
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 14 December 2022
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1054846
This article is part of the Research Topic Biomarkers, Functional Mechanisms, and Therapeutic Potentials in Gastrointestinal Cancers View all 40 articles
Colorectal cancer (CRC) is the most common malignant tumor and one of the most lethal malignant tumors in the world. Despite treatment with a combination of surgery, radiotherapy, and/or systemic treatment, including chemotherapy and targeted therapy, the prognosis of patients with advanced CRC remains poor. Therefore, there is an urgent need to explore novel therapeutic strategies and targets for the treatment of CRC. MicroRNAs (miRNAs/miRs) are a class of short noncoding RNAs (approximately 22 nucleotides) involved in posttranscriptional gene expression regulation. The dysregulation of its expression is recognized as a key regulator related to the development, progression and metastasis of CRC. In recent years, a number of miRNAs have been identified as regulators of drug resistance in CRC, and some have gained attention as potential targets to overcome the drug resistance of CRC. In this review, we introduce the miRNAs and the diverse mechanisms of miRNAs in CRC and summarize the potential targeted therapies of CRC based on the miRNAs.
Colorectal cancer (CRC) is the third most frequent cancer and the second leading cause of cancer death, with an estimated more than 1.9 million new cases and 935,000 deaths worldwide in 2020 (1). The incidence of CRC is steadily increasing year by year in many countries, with a trend of younger age onset (1, 2). At present, the treatment of CRC mainly includes surgery, radiotherapy, chemotherapy, immunotherapy and targeted therapy. However, drug resistance and recurrence after therapy are still the major obstacles to effective anticancer therapy for CRC (3–5), and it is reported that approximately 50% of CRCs are resistant to 5-Fu-based chemotherapy regimens (6). In addition, immunotherapy (checkpoint blockade therapies) is currently considered to be only effective for a proportion (approximately 10-15%) of mismatch-repair-deficient (dMMR) CRC (7–9). The prognosis of CRC was negatively correlated with the progression of tumor stage. Although these treatments can improve the survival rate of CRC patients, the 5-year survival rate for those diagnosed with distant-stage disease remains poor (only approximately 14%) (2). Therefore, it is necessary to find new therapeutic strategies that can effectively treat CRC or improve the drug resistance of CRC to prevent CRC relapse and improve the prognosis of CRC patients.
MicroRNAs (miRNAs/miRs) are a category of short, noncoding, highly conserved and single-stranded RNAs that regulate gene expression at the posttranscriptional level by mRNA degradation or silencing (10–12). MiRNAs are not only critical for regulating normal physiological activities in various biological processes, such as cell development, metabolism, proliferation and apoptosis, but also play an important role in the progression of cancer (13, 14). In 2003, the reduction in miRNAs was reported to be closely related to CRC (15). Since then, research focusing on the effects of miRNAs on CRC has gradually shown that aberrant expression of miRNAs is associated with CRC progression, including tumor formation, metastasis, and drug resistance (16–19). For instance, Li et al. (20)reported that the expression of miR-186-5p in CRC cell lines (HT116, H29, SW620 and LoVo) was lower than that in the normal colonic epithelial cell line NCM460. Moreover, the high expression of miR-186-5p can inhibit the proliferation, epithelial-to-mesenchymal transition (EMT), and metastasis of the CRC cell line LoVo by targeting inhibition of ZEB1. Jin et al. (21) found that the expression of miRNA-30a was significantly increased in CRC tissues compared with normal colorectal tissues, and the expression level of miRNA-30a was inversely correlated with the invasiveness of CRC cell lines. Therefore, miRNAs are expected to be potential novel therapeutic targets for CRC.
Therefore, in this article, we will review the latest research progress on the involvement of miRNAs in the occurrence and development of CRC. This review also discusses the current understanding of CRC drug resistance-related miRNAs and their underlying molecular mechanisms. Furthermore, we highlight the potential of miRNAs as CRC therapeutic targets.
miRNAs are short, noncoding RNAs that are not translated into proteins, with a length of approximately 22 nucleotides (22). The first animal microRNA was lin4, called “small molecule sequential RNA (stRNA)”, which was found in Caenorhabditis elegans in 1993 (23). It was not until 2001 that a large number of functional microRNAs with common characteristics were found in many species, which were named miRNAs (24–26). Until recently, the exploration of the roles and mechanisms of this noncoding RNA in biological activities was in progress. Currently, the majority of studies agree that miRNA is derived from a primary miRNA transcript (pri-miRNA). Firstly, the miRNA gene is transcribed into a large pri-miRNA containing multiple hairpin-loop structures by RNA polymerase II in the nucleus (27). The pri-miRNA typically contains thousands of nucleotides; subsequently, the pri-miRNA is converted into a shorter hairpin-shaped precursor miRNA (pre-miRNA) containing approximately 70 nucleotides by the Drosha enzyme (one type of RNAseIII) and DGCR8 in the nucleus (28–30). Pre-miRNA is transported from the nucleus to the cytoplasm by the Ran/GTP/Exportin-5 complex (31, 32); the pre-miRNA is cleaved and processed to form a double-stranded miRNA of approximately 22 nucleotides in length in the cytoplasm with the help of the Dicer enzyme (another type of RNAseIII), TRBP and PACT (33–35). Finally, one strand of the double-stranded miRNA is bound to AGO2 and loaded onto the RNA-induced silencing complex (RISC) to become the RISC complex, while the other strand is degraded (36). The mature RISC complex binds to target mRNAs with complementary sites, resulting in translational inhibition or degradation of target mRNAs (37) (Figure 1).
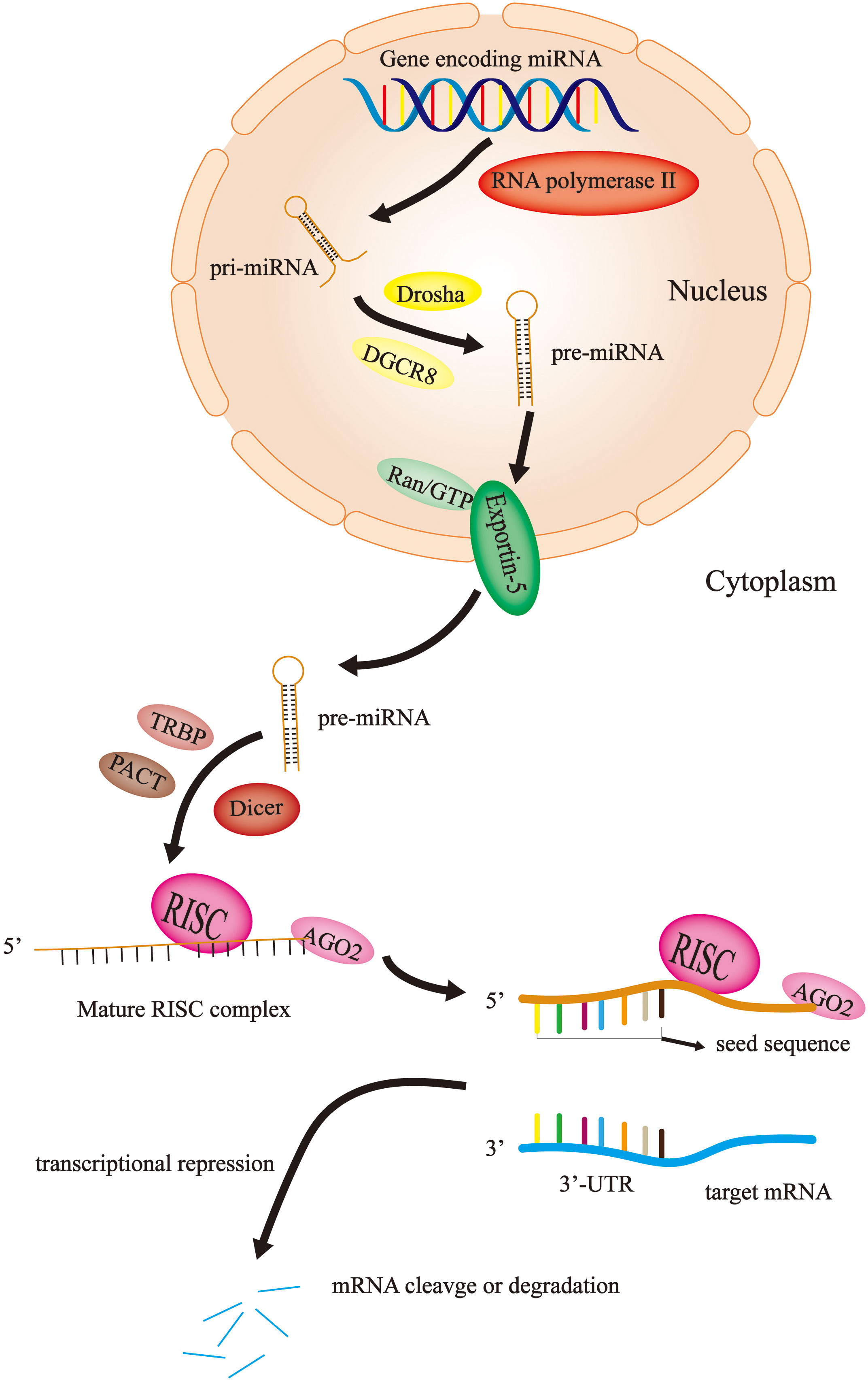
Figure 1 miRNA biosynthesis and mechanism of action. In the nucleus, the miRNA gene is transcribed by RNA polymerase II into pri-miRNA, which become pre-miRNA with the help of Drosha and DGCR8. Then, the Ran/GTP/Exportin-5 complex delivers the pre-miRNA to the cytoplasm. Here, with the assistance of Dicer, TRBP and PACT, the pre-miRNA is disassembled into single strands and combined with AGO2 and RISC to become a mature RISC complex. Finally, the RISC complex binds to the target mRNA to cause mRNA degradation or translational repression.
Initially, it was not thought that these noncoding miRNAs could have a severe impact on human health and even lead to the occurrence and progression of cancer. With the continuous exploration of miRNAs, humans have gradually changed this view. Currently, it is widely assumed that miRNAs carrying RISC recognize the binding site on the 3’-UTR of the target gene mRNA through its seed sequence (nucleotides 2-8 at the 5-terminal). This process produces two effects: transcriptional repression and mRNA cleavage or degradation (38) (Figure 1). In 2002, Calin et al. (39)showed that miRNA genes (miR-15 and miR-16) at 13q14 in chronic lymphocytic leukemia are usually deleted or downregulated by detailed deletion and expression analysis. This study suggests that miRNAs may play an important role in tumorigenesis and tumor progression. Thus, since then, an increasing number of studies have demonstrated that the aberrant expression of miRNAs is associated with cancer progression, including CRC (40–42).
Intestinal microbiome is considered as a key participant in CRC immune regulation and tumor promoting microenvironment, because bacteria from different intestinal sources can induce tumor growth. In recent years, studies have found that the cross-talk between gut microbes and the host in tumor cell metabolism is largely achieved by regulating the level of miRNAs (43, 44). Intestinal microbiota can affect the expression of miRNA, and the abnormal expression of miRNA subsequently activates signal pathways and regulates various aspects of tumor pathobiology in CRC (45). The miRNA in the tumor microenvironment alters the composition of the intestinal microbiota by influencing the gene expression of the microbiota and transferring the metabolites secreted by cancer (46). In conclusion, this interaction will eventually create a favorable microenvironment for tumor cells, including angiogenesis, immune escape and microbiota composition. Meanwhile, the two-way interaction between host and gut microbiome mediated by miRNAs brings new complexity to miRNAs research.
There is abundant evidence that miRNAs and their biogenesis mechanisms are related to the occurrence and development of CRC. Compared to normal tissues, miRNAs are frequently dysregulated in tumors. It suggests that the aberrant expression of miRNAs is closely related to the progression, metastasis and drug resistance of CRC.
The progression and metastasis of CRC are the result of a multistep process involving multiple genetic and epigenetic alterations of oncogenes and tumor suppressor genes over time. In addition, metastasis is also a major cause of poor prognosis in CRC patients. In recent years, substantial data have identified important roles for miRNAs in numerous regulators involved in CRC pathogenesis and metastasis (47, 48). It is being increasingly regarded that miRNA regulation of CRC progression and metastasis occurs by various mechanisms, such as influencing signaling pathways, EMT, and angiogenesis.
There are many signaling pathways mediated by miRNAs in CRC cells that play an important role in the progression and metastasis of CRC (Table 1). As a regulatory pathway regulating colorectal development, hyperactivation of the Wnt/β-catenin pathway is found in more than 90% of CRC cells. The loss or inactivation of adenomatous polyposis coli (APC, a key negative regulator of the canonical Wnt signaling pathway) and the overactivation of the Wnt/β-catenin signaling pathway are considered to be the key processes in the initiation of CRC (83, 84). Zheng et al. (49)aimed to investigate the tumorigenic role of miR-490-3p in CRC. They reported that downregulation of miR-490-3p promoted CRC progression by activating the classic Wnt/β-catenin signaling pathway. MiR-224 has been found to directly target the GSK3β and SFRP2 genes to activate Wnt/β-catenin signaling and direct the nuclear translocation of β-catenin in CRC. Furthermore, miR-224 upregulation promoted CRC cell proliferation. Knockdown of miR-224 attenuated the effects of Wnt/β-catenin on the metastasis and proliferation of CRC cells (50). Similarly, miR-452 enhanced the proliferation and metastasis of CRC cells by activating the Wnt/β-catenin signaling pathway via directly targeting GSK3β. While knockdown of miR-452 was found to restore the expression of GSK3β and inhibit Wnt/β-catenin-mediated cell metastasis and cell proliferation (51). The expression of miR-494 was significantly increased in CRC, with a negative correlation to APC expression in CRC tissues. And up-regulation of this miR-494 has also been found to enhance cell proliferation and tumorigenesis of CRC by suppressing the expression of APC (52). As a member of the BTB/POZ/zinc finger (ZF) family of transcription factors, BCL6 has been shown to target multiple functional signaling pathways, including the Wnt/β-catenin signaling pathway. In addition, Sun et al. (53) suggested that BCL6 was a mediator of miR-144-3p in regulating CRC cell proliferation and cell cycle arrest. They reported that miR-144-3p inhibited cell proliferation and the G1/S phase transition of CRC cells by targeting BCL6 via inhibition of Wnt/β-catenin signaling. As a negative regulator of Wnt/β-catenin signaling, miR-377-3p was shown to inhibit CRC cell growth in vitro and in vivo by targeting XIAP and ZEB2 (54). The PI3K/AKT signaling pathway is an important signaling pathway that controls the growth and metastasis of CRC (85). Jia et al. reported that miR182/-135b promoted CRC progression by targeting ST6GALNAC2 via regulation of the PI3K/AKT pathway (55). miR-7 conferred its tumor-suppressing function in CRC by inhibiting the activation of the PI3K/AKT pathway by downregulating the miR-7 potential target TYRO3 (56). Therefore, targeting these signaling pathways mediated by miRNAs that regulate the progression and metastasis of CRC may become a promising therapeutic strategy for targeting CRC.
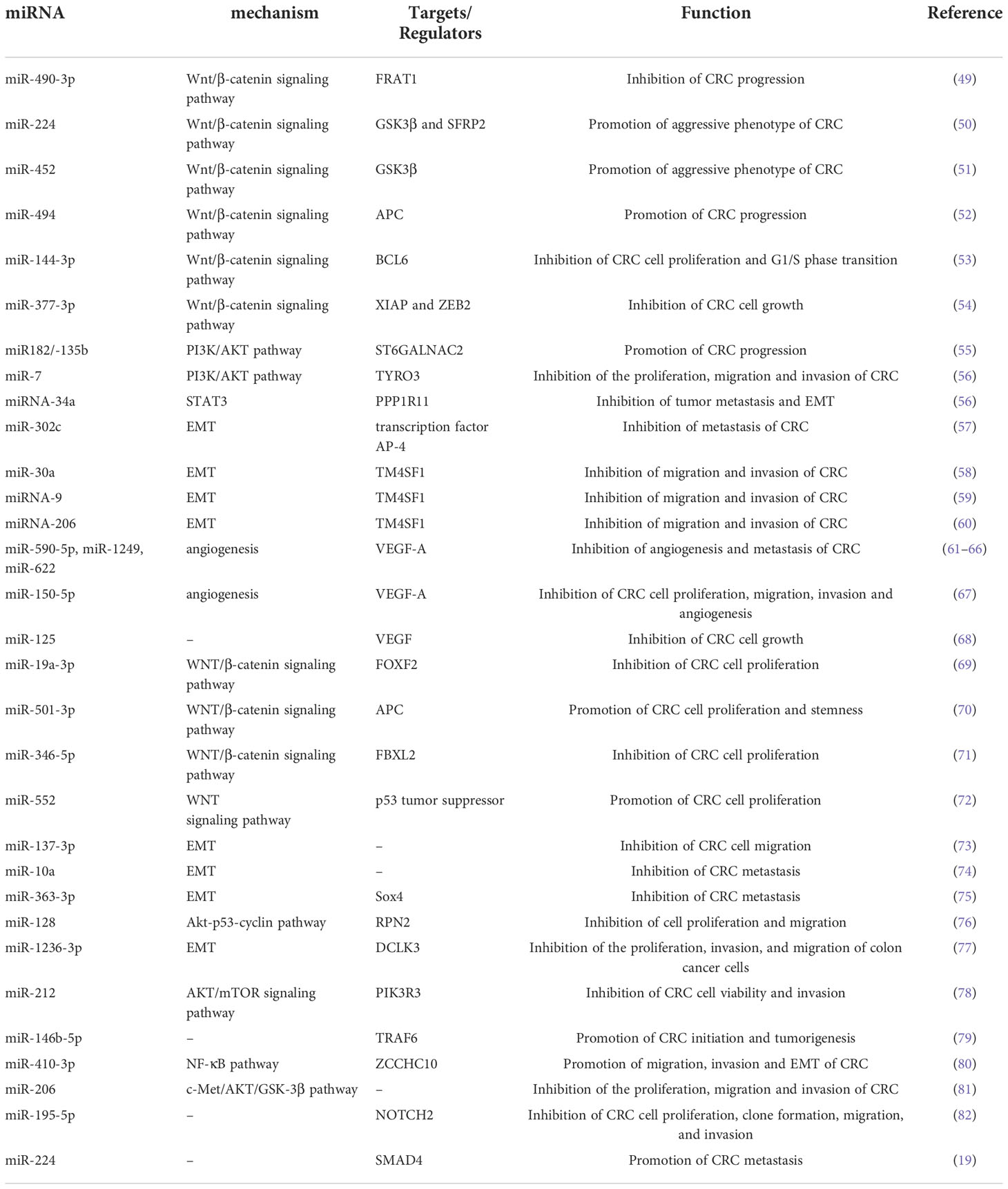
Table 1 A list of representative miRNAs identified in CRC that are associated with the progression/metastasis of CRC.
It is widely accepted that epithelial–mesenchymal transition (EMT) is the transition from epithelial cells to mesenchymal cells during the progression and metastasis of malignancy. And EMT is known to be associated with a wide array of malignant behaviors of CRC, including tumorigenicity and metastasis (82). There is growing evidence that miRNAs play crucial roles in regulating the phenotype of EMT (86). Ding et al. (73) illustrated that miR-137-3p could attenuate CRC cell migration by regulating a KDM1A-dependent EMT process. Li et al. (87) reported that miRNA-34a inhibits tumor metastasis and EMT by reducing the expression of PPP1R11 to prevent the activation of STAT3. Low expression levels of miR-302c was found to significantly correlate with advanced tumor stage, lymph node metastasis and deeper tumor invasion. And miR-302c inhibit EMT and metastasis of CRC by targeting the transcription factor AP-4 (57). Transmembrane 4 L six family member 1 (TM4SF1) is a direct target gene of miRNAs in CRC cells and is involved in the regulation of EMT progression in CRC. MiR−30a, which is involved in the EMT of CRC, was shown to suppress malignant behaviors of CRC, including migration and invasion, by directly targeting oncogenic TM4SF1 (58). Similarly, miRNA-206 and miRNA-9 could directly target TM4SF1, thus suppressing EMT of CRC cells and leading to the suppression of cell proliferation, migration, and invasion in CRC cells (59, 60).
One of the characteristics of tumor growth is angiogenesis, which fosters tumorigenesis and metastasis by supplying oxygen and diffusible nutrients as well as releasing proangiogenic chemicals. Angiogenesis is integral to the development and progression of CRC, and it is crucial to the growth and metastasis of CRC (61). There are many angiogenic factors that control angiogenesis, including vascular endothelial growth factor (VEGF) and hypoxia-inducible factor 1 (HIF-1). The overexpression of miR-145-5p suppressed RHBDD1 through suppression of the EGFR-associated signaling pathway (EGFR/Raf/MEK/ERK cascades), which in turn inhibited the growth, invasion and migration of CRC cells (62). As a key VEGF receptor, VEGF-A is involved in angiogenesis and stimulates the germination of prevascular endothelial cells, which results in the development of new vasculature. MiR-590-5p was found to suppress the angiogenesis and metastasis of CRC by modulating VEGF-A (63). Similarly, miR-1249 and miR-622 can also inhibit CRC angiogenesis by regulating the level of VEGF-A, thereby inhibiting CRC growth and metastasis (64, 65). Additionally, miR-520a serves as a direct target of VEGF-A, and ATAD2 can suppress VEGF-A production by elevating the expression of miR-520a, hence inhibiting angiogenesis in CRC (66). In vitro and in vivo, miR-150-5p inhibited the ability of CRC cells to proliferate, migrate, invade, and undergo angiogenesis. This inhibitory impact could be reversed by transfecting a plasmid encoding the VEGF-A (67). HIF-1 is a critical regulator of VEGF and one of the key molecules that mediates the growth of CRC (88, 89). Additionally, numerous miRNAs have been identified as having a role in the regulation of HIF-1 on the angiogenesis and development of CRC (90, 91). MiR-148a inhibited CRC angiogenesis and reduced the risk of early recurrence of CRC by regulating the level of pERK/HIF-1α (92).
In conclusion, miRNAs play an important regulatory role in the biological behavior of CRC. MiRNAs affect the self-renewal, proliferation, differentiation, metastasis and other biological behaviors of CRC by regulating related molecules of various signaling pathways, EMT or angiogenesis. And miRNAs may have broad prospects in clinical application. Further exploration of key miRNAs and their regulatory mechanisms related to the characteristics of CRC will provide more references for future CRC targeted therapy.
Resistance to anticancer therapy is one of the major barriers to the successful treatment of CRC. It was reported that there were approximately 80% of responders may develop drug resistance (93). The mechanism of drug resistance acquired by CRC has been continuously explored, and it is believed that the drug resistance of CRC is associated with multiple factors, such as the cancer stem cells and tumor microenvironment of CRC (94, 95). However, no single viewpoint can explain the drug resistance of all CRC patients. In recent years, an increasing number of studies have shown that miRNAs are involved in the drug resistance of CRC by regulating autophagy, the cell cycle, important signaling pathways and efflux pumps (Table 2).
Autophagy is a type of programmed cell death different from apoptosis. Autophagy is the process of transporting damaged, denatured or aged proteins and organelles in cells to lysosomes for digestion and degradation. On the one hand, autophagy is a physiological process and a defense mechanism of cells in adverse environments. On the other hand, the occurrence and development of autophagy is also closely related to the drug resistance of tumors (111, 112). In recent years, it has been reported that miRNAs regulate the drug resistance by modulating autophagy (113). Despite tremendous progress in anticancer therapy, 5-FU-containing regimens remain one of the most commonly used and effective treatment regimens for CRC. Furthermore, evidence suggests that miRNAs may be involved in the chemoresistance of CRC cells to 5-FU. Zhao et al. (96) found significantly low expression of miR-145 and p53, whereas the expression of activating transcription factor 4 (ATF4) histone deacetylase 4 (HDAC4) increased considerably by RT-qPCR and Western blot analysis. They further found that ATF4-regulated miR-145 enhanced CRC tumorigenesis and the resistance to 5-FU via regulating the HDAC4/p53 axis. Oxaliplatin (OXA) is another crucial component of the combinatorial chemotherapeutic standard of CRC. And OXA resistance is also another major obstacle to effective chemotherapy in CRC patients. In OXA-resistant cell lines (SW480-OxR and HCT116-OxR), the expression of miR-27b-3p was significantly decreased compared to that in the corresponding parental cells. miR-27b-3p was able to inhibit autophagy in CRC cells by suppressing the expression of ATG10 at the posttranscriptional level. Meanwhile, miR-27b-3p could increase the sensitivity of CRC cells to OXA in vivo and in vitro by regulating the level of autophagy (99). However, there are still few studies on miRNAs affecting CRC drug resistance by regulating autophagy levels, and more high-quality studies are needed in the future to confirm this view.
The cell cycle is the process of DNA replication and cell division and includes four stages: G1, S, G2 and M (114). Studies have shown that impaired cell cycle regulation is the key mechanism that promotes drug resistance in cancer. However, miRNAs may facilitate the resistance of cancer cells to chemotherapy by affecting the cell cycle (115). Zhao et al. (98)found that miR-92b-3p was highly expressed by CRC HCT8/T cells and that knockdown of miR-92b-3p may attenuate the resistance of MDR HCT8/T cells to chemotherapy in vitro and in vivo. They further demonstrated that miR-92b-3p regulated the sensitivity of CRC cells to chemotherapeutic drugs by regulating the cell cycle and apoptosis (via directly targeting cyclin-dependent kinase inhibitor 1C and suppressing its expression). The expression of miRNA-375-3p was significantly decreased in CRC cell lines, and the expression level of miRNA-375-3p was proportional to the sensitivity of CRC cells to 5-FU. It was confirmed to enhance the sensitivity of CRC cells to 5-FU by inducing apoptosis and cycle arrest of CRC cells (116). The expression of miR-577 was found to be significantly increased in 5-fluorouracil (5-FU)-resistant SW480 cells (SW480/5-FU). And Jiang et al. revealed that miR-577 inhibited tumor growth and enhanced 5-FU sensitivity in SW480/5-FU cells by inducing G0/G1 cell cycle arrest in CRC cells (100).
Many signaling pathways have been shown to be involved in tumor chemoresistance. In recent years, there has also been growing evidence that miRNAs regulate the resistance of CRC cells to therapy through signaling pathways. MiRNA-506 was found to be low expressed in OXA-resistant CRC tissues. And the overexpression of miRNA-506 could not only inhibit the growth of CRC cells, but also reverse the resistance of OXA-resistant CRC cells to OXA. Further exploration by Zhou et al. (101) showed that miRNA-506 increased the sensitivity of CRC to OXA therapy by inhibiting MDR1/P-gp expression via downregulation of the Wnt/β-catenin pathway. miR-199b-3p was shown to enhance the drug resistance of CRC cells. Han et al. (102) suggested that suppressing miR-199b-3p could enhance the sensitivity of CRC cetuximab (CTx)-resistant cells to CTx in vitro and in mouse xenograft models by targeting the inhibition of CRIM1 via the Wnt/β-catenin signaling pathway. Wu et al. (103) found that the overexpression of miR-200b-3p could enhance OXA sensitivity in OXA-resistant CRC cells (HT29 and HCT116 cells) and induce growth inhibition and apoptosis of OXA-resistant CRC cells by inhibiting the expression of βIII-tubulin protein. As an important regulator of the PI3K/AKT signaling pathway, PTEN is also a direct target of miRNAs. The expression of miR-454-3p was significantly upregulated in OXA-resistant cells. Interestingly, inhibition of miR-454-3p was observed to sensitize OXA-resistant cells to OXA treatment and enhance OXA-induced cells apoptosis. In the xenograft model, this effect also exists. Meanwhile, Qian et al. (104) further confirmed that miR-454-3p enhanced OXA resistance by targeting PTEN and activating the AKT signaling pathway. And the PI3K/AKT signaling pathway is closely related to the chemosensitivity of CRC cells to 5-FU. Studies have shown that miRNAs affect the sensitivity of CRC cells to 5-FU by targeting the targets of the PI3K/AKT signaling pathway. Liu et al. (105) found that upregulation of miR-135b or miR-182 could enhance the resistance to 5-FU in CRC cells by targeting ST6GALNAC2 via the PI3K/AKT pathway. Similarly, Liu et al. (106) revealed that highly expressed miR-543 enhanced the resistance of CRC cells to 5-FU by downregulating the expression of PTEN, and the low expression of PTEN can activate the PI3K/AKT signaling pathway. As another regulator of the PI3K/AKT signaling pathway, PIK3R1 was confirmed to be a target of miR-455-5p in CRC cells. Lou et al. (107) found that miR-455-5p sensitized CRC cells to 5-FU through CCK-8 and flow cytometry analysis. They further studied the mechanism underlying this phenomenon and revealed that miR-455-5p enhanced the sensitivity of CRC cells to 5-FU by targeting PIK3R1 and DEPDC1. However, these conclusions were supported at the cellular level. And future in vivo studies to confirm these findings are urgently required. Shi et al. (108) selectively enriched cisplatin-resistant CRC cell lines from the SW480 cell line by using cisplatin. By PCR assay, they found that the expression of miR-148a was down-regulated in cisplatin-resistant SW480 cells, while overexpression of miR-148a was able to attenuate cisplatin resistance and inhibit the growth of CRC cells in cisplatin-resistant SW480 cells. Further research found that miR-148a played a role in regulating CRC cisplatin resistance and tumor development by inhibiting the expression of its downstream target Wnt10b and the activity of β-catenin signaling. And this conclusion was also validated in an immunized mouse xenograft model of SW480 resistance. The Notch pathway is a highly conserved signaling pathway that plays a key role in CRC, which contributes to cell proliferation, EMT and chemoresistance (117). MiR-139-5p was downregulated in 5-FU-resistant CRC cell lines (HCT-8/5-FU and HCT-116/5-FU). And the expression of miR-139-5p increased 5-FU-induced apoptosis and sensitized CRC cells to 5-FU by inhibiting NOTCH-1 and its downstream molecules (MRP-1 and BCL-2) (109). Similarly, in 5-FU-resistant cells (SW620 and HT-29 cells), the expression of miR-195-5p was decreased. Jin et al. (97) demonstrated that miR-195-5p reduced the stemness and chemoresistance of CRC cells by inhibiting the Notch signaling pathway.
The most common mechanism for cancer drug resistance is reduced drug accumulation concentrations in cancer cells, and studies have shown that a major cause of this phenomenon is increased drug efflux mediated by the ATP-binding cassette (ABC) efflux pump. ABC efflux pump can directly excrete drugs from cancer cells, affecting drug absorption, distribution, and metabolic clearance, resulting in chemotherapy failure (118, 119). Recently, studies on miRNAs have shown that miRNAs may regulate the drug resistance of CRC cells by affecting the ABC efflux pump. Liu et al. (110)showed that miR-128-3p inhibited EMT and increased intracellular OXA concentrations in OXA-resistant CRC cell lines. MiR-128-3p was found to competitively bind Bmi1 and MRP5 (an ABC efflux pump), resulting in decreased intracellular OXA efflux and enhanced OXA-induced EMT. Therefore, overexpression of miR-128-3p can enhance the therapeutic response of CRC cells to OXA.
Together, these studies provide a rationale for developing miRNA-based therapies to effectively treat drug-resistant CRC cells. However, one important limitation of these findings must be recognized. Most of the current research is at the cellular level and includes a few studies in animal models. There are more clinical research trials to be made before these miRNAs can truly become targeted drugs for reversing CRC resistance.
As discussed above, some miRNAs promote CRC progression, metastasis, or drug resistance, while others show the opposite effect. According to their regulatory effects on tumors, miRNAs are broadly divided into two types: tumor suppressor miRNAs and oncogenic miRNAs. Therefore, the current treatment approaches based on targeting miRNAs include: (1) the upregulation of tumor suppressor miRNAs utilizing miRNA mimics, such as double-stranded synthetic miRNAs and miRNA expression vectors, when tumor suppressor miRNAs are downregulated; (2) inhibiting the expression of oncomiRs by miRNA antagonists, such as antisense oligonucleotides, antagomirs, and miRNA sponges, when oncomiRs are overexpressed (120). An effective treatment regimen requires not only the selection of the right regulatory molecules, but also the selection of appropriate drug delivery strategies. Although targeting miRNAs is a promising therapeutic strategy for the treatment of CRC, naked miRNA-based agents have many shortcomings, such as poor targeting capabilities, short circulation times, and off-target effects. In recent years, nanoparticle carriers have provided unprecedented opportunities for efficiently delivering therapeutics of miRNAs by controlling release kinetics, prolonging circulation time and improving biological distribution to improve the therapeutic efficacy of miRNA-targeting agents with less toxicity compared with other anticancer drugs (121, 122). For instance, miR-145 was shown to be downregulated in colon cancer and to have antiproliferative and proapoptotic effects. Through nanotechnology, Liang et al. (123) studied a PLGA/PEI-mediated miRNA vector delivery system and verified the validity of this method by using a colon cancer xenograft model with a miR-145 vector encoding for the expression of miR-145 (pDNA). The results of this work showed that miR-145 could be efficiently delivered to colon cancer cells and exerted potent antitumor efficacy through a PLGA/PEI/HA vehicle. Similarly, Ibrahim et al. (124) developed a miRNA delivery system by using polyethylenimine (PEI)-mediated delivery of unmodified miRNAs and validated the method in a mouse model of colon carcinoma. The results of this study showed that miRNA replacement therapy for miR-145 and miR-33a could reduce tumor growth. Despite the growing number of studies targeting miRNAs for the treatment of CRC, more studies, especially clinically relevant studies, are needed to demonstrate the clinical significance of therapeutic strategies for targeting miRNAs.
Despite having a strong genetic component, most CRC cases are sporadic and undergo a lengthy (usually several years) process of slow progression from adenoma to cancer (125). The prognosis of CRC is highly related to the stage at diagnosis. The 5-year survival rate of early CRC patients can reach more than 90%, while the 5-year survival rate of advanced CRC patients is approximately 14% (2). Therefore, CRC screening and early diagnosis and treatment can effectively reduce the mortality of CRC. At present, the most frequently used diagnostic tool for CRC is colonoscopy. However, colonoscopy has some limitations in CRC screening, such as invasive procedures, poor population compliance, and high technical requirements for operators. In addition, CRC is a heterogeneous disease with various histologic characteristics, molecular characteristics, and prognosis. Although TNM staging system is a crucial clinical parameter to evaluate the prognosis of CRC patients, the prognosis of patients with CRC still varies considerably at the same stage (126). The drug resistance of CRC patients to current chemotherapy drugs also reflects its heterogeneity. Taken together, these limitations highlight the urgent need of CRC for new non-invasive biomarkers. MiRNA is relatively stable, not easily degraded by rnase, and less affected by high temperature and extreme pH. And it is a crucial regulator of life process, closely related to the tumor (can be actively secreted into the circulatory system by cancer cells) (127). Therefore, circulating miRNAs are a potential biomarker. In 2008, Mitchell et al. first proposed that circulating miRNAs are emerging as promising biomarkers for solid tumors (128). And then in 2009, Ng, EKO et al. (129) reported that circulating miRNA (MiR-92) was significantly increased in the plasma of CRC and might be a potential non-invasive biomarker for the diagnosis of CRC. In 2013, Kanaan, Ziad et al. (130) identified and verified the characteristics of miRNAs in the plasma of healthy controls, colorectal adenomas and CRC patients, and designed two powerful prediction models: a panel of 8 plasma miRNAs (miR-532-3p, miR-331, miR-195, miR-17, miR-142-3p, miR-15b, miR-532 and miR-652) could significantly distinguish colorectal adenomas from healthy people [AUC=0.868 (95% confidence interval [CI]: 0.76-0.98)]. Furthermore, a panel of 3 plasma miRNAs (miR-431, miR-15b and miR-139-3p) could also distinguish patients with stage IV CRC from healthy people [AUC=0.896 (95% CI: 0.78-1.0)]. Noteworthy, a growing number of studies have highlighted that circulating miRNAs, especially the prediction models based on miRNA panels, are a promising tool for early detection, prognosis and treatment selection of CRC in recent 5 years (Figure 2).
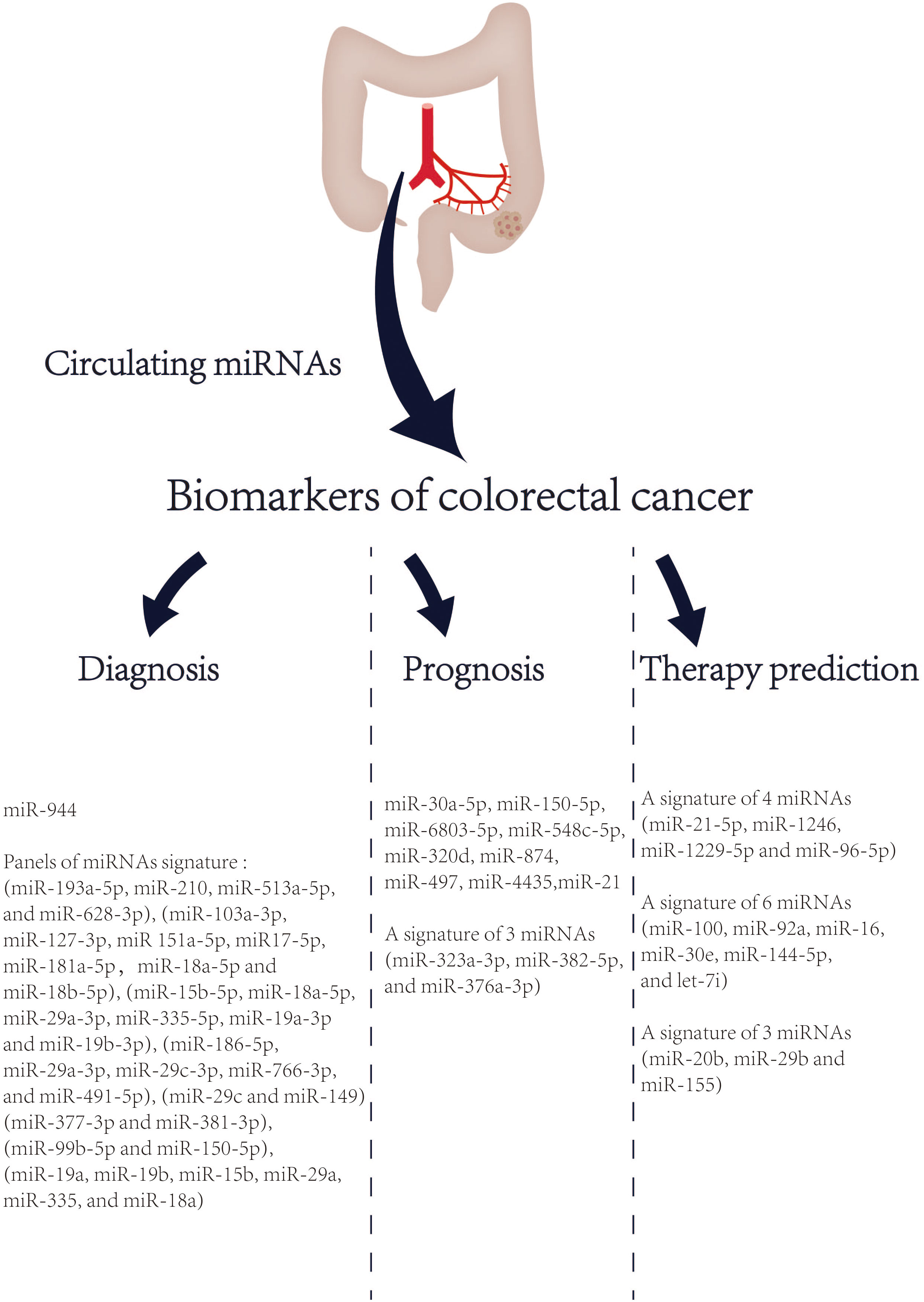
Figure 2 List of circulating miRNAs reported as promising biomarkers in CRC in recent 5 years. References are provided in Table S1.
In summary, miRNAs play important roles in CRC cell proliferation, metastasis, and chemoresistance by regulating CRC-related signaling pathways, EMT, angiogenesis and others. According to the different effects of miRNAs on tumors, miRNAs are also divided into tumor-promoting and tumor-suppressing types, and these two types also provide future directions for targeting miRNAs in the treatment of CRC. However, one of the greatest challenges in developing miRNA-based therapeutics is to design a delivery system that can make miRNA-based therapeutics durable and enable tissue-specific targeting while avoiding potential toxicities and off-target effects. Although there is no lack of scientific evidence that nanostructures containing miRNAs mimetics or antagonists can produce robust antitumor effects with few side effects, most of the current exciting results remain at the level of cell studies, and few relevant clinical studies have been reported. In conclusion, although miRNAs are a promising target for the treatment of CRC, the introduction of miRNA-targeted therapies into clinical practice still requires substantial and in-depth research.
Literature review, data analysis, and manuscript preparation were performed by YY. W-JM and ZW contributed for the study conception, design, and revision. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1054846/full#supplementary-material
5-FU, 5-Fluorouracil; ABC, ATP-binding cassette; AGO2, Argonaute proteins; APC, adenomatous polyposis coli; ATF4, activating transcription factor 4; CRC, Colorectal Cancer; CTx , cetuximab; DGCR8 , DiGeorge syndrome critical region 8; EMT, epithelial-to-mesenchymal transition; HDAC4, histone deacetylase 4; HIF-1, hypoxia-inducible factor 1; OXA, Oxaliplatin ; PACT, protein activator of the interferon-induced protein kinase; RISC, RNA-induced silencing complex; SMAD4, SMAD family member 4; TM4SF1, transmembrane 4 L six family member 1; TRBP, TAR RNA binding protein; VEGF, vascular endothelial growth factor.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin (2020) 70(3):145–64. doi: 10.3322/caac.21601
3. Xu K, Zhan Y, Yuan Z, Qiu Y, Wang H, Fan G, et al. Hypoxia induces drug resistance in colorectal cancer through the HIF-1α/miR-338-5p/IL-6 feedback loop. Mol Ther (2019) 27(10):1810–24. doi: 10.1016/j.ymthe.2019.05.017
4. Ukai S, Sakamoto N, Taniyama D, Harada K, Honma R, Maruyama R, et al. KHDRBS3 promotes multi-drug resistance and anchorage-independent growth in colorectal cancer. Cancer Sci (2021) 112(3):1196–208. doi: 10.1111/cas.14805
5. Luo M, Yang X, Chen H-N, Nice EC, Huang C. Drug resistance in colorectal cancer: An epigenetic overview. Biochim Biophys Acta Rev Cancer (2021) 1876(2):188623. doi: 10.1016/j.bbcan.2021.188623
6. He J, Pei L, Jiang H, Yang W, Chen J, Liang H. Chemoresistance of colorectal cancer to 5-fluorouracil is associated with silencing of the BNIP3 gene through aberrant methylation. J Cancer (2017) 8(7):1187–96. doi: 10.7150/jca.18171
7. Ijsselsteijn ME, Petitprez F, Lacroix L, Ruano D, van der Breggen R, Julie C, et al. Revisiting immune escape in colorectal cancer in the era of immunotherapy. Br J Cancer (2019) 120(8):815–8. doi: 10.1038/s41416-019-0421-x
8. Kim ST, Lee S-J, Lee J, Park SH, Park JO, Lim HY, et al. The impact of microsatellite instability status and sidedness of the primary tumor on the effect of cetuximab-containing chemotherapy in patients with metastatic colorectal cancer. J Cancer (2017) 8(14):2809–15. doi: 10.7150/jca.18286
9. Mo X, Huang X, Feng Y, Wei C, Liu H, Ru H, et al. Immune infiltration and immune gene signature predict the response to fluoropyrimidine-based chemotherapy in colorectal cancer patients. Oncoimmunology (2020) 9(1):1832347. doi: 10.1080/2162402X.2020.1832347
10. Chua BA, van der Werf I, Jamieson C, Signer RAJ. Post-transcriptional regulation of homeostatic, stressed, and malignant stem cells. Cell Stem Cell (2020) 26(2):138–59. doi: 10.1016/j.stem.2020.01.005
11. Gebert LFR, Macrae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol (2019) 20(1):21–37. doi: 10.1038/s41580-018-0045-7
12. Sun L, Wan A, Zhou Z, Chen D, Liang H, Liu C, et al. RNA-Binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut (2021) 70(9):1698–712. doi: 10.1136/gutjnl-2020-320652
13. Chen B, Xia Z, Deng Y-N, Yang Y, Zhang P, Zhu H, et al. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol (2019) 9(1):180212. doi: 10.1098/rsob.180212
14. Tong Z, Cui Q, Wang J, Zhou Y. TransmiR v2.0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res (2019) 47(D1):D253–D8. doi: 10.1093/nar/gky1023
15. Michael MZ, O' Connor SM, Van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res (2003) 1(12):882–91.
16. Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via wnt/β-catenin signaling. Nat Med (2017) 23(11):1331–41. doi: 10.1038/nm.4424
17. Ozawa T, Kandimalla R, Gao F, Nozawa H, Hata K, Nagata H, et al. A MicroRNA signature associated with metastasis of T1 colorectal cancers to lymph nodes. Gastroenterology (2018) 154(4):844–8. doi: 10.1053/j.gastro.2017.11.275
18. Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut (2017) 66(4):654–65. doi: 10.1136/gutjnl-2014-308737
19. Ling H, Pickard K, Ivan C, Isella C, Ikuo M, Mitter R, et al. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut (2016) 65(6):977–89. doi: 10.1136/gutjnl-2015-309372
20. Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys (2018) 640:53–60. doi: 10.1016/j.abb.2018.01.002
21. Jin H, Shi X, Zhao Y, Peng M, Kong Y, Qin D, et al. MicroRNA-30a mediates cell migration and invasion by targeting metadherin in colorectal cancer. Technol Cancer Res Treat (2018) 17:1533033818758108. doi: 10.1177/1533033818758108
22. Kim T, Croce CM. MicroRNA and ER stress in cancer. Semin Cancer Biol (2021) 75:3–14. doi: 10.1016/j.semcancer.2020.12.025
23. Lee RC, Feinbaum RL, Ambros V. The c. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75(5):843–54. doi: 10.1016/0092-8674(93)90529-y
24. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science (2001) 294(5543):853–8. doi: 10.1126/science.1064921
25. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in caenorhabditis elegans. Science (2001) 294(5543):858–62. doi: 10.1126/science.1065062
26. Lee RC, Ambros V. An extensive class of small RNAs in caenorhabditis elegans. Science (2001) 294(5543):862–4. doi: 10.1126/science.1065329
27. Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J (2004) 23(20):4051–60. doi: 10.1038/sj.emboj.7600385
28. Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature (2004) 432(7014):231–5. doi: 10.1038/nature03049
29. Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature (2004) 432(7014):235–40. doi: 10.1038/nature03120
30. Han J, Lee Y, Yeom K-H, Kim Y-K, Jin H, Kim VN. The drosha-DGCR8 complex in primary microRNA processing. Genes Dev (2004) 18(24):3016–27. doi: 10.1101/gad.1262504
31. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev (2003) 17(24):3011–6. doi: 10.1101/gad.1158803
32. Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science (2004) 303(5654):95–8. doi: 10.1126/science.1090599
33. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the dicer complex to Ago2 for microRNA processing and gene silencing. Nature (2005) 436(7051):740–4. doi: 10.1038/nature03868
34. Heyam A, Coupland CE, Dégut C, Haley RA, Baxter NJ, Jakob L, et al. Conserved asymmetry underpins homodimerization of dicer-associated double-stranded RNA-binding proteins. Nucleic Acids Res (2017) 45(21):12577–84. doi: 10.1093/nar/gkx928
35. Park J-E, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature (2011) 475(7355):201–5. doi: 10.1038/nature10198
36. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell (2005) 123(4):631–40. doi: 10.1016/j.cell.2005.10.022
37. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer (2015) 15(6):321–33. doi: 10.1038/nrc3932
38. Kingston ER, Bartel DP. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res (2019) 29(11):1777–90. doi: 10.1101/gr.251421.119
39. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U.S.A. (2002) 99(24):15524–9. doi: 10.1073/pnas.242606799
40. Stang A, Weilert H, Lipp MJ, Oldhafer KJ, Hoheisel JD, Zhang C, et al. MicroRNAs in blood act as biomarkers of colorectal cancer and indicate potential therapeutic targets. Mol Oncol (2021) 15(9):2480–90. doi: 10.1002/1878-0261.13065
41. Li J, Pu W, Sun H-L, Zhou J-K, Fan X, Zheng Y, et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ (2018) 25(9):1612–24. doi: 10.1038/s41418-018-0065-z
42. Tang W, Zhou W, Xiang L, Wu X, Zhang P, Wang J, et al. The p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell proliferation in human colorectal cancer. Nat Commun (2019) 10(1):663. doi: 10.1038/s41467-018-08225-3
43. Yuan C, Steer CJ, Subramanian S. Host-MicroRNA-Microbiota interactions in colorectal cancer. Genes (Basel) (2019) 10(4):270. doi: 10.3390/genes10040270
44. Xing J, Liao Y, Zhang H, Zhang W, Zhang Z, Zhang J, et al. Impacts of MicroRNAs induced by the gut microbiome on regulating the development of colorectal cancer. Front Cell Infect Microbiol (2022) 12:804689. doi: 10.3389/fcimb.2022.804689
45. Li L, Sarver AL, Khatri R, Hajeri PB, Kamenev I, French AJ, et al. Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J Pathol (2014) 234(4):488–501. doi: 10.1002/path.4407
46. Liu S, Da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe (2016) 19(1):32–43. doi: 10.1016/j.chom.2015.12.005
47. Wen X-Q, Qian X-L, Sun H-K, Zheng L-L, Zhu W-Q, Li T-Y, et al. MicroRNAs: Multifaceted regulators of colorectal cancer metastasis and clinical applications. Onco Targets Ther (2020) 13:10851–66. doi: 10.2147/OTT.S265580
48. Ghafouri-Fard S, Hussen BM, Badrlou E, Abak A, Taheri M. MicroRNAs as important contributors in the pathogenesis of colorectal cancer. BioMed Pharmacother (2021) 140:111759. doi: 10.1016/j.biopha.2021.111759
49. Zheng K, Zhou X, Yu J, Li Q, Wang H, Li M, et al. Epigenetic silencing of miR-490-3p promotes development of an aggressive colorectal cancer phenotype through activation of the wnt/β-catenin signaling pathway. Cancer Lett (2016) 376(1):178–87. doi: 10.1016/j.canlet.2016.03.024
50. Li T, Lai Q, Wang S, Cai J, Xiao Z, Deng D, et al. MicroRNA-224 sustains wnt/β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res (2016) 35:21. doi: 10.1186/s13046-016-0287-1
51. Li T, Jian X, He H, Lai Q, Li X, Deng D, et al. MiR-452 promotes an aggressive colorectal cancer phenotype by regulating a wnt/β-catenin positive feedback loop. J Exp Clin Cancer Res (2018) 37(1):238. doi: 10.1186/s13046-018-0879-z
52. Zhang Y, Guo L, Li Y, Feng G-H, Teng F, Li W, et al. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol Cancer (2018) 17(1):1. doi: 10.1186/s12943-017-0753-1
53. Sun N, Zhang L, Zhang C, Yuan Y. miR-144-3p inhibits cell proliferation of colorectal cancer cells by targeting BCL6 via inhibition of wnt/β-catenin signaling. Cell Mol Biol Lett (2020) 25:19. doi: 10.1186/s11658-020-00210-3
54. Huang L, Liu Z, Hu J, Luo Z, Zhang C, Wang L, et al. MiR-377-3p suppresses colorectal cancer through negative regulation on wnt/β-catenin signaling by targeting XIAP and ZEB2. Pharmacol Res (2020) 156:104774. doi: 10.1016/j.phrs.2020.104774
55. Jia L, Luo S, Ren X, Li Y, Hu J, Liu B, et al. miR-182 and miR-135b mediate the tumorigenesis and invasiveness of colorectal cancer cells via targeting ST6GALNAC2 and PI3K/AKT pathway. Dig Dis Sci (2017) 62(12):3447–59. doi: 10.1007/s10620-017-4755-z
56. Qin A, Qian W. MicroRNA-7 inhibits colorectal cancer cell proliferation, migration and invasion via TYRO3 and phosphoinositide 3-kinase/protein b kinase/mammalian target of rapamycin pathway suppression. Int J Mol Med (2018) 42(5):2503–14. doi: 10.3892/ijmm.2018.3864
57. Ma W, Liu B, Li J, Jiang J, Zhou R, Huang L, et al. MicroRNA-302c represses epithelial-mesenchymal transition and metastasis by targeting transcription factor AP-4 in colorectal cancer. BioMed Pharmacother (2018) 105:670–6. doi: 10.1016/j.biopha.2018.06.025
58. Park YR, Kim SL, Lee MR, Seo SY, Lee JH, Kim SH, et al. MicroRNA-30a-5p (miR-30a) regulates cell motility and EMT by directly targeting oncogenic TM4SF1 in colorectal cancer. J Cancer Res Clin Oncol (2017) 143(10):1915–27. doi: 10.1007/s00432-017-2440-4
59. Park YR, Lee ST, Kim SL, Liu YC, Lee MR, Shin JH, et al. MicroRNA-9 suppresses cell migration and invasion through downregulation of TM4SF1 in colorectal cancer. Int J Oncol (2016) 48(5):2135–43. doi: 10.3892/ijo.2016.3430
60. Park YR, Seo SY, Kim SL, Zhu SM, Chun S, Oh J-M, et al. MiRNA-206 suppresses PGE2-induced colorectal cancer cell proliferation, migration, and invasion by targetting TM4SF1. Biosci Rep (2018) 38(5):BSR20180664. doi: 10.1042/BSR20180664
61. Van Cutsem E, Yoshino T, Lenz HJ, Lonardi S, Falcone A, Limón ML, et al. Nintedanib for the treatment of patients with refractory metastatic colorectal cancer (LUME-colon 1): a phase III, international, randomized, placebo-controlled study. Ann Oncol (2018) 29(9):1955–63. doi: 10.1093/annonc/mdy241
62. Niu Y, Zhang J, Tong Y, Li J, Liu B. miR-145-5p restrained cell growth, invasion, migration and tumorigenesis via modulating RHBDD1 in colorectal cancer via the EGFR-associated signaling pathway. Int J Biochem Cell Biol (2019) 117:105641. doi: 10.1016/j.biocel.2019.105641
63. Zhou Q, Zhu Y, Wei X, Zhou J, Chang L, Sui H, et al. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor a axis. Cell Death Dis (2016) 7(10):e2413. doi: 10.1038/cddis.2016.306
64. Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, et al. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis (2019) 10(2):131. doi: 10.1038/s41419-018-1188-3
65. Fang Y, Sun B, Wang J, Wang Y. miR-622 inhibits angiogenesis by suppressing the CXCR4-VEGFA axis in colorectal cancer. Gene (2019) 699:37–42. doi: 10.1016/j.gene.2019.03.004
66. Hong S, Chen S, Wang X, Sun D, Yan Z, Tai J, et al. ATAD2 silencing decreases VEGFA secretion through targeting has-miR-520a to inhibit angiogenesis in colorectal cancer. Biochem Cell Biol (2018) 96(6):761–8. doi: 10.1139/bcb-2018-0081
67. Chen X, Xu X, Pan B, Zeng K, Xu M, Liu X, et al. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging (Albany NY) (2018) 10(11):3421–37. doi: 10.18632/aging.101656
68. Wu Q-B, Chen J, Zhu J-W, Yin X, You H-Y, Lin Y-R, et al. MicroRNA−125 inhibits RKO colorectal cancer cell growth by targeting VEGF. Int J Mol Med (2018) 42(1):665–73. doi: 10.3892/ijmm.2018.3632
69. Yu F-B, Sheng J, Yu J-M, Liu J-H, Qin X-X, Mou B. MiR-19a-3p regulates the forkhead box F2-mediated wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J Gastroenterol (2020) 26(6):627–44. doi: 10.3748/wjg.v26.i6.627
70. Wu F, Xing T, Gao X, Liu F. miR−501−3p promotes colorectal cancer progression via activation of wnt/β−catenin signaling. Int J Oncol (2019) 55(3):671–83. doi: 10.3892/ijo.2019.4852
71. Pan S, Wu W, Ren F, Li L, Li Y, Li W, et al. MiR-346-5p promotes colorectal cancer cell proliferation in vitro and in vivo by targeting FBXL2 and activating the β-catenin signaling pathway. Life Sci (2020) 244:117300. doi: 10.1016/j.lfs.2020.117300
72. Kwak B, Kim DU, Kim TO, Kim H-S, Kim S-W. MicroRNA-552 links wnt signaling to p53 tumor suppressor in colorectal cancer. Int J Oncol (2018) 53(4):1800–8. doi: 10.3892/ijo.2018.4505
73. Ding X, Zhang J, Feng Z, Tang Q, Zhou X. MiR-137-3p inhibits colorectal cancer cell migration by regulating a KDM1A-dependent epithelial-mesenchymal transition. Dig Dis Sci (2021) 66(7):2272–82. doi: 10.1007/s10620-020-06518-6
74. Liu Y, Zhang Y, Wu H, Li Y, Zhang Y, Liu M, et al. miR-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis (2017) 8(4):e2739. doi: 10.1038/cddis.2017.61
75. Hu F, Min J, Cao X, Liu L, Ge Z, Hu J, et al. MiR-363-3p inhibits the epithelial-to-mesenchymal transition and suppresses metastasis in colorectal cancer by targeting Sox4. Biochem Biophys Res Commun (2016) 474(1):35–42. doi: 10.1016/j.bbrc.2016.04.055
76. Zhou T, Wu L, Wang Q, Jiang Z, Li Y, Ma N, et al. MicroRNA-128 targeting RPN2 inhibits cell proliferation and migration through the akt-p53-cyclin pathway in colorectal cancer cells. Oncol Lett (2018) 16(6):6940–9. doi: 10.3892/ol.2018.9506
77. Zhao Y, Zhou H, Shen J, Yang S, Deng K, Li Q, et al. MiR-1236-3p inhibits the proliferation, invasion, and migration of colon cancer cells and hinders epithelial-mesenchymal transition by targeting DCLK3. Front Oncol (2021) 11:688882. doi: 10.3389/fonc.2021.688882
78. Zhang J, Zhang Y, Li X, Wang H, Li Q, Liao X. MicroRNA−212 inhibits colorectal cancer cell viability and invasion by directly targeting PIK3R3. Mol Med Rep (2017) 16(5):7864–72. doi: 10.3892/mmr.2017.7552
79. Shi L, Su Y, Zheng Z, Qi J, Wang W, Wang C. miR-146b-5p promotes colorectal cancer progression by targeting TRAF6. Exp Ther Med (2022) 23(3):231. doi: 10.3892/etm.2022.11155
80. Ma Z-H, Shi P-D, Wan B-S. MiR-410-3p activates the NF-κB pathway by targeting ZCCHC10 to promote migration, invasion and EMT of colorectal cancer. Cytokine (2021) 140:155433. doi: 10.1016/j.cyto.2021.155433
81. Lyu J, Sun Y, Li X, Ma H. MicroRNA-206 inhibits the proliferation, migration and invasion of colorectal cancer cells by regulating the c-Met/AKT/GSK-3β pathway. Oncol Lett (2021) 21(2):147. doi: 10.3892/ol.2020.12408
82. Lin X, Wang S, Sun M, Zhang C, Wei C, Yang C, et al. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol (2019) 12(1):20. doi: 10.1186/s13045-019-0708-7
83. Mizutani A, Yashiroda Y, Muramatsu Y, Yoshida H, Chikada T, Tsumura T, et al. RK-287107, a potent and specific tankyrase inhibitor, blocks colorectal cancer cell growth in a preclinical model. Cancer Sci (2018) 109(12):4003–14. doi: 10.1111/cas.13805
84. Wang Q, Zhou Y, Rychahou P, Harris JW, Zaytseva YY, Liu J, et al. Deptor is a novel target of wnt/β-Catenin/c-Myc and contributes to colorectal cancer cell growth. Cancer Res (2018) 78(12):3163–75. doi: 10.1158/0008-5472.CAN-17-3107
85. Jiang T, Wang H, Liu L, Song H, Zhang Y, Wang J, et al. CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Mol Cancer (2021) 20(1):167. doi: 10.1186/s12943-021-01474-9
86. Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond) (2021) 41(3):199–217. doi: 10.1002/cac2.12138
87. Li H, Rokavec M, Jiang L, Horst D, Hermeking H. Antagonistic effects of p53 and HIF1A on microRNA-34a regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to mesenchymal transition in colorectal cancer cells. Gastroenterology (2017) 153(2):505–20. doi: 10.1053/j.gastro.2017.04.017
88. Fang C, Dai L, Wang C, Fan C, Yu Y, Yang L, et al. Secretogranin II impairs tumor growth and angiogenesis by promoting degradation of hypoxia-inducible factor-1α in colorectal cancer. Mol Oncol (2021) 15(12):3513–26. doi: 10.1002/1878-0261.13044
89. Sui H, Zhao J, Zhou L, Wen H, Deng W, Li C, et al. Tanshinone IIA inhibits β-catenin/VEGF-mediated angiogenesis by targeting TGF-β1 in normoxic and HIF-1α in hypoxic microenvironments in human colorectal cancer. Cancer Lett (2017) 403:86–97. doi: 10.1016/j.canlet.2017.05.013
90. Sabry D, El-Deek SEM, Maher M, El-Baz MAH, El-Bader HM, Amer E, et al. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1α-VEGF signaling pathway. Mol Cell Biochem (2019) 454(1-2):177–89. doi: 10.1007/s11010-018-3462-1
91. Jin F, Yang R, Wei Y, Wang D, Zhu Y, Wang X, et al. HIF-1α-induced miR-23a∼27a∼24 cluster promotes colorectal cancer progression via reprogramming metabolism. Cancer Lett (2019) 440-441:211–22. doi: 10.1016/j.canlet.2018.10.025
92. Tsai H-L, Miao Z-F, Chen Y-T, Huang C-W, Yeh Y-S, Yang IP, et al. miR-148a inhibits early relapsed colorectal cancers and the secretion of VEGF by indirectly targeting HIF-1α under non-hypoxia/hypoxia conditions. J Cell Mol Med (2019) 23(5):3572–82. doi: 10.1111/jcmm.14257
93. Zhou J, Ji Q, Li Q. Resistance to anti-EGFR therapies in metastatic colorectal cancer: underlying mechanisms and reversal strategies. J Exp Clin Cancer Res (2021) 40(1):328. doi: 10.1186/s13046-021-02130-2
94. Lei X, He Q, Li Z, Zou Q, Xu P, Yu H, et al. Cancer stem cells in colorectal cancer and the association with chemotherapy resistance. Med Oncol (2021) 38(4):43. doi: 10.1007/s12032-021-01488-9
95. Chen H, Yao J, Bao R, Dong Y, Zhang T, Du Y, et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer (2021) 20(1):29. doi: 10.1186/s12943-021-01322-w
96. Zhao L, Chen H, Zhang Q, Ma J, Hu H, Xu L. ATF4-mediated microRNA-145/HDAC4/p53 axis affects resistance of colorectal cancer cells to 5-fluorouracil by regulating autophagy. Cancer Chemother Pharmacol (2022) 89(5):595–607. doi: 10.1007/s00280-021-04393-0
97. Jin Y, Wang M, Hu H, Huang Q, Chen Y, Wang G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol (2018) 117:445–53. doi: 10.1016/j.ijbiomac.2018.05.151
98. Zhao F, Yang Z, Gu X, Feng L, Xu M, Zhang X. miR-92b-3p regulates cell cycle and apoptosis by targeting , thereby affecting the sensitivity of colorectal cancer cells to chemotherapeutic drugs. Cancers (Basel) (2021) 13(13):3323. doi: 10.3390/cancers13133323
99. Sun W, Li J, Zhou L, Han J, Liu R, Zhang H, et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics (2020) 10(5):1981–96. doi: 10.7150/thno.37621
100. Jiang H, Ju H, Zhang L, Lu H, Jie K. microRNA-577 suppresses tumor growth and enhances chemosensitivity in colorectal cancer. J Biochem Mol Toxicol (2017) 31(6):e21888. doi: 10.1002/jbt.21888
101. Zhou H, Lin C, Zhang Y, Zhang X, Zhang C, Zhang P, et al. miR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp expression. Cell Prolif (2017) 50(3):e12341. doi: 10.1111/cpr.12341
102. Han H, Li Y, Qin W, Wang L, Yin H, Su B, et al. miR-199b-3p contributes to acquired resistance to cetuximab in colorectal cancer by targeting CRIM1 via wnt/β-catenin signaling. Cancer Cell Int (2022) 22(1):42. doi: 10.1186/s12935-022-02460-x
103. Wu Y-Z, Lin H-Y, Zhang Y, Chen W-F. miR-200b-3p mitigates oxaliplatin resistance via targeting TUBB3 in colorectal cancer. J Gene Med (2020) 22(7):e3178. doi: 10.1002/jgm.3178
104. Qian X-L, Zhou F, Xu S, Jiang J, Chen Z-P, Wang S-K, et al. MiR-454-3p promotes oxaliplatin resistance by targeting PTEN in colorectal cancer. Front Oncol (2021) 11:638537. doi: 10.3389/fonc.2021.638537
105. Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y, et al. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog (2017) 56(12):2669–80. doi: 10.1002/mc.22710
106. Liu G, Zhou J, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep (2019) 39(3):BSR20190249. doi: 10.1042/BSR20190249
107. Lou T, Zhang L, Jin Z, Miao C, Wang J, Ke K. miR-455-5p enhances 5-fluorouracil sensitivity in colorectal cancer cells by targeting PIK3R1 and DEPDC1. Open Med (Wars) (2022) 17(1):847–56. doi: 10.1515/med-2022-0474
108. Shi L, Xi J, Xu X, Peng B, Zhang B. MiR-148a suppressed cell invasion and migration via targeting WNT10b and modulating β-catenin signaling in cisplatin-resistant colorectal cancer cells. BioMed Pharmacother (2019) 109:902–9. doi: 10.1016/j.biopha.2018.10.080
109. Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, et al. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract (2016) 212(7):643–9. doi: 10.1016/j.prp.2016.04.011
110. Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer (2019) 18(1):43. doi: 10.1186/s12943-019-0981-7
111. Gremke N, Polo P, Dort A, Schneikert J, Elmshäuser S, Brehm C, et al. mTOR-mediated cancer drug resistance suppresses autophagy and generates a druggable metabolic vulnerability. Nat Commun (2020) 11(1):4684. doi: 10.1038/s41467-020-18504-7
112. Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol (2019) 247(5):708–18. doi: 10.1002/path.5222
113. Xia C, Li Q, Cheng X, Wu T, Gao P, Gu Y. Insulin-like growth factor 2 mRNA-binding protein 2-stabilized long non-coding RNA taurine up-regulated gene 1 (TUG1) promotes cisplatin-resistance of colorectal cancer via modulating autophagy. Bioengineered (2022) 13(2):2450–69. doi: 10.1080/21655979.2021.2012918
114. Suski JM, Braun M, Strmiska V, Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell (2021) 39(6):759–78. doi: 10.1016/j.ccell.2021.03.010
115. Tormo E, Ballester S, Adam-Artigues A, Burgués O, Alonso E, Bermejo B, et al. The miRNA-449 family mediates doxorubicin resistance in triple-negative breast cancer by regulating cell cycle factors. Sci Rep (2019) 9(1):5316. doi: 10.1038/s41598-019-41472-y
116. Xu F, Ye M-L, Zhang Y-P, Li W-J, Li M-T, Wang H-Z, et al. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci (2020) 111(5):1528–41. doi: 10.1111/cas.14356
117. Li D-D, Zhao C-H, Ding H-W, Wu Q, Ren T-S, Wang J, et al. A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-fluorouracil by reversing notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif (2018) 51(5):e12480. doi: 10.1111/cpr.12480
118. Eckford PDW, Sharom FJ. ABC Efflux pump-based resistance to chemotherapy drugs. Chem Rev (2009) 109(7):2989–3011. doi: 10.1021/cr9000226
119. Degorter MK, Xia CQ, Yang JJ, Kim RB. Drug transporters in drug efficacy and toxicity. Annu Rev Pharmacol Toxicol (2012) 52:249–73. doi: 10.1146/annurev-pharmtox-010611-134529
120. Lai X, Eberhardt M, Schmitz U, Vera J. Systems biology-based investigation of cooperating microRNAs as monotherapy or adjuvant therapy in cancer. Nucleic Acids Res (2019) 47(15):7753–66. doi: 10.1093/nar/gkz638
121. Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Kamm RD, et al. MicroRNA delivery through nanoparticles. J Control Release (2019) 313:80–95. doi: 10.1016/j.jconrel.2019.10.007
122. Fernandez-Piñeiro I, Badiola I, Sanchez A. Nanocarriers for microRNA delivery in cancer medicine. Biotechnol Adv (2017) 35(3):350–60. doi: 10.1016/j.biotechadv.2017.03.002
123. Liang G, Zhu Y, Jing A, Wang J, Hu F, Feng W, et al. Cationic microRNA-delivering nanocarriers for efficient treatment of colon carcinoma in xenograft model. Gene Ther (2016) 23(12):829–38. doi: 10.1038/gt.2016.60
124. Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res (2011) 71(15):5214–24. doi: 10.1158/0008-5472.CAN-10-4645
125. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet (2014) 383(9927):1490–502. doi: 10.1016/S0140-6736(13)61649-9
126. Mattesen TB, Rasmussen MH, Sandoval J, Ongen H, Árnadóttir SS, Gladov J, et al. MethCORR modelling of methylomes from formalin-fixed paraffin-embedded tissue enables characterization and prognostication of colorectal cancer. Nat Commun (2020) 11(1):2025. doi: 10.1038/s41467-020-16000-6
127. Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol (2016) 27(10):1879–86. doi: 10.1093/annonc/mdw279
128. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U.S.A. (2008) 105(30):10513–8. doi: 10.1073/pnas.0804549105
129. Ng EKO, Chong WWS, Jin H, Lam EKY, Shin VY, Yu J, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut (2009) 58(10):1375–81. doi: 10.1136/gut.2008.167817
Keywords: colorectal cancer, MicroRNAs, drug resistance, metastasis, signaling pathways, therapeutic targets
Citation: Yang Y, Meng W-J and Wang Z-Q (2022) MicroRNAs (miRNAs): Novel potential therapeutic targets in colorectal cancer. Front. Oncol. 12:1054846. doi: 10.3389/fonc.2022.1054846
Received: 27 September 2022; Accepted: 29 November 2022;
Published: 14 December 2022.
Edited by:
Qun Zhang, Nanjing Medical University, ChinaCopyright © 2022 Yang, Meng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Jian Meng, bWVuZ3dlbmppYW5AMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.