- 1Department of Urology, Affiliated Drum Tower Hospital, Medical School of Nanjing University, Nanjing, China
- 2Jiangsu Engineering Research Center for MicroRNA Biology and Biotechnology, State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, China
- 3Nanjing Drum Tower Hospital Clinical College of Jiangsu University, Nanjing, China
Cancer metastasis, a typical malignant biological behavior involving the distant migration of tumor cells from the primary site to other organs, contributed majorly to cancer-related deaths of patients. Although constant efforts have been paid by researchers to elucidate the mechanisms of cancer metastasis, we are still far away from the definite answer. Recently, emerging evidence demonstrated that cancer metastasis is a continuous coevolutionary process mediated by the interactions between tumor cells and the host organ microenvironment, and epigenetic reprogramming of metastatic cancer cells may confer them with stronger metastatic capacities. The lymph node served as the first metastatic niche for many types of cancer, and the appearance of lymph node metastasis predicted poor prognosis. Importantly, multiple immune cells and stromal cells station and linger in the lymph nodes, which constitutes the complexity of the lymph node microenvironment. The active cross talk between cancer cells and immune cells could happen unceasingly within the metastatic environment of lymph nodes. Of note, diverse immune cells have been found to participate in the formation of malignant properties of tumor, including stemness and immune escape. Based on these available evidence and data, we hypothesize that the metastatic microenvironment of lymph nodes could drive cancer cells to metastasize to further organs through epigenetic mechanisms.
Introduction
Lymph node metastasis served as the most common metastatic manner for many tumor types, such as bladder cancer, breast cancer, melanoma, and colorectal cancer (1–4). In many cases, the lymphatic system was the first receiving station for the metastatic tumor cells, and the presence of lymph node metastasis has been recognized as an effective predictor for the further metastasis to other organs and the poor prognosis of cancer patients (5, 6). With regard to the mechanisms of lymph node metastasis, multiple studies have been conducted to demonstrate that the properties of lymph node metastasis for some cancers were related to the upregulation of lymphangiogenic growth factors, higher lymphatic invasiveness, and increased density of lymphatic vessels (7, 8). In addition, due to the massive enrichment of diverse immune cells within the lymph nodes, the active or suppressive immune status of the lymph node microenvironment is also found to mediate the elimination or protection of cancer cells within the metastatic niche of lymph nodes (9–11). However, a definite theory to elucidate the mechanism of lymph node metastasis remains unclear.
In recent years, the rapid innovation and progress of tumor lineage tracking technology and cancer sequencing techniques enabled us to reconstruct and detect the phylogenetic and evolutionary process of the distinct clonal populations of cancer cells. Multiple studies have demonstrated that metastases are derived from distinct clonal groups from the primary tumor, including “monoclonal seeding” and “polyclonal seeding.” Tumor heterogeneity is originated from continuous mutations produced mainly during cell division, and the subsequent formation of multiple subpopulations provided the raw material for the secondary environmental selection. A comparative analysis performed on the whole-exome sequence expressed between paired primary tumor and metastasis samples showed that the clonal selection on a major subpopulation from the primary tumor is more limited to a single major clone and harbored fewer metastasis-driven specific mutations than expected across several cancer types (12). However, this clone-selective pressure could be amplified by treatment, which resulted in evolved drug resistance instead of metastasis capacities (13). Nevertheless, these observations are based on retrospective analysis on the static gene expression profile, which largely limited the resolution and accuracy. To overcome this problem, Simeonov et al. developed a novel single-cell pedigree localization and tracking approach based on CRISPR-Cas9 technology to reveal the evolutionary history of single-cell transcriptional states during the process of metastasis in pancreatic cancer (14). Surprisingly, they found that over half of the clones of the primary cancer cells were confined to the primary sites and never metastasize to other sites, while the dominant clones survived in the metastatic sites with an enhanced expression of epithelial-to-mesenchymal transition (EMT) state. In consistency with previous studies, this research emphasized the domination of advantageous clonal selection in the metastasis. More importantly, it pointed out the distinct genetic expression phenotype associated with the highly metastatic cancer cells, which brought us with several new questions: (1) Do the genetic mutations related with the metastatic capacities all derive from the evolutional selection pressure during metastasis? (2) Could the microenvironment of the metastasis sites exert regulatory functions on the disseminated cancer cells to endow them with enhanced malignant biological characteristics? (3) What factors determine the tropism of further metastasis of tumor, and is the dissemination to different organs totally randomized or programmed?
Following these questions, we found quite a few related studies revealing somewhat different answers. In contrast with the mainstream cognition that primary lesion is the dominant origin for further dissemination of metastatic cells to other organs, a range of studies indicated that most seeding of metastatic cells originated from metastatic sites rather than the primary tumor (15, 16), which suggested that the clonal selection from primary tumor may be insufficient to explain the mechanism of cancer metastasis and the cross-seeding between metastases and secondary metastasis from distinct organs may have underestimated functions on the regulation of metastatic cancer cells.
Recently, Zhang et al. utilized the approach of parabiosis and an evolving barcode system to trace the evolution history of metastasis in mouse models of prostate and breast cancer, which are all highly bone-metastatic cancers. Interestingly, they also detected that majority of the metastases in further organs were from the first metastatic bone lesions rather than the primary tumor (17). Of note, this study demonstrated that the bone microenvironment not only simply served as an early transport station for metastatic cells but also exerted crucial epigenetic modulatory functions on the stemness of the cancer cells, which finally enhanced the metastatic capacities of cancer cells and promoted the further dissemination to further organs.
In summary, the process of metastasis is not only simply a unidirectional evolution route under the selective pressure of different metastatic environments, but rather an active two-way interactive process, which finally contributed to the evolution and further dissemination of highly metastatic cancer cell populations. Notably, the microenvironment of the metastatic sites could initiate a process of epigenetic reprogramming on the cancer cells, which explained the enhanced metastatic capacities and increased mortality after the event of first metastasis. As has been mentioned before, the lymph node is the first metastatic organ with the greatest possibility for many cancers, such as bladder cancer and gastric cancer. Correspondingly, the higher possibility of further dissemination and a poorer prognosis are found to be significantly associated with lymph node metastasis. However, there still lacks an exact explanation for this troublesome clinical problem.
The Hypothesis/Theory
We proposed that in those cancers tending to first metastasize to the lymph nodes, the microenvironment of lymph nodes would facilitate the cancer cells to undergo secondary metastasis to multiple further organs through epigenetic regulation or other mechanisms, which most likely mediated the enhancement of the immune-escape ability of tumor cells. In particular, the cross talk between cancer cells and the lymph node metastasis served as a crucial and indispensable part of the tumor evolution during the process of metastasis.
Justification of the Hypothesis
Cancers With Lymph Node Involvement Are More Progressive and Malignant
Multiple studies verified that lymph node metastasis was an independent predictor for the oncological progression and patient survival in many cancers (1, 18–20). Of note, the probability of metastasis into other further organs significantly increased along with the appearance of lymph node metastasis. One of the reasons may be the route of lymphatic vessels facilitates the migration of cancer cells, which accelerated the speed of tumor dissemination. However, there still existed several problems remaining to be resolved. For one thing, under the circumstance of cancer with distant metastases, we are still unclear about the major origin of the metastatic cells in the distant organs. According to traditional understanding, the hematogenous metastasis from the primary tumor should be the “root of all evils” for the distant metastasis. However, recent studies demonstrated the high heterogeneity of the tumor population during the process of metastasis, which indicated the complexity of distribution of tumor cells and their cloning clustering (14–17). Also, emerging evidence proved that the major origin of further metastatic tumor cells on distant organs may not necessarily be the primary tumor, but rather the first metastatic lesions, such as bones (17). More importantly, the microenvironment of the bones enhanced the stemness of tumor cells through epigenetic modulation, which finally contributed to the further metastasis (17). To some extent, the lymph node shares many similar characteristics with bone for tumor metastasis, such serving the most frequent first metastatic niche for multiple cancers and increased probabilities of further metastasis. Taking these factors into consideration, we may naturally come to the hypothesis that the lymph node could also exert crucial functions on the metastatic tumor cells to enhance their metastatic abilities for further dissemination. To elucidate this issue, we may firstly trace the evolutionary process and cloning distribution in the context of lymph node metastasis. Once we find that most clones of tumor cells in the further distant organs are derived from the lymph node metastases rather than the primary tumor, it may be more solid evidence for our hypothesis.
Classical Immune Phenotype Alterations of Metastatic Cells in the Microenvironment of Lymph Nodes
As the most widely distributed immune organ in our body, lymph nodes have unique environmental characteristics. Traditionally, lymph nodes were viewed as killers against invaded cancer cells. Resident or recruited cytotoxic immune cells, such as natural killer cells, CD8+ T cells, and M1-type macrophages, would help eliminate tumor cells through different mechanisms (21–23). However, the fact of high incidence of lymph node metastasis indicated that metastatic tumor cells as well as the microenvironment of the lymph node must encounter significant alterations to help the tumor cells escape the immune attack from immune cells. Recent studies have demonstrated that in the tumor-involved lymph nodes, a range of immune-suppressive cells and biological factors would be recruited or activated, which facilitated the survival and growth of metastatic tumor cells. The mechanisms include the decrease of CD8+ T cells, myeloid-derived suppressor cell (MDSC) expansion, and upregulation of regulatory T cells (24–27). However, the exact alterations of metastatic tumor cells from primary tumor cells remained unclear. Some studies presented changes in expressive levels of immune-checkpoint proteins such as PD-1 and PD-L1 of the lymph node microenvironment and explored its correlations with prognosis (28–30), but these studies are unable to show the changes in tumor cells themselves. To explore the alterations in genetics as well as the biological phenotypes between the primary tumor and the lymphatic metastases, we performed a bioinformatic analysis to compare the differential genes and the enrichment of possible pathways from the Gene Expression Omnibus (GEO) database. The differentiated expressed genes between primary tumor and lymph node metastases were extracted from GSE121738 for bladder cancer and GSE180186 for breast cancer (Figure 1). Surprisingly, we found that the predicted enriched pathways were focused on the negative regulation of immune responses, which revealed that the metastatic cells in the lymph node possessed upregulated immune-escape characteristics (31). From this result, we may naturally speculate that those metastatic cells colonized on the lymph nodes could strive to survive and resist the immune attack from lymph nodes through acquisition of higher immune-escape capacities. More importantly, in this way, these metastatic cells would possess stronger progressive phenotypes, which facilitates them to metastasize into further organs. The possible immune-escape mechanisms may include upregulation of immune-checkpoint proteins, such as PD-L1 and CD47, expression and release of immune-suppressive cytokines or chemokines, such as TGF-β1, IL-10 and so on, and abnormal regulation of non-coding RNA components. Nevertheless, we need to elucidate the origin of the acquisition and whether strengthening of these immune-escape abilities is from the natural cloning selection during metastasis or from the phenotypic regulation of the lymph node environment on cells or both.
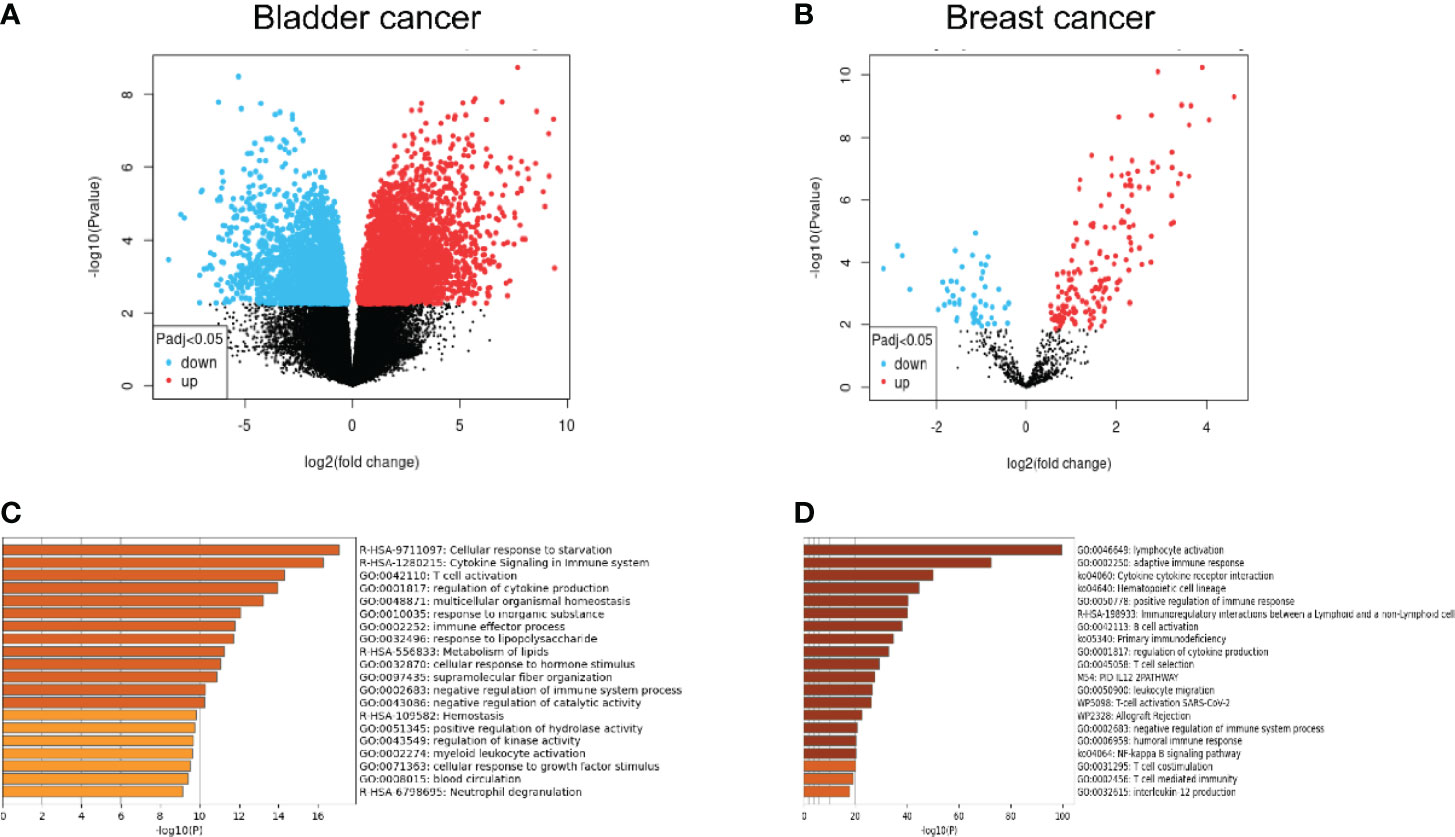
Figure 1 (A, B) Comparative analysis from the microarray data extracted from the GEO database illustrating the significantly upregulated and downregulated genes between lymph node metastases and primary tumor. (C, D) Gene-enrichment analysis showing the predicted enriched pathways mediated by the significantly upregulated genes in the lymph node metastases compared with the primary tumor using Metascape (http://metascape.org/).
Complexity and Heterogeneity of the Lymph Node Microenvironment
In our hypothesis, the complexity of the lymph node microenvironment and the heterogeneity of its composed cell populations are the potential inducers or regulators for cancer second-time evolution with enhanced metastatic capacities. Therefore, it is essential to carefully review the structural and compositional properties of lymph nodes, especially in the context of cancer involvement.
Three main structural elements, including the cortex, paracortex, and medulla, help constitute the anatomical framework of the lymph node, and different types of immune cell populations own their main regional distribution. The cortex, consisting of the follicle, the interfollicular zone, and the subcapsular sinus, is the main resident area for B cell and dendritic cells, which regulated the process of antigen presentation and antibody production, while the paracortex is mainly distributed with T cells and the medulla is covered with macrophages (32, 33).
Generally, immune cells are the “flesh” of the lymph node and the stromal cells are the “skeleton.” Functionally, lymph node stromal cells not only help form the architecture of the lymph node but also play important roles in regulating the normal function of those immune cells mainly by releasing cytokines such as CCL19, CCL21, and IL17 (33, 34). Similar with previously mentioned immune cells, lymph node stromal cells also possess strong heterogeneity and regional distribution difference. Based on the surface biomarker expression and regional location, the lymph node stromal cells could be mainly divided into four subtypes, including lymphatic endothelial cells, blood endothelial cells, fibroblastic reticular cells, and CD31gp38-double negative cells (35).
Besides the cellular component of the lymph node microenvironment, the extracellular matrix is an indispensable constitute for the lymph node. Structural stability and biological sustainability are ensured by the complex network of the lymph node extracellular matrix (36).
To conclude, the lymph node microenvironment is a very complex network involving various cell types and components. However, it is the great cellular heterogeneity and the structural complexity that provide the metastatic cancer cells with more possibilities of multidirectional genetic selective pressure and more opportunities of cell–cell interactions. In the next section, we will discuss several potential mechanisms of regulatory roles for reprogramming metastatic cancers cells in the lymph node microenvironment, especially based on the possible origin from the distinct component of the lymph node.
Potential Mechanisms of Regulatory Roles for Reprogramming Metastatic Cancer Cells in the Lymph Node Microenvironment
Herein, we listed several potential possible regulatory mechanisms which may explain the enhancement of metastatic capacities of cancer cells granted by the microenvironment.
Potential Regulatory Roles Played by Lymph Node Stromal Cells
As has been discussed in the previous section, lymph node stromal cells not only served as major structural participators in the microenvironment of lymph node but also exerted important immune-regulatory functions on the immune cells. A previous study revealed that the lymphatic endothelial cells are the major source to produce sphingosine-1-phosphate (S1P) to mediate the emigration of lymphocytes from the lymph node (37, 38). However, as a pleiotropic bioactive molecule, S1P is involved in multiple pathological processes, including carcinogenesis. Genetic expression could be regulated by S1P through histone deacetylase binding, which may lead to abnormal expression of a series of genes such as p21 (39). Whether this epigenetic modulation could be utilized by lymphatic endothelial cells to regulate cancer cell phenotype alteration warrants further investigations. Also, proliferation capabilities of a range of tumor cells have been reported to be upregulated via the S1P–S1PR pathway (40–42). In addition, invasiveness, which is another key factor for metastasis, has also been found to be enhanced by S1P activation in glioma, which drove the malignant behavior (43). What determines our attention is that S1P-exerted functions are limited not only on the cancer cells but also on the whole immune environment, including M2 differentiation (44, 45) and Treg accumulation (46), and these immunosuppressive cells may also play regulatory roles for cancer cells, which indicated that lymphatic endothelial cells may also reprogram cancer cells through indirect mechanisms.
Under inflammatory conditions, nitrite oxide (NO) was found to be produced by lymphatic endothelial cells and fibroblastic reticular cells as an immune-suppressive pattern to inhibit the overactivation of T cells in the lymph node (47, 48). However, this pathway could also be hijacked by metastatic cancer cells for self-evolution. As a powerful deamination agent, NO was found to cause DNA damage through C–T transformation, which brought great genotoxicity and negatively influenced the DNA repair ability (49). This way, naturally we may hypothesize that the metastatic cancer cells arriving in the lymph node would face greater possibilities of potential gene mutations, which would provide a large-scale gene screening pool under the selective pressure of the lymph node microenvironment and more possibly evolve into a more malignant phenotype. Notably, different concentration levels of NO may reprogram the cancer cells through different oncogenic mechanisms (50). Epigenetic modulations could also be imposed on the cancer cells by the stromal cell-secreted NO. The DNA methylation event and histone acetylation event are the two major epigenetic regulatory mechanisms for cancer cells (51, 52). What is interesting is that NO was proved to be able to increase acetyl events through enhanced production of acetyl CoA induced by deprivation of aconitase 2 (53). Also, the methylation level of DNA CpG islands could be upregulated under the condition with high levels of NO through a direct inhibition of demethylase (54). In all, epigenetic alterations on the cancer cells could be regulated by the lymph node stromal cells. Recent studies have indicated that many crucial oncogenic pathways are mediated by these epigenetic mechanisms such as immune escape ability (55, 56) and stemness (57, 58). However, whether similar epigenetic reprogramming processes would happen in the lymph node microenvironment to drive the enhancement of cancer cell malignancy via NO produced by lymph node stromal cells remains to be elucidated.
When we mentioned the term “lymph node stromal cells” under the context of cancer metastasis, it is natural for us to associate another well-known term, cancer-associated fibroblasts (CAFs), which represents a unique stromal cell subpopulation, originated from preexisting normal stromal cells, which coevolved with the cancer cells in the tumor microenvironment with a common pro-tumor capability (59, 60). Similarly, the lymph node stromal cells seemed to also share these characteristics when they encountered the arrival of metastatic cancer cells. Nevertheless, existing investigations still cannot prove that the stromal cells in the metastatic lymph node micro-environment experienced similar reprogramming. But we may easily infer that a great possibility does exist that the lymph node stromal cells may coevolve with the cancer cells and finally acquire a phenotype similar with CAFs to in turn exert crucial regulatory functions on the cancer cells to facilitate their further invasion and metastasis. However, this issue required further investigations to verify.
Potential Regulatory Roles Played by Lymph Node Immune Cells
As has been discussed before, in addition to the lymph node stromal cells, immune cells are the other important cell population in the lymph node microenvironment. The immune cells mainly include two subpopulations: myeloid cells and lymphoid cells. The myeloid cells are composed of dendritic cells and macrophages while the lymphoid cells include B cells and T cells (22, 61). During the process of lymph node metastasis, innate and adaptive immune responses would be activated for tumor elimination while immune escape could also be induced to facilitate cancer progression. Combining the results of the gene-pathway enrichment analysis above and the common phenomenon of cancer immune escape in the lymph node, we assume that the immune cells in the lymph node may participate in the regulation of the malignancy enhancement of metastatic cancer cells.
PD-L1 is a classical immune-checkpoint protein mainly expressed on the cell membrane of tumor cells. By interacting with PD-1, which is expressed on T cells, PD-L1 induced the apoptosis and dysfunction of T cells and suppressed the anticancer immunity (62, 63). In consistency with our previous findings, many studies have demonstrated that lymph node metastasis in multiple cancers possessed overexpression of PD-L1. In addition, the high expression level of PD-L1 in lymph nodes strongly correlated with the poor prognosis of cancer patients (28, 64, 65). The mechanisms controlling PD-L1 expression covered different levels including genomic alterations, transcription, post-transcription, translation, and post-translation. Cha et al. made a brilliant review summarizing the intricate regulatory networks of PD-L1 expression (66). Aberrant inflammatory pathways have been clarified to contribute to loss of immunosurveillance in the tumor microenvironment, including transcriptionally upregulating the PD-L1 expression. MYC is a well-studied oncogenic transcription factor, which has been found to be able to bind the PD-L1 promoter and subsequently promote PD-L1 mRNA expression (67), while in the microenvironment of lymph node metastases, emerging evidence indicates that MYC expression showed an abnormal upregulation in the metastatic lymph nodes (68–70). Therefore, it is possible that the lymph node microenvironment harnessed specific mechanisms to promote PD-L1 expression of cancer cells through MYC modulation.
As an important part of the immune system, the lymph nodes basically assumed two major rules: one is to eliminate the dangerous foreign “non-self”, and the other is to recognize the “self” and avoid the unnecessary harm to the host. In the context of cancer, interferon-γ (IFN-γ) plays a central role against cancer development with its cytotoxic, pro-apoptotic, and immune-boosting functions. However, in the face of immune attack, tumor cells are able to hijack the IFN-γ pathway to enhance the PD-L1 expression (66, 71, 72), which finally help tumor cells survive the immune killing and survive. Considering the large enrichment of killer immune cells in the lymph node metastasis, metastatic cancer cells would encounter fierce attack in the metastatic niche, which offered great opportunity for them to evolve in this harsh living environment, in part through the upregulation of PD-L1 membrane expression. Moreover, other inflammatory cytokines also participated in the induction of mRNA expression of PD-L1, including TGF-β, TNF-α, and IL-6 (66, 73–75), and these cytokines are also important participators in the lymph node metastases, which may also serve as alternative pathways for the upregulation of PD-L1 in cancer cells and train them to become more powerful metastatic cancer cells for further dissemination in other organs. Seeing that the major origin of these inflammatory mediators is the immune effector cells, we assumed that the PD-L1 upregulation under the pressure of immune attack from immune cells of the lymph node may be a crucial step for the enhancement of immune-escape ability of metastatic cancer cells. However, this hypothesis has not been reproduced or confirmed in previous studies and we still need further experiments to verify this possibility. Also, the upregulation of PD-L1 as well as other immune-checkpoint proteins may not only be dependent on the direct defense mechanism for the cytotoxic immune cells but also derive from an indirect regulatory direction from other immune-suppressive cells in the microenvironment of lymph nodes. Multiple studies suggested that the presence of a high infiltration of Treg cells in the lymph node is significantly associated with cancer lymph node metastasis (76–78). As a powerful subpopulation of immune-suppressive cells, Treg cells would produce immune-suppressive cytokines, such as IL-10 and TGF-β, for controlling the balance of immune function (79, 80). However, these immune-suppressive mediators could also be utilized by metastatic cancer cells for their self-reinforcement. TGF-β and IL-10 could all serve as upstream activation factors for PD-L1 expression (66, 75). Similar functions may also be observed in other immune-suppressive cells, such as M2 macrophages and MDSCs, via future investigations. In a word, during the process of reprogramming for metastatic cancer cells, the driving forces may come from two directions: the direct pro-tumor regulations from immune-suppressive cells and the resisting self-defense mechanisms against the immune-killing cytotoxic cells. This regulatory process could be simultaneous or sequential, but we assume that the difference of strong and weak is dependent on the degree of metastasis.
In addition to the upregulation of membrane immune-checkpoint proteins, the metastatic capacities of cancer cells may also depend on the epigenetic reprogramming of other key genetic targets. TGF-β was identified as a powerful oncogenic cytokine mainly produced and released by tumor cells in the tumor microenvironment (81, 82). For one thing, TGF-β is able to promote Treg cell differentiation and recruitment, suppression of CD8+ T cell function, and MDSC expansion, which focused on the formation of the immune-suppressive microenvironment (83–85). For another, TGF-β could directly promote tumorigenesis through induction of epithelial to mesenchymal transition (EMT) and angiogenesis (86, 87), which both led to easier and faster tumor metastasis. Notably, a higher expression of TGF-β1 in the lymph node is significantly associated with the more malignant phenotype and poor survival across a range of cancer types (88–90). Then, we may naturally question whether the interactions between the lymph node microenvironment and metastatic cancer cells could enhance the regulatory function of TGF-β1 in metastatic cancer cells. MicroRNAs (miRNA) are well-studied small non-coding RNA molecules widely distributed in our body (91). By recognizing and binding with the target mRNA with the complementary sequences, miRNA could then repress the transcription of the specific RNA, which plays major regulatory roles in various physiologic or pathologic processes, including oncology (92, 93). For example, the decrease of the miR-200 family by TGF-β could promote TGF-β expression in return, which formed a positive feedback loop to further enhance the oncogenic function of TGF-β (94, 95). Importantly, a range of crucial tumor-suppressor miRNAs, including the miR-200 family, was also found to show a dramatic downregulation in the triple-negative breast cancer type with lymph node metastasis (96), which indicated that the aberrant expression of specific miRNAs was significantly associated with the enhanced metastatic capacities of cancer cells. Nevertheless, whether the miRNA networks regulate the enhancement of metastatic capabilities of cancer through modulating the TGF-β pathway remained elusive.
Potential Regulatory Roles Played by Other Eenvironmental Factors of the Lymph Node Microenvironment
As has been discussed above, non-coding RNAs (ncRNAs), including miRNA, circular RNAs (circRNA), long non-coding RNAs (lincRNA), and tRNA-derived small RNAs (tsRNA), are pivotal biological molecules exerting powerful epigenetic functions majorly controlling the post-transcription of a range of crucial genes during the process of carcinogenesis (97–99). Multiple studies have demonstrated that tumor cells could influence the lymph node microenvironment through secretion of individual or exosome-capsuled ncRNAs, which further facilitate their dissemination and progression (100–102). However, in the tumor-involved lymph node microenvironment, the ncRNA expression profile of the lymph node stromal cells and immune cells is still unclear to us. Up until now, a series of circular miRNAs, such as miR-20a, miR-203 (103), and miR-10b (104), have been identified as potential biomarkers for lymph node metastasis prediction. However, their exact origin and biological functions are still unknown. Chen et al. performed a comprehensive bioinformatic analysis based on a comparative miRNA array between lymph node metastases and paired primary tumor and found that miR-10a was significantly upregulated in the lymph nodes with a further verification by detection of relative cell lines (105). Then, we may question whether ncRNA secretion from the immune cells or stromal cells into the metastatic cancer cells could also epigenetically regulate their phenotype alterations and further result in their stronger progressive malignancy. Further studies are required via a finer cell clustering before gene sequencing for the lymph node metastasis, which may indicate the alterations of the ncRNA expressive profile under the condition of tumor involvement and further verify the reaction from the lymph node microenvironment to the cancer cells.
Hypoxia represented a specific environmental status with a low oxygen pressure of less than 5–10 mmHg (106). In the context of cancer lymph node metastasis, hypoxia has been found to be a factor that cannot be neglected for driving progression and metastasis (107, 108). The reason for the formation of hypoxia mainly originated from the oxygen overconsumption and enhanced metabolism under the pressure of tumor overgrowth. Importantly, hypoxia along with its induced alterations of a series of key oncogenes is an indispensable contributory factor for lymphangiogenesis (109, 110). HIF-1α, which is the major functional effector in response to hypoxia, could regulate the remodeling of lymph node endothelial cells (111, 112). Nevertheless, HIF-1α could also exert key regulatory functions on the cancer cells. Multiple studies suggested that HIF-1α served as a key transcriptionally regulatory element for PD-L1 upregulation in a range of cancers (66, 113–115). Therefore, great possibilities exist that metastatic cancer cells that colonized on the involved lymph nodes may acquire enhanced immune-escape capacities in response to the hypoxia status in the lymph node through the HIF-1α pathway. Of note, hypoxia and HIF-1α pathway-induced phenotype alterations not only are limited in PD-L1 expression but also may influence other important characteristics and functions of cancer, such as EMT (116, 117) and glycolysis metabolism induction (118, 119).
In conclusion, due to the complexity of the components in the lymph node and the rich changes in face of the tumor involvement, various subpopulations of the cells and other environmental elements of the lymph node microenvironment own the potential to modulate or reprogram the metastatic cancer cells. The enhanced metastatic capability conferred by the lymph node microenvironment may not only depend on the induction of immune escape but also may derive from the other key oncogenic pathway activation and strengthening, such as stronger tumor stemness and migratory and invasive abilities. However, considering the distinct environmental characteristics of the lymph node, we may prefer the hypothesis that the enhanced metastatic capabilities conferred by the lymph node microenvironment largely relied on the stronger immune-escape ability. The possible regulatory roles played by different components of the lymph node microenvironment are summarized in Table 1.
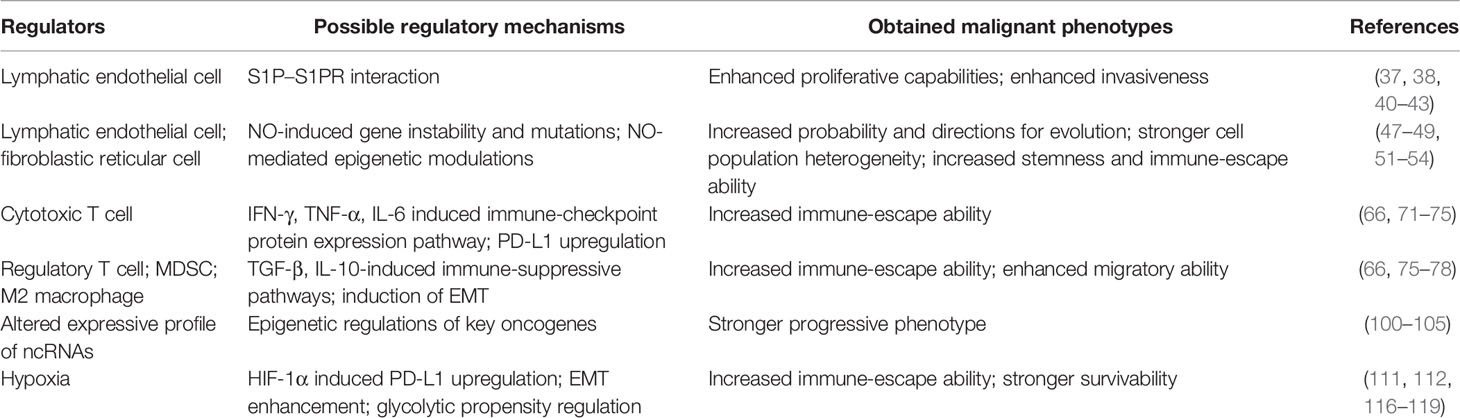
Table 1 Possible regulatory roles played by different components from the lymph node microenvironment for metastatic tumor reprogramming.
Discussion
Traditionally, tumor metastasis into further organs is identified as the terminal step of tumor progression, which served as the major cause of cancer-related death. Rapid development of gene-editing and sequencing technology allowed us to trace the metastasis process more precisely. Growing evidence suggested that tumor metastasis is not a unidirectional and even process originating from the parental niche but a cross-seeding interactive process between multiple metastatic environments. Importantly, further secondary metastasis within further organs is found to be more homologous with the first metastatic niche rather than the primary tumor, and the microenvironment of the first metastatic environment could exert epigenetic reprogramming function on the cancer cells, which strengthened their metastatic capabilities and facilitate them to further metastasize. Lymph node metastasis is commonly viewed as a crucial factor of poor prognosis in cancer patients and served as the first metastatic niche for many tumor types. The unique state of immune cell enrichment and inflammatory status determined that potential training effects could be performed on the cancer cells, involving upregulation of immune-escape capacities. Thus, we hypothesize that in those cancers tending to metastasize to lymph nodes first, the microenvironment of metastatic lymph nodes could invigorate the cancer cells with enhanced metastatic abilities, which contributed to the further metastasis.
As we discussed before, the contributory driving forces for possible reprogramming regulatory mechanisms on the malignancy of metastatic cancer cells could be mainly divided into three branches. The first one is from the lymph node stromal cells. Under normal physiologic conditions, these stromal cells are identified as structural supporters as well as regulators or recruiters of immune cells (33, 120, 121). Nevertheless, their possible epigenetic modulatory roles on the metastatic cancer cells should not be ignored. For one thing, NO, as a powerful genotoxic substance, could be secreted by lymphatic endothelial cells and fibroblastic reticular cells (47, 48), which may create an environment for more possibilities of gene mutations and epigenetic reprogramming in cancer cells. The increased cell heterogeneity provided primitive accumulation of evolutionary materials while the epigenetic modifications offered the required opening and closing of key genes. For another, the S1P–S1PR pathway could be activated by lymphatic endothelial cells to promote the enhancement of proliferative and invasive capabilities of cancer cells, which may explain the reason for the strengthening of second-metastatic ability (40–42). Notably, we presented a possibility that the lymph node stromal cells could experience a phenotype alteration under the invasion of cancer cells, which may form a positive-feedback loop to lead to their further negative regulation on the cancer cells.
The second possible regulatory pathway is originated from the immune cells in the lymph node microenvironment. As is shown in Figure 1, the upregulated genes in lymph node metastatic cells compared with the primary niche demonstrated significant enrichment in a range of immune-related pathways, including the negative regulation of the immune system process, T cell activation, and immune effector process. Considering the highly immune-attack pressure in the lymph node, it is reasonable to hypothesize that the cancer cells may acquire the enhancement of immune-escape capabilities to adapt to or resist the immune killing from the lymph node immune cells and further metastasize into further organs. PD-L1 has been recognized as a significant biomarker for lymph node metastasis in a range of cancers (28, 64, 65). What is intriguing is that the functional cytokines and inflammatory mediators produced by those immune-killing cells may be utilized by cancer cells as upstream factors to in turn upregulate the expression of PD-L1 (66, 71–74). In addition, the infiltration of immune-suppressive cells, such as Treg cell, MDSC, and M2 macrophage, could also exert similar functions (66, 75, 79, 80).
The third possible regulator we hypothesized is the environmental characteristic of hypoxia in the lymph-node microenvironment. HIF-1α, in response to the oxygen concentration in the lymph node microenvironment, could serve as an important transcriptionally regulatory mediator for a series of key phenotypes in the cancer cells, such as immune escape (113–115), EMT (116, 117), and metabolism alteration (118, 119).
Despite that a few specific alterations along with their regulators in the lymph node microenvironment have been discussed and hypothesized previously, it is certain that we still cannot include all the possible regulatory pathways because the co-evolutional process between the metastatic cancer cells and the lymph node microenvironment involves constant and complex cell–cell interactions. Therefore, further investigations are required for elucidating the exact mechanisms. The hypothesis is shown in Figure 2, illustrating the whole process and potential mechanisms.
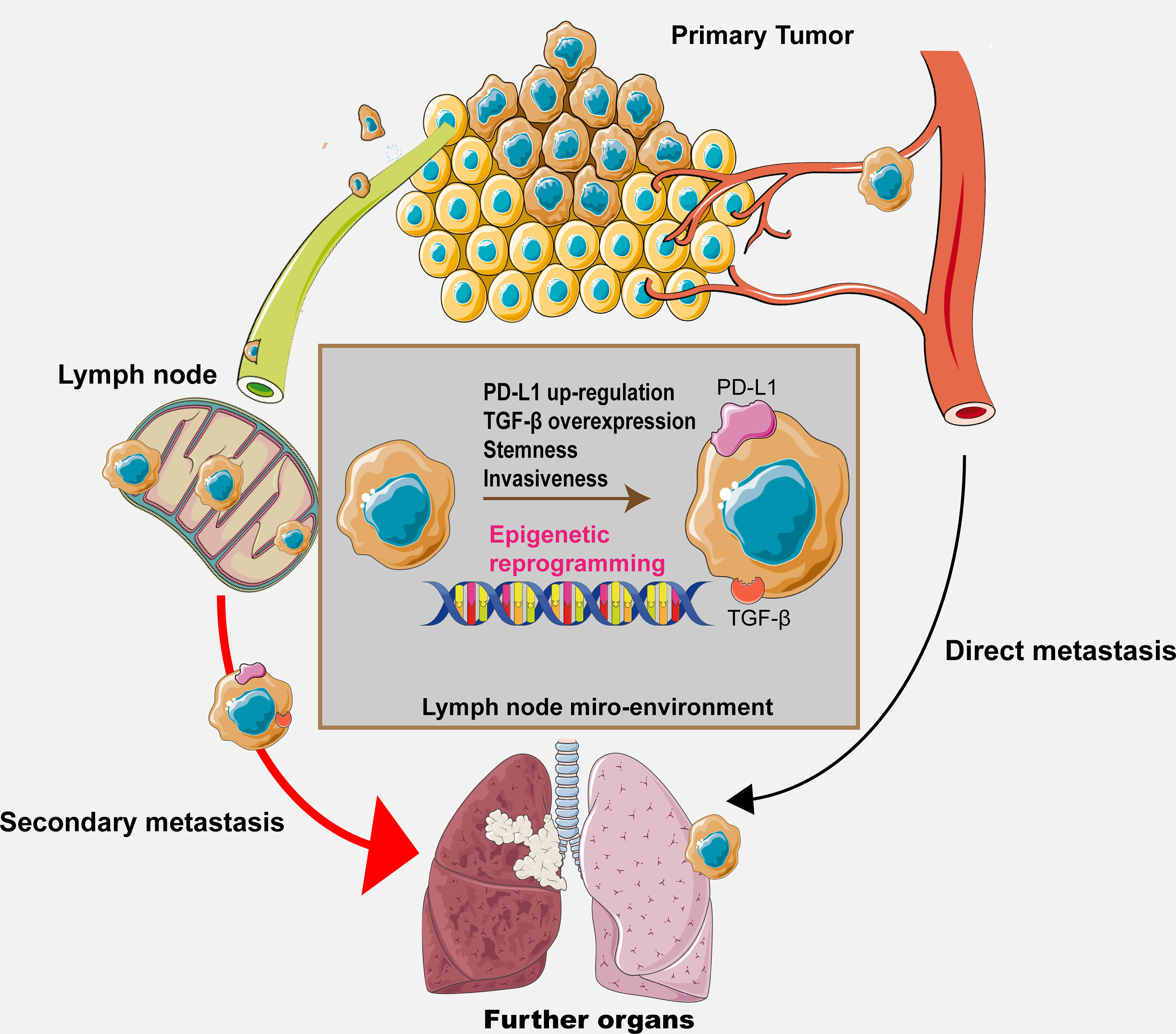
Figure 2 The graphical hypothesis illustrating the regulatory process and potential mechanism throughout the metastasis process.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
RY and HG designed and constructed the scientific idea. ThL extended and systemically analyzed the feasibility of the scientific hypothesis and wrote the paper. TyL, XX, and ZZ participated in the discussion and provided the crucial components of this hypothesis. SbZ and SkZ participated in the discussion and collected the data. NJ, WZ, and RS polished the language and revised the manuscript. FW and BF participated in the discussion and drew the figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81772727, 81772710 and 82172691) and Nanjing Science and Technology Development Key Project (YKK19011).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors give sincere appreciation to all the members from the M3 laboratory of the School of Life Sciences, Nanjing University, for their generous help and inspiring discussion. Also, we express great thanks to Professor Jinbo Chen, Professor Xiongbing Zu, et al., from Xiangya Hospital of Central South University and Professor Anna J Zaczek et al. from the Medical University of Gdansk for their kind and generous sharing of the mRNA-sequencing microarray results in the GEO database, which provided a lot of inspiration.
References
1. Liedberg F, Månsson W. Lymph Node Metastasis in Bladder Cancer. Eur Urol (2006) 49:13–21. doi: 10.1016/j.eururo.2005.08.007
2. Veronesi P, Corso G. Standard and Controversies in Sentinel Node in Breast Cancer Patients. Breast (2019) 48(Suppl 1):S53–6. doi: 10.1016/S0960-9776(19)31124-5
3. Jin M, Frankel WL. Lymph Node Metastasis in Colorectal Cancer. Surg Oncol Clin N Am (2018) 27:401–12. doi: 10.1016/j.soc.2017.11.011
4. Faries MB, Han D, Reintgen M, Kerivan L, Reintgen D, Caracò C. Lymph Node Metastasis in Melanoma: A Debate on the Significance of Nodal Metastases, Conditional Survival Analysis and Clinical Trials. Clin Exp Metastasis (2018) 35:431–42. doi: 10.1007/s10585-018-9898-6
5. Yang J, Lu Z, Li L, Li Y, Tan YL, Zhang DK, et al. Relationship of Lymphovascular Invasion With Lymph Node Metastasis and Prognosis in Superficial Esophageal Carcinoma: Systematic Review and Meta-Analysis. BMC Cancer (2020) 20:176. doi: 10.1186/s12885-020-6656-3
6. Deng JY, Liang H. Clinical Significance of Lymph Node Metastasis in Gastric Cancer. World J Gastroenterol (2014) 20:3967–75. doi: 10.3748/wjg.v20.i14.3967
7. Datta K, Muders M, Zhang H, Tindall DJ. Mechanism of Lymph Node Metastasis in Prostate Cancer. Future Oncol (2010) 6:823–36. doi: 10.2217/fon.10.33
8. Brown M, Assen FP, Leithner A, Abe J, Schachner H, Asfour G, et al. Lymph Node Blood Vessels Provide Exit Routes for Metastatic Tumor Cell Dissemination in Mice. Science (2018) 359:1408–11. doi: 10.1126/science.aal3662
9. Frazao A, Messaoudene M, Nunez N, Dulphy N, Roussin F, Sedlik C, et al. CD16+NKG2Ahigh Natural Killer Cells Infiltrate Breast Cancer-Draining Lymph Nodes. Cancer Immunol Res (2019) 7:208–18. doi: 10.1158/2326-6066.CIR-18-0085
10. Cheng L, Wang Y, Huang L. Exosomes From M1-Polarized Macrophages Potentiate the Cancer Vaccine by Creating a Pro-Inflammatory Microenvironment in the Lymph Node. Mol Ther (2017) 25:1665–75. doi: 10.1016/j.ymthe.2017.02.007
11. Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, et al. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell (2016) 30:324–36. doi: 10.1016/j.ccell.2016.06.003
12. Brown D, Smeets D, Székely B, Larsimont D, Szász AM, Adnet PY, et al. Phylogenetic Analysis of Metastatic Progression in Breast Cancer Using Somatic Mutations and Copy Number Aberrations. Nat Commun (2017) 8:14944. doi: 10.1038/ncomms14944
13. Salehi S, Kabeer F, Ceglia N, Andronescu M, Williams MJ, Campbell KR, et al. Clonal Fitness Inferred From Time-Series Modelling of Single-Cell Cancer Genomes. Nature (2021) 595:585–90. doi: 10.1038/s41586-021-03648-3
14. Simeonov KP, Byrns CN, Clark ML, Norgard RJ, Martin B, Stanger BZ, et al. Single-Cell Lineage Tracing of Metastatic Cancer Reveals Selection of Hybrid EMT States. Cancer Cell (2021) 39:1150–62.e9. doi: 10.1016/j.ccell.2021.05.005
15. Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The Evolutionary History of Lethal Metastatic Prostate Cancer. Nature (2015) 520:353–7. doi: 10.1038/nature14347
16. Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, et al. Minimal Functional Driver Gene Heterogeneity Among Untreated Metastases. Science (2018) 361:1033–7. doi: 10.1126/science.aat7171
17. Zhang W, Bado IL, Hu J, Wan YW, Wu L, Wang H, et al. The Bone Microenvironment Invigorates Metastatic Seeds for Further Dissemination. Cell (2021) 184:2471–86.e20. doi: 10.1016/j.cell.2021.03.011
18. Bochner BH. Lymph Node Dissection for Advanced Bladder Cancer: Is There a Role? Eur Urol Focus (2020) 6:615–6. doi: 10.1016/j.euf.2019.09.008
19. Rausei S, Ruspi L, Galli F, Pappalardo V, Rocco GD, Martignoni F, et al. Seventh Tumor-Node-Metastasis Staging of Gastric Cancer: Five-Year Follow-Up. World J Gastroenterol (2016) 22:7748–53. doi: 10.3748/wjg.v22.i34.7748
20. Kayani B, Zacharakis E, Ahmed K, Hanna GB. Lymph Node Metastases and Prognosis in Oesophageal Carcinoma–a Systematic Review. Eur J Surg Oncol (2011) 37:747–53. doi: 10.1016/j.ejso.2011.06.018
21. O'Neill NA, Eppler HB, Jewell CM, Bromberg JS. Harnessing the Lymph Node Microenvironment. Curr Opin Organ Transplant (2018) 23:73–82. doi: 10.1097/MOT.0000000000000488
22. Pereira ER, Jones D, Jung K, Padera TP. The Lymph Node Microenvironment and its Role in the Progression of Metastatic Cancer. Semin Cell Dev Biol (2015) 38:98–105. doi: 10.1016/j.semcdb.2015.01.008
23. Minute L, Teijeira A, Sanchez-Paulete AR, Ochoa MC, Alvarez M, Otano I, et al. Cellular Cytotoxicity is a Form of Immunogenic Cell Death. J Immunother Cancer (2020) 8:e000325. doi: 10.1136/jitc-2019-000325
24. Hasegawa H, Matsumoto T. Mechanisms of Tolerance Induction by Dendritic Cells. In Vivo Front Immunol (2018) 9:350:350. doi: 10.3389/fimmu.2018.00350
25. Kitamura T, Qian BZ, Pollard JW. Immune Cell Promotion of Metastasis. Nat Rev Immunol (2015) 15:73–86. doi: 10.1038/nri3789
26. Lee HE, Park DJ, Kim WH, Kim HH, Lee HS. High FOXP3+ Regulatory T-Cell Density in the Sentinel Lymph Node is Associated With Downstream non-Sentinel Lymph-Node Metastasis in Gastric Cancer. Br J Cancer (2011) 105:413–9. doi: 10.1038/bjc.2011.248
27. Gillot L, Baudin L, Rouaud L, Kridelka F, Noël A. The Pre-Metastatic Niche in Lymph Nodes: Formation and Characteristics. Cell Mol Life Sci (2021) 78:5987–6002. doi: 10.1007/s00018-021-03873-z
28. Li M, Li A, Zhou S, Xu Y, Xiao YX, Bi R, et al. Heterogeneity of PD-L1 Expression in Primary Tumors and Paired Lymph Node Metastases of Triple Negative Breast Cancer. BMC Cancer (2018) 18:4. doi: 10.1186/s12885-017-3916-y
29. Schneider S, Kadletz L, Wiebringhaus R, Kenner L, Selzer E, Füreder T, et al. PD-1 and PD-L1 Expression in HNSCC Primary Cancer and Related Lymph Node Metastasis - Impact on Clinical Outcome. Histopathology (2018) 73:573–84. doi: 10.1111/his.13646
30. Raniszewska A, Polubiec-Kownacka M, Rutkowska E, Domagala-Kulawik J. PD-L1 Expression on Lung Cancer Stem Cells in Metastatic Lymph Nodes Aspirates. Stem Cell Rev Rep (2019) 15:324–30. doi: 10.1007/s12015-018-9860-7
31. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat Commun (2019) 10:1523. doi: 10.1038/s41467-019-09234-6
32. Lucas ED, Tamburini BAJ. Lymph Node Lymphatic Endothelial Cell Expansion and Contraction and the Programming of the Immune Response. Front Immunol (2019) 10:36. doi: 10.3389/fimmu.2019.00036
33. Harlé G, Kowalski C, Garnier L, Hugues S. Lymph Node Stromal Cells: Mapmakers of T Cell Immunity. Int J Mol Sci (2020) 21:7785. doi: 10.3390/ijms21207785
34. Luther SA, Tang HL, Hyman PL, Farr AG, Cyster JG. Coexpression of the Chemokines ELC and SLC by T Zone Stromal Cells and Deletion of the ELC Gene in the Plt/Plt Mouse. Proc Natl Acad Sci USA (2000) 97:12694–9. doi: 10.1073/pnas.97.23.12694
35. Rodda LB, Lu E, Bennett ML, Sokol C, Wang X, Luther SA, et al. Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity (2018) 48:1014–28.e6. doi: 10.1016/j.immuni.2018.04.006
36. Li X, Zhao J, Kasinath V, Uehara M, Jiang L, Banouni N, et al. Lymph Node Fibroblastic Reticular Cells Deposit Fibrosis-Associated Collagen Following Organ Transplantation. J Clin Invest (2020) 130:4182–94. doi: 10.1172/JCI136618
37. Fujimoto N, He Y, D'Addio M, Tacconi C, Detmar M, Dieterich LC, et al. Single-Cell Mapping Reveals New Markers and Functions of Lymphatic Endothelial Cells in Lymph Nodes. PloS Biol (2020) 18:e3000704. doi: 10.1371/journal.pbio.3000704
38. Pham TH, Baluk P, Xu Y, Grigorova I, Bankovich AJ, Pappu R, et al. Lymphatic Endothelial Cell Sphingosine Kinase Activity is Required for Lymphocyte Egress and Lymphatic Patterning. J Exp Med (2010) 207:17–27. doi: 10.1084/jem.20091619
39. Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of Histone Acetylation in the Nucleus by Sphingosine-1-Phosphate. Science (2009) 325:1254–7. doi: 10.1126/science.1176709
40. Chang CL, Ho MC, Lee PH, Hsu CY, Huang WP, Lee H, et al. S1P(5) is Required for Sphingosine 1-Phosphate-Induced Autophagy in Human Prostate Cancer PC-3 Cells. Am J Physiol Cell Physiol (2009) 297:C451–8. doi: 10.1152/ajpcell.00586.2008
41. Cheng JC, Wang EY, Yi Y, Thakur A, Tsai SH, Hoodless PA, et al. S1P Stimulates Proliferation by Upregulating CTGF Expression Through S1PR2-Mediated YAP Activation. Mol Cancer Res (2018) 16:1543–55. doi: 10.1158/1541-7786.MCR-17-0681
42. Li MH, Sanchez T, Pappalardo A, Lynch KR, Hla T, Ferrer F, et al. Induction of Antiproliferative Connective Tissue Growth Factor Expression in Wilms' Tumor Cells by Sphingosine-1-Phosphate Receptor 2. Mol Cancer Res (2008) 6:1649–56. doi: 10.1158/1541-7786.MCR-07-2048
43. Young N, Van Brocklyn JR. Roles of Sphingosine-1-Phosphate (S1P) Receptors in Malignant Behavior of Glioma Cells. Differential Effects of S1P2 on Cell Migration and Invasiveness. Exp Cell Res (2007) 313:1615–27. doi: 10.1016/j.yexcr.2007.02.009
44. Weigert A, Tzieply N, von Knethen A, Johann AM, Schmidt H, Geisslinger G, et al. Tumor Cell Apoptosis Polarizes Macrophages Role of Sphingosine-1-Phosphate. Mol Biol Cell (2007) 18:3810–9. doi: 10.1091/mbc.e06-12-1096
45. He H, Zhang S, Tighe S, Son J, Tseng SCG. Immobilized Heavy Chain-Hyaluronic Acid Polarizes Lipopolysaccharide-Activated Macrophages Toward M2 Phenotype. J Biol Chem (2013) 288:25792–803. doi: 10.1074/jbc.M113.479584
46. Priceman SJ, Shen S, Wang L, Deng J, Yue C, Kujawski M, et al. S1PR1 is Crucial for Accumulation of Regulatory T Cells in Tumors via STAT3. Cell Rep (2014) 6:992–9. doi: 10.1016/j.celrep.2014.02.016
47. Lukacs-Kornek V, Malhotra D, Fletcher AL, Acton SE, Elpek KG, Tayalia P, et al. Regulated Release of Nitric Oxide by Nonhematopoietic Stroma Controls Expansion of the Activated T Cell Pool in Lymph Nodes. Nat Immunol (2011) 12:1096–104. doi: 10.1038/ni.2112
48. Nörder M, Gutierrez MG, Zicari S, Cervi E, Caruso A, Guzmán CA, et al. Lymph Node-Derived Lymphatic Endothelial Cells Express Functional Costimulatory Molecules and Impair Dendritic Cell-Induced Allogenic T-Cell Proliferation. FASEB J (2012) 26:2835–46. doi: 10.1096/fj.12-205278
49. Nguyen T, Brunson D, Crespi CL, Penman BW, Wishnok JS, Tannenbaum SR, et al. DNA Damage and Mutation in Human Cells Exposed to Nitric Oxide. Vitro Proc Natl Acad Sci USA (1992) 89:3030–4. doi: 10.1073/pnas.89.7.3030
50. Somasundaram V, Basudhar D, Bharadwaj G, No HJ, Ridnour LA, Cheng RYS, et al. Molecular Mechanisms of Nitric Oxide in Cancer Progression, Signal Transduction, and Metabolism. Antioxid Redox Signal (2019) 30:1124–43. doi: 10.1089/ars.2018.7527
51. Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol (2016) 8:a019521. doi: 10.1101/cshperspect.a019521
52. Kulis M, Esteller M. DNA Methylation and Cancer. Adv Genet (2010) 70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2
53. Baseler WA, Davies LC, Quigley L, Ridnour LA, Weiss JM, Hussain SP, et al. Autocrine IL-10 Functions as a Rheostat for M1 Macrophage Glycolytic Commitment by Tuning Nitric Oxide Production. Redox Biol (2016) 10:12–23. doi: 10.1016/j.redox.2016.09.005
54. Hickok JR, Vasudevan D, Antholine WE, Thomas DD. Nitric Oxide Modifies Global Histone Methylation by Inhibiting Jumonji C Domain-Containing Demethylases. J Biol Chem (2013) 288:16004–15. doi: 10.1074/jbc.M112.432294
55. Vos L, Dietrich J, Strieth S, Bootz F, Dietrich D, Franzen A, et al. PD-1, CTLA4, PD-L1 and PD-L2 DNA Methylation in Papillary Thyroid Carcinoma. Immunotherapy (2020) 12:903–20. doi: 10.2217/imt-2020-0037
56. Wang H, Fu C, Du J, Wang H, He R, Yin X, et al. Enhanced Histone H3 Acetylation of the PD-L1 Promoter via the COP1/c-Jun/HDAC3 Axis is Required for PD-L1 Expression in Drug-Resistant Cancer Cells. J Exp Clin Cancer Res (2020) 39:29. doi: 10.1186/s13046-020-1536-x
57. Wong CC, Xu J, Bian X, Wu J, Kang W, Qian Y, et al. In Colorectal Cancer Cells With Mutant KRAS, SLC25A22-Mediated Glutaminolysis Reduces DNA Demethylation to Increase WNT Signaling, Stemness, and Drug Resistance. Gastroenterology (2020) 159:2163–80.e6. doi: 10.1053/j.gastro.2020.08.016
58. Pangeni RP, Yang L, Zhang K, Wang J, Li W, Guo C, et al. G9a Regulates Tumorigenicity and Stemness Through Genome-Wide DNA Methylation Reprogramming in non-Small Cell Lung Cancer. Clin Epigenet (2020) 12:88. doi: 10.1186/s13148-020-00879-5
59. Chen X, Song E. Turning Foes to Friends: Targeting Cancer-Associated Fibroblasts. Nat Rev Drug Discovery (2019) 18:99–115. doi: 10.1038/s41573-018-0004-1
60. Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In Search of Definitions: Cancer-Associated Fibroblasts and Their Markers. Int J Cancer (2020) 146:895–905. doi: 10.1002/ijc.32193
61. Cochran AJ, Huang RR, Lee J, Itakura E, Leong SP, Essner R, et al. Tumour-Induced Immune Modulation of Sentinel Lymph Nodes. Nat Rev Immunol (2006) 6:659–70. doi: 10.1038/nri1919
62. Dermani FK, Samadi P, Rahmani G, Kohlan AK, Najafi R. PD-1/PD-L1 Immune Checkpoint: Potential Target for Cancer Therapy. J Cell Physiol (2019) 234:1313–25. doi: 10.1002/jcp.27172
63. Gou Q, Dong C, Xu H, Khan B, Jin JH, Liu Q, et al. PD-L1 Degradation Pathway and Immunotherapy for Cancer. Cell Death Dis (2020) 11:955. doi: 10.1038/s41419-020-03140-2
64. Zhang M, Sun H, Zhao S, Wang Y, Pu HH, Wang Y, et al. Expression of PD-L1 and Prognosis in Breast Cancer: A Meta-Analysis. Oncotarget (2017) 8:31347–54. doi: 10.18632/oncotarget.15532
65. Schoenfeld AJ, Rizvi H, Bandlamudi C, Sauter JL, Travis WD, Rekhtman N, et al. Clinical and Molecular Correlates of PD-L1 Expression in Patients With Lung Adenocarcinomas. Ann Oncol (2020) 31:599–608. doi: 10.1016/j.annonc.2020.01.065
66. Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell (2019) 76:359–70. doi: 10.1016/j.molcel.2019.09.030
67. Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC Regulates the Antitumor Immune Response Through CD47 and PD-L1. Science (2016) 352:227–31. doi: 10.1126/science.aac9935
68. Pan W, Wang W, Huang J, Lu K, Huang S, Jiang DX, et al. The Prognostic Role of C-MYC Amplification in Schistosomiasis-Associated Colorectal Cancer. Jpn J Clin Oncol (2020) 50:446–55. doi: 10.1093/jjco/hyz210
69. Soave A, Kluwe L, Yu H, Rink M, Gild P, Vetterlein MW, et al. Copy Number Variations in Primary Tumor, Serum and Lymph Node Metastasis of Bladder Cancer Patients Treated With Radical Cystectomy. Sci Rep (2020) 10:21562. doi: 10.1038/s41598-020-75869-x
70. Watson PH, Safneck JR, Le K, Dubik D, Shiu RP. Relationship of C-Myc Amplification to Progression of Breast Cancer From in Situ to Invasive Tumor and Lymph Node Metastasis. J Natl Cancer Inst (1993) 85:902–7. doi: 10.1093/jnci/85.11.902
71. Carbotti G, Barisione G, Airoldi I, Mezzanzanica D, Bagnoli M, Ferrero S, et al. IL-27 Induces the Expression of IDO and PD-L1 in Human Cancer Cells. Oncotarget (2015) 6:43267–80, 6. doi: 10.18632/oncotarget.6530
72. Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep (2017) 19:1189–201. doi: 10.1016/j.celrep.2017.04.031
73. Wang X, Yang L, Huang F, Zhang QY, Liu S, Ma L, et al. Inflammatory Cytokines IL-17 and TNF-α Up-Regulate PD-L1 Expression in Human Prostate and Colon Cancer Cells. Immunol Lett (2017) 184:7–14. doi: 10.1016/j.imlet.2017.02.006
74. Zhang N, Zeng Y, Du W, Zhu JJ, Shen D, Liu ZY, et al. The EGFR Pathway is Involved in the Regulation of PD-L1 Expression via the IL-6/JAK/STAT3 Signaling Pathway in EGFR-Mutated non-Small Cell Lung Cancer. Int J Oncol (2016) 49:1360–8. doi: 10.3892/ijo.2016.3632
75. Ni XY, Sui HX, Liu Y, Ke SZ, Wang YN, Gao FG. TGF-β of Lung Cancer Microenvironment Upregulates B7H1 and GITRL Expression in Dendritic Cells and is Associated With Regulatory T Cell Generation. Oncol Rep (2012) 28:615–21. doi: 10.3892/or.2012.1822
76. Núñez NG, Tosello Boari J, Ramos RN, Richer W, Cagnard N, Anderfuhren CD, et al. Tumor Invasion in Draining Lymph Nodes is Associated With Treg Accumulation in Breast Cancer Patients. Nat Commun (2020) 11:3272. doi: 10.1038/s41467-020-17046-2
77. Solis-Castillo LA, Garcia-Romo GS, Diaz-Rodriguez A, Hernandez DR, Tellez-Rivera E, Rosales-Garcia VH, et al. Tumor-Infiltrating Regulatory T Cells, CD8/Treg Ratio, and Cancer Stem Cells are Correlated With Lymph Node Metastasis in Patients With Early Breast Cancer. Breast Cancer (2020) 27:837–49. doi: 10.1007/s12282-020-01079-y
78. Seifert AM, Eymer A, Heiduk M, Wehner R, Tunger A, Renesse JV, et al. PD-1 Expression by Lymph Node and Intratumoral Regulatory T Cells Is Associated With Lymph Node Metastasis in Pancreatic Cancer. Cancers (Basel) (2020) 12:2756. doi: 10.3390/cancers12102756
79. Tanaka A, Sakaguchi S. Regulatory T Cells in Cancer Immunotherapy. Cell Res (2017) 27:109–18. doi: 10.1038/cr.2016.151
80. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T Cells and Immune Tolerance. Cell (2008) 133:775–87. doi: 10.1016/j.cell.2008.05.009
81. Colak S, Ten Dijke P. Targeting TGF-β Signaling in Cancer. Trends Cancer (2017) 3:56–71. doi: 10.1016/j.trecan.2016.11.008
82. Katsuno Y, Lamouille S, Derynck R. TGF-β Signaling and Epithelial-Mesenchymal Transition in Cancer Progression. Curr Opin Oncol (2013) 25:76–84. doi: 10.1097/CCO.0b013e32835b6371
83. Liu M, Li S, Li MO. TGF-β Control of Adaptive Immune Tolerance: A Break From Treg Cells. Bioessays (2018) 40:e1800063. doi: 10.1002/bies.201800063
84. Zhao Y, Wu T, Shao S, Shi B, Zhao Y. Phenotype, Development, and Biological Function of Myeloid-Derived Suppressor Cells. Oncoimmunology (2015) 5:e1004983. doi: 10.1080/2162402X.2015.1004983
85. Gunderson AJ, Yamazaki T, McCarty K, Fox N, Phillips M, Alice A, et al. Tgfβ Suppresses CD8+ T Cell Expression of CXCR3 and Tumor Trafficking. Nat Commun (2020) 11:1749. doi: 10.1038/s41467-020-15404-8
86. Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci (2019) 20:2767. doi: 10.3390/ijms20112767
87. Kim BN, Ahn DH, Kang N, Yeo CD, Kim YK, Lee KY, et al. TGF-β Induced EMT and Stemness Characteristics are Associated With Epigenetic Regulation in Lung Cancer. Sci Rep (2020) 10(1):10597. doi: 10.1038/s41598-020-67325-7
88. Noordhuis MG, Fehrmann RS, Wisman GB, Yeo CD, Kim YK, Lee KY, et al. Involvement of the TGF-Beta and Beta-Catenin Pathways in Pelvic Lymph Node Metastasis in Early-Stage Cervical Cancer. Clin Cancer Res (2011) 17:1317–30. doi: 10.1158/1078-0432.CCR-10-2320
89. Huang SC, Wei PC, Hwang-Verslues WW, Kuo WH, Jeng YM, Hu CM, et al. TGF-β1 Secreted by Tregs in Lymph Nodes Promotes Breast Cancer Malignancy via Up-Regulation of IL-17rb. EMBO Mol Med (2017) 9:1660–80. doi: 10.15252/emmm.201606914
90. Pelon F, Bourachot B, Kieffer Y, Magagna I, Mermet-Meillon F, Bonnet I, et al. Cancer-Associated Fibroblast Heterogeneity in Axillary Lymph Nodes Drives Metastases in Breast Cancer Through Complementary Mechanisms. Nat Commun (2020) 11:404. doi: 10.1038/s41467-019-14134-w
91. Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol (2017) 1509:1–10. doi: 10.1007/978-1-4939-6524-3_1
92. Lee YS, Dutta A. MicroRNAs in Cancer. Annu Rev Pathol (2009) 4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222
93. Rupaimoole R, Slack FJ. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat Rev Drug Discov (2017) 16:203–22. doi: 10.1038/nrd.2016.246
94. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 Family and miR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat Cell Biol (2008) 10:593–601. doi: 10.1038/ncb1722
95. Suzuki HI. MicroRNA Control of TGF-β Signaling. Int J Mol Sci (2018) 19:1901. doi: 10.3390/ijms19071901
96. Avery-Kiejda KA, Braye SG, Mathe A, Forbes JF, Scott RJ. Decreased Expression of Key Tumour Suppressor microRNAs is Associated With Lymph Node Metastases in Triple Negative Breast Cancer. BMC Cancer (2014) 14:51. doi: 10.1186/1471-2407-14-51
97. Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, et al. tsRNA Signatures in Cancer. Proc Natl Acad Sci U.S.A. (2017) 114:8071–6. doi: 10.1073/pnas.1706908114
98. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene (2018) 37:555–65. doi: 10.1038/onc.2017.361
99. Anastasiadou E, Jacob LS, Slack FJ. Non-Coding RNA Networks in Cancer. Nat Rev Cancer (2018) 18:5–18. doi: 10.1038/nrc.2017.99
100. Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, et al. Exosomal Long Noncoding RNA LNMAT2 Promotes Lymphatic Metastasis in Bladder Cancer. J Clin Invest (2020) 130:404–21. doi: 10.1172/JCI130892
101. He F, Zhong X, Lin Z, Lin J, Qiu M, Li X, et al. Plasma Exo-Hsa_circRNA_0056616: A Potential Biomarker for Lymph Node Metastasis in Lung Adenocarcinoma. J Cancer (2020) 11:4037–46. doi: 10.7150/jca.30360
102. Jadhav KB, Nagraj SK, Arora S. miRNA for the Assessment of Lymph Node Metastasis in Patients With Oral Squamous Cell Carcinoma: Systematic Review and Metanalysis. J Oral Pathol Med (2021) 50):345–52. doi: 10.1111/jop.13134
103. Zhao S, Yao D, Chen J, Ding N. Circulating miRNA-20a and miRNA-203 for Screening Lymph Node Metastasis in Early Stage Cervical Cancer. Genet Test Mol Biomarkers (2013) 17:631–6. doi: 10.1089/gtmb.2013.0085
104. Chen W, Cai F, Zhang B, Barekati Z, Zhong XY. The Level of Circulating miRNA-10b and miRNA-373 in Detecting Lymph Node Metastasis of Breast Cancer: Potential Biomarkers. Tumour Biol (2013) 34:455–62. doi: 10.1007/s13277-012-0570-5
105. Chen W, Tang Z, Sun Y, Zhang Y, Wang X, Shen Z, et al. miRNA Expression Profile in Primary Gastric Cancers and Paired Lymph Node Metastases Indicates That miR-10a Plays a Role in Metastasis From Primary Gastric Cancer to Lymph Nodes. Exp Ther Med (2012) 3:351–6. doi: 10.3892/etm.2011.411
106. Lu X, Kang Y. Hypoxia and Hypoxia-Inducible Factors: Master Regulators of Metastasis. Clin Cancer Res (2010) 16:5928–35. doi: 10.1158/1078-0432.CCR-10-1360
107. Schito L, Rey S, Tafani M, Zhang H, Wong CC, Russo A, et al. Hypoxia-Inducible Factor 1-Dependent Expression of Platelet-Derived Growth Factor B Promotes Lymphatic Metastasis of Hypoxic Breast Cancer Cells. Proc Natl Acad Sci U.S.A. (2012) 109:E2707–16. doi: 10.1073/pnas.1214019109
108. Zhuo W, Jia L, Song N, Lu X, Ding Y, Wang X, et al. The CXCL12-CXCR4 Chemokine Pathway: A Novel Axis Regulates Lymphangiogenesis. Clin Cancer Res (2012) 18:5387–98. doi: 10.1158/1078-0432.CCR-12-0708
109. Semenza GL. Cancer-Stromal Cell Interactions Mediated by Hypoxia-Inducible Factors Promote Angiogenesis, Lymphangiogenesis, and Metastasis. Oncogene (2013) 32:4057–63. doi: 10.1038/onc.2012.578
110. Subarsky P, Hill RP. The Hypoxic Tumour Microenvironment and Metastatic Progression. Clin Exp Metastasis (2003) 20:237–50. doi: 10.1023/a:1022939318102
111. Cursiefen C, Chen L, Borges LP, Jackson D, Cao J, Radziejewski C, et al. VEGF-A Stimulates Lymphangiogenesis and Hemangiogenesis in Inflammatory Neovascularization via Macrophage Recruitment. J Clin Invest (2004) 113:1040–50. doi: 10.1172/JCI20465
112. Ji RC. Macrophages are Important Mediators of Either Tumor- or Inflammation-Induced Lymphangiogenesis. Cell Mol Life Sci (2012) 69:897–914. doi: 10.1007/s00018-011-0848-6
113. Zhao Y, Wang XX, Wu W, Long H, Huang J, Wang Z, et al. EZH2 Regulates PD-L1 Expression via HIF-1α in non-Small Cell Lung Cancer Cells. Biochem Biophys Res Commun (2019) 517:201–9. doi: 10.1016/j.bbrc.2019.07.039
114. Ding XC, Wang LL, Zhang XD, Xu JL, Li PF, Liang H, et al. The Relationship Between Expression of PD-L1 and HIF-1α in Glioma Cells Under Hypoxia. J Hematol Oncol (2021) 14:92. doi: 10.1186/s13045-021-01102-5
115. Chang YL, Yang CY, Lin MW, Wu CT, Yang PC. High Co-Expression of PD-L1 and HIF-1α Correlates With Tumour Necrosis in Pulmonary Pleomorphic Carcinoma. Eur J Cancer (2016) 60:125–35. doi: 10.1016/j.ejca.2016.03.012
116. Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin Switching. J Cell Sci (2008) 121:727–35. doi: 10.1242/jcs.000455
117. Tirpe AA, Gulei D, Ciortea SM, Crivii C, Berindan-Neagoe I. Hypoxia: Overview on Hypoxia-Mediated Mechanisms With a Focus on the Role of HIF Genes. Int J Mol Sci (2019) 20:6140. doi: 10.3390/ijms20246140
118. Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial Metabolism and ROS Generation are Essential for Kras-Mediated Tumorigenicity. Proc Natl Acad Sci USA (2010) 107:8788–93. doi: 10.1073/pnas.1003428107
119. Agrimi G, Brambilla L, Frascotti G, Pisano I, Porro D, Vai M, et al. Deletion or Overexpression of Mitochondrial NAD+ Carriers in Saccharomyces Cerevisiae Alters Cellular NAD and ATP Contents and Affects Mitochondrial Metabolism and the Rate of Glycolysis. Appl Environ Microbiol (2011) 77:2239–46. doi: 10.1128/AEM.01703-10
120. Krishnamurty AT, Turley SJ. Lymph Node Stromal Cells: Cartographers of the Immune System. Nat Immunol (2020) 21:369–80. doi: 10.1038/s41590-020-0635-3
Keywords: lymph node, tumor evolution, epigenetic reprogramming, immune escape, PD-L1, stemness, tumor metastasis
Citation: Li T, Liu T, Zhao Z, Xu X, Zhan S, Zhou S, Jiang N, Zhu W, Sun R, Wei F, Feng B, Guo H and Yang R (2022) The Lymph Node Microenvironment May Invigorate Cancer Cells With Enhanced Metastatic Capacities. Front. Oncol. 12:816506. doi: 10.3389/fonc.2022.816506
Received: 16 November 2021; Accepted: 02 February 2022;
Published: 28 February 2022.
Edited by:
Roger Chammas, University of São Paulo, BrazilCopyright © 2022 Li, Liu, Zhao, Xu, Zhan, Zhou, Jiang, Zhu, Sun, Wei, Feng, Guo and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Yang, ZG9jdG9yeXJAZ21haWwuY29t; Hongqian Guo, ZHIuZ2hxQG5qdS5lZHUuY24=
 Tianhang Li
Tianhang Li Tianyao Liu1
Tianyao Liu1 Zihan Zhao
Zihan Zhao Hongqian Guo
Hongqian Guo Rong Yang
Rong Yang