- 1Department of Medical Oncology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Prenatal Diagnosis Center, Affiliated Hospital of Weifang Medical University, Weifang, China
- 3Acornmed Biotechnology Co., Ltd., Beijing, China
- 4Department of Medical Oncology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 5Department of Radiotherapy and Chemotherapy, Hwamei Hospital, University of Chinese Academy of Sciences, Ningbo, China
- 6Department of Anorectal Surgical, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
Background: Colorectal cancer (CRC) is one of the most prevalent malignances worldwide. However, CRC with situs inversus totalis (SCRC) is extremely rare, and molecular characterization of this disease has never been investigated.
Methods: Tumor tissue samples from 8 patients with SCRC and 33 CRC patients without situs inversus totalis (NSCRC) were subjected to multigene next-generation sequencing.
Results: The most frequently mutated genes in SCRC were APC, TP53, CHEK2, MDC1, GNAQ, KRAS, and SMAD4. A high frequency of SCRC tumors had mutations in DNA damage repair genes. Single amino acid substitutions in the DNA damage repair genes caused by continuous double base substitution was identified in the majority of this population. Furthermore, mutational profiles showed notable differences between the SCRC and NSCRC groups. In particular, CHEK2, MDC1, GNAQ, SMAD4, BRCA1, HLA-B, LATS2, and NLRC5 mutations were more frequently observed in SCRC patients. The mutation loci distributions of KRAS in the SCRC cohort differed from that of the NSCRC cohort. Additionally, differences in the targeted genomic profiles and base substitution patterns were observed between the two groups.
Conclusions: These findings comprehensively revealed a molecular characterization of SCRC, which will contribute to the development of personalized therapy and improved clinical management of SCRC patients.
Introduction
Colorectal cancer (CRC) is the third most prevalent type of cancer and the second leading cause of cancer-associated mortality worldwide (1). In recent years, the incidence and mortality rates for CRC have been steadily increasing (2). CRC patients can achieve promising clinical outcomes with early diagnosis. However, most CRC patients are generally diagnosed at an advanced stage, and over 50% of CRC patients develop liver, lung and lymph node metastases with a high mortality rate (3).
A series of genomic mutations leads to the development and progression of CRC. In recent years, next-generation sequencing (NGS) has revealed a diversity of driver gene mutations and affected signaling pathways in CRC. Numerous studies have demonstrated that the most commonly mutated driver genes in CRC are APC, TP53, KRAS, SMAD4, and PIK3CA (4, 5). Additionally, the Cancer Genome Atlas network comprehensively investigated the mutational landscape and identified some novel genomic alterations in CRC (6). The WNT, TGF-β PI3K, RAS/MAPK, and p53 signaling pathways were found to be the most frequently altered pathways in CRC (6). Recurrent copy number variations included the amplification of ERBB2 and IGF2, and frequent chromosomal rearrangements included the fusion of NAV2 and TCF7L1 (6). Multi-omic results classified the CRC patients into hypermutated (16%) and non-hypermutated (84%) groups, and the hypermutated subgroup was characterized by high microsatellite instability, hypermethylation, MLH1 silencing, and genomic mutations in mismatch repair genes and POLE (6). Moreover, some large cohort studies found statistically significant differences in mutation status between right-sided and left-sided CRC for several genes, including KRAS, BRAF, and FBXW7, as well as for several pathways, including RAS/MAPK, TGF-β, and p53 signaling pathways (5, 7).
Situs inversus is an extremely rare congenital condition, with an overall incidence rate of 0.01% (8). The majority of situs inversus cases are situs inversus totalis, which are characterized by a left-to-right reversal of all the thoracic and abdominal organs. A growing number of people with situs inversus totalis have been identified because of advances in medical imaging technology. A large amount of literature has been published that covers treatments and appropriate operative approaches for CRC patients with situs inversus totalis (SCRC) (9–11). However, the molecular characterization of SCRC has been largely unexplored. In addition, genomic differences between SCRC patients and CRC patients without situs inversus totalis (NSCRC) are currently unknown.
To better understand SCRC disease biology and to elucidate the molecular features of SCRC, we systematically explored the molecular signatures in SCRC by NGS, and further compared these signatures between SCRC and NSCRC patients. These findings provide valuable genomic information for SCRC, which will contribute to promoting treatment strategies and clinical management of SCRC patients.
Materials and Methods
Patients and Sample Collection
A total of 41 patients with CRC from the Affiliated Sir Run Run Shaw Hospital of Zhejiang University School of Medicine and the First Affiliated Hospital of Zhejiang University School of Medicine between January 2014 and January 2020 were enrolled. Eight SCRC and 33 NSCRC patients were included. Clinical stages of patients ranged from I to IV, which was verified based on the American Joint Committee on Cancer staging scheme (8th edition). This study was approved by the ethical committee of the Affiliated Sir Run Run Shaw Hospital of Zhejiang University School of Medicine. Informed consent was obtained from all participants.
NGS
Tissue DNA was extracted utilizing the QIAamp Genomic DNA kit (Germany, QIAGEN) based on the manufacturer’s instructions. Sequencing libraries were generated according to Illumina standard library construction protocols (Illumina Inc.). The libraries were enriched using an Acornmed panel targeting 808 cancer-related genes. The captured libraries were then sequenced on the NovaSeq6000 System (Illumina Inc.). Sequencing reads were aligned to the reference human genome (hg19) using the BWA aligner (version 0.7.12). Base recalibration was conducted using GATK software (version 3.8). Single nucleotide variants (SNVs) and small insertions or deletions were identified using MuTect2 software (version 1.1.7). CONTRA software (version 2.0.8) was used to analyze copy number variant calling. An average coverage depth for tumor tissue was > 5000×. Mutations were identified based on these standards: mutant allele frequency (MAF) ≥ 1.0%, and at least 5 high-quality supporting reads (base quality ≥ 30, mapping quality ≥ 30).
Identification of Potentially Actionable Mutations With OncoKB
Potentially actionable mutations were identified using MSK’s Precision Oncology Knowledge Base (OncoKB) (https://www.oncokb.org/). This clinical support tool distills information from FDA approved regimens, National Comprehensive Cancer Network (NCCN) guidelines, and the published scientific literature. Clinically relevant alterations were classified into one of four levels according to the strength of evidence. Mutations are annotated by the level of evidence supporting the use of certain drugs. The levels of evidence are as follows:
Level 1: FDA-recognized biomarker predictive of response to an FDA-approved drug in this indication.
Level 2: Standard care biomarker recommended by the NCCN or other professional guidelines predictive of response to an FDA-approved drug in this indication.
Level 3A: Compelling clinical evidence supports the biomarker as being predictive of response to a drug in this indication.
Level 3B: Standard care or investigational biomarker predictive of response to an FDA-approved or investigational drug in another indication.
Level 4: Compelling biological evidence supports the biomarker as being predictive of response to a drug.
Statistical Analysis
SPSS 21.0 statistical software (IBM Corp.) was used to conduct our statistical analyses. A Fisher’s exact test was used to determine the associations between gene mutation status and clinical characteristics. Differences in continuous variables were measured using a Student’s t-test. A two-sided P< 0.05 was considered statistically significant.
Results
Patient Features
In the present study, a total of eight SCRC patients and 33 NSCRC patients were included. Among the SCRC patients, the median age at diagnosis was 60 years old (range, 51-75 years old). Seventy-five percent of the patients were female, and none of them were smokers. In the NSCRC cohort, the median age at diagnosis was also 60 years old (range, 28-79 years old). A total of 36.4% of the patients were female, and 24.2% of them were smokers. We further compared the clinical characteristics between the SCRC and NSCRC cohorts. There was no difference in the age between the two groups. However, compared with NSCRC patients, SCRC patients were more likely to be female (P = 0.048). The clinical and pathological characteristics of the patients are listed in Table 1.
Mutational Landscape in SCRC
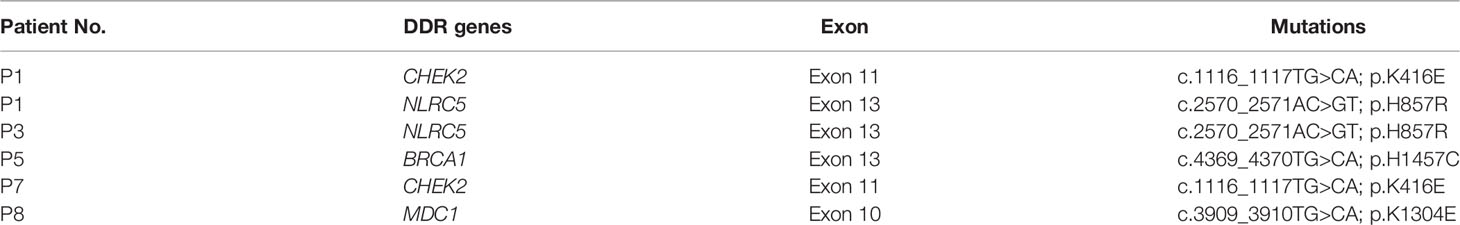
Samples from SCRC patients were profiled by targeted sequencing with an 808 cancer-related gene panel. An average coverage depth > 5000× was obtained for tumor tissue samples. The quality criteria used for mutation identification were a detection threshold of 1.0% and at least 5 high-quality supporting reads. All sequenced samples had at least one genetic alteration. A total of 110 somatic variants from 75 genes were identified. The most commonly mutated genes were APC (50%), TP53 (50%), CHEK2 (50%), MDC1 (50%), GNAQ (38%), KRAS (38%), and SMAD4 (38%) (Figure 1). Copy number variations in ERBB2, MRE11A, FANCM, BRD4, and TOP1 were found (Table S1). Gene rearrangements were identified in one SCRC patient including NTRK1-TPM3. Among the KRAS mutations, only G13D substitutions were observed.
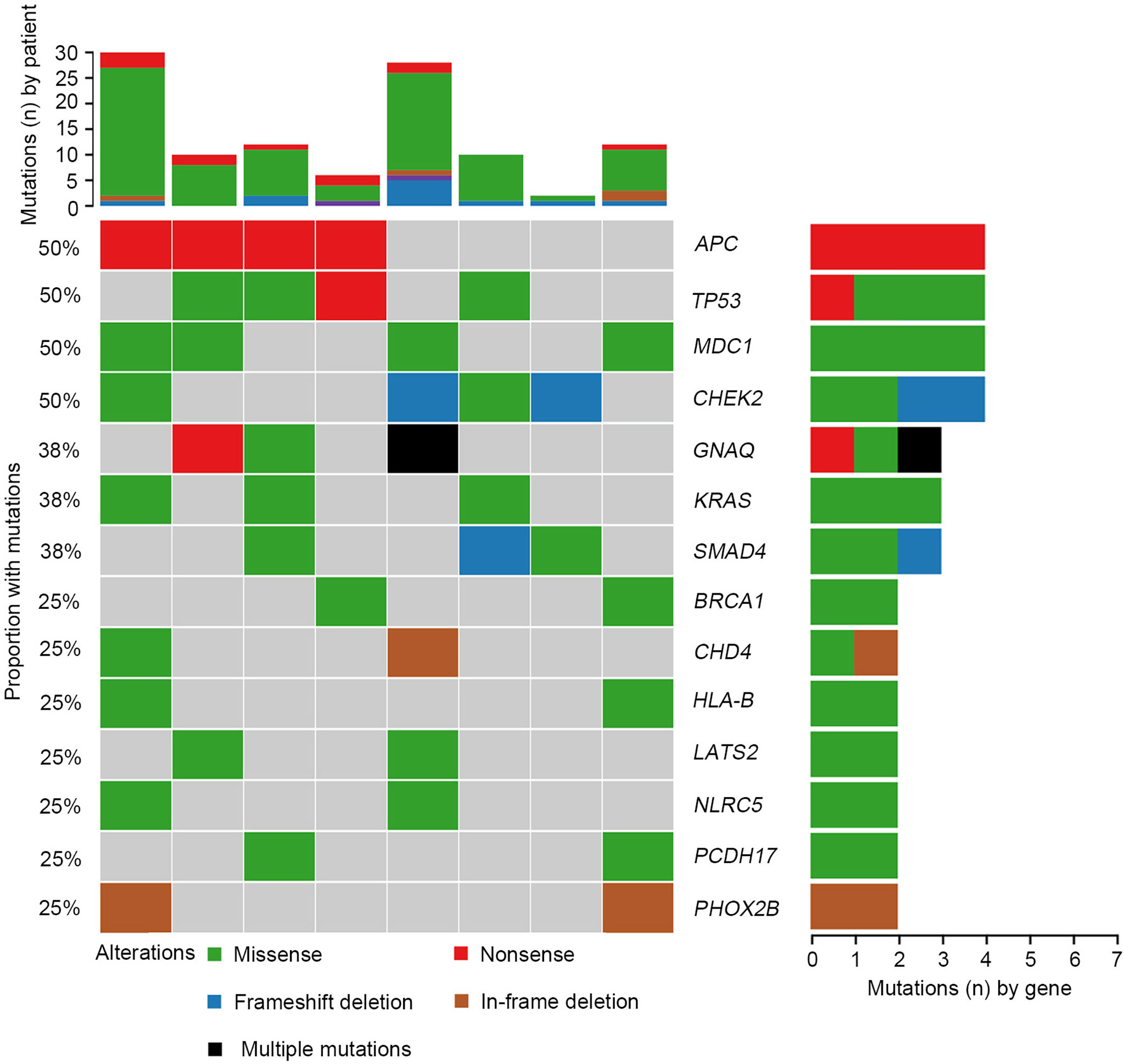
Figure 1 Mutational landscape of colorectal cancer patients with situs inversus totalis (SCRC). Genomic mutations were identified by targeted NGS of the tumor tissues from patients with SCRC. Multiple mutations, mutations numbers greater than two.
We further investigated the affected signaling pathways in SCRC based on the mutation data. The DNA damage repair (DDR), WNT, RAS/MAPK, TGF-β, and p53 signaling pathways were the most commonly altered pathways. Additionally, the Ca2+, epigenetic, IFN-γ, NOTCH, and PI3K signaling pathways were affected in some patients (Table S2, S3). Notably, the DDR pathway was strikingly altered in SCRC (87.5%) patients, and the majority of these patients (71.4%) had a single amino acid substitution in the DDR genes caused by continuous double base substitution (Table 2). To further understand the background of carcinogenesis, the pattern of nucleotide substitution was explored. All SNVs could be classified into transition (Ti) and transversion (Tv) substitutions according to the specific substitutions of pyrimidines and purines. The profile of base substitutions in SCRC showed the frequency of Ti was significantly higher than that of Tv (P = 0.006) (Figure 2A). Additionally, a high frequency of C > T Ti at the CpG dinucleotide was identified in 58.7% of SNVs (Figures 2B, C).
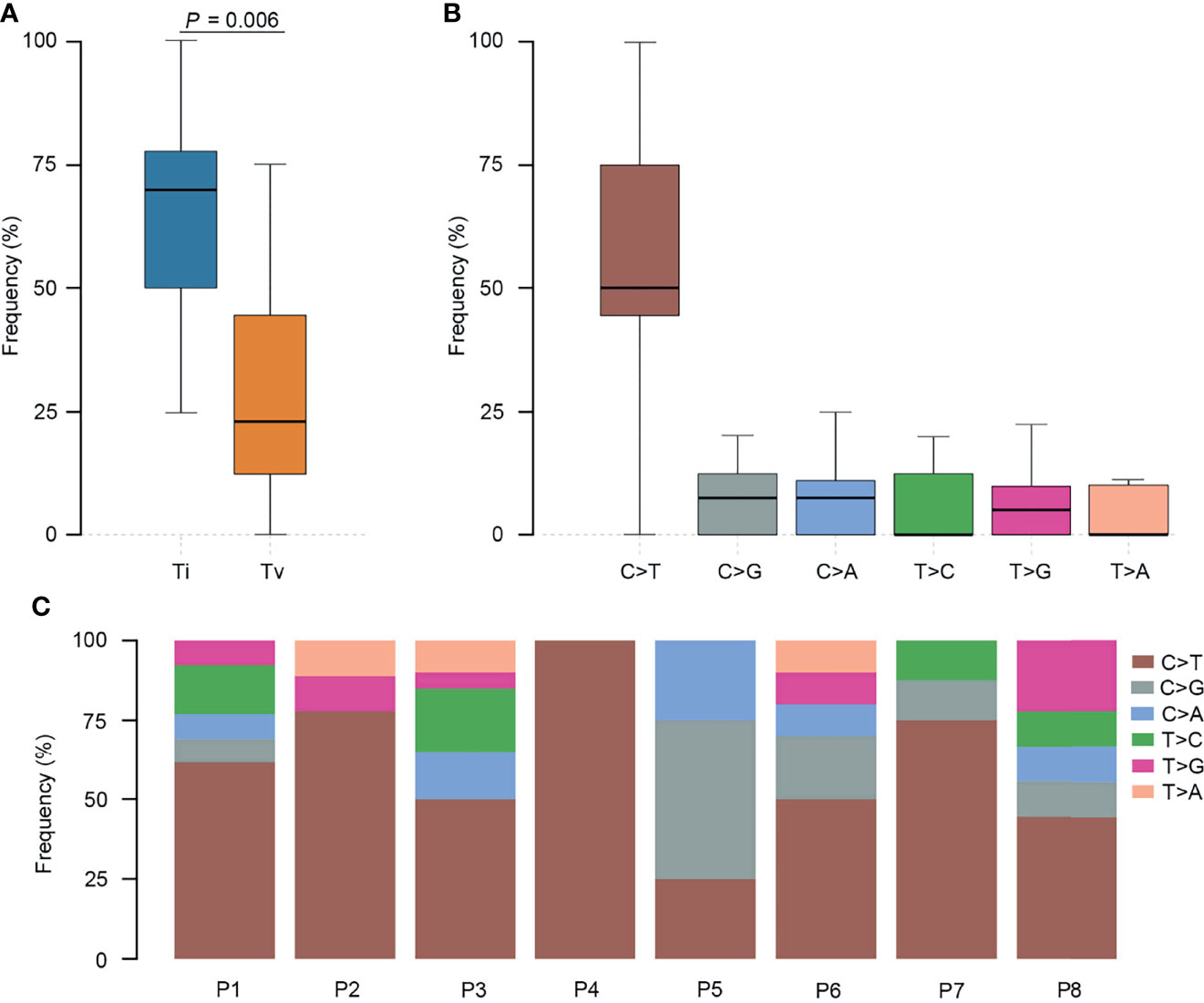
Figure 2 Transition (Ti) and transversion (Tv) profiles for SCRC. (A) Comparison between Ti and Tv ratios in SCRC. (B) The bar plot shows each type of Ti or Tv. (C) The box plot exhibits the ratio of each type of Ti or Tv for each patient.
Comparison of Genomic Profiles Between SCRC and NSCRC Patients
To further elucidate the molecular basis of SCRC tumorigenesis, a comprehensive comparison of genomic profiles between SCRC and NSCRC was conducted. Compared with NSCRC patients, significantly more genomic alterations were observed in CHEK2, MDC1, GNAQ, SMAD4, BRCA1, HLA-B, LATS2, and NLRC5 among SCRC patients. A potential statistical difference was identified in PHOX2B between the two group (P = 0.092) (Figures 3A, B). The incidence of KRAS mutations between the two groups was not significantly different (Figures 3A, B). The mutation loci distribution of KRAS was further evaluated. In the SCRC group, KRAS mutations were only distributed on codon 13 (Figure 4A). However, in the NSCRC group, KRAS mutations were distributed on different codons (Figure 4B). Further analysis indicated that the frequencies of mutation loci distributions of KRAS between the SCRC and NSCRC groups showed a statistically significant difference (P = 0.036) (Figure 4C). Moreover, the profile of base substitutions between the two groups was found to be different. A total of 34.7% of SNVs identified were C > T Ti in NSCRC (Figure 5A), which was lower than that in SCRC (34.7% vs 58.7%). No difference between the frequencies of Ti and Tv in NSCRC was observed (Figure 5B). The proportion of Ti in SCRC tended to be higher than that in NSCRC, although no statistically significant difference was observed (Figure 5C).

Figure 3 Comparison of the genomic landscape between SCRC and NSCRC cohorts. (A) Frequencies of genomic mutations in SCRC and NSCRC cohorts. (B) Corresponding P values comparing the prevalence of a gene’s mutations between different cohorts. Cases with a statistically significant difference (P < 0.05) are highlighted in yellow.
Figure 4 The mutation loci distributions of KRAS in SCRC and NSCRC groups. (A) Distributions of KRAS mutations identified in SCRC. (B) Distribution of KRAS mutations identified in NSCRC. (C) Comparison of the distributions of KRAS mutations between SCRC and NSCRC patients.
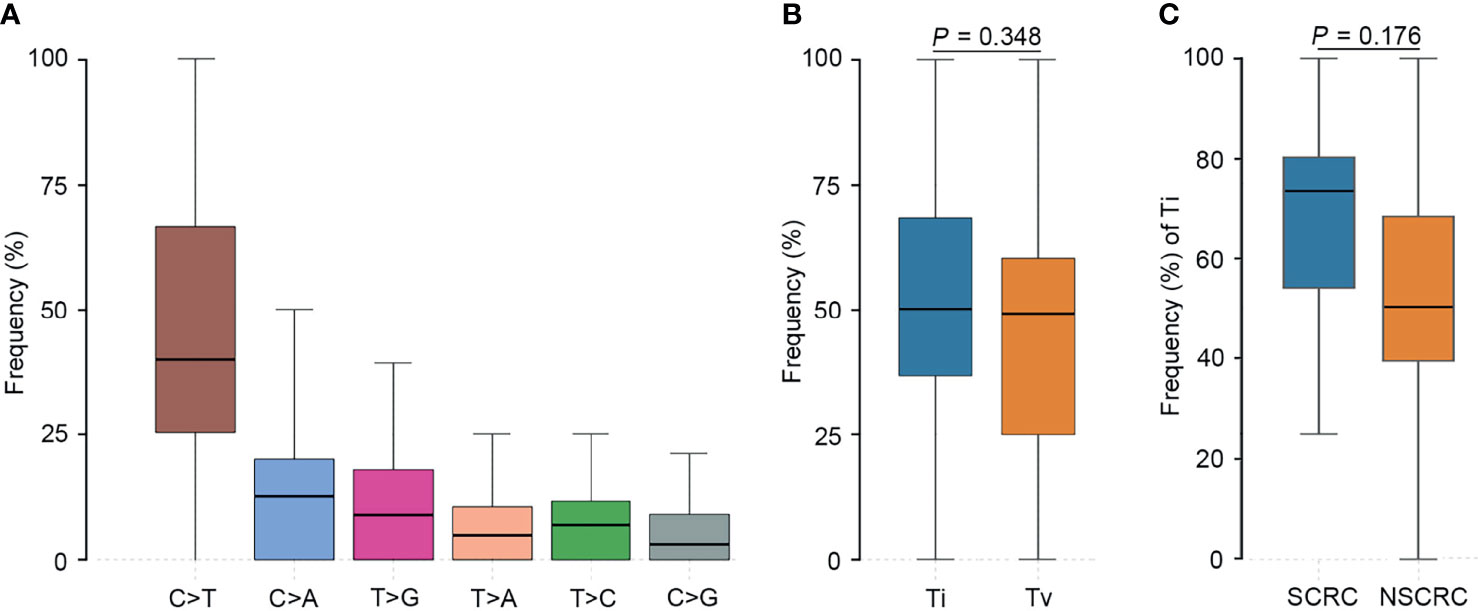
Figure 5 Comparison of the profiles of transition (Ti) and transversion (Tv) between SCRC and NSCRC patients. (A) The bar plot shows each type of Ti or Tv in NSCRC. (B) Comparison between Ti and Tv ratios in NSCRC. (C) Comparison of Ti ratios between the SCRC and NSCRC groups.
When investigating differences in altered signaling pathways, we found that the frequencies of the DDR, Ca2+, and IFN-γ signaling pathways in SCRC patients were significantly higher than those in NSCRC patients. A statistical trend was observed for the TGF-β pathway between the two cohorts (P = 0.060)(Figure 6). Notably, the mutated genes in the DDR signaling pathway between the two cohorts were markedly different. In SCRC, the most commonly altered DDR genes were CHEK2 (50%), MDC1 (50%), BRCA1 (25%), and NLRC5 (25%) (Figure 7A). However, the most frequently mutated DDR gene in NSCRC was POLE (6%). The other DDR mutations occurred in only one patient (Figure 7B). Furthermore, the phenomenon of single amino acid variations in DDR genes caused by continuous double base substitutions was not detected in NSCRC.
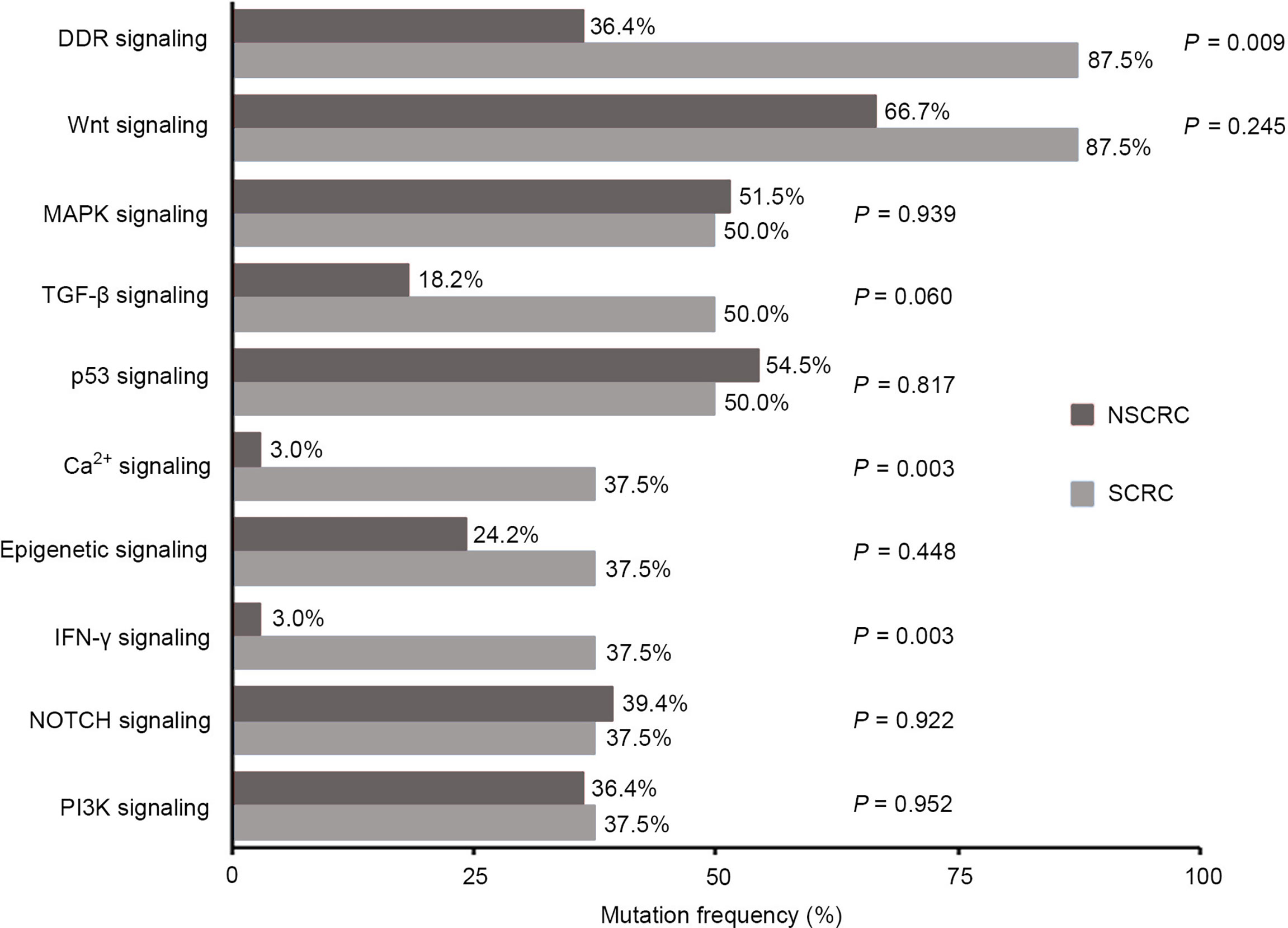
Figure 6 Comparison between SCRC and NSCRC patients of the detection rate of genomic mutations in signaling pathways.
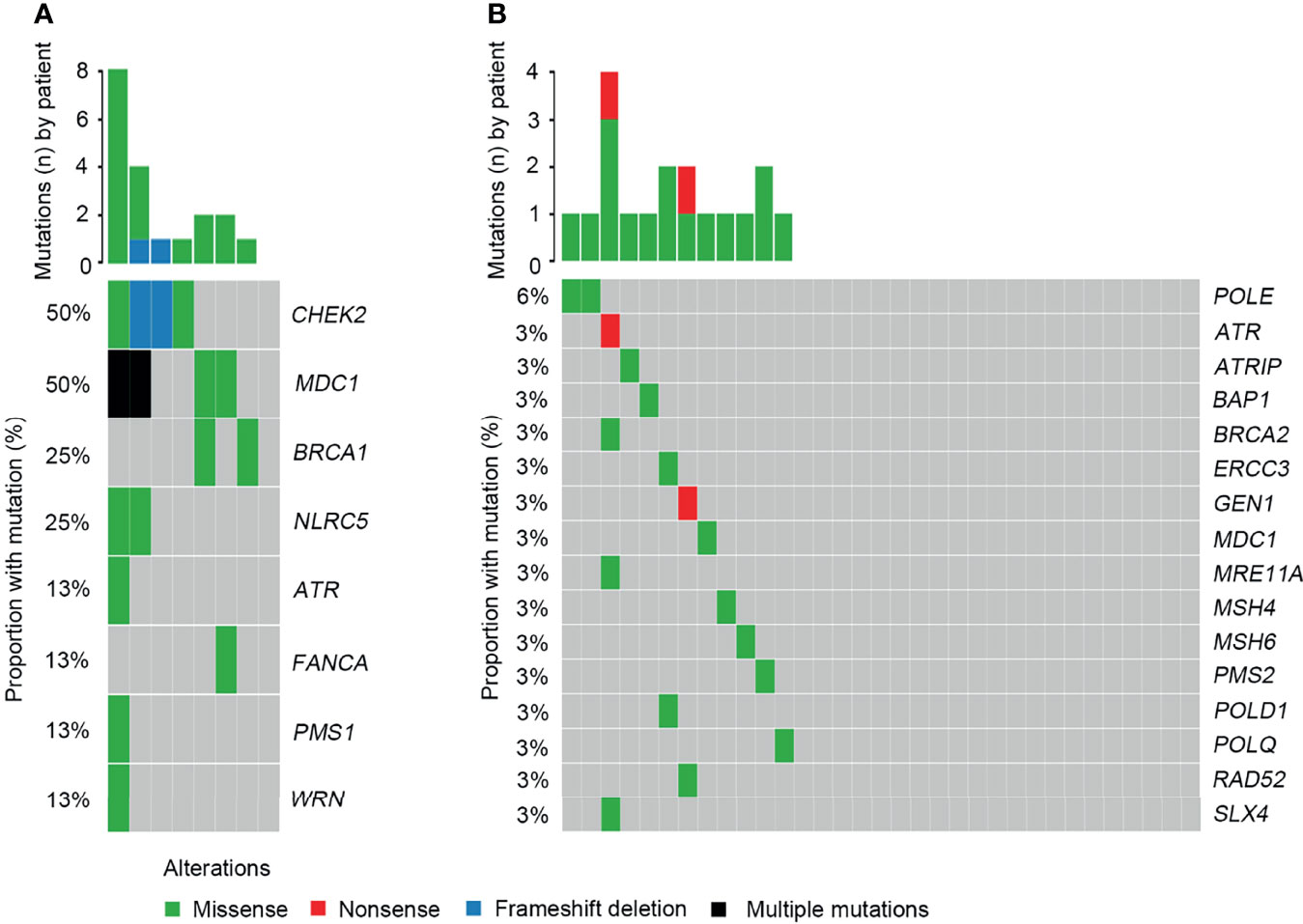
Figure 7 Comparison of the DDR profiles between SCRC and NSCRC patients. (A) The DDR profile in the SCRC group. (B) The DDR profile in the NSCRC group.
According to the OncoKB classification system, the profile of clinically relevant alterations was comprehensively evaluated. Overall, 24 clinically relevant mutations were detected in 87.5% of SCRC patients. Alterations in CHEK2, KRAS, and BRCA1 were the most common targets (Figure 8A and Table S4). One patient harboring a BRAF Y633C mutation was identified. Additionally, a clinically significant NTRK1-TPM3 fusion was identified. Although prior large cohort studies have reported the distributions of NTRK rearrangements in various tumors (12), they had not been uncovered in SCRC. Additionally, 78.8% of NSCRC cases harbored at least one clinically relevant alteration. However, the most frequently identified mutations occurred in KRAS and PIK3CA for this group (Figure 8B and Table S4). BRAF alterations were observed in two patients, including V600E and P655A mutations.
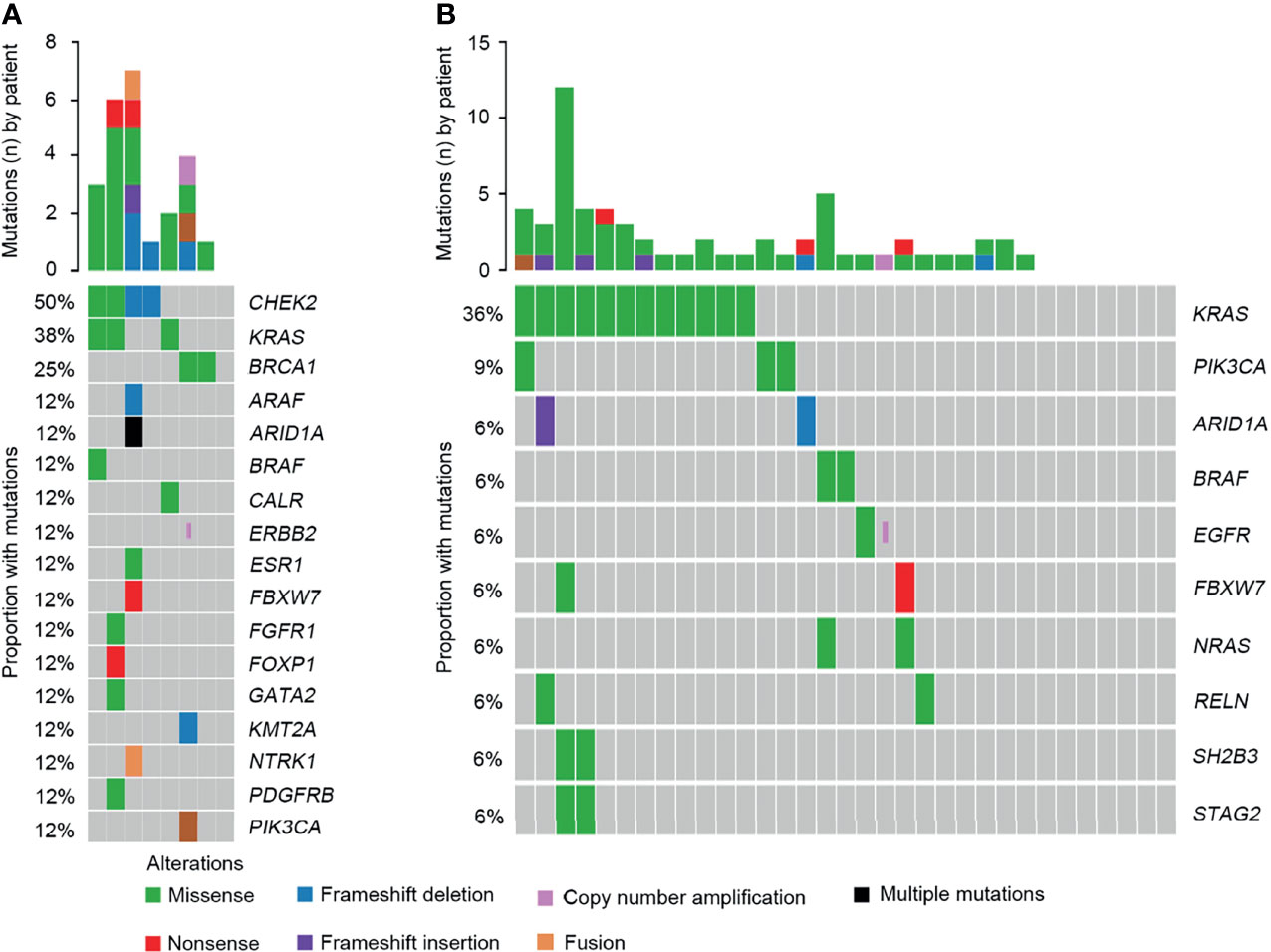
Figure 8 Comparison of the targeted genomic profiles between CRC and NSCRC patients. (A) The targeted genomic profile in the SCRC group. (B) The targeted genomic profile in the NSCRC group.
Discussion
Given the rarity of SCRC, the clinical characteristics and molecular features of SCRC remain largely unknown. Numerous studies indicated that CRC is more common in men than in women (13, 14). However, our study indicated that SCRC patients were more likely to be female, demonstrating that female gender has a larger impact on the prevalence of SCRC. With the advent of high-throughput sequencing technology, efforts have been made to identify the molecular characterization of CRC. However, most of the studies focused on CRC patients with normally positioned structures of the abdominal and thoracic cavities. No reports have described the molecular features of SCRC. For the first time, we investigated the molecular signatures of SCRC, which will be of great significance in understanding the biological characteristics of cancer cells and in promoting the clinical management of this population.
In the present study, we identified a large number of genes with point mutations, insertions, deletions, and copy number variations in SCRC. The most frequently mutated genes were APC, TP53, CHEK2, MDC1, GNAQ, KRAS, and SMAD4. It is known that APC, TP53, KRAS, and SMAD4 were the common driver genes in CRC (4, 5). In addition to these genes, our analysis identified novel driver genes in SCRC, including GNAQ, CHEK2, and MDC1. GNAQ encodes heterotrimeric G protein alpha subunits, which are crucial for G protein-coupled receptor signaling. Numerous studies have demonstrated that GNAQ mutations can lead to constitutive activation of the downstream RAS/MAPK pathway (15, 16), further indicating the important role of the RAS/MAPK pathway in SCRC. CHEK2 encodes a cell cycle checkpoint kinase involved in the DDR process (17). MDC1 encodes a scaffold protein that functions as a platform for the recruitment of different DDR factors, such as RNF8 and 53BP1, to regulate the DDR process (18). Furthermore, we found that 87.5% of SCRC patients carried genomic mutations in DDR genes. The DDR system is mainly involved in the mismatch repair of both DNA single-strand breaks (SSB) and double-strand breaks (DSB) (19). It is necessary for genomic integrity, and alterations in DDR genes frequently result in DDR deficiency. Recently, Mok et al. revealed a higher risk of colorectal cancer in BRCA1 mutation carriers through systematic review and meta-analysis (20). In clinical practice, many clinical trials have demonstrated BRCA1 mutation was used to predict the treatment benefit from Poly (ADP-ribose) polymerase inhibitors (PARPi) (21, 22). CHEK2 is one of the DDR genes which are frequently analyzed because of their important role on maintaining genomic stability. CHEK2 mutation is widely reported in sporadic colorectal cancer and hereditary colorectal cancer (23–25). However, the prevalence of CHEK2 mutation is relatively low in colorectal cancer (25–28). In this study, the frequency of CHEK2 mutation in SCRC is 50%, whereas no CHEK2 mutation is found in NSCRC. These findings suggest that DDR deficiency plays an important role in the development of SCRC. Notably, single amino acid substitutions in the DDR genes caused by continuous double base substitutions in SCRC were a novel observation. Although the mechanism of this observation is unknown, the role and clinical significance in SCRC should be further explored.
Previous studies have reported that genomic mutations in CRC are dominated by C to T Ti at CpG sites, indicating that the deamination of 5-methylcytosine is a key initiating event in cancer-driving mutations (29). In our study, we found this phenomenon in SCRC and NSCRC simultaneously, whereas the ratio of C to T Ti in SCRC was higher than that in NSCRC. Additionally, the frequency of Ti in SCRC was significantly higher than that for Tv, but no difference was found between the frequencies of Ti and Tv in NSCRC. Furthermore, we comprehensively investigated the mutational landscape between SCRC and NSCRC and found that several genes had remarkably different mutation frequencies, including CHEK2, MDC1, GNAQ, SMAD4, BRCA1, HLA-B, LATS2, and NLRC5. In this study, targetable mutations, defined as molecular targets for drugs that can guide treatment decisions (30, 31), were analyzed. We found 87.5% of SCRC patients carried at least one clinically actionable mutation, and the difference was observed in the targetable mutation profile between SCRC and NSCRC patients. Thus, although there was a similarity between the mutational profiles of SCRC and NSCRC, SCRC still displayed a diverse mutational pattern. According to the mutation loci distributions of KRAS, it was found that patients with SCRC carried KRAS codon 13 mutations, whereas the majority (61.5%) of NSCRC patients harbored KRAS codon 12 mutations. In CRC, numerous studies revealed that KRAS codon 13 mutations displayed less tumorigenic activity than codon 12 mutations (32, 33). Moreover, experimental data demonstrated that KRAS codon 12 mutated tumors were more aggressive than codon 13-mutated tumors (34). A recent study by Tahir et al. exhibited that alterations at codon 12 and codon 13 in KRAS could lead to varying metastatic efficiencies and oncogenic transformation in colorectal cancer cell lines (35). In SCRC, only KRAS codon 13 mutations were found, which might lead to low tumorigenic activity in SCRC compared with NSCRC. Additionally, many studies have reported that KRAS codon 12 mutations are associated with a poor prognosis in patients with CRC (36, 37). The prognosis between the two groups may be different because of the differences in mutation loci distributions of KRAS. Therefore, it is critical to systematically investigate and understand the mutational spectrum of SCRC. Future treatments and clinical strategies for SCRC should be based on the specific molecular features.
In recent years, NGS has improved the understanding of the signal transduction cascades during the development and progression of CRC. It is known that the RAS/MAPK, PI3K, Wnt, and p53 signaling pathways are most affected in CRC (6). Besides these signaling pathways, we further revealed that Ca2+ and IFN-γ pathways were frequently affected in SCRC. Although numerous studies have demonstrated that the Ca2+ pathway plays an important role in the development and progression of many cancer types (38, 39), few reports revealed the role in CRC. This could be because of the low incidence of Ca2+ pathway alterations in CRC. In this study, the detection rate of Ca2+ pathway related gene alterations was significantly higher in SCRC compared with that in NSCRC, indicating a crucial role of this pathway in the development of SCRC. Recently, interferons, especially IFN-γ, have been found to act as key regulators in tumor immunotherapy (40). Interferons are mainly generated in response to immune stimuli or inflammation, and are crucial in tumor immunosurveillance (41). In SCRC, the common alterations in the IFN-γ pathway highlight the importance of understanding the interferon response, which may contribute to improving the design of immunotherapy trials in this population. Additionally, the DDR pathway is commonly affected in SCRC and NSCRC simultaneously, however, the detection rate of DDR alterations in SCRC is significantly higher than that in NSCRC (87.5% vs 37.5%). Furthermore, DDR gene alterations were remarkably different between the two groups. Recently, DDR deficiencies have become a novel predictive factor of response to immunotherapies, and mutations in DDR genes increases immunogenicity by enhancing the tumor neoantigen load (42). A number of studies have revealed that patients with DDR alterations benefit from PD-1/PD-L1 blockade in advanced urothelial and non-small cell lung cancer (43, 44). Therefore, the high mutation frequencies of genes related to the IFN-γ and DDR signaling pathways imply that immunotherapies may be an important treatment strategy in SCRC.
To the best of our knowledge, this is the first and largest study to characterize the mutational profile in SCRC, and in addition, to compare these profiles between SCRC and NSCRC patients. We revealed a diverse genomic landscape between the two groups as well as novel mutations for targets for future therapy in SCRC. Additionally, we discovered a feature of single amino acid substitutions in DDR genes caused by continuous double base substitutions, which was the first to be reported for CRC and other cancers. These results suggest that SCRC patients present with some unique molecular characteristics. These observations will contribute to promoting the development of personalized therapy and the clinical management in this population.
Data Availability Statement
The sequencing data are available in the NGDC, GSA database (https://bigd.big.ac.cn/gsa/) under the accession number of PRJCA008881.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethical Committee of the Affiliated Sir Run Run Shaw Hospital of Zhejiang University School of Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YF, HP, and SC designed the study. HZL, LG, XZ, CR, ZS, DW, and HSL enrolled the patients and collected the corresponding clinical information. HC, HW, and FL conducted the data analysis. HZL, HSL LG, HC, and HW wrote the paper. All authors contributed to the article and approved the final version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81702809 and No. 81872238) and the Medical Science and Technology Project of Zhejiang Province (No. 2016ZDB007 and No. 2017ZD021).
Conflict of Interest
HW, FL, and SC are employees of Acornmed Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the patients for their contribution to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.813253/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Patterns and Trends in Colorectal Cancer Incidence and Mortality. Gut (2017) 66(4):683–91. doi: 10.1136/gutjnl-2015-310912
3. Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of Metastatic Competency in Colorectal Cancer. Mol Oncol (2017) 11(1):97–119. doi: 10.1002/1878-0261.12018
4. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational Landscape and Significance Across 12 Major Cancer Types. Nature (2013) 502(7471):333–9. doi: 10.1038/nature12634
5. Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell (2018) 33(1):125–136.e3. doi: 10.1016/j.ccell.2017.12.004
6. Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature (2012) 487(7407):330–7. doi: 10.1038/nature11252
7. Zaidi SH, Harrison TA, Phipps AI, Steinfelder R, Trinh QM, Qu C, et al. Landscape of Somatic Single Nucleotide Variants and Indels in Colorectal Cancer and Impact on Survival. Nat Commun (2020) 11(1):3644. doi: 10.1038/s41467-020-17386-z
8. Spoon JM. Situs Inversus Totalis. Neonatal Netw (2001) 20(1):59–63. doi: 10.1891/0730-0832.20.1.63
9. Choi SI, Park SJ, Kang BM, Lee KY, Lee HC, Lee SH. Laparoscopic Abdominoperineal Resection for Rectal Cancer in a Patient With Situs Inversus Totalis. Surg Laparosc Endosc Percutan Tech (2011) 21(2):e87–90. doi: 10.1097/SLE.0b013e31820b0258
10. Yaegashi M, Kimura T, Sakamoto T, Sato T, Kawasaki Y, Otsuka K, et al. Laparoscopic Sigmoidectomy for a Patient With Situs Inversus Totalis: Effect of Changing Operator Position. Int Surg (2015) 100(4):638–42. doi: 10.9738/INTSURG-D-14-00217.1
11. Kojima Y, Sakamoto K, Tomiki Y, Sugimoto K, Okazawa Y, Makino Y. Laparoscopic Right Colectomy for a Patient With Situs Inversus Totalis. J Surg Case Rep (2019) 2019(3):rjz080. doi: 10.1093/jscr/rjz080
12. Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis Oncol (2018) 2018:PO.18.00183. doi: 10.1200/PO.18.00183
13. Hoffmeister M, Schmitz S, Karmrodt E, Stegmaier C, Haug U, Arndt V, et al. Male Sex and Smoking Have a Larger Impact on the Prevalence of Colorectal Neoplasia Than Family History of Colorectal Cancer. Clin Gastroenterol Hepatol (2010) 8(10):870–6. doi: 10.1016/j.cgh.2010.07.004
14. Hebbar S, Fuggle WJ, Nevill AM, Veitch AM. Colorectal Cancer Incidence and Trend in UK South Asians: A 20-Year Study. Colorectal Dis (2012) 14(6):e319–22. doi: 10.1111/j.1463-1318.2012.02960.x
15. Truong A, Yoo JH, Scherzer MT, Sanchez JMS, Dale KJ, Kinsey CG, et al. Chloroquine Sensitizes GNAQ/11-Mutated Melanoma to MEK1/2 Inhibition. Clin Cancer Res (2020) 26(23):6374–86. doi: 10.1158/1078-0432.CCR-20-1675
16. Li H, Li Q, Dang K, Ma S, Cotton JL, Yang S, et al. YAP/TAZ Activation Drives Uveal Melanoma Initiation and Progression. Cell Rep (2019) 29(10):3200–3211.e4. doi: 10.1016/j.celrep.2019.03.021
17. leiblova P, Stolarova L, Krizova K, Lhota F, Hojny J, Zemankova P, et al. Identification of Deleterious Germline CHEK2 Mutations and Their Association With Breast and Ovarian Cancer. Int J Cancer (2019) 145(7):1782–97. doi: 10.1002/ijc.32385
18. Leimbacher PA, Jones SE, Shorrocks AK, de Marco Zompit M, Day M, Blaauwendraad J, et al. MDC1 Interacts With TOPBP1 to Maintain Chromosomal Stability During Mitosis. Mol Cell (2019) 74(3):571–583.e8. doi: 10.1016/j.molcel.2019.02.014
19. Sjakste N, Riekstiņa U. DNA Damage and Repair in Differentiation of Stem Cells and Cells of Connective Cell Lineages: A Trigger or a Complication? Eur J Histochem (2021) 65(2):3236. doi: 10.4081/ejh.2021.3236
20. Oh M, McBride A, Yun S, Bhattacharjee S, Slack M, Martin JR, et al. BRCA1 and BRCA2 Gene Mutations and Colorectal Cancer Risk: Systematic Review and Meta-Analysis. J Natl Cancer Inst (2018) 110(11):1178–89. doi: 10.1093/jnci/djy148
21. Przybycinski J, Nalewajska M, Marchelek-Mysliwiec M, Dziedziejko V, Pawlik A. Poly-ADP-Ribose Polymerases (PARPs) as a Therapeutic Target in the Treatment of Selected Cancers. Expert Opin Ther Targets (2019) 23(9):773–85. doi: 10.1080/14728222.2019.1654458
22. Mauri G, Arena S, Siena S, Bardelli A, Sartore-Bianchi A. The DNA Damage Response Pathway as a Land of Therapeutic Opportunities for Colorectal Cancer. Ann Oncol (2020) 31(9):1135–47. doi: 10.1016/j.annonc.2020.05.027
23. Vysotskaia V, Kaseniit KE, Bucheit L, Ready K, Price K, Johansen Taber K. Clinical Utility of Hereditary Cancer Panel Testing: Impact of PALB2, ATM, CHEK2, NBN, BRIP1, RAD51C, and RAD51D Results on Patient Management and Adherence to Provider Recommendations. Cancer (2020) 126(3):549–58. doi: 10.1002/cncr.32572
24. Marks EI, Matera R, Olszewski AJ, Yakirevich E, El-Deiry WS, Safran H, et al. Mutations in DNA Repair Genes and Clinical Outcomes of Patients With Metastatic Colorectal Cancer Receiving Oxaliplatin or Irinotecan-Containing Regimens. Am J Clin Oncol (2021) 44(2):68–73. doi: 10.1097/COC.0000000000000785
25. Sutcliffe EG, Stettner AR, Miller SA, Solomon SR, Marshall ML, Roberts ME, et al. Differences in Cancer Prevalence Among CHEK2 Carriers Identified via Multi-Gene Panel Testing. Cancer Genet (2020) 246-247:12–7. doi: 10.1016/j.cancergen.2020.07.001
26. Naseem H, Boylan J, Speake D, Leask K, Shenton A, Lalloo F, et al. Inherited Association of Breast and Colorectal Cancer: Limited Role of CHEK2 Compared With High-Penetrance Genes. Clin Genet (2006) 70(5):388–95. doi: 10.1111/j.1399-0004.2006.00698.x
27. Arai H, Elliott A, Xiu J, Wang J, Battaglin F, Kawanishi N, et al. The Landscape of Alterations in DNA Damage Response Pathways in Colorectal Cancer. Clin Cancer Res (2021) 27(11):3234–42. doi: 10.1158/1078-0432.CCR-20-3635
28. Vargas-Parra G, Del Valle J, Rofes P, Gausachs M, Stradella A, Moreno-Cabrera JM, et al. Comprehensive Analysis and ACMG-Based Classification of CHEK2 Variants in Hereditary Cancer Patients. Hum Mutat (2020) 41(12):2128–42. doi: 10.1002/humu.24110
29. Hsu CW, Sowers ML, Hsu W, Eyzaguirre E, Qiu S, Chao C, et al. How Does Inflammation Drive Mutagenesis in Colorectal Cancer? Trends Cancer Res (2017) 12:111–32.
30. Carr TH, McEwen R, Dougherty B, Johnson JH, Dry JR, Lai Z, et al. Defining Actionable Mutations for Oncology Therapeutic Development. Nat Rev Cancer (2016) 16(5):319–29. doi: 10.1038/nrc.2016.35
31. Shen C, Meric-Bernstam F, Su X, Mendelsohn J, Giordano S. Prevalence of Actionable Mutations and Copy Number Alterations and the Price of a Genomic Testing Panel. Oncotarget (2016) 7(44):71686–95. doi: 10.18632/oncotarget.11994
32. Guerrero S, Casanova I, Farré L, Mazo A, Capellà G, Mangues R. K-Ras Codon 12 Mutation Induces Higher Level of Resistance to Apoptosis and Predisposition to Anchorage-Independent Growth Than Codon 13 Mutation or Proto-Oncogene Overexpression. Cancer Res (2000) 60(23):6750–6.
33. Kwak MS, Cha JM, Yoon JY, Jeon JW, Shin HP, Chang HJ, et al. Prognostic Value of KRAS Codon 13 Gene Mutation for Overall Survival in Colorectal Cancer: Direct and Indirect Comparison Meta-Analysis. Medicine (2017) 96(35):e7882. doi: 10.1097/MD.0000000000007882
34. Ward RL, Todd AV, Santiago F, O'Connor T, Hawkins NJ. Activation of the K-Ras Oncogene in Colorectal Neoplasms is Associated With Decreased Apoptosis. Cancer (1997) 79(6):1106–13. doi: 10.1002/(SICI)1097-0142(19970315)79:6<1106::AID-CNCR8>3.0.CO;2-D
35. Tahir R, Renuse S, Udainiya S, Madugundu AK, Cutler JA, Nirujogi RS, et al. Mutation-Specific and Common Phosphotyrosine Signatures of KRAS G12D and G13D Alleles. J Proteome Res (2021) 20(1):670–83. doi: 10.1021/acs.jproteome.0c00587
36. Jones RP, Sutton PA, Evans JP, Clifford R, McAvoy A, Lewis J, et al. Specific Mutations in KRAS Codon 12 are Associated With Worse Overall Survival in Patients With Advanced and Recurrent Colorectal Cancer. Br J Cancer (2017) 116(7):923–9. doi: 10.1038/bjc.2017.37
37. Hayama T, Hashiguchi Y, Okamoto K, Okada Y, Ono K, Shimada R, et al. G12V and G12C Mutations in the Gene KRAS are Associated With a Poorer Prognosis in Primary Colorectal Cancer. Int J Colorectal Dis (2019) 34(8):1491–6. doi: 10.1007/s00384-019-03344-9
38. Roberts-Thomson SJ, Chalmers SB, Monteith GR. The Calcium-Signaling Toolkit in Cancer: Remodeling and Targeting. Cold Spring Harb Perspect Biol (2019) 11(8):a035204. doi: 10.1101/cshperspect.a035204
39. Marchi S, Pinton P. Alterations of Calcium Homeostasis in Cancer Cells. Curr Opin Pharmacol (2016) 29:1–6. doi: 10.1016/j.coph.2016.03.002
40. Lu C, Klement JD, Ibrahim ML, Xiao W, Redd PS, Nayak-Kapoor A, et al. Type I Interferon Suppresses Tumor Growth Through Activating the STAT3-Granzyme B Pathway in Tumor-Infiltrating Cytotoxic T Lymphocytes. J Immunother Cancer (2019) 7(1):157. doi: 10.1186/s40425-019-0635-8
41. Ni L, Lu J. Interferon Gamma in Cancer Immunotherapy. Cancer Med (2018) 7(9):4509–16. doi: 10.1002/cam4.1700
42. Mouw KW, Goldberg MS, Konstantinopoulos PA, D'Andrea AD. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discovery (2017) 7(7):675–93. doi: 10.1158/2159-8290.CD-17-0226
43. Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, et al. Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From PD-1/PD-L1 Blockade in Advanced Urothelial Cancers. J Clin Oncol (2018) 36(17):1685–94. doi: 10.1200/JCO.2017.75.7740
Keywords: colorectal cancer, SCRC, NSCRC, mutational profile, next-generation sequencing
Citation: Li H, Gong L, Cheng H, Wang H, Zhang X, Rao C, Song Z, Wang D, Lou H, Lou F, Cao S, Pan H and Fang Y (2022) Comprehensive Molecular Profiling of Colorectal Cancer With Situs Inversus Totalis by Next-Generation Sequencing. Front. Oncol. 12:813253. doi: 10.3389/fonc.2022.813253
Received: 11 November 2021; Accepted: 15 March 2022;
Published: 20 April 2022.
Edited by:
Paola Ulivi, Scientific Institute of Romagna for the Study and Treatment of Tumors (IRCCS), ItalyReviewed by:
Francesco Pepe, University of Naples Federico II, ItalyMurali Bashyam, Centre for DNA Fingerprinting and Diagnostics (CDFD), India
Copyright © 2022 Li, Gong, Cheng, Wang, Zhang, Rao, Song, Wang, Lou, Lou, Cao, Pan and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Fang, ZmFuZ3lvbmdAemp1LmVkdS5jbg==; Hongming Pan, cGFuaG9uZ21pbmdAemp1LmVkdS5jbg==; Shanbo Cao, c2hhbmJvY2FvQGFjb3JubWVkLmNvbQ==
†These authors have contributed equally to this work
 Hongsen Li
Hongsen Li Liu Gong1†
Liu Gong1† Huanqing Cheng
Huanqing Cheng Huina Wang
Huina Wang Zhangfa Song
Zhangfa Song Haizhou Lou
Haizhou Lou Hongming Pan
Hongming Pan Yong Fang
Yong Fang