- Department of Medical Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Hepatocellular carcinoma (HCC) is the most common liver cancer and one of the leading causes of cancer-related deaths in the world. Mono-immunotherapy and combination therapy with immune checkpoint inhibitors (ICIs) and multitargeted tyrosine kinase inhibitors (TKIs) or anti-vascular endothelial growth factor (anti-VEGF) inhibitors have become new standard therapies in advanced HCC (aHCC). However, the clinical benefit of these treatments is still limited. Thus, proper biomarkers which can predict treatment response to immunotherapy to maximize clinical benefit while sparing unnecessary toxicity are urgently needed. Contrary to other malignancies, up until now, no acknowledged biomarkers are available to predict resistance or response to immunotherapy for HCC patients. Furthermore, biomarkers, which are established in other cancer types, such as programmed death ligand 1 (PD-L1) expression and tumor mutational burden (TMB), have no stable predictive effect in HCC. Thus, plenty of research focusing on biomarkers for HCC is under exploration. In this review, we summarize the predictive and prognostic biomarkers as well as the potential predictive mechanism in order to guide future research direction for biomarker exploration and clinical treatment options in HCC.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and mostly develops on a background of chronic liver disease (1). Most patients were diagnosed at an advanced stage and/or had underlying chronic liver disease, with no opportunity to receive liver resection and transplantation. Moreover, even diagnosed at an early stage, the recurrence rates remain at about 70% in 5 years after surgery (2). Systemic treatment options for advanced HCC (aHCC) by multitargeted tyrosine kinase inhibitors (TKIs) of sorafenib, lenvatinib, regorafenib, cabozantinib, and ramucirumab have improved aHCC patients’ survival in a certain degree. However, the overall survival (OS) is merely 10.7-13.6 months (3–7), far from clinical expectation. In recent years, immune checkpoint inhibitors (ICIs) including nivolumab and pembrolizumab have shown survival benefits and have been approved by the United States Food and Drug Administration (FDA) for aHCC treatment (8, 9). Since 2020, anti-programmed death 1 (anti-PD-1) antibodies such as camrelizumab and tislelizumab have been successively approved by National Medical Products Administration (NMPA) as second-line treatment regimens for HCC patients (10, 11). The IMbrave150 trial achieved an improvement in OS of up to 19.2 months with atezolizumab and bevacizumab combination therapy, making it the standard first-line treatment for aHCC (12, 13). Regrettably, the objective response rate (ORR) of combination therapy was only about 30% (14). In addition, approximately 5%–30% of patients develop ≥ grade 3 immune-related adverse events (irAEs) (14). Therefore, proper biomarkers used to predict patient clinical response and spare unnecessary toxicity are urgently needed. Although no widely accepted biomarkers have been identified currently, multidimensional analyses of potential biomarkers for immunotherapy of HCC have been under exploration. In this review, we aim to summarize the predictive and prognostic biomarkers from multiple dimensions to guide future biomarker exploration in HCC (Figure 1).
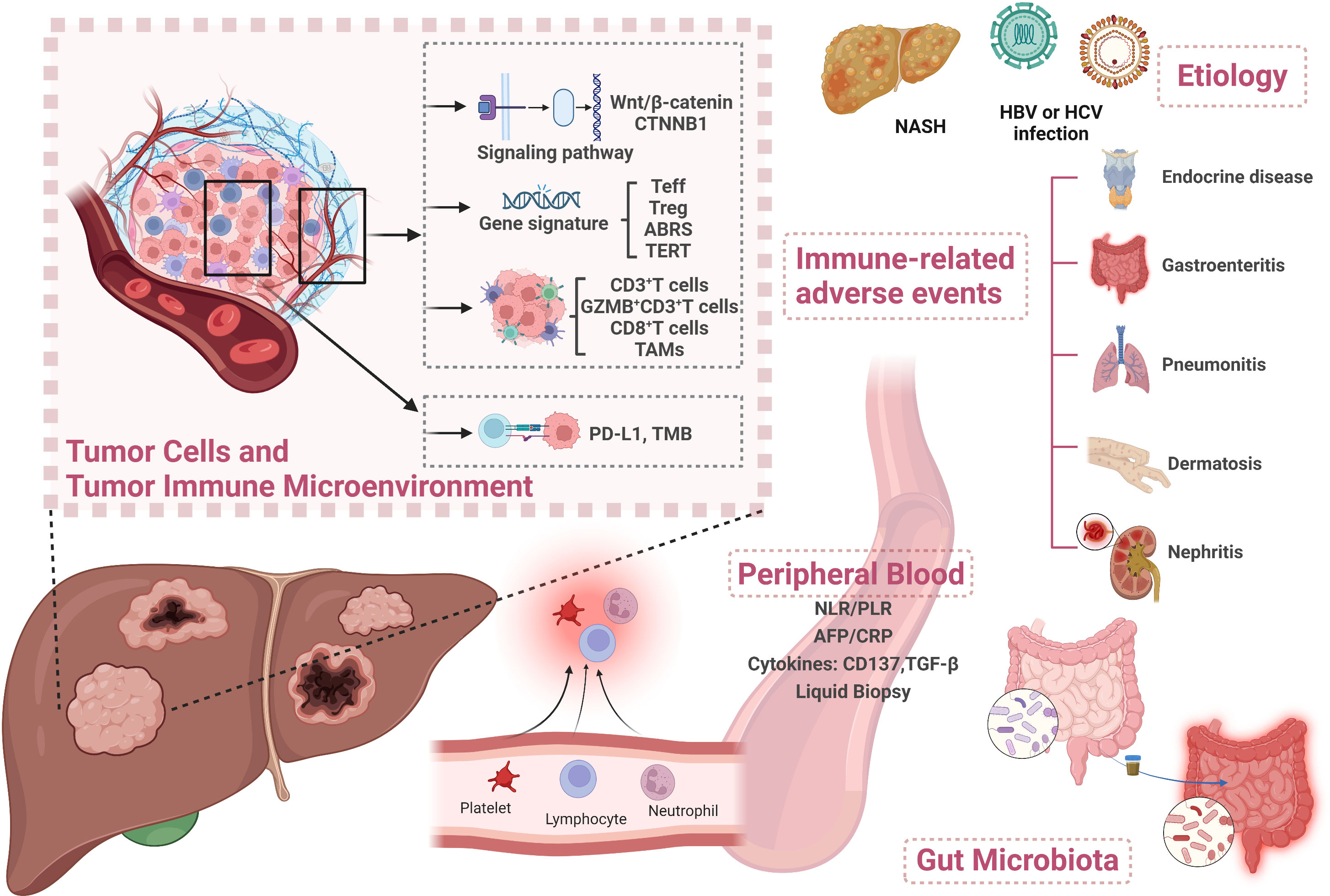
Figure 1 Overview of biomarkers for predicting treatment response to immunotherapy in HCC. Created with BioRender.com. Teff, T effector; ABRS, atezolizumab + bevacizumab response signature; TAM, tumor-associated macrophage; PD-L1, programmed death ligand 1; TMB, tumor mutational burden; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; AFP, alpha-fetoprotein; CRP, C-reactive protein; TGF-β, transforming growth factor-β; HBV, hepatitis B virus; HCV, hepatitis C virus; NASH, non-alcoholic steatohepatitis.
Circulating biomarkers in peripheral blood
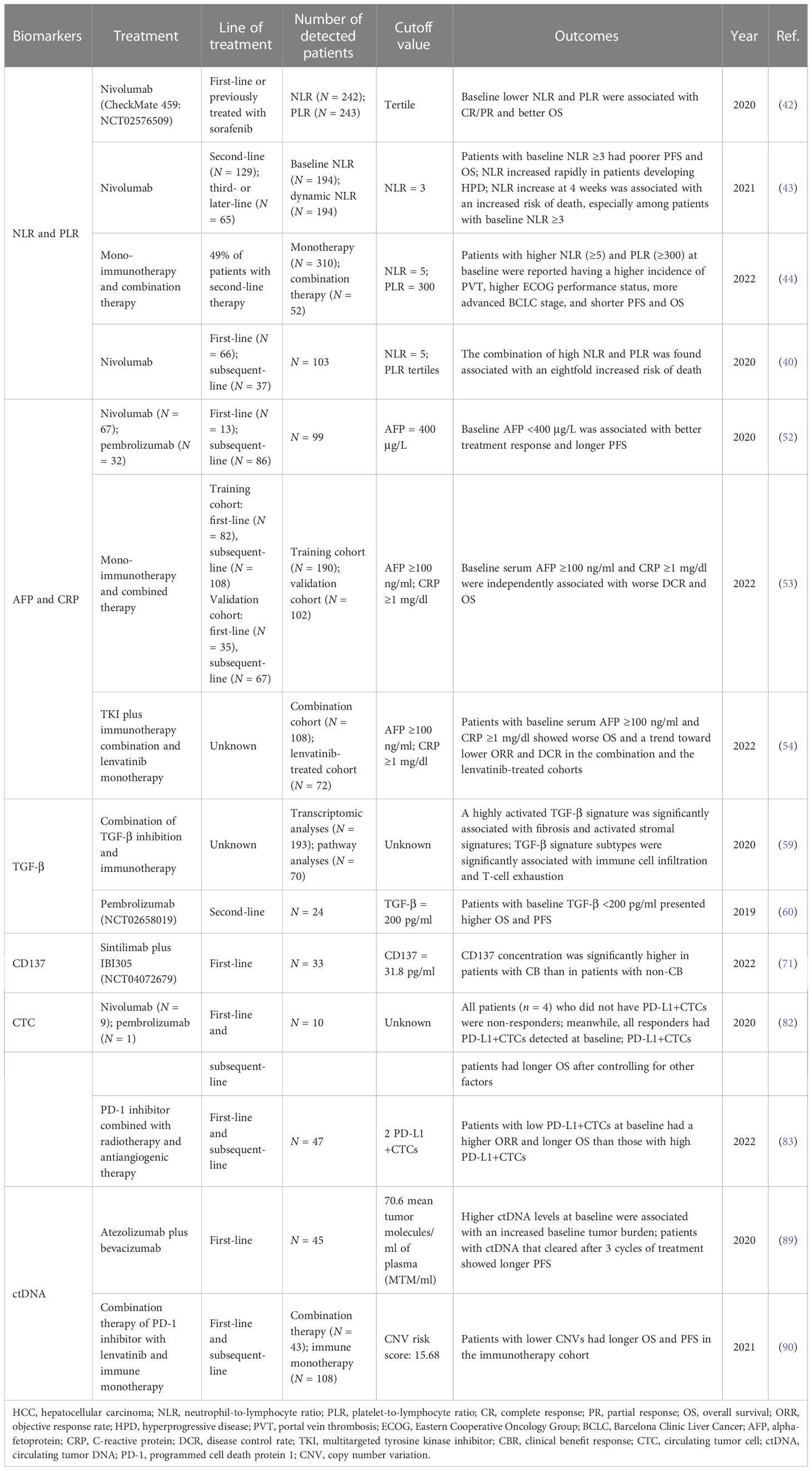
NLR and PLR
Human neutrophils and platelets produce a host of cytokines and growth factors relevant to tumor growth and progression (15–25). Neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) have been reported as predictive factors in several cancer types (26–34). Elevated NLR and PLR were also found to be associated with poor response to transarterial chemoembolization (TACE) and sorafenib treatment in HCC (35–41). As for immunotherapy in HCC, the same predictive effect has also been reported. In a subcohort of 242 patients in the CheckMate 040 trial, patients with NLR in the low tertile showed better OS than those with medium or high tertile (p = 0.015) (42). A similar result was observed in PLR (p = 1.38e−07). Patients with complete response or partial response (CR/PR) had lower PLR than those with progressive disease (PD) (p = 0.05). In another cohort of 194 aHCC patients treated with nivolumab, those with baseline NLR ≥3 presented poorer progression-free survival (PFS) [11.0 vs. 7.1 weeks; HR = 1.52 (95% CI 1.11–2.07), p = 0.01] and OS [61.3 vs. 21.0 weeks; HR = 2.72 (95% CI 1.86–3.99), p < 0.001]. Moreover, a dynamic increase of NLR at 4 weeks was associated with an increased risk of death [HR = 1.79, 95% CI (1.19–2.68)]. Interestingly, in this study, NLR increased at 4 weeks also had a role in predicting hyperprogressive disease (HPD), which may guide treatment plan in an early phase (43). In a cohort of 362 HCC patients treated with mono or combination immunotherapy, patients with higher NLR and PLR at baseline were reported to have a higher incidence of portal vein thrombosis (PVT), higher Eastern Cooperative Oncology Group (ECOG) performance status, and more advanced Barcelona Clinic Liver Cancer (BCLC) stage. Significantly shorter OS and PFS were observed in patients with NLR ≥5 (OS: 7.7 vs. 17.6 months, p < 0.0001; PFS: 2.1 vs. 3.8 months, p = 0.03) and PLR ≥300 (OS: 6.7 vs.16.5 months, p < 0.0001; PFS: 1.8 vs. 3.7 months, p = 0.0006) (44). On the basis of the independent predictive role for OS of NLR and PLR, Schobert et al. found that the combination of high NLR and PLR was associated with an eightfold increased risk of death (40).
In conclusion, several analyses in different trials have demonstrated the strong survival predictive power of NLR and PLR in HCC immunotherapy and their predictive trend in treatment response. As for potential mechanisms, some reported that IL-8 and other tumor growth factors secreted by tumors may promote neutrophil recruitment (45). The increasing circulating and intratumoral neutrophils can further secrete vascular endothelial growth factor (VEGF), thereby causing higher levels of VEGF in the tumors (46) and promoting angiogenesis.
AFP and CRP
Alpha-fetoprotein (AFP) is widely used for the surveillance, diagnosis, and prognostication of HCC. In recent decades, several studies have been conducted to explore its additional roles, such as being employed for defining HCC molecular classes or as biomarkers for HCC treatment (47–51). In a cohort of 99 patients who received nivolumab or pembrolizumab, those with AFP <400 μg/L at the beginning of ICI treatment were more likely to achieve a higher rate of CR or PR than those with AFP ≥400 μg/L (24% vs. 13%). Patients with baseline serum AFP <400 μg/L presented longer PFS (5.4 vs. 2.6 months, p < 0.05) and OS (21.8 vs. 8.7 months, p < 0.0001) (52). Moreover, a simple and easily applicable score called C-reactive protein (CRP) and AFP in Immunotherapy (CRAFITY) constructed by the analysis of 190 aHCC patients who received mono or combination immunotherapy based on CRP and AFP was recently reported (53). In this score, AFP ≥100 ng/ml and CRP ≥1 mg/dl were both assigned 1 point. Patients could achieve either 0, 1, or 2 points depending on the level of these two variables. Results showed that baseline serum AFP ≥100 ng/ml and CRP ≥1 mg/dl were independently associated with worse OS in ICI-treated patients with HCC. The median OS of patients with 0 points (CRAFITY - low) (n = 53), 1 point (CRAFITY - intermediate) (n = 75), and 2 points (CRAFITY - high) (n = 62) were 27.6 vs. 11.3 vs. 6.4 months (p < 0.001). In addition, a high CRAFITY score also predicted a worse radiological response, and the disease control rate (DCR) was 80% vs. 64% vs. 39% for a score of 0, 1, and 2, respectively (p < 0.001). Yang et al. further verified the CRAFITY score in TKI plus immunotherapy and lenvatinib monotherapy cohorts. A high score successfully predicts worse OS and a trend toward worse ORR and DCR (54). This simple prognostic score facilitates early survival evaluation of immunotherapy treatment and is promising to be adopted in clinical application. However, CRP is an acute-phase protein which may increase after injury or infection. Diseases which may increase CRP levels should be considered before score application.
Cytokines
Transforming growth factor-β
Transforming growth factor-β (TGF-β) is known as an immunosuppressive and fibrotic cytokine. Approximately 38% of HCC patients have somatic mutations in the TGF-β pathway (55). High TGF-β levels present more aggressive tumor characteristics and may also cause T-cell exhaustion by upregulating PD-1 signaling in HCC, which demonstrates a specific immunosuppressive role of TGF-β in mediating immunotherapy resistance (56–59). Feun et al. conducted a phase 2 study of pembrolizumab in 29 patients (24 provided plasma samples) with aHCC. In the biomarker analyses, plasma TGF-β levels in responders [those with CR/PR/stable disease (SD)] were lower than those in non-responders (141.9 vs. 1,071.8 pg/ml, p = 0.004). Survival analysis showed that patients with plasma TGF-β ≥200 pg/ml had significantly shorter PFS (2 vs. over 25 months, p = 0.008) and OS (7 vs. over 25 months, p = 0.005), indicating that higher TGF-β levels were associated with poor treatment outcomes (60). This suggests that high plasma TGF-β may be a potential biomarker for poor treatment response and outcome to immunotherapy, which may be related to the tumor microenvironment of decreased T-cell infiltration in tumors shaped by TGF-β (61). However, the role of TGF-β in HCC is still in the exploratory stage, and its predictive value needs to be further confirmed in large-scale studies.
CD137
CD137, also known as 4-1BB or TNF receptor superfamily member 9 (TNFRSF9), is a member of the tumor necrosis factor family and an important costimulatory molecule in the process of T-cell activation, which can enhance the antitumor effects of T cells (62). CD137 is mainly expressed by activated CD4+ and CD8+ T cells (63), it is also found on the surfaces of NK cells, neutrophils, dendritic cells, and monocytes (64, 65). The expression of CD137 in HCC was higher than that in other types of cancer (e.g., small cell lung cancer and colorectal cancer) and was found to be expressed predominantly on exhausted PD-1highCD8+ T cells (66), as well as activated T cells in peripheral blood samples (67, 68). Preclinical studies have found a synergistic antitumor activity between PD-1/programmed death ligand 1 (PD-L1) inhibitors and activation of the CD137 signaling pathway (69). The increased number of CD137+CD8+ T cells in peripheral blood was correlated with longer disease-free survival (DFS) in patients with melanoma who were treated with ipilimumab plus nivolumab (70). A study recently conducted on 50 aHCC patients who received sintilimab (a PD-1 inhibitor) plus IBI305 reported the potential predictive role of serum CD137. Among 33 patients with serum CD137 detected, the CD137 concentration was significantly higher in patients with clinical benefit (CB, patients with CR/PR, or SD ≥12 weeks) than in those with non-CB (patients with PD or SD <12 weeks) (32.8 vs. 19.8 pg/ml, p = 0.034). Markedly longer PFS (14.2 vs. 4.1 months, p < 0.001) and OS (undefined vs. 15.6 months, p = 0.023) were observed in patients with high CD137 concentrations (71). However, relevant studies are mainly small sample research. Its predictive role remains to be further explored.
Liquid biopsy
HCC exhibits significant heterogeneity from genetic aberrations and transcriptional and epigenetic dysregulation. A single biopsy specimen containing a small amount of tumor tissue may not be representative of the whole tumor (72). In recent years, liquid biopsy techniques have been developed to collect samples from patients’ body fluids to obtain phenotypic, genetic, and transcriptomic information about the primary tumor (73). The primary forms of liquid biopsy include circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), microRNA, and extracellular vesicles (74–77).
Circulating tumor cells
CTCs are malignant cells derived from either the primary or metastasis tumor that migrate into the systemic circulation, which represents a heterogeneous population of cells from the tumor. CTCs have been shown to be a reliable predictor of metastatic prostate cancer and breast cancer (78–81). A recent study conducted on HCC patients, of which 10 patients received anti-PD-1 therapy (9 with nivolumab and 1 with pembrolizumab), reported that all patients (n = 4) who did not have PD-L1+CTCs were non-responders (patients with PD or died within 6 months from initiating treatment). Meanwhile, all responders (patients with PR/SD) had PD-L1+CTCs detected at baseline. A longer OS was also found in PD-L1+CTC patients even after controlling for other factors [HR = 3.22 (95%CI 1.33-7.79), p = 0.01] (82). However, in a study of 47 HCC patients receiving a PD-1 inhibitor in combination with antiangiogenic therapy and radiotherapy, patients with low PD-L1+CTCs at baseline had a higher ORR (56.5% vs. 16.7%, p = 0.007) and longer OS (not reached vs. 10.8 months, p = 0.001) than those with high PD-L1+CTCs (83), indicating that CTC is still a controversial biomarker for predicting the treatment response to immunotherapy in HCC. As a result, larger sample studies are required to further explore its predictive value. Meanwhile, an extremely rare frequency of CTCs has been found in the circulation. All of these reasons make the detection of CTCs in the early stage of disease challenging (84).
Circulating tumor DNA
Circulating tumor DNA (ctDNA) can arise in the bloodstream of cancer patients as a result of tumor cell apoptosis or necrosis (85). ctDNA contains cancer-associated molecular characteristics, which allow its discrimination from total normal circulating cellular free DNA (86–88). In a subset of GO30140 arm A of 45 patients, higher ctDNA levels at baseline were associated with an increased baseline tumor burden (p < 0.03). After 3 cycles of treatment, ctDNA turned negative in 70% (CR), 27% (PR), 9% (SD), and 0% (PD) of patients, respectively. Patients with ctDNA cleared after 3 cycles of treatment showed longer PFS compared with those still present (6.5 vs. not reached months, p < 0.00029) (89). Patients with lower copy number variations (CNVs) in cell-free DNA risk score were also found to have longer OS and PFS in the ICI-treated cohort (90). Moreover, tumor mutational burden (TMB) evaluated by ctDNA was reported to be highly consistent with TMB detected by tissues (91), suggesting that ctDNA analysis could be an alternative option to evaluate TMB prior to immunotherapy in aHCC patients to whom tissue biopsy was not recommended if necessary. Table 1 provides a brief overview of the biomarkers in peripheral blood for HCC immunotherapy.
Tumor tissue-related biomarkers
Tumor immune microenvironment
The HCC tumor immune microenvironment (TiME) is acknowledged for its immunosuppressive character. The crosstalk between tumor cells and the immune microenvironment promotes tumor proliferation, invasion, and metastasis (92). Recent advances in basic and translational research have shown that the different manifestations of the tumor microenvironment are closely related to the efficacy of immunotherapy, revealing that the TiME profile may be a valuable potential biomarker of immunotherapy (14).
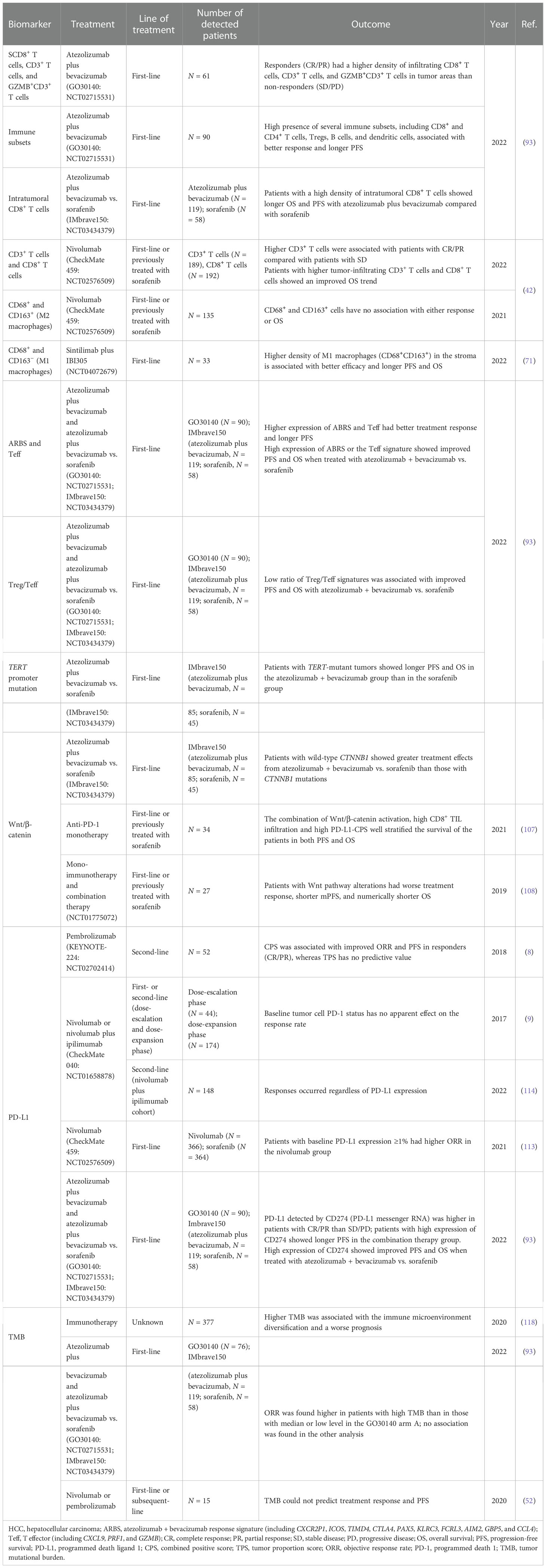
Analyses of patients treated with atezolizumab plus bevacizumab in the GO30140 arm A cohort showed that responders (CR/PR) had a higher density of infiltrating CD8+ T cells, CD3+ T cells, and GZMB+CD3+ T cells in tumor areas than non-responders (SD/PD) (p = 0.007, p = 0.039, and p = 0.044, respectively). The high presence of several immune subsets, including CD8 and CD4 T cells, Tregs, B cells, and dendritic cells, also seemed to be associated with better response and longer PFS (93). In the IMbrave150 cohort, patients with a high density of intratumoral CD8+ T cells showed longer OS [HR = 0.29 (95% CI 0.14–0.61), p = 0.001] and PFS [HR = 0.54 (95% CI 0.29–1.00), p = 0.053] in atezolizumab plus bevacizumab compared with sorafenib. The reason may be that blockade of the PD-1/PD-L1 axis could restore the antitumor immunity induced by CD8+ lymphocytes in tumors (94). The results above demonstrated that patients with pre-existing immunity seemed to possess improved clinical outcomes to combination therapy. An exploratory research in CheckMate 040 analyzed the levels of multiple inflammation biomarkers and their association between treatment response and survival to nivolumab in patients previously treated with or without sorafenib (42). The results showed that patients with CR/PR had higher CD3+ T cells compared with those with SD (p = 0.03). Those with higher tumor-infiltrating CD3+ and CD8+ T cells showed a trend toward improved OS (both p = 0.08).
Macrophages are major components of the TiME, which can be classified into two main subtypes: the classically activated macrophages (M1 macrophages) with pro-inflammatory functions and the alternatively activated macrophages (M2 macrophages) with immunosuppressive functions (95). Several tumor-promoting roles, such as immune suppression, cancer invasion and metastasis, angiogenesis, maintenance of cancer cell stemness, and drug resistance, have been attributed to these tumor-associated macrophages (TAMs), especially M2 macrophages (96, 97). A previous study had shown that high levels of M2 macrophages have been associated with poor prognosis in patients with HCC (98). Although the expression of CD68+ and CD163+ (M2 macrophages) cells has no association with either treatment response or OS in the CheckMate 040 subgroup (42), the other study reported that a higher density of M1 macrophages (CD68+CD163−) in the stroma was associated with better efficacy and longer PFS (M1 macrophages low vs. high: 11.4 vs. 3.0 months, p = 0.024) and OS (M1 macrophages low vs. high: undefined vs. 17.5 months, p = 0.046) (71). Therefore, a better understanding of the mechanisms underlying the function of TAMs is necessary for the development of novel TAM-targeting immunological interventions, which may provide promising therapeutic approaches for HCC patients. Collectively, immunocytes with different functions can directly reflect tumor immune status. This advantage makes it become the main research direction at present. More mechanistic research is expected to be further carried out.
Signaling pathway and gene signature
As important regulatory factors in tumor progression and the immune environment, the correlation between molecular features and treatment efficacy has become the focus of recent research. In the GO30140 group A cohort, pathways and immune subsets were identified by genome-wide differentially expressed genes (DEGs), gene set enrichment analysis (GSEA), and xCell analyses. An atezolizumab + bevacizumab response signature (ABRS) consisting of the top 10 genes from the DEG analyses (namely, CXCR2P1, ICOS, TIMD4, CTLA4, PAX5, KLRC3, FCRL3, AIM2, GBP5, and CCL4) was found consistently higher in patients with CR/PR than in those with SD/PD, as well as the T effector (Teff) signature (CXCL9, PRF1, and GZMB). Patients with a high expression of these markers had longer PFS than those with low expression [ABRS: HR = 0.51 (95% CI 0.30–0.87), p = 0.013; Teff signature: HR = 0.46 (95% CI 0.27–0.78), p = 0.0035], which was further validated in the IMbrave150 cohort. Patients with high ABRS or the Teff signature showed improved PFS [ABRS: HR = 0.49 (95% CI 0.25–0.97), p = 0.041; Teff signature: HR = 0.52 (95% CI 0.28–0.99), p = 0.047] and OS [ABRS: HR = 0.26 (95% CI 0.11–0.58), p = 0.0012; Teff signature: HR = 0.24 (95% CI 0.11–0.5), p = 0.0002] when treated with atezolizumab + bevacizumab compared with sorafenib. Signature analysis in IMbrave150 revealed that a low ratio of Treg/Teff signatures was associated with improved PFS [HR = 0.42 (95% CI 0.22–0.79), p = 0.007] and OS [HR = 0.24 (95% CI 0.11–0.54), p = 0.0006] when treated with atezolizumab + bevacizumab compared with sorafenib. As for the mutation landscape, TERT promoter mutations were observed in 56.2% of patients in the IMbrave150 trial. The benefit of atezolizumab + bevacizumab was more pronounced in patients with TERT-mutant than in the sorafenib group [PFS: HR = 0.61 (95% CI 0.33–1.10), p = 0.047; OS: HR = 0.38 (95% CI 0.16–0.89), p = 7.8 × 10−5] (93).
Hyperactive Wnt/β‐catenin signaling is implicated in the initiation and progression of various types of cancer, which may be related to the exclusion of CD8+ cells in tumor tissues in melanoma cases (99). CTNNB1 which is involved in the Wnt/β-catenin signaling pathway is a prevalent mutation gene in HCC (100). Approximately 11%–41% of liver malignancies harbor CTNNB1-activating mutations (93, 101–104). Several studies have shown that β‐catenin signaling may mediate the immune escape of cancer cells and the resistance to ICIs (99, 105, 106). In the IMbrave150 trial, patients with wild-type CTNNB1 showed greater treatment effect with atezolizumab + bevacizumab than sorafenib [PFS: HR = 0.45 (95% CI 0.27–0.86), p = 0.0086; OS: HR = 0.42 (95% CI 0.19–0.91), p = 3 × 10−4) (93). A small sample cohort of 34 patients who were treated with anti-PD-1 monotherapy with or without previous treatment with sorafenib found that although patients with negative Wnt/β-catenin activation, high CD8+ TIL infiltration, and high PD-L1-CPS showed higher DCR, PFS, and OS in the univariate analysis, no significant difference was presented after the multivariate analysis. However, the combination of these factors well stratified the survival in both PFS (p < 0.0001) and OS (p = 0.0048), suggesting that in patients lacking β-catenin activation, blockade of the PD-1/PD-L1 axis might overcome the presence of exhausted TILs (107). Harding et al. (108) studied 27 HCC patients treated with ICIs (both monotherapy and combination therapy). The results showed that all patients with Wnt pathway alterations had PD at the first interval scan, whereas 9 of 17 non-Wnt pathway-altered patients had durable disease (SD ≥4 months) or better as the best response (p < 0.009). Wnt-activated patients presented shorter mPFS (2.0 vs. 7.4 months, p < 0.0001) and numerically shorter OS (9.1 vs. 15.2 months, p = 0.11). Lin et al. analyzed the same cohort and verified the results (109). The role of Wnt/β-catenin signaling as a biomarker has been verified in a number of studies, which can be further explored as a powerful factor in a prospective prediction model.
PD-L1 expression and tumor mutational burden
PD-L1 has been reported as an indicator of anti-PD-1/PD-L1 treatment in several cancer types (110–112). However, the predictive role of PD-L1 expression in HCC immunotherapy remains controversial. In the KEYNOTE-224 trial, PD-L1 expression calculated by the combined positive score (CPS, cutoff = 1) was found to be associated with improved ORR and PFS in responders (CR/PR), whereas PD-L1 expression calculated by the tumor proportion score (TPS, cutoff = 1%) has no predictive value as CPS (8). In the CheckMate 459 trial, although patients with baseline PD-L1 expression ≥1% had higher ORR in the nivolumab group (28% vs. 12%), no difference was observed in PFS and OS (113). In the dose-escalation and dose-expansion cohort of Checkmate 040, TPS did not have an apparent predictive effect on the response rate (9). The same result was found in the nivolumab and ipilimumab combined cohort of CheckMate 040 (114). When PD-L1 was detected by the expression of CD274 (PD-L1 messenger RNA) in the G030140 arm A group and in the IMbrave150 trial, it was found to be higher in patients with CR/PR than in patients with SD/PD. Patients with high levels of CD274 also showed longer PFS than those with low expression [HR = 0.42 (95% CI 0.25–0.72), p = 0.0011] (93). Patients with high expression of CD274 showed improved PFS and OS in the combination therapy group than in the sorafenib group. In conclusion, the predictive value of PD-L1 is limited in HCC immunotherapy.
Previous studies in melanoma and NSCLC showed that higher TMB was associated with higher tumor responsiveness to PD-1/PD-L1 immunotherapy (115–117). Nonetheless, the value of TMB as an objective biomarker in immunotherapy in HCC remains indefinite. Xie et al. analyzed the HCC cohort with immunotherapy from The Cancer Genome Atlas and found that higher TMB was associated with the immune microenvironment diversification and worse prognosis (118). In the GO30140 biomarker exploration study, ORR was found to be higher in patients with high TMB than in those with median or low levels in 76 patients in arm A (56% vs. 17% vs. 35%), while PFS has no difference in all the groups (high vs. median vs. low = 13.6 vs. 5.9 vs. 7.9 months). However, no association between TMB and treatment response or survival was found in the combination therapy group in the IMbrave150 cohort (93). The same result was found in another anti-PD-1 treatment cohort (52). The potential reason may be attributed to the generally low level of TMB in HCC (median TMB in HCC was only 4.08) (108). Table 2 provides a brief overview of the biomarkers in tumor tissues for HCC immunotherapy.
Gut microbiota
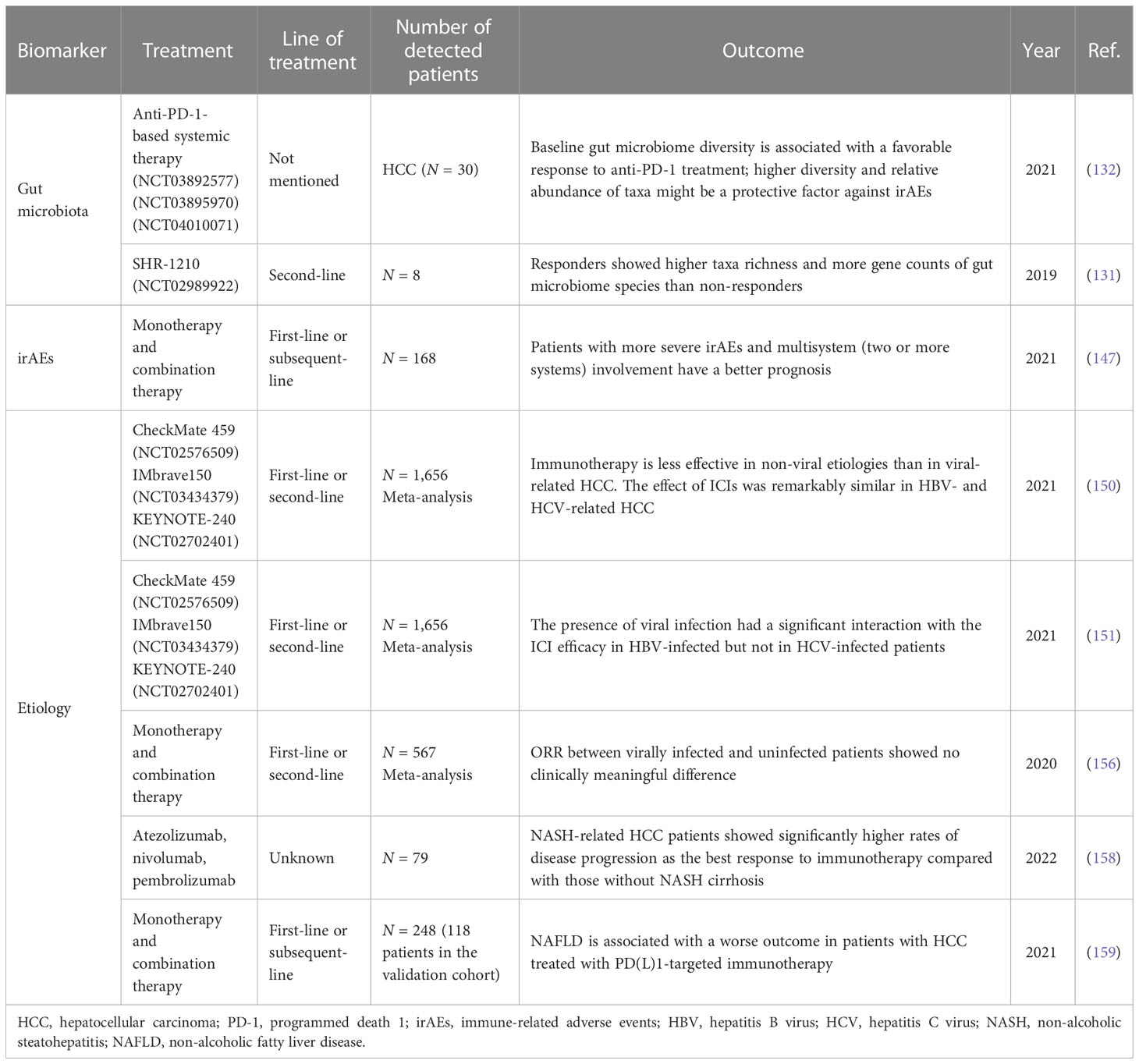
The gut microbiota is known to influence immune responses and even promote carcinogenesis, which supports its potential role as a biomarker in immunotherapy. Accumulated evidence has shown that the gut microbiota may predict immunotherapy efficacy in various cancer types (119–123). Several mechanisms have been reported such as modulating DNA damage, influencing oncogenesis or tumor suppression by metabolic processes (124), and inducing regulatory T-cell expansion and CD8+ T-cell attenuation (125). The above mechanisms finally inhibit antitumor immunity through mediating immune cells and cytokine production (126–130). Several studies have been conducted to explore the role of the gut microbiome in HCC immunotherapy in recent years. Zheng et al. (131) found that fecal samples from HCC patients treated with camrelizumab showed higher taxa richness and more gene counts of gut microbiome species, such as Akkermansia and Ruminococcaceae, than from non-responders. Meanwhile, the dissimilarity of beta diversity became prominent as early as 6 weeks, which indicated that the gut microbiome might be used for early prediction for anti-PD-1 immunotherapy after treatment initiation. However, only 8 patients were enrolled in this study. Zhao et al. (132) conducted an analysis of 65 patients with advanced hepatobiliary cancer receiving anti-PD-1 treatment to explore the potential mechanism. The results showed that the clinical benefit response (CBR) group (patients with CR, PR, or SD ≥6 months) had more taxa enrichment than the non-clinical benefit (NCB) group (patients with SD <6 months or PD). Lachnospiraceae bacterium-GAM79 and Alistipes spMarseille-P5997 were significantly enriched in the CBR group (74 vs. 40 taxa). Patients with a higher abundance of Ruminococcus calidus and Erysipelotichaceae bacterium-GAM147 presented longer PFS and OS. During treatment, the gut microbiome composition in the CBR group remained stable, while in the NCB group, the microbial diversity seemed to decrease. Fecal microbiota transplantation (FMT) from donors who achieved CR/PR for a long duration treated with anti-PD-1 therapy to patients who were refractory to immunotherapy was reported to increase intratumor lymphocyte infiltration in patients with poor efficacy in melanoma (133, 134). A study conducted in a mouse model showed that FMT could significantly enhance the therapeutic efficacy of ICIs in syngeneic tumor models by increasing tumor-infiltrating IFN-γ+CD8+ T cells and the tumor suppression effect (135). All these findings demonstrated that the gut microbiome might be an effective biomarker to predict the clinical response and survival benefit of immunotherapy in HCC. Its predictive prospect is worth anticipating.
Immune-related adverse events
Immunotherapy will inevitably result in irAEs, which are defined as side effects with potential immunological basis and require more frequent monitoring and possible treatment with systemic steroids (136). Mono-immunotherapy conducted by nivolumab in CheckMate 040 and CheckMate 459 resulted in 22%–25% grade 3–5 AEs (9, 137). For pembrolizumab in KEYNOTE-240 and KEYNOTE-224, grade 3–5 AEs were about 52% and 26% (8, 138). As for the combination of immune and targeted therapy, grade 3–5 AEs were about 50%–60% in IMbrave150, GO30140, and ORIENT-32 (12, 139, 140). Several studies conducted in solid malignancies have demonstrated a positive association between irAEs with improved clinical outcomes, such as melanoma, urothelial cancer, renal cell carcinoma, NSCLC, and gastric cancer (141–146). A consistent result was found in a retrospective cohort study of 168 patients with aHCC. In this study, patients with grade ≥3 irAEs demonstrated improved ORR and DCR than those with no irAEs (ORR: 50% vs. 11.3%, p = 0.002; DCR: 87.5% vs. 28.2%, p < 0.001). The median PFS and OS in patients with grade ≥3 irAEs and grade 1–2 irAEs were significantly longer than in patients with no irAEs (PFS: 8.5 vs. 3.6 vs. 1.3 months, p < 0.001; OS: 26.9 vs. 14.0 vs. 4.6 months, p < 0.001). Patients with more severe and multisystem (two or more systems) irAEs have a better prognosis (147). The mechanisms are still unclear. Possibly, patients who experience more serious irAEs could have higher T-cell activity and experience better antitumor outcomes. Other possible mechanisms may rely on the potential similar pathway shared by adverse events and the ICIs. Thus, the occurrence of adverse events may reflect that the relative pathway has been inhibited at a high level, which certainly led to better efficacy (148).
The etiology of hepatocellular carcinoma
Over 90% of HCC cases occur in the setting of chronic liver disease. The major risk factors for HCC are chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), heavy alcohol intake, excess body weight, diabetes, or non-alcoholic fatty liver disease (NAFLD) (1).
The response of HCC induced by various etiologies to immunotherapy may differ. Chun et al. reported that Treg and CD8+ resident memory T cells (TRM) were enriched in HBV-related HCC. Treg and TRM from HBV-related HCC expressed more PD-1 and were functionally more suppressive and exhausted than those from non-viral-related HCC, which could be reversed by anti-PD-1 blockade (149). A meta-analysis included eight systemic therapies cohorts to evaluate the impact of targeted and immune therapies according to different HCC etiologies. Among them, five were TKI/anti-VEGF cohorts (REACH, REACH-2, METIV-HCC, CELESTIAL, and JET-HCC) and three were immunotherapy cohorts (Checkmate 459, IMbrave150, and KEYNOTE-240). Patients with viral-related HCC presented significantly better OS than those with non-viral-related HCC (p = 0.0259) in immunotherapy. Efficacy was similar in HBV- and HCV-related HCC [HR = 0.64 (95% CI 0.49–0.83) vs. HR = 0.68 (95% CI 0.47–0.98)]. No impact of etiology was observed in TKI/anti-VEGF therapies (150). In another meta-analysis, the presence of viral infection had a significant interaction with the ICI efficacy in HBV-infected HCC (pinteraction = 0.016) but not in HCV-infected HCC (pinteraction = 0.081) (151). The potential reason may be that patients with HCV–HCC were rich in Tregs and M2 macrophages and had an upregulated expression of CTLA4 and other immunosuppressive molecules (152–154), and the expression of negative co-stimulatory signals may contribute to treatment resistance. Meanwhile, unlike HBV-related HCC, the function of HCV-specific CD8+ T cells did not recover after PD-1/PD-L1 blockade (155). However, contrary to the above results, Ho et al. found that the ORR between viral-infected and uninfected patients showed no clinical difference when treated with PD-1/PD-L1 inhibitors, which means that viral status is not suitable to be used as a criterion to select patients for immunotherapy (156). Considering that the meta-analyses were not based on individual patient’s data and the trials included were heterogeneous in terms of treatment line and control arm, whether patients with viral infection respond better to immunotherapy than those without infection requires further research.
NAFLD has become an emerging risk factor for HCC over the past decade (157). In a retrospective study with 79 patients [15 patients in the non-alcoholic steatohepatitis (NASH) cirrhosis-related HCC group and 64 patients in the HCC group without NASH cirrhosis], there were significantly higher rates of PD as the best response to immunotherapy in patients with HCC and NASH cirrhosis compared with those without NASH cirrhosis (46.7% vs. 10.9%, p = 0.004) (158). A relevant mechanism research found that the exhausted, unconventionally activated CD8+PD1+ T cells progressively accumulated in NASH-affected livers. However, in mice with NASH but without HCC, preventive CD8+ T-cell depletion significantly decreased the incidence of HCC. Meanwhile, preventive anti-PD-1 treatment in NASH mice increased CD8+PD1+ T cells and also caused a marked increase in cancer incidence, which means CD8+PD1+ T cells from patients with NAFLD or NASH might help induce NASH–HCC, rather than invigorating or executing immune surveillance (159). Collectively, NASH–HCC might be less responsive to immunotherapy, probably owing to NASH-related aberrant T-cell activation causing tissue damage that leads to impaired immune surveillance. Table 3 provides a brief overview of the biomarkers of other types for HCC immunotherapy.
Conclusion
We have concluded an exploratory research on biomarkers as immunotherapy for HCC. As biomarkers detected from peripheral blood, NLR, PLR, and CRAFITY (CRP and AFP in Immunotherapy) score, which are not only easy to be collected but also with high prognostic value supported by a large sample research, are of great significance in the future construction of predictive models. The gene signature and the tumor immune microenvironment have the ability to precisely reflect the pre-existing immunity in baseline tumor tissues, which have shown potential predictive value to drive the clinical activity of immunotherapy in aHCC in the IMbrave150 trial. The gut microbiota and irAEs which were found to be potential biomarkers in immunotherapy are now being further analyzed and are expected to be explored in the future. Fecal microbiota transplantation has been even developed into a combination treatment method and has shown great promise to increase immunotherapy efficacy. CD137 and other cytokines are potential predictive factors that need to be verified in large sample cohorts. Integrative multiparametric approaches that combine peripheral markers, the tumor microenvironment, and immune signatures appear to be the most comprehensive way to assess treatment outcomes and seem to be promising in the future.
Author contributions
XP: conceptualization, data curation, figure preparation, and writing—original draft preparation. CG: writing—review and editing. WZ: writing—review and editing, project administration, and funding acquisition. AZ: conceptualization, writing—original draft preparation, writing—review and editing, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the CAMS Innovation Fund for Medical Sciences (CIFMS, Grant No. 2021-I2M-1-066) and the Translational Research Project of Medical Oncology Key Foundation of Cancer Hospital Chinese Academy of Medical Sciences (CICAMS-MOTRP, Grant No. 2022005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Abbreviations
HCC, hepatocellular carcinoma; aHCC, advanced hepatocellular carcinoma; TKIs, multitargeted tyrosine kinase inhibitors; OS, overall survival; ICIs, immune checkpoint inhibitors; FDA, Food and Drug Administration; PD-1, programmed death 1; NMPA, National Medical Products Administration; ORR, objective response rate; irAEs, immune-related adverse effects; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; TACE, transarterial chemoembolization; CR, complete response; PR, partial response; PD, progressive disease; PFS, progression-free survival; HR, hazard ratio; HPD, hyperprogressive disease; PVT, portal vein thrombosis; ECOG, Eastern Cooperative Oncology Group; BCLC, Barcelona Clinic Liver Cancer; VEGF, vascular endothelial growth factor; AFP, alpha-fetoprotein; CRP, C-reactive protein; SD, stable disease; DCR, disease control rate; TGF-β, transforming growth factor-β; TNFRSF9, TNF receptor superfamily member 9; PD-L1, programmed death ligand 1; DFS, disease-free survival; CB, clinical benefit; CTC, circulating tumor cell; ctDNA, circulating tumor DNA; CNV, copy number variation; TiME, tumor immune microenvironment; TAM, tumor-associated macrophage; DEG, differentially expressed gene; GSEA, gene set enrichment analysis; ABRS, atezolizumab + bevacizumab response signature; Teff, T effector; TMB, tumor mutational burden; CPS, combined positive score; TPS, tumor proportion score; CBR, clinical benefit response; NCB, non-clinical benefit; FMT, fecal microbiota transplantation; HBV, hepatitis B virus; HCV, hepatitis C virus; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
References
1. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers (2021) 7(1):6. doi: 10.1038/s41572-020-00240-3
2. Roayaie S, Obeidat K, Sposito C, Mariani L, Bhoori S, Pellegrinelli A, et al. Resection of hepatocellular cancer ≤ 2 cm: results from two Western centers. Hepatology (2013) 57(4):1426–35. doi: 10.1002/hep.25832
3. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med (2008) 359(4):378–90. doi: 10.1056/NEJMoa0708857
4. Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet (2018) 391(10126):1163–73. doi: 10.1016/s0140-6736(18)30207-1
5. Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet (2017) 389(10064):56–66. doi: 10.1016/s0140-6736(16)32453-9
6. Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med (2018) 379(1):54–63. doi: 10.1056/NEJMoa1717002
7. Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol (2019) 20(2):282–96. doi: 10.1016/s1470-2045(18)30937-9
8. Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol (2018) 19(7):940–52. doi: 10.1016/s1470-2045(18)30351-6
9. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet (2017) 389(10088):2492–502. doi: 10.1016/s0140-6736(17)31046-2
10. Qin S, Ren Z, Meng Z, Chen Z, Chai X, Xiong J, et al. Camrelizumab in patients with previously treated advanced hepatocellular carcinoma: a multicentre, open-label, parallel-group, randomised, phase 2 trial. Lancet Oncol (2020) 21(4):571–80. doi: 10.1016/s1470-2045(20)30011-5
11. Ducreux M, Abou-Alfa G, Ren Z, Edeline J, Li Z, Assenat E, et al. Results from a global phase 2 study of tislelizumab, an investigational PD-1 antibody, in patients with unresectable hepatocellular carcinoma. Ann Oncol (2021) 32:S217. doi: 10.1016/j.annonc.2021.05.005
12. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med (2020) 382(20):1894–905. doi: 10.1056/NEJMoa1915745
13. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. IMbrave150: Updated overall survival (OS) data from a global, randomized, open-label phase III study of atezolizumab (atezo) + bevacizumab (bev) versus sorafenib (sor) in patients (pts) with unresectable hepatocellular carcinoma (HCC). J Clin Oncol (2021) 39(3_suppl):267. doi: 10.1200/JCO.2021.39.3_suppl.267
14. Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol (2022) 19(3):151–72. doi: 10.1038/s41571-021-00573-2
15. Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, Mantovani A. Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer (2020) 20(9):485–503. doi: 10.1038/s41568-020-0281-y
16. Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Maase H. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J Clin Oncol (2009) 27(28):4709–17. doi: 10.1200/jco.2008.18.9498
17. Kuang DM, Zhao Q, Wu Y, Peng C, Wang J, Xu Z, et al. Peritumoral neutrophils link inflammatory response to disease progression by fostering angiogenesis in hepatocellular carcinoma. J Hepatol (2011) 54(5):948–55. doi: 10.1016/j.jhep.2010.08.041
18. Gkolfinopoulos S, Jones RL, Constantinidou A. The emerging role of platelets in the formation of the micrometastatic niche: Current evidence and future perspectives. Front Oncol (2020) 10:374. doi: 10.3389/fonc.2020.00374
19. Durán-Saenz NZ, Serrano-Puente A, Gallegos-Flores PI, Mendoza-Almanza BD, Esparza-Ibarra EL, Godina-González S, et al. Platelet membrane: An outstanding factor in cancer metastasis. Membranes (Basel) (2022) 12(2):182. doi: 10.3390/membranes12020182
20. Haemmerle M, Taylor ML, Gutschner T, Pradeep S, Cho MS, Sheng J, et al. Platelets reduce anoikis and promote metastasis by activating YAP1 signaling. Nat Commun (2017) 8(1):310. doi: 10.1038/s41467-017-00411-z
21. Mendoza-Almanza G, Burciaga-Hernández L, Maldonado V, Melendez-Zajgla J, Olmos J. Role of platelets and breast cancer stem cells in metastasis. World J Stem Cells (2020) 12(11):1237–54. doi: 10.4252/wjsc.v12.i11.1237
22. Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell (2011) 20(5):576–90. doi: 10.1016/j.ccr.2011.09.009
23. Wang Y, Yao R, Zhang D, Chen R, Ren Z, Zhang L. Circulating neutrophils predict poor survival for HCC and promote HCC progression through p53 and STAT3 signaling pathway. J Cancer (2020) 11(13):3736–44. doi: 10.7150/jca.42953
24. Li XF, Chen DP, Ouyang FZ, Chen MM, Wu Y, Kuang DM, et al. Increased autophagy sustains the survival and pro-tumourigenic effects of neutrophils in human hepatocellular carcinoma. J Hepatol (2015) 62(1):131–9. doi: 10.1016/j.jhep.2014.08.023
25. Geh D, Leslie J, Rumney R, Reeves HL, Bird TG, Mann DA. Neutrophils as potential therapeutic targets in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol (2022) 19(4):257–73. doi: 10.1038/s41575-021-00568-5
26. Grenader T, Nash S, Adams R, Kaplan R, Fisher D, Maughan T, et al. Derived neutrophil lymphocyte ratio is predictive of survival from intermittent therapy in advanced colorectal cancer: a post hoc analysis of the MRC COIN study. Br J Cancer (2016) 114(6):612–5. doi: 10.1038/bjc.2016.23
27. Gu X, Gao X, Li X, Qi X, Ma M, Qin S, et al. Prognostic significance of neutrophil-to-lymphocyte ratio in prostate cancer: evidence from 16,266 patients. Sci Rep (2016) 6:22089. doi: 10.1038/srep22089
28. Lin G, Liu Y, Li S, Mao Y, Wang J, Shuang Z, et al. Elevated neutrophil-to-lymphocyte ratio is an independent poor prognostic factor in patients with intrahepatic cholangiocarcinoma. Oncotarget (2016) 7(32):50963–71. doi: 10.18632/oncotarget.7680
29. Sun HL, Pan YQ, He BS, Nie ZL, Lin K, Peng HX, et al. Prognostic performance of lymphocyte-to-monocyte ratio in diffuse large b-cell lymphoma: an updated meta-analysis of eleven reports. Onco Targets Ther (2016) 9:3017–23. doi: 10.2147/ott.S96910
30. Diem S, Schmid S, Krapf M, Flatz L, Born D, Jochum W, et al. Neutrophil-to-Lymphocyte ratio (NLR) and platelet-to-Lymphocyte ratio (PLR) as prognostic markers in patients with non-small cell lung cancer (NSCLC) treated with nivolumab. Lung Cancer (2017) 111:176–81. doi: 10.1016/j.lungcan.2017.07.024
31. Thiagarajan S, Tan JW, Zhou S, Tan QX, Hendrikson J, Ng WH, et al. Postoperative inflammatory marker surveillance in colorectal peritoneal carcinomatosis. Ann Surg Oncol (2021) 28(11):6625–35. doi: 10.1245/s10434-020-09544-w
32. Templeton AJ, Knox JJ, Lin X, Simantov R, Xie W, Lawrence N, et al. Change in neutrophil-to-lymphocyte ratio in response to targeted therapy for metastatic renal cell carcinoma as a prognosticator and biomarker of efficacy. Eur Urol (2016) 70(2):358–64. doi: 10.1016/j.eururo.2016.02.033
33. Mason M, Maurice C, McNamara MG, Tieu MT, Lwin Z, Millar BA, et al. Neutrophil-lymphocyte ratio dynamics during concurrent chemo-radiotherapy for glioblastoma is an independent predictor for overall survival. J Neurooncol (2017) 132(3):463–71. doi: 10.1007/s11060-017-2395-y
34. Walsh SR, Cook EJ, Goulder F, Justin TA, Keeling NJ. Neutrophil-lymphocyte ratio as a prognostic factor in colorectal cancer. J Surg Oncol (2005) 91(3):181–4. doi: 10.1002/jso.20329
35. Guo L, Ren H, Pu L, Zhu X, Liu Y, Ma X. The prognostic value of inflammation factors in hepatocellular carcinoma patients with hepatic artery interventional treatments: A retrospective study. Cancer Manag Res (2020) 12:7173–88. doi: 10.2147/cmar.S257934
36. Limaye AR, Clark V, Soldevila-Pico C, Morelli G, Suman A, Firpi R, et al. Neutrophil-lymphocyte ratio predicts overall and recurrence-free survival after liver transplantation for hepatocellular carcinoma. Hepatol Res (2013) 43(7):757–64. doi: 10.1111/hepr.12019
37. McVey JC, Sasaki K, Firl DJ, Fujiki M, Diago-Uso T, Quintini C, et al. Prognostication of inflammatory cells in liver transplantation: Is the waitlist neutrophil-to-lymphocyte ratio really predictive of tumor biology? Clin Transplant (2019) 33(12):e13743. doi: 10.1111/ctr.13743
38. Bhatti I, Peacock O, Lloyd G, Larvin M, Hall RI. Preoperative hematologic markers as independent predictors of prognosis in resected pancreatic ductal adenocarcinoma: neutrophil-lymphocyte versus platelet-lymphocyte ratio. Am J Surg (2010) 200(2):197–203. doi: 10.1016/j.amjsurg.2009.08.041
39. Okamura Y, Sugiura T, Ito T, Yamamoto Y, Ashida R, Mori K, et al. Neutrophil to lymphocyte ratio as an indicator of the malignant behaviour of hepatocellular carcinoma. Br J Surg (2016) 103(7):891–8. doi: 10.1002/bjs.10123
40. Schobert IT, Savic LJ, Chapiro J, Bousabarah K, Chen E, Laage-Gaupp F, et al. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios as predictors of tumor response in hepatocellular carcinoma after DEB-TACE. Eur Radiol (2020) 30(10):5663–73. doi: 10.1007/s00330-020-06931-5
41. Yuan J, Liang H, Li J, Li M, Tang B, Ma H, et al. Peripheral blood neutrophil count as a prognostic factor for patients with hepatocellular carcinoma treated with sorafenib. Mol Clin Oncol (2017) 7(5):837–42. doi: 10.3892/mco.2017.1416
42. Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng AL, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol (2020) 73(6):1460–9. doi: 10.1016/j.jhep.2020.07.026
43. Choi WM, Kim JY, Choi J, Lee D, Shim JH, Lim YS, et al. Kinetics of the neutrophil-lymphocyte ratio during PD-1 inhibition as a prognostic factor in advanced hepatocellular carcinoma. Liver Int (2021) 41(9):2189–99. doi: 10.1111/liv.14932
44. Muhammed A, Fulgenzi CAM, Dharmapuri S, Pinter M, Balcar L, Scheiner B, et al. The systemic inflammatory response identifies patients with adverse clinical outcome from immunotherapy in hepatocellular carcinoma. Cancers (2021) 14(1):186. doi: 10.3390/cancers14010186
45. Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res (2008) 14(21):6735–41. doi: 10.1158/1078-0432.Ccr-07-4843
46. Motomura T, Shirabe K, Mano Y, Muto J, Toshima T, Umemoto Y, et al. Neutrophil-lymphocyte ratio reflects hepatocellular carcinoma recurrence after liver transplantation via inflammatory microenvironment. J Hepatol (2013) 58(1):58–64. doi: 10.1016/j.jhep.2012.08.017
47. Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc JF, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol (2017) 67(4):727–38. doi: 10.1016/j.jhep.2017.05.014
48. Chan SL, Mo FK, Johnson PJ, Hui EP, Ma BB, Ho WM, et al. New utility of an old marker: serial alpha-fetoprotein measurement in predicting radiologic response and survival of patients with hepatocellular carcinoma undergoing systemic chemotherapy. J Clin Oncol (2009) 27(3):446–52. doi: 10.1200/jco.2008.18.8151
49. Vora SR, Zheng H, Stadler ZK, Fuchs CS, Zhu AX. Serum alpha-fetoprotein response as a surrogate for clinical outcome in patients receiving systemic therapy for advanced hepatocellular carcinoma. Oncologist (2009) 14(7):717–25. doi: 10.1634/theoncologist.2009-0038
50. Shao YY, Lin ZZ, Hsu C, Shen YC, Hsu CH, Cheng AL. Early alpha-fetoprotein response predicts treatment efficacy of antiangiogenic systemic therapy in patients with advanced hepatocellular carcinoma. Cancer (2010) 116(19):4590–6. doi: 10.1002/cncr.25257
51. Hu X, Chen R, Wei Q, Xu X. The landscape of alpha fetoprotein in hepatocellular carcinoma: Where are we? Int J Biol Sci (2022) 18(2):536–51. doi: 10.7150/ijbs.64537
52. Spahn S, Roessler D, Pompilia R, Gabernet G, Gladstone BP, Horger M, et al. Clinical and genetic tumor characteristics of responding and non-responding patients to PD-1 inhibition in hepatocellular carcinoma. Cancers (Basel) (2020) 12(12):3830. doi: 10.3390/cancers12123830
53. Scheiner B, Pomej K, Kirstein MM, Hucke F, Finkelmeier F, Waidmann O, et al. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy - development and validation of the CRAFITY score. J Hepatol (2022) 76(2):353–63. doi: 10.1016/j.jhep.2021.09.035
54. Yang Y, Ouyang J, Zhou Y, Zhou J, Zhao H. The CRAFITY score: A promising prognostic predictor for patients with hepatocellular carcinoma treated with tyrosine kinase inhibitor and immunotherapy combinations. J Hepatol (2022) 77(2):574–6. doi: 10.1016/j.jhep.2022.03.018
55. Chen J, Zaidi S, Rao S, Chen JS, Phan L, Farci P, et al. Analysis of genomes and transcriptomes of hepatocellular carcinomas identifies mutations and gene expression changes in the transforming growth factor-β pathway. Gastroenterology (2018) 154(1):195–210. doi: 10.1053/j.gastro.2017.09.007
56. Chen J, Gingold JA, Su X. Immunomodulatory TGF-β signaling in hepatocellular carcinoma. Trends Mol Med (2019) 25(11):1010–23. doi: 10.1016/j.molmed.2019.06.007
57. Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol (2020) 72(2):215–29. doi: 10.1016/j.jhep.2019.08.017
58. Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J, et al. TGFβ1-mediated SMAD3 enhances PD-1 expression on antigen-specific T cells in cancer. Cancer Discovery (2016) 6(12):1366–81. doi: 10.1158/2159-8290.Cd-15-1347
59. Chen J, Su X. Abstract 4286: TGF-β signature is a therapeutic biomarker for combination immunotherapy for hepatocellular carcinoma. Cancer Res (2020) 80(16_Suppl):4286. doi: 10.1158/1538-7445.Am2020-4286
60. Feun LG, Li YY, Wu C, Wangpaichitr M, Jones PD, Richman SP, et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer (2019) 125(20):3603–14. doi: 10.1002/cncr.32339
61. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature (2018) 554(7693):544–8. doi: 10.1038/nature25501
62. Cheuk AT, Mufti GJ, Guinn BA. Role of 4-1BB:4-1BB ligand in cancer immunotherapy. Cancer Gene Ther (2004) 11(3):215–26. doi: 10.1038/sj.cgt.7700670
63. Reithofer M, Rosskopf S, Leitner J, Battin C, Bohle B, Steinberger P, et al. 4-1BB costimulation promotes bystander activation of human CD8 T cells. Eur J Immunol (2021) 51(3):721–33. doi: 10.1002/eji.202048762
64. Melero I, Johnston JV, Shufford WW, Mittler RS, Chen L. NK1.1 cells express 4-1BB (CDw137) costimulatory molecule and are required for tumor immunity elicited by anti-4-1BB monoclonal antibodies. Cell Immunol (1998) 190(2):167. doi: 10.1006/cimm.1998.1396
65. Heinisch IV, Daigle I, Knöpfli B, Simon HU. CD137 activation abrogates granulocyte-macrophage colony-stimulating factor-mediated anti-apoptosis in neutrophils. Eur J Immunol (2000) 30(12):3441–6. doi: 10.1002/1521-4141(2000012)30:12<3441::Aid-immu3441>3.0.Co;2-l
66. Kim HD, Park S, Jeong S, Lee YJ, Lee H, Kim CG, et al. 4-1BB delineates distinct activation status of exhausted tumor-infiltrating CD8(+) T cells in hepatocellular carcinoma. Hepatology (2020) 71(3):955–71. doi: 10.1002/hep.30881
67. Michel J, Langstein J, Hofstädter F, Schwarz H. A soluble form of CD137 (ILA/4-1BB), a member of the TNF receptor family, is released by activated lymphocytes and is detectable in sera of patients with rheumatoid arthritis. Eur J Immunol (1998) 28(1):290–5. doi: 10.1002/(sici)1521-4141(199801)28:01<290::Aid-immu290>3.0.Co;2-s
68. DSV A, BSK B. Role of 4-1BB in immune responses. Semin Immunol (1998) 10(6):481–9. doi: 10.1006/smim.1998.0157
69. Geuijen C, Tacken P, Wang LC, Klooster R, van Loo PF, Zhou J, et al. A human CD137×PD-L1 bispecific antibody promotes anti-tumor immunity via context-dependent T cell costimulation and checkpoint blockade. Nat Commun (2021) 12(1):4445. doi: 10.1038/s41467-021-24767-5
70. Jacquelot N, Roberti MP, Enot DP, Rusakiewicz S, Ternès N, Jegou S, et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat Commun (2017) 8(1):592. doi: 10.1038/s41467-017-00608-2
71. Zhang W, Gong C, Peng X, Bi X, Sun Y, Zhou J, et al. Serum concentration of CD137 and tumor infiltration by M1 macrophages predict the response to sintilimab plus bevacizumab biosimilar in advanced hepatocellular carcinoma patients. Clin Cancer Res (2022) 28(16):3499–508. doi: 10.1158/1078-0432.Ccr-21-3972
72. Zhu S, Hoshida Y. Molecular heterogeneity in hepatocellular carcinoma. Hepat Oncol (2018) 5(1):Hep10. doi: 10.2217/hep-2018-0005
73. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer (2019) 18(1):114. doi: 10.1186/s12943-019-1043-x
74. Alunni-Fabbroni M, Rönsch K, Huber T, Cyran CC, Seidensticker M, Mayerle J, et al. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: a translational exploratory study from the SORAMIC trial. J Transl Med (2019) 17(1):328. doi: 10.1186/s12967-019-2079-9
75. Moshiri F, Salvi A, Gramantieri L, Sangiovanni A, Guerriero P, De Petro G, et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget (2018) 9(20):15350–64. doi: 10.18632/oncotarget.24601
76. Kelley RK, Magbanua MJ, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer (2015) 15:206. doi: 10.1186/s12885-015-1195-z
77. Julich-Haertel H, Urban SK, Krawczyk M, Willms A, Jankowski K, Patkowski W, et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J Hepatol (2017) 67(2):282–92. doi: 10.1016/j.jhep.2017.02.024
78. Pantel K, Hille C, Scher HI. Circulating tumor cells in prostate cancer: From discovery to clinical utility. Clin Chem (2019) 65(1):87–99. doi: 10.1373/clinchem.2018.287102
79. Chen W, Zhang J, Huang L, Chen L, Zhou Y, Tang D, et al. Detection of HER2-positive circulating tumor cells using the LiquidBiopsy system in breast cancer. Clin Breast Cancer (2019) 19(1):e239–e46. doi: 10.1016/j.clbc.2018.10.009
80. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol (2021) 18(5):297–312. doi: 10.1038/s41571-020-00457-x
81. Munzone E, Botteri E, Sandri MT, Esposito A, Adamoli L, Zorzino L, et al. Prognostic value of circulating tumor cells according to immunohistochemically defined molecular subtypes in advanced breast cancer. Clin Breast Cancer (2012) 12(5):340–6. doi: 10.1016/j.clbc.2012.07.001
82. Winograd P, Hou S, Court CM, Lee YT, Chen PJ, Zhu Y, et al. Hepatocellular carcinoma-circulating tumor cells expressing PD-L1 are prognostic and potentially associated with response to checkpoint inhibitors. Hepatol Commun (2020) 4(10):1527–40. doi: 10.1002/hep4.1577
83. Su K, Guo L, He K, Rao M, Zhang J, Yang X, et al. PD-L1 expression on circulating tumor cells can be a predictive biomarker to PD-1 inhibitors combined with radiotherapy and antiangiogenic therapy in advanced hepatocellular carcinoma. Front Oncol (2022) 12:873830. doi: 10.3389/fonc.2022.873830
84. Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol (2017) 23(31):5650–68. doi: 10.3748/wjg.v23.i31.5650
85. Chakrabarti S, Xie H, Urrutia R, Mahipal A. The promise of Circulating Tumor DNA (ctDNA) in the management of early-stage colon cancer: A critical review. Cancers (2020) 12(10):2808. doi: 10.3390/cancers12102808
86. von Felden J, Craig AJ, Garcia-Lezana T, Labgaa I, Haber PK, D'Avola D, et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene (2021) 40(1):140–51. doi: 10.1038/s41388-020-01519-1
87. Wu X, Li J, Gassa A, Buchner D, Alakus H, Dong Q, et al. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int J Biol Sci (2020) 16(9):1551–62. doi: 10.7150/ijbs.44024
88. Kaseb A, Sánchez N, Sen S, Kelley R, Tan B, Bocobo A, et al. Molecular profiling of hepatocellular carcinoma using circulating cell-free DNA. Clin Cancer Res (2019) 25(20):6107–18. doi: 10.1158/1078-0432.CCR-18-3341. clincanres.3341.2018.
89. Hsu C-H, Lu S, Abbas A, Guan Y, Zhu AX, Aleshin A, et al. Longitudinal and personalized detection of circulating tumor DNA (ctDNA) for monitoring efficacy of atezolizumab plus bevacizumab in patients with unresectable hepatocellular carcinoma (HCC). J Clin Oncol (2020) 38(15_suppl):3531. doi: 10.1200/JCO.2020.38.15_suppl.3531
90. Yang X, Hu Y, Yang K, Wang D, Lin J, Long J, et al. Cell-free DNA copy number variations predict efficacy of immune checkpoint inhibitor-based therapy in hepatobiliary cancers. J Immunother Cancer (2021) 9(5):e001942. doi: 10.1136/jitc-2020-001942
91. Zhao W, Qiu L, Liu H, Xu Y, Zhan M, Zhang W, et al. Circulating tumor DNA as a potential prognostic and predictive biomarker during interventional therapy of unresectable primary liver cancer. J Gastrointest Oncol (2020) 11(5):1065–77. doi: 10.21037/jgo-20-409
92. Santhakumar C, Gane EJ, Liu K, McCaughan GW. Current perspectives on the tumor microenvironment in hepatocellular carcinoma. Hepatol Int (2020) 14(6):947–57. doi: 10.1007/s12072-020-10104-3
93. Zhu AX, Abbas AR, de Galarreta MR, Guan Y, Lu S, Koeppen H, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med (2022) 28(8):1599–611. doi: 10.1038/s41591-022-01868-2
94. Nishida N, Sakai K, Morita M, Aoki T, Takita M, Hagiwara S, et al. Association between genetic and immunological background of hepatocellular carcinoma and expression of programmed cell death-1. Liver Cancer (2020) 9(4):426–39. doi: 10.1159/000506352
95. Tian Z, Hou X, Liu W, Han Z, Wei L. Macrophages and hepatocellular carcinoma. Cell Biosci (2019) 9:79. doi: 10.1186/s13578-019-0342-7
96. Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discovery (2018) 17(12):887–904. doi: 10.1038/nrd.2018.169
97. Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol (2019) 12(1):76. doi: 10.1186/s13045-019-0760-3
98. Dong N, Shi X, Wang S, Gao Y, Kuang Z, Xie Q, et al. M2 macrophages mediate sorafenib resistance by secreting HGF in a feed-forward manner in hepatocellular carcinoma. Br J Cancer (2019) 121(1):22–33. doi: 10.1038/s41416-019-0482-x
99. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature (2015) 523(7559):231–5. doi: 10.1038/nature14404
100. Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology (2015) 149(5):1226–39.e4. doi: 10.1053/j.gastro.2015.05.061
101. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet (2012) 44(6):694–8. doi: 10.1038/ng.2256
102. Takai A, Dang HT, Wang XW. Identification of drivers from cancer genome diversity in hepatocellular carcinoma. Int J Mol Sci (2014) 15(6):11142–60. doi: 10.3390/ijms150611142
103. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell (2017) 169(7):1327–41.e23. doi: 10.1016/j.cell.2017.05.046
104. Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet (2015) 47(5):505–11. doi: 10.1038/ng.3252
105. Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, et al. β-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discovery (2019) 9(8):1124–41. doi: 10.1158/2159-8290.Cd-19-0074
106. Waaler J, Mygland L, Tveita A, Strand MF, Solberg NT, Olsen PA, et al. Tankyrase inhibition sensitizes melanoma to PD-1 immune checkpoint blockade in syngeneic mouse models. Commun Biol (2020) 3(1):196. doi: 10.1038/s42003-020-0916-2
107. Morita M, Nishida N, Sakai K, Aoki T, Chishina H, Takita M, et al. Immunological microenvironment predicts the survival of the patients with hepatocellular carcinoma treated with anti-PD-1 antibody. Liver Cancer (2021) 10(4):380–93. doi: 10.1159/000516899
108. Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, et al. Prospective genotyping of hepatocellular carcinoma: Clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res (2019) 25(7):2116–26. doi: 10.1158/1078-0432.Ccr-18-2293
109. Chen L, Zhou Q, Liu J, Zhang W. CTNNB1 alternation is a potential biomarker for immunotherapy prognosis in patients with hepatocellular carcinoma. Front Immunol (2021) 12:759565. doi: 10.3389/fimmu.2021.759565
110. Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet (2019) 393(10183):1819–30. doi: 10.1016/s0140-6736(18)32409-7
111. Rini BI, Powles T, Atkins MB, Escudier B, McDermott DF, Suarez C, et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open-label, phase 3, randomised controlled trial. Lancet (2019) 393(10189):2404–15. doi: 10.1016/s0140-6736(19)30723-8
112. Abdel-Rahman O. PD-L1 expression and outcome of advanced melanoma patients treated with anti-PD-1/PD-L1 agents: a meta-analysis. Immunotherapy (2016) 8(9):1081–9. doi: 10.2217/imt-2016-0025
113. Yau T, Park JW, Finn RS, Cheng AL, Mathurin P, Edeline J, et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol (2022) 23(1):77–90. doi: 10.1016/s1470-2045(21)00604-5
114. Yau T, Kang YK, Kim TY, El-Khoueiry AB, Santoro A, Sangro B, et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: The CheckMate 040 randomized clinical trial. JAMA Oncol (2020) 6(11):e204564. doi: 10.1001/jamaoncol.2020.4564
115. Chan TA, Wolchok JD, Snyder A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med (2015) 373(20):1984. doi: 10.1056/NEJMc1508163
116. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (2015) 348(6230):124–8. doi: 10.1126/science.aaa1348
117. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell (2017) 171(4):934–49.e16. doi: 10.1016/j.cell.2017.09.028
118. Xie F, Bai Y, Yang X, Long J, Mao J, Lin J, et al. Comprehensive analysis of tumour mutation burden and the immune microenvironment in hepatocellular carcinoma. Int Immunopharmacol (2020) 89(Pt A):107135. doi: 10.1016/j.intimp.2020.107135
119. Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun (2016) 7:10391. doi: 10.1038/ncomms10391
120. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer (2019) 19(3):133–50. doi: 10.1038/s41568-019-0116-x
121. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
122. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
123. Peng Z, Cheng S, Kou Y, Wang Z, Jin R, Hu H, et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res (2020) 8(10):1251–61. doi: 10.1158/2326-6066.Cir-19-1014
124. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
125. Behary J, Amorim N, Jiang XT, Raposo A, Gong L, McGovern E, et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat Commun (2021) 12(1):187. doi: 10.1038/s41467-020-20422-7
126. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science (2021) 371(6536):eabc4552. doi: 10.1126/science.abc4552
127. Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell (2021) 39(10):1317–41. doi: 10.1016/j.ccell.2021.08.006
128. Skelly AN, Sato Y, Kearney S, Honda K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol (2019) 19(5):305–23. doi: 10.1038/s41577-019-0144-5
129. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33(4):570–80. doi: 10.1016/j.ccell.2018.03.015
130. Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology (2021) 160(2):600–13. doi: 10.1053/j.gastro.2020.11.041
131. Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer (2019) 7(1):193. doi: 10.1186/s40425-019-0650-9
132. Mao J, Wang D, Long J, Yang X, Lin J, Song Y, et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J Immunother Cancer (2021) 9(12):e003334. doi: 10.1136/jitc-2021-003334
133. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
134. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science (2021) 371(6529):595–602. doi: 10.1126/science.abf3363
135. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
136. Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med (2018) 378(2):158–68. doi: 10.1056/NEJMra1703481
137. Yau T, Park JW, Finn RS, Cheng AL, Mathurin P, Edeline J, et al. CheckMate 459: A randomized, multi-center phase III study of nivolumab (NIVO) vs sorafenib (SOR) as first-line (1L) treatment in patients (pts) with advanced hepatocellular carcinoma (aHCC). Ann Oncol (2019) 30:v874–v5. doi: 10.1093/annonc/mdz394
138. Finn RS, Ryoo BY, Merle P, Kudo M, Bouattour M, Lim HY, et al. Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE-240: A randomized, double-blind, phase III trial. J Clin Oncol (2020) 38(3):193–202. doi: 10.1200/jco.19.01307
139. Ren Z, Xu J, Bai Y, Xu A, Cang S, Du C, et al. Sintilimab plus a bevacizumab biosimilar (IBI305) versus sorafenib in unresectable hepatocellular carcinoma (ORIENT-32): a randomised, open-label, phase 2-3 study. Lancet Oncol (2021) 22(7):977–90. doi: 10.1016/s1470-2045(21)00252-7
140. Lee MS, Ryoo BY, Hsu CH, Numata K, Stein S, Verret W, et al. Atezolizumab with or without bevacizumab in unresectable hepatocellular carcinoma (GO30140): an open-label, multicentre, phase 1b study. Lancet Oncol (2020) 21(6):808–20. doi: 10.1016/s1470-2045(20)30156-x
141. Indini A, Di Guardo L, Cimminiello C, Prisciandaro M, Randon G, De Braud F, et al. Immune-related adverse events correlate with improved survival in patients undergoing anti-PD1 immunotherapy for metastatic melanoma. J Cancer Res Clin Oncol (2019) 145(2):511–21. doi: 10.1007/s00432-018-2819-x
142. Maher VE, Fernandes LL, Weinstock C, Tang S, Agarwal S, Brave M, et al. Analysis of the association between adverse events and outcome in patients receiving a programmed death protein 1 or programmed death ligand 1 antibody. J Clin Oncol (2019) 37(30):2730–7. doi: 10.1200/jco.19.00318
143. Ishihara H, Takagi T, Kondo T, Homma C, Tachibana H, Fukuda H, et al. Association between immune-related adverse events and prognosis in patients with metastatic renal cell carcinoma treated with nivolumab. Urol Oncol (2019) 37(6):355.e21–.e29. doi: 10.1016/j.urolonc.2019.03.003
144. Haratani K, Hayashi H, Chiba Y, Kudo K, Yonesaka K, Kato R, et al. Association of immune-related adverse events with nivolumab efficacy in non-Small-Cell lung cancer. JAMA Oncol (2018) 4(3):374–8. doi: 10.1001/jamaoncol.2017.2925
145. Masuda K, Shoji H, Nagashima K, Yamamoto S, Ishikawa M, Imazeki H, et al. Correlation between immune-related adverse events and prognosis in patients with gastric cancer treated with nivolumab. BMC Cancer (2019) 19(1):974. doi: 10.1186/s12885-019-6150-y
146. Shankar B, Zhang J, Naqash AR, Forde PM, Feliciano JL, Marrone KA, et al. Multisystem immune-related adverse events associated with immune checkpoint inhibitors for treatment of non-small cell lung cancer. JAMA Oncol (2020) 6(12):1952–6. doi: 10.1001/jamaoncol.2020.5012
147. Ng KYY, Tan SH, Tan JJE, Tay DSH, Lee AWX, Ang AJS, et al. Impact of immune-related adverse events on efficacy of immune checkpoint inhibitors in patients with advanced hepatocellular carcinoma. Liver Cancer (2022) 11(1):9–21. doi: 10.1159/000518619
148. Quandt Z, Young A, Anderson M. Immune checkpoint inhibitor diabetes mellitus: a novel form of autoimmune diabetes. Clin Exp Immunol (2020) 200(2):131–40. doi: 10.1111/cei.13424
149. Lim CJ, Lee YH, Pan L, Lai L, Chua C, Wasser M, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis b virus-related hepatocellular carcinoma. Gut (2019) 68(5):916–27. doi: 10.1136/gutjnl-2018-316510
150. Haber PK, Puigvehí M, Castet F, Lourdusamy V, Montal R, Tabrizian P, et al. Evidence-based management of hepatocellular carcinoma: Systematic review and meta-analysis of randomized controlled trials (2002-2020). Gastroenterology (2021) 161(3):879–98. doi: 10.1053/j.gastro.2021.06.008
151. Liu T, Li Q, Lin Z, Wang P, Chen Y, Fu Y, et al. Viral infections and the efficacy of PD-(L)1 inhibitors in virus-related cancers: Head and neck squamous cell carcinoma and hepatocellular carcinoma. Int Immunopharmacol (2021) 100:108128. doi: 10.1016/j.intimp.2021.108128
152. De Battista D, Zamboni F, Gerstein H, Sato S, Markowitz TE, Lack J, et al. Molecular signature and immune landscape of HCV-associated hepatocellular carcinoma (HCC): Differences and similarities with HBV-HCC. J Hepatocellular Carcinoma (2021) 8:1399–413. doi: 10.2147/jhc.S325959
153. Larrea E, Riezu-Boj JI, Gil-Guerrero L, Casares N, Aldabe R, Sarobe P, et al. Upregulation of indoleamine 2,3-dioxygenase in hepatitis c virus infection. J Virol (2007) 81(7):3662–6. doi: 10.1128/jvi.02248-06
154. Riezu-Boj JI, Larrea E, Aldabe R, Guembe L, Casares N, Galeano E, et al. Hepatitis c virus induces the expression of CCL17 and CCL22 chemokines that attract regulatory T cells to the site of infection. J Hepatol (2011) 54(3):422–31. doi: 10.1016/j.jhep.2010.07.014
155. Nakamoto N, Kaplan DE, Coleclough J, Li Y, Valiga ME, Kaminski M, et al. Functional restoration of HCV-specific CD8 T cells by PD-1 blockade is defined by PD-1 expression and compartmentalization. Gastroenterology (2008) 134(7):1927–37, 37.e1-2. doi: 10.1053/j.gastro.2008.02.033
156. Ho WJ, Danilova L, Lim SJ, Verma R, Xavier S, Leatherman JM, et al. Viral status, immune microenvironment and immunological response to checkpoint inhibitors in hepatocellular carcinoma. J Immunother Cancer (2020) 8(1):e000394. doi: 10.1136/jitc-2019-000394
157. Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol (2019) 16(7):411–28. doi: 10.1038/s41575-019-0145-7
158. Chang J, Ryan JS, Ajmera V, Ting S, Tamayo P, Burgoyne A. Responses to immunotherapy in hepatocellular carcinoma patients with nonalcoholic steatohepatitis cirrhosis. J Clin Oncol (2022) 40(4_suppl):389. doi: 10.1200/JCO.2022.40.4_suppl.389
Keywords: hepatocellular carcinoma, immunotherapy, combination therapy, biomarker, tumor immune microenvironment
Citation: Peng X, Gong C, Zhang W and Zhou A (2023) Advanced development of biomarkers for immunotherapy in hepatocellular carcinoma. Front. Oncol. 12:1091088. doi: 10.3389/fonc.2022.1091088
Received: 06 November 2022; Accepted: 20 December 2022;
Published: 16 January 2023.
Edited by:
Xiong Chen, Nanjing General Hospital of Nanjing Military Command, ChinaReviewed by:
Eleonora Lai, University Hospital and University of Cagliari, ItalyFucun Xie, Peking Union Medical College Hospital (CAMS), China
Copyright © 2023 Peng, Gong, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aiping Zhou, YWlwaW5nX3pob3VAeWVhaC5uZXQ=
 Xuenan Peng
Xuenan Peng Caifeng Gong
Caifeng Gong Wen Zhang
Wen Zhang Aiping Zhou
Aiping Zhou