- 1Department of Laboratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, Wenzhou, China
Aberrant expression of the gene encoding the Ndc80 kinetochore complex component (NUF2) reportedly contributes to the progression of several human cancers. However, the functional roles of NUF2 and their underlying mechanisms in lung adenocarcinoma (LUAD) are largely unknown. The current study aimed to investigate the role of NUF2 in LUAD tumorigenesis. Here, TCGA, ONCOMINE, the Human Protein Atlas, UALCAN, and the results of our cohort were used to analyze the expression of NUF2 in LUAD. A Kaplan–Meier analysis and univariate and multivariate Cox regression analyses were performed to estimate the prognostic values of NUF2 expression in the Cancer Genome Atlas cohort. We studied the effects of NUF2 expression on proliferation, migration, invasion, and tumor growth using LUAD cell lines. Gene set enrichment analysis (GSEA) was used to analyze the pathways and biological function enrichment of NUF2 in LUAD. The ssGSEA database was used to analyze the relationship between NUF2 expression and immune cell infiltration in LUAD. Results revealed elevated expression of NUF2 in LUAD specimens. Patients overexpressing NUF2 had poor prognoses relative to those with low NUF2 expression. Knockdown of NUF2 suppressed the proliferation, migration, invasion, epithelial-mesenchymal transition, and colony formation of LUAD cells. Moreover, NUF2 knockdown induced cell cycle arrest at the G0/G1 phase. Gene Ontology and GSEA analyses suggested that NUF2 may be involved in immunity, proliferation, and apoptosis-related pathways. NUF2 overexpression was positively correlated with differential immune cell infiltration. In conclusion, NUF2 expression was associated with the clinical phenotype of LUAD and hence has potential implications in LUAD treatment.
Introduction
Lung adenocarcinoma (LUAD) is the most prevalent subtype of non-small cell lung cancer (NSCLC) (1). Since its early symptoms are not obvious, LUAD is mostly diagnosed in advanced stages (2). Thus, reliable biomarkers to estimate the key parameters for diagnosis and disease monitoring are urgently needed.
The NDC80 complex, which is composed of NDC80 (also known as HEC1), NUF2, SPC24, and SPC25, is the primary microtubule receptor at the kinetochore. NUF2 is a key regulator of the cell cycle and is overexpressed in several types of cancers, including bladder cancer, renal cell carcinoma, cholangiocarcinoma, and lung cancer (3). Mounting evidence suggests that NUF2 is a valuable prognostic biomarker for detecting breast cancer, hepatocellular carcinoma, and oral cancer (4–6). NUF2, as a part of the aforementioned NDC80 complex, is differentially expressed in tumor tissues and has diagnostic value for LUAD (7). Moreover, this gene combination has a better diagnostic value for LUAD than NUF2 alone. NUF2 overexpression is reportedly associated with poor overall patient survival. Reports have also suggested that NUF2 is a potential prognostic biomarker in NSCLC (8). Taken together, NUF2 appears to play an important role in carcinogenesis, although systematic studies are required to further shed light on its functional and clinical role as a biomarker for LUAD.
Immunotherapy has generated significant interest in the context of NSCLC treatment, especially with the recent success of immune checkpoint inhibitors for treating metastatic stage IV cancers (9, 10). Recently, focus has shifted towards novel strategies that target LAG3 and TIM3, as well as targeting Tregs and their immunosuppressive factors (e.g. TGF-β) into the tumor microenvironment, which have been proposed as more effective in stimulating an anti-tumor immune response (11). The tumor microenvironment is an active research field for tumor diagnosis, treatment targets, and prognostic biomarkers (12). Overall, the study of immune-related therapeutic targets is essential for effective treatment of lung cancer.
In this study, we evaluated the expression of NUF2 in LUAD as described in the following five cohorts: The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx), Gene Expression Omnibus (GEO), ONCOMINE, and UALCAN. Furthermore, we used our independent cohort to identify the expression pattern of NUF2, as well as the related clinicopathological features. We used TCGA to explore the role of NUF2 expression level in LUAD as a pathological and prognostic biomarker. A nomogram integrating clinicopathological indexes and NUF2 expression was established to predict prognosis. Moreover, we examined the association between NUF2 expression and immune cell infiltration. We used NUF2-targeting siRNA to suppress its endogenous expression to examine its role in LUAD. The relationships between NUF2 and its pathways were analyzed using gene set expression analysis (GSEA).
Materials and Methods
Patient and Sample
Specimens of LUAD tissues and para-cancerous tissues were obtained from 61 patients with LUAD, who had undergone surgical operation in the First Affiliated Hospital of Wenzhou Medical University, China, from February 2017 to March 2020. The inclusion criteria for patients were as follows: (I) confirmed histology following pathology review; (II) aged between 18 and 80 years; (III) absence of palliative surgery or neoadjuvant chemo- and/or radiotherapy; (IV) no major organ dysfunction unless caused by the malignant disease; and (V) no history of major neurological or psychiatric disease; (IV) no significant renal dysfunction, cardiovascular or cerebrovascular diseases, hematological or endocrine system diseases, or metabolic illness; and (III) psychiatric illness.
Data Mining
In the first round of verification of NUF2 expression, genomic data of samples from subjects with LUAD (n = 513) and from 59 matched normal lungs were obtained from TCGA-LUAD; 57 paired LUAD and adjacent normal tissue samples were included in TCGA. To evaluate NUF2 expression, tumor tissues were obtained from TCGA, and normal tissues from the TCGA and GTEx databases were combined. Thereafter, we searched the GEO database and downloaded human LUAD-related datasets. Among them, 57 LUAD patients and 11 normal specimens were included in GSE116959, and 52 pairs of LUAD and normal lung specimens were included in GSE115002. To investigate NUF2 expression in other databases, we referred to the ONCOMINE database (https://www.oncomine.org/resource/main.html).
In the second round of verification, differential expression of the NUF2 protein between normal human lung and LUAD tissues was determined using the UALCAN database (http://ualcan.path.uab.edu/) using 111 LUAD and 111 healthy lung tissues.
In the third round of experimental verification, 61 pairs of LUAD tissue and paired tissue samples stored in our laboratory were used. NUF2 mRNA transcription levels were evaluated in tissue and cancer cell lines (HCC827, A549, and SPCA1). Briefly, 500 ng total RNA (for each sample) was reverse-transcribed using the first-strand cDNA synthesis kit (TaKaRa, Kusatsu, Japan) in accordance with the manufacturer’s instructions. cDNAs were then used for RT-qPCR analysis using SYBR Green (TaKaRa). The primers used were as follows: NUF2 forward primer, 5′-TTTTGCCTATCTGCCGGGTG-3′; reverse, 5′-GTCCGCAGAGGATTTATATTGCC-3′. β-actin forward primer: 5′-CCTGGCACCCAGCACAAT-3′; reverse primer, 5′-GCTGATCCACATCTGCTGGAA-3′.
Nomogram Construction and Validation
Next, a nomogram integrating clinical prognostic factors, such as primary therapeutic outcome, tumor status, pathologic stage, an NUF2 expression was constructed.The NUF2 expression profile was used to predict 1-, 3-, and 5-year prognoses of patients with LUAD with the R rms package. Calibration curves were established to examine the accuracy of the nomogram. Finally, C-index was used to evaluate the predictive ability of the model.
Functional Analyses
The associations between NUF2 expression and biological processes, molecular functions, and cellular components were evaluated by Gene Ontology (GO) annotation. The impact of NUF2 on pathway activation and inhibition was evaluated using GSEA, a web service aimed at unmasking potential cancer-related pathways.
Protein-Protein Interaction Network Analysis
STRING (https://string-db.org/), a tool for predicting protein-protein interactions (PPIs), was used to quantify the functional interactions of the NUF2 protein (13). A combined score > 0.4 was set as the cut-off.
Estimation of Relative Abundance of Immune Cell Types
Single-sample gene set enrichment analysis (ssGSEA) is a database that shows the relative abundance of immune cells in LUAD. ssGSEA was used to evaluate the infiltration level of 22 types of immune cells in LUAD samples based on their NUF2 expression profile. Subsequently, differences in the composition of immune cells between low-risk and high-risk patients with NUF2 expression was compared.
Immunohistochemical Staining of the Tissue Microarray
Samples from 7 LUAD patients were stained with rabbit polyclonal antibodies against NUF2 (1:500, #ab244470, Abcam). The staining results were evaluated by two independent pathologists. Staining intensity was classified as 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The amounts of positive tumor cells were classified according to the following percentages: 1+ (≤25%), 2+ (26%–50%), 3+ (51%–75%), and 4+ (>75%). The final expression scores were calculated by multiplying the two variables together.
Cell Line and siRNA Transfection
LUAD cell lines HCC827 and A549 were used to determine the regulation of NUF2 expression. siRNAs targeting NUF2 were transfected into HCC827 and A549 cells using Polyethylenimine (PEI); briefly, 35 × 104 cells/well were seeded in 6-well plates, and incubated with 50 nM siRNA and 4 μg/ml PEI. After 8 h, the medium was replaced with complete medium.
Cell Proliferation Assay
A549 and HCC827 cells and Cell Counting Kit-8 (CCK-8) kit (TaKaRa) were used for the CCK-8 assay. A total of 1500 LUAD cells in full medium were seeded in 96-well plates. The next day, transfections were performed with PEI (4 μg/ml) for 8h. Briefly, the 96-well plates containing the transfected cells were incubated at 37 °C for the indicated time points. CCK-8 reaction solution (10%) was added to the cells and incubated for 1 h at 37°C. The optical density (OD) values at 450 nm were measured with a microplate reader (Thermo Fisher, Waltham, MA, USA) to analyze the number of proliferating cells.
Colony Formation Assay
A549 and HCC827 cells (600 cells/well), transfected with siRNAs, were seeded in 6-well plates. Subsequently, the plates were incubated under standard culture conditions for 14 days and stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 1 h. Finally, cells were imaged under a microscope (Olympus, Tokyo, Japan).
Cell Cycle Analysis
A549 and HCC827 cells were seeded in 6-well plates at 2.0 × 105 cells per well and subjected to various transfections. Thereafter, the cells were harvested and fixed in 75% (v/v) ethanol overnight at −20°C. Subsequently, the cells were resuspended in cold PBS and incubated in the dark for 30 min at room temperature in a buffer containing 25 mg/mL 7-aminoactinomycin (7-AAD Sigma Aldrich, St. Louis, MO, USA) and 40 mg/mL RNase. Subsequently, the cells were analyzed by flow cytometry (FACS Calibur, BD Biosciences, Franklin Lakes, NJ, USA), and the percentage of cells in different phases of the cell cycle was determined.
Transwell Assay
Two days after transfection, A549 and HCC827 cells were plated at a density of 2.5 × 105 in the upper chamber of the transwell plate (8 μm, Corning, Tewksbury, MA, USA) with serum-free RPMI 1640, while culture medium supplemented with 20% fetal bovine serum (FBS) was added to the lower chamber. Cells in the Matrigel (Corning) were fixed with 4% paraformaldehyde, before being stained with 0.1% crystal violet (Beyotime, Shanghai, China), photographed, and counted.
Western Blotting
Total proteins were extracted from human LUAD cells using the RIPA buffer (Beyotime), according to the manufacturer’s protocol. Protein concentration was determined using the BCA kit (Beyotime). Western blotting was performed to determine the expression levels of proteins involved in this study. Primary antibodies specific to N-cadherin and E-cadherin were purchased from Proteintech (Wuhan, China), while anti-vimentin antibody was purchased from Abcam (Cambridge, UK). The secondary antibodies were: 1:2000 goat anti-mouse IgG conjugated with HRP (Beyotime) and 1:2000 goat anti-rabbit IgG conjugated with HRP (Beyotime). The protein bands were imaged using an electrochemiluminescence (ECL) system, and the grey values were measured using by ImageJ software to evaluate relative protein levels normalized to β-actin expression level.
Statistical Analysis
NUF2 levels between LUAD and normal groups were compared using Student’s t-test. Pearson’s chi-square test was performed to assess the relationship between NUF2 expression and clinical parameters. The overall survival (OS) and recurrence-free survival (RFS) differences between the low and high gene expression groups were compared by Kaplan–Meier analysis and log-rank test. Cox proportional hazards model was used to determine the independent prognostic factors related to OS or RFS. Prognostic factors screened by the Cox analysis of OS and RFS were incorporated into T stage, N stage, M stage, pathologic stage, primary therapeutic outcome, residual tumors, tumor status, and NUF2 expression to construct the corresponding line graph. NUF2 levels expression between LUAD and benign lung lesions were compared using the t-test function in R. Group differences in the biomarker levels were assessed with Mann–Whitney test or univariate general linear models, adjusted for age and sex as covariates. For all the analyses, results were considered significant at P < 0.05.
Results
Analysis of NUF2 Expression in Databases
Data from TCGA revealed that NUF2 was overexpressed in the LUAD samples relative to normal or para-cancerous samples (P < 0.001, Figures 1A, B). Based on GSE116959 and GSE115002 datasets, NUF2 expression was remarkably higher than that in the normal lung specimens (P < 0.001, Figures 1C, D). We integrated normal tissue data into GTEx data. As shown in Figure 1E, NUF2 mRNA levels also increased significantly in LUAD compared to those in their corresponding normal controls. Furthermore, we used UALCAN and ONCOMINE database to validate the expression of the NUF2 protein; our results validated the upregulation of NUF2 in LUAD samples (P < 0.001, Figures 1F–H) (14, 15).
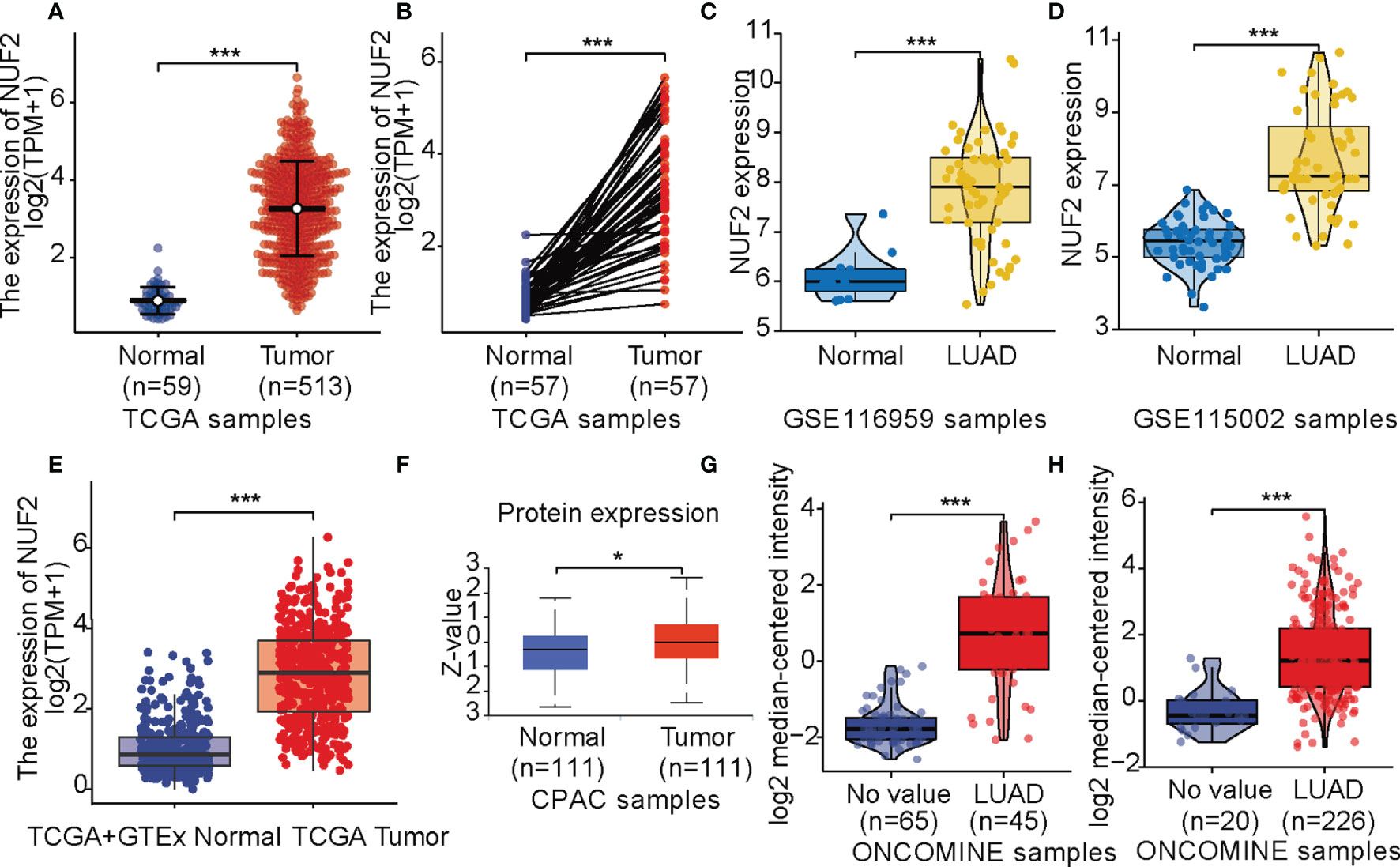
Figure 1 NUF2 expression levels. (A) Expression levels of NUF2 in lung adenocarcinoma (LUAD) and normal tissue from The Cancer Gene Atlas (TCGA). (B) the expression of NUF2 in LUAD and its paired adjacent tissues. (C) mRNA expression of NUF2 in LUAD was obtained from GSE116959. (D) mRNA expression of NUF2 in LUAD was obtained from GSE115002. (E) Expression levels of NUF2 in tumor and normal tissues in TCGA and Genotype-Tissue Expression data. (F) The protein expression of NUF2 in LUAD was obtained from the CPTAC dataset. (G) NUF2 expression in tumor and normal tissues in LUAD from ONCOMINE database (Hou Lung). (H) NUF2 expression in tumor and normal tissues in LUAD from ONCOMINE database (Okayama Lung). ***p < 0.001,*p < 0.05.
NUF2 Validation
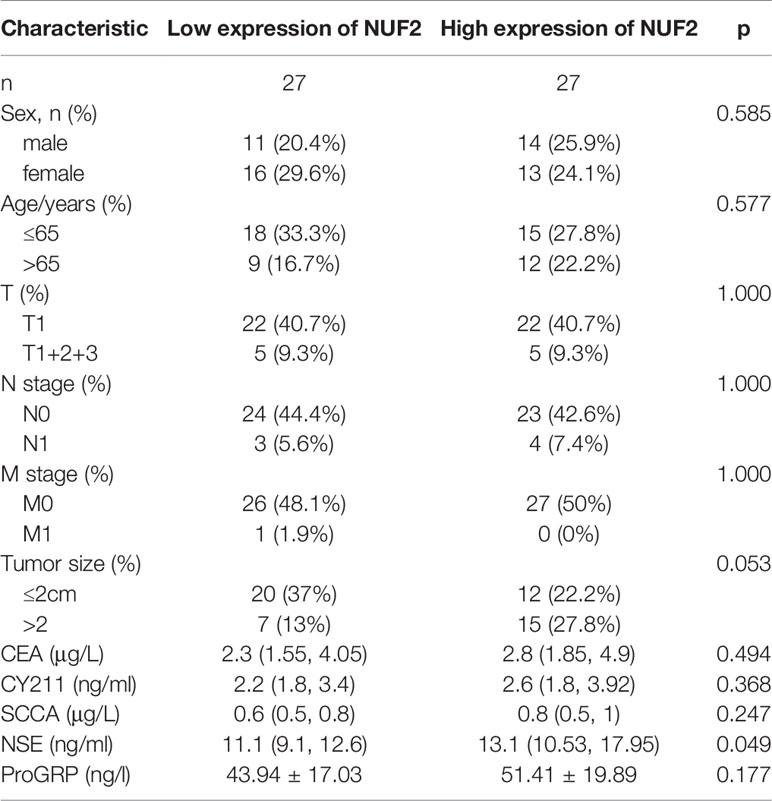
RT-qPCR assay and immunohistochemistry were was performed to assess the expression of NUF2 in LUAD tissues. The expression was elevated in the LUAD samples of our cohort (Figures 2A, B). As shown in Figure 2C, it was demonstrated that the expression of NUF2 in lung cancer tissues was significantly higher than that in the corresponding paired para-tumor tissues, and the subcellular localization of NUF2 protein was in the nucleus and cytoplasm. Furthermore, NUF2 was found to be overexpressed in LUAD cell lines (Figure 2D). As shown in Table 1, the serum concentrations of neuron specific enolase (NSE) in the group expressing high levels of NUF2 were higher than those in the low NUF2 expression group.
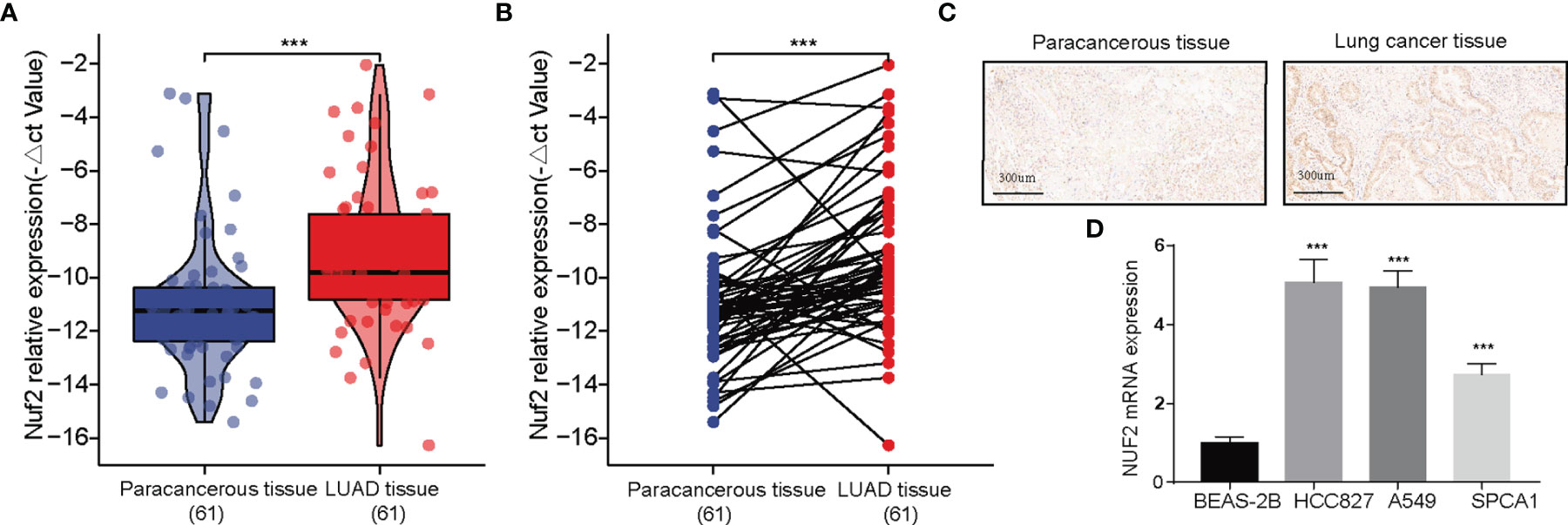
Figure 2 Expression analysis of NUF2 in lung adenocarcinoma (LUAD). (A, B) Qualitative real-time PCR analysis of NUF2 in tumor and adjacent samples from our recruited cohort. (C) Immunohistochemical (IHC) staining sections for NUF2 of lung cancer tissues and cancer-adjacent normal lung tissues. (D) Differential expression of NUF2 in in LUAD cell lines and normal lung epithelial cell line 2B. ***p < 0.001.
Significant Association of NUF2 With Clinicopathological Features
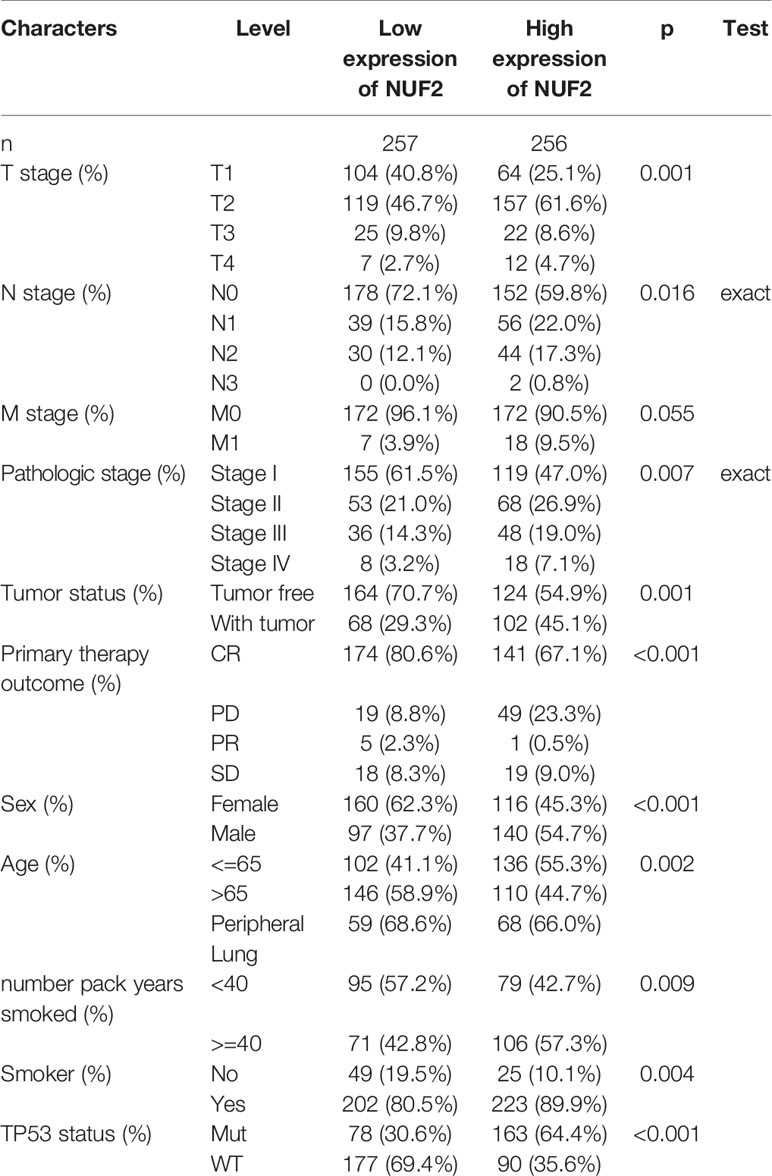
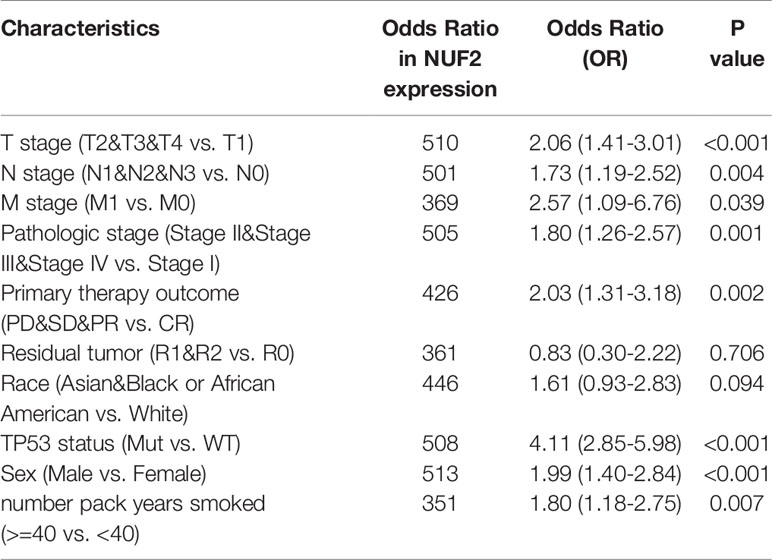
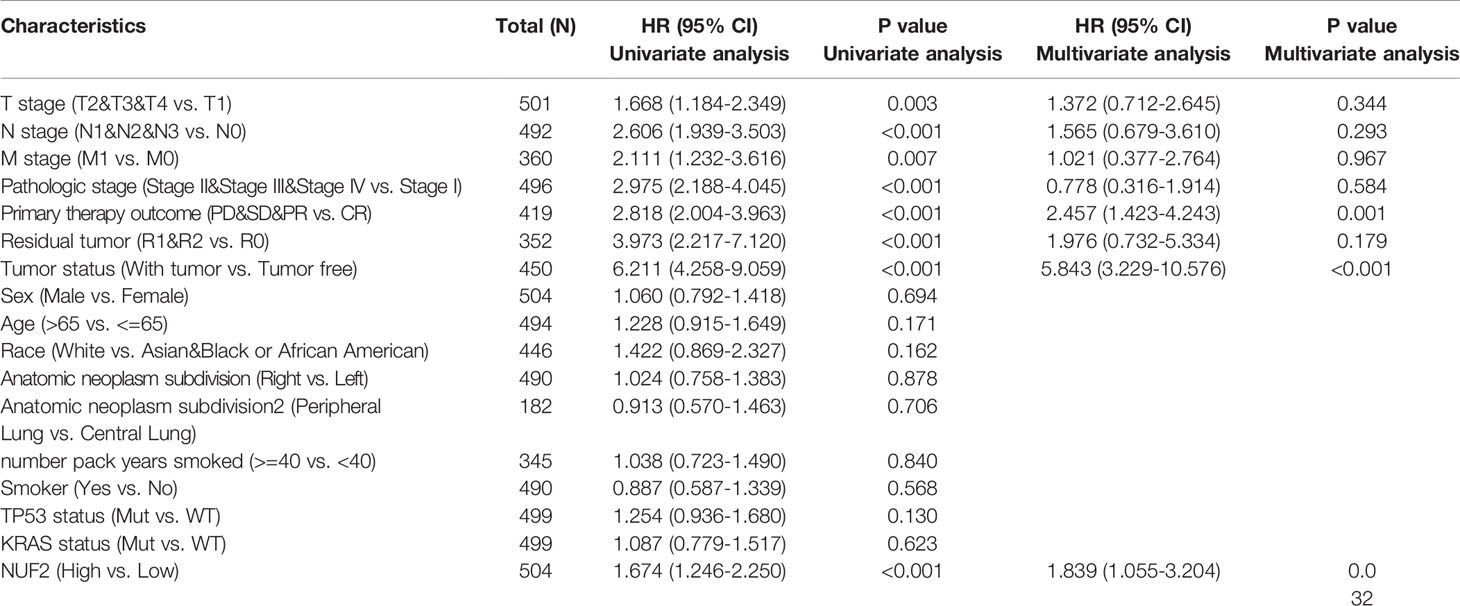
The correlations between NUF2 overexpression and its clinical relevance and prognostic value were analyzed by collating information from the LUAD-TCGA database. NUF2 overexpression was significantly and positively correlated with a higher T stage (T2 Stage vs. T1 Stage, P < 0.001; T4 Stage vs. T1 Stage, P = 0.026), N stage (N1 Stage vs. N0 Stage, P = 0.048; T2 Stage vs. N0 Stage, P = 0.048), M stage (M1 Stage vs. N0 Stage, P = 0.035), pathological stage (Stage II vs. Stage I, P = 0.026; Stage III vs. Stage I, P = 0.017; Stage IV vs. Stage I, P = 0.018), primary therapeutic outcome (PD vs. CR, P < 0.001), sex (Male vs. Female, P < 0.001), smoking status (Yes vs. No, P < 0.001), and TP53 status (Mut vs. WT, P < 0.001) (Table 2 and Figures 3A–H). Logistic regression analysis showed that NUF2 overexpression was observably positively correlated with multiple factors, such as T stage (P < 0.001), N stage (P = 0.004), M stage (P = 0.039), pathologic stage (P = 0.001), and primary therapy outcome (P = 0.002), TP53 status (P < 0.001), Sex (P < 0.001), number of pack years smoked (P = 0.007) (Table 3). As shown in Figures 4A–C, survival analysis indicated that NUF2 overexpression led to a significant reduction in OS (HR = 1.67, P < 0.001), PFI (HR = 1.66, P < 0.001), and DSS (HR = 2.12, P < 0.001). In addition, area under the curve (AUC) was 0.98, providing evidence of the favorable diagnostic ability of NUF2 for LUAD (Figure 4D). Univariate Cox analysis revealed NUF2 (HR = 1.674, P < 0.001) as a high-risk factor for LUAD (Table 3); meanwhile, multivariate Cox analysis highlighted that NUF2 expression (HR = 1.839, P = 0.032) was independently related to OS (Table 4). In addition, based on the above results, we established a clinical nomogram for overall survival by fitting the expression of NUF2 and other clinical parameters. (Figures 4E, F) includes the calibration curves of our nomogram; plots were very close to the ideal line, which indicated the high predictive accuracy.

Figure 3 Correlation of NUF2 expression of and clinicopathologic variables. (A) T stage, (B) N stage, (C) M stage. (D) pathologic stage. (E) primary therapy outcome, (F) sex, (G) smoking. (H) TP53 status.
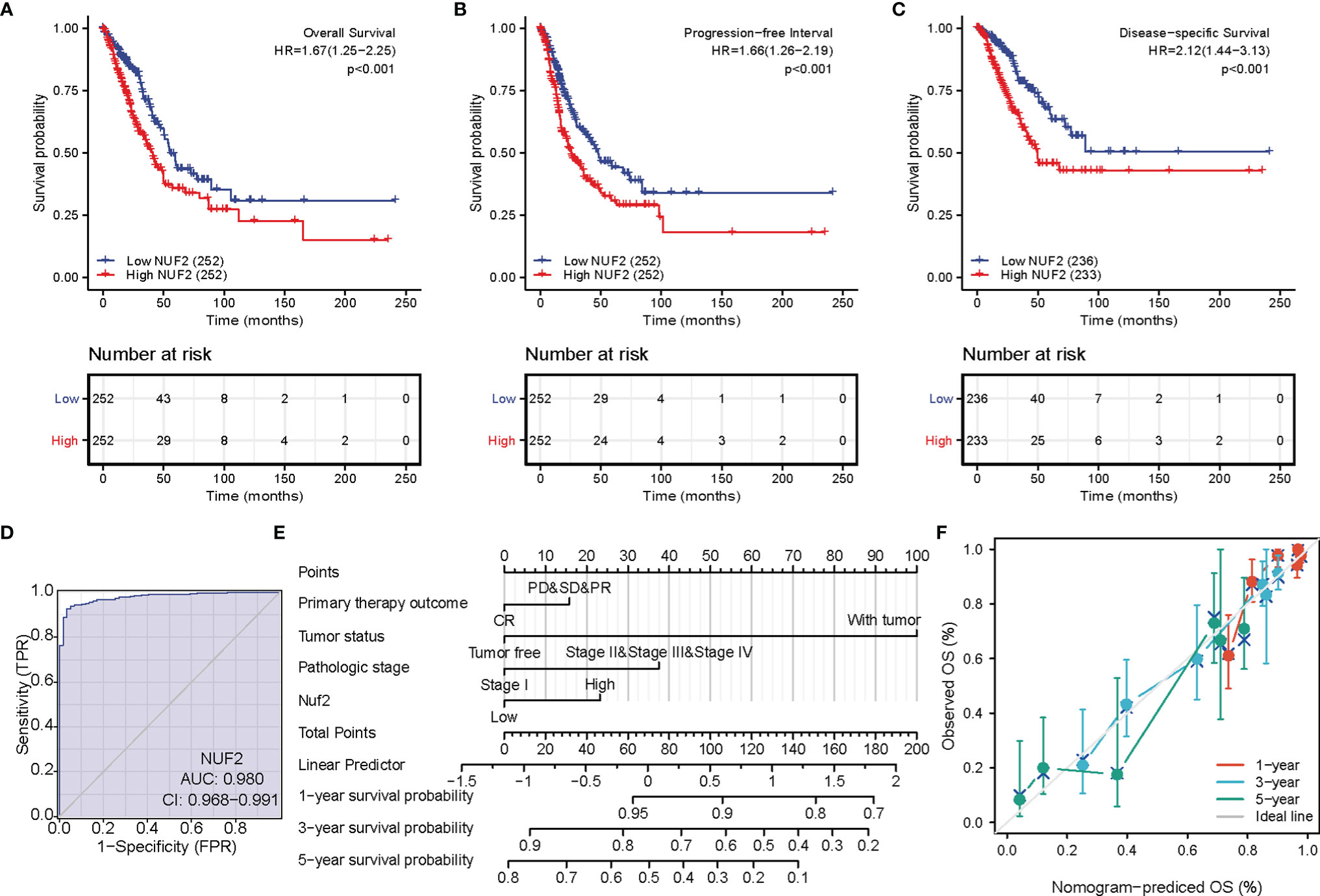
Figure 4 Prognostic value of NUF2 expression in lung adenocarcinoma (LUAD). (A) Survival curves of overall survival. (B) Survival curves of progression-free interval. (C) Survival curves of disease-specific survival. (D) Receiver operating characteristic (ROC) curves of NUF2 in LUAD. (E) Prognostic nomogram that integrated NUF2 expression and other prognostic factors for OS in LUAD from The Cancer Genome Atlas data. (F) A calibration curve at 1-, 3- and 5-year.
Interaction Network of NUF2
Next, we investigated the associations between NUF2 and known and predicted proteins. As shown in Figure 5A, the top 10 predicted partners with their scores were: BUB1 (0.994), CASC5 (0.988), CENPE (0.988), DSN1 (0.992), KIF11 (0.987), MIS12 (0.983), NDC80 (0.999), SPC24 (0.999), SPC25(0.998), and TTK (0.991).
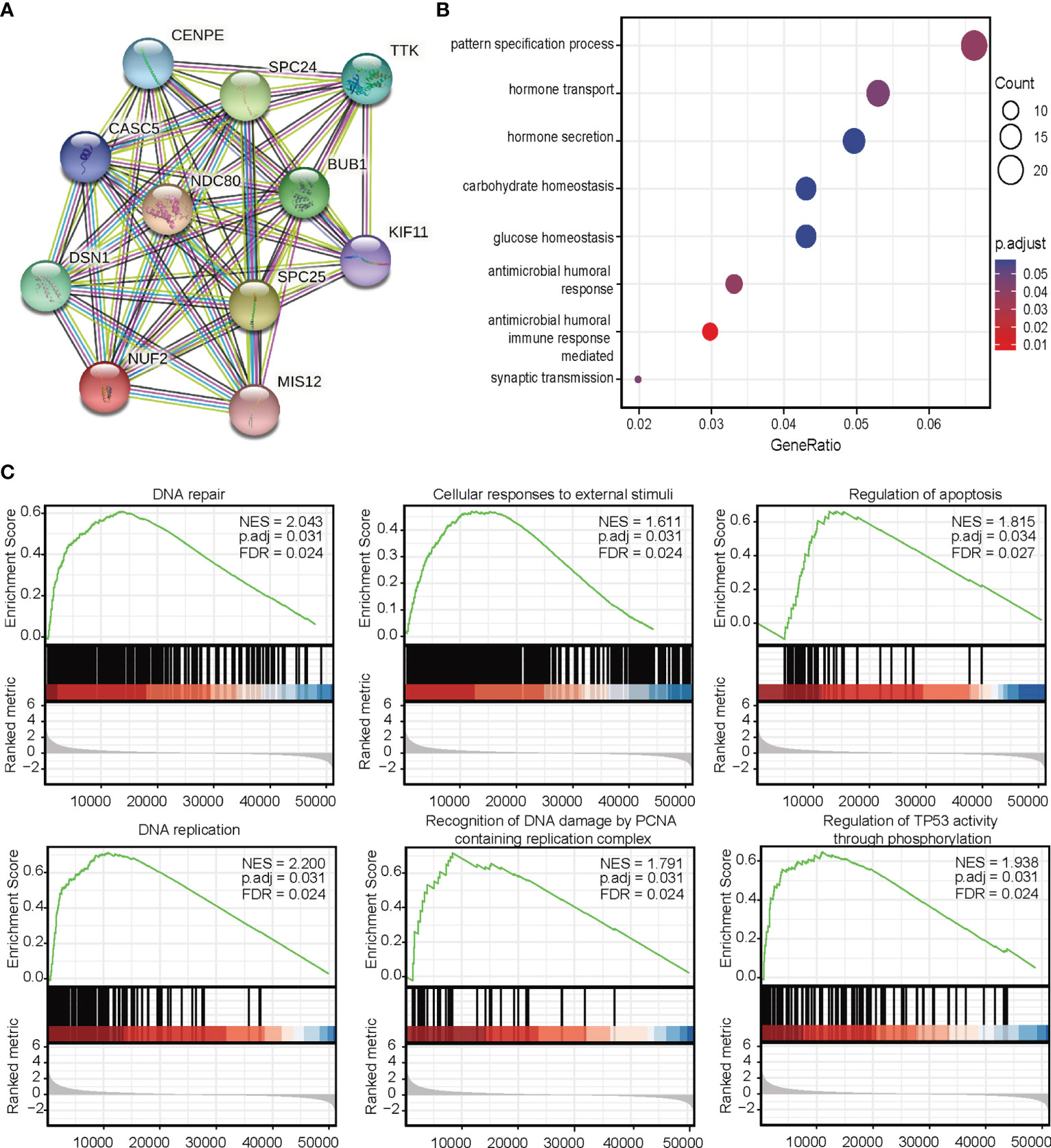
Figure 5 (A) Protein-protein interaction network analysis of NUF2-related genes. (B) Gene Ontology analysis. (C) Gene set expression analysis.
Enrichment Analyses
To predict the function of NUF2, we performed a GO analysis. Functional enrichment analysis of genes in this network showed that they were enriched for immune response (Figure 5B). GSEA indicated an enrichment in DNA repair, cellular responses to external stimuli, regulation of apoptosis, DNA replication, recognition of DNA damage by the PCNA containing replication complex, and regulation of TP53 activity through phosphorylation pathways (Figure 5C).
Relevance of NUF2 Expression in Immune Infiltration
Correlations between NUF2 overexpression and immune cell infiltration level in LUAD tissues were evaluated using the ssGSEA database. The analysis revealed remarkable positive correlation of NUF2 overexpression with Th2 cells, T gamma delta (Tgd), NK CD56dim cells, and T helper cells in LUAD (Figure 6).

Figure 6 Association between the expression level of NUF2 and immune infiltration in the tumor microenvironment. (A) The forest plot shows the correlation between NUF2 expression level and 24 immune cells. (B) the enrichment scores of NUF2 expression in Th2 cells and macrophages; (C) Enrichment scores of NUF2 expression in Tgd cells; (D) Enrichment scores of NUF2 expression in NK CD56dim cells. (E) Enrichment scores of NUF2 expression in T helper cells.
NUF2 Promotes Cell Proliferation, Invasion, and Migration in LUAD
To verify the effect of NUF2 on the biological function of LUAD cells, siRNA targeting NUF2 (si- NUF2) was employed to knock down the expression of NUF2 in HCC827 and A549 cell lines. RT-qPCR results indicated that si-NUF2 effectively inhibited NUF2 expression in LUAD cell lines (Figure 7A). As shown in Figures 7B–E, downregulation of NUF2 expression significantly inhibited cell proliferation. Furthermore, it dramatically reduced the invasion and migration capacities of LUAD cells (Figures 7F, G). Epithelial-mesenchymal transition (EMT) is characterized by the upregulated expression of N-cadherin followed by the downregulated expression of E-cadherin. NUF2 silencing increased E-cadherin expression and decreased that of N-cadherin (Figures 7H, I). These findings indicated that NUF2 may promote EMT. Cell cycle analysis revealed that NUF2 induced cell cycle arrest at G0/G1 (Figures 7J, K). Collectively, these findings suggested that NUF2 may drive tumor progression and act as an oncogene.
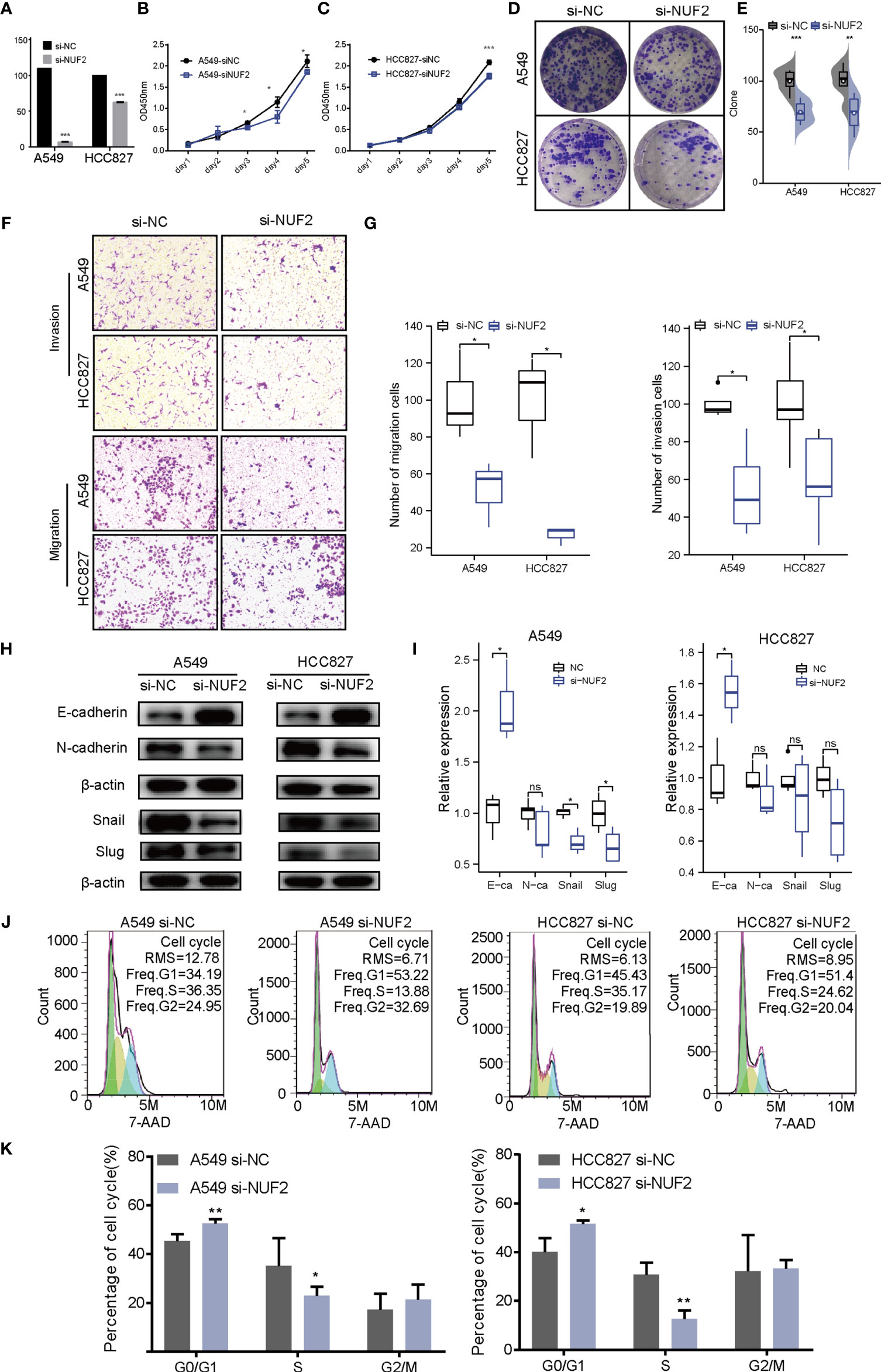
Figure 7 Biological function of NUF2 in lung adenocarcinoma (LUAD) cells. (A) Reverse transcription-quantitative polymerase chain reaction analysis of NUF2 in A549 and HCC827 cells transfected with si-NC or si-NUF2. (B, C) Cell proliferation rates as determined by CCK8 assays in A549 and HCC827 cells transfected with control si-NC or si-NUF2. (D, E) Colony formation assay for evaluating the clonogenic ability of A549 and HCC827 cells. (F, G) The invasive ability of A549 and HCC827 cells was detected by transwell assay. (H, I) Expression of epithelial-mesenchymal transition markers in NUF2-silenced samples as measured using western blotting assays. (J, K) Effect of NUF2 knockdown on cell cycle progression. ***p < 0.001,**p < 0.01,*p < 0.05.
Discussion
Molecular biomarkers can guide the diagnosis, prognosis, and treatment of patients with LUAD. In combination with other genes, NUF2 has been reported to be a biomarker of LUAD, which is consistent with our results. NUF2 overexpression can increase the proliferative ability of liver (16), pancreatic (17), and breast cancer cells (18, 19). Previous studies have analyzed NUF2 in lung adenocarcinoma using multiple omics methods, verifying that NUF2 mRNA is highly expressed in lung adenocarcinoma cell lines. However, the potential biomarkers and important functional genes have not been tested in clinical cohorts and cell function experiments in LUAD. In this study, we performed a more systematic clinical correlation analysis of NUF2. Based on the results of univariate and multivariate Cox analyses, NUF2 was indicated as a high-risk factor for LUAD development, and it could serve as an independent indicator to predict the clinical outcomes of patients with LUAD. Chen et al. analyzed by GO, KEGG, and GSEA enrichment and found that NUF2 is significantly enriched in the cell cycle, especially during DNA replication (8). Previous studies have also found that knockdown of NUF2 induces cell cycle arrest in pancreatic cancer and breast cancer (17, 18). However, in our research, we found that NUF2 has a minimal effect on cell cycle. The differential effect of NUF2 on the cycle may indicate that the NUF2 gene has various effects on epigenetic regulation in different cancer types. Notably, downregulated NUF2 expression was associated with decreased cell proliferation. In addition, the results demonstrated NUF2 to be involved in regulating EMT, which has not been previously reported. Collectively, these results provide direct evidence that NUF2 acts as an oncogene in LUAD.
GSEA results showed that NUF2 is involved in proliferation and apoptosis-related pathways, including proliferating cell nuclear antigen (PCNA) and TP53 pathways. PCNA is related to tumor growth rate; therefore, PCNA expression is used as an important proliferative marker (20). TP53 act as a tumor suppressor gene by regulating apoptosis and the cell cycle as well as mediating DNA damage repair (21). Therefore, we hypothesized that the functions of NUF2 in tumorigenesis are mediated by the PCNA and P53 pathways. By establishing a PPI network, we further identified that these genes play a key role in cell cycle-related meiosis. The PPI was positively associated with top 10 NUF2 co-expressed genes (BUB1, CASC5, CENPE, DSN1, KIF11, MIS12, NDC80, SPC24, SPC25, and TTK). Chen et al. had reported that the increased expression levels of BUB1B and BUB1 was related to the OS in patients with LUAD (22). Incidentally, CENPE encodes centromere-associated protein E, which is a human kinetochore protein that promotes lung adenocarcinoma proliferation (23).
Immune cells present in the tumor-microenvironment play a key role in tumor tissues, with increasing evidence supporting their clinicopathological significance in predicting the survival status of and therapeutic efficacy in cancer patients (24, 25). According to a recent report, various immune cells subtypes, such as Th2, Tgd, NK CD56dim cells, and T helper cells, are vital components of the tumor microenvironment (26). Moreover, studies have reported that Th2 cell infiltration correlates with reduced survival in patients with pancreatic cancer and clear cell renal cell carcinoma. Furthermore, in our study, the Th2 cell infiltration level was significantly higher in the high NUF2 expression group; therefore, we hypothesized that the infiltration level of Th2 accelerates the progression of LUAD.
Despite presenting some credible data, our study did have some limitations. Firstly, it had some inherent limitations due to its retrospective design and small sample size. To further confirm our results, a large-scale prospective study would be required. Secondly, although the report indicated the biological effects related to EMT, the study failed to explore the underlying mechanism of the signaling pathways involving NUF2. Thus, further studies are required to investigate the mechanism responsible for regulating NUF2 expression and its role in LUAD.
In conclusion, to the best of our knowledge, this is the first comprehensive analysis of the expression pattern and clinical significance of NUF2 in LUAD. Our results revealed that NUF2 is overexpressed in LUAD compared to neighboring tissues, and that NUF2 expression is an independent prognostic factor related to OS. Overall, the study provides new evidence of NUF2 being closely linked with the development and progression of LUAD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by This study was approved by the Institutional Ethical Review Committee of the First Affiliated Hospital of Wenzhou Medical University (YS2018001). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
FJ conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft. XH and XY performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft. HZ analyzed the data, prepared figures and/or tables. YW conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft. All authors contributed to the article and approved the submitted version.
Funding
Zhejiang Provincial Research Center for Cancer Intelligent Diagnosis and Molecular Technology (JBZX-202003). Wenzhou Municipal Science and Technology Bureau of China (Y20180113). Zhejiang Provincial Natural Science Foundation (LQ22H200005). Scientific Research Incubation Project of The First Affiliated Hospital of Wenzhou Medical University (FHY2019084).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our gratitude to all the members who participated in discussion and assisted in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.795971/full#supplementary-material
References
1. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell Death-Based Treatment of Lung Adenocarcinoma. Cell Death Dis (2018) 9(2):117. doi: 10.1038/s41419-017-0063-y
2. Dong M, Yang Z, Li X, Zhang Z, Yin A. Screening of Methylation Gene Sites as Prognostic Signature in Lung Adenocarcinoma. Yonsei Med J (2020) 61(12):1013–23. doi: 10.3349/ymj.2020.61.12.1013
3. Jiang X, Jiang Y, Luo S, Sekar K, Koh CKT, Deivasigamani A, et al. Correlation of NUF2 Over-Expression With Poorer Patient Survival in Multiple Cancers. Cancer Res Treat (2021) 53(4):944–61. doi: 10.4143/crt.2020.466
4. Xu W, Wang Y, Wang Y, Lv S, Xu X, Dong X. Screening of Differentially Expressed Genes and Identification of NUF2 as a Prognostic Marker in Breast Cancer. Int J Mol Med (2019) 44(2):390–404. doi: 10.3892/ijmm.2019.4239
5. Wang Y, Tan PY, Handoko YA, Sekar K, Shi M, Xie C, et al. NUF2 is a Valuable Prognostic Biomarker to Predict Early Recurrence of Hepatocellular Carcinoma After Surgical Resection. Int J Cancer (2019) 145(3):662–70. doi: 10.1002/ijc.32134
6. Thang PM, Takano A, Yoshitake Y, Shinohara M, Murakami Y, Daigo Y. Cell Division Cycle Associated 1 as a Novel Prognostic Biomarker and Therapeutic Target for Oral Cancer. Int J Oncol (2016) 49(4):1385–93. doi: 10.3892/ijo.2016.3649
7. Sun ZY, Wang W, Gao H, Chen QF. Potential Therapeutic Targets of the Nuclear Division Cycle 80 (NDC80) Complexes Genes in Lung Adenocarcinoma. J Cancer (2020) 11(10):2921–34. doi: 10.7150/jca.41834
8. Chen M, Li S, Liang Y, Zhang Y, Luo D, Wang W. Integrative Multi-Omics Analysis of Identified NUF2 as a Candidate Oncogene Correlates With Poor Prognosis and Immune Infiltration in Non-Small Cell Lung Cancer. Front Oncol (2021) 11:656509. doi: 10.3389/fonc.2021.656509
9. Theelen W, Peulen HMU, Lalezari F, van der Noort V, de Vries JF, Aerts J, et al. Effect of Pembrolizumab After Stereotactic Body Radiotherapy vs Pembrolizumab Alone on Tumor Response in Patients With Advanced Non-Small Cell Lung Cancer: Results of the PEMBRO-RT Phase 2 Randomized Clinical Trial. JAMA Oncol (2019) 5(9):1276–82. doi: 10.1001/jamaoncol.2019.1478
10. Bang A, Schoenfeld JD, Sun AY. PACIFIC: Shifting Tides in the Treatment of Locally Advanced non-Small Cell Lung Cancer. Transl Lung Cancer Res (2019) 8(Suppl 2):S139–s146. doi: 10.21037/tlcr.2019.09.04
11. Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, et al. Comprehensive Genomic and Immunological Characterization of Chinese non-Small Cell Lung Cancer Patients. Nat Commun (2019) 10(1):1772. doi: 10.1038/s41467-019-09762-1
12. Lin A, Wei T, Meng H, Luo P, Zhang J. Role of the Dynamic Tumor Microenvironment in Controversies Regarding Immune Checkpoint Inhibitors for the Treatment of Non-Small Cell Lung Cancer (NSCLC) With EGFR Mutations. Mol Cancer (2019) 18(1):139. doi: 10.1186/s12943-019-1062-7
13. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING V10: Protein-Protein Interaction Networks, Integrated Over the Tree of Life. Nucleic Acids Res (2015) 43:D447–52. doi: 10.1093/nar/gku1003
14. Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene Expression-Based Classification of Non-Small Cell Lung Carcinomas and Survival Prediction. PloS One (2010) 5(4):e10312. doi: 10.1371/journal.pone.0010312
15. Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-Negative Lung Adenocarcinomas. Cancer Res (2012) 72(1):100–11. doi: 10.1158/0008-5472.can-11-1403
16. Xie X, Jiang S, Li X. Nuf2 Is a Prognostic-Related Biomarker and Correlated With Immune Infiltrates in Hepatocellular Carcinoma. Front Oncol (2021) 11:621373. doi: 10.3389/fonc.2021.621373
17. Hu P, Chen X, Sun J, Bie P, Zhang LD. siRNA-Mediated Knockdown Against NUF2 Suppresses Pancreatic Cancer Proliferation In Vitro and In Vivo. Biosci Rep (2015) 35(1):e00170. doi: 10.1042/bsr20140124
18. Lv S, Xu W, Zhang Y, Zhang J, Dong X. NUF2 as an Anticancer Therapeutic Target and Prognostic Factor in Breast Cancer. Int J Oncol (2020) 57(6):1358–67. doi: 10.3892/ijo.2020.5141
19. Zhai X, Yang Z, Liu X, Dong Z, Zhou D. Identification of NUF2 and FAM83D as Potential Biomarkers in Triple-Negative Breast Cancer. PeerJ (2020) 8:e9975. doi: 10.7717/peerj.9975
20. Zheng W, Xu S. Analysis of Differential Expression Proteins of Paclitaxel-Treated Lung Adenocarcinoma Cell A549 Using Tandem Mass Tag-Based Quantitative Proteomics. Onco Targets Ther (2020) 13:10297–313. doi: 10.2147/ott.s259895
21. Burke MJ, Kostadinov R, Sposto R, Gore L, Kelley SM, Rabik C, et al. Decitabine and Vorinostat With Chemotherapy in Relapsed Pediatric Acute Lymphoblastic Leukemia: A TACL Pilot Study. Clin Cancer Res (2020) 26(10):2297–307. doi: 10.1158/1078-0432.ccr-19-1251
22. Chen C, Guo Q, Song Y, Xu G, Liu L. SKA1/2/3 Serves as a Biomarker for Poor Prognosis in Human Lung Adenocarcinoma. Transl Lung Cancer Res (2020) 9(2):218–31. doi: 10.21037/tlcr.2020.01.20
23. Shan L, Zhao M, Lu Y, Ning H, Yang S, Song Y, et al. CENPE Promotes Lung Adenocarcinoma Proliferation and is Directly Regulated by FOXM1. Int J Oncol (2019) 55(1):257–66. doi: 10.3892/ijo.2019.4805
24. Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity (2019) 51(1):27–41. doi: 10.1016/j.immuni.2019.06.025
25. Xu F, Zhan X, Zheng X, Xu H, Li Y, Huang X, et al. A Signature of Immune-Related Gene Pairs Predicts Oncologic Outcomes and Response to Immunotherapy in Lung Adenocarcinoma. Genomics (2020) 112(6):4675–83. doi: 10.1016/j.ygeno.2020.08.014
Keywords: NUF2, prognosis, immune infiltration, lung adenocarcinoma, survival
Citation: Jiang F, Huang X, Yang X, Zhou H and Wang Y (2022) NUF2 Expression Promotes Lung Adenocarcinoma Progression and Is Associated With Poor Prognosis. Front. Oncol. 12:795971. doi: 10.3389/fonc.2022.795971
Received: 15 October 2021; Accepted: 17 May 2022;
Published: 23 June 2022.
Edited by:
Matiullah Khan, AIMST University, MalaysiaReviewed by:
Kamini Singh, Albert Einstein College of Medicine, United StatesYunchao Huang, Yunnan Cancer Hospital, China
Copyright © 2022 Jiang, Huang, Yang, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumin Wang, d2FuZ3l1bWluMDU3N0B3bXUuZWR1LmNu
†These authors have contributed equally to this work
 Feng Jiang
Feng Jiang Xiaolu Huang1,2†
Xiaolu Huang1,2† Huixin Zhou
Huixin Zhou Yumin Wang
Yumin Wang