- Department of Biotechnology, Assam University, Silchar, India
Background: Several studies have reported the role of circRNAs in the pathogenesis, diagnosis and prognosis of different cancers. This meta-analysis study aimed to evaluate the potential of using circRNAs as prognostic biomarkers of head and neck squamous cell carcinoma (HNSCC).
Methods: 816 relevant articles were retrieved from PubMed and Science Direct databases, out of which 17 met the inclusion criteria. These 17 studies were assessed for quality by the Newcastle-Ottawa Scale (NOS) system, and 9 high quality studies (NOS>7) were included in the meta-analysis. Cochran Q test and the I square (I2) metric were calculated to detect potential heterogeneity among studies. Sensitivity analysis was performed to validate the credibility of outcomes, and publication bias was determined using Begg’s funnel plot and Egger’s test. Hazard ratio (HR) and 95% Confidence Intervals (CIs) were used to evaluate overall survival (OS) of HNSCC patients by univariate and multivariate analyses.
Results: The dysregulated levels of 9 circRNAs (circPVT1, circCORO1C, circ_0000199, circCUX1, circPARD3, circMYC, circ_0102272, circ_0092125 and circ_00072387) were inversely related to OS of HNSCC patients [upregulated circRNA (univariate analysis: HR = 3.40, 95% CI: 2.66-4.36, p < 0.0001, I2 = 0%; multivariate analysis: HR = 3.33, 95% CI: 2.54-4.38, p < 0.0001, I2 = 0%), downregulated circRNA (univariate analysis: HR = 2.83, 95% CI: 1.73-4.65, p < 0.0001, I2 = 57.8%; multivariate analysis: HR = 2.35, 95% CI: 1.42-3.89, p = 0.0009, I2 = 0%)]. The individual HR for these 9 circRNAs indicated inverse relation to OS, validating the overall HR. The dyregulated levels of these circRNAs were also associated with poor clinicopathological outcomes such as primary tumor size, lymph node metastasis, distant metastasis and poor tumor (T), nodes (N), metastases (M); i.e TNM staging, and six of these circRNAs regulated diverse micro RNAs, revealing their role in tumorigenesis and cancer progression.
Conclusion: Nine different circRNAs dysregulated in HNSCC tumors may serve as potential prognostic markers of HNSCC. These markers are associated with reduced OS and poor clinicopathological outcomes of HNSCC patients. They are also involved in the pathogenesis and progression of HNSCC through diverse mechanisms.
Introduction
Head and neck cancers are a group of aggressive and genetically complex cancers arising from diverse anatomical sites of the head and neck (1). According to the GLOBOCAN 2020 database, out of an estimated 19.3 million new cancer cases, there were a cumulative number of 878348 (4.5%) new cases of cancer of the lip and oral cavity, larynx, nasopharynx, orpharynx, and hypopharynx reported in 2020, worldwide (2). The incidence rate of lip and oral cavity cancer (Age Standardized Rate 10.2 per 100,000) follows that of prostate and lung cancer as the leading cancers among males in the lower Human Development Index (HDI) countries, and is the leading cause of cancer-related death among males in India. The high incidence of cancers of the lip and oral cavity cancers in South Central Asia and Melanesia (Papua New Guinea) is largely attributed to the popularity of betel nut chewing in these regions. On the other hand, the high incidence rates in Eastern and Western Europe and in Australia/New Zealand have been attributed to alcohol consumption, tobacco smoking and HPV infection for cancers of the oropharyngeal region, and to ultraviolet radiation from sunlight exposure for lip cancer (2). While most of these cancers, collectively known as head and neck squamous cell carcinoma (HNSCC) are derived from the mucosal epithelium of the oral cavity, pharynx and larynx (1, 3), their heterogeneity at the molecular level has proved to be a major impediment in the identification of drug targets and targeted therapeutics (1). Thus, the 5-year overall survival (OS) rate of HNSCCs has remained approximately at 40–50% without much improvement over the past decade, largely due to poor availability of effective therapeutic options for HNSCC patients with recurrent disease (4). Additionally, locoregional recurrences occur in 15%-50% of patients with HNSCC and is a major factor contributing towards deaths because of the complexities associated with treatment of recurrent HNSCC, which include the effects of prior treatment on tumor cells and the infiltrative and the multifocal nature of HNSCC itself (5).
Majority of HNSCC patients present with advanced stage cancer (3) which is difficult to treat even using a combinatorial approach (1), thereby underscoring the significance of early detection. Careful physical examination and early recognition of oral pre-malignant lesions (OPLs) which present as leukoplakia (white patches) or erythroplakia (red patches), and may progress to invasive cancer, remain the cornerstone of the early detection of HNSCC (3). Indeed, identification of early, asymptomatic oral squamous cell carcinoma (OSCC) during routine oral examinations by community-based dentists and physicians followed by oral brush biopsy could increase the 5 year OS rate to a remarkable 94% (6). Prognosis of the cancer and determination of the likelihood of disease recurrence or progression is equally important. Thus, the recent years have seen the development of significant interest in the formulation of strategies based on specific markers and molecular signatures for early detection and/or clinical prognosis of HNSCC (4, 7), with non-invasive diagnostic testing holding particular appeal (7). Key amongst these are “liquid biopsies,” which involve analysis of body fluid samples such as blood or other accessible fluids like ascites, pleural effusions, saliva, or urine. Most liquid biopsy-based diagnostic tests for solid malignancies are based on blood, serum or plasma specimens, which are less invasive, and have the potential to detect HNSCC, tumor recurrences or metastases at initial stages (7, 8). The primary markers being investigated from liquid biopsies are circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs), tumor antigens, cell free coding and noncoding RNAs, metabolites as well as extracellular vesicles and exosomes (7, 8).
High throughput transcriptome analysis techniques have recently revealed circular RNAs (circRNAs) as potentially useful markers for the prognosis of cancer from body fluids. CircRNAs, also known as competitive endogenous RNAs (ceRNAs), are a large family of covalently closed, single-stranded, stable RNA molecules which regulate gene expression by regulating micro RNAs (miRNAs), binding with RNA-binding proteins (RBPs) or by getting directly translated into proteins. They exist stably and at high levels in body fluids, including plasma, serum, exosomes, and urine, and exhibit distinct expression patterns between patients with cancer and healthy controls (9, 10). Different circRNAs have been identified as functional prognostic biomarkers in lung, breast, colorectal, prostate, gastric, cervical, thyroid, bladder and ovarian cancers, hepatocellular carcinoma and esophageal squamous cell carcinoma (ESCC) (9, 11, 12). The circRNAs were found to be either significantly upregulated or downregulated in the body fluids of cancer patients in comparison to healthy controls with high discriminatory accuracy, and the expression level of most circRNAs were associated with tumour size, tumour differentiation, distant metastasis, and TNM stage (9).
The circular RNA, circPVT1, was proposed to play a pivotal and oncogenic role in the pathogenesis of HNSCC. CircPVT1 was found to be over-expressed in tumors compared to matched non-tumoral tissues, with particular enrichment in patients with TP53 mutations, and circPVT1 upregulation and downregulation were respectively associated with an increase or decrease in the malignant phenotype of HNSCC cell lines (13), indicating its relevance as a prognostic marker. Dysregulated circRNA expression has been reported in different types of HNSCC like ESCC, OSCC, laryngeal squamous cell carcinoma (LSCC), nasopharyngeal cancer and hypopharyngeal cancer (14–18).
In addition to the high heterogeneity of the molecular factors associated with HNSCC, inconsistencies are likely to arise among the findings of different studies on the potential of using circRNA as a biomarker of HNSCC due to variation in population ethnicity, sample size, specimens and control sources, which can become a hurdle to the application of circRNA in clinical practice (19). Therefore, we have performed a meta-analysis in order to investigate the relationship between the dysregulated expression of circRNAs and the prognosis and clinicopathological characteristics of HNSCC with the aim of determining whether circRNAs may be candidate biomarkers for HNSCC prognosis.
Methods
Literature Search Strategy
This study was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist (20). The studies were retrieved from PubMed, and ScienceDirect databases using the search terms “Circular RNA AND Head & Neck Cancer”, “circRNA in Head and Neck Cancer”, “Circular RNA AND Head & Neck Cancer biomarker”, “Circular RNA AND HNSCC”, “circRNA AND HNSCC”, “circRNA AND prognosis of HNSCC”. Studies published from 20th December 2017 to 10th September 2021 were included. The search was advanced using the term ‘AND’ to recover the desired outcomes. The potentially significant records were recovered from this search by all authors by surveying the title and abstract of each article, which were additionally inspected cautiously for meta-analysis eligibility. Furthermore, the reference lists of the retrieved articles were searched physically in order to recover more qualified studies for the meta-analysis. In addition to the articles screened for eligibility in meta-analysis, articles relevant for writing the Introduction, Methods and Discussion sections of this study were also included from the above mentioned and other sources.
Eligibility Criteria
The following inclusion criteria were predetermined for selecting articles eligible for meta-analysis (1) original research articles that reported data on the involvement of aberrantly expressed circRNAs in HNSCC patients; (2) articles reporting the link between circRNAs and survival outcomes in HNSCC patients; (3) studies on pathologically confirmed HNSCC (4) articles reporting the prognostic ability of circRNAs in HNSCC with available HR values and 95% confidence interval (CI); (5) studies that investigated the association between circRNAs and the clinicopathological features of HNSCC. The criteria for exclusion of articles were (1) articles not published in English (2) articles on the role of circRNAs in cancers other than HNSCC (3) reviews, basic studies, comments, meta-analyses, letters or case reports (4) low quality studies and (5) articles reporting the application of circRNAs in HNSCC diagnosis.
Data Extraction and Quality Assessment
All data was extracted from the included articles by two investigators (DR and MN) carefully and independently in order to retain accuracy of the data. The information collected from each study were: (1) basic information including author’s name, year of publication, number of cases, number of controls (2) clinicopathologic information such as circRNAs profiles, altered expression, specimen type, detection method and TNM stage (3) prognostic data including the univariate and multivariate HR values with 95% CI for OS.
Eligible studies retrieved after applying the inclusion and exclusion criteria were further screened for quality by the NOS (Newcastle-Ottawa Scale) system (Table 1). The NOS system is a semi-quantitative system for determining the quality of studies and comprises of three dimensions viz; selection, comparability, and study type, which are further subdivided into eight categories. The NOS ranges between 0 and 9 score, with NOS > 7 indicating high quality studies (35). Thus, only studies with NOS > 7 were finally selected for inclusion in meta-analysis.
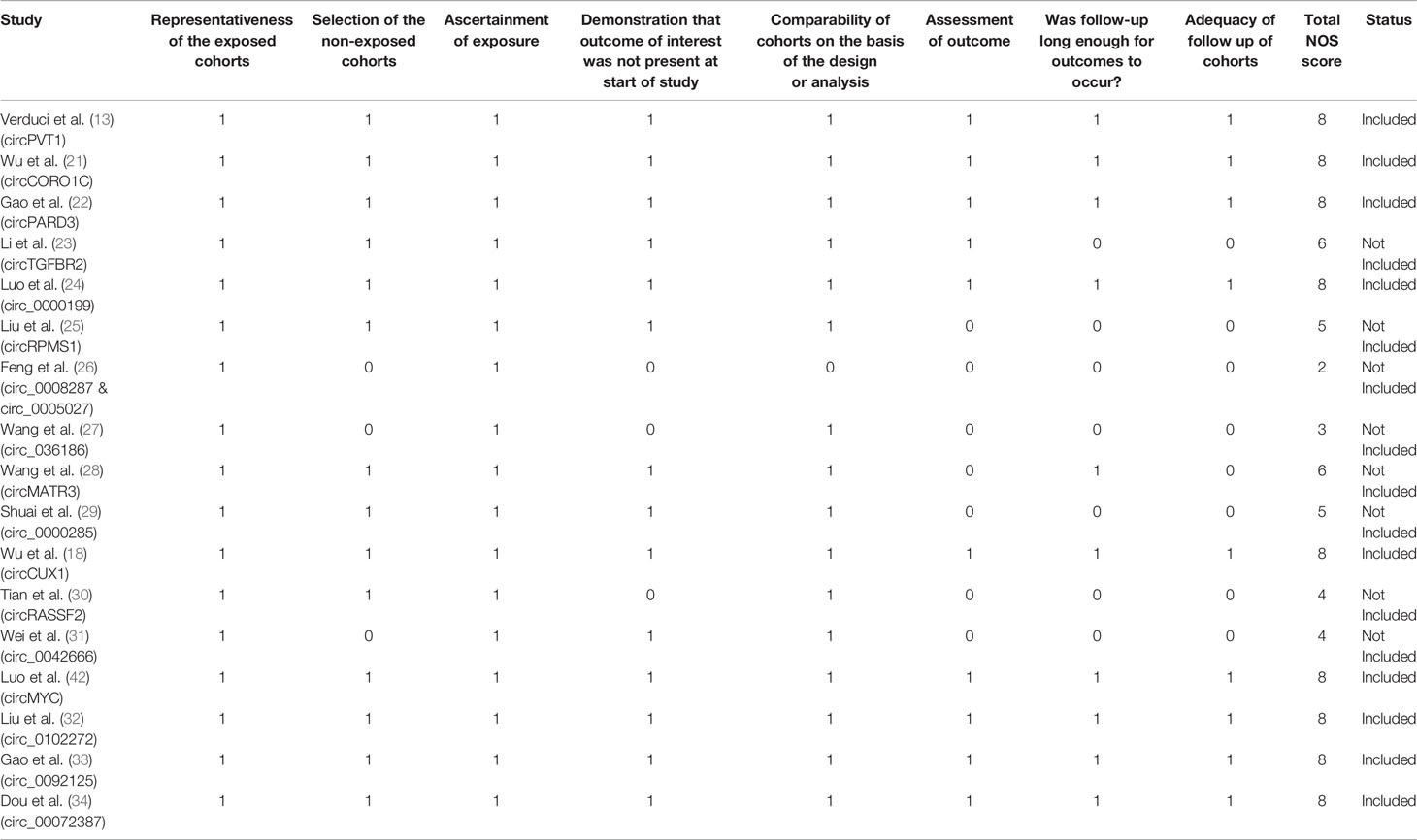
Table 1 Quality assessment of prognostic studies included on basis of the Newcastle-Ottawa Scale (NOS).
Statistical Analysis
Hazard ratio (HR) and 95% CIs were used to evaluate overall survival (OS) of HNSCC patients. Heterogeneity among the included studies for meta-analysis were examined by Cochran’s Q test (36), with P < 0.05 considered to be significant. The I2 statistic (37) was used to evaluate the percentage of the total variation due to heterogeneity, such that I2 values of 0% - 25% suggest no heterogeneity, while values of 25% - 50%, 50% - 75% and 75% - 100% indicate reasonable heterogeneity, huge heterogeneity and outrageous heterogeneity, respectively. In the presence of significant heterogeneity (Q statistic P < 0.05), the random effect model (38) was used to calculate the pooled HR, else the fixed effect model (39) was used. Sensitivity analysis was performed in order to validate the credibility of outcomes by chronological omission of individual studies. All investigations were performed by utilizing the StatsDirect statistical software (Version 2.7.2). Publication bias was determined using Begg’s funnel plot and Egger’s test (40, 41).
Results
Search Results
The methodology employed for literature search is shown in Figure 1. After a thorough search of PubMed and ScienceDirect databases using the mentioned search terms and a manual search through reference lists, a total of 816 records which appeared to be relevant to the role of circRNAs in HNSCC were retrieved. 84 articles were found to be relevant after reading the article titles and abstracts and removal of the duplicate articles (n=398) and irrelevant articles (n=334). These 84 articles were further screened on the basis of the inclusion and exclusion criteria to determine their eligibility for meta-analysis, and 67 articles were eliminated. The remaining 17 articles were further segregated as records reporting upregulated circRNAs and downregulated circRNAs in HNSCC, respectively. These 17 records were screened for quality by the NOS system, leading to the exclusion of 8 studies due to a low NOS score (NOS < 7) (Table 1). Finally, 7 high quality studies reporting upregulated circRNAs and 2 studies reporting downregulated circRNAs were included in the quantitative meta-analysis.
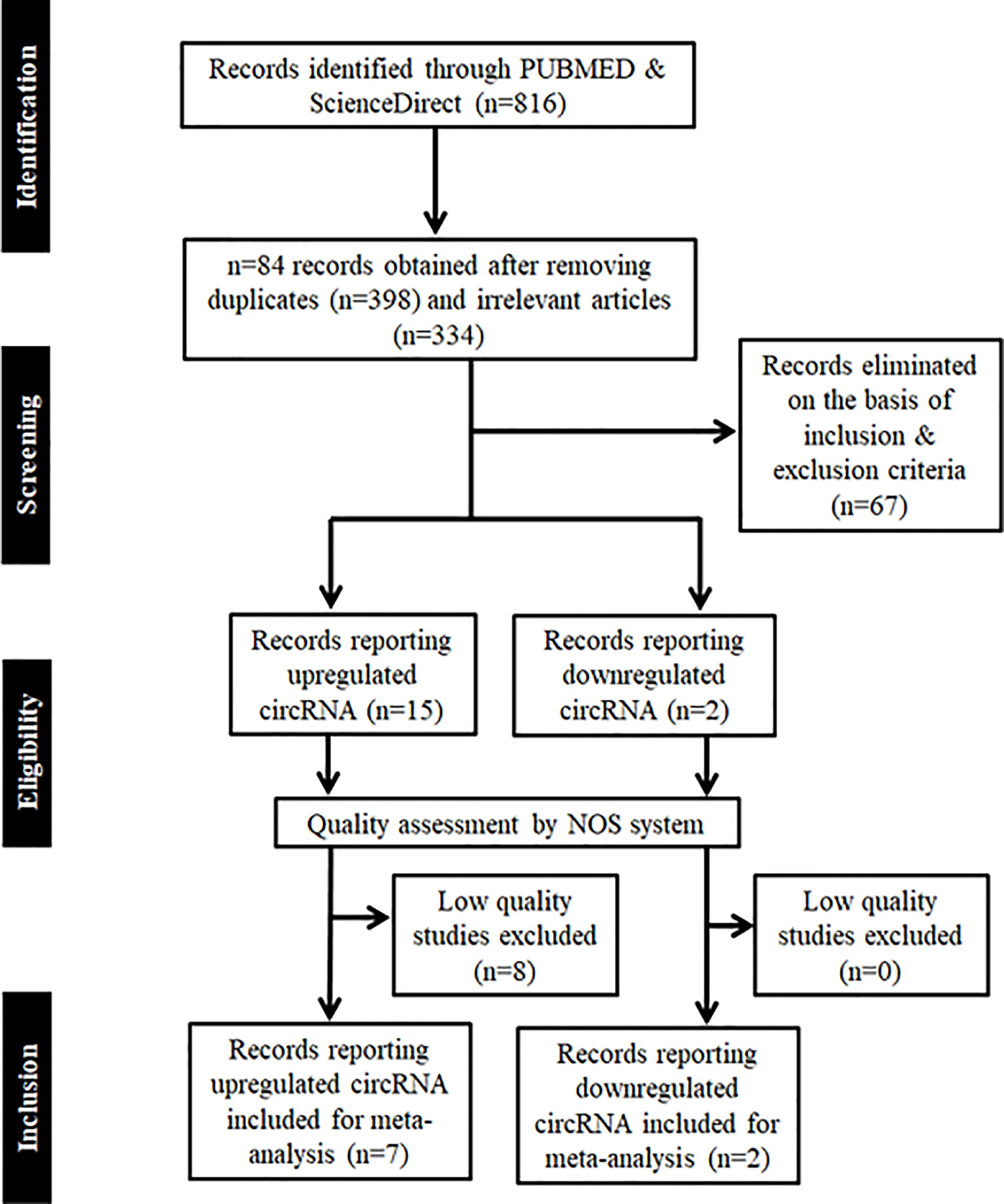
Figure 1 PRISMA flow chart showing the methodology employed for literature retrieval for meta-analysis. Records were screened on the basis of inclusion and exclusion criteria, whose quality was further assessed by NOS system.
In addition, 8 articles were included as references to the methods used for meta-analysis and 25 articles were included for writing the introduction and discussion sections of this article.
Characteristics of Included Studies
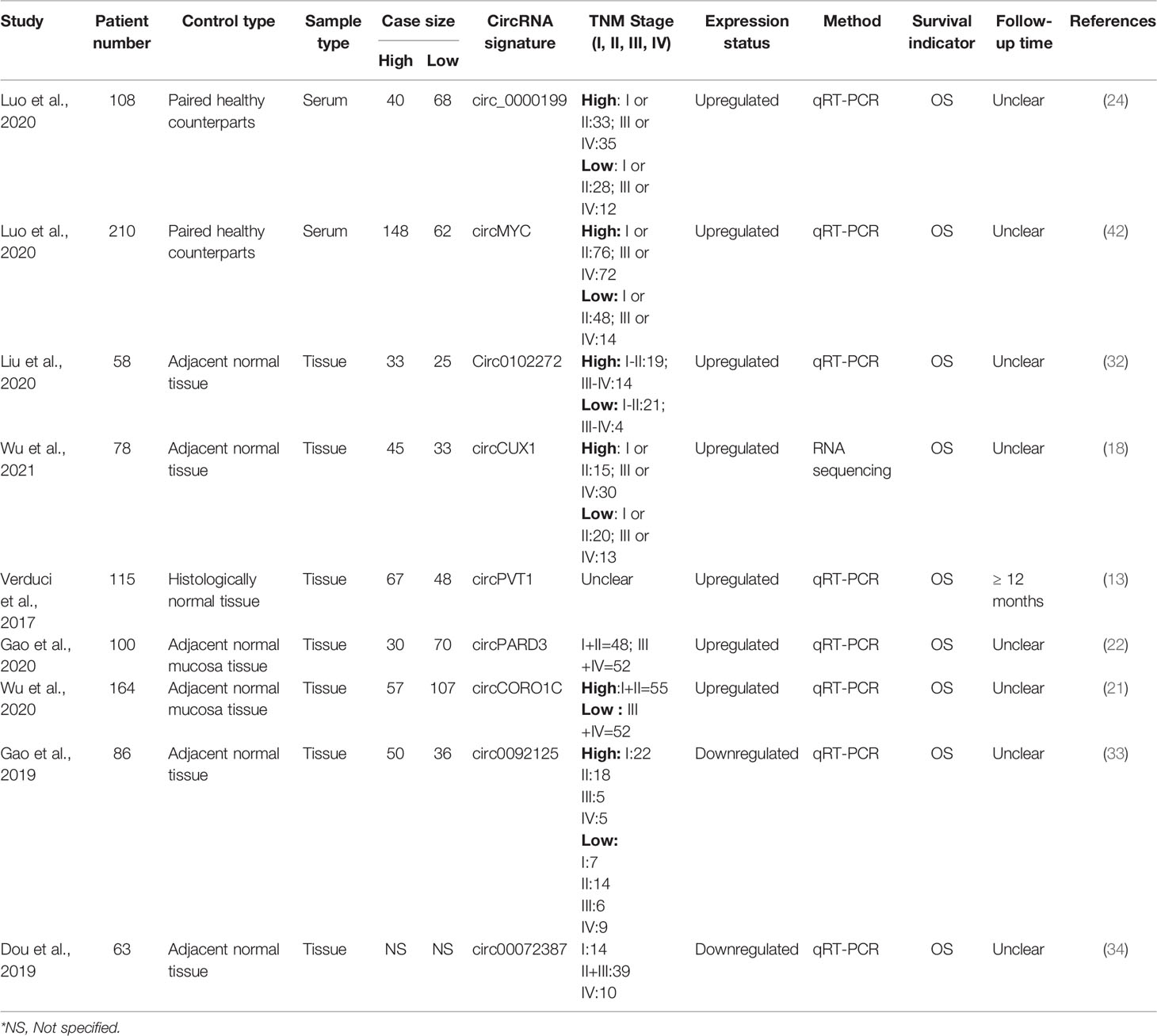
The NOS was used to evaluate the quality of the included studies and the nine included studies received high scores (NOS>7) (Table 1) indicating a high quality of these studies, and ensuring the reliability of the overall result. The baseline data from the 9 included studies is shown in Table 2. Collectively, these studies comprised a total number of 982 patients who were pathologically confirmed as suffering from HNSCC. These circRNAs were detected by the techniques of RNA Sequencing (18) or quantitative polymerase chain reaction, qPCR (13, 21, 22, 24, 32–34, 42). All of these circular RNAs were dysregulated in cancer tissue and were associated with poor clinicopathological outcomes such as primary tumor size, lymph node metastasis, distant metastasis and tumor (T), nodes (N), metastases (M); i.e TNM stage. Upregulated levels of circCUX1 in hypopharyngeal cancer tissue in comparison to adjacent normal tissue correlated significantly with primary tumor size, lymph node metastasis, distant metastasis and TNM stage (18). High circPARD3 levels in LSCC tissue in comparison to adjacent normal mucosa positively correlated with T stage, N stage and clinical stages, and increased progressively from T1 to T4 (22). Similarly, OSCC patients with higher exosomal circ_0000199 had higher tumor recurrence rate and higher mortality than patients with lower levels of exosomal circ_0000199 (24). Wu et al., reported that circCORO1C expression was relatively abundant in LSCC than in adjacent normal mucosa, and was significantly correlated with T stage, N stage and clinical stage (21). In thyroid cancer patients, high levels of circ_0102272 was associated with lymph node metastasis along with advanced clinical stage indicating shorter overall survival in comparison to patients with low expression of circ_0102272 (32). Exosomal circMYC acts as an independent predictor of nasopharyngeal carcinoma patient survival as its high expression is closely associated with tumor size, lymph node metastasis and TNM stage (42). Apart from upregulated circRNAs, the downregulation of two circRNAs (circ_0092125, circ_0072387) were significantly correlated with tumor size, TNM stage, and lymph node metastasis in OSCC patients (33, 34). Kaplan- Meir analysis also revealed a correlation between the upregulation of these seven circRNAs (circPVT1, circCOROC1, circPARD3, circ_0000199, circ_0102272, circMYC and circCUX1) (13, 18, 21, 22, 24, 32, 42) and downregulation of two circRNAs (circ_0092125, circ_0072387) (33, 34) with inferior OS of patients of HNSCC patients.
Meta-Analysis for Overall Survival
Our meta-analysis indicates that the upregulated levels of 7 circRNAs (circ_0000199, circ MYC, circ_0102272, circCUX1, circPVT1, circPARD3 and circCORO1C) were inversely related to OS of HNSCC patients (univariate analysis: HR = 3.40, 95% CI: 2.66-4.36, p < 0.0001, I2 = 0%; multivariate analysis: HR = 3.33, 95% CI: 2.54-4.38, p < 0.0001, I2 = 0%). Also, the individual HR for these 7 circRNAs [circ_0000199 (univariate analysis: HR=3.07, 95% CI: 1.95-4.72; multivariate analysis: HR=3.57, 95% CI: 2.48-6.24), circ MYC (univariate analysis: HR=4.24, 95% CI: 2.08-6.64; multivariate analysis: HR=4.16, 95% CI: 2.52-7.06), circ 0102272 (univariate analysis: HR=2.99, 95% CI: 1.62-9.72; multivariate analysis: HR=2.24, 95% CI: 1.42-7.55), circCUX1 (univariate analysis: HR=3.87, 95% CI: 1.94-6.24; multivariate analysis: HR=2.83, 95% CI: 1.74-5.02), circPVT1 (univariate analysis: HR=2.10, 95% CI: 0.87-4.95), circPARD3 (HR=3.93, 95% CI:1.72-8.99), circCORO1C (HR=3.64, 95% CI: 1.54-8.62), indicated inverse relation to OS of HNSCC patients validating the overall HR calculated for both univariate (Figure 2A) and multivariate (Figure 2B) analyses. The variables used for multivariate analyses were areca nut use (24), TNM stage (18, 24, 42), lymphatic metastasis (18, 24, 32, 42), tumor size (18, 24, 42) and distant metastasis (18). Finally, the statistical results show that the various circRNAs had no significant heterogeneity (I2 = 0%, p > 0.05) and thus, a fixed-effect model was used for both the analyses.
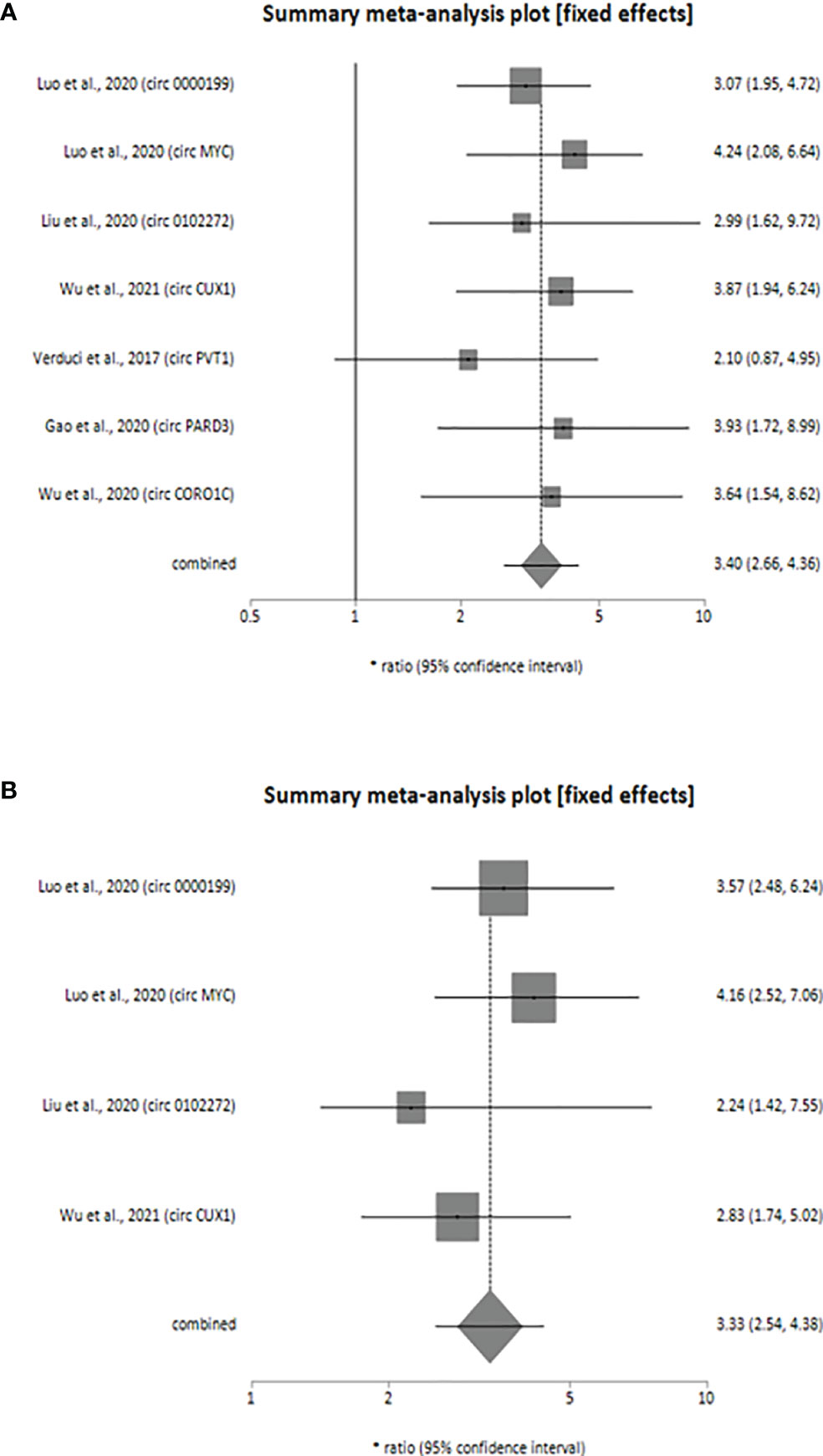
Figure 2 Forest plot showing the combined HR along with individual HR values at 95% CI for the upregulated circRNA profiles in predicting the overall survival of patients with HNSCC (A) univariate analysis, (B) multivariate analysis. Square shapes indicate the effect size of each upregulated circRNA profile, while diamond shapes indicate the overall effect size. The 95% CI of each effect sizes are indicated by respective bars.
Additionally, downregulation of 2 circRNAs (circ_0092125 and circ_00072387) was associated with poor prognosis of HNSCC patients as revealed by our meta-analysis (univariate analysis: HR=2.83, 95% CI: 1.73-4.65, p < 0.0001, I2 = 57.8%; multivariate analysis: HR=2.35, 95% CI: 1.42-3.89, p < 0.0009, I2 = 0%). The variables used for multivariate analyses were TNM stage (33, 34), tumor size (33) and lymph node metastasis (33). Like upregulated circRNAs, the individual HR for these 2 circRNAs [circ_0092125 (univariate analysis: HR=5.25, 95% CI: 2.07-13.30; multivariate analysis: HR=3.02, 95% CI: 1.08-8.45), circ_00072387 (univariate analysis: HR=2.22, 95% CI: 1.23-3.98; multivariate analysis: HR=2.17, 95% CI: 1.37-4.35) also indicated poor prognosis of HNSCC patients, validating the overall HR calculated for both univariate (Figure 3A) and multivariate (Figure 3B) analyses. The statistical results show that the circRNAs had no significant heterogeneity (univariate analysis: I2 = 57.8% p > 0.05, multivariate analysis: I2 = 0%, p > 0.05) and thus, a fixed-effect model was used for both the analyses.
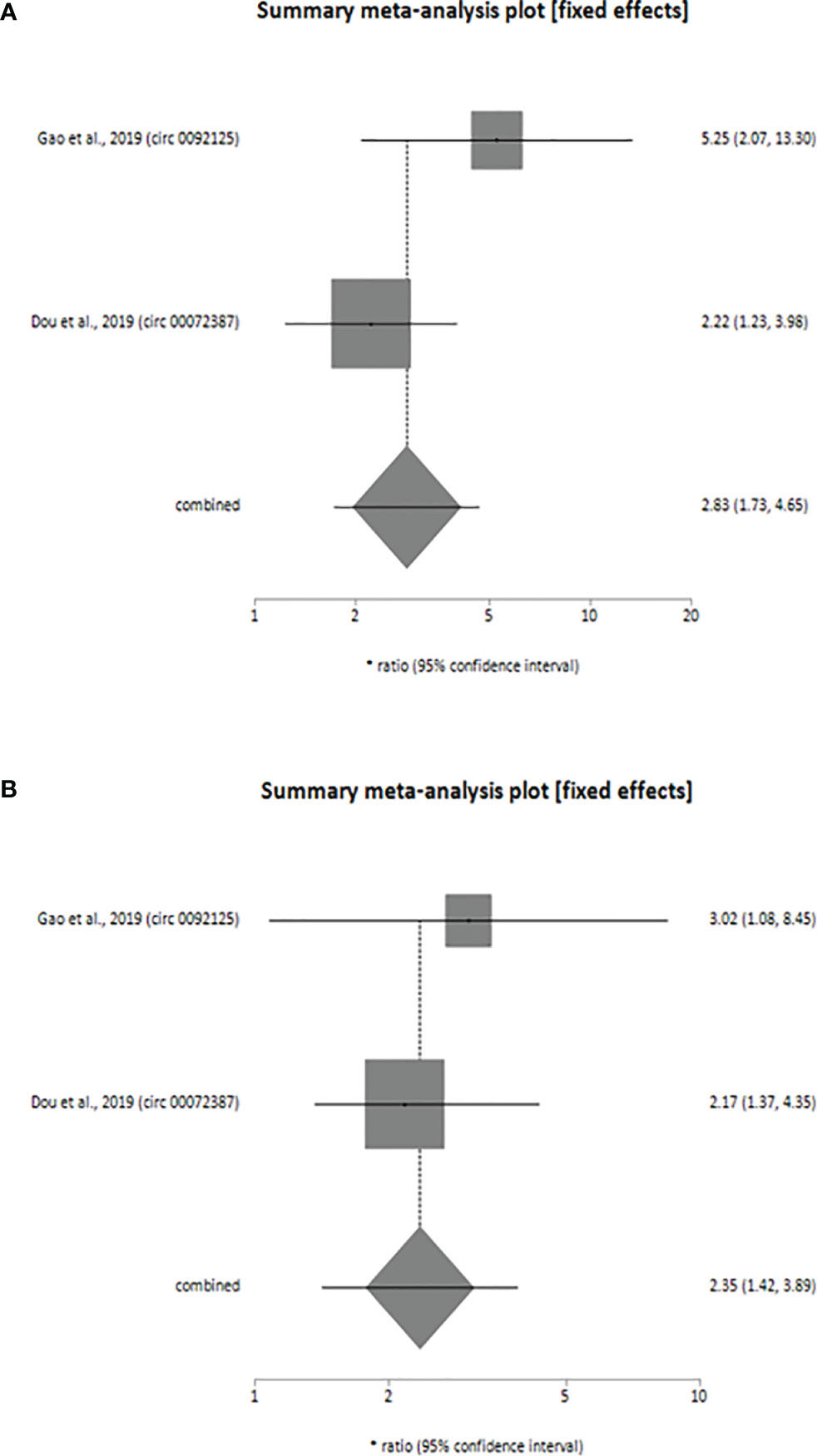
Figure 3 Forest plot showing the combined HR along with individual HR values at 95% CI for the downregulated circRNA profiles in predicting the overall survival of patients with HNSCC (A) univariate analysis, (B) multivariate analysis. Square shapes indicate the effect size of each upregulated circRNA profile, while diamond shapes indicate the overall effect size. The 95% CI of each effect sizes are indicated by respective bars.
Sensitivity Analysis and Publication Bias
Sensitivity analysis was performed for studies reporting upregulated level of circRNAs in HNSCC patients in order to determine the impact of individual studies on the pooled HR obtained by both univariate (Figure 4) and multivariate (Figure 5) analyses, and to validate whether any single study skewed the combined HR significantly. When the included studies of univariate analysis, namely Luo et al., 2020 (circ 0000199), Verduci et al., 2017 (circ PVT1), Luo et al., 2020 (circ MYC), Liu et al., 2020 (circ 0102272), Gao et al., 2020 (circ PARD3), Wu et al., 2020 (circ CORO1C) and Wu et al., 2021 (circ CUX1) were excluded one at a time, the combined HR values obtained (3.57, 3.55, 3.24, 3.44, 3.36, 3.38 and 3.31 respectively) did not indicate significant misrepresentation by any individual study in comparison to the overall HR value (HR = 3.40) (Figure 4). Similarly, sequential omission of included studies of multivariate analysis, namely, Luo et al., 2020 (circ 0000199), Luo et al., 2020 (circ MYC), Liu et al., 2020 (circ 0102272) and Wu et al., 2021 (circ CUX1) resulted in combined HR values (HR = 3.21, 3.06, 3.50 and 3.54 respectively) which did not skew the overall HR value (HR = 3.33) (Figure 5). Sensitivity analysis was not performed for meta-analysis conducted for downregulated circRNAs as only two studies met the inclusion criteria, and individual HR values of both the studies did not skew the combined HR (Figures 3A, B). Results based on Begg’s funnel plots show that there was no significant publication bias in both the analyses for upregulated circRNAs [univariate analysis (Begg’s test: p = 0.13, Egger’s test: p = 0.74), multivariate analysis (Begg’s test: p = 0.08, Egger’s test: p = 0.27) (Figure 6). However, Begg’s funnel plot and Egger’s test value could not be obtained for studies reporting downregulated circRNAs due to the small number of studies (the minimum number of studies required to generate a funnel plot is four).
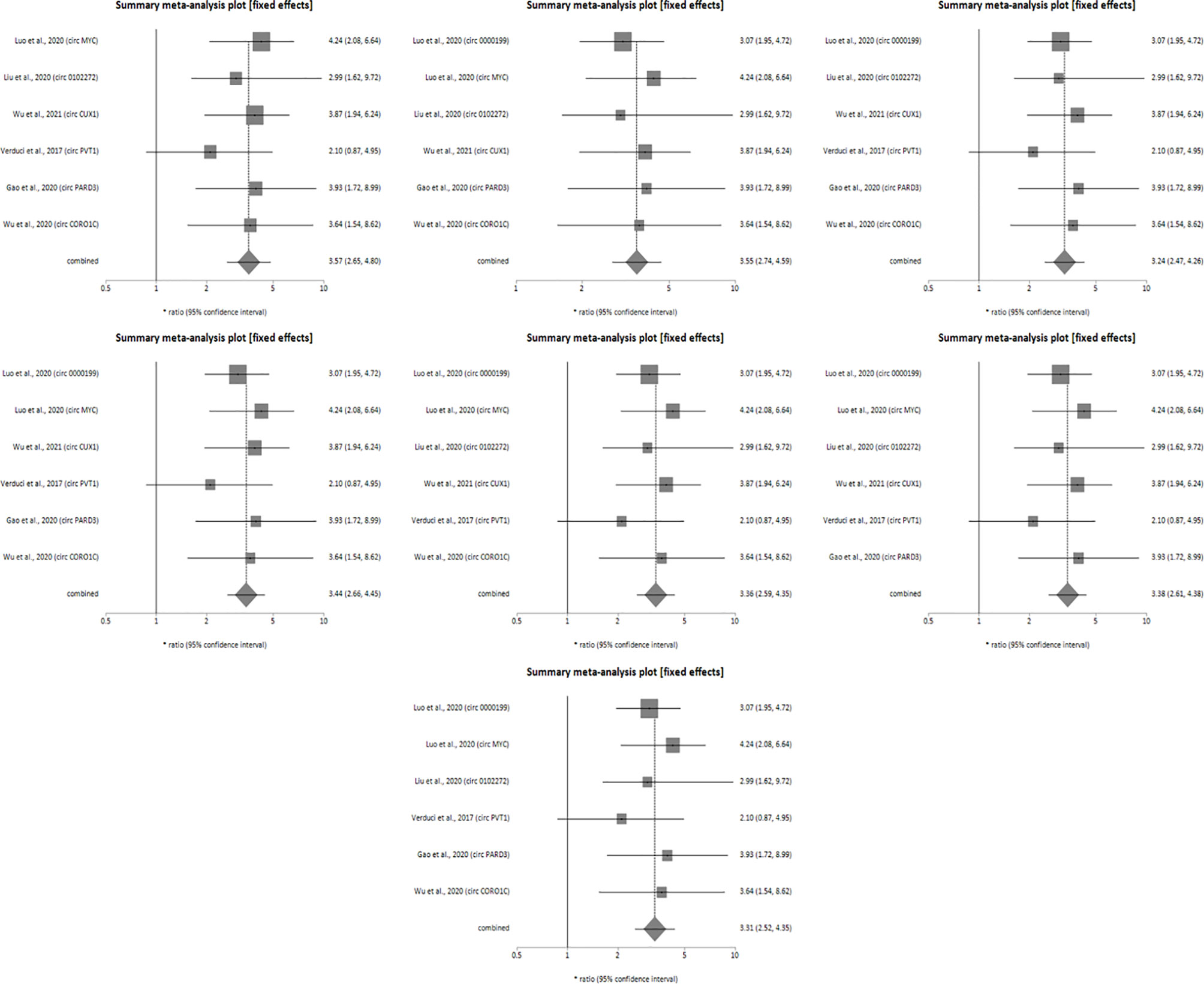
Figure 4 Forest plots of sensitivity analysis showing the combined HR values at 95% CI upon sequential deletion of the studies included in univariate analysis which reported upregulated circRNAs. Square shapes indicate the effect size of each upregulated circRNA profile, while diamond shapes indicate the overall effect size. The 95% CI of each effect sizes are indicated by respective bars.
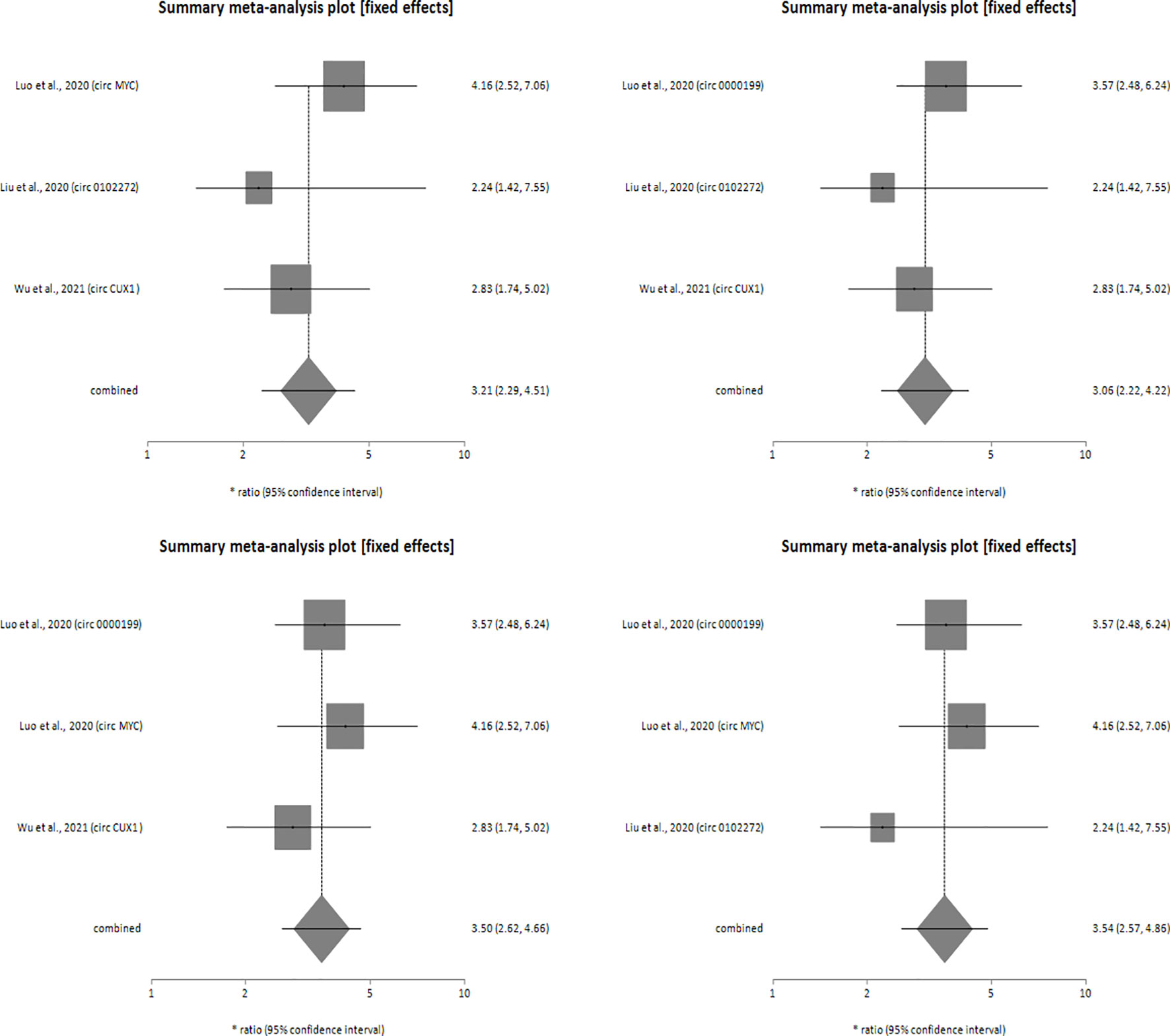
Figure 5 Forest plots of sensitivity analysis showing the combined HR values at 95% CI upon sequential deletion of the studies included in multivariate analysis which reported upregulated circRNAs. Square shapes indicate the effect size of each upregulated circRNA profile, while diamond shapes indicate the overall effect size. The 95% CI of each effect sizes are indicated by respective bars.

Figure 6 Begg’s funnel plot showing publication bias of upregulated (A) univariate analysis & (B) multivariate analysis. Each dot of the Begg’s funnel plot represents a separate study. The two border lines of the plot represent the pseudo 95% confidence intervals against their corresponding standard errors and the middle solid line indicates the overall effect from the meta-analysis.
Discussion
The therapies for HNSCC currently available and administered alone or in combination are radiotherapy, chemotherapy, targeted drugs and immunotherapy. While radiotherapy and chemotherapy remain highly toxic, the efficacy of EGFR inhibitors which are the only approved targeted drugs, is limited by inherent and acquired resistance (1). While newer strategies have contributed towards a significant improvement of the quality of life in the past two decades (43), there is no improvement in OS rates of patients in recent decades. Thus, an improved understanding of the molecular mechanisms underlying HNSCC is necessary to enhance therapeutic efficacy. Besides this, the requirement to identify patients who are more susceptible to metastases and recurrences has led to a pressing need for more compelling biomarkers for the identification of individuals who are at higher risk. This indicates the critical requirement of novel biomarkers to more precisely predict outcome of HNSCCs and help in the decision of therapeutic strategies. Studies indicate that circRNA play a vital role in the regulation of various molecular events which determine the occurrence, progression, and metastasis of various malignant tumors (44). Studies have also demonstrated that circRNAs are relatively stable because of their covalently closed circular structures with neither 5’ caps nor 3’ tails; consequently, most circRNAs are resistant to RNA exonucleases, and are more stable when compared with linear transcripts (45), making them good candidates for biomarkers.
A number of meta-analysis studies have established circRNA as a prognostic biomarker for cancer (19), such as hepatocellular carcinoma (46), colorectal cancer (47) and breast cancer (48). However, to the best of our knowledge, no meta-analysis has been performed to determine the prognostic significance of circRNAs in HNSCCs. In performing this meta-analysis, we included 9 studies with a total of 982 HNSCC cases in order to uncover the connection between the expression of different types of circular RNA and their clinicopathological and prognostic significance in HNSCC. Our results indicate that in HNSCC patients, the high expressions of seven circRNAs (circPVT1, circCOROC1, circPARD3, circ_0000199, circMYC, circ_0102272 and circCUX1) and low expressions of two circRNAs (circ0092125 and circ00072387) in tumors, in comparison to the levels in adjacent normal tissue were associated with poor clinicopathological features such as poor TNM staging, histological grading, lymph node metastasis, and recurrence in HNSCC and showed inferior OS [increased circRNA expression (univariate analysis: HR = 3.40, 95% CI: 2.66-4.36, p < 0.0001, I2 = 0%; multivariate analysis: HR = 3.33, 95% CI: 2.54-4.38, p < 0.0001, I2 = 0%), decreased circRNA expression (univariate analysis: HR = 2.83, 95% CI: 1.73-4.65, p < 0.0001, I2 = 57.8%; multivariate analysis: HR = 2.35, 95% CI: 1.42-3.89, p = 0.0009, I2 = 0%)].
The miRNAs that were targeted by these circRNAs were also identified in six of the nine included studies, revealing the mechanisms through which the circRNAs were involved in tumorigenesis and cancer progression. CircPARD3 acts as a sponge of the tumor-suppressor miR-145-5p in LSCC (22), while circCORO1C sponges let-7c-7p whose normal function is to inhibit the proliferation, migration and invasion of LSCC cells and to promote their apoptosis (21). Circ0000199 and circ0072387 interact simultaneously with the tumor suppressor miR-145-5p & miR-29b-3p and hsa-miR-129-3p, hsa-miR-141-3p, & hsa-miR-29-3p respectively in OSCC cells thereby regulating cell proliferation, growth and differentiation (24, 34). In nasopharyngeal carcinoma, circMYC is reported to interact with miR-20b-5p and let-7e-5p simultaneously, which in turn target argonaute RISC component 1 (AGO1) and cryptochrome circadian regulator 2 (CRY2) (42). Both AGO1 and CRY2 tightly regulate cell development, proliferation and migration (49, 50). Finally, circPVT1 binds to miR-497-5p through a specific binding site and affects its expression, leading to downregulation of miR-497-5p in HNSCC tumors with mutant p53 (13). The downregulation of both circ_00072387 and circ_0092125 was reported to significantly affect tumor size, TNM stage, and lymph node metastasis in OSCC patients by interacting with respective miRNAs (33, 34). Circ_00072387 binds with multiple miRNAs such as miR-129-3p, miR-141-3p, and miR-29-3p which regulate several tumor related pathways viz; MAPK signaling pathway, Ras signaling pathway, and Hippo signaling pathway (34). Similarly, circ_0092125 interacts with miR_1184, miR_1205, and miR_1322 which are reported to be involved in the occurrence and progression of OSCC (33). In nasopharyngeal carcinoma, interaction of circMYC with miR-20b-5p and let-7e-3p regulates tumor development as both these miRNAs exhibit tumor suppressor functions in a variety of cancers (42). Luo et al., 2020 also reported that miR-20b-5p and let-7e-3p jointly target AGO1 and CRY2. Various reports indicate the crucial role played by AGO1 in solid tumors and the involvement of CRY2 in epithelial-mesenchymal transition, Akt signaling and p53 signaling (42).
In addition, some of the included studies also revealed other molecular mechanisms through which the identified circRNAs contributed towards the cancer phenotype. The upregulation of circCUX1 was associated with radiotherapy resistance of hypopharyngeal tumors attributed to the binding of circCUX1 to the 3’UTR-of caspase-1, thereby inhibiting its expression and resulting in a decrease in inflammatory factors, thus leading to tolerance to radiotherapy (18). Similarly, circPARD3 promoted the proliferation, migration, invasion and chemoresistance of LSCC by inhibiting autophagy via the PRKC1-Akt-mTOR pathway (22). CircCORO1C promoted the proliferation, migration and invasion of LSCC cells by specifically upregulating the expression of the target PBX3 gene and affecting the epithelial mesenchymal transition (EMT) process. Indeed, dysregulation of PBX3 has also been reported in prostate, gastric, cervical and liver cancer (21). CircPVT1 was significantly upregulated in tumors with mutant p53 compared to those with wild type p53, and only in HNSCC patients with p53 mutations. In fact, the expression of circPVT1 was found to be regulated at the transcriptional level by the mut-p53/YAP/TEAD complex which resides on the promoter of circPVT1, such that increased expression of mutant p53, YAP and TEAD1 can increase the nascent circPVT1 expression. Simultaneously, circPVT1 might act within a positive self-regulating loop, controlling and enhancing its own nuclear expression, and thus regulating tumorigenesis (13).
Limitations: Our current study has some limitations which should be considered: (a) Based on our inclusion criteria, eight out of nine studies were only from Asian population (b) The sample size and number of the enrolled studies in this analysis were relatively small (c) Studies with good quality diagnostic data could not be retrieved because of which it was not possible to determine the potential of circRNAs as diagnostic markers of HNSCC (d) Begg’s funnel plot and Egger’s test value could not be generated for studies reporting downregulation of circRNAs in HNSCC due to the small number of studies. Thus, more high-quality studies on larger sample sizes and diverse population groups are required in order to firmly establish circRNAs as a good prognostic marker of HNSCC.
Conclusion
Taken together, our meta-analysis indicates that the nine circRNAs (circPVT1, circCOROC1, circPARD3, circ_0000199, circMYC, circ_0102272, circCUX1, circ_0092125 and circ_00072387) are potential prognostic biomarkers of HNSCC.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
MN and DR contributed to the design of study, performed the statistical analyses and drafted the manuscript. YC designed the study, provided intellectual input, approved the protocols to be followed in the study and drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Alsahafi E, Begg K, Amelio I, Raulf N, Lucarelli P, Sauter T, et al. Clinical Update on Head and Neck Cancer: Molecular Biology and Ongoing Challenges. Cell Death Dis (2019) 10(8):1–17. doi: 10.1038/s41419-019-1769-9
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and Neck Squamous Cell Carcinoma. Nat Rev Dis Primers (2020) 6(1):1–22. doi: 10.1038/s41572-020-00224-3
4. Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, et al. Heterogeneity of the Head and Neck Squamous Cell Carcinoma Immune Landscape and its Impact on Immunotherapy. Front Cell Dev Biol (2019) 7:52. doi: 10.3389/fcell.2019.00052
5. Chang J-H, Wu C-C, Yuan KS-P, Wu AT, Wu S-Y. Locoregionally Recurrent Head and Neck Squamous Cell Carcinoma: Incidence, Survival, Prognostic Factors, and Treatment Outcomes. Oncotarget (2017) 8(33):55600. doi: 10.18632/oncotarget.16340
6. Sciubba JJ, Larian B. Oral Squamous Cell Carcinoma: Early Detection and Improved 5-Year Survival in 102 Patients. Gen Dent (2018) 66(6):e11–6.
7. Spector ME, Farlow JL, Haring CT, Brenner JC, Birkeland AC. The Potential for Liquid Biopsies in Head and Neck Cancer. Discov Med (2018) 25(139):251.
8. Kong L, Birkeland AC. Liquid Biopsies in Head and Neck Cancer: Current State and Future Challenges. Cancers (Basel) (2021) 13(8):1874. doi: 10.3390/cancers13081874
9. Wang M, Zhang L, Ren W, Li S, Zhi K, Zheng J, et al. Diagnostic Value of CircRNAs as Potential Biomarkers in Oral Squamous Cell Carcinoma: A Meta-Analysis. Front Oncol (2021) 11. doi: 10.3389/fonc.2021.693284
10. Khan S, Jha A, Panda AC, Dixit A. Cancer-Associated circRNA–miRNA–mRNA Regulatory Networks: A Meta-Analysis. Front Mol Biosci (2021) 8:337. doi: 10.3389/fmolb.2021.671309
11. Li J, Xu Q, Huang Z-J, Mao N, Lin Z-T, Cheng L, et al. CircRNAs: A New Target for the Diagnosis and Treatment of Digestive System Neoplasms. Cell Death Dis (2021) 12(2):1–13. doi: 10.1038/s41419-021-03495-0
12. Brown JR, Chinnaiyan AM. The Potential of Circular RNAs as Cancer Biomarkers. Cancer Epidemiol Prev Biomarkers (2020) 29(12):2541–55. doi: 10.1158/1055-9965.EPI-20-0796
13. Verduci L, Ferraiuolo M, Sacconi A, Ganci F, Vitale J, Colombo T, et al. The Oncogenic Role of Circpvt1 in Head and Neck Squamous Cell Carcinoma Is Mediated Through the Mutant P53/YAP/TEAD Transcription-Competent Complex. Genome Biol (2017) 18(1):1–24. doi: 10.1186/s13059-017-1368-y
14. Kristensen L, Hansen T, Venø M, Kjems J. Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene (2018) 37(5):555–65. doi: 10.1038/onc.2017.361
15. Li B, Wang F, Li X, Sun S, Shen Y, Yang H. Hsa_circ_0008309 May Be a Potential Biomarker for Oral Squamous Cell Carcinoma. Dis Markers (2018) 2018. doi: 10.1155/2018/7496890
16. Guo Y, Yang J, Huang Q, Hsueh C, Zheng J, Wu C, et al. Circular RNAs and Their Roles in Head and Neck Cancers. Mol Cancer (2019) 18(1):1–18. doi: 10.1186/s12943-019-1003-5
17. Shuai M, Huang L. High Expression of Hsa_circRNA_001387 in Nasopharyngeal Carcinoma and the Effect on Efficacy of Radiotherapy. Onco Targets Ther (2020) 13:3965. doi: 10.2147/OTT.S249202
18. Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, et al. N6-Methyladenosine Modification of Circcux1 Confers Radioresistance of Hypopharyngeal Squamous Cell Carcinoma Through Caspase1 Pathway. Cell Death Dis (2021) 12(4):1–13. doi: 10.1038/s41419-021-03558-2
19. Li J, Li H, Lv X, Yang Z, Gao M, Bi Y, et al. Diagnostic Performance of Circular RNAs in Human Cancers: A Systematic Review and Meta-Analysis. Mol Genet Genomic Med (2019) 7(7):e00749. doi: 10.1002/mgg3.749
20. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PloS Med (2009) 6(7):e1000097. doi: 10.1371/journal.pmed.1000097
21. Wu Y, Zhang Y, Zheng X, Dai F, Lu Y, Dai L, et al. Circular RNA Circcoro1c Promotes Laryngeal Squamous Cell Carcinoma Progression by Modulating the let-7c-5p/PBX3 Axis. Mol Cancer (2020) 19:1–18. doi: 10.1186/s12943-020-01215-4
22. Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, et al. circPARD3 Drives Malignant Progression and Chemoresistance of Laryngeal Squamous Cell Carcinoma by Inhibiting Autophagy Through the PRKCI-Akt-mTOR Pathway. Mol Cancer (2020) 19(1):1–22. doi: 10.1186/s12943-020-01279-2
23. Li W, Lu H, Wang H, Ning X, Liu Q, Zhang H, et al. Circular RNA TGFBR2 Acts as a ceRNA to Suppress Nasopharyngeal Carcinoma Progression by Sponging miR-107. Cancer Lett (2021) 499:301–13. doi: 10.1016/j.canlet.2020.11.001
24. Luo Y, Liu F, Guo J, Gui R. Upregulation of circ_0000199 in Circulating Exosomes is Associated With Survival Outcome in OSCC. Sci Rep (2020) 10(1):1–9. doi: 10.1038/s41598-020-70747-y
25. Liu Q, Shuai M, Xia Y. Knockdown of EBV-Encoded circRNA Circrpms1 Suppresses Nasopharyngeal Carcinoma Cell Proliferation and Metastasis Through Sponging Multiple miRNAs. Cancer Manag Res (2019) 11:8023–31. doi: 10.2147/CMAR.S218967
26. Feng C, Li Y, Lin Y, Cao X, Li D, Zhang H, et al. CircRNA-Associated ceRNA Network Reveals ErbB and Hippo Signaling Pathways in Hypopharyngeal Cancer. Int J Mol Med (2019) 43(1):127–42. doi: 10.3892/ijmm.2018.3942
27. Wang WL, Yang Z, Zhang YJ, Lu P, Ni YK, Sun CF, et al. Competing Endogenous RNA Analysis Reveals the Regulatory Potency of circRNA_036186 in HNSCC. Int J Oncol (2018) 53(4):1529–43. doi: 10.3892/ijo.2018.4499
28. Wang Z, Wei P, Wei D, Cao S, Liu H, Chen L, et al. Effect of Up-Regulation of Circmatr3 on the Proliferation, Metastasis, Progression and Survival of Hypopharyngeal Carcinoma. J Cell Mol Med (2020) 24(8):4687–97. doi: 10.1111/jcmm.15134
29. Shuai M, Hong J, Huang D, Zhang X, Tian Y. Upregulation of circRNA_0000285 Serves as a Prognostic Biomarker for Nasopharyngeal Carcinoma and is Involved in Radiosensitivity. Oncol Lett (2018) 16(5):6495–501. doi: 10.3892/ol.2018.9471
30. Tian L, Cao J, Jiao H, Zhang J, Ren X, Liu X, et al. CircRASSF2 Promotes Laryngeal Squamous Cell Carcinoma Progression by Regulating the miR-302b-3p/IGF-1R Axis. Clin Sci (Lond) (2019) 133(9):1053–66. doi: 10.1042/CS20190110
31. Wei Z, Chang K, Fan C. Hsa_circ_0042666 Inhibits Proliferation and Invasion via Regulating miR-223/TGFBR3 Axis in Laryngeal Squamous Cell Carcinoma. BioMed Pharmacother (2019) 119:109365. doi: 10.1016/j.biopha.2019.109365
32. Liu J, Zheng X, Liu H. Hsa_circ_0102272 Serves as a Prognostic Biomarker and Regulates Proliferation, Migration and Apoptosis in Thyroid Cancer. J Gene Med (2020) 22(9):e3209. doi: 10.1002/jgm.3209
33. Gao L, Wang Q-B, Zhi Y, Ren W-H, Li S-M, Zhao C-Y, et al. Down-Regulation of hsa_circ_0092125 is Related to the Occurrence and Development of Oral Squamous Cell Carcinoma. Int J Oral Maxillofac Surg (2020) 49(3):292–7. doi: 10.1016/j.ijom.2019.07.014
34. Dou Z, Li S, Ren W, Wang Q, Liu J, Kong X, et al. Decreased Expression of hsa_circ_0072387 as a Valuable Predictor for Oral Squamous Cell Carcinoma. Oral Dis (2019) 25(5):1302–8. doi: 10.1111/odi.13094
35. Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottowa: Oxford (2000).
36. Cochran WG. The Comparison of Percentages in Matched Samples. Biometrika (1950) 37(3/4):256–66. doi: 10.1093/biomet/37.3-4.256
37. Higgins J, Tompson S, Deeks J, Altman D. A Meta-Analysis on the Effectiveness of Smart-Learning. BMJ (2003) 327(1):557–60. doi: 10.1136/bmj.327.7414.557
38. DerSimonian R, Laird N. Meta-Analysis in Clinical Trials Revisited. Contemp Clin Trials (2015) 45:139–45. doi: 10.1016/j.cct.2015.09.002
39. Mantel NH. Statistical Aspects of the Analysis of Data From Retrospective Studies of Disease. J Natl Cancer Institute (1959) 22:719748.
40. Egger M, Smith GD, Schneider M, Minder C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ (1997) 315(7109):629–34. doi: 10.1136/bmj.315.7109.629
41. Stuck AE, Rubenstein LZ, Wieland D. Bias in Meta-Analysis Detected by a Simple, Graphical Test. Asymmetry Detected in Funnel Plot was Probably Due to True Heterogeneity. BMJ: Br Med J (1998) 316(7129):469. doi: 10.1136/bmj.316.7129.469
42. Luo Y, Ma J, Liu F, Guo J, Gui R. Diagnostic Value of Exosomal circMYC in Radioresistant Nasopharyngeal Carcinoma. Head Neck (2020) 42(12):3702–11. doi: 10.1002/hed.26441
43. Castelli J, Simon A, Lafond C, Perichon N, Rigaud B, Chajon E, et al. Adaptive Radiotherapy for Head and Neck Cancer. Acta Oncol (2018) 57(10):1284–92. doi: 10.1080/0284186X.2018.1505053
44. Zhao Z-J, Shen J. Circular RNA Participates in the Carcinogenesis and the Malignant Behavior of Cancer. RNA Biol (2017) 14(5):514–21. doi: 10.1080/15476286.2015.1122162
45. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
46. Hao Q, Han Y, Xia W, Wang Q, Qian H. Systematic Review and Meta-Analysis of the Utility of Circular RNAs as Biomarkers of Hepatocellular Carcinoma. Can J Gastroenterol Hepatol (2019) 2019. doi: 10.1155/2019/1684039
47. Li C, He X, Zhang L, Li L, Zhao W. A Pair-Wise Meta-Analysis Highlights Circular RNAs as Potential Biomarkers for Colorectal Cancer. BMC Cancer (2019) 19(1):1–11. doi: 10.1186/s12885-019-6136-9
48. Ma Y, Niu X, Yan S, Liu Y, Dong R, Li Y. Circular RNA Profiling Facilitates the Diagnosis and Prognostic Monitoring of Breast Cancer: A Pair-Wise Meta-Analysis. J Clin Lab Anal (2021) 35(1):e23575. doi: 10.1002/jcla.23575
49. Nowak I, Sarshad AA. Argonaute Proteins Take Center Stage in Cancers. Cancers (Basel) (2021) 13(4):788. doi: 10.3390/cancers13040788
Keywords: circular RNA, HNSCC, meta-analysis, overall survival, prognostic markers
Citation: Nath M, Roy D and Choudhury Y (2022) Circular RNAs are Potential Prognostic Markers of Head and Neck Squamous Cell Carcinoma: Findings of a Meta-Analysis Study. Front. Oncol. 12:782439. doi: 10.3389/fonc.2022.782439
Received: 27 September 2021; Accepted: 02 February 2022;
Published: 28 February 2022.
Edited by:
Alvaro Sanabria, University of Antioquia, ColombiaReviewed by:
Carolina Ferraz, Santa Casa of Sao Paulo, BrazilGiovanni Blandino, Hospital Physiotherapy Institutes (IRCCS), Italy
Copyright © 2022 Nath, Roy and Choudhury. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yashmin Choudhury, yashminchoudhury@gmail.com; yashmin.choudhury@aus.ac.in
 Moumita Nath
Moumita Nath Dibakar Roy
Dibakar Roy Yashmin Choudhury
Yashmin Choudhury