- 1Department of Spinal Bone Disease, First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 2Department of Trauma Orthopedic and Hand Surgery, First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 3Center for Genomic and Personalized Medicine, School of Preclinical Medicine, Guangxi Medical University, Nanning, China
- 4Guangxi Key Laboratory for Genomic and Personalized Medicine, Guangxi Key Laboratory of Colleges and Universities, Nanning, China
- 5Guangxi Collaborative Innovation Center for Genomic and Personalized Medicine, Guangxi Medical University, Nanning, China
- 6Department of Bone and Soft Tissue Surgery, The Affiliated Tumor Hospital, Guangxi Medical University, Nanning, China
- 7School of Biomedical Sciences, The University of Western Australia, Perth, WA, Australia
- 8Department of Medical Oncology, First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 9Guangxi Key Laboratory of Regenerative Medicine, Research Centre for Regenerative Medicine, Guangxi Medical University, Nanning, China
A Corrigendum on
Single-Cell Transcriptomics Reveals the Complexity of the Tumor Microenvironment of Treatment-Naive Osteosarcoma
By Liu Y, Feng W, Dai Y, Bao M, Yuan Z, He M, Qin Z, Liao S, He J, Huang Q, Yu Z, Zeng Y, Guo B, Huang R, Yang R, Jiang Y, Liao J, Xiao Z, Zhan X, Lin C, Xu J, Ye Y, Ma J, Wei Q and Mo Z (2021) 11:709210. doi: 10.3389/fonc.2021.709210
In the published article, there was an error in Figure 1D as published. The labels of feature plots in Figure 1D were misleadingly described. The original labels for “osteoblastic OS cells” should switch with “CAFs”, as well as “Osteoclasts” should switch with “Plasmocytes”. The corrected Figure 1D and its caption appear below.
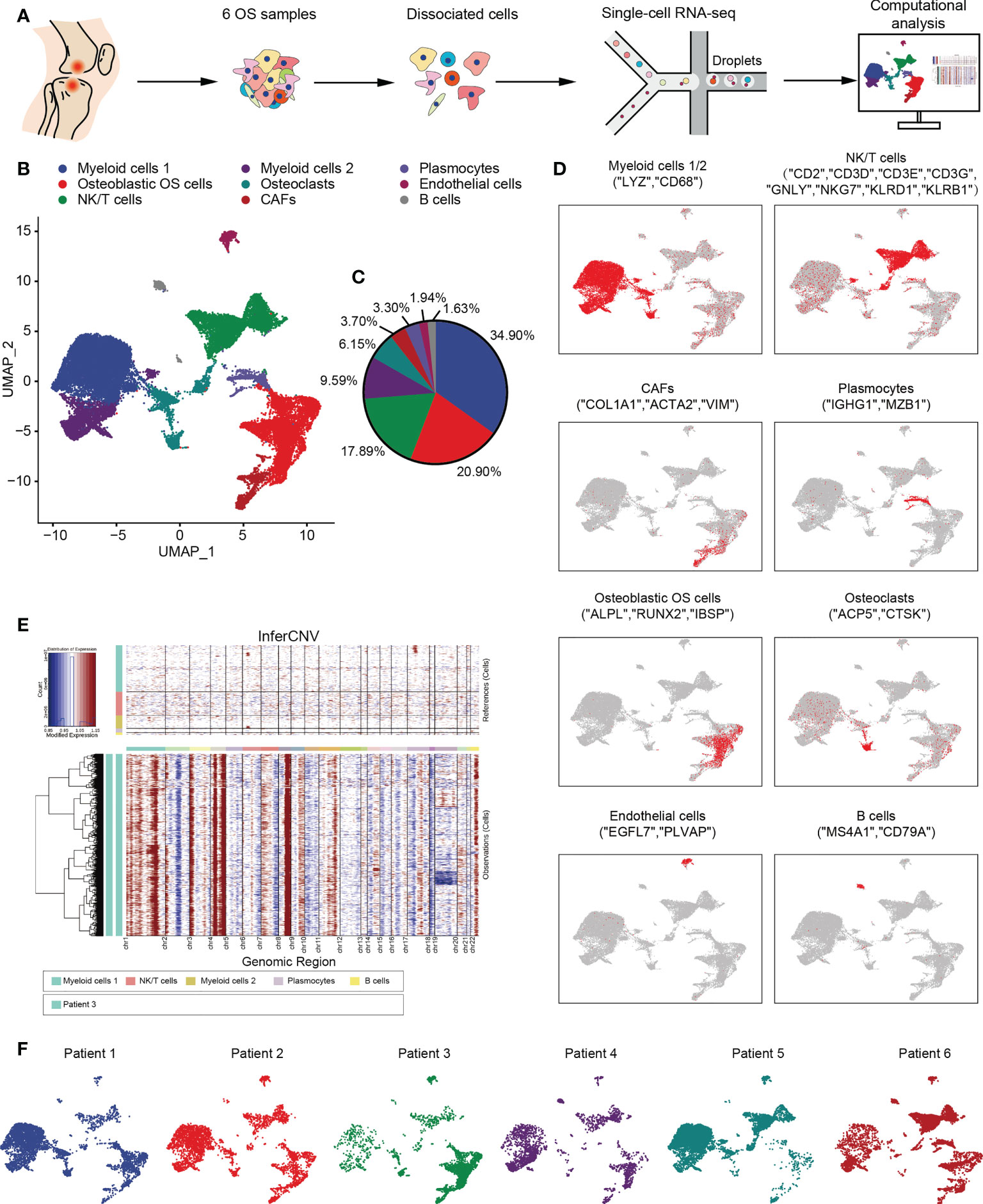
Figure 1 Overview of single cells derived from OS tissues. (A) Workflow depicting collection and processing of specimens of OS tumors for scRNA-seq. (B) UMAP plot of all the single cells, with each color-coded for the 9 major cell types. (C) Pie chart, indicating the cell composition of OS. (D) UMAP plots of the normalized marker expression of the 9 major cell types. (E) The large-scale chromosomal landscape in patient 3 was calculated using reference cells (myeloid cells 1/2, NK/T cells, plasmocytes and B cells); the red color represents an increased copy number, whereas the blue color represents a decreased copy number. (F) UMAP plot of all the single cells, with each cell color-coded for different patients. OS, osteosarcoma; UMAP, uniform manifold approximation and projection; scRNA-seq, single-cell RNA sequencing; NK, natural killer.
In the published article, there was an error in Figure 1E as published. The label of “Patient 1” in Figure 1E was incorrectly described, which should be changed to “Patient 3”. The corrected Figure 1E and its caption appear below.
In the published article, there was an error in Supplementary Figure 1A as published. The labels of “Patient 1” in Supplementary Figure 1A were incorrectly described. The six “Patient 1” in inferCNV of Supplementary Figure 1A should need to be corrected in order.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: single-cell RNA sequencing, tumor microenvironment, naive osteosarcoma, heterogeneity, osteolysis
Citation: Liu Y, Feng W, Dai Y, Bao M, Yuan Z, He M, Qin Z, Liao S, He J, Huang Q, Yu Z, Zeng Y, Guo B, Huang R, Yang R, Jiang Y, Liao J, Xiao Z, Zhan X, Lin C, Xu J, Ye Y, Ma J, Wei Q and Mo Z (2022) Corrigendum: Single-cell transcriptomics reveals the complexity of the tumor microenvironment of treatment-naive osteosarcoma. Front. Oncol. 12:1077067. doi: 10.3389/fonc.2022.1077067
Received: 22 October 2022; Accepted: 24 October 2022;
Published: 15 November 2022.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2022 Liu, Feng, Dai, Bao, Yuan, He, Qin, Liao, He, Huang, Yu, Zeng, Guo, Huang, Yang, Jiang, Liao, Xiao, Zhan, Lin, Xu, Ye, Ma, Wei and Mo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingjun Wei, d2VpcWluZ2p1bkBneG11LmVkdS5jbg==; Jie Ma, bWFqaWUwODZAMTYzLmNvbQ==; Yu Ye, Z3htdWtqY0AxMjYuY29t; Jiake Xu, amlha2UueHVAdXdhLmVkdS5hdQ==
†These authors have contributed equally to this work
 Yun Liu
Yun Liu Wenyu Feng
Wenyu Feng Yan Dai3,4,5†
Yan Dai3,4,5† Mingwei He
Mingwei He Zhaojie Qin
Zhaojie Qin Shijie Liao
Shijie Liao Qian Huang
Qian Huang Zhenyuan Yu
Zhenyuan Yu Yanyu Zeng
Yanyu Zeng Binqian Guo
Binqian Guo Rong Huang
Rong Huang Rirong Yang
Rirong Yang Yonghua Jiang
Yonghua Jiang Jiake Xu
Jiake Xu Jie Ma
Jie Ma Qingjun Wei
Qingjun Wei