- 1The First Clinical Medical College, Lanzhou University, Lanzhou, China
- 2Department of Gastroenterology, The First Hospital of Lanzhou University, Lanzhou, China
- 3Key Laboratory for Gastrointestinal Diseases of Gansu Province, The First Hospital of Lanzhou University, Lanzhou, China
The metastasis of malignant tumors determines patient prognosis. This is the main reason for the poor prognosis of patients with cancer and the most challenging aspect of treating malignant tumors. Therefore, it is important to identify early tumor markers and molecules that can predict patient prognosis. However, there are currently no molecular markers with good clinical accuracy and specificity. Many non-coding RNA (ncRNAs)have been identified, which can regulate the process of tumor development at multiple levels. Interestingly, some ncRNAs are translated to produce functional peptides. Exosomes act as signal carriers, are encapsulated in nucleic acids and proteins, and play a messenger role in cell-to-cell communication. Recent studies have identified exosome peptides with potential diagnostic roles. This review aims to provide a theoretical basis for ncRNA-encoded peptides or proteins transported by exosomes and ultimately to provide ideas for further development of new diagnostic and prognostic cancer markers.
1 Introduction
According to the statistical results of the World Health Organization in 2019, cancer is the first or second leading cause of death before the age of 70 years in 112 of 183 countries. Furthermore, the status of cancer as a leading cause of death continues to rise (1). During the growth and development of the body, oncogenes and tumor suppressor genes accumulate mutations due to repeated stimulation by various factors in vivo and in vitro, thus triggering the process of tumor development and metastasis, most of which are located in genomic regions lacking protein-coding capacity (2). However, currently available tumor biomarkers and therapeutic targets have been identified based on coding genes. Genome-wide association studies have identified many genetic variants associated with tumors and other diseases, two-thirds of which are located in non-coding regions (3, 4).
Non-coding RNAs (ncRNAs) is RNA molecules transcribed by the genome that do not encode a protein. NcRNAs can be divided into two main types: basic structural ncRNA and regulatory ncRNA. Basic structural ncRNAs are composed of ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and small nuclear RNAs (snRNAs). The functions of these ncRNAs are relatively well defined. Regulatory ncRNA including three main types: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) (5, 6). Initially, ncRNAs were considered non-functional byproducts of RNA polymerase II transcripts and existed as “transcriptional noise” in the genome (7). However, there is growing evidence that ncRNAs have various essential functions in cancer cell growth, development, apoptosis, and metabolism (8–11) (Figure S1). The functions of the ncRNA molecules described above are based on their RNA molecules. Recent research on ncRNA molecules in tumors, such as breast, liver, and intestinal cancers has shown that ncRNAs can encode translation to produce functional peptides or protein molecules that participate in the primary developmental processes of various organisms and regulate the development of tumor cells (12–14). Although the functions of some peptides have been demonstrated, the mechanism of their action in cancer remains unclear. Studies have shown that exosomes are essential carriers of intercellular communication and contain a variety of functional molecules, such as mRNA, miRNAs, lncRNAs, circRNAs, and proteins (15–17). Exosome membranes influence the biological behavior of target cells by delivering their contents to them, thus influencing the biological behavior of the latter.
Interestingly, exosomes and peptides may also work together to deliver signaling molecules to modify or modulate receptor cells. This review summarizes the current studies concerning the relationship between exosomes and ncRNA with encoding potential in cancers. This review aimed to provide a theoretical basis for the study of the mechanisms of peptides encoded by ncRNAs.
2 Prediction and validation of novel peptides encoded by ncRNAs
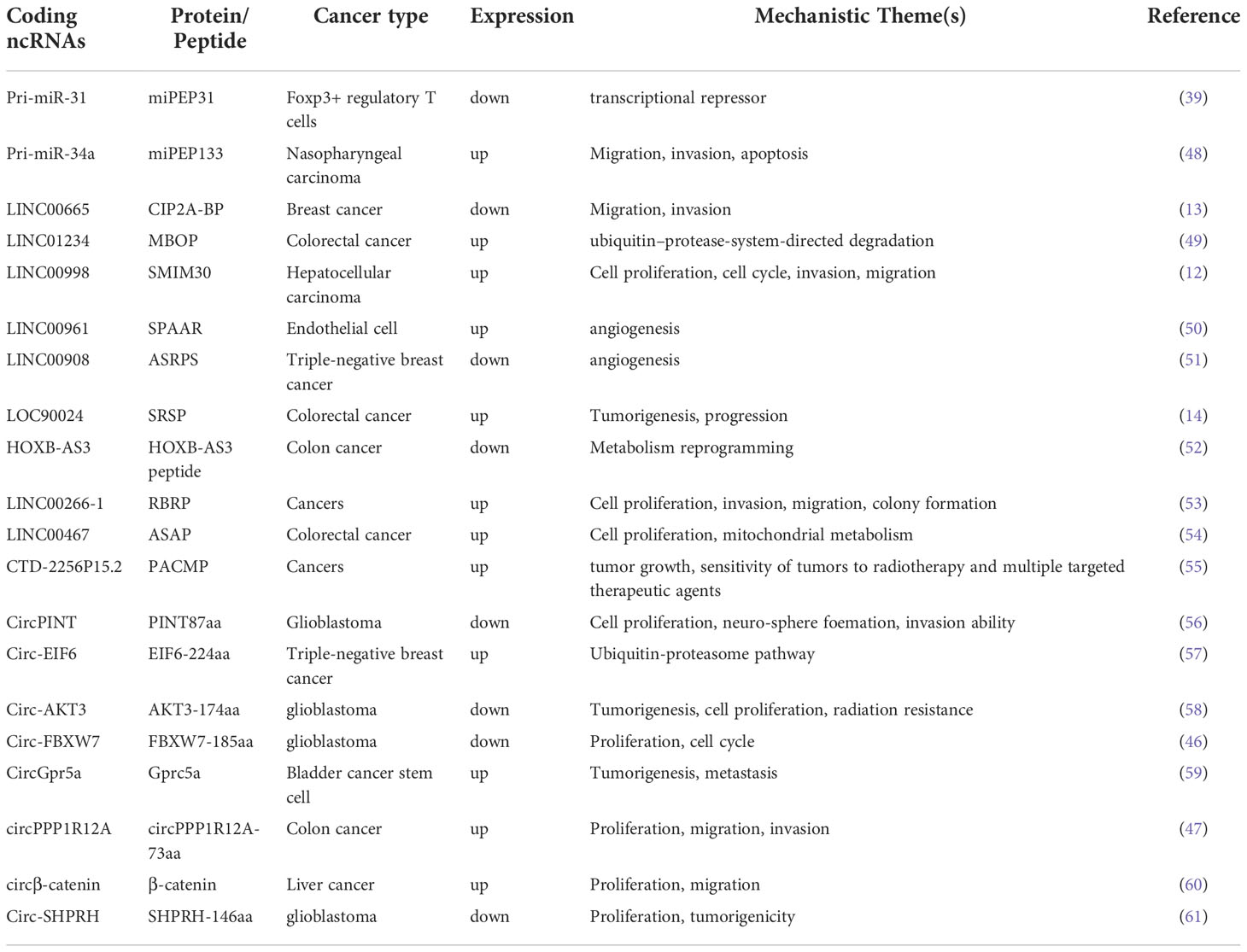
A small percentage of ncRNAs contain a small open reading frame (smORF, < 300 nucleo tide) that can be translated into a peptide. Translation of ncRNAs has been neglected mainly because the peptides or proteins are encoded in a noncanonical mode, and the peptides are unstable and rapidly degraded (18, 19). However, owing to advances in sequencing technology, many studies have found that ncRNAs encode peptides and proteins during cancer development (20). To identify hidden peptides in ncRNAs, there are tools to identify smORFs with coding potential, such as CPC2 (21), ORF finder (22), LGC (23), ORF-RATER (24), CPAT (25), ORFscore (26), PhyloCSF (27), and PLEK (28). In the input box on the CPC2 homepage, you can simply paste or upload the FASTA format of the nucleotide sequences to obtain a report containing coding probability, putative peptide length, Fickett score, and isoelectric point (21). We then entered the DNA sequence in the ORF finder search box to search for potential protein-coding fragments and used SMART BLAST or BLASTP to validate the predicted proteins (22). LGC was the first new algorithm based on the difference in ORF length and GC content between lncRNAs and protein-coding RNAs. LGC outperforms the existing algorithms owing to its higher accuracy, sensitivity, and specificity (23). Based on these online tools (Table 1), we can initially predict whether a ncRNA of interest has coding properties. Using these tools, we will find some ncRNAs with encoding potential, and in the next step, we need to use some methods to verify the presence of these peptides. Mass spectrometry (MS) methods and peptidomics testing of encoded peptides are comparative direct evidence for the translability of ORFs. Build a fusion epitope tags, such as FLAG, GFP, HIS, etc., together with peptides/proteins to use western blot (WB), immunofluorescence to detect the presence of fusion proteins, and antibodies against peptide/protein can also be prepared for detection. CRISPR(clustered regularly interspaced short palindromic repeats)-Cas9 (CRISPR-associated protein 9) can insert the epitope tag into the site of the peptide/protein, and then use WB to detect the presence of the peptide/protein. Through the above experimental methods, we can confirm that the ncRNA can be translated into a peptide.
3 Functional ncRNAs-encoded peptides
3.1 MicroRNAs
MiRNAs are small RNAs approximately 20-25 nucleotides in length that regulate gene expression by explicitly cleaving or inhibiting the translation of target mRNAs (36, 37). The biogenesis of miRNAs undergoes two main steps. MiRNAs are first transcribed into primary transcripts of miRNAs (pri-miRNAs) in the nucleus. Then, miRNA is further processed to form a pre-miRNA for translocation into the cytoplasm and finally sheared by the Dicer enzyme. The mature miRNA binds to the 3’-UTR of the target mRNA to function (37). Usually, after a mature miRNA undergoes processing and excitement, the upstream and downstream sequences of the pri-miRNA become nonfunctional. They do not appear to be translated further to produce peptides or protein molecules (38). However, the current study shows that miRNA-encoded peptides (miPEPs) begin a new phase of gene regulation. Lauressegues et al. (36) found that in Arabidopsis, pri-miR171b and pri-miR165a have the potential to encode the peptides miPEP171b and miPEP165a, which downregulate downstream genes involved in root development. More importantly, Zhou et al. (39) found that pri-miRNA-31 encodes a peptide, miPEP31. MiPEP31 inhibits the transcription of miRNA-31 and downregulates its expression. Additionally, miRNA-31 is a negative regulator of Tregs-cells. MiPEP31 significantly inhibited experimental autoimmune encephalomyelitis by promoting Treg cell differentiation. This study found the peptide encoded by pri-miR-31 provides new approaches for the treatment of autoimmune disease. Current research on miRNAs is unclear as to whether miPEPs exist in all organisms and whether they all have a biological role. However, the relationship between miPEPs and tumors requires further investigation (Figure 1).
3.2 LncRNA
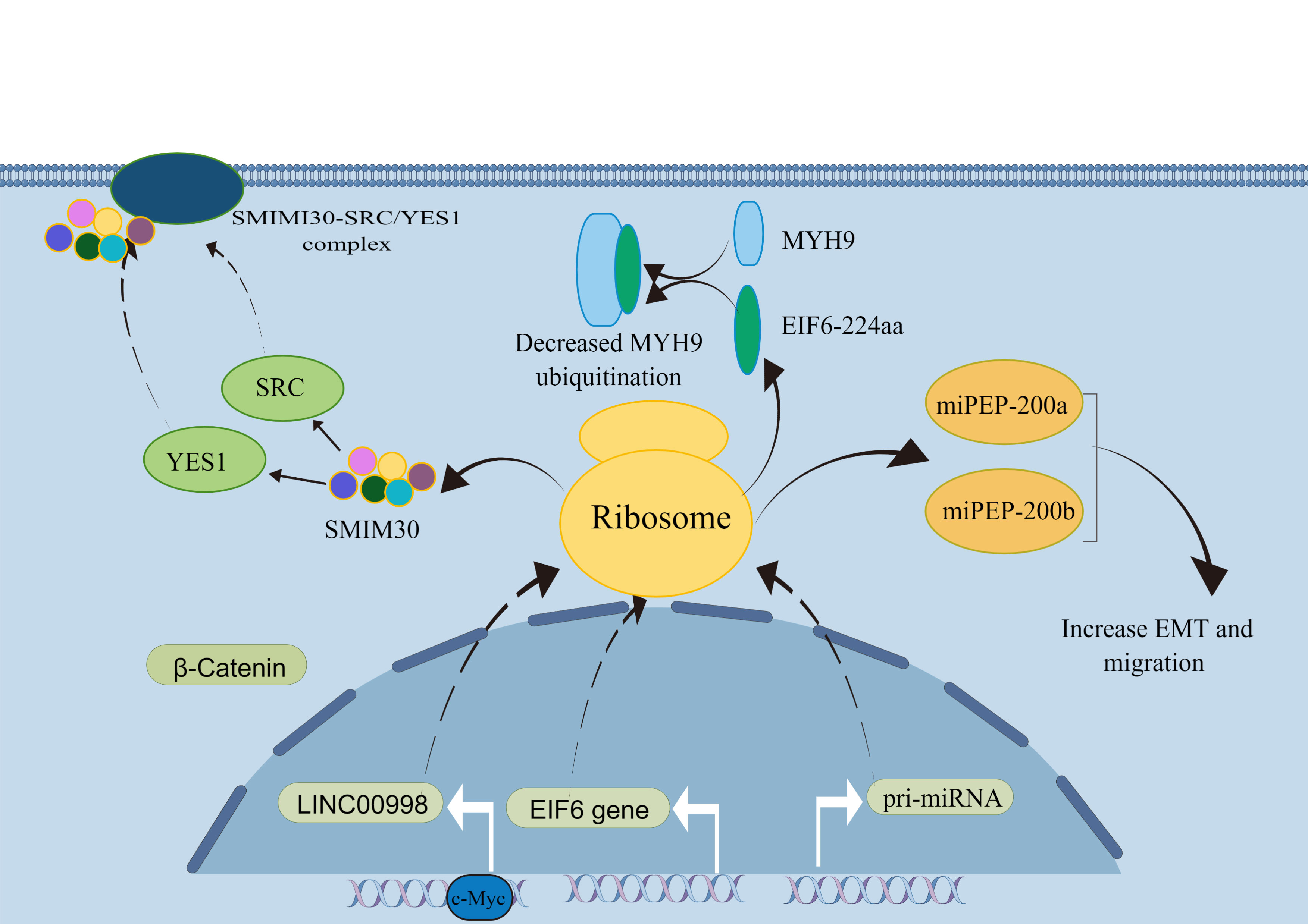
LncRNAs are a class of RNA molecules with transcripts longer than 200 nt transcribed by RNA polymerase II. It has a specific spatial secondary structure and a low interspecific conservation. LncRNA was initially considered as a redundant product of transcription. However, current studies suggest that it can undergo post-transcription modification, such as capping, polyadenylation, and splicing (40–42). Meanwhile, LncRNA generally does not encode proteins, but the number of molecules is much larger than protein-coding genes. LncRNA molecules are usually resided at separate locations on the genome and do not overlap with classical protein-coding genes, with some having small open reading frames (smORFs) (43). Using bioinformatics analysis, Pang et al. (12) identified LINC00998 as having a translational potential. The expression of peptide molecules corresponding to the smORF sequences was identified in vitro and in vivo by constructing fusion plasmids with FLAG tags. A recombinant plasmid fused to GFP was constructed, and SMIM30 was found to be localized to the cell membrane with a significant ability to guide membrane localization. In addition, extensive cellular functional assays revealed that the peptide SMIM30 plays a vital role in promoting the proliferation, invasion, and migration of hepatocellular carcinoma cells. Therefore, peptide SMIM30 may be used as a new prognostic indicator of hepatocellular carcinoma (HCC) and can provide a reference for the treatment of HCC patients. Guo et al. (13) identified LINC00665 in breast cancer that was significantly downregulated in translation after TGF-β treatment, further demonstrating that TGF-β treatment activates the Smad signaling pathway. Smad4 directly induced the expression of the translation repressor protein 4E-BP1, which led to a decrease in the expression of the peptide encoded by LINC00665 through direct binding to the eukaryotic translation initiation factor eIF4E. TGF-β acts as an oncogenic factor in advanced tumors and, to some extent, TGF-β induces metastasis and invasion in triple-negative breast cancer by regulating CIP2A-BP translation. In conclusion, the micropeptide protein CIP2A-BP, encoded by LINC00665, may soon become a potential therapeutic agent for treating triple-negative breast cancer metastasis. Although a limited number of peptides/proteins have been functionally characterized, their mechanisms of action in tumors remain unclear. Therefore, the study of ncRNAs molecules, especially lncRNAs, should not be confined to the RNA level but should also focus on whether they can be translated to produce functional protein molecules that can play a role in regulating basic life activities and disease development.
3.3 CircRNA
CircRNAs are produced from the exon or intron of a precursor messenger RNA by reverse splicing. CircRNAs are immune to digestion and degradation by nucleic acid exonucleases due to the absence of a 5’ end cap structure and a 3’ end tail (44). Currently, most circRNAs act as molecular sponges for miRNAs as the primary mechanism of action. However, in recent years, with the development of mass spectrometry and ribosome mapping, proteins or peptides produced by translation have become involved in the process of tumor development. There are two primary forms of protein translation: classical cap-dependent and non-classical non-cap-dependent. The non-cap mechanism allows cells to respond to a variety of transient stresses (45), and this translation is the mechanism underlying circRNA translation. In 2018, Yang et al. (46) discovered that circRNAs can encode peptides. In gliomas, circ-FBXW7 can encode a peptide FBXW7-185aa of 185 amino acids, which promotes the degradation of the proto-oncogene c-Myc by competitively binding USP28 ubiquitinated proteins, thereby inhibiting the development and progression of malignant gliomas.
Furthermore, high expression of circPPP1R12A was found in gastric cancer, encoding the production of the functional protein circPPP1R12A-73aa. In addition, cellular functional assays confirmed that it was the peptide but not circPPP1R12A that promoted the proliferation, invasion, and migration of gastric cancer cells in vivo and in vitro (47). Whether circRNAs that act through an endogenous competitive RNA mechanism can also act through peptides is interesting. A growing number of studies have confirmed that circRNA-encoded proteins or peptides have different mechanisms of action in tumors. Circ-EIF6 encodes EIF6-224aa which acts as a prognostic and therapeutic target in triple-negative breast cancer. This also suggests that circRNAs or the peptides they encode can be used as tumor markers and potential therapeutic targets (Table 2).
4 Exosomal ncRNAs and cancer
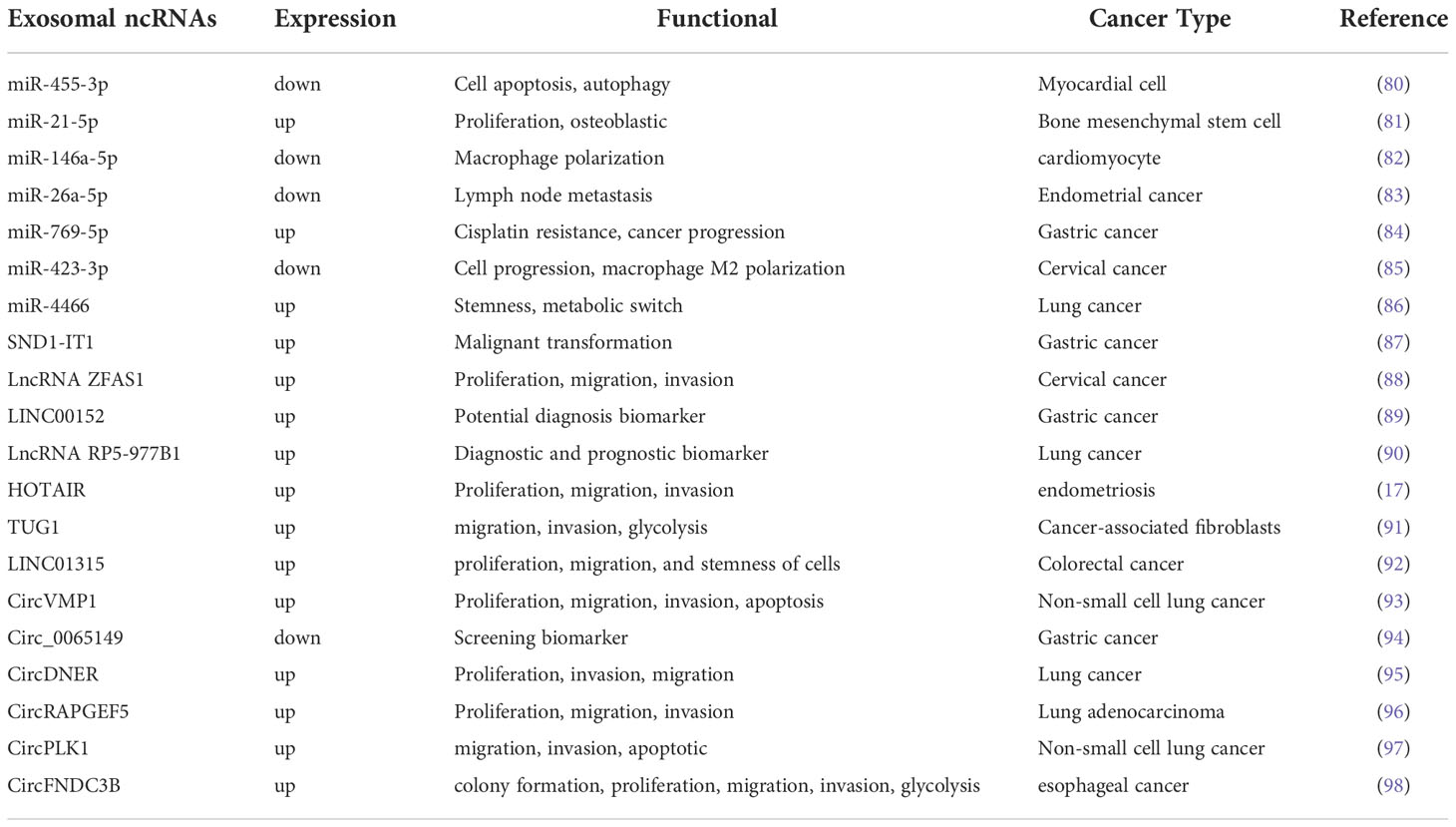
Exosomes are bilayer membrane vesicles of size 30-150nm, which are secreted to the extracellular uptake of recipient cells after serosal fusion, and play an important role in intercellular communication. Exosomes provide the recipient cells with multiple biomolecules, including lipids, proteins, DNA, and ncRNAs, mediating cell activation, phenotypic modification, and reprogramming of cellular functions. Working as a novel mode of intercellular communication, we summarize some of the mechanisms of action of exosome-carrying ncRNAs in tumors (Figure 2).
4.1 Exosomal microRNA and cancer
In patients with ovarian cancer, tumor cells produce exosomes, which are particularly abundant in malignant exudates, such as ascites (62). Exosomal miRNAs can be secreted in various body fluids, such as serum, plasma, urine, and peritoneal fluid, thereby avoiding ribonuclease degradation (63). Exosomal miRNAs in the circulating blood have been reported in most tumors for early cancer screening, disease course monitoring, and outcome assessment. Serum miR-100 levels were significantly lower in glioma patients than in healthy controls. However, they were significantly higher after treatment. Multivariate analysis was performed to identify serum miR-100 as an independent prognostic indicator in glioma patients (64).
Moreover, in colorectal cancer, miR-182 and miR-20a were up-regulated in tissues and plasma compared to those in healthy individuals. The area under the receiver operating characteristic (ROC) for circulating miR-182 at the validation stage was 0.891 (95% CI, 0.821–0.961), and the Area Under Curve (AUC) for circulating miR-20a expression was 0.801 (95% CI, 0.695–0.906) (65). Tumor-derived exosomes contribute to the immune escape of tumor cells by negatively regulating effector T and NK cells (66). To investigate how exosomal miRNAs derived from pancreatic cancer suppress the mRNA expression in dendritic cells, Ding et al. (67) found that their derived exosomes induced immune tolerance in dendritic cells by inhibiting the expression of the target gene RFXAP via miR-212-3p, through exosome stimulation of dendritic cells in pancreatic cancer. Whether exosomes can carry pre-miRNA-encoded peptides to play a role in tumor development and how they achieve this requires further exploration.
4.2 Exosomal LncRNAs and cancer
The bilayer membrane structure of exosomes effectively prevents their contents from being removed or experiencing RNA damage or degradation, further extending the circulating half-life of lncRNAs and enhancing their biological activity (68). Current research indicates that lncRNAs are closely associated with tumor cell invasion, metastasis, and immune system regulation (69). The process of exosome secretion is a multi-step process. Yang et al. (70) found that genes associated with exosome secretion were enriched in the HOTAIR high expression group. HOTAIR controls the docking process by regulating the expression and localization of RAB35. Further, HOTAIR facilitated fusion to complete the final step of exosome secretion by influencing the co-localization of VAMP3 and SNAP23. Moreover, several studies have suggested that exosomal lncRNAs can modulate chemoresistance. The results obtained by Wang et al. (71) showed that HOTTIP is highly expressed in cisplatin-resistant gastric cancer cells, and further studies revealed that exosomal HOTTIP promotes cisplatin resistance by activating HMGA1 in gastric cancer cells. Considering that tumor cells use various intercellular communication mechanisms to adapt to the local microenvironment, manipulate the immune system, and promote metastasis, the involvement of peptide/proteins in intercellular communication between tumor and stromal cells in the local or distant microenvironment via the exosomal pathway should be investigated in depth.
4.3 Exosomal circRNA and cancer
CircRNAs are endogenous RNAs that are structurally stable and conserved in sequence, with no 5’-end cap or 3’-end poly(A) tail (72). CircRNAs are more consistently present in cells, body fluids, and tissues than linear RNA (73). Memczak et al. (74) first proposed that circRNAs could compete with other RNAs for miRNA binding and play a role in post-transcriptional processes, laying the foundation for circRNAs in studying biological functions. Certain circRNAs are highly expressed at a predominant concentration at one locus, suggesting a tissue-specific expression pattern of circRNA (75). CircRNAs are enriched in exosomes and exhibit good stability (76). High-throughput sequencing analysis of exosomal RNA revealed different species of circRNAs in exosomes and cell plasma, indicating that circRNAs are selective in their entry into exosomes (77). However, the mechanism through which circRNAs enter exosomes remains unclear. Exosomal circRNAs may be essential mediators of communication between tumor cells and their microenvironment and play an important role in tumor development.
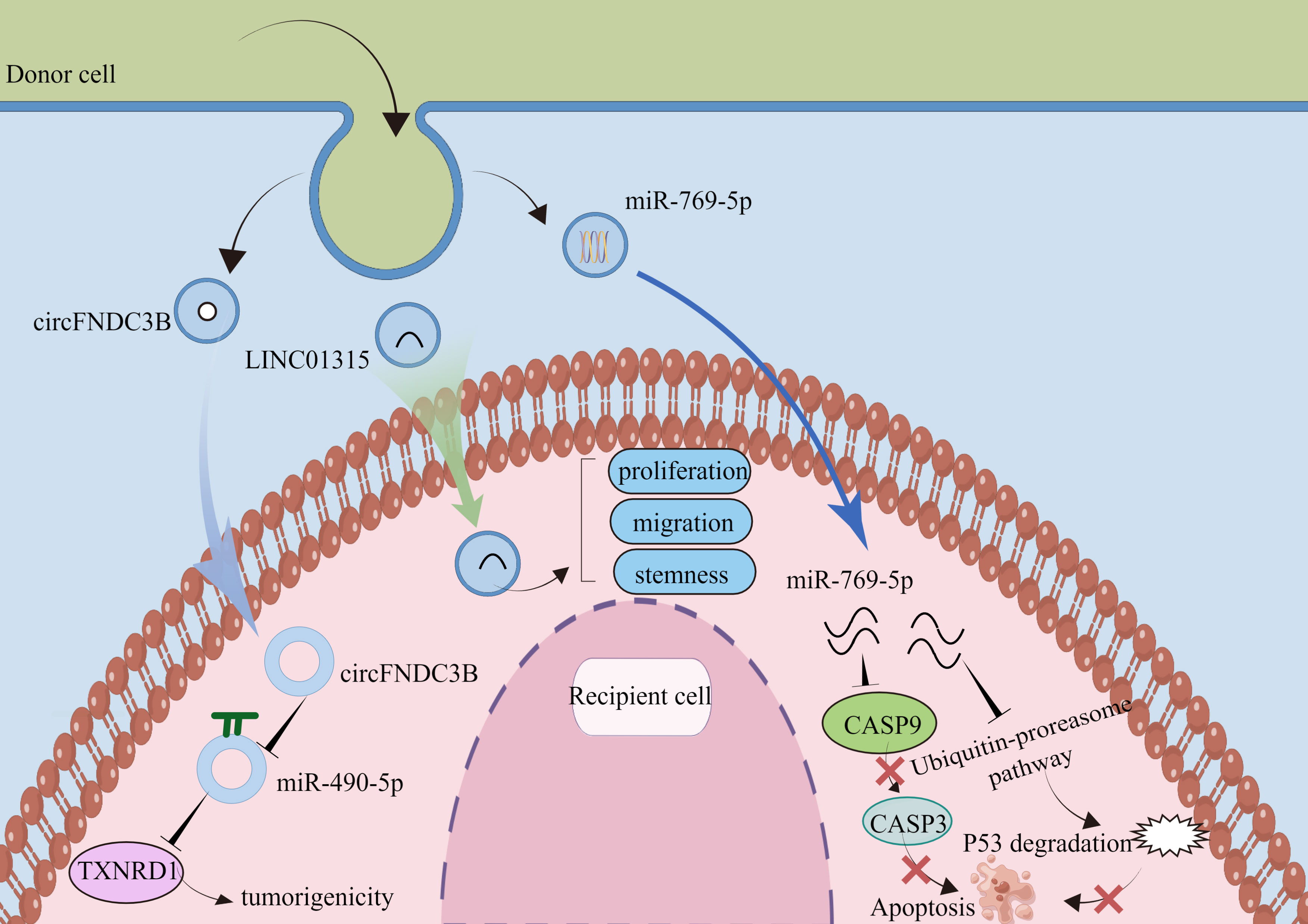
In non-small cell lung cancer (NSCLC), circ-MEMO1 is highly expressed in serum exosomes. Circ-MEMO1 promotes proliferation, cell cycle progression, and glycolysis in NSCLC cells by regulating the miR-101-3p/KRAS axis. High circ-MEMO1 expression predicts poor prognosis for patients with NSCLC. Additionally, it may be a valuable diagnostic marker for NSCLC (78). At the same time, circ-0051443 is expressed at low levels in the tissue and plasma exosomes of patients with hepatocellular carcinoma. Circ-0051443 is transported from normal cells to hepatocellular carcinoma cells by exosomes and inhibits the malignant biological behavior of hepatocellular carcinoma cells by affecting the cell cycle and apoptosis (79). Although a growing number of studies have shown the potential of circRNAs as tumor targets or diagnostic markers, further research is needed on the clinical applications of circRNAs. Exosomal circFNDC3B drove esophageal squamous cell carcinoma progression by miR-490-5p/TXNRD1 axis. Exosomal LINC01315 promotes proliferation, migration, and stemness. Exosomal miR-769-5p by the ubiquitin-proteasome pathway promotes degrading protein p53 (Table 3).
5 Relationship between peptide encoding by ncRNAs and exosome
Although the functions of peptide or proteins have been elucidated in liver, breast, and colorectal cancers, their mode of action remains incompletely understood. In light of recent studies suggesting that exosomes represent a new mode of intercellular communication, it is possible that peptide/proteins together with exosomes are involved in intercellular communication in the local or distant microenvironment. Cai et al. (99) identified 84 peptides in glioma cells and 29 peptides in their exosomes by improving the workflow and criteria for peptide-protein analysis, nine of which were present in both cells and exosomes. These results suggested that the release of specific peptides in or with exosomes may be actively regulated. Notably, tumor cells secrete exosomes that act as a bridge between tumor cells and their microenvironment. Thus, the derived exosomal peptides have the potential to regulate receptor cell function. In addition, exosomes contain a lipid bilayer membrane structure that prevents degradation of the nucleic acids and proteins they encapsulate, while their small size and membrane structure facilitate their uptake by tumor cells. The exosome peptides will therefore have greater function and integrity and great potential as diagnostic and prognostic markers (100).
6 Conclusion
This review summarizes many studies in which ncRNAs can encode peptides and proteins, in which ncRNAs carried by exosomes can act as biomarkers in tumors. Most importantly, the peptides encoded by ncRNAs in exosomes can serve as potential diagnostic markers, also as targets for therapeutic oncology agents. In the past time, peptides have been widely used in medicine. Currently, there are many peptide drugs in preclinical and clinical trials. Peptide drugs have the characteristics of high efficiency, safety and more tolerance, Furthermore, have the advantages of high selectivity and not easy to accumulate in the body. Although the current development of peptide drugs still faces great challenges, we believe that with the development of technology, the shortcomings of peptide drugs will eventually be overcome and the application scope of polypeptide drugs will be expanded. The limitations of this research field should to be point. Our current technology still cannot overcome the discovery of peptides such as low molecular weight and low abundance. Other ncRNAs such as snRNAs, tRNAs and rRNAs Is it possible to encode a new peptide/protein. What function ncRNAs binds to the ribosome but without an ORF remains unclear.
However, in future, several questions remain our to address. For example, what are the molecular mechanisms by which exosome peptides act, and how can the discovered molecular markers be translated into clinical use? Nevertheless, peptides in exosomes have the potential to play a key role in the diagnosis and treatment of tumors.
Author contributions
Conceptualization, JY, ML and XF. Investigation, HZ and QR. Data curation, YZ. Writing—original draft preparation, JY. Writing—review and editing, YW and YNZ. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Fundamental Research Funds for the Central Universities, China (lzujbky-2021-ct17), the Foundation of The First Hospital of Lanzhou University, China (ldyyyn2019-74).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1081997/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Rajagopalan D, Jha S. An epi(c) genetic war: Pathogens, cancer and human genome. Biochim Biophys Acta Rev Cancer (2018) 1869:333–45. doi: 10.1016/j.bbcan.2018.04.003
3. Costa FF. Non-coding RNAs: could they be the answer? Brief Funct Genomics (2011) 10:316–9. doi: 10.1093/bfgp/elq029
4. Ashar FN, Mitchell RN, Albert CM, Newton-Cheh C, Brody JA, Müller-Nurasyid M, et al. A comprehensive evaluation of the genetic architecture of sudden cardiac arrest. Eur Heart J (2018) 39:3961–9. doi: 10.1093/eurheartj/ehy474
5. Rahimian N, Razavi ZS, Aslanbeigi F, Mirkhabbaz AM, Piroozmand H, Shahrzad MK, et al. Non-coding RNAs related to angiogenesis in gynecological cancer. Gynecol Oncol (2021) 161:896–912. doi: 10.1016/j.ygyno.2021.03.020
6. Zhang C, Liu N. Noncoding RNAs in the glycolysis of ovarian cancer. Front Pharmacol (2022) 13:855488. doi: 10.3389/fphar.2022.855488
7. Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol (2007) 14:103–5. doi: 10.1038/nsmb0207-103
8. Song H, Tian D, Sun J, Mao X, Kong W, Xu D, et al. circFAM120B functions as a tumor suppressor in esophageal squamous cell carcinoma via the miR-661/PPM1L axis and the PKR/p38 MAPK/EMT pathway. Cell Death Dis (2022) 13:361. doi: 10.1038/s41419-022-04818-5
9. Wang X, Yu X, Xu H, Wei K, Wang S, Wang Y, et al. Serum-derived extracellular vesicles facilitate temozolomide resistance in glioblastoma through a HOTAIR-dependent mechanism. Cell Death Dis (2022) 13:344. doi: 10.1038/s41419-022-04699-8
10. Yang W, Zhou J, Zhang Z, Zhang K, Xu Y, Li L, et al. Downregulation of lncRNA APCDD1L-AS1 due to DNA hypermethylation and loss of VHL protein expression promotes the progression of clear cell renal cell carcinoma. Int J Biol Sci (2022) 18:2583–96. doi: 10.7150/ijbs.71519
11. Li S, Li L, Li J, Liang X, Song C, Zou Y. miR-203, fine-tunning neuroinflammation by juggling different components of NF-κB signaling. J Neuroinflamm (2022) 19:84. doi: 10.1186/s12974-022-02451-9
12. Pang Y, Liu Z, Han H, Wang B, Li W, Mao C, et al. Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. J Hepatol (2020) 73:1155–69. doi: 10.1016/j.jhep.2020.05.028
13. Guo B, Wu S, Zhu X, Zhang L, Deng J, Li F, et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J (2020) 39:e102190. doi: 10.15252/embj.2019102190
14. Meng N, Chen M, Chen D, Chen XH, Wang JZ, Zhu S, et al. Small protein hidden in lncRNA LOC90024 promotes "Cancerous" RNA splicing and tumorigenesis. Adv Sci (Weinh) (2020) 7:1903233. doi: 10.1002/advs.201903233
15. Ye D, Gong M, Deng Y, Fang S, Cao Y, Xiang Y, et al. Roles and clinical application of exosomal circRNAs in the diagnosis and treatment of malignant tumors. J Transl Med (2022) 20:161. doi: 10.1186/s12967-022-03367-x
16. Zhang F, Isak AN, Yang S, Song Y, Ren L, Feng C, et al. Smartly responsive DNA-miRNA hybrids packaged in exosomes for synergistic enhancement of cancer cell apoptosis. Nanoscale (2022) 14:6612–19. doi: 10.1039/D1NR08539E
17. Zhang L, Yu Z, Qu Q, Li X, Lu X, Zhang H. Exosomal lncRNA HOTAIR promotes the progression and angiogenesis of endometriosis via the miR-761/HDAC1 axis and activation of STAT3-mediated inflammation. Int J Nanomedicine (2022) 17:1155–70. doi: 10.2147/IJN.S354314
18. Malone B, Atanassov I, Aeschimann F, Li X, Großhans H, Dieterich C. Bayesian Prediction of RNA translation from ribosome profiling. Nucleic Acids Res (2017) 45:2960–72. doi: 10.1093/nar/gkw1350
19. Mackowiak SD, Zauber H, Bielow C, Thiel D, Kutz K, Calviello L, et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol (2015) 16:179. doi: 10.1186/s13059-015-0742-x
20. Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of noncoding RNAs and cancer. Cancer Lett (2021) 497:89–99. doi: 10.1016/j.canlet.2020.10.002
21. Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res (2017) 45:W12–w16. doi: 10.1093/nar/gkx428
22. Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH, et al. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics (2010) 26:399–400. doi: 10.1093/bioinformatics/btp688
23. Wang G, Yin H, Li B, Yu C, Wang F, Xu X, et al. Characterization and identification of long non-coding RNAs based on feature relationship. Bioinformatics (2019) 35:2949–56. doi: 10.1093/bioinformatics/btz008
24. Fields AP, Rodriguez EH, Jovanovic M, Ginossar N, Haas BJ, Mertins P, et al. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell (2015) 60:816–27. doi: 10.1016/j.molcel.2015.11.013
25. Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W, et al. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res (2013) 41:e74. doi: 10.1093/nar/gkt006
26. Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J (2014) 33:981–93. doi: 10.1002/embj.201488411
27. Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics (2011) 27:i275–82. doi: 10.1093/bioinformatics/btr209
28. Li A, Zhang J, Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinf (2014) 15:311. doi: 10.1186/1471-2105-15-311
29. Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res (2013) 41:e166. doi: 10.1093/nar/gkt646
30. Chen X, Han P, Zhou T, Guo X, Song X, Li Y, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep (2016) 6:34985. doi: 10.1038/srep34985
31. Zhao J, Wu J, Xu T, Yang Q, He J, Song X, et al. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J Genet Genomics (2018) 45:403–6. doi: 10.1016/j.jgg.2018.07.006
32. Sun P, Li G. CircCode: A powerful tool for identifying circRNA coding ability. Front Genet (2019) 10:981. doi: 10.3389/fgene.2019.00981
33. Mokrejs M, Masek T, Vopálensky V, Hlubucek P, Delbos P, Pospísek M, et al. IRESite–a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res (2010) 38:D131–6. doi: 10.1093/nar/gkp981
34. Mokrejs M, Vopálenský V, Kolenaty O, Masek T, Feketová Z, Sekyrová P, et al. IRESite: the database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res (2006) 34:D125–30. doi: 10.1093/nar/gkj081
35. Wu W, Ji P, Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol (2020) 21:101. doi: 10.1186/s13059-020-02018-y
36. Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Bécard G, et al. Primary transcripts of microRNAs encode regulatory peptides. Nature (2015) 520:90–3. doi: 10.1038/nature14346
37. Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell (2009) 136:669–87. doi: 10.1016/j.cell.2009.01.046
38. Yadav A, Sanyal I, Rai SP, Lata C. An overview on miRNA-encoded peptides in plant biology research. Genomics (2021) 113:2385–91. doi: 10.1016/j.ygeno.2021.05.013
39. Zhou H, Lou F, Bai J, Sun Y, Cai W, Sun L, et al. A peptide encoded by pri-miRNA-31 represses autoimmunity by promoting t(reg) differentiation. EMBO Rep (2022) 23:e53475. doi: 10.15252/embr.202153475
40. Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature (2002) 420:563–73. doi: 10.1038/nature01266
41. Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet (2012) 8:e1003078. doi: 10.1371/journal.pgen.1003078
42. Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, et al. Long noncoding RNAs with snoRNA ends. Mol Cell (2012) 48:219–30. doi: 10.1016/j.molcel.2012.07.033
43. Prabakaran S, Hemberg M, Chauhan R, Winter D, Tweedie-Cullen RY, Dittrich C, et al. Quantitative profiling of peptides from RNAs classified as noncoding. Nat Commun (2014) 5:5429. doi: 10.1038/ncomms6429
44. Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004
45. Borden KLB, Volpon L. The diversity, plasticity, and adaptability of cap-dependent translation initiation and the associated machinery. RNA Biol (2020) 17:1239–51. doi: 10.1080/15476286.2020.1766179
46. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst (2018) 110:304–15. doi: 10.1093/jnci/djx166
47. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via hippo-YAP signaling. Mol Cancer (2019) 18:47. doi: 10.1186/s12943-019-1010-6
48. Kang M, Tang B, Li J, Zhou Z, Liu K, Wang R, et al. Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol Cancer (2020) 19:143. doi: 10.1186/s12943-020-01248-9
49. Tang C, Zhou Y, Sun W, Hu H, Liu Y, Chen L, et al. Oncopeptide MBOP encoded by LINC01234 promotes colorectal cancer through MAPK signaling pathway. Cancers (Basel) (2022) 14:2338. doi: 10.3390/cancers14092338
50. Spencer HL, Sanders R, Boulberdaa M, Meloni M, Cochrane A, Spiroski AM, et al. The LINC00961 transcript and its encoded micropeptide, small regulatory polypeptide of amino acid response, regulate endothelial cell function. Cardiovasc Res (2020) 116:1981–94. doi: 10.1093/cvr/cvaa008
51. Wang Y, Wu S, Zhu X, Zhang L, Deng J, Li F, et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med (2020) 217:jem.20190950. doi: 10.1084/jem.20190950
52. Huang JZ, Chen M, Chen D, Zao XC, Zhu S, Huang H, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell (2017) 68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015
53. Zhu S, Wang JZ, Chen D, He YT, Meng N, Chen M, et al. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun (2020) 11:1685. doi: 10.1038/s41467-020-15403-9
54. Ge Q, Jia D, Cen D, Qi Y, Shi C, Li J, et al. Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J Clin Invest (2021) 131: e152911. doi: 10.1172/JCI152911
55. Zhang C, Zhou B, Gu F, Liu H, Wu H, Yao F, et al. Micropeptide PACMP inhibition elicits synthetic lethal effects by decreasing CtIP and poly(ADP-ribosyl)ation. Mol Cell (2022) 82:1297–312.e8. doi: 10.1016/j.molcel.2022.01.020
56. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9:4475. doi: 10.1038/s41467-018-06862-2
57. Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang H, et al. Circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the wnt/beta-catenin pathway. Mol Ther (2022) 30:415–30. doi: 10.1016/j.ymthe.2021.08.026
58. Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent kinase-1. Mol Cancer (2019) 18:131. doi: 10.1186/s12943-019-1056-5
59. Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S, et al. circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol Ther Nucleic Acids (2018) 13:633–41. doi: 10.1016/j.omtn.2018.10.008
60. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the wnt pathway. Genome Biol (2019) 20:84. doi: 10.1186/s13059-019-1685-4
61. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene (2018) 37:1805–14. doi: 10.1038/s41388-017-0019-9
62. Navabi H, Croston D, Hobot J, Clayton A, Zitvogel L, Jasani B, et al. Preparation of human ovarian cancer ascites-derived exosomes for a clinical trial. Blood Cells Mol Dis (2005) 35:149–52. doi: 10.1016/j.bcmd.2005.06.008
63. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One (2008) 3:e3148. doi: 10.1371/journal.pone.0003148
64. Zhang H, Wang J, Wang Z, Ruan C, Wang L, Guo H, et al. Serum miR-100 is a potential biomarker for detection and outcome prediction of glioblastoma patients. Cancer biomark (2019) 24:43–9. doi: 10.3233/CBM-181416
65. Liu X, Xu T, Hu X, Chen X, Zeng K, Sun L, et al. Elevated circulating miR-182 acts as a diagnostic biomarker for early colorectal cancer. Cancer Manag Res (2018) 10:857–65. doi: 10.2147/CMAR.S158016
66. Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol (2006) 176:1375–85. doi: 10.4049/jimmunol.176.3.1375
67. Ding G, Zhou L, Qian Y, Fu M, Chen J, Chen J, et al. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget (2015) 6:29877–88. doi: 10.18632/oncotarget.4924
68. Huang Y, Luo Y, Ou W, Wang Y, Dong D, Peng X, et al. Exosomal lncRNA SNHG10 derived from colorectal cancer cells suppresses natural killer cell cytotoxicity by upregulating INHBC. Cancer Cell Int (2021) 21:528. doi: 10.1186/s12935-021-02221-2
69. Yarani R, Mirza AH, Kaur S, Pociot F. The emerging role of lncRNAs in inflammatory bowel disease. Exp Mol Med (2018) 50:1–14. doi: 10.1038/s12276-018-0188-9
70. Yang L, Peng X, Li Y, Zhang X, Ma Y, Wu C, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer (2019) 18:78. doi: 10.1186/s12943-019-0990-6
71. Wang J, Lv B, Su Y, Wang X, Bu J, Yao L, et al. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther (2019) 12:11325–38. doi: 10.2147/OTT.S231846
72. Han Z, Chen H, Guo Z, Shen J, Luo W, Xie F, et al. Circular RNAs and their role in exosomes. Front Oncol (2022) 12:848341. doi: 10.3389/fonc.2022.848341
73. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther (2018) 187:31–44. doi: 10.1016/j.pharmthera.2018.01.010
74. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
75. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
76. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol (2013) 200:373–83. doi: 10.1083/jcb.201211138
77. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res (2015) 25:981–4. doi: 10.1038/cr.2015.82
78. Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, et al. Exosomal circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS axis. Front Genet (2020) 11:962. doi: 10.3389/fgene.2020.00962
79. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
80. Wang Y, Shen Y. Exosomal miR-455-3p from BMMSCs prevents cardiac ischemia-reperfusion injury. Hum Exp Toxicol (2022) 41:9603271221102508. doi: 10.1177/09603271221102508
81. You M, Ai Z, Zeng J, Fu Y, Zhang L, Wu X, et al. Bone mesenchymal stem cells (BMSCs)-derived exosomal microRNA-21-5p regulates kruppel-like factor 3 (KLF3) to promote osteoblast proliferation in vitro. Bioengineered (2022) 13:11933–44. doi: 10.1080/21655979.2022.2067286
82. Chen C, Cai S, Wu M, Wang R, Liu M, Cao G, et al. Role of cardiomyocyte-derived exosomal MicroRNA-146a-5p in macrophage polarization and activation. Dis Markers (2022) 2022:2948578. doi: 10.1155/2022/2948578
83. Wang J, Gong X, Yang L, Li L, Gao X, Ni T, et al. Loss of exosomal miR-26a-5p contributes to endometrial cancer lymphangiogenesis and lymphatic metastasis. Clin Transl Med (2022) 12:e846. doi: 10.1002/ctm2.846
84. Jing X, Xie M, Ding K, Xu T, Fang Y, Ma P, et al. Exosome-transmitted miR-769-5p confers cisplatin resistance and progression in gastric cancer by targeting CASP9 and promoting the ubiquitination degradation of p53. Clin Transl Med (2022) 12:e780. doi: 10.1002/ctm2.780
85. Yan X, Zhang S, Jia J, Yang J, Song Y, Duan H, et al. Exosomal MiR-423-3p inhibits macrophage M2 polarization to suppress the malignant progression of cervical cancer. Pathol Res Pract (2022) 235:153882. doi: 10.1016/j.prp.2022.153882
86. Tyagi A, Wu SY, Sharma S, Wu K, Zhao D, Deshpande R, et al. Exosomal miR-4466 from nicotine-activated neutrophils promotes tumor cell stemness and metabolism in lung cancer metastasis. Oncogene (2022) 41:3079–92. doi: 10.1038/s41388-022-02322-w
87. Jin G, Zhang J, Cao T, Chen B, Tian Y, Shi Y, et al. Exosome-mediated lncRNA SND1-IT1 from gastric cancer cells enhances malignant transformation of gastric mucosa cells via up-regulating SNAIL1. J Transl Med (2022) 20:284. doi: 10.1186/s12967-022-03306-w
88. Yang Z, Ma J, Han S, Li X, Guo H, Liu D, et al. ZFAS1 exerts an oncogenic role via suppressing miR-647 in an m(6)A-dependent manner in cervical cancer. Onco Targets Ther (2020) 13:11795–806. doi: 10.2147/OTT.S274492
89. Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol (2015) 36:2007–12. doi: 10.1007/s13277-014-2807-y
90. Min L, Zhu T, Lv B, An T, Zhang Q, Shang Y, et al. Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int J Clin Oncol (2022) 27:1013–24. doi: 10.1007/s10147-022-02129-5
91. Lu L, Huang J, Mo J, Da X, Li Q, Fan M, et al. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett (2022) 27:17. doi: 10.1186/s11658-022-00309-9
92. Li Y, Wu M, Xu S, Huang H, Yan L, Gu Y, et al. Colorectal cancer stem cell-derived exosomal long intergenic noncoding RNA 01315 (LINC01315) promotes proliferation, migration, and stemness of colorectal cancer cells. Bioengineered (2022) 13:10827–42. doi: 10.1080/21655979.2022.2065800
93. Xie H, Yao J, Wang Y, Ni B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Delivery (2022) 29:1257–71. doi: 10.1080/10717544.2022.2057617
94. Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res (2020) 26:1475–82. doi: 10.1007/s12253-019-00716-y
95. Li J, Zhu T, Weng Y, Cheng F, Sun Q, Yang K, et al. Exosomal circDNER enhances paclitaxel resistance and tumorigenicity of lung cancer via targeting miR-139-5p/ITGB8. Thorac Cancer (2022) 13:1381–90. doi: 10.1111/1759-7714.14402
96. Zhou H, Huang X, Yang X, Jiang F, Shao F, Shi W, et al. CircRAPGEF5 promotes the proliferation and metastasis of lung adenocarcinoma through the miR-1236-3p/ZEB1 axis and serves as a potential biomarker. Int J Biol Sci (2022) 18:2116–31. doi: 10.7150/ijbs.66770
97. Li C, Wang G, Ma X, Tao T, Li Q, Yang Y, Sang H, et al. Upregulation of exosomal circPLK1 promotes the development of non-small cell lung cancer through the miR-1294/ high mobility group protein A1 axis. Bioengineered (2022) 13:4185–4200. doi: 10.1080/21655979.2022.2026727
98. Tang B, Zhang Q, Liu K, Huang Y. Exosomal circRNA FNDC3B promotes the progression of esophageal squamous cell carcinoma by sponging miR-490-5p and regulating thioredoxin reductase 1 expression. Bioengineered (2022) 13:13829–13848. doi: 10.1080/21655979.2022.2084484
99. Cai T, Zhang Q, Wu B, Wang J, Li N, Zhang T, et al. LncRNA-encoded microproteins: A new form of cargo in cell culture-derived and circulating extracellular vesicles. J Extracell Vesicles (2021) 10:e12123. doi: 10.1002/jev2.12123
Keywords: non-coding RNA, exosome, peptide, translation, cancer
Citation: Yang J, Liu M, Fang X, Zhang H, Ren Q, Zheng Y, Wang Y and Zhou Y (2022) Advances in peptides encoded by non-coding RNAs: A cargo in exosome. Front. Oncol. 12:1081997. doi: 10.3389/fonc.2022.1081997
Received: 27 October 2022; Accepted: 28 November 2022;
Published: 23 December 2022.
Edited by:
Christos K. Kontos, National and Kapodistrian University of Athens, GreeceReviewed by:
Pinelopi Artemaki, National and Kapodistrian University of Athens, GreeceHanchu Xiong, Zhejiang Provincial People’s Hospital, China
Chao Zhang, Lilly Research Laboratories, United States
Copyright © 2022 Yang, Liu, Fang, Zhang, Ren, Zheng, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongning Zhou, emhvdXluQGx6dS5lZHUuY24=; Yuping Wang, d2FuZ3l1cGluZ0BsenUuZWR1LmNu
†These authors have contributed equally to this work
 Jing Yang1,2,3†
Jing Yang1,2,3† Huiyun Zhang
Huiyun Zhang Ya Zheng
Ya Zheng Yuping Wang
Yuping Wang Yongning Zhou
Yongning Zhou