- 1Department of Hepatobiliary Disease, Dongfang Hospital, School of Medicine, Xiamen University, Fuzhou, China
- 2Department of Gastroenterology, Liuzhou Workers’ Hospital (The Fourth Affiliated Hospital), Guangxi Medical University, Liuzhou, China
- 3Department of Gastroenterology, Fuzhou Second Hospital, Fuzhou, China
- 4Department of Transplantation, People’s Hospital of Guangxi Zhuang Autonomous Region, Guangxi, China
Background: Patients with hepatocellular carcinoma (HCC) have poor prognosis, especially in advanced stages. Targeted therapy is the main treatment for advanced HCC patients, but the optimal targets for HCC remain poorly understood. The main purpose of this study was to identify potential novel prognostic markers and therapeutic targets.
Methods: Firstly, differentially expressed genes (DEGs) in HCC were identified from the Gene Expression Omnibus (GEO) database. The expression, significance in prognosis, and potential mechanisms of DEGs were analyzed using GEPIA, TIMER, HPA, Kaplan Meier Plotter, CBioPortal, miRWalk, TargetScan, and ENCORI databases. Immunohistochemical staining was used to determine the protein expression levels of potential candidate genes.
Results: The mRNA levels of MND1, STXBP6, and CLGN were significantly increased in HCC (p< 0.01). HCC patients with elevated CLGN mRNA levels had poorer overall survival (OS), disease-free survival (DFS), progression-free survival (PFS), and disease-specific survival (DSS) (p < 0.05). Higher MND1 mRNA levels significantly correlated with poorer DFS in HCC patients (p< 0.05). However, there was no significant correlation between STXBP6 expression and prognosis of HCC (p> 0.05). Further analysis revealed that patients with elevated CLGN mRNA expression in advanced pathology stages had poorer prognosis (p< 0.01). In addition, CLGN protein levels were elevated in HCC compared to their levels in normal tissues. The mRNA levels of CLGN had no significant correlation with the abundance of six common tumor infiltrating lymphocytes in HCC (COR < 0.5). Moreover, the mutation rate of CLGN was less than 1% in HCC patients (10/1089). Finally, the expression level of hsa-miR-194-3p in HCC was significantly lower than that in normal tissues (p < 0.05), and prognosis of HCC with low expression of hsa-miR-194 was poor (p < 0.05).
Conclusion: The upregulation of CLGN in HCC is significantly associated with poor patient prognosis, especially in the advanced stages, and may be regulated by hsa-miR-194-3p. These findings suggest that CLGN may be closely related to the progression of HCC, and is a potential therapeutic target and prognostic indicator for patients with advanced HCC.
Introduction
Primary liver cancer remains the sixth most common cancer and the third leading cause of cancer-related deaths worldwide in 2020, according to the latest statistics (1). Hepatocellular carcinoma (HCC) encompasses 75% -85% of liver cancer cases (1). Most HCC is in advanced stages at the time of diagnosis and is resistant to conventional cytotoxic drugs. Currently, systemic therapy for patients with advanced HCC includes molecularly targeted agents, immune checkpoint inhibitors, or a combination of both (2–4). However, a substantial proportion of patients have yet to benefit from systemic therapy (5, 6). Recent studies have identified several novel biomarkers that can predict HCC prognosis. Lu et al. showed that serum α-fetoprotein trajectories were associated with overall survival of patients with intermediate-stage HCC after transarterial chemoembolization (7). However, it is imperative to identify the most effective predictive biomarkers to facilitate individualized and targeted treatments. Therefore, it is necessary to screen more biomarkers to improve patient diagnosis, and prognosis and identify therapeutic targets for precision therapy.
In recent years, bioinformatic databases have provided abundant transcriptomic and clinical data (8, 9), which are convenient for the analysis of relevant markers with potential significance (10–12). In this study, we explored several databases for molecular markers of HCC and investigated their expression levels in clinical specimens.
Methods
Screening of DEGs in HCC patients
Tumor and normal tissue Gene expression datasets GSE54236 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54236) (13) and GSE121248 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121248) were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) (14). D ifferentially expressed genes (DEGs) were analyzed using the online tool GEO2R. Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) (15) and Tumor Immune Estimation Resource database (TIMER, https://cistrome.shinyapps.io/timer/) (16) were used to further verify the DEGs. A p-value of <0.05 was statistically significant.
The significance of DEGs in the prognosis of HCC
GEPIA was used to analyze the relationship between DEGs and overall survival (OS) and disease-free survival (DFS) in HCC patients. Kaplan Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq) (17) was used to verify the relationship between candidate gene expression levels and OS, progression-free survival (PFS), relapse-free survival (RFS), and disease-specific survival (DSS) in HCC patients. Then, Kaplan Meier Plotter was used to explore the relationship between candidate genes and OS in patients with different pathological stages. Patients were trichotomized (T1 vs. T3).
CLGN protein expression in HCC and normal tissues
Ethics Statement: The study was conducted in accordance with the principles of the Declaration of Helsinki (18). The collection of HCC specimens was approved by the Ethics Committee of Liuzhou Workers’ Hospital (No.KY2022051).
Immunohistochemical Staining: Samples from 24 HCC patients who underwent surgical resection at Liuzhou Workers’ Hospital were collected. The acquisition of specimens was approved by the Ethics Committee of Liuzhou Workers’ Hospital.
Five-micrometer sections were obtained from paraffin-embedded tumor and non-tumor specimens. All sections were dewaxed in xylene and rehydrated in alcohol, followed by wet autoclave pretreatment in citrate buffer for antigen retrieval (10 min at 120°C, pH=6.0) and then rinsed with phosphate buffer saline. Immunohistochemical staining with antibody to CLGN (Anti-Calmegin/CLGN Antibody, Rabbit: A05261-1, Boster Biological Technology Co. Ltd.) was performed using the avidin-biotin- peroxidase complex method. The primary antibody (1:200 dilution for CLGN) was applied to the sections and allowed to bind for 1 h at room temperature. The sections were then incubated with biotinylated anti-mouse/rabbit antibody for 30 min and an avidin-biotin-peroxidase reagent for 10 min. After color development with diaminobenzidine, the sections were counterstained with hematoxylin.
In addition, PATHOLOGY module in HPA (https://www.proteinatlas.org/) database was used to verify the expression of CLGN protein in HCC patient specimens and normal liver tissue.
Analysis of mutation and immune cell infiltration levels
The CBioportal database (https://www.cbioportal.org/) (19) was used to analyze the mutation of CLGN in HCC patients. The relationships between the expression levels of CLGN and six immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) in HCC were estimated using the TIMER algorithm. Statistical significance was set at p values of <0.05 and correlation coefficients (COR) of ≥ 0.5.
Screening of regulating microRNAs
MiRWalk (http://mirwalk.umm.uni-heidelberg.de/) (20) and TargetScan (http://www.targetscan.org/vert_72) (21) databases were used to predict microRNAs (miRNAs) that regulate CLGN mRNA expression. The Encyclopedia of RNA Interactomes (ENCORI, http://rna.sysu.edu.cn/encori/mirTarPathways.php) (22) database was used to analyze the expression levels of candidate miRNAs in HCC and normal tissues. Kaplan Meier Plotter was used to analyze the relationship between the levels of candidate miRNAs and prognosis of HCC. A p value of <0.05 was considered statistically significant.
Data processing and visualization
Gene expression data were cleaned and visualized using R (Version 3.6.3) and the R packages ggplot2 and VennDiagram. The cleaning algorithm for the gene expression data included removal of non-coding genes, miRNA, and blank probes; FDR ≤0.05 and FC ≥2; retaining one level from one gene that had several high or, consistently, low expression levels in different specimens. We removed genes that had both high expression and low expression in different specimens. Cytoscape (Version: 3.7.2) and cytoHubba plugin were used to analyze miRNAs with the potential to regulate CLGN mRNA expression.
Results
Significant DEGs in HCC
The DEG analysis revealed 290 genes with significantly high expression and 518 genes with significantly low expression in the GSE54236 (G1) dataset (Figures 1A, C). In the GSE121248 (G2) dataset, 319 genes were significantly upregulated, and 611 genes were significantly under-expressed (Figures 1B, C). There were 121 genes with high expression and 249 genes with low expression in both the datasets (Figure 1C). After removing the genes that have been reported, we found that the expression levels of MND1, STXBP6, and CLGN in HCC were significantly higher than those in normal tissues.
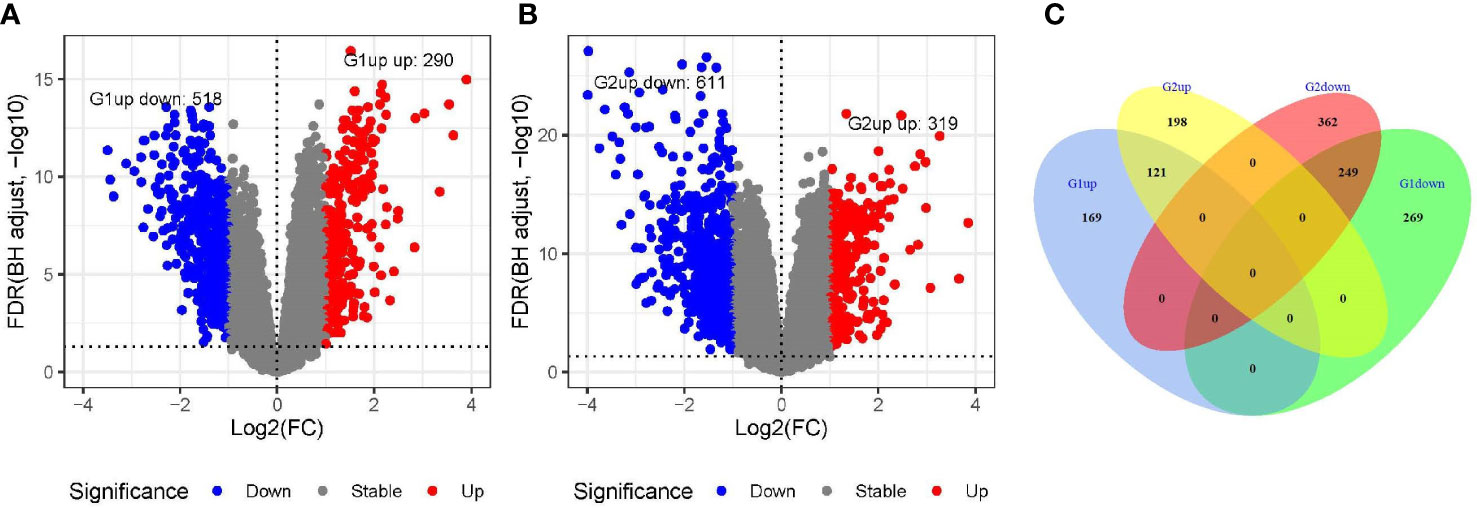
Figure 1 Screening of differentially expressed genes in GSE54236 (G1) and GSE121248 (G2) datasets. (A, B) Gene expression levels in GSE54236 (G1) and GSE121248 (G2) datasets, with blue dots indicating significantly low expression and red dots indicating significantly high expression. (C) The number of genes with high or low expression in both datasets.
The expression levels of MND1, STXBP6, and CLGN mRNA in HCC w ere verified. The results showed that the MND1, STXBP6, and CLGN mRNA levels in HCC were significantly higher than those in normal tissues (p < 0.01, Figures 2A-C). In addition, the mRNA expression of CLGN was high in breast invasive carcinoma, kidney chromophobe, kidney renal papillary cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma prostate adenocarcinoma, and uterine corpus endometrial carcinoma (p < 0.01, Figure 3).
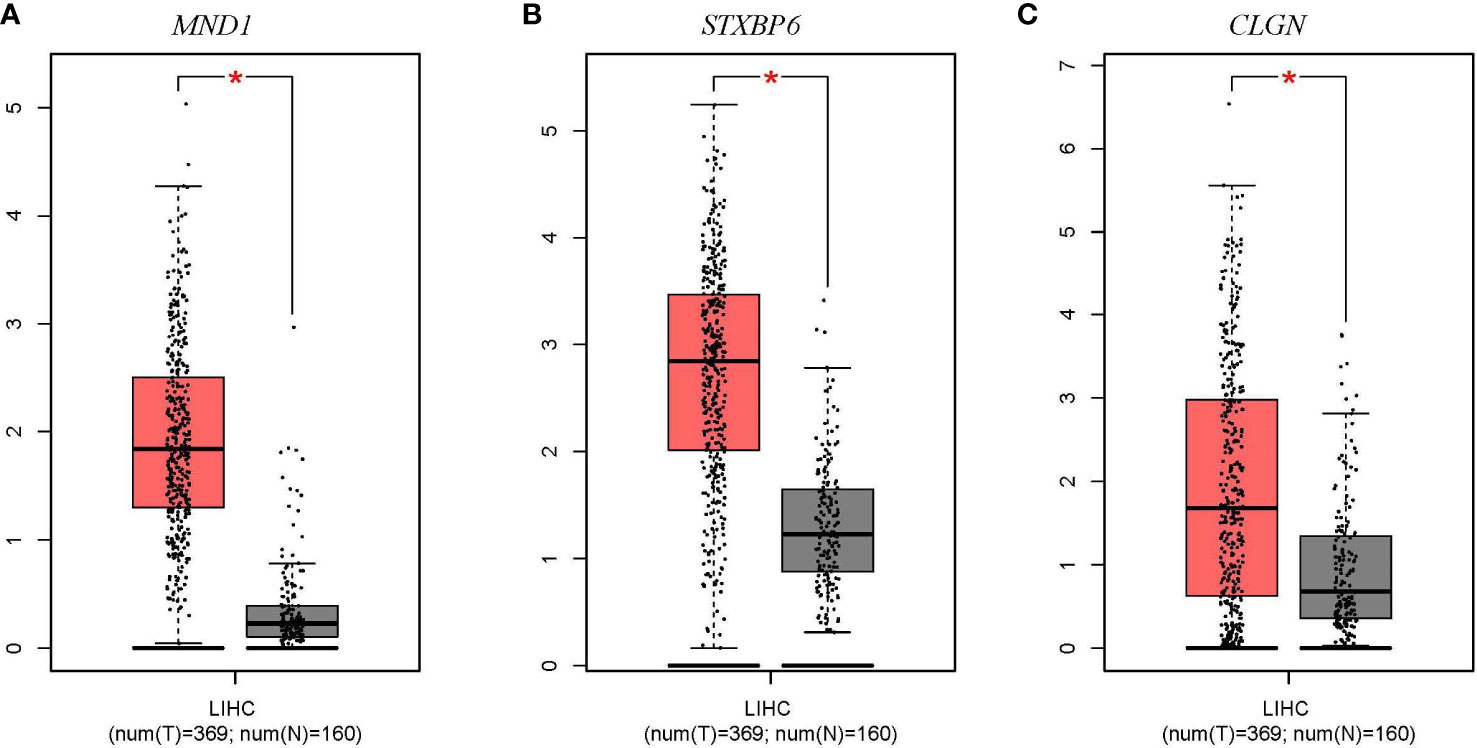
Figure 2 The mRNA expression levels of MND1, STXBP6, and CLGN in HCC (GEPIA). (A-C) The mRNA expression levels of MND1, STXBP6, and CLGN in HCC were significantly higher than in normal tissue. *p < 0.01.
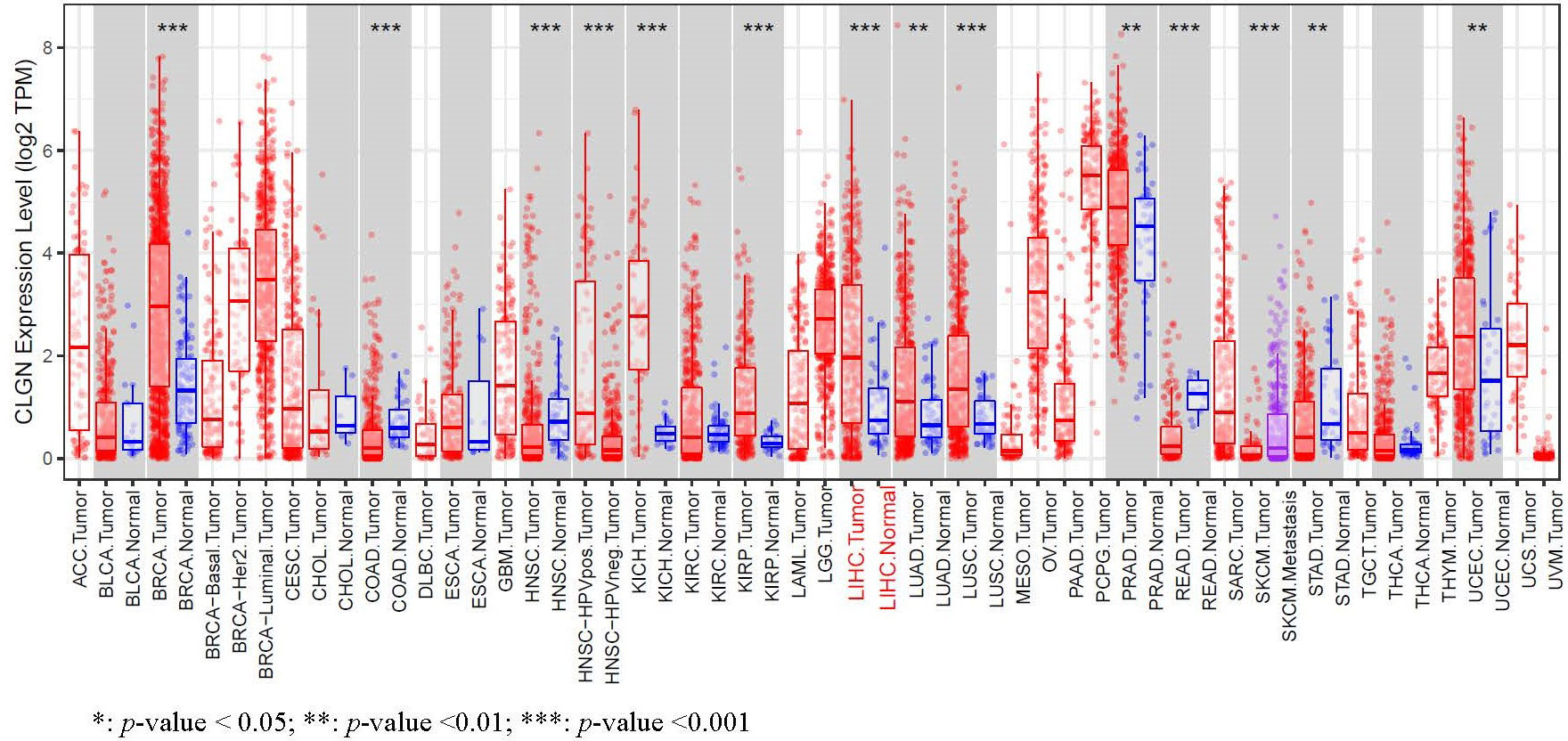
Figure 3 The mRNA expression of levels of CLGN in several common human tumors. The mRNA of CLGN was upregulated in liver and other tumors (TIMER).
Relationship between DEGs and prognosis of HCC
The prognostic value of MND1, STXBP6, and CLGN in HCC patients was explored. HCC patients with higher mRNA levels of CLGN had poorer OS and DFS (p< 0.05, Figures 4A, B). There was no significant correlation between MND1 mRNA levels and OS (p> 0.05; Figure 4C), and patients with higher MND1 mRNA levels had poorer DFS (p< 0.05, Figure 4D). However, there was no significant correlation between the mRNA levels of STXBP6 and OS or DFS in HCC patients (p > 0.05, Figures 4E, F).
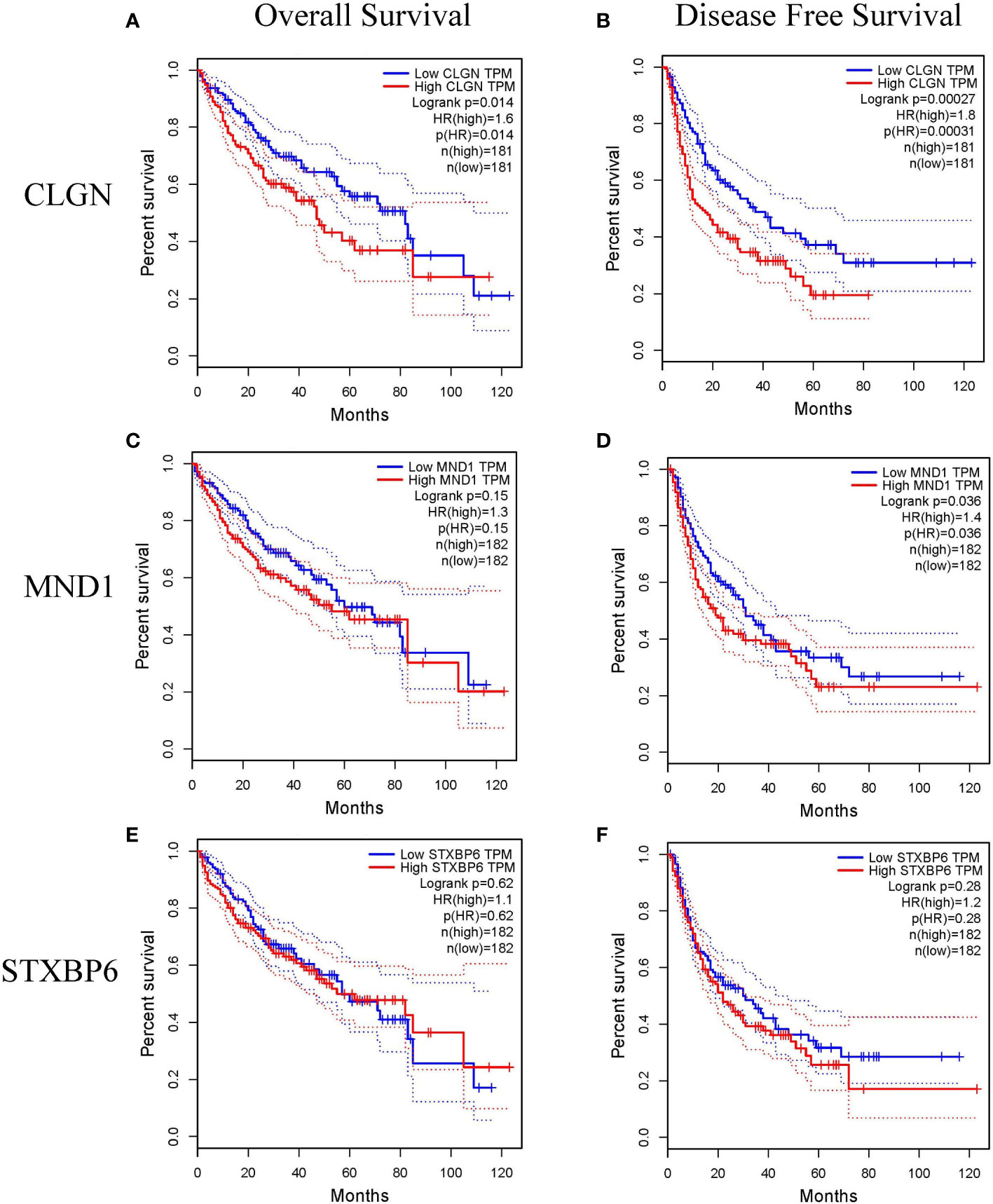
Figure 4 The relationship between the mRNA levels of MND1, STXBP6 and CLGN and the prognosis of HCC (GEPIA). (A, B) HCC patients with higher mRNA levels of CLGN had poorer overall survival and disease-free survival (p< 0.05). (C) No significant relationship between MND1 mRNA levels and patients’ overall survival was observed (p > 0.05). (D) Patients with higher MND1 mRNA levels had poorer disease-free survival (p < 0.05). (E, F) There was no significant correlation between the mRNA levels of STXBP6 and overall survival and disease-free survival in HCC patients (p > 0.05).
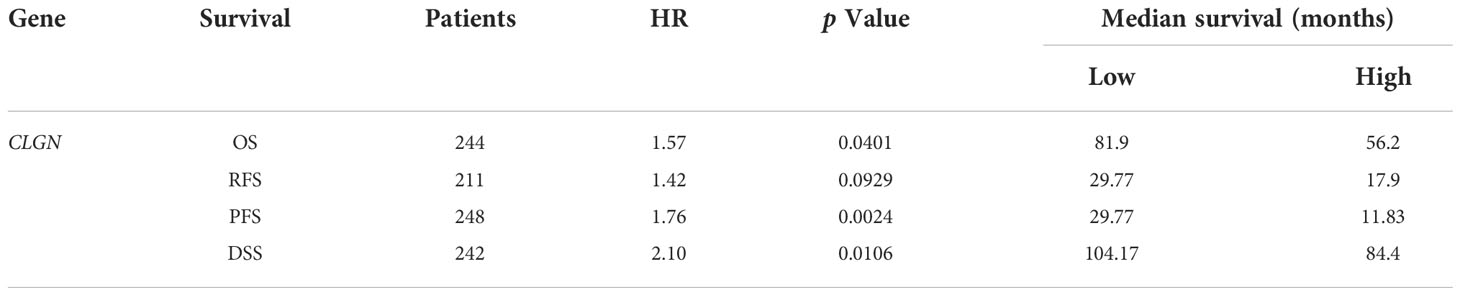
The relationship between CLGN mRNA levels and the prognosis of HCC was also investigated. The results showed that the mRNA levels of CLGN were significantly correlated with OS, PFS, and DSS in HCC patients (p< 0.05, Figures 5A-C and Table 1). However, there was no significant relationship between CLGN mRNA levels and RFS (p> 0.05, Figure 5D and Table 1). Furthermore, the mRNA level of CLGN was significantly correlated with the OS in different pathology stages in HCC patients. Specifically, HCC patients with higher mRNA expression of CLGN in advanced stages (T3, T3+T4) had poorer OS (p < 0.01, Figures 6C, D and Table 2). No significant correlation was found between CLGN mRNA levels and OS in patients with early-stage HCC (T1-T2) (p > 0.05, Figures 6A, B and Table 2).
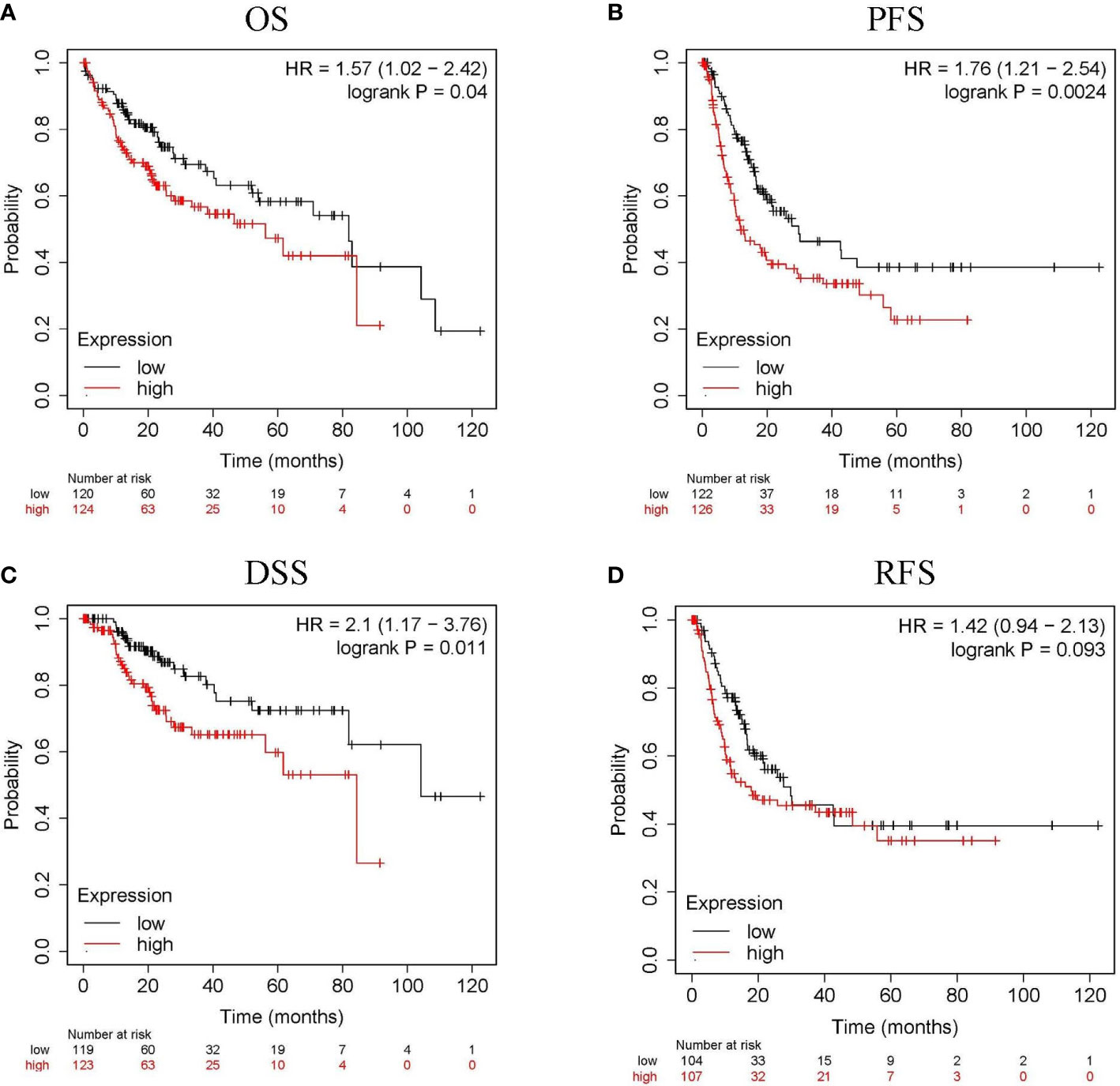
Figure 5 The relationship between CLGN mRNA and prognosis of HCC (Kaplan Meier Plotter). (A–C) HCC patients with higher CLGN mRNA expression have poorer OS, PFS, and DSS, p < 0.05. (D) There was no significant relationship between CLGN mRNA level and RFS, p > 0.05.
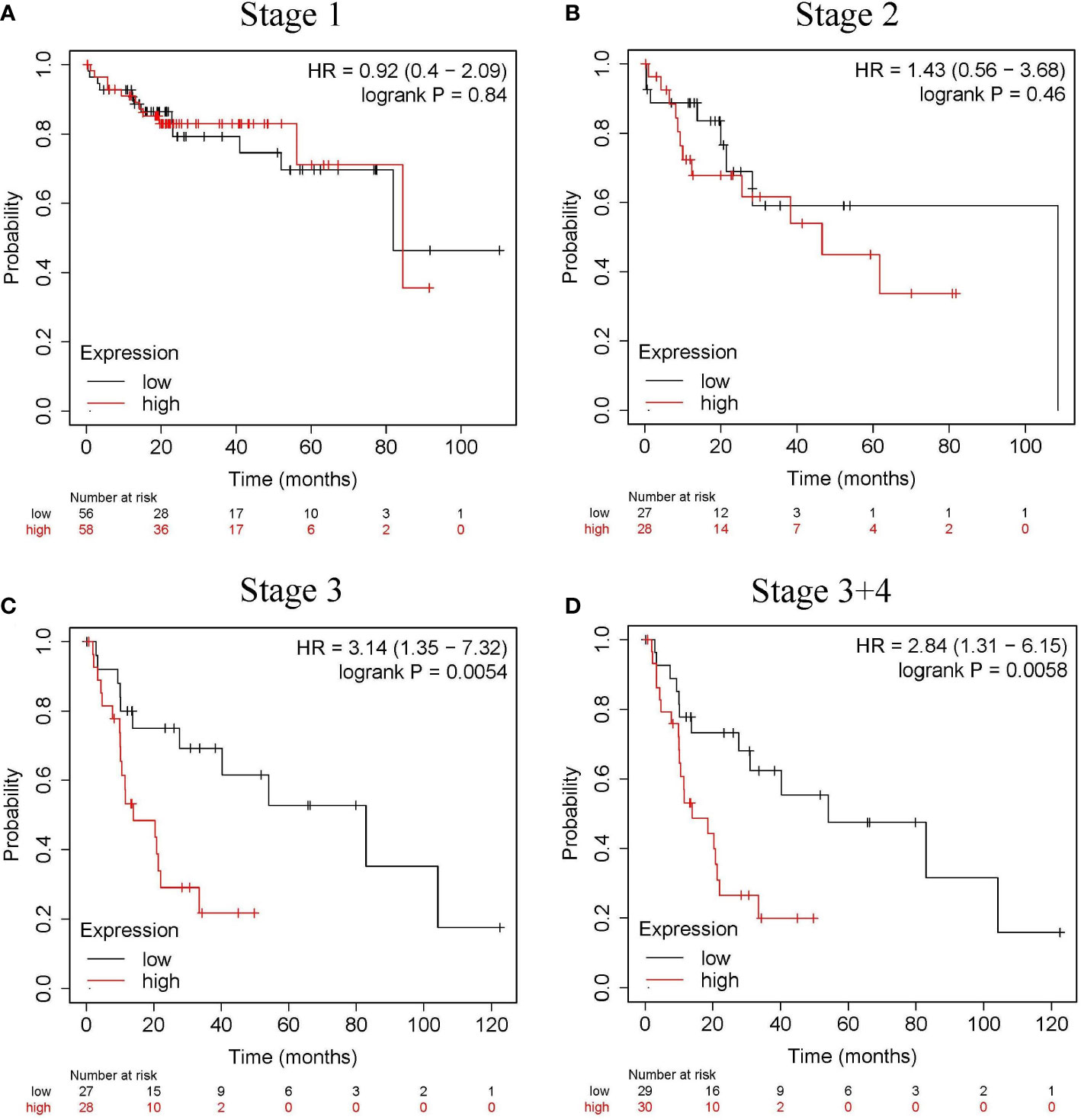
Figure 6 The relationship between the CLGN mRNA level and the OS in different pathology stages in HCC patients. (Kaplan Meier Plotter). (A, B) There was no significant correlation between the CLGN mRNA level and the OS in HCC patients in early stages (T1 and T2), p> 0.05. (C, D) HCC patients in advanced T stages (T3 and T3+T4) have higher CLGN mRNA expression and poorer OS, p < 0.05.
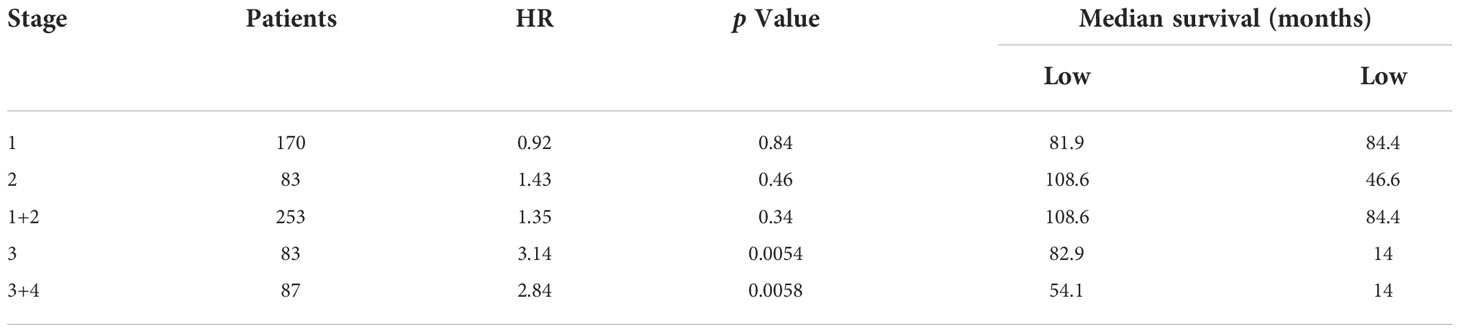
Table 2 Correlation between the CLGN mRNA expression and patient OS at different pathological stages.
CLGN protein expression in clinical specimens
The p rotein expression of CLGN in tumor and normal tissues of clinical specimens from patients with HCC was investigated. Immunohistochemical staining showed that the CLGN protein was significantly regulated in HCC compared with normal liver tissues in 58% (14/24) of cases (Figure 7A). The HPA database results also showed that CLGN protein levels were higher in HCC than in non-tumor tissues (Figure 7B).
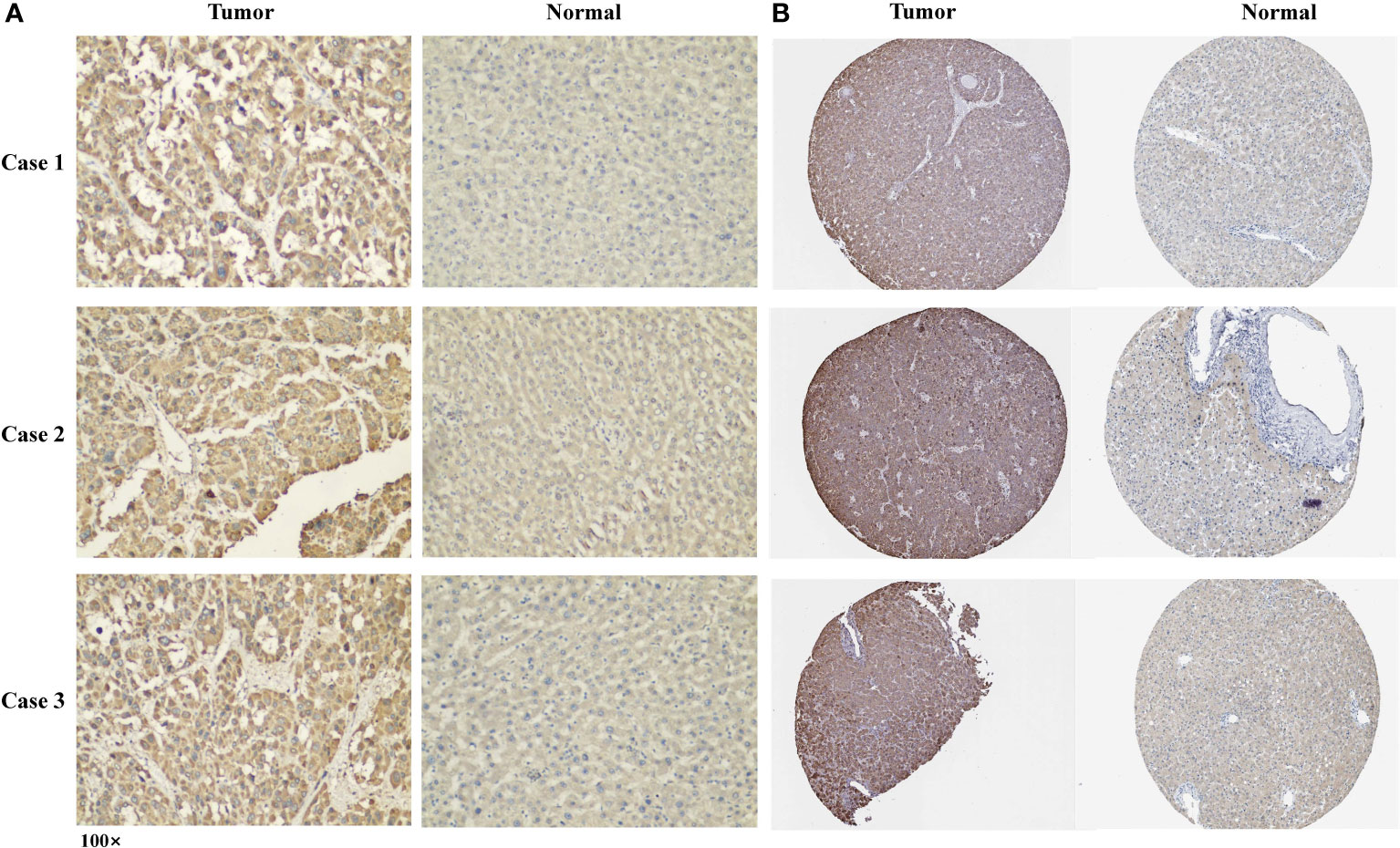
Figure 7 The expression of CLGN protein in HCC and adjacent tissues. (A) Three typical cases of CLGN protein upregulation in HCC in 24 clinical specimens. (B) The expression of CLGN protein in HCC is higher than non-cancerous liver tissue (HPA).
CLGN mutation in HCC and the correlation between CLGN expression and tumor infiltrating lymphocytes
CLGN mutations were analyzed in 1089 patients with HCC from six data sources. The results showed that CLGN was mutated in < 1% (10/1089) of patients (Figure 8A). Furthermore, there was no significant correlation between CLGN expression and the levels of B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, or dendritic cells in HCC (COR < 0.5 (Figure 8B).
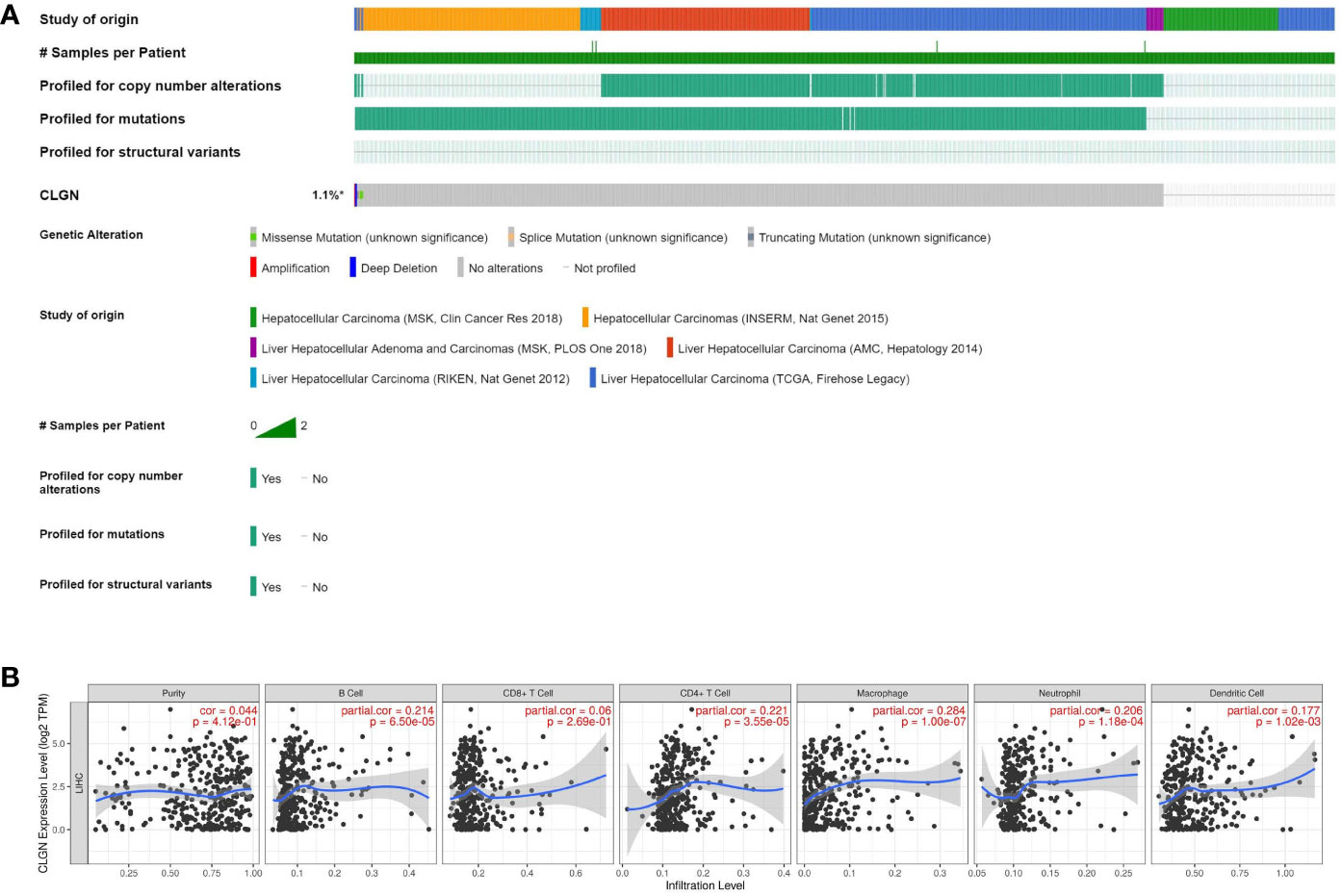
Figure 8 CLGN mutations in HCC patients and the correlation between CLGN expression and tumor infiltrating lymphocytes. (A) CLGN mutations occurred in 10 (< 1%) of 1089 patients from six data sources (CBioportal). (B) There is no significant correlation between the mRNA of CLGN and the infiltrating levels of B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells in HCC, COR < 0.5 (TIMER).
miRNAs that regulate CLGN
Firstly, 4357 and 634 miRNAs that potentially regulate CLGN were predicted in the miRWalk and TargetScan databases, respectively. Duplicate data from the two groups were removed, and their intersection contained 248 potential miRNAs (Supplementary 1). As calculated with Cytoscape, the top 14 miRNAs that were most likely to regulate CLGN mRNA expression were hsa-miR-642b-3p, hsa-miR-7114-5p, hsa-miR-6130, hsa-miR-4738-3p, hsa-miR-520g-3p, hsa-miR-6759-5p, hsa-miR-4446-3p, hsa-miR-6843-3p, hsa-miR-4482-5p, hsa-miR-202-3p, hsa-let-7a-5p, hsa-miR-6080, hsa-miR-2681-5p, and hsa-miR-194-3p (Figure 9A). A further analysis revealed that hsa-miR-194-3p was significantly underexpressed in HCC (Fold Change = 0.72, p < 2.1e-5 and FDR< 0.001, Figure 9B). HCC patients with low hsa-miR-194 expression had poorer OS than those without (p < 0.001, Figure 9C).
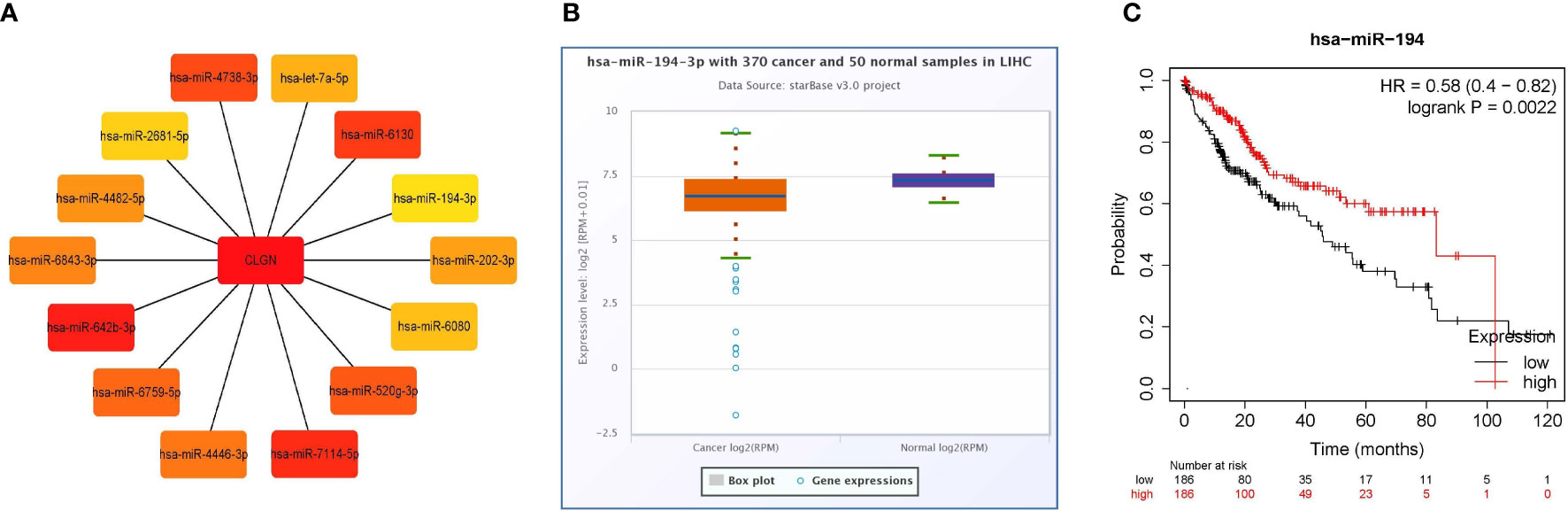
Figure 9 miRNAs that likely regulate the expression of CLGN mRNA (miRWalk and TargetScan). (A) The 14 miRNAs that most closely related to CLGN mRNA. (B) The expression of hsa-miR-194-3p in HCC is significantly lower than in normal tissue, p < 0.05 (ENCORI). (C) HCC patients with lower hsa-miR-194 expression have poorer OS than those without, p < 0.05 (Kaplan Meier Plotter).
Discussion
CLGN (Calmegin) is a testis-specific endoplasmic reticulum chaperone protein (23, 24). Previous studies have shown that CLGN acted as a chaperone for one or more sperm surface proteins to mediate the interaction between sperm and eggs during spermatogenesis (23). Therefore, CLGN may play an important role in spermatogenesis and infertility. A recent study found that CLGN was up-regulated in aldogen-producing adenomas and was related to the generation of aldosterone (25). However, the role of CLGN in several malignant tumors, including HCC, remains unclear.
In this study, it was found that the mRNA and protein of CLGN were highly expressed in HCC compared with normal liver tissues. Furthermore, patients with higher CLGN mRNA levels had poorer prognoses, and CLGN mRNA levels were valuable in predicting the prognosis of patients with advanced pathology. These results suggest that CLGN is associated with malignant progression in HCC patients. Additionally, CLGN was found to be significantly upregulated in invasive breast cancer, chromophobe renal cell carcinoma, papillary renal cell carcinoma lung adenocarcinoma, lung squamous cell carcinoma, prostate adenocarcinoma, and uterine corpus endometrial carcinoma. To the best of our knowledge, no studies have investigated the role of CLGN in these tumors, which is worth of further investigation.
However, the relationship between MND1 and STXBP6 in HCC remains unknown. Notably, Zierhut et al. found that MND1 was necessary for meiosis homologue repair (26). Zhang et al. found that MND1 was upregulated in lung adenocarcinoma and was an independent risk factor for overall survival (27). Furthermore, MND1 was a prognostic biomarker for renal clear cell carcinoma (28). Lenka et al. found that STXBP6 hypermethylation was associated with adverse clinical outcomes in patients with lung cancer (29). A nother recent study showed that the expression level of STXBP6 predicted the response to PD-1/PD-L1 immunotherapy in patients with cancer (30). In this study, we found that MND1 and STXBP6 were upregulated in HCC, but no significant correlation was found between their expression and patient prognosis .
Gene mutations are a common tumorigenic mechanism in liver cancer (31–34). In this study, it was found that CLGN was rarely mutated in HCC patients, which suggests that the pathological effects of CLGN may not be exerted through genetic mutations. The immune microenvironment is closely related to tumor progression (35–37). Therefore, we also analyzed the relationship between CLGN mRNA levels and the abundance of tumor-infiltrating lymphocytes in HCC; however, no significant correlation was found. Based on this, we hypothesized that CLGN might influence the progression of HCC by promoting cell proliferation or inhibiting tumor cell apoptosis, which requires further study in the future.
MicroRNAs (miRNAs) are small non-coding RNAs composed of approximately 20 nucleotides that regulate gene expression by binding to the 3’-UTR of the target mRNA (38). Recent studies have shown that miRNAs were an important regulator of mRNA expression (39–41). hsa-miR-194-3p was significantly underexpressed in HCC, and patients with lower hsa-miR-194 expression had poorer prognosis. A study found that hsa-miR-194-3p might target MECP2 to promote breast cancer progression and reduce sensitivity to rapamycin (42). Recent studies have shown that hsa-miR-194-3p might be associated with colorectal cancer progression (43). However, its specific therapeutic and prognostic value in tumors requires further investigation. In this study, we discovered that hsa-miR-194-3p may also play an important role in HCC progression by regulating CLGN expression.
Our findings suggest that CLGN is a potential prognostic marker for HCC and is associated with HCC progression, however, several limitations are associated with our study. Specifically, the relationship between CLGN expression and patient prognosis has not yet been verified at the center. Additionally, laboratory research on the cancer-promoting mechanisms of CLGN were not available, and the sample size used for immunohistochemistry in this study was small. Therefore, a large-scale verification study is required in the future.
Conclusion
CLGN is upregulated in HCC and significantly correlates with patient prognosis, especially in the advanced stages. In addition, the mRNA levels of CLGN were found to be potentially regulated by hsa-miR-194-3p. These results suggest that CLGN may have important therapeutic and prognostic value in HCC patients.
Data availability statement
The datasets analyzed during the current study are available on Gene Expression Omnibus (GEO) (accession numbers: GSE54236, GSE121248), Gene Expression Profiling Interactive Analysis (GEPIA), Tumor Immune Estimation Resource (TIMER), Kaplan Meier Plotter, The Human Protein Atlas (HPA), cBioPortal, miRWalk, TargetScan, and The Encyclopedia of RNA Interactomes (ENCORI).
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Liuzhou Workers’ Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
ZC: Data analysis and writing of the first draft. BC, YL and ZW: Data analysis, manuscript editing, review, and approval. JW, ZC, and GC: Data analysis, data collection and experiments. DL: Data analysis, manuscript editing and review. All authors have read and approved the final manuscript.
Funding
This study was supported by the Science and Technology Innovation Joint Fund Project of Fujian Province (No. 2019Y9044) and Natural Science Foundation of Fujian Province (No. 2020J011131).
Acknowledgments
We are grateful to the generous data contributors, developers, and maintainers of the public databases.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1081510/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Zhang H, Zhang W, Jiang L, Chen Y. Recent advances in systemic therapy for hepatocellular carcinoma. biomark Res (2022) 10(1):3. doi: 10.1186/s40364-021-00350-4
3. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y
4. Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol (2022) 19(3):151–72. doi: 10.1038/s41571-021-00573-2
5. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet (2017) 389(10088):2492–502. doi: 10.1016/S0140-6736(17)31046-2
6. Pallozzi M, Di Tommaso N, Maccauro V, Santopaolo F, Gasbarrini A, Ponziani FR, et al. Non-invasive biomarkers for immunotherapy in patients with hepatocellular carcinoma: Current knowledge and future perspectives. Cancers (Basel) (2022) 14(19). doi: 10.3390/cancers14194631
7. Lu L, Shen L, Wu Z, Shi Y, Hou P, Xue Z, et al. Trajectories of serum alpha-fetoprotein and intermediate-stage hepatocellular carcinoma outcomes after transarterial chemoembolization: A longitudinal, retrospective, multicentre, cohort study. E Clin Med (2022) 47:101391. doi: 10.1016/j.eclinm.2022.101391
8. Hussen BM, Abdullah ST, Salihi A, Sabir DK, Sidiq KR, Rasul MF, et al. The emerging roles of NGS in clinical oncology and personalized medicine. Pathol Res Pract (2022) 230:153760. doi: 10.1016/j.prp.2022.153760
9. Morganti S, Tarantino P, Ferraro E, D'Amico P, Viale G, Trapani D, et al. Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit Rev Oncol Hematol (2019) 133:171–82. doi: 10.1016/j.critrevonc.2018.11.008
10. Schmidt B, Hildebrandt A. Deep learning in next-generation sequencing. Drug Discovery Today (2021) 26(1):173–80. doi: 10.1016/j.drudis.2020.10.002
11. Zhong Y, Xu F, Wu J, Schubert J, Li MM. Application of next generation sequencing in laboratory medicine. Ann Lab Med (2021) 41(1):25–43. doi: 10.3343/alm.2021.41.1.25
12. Chen F, Wang J, Wu Y, Gao Q, Zhang S. Potential biomarkers for liver cancer diagnosis based on multi-omics strategy. Front Oncol (2022) 12:822449. doi: 10.3389/fonc.2022.822449
13. Villa E, Critelli R, Lei B, Marzocchi G, Camma C, Giannelli G, et al. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. results from a prospective study. Gut (2016) 65(5):861–9. doi: 10.1136/gutjnl-2014-308483
14. Wang SM, Ooi LL, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res (2007) 13(21):6275–83. doi: 10.1158/1078-0432.CCR-06-2236
15. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
16. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77(21):e108–e10. doi: 10.1158/0008-5472.CAN-17-0307
17. Lanczky A, Gyorffy B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J Med Internet Res (2021) 23(7):e27633. doi: 10.2196/27633
18. World Medical A. World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA (2013) 310(20):2191–4. doi: 10.1001/jama.2013.281053
19. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
20. Sticht C, de la Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PloS One (2018) 13(10):e0206239. doi: 10.1371/journal.pone.0206239
21. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife (2015) 4. doi: 10.7554/eLife.05005
22. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res (2014) 42(Database issue):D92–7. doi: 10.1093/nar/gkt1248
23. Ikawa M, Wada I, Kominami K, Watanabe D, Toshimori K, Nishimune Y, et al. The putative chaperone calmegin is required for sperm fertility. Nature (1997) 387(6633):607–11. doi: 10.1038/42484
24. Siep M, Sleddens-Linkels E, Mulders S, van Eenennaam H, Wassenaar E, Van Cappellen WA, et al. Basic helix-loop-helix transcription factor Tcfl5 interacts with the calmegin gene promoter in mouse spermatogenesis. Nucleic Acids Res (2004) 32(21):6425–36. doi: 10.1093/nar/gkh979
25. Itcho K, Oki K, Gomez-Sanchez CE, Gomez-Sanchez EP, Ohno H, Kobuke K, et al. Endoplasmic reticulum chaperone calmegin is upregulated in aldosterone-producing adenoma and associates with aldosterone production. Hypertension (2020) 75(2):492–9. doi: 10.1161/HYPERTENSIONAHA.119.14062
26. Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F. Mnd1 is required for meiotic interhomolog repair. Curr Biol (2004) 14(9):752–62. doi: 10.1016/j.cub.2004.04.030
27. Zhang Q, Shi R, Bai Y, Meng L, Hu J, Zhu H, et al. Meiotic nuclear divisions 1 (MND1) fuels cell cycle progression by activating a KLF6/E2F1 positive feedback loop in lung adenocarcinoma. Cancer Commun (Lond) (2021) 41(6):492–510. doi: 10.1002/cac2.12155
28. Fang J, Zhen J, Gong Y, Ke Y, Fu B, Jiang Y, et al. MND1 functions as a potential prognostic biomarker associated with cell cycle and immune infiltration in kidney renal clear cell carcinoma. Aging (Albany NY) (2022) 14(18):7416–42. doi: 10.18632/aging.204280
29. Lenka G, Tsai MH, Lin HC, Hsiao JH, Lee YC, Lu TP, et al. Identification of methylation-driven, differentially expressed STXBP6 as a novel biomarker in lung adenocarcinoma. Sci Rep (2017) 7:42573. doi: 10.1038/srep42573
30. Liu Y, Huang Z, Wei Y, Zhang M, Li X, Yang S, et al. Identification of STXBP6-IRF1 positive feedback loop in regulation of PD-L1 in cancer. Cancer Immunol Immunother (2021) 70(2):275–87. doi: 10.1007/s00262-020-02678-6
31. Li Y, Li D, Liu Y, Wang S, Sun M, Zhang Z, et al. The positive feedback loop of NHE1-ERK phosphorylation mediated by BRAF(V600E) mutation contributes to tumorigenesis and development of glioblastoma. Biochem Biophys Res Commun (2022) 588:1–7. doi: 10.1016/j.bbrc.2021.11.104
32. Vu T, Datta A, Banister C, Jin L, Yuan G, Samuel T, et al. Serine-threonine kinase receptor-associated protein is a critical mediator of APC mutation-induced intestinal tumorigenesis through a feed-forward mechanism. Gastroenterology (2022) 162(1):193–208. doi: 10.1053/j.gastro.2021.09.010
33. Wen X, Lu F, Liu S. Prognostic value of p53 mutation for poor outcome of Asian primary liver cancer patients: evidence from a cohort study and meta-analysis of 988 patients. OncoTargets Ther (2016) 9:7425–33. doi: 10.2147/OTT.S121594
34. Li J, Yu CP, Li Q, Chang S, Xie LL, Wang S. Large-Scale omics data reveal the cooperation of mutation-circRNA-miRNA-target gene network in liver cancer oncogenesis. Future Oncol (2022) 18(2):163–78. doi: 10.2217/fon-2021-0940
35. Chew V, Toh HC, Abastado JP. Immune microenvironment in tumor progression: characteristics and challenges for therapy. J Oncol (2012) 2012:608406. doi: 10.1155/2012/608406
36. Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J hepatol (2015) 62(6):1420–9. doi: 10.1016/j.jhep.2015.02.038
37. Xing Y, Ruan G, Ni H, Qin H, Chen S, Gu X, et al. Tumor immune microenvironment and its related miRNAs in tumor progression. Front Immunol (2021) 12:624725. doi: 10.3389/fimmu.2021.624725
38. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5
39. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet (2016) 17(5):272–83. doi: 10.1038/nrg.2016.20
40. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res (2009) 19(1):92–105. doi: 10.1101/gr.082701.108
41. Pan X, Wenzel A, Jensen LJ, Gorodkin J. Genome-wide identification of clusters of predicted microRNA binding sites as microRNA sponge candidates. PloS One (2018) 13(8):e0202369. doi: 10.1371/journal.pone.0202369
42. Zhou Q, Guo J, Huang W, Yu X, Xu C, Long X. Linc-ROR promotes the progression of breast cancer and decreases the sensitivity to rapamycin through miR-194-3p targeting MECP2. Mol Oncol (2020) 14(9):2231–50. doi: 10.1002/1878-0261.12700
Keywords: hepatocellular carcinoma (HCC), prognosis, CLGN, MND1, STXBP6, MiR-194-3p
Citation: Cui Z, Wang J, Chen G, Li D, Cheng B, Lai Y and Wu Z (2023) The upregulation of CLGN in hepatocellular carcinoma is potentially regulated by hsa-miR-194-3p and associated with patient progression. Front. Oncol. 12:1081510. doi: 10.3389/fonc.2022.1081510
Received: 27 October 2022; Accepted: 29 November 2022;
Published: 09 January 2023.
Edited by:
Wenyu Lin, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Linbin Lu, Xijing Hospital, ChinaXiaoqiong Duan, Institut National de la Transfusion Sanguine, France
Copyright © 2023 Cui, Wang, Chen, Li, Cheng, Lai and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bianqiao Cheng, Y2JxNzUxMTE1QHNpbmEuY29t; Yanhua Lai, MTM3OTc3MTgxMkBxcS5jb20=; Zhixian Wu, enh3dUB4bXUuZWR1LmNu
†These authors share first authorship
 Zhongyuan Cui
Zhongyuan Cui Jielong Wang1†
Jielong Wang1† Zhixian Wu
Zhixian Wu