of mycoplasmas in causing tumors needs to be strengthened with more laboratory studies.
Prevention of Infection-Associated Cancers
Many integrated approaches may be needfully employed to prevent and control the disease based on different mechanisms linked to the origin of cancers via bacterial infections. The primary approach is to prevent the infection and eliminate the root causes of infection in healthy individuals, which may be achieved via effective vaccination strategies, preventative antibacterial therapy in endemic regions, and/or prevention of persistent infection (372). Secondary prevention may address patients in the pre-clinical or early stages of cancer and prevent tumor progression (373). For example, certain Asian countries have adapted country-wide screening programs to detect stomach cancer (374, 375). Finally, post-therapy monitoring of the patients for relapses is also an efficient method to ensure the quality of life (376, 377).
Conclusion
Bacterial etiology for cancer has been suspected for many years, yet not much proof has been obtained. Many organisms have been studied concerning their role in oncogenesis. This review lists the possible cancer-causing bacteria and the associated molecular processes through which oncogenesis may be achieved. While chronic inflammation and toxic bacterial neoplastic metabolites have remained the major concerns, further research into the molecular mechanisms of these infectious agents in the process of the cancer formation is of importance. Additionally, several factors pose a challenge for confirming the role of these bacteria in oncogenesis, including multiple etiology, variable periods between the onset of infection, and diagnosis of cancer.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This research work was partially supported by Chiang Mai University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
SP and SB acknowledge the infrastructure and support provided by the 1Department of Biotechnology and Bioinformatics, Faculty of Life Sciences, JSS Academy of Higher Education and Research, Mysuru, India. DS acknowledges Adichunchanagiri Institute of Medical Sciences, India for the support. AR thanks the Department of Medical Oncology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India for support. DD and PD would acknowledge the support provided by the Centre of Excellence in Molecular Biology and Regenerative Medicine, Department of Biochemistry, JSS Medical College, JSS Academy of Higher Education and Research, Mysuru, India. PR and SS acknowledge the support provided by Chiang Mai University, Chiang Mai 50200, Thailand.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–86. doi: 10.1002/ijc.29210
2. International Agency for Research on Cancer. (2021). Available at: https://gco.iarc.fr/today/data/factsheets/cancers/39-All-cancers-fact-sheet.pdf.
3. Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S, et al. Smoking and Drinking in Relation to Oral and Pharyngeal Cancer. Cancer Res (1988) 48(11):3282–7.
4. Mons U, Gredner T, Behrens G, Stock C, Brenner H. Cancers Due to Smoking and High Alcohol Consumption. Dtsch Arztebl Int (2018) 115(35–36):571–7. doi: 10.3238/arztebl.2018.0571
5. van Elsland D, Neefjes J. Bacterial Infections and Cancer. EMBO Rep (2018) 19(11):e46632. doi: 10.15252/embr.201846632
6. Al-Hilu SA, Al-Shujairi WH. Dual Role of Bacteria in Carcinoma: Stimulation and Inhibition. Callaway TR, Editor. Int J Microbiol (2020) 2020:4639761. doi: 10.1155/2020/4639761
7. Duong MT-Q, Qin Y, You S-H, Min J-J. Bacteria-Cancer Interactions: Bacteria-Based Cancer Therapy. Exp Mol Med (2019) 51(12):1–15. doi: 10.1038/s12276-019-0297-0
8. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global Burden of Cancer Attributable to Infections in 2018: A Worldwide Incidence Analysis. Lancet Glob Heal (2020) 8(2):e180–90. doi: 10.1016/S2214-109X(19)30488-7
9. Wolff J. The Science of Cancerous Disease From Earliest Time to the Present. Bethesda, MD: Science (1989).
10. Jacqueline C, Tasiemski A, Sorci G, Ujvari B, Maachi F, Missé D, et al. Infections and Cancer: The “Fifty Shades of Immunity” Hypothesis. BMC Cancer (2017) 17(1):257. doi: 10.1186/s12885-017-3234-4
11. Lindahl JF, Grace D. The Consequences of Human Actions on Risks for Infectious Diseases: A Review. Infect Ecol Epidemiol (2015) 5:30048. doi: 10.3402/iee.v5.30048
12. Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol (2007) 35(4):495–516. doi: 10.1080/01926230701320337
13. Balkwill F, Charles KA, Mantovani A. Smoldering and Polarized Inflammation in the Initiation and Promotion of Malignant Disease. Cancer Cell (2005) 7(3):211–7. doi: 10.1016/j.ccr.2005.02.013
14. Garrett WS. Cancer and the Microbiota. Science (2015) 348(6230):80–6. doi: 10.1126/science.aaa4972
15. Coussens LM, Werb Z. Inflammation and Cancer. Nature (2002) 420(6917):860–7. doi: 10.1038/nature01322
16. Piotrowski I, Kulcenty K, Suchorska W. Interplay Between Inflammation and Cancer. Rep Pract Oncol Radiother J Gt Cancer Cent Pozn Polish Soc Radiat Oncol (2020) 25(3):422–7. doi: 10.1016/j.rpor.2020.04.004
17. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
18. Flossmann E, Rothwell PM. Effect of Aspirin on Long-Term Risk of Colorectal Cancer: Consistent Evidence From Randomised and Observational Studies. Lancet (2007) 369(9573):1603–13. doi: 10.1016/S0140-6736(07)60747-8
19. Schistosomes, Liver Flukes and Helicobacter pylori. Available at: https://publications.iarc.fr/Book-And-Report-Series/Iarc-Monographs-On-The-Identification-Of-Carcinogenic-Hazards-To-Humans/Schistosomes-Liver-Flukes-And-Em-Helicobacter-Pylori-Em–1994.
20. Ferreri AJM, Ernberg I, Copie-Bergman C. Infectious Agents and Lymphoma Development: Molecular and Clinical Aspects. J Intern Med (2009) 265(4):421–38. doi: 10.1111/j.1365-2796.2009.02083.x
21. Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter Pylori-Associated Gastritis and Primary B-Cell Gastric Lymphoma. Lancet (Lond Engl) (1991) 338(8776):1175–6. doi: 10.1016/0140-6736(91)92035-Z
22. Arnheim Dahlström L, Andersson K, Luostarinen T, Thoresen S, Ögmundsdottír H, Tryggvadottír L, et al. Prospective Seroepidemiologic Study of Human Papillomavirus and Other Risk Factors In Cervical Cancer. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored by Am Soc Prev Oncol (2011) 20(12):2541–50. doi: 10.1158/1055-9965.EPI-11-0761
23. Koskela P, Anttila T, Bjørge T, Brunsvig A, Dillner J, Hakama M, et al. Chlamydia Trachomatis Infection as a Risk Factor for Invasive Cervical Cancer. Int J Cancer (2000) 85(1):35–9. doi: 10.1002/(SICI)1097-0215(20000101)85:1<35::AID-IJC6>3.0.CO;2-A
24. Taylor ML, Mainous AG 3rd, Wells BJ. Prostate Cancer and Sexually Transmitted Diseases: A Meta-Analysis. Fam Med (2005) 37(7):506–12.
25. Dennis LK, Dawson DV. Meta-Analysis of Measures of Sexual Activity and Prostate Cancer. Epidemiology (2002) 13(1):72–9. doi: 10.1097/00001648-200201000-00012
26. Ugge H, Udumyan R, Carlsson J, Andrén O, Montgomery S, Davidsson S, et al. Acne in Late Adolescence and Risk of Prostate Cancer. Int J Cancer (2018) 142(8):1580–5. doi: 10.1002/ijc.31192
27. Davidsson S, Mölling P, Rider JR, Unemo M, Karlsson MG, Carlsson J, et al. Frequency and Typing of Propionibacterium Acnes in Prostate Tissue Obtained From Men With and Without Prostate Cancer. Infect Agent Cancer (2016) 11(1):26. doi: 10.1186/s13027-016-0074-9
28. Kakegawa T, Bae Y, Ito T, Uchida K, Sekine M, Nakajima Y, et al. Frequency of Propionibacterium Acnes Infection in Prostate Glands With Negative Biopsy Results Is an Independent Risk Factor for Prostate Cancer in Patients With Increased Serum PSA Titers. PloS One (2017) 12(1):e0169984. doi: 10.1371/journal.pone.0169984
29. Dadashi M, Eslami G, Taghavi A, Goudarzi H, Hajikhani B, Goudarzi M, et al. Is Propionibacterium Acnes a Causative Agent in Benign Prostate Hyperplasia and Prostate Cancer? Arch Clin Infect Dis (2018) 13(3):e58947. doi: 10.5812/archcid.58947
30. Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium Nucleatum Associates With Stages of Colorectal Neoplasia Development, Colorectal Cancer and Disease Outcome. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol (2014) 33(8):1381–90. doi: 10.1007/s10096-014-2081-3
31. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
32. Snezhkina AV, Krasnov GS, Lipatova AV, Sadritdinova AF, Kardymon OL, Fedorova MS, et al. The Dysregulation of Polyamine Metabolism in Colorectal Cancer Is Associated With Overexpression of C-Myc and C/Ebpβ Rather Than Enterotoxigenic Bacteroides Fragilis Infection. Oxid Med Cell Longev (2016) 2016:2353560. doi: 10.1155/2016/2353560
33. Lv Y, Ye T, Wang H-P, Zhao J-Y, Chen W-J, Wang X, et al. Suppression of Colorectal Tumorigenesis by Recombinant Bacteroides Fragilis Enterotoxin-2 In Vivo. World J Gastroenterol (2017) 23(4):603–13. doi: 10.3748/wjg.v23.i4.603
34. Barykova YA, Logunov DY, Shmarov MM, Vinarov AZ, Fiev DN, Vinarova NA, et al. Association of Mycoplasma Hominis Infection With Prostate Cancer. Oncotarget (2011) 2(4):289–97. doi: 10.18632/oncotarget.256
35. Abdul-Wahab OMS, Al-Shyarba MH, Mardassi BBA, Sassi N, Al Fayi MSS, Otifi H, et al. Molecular Detection of Urogenital Mollicutes in Patients With Invasive Malignant Prostate Tumor. Infect Agent Cancer (2021) 16(1):6. doi: 10.1186/s13027-021-00344-9
36. Yow MA, Tabrizi SN, Severi G, Bolton DM, Pedersen J, Longano A, et al. Detection of Infectious Organisms in Archival Prostate Cancer Tissues. BMC Cancer (2014) 14(1):579. doi: 10.1186/1471-2407-14-579
37. Saadat S, Karami P, Jafari M, Kholoujini M, Rikhtegaran Tehrani Z, Mohammadi Y, et al. The Silent Presence of Mycoplasma Hominis in Patients With Prostate Cancer. Pathog Dis (2020) 78(7):ftaa037. doi: 10.1093/femspd/ftaa037
38. Parkin DM. The Global Health Burden of Infection-Associated Cancers in the Year 2002. Int J Cancer (2006) 118(12):3030–44. doi: 10.1002/ijc.21731
39. Di Napoli A, Petrino R, Boero M, Bellis D, Chiandussi L. Quantitative Assessment of Histological Changes in Chronic Gastritis After Eradication of Helicobacter Pylori. J Clin Pathol (1992) 45(9):796–8. doi: 10.1136/jcp.45.9.796
40. Eusebi LH, Zagari RM, Bazzoli F. Epidemiology of Helicobacter Pylori Infection. Helicobacter (2014) 19(Suppl 1):1–5. doi: 10.1111/hel.12165
41. An International Association Between Helicobacter Pylori Infection and Gastric Cancer. The EUROGAST Study Group. Lancet (Lond Engl) (1993) 341(8857):1359–62.
42. Forman D, Sitas F, Newell DG, Stacey AR, Boreham J, Peto R, et al. Geographic Association of Helicobacter Pylori Antibody Prevalence and Gastric Cancer Mortality in Rural China. Int J Cancer (1990) 46(4):608–11. doi: 10.1002/ijc.2910460410
43. Fukao A, Komatsu S, Tsubono Y, Hisamichi S, Ohori H, Kizawa T, et al. Helicobacter Pylori Infection and Chronic Atrophic Gastritis Among Japanese Blood Donors: A Cross-Sectional Study. Cancer Causes Control (1993) 4(4):307–12. doi: 10.1007/BF00051332
44. Tsugane S, Kabuto M, Imai H, Gey F, Tei Y, Hanaoka T, et al. Helicobacter Pylori, Dietary Factors, and Atrophic Gastritis in Five Japanese Populations With Different Gastric Cancer Mortality. Cancer Causes Control (1993) 4(4):297–305. doi: 10.1007/BF00051331
46. Correa P, Haenszel W, Cuello C, Zavala D, Fontham E, Zarama G, et al. Gastric Precancerous Process in a High Risk Population: Cohort Follow-Up. Cancer Res (1990) 50(15):4737–40.
47. Sipponen P, Riihelä M, Hyvärinen H, Seppälä K. Chronic Nonatropic (’Superficial’) Gastritis Increases the Risk of Gastric Carcinoma. A Case-Control Study. Scand J Gastroenterol (1994) 29(4):336–40. doi: 10.3109/00365529409094845
48. Guarner J, Mohar A, Parsonnet J, Halperin D. The Association of Helicobacter Pylori With Gastric Cancer and Preneoplastic Gastric Lesions in Chiapas, Mexico. Cancer (1993) 71(2):297–301. doi: 10.1002/1097-0142(19930115)71:2<297::aid-cncr2820710205>3.0.co;2-9
49. Hennings H, Glick AB, Greenhalgh DA, Morgan DL, Strickland JE, Tennenbaum T, et al. Critical Aspects of Initiation, Promotion, and Progression in Multistage Epidermal Carcinogenesis. Proc Soc Exp Biol Med Soc Exp Biol Med (New York NY) (1993) 202(1):1–8. doi: 10.3181/00379727-202-43511A
50. Pignatelli B, Bancel B, Plummer M, Toyokuni S, Patricot L-M, Ohshima H. Helicobacter Pylori Eradication Attenuates Oxidative Stress in Human Gastric Mucosa. Am J Gastroenterol (2001) 96(6):1758–66. doi: 10.1111/j.1572-0241.2001.03869.x
51. Baek HY, Lim JW, Kim H, Kim JM, Kim JS, Jung HC, et al. Oxidative-Stress-Related Proteome Changes in Helicobacter Pylori-Infected Human Gastric Mucosa. Biochem J (2004) 379(2):291–9. doi: 10.1042/bj20031208
52. Lee W-P, Hou M-C, Lan K-H, Li C-P, Chao Y, Lin H-C, et al. Helicobacter Pylori-Induced Chronic Inflammation Causes Telomere Shortening of Gastric Mucosa by Promoting PARP-1-Mediated Non-Homologous End Joining of DNA. Arch Biochem Biophys (2016) 606:90–8. doi: 10.1016/j.abb.2016.07.014
53. Ernst PB, Crowe SE, Reyes VE. How Does Helicobacter Pylori Cause Mucosal Damage? The Inflammatory Response. Gastroenterology (1997) 113(6 Suppl):S35–42; discussion S50. doi: 10.1016/S0016-5085(97)80009-1
54. Peek RMJ, Fiske C, Wilson KT. Role of Innate Immunity in Helicobacter Pylori-Induced Gastric Malignancy. Physiol Rev (2010) 90(3):831–58. doi: 10.1152/physrev.00039.2009
55. Santos MP, Pereira JN, Delabio RW, Smith MAC, Payão SLM, Carneiro LC, et al. Increased Expression of Interleukin-6 Gene in Gastritis and Gastric Cancer. Braz J Med Biol Res = Rev Bras Pesqui medicas e Biol (2021) 54(7):e10687. doi: 10.1590/1414-431x2020e10687
56. Piao J-Y, Lee HG, Kim S-J, Kim D-H, Han H-J, Ngo H-K-C, et al. Helicobacter Pylori Activates IL-6-STAT3 Signaling in Human Gastric Cancer Cells: Potential Roles for Reactive Oxygen Species. Helicobacter (2016) 21(5):405–16. doi: 10.1111/hel.12298
57. Balic JJ, Saad MI, Dawson R, West AJ, McLeod L, West AC, et al. Constitutive STAT3 Serine Phosphorylation Promotes Helicobacter-Mediated Gastric Disease. Am J Pathol (2020) 190(6):1256–70. doi: 10.1016/j.ajpath.2020.01.021
58. Moss SF, Calam J, Agarwal B, Wang S, Holt PR. Induction of Gastric Epithelial Apoptosis by Helicobacter Pylori. Gut (1996) 38(4):498–501. doi: 10.1136/gut.38.4.498
59. Shimoyama T, Fukuda S, Tanaka M, Mikami T, Munakata A, Crabtree JE. CagA Seropositivity Associated With Development of Gastric Cancer in a Japanese Population. J Clin Pathol (1998) 51(3):225–8. doi: 10.1136/jcp.51.3.225
60. Touati E, Michel V, Thiberge J-M, Wuscher N, Huerre M, Labigne A. Chronic Helicobacter Pylori Infections Induce Gastric Mutations in Mice. Gastroenterology (2003) 124(5):1408–19. doi: 10.1016/S0016-5085(03)00266-X
61. Raza Y, Khan A, Farooqui A, Mubarak M, Facista A, Akhtar SS, et al. Oxidative DNA Damage as a Potential Early Biomarker of Helicobacter Pylori Associated Carcinogenesis. Pathol Oncol Res (2014) 20(4):839–46. doi: 10.1007/s12253-014-9762-1
62. Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an Abundant Form of Oxidative DNA Damage, Causes G—-T and A—-C Substitutions. J Biol Chem (1992) 267(1):166–72. doi: 10.1016/S0021-9258(18)48474-8
63. Hahm KB, Lee KJ, Choi SY, Kim JH, Cho SW, Yim H, et al. Possibility of Chemoprevention by the Eradication of Helicobacter Pylori: Oxidative DNA Damage and Apoptosis in H. Pylori Infection. Am J Gastroenterol (1997) 92(10):1853–7.
64. Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, et al. Cag, a Pathogenicity Island of Helicobacter Pylori, Encodes Type I-Specific and Disease-Associated Virulence Factors. Proc Natl Acad Sci USA (1996) 93(25):14648–53. doi: 10.1073/pnas.93.25.14648
65. Sgouras DN, Trang TTH, Yamaoka Y. Pathogenesis of Helicobacter Pylori Infection. Helicobacter (2015) 20 Suppl 1(0 1):8–16. doi: 10.1111/hel.12251
66. Alfarouk KO, Bashir AHH, Aljarbou AN, Ramadan AM, Muddathir AK, AlHoufie STS, et al. The Possible Role of Helicobacter Pylori in Gastric Cancer and Its Management. Front Oncol (2019) 9:75. doi: 10.3389/fonc.2019.00075
67. Yamaoka Y. Mechanisms of Disease: Helicobacter Pylori Virulence Factors. Nat Rev Gastroenterol Hepatol (2010) 7(11):629–41. doi: 10.1038/nrgastro.2010.154
68. Backert S, Tegtmeyer N, Fischer W. Composition, Structure and Function of the Helicobacter Pylori Cag Pathogenicity Island Encoded Type IV Secretion System. Future Microbiol (2015) 10(6):955–65. doi: 10.2217/fmb.15.32
69. Brandt S, Kwok T, Hartig R, König W, Backert S. NF-kappaB Activation and Potentiation of Proinflammatory Responses by the Helicobacter Pylori CagA Protein. Proc Natl Acad Sci USA (2005) 102(26):9300–5. doi: 10.1073/pnas.0409873102
70. Xu S, Wu X, Zhang X, Chen C, Chen H, She F. CagA Orchestrates Eef1a1 and Pkcδ to Induce Interleukin-6 Expression in Helicobacter Pylori-Infected Gastric Epithelial Cells. Gut Pathog (2020) 12(1):31. doi: 10.1186/s13099-020-00368-3
71. Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, Sgouras D, et al. C-Src and C-Abl Kinases Control Hierarchic Phosphorylation and Function of the CagA Effector Protein in Western and East Asian Helicobacter Pylori Strains. J Clin Invest (2012) 122(4):1553–66. doi: 10.1172/JCI61143
72. Higashi H, Nakaya A, Tsutsumi R, Yokoyama K, Fujii Y, Ishikawa S, et al. Helicobacter Pylori CagA Induces Ras-Independent Morphogenetic Response Through SHP-2 Recruitment and Activation. J Biol Chem (2004) 279(17):17205–16. doi: 10.1074/jbc.M309964200
73. Selbach M, Paul FE, Brandt S, Guye P, Daumke O, Backert S, et al. Host Cell Interactome of Tyrosine-Phosphorylated Bacterial Proteins. Cell Host Microbe (2009) 5(4):397–403. doi: 10.1016/j.chom.2009.03.004
74. Bhardwaj V, Noto JM, Wei J, Andl C, El-Rifai W, Peek RM, et al. Helicobacter Pylori Bacteria Alter the P53 Stress Response via ERK-HDM2 Pathway. Oncotarget (2015) 6(3):1531–43. doi: 10.18632/oncotarget.2828
75. Wei J, Noto J, Zaika E, Romero-Gallo J, Correa P, El-Rifai W, et al. Pathogenic Bacterium ≪Em<Helicobacter Pylori≪/Em< Alters the Expression Profile of P53 Protein Isoforms and P53 Response to Cellular Stresses. Proc Natl Acad Sci (2012) 109(38):E2543–50. doi: 10.1073/pnas.1205664109
76. Hatakeyama M. Helicobacter Pylori CagA and Gastric Cancer: A Paradigm for Hit-and-Run Carcinogenesis. Cell Host Microbe (2014) 15(3):306–16. doi: 10.1016/j.chom.2014.02.008
77. Li N, Tang B, Jia Y, Zhu P, Zhuang Y, Fang Y, et al. Helicobacter Pylori CagA Protein Negatively Regulates Autophagy and Promotes Inflammatory Response via C-Met-PI3K/Akt-mTOR Signaling Pathway. Front Cell Infect Microbiol (2017) 7:417:417. doi: 10.3389/fcimb.2017.00417
78. Abu-Taleb AMF, Abdelattef RS, Abdel-Hady AA, Omran FH, El-korashi LA, Abdel-aziz El-hady H, et al. Prevalence of Helicobacter Pylori cagA and iceA Genes and Their Association With Gastrointestinal Diseases. Ahmed AM, Editor. Int J Microbiol (2018) 2018:4809093. doi: 10.1155/2018/4809093
79. Torres J, Pérez-Pérez GI, Leal-Herrera Y, Muñoz O. Infection With CagA+ Helicobacter Pylori Strains as a Possible Predictor of Risk in The Development of Gastric Adenocarcinoma in Mexico. Int J Cancer (1998) 78(3):298–300. doi: 10.1002/(SICI)1097-0215(19981029)78:3<298::AID-IJC6>3.0.CO;2-Q
80. Selbach M, Moese S, Backert S, Jungblut PR, Meyer TF. The Helicobacter Pylori CagA Protein Induces Tyrosine Dephosphorylation of Ezrin. Proteomics (2004) 4(10):2961–8. doi: 10.1002/pmic.200400915
81. Bourzac KM, Botham CM, Karen G. Helicobacter Pylori CagA Induces AGS Cell Elongation Through a Cell Retraction Defect That Is Independent of Cdc42, Rac1, and Arp2/3. Infect Immun (2007) 75(3):1203–13. doi: 10.1128/IAI.01702-06
82. Chen L, Liu Y-C, Zheng Y-Y, Xu J, Zhang Y, Liu W-L, et al. Furanodienone Overcomes Temozolomide Resistance in Glioblastoma Through the Downregulation of CSPG4-Akt-ERK Signaling by Inhibiting EGR1-Dependent Transcription. Phytother Res (2019) 33(6):1736–47. doi: 10.1002/ptr.6363
83. Ducker C, Chow LKY, Saxton J, Handwerger J, McGregor A, Strahl T, et al. De-Ubiquitination of ELK-1 by USP17 Potentiates Mitogenic Gene Expression and Cell Proliferation. Nucleic Acids Res (2019) 47(9):4495–508. doi: 10.1093/nar/gkz166
84. Olea-Flores M, Zuñiga-Eulogio MD, Mendoza-Catalán MA, Rodríguez-Ruiz HA, Castañeda-Saucedo E, Ortuño-Pineda C, et al. Extracellular-Signal Regulated Kinase: A Central Molecule Driving Epithelial-Mesenchymal Transition in Cancer. Int J Mol Sci (2019) 20(12):2885. doi: 10.3390/ijms20122885
85. Garces de los Fayos Alonso I, Liang H-C, Turner SD, Lagger S, Merkel O, Kenner L. The Role of Activator Protein-1 (AP-1) Family Members in CD30-Positive Lymphomas. Cancers (2018) 10(4):93. doi: 10.3390/cancers10040093
86. Takahashi-Yanaga F, Sasaguri T. GSK-3beta Regulates Cyclin D1 Expression: A New Target for Chemotherapy. Cell Signal (2008) 20(4):581–9. doi: 10.1016/j.cellsig.2007.10.018
87. Li X, Liu F, Lin B, Luo H, Liu M, Wu J, et al. Mir−150 Inhibits Proliferation and Tumorigenicity via Retarding G1/S Phase Transition in Nasopharyngeal Carcinoma. Int J Oncol (2017) 50(4):1097–108. doi: 10.3892/ijo.2017.3909
88. Icard P, Fournel L, Wu Z, Alifano M, Lincet H. Interconnection Between Metabolism and Cell Cycle in Cancer. Trends Biochem Sci (2019) 44(6):490–501. doi: 10.1016/j.tibs.2018.12.007
89. Nešić D, Buti L, Lu X, Stebbins CE. Structure of the Helicobacter Pylori CagA Oncoprotein Bound to the Human Tumor Suppressor ASPP2. Proc Natl Acad Sci USA (2014) 111(4):1562–7. doi: 10.1073/pnas.1320631111
90. Lin W-C, Tsai H-F, Kuo S-H, Wu M-S, Lin C-W, Hsu P-I, et al. Translocation of Helicobacter Pylori CagA Into Human B Lymphocytes, the Origin of Mucosa-Associated Lymphoid Tissue Lymphoma. Cancer Res (2010) 70(14):5740–8. doi: 10.1158/0008-5472.CAN-09-4690
91. Vallejo-Flores G, Torres J, Sandoval-Montes C, Arévalo-Romero H, Meza I, Camorlinga-Ponce M, et al. Helicobacter Pylori CagA Suppresses Apoptosis Through Activation of AKT in a Nontransformed Epithelial Cell Model of Glandular Acini Formation. Marmiroli S, Editor. BioMed Res Int (2015) 2015:761501. doi: 10.1155/2015/761501
92. Noto JM, Piazuelo MB, Chaturvedi R, Bartel CA, Thatcher EJ, Delgado A, et al. Strain-Specific Suppression of microRNA-320 by Carcinogenic Helicobacter Pylori Promotes Expression of the Antiapoptotic Protein Mcl-1. Am J Physiol Liver Physiol (2013) 305(11):G786–96. doi: 10.1152/ajpgi.00279.2013
93. Mimuro H, Suzuki T, Nagai S, Rieder G, Suzuki M, Nagai T, et al. Helicobacter Pylori Dampens Gut Epithelial Self-Renewal by Inhibiting Apoptosis, a Bacterial Strategy to Enhance Colonization of the Stomach. Cell Host Microbe (2007) 2(4):250–63. doi: 10.1016/j.chom.2007.09.005
94. Palrasu M, Zaika E, El-Rifai W, Garcia-Buitrago M, Piazuelo MB, Wilson KT, et al. Bacterial CagA Protein Compromises Tumor Suppressor Mechanisms in Gastric Epithelial Cells. J Clin Invest (2020) 130(5):2422–34. doi: 10.1172/JCI130015
95. Buti L, Ruiz-Puig C, Sangberg D, Leissing TM, Brewer RC, Owen RP, et al. CagA–ASPP2 Complex Mediates Loss of Cell Polarity and Favors ≪Em<H. Pylori≪/Em< Colonization of Human Gastric Organoids. Proc Natl Acad Sci (2020) 117(5):2645–55. doi: 10.1073/pnas.1908787117
96. Lang BJ, Gorrell RJ, Tafreshi M, Hatakeyama M, Kwok T, Price JT. The Helicobacter Pylori Cytotoxin CagA is Essential for Suppressing Host Heat Shock Protein Expression. Cell Stress Chaperones (2016) 21(3):523–33. doi: 10.1007/s12192-016-0680-x
97. Liu B, Li X, Sun F, Tong X, Bai Y, Jin K, et al. HP-CagA+ Regulates the Expression of CDK4/CyclinD1 via Reg3 to Change Cell Cycle and Promote Cell Proliferation. Int J Mol Sci (2020) 21(1):224. doi: 10.3390/ijms21010224
98. Choi S, Yoon C, Park MR, Lee D, Kook M-C, Lin J-X, et al. CDX1 Expression Induced by CagA-Expressing ≪Em<Helicobacter Pylori≪/Em< Promotes Gastric Tumorigenesis. Mol Cancer Res (2019) 17(11):2169–83. doi: 10.1158/1541-7786.MCR-19-0181
99. Zhang X-Y, Zhang P-Y, Aboul-Soud MAM. From Inflammation to Gastric Cancer: Role of Helicobacter Pylori. Oncol Lett (2017) 13(2):543–8. doi: 10.3892/ol.2016.5506
100. Murata-Kamiya N, Kurashima Y, Teishikata Y, Yamahashi Y, Saito Y, Higashi H, et al. Helicobacter Pylori CagA Interacts With E-Cadherin and Deregulates the Beta-Catenin Signal That Promotes Intestinal Transdifferentiation in Gastric Epithelial Cells. Oncogene (2007) 26(32):4617–26. doi: 10.1038/sj.onc.1210251
101. Saadat I, Higashi H, Obuse C, Umeda M, Murata-Kamiya N, Saito Y, et al. Helicobacter Pylori CagA Targets PAR1/MARK Kinase to Disrupt Epithelial Cell Polarity. Nature (2007) 447(7142):330–3. doi: 10.1038/nature05765
102. Li M, Chen T, Wang R, Luo J-Y, He J-J, Ye R-S, et al. Plant MIR156 Regulates Intestinal Growth in Mammals by Targeting the Wnt/β-Catenin Pathway. Am J Physiol Physiol (2019) 317(3):C434–48. doi: 10.1152/ajpcell.00030.2019
103. Chang W-L, Yeh Y-C, Sheu B-S. The Impacts of H. Pylori Virulence Factors on the Development of Gastroduodenal Diseases. J BioMed Sci (2018) 25(1):68. doi: 10.1186/s12929-018-0466-9
104. Barden S, Lange S, Tegtmeyer N, Conradi J, Sewald N, Backert S, et al. A Helical RGD Motif Promoting Cell Adhesion: Crystal Structures of the Helicobacter Pylori Type IV Secretion System Pilus Protein CagL. Structure (2013) 21(11):1931–41. doi: 10.1016/j.str.2013.08.018
105. Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, et al. Transgenic Expression of ≪Em<Helicobacter Pylori≪/Em< CagA Induces Gastrointestinal and Hematopoietic Neoplasms in Mouse. Proc Natl Acad Sci (2008) 105(3):1003–8. doi: 10.1073/pnas.0711183105
106. Amieva M, Peek RMJ. Pathobiology of Helicobacter Pylori-Induced Gastric Cancer. Gastroenterology (2016) 150(1):64–78. doi: 10.1053/j.gastro.2015.09.004
107. Sugimoto M, Ohno T, Yamaoka Y. Expression of Angiotensin II Type 1 and Type 2 Receptor mRNAs in the Gastric Mucosa of Helicobacter Pylori-Infected Mongolian Gerbils. J Gastroenterol (2011) 46(10):1177. doi: 10.1007/s00535-011-0433-7
108. Sugimoto M, Ohno T, Graham DY, Yamaoka Y. Helicobacter Pylori Outer Membrane Proteins on Gastric Mucosal Interleukin 6 and 11 Expression in Mongolian Gerbils. J Gastroenterol Hepatol (2011) 26(11):1677–84. doi: 10.1111/j.1440-1746.2011.06817.x
109. Tsugawa H, Suzuki H, Saya H, Hatakeyama M, Hirayama T, Hirata K, et al. Reactive Oxygen Species-Induced Autophagic Degradation of Helicobacter Pylori CagA is Specifically Suppressed in Cancer Stem-Like Cells. Cell Host Microbe (2012) 12(6):764–77. doi: 10.1016/j.chom.2012.10.014
110. Isomoto H, Moss J, Hirayama T. Pleiotropic Actions of Helicobacter Pylori Vacuolating Cytotoxin, VacA. Tohoku J Exp Med (2010) 220(1):3–14. doi: 10.1620/tjem.220.3
111. Nejati S, Karkhah A, Darvish H, Validi M, Ebrahimpour S, Nouri HR. Influence of Helicobacter Pylori Virulence Factors CagA and VacA on Pathogenesis of Gastrointestinal Disorders. Microb Pathog (2018) 117:43–8. doi: 10.1016/j.micpath.2018.02.016
112. Fahimi F, Sarhaddi S, Fouladi M, Samadi N, Sadeghi J, Golchin A, et al. Phage Display-Derived Antibody Fragments Against Conserved Regions of VacA Toxin of Helicobacter Pylori. Appl Microbiol Biotechnol (2018) 102(16):6899–913. doi: 10.1007/s00253-018-9068-4
113. Yahiro K, Akazawa Y, Nakano M, Suzuki H, Hisatune J, Isomoto H, et al. Helicobacter Pylori VacA Induces Apoptosis by Accumulation of Connexin 43 in Autophagic Vesicles via a Rac1/ERK-Dependent Pathway. Cell Death Discov (2015) 1(1):15035. doi: 10.1038/cddiscovery.2015.35
114. Chauhan N, Tay ACY, Marshall BJ, Jain U. Helicobacter Pylori VacA, a Distinct Toxin Exerts Diverse Functionalities in Numerous Cells: An Overview. Helicobacter (2019) 24(1):e12544. doi: 10.1111/hel.12544
115. Gauthier NC, Monzo P, Kaddai V, Doye A, Ricci V, Boquet P. Helicobacter Pylori VacA Cytotoxin: A Probe for a Clathrin-Independent and Cdc42-Dependent Pinocytic Pathway Routed to Late Endosomes. Mol Biol Cell (2005) 16(10):4852–66. doi: 10.1091/mbc.e05-05-0398
116. Ricci V, Galmiche A, Doye A, Necchi V, Solcia E, Boquet P. High Cell Sensitivity to Helicobacter Pylori VacA Toxin Depends on a GPI-Anchored Protein and Is Not Blocked by Inhibition of the Clathrin-Mediated Pathway of Endocytosis. Mol Biol Cell (2000) 11(11):3897–909. doi: 10.1091/mbc.11.11.3897
117. Yahiro K, Wada A, Nakayama M, Kimura T, Ogushi K, Niidome T, et al. Protein-Tyrosine Phosphatase Alpha, RPTP Alpha, Is a Helicobacter Pylori VacA Receptor. J Biol Chem (2003) 278(21):19183–9. doi: 10.1074/jbc.M300117200
118. Fujikawa A, Shirasaka D, Yamamoto S, Ota H, Yahiro K, Fukada M, et al. Mice Deficient in Protein Tyrosine Phosphatase Receptor Type Z Are Resistant to Gastric Ulcer Induction by VacA of Helicobacter Pylori. Nat Genet (2003) 33(3):375–81. doi: 10.1038/ng1112
119. Utt M, Danielsson B, Wadström T. Helicobacter Pylori Vacuolating Cytotoxin Binding to a Putative Cell Surface Receptor, Heparan Sulfate, Studied by Surface Plasmon Resonance. FEMS Immunol Med Microbiol (2001) 30(2):109–13. doi: 10.1111/j.1574-695X.2001.tb01557.x
120. Gupta VR, Patel HK, Kostolansky SS, Ballivian RA, Eichberg J, Blanke SR. Sphingomyelin Functions as a Novel Receptor for Helicobacter Pylori VacA. PloS Pathog (2008) 4(5):e1000073. doi: 10.1371/journal.ppat.1000073
121. Kroemer G, Mariño G, Levine B. Autophagy and the Integrated Stress Response. Mol Cell (2010) 40(2):280–93. doi: 10.1016/j.molcel.2010.09.023
122. Papini E, de Bernard M, Milia E, Bugnoli M, Zerial M, Rappuoli R, et al. Cellular Vacuoles Induced by Helicobacter Pylori Originate From Late Endosomal Compartments. Proc Natl Acad Sci (1994) 91(21):9720–4. doi: 10.1073/pnas.91.21.9720
123. Nakayama M, Hisatsune J, Yamasaki E, Isomoto H, Kurazono H, Hatakeyama M, et al. Helicobacter Pylori VacA-Induced Inhibition of GSK3 Through the PI3K/Akt Signaling Pathway. J Biol Chem (2009) 284(3):1612–9. doi: 10.1074/jbc.M806981200
124. Yudushkin I. Getting the Akt Together: Guiding Intracellular Akt Activity by PI3K. Biomolecules (2019) 9(2):67. doi: 10.3390/biom9020067
125. Kim I-J, Lee J, Oh SJ, Yoon M-S, Jang S-S, Holland RL, et al. Helicobacter Pylori Infection Modulates Host Cell Metabolism Through VacA-Dependent Inhibition of Mtorc1. Cell Host Microbe (2018) 23(5):583–93.e8. doi: 10.1016/j.chom.2018.04.006
126. Gao C, Yuan X, Jiang Z, Gan D, Ding L, Sun Y, et al. Regulation of AKT Phosphorylation by GSK3β and PTEN to Control Chemoresistance in Breast Cancer. Breast Cancer Res Treat (2019) 176(2):291–301. doi: 10.1007/s10549-019-05239-3
127. Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell (2017) 169(3):381–405. doi: 10.1016/j.cell.2017.04.001
128. Vallée A, Lecarpentier Y. Alzheimer Disease: Crosstalk Between the Canonical Wnt/Beta-Catenin Pathway and PPARs Alpha and Gamma. Front Neurosci (2016) 10:459. doi: 10.3389/fnins.2016.00459
129. Singh S, Mishra A, Mohanbhai SJ, Tiwari V, Chaturvedi RK, Khurana S, et al. Axin-2 Knockdown Promote Mitochondrial Biogenesis and Dopaminergic Neurogenesis by Regulating Wnt/β-Catenin Signaling in Rat Model of Parkinson’s Disease. Free Radic Biol Med (2018) 129:73–87. doi: 10.1016/j.freeradbiomed.2018.08.033
130. Badimon L, Casaní L, Camino-Lopez S, Juan-Babot O, Borrell-Pages M. Gsk3β Inhibition and Canonical Wnt Signaling in Mice Hearts After Myocardial Ischemic Damage. PloS One (2019) 14(6):e0218098. doi: 10.1371/journal.pone.0218098
131. Jones KR, Whitmire JM, Merrell DS. A Tale of Two Toxins: Helicobacter Pylori CagA and VacA Modulate Host Pathways That Impact Disease. Front Microbiol (2010) 1:115. doi: 10.3389/fmicb.2010.00115
132. Noto JM, Romero-Gallo J, Piazuelo MB, Peek RM. The Mongolian Gerbil: A Robust Model of Helicobacter Pylori-Induced Gastric Inflammation and Cancer. Methods Mol Biol (2016) 1422:263–80. doi: 10.1007/978-1-4939-3603-8_24
133. Machado AMD, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter Pylori Infection Generates Genetic Instability in Gastric Cells. Biochim Biophys Acta (2010) 1806(1):58–65. doi: 10.1016/j.bbcan.2010.01.007
134. Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, et al. Carcinogenic Bacterial Pathogen Helicobacter Pylori Triggers DNA Double-Strand Breaks and a DNA Damage Response in its Host Cells. Proc Natl Acad Sci USA (2011) 108(36):14944–9. doi: 10.1073/pnas.1100959108
135. Hanada K, Uchida T, Tsukamoto Y, Watada M, Yamaguchi N, Yamamoto K, et al. Helicobacter Pylori Infection Introduces DNA Double-Strand Breaks in Host Cells. Infect Immun (2014) 82(10):4182–9. doi: 10.1128/IAI.02368-14
136. Fukase K, Kato M, Kikuchi S, Inoue K, Uemura N, Okamoto S, et al. Effect of Eradication of Helicobacter Pylori on Incidence of Metachronous Gastric Carcinoma After Endoscopic Resection of Early Gastric Cancer: An Open-Label, Randomised Controlled Trial. Lancet (Lond Engl) (2008) 372(9636):392–7. doi: 10.1016/S0140-6736(08)61159-9
137. Du M-Q, Isaccson PG. Gastric MALT Lymphoma: From Aetiology to Treatment. Lancet Oncol (2002) 3(2):97–104. doi: 10.1016/S1470-2045(02)00651-4
138. Wotherspoon AC. Gastric MALT Lymphoma and Helicobacter Pylori. Yale J Biol Med (1996) 69(1):61–8.
139. Bergman MP, D’Elios MM. Cytotoxic T Cells in H. Pylori-Related Gastric Autoimmunity and Gastric Lymphoma. J BioMed Biotechnol (2010) 2010:104918. doi: 10.1155/2010/104918
140. Zhu Y, Wang C, Huang J, Ge Z, Dong Q, Zhong X, et al. The Helicobacter Pylori Virulence Factor CagA Promotes Erk1/2-Mediated Bad Phosphorylation in Lymphocytes: A Mechanism of CagA-Inhibited Lymphocyte Apoptosis. Cell Microbiol (2007) 9(4):952–61. doi: 10.1111/j.1462-5822.2006.00843.x
141. Umehara S, Higashi H, Ohnishi N, Asaka M, Hatakeyama M. Effects of Helicobacter Pylori CagA Protein on the Growth and Survival of B Lymphocytes, the Origin of MALT Lymphoma. Oncogene (2003) 22(51):8337–42. doi: 10.1038/sj.onc.1207028
142. Du M, Peng H, Singh N, Isaacson PG, Pan L. The Accumulation of P53 Abnormalities is Associated With Progression of Mucosa-Associated Lymphoid Tissue Lymphoma. Blood (1995) 86(12):4587–93. doi: 10.1182/blood.V86.12.4587.bloodjournal86124587
143. Boncristiano M, Paccani SR, Barone S, Ulivieri C, Patrussi L, Ilver D, et al. The Helicobacter Pylori Vacuolating Toxin Inhibits T Cell Activation by Two Independent Mechanisms. J Exp Med (2003) 198(12):1887–97. doi: 10.1084/jem.20030621
144. Torres VJ, VanCompernolle SE, Sundrud MS, Unutmaz D, Cover TL. Helicobacter Pylori Vacuolating Cytotoxin Inhibits Activation-Induced Proliferation Of Human T and B Lymphocyte Subsets. J Immunol (2007) 179(8):5433–40. doi: 10.4049/jimmunol.179.8.5433
145. Kondo T, Oka T, Sato H, Shinnou Y, Washio K, Takano M, et al. Accumulation of Aberrant CpG Hypermethylation by Helicobacter Pylori Infection Promotes Development and Progression of Gastric MALT Lymphoma. Int J Oncol (2009) 35(3):547–57. doi: 10.3892/ijo_00000366
146. Fukuhara N, Nakamura T, Nakagawa M, Tagawa H, Takeuchi I, Yatabe Y, et al. Chromosomal Imbalances are Associated With Outcome of Helicobacter Pylori Eradication in T (11,18)(Q21;Q21) Negative Gastric Mucosa-Associated Lymphoid Tissue Lymphomas. Genes Chromosomes Cancer (2007) 46(8):784–90. doi: 10.1002/gcc.20464
147. Lehours P, Ménard A, Dupouy S, Bergey B, Richy F, Zerbib F, et al. Evaluation of the Association of Nine Helicobacter Pylori Virulence Factors With Strains Involved in Low-Grade Gastric Mucosa-Associated Lymphoid Tissue Lymphoma. Infect Immun (2004) 72(2):880–8. doi: 10.1128/IAI.72.2.880-888.2004
148. Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium Nucleatum: An Emerging Gut Pathogen? Gut Microbes (2011) 2(5):294–8. doi: 10.4161/gmic.2.5.18603
149. Nagy TA, Frey MR, Yan F, Israel DA, Polk DB, Peek RMJ. Helicobacter Pylori Regulates Cellular Migration and Apoptosis by Activation of Phosphatidylinositol 3-Kinase Signaling. J Infect Dis (2009) 199(5):641–51. doi: 10.1086/596660
150. Beevers DG, Lip GYH, Blann AD. Salt Intake and Helicobacter Pylori Infection. J Hypertens (2004) 22(8):1475–7. doi: 10.1097/01.hjh.0000133736.77866.77
151. Shikata K, Kiyohara Y, Kubo M, Yonemoto K, Ninomiya T, Shirota T, et al. A Prospective Study of Dietary Salt Intake and Gastric Cancer Incidence in a Defined Japanese Population: The Hisayama Study. Int J Cancer (2006) 119(1):196–201. doi: 10.1002/ijc.21822
152. Gamboa-Dominguez A, Ubbelohde T, Saqui-Salces M, Romano-Mazzoti L, Cervantes M, Domínguez-Fonseca C, et al. Salt and Stress Synergize H. Pylori-Induced Gastric Lesions, Cell Proliferation, and P21 Expression in Mongolian Gerbils. Dig Dis Sci (2007) 52(6):1517–26. doi: 10.1007/s10620-006-9524-3
153. Randel AH. Pylori Infection: ACG Updates Treatment Recommendations. Am Fam Physician (2018) 97(2):135–7.
154. Wu C-Y, Wu M-S, Kuo KN, Wang C-B, Chen Y-J, Lin J-T. Effective Reduction of Gastric Cancer Risk With Regular Use of Nonsteroidal Anti-Inflammatory Drugs in Helicobacter Pylori-Infected Patients. J Clin Oncol Off J Am Soc Clin Oncol (2010) 28(18):2952–7. doi: 10.1200/JCO.2009.26.0695
155. Doorakkers E, Lagergren J, Engstrand L, Brusselaers N. Eradication of Helicobacter Pylori and Gastric Cancer: A Systematic Review and Meta-Analysis of Cohort Studies. JNCI J Natl Cancer Inst (2016) 108(9):djw132. doi: 10.1093/jnci/djw132
156. Li P, Zhang H, Chen J, Shi Y, Cai J, Yang J, et al. Association Between Dietary Antioxidant Vitamins Intake/Blood Level and Risk of Gastric Cancer. Int J Cancer (2014) 135(6):1444–53. doi: 10.1002/ijc.28777
157. Correa P, Fontham ET, Bravo JC, Bravo LE, Ruiz B, Zarama G, et al. Chemoprevention of Gastric Dysplasia: Randomized Trial of Antioxidant Supplements and Anti-Helicobacter Pylori Therapy. J Natl Cancer Inst (2000) 92(23):1881–8. doi: 10.1093/jnci/92.23.1881
158. Mitchell HM, Ally R, Wadee A, Wiseman M, Segal I. Major Differences in the IgG Subclass Response to Helicobacter Pylori in the First and Third Worlds. Scand J Gastroenterol (2002) 37(5):517–22. doi: 10.1080/00365520252903044
159. Lee S-A, Kang D, Shim KN, Choe JW, Hong WS, Choi H. Effect of Diet and Helicobacter Pylori Infection to the Risk of Early Gastric Cancer. J Epidemiol (2003) 13(3):162–8. doi: 10.2188/jea.13.162
160. Stolte M, Bayerdörffer E, Morgner A, Alpen B, Wündisch T, Thiede C, et al. ≪Em<Helicobacter</em< and Gastric MALT Lymphoma. Gut (2002) 50(suppl 3):iii19–24. doi: 10.1136/gut.50.suppl_3.iii19
161. Choi J. Successful Endoscopic Resection of Gastric Mucosa-Associated Lymphoid Tissue Lymphoma Unresponsive to Helicobacter Pylori Eradication Therapy. Korean J Gastrointest Endosc (2020) 0(0). doi: 10.5946/ce.2020.232
162. Gong EJ, Ahn JY, Jung H-Y, Park H, Ko YB, Na HK, et al. Helicobacter Pylori Eradication Therapy Is Effective as the Initial Treatment for Patients With H. Pylori-Negative and Disseminated Gastric Mucosa-Associated Lymphoid Tissue Lymphoma. Gut Liver (2016) 10(5):706–13.
163. Kuo S-H, Yeh K-H, Wu M-S, Lin C-W, Wei M-F, Liou J-M, et al. First-Line Antibiotic Therapy in Helicobacter Pylori-Negative Low-Grade Gastric Mucosa-Associated Lymphoid Tissue Lymphoma. Sci Rep (2017) 7(1):14333. doi: 10.1038/s41598-017-14102-8
164. Ford AC, Forman D, Hunt RH, Yuan Y, Moayyedi P. Helicobacter Pylori Eradication Therapy to Prevent Gastric Cancer in Healthy Asymptomatic Infected Individuals: Systematic Review and Meta-Analysis of Randomised Controlled Trials. BMJ (2014) 348:g3174. doi: 10.1136/bmj.g3174
165. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global Cancer Statistics. CA Cancer J Clin (2011) 61(2):69–90. doi: 10.3322/caac.20107
166. Smith JS, Bosetti C, Muñoz N, Herrero R, Bosch FX, Eluf-Neto J, et al. Chlamydia Trachomatis and Invasive Cervical Cancer: A Pooled Analysis of the IARC Multicentric Case-Control Study. Int J Cancer (2004) 111(3):431–9. doi: 10.1002/ijc.20257
167. Castellsagué X, Pawlita M, Roura E, Margall N, Waterboer T, Bosch FX, et al. Prospective Seroepidemiologic Study on the Role of Human Papillomavirus and Other Infections in Cervical Carcinogenesis: Evidence From the EPIC Cohort. Int J Cancer (2014) 135(2):440–52. doi: 10.1002/ijc.28665
168. Malhotra M, Sood S, Mukherjee A, Muralidhar S, Bala M. Genital Chlamydia Trachomatis: An Update. Indian J Med Res (2013) 138(3):303–16.
169. Zhu H, Shen Z, Luo H, Zhang W, Zhu X. Chlamydia Trachomatis Infection-Associated Risk of Cervical Cancer: A Meta-Analysis. Med (Baltimore) (2016) 95(13):e3077. doi: 10.1097/MD.0000000000003077
170. Tungsrithong N, Kasinpila C, Maneenin C, Namujju PB, Lehtinen M, Anttila A, et al. Lack of Significant Effects of Chlamydia Trachomatis Infection on Cervical Cancer Risk in a Nested Case-Control Study in North-East Thailand. Asian Pac J Cancer Prev (2014) 15(3):1497–500. doi: 10.7314/APJCP.2014.15.3.1497
171. Zereu M, Zettler CG, Cambruzzi E, Zelmanowicz A. Herpes Simplex Virus Type 2 and Chlamydia Trachomatis in Adenocarcinoma of the Uterine Cervix. Gynecol Oncol (2007) 105(1):172–5. doi: 10.1016/j.ygyno.2006.11.006
172. Kiviat NB, Paavonen JA, Brockway J, Critchlow CW, Brunham RC, Stevens CE, et al. Cytologic Manifestations of Cervical and Vaginal Infections. I. Epithelial and Inflammatory Cellular Changes. JAMA (1985) 253(7):989–96.
173. Paavonen J. ≪Em<Chlamydia Trachomatis≪/Em< and Cancer. Sex Transm Infect (2001) 77(3):154–6. doi: 10.1136/sti.77.3.154
174. Scidmore MA, Fischer ER, Hackstadt T. Restricted Fusion of Chlamydia Trachomatis Vesicles With Endocytic Compartments During the Initial Stages of Infection. Infect Immun (2003) 71(2):973–84. doi: 10.1128/IAI.71.2.973-984.2003
175. Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a Chlamydial Protease-Like Activity Factor Responsible for the Degradation of Host Transcription Factors. J Exp Med (2001) 193(8):935–42. doi: 10.1084/jem.193.8.935
176. WHO. Available at: https://monographs.iarc.who.int/home/iarc-monographs-general-information/.
177. Xia M, Bumgarner RE, Lampe MF, Stamm WE. Chlamydia Trachomatis Infection Alters Host Cell Transcription in Diverse Cellular Pathways. J Infect Dis (2003) 187(3):424–34. doi: 10.1086/367962
178. Knowlton AE, Brown HM, Richards TS, Andreolas LA, Patel RK, Grieshaber SS. Chlamydia Trachomatis Infection Causes Mitotic Spindle Pole Defects Independently From its Effects on Centrosome Amplification. Traffic (2011) 12(7):854–66. doi: 10.1111/j.1600-0854.2011.01204.x
179. Sun HS, Wilde A, Harrison RE. Chlamydia Trachomatis Inclusions Induce Asymmetric Cleavage Furrow Formation and Ingression Failure in Host Cells. Mol Cell Biol (2011) 31(24):5011–22. doi: 10.1128/MCB.05734-11
180. Grieshaber SS, Grieshaber NA, Miller N, Hackstadt T. Chlamydia Trachomatis Causes Centrosomal Defects Resulting in Chromosomal Segregation Abnormalities. Traffic (2006) 7(8):940–9. doi: 10.1111/j.1600-0854.2006.00439.x
181. Fan T, Lu H, Hu H, Shi L, McClarty GA, Nance DM, et al. Inhibition of Apoptosis in Chlamydia-Infected Cells: Blockade of Mitochondrial Cytochrome C Release and Caspase Activation. J Exp Med (1998) 187(4):487–96. doi: 10.1084/jem.187.4.487
182. Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a Human Protein Homologous to C. Elegans CED-4, Participates in Cytochrome C-Dependent Activation of Caspase-3. Cell (1997) 90(3):405–13. doi: 10.1016/s0092-8674(00)80501-2
183. Shi L, Chen G, MacDonald G, Bergeron L, Li H, Miura M, et al. Activation of an Interleukin 1 Converting Enzyme-Dependent Apoptosis Pathway by Granzyme B. Proc Natl Acad Sci USA (1996) 93(20):11002–7. doi: 10.1073/pnas.93.20.11002
184. Cai J, Yang J, Jones DP. Mitochondrial Control of Apoptosis: The Role of Cytochrome C. Biochim Biophys Acta (1998) 1366(1–2):139–49. doi: 10.1016/S0005-2728(98)00109-1
185. Erhardt P, Cooper G. Activation of the CPP32 Apoptotic Protease by Distinct Signaling Pathways With Differential Sensitivity to Bcl-Xl. J Biol Chem (1996) 271(30):17601–4. doi: 10.1074/jbc.271.30.17601
186. Bliska JB, Galán JE, Falkow S. Signal Transduction in the Mammalian Cell During Bacterial Attachment and Entry. Cell (1993) 73(5):903–20. doi: 10.1016/0092-8674(93)90270-Z
187. Birkelund S, Johnsen H, Christiansen G. Chlamydia Trachomatis Serovar L2 Induces Protein Tyrosine Phosphorylation During Uptake by HeLa Cells. Infect Immun (1994) 62(11):4900–8. doi: 10.1128/iai.62.11.4900-4908.1994
188. Fawaz FS, van Ooij C, Homola E, Mutka SC, Engel JN. Infection With Chlamydia Trachomatis Alters the Tyrosine Phosphorylation and/or Localization of Several Host Cell Proteins Including Cortactin. Infect Immun (1997) 65(12):5301–8. doi: 10.1128/iai.65.12.5301-5308.1997
189. Gurumurthy RK, Mäurer AP, Machuy N, Hess S, Pleissner KP, Schuchhardt J, et al. A Loss-of-Function Screen Reveals Ras- and Raf-Independent MEK-ERK Signaling During Chlamydia Trachomatis Infection. Sci Signal (2010) 3(113):ra21. doi: 10.1126/scisignal.2000651
190. Vignola MJ, Kashatus DF, Taylor GA, Counter CM, Valdivia RH. Cpla2 Regulates the Expression of Type I Interferons and Intracellular Immunity to Chlamydia Trachomatis. J Biol Chem (2010) 285(28):21625–35. doi: 10.1074/jbc.M110.103010
191. Abdul-Sater AA, Saïd-Sadier N, Lam VM, Singh B, Pettengill MA, Soares F, et al. Enhancement of Reactive Oxygen Species Production and Chlamydial Infection by the Mitochondrial Nod-Like Family Member NLRX1. J Biol Chem (2010) 285(53):41637–45. doi: 10.1074/jbc.M110.137885
192. Wang F, Zhang H, Lu X, Zhu Q, Shi T, Lu R, et al. Chlamydia Trachomatis Induces Autophagy by P62 in HeLa Cell. World J Microbiol Biotechnol (2021) 37(3):50. doi: 10.1007/s11274-021-03014-5
193. Zou Y, Lei W, Su S, Bu J, Zhu S, Huang Q, et al. Chlamydia Trachomatis Plasmid-Encoded Protein Pgp3 Inhibits Apoptosis via the PI3K-AKT-Mediated MDM2-P53 Axis. Mol Cell Biochem (2019) 452(1):167–76. doi: 10.1007/s11010-018-3422-9
194. Lei W, Li Q, Su S, Bu J, Huang Q, Li Z. Chlamydia Trachomatis Plasmid-Encoded Protein Porf5 Protects Mitochondrial Function by Inducing Mitophagy and Increasing HMGB1 Expression. Pathog Dis (2017) 75(9). doi: 10.1093/femspd/ftx111
195. Al-Zeer MA, Xavier A, Abu Lubad M, Sigulla J, Kessler M, Hurwitz R, et al. Chlamydia Trachomatis Prevents Apoptosis Via Activation of PDPK1-MYC and Enhanced Mitochondrial Binding of Hexokinase II. EBioMedicine (2017) 23:100–10. doi: 10.1016/j.ebiom.2017.08.005
196. Greene W, Zhong G. Inhibition of Host Cell Cytokinesis by Chlamydia Trachomatis Infection. J Infect (2003) 47(1):45–51. doi: 10.1016/S0163-4453(03)00039-2
197. Greene W, Xiao Y, Huang Y, McClarty G, Zhong G. Chlamydia-Infected Cells Continue to Undergo Mitosis and Resist Induction of Apoptosis. Infect Immun (2004) 72(1):451–60. doi: 10.1128/IAI.72.1.451-460.2004
198. Johnson KA, Tan M, Sütterlin C. Centrosome Abnormalities During a Chlamydia Trachomatis Infection Are Caused by Dysregulation of the Normal Duplication Pathway. Cell Microbiol (2009) 11(7):1064–73. doi: 10.1111/j.1462-5822.2009.01307.x
199. Voos W, Röttgers K. Molecular Chaperones as Essential Mediators of Mitochondrial Biogenesis. Biochim Biophys Acta (2002) 1592(1):51–62. doi: 10.1016/S0167-4889(02)00264-1
200. Campanella C, Marino Gammazza A, Mularoni L, Cappello F, Zummo G, Di Felice V. A Comparative Analysis of the Products of GROEL-1 Gene From Chlamydia Trachomatis Serovar D and the HSP60 Var1 Transcript From Homo Sapiens Suggests a Possible Autoimmune Response. Int J Immunogenet. (2009) 36(1):73–8. doi: 10.1111/j.1744-313X.2008.00819.x
201. Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, Jang JH, et al. Global Profiling of the Cell Surface Proteome of Cancer Cells Uncovers an Abundance Of Proteins With Chaperone Function. J Biol Chem (2003) 278(9):7607–16. doi: 10.1074/jbc.M210455200
202. Cappello F, Conway de Macario E, Di Felice V, Zummo G, Macario AJL. Chlamydia Trachomatis Infection and Anti-Hsp60 Immunity: The Two Sides of the Coin. PloS Pathog (2009) 5(8):e1000552. doi: 10.1371/journal.ppat.1000552
203. Bavoil P, Stephens RS, Falkow S. A Soluble 60 Kilodalton Antigen of Chlamydia Spp. Is a Homologue of Escherichia Coli GroEL. Mol Microbiol (1990) 4(3):461–9.
204. Vabulas RM, Ahmad-Nejad P, da Costa C, Miethke T, Kirschning CJ, Häcker H, et al. Endocytosed HSP60s Use Toll-Like Receptor 2 (TLR2) and TLR4 to Activate the Toll/Interleukin-1 Receptor Signaling Pathway in Innate Immune Cells. J Biol Chem (2001) 276(33):31332–9. doi: 10.1074/jbc.M103217200
205. Sasu S, LaVerda D, Qureshi N, Golenbock DT, Beasley D. Chlamydia Pneumoniae and Chlamydial Heat Shock Protein 60 Stimulate Proliferation of Human Vascular Smooth Muscle Cells via Toll-Like Receptor 4 and P44/P42 Mitogen-Activated Protein Kinase Activation. Circ Res (2001) 89(3):244–50. doi: 10.1161/hh1501.094184
206. Kirchhoff SR, Gupta S, Knowlton AA. Cytosolic Heat Shock Protein 60, Apoptosis, and Myocardial Injury. Circulation (2002) 105(24):2899–904. doi: 10.1161/01.CIR.0000019403.35847.23
207. Beatty WL, Byrne GI, Morrison RP. Morphologic and Antigenic Characterization of Interferon Gamma-Mediated Persistent Chlamydia Trachomatis Infection In Vitro. Proc Natl Acad Sci USA (1993) 90(9):3998–4002. doi: 10.1073/pnas.90.9.3998
208. Hjelholt A, Christiansen G, Johannesson TG, Ingerslev HJ, Birkelund S. Tubal Factor Infertility is Associated With Antibodies Against Chlamydia Trachomatis Heat Shock Protein 60 (HSP60) But Not Human HSP60. Hum Reprod (2011) 26(8):2069–76. doi: 10.1093/humrep/der167
209. Madeleine MM, Anttila T, Schwartz SM, Saikku P, Leinonen M, Carter JJ, et al. Risk of Cervical Cancer Associated With Chlamydia Trachomatis Antibodies by Histology, HPV Type and HPV Cofactors. Int J Cancer (2007) 120(3):650–5. doi: 10.1002/ijc.22325
210. Carratelli CR, Rizzo A, Catania MR, Gallè F, Losi E, Hasty DL, et al. Chlamydia Pneumoniae Infections Prevent the Programmed Cell Death on THP-1 Cell Line. FEMS Microbiol Lett (2002) 215(1):69–74. doi: 10.1111/j.1574-6968.2002.tb11372.x
211. Anttila T, Saikku P, Koskela P, Bloigu A, Dillner J, Ikäheimo I, et al. Serotypes of Chlamydia Trachomatis and Risk for Development of Cervical Squamous Cell Carcinoma. JAMA (2001) 285(1):47–51. doi: 10.1001/jama.285.1.47
212. Huh WK. Human Papillomavirus Infection: A Concise Review of Natural History. Obstet Gynecol (2009) 114(1):139–43. doi: 10.1097/AOG.0b013e3181ab6878
213. Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human Papillomavirus Testing in the Prevention of Cervical Cancer. J Natl Cancer Inst (2011) 103(5):368–83. doi: 10.1093/jnci/djq562
214. Insinga RP, Perez G, Wheeler CM, Koutsky LA, Garland SM, Leodolter S, et al. Incidence, Duration, and Reappearance of Type-Specific Cervical Human Papillomavirus Infections in Young Women. Cancer Epidemiol Biomarkers & Prev (2010) 19(6):1585–94. doi: 10.1158/1055-9965.EPI-09-1235
215. Discacciati MG, Gimenes F, Pennacchi PC, Faião-Flores F, Zeferino LC, Derchain SM, et al. MMP-9/RECK Imbalance: A Mechanism Associated With High-Grade Cervical Lesions and Genital Infection by Human Papillomavirus and Chlamydia Trachomatis. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored by Am Soc Prev Oncol (2015) 24(10):1539–47. doi: 10.1158/1055-9965.EPI-15-0420
216. Lau C-Y, Qureshi AK. Azithromycin Versus Doxycycline for Genital Chlamydial Infections: A Meta-Analysis Of Randomized Clinical Trials. Sex Transm Dis (2002) 29(9):497–502. doi: 10.1097/00007435-200209000-00001
217. STI Treatment Guidelines 2021. (2021). Available at: https://www.cdc.gov/std/treatment-guidelines/chlamydia.htm.
218. Nozawa A, Oshima H, Togawa N, Nozaki T, Murakami S. Development of Oral Care Chip, a Novel Device for Quantitative Detection of the Oral Microbiota Associated With Periodontal Disease. PloS One (2020) 15(2):e0229485. doi: 10.1371/journal.pone.0229485
219. Abed J, Maalouf N, Manson AL, Earl AM, Parhi L, Emgård JEM, et al. Colon Cancer-Associated Fusobacterium Nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front Cell Infect Microbiol (2020) 10:400:400. doi: 10.3389/fcimb.2020.00400
220. Han YW. Can Oral Bacteria Cause Pregnancy Complications? Women’s Health (Lond Engl) United States; (2011) 7:401–4. doi: 10.2217/WHE.11.37
221. Han YW. Oral Health and Adverse Pregnancy Outcomes - What’s Next? J Dent Res (2011) 90(3):289–93. doi: 10.1177/0022034510381905
222. Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High Occurrence of Fusobacterium Nucleatum and Clostridium Difficile in the Intestinal Microbiota of Colorectal Carcinoma Patients. Braz J Microbiol [publication Braz Soc Microbiol (2015) 46(4):1135–40. doi: 10.1590/S1517-838246420140665
223. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium Nucleatum Infection is Prevalent in Human Colorectal Carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
224. McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is Associated With Colorectal Adenomas. PloS One (2013) 8(1):e53653. doi: 10.1371/journal.pone.0053653
225. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium Nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol (2015) 1(5):653–61. doi: 10.1001/jamaoncol.2015.1377
226. Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium Nucleatum With Clinical and Molecular Features in Colorectal Serrated Pathway. Int J Cancer (2015) 137(6):1258–68. doi: 10.1002/ijc.29488
227. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium Nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κb, and Up-Regulating Expression of MicroRNA-21. Gastroenterology (2017) 152(4):851–66.e24. doi: 10.1053/j.gastro.2016.11.018
228. Li Y-Y, Ge Q-X, Cao J, Zhou Y-J, Du Y-L, Shen B, et al. Association of Fusobacterium Nucleatum Infection With Colorectal Cancer in Chinese Patients. World J Gastroenterol (2016) 22(11):3227–33. doi: 10.3748/wjg.v22.i11.3227
229. Wong SH, Kwong TNY, Chow T-C, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of Faecal Fusobacterium Improves Faecal Immunochemical Test in Detecting Advanced Colorectal Neoplasia. Gut (2017) 66(8):1441–8. doi: 10.1136/gutjnl-2016-312766
230. Gethings-Behncke C, Coleman HG, Jordao HWT, Longley DB, Crawford N, Murray LJ, et al. Fusobacterium Nucleatum in the Colorectum and Its Association With Cancer Risk and Survival: A Systematic Review and Meta-Analysis. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored by Am Soc Prev Oncol (2020) 29(3):539–48. doi: 10.1158/1055-9965.EPI-18-1295
231. Shet UK, Oh H-K, Kim H-J, Chung H-J, Kim Y-J, Kim O-S, et al. Quantitative Analysis of Periodontal Pathogens Present in the Saliva of Geriatric Subjects. J Periodontal Implant Sci (2013) 43(4):183–90. doi: 10.5051/jpis.2013.43.4.183
232. Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, et al. Co-Occurrence of Anaerobic Bacteria in Colorectal Carcinomas. Microbiome (2013) 1(1):16. doi: 10.1186/2049-2618-1-16
233. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic Analysis of Faecal Microbiome as a Tool Towards Targeted Non-Invasive Biomarkers for Colorectal Cancer. Gut (2017) 66(1):70–8. doi: 10.1136/gutjnl-2015-309800
234. Eisele Y, Mallea PM, Gigic B, Stephens WZ, Warby CA, Buhrke K, et al. Fusobacterium Nucleatum and Clinicopathologic Features of Colorectal Cancer: Results From the ColoCare Study. Clin Colorect Cancer (2021) 20(3):e165–72. doi: 10.1016/j.clcc.2021.02.007
235. Chen T, Li Q, Zhang X, Long R, Wu Y, Wu J, et al. TOX Expression Decreases With Progression of Colorectal Cancers and Is Associated With CD4 T-Cell Density and Fusobacterium Nucleatum Infection. Hum Pathol (2018) 79:93–101. doi: 10.1016/j.humpath.2018.05.008
236. Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, et al. Fusobacterium Nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol Res (2018) 6(11):1327–36. doi: 10.1158/2326-6066.CIR-18-0174
237. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors From Immune Cell Attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
238. Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, et al. Fusobacterium Nucleatum Supresses Anti-Tumor Immunity by Activating CEACAM1. Oncoimmunology (2019) 8(6):e1581531. doi: 10.1080/2162402X.2019.1581531
239. Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, et al. Fusobacterium Nucleatum Promotes Colorectal Cancer by Inducing Wnt/β-Catenin Modulator Annexin A1. EMBO Rep (2019) 20(4):e47638. doi: 10.15252/embr.201847638
240. Wu Y, Wu J, Chen T, Li Q, Peng W, Li H, et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis in Mice via a Toll-Like Receptor 4/P21-Activated Kinase 1 Cascade. Dig Dis Sci (2018) 63(5):1210–8. doi: 10.1007/s10620-018-4999-2
241. Ma N, Liu Q, Hou L, Wang Y, Liu Z. MDSCs are Involved in the Protumorigenic Potentials of GM-CSF in Colitis-Associated Cancer. Int J Immunopathol Pharmacol (2017) 30(2):152–62. doi: 10.1177/0394632017711055
242. Tang B, Wang K, Jia Y-P, Zhu P, Fang Y, Zhang Z-J, et al. Fusobacterium Nucleatum-Induced Impairment of Autophagic Flux Enhances the Expression of Proinflammatory Cytokines via ROS in Caco-2 Cells. PloS One (2016) 11(11):e0165701. doi: 10.1371/journal.pone.0165701
243. Chaushu S, Wilensky A, Gur C, Shapira L, Elboim M, Halftek G, et al. Direct Recognition of Fusobacterium Nucleatum by the NK Cell Natural Cytotoxicity Receptor NKp46 Aggravates Periodontal Disease. PloS Pathog (2012) 8(3):e1002601. doi: 10.1371/journal.ppat.1002601
244. Ikegami A, Chung P, Han YW. Complementation of the fadA Mutation in Fusobacterium Nucleatum Demonstrates That The Surface-Exposed Adhesin Promotes Cellular Invasion and Placental Colonization. Infect Immun (2009) 77(7):3075–9. doi: 10.1128/IAI.00209-09
245. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
246. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
247. Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in Colonic Flora and Molecular Features of Colorectal Carcinoma. Cancer Res (2014) 74(5):1311–8. doi: 10.1158/0008-5472.CAN-13-1865
248. Kaplan CW, Lux R, Haake SK, Shi W. The Fusobacterium Nucleatum Outer Membrane Protein RadD is an Arginine-Inhibitable Adhesin Required for Inter-Species Adherence and the Structured Architecture of Multispecies Biofilm. Mol Microbiol (2009) 71(1):35–47. doi: 10.1111/j.1365-2958.2008.06503.x
249. Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, et al. Fap2 of Fusobacterium Nucleatum is a Galactose-Inhibitable Adhesin Involved in Coaggregation, Cell Adhesion, and Preterm Birth. Infect Immun (2015) 83(3):1104–13. doi: 10.1128/IAI.02838-14
250. Keku TO, McCoy AN, Azcarate-Peril AM. Fusobacterium Spp. And Colorectal Cancer: Cause or Consequence? Trends Microbiol (2013) 21(10):506–8. doi: 10.1016/j.tim.2013.08.004
251. Sun C-H, Li B-B, Wang B, Zhao J, Zhang X-Y, Li T-T, et al. The Role of Fusobacterium Nucleatum in Colorectal Cancer: From Carcinogenesis to Clinical Management. Chronic Dis Transl Med (2019) 5(3):178–87. doi: 10.1016/j.cdtm.2019.09.001
252. Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, et al. Invasive Fusobacterium Nucleatum Activates Beta-Catenin Signaling in Colorectal Cancer via a TLR4/P-PAK1 Cascade. Oncotarget (2017) 8(19):31802–14. doi: 10.18632/oncotarget.15992
253. Kaller M, Hermeking H. Interplay Between Transcription Factors and MicroRNAs Regulating Epithelial-Mesenchymal Transitions in Colorectal Cancer. Adv Exp Med Biol (2016) 937:71–92. doi: 10.1007/978-3-319-42059-2_4
254. Sheen MR, Fields JL, Northan B, Lacoste J, Ang L-H, Fiering S. Replication Study: Biomechanical Remodeling of the Microenvironment by Stromal Caveolin-1 Favors Tumor Invasion and Metastasis. Elife (2019) 146(1):148–63. doi: 10.7554/eLife.45120.sa2
255. Chen H-N, Yuan K, Xie N, Wang K, Huang Z, Chen Y, et al. PDLIM1 Stabilizes the E-Cadherin/β-Catenin Complex to Prevent Epithelial-Mesenchymal Transition and Metastatic Potential of Colorectal Cancer Cells. Cancer Res (2016) 76(5):1122–34. doi: 10.1158/0008-5472.CAN-15-1962
256. Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, et al. Novel Evidence for an Oncogenic Role of microRNA-21 in Colitis-Associated Colorectal Cancer. Gut (2016) 65(9):1470–81. doi: 10.1136/gutjnl-2014-308455
257. Chi Y, Zhou D. MicroRNAs in Colorectal Carcinoma–From Pathogenesis to Therapy. J Exp Clin Cancer Res (2016) 35:43. doi: 10.1186/s13046-016-0320-4
258. Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium Nucleatum With Immunity and Molecular Alterations in Colorectal Cancer. World J Gastroenterol (2016) 22(2):557–66. doi: 10.3748/wjg.v22.i2.557
259. Engevik MA, Danhof HA, Ruan W, Engevik AC, Chang-Graham AL, Engevik KA, et al. Fusobacterium Nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. MBio (2021) 12(2):e02706-20. doi: 10.1128/mBio.02706-20
260. Kang W, Jia Z, Tang D, Zhang Z, Gao H, He K, et al. Fusobacterium Nucleatum Facilitates Apoptosis, ROS Generation, and Inflammatory Cytokine Production by Activating AKT/MAPK and NF-κb Signaling Pathways in Human Gingival Fibroblasts. Oxid Med Cell Longev (2019) 2019:1681972. doi: 10.1155/2019/1681972
261. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell (2017) 170(3):548–63. doi: 10.1016/j.cell.2017.07.008
262. Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, et al. Fusobacterium Nucleatum Promotes Chemoresistance to 5-Fluorouracil by Upregulation of BIRC3 Expression in Colorectal Cancer. J Exp Clin Cancer Res (2019) 38(1):1–13. doi: 10.1186/s13046-018-0985-y
263. Stergiopoulou T, Walsh TJ. Fusobacterium Necrophorum Otitis and Mastoiditis in Infants and Young Toddlers. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol (2016) 35(5):735–40. doi: 10.1007/s10096-016-2612-1
264. Yarden-Bilavsky H, Raveh E, Livni G, Scheuerman O, Amir J, Bilavsky E. Fusobacterium Necrophorum Mastoiditis in Children - Emerging Pathogen in an Old Disease. Int J Pediatr Otorhinolaryngol (2013) 77(1):92–6. doi: 10.1016/j.ijporl.2012.10.003
265. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The Bacteroides Fragilis Toxin Gene is Prevalent in the Colon Mucosa of Colorectal Cancer Patients. Clin Infect Dis an Off Publ Infect Dis Soc Am (2015) 60(2):208–15. doi: 10.1093/cid/ciu787
266. Sears CL, Geis AL, Housseau F. Bacteroides Fragilis Subverts Mucosal Biology: From Symbiont to Colon Carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/JCI72334
267. Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides Fragilis Enterotoxin Induces C-Myc Expression and Cellular Proliferation. Gastroenterology (2003) 124(2):392–400. doi: 10.1053/gast.2003.50047
268. Wu S, Lim K-C, Huang J, Saidi RF, Sears CL. Bacteroides Fragilis Enterotoxin Cleaves the Zonula Adherens Protein, E-Cadherin. Proc Natl Acad Sci (1998) 95(25):14979–84. doi: 10.1073/pnas.95.25.14979
269. Cheng WT, Kantilal HK, Davamani F. The Mechanism of Bacteroides Fragilis Toxin Contributes to Colon Cancer Formation. Malays J Med Sci (2020) 27(4):9–21. doi: 10.21315/mjms2020.27.4.2
270. Wick EC, Rabizadeh S, Albesiano E, Wu X, Wu S, Chan J, et al. Stat3 Activation in Murine Colitis Induced by Enterotoxigenic Bacteroides Fragilis. Inflamm Bowel Dis (2014) 20(5):821–34. doi: 10.1097/MIB.0000000000000019
271. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota Organization Is a Distinct Feature of Proximal Colorectal Cancers. Proc Natl Acad Sci USA (2014) 111(51):18321–6. doi: 10.1073/pnas.1406199111
272. Pierce JV, Bernstein HD. Genomic Diversity of Enterotoxigenic Strains of Bacteroides Fragilis. PloS One (2016) 11(6):e0158171. doi: 10.1371/journal.pone.0158171
273. Liu Q-Q, Li C-M, Fu L-N, Wang H-L, Tan J, Wang Y-Q, et al. Enterotoxigenic Bacteroides Fragilis Induces the Stemness in Colorectal Cancer via Upregulating Histone Demethylase JMJD2B. Gut Microbes (2020) 12(1):1788900. doi: 10.1080/19490976.2020.1788900
274. Yu H, Pardoll D, Jove R. STATs in Cancer Inflammation and Immunity: A Leading Role for STAT3. Nat Rev Cancer (2009) 9(11):798–809. doi: 10.1038/nrc2734
275. Klampfer L. Cytokines, Inflammation and Colon Cancer. Curr Cancer Drug Targets (2011) 11(4):451–64. doi: 10.2174/156800911795538066
276. Zhang Z, Zheng F, Yu Z, Hao J, Chen M, Yu W, et al. XRCC5 Cooperates With P300 to Promote Cyclooxygenase-2 Expression and Tumor Growth In Colon Cancers. PloS One (2017) 12(10):e0186900. doi: 10.1371/journal.pone.0186900
277. Sheng H, Shao J, Washington MK, DuBois RN. Prostaglandin E2 Increases Growth and Motility of Colorectal Carcinoma Cells. J Biol Chem (2001) 276(21):18075–81. doi: 10.1074/jbc.M009689200
278. Kakiuchi Y, Tsuji S, Tsujii M, Murata H, Kawai N, Yasumaru M, et al. Cyclooxygenase-2 Activity Altered the Cell-Surface Carbohydrate Antigens on Colon Cancer Cells and Enhanced Liver Metastasis. Cancer Res (2002) 62(5):1567–72.
279. Purcell RV, Pearson J, Frizelle FA, Keenan JI. Comparison of Standard, Quantitative and Digital PCR in the Detection of Enterotoxigenic Bacteroides Fragilis. Sci Rep (2016) 6:34554. doi: 10.1038/srep34554
280. Formica V, Cereda V, Nardecchia A, Tesauro M, Roselli M. Immune Reaction and Colorectal Cancer: Friends or Foes? World J Gastroenterol (2014) 20(35):12407–19. doi: 10.3748/wjg.v20.i35.12407
281. Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-Regulation of Cyclooxygenase 2 Gene Expression in Human Colorectal Adenomas and Adenocarcinomas. Gastroenterology (1994) 107(4):1183–8. doi: 10.1016/0016-5085(94)90246-1
282. Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, et al. Enterotoxigenic Bacteroides Fragilis Promotes Intestinal Inflammation and Malignancy by Inhibiting Exosome-Packaged miR-149-3pf. Gastroenterology (2021) 161(5):1552–66.e12. doi: 10.1053/j.gastro.2021.08.003
283. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides Fragilis Toxin Coordinates a Pro-Carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe (2018) 23(2):203–14.e5. doi: 10.1016/j.chom.2018.01.007
284. Hsu C-J, Wu M-H, Chen C-Y, Tsai C-H, Hsu H-C, Tang C-H. AMP-Activated Protein Kinase Activation Mediates CCL3-Induced Cell Migration and Matrix Metalloproteinase-2 Expression in Human Chondrosarcoma. Cell Commun Signal (2013) 11(1):1–15. doi: 10.1186/1478-811X-11-68
285. Xie X, Jiang D, Zhou X, Ye X, Yang P, He Y. Recombinant Bacteroides Fragilis Enterotoxin-1 (rBFT-1) Promotes Proliferation of Colorectal Cancer via CCL3-Related Molecular Pathways. Open Life Sci (2021) 16(1):408–18. doi: 10.1515/biol-2021-0043
286. Takesue Y, Kusachi S, Mikamo H, Sato J, Watanabe A, Kiyota H, et al. Antimicrobial Susceptibility of Common Pathogens Isolated From Postoperative Intra-Abdominal Infections in Japan. J Infect Chemother Off J Japan Soc Chemother (2018) 24(5):330–40. doi: 10.1016/j.jiac.2018.02.011
287. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
288. THE GLOBAL CANCER OBSERVATORY. Available at: https://gco.iarc.fr/tomorrow/en.
289. Lian W-Q, Luo F, Song X-L, Lu Y-J, Zhao S-C. Gonorrhea and Prostate Cancer Incidence: An Updated Meta-Analysis of 21 Epidemiologic Studies. Med Sci Monit (2015) 21:1902–10. doi: 10.12659/MSM.893579
290. Dennis LK, Lynch CF, Torner JC. Epidemiologic Association Between Prostatitis and Prostate Cancer. Urology (2002) 60(1):78–83. doi: 10.1016/S0090-4295(02)01637-0
291. Chung SD, Lin YK, Huang CC, Lin HC. Increased Risk of Prostate Cancer Following Sexually Transmitted Infection in an Asian Population. Epidemiol Infect (2013) 141(12):2663–70. doi: 10.1017/S0950268813000459
292. Källström H, Liszewski MK, Atkinson JP, Jonsson AB. Membrane Cofactor Protein (MCP or CD46) is a Cellular Pilus Receptor for Pathogenic Neisseria. Mol Microbiol (1997) 25(4):639–47. doi: 10.1046/j.1365-2958.1997.4841857.x
293. Merz AJ, Enns CA, So M. Type IV Pili of Pathogenic Neisseriae Elicit Cortical Plaque Formation in Epithelial Cells. Mol Microbiol (1999) 32(6):1316–32. doi: 10.1046/j.1365-2958.1999.01459.x
294. Binnicker MJ, Williams RD, Apicella MA. Gonococcal Porin IB Activates NF-kappaB in Human Urethral Epithelium and Increases The Expression of Host Antiapoptotic Factors. Infect Immun (2004) 72(11):6408–17. doi: 10.1128/IAI.72.11.6408-6417.2004
295. Follows SA, Murlidharan J, Massari P, Wetzler LM, Genco CA. Neisseria Gonorrhoeae Infection Protects Human Endocervical Epithelial Cells From Apoptosis via Expression of Host Antiapoptotic Proteins. Infect Immun (2009) 77(9):3602–10. doi: 10.1128/IAI.01366-08
296. Haverkamp J, Charbonneau B, Ratliff TL. Prostate Inflammation and its Potential Impact on Prostate Cancer: A Current Review. J Cell Biochem (2008) 103(5):1344–53. doi: 10.1002/jcb.21536
297. Wang W, Bergh A, Damber J-E. Morphological Transition of Proliferative Inflammatory Atrophy to High-Grade Intraepithelial Neoplasia and Cancer in Human Prostate. Prostate (2009) 69(13):1378–86. doi: 10.1002/pros.20992
298. Lu P, Wang S, Lu Y, Neculai D, Sun Q, van der Veen S. A Subpopulation of Intracellular Neisseria Gonorrhoeae Escapes Autophagy-Mediated Killing Inside Epithelial Cells. J Infect Dis (2019) 219(1):133–44. doi: 10.1093/infdis/jiy237
299. Jones A, Jonsson A-B, Aro H. Neisseria Gonorrhoeae Infection Causes a G1 Arrest in Human Epithelial Cells. FASEB J Off Publ Fed Am Soc Exp Biol (2007) 21(2):345–55. doi: 10.1096/fj.06-6675com
300. Klein EA, Silverman R. Inflammation, Infection, and Prostate Cancer. Curr Opin Urol (2008) 18(3):315–9. doi: 10.1097/MOU.0b013e3282f9b3b7
301. Vielfort K, Söderholm N, Weyler L, Vare D, Löfmark S, Aro H. Neisseria Gonorrhoeae Infection Causes DNA Damage and Affects the Expression of P21, P27 and P53 in Non-Tumor Epithelial Cells. J Cell Sci (2013) 126(1):339–47. doi: 10.1242/jcs.117721
302. Weyler L, Engelbrecht M, Mata Forsberg M, Brehwens K, Vare D, Vielfort K, et al. Restriction Endonucleases From Invasive Neisseria Gonorrhoeae Cause Double-Strand Breaks and Distort Mitosis in Epithelial Cells During Infection. PloS One (2014) 9(12):e114208. doi: 10.1371/journal.pone.0114208
303. Sutcliffe S, Giovannucci E, De Marzo AM, Leitzmann MF, Willett WC, Platz EA. Gonorrhea, Syphilis, Clinical Prostatitis, and the Risk of Prostate Cancer. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored by Am Soc Prev Oncol (2006) 15(11):2160–6. doi: 10.1158/1055-9965.EPI-05-0913
304. Cheng I, Witte JS, Jacobsen SJ, Haque R, Quinn VP, Quesenberry CP, et al. Prostatitis, Sexually Transmitted Diseases, and Prostate Cancer: The California Men’s Health Study. PloS One (2010) 5(1):e8736. doi: 10.1371/journal.pone.0008736
305. Gottlieb SL, Ndowa F, Hook EW, Deal C, Bachmann L, Abu-Raddad L, et al. Gonococcal Vaccines: Public Health Value and Preferred Product Characteristics; Report of a WHO Global Stakeholder Consultation, January 2019. Vaccine (2020) 38(28):4362–73. doi: 10.1016/j.vaccine.2020.02.073
306. Wang Y-C, Chung C-H, Chen J-H, Chiang M-H, Ti-Yin, Tsao C-H, et al. Gonorrhea Infection Increases the Risk of Prostate Cancer in Asian Population: A Nationwide Population-Based Cohort Study. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol (2017) 36(5):813–21. doi: 10.1007/s10096-016-2866-7
307. Cohen RJ, Shannon BA, McNeal JE, Shannon T, Garrett KL. Propionibacterium Acnes Associated With Inflammation in Radical Prostatectomy Specimens: A Possible Link to Cancer Evolution? J Urol (2005) 173(6):1969–74. doi: 10.1097/01.ju.0000158161.15277.78
308. Sfanos KS, Isaacs WB. An Evaluation of PCR Primer Sets Used for Detection of Propionibacterium Acnes in Prostate Tissue Samples. Prostate (2008) 68(14):1492–5. doi: 10.1002/pros.20820
309. Mak TN, Yu S-H, De Marzo AM, Brüggemann H, Sfanos KS. Multilocus Sequence Typing (MLST) Analysis of Propionibacterium Acnes Isolates From Radical Prostatectomy Specimens. Prostate (2013) 73(7):770–7. doi: 10.1002/pros.22621
310. Valanne S, McDowell A, Ramage G, Tunney MM, Einarsson GG, O’Hagan S, et al. CAMP Factor Homologues in Propionibacterium Acnes: A New Protein Family Differentially Expressed by Types I and II. Microbiology (2005) 151(Pt 5):1369–79. doi: 10.1099/mic.0.27788-0
311. Squaiella CC, Ananias RZ, Mussalem JS, Braga EG, Rodrigues EG, Travassos LR, et al. In Vivo and In Vitro Effect of Killed Propionibacterium Acnes and its Purified Soluble Polysaccharide on Mouse Bone Marrow Stem Cells and Dendritic Cell Differentiation. Immunobiology (2006) 211(1–2):105–16. doi: 10.1016/j.imbio.2005.10.013
312. Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, et al. The Complete Genome Sequence of Propionibacterium Acnes, a Commensal of Human Skin. Science (2004) 305(5684):671–3. doi: 10.1126/science.1100330
313. Holland C, Mak TN, Zimny-Arndt U, Schmid M, Meyer TF, Jungblut PR, et al. Proteomic Identification of Secreted Proteins of Propionibacterium Acnes. BMC Microbiol (2010) 10(1):1–11. doi: 10.1186/1471-2180-10-230
314. Drott JB, Alexeyev O, Bergström P, Elgh F, Olsson J. Propionibacterium Acnes Infection Induces Upregulation of Inflammatory Genes and Cytokine Secretion in Prostate Epithelial Cells. BMC Microbiol (2010) 10:126. doi: 10.1186/1471-2180-10-126
315. Radej S, Płaza P, Olender A, Szewc M, Bar K, Maciejewski R. Infiltrating Treg and Th17 Cells of the Prostate Hypertrophy Gland Associated With Propionibacterium Acnes Infection. Res Rep Urol (2020) 12:593–7. doi: 10.2147/RRU.S284066
316. Liu J, Duan Y, Cheng X, Chen X, Xie W, Long H, et al. IL-17 Is Associated With Poor Prognosis and Promotes Angiogenesis via Stimulating VEGF Production of Cancer Cells in Colorectal Carcinoma. Biochem Biophys Res Commun (2011) 407(2):348–54. doi: 10.1016/j.bbrc.2011.03.021
317. Saleh R, Elkord E. FoxP3(+) T Regulatory Cells in Cancer: Prognostic Biomarkers and Therapeutic Targets. Cancer Lett (2020) 490:174–85. doi: 10.1016/j.canlet.2020.07.022
318. Shannon BA, Cohen RJ, Garrett KL. The Antibody Response to Propionibacterium Acnes Is an Independent Predictor of Serum Prostate-Specific Antigen Levels in Biopsy-Negative Men. BJU Int (2008) 101(4):429–35. doi: 10.1111/j.1464-410X.2007.07214.x
319. Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, et al. Prevalence of Propionibacterium Acnes in Diseased Prostates and Its Inflammatory and Transforming Activity on Prostate Epithelial Cells. Int J Med Microbiol (2011) 301(1):69–78. doi: 10.1016/j.ijmm.2010.08.014
320. Talib WH, Saleh S. Propionibacterium Acnes Augments Antitumor, Anti-Angiogenesis and Immunomodulatory Effects of Melatonin on Breast Cancer Implanted in Mice. PloS One (2015) 10(4):e0124384. doi: 10.1371/journal.pone.0124384
321. Severi G, Shannon BA, Hoang HN, Baglietto L, English DR, Hopper JL, et al. Plasma Concentration of Propionibacterium Acnes Antibodies and Prostate Cancer Risk: Results From an Australian Population-Based Case-Control Study. Br J Cancer (2010) 103(3):411–5. doi: 10.1038/sj.bjc.6605757
322. Johansson M, Denardo DG, Coussens LM. Polarized Immune Responses Differentially Regulate Cancer Development. Immunol Rev (2008) 222:145–54. doi: 10.1111/j.1600-065X.2008.00600.x
323. Savli H, Szendröi A, Romics I, Nagy B. Gene Network and Canonical Pathway Analysis in Prostate Cancer: A Microarray Study. Exp Mol Med (2008) 40(2):176–85. doi: 10.3858/emm.2008.40.2.176
324. Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Inflammation and Cancer: How Hot Is the Link? Biochem Pharmacol (2006) 72(11):1605–21. doi: 10.1016/j.bcp.2006.06.029
325. Shariat SF, Karam JA, Margulis V, Karakiewicz PI. New Blood-Based Biomarkers for the Diagnosis, Staging and Prognosis of Prostate Cancer. BJU Int (2008) 101(6):675–83. doi: 10.1111/j.1464-410X.2007.07283.x
326. Buettner R, Mora LB, Jove R. Activated STAT Signaling in Human Tumors Provides Novel Molecular Targets for Therapeutic Intervention. Clin Cancer Res an Off J Am Assoc Cancer Res (2002) 8(4):945–54.
327. Korniluk A, Koper O, Kemona H, Dymicka-Piekarska V. From Inflammation to Cancer. Ir J Med Sci (2017) 186(1):57–62. doi: 10.1007/s11845-016-1464-0
328. Wierstra I. FOXM1 (Forkhead Box M1) in Tumorigenesis: Overexpression in Human Cancer, Implication in Tumorigenesis, Oncogenic Functions, Tumor-Suppressive Properties, and Target of Anticancer Therapy. Adv Cancer Res (2013) 119:191–419. doi: 10.1016/B978-0-12-407190-2.00016-2
329. Huggins C, Hodges CV. Studies on Prostatic Cancer: I. The Effect of Castration, of Estrogen and of Androgen Injection on Serum Phosphatases in Metastatic Carcinoma of the Prostate. CA Cancer J Clin (1972) 22(4):232–40. doi: 10.3322/canjclin.22.4.232
330. Klap J, Schmid M, Loughlin KR. The Relationship Between Total Testosterone Levels and Prostate Cancer: A Review of the Continuing Controversy. J Urol (2015) 193(2):403–14. doi: 10.1016/j.juro.2014.07.123
331. Olsson J, Drott JB, Laurantzon L, Laurantzon O, Bergh A, Elgh F. Chronic Prostatic Infection and Inflammation by Propionibacterium Acnes in a Rat Prostate Infection Model. PloS One (2012) 7(12):e51434. doi: 10.1371/journal.pone.0051434
332. Shinohara DB, Vaghasia AM, Yu S-H, Mak TN, Brüggemann H, Nelson WG, et al. A Mouse Model of Chronic Prostatic Inflammation Using a Human Prostate Cancer-Derived Isolate of Propionibacterium Acnes. Prostate (2013) 73(9):1007–15. doi: 10.1002/pros.22648
333. Khalili M, Mutton LN, Gurel B, Hicks JL, De Marzo AM, Bieberich CJ. Loss of Nkx3.1 Expression in Bacterial Prostatitis: A Potential Link Between Inflammation and Neoplasia. Am J Pathol (2010) 176(5):2259–68. doi: 10.2353/ajpath.2010.080747
334. Achermann Y, Goldstein EJC, Coenye T, Shirtliff ME. Propionibacterium Acnes: From Commensal to Opportunistic Biofilm-Associated Implant Pathogen. Clin Microbiol Rev (2014) 27(3):419–40. doi: 10.1128/CMR.00092-13
335. Barile MF, Leventhal BG. Possible Mechanism for Mycoplasma Inhibition of Lymphocyte Transformation Induced by Phytohaemagglutinin. Nature (1968) 219(5155):751–2. doi: 10.1038/219751a0
336. Dmochowski L, Taylor HG, Grey CE, Dreyer DA, Sykes JA, Langford PL, et al. Viruses and Mycoplasma (PPLO) in Human Leukemia. Cancer (1965) 18(10):1345–68. doi: 10.1002/1097-0142(196510)18:10%3C1345::AID-CNCR2820181021%3E3.0.CO
337. Murphy WH, Bullis C, Dabich L, Heyn R, Zarafonetis CJD. Isolation of Mycoplasma From Leukemic and Nonleukemic Patients2. JNCI J Natl Cancer Inst (1970) 45(2):243–51. doi: 10.1093/jnci/45.2.243
338. Huang S, Li JY, Wu J, Meng L, Shou CC. Mycoplasma Infections and Different Human Carcinomas. World J Gastroenterol (2001) 7(2):266–9. doi: 10.3748/wjg.v7.i2.266
339. Rogers MB. Mycoplasma and Cancer: In Search of the Link. Oncotarget (2011) 2(4):271–3. doi: 10.18632/oncotarget.264
340. Razin S. Medical Microbiology, 4th. Barone S, editor. Galveston (TX): University of Texas Medical Branch at Galveston (1996).
341. Zarei O, Rezania S, Mousavi A. Mycoplasma Genitalium and Cancer: A Brief Review. Asian Pacif J Cancer Prev (2013) 14(6):3425–8. doi: 10.7314/APJCP.2013.14.6.3425
342. Ainsworth JG, Easterbrook PJ, Clarke J, Gilroy CB, Taylor-Robinson D. An Association of Disseminated Mycoplasma Fermentans in HIV-1 Positive Patients With non-Hodgkin’s Lymphoma. Int J STD AIDS (2001) 12(8):499–504. doi: 10.1258/0956462011923589
343. Atallah S, Berçot B, Laurence V, Hoffmann C. Association of Mycoplasma Hominis and Head and Neck Cancer With Unknown Primary. Eur Ann Otorhinolaryngol Head Neck Dis (2020) 137(1):69–71. doi: 10.1016/j.anorl.2019.05.020
344. Klein C, Samwel K, Kahesa C, Mwaiselage J, West JT, Wood C, et al. Mycoplasma Co-Infection Is Associated With Cervical Cancer Risk. Cancers (2020) 12:1093. doi: 10.3390/cancers12051093
345. Sfanos KS, De Marzo AM. Prostate Cancer and Inflammation: The Evidence. Histopathology (2012) 60(1):199–215. doi: 10.1111/j.1365-2559.2011.04033.x
346. Khan S, Zakariah M, Palaniappan S. Computational Prediction of Mycoplasma Hominis Proteins Targeting in Nucleus of Host Cell and Their Implication in Prostate Cancer Etiology. Tumor Biol (2016) 37(8):10805–13. doi: 10.1007/s13277-016-4970-9
347. Karan D, Dubey S. From Inflammation to Prostate Cancer: The Role of Inflammasomes. Ather MH, Editor. Adv Urol (2016) 2016:3140372. doi: 10.1155/2016/3140372
348. Caini S, Gandini S, Dudas M, Bremer V, Severi E, Gherasim A. Sexually Transmitted Infections and Prostate Cancer Risk: A Systematic Review and Meta-Analysis. Cancer Epidemiol (2014) 38(4):329–38. doi: 10.1016/j.canep.2014.06.002
349. Sethi S, Singh G, Samanta P, Sharma M. Mycoplasma Genitalium: An Emerging Sexually Transmitted Pathogen. Indian J Med Res (2012) 136(6):942–55.
350. Frølund M, Wikström A, Lidbrink P, Abu Al-Soud W, Larsen N, Harder CB, et al. The Bacterial Microbiota in First-Void Urine From Men With and Without Idiopathic Urethritis. PloS One (2018) 13(7):e0201380. doi: 10.1371/journal.pone.0201380
351. Namiki K, Goodison S, Porvasnik S, Allan RW, Iczkowski KA, Urbanek C, et al. Persistent Exposure to Mycoplasma Induces Malignant Transformation of Human Prostate Cells. PloS One (2009) 4(9):e6872. doi: 10.1371/journal.pone.0006872
352. Benedetti F, Curreli S, Zella D. Mycoplasmas–Host Interaction: Mechanisms of Inflammation and Association With Cellular Transformation. Microorganisms (2020) 8:1351. doi: 10.3390/microorganisms8091351
353. Borchsenius SN, Vishnyakov IE, Chernova OA, Chernov VM, Barlev NA. Effects of Mycoplasmas on the Host Cell Signaling Pathways. Pathogens (2020) 9:308. doi: 10.3390/pathogens9040308
354. Qin L, Chen Y, You X. Subversion of the Immune Response by Human Pathogenic Mycoplasmas. Front Microbiol (2019) 10:1934. doi: 10.3389/fmicb.2019.01934
355. Zou S, Fang L, Lee M-H. Dysbiosis of Gut Microbiota in Promoting the Development of Colorectal Cancer. Gastroenterol Rep (2018) 6(1):1–12. doi: 10.1093/gastro/gox031
356. Cimolai N. Do Mycoplasmas Cause Human Cancer? Can J Microbiol (2001) 47(8):691–7. doi: 10.1139/w01-053
357. Tsai S, Wear DJ, Shih JW, Lo SC. Mycoplasmas and Oncogenesis: Persistent Infection and Multistage Malignant Transformation. Proc Natl Acad Sci U S A. (1995) 92(22):10197–201. doi: 10.1073/pnas.92.22.10197
358. Xu Y, Li H, Chen W, Yao X, Xing Y, Wang X, et al. Mycoplasma Hyorhinis Activates the NLRP3 Inflammasome and Promotes Migration and Invasion of Gastric Cancer Cells. PloS One (2013) 8(11):e77955–5. doi: 10.1371/journal.pone.0077955
359. Feng SH, Tsai S, Rodriguez J, Lo SC. Mycoplasmal Infections Prevent Apoptosis and Induce Malignant Transformation of Interleukin-3-Dependent 32D Hematopoietic Cells. Mol Cell Biol (1999) 19(12):7995–8002. doi: 10.1128/MCB.19.12.7995
360. Razin S, Yogev D, Naot Y. Molecular Biology and Pathogenicity of Mycoplasmas. Microbiol Mol Biol Rev (1998) 62(4):1094–156. doi: 10.1128/MMBR.62.4.1094-1156.1998
361. Zhang B, Shih JW, Wear DJ, Tsai S, Lo SC. High-Level Expression of H-Ras and C-Myc Oncogenes in Mycoplasma-Mediated Malignant Cell Transformation. Proc Soc Exp Biol Med Soc Exp Biol Med (New York NY) (1997) 214(4):359–66. doi: 10.3181/00379727-214-44104
362. Zhang S, Tsai S, Lo S-C. Alteration of Gene Expression Profiles During Mycoplasma-Induced Malignant Cell Transformation. BMC Cancer (2006) 6:116. doi: 10.1186/1471-2407-6-116
363. Benedetti F, Cocchi F, Latinovic OS, Curreli S, Krishnan S, Munawwar A, et al. Role of Mycoplasma Chaperone DnaK in Cellular Transformation. Int J Mol Sci (2020) 21:1311. doi: 10.3390/ijms21041311
364. Zella D, Curreli S, Benedetti F, Krishnan S, Cocchi F, Latinovic OS, et al. Mycoplasma Promotes Malignant Transformation In Vivo, and its DnaK, a Bacterial Chaperone Protein, has Broad Oncogenic Properties. Proc Natl Acad Sci USA (2018) 115(51):E12005–14. doi: 10.1073/pnas.1815660115
365. Goodison S, Nakamura K, Iczkowski KA, Anai S, Boehlein SK, Rosser CJ. Exogenous Mycoplasmal P37 Protein Alters Gene Expression, Growth and Morphology of Prostate Cancer Cells. Cytogenet Genome Res (2007) 118(2–4):204–13. doi: 10.1159/000108302
366. Ketcham CM, Anai S, Reutzel R, Sheng S, Schuster SM, Brenes RB, et al. P37 Induces Tumor Invasiveness. Mol Cancer Ther (2005) 4(7):1031–8. doi: 10.1158/1535-7163.MCT-05-0040
367. Logunov DY, Scheblyakov DV, Zubkova OV, Shmarov MM, Rakovskaya IV, Gurova KV, et al. Mycoplasma Infection Suppresses P53, Activates NF-kappaB and Cooperates With Oncogenic Ras in Rodent Fibroblast Transformation. Oncogene (2008) 27(33):4521–31. doi: 10.1038/onc.2008.103
368. Zhang S, Wear DJ, Lo S. Mycoplasmal Infections Alter Gene Expression in Cultured Human Prostatic and Cervical Epithelial Cells. FEMS Immunol Med Microbiol (2000) 27(1):43–50. doi: 10.1111/j.1574-695X.2000.tb01410.x
369. Reddel RR, Salghetti SE, Willey JC, Ohnuki Y, Ke Y, Gerwin BI, et al. Development of Tumorigenicity in Simian Virus 40-Immortalized Human Bronchial Epithelial Cell Lines. Cancer Res (1993) 53(5):985–91.
370. Pehlivan M, Itirli G, Onay H, Bulut H, Koyuncuoglu M, Pehlivan S. Does Mycoplasma Sp. Play Role in Small Cell Lung Cancer? Lung Cancer (2004) 45(1):129–30. doi: 10.1016/j.lungcan.2004.01.007
371. Pehlivan M, Pehlivan S, Onay H, Koyuncuoglu M, Kirkali Z. Can Mycoplasma-Mediated Oncogenesis be Responsible for Formation of Conventional Renal Cell Carcinoma? Urology (2005) 65(2):411–4. doi: 10.1016/j.urology.2004.10.015
372. De Flora S, Crocetti E, Bonanni P, Ferro A, Vitale F. Incidence of Infection-Associated Cancers in Italy and Prevention Strategies. Epidemiol Prev (2015) 39(4 Suppl 1):14–20.
373. De Flora S, La Maestra S. Epidemiology of Cancers of Infectious Origin and Prevention Strategies. J Prev Med Hyg (2015) 56(1):E15–20.
374. Tan YK, Fielding JWL. Early Diagnosis of Early Gastric Cancer. Eur J Gastroenterol Hepatol (2006) 18(8):821–9. doi: 10.1097/00042737-200608000-00004
375. Leung WK, Wu M, Kakugawa Y, Kim JJ, Yeoh K, Goh KL, et al. Screening for Gastric Cancer in Asia: Current Evidence and Practice. Lancet Oncol (2008) 9(3):279–87. doi: 10.1016/S1470-2045(08)70072-X
376. De Flora S, Ferguson LR. Overview of Mechanisms of Cancer Chemopreventive Agents. Mutat Res (2005) 591(1–2):8–15. doi: 10.1016/j.mrfmmm.2005.02.029
Keywords: oncogenesis, chronic inflammation, Helicobacter pylori, bacteria, carcinogen
Citation: Prasad SK, Bhat S, Shashank D, C. R. A, R. S, Rachtanapun P, Devegowda D, Santhekadur PK and Sommano SR (2022) Bacteria-Mediated Oncogenesis and the Underlying Molecular Intricacies: What We Know So Far. Front. Oncol. 12:836004. doi: 10.3389/fonc.2022.836004
Received: 15 December 2021; Accepted: 22 February 2022;
Published: 04 April 2022.
Edited by:
Marie R. Webster, Lankenau Institute for Medical Research, United StatesReviewed by:
Francesca Benedetti, University of Maryland, United StatesCopyright © 2022 Prasad, Bhat, Shashank, C. R., R., Rachtanapun, Devegowda, Santhekadur and Sommano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasanna K. Santhekadur, cHJhc2FubmFrdW1hcnNAanNzdW5pLmVkdS5pbg==; Devananda Devegowda, ZGV2YW5hbmRkQGpzc3VuaS5lZHUuaW4=; Sarana Rose Sommano, c2FyYW5hLnNAY211LmFjLnRo
†These authors have contributed equally to this work and share first authorship
 Shashanka K. Prasad
Shashanka K. Prasad Smitha Bhat
Smitha Bhat Dharini Shashank2
Dharini Shashank2 Sindhu R.
Sindhu R. Devananda Devegowda
Devananda Devegowda Prasanna K. Santhekadur
Prasanna K. Santhekadur Sarana Rose Sommano
Sarana Rose Sommano