
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 11 March 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.763464
This article is part of the Research Topic Women in Molecular and Cellular Oncology: 2021 View all 15 articles
Steroid hormones govern the complex, cyclic changes of the endometrium, predominantly through their receptors. An interplay between steroid hormones and epigenetic mechanisms controls the dynamic endometrial gene regulation. Abnormalities in expression of genes and enzymes associated with steroid hormone signaling, contribute to a disturbed hormonal equilibrium. Limited evidence suggests the involvement of TET (Ten Eleven Translocation)-mediated DNA hydroxymethylation in endometrial cancer, with some data on the use of TET1 as a potential prognostic and diagnostic biomarker, however the mechanisms guiding it and its regulation remains unexplored. This study aims to explore the changes in the expressions of TETs and steroid hormone receptors in response to estrogen and progesterone in endometrial cancer cells. Gene expression was examined using real-time PCR and protein expression was quantified using fluorescent western blotting in endometrial cancer cell lines (AN3 and RL95-2). Results indicate that TET1 and TET3 gene and protein expression was cell-specific in cancer cell-lines. Protein expression of TET1 was downregulated in AN3 cells, while TET1 and TET3 expressions were both upregulated in RL95-2 cells in response to estrogen-progesterone. Further, a decreased AR expression in AN3 cells and an increased ERα and ERβ protein expressions in RL95-2 cells was seen in response to estrogen-progesterone. PR gene and protein expression was absent from both cancer cell-lines. Overall, results imply that expressions of steroid hormones, steroid-hormone receptors and TETs are co-regulated in endometrial cancer-cells. Further studies are needed to interpret how these mechanisms fit in with DNMTs and DNA methylation in regulating endometrial biology. Understanding the role of TETs and hydroxymethylation in steroid hormone receptor regulation is crucial to comprehend how these mechanisms work together in a broader context of epigenetics in the endometrium and its pathologies.
A two-way communication between epigenetic mechanisms and steroid hormones is crucial for the healthy functioning of the endometrium. Estrogen and progesterone, secreted by the ovaries, execute their functions predominantly via steroid hormone receptors - estrogen receptor (ER) and progesterone receptor (PR). Transcriptional regulation of steroid hormone receptors in the endometrium is partly controlled by epigenetic factors like DNA methylation and hydroxymethylation (1–5). DNA methylation yields 5-methylcytosine (5mC), making for one of the most important forms of epigenetic modification in the mammalian DNA (6). However, the modification of DNA from 5C (5-Cytosine) to 5mC can be actively or passively reversed via the process of DNA de-methylation. The DNA de-methylation cascade consists of the initial oxidation of 5mC into 5-hydroxymethylcytosine (5hmC) followed by a series of additional oxidation steps (7, 8). 5hmC is identified as an independent epigenetic modification that can alter gene expression and might be important in epigenetic reprogramming (8). The active de-methylation process is catalysed by ten-eleven translocation (TET) enzymes, making them an essential component in epigenetic machinery. Dysregulation of TETs and subsequent 5hmC marks have been implicated in endometrial diseases such as endometrial cancer and endometriosis. (9, 10). Knockout study models have previously been used to establish the function of TETs in various tissues and cells including the maintenance of reproductive axis and epigenetic reprogramming (11–15). DNA methylation is known to be involved in maintaining successful steroid hormone signaling by regulating steroid hormone receptors (16). On the other hand, estrogen and progesterone can influence mRNA and protein expression of DNA Methyltransferases (DNMTs), thereby affecting methylation patterns (17–19).
In the normal endometrium, increasing estrogen levels during the proliferative phase, lead to an increase in the expression of estrogen (ER), progesterone (PR) and androgen receptors (AR) (20). This is followed by an antagonistic progesterone action which is mediated by the increased levels of progesterone receptors (21, 22). The interplay between estrogen and progesterone implies that while estrogen action aids in upregulating steroid receptors in the endometrium, progesterone action downregulates them (20). Since the maintenance of this steroid hormone equilibrium is essential to endometrial biology, abnormal regulation of steroid hormone receptor expression can contribute to endometrial pathologies (23–28). Previously, it has been suggested that TETs and DNMTs could potentially be inversely regulated by steroid hormones, with epithelial cells being more sensitive and responsive to steroid hormone treatments (29). This study is aimed at mimicking the hormonal influences seen during the menstrual cycle in vitro, to explore the mechanisms involved in the regulation of TETs and steroid hormone receptors in endometrial cancer cells. Four steroid hormone receptors– Estrogen receptors alpha (ERα) and beta (ERβ), Progesterone receptor (PR) and Androgen Receptor (AR) along with TETs were examined to assess the role of steroid hormones in their transcriptional and translational regulation.
Endometrial adenocarcinoma cell lines, AN3 (ATCC® HTB-111™) and RL95-2 (ATCC® CRL-1671™) were used for this study. All cells were cultured either in phenol-free DMEM or RPMI medium, supplemented with 10% charcoal stripped FBS (CS-FBS) as well as 1% of penicillin-streptomycin antibiotic (ThermoFisher Scientific, USA). The cells were then cultured in a humidified atmosphere with 5% CO2 at 37°C until confluent. Cells were then plated in twelve-well culture plates and upon 80% confluence, they were primed with 0.01μM of β-estradiol (E treatment) for 24h. Followed by the addition of progesterone (1μM) to the estrogen primed (EP treatment) wells for 24, 48 and 72h. Ethanol at a concentration of less than 0.01% was used as control (C treatment). The treatment solutions were prepared using commercially available powdered concentrates (Sigma- Aldrich, USA) and dissolved in analytical grade ethanol. The final concentrations were prepared in culture media and stored at -80°C until further use.
Trizol® reagent (Life Technologies, NZ) was used to extract Total RNA. 1ml of Trizol® was added per well and cells were detached using a cell scrapper. The cells were homogenized and treated according to the manufacturer’s instructions. Using the protocol provided, chloroform (0.2ml/1ml of Trizol®) was added to the samples and vigorously shaken and incubated for 3minutes at room temperature. After a 15minute centrifugation (12000xg) at 4°C, isopropanol (0.5ml/1ml of Trizol®) was added to the aqueous phase and incubated for 20minutes on ice. Followed by another centrifugation under similar conditions, the RNA pellet was obtained and washed in 70% ethanol with additional 10minute centrifugations, twice. The pellet was air dried at room temperature and suspended in DEPC treated water. The concentration and quality of RNA was assessed using the NanoPhotometer® (Implen, Germany). An OD260/280 ratio of 1.8 to 2.0 was considered quality RNA.
As directed by the manufacturer’s instruction manual, 1µg of RNA was treated with 1µl of 10xDNase and DNAse Buffer each and made up to 10µl with DEPC-water. After a 15minute incubation at room temperature, 1µl of EDTA was added to each reaction tube and incubated at 65°C for 10minutes. Reverse transcription into single-stranded cDNA was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). According to the manufacturer’s instructions, each tube was mixed with Reverse Transcriptase Buffer, Random primers, Deoxynucleotide Mix and Reverse Transcriptase and made it up to a total reaction volume of 20μl. Using the BioRad DNA Engine® Peltier Thermal Cycler under the following conditions: 10minutes at 25°C, 120minutes at 37°C and 5minutes at 85°C, reverse transcription was performed. The resulting cDNA was diluted with nuclease-free water (1:10) and used for real-time PCR. PCR analysis was performed and conducted using QuantStudio (Applied Biosystems, USA) as previously described (29). Primers for TET1, TET2, TET3, RPL13a, YWHAZ and RPLO (Table 1) were obtained from Primer Bank (30–33). Primers for ERα, ERβ and PGR were the PrimeTime predesigned qPCR Assays (IDT) (Table 1). Primer (Table 1) for AR was obtained from a previously published study by Kamal et al. (34). Gene expression analysis was done using the comparative CT method (ΔΔCT method) (35). All the results were normalized to the geomeans of the three reference genes- YWHAZ, RPL13a and RPLO as described previously (29).
AN3 and RL95-2 cells were extracted from culture plates using RIPA lysis and extraction buffer (ThermoFisher Scientific, USA). According to the instructions provided, culture media was aspirated and 1ml/well of cold RIPA buffer was added to lyse the cells. Halt™ Protease Inhibitor Cocktail (EDTA-Free (100X)) (Thermo Fisher Scientific) was also added (20µL per 1mL of RIPA lysis buffer). The plate was then incubated on ice for 5minutes with intermittent swirling for uniform spreading of the buffer. The lysate was gathered using a cell scraper and transferred to a 2ml tube. The samples were then centrifuged at 14000xg for 15 minutes and the supernatant protein was collected and stored at -80°C for further analysis.
Western blot analysis was performed to evaluate the expression of TETs and steroid hormone receptor proteins post-treatment. Protein extracted from the cells were loaded on a 3-8% NuPage™ Tris Acetate gel (Invitrogen, USA) and transferred onto a 0.45 micron pored fluorescent polyvinyl difluoride membrane (Fl-PVDF), (Millipore, USA). The protocol for a wet transfer was followed according to the manufacturer’s instructions using 20X NuPage™ Transfer Buffer (Invitrogen, USA). Once the proteins were transferred onto the membrane, it was stained and washed with Revert™ total protein stain and wash solution respectively, (Licor Biosciences, USA). The membrane was imaged at 700nm Odyssey® imaging system and blocked using Intercept Blocking Buffer (Licor Biosciences, USA) for an hour at room temperature. Primary antibodies were diluted according to Table 2 in the blocking buffer. Following which, the membrane was incubated in primary antibody and left overnight at 4°C and conjugated with secondary antibodies the next day. The membrane was washed thrice using 1XTBST (Tris-buffered saline with Tween-20) with 5minute intervals and incubated with the secondary antibody for 1hr at room temperature. The membrane was washed, dried, and imaged for 10minutes at 800nm channel using Odyssey® imaging system (Licor Biosciences, USA). All the primary antibodies (Table 2) used for this experiment were from Thermo Fisher Scientific and the secondary antibodies were from Licor Biosciences - donkey anti-mouse (P/N: 926-32212) or goat anti-rabbit (Catalog# P/N: 926-32211) IRDye® 800CW depending on primary antibody reactivity. Protein expression for all samples were normalized to the total protein stain for each blot. Target protein bands were normalized against the total protein transferred per lane. Total protein signal (TPS) was used to calculate the proteins in each lane and the normalization factor. The formulas used for each calculation are below:
The normalized signal for each sample was calculated to be used for relative quantitative comparison. The x-axis demonstrated the fold change that was normalized to the control and was plotted against the treatment stage (y-axis) for each sample.
GraphPad Prism 8 (GraphPad Software, La Jolla, CA) and IBM SPSS version 27.0 (Armonk, NY) were used to analyze the data obtained. Statistical tests included one-way analysis of variance (ANOVA) and paired t-test to determine significance (P<0.05 was considered statistically significant; P ≤ 0.1 was considered as approaching significance). All the Graphs were generated using GraphPad Prism 8 (GraphPad Software, CA).
TET1 was significantly downregulated (p=0.0479) post the initial 24 hour estrogen treatment, followed by a significant increase in response to a combined estrogen-progesterone treatment for 24 hours (p=0.0361). Prolonged exposure to combined estrogen-progesterone for 48 and 72 hours resulted in a significant reduction of TET1 gene expression (p=0.0302). TET2 and TET3 did not display any significant in response to any treatments, although there was a significant increase in TET2 between combined estrogen-progesterone treatment from 24 to 48 hours (p=0.0276). No significant changes in steroid hormone receptor expression were observed in AN3 cells in response to any treatments.
TET1, 2 and 3 proteins in AN3 cells exhibited no changes during estrogen only treatment as observed in Figure 2. However, differential expression was observed when treated with combined estrogen-progesterone for 24, 48 and 72 hours. TET1 protein expression displayed a decreasing trend when exposed to 72 hours of combined estrogen-progesterone treatment (p=0.1). Conversely, TET3 protein expression displayed a trend toward increasing upon 72 hours of estrogen-progesterone treatment (p=0.1). Furthermore, there was a trend towards an increase in TET2 protein expression in response to 24-hour estrogen-progesterone treatment, approaching significance (p=0.1). Protein expression for steroid hormone receptors revealed no significant changes in ERβ expression. However, AR protein expression was consistently downregulated during treatment with estrogen-progesterone for 24 (p=0.08), 48 (p=0.059) and 72 (p=0.09) hours (Figure 3). No bands for ERα, PRA and PRB proteins were not detectable by western blotting in AN3 cells under any treatments.
Gene expressions for all three TETs varied significantly across different treatments (p< 0.0001) in RL95-2 cells. However, no statistical significance was found between individual treatment groups in comparison to the control. Gene expression for ERα, PR or AR were not detected in RL95-2 cells. However, ERβ gene expression was prominent in hormone treated RL95-2 cells with significant changes between control and treatments (p<0.0001), as revealed by one way ANOVA with no significant differences between individual treatment groups (Figure 4).
Protein expression of TETs varied across different treatments as shown in Figure 5. TET1 (p=0.01) expression was significantly decreased and a similar trend towards a decrease in TET3 (p=0.1) expression was also seen in response to estrogen only treatment. There was a significant increase in TET3 (p=0.019) and a trend towards an increase in TET1 (p=0.1), when treated with estrogen-progesterone for 72 hours. In response to 48 hours of estrogen-progesterone treatment, a significant increase in TET3 (p=0.02) and a trend towards reduction in TET1 (p=0.1) expression was observed. TET2 protein expression was significantly upregulated during estrogen only treatment (*p=0.059). The protein expression of ERs revealed a differential and treatment dependent regulation as shown in Figure 6. There was a trend towards an increase in ERα protein (p=0.1) expression in response to 24 hours of estrogen-progesterone treatment, and ERβ expression (p=0.1) in response to estrogen only treatment which stayed consistent across treatments with significant increase seen in response to 72 hours of a combined estrogen-progesterone treatment (p=0.038). Very faint bands for AR were observed with no significant differences between treatments. PRA and PRB were not detected in RL95-2 cells even in response to treatment.
DNA methylation and hydroxymethylation are crucial components of the epigenetic machinery. The aim of this study was to evaluate the contribution of steroid hormones in the transcriptional and translational regulation of TETs and steroid hormone receptors in endometrial cancer cells. The results indicate that the gene and protein expressions of TETs and steroid hormone receptors and their response to steroid hormones is cell-specific and differ between AN3 and RL95 cells.
Endometrial pathologies such as endometrial cancer are steroid dependent disorders. Steroid hormones guide the fluctuating epigenetic patterns, allowing genes to be expressed or repressed during the menstrual cycle (16). In AN3 cells, TET1 transcription was significantly downregulated when exposed to estrogen for 24 hours. This was then marked by a significant increase when exposed to a combined estrogen-progesterone treatment for 24 hours. However, a further prolonged estrogen-progesterone treatment for 48 and 72 hours resulted in a significant decrease in TET1 mRNA expression (Figure 1). TET1 gene expression in AN3 cells, was responsive to the slightest change in treatment, which could indicate its sensitivity to subtle hormonal changes.
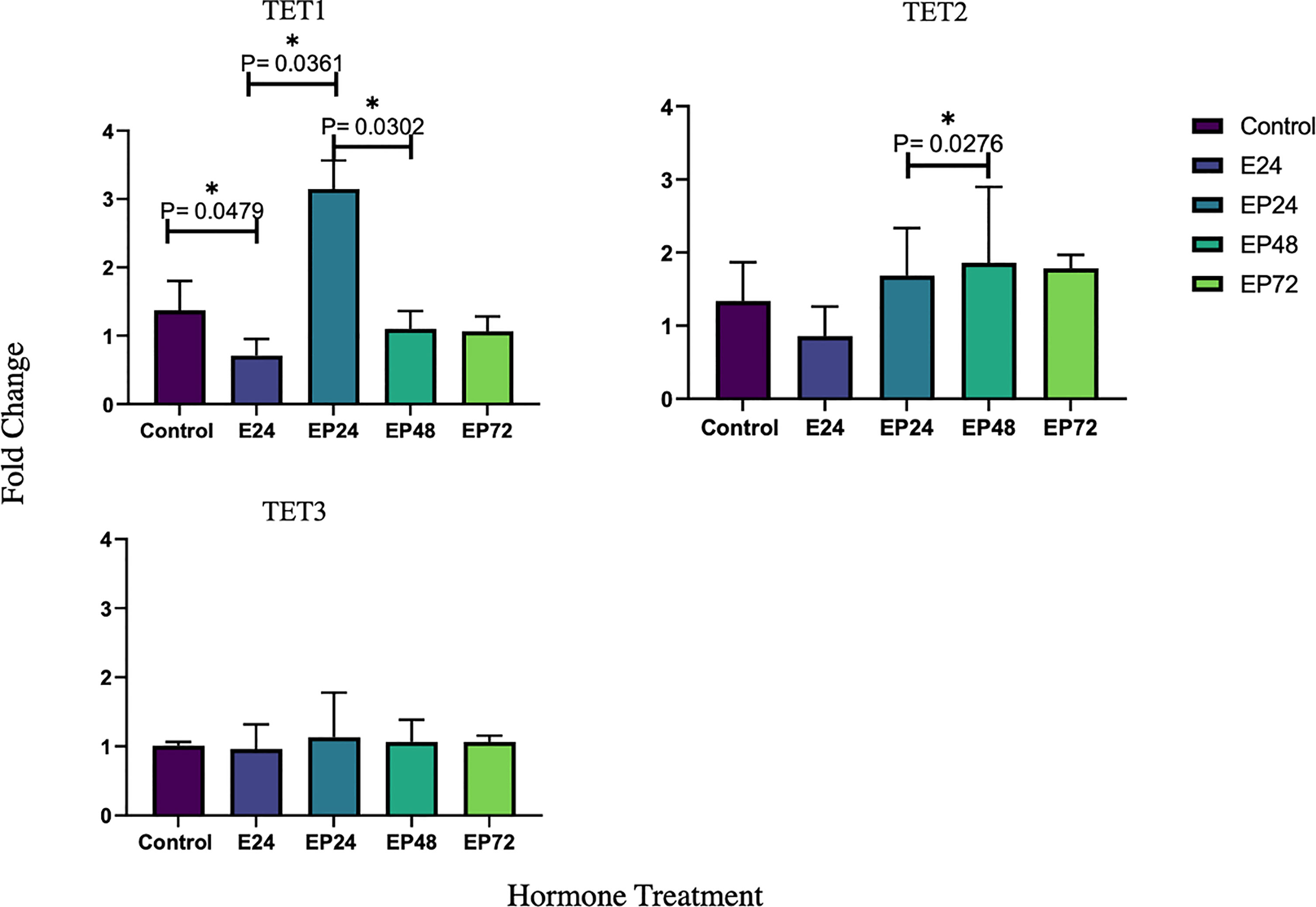
Figure 1 Relative TET1 TET2 and TET3 mRNA expression in response to steroid hormone treatment in AN3 cells. The y-axis shows the fold change of mRNA levels following different treatments compared with control, all results corrected against geo-mean expressions of three reference genes - YWHAZ, RPLO and RPL13a. The x-axis shows different treatment groups. E24 = 24h Estrogen; EP24, EP48 and EP72 = both Estrogen + Progesterone for 24, 48 and 72h. Data are presented as mean ± SEM *p < 0.05. P ≤ 0.1 was considered as approaching significance. The experimental setup included three independent sets of cell culture experiments (n = 3) and triplicates of each sample for the RT-PCR.
TET1 protein expression parallels the gene expression and is significantly downregulated when treated with estrogen-progesterone for 72 hours. According to the results of our previous study, TET1 mRNA was upregulated during the mid-secretory phase in healthy endometrial tissues and in response to progesterone treatment in epithelial cells, in vitro (29). The decreased protein expression during 72 hours of estrogen-progesterone treatment, suggests a potential aberrant regulation of TET1 in AN3 cells. Data by other studies report similar findings with decreased TET1 mRNA and protein expression in endometrial cancer tissues compared to normal (9, 36). It has been suggested that overexpression of DNMT3a and DNMT3b contribute to hypermethylation of ERα and PR, subsequently silencing these genes in endometrial cancer (37). TETs mediate epigenetic alterations via DNA de-methylation, a process where they actively remove the methyl group, to activate gene expression (38). Downregulation of TET1 gene and protein expression, could be associated with the abnormal inactivation of ERα and PR seen in endometrial cancer tissues (37, 39, 40). While other studies have reported mRNA expression of ERa in AN3 cells at the basal level (41), neither gene nor protein expression of ERa or PR in AN3 cells in the present study. The discrepancy in the ERa gene expression in both the studies could be attributed to the differences in the treatment protocol used. The data obtained from this study indicates that the downregulation of TET1 in response to estrogen and progesterone could potentially be contributing to epigenetic deregulation and warrants the need for more studies to investigate its role in endometrial cancer.
TET2 has been previously implicated in various types of malignancies (11, 42–46). Data from the present study imply that TET2 protein expression is upregulated when exposed to a combined estrogen-progesterone treatment for 24 hours (Figure 2). Further, it is also seen that mRNA expression of TET2 remains upregulated upon continued exposure to estrogen-progesterone treatment for 48 hours (Figure 1) in AN3 cells. TET2 expression has been shown to be significantly reduced in severe endometrial cancer and cervical squamous cell carcinoma tissues compared to their normal counterparts (9, 47). So far to our knowledge, there are no studies that have evaluated any cell specific changes in relation to malignancy or hormonal treatment in endometrium. Our previous study reported an upregulation of TET2 expression in non-estrogen primed, non-cancerous endometrial epithelial cells, in response to progesterone (29). Further studies are needed to fine tune the mechanisms by which TET2 might be deregulated in endometrial cancer.
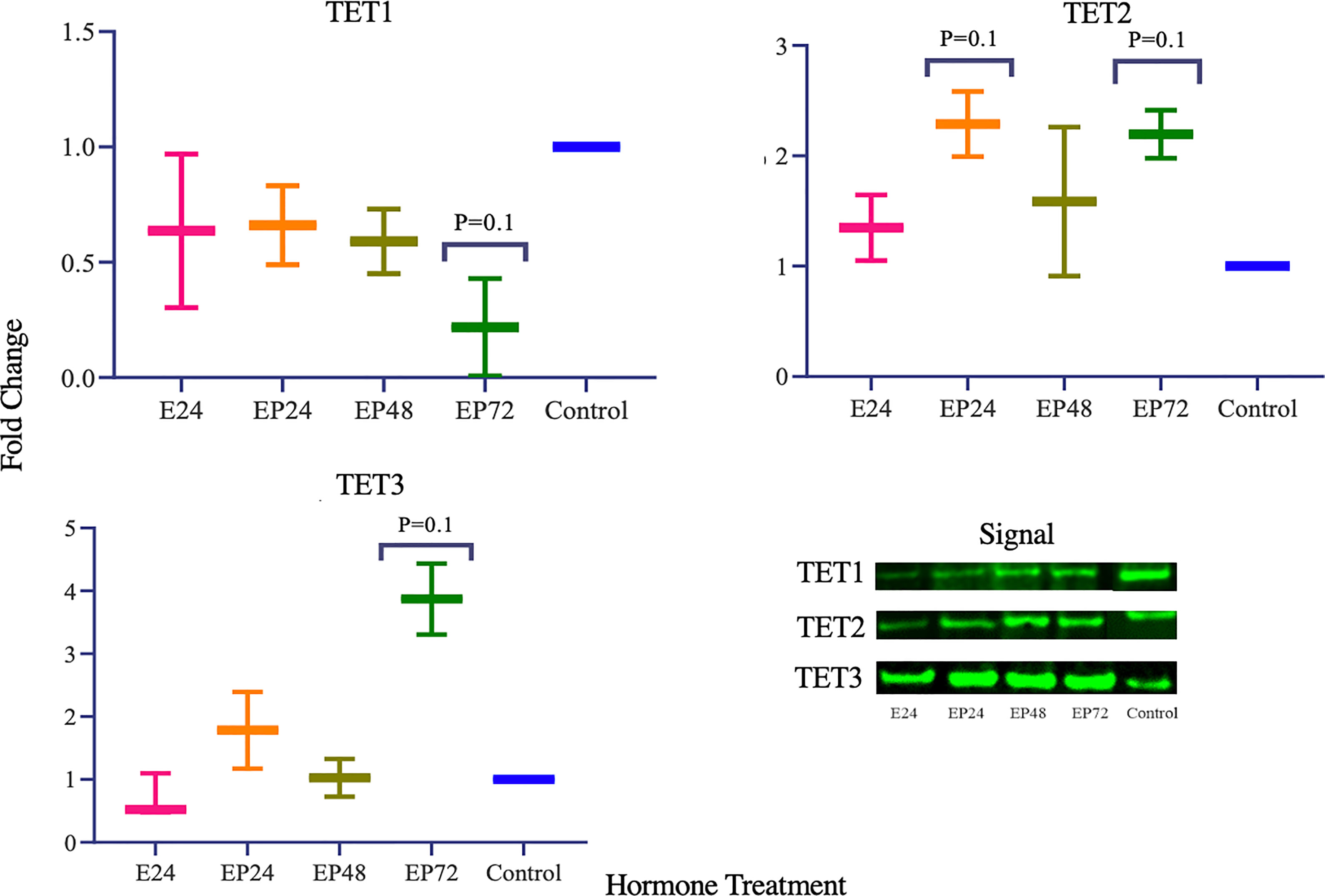
Figure 2 TET protein expression in response to different steroid hormone treatments in AN3 cells. A representative blot image for the particular weight band is shown next to the graph. The y-axis shows the fold change of protein levels following different treatments compared to control and x-axis shows the different treatment groups. E24, 24h Estrogen; EP24, EP48 and EP72 = both Estrogen + Progesterone for 24, 48 and 72h. Data are presented as mean ± SEM, *p < 0.05 (P ≤ 0.1 is considered as approaching significance). The experimental setup included three independent sets of cell culture experiments (n = 3) with three technical replicates for each sample.
While no hormonal effects on TET3 mRNA expression were observed, TET3 protein was significantly increased during 72 hours of estrogen-progesterone treatment (Figure 2). This is in agreement with Cieselski et al., who also reported an increased TET3 mRNA expression in endometrial cancer tissue biopsies (9). TET3 has been reported to be crucial in the maintenance of stem cell identity, DNA repair and overall genome stability in various tissues (48–51). Aberrations in stem cells have been implicated in the origin and progression of endometrial cancer (52, 53). Increased TET3 protein expression in cancer, could potentially indicate its involvement in abnormal stem cell regulation, contributing to progression, invasiveness and metastasis. Furthermore, reduced mRNA expression of TET2 and TET3 have been implicated in the induction of epithelial-mesenchymal transition in melanoma (54). This study, indicates an increased expression of TET2 and TET3 in the combined estrogen-progesterone treated samples in AN3 cells. The difference in results could be attributed to the type of cell and treatment protocols used. Collectively, it is implied that TET2 and TET3 could be involved in the differential regulation seen in cancers, however its exact association still needs to be explored further.
ER and PR have been extensively studied in endometrial pathologies such as cancer and endometriosis. AR, however, is a lesser explored steroid hormone receptor in endometrial biology. Our data imply that in AN3 cells, AR protein levels were significantly downregulated when treated with estrogen-progesterone together for 24, 48 and 72 hours (Figure 3). AR is a known anti-estrogen, which means that it has the ability to counteract the proliferative activity of estrogen (55). In normal epithelial cells, a differential and increased expression of AR is reported using immunohistochemistry during the secretory phase of the endometrium (56). Downregulation of AR levels are seen in the estrogen-progesterone treated samples, could be related to the decreased TET1 mRNA and protein expression. An association between TET1 and AR has also been suggested by Dhiman et al. (57). Their study reports that TET1, AR and thymine DNA glycosylase are co-recruited to the transcription start site of the Androgen Responsive Elements (AREs) to influence gene regulation in human prostate cells (57). Thus, suggesting that TET1 could be potentially involved in the transcriptional activation of AR in endometrial biology. Moreover, it has been suggested that AR suppresses tumor growth in ER positive breast malignancies (58). The findings of this study imply that the downregulation of AR and absence of PR gene expression in AN3 cells, could be contributing to the uncontrolled proliferation, seen in endometrial cancer cells. This study suggests that steroid hormones regulate the crosstalk between TETs and steroid hormone receptors in endometrial biology. Understanding this regulation more robustly in the endometrium, could help provide novel targets for therapeutic interventions for associated pathologies.
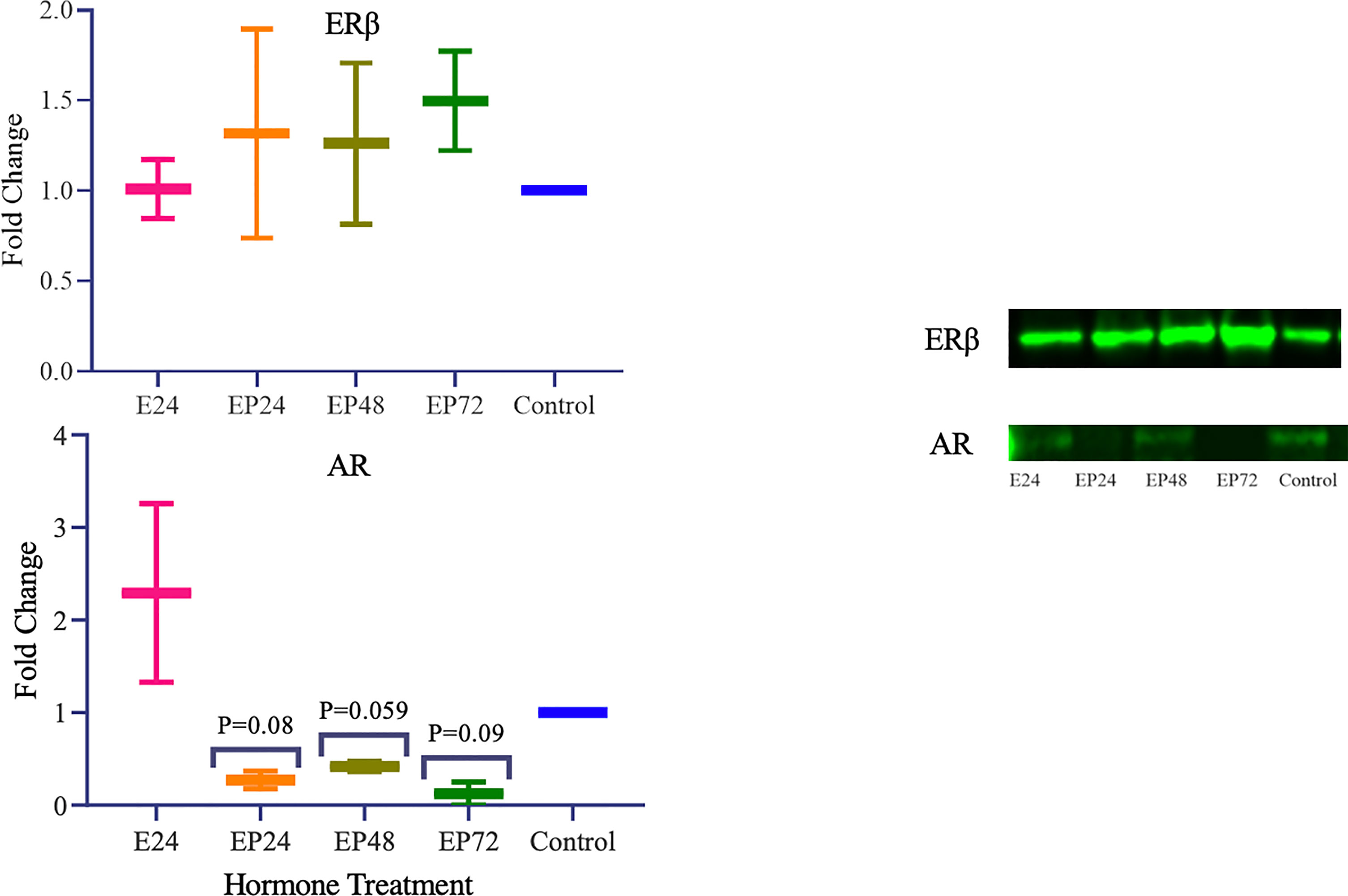
Figure 3 Steroid hormone protein expression in response to different steroid hormone treatments in AN3 cells. A representative blot image for the particular weight band is shown next to the graph. The y-axis shows the fold change of protein levels following different treatments compared to control and x-axis shows the different treatment groups. E24 = 24h Estrogen; EP24, EP48 and EP72 = both Estrogen + Progesterone for 24, 48 and 72h. Data are presented as mean ± SEM. P ≤ 0.1 is considered as approaching significance. The experimental setup included three independent sets of cell culture experiments (n =3) with three technical replicates for each sample.
In RL95-2 cells, one way ANOVA analysis suggest a significant influence of hormones on TET mRNA expressions. However, no significant differences between individual treatments were observed. Gene expression for TET1, TET2 and TET3 were highest when exposed to combined estrogen-progesterone treatments for 24 hours and 48 hours (Figure 4). Protein data indicated a differential expression of TETs when exposed to different treatments. TET1 and TET3 levels were significantly increased in response to 72 hour combined estrogen-progesterone treatment (Figure 5). mRNA and Protein upregulation of TET1 is also demonstrated in another study suggesting that the hypoxic, chronic inflammatory environment seen in endometrial cancer, can up-regulate TET1 expression and induce its downstream gene transcription (59). Upon exposure to 24 hours of estrogen, TET1 and TET3 were significantly downregulated whereas TET2 was upregulated (Figure 5), implying a potential difference in the regulation and function of TETs. TET2 has been reported to serve as a co-activator of ERα by de-methylating and maintaining low CpG methylation levels in breast cancer cell lines (60). This could potentially explain the significant upregulation of ERα when treated with estrogen-progesterone for 24 hours (Figure 6). Additionally, the upregulation of ERβ could be correlated to the increased protein expression of TET1 and TET3 in response to 72 hours of estrogen-progesterone treatment. This finding suggests a possible interplay between TETs, regulated by hormones and influencing ERα and ERβ expression in RL95-2 cells.
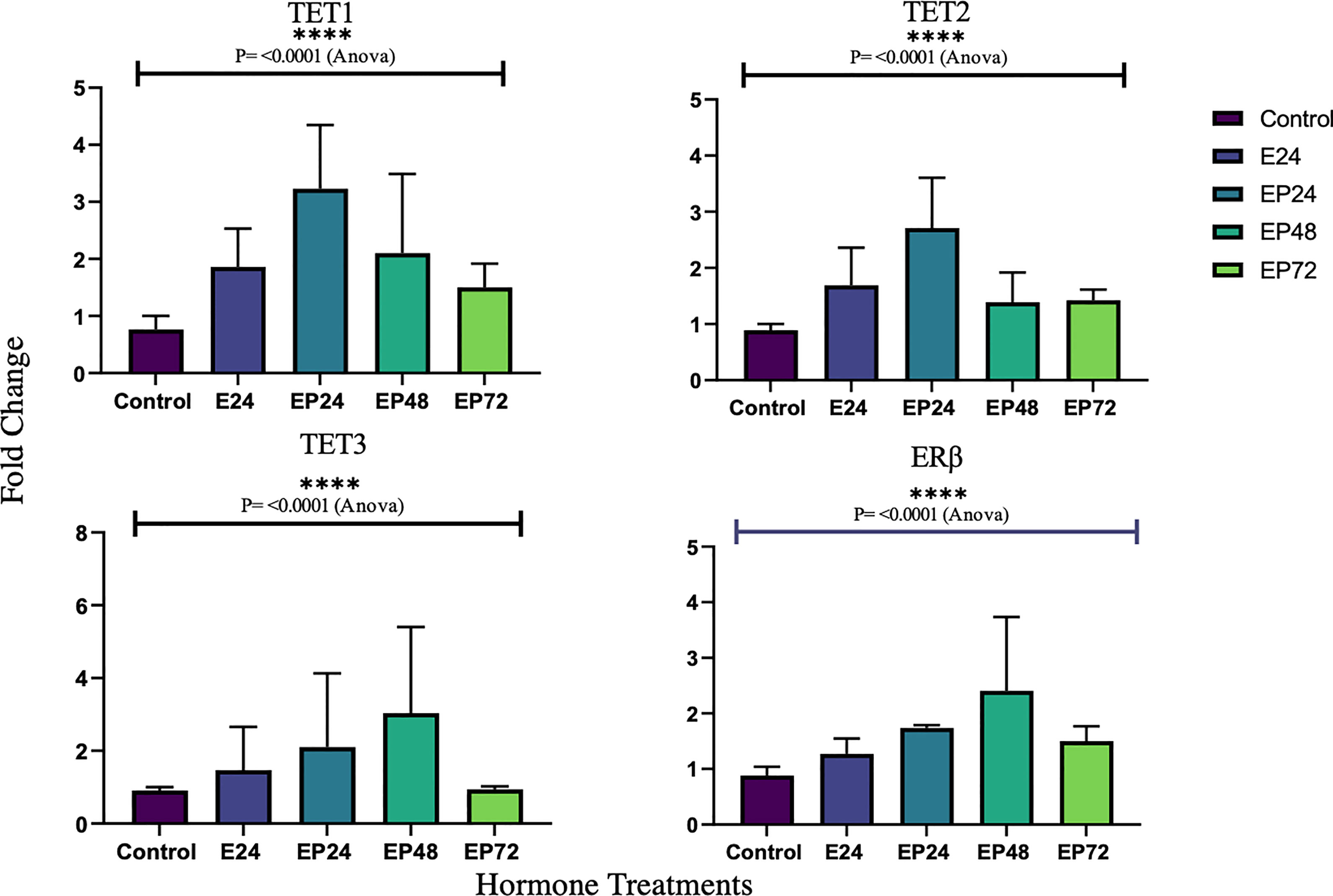
Figure 4 Relative TET and ERb mRNA expression in response to different steroid hormone treatments in RL95-2 cells. The y-axis shows the fold change of mRNA levels following different treatments compared with control, all results corrected against geo-mean expressions of three reference genes - YWHAZ, RPLO and RPL13a. The x-axis shows different treatment groups E24 = 24h Estrogen; EP24, EP48 and EP72 = both Estrogen + Progesterone for 24, 48 and 72h. Data are presented as mean ± SEM; ***P < 0.001; P ≤ 0.1 was considered as approaching significance. One way ANOVA test revealed significant variations in TET and ERb expression across treatments (****P<0.0001). The experimental setup included three independent sets of cell culture experiments (n =3) and triplicates of each sample for the RT-PCR.
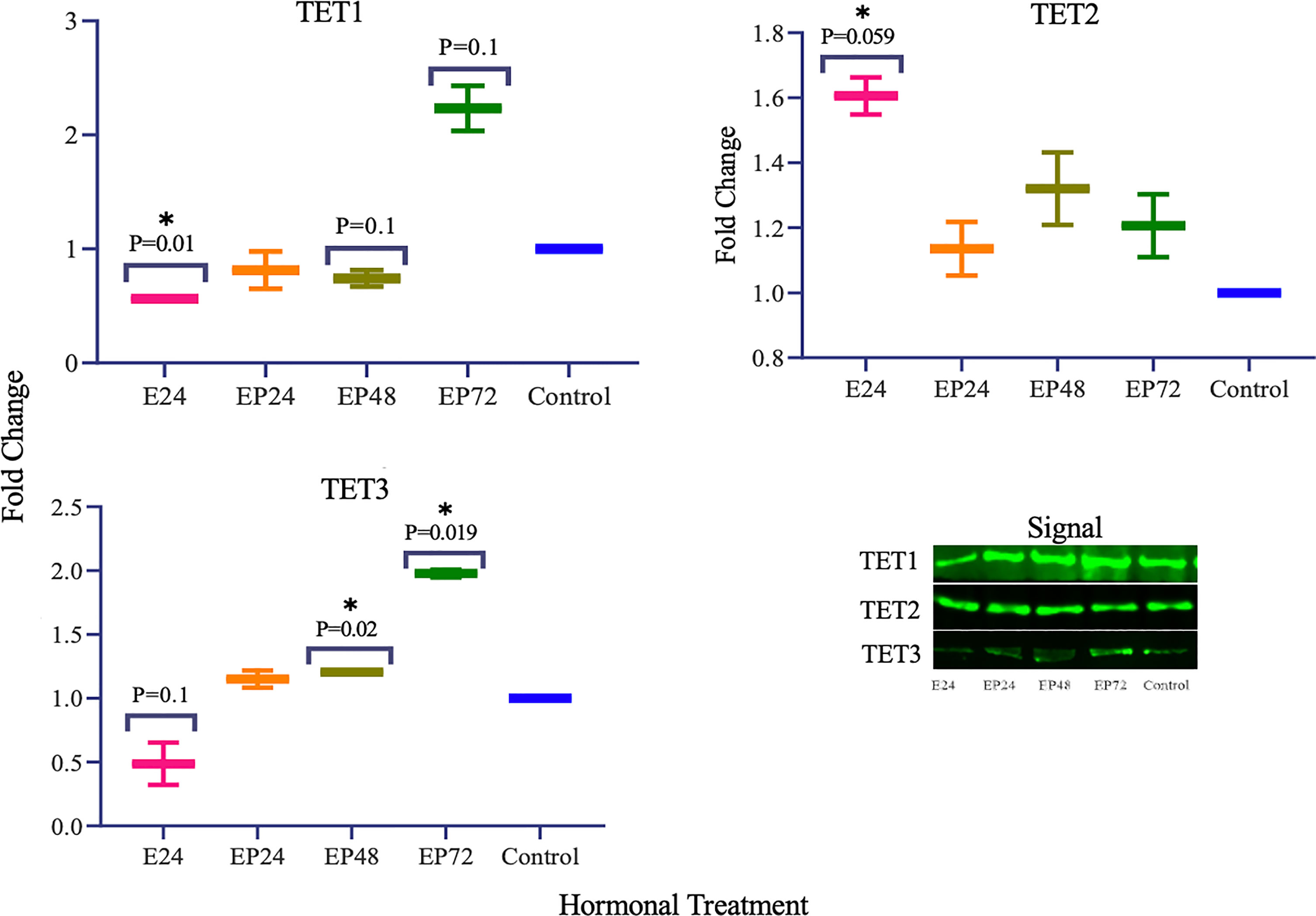
Figure 5 TET protein expression in response to different steroid hormone treatments in RL95-2 cells. A representative blot image for the particular weight band is shown next to the graph. The y-axis shows the fold change of protein levels following different treatments compared to control and x-axis shows the different treatment groups. E24 = 24h Estrogen; EP24, EP48 and EP72 = both Estrogen + Progesterone for 24, 48 and 72h. Data are presented as mean ± SEM, *p < 0.05, P ≤ 0.1 is considered as approaching significance. The experimental setup included three independent sets of cell culture experiments (n =3) with three technical replicates for each sample.
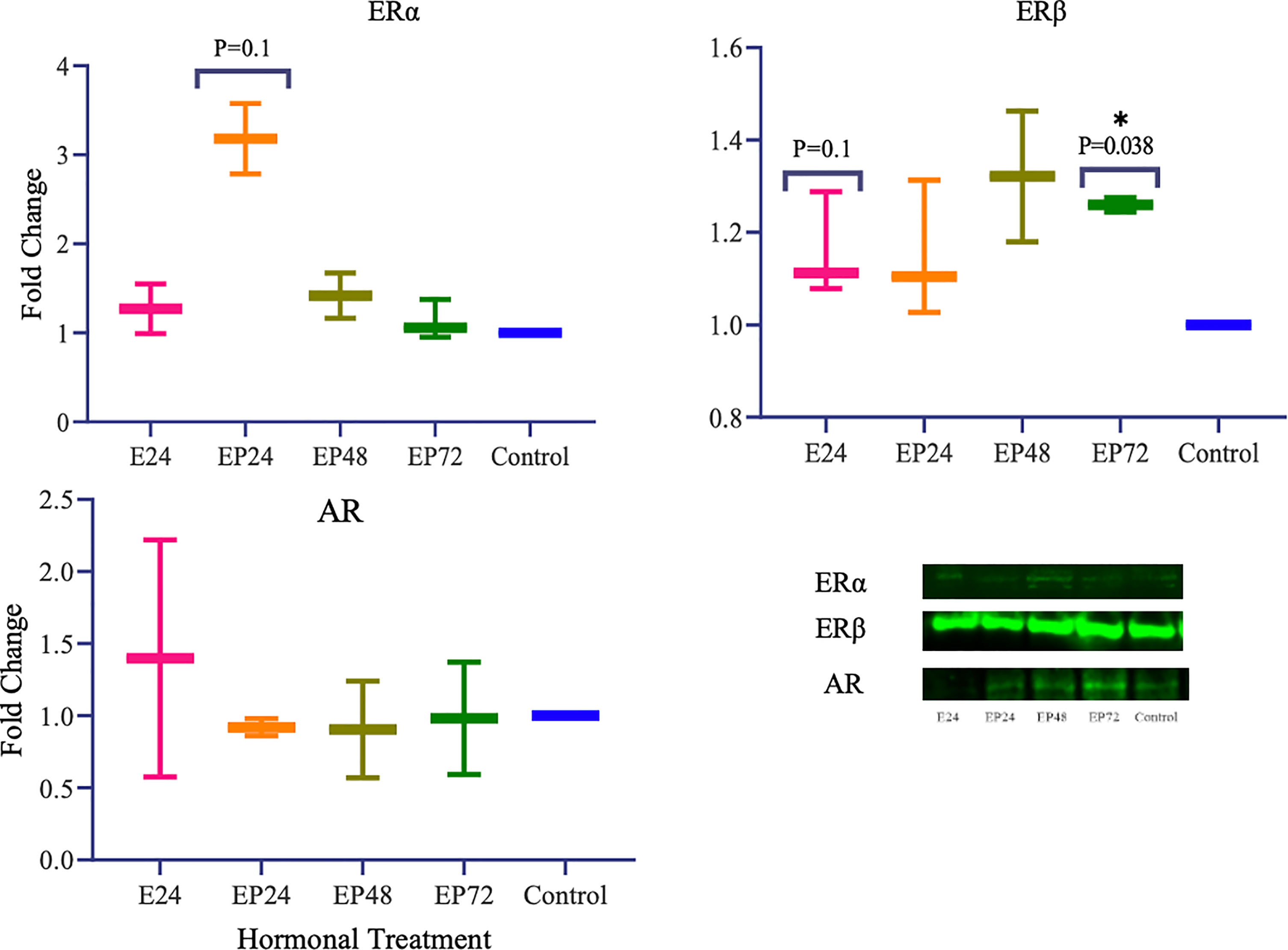
Figure 6 Steroid hormone protein expression in response to different steroid hormone treatments in RL95-2 cells. A representative blot image for the particular weight band is shown next to the graph. The y-axis shows the fold change of protein levels following different treatments compared to control and x-axis shows the different treatment groups. E24 = 24h Estrogen; EP24, EP48 and EP72 = both Estrogen + Progesterone for 24, 48 and 72h. Data are presented as mean ± SEM, *p < 0.05, P ≤ 0.1 is considered as approaching significance. The experimental setup included three independent sets of cell culture experiments (n = 3) with three technical replicates for each sample.
Conflicting studies on the mRNA expression of ERα and ERβ have been reported in endometrial cancer. While some suggest lower ERα expression (37, 61), others report higher ERα expression in comparison to ERβ in endometrial cancer tissues (62, 63). The results of this study indicate a differential steroid hormone receptor regulation between RL95-2 and AN3 cells. RL95-2 cells have an increased expression of ERα and ERβ protein during treatments (Figure 6). Whereas in AN3 cells, no mRNA data for either were observed and only ERβ protein bands were seen with no statistically significant differences. The results of this study partially agrees with Sun et al., who report significantly increased ERα mRNA and protein expression in AN3 and RL95-2 cells in comparison to other endometrial cancer cell lines (41). They also suggest an increased gene expression of estrogen related receptor alpha (ERRα), an orphan nuclear receptor known to mediate the effects of estrogen, in AN3 and RL95-2 cells (41). Therefore implying the need to study the involvement of orphan nuclear receptors in estrogen signaling and action, as well as understanding their association with TETs is imperative. This is also consistent with findings in other endometrial cancer cells that suggest, TET1 increases estrogen sensitivity by upregulating mRNA expression of orphan nuclear receptor - GPER, in ishikawa and HEC-1-A cells (64).
PR was not expressed in RL95-2 cells, either at the mRNA level or at the protein level, which is a finding reported in another study as well (65). AR however, was not expressed at the mRNA level but was seen at the protein level, however the differences were not significant (Figure 6). Previously, AR protein expression has been reported in endometrial carcinomas with conflicting data on the level of expression. While Sasaki et al., demonstrated hypermethylation mediated AR gene silencing, Ito et al., suggested increased AR expression in endometrial carcinoma tissues (56, 66). Recent data imply that AR positivity is seen in a subset of endometrial carcinomas and is expressed conversely to ERs (55). A similar correlation can be drawn from the findings of this study, wherein increased ERα and ERβ is associated with reduced AR protein expression in RL95-2 cells. Since AR is anti-proliferative, its use as a potential target for curbing uncontrollable cellular hypertrophy is implied however this might not be the case for all tumors and more studies on tumor endocrinology is needed. A better understanding of the steroid hormone regulation and epigenetic axis in female cancers could help in the development of targeted transcriptional endocrine therapies.
In this study, a degree of variation between mRNA and protein expression of TETs and steroid hormone receptors in both, AN3 and RL95-2 cells was observed. The correlation between transcription and translation is complex and depends on several biological and technical factors. It has been suggested that the physical properties of transcription, can alter the translation efficiency at various levels contributing to a discrepancy between mRNA and protein data (67). The other most important and highly variable factor, influencing mRNA-protein correlation is the individual half-lives of proteins (68). For instance, it has been reported that long term estrogen exposure, increases ERα half-life, maintaining protein stability and slowing rate of proteolysis, which could explain the presence of ERα in RL95-2, despite no mRNA expression being observed (69). Subsequently, post-translational and post transcriptional modifications and delayed synthesis between mRNA and protein, could also result in a poor mRNA-protein correlation (70, 71).
In summary, endometrial cancer is complex and involves abnormal steroid hormone signaling. This study evaluates steroid hormone regulation of TETs and steroid hormone receptors in vitro and also highlights the importance of evaluating different cancer cell lines independently, to understand the mechanisms of hormone action. It is proposed that differential protein expression of TETs during different hormonal treatments could be involved in the regulation of ERα and ERβ in RL95-2 and AN3 cells. The downregulation AR in AN3 cell line could be explored further as a potential target for hormone therapy. However, for a more comprehensive understanding of the association between TETs and steroid hormone receptors, additional studies including endometrial cancer tissues and primary cells, need to be undertaken. Overall, this study provides a preliminary account, indicating that TETs, steroid hormones and their receptors might be co-regulated to maintain hormone signaling in the endometrium. Future studies involving the assessment of 5-hmC levels and gene promoter sequencing might help in determining the epigenomic regulation of steroid hormone receptors in endometrial cancer cells more definitively.
The protocol used in this study, included a limited 24h of estrogen treatment prior to the addition of a combined estrogen and progesterone. This was done to mimic a snapshot of the molecular events that occur in utero during the early proliferative stage. The crucial estrogen priming process, enriches the endometrium with steroid hormone receptors preparing it for a successful progesterone action during the secretory phase. Due to the challenging nature of the tissue and complexity of the experiments, it was not possible to include multiple time points for estrogen priming at this stage but is suggested in the scope for future studies.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
VM: performed major experiments and primary contributor of manuscript writing and editing manuscript. PG: reviewed manuscript and minor experiments. LJ: reviewed manuscript and minor experiments. AP: conceived the idea, provided perceptive comments on drafts, and approved the content of the manuscript. All authors contributed to the article and approved the submitted version.
The research project was funded and supported by Auckland Medical Research Foundation (AMRF, grant number: 1116005). VM was supported by Liggins Institute Departmental Fund and AP was supported by Sidney Taylor Trust, University of Auckland.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine; AR, Androgen Receptor; AREs, Androgen Responsive Elements; CS-FBS, Charcoal stripped FBS; C treatment, Control; DNMTs, DNA Methyltransferases; E treatment, Estradiol treatment; EP treatment, Estrogen + progesterone treatment; Era, Estrogen receptor alpha; ERβ, Estrogen receptor beta; ER, Estrogen receptors; ERRα, Estrogen related receptor alpha; PR, Progesterone receptor; TDG, thymine DNA glycosylase; TET, Ten-eleven translocation.
1. Mertens HJ, Heineman MJ, Theunissen PH, de Jong FH, Evers JL. Androgen, Estrogen and Progesterone Receptor Expression in the Human Uterus During the Menstrual Cycle. Eur J Obstetrics Gynecology Reprod Biol (2001) 98:58–65. doi: 10.1016/S0301-2115(00)00554-6
2. Munro SK, Farquhar CM, Mitchell MD, Ponnampalam AP. Epigenetic Regulation of Endometrium During the Menstrual Cycle. Mol Hum Reprod (2010) 16:297–310. doi: 10.1093/molehr/gaq010
3. Dyson MT, Roqueiro D, Monsivais D, Ercan CM, Pavone ME, Brooks DC, et al. Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis. PloS Genet (2014) 10:e1004158. doi: 10.1371/journal.pgen.1004158
4. Hsiao K-Y, Wu M-H, Tsai S-J. Epigenetic Regulation of the Pathological Process in Endometriosis. Reprod Med Biol (2017) 16:314–9. doi: 10.1002/rmb2.12047
5. Hewitt SC, Korach KS. Estrogen Receptors: New Directions in the New Millennium. Endocr Rev (2018) 39(5):664–75. doi: 10.1210/er.2018-00087
6. Spruijt CG, Vermeulen M. DNA Methylation: Old Dog, New Tricks? Nat Struct Mol Biol (2014) 21:949–54. doi: 10.1038/nsmb.2910
7. Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science (2009) 324:929–30. doi: 10.1126/science.1169786
8. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner Tet1. Science (2009) 324:930–5. doi: 10.1126/science.1170116
9. Ciesielski P, Jóźwiak P, Wójcik-Krowiranda K, Forma E, Cwonda Ł, Szczepaniec S, et al. Differential Expression of Ten-Eleven Translocation Genes in Endometrial Cancers. Tumor Biol (2017) 39:101042831769501. doi: 10.1177/1010428317695017
10. Szczepańska M, Wirstlein P, Zawadzka M, Wender-Ożegowska E, Jagodziński PP. Alternation of Ten-Eleven Translocation 1, 2, and 3 Expression in Eutopic Endometrium of Women With Endometriosis-Associated Infertility. Gynecological Endocrinol (2018) 34:1084–90. doi: 10.1080/09513590.2018.1490403
11. Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, et al. Combined Deficiency of Tet1 and Tet2 Causes Epigenetic Abnormalities But is Compatible With Postnatal Development. Dev Cell (2013) 24:310–23. doi: 10.1016/j.devcel.2012.12.015
12. Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, et al. The Role of Tet3 DNA Dioxygenase in Epigenetic Reprogramming by Oocytes. Nature (2011) 477:606–10. doi: 10.1038/nature10443
13. Kurian JR, Louis S, Keen KL, Wolfe A, Terasawa E, Levine JE. The Methylcytosine Dioxygenase Ten-Eleven Translocase-2 (Tet2) Enables Elevated GnRH Gene Expression and Maintenance of Male Reproductive Function. Endocrinology (2016) 157:3588–603. doi: 10.1210/en.2016-1087
14. Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the Mammalian Zygote is Linked With Epigenetic Reprogramming. Nat Commun (2011) 2:241. doi: 10.1038/ncomms1240
15. Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in Erasure of Genomic Imprinting. Nature (2013) 504:460–4. doi: 10.1038/nature12805
16. Kukushkina V, Modhukur V, Suhorutšenko M, Peters M, Mägi R, Rahmioglu N, et al. DNA Methylation Changes in Endometrium and Correlation With Gene Expression During the Transition From Pre-Receptive to Receptive Phase. Sci Rep (2017) 7:3916. doi: 10.1038/s41598-017-03682-0
17. Jin X, Li Y, Guo Y, Jia Y, Qu H, Lu Y, et al. Erα is Required for Suppressing OCT4-Induced Proliferation of Breast Cancer Cells via DNMT1/ISL1/ERK Axis. Cell Proliferation (2019) 52:e12612. doi: 10.1111/cpr.12612
18. Yamagata Y, Asada H, Tamura I, Lee L, Maekawa R, Taniguchi K, et al. DNA Methyltransferase Expression in the Human Endometrium: Down-Regulation by Progesterone and Estrogen. Hum Reprod (2009) 24:1126–32. doi: 10.1093/humrep/dep015
19. Zhao Z, Fan L, Fortress AM, Boulware MI, Frick KM. Hippocampal Histone Acetylation Regulates Object Recognition and the Estradiol-Induced Enhancement of Object Recognition. J Neurosci (2012) 32:2344–51. doi: 10.1523/JNEUROSCI.5819-11.2012
20. Jabbour HN, Kelly RW, Fraser HM, Critchley HOD. Endocrine Regulation of Menstruation. Endocr Rev (2006) 27:17–46. doi: 10.1210/er.2004-0021
21. Jacobsen BM, Horwitz KB. Progesterone Receptors, Their Isoforms and Progesterone Regulated Transcription. Mol Cell Endocrinol (2012) 357(1-2):18–29. doi: 10.1016/j.mce.2011.09.016
22. Vienonen A, Miettinen S, Bläuer M, Martikainen PM, Tomás E, Heinonen PK, et al. Expression of Nuclear Receptors and Cofactors in Human Endometrium and Myometrium. J Soc Gynecologic Invest (2004) 11:104–12. doi: 10.1016/j.jsgi.2003.09.003
23. Bulun S, Cheng Y-H, Pavone M, Xue Q, Attar E, Trukhacheva E, et al. Estrogen Receptor-β, Estrogen Receptor-α, and Progesterone Resistance in Endometriosis. Semin Reprod Med (2010) 28:036–43. doi: 10.1055/s-0029-1242991
24. Bulun SE, Cheng Y-H, Yin P, Imir G, Utsunomiya H, Attar E, et al. Progesterone Resistance in Endometriosis: Link to Failure to Metabolize Estradiol. Mol Cell Endocrinol (2006) 248:94–103. doi: 10.1016/j.mce.2005.11.041
25. Kim JJ, Kurita T, Bulun SE. Progesterone Action in Endometrial Cancer, Endometriosis, Uterine Fibroids, and Breast Cancer. Endocr Rev (2013) 34:130–62. doi: 10.1210/er.2012-1043
26. Ma X, Gao X. Epigenetic Modifications and Carcinogenesis of Human Endometrial Cancer. Austin J Clin Pathol Austin J Clin Pathol (2014) 1:1014–3.
27. Sasaki M, Kaneuchi M, Fujimoto S, Tanaka Y, Dahiya R. Hypermethylation can Selectively Silence Multiple Promoters of Steroid Receptors in Cancers. Mol Cell Endocrinol (2003) 202:201–7. doi: 10.1016/S0303-7207(03)00084-4
28. Tao MH, Freudenheim JL. DNA Methylation in Endometrial Cancer. Epigenetics (2010) 5:491–8. doi: 10.4161/epi.5.6.12431
29. Mahajan V, Osavlyuk D, Logan PC, Amirapu S, Ponnampalam AP. Expression and Steroid Hormone Regulation of TETs and DNMTs in Human Endometrium. Reproduction (2020) 1:247–57. doi: 10.1530/REP-19-0562
30. Spandidos A, Wang X, Wang H, Seed B. PrimerBank: A Resource of Human and Mouse PCR Primer Pairs for Gene Expression Detection and Quantification. Nucleic Acids Res (2010) 38:D792–9. doi: 10.1093/nar/gkp1005
31. Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A Comprehensive Collection of Experimentally Validated Primers for Polymerase Chain Reaction Quantitation of Murine Transcript Abundance. BMC Genomics (2008) 9:633. doi: 10.1186/1471-2164-9-633
32. Wang X, Spandidos A, Wang H, Seed B. PrimerBank: A PCR Primer Database for Quantitative Gene Expression Analysis 2012 Update. Nucleic Acids Res (2012) 40:D1144–9. doi: 10.1093/nar/gkr1013
33. Wang X, Seed B. A PCR Primer Bank for Quantitative Gene Expression Analysis. Nucleic Acids Res (2003) 31:154e–154. doi: 10.1093/nar/gng154
34. Kamal AM, Bulmer JN, Decruze SB, Stringfellow HF, Martin-Hirsch P, Hapangama DK. Androgen Receptors are Acquired by Healthy Postmenopausal Endometrial Epithelium and Their Subsequent Loss in Endometrial Cancer is Associated With Poor Survival. Br J Cancer (2016) 114:688–96. doi: 10.1038/bjc.2016.16
35. Schmittgen TD, Livak KJ. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat Protoc (2008) 3:1101–8. doi: 10.1038/nprot.2008.73
36. Liu NT, Perng CL, Chou YC, Ko PS, Lin YJ, Lin YV, et al. Loss of Ten-Eleven Translocation 1 (TET1) Expression as a Diagnostic and Prognostic Biomarker of Endometrial Carcinoma. PloS One (2021) 16:e0259330. doi: 10.1371/journal.pone.0259330
37. He D, Wang X, Zhang Y, Zhao J, Han R, Dong Y. DNMT3A/3B Overexpression Might be Correlated With Poor Patient Survival, Hypermethylation and Low Expression of ESR1/PGR in Endometrioid Carcinoma: An Analysis of the Cancer Genome Atlas. Chin Med J (2019) 132:161–70. doi: 10.1097/CM9.0000000000000054
38. Kovács T, Szabó-Meleg E, Ábrahám IM. Estradiol-Induced Epigenetically Mediated Mechanisms and Regulation of Gene Expression. Int J Mol Sci (2020) 21(9):3177. doi: 10.3390/ijms21093177
39. Qu W, Zhao Y, Wang X, Qi Y, Zhou C, Hua Y, et al. Culture Characters, Genetic Background, Estrogen/Progesterone Receptor Expression, and Tumorigenic Activities of Frequently Used Sixteen Endometrial Cancer Cell Lines. Clin Chim Acta (2019) 489:225–32. doi: 10.1016/j.cca.2018.08.013
40. Zhou X, Wang Z, Zhao Y, Podratz K, Jiang S. Characterization of Sixteen Endometrial Cancer Cell Lines. Cancer Res (2007) 67:3870.
41. Sun P, Mao X, Gao M, Huang M, Chen L, Ruan G, et al. Novel Endocrine Therapeutic Strategy in Endometrial Carcinoma Targeting Estrogen-Related Receptor α by XCT790 and siRNA. Cancer Manage Res (2018) 10:2521–35. doi: 10.2147/CMAR.S168043
42. Bronowicka-Kłys DE, Roszak A, Pawlik P, Sajdak S, Sowińska A, Jagodziński PP. Transcript Levels of Ten-Eleven Translocation Type 1-3 in Cervical Cancer and non-Cancerous Cervical Tissues. Oncol Lett (2017) 13:3921–7. doi: 10.3892/ol.2017.5930
43. Delhommeau F, Dupont S, Valle VD, James C, Trannoy S, Massé A, et al. Mutation in TET2 in Myeloid Cancers. New Engl J Med (2009) 360:2289–301. doi: 10.1056/NEJMoa0810069
44. Li R, Zhou Y, Cao Z, Liu L, Wang J, Chen Z, et al. TET2 Loss Dysregulates the Behavior of Bone Marrow Mesenchymal Stromal Cells and Accelerates Tet2-/–Driven Myeloid Malignancy Progression. Stem Cell Rep (2018) 10:166–79. doi: 10.1016/j.stemcr.2017.11.019
45. Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event During Human Lymphomagenesis. Cancer Cell (2011) 20:25–38. doi: 10.1016/j.ccr.2011.06.003
46. Scourzic L, Mouly E, Bernard OA. TET Proteins and the Control of Cytosine Demethylation in Cancer. Genome Med (2015) 7:9. doi: 10.1186/s13073-015-0134-6
47. Zhang LY, Han CS, Li PL, Zhang XC. 5-Hydroxymethylcytosine Expression is Associated With Poor Survival in Cervical Squamous Cell Carcinoma. Jpn J Clin Oncol (2016) 46:427–34. doi: 10.1093/jjco/hyw002
48. Guan W, Guyot R, Samarut J, Flamant F, Wong J, Gauthier KC. Methylcytosine Dioxygenase TET3 Interacts With Thyroid Hormone Nuclear Receptors and Stabilizes Their Association to Chromatin. Proc Natl Acad Sci U States America (2017) 114:8229–34. doi: 10.1073/pnas.1702192114
49. Jiang D, Wei S, Chen F, Zhang Y, Li J. TET3-Mediated DNA Oxidation Promotes ATR-Dependent DNA Damage Response. EMBO Rep (2017) 18:781–96. doi: 10.15252/embr.201643179
50. Santiago M, Antunes C, Guedes M, Iacovino M, Kyba M, Reik W, et al. Tet3 Regulates Cellular Identity and DNA Methylation in Neural Progenitor Cells. Cell Mol Life Sci (2019) 77:2871. doi: 10.1007/s00018-019-03335-7
51. Zhang J, Hao L, Wei Q, Zhang S, Cheng H, Zhai Y, et al. TET3 Overexpression Facilitates DNA Reprogramming and Early Development of Bovine SCNT Embryos. Reproduction (2020) 160:379–91. doi: 10.1530/REP-20-0021
52. Giannone G, Attademo L, Scotto G, Genta S, Ghisoni E, Tuninetti V, et al. Endometrial Cancer Stem Cells: Role, Characterization and Therapeutic Implications. Cancers (2019) 11:1820. doi: 10.3390/cancers11111820
53. Tempest N, Maclean A, Hapangama DK. Endometrial Stem Cell Markers: Current Concepts and Unresolved Questions. Int J Mol Sci (2018) 19(10):3240. doi: 10.3390/ijms19103240
54. Gong F, Guo Y, Niu Y, Jin Y, Zhang J, Shi X, et al. Epigenetic Silencing of TET2 and TET3 Induces an EMT-Like Process in Melanoma. Oncotarget (2017) 8:315–28. doi: 10.18632/oncotarget.13324
55. Zadeh SL, Duska LR, Mills AM. Androgen Receptor Expression in Endometrial Carcinoma. Int J Gynecological Pathol (2018) 37:167–73. doi: 10.1097/PGP.0000000000000401
56. Ito K, Suzuki T, Akahira J, Moriya T, Kaneko C, Utsunomiya H, et al. Expression of Androgen Receptor and 5?-Reductases in the Human Normal Endometrium and its Disorders. Int J Cancer (2002) 99:652–7. doi: 10.1002/ijc.10394
57. Dhiman VK, Attwood K, Campbell MJ, Smiraglia DJ. Hormone Stimulation of Androgen Receptor Mediates Dynamic Changes in DNA Methylation Patterns at Regulatory Elements. Oncotarget (2015) 6:42575–89. doi: 10.18632/oncotarget.6471
58. Hickey TE, Selth LA, Chia KM, Laven-Law G, Milioli HH, Roden D, et al. The Androgen Receptor is a Tumor Suppressor in Estrogen Receptor–Positive Breast Cancer. Nat Med (2021) 27:310–20. doi: 10.1038/s41591-020-01168-7
59. Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y, et al. Role of Tet Proteins in 5mc to 5hmc Conversion, ES-Cell Self-Renewal and Inner Cell Mass Specification. Nature (2010) 466:1129–33. doi: 10.1038/nature09303
60. Wang L, Ozark PA, Smith ER, Zhao Z, Marshall SA, Rendleman EJ, et al. TET2 Coactivates Gene Expression Through Demethylation of Enhancers. Sci Adv (2018) 4:6986. doi: 10.1126/sciadv.aau6986
61. Yu HC, Lin CY, Chang WC, Shen BJ, Chang WP, Chuang CM. Increased Association Between Endometriosis and Endometrial Cancer: A Nationwide Population-Based Retrospective Cohort Study. Int J Gynecological Cancer (2015) 25:447–52. doi: 10.1097/IGC.0000000000000384
62. Levine DA. Integrated Genomic Characterization of Endometrial Carcinoma. Nature (2013) 497:67–73. doi: 10.1038/nature12113
63. Rodriguez AC, Blanchard Z, Maurer KA, Gertz J. Estrogen Signaling in Endometrial Cancer: A Key Oncogenic Pathway With Several Open Questions. Hormones Cancer (2019) 10(2-3):51–63. doi: 10.1007/s12672-019-0358-9
64. Lv QY, Xie BY, Yang BY, Ning CC, Shan WW, Gu C, et al. Increased TET1 Expression in Inflammatory Microenvironment of Hyperinsulinemia Enhances the Response of Endometrial Cancer to Estrogen by Epigenetic Modulation of GPER. J Cancer (2017) 8:894–902. doi: 10.7150/jca.17064
65. Yan X, Zhang H, Ke J, Zhang Y, Dai C, Zhu M, et al. Progesterone Receptor Inhibits the Proliferation and Invasion of Endometrial Cancer Cells by Up Regulating Krüppel-Like Factor 9. Trans Cancer Res (2020) 9:2220–30. doi: 10.21037/tcr.2020.03.53
66. Sasaki M, Oh BR, Dharia A, Fujimoto S, Dahiya R. Inactivation of the Human Androgen Receptor Gene is Associated With CpG Hypermethylation in Uterine Endometrial Cancer. Mol Carcinog (2000) 29:59–66. doi: 10.1002/1098-2744(200010)29:2<59::AID-MC2>3.0.CO;2-6
67. Maier T, Güell M, Serrano L. Correlation of mRNA and Protein in Complex Biological Samples. FEBS Lett (2009) 583(24):3966–73. doi: 10.1016/j.febslet.2009.10.036
68. Wu G, Nie L, Zhang W. Integrative Analyses of Posttranscriptional Regulation in the Yeast Saccharomyces Cerevisiae Using Transcriptomic and Proteomic Data. Curr Microbiol (2008) 57:18–22. doi: 10.1007/s00284-008-9145-5
69. Valley CC, Solodin NM, Powers GL, Ellison SJ, Alarid ET. Temporal Variation in Estrogen Receptor-α Protein Turnover in the Presence of Estrogen. J Mol Endocrinol (2008) 40:23–34. doi: 10.1677/JME-07-0067
70. Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship Between Differentially Expressed mRNA and mRNA-Protein Correlations in a Xenograft Model System. Sci Rep (2015) 5:1–9. doi: 10.1038/srep10775
Keywords: gene expression, steroid hormones and receptors, endometrial cancer cells, ten eleven translocation (TET proteins), DNA hydroxymethylation (5hmC)
Citation: Mahajan V, Gujral P, Jain L and Ponnampalam AP (2022) Differential Expression of Steroid Hormone Receptors and Ten Eleven Translocation Proteins in Endometrial Cancer Cells. Front. Oncol. 12:763464. doi: 10.3389/fonc.2022.763464
Received: 24 August 2021; Accepted: 16 February 2022;
Published: 11 March 2022.
Edited by:
Petranel T. Ferrao, Independent Researcher, Adelaide, SA, AustraliaReviewed by:
Flávia Karina Delella, São Paulo State University, BrazilCopyright © 2022 Mahajan, Gujral, Jain and Ponnampalam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna P. Ponnampalam, YS5wb25uYW1wYWxhbUBhdWNrbGFuZC5hYy5ueg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.