
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 10 February 2022
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.812655
 Zilong Yan1,2
Zilong Yan1,2 Qingen Da1,3
Qingen Da1,3 Zhangfu Li1,2
Zhangfu Li1,2 Qirui Lin2
Qirui Lin2 Jing Yi2
Jing Yi2 Yanze Su2
Yanze Su2 Guanyin Yu4
Guanyin Yu4 Qingqi Ren2
Qingqi Ren2 Xu Liu2
Xu Liu2 Zewei Lin2
Zewei Lin2 Jianhua Qu2*
Jianhua Qu2* Weihua Yin4
Weihua Yin4 Jikui Liu2*
Jikui Liu2*NIMA-related kinase 7 (NEK7) is a serine/threonine kinase involved in cell cycle progression via mitotic spindle formation and cytokinesis. It has been related to multiple cancers, including breast cancer, hepatocellular cancer, lung cancer, and colorectal cancer. Moreover, NEK7 regulated the NLRP3 inflammasome to activate Caspase-1, resulting in cell pyroptosis. In the present study, we investigated whether NEK7 is involved in cell pyroptosis of hepatocellular carcinoma (HCC). Interestingly, we found that NEK7 was significantly related to expression of pyroptosis marker GSDMD in HCC. We found that NEK7 expression was significantly correlated with GSDMD expression in bioinformatics analysis, and NEK7 expression was significantly co-expressed with GSDMD in our HCC specimens. Cell viability, migration, and invasion capacity of HCC cell lines were inhibited, and the tumor growth in the xenograft mouse model was also suppressed following knockdown of NEK7 expression. Mechanistic studies revealed that knockdown of NEK7 in HCC cells significantly upregulated the expression of pyroptosis markers such as NLRP3, Caspase-1, and GSDMD. Coculture of HCC cells stimulated hepatic stellate cell activation by increasing p-ERK1/2 and α-SMA. Knockdown of NEK7 impaired the stimulation of HCC cells. Therefore, downregulation of NEK7 inhibited cancer–stromal interaction by triggering cancer cell pyroptosis. Taken together, this study highlights the functional role of NEK7-regulated pyroptosis in tumor progression and cancer–stromal interaction of HCC, suggesting NEK7 as a potential target for a new therapeutic strategy of HCC treatment.
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and the second most common cause of cancer deaths worldwide (1), with a 5-year overall survival rate of 12% and a median survival of 23 months for Chinese HCC patients (2). Early-stage HCC may be treated with surgical resection and systemic therapy. However, treatment is associated with high overall relapse rates, and curative resection is not feasible after tumor progressed (3). More unfortunately, >70% of patients with HCC are diagnosed at the late stage (4, 5). As curative resection and chemotherapy are not feasible, it is urgent to explore non-surgical therapeutic approaches for new biomarkers and effective therapeutic strategies (6).
NIMA-related kinases (NEKs) of NEK1 to NEK11 are serine/threonine kinases that drive cell-cycle dynamics by modulating mitotic spindle formation and cytokinesis (7). Among the NEK family members, the smallest NEK7 and NEK6 share more than 85% sequence identity (8). Previous studies reported overexpression of NEK7 in human cancer, including head and neck (9), breast (10), pancreas (11), liver (12), and colorectum and lung (13). Moreover, the biological role of NEK7 was not limited to cell proliferation capacity; involvement of NEK7 in tumorigenesis and metastasis was also proved (9–12).
Recent studies have shown that ROS, potassium efflux, and other factors activate Caspase-1 by regulating the interaction of NEK7 and NLRP3 inflammasome, leading to cell pyroptosis (14–16). Pyroptosis is an inflammatory form of programmed cell death, mainly mediated by the inflammasome to activate a variety of cysteine proteases including Caspase-1/4/5/11, resulting in pyroptosis execution of protein Gasdermin D (GSDMD) cleavage. The pore-forming activity of GSDMD leads to cell perforation, changing the osmotic pressure of the cell membrane and losing its integrity, and then mediating the maturation and secretion of IL-1β and IL-18 (17, 18). Accumulating studies of NEK7 or pyroptosis were reported individually in breast cancer, colorectal cancer, lung cancer, and liver cancer (12, 13, 19–21). However, the relationship between NEK7 and pyroptosis in HCC and the biological role and underlying mechanisms of pyroptosis in HCC remain unclear.
In the present study, by using bioinformatic databases, cell lines, and tumor samples, we investigated the correlation between NEK7 expression and GSDMD expression in human HCC. We detected a significant correlation of NEK7 in the migration and invasion ability of HCC cell lines. Moreover, NEK7 inhibited hepatic stellate cell (HSC) activation and cancer–stromal interaction through regulation of cancer cell pyroptosis. In a splenic xenograft mouse model, knockdown of NEK7 expression suppressed HCC primary tumor formation. Therefore, the potential of NEK7-targeted treatment for cancer–stromal interaction of HCC may be possible.
The HCC specimens were obtained from 12 patients who underwent hepatectomy for liver cancer in Peking University Shenzhen Hospital from 2011 to 2014. For immunohistochemistry staining, tissues were embedded, sliced, and stained at the Pathology Department of Peking University Shenzhen Hospital. The tissues were snap frozen in liquid nitrogen immediately after resection and stored at −80°C until RNA extraction. Paired HCC tumor and tumor-adjacent normal tissues were used to examine mRNA expression levels in our study. Experiments involving human samples have been approved by the committee for ethical review of research involving human subjects at Peking University and Peking University Shenzhen Hospital. All patients signed informed consent for participation in the study. The clinicopathological characteristics of all patients in this study are summarized in Table 1.
Immunohistochemistry (IHC) staining was performed in tissue sections as described previously (22). Sections were incubated with goat anti-NEK7 (ab166776, Abcam, Cambridge, MA, USA) and anti-GSDMD (ab219800, Abcam) overnight at 4°C and stained with a second antibody by using an SP-POD Kit according to the manufacturer’s instructions (#SP0041, Solarbio, Beijing, China). Multiplexed IHC (mIHC) was performed using a multiple fluorescent immunohistochemical staining kit (abs50012, Absin, Shanghai, China) according to the manufacturer’s instructions (23). Sections were observed using an optical microscope (BX53, Olympus, Tokyo, Japan).
The following HCC cell lines were used in this study: Hep3B, Huh7, HepG2, MHCC97L, MHCC97H, and L02 (Chinese Academy of Medical Sciences, Beijing, China). All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) and supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U/ml penicillin, and 100 U/ml streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere with 5% CO2. For inducing pyroptosis, cells (2 × 105 cells/well in 6-well culture plates) were treated with 1 μg/ml lipopolysaccharide (L2630, Sigma-Aldrich) for 4 h and then cells were treated with 10 μM nigericin (HY-100381, MedChemExpress, Princeton, NJ, USA) for 2 h before subsequent analyses (24).
The medium of each treatment group was collected and centrifuged at 1,000 g for 20 min at 4°C to remove impurities and cell debris, according to the instructions of Human IL-1 Beta ELISA Kit (ELH-IL1b, RayBio, Peachtree Corners, GA, USA). The absorbance of the microplate reader was set to 450 nm, and the standard curve was generated using a standard product to determine the concentration of each culture of IL-1 in the supernatant. Three separate preparations were used, and each experiment was carried out in triplicate wells and repeated 3 times.
The lactate dehydrogenase (LDH) assay was performed according to the LDH Cytotoxicity Assay Kit (C0016, Beyotime, Shanghai, China). The cells were seeded on 96-well plates for the logarithmic growth phase and divided into a negative control group, maximum active well, and experiment-indicated groups. The LDH release reagent was added 1 h before the scheduled time. The 96-well plate was centrifuged at 400 g for 5 min. Then, 120 μl of the supernatant was aspirated into a new 96-well plate, before measuring the absorbance at a wavelength of 490 nm using a microplate reader to calculate the LDH content. Each experiment was performed in triplicate and repeated three times.
Total RNA was extracted from cultured cells using a High Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany) and DNase I (Roche Diagnostics, Sigma-Aldrich) treatment according to the manufacturer’s instructions. RNA was directly reverse transcribed using the HiScript® II 1st Strand cDNA Synthesis Kit (MR101-01, Vazyme, Nanjing, China) according to the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed using the AceTaq® qPCR SYBR Green Master Mix (Q121-03, Vazyme, China). We designed several specific primer sequences (Gene Pharma, China). For PCR analysis, three independent experimental repeats were performed, and data are presented as mean ± standard error of mean. Primer sequences are listed in Table 2. mRNA expression levels are presented as relative expression normalized to β-actin.
Cells were prepared at 4°C in RIPA buffer (P0013J, Beyotime, China) containing proteinase inhibitor cocktail (B14001, Bimake, Houston, TX, USA) and phosphatase inhibitor cocktail (B15001, Bimake). Proteins were separated on 4%–20% precast mini polyacrylamide gels (SurePAGE™, GenScript, Nanjing, China) and transferred to PVDF membranes. The membranes were incubated overnight at 4°C with anti-NEK7 (ab95873, Abcam), anti-GSDMD (ab219800, Abcam), anti-cleaved N-terminal GSDMD ab215203, Abcam), anti-NLRP3 (ab263899, Abcam), anti-cleavage caspase-1 (4199S, Cell Signaling Technology, Danvers, MA, USA), anti-ERK1/2 (9102S, Cell Signaling Technology), anti-p-ERK1/2 (9101S, Cell Signaling Technology), anti-α-SMA (A17910, ABclonal, Woburn, MA, USA), and anti-β-actin (66009-1-Ig, Proteintech, Rosemont, IL, USA). Membranes were then probed with appropriate secondary antibodies (Cell Signaling Technology). Immunoblot signals were detected by a Millipore chemical developer (Millipore Sigma, Burlington, MA, USA).
The invasiveness and migration capacity of cancer cells were assessed by determining the number of cells invading or migrating across transwell chambers as previously described (25). Cancer cells were labeled with CellTracker Green CMFDA (40721ES50, Yeasen, Shanghai, China) according to the manufacturer’s instructions. For invasion assays, cells (1 × 105 cells/250 μl) were seeded in the upper transwell chamber (8-μm pore size; Becton Dickinson, Franklin Lakes, NJ, USA) containing 100 ml of reconstituted matrigel-coated membrane (20 μg/well, BD Biosciences, Bedford, MA, USA) at 24 h after knockdown of NEK7. Cells were incubated for 48 h, and the number of invaded cancer cells was counted. Cell migration assays were performed using the same protocol as the invasion assay without a matrigel-coated membrane. Cells were allowed to migrate and counted 24 h after cell seeding into the upper chamber. In both assays and at each time point, invaded or migrated cells at the bottom of the chamber were determined in five random fields at ×100 magnification using a fluorescent microscope (Leica, DMi8, Wetzlar, Germany). Each experiment was performed in triplicate and repeated at least three times.
The migration capacity of coculture was assessed by wound healing assay. Cells were dyed with Cell Tracker Green or Red, then seeded in a culture insert (ibidi, 81176, Martinsried, Germany). After cell attachment, the culture insert was removed and cells were allowed to migrate. At each time point, the wound closure was measured (BX53, Olympus, Japan). Graphs show the quantification of closure length (reference to 0 h). For direct coculture of cancer cells and HSCs, cancer cells and HSCs were dyed with Cell Tracker Green or Red, mixed, and then seeded in the same culture insert. At the indicated time point, migrated cell number and distance were measured by using a fluorescent microscope (Leica, DMi8, Germany). Each experiment was performed in triplicate and repeated three times.
Cells (1 × 103 cells/well) were seeded in 96-well plates. Cell viability was examined using the CellTiter-Lumi™ Plus Cell Viability Assay Kit (C0068M, Beyotime, China) following the manufacturer’s instructions. Background was subtracted using values from wells containing only culture medium.
Two NEK7 small-hairpin (shRNA, shNEK7-1: ccggatatgggctataataca, shNEK7-2: ctccgacagttagttaatatg) vectors (GenePharma, Shanghai, China) were transfected into MHCC97L cells according to the manufacturer’s instructions. Non-targeting shRNA (GenePharma) was used as control. Puromycin (S7417, Selleck Chemicals, Houston, TX, USA) was used to select NEK7-stably expressing clones. Selection was performed for more than 3 weeks. shRNA-mediated NEK7 knockdown was confirmed by quantitative RT-PCR and Western blot.
BALB/c athymic female nude mice were purchased (GemPharmatech, Nanjing, China) and transported to our institution at 4 weeks of age. 10 mice were randomized divided into 2 groups. Mice were raised in a specific pathogen-free animal room (SPF) of Shenzhen Peking University, Hong Kong University of Science and Technology Medical Center. The Animal Experimentation Ethics Committee approved our study and ensured that all experiments conformed to all relevant regulatory standards. After 1 week of accustomization, 1 × 106 MHCC97L cells were injected into the spleen of mice and liver metastasis was quantified after 6 weeks of cell implantation. Mice were cervically dislocated after anesthesia of 2% inhalable isoflurane, and the liver metastases were collected for the following evaluation.
The chi-squared test was performed to assess relationships between NEK7 protein expression and clinicopathological features. For results of in vitro experiments, values are expressed as the mean ± SEM. Comparisons of RT-QPCR were carried out using the Student’s t-test. Comparisons of RNA expression in bioinformatic analysis were performed using the Wilcox test and Kruskal–Wallis test. Kaplan–Meier analyses were compared using the log-rank test. p < 0.05 was used to define statistical significance. Statistical analysis was conducted using R language and GraphPad Prism.
By analyzing UALCAN (http://ualcan.path.uab.edu/index.html) (26), we found that expressions of NEK7 and GSDMD were significantly upregulated in tumor tissues compared to tumor-adjacent normal tissues of liver hepatocellular carcinoma (Figure 1A). The expression of NEK7 in liver hepatocellular carcinoma was correlated with cancer stage, tumor grade, nodal metastasis, and TP53 mutant and histological subtypes (Figures 1B–E and S1A). Based on the bioinformatic analysis of the datasets from The Cancer Genome Atlas (TCGA, https://www.cancer.gov/tcga) database (27), we found that NEK7 expression was highly correlated with expression levels of Kras and GSDMD in HCC tumor tissues (Figures 1F, G and S1B). In primary tumors, heterogenetic expressions of NEK7 and GSDMD including negative, weak, and strong were observed using Human Protein Atlas (www.protenatlas.org) (28). Meanwhile, we found a co-expression relationship between NEK7 and GSDMD in human specimens (Figure 1G). Given the results of NEK7 and GSDMD in HCC, we speculated that the expression of NEK7 may regulate HCC cell pyroptosis and play a crucial role in cancer progression.
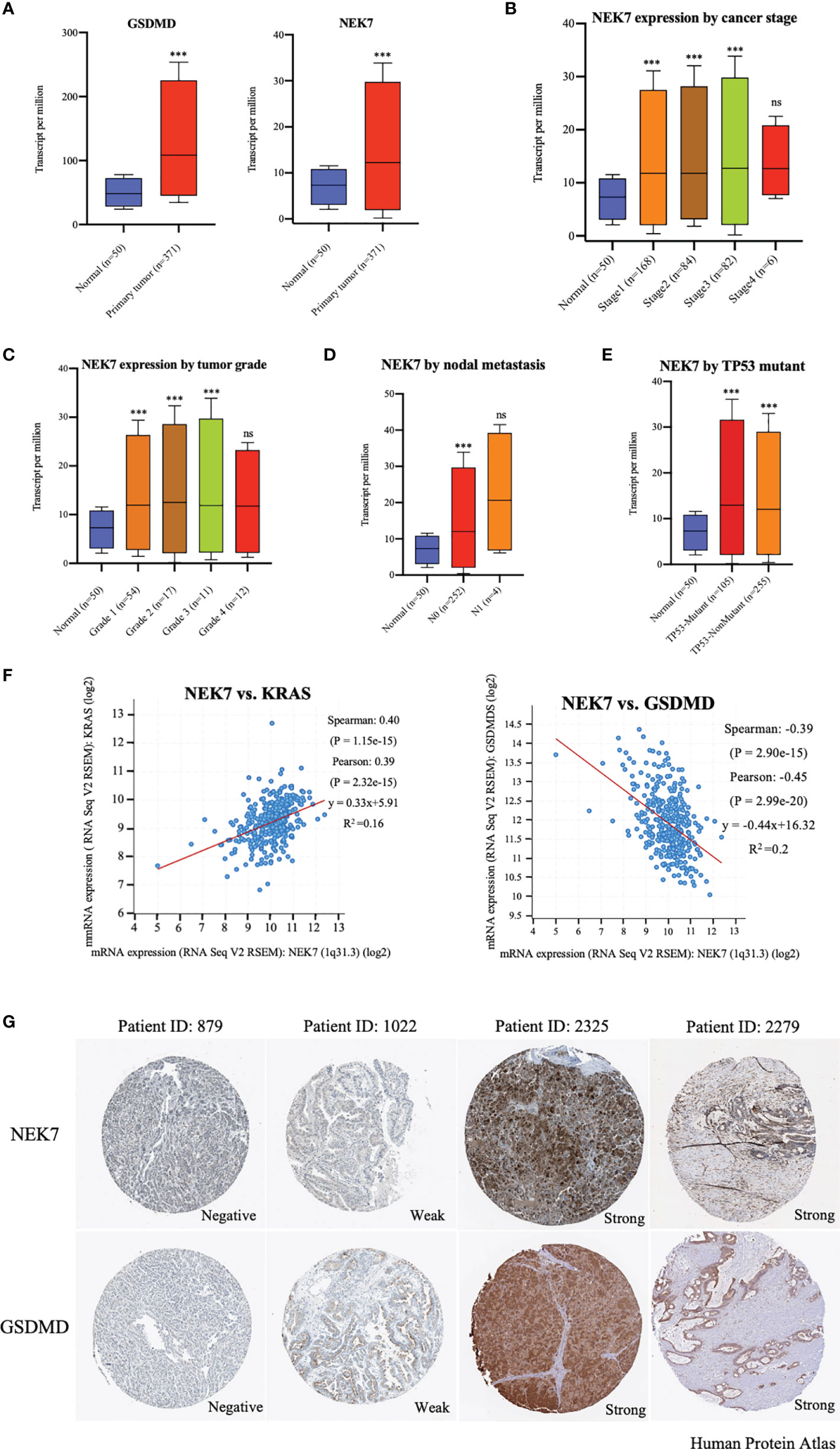
Figure 1 Expression of NEK7 correlated with GSDMD in hepatocellular carcinoma in bioinformatics analysis. (A) Expressions of NEK7 and GSDMD were significantly upregulated in tumor tissues compared to tumor-adjacent normal tissues. ***p < 0.001. (B–E) Expression of NEK7 significantly correlated with liver hepatocellular carcinoma cancer stage, tumor grade, nodal metastasis, and TP53 mutant. ***p < 0.001. ns, no significance. (F) Expression of NEK7 significantly correlated with Kras and GSDMD in HCC tissues. (G) Heterogenetic expressions of NEK7 and GSDMD in HCC tissues. NEK7 and GSDMD were co-expressed in the same tumor samples.
By using fresh frozen samples of tumor and paired tumor-adjacent normal tissues from our institution, we found that NEK7 and GSDMD were significantly upregulated in tumor tissues compared to normal tissues (Figures 2A, B). Then, we examined the expressions of NEK7 and GSDMD in normal liver tissue and HCC tissues. As negative expressions of NEK7 and GSDMD were found in normal liver tissue, we confirmed co-localization between NEK7 and GSDMD in multiple fluorescent immunohistochemical stainings (Figures 2C and S2A) and serial sections (Figure 2D) using HCC specimens from our institution. Moreover, we investigated mRNA (Figures 2E–G) and protein (Figure 2H) expressions of NEK7, as well as pyroptosis marker NLRP3 and GSDMD expression of HCC cell lines.
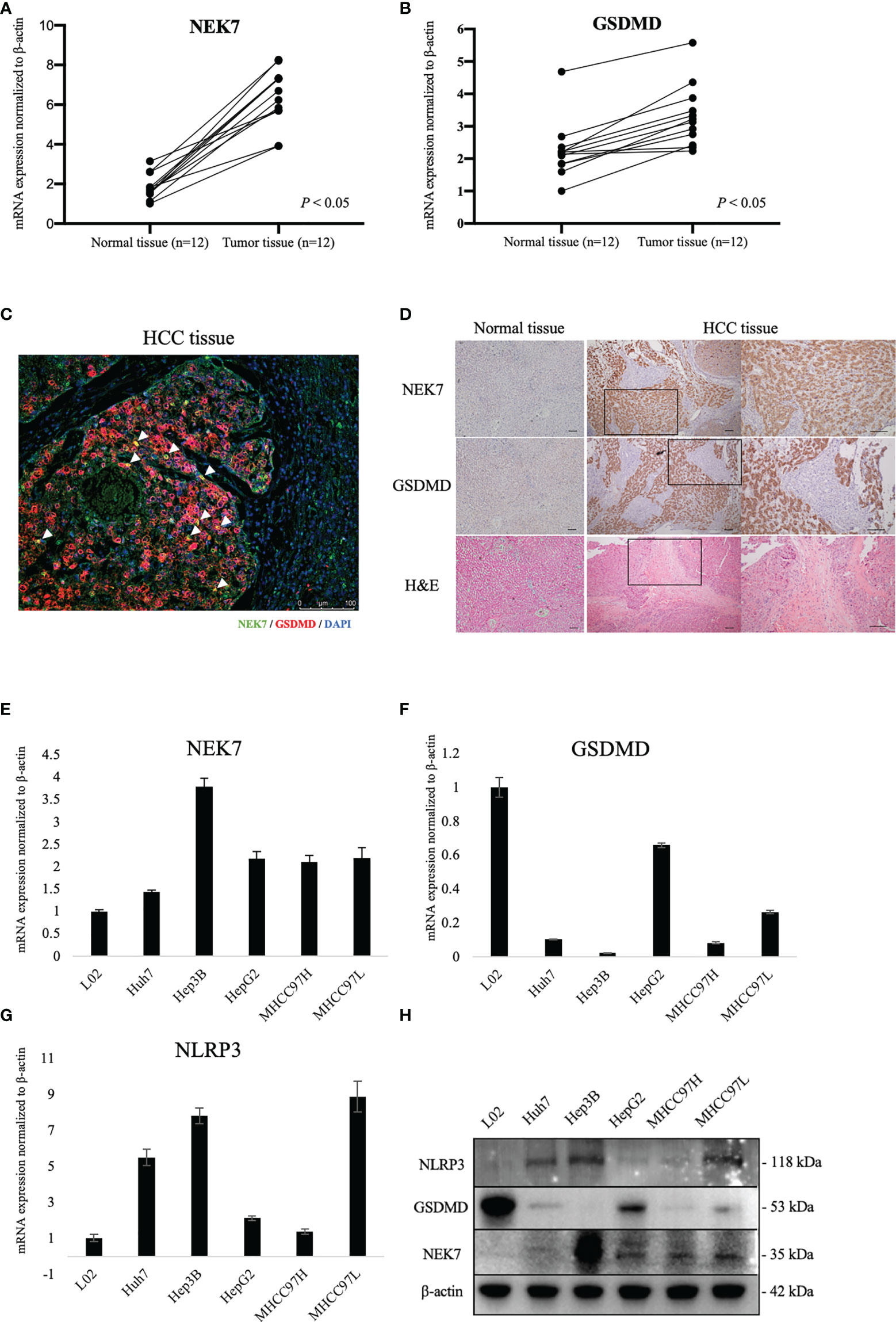
Figure 2 Expression of NEK7 and GSDMD in hepatocellular carcinoma tissues and cell lines. (A, B) mRNA expressions of NEK7 and GSDMD were significantly upregulated in tumor tissues compared to tumor-adjacent normal tissues. (C) mIHC staining of NEK7 (green) and GSDMD (red) on HCC specimens. Scale bar = 100 μm. (D) Co-locolization between NEK7 and GSDMD was observed on serial sections of HCC specimens. Scale bars = 100 μm. (E) NEK7 mRNA expression in HCC cell lines. (F) GSDMD mRNA expression in HCC cell lines. (G) NLRP3 mRNA expression in HCC cell lines. mRNA expression was normalized by β-actin expression. Error bar, error value in triplicate. (H) Protein expressions of NEK7, GSDMD, and NLRP3 in HCC cell lines.
To explore the biological role of NEK7 in HCCs, we investigated the effect of NEK7 knockdown on the invasiveness and migration activities of MHCC97L and HepG2 cells due to their overexpression of NEK7 and GSDMD. Downregulation of NEK7 expression in HepG2 and MHCC97L cells was achieved stably and efficiently by using short hairpin RNA transfection (Figures 3A, B). Cells were labeled with green fluorescence, and then transwell migration and invasion assay were performed. In result, the migration and invasion abilities of cancer cells were significantly decreased after NEK7 knockdown compared to the control group (Figure 3C). Meanwhile, cell proliferation was significantly decreased following downregulation of NEK7 expression (Figure 3D).
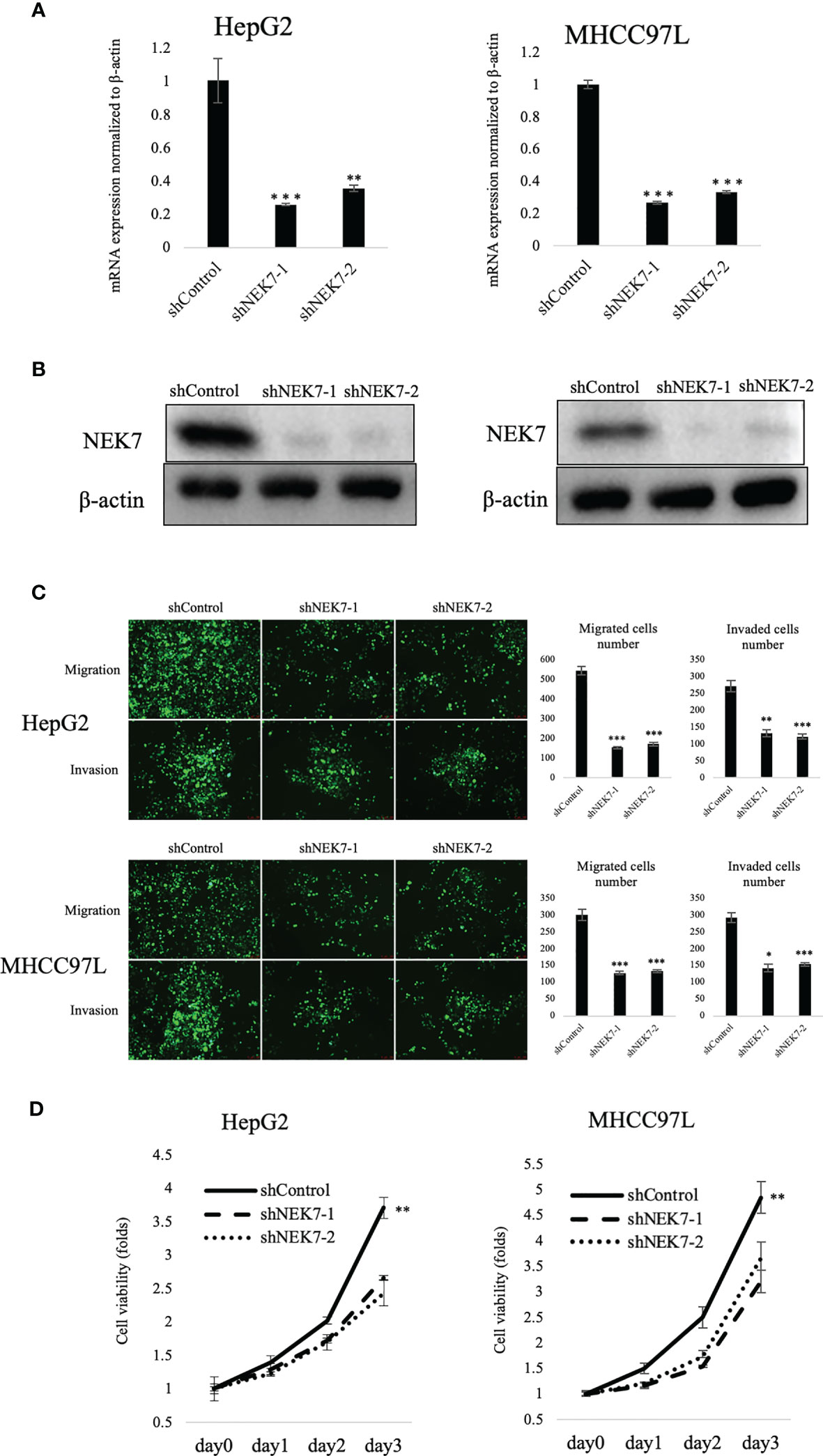
Figure 3 Knockdown of NEK7 reduces migration, invasion, and cell viability of HCCs. (A, B) qRT-PCR and Western blot of NEK7 mRNA and protein levels in cells transfected with shRNAs targeting NEK7 or negative control. ***p < 0.001. (C) shControl or shNEK7 cells were dyed with CellTracker Green; migration and invasion assays were performed for 24 and 48 h. Graphs show the quantification of cells calculated from five fields. Original magnification: ×100. Scale bars = 100 μm. *p < 0.05, **p < 0.01, ***p < 0.001. (D) Cell viability of cells was determined by CellTiter-Lumi™ Plus cell viability assay. **p < 0.01.
We next investigated the effect of NEK7 in the cancer–stromal interaction of HCC (29, 30).
Lipopolysaccharide (LPS) combined with nigericin was reported to induce pyroptosis (24, 31); our results showed that treatment of LPS and nigericin could induce HCC cell pyroptosis by remarkably upregulating the expression of active Caspase-1 and N-terminal GSDMD which are commonly used markers of pyroptosis. Compared to control, independent knockdown of NEK7 expression slightly upregulated the expressions of pyroptosis-related genes NLRP3, active Caspase-1, and N-terminal GSDMD (Figure 4A). The occurrence of pyroptosis was confirmed by investigating the IL-1β and LDH release (Figures 4B, C). Then we conducted a wound healing assay to investigate the cancer–stromal interaction by using cancer cells and human hepatic stellate cell line LX-2 cells. We found that knockdown of NEK7 in cancer cells inhibited the cancer–stromal interaction by repression of cell migratory toward LX-2 cells (Figure 4D).
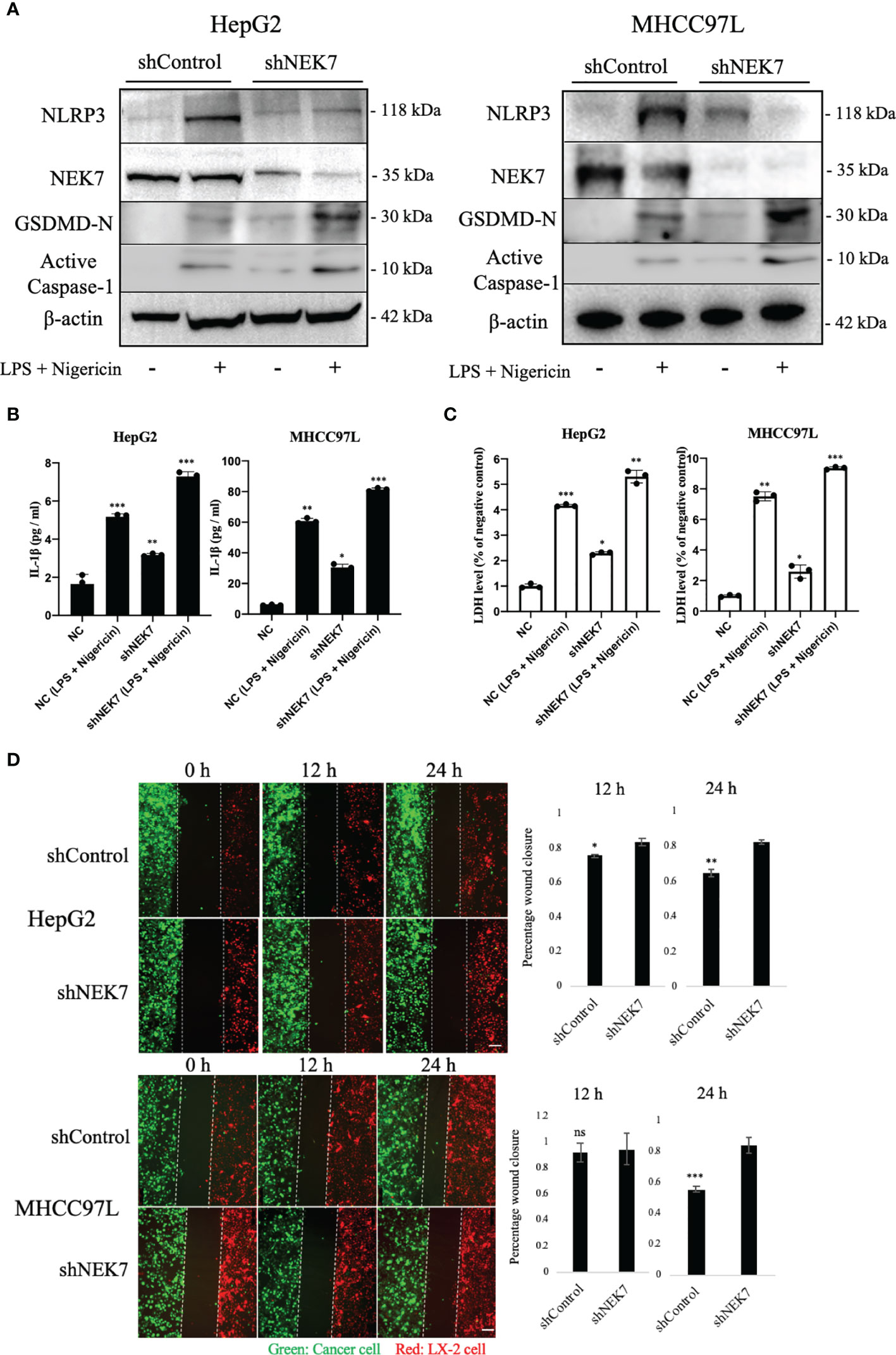
Figure 4 NEK7 regulated HCC pyroptosis and inhibited its migratory toward HSC. (A) Protein levels of NLRP3, GSDMD, NEK7, and Cleaved caspase-1 in HCCs transfected with shRNAs targeting NEK7 or negative control. LPS and nigericin treatment was positive control for cell pyroptosis. (B) IL-1β release of HCC supernatant. (C) LDH release of HCC supernatant. (D) shControl or shNEK7 cells were dyed with CellTracker Green, LX-2 cells were dyed with CellTracker Red, and migration was performed for 12 and 24 h. Original magnification: ×100. Scale bars = 100 μm. *p < 0.05, **p < 0.01, ***p < 0.001.
Cancer–stromal interaction plays an important role in HCC progression (32). Cancer cells activated HSC and promoted tumor-stromal interaction (33, 34). By using the transwell indirect coculture system (Figure 5A), we confirmed that coculture of cancer cells significantly stimulated LX-2 activation by upregulating the expressions of α-SMA and p-ERK1/2 (Figure 5B). α-SMA and p-ERK1/2 were reported as markers of HSC activation (32, 35); coculture of shNEK7-cells impaired the activation of α-SMA or p-ERK1/2 in LX-2 cells compared to the positive control (Figure 5B). Next, we mixed cancer cells and HSCs directly and monitored the migration of both cancer cells and HSCs. Knockdown of NEK7 significantly reduced migrated cell numbers and maximum migrated distance of both cancer cells and HSCs (Figure 5C). As knockdown of NEK7 alone was sufficient to induce pyroptosis by mediating GSDMD-N and IL-1β release, these results suggested that NEK7-regulated HCC cell pyroptosis inhibited HSC activation and cancer–stromal interaction.
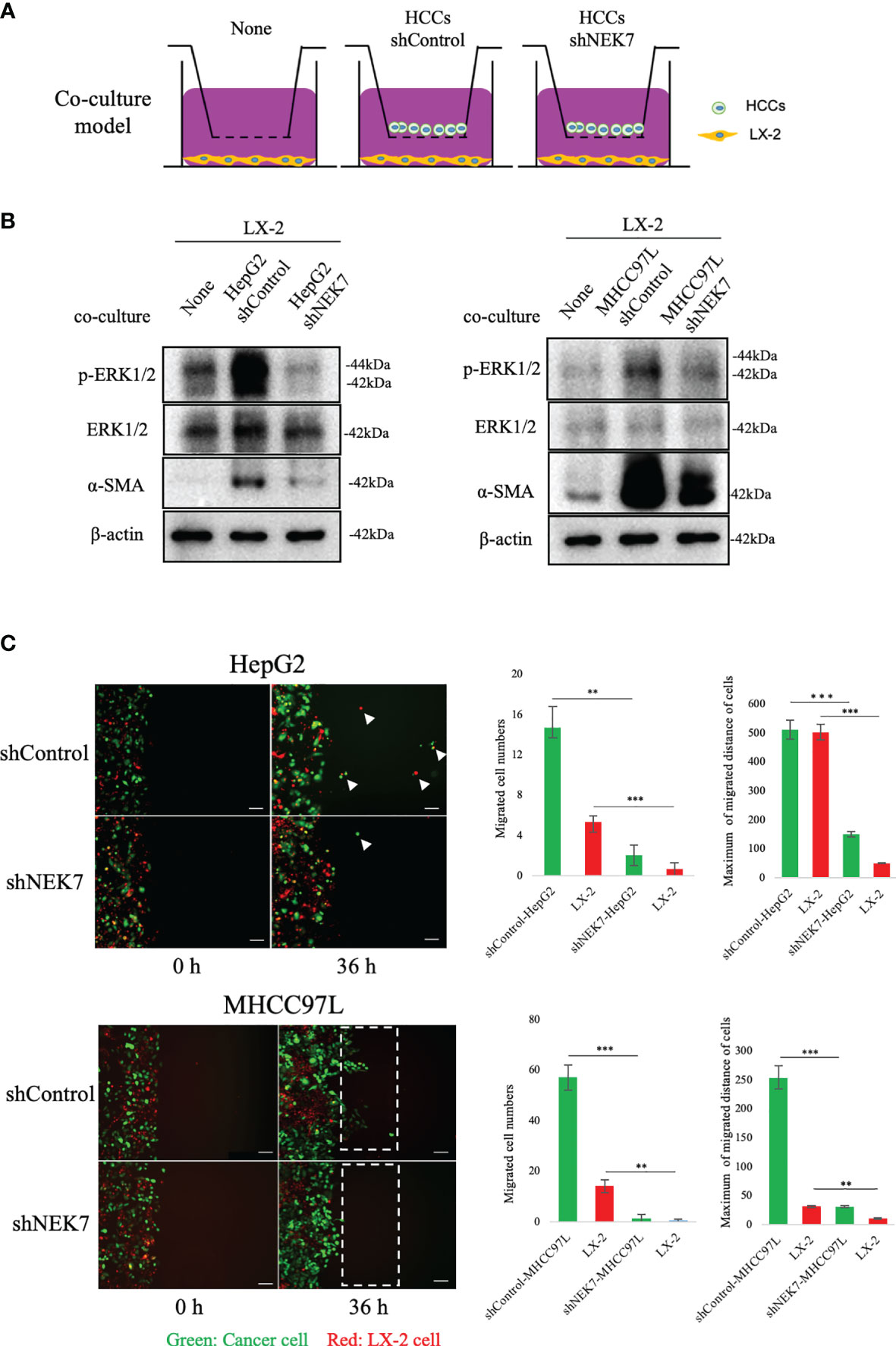
Figure 5 NEK7 promoted cancer–stromal interaction by activating HSC. (A) In indirect coculture experiments, first HSCs were seeded, and 24 h later, medium was replaced and transwell chambers (3-μm pores; Becton Dickinson) were placed in 24-well dishes, and then cells were seeded into the transwell chambers. After incubation for 2 days, HSCs in the lower chamber were collected and total protein was extracted. (B) Western blot of ERK1/2, p-ERK1/2, and α-SMA levels in HSCs, alone, or after coculture with HCCs. (C) shControl or shNEK cells direct the coculture model, coculture promoted the migration of both cancer cells and HSCs, meanwhile downregulation of NEK7 impaired the migration ability. Original magnification: ×100. Scale bars = 100 μm. **p < 0.01, ***p < 0.001.
We next investigated the effect of NEK7 downregulation on HCC progression in a splenic xenograft mouse model. ShControl or shNEK7 MHCC97L cells were splenic transplanted into nude mice, and liver tumor growth was evaluated after 6 weeks of cell transplantation (Figure 6A). Compared to the control group, mice with NEK7-depleted cells exhibited decreased liver weight and volume (Figures 6B–D). Moreover, histological analysis revealed that cancer cells that colonized into the liver parenchyma were suppressed in mice injected with NEK7 knockdown cancer cells. Meanwhile, decreased expression of Ki67 and p-ERK1/2 indicates that knockdown of NEK7 suppressed the tumor formation capacity of HCCs (Figure 6E). Taken together, these results indicated that downregulation of NEK7 reduced the colonization and proliferation of HCCs and inhibited tumor formation in vivo.
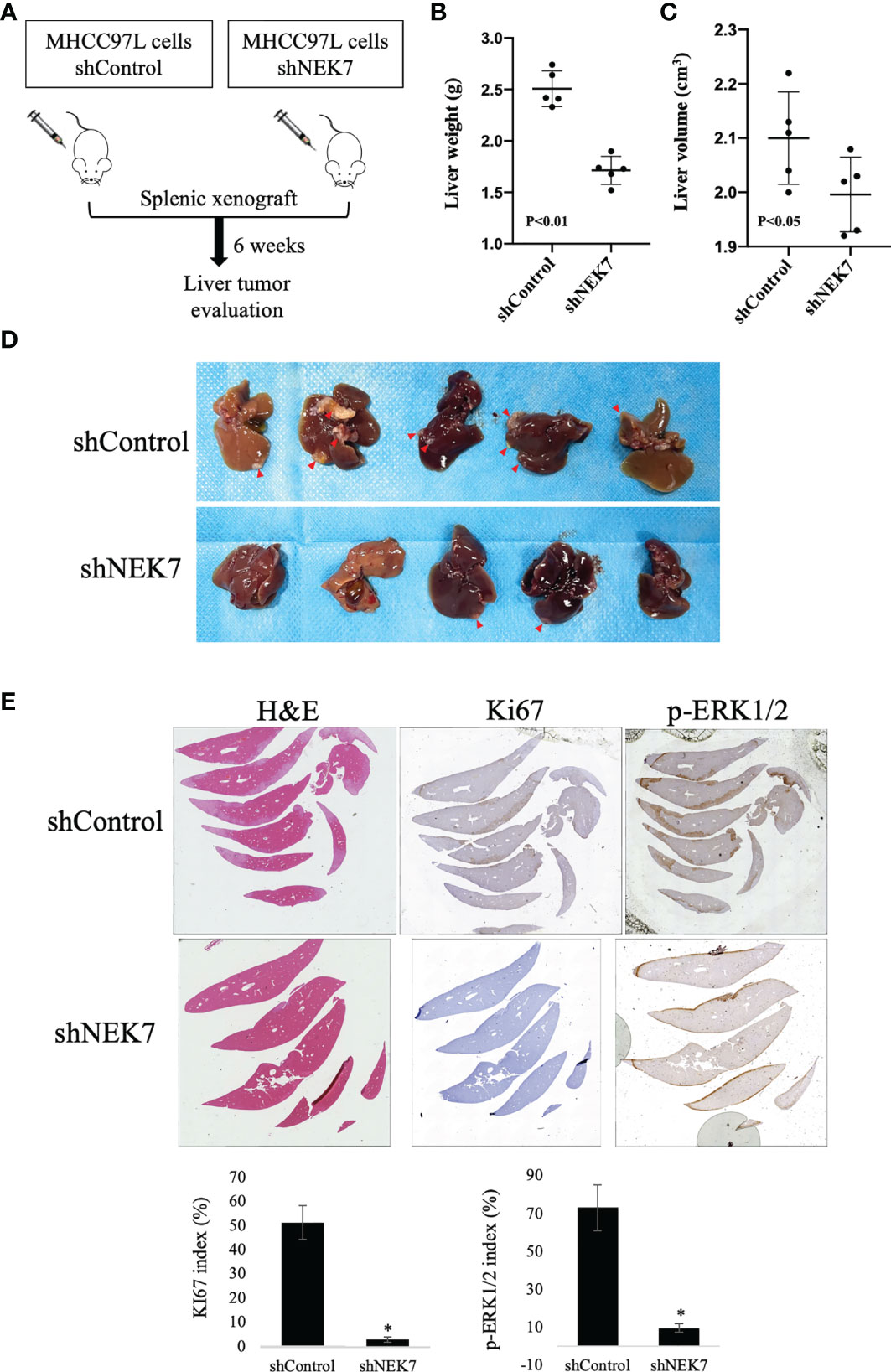
Figure 6 NEK7 decreased HCC tumor formation in a splenic xenograft mouse model. (A) Scheme of the xenograft experiment. Female nude mice were intrasplenic transplanted with MHCC97L cells and randomized divided into 2 groups (n = 5/group). 6 weeks after implantation, mice were sacrificed and liver tumors were harvested. (B, C) Downregulation of NEK7 decreased liver weight (p < 0.01) and volume (p < 0.05) of mice. (D) Gross pathology showed that knockdown of NEK7 significantly reduced liver metastasis formation (arrowheads: metastasis lesions). (E) H&E and IHC staining revealed decreased expression of Ki67 and p-ERK1/2 in livers of shNEK7 xenograft mice. *P < 0.05.
In the present study, we examined a NEK7 expression pattern with GSDMD in l HCC and investigated the functional impact of NEK7-regulated pyroptosis on tumor progression. Our results showed that NEK7 expression was correlated with pyroptosis marker GSDMD. NEK7 was associated with the migratory, invasive, and proliferative capacities of HCCs. A coculture experiment indicated that NEK7-regulated cancer cell pyroptosis inhibited stimulation of HSC activation and tumor-stromal interaction of cancer cells. In vivo experiments using a splenic injection model revealed that downregulation of NEK7 inhibited HCC tumor formation. Taken together, these results suggest that NEK7-regulated pyroptosis may play an important role in HCC progression; targeting pyroptosis by inhibiting NEK7 could be an effective therapeutic strategy for HCC progression and cancer–stromal interaction.
Pyroptosis plays a dual role in the pathogenesis of tumors (36). On the one hand, the multiple signaling pathways and inflammatory chemokines released during pyroptosis are contributed to tumorigenesis and chemoresistance (37, 38). On the other hand, as a type of cell death, pyroptosis can inhibit the occurrence and development of tumors (37, 39). Wei et al. found a different result of the NLRP3 inflammasome in HCC: on the one hand, the NLPR3 expression was downregulated in HCC tissues compared to non-cancerous liver tissues, and its expression was negatively correlated with the pathological grade and clinical stage of HCC (19); on the other hand, in the comparison of normal liver tissue and liver cancer tissue, NLRP3 expression in normal liver tissue was low, while that in cancer tissues was upregulated. Furthermore, they found an anticancer effect of 17β-estradiol, by trigger pyroptosis via activation of the NLRP3 inflammasome (40). Another inflammasome signaling relevant gene, IRF2, was reported to regulate caspase-1 and GSDMD transcript expression in macrophages, endothelial cells, and multiple tissues (41). Moreover, LPS induces GSDMD expression change in varying degrees by mediating caspase-4 or caspase-11 (42, 43). An FDA-approved drug, disulfiram, protected against LPS-induced GSDMD pore formation and IL-1β leaking, while the expression of GSDMD was inhibited following treatment of disulfiram (44). Our data demonstrated a knockdown of NEK7 inhibited GSDMD expression, and this effect was strengthened in the presence of LPS and nigericin treatment. However, the regulation of GSDMD and cleaved-GSDMD is a complex process far from being clearly understood.
Cancer–stromal interactions reported a key regulator of HCC progression (32). Cancer cells are able to promote HSC activation and proliferation. Meanwhile, activated HSCs in turn generate a variety of cytokines, growth factors, and ECM proteins, which contribute to forming a microenvironment favorable for tumor growth and promoting tumor cell progression, metastasis, and chemotherapy resistance (33). Activated HSCs are myofibroblast-like cells that express α-smooth muscle actin (α-SMA) and become proliferative and contractile (45). By coculturing with HSCs, cancer cells enhanced proliferation and migration through ERK and NF-κB signaling pathways (34). Co-inoculation of activated primary HSCs with tumor cells in vivo leads to enhanced tumor growth and proliferation of HCC (46). Currently, therapeutic approaches are used to target tumor cells of l HCC; recent studies have instead focused on potentially targeting the stromal cell therapeutic approach to HCC treatment (47). We provide new insights into and perspectives for effective therapeutic HSC targeting, which exhibited a brand-new strategy through tumor cell pyroptosis.
In conclusion, NEK7 increased the cell migration and invasion capacity of HCC. Downregulation of NEK7 suppressed cancer–stromal interaction by inducing cancer cell pyroptosis. NEK7 promoted HCC cell growth in vitro and in vivo. Our findings suggest that NEK7 plays an important role in cancer–stromal interaction of HCC. Investigation of NEK7-regulated pyroptosis could move forward for revealing the genomic alteration of HCC. Moreover, anti-NEK7 drugs may have potential to regulate NLRP3 to abolish the inflammation response and NLRP3-related diseases (48, 49). Therefore, NEK7 targeting therapy might be a potential new strategy for HCC treatment.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The research study was approved by the Ethics Committee of Human Experimentation at the Peking University Shenzhen Hospital (Shenzhen, China). All patients studied signed informed consent for participation. The patients/participants provided their written informed consent to participate in this study. The animal study was approved by the Animal Ethics Committee of Peking University Shenzhen Hospital. All animal procedures and care were conducted in accordance with institutional guidelines and in compliance with national and international laws and policies.
ZY designed the study, conducted experiments, acquired and analyzed data, and wrote the manuscript. QD, ZFL analyzed bioinformatic data. JY, YS, QL and ZWL discussed and revised the manuscript. WY, GY, XL and QR conducted evaluation of pathology study. JQ and JL was responsible for the conception and supervision of the study and wrote the manuscript. All authors corrected drafts and approved the final version of the manuscript.
This study was supported by the Sanming Project of Medicine in Shenzhen (No. SZSM201612021) and China Postdoctoral Science Foundation (No. 2020M682841).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Fangting Zhang, Minghua Li, and Xia Long (Central Laboratory, Peking University Shenzhen Hospital) for the skillful technical assistance and laboratory management.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.812655/full#supplementary-material
Supplementary Figure 1 | Bioinformatics analysis of NEK7 in hepatocellular carcinoma. (A) Expression of NEK7 significantly correlated to liver hepatocellular carcinoma cancer histological subtypes. ***P < 0.001, *P < 0.05. (B) Correlation between NEK7 expression and NLRP3, IL-18, GSDMB, GSDMC expression in HCC tissues. (C) Co-expression of NEK7 (green) and GSDMD (red) by multiple fluorescent immunohistochemical staining of HCC specimen. Scale bars = 100 μm.
Supplementary Figure 2 | co-expression of NEK7 and GSDMD in HCC specimens. (A) mIHC staining of NEK7 and GSDMD on HCC specimens. (B) Co-expression of NEK7 and GSDMD was observed on serial sections of HCC specimens.
HCC, hepatocellular carcinoma; NEK7, NIMA-related protein kinase 7; NLRP3, NLR family pyrin domain containing 3; HSC, hepatic stellate cell; mIHC, multiplexed immunohistochemistry; LDH, lactate dehydrogenase; IL-1β, interleukin 1β; IL-18, interleukin 18; LPS, lipopolysaccharide; IHC, immunohistochemical; qRT-PCR, real-time quantitative polymerase chain reaction; FBS, fetal bovine serum; NC, negative control; shRNA, short hairpin RNA; TCGA, Cancer Genome Atlas Program; GSDMD, Gasdermin D; α-SMA, α-smooth muscle actin.
1. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A Global View of Hepatocellular Carcinoma: Trends, Risk, Prevention and Management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y
2. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. Changing Cancer Survival in China During 2003–15: A Pooled Analysis of 17 Population-Based Cancer Registries. Lancet Glob Health (2018) 6(5):e555–67. doi: 10.1016/S2214-109X(18)30127-X
3. Bruix J, Reig M, Sherman M. Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology (2016) 150(4):835–53. doi: 10.1053/j.gastro.2015.12.041
4. Crocetti L, Bargellini I, Cioni R. Loco-Regional Treatment of HCC: Current Status. Clin Radiol (2017) 72(8):626–35. doi: 10.1016/j.crad.2017.01.013
5. Wang EA, Stein JP, Bellavia RJ, Broadwell SR. Treatment Options for Unresectable HCC With a Focus on SIRT With Yttrium-90 Resin Microspheres. Int J Clin Pract (2017) 71(11):e12972. doi: 10.1111/ijcp.12972
6. Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk Factors and Prevention of Hepatocellular Carcinoma in the Era of Precision Medicine. J Hepatol (2018) 68(3):526–49. doi: 10.1016/j.jhep.2017.09.016
7. O’Connell MJ, Krien MJE, Hunter T. Never Say Never. The NIMA-Related Protein Kinases in Mitotic Control. Trends Cell Biol (2003) 13(5):221–8. doi: 10.1016/S0962-8924(03)00056-4
8. Kimura M, Okano Y. Identification and Assignment of the Human NIMA-Related Protein Kinase 7 Gene (NEK7) to Human Chromosome 1q31.3. Cytogenet Genome Res (2001) 94(1-2):33–8. doi: 10.1159/000048779
9. Saloura V, Cho H-S, Kiyotani K, Alachkar H, Zuo Z, Nakakido M, et al. WHSC1 Promotes Oncogenesis Through Regulation of NIMA-Related Kinase-7 in Squamous Cell Carcinoma of the Head and Neck. Mol Cancer Res (2015) 13(2):293–304. doi: 10.1158/1541-7786.MCR-14-0292-T
10. Eisa NH, Jilani Y, Kainth K, Redd P, Lu S, Bougrine O, et al. The Co-Chaperone UNC45A Is Essential for the Expression of Mitotic Kinase NEK7 and Tumorigenesis. J Biol Chem (2019) 294(14):5246–60. doi: 10.1074/jbc.RA118.006597
11. Yan Z, Qu J, Li Z, Yi J, Su Y, Lin Q, et al. NEK7 Promotes Pancreatic Cancer Progression And Its Expression Is Correlated With Poor Prognosis. Front Oncol (2021) 11:705797. doi: 10.3389/fonc.2021.705797
12. Zhou L, Wang Z, Xu X, Wan Y, Qu K, Fan H, et al. Nek7 is Overexpressed in Hepatocellular Carcinoma and Promotes Hepatocellular Carcinoma Cell Proliferation In Vitro and In Vivo. Oncotarget (2016) 7(14):18620–30. doi: 10.18632/oncotarget.7620
13. Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, et al. Frequent Alterations in the Expression of Serine/Threonine Kinases in Human Cancers. Cancer Res (2006) 66:8147–54. doi: 10.1158/0008-5472.CAN-05-3489
14. Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, et al. NLRP3 Activation and Mitosis Are Mutually Exclusive Events Coordinated by NEK7, a New Inflammasome Component. Nat Immunol (2016) 17(3):250–8. doi: 10.1038/ni.3333
15. Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, et al. Genome-Wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J Biol Chem (2016) 291(1):103–9. doi: 10.1074/jbc.C115.700492
16. Domingo-Fernández R, Coll RC, Kearney J, Breit S, O’Neill LAJ. The Intracellular Chloride Channel Proteins CLIC1 and CLIC4 Induce IL-1β Transcription and Activate the NLRP3 Inflammasome. J Biol Chem (2017) 292(29):12077–87. doi: 10.1074/jbc.M117.797126
17. Fink SL, Cookson BT. Caspase-1-Dependent Pore Formation During Pyroptosis Leads to Osmotic Lysis of Infected Host Macrophages. J Immunol (2006) 8:1812–25. doi: 10.1111/J.1462-5822.2006.00751.x
18. Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, et al. The Pyroptosome: A Supramolecular Assembly of ASC Dimers Mediating Inflammatory Cell Death via Caspase-1 Activation. Cell Death Differ (2007) 14:1590–604. doi: 10.1038/sj.cdd.4402194
19. Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X, et al. Deregulation of the NLRP3 Inflammasome in Hepatic Parenchymal Cells During Liver Cancer Progression. Lab Investig (2014) 94:52–62. doi: 10.1038/labinvest.2013.126
20. Allen IC, TeKippe EM, Woodford R-MT, Uronis JM, Holl EK, Rogers AB, et al. The NLRP3 Inflammasome Functions as a Negative Regulator of Tumorigenesis During Colitis-Associated Cancer. J Exp Med (2010) 207(5):1045–56. doi: 10.1084/jem.20100050
21. Gao J, Qiu X, Xi G, Liu H, Zhang F, Lv T, et al. Downregulation of GSDMD Attenuates Tumor Proliferation via the Intrinsic Mitochondrial Apoptotic Pathway and Inhibition of EGFR/Akt Signaling and Predicts a Good Prognosis in Non-Small Cell Lung Cancer. Oncol Rep (2018) 40:1971–84. doi: 10.3892/or.2018.6634
22. Zheng B, Qu J, Ohuchida K, Feng H, Chong SJF, Yan Z, et al. LAMA4 Upregulation Is Associated With High Liver Metastasis Potential and Poor Survival Outcome of Pancreatic Cancer. Theranostics (2020) 10. doi: 10.7150/thno.47001
23. Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed Immunohistochemistry, Imaging, and Quantitation: A Review, With an Assessment of Tyramide Signal Amplification, Multispectral Imaging and Multiplex Analysis. Methods (2014) 70:46–58. doi: 10.1016/j.ymeth.2014.08.016
24. Hörauf J-A, Kany S, Janicova A, Xu B, Vrdoljak T, Sturm R, et al. Short Exposure to Ethanol Diminishes Caspase-1 and ASC Activation in Human HepG2 Cells In Vitro. Int J Mol Sci (2020) 21:3196. doi: 10.3390/ijms21093196
25. Yan Z, Ohuchida K, Zheng B, Okumura T, Takesue S, Nakayama H. CD110 Promotes Pancreatic Cancer Progression and Its Expression Is Correlated With Poor Prognosis. J Cancer Res Clin Oncol (2019) 145(5):1147–64. doi: 10.1007/s00432-019-02860-z
26. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
27. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov (2012) 2. doi: 10.1158/2159-8290.CD-12-0095
28. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-Based Map of the Human Proteome. Science (80-) (2015) 347:1260419. doi: 10.1126/science.1260419
29. Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, et al. Inflammatory Caspases Are Innate Immune Receptors for Intracellular LPS. Nature (2014) 514(7521):187–92. doi: 10.1038/nature13683
30. Zhang C, Zhao C, Chen X, Tao R, Wang S, Meng G, et al. Induction of ASC Pyroptosis Requires Gasdermin D or Caspase-1/11-Dependent Mediators and Ifnβ From Pyroptotic Macrophages. Cell Death Dis (2020) 11(6):470. doi: 10.1038/s41419-020-2664-0
31. He W, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin D Is an Executor of Pyroptosis and Required for Interleukin-1β Secretion. Cell Res (2015) 25(12):1285–98. doi: 10.1038/cr.2015.139
32. Ogunwobi OO, Harricharran T, Huaman J, Galuza A, Odumuwagun O, Tan Y, et al. Mechanisms of Hepatocellular Carcinoma Progression. World J Gastroenterol (2019) 25(19):2279–93. doi: 10.3748/wjg.v25.i19.2279
33. Yang JD, Nakamura I, Roberts LR. The Tumor Microenvironment in Hepatocellular Carcinoma: Current Status and Therapeutic Targets. Semin Cancer Biol (2011) 21:35–43. doi: 10.1016/j.semcancer.2010.10.007
34. Amann T, Bataille F, Spruss T, Mühlbauer M, Gäbele E, Schölmerich J, et al. Activated Hepatic Stellate Cells Promote Tumorigenicity of Hepatocellular Carcinoma. Cancer Sci (2009) 100:646–53. doi: 10.1111/j.1349-7006.2009.01087.x
35. Dewidar, Meyer, Dooley, Meindl-Beinker. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis—Updated 2019. Cells (2019) 8:1419. doi: 10.3390/cells8111419
36. Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J, et al. The Role of Pyroptosis in Cancer: Pro-Cancer or Pro-“Host”? Cell Death Dis (2019) 10:650. doi: 10.1038/s41419-019-1883-8
37. Thi HTH, Hong S. Inflammasome as a Therapeutic Target for Cancer Prevention and Treatment. J Cancer Prev (2017) 22:62–73. doi: 10.15430/JCP.2017.22.2.62
38. Zhou C-B, Fang J-Y. The Role of Pyroptosis in Gastrointestinal Cancer and Immune Responses to Intestinal Microbial Infection. Biochim Biophys Acta Rev Cancer (2019) 1872:1–10. doi: 10.1016/j.bbcan.2019.05.001
39. Nagarajan K, Soundarapandian K, Thorne RF, Li D, Li D. Activation of Pyroptotic Cell Death Pathways in Cancer: An Alternative Therapeutic Approach. Transl Oncol (2019) 12:925–31. doi: 10.1016/j.tranon.2019.04.010
40. Wei Q, Zhu R, Zhu J, Zhao R, Li M. E2-Induced Activation of the NLRP3 Inflammasome Triggers Pyroptosis and Inhibits Autophagy in HCC Cells. Oncol Res Featur Preclin Clin Cancer Ther (2019) 27:827–34. doi: 10.3727/096504018X15462920753012
41. Kayagaki N, Lee BL, Stowe IB, Kornfeld OS, O’Rourke K, Mirrashidi KM, et al. IRF2 Transcriptionally Induces GSDMD Expression for Pyroptosis. Sci Signal (2019) 12. doi: 10.1126/scisignal.aax4917
42. Wang K, Sun Q, Zhong X, Zeng M, Zeng H, Shi X, et al. Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis. Cell (2020) 180:941–55.e20. doi: 10.1016/j.cell.2020.02.002
43. Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, et al. GsdmD P30 Elicited by Caspase-11 During Pyroptosis Forms Pores in Membranes. Proc Natl Acad Sci (2016) 113:7858–63. doi: 10.1073/pnas.1607769113
44. Hu JJ, Liu X, Xia S, Zhang Z, Zhang Y, Zhao J, et al. FDA-Approved Disulfiram Inhibits Pyroptosis by Blocking Gasdermin D Pore Formation. Nat Immunol (2020) 21:736–45. doi: 10.1038/s41590-020-0669-6
45. Flier JS, Underhill LH, Friedman SL. The Cellular Basis of Hepatic Fibrosis – Mechanisms and Treatment Strategies. N Engl J Med (1993) 328:1828–35. doi: 10.1056/NEJM199306243282508
46. Zhao W, Zhang L, Yin Z, Su W, Ren G, Zhou C, et al. Activated Hepatic Stellate Cells Promote Hepatocellular Carcinoma Development in Immunocompetent Mice. Int J Cancer (2011) 129:2651–61. doi: 10.1002/ijc.25920
47. Ruan Q, Wang H, Burke LJ, Bridle KR, Li X, Zhao C, et al. Therapeutic Modulators of Hepatic Stellate Cells for Hepatocellular Carcinoma. Int J Cancer (2020) 147:1519–27. doi: 10.1002/ijc.32899
48. Xu J, Lu L, Li L. NEK7: A Novel Promising Therapy Target for NLRP3-Related Inflammatory Diseases. Acta Biochim Biophys Sin (Shanghai) (2016) 48:966–8. doi: 10.1093/abbs/gmw080
Keywords: NEK7, hepatocellular carcinoma, pytoptosis, cancer–stromal interaction, migration, invasion
Citation: Yan Z, Da Q, Li Z, Lin Q, Yi J, Su Y, Yu G, Ren Q, Liu X, Lin Z, Qu J, Yin W and Liu J (2022) Inhibition of NEK7 Suppressed Hepatocellular Carcinoma Progression by Mediating Cancer Cell Pyroptosis. Front. Oncol. 12:812655. doi: 10.3389/fonc.2022.812655
Received: 18 November 2021; Accepted: 07 January 2022;
Published: 10 February 2022.
Edited by:
Mark De Ridder, Vrije University Brussel, BelgiumReviewed by:
Ting Zhang, First Affiliated Hospital of Chengdu Medical College, ChinaCopyright © 2022 Yan, Da, Li, Lin, Yi, Su, Yu, Ren, Liu, Lin, Qu, Yin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Qu, cXVqaWFuaHVhLmplbnNlbkBnbWFpbC5jb20=; Jikui Liu, bGl1ODkyOUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.