
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 24 February 2022
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.790916
This article is part of the Research TopicExtracellular Vesicle-derived Non-coding RNAs (EV-ncRNAs) and Their Multifaceted Roles in Cancer BiologyView all 7 articles
Hepatocellular carcinoma (HCC) is the sixth highest-incidence cancer and the 4th most deadly cancer all over the world, with a high fatality and low diagnostic rate. Nowadays, Excessive alcohol consumption, type-2 diabetes, smoking and obesity have become some primary risk factors of HCC. As intercellular messenger transporting information cargoes between cells, exosomes are a type of extracellular vesicles (EVs) released by most types of cells including tumor cells and non-tumor cells and play a pivotal role in establishing an HCC microenvironment. Exosomes, and more generally EVs, contain different molecules, including messenger RNAs (mRNAs), non-coding RNAs (ncRNAs), proteins, lipids and transcription factors. The three main ncRNAs in exosomes are microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs). NcRNAs, identified as essential components, are selectively sorted into exosomes and exosomal ncRNAs show great potential in regulating tumor development, including proliferation, invasion, angiogenesis, metastasis, immune escape and drug resistance. Here, we chiefly review the formation and uptake of exosomes, classification of exosomal ncRNAs and current research on the roles of exosomal ncRNAs in HCC progression. We also explored their clinical applications as new diagnostic biomarkers and therapeutic avenues in HCC.
As a major adult liver malignancy, Hepatocellular carcinoma (HCC) accounts for 75-85% of all primary liver cancers with easy metastasis, poor prognosis and low survival rate (1). Hepatitis B virus (HBV) infection is the main reason of liver cancer in China, while hepatitis C virus (HCV) infection is more frequent in Western countries (2). Other etiologies including type-2 diabetes, smoking and obesity, alcoholic liver disease, non-alcoholic fatty liver disease (NAFLD), non-alcoholic steatohepatitis (NASH) can result in HCC (3–5). HCC occurs largely on the basis of liver cirrhosis (LC) (4, 6, 7). In the past few years, accumulating evidence has demonstrated the regulatory roles of diverse categories of non-coding RNAs (ncRNAs) in liver carcinogenesis associated with many etiologies, including HBV, HCV and NAFLD. For instance, HCV infection could suppress miR-122 and miR-122 targets to induce liver carcinogenesis. Dysregulated expression of miRNA has been connected with NAFLD-associated HCC development (8). In China, 5-year survival rates of HCC have been reported to be as low as 12% (9), mainly ascribed to its insidious onset, lack of obvious symptoms in the early stage, rapid progression and difficulty of early effective diagnosis through current non-invasive clinical screening technology like serum alpha-fetoprotein (AFP) and imaging examination (10). As a result, when diagnosed, most patients with HCC are at an advanced stage. They usually accompany with multiple intrahepatic or distant metastasis and miss the opportunity of radical treatments such as surgical resection or orthotopic liver transplantation. Therapeutic strategies for liver cancer include radical surgical resection, radiofrequency ablation (RFA), systemic chemotherapy, liver transplantation and transcatheter arterial chemoembolization (TACE). However, due to poor liver function, current treatment choices for advanced HCC are limited to palliative treatments or strive for second-stage surgery through chemotherapy, interventional therapy and targeted therapy (11). Sorafenib was first identified as an oral multitargeted tyrosine kinase inhibitor drug for systemic chemotherapy in advanced HCC patients with metastasis and can prominently extend the median survival of patients. However, drug resistance gradually emerges during the later stage of treatment, which become a major difficulty in the treatment of liver cancer (12). Therefore, it is an urgent need to improve the early diagnostic rate and explore effective treatment for HCC.
EVs can be divided into two main types: exosomes and ectosomes or microvesicles, on the basis of their differential modes of biogenesis and size (13, 14). Exosomes correspond to a series of nano-extracellular vesicles with a phospholipid bilayer membrane (15, 16) with 30–100 nm in diameter and 1.13–1.19 g/mL in density (16–19). Microvesicles or ectosomes, in comparison to exosomes, range in size from exosome-like EVs of 50nm to EVs as large as 10μm in diameter (13). Observed by transmission electron microscopy (TEM), exosomes present distinctive cup-shaped or disc-shaped morphology (20). Exosomes are distributed in the majority of body fluids, including blood, urine, breast milk, saliva, tears, cerebrospinal fluid, bronchial lavage fluid, amniotic fluid and synovial fluid (21–23). These nanostructures usually contain proteins, lipid molecules, nucleic acids and other inorganic substances such as Ca2+ (16, 20). Nucleic acids in exosomes mainly comprise messenger RNAs (mRNAs), long noncoding RNAs (lncRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs/miRs), transfer RNAs (tRNAs), circular RNAs (circRNAs), cell free DNAs (cfDNAs) and mitochondrial DNAs (mtDNAs) (24). Many types of cells may induce the release of exosomes such as tumor cells, endothelial cells, mesenchymal cells, B lymphocytes, T cells, dendritic cells and mast cells (25). In addition, extensive evidence have confirmed that exosomes might play a central part in the initiation, development, diagnosis and therapy of liver cancer (26). Furthermore, the biologically active contents of exosomes have provided novel approaches for the establishment of efficient clinical diagnostics and therapeutic strategies (27). Despite several breakthroughs have been made in exploring the exosomes, further studies are required to fully elucidate the specific biological role of exosomes.
NcRNAs are a series of RNAs unable to encode protein which are divided into lncRNAs (>200 nucleotides) and miRNAs (19–25 nucleotides) according to their length. NcRNAs regulate gene expression post-transcriptionally by translational inhibition or degradation of target mRNA (28, 29). A growing body of studies have demonstrated that exosomal ncRNAs are involved in the potential regulation of tumor metastasis, angiogenesis, immune escape, metabolic reprogramming and drug resistance. Exosomal ncRNAs become endogenous tumor regulators in the body (30), and their dysregulation directly associated with the onset and progression of carcinogenesis (31). More importantly, ncRNAs inside the exosome are stable because the double-layer lipid membrane of exosomes can protect ncRNAs from the ribonuclease-mediated degradation. Thus, the integrity and function of the RNAs are not changed (32). It has been found that there is an obvious ncRNA change in HCC patients’ exosomes that is associated with HCC development, angiogenesis and metastasis in previous studies of ncRNAs (25). This review, we aim to address the biology of exosomes, how exosomal ncRNAs contribute to the development of HCC with a major focus on their potential clinical applications in diagnosis and therapy.
EVs were firstly discovered in mature sheep reticulocytes by Johnstone and coworkers in 1983 and named them as exosomes. They were originally thought of as “fragments” of cells (33). In 1996, It has been confirmed by Raposo and coworkers that exosomes play a crucial role in B cell antigen presentation and cause T cell responses (34). Therefore, exosomes have seen a progressive increase in popularity in recent years and the imperative role of exosomes in tumor diagnosis and treatment has gradually attracted people’s attention. However, the term “exosomes” has been often used in a less restrictive and not univocal manner with respect to the original definition centered on biogenesis (16, 35, 36). This required a great commitment of many scientists and of the International Society for Extracellular Vesicles (ISEV) itself to make order in the nomenclature and identify the minimal information for studies of EVs to avoid irreproducibility in the results (16, 36).
Exosomes are endosome-derived and originate inside multivesicular bodies (MVBs) secreted in physiological as well as pathological conditions (37). Exosomes (Figure 1) are in the form of intraluminal vesicles (ILVs) when early endosomes get maturation into late endosomes by specific signals stimulation (14). Their secretory process contains a series of procedures, start with the development of endocytic vesicles by inward budding of MVBs, then late endosomes moving along microtubules, fusing with the plasma membrane and at last releasing to the extracellular space (38, 39). In recent years, it has been increasingly confirmed that exosomes played a central part in modulating the crosstalk between tumor cells and stromal cells in the tumor microenvironment (TME) and participating in modulating various of biological processes related to cancer, such as cell proliferation, angiogenesis, metastasis and drug resistance (40, 41).
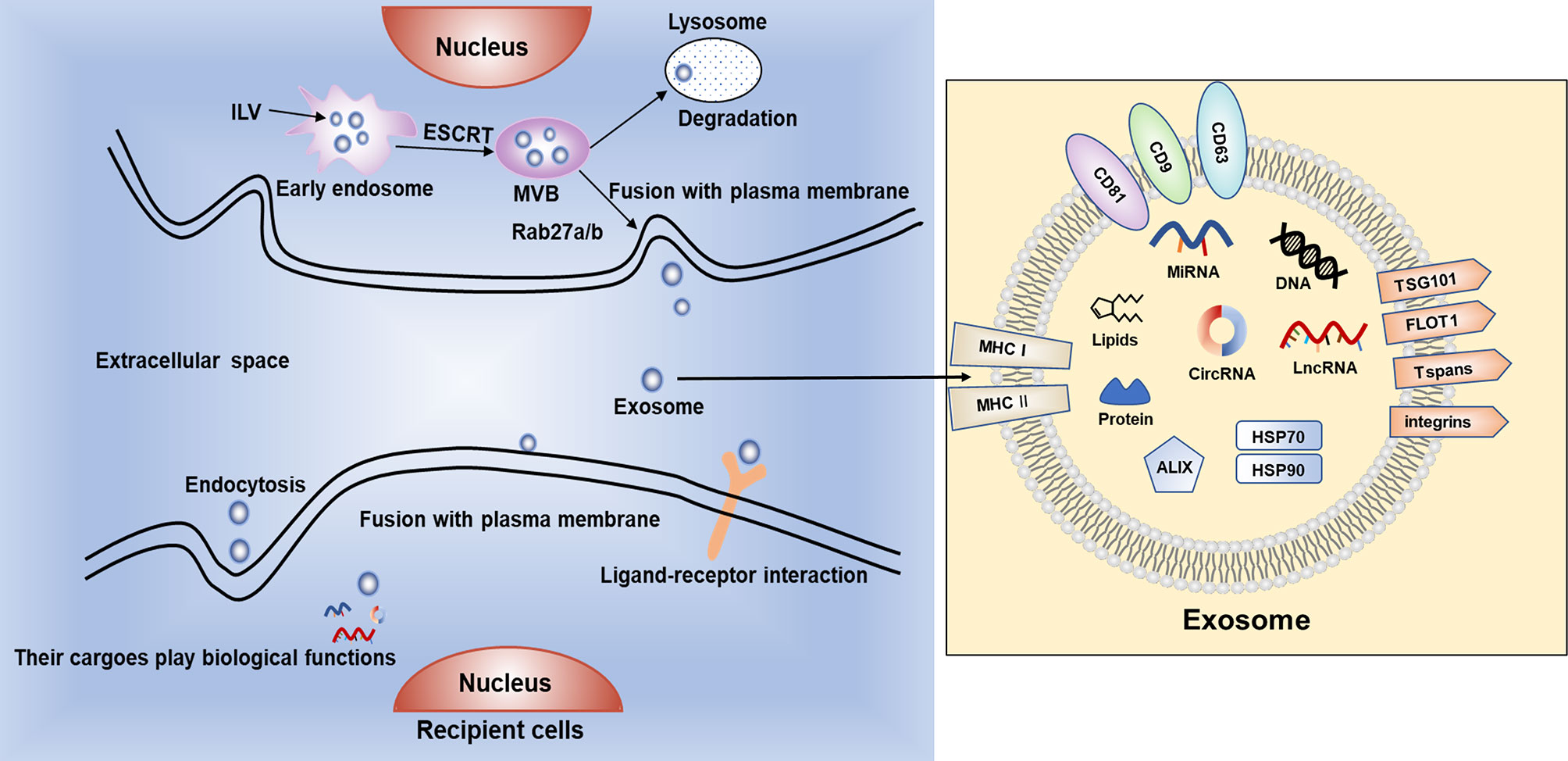
Figure 1 The biogenesis, secretion and uptake of exosomes. Exosomes contain bioactive cargoes including RNAs, DNAs, proteins and lipids. With the help of endosomal sorting complex required for transport (ESCRT), intraluminal vesicles (ILVs) are produced when early endosomes develop to multivesicular bodies (MVBs). Then, MVBs can be degraded by lysosomes or fuse with cell membrane and release exosomes into extracellular space. Exosomes can be ingested by recipient cells by endocytosis or fusing with plasma membrane, or through ligand–receptor interaction to exert various biological functions.
Exosomes are originally endocytic and usually contain proteins associated with endosomes including ALG-2 interacting protein X (ALIX), major histocompatibility complex class I and class II molecules (MHC I/II), Rab GTPases, tumor susceptibility gene 101 protein (TSG101), Annexins, flotillin (FLOT1), integrins and tetraspanins (Tspans) such as CD9, CD63, CD81 (42). The mechanism of exosomal biogenesis is mostly mediated by endosomal sorting complex required for transport (ESCRT). The ESCRT system mainly comprises of 4 core components, named ESCRT 0, I, II and III respectively. ESCRT-0 and ESCRT-I take charge of clustering cargoes and recruiting ubiquitylated transmembrane cargoes. ESCRT-II participates in initiating the process of inward budding, and ESCRT-III is thought to be associated with vesicle scission and membrane budding (43–45). These multiple protein complexes work synergistically with VTA1, VPS4 and ALIX to help protein ubiquitination and classify the proteins as ILVs (46). Additionally, Ca2+ can be used to regulate the fusion process of secretory lysosomes with plasma membrane and increased level of intracellular Ca2+ can induce the secretion of exosomes (47, 48).
Once released in the extracellular environment, exosomes can interact with recipient cells by recognition and conjugation to membrane-bound receptors, thus activating specific signal pathways in target cells. Some of the widely discussed mechanisms for exosome uptake are phagocytosis, macropinocytosis, clathrin-mediated endocytosis (CME), caveolin-dependent endocytosis (CDE), and plasma membrane fusion. The exact mechanism of exosome uptake by recipient cells is yet to be elucidated (49). The cargoes can induce a series of phenotypic changes in recipient cells (50). It has been reported that tumor cell-derived exosomes (TDEs) can alter the behavior of peripheral stromal cells, eventually creating an appropriate microenvironment for tumor growth (51). TDEs also involve in eukaryotic intercellular crosstalk, tumor angiogenesis, treatment resistance and tumor metastasis (52–54). Studies have shown that high levels of exosomes can be released by tumor cells and the ingredients of exosomes change under different pathological and physiological conditions (55, 56).
To data, several conventional techniques have been employed to separate exosomes, including ultracentrifugation-based separation, immunological separation, ultrafiltration separation, size-exclusion chromatography (SEC), and polymer-based precipitation separation (57). Among these techniques, the ultracentrifugation-based separation technique is the most commonly used technique for separating exosomes. However, the ultracentrifugation-based separation technique is not suitable for clinical diagnosis because of the time consuming (>4h), low recovery rate (5–25%) and poor repeatability. With the rapid development of microfluidics, microfluidics-based techniques have received increasing attention. Microfluidics-based techniques for exosome separation rely on the physical and chemical features of exosomes, such as size, density, and surface antigens present (58). However, it is difficult to completely isolate exosomes from blood samples because there are many biomolecules or biological nanoparticlesin plasma/serum that have similar size and/or density to EVs, including lipoproteins, subcellular organelles, cellular debris and viral particles (59). Among them, low/very low/high density lipoproteins (LDL, VLDL, HDL) represent ones of the most common contaminants in EV isolation as there are∼1016 lipoproteins/ml in plasma (60). The separation of exosomes from serum requires successful separation of exosomes from lipoproteins. By optimizing the pore size, increasing bed volume and bulk elution volume of column resin, Cui and coworkers improved SEC and developed dichotomic SEC. This method could isolate exosomes from serum with high purity and particle recovery rate, which is expected to accelerate the clinical research and achievement transformation of EVs (61).
Simultaneously, the conventional detection techniques for analyzing and quantifying exosomes include scanning electron microscopy (SEM), transmission electron microscopy (TEM), atomic force microscopy (AFM), enzyme-linked immunosorbent assay (ELISA), dynamic light scattering (DLS), and nanoparticle tracking analysis (NTA). On the other hand, microfluidics-based exosome detection methods have the advantages of high-throughput capacity, high sensitivity, low reagent consumption and portability, which include fluorescence imaging, colorimetric detection, optical special effects methods, magnetic detection and electro-chemical detection. Microfluidics combined with conventional exosome separation techniques are expected to be a research hotspot in separation and detection of exosomes (58).
Exosomal ncRNAs could be extracted from exosomes by RNA extracting kits or Trizol after separating exosomes and quantified by next-generation sequencing (NGS) (62), microarray (63), real-time quantitative polymerase chain reaction (qPCR) and digital PCR (64).
The first discovery of miRNA was reported by Lee and coauthors in 1993 that small RNA molecules were encoded by gene lin-4 to regulate protein lin-14 expression in Caenorhabditis elegans (65). MiRNAs are small single-stranded RNA molecules without the ability of encoding proteins. They usually have a length of approximately 20–24 nucleotides, which modulate gene expression post-transcriptionally by complementary binding to 3′untranslated regions (UTR) of target mRNA’s and guide degradation of target mRNAs (66–68). MiRNAs may have carcinogenic effects or play the role of tumor suppressors under different conditions (69). Alteration in miRNA expression levels have been recognized as critical factors for many aspects of tumorigenesis and development such as tumor angiogenesis, immune disturbance, metastasis and so on (66–68). MiRNAs in exosomes have been considered as critical biologically active content of exosomes and this has become a hotspot in recent years. Research in recent years has showed that miRNAs in exosomes can influence a lot of aspects of liver cancer physiologically and pathologically. What’s more, exosomal miRNAs can not only reflect aberrant intrahepatic regulation but also can affect other organs and their microenvironment. More importantly, previous studies have revealed that serum miRNAs are enriched in the exosomes and can be protected from degradation by RNase in circulation (70). Taken together, exosomal miRNAs have the potential diagnostic and therapeutic value (Figure 2).
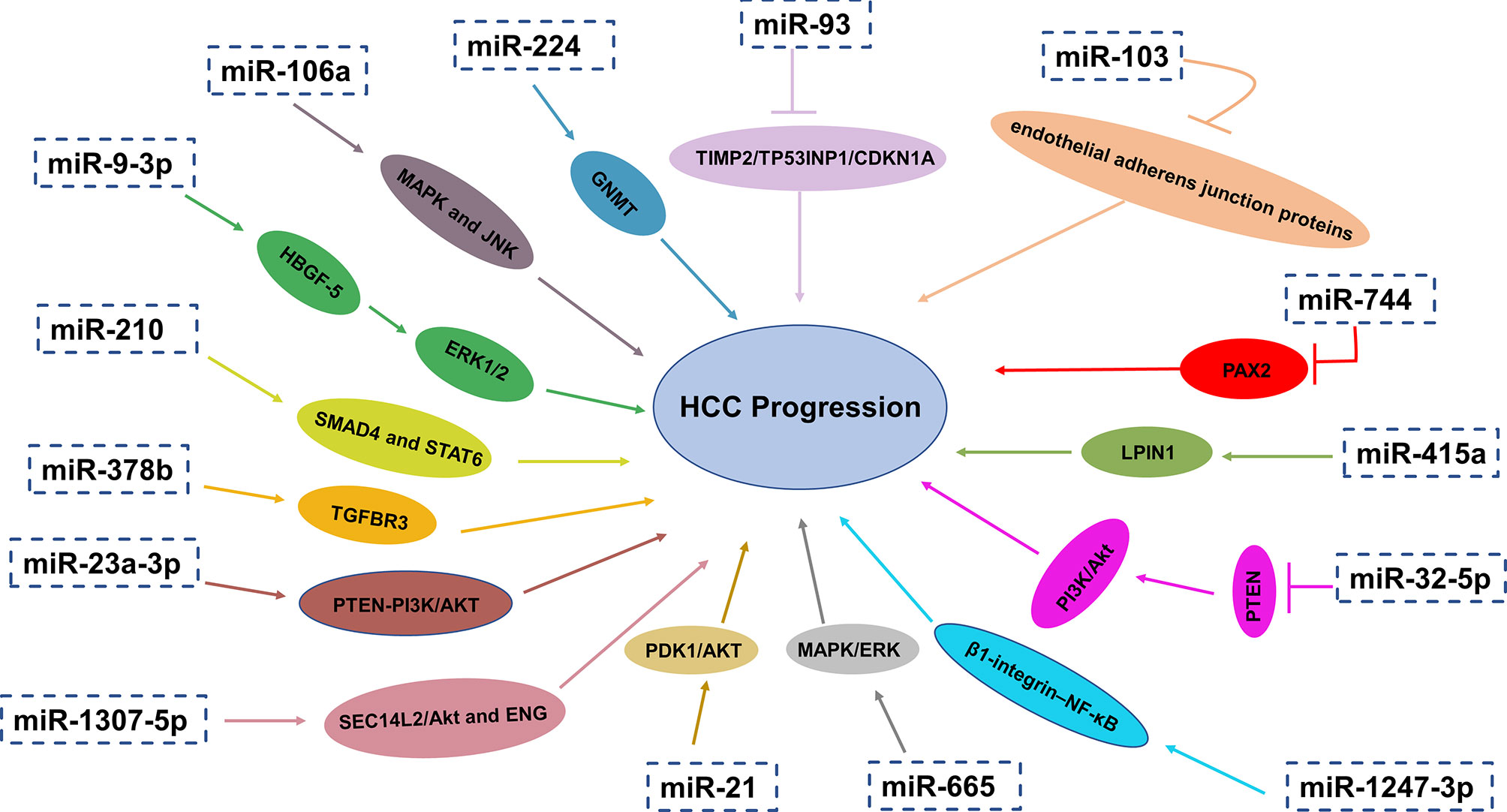
Figure 2 Exosomal miRNAs play a crucial role in HCC progression through different pathways and mechanisms.
Exosomal miR-210 was upregulated in HCC patients and related to microvessel density (MVD). Exosomal miR-210 could promote proliferation, migration and tube formation of endothelial cells in HCC cells via targeting SMAD4 and STAT6 in endothelial cells, which are critical for angiogenesis (71). In addition, high levels of exosomal miR-378b facilitates the angiogenesis and progression of HCC cells, which may target and negatively associated with TGFBR3, offering therapeutic strategies for HCC treatment (72). On the contrary, in the serum exosomes of patients with liver cancer, the expression of miR-451a was remarkably reduced and low level of miR-451a was associated with later clinical stages and high T classification. As a tumor suppressor, exosomal miR-451a targets LPIN1 to suppress hepatocellular tumorigenesis by regulating tumor cell apoptosis and angiogenesis (73).
Recent study by Wang and coauthors demonstrated that the level of exosomal miR-744 in liver cancer tissues and serum related to the propagation and the sorafenib resistance of HCC cells was also remarkably decreased. They also reported that miR-744 directly targeted PAX2. Compared with adjacent normal tissues, PAX2 in HCC tissues increased significantly. Intriguingly, the proliferation and drug resistance of liver cancer cells were dramatically inhibited when treated with miR-744-enriched exosomes (74).
Fu and coauthors have discovered that exosomal miR-32-5p was significantly overexpressed in HCC and neighboring tissues while PTEN was significantly lower-expressed. Furthermore, upregulated expression of miR-32-5p and downregulated expression of PTEN had a positively correlation to unfavorable prognosis of patients with HCC. They also found that miR-32-5p, targeting PTEN, caused multiple drug resistance through the activation of PI3K/Akt pathway and going through epithelial-mesenchymal transition and angiogenesis. However, how miR-32-5p converts sensitive cells to resistant cells is still confusing (75). In another study, exosomal miRNA-21 could convert normal hepatic stellate cells (HSCs) into cancer-associated fibroblasts (CAFs) via directly targeting PTEN, resulting in activating PDK1/AKT signaling pathway in HCC. In addition, by secreting angiogenic cytokines such as VEGF, TGF-β, bFGF, MMP2 and MMP9, activated CAFs further promoted cancer progression. Furthermore, high expression of exosomal miRNA-21 was linked with low survival rate, which has guiding significance for effective prevention and treatment strategies (76).
Epithelial-mesenchymal transition (EMT) is a conservative process in which epithelial cells transition to a mesenchymal cell state, resulting in decreased intercellular adhesion and increased motility. Accumulative evidence indicates that inappropriate activation of EMT is associated with malignant transformation and the initiation of metastasis (77). It has been demonstrated that TDEs carry a pro-EMT programme including hypoxia-inducible factor 1 alpha (HIF1a), transforming growth factor beta (TGFb), caveolin-1 and b-catenin,which enhances the ability of recipient cells to invade and migrate and facilitate matrix remodelling and the formation of premetastatic niche (78).
Pulmonary metastasis is the most common remote invasive progression and one of the leading reasons of deaths related to HCC (78, 79). The study from Fang T confirmed that through reducing the expression of its target B4GALT3, the miR-1247-3p derived from tumor exosomes transform fibroblasts to CAFs. Consequently, CAFs in the lung metastatic niche are activated by activating β1-integrin–NF-κB signaling pathway. Additionally, CAFs promote tumorigenicity, chemoresistance, stemness and EMT of tumor cells by increasing secretion of IL-6 and IL-8. What’s more, upregulation of exosomal miR-1247-3p is positively related with lung metastasis in liver cancer patients (80). Also, according to the research proposed by Fang and coworkers, the level of exosomal miR-103 in HCC cells was upregulated. Exosomal miR-103 could be delivered into endothelial cells to reduce the integrity of endothelial junction and increase the permeability of vessels by targeting zonula occludens-1 (ZO-1), VE-Cadherin and p120-catenin. As a result, the transendothelial infiltration of HCC cells increased and metastasis occurred (81). In addition to the aforementioned, exosomal miR-1307-5p was reported to promote metastasis and help predict metastasis in HCC, which was dramatically overexpressed in the metastasis group (p = 0.04), as well as in the vascular invasion and tumor recurrence groups. SEC14L2/Akt and ENG-related signaling pathways were identified as downstream pathways of miR-1307-5p for promoting the EMT in HCC by comprehensive bioinformatics analysis (82).
To the best of our knowledge, the effective treatment of liver cancer is living donor liver transplantation (LDLT), facing the problem of recurrence after transplantation. According to Nakano and coworkers, elevated levels of AFP (>24.1ng/ml) and exosomal miR-92b (>23.4-fold change) in HCC patients before LDLT is one of risk factors for early recurrence of HCC after transplantation and the accuracy of prediction was great with the area under the curve (AUC) values reaching 0.760. It is demonstrated by ROC curve (receiver operating characteristic curve) that there is a great predictive accuracy of posttransplant miR-92b (>41.8-fold change) for early recurrence of HCC after transplantation (AUC= 0.925). But exosomal miR-92b combined with AFP are not enough to predict the recurrence of advanced HCC after transplantation. MiR-92b in exosomes may have effects on the activity of NK cells by inhibiting CD69 expression and NK cell-mediated cytotoxicity, which is crucial for tumor immune surveillance. Therefore, exosomal miR-92b may function as prospective biomarkers for early predicting HCC recurrence after transplantation (83). Under Endoplasmic reticulum stress, miR-23a-3p derived from exosomes in HCC could increase PD-L1 expression in macrophages through PTEN-PI3K/AKT pathway, thereby suppressing T cell function (84).
MiR-93 in the exosomes of HCC patients was overexpressed, which was associated with the clinical characteristics of HCC such as the size and stage of tumor stage as well as survival rate. By targeting TIMP2/TP53INP1/CDKN1A, exosomal miR-93 may increase the ability of proliferation and invasion of liver cancer. It is also revealed that the expression level of exosomal miR-93 is a reliable biomarker for HCC patients related to tumor size and advanced TNM stage. Liver cancer patients with higher exosomal miR-93 have poorer prognosis. In addition, they also found that the 2-year overall survival rate of liver cancer patients who had higher serum exosome miR-93 expression was lower than that of HCC patients with lower expression of serum exosome miR-93 (85). One previous study investigated the levels of miR-665 in the serum exosomes of HCC patients and the possibility as a promising diagnostic biomarker and prognostic factors for HCC was assessed. Researchers demonstrated that the level of miR-665 in serum exosomes of HCC group was obviously higher than that of normal control group (NC), which can stimulate HCC cells proliferation and the growth of tumors through MAPK/ERK pathway. The overexpression of miR-665 in exosomes had a positive relation with local invasion, tumor size and clinical stage and had an inverse relation with survival time (86).
Cui and coauthors found that compared to the normal hepatic cell line, the level of exosomal miR-224 was remarkably elevated in HCC lines, especially in HCC patients who have larger tumors and later stages. HCC patients with increased levels of miR-224 in the exosomes had low overall survival time according to Kaplan-Meier survival curves. The feasibility of miR-224 in exosomes to differentiating HCC patients from healthy controls was evaluated with an AUC of 0.910. Exosomal miR-224 could directly target glycine N-methyltransferase (GNMT), increasing the proliferation and invasion of liver cancer cells. Thus, miR-224 in the serum exosomes can serve as a potential diagnostic biomarker and an independent prognostic factor for HCC (87). However, its capability of differentiating liver diseases needs further evaluation. Additionally, the level of exosomal miR-148a and miR-1246 were remarkably higher from liver cancer patients than that of normal individuals and patients with liver cirrhosis. Exosomal miR-148a could discriminate HCC from LC (AUC=0.891), significantly higher than AFP (AUC=0.712), compared with miR-1246 (AUC=0.785). AFP combined with miR-122 and miR-148a was the greatest diagnostic group (AUC=0.947). The sensitivity and specificity of it were 87.0% and 90.0% respectively (88). Tang and coauthors have discovered that compared with normal donors, the level of miR-9-3p in the serum exosomes of liver cancer patients was significantly downregulated (p<0.01). Exosomal miR-9-3p targets the expression of fibroblast growth factor 5 (FGF5) to suppress HCC cell proliferation and the activation of the extracellular signal-regulated protein kinase1 and 2(ERK1/2) signaling pathway. The diagnostic ability of exosomal miR-9-3p was further evaluated with 67.31% sensitivity and 61.21% specificity (89).
Liu and coworkers have reported that the level of exosomal miR-125b in patients with HCC is remarkably lower than that in patients with chronic hepatitis B (CHB) and LC. Additionally, the low expression of serum exosomal miR-125b is closely related to tumor differentiation, tumor number and TNM stage. The Kaplan-Meier curve of time to relapse (TTR) and overall survival (OS) shows that the expression level of exosomal miR-125b was positively related with HCC patient’s TTR and OS, which can independently predict TTR and OS in patients with liver cancer (90). Furthermore, it is reported by Min and coauthors that levels of serum exosomal miR-638 from patients with HCC were remarkably lower than that of healthy controls, which predicts poor prognosis in HCC. Therefore, serum exosomal miR-638 was an innovative cancer biomarker and an independent prognostic factor for HCC patients. Nevertheless, the underlying molecular mechanisms and potential targeted genes related to miR-638 in HCC continue to be elusive to date and need to be further investigated (91).
Moreover, the levels of exosomal miR-106a, miR-122, miR-125b, miR-145, miR-192, miR-194, miR-17-5p and miR-29a were dramatically increased in liver cancer patients. What’s more, these exosomal miRNAs apart from miR-145 could distinguishing HCC from healthy subjects with an AUC of 0.650 to 0.850. What’s more, it was reported that the elevated levels of exosomal miR-106a, miR-17-5p and miR-194 were associated with larger tumor size (>3 cm). In further, exosomal miR-106a facilitates tumorigenesis via modulating MAPK and JNK pathways, serving as a promising serum prognostic biomarker and providing a novel insight into the therapy of HCC (92). Further effort is required to validate the specificity and sensitivity of these biomarkers in prospective studies with larger patient cohorts and standardized methodology. Currently, several miRNA-based therapies for different cancers have entered into clinical trials.
Long non-coding RNA (lncRNA) is a non-protein transcript longer than 200 nucleotides in length (93). LncRNAs can be found in the nucleus and cytoplasm, and exert different functions according to their subcellular localizations. lncRNAs may be involved in transcriptional regulation of gene expression and mRNA splicing in the nucleus. While in the cytoplasm, they could influence mRNA stability and regulate protein function (94). LncRNAs play its regulatory role mainly through multiple molecular mechanisms including binding with DNA to modulate gene transcription, acting as the competing endogenous RNA (ceRNA) or miRNA sponges to regulate gene expression at posttranscriptional level, associating with proteins, and encoding functional small peptides (95). LncRNAs are involved in various biological processes, such as cell proliferation, apoptosis, metastasis and regulate the tumor microenvironment in HCC, eventually causing tumor development (96). Exosomal lncRNAs obtained by the recipient cell will play its cancer-related role in the recipient cell to regulate cancer progression (Table 1).
Lnc-EPC1-4 overexpression inhibits cell proliferation and stimulates apoptosis, suggesting its functional role as a tumor suppressor gene. In contrast, knockdown of lnc-FAM72D-3 can reduce cell viability and promote cell apoptosis, indicating that lnc-FAM72D-3 acts as an oncogene in HCC (97). It has been proven previously that exosomal lncRNA TUC339 is rich in HCC patients and can promote cell proliferation and decrease cell adhesion to the extracellular matrix (ECM), which leads to the spread of HCC (98). Macrophages are imperative in the innate immune defense and have received increasing attention in the tumor microenvironment. In response to tumor-derived stimuli, macrophages can be polarized into the classical (M1) or alternative (M2) phenotypes. M1 macrophages exhibit anti-tumor activity, whereas M2 macrophages can promote tumorgenesis and tumor progression (112). Recently, Li and coauthors found that exosomal lncRNA TUC339 can be transferred to neighboring macrophages where it regulates the M1/M2 polarization and inhibits the anti-tumor immune response in vitro (99). It has also been shown that exosomal lncRNA H19 promoted the development of HCC including proliferation, invasion and migration and suppressed HCC cells apoptosis treated with propofol through upregulating LIM domain kinase 1 (LIMK1) via sponging miR-520a-3p (100). In another study, exosomal linc-FAM138B were reduced and inhibited the proliferation, migration and invasion of HCC cells via regulating miR-765 (101). On the contrary, exosomal lncRNA FAL1 was elevated in patients with HCC, which functioned as an oncogene and promoted the proliferation and migration of HCC cells through binding to miR-1236 competitively, and subsequently increased the expression of its target gene AFP and ZEB1 (102).
Other groups reported that exosomal linc-VLDLR could induce resistance to chemotherapeutic drugs (camptothecin and doxorubicin) and sorafenib targeted therapy in HCC. Exosomal linc-VLDLR derived from HCC cell lines were overexpressed when exposed to camptothecin, doxorubicin and sorafenib (103). The same research team also discovered another lncRNA, linc-ROR that contributes to doxorubicin and sorafenib resistance in HCC. The underlying mechanism is that TGF-β could enrich linc-ROR in exosomes from HCC, leading to an increase in the number of CD133+ liver cancer stem cells and reduced chemotherapy-induced cell death through inhibiting p53 (104). Although exosomal lncRNAs haven’t been investigated exhaustively in HCC, the above-mentioned exosomal lncRNAs promote the development of exosomal lncRNAs as a biomarker, drug resistance and therapeutic target in liver cancer.
Compared with patients with CHB, the expression of exosomal lncRNA LINC00635 and ENSG00000258332.1 of HCC patients are both higher with the AUC of 0.750 and 0.719 respectively (105). Similarly, elevated serum exosomal lncRNA HEIH levels can also distinguish HCV-related HCC patients from chronic hepatitis C (CHC) or HCV-induced cirrhosis patients (106). In the recent studies, Huang and coauthors reported that lncRNA 85 promotes proliferation, apoptosis and metastasis of HCC cells by modulating the expression of target genes associated with miR-324-5p. LncRNA 85 was highly expressed in both AFP+ and AFP- HCC-exosomes. The specificity and sensitivity of lncRNA 85 in diagnosing AFP+ HCC were 76.7% and 80.5% respectively (cut-off = 1.650) while specificity and sensitivity were 76.7% and 80.0% respectively (cut-off = 1.645) in AFP− HCC. All these findings indicated that lncRNA 85 is capable of serving as a promising biomarker in diagnosing HCC and contributing to improving the diagnostic sensitivity of AFP-HCC (107).
Exosomal lnc RNA SENP3-EIF4A1 was obviously decreased in liver cancer tissues, which can distinguish HCC patients from healthy people with the AUC reaching 0.8028. Exosomal SENP3-EIF4A1 released by normal cells was transported to liver cancer cells to induce apoptosis and attenuate the migration and invasion abilities of HCC cells and thus reverse their malignant biological behavior (P<0.05). In addition, it was able to suppress the tumor growth in vivo and regulate ZFP36 expression by binding with miR-9-5p competitively. To conclude, exosomal SENP3-EIF4A1 is a novel beneficial biomarker for clinical detection of HCC, and can spread from normal cells to HCC cells through exosomes, thereby in vivo and in vitro inhibiting HCC progression (108). Exosomal LINC00161 was upregulated in HCC patients compared with the controls, which may be another innovative biomarker for HCC with remarkable stability and specificity (109).
Although the treatment for HCC have been greatly improved, the prognosis of HCC is still poor. Lee and colleagues (110) provided strong evidence that exosomal lncRNA-ATB is a new independent prognostic marker and therapeutic target for HCC. The serum exosomal lncRNA CRNDE expression levels were significantly increased in patients with HCC compared with normal controls. High serum exosomal lncRNA CRNDE expression was positively associated HCC progression and poorer prognosis (111).
Identified as a novel member of RNAs, circular RNAs (circRNAs) originate from pre-mRNA back splicing with considerable stability and conservation (113–116). Different from linear RNA, circRNAs have a closed loop structure without a poly A tail or 5’−3’ polarity, which are insusceptible to RNA exonuclease or RNase R-induced degradation (117). It has been revealed that exosomal circRNAs are closely related to cellular proliferation, invasion, migration, metastasis and drug resistance (118) and due to high abundance and relative stability, the combined use of exosomes and circRNAs may become the promising diagnostic and prognostic markers in cancers (119, 120).
It was first reported that circRNAs in exosomes is rich and stable on the basis of the RNA-Seq analysis from MHCC-LM3 liver cancer cells and cell-derived exosomes. By analysis of RNA-seq, we found that the ratio of circRNAs levels to linear RNA levels in exosomes was approximately 6 times higher than in producing cells. In further, the expression levels of circRNAs were almost unchanged after incubation for 24h at room temperature. These results show that exosomal circRNAs are extremely plentiful and remain stable (121). In recent years, there has been lots of research on exosomal lncRNAs and miRNAs in cancer, but relatively little attention has been paid to exosomal circRNAs. However, in recent years, the clinical application of exosomal circRNAs is in an era of rapid development (Table 2).
CircRNA-100,338 was elevated in the exosomes from liver cancer cells with potential metastasis, which involved in regulating angiogenesis and metastasis of HCC. It was revealed that the invasive abilities of liver cancer cells can be increased or reduced by upregulation or downregulation of exosomal circRNA-100,338. RNA pull-down assay showed that circRNA-100338 can bind RNA-binding proteins including NOVA2, which was reported to regulate vascular development and lumen formation. They also found that HCC patients undergoing radical hepatectomy had continuous upregulated level of exosomal circRNA-100,338 may be at risk of poor survival and lung metastasis (122). Wang recently proved that the overexpression of exosomal circPTGR1 was linked to the clinical stage and prognosis of HCC. Furthermore, downregulation of exosomal circPTGR1 remarkably inhibited the invasion and migration of HCC cell lines. There are evidence demonstrating that the exosome circPTGR1 competed with Met to target MIR449a, causing dysregulation of the TME and promoting liver cancer metastasis (119).
Several studies in recent years have proven that circRNAs can absorb miRNAs by stable complementary binding and sponge miRNAs to modulate gene expression (134, 135). For example, exosomal circRNAs derived from adipocytes play a momentous role in promoting the tumor growth of liver cancer via absorbing miR-34a and activating the USP7/Cyclin A2 pathway (126). What’s more, Dai found that exosomal circRNA 100284 acted as a sponge for miR-217 to regulate cell cycle and inhibited cell proliferation via inducing the G2/M phase of cell cycle arrest and targeting EZH2 in HCC (123, 124). Exosomal CDR1-AS could promote HCC progression. In HCC cells, upregulated CDR1-AS accelerated cell proliferation and migration by sponging of miR-1270, increasing the expression of alpha-fetoprotein (125). It is worth noting that not only exosomal circRNAs derived from tumors, but also exosomal circRNAs derived from adipocytes, promote tumor growth. For example, the expression level of exosomal circ-deubiquitination (Circ-DB) was high in HCC patients with higher body fat ratios. Circ-DB derived from adipocytes promoted HCC proliferation and reduces DNA damage via functioning as a spong of miR-34a, thereby activating the USP7/Cyclin A2 signaling pathway (126). Furthermore, circ0051443, secreted by normal hepatocytes and transferred to adjacent HCC cells, was decreased in plasma exosomes of HCC patients, causing tumor cell apoptosis and cell cycle arrest through sponging of miR331-3p and upregulation of BAK1 expression (127). However, the specific functions of exosome circRNAs are under intense investigation.
As the largest immune organ in human body, liver plays a protective role by promoting immunotolerance under physiologic conditions (136). More recently, immunotherapy has emerged as a promising therapeutic option for advanced HCC patients. Several novel immunotherapeutic methods, including the use of immune checkpoint inhibitors, new types of immune cell adoption, such as chimeric antigen receptor T cell (CAR-T), TCR gene-modified T cells and stem cells, and microRNAs have been used in clinical trials for the treatment of HCC (137). Zhang and colleagues found that circUHRF1 was overexpressed in HCC tissues and HCC-derived exosomes and inhibited NK cell function by upregulating the expression of TIM-3 via degradation of miR-449c-5p. Furthermore, exosomal circUHRF1 may drive resistance to anti-PD1 immunotherapy, providing a potential therapeutic strategy for patients with HCC (129).
TACE is the treatment of choice for intermediate-stage HCC, including unresectable multinodular HCC without extrahepatic spread. However, it is difficult and subjective to decide whether to repeat or stop TACE (138). It has been proved that circ-G004213 in exosomes could promote cisplatin sensitivity via regulation of miR-513b-5p/PRPR39 and was positively associated with the prognosis of HCC patients following TACE. Exosomal circ-G004213 may be an indicator for predicting the efficacy of TACE in patients with HCC (128).
Glycolysis is a hallmark feature of cancer especially cancer cells preferentially use glycolysis to produce glucose-dependent ATP (139). Lactate is an important metabolite in cancer metabolic reprogramming, and its large accumulation causes extracellular pH in the TME being acidified, ranging from 6.0 to 6.5 (140). Besides, TDEs are strongly connected with the TME. For example, exosomal circFBLIM1 was highly expressed in HCC and made contributions to the progression and glycolysis of HCC by sponging miR-338 and upregulating LRP6. The circFBLIM1/miR-338/LRP6 axis may provide a new therapeutic approach for HCC treatment (130).
Exosomal circ-0006602 was generally upregulated in HCC and combined with AFP can greatly improve the accuracy of early diagnosis of HCC, having the potential to become non-invasive biomarker for the early diagnosis and screening of liver cancer (131). Similarly, exosomal circ 0070396 expressed dramatically higher in HCC samples compared with corresponding controls and displayed a better diagnostic performance than AFP and their combination provided higher diagnostic value. Moreover, the exosomal circ 0070396 expression was positively correlated with HCC progression (132).
Additionally, the levels of circulating exosomal circAKT3 in patients with HCC were significantly higher than in healthy subjects. High expression of circulating exosomal circAKT3 is associated with a higher risk of HCC recurrence and mortality. Follow-up of patients with HCC have high circAKT3 expression after surgery may help to reduce the risk of recurrence and improve prognosis (133).
Extensive evidence has revealed ncRNAs in exosomes function as diagnostic and prognostic markers in different malignancies, including lung cancer, gastric cancer, breast cancer, liver cancer, colorectal cancer and so on. In recent years, tumor-associated cargoes in exosomes, especially with regard to ncRNAs have been a research hotspot. In this review, we talk about the clinical applications of exosomal ncRNAs in liver cancer.
Exosomes are nanometer-sized membrane vesicle structure secreted from most cell types which regulate the activation of corresponding signaling pathways in recipient (target) cells. Exosomes serve as mediators in intercellular communications both locally and systemically in the tumor microenvironment by transferring information cargo, such as proteins, lipids and diverse forms of RNAs, contributing to the progression of HCC. Because ncRNAs are stable in serum exosomes and exhibit distinct expression profiles that reflect the characteristics of cancer cells, they could function as noninvasive and sensitive ideal biomarkers for the early diagnosis and prognosis of liver cancer. The study of exosomes is still in its infancy. With the further development of the research, its role in the molecular pathogenesis of liver cancer will be clarified. Moreover, exosomes can not only be used as vaccines or drug carriers, but also are expected to transfer target genes into exosomes through genetic engineering technology for gene therapy of liver cancer. Given the above, exosomal ncRNAs provide emerging diagnostic methods and treatments for HCC.
As we further understand the nature of exosomes, diagnostic and therapeutic techniques related to exosomes are being constantly improved. Increasing emphasis has been given to exosomal microRNAs and many exosomal microRNAs have been explored in the recent years. As noninvasive, specific and sensitive tumor markers, exosomal lncRNAs have attracted more and more attention. Liquid biopsy of serum or plasma specimens is usually used for exosomal lncRNAs detection, which has the advantages of non-invasive, repeatable detection and real-time monitoring. Combined with imaging data and traditional tumor marker detection, it is expected to realize early diagnosis, recurrence monitoring, prognosis judgment and efficacy evaluation of tumors. Exosomal lncRNAs and circRNAs have a great research prospect in cancers, but a lot of lncRNAs and circRNAs have not been explored in liver cancer. At present, there are relatively few studies on the association between exosomal ncRNAs and immunotherapy in HCC. The following research can focus on this aspect to open a new chapter in liver cancer immunotherapy. In addition, there is a lack of standard methods for exosome isolation and purification. To solve the present problem, first of all, it is necessary to identify, isolate and quantify exosomes accurately, efficiently and selectively. What’s more, the detection technology of lncRNAs and the specificity of exosomal ncRNAs as biomarker of HCC needs to be improved. Both human tumor cells and normal cells can secrete exosomes, but there is no specific marker to distinguish the source of exosomes at present. Therefore, how to avoid the interference caused by exosomes secreted by normal cells and separate tumor-specific exosomes is also a major challenge. Many experiments are done in mice or in vitro and the application of exosomal lncRNA in the treatment of liver cancer needs to be further verified in vivo.
RW conceptualized the review. WW was the major contributors in writing the manuscript. WW designed the figures. L-PH prepared the tables. HS and X-YC critically reviewed and edited the manuscript. All the authors read and approved the final manuscript.
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81772995 and 81472266); the Excellent Youth Foundation of Jiangsu Province, China (No. BK20140032); Jiangsu Province’s Key Provincial Talents Program (No. ZDRCA2016090).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Locarnini S, Hatzakis A, Chen D-S, Lok A. Strategies to Control Hepatitis B: Public Policy, Epidemiology, Vaccine and Drugs. J Hepatol (2015) 62:S76–86. doi: 10.1016/j.jhep.2015.01.018
3. Mittal S, El-Serag HB. Epidemiology of Hepatocellular Carcinoma: Consider the Population. J Clin Gastroenterol (2013) 47(Suppl):S2–6. doi: 10.1097/MCG.0b013e3182872f29
4. Forner A, Llovet JM, Bruix J. Hepatocellular Carcinoma. Lancet (2012) 379(9822):1245–55. doi: 10.1016/S0140-6736(11)61347-0
5. Clark T, Maximin S, Meier J, Pokharel S, Bhargava P. Hepatocellular Carcinoma: Review of Epidemiology, Screening, Imaging Diagnosis, Response Assessment, and Treatment. Curr Probl Diagn Radiol (2015) 44(6):479–86. doi: 10.1067/j.cpradiol.2015.04.004
6. Wallace MC, Preen D, Jeffrey GP, Adams LA. The Evolving Epidemiology of Hepatocellular Carcinoma: A Global Perspective. Expert Rev Gastroenterol Hepatol (2015) 9(6):765–79. doi: 10.1586/17474124.2015.1028363
7. Petruzziello A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) Related Hepatocellular Carcinoma. Open Virol J (2018) 12:26–32. doi: 10.2174/1874357901812010026
8. Wong CM, Tsang FH, Ng IO. Non-Coding RNAs in Hepatocellular Carcinoma: Molecular Functions and Pathological Implications. Nat Rev Gastroenterol Hepatol (2018) 15(3):137–51. doi: 10.1038/nrgastro.2017.169
9. Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu X, et al. Liver Cancer Incidence and Mortality in China: Temporal Trends and Projections to 2030. Chin J Cancer Res (2018) 30:571–9. doi: 10.21147/j.issn.1000-9604.2018.06.01
10. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-Analysis. Gastroenterology (2018) 154(6):1706–18.e1701. doi: 10.1053/j.gastro.2018.01.064
11. Kanda T, Ogasawara S, Chiba T, Haga Y, Omata M, Yokosuka O. Current Management of Patients With Hepatocellular Carcinoma. World J Hepatol (2015) 7(15):1913–20. doi: 10.4254/wjh.v7.i15.1913
12. Kuczynski EA, Lee CR, Man S, Chen E, Kerbel RS. Effects of Sorafenib Dose on Acquired Reversible Resistance and Toxicity in Hepatocellular Carcinoma. Cancer Res (2015) 75:2510–9. doi: 10.1158/0008-5472.CAN-14-3687
13. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and Function of Extracellular Vesicles in Cancer. Pharmacol Ther (2018) 188:1–11. doi: 10.1016/j.pharmthera.2018.02.013
14. van Niel G, D’Angelo G, Raposo G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat Rev Mol Cell Biol (2018) 19(4):213–28. doi: 10.1038/nrm.2017.125
15. Muhsin-Sharafaldine MR, Saunderson SC, Dunn AC, Faed JM, Kleffmann T, McLellan AD. Procoagulant and Immunogenic Properties of Melanoma Exosomes, Microvesicles and Apoptotic Vesicles. Oncotarget (2016) 7:56279–94. doi: 10.18632/oncotarget.10783
16. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J Extracell Vesicles (2018) 7(1):1535750. doi: 10.1080/20013078.2018.1535750
17. Shen H, Yao X, Li H, Li X, Zhang T, Sun Q, et al. Role of Exosomes Derived From miR-133b Modified MSCs in an Experimental Rat Model of Intracerebral Hemorrhage. J Mol Neurosci (2018) 64(3):421–30. doi: 10.1007/s12031-018-1041-2
18. Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a Liquid Biopsy for Lung Cancer. Lung Cancer (2017) 116:46–54. doi: 10.1016/j.lungcan.2017.12.012
19. Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, et al. Mesenchymal Stem Cell: An Efficient Mass Producer of Exosomes for Drug Delivery. Adv Drug Deliv Rev (2012) 65(3):336–41. doi: 10.1016/j.addr.2012.07.001
20. Jiao Y, Xu P, Shi H, Chen D, Shi H. Advances on Liver Cell-Derived Exosomes in Liver Diseases. J Cell Mol Med (2021) 25(1):15–26. doi: 10.1111/jcmm.16123
21. Oosthuyzen W, Sime NE, Ivy JR, Turtle EJ, Street JM, Pound J, et al. Quantification of Human Urinary Exosomes by Nanoparticle Tracking Analysis. J Physiol (2013) 591:58335842. doi: 10.1113/jphysiol.2013.264069
22. Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, et al. Identification and Proteomic Profiling of Exosomes in Human Cerebrospinal Fluid. J Transl Med (2012) 10:5. doi: 10.1186/1479-5876-10-5
23. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of Serum Exosomal microRNA-21 in Human Hepatocellular Carcinoma. J Biomedicine Biotechnol (2014) 2014:864894. doi: 10.1155/2014/864894
24. Bu H, He D, He X, Wang K. Exosomes: Isolation, Analysis, and Applications in Cancer Detection and Therapy. Chembiochem (2018) 20:451–61. doi: 10.1002/cbic.201800470
25. Li C, Xu X. Biological Functions and Clinical Applications of Exosomal Non-Coding RNAs in Hepatocellular Carcinoma. Cell Mol Life Sci (2019) 76(21):4203–19. doi: 10.1007/s00018-019-03215-0
26. Abudoureyimu M, Zhou H, Zhi Y, Wang T, Feng B, Wang R, et al. Recent Progress in the Emerging Role of Exosome in Hepatocellular Carcinoma. Cell Prolif (2018) 52:e12541. doi: 10.1111/cpr.12541
27. Cobelli NJ, Leong DJ, Sun HB. Exosomes: Biology, Therapeutic Potential, and Emerging Role in Musculoskeletal Repair and Regeneration. Ann N Y Acad Sci (2017) 1410:57–67. doi: 10.1111/nyas.13469
28. Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
29. Calin GA, Croce CM. MicroRNA-Cancer Connection: The Beginning of a New Tale. AACR Educ Book (2006) 66:7390–4. doi: 10.1158/AACR.EDB-CAN-06-0800
30. Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, et al. The Emerging Role of Exosome-Derived Non-Coding RNAs in Cancer Biology. Cancer Lett (2018) 414:107–15. doi: 10.1016/j.canlet.2017.10.040
31. Xie Y, Dang W, Zhang S, Yue W, Yang L, Zhai X, et al. The Role of Exosomal Noncoding RNAs in Cancer. Mol 535 Cancer (2019) 18(1):37. doi: 10.1186/s12943-019-0984-431
32. Sukowati CHC, Cabral LKD, Tiribelli C, Pascut D. Circulating Long and Circular Noncoding RNA as Non-Invasive Diagnostic Tools of Hepatocellular Carcinoma. Biomedicines (2021) 9(1):90. doi: 10.3390/biomedicines9010090
33. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle Formation During Reticulocyte Maturation. Association of Plasma Membrane Activities With Released Vesicles (Exosomes). J Biol Chem (1987) 262(19):9412–20. doi: 10.1016/S0021-9258(18)48095-7
34. Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B Lymphocytes Secrete Antigen-Presenting Vesicles. J Exp Med (1996) 183(3):1161–72. doi: 10.1084/jem.183.3.1161
35. Gould SJ, Raposo G. As We Wait: Coping With an Imperfect Nomenclature for Extracellular Vesicles. J Extracell Vesicles (2013) 2:10.3402/jev.v2i0.20389. doi: 10.3402/jev.v2i0.20389
36. Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. Minimal Experimental Requirements for Definition of Extracellular Vesicles and Their Functions: A Position Statement From the International Society for Extracellular Vesicles. J Extracell Vesicles (2014). doi: 10.3402/jev.v3.26913
37. Mittelbrunn M, Sanchez-Madrid F. Intercellular Communication: Diverse Structures for Exchange of Genetic Information. Nat Rev Mol Cell Biol (2012) 13(5):328–35. doi: 10.1038/nrm3335
38. French KC, Antonyak MA, Cerione RA. Extracellular Vesicle Docking at the Cellular Port: Extracellular Vesicle Binding and Uptake. Semin Cell Dev Biol (2017) 67:48–55. doi: 10.1016/j.semcdb.2017.01.002
39. Kourembanas S. Exosomes: Vehicles of Intercellular Signaling, Biomarkers, and Vectors of Cell Therapy. Annu Rev Physiol (2015) 77:13–27. doi: 10.1146/annurev-physiol-021014-071641
40. Atienzar-Aroca S, Flores-Bellver M, Serrano-Heras G, Martinez-Gil N, Barcia JM, Aparicio S, et al. Oxidative Stress in Retinal Pigment Epithelium Cells Increases Exosome Secretion and Promotes Angiogenesis in Endothelial Cells. J Cell Mol Med (2016) 20:1457–66. doi: 10.1111/jcmm.12834
41. Cheng Y, Dai X, Yang T, Zhang N, Liu Z, Jiang Y. Low Long Noncoding RNA Growth Arrest-Specific Transcript 5 Expression in the Exosomes of Lung Cancer Cells Promotes Tumor Angiogenesis. J Oncol (2019) 2019:2476175. doi: 10.1155/2019/2476175
42. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc Natl Acad Sci (2016) 113:E968–77. doi: 10.1073/pnas.1521230113
43. Schmidt O, Teis D. The ESCRT Machinery. Curr Biol (2012) 22(4):R116–20. doi: 10.1016/j.cub.2012.01.028
44. Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark. Cellular functions H. And Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem Sci (2016) 42(1):42–56. doi: 10.1016/j.tibs.2016.08.016
45. Schoneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-Topology Membrane Scission by the ESCRT Proteins. Nat Rev Mol Cell Biol (2016) 18(1):5–17. doi: 10.1038/nrm.2016.121
46. Colombo M, Raposo G, Thery. Biogenesis C. Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol (2014) 30(1):255–89. doi: 10.1146/annurev-cellbio-101512-122326
47. Savina A, Furlan M, Vidal M, Colombo MI. Exosome Release is Regulated by a Calcium-Dependent Mechanism in K562 Cells. J Biol Chem (2003) 278(22):20083–90. doi: 10.1074/jbc.M301642200
48. Tiwari N, Wang CC, Brochetta C, Ke G, Vita F, Qi Z, et al. VAMP-8 Segregates Mast Cell-Preformed Mediator Exocytosis From Cytokine Trafficking Pathways. Blood (2008) 111(7):3665–74. doi: 10.1182/blood-2007-07-103309
49. Jadli AS, Ballasy N, Edalat P, Patel VB. Inside (Sight) of Tiny Communicator: Exosome Biogenesis, Secretion, and Uptake. Mol Cell Biochem (2020) 467:77– 94. doi: 10.1007/s11010-020-03703-z
50. Mulcahy LA, Pink RC, Carter DR. Routes and Mechanisms of Extracellular Vesicle Uptake. J Extracell Vesicles (2014). doi: 10.3402/jev.v3.24641
51. van der Vos KE, Abels ER, Zhang X, Lai C, Carrizosa E, Oakley D, et al. Directly Visualized Glioblastoma-Derived Extracellular Vesicles Transfer RNA to Microglia/Macrophages in the Brain. Neuro-Oncology (2015) 18(1):58–69. doi: 10.1093/neuonc/nov244
52. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome Mediated Communication Within the Tumor Microenvironment. J Control Release (2015) 219:278294. doi: 10.1016/j.jconrel.2015.06.029
53. Yu DD, Wu Y, Shen HY, Lv MM, Chen WX, Zhang XH, et al. Exosomes in Development, Metastasis and Drug Resistance of Breast Cancer. Cancer Sci (2015) 106:959964. doi: 10.1111/cas.12715
54. Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, et al. Radiation-induced Mir-208a Increases the Proliferation and Radioresistance by Targeting P21 in Human Lung Cancer Cells. J Exp Clin Cancer Res (2016) 35:7. doi: 10.1186/s13046-016-0285-3
55. Soung YH, Nguyen T, Cao H, Lee J, Chung J. Emerging Roles of Exosomes in Cancer Invasion and Metastasis. BMB Rep (2016) 49:1825. doi: 10.5483/BMBRep.2016.49.1.239
56. Riches A, Campbell E, Borger E, Powis S. Regulation of Exosome Release From Mammary Epithelial and Breast Cancer Cells - A New Regulatory Pathway. Eur J Cancer (2014) 50:10251034. doi: 10.1016/j.ejca.2013.12.019
57. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics (2017) 7(3):789–804. doi: 10.7150/thno.18133
58. Lin S, Yu Z, Chen D, Wang Z, Miao J, Li Q, et al. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small (2020) 16(9):e1903916. doi: 10.1002/smll.201903916
59. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat Cell Biol (2019) 21(1):9–17. doi: 10.1038/s41556-018-0250-9
60. Simonsen JB. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ Res (2017) 121(8):920–2. doi: 10.1161/CIRCRESAHA.117.31176
61. Guo J, Wu C, Lin X, Zhou J, Zhang J, Zheng W, et al. Establishment of a Simplified Dichotomic Size-Exclusion Chromatography for Isolating Extracellular Vesicles Toward Clinical Applications. J Extracell Vesicles (2021) 10(11):e12145. doi: 10.1002/jev2.12145
62. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin Cancer Res (2017) 23(17):5311–9. doi: 10.1158/1078-0432.CCR-17-0577
63. Chen J, Wang S, Jia S, Ding G, Jiang G, Cao L. Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profile in Pancreatic Cancer Derived Exosomes Treated Dendritic Cells by Microarray Analysis. J Cancer (2018) 9(1):21–31. doi: 10.7150/jca.21749
64. Bellingham SA, Shambrook M, Hill AF. Quantitative Analysis of Exosomal miRNA via qPCR and Digital PCR. Methods Mol Biol (2017) 1545:55–70. doi: 10.1007/978-1-4939-6728-5_5
65. Lee RC, Feinbaum RL, Ambros VJ. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell (1994) 75(5):843–54. doi: 10.1016/0092-8674(93)90529-Y
66. Ruan K, Fang X, Ouyang G. MicroRNAs: Novel Regulators in the Hallmarks of Human Cancer. Cancer Lett (2009) 285(2):116–26. doi: 10.1016/j.canlet.2009.04.031
67. Di Leva G, Garofalo M, Croce CM. MicroRNAs in Cancer. Annu Rev Pathol Mech Dis (2013) 9(1):287–314. doi: 10.1146/annurev-pathol-012513-104715
68. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in Cancer: Biomarkers, Functions and Therapy. Trends Mol Med (2014) 20(8):460–9. doi: 10.1016/j.molmed.2014.06.005
69. Kozomara A, Birgaoanu M, Griffiths-Jones S. Mirbase: From microRNA Sequences to Function. Nucleic Acids Res (2018) 47(D1):D155–62. doi: 10.1093/nar/gky1141
70. Lima LG, Chammas R, Monteiro RQ, Moreira ME, Barcinski MA. Tumor-Derived Microvesicles Modulate the Establishment of Metastatic Melanoma in a Phosphatidylserine-Dependent Manner. Cancer Lett (2009) 283:168–75. doi: 10.1016/j.canlet.2009.03.041
71. Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol Ther Nucleic Acids (2018) 11:243–52. doi: 10.1016/j.omtn.2018.02.014
72. Chen W, Huang L, Liang J, Ye Y, He S, Niu J. Hepatocellular Carcinoma Cells-Derived Exosomal microRNA-378b Enhances Hepatocellular Carcinoma Angiogenesis. Life Sci (2021) 273(6):119184. doi: 10.1016/j.lfs.2021.119184
73. Zhao S, Li J, Zhang G, Wang Q, Wu C, Zhang Q, et al. Exosomal miR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting Lpin1. Cell Physiol Biochem (2019) 53(1):19–35. doi: 10.33594/000000118
74. Wang G, Zhao W, Wang H, Qiu G, Jiang Z, Wei G, et al. Exosomal MiR-744 Inhibits Proliferation and Sorafenib Chemoresistance in Hepatocellular Carcinoma by Targeting Pax2. Med Sci Monit (2019) 25:7209–17. doi: 10.12659/MSM.919219
75. Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, et al. Exosomal microRNA-32-5p Induces Multidrug Resistance in Hepatocellular Carcinoma via the PI3K/Akt Pathway. J Exp Clin Cancer Res (2018) 37(1):52. doi: 10.1186/s13046-018-0677-7
76. Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular Carcinoma-Derived Exosomal miRNA-21 Contributes to Tumor Progression Byconverting Hepatocyte Stellate Cells to Cancer-Associated Fibroblasts. J Exp Clin Cancer Res (2018) 37(1):324. doi: 10.1186/s13046-018-0965-2
77. Campbell K. Contribution of Epithelial-Mesenchymal Transitions to Organogenesis and Cancer Metastasis. Curr Opin Cell Biol (2018) 55:30–5. doi: 10.1016/j.ceb.2018.06.008
78. Uka K, Aikata H, Takaki S, Shirakawa H, Jeong SC, Yamashina K, et al. Clinical Features and Prognosis of Patients With Extrahepatic Metastases From Hepatocellular Carcinoma. World J Gastroenterol (2007) 13:414–20. doi: 10.3748/wjg.v13.i3.414
79. Wang H, Chen L. Tumor Microenviroment and Hepatocellular Carcinoma Metastasis. J Gastroenterol Hepatol (2013) 28(Suppl. S1):43–8. doi: 10.1111/jgh.12091
80. Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-Derived Exosomal miR-1247-3p Induces Cancer-Associated Fibroblast Activation to Foster Lung Metastasis of Liver Cancer. Nat Commun (2018) 9(1):191. doi: 10.1038/s41467-017-02583-0
81. Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, et al. Hepatoma Cell-Secreted Exosomal microRNA-103 Increases Vascular Permeability and Promotes Metastasis by Targeting Junction Proteins. Hepatology (2018) 68(4):1459–75. doi: 10.1002/hep.29920
82. Eun JW, Seo CW, Baek GO, Yoon MG, Ahn HR, Son JA, et al. Circulating Exosomal MicroRNA-1307-5p as a Predictor for Metastasis in Patients With Hepatocellular Carcinoma. Cancers (Basel) (2020) 12(12):3819. doi: 10.3390/cancers12123819
83. Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, et al. Circulating Exosomal miR-92b: Its Role for Cancer Immunoediting and Clinical Value for Prediction of Posttransplant Hepatocellular Carcinoma Recurrence. Am J Transplant (2019) 19(12):3250–62. doi: 10.1111/ajt.15490
84. Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, et al. Endoplasmic Reticulum Stress Promotes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-Regulate PD-L1 Expression in Macrophages. Hepatology (2019) 70(1):241–58. doi: 10.1002/hep.30607
85. Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 Promotes Proliferation and Invasion in Hepatocellular Carcinoma by Directly Inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun (2018) 502(4):515–21. doi: 10.1016/j.bbrc.2018.05.208
86. Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, et al. Exosomal miR-665 as a Novel Minimally Invasive Biomarker for Hepatocellular Carcinoma Diagnosis and Prognosis. Oncotarget (2017) 8(46):80666–78. doi: 10.18632/oncotarget.20881
87. Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, et al. Mechanism of Exosomal microRNA-224 in Development of Hepatocellular Carcinoma and its Diagnostic and Prognostic Value. World J Gastroenterol (2019) 25(15):1890–8. doi: 10.3748/wjg.v25.i15.1890
88. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum Exosomal microRNAs Combined With Alpha-Fetoprotein as Diagnostic Markers of Hepatocellular Carcinoma. Cancer Med (2018) 7(5):1670–9. doi: 10.1002/cam4.1390
89. Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, et al. Exosomal miR-9-3p Suppresses HBGF-5 Expression and is a Functional Biomarker in Hepatocellular Carcinoma. Minerva Med (2018) 109(1):15–23. doi: 10.23736/S0026-4806.17.05167-9
90. Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, et al. Serum Exosomal miR-125b is a Novel Prognostic Marker for Hepatocellular Carcinoma. OncoTargets Ther (2017) 10:3843–51. doi: 10.2147/OTT.S140062
91. Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased Levels of Serum Exosomal miR-638 Predict Poor Prognosis in Hepatocellular Carcinoma. J Cell Biochem (2018) 119(6):4711–6. doi: 10.1002/jcb.26650
92. Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and Validation of Serum Exosomal microRNAs as Diagnostic and Prognostic Biomarkers for Hepatocellular Carcinoma. J Cell Biochem (2019) 120(1):135–42. doi: 10.1002/jcb.27165
93. Wilusz JE, Sunwoo H, Spector DL. Long Noncoding RNAs: Functional Surprises From the RNA World. Genes Dev (2009) 23:1494–504. doi: 10.1101/gad.1800909
94. Quinn JJ, Chang HY. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat Rev Genet (2016) 17(1):47–62. doi: 10.1038/nrg.2015.10
95. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
96. Li C, Yang J, Liu C, Wang X, Zhang L. Long Non-Coding RNAs in Hepatocellular Carcinoma: Ordering of the Complicated lncRNA Regulatory Network and Novel Strategies for HCC Clinical Diagnosis and Treatment. Pharmacol Res (2020) 158:104848. doi: 10.1016/j.phrs.2020.104848
97. Yao Z, Jia C, Tai Y, Liang H, Zhong Z, Xiong Z, et al. Serum Exosomal Long Noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as Diagnostic Biomarkers for Hepatocellular Carcinoma. Aging (Albany NY) (2020) 12(12):11843–63. doi: 10.18632/aging.103355
98. Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer (2013) 4(7–8):261–72. doi: 10.1177/1947601913499020
99. Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by Hcc-Derived Exosomal lncrna Tuc339. Int J Mol Sci (2018) 19(10):2958. doi: 10.3390/ijms19102958
100. Wang D, Xing N, Yang T, Liu J, Zhao H, He J, et al. Exosomal lncRNA H19 Promotes the Progression of Hepatocellular Carcinoma Treated With Propofol via miR-520a-3p/LIMK1 Axis. Cancer Med (2020) 9(19):7218–30. doi: 10.1002/cam4.3313
101. Zhuo C, Yi T, Pu J, Cen X, Zhou Y, Feng S, et al. Exosomal Linc-FAM138B From Cancer Cells Alleviates Hepatocellular Carcinoma Progression via Regulating miR-765. Aging (Albany NY) (2020) 12(24):26236–47. doi: 10.18632/aging.202430
102. Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 Promotes Cell Proliferation and Migration by Acting as a CeRNA of miR-1236 in Hepatocellular Carcinoma Cells. Life Sci (2018) 197:122–9. doi: 10.1016/j.lfs.2018.02.006
103. Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of Extracellular Vesicle Long Noncoding RNA (linc-VLDLR) in Tumor Cell Responses to Chemotherapy. Mol Cancer Res (2014) 12:1377–87. doi: 10.1158/1541-7786.MCR-13-0636
104. Takahashi K, Yan I, Kogure T, Haga H, Patel T. Extracellular Vesicle-Mediated Transfer of Long Non-Coding RNA ROR Modulates Chemosensitivity in Human Hepatocellular Cancer. FEBS Open Bio (2014) 4:458–46. doi: 10.1016/j.fob.2014.04.007
105. Xu H, Chen Y, Dong X, Wang X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev (2018) 27(6):710–6. doi: 10.1158/1055-9965.epi-17-0770
106. Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNAHEIH in Serum and Exosomes as a Potential Biomarker in the HCV-Related Hepatocellular Carcinoma. Cancer Biomark (2018) 21(3):651–9. doi: 10.3233/CBM-170727
107. Huang X, Sun L, Wen S, Deng D, Wan F, He X, et al. RNA Sequencing of Plasma Exosomes Revealed Novel Functional Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Sci (2020) 111(9):3338–49. doi: 10.1111/cas.14516
108. Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, et al. Exosome-Transmitted Long Non-Coding RNA SENP3-EIF4A1 Suppresses the Progression of Hepatocellular Carcinoma. Aging (Albany NY) (2020) 12(12):11550–67. doi: 10.18632/aging.103302
109. Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, et al. Serum and Exosome Long Non Coding RNAs as Potential Biomarkers for Hepatocellular Carcinoma. J Cancer (2018) 9(15):2631–9. doi: 10.7150/jca.24978
110. Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, et al. Circulating Exosomal Noncoding RNAs as Prognostic Biomarkers in Human Hepatocellular Carcinoma. Int J Cancer (2019) 144(6):1444–52. doi: 10.1002/ijc.31931
111. Wang T, Zhu H, Xiao M, Zhou S. Serum Exosomal Long Noncoding RNA CRNDE as a Prognostic Biomarker for Hepatocellular Carcinoma. J Clin Lab Anal (2021) 35(11):e23959. doi: 10.1002/jcla.23959
112. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long Non-Coding RNA Cox-2 Prevents Immune Evasion and Metastasis of Hepatocellular Carcinoma by Altering M1/M2 Macrophage Polarization. J Cell Biochem (2018) 119(3):2951–63. doi: 10.1002/jcb.26509
113. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
114. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-Type Specific Features of Circular RNA Expression. PloS Genet (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
115. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
116. Yu B, Shan G. Functions of Long Noncoding RNAs in the Nucleus. Nucleus (2016) 7:155–66. doi: 10.1080/19491034.2016.1179408
117. Chen LL, Yang L. Regulation of circRNA Biogenesis. RNA Biol (2015) 12:381388. doi: 10.1080/15476286.2015.1020271
118. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: Biogenesis, Effect and Application in Human Diseases. Mol Cancer (2019) 18(1):116. doi: 10.1186/s12943-019-1041-z
119. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three Isoforms of Exosomal Circptgr1 Promote Hepatocellular Carcinoma Metastasis via the Mir449a-MET Pathway. EBioMedicine (2019) 40:432445. doi: 10.1016/j.ebiom.2018.12.062
120. Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, et al. Circular RNA Circ-RanGAP1 Regulates VEGFA Expression by Targeting miR-877-3p to Facilitate Gastric Cancer Invasion and Metastasis. Cancer Lett (2020) 471:38–48. doi: 10.1016/j.canlet.2019.11.038
121. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25:981–4. doi: 10.1038/cr.2015.82
122. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 Promotes Hepatocellular Carcinoma Metastasis via Enhancing Invasiveness and Angiogenesis. J Exp Clin Cancer Res (2020) 39(1):20. doi: 10.1186/s13046-020-1529-9
123. Xue J, Liu Y, Luo F, Lu X, Xu H, Liu X, et al. Circ100284, via miR-217 Regulation of EZH2, is Involved in the Arseniteaccelerated Cell Cycle of Human Keratinocytes in Carcinogenesis. Biochim Biophys Acta Mol Basis Dis (2017) 1863(2017):753–63. doi: 10.1016/j.bbadis.2016.12.018
124. Zhang M, Li M, Li N, Zhang Z, Liu N, Han X, et al. miR-217 Suppresses Proliferation, Migration, and Invasion Promoting Apoptosis via Targeting MTDH in Hepatocellular Carcinoma. Oncol Rep (2017) 37:1772–8. doi: 10.3892/or.2017.5401
125. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as Functions as a Competitive Endogenous RNA to Promote Hepatocellular Carcinoma Progression. Aging (Albany NY) (2019) 11(19):8183–203. doi: 10.18632/aging.102312
126. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA Secreted From Adipocytes Promotes Thegrowth of Hepatocellular Carcinoma by Targeting Deubiquitination-Related USP7. Oncogene (2013) 38(15):2844–59. doi: 10.1038/s41388-018-0619-z
127. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-Transmitted Circular RNA Hsa_Circ_0051443 Suppresses Hepatocellular Carcinoma Progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
128. Qin L, Zhan Z, Wei C, Li X, Zhang T, Li J. Hsa-circRNA-G004213 Promotes Cisplatin Sensitivity by Regulating miR-513b-5p/PRPF39 in Liver Cancer. Mol Med Rep (2021) 23(6):421. doi: 10.3892/mmr.2021.12060
129. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer Cell-Derived Exosomal circUHRF1 Induces Natural Killer Cell Exhaustion and may Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5
130. Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal Circfblim1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3564
131. Guo S, Hu C, Zhai X, Sun D. Circular RNA 0006602 in Plasma Exosomes: A New Potential Diagnostic Biomarker for Hepatocellular Carcinoma. Am J Transl Res (2021) 13(6):6001–15.
132. Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin A, et al. The Diagnostic Value of Plasma Exosomal Hsa_Circ_0070396 for Hepatocellular Carcinoma. biomark Med (2021) 15(5):359–71. doi: 10.2217/bmm-2020-0476
133. Luo Y, Liu F, Gui R. High Expression of Circulating Exosomal circAKT3 Is Associated With Higher Recurrence in HCC Patients Undergoing Surgical Treatment. Surg Oncol (2020) 33:276–81. doi: 10.1016/j.suronc.2020.04.021
134. Wilusz JE, Sharp PA. A Circuitous Route to Noncoding RNA. Science (2013) 340:440–1. doi: 10.2307/41942637
135. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
136. Kubes P, Jenne C. Immune Responses in the Liver. Annu Rev Immunol (2018) 36:247–77. doi: 10.1146/annurev-immunol-051116-052415
137. Wang L, Wang FS. Clinical Immunology and Immunotherapy for Hepatocellular Carcinoma: Current Progress and Challenges. Hepatol Int (2019) 13(5):521–33. doi: 10.1007/s12072-019-09967-y
138. Chang Y, Jeong SW, Young Jang J, Jae Kim Y. Recent Updates of Transarterial Chemoembolilzation in Hepatocellular Carcinoma. Int J Mol Sci (2020) 21(21):8165. doi: 10.3390/ijms21218165
139. Lunt SY, Vander Heiden MG. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu Rev Cell Dev Biol (2011) 27:441–64. doi: 10.1146/annurev-cellbio-092910-154237
Keywords: exosomal non-coding RNAs, hepatocellular carcinoma, exosome, extracellular vesicles, diagnostic biomarkers, therapeutic target
Citation: Wang W, Hao L-P, Song H, Chu X-Y and Wang R (2022) The Potential Roles of Exosomal Non-Coding RNAs in Hepatocellular Carcinoma. Front. Oncol. 12:790916. doi: 10.3389/fonc.2022.790916
Received: 07 October 2021; Accepted: 26 January 2022;
Published: 24 February 2022.
Edited by:
Lipeng Qiu, Jiangsu University, ChinaReviewed by:
Giovanna Lucia Liguori, Institute of Genetics and Biophysics (CNR), ItalyCopyright © 2022 Wang, Hao, Song, Chu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Wang, d2FuZ3J1aTIxOEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.