
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 09 December 2022
Sec. Radiation Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1061024
This article is part of the Research Topic Interpretable and Explainable Machine Learning Models in Oncology View all 6 articles
 Yi Luo1*
Yi Luo1* Kyle C. Cuneo2
Kyle C. Cuneo2 Theodore S. Lawrence2
Theodore S. Lawrence2 Martha M. Matuszak2
Martha M. Matuszak2 Laura A. Dawson3
Laura A. Dawson3 Dipesh Niraula1
Dipesh Niraula1 Randall K. Ten Haken2
Randall K. Ten Haken2 Issam El Naqa1
Issam El Naqa1Background: Imbalanced outcome is one of common characteristics of oncology datasets. Current machine learning approaches have limitation in learning from such datasets. Here, we propose to resolve this problem by utilizing a human-in-the-loop (HITL) approach, which we hypothesize will also lead to more accurate and explainable outcome prediction models.
Methods: A total of 119 HCC patients with 163 tumors were used in the study. 81 patients with 104 tumors from the University of Michigan Hospital treated with SBRT were considered as a discovery dataset for radiation outcomes model building. The external testing dataset included 59 tumors from 38 patients with SBRT from Princess Margaret Hospital. In the discovery dataset, 100 tumors from 77 patients had local control (LC) (96% of 104 tumors) and 23 patients had at least one grade increment of ALBI (I-ALBI) during six-month follow up (28% of 81 patients). Each patient had a total of 110 features, where 15 or 20 features were identified by physicians as expert knowledge features (EKFs) for LC or I-ALBI prediction. We proposed a HITL based Bayesian network (HITL-BN) approach to enhance the capability of selecting important features from imbalanced data in terms of accuracy and explainability through humans’ participation by integrating feature importance ranking and Markov blanket algorithms. A pure data-driven Bayesian network (PD-BN) method was applied to the same discovery dataset of HCC patients as a benchmark.
Results: In the training and testing phases, the areas under receiver operating characteristic curves of the HITL-BN models for LC or I-ALBI prediction during SBRT are 0.85 (95% confidence interval: 0.75-0.95) or 0.89 (0.81-0.95) and 0.77 or 0.78, respectively. They significantly outperformed the during-treatment PD-BN model in predicting LC or I-ALBI based on the discovery cross-validation and testing datasets from the Delong tests.
Conclusion: By allowing the human expert to be part of the model building process, the HITL-BN approach yielded significantly improved accuracy as well as better explainability when dealing with imbalanced outcomes in the prediction of post-SBRT treatment response of HCC patients when compared to the PD-BN method.
Hepatocellular cancer (HCC) is the third leading cause of cancer death worldwide. In 2020, the American Society of Clinical Oncology (ASCO) estimated that 830,180 people around the world died from the disease. While radiotherapy is designed to achieve tumor local control (LC) in HCC patients, it may also lead to radiation-induced toxicities (RITs). As a relatively newer radiation treatment technique, stereotactic body radiation therapy (SBRT) uses focused beams of radiation aimed at the tumor from many different angles given in one to five treatment fractions. Thus, the aim of SBRT is to cure tumors in the meanwhile decreasing the radiation to nearby healthy tissues. While it is more effective for tumor LC and RITs reduction compared to conventional approaches, stringent dose volume constraints of SBRT require the treatment planning to be highly personalized to meet its intended goals (1).
In HCC SBRT, LC can be evaluated radiologically from a lesion that is no longer arterially enhancing and has not spread to neighboring lymph nodes without any failures within the irradiated area over long-term follow-up (2). The impact of RITs to baseline liver function of HCC patients before and after SBRT can be evaluated by albumin-bilirubin (ALBI) grades for personalized standard or adaptive implementation (3–5). Specifically, physicians are concerned whether patients’ ALBI grades will increase at least by one grade or not during 6-month follow-up, which is denoted as I-ALBI. Thus, we considered I-ALBI as another relevant SBRT outcome in addition to tumor LC in this study. The literature on outcomes prediction models for HCC patients with SBRT and their explainability capability remains limited and challenging (6, 7). The purpose of this study is to fill these gaps by developing accurate and explainable LC or I-ALBI prediction models for HCC patients with SBRT.
In clinical practice, oncology datasets usually have high dimensional features with limited sample size making susceptible to spurious correlations including the Simpson paradox (8). The dataset of HCC patients with SBRT in this study is not an exception. Machine learning (ML) is defined as the task of extracting information from possibly high-dimensional and noisy data to give some guarantees of performance on unseen data. However, extracting the structure based on the proximity between empirical and population densities becomes challenging in the higher dimensions, since the distance between objects may be heavily dominated by noise, and the associated optimization process has an exponential dependency on these dimensions (9). Then, feature selection is designed to help conventional ML approaches handle high-dimensional datasets. For example, in our previous study on personalized adaptive radiotherapy for non-small-cell lung cancer patients, a pure data-driven Bayesian network (PD-BN) approach is developed including feature selection and BN structure building two steps. While Markov blanket (MB) algorithms were employed in the first step to identify the most important features from high-dimensional oncology datasets, Tabu Search was used in the second step to learn network structure based on the selected features. In addition to unraveling the biophysical relationships among lung cancer patients’ personal characteristics, radiation treatment, and outcomes, the PD-BNs can predict lung tumor LC or/and RITs and identify the best treatment strategies before and during the radiotherapy to improve patients’ therapeutic satisfaction (10–12).
Initially proposed by Pearl (13), the concept of variable X’s MB is to identify its optimal feature subset containing strongly relevant and non-redundant features, such as the variable’s parents, children, and spouses as shown in the shadow area of Figure 1. Given these features in the subset, the variable is independent to other features outside it. Due to its capability of fully explaining a target variable, the MB has the potential of selecting the features that have strong relevance to an outcome for building its prediction models. Then the MB algorithms such as incremental association MB (14) and its variants (15) were successfully employed in the feature selection process of our previous PD-BN approach to develop accurate and interpretable outcome prediction models.
However, in addition to high dimensional features with limited sample size, oncology datasets usually have imbalanced outcomes, such as HCC patients’ LC or I-ALBI in this study. The prediction of these treatment outcomes can be modeled as a binary classification problem under supervised machine learning. Class imbalance occurs when the minority group, such as non-LC or I-ALBI, contains significantly fewer events samples than the majority group, such as LC or non-I-ALBI. Learning these imbalanced outcomes from high-dimensional datasets can be very difficult (15, 16), and non-standard machine learning methods are often guaranteed to achieve desirable results (14). Moreover, features selected from the above theoretically sound MB algorithms to have a strong relevance with an outcome may not be able to build the BN-based outcome prediction model with high accuracy, since accuracy and explainability are two different criteria for feature selection (17). Then, the PD-BN based outcome prediction models have a limited prediction performance in this case. Furthermore, the developed PD-BNs are not necessarily following physicians’ common practice knowledge, and unconfirmed biophysical interactions explored from the PD-BN approach can barely gain physicians’ trust for application in routine clinical decision making. Therefore, the goal of this study is to develop a new ML approach in handling imbalanced oncology data to improve the accuracy and explainability of predicting HCC patients’ outcomes with SBRT.
Building accurate and explainable outcome prediction models from high-dimensional imbalanced data is a complex process that requires nontrivial understanding of complex ML algorithms (18). Humans are typically involved in unstructured manner at various points in the processes of the model development, model training, and testing of the underlying ML algorithm implementation. Human-in-the-loop ML (HITL-ML) approaches are proposed to rather define a new type of structured interactions between humans and machine learning algorithms. Being developed initially from reinforcement learning, preference learning, and active learning, the HITL-ML is a hybrid of data-driven and knowledge-driven approach that integrates a priori expert knowledge (EK) into ML frameworks to overcome issues related to model bias and uncertainty (19). In addition to making ML more accurate or to obtain the desired accuracy faster, the HITL-ML approach makes humans more effective and efficient (18). Especially, it is useful in handling imbalanced data (20, 21). Due to the transparency of the BN for potential clinical causal inferences, in this study we develop an HITL-BN approach to build HCC SBRT outcome prediction models from imbalanced oncology data by incorporating EK features and allowing human agents to participate in the BN feature selection process. The accuracy and explainability of HITL-BN based outcome prediction models are evaluated and compared to the PD-BN based models that do not involve human agents.
The rest of paper is organized as follows. Section 2 introduces the properties of our dataset and the details of the HITL-BN approach. Section 3 shows and compares outcome prediction models developed from the PD-BN and HITL-BN approaches. Section 4 discusses the accuracy and explainability of our new approach and verifies the relationships among biophysical features in developed HITL-BNs based on related literatures. Section 5 concludes our paper.
Our study uses 81 HCC patients with SBRT on prospective protocols under institutional review board (IRB) approval from University of Michigan Hospital (Michigan Medicine). Since each patient may have one or more tumors, there are totally 104 tumors in our discovery dataset. In this study, two or more tumors in an HCC patient are assumed to be independent from each other for the sake of simplicity. There are 23 patients with I-ALBI during six-month follow up, and 100 tumors from 77 patients achieved LC. Each patient has 97 features, including dosimetric information, clinical factors, pre- and during-treatment labs and cytokines as summarized in Table 1. The change of a lab or cytokine value during treatment was calculated from the difference between its post treatment (or three months after treatment) and pre-treatment (or baseline) values, and it is formulated by adding prefix “D_” to its name in our study. To avoid confusion in outcome prediction, biophysical features related to LC or I-ALBI were specified and manually designated by a human expert. For example, “gross tumor volume (GTV)” is considered for predicting LC instead of I-ALBI. The number of features for LC or I-ALBI prediction before and during SBRT are listed in Table 2. For independent external validation, we tested our developed models on a dataset from the Princess Margaret Hospital, which included 59 tumors from 38 HCC patients.

Table 2 The number of features associated with each of 104 tumors before and during treatment for I-ALBI or LC prediction.
Physicians’ acquired knowledge and experience from treating HCC SBRT patients were collected and treated as expert knowledge (EK) for SBRT outcomes prediction in this study. The EK features (EKFs) related to I-ALBI prediction comprise “LIVER_GTV_Mean_Dose”, “pre_ICGR15”, “D_ICGR15”, “Child_Pugh”, “Cirrhosis”, “Liver_GTV_Volume”, “pre_HGF”, “D_HGF”, “Age”, “Sex”, “pre_CD40_L”, “D_CD40_L”. The EKFs for LC prediction include “Child_Pugh”, “GTV”, “Total_BED”, “LIVER_GTV_Mean_Dose”, “Prior_Liver_Occurrences”, “pre_MELD”, “pre_Bilirubin”, “pre_Albumin”, “Tumor_Size”, “GTV_Mean_Dose_LQ”, “pre_ICGR15”, “pre_HGF”, and “pre_TGF_Beta”. Except the above EKFs related to two different outcomes, the rest of features in Table 1 were denoted as non-EK features (non-EKFs) in this study.
Gained from many years of experience, reading articles, training, peers’ interaction, EK has a potential of bypassing otherwise complex systems and providing parsimonious solutions that focus on key aspects of a given situation. By incorporating EKFs into the feature selection of the PD-BN approach, we previously developed a situational awareness BN (SA-BN) method to predict the radiation outcomes of lung cancer patients (29). With the SA-BNs, the physicians not only are able to know patients’ situation and predict LC and potential RITs starting from their acquired knowledge, but also can evaluate the best treatment strategies to maximize the LC and minimize the RITs before and during the course of radiotherapy. Focusing on improving the PD-BN based models’ explainability, the SA-BN method has limitations in alleviating the impact of high-dimensional imbalanced data on the PD-BN approach’s feature selection process to improve the accuracy of its associated outcome prediction models (29). However, the tighter confidence intervals of prediction performance and well-known biophysical relationships in the SA-BNs indicated that the EK has potential to improve the accuracy and explainability of outcome prediction models. Then, the EK methodology was employed in the HITL-BN approach to guide feature selection from imbalanced HCC SBRT data.
Selecting an ML approach for outcome prediction often involves a trade-off between prediction accuracy and explainability (30). While some ML approaches may lead to relatively more accurate outcome prediction models, other ML methods can result in more explainable ones. According to the explainability of their associated outcome prediction models, the ML approaches can be generally classified into explainable ML (EML) and unexplainable ML (UML) methods. The former includes Decision Trees, Logistic Regression and its variants, Naïve BNs, BNs, etc., and the latter comprises Random Forests (RFs), Support Vector Machines (SVMs), Gradient Boosting Machines (GBMs), Deep Learning (DL), etc. Although the EML-based outcome prediction models generally have relatively lower prediction accuracy compared to the UML-based models, they can be used to identify the most relevant features in explaining an outcome. On the other hand, while the UML-based outcome prediction models have difficulties in interpreting the relationships between specific features and the outcome, a list of ranked features can be generated from each of them based on features’ importance in terms of outcome prediction (31). However, the ranking lists generated from different UML approaches may not be the same, resulting in different important features selected from the top rank of these lists for outcome prediction. An integrated feature ranking list is developed in this study by combining these lists based on the performance of its associated UML-based outcome prediction models to achieve robust feature selection as introduced in the next section.
The selected features from the EML and UML approaches are generally different, even though they are evaluated from one single dataset. The former and latter have the potential of improving an outcome prediction model’s explainability and accuracy respectively. While the MB algorithm and network structure learning were considered as a computational agent to improve the prediction model’s explainability by exploring EKFs and non-EKFs that have strong relevance to an outcome, the integrated feature ranking list was treated as another computational agent to enhance the prediction model’s accuracy by investigating each feature’s importance in terms of outcome prediction. Then, the HITL-BN approach can improve its capability of learning from the imbalanced HCC SBRT data by allowing human agents to interact these two computational agents during the process of feature selection.
As stated previously, the UML approaches include RF, SVM, GBM, DL, etc., and each of them can generate a feature ranking list in terms of importance in outcome prediction from all features including EKFs and non-EKFs in a dataset. Let K be the total kinds of these UML approaches, k be the index of these approaches (k =1, 2, 3, …, K), Lk be a feature ranking list obtained from UML approach k (k =1, 2, 3, …, K) with the most important feature for outcome prediction at the top of the list, and AUCk be the performance of an outcome prediction model developed from UML approach k based on cross validation in the discovery dataset (k =1, 2, 3, …, K). Let J be the total number of features in the discovery dataset, j be the index randomly assigned to them (j = 1, 2, 3, …, J), Nj(Lk) be the rank of feature j in ranking list Lk (j = 1, 2, 3, …, J, k =1, 2, 3, …, K). The rankings Nj(Lk) of the feature in different lists Lk may not be the same, and the performance AUCk of UML approaches for outcome prediction could be different. It is assumed that a robust feature ranking list can be developed by integrating all these ranking lists based on their corresponding UML approaches’ prediction performance. Let L* be an integrated feature ranking list based on K UML approaches, be the weighted ranking score (WRS) of feature j to determine its ranking in list L* , and its value can be evaluated from the following equation by integrating its ranks Nj(Lk) in different ranking lists Lk
Then, the ranking list L* in terms of features’ importance in outcome prediction can be obtained from sorting all the features based on their WRSj , where the feature with the minimal score value is ranked at the top of the list.
Including feature selection and BN structure learning processes, HITL-BN based outcome prediction models are mainly developed based on the integrated ranking list. Let I be the total number of EKFs in list L* with I<J , i represent the order of an EKF within all EKFs (i = 1, 2, 3, …, I). An initial HITL-BN is developed from the top n percent of features in the list. The value of n depends on the total number N of features in a dataset and appropriate feature dimension D to satisfy the MB algorithms’ faithfulness assumption, and we assumed n = 100* . Suppose the top n percent of features in list includes i EKFs (i≤D ), an initial HITL-BN based outcome prediction model can be denoted as HITL-BN(i). Since some EKFs in the top rank of list L* may be redundant or less relevance to an outcome compared to other ones, the most relevant EKFs can be identified from the outcome’s MB. Given the selected EKFs, important non-EKFs in the top rank of the list to improve the outcome prediction should also be strongly related to these EKFs, which can be identified from each of their MBs. Thus, important EKFs and non-EKFs can be selected from the top rank of the list with balanced accuracy and explainability for HITL-BN(i) development. Note that the structure learning of HITL-BN(i) is the same as that of PD-BNs, where less important EKFs and non-EKFs are eliminated from the network to maximize its prediction performance. The rest of our HITL-BN approach is to repeatedly evaluate whether a next EKF and additional non-EKFs before it in list L* can improve the accuracy of previous outcome prediction models or not.
Let ri be the rank of i-th EKF in list L* (i =1, 2, 3, …, I). As the evaluation moves from i-th EKF to i+1-th EKF in the integrated feature ranking list, the set of additional indices between them is denoted as ri, i+1 . Let S(ri, i+1) represents the set of non-EKFs associated with ri, i+1 , and the number of non-EKFs in the set could be zero when two EKFs are consecutive in the list. If set S(ri, i+1) is not empty, the importance of these non-EKFs for the outcome prediction depends on whether they have strong relevance with selected EKFs, including the EKFs in HITL-BN(i) and i+1-th EKF. Let MBs (S(ri,i+1) ) be these EKFs’ MBs based on non-EKFs in S(ri,i+1) , and the set of selected non-EKFs from these MBs is indicated as . Then HITL-BN(i+1) can be developed based on i+1-th EKF, non-EKFs in together with all the features in HITL-BN(i) by employing PD-BN’s structure learning process. The process continues along list until the performance of prediction model cannot be improved or meet a target prediction performance. The details of the HITL-BN approach to generate an accurate and explainable outcome prediction model are described in Figure 2.
As a comparison of the HITL-BN approach, PD-BN models for I-ALBI or LC prediction were developed based on our HCC SBRT patients as shown in Figures 3 or 4. Numerical experiments in this study were conducted in an R environment, where function “inter-MB” in R package “bnlearn” was employed as the MB algorithm for feature selection and function “boot.strength” in the same R package was used for BN structure learning. Figures 3A or 3D shows pre- or during-treatment PD-BN model for I-ALBI prediction developed from the discovery dataset. While the PD-BN method selected biophysical features “pre_Bilirubin”, “pre_Cirrhosis”, “Portal_Vein_Thrombosis”, “pre_Creatinie”, “pre_CD40_L”, “pre_HGF”, and “Liver_GTV_DC_LQ_EQD2” for pre-treatment I-ALBI prediction, additional variables “D_Protime_INR”, “D_Bilirubin”, and “D_ICGR15” were chosen for during-treatment I-ALBI prediction. The prediction performances of the former and the latter based on the discovery dataset are 0.78 (95%CI: 0.67-0.83) and 0.82 (95%CI: 0.74-0.88) as described in Figures 3B and 3E respectively. The prediction performance of the former or the latter based on the testing dataset is 0.68 or 0.73 as illustrated by Figures 3C or 3F.
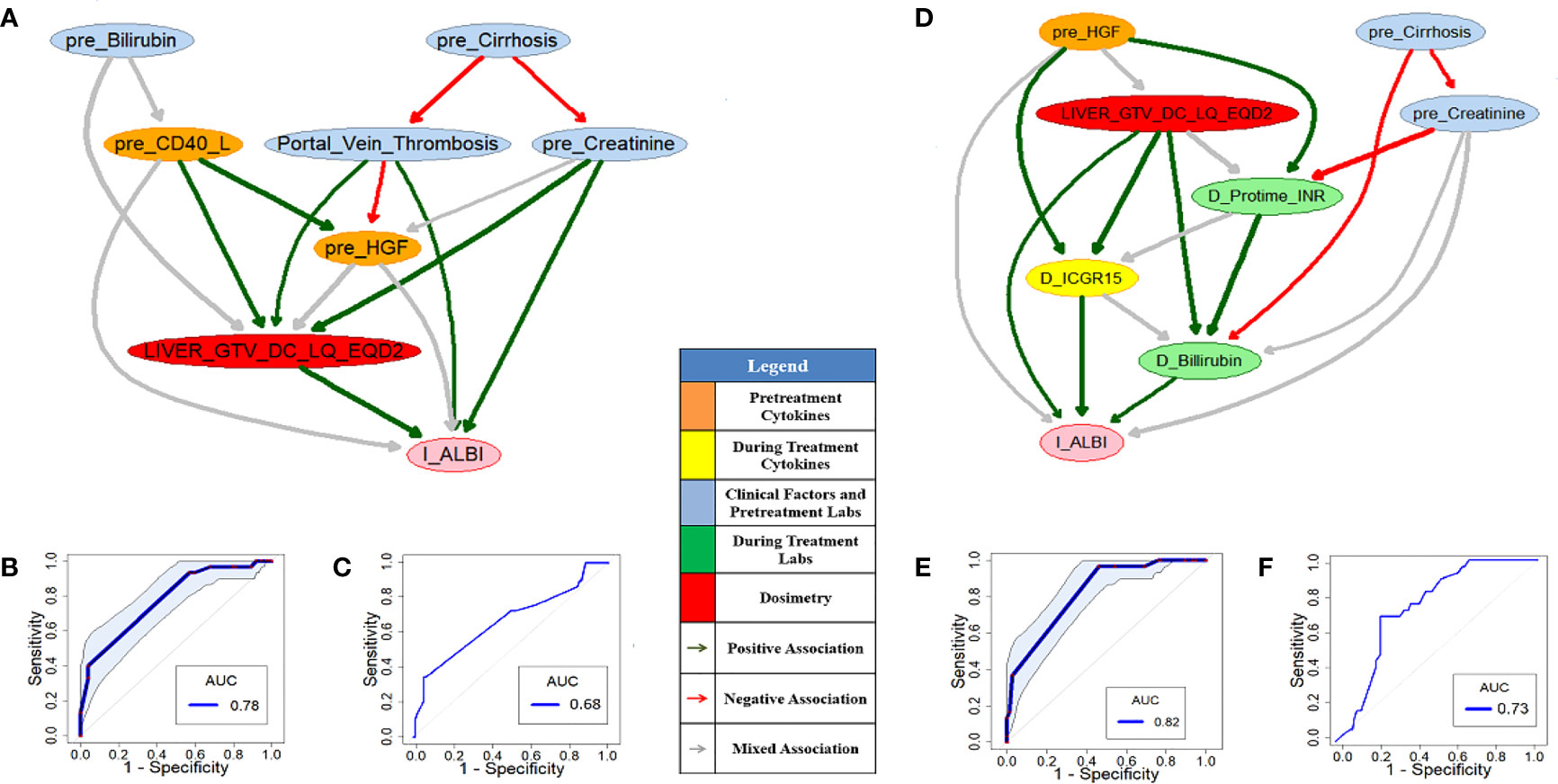
Figure 3 Pre- (A) and during-treatment (D) PD-BNs for I-ALBI prediction. The prediction performance of pre- (B) and during-treatment (E) PD-BNs based on the discovery dataset. The prediction performance of pre- (C) and during-treatment (F) PD-BNs based on the testing dataset.
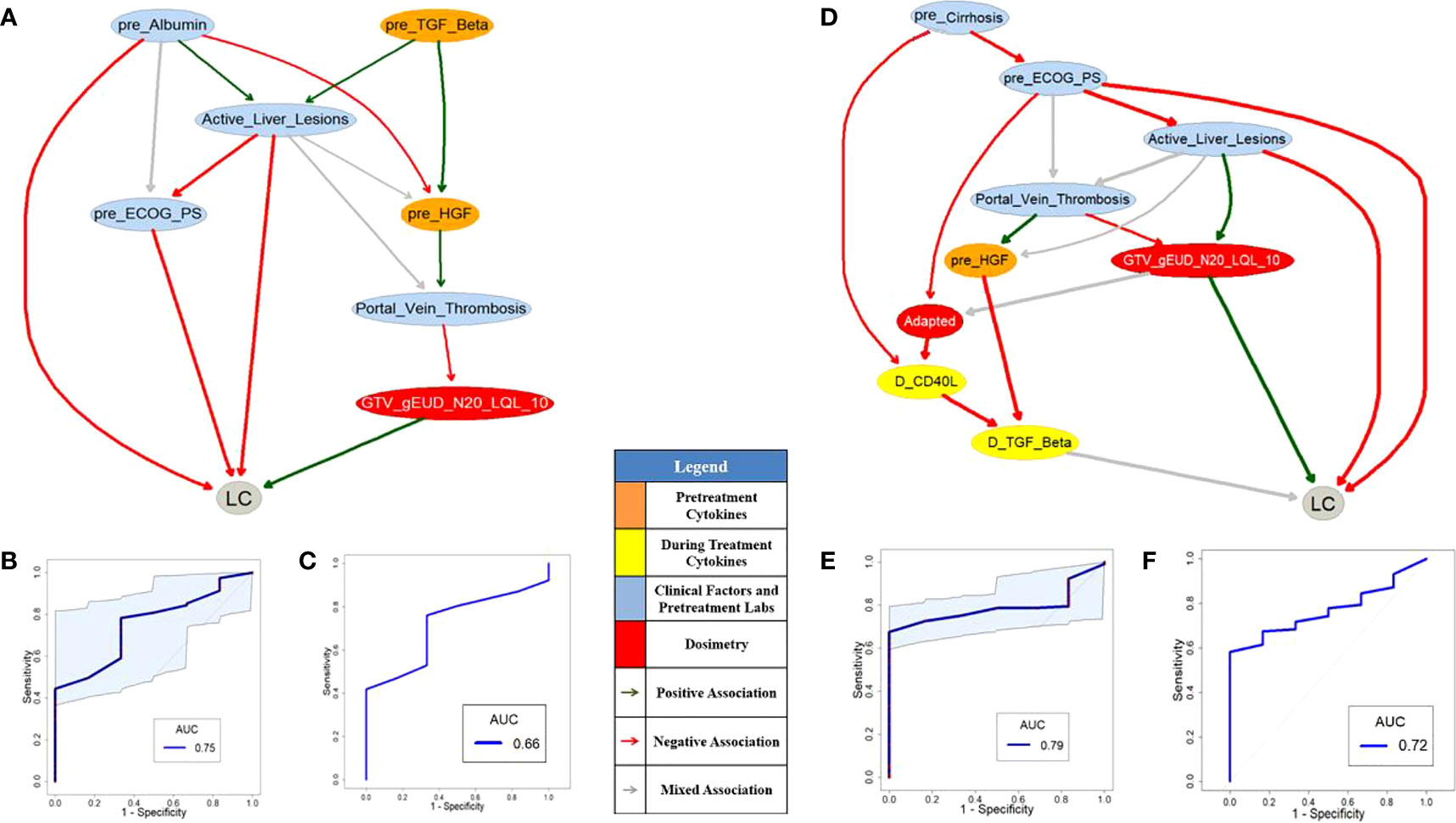
Figure 4 Pre- (A) and during-treatment (D) PD-BNs for LC prediction. The prediction performance of pre- (B) and during-treatment (E) PD-BNs based on the discovery dataset. The prediction performance of pre- (C) and during-treatment (F) PD-BNs based on the testing dataset.
Figures 4A or 4D shows pre- or during-treatment PD-BN for LC prediction generated from the discovery dataset. While the PD-BN method selected features “pre_Albumin”, “Active_Liver_Lesions”, “Portal_Vein_Thrombosis”, “pre_ECOG_PS”, “pre_TGF_Beta”, “pre_HGF”, and “GTV_gEUD_N20_LQL_10” for pre-treatment LC prediction, additional variables “pre_Cirrhosis”, “Adapted”, “D_CD40_L”, and “D_TGF_Beta” were chosen for during-treatment LC prediction. The prediction performances of the former and the latter based on the discovery dataset are 0.75 (95%CI: 0.60-0.86) and 0.79 (95%CI: 0.69-0.89) as shown in Figures 4B and 4E respectively. The prediction performance of the former or the latter based on the testing dataset is 0.66 or 0.72 as illustrated by Figures 4C and 4F.
We conducted numerical experiments to develop or test HITL-BN models for I-ALBI or LC prediction based on the discovery and testing datasets in the same R environment as that of developing or testing the PD-BN models. Two UML approaches, the RF and GBM (K=2), were employed in this study to generate an integrated feature ranking list for a HITL-BN based outcome prediction model development before or during treatment. RF and GBM share similar tree/graph structure learning to BN. Packages ‘randomForestSRC’ and ‘gbm’ were used to identify feature ranking lists from the former and latter approaches based on the discovery dataset respectively. After evaluating the two UML approaches’ prediction performances, each feature’s WRS was computed based on its ranks in two different ranking lists and the corresponding UML approaches’ prediction performances from Equation (1). Then, an integrated feature ranking list to rank all the features in the discovery dataset for I-ALBI or LC prediction before or during SBRT can be generated from their WRSs.
Figures 5A or 5D shows pre- or during-treatment HITL-BN for I-ALBI prediction developed from the discovery dataset. While the HITL-BN approach selected features “Sex”, “Age”, “pre_Na”, “pre_Cirrhosis”, “pre_Alkphos”, “pre_Billirubin”, “pre_ICGR15”, and “LIVER_GTV_DC_LQ_EQD2” for pre-treatment I-ALBI prediction, additional variables “D_MELD”, “D_Albumin”, and “D_ICGR15” were chosen for during-treatment I-ALBI prediction. Tables 3 and 4 show the integrated feature ranking lists of all features according to their WRSs for I-ALBI prediction before and during SBRT respectively. The features in PD-BNs as shown in Figure 3 are highlighted with italic font in these tables, and the features in HITL-BNs as illustrated in Figure 5 are emphasized with bold font in them. Especially, the features marked with italic and bold fonts come from both the PD-BN and HITL-BN.
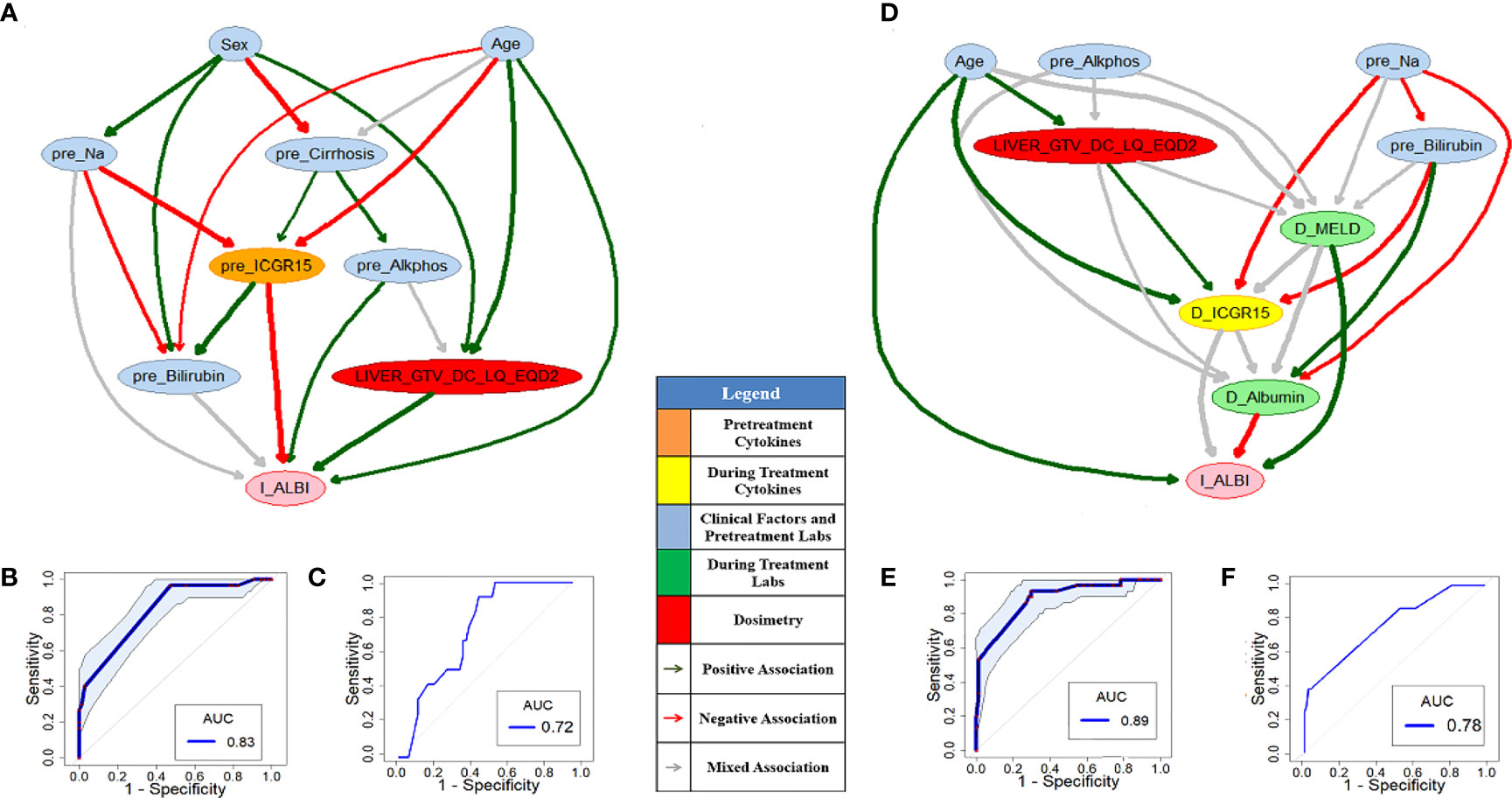
Figure 5 Pre- (A) and during-treatment (D) HITL-BNs for I-ALBI prediction. The prediction performance of pre- (B) and during-treatment (E) HITL-BNs based on the discovery dataset. The prediction performance of pre- (C) and during-treatment (F) HITL-BNs based on the testing dataset.
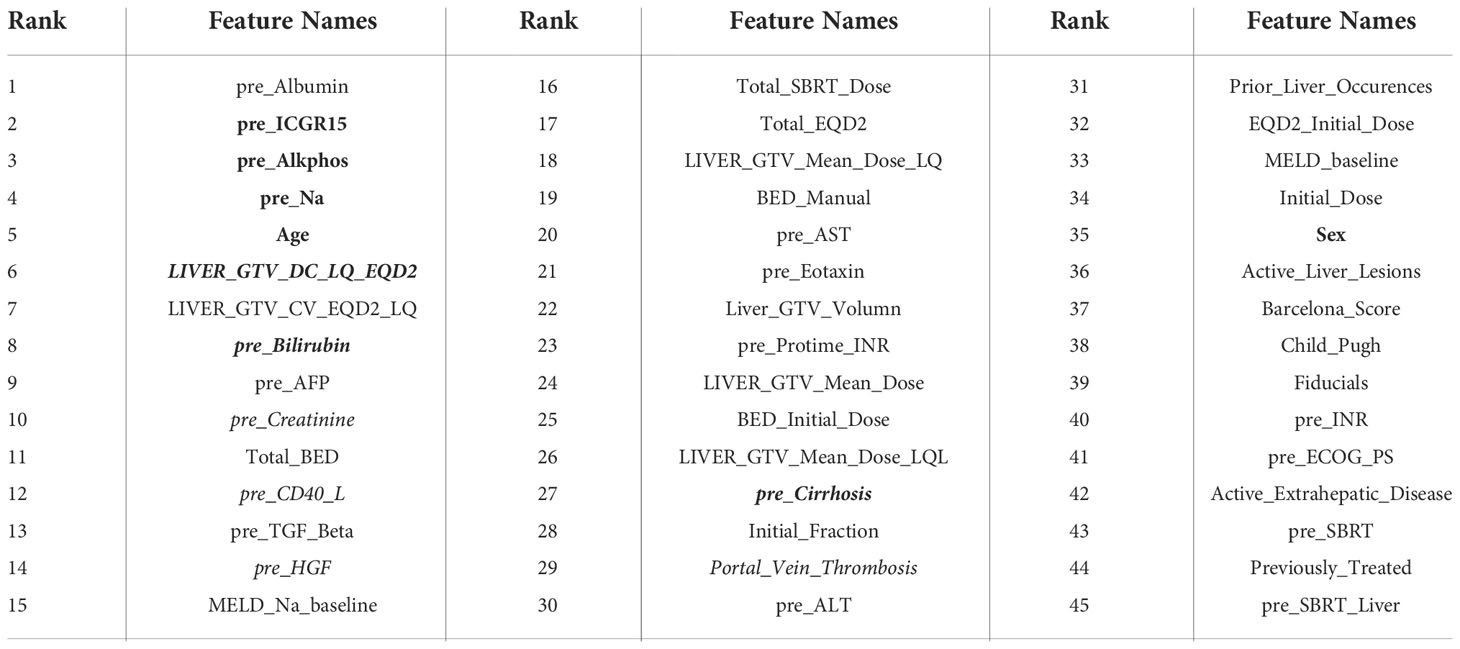
Table 3 The rank of features in an integrated feature ranking list for pre-treatment I-ALBI prediction.

Table 4 The rank of features in an integrated feature ranking list for during-treatment I-ALBI prediction.
The performances AUCs of pre- and during-treatment HITL-BNs for I-ALBI prediction based on the discovery dataset are 0.83 (95%CI: 0.75-0.89) and 0.89 (95%CI: 0.81-0.95) as shown in Figures 5B, E respectively. While the performance of the former is not significantly better than that of pre-treatment PD-BN as illustrated in Figure 3A, the latter significantly outperforms during-treatment PD-BN as shown in Figure 3D based on the DeLong test with p-value=0.0253. For the testing dataset, the performance of pre- or during-treatment HITL-BN for I-ALBI prediction is 0.72 or 0.78 as illustrated by Figures 5C or 5F, and the latter significantly outperforms during-treatment PD-BN from the Delong test with p-value=0.0318.
Figures 6A or 6D shows pre- or during-treatment HITL-BN for LC prediction developed from the discovery dataset. While the HITL-BN for LC prediction approach selected features “Prior_Liver_Occurences”, “GTV”, “MELD_baseline”, “pre_TGF_Beta”, “pre_HGF”, “GTV_gEUD_LQ”, and “LIVER_GTV_Mean_Dose” for pre-treatment LC prediction, additional variables “MELD_Na_baseline”, “pre_Billirubin”, “pre_ICGR15”, “GTV_Mean_Dose_LQ”, “D_Protime_INR”, and “D_TGF_Beta” were chosen for during-treatment LC prediction. Tables 5 and 6 show the ranking lists of all features according to their WRSs for LC prediction before and during SBRT respectively. The features from the PD-BNs as shown in Figure 4 are highlighted with italic font in these tables, and the features from the HITL-BNs are emphasized with bold font in them. Especially, the features marked with italic and bold fonts come from both the PD-BN and HITL-BN.
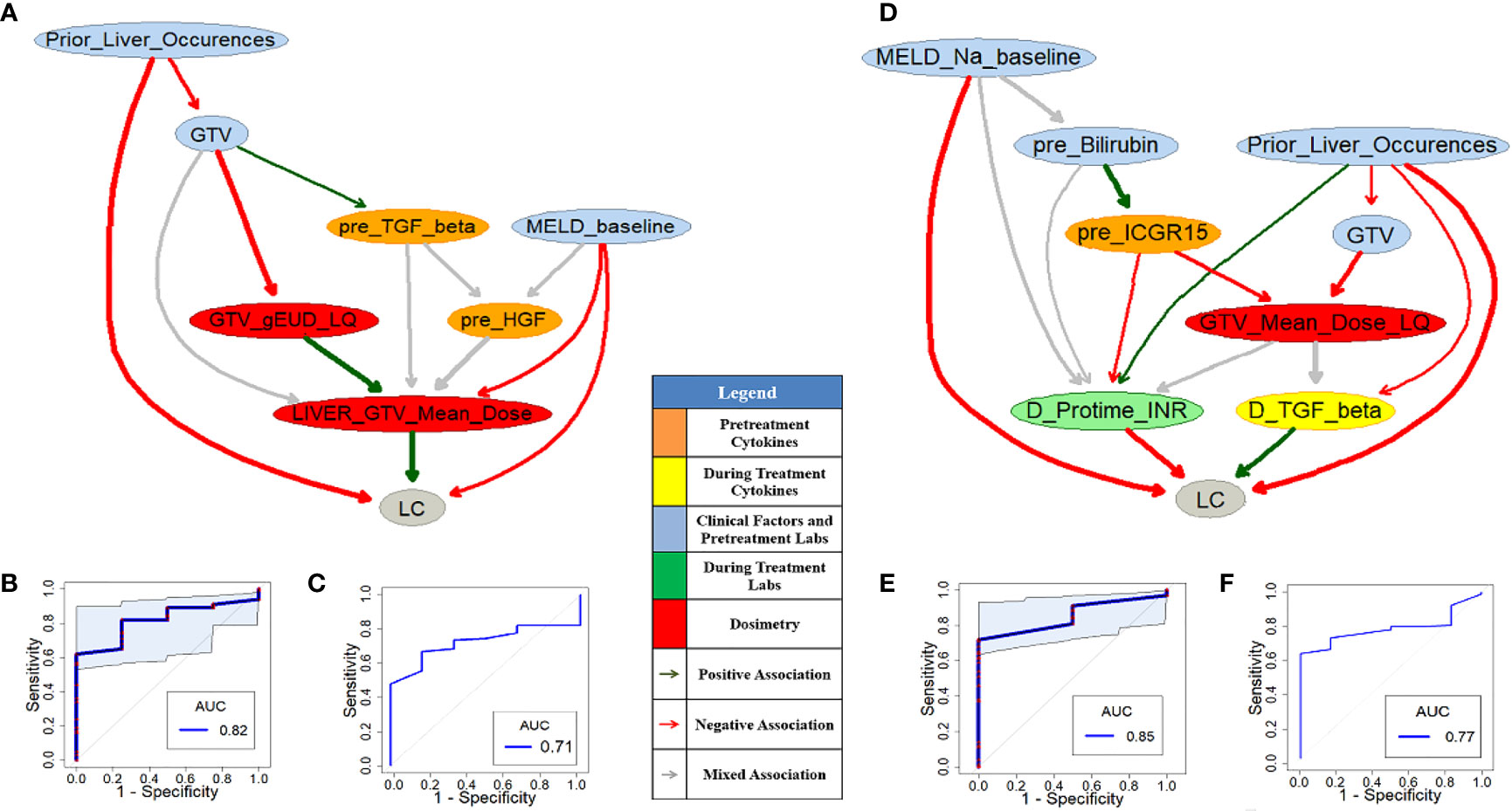
Figure 6 Pre- (A) and during-treatment (D) HITL-BNs for LC prediction. The prediction performance of pre- (B) and during-treatment (E) HITL-BNs based on the discovery dataset. The prediction performance of pre- (C) and during-treatment (F) HITL-BNs based on the testing dataset.
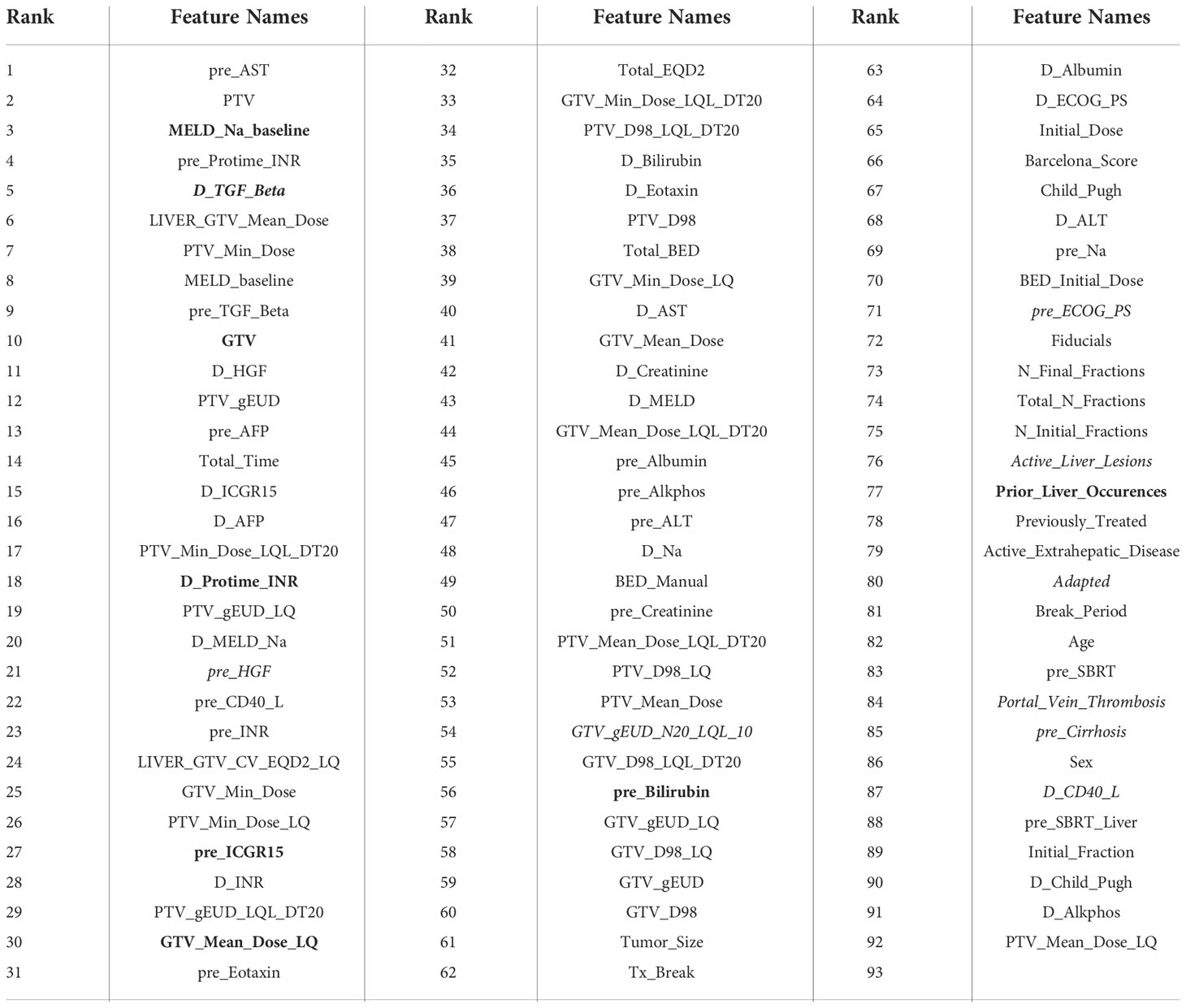
Table 6 The rank of features in an integrated feature ranking list for during-treatment LC prediction.
The performances of pre- and during-treatment HITL-BNs for LC prediction based on the discovery dataset are 0.82 (95%CI: 0.67-0.93) and 0.85 (95%CI: 0.75-0.95) as shown in Figures 6B and 6E and 6E respectively. While the performance of the former is not significantly better than that of pre-treatment PD-BN as illustrated in Figure 4A, the latter significantly outperforms the during-treatment PD-BN as shown in Figure 4D based on the DeLong test with p-value=0.0367. For the testing dataset, the performance of pre- or during-treatment HITL-BN for LC prediction is 0.71 or 0.77 as illustrated by Figure 6C or 6F, and the latter significantly outperforms the during-treatment PD-BN from the Delong test with p-value=0.0406. The results of our numerical experiments are summarized in Table 7.
Developed from our previous PD-BN method, the HITL-BN approach also includes feature selection and BN structure learning. To handle imbalanced data, the HITL-BN approach allows human agents to integrate the EML-based and UML-based feature selections in identifying important EKFs and non-EKFs in terms of outcome prediction. Tables 3 and 4 show that EKFs and non-EKFs obtained from the HITL-BNs (highlighted by bold font) for I-ALBI prediction are generally ranking higher than those from the PD-BNs (emphasized by italic font) before and during SBRT respectively. Also, a similar situation can be found from Tables 5 and 6 for LC prediction. These findings not only echo that the HITL-BNs outperform the PD-BNs for I-ALBI or LC prediction before and during SBRT as shown in Figures 3 and 5 or Figures 4 and 6, but also indicate that the HITL-BN approach can increase the capability of feature selection from imbalanced data. Since the properties of imbalanced outcomes in the testing dataset are not the same as those of the training dataset, the prediction performance of the HITL-BN based outcome prediction models of the former is expectedly less than that of the latter.
The reasons for the improvement of accuracy and explainability of HITL-BN based outcome prediction models in handling the imbalanced proportion of tumors with and without LC or I-ALBI in our HCC SBRT patient dataset could be twofold. First, since traditional ML approaches for crowdsourcing labeled training examples are not effective at locating the scarce minority class examples (32), they have difficulties in handling the high-skewed domain in the real-world, and their associated outcome prediction models may have low accuracy. Active learning is designed to select representative subsets of unlabeled datasets for manual labeling, and an ML algorithm can achieve accuracy with fewer training labels if it is allowed to choose the data from which it learns (14, 33). Originating from active learning, our HITL-BN approach intends to manually label important EKFs and non-EKFs based on their strong relevance to an outcome or/and their importance in the outcome prediction, which is intended to improve the prediction of the imbalanced LC classes and I-ALBI classes. Secondly, while EKFs play an important role in the HITL-BN approach due to its explainability to gain physicians’ trust in clinical decision making, not all of them are ranked at the top of an integrated feature ranking list. They are evenly distributed into the ranking list as shown in Tables 3-6. Only the top-ranked EKFs that are strongly related to the outcome were selected to build an initial HITL-BN. In the meantime, top-ranked non-EKFs have potential to improve the accuracy of the initial HITL-BN model as well. However, given the selected EKFs in the initial model, only the non-EKFs with strong relevance to these EKFs can improve its prediction performance. Our HITL-BN approach is designed to determine the important EKFs or/and non-EKFs from integrating the EML-based and UML-based feature selection methods and maximizing the prediction performance of the developed BNs through feedback. The focused, interactive, incremental process to improve the accuracy and explainability of an outcome prediction model can be considered as an extension of cost-sensitive learning, which is one of traditional methods for class imbalance (14, 34, 35).
As some EKFs may be missing or not available in clinical practice, the HITL-BN approach can skip these EKFs or investigate the EKFs that physicians are most interested in along the integrated ranking list for the outcome prediction model development. The purpose of this study is to verify whether the HITL-BN approach can significantly improve the performance of HCC SBRT patients’ outcome prediction models or not based on imbalanced data compared to the PD-BN method. The HITL-BN approach based on two UML approaches with RF and GBM had been implemented in our numerical experiments to test the hypophysis. Our choices of these two because they resemble BN in terms of graph/tree structures. However, if the number of UML algorithms increases, whether the predictive power of the HITL-BN based outcome prediction models could be improved or not and how much it can be improved are interesting research topics that beyond our current scope and we would like to explore in the next step.
Our numerical experiments on developing the HITL-BN based outcome prediction models for HCC SBRT patients have shown that human intelligence can positively augment machine intelligence, and the assistance of human agents involved in the learning phase can enhance the capability of learning from imbalanced data. However, our study still has limitations in terms of small sample size and the assumption of two or more independent lesions in an HCC patient. In the next steps, in addition to developing more robust HITL-BN approaches by removing the within patient tumor independence assumption and conducting further external independent validations, we plan to explore an interactive human-computer interface via the HITL-BN approach to conduct prospective personalized SBRT trials for improving HCC patients’ radiation treatment outcomes.
In addition to outperforming the PD-BN based outcome prediction models in terms of accuracy, the HITL-BN based outcome prediction models also have a better explainability due to the incorporation of the EKFs in their model buildings. The biophysical pathways displayed in our HITL-BNs for I-ALBI prediction before SBRT are supported by cited literatures. Since a longitudinal increase in the ALBI score is closely associated with non-malignancy-related mortality and quality of life (36), the incorporation of mid-treatment change in ALBI in addition to baseline ALBI improves the ability to predict treatment-related toxicity in patients with HCC receiving SBRT (13). Then change in albumin–bilirubin score (ALBI) score at three months after SBRT were used in many studies to capture acute toxicity occurring <90 days after SBRT (37). Studies showed that repeated SBRT in patients with advanced liver cirrhosis seems to exhibit higher hepatic toxicity (38), and the severity of hepatic cirrhosis is a major prognostic factor for radiation induced liver disease (39). Also, researchers found out that direct total bilirubin and total bilirubin are not related to delivery dose, and age is a significant predictive factor for radiation-induced liver injury based on univariate analysis of clinical factors (39). Moreover, an elevation in alkaline phosphatase (alkphos) of at least 5-fold and/or that of bilirubin of at least 3-fold compared to either the upper normal limit or the pretreatment level corresponding to grade 3 or higher hepatic toxicity without disease progression within 3 months after SBRT is one of the conditions to define radiation-induced liver disease (40).
The following findings from literatures support the biophysical pathways displayed in the HITL-BN for I-ALBI prediction during SBRT. Increasing mean liver dose was associated with larger increases in toxicities (41). As the percentage of retained ICG at 15 minutes, ICGR15’s normal value would be in the range of 4–10% (42). While baseline values of ICGR15 may be associated with the development of radiation induced liver disease, the change of ICGR15 after radiation therapy appears to be most indication of the toxicity (43, 44). There may exist prognostic significance of baseline serum sodium value (pre_Na) in HCC patients complicating with liver cirrhosis, and lower serum sodium concentration is a useful predictor for these patients (45). The time course of changes of the liver function after SBRT was analyzed in patients treated for non-resectable HCC. Albumin was the only blood test that changed systematically during a three-month period, and it stabilized thereafter, which indicates the decrease in albumin reflects a minor radiation-induced liver disease (46). Model for end-stage liver disease (MELD) is a scoring system used to predict three-month mortality in patients with advanced liver disease (47). An increase in MELD score is associated with a decrease in residual liver function or deterioration in liver function (48).
Moreover, our HITL-BNs to predict LC before and during SBRT are endorsed by the following recorded observations. Higher treatment dose was associated with improved freedom from local progression (41) (49). Larger GTV volume was significantly associated with a higher risk of death (39). While increased TGF-beta signaling has demonstrated radiation resistance (50), study shows that inhibition of TGF-beta stops disease progression in liver metastases from colon cancer (51) (52). Incorporation of ICGR15 variables significantly improves the prediction of post-SBRT liver function. The use of ICGR15 can facilitate the delivery of the maximum safe dose of radiation for patients with hepatocellular carcinoma and has the potential to improve uncomplicated tumor control and survival (43). Prolonged prothrombin time (Protime) is the most important score when determining the incidence of radiation-induced liver disease during SBRT in patients with CP-A score 6 (53). International Normalized Ratio (INR) is derived from Protime which is calculated as a ratio of the patient’s Protime to a control Protime standardized for the potency of the thromboplastin reagent developed by the World Health Organization. The MELD is used to prioritize patients for liver transplantation and includes results for creatinine, bilirubin, and Protime expressed as international normalized ratio (Protime-INR) (54). Evidence was provided that the Protime-INR was identified as the most important methodologies may influence the MELD (54). While lower MELD scores were associated with improved survival following SBRT (55), a mathematical equation based on MELD and sodium, named the MELD-Na score, is a feasible and independent prognostic predictor for both short- and long-term outcome predictions in patients with hepatocellular carcinoma (56). It turns out that some features related to radiation induced liver disease such as TGF-Beta, MELD-Na, Bilirubin, etc. appeared in the HITL-BN for LC prediction, and its reason may be related to the fact that liver SBRT was conducted by limiting the toxicity from therapy and not compromising the primary objective of local control.
In this study, we have developed a new HITL-BN approach for HCC patients’ I-ALBI or LC prediction before and during SBRT based on previous PD-BN method. In addition to incorporating EK into its feature selection process, the HITL-BN approach allows humans to participate in an outcome prediction model building process for better handling of imbalanced HCC SBRT data. Especially, we created a novel feature selection mechanism for the HITL-BN approach by integrating the prediction strength of multiple UML methods and the explainable capability of the theoretically sound MB algorithms. Numerical experiments show that the HITL-BN based outcome prediction models significantly outperform the PD-BN based models during SBRT in terms of accuracy and explainability. In addition to gaining physicians’ trust in clinical decision making, the HITL-BN approach has the potential of becoming an important component of future human-computer interface to bridge physicians and advanced ML techniques in improving HCC patients’ treatment outcomes. Our approach can be applied to the outcome prediction of treating other types of cancer, but it still needs to be validated in external further independent datasets.
The raw data supporting the conclusions of this article will be made available upon request from authors and per institutional guidelines.
KC, TL, MM, RT, IE, and YL conceived of the presented idea. YL developed the theory and performed the computations. IE and DN verified the analytical methods. LD provided the testing dataset. All authors discussed the results and contributed to the final manuscript.
This work was supported in part by the National Institutes of Health P01 CA059827, R37-CA222215 and R01-CA233487.
The authors would like to thank Princess Margaret Hospital for providing the testing dataset to independently evaluate our models.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Cai J, Wijesooriya K, Benedict S. SBRT I: Overview of simulation, planning, and delivery. Med Phys (2014) 41(6):412–3. doi: 10.1118/1.4889122
2. Matoba M, Tsuchiya H, Kondo T, Ota K. Stereotactic body radiotherapy delivered with IMRT for oligometastatic regional lymph node metastases in hepatocellular carcinoma: a single-institutional study. J Radiat Res (2020) 61(5):776–83. doi: 10.1093/jrr/rraa067
3. Demirtas CO, 'Alessio AD, Rimassa L, Sharma R, Pinato DJ. ALBI grade: Evidence for an improved model for liver functional estimation in patients with hepatocellular carcinoma. Jhep Rep (2021) 3(5):100347. doi: 10.1016/j.jhepr.2021.100347
4. Johnson PJ, Berhane S, Kagebayashi C, Satomura S, Teng M, Reeves HL, et al. Assessment of liver function in patients with hepatocellular carcinoma: A new evidence-based approach-the ALBI grade. J Clin Oncol (2015) 33(6):550–U45. doi: 10.1200/JCO.2014.57.9151
5. Jackson WC, Hartman HE, Gharzai LA, Maurino C, Karnak D, Mendiratta-Lala M, et al. A mid-treatment increase in albi score is strongly associated with treatment related toxicity following liver radiation therapy. Int J Radiat Oncol (2019) 105(1):S206–S7. doi: 10.1016/j.ijrobp.2019.06.277
6. El Naqa I, Johansson A, Owen D, Cuneo K, Cao Y, Matuszak M, et al. Modeling of normal tissue complications using imaging and biomarkers after radiation therapy for hepatocellular carcinoma. Int J Radiat Oncol (2018) 100(2):335–43. doi: 10.1016/j.ijrobp.2017.10.005
7. Pursley J, El Naqa I, Sanford NN, Noe B, Wo JY, Eyler CE, et al. Dosimetric analysis and normal-tissue complication probability modeling of child-pugh score and albumin-bilirubin grade increase after hepatic irradiation. Int J Radiat Oncol (2020) 107(5):986–95. doi: 10.1016/j.ijrobp.2020.04.027
8. Naqa I. Perspectives on making big data analytics work for oncology. Methods (2016) 111:32–44. doi: 10.1016/j.ymeth.2016.08.010
9. Bach F. Learning theory from first principles. In: Draft of a book, vol. 6. (2021). p. 2021. The MIT Press.
10. Luo Y, El Naqa I, McShan DL, Ray D, Lohse I, Matuszak MM, et al. Unraveling biophysical interactions of radiation pneumonitis in non-small-cell lung cancer via Bayesian network analysis. Radiother Oncol (2017) 123(1):85–92. doi: 10.1016/j.radonc.2017.02.004
11. Luo Y, McShan D, Ray D, Matuszak M, Jolly S, Lawrence T, et al. Development of a fully cross-validated Bayesian network approach for local control prediction in lung cancer. IEEE Trans Radiat Plasma Med Sci (2019) 3(2):232–41. doi: 10.1109/TRPMS.2018.2832609
12. Luo Y, McShan DL, Matuszak MM, Ray D, Lawrence TS, Jolly S, et al. A multiobjective Bayesian networks approach for joint prediction of tumor local control and radiation pneumonitis in nonsmall-cell lung cancer (NSCLC) for response-adapted radiotherapy. Med Phys (2018) 45(8):3980–95. doi: 10.1002/mp.13029
13. Pearl J. Probabilistic reasoning in intelligent systems: Networks of plausible inference. (1988). Morgan Kaufmann. doi: 10.1016/C2009-0-27609-4
14. Johnson JM, Khoshgoftaar TM. Survey on deep learning with class imbalance. J Big Data (2019) 6(1):27. doi: 10.1186/s40537-019-0192-5
15. Bauder RA, Khoshgoftaar TM. The effects of varying class distribution on learner behavior for medicare fraud detection with imbalanced big data. Health Inf Sci Syst (2018) 6(1):9. doi: 10.1007/s13755-018-0051-3
16. Bauder RA, Khoshgoftaar TM, Hasanin T eds. An empirical study on class rarity in big data. 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA) (2018). Orlando, FL, USA.
17. Yu K, Wu XD, Ding W, Mu Y, Wang H. Markov Blanket feature selection using representative sets. IEEE Trans Neural Networks Learn Syst (2017) 28(11):2775–88. doi: 10.1109/TNNLS.2016.2602365
18. Mosqueira-Rey E, Hernández-Pereira E, Alonso-Ríos D, Bobes-Bascarán J, Fernández-Leal Á. Human-in-the-loop machine learning: a state of the art. Artif Intell Rev (2022). doi: 10.1007/s10462-022-10246-w
19. Wu XJ, Xiao LW, Sun YX, Zhang JH, Ma TL, He L. A survey of human-in-the-loop for machine learning. Future Generation Comput Systems-the Int J Escience (2022) 135:364–81. doi: 10.1016/j.future.2022.05.014
20. Chen J, Lim CP, Tan KH, Govindan K, Kumar A. Artificial intelligence-based human-centric decision support framework: an application to predictive maintenance in asset management under pandemic environments. Ann operations Res (2021) 306:1–24. doi: 10.1007/s10479-021-04373-w
21. Wang Y, Gan W, Yang J, Wu W, Yan J eds. Dynamic curriculum learning for imbalanced data classification. 2019 IEEE/CVF International Conference on Computer Vision (ICCV) (2019). Seoul, Korea (South).
22. Jackson WC, Suresh K, Maurino C, Feng M, Cuneo KC, Ten Haken RK, et al. A mid-treatment break and reassessment maintains tumor control and reduces toxicity in patients with hepatocellular carcinoma treated with stereotactic body radiation therapy. Radiother Oncol (2019) 141:101–7. doi: 10.1016/j.radonc.2019.07.027
23. Millar WT, Hopewell JW, Paddick I, Lindquist C, Nordstron H, Lidberg P, et al. The role of the concept of biologically effective dose (BED) in treatment planning in radiosurgery. Physica Medica-European J Med Phys (2015) 31(6):627–33. doi: 10.1016/j.ejmp.2015.04.008
24. Jackson WC, Tang M, Maurino C, Mendiratta-Lala M, Parikh ND, Matuszak MM, et al. Individualized adaptive radiation therapy allows for safe treatment of hepatocellular carcinoma in patients with child-Turcotte-Pugh b liver disease. Int J Radiat Oncol (2021) 109(1):212–9. doi: 10.1016/j.ijrobp.2020.08.046
25. Feng M, Suresh K, Schipper MJ, Bazzi L, Ben-Josef E, Matuszak MM, et al. Individualized adaptive stereotactic body radiotherapy for liver tumors in patients at high risk for liver damage a phase 2 clinical trial. JAMA Oncol (2018) 4(1):40–7. doi: 10.1001/jamaoncol.2017.2303
26. Stenmark MH, Cao Y, Wang HS, Jackson A, Ben-Josef E, Ten Haken RK, et al. Estimating functional liver reserve following hepatic irradiation: Adaptive normal tissue response models. Radiother Oncol (2014) 111(3):418–23. doi: 10.1016/j.radonc.2014.04.007
27. Cousins MM, Morris E, Maurino C, Devasia TP, Karnak D, Ray D, et al. TNFR1 and the TNF alpha axis as a targetable mediator of liver injury from stereotactic body radiation therapy. Trans Oncol (2021) 14(1):100950.doi: 10.1016/j.tranon.2020.100950
28. Cuneo KC, Devasia T, Sun YL, Schipper MJ, Karnak D, Davis MA, et al. Serum levels of hepatocyte growth factor and CD40 ligand predict radiation-induced liver injury. Trans Oncol (2019) 12(7):889–94. doi: 10.1016/j.tranon.2019.04.003
29. Luo Y, Jolly S, Palma D, Lawrence TS, Tseng HH, Valdes G, et al. A situational awareness Bayesian network approach for accurate and credible personalized adaptive radiotherapy outcomes prediction in lung cancer patients. Phys Med (2021) 87:11–23. doi: 10.1016/j.ejmp.2021.05.032
30. Wu L, Huang R, Tetko IV, Xia Z, Xu J, Tong W. Trade-off predictivity and explainability for machine-learning powered predictive toxicology: An in-depth investigation with Tox21 data sets. Chem Res Toxicol (2021) 34(2):541–9. doi: 10.1021/acs.chemrestox.0c00373
31. Wojtas MA, Chen K. Feature importance ranking for deep learning. In: Proceedings of the 34th international conference on neural information processing systems. Vancouver, BC, Canada: Curran Associates Inc (2020). p. 429.
32. Lin CH, Weld DS eds. Active learning with unbalanced classes and example-generation queries. HCOMP (2018). Zürich, Switzerland.
33. Settles B. Active learning literature survey. In: Computer sciences technical report. Madison: University of Wisconsin (2009). p. 1648.
34. Li Z, Sharma P, Lu XH, Cheung J, Reddy S. Using interactive feedback to improve the accuracy and explainability of question answering systems post-deployment. Findings of the Association for Computational Linguistics: ACL (2022), p. 926–37. Dublin, Ireland. arXiv:2204.03025.
35. Kumar P, Bhatnagar R, Gaur K, Bhatnagar A. Classification of imbalanced Data:Review of methods and applications. IOP Conf Series: Materials Sci Eng (2021) 1099(1):012077. doi: 10.1088/1757-899X/1099/1/012077
36. Sakamaki A, Takamura M, Sakai N, Watanabe Y, Arao Y, Kimura N, et al. Longitudinal increase in albumin-bilirubin score is associated with non-malignancy- related mortality and quality of life in patients with liver cirrhosis. PloS One (2022) 17(2):e0263464. doi: 10.1371/journal.pone.0263464
37. Mathew AS, Atenafu EG, Owen D, Maurino C, Brade A, Brierley J, et al. Long term outcomes of stereotactic body radiation therapy for hepatocellular carcinoma without macrovascular invasion. Eur J Cancer (2020) 134:41–51. doi: 10.1016/j.ejca.2020.04.024
38. Huang Y, Chen SW, Fan CC, Ting LL, Kuo CC, Chiou JF. Clinical parameters for predicting radiation-induced liver disease after intrahepatic reirradiation for hepatocellular carcinoma. Radiat Oncol (2016) 11(1):89. doi: 10.1186/s13014-016-0663-1
39. Liang SX, Zhu XD, Xu ZY, Zhu J, Zhao JD, Lu HJ, et al. Radiation-induced liver disease in three-dimensional conformal radiation therapy for primary liver carcinoma: the risk factors and hepatic radiation tolerance. Int J Radiat Oncol Biol Phys (2006) 65(2):426–34. doi: 10.1016/j.ijrobp.2005.12.031
40. Lee S, Kim H, Ji Y, Cho B, Kim SS, Jung J, et al. Evaluation of hepatic toxicity after repeated stereotactic body radiation therapy for recurrent hepatocellular carcinoma using deformable image registration. Sci Rep (2018) 8(1):16224. doi: 10.1038/s41598-018-34676-1
41. Jackson WC, Hartman HE, Gharzai LA, Maurino C, Karnak DM, Mendiratta-Lala M, et al. The potential for midtreatment albumin-bilirubin (ALBI) score to individualize liver stereotactic body radiation therapy. Int J Radiat Oncol (2021) 111(1):127–34. doi: 10.1016/j.ijrobp.2021.04.012
42. Koay EJ, Owen D, Das P. Radiation-induced liver disease and modern radiotherapy. Semin Radiat Oncol (2018) 28(4):321–31. doi: 10.1016/j.semradonc.2018.06.007
43. Suresh K, Owen D, Bazzi L, Jackson W, Ten Haken RK, Cuneo K, et al. Using indocyanine green extraction to predict liver function after stereotactic body radiation therapy for hepatocellular carcinoma. Int J Radiat Oncol (2018) 100(1):131–7. doi: 10.1016/j.ijrobp.2017.09.032
44. Yoon HI, Koom WS, Lee IJ, Jeong K, Chung Y, Kim JK, et al. The significance of ICG-R15 in predicting hepatic toxicity in patients receiving radiotherapy for hepatocellular carcinoma. Liver Int (2012) 32(7):1165–71. doi: 10.1111/j.1478-3231.2012.02784.x
45. Nishikawa H, Kita R, Kimura T, Ohara Y, Sakamoto A, Saito S, et al. Hyponatremia in hepatocellular carcinoma complicating with cirrhosis. J Cancer (2015) 6(5):482–9. doi: 10.7150/jca.11665
46. Dreher C, Hoyer KI, Fode MM, Habermehl D, Combs SE, Hoyer M. Metabolic liver function after stereotactic body radiation therapy for hepatocellular carcinoma. Acta Oncol (2016) 55(7):886–91. doi: 10.3109/0284186X.2015.1137352
47. Tang JY, Ohri N, Kabarriti R, Aparo S, Chuy J, Goel S, et al. Model for end-stage liver disease and sodium velocity predicts overall survival in nonmetastatic hepatocellular carcinoma patients. Can J Gastroenterol Hepatol (2018) 2018:5681979. doi: 10.1155/2018/5681979
48. Botta F, Giannini E, Romagnoli P, Fasoli A, Malfatti F, Testa E, et al. MELD scoring system is useful for predicting prognosis in patients with liver cirrhosis and is correlated with residual liver function: a European study. Gut (2003) 52(1):134–9. doi: 10.1136/gut.52.1.134
49. Bibault JE, Dewas S, Vautravers-Dewas C, Hollebecque A, Jarraya H, Lacornerie T, et al. Stereotactic body radiation therapy for hepatocellular carcinoma: Prognostic factors of local control, overall survival, and toxicity. PloS One (2013) 8(10):e77472. doi: 10.1371/journal.pone.0077472
50. Wu CT, Hsieh CC, Yen TC, Chen WC, Chen MF. TGF-beta1 mediates the radiation response of prostate cancer. J Mol Med (Berl) (2015) 93(1):73–82. doi: 10.1007/s00109-014-1206-6
51. Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet (2015) 47(4):320–U62. doi: 10.1038/ng.3225
52. Onderdonk BE, Chmura SJ. The yin and yang of cytoreductive SBRT in oligometastases and beyond. Front Oncol (2019) 9. doi: 10.3389/fonc.2019.00706
53. Wang PM, Chung NN, Hsu WC, Chang FL, Jang CJ, Scorsetti M. Stereotactic body radiation therapy in hepatocellular carcinoma: Optimal treatment strategies based on liver segmentation and functional hepatic reserve. Rep Pract Oncol Radiother (2015) 20(6):417–24. doi: 10.1016/j.rpor.2015.03.005
54. Tripodi A, Chantarangkul V, Primignani M, Fabris F, Dell'Era A, Sei C, et al. The international normalized ratio calibrated for cirrhosis (INR(liver)) normalizes prothrombin time results for model for end-stage liver disease calculation. Hepatology (2007) 46(2):520–7. doi: 10.1002/hep.21732
55. Chan J, Behr S, Pai J, Feng MUS, Chang A, Haas-Kogan DA, et al. Stereotactic body radiotherapy for hepatocellular carcinoma in patients with poor liver function. J Clin Oncol (2018) 36(4):397–397. doi: 10.1200/JCO.2018.36.4_suppl.397
Keywords: accuracy and explainability, Bayesian networks, human-in-the-loop, hepatocellular cancer, outcome prediction, stereotactic body radiotherapy
Citation: Luo Y, Cuneo KC, Lawrence TS, Matuszak MM, Dawson LA, Niraula D, Ten Haken RK and El Naqa I (2022) A human-in-the-loop based Bayesian network approach to improve imbalanced radiation outcomes prediction for hepatocellular cancer patients with stereotactic body radiotherapy. Front. Oncol. 12:1061024. doi: 10.3389/fonc.2022.1061024
Received: 04 October 2022; Accepted: 01 November 2022;
Published: 09 December 2022.
Edited by:
Tonghe Wang, Emory University, United StatesReviewed by:
Kevin Camphausen, National Cancer Institute (NIH), United StatesCopyright © 2022 Luo, Cuneo, Lawrence, Matuszak, Dawson, Niraula, Ten Haken and El Naqa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Luo, WWkuTHVvQG1vZmZpdHQub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.