
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 23 March 2022
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.769709
This article is part of the Research TopicEmerging Therapeutic Targets, Potential Diagnostic or Prognostic markers for Colorectal CancerView all 28 articles
 Qi-Wen Wang1†
Qi-Wen Wang1† Xin-Yuan Wang1†
Xin-Yuan Wang1† Qing-Wei Zhang1
Qing-Wei Zhang1 Jin-Nan Chen1
Jin-Nan Chen1 Yu-Jie Zhou1
Yu-Jie Zhou1 Zhao-Rong Tang2
Zhao-Rong Tang2 Rui-Lan Wang3
Rui-Lan Wang3 Haoyan Chen4
Haoyan Chen4 Huimin Chen1*
Huimin Chen1* Xiao-Bo Li1*
Xiao-Bo Li1*Background: Follow-up guidelines for serrated polyps (SPs) are mainly based on factors such as histology and size with limited evidence. The underlying genomic mechanism of SPs in relation to recurrence risks is utterly unknown.
Methods: We applied targeted next-generation sequencing (NGS) approach on two groups of SPs [polyp-relapsed SPs (PRSPs) vs. polyp-free SPs (PFSPs)] based on the surveillance outcomes to compare differences of DNA variants in 71 colorectal cancer-associated genes. A multicenter validation cohort was established longitudinally from 2016 to 2019 to confirm the relevant results.
Results: Among the 96 NGS samples, at least one mutant after filtration was detected in 90 samples (94%). Molecular profiling presented BRAF, KRAS, and APC as top 3 mutated genes. FBXW7, MSH2, and ERBB2 might be recurrence-relevant, while DMD, BRCA1, and BRCA2 might be negatively correlated with recurrence. Notably, ERBB2 mutants (R678Q and V842I) (n = 5) had higher risks of polyp recurrence than the wild types (n = 85), with a median polyp-free interval of 15 months compared to 26 months [P < 0.001; hazard ratio (HR) = 4.9; 95% confidence interval (CI) = 1.9–12.8]. Furthermore, a multicenter cohort composed by 321 SPs verified that ERBB2-mutated SPs had increased risks of polyp recurrence (P < 0.001; HR = 3.7; 95% CI = 2.3–6.0) and advanced neoplastic lesion (ANL) recurrence (P < 0.001; HR = 10.0; 95% CI = 2.7–36.9) compared with wild-type SPs, respectively.
Conclusions: Our results are emphasizing that SP individuals with ERBB2 mutants are at higher risks of subsequent colorectal neoplasms. ERBB2 mutants might work as facilitated markers for prediction of high-risk SPs and might implicate a potential mechanism in the serrated pathway to colorectal carcinoma (CRC).
Serrated polyps (SPs) are the second most common type of colorectal polyps with a distinct histological appearance of saw-toothed colonic crypts. According to the 2019 World Health Organization’s criteria, SPs are histologically classified as hyperplastic polyps (HPs), sessile serrated lesions (SSLs), or traditional serrated adenomas (TSAs) (1). Unlike the canonical adenoma-carcinoma sequence, SPs are proven to be early precursors to about 15%–30% colorectal carcinoma (CRC) through serrated pathway, frequently associated with BRAF or KRAS oncogenic mutations, microsatellite instability (MSI), and high CpG island methylator phenotype (CIMP) (2).
There is no consensus in the literature on which SPs are clinically relevant. A wide risk variation exists among SPs, as some case reports present SP rapid progression to invasive cancer within months (3), while other findings suggest a mean interval of 15 years for malignant transformation (4). Several longitudinal studies have investigated the relationship between the future risks of colorectal neoplasms and the clinicopathological characteristics of SPs, such as histology, size, anatomic location, or numbers (5–7). Evidence indicates that the detection of large SPs at the first endoscopy is more likely to have metachronous advanced neoplasms than those with no SPs (8), and it is an independent risk factor for subsequent CRC even with stronger association than that for advanced adenomas (9, 10). Cross-sectional reports have also substantially manifested that over-representation of malignancy hallmarks in SPs including BRAF mutation, MLH1 methylation, MUC5AC demethylation, and CIMP proposes possibly higher risks of CRC compared to conventional adenomas (11–13). Nevertheless, little is known about the genomic aberrations of SPs in relation to recurrence risks.
Therefore, we applied targeted next-generation sequencing (NGS) approach on groups of polyp-relapsed SPs (PRSPs) and polyp-free SPs (PFSPs) based on the surveillance outcomes to compare the intergroup differences of variants in 71 colorectal cancer-associated genes. In order to get insight about the value of genomic variants in predicting polyps’ recurrence and in planning colonoscopic surveillance, we further validated our results in a multicenter validation cohort established longitudinally from 2016 to 2019.
For the targeted NGS cohort, 96 candidates were enrolled at the Department of Gastroenterology and Hepatology, Ren-ji Hospital, Shanghai, between 2016 and 2019. In total, 93 colorectal SPs including 49 PRSPs and 44 PFSPs and 3 normal colon mucosae were retrieved from the tissue bank. A multicenter validation cohort was established longitudinally in Shanghai Renji Hospital, Chongqing Traditional Chinese Medicine Hospital and Sichuan Provincial Corps Hospital of Chinese People’s Armed Forces to confirm the relevant results. The overall schematic of the cohort selection is shown in Figure 1.
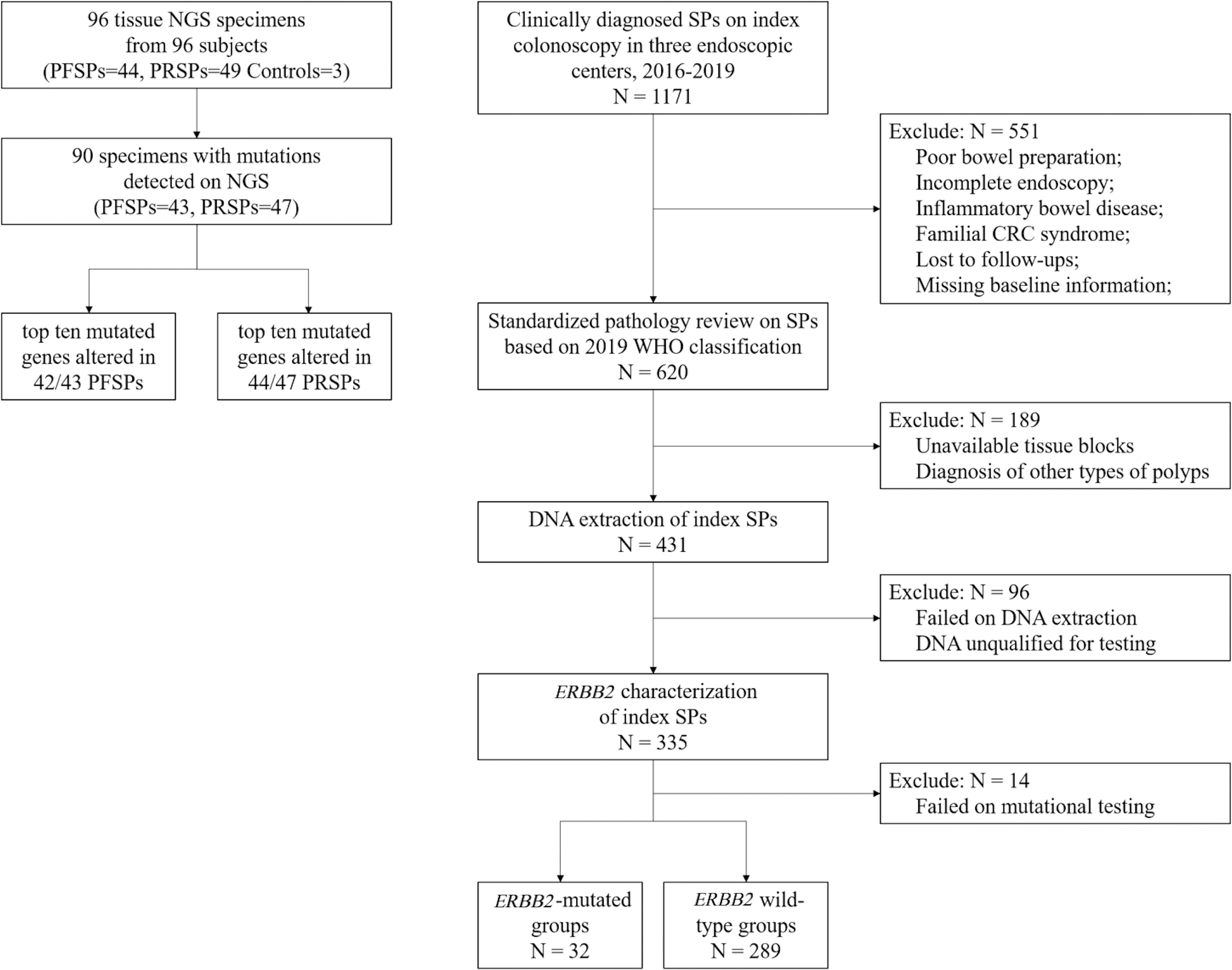
Figure 1 Cohort flowchart. NGS, next generation sequencing; PRSPs, polyp-relapsed serrated polyps; PFSPs, polyp-free serrated polyps; SPs, serrated polyps; CRC, colorectal cancer.
The eligibility criteria of all the subjects in our study were as follows: age ≥18 years; received an index colonoscopy from 2016 to 2019 with a clinical diagnosis of SPs; fulfilled adequate bowel preparation and cecum reach. Subjects were ineligible if they had inflammatory bowel disease or familial CRC syndromes at index colonoscopy or were lost to follow-ups or baseline information. All parts of the colon were scrupulously examined, and all the polyps were completely removed on the index colonoscopy. All the subjects received surveillance colonoscopy annually with available demographics and clinicopathological medical data. The histology of all samples including index and recurrent polyps was reevaluated from pathology reports and biopsies by two pathologists to confirm the diagnosis according to the 2019 WHO classification. Written informed consent was obtained from each patient before the specimen collection. Approval of this study was achieved by the Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University.
Since CRC was a relatively infrequent event among the subjects qualified with available medical records and regular surveillance in our study, we used risk of polyp recurrence as a surrogate marker. We defined PRSPs as the SPs with recurrence of polyps during surveillance colonoscopy, and PFSPs as the SPs free of polyps during surveillance colonoscopy. Index colonoscopy was referred to as a baseline colonoscopy for participants. Index polyps were the polyps diagnosed at index colonoscopy. We acknowledged a polyp recurrence when polyps were diagnosed during surveillance colonoscopy after the index colonoscopy. Recurrent polyps were classified into SPs (including HPs), non-advanced adenomas (NAs), or advanced neoplastic lesions (ANLs). ANLs included advanced adenomas (AAs) and cancers. AAs were considered as advanced adenomas that measure ≥10 mm in size, with villous or tubulovillous component, high-grade dysplasia, intramucosal carcinoma, or any combination thereof (14). Patients’ polyp-free intervals were calculated according to the time interval from the date of the index colonoscopy to the first diagnosis of polyp recurrence or until the date of last negative surveillance colonoscopy.
Genomic DNA was extracted from 10-μm formalin-fixed, paraffin-embedded (FFPE) tissue samples using the QIAamp DNA FFPE Tissue Kit (Qiagen Inc., Chatsworth, CA, USA), following the manufacturer’s instructions. DNA quantity and quality were checked using Qubit dsDNA HS assay on the Qubit Fluorometer (Thermo Fisher Scientific, Waltham, MA). DNA libraries were constructed from 100 to 250 ng DNA samples using QIAseq Targeted DNA Human Colorectal Cancer Panel (#DHS-002Z, Qiagen Inc., Chatsworth, CA, USA) including 71 most commonly mutated genes in human CRCs. All DNA fragments were tagged with a unique molecular index (UMI; a 12-nucleotide random sequence).
Libraries were sequenced via the Illumina NextSeq instrument with 75~155-bp paired-end reads. Sequence reads were aligned to the hg19 human genome build. Sufficient sequencing quality was guaranteed from all samples with the 30× minimum total read depth. Variant detection, annotation, scoring, and further filtering were implemented by Biomedical Genomics Workbench version 5 (Qiagen, Valencia, CA, USA) with the gene panel-specific plugin QIAseq Targeted Panel Analysis.
The plugin used five different filters for removal of called variants. The confidence filter only retained variants that were not residing in the top 5% of the most exonically variable 100-base windows in healthy public genome database (four reference databases: Allele Frequency Community; 1000 Genomes Project; ExAC; NHLBI ESP exomes). Besides, the confidence filter removed all variants below a call quality of 20 and with a prevalence of 0.5% in the healthy population. The genetic analysis filter only kept variant UMI level allele fraction (VMF) that ranged from 1% to 45% for each tested region, described to be pathogenic and/or likely pathogenic or loss of function-associated, which causes frameshift, missense, etc. An R Bioconductor package, maftools, was used for integrative analysis of somatic variants (15). The ggplot2 package was applied to draw the distribution map of mutation.
DNA samples from the validation cohort were tested by ERBB2 mutation status (R678Q, V842I) using Sanger sequencing. The two designed fragments of ERBB2 domain mutations from the DNA of the patients were amplified by multiplex PCR using primers as follows: ERBB2 R678Q forward (5’-GTTGGCATTCTGCTGGTCGT-3’) and reverse (5’-AGCAGTCTCCGCATCGTGTA-3’); ERBB2 V842I forward (5’-GCTAGGATGGGGACTCTTGC-3’) and reverse (5’-CCCCCATCTGCATGGTACTC-3’). The obtained reaction products were confirmed successful amplification by electrophoresis and then subjected to direct sequencing and analyzed on a 3730xl DNA Analyzer (Applied Biosystems). Sequencing results were compared with the reference DNA sequence and were interpreted by two separate approaches to improve the mutation detection: electronically with a set threshold of 10% and by visual inspection of the electropherogram by two researchers using Sequencher 5.0 software.
R (Version 4.0.3) was used for statistical analysis. Continuous parameters expressed as mean ± SD were analyzed by one-way analysis of variance (ANOVA) or independent Student’s t-test, while comparison between categorial data was evaluated by chi-square test or Fisher’s exact test. Cox regression models were applied for the time-to-event outcome analysis. Kaplan–Meier curves were reported by log-rank test for the variants detected to assess the polyps-free probability. Hazard ratios (HRs) and 95% confidence intervals (CIs) were reported in Cox regression models, and P values from a likelihood ratio test less than 0.05 were considered statistically significant. All the P values were two-tailed.
The baseline information of the sequencing samples (n = 96) was listed in Supplementary Table S1. The ANNOVAR package was used to annotate the variants with all available public population information (16). The identified variants were classified as benign or pathogenic according to ClinVar database (17). In total, 90/96 samples (94%) (including 47 PRSPs and 43 PFSPs) displayed at least one pathogenic or likely pathogenic variant that was categorized as missense, nonsense mutation, or splice site according to the frequency-based analysis (Figure 2). The final filtered NGS cohort (n = 90) was listed in Table 1.
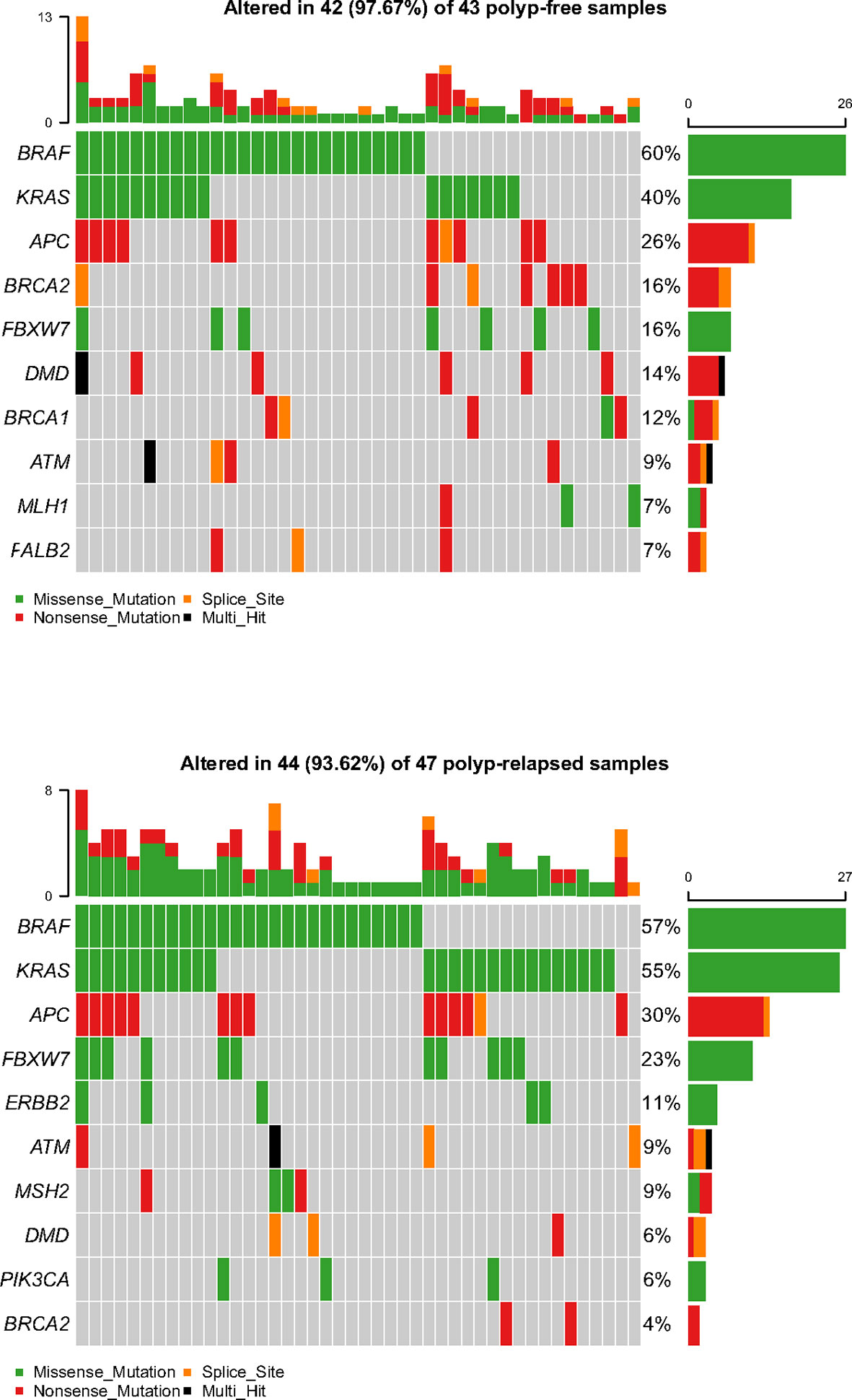
Figure 2 Representative spectrum of frequently mutated genes in polyp-relapsed and polyp-free colorectal serrated polyps after variant filtration.
We compared clinical characteristics between PRSPs and PFSPs. There were no significant differences in patients’ gender and age. SPs in PRSPs were significantly larger than that in PFSPs (P < 0.001). No differences were detected for polyp shape or location or dysplasia grade. Higher percentages of SSLs and TSAs were observed in PRSPs compared to PFSPs with remarkable differences (P < 0.001). The presence of three or more synchronous polyps or at least one ANL on index colonoscopy was not statistically different in the two groups. During the median polyp-free period of 26 months, three individuals in total developed ANLs during the surveillance colonoscopy. Polyp-free intervals {median months [interquartile range (IQR)]} were remarkably longer in PFSPs than those in PRSPs [25 (15) vs. 30 (16), P = 0.006].
Molecular profiling of PRSPs vs. PFSPs revealed that the three most common mutations were BRAF (57% vs. 60%), KRAS (55% vs. 40%), and APC (30% vs. 26%). FBXW7 (23% vs.16%, P = 0.399), MSH2 (9% vs. 2%, P = 0.413), and ERBB2 (11% vs. 0%, P = 0.078) had a larger proportion in PRSPs than PFSPs, which might be recurrence-relevant genes. While DMD (6% vs. 14%, P = 0.399), BRCA1 (2% vs. 12%, P = 0.167), and BRCA2 (4% vs. 16%, P = 0.122) might be negatively correlated with recurrence (Figure 3).
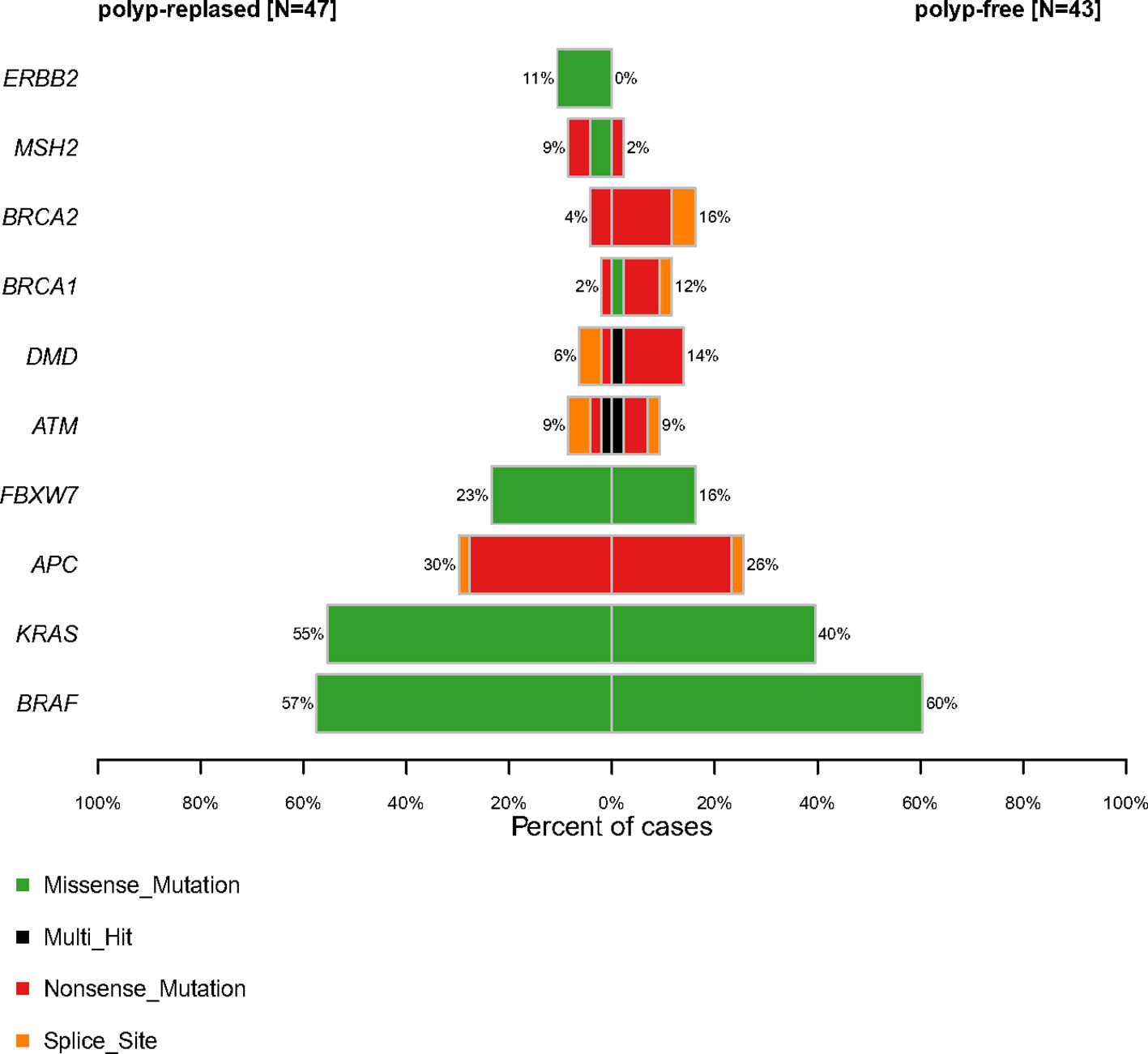
Figure 3 Comparative profiles of top 10 genomic alterations in polyp-relapsed and polyp-free colorectal serrated polyps.
The top 10 mutated genes in the NGS cohort were all assessed for polyp-free intervals by Kaplan–Meier plot analysis (n = 90). The findings illustrated that only ERBB2 mutations (n = 5) displayed a significant correlation with polyp recurrence, displaying a shorter median polyp-free interval of 15 months compared to 26 months than the wild types (n = 85) (P < 0.001; HR = 4.9; 95% CI = 1.9–12.8). Among the ERBB2 somatic point mutations in five samples, three were in the protein kinase domain (V842I) and two were non-activating mutations (R678Q) (18) (Supplementary Table S2 and Figure 4).
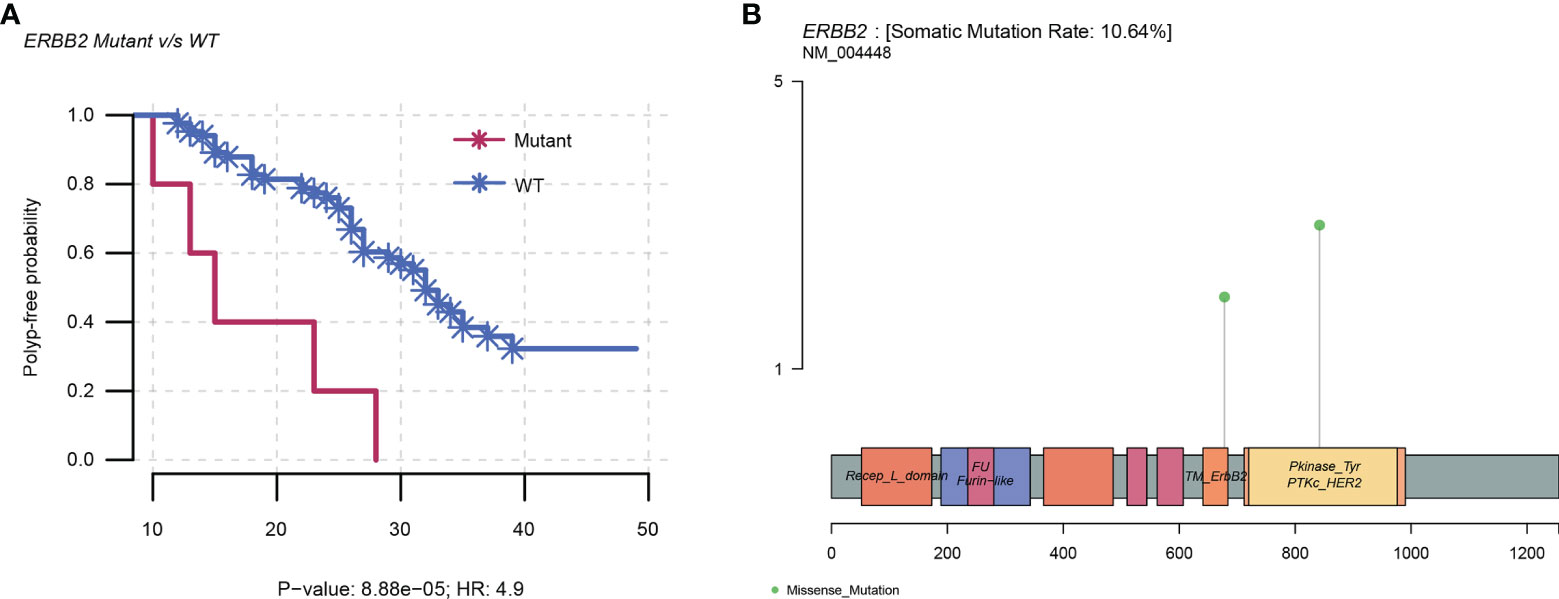
Figure 4 ERBB2 mutations in the NGS cohort. (A) Kaplan–Meier plot showing an increased risk of polyp recurrence over time in the ERBB2 mutants than in the wild types, with a median polyp-free interval of 15 months compared to 26 months (P < 0.001; HR = 4.9; 95% CI = 1.9–12.8). (B) Linear structure of ERBB2 illustrating the region and frequency of the variants (R678Q and V842I). WT, wild type; HR, hazard ratio.
The clinical relevance of ERBB2 genes in serrated lesions with risks of polyp recurrence has not been documented yet. To further validate the presence of ERBB2 mutations in SPs as a target of interest, we developed a separate validation cohort of 321 SPs from 308 patients recruited from three endoscopic centers (263 from Shanghai Ren-ji Hospital, 58 from the others). All the samples were screened for ERBB2 mutation status (R678Q and V842I). The characteristics were shown in Table 2, with no statistically significant difference found among the 3 centers in baseline information and mutation frequencies (Supplementary Table S3).
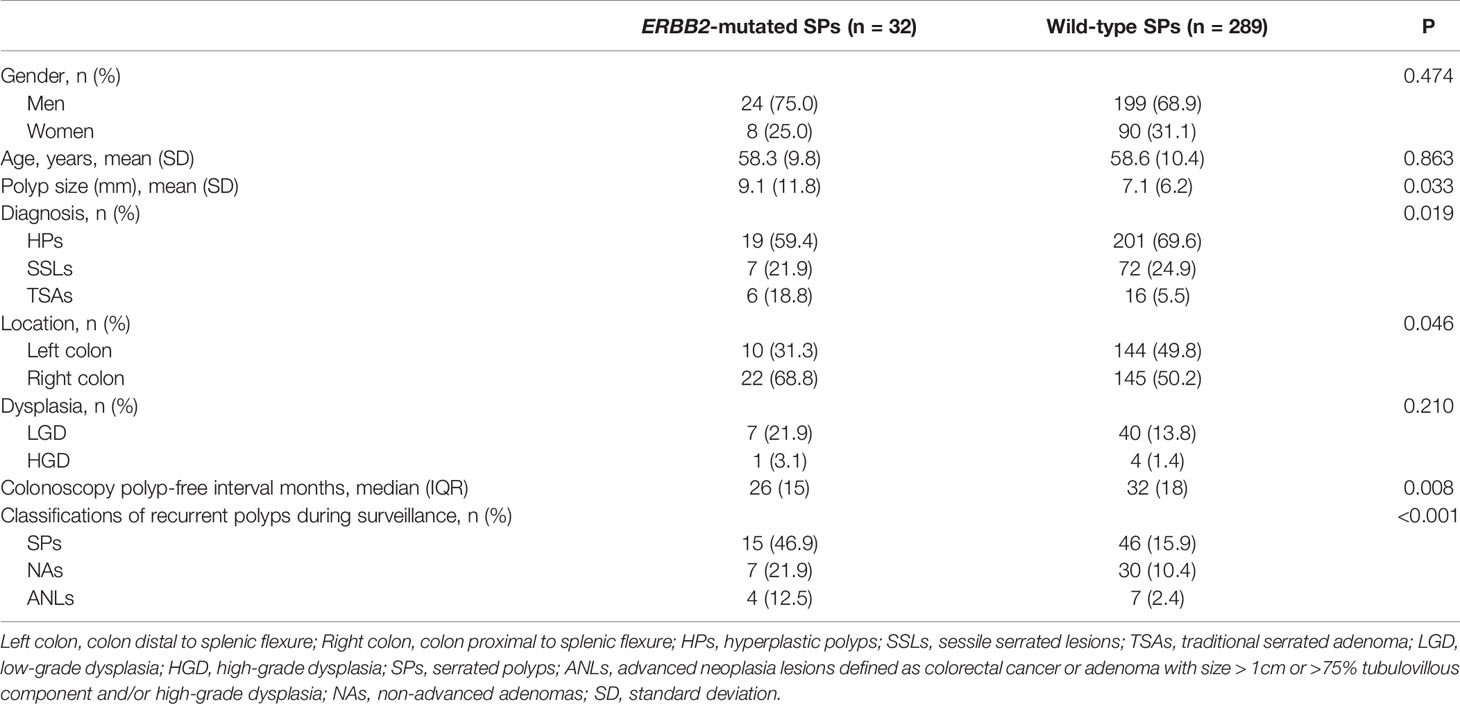
Table 2 Baseline characteristics of the validation cohort of SPs according to findings on index colonoscopy (total n = 321).
ERBB2 mutations were detected in 32/321 SPs (10.0%), with 18/32 R678Q (56.3%) and 14/32 V842I (43.7%) mutants. There were no differences in terms of gender and age between ERBB2-mutated and wild-type patients. The ERBB2-mutated group tended to be more right-sided (68.8% vs. 50.2%, P = 0.046) and significantly larger than the wild-type group (9.1 ± 11.8 vs. 7.1 ± 6.2, P = 0.033). TSAs were more present in the ERBB2-mutated group (18.8%) than in the wild-type group (5.5%).
In the validation cohort, Kaplan–Meier method and multivariate Cox regression were used to assess the association between ERBB2 mutations and polyp recurrence. Cox regression analysis showed that age, pathology of SSLs, and ERBB2 mutations independently predicted polyp recurrence. ERBB2-mutated SPs displayed increased risks of polyp recurrence, with a median polyp-free interval of 26 months compared to 32 months in wild-type SPs (P < 0.001; HR = 3.7; 95% CI = 2.3–6.0). ERBB2 mutations were further associated with reduced ANL-free intervals in SPs with statistical significance (P < 0.001; HR = 10.0; 95% CI = 2.7–36.9) (Figure 5). ERBB2 mutants displayed increased risks of polyp recurrence in Shanghai Renji center (P < 0.001), while no mutational factor was associated with polyp recurrence in patients from Chongqing TCM or Sichuan centers possibly due to the small sample size (Supplementary Figure S1).
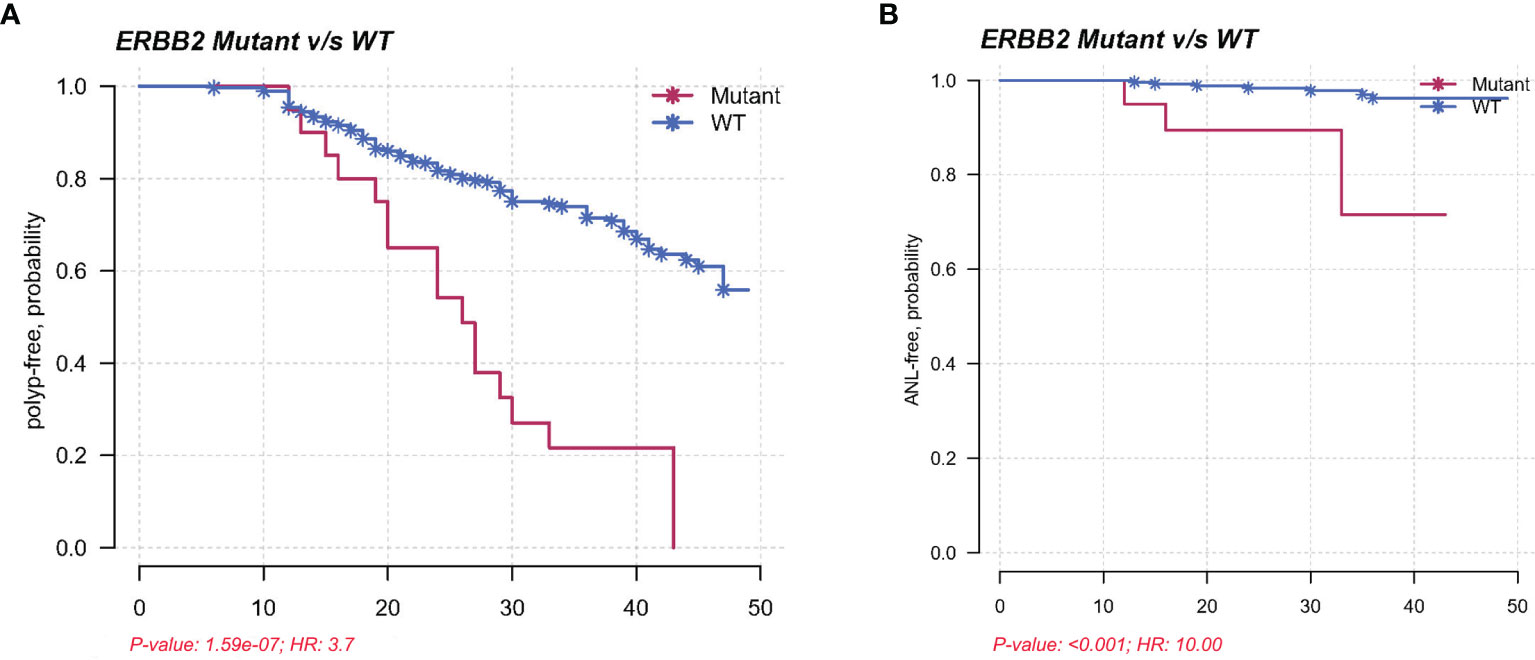
Figure 5 (A) Kaplan–Meier plot demonstrating a shorter polyp-free duration in the ERBB2 mutants than in the wild types in a validation cohort, with a median interval of 26 months compared to 32 months (P < 0.001; HR = 3.7; 95% CI = 2.3–6.0). (B) Kaplan–Meier plot showing reduced ANL-free intervals in ERBB2-mutated SPs in a validation cohort (P < 0.001; HR = 10.0; 95% CI = 2.7–36.9).
In general, it is consistently suggested that patients with SSLs and TSAs yielding increased risks of metachronous ANLs compared with persons without polyps should have more aggressive surveillance. To distinguish patients impaired from pathology of SSLs and TSAs, we stratified the patients according to histological diagnosis. The Kaplan–Meier curves illustrated that patients with ERBB2-mutated HPs and SSLs had strikingly shorter polyp-free intervals compared with corresponding wild-type groups, respectively (P < 0.001; P = 0.036), while in patients with TSAs, no difference in recurrent outcome was observed (Supplementary Figure S2).
It has not been widely appreciated until 2003–2005 that distinct from conventional sequence, SPs are integrated into clinical practice as malignant precursors of CRCs through a serrated tumorigenic pathway. Clinical surveillance recommendations for SP patients on colonoscopy are difficult to set up uniformly on account of unknown underlying genetic mechanisms behind risks of subsequent CRCs (19–22). Controversy exists around the effectiveness of colonoscopy even based on the foremost guidelines, as unexpected interval cancers are sporadically developed within serial surveillance colonoscopies (23). Research regarding the remarkable resemblance of interval cancers to SPs in right-sided preference and epigenetic features such as MSI and CIMP (24, 25) has aroused extensive concern on identification of high-risk SPs in the clinic. Though various studies have documented generally accepted features of high-risk serrated lesions such as SSLs with size larger than 10 mm or SLs harboring dysplasia including TSAs that require intensive surveillance colonoscopy (5–7, 19), they are based on limited evidence. A recent study firstly provided longitudinal evidence on molecular markers (including BRAF V600E, CIMP, and MLH1 methylation) of SPs, suggesting that epigenetic defect of MLH1 methylation was in relation to subsequent advanced neoplasms (26). However, somatic mutations have not been screened yet.
In the present study, we firstly investigated the genetic alterations of SPs in relation to polyp recurrence by NGS. The mutational profile analyses demonstrated pathogenic variants in genes as previously reported, such as BRAF V600E and KRAS codon12/13 but also illustrated genes less recorded such as FBXW7, ATM, and DMD. This work provided several interesting findings. First, BRAF, KRAS, and APC were the key drivers of serrated tumorigenesis but not the useful markers to identify high-risk SPs. Second, higher prevalence of FBXW7, MSH2, and ERBB2 mutants in PRSPs rather than PFSPs might suggest their association with higher risks of developing colorectal polyps. Third, lower incidences of DMD, BRCA1, and BRCA2 mutants in PRSPs rather than PFSPs delineated the unlikelihood of their contribution to interval cancer. Finally, ERBB2 mutants demonstrated a significant relationship with colorectal advanced neoplasm recurrence, implicating its value as an important marker of high-risk SPs.
In line with previous studies, our findings recognized BRAF V600E or KRAS mutations as the triggering event of the serrated pathway (27). The overall rate of BRAF V600E (53/90, 59%) was within the range reported in the existing literatures from 50% to 83% (28, 29). The high level of mutant BRAF in SPs is becoming growingly relevant as a poor prognostic factor (30). However, our result indicated that BRAF V600E was not related to the future risks of neoplasms, consistent with the result of Hua et al. (26). The prevalence of KRAS mutations is discrete, ranging from 15% to 75% in colorectal polyps in previous studies (31, 32). The frequency of KRAS mutations in our NGS analysis was 48% (43/90). Interestingly, APC was the third most mutated gene among the SPs. APC gene typically acts as a driver gene in adenoma-to-carcinoma sequence through aberrant Wnt signaling (33). The potential role of APC mutations in the serrated pathway remains controversial. It is generally believed that Wnt pathway of SPs is mainly activated by a number of alternative mechanisms such as PTPRK-RSPO3 fusions and RNF43 mutations other than APC (28, 34). Although recently, research is suggesting that APC mutations are likely the main pathogenic reason for WNT signaling activation in serrated pathway based on their high frequency (35, 36).
ERBB2 is a member of the ErbB receptor tyrosine kinase (RTK) family that transduces downstream signaling pathway, such as Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/protein kinase B (AKT) axis through the active form of homodimer or heterodimer complexes with other RTKs (37). ERBB2 overexpression occurs in many kinds of human cancers such as breast and ovarian cancers (38), and its association with poor prognosis in these cancers has been widely proven (39, 40). Moreover, the use of NGS has revealed the presence of ERBB2 sequence mutations in human tumors over recent 10 years. GENIE consortium data published in 2017 have exhibited that ERBB2 is altered in 4.69% of 2,081 CRC patients with R678Q and V842I present in 0.48% and 0.38% of all CRC patients, respectively (41). Given the nearly 5% frequency of ERBB2 alterations in CRCs based on the current literatures, ERBB2 becomes a promising target in anti-ERBB2-targeted therapies and prognoses. Whereas the presence of ERBB2 point mutations in colorectal serrated precursors has not been published before. Our findings have yielded important insights into the functional consequences of ERBB2 mutants. In our study, ERBB2 was mutated in 5.56% of 90 NGS subjects, and it was altered in 9.97% of 321 validation subjects with no statistical differences. Given the current identification of ERBB2 mutations in high-risk SPs, it could help shed some light on better understanding of polyp development and recurrence and could mark the significance that ERBB2 mutations may serve as an independent biomarker for guidance of clinical surveillance strategy. Patients with ERBB2 mutation-positive SPs may be recommended to receive more aggressive follow-up colonoscopy.
Our study has several strengths. First, SPs in this study are reclassified both historically and clinically. The historical reclassification was based on the newest 2019 WHO definitions by two independent pathologists, while the clinical subgroups were defined according to the follow-up outcomes. Second, this is the first cohort study exploring the correlation between high-risk genomic aberrations of SPs and recurrent outcome over time by using a targeted colorectal NGS panel. Prior literature only identified several endoscopic, histologic, or epigenetic features of serrated precursors that were strongly related to subsequent high-risk adenomas or CRCs identified during surveillance colonoscopy. Finally, the correlation between the ERBB2 mutational targets and colorectal polyp recurrence is first identified in the NGS cohort and was ulteriorly validated in a multicenter cohort. ERBB2 mutations are qualified as a clinically relevant biomarker to predict risks of future ANLs that have HRs >3.
This study has likewise some limitations. The retrospective study contained only a small number of SPs, and the follow-up duration was relatively short. Sanger sequencing platform had the limitation for reliable detection that required more than 10% fraction of mutated allele fraction. A sensitive digital PCR-based ERBB2 assay should be established in our future work to accurately evaluate its performance. The exploration of relevant factors of recurrence was limited in genomic features, while epigenetic characteristics of SPs were not dug out. Hence, the prognostic application of ERBB2 mutants in serrated pathway should be confirmed prospectively and extensively in the future.
In summary, for the first time, we have identified distinct genomic features of SPs in relation to subsequent polyp recurrence. This is also the first study detecting ERBB2 mutants in SPs. The clinical relevance of ERBB2 mutants with higher risks of subsequent colorectal neoplasms suggests their prospects as molecular markers for high-risk SP identification.
The original contributions presented in the study are publicly available. These data can be found here: https://ngdc.cncb.ac.cn/gsa-human, PRJCA007048 and GSA-Human: HRA001502.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Renji Hospital, School of Medicine, Shanghai Jiao Tong University. The patients/participants provided their written informed consent to participate in this study.
Guarantor of the paper: X-BL and H-MC. Specific author contributions: Q-WW and X-YW performed the experiments, analyzed data, and wrote the original draft. H-YC revised the article. Q-WZ, J-NC, and Y-JZ collected clinical and pathological information. Z-RT and R-LW provided tissue specimens. X-BL and H-MC conceived and supervised the study. All the authors revised and approved the final article.
The project described was supported by a funding grant from the National Natural Science Foundation of China (NSFC) (General Program: 81772519) and by a grant from the Excellent Youth Program of Shanghai Municipal Commission of Health and Family Planning (2018YQ29). There was no influence from the NSFC or Shanghai Municipal Commission of Health and Family Planning on study design or conduct, data collection and management, analysis, interpretation, preparation and review or approval of the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.769709/full#supplementary-material
Supplementary Figure 1 | (A) Kaplan-Meier plot showing no mutational factor associated with polyp recurrence in patients from Chongqing TCM (P = 0.086). (B) ERBB2 mutants displaying increased risks of polyp recurrence, with a median polyp-free interval of 27 months compared to 40 months in wild types from Shanghai Renji center (P<0.001).
Supplementary Figure 2 | Kaplan-Meier plot demonstrating a shorter polyp-free duration in the ERBB2 mutated HPs (A)/SSLs (B) than in the corresponding wild-type groups in the validation cohort, with a median interval of 27/23 months compared to 43/34 months (P < 0.001; P = 0.036). (C) Kaplan-Meier plot showing no difference in recurrent outcome in validation cohort of TSAs (P=0.345).
Supplementary Table 1 | Baseline characteristics of FFPE sections for sequencing (total n=96).
Supplementary Table 2 | Summary of clinicopathological features in the NGS cases with ERBB2 mutants (n=5).
Supplementary Table 3 | Baseline characteristics of the validation cohort of SPs classified by clinical centers (total n=321).
1. Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO Classification of Tumours of the Digestive System. Histopathology (2020) 76:182–8. doi: 10.1111/his.13975
2. Dehghanizadeh S, Khoddami V, Mosbruger TL, Hammoud SS, Edes K, Berry TS, et al. Active BRAF-V600E Is the Key Player in Generation of a Sessile Serrated Polyp-Specific DNA Methylation Profile. PloS One (2018) 13:e0192499. doi: 10.1371/journal.pone.0192499
3. Amemori S, Yamano HO, Tanaka Y, Yoshikawa K, Matsushita HO, Takagi R, et al. Sessile Serrated Adenoma/Polyp Showed Rapid Malignant Transformation in the Final 13 Months. Dig Endosc (2020) 32(6):979–83. doi: 10.1111/den.13572
4. Lash RH, Genta RM, Schuler CM. Sessile Serrated Adenomas: Prevalence of Dysplasia and Carcinoma in 2139 Patients. J Clin Pathol (2010) 63:681–6. doi: 10.1136/jcp.2010.075507
5. Anderson JC, Robinson CM, Butterly LF. Increased Risk of Metachronous Large Serrated Polyps in Individuals With 5- to 9-Mm Proximal Hyperplastic Polyps: Data From the New Hampshire Colonoscopy Registry. Gastrointest Endosc (2020) 92:387–93. doi: 10.1016/j.gie.2020.04.034
6. Park SK, Kim HS, Yang HJ, Jung YS, Park JH, Sohn CI, et al. Coexistent Adenoma and Serrated Polyps on Index Colonoscopy and the Risk of Metachronous Advanced Colorectal Neoplasia. Endosc Int Open (2019) 7:E1748–54. doi: 10.1055/a-1019-2976
7. Anderson JC, Butterly LF, Robinson CM, Weiss JE, Amos C, Srivastava A. Risk of Metachronous High-Risk Adenomas and Large Serrated Polyps in Individuals With Serrated Polyps on Index Colonoscopy: Data From the New Hampshire Colonoscopy Registry. Gastroenterology (2018) 154:117–27.e2. doi: 10.1053/j.gastro.2017.09.011
8. Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, et al. Serrated Lesions of the Colorectum: Review and Recommendations From an Expert Panel. Am J Gastroenterol (2012) 107:1315–29; quiz 1314, 1330. doi: 10.1038/ajg.2012.161
9. Erichsen R, Baron JA, Hamilton-Dutoit SJ, Snover DC, Torlakovic EE, Pedersen L, et al. Increased Risk of Colorectal Cancer Development Among Patients With Serrated Polyps. Gastroenterology (2016) 150:895–902.e5. doi: 10.1053/j.gastro.2015.11.046
10. He X, Hang D, Wu K, Nayor J, Drew DA, Giovannucci EL, et al. Long-Term Risk of Colorectal Cancer After Removal of Conventional Adenomas and Serrated Polyps. Gastroenterology (2020) 158:852–61.e4. doi: 10.1053/j.gastro.2019.06.039
11. Bettington M, Walker N, Rosty C, Brown I, Clouston A, McKeone D, et al. Clinicopathological and Molecular Features of Sessile Serrated Adenomas With Dysplasia or Carcinoma. Gut (2017) 66:97–106. doi: 10.1136/gutjnl-2015-310456
12. Renaud F, Mariette C, Vincent A, Wacrenier A, Maunoury V, Leclerc J, et al. The Serrated Neoplasia Pathway of Colorectal Tumors: Identification of MUC5AC Hypomethylation as an Early Marker of Polyps With Malignant Potential. Int J Cancer (2016) 138:1472–81. doi: 10.1002/ijc.29891
13. Meester RGS, van Herk MMAGC, Lansdorp-Vogelaar I, Ladabaum U. Prevalence and Clinical Features of Sessile Serrated Polyps: A Systematic Review. Gastroenterology (2020) 159:105–18.e25. doi: 10.1053/j.gastro.2020.03.025
14. Ng SC, Ching JY, Chan VC, Wong MC, Tang R, Wong S, et al. Association Between Serrated Polyps and the Risk of Synchronous Advanced Colorectal Neoplasia in Average-Risk Individuals. Aliment Pharmacol Ther (2015) 41:108–15. doi: 10.1111/apt.13003
15. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res (2018) 28:1747–56. doi: 10.1101/gr.239244.118
16. Wang K, Li M, Hakonarson H. ANNOVAR: Functional Annotation of Genetic Variants From High-Throughput Sequencing Data. Nucleic Acids Res (2010) 38:e164. doi: 10.1093/nar/gkq603
17. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: Public Archive of Relationships Among Sequence Variation and Human Phenotype. Nucleic Acids Res (2014) 42:D980–5. doi: 10.1093/nar/gkt1113
18. Li Z, Shao C, Liu X, Lu X, Jia X, Zheng X, et al. Oncogenic ERBB2 Aberrations and KRAS Mutations Cooperate to Promote Pancreatic Ductal Adenocarcinoma Progression. Carcinogenesis (2020) 41:44–55. doi: 10.1093/carcin/bgz086
19. East JE, Atkin WS, Bateman AC, Clark SK, Dolwani S, Ket SN, et al. British Society of Gastroenterology Position Statement on Serrated Polyps in the Colon and Rectum. Gut (2017) 66:1181–96. doi: 10.1136/gutjnl-2017-314005
20. Crockett SD, Nagtegaal ID. Terminology, Molecular Features, Epidemiology, and Management of Serrated Colorectal Neoplasia. Gastroenterology (2019) 157:949–66.e4. doi: 10.1053/j.gastro.2019.06.041
21. Gupta V, East JE. Optimal Endoscopic Treatment and Surveillance of Serrated Polyps. Gut Liver (2020) 14:423–9. doi: 10.5009/gnl19202
22. Lieberman DA. American Gastroenterological A. Colon Polyp Surveillance: Clinical Decision Tool. Gastroenterology (2014) 146:305–6. doi: 10.1053/j.gastro.2013.11.029
23. Samadder NJ, Curtin K, Tuohy TM, Pappas L, Boucher K, Provenzale D, et al. Characteristics of Missed or Interval Colorectal Cancer and Patient Survival: A Population-Based Study. Gastroenterology (2014) 146:950–60. doi: 10.1053/j.gastro.2014.01.013
24. Lee YM, Huh KC. Clinical and Biological Features of Interval Colorectal Cancer. Clin Endosc (2017) 50:254–60. doi: 10.5946/ce.2016.115
25. Richter JM, Pino MS, Austin TR, Campbell E, Szymonifka J, Russo AL, et al. Genetic Mechanisms in Interval Colon Cancers. Dig Dis Sci (2014) 59:2255–63. doi: 10.1007/s10620-014-3134-2
26. Hua X, Newcomb PA, Chubak J, Malen RC, Ziebell R, Kamineni A, et al. Associations Between Molecular Characteristics of Colorectal Serrated Polyps and Subsequent Advanced Colorectal Neoplasia. Cancer Causes Control (2020) 31:631–40. doi: 10.1007/s10552-020-01304-1
27. Lin SH, Raju GS, Huff C, Raju GS, Huff C, Ye Y, Gu J, Chen JS, et al. The Somatic Mutation Landscape of Premalignant Colorectal Adenoma. Gut (2018) 67:1299–305. doi: 10.1136/gutjnl-2016-313573
28. Yan HHN, Lai JCW, Ho SL, Leung WK, Law WL, Lee JFY, et al. RNF43 Germline and Somatic Mutation in Serrated Neoplasia Pathway and Its Association With BRAF Mutation. Gut (2017) 66:1645–56. doi: 10.1136/gutjnl-2016-311849
29. Chan AW, Pan Y, Tong JH, Lung RW, Kwan JS, Chow C, et al. Receptor Tyrosine Kinase Fusions Act as a Significant Alternative Driver of the Serrated Pathway in Colorectal Cancer Development. J Pathol (2020) 251:74–86. doi: 10.1002/path.5418
30. Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, et al. Identification of a Poor-Prognosis BRAF-Mutant-Like Population of Patients With Colon Cancer. J Clin Oncol (2012) 30:1288–95. doi: 10.1200/JCO.2011.39.5814
31. Fu X, Qiu Y, Zhang Y. Screening, Management and Surveillance for the Sessile Serrated Adenomas/Polyps. Int J Clin Exp Pathol (2014) 7:1275–85.
32. Yi C, Huang Y, Yu X, Li X, Zheng S, Ding K, et al. Clinicopathologic Distribution of KRAS and BRAF Mutations in a Chinese Population With Colorectal Cancer Precursor Lesions. Oncotarget (2016) 7:17265–74. doi: 10.18632/oncotarget.7504
33. Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet (2001) 10:721–33. doi: 10.1093/hmg/10.7.721
34. Sekine S, Yamashita S, Tanabe T, Hashimoto T, Yoshida H, Taniguchi H, et al. Frequent PTPRK-RSPO3 Fusions and RNF43 Mutations in Colorectal Traditional Serrated Adenoma. J Pathol (2016) 239:133–8. doi: 10.1002/path.4709
35. Borowsky J, Dumenil T, Bettington M, Pearson SA, Bond C, Fennell L, et al. The Role of APC in WNT Pathway Activation in Serrated Neoplasia. Mod Pathol (2018) 31:495–504. doi: 10.1038/modpathol.2017.150
36. Okamura T, Hashimoto T, Naka T, Yoshida T, Tanabe N, Ogawa R, et al. Clinicopathologic and Molecular Characteristics of Familial Adenomatous Polyposis-Associated Traditional Serrated Adenoma. Am J Surg Pathol (2020) 44:1282–9. doi: 10.1097/PAS.0000000000001502
37. Wang Z. ErbB Receptors and Cancer. Methods Mol Biol (2017) 1652:3–35. doi: 10.1007/978-1-4939-7219-7_1
38. Cenaj O, Ligon AH, Hornick JL, Sholl LM. Detection of ERBB2 Amplification by Next-Generation Sequencing Predicts HER2 Expression in Colorectal Carcinoma. Am J Clin Pathol (2019) 152:97–108. doi: 10.1093/ajcp/aqz031
39. Adam L, San Lucas FA, Fowler R, Yu Y, Wu W, Liu Y, et al. DNA Sequencing of Small Bowel Adenocarcinomas Identifies Targetable Recurrent Mutations in the ERBB2 Signaling Pathway. Clin Cancer Res (2019) 25:641–51. doi: 10.1158/1078-0432.CCR-18-1480
40. Luo SP, Wu QS, Chen H, Wang XX, Chen QX, Zhang J, et al. Validation of the Prognostic Significance of the Prognostic Stage Group According to the Eighth Edition of American Cancer Joint Committee on Cancer Staging System in Triple-Negative Breast Cancer: An Analysis From Surveillance, Epidemiology, and End Results 18 Database. J Surg Res (2020) 247:211–9. doi: 10.1016/j.jss.2019.09.072
Keywords: serrated polyps, serrated pathway, colorectal cancer, ERBB2, recurrence
Citation: Wang Q-W, Wang X-Y, Zhang Q-W, Chen J-N, Zhou Y-J, Tang Z-R, Wang R-L, Chen H, Chen H and Li X-B (2022) ERBB2 Mutations as Potential Predictors for Recurrence in Colorectal Serrated Polyps by Targeted Next-Generation Sequencing. Front. Oncol. 12:769709. doi: 10.3389/fonc.2022.769709
Received: 02 September 2021; Accepted: 22 February 2022;
Published: 23 March 2022.
Edited by:
Nadia M. Hamdy, Ain Shams University, EgyptReviewed by:
Jun Li, The University of Sydney, AustraliaCopyright © 2022 Wang, Wang, Zhang, Chen, Zhou, Tang, Wang, Chen, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Bo Li, bHhiXzE5NjlAMTYzLmNvbQ==; Huimin Chen, aHVpbWluLmNoYW5AZm94bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.