- 1Department of Pathology, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Department of Oncology, The Second Xiangya Hospital, Central South University, Changsha, China
- 3Department of Respiratory and Critical Care Medicine, The Second Xiangya Hospital, Central South University, Changsha, China
- 4State Key Laboratory of Translational Medicine and Innovative Drug Development, Jiangsu Simcere Diagnostics Co., Ltd., Nanjing, China
- 5Department of Medicine, Nanjing Simcere Medical Laboratory Science Co., Ltd., Nanjing, China
- 6Department of Thoracic Surgery, The First Affiliated Hospital of China Medical University, Shenyang, China
The fusions of receptor tyrosine kinase (RTK) involving anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1), and neurotrophic receptor tyrosine kinase (NTRK) represent the potential targets of therapeutic intervention for various types of solid tumors. Here, the genomic features of 180 Chinese solid tumor patients with ALK, ROS1, and NTRK fusions by next generation sequencing (NGS) were comprehensively characterized, and the data from 121 patients in Memorial Sloan Kettering Cancer Center (MSKCC) database were used to compare. We found that ALK, ROS1, and NTRK fusions were more common in younger female patients (p<0.001) and showed a higher expression of programmed death ligand 1 (PD-L1). The gene-intergenic fusion and the fusion with rare formation directions accounted for a certain proportion in all samples and 62 novel fusions were discovered. Alterations in TP53 and MUC16 were common in patients with RTK fusions. The mutational signatures of patients were mainly distributed in COSMIC signature 1, 2, 3, 15 and 30, while had a higher frequency in copy number variations (CNVs) of individual genes, such as IL-7R. In the MSKCC cohort, patients with fusions and CNVs showed shorter overall survival than those with only fusions. Furthermore, the differentially mutated genes between fusion-positive and -negative patients mainly concentrated on MAPK signaling and FOXO signaling pathways. These results may provide genomic information for the personalized clinical management of solid tumor patients with ALK, ROS1, and NTRK fusions in the era of precision medicine.
Introduction
Chromosomal inversions, deletions or translocations leading to the constitutive activation of receptor tyrosine kinase (RTK) drive tumorigenesis across different malignancies (1, 2). The prevalence of RTK fusions involving anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1), and neurotrophic receptor tyrosine kinase (NTRK) ranges from 0.3% to 5% in solid tumors and tyrosine kinase inhibitors (TKIs) are the standard treatment modality for the first-line setting of patients with advanced cancer harboring such fusions (3–5). Currently, multiple ALK fusion partners have been identified, of which echinoderm microtubule-associated protein-like 4 (EML4) is the most frequent, with nine variants occurring in nearly 80% of all the ALK fusion cases of non-small cell lung cancer (NSCLC) (6–8). Meanwhile, many different 5´ gene partners have been identified in the fusion with 3´ regions of ROS1. These fusions are discovered in adult glioblastoma, paediatric glioma, NSCLC, and inflammatory myofibroblastic tumor (IMTs) (4, 9). Additionally, approximately 80 NTRK fusion partners have also been described (10, 11). Although the frequency of NTRK fusions is low, they are ubiquitous in rare cancer types, such as mammary analog secretory carcinoma and infantile fibrosarcoma (12–15).
In recent years, a lot of clinical trials on treatments targeting specific molecular mechanisms like ALK, ROS1, and NTRK fusions have been conducted. Small molecule inhibitors for ALK, ROS1, and NTRK fusions, such as crizotinib, brigatinib, lorlatinib, entrectinib and larotrectinib, have been approved by the US Food and Drug Administration (FDA) for different cancer types (16–20). Despite the potential benefit from identifying these fusions, it remains unclear whether the tumors with ALK, ROS1, and NTRK fusions represent a distinct, although rare, disease subtype that should be detected early for targeted therapy. Herein, a comprehensive study was carried out to characterize the molecular and clinicopathological characteristics, and prognosis of solid tumor patients with ALK, ROS1, and NTRK fusions.
Materials and Methods
Sample Collection
In this study, the sequencing data of 7,537 solid tumor samples from the database of Simcere Diagnostics, Co. Ltd. (Nanjing, China) for genomic profiling between June 2019 and November 2020 were retrospectively analyzed, including lung cancer (n=3001), liver cancer (n=762), soft tissue sarcoma (n=281), bile duct carcinoma (n=232), esophageal cancer (n=155), breast cancer (n=154), melanoma (n=125), gallbladder carcinoma (n=121), bone tumor (n=59) and other unspecified tumors (n=2647) (Table 1). All patients signed written informed consents. The formalin fixed, paraffin-embedded (FFPE) tissue samples were selected for analysis, and peripheral blood samples were collected as the control. Here, 121 ALK, ROS1, and NTRK fusion-positive cancer patients from the MSK-IMPACT Clinical Sequencing Cohort (MSKCC, Nat Med 2017), which was composed of 10,945 samples, were used as the compared cohort.
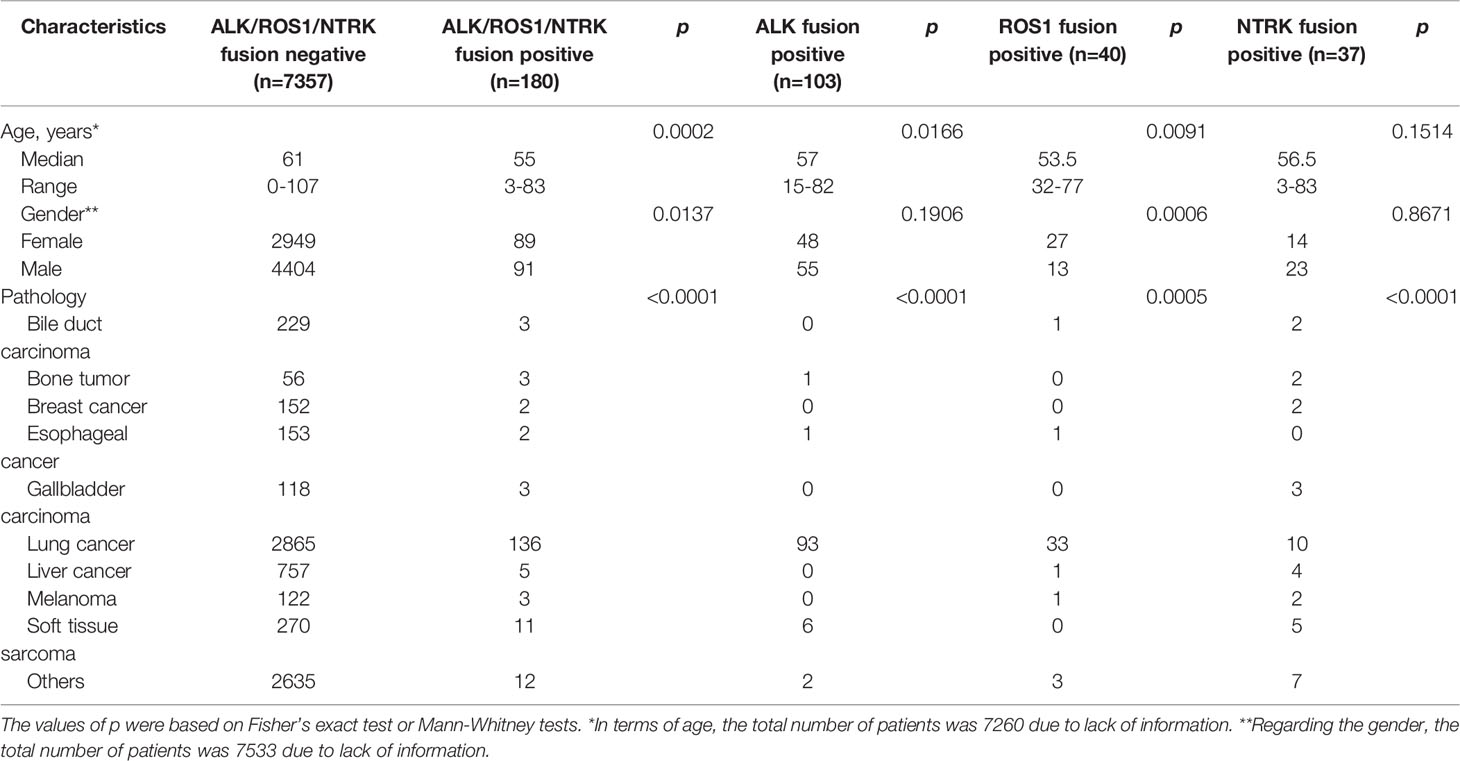
Table 1 Patients’ characteristics according to the presence or absence of ALK, ROS1, and NTRK fusions.
DNA Extraction, Library Construction and Sequencing
DNA was extracted from unstained FFPE sections with more than 20% tumor cells according to the manufacturer’s protocol. Library construction was performed using the KAPA Library Preparation kit. The concentration of the library was assessed using the Invitrogen Qubit4.0, and the inserted size was examined on the Agilent 4200 TapeStation. Next generation sequencing (NGS) was performed on the Illumina Novaseq 6000 system at an average depth of 1000X with a panel of 539 cancer-related genes (Supplementary Table 1). Genomic alterations, including single nucleotide variants (SNVs), copy number variations (CNVs), small insertions, deletions and gene arrangements were covered. The tumor mutation burden (TMB) and microsatellite status (MSI) were also calculated by NGS.
Statistical Analysis
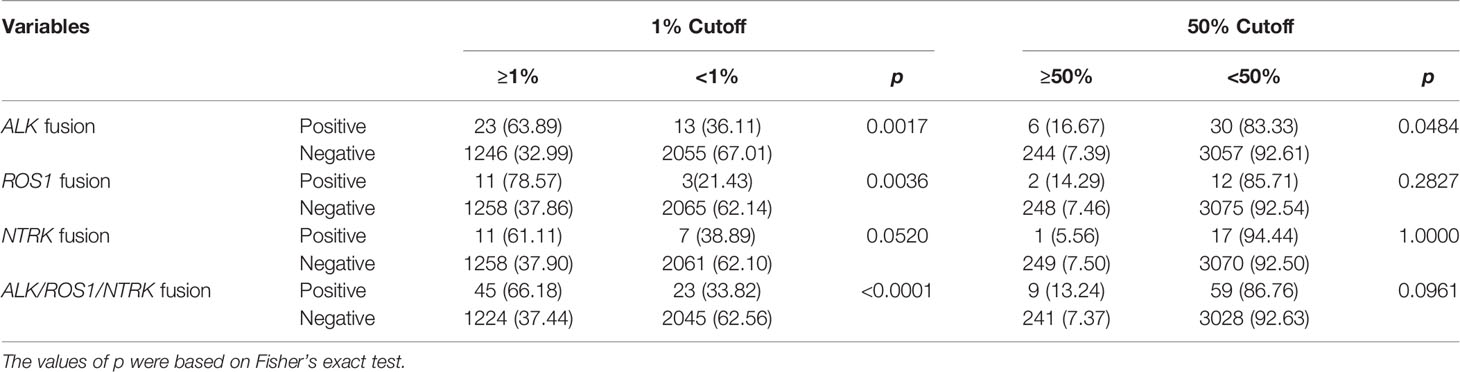
The Fisher’s exact test and Mann-Whitney test were used to assess the association of ALK, ROS1, and NTRK fusions with age, gender, and cancer types. To assess the probability of gene fusions in various cancer types, odds ratios (ORs) and relative 95% confidence intervals (CIs) were calculated. The overall survival (OS) was analyzed using the Kaplan-Meier method, and survival curves (mutational signature, SNVs, and CNVs) were compared using the log-rank test. Fisher’s exact test was used to evaluate the association of genomic characteristics with the proportion of PD-L1 expression, with 1% and 50% as the cutoff value. All statistical tests were two-sided, and p < 0.05 was considered statistically significant.
Results
Patient Characteristics
There were 103 (1.37%) cases harboring ALK rearrangements in 7,537 solid tumor patients, including lung cancer (n=93), soft tissue sarcoma (n=6), bone tumor (n=1), esophagus cancer (n=1) and other unspecified tumors (n=2). ROS1 rearrangements were detected in 40 cases (0.53%), among whom 33 cases suffered from lung cancer (Table 1). 37 cases (0.49%) harbored NTRK fusions, including 9 cases of NTRK1 fusions, 2 cases of NTRK2 fusions, and 26 cases of NTRK3 fusions.
Through NGS, a total of 180 patients were found to harbor ALK/ROS1/NTRK fusions and were set as ALK/ROS1/NTRK fusion-positive group (n=180), while those without ALK/ROS1/NTRK fusions were as ALK/ROS1/NTRK fusion-negative group (n=7357). As shown in Table 1, ALK/ROS1/NTRK fusions were more common in young patients (p<0.001). However, no age bias was presented in patients with NTRK fusion (p>0.05). There was a higher ALK/ROS1/NTRK fusion-positive frequency in females than males (p=0.0137), and subgroup analysis further showed that the ROS1 fusion-positive rate in females was significantly higher than that in males (p=0.0006), but not ALK fusion (p=0.1906) and NTRK fusion (p=0.8671). The incidence of RTK fusions in soft tissue sarcoma and bone tumor was significantly higher than that in liver cancer (p<0.05). Meanwhile, the rates of RTK fusions in bile duct carcinoma and liver cancer was much lower than that in lung cancer (p<0.05, OR=0.285) (Supplemental Figure 1).
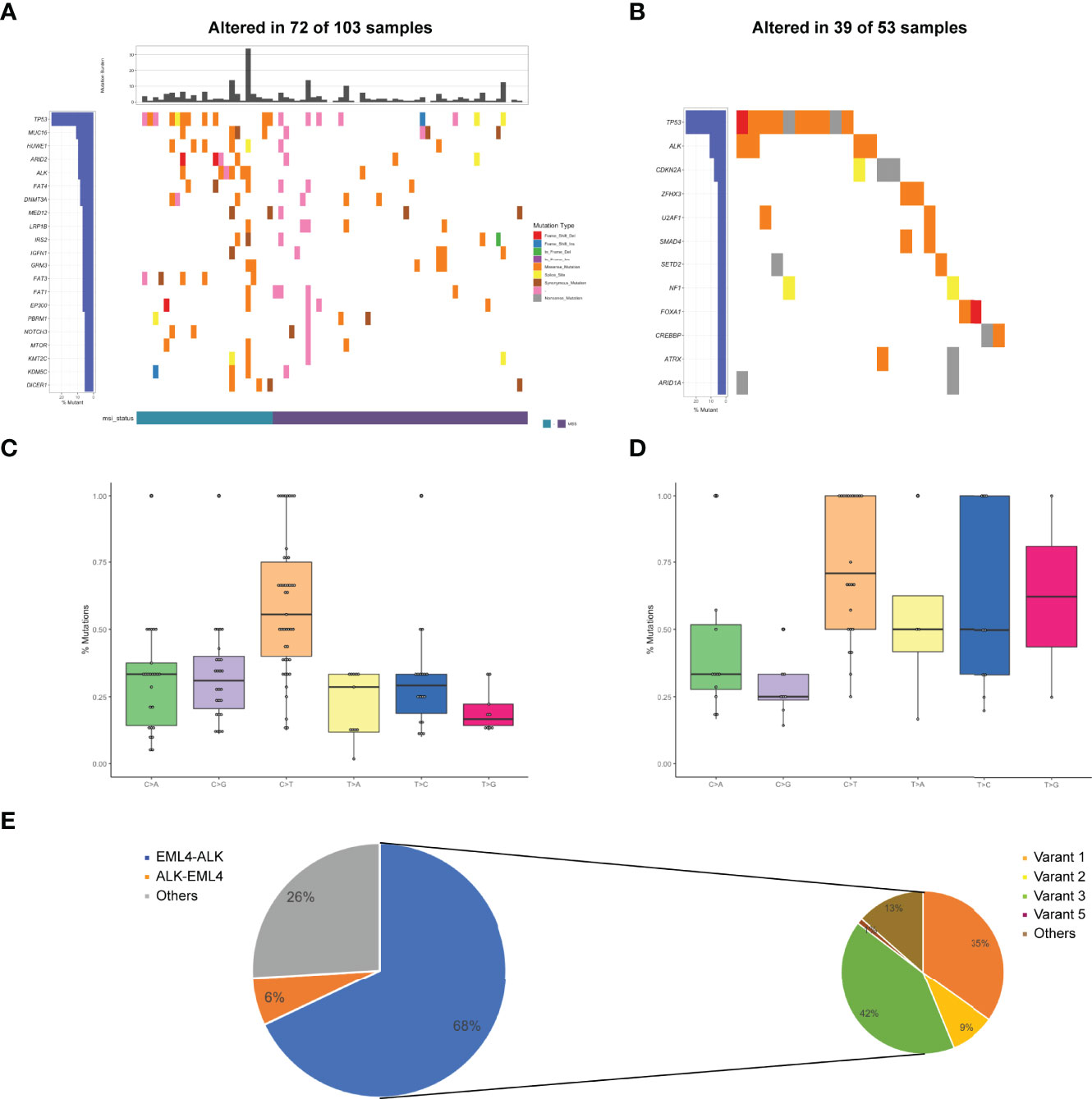
Figure 1 Mutational profiles and partners of ALK fusion-positive patients. (A) The oncoprint of the somatic SNVs in 103 patients harboring ALK fusion in our study. (B) The oncoprint of the somatic SNVs in 53 patients harboring ALK fusion in the MSKCC database. (C) Mutational signatures of ALK fusion-positive patients in our cohort. (D) Mutational signatures of ALK fusion-positive patients in the MSKCC cohort. (E) Distribution of ALK fusion partners and EML4-ALK variants. MSI, microsatellite instability.
Molecular Features of ALK Fusion-Positive Tumors
Of 103 ALK fusion-positive samples, a total of 491 variants were identified, including frame InDel, missense mutations, nonsense mutations and splicing mutations. TP53 alterations (26%) were the most common, followed by MUC16 (11%), HUWE1 (10%), ARID2 (10%), and ALK (10%). Other genomic alterations included NOTCH3 (6%), MTOR (6%), KMT2C (6%), KDM5C (6%), and DICER1 (6%) (Figure 1A). The median TMB was 2.21 mut/Mb (0-33.82 mut/Mb). Although MSI status was available in 47% of patients, there was a higher proportion of microsatellite stability (MSS) in the tumors bearing ALK fusions. In the MSKCC cohort, totally 94 mutations occurred in 53 ALK fusion-positive cases, suggesting TP53 and ALK were the most frequently altered genes (Figure 1B).
Analysis of mutational signatures showed that C>T transition were the most common, followed by C>A and C>G transitions (Figure 1C). The probability of T>G and T>A transitions was the lowest, consistent with COSMIC signature 1 identified in most cancer samples. Accordingly, our results were highly in accordance with MSKCC findings that C>T transition was the most frequently mutation (Figure 1D). Additionally, the breakpoints corresponding to ALK fusion in the sequencing data of these patients were also identified. Most of breakpoints were located at the intron between exon 19 and exon 20 of ALK gene. In the ALK cohort, EML4-ALK fusion accounted for 67%, ALK-EML4 fusion for 6%, and others for the remaining 27%. Coexistence of these fusions was present in 26 patients (25%). 89 out of 103 patients had an EML4-ALK fusion, with variant 1 (v1, E13:A20), variant 2 (v2, E20:A20), variant 3 (v3, E6:A20) and variant 5 (v5, E2:A20) detected in 31, 8, 37 and 1 patients, respectively (Figure 1E). Sixteen novel ALK fusion partners identified were shown in Supplementary Table 2.
Notably, in our cohort, the mutations at the site of ALK resistance were detected. The gatekeeper L1196M (4/103) was present in crizotinib-resistant cases, while the solvent-front G1202R mutation (2/103) was highly resistant to crizotinib, as well as to next-generation ALK inhibitors (21).
Molecular Features of ROS1 Fusion-Positive Tumors
Genomic alterations in ROS1 fusion-positive samples (n=40) were shown in Figure 2A. The median TMB was 2.94 mut/Mb with a range of 0-25 mut/Mb. 61% of ROS1 fusion-positive tumors harboring MSI data showed MSS, while only one case showed MSI-L. The frequency of TP53 mutations was obviously the highest (54%), followed by MUC16 (29%), LRP18 (14%), FAT1 (14%), CARD11 (14%), and ARID18 (14%) mutations. We further compared our results with the MSKCC cohort that included 43 ROS1 fusion-positive cases harboring 155 mutations. TP53 was the most frequently altered gene in the MSKCC cohort, followed by MLL2 instead of MUC16 (Figure 2B). Analysis of their mutational signatures showed that C>T transition was the most prevalent, followed by T>A and T>C transitions (Figure 2C). The T>G transition showed the lowest frequency. This pattern was also consistent with COSMIC signature 1. Moreover, C>T transition also occurred most frequently in the MSKCC cohort (Figure 2D).
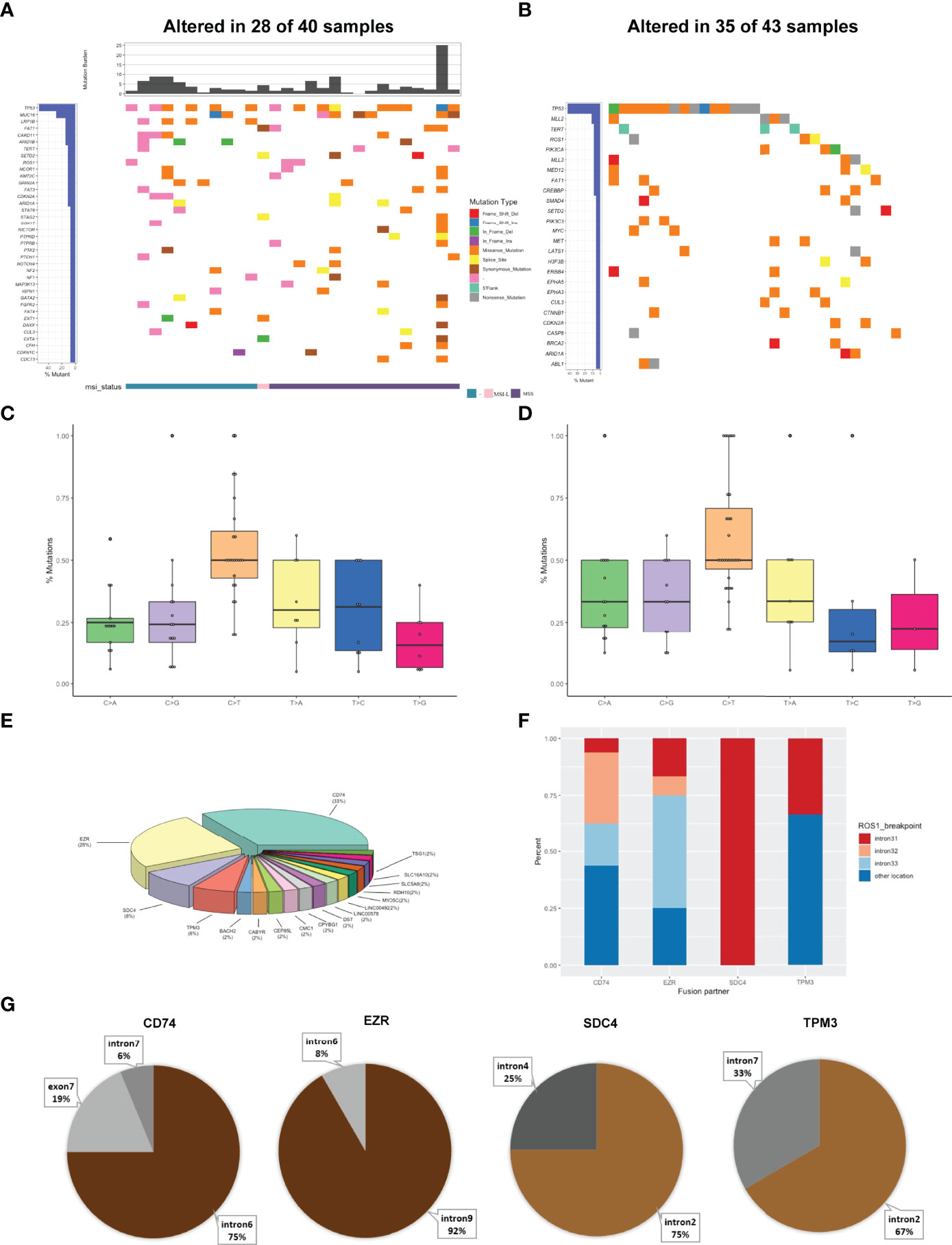
Figure 2 Mutational profiles and partners of ROS1 fusion-positive patients. (A) The oncoprint of the somatic SNVs in 40 patients harboring ROS1 fusion in our study. (B) The oncoprint of the somatic SNVs in 43 patients harboring ROS1 fusion in the MSKCC database. (C) Mutational signatures of ROS1 fusion-positive patients in our cohort. (D) Mutational signatures of ROS1 fusion-positive patients in the MSKCC cohort. (E) Distribution of ROS1 fusion partners. (F) Distribution of fusion breakpoint positions in the most common ROS1 fusions including CD74-ROS1, EZR-ROS1, SDC4-ROS1, and TPM3-ROS1. (G) Distribution of breakpoint locations for ROS1 fusion partner genes, including CD74, EZR, SDC4, and TPM3.
Then we identified the breakpoints and partner genes of the ROS1 fusion in the sequencing data of these patients. In our cohort, CD74 was the most common ROS1 fusion partner (33%), followed by EZR (25%), SDC4 (8%), TPM3 (6%) and more (Figure 2E). ROS1 fusions were formed via intra chromosomes, most frequently occurring in ROS1 introns 31, 32, 33, while less frequently in other exons and introns (Figure 2F). Meanwhile, ROS1 most frequently fused to intron 6 of CD74, intron 9 of EZR, intron 2 of SDC4, and intron 7 of TPM3 (Figure 2G). We also identified 11 novel ROS1 fusion partners (Supplementary Table 2). Mutations resulting in substitutions at solvent-front residues (G2032R) of ROS1 were identified in one CD74-ROS1 fusion case. The G2032R mutation had been reported to introduce steric hindrance and diminish high-affinity crizotinib binding (22).
Molecular Features of NTRK Fusion-Positive Tumors
Among 37 NTRK fusion-positive cases (0.49%, 37/7537), 479 variants were totally identified in our cohort (Figure 3A). The median TMB was 4.41 mut/Mb, with the peak value of 93.38 mut/Mb. In the cases with MSI data, only one case bearing rearrangements was MSI-H and the others were MSS.
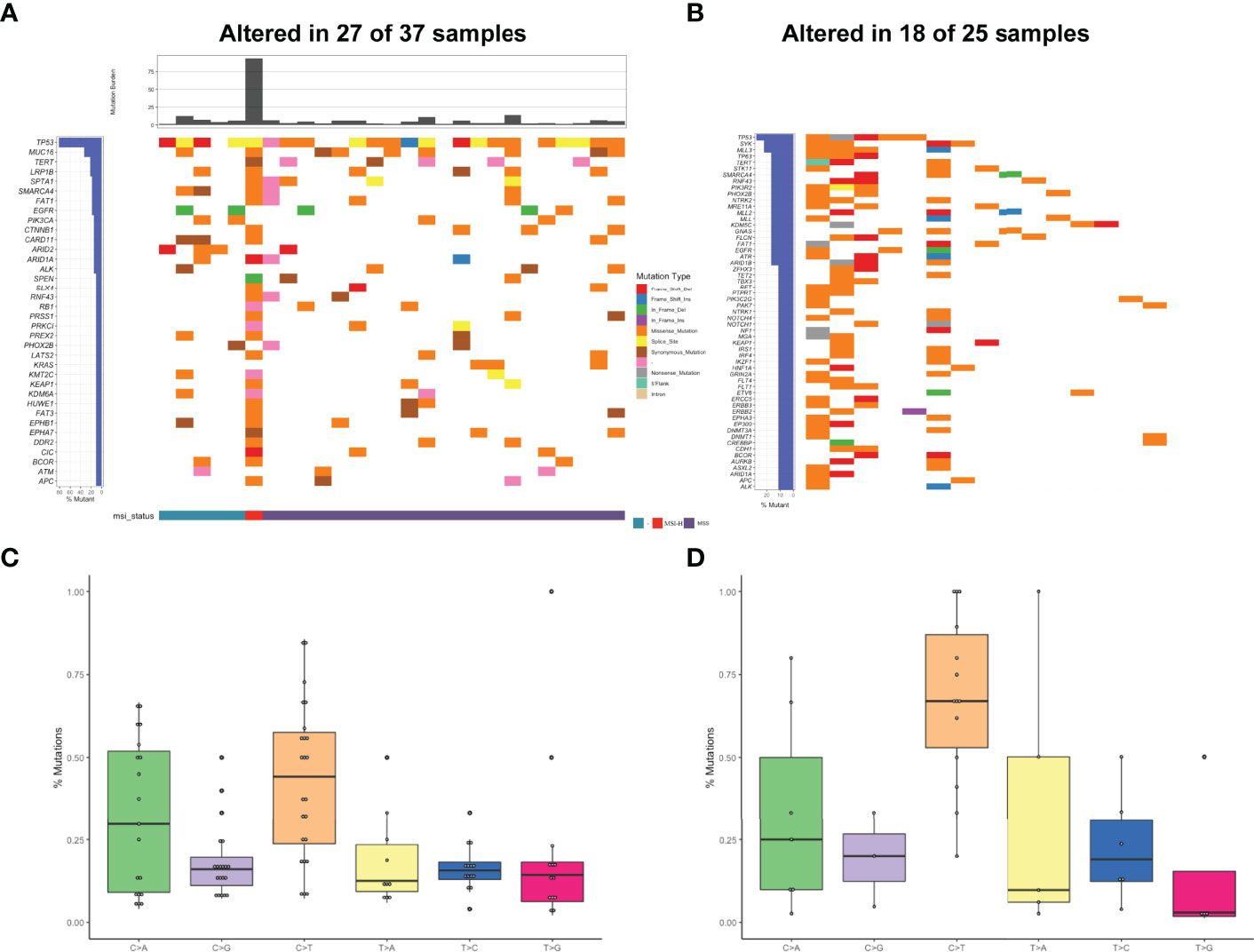
Figure 3 Mutational profiles and partners of NTRK fusion-positive patients. (A) The oncoprint of the somatic SNVs in 37 patients harboring NTRK fusion in our study. (B) The oncoprint of the somatic SNVs in 25 patients harboring NTRK fusion in the MSKCC database. (C) Mutational signatures of NTRK fusion positive patients in our cohort. (D) Mutational signatures of NTRK fusion-positive patients in the MSKCC cohort.
The heatmap of somatic mutations showed that TP53 was the most altered gene (81%), followed by MUC16 (33%), TERT (22%), LRP1B (22%), SPTA1 (19%), SMARCA4 (19%), FAT1 (19%), and EGFR (19%) mutations. By analysis of the MSKCC cohort that comprised 25 NTRK fusion-positive cases harboring 299 mutations, TP53 was also found to be the most frequently altered gene, followed by SYK but not MUC16 (Figure 3B). Analysis of the mutational signatures showed that C>T transition occurred most frequently, followed by C>A transition (Figure 3C). The other transitions were at a low frequency. As shown in Figure 3D, the frequency of C>T transition in the MSKCC cohort was the highest, even higher than ours, which might be associated with different ethnicities and diets.
The positive rates of NTRK fusions were generally low in a wide range of cancers and tended to be enriched among rare cancers. By analyzing its partner genes, we found the proportion of NTRK3 partner genes was the highest (76%), followed by NTRK1 (17%), and NTRK2 (7%). Meanwhile, 35 novel NTRK fusions were identified (Supplementary Table 2). Notably, G709C mutation resulting in amino acid substitutions was identified in the QKI-NTRK2 fusion case, which involved the regions of the xDFG motif and was paralogous to G1269 (ALK) substitutions (23).
Classification of ALK, ROS1, and NTRK Fusion Events
As shown in Figure 4A, a total of 225 gene fusions were identified in 180 samples, in which coexistence was present in 41 cases. These fusions occurred mostly in chromosomes, with a few occurring between adjacent chromosomes (Figure 4B). These fusions were classified into two categories based on the genome annotation containing the breakpoint regions (Figure 4C): gene-gene (91.1%) and gene-intergenic (8.9%). The gene-intergenic fusions accounted for 3.1% of ALK fusions, 6.3% of ROS1 fusions, and 28.2% of NTRK fusions. Based on the gene breakpoint regions, we discovered 14% fusions harboring rare fusion directions, namely “upstream-upstream-breakpoint” cases (8%) and “downstream-downstream-breakpoint” cases (6%) (Figure 4D). Due to lack of chimeric transcripts, they were set aside in most fusion analyses as being unlikely to be functionally relevant.
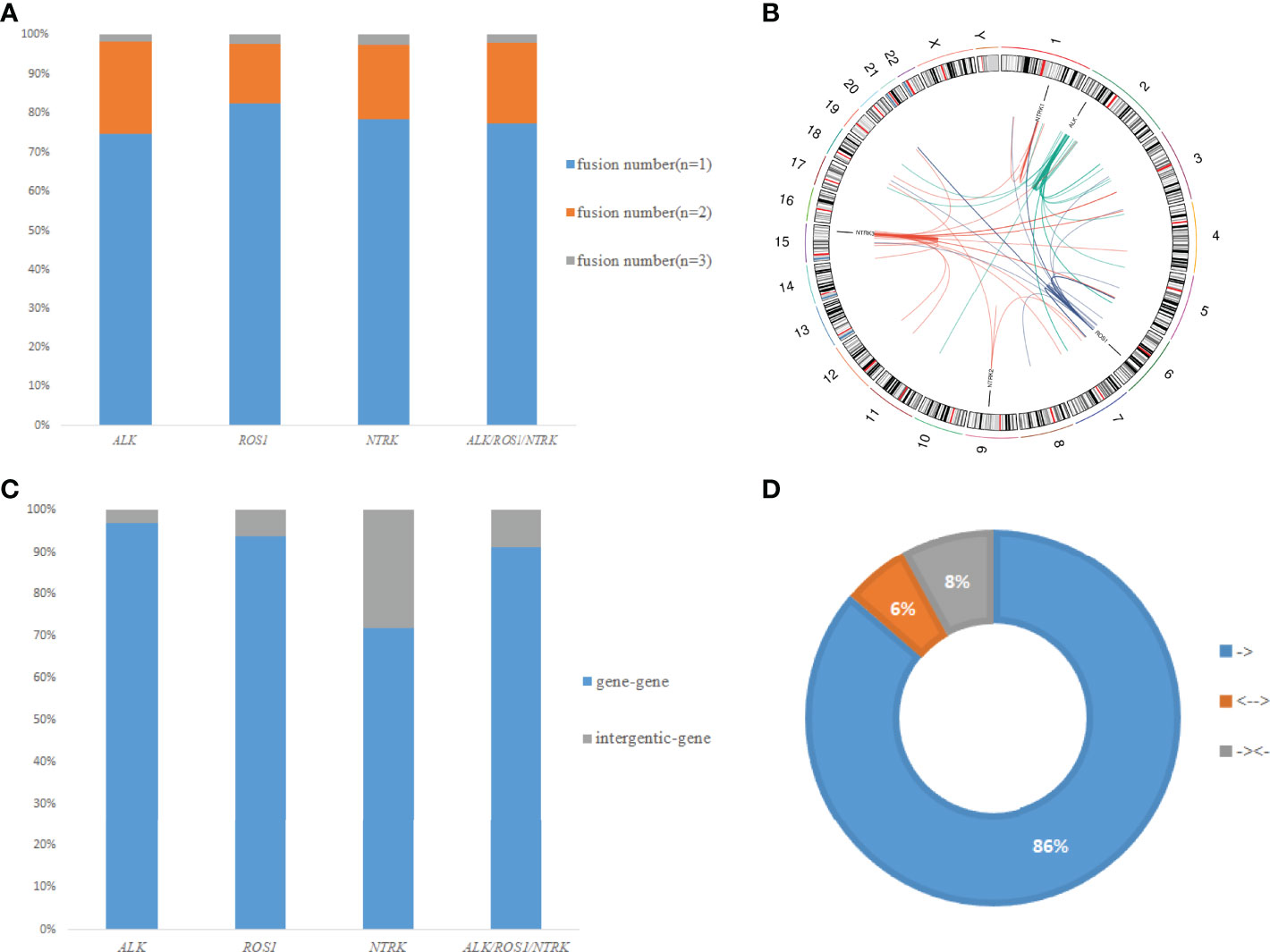
Figure 4 Classification of fusion events. (A) Distribution of different fusion numbers (n=1, 2, 3) in our study. (B) A circos plot of 225 gene fusions identified in all patients. (C) Distribution of different fusion types (gene-gene and gene-intergenic). (D) Distribution of fusions with different formation directions.
Impacts of ALK, ROS1, and NTRK Positivity on the Prognosis
In this study, we used Signature Multivariate Analysis (SigMA), a computational tool, to call the mutational signatures, which could accurately detect the mutational signatures associated with homologous recombination deficiency from targeted gene panels (24). Using deconstructSigs R package to extract the mutational signatures, we identified the presence of signature 1 (Sig 1) in 18.2% (22/121), signature 3 (Sig 3) in 11.6% (14/121), signature 30 (Sig 30) in 9.9% (12/121), and signature 2 in 9% (11/121) of the patients in our cohort (Figure 5A). Likewise, Sig 1, signature 7 (Sig 7), signature 15 (Sig 15), and Sig 30 were detected in 25.8% (24/93), 6.5% (6/93), 8.6% (8/93), and 6.5% (6/93) of the MSKCC samples, respectively (Figure 5B).
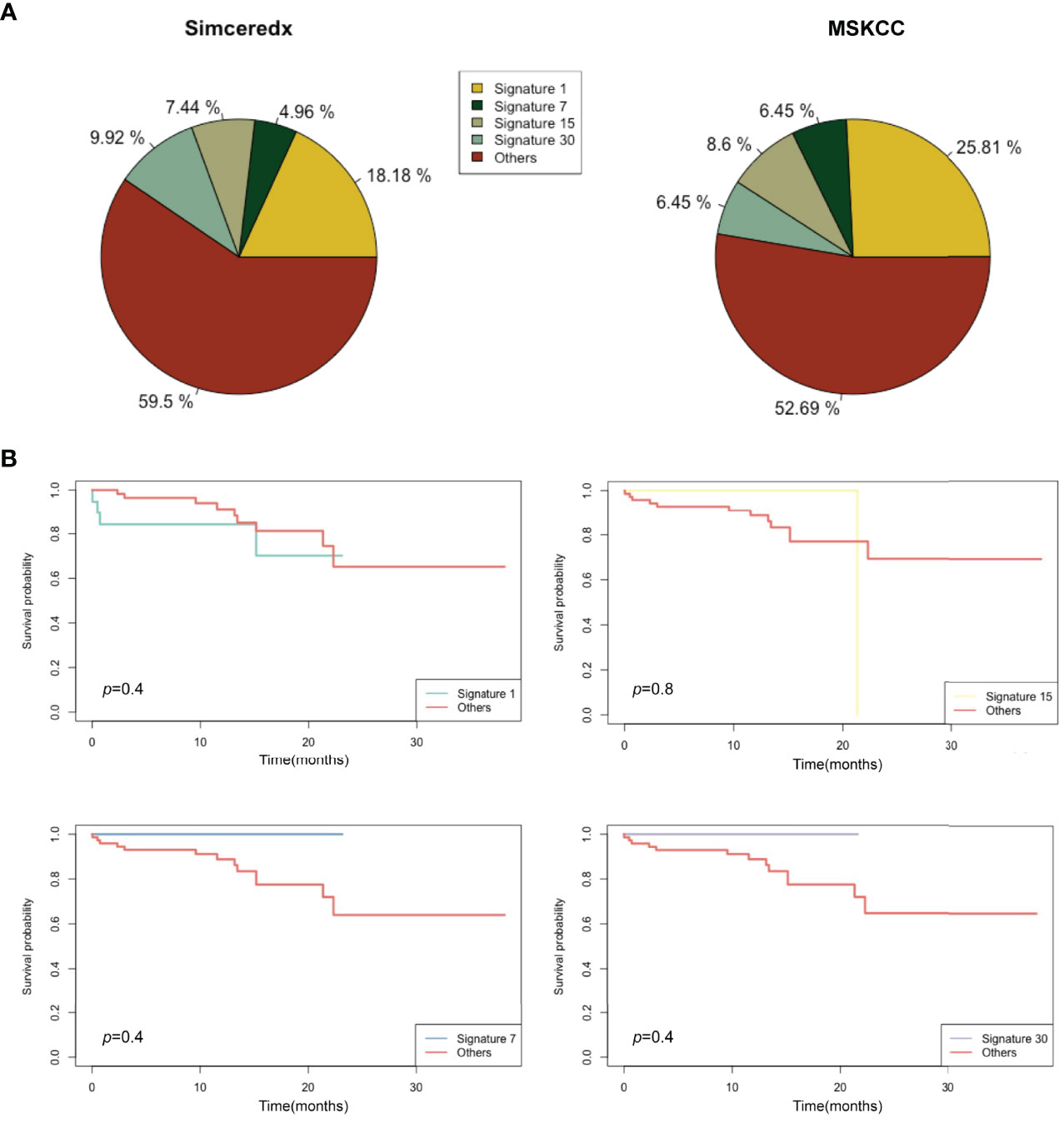
Figure 5 Mutational signatures of ALK/ROS1/NTRK fusion-positive patients. (A) Distribution of mutational signatures in all patients harboring ALK/ROS1/NTRK fusions in our study and that from the MSKCC database. (B) Kaplan-Meier graph for survival probability according to Sig 1, Sig 7, Sig 15, and Sig 30 status.
A previous study indicated that Sig 3 positivity was indicative of clinical benefits (25), so we analyzed the association of Sig 1, Sig 7, Sig 15, and Sig 30 positive patients with clinical benefits. Sig 1 and Sig 15 were apparently not associated with prolonged OS (Figure 5B). Sig 7 and Sig 30 positive patients showed slightly longer OS than others, but without statistical significance. Interestingly, some fusion samples were absent of SNVs. On this basis, we examined whether fusion-positive samples without SNVs could indicate clinical benefits. Unfortunately, the absence of SNVs did not make a significant difference in OS (Supplemental Figure 2A).
CNVs in Patients With ALK, ROS1, and NTRK Fusions
CNVs were found in 50% of 180 samples. About 11% of the patients in our cohort harbored MYC CNVs, which may be a candidate for tumor genesis and progression (26). In addition, CNVs of CDKN2A, CDKN2B, MCL1, MDM2, and IRS2 have been reported to be associated with prognosis (27–31). CNVs of these genes were also found in fusion-positive samples from the MSKCC database (Figure 6). Interestingly, we found that CNVs of IL7R showed a high frequency. Moreover, the CNVs in fusion-positive samples were related to poor prognosis (p=0.01) (Supplemental Figure 2B), which needed more data to verify.
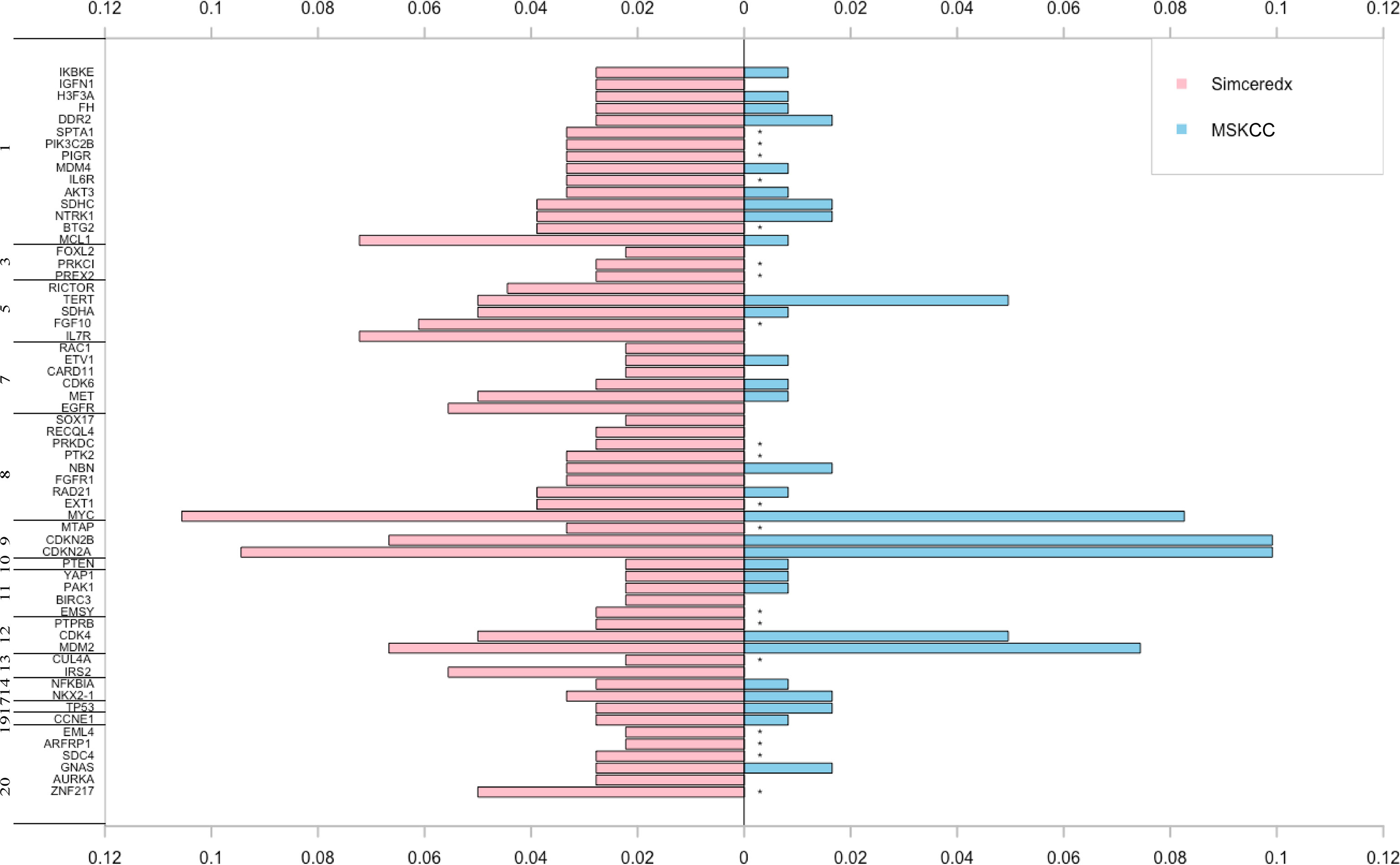
Figure 6 The pink and blue bars represent CNV events occurring in ALK/ROS1/NTRK fusion-positive patients in our cohort and MSKCC cohort, respectively. *represents the genes not covered in the MSKCC panel.
PD-L1 Expression in ALK, ROS1, and NTRK Fusion-Positive Tumors
Over-expression of ALK fusion protein increased PD-L1 expression, while anti-PD-1 antibody (immunotherapy) was effective in both crizotinib sensitive and resistant NSCLC cells (32). Hence, we examined the expression of PD-L1 in our cohort. A total of 3337 patients were eligible after excluding those without PD-L1 expression. PD-L1 immunohistochemistry testing was performed using the SP263 antibody. In our cohort, PD-L1 expression was higher in tumors with ALK (p=0.0017) and ROS1 (p=0.0036) fusions than fusion-negative tumors at 1% cutoff. However, PD-L1 expression between NTRK fusion-positive and -negative tumors showed no statistical difference (p=0.052). RTK fusions including ALK, ROS1, and NTRK exhibited a higher PD-L1 expression than fusion-negative tumors (p<0.0001). Using ≥50% cutoff, a higher PD-L1 positivity was also observed in tumors with ALK (p=0.0484), but not ROS1 (p=0.2827), NTRK (p=1), or RTK fusions (p=0.0961) (Table 2). These results indicated that PD-L1 expression from different RTK fusion-positive tumors may have different predictive values for benefiting from immune checkpoint inhibitors (ICIs) in solid tumors.
Aberrations in Relevant Signaling Pathways
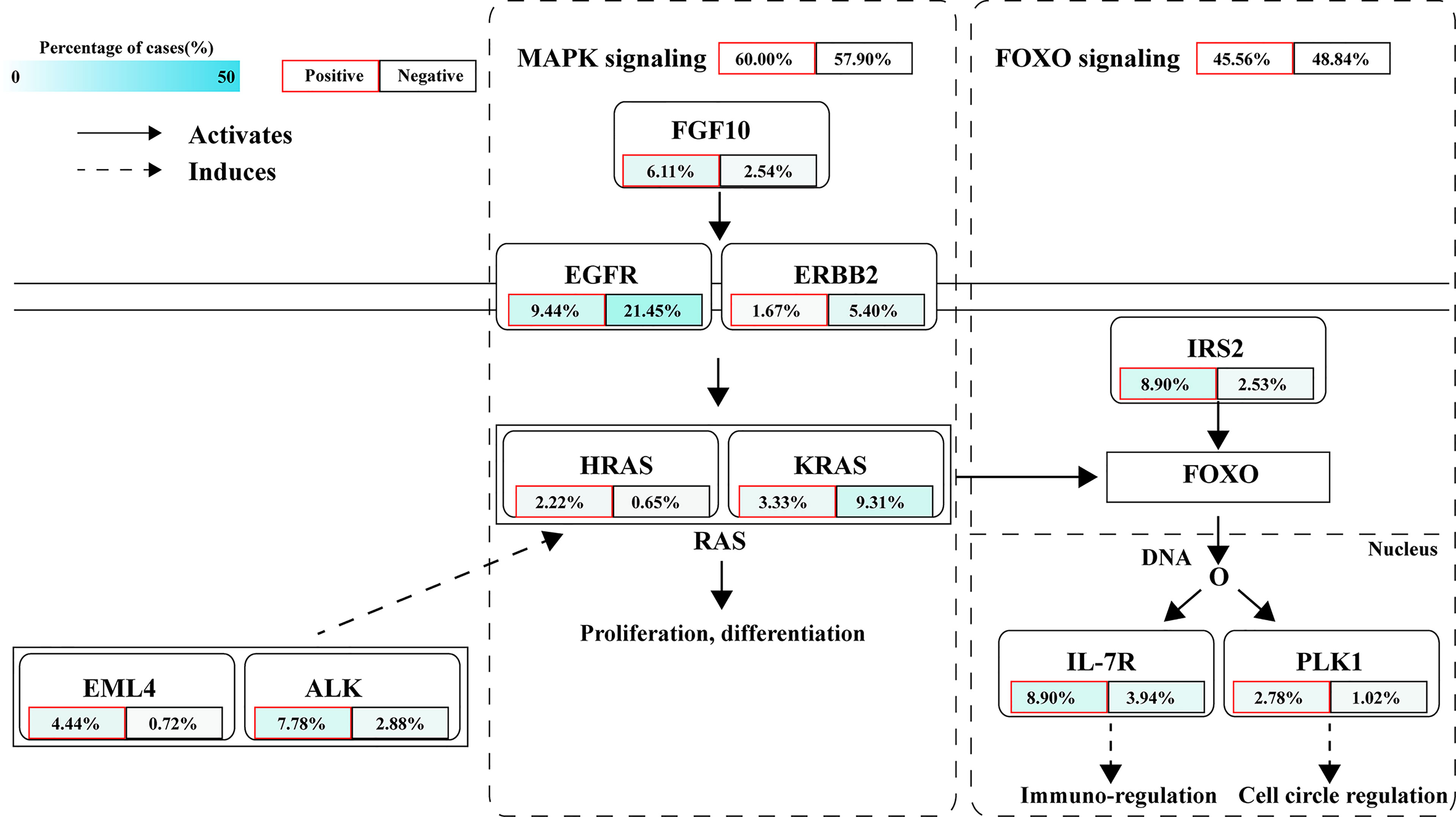
The signaling pathway analysis of NTRK/ROS1/ALK fusion-positive and -negative patients exhibited significant dysregulations in well-defined pathways, namely MAPK and FOXO pathways (Figure 7). According to prior reports, MAPK pathway regulates cell proliferation, differentiation, apoptosis, and migration, while FOXO signaling pathway is related to cell cycle, apoptosis, autophagy, metabolism, oxidation, immune response, and stem cell maintenance (33, 34). We found that the MAPK signaling pathway was altered in 60% of fusion-positive patients and 57.9% of negative patients. Fusion-positive patients had a higher frequency of mutations in EML4, ALK, FGF10, and HRAS, while the rates of EGFR, ERBB2, and KRAS were higher in fusion-negative patients. Differential frequencies of IRS2, IL7R, and PLK1 mutations resulted in dysregulation of the FOXO1 signaling pathway between these two groups.
Discussion
By analyzing the genomic landscape of patients with ALK, ROS1, and NTRK fusions, a relatively high frequency of TP53 mutation, MSS status, and different TMB levels (NTRK>ALK/ROS1) were found, supported by previous studies (7, 9, 35, 36). In our cohort, the frequency of MUC16 mutations was secondary to TP53. MUC16 mutations appeared to be associated with the therapeutic and prognostic factors and were expected to be a biomarker to guide immunotherapy (37, 38). In terms of mutational signatures, ALK, ROS1, and NTRK fusion-positive patients showed similar point mutant characteristics, and the C>T transition was most common, followed by C>A transition. This pattern was consistent with COSMIC Sig 1 that had been found in most cancer types (39). Furthermore, survival curves suggested that Sig 7 and Sig 30 may be associated with a favorable prognosis in some way, which needed more data to verify.
Accumulating evidence has suggested that CNVs might be a potential biomarker or prognostic factor for tumor treatment. Apart from the genes with a high frequency of copy number amplification, such as MYC and MDM2, we identified some genes with copy number loss, such as CDKN2A and CDKN2B. CDKN2A/B deletions were independent prognostic markers for both adult and paediatric lymphoblastic leukaemia (40). MDM2 amplification was associated with poor clinical outcomes and significantly increased tumor growth rates with anti-PD-1/PD-L1 immunotherapy (41). This information is important for guiding clinical treatment. We observed that fusion-positive patients without CNVs had a favorable prognosis. Notably, the pathogenic IL-7R CNVs exist in 7% of Chinese patients with fusions, which is higher than that in the Western population (0%). IL7R was previously reported to be amplified in various cancers, with the function of mediating potential tumor promotion, and high levels of IL-7R may be associated with poor prognosis (42). Currently, the risk factors for IL-7R-mutant fusions are unknown. Future studies should focus on how diet and ethnic differences increase the risk of IL-7R mutations.
Of the 180 fusion-positive samples by NGS, EML4 and CD74 were the most common ALK and ROS1 fusion partners, respectively. EML4-ALK occurred mainly in the forms of three variants: variant 1, variant 2, and variant 3 (43, 44). Diverse ROS1 fusion partners were identified, and the top four fusion partners were CD74, EZR, SDC4, and TPM3. As the most common ROS1 fusion partner, CD74 had a frequency similar to the previous ones (9, 45). There were no high-frequency partner genes occurring in NTRK fusions, which might be related to the high incidence of NTRK fusions in rare tumors. We also detected some novel ALK/ROS1/NTRK fusion partners, such as LPIN1 and SMARCC1 (ALK), SLC16A10 and CRYBG1 (ROS1), SDK1 and GYPA (NTRK3). These results suggested that the NGS-based evaluation for ALK/ROS1/NTRK fusions was accurate and comprehensive. Compared with traditional methods, such as IHC, FISH, and Sanger sequencing, NGS had unique advantages in detecting unknown fusion partners and identifying accurate breakpoints. However, the rare fusions remain clinically interesting, further studies are needed to confirm these observations in preclinical and clinical studies.
A previous study reported the impacts of gene-intergenic and intergenic-intergenic fusions on the upregulation of their target genes (46). Therefore, we classified the fusions in our cohort into two categories: gene-gene fusion and gene-intergenic fusion. Neither intergenic sequence-ALK nor coexistence of fusions showed a significant effect on the benefit from crizotinib treatment (47). However, a substantial portion of chimeric transcripts was produced by gene-intergenic fusions. The impact of such intergenic breakpoints on transcriptome has been unclear. Meanwhile, these fusions with rare fusion directions mostly coexisted with classic fusions, and their clinical significance was currently unknown, even though a portion of them harboring kinase domains. Future research may focus on investigating the clinical role of gene-intergenic fusions and fusions with rare fusion directions in cancers.
PD-L1 protein expression in tumor cells emerged as the first potential predictive biomarker for sensitivity to ICIs (48). In our cohort, 44.27% of the patients had clinically relevant information in PD-L1 expression. Consistent with the literature, we observed a significantly higher expression of PD-L1 in the ALK fusion-positive cohort. Of note, the expression of PD-L1 in the ROS1 or NTRK fusion-positive cohort was similar to that in the fusion-negative cohort. However, other data suggested that immune escape may confer a higher PD-L1 expression in NSCLC patients with an aggressive tumor phenotype, leading to a poor prognosis with TKI therapy (40). Moreover, the differentially mutated genes between fusion-positive and fusion-negative samples were mainly enriched in the MAPK and FOXO signaling pathways. The mutational frequency of individual gene varied greatly between fusion-positive and fusion-negative samples, but with similar mutational frequency in the whole signaling pathways.
In conclusion, we characterized the genomic landscape of solid tumor patients with ALK, ROS1, and NTRK fusions and 62 novel fusions were discovered, which may provide more clinically actionable targets for cancer therapy to a great extent. Although the gene-intergenic fusion and fusion with rare fusion directions accounted for a certain proportion of all fusion samples, the clinical significance of these fusions remained to be unclear, thus RTK-targeted therapy should be explored further in solid tumors in the future. Notably, the frequency of CNVs was high and associated with a poor prognosis in fusion-positive patients, highlighting the importance of CNVs as a potential biomarker or prognostic factor for cancer therapy. PD-L1 high-expression was more common in the ALK fusion-positive cohort than that in the fusion-negative cohort, leading us to hypothesize that ICIs might bring clinical benefits to the solid tumor patients harboring RTK fusions. Collectively, all these findings may provide genomic information for personalized clinical management of patients with ALK, ROS1, and NTRK fusions in the era of precision medicine.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank, submission #2562890.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YD, CS, YL and YS contributed to conception and design of the study. YD wrote the first draft of the manuscript. PL, WH, LY wrote sections of the manuscript. YN, XM and FD organized the database and performed the statistical analysis. CS, YL and YS contributed to edit and review the manuscript. All authors approved the submitted version.
Funding
This study was supported by Hunan Principal Project “The molecular mechanism of the reversion of glioma differentiation induced by Toll-like receptor 4 activation of Notch signal pathway” (ID: 2019JJ40422).
Conflict of Interest
Authors YN, XM, FD, and CS were employed by Jiangsu Simcere Diagnostics Co., Ltd. And Nanjing Simcere Medical Laboratory Science Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.813158/full#supplementary-material
References
1. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The Landscape of Kinase Fusions in Cancer. Nat Commun (2014) 5:4846. doi: 10.1038/ncomms5846
2. Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine Kinase Gene Rearrangements in Epithelial Malignancies. Nat Ret Cancer (2013) 13:772–87. doi: 10.1038/nrc3612
3. Chia PL, Mitchell P, Dobrovic A, John T. Prevalence and Natural History of ALK Positive Non-Small-Cell Lung Cancer and the Clinical Impact of Targeted Therapy With ALK Inhibitors. Clin Epidemiol (2014) 6:423–32. doi: 10.2147/CLEP.S69718
4. Drilon A, Jenkins C, Iyer S, Schoenfeld A, Keddy C, Davare MA. ROS1-Dependent Cancers- Biology, Diagnostics and Therapeutics. Nat Ret Clin Oncol (2021) 18:35–55. doi: 10.1038/s41571-020-0408-9
5. Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis Oncol (2018) 2018:PO.18.00183. doi: 10.1200/PO.18.00183
6. Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK Variants: Biological and Molecular Properties, and the Implications for Patients. Cancers (2017) 9:118. doi: 10.3390/cancers9090118
7. Liu S, Huang T, Liu M, He W, Zhao Y, Yang L, et al. The Genomic Characteristics of ALK Fusion Positive Tumors in Chinese NSCLC Patients. Front Oncol (2020) 10:726. doi: 10.3389/fonc.2020.00726
8. Noh KW, Lee MS, Lee SE, Song JY, Shin HT, Kim YJ, et al. Molecular Breakdown: A Comprehensive View of Anaplastic Lymphoma Kinase (ALK)-Rearranged Non-Small Cell Lung Cancer. J Pathol (2017) 243:307–19. doi: 10.1002/path.4950
9. Cui M, Han Y, Li P, Zhang J, Ou Q, Tong X, et al. Molecular and Clinicopathological Characteristics of ROS1-Rearranged Non-Small-Cell Lung Cancers Identified by Next-Generation Sequencing. Mol Oncol (2020) 14:2787–95. doi: 10.1002/1878-0261.12789
10. Farago AF, Taylor MS, Doebele RC, Zhu VW, Kummar S, Spira AI, et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis Oncol (2018) 2018:PO.18.00037. doi: 10.1200/PO.18.00037
11. Kummar S, Lassen UN. TRK Inhibition: A New Tumor-Agnostic Treatment Strategy. Target Oncol (2018) 13:545–56. doi: 10.1007/s11523-018-0590-1
12. Assi T, Rassy E, Nassereddine H, Farhat F, Karak FE, Kattan J, et al. TRK Inhibition in Soft Tissue Sarcomas: A Comprehensive Review. Semin Oncol (2020) 47:73–84. doi: 10.1053/j.seminoncol.2020.02.009
13. Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary Analogue Secretory Carcinoma of Salivary Glands, Containing the ETV6-NTRK3 Fusion Gene: A Hitherto Undescribed Salivary Gland Tumor Entity. Am J Surg Pathol (2010) 34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc
14. Bourgeois JM, Knezevich SR, Mathers JA, Sorensen PH. Molecular Detection of the ETV6-NTRK3 Gene Fusion Differentiates Congenital Fibrosarcoma From Other Childhood Spindle Cell Tumors. Am J Surg Pathol (2000) 24:937–46. doi: 10.1097/00000478-200007000-00005
15. Zhao R, Yao F, Xiang C, Zhao J, Shang Z, Guo L, et al. Identification of NTRK Gene Fusions in Lung Adenocarcinomas in the Chinese Population. J Pathol Clin Res (2021) 7:375–84. doi: 10.1002/cjp2.208
16. Rodig SJ, Shapiro GI. Crizotinib, a Small-Molecule Dual Inhibitor of the C-Met and ALK Receptor Tyrosine Kinases. Curr Opin Investig Drugs (2010) 11:1477–90.
17. Markham A. Brigatinib: First Global Approval. Drugs (2017) 77:1131–5. doi: 10.1007/s40265-017-0776-3
18. Kuang S, Leighl NB. Lorlatinib in ALK-Rearranged Lung Cancer. Cancer Cell (2021) 39:25–7. doi: 10.1016/j.ccell.2020.12.017
19. Marcus L, Donoghue M, Aungst S, Myers CE, Helms WS, Shen G, et al. FDA Approval Summary: Entrectinib for the Treatment of NTRK Gene Fusion Solid Tumors. Clin Cancer Res (2021) 27:928–32. doi: 10.1158/1078-0432.CCR-20-2771
20. Scott LJ. Larotrectinib: First Global Approval. Drugs (2019) 79:201–6. doi: 10.1007/s40265-018-1044-x
21. McCoach CE, Le AT, Gowan K, Jones K, Schubert L, Doak A, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-Small Cell Lung Cancer. Clin Cancer Res (2018) 24:3334–47. doi: 10.1158/1078-0432.CCR-17-2452
22. Wu X, Fu Y, Wang Y, Wan S, Zhang J. Gaining Insight Into Crizotinib Resistance Mechanisms Caused by L2026M and G2032R Mutations in ROS1 via Molecular Dynamics Simulations and Free-Energy Calculations. J Mol Model (2017) 23:141. doi: 10.1007/s00894-017-3314-z
23. Drilon A. TRK Inhibitors in TRK Fusion-Positive Cancers. Ann Oncol (2019) 30:viii23–30. doi: 10.1093/annonc/mdz282
24. Gulhan DC, Lee JJ, Melloni GEM, Cortés-Ciriano I, Park PJ. Detecting the Mutational Signature of Homologous Recombination Deficiency in Clinical Samples. Nat Genet (2019) 51:912–9. doi: 10.1038/s41588-019-0390-2
25. Farkkila A, Gulhan DC, Casado J, Jacobson CA, Nguyen H, Kochupurakkal B, et al. Immunogenomic Profiling Determines Responses to Combined PARP and PD-1 Inhibition in Ovarian Cancer. Nat Commun (2020) 11:1459. doi: 10.1038/s41467-020-15315-8
26. Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, et al. MYC Protein Interactors in Gene Transcription and Cancer. Nat Rev Cancer (2021) 21:579–91. doi: 10.1038/s41568-021-00367-9
27. Chen WS, Bindra RS, Mo A, Hayman T, Husain Z, Contessa JN, et al. CDKN2A Copy Number Loss Is an Independent Prognostic Factor in HPV-Negative Head and Neck Squamous Cell Carcinoma. Front Oncol (2018) 8:95. doi: 10.3389/fonc.2018.00095
28. Zhao Y, Li Y, Lu H, Chen J, Zhang Z, Zhu ZZ. Association of Copy Number Loss of CDKN2B and PTCH1 With Poor Overall Survival in Patients With Pulmonary Squamous Cell Carcinoma. Clin Lung Cancer (2011) 12:328–34. doi: 10.1016/j.cllc.2011.02.007
29. Yin J, Li Y, Zhao H, Qin Q, Li X, Huang J, et al. Copy-Number Variation of MCL1 Predicts Overall Survival of Non-Small-Cell Lung Cancer in a Southern Chinese Population. Cancer Med (2016) 5:2171–9. doi: 10.1002/cam4.774
30. Sawada R, Maehara R, Oshikiri T, Nakamura T, Itoh T, Kodama Y, et al. MDM2 Copy Number Increase: A Poor Prognostic, Molecular Event in Esophageal Squamous Cell Carcinoma. Hum Pathol (2019) 89:1–9. doi: 10.1016/j.humpath.2019.04.002
31. Huang F, Chang H, Greer A, Hillerman S, Reeves KA, Hurlburt W, et al. IRS2 Copy Number Gain, KRAS and BRAF Mutation Status as Predictive Biomarkers for Response to the IGF-1r/IR Inhibitor BMS-754807 in Colorectal Cancer Cell Lines. Mol Cancer Ther (2015) 14:620–30. doi: 10.1158/1535-7163.MCT-14-0794-T
32. Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin Cancer Res (2015) 21:4014–21. doi: 10.1158/1078-0432.CCR-15-0016
33. Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL. ERK/MAPK Signalling Pathway and Tumorigenesis. Exp Ther Med (2020) 19:1997–2007. doi: 10.3892/etm.2020.8454
34. Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int J Biol Sci (2017) 13:815–27. doi: 10.7150/ijbs.20052
35. Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, et al. ALK, ROS1, and NTRK Rearrangements in Metastatic Colorectal Cancer. J Natl Cancer Inst (2017) 109:1–10. doi: 10.1093/jnci/djx089
36. Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular Characterization of Cancers With NTRK Gene Fusions. Mod Pathol (2019) 32:147–53. doi: 10.1038/s41379-018-0118-3
37. Aithal A, Rauth S, Kshirsagar P, Shah A, Lakshmanan I, Junker WM, et al. MUC16 as a Novel Target for Cancer Therapy. Expert Opin Ther Targets (2018) 22:675–86. doi: 10.1080/14728222.2018.1498845
38. Li X, Pasche B, Zhang W, Chen K. Association of MUC16 Mutation With Tumor Mutation Load and Outcomes in Patients With Gastric Cancer. JAMA Oncol (2018) 4:1691–8. doi: 10.1001/jamaoncol.2018.2805
39. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of Mutational Processes in Human Cancer. Nature (2013) 500:415–21. doi: 10.1038/nature12477
40. Zhang W, Kuang P, Liu T. Prognostic Significance of CDKN2A/B Deletions in Acute Lymphoblastic Leukaemia: A Meta-Analysis. Ann Med (2019) 51:28–40. doi: 10.1080/07853890.2018.1564359
41. Fang W, Zhou H, Shen J, Li J, Zhang Y, Hong S, et al. MDM2/4 Amplification Predicts Poor Response to Immune Checkpoint Inhibitors: A Pan-Cancer Analysis. ESMO Open (2020) 5:e000614. doi: 10.1136/esmoopen-2019-000614
42. Barata JT, Durum SK, Seddon B. Flip the Coin: IL-7 and IL-7R in Health and Disease. Nat Immunol (2019) 20:1584–93. doi: 10.1038/s41590-019-0479-x
43. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the Transforming EML4-ALK Fusion Gene in Non-Small-Cell Lung Cancer. Nature (2007) 448:561–6. doi: 10.1038/nature05945
44. Gristina V, La Mantia M, Iacono F, Galvano A, Russo A, Bazan V. The Emerging Therapeutic Landscape of ALK Inhibitors in Non-Small Cell Lung Cancer. Pharmaceuticals (2020) 13:474. doi: 10.3390/ph13120474
45. Huang RSP, Haberberger J, Sokol E, Schrock AB, Danziger N, Madison R, et al. Clinicopathologic, Genomic and Protein Expression Characterization of 356 ROS1 Fusion Driven Solid Tumors Cases. Int J Cancer (2021) 148:1778–88. doi: 10.1002/ijc.33447
46. Yun JW, Yang L, Park HY, Lee CW, Cha H, Shin HT, et al. Dysregulation of Cancer Genes by Recurrent Intergenic Fusions. Genome Biol (2020) 21:166. doi: 10.1186/s13059-020-02076-2
47. Cai C, Tang Y, Li Y, Chen Y, Tian P, Wang Y, et al. Distribution and Therapeutic Outcomes of Intergenic Sequence-ALK Fusion and Coexisting ALK Fusions in Lung Adenocarcinoma Patients. Lung Cancer (2021) 152:104–8. doi: 10.1016/j.lungcan.2020.12.018
48. Davis AA, Patel VG. The Role of PD-L1 Expression as a Predictive Biomarker: An Analysis of All US Food and Drug Administration (FDA) Approvals of Immune Checkpoint Inhibitors. J Immunother Cancer (2019) 7:278. doi: 10.1186/s40425-019-0768-9
Keywords: ALK, ROS1, NTRK, gene fusion, next generation sequencing, mutational signature, copy number variants, programmed death ligand 1
Citation: Dai Y, Liu P, He W, Yang L, Ni Y, Ma X, Du F, Song C, Liu Y and Sun Y (2022) Genomic Features of Solid Tumor Patients Harboring ALK/ROS1/NTRK Gene Fusions. Front. Oncol. 12:813158. doi: 10.3389/fonc.2022.813158
Received: 11 November 2021; Accepted: 19 April 2022;
Published: 16 June 2022.
Edited by:
Ondrej Slaby, Central European Institute of Technology (CEITEC), CzechiaReviewed by:
Pasqualino De Antonellis, University of Toronto, CanadaGuan Wang, Sichuan University, China
Copyright © 2022 Dai, Liu, He, Yang, Ni, Ma, Du, Song, Liu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Sun, c3VueWl4aWFuZ3lhMkAxMjYuY29t; Yang Liu, bGl1eWRvY3RvcjkxMUAxNjMuY29t; Chao Song, Y2hhby5zb25nQHNpbWNlcmVkeC5jb20=
 Yinghuan Dai1
Yinghuan Dai1 Chao Song
Chao Song Yang Liu
Yang Liu Yi Sun
Yi Sun