- 1School of Biomedical Sciences and Pharmacy, College of Health, Medicine and Wellbeing, University of Newcastle, Callaghan, NSW, Australia
- 2Hunter Medical Research Institute, New Lambton Heights, NSW, Australia
- 3Translational Research Institute, Henan Provincial People’s Hospital, Academy of Medical Science, Zhengzhou University, Zhengzhou, China
- 4NSW Health Pathology, John Hunter Hospital, New Lambton Heights, NSW, Australia
Triple negative breast cancer (TNBC) is a highly aggressive subtype with a high rate of metastasis, early distant recurrence and resistance to therapy leading to worse survival than other breast cancer subtypes. There are no well-established biomarkers that can determine women who will do better and those who are likely to have poorer outcomes with TNBC, nor are there targeted therapies. Thus, the identification of prognostic and/or predictive biomarkers will enable tailored therapies based on their likelihood of disease outcomes and may prevent over- and under-diagnosis. Previous studies from our laboratory have identified four genes (ANP32E, DSC2, ANKRD30A and IL6ST/gp130) that are specific to TNBC and were associated with lymph node metastasis (LNmets), the earliest indicator of tumor progression via distal spread. This study aimed to validate these findings using absolute quantitation by digital droplet PCR (ddPCR) and to determine relationships with clinicopathological features and survival. Our analysis confirmed all four genes displayed significant expression differences between TNBC cases and non-TNBC cases. Moreover, low IL6ST expression was significantly associated with grade 3 disease, hormone receptor negativity and earlier age at diagnosis; low ANKRD30A expression was associated with tumor size; and high ANP32E expression was significantly associated with grade and the number of positive lymph nodes. Individually, three of the four genes were associated with relapse-free survival in TNBC and in combination, all four genes were significantly associated with TNBC survival, but not in hormone receptor-positive cases. Collectively our results suggest that the four genes may have utility in TNBC prognostication.
Introduction
Triple negative breast cancer (TNBC) is one of the most aggressive subtypes due to it not being amenable to targeted therapies including Tamoxifen and Herceptin (trastuzumab) and it is associated with rapid metastasis, higher risk of recurrence and poorer survival outcomes, when compared to receptor positive breast cancer subtypes (1, 2). Because of the lack of targeted treatment, the current treatment options are limited to chemotherapy and surgery. There are no well-established prognostic biomarkers in TNBC that can be used in disease prognosis. Therefore, identification of prognostic biomarkers to improve treatment regimens and that can potentially be targets for therapy in this breast cancer subtype are urgently required.
Previous studies from our laboratory have identified four genes that are differentially expressed in invasive ductal carcinoma (IDCs) compared to normal adjacent tissues (NATs) in TNBC as well as being differentially expressed in TNBC compared to non-TNBC. These results were validated in two independent cohorts, including a large cohort sourced from The Cancer Genome Atlas (TCGA) using the same method (cDNA microarrays) (3). Ankyrin repeat domain 30A (ANKRD30A) and interleukin 6 signal transducer (IL6ST) were downregulated in TNBC compared to non-TNBC, whereas desmocollin-2 (DSC2) and acidic nuclear phosphoprotein 32 family member E (ANP32E) were upregulated in TNBC compared to non-TNBC. ANKRD30A is a breast differentiation antigen responsible for protein-protein interactions and other cellular functions (4). IL6ST also known as glycoprotein 130 (gp130) is a signal transducer for the interleukin family of cytokines such as IL6, CNTF, LIF and OSM; and an activator of JAK/STAT and MAPK/PI3K/ERK signaling pathways (5). DSC2 is one of the main components of desmosomes, which aid in cell-cell attachments as well as play a key role in cell growth and apoptosis (6). ANP32E is a histone chaperone that has the ability to strip H2A.Z away from DNA, allowing chromatin remodeling and thus altering gene expression (7). Although some of these genes have been implicated in other types of breast cancer, the relationship of these genes to prognosis in TNBC is currently unknown. Moreover, our previous results need to be verified using a different method in order to move these results forward to the clinic.
Amplification by digital droplet PCR (ddPCR) offers several advantages over conventional qRT-PCR; it can be used to calculate the absolute concentration of cDNA in a sample without the need for any standards, as the cDNA within a sample is partitioned into thousands of droplets, amplified and counted directly by Poisson statistics. Because of the sample partitioning and endpoint quantitation used in ddPCR, PCR amplification is independent of reaction efficiency as well as being less susceptible to Taqman polymerase inhibitors compared to that in qPCR in which PCR amplification is dependent on the concentration of inhibitors in the entire sample. Thus ddPCR provides accurate, precise and reproducible data (8, 9) and also can be used for low concentration samples with increased precision (10). Moreover, ddPCR can be utilized to perform multiplexing to detect more than one target in the reaction, reducing the time and cost of the experiment (11).
The aim of this study was to verify the differential expression of ANKRD30A, IL6ST, DSC2 and ANP32E in TNBC compared to NATs and between TNBC and non-TNBC using an independent method (ddPCR) and to define their relationship with clinicopathological features and survival outcomes. We have shown that these genes were significantly different between TNBC cases and non-TNBC cases. Individually, three of the four genes were associated with relapse-free survival in TNBC and when combined, the four genes were significantly associated with survival in TNBC, but not in hormone receptor-positive cases. Thus, ANKRD30A, IL6ST, DSC2 and ANP32E may represent novel prognostic markers for the TNBC subtype.
Materials and Methods
Cohorts
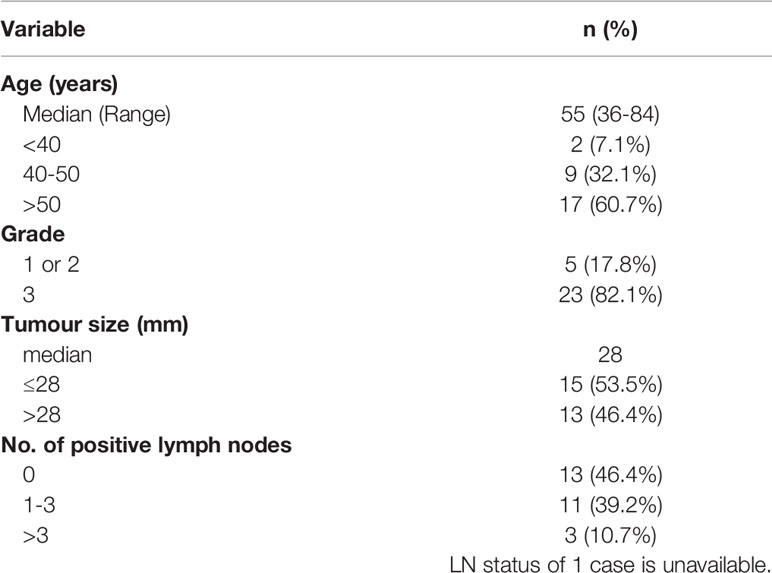
Two cohorts were used in this study. The first cohort consisted of a total of 28 invasive ductal carcinomas of the TNBC subtype, with 13 matched LNmets and 2 unmatched LNmets as well as 8 matched normal adjacent tissue (NAT) and 1 unmatched NAT. All samples were formalin fixed paraffin embedded (FFPE) and obtained by 1.5mm punch biopsy from the archives of NSW Health Pathology, John Hunter Hospital, Newcastle, Australia. This cohort has been described previously (3). Areas of IDC, LNmet and NAT were identified and confirmed by a pathologist. The clinical characteristics of the patients used in this study are shown in Table 1.
A second cohort with a total of 13 TNBCs and 105 non-TNBCs for comparisons with a non-TNBC cohort. The samples were fresh frozen IDC and were provided by the Australian Breast Cancer Tissue Bank (Westmead, NSW, Australia), which have been previously described (12). The cohort characteristics are described in Table 2.
This study complies with the Helsinki Declaration with ethical approval from the Hunter New England Human Research Ethics Committee (Approval number: 09/05/20/5.02). In accordance with the National Statement on Ethical Conduct in Research Involving Humans, a waiver of consent was granted for cases from NSW Health Pathology, whilst all other cases have consented to their tissue and clinical information being used for research.
RNA Extraction and Quantification
RNA extraction of the whole biopsy samples was previously described (13). All samples were stored at -80°C. These were quantitated before cDNA synthesis using the Qubit™ RNA BR (broad range) Assay Kit. The extracted FFPE DNA stored at -20°C was quantitated using the Qubit® dsDNA HS Assay Kit.
Reverse Transcription
Either 75ng RNA from fresh frozen tissues or 125 ng RNA from FFPE tissues was used for cDNA synthesis, the latter amount increased to counter the highly degraded nature of the RNA in FFPE samples. cDNA synthesis was performed as using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Mulgrave, VIC, Australia) to generate complementary DNA (cDNA) according to the manufacturers’ instructions.
Digital Droplet PCR (ddPCR)
A total reaction volume of 25 μl was prepared according to the manufacturers’ instructions (Bio-Rad). The amount of cDNA added to the PCR reaction depended on whether it was extracted from fresh frozen or FFPE tissues, due to the differing amplification efficiencies: 12.5 ng cDNA equivalent to RNA input from FFPE tissues was used in the reaction; while 3.75 ng cDNA equivalent to RNA input that had been reverse transcribed from fresh frozen tissues was used. TaqMan Gene Expression Assays (Life Technologies) for ANKRD30A (Hs00369567_m1), IL6ST (Hs00174360_m1), ANP32E (Hs01064731_m1) and DSC2 (Hs00951428_m1) were used for digital droplet PCR. Droplets were generated in an Automated Droplet QX200 Generator (1864101, Bio-Rad) according to the manufacturers’ instructions. PCR amplification of the cDNA within the droplets was performed using the C1000 thermal cycler (Bio-Rad) according to the manufacturers’ instructions.
After PCR amplification of the target cDNA within the droplets, the sample plate was placed in the droplet reader (QX200 Droplet Reader, 1864003, Bio-Rad) which counts each droplet individually using a fluorescent detection system which is set to detect FAM or HEX/VIC and classifies them as positive or negative droplets based on endpoint fluorescent amplitude. Positive droplets containing at least one copy of the target cDNA molecule have an increased fluorescence compared to negative droplets. The number of positive and negative droplets read by droplet reader was then used by Quantasoft software (Bio-Rad) to calculate the absolute quantity of DNA per sample in copies/μl where it first determines the fraction of positive droplets and after combination with a Poisson algorithm, provides the original concentration of the target template. Based on Poisson statistics, the average copies (of target) per droplet (CPD) was calculated as: CPD = −ln(1−p); where p = fraction of positive droplet. CPD can then be converted into the concentration of target (copies/μl) in the initial sample as shown below:
where Vdroplet is the average droplet volume (μL).
Statistical Analysis
All statistical analysis was performed using GraphPad Prism 7 (San Diego, California, USA). The normality of the distribution was tested using the D’Agostino & Pearson normality test. As some of the groups were not normally distributed, a Kruskal-Wallis test was used to determine if differences in the expression of DSC2, ANP32E and IL6ST were statistically significant between unmatched groups. A two-tailed Mann Whitney test was used to assess if there was a statistically significant difference in the expression of DSC2, ANP32E, IL6ST and ANKRD30A between two groups. To assess if the differential expression was associated with clinical features including age, tumor size, grade and lymph node positivity, a chi-squared test was used. A p value ≤ 0.05 was considered to be statistically significant.
Kaplan-Meier (KM) Plotter Database Analysis
KM plotter is a publicly available online database with gene expression and survival information data downloaded from Gene Expression Omnibus (GEO), European genome-phenome Archive (EGA) and The Cancer Genome Atlas (TCGA). This database was used to perform relapse free survival analysis in TNBC and non-TNBC cases. Each gene of interest was entered into the database to obtain KM survival curves plots and number at risk. The following probe IDs were used for this analysis: ANKRD30A (223864_at), IL6ST (204863_s_at, 204864_s_at, 211000_s_at, 212195_at, 212196_at), DSC2 (204750_s_at, 204751_x_at, 226817_at) and ANP32E (208103_s_at, 221505_at). A total of 255 TNBC cases in the database were available for RFS analysis on ANP32E and IL6ST, while only 161 TNBC cases were available for RFS analysis with the DSC2 and ANKRD30A microarray probes. No gene expression microarray data for DSC2 and ANKRD30A were available in the rest of the 94 TNBC samples. The automatically generated best cut-off (more accurate than median) was chosen to classify the expression of genes into high and low values. The best cut-off provided in the KM plotter is the best performing cut-off with the most statistically significant p-value (Cox regression analysis) from all the possible cut-offs computed automatically by the database between the lower and upper quartiles. The publicly available microarray datasets including E-MTAB-365, E-TABM-43 and GSE (Gene Expression Omnibus Series) in the software were selected to generate the Kaplan-Meier plots in this study. A log rank p-value of 0.05 was considered statistically significant. The hazard ratio, 95% confidence interval and number at risk were obtained using the database.
Results
Verification of Differential mRNA Expression of ANP32E, DSC2, IL6ST and ANKRD30A Between Normal and Tumour Samples by ddPCR
The mRNA expression of ANP32E, DSC2, IL6ST and ANKRD30A was quantified using ddPCR to verify the differential mRNA expression of these genes in IDC (n = 28) compared to NAT (n = 9) as well as LNmets (n = 15). This was performed in the first cohort, which was previously used for gene expression analysis in a recent publication by our group (3). The sample number used in this study varied from the numbers in the previous study as some samples were excluded due to the low RNA amount. In concordance with our previous findings (3), the expression of IL6ST was significantly downregulated in IDCs (median fold change = -0.2) and LNmets (median fold change = -0.2) compared to NATs (p=0.0171 and 0.0020, respectively). The expression of DSC2 (median fold change = 1.3) and ANP32E (median fold change = 1.8) were upregulated in IDCs compared to NATs, however, the increase in expression was not statistically significant (p = 0.4537, p = 0.1743, respectively) (Figure 1). Additionally, the expression of these genes was increased in LNmets when compared to NAT (median fold change for ANP32E = 1.5), but this difference was not significant (p ≥ 0.999 and 0.9864, respectively). ANKRD30A showed very low to no expression in IDCs and LNmets compared to NATs and was undetectable in the majority of samples (Figure 1).
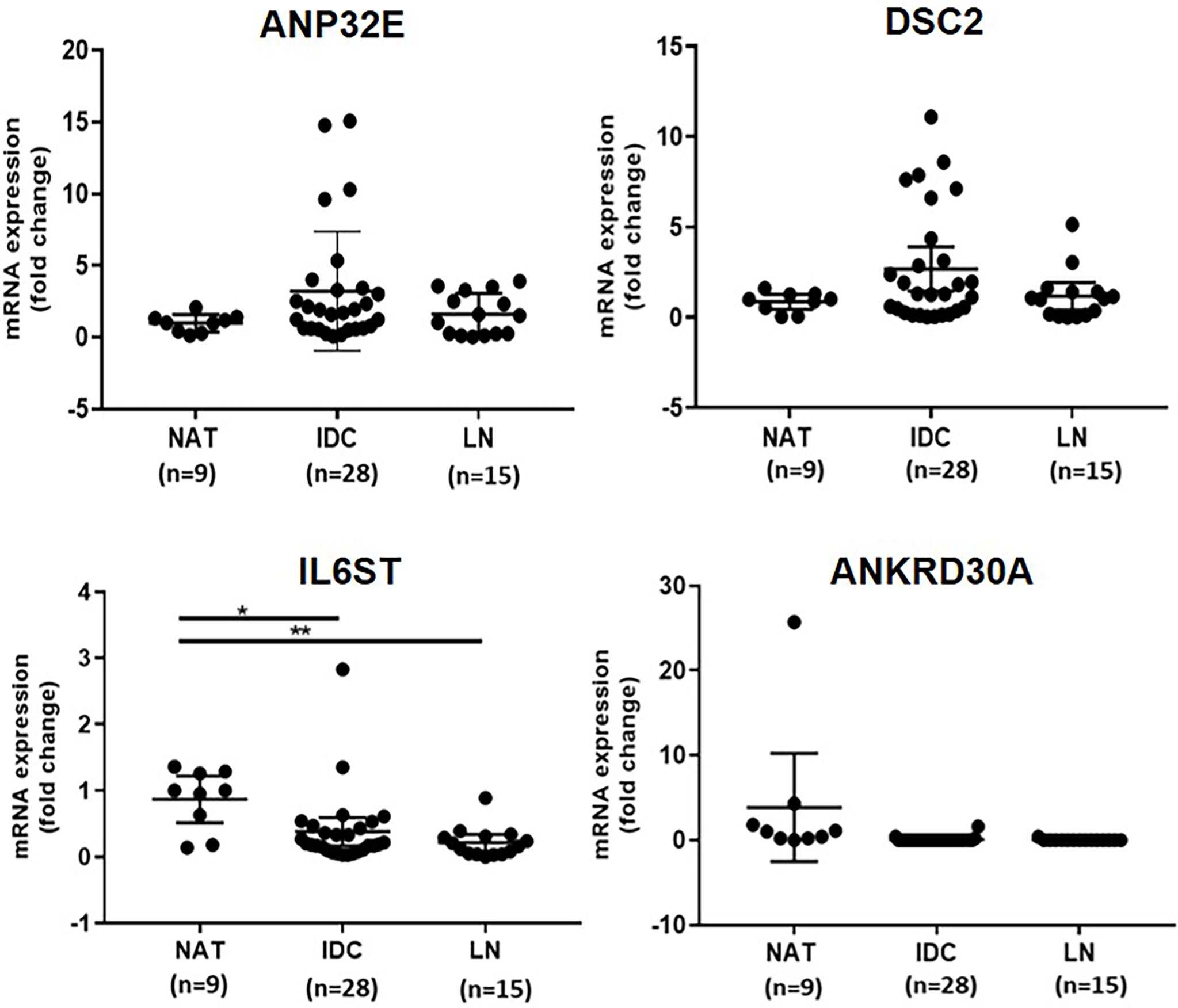
Figure 1 Verification of differential expression of ANP32E, DSC2, IL6ST and ANKRD30A genes at the mRNA level in tumour compared to normal tissue in the first cohort. The mRNA expression of ANP32E, DSC2, IL6ST and ANKRD30A genes obtained as copies per μl using ddPCR. Results show the fold change expression of four genes in IDCs and LNmets compared to NATs. Values are presented as the median ± interquartile range. A Kruskal-Wallis test followed by Dunn’s test for multiple comparisons was performed to determine the statistical significance of the expression. A p value ≤ 0.05 was considered significant. Asterisks in the figure represents statistical significance (*p ≤ 0.05; **p ≤ 0.01). NAT, Normal adjacent tissue; IDC, Invasive ductal carcinoma and LNmet, Lymph node metastasis.
Verification of Differential mRNA Expression of ANP32E, DSC2, IL6ST and ANKRD30A Between TNBC and Non-TNBC by ddPCR
Next, the differential expression of ANP32E, DSC2, IL6ST and ANKRD30A in TNBC compared to non-TNBC was verified using digital droplet PCR. For this analysis, 13 TNBC and 105 non-TNBC samples (second cohort) were used. This was to confirm the differential expression of these genes identified in our previous study at the mRNA level using a different method. In concordance with our previous finding (3), ANP32E (median fold change = 2.1) and DSC2 (median fold change = 3.5) were significantly upregulated in TNBC compared to non-TNBC (p = 0.0009, p<0.0001, respectively), whereas IL6ST (median fold change = -0.2) and ANKRD30A (median fold change = 0.00) were significantly downregulated in TNBC compared to non-TNBC (p <0.0001, p < 0.0001, respectively) (Figure 2).
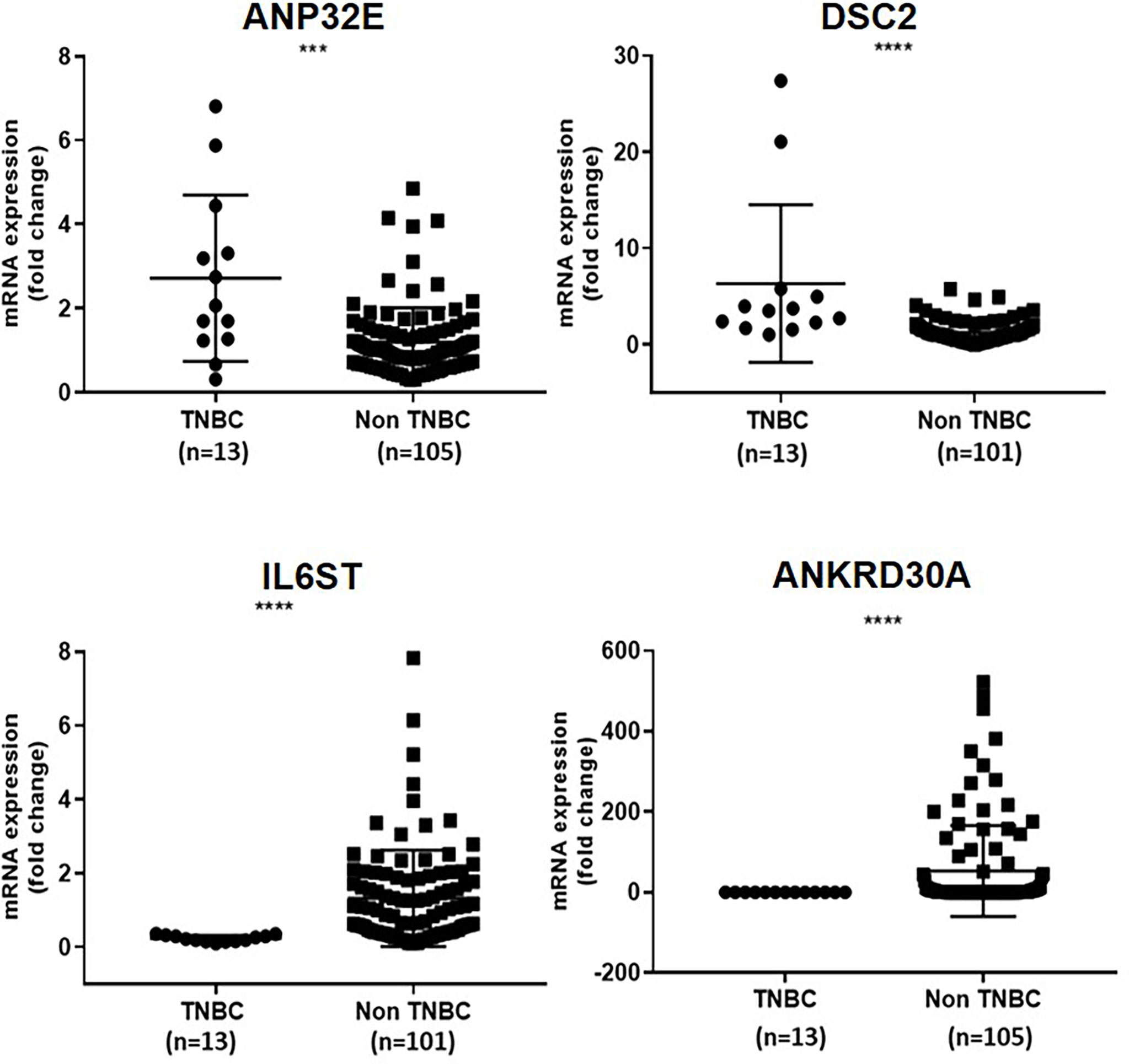
Figure 2 Verification of differential expression of ANP32E, DSC2, IL6ST and ANKRD30A genes in TNBC compared to non-TNBC at the mRNA level in the second cohort. The mRNA expression of ANP32E, DSC2, IL6ST and ANKRD30A genes obtained as copies per μl using ddPCR. Results are shown as fold change expression of target genes in TNBC compared to non-TNBC. Values are presented as the median ± interquartile range. A Mann Whitney test was used to determine the statistical significance of the expression in TNBC. A p value ≤ 0.05 was considered significant. Asterisks in the figure represent statistical significance (***p ≤ 0.001, ****p ≤ 0.0001).
Association of mRNA Expression of ANP32E, DSC2 and IL6ST With Clinicopathological Features
To assess whether the mRNA expression of ANP32E, DSC2 and IL6ST in TNBC were correlated with clinicopathological features, a chi-square test was performed for all genes except ANKRD30A (due to the low number of samples with detectable expression) in relation to age, grade, tumor size, and number of positive lymph nodes. The samples were divided into high (n = 14), and low (n = 14) mRNA expression based on the median expression of each gene within the first cohort. However, the differential mRNA expression of DSC2, ANP32E and IL6ST genes showed no correlation with the clinical characteristics in the first cohort (Table 3).
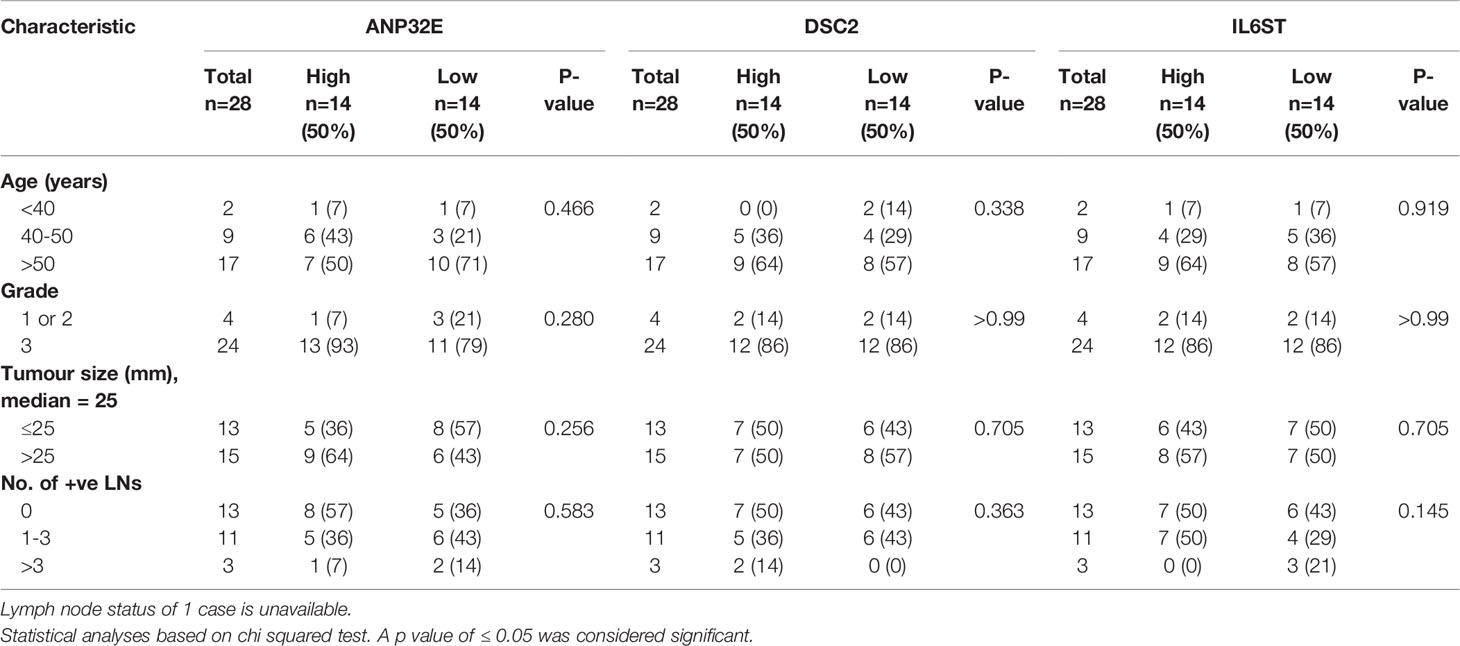
Table 3 Association of ANP32E, DSC2 and IL6ST mRNA expression with clinicopathological features in TNBC cases from the first cohort.
In the second cohort, which contained both receptor positive and negative IDC cases, a chi-square test was performed to determine whether the high or low mRNA expression of the four genes was associated with age, grade, tumor size, number of positive lymph nodes, hormone receptor positivity and TNBC status. ANP32E was significantly associated with grade (p = 0.0017), number of positive lymph nodes (p = 0.0304); and ER, PR and TNBC status (p = 0.0282, 0.0384 and 0.0081, respectively). DSC2 expression was significantly associated with ER, PR and TNBC status (p = 0.001, 0.018 and 0.001, respectively). IL6ST expression showed a significant association with age (p = 0.05), tumor grade (p < 0.0001) as well as with ER, PR, HER2 and TNBC status (p < 0.0001, < 0.0001, 0.0033 and 0.0001, respectively). ANKRD30A expression was significantly associated with tumor size, ER, PR and TNBC status (p = 0.0161, 0.0007, 0.0009, 0.0012, respectively) (Table 4).
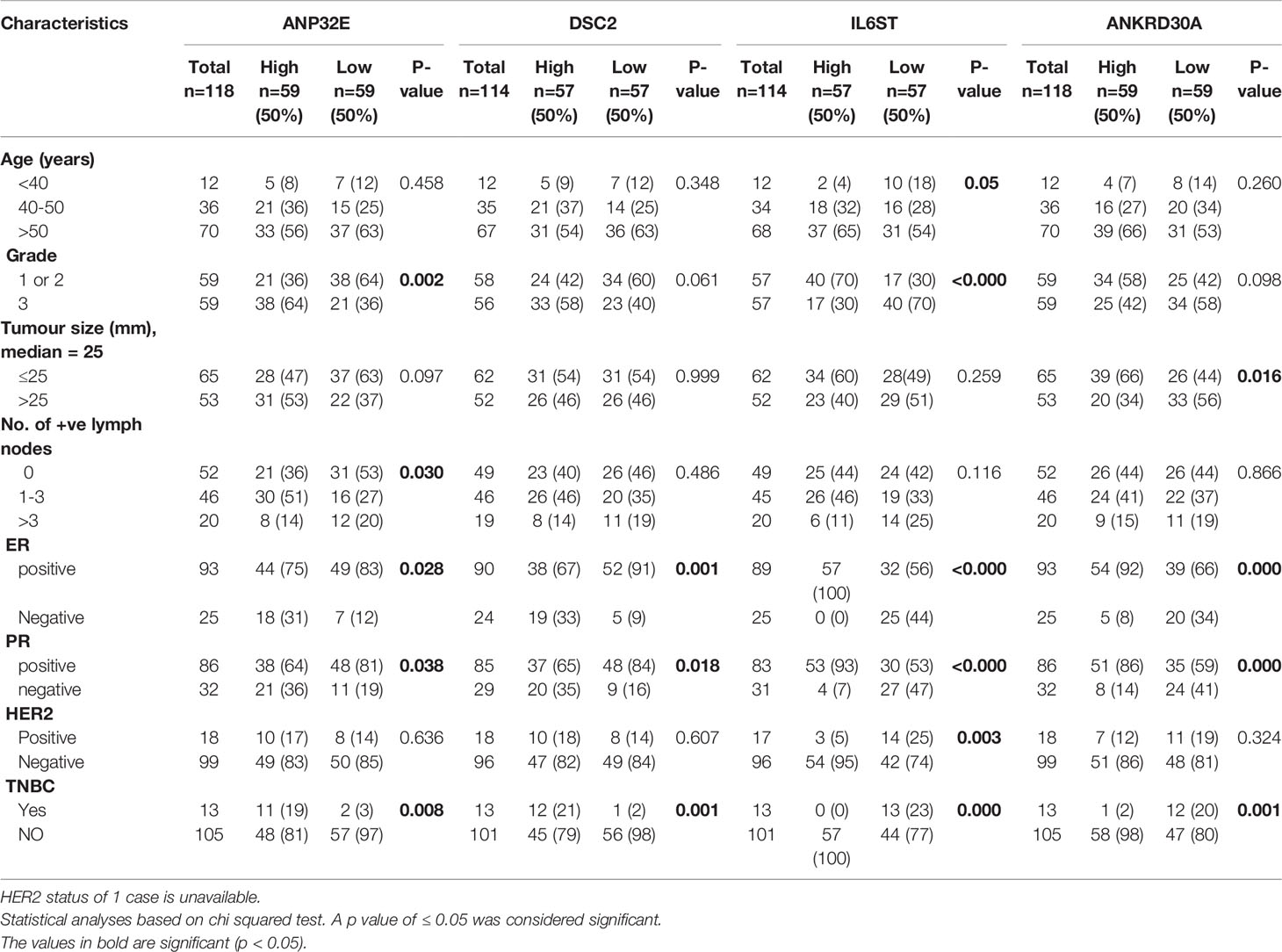
Table 4 Association of ANP32E, DSC2, IL6ST and ANKRD30A mRNA expression with clinicopathological features in breast cancer cases from the second cohort.
Association of mRNA Expression of ANP32E, DSC2, IL6ST and ANKRD30A With Survival
Next, the impact of each of the four genes on survival was assessed after segmenting cases into high and low mRNA expression. Due to the low number of TNBC samples in the first and second cohorts, relapse-free survival (RFS) analysis was performed on the four genes in a larger cohort of TNBC cases using the KM plotter online database, which contains expression values from publicly available microarray data. A total of 255 TNBC cases in the database were available for RFS analysis on ANP32E and IL6ST, while only 161 TNBC cases were available for RFS analysis with the DSC2 and ANKRD30A microarray probes. No gene expression microarray data for DSC2 and ANKRD30A were available in the other 94 TNBC samples. High and low expression of these genes were split based on the automatically generated best cut-off value as described the methods. All the microarray probes that were specifically related to the gene of interest were selected. High expression of ANP32E (p = 0.0092) was significantly associated with decreased RFS while the high expression of DSC2 (p = 0.26) showed a non-significant trend of increased RFS. Low expression of IL6ST (p = 0.011) and ANKRD30A (p = 0.027) was significantly associated with decreased RFS (Figure 3).
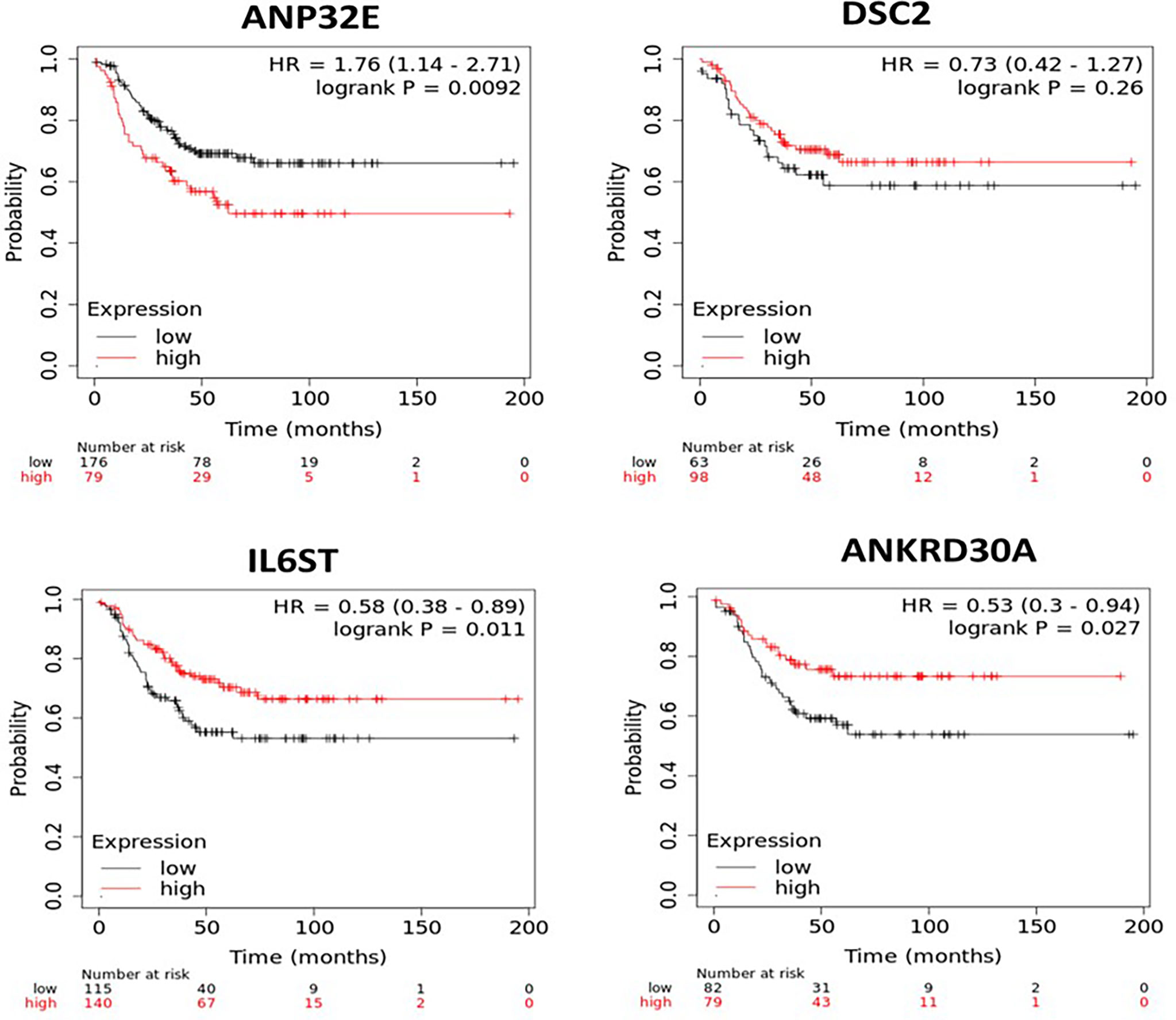
Figure 3 Kaplan-Meier survival curves for patients with high or low mRNA expression of ANP32E, DSC2, IL6ST and ANKRD30A in TNBC samples. The survival analysis was performed in KM plotter using publicly available TNBC microarray data that is available within the database. TNBC samples in the KM plotter database were divided into high or low expression groups based on auto best cut-off expression value of each gene and compared by Kaplan-Meier survival analysis. A Log-rank p-value ≤ 0.05 was considered significant. HR, hazard ratio. The red curve represents high expression, and the black curve represents low expression of the indicated gene.
To determine if the combined expression of the four gene panel was associated with survival, relapse free survival analysis of the four gene panel including ANKRD30A, IL6ST, ANP32E and DSC2 was performed using the KM plotter database in both TNBC and non-TNBC samples. For this, the mean expression of the four genes was selected for survival curve analysis with inverted expression of the two low expressed genes: ANKRD30A and IL6ST and not inverted for the two highly expressed genes: ANP32E and DSC2 in both TNBC and non-TNBC samples. A total of 161 TNBC cases and 467 non-TNBC cases was available for four gene panel survival analysis in the KM plotter database. High expression of the four gene panel was significantly associated with low RFS compared to its low expression in TNBC. In contrast, the differential expression of the 4-gene panel in non-TNBC cases (ER+/PR+/HER2+-) was not significantly associated with RFS (Figure 4).
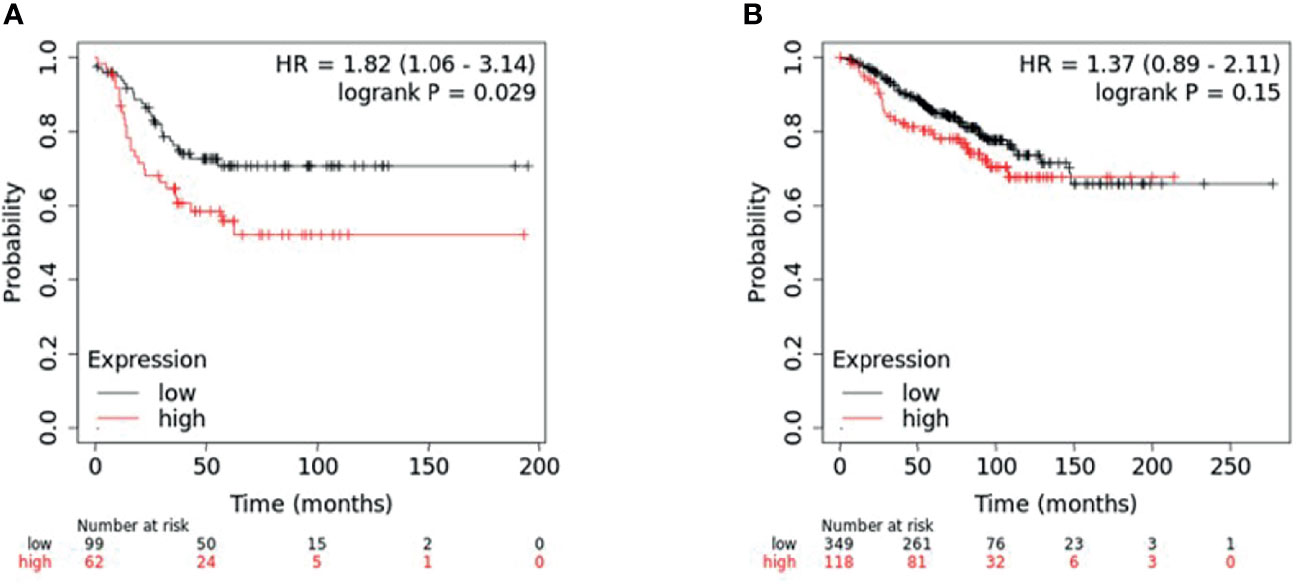
Figure 4 KM plotter survival analysis for patients with differential expression of four-gene panel including ANP32E, DSC2, IL6ST, and ANKRD30A. Survival analysis was performed in KM plotter using publicly available microarray data that is available within the database. (A) TNBC samples (n = 161) and (B) non-TNBC (n = 467) were divided into high or low expression groups based on the median expression of the four-gene panel (with inverted expression of the two low expressed genes: ANKRD30A and IL6ST and not inverted for the two highly expressed genes: ANP32E and DSC2) and compared by Kaplan-Meier survival analysis. A Log-rank p-value ≤ 0.05 was considered significant. HR, hazard ratio. The red curve represents high expression, and the black curve represents low expression.
To determine if the combined expression four gene panel was associated with survival, relapse free survival analysis was performed on the 6 TNBC subtypes (14). The four gene panel was associated with worse RFS (when compared to tumours in the low expression group), in the basal-like 1 and 2 (BL1, BL2) subtypes (HR=2.27, p=0.015; HR=2.52, p=0.013 respectively) and the mesenchymal subtype (HR=1.8; p=0.034). In contrast, this four gene panel was associated with better RFS in the luminal androgen receptor (LAR) subtype (HR=0.56, p=0.031) (Figure 5). Taken together, these results suggest that the four gene panel can predict distinct survival outcomes amongst the distinct TNBC subytpes, implying that this signature would be useful for most TNBC cases. However, it should be noted that the number of cases in these survival analyses is low, particularly in the mesenchymal stem-like and basal-like 2 subgroups, and should be interpreted with caution.
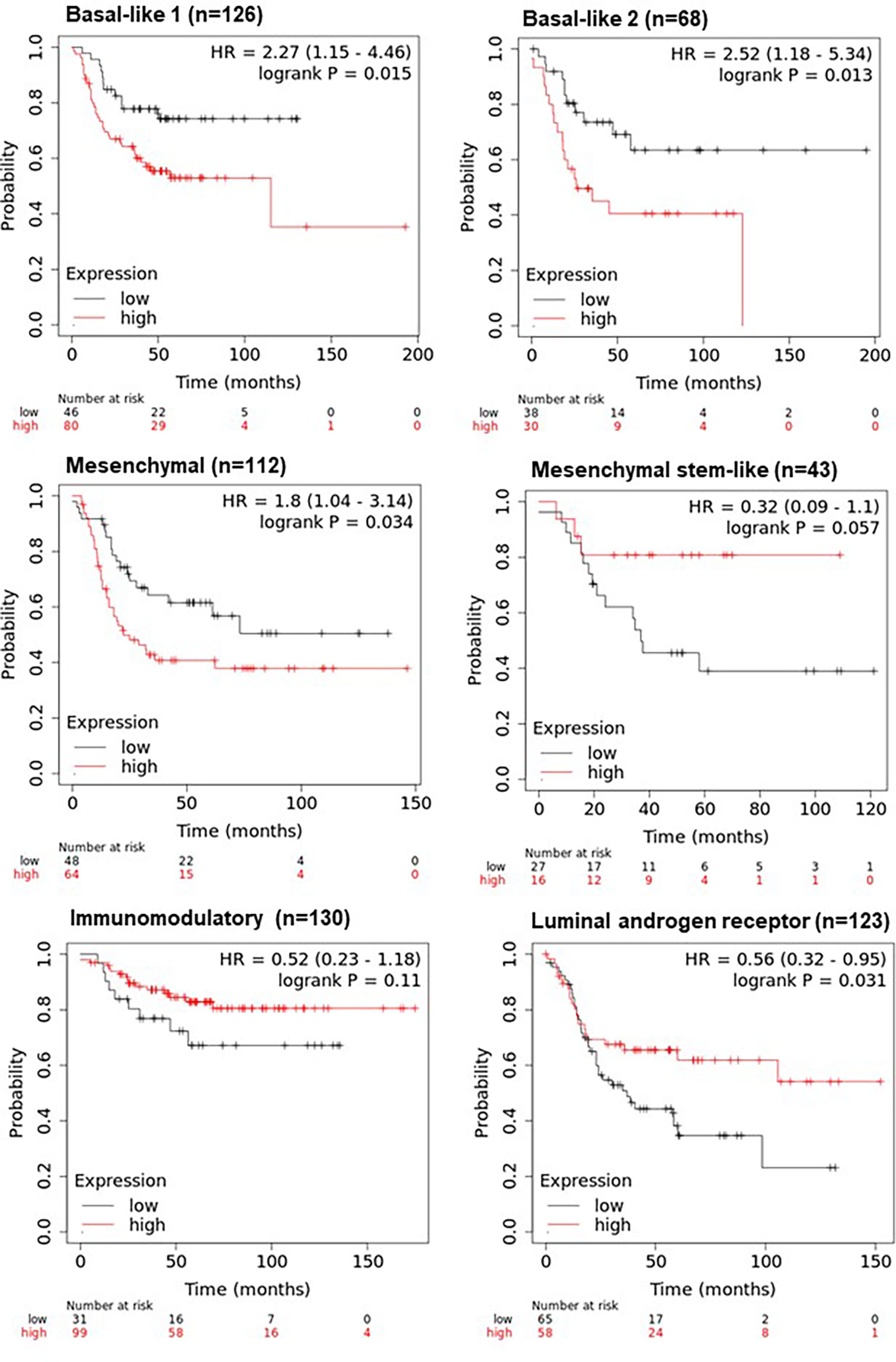
Figure 5 KM plotter survival analysis of TNBC subtypes using the four-gene panel including ANP32E, DSC2, IL6ST, and ANKRD30A. Survival analysis was performed in KM plotter using publicly available microarray data that is available within the database according to the TNBC subtypes of basal-like 1, basal-like 2, mesenchymal, mesenchymal stem-like, immunomodulatory and the luminal androgen receptor subtype. Cases were divided into high or low expression groups based on the median expression of the four-gene panel (with inverted expression of the two low expressed genes: ANKRD30A and IL6ST and not inverted for the two highly expressed genes: ANP32E and DSC2) and compared by Kaplan-Meier survival analysis. A Log-rank p-value ≤ 0.05 was considered significant. HR, hazard ratio. The red curve represents high expression, and the black curve represents low expression.
Discussion
TNBC is a highly aggressive subtype of breast cancer with decreased survival rates compared to other subtypes of breast cancer (1, 2). Because of the absence of well-established prognostic and predictive biomarkers, there are no targeted treatment options available to improve patient survival. Previous studies from our laboratory have indicated that ANKRD30A, IL6ST, ANP32E and DSC2 are differentially expressed in TNBC when compared to receptor positive disease, using cDNA microarrays. In this study, the expression of ANKRD30A, IL6ST, ANP32E and DSC2 were analyzed using ddPCR to verify these results and to determine whether their expression was associated with clinical features and survival.
IL6ST/gp130 was significantly downregulated in IDCs and LNmets compared to NATs as well as in TNBC compared to non-TNBCs, verifying the results of our previous study (3). Additionally, its low expression level was significantly associated with grade 3 disease, hormone receptor negativity, earlier age at diagnosis and worse relapse free survival. Taken together, this implies that its downregulation may increase the aggressiveness of the disease and that its high expression is a marker of good prognosis in TNBC. Similar to this study, IL6ST has been shown to be downregulated in basal-like breast cancer compared to luminal A/B breast cancer subtypes and its lower expression was associated with poor overall survival in TNBC patients (15, 16). The loss of IL6ST is involved in pathways related to lymphovascular invasion in breast cancer patients (17) and its function has been associated with other physiologies such as myocardial and hematological development where embryos of mice deficient for IL6ST gradually die between 12.5 days post-coitum (18). IL6ST is known to be involved in the JAK/STAT pathway (19, 20) including the activation of STAT3 and STAT1, as well as the PI3K/AKT and RAS/MAPK pathways. Therefore, its loss in TNBC may disrupt the JAK/STAT pathway resulting in transcriptional mis-regulation of associated genes and promoting tumor proliferation, migration and invasion. However, further study of IL6ST in a larger cohort is needed to understand its prognostic role in TNBC.
ANKRD30A showed significantly lower expression in TNBC compared to non-TNBC cases and its lower expression was associated with ER, PR status and tumor size; as well as worse RFS, suggesting a role in disease aggressiveness. Similar to the findings reported herein, another study have also found its downregulation in TNBC tissues (21, 22) and its expression was associated with ER status (23).Notably, ANKRD30A possesses an estrogen response element in the promoter region which may be regulated through estrogen receptor signaling (24), suggesting a possible mechanism for its low expression in TNBC. Additionally, the downregulation of Long Non-coding RNA LINC00993 (Long Intergenic Non-Protein Coding RNA 993) was significantly associated with the downregulated expression of the nearest coding gene, ANKRD30A in a microarray study of TNBC (21). It should be noted that many genomic regions can be co-regulated. Hence, the association may well be coincidental rather than causative. Interestingly, the lncRNA LINC00993 was identified to act as a tumor suppressor in TNBC which suppresses the growth of tumor cells both in vivo and in vitro (25). ANKRD30A is classed as a transcription factor due to the presence of bZIPsite and bipartite nuclear localization signal motif, hence it may be involved in regulating the expression of LINC00993. Thus, this implies that downregulation of ANKRD30A may have a significant role in tumor progression in TNBC.
ANP32E was significantly upregulated in TNBC compared to non-TNBC. Furthermore, its high expression was significantly associated with grade, the number of positive lymph nodes and worse RFS. ANP32E knockdown has been shown to inhibit the proliferation, migration and metastasis of breast cancer cells (26). It has also been shown to be highly expressed in primary breast cancers with a high propensity of metastasizing to the lungs (27). One study showed that ANP32E promotes G1/S progression by increasing the expression of E2F1, thus inducing proliferation in TNBC cells (28). The same study also showed that ANP32E is highly expressed at protein level in TNBC cases compared to non-TNBC cell lines and tumors. Taken together, these data support a role for ANP32E in cancer progression. However, further studies in a larger cohort are needed to understand its prognostic role in TNBC.
DSC2 was significantly upregulated in TNBC compared to non-TNBC but was not associated with clinicopathological features or survival outcomes. Microarray gene expression analysis of 23 breast cancer metastases showed that the upregulation of DSC2 expression may contribute to lung metastasis as a part of a 6-gene signature (27). Moreover, a study validated that the overexpression of five genes including DSC2 worsens lung metastasis-free survival in geminin-overexpressing TNBC cells (29). In contrast, a reduction in DSC2 expression has been identified in other cancer types such as colorectal, lung and esophageal squamous cell carcinoma (30–32). Conversely, a reduction in the expression of DSC2 has been found in other cancers including colon cancer (33) and urothelial carcinoma tissues, where its downregulation was associated with rapid migration and invasion (34). Currently, there have been no other studies of DSC2 expression and its role in TNBC.
Perhaps one of the most important findings was that when combined, the four-gene panel was strongly associated with RFS in TNBC, but not in hormone receptor positive breast cancers. Additionally, the four-gene panel provided a better survival curve discrimination when compared to each of the individual genes. The signature was further associated with RFS in 4/6 TNBC subtypes (Figure 5), indicating that this signature may be useful in predicting survival in the majority of TNBC cases. Interestingly, high expression of the gene panel was associated with better RFS in the LAR subtype, and worse RFS in the BL1, BL2 and mesenchymal subtypes. Given that xenografts developed from cell lines representative of these distinct subtypes show distinct responses to chemotherapeutic agents (14), the four gene panel defined in this study may have significant clinical utility, but these results would need to be validated in an independent cohort given the small sample size. Another key consideration is whether the mRNA expression of these four genes is correlated with their protein expression, the functional unit of the gene. There has been one published study showing that ANP32E is highly expressed at the mRNA and protein level in TNBC compared to non-TNBC tissues (28). However, for the other three genes, the correlation with protein expression has not been examined in breast cancer tissues to the best of our knowledge.
Taken together, these studies have verified the differential expression of ANKRD30A, IL6ST, ANP32E and DSC2 in TNBC compared to non-TNBC and determined their association with clinical features and survival. Additionally, the four-gene panel may serve as a specific prognostic tool in TNBC management.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Hunter New England Human Research Ethics Committee. The ethics committee waived the requirement of written informed consent for participation.
Author Contributions
KA-K: conceptualization. MP and KA-K: data collection, curation, and methodology. MP and KA-K: writing—original draft preparation. MP, RT, RS, KA-K: review and editing. RT, RS, and KA-K: supervision. RS and KA-K: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Breast Cancer Foundation and the Hunter Medical Research Institute, through donations from the Hunter Breast Cancer Foundation. MP was supported by a University Postgraduate Award. KA-K is supported by the Cancer Institute NSW (Career Development Fellowship; CDF181205).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Pathologist Dr. Ricardo Vilain for the validation of all tumor and normal tissue specimens used in this study, as well as Ms Sarah Nielsen and Dr. Mythily Mariasegaram for their assistance with archival specimens.
References
1. Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive Analysis of Estrogen Receptor (ER)-Negative, Progesterone Receptor (PR)-Negative, and HER2-Negative Invasive Breast Cancer, the So-Called Triple-Negative Phenotype: A Population-Based Study From the California Cancer Registry. Cancer (2007) 109(9):1721–8. doi: 10.1002/cncr.22618
2. Gonçalves H Jr., Guerra MR, Duarte JR, Cintra VA, Fayer IV, Brum MT, et al. Survival Study of Triple-Negative and Non-Triple-Negative Breast Cancer in a Brazilian Cohort. Clin Med Insights Oncol (2018) 12:1179554918790563. doi: 10.1177/1179554918790563
3. Mathe A, Wong-Brown M, Morten B, Forbes JF, Braye SG, Avery-Kiejda KA, et al. Novel Genes Associated With Lymph Node Metastasis in Triple Negative Breast Cancer. Sci Rep (2015) 5(1):15832. doi: 10.1038/srep15832
4. Jäger D, Stockert E, Güre AO, Scanlan MJ, Karbach J, Jäger E, et al. Identification of a Tissue-Specific Putative Transcription Factor in Breast Tissue by Serological Screening of a Breast Cancer Library. Cancer Res (2001) 61(5):2055.
5. Reihmane D, Dela F. Interleukin-6: Possible Biological Roles During Exercise. Eur J Sport Sci (2014) 14(3):242–50. doi: 10.1080/17461391.2013.776640
6. Fang WK, Liao LD, Li LY, Xie YM, Xu XE, Zhao WJ, et al. Down-Regulated Desmocollin-2 Promotes Cell Aggressiveness Through Redistributing Adherens Junctions and Activating Beta-Catenin Signalling in Oesophageal Squamous Cell Carcinoma. J Pathol (2013) 231(2):257–70. doi: 10.1002/path.4236
7. Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, et al. Anp32e, a Higher Eukaryotic Histone Chaperone Directs Preferential Recognition for H2A.Z. Cell Res (2014) 24(4):389–99. doi: 10.1038/cr.2014.30
8. Taylor SC, Laperriere G, Germain H. Droplet Digital PCR Versus qPCR for Gene Expression Analysis With Low Abundant Targets: From Variable Nonsense to Publication Quality Data. Sci Rep (2017) 7(1):2409. doi: 10.1038/s41598-017-02217-x
9. Dingle TC, Sedlak RH, Cook L, Jerome KR, et al. Tolerance of Droplet-Digital PCR vs Real-Time Quantitative PCR to Inhibitory Substances. Clin Chem (2013) 59(11):1670–2. doi: 10.1373/clinchem.2013.211045
10. Ma J, Li N, Guarnera M, Jiang F. Quantification of Plasma miRNAs by Digital PCR for Cancer Diagnosis. Biomark Insights (2013) 8:127–36. doi: 10.4137/BMI.S13154
11. Dobnik D, Štebih D, Blejec A, Morisset D, Žel J. Multiplex Quantification of Four DNA Targets in One Reaction With Bio-Rad Droplet Digital PCR System for GMO Detection. Sci Rep (2016) 6(1):35451. doi: 10.1038/srep35451
12. Avery-Kiejda KA, Morten B, Wong-Brown MW, Mathe A, Scott RJ. The Relative mRNA Expression of P53 Isoforms in Breast Cancer Is Associated With Clinical Features and Outcome. Carcinogenesis (2014) 35(3):586–96. doi: 10.1093/carcin/bgt411
13. Avery-Kiejda KA, Braye SG, Mathe A, Forbes JF, Scott RJ. Decreased Expression of Key Tumour Suppressor microRNAs Is Associated With Lymph Node Metastases in Triple Negative Breast Cancer. BMC Cancer (2014) 14:51. doi: 10.1186/1471-2407-14-51
14. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of Human Triple-Negative Breast Cancer Subtypes and Preclinical Models for Selection of Targeted Therapies. J Clin Invest (2011) 121(7):2750–67. doi: 10.1172/JCI45014
15. Fertig EJ, Lee E, Pandey NB, Popel AS. Analysis of Gene Expression of Secreted Factors Associated With Breast Cancer Metastases in Breast Cancer Subtypes. Sci Rep (2015) 5(1):12133. doi: 10.1038/srep12133
16. Jia R, Weng Y, Li Z, Liang W, Ji Y, Liang Y, et al. Bioinformatics Analysis Identifies IL6ST as a Potential Tumor Suppressor Gene for Triple-Negative Breast Cancer. Reprod Sci (2021) 28(8):2331–41. doi: 10.1007/s43032-021-00509-2
17. Klahan S, Wong HS, Tu SH, Chou WH, Zhang YF, Ho TF, et al. Identification of Genes and Pathways Related to Lymphovascular Invasion in Breast Cancer Patients: A Bioinformatics Analysis of Gene Expression Profiles. Tumour Biol (2017) 39(6):1010428317705573. doi: 10.1177/1010428317705573
18. Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, et al. Targeted Disruption of Gp130, a Common Signal Transducer for the Interleukin 6 Family of Cytokines, Leads to Myocardial and Hematological Disorders. Proc Natl Acad Sci USA (1996) 93(1):407–11. doi: 10.1073/pnas.93.1.407
19. Simpson RJ, Hammacher A, Smith DK, Matthews JM, Ward LD. Interleukin-6: Structure-Function Relationships. Protein Sci: Publ Protein Soc (1997) 6(5):929–55. doi: 10.1002/pro.5560060501
20. Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-Type Cytokine Signalling Through the Gp130/Jak/STAT Pathway. Biochem J (1998) 334(Pt 2):297–314. doi: 10.1042/bj3340297
21. Chen C, Li Z, Yang Y, Xiang T, Song W, Liu S. Microarray Expression Profiling of Dysregulated Long Non-Coding RNAs in Triple-Negative Breast Cancer. Cancer Biol Ther (2015) 16(6):856–65. doi: 10.1080/15384047.2015.1040957
22. Zombori T, Cserni G. Immunohistochemical Analysis of the Expression of Breast Markers in Basal-Like Breast Carcinomas Defined as Triple Negative Cancers Expressing Keratin 5. Pathol Oncol Res (2018) 24(2):259–67. doi: 10.1007/s12253-017-0246-y
23. Theurillat J-P, Zürrer-Härdi U, Varga Z, Storz M, Probst-Hensch NM, Seifert B, et al. NY-BR-1 Protein Expression in Breast Carcinoma: A Mammary Gland Differentiation Antigen as Target for Cancer Immunotherapy. Cancer Immunol Immunother (2007) 56(11):1723–31. doi: 10.1007/s00262-007-0316-1
24. Theurillat JP, Zürrer-Härdi U, Varga Z, Barghorn A, Saller E, Frei C, et al. Distinct Expression Patterns of the Immunogenic Differentiation Antigen NY-BR-1 in Normal Breast, Testis and Their Malignant Counterparts. Int J Cancer (2008) 122(7):1585–91. doi: 10.1002/ijc.23241
25. Guo S, Jian L, Tao K, Chen C, Yu H, Liu S. Novel Breast-Specific Long Non-Coding RNA LINC00993 Acts as a Tumor Suppressor in Triple-Negative Breast Cancer. Front Oncol (2019) 9(1325). doi: 10.3389/fonc.2019.01325
26. Li P, Xu T, Zhou X, Liao L, Pang G, Luo W, et al. Downregulation of miRNA-141 in Breast Cancer Cells Is Associated With Cell Migration and Invasion: Involvement of ANP32E Targeting. Cancer Med (2017) 6(3):662–72. doi: 10.1002/cam4.1024
27. Landemaine T, Jackson A, Bellahcene A, Rucci N, Sin S, Abad BM, et al. A Six-Gene Signature Predicting Breast Cancer Lung Metastasis. Cancer Res (2008) 68(15):6092–9. doi: 10.1158/0008-5472.CAN-08-0436
28. Xiong Z, Ye L, Zhenyu H, Li F, Xiong Y, Lin C, et al. ANP32E Induces Tumorigenesis of Triple-Negative Breast Cancer Cells by Upregulating E2F1. Mol Oncol (2018) 12(6):896–912. doi: 10.1002/1878-0261.12202
29. Sami E, Bogan D, Molinolo A, Koziol J, ElShamy WM. The Molecular Underpinning of Geminin-Overexpressing Triple-Negative Breast Cancer Cells Homing Specifically to Lungs. Cancer Gene Ther (2021). doi: 10.1038/s41417-021-00311-x
30. Kolegraff K, Nava P, Helms MN, Parkos CA, Nusrat A. Loss of Desmocollin-2 Confers a Tumorigenic Phenotype to Colonic Epithelial Cells Through Activation of Akt/β-Catenin Signaling. Mol Biol Cell (2011) 22(8):1121–34. doi: 10.1091/mbc.e10-10-0845
31. Fang W-K, Gu W, Li E-M, Wu Z-Y, Shen Z-Y, Shen J-H, et al. Reduced Membranous and Ectopic Cytoplasmic Expression of DSC2 in Esophageal Squamous Cell Carcinoma: An Independent Prognostic Factor. Hum Pathol (2010) 41(10):1456–65. doi: 10.1016/j.humpath.2010.04.003
32. Cui T, Chen Y, Yang L, Mireskandari M, Knösel T, Zhang Q, et al. Diagnostic and Prognostic Impact of Desmocollins in Human Lung Cancer. J Clin Pathol (2012) 65(12):1100. doi: 10.1136/jclinpath-2011-200630
33. Khan K, Hardy R, Haq A, Ogunbiyi O, Morton D, Chidgey M. Desmocollin Switching in Colorectal Cancer. Br J Cancer (2006) 95(10):1367–70. doi: 10.1038/sj.bjc.6603453
Keywords: triple negative breast cancer (TNBC), metastasis, gene expression, ddPCR, prognostic
Citation: Pariyar M, Thorne RF, Scott RJ and Avery-Kiejda KA (2022) Verification and Validation of a Four-Gene Panel as a Prognostic Indicator in Triple Negative Breast Cancer. Front. Oncol. 12:821334. doi: 10.3389/fonc.2022.821334
Received: 24 November 2021; Accepted: 25 February 2022;
Published: 21 March 2022.
Edited by:
San-Gang Wu, First Affiliated Hospital of Xiamen University, ChinaReviewed by:
Victoria Seewaldt, City of Hope, United StatesAnuraag Shrivastav, University of Winnipeg, Canada
Copyright © 2022 Pariyar, Thorne, Scott and Avery-Kiejda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kelly A. Avery-Kiejda, a2VsbHkua2llamRhQG5ld2Nhc3RsZS5lZHUuYXU=
 Mamta Pariyar
Mamta Pariyar Rick F. Thorne
Rick F. Thorne Rodney J. Scott
Rodney J. Scott Kelly A. Avery-Kiejda
Kelly A. Avery-Kiejda