
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 14 December 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1049928
This article is part of the Research Topic Epigenetic Insights into Diagnostic and Therapeutic Applications View all 6 articles
The biological functions of N6-methyladenosine (m6A) modification of mRNA have recently received a great deal of attention. In previous studies, m6A methylation modification has been shown to regulate mRNA fate and to be crucial for the progression and development of tumors. BTG2 (B-cell translocation gene 2) is a member of BTG/TOB anti-proliferative protein family. BTG2 could inhibit cell proliferation and migration and regulate the cell cycle progression. In this study, we confirm that BTG2 is frequently down-regulated in renal cell carcinoma (RCC) tissues and its low expression is associated with unfavorable prognosis and decreased m6A level. Moreover, we found that m6A methylation modifies the 5’UTR of BTG2 to promote its mRNA stability by binding to IGF2BP2. It has been shown that CRISPR/dCas13b-METLL3 can specifically increase BTG2 m6A modification to significantly increase its m6A and expression levels. Then m6A hypermethylation in BTG2 mRNA could dramatically inhibit RCC cells proliferation and migration, and induce cells apoptosis. Taken together, our data show that BTG2 functions as a tumor suppressor and is frequently silenced via m6A modification in RCC.
Renal cell carcinoma (RCC) is one of the common malignant tumors of the urinary system, and its incidence is 5% in men and 3% in women, and its incidence is gradually increasing (1, 2). Its etiology is not clear, and its pathogenesis may be closely related to genetic factors, smoking, obesity, diet, hypertension and so on. Genetic mutations are an important cause of RCC and can be detected early (3). With the development of gene therapy and the in-depth study on the mechanism of various tumor suppressor genes, it brings new directions and ideas for the treatment of tumors to find targets from gene level.
RNA modifications play an essential role in regulating gene expression post-transcriptionally. Eukaryotes have the most abundant internal modification, m6A, which constitutes 0.1-0.3% of total adenosine residues (4, 5). It is located in the non-coding 3’term, near the stop codon and long internal exons, and is related to RNA stability, splicing, intracellular distribution, and translation (6, 7). The cellular m6A state is controlled by a group of genes called “writers” (WTAP, METTL3 and METTLL4), “erasers” (FTO and ALKBH5), and “readers” (YTHDF1/2/3, IGF2BP2/3, YTHDCL and YTHDC2) (8–13). N6-methyladenosine RNA modifications on the sixth nitrogen atom of adenine, one of the most important nitrogen atoms in RNA, have become one of the hottest topics in various human diseases including hypertension (14), cardiac hypertrophy (15), viral infection (16), diabetes (17) and cancers (18, 19). Yet, the expression patterns of RNA m6A methylation modification and their underlying mechanisms remain largely unknown in RCC.
B-cell translocation gene 2 (BTG2) belongs to the anti-proliferation gene family and is an early growth response gene. Many studies have shown that BTG2 participates in the regulation of cell growth, differentiation and apoptosis (20, 21). At the same time, the expression of BTG2 in many types of cancer cells has varying degrees of reduction or even direct deletion, while the expression level of BTG2 gene in many corresponding normal cells is relatively higher. This suggests that abnormal expression of BTG2 is closely related to tumor formation, suggesting that tumor formation may be related to BTG2 expression (22–24). BTG2 expression level has been found to be correlated with the clinical characteristics of the tumor in breast cancer samples. The lower the level of BTG2 expression, the higher the risk of lymphatic invasion and vascular invasion in breast cancer patients, and the higher the risk of tumor metastasis. Low expression of BTG2 is associated with tumor recurrence. And low expression of BTG2 is associated with high tumor recurrence rate and low overall survival rate (25). However, the role of BTG2 in the growth and migration of RCC remains unclear.
In this study, we observed RCC tissues exhibited significantly reduced BTG2 m6A level and expression level relative to normal tissues. Preliminary experimental results showed that methylation of 5’UTR in BTG2 mRNA can regulate its mRNA stability by recruiting m6A reader protein IGF2BP2. Further, upregulation of BTG2 m6A methylation level can inhibit RCC cells proliferation and migration, and promotes RCC cells apoptosis by dCas13b-METTL3.
BTG2 has been found to be expressed at low levels in a variety of cancer, which has potential role in the development and subsequent progression of tumors. However, there is limited evidence regarding the regulatory pathways that control the expression levels of BTG2 in RCC. According to The Cancer Genome Atlas (TCGA) database, we first found that BTG2 expression levels are downregulated in RCC tissues compared with adjacent normal tissues (Figures 1A, B). We then examined the impact of BTG2 expression on patient survival using TCGA dataset. The results showed that there was a significantly negative correlation between low expression of BTG2 and poor survival rates in patients (Figure 1C). In addition, we also performed IHC and RT-qPCR experiments on RCC and normal tissues to determine the expression levels of BTG2 protein and mRNA. The results showed significant downregulation of BTG2 in tumor tissues compared to adjacent normal tissues (Figures 1D–F). The expression levels of METTL3 and IGF2BP2 were also significantly decreased in RCC tumor tissues compared to normal tissues (Supplementary Figure S1).
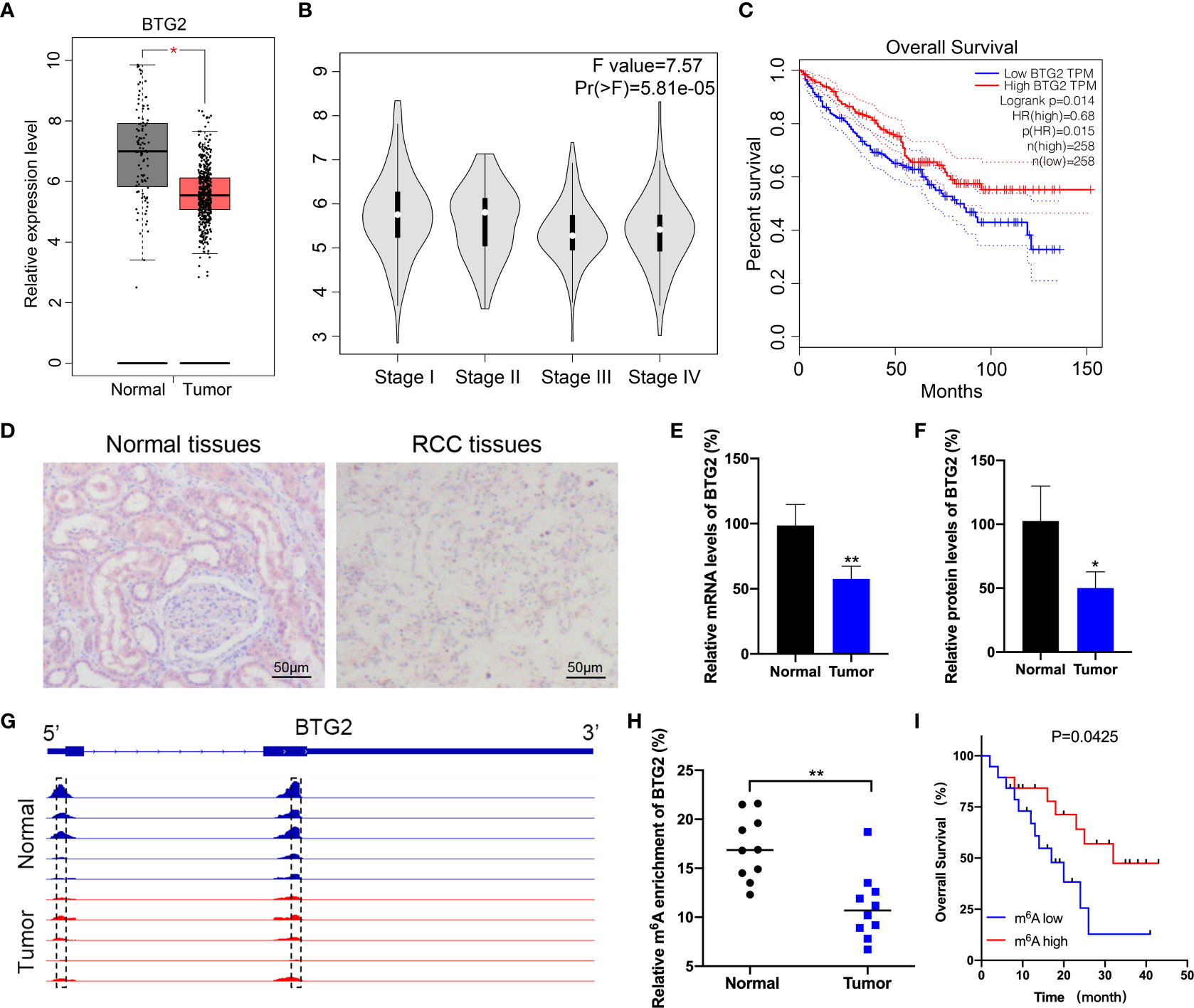
Figure 1 BTG2 expression and m6A methylation level in RCC patients. (A) An analysis of BTG2 mRNA expression in RCC tumor tissues (red plot) and normal tissues (grey plot) is shown in the box plot below based on TCGA database. (B) The expression levels of BTG2 in different stages of RCC based on TCGA database. (C) The survival plot for RCC patients stratified by expression of BTG2. (D) The protein expression levels of BTG2 in tumor tissues and normal tissues analyzed by IHC. (E) The mRNA expression levels of BTG2 in tumor tissues and normal tissues analyzed by RT-qPCR. (F) The protein expression levels of BTG2 in tumor tissues and normal tissues analyzed by western blot. (G) The m6A peaks within BTG2 mRNA in tumor tissues and normal tissues analyzed by IGV. (H) The m6A enrichment of BTG2 in tumor tissues and normal tissues analyzed by MeRIP-qPCR. (I) The survival plot for RCC patients stratified by m6A level of BTG2. * P<0.05, ** P<0.01.
It is evident that m6A RNA methylation plays an important role in tumorigenesis by inhibiting tumor suppressor genes. Based on previous MeRIP-seq results, we examined the m6A methylation level of BTG2 in RCC tissues to investigate the role of m6A methylation modification in RCC progression. The data revealed that the m6A peaks on BTG2 mRNA were located in its 5′UTR and CDS region. And compared to the normal tissue, tumor tissues showed lower peak enrichment (Figure 1G). MeRIP-qPCR results validated that the m6A methylation level of BTG2 was significantly decreased in tumor tissues compared to normal tissues (Figure 1H). Based on the Kaplan-Meier survival analysis, RCC patients with lower m6A levels of BTG2 had a shorter overall survival (OS) (Figure 1I). Overall, BTG2 expression and m6A methylation levels were correlated with overall survival of RCC patients.
Using Western blot and RT-qPCR, we first detected BTG2 expression in RCC cells to explore possible mechanisms involved in m6A regulation of BTG2 expression. As compared to normal renal tubule HK-2 cell line, BTG2 expression was significantly decreased in other RCC cell lines except for OSRC cell (Figures 2A, 2B and Supplementary Figure S2). Further, based on MeRIP-qPCR results, BTG2 m6A enrichment in 786O and 769P RCC cells were significantly decreased in comparison with HK-2 cells (Figure 2C). Then we examined the mechanisms that influence BTG2 expression in RCC by the m6A methylation modification. As shown by RT-qPCR results, Mettl3-overexpressing 786O and 769P cells exhibited higher expression of BTG2 (Figure 2D). In addition, the m6A modification levels of BTG2 were also significantly increased in 786O and 769P cells with overexpression of Mettl3 (Figures 2E, F). To block transcription, we treated control cells and 786O cells overexpressing Mettl3 with Act-D. In RNA stability assays, Mettl3 overexpression prolonged BTG2 mRNA half-life (Figure 2G). Totally, our results showed that the m6A methylation modification in BTG2 mRNA might delay its degradation.
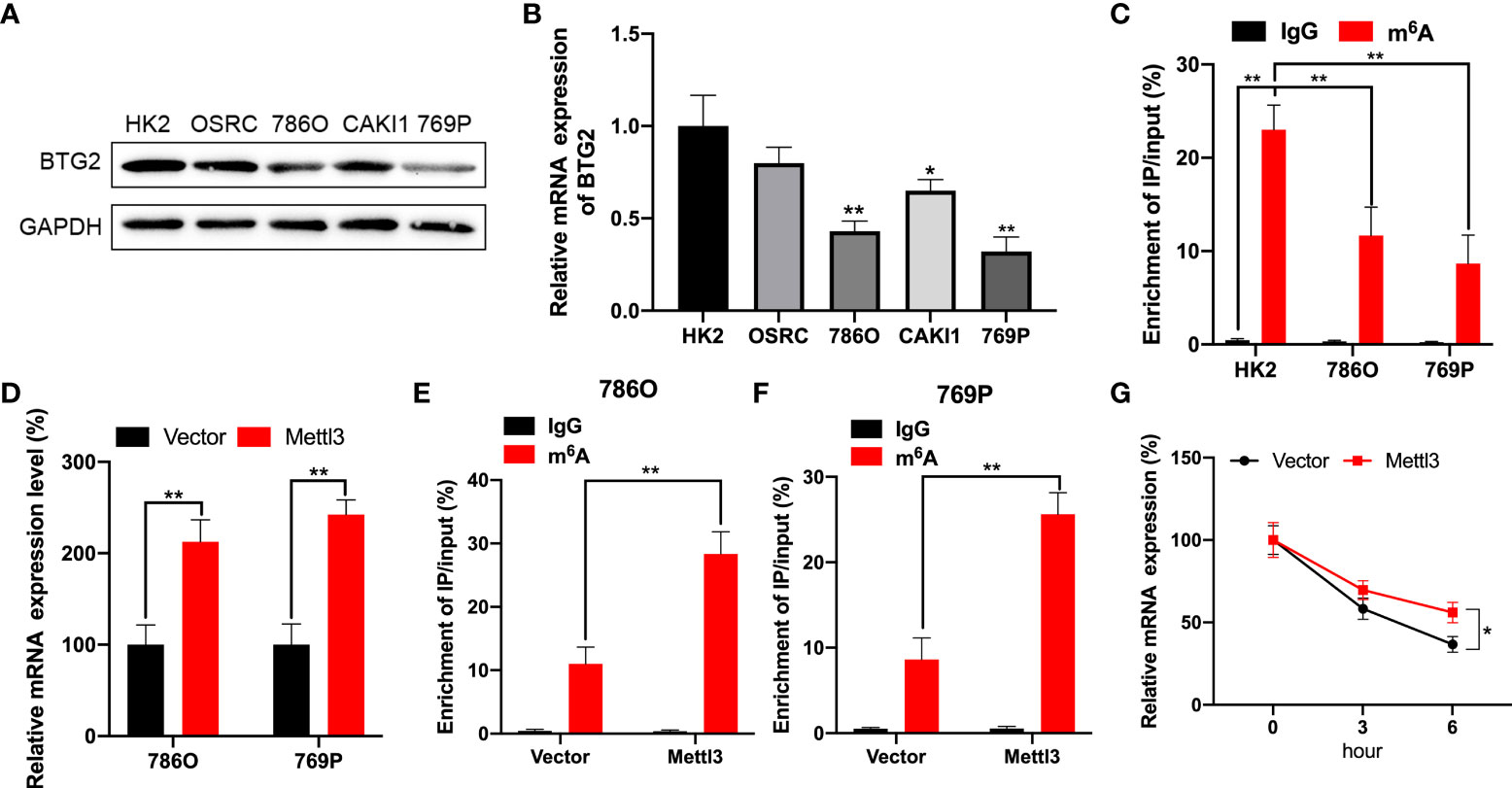
Figure 2 BTG2 expression and m6A methylation level in RCC cells. (A) The protein expression levels of BTG2 in RCC cells were detected using western blot. (B) The mRNA expression levels of BTG2 in RCC cells were detected using RT-qPCR. (C) The m6A methylation levels of BTG2 in RCC cells were detected using MeRIP-qPCR. (D) The mRNA expression levels of BTG2 in 786O and 769P cells after overexpressing Mettl3 were detected using RT-qPCR. E-F. The m6A methylation levels of BTG2 in 786O cell (E) and 769P cells (F) after overexpressing Mettl3 were detected using MeRIP-qPCR. (G) The mRNA expression levels of BTG2 in 786O cells after overexpressing Mettl3 and treatment with ActD for indicated times were detected using RT-qPCR. * P<0.05, ** P<0.01.
Based on MeRIP-seq data, there were two differentially methylated m6A peaks (DMMs) in BTG2 mRNA’s 5′UTR and CDS regions (Figures 1G, 3A). Then, RNA fragments isolated from RCC cell are immunoprecipitated with anti- m6A antibody to characterize BTG2 mRNA methylation modification. According MeRIP-qPCR results, we found that the 5′UTR region in BTG2 mRNA had the highest m6A modification level, followed by the 3′UTR and CDS (Figure 3B). In addition, Mettl3-overexpressing cells showed increased enrichment of m6A in BTG2 5′UTR, which suggests that m6A modification in the 5′UTR is more dynamic than in the CDS region (Figure 3B). In order to investigate the possible roles of m6A methylation in the BTG2 mRNA CDS region, we used luciferase reporters containing the WT or MUT BTG2 CDS in RCC cells. The results showed that the luciferase activity of BTG2-CDS-MUT was similar between Mettl3-overexpressing and control groups compared to BTG2-CDS-WT (Figure 3C). In summary, these results indicate that m6A methylation level in the CDS region of BTG2 mRNA does not correlate with m6A modification, which promotes its mRNA stability.
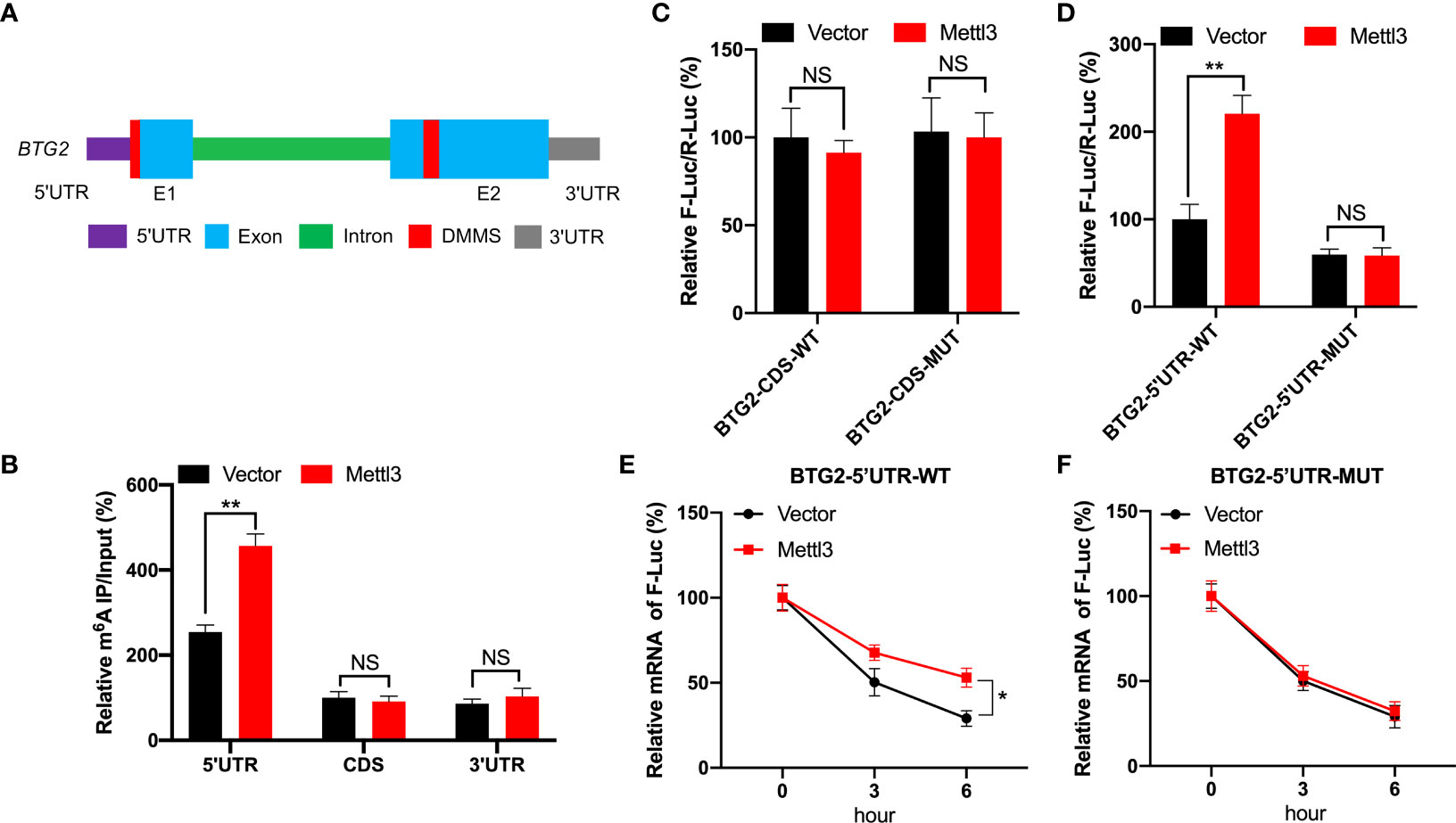
Figure 3 BTG2 is regulated by m6A modification through methylation sites. (A) Diagram showing DMMs positions within BTG2 mRNA. (B) The MeRIP-qPCR assay was used to analyze the m6A enrichment in different regions of BTG2 mRNA in control or Mettl3 overexpression 786O cells. (C) F-Luc/R-Luc activity in control and Mettl3-overexpressing 786O cells transfected with pmirGLO-BTG2-CDS-WT or pmirGLO-BTG2-CDS-MUT. (D) F-Luc/R-Luc activity in control and Mettl3-overexpressing 786O cells transfected with pmirGLO-BTG2-5’UTR-WT or pmirGLO-BTG2-5’UTR-MUT. E-F. The mRNA expression levels of pmirGLO-BTG2-5’UTR-WT (E) or pmirGLO-BTG2-5’UTR-MUT (F) in 786O cells after overexpressing Mettl3 and treatment with ActD for indicated times. * P<0.05, ** P<0.01, NS means No significance.
Therefore, we tested whether BTG2 mRNA stability is regulated by m6A methylation modification in the 5′UTR region. In Mettl3-overexpressing RCC cells, BTG2-5′UTR-WT produced significantly higher luciferase activity than control group by the dual-luciferase assay (Figure 3D). While in Mettl3-overexpressing RCC cells, BTG2-5′UTR-MUT didn’t have a significant upregulation of luciferase activity than control group (Figure 3D). Moreover, the stability of BTG2-5′UTR-WT mRNA in Mettl3-overexpressing cells was increased over control cells (Figure 3E), while Mettl3-overexpressing cells and control cells didn’t have different mRNA half-lives of BTG2-5′UTR-MUT (Figure 3F). In conclusion, BTG2 mRNA stability is mediated by m6A methylation modifications in the mRNA 5′UTR region.
We further investigated mechanisms involved in m6A-regulated BTG2 mRNA stability. Several studies have shown that m6A modification can affect mRNA stability via readers such as IGF2BP1∼3 (26, 27). Based on the TCGA database, IGF2BP2 was evidently downregulated in RCC tumour samples compared to normal samples (Figure 4A). Consequently, we tested whether IGF2BP2 enhances BTG2 mRNA stability in a m6A-dependent manner. Through RIP-qPCR assays, we first detected that IGF2BP2 binds to BTG2 m6A modification sites. It was found that IGF2BP2 strongly binds to BTG2 mRNA in RCC cells (Figure 4B). Furthermore, compared to control cells, Mettl3-overexpressing cells had significantly higher binding between IGF2BP2 and BTG2 (Figure 4B). Then we overexpressed IGF2BP2 in RCC cells. It was found that IGF2BP2 could increase BTG2 mRNA levels (Figure 4C). Moreover, IGF2BP2 can increase mRNA stability in BTG2-5′UTR-WT (Figure 4D), but not in BTG2-5′UTR-MUT (Figure 4E). In our study, IGF2BP2 was proved to be implicated in m6A methylation-dependent BTG2 mRNA stabilization.
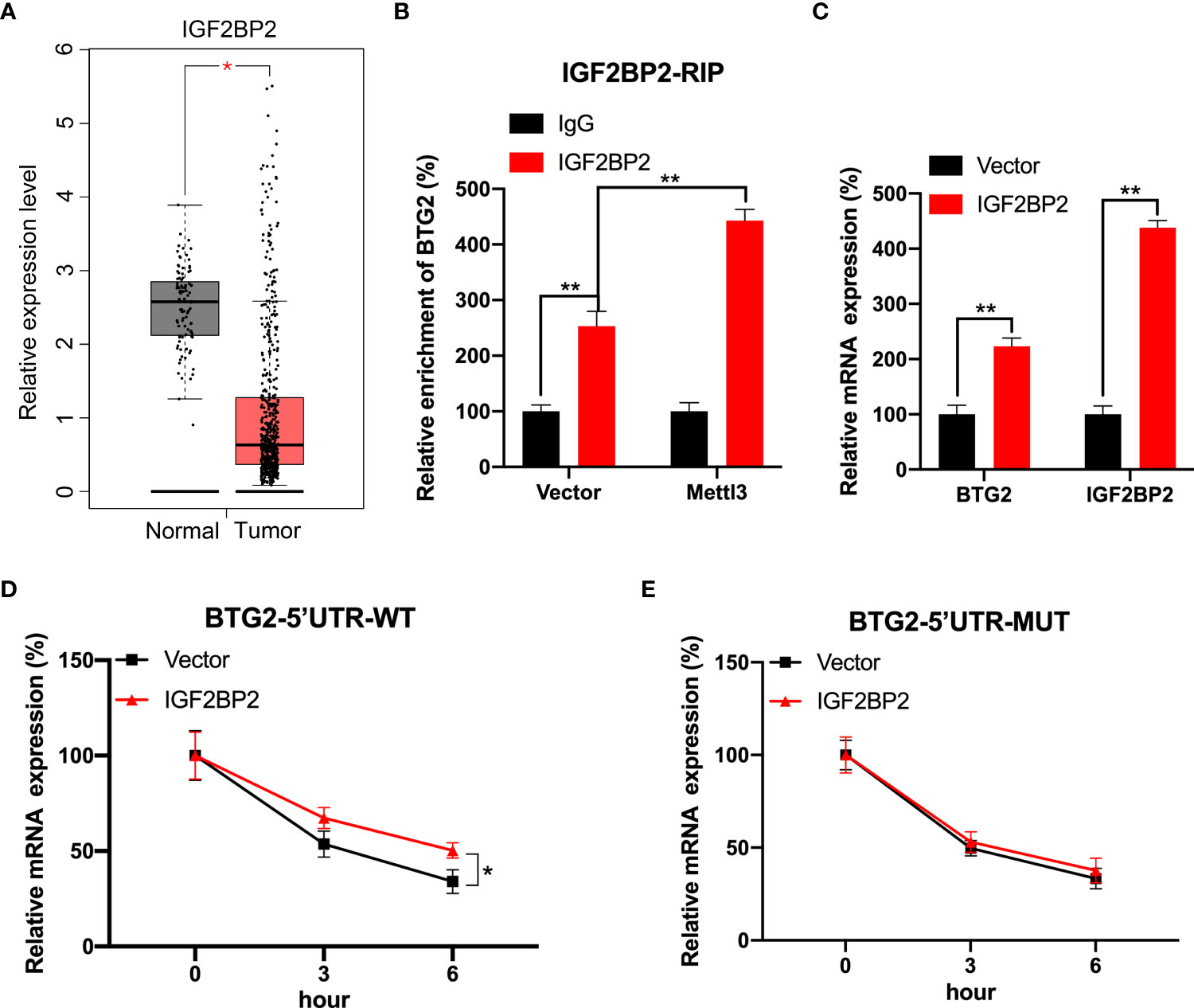
Figure 4 IGF2BP2 involved in m6A-regulated expression of BTG2. (A) An analysis of IGF2BP2 expression level in RCC tumor tissues (red plot) and normal tissues (grey plot) is shown in the box plot below based on TCGA database. (B) An analysis of BTG2 mRNA in control or Mettl3-overexpressing 786O cells using IGF2BP2 RIP-qPCR. (C) The expression levels of BTG2 and IGF2BP2 in control or IGF2BP2-overexpressing 786O cells using RT-qPCR. D-E. The mRNA expression levels of pmirGLO-BTG2-5’UTR-WT (D) or pmirGLO-BTG2-5’UTR-MUT (E) in 786O cells after overexpressing IGF2BP2 and treatment with ActD for indicated times. * P<0.05, ** P<0.01.
In order to specifically methylate the m6A of BTG2, we combined a catalytically dead Cas13 enzyme with METTL3 (dCas13b-M3) (28). To target the BTG2 mRNA, guide RNAs were positioned around the m6A site in BTG2 mRNA (Figure 5A). As a first step, MeRIP-qPCR was performed on 786O and 769P cells to confirm that dCas13b-M3 induced m6A methylation of BTG2 (Figure 5B). Furthermore, we found that dCas13b-M3 targeting BTG2 significantly increased BTG2 mRNA and protein levels in 786O and 769P cells (Figures 5C, D). It may be due to dCas13b-M3 targets for BTG2 can significantly enhance BTG2 mRNA binding to IGF2BP2 (Figure 5E). Then the effects of dCas13b-M3 with control or gRNA for BTG2 mRNA were compared in order to determine whether m6A-mediated mRNA stability relates to dCas13b-M3-induced methylation upregulation. Based on the results of our study, targeted m6A methylation of BTG2 significantly lengthened its mRNA half-life, suggesting that dCas13b-M3 methylated m6A at 5′UTR region to increase its mRNA stability (Figure 5F).
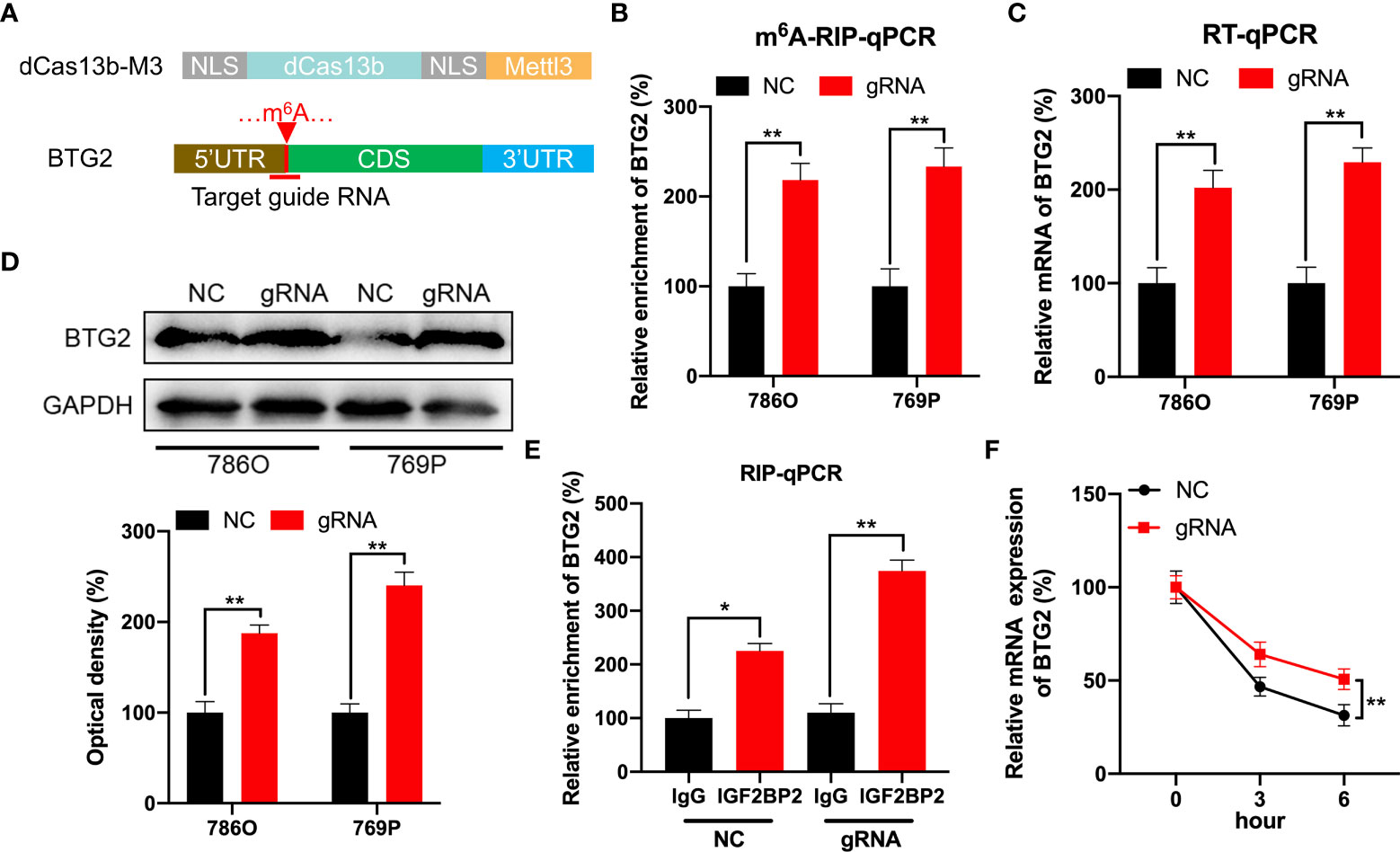
Figure 5 CRISPR/dCas13b-METTL3 specifically targets m6A methylation of BTG2 to regulate its expression. (A) Positions of m6A sites within BTG2 mRNA along with target guide RNA regions. (B–D). The m6A (B), mRNA (C) and protein levels (D) of BTG2 in RCC cells transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2, respectively. (E) A RIP-qPCR analysis of BTG2 mRNA is performed by using antibodies against IGF2BP2 on 786O cells transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2, respectively. (F) The mRNA expression levels of BTG2 in 786O cells transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2 and treatment with ActD for indicated times. * P<0.05, ** P<0.01.
Further investigation to detect the role of BTG2 m6A hypermethylation might regulate RCC cell homeostasis was performed by detecting cell proliferation, migration and apoptosis in RCC cells. According to our results, BTG2 gRNA significantly decreased cell proliferation and promoted apoptosis in comparison with non-targeted control gRNA transfected with dCas13b-M3 in 786O and 769P cells (Figures 6A–C). According to wound healing assays and cell migration assays, hypermethylation of BTG2 inhibited cell migration in 786O and 769P cells (Figures 6D–I). These results showed that BTG2 could inhibit the RCC cells proliferation and migration, and promote RCC cells apoptosis through m6A methylation modification.
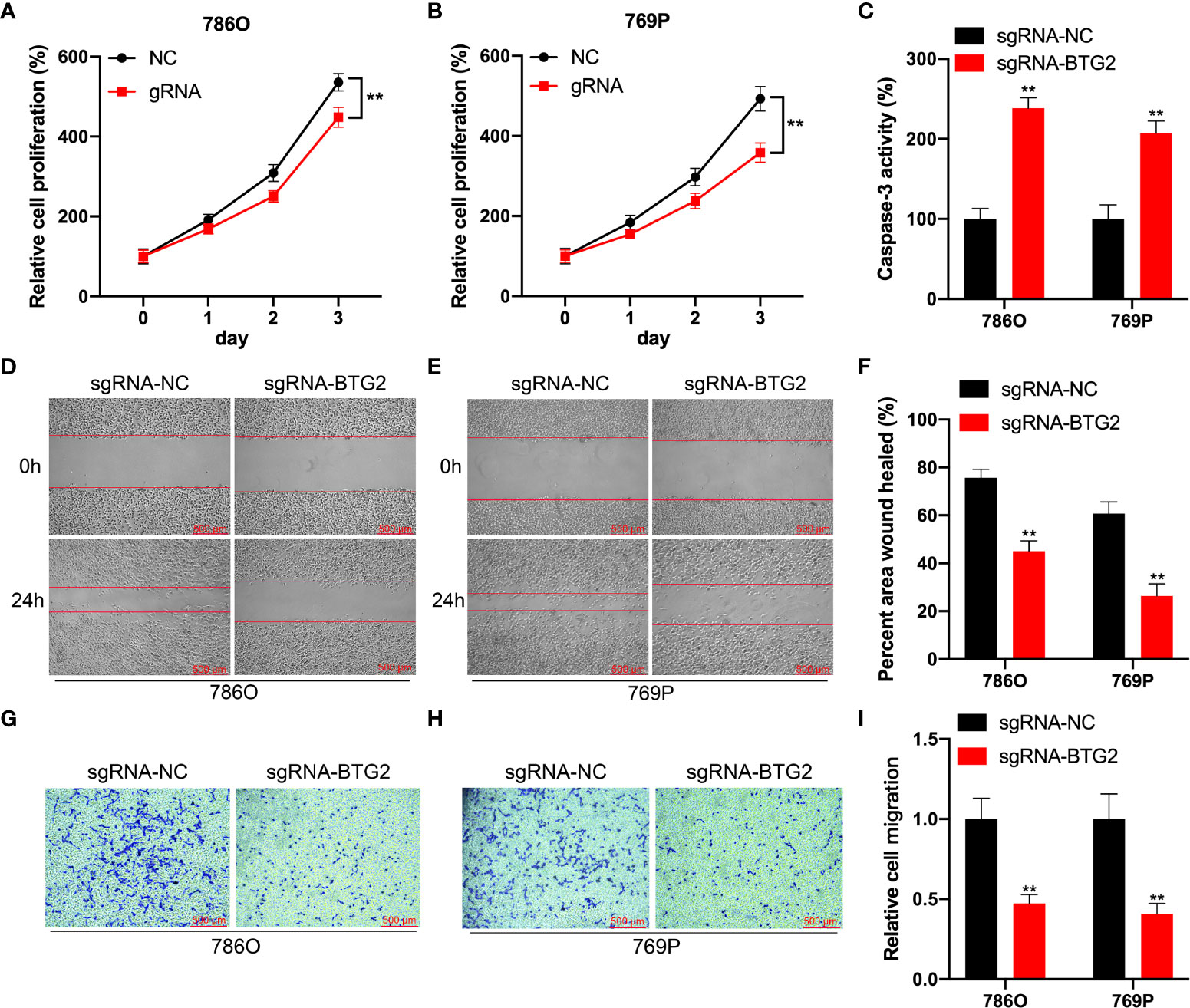
Figure 6 Specifical m6A methylation of BTG2 inhibits RCC cells proliferation and promotes cell apoptosis by CRISPR/dCas13b-METTL3. (A, B). The cell proliferation of 786O (A) and 769P (B) cells transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2 was detected by CCK8 assay. (C) The cell apoptosis of 786O (A) and 769P (B) cells transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2. (D-F) Wound-healing assays for migration ability of 786O (D) and 769P (E) cells after transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2. (G-I). Transwell assays for migration ability of 786O (D) and 769P (E) cells after transfected with dCas13b-M3 combined with gRNA control or gRNA for BTG2. ** P<0.01.
Renal cell carcinoma includes many histological subtypes with different biological manifestations and prognosis. The most basic histological subtypes include clear cell carcinoma of the kidney (75-80%), papillary carcinoma (10-15%), and chromocytoma (4-6%). Among them, renal clear cell carcinoma is the most common histological type, and is the most aggressive tumor subtype with the worst prognosis among all renal cell carcinoma subtypes. Surgical resection is the main treatment in clinical practice. For patients with advanced renal cancer who are not suitable for surgery, radiotherapy, chemotherapy and hormone therapy are the alternative treatment methods. However, clinical practice has proved that patients with renal cancer have low sensitivity to radiotherapy, chemotherapy and hormone therapy. Molecular targeted drug therapy is the main treatment for advanced renal cancer and recurrent renal cancer in recent years.
A reversible and dynamic RNA modification, m6A is a result of both a methyltransferase and demethylase protein. There is evidence that m6A modification plays a role in multiple cellular processes, such as heat shock (29), UV light (30), hypoxic stress (31), and oxidative stress (32). It has been reported that m6A modification is associated with the development of tumours, but their involvement in RCC is less clear. BTG2 has been found to be expressed at low levels in a variety of cancer cells, which has potential in the development and subsequent progression of tumors. Studies have shown that the expression level of BTG2 is low in breast cancer cell lines and is associated with tumor grade, size, metastasis, recurrence and breast cancer survival (25). Another study also reported low levels of BTG2 expression in renal cell carcinoma (33). Our study also showed a significant difference in the expression level of BTG2 gene in normal renal tissues and tumor tissues. However, it remains unclear whether m6A methylation modification influences BTG2 expression to alter epigenetic remodeling or contribute to malignant characteristics of RCC.
The findings of this study indicated that BTG2 suppresses RCC tumor growth. First, we found that the expression of BTG2 was repressed in RCC tumour tissues compared to normal tissues based on TCGA database. And patient outcomes were also adversely by BTG2 downregulation. Furthermore, we found that BTG2 inactivation might result from reduced m6A methylation modifications in the 5′UTR of mRNA. And Mettl3 regulates BTG2’s mRNA stability positively, thereby increasing the BTG2 m6A and expression levels. Nearly all stages in the lifecycle of RNA can be regulated by the m6A modification, including RNA processing, nuclear export, and translation (34, 35). The results of luciferase reporter system showed that m6A modification in 5′UTR region positively regulated m6A methylation level of BTG2 instead of CDS region. Moreover, we demonstrated that BTG2 mRNA stability was regulated by IGF2BP2 in RCC cells. It has also been shown that IGF2BPs could bind the GG(m6A)C sequence of mRNA to increase its stability and storage (26). As a result, our findings provide a fundamentally new understanding of the role of IGF2BP2 in RCC through its involvement in regulating the mRNA stability of BTG2.
Furthermore, CRIPSR/dCas13b-METTL3 was used specifically to methylate the m6A of BTG2 mRNA (28). It is also a newly developed method which can manipulate cell homeostasis by targeting specific methylation events in mRNA transcripts. The results showed that there was a 2-fold increase in m6A methylation level and a significant increase in BTG2 expression with this system. And dCas13b-M3 decreased RCC cells proliferation and migration and induced RCC cells apoptosis.
In summary, we demonstrate that m6A-mediated mRNA stability depends on methylation of 5′UTR regions in BTG2 mRNA to downregulate its expression. Our study further demonstrated that m6A regulates RCC cells growth and migration in RCC by regulating BTG2 expression. It is important to note that our results demonstrate that m6A methylation modification is instrumental in regulating the progression of RCC. Finally, METTL3/IGF2BP2/BTG2, a new regulatory complex, might provide novel insight into the pathogenesis and development of RCC.
We obtained renal cell carcinoma samples and adjacent nonmalignant renal tissues with informed consent from Shengli OilFiled Central Hospital’s (SOFCH) Urology Department. SOFCH’s Institutional Ethical Review Board approved this study in accordance with the Helsinki declaration. To isolate RNA from the samples, they were collected immediately after surgical removal and frozen in liquid nitrogen after rapid freezing.
In accordance with the manufacturer’s instructions, RIP assays were performed with the Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore). A protease inhibitor cocktail and RNase inhibitor were included in the complete radioimmunoprecipitation assay buffer for lysing the cells. Protein A/G magnetic beads were pre-bound to antibodies (5 g) for 2 h before being incubated overnight with 100 L of cell lysate at 4°C. By incubating the beads in 400 μL of elution buffer for 2 h, eluting with ethanol, and dissolving in RNase-free water, the RNA was eluted from the beads. Real-time PCR was used to determine fragment enrichment.
Following the instructions of Vazyme Biotech, Nanjing, China, total RNA was extracted from tissues and cell lines using the RNA-easy Isolation Reagent. Anti-m6A antibody-coupled beads were used to incubate fragmented RNA. IGF2BP2 was used to immunoprecipitate the m6A-containing RNA and then it was eluted from the beads. The RT-qPCR was performed with gene-specific primers on both input control and m6A-IP samples. A HiScript III RT SuperMix for qPCR was used to synthesize the cDNA (Vazyme Biotech, Nanjing, China). With universal SYBR Green qPCR Master Mix (Vazyme Biotech, Nanjing, China) and spectrophotometry (ABI Prism 7500TM instrument, Applied Biosystems), qRT-PCR was performed. The primers used in this study were listed in Supplement Table S1.
KeyGEN Bio TECH protein extraction kit (KGP1100) was used to extract protein from cells and separate it on 10% SDS-PAGE and transfer it to nitrocellulose membrane. According to previous instructions, blots were then immunostained with primary antibodies and secondary antibodies. The antibodies were as follows: BTG2 (1:1000; Abcam, United States) and GAPDH (1:10000; Proteintech, United States).
Image density was quantified using ImageJ analysis software (NIH, Bethesda, MD, USA). And we are using a ratio to GAPDH.
Addgene provided the CRISPR dCas13b plasmids and Cas13b-gRNA plasmids. A number of designed gRNAs and the dCas13b-METTL3 vector have been constructed by Synbio Technologies. The overexpression plasmids were generated using the CDS of METTL3 or IGF2BP2 cloned into pcDNA3.1. The vector control used for analysis was pcDNA3.1.
This study used cells from the RCC cell lines. Cell lines for this study was purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States) and the National Infrastructure of Cell Line Resource, China. DMEM with 10% fetal bovine serum was routinely used for cell culture, supplemented with 5% CO2 in a 37°C environment (Invitrogen, Carlsbad, CA, United States). Lipo3000 (Invitrogen) was used for transfection of all plasmids as per the manufacturer’s instructions, and 1 μg of plasmids was used in each experiment.
In order to determine the effect of BTG2 expression, BTG2’s wild type or mutant 5’UTR was inserted at the end of the F-luc coding sequence. In both wild type and METTL3 overexpression cells, the pmirGLO-BTG2-5’UTR-WT and pmirGLO- BTG2-5’UTR-Mut were transfected for 24 hours. We analyzed firefly luciferase (F-luc) and renilla luciferase (R-luc) by using the Dual-Glo Luciferase Assay system (Promega) according to instructions. The activity of Renilla Luciferase (R-luc) was used to normalize the activity of Firefly Luciferase (F-luc) to evaluate reporter transcription. The experiments were repeated three times and the results were similar each time.
Cells transfected with different plasmids were stabilized with actinomycin D (Act-D, catalog #A9415, Sigma, U.S.A.) at 50 mg/ml during incubation. To conduct real-time PCR, RNA was isolated from the cells at the indicated times. BTG2 mRNA half-life was calculated by using ln2/slope and normalized using GAPDH.
Detection of cell proliferation was carried out using the Cell Counting Kit 8 (CCK8) assay (Transgen, China). In 96-well plates, 3×103 cells were seeded per well. A total of 24h, 48h, 72h, and 96h of cell culture was followed by 3h of incubation with CCK8 at 37°C. After that, 450 nm absorbance was measured using a microplate reader. Cells transfected with vectors were inoculated on a
12-well plate (2X105 cells/well) with 70%–80% confluency. After 48 h, the cell was detected by the caspase-3/ELISA (enzyme-linked immunosorbent assay) (Hcusabio, China). The caspase-3 enzyme is a marker for inflammation and apoptosis signaling, as it can regulate the destruction of DNA or cytoskeletal proteins. Each test was performed at least three times.
The wound healing migration experiments were carried out exactly as indicated. Cells were planted at a density of 5x105 cells/well in a six-well chamber slide. A sterile 10 μL pipette tip was used to scratch the center of the slides. After 24 hours of incubation, photos were taken to assess the gap’s closure. The wound closure was measured using the following formulae:
At least three independent experiments were used to gather data. Data are reported as mean + standard deviation. A two-tailed unpaired Student’s t-test was used between two groups, along with one-way or two-way ANOVA and Bonferroni testing for multiple comparisons. A two-sided test was used for all statistical analyses, using SPSS 16.0 for Windows. In order to be considered statistically significant, the p-value must be 0.05 or less. *p < 0.05, **p < 0.01; NS, no significant.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Shengli OilField Central Hospital's (SOFCH) Urology Department. The patients/participants provided their written informed consent to participate in this study.
FQ, WL, and BT performed the experiments and data analysis. JZ and YM prepared diagrams and wrote the manuscript. FM, CC and BZ designed the project. JY and XL supervised the project and provided financial support. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1049928/full#supplementary-material
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin (2017) 67(1):7–30. doi: 10.3322/caac.21387
2. King SC, Pollack LA, Li J, King JB, Master VA. Continued increase in incidence of renal cell carcinoma, especially in young patients and high grade disease: United states 2001 to 2010. J Urol (2014) 191(6):1665–70. doi: 10.1016/j.juro.2013.12.046
3. Lipworth L, Tarone RE, McLaughlin JK. The epidemiology of renal cell carcinoma. J Urol (2006) 176(6 Pt 1):2353–8. doi: 10.1016/j.juro.2006.07.130
4. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
5. Zhu W, Wang JZ, Xu Z, Cao M, Hu Q, Pan C, et al. Detection of N6−methyladenosine modification residues (Review). Int J Mol Med (2019) 43(6):2267–78. doi: 10.3892/ijmm.2019.4169
6. Csepany T, Lin A, Baldick CJ Jr, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem (1990) 265(33):20117–22. doi: 10.1016/S0021-9258(17)30477-5
7. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
8. Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, et al. Regulation of Co-transcriptional pre-mRNA splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell (2019) 76(1):70–81.e9. doi: 10.1016/j.molcel.2019.07.005
9. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A (2016) 113(14):E2047–56. doi: 10.1073/pnas.1602883113
10. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell (2014) 15(6):707–19. doi: 10.1016/j.stem.2014.09.019
11. Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-Methylation-dependent RNA processing controls the speed of the circadian clock. Cell (2013) 155(4):793–806. doi: 10.1016/j.cell.2013.10.026
12. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell (2016) 62(3):335–45. doi: 10.1016/j.molcel.2016.03.021
13. Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, et al. The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature (2018) 555(7695):256–9. doi: 10.1038/nature25784
14. Paramasivam A, Vijayashree Priyadharsini J, Raghunandhakumar S. N6-adenosine methylation (m6A): A promising new molecular target in hypertension and cardiovascular diseases. Hypertens Res (2020) 43(2):153–4. doi: 10.1038/s41440-019-0338-z
15. Dorn LE, Lasman L, Chen J, Xu X, Hund TJ, Medvedovic M, et al. The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation (2019) 139(4):533–45. doi: 10.1161/CIRCULATIONAHA.118.036146
16. Williams GD, Gokhale NS, Horner SM. Regulation of viral infection by the RNA modification N6-methyladenosine. Annu Rev Virol (2019) 6(1):235–53. doi: 10.1146/annurev-virology-092818-015559
17. Yang Y, Shen F, Huang W, Qin S, Huang JT, Sergi C, et al. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J Clin Endocrinol Metab (2019) 104(3):665–73. doi: 10.1210/jc.2018-00619
18. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell (2017) 31(4):591–606.e6. doi: 10.1016/j.ccell.2017.02.013
19. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell (2017) 31(1):127–41. doi: 10.1016/j.ccell.2016.11.017
20. Li YJ, Dong BK, Fan M, Jiang WX. BTG2 inhibits the proliferation and metastasis of osteosarcoma cells by suppressing the PI3K/AKT pathway. Int J Clin Exp Pathol (2015) 8(10):12410–8. doi: 10.3389/fmolb.2021.675683
21. Chen Y, Chen C, Zhang Z, Xiao H, Mao B, Huang H, et al. Expression of b-cell translocation gene 2 is associated with favorable prognosis in hepatocellular carcinoma patients and sensitizes irradiation-induced hepatocellular carcinoma cell apoptosis in vitro and in nude mice. Oncol Lett (2017) 13(4):2366–72. doi: 10.3892/ol.2017.5685
22. Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, et al. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol (2007) 27(22):7791–801. doi: 10.1128/MCB.01254-07
23. Lim IK. TIS21 (/BTG2/PC3) as a link between ageing and cancer: Cell cycle regulator and endogenous cell death molecule. J Cancer Res Clin Oncol (2006) 132(7):417–26. doi: 10.1007/s00432-006-0080-1
24. Winkler GS. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol (2010) 222(1):66–72. doi: 10.1002/jcp.21919
25. Takahashi F, Chiba N, Tajima K, Hayashida T, Shimada T, Takahashi M, et al. Breast tumor progression induced by loss of BTG2 expression is inhibited by targeted therapy with the ErbB/HER inhibitor lapatinib. Oncogene (2011) 30(27):3084–95. doi: 10.1038/onc.2011.24
26. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
27. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
28. Wilson C, Chen PJ, Miao Z, Liu DR. Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol (2020) 38(12):1431–40. doi: 10.1038/s41587-020-0572-6
29. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature (2015) 526(7574):591–4. doi: 10.1038/nature15377
30. Robinson M, Shah P, Cui YH, He YY. The role of dynamic m(6) a RNA methylation in photobiology. Photochem Photobiol (2019) 95(1):95–104. doi: 10.1111/php.12930
31. Fry NJ, Law BA, Ilkayeva OR, Holley CL, Mansfield KD. N(6)-methyladenosine is required for the hypoxic stabilization of specific mRNAs. RNA (2017) 23(9):1444–55. doi: 10.1261/rna.061044.117
32. Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, et al. Dynamic m(6)A methylation facilitates mRNA triaging to stress granules. Life Sci Alliance (2018) 1(4):e201800113. doi: 10.26508/lsa.201800113
33. Struckmann K, Schraml P, Simon R, Elmenhorst K, Mirlacher M, Kononen J, et al. Impaired expression of the cell cycle regulator BTG2 is common in clear cell renal cell carcinoma. Cancer Res (2004) 64(5):1632–8. doi: 10.1158/0008-5472.CAN-03-1687
34. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
Keywords: BTG2, m6A, RCC, dCas13b, Mettl3
Citation: Qi F, Liu W, Tan B, Zhang J, Ma Y, Cao C, Ma F, Zhu B, Yang J and Liu X (2022) BTG2 suppresses renal cell carcinoma progression through N6-methyladenosine. Front. Oncol. 12:1049928. doi: 10.3389/fonc.2022.1049928
Received: 21 September 2022; Accepted: 01 November 2022;
Published: 14 December 2022.
Edited by:
Samantha Pattenden, University of North Carolina at Chapel Hill, United StatesReviewed by:
Linhua Liu, Guangdong Medical University, ChinaCopyright © 2022 Qi, Liu, Tan, Zhang, Ma, Cao, Ma, Zhu, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyun Liu, bHh5bHh5OTI4QDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.