
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 22 December 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1051497
Claudins are a tetraspan membrane protein multigene family that plays a structural and functional role in constructing tight junctions. Claudins perform crucial roles in maintaining cell polarity in epithelial and endothelial cell sheets and controlling paracellular permeability. In the last two decades, increasing evidence indicates that claudin proteins play a major role in controlling paracellular permeability and signaling inside cells. Several types of claudins are dysregulated in various cancers. Depending on where the tumor originated, claudin overexpression or underexpression has been shown to regulate cell proliferation, cell growth, metabolism, metastasis and cell stemness. Epithelial-to-mesenchymal transition is one of the most important functions of claudin proteins in disease progression. However, the exact molecular mechanisms and signaling pathways that explain why claudin proteins are so important to tumorigenesis and progression have not been determined. In addition, claudins are currently being investigated as possible diagnostic and treatment targets. Here, we discuss how claudin-related signaling pathways affect tumorigenesis, tumor progression, and treatment sensitivity.
Claudins (CLDNs) are a group of indispensable membrane tight junction (TJ) proteins that range in size from 17 to 27 kDa and include up to 27 members in mammals that exhibit a high degree of sequence homology (1). CLDNs were first identified in 1998 in a purified junctional fraction from chicken liver (2). According to hydrophilicity studies, claudin proteins comprise four transmembrane helices, two extracellular loops, an internal C-terminus, and a very short internal N-terminal region (2). The C-terminus contacts cytoplasmic proteins via a PDZ motif. Here, the first loop regulates paracellular charge selectivity, and the second loop of CLDN3 and CLDN4 is the receptor for a bacterial toxin (3–5). All known epithelial tissues exhibit tissue- and/or cell-specific expression of CLDNs. For instance, CLDN18.1 and CLDN18.2 are both particularly expressed in the stomach and lung alveolar epithelium, respectively (6, 7). CLDN3 is primarily expressed in alveolar epithelial type II cells (8). CLDN2, CLDN3, CLDN7, and CLDN15 are all found in high concentrations in intestinal epithelial cells (9, 10). CLDNs are generally located in the cell membrane and, under specific conditions, can be localized to the cytoplasm in both normal and neoplastic tissues. This cytoplasmic localization may be associated with CLDN phosphorylation and vesicle trafficking (11–14). The extracellular loops of CLDNs play an essential role in TJ formation by interacting with each other to seal the cellular sheet, thereby playing a role in regulating paracellular permeability and maintaining cell polarity (15). Growing evidence indicates that CLDNs are also involved in signal transduction by interacting with multiple proteins, regulating cell proliferation, cell growth, metabolism, metastasis and cell stemness. In addition, mutation of some CLDNs has been causally associated with human diseases, and CLDNs have been found to be deregulated in various cancers. This review aims to provide an overview of the potential signaling pathways and mechanisms of CLDNs (Table 1) in the occurrence, metastasis, and treatment of tumors.
CLDN1 is one of the most-studied CLDNs in cancer (Figure 1). CLDN1 promotes the growth and progression of various tumors, including breast cancer, melanoma, oral squamous cell carcinoma, thyroid cancer, ovarian cancer, colon cancer, gastric cancer, hepatocellular carcinoma, and pancreatic cancer. CLDN1 is thought to be a tumor suppressor in lung and prostate cancers (57). In breast cancer, CLDN1 expression levels vary in pathology subtypes (58). In most breast cancers, downregulation of CLDN1 is more frequently correlated with higher invasiveness and poor prognosis (59). The basal-like subtype of breast cancer, which is typically associated with a poor prognosis, is one subtype where CLDN1 expression is elevated (60). At the molecular level, CLDN1 is regulated by disintegrin and metalloproteinase-15 (ADAM15) via PI3K/Akt/mTOR signaling in breast cancer (16). In addition, CLDN1 expression is also be upregulated through the ERK signaling pathway in the MCF7 cell line (17).
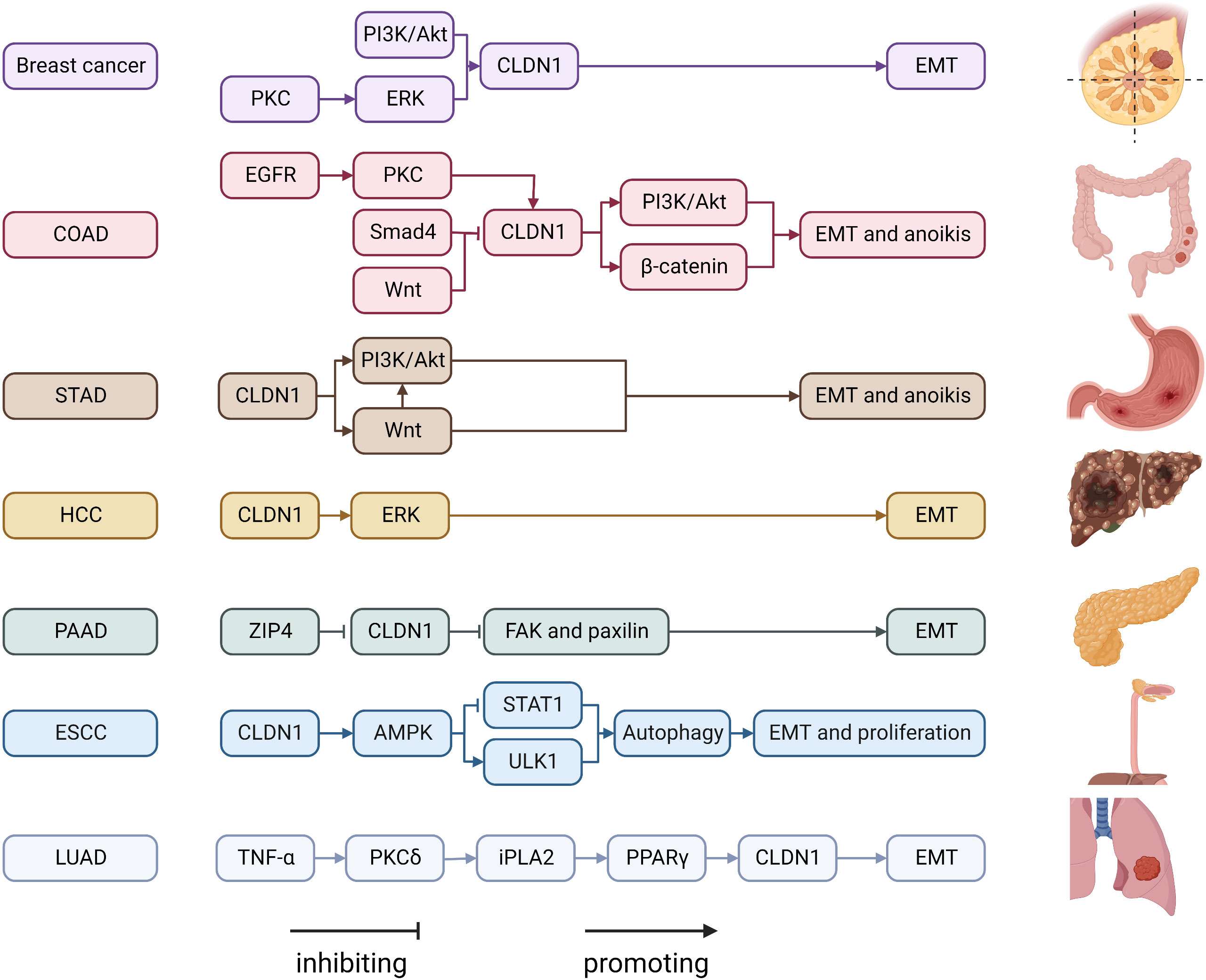
Figure 1 Regulatory mechanisms of CLDN1 expression in cancers. COAD, Colon adenocarcinoma; ESCC, esophageal squamous cell carcinoma; FAK, focal adhesion kinase; HCC, hepatocellular carcinoma; iPLA2, calcium-independent phospholipase A2; LUAD, lung adenocarcinoma; PAAD, pancreatic adenocarcinoma; PGE2, Prostaglandin E2; PI3K, phospatidylinositol-3 kinase; PKC, protein kinase C; PPAR-γ, Peroxisome proliferator-activated receptor-γ; STAD, stomach adenocarcinoma; STAT1, signal transducer and activator of transcription 1; TNF α, tumor necrosis factor α; ULK1, unc-51 like autophagy activating kinase 1; ZIP4, solute carrier family 39 member 4.
The role of CLDN1 expression in colon cancer is debated. While some studies link increasing CLDN1 expression to the growth and spread of colon cancer, others believe that low CLDN1 levels are a standalone predictor of a poor prognosis (61, 62). This disparity could be caused by Resnick’s (62) work mistakenly identifying nuclear translocation of CLDN1 as low expression of CLDN1, when in fact CLDN1 expression was high in metastatic colon cancer. The migration ability of colon cancer cells is enhanced by the upregulation and internalization of CLDN1 via the EGFR/PKC/CLDN1 signaling pathway (13). Additionally, CLDN1 can upregulate ZEB-1, which reduces E-cadherin expression in colon cancer cells, increases their invasive activity and decreases anoikis (19). Previous studies demonstrated that the Wnt signaling pathway promoted CLDN1 expression and enhanced colon cancer cell proliferation, invasion and metastasis (63–67). By increasing CLDN1 gene expression, the Wnt signaling pathway inhibitor FH535 prevented colorectal cancer cells from proliferating and migrating (18). In stomach adenocarcinoma, CLDN1 is an oncogenic molecule. CLDN1’s malignant potential may be attributed in part to its regulation of the Wnt signaling pathway (21) with its knockdown significantly inhibiting cell proliferation, migration, and invasion while increasing apoptosis (68). CLDN1 is highly expressed in hepatocellular carcinoma and acts as a promoter of epithelial-to-mesenchymal transition (EMT) through the c-Abl-Ras-Raf-1-ERK1/2 signaling axis (22, 69, 70). It has been demonstrated that CLDN1 downregulation in pancreatic cancer results in the phosphorylation of FAK and paxillin, which further promotes cell invasion, migration, and tumor metastasis (23). Given that CLDN1 is mostly found in the nucleus of esophageal squamous cell carcinoma (ESCC) and has been shown to be abnormally elevated, CLDN1 promotes ESCC growth and metastasis by upregulating the expression of ULK1 via the AMPK/STAT1 signaling pathway (14). Low CLDN1 expression in response to TNF-α is regulated by the PKC/iPLA2/PGE2/PPAR signaling cascade and is associated with lung adenocarcinoma rather than lung squamous cell carcinoma, whereas increased CLDN1 expression is associated with a better prognosis and tumor suppressive activity (24, 71, 72). CLDN1 is involved in a variety of signaling pathways that are particularly involved in invasion and migration. Numerous CLDN1-targeting antibodies, including 3A2 and 6F6, have been demonstrated to be tumor suppressive (73, 74). As a result, the molecules in the pathway that interact with CLDN1 can be investigated further as potential drug targets.
According to a growing body of evidence, dysregulated CLDN2 expression affects processes underlying carcinogenesis and metastasis formation, such as proliferation, migration, and EMT (29, 75–80). Increased CLDN2 expression is mediated by the EGF/Ras/Raf/MEK/ERK/c-Fos pathway and the PI3K/Akt/NF-B pathway (25, 27, 75, 81–85). Loss of CLDN2 promotes the dissociation of the CLDN2/ZO1/ZONAB complex, which then induces ZONAB translocation to the nucleus and enrichment at the NDRG1 promotor to activate its transcription, thereby preventing the growth and spread of colorectal cancer (26, 86–91). Through its PDZ binding motif, CLDN2 interacts with YAP in renal clear cell carcinoma and prevents YAP nuclear localization and activation (28). Osteosarcoma tissue exhibits reduced levels of CLDN2 expression, thereby inhibiting the Ras/Raf/MEK/ERK signaling pathway via afadin and eventually reducing the capacity of osteosarcoma cells to migrate (29). However, the effective development of breast cancer metastases is made possible by signaling downstream from a CLDN2/Afadin complex (30). Notably, increased intracellular accumulation of anticancer agents and decreased expression of ABCC2 were observed in CLDN2-downregulated lung adenocarcinoma cells (31). Hence, inhibiting CLDN2-related signaling pathways could represent a potential novel target for therapeutic intervention. In fact, flavonoids, such as quercetin, chrysin, kaempferol, and luteolin as well as inhibitors of intracellular signaling reduce chemoresistance, invasion, and proliferation in a variety of malignancies (82, 92). VPDSM and DSMKF, short peptides that mimic CLDN2’s second extracellular loop (ECL2), may directly act on the CLDN2 protein and lead to CLDN2 internalization, whereas CLDN2 expression is reduced via the clathrin-dependent endocytosis pathway (93, 94). In xenografted mice, CLDN2 ECL-targeting antibody 1A2 suppresses tumor growth in a manner similar to CLDN1-targeting antibody 3A2 (95). Inhibitors targeting CLDN2-related intracellular signal transduction molecules inhibit cancer development. The idea of reducing CLDN2 expression in cells by mimicking the short peptide of CLDN2’s second extracellular loop (ECL2) to improve tumor cell sensitivity to chemotherapy drugs provides us with new inspiration. We consider whether this drug design concept can be applied to other CLDNs.
Numerous human tumors and their subsequent metastasis have been linked to the dysregulation of CLDN3. CLDN3 regulation involves a variety of signaling pathways. Studies have revealed that SCF/c-kit signaling exclusively enhances CLDN3 expression in colorectal cancer, mainly by activating the JNK pathway, and that the SCF/c-kit-JNK-AP-1 signaling axis is crucial for controlling CLDN3 expression (33). EGF-induced CLDN3 upregulation via the ERK pathway and PI3K-Akt pathway is associated with increased malignancy in HT-29 colon cancer cells (32). Another contentious finding, however, demonstrated that CLDN3 is the most abundant cell-adhesion protein in the normal colon and that its absence promotes Wnt signaling, which is subsequently excessively triggered by IL-6/gp130/STAT3 signaling (34). Contradictory findings from these two studies on the involvement of CLDN3 in colorectal cancer indicate that we still know very little about the role of CLDN3 in colorectal cancer. In addition to what is already known, CLDN3 overexpression or silencing alter the structure and function of TJs, hence disrupting intestinal homeostasis. CLDN3 expression is increased in lung adenocarcinoma compared with lung squamous cell carcinoma (96). Through Wnt/β-catenin signaling pathway inhibition, CLDN3 prevents EMT in lung squamous cell carcinoma (36). However, CLDN3 overexpression in lung cancer can increase malignant potential by activating the EGF-activated MEK/ERK and PI3K-Akt pathways (35). The reasons for this discrepancy are unclear but may be attributed to the different properties of tumorigenicity. Type II alveolar epithelial cells are the primary lung cells that express CLDN3 (97). Lung adenocarcinoma is thought to develop from type II alveolar epithelial cells, whereas lung adenosquamous carcinoma and lung squamous cell carcinoma develop from different epithelial cell components (98). For primary hepatocellular carcinoma patients, downregulation of the CLDN3-Wnt/β-catenin-EMT axis is significantly correlated with survival differences (37). CLDN3 is associated with drug resistance and prognosis in a number of tumors, and its role involves multiple signaling pathways, including the PI3K signaling pathway, ERK signaling pathway, and Wnt signaling pathway. CLDN3 and CLDN4 have been identified as useful biomarkers in the diagnosis of gynecological malignancies and can be combined with other markers such as CA125. CLDN3 and CLDN4 have been demonstrated to be effective Clostridium perfringens enterotoxin (CPE) targets in the treatment of gynecological malignancies (99).CPE’s C-terminal domain(c-CPE) can bind to the second extracellular loop of a variety of claudin proteins to form hexameric pores at the membrane surface, resulting in calcium influx and cell death (100).CLDN3, 4, 5, 6, 7, 8, 9 and 14 are examples of claudin proteins that have been found to bind to CPE. As a result, Claudius exhibit considerable potential as a cancer therapy targets.
CLDN4 expression is dysregulated in multiple cancers, including esophageal, gastric, breast, lung, biliary, ovarian, endometrial, bladder, uterine, renal, nasopharyngeal, and prostate cancers (101–112). EGF enhances the degradation of CLDN4 at the posttranslational level via MEK/ERK signaling and PI3K/Akt signaling (113). Furthermore, PI3K/Akt signaling is activated by reduced CLDN4 expression (38, 114). TGF-β and SMAD2/3/4 signaling regulate CLDN4 promoter activity, and expression levels differ between cancers (115, 116). CLDN4, which does not form TJs, participates in intracellular signaling (117). EphA2 and PKC phosphorylation of CLDN4 may weaken TJs, release CLDN4 from TJs, and boost CLDN4’s ability to bind to YAP and ZO-1 to create a nuclear translocating complex, which would activate YAP and EMT (39, 118). CLDN4 upregulation also activates Wnt/β-catenin signaling and triggers the progression of hepatocellular carcinoma and ovarian cancer (40–42). In uterine corpus endometrial carcinoma, CLDN4 induces tumorigenesis by activating the PI3K/AKT pathway (43). CLDN4 is commonly overexpressed in many epithelial cancers. As previously stated, CLDN4 has sparked considerable interest due to its role as a natural CPE receptor. A C-terminal fragment containing only CPE (c-CPE) can bind specifically to claudin protein and increase paracellular permeability by disrupting TJs, improving drug delivery across tissue membranes without disrupting plasma membrane integrity and causing cytotoxicity (119). This finding indicates that CPE represents an effective drug carrier and is less toxic than natural CPE, making it more suitable for cancer treatment. Currently, some drugs designed with c-CPE characteristics have been used in clinical research with good results, such as Doxorubicin-loaded C-SNPs (DOX-C-SNPs) and poly(lactic-co-glycolic-acid) (PLGA) nanoparticles (NPs) modified with the carboxy-terminal binding domain of CPE (c-CPE-NPs) (120, 121) c-CPE 194 is a c-CPE mutant that only binds to Claudin-4 and improves the efficacy of anticancer agents (122). Furthermore, many studies have shown that combining c-CPE and chemotherapy drugs can increase tumor cell sensitivity to chemotherapy drugs (123). A monoclonal antibody that recognized the ECL1 of CLDN3 and CLDN4, KM3907 (IgG2a), induced antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity in vitro and significantly inhibited tumor formation in SCID mice (124). Finally, CLDN4 exhibits significant therapeutic and diagnostic utility.
CLDN6 is described as a tumor suppressor gene and helps maintain cell stemness. CLDN6 expression levels are altered in various types of cancer (Figure 2). CLDN6 is well studied in breast cancer. Compared with adjacent normal tissues, CLDN6 expression levels are significantly reduced in breast cancer tissues (125, 126). It has been proposed that CLDN6 silencing is associated with DNA methyltransferase 1 (DNMT1)-induced DNA methylation, which is controlled by the TGF-β/SMAD2 pathway (127). By bringing MeCP2 to the CLDN6 promoter, deacetylating H3 and H4, and changing the chromatin structure, DNA methylation inhibits the production of CLDN6 (128). Silencing the CLDN6 gene causes increased growth and migratory ability, as well as increased MMP-2 expression and activity, which may be mediated by the MAPK pathway (129). CLDN6 expression is linked to apoptosis signal-regulating kinase 1 (ASK1) expression, and restoring CLDN6 expression in breast cancer cells reduces ASK1 phosphorylation, activates the downstream target proteins JNK and p38 kinase, and induces apoptosis (130–133). 17β-Estradiol (E2) enhances CLDN6 expression via estrogen receptor α (ERα) and estrogen receptor β (ERβ) (44, 134). In addition, CLDN6, which triggers the beclin1-dependent autophagic cascade, is a mediator of the inhibitory effect of ERβ on breast cancer cells migration and inveations (44). The results from studies on the sensitivity of conventional therapeutic drugs have demonstrated that the reinstatement of CLDN6 expression in breast cancer cells increases chemoresistance to Adriamycin, an anticancer medication that is frequently used to treat breast cancer by activating the AF-6/ERK signaling pathway and upregulating cancer stem cell properties (135, 136). CLDN6-induced chemoresistance in breast cancer is mediated by glutathione S-transferase-p1 (GSTP1), which is regulated by p53 (137). CLDN6 binds to the transcriptional coactivator with a PDZ-binding motif (TAZ) and reduces the amount of TAZ. This activity inhibits the transcription of c-MYC, which slows the growth of breast cancer cells (138). Under hypoxic conditions, the accumulation of hypoxia-inducible factor 1 (HIF-1) promotes CLDN6 transcription, and increased CLDN6 weakens HIF-1α protein stability by reducing SUMO-specific protease 1 expression and preventing the deSUMOylation of HIF-1α, ultimately leading to HIF-1α degradation and breast cancer metastasis suppression (139).
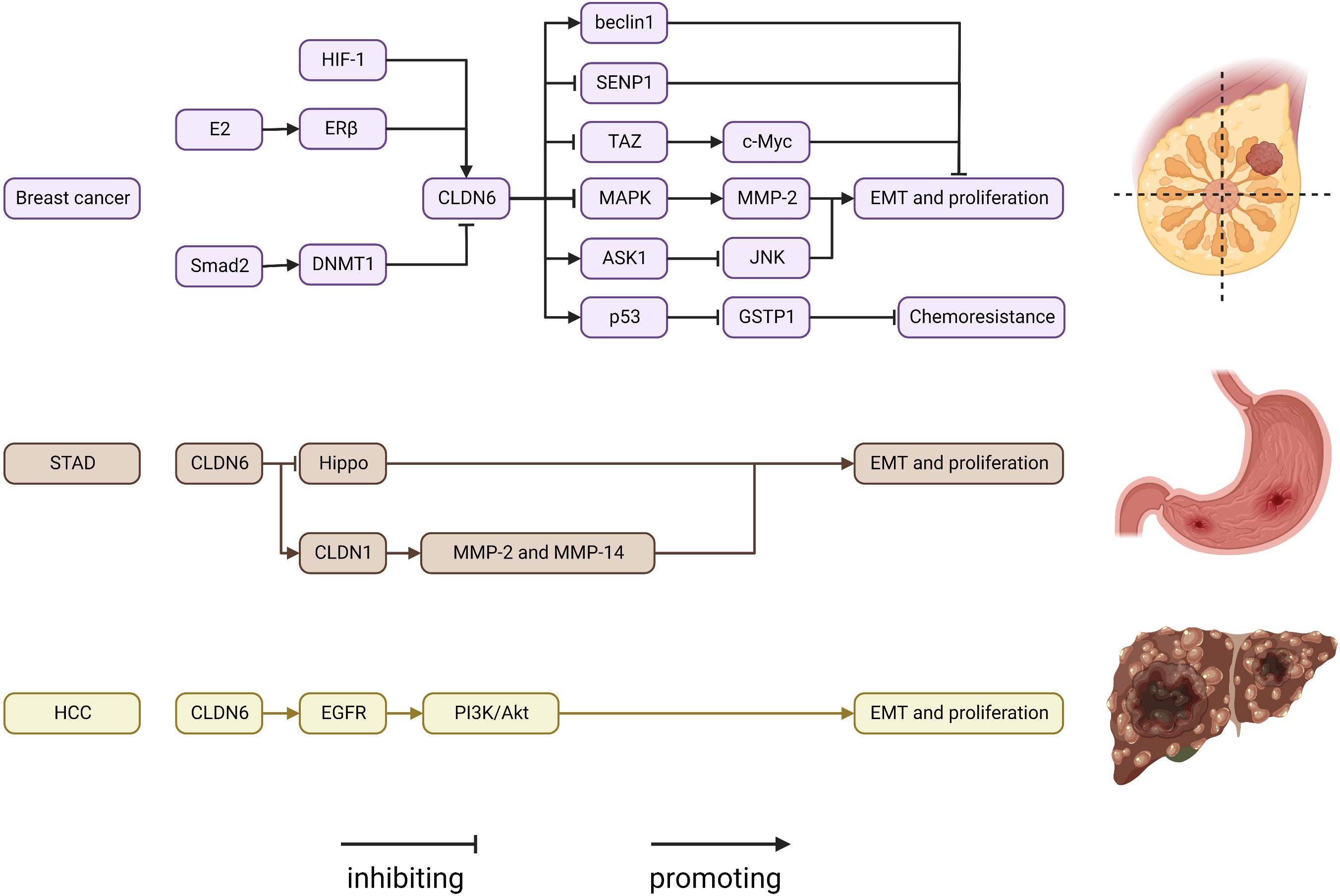
Figure 2 Regulatory mechanisms of CLDN6 expression in cancers. ASK1, apoptosis signal-regulating kinase 1; CLDN, claudin; DNMT1, DNA methyltransferase 1; E2, 17β-estradiol; EGFR, epidermal growth factor receptor; ERβ, estrogen receptor β; GSTP1, glutathione S-transferase-p1; HIF-1, hypoxia-inducible factor 1; MMP, matrix metalloproteinase; SENP1, SUMO-specific protease 1; STAD, stomach adenocarcinoma; TAZ, transcriptional coactivator with PDZ-binding motif.
Compared to histologically normal gastric tissues, CLDN6 expression is noticeably reduced in the majority of gastric cancer tissues; however, some have remarkably greater expression levels (140–143). Additionally, CLDN6 interacts with LATS1/2 to reduce LATS1/2 and YAP1 phosphorylation, influence YAP1 entry into the nucleus, and enhance the invasive ability of gastric cancer cells (45, 144). Through the expression of CLDN1 on the membrane, CLDN6 can also cause MMP-2 activation, which encourages cell migration and invasion (145). However, another study found the converse to be true; specifically, CLDN6 overexpression caused differentiation in BGC-823 cells by blocking the JNK pathway with no impact on in vitro apoptosis, proliferation, invasiveness, or migration (143). The different gastric cancer cell lines that were used might be the cause of the inconsistent results. CLDN6 is highly expressed in hepatocellular carcinoma, and can promote EMT in hepatoma cells (146). In addition, CLDN6 overexpression may act as an oncogene and enhance HepG2 cell migration, invasion, and proliferation via EGFR/AKT/mTOR signaling in hepatocellular carcinoma (46). Chimeric antigen receptor (CAR)-T/NK and antibody−drug conjugates are two cutting-edge and effective therapy options for cancer. CLDN6 was used as a promising target in the development of treatments. AMG 794 for nonsquamous non-small cell lung cancer or epithelial ovarian cancer (NCT05317078) as well as DS-9606a for advanced solid tumors are currently being assessed in two phase 1 studies (NCT05394675) (147, 148). Additionally, research on CLDN6-CAR-NK-cell treatment for advanced solid tumors (NCT05410717) is ongoing (149). The results from the aforementioned CLDN6 studies are anticipated.
CLDN7 is either downregulated or disrupted in a variety of tumors. CLDN7 downregulation may lead to epithelial damage and TJ structural changes, which are associated with the etiology and progression of malignancies (150–154). However, CLDN7 was strongly expressed in benign bronchial epithelial cells but substantially expressed or completely absent in lung cancer cells. This finding suggests that CLDN7 may act as a tumor suppressor in lung cancers. CLDN7 transfection induces the downregulation of ERK1/2 phosphorylation, leading to MAPK/ERK pathway inhibition. MAPK/ERK pathway inhibition causes the formation of shorter foot processes, thereby decreasing cellular motility (47). CLDN7 is phosphorylated by protein kinase C at serine 204, and CLDN7 phosphorylation increases chemosensitivity to cisplatin treatment by activating the caspase pathway in lung cancer cells (48). CLDN7 expression in salivary adenoid cystic carcinoma was lower than that in paired normal tissues, and CLDN7 knockdown promotes cell proliferation and metastasis through the Wnt/β-catenin signaling pathway in SACC-LM cells (49). CLDN7 expression is downregulated as colorectal cancer tissue differentiation grade decreases, the invasion and migration abilities of colorectal cancer cells are subsequently enhanced as a result of modulation of CLDN7 expression (155, 156). The stemness properties of colorectal cancer are enhanced by CLDN7, which mediates Wnt/β-catenin pathway activation by SOX-9 (50, 157, 158). CLDN7 expression is involved in a variety of activities and is a crucial target for preventing tumor proliferation and invasion. Early detection, treatment, and prognosis of cancer may be improved through research focusing on CLDN7.
Multiple groups have reported that the CLDN10 expression level is associated with the progression of a variety of tumors and may serve as a potential prognostic biomarker (159–164). CLDN10 is highly expressed in osteosarcoma cells compared with fetal osteoblast cells, and CLDN10 overexpression in osteosarcoma cells enhances the JAK1/STAT1 signaling pathway to significantly promote proliferation and motility (51). However, the underlying mechanisms have received little attention, and more research is needed.
Tumor-related mechanisms in signaling events related to CLDN17 have been less studied. CLDN17 overexpression is strongly linked to cancer metastasis and survival rate in patients with hepatocellular carcinoma and promotes cell migration and invasion in the hepatocyte line HL7702. Tyk2/STAT3 signaling may be one of the most important mechanisms (52). In contrast, CLDN17 may serve as a negative regulator in oral cancer by blocking invasion and migration via the EMT process (165). More research is needed to determine how CLDN17 contributes to the pathogenesis of cancer.
CLDN18 has received the most attention in recent years (Figure 3). It has two tissue-specific isoforms, each with its own promoter: exon 1 and common exons 2–5. CLDN18.1 is exclusively expressed in lung epithelium, whereas CLDN18.2 is only found in the stomach and bone. CLDN18.1 is aberrantly localized in many cancers, especially pancreatic and gastric cancers (7, 55, 166–170).
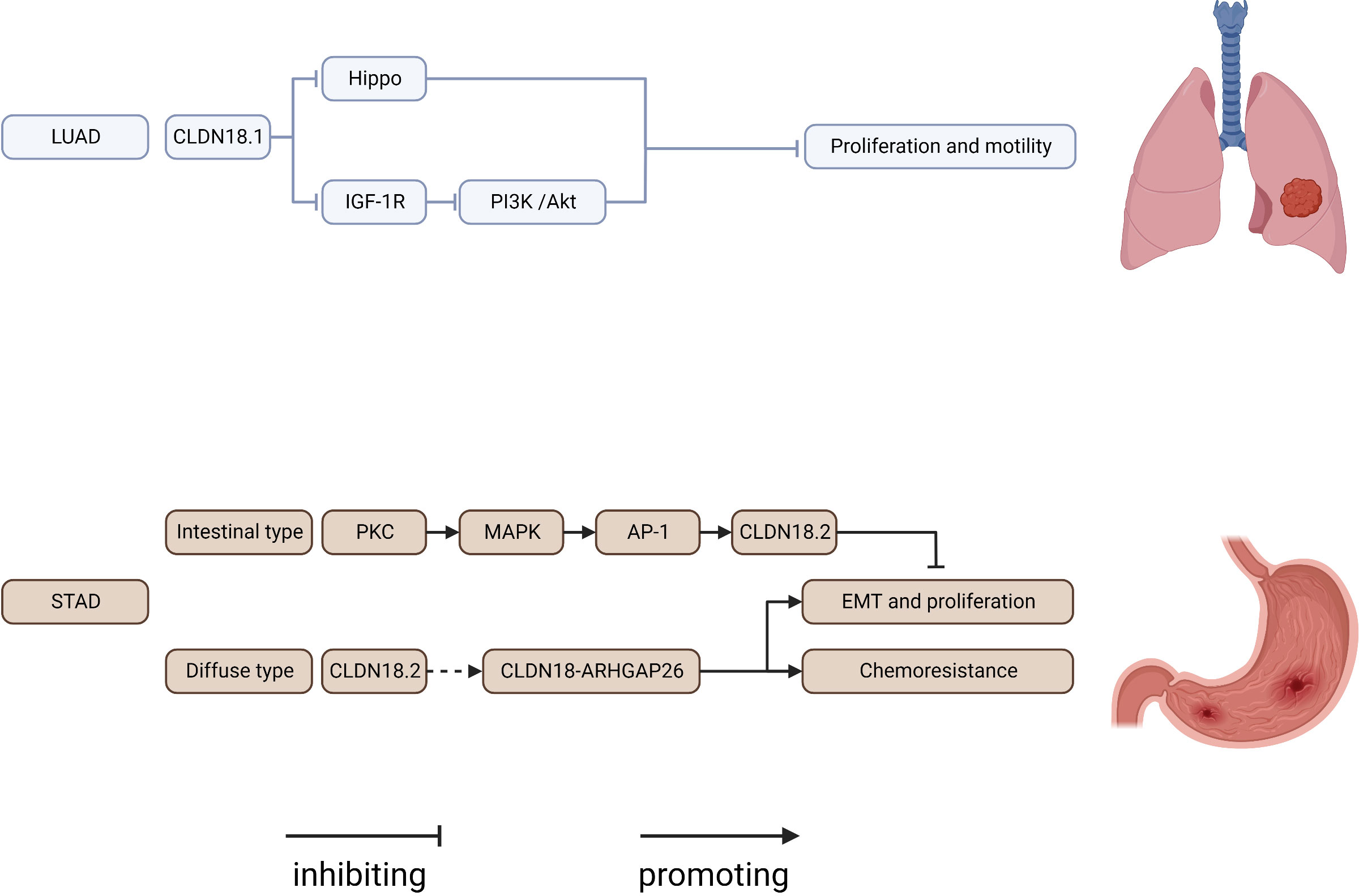
Figure 3 Regulatory mechanisms of CLDN18 expression in cancers. AP-1, AP1 transcription factor; IGF-1R, insulin-like growth factor 1 receptor; LUAD, lung adenocarcinoma; PI3K, phospatidylinositol-3 kinase; PKC, protein kinase C; STAD, stomach adenocarcinoma.
CLDN18.1 knockout resulted in increases in lung size and cellularity due to alveolar epithelial type II cell proliferation in mice. Furthermore, most CLDN18 knockout mice develop lung adenocarcinomas spontaneously between the ages of 18 and 20 months (53, 171–173). CLDN18.1 inhibits two oncogenic pathways, Yap/Taz and IGF-1R signaling, resulting in the suppression of cell motility and increased cell death in anoikis by inhibiting PDK1 and subsequent inhibition of Akt phosphorylation (53, 54, 174).
CLDN18.2 represents a promising new therapeutic target for pancreatic and stomach cancer. Downregulation of CLDN18.2 is linked to the intestinal phenotype of gastric cancers, and CLDN18.2 expression in gastric cells is upregulated via a PKC/MAPK/AP-1-dependent pathway (55). CLDN18.2 is highly expressed in gastric signet-ring cell carcinoma (175). According to reports, genomically stable gastric cancers frequently have chromosomal rearrangements of the CLDN18 and ARHGAP genes, primarily CLDN18-ARHGAP26/6 fusions. Importantly, after CLDN18-ARHGAP26 was introduced, considerable resistance to 5-fluorouracil and oxaliplatin was observed in a number of gastric cancer cell lines, which may account for the poor drug response of patients harboring this fusion product (176–180). The relationship between CLDN18.2 expression and the prognosis of gastric cancers remains controversial. Past research indicated that CLDN18.2 was a good marker of poor survival in gastric cancer and that the downregulation of CLDN18 may represent an early event of gastric carcinogenesis with an intestinal phenotype (181, 182). However, resent research revealed that CLDN18.2 expression was not related to prognosis among patients with gastric cancer (183–186). The diverse ethnic makeup of the cohort, intra-tumor variation and the limitations of a limited sample size can all be used to explain this variation in CLDN18 expression rate. The use of different CLDN18 expression detection antibodies can also result in variations in CLDN18 expression rates. For the creation of comparable data in future research, uniform immunostaining and grading methodologies are therefore crucial. CLDN18.2 is ectopically highly expressed in pancreatic cancer, and activation of the PKC pathway might be involved (56, 187). CLDN18 has been considered a potential marker and therapeutic target in gastric-type mucinous carcinoma, esophageal adenocarcinoma, mucinous ovarian cancer, ductal pancreatic adenocarcinoma, and intrahepatic cholangiocarcinoma (170, 188–190). The creation of new medications and cutting-edge therapies for CLDN18.2 has recently become a popular area of study. Zolbetuximab (IMAB362), a monoclonal antibody targeting CLDN18.2, extends progression-free and overall survival in CLDN182-prositive patients with gastric and gastroesophageal adenocarcinoma (191–193). In addition, patients with advanced gastric cancer also benefited from CLDN18.2-targeted CAR-T cells (CT041) in a phase 1 trial (194). We are eagerly waiting for the results of a number of ongoing prospective clinical studies assessing CLDN18.2 in solid tumors. In conclusion, CLDN18 is a very good target for determining what is wrong with a tumor and treating it.
Since the discovery of the claudin protein, a great deal has been learned regarding its structure and function. On the one hand, the claudin family of membrane proteins interacts with the zonula occludens (ZO) family, junctional adhesion molecules (JAMs), and membrane lipids to play a central role in the structure and function of tight junctions. In MDCK II cells, Claudin-1/2/3/4/7 quintuple-KO (claudin quintuple-KO) cells lack TJ chains, but the adjacent plasma membranes remain tightly attached to each other, showing only disruption of the permeability barrier to ions and small molecules. However, when JAM-A was further removed from claudin quintuple-KO MDCK II cells, the intercellular space was enlarged, and the macromolecular permeability barrier was disrupted (195).These phenomena indicate that claudin proteins play a decisive role in the formation of TJs and affect the function of the permeability barrier together with cell membrane molecules, such as JAMs. When the expression of claudin protein is altered or translocated, the structure or function of TJs is damaged, and barrier permeability changes, resulting in homeostasis dysregulation. TJ results and functional changes also weaken the cell anchor, making tumor cells more prone to invasion and migration. On the other hand, Claudin proteins are an important component of cell signaling that is involved in several classic tumor-related signaling pathways and affects every aspect of tumor biology and all stages of tumor development. In addition, the tissue-specific expression of CLDNs and the disruption of the tight junction structure in malignant tissues suggest that CLDNs may represent ideal diagnostic and therapeutic targets (e.g., CLDN18.2 for gastric cancer, c-CPE for breast cancer and pancreatic cancer). As more research is conducted, we will learn more about their role in tumorigenesis, which will lead to the development of novel cancer treatments and prevention strategies.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
National Natural Science Foundation of China (No. 82072688, No. 81902437). Foundation of Science & Technology Department of Sichuan Province (No. 2020YJ3357, No. 2021YJ0475). Medical Science and Technique Project of Health Commission of Sichuan Province (No. 21PJ043).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.

1. Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, et al. Predicted expansion of the claudin multigene family. FEBS Lett (2011) 585(4):606–12. doi: 10.1016/j.febslet.2011.01.028
2. Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol (1998) 141(7):1539–50. doi: 10.1083/jcb.141.7.1539
3. Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol (2002) 283(1):C142–147. doi: 10.1152/ajpcell.00038.2002
4. Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tsukita S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett (2000) 476(3):258–61. doi: 10.1016/S0014-5793(00)01744-0
5. Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol (1999) 147(6):1351–63. doi: 10.1083/jcb.147.6.1351
6. Koval M. Claudin heterogeneity and control of lung tight junctions. Annu Rev Physiol (2013) 75:551–67. doi: 10.1146/annurev-physiol-030212-183809
7. Niimi T, Nagashima K, Ward JM, Minoo P, Zimonjic DB, Popescu NC, et al. Claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol Cell Biol (2001) 21(21):7380–90. doi: 10.1128/mcb.21.21.7380-7390.2001
8. Guo R, Li Y, Han M, Liu J, Sun Y. Emodin attenuates acute lung injury in cecal-ligation and puncture rats. Int Immunopharmacol (2020) 85:106626. doi: 10.1016/j.intimp.2020.106626
9. Holmes JL, Van Itallie CM, Rasmussen JE, Anderson JM. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr Patterns (2006) 6(6):581–8. doi: 10.1016/j.modgep.2005.12.001
10. Garcia-Hernandez V, Quiros M, Nusrat A. Intestinal epithelial claudins: expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci (2017) 1397(1):66–79. doi: 10.1111/nyas.13360
11. Blackman B, Russell T, Nordeen SK, Medina D, Neville MC. Claudin 7 expression and localization in the normal murine mammary gland and murine mammary tumors. Breast Cancer Res (2005) 7(2):R248–255. doi: 10.1186/bcr988
13. Kim H, Kim SH, Hwang D, An J, Chung HS, Yang EG, et al. Extracellular pyruvate kinase M2 facilitates cell migration by upregulating claudin-1 expression in colon cancer cells. Biochem Cell Biol (2020) 98(2):219–26. doi: 10.1139/bcb-2019-0139
14. Wu J, Gao F, Xu T, Li J, Hu Z, Wang C, et al. CLDN1 induces autophagy to promote proliferation and metastasis of esophageal squamous carcinoma through AMPK/STAT1/ULK1 signaling. J Cell Physiol (2020) 235(3):2245–59. doi: 10.1002/jcp.29133
15. Van Itallie CM, Anderson JM. Claudin interactions in and out of the tight junction. Tissue Barriers (2013) 1(3):e25247. doi: 10.4161/tisb.25247
16. Mattern J, Roghi CS, Hurtz M, Knäuper V, Edwards DR, Poghosyan Z. ADAM15 mediates upregulation of claudin-1 expression in breast cancer cells. Sci Rep (2019) 9(1):12540. doi: 10.1038/s41598-019-49021-3
17. Blanchard AA, Ma X, Wang N, Hombach-Klonisch S, Penner C, Ozturk A, et al. Claudin 1 is highly upregulated by PKC in MCF7 human breast cancer cells and correlates positively with PKCϵ in patient biopsies. Transl Oncol (2019) 12(3):561–75. doi: 10.1016/j.tranon.2018.12.011
18. Tu X, Hong D, Jiang Y, Lou Z, Wang K, Jiang Y, et al. FH535 inhibits proliferation and migration of colorectal cancer cells by regulating CyclinA2 and Claudin1 gene expression. Gene (2019) 690:48–56. doi: 10.1016/j.gene.2018.12.008
19. Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S, et al. Claudin-1 up-regulates the repressor ZEB-1 to inhibit e-cadherin expression in colon cancer cells. Gastroenterology (2011) 141(6):2140–53. doi: 10.1053/j.gastro.2011.08.038
20. Shiou SR, Singh AB, Moorthy K, Datta PK, Washington MK, Beauchamp RD, et al. Smad4 regulates claudin-1 expression in a transforming growth factor-beta-independent manner in colon cancer cells. Cancer Res (2007) 67(4):1571–9. doi: 10.1158/0008-5472.CAN-06-1680
21. Huang J, Zhang L, He C, Qu Y, Li J, Zhang J, et al. Claudin-1 enhances tumor proliferation and metastasis by regulating cell anoikis in gastric cancer. Oncotarget (2015) 6(3):1652–65. doi: 10.18632/oncotarget.2936
22. Mahati S, Bolati D, Yang Y, Mao R, Zhang H, Bao Y. TMPRSS4 promotes cancer stem cell traits by regulating CLDN1 in hepatocellular carcinoma. Biochem Biophys Res Commun (2017) 490(3):906–12. doi: 10.1016/j.bbrc.2017.06.139
23. Liu M, Yang J, Zhang Y, Zhou Z, Cui X, Zhang L, et al. ZIP4 promotes pancreatic cancer progression by repressing ZO-1 and claudin-1 through a ZEB1-dependent transcriptional mechanism. Clin Cancer Res (2018) 24(13):3186–96. doi: 10.1158/1078-0432.CCR-18-0263
24. Iitaka D, Moodley S, Shimizu H, Bai XH, Liu M. PKCδ-iPLA2-PGE2-PPARγ signaling cascade mediates TNF-α induced claudin 1 expression in human lung carcinoma cells. Cell Signal (2015) 27(3):568–77. doi: 10.1016/j.cellsig.2014.12.015
25. Ahmad R, Kumar B, Pan K, Dhawan P, Singh AB. HDAC-4 regulates claudin-2 expression in EGFR-ERK1/2 dependent manner to regulate colonic epithelial cell differentiation. Oncotarget (2017) 8(50):87718–36. doi: 10.18632/oncotarget.21190
26. Wei M, Zhang Y, Yang X, Ma P, Li Y, Wu Y, et al. Claudin-2 promotes colorectal cancer growth and metastasis by suppressing NDRG1 transcription. Clin Transl Med (2021) 11(12):e667. doi: 10.1002/ctm2.667
27. Mankertz J, Amasheh M, Krug SM, Fromm A, Amasheh S, Hillenbrand B, et al. TNFalpha up-regulates claudin-2 expression in epithelial HT-29/B6 cells via phosphatidylinositol-3-kinase signaling. Cell Tissue Res (2009) 336(1):67–77. doi: 10.1007/s00441-009-0751-8
28. Kumar B, Ahmad R, Giannico GA, Zent R, Talmon GA, Harris RC, et al. Claudin-2 inhibits renal clear cell carcinoma progression by inhibiting YAP-activation. J Exp Clin Cancer Res (2021) 40(1):77. doi: 10.1186/s13046-021-01870-5
29. Zhang X, Wang H, Li Q, Li T. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int (2018) 18:160. doi: 10.1186/s12935-018-0662-4
30. Tabariès S, McNulty A, Ouellet V, Annis MG, Dessureault M, Vinette M, et al. Afadin cooperates with claudin-2 to promote breast cancer metastasis. Genes Dev (2019) 33(3-4):180–93. doi: 10.1101/gad.319194.118
31. Maruhashi R, Akizuki R, Sato T, Matsunaga T, Endo S, Yamaguchi M, et al. Elevation of sensitivity to anticancer agents of human lung adenocarcinoma A549 cells by knockdown of claudin-2 expression in monolayer and spheroid culture models. Biochim Biophys Acta Mol Cell Res (2018) 1865(3):470–9. doi: 10.1016/j.bbamcr.2017.12.005
32. de Souza WF, Fortunato-Miranda N, Robbs BK, de Araujo WM, de-Freitas-Junior JC, Bastos LG, et al. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-akt as modulators of EGFR signaling. PloS One (2013) 8(9):e74994. doi: 10.1371/journal.pone.0074994
33. Wang Y, Sun T, Sun H, Yang S, Li D, Zhou D. SCF/C-Kit/JNK/AP-1 signaling pathway promotes claudin-3 expression in colonic epithelium and colorectal carcinoma. Int J Mol Sci (2017) 18(4):765. doi: 10.3390/ijms18040765
34. Ahmad R, Kumar B, Chen Z, Chen X, Müller D, Lele SM, et al. Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer malignancy by hyperactivating wnt/β-catenin signaling. Oncogene (2017) 36(47):6592–604. doi: 10.1038/onc.2017.259
35. Zhang L, Wang Y, Zhang B, Zhang H, Zhou M, Wei M, et al. Claudin-3 expression increases the malignant potential of lung adenocarcinoma cells: role of epidermal growth factor receptor activation. Oncotarget (2017) 8(14):23033–47. doi: 10.18632/oncotarget.14974
36. Che J, Yue D, Zhang B, Zhang H, Huo Y, Gao L, et al. Claudin-3 inhibits lung squamous cell carcinoma cell epithelial-mesenchymal transition and invasion via suppression of the wnt/β-catenin signaling pathway. Int J Med Sci (2018) 15(4):339–51. doi: 10.7150/ijms.22927
37. Jiang L, Yang YD, Fu L, Xu W, Liu D, Liang Q, et al. CLDN3 inhibits cancer aggressiveness via wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget (2014) 5(17):7663–76. doi: 10.18632/oncotarget.2288
38. Luo J, Wang H, Chen H, Gan G, Zheng Y. CLDN4 silencing promotes proliferation and reduces chemotherapy sensitivity of gastric cancer cells through activation of the PI3K/Akt signalling pathway. Exp Physiol (2020) 105(6):979–88. doi: 10.1113/EP088112
39. Owari T, Sasaki T, Fujii K, Fujiwara-Tani R, Kishi S, Mori S, et al. Role of nuclear claudin-4 in renal cell carcinoma. Int J Mol Sci (2020) 21(21):8340. doi: 10.3390/ijms21218340
40. Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B, et al. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate wnt/β-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis (2018) 9(9):851. doi: 10.1038/s41419-018-0937-7
41. Kuang L, Li L. E74-like factor 3 suppresses microRNA-485-5p transcription to trigger growth and metastasis of ovarian cancer cells with the involvement of CLDN4/Wnt/β-catenin axis. Saudi J Biol Sci (2021) 28(8):4137–46. doi: 10.1016/j.sjbs.2021.04.093
42. Hicks DA, Galimanis CE, Webb PG, Spillman MA, Behbakht K, Neville MC, et al. Claudin-4 activity in ovarian tumor cell apoptosis resistance and migration. BMC Cancer (2016) 16(1):788. doi: 10.1186/s12885-016-2799-7
43. Wang P, Liu T, Zhao Z, Wang Z, Liu S, Yang X. SPTBN2 regulated by miR-424-5p promotes endometrial cancer progression via CLDN4/PI3K/AKT axis. Cell Death Discov (2021) 7(1):382. doi: 10.1038/s41420-021-00776-7
44. Song P, Li Y, Dong Y, Liang Y, Qu H, Qi D, et al. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J Exp Clin Cancer Res (2019) 38(1):354. doi: 10.1186/s13046-019-1359-9
45. Yu S, Zhang Y, Li Q, Zhang Z, Zhao G, Xu J. CLDN6 promotes tumor progression through the YAP1-snail1 axis in gastric cancer. Cell Death Dis (2019) 10(12):949. doi: 10.1038/s41419-019-2168-y
46. Huang L, Zhao C, Sun K, Yang D, Yan L, Luo D, et al. Downregulation of CLDN6 inhibits cell proliferation, migration, and invasion via regulating EGFR/AKT/mTOR signalling pathway in hepatocellular carcinoma. Cell Biochem Funct (2020) 38(5):541–8. doi: 10.1002/cbf.3489
47. Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res (2011) 317(13):1935–46. doi: 10.1016/j.yexcr.2011.05.019
48. Hoggard J, Fan J, Lu Z, Lu Q, Sutton L, Chen YH. Claudin-7 increases chemosensitivity to cisplatin through the upregulation of caspase pathway in human NCI-H522 lung cancer cells. Cancer Sci (2013) 104(5):611–8. doi: 10.1111/cas.12135
49. Ji H, Ding X, Zhang W, Zheng Y, Du H, Zheng Y, et al. Claudin-7 inhibits proliferation and metastasis in salivary adenoid cystic carcinoma through wnt/β-catenin signaling. Cell Transplant (2020) 29:963689720943583. doi: 10.1177/0963689720943583
50. Xu C, Ding YH, Wang K, Hao M, Li H, Ding L. Claudin-7 deficiency promotes stemness properties in colorectal cancer through Sox9-mediated wnt/β-catenin signalling. J Transl Med (2021) 19(1):311. doi: 10.1186/s12967-021-02983-3
51. Zhang X, Wang X, Wang A, Li Q, Zhou M, Li T. CLDN10 promotes a malignant phenotype of osteosarcoma cells via JAK1/Stat1 signaling. J Cell Commun Signal (2019) 13(3):395–405. doi: 10.1007/s12079-019-00509-7
52. Sun L, Feng L, Cui J. Increased expression of claudin-17 promotes a malignant phenotype in hepatocyte via Tyk2/Stat3 signaling and is associated with poor prognosis in patients with hepatocellular carcinoma. Diagn Pathol (2018) 13(1):72. doi: 10.1186/s13000-018-0749-1
53. Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu FX, et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest (2018) 128(3):970–84. doi: 10.1172/JCI90429
54. Shimobaba S, Taga S, Akizuki R, Hichino A, Endo S, Matsunaga T, et al. Claudin-18 inhibits cell proliferation and motility mediated by inhibition of phosphorylation of PDK1 and akt in human lung adenocarcinoma A549 cells. Biochim Biophys Acta (2016) 1863(6 Pt A):1170–8. doi: 10.1016/j.bbamcr.2016.02.015
55. Yano K, Imaeda T, Niimi T. Transcriptional activation of the human claudin-18 gene promoter through two AP-1 motifs in PMA-stimulated MKN45 gastric cancer cells. Am J Physiol Gastrointest Liver Physiol (2008) 294(1):G336–343. doi: 10.1152/ajpgi.00328.2007
56. Ito T, Kojima T, Yamaguchi H, Kyuno D, Kimura Y, Imamura M, et al. Transcriptional regulation of claudin-18 via specific protein kinase c signaling pathways and modification of DNA methylation in human pancreatic cancer cells. J Cell Biochem (2011) 112(7):1761–72. doi: 10.1002/jcb.23095
57. Bhat AA, Syed N, Therachiyil L, Nisar S, Hashem S, Macha MA, et al. Claudin-1, a double-edged sword in cancer. Int J Mol Sci (2020) 21(2):569. doi: 10.3390/ijms21020569
58. Ricardo S, Gerhard R, Cameselle-Teijeiro JF, Schmitt F, Paredes J. Claudin expression in breast cancer: high or low, what to expect? Histol Histopathol (2012) 27(10):1283–95. doi: 10.14670/hh-27.1283
59. Blanchard AA, Zelinski T, Xie J, Cooper S, Penner C, Leygue E, et al. Identification of claudin 1 transcript variants in human invasive breast cancer. PloS One (2016) 11(9):e0163387. doi: 10.1371/journal.pone.0163387
60. Blanchard AA, Skliris GP, Watson PH, Murphy LC, Penner C, Tomes L, et al. Claudins 1, 3, and 4 protein expression in ER negative breast cancer correlates with markers of the basal phenotype. Virchows Arch (2009) 454(6):647–56. doi: 10.1007/s00428-009-0770-6
61. Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest (2005) 115(7):1765–76. doi: 10.1172/JCI24543
62. Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol (2005) 18(4):511–8. doi: 10.1038/modpathol.3800301
63. Choi BR, Gwak J, Kwon HM, Oh S, Kim KP, Choi WH, et al. Oligodeoxyribozymes that cleave beta-catenin messenger RNA inhibit growth of colon cancer cells via reduction of beta-catenin response transcription. Mol Cancer Ther (2010) 9(6):1894–902. doi: 10.1158/1535-7163.MCT-10-0056
64. Roh H, Green DW, Boswell CB, Pippin JA, Drebin JA. Suppression of beta-catenin inhibits the neoplastic growth of APC-mutant colon cancer cells. Cancer Res (2001) 61(17):6563–8.
65. Renard CA, Labalette C, Armengol C, Cougot D, Wei Y, Cairo S, et al. Tbx3 is a downstream target of the wnt/beta-catenin pathway and a critical mediator of beta-catenin survival functions in liver cancer. Cancer Res (2007) 67(3):901–10. doi: 10.1158/0008-5472.CAN-06-2344
66. Bhat AA, Sharma A, Pope J, Krishnan M, Washington MK, Singh AB, et al. Caudal homeobox protein cdx-2 cooperates with wnt pathway to regulate claudin-1 expression in colon cancer cells. PloS One (2012) 7(6):e37174. doi: 10.1371/journal.pone.0037174
67. Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res (2001) 12(11-12):469–76. doi: 10.3727/096504001108747477
68. Shiozaki A, Shimizu H, Ichikawa D, Konishi H, Komatsu S, Kubota T, et al. Claudin 1 mediates tumor necrosis factor alpha-induced cell migration in human gastric cancer cells. World J Gastroenterol (2014) 20(47):17863–76. doi: 10.3748/wjg.v20.i47.17863
69. Stebbing J, Filipović A, Giamas G. Claudin-1 as a promoter of EMT in hepatocellular carcinoma. Oncogene (2013) 32(41):4871–2. doi: 10.1038/onc.2012.591
70. Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene (2013) 32(41):4873–82. doi: 10.1038/onc.2012.505
71. Chao YC, Pan SH, Yang SC, Yu SL, Che TF, Lin CW, et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med (2009) 179(2):123–33. doi: 10.1164/rccm.200803-456OC
72. Paschoud S, Bongiovanni M, Pache JC, Citi S. Claudin-1 and claudin-5 expression patterns differentiate lung squamous cell carcinomas from adenocarcinomas. Mod Pathol (2007) 20(9):947–54. doi: 10.1038/modpathol.3800835
73. Cherradi S, Ayrolles-Torro A, Vezzo-Vié N, Gueguinou N, Denis V, Combes E, et al. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J Exp Clin Cancer Res (2017) 36(1):89. doi: 10.1186/s13046-017-0558-5
74. Hashimoto Y, Tada M, Iida M, Nagase S, Hata T, Watari A, et al. Generation and characterization of a human-mouse chimeric antibody against the extracellular domain of claudin-1 for cancer therapy using a mouse model. Biochem Biophys Res Commun (2016) 477(1):91–5. doi: 10.1016/j.bbrc.2016.06.025
75. Dhawan P, Ahmad R, Chaturvedi R, Smith JJ, Midha R, Mittal MK, et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene (2011) 30(29):3234–47. doi: 10.1038/onc.2011.43
76. Kinugasa T, Huo Q, Higashi D, Shibaguchi H, Kuroki M, Tanaka T, et al. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res (2007) 27(6a):3729–34.
77. Moldvay J, Jäckel M, Páska C, Soltész I, Schaff Z, Kiss A. Distinct claudin expression profile in histologic subtypes of lung cancer. Lung Cancer (2007) 57(2):159–67. doi: 10.1016/j.lungcan.2007.02.018
78. Xin S, Huixin C, Benchang S, Aiping B, Jinhui W, Xiaoyan L, et al. Expression of Cdx2 and claudin-2 in the multistage tissue of gastric carcinogenesis. Oncology (2007) 73(5-6):357–65. doi: 10.1159/000135351
79. Jung H, Jun KH, Jung JH, Chin HM, Park WB. The expression of claudin-1, claudin-2, claudin-3, and claudin-4 in gastric cancer tissue. J Surg Res (2011) 167(2):e185–191. doi: 10.1016/j.jss.2010.02.010
80. Aung PP, Mitani Y, Sanada Y, Nakayama H, Matsusaki K, Yasui W. Differential expression of claudin-2 in normal human tissues and gastrointestinal carcinomas. Virchows Arch (2006) 448(4):428–34. doi: 10.1007/s00428-005-0120-2
81. Venugopal S, Anwer S, Szászi K. Claudin-2: Roles beyond permeability functions. Int J Mol Sci (2019) 20(22):5655. doi: 10.3390/ijms20225655
82. Sonoki H, Sato T, Endo S, Matsunaga T, Yamaguchi M, Yamazaki Y, et al. Quercetin decreases claudin-2 expression mediated by up-regulation of microRNA miR-16 in lung adenocarcinoma A549 cells. Nutrients (2015) 7(6):4578–92. doi: 10.3390/nu7064578
83. Suzuki T, Yoshinaga N, Tanabe S. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J Biol Chem (2011) 286(36):31263–71. doi: 10.1074/jbc.M111.238147
84. Ikari A, Sato T, Watanabe R, Yamazaki Y, Sugatani J. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-fos pathway in lung adenocarcinoma A549 cells. Biochim Biophys Acta (2012) 1823(6):1110–8. doi: 10.1016/j.bbamcr.2012.04.005
85. Wang YB, Shi Q, Li G, Zheng JH, Lin J, Qiu W. MicroRNA-488 inhibits progression of colorectal cancer via inhibition of the mitogen-activated protein kinase pathway by targeting claudin-2. Am J Physiol Cell Physiol (2019) 316(1):C33–c47. doi: 10.1152/ajpcell.00047.2018
86. Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci (2013) 14(9):18148–80. doi: 10.3390/ijms140918148
87. Dhawan P, Ahmad R, Srivastava AS, Singh AB. Cancer stem cells and colorectal cancer: an overview. Curr Top Med Chem (2011) 11(13):1592–8. doi: 10.2174/156802611796117694
88. Paquet-Fifield S, Koh SL, Cheng L, Beyit LM, Shembrey C, Mølck C, et al. Tight junction protein claudin-2 promotes self-renewal of human colorectal cancer stem-like cells. Cancer Res (2018) 78(11):2925–38. doi: 10.1158/0008-5472.CAN-17-1869
89. Buchert M, Papin M, Bonnans C, Darido C, Raye WS, Garambois V, et al. Symplekin promotes tumorigenicity by up-regulating claudin-2 expression. Proc Natl Acad Sci USA (2010) 107(6):2628–33. doi: 10.1073/pnas.0903747107
90. Mezheyeuski A, Strell C, Hrynchyk I, Guren TK, Dragomir A, Doroshenko T, et al. Treatment-related survival associations of claudin-2 expression in fibroblasts of colorectal cancer. Virchows Arch (2018) 472(3):395–405. doi: 10.1007/s00428-017-2263-3
91. Tabariès S, Annis MG, Lazaris A, Petrillo SK, Huxham J, Abdellatif A, et al. Claudin-2 promotes colorectal cancer liver metastasis and is a biomarker of the replacement type growth pattern. Commun Biol (2021) 4(1):657. doi: 10.1038/s42003-021-02189-9
92. Sonoki H, Tanimae A, Endo S, Matsunaga T, Furuta T, Ichihara K, et al. Kaempherol and luteolin decrease claudin-2 expression mediated by inhibition of STAT3 in lung adenocarcinoma A549 cells. Nutrients (2017) 9(6):597. doi: 10.3390/nu9060597
93. Nasako H, Akizuki R, Takashina Y, Ishikawa Y, Shinoda T, Shirouzu M, et al. Claudin-2 binding peptides, VPDSM and DSMKF, down-regulate claudin-2 expression and anticancer resistance in human lung adenocarcinoma A549 cells. Biochim Biophys Acta Mol Cell Res (2020) 1867(4):118642. doi: 10.1016/j.bbamcr.2019.118642
94. Ikari A, Taga S, Watanabe R, Sato T, Shimobaba S, Sonoki H, et al. Clathrin-dependent endocytosis of claudin-2 by DFYSP peptide causes lysosomal damage in lung adenocarcinoma A549 cells. Biochim Biophys Acta (2015) 1848(10 Pt A):2326–36. doi: 10.1016/j.bbamem.2015.07.003
95. Hashimoto Y, Hata T, Tada M, Iida M, Watari A, Okada Y, et al. Safety evaluation of a human chimeric monoclonal antibody that recognizes the extracellular loop domain of claudin-2. Eur J Pharm Sci (2018) 117:161–7. doi: 10.1016/j.ejps.2018.02.016
96. Moldvay J, Fábián K, Jäckel M, Németh Z, Bogos K, Furák J, et al. Claudin-1 protein expression is a good prognostic factor in non-small cell lung cancer, but only in squamous cell carcinoma cases. Pathol Oncol Res (2017) 23(1):151–6. doi: 10.1007/s12253-016-0115-0
97. Mitchell LA, Overgaard CE, Ward C, Margulies SS, Koval M. Differential effects of claudin-3 and claudin-4 on alveolar epithelial barrier function. Am J Physiol Lung Cell Mol Physiol (2011) 301(1):L40–49. doi: 10.1152/ajplung.00299.2010
98. Oghiso Y, Yamada Y. Immunohistochemical study on cellular origins of rat lung tumors induced by inhalation exposures to plutonium dioxide aerosols as compared to those by X-ray irradiation. J Radiat Res (2002) 43(3):301–11. doi: 10.1269/jrr.43.301
99. Uthayanan L, El-Bahrawy M. Potential roles of claudin-3 and claudin-4 in ovarian cancer management. J Egypt Natl Canc Inst (2022) 34(1):24. doi: 10.1186/s43046-022-00125-4
100. Mitchell LA, Koval M. Specificity of interaction between clostridium perfringens enterotoxin and claudin-family tight junction proteins. Toxins (Basel) (2010) 2(7):1595–611. doi: 10.3390/toxins2071595
101. Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res (2005) 65(16):7378–85. doi: 10.1158/0008-5472.CAN-05-1036
102. Hwang TL, Changchien TT, Wang CC, Wu CM. Claudin-4 expression in gastric cancer cells enhances the invasion and is associated with the increased level of matrix metalloproteinase-2 and -9 expression. Oncol Lett (2014) 8(3):1367–71. doi: 10.3892/ol.2014.2295
103. Sheehan GM, Kallakury BV, Sheehan CE, Fisher HA, Kaufman RP Jr., Ross JS. Loss of claudins-1 and -7 and expression of claudins-3 and -4 correlate with prognostic variables in prostatic adenocarcinomas. Hum Pathol (2007) 38(4):564–9. doi: 10.1016/j.humpath.2006.11.007
104. Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer (2006) 6:186. doi: 10.1186/1471-2407-6-186
105. Suren D, Yildirim M, Kaya V, Elal R, Selcuk OT, Osma U, et al. Expression patterns of claudins 1, 4, and 7 and their prognostic significance in nasopharyngeal carcinoma. J buon (2015) 20(1):212–7.
106. Pan XY, Wang B, Che YC, Weng ZP, Dai HY, Peng W. Expression of claudin-3 and claudin-4 in normal, hyperplastic, and malignant endometrial tissue. Int J Gynecol Cancer (2007) 17(1):233–41. doi: 10.1111/j.1525-1438.2006.00748.x
107. Maeda T, Murata M, Chiba H, Takasawa A, Tanaka S, Kojima T, et al. Claudin-4-targeted therapy using clostridium perfringens enterotoxin for prostate cancer. Prostate (2012) 72(4):351–60. doi: 10.1002/pros.21436
108. Kwon MJ, Kim SH, Jeong HM, Jung HS, Kim SS, Lee JE, et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab Invest (2011) 91(11):1652–67. doi: 10.1038/labinvest.2011.117
109. Lódi C, Szabó E, Holczbauer A, Batmunkh E, Szíjártó A, Kupcsulik P, et al. Claudin-4 differentiates biliary tract cancers from hepatocellular carcinomas. Mod Pathol (2006) 19(3):460–9. doi: 10.1038/modpathol.3800549
110. Lechpammer M, Resnick MB, Sabo E, Yakirevich E, Greaves WO, Sciandra KT, et al. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod Pathol (2008) 21(11):1320–9. doi: 10.1038/modpathol.2008.116
111. Székely E, Törzsök P, Riesz P, Korompay A, Fintha A, Székely T, et al. Expression of claudins and their prognostic significance in noninvasive urothelial neoplasms of the human urinary bladder. J Histochem Cytochem (2011) 59(10):932–41. doi: 10.1369/0022155411418829
112. Kwon MJ, Kim SS, Choi YL, Jung HS, Balch C, Kim SH, et al. Derepression of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with loss of repressive histone modifications. Carcinogenesis (2010) 31(6):974–83. doi: 10.1093/carcin/bgp336
113. Ogawa M, Kojima T, Someya M, Nomura K, Takasawa A, Murata M, et al. Epidermal growth factor modulates claudins and tight junctional functions in ovarian cancer cell lines. Histochem Cell Biol (2012) 138(2):323–38. doi: 10.1007/s00418-012-0956-x
114. Lin X, Shang X, Manorek G, Howell SB. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PloS One (2013) 8(6):e67496. doi: 10.1371/journal.pone.0067496
115. Rachakonda G, Vu T, Jin L, Samanta D, Datta PK. Role of TGF-β-induced claudin-4 expression through c-jun signaling in non-small cell lung cancer. Cell Signal (2016) 28(10):1537–44. doi: 10.1016/j.cellsig.2016.07.006
116. Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res (2003) 63(19):6265–71.
117. Fredriksson K, Van Itallie CM, Aponte A, Gucek M, Tietgens AJ, Anderson JM. Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks. PloS One (2015) 10(3):e0117074. doi: 10.1371/journal.pone.0117074
118. Nakashima C, Yamamoto K, Kishi S, Sasaki T, Ohmori H, Fujiwara-Tani R, et al. Clostridium perfringens enterotoxin induces claudin-4 to activate YAP in oral squamous cell carcinomas. Oncotarget (2020) 11(4):309–21. doi: 10.18632/oncotarget.27424
119. Kojima T, Kondoh M, Keira T, Takano KI, Kakuki T, Kaneko Y, et al. Claudin-binder c-CPE mutants enhance permeability of insulin across human nasal epithelial cells. Drug Deliv (2016) 23(8):2703–10. doi: 10.3109/10717544.2015.1050530
120. Cocco E, Deng Y, Shapiro EM, Bortolomai I, Lopez S, Lin K, et al. Dual-targeting nanoparticles for In vivo delivery of suicide genes to chemotherapy-resistant ovarian cancer cells. Mol Cancer Ther (2017) 16(2):323–33. doi: 10.1158/1535-7163.MCT-16-0501
121. Shim MK, Na J, Cho IK, Jang EH, Park J, Lee S, et al. Targeting of claudin-4 by clostridium perfringens enterotoxin-conjugated polysialic acid nanoparticles for pancreatic cancer therapy. J Control Release (2021) 331:434–42. doi: 10.1016/j.jconrel.2021.01.031
122. Kono T, Kondoh M, Kyuno D, Ito T, Kimura Y, Imamura M, et al. Claudin-4 binder c-CPE 194 enhances effects of anticancer agents on pancreatic cancer cell lines via a MAPK pathway. Pharmacol Res Perspect (2015) 3(6):e00196. doi: 10.1002/prp2.196
123. Hashimoto Y, Tachibana K, Krug SM, Kunisawa J, Fromm M, Kondoh M. Potential for tight junction protein-directed drug development using claudin binders and angubindin-1. Int J Mol Sci (2019) 20(16):4016. doi: 10.3390/ijms20164016
124. Kato-Nakano M, Suzuki M, Kawamoto S, Furuya A, Ohta S, Nakamura K, et al. Characterization and evaluation of the antitumour activity of a dual-targeting monoclonal antibody against claudin-3 and claudin-4. Anticancer Res (2010) 30(11):4555–62.
125. Wu Q, Liu Y, Ren Y, Xu X, Yu L, Li Y, et al. Tight junction protein, claudin-6, downregulates the malignant phenotype of breast carcinoma. Eur J Cancer Prev (2010) 19(3):186–94. doi: 10.1097/CEJ.0b013e328337210e
126. Jia H, Chai X, Li S, Wu D, Fan Z. Identification of claudin-2, -6, -11 and -14 as prognostic markers in human breast carcinoma. Int J Clin Exp Pathol (2019) 12(6):2195–204.
127. Lu Y, Wang L, Li H, Li Y, Ruan Y, Lin D, et al. SMAD2 inactivation inhibits CLDN6 methylation to suppress migration and invasion of breast cancer cells. Int J Mol Sci (2017) 18(9):1863. doi: 10.3390/ijms18091863
128. Liu Y, Jin X, Li Y, Ruan Y, Lu Y, Yang M, et al. DNA Methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J Exp Clin Cancer Res (2016) 35(1):120. doi: 10.1186/s13046-016-0396-x
129. Ren Y, Wu Q, Liu Y, Xu X, Quan C. Gene silencing of claudin−6 enhances cell proliferation and migration accompanied with increased MMP−2 activity via p38 MAPK signaling pathway in human breast epithelium cell line HBL−100. Mol Med Rep (2013) 8(5):1505–10. doi: 10.3892/mmr.2013.1675
130. Lin D, Guo Y, Li Y, Ruan Y, Zhang M, Jin X, et al. Bioinformatic analysis reveals potential properties of human claudin-6 regulation and functions. Oncol Rep (2017) 38(2):875–85. doi: 10.3892/or.2017.5756
131. Guo Y, Xu X, Liu Z, Zhang T, Zhang X, Wang L, et al. Apoptosis signal-regulating kinase 1 is associated with the effect of claudin-6 in breast cancer. Diagn Pathol (2012) 7:111. doi: 10.1186/1746-1596-7-111
132. Guo Y, Lin D, Zhang M, Zhang X, Li Y, Yang R, et al. CLDN6-induced apoptosis via regulating ASK1-p38/JNK signaling in breast cancer MCF-7 cells. Int J Oncol (2016) 48(6):2435–44. doi: 10.3892/ijo.2016.3469
133. Wu Q, Liu X, Liu YF, Lu Y, Wang LP, Zhang XW, et al. Inhibition of p38 activity reverses claudin-6 induced cell apoptosis, invasion, and migration. Chin Med J (Engl) (2013) 126(18):3539–44.
134. Yafang L, Qiong W, Yue R, Xiaoming X, Lina Y, Mingzi Z, et al. Role of estrogen receptor-α in the regulation of claudin-6 expression in breast cancer cells. J Breast Cancer (2011) 14(1):20–7. doi: 10.4048/jbc.2011.14.1.20
135. Yang M, Li Y, Ruan Y, Lu Y, Lin D, Xie Y, et al. CLDN6 enhances chemoresistance to ADM via AF-6/ERKs pathway in TNBC cell line MDAMB231. Mol Cell Biochem (2018) 443(1-2):169–80. doi: 10.1007/s11010-017-3221-8
136. Bosch A, Eroles P, Zaragoza R, Viña JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat Rev (2010) 36(3):206–15. doi: 10.1016/j.ctrv.2009.12.002
137. Yang M, Li Y, Shen X, Ruan Y, Lu Y, Jin X, et al. CLDN6 promotes chemoresistance through GSTP1 in human breast cancer. J Exp Clin Cancer Res (2017) 36(1):157. doi: 10.1186/s13046-017-0627-9
138. Qu H, Qi D, Wang X, Dong Y, Jin Q, Wei J, et al. CLDN6 suppresses c-MYC-Mediated aerobic glycolysis to inhibit proliferation by TAZ in breast cancer. Int J Mol Sci (2021) 23(1):129. doi: 10.3390/ijms23010129
139. Jia Y, Guo Y, Jin Q, Qu H, Qi D, Song P, et al. A SUMOylation-dependent HIF-1α/CLDN6 negative feedback mitigates hypoxia-induced breast cancer metastasis. J Exp Clin Cancer Res (2020) 39(1):42. doi: 10.1186/s13046-020-01547-5
140. Lin Z, Zhang X, Liu Z, Liu Q, Wang L, Lu Y, et al. The distinct expression patterns of claudin-2, -6, and -11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagn Pathol (2013) 8:133. doi: 10.1186/1746-1596-8-133
141. Kohmoto T, Masuda K, Shoda K, Takahashi R, Ujiro S, Tange S, et al. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer (2020) 23(3):403–17. doi: 10.1007/s10120-019-01014-x
142. Gao F, Li M, Xiang R, Zhou X, Zhu L, Zhai Y. Expression of CLDN6 in tissues of gastric cancer patients: Association with clinical pathology and prognosis. Oncol Lett (2019) 17(5):4621–5. doi: 10.3892/ol.2019.10129
143. Lu YZ, Li Y, Zhang T, Han ST. Claudin-6 is down-regulated in gastric cancer and its potential pathway. Cancer biomark (2020) 28(3):329–40. doi: 10.3233/CBM-201554
144. Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, Fortoul TI, Montaño LF, Rendon-Huerta EP. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest (2011) 29(1):1–11. doi: 10.3109/07357907.2010.512594
145. Torres-Martínez AC, Gallardo-Vera JF, Lara-Holguin AN, Montaño LF, Rendón-Huerta EP. Claudin-6 enhances cell invasiveness through claudin-1 in AGS human adenocarcinoma gastric cancer cells. Exp Cell Res (2017) 350(1):226–35. doi: 10.1016/j.yexcr.2016.11.025
146. Lu Y, Dang Q, Bo Y, Su X, Wang L, Sun J, et al. The expression of CLDN6 in hepatocellular carcinoma tissue and the effects of CLDN6 on biological phenotypes of hepatocellular carcinoma cells. J Cancer (2021) 12(18):5454–63. doi: 10.7150/jca.55727
147. Amgen. A safety, tolerability, and pharmacokinetics study of AMG 794 with claudin 6-positive non-squamous non-small cell lung cancer or epithelial ovarian cancer . Available at: https://ClinicalTrials.gov/show/NCT05317078.Published2025 (Accessed December 7).
148. Daiichi Sankyo I. A study of DS-9606a in patients with advanced solid tumors (2023). Available at: https://ClinicalTrials.gov/show/NCT05394675 (Accessed November 30).
149. Second Affiliated Hospital of Guangzhou Medical U. CLDN6-CAR-NK cell therapy for advanced solid tumors (2024). Available at: https://ClinicalTrials.gov/show/NCT05410717 (Accessed June 1).
150. Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, et al. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol (2006) 37(5):569–77. doi: 10.1016/j.humpath.2005.12.018
151. Hsueh C, Chang YS, Tseng NM, Liao CT, Hsueh S, Chang JH, et al. Expression pattern and prognostic significance of claudins 1, 4, and 7 in nasopharyngeal carcinoma. Hum Pathol (2010) 41(7):944–50. doi: 10.1016/j.humpath.2010.01.005
152. Oshima T, Kunisaki C, Yoshihara K, Yamada R, Yamamoto N, Sato T, et al. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol Rep (2008) 19(4):953–9. doi: 10.3892/or.19.4.953
153. Bernardi MA, Logullo AF, Pasini FS, Nonogaki S, Blumke C, Soares FA, et al. Prognostic significance of CD24 and claudin-7 immunoexpression in ductal invasive breast cancer. Oncol Rep (2012) 27(1):28–38. doi: 10.3892/or.2011.1477
154. Li Y, Gong Y, Ning X, Peng D, Liu L, He S, et al. Downregulation of CLDN7 due to promoter hypermethylation is associated with human clear cell renal cell carcinoma progression and poor prognosis. J Exp Clin Cancer Res (2018) 37(1):276. doi: 10.1186/s13046-018-0924-y
155. Hou Y, Hou L, Liang Y, Zhang Q, Hong X, Wang Y, et al. The p53-inducible CLDN7 regulates colorectal tumorigenesis and has prognostic significance. Neoplasia (2020) 22(11):590–603. doi: 10.1016/j.neo.2020.09.001
156. Wang K, Li T, Xu C, Ding Y, Li W, Ding L. Claudin-7 downregulation induces metastasis and invasion in colorectal cancer via the promotion of epithelial-mesenchymal transition. Biochem Biophys Res Commun (2019) 508(3):797–804. doi: 10.1016/j.bbrc.2018.10.049
157. Xu C, Wang K, Ding YH, Li WJ, Ding L. Claudin-7 gene knockout causes destruction of intestinal structure and animal death in mice. World J Gastroenterol (2019) 25(5):584–99. doi: 10.3748/wjg.v25.i5.584
158. Wang K, Ding Y, Xu C, Hao M, Li H, Ding L. Cldn-7 deficiency promotes experimental colitis and associated carcinogenesis by regulating intestinal epithelial integrity. Oncoimmunology (2021) 10(1):1923910. doi: 10.1080/2162402X.2021.1923910
159. Li Z, Xuan W, Huang L, Chen N, Hou Z, Lu B, et al. Claudin 10 acts as a novel biomarker for the prognosis of patients with ovarian cancer. Oncol Lett (2020) 20(1):373–81. doi: 10.3892/ol.2020.11557
160. Rao X, Jiang J, Liang Z, Zhang J, Zhuang Z, Qiu H, et al. Down-regulated CLDN10 predicts favorable prognosis and correlates with immune infiltration in gastric cancer. Front Genet (2021) 12:747581. doi: 10.3389/fgene.2021.747581
161. Yang W, Li L, Zhang K, Ma K, Gong Y, Zhou J, et al. CLDN10 associated with immune infiltration is a novel prognostic biomarker for clear cell renal cell carcinoma. Epigenomics (2021) 13(1):31–45. doi: 10.2217/epi-2020-0256
162. Zhou Y, Xiang J, Bhandari A, Guan Y, Xia E, Zhou X, et al. CLDN10 is associated with papillary thyroid cancer progression. J Cancer (2018) 9(24):4712–7. doi: 10.7150/jca.28636
163. Xiang Z, Zhong C, Chang A, Ling J, Zhao H, Zhou W, et al. Immune-related key gene CLDN10 correlates with lymph node metastasis but predicts favorable prognosis in papillary thyroid carcinoma. Aging (Albany NY) (2020) 12(3):2825–39. doi: 10.18632/aging.102780
164. Cheung ST, Leung KL, Ip YC, Chen X, Fong DY, Ng IO, et al. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res (2005) 11(2 Pt 1):551–6. doi: 10.1158/1078-0432.551.11.2
165. Xu YN, Deng MS, Liu YF, Yao J, Xiao ZY. Tight junction protein CLDN17 serves as a tumor suppressor to reduce the invasion and migration of oral cancer cells by inhibiting epithelial-mesenchymal transition. Arch Oral Biol (2022) 133:105301. doi: 10.1016/j.archoralbio.2021.105301
166. Linares GR, Brommage R, Powell DR, Xing W, Chen ST, Alshbool FZ, et al. Claudin 18 is a novel negative regulator of bone resorption and osteoclast differentiation. J Bone Miner Res (2012) 27(7):1553–65. doi: 10.1002/jbmr.1600
167. Kyuno D, Yamaguchi H, Ito T, Kono T, Kimura Y, Imamura M, et al. Targeting tight junctions during epithelial to mesenchymal transition in human pancreatic cancer. World J Gastroenterol (2014) 20(31):10813–24. doi: 10.3748/wjg.v20.i31.10813
168. Wheal A, Jenkins R, Mikami Y, Das N, Hirschowitz L. Primary mucinous carcinoma of the fallopian tube: Case report and review of literature. Int J Gynecol Pathol (2017) 36(4):393–9. doi: 10.1097/PGP.0000000000000330
169. Arnold A, Daum S, von Winterfeld M, Berg E, Hummel M, Rau B, et al. Prognostic impact of claudin 18.2 in gastric and esophageal adenocarcinomas. Clin Transl Oncol (2020) 22(12):2357–63. doi: 10.1007/s12094-020-02380-0
170. Kiyokawa T, Hoang L, Pesci A, Alvarado-Cabrero I, Oliva E, Park KJ, et al. Claudin-18 as a promising surrogate marker for endocervical gastric-type carcinoma. Am J Surg Pathol (2021) 46(5):628–63. doi: 10.1097/PAS.0000000000001847
171. Li G, Flodby P, Luo J, Kage H, Sipos A, Gao D, et al. Knockout mice reveal key roles for claudin 18 in alveolar barrier properties and fluid homeostasis. Am J Respir Cell Mol Biol (2014) 51(2):210–22. doi: 10.1165/rcmb.2013-0353OC
172. LaFemina MJ, Sutherland KM, Bentley T, Gonzales LW, Allen L, Chapin CJ, et al. Claudin-18 deficiency results in alveolar barrier dysfunction and impaired alveologenesis in mice. Am J Respir Cell Mol Biol (2014) 51(4):550–8. doi: 10.1165/rcmb.2013-0456OC
173. Kotton DN. Claudin-18: unexpected regulator of lung alveolar epithelial cell proliferation. J Clin Invest (2018) 128(3):903–5. doi: 10.1172/JCI99799
174. Luo J, Chimge NO, Zhou B, Flodby P, Castaldi A, Firth AL, et al. CLDN18.1 attenuates malignancy and related signaling pathways of lung adenocarcinoma in vivo and in vitro. Int J Cancer (2018) 143(12):3169–80. doi: 10.1002/ijc.31734
175. Xu B, Liu F, Liu Q, Shi T, Wang Z, Wu N, et al. Highly expressed Claudin18.2 as a potential therapeutic target in advanced gastric signet-ring cell carcinoma (SRCC). J Gastrointest Oncol (2020) 11(6):1431–9. doi: 10.21037/jgo-20-344
176. Ushiku T, Ishikawa S, Kakiuchi M, Tanaka A, Katoh H, Aburatani H, et al. RHOA mutation in diffuse-type gastric cancer: a comparative clinicopathology analysis of 87 cases. Gastric Cancer (2016) 19(2):403–11. doi: 10.1007/s10120-015-0493-0
177. Tanaka A, Ishikawa S, Ushiku T, Yamazawa S, Katoh H, Hayashi A, et al. Frequent CLDN18-ARHGAP fusion in highly metastatic diffuse-type gastric cancer with relatively early onset. Oncotarget (2018) 9(50):29336–50. doi: 10.18632/oncotarget.25464
178. Nakayama I, Shinozaki E, Sakata S, Yamamoto N, Fujisaki J, Muramatsu Y, et al. Enrichment of CLDN18-ARHGAP fusion gene in gastric cancers in young adults. Cancer Sci (2019) 110(4):1352–63. doi: 10.1111/cas.13967
179. Zhang WH, Zhang SY, Hou QQ, Qin Y, Chen XZ, Zhou ZG, et al. The significance of the CLDN18-ARHGAP fusion gene in gastric cancer: A systematic review and meta-analysis. Front Oncol (2020) 10:1214. doi: 10.3389/fonc.2020.01214
180. Shu Y, Zhang W, Hou Q, Zhao L, Zhang S, Zhou J, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun (2018) 9(1):2447. doi: 10.1038/s41467-018-04907-0
181. Sanada Y, Oue N, Mitani Y, Yoshida K, Nakayama H, Yasui W. Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol (2006) 208(5):633–42. doi: 10.1002/path.1922
182. Jun KH, Kim JH, Jung JH, Choi HJ, Chin HM. Expression of claudin-7 and loss of claudin-18 correlate with poor prognosis in gastric cancer. Int J Surg (2014) 12(2):156–62. doi: 10.1016/j.ijsu.2013.11.022
183. Baek JH, Park DJ, Kim GY, Cheon J, Kang BW, Cha HJ, et al. Clinical implications of Claudin18.2 expression in patients with gastric cancer. Anticancer Res (2019) 39(12):6973–9. doi: 10.21873/anticanres.13919
184. Pellino A, Brignola S, Riello E, Niero M, Murgioni S, Guido M, et al. Association of CLDN18 protein expression with clinicopathological features and prognosis in advanced gastric and gastroesophageal junction adenocarcinomas. J Pers Med (2021) 11(11):1095. doi: 10.3390/jpm11111095
185. Li J, Zhang Y, Hu D, Gong T, Xu R, Gao J. Analysis of the expression and genetic alteration of CLDN18 in gastric cancer. Aging (Albany NY) (2020) 12(14):14271–84. doi: 10.18632/aging.103457
186. Ungureanu BS, Lungulescu CV, Pirici D, Turcu-Stiolica A, Gheonea DI, Sacerdotianu VM, et al. Clinicopathologic relevance of claudin 18.2 expression in gastric cancer: A meta-analysis. Front Oncol (2021) 11:643872. doi: 10.3389/fonc.2021.643872
187. Tanaka M, Shibahara J, Fukushima N, Shinozaki A, Umeda M, Ishikawa S, et al. Claudin-18 is an early-stage marker of pancreatic carcinogenesis. J Histochem Cytochem (2011) 59(10):942–52. doi: 10.1369/0022155411420569
188. Sahin U, Koslowski M, Dhaene K, Usener D, Brandenburg G, Seitz G, et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res (2008) 14(23):7624–34. doi: 10.1158/1078-0432.CCR-08-1547
189. Li WT, Jeng YM, Yang CY. Claudin-18 as a marker for identifying the stomach and pancreatobiliary tract as the primary sites of metastatic adenocarcinoma. Am J Surg Pathol (2020) 44(12):1643–8. doi: 10.1097/PAS.0000000000001583
190. Shinozaki A, Shibahara J, Noda N, Tanaka M, Aoki T, Kokudo N, et al. Claudin-18 in biliary neoplasms. its significance in the classification of intrahepatic cholangiocarcinoma. Virchows Arch (2011) 459(1):73–80. doi: 10.1007/s00428-011-1092-z
191. Kyuno D, Takasawa A, Takasawa K, Ono Y, Aoyama T, Magara K, et al. Claudin-18.2 as a therapeutic target in cancers: cumulative findings from basic research and clinical trials. Tissue Barriers (2022) 2021:1967080. doi: 10.1080/21688370.2021.1967080
192. Sahin U, Türeci Ö, Manikhas G, Lordick F, Rusyn A, Vynnychenko I, et al. FAST: a randomised phase II study of zolbetuximab (IMAB362) plus EOX versus EOX alone for first-line treatment of advanced CLDN18.2-positive gastric and gastro-oesophageal adenocarcinoma. Ann Oncol (2021) 32(5):609–19. doi: 10.1016/j.annonc.2021.02.005
193. Türeci O, Sahin U, Schulze-Bergkamen H, Zvirbule Z, Lordick F, Koeberle D, et al. A multicentre, phase IIa study of zolbetuximab as a single agent in patients with recurrent or refractory advanced adenocarcinoma of the stomach or lower oesophagus: the MONO study. Ann Oncol (2019) 30(9):1487–95. doi: 10.1093/annonc/mdz199
194. Qi C, Gong J, Li J, Liu D, Qin Y, Ge S, et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results. Nat Med (2022) 28(6):1189–98.
Keywords: claudin, signaling pathways, cancer, molecular targeted therapy, tumor diagnosis
Citation: Wang D-W, Zhang W-H, Danil G, Yang K and Hu J-K (2022) The role and mechanism of claudins in cancer. Front. Oncol. 12:1051497. doi: 10.3389/fonc.2022.1051497
Received: 22 September 2022; Accepted: 28 November 2022;
Published: 22 December 2022.
Edited by:
Yinan Zhang, Nanjing University of Chinese Medicine, ChinaReviewed by:
Dario Mizrachi, Brigham Young University, United StatesCopyright © 2022 Wang, Zhang, Danil, Yang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Kun Hu, aHVqa3djaEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.