
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 11 January 2023
Sec. Gynecological Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1064722
This article is part of the Research Topic New Molecular Approaches to Improve Gynecological Cancer Management View all 11 articles
Objective: To explore the ability of PAX1 methylation (PAX1m) to predict the pathological upgrade of cervical intraepithelial neoplasia (CIN) before cold knife conization (CKC).
Methods: A total of 218 women that underwent colposcopy-directed biopsy (CDB) pathology for the confirmation of CIN2 and CIN3 between December 2020 to September 2021 were enrolled in this study. The methylation levels of PAX1 (ΔCpPAX1) were determined by quantitative methylation-specific polymerase chain reaction (qMSP). Receiver operating characteristic curve was used to identify the optimal cut-off value of ΔCpPAX1 for predicting the pathological upgrade of disease.
Results: In the CDB-confirmed CIN2 group, 36% of CIN2 was found to have pathologically upgraded to CIN3 and 30% regressed to low-grade squamous intraepithelial lesion (LSIL) and below, and none of CIN2 upgraded to early-stage cervical cancer (ESCC) after CKC. In the CDB-confirmed CIN3 group, 19.5% (23/118) of CDB-confirmed CIN3 were pathologically upgraded to ESCC after CKC. Regardless of CIN2 or CIN3, the ΔCpPAX1 level of women with upgraded pathology after CKC was significantly lower than that of women with degraded pathology. The optimal △CpPAX1 cut-off value in predicting CIN3 to be upgraded to ESCC after CKC was 6.360 and the area under the curve (AUC) was 0.814, with similar sensitivity (78.3%) and higher specificity (84.2%) than cytology≥LSIL (Se:78.3%;Sp:58.9%) and HPV16/18 positive (Se:73.9%;Sp:46.3%) patients.
Conclusions: PAX1m could be a promising auxiliary marker in predicting the pathological upgrade of CIN before CKC. We found that if the △Cp PAX1 cut-off value is lower than 6.360, it is highly suggestive of invasive cervical cancer.
Persistent infection of high-risk human papillomavirus (hr-HPV) is an important risk factor for the development of cervical intraepithelial neoplasia (CIN) and cervical cancer. In 2020, the World Health Organization (WHO) classification of female genital tumours was updated from the original three-level classification of cervical intraepithelial neoplasia (CIN1, CIN2, CIN3) to a two-level classification that included low-grade squamous intraepithelial lesions (LSIL/CIN1) and high-grade squamous intraepithelial lesions (HSIL/CIN2 and CIN3) (1). HSIL is recognized as the true precancer with a higher risk of progression. However, it is difficult to determine whether or when a patient with HSIL will progress to invasive cervical cancer from an individual perspective, in fact, some patients may already have occult cervical cancer when diagnosed with HSIL. HSIL are primarily treated with cervical conization, including cervical cold knife conization (CKC) or loop electrode excision procedure (LEEP). On the other hand, most CIN2 lesions (60%), particularly in young women (<30 years), regress spontaneously, indicating that active surveillance, rather than immediate intervention, is justified, especially if patients adhere to monitoring (2). However, there are some limitations in the consistency between pathological assessment via colposcopy-directed biopsy(CDB) and final pathological diagnosis after conization, with an upgrade rate of 23.1% and degrade rate of 33.6% post-conization pathology (3).
To date, there are no accurate tests to determine whether CIN lesions have a tendency to regress or progress. The HPV genotype present in affected patients could not provide additional information to predict high-grade disease progression (4). Although the proportion of severe lesions caused by HPV16/18 has increased over time, its potential for progression remains uncertain (5, 6). Gene methylation is a kind of epigenetic modification that can contribute to the accumulation of mutated genes over time and methylation may play an important role in tumor genesis and progression. As such, using methylation as a marker due to its high sensitivity for cancer has potential as a primary screening tool. It may also be used for the management of women with CIN lesions to prevent overtreatment of CIN2/CIN3 lesions (7).
In particular, the efficacy of paired boxed gene 1 (PAX1) methylation (PAX1m) as a biomarker for the detection of CIN3 or worse (CIN3+) has been demonstrated in various studies (8–10). PAX1m can be used as a triage method for women with atypical squamous cells of undetermined significance (ASCUS) and has shown better diagnostic performance than HPV-DNA in predicting CIN2+ (11). Besides, PAX1m has a comparable clinical performance to cytology and better accuracy and specificity than HPV16/18 as a triage tool for detecting CIN3+ in women with hr-HPV (12). PAX1m has also been reported to predict the efficacy of concurrent chemo-radiotherapy in cervical cancer (13), and is a potential biomarker for monitoring the prognosis of cervical adenocarcinoma (14). However, few studies have evaluated PAX1 gene methylation before conization, it has been previously reported that PAX1m would be a suitable alternative method to conventional options and it has the ability to predict the outcome of conization in CIN3 cases (15). However, the role of PAX1m in predicting the pathological upgrade of CIN2 is unclear. In this study, we aim to investigate the predictive value of PAX1m status in determining the upgrade tendency of CIN2 and CIN3. This information would help patients and doctors make more individualized treatment decisions.
In total, 247 women with pathologically confirmed HSIL by CDB were included in this study at the Peking University People’s Hospital between December 2020 to September 2021. The exclusion criteria were as follows: (1) CDB revealed the presence of squamous cell cancer, adenocarcinoma in situ (AIS), or adenocarcinoma, (2) inability to undergo CKC, (3) inadequate DNA concentration in cell samples, (3) HSIL in patients who were also pregnant, had immune system diseases, or receiving immunosuppressive therapy, (4) patients that had a history of cervical disease treatment, hysterectomy, or chemoradiotherapy. Of the 247 women with CDB-confirmed HSIL, 16 cases of CIN2 and 4 of CIN3 chose observational follow-up rather than CKC and 9 cases were determined to have AIS. Therefore, a total of 218 women with pathologically confirmed HSIL by CDB were included in this study.
Exfoliated cervical cell samples were collected after biopsy pathology had confirmed HSIL within 7 days before the CKC procedure. Briefly, a vaginal speculum was placed to expose the cervix and cervical exfoliation was performed at the squamocolumnar junction of the cervix using a sampling brush. The sampling brush was then placed into a 20mL PreservCyt1 solution (Hologic, Marlborough Mass, USA, DOC sample) for testing. All specimens were tested for cytology, HPV detection, and PAX1 methylation. We informed patients of the research programs and obtained written consent before CKC. The study was approved by the Institutional Review Board of Peking University People’s Hospital (2020PHB298-01).
Cervical exfoliated cells were centrifuged and stored in phosphate-buffered saline at -20°C. Genomic DNA was extracted using standard protocols and then converted to bisulfite form using the EZ DNA Methylation-Gold kits (Zymo Research, Irvine, CA, USA). Quantitative methylation-specific PCR (qMSP) was performed using a Light Cycler LC480 system (Roche Applied Science, Penzberg, Germany) to determine the methylation level of PAX1 according to the manufacturer’s instructions (Hoomya Ltd, Hunan, P.R China). Type II collagen gene (COL2A) was used as an internal reference. The △Cp is the difference between the △Cp values for PAX1 and COL2A. The methylation level (△Cp) was assessed by the following formula: △Cp=Cp target gene - Cp Col2A (16). A smaller △CpPAX1 value denotes a higher degree of PAX1 methylation detected in the collected samples.
Type-specific HR-HPV viral genotyping was simultaneously measured using a BioPerfectus Multiplex Real-Time PCR(BMRT) assay. BMRT is a PCR-based assay for the detection of high-risk HPV strains and it was performed using a fluorescence-based multiplex HPV DNA genotyping kit (Bioperfectus Ltd, Jiangsu, P.R. China). This assay can detect 14 high-risk HPV subtypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) and 7 medium- and low-risk subtypes. For this study, all types specifically refer to high-risk HPV.
Colposcopic impressions were made according to the American Society for Colposcopy and Cervical Pathology (ASCCP) standard, multiple biopsies targeting all areas with acetowhitening, metaplasia, or higher abnormalities are recommended. At least 2-4 targeted biopsies from distinct acetowhite lesions should be taken (17). A circular knife cut is made 3 mm peripheral to the abnormal transformation zone (ATZ). The knife is angled toward the endocervical canal and cuts deeper into the stroma, the depth of excision depends on the type of TZ according to 2011 Colposcopic Terminology of International Federation for Cervical Pathology and Colposcopy (IFCPC) (18). Cervical biopsy and CKC specimens were histologically examined and classified according to the 2020 WHO classification of female genital tumours (1), which reported as HSIL(CIN2) and HSIL (CIN3), p16 immunohistochemistry was used only in morphologically ambiguous cases when HSIL is suspected according to the guidance provided by the Lower Anogenital Squamous Terminology (LAST) Project (19). The highest pathological grade was taken as the final pathological diagnosis. Pathological upgrade of disease is defined as CIN2CDB (CDB-confirmed CIN2) →CIN3CKC (CKC-confirmed CIN3) and pathology-confirmed CIN3CDB (CDB-confirmed CIN3) →ESCCCKC (CKC-confirmed early-stage cervical cancer). The cervical lesions were diagnosed by two professional pathologists.
The samples were characterized using descriptive statistics in two groups of biopsy diagnosis. The Mann-Whitney test was utilized to analyze the differences between △CpPAX1 levels. We used restricted cubic spline models fitted for logistic odds ratio with 3 knots of PAX1m using statistical software (rms in R, version 4.1.2) (Supplementary Figure 1). We divided the △CpPAX1 into 3 groups to form grade variables and assigned the group names of Low (1:△Cp >15), Moderate (2: 9 ≤ △Cp ≤ 15), and High (3:△Cp ≤ 9). Logistic regression was used to evaluate the odds ratio (OR) and control for confounding factors (e.g., HPV, cytology) (model 1). The receiver operating characteristic (ROC) curve was used to identify the optimal cut-off value of PAX1m for predicting pathological upgrade of disease. Sensitivity (Se), specificity (Sp), positive predictive value (PPV), and negative predictive value (NPV) were calculated. Confidence intervals for Se and Sp are Clopper-Pearson confidence intervals. Confidence intervals for the predictive values are the standard logit confidence intervals (20). SPSS software version (Version 26.0, SPSS, Inc, Chicago, IL) were used for statistical analysis. All differences were considered two-sided and statistically significant at P < 0.05.
Among the 218 women with CDB-confirmed HSIL, 100 cases were CIN2CDB and 118 cases were CIN3CDB. The mean age was 40.7 ± 10.8 years (22-69). The median △CpPAX1 was 19.3 (10.5-20.9) and 8.9 (6.0-14.0) in CIN2CBD and CIN3 CBD, respectively, which was significantly different (Figure 1A). In the CIN2CBD group, 36% of CIN2 was pathologically upgraded to CIN3 and 30% regressed to LSIL and below, none of the CIN2 cases upgraded to SCC after CKC, and the positive margin rate was only 2%. However, in the CIN3CBD group, 19.5% (23/118) of CDB-confirmed CIN3 were pathologically upgraded to ESCC after CKC. The detailed characteristics are presented in Table 1.
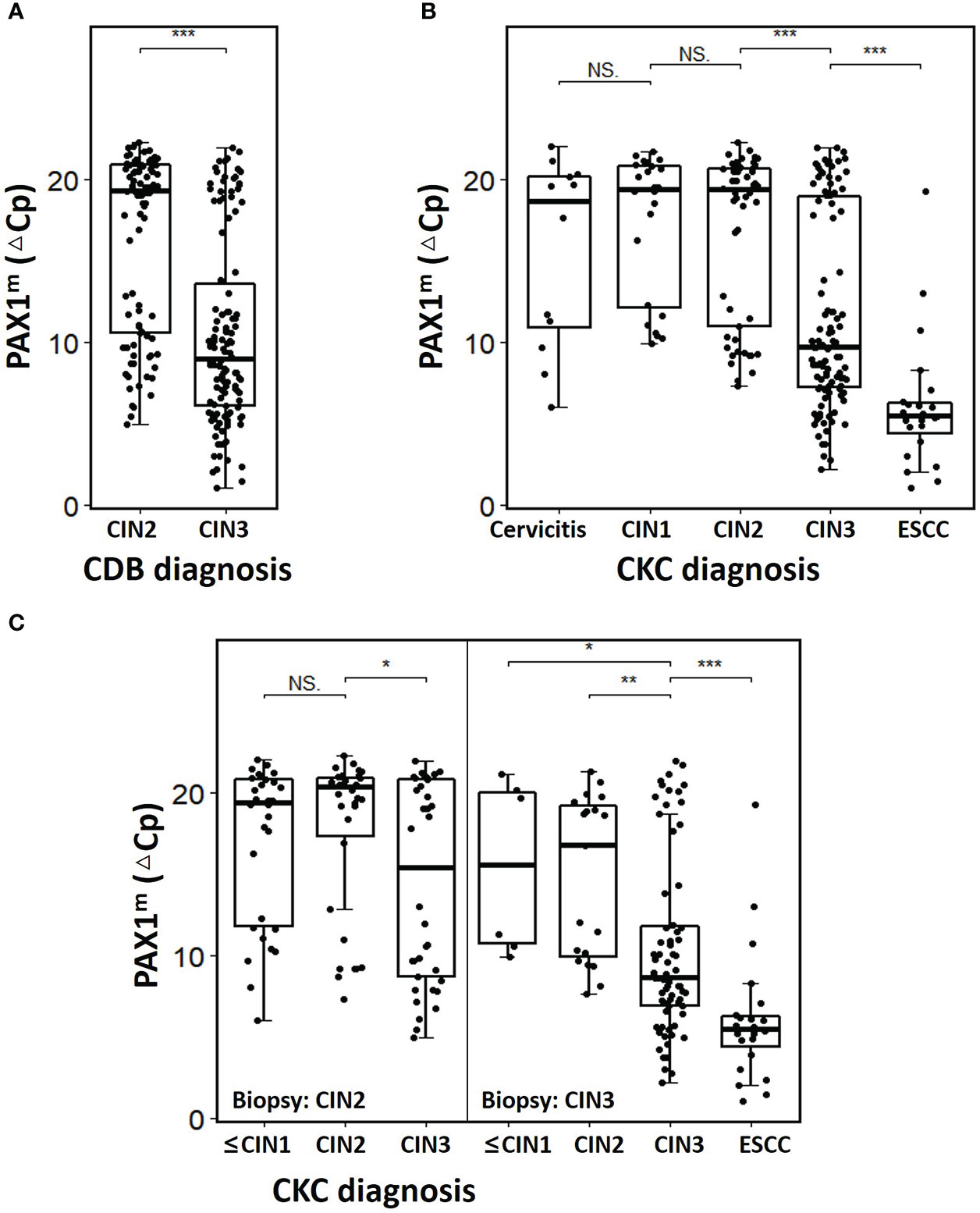
Figure 1 The PAX1m distribution of cervical lesions diagnosed by CDB and CKC. (A) Comparison of ΔCpPAX1 levels of CIN2 and CIN3 diagnosis by CDB; (B) Comparison of ΔCpPAX1 levels of different cervical lesions diagnosed by CKC; (C) Comparison of ΔCpPAX1 levels changing in upgraded, maintained, or degraded lesions after CKC. The middle line is the median; the box shows the inter-quartile range (IQR), and the whiskers extend to, at most, 1.5 times the IQR. *p<0.05, **p<0.01, ***p<0.001, NS: not significant.
After conization, the△CpPAX1 level of ESCCCKC was significantly lower than that of CIN3CKC, and that of CIN3CKC was lower than CIN2CKC, with statistically significant differences (p<0.001) (Figure 1B). Within the CIN2CDB group, there was no difference in △CpPAX1 between CIN2CDB that had degraded to ≤CIN1CKC and those that had maintained at CIN2CKC, but there was a significant difference between those that had maintained at CIN2CKC and those that upgraded to CIN3CKC. Among the CIN3CDB group, △CpPAX1levels were significantly lower in those that had upgraded to ESCCCKC than those that had maintained at CIN3CKC and downgraded to ≤CIN2CKC (p<0.001) (Figure 1C). Regardless of CIN2 or CIN3 status, △CpPAX1 level of women with upgraded pathology after CKC was significantly lower than that of women with degraded pathology (detailed information see Supplementary Table 1).
When analyzing PAX1m at different thresholds, it was found that the risk of CIN3+ increased significantly when ΔCpPAX1 < 7, while the trend turns flat when ΔCpPAX1 > 15 (Supplementary Figure 1). We divided the different threshold levels of PAX1m according to high-, medium-, and low-risk for CIN2+, CIN3+, and ESCC. When ΔCpPAX1 was 6.4 (1.1-9.0), the OR values of CIN2+, CIN3+, and ESCC were 12.52 (2.85-55.00), 20.61 (8.08-52.57), and 34.07 (4.45-261.08), respectively. In order to adjust variables that would have effects on PAX1m, we established PAX1mModel 1(shown in the “Methods” section), and further confirmed that the risk of ESCC, CIN3+, and CIN2+ was still high, and the OR values were 24.85 (3.17-194.67), 19.27 (7.39-50.22), and 11.98 (2.68-53.65), respectively (Table 2), indicating that if ΔCpPAX1 less than 6.4, it should be alert for the occurrence of high-grade lesions or even cervical cancer.
The optimal ΔCpPAX1 cut-off value in predicting whether CIN3 would upgrade to ESCC after CKC was 6.360 and the area under the curve (AUC) was 0.814 (95% CI: 0.714–0.915), with similar sensitivity (78.3%) but higher specificity (84.2%) than cytology≥ LSIL (Se:78.3%;Sp:58.9%) and HPV16/18 positive (Se:73.9%;Sp:46.3%). The optimal ΔCpPAX1 cut-off value in predicting whether CIN2 would upgrade to CIN3 was 10.830 and the AUC was 0.636 (95% CI: 0.515–0.756), with lower sensitivity (44.4%) but higher specificity (82.8%), compared with cytology>ASCUS (Se:44.4%;Sp:45.3%) and HPV positive (Se:97.2%;Sp:9.4%) (Figure 2 and Table 3).
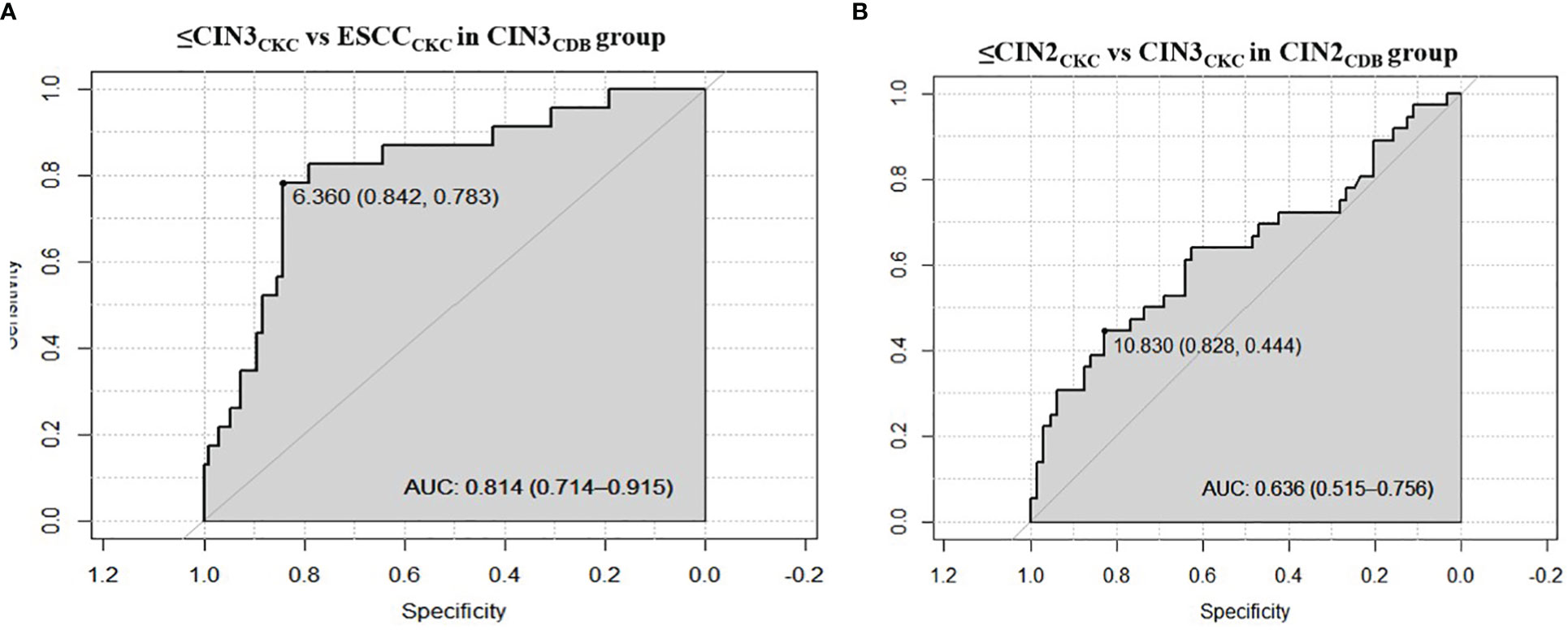
Figure 2 Receiver operating characteristic curves of performance of PAX1 methylation. (A) The optimal ΔCpPAX1cut-off value in predicting whether CIN3 would be upgraded to ESCC after CKC; (B) The optimal ΔCpPAX1cut-off value in predicting whether CIN2 would be upgraded to CIN3.
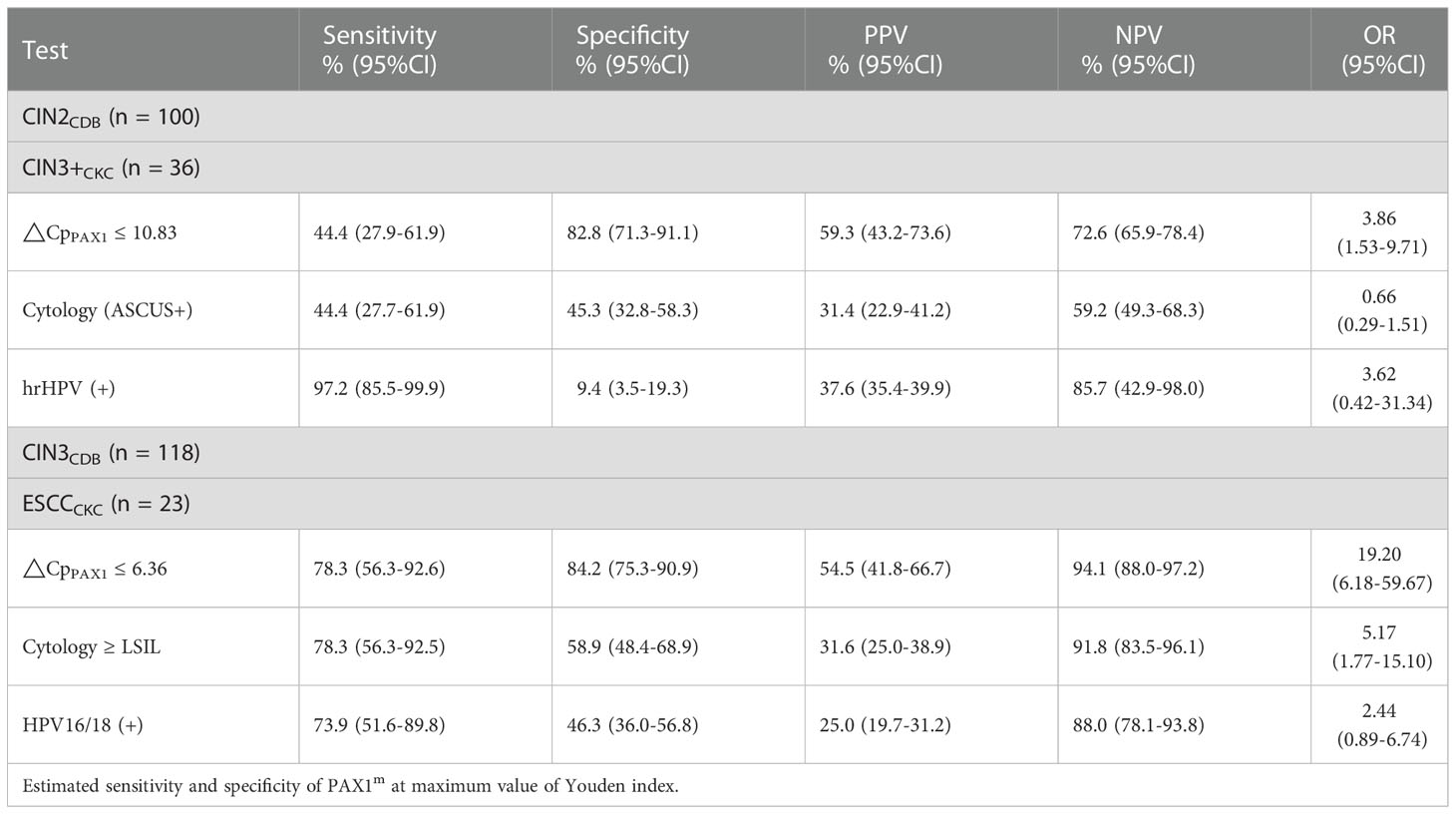
Table 3 The Performance of ΔCpPAX1 in predicting CIN2 upgrade to CIN3, and CIN3 upgrade to ESCC after conization.
In our study, CDB-confirmed CIN2 and CIN3 were stratified to predict pathological progression after CKC. We found that 19.5% (23/118) of the CDB-confirmed CIN3 cases were pathologically upgraded to ESCC after CKC, which was higher than previously reported (15), indicating that CDB alone is insufficient for the diagnosis of microinvasive cervical cancer (21). In the CDB-confirmed CIN2 group, 36% of the CIN2 cases were pathologically upgraded to CIN3 and 30% regressed to LSIL and below, however, none of the CIN2 cases were pathologically upgraded to SCC after CKC. Using accurate tests to determine whether CIN lesions have a tendency to regress or progress is crucial for subsequent disease management.
Prognostic testing for CIN could dramatically alter the treatment algorithm. Underdiagnosis leads to multiple follow-up visits, and either delayed or progressed the lesion, exacerbating the potential harm to patients. Alternatively, overdiagnosis can result in unnecessary or premature treatment, especially in younger women, as inappropriate treatment significantly increases the risk of adverse outcomes in subsequent pregnancies (22). The majority of HSILs require surgery for the purpose of completely removing lesions, as well as prevent cancer, a small number of special conditions or periods (such as young or pregnant women) can be given a short-term close follow-up (23). As recommended by the ASCCP2019 guidelines, CIN2 and CIN3 should be managed separately, and for patients with CIN 2 that are more concerned about the effects of treatment on a future pregnancy outweigh their concerns about cancer, observation without treatment is acceptable (24).
However, CIN2 and CIN3 diagnosed by CDB have limitations to some extent. For example, the diagnosis of CIN2 has historically been a gray area in pathology and it is difficult for pathologists to reproduce which might be overcalled CIN1 or under-called CIN3 (25). Some pathologists even use “CIN1–2 or CIN2–3” to equivocate the classification. Based on difficulties associated with receiving an accurate diagnosis, it is challenging to determine whether or when a patient with CIN3 will progress to invasive cervical cancer from an individual perspective. In fact, some patients may already have occult cervical cancer when diagnosed with HSIL. The rate of progression to invasive cancer after conization have been reported to be about 0.3%-15% (26–28). In our study, although none of the CIN2 cases progressed to invasive cancer, 36% of the women within the afore mentioned group did progress to CIN3, and progression to invasive cancer in the CIN3 group was as high as 20%. Relying on HPV testing alone cannot accurately predict the progression of cancer satisfactorily. A better prognostic risk evaluation for CIN2 and CIN3 is needed. The integration of molecular markers in cervical cancer screening, such as DNA methylation, might help avoid unnecessary referrals and repeatedly performing diagnostic procedures, which is a waste medical resources and generate needless worry for the patient and her family (29).
The PAX1 gene is located on chromosome 20p11 and consists of a paired domain (PD) and an octapeptide domain (OP). The expression PAX1 is associated with embryogenesis, especially the development of the skeleton, thymus, and the parathyroid glands (30, 31). In 2008, Lai et al. first reported that abnormal methylation of PAX1 was associated with cervical cancer, and that the PAX1 gene was found to be silenced by hyper-methylation and under-expressed in cervical cancer biopsies (8). PAX1 can regulate cell division and differentiation, and methylation and silencing of PAX1 is closely related to the progression of precancerous lesions into cervical cancer (32). It has been reported that the disruption between kinases and phosphatases caused by PAX1 methylation is involved in cervical carcinogenesis (33). An increasing number of studies have confirmed PAX1 methylation as a promising biomarker for cervical cancer based on its ability to discriminate between high‐grade cervical lesions and normal tissues, resulting in a reduced necessity for colposcopy referral and biopsy (9, 10, 34). The current study demonstrated that the ΔCpPAX1 level of CIN3 determined by CDB was lower than that of CIN2 and the ΔCpPAX1 level of ESCC was lower than that of both CIN2 and CIN3. Regardless of CIN2 or CIN3 status, the ΔCpPAX1 level of women with upgraded pathology after conization was significantly lower than that of women with degraded pathology. We further stratified the PAX1m level by different thresholds and found that the risk of CIN3+ increased significantly when ΔCpPAX1 < 7. The optimal ΔCpPAX1 cut-off value in predicting whether CIN3 would be upgraded to ESCC, and whether CIN2 would be upgraded to CIN3 after CKC was 6.360 and 10.830, respectively, and is more specific than using with cytology or HPV abnormalities. This concept implies that if biopsy pathology indicates CIN2 status with a ΔCpPAX1 less than 10.83, then the actual pathology is more likely to be upgraded to CIN3, so it is necessary to be more careful if observation without treatment was selected. If biopsy pathology indicates CIN3 with a ΔCpPAX1 less than 6.36, then cervical conization is inevitable because of the increased risk of pathological upgrade to early-stage cervical cancer. On the other hand, women with negative PAX1 methylation do not need immediate colposcopy or conization because of there being a relatively low short-term progression risk for cancer. For young women with CIN2 who have fertility requirements, this approach seems to be particularly important, since only hypermethylated lesions require treatment and the risk of preterm abortion due to treatment could be reduced.
This study has several limitations. First, since our research has not reached the follow-up endpoint, residual and recurrence of lesions have not been discussed here, and continued follow-up is needed in the future. Secondly, the sample size was not large enough and a larger longitudinal study is necessary to validate the natural history of CIN2 and CIN3 progression in relation to DNA methylation. Thirdly, only hospitalized patients with CKC were included in this study who can be followed up well, however, those patients for LEEP from outpatient were not included due to unstable follow-up. In addition, further studies are needed to explore PAX1m levels after treatment and to compare PAX1m changes before and after CKC. At last, further studies are also needed to determine the ideal interval of monitoring using PAX1m to avoid underdiagnosis and overdiagnosis.
In this exploratory study, we found that PAX1 methylation could be a promising auxiliary marker in the prediction of pathological upgrade risk in patients with CIN2 or CIN3 before conization, especially if △CpPAX1 cut-off value is lower than 6.360, as we found this to be highly suggestive of invasive cervical cancer. Using PAX1 methylation as a monitoring tool could help prevent inappropriate conservative observation or ablation therapy. Further validation and prospective clinical trials are needed to confirm these findings in the future.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Peking University People’s Hospital Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
Data collection, study conception, and study design was performed by ML. Personnel organization, data collection, and the cold knife conization procedure was performed by CZ. Sample collection was performed by YZ, JL, HL, ZT, JW and YG. Supervision of the research program and manuscript review and guidance during the study was provided by LW. The first draft of the manuscript was written by ML and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
This work was supported by The National Key Research and Development Program of China (2021YFC2701200) (2021YFC2701202) and Peking University People’s Hospital Scientific Research Development Funds (RDL2020-02) (RDL2022-34).
Thanks to Hong Tao for statistics and interpretation of data, and Ching-Tung Yeh as well as Yu-ligh Liou for giving guidance and advice on statistics and interpretation of data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1064722/full#supplementary-material
Supplementary Figure 1 | Restricted cubic splines of odds ratio for CIN3+CKC according to ΔCpPAX1 levels in all participants.
1. World Health Organization. Female genital tumours. IARCpress. WHO classification of tumours. 5th Edition Volume 4[EB/OL]. (2020-09-09)[2021-01-25]. Available at: https://tumourclassification.iarc.who.int/9789283245049.
2. Tainio K, Athanasiou A, Tikkinen K, Aaltonen R, Cardenas J, HernandesGlazer-Livson S, et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: systematic review and meta-analysis. BMJ (2018) 360:k499. doi: 10.1136/bmj.k499
3. Jung Y, Lee AR, Lee SJ, Lee YS, Park DC, Park EK. Clinical factors that affect diagnostic discrepancy between colposcopically directed biopsies and loop electrosurgical excision procedure conization of the uterine cervix. Obstet Gynecol Sci (2018) 61(4):477–88. doi: 10.5468/ogs.2018.61.4.477
4. Louvanto K, Aro K, Nedjai B, Butzow R, Jakobsson M, Kalliala I, et al. Methylation in predicting progression of untreated high-grade cervical intraepithelial neoplasia. Clin Infect Dis (2020) 70(12):2582–90. doi: 10.1093/cid/ciz677
5. Guan P, Howell-Jones R, Li N, Bruni L, de Sanjose S, Franceschi S, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer (2012) 131(10):2349–59. doi: 10.1002/ijc.27485
6. Moscicki AB, Ma Y, Wibbelsman C, Darragh TM, Powers A, Farhat S, et al. Rate of and risks for regression of cervical intraepithelial neoplasia 2 in adolescents and young women. Obstet Gynecol (2010) 116(6):1373–80. doi: 10.1097/AOG.0b013e3181fe777f
7. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer (2014) 14(6):395–405. doi: 10.1038/nrc3728
8. Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer (2008) 123(1):161–7. doi: 10.1002/ijc.23519
9. Luan T, Hua Q, Liu X, Xu P, Gu Y, Qian H, et al. PAX1 methylation as a potential biomarker to predict the progression of cervical intraepithelial neoplasia: A meta-analysis of related studies. Int J Gynecol Cancer (2017) 27(7):1480–8. doi: 10.1097/IGC.0000000000001011
10. Xu J, Xu L, Yang B, Wang L, Lin X, Tu H. Assessing methylation status of PAX1 in cervical scrapings, as a novel diagnostic and predictive biomarker, was closely related to screen cervical cancer. Int J Clin Exp Pathol (2015) 8(2):1674–81.
11. Li SR, Wang ZM, Wang YH, Wang XB, Zhao JQ, Xue HB, et al. Value of PAX1 methylation analysis by MS-HRM in the triage of atypical squamous cells of undetermined significance. Asian Pac J Cancer Prev (2015) 16(14):5843–6. doi: 10.7314/apjcp.2015.16.14.5843
12. Chang CL, Ho SC, Su YF, Juan YC, Huang CY, Chao AS, et al. DNA Methylation marker for the triage of hrHPV positive women in cervical cancer screening: Real-world evidence in Taiwan. Gynecol Oncol (2021) 161(2):429–35. doi: 10.1016/j.ygyno.2021.02.011
13. Li X, Zhou X, Zeng M, Zhou Y, Zhang Y, Liou YL, et al. Methylation of PAX1 gene promoter in the prediction of concurrent chemo-radiotherapy efficacy in cervical cancer. J Cancer (2021) 12(17):5136–43. doi: 10.7150/jca.57460
14. Zhao Z, Zhang X, Zhao X, Cai J, Wu NY, Wang J. SOX1 and PAX1 are hypermethylated in cervical adenocarcinoma and associated with better prognosis. BioMed Res Int (2020) 2020:3981529. doi: 10.1155/2020/3981529
15. Fu K, Lei M, Wu LS, Shi JC, Yang SY, Yang WQ, et al. Triage by PAX1 and ZNF582 methylation in women with cervical intraepithelial neoplasia grade 3: A multicenter case-control study. Open Forum Infect Dis (2022) 9(5):c13. doi: 10.1093/ofid/ofac013
16. Kan YY, Liou YL, Wang HJ, Chen CY, Sung LC, Chang CF, et al. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer (2014) 24(5):928–34. doi: 10.1097/IGC.0000000000000155
17. Wentzensen N, Massad LS, Mayeaux EJ, Khan MJ, Waxman AG, Einstein MH, et al. Evidence-based consensus recommendations for colposcopy practice for cervical cancer prevention in the united states. J Low Genit Tract Dis (2017) 21(4):216–22. doi: 10.1097/LGT.0000000000000322
18. Bornstein J, Bentley J, Bosze P, Girardi F, Haefner H, Menton M, et al. 2011 colposcopic terminology of the international federation for cervical pathology and colposcopy. Obstet Gynecol (2012) 120(1):166–72. doi: 10.1097/AOG.0b013e318254f90c
19. Darragh TM, Colgan TJ, Cox JT, Heller DS, Henry MR, Luff RD, et al. The Lower Anogenital SquamousTerminology standardization project for HPV-associated lesions:background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy andCervical Pathology. J Low Genit Tract Dis (2012) 16:20542.
20. Mercaldo ND, Lau KF, Zhou XH. Confidence intervals for predictive values with an emphasis to case-control studies. Stat Med (2007) 26(10):2170–83. doi: 10.1002/sim.2677
21. Xiao FY, Wang Q, Zheng RL, Chen M, Su TT, Sui L. [Diagnosis and treatment value of colposcopy and loop electrosurgical excision procedure in microinvasive cervical cancer: analysis of 135 cases]. Zhonghua Fu Chan Ke Za Zhi (2016) 51(3):186–91. doi: 10.3760/cma.j.issn.0529-567X.2016.03.005
22. Kyrgiou M, Athanasiou A, Kalliala I, Paraskevaidi M, Mitra A, Martin-Hirsch PP, et al. Obstetric outcomes after conservative treatment for cervical intraepithelial lesions and early invasive disease. Cochrane Database Syst Rev (2017) 11(11):CD012847. doi: 10.1002/14651858.CD012847
23. Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, et al. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet Gynecol (2013) 121(4):829–46. doi: 10.1097/AOG.0b013e3182883a34
24. Perkins RB, Guido RS, Castle PE, Chelmow D, Einstein MH, Garcia F, et al. 2019 ASCCP risk-based management consensus guidelines for abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis (2020) 24(2):102–31. doi: 10.1097/LGT.0000000000000525
25. Carreon JD, Sherman ME, Guillen D, Solomon D, Herrero R, Jeronimo J, et al. CIN2 is a much less reproducible and less valid diagnosis than CIN3: results from a histological review of population-based cervical samples. Int J Gynecol Pathol (2007) 26(4):441–6. doi: 10.1097/pgp.0b013e31805152ab
26. McIndoe WA, McLean MR, Jones RW, Mullins PR. The invasive potential of carcinoma in situ of the cervix. Obstet Gynecol (1984) 64(4):451–8.
27. Milojkovic M. Residual and recurrent lesions after conization for cervical intraepithelial neoplasia grade 3. Int J Gynaecol Obstet (2002) 76(1):49–53. doi: 10.1016/s0020-7292(01)00523-9
28. Temkin SM, Hellmann M, Lee YC, Abulafia O. Dysplastic endocervical curettings: a predictor of cervical squamous cell carcinoma. Am J Obstet Gynecol (2007) 196(5):461–9. doi: 10.1016/j.ajog.2006.11.018
29. Lai HC, Ou YC, Chen TC, Huang HJ, Cheng YM, Chen CH, et al. PAX1/SOX1 DNA methylation and cervical neoplasia detection: a Taiwanese gynecologic oncology group (TGOG) study. Cancer Med (2014) 3(4):1062–74. doi: 10.1002/cam4.253
30. Paixao-Cortes VR, Salzano FM, Bortolini MC. Origins and evolvability of the PAX family. Semin Cell Dev Biol (2015) 44:64–74. doi: 10.1016/j.semcdb.2015.08.014
31. Yamazaki Y, Urrutia R, Franco LM, Giliani S, Zhang K, Alazami AM, et al. PAX1 is essential for development and function of the human thymus. Sci Immunol (2020) 5(44):eaax1036. doi: 10.1126/sciimmunol.aax1036
32. Da SM, De Albuquerque B, Allyrio T, De Almeida VD, Cobucci R, Bezerra FL, et al. The role of HPV-induced epigenetic changes in cervical carcinogenesis (Review). BioMed Rep (2021) 15(1):60. doi: 10.3892/br.2021.1436
33. Su PH, Lai HC, Huang RL, Chen LY, Wang YC, Wu TI, et al. Paired box-1 (PAX1) activates multiple phosphatases and inhibits kinase cascades in cervical cancer. Sci Rep (2019) 9(1):9195. doi: 10.1038/s41598-019-45477-5
Keywords: cervical intraepithelial neoplasia, cold knife conization, PAX1, methylation, pathological upgrade
Citation: Li M, Zhao C, Zhao Y, Li J, Wang J, Luo H, Tang Z, Guo Y and Wei L (2023) The role of PAX1 methylation in predicting the pathological upgrade of cervical intraepithelial neoplasia before cold knife conization. Front. Oncol. 12:1064722. doi: 10.3389/fonc.2022.1064722
Received: 08 October 2022; Accepted: 22 December 2022;
Published: 11 January 2023.
Edited by:
Gabriella Lillsunde Larsson, Örebro University, SwedenReviewed by:
Stephanie M. McGregor, University of Wisconsin-Madison, United StatesCopyright © 2023 Li, Zhao, Zhao, Li, Wang, Luo, Tang, Guo and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihui Wei, d2VpbGhAYmptdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.